Abstract
In the endoplasmic reticulum (ER), ER oxidoreductin 1 (ERO1) catalyzes intramolecular disulfide-bond formation within its substrates in coordination with protein-disulfide isomerase (PDI) and related enzymes. However, the molecular mechanisms that regulate the ERO1–PDI system in plants are unknown. Reduction of the regulatory disulfide bonds of the ERO1 from soybean, GmERO1a, is catalyzed by enzymes in five classes of PDI family proteins. Here, using recombinant proteins, vacuum-ultraviolet circular dichroism spectroscopy, biochemical and protein refolding assays, and quantitative immunoblotting, we found that GmERO1a activity is regulated by reduction of intramolecular disulfide bonds involving Cys-121 and Cys-146, which are located in a disordered region, similarly to their locations in human ERO1. Moreover, a GmERO1a variant in which Cys-121 and Cys-146 were replaced with Ala residues exhibited hyperactive oxidation. Soybean PDI family proteins differed in their ability to regulate GmERO1a. Unlike yeast and human ERO1s, for which PDI is the preferred substrate, GmERO1a directly transferred disulfide bonds to the specific active center of members of five classes of PDI family proteins. Of these proteins, GmPDIS-1, GmPDIS-2, GmPDIM, and GmPDIL7 (which are group II PDI family proteins) failed to catalyze effective oxidative folding of substrate RNase A when there was an unregulated supply of disulfide bonds from the C121A/C146A hyperactive mutant GmERO1a, because of its low disulfide-bond isomerization activity. We conclude that regulation of plant ERO1 activity is particularly important for effective oxidative protein folding by group II PDI family proteins.
Keywords: endoplasmic reticulum (ER), protein-disulfide isomerase, protein folding, disulfide, oxidation-reduction (redox), ER oxidoreductin 1 (ERO1), GmERO1a, intrinsically disordered region, isomerization
Introduction
The folding of most proteins synthesized in the endoplasmic reticulum (ER)5 involves intramolecular disulfide-bond formation catalyzed by protein-disulfide isomerase (PDI) (EC 5.3.4.1) and other PDI family proteins (1–4). PDI, composed of four thioredoxin-folded domains (designated a, b, b′, and a′), catalyzes the formation, isomerization, and reduction of disulfide bonds via one or both of the CXXC active centers, which are located in domains a and a′ and assist in oxidative protein folding (Fig. 1). Disulfides in the active centers of PDI family proteins are reduced by catalyzing the oxidation of substrate dithiol groups. The reduced active center of PDI must then be re-oxidized prior to catalyzing the next oxidation reaction.
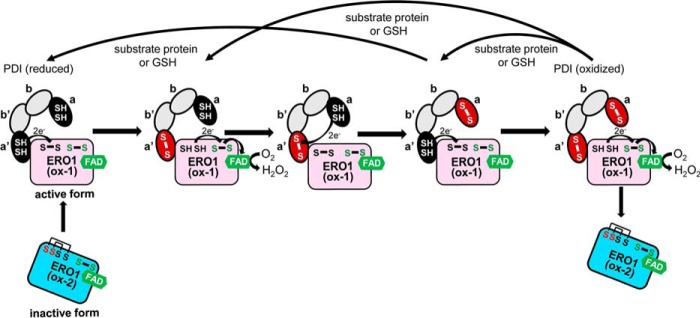
Figure 1.
Model of inter- and intramolecular electron transfer cascade between ERO1 and PDI in humans. In the oxidation of PDI by ERO1, ERO1 transfers electrons from the active center of PDI domain a′ to O2. In PDI, electrons are transferred intramolecularly from domain a to domain a′. The black ellipses indicate the reduced domains a or a′ of PDI, and the red ellipses indicate the oxidized domains. The gray ellipses indicate PDI domain b or b′. For ERO1, the two SH and S–S (black text) indicate the thiols and disulfide of the shuttle cysteine pair, and the S–S (green text) indicates the disulfide of the active cysteine pair. In inactive form (ox-2) of ERO1, the regulatory cysteines (red) formed disulfide bonds with the shuttle cysteines, which are reduced in active form (ox-1).
Yeast ER oxidoreductin 1p (ERO1p) (5, 6), mammalian ERO1 (ERO1α and ERO1β) (7, 8), and plant ERO1 enzymes (9–11) reportedly oxidize PDI family proteins, and the respective enzymatic properties have been characterized. ERO1p, ERO1α, and ERO1β preferentially oxidize PDI but not other PDI family proteins (ERp57, ERp72, P5, and ERp46) (12, 13). By contrast, plant ERO1 (soybean ERO1: GmERO1a) exhibits broad substrate specificity, oxidizing five PDI family proteins: GmPDIL-1, GmPDIM, GmPDIS-1, GmPDIS-2, and GmPDIL7 (10). The two active centers of PDI are known to undergo asymmetric oxidation catalyzed by ERO1s. In the yeast PDI ortholog (Pdi1p), the active center of domain a is preferentially oxidized by ERO1p (12). By contrast, human ERO1α oxidizes the active center of PDI domain a′ (14–16).
Such asymmetric active-center oxidation by PDIs provides a major clue as to how the enzymes catalyze both oxidation and isomerization/reduction reactions. It was suggested that yeast Pdi1p domain a catalyzes substrate oxidation, whereas domain a′ catalyzes isomerase or reductase reactions (14). Asymmetric oxidation of human PDI results from docking of ERO1α to PDI domain b′, which is also a substrate-binding site and aligns the active centers of ERO1α and PDI a′ (17). Because the a active center of human PDI can be oxidized via intramolecular thiol-disulfide exchange with the a′ active center, PDI domain a can also oxidize substrates.
Previously, we found that GmERO1a preferentially oxidizes the a′ active center of GmPDIM, a plant ortholog of mammalian P5 (10). Because the GmPDIM a active center can be oxidized via intramolecular thiol-disulfide exchange with the a′ active center, both domains a and a′ catalyze substrate oxidation. GmPDIM oxidizes both the a and a′ active centers of GmPDIL-2, which facilitates oxidative folding of substrate proteins. However, the specificity of GmERO1a for active centers of other PDI family proteins, including the PDI ortholog, has not been characterized to date.
The active center of ERO1s, known as the “shuttle cysteine pair,” oxidizes PDI and transfers the resulting electrons to oxygen molecules in concert with flavin adenine dinucleotide (FAD) via another active center known as the “active cysteine pair,” generating one hydrogen peroxide (H2O2) molecule per disulfide bond (18–23). Hence, ERO1-catalyzed oxidation results in continual consumption of cellular glutathione (GSH) and accumulation of cytotoxic reactive oxygen species from H2O2 as a side product (19). As H2O2 is cytotoxic when overproduced, the oxidation mediated by ERO1s and reducing power (import of GSH into the ER lumen) are tightly regulated to link disulfide-bond formation with the overall ER redox status (24, 25). The activity of yeast ERO1p is regulated by reduction of the disulfide bond between the thiols of noncatalytic regulatory cysteine residues (Cys-150 and Cys-295) in the mechanism of oxidizing power control in the ER. Reduction of this disulfide bond induces conformational changes that enable the transfer of electrons from the thiols of the shuttle cysteine residues to those of the active-cysteine residues. Human ERO1α is also regulated via reduction of the disulfide bond between thiols of shuttle Cys-94 and noncatalytic Cys-131 and shuttle Cys-99 and noncatalytic Cys-104, which directly blocks the pair of shuttle cysteines (26). A similar regulatory mechanism was reported for ERO1β (27). In ERO1α, another regulatory disulfide bond formed between Cys-208 and Cys-241 limits access to the FAD moiety by O2 molecules (28). Such regulatory reduction of disulfide bonds is catalyzed by reduced PDI and other PDI family proteins (29). A third regulation system involving the formation of a dynamic mixed–disulfide complex with Cys-166 and Cys-165 in both the human ERO1 isoforms and PDI has been reported (30).
Several systems in mammalian cells function to quench H2O2 produced by the PDI/ERO1 pathway and thus mitigate H2O2 cytotoxicity. Peroxiredoxin IV (PRDX4) (31–34), PDI peroxidases (GPx7 and GPx8) (35), and vitamin K epoxide reductase use H2O2 to oxidize PDI (36). These reaction systems are thus also useful as adjuncts to the PDI/ERO1 pathway. In addition, these enzymes exhibit a relatively broad specificity for PDI family proteins. Erp46 and P5 are oxidized by PRDX4, and ERp72, ERp57, and P5 are oxidized by GPx7/8 (i.e. oxidizing equivalents are supplied to multiple PDI family proteins by PRDX4 and Gpx7/8 using H2O2 generated by ERO1α). A type II transmembrane GSH peroxidase–like protein located in the ER and Golgi apparatus of plants has also been described (37). However, whether this enzyme functions as an adjunct to the PDI/ERO1 system in the ER like GPx7/8 has not been determined. In addition, the mechanism regulating the ERO1/PDI system in plants remains to be elucidated.
In this study, we examined the mechanism by which plant ERO1 activity is regulated and the relationship of this mechanism to oxidative protein folding. We found that plant ERO1 activity is regulated by the reduction of intramolecular disulfide bonds and that this regulatory mechanism is essential for effective oxidative protein folding mediated by various PDI family proteins. Based on these data, we propose that the regulation of ERO1 activity functions as a safety mechanism that maintains the integrity of the oxidative protein folding process.
Results
Plant ERO1 activity is regulated by redox of intramolecular disulfide bonds
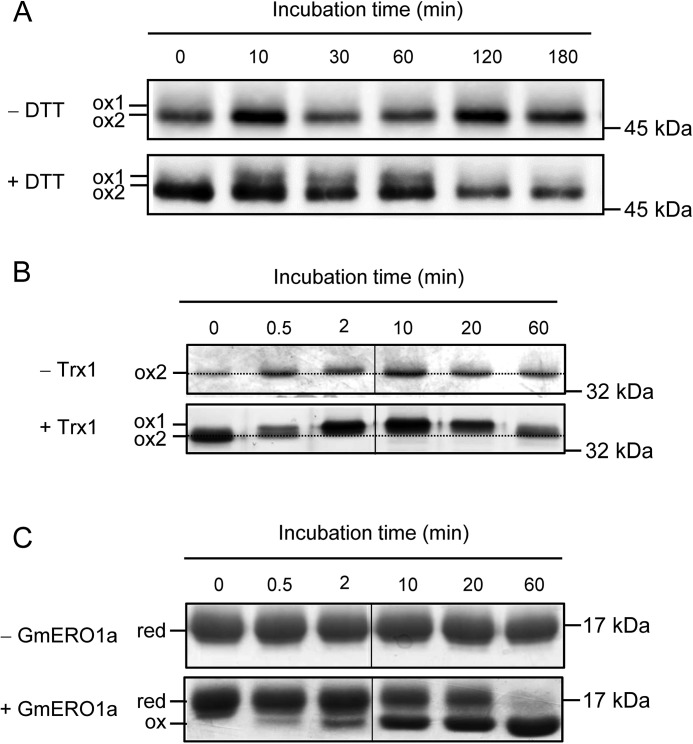
Both human and yeast ERO1 are activated in vivo under reducing conditions created by the addition of dithiothreitol (DTT). The active form of ERO1 is detectable as a band known as “ox-1” by SDS-PAGE under nonreducing conditions (38). Incubation of ERO1 with Escherichia coli thioredoxin 1 (Trx1), which is known to be a good substrate for ERO1s, reportedly generates ox-1 in vitro (24). Therefore, to determine whether plant ERO1 activity is regulated in a similar manner in yeast and humans, we examined the redox status of GmERO1a in cultured soybean cells treated with DTT, and also in vitro during the reaction with Trx1. After treatment of soybean culture cells with DTT, a new GmERO1 band (ox-1) appeared on SDS-PAGE, with mobility on the gel lower than that of original GmERO1 (ox-2) (Fig. 2A). Upon incubation of recombinant GmERO1a with Trx1, GmERO1a incubated for 2 min migrated slower through the gel than the un-incubated control, indicating the reduction of intramolecular disulfide bonds (Fig. 2B). As Trx1 became fully oxidized by 60 min (Fig. 2C), GmERO1a migrated faster, indicating re-formation of the intramolecular disulfide bonds in GmERO1a. These results strongly suggested that activity of plant ERO1, similar to yeast and human ERO1, is regulated via reduction of intramolecular disulfide bonds.
Figure 2.
Change in GmERO1a redox status. A, cultured soybean cells (DG330) were incubated with or without 1 mm DTT at 25 °C. Change in GmERO1a redox status was analyzed by nonreducing SDS-PAGE and Western blotting using anti-GmERO1 serum after N-ethylmaleimide treatment of the proteins extracted from cultured soybean cells. B and C, change in GmERO1a (B) and Trx1 (C) redox status during oxidation catalyzed by GmERO1a was analyzed by nonreducing SDS-PAGE after alkylation with N-ethylmaleimide (B) and 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (C) treatment. Proteins were stained with silver-staining (B) or Coomassie Brilliant Blue G-250 (C). B, dotted line indicates the position of ox-2 band. C, red, reduced Trx1; ox, oxidized Trx1.
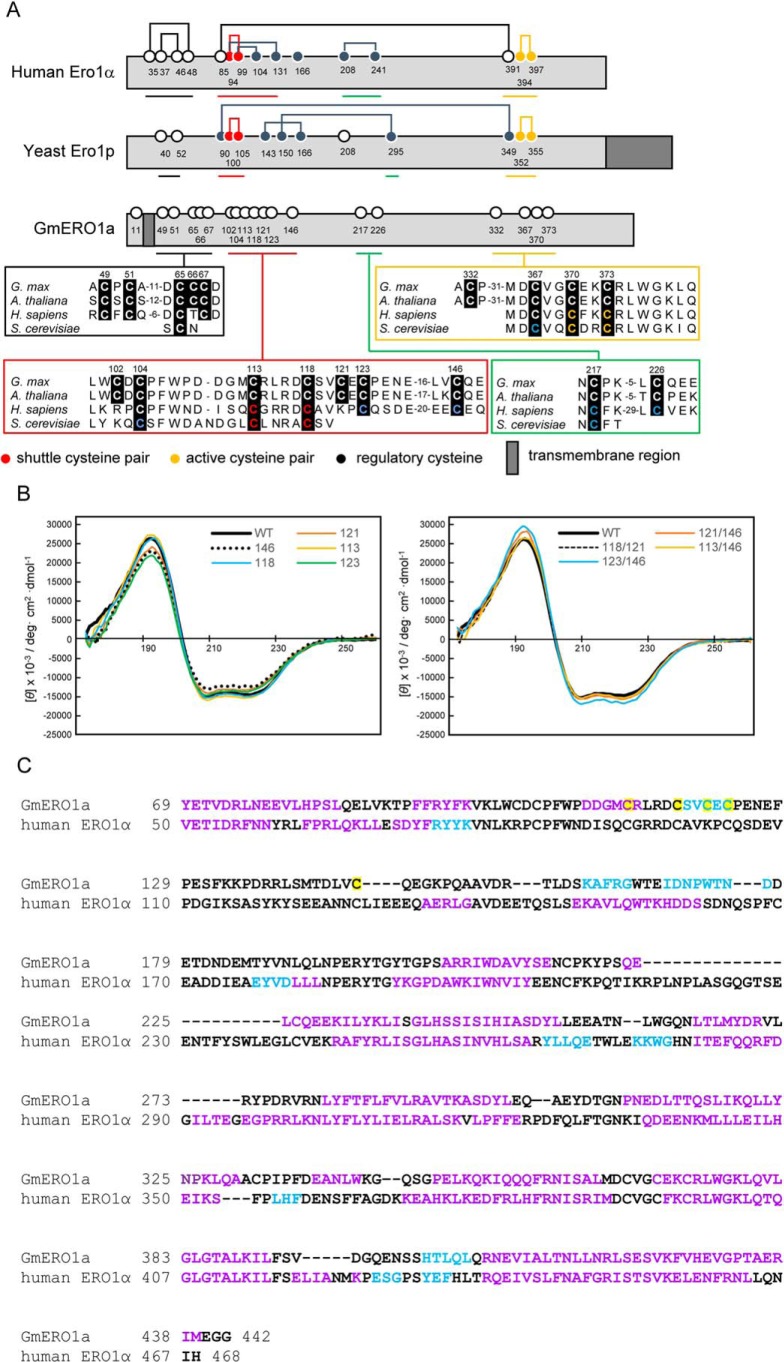
Next, we tried to identify the cysteine residues involved in the regulation of GmERO1a activity. Alignment of the amino acid sequences of human ERO1α, yeast ERO1p, and plant ERO1 (GmERO1a) revealed that active-center cysteines (shuttle cysteine pair and active cysteine pair) are conserved (Fig. 3A). In adding to the active-center cysteines, several regulatory cysteines in human ERO1α were also conserved in plants. In studies of the crystal structures, both the shuttle cysteines (Cys-94 and Cys-99) and regulatory cysteines (Cys-104 and Cys-131) of human ERO1α were found to be located in a single intrinsically flexible loop region (22). Unfortunately, no crystals of recombinant GmERO1a were obtained, despite repeated attempts. Therefore, vacuum-UV CD (VUVCD) spectra of recombinant GmERO1a were collected over the wavelength range 172–260 nm to compare the secondary structure of GmERO1a with that of human ERO1α (Fig. 3B). The spectrum exhibited two negative peaks around 222 and 208 nm and one positive peak around 190 nm, indicating that the recombinant GmERO1a had high α-helix content. The secondary structural contents of recombinant GmERO1a were estimated as 51.1% α-helix and 5.5% β-strand and the number of α-helix segments to be 20 (Table S1). These estimated contents by VUVCD analysis are similar to those of human ERO1α obtained from the crystal structure (48.9% α-helix and 6.4% β-strand) (Table S1). The positions of secondary structure of GmERO1 on the amino acid sequence are predicted by combining a neural network (NN) analysis treating the amino acid sequence with the secondary structure data obtained from the VUVCD analysis (VUVCD–NN method) (Fig. 3C). Conserved putative shuttle cysteines (Cys-113 and Cys-118) and adjacent putative regulatory cysteines (Cys-121, Cys-123, and Cys-146) were located in a disordered region of GmERO1a, analogous to the context of shuttle and regulatory cysteines in human ERO1α.
Figure 3.
Secondary structure of GmERO1a. A, schematic illustration and alignment of amino acid sequences around the cysteine residues of GmERO1a with human ERO1α and yeast ERO1p. Numbered circles indicate positions of cysteine residues. Lines connecting circles indicate disulfide bonds. B, VUVCD spectra of recombinant wildtype (WT) GmERO1a and mutants in which cysteine residues were substituted with alanine residues. Colored lines in each spectrum indicate mutants in which the indicated cysteine residues were replaced with alanine residues. The number on each spectrum indicates the cysteine residue replaced with alanine. C, alignment of secondary structures of GmERO1a and human ERO1α. Secondary structures of GmERO1a were predicted from the VUVCD–NN method. Secondary structural data of human ERO1α were obtained from the crystal structure (Protein Data Bank code 3AHR). α-Helix, β-strand, and coil regions are indicated in purple, light blue, and black in the amino acid sequences, respectively. Cys-113, Cys-118, Cys-121, Cys-123, and Cys-146 are highlighted in yellow.
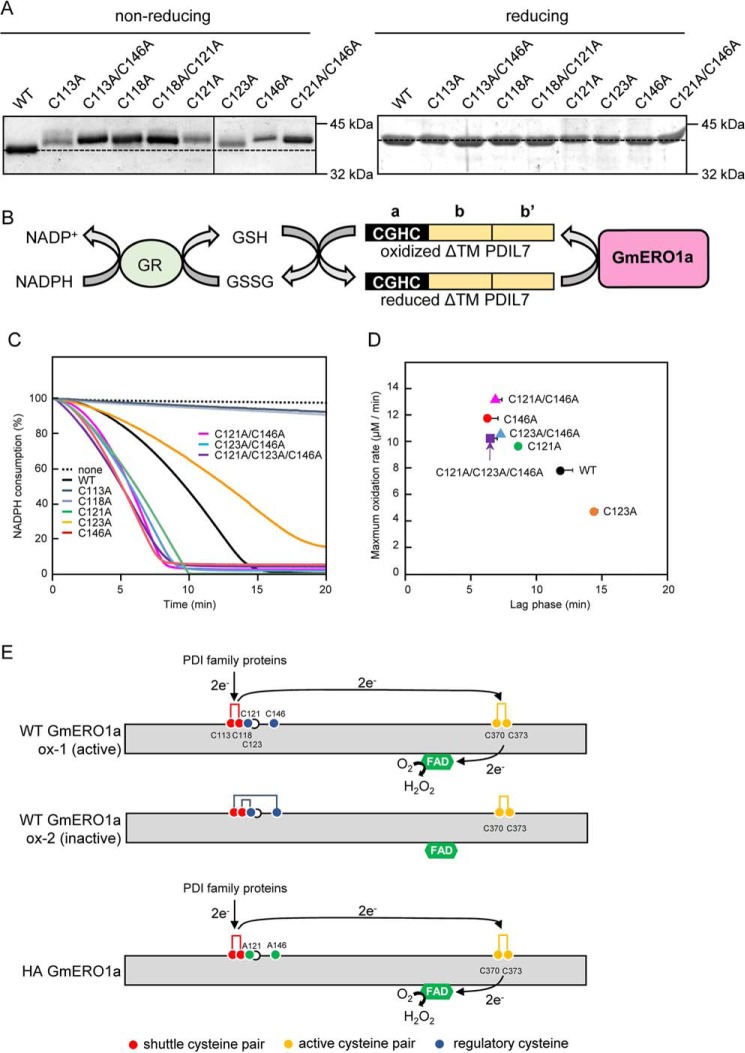
To assess the function of the putative shuttle, active and regulatory cysteines in GmERO1a, we prepared recombinant GmERO1a mutants in which alanine residues were substituted for Cys-113 and Cys-118 (putative shuttle cysteine residues), Cys-370 (putative active-cysteine residue), and Cys-121, Cys-123, and Cys-146 (putative regulatory cysteine residues). All mutants were expressed as soluble proteins in E. coli cells. However, the C370A mutant was not used in the experiments because this protein became insoluble and lost during the purification process. The other mutants were purified as stable, soluble proteins. These proteins were analyzed by SDS-PAGE under reducing and nonreducing conditions (Fig. 4A). The nonreducing SDS-polyacrylamide gel indicates the oxidation state of the protein as purified, which could include a number of different disulfide bond states. Under nonreducing conditions, all of the GmERO1a mutants migrated slower on SDS-PAGE than WT GmERO1a, suggesting that Cys-113, Cys-118, Cys-121, Cys-123, and Cys-146 form intramolecular disulfide bonds. Although there are slight differences in the VUVCD spectra among the mutants, the secondary structures of all mutants were basically similar to those of WT GmERO1a (Fig. 3B and Fig. S1 and Table S1), indicating that the mutations did not affect proper folding of the polypeptides.
Figure 4.
Identification of cysteine residues involved in the activity of GmERO1a. A, WT GmERO1a and mutants in which cysteine residues were substituted with alanine residues were separated by nonreducing (left) or reducing SDS-PAGE (right) on the same gel and stained with Coomassie Brilliant Blue G-250. Dotted line indicates the position of the WT GmERO1a band. B, schematic representation of the coupling reaction of oxidation of recombinant GmPDIL-7–truncated N-terminal transmembrane region (ΔTM PDIL-7) by GmERO1a and the reduction of GSSG by GSH-disulfide reductase (GR) in the presence of GSH and NADPH. C, oxidation of ΔTM PDIL-7 by WT GmERO1a and cysteine mutants. Oxidation reactions were carried out without (dotted line) or with 1 μm WT GmERO1a (black line), C113A mutant (blue-gray line), C118A mutant (gray line), C121A mutant (green line), C123A mutant (orange line), C146A mutant (red line), C121A/C146A mutant (pink line), C123A/C146A mutant (blue line), or C121A/C123A/C146A mutant (purple line) in the presence of 3 μm ΔTM GmPDIL7, 3 mm GSH, 120 μm NADPH, and 1 unit/ml GSH reductase. Rate of ΔTM GmPDIL7 oxidation was monitored as the decrease in NADPH. Schematic representation of ΔTM GmPDIL7 is shown at the top of the graph. CGHC is the active center located in domain a. D, maximum oxidation rate (y axis) was calculated from the maximum slope of oxidation curves shown in C. Lag phase (x axis) was time until achieving maximum oxidation rate. Black circle, WT GmERO1a; green circle, C121A; orange circle, C123A; red circle, C146A; pink triangle, C121A/C146A ; blue triangle, C123A/C146A; purple square, C121A/C123A/C146A. Data are presented as the mean ± S.E. of n = 3–4 experiments. E, model of disulfide connectivity and intramolecular electron transfer in GmERO1a. Active WT GmERO1a (ox-1) contains a shuttle disulfide bond between Cys-113 and Cys-118, whereas Cys-121 and Cys-146 are reduced. The electrons obtained from PDI family proteins are transferred from the shuttle cysteine pairs to FAD via the active cysteine pair (Cys-370 and Cys-373). However, the inactive form (ox-2) possesses Cys-113–Cys-146 and Cys-118–Cys-121 disulfide bonds, which are not functional for intramolecular electron transfer. C121A/C146A mutant (HA GmRO1a) constitutively contains the shuttle disulfide bond.
To assess the function of Cys-113, Cys-118, Cys-121, Cys-123, and Cys-146, we assayed the ability of the mutant GmERO1a enzymes to oxidize GmPDIL7, a soybean PDI family protein with a single redox active-site CGHC in domain a that is readily oxidized by recombinant WT GmERO1a in vitro (39). Oxidation of GmPDIL7 by GmERO1a was assayed by experiments coupled with GSH disulfide reductase, and NADPH reduced (NADPH) consumption (Fig. 4B). The reaction was performed in the presence of GSH, as a substrate for oxidation by GmPDIL7 oxidized by GmERO1a. As expected, the GmERO1a C113A and C118A mutants did not consume NADPH because these cysteines constitute the shuttle cysteine pair essential for oxidation of the substrate (Fig. 4C). A pronounced lag phase was observed in NADPH consumption by WT GmERO1a, which we interpreted to reflect activation of the GmERO1a catalytic activity. The rate of NADPH consumption reached a maximum 12 min after initiation of the reaction (Fig. 4, C and D). The C121A and C146A GmERO1a mutants exhibited enhanced catalytic activity, with 1.2- and 1.5-fold increases in the maximal rate of NADPH consumption relative to the WT enzyme (Fig. 4D). The C121A/C146A double mutant exhibited a higher maximal rate of NADPH consumption (1.7-fold higher than that of WT GmERO1a). Additionally, the time required to reach the maximum NADPH consumption rate was shortened to 8.5, 6.5, and 7 min in the C121A, C146A, and C121A/C146A mutants (Fig. 4D). These results suggest that both Cys-121 and Cys-146 normally form regulatory disulfide bonds with other cysteines (perhaps with shuttle cysteines Cys-113 and Cys-118), thus suppressing GmERO1a activity (Fig. 4E). Interestingly, the C123A mutant exhibited a 1.6-fold decrease in the maximal rate of NADPH consumption relative to the WT enzyme, and the time required to reach the maximum NADPH consumption rate was prolonged to 14 min. However, mutation of Cys-123 had no other effect on either the maximum NADPH consumption rate or the lag phase observed with the C146A and C121A/C146A mutants. Taken together, these results suggest that the activity of GmERO1a is fine-tuned by the formation or reduction of disulfide bonds between multiple combinations of cysteines.
PDI family proteins convert GmERO1a from the ox-2 to ox-1 form to differing extents
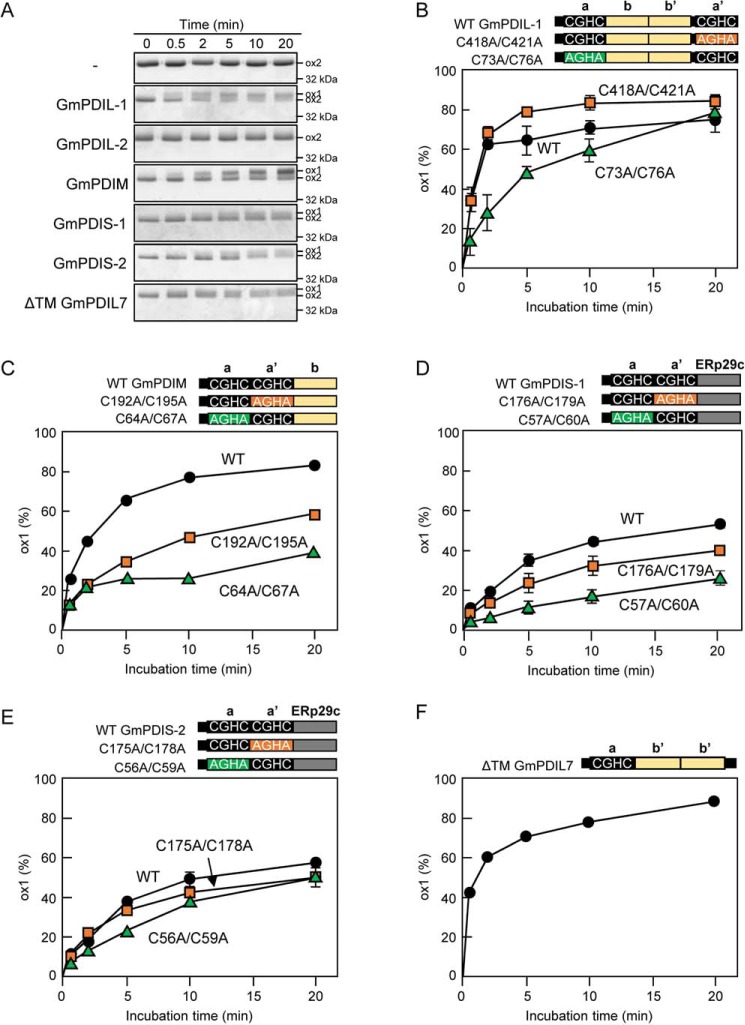
PDI regulates the activities of yeast and human ERO1 in a feedback manner. In the reduced state, yeast and human PDIs can reduce the regulatory disulfides, and thereby activate yeast ERO1p and human ERO1α/β (40, 41). In the oxidized state, human PDI can re-oxidize the regulatory disulfides, and thereby inhibit the activity of ERO1α (42). Based on those observations and our own experimental results, we hypothesized that the activation of WT GmERO1a, which involves Cys-121 and Cys-146, depends upon the presence of PDI. We therefore examined the ability of various PDI family proteins to reduce the regulatory disulfides of GmERO1a. The majority of recombinant GmERO1a molecules purified from E. coli exists in the ox-2 (oxidized) state. No transition of GmERO1a from the ox-2 to the reduced and activated ox-1 form was observed in the presence of GSH but in the absence of the PDI family protein (Fig. 5A). As expected, incubation of GmERO1a with GmPDIL-1 in the presence of GSH resulted in the appearance of the ox-1 form by 0.5 min. The ox-1 level increased with continued incubation as the ox-2 level decreased. These results indicated that GmPDIL-1, reduced by GSH, converted GmERO1a from the ox-2 to ox-1 form. In contrast to GmPDIL-1, GmPDIL-2 did not convert GmERO1a from the ox-2 to ox-1 form. GmPDIM, GmPDIS-1, GmPDIS-2, and GmPDIL7 did, however, convert GmERO1a from the ox-2 to ox-1 form.
Figure 5.
Conversion of ox-2 GmERO1a to the ox-1 form by reduced PDI family proteins. A, GmERO1a (5 μm) was incubated with 2 μm reduced GmPDIL-1, GmPDIL-2, GmPDIM, GmPDIS-1, GmPDIS-2, or ΔTM GmPDIL7 in the presence of 10 mm GSH at 25 °C, then treated with N-ethylmaleimide, and subjected to nonreducing SDS-PAGE. Proteins were stained with Coomassie Brilliant Blue G-250. B–F, schematic representations of WT PDI family proteins and respective mutants are shown. Numbers in the designations of mutants indicate cysteine residues substituted with alanine residues. GmERO1a and each PDI family protein (circles) or respective domain a (green triangles) or domain a′ (orange squares) active-center cysteine mutant was incubated and separated as described in A. The percentage of GmERO1a in the ox-1 form was calculated based on the ox-1 and ox-2 band intensities. Data are presented as the mean ± S.E. of n = 3 experiments.
The rate of ox-2 to ox-1 conversion differed between PDI family proteins. GmPDIL-1 exhibited the highest ox-2 to ox-1 GmERO1a conversion rate, reaching a plateau within 10 min (Fig. 5B). GmPDIL-1 converted ∼80% of GmERO1a to the ox-1 form within 5 min. To determine the contribution of the active centers in domains a and a′ of GmPDIL-1, we examined the conversion activity of the active-center mutants. Both the C418A/C421A and C73A/C76A active-center mutants converted GmERO1a from the ox-2 to ox-1 form, indicating that both active centers of can convert GmERO1a (Fig. 5B and Fig. S2A). However, the active centers differed in terms of conversion activity; the C418A/C421A mutant converted GmERO1a to the ox-1 form more rapidly and to a greater percentage than either WT GmPDIL-1 or the C73A/C76A mutant.
Compared with GmPDIL-1, GmPDIM catalyzed conversion of the ox-2 form of GmERO1a to the ox-1 form at a lower rate (Fig. 5C). The reaction plateaued with ∼80% of GmERO1a converted to the ox-1 form. The C192A/C195A GmPDIM mutant converted GmERO1a to the ox-1 form more rapidly and to a greater percentage than the C64A/C67A mutant (Fig. 5C and Fig. S2B).
Compared with GmPDIL-1 and GmPDIM, GmPDIS-1 and GmPDIS-2 exhibited lower activity in converting GmERO1a from the ox-2 to ox-1 form (Fig. 5, D and E). The rates of conversion of GmPDIS-1 and GmPDIS-2 were lower than that of GmPDIL-1, with only 50% of GmERO1a converted to the ox-1 form even after 20 min. The C176A/C179A GmPDIS-1 mutant exhibited higher activity than the C57A/C60A GmPDIS-1 mutant, suggesting that domain a has higher conversion activity than domain a′ (Fig. 5D and Fig. S2C). The conversion activity of the C175A/C178A GmPDIS-2 mutant is comparable with the WT and slightly higher than that of the C56A/C59A (Fig. 5E and Fig. S2D).
GmPDIL-7, which has one active site, also converted GmERO1a from the ox-2 to ox-1 form in a manner comparable with that of GmPDIL-1 (Fig. 5, A and F), converting >80% of GmERO1a to the ox-1 form within 10 min.
Next, using semi-quantitative Western blot analysis, we determined the content and molar ratios of each of the PDI family proteins and GmERO1 in immature soybean cotyledons (Table 1). The GmPDIL-1 was markedly higher than that of other PDI family proteins, with a molar ratio of GmPDIL-1 to the total amount of other PDI family proteins of 3.4. Previously, we reported that a wheat ortholog of GmPDIL-1 (TaPDIL1) exhibited the highest content among PDI family proteins in the wheat caryopses throughout the grain-filling period (43). Furthermore, the content of GmERO1 was extremely low compared with that of other PDI family proteins (Table 1). The molar ratio of total PDI family protein content to that of GmERO1 was 178. Collectively, these data suggest that the redox status of GmPDIL-1 plays a crucial role in the regulation of GmERO1a activity in the ER.
Table 1.
Amounts of PDI family proteins and GmERO1 in the immature soybean cotyledon
The intensities of the bands immunostained with antiserum against each PDI family proteins or GmERO1 were semi-quantified. Data are represented as mean ± S.E. of n = 3.
| GmPDIL-1 | GmPDIL-2 | GmPDIM | GmPDIS-1 | GmPDIS-2 | GmPDIL7 | GmERO1a | |
|---|---|---|---|---|---|---|---|
| nmol/g protein | 160.1 ± 44.3 | 0.8 ± 0.1 | 6.9 ± 0.9 | 28.7 ± 5.5 | 6.2 ± 2.7 | 4.2 ± 0.4 | 1.2 ± 0.5 |
| Molar ratio to GmERO1a | 136.8 | 0.7 | 5.9 | 24.5 | 5.3 | 3.6 | 1.0 |
GmERO1a oxidizes PDI family proteins asymmetrically
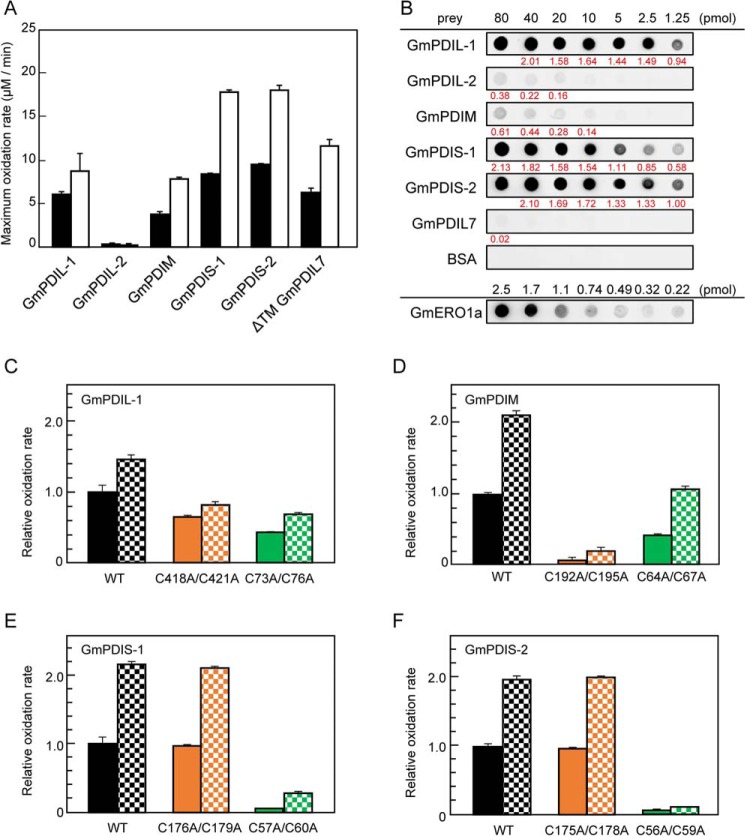
Previously, we have found that plant ERO1 has a broader substrate specificity than human and yeast ERO1, which preferentially oxidize PDI (10, 39). GmERO1a oxidizes five soybean PDI family proteins (at different rates): GmPDIL-1, GmPDIM, GmPDIS-1, GmPDIS-2, and GmPDIL7 (Fig. 6A, black bars), but not GmPDIL-2, which has a PDI-like domain organization identical to GmPDIL-1 and exhibits high oxidative refolding activity in the presence of GSH redox buffer (44). Such differences in rates of oxidation of PDI family proteins by GmERO1a likely reflect differences in the abilities of PDI family proteins to activate GmERO1a. To confirm this, we determined the oxidation rates of PDI family proteins by the C121A/C146A-hyperactive GmERO1a. The C121A/C146A-hyperactive GmERO1a oxidized GmPDIL-1, GmPDIM, GmPDIS-1, GmPDIS-2, and GmPDIL7 at higher reaction rates than WT GmERO1a (Fig. 6A, white bars), but the relative rates for the different PDI proteins were similar to the pattern observed with WT GmERO1a. Also, like WT GmERO1a, C121A/C146A-hyperactive GmERO1a did not oxidize GmPDIL-2. For the human proteins, crystal structure and domain-swapping analyses demonstrated that replacement of PDI domain b′ with domain b′ of ERp57, which has a PDI-like domain organization but low reactivity with ERO1α, substantially altered the affinity and reactivity for ERO1α (17, 22, 45). Therefore, it was expected that the differences in rates of oxidation of PDI family proteins by activated GmERO1a were associated with differences in the affinity of GmERO1a for the various PDI family proteins. Accordingly, we examined the affinity of GmERO1a for soybean PDI family proteins using far-Western blot analysis with recombinant WT GmERO1a as the bait and each of the PDI family proteins as prey. GmERO1a bound to all of the PDI family proteins, including GmPDIL-2, but with different affinities (Fig. 6B). The rates of oxidation by GmERO1a and affinities for PDI family proteins showed poor correlation, with exception of GmPDIL7, which was found to be a superior substrate of GmERO1a, but exhibited very weak affinity for GmERO1a.
Figure 6.
Substrate specificity of GmERO1a for PDI family proteins. A, oxidation of each PDI family protein by WT GmERO1a (black bars) or C121A/C146A-hyperactive mutant GmERO1a (HA) (white bars), determined as described in the legend for Fig. 4B. B, GmERO1a binds to PDI family proteins with different affinities. Far-Western blot analysis was performed with each PDI family protein as prey and GmERO1a as bait. Bound GmERO1a was detected using anti-GmERO1a serum. At the bottom lane (GmERO1a), 2.5, 1.7, 1.1, 0.74, 0.49, 0.32, or 0.22 pmol GmERO1a was dotted as a positive control. The amount of GmERO1a bound on each spot of PDI family proteins was calculated based on the dot intensities and GmERO1a standard dots using ImageJ software. All experiments were performed on the same nitrocellulose membrane. Value written in red below each dot indicates the amount (picomoles) of GmERO1a bound on each dot. C–F, relative ratio of maximum oxidation rates WT GmPDIL-1 (C), GmPDM (D), GmPDIS-1 (E), and GmPDIS-2 (F) and respective active-cysteine residue mutants by WT (solid bars) or HA GmERO1a (checkered bars). Numbers in the designations of mutants indicate cysteine residues substituted with alanine residues. The rates of oxidation of PDI family proteins were monitored as amount of NADPH consumed in the presence of 1 μm WT or HA GmERO1a and 3 μm of each PDI family protein or respective mutant in the presence of 3 mm GSH, 120 μm NADPH, and 1 unit/ml GSH reductase. Data are presented as the mean ± S.E. of n = 3 experiments.
Human ERO1α/β oxidizes domain a′ of human PDI more preferentially than it oxidizes domain a (13, 15). This asymmetric oxidation of PDI domains is thought to be related to the opposing functional roles of domains a and a′ as a disulfide isomerase and disulfide oxidase, respectively. We determined the rates of oxidation of the active-cysteine mutants of PDI family proteins by WT and C121A/C146A-hyperactive GmERO1a. Both GmPDIL-1 (C418A/C421A) and GmPDIL-1 (C73A/C76A) were oxidized at almost the same rate (Fig. 6C), suggesting that GmERO1a oxidizes both domains a and a′ of GmPDIL-1. However, domain oxidation of GmPDIM, GmPDIS-1, and GmPDIS-2 was asymmetric. GmERO1a oxidized GmPDIM (C64A/C67A) much more effectively than it oxidized GmPDIM (C192A/C195A) (Fig. 6D), indicating that domain a′ is more readily oxidized than domain a. In contrast to GmPDIM, domain a of GmPDIS-1 and GmPDIS-2 was preferred by GmERO1a because GmERO1a oxidized GmPDIS-1 (C176A/C179A) and GmPDIS-2 (C175A/C178A) much more effectively than GmPDIS-1 (C57A/C60A) and GmPDIS-2 (C56A/C59A) (Fig. 6, E and F). GmPDIS-1 (C176A/C179A) and WT GmPDIS-1 and GmPDIS-2 (C175A/C178A) and WT GmPDIS-2 were oxidized by GmERO1a at almost the same rate. GmPDIM (C64A/C67A) was oxidized more slowly than WT GmPDIM, suggesting that the active center of domain a of GmPDIM plays an important role for oxidation of domain a′.
In human PDI, the redox reaction is affected by differences in the reduction potential E0 of the active-center disulfides (15, 46). However, there was no significant difference in the E0 values of soybean PDI family protein-active centers (Table 2). The domain specificity of GmERO1a is therefore probably associated with the interactions between the various PDI family proteins and GmERO1a.
Table 2.
Redox potential of PDI family proteins and their active centers
Data are represented as the mean of triplicate experiments.
Loss of regulation of GmERO1a diminishes the effectiveness of oxidative folding catalyzed by PDI family proteins
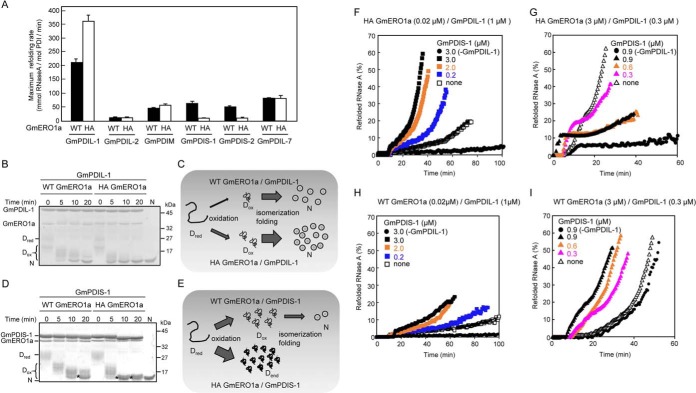
Maintenance of the appropriate redox status in the active centers of PDI family proteins is critical for effective oxidative folding of unfolded proteins. Feedback regulation of ERO1 activity is thought to play a critical role in the maintenance of proper redox status (24). We therefore examined the PDI-catalyzed oxidative folding of RNase A as a model substrate in the presence of C121A/C146A-hyperactive GmERO1a lacking feedback regulation activity. The maximum oxidative refolding rate of RNase A by GmPDIL-1 in the presence of C121A/C146A-hyperactive GmERO1a increased to 1.7 times the rate observed in the presence of WT GmERO1a (Fig. 7A). GmPDIL-2, which is not oxidized by GmERO1a, could not mediate the refolding of RNase A. The refolding activities of GmPDIM, GmPDIS-1, GmPDIS-2, and GmPDIL-7 were significantly lower than that of GmPDIL-1 in the presence of WT GmERO1a. As the rates at which GmERO1a oxidized GmPDIS-1, GmPDIS-2, GmPDIM, and GmPDIL-7 were equal to or higher than that of GmPDIL-1 (Fig. 5A), and the very low refolding activities of these PDI family proteins in the presence of GmERO1 are likely associated with their low activities in terms of isomerizing disulfide bonds into the native configuration. Other noteworthy results were also obtained in the presence of C121A/C146A-hyperactive GmERO1a. Minimal oxidative refolding of RNase A catalyzed by GmPDIS-1 and GmPDIS-2 was detected in the presence of C121A/C146A-hyperactive GmERO1a (Fig. 7A).
Figure 7.
Effect of the loss of regulation of GmERO1a activity on oxidative folding catalyzed by PDI family proteins. A, reduced and denatured RNase A (8 μm) was incubated with 3 μm of each PDI family protein in the presence of 1 μm WT GmERO1a (black bars) or C121A/C146A-hyperactive (HA) mutant GmERO1a (white bars) at 25 °C, after which the recovered RNase A activity was assayed. Data are represented as the mean ± S.E. of n = 3 experiments. B and D, formation of disulfide bonds in reduced and denatured RNase A during refolding catalyzed by GmPDIL-1 (B) or GmPDIS-1 (D) described in A. The reaction was quenched with 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid. Proteins in the reaction mixture were analyzed by nonreducing SDS-PAGE. Dred, reduced and denatured RNase A; Dox, denatured RNase A with nonnative disulfides; asterisks, denatured RNase A with dead-end disulfide bonds; N, native RNase A. C, model of RNase A refolding catalyzed by GmPDIL-1 in the presence of WT or HA GmERO1a. GmPDIL-1 transfers nonnative disulfide bonds to Dred in the presence of WT or HA GmERO1a. More Dox is generated in the presence of HA GmERO1a than in the presence of WT GmERO1a. Because Dox is rapidly folded into the native form, accompanied by the isomerization of disulfide bonds catalyzed by GmPDIL-1, more N is formed in the presence of HA GmERO1a than in the presence of WT GmERO1a. E, model of RNase A refolding catalyzed by GmPDIS-1 in the presence of WT or HA GmERO1a. Denatured RNase A with dead-end disulfide bonds (Dend) is generated by GmPDIS-1 in the presence of HA GmERO1a than in the presence of WT GmERO1a. F–I, cooperative refolding of RNase A catalyzed by GmPDIL-1 and GmPDIS-1 in the presence of a nonregulated (F and G) or regulated (H and I) supply of oxidizing equivalents. Reduced and denatured RNase A (8 μm) was incubated without (−GmPDIL-1) or with 1 μm (F and H) or 0.3 μm GmPDIL-1 (G and I) and GmPDIS-1 at the indicated concentrations in the presence of 0.02 μm HA (F), 3 μm HA (G), or 0.02 μm WT (H), or 3 μm WT (I) GmERO1a.
We then sought the cause of the lower activities of some PDI family proteins, such as GmPDIS-1, in the presence of the C121A/C146A-hyperactive GmERO1a. Oxidative refolding of unfolded proteins is generally thought to proceed via a two-step process, consisting of the introduction of nonnative disulfide bonds in the substrate followed by isomerization of the disulfide bonds to restore the native conformation. To identify the rate-determining step in the oxidative refolding by PDI family proteins, we used nonreducing SDS-PAGE to analyze the folding intermediates. These experiments focused on GmPDIL-1 and GmPDIS-1, because GmPDIL-1 is the plant ortholog of PDI, and immunoprecipitation experiments revealed that GmPDIL-1 associates with GmPDIS-1 in the ER and in vitro (Fig. S3). Incubation of denatured and reduced RNase A and GmPDIL-1 for various times in the presence of WT GmERO1a resulted in the oxidation of most RNase A molecules to form an intermediate with nonnative disulfide bonds, leading to the appearance of a weakly-staining band at the same position as native RNase A within 5 min (Fig. 7B). With incubation between 10 and 20 min, this band increased in intensity, with an accompanying decrease in that of the intermediates. As 80% of the RNase activity was recovered within 10 min (Fig. S4A), the band migrating at the same position as native RNase A was identified as refolded RNase A. In the presence of C121A/C146A-hyperactive GmERO1a, a clearly distinguishable native RNase A band appeared within 5 min (Fig. S4B), and most RNase A molecules were folded by 10 min (Fig. S4A), indicating that the more rapid oxidation of GmPDIL-1 by C121A/C146A-hyperactive GmERO1a accelerated the overall oxidative folding of RNase A. The C418A/C421A and C73A/C76A active-center cysteine mutants also refolded RNase A in the presence of GmERO1a (Fig. S4, A and B), suggesting that the active centers in domains a and a′ are capable of catalyzing the refolding of RNase A using oxidation equivalents supplied by both WT and C121A/C146A-hyperactive GmERO1a. These results suggest that the first-step reaction (introduction of nonnative disulfide bonds) is the rate-limiting step in the overall oxidative folding process catalyzed by GmPDIL-1 (Fig. 7C). The ability of GmPDIL-1 to isomerize disulfide bonds into their native conformation is very strong. Even in the presence of 10 molar eq of C121A/C146A-hyperactive GmERO1a against GmPDIL-1, the rate of RNase A refolding catalyzed by GmPDIL-1 remained constant, although the lag time before the commencement of RNase A refolding declined (Fig. S5B).
GmPDIS-1 introduced nonnative disulfide bonds in all RNase A molecules within 5 min; by 5–10 min, oxidized intermediates and a few native RNase A molecules were generated (Fig. 7D). In the presence of C121A/C146A-hyperactive GmERO1a, nonnative disulfide bonds were introduced in all RNase A molecules within 5 min. After 10 min, the RNase A migrated as a single band immediately above that of native RNase A. As no recovery of RNase A activity was detected in the reaction consisting of GmPDIS-1 in the presence of C121A/C146A-hyperactive GmERO1a (Fig. 7A and Fig. S4C), the band migrating immediately above native RNase A was determined to represent dead-end products containing nonnative disulfide bonds. In contrast to the case of GmPDIL-1, these results suggest that the second-step reaction (isomerization of disulfide bonds into the native conformation) is the rate-limiting step in the oxidative folding process catalyzed by GmPDIS-1 (Fig. 7E). Continuous oxidation of GmPDIS-1 by nonregulated GmERO1a results in the introduction of nonnative disulfide bonds in the substrate that cannot be isomerized into the native conformation by GmPDIS-1, resulting in the accumulation of dead-end products. These data indicate that the feedback regulation of GmERO1a plays an important role in oxidative folding catalyzed by PDI family proteins such as GmPDIS-1.
Because GmPDIL-1 and GmPDIS-1 associate in the ER, we examined the impact of dysregulation of GmERO1a-associated cooperative refolding with GmPDIL-1 and GmPDIS-1. At low molar ratios of C121A/C146A-hyperactive GmERO1a to GmPDIL-1 and RNase A (0.02 to 1 and 8 μm), the maximum oxidative folding rate of RNase A increased with the addition of GmPDIS-1 in a GmPDIS-1 concentration-dependent manner (Fig. 7F). The effect of GmPDIS-1 addition was synergistic, suggesting that GmPDIS-1 compensates for the insufficient formation of nonnative disulfide bonds in RNase A by GmPDIL-1. However, at high molar ratios of C121A/C146A-hyperactive GmERO1a to GmPDIL-1 and RNase A (3 to 1 and 8 μm), adding GmPDIS-1 inhibited oxidative refolding (Fig. 7G). In the presence of WT GmERO1a, by contrast, the addition of GmPDIS-1 accelerated oxidative refolding at both low and high molar ratios of WT GmERO1a to GmPDIL-1 and RNase A (Fig. 7, H and I).
Discussion
In this study, we demonstrated that the activity of plant ERO1 (GmERO1a) is regulated by the reduction and/or exchange of disulfide bonds involving residues Cys-121 and Cys-146. The location of these cysteine residues in GmERO1a is similar to that of the regulatory cysteines (Cys-104 and Cys-131) in human ERO1α, near the shuttle cysteine pair. Analyses of the crystal structures of constitutively active and inactive human ERO1α mutants revealed that the regulatory cysteines are located within an intrinsically flexible loop exhibiting electron shuttle activity that is finely tuned by intra-loop disulfide-bond rearrangement (22). As the VUVCD–NN method predicted that Cys-146 of GmERO1a is located in the intrinsically flexible loop, GmERO1a activity could be regulated in a manner similar to human ERO1α. The additional Cys-208–Cys-241 disulfide bond in human ERO1α functions to “clamp” the two helices that seal the flavin cofactor FAD. The a′-active center of PDI unclamps this seal by forming a Cys-208- and Cys-241–dependent mixed–disulfide complex with ERO1α to facilitate entry of O2 molecules into the flavoprotein-active center (28, 47). In GmERO1a, Cys-217 and Cys-226 are located in positions similar to Cys-208 and Cys-241 of human ERO1α. Indeed, a brief but nonnegligible lag phase in the GmPDIL-1–catalyzed oxidative folding of RNase A was observed in the presence of the C121A/C146A-hyperactive GmERO1a, suggesting the presence of a regulation mechanism that does not involve Cys-121/Cys-146. We also found that substitution of Cys-123 with an Ala residue diminished the oxidation activity of GmERO1a and prolonged the lag phase until the oxidation rate reached a maximum. Moilanen et al. (30) identified a third regulatory disulfide bond involving human ERO1α Cys-166 and ERO1β Cys-165, which forms a mixed–disulfide-linked complex with the a′ active-center cysteine of PDI in the inactive state. Substitution of ERO1α Cys-166 or ERO1β Cys-165 with an Ala residue reportedly causes activity of human ERO1s to decline over the course of the assay, with a concomitant shortening of the lag phase (i.e. enhancement of the activation rate). As the Cys-123 mutation in C121A/C146A-hyperactive GmERO1a had no effect on either activity or activation rate, this suggests that Cys-123 stabilizes the active form of WT GmERO1a.
Analyses of the interactions between GmERO1a and PDI family proteins revealed that PDI family proteins also regulate GmERO1a. However, GmPDIL-2 (which is not a substrate of GmERO1a) did not regulate GmERO1a, in contrast to the observation that human PDI family proteins, which are not good substrates of ERO1α, function as potent regulators of ERO1α/β (41). Furthermore, the activity of GmPDIL-1, GmPDIM, GmPDIS-1, GmPDIS-2, and GmPDIL7 as regulators of GmERO1a differed. The specific characteristics of PDI family proteins that lead to differential activation of GmERO1a may be necessary to elicit the oxidative folding enzymatic activity of each PDI family protein.
GmERO1a oxidizes both the a and a′ domains of GmPDIL-1. GmERO1a also preferentially oxidizes the C-terminal a′ domain of GmPDIM and the N-terminal a domains of GmPDIS-1 and GmPDIS-2. This broad specificity and the differences in domain preference of GmERO1a for PDI family proteins appear to be important in the intra- and intermolecular relay of electrons between GmERO1a, the PDI family protein, and the substrate during oxidative folding, although the underlying structural details remain unclear. We previously found that GmPDIL-2 and GmPDIM associate in the ER and that GmPDIL-2 synergistically accelerates oxidative refolding in vitro using oxidizing equivalents obtained by GmPDIM from GmERO1a (10). In this process, a disulfide bond introduced in the active center of domain a′ of GmPDIM by GmERO1a is transferred to the active center of the GmPDIM domain a and both domains a and a′ of GmPDIL-2. In this study, we demonstrated that GmPDIS-1 associates with GmPDIL-1 in the ER and accelerates oxidative refolding catalyzed by GmPDIL-1 in such a way as to compensate for the lack of disulfide bonds.
In this study, we showed that plant PDI family proteins are categorized into two groups based on the rate-limiting enzymatic activity (e.g. formation of nonnative disulfide bonds in the substrate protein or isomerization of disulfide bonds into the native conformation). Formation of nonnative disulfide bonds in the substrate protein is the rate-limiting activity of group I PDI family proteins (GmPDIL-1 and GmPDIL-2), whereas isomerization of disulfide bonds into the native conformation is the rate-limiting activity of group II PDI family proteins (GmPDIM, GmPDIS-1, GmPDIS-2, and GmPDIL7). Our folding experiments involving the C121A/C146A-hyperactive mutant GmERO1a indicated that dead-end products containing nonnative disulfide bonds that cannot be isomerized to the native conformation are generated by group II PDI family proteins. The generation of these dead-end products is prevented by feedback regulation of GmERO1a activity. The rapid generation of a supply of oxidizing equivalents by C121A/C146A-hyperactive GmERO1a was found to accelerate the rate of oxidative folding by the group I PDI family protein GmPDIL-1. In addition, the rate of oxidative folding catalyzed by GmPDIL-1 was increased by interaction with GmPDIS-1, which associates with GmPDIL-1 in the presence of WT GmERO1a or C121A/C146A-hyperactive GmERO1a at a molar ratio similar to that occurring in vivo. However, levels of C121A/C146A-hyperactive GmERO1a above this threshold inhibit synergistic folding catalyzed by GmPDIL-1 and GmPDIS-1, demonstrating the importance of feedback regulation of GmERO1a for group II PDI family proteins. These data also importantly suggest that GmPDIL-1 can activate GmERO1a and the increase supply of oxidizing equivalents via GmPDIS-1 as required in vivo by feedback. Decreased activation of GmERO1a by GmPDIS-1 and GmPDIS-2 probably prevents excessive oxidation of their active-center cysteines by GmERO1a.
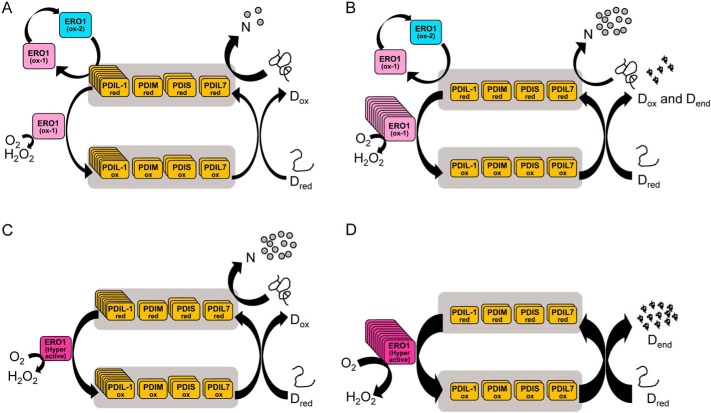
In eukaryotes, the PDI/ERO1 system is a major supplier of disulfides necessary for the oxidative folding of nascent proteins in the ER. In humans, electron transfer between PDI family proteins occurs in a hierarchical cascade from PDI, the main substrate of Ero1α (13). Here, we demonstrated that plant ERO1 (GmERO1a) has broad substrate specificity, and can oxidize not only the PDI ortholog (GmPDIL-1), but also other PDI family proteins (GmPDIM, GmPDIS-1, GmPDIS-2, and GmPDIL7). These findings suggest that, in the plant system, the electron transfer network of the ERO1/PDI family proteins is distinct from the human system. The oxidation power of this system is taken from O2 by ERO1, resulting in the generation of H2O2, which is highly toxic to the cells. Regulation of ERO1 activity is thus thought to be necessary to protect cells from excessive H2O2 generation (48–50). In this study, we demonstrated that the presence of an unregulated supply of disulfide bonds provided by the hyperactive plant ERO1 led to inhibition of folding. This implies that feedback regulation of ERO1 plays important roles not only in regulating the redox status of the active center of PDI family proteins, especially group II PDI proteins, but also in protection from H2O2 generation. In addition, we showed that the concentration ratio between PDI family members and ERO1 is important for efficient oxidative protein folding. Fig. 8 provides a framework for understanding oxidative protein folding in the ER of plants, at different abundance ratios of ERO1 to PDI family proteins, and for either WT or hyperactive ERO1. At a proper molar ratio of WT ERO1 to PDI family proteins, oxidized PDI family proteins introduce disulfide bonds in unfolded protein (Dred) producing protein with nonnative disulfide bonds (Dox), and reduced PDI family proteins rearrange the disulfide bonds to the native ones (N) (Fig. 8A). However, when a molar ratio of WT ERO1 to PDI family proteins is improperly high, too many disulfide bonds beyond the isomerization capacity of PDI family proteins are supplied by ERO1, resulting in production of denatured proteins with dead-end disulfides (Dend) simultaneously with production of native proteins (Fig. 8B). At a proper molar ratio of hyperactive ERO1 to PDI family proteins, disulfide bonds within the limit of isomerization capacity of PDI family proteins are supplied to Dred by hyperactive ERO1 via PDI family proteins, and the disulfide bonds are rearranged to the native ones efficiently (Fig. 8C). However, at an improperly high molar ratio of hyperactive ERO1 to PDI family proteins, too many disulfide bonds are supplied by hyperactive ERO1, resulting in most of the Dred becoming Dend (Fig. 8D). The properties of plant PDI family proteins and the regulation of plant ERO1 revealed in this work should be useful for understanding oxidative protein folding in other eukaryotes as well.
Figure 8.
Model of oxidative protein folding by the multiple PDI family proteins/ERO1 system in plants. A, oxidative protein folding at a proper molar ratio of WT ERO1 to PDI family proteins. Oxidized PDI family proteins introduce disulfide bonds in unfolded protein (Dred) producing denatured protein with nonnative disulfide bonds (Dox), and reduced PDI family proteins rearrange the disulfide bonds to the native ones, producing correctly folded native protein (N). Reduced PDI family proteins convert inactive ERO1 (ox-2) to the active form (ox-1). B, oxidative protein folding at an improperly high molar ratio of WT ERO1 to PDI family proteins. Oxidized PDI family proteins introduce disulfide bonds in Dred, and reduced PDI family proteins produce N. However, too many disulfide bonds beyond isomerization capacity of PDI family proteins are supplied by ERO1, resulting in part of Dred becoming denatured proteins with dead-end disulfides (Dend). C, oxidative protein folding at a proper molar ratio of hyperactive ERO1 to PDI family proteins. Many disulfide bonds within the limit of isomerization capacity of PDI family proteins are supplied by hyperactive ERO1 to Dred, resulting in Dred becoming N effectively. D, oxidative protein folding at an improperly high molar ratio of hyperactive ERO1 to PDI family proteins. Too many disulfide bonds are supplied by hyperactive ERO1, resulting in most of Dred becoming Dend. The number of copies of boxes are reflecting the relative concentration of the proteins.
Experimental procedures
Preparation of recombinant proteins
Recombinant GSH S-transferase (GST)–GmERO1a, GST–GmPDIL-1, GST–GmPDIL-2, GST–GmPDIM, GST–GmPDIS-1, GST–GmPDIS-2, and GST–GmPDIL-7 were expressed as described previously (10, 39, 44, 51, 52). Expression plasmids encoding the cysteine mutants of these proteins were constructed as follows. The appropriate DNA fragments were amplified by PCR using the primers listed in Table S2 and then subcloned into the pGEX6p-2 vector (GE Healthcare). BL21 (DE3) cells (Takara Bio, Inc.) transformed with (GST–GmERO1a and its cysteine mutants) or without (GST–PDI family proteins and respective cysteine mutants) pTf16 (Takara Bio, Inc.) were used for the expression of recombinant proteins. Expression of recombinant proteins was induced in the presence of 0.5 mm isopropyl β-d-thiogalactopyranoside, extracted by sonication, and purified by GSH–Sepharose 4B column chromatography (GE Healthcare) followed by digestion with PreScission protease (GE Healthcare) and further purification by TSK gel G3000SW column chromatography (Tosoh Co., Ltd.). Purified GmERO1a and respective mutants were incubated in Laemmli's SDS-loading buffer (38) with and without reducing reagent containing 25 mm N-ethylmaleimide at 37 °C for 2 h and then subjected to nonreducing SDS-PAGE. Proteins were stained with Coomassie Brilliant Blue G-250. For expression and purification of Trx1, an expression vector for Trx1 was constructed as follows. The cDNA was amplified from total RNA extracted from E. coli (TOP-10F) (Invitrogen) using the RT-PCR primers shown in Table S2. The vector was then subcloned into the pET-46 Ek/LIC expression vector (Novagen). Expression of recombinant Trx1 was induced in LB medium containing 0.5 mm isopropyl β-d-thiogalactopyranoside and 100 μg/ml ampicillin at 37 °C for 3 h. Recombinant Trx1 was purified using nickel-nitrilotriacetic acid chromatography (Invitrogen) followed by gel-filtration chromatography on a TSK gel G3000SW column (Tosoh).
Trx1 oxidation assays
Recombinant Trx1 was reduced using DTT and desalted using a Zeba Spin Desalting column (Thermo Fisher Scientific). Reactions with Trx1 were initiated by adding 1 μm GmERO1a to 50 μm reduced Trx1 in 50 mm Tris-HCl (pH 7.5) at 25 °C. At the indicated times, reactions were quenched by addition of SDS-PAGE sample buffer containing 1 mm 4′-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid and incubated for 2 h at 37 °C. After quenching, samples were separated on 15% polyacrylamide gels and stained with Coomassie Brilliant Blue G-250. GmERO1a redox status was quenched by addition of SDS-PAGE sample buffer containing 25 mm N-ethylmaleimide and incubated for 2 h at 37 °C. After quenching, samples were separated on 15% polyacrylamide gels and stained with a silver-staining kit (Wako Co.)
Cultivation of soybean cells
Soybean DG330 (rpc00051) cells were provided by RIKEN BioResource Center which is participating in the National Bio-Resource Project of the MEXT, Japan (53). Cells were cultured, propagated, and passaged in a 200-ml Erlenmeyer flask containing 80 ml of 0.25% gellan gum-solidified Murashige and Skoog medium at 25 °C under 5000 1× continuous light.
VUVCD spectroscopy
The VUVCD measurements of GmERO1a and its cysteine mutants were measured over the wavelength range of 172–260 nm by the VUVCD spectrophotometer at the Hiroshima Synchrotron Radiation Center (HiSOR) (54). Details regarding the optical and electronic setup of this spectrophotometer are described elsewhere (54, 55). Calibration of the ellipticity amplitude and wavelength position was confirmed by monitoring the CD spectra of an aqueous solution of ammonium d-camphor-10-sulfonate. The spectra were recorded with a 1.0-mm slit using a 4-s time constant and 20 nm/min scan speed, and final spectra were obtained by averaging eight accumulations and subtracting signals associated with the buffer solution. The secondary structure contents and numbers of segments of GmERO1a and its cysteine mutants were estimated by the SELCON3 program (56) with the VUVCD spectra and secondary structure dataset of 31 reference proteins (57, 58). The positions of the secondary structures on the amino acid sequences of GmERO1a and its mutants were predicted by combining VUVCD analysis with the NN algorithm. This combination method was detailed previously (54, 59).
GmERO1a mutant oxidation activity assays
Oxidation activity was assessed using a coupled assay following the decrease in absorbance at 340 nm due to the consumption of NADPH (Oriental Yeast Co., Ltd.) by GSH reductase (Sigma), as described previously (10, 35, 60). A molar extinction coefficient for NADPH of 6,200 m−1 cm−1 was used for calculations. GmERO1a (or respective mutant) (1 μm) was incubated with each recombinant PDI family protein (3 μm) in 100 mm HEPES buffer (pH 7.5) containing 2 mm CaCl2, 150 mm NaCl, 3 mm GSH, 120 μm NADPH, and 1 unit/ml GSH reductase (Sigma) at 25 °C.
Far-Western blot analysis
Far-Western blot analyses were carried out as described previously (10). Briefly, purified recombinant PDI family protein and BSA (as prey proteins) were spotted onto a nitrocellulose membrane (GenScript Biotech Corp.). The membrane was blocked with 20 mm Tris-HCl (pH 8) containing 150 mm NaCl, 0.05% Tween 20, and 5% nonfat dry milk (blocking solution) at 4 °C for 16 h and incubated in 0.2 μm GmERO1a as bait protein in blocking solution without nonfat dry milk (TBST) for 3 h at 4 °C. The membrane was incubated with anti-GmERO1a antiserum (10) and then horseradish peroxidase-conjugated anti-guinea pig IgG as the secondary antibody (Promega Corp.) diluted with blocking solution as the secondary antibody. Blots were washed four times with TBST for 20 min/wash and developed using Western Lightning chemiluminescence reagent (PerkinElmer Life Sciences).
Determination of the redox state of GmERO1a reacting with PDI family proteins
Recombinant PDI family proteins and respective mutants were reduced using DTT and desalted using Zeba Spin Desalting columns (Thermo Fisher Scientific). GmERO1a (5 μm) was incubated with 2 μm each reduced PDI family protein and 10 mm GSH in 50 mm Tris-HCl buffer (pH 7.5) at 25 °C. At the indicated times, reactions were quenched for 2 h at 37 °C by addition of Laemmli's SDS-loading buffer containing 25 mm N-ethylmaleimide. Proteins were separated on 10% polyacrylamide gels under nonreducing conditions and stained with Coomassie Brilliant Blue G-250. Staining intensity for each band was quantified using ImageJ software (National Institutes of Health).
Quantitative Western blot analysis
Soybean (Glycine max (L.) Merrill cv. Jack) seeds were planted in 5-liter pots and grown in a controlled-environment chamber at 25 °C under 16-h day/8-h night cycles. Seeds (230 mg) were collected from soybean plants, and the cotyledons were then isolated. Tissues were frozen in liquid nitrogen and then ground with a micro-pestle SK-100 (Tokken). Proteins were extracted by boiling for 5 min in Laemmli's SDS-loading buffer containing a 1% mixture of protease inhibitors (Sigma). The protein concentration in the samples was measured using an RC/DC protein assay kit (Bio-Rad). Extracted proteins were separated along with known amounts of recombinant PDI family proteins and GmERO1 on the same gel by SDS-PAGE and then blotted onto polyvinylidene difluoride membranes (Bio-Rad). The blotted proteins were then immunostained using specific rabbit or guinea pig antiserum as primary antibodies and horseradish peroxidase-conjugated anti-rabbit IgG (Santa Cruz Biotechnology, Inc.) or anti-guinea pig IgG as the secondary antibody. Signals were visualized using Western Lightning Plus-ECL Enhanced Chemiluminescence Substrate (PerkinElmer Life Sciences) and an Image Quant LAS4010 imager (GE Healthcare). The amount of each protein was calculated based on the intensities of the sample and corresponding recombinant protein standard bands using ImageJ software.
Determination of redox equilibrium constants of PDI family proteins
Redox equilibrium constants of PDI family proteins and their active-center mutants (GmPDIL-1: C418A/C421A and C73A/C76A; GmPDIS-1: C176A/C179A and C57A/C60A; and GmPDIS-2: C175A/C178A) were determined with WT proteins as described previously (10). Briefly, PDI family proteins or respective mutants were incubated with 0.1 mm GSSG and 0.015–28 mm GSH at 25 °C for 1 h in 0.1 m sodium phosphate buffer (pH 7.0) containing 1 mm EDTA and 150 mm NaCl and then incubated under N2 at 25 °C for 1 h. Free thiol groups were modified by incubation on methoxypolyethylene glycol-maleimide (Fluka Sigma) at 25 °C for 30 min. Proteins were separated by SDS-PAGE and stained with Coomassie Brilliant Blue G-250. The reduced, oxidized, and intermediate fractions were quantified using ImageJ software (National Institutes of Health). Values for the completely oxidized or reduced states were regarded as 0 or 100%, respectively, and all intermediate states were recalibrated. The Keq value was calculated by fitting the recalibrated fraction of the apparent reduced form to the following equation: R = ([GSH]2/[GSH disulfide (GSSG)])/{Keq + ([GSH]2/[GSSG])}, where R represents the relative ratio of the reduced forms. The protein equilibrium redox potential was calculated using the Nernst equation, (E′0 = E′0(GSH/GSSG) − (RT/nF) × lnKeq), using a GSH standard potential E′0(GSH/GSSG) of −0.240 V at pH 7.0 and 25 °C.
RNase A refolding assays
Thiol oxidative refolding activity was assayed as described previously (61, 62). Briefly, each reaction mixture contained 100 mm HEPES buffer (pH 7.5), 150 mm NaCl, 2 mm CaCl2, GmERO1a, 2 mm cCMP, reduced and denatured RNase A, and recombinant PDI family proteins and respective mutants. The mixtures were incubated at 25 °C, and the formation of active RNase A was assessed spectrophotometrically by monitoring the hydrolysis of the RNase A substrate cCMP at 296 nm.
Gel-based RNase A refolding experiments
Analyses were carried out as described previously (10). In brief, the refolding reactions were performed by the addition of 1 μm GmERO1a or C121A/C146A-hyperactive GmERO1a in 100 mm HEPES buffer (pH 7.5), 150 mm NaCl, 2 mm CaCl2, and 3 μm recombinant PDI family proteins and respective mutants along with 8 μm denatured and reduced RNase A. At the indicated time points, free thiols were blocked by the addition of Laemmli's SDS-loading buffer containing 8 mm 4-acetamido-4-maleimidylstilbene-2,2-disulfonic acid. Proteins were then separated by SDS-PAGE on a 15% polyacrylamide gel without reducing reagent. Proteins were detected by Coomassie Blue staining.
Statistical analysis
All data are shown as the mean ± S.E. of at least three replicates. Welch's t test (two-tailed, unpaired) or the Tukey-Kramer test was used for comparison of data from more than three experiments.
Author contributions
M. M. and R. U. conceptualization; M. M., A. O., K. M., K. G., T. M., Y. N., A. H., K. K., and R. U. data curation; M. M., A. O., K. M., K. G., T. M., Y. N., A. H., K. K., Y. T., and R. U. formal analysis; M. M. and R. U. supervision; M. M., A. O., and R. U. funding acquisition; M. M., A. O., K. M., K. G., Y. N., K. K., Y. T., and R. U. validation; M. M., A. O., K. M., K. G., T. M., Y. N., A. H., K. K., Y. T., and R. U. investigation; M. M., A. O., K. M., K. G., Y. N., A. H., K. K., Y. T., and R. U. visualization; M. M., A. O., K. M., K. G., T. M., Y. N., A. H., K. K., Y. T., and R. U. methodology; M. M., A. O., K. M., K. G., Y. N., A. H., K. K., Y. T., and R. U. writing-original draft; M. M. and R. U. project administration; M. M., A. O., K. M., K. G., and R. U. writing-review and editing.
Supplementary Material
This work was supported by Grants-in-aid for Scientific Research 18658055 and 26660111, Grant-in-aid for JSPS Fellows 16J05933 from the Japan Society for the Promotion of Science, a grant from the Program for Promotion of Basic Research Activities for Innovative Biosciences, a grant from the Fuji Foundation for Protein Research, and a grant from the Takano Life Science Research Foundation.The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1 and S2 and Figs. S1–S5.
- ER
- endoplasmic reticulum
- ERO1
- ER oxidoreductin 1
- PDI
- protein-disulfide isomerase
- H2O2
- hydrogen peroxide
- PRDX4
- peroxiredoxin IV
- Trx1
- thioredoxin 1
- VUVCD
- vacuum-ultraviolet circular dichroism
- NN
- neural network
- GST
- GSH S-transferase.
References
- 1. Ellgaard L., and Ruddock L. W. (2005) The human protein-disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 6, 28–32 10.1038/sj.embor.7400311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kozlov G., Määttänen P., Thomas D. Y., and Gehring K. (2010) A structural overview of the PDI family of proteins. FEBS J. 277, 3924–3936 10.1111/j.1742-4658.2010.07793.x [DOI] [PubMed] [Google Scholar]
- 3. Wallis A. K., and Freedman R. B. (2013) Assisting oxidative protein folding: how do protein disulphide-isomerases couple conformational and chemical processes in protein folding? Top. Curr. Chem. 328, 1–34 10.1007/128_2011_171 [DOI] [PubMed] [Google Scholar]
- 4. Urade R. (2019) Oxidative protein folding in the plant endoplasmic reticulum. Biosci. Biotechnol. Biochem. 83, 781–793 10.1080/09168451.2019.1571900 [DOI] [PubMed] [Google Scholar]
- 5. Frand A. R., and Kaiser C. A. (1998) The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol. Cell 1, 161–170 10.1016/S1097-2765(00)80017-9 [DOI] [PubMed] [Google Scholar]
- 6. Pollard M. G., Travers K. J., and Weissman J. S. (1998) Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol. Cell 1, 171–182 10.1016/S1097-2765(00)80018-0 [DOI] [PubMed] [Google Scholar]
- 7. Cabibbo A., Pagani M., Fabbri M., Rocchi M., Farmery M. R., Bulleid N. J., and Sitia R. (2000) ERO1-L, a human protein that favors disulfide bond formation in the endoplasmic reticulum. J. Biol. Chem. 275, 4827–4833 10.1074/jbc.275.7.4827 [DOI] [PubMed] [Google Scholar]
- 8. Pagani M., Fabbri M., Benedetti C., Fassio A., Pilati S., Bulleid N. J., Cabibbo A., and Sitia R. (2000) Endoplasmic reticulum oxidoreductin 1-Lβ (ERO1-Lβ), a human gene induced in the course of the unfolded protein response. J. Biol. Chem. 275, 23685–23692 10.1074/jbc.M003061200 [DOI] [PubMed] [Google Scholar]
- 9. Onda Y., Kumamaru T., and Kawagoe Y. (2009) ER membrane-localized oxidoreductase Ero1 is required for disulfide bond formation in the rice endosperm. Proc. Natl. Acad. Sci. U.S.A. 106, 14156–14161 10.1073/pnas.0904429106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Matsusaki M., Okuda A., Masuda T., Koishihara K., Mita R., Iwasaki K., Hara K., Naruo Y., Hirose A., Tsuchi Y., and Urade R. (2016) Cooperative protein folding by two protein thiol disulfide oxidoreductases and ERO1 in soybean. Plant Physiol. 170, 774–789 10.1104/pp.15.01781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fan F., Zhang Y., Huang G., Zhang Q., Wang C.-C., Wang L., and Lu D. (2019) AtERO1 and AtERO2 exhibit differences in catalyzing oxidative protein folding in the endoplasmic reticulum. Plant Physiol. 180, 2022–2033 10.1104/pp.19.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vitu E., Kim S., Sevier C. S., Lutzky O., Heldman N., Bentzur M., Unger T., Yona M., Kaiser C. A., and Fass D. (2010) Oxidative activity of yeast Ero1p on protein-disulfide isomerase and related oxidoreductases of the endoplasmic reticulum. J. Biol. Chem. 285, 18155–18165 10.1074/jbc.M109.064931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Araki K., Iemura S., Kamiya Y., Ron D., Kato K., Natsume T., and Nagata K. (2013) Ero1-α and PDIs constitute a hierarchical electron transfer network of endoplasmic reticulum oxidoreductases. J. Cell Biol. 202 861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kulp M. S., Frickel E.-M., Ellgaard L., and Weissman J. S. (2006) Domain architecture of protein-disulfide isomerase facilitates its dual role as an oxidase and an isomerase in Ero1p-mediated disulfide formation. J. Biol. Chem. 281, 876–884 10.1074/jbc.M511764200 [DOI] [PubMed] [Google Scholar]
- 15. Chambers J. E., Tavender T. J., Oka O. B., Warwood S., Knight D., and Bulleid N. J. (2010) The reduction potential of the active center disulfides of human protein-disulfide isomerase limits oxidation of the enzyme by Ero1α. J. Biol. Chem. 285, 29200–29207 10.1074/jbc.M110.156596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Araki K., and Nagata K. (2011) Functional in vitro analysis of the ERO1 protein and protein-disulfide isomerase pathway. J. Biol. Chem. 286, 32705–32712 10.1074/jbc.M111.227181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Masui S., Vavassori S., Fagioli C., Sitia R., and Inaba K. (2011) Molecular bases of cyclic and specific disulfide interchange between human ERO1α protein and protein-disulfide isomerase (PDI). J. Biol. Chem. 286, 16261–16271 10.1074/jbc.M111.231357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gross E., Kastner D. B., Kaiser C. A., and Fass D. (2004) Structure of Ero1p, source of disulfide bonds for oxidative protein folding in the cell. Cell 117, 601–610 10.1016/S0092-8674(04)00418-0 [DOI] [PubMed] [Google Scholar]
- 19. Gross E., Sevier C. S., Heldman N., Vitu E., Bentzur M., Kaiser C. A., Thorpe C., and Fass D. (2006) Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc. Natl. Acad. Sci. U.S.A. 103, 299–304 10.1073/pnas.0506448103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tu B. P., and Weissman J. S. (2002) The FAD- and O2-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol. Cell 10, 983–994 10.1016/S1097-2765(02)00696-2 [DOI] [PubMed] [Google Scholar]
- 21. Tu B. P., Ho-Schleyer S. C., Travers K. J., and Weissman J. S. (2000) Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science 290, 1571–1574 10.1126/science.290.5496.1571 [DOI] [PubMed] [Google Scholar]
- 22. Inaba K., Masui S., Iida H., Vavassori S., Sitia R., and Suzuki M. (2010) Crystal structures of human Ero1α reveal the mechanisms of regulated and targeted oxidation of PDI. EMBO J. 29, 3330–3343 10.1038/emboj.2010.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tavender T. J., and Bulleid N. J. (2010) Molecular mechanisms regulating oxidative activity of the Ero1 family in the endoplasmic reticulum. Antioxid. Redox Signal. 13, 1177–1187 10.1089/ars.2010.3230 [DOI] [PubMed] [Google Scholar]
- 24. Sevier C. S., Qu H., Heldman N., Gross E., Fass D., and Kaiser C. A. (2007) Modulation of cellular disulfide-bond formation and the ER redox environment by feedback regulation of Ero1. Cell 129, 333–344 10.1016/j.cell.2007.02.039 [DOI] [PubMed] [Google Scholar]
- 25. Ponsero A. J., Igbaria A., Darch M. A., Miled S., Outten C. E., Winther J. R., Palais G., D'Autréaux B., Delaunay-Moisan A., and Toledano M. B. (2017) Endoplasmic reticulum transport of glutathione by Sec61 is regulated by Ero1 and Bip. Mol. Cell 67, 962–973.e5 10.1016/j.molcel.2017.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Appenzeller-Herzog C., Riemer J., Christensen B., Sørensen E. S., and Ellgaard L. (2008) A novel disulphide switch mechanism in Ero1α balances ER oxidation in human cells. EMBO J. 27, 2977–2987 10.1038/emboj.2008.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang L., Zhu L., and Wang C.-C. (2011) The endoplasmic reticulum sulfhydryl oxidase Ero1β drives efficient oxidative protein folding with loose regulation. Biochem. J. 434, 113–121 10.1042/BJ20101357 [DOI] [PubMed] [Google Scholar]
- 28. Ramming T., Okumura M., Kanemura S., Baday S., Birk J., Moes S., Spiess M., Jenö P., Bernèche S., Inaba K., and Appenzeller-Herzog C. (2015) A PDI-catalyzed thiol–disulfide switch regulates the production of hydrogen peroxide by human ERO1. Free Radic. Biol. Med. 83, 361–372 10.1016/j.freeradbiomed.2015.02.011 [DOI] [PubMed] [Google Scholar]
- 29. Kim S., Sideris D. P., Sevier C. S., and Kaiser C. A. (2012) Balanced Ero1 activation and inactivation establishes ER redox homeostasis. J. Cell Biol. 196, 713–725 10.1083/jcb.201110090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moilanen A., Korhonen K., Saaranen M. J., and Ruddock L. W. (2018) Molecular analysis of human Ero1 reveals novel regulatory mechanisms for oxidative protein folding. Life Sci. Alliance 1, e201800090 10.26508/lsa.201800090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zito E., Melo E. P., Yang Y., Wahlander Å., Neubert T. A., Ron D. (2010) Oxidative protein folding by an endoplasmic reticulum-localized peroxiredoxin. Mol. Cell 40, 787–797 10.1016/j.molcel.2010.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tavender T. J., Springate J. J., and Bulleid N. J. (2010) Recycling of peroxiredoxin IV provides a novel pathway for disulphide formation in the endoplasmic reticulum. EMBO J. 29, 4185–4197 10.1038/emboj.2010.273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Konno T., Pinho Melo E., Lopes C., Mehmeti I., Lenzen S., Ron D., and Avezov E. (2015) ERO1-independent production of H2O2 within the endoplasmic reticulum fuels Prdx4-mediated oxidative protein folding. J. Cell Biol. 211, 253–259 10.1083/jcb.201506123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kojima R., Okumura M., Masui S., Kanemura S., Inoue M., Saiki M., Yamaguchi H., Hikima T., Suzuki M., Akiyama S., and Inaba K. (2014) Radically different thioredoxin domain arrangement of ERp46, an efficient disulfide bond introducer of the mammalian PDI family. Structure 22, 431–443 10.1016/j.str.2013.12.013 [DOI] [PubMed] [Google Scholar]
- 35. Nguyen V. D., Saaranen M. J., Karala A. R., Lappi A. K., Wang L., Raykhel I. B., Alanen H. I., Salo K. E., Wang C. C., and Ruddock L. W. (2011) Two endoplasmic reticulum PDI peroxidases increase the efficiency of the use of peroxide during disulfide bond formation. J. Mol. Biol. 406, 503–515 10.1016/j.jmb.2010.12.039 [DOI] [PubMed] [Google Scholar]
- 36. Rutkevich L. A., and Williams D. B. (2012) Vitamin K epoxide reductase contributes to protein disulfide formation and redox homeostasis within the endoplasmic reticulum. Mol. Biol. Cell 23, 2017–2027 10.1091/mbc.e12-02-0102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Attacha S., Solbach D., Bela K., Moseler A., Wagner S., Schwarzländer M., Aller I., Müller S. J., and Meyer A. J. (2017) Glutathione peroxidase-like enzymes cover five distinct cell compartments and membrane surfaces in Arabidopsis thaliana. Plant Cell Environ. 40, 1281–1295 10.1111/pce.12919 [DOI] [PubMed] [Google Scholar]
- 38. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 39. Okuda A., Matsusaki M., Masuda T., and Urade R. (2017) Identification and characterization of GmPDIL7, a soybean ER membrane-bound protein-disulfide isomerase family protein. FEBS J. 284, 414–428 10.1111/febs.13984 [DOI] [PubMed] [Google Scholar]
- 40. Niu Y., Zhang L., Yu J., Wang C. C., and Wang L. (2016) Novel roles of the non-catalytic elements of yeast protein-disulfide isomerase in its interplay with endoplasmic reticulum oxidoreductin 1. J. Biol. Chem. 291, 8283–8294 10.1074/jbc.M115.694257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang L., Niu Y., Zhu L., Fang J., Wang X., Wang L., and Wang C. C. (2014) Different interaction modes for protein-disulfide isomerase (PDI) as an efficient regulator and a specific substrate of endoplasmic reticulum oxidoreductin-1a (Ero1a). J. Biol. Chem. 289, 31188–31199 10.1074/jbc.M114.602961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oka O. B., Yeoh H. Y., and Bulleid N. J. (2015) Thiol-disulfide exchange between the PDI family of oxidoreductases negates the requirement for an oxidase or reductase for each enzyme. Biochem. J. 469, 279–288 10.1042/BJ20141423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kimura S., Higashino Y., Kitao Y., Masuda T., and Urade R. (2015) Expression and characterization of protein-disulfide isomerase family proteins in bread wheat. BMC Plant Biol. 15, 73 10.1186/s12870-015-0460-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kamauchi S., Wadahama H., Iwasaki K., Nakamoto Y., Nishizawa K., Ishimoto M., Kawada T., and Urade R. (2008) Molecular cloning and characterization of two soybean protein-disulfide isomerases as molecular chaperones for seed storage proteins. FEBS J. 275, 2644–2658 10.1111/j.1742-4658.2008.06412.x [DOI] [PubMed] [Google Scholar]
- 45. Araki K., and Inaba K. (2012) Structure, mechanism, and evolution of ero1 family enzymes. Antioxid. Redox Signal. 16, 790–799 10.1089/ars.2011.4418 [DOI] [PubMed] [Google Scholar]
- 46. Baker K. M., Chakravarthi S., Langton K. P., Sheppard A. M., Lu H., and Bulleid N. J. (2008) Low reduction potential of Ero1α regulatory disulphides ensures tight control of substrate oxidation. EMBO J. 27, 2988–2997 10.1038/emboj.2008.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kanemura S., Okumura M., Yutani K., Ramming T., Hikima T., Appenzeller-Herzog C., Akiyama S., and Inaba K. (2016) Human ER oxidoreductin-1α (Ero1α) undergoes dual regulation through complementary redox interactions with protein-disulfide isomerase. J. Biol. Chem. 291, 23952–23964 10.1074/jbc.M116.735662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ramming T., Hansen H. G., Nagata K., Ellgaard L., and Appenzeller-Herzog C. (2014) GPx8 peroxidase prevents leakage of H2O2 from the endoplasmic reticulum. Free Radic. Biol. Med. 70, 106–116 10.1016/j.freeradbiomed.2014.01.018 [DOI] [PubMed] [Google Scholar]
- 49. Hansen H. G., Schmidt J. D., Søltoft C. L., Ramming T., Geertz-Hansen H. M., Christensen B., Sørensen E. S., Juncker A. S., Appenzeller-Herzog C., and Ellgaard L. (2012) Hyperactivity of the Ero1α oxidase elicits endoplasmic reticulum stress but no broad antioxidant response. J. Biol. Chem. 287, 39513–39523 10.1074/jbc.M112.405050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hansen H. G., Søltoft C. L., Schmidt J. D., Birk J., Appenzeller-Herzog C., and Ellgaard L. (2014) Biochemical evidence that regulation of Ero1β activity in human cells does not involve the isoform-specific cysteine. Biosci. Rep. 34, e00103 10.1042/BSR20130124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wadahama H., Kamauchi S., Ishimoto M., Kawada T., and Urade R. (2007) Protein disulfide isomerase family proteins involved in soybean protein biogenesis. FEBS J. 274, 687–703 10.1111/j.1742-4658.2006.05613.x [DOI] [PubMed] [Google Scholar]
- 52. Wadahama H., Kamauchi S., Nakamoto Y., Nishizawa K., Ishimoto M., Kawada T., and Urade R. (2008) A novel plant protein-disulfide isomerase family homologous to animal P5- Molecular cloning and characterization as a functional protein for folding of soybean seed-storage proteins. FEBS J. 275, 399–410 10.1111/j.1742-4658.2007.06199.x [DOI] [PubMed] [Google Scholar]
- 53. Asano S., and Otobe K. (2011) Production of phytochemicals by using habituated and long-term cultured cells. Plant Biotechnol. 28, 51–62 10.5511/plantbiotechnology.10.1109a [DOI] [Google Scholar]
- 54. Matsuo K., and Gekko K. (2013) Construction of a synchrotron-radiation vacuum-ultraviolet circular-dichroism spectrophotometer and its application to the structural analysis of biomolecules. Bull. Chem. Soc. Jpn. 6, 675–689 [Google Scholar]
- 55. Matsuo K., Sakai K., Matsushima Y., Fukuyama T., and Gekko K. (2003) Optical cell with a temperature-control unit for a vacuum-ultraviolet circular dichroism spectrophotometer. Anal. Sci. 19, 129–132 10.2116/analsci.19.129 [DOI] [PubMed] [Google Scholar]
- 56. Sreerama N., and Woody R. W. (2000) Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 10.1006/abio.2000.4880 [DOI] [PubMed] [Google Scholar]
- 57. Matsuo K., Yonehara R., and Gekko K. (2004) Secondary-structure analysis of proteins by vacuum-ultraviolet circular dichroism spectroscopy. J. Biochem. 135, 405–411 10.1093/jb/mvh048 [DOI] [PubMed] [Google Scholar]
- 58. Matsuo K., Yonehara R., and Gekko K. (2005) Improved estimation of the secondary structures of proteins by vacuum-ultraviolet circular dichroism spectroscopy. J. Biochem. 138, 79–88 10.1093/jb/mvi101 [DOI] [PubMed] [Google Scholar]
- 59. Matsuo K., and Gekko K. (2013) Circular-dichroism and synchrotron-radiation circular-dichroism spectroscopy as tools to monitor protein structure in a lipid environment. Methods Mol. Biol. 974, 151–176 10.1007/978-1-62703-275-9_8 [DOI] [PubMed] [Google Scholar]
- 60. Sato Y., Kojima R., Okumura M., Hagiwara M., Masui S., Maegawa K., Saiki M., Horibe T., Suzuki M., and Inaba K. (2013) Synergistic cooperation of PDI family members in peroxiredoxin 4-driven oxidative protein folding. Sci. Rep. 3, 2456 10.1038/srep02456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Creighton T. E. (1977) Kinetics of refolding of reduced ribonuclease. J. Mol. Biol. 113, 329–341 10.1016/0022-2836(77)90145-0 [DOI] [PubMed] [Google Scholar]
- 62. Lyles M. M., and Gilbert H. F. (1991) Catalysis of the oxidative folding of ribonuclease A by protein-disulfide isomerase: pre-steady-state kinetics and the utilization of the oxidizing equivalents of the isomerase. Biochemistry 30, 619–625 10.1021/bi00217a005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.