Abstract
The interaction between legumes and bacteria of rhizobia type results in a beneficial symbiotic relationship characterized by the formation of new root organs, called nodules. Within these nodules the bacteria, released in plant cells, differentiate into bacteroids and fix atmospheric nitrogen through the nitrogenase activity. This mutualistic interaction has evolved sophisticated signaling networks to allow rhizobia entry, colonization, bacteroid differentiation and persistence in nodules. Nodule cysteine rich (NCR) peptides, reactive oxygen species (ROS), reactive nitrogen species (RNS), and toxin–antitoxin (TA) modules produced by the host plants or bacterial microsymbionts have a major role in the control of the symbiotic interaction. These molecules described as weapons in pathogenic interactions have evolved to participate to the intracellular bacteroid accommodation by escaping control of plant innate immunity and adapt the functioning of the nitrogen-fixation to environmental signalling cues.
Keywords: legumes, symbiosis, bacteroid, reactive oxygen species, nitric oxide, nitrogen- fixation, nodule-specific cysteine rich peptides, toxin–antitoxin
Introduction
The nitrogen-fixing symbiosis (NFS) results from the relationship between plants of the legume family and soil bacteria referred to as rhizobia. After a recognition step, bacteria infect legume roots, induce the formation of specialized root organs, the nodules, and colonize nodule cells by endocytosis to form structures called symbiosomes (Ferguson et al., 2010). Inside symbiosomes, bacteria differentiate into bacteroids that can convert atmospheric dinitrogen (N2) into ammonia (NH4 +), via the nitrogenase activity. NH4 + is then transferred to the whole plant through either amino acids or ureide compounds (Oldroyd and Downie, 2008; Masson-Boivin et al., 2009). NFS provides substantial agronomic and environmental benefits such as the substitution to nitrogen (N) fertilizer inputs to increase the plant yields (Vitousek et al., 2013).
The setting of NFS depends on a signal exchange. An initial plant defense response is observed during the first hours of the interaction with the rhizobium, and then is actively suppressed after the recognition. How rhizobia are recognized as symbionts rather than pathogens by the host plant is well described (Jones et al., 2007; Soto et al., 2009), and the strategies of the plants to adjust their own defense systems to enable rhizobia entry, colonization, and differentiation are detailed in several reviews (Oldroyd et al., 2011; Oldroyd, 2013). Recent reports support the hypothesis that the regulation of immune response does not end at the recognition stage, but rather continue to allow rhizobial long-term accommodation inside the plant cells (Cao et al., 2017; Zipfel and Oldroyd, 2017; Berrabah et al., 2018; Yu et al., 2019). Multiple compounds such as nodule-specific cysteine rich (NCR) peptides, reactive oxygen species (ROS), reactive nitrogen species (RNS) and toxin–antitoxin (TA) modules have been shown to control the setup and the functioning of the interaction between the two partners The purpose of the present review is to provide an overview of the role of these compounds, described as weapons in pathogenic interactions, in the intracellular bacteroid accommodation (rhizobial colonization, differentiation, and control of plant innate immunity for persistence) and the adjustment of the nitrogen-fixation activity to environmental signalling cues.
Reactive Oxygen Species (ROS) in Bacterial Colonization of the Plant Cell and Bacteroid Persistence in the Nodule
ROS are involved in adaptation to environmental perturbations (Apel and Hirt, 2004; Waszczak et al., 2018). They are also essential for promoting normal cellular processes in bacteria and plants (Mittler, 2017). The level of ROS in cells depends on the tight regulation of a complex array of ROS generating systems and detoxification mechanisms, and antioxidant metabolites like glutathione and ascorbate. The balance between ROS production and detoxification regulates the cellular redox homeostasis in plants as well as in bacteria (Apel and Hirt, 2004).
In plants, the respiratory burst oxidase homologs (RBOH) proteins (also called NADPH oxidase) emerged as the major sources of apoplastic ROS ( Figure 1 ) and key players in the redox signaling during pathogen infection and other processes (Kadota et al., 2015; Liu and He, 2016; Montiel et al., 2016). Some members of this multigenic family are differentially expressed in Medicago truncatula nodule tissues and play different roles from the establishment of the symbiotic interaction to the functioning of mature nodule (Marino et al., 2011; Montiel et al., 2018). The reduction in the N2-fixation capacity in transgenic roots knocked-down for MtRbohA was the first evidence of a RBOH involvement in nodule functioning (Marino et al., 2011)( Table 1 ). Authors suggested that MtRBOHA activity contributes to the communication between the plant and the microsymbiont. Hydrogen peroxide production was visualized in M. truncatula infection zone and regulates genes involved in the nodulation process (Andrio et al., 2013). Similar results were obtain in Phaseolus vulgaris using knocked-down PvRbohA gene (Arthikala et al., 2017). Moreover, the roots overexpression of PvRbohB increases the number of bacteroids in the symbiosomes and improves biological N2-fixation in P. vulgaris (Arthikala et al., 2014). In contrast, mutations of NAD1 gene (Nodules with Activated Defence) in M. truncatula activate a strong defence response after rhizobia are released from infection threads into plant cells, leading to necrotic cell death of symbiotic cells (Wang et al., 2016). The knock-out of either MtRbohB, MtRbohC, or MtRbohD in the nad1 mutant reverts this cell death phenotype indicating that nodule innate immunity is notably mediated by RBOH activity (Yu et al., 2018; Yu et al., 2019). These data provide evidences that MtRBOH-mediated ROS production has positive and negative functions in the reception of the microsymbionts in the nodule cells.
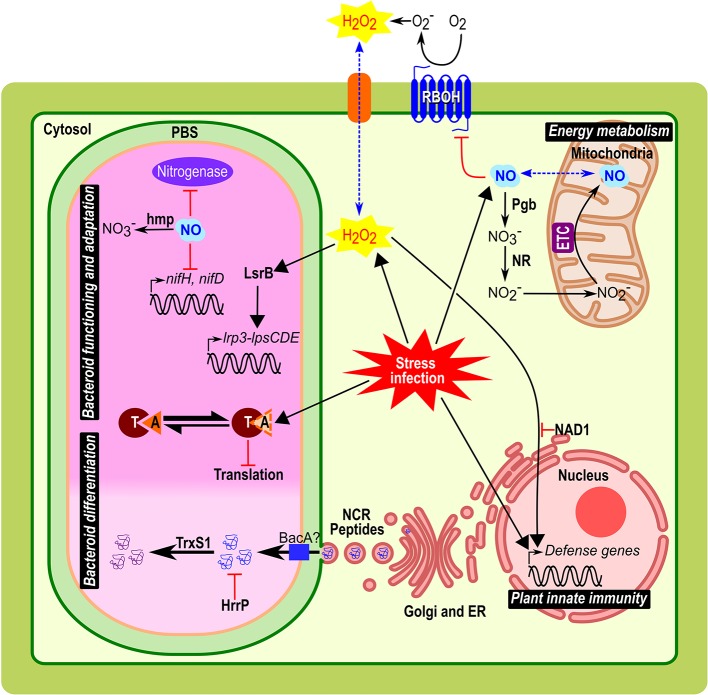
Figure 1.
Implication and connection of ROS, NO, NCR peptides, and TA modules in symbiosomes from Medicago root nodules. Biological role of these compounds during bacteroid differentiation, nodule functioning and adaptation, plant innate immunity, and energy metabolism are represented. Plant host cells infected by bacteria/bacteroid, implied various stress responses such as oxidative/nitrosative stress, acidic pH, microoxia, and exposure to NCRs. In the symbiosome, the clear part corresponds to the infection zone and the dark pink to the fixation zone with bacteria differentiated in bacteroid. Black arrows indicate metabolism reaction or downstream signal transduction pathways; red arrows indicate regulation mechanism (activation with arrowhead or repression with bar-headed lines). Blue dotted arrow indicates a diffusion through the membrane. Abbreviations: PBS, peribacteroid space; NR, nitrate reductase; Pgb, Phytoglobin; RBOH, respiratory burst oxidase homologs; O2 −, superoxide radical; H2O2, hydrogen peroxide; ETC, electron transfer chain; NO, nitric oxide; ER; endoplasmic reticulum; NCR peptides, nodule-specific cysteine-rich peptides; Hmp, flavohemoglobin; NAD1, Nodules with Activated Defence 1; TrxS1, Thioredoxine S1; HrrP, Host-range restriction peptidase; LsrB, LysR transcription factor; T, toxin; A, antitoxin.
Table 1.
Non-exhaustive summary of genes involved in ROS, NCR, NO, and TA modules pathways within legume nodule cells.
| Mutant/transgenic line | Origin | Proteic activity | Symbiotic function | Reference | ||
|---|---|---|---|---|---|---|
| ROS | nad1 | M. truncatula | Nodule activated defense protein -uncharacterized | Nodule innate immunity, Bacteroid differenciation/survival, N2-fixation | Wang et al. (2016) | |
| RbohA: RNAi | N2-fixation | Marino et al. (2011) | ||||
| RbohA: RNAi | P. vulgaris | Respiratory burst oxidase homolog - ROS production | Bacterial infection, Nodule formation, Bacteroid survival, N2-fixation | Arthikala et al. (2017) | ||
| RbohB: RNAi | Bacterial infection, Nodule formation, N2-fixation | Montiel et al. (2012) | ||||
| RbohB: OE | Bacterial infection, Nodule formation, Bacteroid differenciation, N2-fixation | Arthikala et al. (2014) | ||||
| gshB | S. meliloti | ROS detoxification enzymes | Nodule formation, N2-fixation | Harrison et al. (2005) | ||
| trxL | N2-fixation | Castro-Sowinski et al. (2007) | ||||
| grx1 | N2-fixation | Benyamina et al. (2013) | ||||
| grx2 | Nodule formation, N2-fixation | |||||
| sodA | N2-fixation, Bacteroid differenciation | Santos et al. (2000) | ||||
| katA/katC; katB/katC | Nodule formation, Infection, N2-fixation, Bacteroid differenciation | Jamet et al. (2003) | ||||
| lsrB | LysR transcription factor | Infection, Bacteroid differenciation/survival, N2-fixation | Luo et al. (2005); Tang et al. (2013); Tang et al., (2014); Tang et al., (2017) | |||
| NCR | dnf7-2 deletion mutant | M. truncatula | Antimicrobial peptide NCR169 | Bacteroid survival/persistence | Horváth et al. (2015) | |
| dnf4 deletion mutant | M. truncatula | Antimicrobial peptide NCR211- Symbiont Specificity | Bacteroid survival/persistence | Kim et al. (2015) | ||
| NFS1-/- (NCRα-β) | M. truncatula | Antimicrobial peptide - Symbiont Specificity | Bacteroid survival, Senescence | Yang et al. (2017) | ||
| Trx s1: RNAi | M. truncatula | Thioredoxin-NCR reduction | Bacteroid differenciation | Ribeiro et al. (2017) | ||
| Trx s1: OE | ||||||
| bacA | S. meliloti | ABC transporter- Symbiont protection against NCRs | Bacteroid differenciation | Haag et al. (2011) | ||
| bclA | B. japonicum | Bacteroid differenciation/survival | Guefrachi et al. (2015) | |||
| hrrP | S. meliloti | M16A family metallopeptidase- Escape NCR control | Bacteroid fitness | Price et al. (2015) | ||
| NO | Hb1: RNAi | L. japonicus | Leghemoglobin- degradation of nitric oxide | N2-fixation | Ott et al. (2005) | |
| Hb1: OE | Phytoglobin- degradation of nitric oxide | N2-fixation | Shimoda et al. (2009) | |||
| Hb1: OE | A. firma | |||||
| hmp | S. meliloti | Flavohemoprotein- NO degradation | Bacteroid survival, N2-fixation, Senesence | Cam et al. (2012); Meilhoc et al. (2013); Blanquet et al. (2015) | ||
| hmp++ | ||||||
| norB | Blanquet et al. (2015) | |||||
| nnrS1 | ||||||
| TA modules | vapC-4 (ntrR) | S. meliloti | VapB (antidote), VapC (site-specific RNase) | Nodule formation | Dusha et al. (1989) | |
| N2-fixation, Senescence | Oláh et al. (2001) | |||||
| vapB-5 | Nodule formation, Bacteroid differenciation | Lipuma et al. (2014) | ||||
| vapC-5 | N2-fixation, Bacteroid survival, Senescence | |||||
| bat/bto = vapBC | B. japonicum | Nodule formation, N2-fixation | Miclea et al. (2010) | |||
Genes studied have a rhizobial (orange) or a plant (light green) origin. The nitrogen-fixing phenotype of the mutant or transgenic line is depicted in green if defective or in pink if improved in the column symbiotic function.
To cope with the plant ROS production, the microsymbiont contains a number of antioxidants and ROS-scavenging enzymes to preserve the bacteroids against ROS damages (Puppo et al., 2005; Becana et al., 2010). Analysis of bacterial mutants deficient in glutathione synthetase (gshB), thioredoxin (trxL), glutaredoxins (grx1, or grx2), superoxide dismutase (sodA), and catalases (double mutants katA/katC or katB/katC) showed that the alteration in antioxidant pools as well as the mutation of ROS detoxification enzymes impact the formation of nodules, decrease the N2-fixing capacity and induce a premature nodule senescence ( Table 1 )(Santos et al., 2000; Jamet et al., 2003; Harrison et al., 2005; Castro-Sowinski et al., 2007; Benyamina et al., 2013). Besides, nodules induced by a Sinorhizobium meliloti deletion mutant of lsrB, which encodes a LysR transcription factor acting as a ROS regulator, showed premature senescence with impaired bacteroid differentiation (Luo et al., 2005; Tang et al., 2013). LsrB was found to induce the expression of the lrp3-lpsCDE operon involved in lipopolysaccharide biosynthesis required for infection or bacteroid survival in host cells ( Figure 1 )(Tang et al., 2014) and that of γ-glutamylcysteine synthetase, involved in glutathione synthesis (Tang et al., 2017).
Nodule-Specific Cysteine Rich (NCR) Peptides and Terminal Bacteroid Differentiation
NCR peptides have been specifically found in the Inverted Repeat-Lacking Clade (IRLC) legumes such as Medicago spp., and in Dalbergoid legumes such as Aeschynomene spp., where bacteria are terminally differentiated to polyploïd non-dividing bacteroids (Mergaert et al., 2003; Mergaert et al., 2006; Alunni and Gourion, 2016). They encode highly divergent peptides, which resemble defensin-type antimicrobial peptides involved in plant and animal innate immunity (Mergaert et al., 2003). Indeed, some NCR peptides have a strong in vitro antimicrobial activity when applied to free-living bacteria (Van de Velde et al., 2010; Maróti and Kondorosi, 2014; Farkas et al., 2017).
Almost all NCR genes are exclusively expressed in the infected cells of nodules and their products are targeted to the symbiosome through the endoplasmic reticulum secretory system ( Figure 1 )(Wang et al., 2010; Guefrachi et al., 2014). Challenge of cultured bacteria with synthetic NCR peptides and ectopic expression of NCR peptides in legumes devoid of NCR genes cause features of bacteroid differentiation, demonstrating that these NCR peptides are sufficient to induce the irreversible differentiation (Van de Velde et al., 2010). The number of NCR genes is remarkably variable (from 7 in Glycyrrhiza uralensis to over 700 members in M. truncatula), and a positive correlation was found between the size of the NCR peptide family in the plant genome and the degree of bacteroid elongation (Montiel et al., 2017). Despite the large size of NCR peptide family in M. truncatula suggesting an extensive redundancy, NCR169, NCR211, and NFS1 are essential and the corresponding plant mutants are unable to establish a functional NFS ( Table 1 ) (Horváth et al., 2015; Kim et al., 2015; Yang et al., 2017). Both NFS1 and NCR211 exemplify NCR peptides that control the survival of fully differentiated bacteroids instead of triggering the terminal differentiation of bacteroids. NFS1 controls the discrimination mechanisms against incompatible microsymbionts (Yang et al., 2017), provoking bacterial cell death and early nodule senescence in an allele-specific and rhizobial strain-specific manner, while NCR211 is required for bacteroid persistence inside symbiotic cells (Kim et al., 2015).
To survive exposure to toxic NCR peptides S. meliloti requires the integrity of the BacA ABC-transporter. A bacA mutant strain is unable to differentiate and rapidly dies after its release from infection threads ( Figure 1 ) (Haag et al., 2011). Similarly, the BacA-like peptide transporter BclA of Bradyrhizobium japonicum is essential for bacteroid differentiation and survival in Aeschynomene nodule, which suggests that the NCR peptides uptake may be a common mechanism used by different rhizobia to counteract the toxic effect of peptides (Guefrachi et al., 2015). In the symbiosis between Sinorhizobium freedi and G. uralensis alternatively, bacteroid differentiation occurs via a bacA-independent pathway and is rather associated with LPS modification of the bacteroid outer membrane (Crespo-Rivas et al., 2016). Additionally, a S. meliloti natural strain can escape the control of NCR peptides and proliferate in nodules using the plasmid encoded host-range restriction peptidase Hrrp, which is able to digest NCR peptides in vitro ( Figure 1 and Table 1 )(Price et al., 2015). The expression of hrrp increases the fitness of rhizobial strains while inhibiting N2-fixation in some plant ecotypes (Price et al., 2015).
Another layer of regulation may come from posttranslational modifications of NCR peptides (Marx et al., 2016). In particular, a nodule-specific thioredoxin, TrxS1, capable to reduce NCR peptides and targeted to symbiosomes, has been shown to be required for bacteroid differentiation, suggesting that NCR redox state is important in planta ( Figure 1 and Table 1 )(Ribeiro et al., 2017). In this context, the redox control of the bacteroid differentiation probably occurs through the NCR peptide activity suggesting a crosstalk between the different regulators described in this review.
Considered together, these data indicate that the symbiosis efficiency of terminally differentiated bacteria is the outcome of a tight balance between the effects of NCR peptides and the ability of rhizobia to resist them. The rupture of this balance can lead to the activation of the plant innate immunity (Yu et al., 2019).
Nitric Oxide (NO) in Functional Nodules: Microoxia, Energetic Metabolism
NO production was observed in functional nodules of Lotus japonicus and M. truncatula, mainly in the N2-fixation zone (Baudouin et al., 2006; Shimoda et al., 2009), and in the nodule senescence zone (Cam et al., 2012; Fukudome et al., 2018). Although the origin and the biological significance of NO production in nodules has been thoroughly analyzed over the last few years (Boscari et al., 2013; Hichri et al., 2015; Berger et al., 2019), there are still many questions to be clarified concerning the relative importance of the signaling/metabolic functions of NO versus its toxic action on host plant and symbiont.
Functional nodules are characterized by a microoxic environment to protect the bacterial nitrogenase from irreversible denaturation by oxygen (O2) which requires the setup of an O2 barrier in the outer cell layers of the nodule and the synthesis of leghemoglobin (Lb) (Appleby, 1992). In plant roots submitted to hypoxia, a “Phytoglobin-NO respiration” has been shown to use nitrite as a final electron acceptor instead of O2 to be reduced to NO by the mitochondrial electron transfer chain (ETC), which allows cell energy status retention ( Figure 1 ) (Igamberdiev and Hill, 2009; Gupta and Igamberdiev, 2011). Accumulated data support the existence of such a Phytoglobin-NO respiration in M. truncatula and Medicago sativa nodules, in which both nitrate reductase (NR) and ETC are involved in NO production and in the maintenance of the nodule energy state (Horchani et al., 2011; Berger et al., 2018).
Despite its role in acclimation to microoxic environment, NO is also a potent inhibitor of nitrogenase activity (Trinchant and Rigaud, 1982; Sasakura et al., 2006; Kato et al., 2010). In nodules of soybean plants subjected to flooding, the increase in NO production is associated with the repression of bacterial nifH and nifD ( Figure 1 ), and this inhibition is partially reversed by the application of the NO scavenger cPTIO, which illustrates the inhibitory role of NO on the expression of nitrogenase genes (Sánchez et al., 2010). Furthermore, using both pharmacological approach, with NO-donors and scavengers, and molecular approach with transgenic plants with modified NO levels, several studies report that NO inhibits in vivo N2-fixing activity in soybean, L. japonicus, and M. truncatula nodules ( Table 1 )(Shimoda et al., 2009; Kato et al., 2010; Cam et al., 2012).
The biological activity of NO is mediated through redox-dependent protein modifications such as metal-nitrosylation, S-nitrosation and Tyr-nitration (Stamler et al., 2001; Besson-Bard et al., 2008). In M. truncatula mature nodules, 80 proteins have been reported to be S-nitrosated, most of them involved in primary metabolism, energy regeneration and nitrogen assimilation (Puppo et al., 2013). In this context, M. truncatula glutathione peroxidase 1 and glutamine synthetase 1a were shown to be regulated by NO through S-nitrosation and Tyr-nitration modifications (Melo et al., 2011; Castella et al., 2017).
Beside the nodule metabolism regulation, a participation of NO to the life-time of the symbiotic interaction was also observed (Cam et al., 2012). Increased NO level in nodule obtained either by using S. meliloti mutant strains deficient in the degradation of NO (hmp, norB, nnrS1)( Table 1 ), or by treating nodules with NO donors (Cam et al., 2012; Meilhoc et al., 2013; Blanquet et al., 2015) leads to premature nodule senescence. Conversely, by using S. meliloti mutant strains that over-expressed hmp, a decrease in NO level was observed correlated to a delay of nodule senescence ( Table 1 )(Cam et al., 2012). Therefore, NO concentration should be tightly controlled, in time and space, in both partners to avoid its toxic effects and to fulfil its signaling and metabolic functions during nodule functioning and under environmental stresses (Berger et al., 2019).
Toxin-Antitoxin (TA) Systems in Bacteroid Adaptation in Infected Plant Cells
TA systems are key players of intracellular survival of invading bacteria during eukaryote interactions (Lobato-Márquez et al., 2016). TA genes encode a stable toxin and its cognate antitoxin. Depending on the antitoxin nature (RNA or protein) and its mode of action, TA modules are classified into six different types (I–VI). The type II, where both toxin and antitoxin are small proteins forming a stable complex, is the most abundant type in pathogens, particularly exposed to diverse micro-environments during host interaction (Ramage et al., 2009; Georgiades and Raoult, 2011). Due to the self-poisoning effect of the toxin, TA modules could be considered as intracellular molecular timebombs. TA expression is tightly regulated to allow either growth arrest and bacterial adaptation or cell death (Hayes and Kędzierska, 2014). Under various stress conditions, the antitoxin is degraded by bacterial proteases leading to the deregulation of the TA operon and delivery of the toxin which targets specific cellular functions (DNA replication, translation) (Gerdes et al., 2005). In phytopathogenic bacteria, TA have been recently demonstrated as involved in virulence and biofilm formation during plant infection (Shidore and Triplett, 2017; Martins et al., 2018).
Among the 29 chromosomal type II TA systems of S. meliloti, eleven belong to the VapBC family; VapB being the antitoxin and VapC the toxin, acting as a site-specific RNase ( Table 1 ). The importance of two vapBC operons, vapBC-4 (ntrPR) and vapBC-5, has been shown in S. meliloti during symbiotic interaction with Medicago sp. (Dusha et al., 1989; Lipuma et al., 2014). NtrPR was identified on the capacity of the toxin ntrR mutant (for nitrogen regulator) to form more nodules on alfalfa roots in the presence of exogenous ammonium (Dusha et al., 1989). This suggests that NtrR toxin is involved in the nodulation efficiency depending on the level of nitrogen supply. This module plays also a role in mature nodules in a nitrogen-tolerant manner, as ntrR-induced nodules have an enhanced N2-fixation capacity and an increased plant yield (Oláh et al., 2001). Regarding VapBC-5 module, the vapC-5 toxin mutant improves the symbiotic interaction with alfalfa (increase in N2-fixation capacity and plant yield) associated to a delay in nodule senescence (Lipuma et al., 2014).
These vapC mutants have no free-living phenotypes. Therefore, TA modules might play a role in the bacterial adaptation to infection stresses (metabolic shifting, acidic pH, microoxia, ROS, antimicrobial peptides, stresses known to activate pathogen TA modules (Lobato-Márquez et al., 2016)) ( Figure 1 ). Thus, in a wild-type context, NtrPR and VapBC-5 modules likely limit the symbiotic interaction upon specific plant signals and/or contribute to the nodule senescence onset. The high number of TA systems in S. meliloti genome could be due to functional redundancy or to different roles independent of the NFS. Indeed, Milunovic et al. (2014) showed that the deletion of four TA operons from the pSyma and pSymb plasmids induces a cell toxicity phenotype in free living, with no symbiotic effect during alfalfa interaction (Milunovic et al., 2014). In contrast, in B. japonicum USDA110, the complete deletion of the bat/bto TA resulted in a limited production of soybean nodules associated to a reduced plant yield (Miclea et al., 2010). Such a phenotype suggests that this system might play a positive role on the symbiotic interaction with soybean, although this could also be linked to the pleiotropic effects observed for this deletion mutant in free-living conditions.
Concluding Remarks
The evidences presented in this review show the importance of ROS, NO, NCR peptides, and TA modules in the intracellular bacteroid accommodation and the N2-fixation activity regulation. These molecules, considered in certain situations as cellular weapons, are necessary not only in the nodule functioning, but also in the rupture of the symbiosis under unfavourable conditions such as deficient bacterial symbionts, adverse environmental conditions or cellular aging. The importance of these regulatory elements is now clearly demonstrated, but their mode of action still remains to be fully deciphered. Identification of the molecular pathways involved in the regulation of the bacterial intracellular life during NFS will be helpful to dissect the crosstalk between these different regulatory elements. Evidences exist of the connection between ROS, NO, and NCR in plant cells to balance the plant immune response, to regulate the rhizobial differentiation and control the switch from bacteroid persistence to cell death. Among these recent findings it can be noted the involvement of three RBOH in the activation of immunity in Medicago nodules and the regulation of bacteroid differentiation via TrxS1-dependent redox regulation of some NCRs in planta (Ribeiro et al., 2017)( Figure 1 ). Furthermore, it was previously shown that NO could inhibit NADPH oxidase activity by post-translational modification ( Figure 1 )(Yun et al., 2011).
Similarly, the connection between TA, NO, ROS, and NCR produced by both partners represents a field of future interest to identify the signals involved in TA activation. The delayed senescence of nodules induced by the vapC-5 toxin mutants conducted to higher the expression of NCR001 gene compared to control Rhizobium strain (Lipuma et al., 2014). Finally, a better understanding of these regulatory processes may give promising strategies to improve the NFS and reduce the use of fertilizers.
Author Contributions
CS and AB conceived the idea of the review. All the authors were involved in the manuscript writing.
Funding
This work was supported by the “Institut National de la Recherche Agronomique,” the “Centre National de la Recherche Scientifique,” the University of Nice-Sophia-Antipolis, and the French Government (National Research Agency, ANR) through the LABEX SIGNALIFE program (reference # ANR-11-LABX-0028-01) and the STAYPINK project (ANR-15-CE20-0005).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Alunni B., Gourion B. (2016). Terminal bacteroid differentiation in the legume–rhizobium symbiosis: nodule-specific cysteine-rich peptides and beyond. New Phytol. 211, 411–417. 10.1111/nph.14025 [DOI] [PubMed] [Google Scholar]
- Andrio E., Marino D., Marmeys A., de Segonzac M. D., Damiani I., Genre A. (2013). Hydrogen peroxide-regulated genes in the Medicago truncatula–Sinorhizobium meliloti symbiosis. New Phytol. 198, 179–189. 10.1111/nph.12120 [DOI] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Appleby C. A. (1992). The origin and functions of haemoglobin in plants. Sci. Prog. 76, 365–398. 10.2307/43421309 [DOI] [Google Scholar]
- Arthikala M.-K., Montiel J., Sánchez-López R., Nava N., Cárdenas L., Quinto C. (2017). Respiratory burst oxidase homolog gene a is crucial for rhizobium infection and nodule maturation and function in common bean. Front. Plant Sci. 8, 2003. 10.3389/fpls.2017.02003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthikala M.-K., Sánchez-López R., Nava N., Santana O., Cárdenas L., Quinto C. (2014). RbohB, a Phaseolus vulgaris NADPH oxidase gene, enhances symbiosome number, bacteroid size, and nitrogen fixation in nodules and impairs mycorrhizal colonization. New Phytol. 202, 886–900. 10.1111/nph.12714 [DOI] [PubMed] [Google Scholar]
- Bao H., Zhang Z., Cao Y. (2019). Immune Signal. Pathw. Dur. Termin. bacteroid Differ. nodules. Trends Plant Sci. 24, 299–302. 10.1016/j.tplants.2019.01.010 [DOI] [PubMed] [Google Scholar]
- Baudouin E., Pieuchot L., Engler G., Pauly N., Puppo A. (2006). Nitric oxide is formed in Medicago truncatula-Sinorhizobium meliloti functional nodules. Mol. Plant Microbe Interact. 19, 970–975. 10.1094/MPMI-19-0970 [DOI] [PubMed] [Google Scholar]
- Becana M., Matamoros M. A., Udvardi M., Dalton D. A. (2010). Recent insights into antioxidant defenses of legume root nodules. New Phytol. 188, 960–976. 10.1111/j.1469-8137.2010.03512.x [DOI] [PubMed] [Google Scholar]
- Benyamina S. M., Baldacci-Cresp F., Couturier J., Chibani K., Hopkins J., Bekki A. (2013). Two Sinorhizobium meliloti glutaredoxins regulate iron metabolism and symbiotic bacteroid differentiation. Environ. Microbiol. 15, 795–810. 10.1111/j.1462-2920.2012.02835.x [DOI] [PubMed] [Google Scholar]
- Berger A., Boscari A., Frendo P., Brouquisse R. (2019). Nitric oxide signaling, metabolism and toxicity in nitrogen-fixing symbiosis. J. Exp. Bot. 70, 4505–4520. 10.1093/jxb/erz159 [DOI] [PubMed] [Google Scholar]
- Berger A., Brouquisse R., Pathak P. K., Hichri I., Bhatia S., Boscari A. (2018). Pathways of nitric oxide metabolism and operation of phytoglobins in legume nodules: missing links and future directions. Plant Cell Environ. 41, 2057–2068. 10.1111/pce.13151 [DOI] [PubMed] [Google Scholar]
- Berrabah F., Ratet P., Gourion B. (2018). Legume nodules: massive infection in the absence of defense induction. Mol. Plant-Microbe Interact. 32, 35–44. 10.1094/MPMI-07-18-0205-FI [DOI] [PubMed] [Google Scholar]
- Besson-Bard A., Pugin A., Wendehenne D. (2008). New insights into nitric oxide signaling in plants. Annu. Rev. Plant Biol. 59, 21–39. 10.1146/annurev.arplant.59.032607.092830 [DOI] [PubMed] [Google Scholar]
- Blanquet P., Silva L., Catrice O., Bruand C., Carvalho H., Meilhoc E. (2015). Sinorhizobium meliloti controls nitric oxide–mediated post-translational modification of a medicago truncatula nodule protein. Mol. Plant-Microbe Interact. 28, 1353–1363. 10.1094/MPMI-05-15-0118-R [DOI] [PubMed] [Google Scholar]
- Boscari A., Meilhoc E., Castella C., Bruand C., Puppo A., Brouquisse R. (2013). Which role for nitric oxide in symbiotic N2-fixing nodules: toxic by-product or useful signaling/metabolic intermediate?. Front. Plant Sci. 4, 1–6. 10.3389/fpls.2013.00384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam Y., Pierre O., Boncompagni E., Hérouart D., Meilhoc E., Bruand C. (2012). Nitric oxide (NO): a key player in the senescence of Medicago truncatula root nodules. New Phytol. 196, 548–560. 10.1111/j.1469-8137.2012.04282.x [DOI] [PubMed] [Google Scholar]
- Cao Y., Halane M. K., Gassmann W., Stacey G. (2017). The role of plant innate immunity in the legume-rhizobium symbiosis. Annu. Rev. Plant Biol. 68, 535–561. .org/ 10.1146/annurev-arplant-042916-041030 [DOI] [PubMed] [Google Scholar]
- Castella C., Mirtziou I., Seassau A., Boscari A., Montrichard F., Papadopoulou K. (2017). Post-translational modifications of Medicago truncatula glutathione peroxidase 1 induced by nitric oxide. Nitric. Oxide-Biol. Chem. 68, 125–136. 10.1016/j.niox.2017.02.004 [DOI] [PubMed] [Google Scholar]
- Castro-Sowinski S., Matan O., Bonafede P., Okon Y. (2007). A thioredoxin of Sinorhizobium meliloti CE52G is required for melanin production and symbiotic nitrogen fixation. Mol. Plant-Microbe Interact. 20, 986–993. 10.1094/MPMI-20-8-0986 [DOI] [PubMed] [Google Scholar]
- Crespo-Rivas J. C., Guefrachi I., Mok K. C., Villaécija-Aguilar J. A., Acosta-Jurado S., Pierre O. (2016). Sinorhizobium fredii HH103 bacteroids are not terminally differentiated and show altered O-antigen in nodules of the inverted repeat-lacking clade legume glycyrrhiza uralensis. Environ. Microbiol. 18, 2392–2404. 10.1111/1462-2920.13101 [DOI] [PubMed] [Google Scholar]
- Dusha I., Bakos A., Kondorosi A., de Bruijn F. J., Schell J. (1989). The Rhizobium meliloti early nodulation genes (nodABC) are nitrogen-regulated: isolation of a mutant strain with efficient nodulation capacity on alfalfa in the presence of ammonium. Mol. Gen. Genet. 219, 89–96. .org/ 10.1007/BF00261162 [Google Scholar]
- Farkas A., Maróti G., Kereszt A., Kondorosi É. (2017). Comparative analysis of the bacterial membrane disruption effect of two natural plant antimicrobial peptides. Front. Microbiol. 8, 51. .org/ 10,3389/fmicb. 2017.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B. J., Indrasumunar A., Hayashi S., Lin M.-H., Lin Y.-H., Reid D. E. (2010). Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant Biol. 52, 61–76. 10.1111/j.1744-7909.2010.00899.x [DOI] [PubMed] [Google Scholar]
- Fukudome M., Watanabe E., Osuki K.-I., Imaizumi R., Aoki T., Becana M., et al. (2018). Stably-transformed lotus japonicus plants overexpressing phytoglobin LjGlb1-1 show decreased nitric oxide levels in roots and nodules as well as delayed nodule senescence stably-transformed lotus japonicus plants overexpressing phytoglobin LjGlb1-1 show D. 60, 1–21. [DOI] [PubMed]
- Georgiades K., Raoult D. (2011). Genomes of the most dangerous epidemic bacteria have a virulence repertoire characterized by fewer genes but more toxin-antitoxin modules. PloS One 6, e17962–e17962. 10.1371/journal.pone.0017962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Christensen S. K., Løbner-Olesen A. (2005). Prokaryotic toxin–antitoxin stress response loci. Nat. Rev. Microbiol. 3, 371–382. 10.1038/nrmicro1147 [DOI] [PubMed] [Google Scholar]
- Guefrachi I., Nagymihaly M., Pislariu C. I., de Velde W., Ratet P., Mars M. (2014). Extreme specificity of NCR gene expression in Medicago truncatula. BMC Genomics 15, 712. 10.1186/1471-2164-15-712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guefrachi I., Pierre O., Timchenko T., Alunni B., Barrière Q., Czernic P. (2015). Bradyrhizobium BclA is a peptide transporter required for bacterial differentiation in symbiosis with Aeschynomene legumes. Mol. Plant-Microbe Interact. 28, 1155–1166. 10.1094/MPMI-04-15-0094-R [DOI] [PubMed] [Google Scholar]
- Gupta K. J., Igamberdiev A. U. (2011). The anoxic plant mitochondrion as a nitrite: NO reductase. Mitochondrion 11, 537–543. 10.1016/j.mito.2011.03.005 [DOI] [PubMed] [Google Scholar]
- Haag A. F., Baloban M., Sani M., Kerscher B., Pierre O., Farkas A. (2011). Protection of Sinorhizobium against host cysteine-rich antimicrobial peptides is critical for symbiosis. PloS Biol. 9, e1001169. 10.1371/journal.pbio.1001169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J., Jamet A., Muglia C. I., de Sype G., Aguilar O. M., Puppo A. (2005). Glutathione plays a fundamental role in growth and symbiotic capacity of Sinorhizobium meliloti. J. Bacteriol. 187, 168–174. 10.1128/JB.187.1.168-174.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F., Kędzierska B. (2014). Regulating toxin-antitoxin expression: controlled detonation of intracellular molecular timebombs. Toxins (Basel). 6, 337–358. 10.3390/toxins6010337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hichri I., Boscari A., Castella C., Rovere M., Puppo A., Brouquisse R. (2015). Nitric oxide: a multifaceted regulator of the nitrogen-fixing symbiosis. J. Exp. Bot. 66, 2877–2887. 10.1093/jxb/erv051 [DOI] [PubMed] [Google Scholar]
- Horchani F., Prévot M., Boscari A., Evangelisti E., Meilhoc E., Bruand C. (2011). Both plant and bacterial nitrate reductases contribute to nitric oxide production in Medicago truncatula nitrogen-fixing nodules. Plant Physiol. 155, 1023–1036. 10.1104/pp.110.166140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth B., Domonkos Á., Kereszt A., Szűcs A., Ábrahám E., Ayaydin F. (2015). Loss of the nodule-specific cysteine rich peptide, NCR169, abolishes symbiotic nitrogen fixation in the Medicago truncatula dnf7 mutant. Proc. Natl. Acad. Sci. 112, 15232–15237. 10.1073/pnas.1500777112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igamberdiev A. U., Hill R. D. (2009). Plant mitochondrial function during anaerobiosis. Ann. Bot. 103, 259–268. 10.1093/aob/mcn100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet A., Sigaud S., de Sype G., Puppo A., Hérouart D. (2003). Expression of the bacterial catalase genes during Sinorhizobium meliloti-Medicago sativa symbiosis and their crucial role during the infection process. Mol. Plant-Microbe Interact. 16, 217–225. 10.1094/MPMI.2003.16.3.217 [DOI] [PubMed] [Google Scholar]
- Jones K. M., Kobayashi H., Davies B. W., Taga M. E., Walker G. C. (2007). How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 5, 619–633. 10.1038/nrmicro1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y., Shirasu K., Zipfel C. (2015). Regulation of the NADPH oxidase rbohd during plant immunity. Plant Cell Physiol. 56, 1472–1480. 10.1093/pcp/pcv063 [DOI] [PubMed] [Google Scholar]
- Kato K., Kanahama K., Kanayama Y. (2010). Involvement of nitric oxide in the inhibition of nitrogenase activity by nitrate in Lotus root nodules. J. Plant Physiol. 167, 238–241. 10.1016/j.jplph.2009.08.006 [DOI] [PubMed] [Google Scholar]
- Kim M., Chen Y., Xi J., Waters C., Chen R., Wang D. (2015). An antimicrobial peptide essential for bacterial survival in the nitrogen-fixing symbiosis. Proc. Natl. Acad. Sci. U. S. A. 112, 15238–15243. 10.1073/pnas.1500123112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipuma J., Cinege G., Bodogai M., Oláh B., Kiers A., Endre G. (2014). A vapBC-type toxin–antitoxin module of Sinorhizobium meliloti influences symbiotic efficiency and nodule senescence of Medicago sativa. Environ. Microbiol. 16, 3714–3729. 10.1111/1462-2920.12608 [DOI] [PubMed] [Google Scholar]
- Liu Y., He C. (2016). Regulation of plant reactive oxygen species (ROS) in stress responses: learning from AtRBOHD. Plant Cell Rep. 35, 995–1007. 10.1007/s00299-016-1950-x [DOI] [PubMed] [Google Scholar]
- Lobato-Márquez D., Díaz-Orejas R., García-del Portillo F. (2016). Toxin-antitoxins and bacterial virulence. FEMS Microbiol. Rev. 40, 592–609. 10.1093/femsre/fuw022 [DOI] [PubMed] [Google Scholar]
- Luo L., Yao S.-Y., Becker A., Rüberg S., Yu G.-Q., Zhu J.-B. (2005). Two new Sinorhizobium meliloti LysR-type transcriptional regulators required for nodulation. J. Bacteriol. 187, 4562–4572. 10.1128/JB.187.13.4562-4572.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino D., Andrio E., Danchin E. G. J., Oger E., Gucciardo S., Lambert A. (2011). A Medicago truncatula NADPH oxidase is involved in symbiotic nodule functioning. New Phytol. 189, 580–592. 10.1111/j.1469-8137.2010.03509.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maróti G., Kondorosi E. (2014). Nitrogen-fixing Rhizobium-legume symbiosis: are polyploidy and host peptide-governed symbiont differentiation general principles of endosymbiosis?. Front. Microbiol. 5, 326. 10.3389/fmicb.2014.00326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins P. M. M., Merfa M. V., Takita M. A., De Souza A. A. (2018). Persistence in Phytopathogenic bacteria: do we know enough?. Front. Microbiol. 9, 1099. .org/ 10,3389/fmicb. 2018.01099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx H., Minogue C. E., Jayaraman D., Richards A. L., Kwiecien N. W., Siahpirani A. F. (2016). A proteomic atlas of the legume Medicago truncatula and its nitrogen-fixing endosymbiont Sinorhizobium meliloti. Nat. Biotechnol. 34, 1198. .org/ 10.1038/nbt.3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson-Boivin C., Giraud E., Perret X., Batut J. (2009). Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes?. Trends Microbiol. 17, 458–466. 10.1016/j.tim.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Meilhoc E., Blanquet P., Cam Y., Bruand C. (2013). Control of NO level in rhizobium-legume root nodules: not only a plant globin story. Plant Signal. Behav. 8, e25923. 10.4161/psb.25923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo P. M., Silva L. S., Ribeiro I., Seabra A. R., Carvalho H. G. (2011). Glutamine synthetase is a molecular target of nitric oxide in root nodules of Medicago truncatula and is regulated by tyrosine nitration. Plant Physiol. 157, 1505–1517. 10.1104/pp.111.186056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert P., Nikovics K., Kelemen Z., Maunoury N., Kondorosi A., Kondorosi E. (2003). A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol. 132, 161–173. 10.1104/pp.102.018192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert P., Uchiumi T., Alunni B., Evanno G., Cheron A., Catrice O. (2006). Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium–legume symbiosis. Proc. Natl. Acad. Sci. U. S. A. 103, 5230–5235. 10.1073/pnas.0600912103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miclea P. S., Péter M., Végh G., Cinege G., Kiss E., Váró G. (2010). Atypical transcriptional regulation and role of a new toxin-antitoxin-like module and its effect on the lipid composition of bradyrhizobium japonicum. Mol. Plant-Microbe Interact. 23, 638–650. 10.1094/MPMI-23-5-0638 [DOI] [PubMed] [Google Scholar]
- Milunovic B., George C., Morton R. A., Finan T. M., (2014). Cell growth inhibition upon deletion of four toxin-antitoxin loci from the megaplasmids of sinorhizobium meliloti. J. Bacteriol. 196, 811–824. 10.1128/JB.01104-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. (2017). ROS Are Good. Trends Plant Sci. 22, 11–19. 10.1016/j.tplants.2016.08.002 [DOI] [PubMed] [Google Scholar]
- Montiel J., Arthikala M. K., Cárdenas L., Quinto C. (2016). Legume NADPH oxidases have crucial roles at different stages of nodulation. Int. J. Mol. Sci. 17, 1–12. 10.3390/ijms17050680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel J., Downie J. A., Farkas A., Bihari P., Herczeg R., Bálint B. (2017). Morphotype of bacteroids in different legumes correlates with the number and type of symbiotic NCR peptides. Proc. Natl. Acad. Sci. 114, 5041LP–505046. 10.1073/pnas.1704217114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel J., Fonseca-García C., Quinto C. (2018). Phylogeny and Expression of NADPH Oxidases during Symbiotic Nodule Formation. Agric. 8, 179. 10.3390/agriculture8110179 [DOI] [Google Scholar]
- Montiel J., Nava N., Cárdenas L., Sánchez-López R., Arthikala M.-K., Santana O., et al. (2012). A Phaseolus vulgaris NADPH Oxidase Gene is Required for Root Infection by Rhizobia. Plant Cell Physiol. 53, 1751–1767. 10.1093/pcp/pcs120 [DOI] [PubMed] [Google Scholar]
- Oláh B., Kiss E., Györgypál Z., Borzi J., Cinege G., Csanádi G. (2001). Mutation in the ntrR gene, a member of the vap gene family, increases the symbiotic efficiency of Sinorhizobium meliloti. Mol. Plant-Microbe Interact. 14, 887–894. 10.1094/MPMI.2001.14.7.887 [DOI] [PubMed] [Google Scholar]
- Oldroyd G. E. D. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252. 10.1038/nrmicro2990 [DOI] [PubMed] [Google Scholar]
- Oldroyd G. E. D., Downie A. (2008). Bias and precision of cholesterol analysis by physician’s office analyzers. Annu. Rev. Plant Biol. 59, 519–546. 10.1146/annurev.arplant.59.032607.092839 [DOI] [PubMed] [Google Scholar]
- Oldroyd G. E. D., Murray J. D., Poole P. S., Downie J. A. (2011). The Rules of Engagement in the Legume-Rhizobial Symbiosis. Annu. Rev. Genet. 45, 119–144. 10.1146/annurev-genet-110410-132549 [DOI] [PubMed] [Google Scholar]
- Ott T., Van Dongen J. T., Günther C., Krusell L., Desbrosses G., Vigeolas H. (2005). Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr. Biol. 15, 531–535. 10.1016/j.cub.2005.01.042 [DOI] [PubMed] [Google Scholar]
- Price P. A., Tanner H. R., Dillon B. A., Shabab M., Walker G. C., Griffitts J. S. (2015). Rhizobial peptidase HrrP cleaves host-encoded signaling peptides and mediates symbiotic compatibility. Proc. Natl. Acad. Sci. 112, 15244 LP– 15249. 10.1073/pnas.1417797112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puppo A., Groten K., Bastian F., Carzaniga R., Soussi M., Lucas M. M. (2005). Legume nodule senescence: Roles for redox and hormone signalling in the orchestration of the natural aging process. New Phytol. 165, 683–701. 10.1111/j.1469-8137.2004.01285.x [DOI] [PubMed] [Google Scholar]
- Puppo A., Pauly N., Boscari A., Mandon K., Brouquisse R. (2013). Hydrogen peroxide and nitric oxide: key regulators of the legume – Rhizobium and mycorrhizal symbioses. Antioxid. Redox Signal. 18, 2202–2219. 10.1152/jn.00838.2004 [DOI] [PubMed] [Google Scholar]
- Ramage H. R., Connolly L. E., Cox J. S. (2009). Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PloS Genet. 5, e1000767. 10.1371/journal.pgen.1000767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro C. W., Baldacci-Cresp F., Pierre O., Larousse M., Benyamina S., Lambert A. (2017). Regulation of differentiation of nitrogen-fixing bacteria by microsymbiont targeting of plant Thioredoxin s1 . Curr. Biol. 27, 250–256. 10.1016/j.cub.2016.11.013 [DOI] [PubMed] [Google Scholar]
- Sánchez C., Gates A. J., Meakin G. E., Uchiumi T., Girard L., Richardson D. J. (2010). Production of nitric oxide and nitrosylleghemoglobin complexes in soybean nodules in response to flooding. Mol. Plant Microbe Interact. 23, 702–711. 10.1094/MPMI-23-5-0702 [DOI] [PubMed] [Google Scholar]
- Santos R., Hérouart D., Puppo A., Touati D. (2000). Critical protective role of bacterial superoxide dismutase in Rhizobium–legume symbiosis. Mol. Microbiol. 38, 750–759. 10.1046/j.1365-2958.2000.02178.x [DOI] [PubMed] [Google Scholar]
- Sasakura F., Uchiumi T., Shimoda Y., Suzuki A., Takenouchi K., Higashi S. (2006). A class 1 hemoglobin gene from Alnus firma functions in symbiotic and nonsymbiotic tissues to detoxify nitric oxide. Mol. Plant Microbe Interact. 19, 441–450. 10.1094/MPMI-19-0441 [DOI] [PubMed] [Google Scholar]
- Shidore T., Triplett L. R. (2017). Toxin-Antitoxin systems : implications for plant disease. Annu. Rev. Phytopathol. 55, 7, 1,–7. 19,. .org/ 10.1146/annurev-phyto-080516-035559 [DOI] [PubMed] [Google Scholar]
- Shimoda Y., Shimoda-Sasakura F., Kucho K., Kanamori N., Nagata M., Suzuki A. (2009). Overexpression of class 1 plant hemoglobin genes enhances symbiotic nitrogen fixation activity between Mesorhizobium loti and Lotus japonicus. Plant J. 57, 254–263. 10.1111/j.1365-313X.2008.03689.x [DOI] [PubMed] [Google Scholar]
- Soto M. J., Domínguez-Ferreras A., Pérez-Mendoza D., Sanjuán J., Olivares J. (2009). Mutualism versus pathogenesis: the give-and-take in plant-bacteria interactions. Cell. Microbiol. 11, 381–388. 10.1111/j.1462-5822.2009.01282.x [DOI] [PubMed] [Google Scholar]
- Stamler J. S., Lamas S., Fang F. C. (2001). Nitrosylation: the prototypic redox-based signaling mechanism. Cell 106, 675–683. 10.1016/S0092-8674(01)00495-0 [DOI] [PubMed] [Google Scholar]
- Tang G., Lu D., Wang D., Luo L. (2013). Sinorhizobium meliloti lsrB is involved in alfalfa root nodule development and nitrogen-fixing bacteroid differentiation. Chin. Sci. Bull. 58, 4077–4083. 10.1007/s11434-013-5960-6 [DOI] [Google Scholar]
- Tang G., Wang Y., Luo L. (2014). Transcriptional regulator LsrB of Sinorhizobium meliloti positively regulates the expression of genes involved in lipopolysaccharide biosynthesis. Appl. Environ. Microbiol. 80, 5265–5273. 10.1128/AEM.01393-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G., Xing S., Wang S., Yu L., Li X., Staehelin C., (2017). Regulation of cysteine residues in LsrB proteins from Sinorhizobium meliloti under free-living and symbiotic oxidative stress. Environ. Microbiol. 19, 5130–5145. 10.1111/1462-2920.13992 [DOI] [PubMed] [Google Scholar]
- Trinchant J. C., Rigaud J. (1982). Nitrite and nitric oxide as inhibitors of nitrogenase from soybean bacteroids. Appl. Environ. Microbiol. 44, 1385–1388. Available at: http://www.ncbi.nlm.nih.gov/pubmed/16346155%0Ahttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid = PMC242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Velde W., Zehirov G., Szatmari A., Debreczeny M., Ishihara H., Kevei Z. (2010). Plant peptides govern terminal differentiation of bacteria in symbiosis. Science (80-.), 1122–1126. 10.1126/science.1184057 [DOI] [PubMed] [Google Scholar]
- Vitousek P. M., Menge D. N. L., Reed S. C., Cleveland C. C. (2013). Biological nitrogen fixation: rates, patterns and ecological controls in terrestrial ecosystems. Philos. Trans. R. Soc Lond. B. Biol. Sci. 368, 20130119. 10.1098/rstb.2013.0119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Yu H., Luo L., Duan L., Cai L., He X. , et al, (2016). Nodules with activated defense 1 is required for maintenance of rhizobial endosymbiosis in Medicago truncatula. New Phytol. 212, 176–191. 10.1111/nph.14017 [DOI] [PubMed] [Google Scholar]
- Wang D., Griffitts J., Starker C., Fedorova E., Limpens E., Ivanov S. (2010). A nodule-specific protein secretory pathway required for nitrogen-fixing symbiosis. Science (80-.), 1126–1129. 10.1126/science.1184096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waszczak C., Carmody M., Kangasjärvi J. (2018). Reactive Oxygen Species in Plant Signaling. Annu. Rev. Plant Biol. 69, 209–236. 10.1146/annurev-arplant-042817-040322 [DOI] [PubMed] [Google Scholar]
- Yang S., Wang Q., Fedorova E., Liu J., Qin Q., Zheng Q. (2017). Microsymbiont discrimination mediated by a host-secreted peptide in Medicago truncatula. Proc. Natl. Acad. Sci. 114, 6848–6853. 10.1073/pnas.1700460114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Bao H., Zhang Z., Cao Y. (2019). Immune signaling pathway during terminal bacteroid differentiation in nodules. Trends Plant Sci. 24, 299–302. 10.1016/j.tplants.2019.01.010 [DOI] [PubMed] [Google Scholar]
- Yu H., Xiao A., Dong R., Fan Y., Zhang X., Liu C. (2018). Rapid report suppression of innate immunity mediated by the CDPK-Rboh complex is required for rhizobial colonization in Medicago truncatula nodules. New Phytol. 220, 425–434. 10.1111/nph.15410 [DOI] [PubMed] [Google Scholar]
- Yun B.-W., Feechan A., Yin M., Saidi N. B. B., Le Bihan T., Yu M. (2011). S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478, 264. 10.1038/nature10427 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Oldroyd G. E. D. (2017). Plant signalling in symbiosis and immunity. Nature 543, 328–336. 10.1038/nature22009 [DOI] [PubMed] [Google Scholar]