Abstract
Protein CGL2373 from Corynebacterium glutamicum was previously proposed to be a member of the polyketide_cyc2 family, based on amino-acid sequence and secondary structure features derived from NMR chemical shift assignments. We report here the solution NMR structure of CGL2373, which contains three α-helices and one antiparallel β-sheet and adopts a helix-grip fold. This structure shows moderate similarities to the representative polyketide cyclases, TcmN, WhiE, and ZhuI. Nevertheless, unlike the structures of these homologs, CGL2373 structure looks like a half-open shell with a much larger pocket, and key residues in the representative polyketide cyclases for binding substrate and catalyzing aromatic ring formation are replaced with different residues in CGL2373. Also, the gene cluster where the CGL2373-encoding gene is located in C. glutamicum contains additional genes encoding nucleoside diphosphate kinase, folylpolyglutamate synthase, and valine-tRNA ligase, different from the typical gene cluster encoding polyketide cyclase in Streptomyces. Thus, although CGL2373 is structurally a polyketide cyclase-like protein, the function of CGL2373 may differ from the known polyketide cyclases and needs to be further investigated. The solution structure of CGL2373 lays a foundation for in silico ligand screening and binding site identifying in future functional study.
Keywords: Polyketide_cyc2, START-like, PYR1-like, Bet v1, aromatic ring
1. INTRODUCTION
Corynebacterium glutamicum is a non-pathogenic soil bacterium, and an important bioindustrial bacterium used for producing amino acids, nucleotides and vitamins, which is facilitated by some traits of C. glutamicum such as rapid growth rate and relatively few growth requirements.1 C. glutamicum has a circular chromosome and a plasmid for encoding more than three thousands proteins.2 The protein CGL2373 (UniProt ID: Q8NN40_CORGL) encoded in C. glutamicum genome was proposed to belong to the polyketide_cyc2 family (Pfam ID: PF10604), based on amino-acid sequence and secondary structure features derived from NMR study.3 The polyketide_cyc2 family includes polyketide cyclases for polyketide synthesis in bacteria. All known structures of polyketide cyclases adopt a helix-grip fold with an interior pocket,4–7 similar to the structures of the START (StAR-related transfer)-like proteins (Pfam ID: PF01852) and the Bet v1 (Betula verrucosa birch pollenallergen)-like proteins (Pfam ID: PF00407).
Polyketide cyclase plays an important role in the biosynthesis of aromatic polyketides, a class of natural products among which many are the sources of antibiotics or anticancer drugs.8,9 Polyketide cyclase is a part of the multi-enzyme complex of type II polyketide synthase (PKS), and catalyze the region-specific formation of aromatic rings in polyketide synthesis.10 Several representative polyketide cyclases have been structurally and functionally characterized, which include three monodomain cyclases (TcmN, ZhuI, WhiE) and two di-domain cyclases (StfQ and BexL).4–7 The individual cyclase domain in the five polyketide cyclases all adopts a similar helix-grip fold with an interior pocket. However, they show different residue composition in the interior pocket, which controls the region-specific cyclization and determines the selective specificity for the chain length of the substrate. Consistently, these known polyketide cyclases show different functional features. For instance, TcmN and WhiE both catalyze C9−C14 first-ring cyclization, but show difference in the chain lengths of the respective natural substrates (20 and 24 carbons, respectively).4,6 On the other hand, ZhuI is a C7−C12 first-ring cyclase for substrate chains of variable lengths (C16 and C18−C20).5,11 To our knowledge, most polyketide cyclases that have been investigated for their structure and function are from Streptomyces, while the structure and function of polyketide cyclases from Corynebacterium are still unknown.
We reported the chemical shift assignments of 1H, 13C and 15N-labeled CGL2373 recently,3 and its solution NMR structure has been previously deposited in the Protein Data Bank (PDB ID: 2M47). Here, we describe the details of the NMR structure determination of CGL2373 and make a comparison between CGL2373 and the representative polyketide cyclases in the aspects of sequence and structure. Although the overall structures are similar, structural details are largely different between CGL2373 and the polyketide cyclases. Especially, key residues in the polyketide cyclases for binding substrate and catalyzing ring formation are replaced with other types of residues in CGL2373. Thus, the function of CGL2373 may remarkably differ from the representative polyketide cyclases.
2. MATERIALS AND METHODS
2.1. Protein sample preparation
The U-13C,15N-labeled (NC) recombinant CGL2373 protein with a C-terminal His-tag (LEHHHHHH) was prepared as described previously.3 The purified protein at a concentration of 0.8 mM in the buffer containing 90 % H2O/10 % D2O (v/v), 20 mM MES, 100 mM NaCl, 5 mM CaCl2, 10 mM DTT, 0.05 mM DSS and 0.02 mM NaN3 at pH 6.5 was used for NMR experiments.
2.2. Rotational correlation time (τc) estimate
1D 15N-edited T1 and T2 (CPMG) experiments using NC sample were recorded on a Varian Inova 600 MHz at 298 K for determining the rotational correlation time (τc) of CGL2373. Longitudinal T1 relaxation delays were 100, 200, 300, 400, 600, 800, 1000, 1500, 1700, and 2000 ms; transverse T2 relaxation delays were 10, 30, 50, 70, 90, 110, 130, 170, 210, and 250 ms; both experiments had 1.5 s recycle delays. T1 and T2 relaxation times were obtained through integrating intensity from 8.5 to 10.5 ppm. An τc value of 11.5 ns for NC CGL2373 was derived from the T1 and T2 measurements following the literature equation.12 Based on a linear fitting of τc versus molecular weight (MW) for a series of standard proteins,13 the MW of NC CGL2373 under the NMR conditions was estimated to be 20.2 kDa (Supporting Information Figure S1).
2.3. Structure calculation
The chemical shift assignments of CGL2373 deposited in the BioMagResDB (BMRB) with the accession number 18989 were reported previously.3 For structure determination, NOE-based inter-proton distance restraints of CGL2373 were automatically determined using CYANA 3.0. Input for CYANA included chemical shift assignments, NOESY peak lists from four NOESY spectra with peak intensities, the restraints for backbone phi (φ) and psi (ψ) torsion angle derived from chemical shifts of backbone atoms using the TALOS+ software program.14 Manual and iterative refinements of NOESY peak picking lists were guided through assessing “goodness of fit” between calculated structures and NOESY peak lists using NMR RPF quality.15 Towards the end of the iterative process of structure calculation, hydrogen bond constraints for the NH and CO distances were introduced based on identification of the proximity of potential donors and receptors in earlier structure calculations. The 20 lowest energy structures out of 100 structures calculated by CYANA 3.0 were further refined through restrained molecular dynamics in explicit water CNS 1.2 and the PARAM19 force field, using the final NOE-derived distance restraints and TALOS-derived dihedral angle restraints. The final ensemble of 20 structures has been deposited to the PDB (ID: 2M47). Structural statistics and global structure quality factors were computed using PSVS16 version 1.5 (Table 1).
Table 1.
Structure statistics for CGL2372 from C. glutamicum
| Conformationally-restricting constraintsa | |
|---|---|
| NOE-based distance constraints | |
| Total | 1571 |
| Intra-residue (i=j) | 380 |
| Sequential (|i-j|=1) | 489 |
| Medium-range (1<|i-j|<5) | 238 |
| Long-range (|i-j|≥5) | 464 |
| NOE constraints per restrained residueb | 10.1 |
| Hydrogen bond constraints | |
| Long-range (|i-j |≥5)/total | 62/108 |
| Dihedral angle constraints | 214 |
| Total number of restricting constraintsb | 1893 |
| Total number of restricting constraints per restrained residueb | 12.1 |
| Restricting long-range constraints per restrained residueb | 3.4 |
| Number of structures used | 20 |
| Residue constraint violationsa, c | |
| Distance violations per structure | |
| 0.1–0.2Å | 7.2 |
| 0.2–0.5 Å | 1.4 |
| >0.5Å | 0 |
| RMS of distance violation/constraint (Å) | 0.01 |
| Maximum distance violation (Å)d | 0.42 |
| Dihedral angle violations per structure | |
| 1–10° | 17.8 |
| >10° | 0 |
| Average RMS dihedral angle violation/constraint (degree) | 0.80 |
| Maximum dihedral angle violation (degree)d | 8.10 |
| RMSD Valuese | |
| Backbone/Heavy atoms (Å) | 1.3/1.8 |
| Ramachandran plot summary from Richardson’s lab | |
| Most favored regions (%) | 96.0 |
| Allowed regions (%) | 4 |
| Disallowed regions (%) | 0.0 |
| Structure quality Factors (mean/Z-score)f | |
| Verify3D | 0.27/−3.05 |
| Prosall (-ve) | 0.33/−1.32 |
| Procheck G-factore (φ-Ψ) | −0.35/−1.06 |
| Procheck G-factore (all dihedral angles) | −0.27/−1.60 |
| Molprobity clash score | 12.18/−0.56 |
| RPF Scoresg | |
| Recall/Precision | 0.99/0.89 |
| F-measure/DP-score | 0.93/0.77 |
Calculated using PSVS 1.5 program. Residues (1–163) were analyzed.
There are 156 residues with conformationally restricting constraints.
Calculated for all constraints for the given residues, using sum over r–6.
Largest constraint violation among all the reported structures
Ordered residues ranges (with the sum of ϕ and ψ order parameters >1.8): 6–24, 36–46, 48–55, 57–75, 78–84, 88–111, 114–121, and 124–156.
With respect to mean and standard deviation for a set of 252 X-ray structures < 500 residues, of resolution ≤ 1.80 Å, R-factor ≤ 0.25 and R-free ≤ 0.28; a positive value indicates a ‘better’ score.
RPF scores reflected the goodness-of-fit of the final ensemble of structures including disordered residues to the NMR data.
3. RESULTS AND DISCUSSION
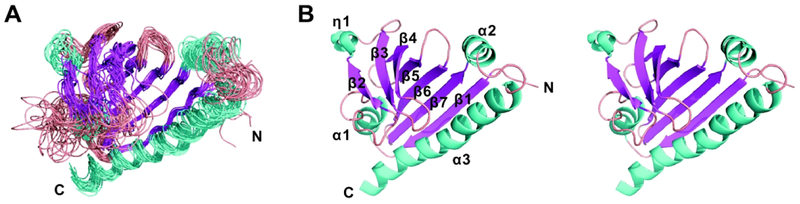
The MW of NC CGL2373 was estimated to be 20.2 kDa based on the determined overall τc value of 11.5 ns (Supporting Information Figure S1), close to its theoretical MW (MW = 19.9 kDa for NC CGL2373 with the C-terminal His-tag), suggesting that CGL2373 exists predominantly as a monomer under the NMR conditions. The overall NMR structure of CGL2373 adopts a helix-grip fold, consisting of a seven-stranded antiparallel β-sheet surrounding a long C-terminal α-helix and two small helices flanking the β-sheet (Figure 1A). Additionally, there is a 310-helix formed at the backside of the β-sheet. The RMSD values for backbone atoms and heavy atoms in the ordered regions were 1.3 and 1.8 Å, respectively (Table 1). Secondary structure elements distribute as α-helices (α1, 17–24; α2, 112–119; α3, 127–155), β-strands (β1, 5–13; β2, 37–41; β3, 53–59; β4, 64–75; β5, 79–84; β6, 89–98; β7, 101–110), and 310-helix (η1, 43–45), with the arranging order of N–β1–α1–β2–η1–β3–β4–β5–β6–β7–α2–α3–C (Figure 1B). The α-helices and β-strands together form a large pocket with a volume of 3302 Å3 in the center of the protein as calculated using the CASTp 3.0 server.17 The α1-β2 and β3-β4 loops are very flexible, making the opening extent of the pocket vary greatly among the 20 conformers of CGL2373 structure (Figure 1A). Surface representation of CGL2373 structure looks like a half-open shell, and electrostatic surface potential analysis shows that the pocket is largely neutral and is only slightly charged (Supporting Information Figure S2A). The Among the 37 residues involved in forming the pocket, 21 residues contain hydrophobic side chain and 16 residues contain polar side chain (Supporting Information Figure S2B). Only 4 residues with charged side chain (one Arg, one Asp and two Glu) are located in the pocket. The properties of the pocket are consistent with the potential function of CGL2373 that involves polycyclic ligand binding. The amphipathic composition of the pocket residues would provide appropriate hydrophobic and hydrogen bond interaction for specific ligands.
Figure 1.
Solution NMR structure of CGL2373 from Corynebacterium glutamicum. A, Superposition of the backbone traces of the 20 conformers of CGL2373 with the lowest energy. B, The stereo-view of cartoon model of CGL2373 structure. The α-helices, β-strands, and loop regions are colored in cyan, magenta, and salmon, respectively.
BLAST search using CGL2373 sequence obtained 500 homologous sequences with alignment scores over 89.7 and sequence identity over 36.4%. However, the obtained sequences are annotated as either SRPBCC (START/RHO_α_C/PITP/Bet_v1/CoxG/CalC) family protein or polyketide cyclase, and gave no detailed functional and structural information. Structure similarity analysis using Dali18 identified 113 peptide chains with Z-scores over 11.0, which can be classified into three groups including polyketide cyclases,4–7 plant receptors for abscisic acid (ABA),19–21 and phenolic oxidative coupling proteins.22 Representative proteins with published papers focusing on function and structure are summarized in Supporting Information Table S1
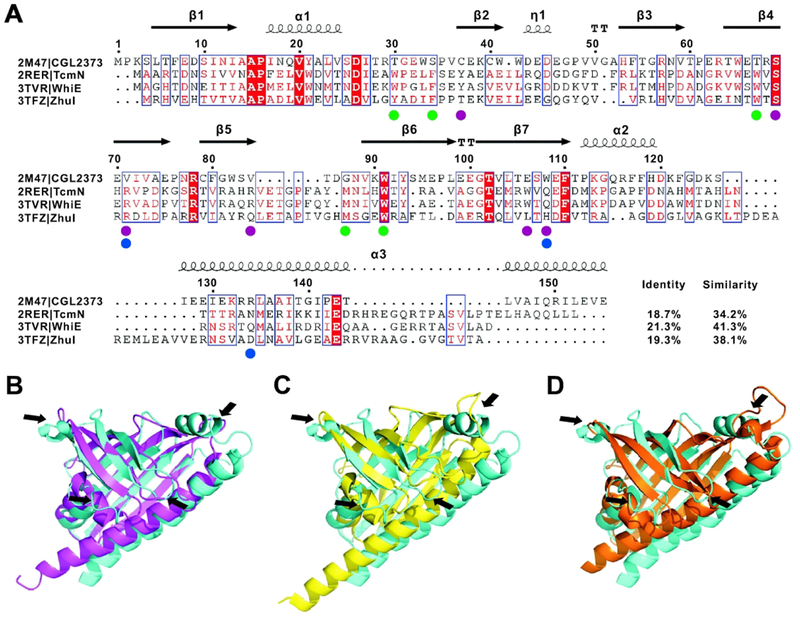
CGL2373 shares sequence identity/similarity of 18.7/34.2%, 21.3/41.3% and 19.3/38.1% with three representative monodomain cyclases, TcmN, WhiE and ZhuI, respectively (Figure 2A). The overall structures are similar, but at last three major differences can be found between CGL2373 and the three cyclases, including the lack of an α-helix between α1 and β2, the forming of a 310-helix between β2 and β3, and the forming of an α-helix between β7 and α3 in CGL2373 (Figure 2B). Besides, the conformation of β3–β4 loop of CGL2373 is significantly different from the counterparts in the three cyclases. The lack of rigid α-helix structure between α1 and β2 and the different conformation of β3–β4 loop may result from the extremely weak interaction of α1–β2 and β3–β4 loops with the α3 helix, leading to an unclosed side surface and markedly larger pocket of CGL2373 compared with TcmN, WhiE, and ZhuI, which contain ligand-binding pockets with the volumes of 1269 Å3, 1304 Å3, and 1066 Å3, respectively (Supporting Information Figure S2A). Thus, it is possible that the potential ligands of CGL2373 are bigger than those of these three cyclases in size. Meanwhile, it’s worth noting that most of the key residues in the three cyclases for binding substrate and catalyzing ring formation, such as Y35, R69 and R82 of TcmN and WhiE,4,6 and R66, H109 and D146 of ZhuI,5 are replaced with residues having different types of side chains in CGL2373 (Figure 2A). Therefore, CGL2373 may not have aromatase/cyclase activity, or if it has, that its catalytic mechanism must be different from the known polyketide cyclases. Similarly, the low sequence identity and notable conformational differences with the ABA receptors, PYR121, PYL120 and PYL1019, as well as lack of key residues for ABA binding (Supporting Information Figure S3), suggest that CGL2373 may also not an ABA binding protein. Because the ligand-binding mechanism of HYP-1 has not been well demonstrated, detailed comparison between HYP-1 and CGL2373 for potential similar function was not carried out.
Figure 2.
Comparison of CGL2373 with three known polyketide cyclases including TcmN, WhiE and ZhuI. A, Sequence alignment of CGL2373 with the three polyketide cyclases. Alignment was rendered using ESPript 3.0 with default settings for similarity calculations. Secondary structural elements of CGL2373 are indicated above the amino-acid residue number in the sequence. Identical (white letters filled with red color) and similar (red letters with blue box) amino acids are denoted. The residues essential for the cyclase activity of TcmN and WhiE are labeled with violet circles below the sequence, while the residues essential for the activity of ZhuI are labeled with blue circles. The residues labeled with green circles are involved in substrate binding for TcmN and WhiE. B, C, and D, Superposition of the structure of CGL2373 (cyan) with those of TcmN (magenta), WhiE (yellow), and ZhuI (orange), respectively. Black arrows denote the remarkable differences between CGL2373 structure and those of the three cyclases.
The gene cluster where CGL2373 gene is located contains additional genes encoding nucleoside diphosphate kinase (NDPK, CGL2370), predicted acetyltransferase (CGL2372), folylpolyglutamate synthase (FPGS, CGL2375), valine-tRNA ligase (CGL2376), and two hypothetical membrane proteins (CGL2371 and CGL2374). The substrates of NDPK include nucleoside diphosphates (NDP),23 and the substrates of valine-tRNA ligase and FPGS include adenosine triphosphate (ATP),24,25 all of which have polycyclic structures. Moreover, the substrates of FPGS include folic acid, also a polycyclic molecule. Because the expression of neighboring genes in bacteria is usually co-regulated, and their functions are associated with each other to some extent, thus, the neighborhood members of CGL2373 may provide some clues for its function.
In summary, the solution NMR structure of CGL2373 was determined to adopt a typical helix-grip fold, but the details of the structure are different from those of the well known polyketide cyclases, implying that CGL2373 may not function similarly to the known cyclases. The biological function of CGL2373 requires further investigation, and the solution structure reported here will be helpful for in silico ligand screening and binding site identifying in future.
Supplementary Material
ACKNOWLEDGMENTS
We thank for grant supports from the National Natural Science Foundation of China (Grant Number: 21575155 and 21703283), and the Protein Structure Initiative-Biology Program by the National Institute of General Medical Sciences (Grant Numbers: U54-GM094597).
REFERENCES
- 1.Yukawa H, Natsuishi BB. Corynebacterium glutamicum: biology and biotechnology. Heidelberg: Springer; 2013. [Google Scholar]
- 2.Ikeda M, Nakagawa S. The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl Microbiol Biotechnol. 2003;62:99–109. [DOI] [PubMed] [Google Scholar]
- 3.Liang C, Hu R, Ramelot TA, Kennedy MA, Li X, Yang Y, Zhu J, Liu M. Chemical shift assignments of polyketide cyclase_like protein CGL2373 from Corynebacterium glutamicum. Biomol NMR Assign. 2017;11:289–292. [DOI] [PubMed] [Google Scholar]
- 4.Ames BD, Korman TP, Zhang W, Smith P, Vu T, Tang Y, Tsai SC. Crystal structure and functional analysis of tetracenomycin ARO/CYC: implications for cyclization specificity of aromatic polyketides. Proc Natl Acad Sci U S A. 2008;105:5349–5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ames BD, Lee MY, Moody C, Zhang W, Tang Y, Tsai SC. Structural and biochemical characterization of ZhuI aromatase/cyclase from the R1128 polyketide pathway. Biochemistry. 2011;50:8392–8406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee MY, Ames BD, Tsai SC. Insight into the molecular basis of aromatic polyketide cyclization: crystal structure and in vitro characterization of WhiE-ORFVI. Biochemistry. 2012;51:3079–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caldara-Festin G, Jackson DR, Barajas JF, Valentic TR, Patel AB, Aguilar S, Nguyen M, Vo M, Khanna A, Sasaki E, Liu HW, Tsai SC. Structural and functional analysis of two di-domain aromatase/cyclases from type II polyketide synthases. Proc Natl Acad Sci U S A. 2015;112:E6844–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hopwood DA. Genetic Contributions to Understanding Polyketide Synthases. Chem Rev. 1997;97:2465–2498. [DOI] [PubMed] [Google Scholar]
- 9.Das A, Khosla C. Biosynthesis of aromatic polyketides in bacteria. Acc Chem Res. 2009;42:631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou H, Li Y, Tang Y. Cyclization of aromatic polyketides from bacteria and fungi. Nat Prod Rep. 2010;27:839–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Y, Lee TS, Khosla C. Engineered biosynthesis of regioselectively modified aromatic polyketides using bimodular polyketide synthases. PLoS Biol. 2004;2:E31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. [DOI] [PubMed] [Google Scholar]
- 13.Rossi P, Swapna GV, Huang YJ, Aramini JM, Anklin C, Conover K, Hamilton K, Xiao R, Acton TB, Ertekin A, Everett JK, Montelione GT. A microscale protein NMR sample screening pipeline. J Biomol NMR. 2010;46:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang YJ, Powers R, Montelione GT. Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics. J Am Chem Soc. 2005;127:1665–1674. [DOI] [PubMed] [Google Scholar]
- 16.Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. [DOI] [PubMed] [Google Scholar]
- 17.Tian W, Chen C, Lei X, Zhao J, Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46:W363–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holm L, Laakso LM. Dali server update. Nucleic Acids Res. 2016;44:W351–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hao Q, Yin P, Li W, Wang L, Yan C, Lin Z, Wu JZ, Wang J, Yan SF, Yan N. The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins. Mol Cell. 2011;42:662–672. [DOI] [PubMed] [Google Scholar]
- 20.Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, Miyauchi Y, Takahashi M, Zhi Y, Fujita Y, Yoshida T, Kodaira KS, Yamaguchi-Shinozaki K, Tanokura M. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. [DOI] [PubMed] [Google Scholar]
- 21.Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Marquez JA. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009;462:665–668. [DOI] [PubMed] [Google Scholar]
- 22.Michalska K, Fernandes H, Sikorski M, Jaskolski M. Crystal structure of Hyp-1, a St. John’s wort protein implicated in the biosynthesis of hypericin. J Struct Biol. 2010;169:161–171. [DOI] [PubMed] [Google Scholar]
- 23.Lascu I, Gonin P. The catalytic mechanism of nucleoside diphosphate kinases. J Bioenerg Biomembr. 2000;32:237–246. [DOI] [PubMed] [Google Scholar]
- 24.Fukai S, Nureki O, Sekine S, Shimada A, Tao J, Vassylyev DG, Yokoyama S. Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNA(Val) and valyl-tRNA synthetase. Cell. 2000;103:793–803. [DOI] [PubMed] [Google Scholar]
- 25.Shane B Folylpolyglutamate synthesis and role in the regulation of one-carbon metabolism. Vitam Horm. 1989;45:263–335. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.