Abstract
Cross-linking of high-affinity immunoglobulin E (IgE) results in the life-threatening allergic reaction, anaphylaxis. Yet the cellular mechanisms that induce B cells to produce IgE in response to allergens remain poorly understood. T follicular helper (Tfh) cells direct the affinity and isotype of antibodies produced by B cells. Although Tfh cell-derived interleukin-4 (IL-4) is necessary for IgE production, it is not sufficient. We report a rare population of IL-13-producing Tfh cells present in mice and humans with IgE to allergens, but not when allergen-specific IgE was absent or only low-affinity. These “Tfh13” cells have an unusual cytokine profile (IL-13hiIL4hiIL-5hiIL-21lo) and co-express the transcription factors BCL6 and GATA3. Tfh13 cells are required for production of high-but not low-affinity IgE and subsequent allergen-induced anaphylaxis. Blocking Tfh13 cells may represent an alternative therapeutic target to ameliorate anaphylaxis.
Keywords: T Follicular Helper Cells, IgE, IL-13, IL-4, GATA3, Anaphylaxis, Allergen, Helminth, DOCK8
One Sentence Summary:
Characterization of a T cell subset that promotes the form of IgE required for allergic anaphylaxis.
Anaphylaxis is a severe form of allergic reaction precipitated by degranulation of immunoglobulin E (IgE)-laden mast cells after allergen recognition (1). Studies from food allergic patients and murine models indicate that high-affinity, but not low-affinity, IgE induces mast-cell degranulation and anaphylaxis (2–4) and that the nature of the B cell that switches to low-versus high-affinity IgE differs (5, 6). Unlike other antibody isotypes, how B cells are instructed to make affinity-matured IgE remains unclear.
Early work on IgE regulation demonstrated that the deletion of T helper 2 (Th2) lineagedefining transcription factors (TFs) such as STAT6 or GATA3 or the prototypical Th2 cytokine interleukin-4 (IL-4) reduced total IgE. Thus, Th2 cells were proposed to control the IgE response (7–9). However, more recent work has demonstrated that IL-4+ T follicular helper (Tfh) cells, not Th2 cells, are required for IgE production (10–15). Tfh cells are the primary helper T cell subset responsible for directing the affinity, longevity, and isotype of antibody produced by B cells. Recent work has supported this functional Th2–Tfh lineage distinction by identifying a distinctive Il4 enhancer locus bound by BATF in Tfh cells that is distinct from the Th2 DNA regulatory element for IL-4, IL-5, and IL-13 bound by GATA3 (16–19). Therefore, it has been argued that GATA3, IL-5, and IL-13, are restricted to Th2 cells and type 2 innate lymphoid cells (ILC2s) (14, 20).
IL-4 is a B cell survival factor expressed by Tfh cells during a variety of immune responses, including those in which IgE is not made (21–23). This suggests that IL-4 from Tfh cells is necessary but not sufficient for the induction of IgE. We hypothesized that IL-4+ Tfh cells induce direct switching of B cells to low-affinity IgE during certain type 2 immune responses, but a distinct Tfh population producing additional signals regulates high-affinity IgE during allergen responses.
Using a murine model of a rare monogenic form of IgE-mediated allergy, dedicator of cytokinesis 8 (Dock8) deficiency, we discovered a subset of Tfh cells associated with high-affinity IgE production. These “Tfh13” cells produced IL-13 along with IL-4 but downregulated IL-21. Accordingly, they expressed GATA3 in addition to the Tfh transcription factor BCL6. We found the same Tfh13 cells in WT mice immunized with multiple allergens, but not other stimuli that failed to induce high-affinity IgE, including bacterial products or helminth infection. Circulating Tfh13 cells were also found in patients with IgE to aeroallergens or peanut(PN). Conditional deletion of Tfh13 cells or IL-13 in Tfh cells abrogated the generation of high-affinity anaphylactic IgE to allergens. Thus, our study identifies the context in which a rare subset of Tfh cells are elicited and uncovers their critical role in the induction of anaphylactic IgE to allergens.
Dock8 deficiency reveals the presence of a distinct Tfh cell population associated with a hyper-IgE state
Patients with mutations in DOCK8 are immunodeficient, but, paradoxically, they present with hyper-IgE syndrome (HIES) and associated food allergies and asthma. The precise reasons for HIES in this condition are not yet understood (24, 25). DOCK8 was originally described as a guanine nucleotide exchange factor that regulates the actin cytoskeleton, but recent evidence reveals diverse roles of DOCK8 in nearly every cell of the immune system (25). We generated both total and immune cell-specific knockouts of Dock8 in mice to study the cellular mechanisms of IgE induction.
IgE antibodies are a characteristic component of type 2 immunity, which is induced in response to allergens and helminths. In contrast, type 1 responses, induced by viral and certain bacterial infections, do not classically elicit the production of IgE. To determine whether Dock8 deficiency promotes an aberrant hyper-IgE response to type 1 immunization, we immunized mice with lipopolysaccharide (LPS) along with the model antigen 4-hydroxy-3-nitrophenylacetyl (NP) conjugated to ovalbumin (NP-OVA), henceforth called LPS+OVA. The hapten NP allows measurement of antigen specificity and affinity. Using conditional Dock8-knockout mice, we discovered that isolated loss of Dock8 in T cells (T-Dock8−/−), but not in B cells or dendritic cells (DCs), recapitulated the hyper-IgE phenotype seen in patients (Fig. 1A). The hyper-IgE phenotype was not present in complete Dock8-knockout mice, which was not surprising given their DCdependent defect in Tfh cell induction (26). The use of T-Dock8−/− mice bypassed the effect of Dock8 deficiency on DC migration (fig. S1A) and B cell development (fig. S1B). T cell-specific deletion of Dock8 was confirmed by immunoblot (fig. S1C) and via known T cell-intrinsic phenotypes of Dock8, including reduced T cell frequencies (fig. S1D) (27). However, Dock8−/− OVA-specific CD4+ T cells (OT-II cells) demonstrated comparable in vivo proliferation and Tfh cell differentiation to Dock8WT OT-II cells to LPS+OVA immunization (fig. S1E and F).
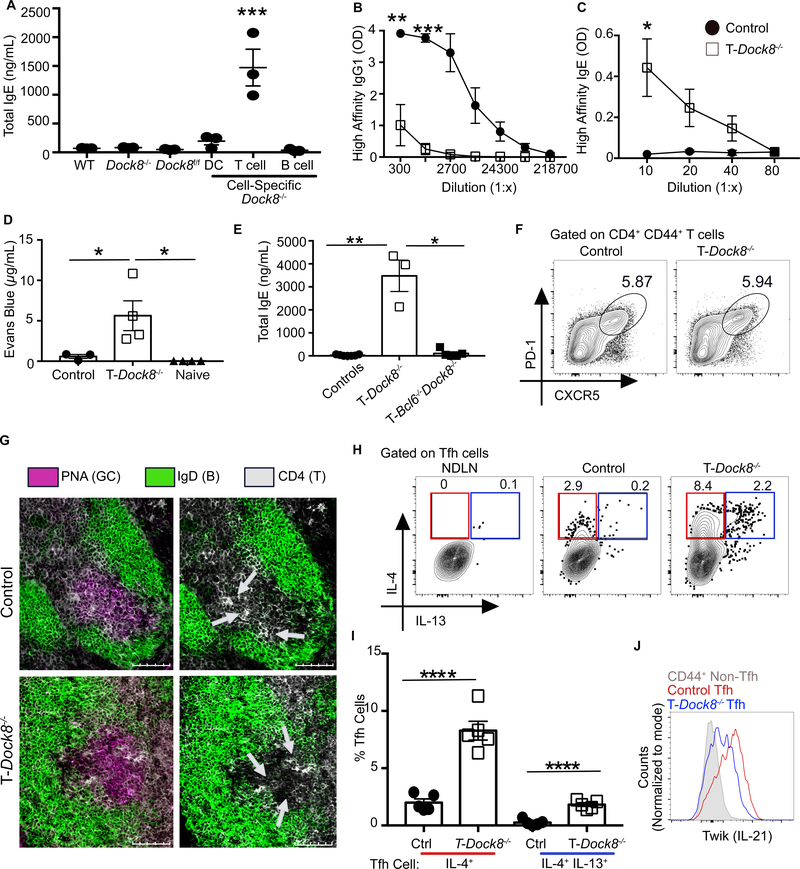
Fig. 1. DOCK8 deficiency reveals the presence of a specific Tfh cell population associated with a hyper-IgE state.
(A) WT, Dock8−/−, Dock8fl/fl, Cd11cCreDock8fl/fl (DC-Dock8−/−), Cd4CreDock8fl/fl (T-Dock8−/−), and Cd19CreDock8fl/fl (B-Dock8−/−) mice were immunized intranasally (i.n.) with LPS and NP16-OVA (LPS+OVA). Day 12 total serum IgE was measured by ELISA. (B and C) T-Dock8−/− or control mice (Dock8fl/fl) were immunized and boosted with LPS+OVA. Day 8 post-boost sera were analyzed by ELISA for (B) NP4-specific IgG1 and (C) NP4-specific IgE. OD, optical density. (D) PCA assay performed by transferring day 8 post-boost sera into naïve recipients and challenging with NP7-BSA and 1% Evans blue. Dye extravasation quantification is shown. (E) Day 12 serum IgE from T-Dock8−/−, T-Bcl6−/−Dock8−/−, or control mice (Dock8fl/fl and Bcl6fl/fl Dock8fl/fl) immunized i.n. with LPS+OVA. (F) Day 8 Tfh cell frequencies are depicted as representative flow cytometry contour plots. (G) Immunofluorescent images of day 9 MedLN GCs from immunized T-Dock8−/− or control Dock8fl/fl mice stained for IgD (green) and CD4 (white) with or without (left and right, respectively) peanut agglutinin (PNA, purple) are shown. Scale bars: 100 μM. Arrows indicate clusters of CD4+ T cells in the GC. (H and I) Intracellular expression of IL-4 and IL-13 by day 8 Tfh cells (gated as in fig. S4A) depicted as (H) flow cytometry plots and (I) bar graphs. NDLN, nondraining lymph node. (J) IL-21 reporter expression in Tfh cells from IL-21 TWIK TDock8−/− or control Dock8fl/fl reporter mice on day 8 after-immunization depicted as histogram overlay. In (A), (D), (E), and (I), each symbol indicates an individual mouse. Numbers in flow plots indicate percentages. Error bars indicate SEM. Statistical tests: analysis of variance (ANOVA) (A and D); Student’s t test (B, C and I); Kruskal–Wallis H test (E). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. Data representative of at least two independent experiments with three to seven mice per group.
In addition to atypical total IgE production, type 1 immunization in T-Dock8−/− mice resulted in reduced NP-OVA-specific IgG antibodies and elevated NP-OVA-specific IgE antibodies (fig. S2A–C). Additionally, there was reduced high-affinity NP-specific IgG1 and elevated high-affinity NP-specific IgE (Fig. 1B and C). Previously immunized T-Dock8−/− mice challenged with NP-conjugated bovine serum albumin (BSA) showed robust acute mast-cell degranulation and systemic tissue edema (fig. S2, D and E) (28). The anaphylactic capacity of IgE from T-Dock8−/− mice was confirmed using a passive cutaneous anaphylaxis (PCA) assay (Fig. 1D) (6, 28). T-Dock8−/− mice also spontaneously developed high levels of serum IgE as they aged (fig. S2, F and G). Thus, T-Dock8−/− mice appear to recapitulate the hyper-IgE and dysgammaglobulinemic presentation of DOCK8-deficient patients.
DOCK8 deficiency has been reported to inhibit FOXP3+ regulatory T cell (Treg) function (29, 30). However, the inducible deletion of Dock8 in Tregs did not develop high-affinity IgE in response to LPS+OVA immunization (fig. S2H). Further, Dock8-deficient Tregs were competent in suppressing T cell activation in vitro (fig. S2I). To determine whether Tfh cells in T-Dock8−/− mice were responsible for the hyper-IgE response to type 1 immunization, we crossed T-Dock8−/− mice to Bcl6fl/fl mice to generate T-Bcl6−/−Dock8−/− (Cd4CreBcl6fl/flDock8fl/fl) mice. In contrast to T-Dock8−/− mice, T-Bcl6−/−Dock8−/− mice did not develop a hyper-IgE response, suggesting that the hyper-IgE phenotype in T-Dock8−/− mice is dependent on Tfh cells but not on other cell types (e.g. CD4+ DCs or CD8+ T cells) that may have lost Dock8 expression as a result of Cd4Cre mediated deletion (Fig. 1E). Finally, T-Dock8−/− mice immunized with the model type 2 allergen Alternaria alternata along with NP-OVA (henceforth called Alt+OVA) showed high-affinity and total IgE titers similar to control mice (fig. S2, J and K). Thus, DOCK8 in Tfh cells blocks inappropriate induction of IgE during type 1 immune responses. Our analysis of Tfh cells showed no difference in frequency or expression of programmed cell death 1 (PD-1) or CXCR5 between control and T-Dock8−/− mice post LPS+OVA immunization (Fig. 1F and fig. S3, A and B). Germinal center (GC) structure, Tfh cell localization, and GC B cell frequencies were also comparable between control and T-Dock8−/− mice (Fig. 1G and fig. S3, C to E). However, in contrast to control mice, there was a significantly greater population of IL-4-producing Tfh cells as well as an unexpected population of IL-4 and IL-13 co-producing Tfh cells in T-Dock8−/− mice (Fig. 1, H and I and fig. S4A). A fraction of these IL-4+IL-13+ Tfh cells, which we call Tfh13 cells, also produced the canonical type 2 cytokine IL-5 (fig. S4B). Tfh13 cells induced in TDock8−/− mice expressed the lineage-defining Tfh TF BCL6 at levels similar to IL-4+ Tfh cells induced in control mice (fig. S4C).
The cytokine IL-21 has been associated with the negative regulation of IgE and promoting IgG1 in mice (31, 32, 33) as well as in humans in the presence of IL-13 (34). Hence, we assessed IL-21 levels in Tfh cells in T-Dock8−/− mice by crossing them with IL-21 TWIK reporter mice (21). IL-21 production by Tfh cells was reduced in T-Dock8−/− mice relative to control mice (Fig. 1J and fig. S4, D and E). Tfh cells from T-Dock8−/− mice expressed more of the canonical Th2 TF GATA3 compared to control Tfh cells and non-Tfh effector cells (fig. S4F). Aged T-Dock8−/− mice that developed the spontaneous hyper-IgE phenotype also had elevated frequencies of Tfh13 cells (Fig. S4G). Thus, a rare population of Tfh cells that expressed GATA3 and unexpectedly produced IL-5 and IL-13 in addition to IL-4, while secreting less IL-21, was associated with the hyper-IgE state in T cell-specific Dock8 deficiency.
Tfh13 cells are induced in WT mice during allergic sensitization
We next asked whether Tfh13 cells are also induced in genetically unmanipulated WT mice during allergic sensitization, which also generates high-affinity, anaphylactic IgE. WT mice immunized with Alt+OVA, but not those immunized with LPS+OVA, produced high-affinity IgE that was anaphylactic (Fig. 2, A and B). Alt+OVA immunization induced less high-affinity IgG1 compared with LPS+OVA (fig. S5A). IgE induction in Alternaria immunization was dependent on Tfh cells, as Cd4CreBcl6fl/fl (T-Bcl6−/−) mice did not generate high-affinity or total IgE (Fig. 2C and fig. S5, B and C). However, eosinophilia in these mice was similar to controls, indicating that the type 2 cellular response was intact but could not compensate for Tfh cell loss in IgE induction, consistent with published studies (fig. S5D) (10, 13).
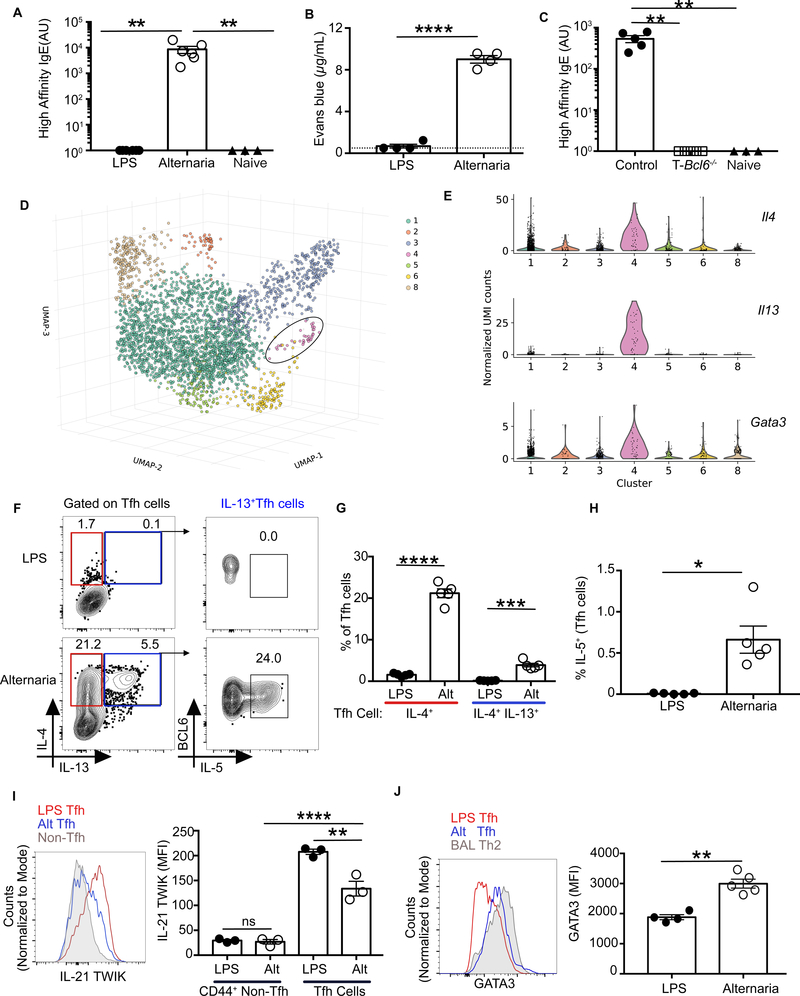
Fig. 2. Tfh13 cells are induced in WT mice during allergic sensitization.
WT mice were immunized and boosted i.n. with LPS or Alternaria extract and NP16-OVA. (A) Day 8 sera from boosted mice were analyzed for high-affinity IgE by ELISA with NP7-BSAcoated plates. (B) Evans blue dye extravasation quantification after PCA with day 8 post-boost sera and NP7-BSA challenge. (C) Cd4CreBcl6fl/fl (T-Bcl6−/−) or control Bcl6fl/fl mice were immunized and boosted with Alternaria extract and NP16-OVA. Eight days later, high-affinity IgE was quantitated using NP4-BSA by ELISA. (D) 3D uniform manifold approximation and projection (UMAP) embedding of the single–cell expression profiles of n=3002 single Tfh cells sorted from WT C57BL/6 mice immunized i.n with Alternaria extract and NP-OVA. Leiden community detection on the cell-cell k=10 nearest neighbor graph segregates cells into seven clusters, five of which were identifiable on the basis expression of previously known markers: 1, Tfh2; 2, type 1 IFN T cell population; 3, proliferating T cells; 4, Tfh13 cells and 6, Tfr cells. The circle identifies cluster 4, a cluster of Il13+ Tfh cells (n=39). (E) Violin plots showing the expression of distinctive marker genes of Il13+ Tfh cluster (Il4, Il13, Gata3). (F to H) Intracellular expression of IL-4, IL-13, and IL-5 in day 8 MedLN Tfh cells induced after primary immunization with LPS or Alternaria and NP-OVA depicted as flow cytometry (F) plots and bar graphs for IL4 and IL-13 (G) and IL-5 (H). (I) IL-21 expression in Tfh cells from IL-21-TWIK reporter mice at day 8 after immunization, depicted as histogram overlay (left) and bar graphs (right). (J) GATA3 expression in Th2 cells from bronchioalveolar lavage fluid of mice immunized with Alternaria and NP-OVA, and Tfh cells from MedLN of mice immunized with LPS or Alternaria and NPOVA. Data are depicted as histogram overlay (left) and bar graphs (right). Each symbol indicates an individual mouse. Numbers in flow plots indicate percentages. Error bars indicate SEM. Dotted lines in bar graphs represent background readings of sera from naïve mice. Statistical tests: Kruskal-Wallis H test (A and C); Student’s t test (B, G and J); ANOVA (I); Mann–Whitney U test (H) *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; ns, not significant. Data representative of two or three independent experiments with three to five mice per group (A to C, and F to J). Data representative of three biological replicates (D and E).
To determine whether Tfh13 cells could be identified as a transcriptionally distinct population in WT mice, we performed single-cell RNA sequencing (scRNA-seq) on sorted Tfh cells after Alt+OVA immunization (n=3 mice). After data processing (fig. S6, and tables S1 to S3), CXCR5+PD1+ T cells formed seven clusters with distinct transcriptional signatures (Fig. 2D and fig. S7A). We could readily identify a Tfh2 cell population (cluster 1), a Tfh cell population with a type I interferon signature (cluster 2), a T follicular regulatory (Tfr) population (cluster 6), and a Tfh13 cell population (cluster 4). Although all clusters similarly expressed Tfh cell markers such as Batf, Cd40lg, Icos, and Pdcd1, Tfh13 cells uniquely expressed high levels of Il4, Il13, and Gata3 (Fig. 2E). Pairwise analysis between the Tfh2 and Tfh13 clusters identified additional genes that discriminate between these two populations, demonstrating that Tfh13 cells are a transcriptionally distinct population (fig. S7B).
We also identified Tfh13 cells by flow cytometry in Alternaria-immunized WT mice. Although the overall magnitude of Tfh cell and GC B cell induction was comparable between LPS-and Alternaria-immunized mice (fig. S8, A to C), a significant Tfh13 population was induced by Alt+OVA but not LPS+OVA (Fig. 2F–G and S8D–F). Related cell types induced during type 2 immune responses—Th2 effector cells and IL-4-single-positive Tfh cells (Tfh2 cells)—were also more abundant with Alt+OVA immunization. However, these populations were distinguished from Tfh13 cells via flow cytometric staining of BCL6, PD-1, and CXCR5 (fig. S8, G to I). Il13 transcripts were detected in unstimulated Tfh cells sorted from mice immunized with Alternaria but not LPS, whereas Il4 transcripts were present in Tfh cells from both conditions (fig. S9, A and B). Like Tfh13 cells in T-Dock8−/− mice, Tfh13 cells in Alt+OVA-immunized WT mice also produced IL-5 (Fig. 2F and 2H). As IL-5 promotes eosinophilia (10, 14, 35) and the majority of IL-5+ T cells in the lymph node (LN) were Tfh cells, we accordingly observed eosinophil infiltration of the LNs in Alt+OVA-immunized mice (fig. S9, C to E).
Alternaria-induced Tfh cells produced substantial levels of IL-21 compared with non-Tfh effector CD4+ T cells, but reduced levels of IL-21 compared with LPS+OVA-induced Tfh cells (Fig. 2I). Tfh cells from Alt+OVA immunization also demonstrated significant GATA3 expression, albeit less than differentiated Th2 cells from the lungs (Fig. 2J). However, canonical Tfh TFs such as BCL6, BATF, IRF4, and TCF1 (36) were expressed at equivalent levels in both immunizations (fig. S10). Thus, Tfh13 cells are also induced in WT mice, but only during type 2 high-affinity IgE responses.
Tfh13 cells are a distinct T cell subset
Given the similar phenotype of Tfh13 cells and conventional Th2 effector cells (i.e. IL-4, IL-5, IL13, and GATA3 expression), we evaluated whether the two populations were transcriptionally distinct. We performed scRNA-seq on Il4-reporter-positive activated CD4+ T cells from Alt+OVA-immunized 4Get/Il4-reporter mice (n=3) (37). The data were processed as before (fig. S6, and tables S1 to S3), followed by selection of Il13-expressing cells. The Il13-expressing T cells formed three transcriptionally distinct clusters (Fig. 3A). Among these clusters, we could identify a population of Th2 effectors (cluster 1), Tfh13 cells (cluster 2), and Il4+ T cells expressing high levels of proliferation markers such as Top2a and Mki67 (cluster 3). Pairwise analysis between Th2 effector and Tfh13 cells confirmed differential expression of T effector versus Tfh cell markers, including Prdm1 (BLIMP1), Bcl6, Il21, and Areg (Fig. 3B and fig S11A). We verified select targets at the protein level (Fig. 3, C to G). We also analyzed the scRNA-seq data from Bcl6-expressing cells and identified two distinct clusters, one with a transcriptional profile consistent with Tfh13 cells (cluster 2) and the other consistent with Tfh2 cells (cluster1) (fig. S11, B and C).
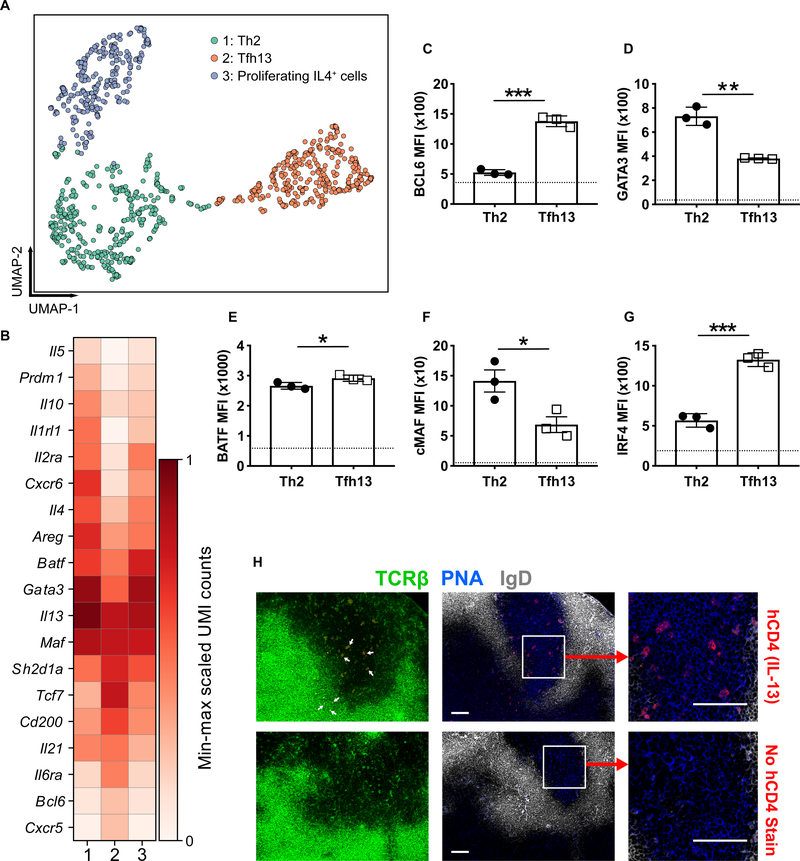
Fig. 3. Tfh13 cells are a distinct T cell subset.
4Get/Il4-reporter mice were immunized i.n. with Alternaria extract and NP19-OVA. GFP(Il4)+CD44+CD4+ T cells were sorted for sc-RNA-seq. Cells expressing one or more Il13 transcripts were isolated and subjected to dimensionality reduction and clustering. (A) A 2D UMAP embedding of n=1,040 Il13+ cells. Low-resolution (0.2) Leiden community detection on the k=10 cell–cell nearest neighbor graph reveals three subpopulations comprising 405, 340, and 295 cells, respectively. Populations 1, 2, and 3 were putatively identified as Th2 cells, Tfh13 cells, and proliferating IL-4+ cells, respectively. (B) A matrix plot showing the expression of key marker genes that are similarly and differentially expressed between Il13+ Th2 and Tfh13 cells. Color scale represents log-transformed normalized unique molecular identifier (UMI) counts scaled between 0 and 1, separately for each gene. (C to H) Smart13 (Il13) reporter mice were immunized with Alternaria extract and NP-OVA. Day 3 after boost, analysis of transcription factors (C) BCL6, (D) GATA3, (E) BATF, (F) cMAF, and (G) IRF4 was performed by intracellular staining of Tfh13 cells and IL-13+ Th2 cells (gated as in fig. S12A). Dotted lines indicate mean fluorescence intensity (MFI) of naïve CD4+ T cells. (H) Microscopic images of MedLN GCs from immunized reporter mice stained for human CD4 (IL-13) (red), TCRβ (green), IgD (white), and PNA (blue) are shown. Arrows in the leftmost panel indicate TCRβ and hCD4 costaining. Scale bars: 100 μM. Statistical tests: Student’s t test (C to G). *P<0.05, **P<0.01, ***P<0.001. Data representative of three biological replicates (A and B). Data representative of two independent experiments (C to H) with three mice.
To determine the location of Tfh13 cells in the LN, we used the Smart13 reporter, which reports Il13 via the expression of human CD4 (hCD4) (14). hCD4 expression was concordant with IL-13 intracellular cytokine staining (fig. S12, A and B). With immunofluorescence, Il13 reporter+ T cells could be visualized in both the GC and T cell zone, corresponding to Tfh13 cells and IL13+ T effectors, respectively (Fig. 3H). Altogether, by transcriptional profile, protein expression, and subanatomic location, we show that Tfh13 cells are a distinct subset from Th2 effector and Tfh2 cells.
Tfh13 cells are induced to multiple allergens in mice and humans
Two other common allergens, house dust mite extract (HDM) and PN, were then assessed for their ability to induce Tfh13 cells. HDM-induced allergic airway responses lead to Tfh and GC B cell induction in the mediastinal LN (MedLN) (38). Similar to Alt+OVA immunization, HDM+OVA immunization elicited Tfh13 cells and antigen-specific IgE (Fig. 4, A and B and fig. S13, A and B). Peanut administered with the mucosal adjuvant cholera toxin (CT) is a commonly used model of IgE-mediated food allergy (39). There was a significant induction of mesenteric LN Tfh13 cells and PN-specific IgE in response to PN+CT but not PN alone (Fig. 4, C and D and fig. S13, C and D).
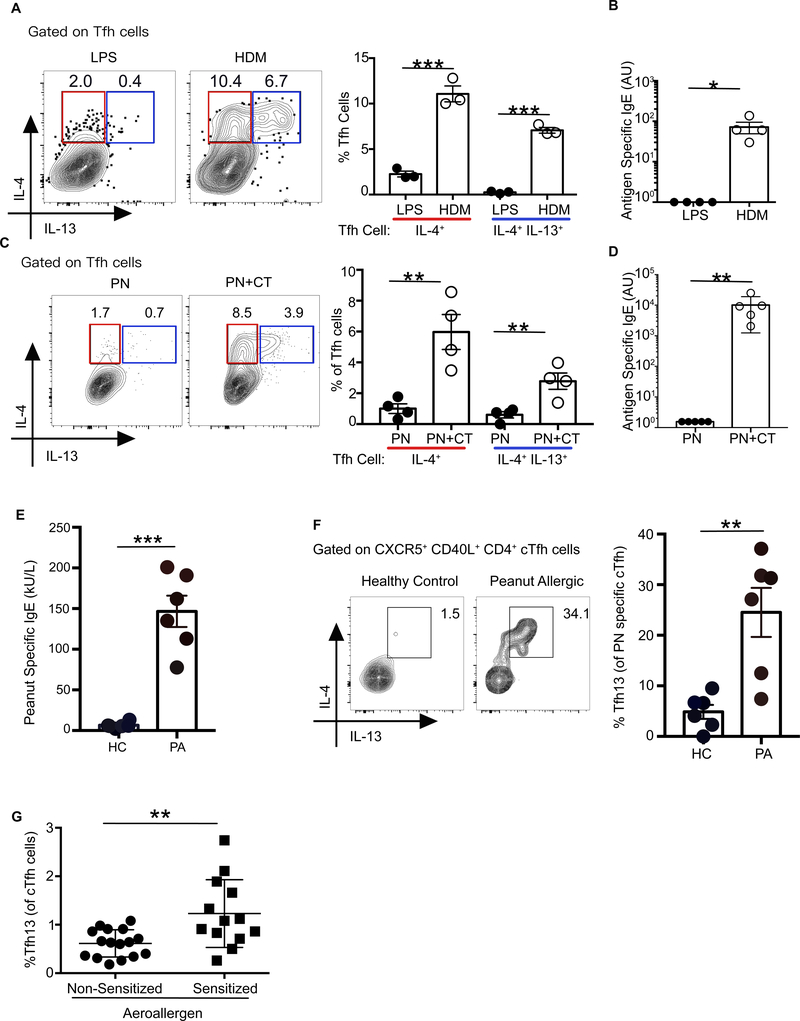
Fig. 4. Tfh13 cells are induced to multiple allergens in mice and humans.
WT C57BL/6 mice were immunized i.n with HDM and NP16-OVA and boosted with HDM and NP16-OVA twice, or with LPS and NP16-OVA as described in Fig 1. (A) Intracellular expression of IL-4 and IL-13 from day 8 MedLN Tfh cells induced after primary immunization is depicted as representative flow cytometry plots (left) and summary bar graphs (right). (B) Day 8 sera from boosted mice were analyzed for NP16-OVA-specific IgE by ELISA. (C and D) WT mice were intragastrically immunized with ground peanut (PN) alone with or without CT in 0.2 M NaHCO3 and boosted with the same immunization up to four times at weekly intervals. (C) Intracellular expression of IL-4 and IL-13 in day 8 mesenteric lymph node (MesLN) Tfh cells induced after primary immunization is depicted as representative flow cytometry plots (left) and summary bar graphs (right). (D) Day 8 sera from boosted mice were analyzed for crude PN extract-specific IgE by ELISA. (E) PN-specific serum IgE ELISA performed with sera from PN-allergic patients (PA) or healthy controls (HC). (F) Cytokine profiles of PN-specific circulating Tfh (cTfh) cells (gated as in fig. S14) obtained from PA or HC. (G) IL-4+ IL-13+ cTfh cells from aeroallergen sensitized or control non-sensitized individuals. Each symbol indicates an individual mouse or subject. Numbers in flow plots indicate percentages. Error bars indicate SEM. Statistical tests: Student’s t test (A, C, and E to G); Mann–Whitney U test (B and D) *P<0.05, **P<0.01, ***P<0.001. Data representative of at least two independent experiments (A to D) with three to five mice per group. (E and F) Data from healthy controls n=6 or PN allergic patients (n=6). (G) Data from aeroallergen sensitized allergic patients (n=13) or non-sensitized individuals (n=16).
We next explored whether Tfh13 cells were present in humans with IgE-mediated allergy by examining “circulating” Tfh (cTfh) cells from a cohort of PN-allergic individuals (40) with PNspecific IgE (Fig. 4E). We selected PN-allergic or healthy individuals whose peripheral blood T cells responded in vitro to PN extract by upregulating CD40L (fig. S14). IL-4+IL13+ Tfh13 cells could be identified within the cTfh cell compartment of PN-allergic patients but not healthy controls (Fig. 4F). We also examined cTfh cells from patients with IgE to aeroallergens (table S4). Compared with non-sensitized individuals, aeroallergen-sensitized individuals had a significantly higher frequency of Tfh13 cells (Fig. 4G). Thus, Tfh13 cells are induced in both mice and humans with antigen-specific IgE responses to multiple allergens.
Tfh13 cells and high-affinity IgE are not induced to helminth infections
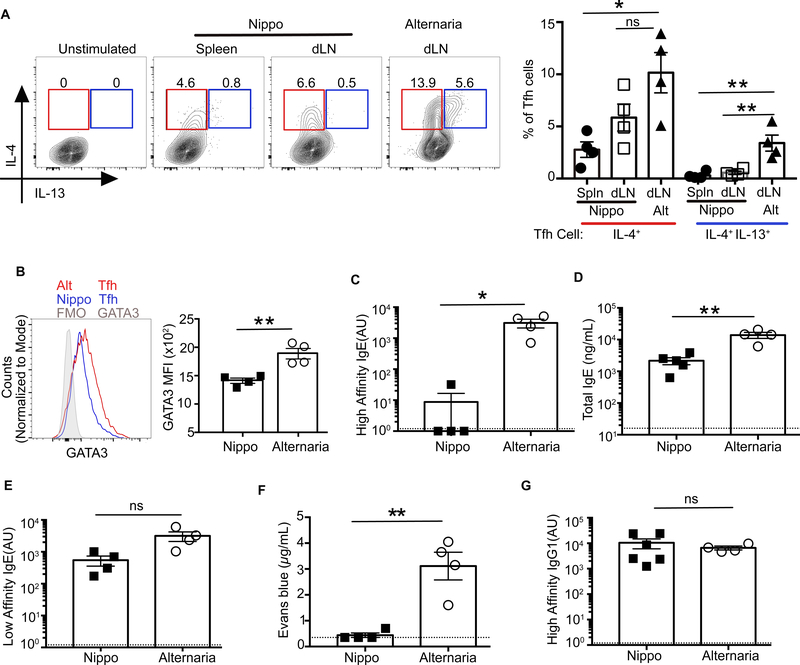
Although Tfh cells are required for IgE production in both allergen and helminth models, it is unclear whether the Tfh cell phenotype in these type 2 immune responses is the same. Previous studies of Tfh cells during helminth-induced type 2 responses have reported IL-4 but not IL-5 or IL-13 production (14, 21). Helminth infections elicit a strong type 2 response characterized by the induction of ILC2s, Th2 cells, eosinophilia, and IgE. Although substantial IgE is produced, little is affinity-matured (6, 41). This suggests that IL-4 production by Tfh cells may be necessary for any IgE induction, but insufficient for the production of high-affinity IgE. We analyzed Tfh cells in mice infected with the helminth, Nippostrongylus brasiliensis, and co-immunized with NPOVA (Nippo+OVA). Nippo+OVA induced a strong IL-4-producing Tfh population in the draining LN and spleen. However, in contrast to Alt+OVA, Tfh13 cells were absent (Fig. 5A). We hypothesized that the absence of Tfh13 cell in N. brasiliensis infection was the result of low GATA3 expression in Tfh cells. Indeed, Tfh cells in N. brasiliensis infection did not upregulate GATA3 (Fig. 5B).
Fig. 5. Tfh13 cells and high-affinity IgE are not induced to helminth infections.
WT C57BL/6 mice were either immunized and boosted with Alternaria extract and NP16-OVA (Alt+OVA), or infected with N. brasiliensis and co-immunized with NP16-OVA and boosted (Nippo+OVA). (A) IL-4 and IL-13 expression in day 8 Tfh cells from N. brasiliensis or Alternaria immunization is shown as flow cytometry plots (left) or as summary bar graphs (right). (B) GATA3 expression in day 8 splenic Tfh cells induced by N. brasiliensis or MedLN Tfh cells from Alternaria immunization. (C to E) Day 8 post-boost sera from mice immunized with Nippo+OVA or Alt+OVA were analyzed by ELISA for (C) high-affinity IgE using NP7-BSA, (D) total IgE, and (E) low-affinity IgE using NP40-BSA. (F) Evans blue dye quantification from PCA assay (with i.v. NP7-BSA challenge) using day 8 post-boost sera from Nippo+OVA or Alt+OVA immunized mice. (G) ELISA for high-affinity IgG1 using NP7-BSA performed on day 8 post-boost sera from mice immunized with Nippo+OVA or Alt+OVA. Each symbol indicates an individual mouse. Numbers in flow plots indicate percentages. Error bars indicate SEM. Dotted lines in bar graphs represent background readings of sera from naïve mice. Statistical tests: ANOVA (A); Student’s t test (B to D to F); Mann-Whitney U test (C and G) *P<0.05, **P<0.01. Data representative of two independent experiments with four to six mice per group.
Nippo+OVA immunization therefore provided a model in which Tfh13 cells were absent but IL-4+IL-13− Tfh2 cells remained present. In the setting where only Tfh2 cells were present, there was strong induction of total IgE, but little was of high affinity (Fig. 5, C and D). However, low-affinity IgE levels were comparable between Nippo+OVA and Alt+OVA immunization (Fig. 5E). This suggests that Tfh2 cells may promote low-affinity IgE but not high-affinity IgE. Serum from Nippo+OVA mice did not contain anaphylactic IgE (Fig. 5F). Similar IgG1 responses in both Nippo+OVA and Alt+OVA models were observed (Fig. 5G). Thus, Tfh13 cells, are uniquely induced during type 2 allergen but not helminth responses and may be responsible for the production of anaphylactic IgE.
Loss of Tfh13 cells abrogates production of high-affinity IgE
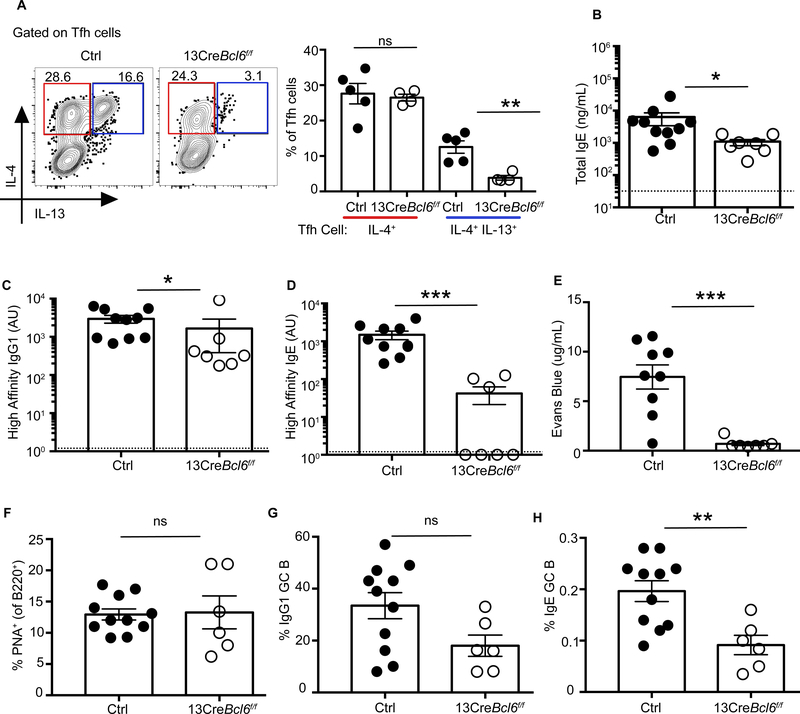
To test whether Tfh13 cells are necessary to induce high-affinity IgE, we bred Il13Cre (14) to Bcl6fl//fl (13CreBcl6fl/fl) mice to specifically delete Tfh13 cells. In 13CreBcl6fl/fl mice, Il13-expressing cells lost Bcl6 expression and could not differentiate into Tfh13 cells. 13CreBcl6fl/fl mice had severely reduced Tfh13 cells but retained similar frequencies of IL-4+ Tfh2 or IL-13+ Th2 cells to Alt+OVA (Fig. 6A and fig. S15A). 13CreBcl6fl/fl mice demonstrated minimal reduction of total IgE and high-affinity IgG1 (Fig. 6, B and C); however, high-affinity IgE was reduced by one to three logs and serum from 13CreBcl6fl/fl mice failed to elicit anaphylaxis (Fig. 6D–E). Although total GC and IgG1 GC B cell frequencies were comparable, IgE GC B cell frequencies (gated as in fig. S15B) were significantly reduced in mice lacking Tfh13 cells (Fig. 6F to H). Thus, although IL-4+ Tfh2 cells may be sufficient to promote total IgE and affinity-matured IgG1 responses, the induction of high-affinity IgE critically depends on Tfh13 cells.
Fig. 6. Loss of Tfh13 cells abrogates the production of high-affinity IgE.
13CreBcl6fl/fl or control Bcl6fl/fl mice were immunized and boosted with Alternaria extract and NP19-OVA. (A) Intracellular expression of IL-4 and IL-13 from day 8 MedLN Tfh cells induced after primary immunization is depicted as representative flow cytometry plots (left) and summary bar graphs (right). (B to E) Day 8 post-boost sera were analyzed for (B) total IgE, (C) NP7-specific high-affinity IgG1 (D), NP7-specific high-affinity IgE, and (E) anaphylactic ability by PCA after i.v. challenge with NP7-BSA. (F to H) Day 7 post-boost MedLN cells were analyzed for: (F) B220+PNA+ GC B cells, (G) percentage of GC B cells expressing IgG1, and (H) percentage of GC B cells expressing IgE. Each symbol indicates an individual mouse. Numbers in flow plots indicate percentages. Error bars indicate SEM. Statistical tests: Student’s t test (A and F to H); Mann–-Whitney U test (B to E). *P<0.05, **P<0.01. Dotted lines in bar graphs represent background readings of sera from naïve mice. Data are either representative of two independent experiments (A) or from two pooled experiments (B to H) with three to six mice per group.
Tfh cell-derived IL-13 is required for anaphylactic IgE production
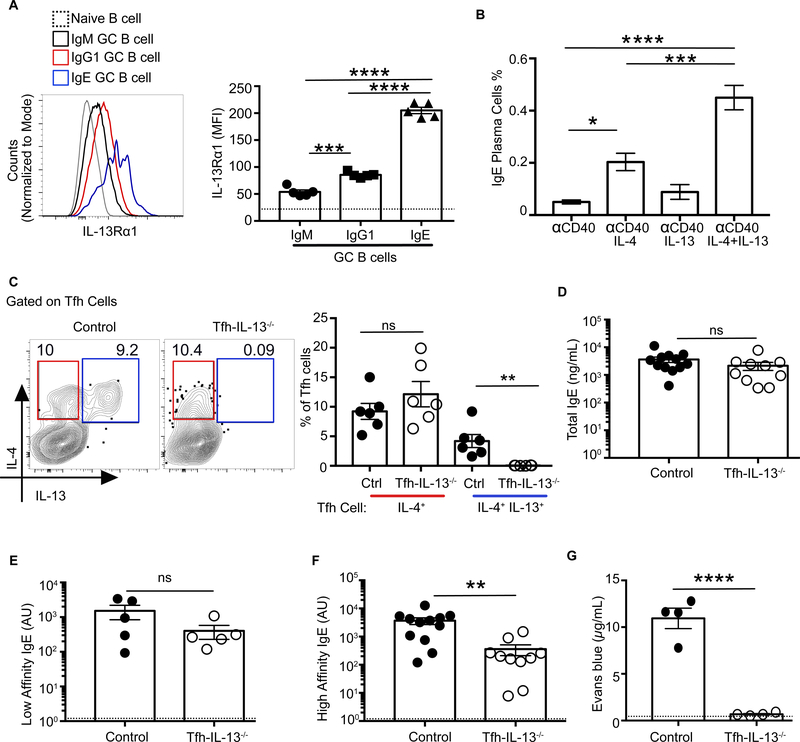
The loss of IgE+ GC B cells in 13CreBcl6fl/fl mice suggested that GC B cells might be able to respond to IL-13. Indeed, after allergic sensitization, GC B cells significantly upregulated IL13Rα1 (fig. S16). IgG1 GC B cells had significantly higher expression of IL-13Rα1 compared with IgM GC B cells, and IgE GC B cells expressed the highest levels (Fig. 7A). Further, IL-13 worked synergistically with IL-4 in ex vivo anti-CD40-stimulated B cells from Alternaria-immunized mice to increase IgE+ plasma cells (Fig. 7B), suggesting that Tfh13-derived IL-4 and IL-13 may together promote IgE in vivo.
Fig. 7. Tfh cell-derived IL-13 is required for anaphylactic IgE production.
(A) Expression of IL-13Rα1 on MedLN GC B cells at day 9 after immunization with Alternaria and NP-OVA is shown as histogram overlay (left) and summary bar graphs (right). Gating strategy as in fig. S15B. (B) Day 8 MedLN lymphocytes from Alternaria and NP-OVA immunized WT mice were cultured with α-CD40 and IL-4 and/or IL-13 for 3 days, and IgE plasma cells were quantified by staining the cells for intracellular IgE and CD138. (C to G) Mixed bone marrow chimeric mice were generated from Il-13−/− and Cd4Cre Bcl6fl/fl donor bone marrow (Tfh-IL-13−/− chimeric mice) or WT and Cd4Cre Bcl6fl/fl donor bone marrow (control chimeric mice). Chimeric mice were immunized and boosted with Alternaria and NP20-OVA. (C) IL-4 and IL-13 expression by day 8 Tfh cells post-boost from Tfh-IL-13−/− or control chimeras is shown as flow cytometry plots (left) or as summary bar graphs (right). (D to F) Day 8 after-boost sera from Tfh-Il-13−/− or control chimeras were analyzed by ELISA for (D) total IgE, (E) low-affinity IgE using NP-40 BSA, and (F) high-affinity IgE using NP7-BSA. (G) Evans blue dye quantification after PCA assay with day 9 after-boost sera from Tfh-Il-13−/− or control chimeras after i.v. challenge with NP7BSA. Each symbol indicates an individual mouse. Error bars indicate SEM. Dotted lines in bar graphs represent (A) IL-13Rα1 expression on naïve B cells or (E and F) background readings of sera from naïve mice. Statistical tests: ANOVA (A and B); Student’s t test (C and G); Mann-Whitney U test (D to F). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. Data are either from two pooled experiments (D and F) or are representative of at least two independent experiments with three to seven mice per group.
Although the loss of IL-13 or its receptor impairs total IgE production (42–44), its role in promoting high-affinity IgE is unclear. To address whether IL-13 produced by Tfh13 cells was necessary for anaphylactic IgE induction, we generated mixed bone marrow chimeras using Cd4CreBcl6fl/fl and Il13−/− mice as donors. Tfh cells in these chimeras did not produce IL-13 but still made IL-4 to Alt+OVA immunization (Fig. 7C). Like 13CreBcl6fl/fl mice, Tfh-Il13−/− chimeric mice produced comparable titers of total and low-affinity IgE to that of controls (Fig. 7, D and E) but had impaired high-affinity IgE (Fig. 7F). The residual high-affinity IgE induced in Tfh-Il13−/− mice was unable to elicit a PCA response (Fig. 7G). Thus, two complementary experimental approaches revealed that Tfh13 cells and the IL-13 they produce are both required for the induction of anaphylactic IgE to allergens.
Discussion
The signals that instruct B cells to make high-affinity IgE remain unresolved. Our work demonstrated that a rare Tfh13 cell population co-expressing IL-4, IL-5, IL-13, BCL6, and GATA3 was responsible for the production of high-affinity anaphylactic IgE. Eliminating Tfh13 cells or Tfh cell-derived IL-13 during allergen immunization resulted in the abrogation of high-affinity anaphylactic IgE while leaving low-affinity IgE intact. Using scRNA-seq, we confirmed that Tfh13 cells are transcriptionally distinct from Tfh2 and Th2 cells. Previous work demonstrates that Th2 and Tfh cells can interconvert during type 2 responses (45, 46). A transitional stage during Tfh development may exist between T effector and Tfh cells during multiple types of immune responses, but there is little reason to believe that a shared developmental stage indicates functional homology (23, 47). We observe functionally discrete Th2 and Tfh13 populations after allergen immunization.
Our data from T-Dock8−/− mice provides insight into possible signaling pathways regulating Tfh13 cells. In humans, DOCK8 has been shown to promote nuclear translocation of STAT3 upon IL-6 or IL-21 stimulation (48, 49). Stat3-deficient mice demonstrate increased GATA3 expression in Tfh cells (50). These studies, along with data, suggest a T cell-intrinsic pathway by which DOCK8 may promote STAT3-dependent suppression of GATA3, thereby blocking inappropriate Tfh-induced IgE during type 1 immune responses.
On the basis of previous studies of IgE-producing B cells and the present work, we propose a two-tiered model of IgE induction that allows for different forms of IgE to be produced depending on the nature of the Tfh cell generated during the type 2 response. Switching to IgE can proceed either directly from IgM or sequentially via IgG1 (6). Directly switched IgE is often of low affinity with poor anaphylactic capabilities and can even protect against anaphylaxis by competing for FceR occupancy (5, 6, 41, 51–54). This is the dominant pathway to IgE during a helminth infection (51, 53). In contrast, allergens are associated with high-affinity anaphylaxis-inducing IgE. During allergic sensitization in both mice and humans, IgE-producing plasma cells show evidence of sequential switching from affinity-matured IgG+ GC B cells (5, 6, 55, 56). We propose that Tfh2 cells instruct the switching of IgM to IgE plasma cells via BATF-driven IL-4, resulting in low-affinity IgE antibodies. These Tfh2 cells, which do not express GATA3, cannot make IL-13. In contrast, allergens induce GATA3+ Tfh13 cells, which instruct sequential switching of IgG1+ B cells, driving high-affinity IgE production and anaphylaxis. This is consistent with our data showing that IgG1 GC B cells express IL-13Rα1 and that loss of Tfh13 cells impairs both high-affinity IgE and GC IgE B cell induction. However, loss of IL-13 does not impair IgE responses in instances where low-affinity IgE is made, such as in helminth infections (43), which is consistent with our findings and the two-tiered IgE model. As loss of IL-4 abrogates all IgE (53), IL-13 likely works synergistically with IL-4 to promote high-affinity IgE by stably engaging the type II IL-4 receptor complex leading to sustained STAT6 activation (57, 58).
Our study defines the role of Tfh13 cells in eliciting anaphylactic IgE to allergens, identifying specific molecular targets that could be leveraged diagnostically and therapeutically for allergies. Identification of Tfh13 cells answers the long-standing question of how, under rare circumstances, anaphylaxis-inducing IgE is produced by high-affinity B cells. It reconciles conflicting literature, including discrepancies between murine and human studies, on whether IL13 and GATA3 are Tfh-relevant effector molecules. Human Tfh cells expressing GATA3, IL-13, and IL-4 have been identified (15, 59, 60) and Il13 remains one of the most replicated genetic associations with elevated IgE, food allergy, atopy, and asthma in humans (61–63). Although this role is typically ascribed to effector Tfh2 cells, Tfh13 cells likely drive the humoral arm of the allergic response. It will be interesting to see whether monotherapies targeting IL-13 in patients affect more than tissue-restricted pathology, specifically allergen-reactive IgE levels. Similarly, recent clinical trials of a GATA3 deoxyribozyme to inhibit GATA3 activity in Th2 cells in asthmatics showed efficacy in reducing eosinophilia (64). The effect of these antisense molecules on Tfh cells and IgE has not been considered given that GATA3 has not typically been associated with Tfh cells (16–19). Thus, the identification of GATA3-expressing Tfh13 cells changes the Th2 paradigm for IgE responses.
Materials and methods
Human subjects
Peanut allergy study: Peanut-allergic subjects were selected from a cohort previously described (40). The clinical study (CoFAR6) provided samples for this study, which is registered with ClinicalTrials.gov with ID NCT01904604. Informed consent was obtained from all subjects or parents/guardians, and all procedures were approved by the institutional review boards at each of the five clinical sites. Peanut allergy was confirmed by double-blind placebo controlled oral challenge with peanut. Controls were sensitized but did not react to peanut challenge. All individuals were avoiding peanut at time of blood draw. Aeroallergen study: Patients 7 to 14 years old, with and without allergies and asthma, were recruited from the pediatric pulmonary and pediatric primary care clinics at Connecticut Children’s Medical Center (CCMC). The allergic phenotype was confirmed with a positive ImmunoCAP clinical laboratory test or positive skin prick testing to at least one environmental allergen (Dermatophagoides farinae, Dermatophagoides pteronyssinus, Alternaria alternata, Cladosporium herbarum, dog, cat, mugwort, mouse, rat, or cockroach). Participants with self-reported food allergies were excluded from analysis (n=1). Clinical details are provided in table S4. CCMC IRB# 15–007
Mice
C57BL/6 and congenic C57BL/6-Ly5.1 [B6.SJL-PtprcaPepcb/BoyCrl] WT mice were purchased from Charles River Laboratories (Wilmington, MA). Cd4cre [Tg(Cd4-cre)1Cwi/BfluJ], Cd11ccre (Itgax-Cre) [B6.Cg-Tg(Itgax-Cre)1–1Reiz/J], Cd19cre [ B6.129P2(C)-Cd19tm1(cre)Cgn/J], Il13cre [C.129S4(B6)-Il13tm1(YFP/cre)Lky/J], Foxp3EGFP-cre-ERT2 [Foxp3tm9(EGFP/cre/ERT2)Ayr/J], Bcl6flox [B6.129S(FVB)-Bcl6tm1.1Dent/J] Smart13[B6.129S4(C)- Il13tm2.1Lky/J], and OT-II [B6.Cg-Tg(TcraTcrb)425Cbn/J] mice were all purchased from Jackson Laboratories (Bar Harbor, ME). Bone marrow from Il13−/− mice (42) in a C57BL6/J background were kindly provided by Dr. E. Gelfand (National Jewish Health, CO). IL-4 4Get reporter mice [C.129Il4tm1.1Lky/J] were back crossed to C57BL/6 background. OT-II mice were crossed onto the CD45.1 or Dock8−/− mice. Dock8−/− and Dock8fl/fl mice were generated as described previously (26). To generate T-Dock8−/−, DC-Dock8−/− mice or B-Dock8−/− mice, Dock8fl/fl were crossed with Cd4Cre, Cd11ccre, or Cd19Cre mice, respectively. IL-13cre mice were in Balb/c background and were backcrossed to Bcl6flox (C57BL6/J background) for three generations and intercrossed to generate Il-13cre/cre Bcl6flox/flox mice in a mixed background and were used for experiments; Cre negative littermates were used as controls. All protocols used in this study were approved by the Institutional Animal Care and Use Committee at the Yale University School of Medicine.
Immunizations
Type 1 LPS model: Mice were immunized intranasally (i.n) with 2 μg of LPS (Sigma) and 25 μg of 4-hydroxy-3-nitrophenylacetyl conjugated ovalbumin (NP16-OVA) (LGC Biosearch Technologies) for primary immunizations. Mice were boosted twice (i.n) with 10 μg NP16-OVA in weekly intervals 10 to 12 days after primary immunization. Type 2 models: Mice were immunized with 10 μg Alternaria (Greer; Lot# 322776) or house dust mite extract (Greer; Lot# 248041) with 25 μg NP16-OVA (i.n) for primary immunizations. Mice were boosted twice (i.n) with 2 μg of the extract and 10 μg NP16-OVA in weekly intervals 10 to 12 days after primary immunization. We observed lot-to-lot variability in house dust mite extract’s ability to elicit Tfh13 cells and antigen-specific IgE. Peanut model: Mice were immunized weekly by oral gavage with 5 mg of ground raw blanched peanut (Western Mixers Produce & Nuts) with or without 10 μg of cholera toxin (List Biologicals) in 200 μl of 0.2 M sodium bicarbonate buffer per mouse. N. brasiliensis and NP-OVA model: Mice were injected with 100 μg NP16-OVA mixed with N. brasiliensis (500 third-stage larvae/mouse) subcutaneously. Mice subsequently were boosted intraperitoneally twice with 50 μg of NP16-OVA in weekly intervals 10 to12 days after primary immunization. For some experiments, NP19-OVA or NP20-OVA were used depending on the lot.
Flow cytometry and cell sorting
Single-cell suspensions from lymph nodes and spleen were prepared and stained as described previously (21, 26). For intracellular cytokine staining (ICS), cells were restimulated with phorbol 12-myristate 13-acetate (PMA) (50 ng/ml) and ionomycin (1 μg/ml) in 96-well U bottom plates in complete IMDM media for 4 h and Golgiplug (BD Biosciences) was added for the last 3 h. ICS was performed using BD ICS kit as per manufacturer’s instructions with overnight incubation (4°C) of permeabilized cells with antibodies. Intracellular staining of transcription factors (TFs) was performed using Foxp3 staining kit (eBiosciences) according to manufacturer’s instructions or by fixing the cells with fixation buffer (BioLegend) for 20 min followed by permeabilization with ice-cold methanol (20 min) and staining for TFs. All TFs were stained at RT for 40 min. Antibodies are listed in Table S5. All flow cytometry samples were acquired on LSRII (BD Biosciences) or MACSQuant (Miltenyi) flow cytometers and analyzed by FlowJo software (Version 10.4.2, TreeStar). Cell sorting was performed using FACS ARIA III (BD Biosciences) on Tfh cells or (IL-4)4Get+ CD44+ CD4+ T cells.
Staining for IgE B cell
The protocol was adapted from (65). Cells obtained from mediastinal lymph nodes (MedLNs) were incubated on ice for 30 min with viability dye, FC-Block (2.4G2), unlabeled anti-IgE (RME-1) to saturate surface and CD23 or Fc receptor bound IgE. Surface staining was performed after this step. Cells were then fixed with and permeabilized with BD intracellular staining kit and stained with fluorochrome conjugated anti-IgE (using same clone that was used to saturate surface IgE), anti-IgG1, and anti-IL-13Rα1 on ice for 30 to 40 min. All incubations prior to fixing cells were done with staining buffer (1X PBS containing 2% FBS and 1 mM EDTA).
Human PBMC stimulation
Peanut study: Peripheral blood mononuclear cells (PBMCs) from healthy control and peanut allergic individuals were stimulated with 100 μg/ml crude peanut extract and stained for Tfh markers and cytokines as described (40). Aeroallergen study: PBMCs were isolated with gradient centrifugation using Lymphoprep (StemCell Technologies) and the cells were frozen until the use. CD4+ T cells were isolated from PBMCs using EasySep™ Human CD4+ T Cell Enrichment Kit (StemCell Technologies) following the manufacturer’s protocol, and the purity was confirmed by flow cytometry. The enriched CD4+ T cells were incubated overnight, 1×106 cells/ml in complete IMDM (IMDM supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin/streptomycin, 2 mM L-glutamine, 10 mM HEPES, 0.1 mM non-essential amino acids, and 1 mM sodium pyruvate) at 37°C + 5% CO2. Cells were stimulated for production of cytokines with PMA (50 ng/ml) + ionomycin (1 μg/ml) in the presence of Brefeldin A (GolgiPlug™ from BD Biosciences) for 6 h. After stimulation, cells were stained for analysis by flow cytometry. CD4+ T cells were incubated with Ghost Dye™ Violet 510 (TonboBiosciences) to stain dead cells for 10 min at room temperature. Fluorochrome-conjugated surface antibodies were incubated for 15 min at room temperature. Cell-surface antibodies against CD3 (UCHT1), CD4 (RPA-T4), and CD45RA (HI100) were all from TonboBiosciences. Anti-CXCR5/CD185 (RF8B2,) was from BD Biosciences, and anti-PD-1/CD279 (EH12.2H7) from BioLegend. Cells were then fixed and permeabilized with Fixation/Permeabilization Solution Kit with BD GolgiPlug™ from BD Biosciences. Intracellular staining was performed for 30 min at 4°C using anti-IL-4 (MP4–25D2), and anti-IL-13 (JES10–5A2) antibodies from BioLegend. FACS analysis was performed on a BD LSR Symphony (BD Biosciences) and data were analyzed with FlowJo software (TreeStar, Ashland, OR).
Enzyme-linked immunosorbent assay (ELISA)
Sera from immunized mice were collected on day 12 post primary and/or 8 to 9 d days post boost. ELISA was performed as described elsewhere (66). Serum samples were analyzed by ELISA for measurement of total, NP-specific, peanut-specific or OVA-specific antibodies. Briefly, for antigen-specific antibodies, 20 μg/ml of NP16-OVA, NP4-BSA or NP7-BSA, NP40-BSA, or crude peanut extract (Lot: 287729, Greer Laboratories), in carbonate buffer was coated on 96-well MaxiSorp plates (Thermo Fisher Scientific) overnight. For total antibodies ELISA, 2 μg/ml anti-mouse IgE (BD Pharmingen) capture antibodies in phosphate buffered saline was coated on 96well MaxiSorp plates overnight. Plates were blocked with 1% BSA in PBS at 37°C for 1 h followed by the addition of serially diluted serum with a 2-h incubation at 37°C. Antigen-specific or total antibody or each isotype were detected with anti-mouse IgE-HRP, anti-IgG1-HRP, or anti-IgG2cHRP (Southern Biotech) and incubated at 37°C. For total antibodies, a starting concentration of 100 ng/ml purified mouse IgE (BD Biosciences) was used as a standard. NP4-BSA or NP7-BSA was used to detect high-affinity IgE depending on the lot. Serum from mice immunized with NPOVA and alum, or complete Freund’s adjuvant or Alternaria extract or peanut with cholera toxin were used as reference standards to calculate arbitrary units for IgG1, IgG2c, and IgE, respectively. Plates were developed with stabilized chromogen tetramethylbenzidine (Life Sciences) and stopped with 3 N hydrochloric acid before reading at 450 nm on a microplate reader (Molecular Devices). Mouse mast cell protease 1 (MMCP-1) ELISA was performed with MMCP-1 ELISA kit (Invitrogen) according to the manufacturer’s instructions.
Passive cutaneous anaphylaxis (PCA)
PCA was performed as described earlier (6). Briefly, ~20 μl of serum from immunized and boosted mice were injected into ear pinnae of naïve B6 recipients. Twenty-four hours later, the recipient mice were challenged with 100 μg NP7-BSA together with 1% Evans blue (i.v.). Thirty minutes later, the mice were euthanized and ear pinnae were harvested and incubated in formamide for 48 to 72 h at 56°C to release the dye. The extent of vascular leakage was measured spectrophotometrically at 650 nm using Evans blue as the standard.
Microscopy
Mediastinal lymph nodes were snap frozen in OCT tissue-freezing solution and stored at −80°C. Tissues were cut into 7-μm sections and processed as described previously (21). Tissues were stained with anti-IgD (11–26.c.2a), PNA (Vector Labs), anti-CD4 (RM4–5); anti-human CD4 (RPA-T4); and anti-PD-1 (RMP1–30). Images were obtained from a laser-scanning confocal microscope (Leica TCS SP5; Lens: Leica HCX PL APO 20x/0.70 Oil) or immunofluorescence microscope (Nikon Eclipse Ti; Camera: Retiga 2000R; Lens: Nikon Plan Apo DIC N1 10X/0.45 and Nikon Plan Fluor DIC N2 20X/0.5). Adobe photoshop or ImageJ software was used for image analysis and the measurement of GC size and T cell counting.
Cytokine ELISPOT assay
MultiScreen HTS plates were coated overnight at 4°C with anti-IL-21 (IL-21 Elispot kit, eBioscience). Sorted cells were cultured (2.5×104 cells/well) with PMA (50 ng/ml) and ionomycin (1 μg/ml) for 36 h at 37°C followed by the addition of primary then secondary detection antibodies. Spots were developed with Vector Blue (Vector Laboratories) and quantified using an ImmunoSpot analyzer (Cellular Technology Limited).
Quantitative PCR (qPCR)
RNA from sorted Tfh cells was isolated using the QIAGEN RNeasy Micro Kit in accordance with the manufacturer’s protocol. cDNA was prepared using the Power SYBR™ Green Cells-to-CT™ Kit (ThermoFisher) in accordance with the manufacturer’s protocol. Real-time PCR was performed using KAPA SYBR Fast Master Mix (Kapa Biosystems) and Low ROX (Kapa Biosystems) and run on the QuantStudio3 (Applied Biosystems). cDNA expression was analyzed by the ΔCt (change in cycle threshold) method normalized to values of Hprt obtained in parallel reactions during each cycle. The following primers were used: Il4: forward 5′-AGATCATCGGCATTTTGAACG-3′, reverse 5′-TTTGGCACATCCATCTCCG-3′; Il13: forward 5′-GCTTATTGAGGAGCTGAGCAACA-3′, reverse 5′-GGCCAGGTCCACACTCCATA-3′; Hprt: forward 5′-CTGGTGAAAAGGACCTCTCG-3′, reverse 5′-TGAAGTACTCATTATAGTCAAGGGCA-3′.
Single-cell labeling, capture, library preparation, and RNA sequencing
TCRβ+ CD4+ CD44+ PD1+ CXCR5+ Tfh cells from WT mice or IL-4+ (GFP) TCRβ+ CD4+ CD44+ cells from Il4 “4Get” reporter mice were sorted 7 days after immunization with Alternaria extract (10 μg) and NP19-OVA (25 μg). Cells were stimulated with PMA and ionomycin for 30 min. For both single-cell RNA sequencing (scRNA-seq) experiments, cell suspensions from each mouse sample were labeled with cell hashing antibodies (BioLegend TotalSeq anti-mouse Hashtag #3–6) according to the manufacturer’s protocol (BioLegend protocol #5009). Cells were washed and suspended in PBS containing 0.04% BSA and immediately processed as follows. Cells were counted on Countess II automated cell counter (ThermoFisher), and 12,000 cells (4,000 cells from each hashtagged mouse) were loaded onto one lane of a 10X Chromium microfluidic chip. Single cell capture, barcoding and library preparation were performed using the 10X Chromium platform, version 2 chemistry for the first experiment and version 3 chemistry for the second, and according to the manufacturer’s protocol (#CG00052) with modifications for generating the hashtag library (67) BioLegend protocol#5009. cDNA and libraries were checked for quality on Agilent 4200 Tapestation, quantified by KAPA qPCR, and pooled using a ratio of 95% gene expression library and 5% hashtag library before sequencing, one on a single lane of an Illumina HiSeq4000 and the other on 16.67% of lane of an Illumina NovaSeq 6000 S2 flow cell, both targeting 6,000 barcoded cells with an average sequencing depth of 50,000 reads per cell.
scRNA-seq data processing, quality control, and analysis
Illumina base call (bcl) files for the 10X V2 and V3 libraries were converted to FASTQ files using Cell Ranger mkfastq versions 2.2.0 and 3.02, respectively (10X Genomics). CellRanger count versions 2.2.0 and 3.0.2 (10X Genomics) were used to align reads from each library to the same mm10 reference genome (GRCm38.84, 10X Genomics reference version 2.1.0), generate digital counts matrices, and call viable cells (n=3,956 and n=7,674, respectively) (68). Processing and mapping statistics for both libraries are listed in table S1 and S2.
All of the following analyses were performed the using Scanpy Python package (version 1.3) (69). Biological replicate identities for each cell were captured by the use of hashtag oligo antibodies (HTO) as described in (67). HTO library FASTQs were processed through the CITESeq-Count tool (version 1.3.3, https://github.com/Hoohm/CITE-seq-Count), producing cell by HTO count matrices for both libraries. Biological replicate labels for each cell were inferred using the HTO count matrices as prescribed in the HTODemux function of the Seurat package (version 2.3.3, https://github.com/satijalab/seurat). Cells which were determined to be HTO multiplets (e.g. doublets, triplets) were excluded from further analysis. Cells were further excluded from downstream analysis based on filtering by the following criteria: UMI counts per cell, gene count per cell, fraction of transcripts mapped to mitochondrial genes, cumulative hemoglobin expression. This quality control process (shown in fig. S6, A and B) yielded n=3,811 and n=4,096 filtered cells for the Tfh cell and the IL-4+ CD4 T cell libraries, respectively, with specific exclusion criteria listed in Table S3. Genes were removed if they were expressed in fewer than three cells, resulting in filtered gene expression matrices 3,811 filtered cells by 12,861 filtered genes and 4096 filtered cells by 14,542 filtered genes, respectively.
The expression matrix of the Tfh cell-sorted library was normalized such that the median UMI counts in each cell was equal to the median UMI count across the dataset and then log transformed and scaled to zero mean and unit variance on a per-gene basis. 2D and 3D UMAP embeddings of the scaled, log-transformed single-cell expression profiles were produced as follows: (i) the 1200 most highly variable genes (HVGs), as measured by dispersion, were selected; (ii) using the scaled, transformed expression at these HVGs, computing the first 50 principal components (PCs); (iii) these PCs are used to construct a k=10 nearest neighbor graph (k-NN graph, where distance is measured using cosine distance); (iv) the UMAP embeddings were computed from this k-NN graph (70). We observe that cells from each of the biological replicates are well-mixed in the UMAP embeddings (fig. S6C) and each replicate contributes roughly equally to all clusters (fig. S6M). Cluster labels were assigned via the Leiden community detection algorithm on this k-NN graph, yielding 11 clusters (shown in fig. S6E) (71). Marker genes were identified via a “one-versus-rest” methodology comparing mean expression of every gene within each cluster which were used to assign putative subtypes. Representative markers used to identify populations of interest and exclude other populations are shown in fig. S6K. These marker genes were used to assign cellular identities to clusters. Specifically, we isolated clusters 1 to 6 and 8, consisting of n=3002 cells.
The expression matrix of the Il4/GFP+ sorted cell library was processed similarly as shown in fig. S6 B, D, F, and N. After a preliminary 2D embedding and cluster analysis, a small population of B cells was removed (fig. S6L) and the remaining n=3863 T cells were reanalyzed. To explore the specific subpopulations of IL-13+, all cells expressing one or more Il13 transcripts were isolated and analyzed separately. Six hundred HVGs were selected and we were careful to exclude mitochondrial and ribosomal genes, sex-linked genes such as Xist, and cell-cycle and proliferation genes from these HVGs. PCs were computed using the log-transformed, normalized expression signatures at these HVGs and the first ten PCs were used to construct a k=10 cosine distance kNN and 2D UMAP embedding. Low-resolution (0.2) Leiden community detection was used to identify three subpopulations of IL-13+ cells. All cells expressing one or more Bcl6 transcript were analyzed identically (as shown in fig. S6). Leiden community detection with resolution 0.1 was used to identify two subpopulations of Bcl6+ cells.
Differential expression analysis between pairs of clusters was performed using edgeR (version 3.24.3), where each cell acts as a single sample (72, 73). For each comparison, raw counts for cells in the two clusters of interest were isolated and processed using glmQLFTest assuming a two-group design matrix according to the edgeR User’s Guide. There are several tools designed to identify differential genes in single cell transcriptomic data; recent studies have shown edgeR and DESeq2 to balance precision and recall in DEG analysis, and to perform similar to if not better than DEG analysis tools designed for scRNA-seq (74).
Immunoblotting
CD4+ T cells were isolated using CD4 T cell purification kit through negative selection (Stem Cell Technologies). Three million cells were lysed in immunoprecipitation assay buffer with protease inhibitors (Roche) and cell lysates were run on SDS–PAGE and transferred onto nitrocellulose membrane. After blocking with 5% non-fat milk in Tris-buffered saline with 5% tween for 1 h at room temperature, membranes were incubated with anti-mouse DOCK8 antibody (Takara) at 4°C overnight. Goat anti-rabbit IgG(H+L)-HRP polyclonal detection antibody (1:5000) (Invitrogen) was incubated with membrane before assessing chemiluminescence signal with ECL substrate on ChemiDoc Imaging System (Bio-Rad). The housekeeping protein β-actin was used to normalize protein concentrations.
In vitro culture
Mediastinal lymph node cells were isolated from mice immunized with Alternaria extract (10 μg) and NP20-OVA (25 μg). Fifty thousand cells/well were cultured in 96-well U-bottom plates with 1 μg CD40 (HM40–3) in the presence of 2 ng of IL-4 (Peprotech) and 50 ng of IL-13 (R&D) for 3 daysd in complete RPMI 1640. IgE plasma cells were analyzed by flow cytometry.
Treg cell suppression assay
Naïve CD45.1+ CD4+ T cells were isolated by negative selection with magnetic beads (Miltenyi) and labelled with carboxyfluorescein succinimidyl ester (CFSE) (Life Technologies) and used as responders. Regulatory T cells (Treg), CD45.2+ CD4+ CD25+ from WT or Dock8−/− mice were isolated by negative selection followed by positive selection with Treg isolation kit (Miltenyi). The cells were plated at a concentration of 3×104 cells per well at ratio of 1 Treg:2 responder T cells. Cells were stimulated with 0.5 μg/ml of α-CD3 and 2 μg/ml of α-CD28. After incubation at 37°C for 72 h, proliferation of responder cells was assessed by flow cytometry and compared to no-Treg controls as an assessment of Treg suppressive abilities.
Mixed bone marrow chimera
Recipient WT CD45.1+ mice were irradiated with two doses of 600 rad 3 h apart. One or two hours after the second irradiation, 5×105 bone marrow cells from WT or Il13−/− mice mixed with 5×105 bone marrow cells from Cd4CreBcl6fl/fl mice were adoptively transferred by i.v. injection into irradiated recipient mice. All experiments with bone marrow chimeric mice were performed 1016 weeks after bone marrow transplant.
Dendritic cell migration assay
Mice were immunized intranasally with 50 μg of OVA-Alexa Fluor 647 (Molecular Probes) and 1 μg of LPS (Invivogen). MedLNs were harvested 18 h after immunization, minced, and digested with collagenase IV (1 mg/ml; Sigma-Aldrich) for 40 min at 37°C. Single-cell suspensions were prepared, stained, and then analyzed on an LSRII (BD Biosciences) or MACSQuant (Miltenyi Biotec) flow cytometer.
In vivo T cell proliferation and Tfh differentiation
To assess proliferation, CD4+ T cells from OT-II Dock8−/− (CD45.2+) or OT-II WT (CD45.1.2+) mice were prepared from the spleen and lymph nodes by negative selection using the EasySep CD4 T Cell Isolation kit (STEMCELL Technologies) according to the manufacturer’s instructions. For T cell proliferation assays, OTII cells were labeled with 1 μM carboxyfluorescein diacetate succinimidyl ester (CFSE) before transfer in to CD45.1+ recipient mice and CFSE dilution was used to assess proliferation. For Tfh cell identification, OT-II cells were not labeled with CFSE and were gated using CD44+CXCR5+PD1+. A total of 2×105 purified OTII cells were transferred into mice by retro-orbital injection. Mice were intranasally immunized 24 h later with 10 μg of NP-OVA (Biosearch) and 2 μg of LPS (Sigma). MedLNs were harvested 3 days (for T cell proliferation) or 6 days after immunization (for Tfh cell analyses). Single–cell suspensions were prepared, stained, and then analyzed on an LSRII flow cytometer (BD Biosciences).
Statistical analysis
ANOVA or Student’s t-test were performed on normally distributed data (analyzed by Shapiro–Wilk test). Mann–Whitney U or Kruskal–Wallis H tests were used otherwise. A P-value of <0.05 was considered significant. Data were analyzed with Prism 7 (GraphPad Software).
Supplementary Material
Acknowledgments:
We thank M. Firla for technical assistance, M. Wimsatt for the illustration, E. Gelfand (National Jewish Health, CO) for the Il13−/− bone marrow, C. Allen (UCSF) for IgE GC B cell staining protocol, R.D. Chow (Yale University) for discussions on analysis of scRNAseq data and The Single Cell Biology Laboratory led by P. Robson at The Jackson Laboratory for Genomic Medicine, for help with scRNA-seq experiments.
Funding: This work was supported by Food Allergy Research and Ira and Diana Riklis Family Research Award in Food Allergy and R01 AI136942 and R01 AI108829 (to S.C.E.) and by R21 AI135221 and R21 AI133440 (to A.W.). U.G. is a recipient of Gershon-Trudeau Fellowship from Immunobiology at Yale University. B.Z. is a recipient of Ph.D. fellowship awarded by Agency for Science, Technology, and Research, Singapore. T.S. is a recipient of Robert E. Leet and Clara Guthrie Patterson Trust Mentored Research Award.
Footnotes
Author contributions: U.G. and S.C.E. designed the study. U.G., J.S.C., M.A.C., J.G., B.Z., W.F.F., D.C., Y.L., W.S., J.J., J.A.G., L.X., J.S.W., M.C.B., A.W., and S.C.E. performed and/or analyzed experiments. U.G., J.S.C., A.W., and S.C.E wrote the manuscript. T.S., M.C.B., J.E.C., J.S.W., A.W., and S.C.E. provided resources, reagents and funding. A.W., and S.C.E supervised the study.
Competing interests: None.
Data and materials availability: The accession number for the RNA-seq datasets is GSE132798. The data is available in the Gene Expression Omnibus Database. All other data needed to evaluate the conclusions in this paper are present either in the main text or the supplementary materials.
References:
- 1.Oettgen HC, Geha RS, IgE regulation and roles in asthma pathogenesis. The Journal of allergy and clinical immunology 107, 429–441(2001); published online EpubMar ( 10.1067/mai.2001.113759). [DOI] [PubMed] [Google Scholar]
- 2.Mita H, Yasueda H, Akiyama K, Affinity of IgE antibody to antigen influences allergen-induced histamine release. Clin. Exp. Allergy : journal of the British Society for Allergy and Clinical Immunology 30, 15831589 (2000); published online EpubNov ( [DOI] [PubMed] [Google Scholar]
- 3.Suzuki R, Leach S, Liu W, Ralston E, Scheffel J, Zhang W, Lowell CA, Rivera J, Molecular editing of cellular responses by the high-affinity receptor for IgE. Science 343, 1021–1025 (2014); published online EpubFeb 28 ( 10.1126/science.1246976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Lin J, Bardina L, Goldis M, Nowak-Wegrzyn A, Shreffler WG, Sampson HA, Correlation of IgE/IgG4 milk epitopes and affinity of milk-specific IgE antibodies with different phenotypes of clinical milk allergy. The Journal of allergy and clinical immunology 125, 695 702 e691–702 e696 (2010); published online EpubMar ( 10.1016/j.jaci.2009.12.017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He JS, Subramaniam S, Narang V, Srinivasan K, Saunders SP, Carbajo D, Wen-Shan T, Hamadee N. Hidayah, Lum J, Lee A, Chen J, Poidinger M, Zolezzi F, Lafaille JJ, de Lafaille M. A. Curotto, IgG1 memory B cells keep the memory of IgE responses. Nat Commun 8, 641 (2017); published online EpubSep 21 ( 10.1038/s41467-017-00723-0). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong H, Dolpady J, Wabl M, de Lafaille M. A. Curotto, Lafaille JJ, Sequential class switching is required for the generation of high affinity IgE antibodies. The Journal of experimental medicine 209, 353–364 (2012); published online EpubFeb 13 ( 10.1084/jem.20111941). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimoda K, van Deursen J, Sangster MY, Sarawar SR, Carson RT, Tripp RA, Chu C, Quelle FW, Nosaka T, Vignali DA, Doherty PC, Grosveld G, Paul WE, Ihle JN, Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature 380, 630–633 (1996); published online EpubApr 18 ( 10.1038/380630a0). [DOI] [PubMed] [Google Scholar]
- 8.Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, Nakanishi K, Yoshida N, Kishimoto T, Akira S, Essential role of Stat6 in IL-4 signalling. Nature 380, 627–630 (1996); published online EpubApr 18 ( 10.1038/380627a0). [DOI] [PubMed] [Google Scholar]
- 9.Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, Killeen N, Urban JF Jr., Guo L, Paul WE, Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nature immunology 5, 1157–1165 (2004); published online EpubNov ( 10.1038/ni1128). [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi T, Iijima K, Dent AL, Kita H, Follicular helper T cells mediate IgE antibody response to airborne allergens. The Journal of allergy and clinical immunology 139, 300–313 e7 (2017); published online EpubJan ( 10.1016/j.jaci.2016.04.021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meli AP, Fontes G, Soo C. Leung, King IL, T Follicular Helper Cell-Derived IL-4 Is Required for IgE Production during Intestinal Helminth Infection. Journal of immunology, (2017); published online EpubMay 22 ( 10.4049/jimmunol.1700141). [DOI] [PubMed] [Google Scholar]
- 12.Noble A, Zhao J, Follicular helper T cells are responsible for IgE responses to Der p 1 following house dust mite sensitization in mice. Clin. Exp.Allergy : journal of the British Society for Allergy and Clinical Immunology 46, 1075–1082 (2016); published online EpubAug ( 10.1111/cea.12750). [DOI] [PubMed] [Google Scholar]
- 13.Dolence JJ, Kobayashi T, Iijima K, Krempski J, Drake LY, Dent AL, Kita H, Airway exposure initiates peanut allergy by involving the IL-1 pathway and T follicular helper cells in mice. The Journal of allergy and clinical immunology 142, 1144–1158. e8 (2018); published online EpubOct ( 10.1016/j.jaci.2017.11.020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang HE, Reinhardt RL, Bando JK, Sullivan BM, Ho IC, Locksley RM, Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nature immunology 13, 58–66 (2012); published online EpubJan ( 10.1038/ni.2182). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morita R, Schmitt N, Bentebibel SE, Ranganathan R, Bourdery L, Zurawski G, Foucat E, Dullaers M, Oh S, Sabzghabaei N, Lavecchio EM, Punaro M, Pascual V, Banchereau J, Ueno H, Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity 34, 108–121 (2011); published online EpubJan 28 ( 10.1016/j.immuni.2010.12.012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harada Y, Tanaka S, Motomura Y, Harada Y, Ohno S, Ohno S, Yanagi Y, Inoue H, Kubo M, The 3’ enhancer CNS2 is a critical regulator of interleukin-4-mediated humoral immunity in follicular helper T cells. Immunity 36, 188–200 (2012); published online EpubFeb 24 ( 10.1016/j.immuni.2012.02.002). [DOI] [PubMed] [Google Scholar]
- 17.Sahoo A, Alekseev A, Tanaka K, Obertas L, Lerman B, Haymaker C, Clise-Dwyer K, McMurray JS, Nurieva R, Batf is important for IL-4 expression in T follicular helper cells. Nat Commun 6, 7997 (2015); published online EpubAug 17 ( 10.1038/ncomms8997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vijayanand P, Seumois G, Simpson LJ, Abdul-Wajid S, Baumjohann D, Panduro M, Huang X, Interlandi J, Djuretic IM, Brown DR, Sharpe AH, Rao A, Ansel KM, Interleukin-4 production by follicular helper T cells requires the conserved Il4 enhancer hypersensitivity site V. Immunity 36, 175–187 (2012); published online EpubFeb 24 ( 10.1016/j.immuni.2011.12.014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams A, Lee GR, Spilianakis CG, Hwang SS, Eisenbarth SC, Flavell RA, Hypersensitive site 6 of the Th2 locus control region is essential for Th2 cytokine expression. Proceedings of the National Academy of Sciences of the United States of America 110, 6955–6960 (2013); published online EpubApr 23 ( 10.1073/pnas.1304720110). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu J, T helper 2 (Th2) cell differentiation, type 2 innate lymphoid cell (ILC2) development and regulation of interleukin-4 (IL-4) and IL-13 production. Cytokine 75, 14–24 (2015); published online EpubSep ( 10.1016/j.cyto.2015.05.010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinstein JS, Herman EI, Lainez B, Licona-Limon P, Esplugues E, Flavell R, Craft J, TFH cells progressively differentiate to regulate the germinal center response. Nature immunology 17, 1197–1205 (2016); published online EpubOct ( 10.1038/ni.3554). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yusuf I, Kageyama R, Monticelli L, Johnston RJ, Ditoro D, Hansen K, Barnett B, Crotty S, Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150). Journal of immunology 185, 190–202 (2010); published online EpubJul 01 ( 10.4049/jimmunol.0903505). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Yan X, Zhong B, Nurieva RI, Wang A, Wang X, Martin-Orozco N, Wang Y, Chang SH, Esplugues E, Flavell RA, Tian Q, Dong C, Bcl6 expression specifies the T follicular helper cell program in vivo. The Journal of experimental medicine 209, 1841–1852, (2012); published online EpubSep 24 ( 10.1084/jem.20120219). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su HC, Jing H, Zhang Q, DOCK8 deficiency. Ann N Y Acad Sci 1246, 26–33 (2011); published online EpubDec ( 10.1111/j.1749-6632.2011.06295.x). [DOI] [PubMed] [Google Scholar]
- 25.Su HC, Jing H, Angelus P, Freeman AF, Insights into immunity from clinical and basic science studies of DOCK8 immunodeficiency syndrome. Immunological reviews 287, 9–19 (2019); published online EpubJan ( 10.1111/imr.12723). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krishnaswamy JK, Gowthaman U, Zhang B, Mattsson J, Szeponik L, Liu D, Wu R, White T, Calabro S, Xu L, Collet MA, Yurieva M, Alsen S, Fogelstrand P, Walter A, Heath WR, Mueller SN, Yrlid U, Williams A, Eisenbarth SC, Migratory CD11b(+) conventional dendritic cells induce T follicular helper cell-dependent antibody responses. Sci Immunol 2, (2017); published online EpubDec 1 ( 10.1126/sciimmunol.aam9169). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Randall KL, Lambe T, Johnson AL, Johnson A, Treanor B, Kucharska E, Domaschenz H, Whittle B, Tze LE, Enders A, Crockford TL, Bouriez-Jones T, Alston D, Cyster JG, Lenardo MJ, Mackay F, Deenick EK, Tangye SG, Chan TD, Camidge T, Brink R, Vinuesa CG, Batista FD, Cornall RJ, Goodnow CC, Dock8 mutations cripple B cell immunological synapses, germinal centers and long-lived antibody production. Nature immunology 10, 1283–1291 (2009); published online EpubDec 1 ( 10.1038/ni.1820). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finkelman FD, Rothenberg ME, Brandt EB, Morris SC, Strait RT, Molecular mechanisms of anaphylaxis: lessons from studies with murine models. The Journal of allergy and clinical immunology 115, 449–457; (2005); published online EpubMar ( 10.1016/j.jaci.2004.12.1125). [DOI] [PubMed] [Google Scholar]
- 29.Janssen E, Morbach H, Ullas S, Bannock JM, Massad C, Menard L, Barlan I, Lefranc G, Su H, Dasouki M, Al-Herz W, Keles S, Chatila T, Geha RS, Meffre E, Dedicator of cytokinesis 8-deficient patients have a breakdown in peripheral B-cell tolerance and defective regulatory T cells. The Journal of allergy and clinical immunology 134, 1365–1374 (2014); published online EpubDec ( 10.1016/j.jaci.2014.07.042). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh AK, Eken A, Hagin D, Komal K, Bhise G, Shaji A, Arkatkar T, Jackson SW, Bettelli E, Torgerson TR, Oukka M, DOCK8 regulates fitness and function of regulatory T cells through modulation of IL2 signaling. JCI Insight 2, (2017); published online EpubOct 5 ( 10.1172/jci.insight.94275). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zotos D, Coquet JM, Zhang Y, Light A, D’Costa K, Kallies A, Corcoran LM, Godfrey DI, Toellner KM, Smyth MJ, Nutt SL, Tarlinton DM, IL-21 regulates germinal center B cell differentiation and proliferation through a B cell-intrinsic mechanism. The Journal of experimental medicine 207, 365–378 (2010); published online EpubFeb 15 ( 10.1084/jem.20091777). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozaki K, Spolski R, Feng CG, Qi CF, Cheng J, Sher A, Morse HC 3rd, Liu C, Schwartzberg PL, Leonard WJ, A critical role for IL-21 in regulating immunoglobulin production. Science 298, 1630–1634 (2002); published online EpubNov 22 ( 10.1126/science.1077002). [DOI] [PubMed] [Google Scholar]
- 33.Suto A, Nakajima H, Hirose K, Suzuki K, Kagami S, Seto Y, Hoshimoto A, Saito Y, Foster DC, Iwamoto I, Interleukin 21 prevents antigen-induced IgE production by inhibiting germ line C(epsilon) transcription of IL-4-stimulated B cells. Blood 100, 4565–4573 (2002); published online EpubDec 15 ( 10.1182/blood-2002-04-1115). [DOI] [PubMed] [Google Scholar]
- 34.Wood N, Bourque K, Donaldson DD, Collins M, Vercelli D, Goldman SJ, Kasaian MT, IL-21 effects on human IgE production in response to IL-4 or IL-13. Cellular immunology 231, 133–145 (2004); published online EpubSep-Oct ( 10.1016/j.cellimm.2005.01.001). [DOI] [PubMed] [Google Scholar]
- 35.Takatsu K, Kouro T, Nagai Y, Interleukin 5 in the link between the innate and acquired immune response. Adv Immunol 101, 191–236 (2009) 10.1016/S0065-2776(08)01006-7). [DOI] [PubMed] [Google Scholar]
- 36.Kubo M, T follicular helper and TH2 cells in allergic responses. Allergol Int 66, 377–381 (2017); published online EpubJul ( 10.1016/j.alit.2017.04.006). [DOI] [PubMed] [Google Scholar]
- 37.Mohrs M, Shinkai K, Mohrs K, Locksley RM, Analysis of type 2 immunity in vivo with a bicistronic IL4 reporter. Immunity 15, 303–311 (2001); published online EpubAug ( [DOI] [PubMed] [Google Scholar]
- 38.Debeuf N, Haspeslagh E, van Helden M, Hammad H, Lambrecht BN, Mouse Models of Asthma. Curr Protoc Mouse Biol 6, 169–184 (2016); published online EpubJun 1 ( 10.1002/cpmo.4). [DOI] [PubMed] [Google Scholar]
- 39.Li XM, Serebrisky D, Lee SY, Huang CK, Bardina L, Schofield BH, Stanley JS, Burks AW, Bannon GA, Sampson HA, A murine model of peanut anaphylaxis: T-and B-cell responses to a major peanut allergen mimic human responses. The Journal of allergy and clinical immunology 106, 150–158 (2000); published online EpubJul ( 10.1067/mai.2000.107395). [DOI] [PubMed] [Google Scholar]
- 40.Chiang D, Chen X, Jones SM, Wood RA, Sicherer SH, Burks AW, Leung DYM, Agashe C, Grishin A, Dawson P, Davidson WF, Newman L, Sebra R, Merad M, Sampson HA, Losic B, Berin MC, Single-cell profiling of peanut-responsive T cells in patients with peanut allergy reveals heterogeneous effector TH2 subsets. The Journal of allergy and clinical immunology 141, 2107–2120 (2018); published online EpubJun ( 10.1016/j.jaci.2017.11.060). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Jackson KJ, Chen Z, Gaeta BA, Siba PM, Pomat W, Walpole E, Rimmer J, Sewell WA, Collins AM, IgE sequences in individuals living in an area of endemic parasitism show little mutational evidence of antigen selection. Scand J Immunol 73, 496–504 (2011); published online EpubMay ( 10.1111/j.1365-3083.2011.02525.x). [DOI] [PubMed] [Google Scholar]
- 42.McKenzie GJ, Emson CL, Bell SE, Anderson S, Fallon P, Zurawski G, Murray R, Grencis R, McKenzie AN, Impaired development of Th2 cells in IL-13-deficient mice. Immunity 9, 423–432 (1998); published online EpubSep ( [DOI] [PubMed] [Google Scholar]
- 43.McKenzie GJ, Fallon PG, Emson CL, Grencis RK, McKenzie AN, Simultaneous disruption of interleukin (IL)-4 and IL-13 defines individual roles in T helper cell type 2-mediated responses. The Journal of experimental medicine 189, 1565–1572 (1999); published online EpubMay 17 ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramalingam TR, Pesce JT, Sheikh F, Cheever AW, Mentink-Kane MM, Wilson MS, Stevens S, Valenzuela DM, Murphy AJ, Yancopoulos GD, Urban JF Jr., Donnelly RP, Wynn TA, Unique functions of the type II interleukin 4 receptor identified in mice lacking the interleukin 13 receptor alpha1 chain. Nature immunology 9, 25–33 (2008); published online EpubJan ( 10.1038/ni1544). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ballesteros-Tato A, Randall TD, Lund FE, Spolski R, Leonard WJ, Leon B, T Follicular Helper Cell Plasticity Shapes Pathogenic T Helper 2 Cell-Mediated Immunity to Inhaled House Dust Mite. Immunity 44, 259–273 (2016); published online EpubFeb 16 ( 10.1016/j.immuni.2015.11.017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zaretsky A. Glatman, Taylor JJ, King IL, Marshall FA, Mohrs M, Pearce EJ, T follicular helper cells differentiate from Th2 cells in response to helminth antigens. The Journal of experimental medicine 206, 991–999 (2009); published online EpubMay 11 ( 10.1084/jem.20090303). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu KT, Johnson TA, Sun HW, Vahedi G, Hakim O, Handon R, Schwartzberg PL, Hager GL, O’Shea JJ, Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity 35, 919–931 (2011); published online EpubDec 23 ( 10.1016/j.immuni.2011.11.012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keles S, Charbonnier LM, Kabaleeswaran V, Reisli I, Genel F, Gulez N, Al-Herz W, Ramesh N, Perez-Atayde A, Karaca NE, Kutukculer N, Wu H, Geha RS, Chatila TA, Dedicator of cytokinesis 8 regulates signal transducer and activator of transcription 3 activation and promotes TH17 cell differentiation. The Journal of allergy and clinical immunology 138, 1384–1394.e2 (2016); published online EpubNov ( 10.1016/j.jaci.2016.04.023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kearney CJ, Randall KL, Oliaro J, DOCK8 regulates signal transduction events to control immunity. Cell Mol Immunol 14, 406–411 (2017); published online EpubMay ( 10.1038/cmi.2017.9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu H, Xu LL, Teuscher P, Liu H, Kaplan MH, Dent AL, An Inhibitory Role for the Transcription Factor Stat3 in Controlling IL-4 and Bcl6 Expression in Follicular Helper T Cells. Journal of immunology 195, 2080–2089 (2015); published online EpubSep 1 ( 10.4049/jimmunol.1500335). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Erazo A, Kutchukhidze N, Leung M, Christ AP, Urban JF Jr., de Lafaille M. A. Curotto, Lafaille JJ, Unique maturation program of the IgE response in vivo. Immunity 26, 191–203 (2007); published online EpubFeb ( 10.1016/j.immuni.2006.12.006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yazdanbakhsh M, Kremsner PG, van Ree R, Allergy, parasites, and the hygiene hypothesis. Science 296, 490–494 (2002); published online EpubApr 19 ( 10.1126/science.296.5567.490). [DOI] [PubMed] [Google Scholar]
- 53.He JS, Narayanan S, Subramaniam S, Ho WQ, Lafaille JJ, de Lafaille M. A. Curotto, Biology of IgE production: IgE cell differentiation and the memory of IgE responses. Curr Top Microbiol Immunol 388, 119 (2015) 10.1007/978-3-319-13725-4_1). [DOI] [PubMed] [Google Scholar]
- 54.Fitzsimmons CM, Falcone FH, Dunne DW, Helminth Allergens, Parasite-Specific IgE, and Its Protective Role in Human Immunity. Front Immunol 5, 61 (2014) 10.3389/fimmu.2014.00061). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Looney TJ, Lee JY, Roskin KM, Hoh RA, King J, Glanville J, Liu Y, Pham TD, Dekker CL, Davis MM, Boyd SD, Human B-cell isotype switching origins of IgE. The Journal of allergy and clinical immunology 137, 579–586.e7 (2016); published online EpubFeb ( 10.1016/j.jaci.2015.07.014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoh RA, Joshi SA, Liu Y, Wang C, Roskin KM, Lee JY, Pham T, Looney TJ, Jackson KJL, Dixit VP, King J, Lyu SC, Jenks J, Hamilton RG, Nadeau KC, Boyd SD, Single B-cell deconvolution of peanut-specific antibody responses in allergic patients. The Journal of allergy and clinical immunology 137, 157–167 (2016); published online EpubJan ( 10.1016/j.jaci.2015.05.029). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCormick SM, Heller NM, Commentary: IL-4 and IL-13 receptors and signaling. Cytokine 75, 38–50 (2015); published online EpubSep ( 10.1016/j.cyto.2015.05.023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.LaPorte SL, Juo ZS, Vaclavikova J, Colf LA, Qi X, Heller NM, Keegan AD, Garcia KC, Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell 132, 259272 (2008); published online EpubJan 25 ( 10.1016/j.cell.2007.12.030). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pattarini L, Trichot C, Bogiatzi S, Grandclaudon M, Meller S, Keuylian Z, Durand M, Volpe E, Madonna S, Cavani A, Chiricozzi A, Romanelli M, Hori T, Hovnanian A, Homey B, Soumelis V, TSLPactivated dendritic cells induce human T follicular helper cell differentiation through OX40-ligand. The Journal of experimental medicine 214, 1529–1546 (2017); published online EpubMay 01 ( 10.1084/jem.20150402). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim CJ, Lee CG, Jung JY, Ghosh A, Hasan SN, Hwang SM, Kang H, Lee C, Kim GC, Rudra D, Suh CH, Im SH, The Transcription Factor Ets1 Suppresses T Follicular Helper Type 2 Cell Differentiation to Halt the Onset of Systemic Lupus Erythematosus. Immunity 49, 1034–1048.e8 (2018); published online EpubDec 18 ( 10.1016/j.immuni.2018.10.012). [DOI] [PubMed] [Google Scholar]
- 61.Ashley SE, Tan HT, Peters R, Allen KJ, Vuillermin P, Dharmage SC, Tang MLK, Koplin J, Lowe A, Ponsonby AL, Molloy J, Matheson MC, Saffery R, Ellis JA, Martino D, t. HealthNuts, Genetic variation at the Th2 immune gene IL13 is associated with IgE-mediated paediatric food allergy. Clin. Exp. Allergy : journal of the British Society for Allergy and Clinical Immunology 47, 1032–1037 (2017); published online EpubAug ( 10.1111/cea.12942). [DOI] [PubMed] [Google Scholar]
- 62.Pykalainen M, Kinos R, Valkonen S, Rydman P, Kilpelainen M, Laitinen LA, Karjalainen J, Nieminen M, Hurme M, Kere J, Laitinen T, Lahesmaa R, Association analysis of common variants of STAT6, GATA3, and STAT4 to asthma and high serum IgE phenotypes. The Journal of allergy and clinical immunology 115, 80–87 (2005); published online EpubJan ( 10.1016/j.jaci.2004.10.006). [DOI] [PubMed] [Google Scholar]
- 63.Huebner M, Kim DY, Ewart S, Karmaus W, Sadeghnejad A, Arshad SH, Patterns of GATA3 and IL13 gene polymorphisms associated with childhood rhinitis and atopy in a birth cohort. The Journal of allergy and clinical immunology 121, 408–414 (2008); published online EpubFeb ( 10.1016/j.jaci.2007.09.020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krug N, Hohlfeld JM, Buhl R, Renz J, Garn H, Renz H, Blood eosinophils predict therapeutic effects of a GATA3-specific DNAzyme in asthma patients. The Journal of allergy and clinical immunology, (2017); published online EpubMar 22 ( 10.1016/j.jaci.2017.02.024). [DOI] [PubMed] [Google Scholar]
- 65.Yang Z, Sullivan BM, Allen CD, Fluorescent in vivo detection reveals that IgE(+) B cells are restrained by an intrinsic cell fate predisposition. Immunity 36, 857–872 (2012); published online EpubMay 25 ( 10.1016/j.immuni.2012.02.009). [DOI] [PubMed] [Google Scholar]
- 66.He JS, Meyer-Hermann M, Xiangying D, Zuan LY, Jones LA, Ramakrishna L, de Vries VC, Dolpady J, Aina H, Joseph S, Narayanan S, Subramaniam S, Puthia M, Wong G, Xiong H, Poidinger M, Urban JF, Lafaille JJ, de Lafaille M. A. Curotto, The distinctive germinal center phase of IgE+ B lymphocytes limits their contribution to the classical memory response. The Journal of experimental medicine 210, 2755–2771 (2013); published online EpubNov 18 ( 10.1084/jem.20131539). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stoeckius M, Zheng S, Houck-Loomis B, Hao S, Yeung BZ, Mauck WM 3rd, Smibert P, Satija R, Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol 19, 224 (2018); published online EpubDec 19 ( 10.1186/s13059-018-1603-1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH, Massively parallel digital transcriptional profiling of single cells. Nat Commun 8, 14049 (2017); published online EpubJan 16 ( 10.1038/ncomms14049). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wolf FA, Angerer P, Theis FJ, SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15 (2018); published online EpubFeb 6 ( 10.1186/s13059-017-1382-0). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, Newell EW, Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol, (2018); published online EpubDec 3 ( 10.1038/nbt.4314). [DOI] [PubMed] [Google Scholar]
- 71.Traag VA, Waltman L, van Eck NJ, From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep 9, 5233 (2019); published online EpubMar 26 ( 10.1038/s41598-019-41695-z). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010); published online EpubJan 1 ( 10.1093/bioinformatics/btp616). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lun AT, Chen Y, Smyth GK, It’s DE-licious: A Recipe for Differential Expression Analyses of RNAseq Experiments Using Quasi-Likelihood Methods in edgeR. Methods in molecular biology 1418, 391–416 (2016) 10.1007/978-1-4939-3578-9_19). [DOI] [PubMed] [Google Scholar]
- 74.Dal Molin A, Baruzzo G, Di Camillo B, Single-Cell RNA-Sequencing: Assessment of Differential Expression Analysis Methods. Front Genet 8, 62 (2017) 10.3389/fgene.2017.00062). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.