Abstract
Assigning functions to the vast array of proteins present in eukaryotic cells remains challenging. To identify relationships between proteins, and thereby enable functional annotations of proteins, we determined changes of abundance of 10,323 human proteins in response to 294 biological perturbations using isotope-labelling mass spectrometry. We applied the machine learning algorithm treeClust to reveal functional associations between co-regulated human proteins from ProteomeHD, a compilation of our own data and datasets from the Proteomics Identifications (PRIDE) database. This produced a co-regulation map of the human proteome. Co-regulation was able to capture relationships between proteins that do not physically interact or co-localize. For example, co-regulation of the peroxisomal membrane protein PEX11β with mitochondrial respiration factors led us to discover an organelle interface between peroxisomes and mitochondria in mammalian cells. We also predicted the function of microproteins that are difficult to study with traditional methods. The co-regulation map can be explored at www.proteomeHD.net.
Functional genomics methods often use a “guilt-by-association” strategy to determine the functions of genes and proteins on a system-wide scale. For example, high-throughput measurements of protein-protein interactions 1–3 and subcellular localization 4–6 have delivered insights into proteome organisation. One limitation of these techniques is that using multiple methods and cross-reacting antibodies may introduce artifacts. Moreover, not all proteins that function in the same biological process also interact physically or co-localize. Those types of relationships are identified using assays with phenotypic readouts, such as genetic interactions 7 or metabolic profiles 8, but have yet to be applied on a genome scale for human proteins.
One of the oldest functional genomics methods is gene expression profiling 9. Genes with correlated activity may participate in similar cellular functions, and coexpression with known genes can be exploited to infer functions of uncharacterized genes 10–12. However, predicting gene function from coexpression can result in inaccurate results 13,14. One reason for this is that gene activity is measured at the mRNA level, which neglects the contribution of protein synthesis and degradation to gene expression control. The precise extent to which protein levels depend on mRNA abundances may differ among genes 15. Further, fundamental differences between mRNA levels and protein expression have emerged. For example, many genes coexpress mRNAs due to their chromosomal proximity, rather than any functional similarity 13,16,17. This non-functional mRNA coexpression results from stochastic transitions between active and inactive chromatin that affect loci genome-wide 16–18, and transcriptional interference from nearby genes 17,19. Importantly, coexpression of spatially close, but functionally unrelated genes, is buffered at the protein level 13,17. Genetic variation affects protein abundance far less than it affects mRNA levels 20, including variations in gene copy numbers 21,22. Therefore protein expression profiling is superior to mRNA expression profiling for prediction of gene function 13,14.
Proteome-level expression profiling underpins protein covariation analysis. For example, protein covariation can be used to infer the composition of protein complexes and organelles 23–31. Most studies to date have focused on relatively small sets of proteins or a few biological conditions, or analysed specific cellular structures. In addition, the scale of coexpression analyses has been limited by the set of statistical tools available. Coexpressed genes are commonly identified using Pearson’s correlation, which is restricted to linear correlations and susceptible to outliers. Machine-learning may offer better sensitivity and specificity. Here we applied large scale quantitative proteomics and machine learning to produce a protein covariation dataset that will enable assignment of functions to human proteins.
Results
ProteomeHD captures protein perturbations
To turn protein covariation analysis into a system-wide, generally applicable method, we created ProteomeHD. In contrast to previous drafts of the human proteome 5,6,32,33, ProteomeHD does not catalogue the proteome of specific tissues or subcellular compartments. Instead, ProteomeHD catalogues the transitions between different proteome states, i.e. changes in protein abundance or localization resulting from cellular perturbations. HD, or high-definition, refers to two aspects of the dataset. First, all experiments are quantified using SILAC (stable isotope labelling by amino acids in cell culture) 34. SILAC essentially eliminates sample processing artifacts and is especially accurate when quantifying small fold-changes. This is crucial to detect subtle, system-wide effects of a perturbation on the protein network. Second, HD refers to the number of observations (pixels) available for each protein. As more perturbations are analysed, regulatory patterns become more refined and can be detected more accurately.
To assemble ProteomeHD we processed the raw data from 5,288 individual mass-spectrometry runs into one coherent data matrix, which covers 10,323 proteins (from 9,987 genes) and 294 biological conditions (Supplementary Table 1). 80 of these conditions, including 43 previously unpublished experiments, were performed in our laboratory and the remaining data were collected from the Proteomics Identifications (PRIDE) 35 repository (Fig. 1a, see Supplementary Table 2 for a complete list of conditions and PRIDE identifiers). These data cover a wide array of quantitative proteomics experiments, such as perturbations with drugs and growth factors, genetic perturbations, cell differentiation studies and comparisons of cancer cell lines. All experiments are comparative studies using SILAC 34, which do not report absolute protein concentrations but report instead fold-changes in response to perturbation. About 60% of the experiments included in ProteomeHD analysed whole-cell samples. The remaining measurements were performed on samples that had been fractionated after perturbation, e.g. to enrich for chromatin-based or secreted proteins (Fig. 1a). This allows for the detection of low-abundance proteins that may not be detected in whole-cell lysates.
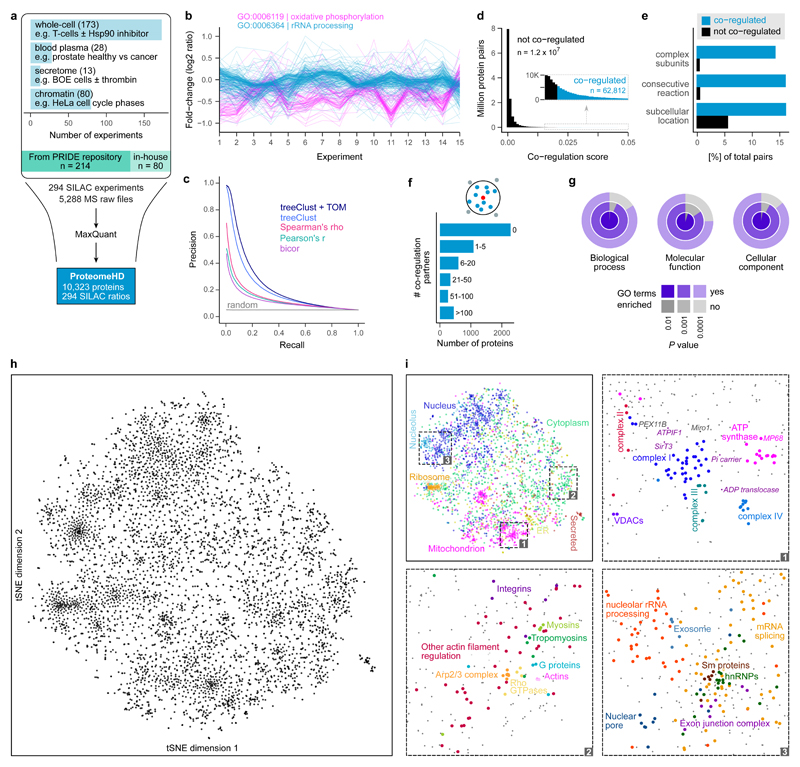
Figure 1. Co-regulation map shows associations between human proteins.
(a) Assembly of ProteomeHD, which quantifies the protein response to 294 perturbations using SILAC 34. Most measurements document protein abundance changes in whole-cell samples, but in some cases subcellular fractions were enriched to detect low-abundance proteins. Data were collected from PRIDE 35 and produced in-house. (b) A random set of experiments from ProteomeHD, showing that groups of proteins with related functions, e.g. Gene Ontology 41 (GO) biological processes, display similar expression changes. Note that the fold-changes are often very small. (c) Precision - recall analysis showing that the treeClust 37,38 algorithm outperforms three correlation-based coexpression measures. Applying the topological overlap measure (TOM) improves performance further. Annotations in Reactome 36 were used as gold standard. (d) Co-regulation scores for all protein pairs are obtained by combining treeClust with TOM. The score distribution is highly skewed. Where an arbitrary threshold is required, the highest-scoring 0.5% of pairs (N = 62,812) are considered “co-regulated”. (e) Co-regulated protein pairs are strongly enriched for subunits of the same protein complex, enzymes catalysing consecutive metabolic reactions and proteins with identical subcellular localization. (f) Most proteins are co-regulated with no or few other proteins, but many have more than 5 co-regulated partners. (g) Considering proteins that are co-regulated with ≥10 proteins, these groups of co-regulated proteins are almost always enriched in one or more GO terms. (h) The global co-regulation map of ProteomeHD created using t-Distributed Stochastic Neighbor Embedding (t-SNE). Distances between proteins indicate how similar their expression patterns are. See www.proteomeHD.net for an interactive version of the map. n = 5,013 proteins. (i) The co-regulation map broadly corresponds to subcellular compartments, and more detailed functional associations can be observed at higher resolution, as exemplified in subpanels 1-3.
Protein coverage in ProteomeHD
On average, each of the 10,323 human proteins in ProteomeHD was quantified on the basis of 28.4 peptides with a sequence coverage of 49% (Supplementary Fig. 1). Not every protein is quantified in every condition. The 294 input experiments quantify 3,928 proteins on average. Each protein is quantified in 112 biological conditions on average (Supplementary Fig. 1). Coexpression studies usually discard transcripts detected in less than half of the samples. However, because ProteomeHD is considerably larger than a typical coexpression analysis, we lowered this arbitrary cut-off to include proteins for downstream analysis if they were quantified in about a third of the conditions. We focused our co-regulation analysis on the 5,013 proteins that were quantified in at least 95 of the 294 perturbation experiments. These 5,013 proteins were quantified in at least 190 conditions; 43% were quantified in more than 200 conditions (Supplementary Fig. 1).
Machine-learning captures protein associations
Proteins that function together have similar patterns of up- and down regulation across the many conditions and samples in ProteomeHD. For example, the patterns of proteins belonging to two well-known biological processes, oxidative phosphorylation and rRNA processing, can be clearly distinguished, even though most expression changes are below 2-fold (Fig. 1b). Therefore, we reasoned that it should be possible to reveal the function of unknown proteins by associating their regulatory patterns with those of well-characterized proteins.
Pearson’s correlation coefficient (PCC) is applied to determine the extent of coexpression between two genes. Since PCC is very sensitive to outlier measurements, Spearman’s rank correlation (rho) or Biweight midcorrelation (bicor) are sometimes used as more robust alternatives. We calculated all three correlation coefficients for 12,562,578 pairwise combinations of the 5,013 protein subset of ProteomeHD. To assess which metric works best for ProteomeHD we performed a precision-recall analysis, using functional protein - protein associations from Reactome 36 as the gold standard. This showed no substantial differences between the correlation measures, although Spearman’s rho performs slightly better than the others (Fig. 1c).
Next we evaluated a coexpression measure based on unsupervised machine-learning. We used the treeClust algorithm developed by Buttrey and Whitaker, which infers dissimilarities based on decision trees 37,38. treeClust runs data through a set of decision trees, which it creates without explicitly provided training data, and essentially counts how often two proteins end up in the same leaves. This results in pairwise protein - protein dissimilarities (not clusters of proteins). Importantly, we found that treeClust dissimilarities were superior to PCC, rho and bicor for prediction of functional relationships between proteins in ProteomeHD (Fig. 1c).
Finally, we applied a topological overlap measure (TOM) 39,40 to the treeClust similarities, which further enhanced performance by approximately 10% as judged by the area under the precision-recall curve (Fig. 1c). The TOM is typically used to improve the robustness of correlation networks by re-weighting connections between two nodes according to how many shared neighbors they have. The TOM-optimised treeClust results form our “co-regulation score”. This score is continuous and reflects how similar two proteins behave across ProteomeHD, i.e. the higher the score the more strongly co-regulated two proteins are. However, for some questions a simplified categorical interpretation is more straightforward. In these cases we arbitrarily consider the top-scoring 0.5% of proteins pairs as “co-regulated”. In this way, we identified 62,812 co-regulated protein pairs (Fig. 1d, Supplementary Table 3). Analysing the same data with Pearson’s correlation, and selecting the top 0.5% pairs would correspond to a cut-off of PCC > 0.69, which is generally considered a strong correlation.
We tested whether co-regulation corresponds to co-function. We found that co-regulated protein pairs are enriched for subunits of the same protein complex, enzymes catalysing linked metabolic reactions and proteins in the same subcellular compartments (Fig. 1e). Most proteins are co-regulated with at least one other protein, and about a third have more than five co-regulation partners (Fig. 1f). For 99% of the tested proteins that had ≥ 10 co-regulated pairs, the group of their co-regulation partners is enriched in at least one Gene Ontology 41 biological process (Fig. 1g).
treeClust improves protein co-regulation analysis
While decision trees are well-understood building blocks of many established machine-learning algorithms, it was unclear which type of information treeClust was capturing from our dataset. For example, treeClust scores could reflect whether two proteins are detected in the same set of samples. Protein co-occurrence can be measured using the Jaccard similarity coefficient, which has previously been exploited to identify protein-protein associations 28. We compared treeClust scores to this Jaccard index (Supplementary Fig. 2). In addition, we forced treeClust to learn dissimilarities solely based on co-occurrence by using a “binary” version of ProteomeHD, in which all SILAC ratios were turned into ones and all missing values were turned into zeroes. We found that the Jaccard index and “binary” treeClust detect functionally related proteins equally well, but with much lower precision than standard treeClust (Supplementary Fig. 2). This suggests that protein co-regulation, that is coordinated changes in protein abundance, rather than solely co-detection underpins the superior performance of treeClust.
Nevertheless, it remained unclear which type of quantitative relationships treeClust can identify, and how it is affected by missing values, outliers and noise. To address this, we created a series of synthetic datasets that allowed us to systematically assess the properties of treeClust dissimilarities. For example, we created a synthetic dataset consisting of 100 variables (“experiments”, “samples” or “biological conditions”) and 200 observations (“proteins”). The dataset is built in such a way that 99.5% of the resulting pairwise “protein - protein” associations are random, i.e. values for both proteins are random samples of a normal distribution (Fig. 2a). The remaining 0.5% pairs are designed to have a clearly defined, linear relationship across the 100 “experiments”. We modified various properties of such synthetic data and assessed how they affect treeClust. For example, we found that treeClust exclusively detects linearly correlated pairs, in contrast to correlation metrics which also detect exponential and logistic relationships (Supplementary Fig. 3a,b). Moreover, treeClust only partially separates anti-correlated from random associations, suggesting that low treeClust similarities indicate a lack of correlation rather than anti-correlation (Supplementary Fig. 3c).
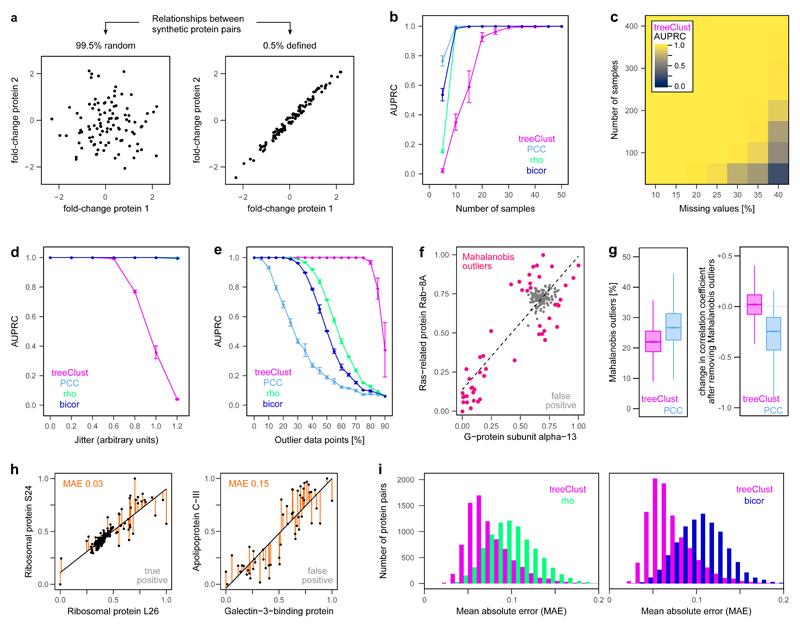
Figure 2. treeClust improves co-regulation analysis through robust selectivity for close linear relationships.
(a) To benchmark treeClust we created a series of synthetic datasets with defined properties. For example, one dataset contains 100 variables and 200 proteins, designed such that out of all possible 19,900 combinations between these proteins, 0.5% have a defined linear relationship, while the remaining 99.5% of pairs have not. We modify the properties of the synthetic data and assess their effect using Precision-Recall (PR) analysis. (b) Impact of sample number on treeClust, Pearson (PCC), Spearman (rho) or Biweight Midcorrelation (bicor). Shown is the average area under the PR curve (AUPRC) of three replicates, error bars indicate the standard error of the mean. n = 500 synthetic proteins. (c) Combinatorial impact of sample number and missing values on treeClust performance. n = 1,000 synthetic proteins. (d) Impact of lowering the goodness-of-fit by increasing the difference between variables (jitter; see Supplementary Fig. 5a for example scatter plots). n = 50 synthetic samples, 500 proteins. (e) Impact of increasing the percentage of outlier measurements. n = 100 synthetic samples, 500 proteins. (f) Real protein pair in ProteomeHD with outliers detected via their Mahalanobis distance. Note in this example outliers drive high PCC even though no general correlation exists. Fold-changes have been scaled to lie between 0 and 1. (g) Co-regulated protein pairs across ProteomeHD were divided into those detected only by treeClust (n = 8,786) or only by PCC (n = 9,593). The latter group contains more outliers. Removing these outliers decreases the PCC of PCC-specific pairs, suggesting their original high PCC was driven by the outliers. Lower and upper hinges correspond to the first and third quartiles, and lower and upper whiskers extend to the smallest or largest value no further than 1.5 interquartile ranges from the hinge, respectively. (h) Two examples pairs from ProteomeHD to illustrate different goodness-of-fit, quantified via the mean absolute error (MAE). Note that only the left pair represents a genuine interaction. (i) Systematic comparison of MAEs across ProteomeHD, from co-regulated pairs detected by treeClust but not rho or bicor (magenta), or by rho or bicor but not treeClust (green and blue, respectively).
We found that treeClust requires a larger dataset than correlation metrics to reach optimal performance, including more experiments (Fig. 2b), more proteins and a higher proportion of defined relationships (Supplementary Fig. 4a-c). By randomly introducing missing values we showed that their impact on treeClust performance depends on the size of the dataset (Fig. 2c, Supplementary Fig. 4d-f). Importantly, the subset of ProteomeHD we used for co-regulation analysis consists of 294 samples and 5,013 proteins, has 35% missing values, and is therefore well within the identified size margins of optimal treeClust performance. In addition, by increasing the dispersion of values around the linear associations (Supplementary Fig. 5a) we found that treeClust specifically detects very close-fitting linear associations, in contrast to correlation metrics (Fig. 2d). Finally, introducing outliers in synthetic data (Supplementary Fig. 5b) showed that treeClust is exceptionally robust against outlier measurements (Fig. 2e).
We then tested which of these properties underpins treeClust’s superior performance on ProteomeHD. For this we analysed co-regulated protein pairs that were detected either by treeClust or by PCC, but not by both (Supplementary Fig. 6a). We observed that outlier measurements lead to PCC detecting many false-positive associations (Fig. 2f,g), while missing many true-positive ones (Supplementary Fig. 6d-f). However, the moderate number of outliers in ProteomeHD has a minor impact on rho and bicor coefficients (Supplementary Fig. 6b-f). Protein pairs that are exclusively detected by rho and bicor, which are largely false-positives (Supplementary Fig. 6a), tend to have a poor goodness-of-fit (Fig. 2h,i). This goodness-of-fit difference is not as pronounced between treeClust and PCC (Supplementary Fig. 7). Taken together, this suggests that treeClust outperforms PCC mainly due to superior outlier handling, whereas its improvement over rho and bicor is predominantly due to treeClust taking into account the “goodness-of-fit” of an association.
The selectivity of treeClust for strong linear relationships implies that it may miss potentially important non-linear associations in ProteomeHD. However, we failed to detect any exponential or logistic associations, suggesting that the vast majority of the interactions in ProteomeHD are of linear nature (Supplementary Fig. 8).
A co-regulation map of the human proteome
treeClust outputs how strongly or weakly each protein is co-regulated with any other protein. In principle, these outputs could be displayed as a scale-free protein interaction network with edges indicating co-regulation (Supplementary Fig. 9). However, such a graph would not be informative due to the size of our co-regulation data (62,812 top-scoring links between 5,013 proteins).
Instead we visualized the protein - protein co-regulation matrix using t-Distributed Stochastic Neighbor Embedding (t-SNE) 42. This produces a two-dimensional proteome co-regulation map in which the distance between proteins indicates how similar they responded to the various perturbations in ProteomeHD (Fig. 1h, Supplementary Table 4). Notably, t-SNE takes all pairwise co-regulation scores into account, rather than focussing on a small number of links above an arbitrary threshold.
Our t-SNE map shows that protein co-regulation is closely related to co-function. For instance, the map reflects the subcellular organization of the cell (Fig. 1i). It broadly separates organelles and separates the nucleolus from the nucleus. Zooming in, the five protein complexes of the respiratory chain are almost resolved (Fig. 1i, section 1). It is possible to discern the phosphate and ADP carriers that transport the substrates for ATP synthesis through the inner mitochondrial membrane, and ATPIF1 - a short-lived, post-transcriptionally controlled key driver of oxidative phosphorylation in mammals 43. Similarly, cytoskeleton proteins such as actins and myosins are next to their regulators, including Rho GTPases and the Arp2/3 complex (Fig. 1i, section 2). Groups of proteins involved in RNA biology, from nucleolar rRNA processing to mRNA splicing and export (Fig. 1i, section 3) are correctly together. This map is generated solely on the basis of protein abundance changes in ProteomeHD without any curated information.
Proteome map complements orthogonal genomics methods
We checked whether protein co-regulation can predict associations that are not detected by other methods by comparing co-regulation with four alternative large-scale resources: IntAct, BioGRID, STRING and BioPlex. The first three are meta-resources that compile curated sets of protein - protein interactions (PPIs) from the results of thousands of individual studies. Since meta-resources generally map interactions to gene loci rather than proteins, we disregarded protein isoforms for this comparison and focused on co-regulated genes.
Our co-regulation map covers fewer distinct genes than other resources, but only STRING captures on average more interactions per gene (Fig. 3a). Based on the 2,565 genes covered by both approaches, around 39% of the gene pairs identified as co-regulated had previously been linked in STRING (Fig. 3b). This suggests that co-regulation can confirm existing links and identify new links. Conversely, only 7% of STRING PPIs are co-regulated, which may reflect the diverse set of associations in STRING. Notably, the overlap between the resources depends on the stringency setting: considering fewer, more stringent STRING interactions decreases the number of co-regulated genes and increases STRING PPIs identified as co-regulated (Fig. 3b). An equivalent trend would be observed when modulating the co-regulation cut-off. STRING associations are based on multiple types of evidence, of which “mRNA coexpression” unsurprisingly shows the highest individual overlap with protein co-regulation results (Fig. 3c).
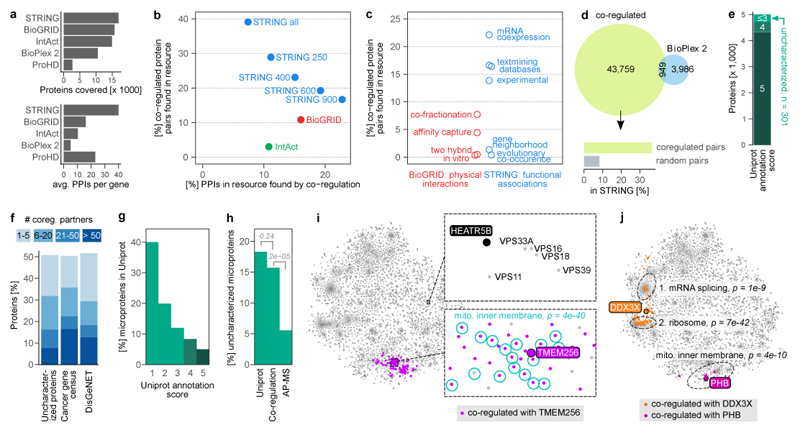
Figure 3. Protein co-regulation predicts functions of unknown proteins.
(a) Coverage of protein - protein interactions (PPIs) in comparison to other resources. Top barchart shows the number of genes covered, i.e. having at least one PPI above cut-off. STRING cut-off used: medium (400). Bottom chart shows the average number of PPIs of covered genes. The co-regulation map (ProHD) covers fewer genes than STRING, BioGRID, IntAct and BioPlex 2, but covers many associations between those genes. (b) Overlap between PPIs discovered by protein co-regulation and PPIs already present in large-scale annotation resources that cover both physical (BioGrid and IntAct) and functional (STRING 69) associations. Multiple association score cut-offs were considered for STRING. These three resources integrate data from many small and large-scale studies. (c) Coverage of co-regulated protein pairs in BioGRID and STRING broken down by the type of functional genomics evidence available in each resource. (d) Number of co-regulation links compared to PPIs found for the same set of genes by BioPlex 2.0 2, one of the largest PPI datasets reported to date by a single study. Associations unique to co-regulation are strongly enriched for links in STRING, compared to random gene pairs. (e) Out of the 5,013 proteins in the co-regulation map, 301 have a UniProt annotation score ≤3 and are thus defined as uncharacterized. (f) Connectivity of either uncharacterized proteins or proteins encoded by disease genes to well-characterized proteins (annotation score ≥4). 51% of uncharacterized proteins have at least one co-regulation partner, 32% have more than five. (g) Barchart showing the percentage of all 20,408 human UniProt (SwissProt) proteins that are microproteins, i.e. have a molecular weight < 15 kDa. Note that microproteins are heavily enriched among less well-characterized proteins. (h) 18% of 5,187 uncharacterized proteins in UniProt are microproteins, compared to 16% of the 153 uncharacterized proteins in the co-regulation map and 6% of 1,422 uncharacterized proteins in state-of-the-art AP-MS experiments, represented by BioPlex. P-values are from one-sided Fisher’s Exact test. (i) The uncharacterized microprotein TMEM256 has many co-regulation partners (n = 130), which are enriched for GO term “mitochondrial inner membrane” (n = 42) among others. Bonferroni-adjusted P-value is from a hypergeometric test. The uncharacterized HEATR5B protein has no co-regulation partners above the default threshold, but its position in the map nevertheless indicates a potential function. (j) For multifunctional proteins, co-regulation can reveal a mix of their functions (DDX3X; n = 14 of 81 co-regulated proteins annotated with GO term “mRNA splicing, via spliceosome, n = 27 with GO term “cytosolic ribosome”), or their main function only (prohibitin, PHB; n = 9 of 11 co-regulated proteins annotated as “mitochondrial inner membrane”). Three representative GO terms are shown.
Next, we compared ProteomeHD-informed co-regulation with physical PPIs catalogued in IntAct and BioGRID. We find that 11% of co-regulated gene pairs have a documented physical protein-protein interaction in BioGRID, and 3% are found in the smaller IntAct database (Fig. 3b).
Finally, we compared our co-regulation approach to a functional genomics project named BioPlex 2.0, which is the most comprehensive affinity purification–mass spectrometry (AP-MS) study reported to date 2. BioPlex reports 4,935 physical interactions between the proteins used in our study, of which 19% are also co-regulated (Fig. 3d). An additional 43,759 potential links between these proteins are identified uniquely by co-regulation. These are strongly enriched for functional protein associations found in STRING, compared to a random set of protein pairs (Fig. 3d).
These comparisons indicate that protein co-regulation identifies protein - protein associations in a way that is reliable and complementary to existing functional genomics methods. Proteins can interact physically without being co-regulated, and vice versa. In summary, protein co-regulation complements other approaches by revealing additional associations and by providing independent evidence for previously detected associations.
ProteomeHD contains difficult-to-characterise proteins
The co-regulation map contains 301 proteins that we defined as uncharacterized proteins because they have a UniProt 44 annotation score of 3 or less (Fig. 3e). Of these, 51% are co-regulated with at least one fully characterized protein (a protein with a UniProt annotation score of 4 or 5) and a median of 9 (Fig. 3f), making it possible to predict their potential function in a “guilt by association” approach. We observed a similar number of fully characterised proteins as co-regulation partners for genes that cause cancer when mutated (listed in the cancer gene census 45), and for genes implicated in a broad range of human diseases (listed in DisGeNET 46) (Fig. 3f). Therefore, protein co-regulation may also be helpful for functional analysis of human disease genes.
Many uncharacterized proteins are small; proteins smaller than 15 kDa constitute 18% of uncharacterized versus 5% of characterized human proteins. 40% of proteins with a UniProt annotation score of 1 are smaller than 15 kDa (Fig. 3g). Of note, hundreds or thousands of these so-called microproteins have been overlooked by genome annotation efforts 47. Microproteins can regulate fundamental biological processes 48, but their size makes it difficult to identify interaction partners 47,49 or to target them in mutagenesis screens 47. Microprotein sequences also tend to be less conserved than those of longer protein-coding genes 50.
We reasoned that our perturbation proteomics approach might be less biased by protein size than methods involving extensive genetic or biochemical sample processing. Indeed, we find that 16% of the uncharacterized proteins in the co-regulation map are smaller than 15 kDa (Fig. 3h). This is a significant difference to BioPlex’s cutting-edge AP-MS data, in which microproteins drop to 6% (p < 2e-5 in a one-tailed Fisher’s Exact test).
The fact that microproteins are not underrepresented in ProteomeHD does not automatically mean that their detection and characterisation is as robust as that of larger proteins. However, the average microprotein in the co-regulation map has been identified by 12.2 peptides, many of which overlap and together result in an average sequence coverage of 76.4% (Supplementary Fig. 10a, d). While in a typical SILAC experiment proteins are considered to be quantifiable from upwards of two independent observations (SILAC ratio counts), microproteins in the co-regulation map are quantified with an average of 9 ratio counts per experiment, totalling a median of 671 ratio counts across ProteomeHD (Supplementary Fig. 10b, c). This indicates that microprotein quantitation in ProteomeHD is robust. Surprisingly, we find that microproteins have more co-regulation partners than larger proteins, and the same is true for their connectivity in STRING (Supplementary Fig. 10f). Within STRING, the majority of microprotein interactions are derived from curated annotations rather than high-throughput efforts such as RNA coexpression and text mining (Supplementary Fig. 10g). Note that, based on BioGRID, microproteins engage in fewer physical PPIs than larger proteins. This may be the result of an experimental bias (microproteins may dissociate more easily during purification and are more difficult to detect) or reflect a biological property (microproteins may have fewer physical interaction partners). In either case, co-regulation offers itself as a powerful alternative approach to study microprotein functions in a systematic way.
Functional annotation of proteins by co-regulation
We created the website www.proteomeHD.net to enable users to search for a protein of interest, showing its position in the co-regulation map together with any co-regulation partners (Supplementary Fig. 11). The online map is interactive and zoomable, making it easy to explore the neighborhood of a query protein. The co-regulation score cut-off can be adjusted and statistical enrichment of Gene Ontology 41 terms among the co-regulated proteins is automatically calculated.
For example, protein co-regulation can be used to predict the potential function of uncharacterized microproteins such as the mitochondrial proteolipid MP68. MP68 is co-regulated with subunits of the ATP synthase complex, suggesting a function in ATP production (Fig. 1i, section 1). Despite being only 6.8 kDa small, its presence in the co-regulation map is documented by 8 distinct peptides that were observed a total of 398 times across 142 experiments (Supplementary Fig. 10e). Intriguingly, MP68 co-purifies biochemically with the ATP synthase complex, but only in buffers containing specific phospholipids 51,52, and knockdown of MP68 decreases ATP synthesis in HeLa cells 53.
Virtually nothing is known about the 12 kDa microprotein TMEM256, although sequence analysis suggests it may be a membrane protein. Its position in the co-regulation map (Fig. 3i) and GO analysis of its co-regulation partners indicates that it likely localizes to the inner mitochondrial membrane (GO:0005743, Bonferroni adj. p < 5e-40), where it may participate in oxidative phosphorylation (GO:0006119, p < 3e-35).
Some proteins have no co-regulation partners above the default score cut-off, but can still be functionally annotated through the co-regulation map. The uncharacterized 224 kDa protein HEATR5B, for example, is located in an area related to vesicle biology (Fig. 3i). Its immediate neighbours are five subunits of the HOPS complex, which mediates the fusion of late endosome to lysosomes. The position in the map shows that the HOPS complex is the closest fit to HEATR5B’s regulation pattern, but they are not as similar as the top-scoring pairs in our overall analysis. If the co-regulation score cut-off is lowered, HOPS subunits and other endolysosomal proteins are eventually identified as co-regulated with HEATR5B, with concomitant enrichment of the related GO terms. This suggests that HEATR5B may not itself be a HOPS subunit, but could have a related vesicle-based function. Notably, a biochemical fractionation profiling approach also predicted HEATR5B to be a vesicle protein 54.
Multifunctional proteins appear to fall into two categories in terms of co-regulation behavior. Prohibitin, for example, functions both as a mitochondrial scaffold protein and a nuclear transcription factor 55. However, only the mitochondrial function is represented in the co-regulation map (Fig. 3j). This could indicate that its nuclear activity is not relevant in the biological conditions covered by ProteomeHD, or that only a small intracellular pool of prohibitin is nuclear, so that changes in its nuclear abundance are insignificant in comparison to the mitochondrial pool. In contrast, the helicase DDX3X shuttles between nucleus and cytoplasm, functioning both as nuclear mRNA processing factor and cytoplasmic regulator of translation 56. In the co-regulation map, DDX3X sits between the areas related to these two activities and is significantly co-regulated both with proteins involved in nuclear RNA biology and with translation factors (Fig. 3j). Therefore, DDX3X is a multifunctional protein whose separate activities result in a mixed regulatory pattern.
The protein co-regulation data presented here has been integrated into the recently released 11th version of STRING 57 (https://string-db.org/). In STRING’s human protein - protein association network, links between proteins inferred from co-regulation in ProteomeHD are shown as network edges of the “coexpression” type (Supplementary Fig. 12). Therefore, STRING is an alternative source for users wishing to explore protein co-regulation in conjunction with other types of association evidence.
PEX11β and peroxisome-mitochondria interactions
Some well-characterized proteins have unexpected co-regulation partners. For example, PEX11β is a key regulator of peroxisomal membrane dynamics and division 58. However, PEX11β’s co-regulation partners are not peroxisomal proteins but subunits of the mitochondrial ATP synthase and other components of the electron transport chain (Fig. 1i, section 1). These proteins are located to the inner mitochondrial membrane, making a physical interaction with PEX11β unlikely. However, peroxisomes and mitochondria in mammals are intimately linked cooperating in fatty acid β-oxidation and ROS homeostasis 59. How these organelles communicate or mediate metabolite flux has been elusive. Live cell imaging revealed that expression of PEX11β-EGFP in mammalian cells induced the formation of peroxisomal membrane protrusions, which interact with mitochondria (Fig. 4, Supplementary movies 1-3). Interactions of elongated peroxisomes with mitochondria were more frequent than those of spherical organelles, but both interactions were long-lasting (Fig. 4n,o). This indicates that peroxisome elongation can facilitate organelle interaction, but once organelles are tethered, the duration of contacts is similar between different morphological forms. Miro1 (RHOT1), a membrane adaptor for the microtubule-dependent motors kinesin and dynein 60, is also co-regulated with PEX11β (Fig. 1i, section 1). We and others recently showed that Miro1 distributes to mitochondria and peroxisomes 61,62 indicating that it coordinates mitochondrial and peroxisomal dynamics with local energy turnover. Peroxisome-targeted Miro1 (Myc-Miro-PO) can be used as a tool to exert pulling forces at peroxisomal membranes, which results in the formation of membrane protrusions in certain cell types 63 (Supplementary Fig. 13). We show here that silencing of PEX11β inhibits membrane elongation by Myc-Miro-PO, confirming that PEX11β is required for the formation of peroxisomal membrane protrusions (Supplementary Fig. 13). These findings are in agreement with studies in plants, where AtPEX11a has been reported to mediate the formation of peroxisomal membrane extensions in response to ROS 64. In yeast, peroxisome-mitochondria contact sites are established by ScPex11 and ScMdm34, a component of the ERMES complex 65. Additional tethering functions for the yeast mitofusin Fzo1 and ScPex34 in peroxisome–mitochondria contacts have recently been revealed 66. Importantly, the study also demonstrated a physiological role for peroxisome–mitochondria contact sites in linking peroxisomal β-oxidation and mitochondrial ATP generation by the citric acid cycle 66. We conclude that PEX11β and Miro1 contribute to peroxisome membrane protrusions, which present a new mechanism of interaction between peroxisomes and mitochondria in mammals. They likely function in the metabolic cooperation and crosstalk between both organelles, and may facilitate transfer of metabolites such as acetyl-CoA and/or ROS homeostasis during mitochondrial ATP production. These findings now enable future studies on the precise functions of peroxisome membrane protrusions in mammalian cells and the role of PEX11β.
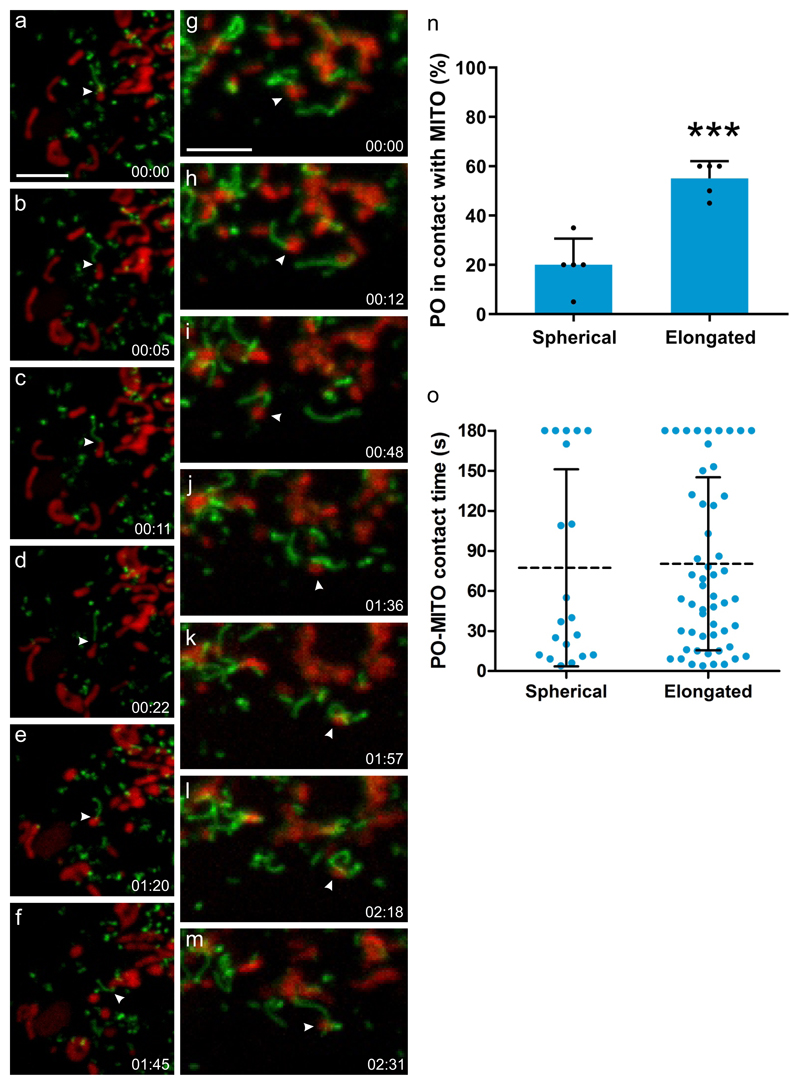
Figure 4. PEX11β and peroxisomal membrane interactions with mitochondria.
(a-m) COS-7 cells were transfected with PEX11β-EGFP, mitochondria were stained with Mitotracker (red) and cells observed live using a spinning disc microscope. PEX11β, a membrane shaping protein, induces the formation of tubular membrane protrusions from globular peroxisomes. We show here that those membrane protrusions can interact with mitochondria. Results are representative of three independent experiments. (a-f) shows a peroxisome which interacts with a mitochondrion via its membrane protrusion (arrowhead), and follows it, occasionally detaching and re-establishing contact before interacting with another mitochondrion (see Supplementary Movie 1). (g-m) shows a mitochondrion (arrowhead) which interacts with a peroxisome via a peroxisomal membrane protrusion. It then detaches and moves away to interact with another peroxisome, which wraps its protrusion around it, before interacting with another mitochondrion (see Supplementary Movie 2). (n) Quantification of interactions between spherical or elongated peroxisomes (PO) with mitochondria (MITO). The average result of 3 independent experiments is shown, error bars indicate the mean +/- standard deviation. (o) Quantification of contact time. Note that elongated PO interact more frequently with MITO than spherical PO, but for similar time periods. PO-MITO interactions are generally long-lasting (see Supplementary Movie 3) (n=200 peroxisomes from 5 different cells). Dotted line indicates the mean, error bars indicate standard deviation. *** P = 0.0003 from a two-tailed unpaired t test; Time (min:sec). Scale bars, 5 µm.
Proteomics for expression profiling
To compare the impact of mRNA and protein abundances on expression profiling we first focussed on 59 SILAC ratios in ProteomeHD that measured abundance changes across a panel of lymphoblastoid cell lines 20. For these samples, corresponding mRNA abundance changes have been determined using RNA-sequencing 67. Repeating treeClust learning on the basis of these data, we observed that protein coexpression predicts functional associations with far higher precision than mRNA coexpression (Fig. 5a). Similar results have recently been reported for a panel of human cancer samples 13.
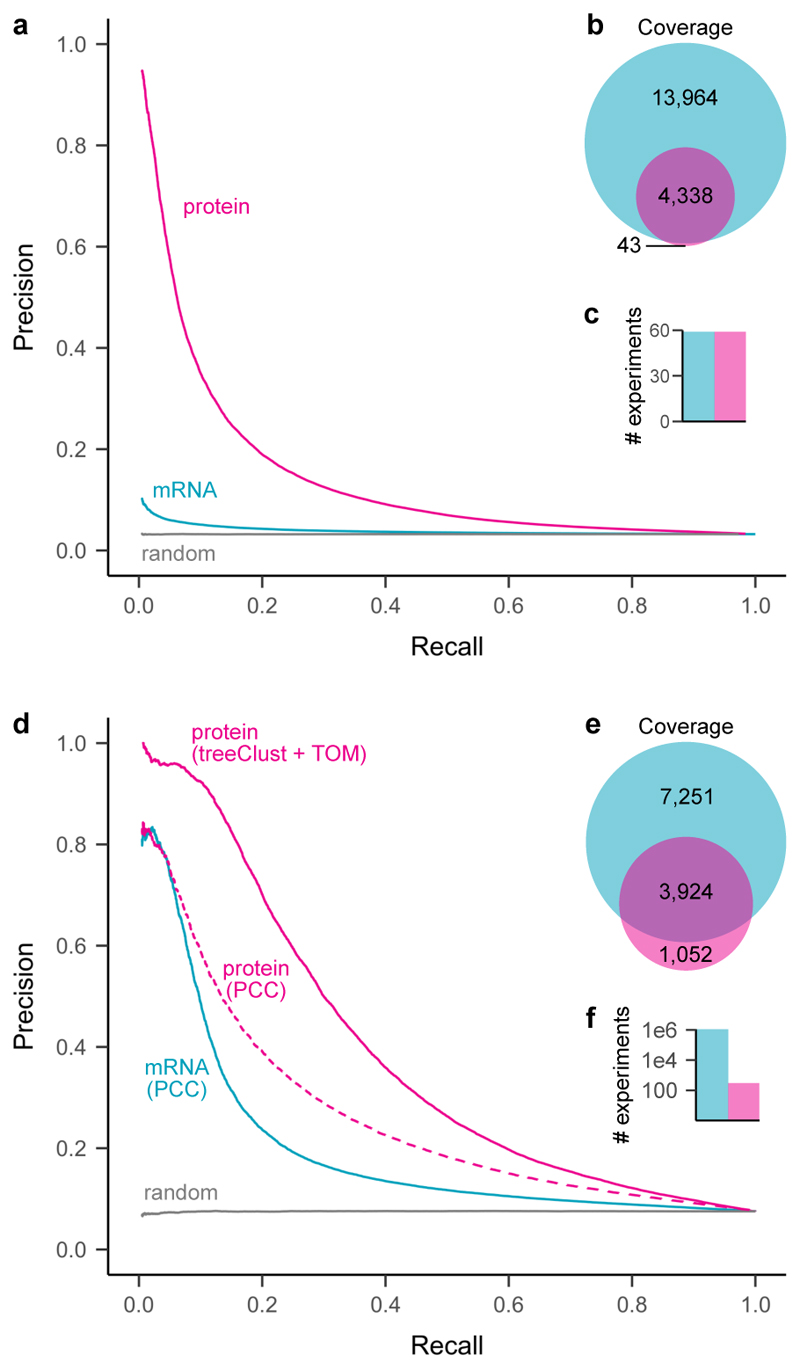
Figure 5. Protein co-regulation enables higher precision but lower coverage than mRNA coexpression.
(a) Precision-recall analysis of treeClust machine-learning on a subset of ProteomeHD, that is 59 samples for which matching RNA-seq data were available from a separate study 67.
Reactome pathways were used as gold standard for true functional associations (proteins found in same pathway) and false associations (never found in same pathway). Only annotated genes covered by both datasets were considered for PR analysis (n = 2,901). (b) Venn diagram showing number of genes covered by each analysis. (c) Barchart showing number of experiments the curves are based on. (d) Similar precision-recall analysis of treeClust machine-learning on the full ProteomeHD database, in comparison to Pearson correlation obtained by STRING 69 on the basis of one million human mRNA profiling samples deposited in the NCBI Gene Expression Omnibus 68 ("mRNA / PCC"). Protein co-regulation outperforms mRNA correlation despite being based on orders-of-magnitude less data. This is partially due to the use of machine-learning, as predicting associations from ProteomeHD using PCC decreases performance markably ("protein / PCC"). Only annotated genes covered by both datasets were considered for the PR analysis (n = 2,743). (e, f) same as (b, c).
Such analyses show that in a direct gene-by-gene, sample-by-sample comparison, protein expression levels are better indicators for gene function than mRNA expression. However, the amount of transcriptomics data published to date vastly exceeds that of proteomics studies. For example, the NCBI GEO repository currently holds mRNA expression profiling data from more than one million human samples 68. This raises the possibility that the sheer quantity of available transcriptomics data could overcome their reduced reflection of functional links and, in combined form, perform better than protein-based measurements. To test this we compared the ProteomeHD co-regulation score with Pearson correlation coefficients obtained by STRING, which leverages the vast amount of mRNA expression experiments deposited in GEO 69. Remarkably, precision-recall analysis shows that the protein co-regulation score still outperforms mRNA coexpression, despite being based on only 294 SILAC ratios (Fig. 5b). Much of this improvement is due to the robustness of treeClust machine-learning, as Pearson’s correlation coefficients derived from the same ProteomeHD data work only moderately better than mRNA correlation (Fig. 5b). While only gene pairs with both mRNA and protein expression measurements were considered for the precision-recall analysis, the transcriptomics and proteomics datasets individually covered 17,436 and 4,976 genes, respectively (Fig. 5b). Therefore, mRNA profiling outperforms protein profiling in terms of gene coverage. In addition, transcriptomics remains the only expression profiling approach suitable for non-coding RNAs.
TreeClust for TMT-based proteomics data
We assessed if treeClust could also improve co-regulation analysis of other isotope-labelled proteomics approaches. For this we applied treeClust to a dataset from Lapek et al 14, which used TMT-based proteomics to monitor protein abundance changes across 41 cancer cell lines. Indeed, we found treeClust to outperform correlation metrics (Supplementary Fig. 14a). Moreover, a t-SNE co-regulation map obtained for Lapek et al’s cancer proteomics dataset contains the complete set of ~6,200 proteins, rather than the 3,024 proteins that correlated with another protein above the author-specified cut-off (Supplementary Fig. 14b).
Discussion
ProteomeHD combined with machine learning adds big-data protein co-regulation analyses to the repertoire of functional genomics methods. A key difference between our approach and previous gene coexpression studies is the application of two machine-learning algorithms, treeClust 37 and t-SNE 42. Inferring protein associations through treeClust learning is more robust and more sensitive than a traditional correlation-based approach, enabling an increase in accuracy with which functionally relevant interactions can be identified from the same dataset. Protein-protein associations visualized by t-SNE can be explored in a hierarchical manner, with larger distances indicating weaker co-regulation. This may be useful for studying connections between related protein complexes (Fig. 1i) or to reveal broad functional clues for uncharacterized proteins for which no detailed predictions are available, such as the HEATR5B protein assigned to the vesicle area of the co-regulation map (Fig. 3i). Our web application at www.proteomeHD.net is designed to support researchers in exploring co-regulation data at multiple scales, to validate existing hypotheses or create new ones.
Only 300 quantitative proteomics measurements sufficed in conjunction with machine learning to establish functional connections between many human genes, which may be of considerable interest for proteome annotation in less studied or difficult to study organisms. Accuracy and coverage could be increased further by adding additional proteomics data. To test this we randomly removed 5%, 10% or 15% of the data points in ProteomeHD. This decreases performance proportionally to the amount of removed data (Supplementary Fig. 15), suggesting that ProteomeHD has not reached saturation and expanding it will further enhance its performance. One possibility would be to incorporate other types of proteomics experiments, such as affinity-purifications or indeed the entire PRIDE 35 repository. However, there is a benefit of restricting ProteomeHD to perturbation experiments. It supports a biological interpretation of protein associations derived from it: two co-regulated proteins are part of the same cellular response to changing biological conditions, even though the precise molecular nature of the connection remains unknown. In this way, protein co-regulation analysis is analogous to genetic interaction screening. This also sets protein co-regulation apart from indiscriminate protein covariation or co-occurrence analyses, which find protein links in a mix of proteomics data and therefore give no insight into the possible biological connection.
In conclusion, protein coexpression analysis identifies functional connections between proteins with an accuracy and sensitivity that is substantially higher than traditional mRNA coexpression analysis. This may be particularly important for constitutively active genes, which constitute about half of human genes 33 and are primarily controlled at the protein level 70. With an ever increasing amount of protein expression data being made available, protein coexpression analysis has huge potential for gene function annotation.
Online Methods
Data selection for ProteomeHD
MS raw data were produced in-house or downloaded from the PRIDE repository 35. Only experiments fulfilling the following inclusion criteria were considered:
(1) Comparative proteomics experiments, i.e. relative protein quantitations of two or more biological states. For example, cells treated with an inhibitor vs. mock control. (2) Biological - not biochemical - comparisons, i.e. fold-changes must have been brought about in vivo, not by differential biochemical purification. For example, SILAC-labelled cells were treated with inhibitor or mock control, harvested and combined, and chromatin was enriched on the combined sample. In such cases any observed fold-change reflects the response to the inhibitor in the living cell, for example a protein re-localising from cytoplasm onto chromatin. We did not consider experiments that compared, for example, a whole-cell lysate with a chromatin-enriched fraction, as this would measure the impact of the biochemical enrichment rather than a biological event. (3) Quantitation by “stable isotope labeling by amino acids in cell culture” (SILAC) 34. (4) Samples of human origin.
In addition to these conceptual considerations, the following restrictions were imposed by the data processing pipeline: (5) The SILAC mass shift introduced by heavy arginine must be distinct from heavy lysine. (6) Raw data acquired on an Orbitrap mass spectrometer. (7) Samples alkylated with iodoacetamide, resulting in carbamidomethylation of cysteines.
In total, we considered 294 experiments (SILAC ratios) from 31 projects. A full list of these is provided in Supplementary Table 2, which also includes the PRIDE identifiers of all previously published datasets.
In-house data collection
80 experiments were performed in-house and analyzed chromatin-enriched samples. Of these, 65 measured the effect of growth factors, radiation and other perturbations on interphase chromatin, which was prepared using Chromatin Enrichment for Proteomics (ChEP) 71. About half of these experiments had previously been published 24. Another 15 experiments documented perturbations specifically on freshly replicated chromatin, which was prepared using Nascent Chromatin Capture (NCC) 72. All mass spectrometry raw files generated in-house have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository 35 with the dataset identifier PXD008888.
MS raw data processing
The 5,288 MS raw files were processed using MaxQuant 1.5.2.8 73 on a Dell PowerEdge R920 server. The following default MaxQuant search parameters were used: MS1 tolerance for the first Andromeda search: 20 ppm, MS1 tolerance for the main Andromeda search: 4.5 ppm, FTMS MS2 match tolerance: 20 ppm, ITMS MS2 match tolerance: 0.5 Da, Variable modifications: acetylation of protein N-termini, oxidation of methionine, Fixed modifications: carbamidomethylation of cysteine, Decoy mode set to reverse, Minimum peptide length: 7 and Max missed cleavages set to 2. The following non-default settings were used: In group-specific parameters, match type was set to “No matching”. In global parameters, “Re-quantify” was enabled, minimum ratio count was set to 1 and “Discard unmodified counterpart peptide” was disabled. Also in global parameters, writing of large tables was disabled. SILAC labels were set as group-specific parameters as indicated in Supplementary Table 2. Canonical and isoform protein sequences were downloaded from UniProt 44 on 28th May 2015, considering only reviewed SwissProt entries that were part of the human proteome. Unprocessed MaxQuant result tables, including peptide evidence data, have been deposited into the PRIDE repository PXD008888.
Protein fold-changes were extracted from the MaxQuant proteinGroups file returned by MaxQuant. Non-normalized SILAC ratios were considered for downstream analysis, log2 transformed and median-normalised. From triple labelling experiments, the heavy/light and medium/light ratios - but not the heavy/medium ratios - were considered. Proteins detected in less than 4 experiments were discarded, as were proteins labeled as contaminants, reverse hits and those only identified by a modification site. The resulting data matrix, ProteomeHD, can be downloaded as Supplementary Table 1.
Calculation of treeClust dissimilarities
It is common in gene coexpression studies to remove genes that were detected in less than half of the samples from the analysis. However, given the unusually large size of ProteomeHD we chose a different arbitrary cut-off, excluding proteins that were detected in less than 95 (about a third) of the 294 experiments. For the remaining 5,013 proteins in ProteomeHD we used the treeClust 37 R package to calculate all 12,562,578 pairwise dissimilarities. Note that treeClust was designed not only to measure inter-point dissimilarities but also to perform clustering 37,38. However, in this study we use it only to calculate dissimilarities, via the treeClust.dist function. The dissimilarity specifier was set to d.num = 2, so that dissimilarities are weighted according to tree quality. We optimised two hyperparameters of treeClust and rpart, which is the routine treeClust uses to create decision trees. These were treeClust’s serule argument, which defines to extent to which trees are pruned, and rpart’s complexity (cp) parameter, which describes the improved fit required to attempt a split. A grid search was performed against the Reactome gold standard (see below) and the area under precision - recall curves was used to identify optimal parameter settings. They were determined to be serule = 1.8 and cp = 0.105, providing approximately a 10% performance improvement over treeClust’s default settings.
Protein co-regulation scores
To calculate the final pairwise co-regulation scores, treeClust dissimilarities were transformed further. First, they were turned into similarities, i.e. 1 - treeClust dissimilarity. Using the WGCNA 74,75 R package, we then performed a sigmoid transformation of these treeClust similarities, creating an adjacency matrix. The settings of parameters mu and alpha for this transformation were optimised in a grid search against the Reactome gold standard, using the area under precision - recall curves as readout. In a third step, the adjacency matrix was transformed into a topological overlap matrix using WGCNA’s TOMsimilarity function, with the TOMDenom parameter set to “mean”. These TOM similarities are the co-regulation scores used throughout our analysis. Co-regulation scores for all of our 12,562,578 protein pairs can be downloaded from the PRIDE repository PXD008888.
While the co-regulation score is continuous, some analyses benefitted from a simplified categorical approach. For these cases we arbitrarily defined the highest-scoring 0.5% of protein pairs as “co-regulated pairs” and the remaining 99.5% of pairs as “not co-regulated pairs”. A list of all 62,812 co-regulated protein pairs is available as Supplementary Table 3. A network of the top 0.5% co-regulated protein pairs can be explored interactively on NDEx 76 (DOI: http://doi.org/10.18119/N9N30Q).
Reactome gold standard
A gold standard set of reference proteins was defined using Reactome 36. Bona fide functionally associated protein pairs (true positives) were defined as protein pairs found in the same “detailed” Reactome pathway. This was inferred from the file UniProt2Reactome.txt (available at https://reactome.org/download-data), where each protein is annotated to the lowest level subset of Reactome pathways. To make sure that only closely related protein pairs were assigned the “true positive” label, we excluded two pathways that were composed of > 200 proteins. We defined protein pairs that are not functionally associated (false positives) as proteins that are never in the same Reactome pathway, at any annotation level. This was inferred from UniProt2Reactome_All_Levels.txt (also available at https://reactome.org/download-data), a file that maps proteins to all levels of the Reactome pathway hierarchy. A copy of this gold standard is available in the Github repository noted above.
Comparison of treeClust and correlation metrics
Pearson’s correlation coefficients (PCC) and Spearman’s rank correlation coefficients (rho) were obtained using the cor function in R, for the same protein pairs covered by the treeClust analysis. Biweight mid-correlation coefficients (bicor) were calculated with default settings using the R package WGCNA 75,77. Changing the maxPOutliers parameter of the bicor function did not improve performance. Precision - recall (PR) analysis was performed with the ROCR package 78 using true and false positive pairs compiled from annotation in Reactome (see paragraph Reactome gold standard). The random classifier was created by scrambling co-regulation scores. AU insert a callout to datasets used and outputs (Supplementary Note?)
Generation of synthetic datasets
Synthetic datasets were generated using a custom function in R (available in our GitHub repository, https://github.com/Rappsilber-Laboratory/treeClust-benchmarking). The function populates a table with values that are randomly sampled from a normal distribution, but includes a user-specified number of observations that have a defined linear relationship with each other. The following properties of the thus created datasets can be manipulated: number of variables (i.e. samples or experiments), number of observations (i.e. proteins), percentage of protein pairs that should have a linear relationship, percentage of outlier data, percentage of missing values and the extent of scatter around the regression line (i.e. biological or measurement noise). Outlier data points are created by random sampling from a broader normal distribution than the rest of the data. In addition to positive linear relationships (y ~ x), we tested relationships that were exponential (y ~ ex), logistic (y ~ 4 / (1 + e-5x)) and quadratic (y ~ x2), as well as linearly anti-correlated (y ~ -x).
Performance evaluation using synthetic data
PR analyses were performed for synthetic data as described for ProteomeHD data above, except that true positive (linear or nonlinear) and false positive (random) associations were known for synthetic data without the need for a gold standard. To test the impact of various data characteristics, synthetic datasets were generated in triplicate and the results were shown as the average area under the PR curves, with error bars indicating the standard error of the mean. No replicates were used for the combinatorial testing of two dataset characteristics.
Model fitting on ProteomeHD data
Base R functions were used to fit and analyse linear models for pairs of proteins in ProteomeHD. Fold-changes of each protein pair were rescaled to fall between 0 and 1 before fitting the model. Outliers were defined as data points with absolute studentized residuals or a Mahalanobis distance larger than 2. Non-linear models were fit using nonlinear least squares. Exponential models (y ~ a + exp(b)x) and logistic models (y ~ a / (1 + e-b(x-c))) were said to outperform the corresponding linear model (y ~ a + bx) if their residual sum of squares (RSS) was at least 10% smaller.
t-SNE visualization
To visualize ProteomeHD as a 2D co-regulation map, co-regulation scores were subjected to t-Distributed Stochastic Neighbor Embedding (t-SNE) 42 using the Rtsne 79 package for R. The theta parameter was set to zero to calculate the exact embedding. The perplexity parameter was set to 50, up from the default of 30, to account for the large size of the co-regulation dataset. 1,500 iterations were performed. However, visual comparison of the t-SNE maps showed that these parameter adaptations provided only a marginal improvement over the default settings. Organelles were labelled based on subcellular locations assigned by UniProt 44 to these proteins, zoom regions were annotated manually based on available literature. Plot coordinates and annotations are available as Supplementary Table 4.
Network visualizations
In addition to t-SNE, the protein co-regulation matrix was also visualized as an undirected, weighted network using the igraph 80 and GGally 81 packages in R. The network contains the same 5,013 proteins as the co-regulation map, but only considers links above the arbitrary co-regulation threshold, i.e. between the top-scoring 0.5% of protein pairs. For these pairs, the network edges are weighted by the co-regulation score. A set of common network layout algorithms were deployed through the sna (social network analysis) 82 R package.
Testing for co-functionality among of co-regulated proteins
To test if protein co-regulation reflects co-function we defined three sets of “functionally related” protein pairs (subunits of the same protein complexes, enzymes catalyzing consecutive metabolic reactions and proteins with identical subcellular localization) as previously described 17.
To test larger groups (not pairs) of co-regulated proteins for functional enrichment, we analyzed enrichment of Gene Ontology terms using the topGO 83 R package. For each protein we tested the group of its co-regulation partners for GO term enrichment. Because some proteins are co-regulated with no or very few other proteins, we restricted the analysis to proteins that are co-regulated with at least 10 proteins. The three aspects (Biological process, Molecular function, Cellular component) of GO were downloaded from QuickGO 84 with taxon set to human and qualifier to null. Rather than the whole proteome, only proteins that were included in the treeClust analysis and had GO annotations were used as the gene “universe” or background for the topGO analysis. Enrichment of GO terms among protein co-regulation groups was tested considering GO graph structure and using a Fisher’s exact test.
Annotation of the co-regulation map
Proteins localizing to specific subcellular compartments were downloaded from UniProt 44 using the following tags: Nucleus (SL-0191), Nucleolus (SL-0188), Endoplasmic reticulum (SL-0095), Mitochondrion (SL-0173), Cytoplasm (SL-0086), Secreted (SL-0243). Proteins and protein complexes in zoom regions (Fig. 1i) were annotated individually based on the available literature.
Creating the www.proteomeHD.net framework
The ProteomeHD online application was written in Python Flask web framework. The interactive plots are generated using Bokeh visualization library for Python (https://github.com/bokeh/bokeh). The Gene Ontology and KEGG enrichment statistics are obtained from a STRING 69 server using an API call with maximally top 100 proteins co-regulated with the query. Only significantly enriched terms (hypergeometric test, Bonferroni adjusted P value < 0.1) are displayed.
Comparison to orthogonal methods
Physical protein-protein-interactions (PPIs) detected by a comprehensive range of small- and large-scale methods were assessed using BioGRID 85, version 3.4.152. Data from IntAct 86 were used as a smaller but curated resource of physical PPIs. Functional protein associations mapped by a large range of methods and publications were inferred from STRING 69, version 10.5. Note that the protein co-regulation scores described here are only used by STRING starting with version 11 57. BioPlex 2.0 2 served as an example for physical interactions mapped by a single project.
Annotation of uncharacterized and disease genes
Proteins were defined as “uncharacterized” on the basis of having an annotation score ≤ 3 in UniProt 44. The UniProt annotation score is a heuristic measure of the annotation state of a protein, expressed as a 5-point system (www.uniprot.org/help/annotation_score). The score combines various types and layers of UniProt annotation, and weights manually curated evidence higher than automated annotation. It may not always agree with the state of “characterization” that field experts would assign to the same protein. However, as an unbiased, data-driven approach we believe the UniProt annotation score is better suited to systematically identify uncharacterized proteins than manual annotation could be. Even with a systematic way of measuring the degree of annotation, the definition of what constitutes an “uncharacterised” protein is an arbitrary one. We chose “3 points or less” as the “uncharacterized” cut-off, because the available information for such proteins tends to be very vague, e.g. a sequence-based prediction as “multi-pass membrane protein”. In contrast, we found that the biological function of most 4-star proteins could be established reasonably well from the available literature.
The Cancer Gene Census, i.e. genes that can cause cancer when mutated, was curated by COSMIC (Catalogue Of Somatic Mutations In Cancer, version 81) 45. DisGeNET was used as a comprehensive, curated list of human gene - disease associations 46.
Comparison of mRNA and protein expression profiling
For the comparison of matched samples and proteins we considered mRNA and protein expression changes across 59 lymphoblastoid cell lines (Fig. 5a). The protein fold-changes are part of ProteomeHD and were originally published by Battle and colleagues 20. RNA-sequencing data for the same cell lines and proteins were also previously reported 67. We used the RNA-sequencing data to calculate mRNA fold-changes relative to a 60th cell line, which was the same cell line used as a SILAC reference for the protein expression data. The combined mRNA and protein dataset has been described in more detail elsewhere 17. Fold-changes for genes covered by both the transcriptomics and proteomics analysis were subjected to treeClust learning (default parameters) and PR curves were obtained as described above.
For a more comprehensive comparison we considered protein associations predicted using treeClust learning or PCC on the basis of all 294 SILAC ratios in ProteomeHD (Fig. 5b). This was compared to mRNA associations inferred by PCC on the basis of all human mRNA expression data processed by STRING. STRING’s state-of-the-art mRNA coexpression analysis pipeline considers all microarray and RNA-sequencing data deposited in the GEO repository 68, resulting in one of the largest mRNA coexpression analyses available to date 69,87. Note that for this comparison we did not use the STRING coexpression score, which is calibrated against the KEGG database, but the original uncalibrated Pearson’s correlations, which were kindly provided by Damian Szklarczyk. STRING PCCs are calculated separately for one- and two-channel microarrays and RNA-sequencing experiments. We used the average of these for the precision - recall analysis, which performed better than any individual experiment type.
Validation of treeClust and t-SNE on the cancer proteomics dataset
Lapek et al measured the abundances for 6,911 proteins in 41 different breast cancer cell lines 14. These data are available as Supplementary Table 2 (tab 3) of their report. As described by Lapek et al, we converted the protein intensities into log2 fold-changes over the median intensity measured for each protein across all cell lines. We then calculated Pearson’s, Spearman’s rank and bicor correlations for all possible protein pairs, as for ProteomeHD. The Spearman’s correlation coefficients obtained in this way are identical to the ones obtained by Lapek et al using the cor.prob function (Supplementary Table 6 in their report 14). We also determined treeClust co-regulation scores for all protein pairs. However, treeClust can only grow one decision tree per input variable, i.e. 41 in this dataset, which would be too few for it to perform properly. To circumvent this, we forced treeClust to generate 1,000 decision trees by applying it iteratively. We created 100 treeClust forests, each generated with a random subset of 10 of the 41 variables, and used the average co-regulation score for downstream analysis. Precision-recall analysis using a Reactome gold standard and t-SNE visualization were performed as described above. The CORUM protein complexes displayed in Lapek et al’s Figure 2, reported in their Supplementary Table 7 14, were color-coded in the co-regulation map.
Comparison of protein co-regulation and co-occurrence
Two different approaches were used to measure protein co-occurrence in ProteomeHD. First, the Jaccard / Tanimoto similarity coefficient 88 was calculated using the Jaccard package for R. Second, a binary version of ProteomeHD was created, where all SILAC ratios were represented by 1s (“protein quantified”) and all missing values were turned to 0s (“protein not quantified”). Subsequently, treeClust dissimilarities were re-calculated based on this binary version of ProteomeHD. The performance of these different metrics was assessed by a precision - recall analysis as described above.
Plasmids, siRNA, and antibodies
For cloning of peroxisome-targeted Miro1, the C-terminal TMD and tail of Myc-Miro1 (kindly provided by P. Aspenström, Karolinska Institute, Sweden) was exchanged by a PEX26/ALDP fragment previously shown to target proteins to the peroxisome membrane 63. PEX11β-EGFP was kindly provided by G. Dodt (Univ. of Tuebingen, Germany). PEX11β siRNA (AUU AGG GUG AGA AUA GAC AGG AUGG) (Eurofins) was previously verified 89. Control siRNA (si-GENOME nontargeting siRNA pool #2) was obtained from GE Healthcare (D-001206-14-05). Antibodies used were as follows: rabbit polyclonal antibody against PEX14 (1:1400, kindly provided by D. Crane, Griffith University, Australia); mouse monoclonal antibody 9E10 against the Myc epitope (1:200, Santa Cruz Biotechnology, Inc., sc-40), rabbit monoclonal antibody against PEX11β (1:1000, Abcam, ab181066); rabbit polyclonal antibody against GAPDH (1:2000, ProSci3783). Secondary anti-IgG antibodies against rabbit (Alexa 594, 1:1000, Molec. Probes/Life Technol. A21207) and mouse (Alexa 488, 1:400, Molec. Probes/Life Technol. A21202) were obtained from ThermoFisher Scientific. HRP-coupled donkey polyclonal antibody against rabbit IgG (1:5000) was obtained from Biorad (172-1013).
Cell culture and transfection
COS-7 cells (African green monkey kidney cells; ATCC CRL-1651), and PEX5 deficient fibroblasts (kindly provided by H. Waterham, AMC, University of Amsterdam, NL) were cultured in DMEM (high glucose, 4.5 g/L) supplemented with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C (5% CO2, 95% humidity) (HERACell 240i CO2 incubator). COS-7 cells were transfected using diethylaminoethyl-dextran (Sigma-Aldrich). dPEX5 fibroblasts have enlarged peroxisomes, which facilitates the visualization of membrane extensions. For transfection of dPEX5 fibroblasts, the Neon® Transfection System (Thermo Fisher Scientific) was used following the manufacturer’s protocol. Briefly, cells (seeded 24h before transfection) were washed once with PBS and trypsinized using TrypLE Express. Trypsinized cells were resuspended in complete medium, pelleted by centrifugation, and washed with PBS. The cells were once again centrifuged and carefully resuspended in 110 μl buffer R. For each condition, 4 × 105 cells were mixed with the DNA construct (5 μg) or with 100 nM siRNA. Cells were microporated using a 100 μl Neon tip with the following settings: 1400 V, 20 ms, one pulse. Microporated cells were immediately seeded into plates with prewarmed complete medium (without antibiotics) and incubated at 37°C with 5% CO2 and 95% humidity. The efficiency of silencing was monitored by immunoblotting of cell lysates and confirmed as previously reported 89.
Immunofluorescence and microscopy
Cells grown on glass coverslips were processed for immunofluorescence 24h after transfection. Cells were fixed for 20 min with 4% paraformaldehyde in PBS (pH 7.4), permeabilized with 0.2% Triton X-100, and blocked with 1% BSA, each for 10 min. Incubation with primary and secondary antibodies took place for 1h each in a humid chamber. Coverslips were washed with ddH2O to remove PBS and mounted with Mowiol medium on glass slides. All immunofluorescence steps were performed at room temperature and cells were washed three times with PBS between each individual step. Cell imaging was performed using an IX81 microscope (Olympus) equipped with an UPlanSApo 100×/1.40 oil objective (Olympus). Digital images were taken with a CoolSNAP HQ2 CCD camera and adjusted for contrast and brightness using the Olympus Soft Imaging Viewer software and MetaMorph 7 (Molecular Devices). For live-cell imaging, COS-7 cells were plated in 3.5 cm diameter glass bottom dishes (Cellvis). MitoTracker Red CMXRos (Life Technologies) at 100 nM was used for visualisation of mitochondria. Live-cell imaging data was collected using an Olympus IX81 microscope equipped with a Yokogawa CSUX1 spinning disk head, CoolSNAP HQ2 CCD camera, 60 x/1.35 oil objective. Digital images were taken and processed using VisiView software (Visitron Systems, Germany). Prior to image acquisition, a controlled temperature chamber was set-up on the microscope stage at 37ºC, as well as an objective warmer. During image acquisition, cells were kept at 37ºC and in CO2–independent medium (HEPES buffered). 200 stacks of 9 planes (0.5 µm thickness, 100 ms exposure) were taken in a continuous stream. All conditions and laser intensities were kept between experiments.
Quantification and statistical analysis of peroxisome morphology and interaction
Analysis of statistical significance was performed using GraphPad Prism 5 software. A two-tailed unpaired t test was used to determine statistical difference against the indicated group. *P < 0.05, **P < 0.01, ***P < 0.001. For analysis of peroxisome morphology, a minimum of 150 cells were examined per condition, and organelle parameters (e.g. membrane protrusions) were microscopically assessed in at least three independent experiments. The analysis was made blind and in different areas of the coverslip. Organelle interaction and contact time were analysed manually from live-cell imaging data using MetaMorph 7 (Molecular Devices). A region of interest (ROI) was drawn in different areas of the cell. Spherical and elongated peroxisomes within the ROI were tracked over the whole time course, and the frequency and duration of contacts monitored. Multiple interactions of the same peroxisome with mitochondria were treated as separate events. Data are presented as mean ± SD.
Statistics
Statistical analyses were performed using R and Prism 5 (GraphPad Software, Inc.). Statistical significance of GO term enrichment was calculated using the topGO 83 R package. Error bars show the standard error of the mean or the standard deviation as indicated in the figure legends. One-sided Fisher’s Exact tests and two-tailed unpaired t-tests were used as indicated in the figure legends.
Supplementary Material
Acknowledgements
We are grateful to Damian Szklarczyk for providing the mRNA Pearson correlation data used by STRING and the STRING team for testing our coregulation data and adding it as novel evidence type to STRING 11. We also thank Karen Wills, Kyosuke Nakamura, Constance Alabert and Anja Groth for contributing chromatin enrichment experiments, and Afsoon S. Azadi for support with live-cell-imaging. This work was supported by the Wellcome Trust through a Senior Research Fellowship to J.R. (grant number 103139) and by the Biotechnology and Biological Sciences Research Council (BB/N01541X/1, BB/R016844/1; to M.S.) and H2020-MSCA-ITN-2018 812968 PERICO (to M.S.). The Wellcome Centre for Cell Biology is supported by core funding from the Wellcome Trust (grant number 203149).
Footnotes
Author Contributions
G. K. and J. R. conceived the project. G. K. and P.G. conducted the data analysis. P. G. created the web application. T. A. S., J. B. P. and M. S. conducted the Pex11β analysis. All authors contributed to writing the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Data availability
All mass spectrometry raw files generated in-house have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository35 with the dataset identifier PXD008888. The co-regulation map is on our website www.proteomeHD.net, pair-wise co-regulation scores are available also through STRING (https://string-db.org). A network of the top 0.5% co-regulated protein pairs can be explored interactively on NDEx (DOI: http://doi.org/10.18119/N9N30Q).
Code availability
Data analysis was performed in R 3.5.1. R scripts and input files required to reproduce the results of this manuscript are available in the following GitHub repository: https://github.com/Rappsilber-Laboratory/ProteomeHD. R scripts related specifically to the benchmarking of the treeClust algorithm using synthetic data are available in the following GitHub repository: https://github.com/Rappsilber-Laboratory/treeClust-benchmarking. The R package data.table was used for fast data processing. Figures were prepared using ggplot2, gridExtra, cowplot and viridis.
Reporting Summary
Further information on research design is available in the Life Sciences Reporting Summary linked to this article.
References
- 1.Havugimana PC, et al. A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huttlin EL, et al. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545:505–509. doi: 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rolland T, et al. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foster LJ, et al. A mammalian organelle map by protein correlation profiling. Cell. 2006;125:187–199. doi: 10.1016/j.cell.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 5.Christoforou A, et al. A draft map of the mouse pluripotent stem cell spatial proteome. Nat Commun. 2016;7 doi: 10.1038/ncomms9992. 8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thul PJ, et al. A subcellular map of the human proteome. Science. 2017;356 doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 7.Costanzo M, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353 doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mülleder M, et al. Functional Metabolomics Describes the Yeast Biosynthetic Regulome. Cell. 2016;167:553–565.e12. doi: 10.1016/j.cell.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 10.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 11.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 12.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, et al. Proteome Profiling Outperforms Transcriptome Profiling for Coexpression Based Gene Function Prediction. Mol Cell Proteomics. 2017;16:121–134. doi: 10.1074/mcp.M116.060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lapek JD, Jr, et al. Detection of dysregulated protein-association networks by high-throughput proteomics predicts cancer vulnerabilities. Nat Biotechnol. 2017;35:983–989. doi: 10.1038/nbt.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Beyer A, Aebersold R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 16.Batada NN, Urrutia AO, Hurst LD. Chromatin remodelling is a major source of coexpression of linked genes in yeast. Trends Genet. 2007;23:480–484. doi: 10.1016/j.tig.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Kustatscher G, Grabowski P, Rappsilber J. Pervasive coexpression of spatially proximal genes is buffered at the protein level. Mol Syst Biol. 2017;13:937. doi: 10.15252/msb.20177548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebisuya M, Yamamoto T, Nakajima M, Nishida E. Ripples from neighbouring transcription. Nat Cell Biol. 2008;10:1106–1113. doi: 10.1038/ncb1771. [DOI] [PubMed] [Google Scholar]
- 20.Battle A, et al. Genomic variation. Impact of regulatory variation from RNA to protein. Science. 2015;347:664–667. doi: 10.1126/science.1260793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geiger T, Cox J, Mann M. Proteomic changes resulting from gene copy number variations in cancer cells. PLoS Genet. 2010;6:e1001090. doi: 10.1371/journal.pgen.1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stingele S, et al. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol. 2012;8:608. doi: 10.1038/msb.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu L, et al. Variation and genetic control of protein abundance in humans. Nature. 2013;499:79–82. doi: 10.1038/nature12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kustatscher G, et al. Proteomics of a fuzzy organelle: interphase chromatin. EMBO J. 2014;33:648–664. doi: 10.1002/embj.201387614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y, et al. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell. 2014;158:1415–1430. doi: 10.1016/j.cell.2014.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kustatscher G, Grabowski P, Rappsilber J. Multiclassifier combinatorial proteomics of organelle shadows at the example of mitochondria in chromatin data. Proteomics. 2016;16:393–401. doi: 10.1002/pmic.201500267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams EG, et al. Systems proteomics of liver mitochondria function. Science. 2016;352 doi: 10.1126/science.aad0189. aad0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta S, Turan D, Tavernier J, Martens L. The online Tabloid Proteome: an annotated database of protein associations. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh SA, et al. Co-regulation proteomics reveals substrates and mechanisms of APC/C-dependent degradation. EMBO J. 2014;33:385–399. doi: 10.1002/embj.201385876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andersen JS, et al. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 31.Kirchner M, et al. Computational protein profile similarity screening for quantitative mass spectrometry experiments. Bioinformatics. 2010;26:77–83. doi: 10.1093/bioinformatics/btp607. [DOI] [PubMed] [Google Scholar]
- 32.Wilhelm M, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 33.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 34.Ong S-E, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 35.Vizcaíno JA, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–56. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fabregat A, et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016;44:D481–7. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buttrey SE, Whitaker LR. treeClust: an R package for tree-based clustering dissimilarities. The R Journal. 2015;7:227–236. [Google Scholar]
- 38.Buttrey SE, Whitaker LR. NPS Technical Report Archive. 2016. A scale-independent, noise-resistant dissimilarity for tree-based clustering of mixed data. https://calhoun.nps.edu/handle/10945/48615. [Google Scholar]
- 39.Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabási AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 40.Yip AM, Horvath S. Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinformatics. 2007;8:22. doi: 10.1186/1471-2105-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.The Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017;45:D331–D338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Der Maaten L, Hinton G. Visualizing High-Dimensional Data Using t-SNE. J Mach Learn Res. 2008;9:26. [Google Scholar]
- 43.García-Aguilar A, Cuezva JM. A Review of the Inhibition of the Mitochondrial ATP Synthase by IF1 in vivo: Reprogramming Energy Metabolism and Inducing Mitohormesis. Front Physiol. 2018;9:1322. doi: 10.3389/fphys.2018.01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forbes SA, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piñero J, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45:D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet. 2014;15:193–204. doi: 10.1038/nrg3520. [DOI] [PubMed] [Google Scholar]
- 48.D’Lima NG, et al. A human microprotein that interacts with the mRNA decapping complex. Nat Chem Biol. 2017;13:174–180. doi: 10.1038/nchembio.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chu Q, et al. Identification of Microprotein-Protein Interactions via APEX Tagging. Biochemistry. 2017 doi: 10.1021/acs.biochem.7b00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slavoff SA, et al. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol. 2013;9:59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyer B, Wittig I, Trifilieff E, Karas M, Schägger H. Identification of two proteins associated with mammalian ATP synthase. Mol Cell Proteomics. 2007;6:1690–1699. doi: 10.1074/mcp.M700097-MCP200. [DOI] [PubMed] [Google Scholar]
- 52.Chen R, Runswick MJ, Carroll J, Fearnley IM, Walker JE. Association of two proteolipids of unknown function with ATP synthase from bovine heart mitochondria. FEBS Lett. 2007;581:3145–3148. doi: 10.1016/j.febslet.2007.05.079. [DOI] [PubMed] [Google Scholar]
- 53.Fujikawa M, Ohsakaya S, Sugawara K, Yoshida M. Population of ATP synthase molecules in mitochondria is limited by available 6.8-kDa proteolipid protein (MLQ) Genes Cells. 2014;19:153–160. doi: 10.1111/gtc.12121. [DOI] [PubMed] [Google Scholar]
- 54.Borner GHH, et al. Multivariate proteomic profiling identifies novel accessory proteins of coated vesicles. J Cell Biol. 2012;197:141–160. doi: 10.1083/jcb.201111049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Signorile A, Sgaramella G, Bellomo F, De Rasmo D. Prohibitins: A Critical Role in Mitochondrial Functions and Implication in Diseases. Cells. 2019;8 doi: 10.3390/cells8010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brennan R, et al. Investigating nucleo-cytoplasmic shuttling of the human DEAD-box helicase DDX3. Eur J Cell Biol. 2018;97:501–511. doi: 10.1016/j.ejcb.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 57.Szklarczyk D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schrader M, Costello JL, Godinho LF, Azadi AS, Islinger M. Proliferation and fission of peroxisomes - An update. Biochim Biophys Acta. 2016;1863:971–983. doi: 10.1016/j.bbamcr.2015.09.024. [DOI] [PubMed] [Google Scholar]
- 59.Schrader M, Costello J, Godinho LF, Islinger M. Peroxisome-mitochondria interplay and disease. J Inherit Metab Dis. 2015;38:681–702. doi: 10.1007/s10545-015-9819-7. [DOI] [PubMed] [Google Scholar]
- 60.Devine MJ, Birsa N, Kittler JT. Miro sculpts mitochondrial dynamics in neuronal health and disease. Neurobiol Dis. 2016;90:27–34. doi: 10.1016/j.nbd.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 61.Costello JL, et al. Predicting the targeting of tail-anchored proteins to subcellular compartments in mammalian cells. J Cell Sci. 2017;130:1675–1687. doi: 10.1242/jcs.200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okumoto K, et al. New splicing variants of mitochondrial Rho GTPase-1 (Miro1) transport peroxisomes. J Cell Biol. 2018;217:619–633. doi: 10.1083/jcb.201708122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Castro IG, et al. A role for Mitochondrial Rho GTPase 1 (MIRO1) in motility and membrane dynamics of peroxisomes. Traffic. 2018;19:229–242. doi: 10.1111/tra.12549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodríguez-Serrano M, Romero-Puertas MC, Sanz-Fernández M, Hu J, Sandalio LM. Peroxisomes Extend Peroxules in a Fast Response to Stress via a Reactive Oxygen Species-Mediated Induction of the Peroxin PEX11a. Plant Physiol. 2016;171:1665–1674. doi: 10.1104/pp.16.00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mattiazzi Ušaj M, et al. Genome-Wide Localization Study of Yeast Pex11 Identifies Peroxisome-Mitochondria Interactions through the ERMES Complex. J Mol Biol. 2015;427:2072–2087. doi: 10.1016/j.jmb.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shai N, et al. Systematic mapping of contact sites reveals tethers and a function for the peroxisome-mitochondria contact. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03957-8. 1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pickrell JK, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barrett T, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Szklarczyk D, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jovanovic M, et al. Immunogenetics. Dynamic profiling of the protein life cycle in response to pathogens. Science. 2015;347:1259038. doi: 10.1126/science.1259038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kustatscher G, Wills KLH, Furlan C, Rappsilber J. Chromatin enrichment for proteomics. Nat Protoc. 2014;9:2090–2099. doi: 10.1038/nprot.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alabert C, et al. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol. 2014;16:281–293. doi: 10.1038/ncb2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 74.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 75.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pratt D, et al. NDEx, the Network Data Exchange. Cell Syst. 2015;1:302–305. doi: 10.1016/j.cels.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Langfelder P, Horvath S. Fast R Functions for Robust Correlations and Hierarchical Clustering. J Stat Softw. 2012;46 [PMC free article] [PubMed] [Google Scholar]
- 78.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 79.Krijthe JH. Rtsne: T-Distributed Stochastic Neighbor Embedding using Barnes-Hut Implementation. 2015 https://github.com/jkrijthe/Rtsne.
- 80.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal. 2006:1695. Complex Systems. [Google Scholar]
- 81.Schloerke B, et al. GGally: Extension to ‘ggplot2’. 2018
- 82.Butts CT. sna: Tools for Social Network Analysis. 2016
- 83.Alexa A, Rahnenfuhrer J. R package version 2.30.0. 2016. topGO: enrichment analysis for gene ontology. [Google Scholar]
- 84.Binns D, et al. QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics. 2009;25:3045–3046. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stark C, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–9. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Orchard S, et al. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–63. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Szklarczyk D, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–52. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jaccard P. Distribution de la flore alpine dans le bassin des Dranses et dans quelques régions voisines. Bull Soc Vaud sci nat. 1901;37:241–272. [Google Scholar]
- 89.Costello JL, et al. ACBD5 and VAPB mediate membrane associations between peroxisomes and the ER. J Cell Biol. 2017;216:331–342. doi: 10.1083/jcb.201607055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.