Abstract
Staphylococcus aureus is one of the major causes of nosocomial infections. This organism produces powerful toxins and cause superficial lesions, systemic infections, and several toxemic syndromes. A total of 109 S. aureus strains isolated from a variety of infections like ocular diseases, wound infection, and sputum were included in the study. Minimum inhibitory concentration (MIC) was determined against 8 antimicrobials. PCR determined the presence of 16S rRNA, nuc, mecA, czrC, qacA/B, pvl, and toxin genes in S. aureus isolates. Pulse-field gel electrophoresis (PFGE), multi-locus sequence typing (MLST), SCCmec, spa-, and agr-typing and serotyping determined the diversity among them. All isolates of S. aureus were resistant to two or more than two antibiotics and generated 32 resistance patterns. These isolates were positive for 16S rRNA and S. aureus-specific nuc gene, but showed variable results for mecA, czrC, and qacA/B and pvl genes. Of the 32 methicillin-resistant S. aureus (MRSA), 13 strains carried SCCmec type V, seven type IV, two type III, and nine carried unreported type UT6. Of the 109 strains, 98.2% were positive for hlg, 94.5% for hla, 86.2% for sei, 73.3% for efb, 70.6% for cna, 30.2% for sea, and 12.8% for sec genes. Serotypes VII and VI were prevalent among S. aureus strains. PFGE analysis grouped the 109 strains into 77 clusters. MLST classified the strains into 33 sequence types (ST) and eight clonal complexes (CCs) of which 12 were singletons, and two belong to new allelic profiles. Isolates showed 46 spa-types that included two new spa-types designated as t14911 and t14912. MRSA and methicillin-susceptible S. aureus (MSSA) isolates were diverse in terms of antibiotic resistance pattern, toxin genotypes, SCCmec types, serotypes and PFGE, MLST, and spa-types. However, few isolates from eye infection and wound infection belong to CC239, ST239, and spa-type t037/t657. The study thus suggests that S. aureus strains are multidrug resistant, virulent, and diverse irrespective of sources and place of isolation. These findings necessitate the continuous surveillance of multidrug-resistant and virulent S. aureus and monitoring of the transmission of infection.
Keywords: antibiotic susceptibility, virulence, MLST, spa-typing, PFGE, biofilm, Staphylococcus aureus
Introduction
Staphylococcus aureus commensal to human skin and mucous membranes could cause nosocomial (Lindsay and Holden, 2004) and systemic infections (Jarraud et al., 2002). The isolation of methicillin-resistant S. aureus (MRSA) from ocular infections varies from 3 to 30% in a hospital in India and other countries (Shanmuganathan et al., 2005; Freidlin et al., 2007). MRSA strains belonging to ST5, ST72, and ST88 and isolated from severe eye infections in India were resistant to all antibiotics except tetracycline, chloramphenicol, and cefazolin (Nadig et al., 2012). Godebo et al. (2013) showed that 94.5% of S. aureus isolated from wound infection were resistant to penicillin, 91.8% to ampicillin, and 76.7% to oxacillin.
Several studies have shown the presence of toxin genes among MRSA. The presence of the sea gene in MRSA varies from country to country (Mehrotra et al., 2000; Kim et al., 2006; Wang et al., 2013). However, hla gene was present in all isolates (Shukla et al., 2010). MRSA isolated from conjunctivitis in Nigeria belonging to ST88 and SCCmec type IV were positive for pvl gene (Ghebremedhin et al., 2009). However, pvl gene positive methicillin-susceptible S. aureus (MSSA) strains belonged to ST30 (D’Souza et al., 2010). S. aureus carrying the pvl gene and belonging to ST239, ST5, and ST88 was reported from a teaching hospital in China (Liu et al., 2009). MSSA belonging to ST121 and spa-type 287 isolated from community-acquired pneumonia in young patients carried the virulence genes (cna and bbp) and pvl (Baranovich et al., 2010). The role of virulence genes in S. aureus pathogenesis may vary from one infection type to another type of infections. Dhawan et al. (2015) reported the isolation of SCCmec type IV and V clones of MRSA in an Indian hospital. Several other workers also showed a decrease in SCCmec III MRSA isolation but increased SCCmec IV and V MRSA isolation (Hsu et al., 2005; D’Souza et al., 2010). Multidrug-resistant isolates belonging to ST239 and SCCmec type III were slowly replaced by multidrug-susceptible ST22 (SCCmec type IV) and ST772 (SCCmec type V) in hospitals (D’Souza et al., 2010).
Several molecular biology techniques like multi-locus sequence typing (MLST), pulse-field gel electrophoresis (PFGE), SCCmec typing, and spa-typing have been used to study epidemiology and clonal diversity of S. aureus (Maslow et al., 1993; Norazah et al., 2001; Ghaznavi-Rad et al., 2011). However, not a single technique alone could discriminate the bacteria because of differences in the degree of typeability, reproducibility, and discriminatory power (Tenover et al., 1994). Overall analysis of different typing techniques can provide information on diversity of the isolates that can be useful for outbreak investigations. In India, S. aureus is rated as one of the major pathogen causing a variety of infections and showing resistance to several antibiotics; however, not much information is available on their antibiotic susceptibility, virulence profile, and genomic diversity. In this study, our aim was to determine the antibiotic susceptibility pattern, virulence profiles, and genomic diversity among MRSA and MSSA isolated from patients with a variety of infections, including ocular diseases and collected from different parts of India from 2007 to 2015. Genetic, serotype, and phenotypic data were used to determine whether isolates from a variety of infections had similar characteristics.
Materials and Methods
Bacterial Strains
A total of 109 S. aureus strains isolated from patients visited/admitted to hospitals with infections in different part of India between July 2007 and November 2015 were included in the study. These isolates were from LV Prasad Eye Institute, Bhubaneswar (n = 54), comprised of microbial keratitis (n = 18), eyelid abscess (n = 8), endophthalmitis (n = 5), Steven Johnson syndrome with bacterial keratitis (n = 9), suture-related infections (n = 3), and other ocular infection (n = 5); LV Prasad Eye Institute, Hyderabad (n = 10) comprised of cornea scrapping (n = 5), pus from eye (n = 4), and suture-related infections (n = 1); Institute of Medical Sciences, Banaras Hindu University, Varanasi (n = 21) comprised of wound infection (n = 16) and unknown sources (n = 5); All India Institute of Medical Sciences, New Delhi (wound infection n = 10); and University College of Medical Sciences, Delhi (wound infection n = 9). Also, five isolates were from the conjunctiva of the asymptomatic healthy volunteers LV Prasad Eye Institute, Bhubaneswar. We conducted the study following the guidelines mentioned in the Declaration of Helsinki. We identified all the 109 isolates by using biochemical tests including Gram staining, catalase production, fermentation of glucose and mannitol, and ID32 STAPH strips using ATBTM NEW v.1.0.0 software on an ATBTM reader (bioMerieux, France) (Panda et al., 2014). The amplification of the S. aureus nuc gene confirmed the identity of isolates (Hirotaki et al., 2011). We used S. aureus ATCC 25293 and S. aureus ATCC 29213 as quality control strains for antibiotic susceptibility testing, and S. aureus ATCC 25923 and ATCC 43300 as a reference for serotyping, PFGE, MLST, and spa-typing.
Coagulase Gene Typing
Coagulation-inhibition test with coagulase type I–VIII-specific antisera (staphylococcal coagulase antiserum kit; Denka Seiken, Inc., Tokyo, Japan) was conducted to determine the coagulase type of S. aureus following the manufacturer’s instructions (Goh et al., 1992). Briefly, a single colony for each test strain was suspended in BHI broth (Becton Dickinson Co.) and incubated at 37°C for overnight. Then centrifuged the culture and 0.1 ml of the supernatant used as test antigen. Distributed an aliquot (0.1 ml) of the test antigen into ten tubes followed by addition of 0.1-ml aliquots of anticoagulase types I–VIII sera to first eight tubes, except 9th and 10th tubes which were used as positive and negative controls and incubated at 37°C for 1 h. After that, 0.2 ml of diluted rabbit plasma was added to each tube and incubated at 37°C for 1 h. Visual inspection judged the coagulation of plasma after 2, 4, 24, and 48 h and accordingly, strains were typed based on results obtained with staphylocoagulase reaction showing coagulation inhibition.
Minimum Inhibitory Concentration (MIC) Determination
Minimum inhibitory concentrations (MICs) of oxacillin, chloramphenicol, vancomycin, tetracycline, gentamicin, erythromycin, clindamycin, and trimethoprim were determined by broth microdilution methodology as recommended by the CLSI breakpoints. The 96-well plates were incubated at 37°C and were read for turbidity after 24 h.
Polymerase Chain Reaction (PCR) Assays
The presence of genes encoding for methicillin resistance (mecA), the nuclease (nuc), Panton-Valentine leukocidin (pvl), cadmium resistance (czrC), and quaternary ammonium resistance (qacA/B) was determined by hexaplex PCR (Panda et al., 2014). PCR identified the presence of msrA, ermA, ermC (erythromycin resistance), tetK (tetracycline resistance) genes (Duran et al., 2012). Also, PCR determined the presence of gene encoding for resistance to aminoglycosides [aac (6’)/aph (2), aph (3’-III)] by the method described earlier (Schmitz et al., 1999). The presence of catpC221, catpC223, and catpC194 (chloramphenicol resistance) was determined by PCR as described by Argudín et al. (2011). The mphC (clindamycin resistance) gene was detected by PCR method described earlier (Panda et al., 2016).
SCCmec Typing
Two PCRs, MPCR1 and MPCR2 were used to detect the presence of mec complex, ccr complex, and SCCmec type among S. aureus (Kondo et al., 2007).
Virulence Gene Profile and Accessory Gene Regulator (Agr) Typing
PCR determined the presence of Staphylococcal enterotoxin (SE) genes encoding for seA, seC, and seI (Monday and Bohach, 1999; Jarraud et al., 2002). Also, the presence of hemolysin genes, hlA and hlG, was determined by PCR (Mitchell et al., 2010; Paniagua-Contreras et al., 2012). PCR was used to detect the presence of collagen adhesion (cna) and extracellular fibrinogen binding protein (efb) among S. aureus strains (Zecconi et al., 2006). The presence of intracellular adhesion genes (icaA, icaD) was determined by PCR as described by Arciola et al. (2001). PCR amplification was carried out to determine the presence of agr alleles using group-specific primers as described by Gilot et al. (2002).
Pulsed-Field Gel Electrophoresis (PFGE)
Pulsed-field gel electrophoresis of S. aureus genomic DNA digested with SmaI (NEB) was carried out by the protocol described for S. aureus by Centre for Disease Control and Prevention. The dendrogram of similarity showing the clustering of the isolates according to banding patterns was generated with Bionumerics software, version 7.1 (Applied Maths, Belgium) using the Dice index and the un-weighted pair group method with arithmetic average (UPGMA) with 0.5% optimization and 1% position tolerance. Isolates showing similarity coefficient of up to 80% were considered belonging to similar pulsotype (Van Belkum et al., 2007).
Multi-Locus Sequence Typing (MLST)
The internal fragments of seven housekeeping genes, viz., arcC, gmk, aroE, glpF, pta, tpi, and yqil were amplified by PCR method described earlier (Enright and Spratt, 1999). The amplified products were purified (ExoSAP; Affymetrix, Cleveland, OH, United States) and both strands sequenced using an ABI sequencer model 3500 (Life Technologies, Marsiling, Singapore) at the sequencing facility of the Institute of Life Sciences (Bhubaneswar, India). The nucleotide sequences were aligned using Mega 5.2 software. After manually comparing with reported alleles, STs were assigned accordingly. Sequencing was performed in biological duplicates to confirm the presence of novel alleles.
The advanced cluster analysis was performed to define the clonal complexes (CCs) by using Bionumerics software, version 7.1 (Applied Maths, Belgium). A minimum spanning tree (MST) was constructed using the MLST data and partitions were created to form clusters. The similarity in at least six alleles grouped isolates of S. aureus in one CC. The central ST of each separation was used to designate a CC.
Spa-Typing
PCR amplified the polymorphic X region of Staphylococcus protein A (spa) gene following the conditions mentioned earlier (Nelson et al., 2007). Amplified products were purified, and both strands were sequenced using an ABI sequencer model 3500 (Life Technologies, Marsiling, Singapore) at the sequencing facility of the Institute of Life Sciences (Bhubaneswar, India). The nucleotide sequences were aligned using Mega 5.2 software. Repeat succession in the polymorphic X-region assigned the spa-types, and accordingly the MST was generated using Bionumerics 7 software (Applied Maths, Belgium) using gap creation cost 250%, gap extension cost 50%, duplicate production cost 25%, duplicate expansion cost 25%, and maximum duplication three repeats.
Statistical Analysis
We performed principal coordinates analysis (PCoA) and discriminant analysis (DA) using PAST program v2.17 for the antibiotic resistance genes and virulence genes in MRSA and MSSA isolates with regard to sources of isolation (Hammer et al., 2001). We carried out the DA using default values to confirm the hypothesis of whether MRSA and MSSA isolates are different.
Results
Hexaplex PCR
All the isolates of S. aureus were positive for 16S rRNA and S. aureus-specific nuc genes. Hexaplex PCR discriminates between MSSA and MRSA isolates. Thirty-one of 109 (29.4%) methicillin-resistant strains were positive for the mecA gene, and 77 (70.6%) methicillin sensitive isolates were negative for the mecA gene. One of the methicillin-resistant strains of S. aureus was negative for the mecA gene. Among 109 isolates, 43 (39.4%) isolates comprising 23 of the 77 (29.9%) MSSA and 20 of the 31 (64.5%) MRSA isolates were positive for pvl gene. Of the 31 MRSA isolates, two (6.5%) strains were positive for the czrC gene and four (12.9%) isolates were positive for qacA/B gene, and remaining isolates were negative for both czrC and qacA/B genes (data not shown).
Coagulase Serotyping
Serotyping classified S. aureus isolates into I–VIII serotypes by using coagulase typing scheme. Twelve of the 109 (11%) strains belong to serotype I, 11 (10%) to serotype II, nine (8%) to serotype III, 14 (12.8%) to serotype IV, 12 (11%) to serotype V, 19 (17.4%) to serotype VI, 20 (18.3%) to serotype VII, and 12 (11%) to serotype VIII, respectively. Nine of 31 (29%) MRSA belong to serotype VI and 17 of 78 (21.8%) and MSSA isolates belong to serotype VII (Table 1). Nine of the 24 (37.5%) isolates from wound infection belong to serotype VI and 16 of 64 (25%) isolates from eye infection belonged to serotype VII.
TABLE 1.
Antibiotic resistance patterns and presence of antibiotic resistance genes in Staphylococcus aureus isolates from different parts of India.
| Phenotypic antibiotic resistance pattern | Number of isolates showing presence of gene(s) encoding for | |||||||||||||||||
| MRSA | MSSA | mecA | aac(6’)/ aph(2) | aph (3’III) | msrA | ermA | ermC | mphC | tetK | tetL | tetM | cat::pC221 | cat::pC223 | cat::pC194 | dfrA | dfrB | dfrG | |
| OX, CHL, TET, GEN, ERY, CL, TMP | 10 | 0 | 10 | 10 | 10 | – | 10 | 10 | 10 | 10 | 10 | 10 | 10 (3) | – | 10 | 10 | 10 | 10 |
| OX, CHL, ERY, TMP | 0 | 1 | – | 1 | – | 1 | – | 1 (1) | 1 | – | – | 1 | 1 (1) | – | 1 | – | – | 1 |
| CHL, ERY, TMP | 0 | 11 | – | 11 | 11 | – | 11 (3) | 11 | 11 | – | 11 | 11 | 11 (3) | 11 | – | 11 | 11 | 11 |
| CHL, TMP | 0 | 6 | – | 6 | – | – | – | + | – | – | 6 | 6 | 6 (2) | – | 6 | – | 6 | 6 |
| OX, CHL, TET, ERY, TMP | 3 | 0 | 3 | 3 | 3 | 3 | 3(1) | 3 | 3 | 3 | 3 | 3 | 3 (1) | – | – | – | 3 | 3 |
| OX, CHL, GEN, ERY, TMP | 11 | 0 | 11 | 11 | 11 | 11 | 11 (5) | 11 | 11 | – | 11 | 11 | 11 (2) | 11 | 11 | – | 11 | 11 |
| OX, CHL, TET, GEN, ERY, TMP | 5 | 0 | 5 | 5 (1) | 5 | 5 | – | 5 | 5 | 5 | 5 | 5 | 5 (1) | 5 | – | 5 | 5 | 5 |
| ERY, CL, TMP | 0 | 1 | – | – | 1 | 1 | 1 | 1 | – | – | – | – | – | – | – | – | 1 | – |
| CHL, ERY, CL, TMP | 0 | 2 | – | 2 | 2 | – | – | 2(1) | 2(1) | – | – | 2 | 2 (1) | 2 | – | 2 | 2 | 2 |
| CHL, ERY, CL | 0 | 13 | – | 13 | 13 | 13 | 13(9) | 13 | 13(1) | 13 | 13 | 13 | 13 (3) | 13 | – | 13 | 13 | 13 |
| CHL | 0 | 3 | – | – | – | 3 | – | 3 | – | 3 | 3 | 3 | 3 (1) | 3 | – | 3 | 3 | 3 |
| CHL, TET, GEN, ERY, CL, TMP | 0 | 3 | – | – | 3 | 3 | – | 3 | –(2) | 3 | – | 33 | 3 | – | 3 | 3 | 3 | |
| CHL, TET, GEN, TMP | 0 | 1 | – | 1 | 1 | – | – | 1 | – | 1 | – | – | 1 (1) | – | – | – | – | – |
| CHL, GEN, ERY, CL, TMP | 0 | 2 | – | 2 (1) | 2 | – | – | 2(2) | 2(2) | 2 | 2 | 2 | 2 | 2 | – | – | 2 | 2 |
| CHL, GEN, ERY, CL, TMP | 0 | 7 | – | 7 (2) | 7 | 7(3) | – | – | −(3) | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 |
| CHL, TET, GEN, ERY, CL, TMP | 0 | 1 | – | – | 1 | – | – | – | 1(1) | 1 | – | – | 1 | 1(1) | – | 1 | 1 | 1 |
| CHL, GEN, ERY | 0 | 3 | – | 3 | 3 | 3 (1) | – | 3 (1) | – | – | 3 | 3 | 3 | 3 | – | 3 | 3 | 3 |
| CHL, TET, GEN, ERY | 0 | 3 | – | 3 (2) | 3 | 3 | – | 3(1) | – | 3 | 3 | 3 | 3 (1) | 3 | – | 3 | 3 | 3 |
| GEN, ERY, CL, TMP | 0 | 1 | – | 1 (1) | 1 | – | – | 1 | 1 | 1 | – | 1 | 1 | 1 | – | 1 | 1 | 1 |
| GEN, ERY | 0 | 2 | – | – | 2(1) | 2 | – | 2(1) | – | – | 2 | 2 | 2 | 2 | – | 2 | 2 | 2 |
| ERY | 0 | 4 | – | 4 | 4 | 4 | – | 4(2) | – | 4 | – | 4 | 4 | 4 | – | 4 | 4 | 4 |
| ERY, TMP | 0 | 3 | – | 3 | 3 | 3 | – | 3(3) | – | – | 3 | 3 | 3 | 3 | – | 3 | 3 | 3 |
| OX, CHL, GEN, ERY | 1 | 0 | 1 | – | 1 | – | – | – (1) | – | 1 | – | 1 | 1 | 1 | – | 1 | 1 | 1 |
| CHL, TET, ERY, TMP | 0 | 2 | – | 2 | 2 | – | – | 2 (1) | – | 2 | 2 | 2 | 2 (1) | 2 | – | 2 | 2 | 2 |
| CHL, ERY, CL | 0 | 1 | – | – | 1 | 1 | – | 1(1) | 1 | – | 1 | 1 | 1 | 1 | – | 1 | 1 | 1 |
| GEN, ERY, TMP, CHL | 0 | 1 | – | 1 | 1 | 1 | – | 1 | – | – | – | 1 | 1 (1) | – | – | – | 1 | 1 |
| CHL, CL, TMP | 0 | 2 | – | 2 | 2 | 2 | – | 2 | 2 | – | 2 | 2 | 2 | 2 | 2 | – | 2 | 2 |
| TET, TMP | 0 | 1 | – | 1 | 1 | 1 | – | 1 | – | 1 | 1 | 1 | 1 | 1 | – | – | 11 | |
| ERY, CL, CHL | 0 | 1 | – | 1 | 1 | 1 | – | 1 | 1(1) | 1 | 1 | 1 | 1 (1) | 1 | – | 1 | 1 | 1 |
| TET, GEN, CL, ERY | 0 | 2 | – | – | 2 | 2 | – | 2 | −(1) | – | – | 2 | 2 | 2 | – | 2 | 2 | 2 |
| OX, CHL, ERY, CL, TMP | 1 | 0 | 1 | 1 | 1 | 1 | – | 1 | −(1) | 1 | 1 | 1 | 1 | 1 | – | – | 1 | 1 |
| CHL, TET, GEN | 0 | 1 | – | 1 | 1 | – | – | – | – | – | 1 (1) | 1 | 1 | 1 | – | – | 1 | 1 |
MRSA: methicillin-resistant Staphylococcus aureus; MSSA: methicillin-susceptible Staphylococcus aureus, OX: oxacillin, GEN: gentamicin, ERY: erythromycin, TET: tetracycline; CL: clindamycin; CHL: chloramphenicol; TMP: trimethoprim. Isolates showing phenotypic resistance to given antibiotic(s) are shown in bold. Number in brackets indicate phenotypic sensitive isolates.
Antibiotic Resistance Genes
One hundred two of the 109 S. aureus isolates were multidrug resistant showing resistance to two or more antibiotics. All the strains were susceptible to vancomycin when tested by broth microdilution assay. Thirty-one isolates of S. aureus were resistant to oxacillin and carried the mecA gene; however, one isolate of S. aureus resistant to oxacillin was negative by PCR for the mecA gene. The remaining 77 isolates were sensitive to oxacillin and negative by PCR for the mecA gene (Table 1).
Ninety-five isolates of S. aureus resistant to chloramphenicol carried cat: pC221 gene; however, 86 isolates carried cat: pC223 and 37 isolates carried cat: pC194 gene, respectively. Twenty isolates carried all the three genes tested; however, 83 isolates were positive for cat: pC221 and cat: pC223 and 37 isolates for cat: pC221 and cat: pC194 genes, respectively (Table 1). One of the isolates sensitive to chloramphenicol was negative by PCR for all three genes. In contrast, 15 strains of S. aureus susceptible to chloramphenicol were positive for cat: pC221 and 14 for cat: pC223 genes, respectively.
Twenty-nine isolates were phenotypically resistant to tetracycline of which 29 isolates were positive for tetK, 25 for tetL, and 28 for tetM genes. Twenty-five isolates carried all the three genes tested; however, three strains carried tetK and tetM genes and one isolate tetL and tetM genes. In contrast, 76 isolates sensitive to tetracycline were positive for the tetM gene, 66 for tetL, and 29 for tetK genes. Among them, 27 isolates carried all the three genes, six had tetK and tetM, and 39 strains had tetL and tetM genes, respectively. One isolate sensitive to tetracycline was negative by PCR for all three genes tested (Table 1).
A total of 54 isolates were resistant to gentamicin of which 45 isolates were positive for aac(6’)/aph(2’) and aph (3’-III) genes and nine isolates for aph (3’-III) gene only. In contrast, 43 gentamycin sensitive isolates showed positive results for aac(6’)/aph(2’) and aph (3’-III), seven isolates for aac(6’)/aph(2’), and two isolates for aph (3’-III) genes. However, 56 isolates sensitive to gentamicin were negative by PCR for aac(6’)/aph(2’) and aph (3’-III) genes (Table 1).
Of the 91 isolates of S. aureus showing resistance to macrolides carried erythromycin resistance genes. Twenty-eight isolates carried all the erythromycin resistance genes, namely, msrA, ermA, and ermC. Fifty-one isolates were positive for two genes, of which 30 isolates carried msrA and ermC genes, and 21 strains had ermA and ermC genes. Besides, 12 isolates were positive for a single gene of which five isolates carried the ermC gene, and seven isolates had msrA gene. In contrast, two of the 10 erythromycin sensitive isolates carried msrA and ermC genes, four strains possess msrA and ermC genes, and three isolates had the ermC gene. Of the 64 isolates carrying the mphC gene, 22 isolates were phenotypically resistant to clindamycin (Table 1). None of the 17 strains showing sensitivity to erythromycin carried any of the erythromycin resistance genes. One of the resistant isolate not carrying any of the erythromycin resistant genes is likely to be mediated by an as-yet-unknown mechanism.
Similarly, 74 isolates were resistant to trimethoprim of which 45 isolates were positive for dfrA, dfrB, and dfrG genes, 27 strains for dfrB and dfrG genes, and one isolate each for dfrB and dfrG genes, respectively. In contrast, 34 isolates sensitive to trimethoprim were also positive for dfrA, dfrB, and dfrG genes; however, one strain was positive for the dfrG gene (Table 1).
D-Test and Macrolide Resistance
Ninety of 109 (89.9%) S. aureus isolates that exhibited erythromycin resistance were evaluated for MLSB resistance phenotype, namely, iMLSB, cMLSB and MSB. Seventy eight of 90 (79.5%) isolates were erythromycin-resistant but clindamycin susceptible were tested for D-test. We found 14 isolates (10 MRSA and four MSSA) showed iMLSB phenotype, and 12 (two MRSA and 10 MSSA) had MSB phenotype. Seven erythromycin-resistant isolates comprising six MRSA and one MSSA had cMLSB phenotype. The remaining 45 isolates (14 MRSA and 31 MSSA) did not show any MLSB phenotypes.
Among MRSA and MSSA showing cMLSB resistance phenotype, three of six MRSA isolates possessed the ermA and ermC genes and one each possessed ermC gene, msrA, ermC, mphC genes, and ermC and mphC genes. One MSSA isolate was positive for msrA, ermA, and ermC genes. On the hand, one of the two MRSA isolates showing MSB phenotype had msrA, ermC genes and other strain had msrA, ermC, and mphC genes (Table 2). Of the 12 MSSA, six isolates contained msrA and ermC genes, one isolate each contained ermC and ermA genes, respectively, two strains had mphC gene. The remaining isolates did not carry any of the genes tested. Of the 10 MRSA, six isolates with iMLSB phenotype had ermC gene. One isolate each carried msrA, ermA, and ermC genes, ermA, ermC genes, msrA, ermC, and mphC genes, respectively. The remaining one isolate did not possess any of the resistance genes. Of the four MSSA isolates that showed iMLSB phenotype, three strains were positive for msrA, ermC genes, and one isolate was positive for ermC gene (Table 2).
TABLE 2.
Result of D-test obtained with MRSA and MSSA isolates showing presence of erythromycin resistance genes and its correlation with MLSB phenotypes among Staphylococcus aureus.
| Erythromycin resistance and MSB phenotypes | Phenotype (%) | Gene combinations | |||||||||
| msrA | ermA | ermC | mphC | msrA, ermC | ermA, ermC | ermC, mphC | msrA, ermA, ermC | msrA, ermC, mphC | msrA, ermA, ermC, mphC | ||
| MRSA (n=32) | |||||||||||
| ER-S, CL-S | 10 (31.25%) | 0 | 0 | 0 | 0 | 3 (30%) | 0 | 1 (10%) | 1 (10%) | 3 (30%) | 1 (10%) |
| ER-R, CL-S (MSB phenotype) | 2 (6.25%) | 0 | 0 | 0 | 0 | 1 (50%) | 0 | 0 | 0 | 1 (50%) | 0 |
| ER-R, CL-R (cMLSB phenotype) | 6 (18.75%) | 0 | 0 | 1 (16.6 %) | 0 | 0 | 3 (50%) | 1 (16.6%) | 0 | 1 (16.6%) | 0 |
| ER-R, CL-D (iMLSB phenotype) | 10 (31.25%) | 0 | 0 | 6 (60%) | 0 | 0 | 1 (10%) | 0 | 1 (10%) | 1 (10%) | 0 |
| MSSA (n=77) | |||||||||||
| ER-S, CL-S | 52 (67.5%) | 3 (5.7%) | 1 (1.9%) | 16 (30.7 %) | 1 (1.9%) | 20 (38.4%) | 0 | 4 (7.6%) | 0 | 4 (7.6%) | 0 |
| ER-R, CL-S (MSB phenotype) | 12 (15.5%) | 0 | 1 (8.3%) | 1 (8.3%) | 2 (16.6 %) | 6 (50%) | 0 | 0 | 0 | 0 | 0 |
| ER-R, CL-R (cMLSB phenotype) | 1 (1.29%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (100%) | 0 | 0 |
| ER-R, CL-D (iMLSB phenotype) | 4 (5.19%) | 0 | 0 | 1 (25%) | 0 | 3 (75%) | 0 | 0 | 0 | 0 | 0 |
S: sensitive; R: resistance; ER: erythromycin; CL: clindamycin.
Of the 109 S. aureus isolates tested for the presence of MLSB resistance genes, 102 isolates carried one or more erm genes. Three strains carried all the erythromycin resistance genes, namely, msrA, ermA, and ermC. Fifty-one isolates were positive for two genes, of which 46 isolates carried msrA and ermC genes, and five had ermA and ermC genes. Besides, 37 isolates were positive for a single gene of which 34 isolates carried the ermC gene, two isolates had ermA gene, and three isolates had the msrA gene (Table 2). In contrast, four of the 13 erythromycin-sensitive isolates carried msrA and ermC genes. One strain each had the ermC gene and msrA gene. The remaining isolates did not carry any resistance genes. Twelve of the 21 mphC gene-positive isolates showed phenotypic resistant to clindamycin. The remaining nine isolates were sensitive to clindamycin (Table 2). Eight erythromycin-resistant strains did not carry any of the erythromycin-resistant genes is likely to be mediated by an as-yet-unknown mechanism.
SCCmec Typing
The presence of the mec complex and ccr complex classified S. aureus strains into different SCCmec types. Thirty-one MRSA isolates showed four known SCCmec types of which 13 (40.6%) belong to type V, nine (28.1) belong to type UT6, seven (21.9%) belong to type IV, and two (6.3%) belong to type III (Table 3). One isolate showing phenotypic resistance to methicillin but negative for mecA gene carried C1 type of ccr complex but lack mec complex. Of the 32 methicillin-sensitive isolates lacking the mec complex, 14 isolates carried ccrA1B1, one strain possesses ccrA4B4, and 17 isolates had ccrA3B3 and ccrA4B4 type of ccr complex, respectively (Table 3).
TABLE 3.
Distribution of SCCmec types among S. aureus strains isolated from wound and ocular infection.
| Distribution of SCCmec types among S. aureus strains isolated from wound and ocular infection | ||||||
| SCCmec type | Recombinase complex | mecA complex | Source of infection | Total no. of isolates (n = 109) | ||
| Wound (n = 34) | Ocular (n = 69) | Unknown (n = 6) | ||||
| III | ccrC1, ccrAB3 | Class A | 2 | 0 | 0 | 2(1.8%) |
| IV | ccrAB2 | Class B | 7 | 0 | 0 | 7(6.4%) |
| V | ccrC1 | Class C2 | 5 | 4 | 4 | 13(11.9%) |
| UT6 | ccrC1 | Class A | 5 | 3 | 1 | 9(8.2%) |
| Untypable-1 | ccrC1 | – | 1 | 0 | 0 | 1(0.91%) |
| Untypable-2 | ccrAB4 | – | 0 | 1 | 0 | 1(0.91%) |
| Untypable-3 | ccrAB1 | – | 0 | 14 | 0 | 14(12.8%) |
| Untypable-4 | ccrAB2 | – | 0 | 1 | 0 | 1(0.91%) |
| Untypable-5 | ccrAB3 | – | 0 | 1 | 0 | 1(0.91%) |
Toxin Gene Profiles
Of the 109 isolates, 34 (31.2%) isolates harbored sea gene, 14 (12.8%) isolates sec gene, 93 (85.3%) isolates sei gene, 76 (69.7%) cna gene, 101 (92.6%) isolates hla gene, 107 (98%) isolates hlg gene, and 84 (77%) isolates carried efb gene, respectively. All the isolates, except one isolate, was positive for the hlg, and carried multiple virulence genes (Table 4).
TABLE 4.
Source, clonal complex, sequence-, spa-, SSCmec-, and agr-types and virulence profiles of S. aureus isolated from different parts of India.
| Source (isolate number) | CC/ST, spa-type | SCCmec type | agr type | pvl gene | icaA/icaD | Serotypes | Virulence pattern |
| MRSA (n = 32) | |||||||
| Wound infection (2095) | 239/239, t037 | III | 1 | – | +/+ | II | sei-cna-hla-hlg-efb |
| Wound infection (2103) | +/+ | IV | sea-sei-cna-hla-hlg-efb | ||||
| Wound infection (2656) | 239/239,t037 | UT6 | 1 | – | +/+ | IV | sea-sei-cna-hla-hlg-efb |
| Wound infection (22/248) | +/+ | III | sea-cna-hla | ||||
| Wound infection (UC650) | +/+ | IV | sea-cna-hla-hlg-efb | ||||
| Wound infection (UC858) | −/− | V | sea-hla-hlg-efb | ||||
| Wound infection (UC1079) | 239/239,t2952 | UT6 | 1 | – | +/+ | I | sea-sei-cna-hla-hlg-efb |
| Wound infection (2658) | 239/241,t037 | UT6 | 1 | – | +/+ | IV | sei-cna-hlg-efb |
| Eye infection (P844628, N307002) | 239/239, t037 | UT6 | 1 | – | +/+ | IV | sei-cna-hla-hlg |
| ± | |||||||
| Eye infection (P853836) | 239/239, t037 | UT6 | 1 | – | ± | V | sea-cna-hla-hlg-efb |
| Wound infection (2380,2452) | 772/772, t657 | V | None | + | +/+ | VI | sea-sec-sei-cna-hla-hlg-efb |
| +/+ | |||||||
| Wound infection (UC609) | 772/772, t657 | V | 2 | + | +/+ | VI | sea-sec-sei-cna-hla-hlg-efb |
| Wound infection (22/252) | 772/Unk, t657 | V | None | + | −/− | VI | sea-sec-sei-cna-hla-hlg-efb |
| Eye infection (845) | 772/772, t345 | V | 3 | + | −/+ | I | sea-sec-sei-cna-hla-hlg-efb |
| Eye infection (1295) | 2884/88, t2526 | V | 2 | + | +/+ | III | sei-hla-hlg-efb |
| Eye infection (1690) | 5/5, t442 | V | 1 | – | +/+ | IV | sei-hla-hlg-efb |
| Eye infection (1820) | 772/772, t657 | V | 1 | + | +/+ | VII | sea-sec-sei-cna-hla-hlg-efb |
| Unknown (1189) | 772/772, t657 | V | 2 | + | +/+ | VI | sec-sei-cna-hla-hlg-efb |
| Unknown (1192,1249) | 772/772, t345 | V | 2 | + | +/+ | VII | sea-sei-cna-hla-hlg-efb |
| +/+ | VI | sec-sei-cna-hla-hlg-efb | |||||
| Unknown (2654) | 772/772, t345 | V | 1 | + | +/+ | VI | sea-sei-cna-hla-hlg-efb |
| Wound infection (284) | Singleton 4/2642, t064 | V | 1 | – | +/+ | IV | hla-hlg-efb |
| Wound infection (221) | 30/30, t012 | IV | 3 | + | +/+ | VI | sei-cna-hla-hlg-efb |
| Wound infection (27/231) | 30/503, t012 | IV | 3 | + | +/+ | VII | sei-cna-hla-hlg |
| Wound infection (296) | 22/22, t005 | IV | 1 | + | +/+ | I | sec-sei-cna-hla-hlg |
| Wound infection (293) | 22/1414, t1328 | IV | 1 | + | +/+ | I | sei-cna-hla-hlg |
| Wound infection (UC104) | 22/22, Unk | IV | 1 | + | +/+ | II | sei-cna-hla-hlg-efb |
| Wound infection (UC101) | 22/22, t091 | +/+ | |||||
| Wound infection (UC463) | 22/22, t309 | IV | 1 | + | −/− | III | sec-sei-cna-hla-hlg-efb |
| Wound infection (2518)∗ | 121/120, t272 | NT | + | +/+ | VI | sea-sei-cna-hla-hlg-efb | |
| MSSA (n = 77) | |||||||
| Wound infection (2130) | 772/772, t345 | 2 | + | +/+ | VI | sec-cna-hla-hlg-efb | |
| Wound infection (2164) | 772/772, t1839 | UT∗ | None | + | +/+ | VI | sea-sec-sei-cna-hla-hlg-efb |
| Wound infection (2493) | 772/1, t386 | 4 | + | +/+ | VI | sei-cna-hla-hlg-efb | |
| Eye infection (N309852) | 772/1, t098 | 3 | – | +/+ | VII | sea-cna-hla-hlg | |
| Eye infection (518) | 772/1, t693 | UT∗ | 3 | – | +/+ | VII | sea-sei-cna-hla-hlg-efb |
| Eye infection (535,1636) | 772/1, t127 | UT∗ | 3 | – | +/+ | VII | sea-sei-cna-hlg-efb |
| +/+ | V | ||||||
| Eye infection (831) | 772/1, t127 | UT∗ | 3 | – | +/+ | II | sea-sei-cna-hla-hlg-efb |
| Eye infection (1361) | 772/1, t128 | UT∗ | 3 | – | +/+ | VII | sea-sec-sei-hla-hlg-efb |
| Eye infection (1321) | 772/1, t177 | UT∗ | 3 | – | +/+ | VII | sea-sei-cna-hla-hlg-efb |
| Eye infection (1476) | 772/1, t127 | 3 | – | +/+ | VIII | sea-sei-cna-hla-hlg-efb | |
| Eye infection (1881) | +/+ | I | |||||
| Eye infection (1503) | 772/1, t127 | 3 | – | +/+ | VI | sei-cna-hla-hlg-efb | |
| Eye infection (975) | 772/1, t8078 | 3 | – | +/+ | VI | sei-hla-hlg-efb | |
| Eye infection (1214) | 772/772, t657 | 3 | + | +/+ | VI | sea-sec-sei-cna-hla-hlg | |
| Healthy conjunctiva (N11OD) | 772/1, t948 | UT∗ | None | – | +/+ | I | sea-sei-cna-hla-hlg-efb |
| Healthy conjunctiva (N12OD) | 772/1, t948 | 3 | – | +/+ | IV | sea-cna-hla-hlg-efb | |
| Wound infection (2151) | 30/714, t021 | 3 | + | +/+ | VI | sei-cna-hla-hlg-efb | |
| Wound infection (2413) | 30/1482, t386 | 3 | + | +/+ | IV | sei-cna-hla-hlg-efb | |
| Eye infection (1196) | 30/938, t021 | 3 | + | +/+ | IV | sei-cna-hla-hlg-efb | |
| Eye infection (1850) | +/+ | V | |||||
| Wound infection (2488) | 121/121, t159 | 4 | – | +/+ | II | sei-cna-hla-hlg-efb | |
| Eye infection (P832812) | 121/121, t3204 | 4 | + | +/+ | V | cna-hla-hlg | |
| Eye infection (P706434) | 121/1964, t272 | 4 | – | +/+ | V | sei-cna-hla-hlg | |
| Eye infection (917) | 121/2160, t159 | 4 | + | +/+ | V | cna-hla-hlg-efb | |
| Unknown (2657) | 2884/2884, t4104 | 3 | + | +/+ | III | hla-hlg-efb | |
| Eye infection (149) | 2884/88, t5562 | 3 | + | −/+ | VI | sei-hla-hlg-efb | |
| Eye infection (1764Y) | 2884/88, t448 | 3 | + | +/+ | VIII | sea-sei-hla-hlg-efb | |
| Eye infection (504, 1035, 1271) | 5/5, t442 | 2 | – | +/+ | II | sei-cna-hla-hlg-efb | |
| +/+ | sei-hla-hlg-efb | ||||||
| +/+ | sei-hlg | ||||||
| Eye infection (N303284) | None | – | +/+ | I | sei-cna-hla-hlg | ||
| Eye infection (843) | 2 | – | +/+ | VIII | sei-hlg-efb | ||
| Eye infection (1042) | 2 | – | +/+ | VII | sei-hla-hlg-efb | ||
| Eye infection (1766, 1862) | 1 | – | +/+ | VIII | sei-hla-hlg | ||
| +/+ | sei-hla-hlg-efb | ||||||
| Eye infection (1867) | 1 | – | +/+ | VII | sei-hla-hlg-efb | ||
| Eye infection (1103) | 5/5, t14912 | 2 | – | +/+ | V | sei-hla-hlg-efb | |
| Eye infection (1306) | 5/83, t442 | 2 | – | +/+ | II | sei-hla-hlg-efb | |
| Eye infection (1424) | 5/5, 8179 | 2 | – | −/ + | VI | sei-hla-hlg-efb | |
| Healthy conjunctiva (N9OD) | 5/5, t010 | 2 | – | +/+ | VII | sei-hla-hlg-efb | |
| Wound infection (17/201) | 813/813, t10579 | 1 | – | +/+ | VII | sei-cna-hla-hlg | |
| Wound infection (262) | 813/291, t1149 | 1 | – | +/+ | VII | hlg | |
| Eye infection (186) | 22/22, t310 | 1 | + | +/+ | II | sei-cna-hla-hlg-efb | |
| Healthy conjunctiva (N61OD) | 22/22, t948 | UT∗ | 1 | + | +/+ | VII | sea-sei-hla-hlg-efb |
| Eye infection (481) | Singleton 1/580, t14911 | None | – | −/ + | V | sei-cna-hla-hlg-efb | |
| Eye infection (N297214) | Singleton 2/45, t302 | 1 | – | +/+ | VII | cna-hla-hlg | |
| Wound infection (2417) | Singleton 3/Unk, t021 | None | – | −/− | VI | sei-hla-hlg-efb | |
| Eye infection (1525,1545) | Singleton 5/72, t148 | 1 | – | −/ + | VI | sei-hla-hlg | |
| None | – | −/ + | V | sei-cna-hla-hlg-efb | |||
| Wound infection (1/229, 861) | Singleton 6/789, t091 | 1 | – | −/− | III | sei-cna-hla-hlg | |
| None | – | +/+ | III | sei-cna-hla-hlg-efb | |||
| Wound infection (379) | Singleton 6/789, t2505 | None | – | +/+ | III | sei-cna-hla-hlg-efb | |
| Eye infection (1603) | Singleton 6/789, t091 | 1 | – | +/+ | V | sei-hla-hlg-efb | |
| Eye infection (1320) | Singleton 7/6, t657 | 1 | – | +/+ | III | sei-cna-hla-hlg-efb | |
| Eye infection (1428) | Singleton 7/6, t4285 | 1 | – | −/ + | VIII | sea-sei-cna-hla-hlg-efb | |
| Eye infection (1698) | Singleton 7/6, t12406 | 1 | – | +/+ | VIII | sea-sei-cna-hla-hlg-efb | |
| Healthy conjunctiva (N21OS) | Singleton 8/15, t084 | 2 | – | +/+ | IV | sei-hla-efb | |
| Wound infection (2508) | Singleton 9/2885, t15579 | 4 | + | +/+ | III | sei-cna-hla-hlg-efb | |
| Wound infection (2653) | Singleton 10/672, t3841 | 2 | – | ± | I | sei-hla-hlg-efb | |
| Eye infection (N259615, N289378, 1049, 1506) | Singleton 10/672, t3841 | 1 | – | +/+ | I | sei-cna-hla-hlg | |
| None | – | +/+ | I | cna-hla-hlg | |||
| 1 | – | +/+ | VII | sei-hla-hlg-efb | |||
| 1 | – | +/+ | VIII | sei-hla-hlg-efb | |||
| Eye infection (188, 1164, 1355, 1670) | Singleton 10/672, t1309 | I | – | +/+ | I | sei-hla-hlg-efb | |
| I | – | −/ + | II | sei-hla-hlg-efb | |||
| I | – | +/+ | I | sei-cna-hla-hlg-efb | |||
| 1 | – | +/+ | VIII | sei-cna-hla-hlg | |||
| Eye infection (884,1333) | Singleton 11/2233, t2663 | 3 | + | +/+ | VII | sei-cna-hlg-efb | |
| +/+ | |||||||
| Eye infection (1716OD, 1758) | 3 | + | +/+ | IV | sei-cna-hla-hlg | ||
| +/+ | |||||||
| Eye infection (1716OS, 1769) | 3 | + | −/+ | VIII | sei-cna-hla-hlg | ||
| 4 | +/+ | ||||||
| Eye infection (915, 1366, 1729) | 3 | + | +/+ | VII | sei-cna-hla-hlg-efb | ||
| UT∗ | 3 | + | −/+ | VII | sei-hla-hlg | ||
| 3 | – | +/+ | VIII | sea-sei-cna-hla-hlg-efb |
MRSA: methicillin-resistant Staphylococcus aureus; MSSA: methicillin-sensitive Staphylococcus aureus; CC: clonal complex; ST: sequence type; SSC: Staphylococcal cassette chromosome; agr: accessory gene regulator; Unk: unknown; UT: untypeable; NT: non-typeable; ∗Isolates with ccrAIB1 complex but lack mec complex; pvl: Panton-valentine leucocidin; icaA: intracellular adhesion gene A; icaD: intracellular adhesion gene D; sea: staphylococcal enterotoxin A; sec: staphylococcal enterotoxin C; sei: staphylococcal enterotoxin I; cna: collagen adhesion; hlyA: hemolysin A; hlyG: hemolysin G; efb: extracellular fibronectin binding protein.
Ninety-one isolates comprising 26 MRSA and 65 MSSA were positive for both icaA and icaD genes, but five strains containing three MRSA and two MSSA were negative for both icaA and icaD genes. Two of the three MRSA isolates were positive for icaA gene, and another strain was positive for icaD gene. Similarly, nine of the 10 MSSA isolates were positive for icaD gene and one isolate for icaA gene, respectively (Table 4).
Also, a total of 25 toxin genes combinations was obtained with 109 strains belonging to 77 PFGE patterns, 32 sequence types (STs), 46 spa-types, and five agr-types. Twenty-three isolates belonging to five MRSA and 18 MSSA showed a toxin pattern comprising sei-cna-hla-hlg-efb genes. On the other hand, five MRSA and two MSSA showed another virulence pattern composed of sea-sec-sei-cna-hla-hlg-efb genes. The remaining isolates showed 23 different virulence gene patterns (Table 4).
Agr-Typing
Of 109 S. aureus strains, 40 (36.7%) isolates belong to agr-I, 31 (28.4%) isolates to agr-III, 18 (16.5%) to agr-II, and seven (6.4%) belong to agr-IV; however, 13 (11.9%) isolates were not typeable by the method employed (Table 4). Of the 32 MRSA isolates, 20 (62.5%) belong to agr-I, five (15.6%) to agr-II, three (9.4%) to agr-III, and remaining isolates were untypeable. On the other hand, 28 of 77 (36.4%) MSSA isolates belong to agr-III, 20 (25.9%) to agr-I, 13 (16.9%) to agr-II, seven (9%) to agr-IV, and nine (11.7%) isolates were untypeable. There was a good correlation between virulence patterns and specific molecular types (Table 4). The sea-sei-cna-hla-hlg-efb was the dominant virulence pattern shown by MRSA belonged to SCCmec type UT6, and agr type I, followed by sei-hla-hlg-efb and sei-cna-hla-hlg-efb pattern showed by MSSA isolates belonged to agr type I and III, respectively (Supplementary Table S1).
Spa-Typing
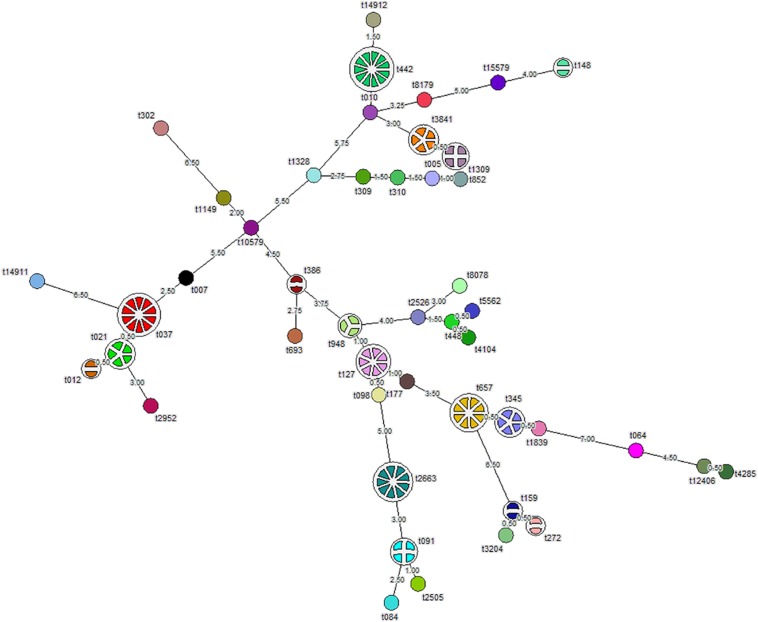
Analysis of the aligned sequence of the polymorphic X region of spa gene using the spa-typing plug-in tool of Bionumerics 7 software showed 46 spa-types (Figure 1). MST analysis classified the strains into six major clusters, seven minor clusters, and 30 singletons. We designated cluster as a minor cluster that contained less than five but more than two strains. Of the 109 S. aureus isolates, 11 (10%) isolates belong to spa-type t442, 10 (9%) to t037, nine (8.2%) to t2663, eight (7.3%) to t657, six (5.5%) to t127, five (4.5%) isolates each to t345 and t3841, and four isolates each belong to t021, t091, and t1309. In addition, four (3.6%) isolates belong to t1309, three (2.7%) isolates belong to t948, and two (1.8%) each belong to t148, t386, t012, t159, and t272, respectively. Moreover, one isolate each of 30 strains belong to single spa-types, namely, t15579, t8179, t14912, t010, t852, t005, t310, t309, t1328, t302, t1149, t10579, t007, t14911, t2952, t693, t2526, t8078, t5562, t448, t4104, t177, t098, t084, t2505, t3204, t1839, t064, t12406, and t4285 (Figure 1). Whereas 10 of 32 (31.2%) MRSA isolates belong to t037, 11 of 77 (14.3%) MSSA isolates belong to t442. S. aureus strain ATCC 25923 showed spa-type t948 along with three test isolates. We found two novel spa-types, namely, t14911 and t14912 among S. aureus strains after submission of nucleotide sequences to the Ridom spa server. Spa-type t14912 showed a close association with major spa-type t442, but 14911 spa-type was diverse and unrelated. One of the isolates was not assigned any spa-type (Figure 1).
FIGURE 1.
Minimum spanning tree (MST) showing 109 S. aureus isolates typed by spa-typing. Each node represents one spa-type, and the corresponding spa-type is given beside the node. The number of disks in a node indicates the number of isolates having a particular spa-type. The number provided on the string depicts the phylogenetic distance between two nodes. Length ≤ 1 is represented by dotted lines and more than one by solid lines.
Multi-Locus Sequence Typing (MLST)
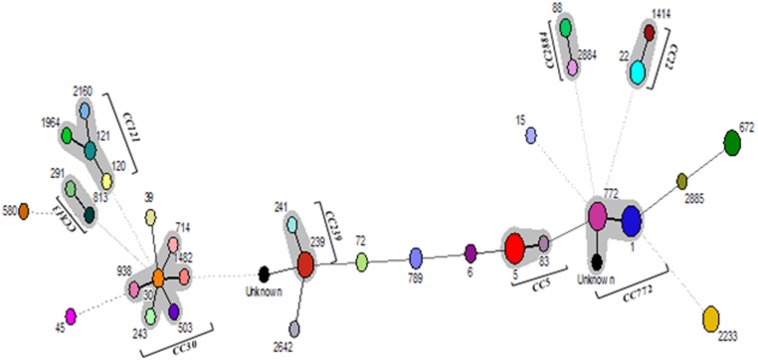
Multi-locus sequence typing of 109 S. aureus isolates showed 32 STs, eight CCs, and 12 singletons (Figure 2). The major ST comprised of ST1 (12.8%), ST5 (11.9%), ST772 (11%) followed by ST239 (9.2%), ST672 (8.3%), and ST2233 (8.3%). Also, we found two new allelic profiles designated as unknown not reported earlier among S. aureus strains (Supplementary Table S2). Of the eight CCs, CC5 contained 14 isolates, CC22 had eight isolates, CC30 had six isolates, CC121 had five isolates, CC239 had 11 isolates, CC772 had 26 isolates, CC813 had two isolates, and CC2884 contained four isolates, respectively. Of the major CCs, CC30 contained five STs, namely, ST30, ST503, ST714, ST938, and ST1482, CC121 contained four STs, namely, ST120, ST121, ST1964, ST2160, and CC772 had three STs, namely, ST772, ST1, and new unknown ST (Figure 2). Seven of the 32 (21.8%) of MRSA strains belong to ST239, spa-type t037, and SCCmec type UT6. However, 14 (18.2%) of MSSA strains possessing ST1 belong to different spa-types, namely, t127, t948, t177, t693, t098, and t386, of which few strains carry ccr complex but devoid of mec complex (Table 4). However, few isolates from eye infection and wound infection belong to CC239, ST239, and spa-type t037/t657. Reference strain of S. aureus ATCC25923 belonged to ST30 and CC30.
FIGURE 2.
Minimum spanning tree (MST) showing the relationship between different STs assigned by the analysis of MLST data. Each node represents one sequence type, and the corresponding ST is given beside the node. The size of each node is directly proportional to the number of isolates included in that ST. Bold lines connect types that are identical for six loci, solid lines connecting types identical for ≥four but ≤six locus, and dotted lines connecting STs differing from each other by ≥four genes out of seven gene locus.
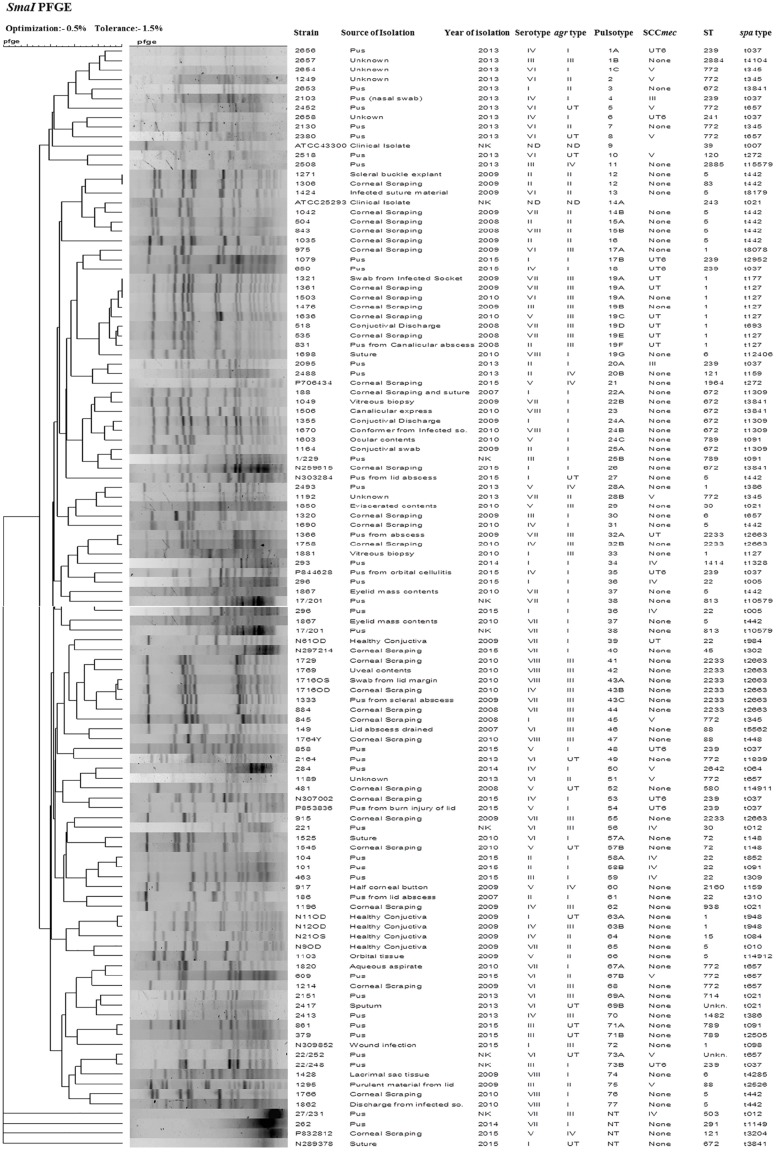
Pulsed-Field Gel Electrophoresis
SmaI-digested genomic DNA of S. aureus yielded bands classifying the 109 strains into 77 pulsotypes that includes two identical pairs (12 and 19A), three major clusters (1, 3, and 19), 17 minor clusters (14, 15, 17, 19, 20, 22, 24, 25, 28, 32, 57, 58, 63, 67, 69, 71, and 73), and 56 singletons. Four isolates were untypeable by the method employed. We found a total of 24 PFGE patterns among 32 MRSA isolates, of which one isolate was untypeable. Similarly, 77 MSSA isolates showed 53 PFGE patterns, of which three MSSA isolates were untypeable (Figure 3). MSSA isolates belonging to the major pulsotype 19 contained seven subtypes 19A, 19B, 19C, 19D, 19E, 19F, and 19G. These isolates were mostly from ocular infection and belong to ST1, agr type III, except one subtype 19G which belongs to ST6 and agr type I. S. aureus strain ATCC 25923 showed pulsotype 14. A dendrogram was generated using Bionumerics 7 software and percentage similarity with a cut-off of 80% and dice coefficients.
FIGURE 3.
Dendrogram representation (Dice coefficient) for macro-restriction banding patterns of S. aureus strains isolated from different sources with ATCC reference strains, generated by pulsed-field gel electrophoresis of total chromosomal DNA digested with SmaI restriction enzyme and correlation between their pulsotype, ST, spa-type, SCCmec type, and agr type with information regarding their source and year of isolation.
Statistical Analysis
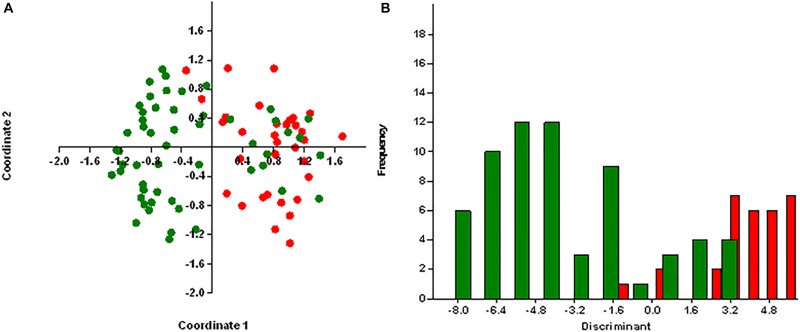
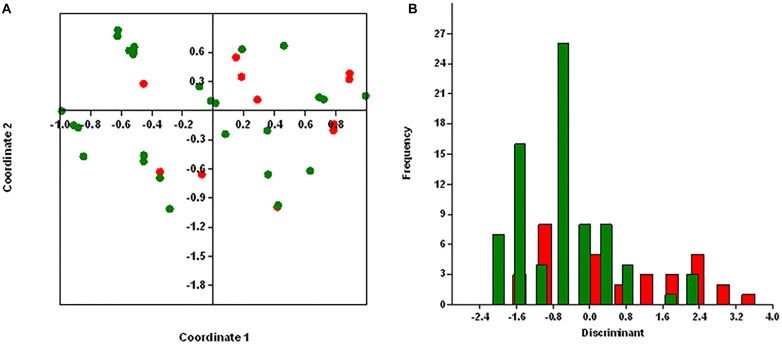
Principal coordinates analysis segregates MRSA and MSSA isolates, except for few isolates with 25.75% of explained variance for antibiotic resistance genes (Figure 4A) and 26% for virulence genes (Figure 5A). We used axis one for the highest percentage of representation. DA graph showed that MRSA isolates grouped within more positive values, whereas MSSA isolates grouped within negative values for both antibiotic resistance genes and virulence genes (Figures 4B, 5B). Predominant biomarkers were determined by calculating the coefficient of discriminant function and considered when the value was equal to 0.5 or >0.5. For antibiotic resistance genes, MRSA isolates are discriminating in the biomarker of resistance to ermA (0.8407), mphC (2.0167), tetK (2.3495), tetL (2.0604), and dfrA (1.3116), whereas the MSSA isolates were discriminating in resistance to aac(6’)/aph(2) (−0.351), aph3 (−2.7179), ermC (−0.8473), tetM (−0.522), cat:pC221 (−2.421), cat:pC223 (−6.601), dfrB (−0.443), and dfrG (−0.603). For virulence genes, MRSA isolates are discriminating in the biomarker of resistance to icaA (0.67169), seA (0.68593), seC (2.3245), cnA (0.90744), and hlA (0.54797). On the other hand, MSSA isolates were discriminating in resistance to icaD (−2.1945), seI (−0.58795), and hlG (−1.4999). PCoA and discriminant function of antibiotic resistance and virulence genes of S. aureus isolates with source and place of isolation was heterologous and complex (data not shown).
FIGURE 4.
Results obtained by statistical analysis using antibiotic resistance genes. (A) Principle component analysis (PCoA) of methicillin-resistant S. aureus (MRSA, red) and methicillin-susceptible S. aureus (MSSA, green). (B) Discriminant analysis of MRSA (red) and MSSA (green). Negative values belong to MSSA isolates and positive values to MRSA isolates.
FIGURE 5.
Results obtained by statistical analysis using virulence genes. (A) Principle component analysis (PCoA) of methicillin-resistant S. aureus (MRSA, red) and methicillin-susceptible S. aureus (MSSA, green). (B) Discriminant analysis of MRSA (red) and MSSA (green). Negative values belong to MSSA isolates and positive values to MRSA isolates.
Discussion
We used hexaplex PCR for detection of MRSA and MSSA isolates along with the presence of mecA, pvl, czrC, and qacA/B genes. We found a good correlation between oxacillin resistance and the presence of the mecA gene. However, one isolate showing resistance to oxacillin and lack mecA gene indicate the occurrence of different mechanism of methicillin resistance. Twenty of 31 MRSA and 23 of 77 MSSA isolates were positive for pvl gene indicating the prevalence of pvl gene among MRSA strains from the wound and eye infections. This finding is in contrast to those who did not find such correlation among clinical isolates (Shittu et al., 2011); therefore, it cannot be used as a reliable marker for MRSA. The presence of czrC and qacA/B genes among the number of MRSA isolates indicates their possible association with the mecA gene; however, further investigation is required to authenticate these findings.
Coagulase gene typing has been used to characterize S. aureus strains. Hwang and Kim (2007) showed the presence of coagulase serotype II among 54.4% MRSA and serotype VII among 30.9% MSSA. In contrast, we found serotype VII was present among 22% of MSSA isolates and serotype VI in 28.1% of MRSA isolates. These observations thus suggest that there is a difference in the presence of serotypes with regard to MRSA and MSSA.
Genetic determinants study among S. aureus showed a good correlation between resistance to aminoglycosides, chloramphenicol, clindamycin, erythromycin, trimethoprim, and tetracycline, and the presence of corresponding resistance genes. In this study, we found 85.3% strains showing resistance to chloramphenicol carried the pC221 gene; however, some of these strains also carried either pC223 or pC194 or both genes. Although one of 109 strains sensitive to chloramphenicol did not carry any of these genes, 13.8% strains showing sensitivity to chloramphenicol carried either pC221 or pC223 genes. These observations thus suggest that chloramphenicol sensitive strains carrying antibiotic resistance genes can develop resistance against this drug on exposure.
The aminoglycoside-modifying enzyme, encoded by aac (6’)-aph(2”) gene, is responsible for resistance against aminoglycosides (Vanhoof et al., 1994). Besides, two other genes encoding for aph(3.III) and ant(4, IV) are accountable for aminoglycoside resistance, but their frequency is less compared to aac(6’)-aph(2”) among staphylococci (Busch-SØRensen et al., 1996). In this study, we found 41.3% S. aureus possesses both aac(6’)-aph(2”) and aph (3, III) genes and 8.3% contained aph (3, III) gene and showed phenotypic resistance to gentamycin. These findings thus suggest that there are strains which harbor aminoglycoside resistance genes other than aac(6’)-aph(2”) and few strains had aph (3, III) only. At least 47.7% strains of S. aureus that were sensitive to aminoglycosides contain either aph (3, III) or aac(6’)-aph(2”) or both; however, three strains susceptible to gentamycin lack resistance genes. These findings are in contrast to those workers who reported that all aminoglycoside-resistant strains carried aac(6’)-aph(2”) (Price et al., 1981; Lovering et al., 1988; Dornbusch et al., 1990; Vanhoof et al., 1994; Martineau et al., 2000). The presence of aminoglycoside resistance gene among gentamycin sensitive isolates of S. aureus indicates that there is likely hood development of aminoglycoside resistance among S. aureus upon exposure to these drugs.
Similarly, 83.4% strains of S. aureus resistant to erythromycin harbored any of the four genes, namely, ermA, ermB, ermC, and msrA; however, an strain sensitive to erythromycin did not carry any of the genes. Previously, it was reported that the ermA gene is dominant among erythromycin resistance genes in S. aureus (Kaur and Chate, 2015). In contrast, we found the presence of the ermC gene in 83.4% strains compared to 49% of ermA gene. Kaur and Chate (2015) reported that majority of MRSA strains showed constitutive MLSB (cMLSB) resistance; however, two isolates had inducible MLSB (iMLSB) phenotype. In this study, 64.5% MRSA and 37.1% MSSA strains belong to iMLSB phenotype; however, 35.4% of MRSA and 43.5% of MSSA strains belong to cMLSB phenotype. This difference could be due to less number of MRSA isolates used in the study, and MSSA isolates were multidrug resistant. Seventeen strains showing sensitivity to erythromycin harbored one of the resistance genes, and one of the strains resistant to erythromycin did not possess any of the resistance genes to indicate that these strains are likely to develop resistance and mediated by an unknown mechanism.
About trimethoprim resistance, 67.8% strains harbored any of the three genes, namely, dfrA, dfrB, and dfrG. The remaining strains showing sensitivity to trimethoprim also carried all or one of the three genes. In this study, 73 of 74 trimethoprim resistance strains possess dfrG and dfrB genes; 45 strains carried the dfrA gene. These findings are in contrast to those who reported the presence of the dfrG gene in 92% strains, dfrA in 7% strains, and one strain carried a dfrB among trimethoprim resistance strains in a travel-associated skin and soft tissue infection study in Europe (Nurjadi et al., 2015).
Like other antibiotic resistance, 26.6% phenotypic resistance strains carried one or all the three tetracycline resistance genes, namely, tetK, tetL, and tetM. One of the strains sensitive to tetracycline was devoid of carrying any genes. However, the majority (69.7%) strain showing sensitivity to tetracycline carried one or all three resistance genes indicating that these isolates could develop resistance after exposure to an antibiotic. From this study, it is clear that erythromycin and gentamicin were least active; however, vancomycin and clindamycin were the most effective drugs. These results corroborate the finding of Pai et al. (2010), who also reported that vancomycin and clindamycin are the most effective drugs.
SCCmec type V was predominant type among MRSA strains followed by SCCmec type UT6, IV, and III, respectively. This finding is similar to Nadig et al. (2012), who also reported the prevalence of SCCmec type V among isolates from eye infections. To our knowledge, we are the first to inform of the presence of SCCmec type UT6 among S. aureus from India. The combination of SCCmec IV, V, and pvl gene was reported as the genetic markers for a community-associated MRSA (Bhutia et al., 2015). Similarly, our study showed the presence of SCCmec V (40.6%), IV (21.9%), and pvl (64.5%); therefore it can be used as a marker for hospital-associated infections. However, new UT6 SCCmec type is emerging in India. Many untypeable strains carried ccr complex but no mec complex. This observation thus suggests the ability of such strains to acquire mec complex and became a known or unknown SCCmec type.
A total of 25 unique toxin combination was found among S. aureus strains, of which at least one toxin gene was present in a given strain. Sotto et al. (2008) reported the presence of sei and sea genes in S. aureus isolated from diabetic foot ulcer. Similarly, we found the presence of sei and sea genes in both MRSA and MSSA strains. Although we noted the high percentage of hlg (98%) and hla (92.6%) among in S. aureus comprising both MSSA and MRSA, other workers reported the presence of these genes in mupirocin resistant in MRSA isolates in China (Liu et al., 2012). Moreover, the distribution of virulence genes with regards to source and place of isolation was complex. Gowrishankar et al. (2016) reported the isolation of 84% MRSA strains carrying icaADBC genes from patients with pharyngitis. Also, in this study, 81.3% MRSA and 84.4% MSSA carrying icaA/icaD genes were isolated from the eye and wound infections (Supplementary Table S3). Absence of icaA/icaD genes in S. aureus strains was similar to those of the previous report (Agarwal and Jain, 2013).
Several molecular genotyping tools are used to trace the origin of the strain, and distribution of CC with regard to methicillin-resistant, methicillin-sensitive, sources and place of isolation. We determined the population structure of S. aureus isolated from ocular and wound infections from different parts of India using MLST, agr-typing, spa-typing, and PFGE.
Multi-locus sequence typing analysis showed the presence of six major ST(s) comprising ST1, ST5, ST772, ST239, ST672, and ST2233, respectively. While ST239-MRSA-UT6 was the typical type among MRSA isolates from wound infection, ST772-MRSA-V were from eye infections (Nadig et al., 2012). Similarly, ST772-SCCmec-V were reported slowly replacing multidrug resistant ST239-SCCmec-III in Asian studies (D’Souza et al., 2010). This finding is in contrast to Suzuki et al. (2012), who reported the presence of ST5 and ST764 among MRSA strains from the infected eye and healthy conjunctiva sacs. Also, Mohammadi et al. (2014) showed emergence of SCCmec-III with variable antimicrobial resistance profiles in Iran. We found ST772-MRSA-V with spa-type t345 and t657 belonging to dominant CC772 among wound infection isolates. Besides, we reported two new spa-types among S. aureus strains from India.
There were eight CCs, namely, CC30, CC121, CC772, CC813, CC239, CC28841, CC22, and CC5 present among S. aureus represents different PFGE clusters. CC30 and CC121 comprising different STs were almost equally distributed among MRSA and MSSA isolates. Whereas CC772-ST772 was dominant among MRSA, CC772-ST1 was prevalent among MSSA isolates. Similarly, CC239-ST239 and CC22-ST22 were prevalent among MRSA isolates and CC5-ST5, CC813-ST813, and CC28841-ST28841 were more commonly found in MSSA isolates. The prevalent CC among Varanasi isolates (mostly wound infection) were CC772 followed by CC239 besides the presence of CC30, and CC121. However, isolates from wound infection from Delhi showed varied results. Whereas AIIMS isolates showed CC CC30, CC22, and CC813, UCMS isolates showed the presence of CC239 and CC22 CCs. Interestingly isolates from Hyderabad (eye infection) had CC239 but isolates from Bhubaneswar (eye infection) showed the presence of CC772, CC5, CC2884, and CC30.
Mobasherizadeh et al. (2019) reported the prevalence of CC5 and CC30 and other CCs among MRSA isolates isolated from nasal carriage in Iranian hospitals. Similarly, CC8, CC121, CC1, CC45, and CC5 were reported in MRSA isolates from Malaysia (Ghasemzadeh-Moghaddam et al., 2011). These observations indicate the existence of different CCs in India and Asian countries. MLST and spa-typing was better than PFGE and toxin genotyping a finding unusual from those who reported a good correlation between various typing schemes. Overall, there was diversity in genotypes, antimicrobial resistance, and virulence determinants among MRSA and MSSA strains.
From this study, it is clear that S. aureus strains sensitive to antibiotics but carried antibiotic resistance genes could develop resistance upon exposure to antibiotic(s), and vancomycin and clindamycin were the most effective drugs. ST239-SCCmec UT6/t035 were dominant clones among S. aureus. There was diversity in genotypes, antimicrobial resistance, and virulence determinants among MRSA and MSSA strains, therefore suggests continuous surveillance of multidrug-resistant strains circulating in the community/hospitals in India, to take adequate measures to control the infection.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Review Board (IRB) of LV Prasad Eye Institute (LEC/08/110/2009) and by the Institute Ethics Sub-Committee (IESC) of All India Institute of Medical Sciences, New Delhi (IESC/T-34/2013), and the data were analyzed anonymously and reported. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
SA, SJ, SS, and DS conceived the experiments. SA, SJ, and SP conducted the experiments. SA, SJ, SP, SS, BD, GN, NS, and DS analyzed the results. KN performed statistical analysis. SA, SJ, and DS wrote the manuscript. All authors reviewed and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
SA and SJ are grateful to the Institute of Life Sciences, Bhubaneswar and Department of Science and Technology, New Delhi, respectively for providing Senior Research Fellowship.
Footnotes
Funding. This study was supported by the Department of Science and Technology (Grant No. SR/SO/HS-117 to DS). This study, in part, was also supported by the fund contributed by the Department of Biotechnology, New Delhi, to the Institute of Life Sciences, Bhubaneswar. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02763/full#supplementary-material
References
- Agarwal A., Jain A. (2013). Glucose & sodium chloride induced biofilm production & ica operon in clinical isolates of staphylococci. Indian J. Med. Res. 138:262. [PMC free article] [PubMed] [Google Scholar]
- Arciola C. R., Baldassarri L., Montanaro L. (2001). Presence of icaA and icaD genes and slime production in a collection of Staphylococcal strains from catheter-associated infections. J. Clin. Microbiol. 39 2151–2156. 10.1128/JCM.39.6.2151-2156.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argudín M., Tenhagen B.-A., Fetsch A., Sachsenröder J., Käsbohrer A., Schroeter A., et al. (2011). Virulence and resistance determinants of German Staphylococcus aureus ST398 isolates from nonhuman sources. Appl. Environ. Microbiol. 77 3052–3060. 10.1128/AEM.02260-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranovich T., Zaraket H., Shabana I., Nevzorova V., Turcutyuicov V., Suzuki H. (2010). Molecular characterization and susceptibility of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from hospitals and the community in Vladivostok, Russia. Clin. Microbiol. Infect. 16 575–582. 10.1111/j.1469-0691.2009.02891.x [DOI] [PubMed] [Google Scholar]
- Bhutia K. O., Singh T., Adhikari L., Biswas S. (2015). Molecular characterization of community-& hospital-acquired methicillin-resistant & methicillin-sensitive Staphylococcus aureus isolates in Sikkim. Indian J. Med. Res. 142:330. 10.4103/0971-5916.166600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch-SØRensen C., SØNmezoglu M., Frimodt-Moller N., Hojbjerg T., Miller G. H., Espersen F. (1996). Aminoglycoside resistance mechanisms in Enterobacteriaceae and Pseudomonas spp. from two Danish hospitals: correlation with type of aminoglycoside used. Apmis 104 763–768. 10.1111/j.1699-0463.1996.tb04940.x [DOI] [PubMed] [Google Scholar]
- Dhawan B., Rao C., Udo E., Gadepalli R., Vishnubhatla S., Kapil A. (2015). Dissemination of methicillin-resistant Staphylococcus aureus SCCmec type IV and SCCmec type V epidemic clones in a tertiary hospital: challenge to infection control. Epidemiol. Infect. 143 343–353. 10.1017/S095026881400065X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornbusch K., Miller G., Hare R., Shaw K., Group E. S. (1990). Resistance to aminoglycoside antibiotics in gram-negative bacilli and staphylococci isolated from blood. Report from a European collaborative study. J. Antimicrob. Chemother. 26 131–144. 10.1093/jac/26.1.131 [DOI] [PubMed] [Google Scholar]
- D’Souza N., Rodrigues C., Mehta A. (2010). Molecular characterization of methicillin-resistant Staphylococcus aureus with emergence of epidemic clones of sequence type (ST) 22 and ST 772 in Mumbai, India. J. Clin. Microbiol. 48 1806–1811. 10.1128/JCM.01867-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran N., Ozer B., Duran G. G., Onlen Y., Demir C. (2012). Antibiotic resistance genes & susceptibility patterns in staphylococci. Indian J. Med. Res. 135:389. [PMC free article] [PubMed] [Google Scholar]
- Enright M. C., Spratt B. G. (1999). Multilocus sequence typing. Trends Microbiol. 7 482–487. 10.1016/S0966-842X(99)01609-1 [DOI] [PubMed] [Google Scholar]
- Freidlin J., Acharya N., Lietman T. M., Cevallos V., Whitcher J. P., Margolis T. P. (2007). Spectrum of eye disease caused by methicillin-resistant Staphylococcus aureus. Am. J. Ophthalmol. 144 313–315. 10.1016/j.ajo.2007.03.032 [DOI] [PubMed] [Google Scholar]
- Ghasemzadeh-Moghaddam H., Ghaznavi-Rad E., Sekawi Z., Yun-Khoon L., Aziz M. N., Hamat R. A., et al. (2011). Methicillin-susceptible Staphylococcus aureus from clinical and community sources are genetically diverse. Int. J. Med. Microbiol. 301 347–353. 10.1016/j.ijmm.2010.10.004 [DOI] [PubMed] [Google Scholar]
- Ghaznavi-Rad E., Goering R., Shamsudin M. N., Weng P., Sekawi Z., Tavakol M., et al. (2011). mec-associated dru typing in the epidemiological analysis of ST239 MRSA in Malaysia. Eu. J. Clin Microbiol. Infect Dis. 30 1365–1369. 10.1007/s10096-011-1230-1 [DOI] [PubMed] [Google Scholar]
- Ghebremedhin B., Olugbosi M., Raji A., Layer F., Bakare R., König B., et al. (2009). Emergence of a community-associated methicillin-resistant Staphylococcus aureus strain with a unique resistance profile in Southwest Nigeria. J. Clin. Microbiol. 47 2975–2980. 10.1128/JCM.00648-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilot P., Lina G., Cochard T., Poutrel B. (2002). Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J. Clin. Microbiol. 40 4060–4067. 10.1128/JCM.40.11.4060-4067.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godebo G., Kibru G., Tassew H. (2013). Multidrug-resistant bacterial isolates in infected wounds at Jimma University Specialized Hospital, Ethiopia. Annals Clin. Microbiol. Antimicrob. 12:17. 10.1186/1476-0711-12-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh S.-H., Byrne S., Zhang J., Chow A. (1992). Molecular typing of Staphylococcus aureus on the basis of coagulase gene polymorphisms. J. Clin. Microbiol. 30 1642–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar S., Kamaladevi A., Balamurugan K., Pandian S. K. (2016). In vitro and in vivo biofilm characterization of methicillin-resistant Staphylococcus aureus from patients associated with pharyngitis infection. Biomed. Res. Int. 2016 1–14. 10.1155/2016/1289157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Ø, Harper D., Ryan P. (2001). Paleontological statistics software: package for education and data analysis. Palaeontol. Electron. 4:9. [Google Scholar]
- Hirotaki S., Sasaki T., Kuwahara-Arai K., Hiramatsu K. (2011). Rapid and accurate identification of human-associated staphylococci by use of multiplex PCR. J. Clin. Microbiol. 49 3627–3631. 10.1128/JCM.00488-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu L.-Y., Koh T.-H., Singh K., Kang M.-L., Kurup A., Tan B.-H. (2005). Dissemination of multisusceptible methicillin-resistant Staphylococcus aureus in Singapore. J. Clin. Microbiol. 43 2923–2925. 10.1128/JCM.43.6.2923-2925.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S.-M., Kim T.-U. (2007). Changes in coagulase serotype of Staphylococcus aureus isolates in Busan, 1994-2005. Korean J. Microbiol. 43 346–350. [Google Scholar]
- Jarraud S., Mougel C., Thioulouse J., Lina G., Meugnier H., Forey F., et al. (2002). Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. immun. 70 631–641. 10.1128/IAI.70.2.631-641.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur D. C., Chate S. S. (2015). Study of antibiotic resistance pattern in methicillin resistant Staphylococcus aureus with special reference to newer antibiotic. J. Glob. Infect. Dis. 7 78–84. 10.4103/0974-777X.157245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-S., Song W., Kim H.-S., Cho H. C., Lee K. M., Choi M.-S., et al. (2006). Association between the methicillin resistance of clinical isolates of Staphylococcus aureus, their staphylococcal cassette chromosome mec (SCCmec) subtype classification, and their toxin gene profiles. Diagn. Microbiol. Infect. Dis. 56 289–295. 10.1016/j.diagmicrobio.2006.05.003 [DOI] [PubMed] [Google Scholar]
- Kondo Y., Ito T., Ma X. X., Watanabe S., Kreiswirth B. N., Etienne J., et al. (2007). Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51 264–274. 10.1128/AAC.00165-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay J. A., Holden M. T. (2004). Staphylococcus aureus: superbug, super genome? Trends Microbiol. 12 378–385. 10.1016/j.tim.2004.06.004 [DOI] [PubMed] [Google Scholar]
- Liu Q., Han L., Li B., Sun J., Ni Y. (2012). Virulence characteristic and MLST-agr genetic background of high-level mupirocin-resistant, MRSA isolates from Shanghai and Wenzhou, China. PLoS One 7:e37005. 10.1371/journal.pone.0037005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wang H., Du N., Shen E., Chen H., Niu J., et al. (2009). Molecular evidence for spread of two major methicillin-resistant Staphylococcus aureus clones with a unique geographic distribution in Chinese hospitals. Antimicrob. Agents Chemother. 53 512–518. 10.1128/AAC.00804-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovering A., Bywater M., Holt H., Champion H., Reeves D. (1988). Resistance of bacterial pathogens to four aminoglycosides and six other antibacterials and prevalence of aminoglycoside modifying enzymes, in 20 UK centres. J. Antimicrob. Chemother. 22 823–839. 10.1093/jac/22.6.823 [DOI] [PubMed] [Google Scholar]
- Martineau F., Picard F. J., Lansac N., Ménard C., Roy P. H., Ouellette M., et al. (2000). Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 44 231–238. 10.1128/aac.44.2.231-238.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslow J. N., Mulligan M. E., Arbeit R. D. (1993). Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin. Infect. Dis. 17 153–162. 10.1093/clinids/17.2.153 [DOI] [PubMed] [Google Scholar]
- Mehrotra M., Wang G., Johnson W. M. (2000). Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J. Clin. Microbiol. 38 1032–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell G., Séguin D. L., Asselin A.-E., Déziel E., Cantin A. M., Frost E. H., et al. (2010). Staphylococcus aureus sigma B-dependent emergence of small-colony variants and biofilm production following exposure to Pseudomonas aeruginosa 4-hydroxy-2-heptylquinoline-N-oxide. BMC Microbiol. 10:33. 10.1186/1471-2180-10-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobasherizadeh S., Shojaei H., Azadi D., Havaei A. A., Rostami S. (2019). Molecular characterization and genotyping of methicillin-resistant Staphylococcus aureus in nasal carriage of healthy Iranian children. J. Med. Microbiol. 68 374–378. 10.1099/jmm.0.000924 [DOI] [PubMed] [Google Scholar]
- Mohammadi S., Sekawi Z., Monjezi A., Maleki M.-H., Soroush S., Sadeghifard N., et al. (2014). Emergence of SCCmec type III with variable antimicrobial resistance profiles and spa types among methicillin-resistant Staphylococcus aureus isolated from healthcare- and community-acquired infections in the west of Iran. Int. J. Infect. Dis. 25 152–158. 10.1016/j.ijid.2014.02.018 [DOI] [PubMed] [Google Scholar]
- Monday S. R., Bohach G. A. (1999). Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J. Clin. Microbiol. 37 3411–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadig S., Velusamy N., Lalitha P., Kar S., Sharma S., Arakere G. (2012). Staphylococcus aureus eye infections in two Indian hospitals: emergence of ST772 as a major clone. Clin. Ophthalmol. 6 165–173. 10.2147/OPTH.S23878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J. L., Rice K. C., Slater S. R., Fox P. M., Archer G. L., Bayles K. W., et al. (2007). Vancomycin-intermediate Staphylococcus aureus strains have impaired acetate catabolism: implications for polysaccharide intercellular adhesin synthesis and autolysis. Antimicrob. Agents Chemother. 51 616–622. 10.1128/AAC.01057-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norazah A., Liew S., Kamel A., Koh Y., Lim V. (2001). DNA fingerprinting of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis (PFGE): comparison of strains from 2 Malaysian hospitals. Singapore Med. J. 42 015–019. [PubMed] [Google Scholar]
- Nurjadi D., Schäfer J., Friedrich-Jänicke B., Mueller A., Neumayr A., Calvo-Cano A., et al. (2015). Predominance of dfrG as determinant of trimethoprim resistance in imported Staphylococcus aureus. Clin. Microbiol. Infect. 21 1095.e5–1095.e9. 10.1016/j.cmi.2015.08.021 [DOI] [PubMed] [Google Scholar]
- Pai V., Rao V. I., Rao S. P. (2010). Prevalence and antimicrobial susceptibility pattern of methicillin-resistant Staphylococcus aureus [MRSA] isolates at a tertiary care hospital in Mangalore South India. J. Lab. Physicians 2:82. 10.4103/0974-2727.72155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S., Kar S., Choudhury R., Sharma S., Singh D. V. (2014). Development and evaluation of hexaplex PCR for rapid detection of methicillin, cadmium/zinc and antiseptic-resistant staphylococci, with simultaneous identification of PVL-positive and-negative Staphylococcus aureus and coagulase negative staphylococci. FEMS Microbiol. Lett. 352 114–122. 10.1111/1574-6968.12383 [DOI] [PubMed] [Google Scholar]
- Panda S., Kar S., Sharma S., Singh D. V. (2016). Multidrug-resistant Staphylococcus haemolyticus isolates from infected eyes and healthy conjunctivae in India. J. Glob. Antimicrob. Res. 6 154–159. 10.1016/j.jgar.2016.05.006 [DOI] [PubMed] [Google Scholar]
- Paniagua-Contreras G., Sáinz-Espuñes T., Monroy-Pérez E., Rodríguez-Moctezuma J. R., Arenas-Aranda D., Negrete-Abascal E., et al. (2012). Virulence markers in Staphylococcus aureus strains isolated from hemodialysis catheters of Mexican patients. Adv. Microbiol. 2:476. 10.4236/aim.2012.24061 24424465 [DOI] [Google Scholar]
- Price K., Kresel P., Farchione L., Siskin S., Karpow S. (1981). Epidemiological studies of aminoglycoside resistance in the USA. J. Antimicrob. Chemother. 8(Suppl. A), 89–105. 10.1093/jac/8.suppl-A.89 [DOI] [PubMed] [Google Scholar]
- Schmitz F.-J., Fluit A. C., Gondolf M., Beyrau R., Lindenlauf E., Verhoef J., et al. (1999). The prevalence of aminoglycoside resistance and corresponding resistance genes in clinical isolates of staphylococci from 19 European hospitals. J. Antimicrob. Chemother. 43 253–259. 10.1093/jac/43.2.253 [DOI] [PubMed] [Google Scholar]
- Shanmuganathan V., Armstrong M., Buller A., Tullo A. (2005). External ocular infections due to methicillin-resistant Staphylococcus aureus (MRSA). Eye 19 284–291. 10.1038/sj.eye.6701465 [DOI] [PubMed] [Google Scholar]
- Shittu A. O., Okon K., Adesida S., Oyedara O., Witte W., Strommenger B., et al. (2011). Antibiotic resistance and molecular epidemiology of Staphylococcus aureus in Nigeria. BMC Microbiol. 11:92. 10.1186/1471-2180-11-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S. K., Karow M. E., Brady J. M., Stemper M. E., Kislow J., Moore N., et al. (2010). Virulence genes and genotypic associations in nasal carriage, community-associated methicillin-susceptible and methicillin-resistant USA400 Staphylococcus aureus isolates. J. Clin. Microbiol. 48 3582–3592. 10.1128/JCM.00657-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotto A., Lina G., Richard J.-L., Combescure C., Bourg G., Vidal L., et al. (2008). Virulence potential of Staphylococcus aureus strains isolated from diabetic foot ulcers: a new paradigm. Diabetes Care 31 2318–2324. 10.2337/dc08-1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Hayashi S., Ohashi Y. (2012). Genotypic characterization of staphylococcus aureus isolates from eyes with keratitis. Invest. Ophthalmol. Vis. Sci. 53 6139–6139. [Google Scholar]
- Tenover F. C., Arbeit R., Archer G., Biddle J., Byrne S., Goering R., et al. (1994). Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J. Clin. Microbiol. 32 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Belkum A., Tassios P., Dijkshoorn L., Haeggman S., Cookson B., Fry N., et al. (2007). Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13 1–46. 10.1111/j.1469-0691.2007.01786.x [DOI] [PubMed] [Google Scholar]
- Vanhoof R., Godard C., Content J., Nyssen H. (1994). Detection by polymerase chain reaction of genes encoding aminoglycoside-modifying enzymes in methicillin-resistant Staphylococcus aureus isolates of epidemic phage types. J. Med. Microbiol. 41 282–290. 10.1099/00222615-41-4-282 [DOI] [PubMed] [Google Scholar]
- Wang L., Hu Z., Hu Y., Tian B., Li J., Wang F., et al. (2013). Molecular analysis and frequency of Staphylococcus aureus virulence genes isolated from bloodstream infections in a teaching hospital in Tianjin. China Genet. Mol. Res. 12 646–654. 10.4238/2013.March.11.12 [DOI] [PubMed] [Google Scholar]
- Zecconi A., Cesaris L., Liandris E., Dapra V., Piccinini R. (2006). Role of several Staphylococcus aureus virulence factors on the inflammatory response in bovine mammary gland. Microb. Pathogen. 40 177–183. 10.1016/j.micpath.2006.01.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.