Abstract
Background:
Adrenocortical carcinoma (ACC) is an aggressive malignancy with a low but variable overall survival rate. The role of long, noncoding RNAs (lncRNAs) in ACC is poorly understood. Thus, in this study we performed lncRNA expression profiling in ACCs, adrenocortical adenomas (ACA), and normal adrenal cortex (NAC).
Methods:
LncRNA expression profile using ArrayStar Human LncRNA/mRNA Expression Microarray V3.0 was analyzed in samples from 11 ACA, 9 ACC, and 5 NAC. Differentially expressed lncRNAs were validated using TaqMan, real-time quantitative PCR with additional samples. The dataset from the ACC Cancer Genome Atlas (TCGA) project was used to evaluate the prognostic utility of lncRNAs.
Results:
Unsupervised hierarchical clustering showed distinct clustering of ACC samples compared with NAC and ACA samples by lncRNA expression profiles. A total of 874 lncRNAs were differentially expressed between ACC and NAC. LINC00271 expression level was associated with prognosis, patients with low LINC00271 expression survived a shorter time than patients with high LINC00271 expression. Low LINC00271 expression was positively associated with WNT signaling, cell cycle, and chromosome segregation pathways.
Conclusions:
ACC has a distinct lncRNA expression profile. LINC00271 is downregulated in ACC and appears to be involved in biologic pathways commonly dysregulated in ACC.
Introduction
Adrenocortical carcinoma (ACC) is a rare and aggressive malignancy with an annual incidence of 0.7–.0 cases per million people and a five-year overall survival rate ranging from 32% to 47%).1,2 Furthermore, even after complete tumor resection, over half of the patients develop recurrent disease.3 Patients with locally advanced and metastatic ACC often undergo therapy with a regimen including adrenolytic mitotane plus combination chemotherapy with etoposide, doxorubicin, and cisplatin. Unfortunately, this regimen has very limited therapeutic benefit.4 The role of adjuvant therapy for ACC is controversial because of questionable therapeutic benefit of current agents and the heterogenous prognosis.3 Understanding the mechanism behind the initiation and progression of ACC could help in identifying diagnostic and prognostic markers, and therapeutic targets.
Several genomic studies of ACC have reported a distinct, ACC genome-wide gene expression and alteration profiles of micro-RNA expression, methylation, and copy number compared with adrenal cortical adenomas (ACAs) and normal adrenal cortex (NAC).5–10 These studies have led to the molecular classification of ACC that is relevant for predicting prognosis. Recently, long, noncoding RNAs (lncRNAs) have been suggested to be dysregulated in ACC.11 LncRNAs are RNA transcripts longer than 200 nucleotides that do not encode protein and are localized in the cell nucleus or cytoplasm.12 The expression of lncRNAs is more tissue-specific than protein-coding genes, and they function as decoys, scaffolds, and enhancer RNAs and are involved in chromatin remodeling, as well as transcriptional and post-transcriptional regulation.13
To our knowledge, the study by Glover and colleagues has been the only study that has investigated lncRNA expression profile in ACCs, ACAs, and NAC.11 These investigators reported that the greatest number of differentially expressed lncRNAs were between ACAs and NAC, with almost 3-fold less lncRNAs being differentially expressed between ACCs and NAC. This finding suggested that changes in lncRNA expression could be an early event in the pathogenesis of both ACC and ACAs. This finding, however, is in contrast to the results of previous, genome-wide analyses that demonstrated a multistep progression in ACC, with increasing genomic changes from NAC to ACA to ACC.8 Therefore, to further our knowledge of the role of lncRNA in ACCs, we performed lncRNA expression profiling using lncRNA microarrays to identify differentially expressed lncRNAs in ACCs compared with NACs and ACAs. We also investigated whether lncRNA expression levels were associated with overall survival times of ACCs.
Materials and methods
Tissue samples
Patient tumor tissues were procured after informed consent for genetic studies on a procurement clinical protocol pproved by our Institutional Review Board( and ). The tissues were immediately snap frozen in liquid nitrogen and stored at −80°C. For this study, we used 11 ACA samples and nine ACC samples. Five normal NACs were obtained at the time of organ donation harvesting. These 25 tissue samples were used for lncRNA microarray profiling. In addition to these samples, an additional 10 ACC samples were included in the quantitative RT-PCR (qRT-PCR) validation (Table 1). Tumors were classified as benign when the Weiss criteria scores were less than 3 (all the benign samples included had a Weiss score of 0), and tumors were classified as ACC, when the Weiss criteria scores were more than or equal to 3.14 Only samples with at least 80% tumor cells were included for analysis.
Table 1.
Clinical features of ACA and ACC patients
| АСА* | ACC† included in microarrav | ACC in validation cohort | |
|---|---|---|---|
| Number of patients | 11 | 9 | 10 |
| Age (average ± SD) | 46 years ± 19 | 52 years ± 15 | 47 years ±14 |
| Sex (female/male) | 9/2 | 7/2 | 6/4 |
| Tumor size (average ± SD) | 3.8 cm ± 1.8 | 6.7 cm ± 5.9 | 5.4 cm ± 2.2 |
| Functional | 55% | 44% | 30% |
| Syndrome‡ | |||
| Adrenal | 3 | 4 | 3 |
| hypercortisolism | |||
| Primary | 3 | 1 | 0 |
| hyperaldosteronism | |||
| Nonfunctioning | 6 | 4 | 7 |
ACA, adrenocortical adenoma
ACC, adrenocortical carcinoma
Functional status at initial presentation
RNA extraction
Total RNA was extracted from fresh frozen tissue samples using an RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Englewood, CO, USA). Only samples with a minimum RNA integrity number of seven were included for analysis.
Microarray profiling
The ArrayStar Human LncRNA/mRNA Expression Microarray Version 3.0 (ArrayStar, Inc., Rockville, MD, USA) used for lncRNA profiling includes 30,586 lncRNA probes and 26,109 coding transcripts,. RNA labeling, microarray hybridization, slide washing, and scanning were performed based on the standard protocols of the ArrayStar. Agilent Feature Extraction software (version 11.0.1.1) which was used to analyze acquired array images. The microarray specifications and derived data are accessible through National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) accession number GSE124531.
TaqMan real-time quantitative PCR
RNA was reverse transcribed using the High Capacity, cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA). TaqMan qRT-PCR was performed using the 7900HT, fast, real-time PCR systems (Applied Biosystems). The reaction contained cDNA, TaqMan 2×universal PCR master mix, and TaqMan gene expression assays primers (Applied Biosystems). LncRNAs were selected for validation based on three criteria: 1) availability of validated TaqMan gene expression primer/probe assays, 2) possible role in cancer, and 3) magnitude of differential expression. The gene expression assays used were: HOTTIP (Hs03649396_m1), CHL1 (Hs04332026_m1), HOXA11-AS1 (Hs_03454334_g1), CRNDE (HS04404483_m1), LINC00271 (Hs03657384_m1), FAM211A-AS1 (Hs03678558_g1), TBXAS1 (Hs01096058_s1) and GAPDH (Hs99999905_m1).
Comparative genomic hybridization (CGH) array analysis
We used our previously published, genome-wide, CGH array data in a cohort of NAC, ACA, and ACC.8 The LINC100271 site was scanned manually for its copy number status using Nexus software.
Statistical and data analysis
LncRNA expression profiles of ACC samples were compared with NAC and ACA samples. The Gaussian linear model was used to calculate P-values, and false discovery rates (FDRs) were calculated using the Benjamini-Hochberg method for each lncRNA. LncRNAs with a log2 fold change ≥2 and an FDR <0.05 were defined as differentially expressed lncRNAs. Differentially expressed lncRNAs were mapped to their associated gene names, and then a gene set enrichment analysis (GSEA) was performed on these genes. An in-house. R package, OmicPath (v 0.1) was used to perform the GSEA to discover potential KEGG pathway associations for each set of differentially expressed lncRNAs. Pathways with a P-value < 0.05 were considered statistically significant. Survival curves were plotted using the Kaplan-Meier methods, and differences in survival rates were determined using the log-rank test. These statistical analyses were done with GraphPad Software with P < 0.05 considered statistically significant.
The ACC cohort from the project database of the Cancer Genome Atlas (TCGA) (https://tcga-data.nci.nih.gov/tcga/) which included 79 patients with HOTTIP, CHL1, HOXA11-AS1, CRNDE, LINC00271, FAM211A-AS1 and TBXAS1 expression data, as well as follow-up information, were used to study the prognostic importance of lncRNAs. For the overall survival analysis, two groups were defined based on the lncRNA expression levels in the primary tumor. Those with a lncRNA level ranked in the top half were classified into the high expression group and the rest into the low expression group based on the median value.
The gene expression profiles of ACC samples deposited in the TCGA project database were analyzed to compare expression patterns in tumors with high (n = 39) vs. low LINC100271 expression (n = 40). The downloaded data consisted of quantified gene expression data that were further processed using the DESeq2 package.15 The differentialy expressed genes were annotated, and GSEA analysis was performed using the clusterProfiler package.16
Results
Differentially expressed lncRNAs in ACC versus NAC
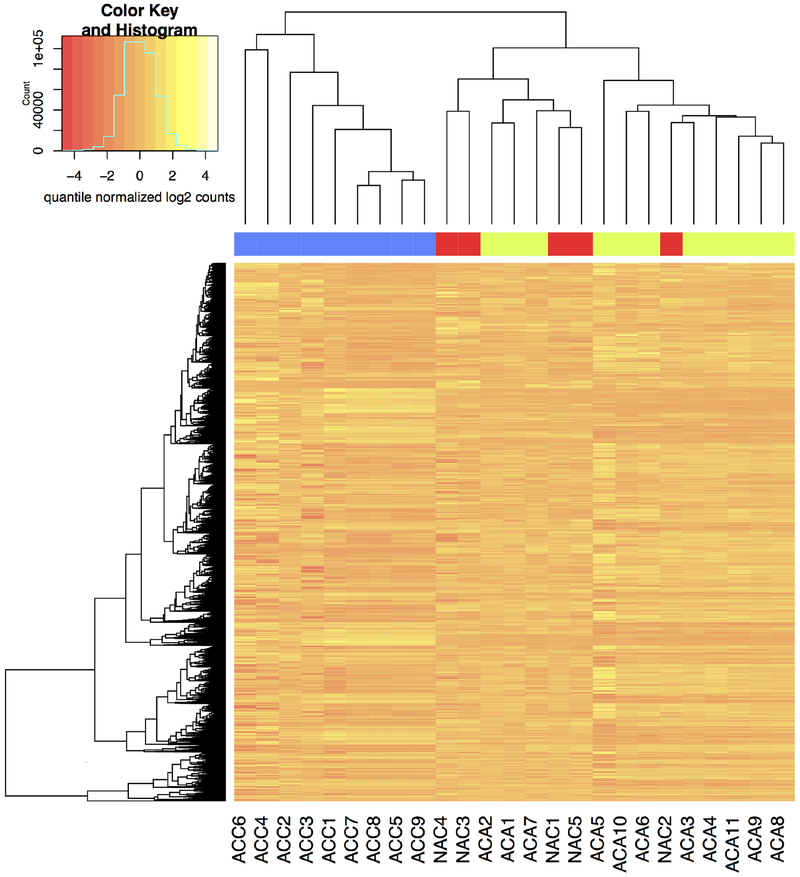
Unsupervised hierarchical and heat map clustering showed distinct clustering of ACC samples compared with NAC and ACA samples (Fig. 1). In these samples, 874 lncRNAs were differentially expressed in ACC compared with NAC, of which 409 were upregulated, and 465 were downregulated. The 874 differentially expressed lncRNAs corresponded to 330 annotated lncRNA genes. Among the upregulated lncRNAs, the greatest log2 fold change was 8.5 for an unannotated lncRNA gene, and RAD50 was the geratest upregulated, annotated lncRNA gene with a log2 fold change of 6.1. Among the downregulated lncRNAs, the greatest log2 fold change was 8.3 for an unannotated lncRNA gene and 6.4 for HAND2, the greatest downregulated annotated lncRNA gene; of these, 183 differently expressed lncRNAs had established functions in cancer development and cancer progression. Selected carcinogenesis-related lncRNAs are summarized in Table 2.
Fig 1.
Unsupervised hierarchical clustering and heat map of lncRNA expression between adrenocortical carcinoma (ACC), adrenocortical adenoma (ACA), and normal adrenal cortex (NAC). Each column represents a sample, and each row represents a lncRNA. High relative expression is indicated in yellow and low relative expression in red.
Table 2.
Selected carcinogenesis-related differentially expressed lncRNAs between ACC and NAC
| Sequence name | Gene symbol | Regulation | P-value | Log2 fold change | Chromosome | Relationship |
|---|---|---|---|---|---|---|
| ENST00000534886 | SRRM4 | Up | 0.001 | 5.14 | Chrl2 | Intron sense-overlapping |
| ENST00000472494 | HOTTIP | Up | 9.11 × 10−5 | 5.05 | Chr7 | Bidirectional |
| ENST00000514846 | GRK6 | Up | 9.92 × 10−6 | 4.75 | Chr5 | Natural antisense |
| NR_002795 | HOXA11 | Up | 4.61 × 10−5 | 4.05 | Chr7 | Bidirectional |
| NR 045572 | CHL1 | Up | 3.45× 10−4 | 4.16 | Chr3 | Exon sense-overlapping |
| ENST00000558031 | CRNDE | Up | 1.30 × 10−5 | 2.45 | Chrl6 | Intergenic |
| ENST00000502941 | HAND2 | Down | 1.52× 10−7 | 6.35 | Chr4 | Bidirectional |
| ENST00000450445 | BNC2 | Down | 1.36× 10−6 | 5.01 | Chr9 | Intronic anti sense |
| ENST00000417354 | DNM3 | Down | 4.46× 10−6 | 3.50 | Chr1 | Intronic anti sense |
| NR_029394 | TBXAS1 | Down | 2.15× 10−4 | 2.51 | Chr7 | Exon sense-overlapping |
| NR_026805 | LINC00271 | Down | 3.99 × 10−6 | 2.50 | Chr6 | Bidirectional |
| NR_027158.1 | FAM211A-AS1 | Down | 2.96× 10−3 | 2.06 | Chrl7 | Intronic anti sense |
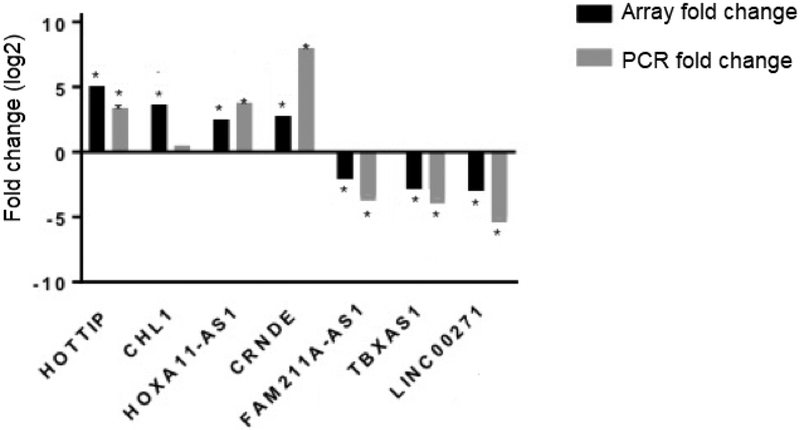
To test the validity of the microarray findings, seven lncRNAs (HOTTIP, CHL1, HOXA11-AS1, CRNDE, LINC00271, FAM211A-AS1 and TBXAS1) were selected among the carcinogenesis-related, differentially expressed lncRNAs, and their expression was analyzed by TaqMan qRT-PCR. The validation cohort included 19 ACC samples and 5 NAC samples. HOTTIP, HOXA11-AS1 and CRNDE were overexpressed in ACC (P < 0.05) and confirmed by TaqMan qRT-PCR in the validation cohort (P < 0.05; Fig. 2). Expression of LINC00271, FAM211A-AS1 and TBXAS1 was downregulated in ACC (P < 0.05) and also by TaqMan qRTPCR (P < 0.05) (Fig 2). The microarray result for CHL1 was not confirmed in the validation cohort. Upregulated expression of CHL1 was identified in the microarray analysis (P < 0.05) while CHL1 was found not to be upregulated by TaqMan qRT-PCR in the validation cohort.
Fig 2.
TaqMan qRT-PCR validation of lncRNA microarray analysis. Fold-change in comparison of ACCversusNAC, *P < 0.05.
Differentially expressed lncRNAs in ACC versus ACA
When comparing ACC with ACA, 1076 lncRNAs were differentially expressed, of which 780 were upregulated, and 296 were downregulated. The 1,076, differentially expressed lncRNAs corresponded to 376 annotated lncRNA genes. Among the upregulated lncRNAs, the greatest log2 fold change was 8.2 for an unannotated lncRNA and 7.0 for NKAIN4, the greatest upregulated annotated lncRNA. Among the downregulated lncRNAs, the greatest log2 fold change was 7.1 for an unannotated lncRNA gene and 6.9 for SSTR5, the greatest downregulated annotated lncRNA.
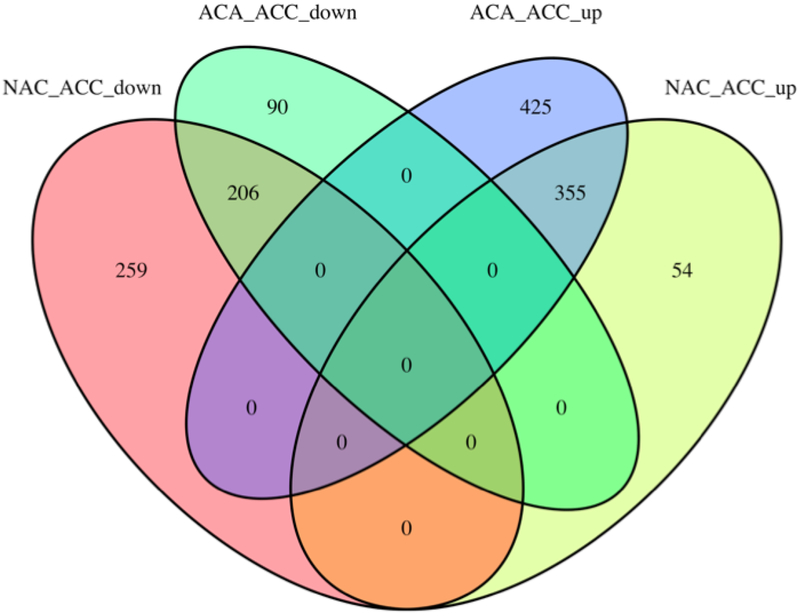
There was overlap in 206 lncRNAs as they were downregulated in ACC compared to NAC and in ACC compared to ACA, and 355 lncRNAs overlapped as they were upregulated in ACC compared to NAC and ACA (Fig. 3).
Fig 3.
Venn diagram showing the number of overlapping up- or downregulated lncRNAs in the different comparisons.
Differentially expressed lncRNAs in ACA versus NAC
Unsupervised hierarchical and heat map clustering showed that NAC samples clustered together with ACA samples (Fig. 1). Only10n lncRNAs were differentially expressed in ACA compared with NAC.
Functional pathway analysis
KEGG pathway analysis of the differentially expressed and annotated lncRNAs in ACC compared with NAC and in ACC compared with ACA was performed to understand the biologic relevance of these lncRNAs. Twenty-one pathways were significantly enriched in ACC versus ACA and 29 pathways were significantly enriched in ACC versus NAC (Tables 3 and 4). Twelve of the altered 21 pathways were common to the comparison of ACC versus ACA and ACC versus NAC. The KEGG pathways common to both comparisons included ‘Transcriptional misregulation in cancer’ and ‘ECM-receptor interaction’.
Table 3.
Statistically significant different KEGG pathways in ACC versus ACA
| Pathways | Genes | P-value |
|---|---|---|
| Pathways in cancer | ADCY2, RALBP1, CSF2RA, DAPK1, FGF13, GSK3B, BIRC5, ITGA3, MMP9, PTGER3, SLC2A1, TGFB2, PAX8, RUNX1 | 1.791e-3 |
| Vascular smooth muscle contraction | KCNMB2, ADCY2, KCNMA1, AVPRIA, PRKCQ, PRKG1 | 3.251e-3 |
| Glucagon signaling pathway | ADCY2, PRKAG2, PGAM2, PHKA2, SLC2A1 | 5.823e-3 |
| Malaria | ITGAL, TGFB2, THBS4 | 7.960e-3 |
| Transcriptional misregulation in cancer | HOXAW, HOXA11, MMP9, PAX8, HMGA2, HIST1H3G, RUNX1 | 8.716e-3 |
| Insulin secretion | KCNMB2,ADCY2, KCNMA1, SLC2A1 | 1.209e-2 |
| Circadian rhythm | ADCY2, PRKG1, PTGER3 | 1.266 e-2 |
| Salivary secretion | NPAS2,FRKAG2 | 1.346 e-2 |
| Cell cycle | ADCY2, KCNMA1, LYZ, PRKG1 | 1.455 e-2 |
| Colorectal cancer | E2F5, GSK3B, MAD2L1, RBL2, TGFB2 | 1.522 e-2 |
| FoxO signaling pathway | GSK3B, BIRC5, TGFB2 | 1.786 e-2 |
| Glycolysis / Gluconeogenesis | S1PR1, PRKAG2, RBL2, ВШРЗ, TGFB2 | 2.149 e-2 |
| Ubiquitin mediated proteolysis | PGAM2, ADPGK, FBP2 | 2.307 e-2 |
| Adipocytokine signaling pathway | UBE2S, UBE2D4, SIAH1, UBE2G2, ITCH | 2.367 e-2 |
| Signaling pathways regulating pluripotency of stem cells | PRKAG2, PRKCQ, SLC2A1 | 2.661 e-2 |
| Bladder cancer | ESRRB, GSK3B, PAX6, P0U5FIB, PCGF1 | 2.762 e-2 |
| Insulin resistance | DAPK1, MMP9 | 2.841 e-2 |
| RNA degradation | GSK3B, PRKAG2, PRKCQ, SLC2A1 | 3.180 e-2 |
| ECM-receptor interaction | LSM1, EXOSCIO, BTG1 | 3.605 e-2 |
| Hypertrophic cardiomyopathy (HCM) | SV2C,ITGA3,THBS4 | 4.385e-2 |
Note Pathways common to the comparison of ACC versus ACA and ACC versus NAC are written in bold type
Table 4.
Statistically significantly different KEGG pathways in ACC versus NAC
| Pathways | Genes | P-value |
|---|---|---|
| ECM-receptor interaction | COL6A2, SV2C, ITGA3, ITGA9, THBS2 | 5.329e-4 |
| Circadian rhythm | NPAS2, PRKAG2, BHLHE40 | 5.596e-4 |
| Vascular smooth muscle contraction | MRVI1, KCNMA1, AVPR1A, PRKACB, PRKCQ, PRKG1 | 7.222e-4 |
| Adipocytokine signaling pathway | IKBKB, PRKAG2, PRKCQ, SLC2A1 | 1.759e-3 |
| Transcriptional misregulation in cancer | HOXA11, MEIS1, MMP9, UTY, PAX8, HMGA2, HIST1H3G | 1.785e-3 |
| Cocaine addiction | GRIN3B, GRM3, PRKACB | 3.175e-3 |
| Salivary secretion | KCNMA1, LYZ, PRKACB, PRKG1 | 5.0121e-3 |
| Glucagon signaling pathway | PRKAG2, PGAM2, PRKACB, SLC2A1 | 8.511e-3 |
| Glycolysis / Gluconeogenesis | ADHIA, PGAM2, ADPGK | 9.687e-3 |
| Insulin resistance | IKBKB, PRKAG2, PRKCQ, SLC2A1 | 1.161e-2 |
| Nicotine addiction | CHRNA4, GRIN3B | 1.345e-2 |
| Proteasome | PSMA3, PSMD7 | 1.740e-2 |
| Platelet activation | LYN, PRKACB, PRKG1, TBXAS1 | 1.817e-2 |
| Hypertrophic cardiomyopathy (HCM) | ITGA3, ITGA9, PRKAG2 | 1.998e-2 |
| Hedgehog signaling pathway | CDON, PRKACB | 2.074e-2 |
| Endocrine and other factor-regulated calcium reabsorption | DNM3, PRKACB | 2.074e-2 |
| Insulin secretion | KCNMA1, PRKACB, SLC2A1 | 2.161e-2 |
| Neuroactive ligand-receptor interaction | CHRNA4, GRIN3B, GRM3, AVPRIA, RXFP1, SSTR5, THRB | 2.227e-2 |
| Dilated cardiomyopathy | ITGA3, ITGA9, PRKACB | 2.602e-2 |
| Morphine addiction | GRK6, PDE4D, PRKACB | 2.697e-2 |
| NF-kappa В signaling pathway | IKBKB, LYN, PRKCQ | 2.891e-2 |
| Circadian entrainment | PRKACB, PRKG1, CACNAIH | 3.094e-2 |
| Regulation of lipolysis in adipocytes | PRKACB, PRKG1 | 3.273e-2 |
| Long-term depression | LYN, PRKG1 | 3.900e-2 |
| Focal adhesion | COL6A2, ITGA3, ITGA9, PAK3, THBS2 | 4.064e-2 |
| T cell receptor signaling pathway | IKBKB, PAK3, PRKCQ | 4.110e-2 |
| Longevity regulating pathway - multiple species | PRKAG2, PRKACB | 4.584e-2 |
| Renin secretion | KCNMA1, PRKACB | 4.584e-2 |
| Renal cell carcinoma | PAK3, SLC2A1 | 4.946e-2 |
Note Pathways common to the comparison of ACC versus ACA and ACC versus NAC are written in bold type
Prognostic lncRNAs in ACC
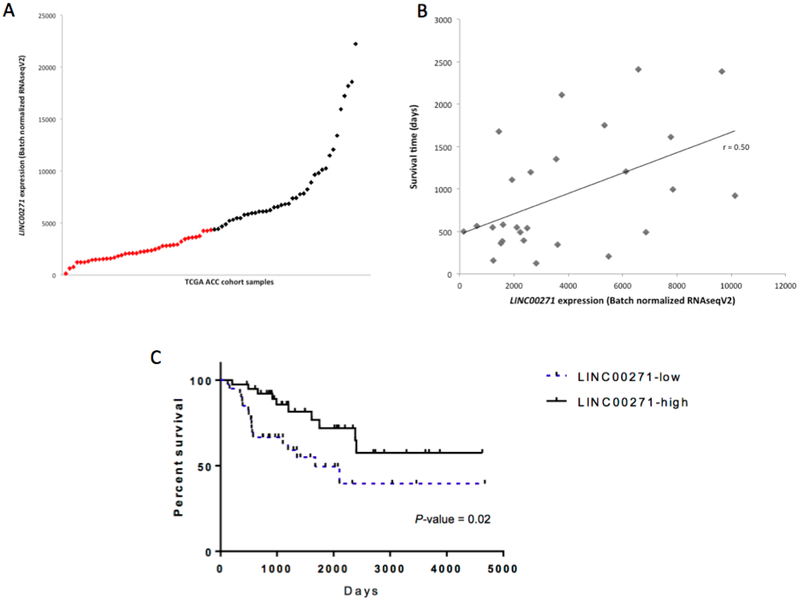
Using the survival data of the ACC TCGA cohort, the prognostic significance of HOTTIP, HOXA11-AS1, CRNDE, LINC00271, FAM211A-AS1 and TBXAS1 was analyzed. Only LINC00271 expression (Fig. 4A) was found to be associated with prognosis. LINC00271 expression levels were positively associated with survival time (Fig. 4B). Median survival time for the low-LINC00271 expression group (n = 40) was 4.9 years, whereas it was not reached for the high-LINC00271 expression group (n = 39) (P < 0.019) (Fig 4C). Student’s t-tests demonstrated that LINC00271 expression levels of stage I tumors were greater than those of stage IV tumors (P < 0.006).
Fig 4.
LINC00271 expression and prognosis. A, Distribution of LINC00271 expression of ACC samples from the TCGA dataset. Red points were defined as low-LINC00271 expression group and black points were defined as high-LINC00271 expression group. B, LINC00271 expression is positively correlated to survival time (Pearson correlation coefficient = 0.50). C, Kaplan-Meier plot of overall survival in the TCGA the ACCcohort is shown according to LINC00271 expression level (low vs. high).
Identification of LINC00271-associated biologic pathways by Gene Set Enrichment Analysis
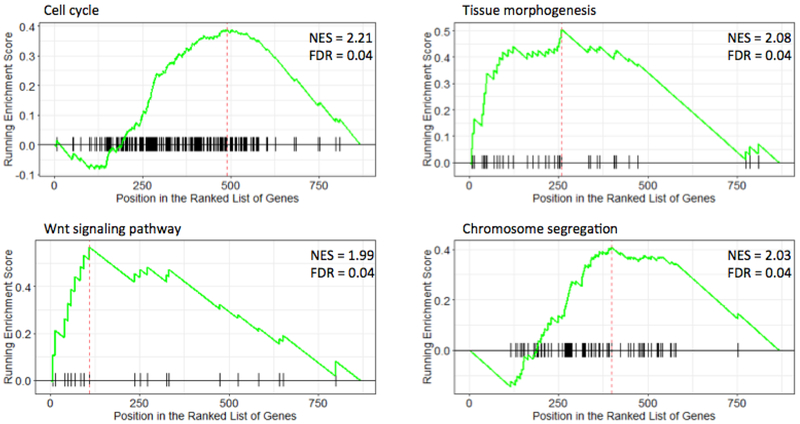
To identify LINC00271-associated biologic pathways, GSEA was performed using high throughput, RNA-sequencing data from the TCGA ACC cohort. Among the GO gene sets, the WNT signaling pathway, cell cycle, chromosome segregation, and tissue morphogenesis were found to be statistically associated with low LINC00271 expression in the ACC TCGA cohort (Fig. 5), suggesting that LINC00271 may be involved in ACC development and/or progression through the above cancer-associated signaling pathways.
Fig 5.
LINC00271-associated biologic signaling pathways. Based on the TCGA dataset, GSEA showed that genes associated with WNT signaling pathway, cell cycle, chromosome segregation and tissue morphogenesis were statistically enriched in lower LINC00271 versus greater LINC00271 expressing ACCs. FDR, false discovery rate; NES, normalized enrichment score.
LINC00271 copy number alterations
We performed an analysis of the LINC00271 chromosomal locus 6q23.3 using genome-wide. CGH array data that were generated previously in a cohort of NAC, ACA, and ACC10 to examine whether the LINC00271 site demonstrated any copy number alterations to explain its downregulated expression in ACC. One of 11 NAC samples demonstrated a deletion at 6q23.3, whereas 2 of 18 ACA samples demonstrated deletions at 6q23.3, and two other ACA samples demonstrated amplifications at 6q23.3. The LINC00271 locus appeared to be the most unstable in ACCs, with 4 of 19 ACC samples demonstrating deletions, and 4 of 19 ACC samples demonstrating amplifications of 6q23.3.
Discussion
This study demonstrated that NAC, ACA, and ACC have distinct lncRNA expression profiles, and that LINC00271, 2whhich aappeared to be involved in biologic pathways commonly dysregulated in ACC, may be a prognostic marker in ACC.
When compared with NAC, 874 lncRNAs were differentially expressed in ACC, 1076 lncRNAs were differentially expressed in ACA compared with ACC, and only t10 lncRNAs were differentially expressed in ACA vs. NAC. Previously, Glover and colleagues demonstrated that the greatest number of differentially expressed lncRNAs in their study was between ACA and NAC (2655 lncRNAs), while 956 lncRNAs were differentially expressed between ACC and NAC, and 85 lncRNAs were differentially expressed between ACC and ACA.11 The data of these investiagotrs suggested that changes in lncRNA expression could be an early part in the pathogenesis of both ACC and ACAs.In contrast, our results are not entirely consistent with their findings, because we found only 10lncRNAs that were differentially expressed between ACA and NAC.;this finding is in line with the multistep hypothesis in tumorigenesis that is present in most human cancers - progressive genetic/genomic alterations increasing/accumulating from NAC to ACA to ACC as described previously in our integrated, genome-wide gene expression, gene methylation, microRNA expression, and CGH analysis in human samples form NACs, ACAs, and ACCs.8 The multistep progression from NAC to ACA to ACC is further supported by our finding of 296 lncRNAs differentially expressed between ACA versus ACC and 465 lncRNAs differentially expressed between ACC vs. NAC. Overall, we found less differently expressed lncRNAs in adrenocortical neoplasmscompared to the Glover et al. study11, but we used a more stringent cut-off in fold-change to identify differentially expressed lncRNAs, and the NAC samples used in our study were not adjacent normal tissue to ACAs.
In the current study, the TCGA ACC dataset was used to screen for prognostic significance of differentially expressed lncRNAs. LINC00271 was found to be associated with malignancy; patients with low LINC00271 expression levels survived a significantly lesser time than patients with high LINC00271 expression levels. Previously, a statidtifvally lesser expression of LINC00271 has been described in invasive breast carcinoma, lung adenocarcinoma, kidney renal papillary cell carcinoma, head and neck squamous cell carcinoma, and papillary thyroid cancer.17 In addition, LINC00271 has been found to be an independent risk factor for extrathyroidal extension, lymph node metastasis, advanced tumor stage III/IV, and recurrence in papillary thyroid cancer.17 GSEA revealed that genes associated with cell adhesion molecules, the TP53 signaling pathway, the JAK/STAT signaling pathway, and the cell cycle were statistically enriched in papillary thyroid cancer with a low LINC00271 expression versus papillary thyroid cancer with greater LINC00271 expression. We also found that genes associated with cell cycle were associated with low LINC00271 expression in the TCGA ACC cohort. Further LINC00271 expression was positively associated with gthe WNT signaling pathway and chromosome segregation which are biologic pathways commonly dysregulated in ACC.18,19 Thus, our findings and other investigators studies suggest that LINC00271 could contribute to abnormal activation of these pathways in a tumor suppressor manner, however, further mechanistic studies are needed to test this hypothesis.
Studies have suggested that genes with causal roles in tumorigenesis are often located in chromosomal areas with alterations in copy number.20,21 Gene expression levels are directly dependent on chromosomal aneuploidies in carcinomas.22 The strongest correlations have been found between genomic copy number and average, chromosome-wide expression levels, but the expression of individual genes has also been associated with genomic copy numbers.23 LncRNAs expression levels have been positively correlated with alterations in copy number as well.24,25 Therefore, we investigated whether alterations in copy number were present at the LINC00271 chromosomal locus 6q23.3. This region had the greatest alteration in ACC samples with 21% of samples demonstrating amplifications and another 21% demonstrating deletions, while only 11% of ACA samples had amplifications and another 11% deletions of 6q23.3. The instability of 6q23.3 might explain the dysregulated expression of LINC00271 in ACC.
In conclusion, ACC has a distinct lncRNA expression profile, and LINC00271 downregulation is appears to be associated with malignancy and may be involved in biologic pathways commonly dysregulated in ACC.
Funding/Support
This work was supported by the intramural research program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health (1ZIABC011275–09).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Presented at the 40th Annual Meeting of the American Association of Endocrine Surgeons, Los Angeles, CA, USA, April 7–9, 2019
COI/Disclosure
The authors report no proprietary or commercial interest in any product mentioned or concept discussed in this article.
References
- 1.Bilmoria KY, Shen WT, Elaraj D, Bentrem DJ, Winchester DJ, Kebebew E, et al. Adrenocortical carcinoma in the United States: treatment utilization and prognostic factors. Cancer 2008;113:3130–6. [DOI] [PubMed] [Google Scholar]
- 2.Kerkhofs TM, Verhoeven RH, Van der Zwan JM, Dieleman J, Kerstens MN, Links TP, et al. Adrenocortical carcinoma: a population-based study on incidence and survival in the Netherlands since 1993. Eur J Cancer 2013;49:2579–86. [DOI] [PubMed] [Google Scholar]
- 3.Grubbs EG, Callender GG, Xing Y, Perrier ND, Evans DB, Phan AT, et al. Recurrence of adrenal cortical carcinoma following resection: surgery alone can achieve results equal to surgery plus mitotane. Ann Surg Oncol 2010;17:263–70. [DOI] [PubMed] [Google Scholar]
- 4.Fassnacht M, Terzolo M, Allolio B, Baudin E, Haak H, Berruti A, et al. Combination chemotherapy in advanced adrenocortical carcinoma. N Engl J Med 2012;366:2189–97. [DOI] [PubMed] [Google Scholar]
- 5.Giordano TJ, Kuick R, Else T, Gauger PG, Vinco M, Bauersfeld J, et al. Molecular classification and prognostication of adrenocortical tumors by transcriptome profiling. Clin Cancer Res 2009;15:668–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soon PS, Gill AJ, Benn DE, Clarkson A, Robinson BG, McDonald KL, et al. Microarray gene expression and immunohistochemistry analyses of adrenocortical tumors identify IGF2 and Ki-67 as useful in differentiating carcinomas from adenomas. Endocr Relat Cancer 2009;16:573–83. [DOI] [PubMed] [Google Scholar]
- 7.Rechache NS, Wang Y, Stevenson HS, Killian JK, Edelman DC, Merino M, et al. DNA methylation profiling identifies global methylation differences and markers of adrenocortical tumors. J Clin Endocrinol Metab 2012;97:E1004–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gara SK, Wang Y, Patel D, Liu-Chittenden Y, Jain M, Boufraqech M, et al. Integrated genome-wide analysis of genomic changes and gene regulation in human adrenocortical tissue samples. Nucleic Acids Res 2015;43:9327–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Assie G, Letouze E, Fassnacht M, Jouinot A, Luscap W, Barreau O., et al. Integrated genomic characterization of adrenocortical carcinoma. Nat Genet 2014;46:607–12. [DOI] [PubMed] [Google Scholar]
- 10.Zheng S, Cherniack AD, Dewal N, Moffitt RA, Danilova L, Murray BA, et al. Comprehensive pan-genomic characterization of adrenocortical carcinoma. Cancer Cell 2016;29:723–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glover AR, Zhao JT, Ip JC, Lee JC, Robinson BG, Gill AJ, et al. Long noncoding RNA profiles of adrenocortical cancer can be used to predict recurrence. Endocr Relat Cancer 2015;22:99–109. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Chang HY. Physiological roles of long noncoding RNAs: insight from knockout mice. Trends Cell Biol 2014;24:594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics 2016;14:42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiss LM, Medeiros LJ, Vickery AL Jr. Pathologic features of prognostic significance in adrenocortical carcinoma. Am J Surg Pathol 1989;13:202–6. [DOI] [PubMed] [Google Scholar]
- 15.Love M, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma B, Liao T, Wen D, Dong C, Zhou L, Yang S, et al. Long intergenic non-coding RNA 271 is predictive of a poorer prognosis of papillary thyroid cancer. Sci Rep 2016;6:36973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roshani L, Fujioka K, Auer G, Kjellman M, Lagercrantz S, Larsson C. Aberrations of centrosomes in adrenocortical tumors. Int J Oncol 2002;20:1161–5. [PubMed] [Google Scholar]
- 19.Ragazzon B, Libe R, Gaujoux S, Assie G, Fratticci A, Launay P, et al. Transcriptome analysis reveals that p53 and b-catenin alterations occur in a group of aggressive adrenocortical cancers. Cancer Res 2010;70:8276–8281. [DOI] [PubMed] [Google Scholar]
- 20.Kim TM, Xi R, Luquette LJ, Park RW, Johnson MD, Park PJ. Functional genomic analysis of chromosomal aberrations in a compendium of 8000 cancer genomes. Genome Res 2013;23:217–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet 2013;45:1134–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grade M, Hormann P, Becker S, Hummon AB, Wangsa D, Varma S, et al. Gene expression profiling reveals a massive, aneuploidy-dependent transcriptional deregulation and distinct differences between lymph node-negative and lymph node-positive colon carcinomas. Cancer Res. 2007;67:41–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ried T, Hu Y, Difilippantonio MJ, Ghadimi BM, Grade M, Camps J. The consequences of chromosomal aneuploidy on the transcriptome of cancer cells. Biochim Biophys Acta 2012;1819:784–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H, Xu J, Hong J, Tang R, Zhang X, Fang JY. Long noncoding RNA profiles identify five distinct molecular subtypes of colorectal cancer with clinical relevance. Mol Oncol 2014;8:1393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng Y, Luo S, Zhang X, Zou C, Yuan H, Liao G, et al. A pan-cancer atlas of cancer hallmark-associated candidate driver lncRNAs. Mol Oncol 2018;12:1980–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]