Abstract
Herpes simplex virus spread between epithelial cells is mediated by virus tegument and envelope protein complexes including gE/gI and pUL51/pUL7. pUL51 interacts with both pUL7 and gE/gI in infected cells. We show that amino acids 30–90 of pUL51 mediate interaction with pUL7. We also show that deletion of amino acids 167–244 of pUL51, or ablation of pUL7 expression both result in failure of gE to concentrate at junctional surfaces of Vero cells. We also tested the hypothesis that gE and pUL51 function on the same pathway for cell-to-cell spread by analyzing the phenotype of a double gE/UL51 mutant. In HaCaT cells, pUL51 and gE function on the same spread pathway, whereas in Vero cells they function on different pathways. Deletion of the gE gene strongly enhanced virus release to the medium in Vero cells, suggesting that the gE-dependent spread pathway may compete with virion release to the medium.
Introduction
Assembly of mature, multi-layer herpesvirions occurs by budding of capsids into a cytoplasmic membrane compartment followed by trafficking of the enveloped virion to the cell surface for release, or to junctional surfaces for cell-to-cell spread (CCS) (reviewed in (1)). The identity of the cytoplasmic membrane compartment used for final envelopment apparently differs between herpesvirus species, but is derived by modification of host cell structures. Human cytomegalovirus (HCMV) for example, undergoes cytoplasmic envelopment in a discrete assembly compartment constructed by massive reorganization of host Golgi and endosomal membranes (2–8). HSV-1, on the other hand, undergoes cytoplasmic envelopment in multiple locations in the cytoplasm. The nature of the enveloping membrane for HSV-1 is not entirely clear. Secondary envelopment at the trans-Golgi network (TGN) has been proposed based on membrane composition of the mature virion, association of capsids with membranes containing TGN markers (9, 10). Secondary envelopment at an endosomal compartment is supported by the presence endocytosed horseradish peroxidase in the lumen of enveloping membrane and co-localization of capsids with transferrin receptor (11).
The herpesvirus tegument is a loosely ordered protein layer that lies between the capsid and the envelope (12). It consists of at least 20 virus-encoded proteins (reviewed in (13)). Tegument proteins are critical for multiple functions late in the virus replication cycle, including assembly of the mature virus particle and trafficking of virus particles for CCS. Interestingly, these functions are not delegated among different sets of proteins, but rather are dual functions of many and, perhaps, most tegument proteins.
The HSV-1 UL51 gene encodes a 244 a.a. palmitoylated tegument protein (14, 15). A complete deletion of any alphaherpesvirus UL51 gene has not yet been constructed because the UL51 protein coding sequence contains promoter/regulatory sequences for the UL52 gene that encodes one of the helicase/primase subunits of the viral DNA replication apparatus. Alphaherpesvirus UL51 gene function has, therefore been explored by the use of partial deletions that remove most of the protein coding sequence (16–18) or by insertion of stop codons a short distance downstream of the initiation codon (19). There are apparent minor differences in the phenotypes obtained with these different approaches, but all of them suggest pUL51 has cell-specific functions in both virion assembly and CCS. Single-step growth in these various mutant viruses is depressed up to 100-fold in some cell lines, including Vero (16–19), and this growth defect has been correlated with accumulation of unenveloped, and sometimes membrane-associated capsids in the cytoplasm (16, 19). This suggests that one function of pUL51 is to facilitate curvature or closure of membrane around the capsid/tegument complex in cytoplasmic assembly. Interestingly, however, single-step growth defects were not observed for an HSV-1 partial deletion mutant on HEp-2 cells (18), suggesting that the pUL51 assembly function can be complemented by host cell factors in some cell types.
pUL51 of HSV-1 forms a complex with another viral tegument protein, pUL7 (19, 20). This complex is necessary for incorporation of pUL7 into the mature virion (20). A double mutant made by stop codon insertion into UL51 and deletion of UL7 showed a defect in single step growth in Vero and HaCaT cells that was no greater than the defects of the individual deletions, suggesting that pUL51 and pUL7 function on the same pathway, and probably as a complex in assembly (19). The UL51 and UL7 genes are conserved among herpesviruses, and deletion of their homologs in HCMV (UL71 and UL103, respectively) causes defects in formation of the assembly compartment and cytoplasmic envelopment, and results in formation of smaller infection foci, suggesting some conservation of function as well (21–23).
All alphaherpesvirus UL51 mutants reported so far show profound defects in plaque formation, even in cells in which no single-step growth defect is observed, suggesting that pUL51 plays a critical role in spread between epithelial cells (16–19). Since the pUL51/pUL7 complex is not displayed on the cell surface, its most likely function in CCS is facilitating delivery of virions to junctional surfaces of cells.
The UL51 gene product is one of several alphaherpesvirus gene products that play an important role in epithelial cell-cell spread (CCS). The best described of these is the gE/gI complex. gI and gE are encoded on the adjacent US7 and US8 genes respectively and, upon translation at the ER, form a stable complex (24, 25). The gE/gI complex is an abundant component of the mature virion envelope, where it functions in immune evasion by interfering with effector functions of IgG by binding to the Fc region (26, 27). In addition to its immune evasion function, the gE/gI complex facilitates CCS in epithelial cells by a mechanism that has been reported to require its localization to adherens junctions where it co-localizes with β-catenin (28–33). Failure to express gE, or failure of gE/gI to localize to junctions results in formation of small plaques in the presence of neutralizing antibody, and in diminished numbers of virus particles found between cells (34). The mechanism by which gE/gI facilitates spread is not clear, but its spread function can be inhibited by overexpression of the soluble ectodomain of gE, suggesting that an interaction with adjacent cells may be required (35).
The localization and CCS function of gE are regulated by viral tegument proteins. A complex of pUL11, pUL16, and pUL21 can form on the cytoplasmic tail of gE, and expression of these proteins is required for gE cell-surface localization (36–39). Failure to form this complex is associated with defects in CCS and syncytium formation, suggesting that gE/gI CCS function depends on these tegument proteins (38). A similar set of observations implicates pUL51 in gE CCS function. A UL51 mutant produced by fusion with EGFP at the C-terminus causes failure of gE to accumulate at cellular junctions in Vero cells, and this is correlated with inhibition of syncytium formation and a severe defect in CCS in Vero cells (18). pUL51 also interacts in the infected cell with gE (18). However, pUL51 may have other functions in CCS, since a UL51 deletion has a far more powerful inhibitory effect on CCS than a deletion of gE (18).
All of these observations suggest the existence of an epithelial CCS pathway in which delivery of the gE/gI complex to cellular junctions is a central event, regulated by interaction between the cytoplasmic tails of gE, gI, or both with tegument protein complexes. The hypothesis that the pUL51/pUL7 complex, the pUL11/pUL16/pUL21 complex, and the gE/gI complex operate on the same pathway for CCS predicts that multiple mutants that affect formation of multiple complexes will have a phenotype no more severe than that of the single mutants. Here we show that this prediction is borne out for a UL51/gE double mutant in HaCaT cells, but not in Vero cells, and that the behavior of gE with respect to junctional localization is cell type dependent as well.
Results
pUL51 sequences that are necessary and sufficient for complex formation with pUL7 are located between a.a. 30 and 90
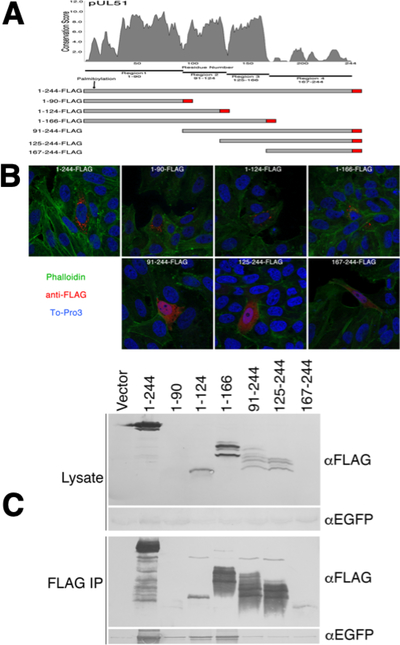
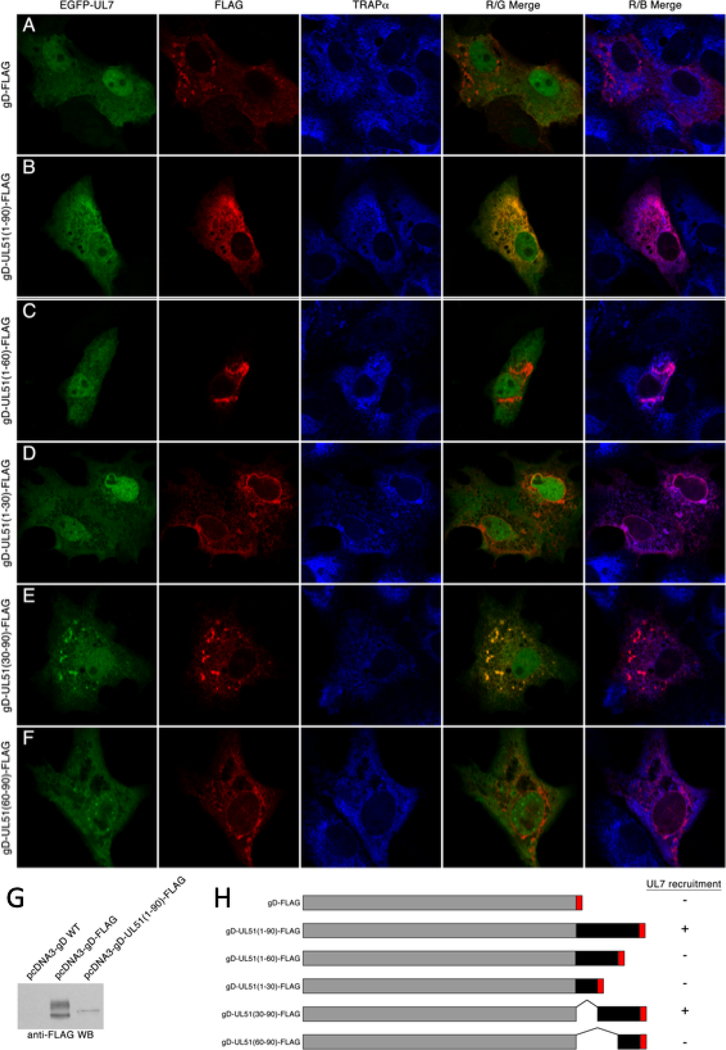
pUL51 is composed of two sequence regions - an N-terminal region from amino acids 1–166 that contains sequence that is well conserved among pUL51 homologs and that is predicted to be globular, and a C-terminal region from amino acid 167 to the end of the protein that poorly conserved and is predicted to be disordered. It has been previously shown that interaction with pUL7 can be mediated by sequences corresponding roughly to the globular domain of pUL51 from residues 29–170 (19, 20). The N-terminal region can be further subdivided into four conserved regions comprising a.a. 1–20, 21–90, 91–124, and 125–166 (Figure 1A) (20). To see whether a smaller pUL7 interaction region could be defined, we created pcDNA3-based plasmid constructs that express N- and C-terminal truncations of a FLAG-tagged pUL51 protein coding sequence with boundaries corresponding to the pUL51 conserved regions (Figure 1A). We first tested the subcellular localization of these truncated proteins. Although pUL51 does not have a transmembrane domain it is tightly associated with membranes in infected and transfected cells due to palmitoylation at C9 near the N-terminus (15). As expected, only the truncation constructs that included the pUL51 N-terminus showed a punctate cytoplasmic distribution with variation in the number and distribution of the puncta due to association with cytoplasmic membranes (Figure 1B). Those that did not include the N-terminus were invariably distributed diffusely in the cytoplasm and nucleus. We then tested those truncated proteins for ability to interact with EGFP-pUL7 in co-immunoprecipitation assays (Figure 1C). Expression of the truncated pUL51 constructs was variable, probably due to differing stability of the truncated proteins, and the shortest truncations (1–90 and 167–244) were detectable only in immunoprecipitated samples. Only constructs that contained residues 1–90 of pUL51 reproducibly co-immunoprecipitated EGFP-pUL7 above the vector background, suggesting that these residues were necessary and sufficient for pUL7 interaction. The pull-down by pUL51(1–90), however, was weak, probably due to the very low expression of this construct. Therefore, to confirm and extend this result, we created in-frame fusion constructs in which pUL51-FLAG truncations were fused to the C-terminus of the viral glycoprotein D (gD) and expressed them via transfection with EGFP-pUL7 (Figure 2). Consistent with our previous report (20), expression of EGFP-pUL7 in combination with the control gD-FLAG, resulted in diffuse localization concentrated in the cell nucleus in all transfected cells (Figure 2A). Co-expression with gD-pUL51(1–90)-FLAG, in contrast, resulted in recruitment of EGFP-pUL7 to cytoplasmic membranes where the two proteins colocalized (Figure 2B). While co-expression of EGFP-pUL7 and gD-pUL51(1–90)-FLAG always results in strong co-localization of the two proteins on cytoplasmic membranes, EGFP-pUL7 was rarely completely recruited to pUL51-labeled membranes – some was found diffusely in the nucleoplasm in most cells. This may reflect differences in expression levels of the constructs. We made a set of further truncations of the pUL51 N-terminal region and found that truncations that included amino acids 30–90 of pUL51 (Figure 2B, E, and H) could recruit EGFP-pUL7 to membranes, whereas truncations that lacked any of amino acids 30–90 (Figure 2C, D, F, and H) did not, suggesting that conserved region 2 of pUL51 is necessary and sufficient for interaction with pUL7.
Figure 1.
Localization and pUL7 interactions of pUL51 truncations. (A) Conservation plot of pUL51 protein coding sequence and schematic diagram of FLAG-tagged pUL51 truncations. The plot shows conservation of biochemical properties of amino acids using all available herpesvirus pUL51 homologous sequences aligned using the program MUSCLE (61). Each residue position receives a conservation score, and scores were averaged over a sliding 5 amino acid residue window. The schematic underneath the plot shows the boundaries of the pUL51 truncations used in this study. The FLAG tag is indicated in red. (B) Localizations of pUL51 truncations in transfected Vero cells. Nuclei are stained with To-Pro3 (blue), actin stress fibers are stained with phalloidin (green) and pUL51 truncations are detected with mouse anti-FLAG (red). Representative images from three independent experiments in which >50 transfected cells were observed are shown. (C) Co-immunoprecipitation of EGFP-pUL7 with pUL51 truncations. Lysates and immunoprecipitates from Vero cells co-transfected with the indicated pUL51 constructs and with pEGFP-pUL7 plasmid are shown. The top two panels show lysate proteins detected by immunoblot using either anti-FLAG or anti- EGFP. The bottom two panels show FLAG immunoprecipitates detected with either anti-FLAG or anti-EGFP. One of two independent experiments is shown.
Figure 2.
Interaction of pUL7 with gD-UL51 truncation fusions. (A-F) Digital images of cells that are immunofluorescently stained and detected by confocal microscopy are shown. The gD fusion construct used is indicated to the left of each panel. EGFP-pUL7 is shown in green, FLAG staining in red, and staining for the ER marker TRAPα is shown in blue. Red/green merge images show co-localization between gD fusions and EGFP-pUL7. Red/blue merged images show co-localization between gD fusions and TRAPα. Representative images from two independent experiments in which >50 transfected cells were observed are shown. (G) Immunoblot detection of gD fusions using anti-FLAG antibody. (H) Summary schematic showing the structures of the gD fusions used for this experiment, and their ability to recruit EGFP-pUL7 to membranes.
Interestingly, the gD-pUL51(1–90)-FLAG fusion construct localized differently than the control gD-FLAG. gD-FLAG is found on membranes distributed throughout the cytoplasm and on the cell surface (Figure 2A). gD-UL51(1–90)-FLAG is found in a reticular distribution in the cytoplasm where it co-localizes completely with the ER marker TRAPα (Figure 2B). Furthermore, whereas gD-FLAG migrates in SDS-PAGE as a heterogeneous collection of bands typical of a glycoprotein that has matured through the Golgi, gD-UL51(1–90)-FLAG migrates largely as a single species typical of immature glycoproteins (Figure 2G). All gD fusions that contained amino acids 1–30 of pUL51 colocalized with TRAPα (Figure 2B, C and D), whereas those that lacked these amino acids did not (Figure 2E and F). These results suggested that the fusion of gD to the N-terminal 30 amino acids of pUL51 results in retention of gD in the ER.
Deletion of the C-terminal third of pUL51 completely ablates its CCS function
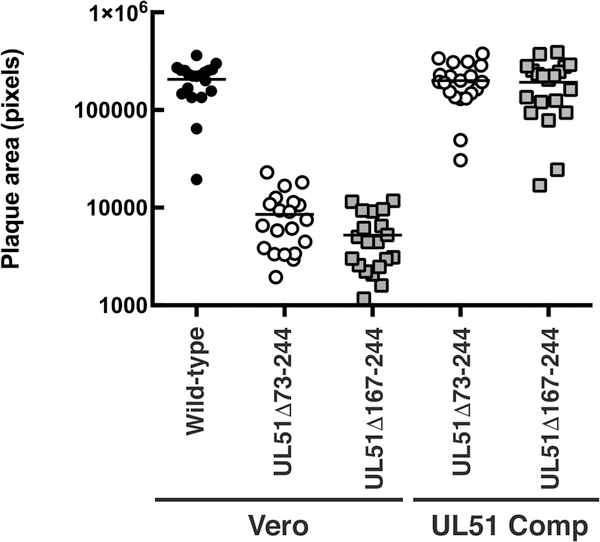
We have previously described the construction and characterization of a recombinant HSV-1 virus in which amino acids 73–244 of pUL51 were deleted, and found that it showed significant defects in both virus production, virus release to the medium, and cell-to-cell spread in Vero cells (18). In a subsequent study, we reported construction of a virus carrying a smaller partial deletion that does not express the poorly conserved C-terminal third of pUL51 (a.a. 167–244) (Figure 3A line 5) (20). In that study, we noted that the virus carrying this smaller deletion, while retaining the interaction between pUL51 and pUL7, appeared to be as deficient in plaque formation as the UL51Δ73–244 mutant (Figure 3A line 3). In order to confirm and quantitate this effect, we performed plaque size assays in the presence of neutralizing antibody (Figure 4). On Vero cells, both the UL51Δ73–244 and UL51Δ167–244-FLAG viruses formed plaques significantly smaller than the wild type control (P<0.001 for both comparisons), and these plaques were ~30-fold smaller than those formed by wild-type virus. The spread defect observed for both deletion viruses could be completely complemented by plaque formation on wild-type pUL51-expressing complementing cells, demonstrating that the defect was solely due to the mutation in the UL51 gene.
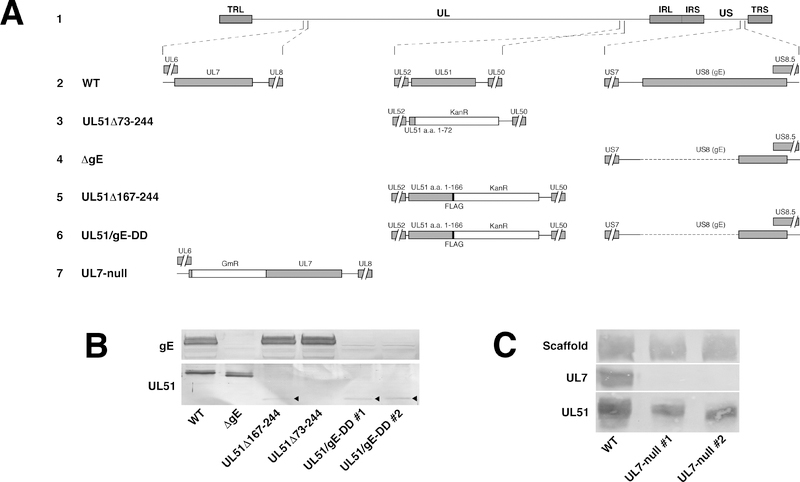
Figure 3.
Schematic diagrams of recombinant viruses used in this study and expression analysis of deletion viruses. (A) Schematic diagram of the HSV-1(F) genome (line 1) and of the recombinant viruses constructed for this study. The positions of the UL7, UL51 and US8 (gE) loci with respect to the unique long (UL) and unique short (US) sequences and the inverted repeats that flank them (TRL and IRL, and IRS and TRS, respectively), are shown in line 1. Expansions of the arrangements of the wild-type sequences in the regions of UL7, UL51 and US8 are shown in line 2. Lines 3 through 7 show altered sequences in recombinant viruses used in this study (Line 3) UL51Δ73–244 carries a stop codon and Kanamycin resistance cassette (KanR) in place of the sequences coding for amino acids 73–244 of pUL51 and has been previously described (18). (Line 4) The gE-null virus has a deletion of the sequence coding for amino acids 1–335 and has been previously described (18). (Line 5) In UL51Δ167–244-FLAG, sequences coding for amino acids 167–244 of pUL51 are replaced by an in-frame FLAG epitope tag, a stop codon and Kanamycin resistance cassette. This virus has been previously described (20). (Line 6) The UL51/gE-DD virus contains the sequence alterations for both the UL51Δ167–244-FLAG ad gE-null viruses, and was constructed by altering the UL51 locus in the previously constructed gE-null virus. (Line 7) The UL7-null virus contains a gentamycin resistance cassette (GmR) insertion immediately following the stop codon for the UL6 coding sequence. This change places a stop codon in the GmR cassette in frame with the UL7 coding sequence and is predicted to result in expression of only the first seven amino acids of pUL7. (B).
Figure 4.
UL51 deletions show similar UL51-dependent cell-to-cell spread defects. Confluent Vero or UL51 complementing cell monolayers were infected at low multiplicity with wild-type (black circles) UL51Δ73–244 (open circles) or UL51Δ167–244 (gray squares), and after two days of incubation in the presence of neutralizing antibody, plaques were immunostained, and plaque areas measured. Statistical significance of differences between samples was measured using one-way ANOVA and Fisher’s least significant difference test.
Mutation of either pUL51 or pUL7 impairs accumulation of gE at cell junctions
pUL51 and gE have both been shown to play important roles in epithelial cell-to-cell spread, and we have previously demonstrated a physical interaction between them that suggests that their functions in cell-to-cell spread might be coupled (18). It has also been previously reported that gE in infected cells concentrates at cellular junctions where it co-localizes with β-catenin, suggesting a localization to adherens junctions (30), and that mutation of pUL51 impairs localization of gE to junctional surfaces of Vero cells (18). We have also shown that pUL51 forms a complex with pUL7, and that pUL7 also functions in cell-to-cell spread (20).
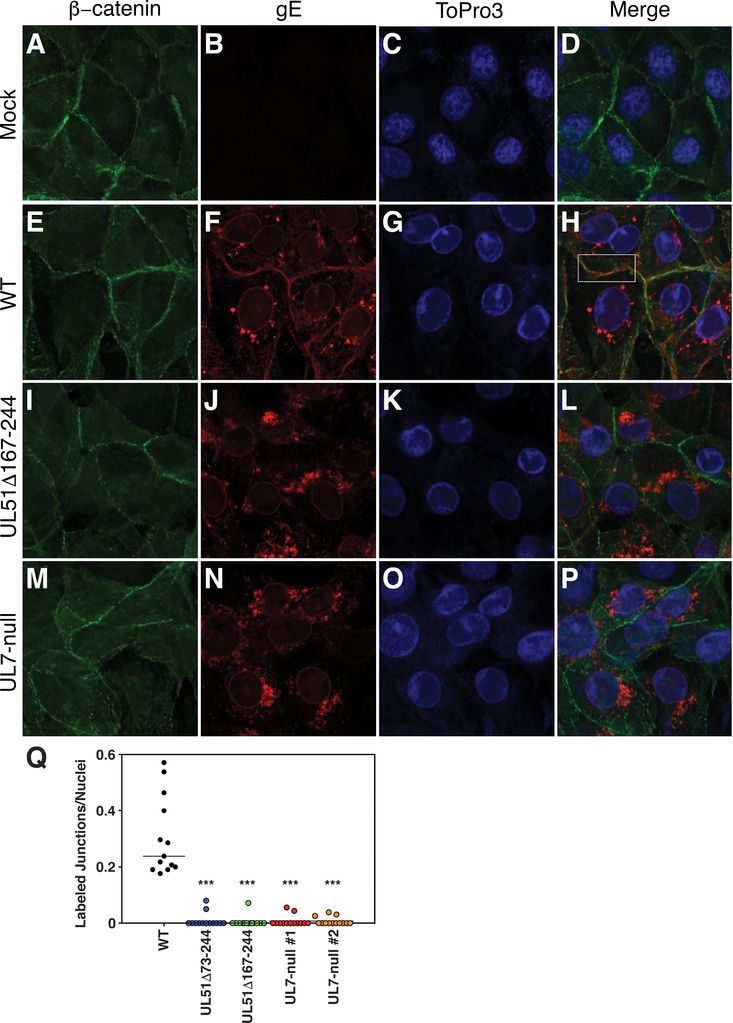
In order to determine whether the UL51Δ167–244 mutant or a UL7 null mutation also disrupt gE localization, we first generated a UL7-null virus (Figure 3A line 7), and demonstrated that it expresses no detectable pUL7 (Figure 3C). We then infected cells with wild-type, UL51 mutant, or UL7 mutant viruses for 12 hours, and then visualized gE and β-catenin localization (Figure 5). Although Vero cells form adherens junctions (40), they do not form polarized epithelia and do not form a continuous belt of β-catenin staining. Rather, we observed β-catenin staining in numerous small patches at the junctional surfaces of uninfected cells (Figure 5A and D) that visually define those junctional surfaces. We observed no difference in the staining intensity or distribution of β-catenin twelve hours after infection with any of the viruses (Figure 5 A, E, I, and M).
Figure 5.
gE mislocalization in UL51 and UL7 mutant-infected cells. (A-P) Vero cells that had been infected with the indicated viruses for 12 h were fixed and immunostained for β-catenin (green), gE (red) or TO-PRO-3 to stain DNA (blue). The white outlined box in panel H indicates apposition of two cells with minimal co-localization of gE and β-catenin. (Q) Quantitation of junctional gE staining. For 20 images, each containing at least 20 cells, apposing surfaces of adjacent cells that stained for gE were counted and divided by the total number of nuclei in the image. For example, in panel F, four apposing surfaces that stain for gE would be counted and divided by the seven nuclei in the image. Each point represents one such image. Statistical significance of differences between samples was measured using one-way ANOVA.
As previously reported, in wild-type virus-infected cells, gE concentrates at the nuclear membrane, on aggregates of membranes in the cytoplasm, and at the junctional surfaces of cells (18). Here, we observed that, although gE and β-catenin both concentrated at junctional surfaces, they were not uniformly distributed and there was very little co-localization between patches of β-catenin and gE staining (Figure 5H, boxed area). Both the UL51Δ167–244 and UL7-null mutant viruses showed almost no gE immunofluorescent staining at junctional surfaces (Figure 5I–P), demonstrating that both deletions impair gE junctional localization in infected cells. Quantitation of gE junctional staining (Figure 5Q) showed that both UL51 deletion and UL7-null viruses showed a similar degree of inhibition.
pUL51 and gE double deletions have different spread phenotypes in HaCaT and Vero cells
The importance of both gE and pUL51 in epithelial cell-to-cell spread, the physical interaction between gE and pUL51, and the effect of pUL51 deletion on gE localization suggested the hypothesis that pUL51 and gE function on the same pathway for cell-to-cell spread and that function of gE in spread might be partially or fully dependent upon pUL51. To test this hypothesis, we created a pair of independently constructed recombinant mutant viruses deficient in pUL51 spread function and in gE expression by combining the UL51Δ167–244 mutation with a deletion in the US8 gene that abrogates expression of gE (Figure 3A, Line 6). Immunoblot analysis showed that these viruses express, as expected, a truncated, FLAG-tagged pUL51, and fail to express detectable gE (Figure 3B). Also as noted previously, the truncated pUL51 is expressed at a considerably lower level than the wild-type protein (18). Previous results evaluating the effects of pUL51 deletion showed cell-specific effects on virus replication, spread and release to the medium (18). Specifically, we observed that both growth and spread defects were considerably greater in Vero than in HEp-2 cells, suggesting that the functions of pUL51 in Vero and human epithelial cells might differ. We therefore evaluated the growth and spread phenotypes of these viruses on Vero cells and on the human HaCaT keratinocyte cell line.
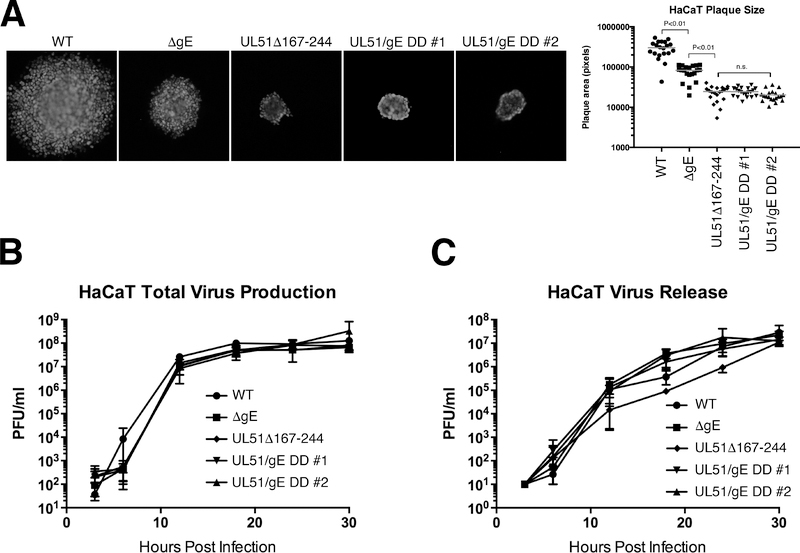
In HaCaT cells, as previously reported, deletion of gE significantly (P<0.01) inhibits cell-to-cell spread of virus in the presence of neutralizing antibodies so that mutant virus plaques are roughly three-fold smaller than the wild-type control (Figure 6A) (30). Deletion of a.a. 167–244 of pUL51 had a more dramatic effect on spread, so that plaques were roughly 10-fold smaller than wild type. The double deletion plaques, however, were no smaller than the plaques formed by the UL51 single deletion, suggesting that, in HaCaT cells, UL51 and gE function on the same cell-to-cell spread pathway. The effects of the single and double deletions on plaque size are completely due to effects on cell-to-cell spread, since neither the UL51Δ167–244 alone, the gE deletion alone, nor the double deletions had any significant effect on single-step virus growth or release of virus to the medium (Figure 6B and C).
Figure 6.
Growth, spread and release of UL51 and gE mutants on HaCaT cells. (A) Double mutants show no enhanced spread defect compared to a UL51 single mutant. Digital images are shown of representative plaques formed by 48 hours on HaCaT cells in the presence of neutralizing antibody. The graph at the right plots areas of 20 randomly chosen plaques for each virus. Statistical significance of differences between samples was measured using one-way ANOVA. (B) Single-step growth of single and double mutant viruses on HaCaT cells. Each point is the mean of three independent experiments, and error bars indicate the range of values. (C) Virus released to the medium in the single-step growth experiments shown in (B).
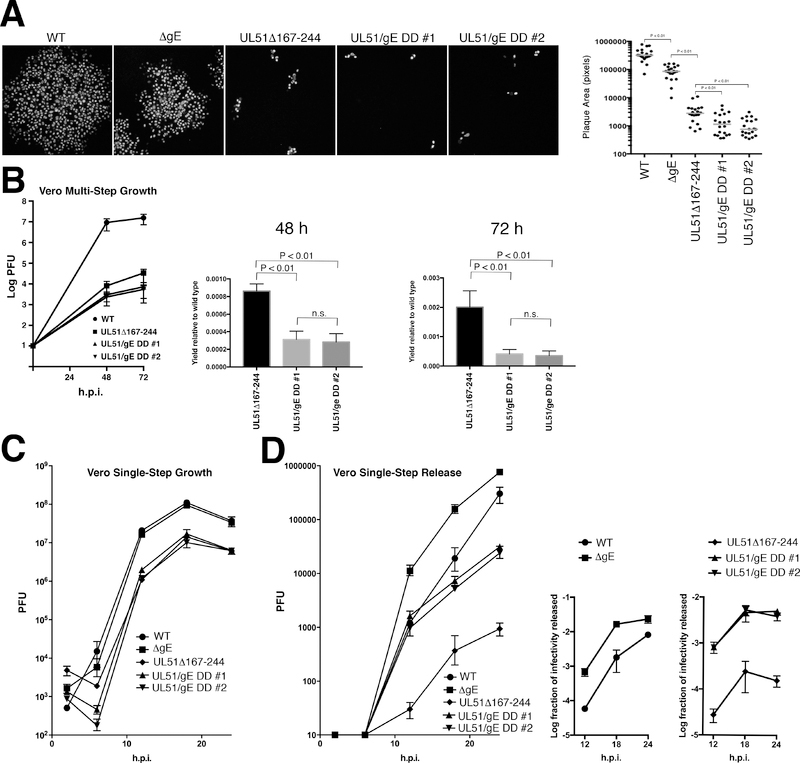
The result in Vero cells was different in several interesting ways (Figure 7). As previously reported, the effects of the UL51 single deletion on plaque formation in the presence of neutralizing antibody were far more dramatic than on HaCaT cells, such that the plaques formed by UL51 deletion viruses consisted of only a few tens of cells (Figure 7A). Over three experiments, the UL51 deletion caused a roughly 40-fold reduction, and the gE deletion caused a roughly three-fold reduction in plaque size (Figure 7A, graph). Second, the double mutants formed significantly smaller plaques (P < 0.01) than either of the single mutants, and the defect (about 120-fold) was the product of multiplication of the effects of the single deletions. In order to confirm this, a multi-step growth experiment was used for comparison of the UL51 single mutant and the double deletions (Figure 7B). As expected, the UL51Δ167–244 single deletion was associated with a dramatic defect in multi-step growth such that by 72 hours, virus yield is diminished by three log orders of magnitude compared to the wild-type control. The two double deletions were further impaired such that by 72 hours, virus yield was diminished 5-fold compared to the UL51Δ167–244 single deletion. The enhanced impairment in cell-to-cell spread seen in the double mutants was not due to an enhanced defect in virus replication. The double deletions replicated no worse in single-step growth than the UL51Δ167–244 single deletion (Figure 7C).
Figure 7.
Growth, spread and release of UL51 and gE mutants on Vero cells. (A) Double mutants show enhanced spread defect compared to single mutants. Digital images are shown of representative plaques formed by 48 hours on Vero cells in the presence of neutralizing antibody. The graph at the right plots areas of 20 randomly chosen plaques for each virus. Statistical significance of differences between samples was measured using one-way ANOVA and Fisher’s least significant difference test. There was no significant difference between the two isolates of the double deletions.
(B) Double mutants show an enhanced spread defect compared to the single UL51 mutant in multi-step growth. The left-hand graph shows the multi-step growth curve. Each point is the mean of three independent experiments, and error bars indicate the range of values. The two right hand graphs compare the values obtained for the UL51Δ167–244 mutant and for the two double mutants at 48 and 72 hours post infection. Error bars indicate the standard deviation. Statistical significance of differences between samples was measured using one-way ANOVA. (C) Single-step growth of single and double mutant viruses on Vero cells. Each point is the mean of three independent experiments, and error bars indicate the range of values. There were no significant differences between WT and ΔgE, and between UL51Δ167–244 and the two double mutant isolates. (D) Virus released to the medium in the single-step growth experiments shown in (C). The graph on the left shows the PFU released to the medium over time. The two smaller graphs to the right show the efficiency of release to the medium calculated as PFU in medium divided by total culture PFU for each virus at each time point. Statistical significance of differences between samples at each time point was measured using one-way ANOVA. Differences between WT and ΔgE and between UL51Δ167–244 and the double mutant viruses were significant (P < 0.001) at all time points. There were no significant differences between the two double mutant isolates.
In order to determine whether the double mutant spread defect was due to a generalized virus release defect, the titer of culture supernatant virus was also measured during the single-step growth experiment. Similar to what was previously reported for a larger UL51 deletion, the UL51Δ167–244 single deletion showed an impairment in virus release in Vero cells compared to the wild-type control (18). Surprisingly, the double mutants both showed enhanced efficiency of virus release to the medium compared to the UL51 single deletion, and the single gE deletion showed enhanced release efficiency compared to the wild-type control (Figure 7D). Comparing wild-type virus to the gE deletion, the effect was most dramatic (greater than one log order of magnitude) at the earliest time points, and diminished as the efficiency of wild-type virus release rose over time. For the UL51 mutants, the double deletions showed greater than a log order of magnitude enhancement of release compared to the single UL51 mutant at all time points tested.
Discussion
The mechanism of herpesvirus cell-to-cell spread is still poorly understood, but several lines of evidence suggest that it reflects specific trafficking of virus particles to junctional surfaces of cells where virus may infect an adjacent cell in a space that is sterically protected from neutralizing antibody and that is confined enough that essential interactions with the host cell (e.g., envelope protein receptor interactions) may occur rapidly and efficiently. This model accounts for the necessity of the HSV entry apparatus for CCS (41–44), and suggests that viral and cellular factors will participate in sorting of virions to junctional surfaces of cells.
Identification of viral factors with roles in epithelial CCS has been problematic because all have additional functions in virion assembly. The phenotypes of complete deletion mutations, therefore, are complex, and exploring CCS mechanisms with such mutants is difficult because an earlier step or steps in virion assembly and egress is also inhibited. Exploring the CCS function of such factors would be greatly aided by characterization of mutants with specific defects in that function. UL51Δ167–244 mutant described here has no detectable defect in virus production or release to the medium in HaCaT cells, but is strongly impaired for CCS, making it a valuable tool for further studies of the CCS mechanism in these cells.
pUL51 and pUL7 have both virion assembly and cell-to-cell spread functions (16–19). The phenotype of single and double deletions of these genes suggest that they function on the same pathways for assembly and spread, and it is likely that they function as a complex (19). We have previously shown that a truncated pUL51 that is missing amino acids 167–244 could interact with pUL7, and Albecka et al. showed that pUL51 sequences necessary for interaction with pUL7 were located from a.a. 29–170 (19, 20). We show here that residues 30–90 of pUL51 are both necessary and sufficient for interaction with pUL7. We also observed that fusion of gD to UL51 truncations that contain amino acids 1–30 results in retention of gD in the ER suggesting the existence of an ER retention signal near the N-terminus of pUL51. Residues 1–30 of pUL51 do not contain a dibasic motif suggesting that ER retention either occurs indirectly, by way of interaction with some protein that does contain a retrieval motif, or by some other mechanism. Interestingly, native pUL51 is retained on cytoplasmic membranes by palmitoylation at cysteine 9 (15). While most palmitoyl transferase enzymes are found on Golgi membranes, some are localized to the ER (45), and it is possible that the gD fusions that contain amino acids 1–30 are palmitoylated, and that this may contribute to their retention in the ER.
Localization of gE to junctional surfaces has been previously shown to be necessary for gE function in epithelial CCS in HaCaT cells (31). gE co-localizes with β-catenin in HaCaT cells, suggesting localization to adherens junctions (30). Here we have observed that, in Vero cells infected with wild-type virus, gE concentrates preferentially at the junctional surfaces of cells, but does not co-localize with β-catenin (Figure 3). This may suggest either that, in Vero cells, gE targets to junctional complexes that are not adherens junctions (e.g., desmosomes), or that it recognizes some other structural feature specific to junctional surfaces.
We have previously shown that pUL51 interacts with gE in infected cells, and that expression of a pUL51-EGFP fusion causes mislocalization of gE (18). Here we show that in cells infected with UL51 partial deletion or UL7-null mutants, gE does not concentrate at junctional surfaces of cells. Our results, therefore, suggest that one function of pUL51 and pUL7 in CCS is to facilitate junctional localization or retention of the gE/gI complex. pUL51 is a cytoplasmically-oriented membrane-associated protein (15) and pUL7 is also located in the cytosol (20, 46). It seems plausible, therefore, that the pUL51/UL7 complex facilitates gE localization by marking gE-containing vesicles for trafficking from the Golgi apparatus or trafficking endosomes to the junctional cell surface. Alternatively, pUL51 might facilitate retention of gE at junctional surfaces by preventing its retrieval and recycling. It is tempting to speculate that this trafficking function requires the interaction between pUL51 and gE/gI.
In one intuitively attractive model, epithelial CCS results from trafficking of nascent virions for release at the basolateral surfaces of cells in a polarized epithelium. The belt of tight and adherens junctions just below the apical surface of the epithelium would then provide protection from immune effectors. This model suggests that CCS trafficking mechanisms should be most important in cells that form polarized monolayers, and less important or completely ineffective in non-polarized cells. Our results show nonetheless that the effects of UL51 mutations that affect CCS are far more potent in non-polarized Vero cell monolayers than in confluent HaCaT monolayers. This suggests the possibility that targeting of virions for CCS is more precise than simple sorting to basolateral cell surfaces. Virions may be targeted to specific junctional complexes that form regardless of whether the cell monolayer is effectively polarized. We observed that gE is not uniformly distributed at junctions between Vero cells, but rather is distributed in patches that do not correspond to concentrations of β-catenin (Figure 5). Whether these concentrations of gE correspond to concentrations of other host cell junctional components is not yet clear.
In addition to the viral envelope proteins that are essential for entry, epithelial CCS in HSV is affected to a greater or lesser degree by mutations in tegument proteins (pUL51, pUL7, pUL21, pUL16, pUL11, and pUL49 (VP22)) (18, 19, 38, 47). All of these proteins that function in CCS have been shown to interact directly or indirectly with gE (18, 37, 38, 48, 49), suggesting that the cytoplasmic domain of the gE/gI complex would be implausibly crowded if all of these interactions occurred simultaneously. Furthermore, several of these proteins (pUL51, pUL7, and the complex of pUL11, pUL16 and pUL21) have been shown to be required for proper localization of gE to junctional surfaces of cells (18, 37). It is possible that some of these interactions are sequential rather than simultaneous and are important for different stages of gE trafficking to and retention at junctional surfaces of cells.
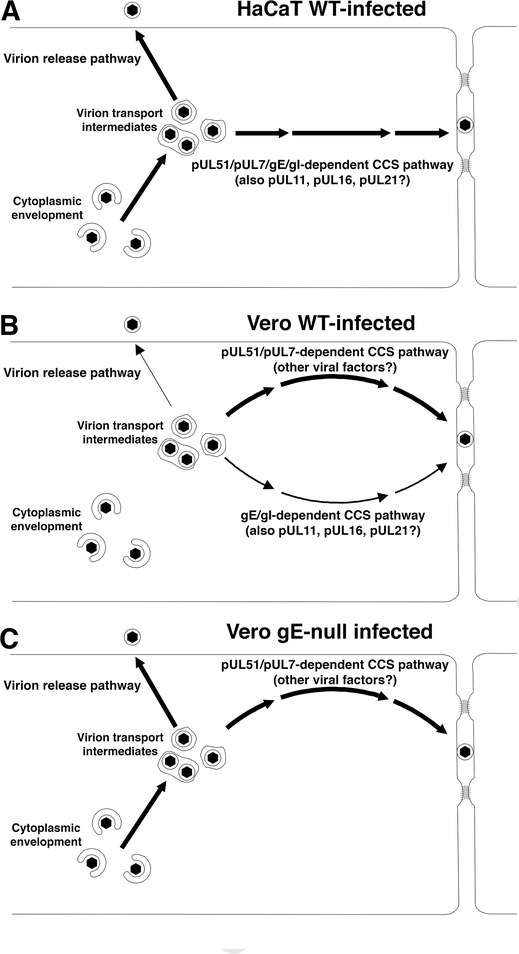
The previously published evidence that pUL51 and gE interact in infected cells, and the evidence presented here that pUL51 and pUL7 expression is necessary for localization of gE to junctional surfaces of cells, suggested a simple model in which gE/gI and the pUL51/pUL7 complex function on the same CCS pathway. In this model, pUL51/pUL7 is required for delivery or retention of gE at cell junctions, and gE would be non-functional in the absence of pUL51/pUL7 due to improper localization in the cell. This model predicts that a pUL51/gE double deletion would have a CCS phenotype no more severe than the worst of the two single mutations. This prediction was borne out in HaCaT cells (Figure 6), suggesting that pUL51 is necessary for gE/gI CCS function and that both proteins function on a single pathway for delivery of virions for CCS as depicted in Figure 8A. Nonetheless, it appears that pUL51 must contribute more to CCS in HaCaT cells than facilitating gE function, since the UL51 mutation had a more powerful inhibitory effect on CCS than a gE deletion. The nature of this additional function is unclear, but does not reflect defects in virus replication or release to the medium.
Figure 8.
Schematic diagrams of spread and release pathways in HaCaT (A) and Vero (B and C) cells infected with wild-type (A and B) or gE-null viruses (C). Thickness of arrows indicates the likely relative usage of pathways based on the effect of mutations that affect each one. In WT virus-infected HaCaT cells (A), virus is trafficked for CCS on a pathway that depends upon both gE and pUL51. Neither gE nor pUL51 participates in virus release to the medium. In WT virus-infected Vero cells (B), gE and pUL51 operate independently in CCS. The pUL51-dependent pathway is evidently more important for CCS (as indicated by the thicker pathway arrow), and pUL51 participates in the virus release pathway as well. We depict the gE and pUL51 pathways with two completely different pathway arrows, but the gE-dependent and pUL51-dependent CCS pathways may share other viral and cellular factors. In Vero cells infected with gE-null virus (C), The gE-dependent CCS pathway is abrogated, diminishing CCS, but the virion release pathway is enhanced.
In contrast to their behavior in HaCaT cells, it appears that gE and pUL51 may function independently in CCS in Vero cells (Figure 8B). The CCS phenotype of the double mutant in Vero cells was significantly more severe than the phenotype of a UL51 deletion alone, and was roughly the product of multiplication of the effects of the two single mutations. This suggests that there are genetically distinct gE- and pUL51-dependent pathways for spread in Vero cells. Although Figure 8B depicts these as entirely separate pathways, it seems likely that they have substantial overlap in the other viral and cellular factors and cellular organelles that are involved. The independence of gE and pUL51 CCS function in Vero cells was surprising, since gE does not concentrate at junctional surfaces in Vero cells infected with the UL51 deletion mutant (Figure 5). This suggests the possibility that the gE/gI CCS function in Vero cells is not dependent on concentration at junctional surfaces. The previously published evidence suggesting that junctional gE localization is necessary for gE function in CCS was based on correlation between junctional localization and CCS for gE cytoplasmic tail partial deletions observed in HaCaT and ARPE-19 cells (31). Whether the same result might be obtained in Vero cells is an interesting subject for further investigation. Further evidence that pUL51 and other tegument factors that affect CCS may not always function on the same pathway comes from the behavior of a double UL51/UL11 deletion in PRV (16). This double deletion showed a more severe plaque formation defect than either of the single mutants despite having a single step growth defect no more severe than the UL11 deletion.
In both HaCaT and Vero cells, a UL51 mutation has a more powerful inhibitory effect on CCS than a gE deletion. This suggests that, in both cell types, pUL51 very likely has an additional, gE-independent CCS function. The nature of this function in HaCaT cells is unclear, but the observation that pUL51 inhibits both CCS and virus release to the medium in Vero cells suggests a role for the pUL51/pUL7 complex in virion export from the cell on either pathway. Our results are consistent with a model for virion trafficking in Vero cells in which pUL51 (and perhaps pUL7) are required for transition of virion transport intermediates onto a general pathway for export from the cell. Once on that pathway, these intermediates arrive at a sorting point for release to the medium or transport to cell junctional surfaces for CCS. At that sorting point, the virions may take one of three pathways: (i) a gE-dependent pathway for CCS; (ii) a pUL51/pUL7-dependent pathway for CCS; or (iii) a pathway for release to the medium. Our data further suggest that the gE-dependent pathway for spread, and the pathway for release to the medium may be competitive, since deletion of gE and consequent elimination of the gE-dependent CCS pathway, results in more efficient release of virus to the medium (Figure 8C).
The molecular basis for cell-specific differences in CCS is unknown, but HSV likely uses a wide range of cellular trafficking factors to move virions from the site of assembly to the plasma membrane. While many of these may be common to virion release and spread pathways, some cellular factors that have a specific function in HSV epithelial CCS have been identified. Inhibition or knock-out of protein tyrosine phosphatase 1b diminishes HSV CCS of HaCaT and mouse embryonic fibroblasts without affecting virus replication or release (50), suggesting that it facilitates spread. The cellular proteins extended synaptotagmins 1 and 3, in contrast, negatively regulate CCS and virus release in Vero cells (51). Furthermore, pUL51 function in CCS in HaCaT and Vero cells is specifically impaired by a mutation that prevents phosphorylation at S184, suggesting that cellular kinases might also differentially regulate CCS (52). Differing levels of expression or activity of these proteins in different cell types might contribute to the differences observed here.
The existence of cell-specific pathways for CCS is not surprising, given that HSV may infect multiple cell-types, in various manifestations of disease, including epidermal keratinocytes, mucoepithelial cells, corneal epithelial cells, and neurons. HaCaT cells are considered a relevant model of skin keratinocytes, since they are derived from these cells, and retain the ability to differentiate into a stratified squamous epithelium in appropriate conditions (53, 54). Whether the pathways of spread observed in Vero cells are typical of any of the other cell types important for HSV pathogenesis is a subject for further exploration. The observation that multiple deletions of spread genes can produce profound, additive spread defects in some cell types suggests that such multiple deletions might form the basis for development of safe and effective anti-HSV vaccines.
Materials and Methods
Cells and Viruses
Vero, HaCaT, and UL51 complementing cells were maintained as previously described (18). All viruses were derived from the HSV-1 strain (F) BAC described in (55). The properties of HSV-1(F) have been previously described (56, 57). The construction of the UL51Δ73–244, UL51Δ167–244, and gE-null viruses has been described previously (18).
Plasmid Constructs
The pcDNA3 pUL51-FLAG expression construct was previously described (20). Inserts for construction of plasmids that express pUL51 truncations were constructed by amplification from pcDNA3 pUL51-FLAG using the primers shown in Table 1. PCR products were digested with EcoRI and Xhol restriction enzymes and then ligated into EcoRI-Xhol-digested pcDNA3.
Table 1.
Primer sequences for cloning of pUL51 truncations
| Insert | Forward Primer | Reverse Primer |
|---|---|---|
| pUL51 truncations in pcDNA3 |
||
| pUL51 (1–90)-FLAG | GTACGAATTCCATGGCTTCTCTTCTCGGGGCTATATG | GGCCCTCGAGTCACTTATCGTCATCGTCTTTGTAGTCGGGGTGGTGGCGCGC |
| pUL51 (1–124)-FLAG | GTACGAATTCCATGGCTTCTCTTCTCGGGGCTATATG | GGCCCTCGAGTCACTTATCGTCATCGTCTTTGTAGTCCATGTACATCTGCAGGATGGTGG |
| pUL51 (1–166)-FLAG | GTACGAATTCCATGGCTTCTCTTCTCGGGGCTATATG | GGCCCTCGAGTCACTTATCGTCATCGTCTTTGTAGTCGAAGGCGGAGAGGCCAAGGG |
| pUL51 (91–244)-FLAG | GTACGAATTCCATGGGGCTCGAGGCCCCCAC | GGCCCTCGAGTCACTTATCGTCATCGTCTTTGTAGTCTTGACCCAAAACACACGGAGCTG |
| pUL51 (125–244)-FLAG | GTACGAATTCCATGGGGGTTGCCGGGGGAACC | GGCCCTCGAGTCACTTATCGTCATCGTCTTTGTAGTCTTGACCCAAAACACACGGAGCTG |
| pUL51 (167–244)-FLAG | GTACGAATTCCATGGGGGTTGCCGGGGGAACC | GGCCCTCGAGTCACTTATCGTCATCGTCTTTGTAGTCTTGACCCAAAACACACGGAGCTG |
| pUL51 truncations fused to gD |
||
| pUL51(1–30)-FLAG | CCAGCCCTTGTTTTACATGGCTTCTCTTCTCGGGGC | TCATCGTCTTTGTAGTCCTCCGAGGGCGGAACGGC |
| pUL51(1–60)-FLAG | CCAGCCCTTGTTTTACATGGCTTCTCTTCTCGGGGC | TCATCGTCTTTGTAGTCGTCCAGGGACCCCAGGGC |
| pUL51(1–90)-FLAG | CCAGCCCTTGTTTTACATGGCTTCTCTTCTCGGGGC | CTTATCGTCATCGTCTTTGTAGTCGGGGTGGTGGCGCGC |
| pUL51(30–90)-FLAG | CCAGCCCTTGTTTTACATGGCGGAGCCGCGGCTG | CTTATCGTCATCGTCTTTGTAGTCGGGGTGGTGGCGCGC |
| pUL51(60–90)-FLAG | CCAGCCCTTGTTTTACATGGACACCCGACGCCTCGTGAAG | CTTATCGTCATCGTCTTTGTAGTCGGGGTGGTGGCGCGC |
pRR1407, for expression of C-terminally FLAG-tagged gD was constructed by amplification of gD coding sequences from HSV-1(F) using the forward primer (5’-GATCAAGCTTGGTGCGTTCCGGTATGGGGG-3’) and the reverse primer (5’-CATGCTCGAGCTACTTATCGTCATCGTCTTTGTAGTCGGATCCCCCGGGGAATTCG TAAAACAAGGGCTGGTGCGAGG-3’), digestion of the resulting PCR product with Hindlll and Xhol, and ligation into Hindlll/Xhol-cut pcDNA3. pRR1407 was then used as the backbone for Gibson assembly cloning of all gD-UL51-FLAG fusions using the New England Biolabs Gibson Assembly Master Mix according to the manufacturer’s instructions. Each plasmid was assembled from two PCR products; one comprised of vector and gD-FLAG, and the other amplified from pcDNA3 pUL51-FLAG. The sequences of primers used for these amplifications are shown in Table 1.
The plasmid that expresses EGFP-UL7 was constructed by amplification of UL7 coding sequences from HSV-1 (F) DNA using the forward primer (5’-GCTCAAGCTTCGATGGCCGCCGCGACG-3’) and the reverse primer (5’-TCGAGAATTCAACAAAACTGATAAAACAGCGACGACG-3’), digestion of the resulting PCR product with Hindlll and EcoRI, and ligation into Hindlll-EcoRI-cut pEGFP-C1 (Clontech).
Construction of Recombinant Viruses
A double mutant recombinant carrying the UL51Δ167–244 and gE-null deletions was constructed using the gE-null BAC as the parent and modifying it to carry the UL51 partial deletion as previously described (18). Two independent isolates were constructed, and were recovered by transfection of the BAC DNAs into UL51 complementing cells. A UL7-null recombinant was constructed by insertion of a gentamicin resistance (GmR) cassette into the UL7 protein coding sequence immediately following the stop codon for the UL6 coding sequence. lnsertion of the GmR cassette places a stop codon in-frame with the UL7 reading frame 10 nucleotides following the insertion junction. The primers used for insertion of the GmR cassette were 5’-GGGGGCATCGGGCACCGGGATGGCCGCCGCGACGGCCGACGATGAgctcggatccta gggataacaggg-3’ and 5’-CGATGGCCTGCTTGAGGATGGTGGCGGCCGACCCCtctagaggccgcggcgttg-3’. ln each case, lowercase nucleotides correspond to sequence from the inserted GmR cassette. Virus construction was performed as described by Tischer et al., using the HSV-1 strain F BAC in bacterial strain GS1783 (kind gift of Greg Smith) (58). The UL7-null recombinant virus was rescued and amplified on Vero cells. Proper structure of the recombinant BAC and virus at the UL7 locus were confirmed by PCR amplification and sequencing of the gene region.
Plaque size assay
Plaque size assays were performed in the presence of neutralizing antibody as previously described (18). Since plaque areas are not always normally distributed, statistical analysis was performed by one-way ANOVA using the non-parametric Fisher’s least significant difference test as implemented in Prism 7 (GraphPad Software, Inc.).
Single-step growth measurement
Measurement of replication of HSV-1(F), UL51Δ167–244 and double mutant viruses on Vero and HaCaT cells after infection at high multiplicity (5 PFU/cell) was performed as previously described (57). Virus release efficiency was calculated as PFU in the culture medium at 24 (Vero) or 48 (HEp-2) h.p.i./PFU produced in the total culture at that time point.
Immunoblotting
Nitrocellulose sheets bearing proteins of interest were blocked in 5% non-fat milk plus 0.2% Tween 20 for at least 2 h. The membranes were probed either with a 1:1000 dilution of mouse M2 anti-FLAG (SIGMA), a 1:5000 dilution of rabbit anti-EFP (gift of Craig Ellermeier), a 1:1000 dilution of a rabbit polyclonal antiserum raised against a UL51-GST fusion protein (18), a 1:1000 dilution of a rabbit polyclonal antiserum raised against gE (kind gift of H. Friedman), a 1:1000 dilution of rabbit polyclonal antiserum raised against pUL7 (kind gift of Yasushi Kawaguchi), or a 1:2000 dilution of mouse monoclonal antibody directed against the HSV-1 scaffold protein (AbD Serotec) followed by reaction with alkaline phosphatase-conjugated secondary antibody (Sigma).
Indirect immunofluorescence
Immunofluorescent detection of pUL51-FLAG or gD-pUL51-FLAG constructs was performed as previously described using 1:1000 mouse M2 anti-FLAG monoclonal antibody (SIGMA) (20). DNA was fluorescently stained using TO-PRO-3 (Invitrogen) at 1 μM and filamentous actin was stained using 1 μM phalloidin-488 (Invitrogen) during the secondary antibody incubation. HSV-1 gE and cellular β-catenin were detected using 1:1000 diluted rabbit polyclonal antiserum raised against gE and 1:1000 diluted mouse monoclonal anti-β-catenin (Invitrogen), respectively (59, 60).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Erika Feutz, Department of Epidemiology, University of Washington, Seattle, WA.
Hilary McLeland-Wieser, Department of Environmental and Occupational Health, Milken Institute School of Public Health, The George Washington University, Washington, DC.
Junlan Ma, Queensland University of Technology, Brisbane, QLD, Australia.
Richard J. Roller, Department of Microbiology and Immunology, Carver College of Medicine, University of Iowa, Iowa City, IA.
References
- 1.Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat Rev Micro 9:382–394. [DOI] [PubMed] [Google Scholar]
- 2.Cepeda V, Esteban M, Fraile-Ramos A. 2010. Human cytomegalovirus final envelopment on membranes containing both trans-Golgi network and endosomal markers. Cell Microbiol 12:386–404. [DOI] [PubMed] [Google Scholar]
- 3.Das S, Ortiz DA, Gurczynski SJ, Khan F, Pellett PE. 2014. Identification of human cytomegalovirus genes important for biogenesis of the cytoplasmic virion assembly complex. J Virol 88:9086–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das S, Pellett PE. 2011. Spatial relationships between markers for secretory and endosomal machinery in human cytomegalovirus-infected cells versus those in uninfected cells. J Virol 85:5864–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Homman-Loudiyi M, Hultenby K, Britt W, Soderberg-Naucler C. 2003. Envelopment of human cytomegalovirus occurs by budding into Golgi-derived vacuole compartments positive for gB, Rab 3, trans-golgi network 46, and mannosidase II. J Virol 77:3191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchez V, Greis KD, Sztul E, Britt WJ. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J Virol 74:975–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schauflinger M, Villinger C, Mertens T, Walther P, von Einem J. 2013. Analysis of human cytomegalovirus secondary envelopment by advanced electron microscopy. Cell Microbiol 15:305–14. [DOI] [PubMed] [Google Scholar]
- 8.Severi B, Landini MP, Govoni E. 1988. Human cytomegalovirus morphogenesis: an ultrastructural study of the late cytoplasmic phases. Arch Virol 98:51–64. [DOI] [PubMed] [Google Scholar]
- 9.Sugimoto K, Uema M, Sagara H, Tanaka M, Sata T, Hashimoto Y, Kawaguchi Y. 2008. Simultaneous tracking of capsid, tegument, and envelope protein localization in living cells infected with triply fluorescent herpes simplex virus 1. J Virol 82:5198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turcotte S, Letellier J, Lippe R. 2005. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J Virol 79:8847–8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollinshead M, Johns HL, Sayers CL, Gonzalez-Lopez C, Smith GL, Elliott G. 2012. Endocytic tubules regulated by Rab GTPases 5 and 11 are used for envelopment of herpes simplex virus, vol 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laine RF, Albecka A, van de Linde S, Rees EJ, Crump CM, Kaminski CF. 2015. Structural analysis of herpes simplex virus by optical super-resolution imaging. Nat Commun 6:5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Owen DJ, Crump CM, Graham SC. 2015. Tegument Assembly and Secondary Envelopment of Alphaherpesviruses. Viruses 7:5084–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daikoku T, Ikenoya K, Yamada H, Goshima F, Nishiyama Y. 1998. Identification and characterization of the herpes simplex virus type 1 UL51 gene product. J Gen Virol 79:3027–3031. [DOI] [PubMed] [Google Scholar]
- 15.Nozawa N, Daikoku T, Koshizuka T, Yamauchi Y, Yoshikawa T, Nishiyama Y. 2003. Subcellular Localization of Herpes Simplex Virus Type 1 UL51 Protein and Role of Palmitoylation in Golgi Apparatus Targeting. J Virol 77:3204–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klupp BG, Granzow H, Klopfleisch R, Fuchs W, Kopp M, Lenk M, Mettenleiter TC. 2005. Functional Analysis of the Pseudorabies Virus UL51 Protein. J Virol 79:3831–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nozawa N, Kawaguchi Y, Tanaka M, Kato A, Kato A, Kimura H, Nishiyama Y. 2005. Herpes Simplex Virus Type 1 UL51 Protein Is Involved in Maturation and Egress of Virus Particles. J Virol 79:6947–6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roller RJ, Haugo AC, Yang K, Baines JD. 2014. The Herpes Simplex Virus 1 UL51 Gene Product Has Cell Type-Specific Functions in Cell-to-Cell Spread. J Virol 88:4058–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albecka A, Owen DJ, Ivanova L, Brun J, Liman R, Davies L, Ahmed MF, Colaco S, Hollinshead M, Graham SC, Crump CM. 2017. Dual Function of the pUL7-pUL51 Tegument Protein Complex in Herpes Simplex Virus 1 Infection. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roller RJ, Fetters R. 2015. The Herpes Simplex Virus 1 UL51 Protein Interacts with the UL7 Protein and Plays a Role in Its Recruitment into the Virion. Journal of Virology 89:3112–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahlqvist J, Mocarski E. 2011. Cytomegalovirus UL103 Controls Virion and Dense Body Egress. Journal of Virology 85:5125–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schauflinger M, Fischer D, Schreiber A, Chevillotte M, Walther P, Mertens T, von Einem J. 2011. The tegument protein UL71 of human cytomegalovirus is involved in late envelopment and affects multivesicular bodies. J Virol 85:3821–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Womack A, Shenk T. 2010. Human cytomegalovirus tegument protein pUL71 is required for efficient virion egress. MBio 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson DC, Feenstra V. 1987. Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J Virol 61:2208–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson DC, Frame MC, Ligas MW, Cross AM, Stow ND. 1988. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J Virol 62:1347–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lubinski J, Nagashunmugam T, Friedman HM. 1998. Viral interference with antibody and complement. Semin Cell Dev Biol 9:329–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ndjamen B, Farley AH, Lee T, Fraser SE, Bjorkman PJ. 2014. The herpes virus Fc receptor gE-gI mediates antibody bipolar bridging to clear viral antigens from the cell surface. PLoS Pathog 10:e1003961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balan P, Davis-Poynter N, Bell S, Atkinson H, Browne H, Minson T. 1994. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. Journal of General Virology 75:1245–1258. [DOI] [PubMed] [Google Scholar]
- 29.Dingwell KS, Brunetti CR, Hendricks RL, Tang Q, Tang M, Rainbow AJ, Johnson DC. 1994. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. Journal of Virology 68:834–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dingwell KS, Johnson DC. 1998. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. Journal of Virology 72:8933–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farnsworth A, Johnson DC. 2006. Herpes Simplex Virus gE/gI Must Accumulate in the trans-Golgi Network at Early Times and Then Redistribute to Cell Junctions To Promote Cell-Cell Spread. J Virol 80:3167–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMillan TN, Johnson DC. 2001. Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. Journal of Virology 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wisner T, Brunetti C, Dingwell K, Johnson DC. 2000. The Extracellular Domain of Herpes Simplex Virus gE Is Sufficient for Accumulation at Cell Junctions but Not for Cell-to-Cell Spread. J Virol 74:2278–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson DC, Webb M, Wisner TW, Brunetti C. 2001. Herpes Simplex Virus gE/gI Sorts Nascent Virions to Epithelial Cell Junctions, Promoting Virus Spread. J Virol 75:821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins WJ, Johnson DC. 2003. Herpes Simplex Virus gE/gI Expressed in Epithelial Cells Interferes with Cell-to-Cell Spread. J Virol 77:2686–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chadha P, Han J, Starkey JL, Wills JW. 2012. Regulated interaction of tegument proteins UL16 and UL11 from herpes simplex virus. J Virol 86:11886–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han J, Chadha P, Meckes DG, Baird NL, Wills JW. 2011. Interaction and Interdependent Packaging of Tegument Protein UL11 and Glycoprotein E of Herpes Simplex Virus. Journal of Virology 85:9437–9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han J, Chadha P, Starkey JL, Wills JW. 2012. Function of glycoprotein E of herpes simplex virus requires coordinated assembly of three tegument proteins on its cytoplasmic tail. Proceedings of the National Academy of Sciences 109:19798–19803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeh PC, Han J, Chadha P, Meckes DG Jr., Ward MD, Semmes OJ, Wills JW. 2011. Direct and specific binding of the UL16 tegument protein of herpes simplex virus to the cytoplasmic tail of glycoprotein E. J Virol 85:9425–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krummenacher C, Baribaud I, Eisenberg RJ, Cohen GH. 2003. Cellular localization of nectin-1 and glycoprotein D during herpes simplex virus infection. J Virol 77:8985–8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai W, Gu B, Person S. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. Journal of Virology 62:2596–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forrester A, Farrell H, Wilkinson G, Kaye J, Davis-Poynter N, Minson T. 1992Construction and properties of a mutant herpes simplex virus type 1 with glycoprotein H coding sequences deleted. Journal of Virology 66:341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ligas MW, Johnson DC. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by ß galactosidase sequences binds to but is unable to penetrate into cells. Journal of Virology 62:1486–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roop C, Hutchinson L, Johnson DC. 1993. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells and its particles lack glycoprotein H. Journal of Virology 67:2285–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gorleku OA, Barns AM, Prescott GR, Greaves J, Chamberlain LH. 2011. Endoplasmic reticulum localization of DHHC palmitoyltransferases mediated by lysine-based sorting signals. J Biol Chem 286:39573–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nozawa N, Daikoku T, Yamauchi Y, Takakuwa H, Goshima F, Yoshikawa T, Nishiyama Y. 2002. Identification and characterization of the UL7 gene product of herpes simplex virus type 2. Virus Genes 24:257–66. [DOI] [PubMed] [Google Scholar]
- 47.Duffy C, LaVail JH, Tauscher AN, Wills EG, Blaho JA, Baines JD. 2006. Characterization of a UL49-Null Mutant: VP22 of Herpes Simplex Virus Type 1 Facilitates Viral Spread in Cultured Cells and the Mouse Cornea. Journal of Virology 80:8664–8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maringer K, Stylianou J, Elliott G. 2012. A network of protein interactions around the herpes simplex virus tegument protein VP22. J Virol 86:12971–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stylianou J, Maringer K, Cook R, Bernard E, Elliott G. 2009. Virion Incorporation of the Herpes Simplex Virus Type 1 Tegument Protein VP22 Occurs via Glycoprotein E-Specific Recruitment to the Late Secretory Pathway. Journal of Virology 83:5204–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carmichael JC, Yokota H, Craven RC, Schmitt A, Wills JW. 2018. The HSV-1 mechanisms of cell-to-cell spread and fusion are critically dependent on host PTP1B. PLoS Pathog 14:e1007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El Kasmi I, Khadivjam B, Lackman M, Duron J, Bonneil E, Thibault P, Lippe R. 2018. Extended Synaptotagmin 1 Interacts with Herpes Simplex Virus 1 Glycoprotein M and Negatively Modulates Virus-Induced Membrane Fusion. J Virol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kato A, Oda S, Watanabe M, Oyama M, Kozuka-Hata H, Koyanagi N, Maruzuru Y, Arii J, Kawaguchi Y. 2018. Roles of the Phosphorylation of Herpes Simplex Virus 1 UL51 at a Specific Site in Viral Replication and Pathogenicity. J Virol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. 1988. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol 106:761–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schoop VM, Mirancea N, Fusenig NE. 1999. Epidermal organization and differentiation of HaCaT keratinocytes in organotypic coculture with human dermal fibroblasts. J Invest Dermatol 112:343–53. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka M, Kagawa H, Yamanashi Y, Sata T, Kawaguchi Y. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J Virol 77:1382–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ejercito PM, Kieff ED, Roizman B. 1968. Characteristics of herpes simplex virus strains differing in their effect on social behavior of infected cells. Journal of General Virology 2:357–364. [DOI] [PubMed] [Google Scholar]
- 57.Roller RJ, Zhou Y, Schnetzer R, Ferguson J, DeSalvo D. 2000. Herpes simplex virus type 1 Ul34 gene product is required for viral envelopment. Journal of Virology 74:117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tischer BK, Smith GA, Osterrieder N. 2010. En Passant Mutagenesis: A Two Step Markerless Red Recombination System, p 421–430. In Braman J (ed), In Vitro Mutagenesis Protocols: Third Edition doi: 10.1007/978-1-60761-652-8_30. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 59.Bjerke SL, Cowan JM, Kerr JK, Reynolds AE, Baines JD, Roller RJ. 2003. Effects of charged cluster mutations on the function of herpes simplex virus type 1 UL34 protein. J Virol 77:7601–7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reynolds AE, Ryckman BJ, Baines JD, Zhou Y, Liang L, Roller RJ. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J Virol 75:8803–8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]