Summary
Immunology research in the last 50 years has made huge progress in understanding the mechanisms of anti‐bacterial defense of deep, normally sterile, tissues such as blood, spleen and peripheral lymph nodes. In the intestine, with its dense commensal microbiota, it seems rare that this knowledge can be simply translated. Here we put forward the idea that perhaps it is not always the theory of immunology that is lacking to explain mucosal immunity, but rather that we have overlooked crucial parts of the mucosal immunological language required for its translation: namely intestinal and bacterial physiology. We will try to explain this in the context of intestinal secretory antibodies (mainly secretory IgA), which have been described to prevent, to alter, to not affect, or to promote colonization of the intestine and gut‐draining lymphoid tissues, and where effector mechanisms have remained elusive. In fact, these apparently contradictory outcomes can be generated by combining the basic premises of bacterial agglutination with an understanding of bacterial growth (i.e. secretory IgA‐driven enchained growth), fluid handling and bacterial competition in the gut lumen.
Keywords: immunoglobulin A, microbiota, modeling, physiology, population dynamics
We put forward the idea that extensive variations in intestinal and bacterial physiology can explain the apparently contradictory functions of intestinal secretory antibodies (i.e. altering, promoting, or inhibiting colonization of the intestine and gut‐draining lymphoid tissues).
Abbreviation
- sIgA
secretory immunoglobulin A
Introduction
Secretory immunoglobulin A (sIgA) is the main component of the mucosal adaptive immune system that is present in the gut lumen and can therefore directly interact with the colonizing microbiota. Secretory IgA is produced mainly as an antibody dimer by plasma cells that populate the gut lamina propria,1 and is actively secreted into the gut lumen by the poly‐immunoglobulin receptor. After transcytosis through epithelial cells,2 this receptor is cleaved close to the cell membrane and the extracellular portion, known as secretory component, remains covalently linked to sIgA. Under certain conditions, sIgA can also be taken up from the gut lumen by epithelial cells, using a variety of carbohydrate‐binding receptors.3 However, many aspects of sIgA biology and interaction with gut bacteria remain controversial. Several excellent recent reviews describe the current knowledge on sIgA induction and specificity.4, 5, 6, 7, 8 In this review, we will focus specifically on the mechanisms by which sIgA can influence intestinal bacterial abundance, phenotype and evolution, with a predominant focus on high‐affinity antibody responses.
In our framework, we specifically focus on a subset of sIgA responses that are induced by monocolonization, infection or vaccination, or that mimic these by adoptive transfer, i.e. situations where the inducing bacterial antigen is known. Typically, these responses have been found to be T‐cell‐dependent and show selectivity for the inducing species.9, 10 However, even confining ourselves only to this narrow definition, the literature contains three apparently different outcomes of such sIgA–bacteria interaction. (i) Host protection and bacterial clearance – in the case of sIgA induced by oral vaccination against Salmonella or Escherichia coli, these antibodies are associated with protection from tissue invasion and faster clearance from the gut.11, 12, 13 (ii) Bacterial adaptation – passive immunization with a monoclonal IgA specific for Bacteroides thetaiotaomicron capsular polysaccharide 4 resulted in selection of phase variants producing a different capsular subtype.9 (iii) Promotion of bacterial colonization – Bacteroides fragilis‐induced sIgA was associated with increased competitiveness of sIgA‐bound B. fragilis in the gut lumen.14
How can similar sIgA responses prevent disease and promote clearance in one case, but benefit colonization in others? The answer appears extremely difficult to find if considering sIgA in isolation, and might give rise to the idea that there are several functionally different types of sIgA. An alternative explanation is that the intestinal sIgA, much like serum antibody isotypes, is actually a constant, but that it operates in the context of the complex, variable flow of intestinal fluid and in a densely colonized and highly dynamic bacterial ecosystem. Intestinal fluid flow and mixing affect how bacteria encounter each other, as well as how quickly they are washed along the intestinal tube and are cleared in feces. Water handling not only resorbs water for the host, it also concentrates nutrients, secreted molecules and secondary metabolites, as material travels along the intestine. The bacterial population of interest in the gut has to grow relatively fast to continuously colonize this flowing ecosystem, and is highly responsive to the concentration of nutrients and potential toxic substances. If sIgA alters how the targeted species is affected by the flow of gut content, how the bacteria are able to interact with host tissues, or how the targeted species interacts with other species or strains in the gut lumen affecting their growth rate, then highly complex outcomes suddenly become plausible. We will therefore begin by highlighting our current understanding of intestinal fluid handling and of bacterial physiology in the intestine, and will subsequently suggest a conceptual framework to provide possible explanations for some of the observed functions of sIgA. We would like to stress that although evidence exists for these mechanisms, and in many cases, these mechanisms are quantitatively sufficient to explain the observed phenomena, we are presenting a simplification and other mechanisms of sIgA function are not excluded.
Host intestinal physiology and the microbiota
Microbial physiology
Although the behavior of the gut microbiota as a whole is clearly extremely complex, the abundance of any individual species can be distilled down to depending on four essential parameters: its growth rate, its death rate, its immigration rate from higher in the intestinal tract and its clearance rate in the intestinal flow. If sIgA is to affect the population density of a bacterial species in the intestine, it must do so by affecting at least one of these parameters.
For microbiota composition to remain constant over time, the average net replication rate of a species over a day (i.e. growth minus death) must be exactly balanced by the clearance of each species due to flow. Generally speaking, the growth rate of a species will be determined by the concentrations of nutrients that it can use and of substances that inhibit its metabolism, as well as environmental variables such as pH and osmolality.15, 16 Recently, microbiota growth rates have been estimated, using snapshot metagenomics approaches, to range from between one and ten replications per day,17, 18 with a differential rate depending on the phyla.19 Of note, as bacteria grow, they add new membrane, cell wall and/or capsule to their outer surface, which becomes available for antibody binding and crosslinking, allowing crosslinking to occur even if antibody concentrations are super‐saturating.
Death rates will be affected by the presence of toxic substances to which the species is susceptible (e.g. bile acids, antimicrobial peptides, antimicrobials produced by other species in the microbiota)20, 21 and phage (either reactivation of dormant prophage or lytic infection).22, 23 It is worth noting that dead bacteria may act as a sink for specific antibodies. Further, massive bacterial death in the intestine is an event strongly associated with dysbiosis and opening or stabilizing of niches for pathogen invasion.24
In the context of a complete microbiota, factors influencing growth and death rates rapidly become extremely complex with extensive cross‐feeding, removal of end‐product inhibitors, and inter‐ and intra‐species aggression. Nevertheless, it is clear that a major influence on bacterial physiology in the gut is intestinal inflammation. This profoundly alters nutrient availability, intestinal secretions and fluid handling, with knock‐on effects on concentrations of bile acids, pH and the flow rate of the intestinal content. The stress exerted by inflammation on intestinal bacteria is also a major signal for the reactivation of temperate phages. Therefore, if sIgA provides protection from invasion of the intestinal epithelium, and decreases inflammatory signaling, one expects major effects on the growth and clearance rates of gut bacteria.
Another frequently overlooked aspect of mucosal immunology is that bacterial communities evolve rapidly in response to the particular conditions of their hosts.18 In E. coli the mutation rate per genome per generation is roughly 1/1000,25 with a typical 5 × 106‐bp genome. When a few hundred E. coli are present during a tissue infection, the chance to pick up a beneficial mutation remains low. However, where E. coli counts reach 5 × 109 colony‐forming units/g of intestinal content, as can happen during dysbiosis, the E. coli in 1 g of intestinal content will scan every possible point mutation every generation. Mobile elements generate genetic change at even faster rates. On top of this, the very dense but metabolically active conditions of the upper large intestine are ideal for horizontal gene transfer, by either phage or plasmids. Therefore, rapid evolution is inevitable. This is another potential aspect of bacterial physiology that could be affected by sIgA, with major effects for the targeted strain and its interactions with the rest of the microbiota.
Gut physiology and flow
The high flow rate of gut content is one of the main challenges in maintaining a stable gut microbiota composition. Although gut content is constantly flowing from mouth to anus, the presence of a sigmoid colon in humans allows us to collect fecal material before ejection and defecate more or less once daily.26 This marks a striking contrast with other mammals such as adult mice that excrete feces in the order of 120 pellets per day.27 Every day, the human digestive system manages around 2 l of fluid coming from ingesta, with a further 7 l added from intestinal secretions (saliva, stomach acid, pancreatic secretions, bile and other small intestinal secretions).28 Gut content enters the proximal colon at a high rate of 1·5–2 l/day, roughly equivalent to a flow velocity of > 30 μm/s.29 This velocity is considerably faster than the swimming speed of most bacteria.30
Several explanations exist for the maintenance of a dense colonic microbiota despite this flow. (i) Mucus‐resident bacteria sticking to the colon walls are often referred to as important for replenishing the luminal bacterial population. However, a quantitative consideration of their contribution indicates that this is far from sufficient to maintain the luminal microbiota.15, 31 To make this clearer, to a human‐sized bacterium (scaling up by 2 × 106‐fold) the ascending colon would be a tube with a 100 km diameter, with the content flowing at around 100 km/hr. (ii) Influx of bacteria from the small intestine could contribute, but can be largely excluded, because ileal effluent contains comparably low bacterial numbers and typically different species from the colonic microbiota.32, 33 (iii) The anatomy and mixing activity in the upper large intestine can provide a plausible mechanism. The upper large intestine in rodents is dominated by the cecum, which forms a large blind‐ended vessel. Here dietary fiber from the small intestine and host glycans and cell debris are plentiful34 and the majority of bacterial growth occurs in this region. In a large chamber‐like cecum, material flowing into the cecum from the small intestine becomes mixed with the content by peristalsis and later a fraction of this content is expelled into the colon. This generates a system similar to a chemostat, where final loss of bacteria into the fecal stream occurs by periodic partial emptying of the cecum, before re‐filling from the ileum. In humans, where the cecum has shrunk, the ascending colon rather represents this. In vitro data and modelling based on transit time in humans35 showed that peristaltic contractions in the ascending colon can generate complex mixing, which includes substantial back‐flushing of gut content close to the walls, effectively mimicking the model where random sampling moves material into the definitive fecal stream for elimination. It is this random sampling process that determines the rate of clearance of a bacterial species in the fecal stream and that can be modified by sIgA binding to a bacterial species, as we will discuss below.
Secretory IgA‐mediated clumping: classical agglutination and enchained growth
Secretory IgA is also known to protect from non‐toxigenic bacterial infections, such as non‐typhoidal salmonellosis,12 through a process referred to as immune exclusion.36 This has been variously attributed to classical agglutination36 (Fig. 1a), to trapping bacteria in mucus, or to altering bacterial motility.37 However, our recent work has indicated that in the murine model of non‐Typhoidal Salmonellosis, bacterial clumping by vaccine‐induced sIgA is the dominant protective mechanism.11 Intravital microscopy demonstrated that clumped bacteria could not approach the cecum wall, preventing the activity of type III secretion systems required for Salmonella virulence.38
Figure 1.
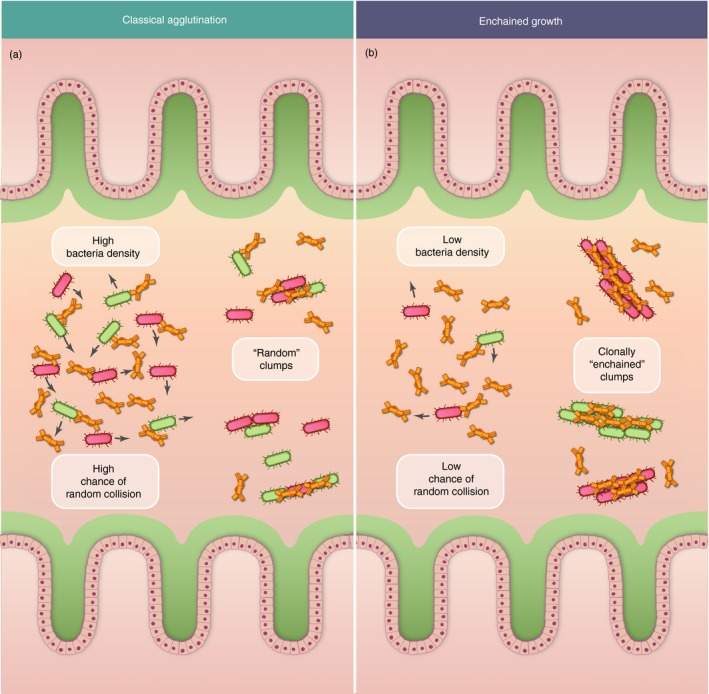
Classical agglutination by secretory immunoglobulin A (sIgA) (a) is efficient at high bacterial densities while only sIgA‐driven enchained growth is possible at low densities (b). Two antigenically identical clones of the same bacterial strain are depicted in red and green, and specific antibody concentrations are saturating in both cases, such that all bacteria of this strain will be sIgA coated.
Close examination of the bacterial and intestinal physiology at play during the formation of these clumps quickly revealed that classical agglutination was an insufficient explanation. In fact, the way in which sIgA interacts with bacteria to form clumps depends on two phenomena: (i) sIgA with sufficient affinity to bind to the bacterial surface must be present, and (ii) bacteria displaying the cognate antigen on their surfaces must come into contact. Classical agglutination relies on random collisions of such antigenically identical bacteria. The efficiency of this process depends on the square of the population density, i.e. is very inefficient at low population densities. A typical bacterial food poisoning starts with the ingestion of 105 bacteria, which are diluted into several liters of gut content in a human, or several milliliters in a mouse, generating a density where random encounters are so rare as to be useless for clump formation. Recent work has suggested that sIgA with moderate affinity can bind broadly to multiple microbiota species39 or can be bound by bacterial superantigens,40 raising the possibility that classical agglutination may generate mixed clumps, partially overcoming this limit. However, such diverse bacterial clumps have not yet been observed, and differences in affinity for epitopes on the different species involved makes this potentially a complex process to envisage.
An alternative way to overcome the quantitative limits of classical agglutination occurs every time a bacterium divides, when two identical cells are always in contact. The sIgA can efficiently crosslink at the point of septation driving ‘enchained growth’ (Fig. 1b). This prevents the two daughter cells from dispersing and through several rounds of division builds up large clumps with a clonal structure. As discussed above, an invading pathogenic bacterium needs to grow to be maintained in the flowing gut ecosystem, and needs to grow fast to expand in this ecosystem during dysbiosis. Therefore, enchained growth will effectively always occur when you have a specific sIgA response present. At densities of over 108 bacteria per ml, bacterial movement due to swimming and complex mixing starts to efficiently drive classical agglutination, such that both classical agglutination and enchained growth will occur simultaneously. In fact, the interaction of these two effects during bacterial growth in the gut can be explicitly modelled from first principles using empirically determined properties of intestinal physiology (see Box 1A). Fitting only the probability of sIgA crosslinking gives a good fit to experimental data generated from the murine model of non‐typhoidal salmonellosis.11
Box 1. IgA–microbe interaction in the context of host physiology: modeling a dynamic system.
Mathematical models can help to bring new insights into complex biological processes and help to identify key parameters that drive them in one direction or the other. Although these can seem daunting, the principles that they represent are actually very simple. Together with experimental testing of hypotheses arising from the models, this begins to detail the intricate relationship between secretory immunoglobulin A (sIgA) and the evolutionary ecology of the intestinal microbiota in ways that move beyond our ability to internally visualize stepwise processes. In recent publications, we used three distinct models to describe IgA–bacteria interaction. This is far from the ‘hit‐and‐kill’ function of serum antibodies. Here we explain their consequences and interrelationships.
Dominant clumping mechanism depends on bacterial population density: 11 Based on the premise that clumped Salmonella can no longer participate in driving host pathology (‘immune exclusion’), we proposed a simple deterministic mathematical model. This model describes the concentration of planktonic bacteria in the gut lumen in the presence of sIgA capable of crosslinking bacteria. It assesses the contribution of enchained growth and classical agglutination to clumping. In this model both classical agglutination and enchained growth depended on IgA crosslinking efficiency, but enchained growth depended on the bacterial growth rate (which is density‐ and size‐independent), whereas classical agglutination depended on the bacterial collision rate (which depends on mixing of intestinal content, bacterial motility, size and population density). We approximated the complex mixing of intestinal content by turbulent mixing. The model quantitatively predicted experimental data that classical agglutination is inefficient at Salmonella Typhimurium densities <108 colony‐forming units/g, as the probability of collision is too low. This is roughly the density of a species present at 1% representation in the fecal microbiota, but is also a typical colonization level for many intestinal pathogens including non‐Typhoidal Salmonella species. Although this model was developed in the context of non‐Typhoidal salmonellosis, it is easily generalizable to quantify the efficiency of clumping of any gut microorganism by sIgA. Therefore, clumping by enchained growth is dominant for low‐abundance but rapidly growing species, with classical agglutination coming into play when species population densities exceed 108 CFU/g.
Enchained growth results in clonal extinction: 11, 24 A second model from the same paper predicted the rate of clonal extinction from a mouse gut luminal bacterial population. This model was dependent on how efficiently sIgA‐mediated enchained growth retained a single clone in an individual clump, and modelled each partial emptying of the cecum as a binomial sampling process. Our model estimated the evenness of distribution of neutrally tagged bacterial clones over time, with evenness being close to 1 if all the barcoded clones where retained or near to 0 if most barcoded bacteria clones become randomly extinct. It relies on three important parameters: (i) probability to colonize the cecum, (ii) bacterial growth rate and (iii) clearance rate in the fecal stream. We estimated values for these parameters empirically and produced families of stochastic simulations under two extreme scenarios (i) no enchained growth (no clonal clumps) or (ii) ‘perfect’ enchained growth (all bacteria form clonal clumps). Experimental data from naive mice were accurately represented by the ‘no clumping’ simulation, consistent with no effect of ‘natural’ sIgA against Salmonella. In contrast, vaccinated mouse experimental data clearly falls between the extremes of zero and perfect enchained growth, consistent with imperfect enchained growth. It is worth mentioning that when very high bacterial inocula are given (i.e. when classical agglutination is dominant) the clonal extinction rate is barely affected by the presence of sIgA up to 24 hr post‐colonization. Therefore, sIgA‐driven enchained growth can drive clonal extinction even without changing the overall growth and clearance rates of the population: instead, it changes the probability of clearance of an individual clone. As clonal extinction is equivalent to selective pressure, enchained growth also represents a major selective pressure on targeted intestinal bacteria.
Bacterial growth rate modifies enchained growth: 53 A third model, published recently, posits the concept that sIgA‐mediated enchained growth can generate a selective pressure for low growth rates. We used several types of models to estimate the expected size distribution of a bacterial clump as a product of enchained growth. Main parameters included in the models were bacterial replication rate, clump breaking rate (rate by which bacteria detach from the clump), and clearance rate by fecal stream. Interestingly, the model predicts that at higher replication rates (such as those observed in pathogens and pathobionts), bacteria divide before the link between daughter bacteria breaks, therefore producing enchained growth and increasing the number of cells present in the clump. On the other hand, at low growth rates the sIgA link between two daughter bacteria can break apart before the next round of division starts, leading to a low chance of observing large clumps. These predictions matched experimentally observed clump sizes. It is important to remember, as described in section B, that remaining in a clump can lead to clonal extinction of the enchained, fast‐growing strain. These phenomena have great implications for how the microbiota community can regulate itself (even in the presence of high‐affinity, clump‐forming sIgA) during recovery to homeostasis after a perturbation.
These examples clearly show the potential of using mathematical models to dissect complex processes such as sIgA–bacteria interactions. Further experimental and theoretical studies should take into consideration more complex scenarios of sIgA–microbiota interplay, such as the interaction between sIgA function and mucous structure and dynamics along the intestinal tract, or/and how sIgA function may be affected by circadian variations in flow and nutrients.
Determinants and consequences of clumping
In the context of a Salmonella infection, sIgA‐mediated defense is a logical response to prevent disease caused by a bona fide pathogen. However, this situation seems to be more complex in the context of the host microbiota. For example, a significant fraction of the microbiota is constitutively coated with sIgA. This sIgA‐coating has been associated with a more pathogenic subset of the microbiota,41 in line with the concept that sIgA may control bacterial pathogenicity. On the other hand some experimental data suggests that sIgA binding to B. fragilis promotes colonization.14 Further, polyclonal binding of sIgA to a range of microbiota species has been observed without obvious positive or negative effects on bacterial abundance.39 Absence of sIgA as the result of selective sIgA deficiency is a common and mild immunodeficiency in humans, and although a significant part of sIgA function appears to be compensated for by secretory IgM,42, 43 this disease is associated with loss of some normally sIgA‐coated species. So, how do these observations fit with our model of sIgA‐driven clumping? To understand this, we need to recognize the major determinants of sIgA‐driven clumping, and its consequences in interacting with the host/bacterial physiology.
Determinants of clumping
As described before, our framework focuses on sIgA induced by monocolonization, infection or vaccination, which are typically high‐avidity and T‐cell‐dependent and can mediate enchained growth/classical agglutination.11 Although there have been some descriptions of natural (germline) sIgA that bind several members of the gut microbiota, this binding has usually low‐ or moderate‐affinity39 and may originate from non‐specific B‐cell stimulation by bacterial superantigens.40
In addition, we should note that the ability of a specific sIgA to crosslink two bacteria, given that these two bacteria are in contact, depends on considerably more than the affinity of the antibody‐binding sites. The antigen must be exposed at the surface and sufficiently abundant that binding sites can be found on both cells, and ideally that multiple antibodies can form crosslinks. The antigen also needs to be strongly anchored to the bacterial surface. Notably, some major surface antigens, such as the enterobacterial common antigen,44 or some bacterial capsules are only very weakly associated to the bacterial surface and crosslinking these will most likely result in the antigen dissociating from the surface, rather than the two cells sticking together.
It should be noted that many bacterial surface antigens are in fact glycans (e.g. O‐antigens, teichoic acids, capsules), which are often very long, flexible, hydrophilic molecules that pose significant challenges for generation of high‐affinity antibodies.45 However, these structures are usually highly repetitive polymers, providing potential for high‐avidity interactions. They are also massively produced during bacterial growth; therefore, new antibody‐binding sites for crosslinks are being generated even at supersaturating antibody concentrations.
Critical consequences of clumping: 1. Increased clonal extinction rate
In experiments with barcoded bacteria in the gut, we observed a great reduction in clonal diversity of the strain targeted by vaccine‐induced sIgA. As the bacterial growth rate is unaltered between clumped and non‐clumped bacteria, this observation could be explained by invoking an elevated clearance by flow or direct killing (See Box 1‐B). However, there is no evidence that sIgA has bactericidal activity in our study,11 despite isolated reports of in vitro effects.37, 46 Although sIgA binding prevents bacterial motility, experiments with non‐flagellated mutants determined that this was not contributing to increased clearance.11 In fact, the reality was more subtle.
Consider a population of bacteria derived from several thousand individual clones. If these are planktonic, each fluid sampling event that moves bacteria from the well‐mixed upper large intestine into the fecal stream for evacuation will take a well‐mixed sample of progeny of all clones. It is highly unlikely that a single clone will go extinct, at least over short time‐scales. However, clumps of bacteria will be sampled en bloc. Here a major difference derives from whether the clumps were formed by classical agglutination (i.e. composed of various random clones) or by enchained growth (i.e. single‐clone composition). Enchained clumps will tend to result in a single clone being sampled and eliminated en bloc, driving rapid clonal extinction, even when clumped bacteria are shed at the same rate as planktonic bacteria (see Box 1‐A and Fig. 2a). Conversely, randomly agglutinated clumps could be expected to behave like planktonic bacterial clones. Nevertheless, we could also imagine a generally faster flow‐through of larger versus smaller particles due to the complex fluid mixing in the upper large intestine and the tendency of planktonic bacteria to associate with mucus.47 Hence, sIgA‐mediated clumping by enchained growth, and potentially also via agglutination can increase the clearance rate of bacteria in the fecal stream. Efficient en bloc elimination of sIgA‐clumped bacteria versus free‐swimming bacteria exerts a strong selective pressure.
Figure 2.
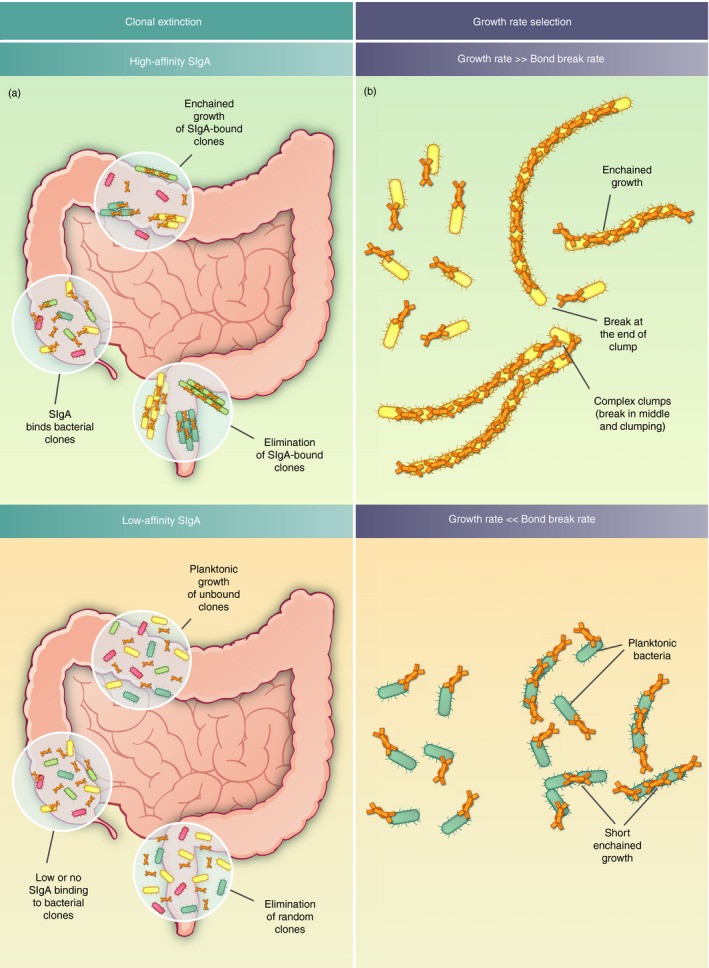
Consequences of bacterial clumping in the gut lumen. (a) Increased clonal extinction due to enchained growth in flowing gut content Bacterial clones in green, brown and yellow form clumps by secretory immunoglobulin A (sIgA) ‐driven enchained growth; are sampled into the final fecal stream and are eliminated (top left panel). Conversely, clones under low‐affinity sIgA pressure are planktonic and are unlikely to be completely eliminated by random sampling into the fecal stream (bottom left panel) (b). Relationship between growth rate, sIgA‐crosslink breaking rate and clump size could drive growth rate selection. It should be realized that sIgA crosslinks have a finite half‐life. Fast‐growing bacteria are more likely to undergo enchained growth, as their growth rate is faster than the breaking rate of the sIgA–bacteria complex. On the contrary, slow‐growing bacteria can escape the selective pressure of sIgA by having a growth rate far slower than the bond‐breaking rate.
We can expect the selective pressure from sIgA to foster strains with variation in surface antigen identity and indeed the majority of commensal bacteria are professionals at phase variation48, 49, 50 (of note, this is likely driven by phage predation,51, 52 but can be repurposed for antibody evasion). Indeed one of the earliest observations of an effect of a monoclonal sIgA against B. thetaiotaomicron was to drive phase variation away from the targeted surface antigen.9
Critical consequences of clumping: 2. Growth rate selection
An intriguing behavior emerges when the selective pressure from clumping is combined with our observations of sIgA–bacteria binding dynamics.53 Despite apparently nanomolar‐affinity T‐dependent antibody responses for experimental systems with vaccine‐induced sIgA against Salmonella, the probability of crosslinking remains quite low in the gut lumen (in the order of 40% of encounters result in crosslinking11) and the links break roughly every 2 hr.53 Whereas this is clearly sufficient to drive massive clumping and protection during pathogenic bacterial overgrowth, 2 hr is expected to be shorter than the division time of most bacteria in the microbiota during health (estimated to be between 2·5 and 24 hr17, 19). In fact, as long as crosslink breakage happens more quickly than growth, you will only rarely generate a clump larger than two bacteria, as the crosslink will disintegrate faster than the clump could grow (Fig. 2b).
Hence, the proposed low‐affinity (and therefore fast‐breaking) bonds formed by T‐independent sIgA coating of the microbiota are unlikely to result in extensive enchained growth in conditions of health. If during dysbiosis a sIgA‐targeted strain begins to grow faster than the crosslinks break, enchained clumps will be formed and a selective pressure will be generated. An alternative to escape this pressure (besides phase variation) is to reduce the growth rate, and in fact for a given antibody crosslinking strength, there is an optimal growth rate for generating planktonic cells.11, 53 This makes it theoretically possible for sIgA to select for slower growth: potentially a highly useful selection in recovering a diverse microbiota after a period of dysbiosis or during preliminary establishment of the microbiota.54, 55 Moreover, this potentially gives sIgA the ability to specifically exert a selective pressure on bacteria demonstrating the pathogen‐associated behavior of rapid growth.
Critical consequences of clumping: 3. Decreased horizontal gene transfer
A third and unexpected consequence of sIgA‐driven enchained growth is the inhibition of horizontal gene transfer by either contact‐dependent plasmid transfer or by temperate phage transduction. In fact, two mechanisms are at play.
First, as sIgA‐driven enchained growth locks individual clones of a species into physically separated clumps, these clones are much less likely to come into contact than in their planktonic state, and are therefore less likely to undergo contact‐dependent exchange of plasmids. This could be directly quantified in vivo and in situations where enchained growth, rather than classical agglutination, dominated the clumping mechanisms, could delay transfer by 24–48 hr. Interestingly, the same antibodies have no effect on transfer when plasmid donor and recipient clones are mixed at high‐density in vitro, i.e. where classical agglutination is the dominant clumping mechanism.
Second, the main driver of temperate phage reactivation in the gut lumen is inflammation.56 Secretory IgA‐driven clumping, via either classical agglutination or enchained growth, leads to inhibition of virulence mechanisms that require contact with gut epithelial cells. This significantly reduces inflammation driven by opportunistic pathogens or pathobionts, and so reduces the stimulus for temperate phage reactivation, often driven by SOS response. It is noteworthy that sIgA can also target bacteriophages in the gut,57 so indirectly affecting bacteria–phage interaction. This adds an extra piece to the puzzle of the effect of sIgA on bacterial evolution and on microbiota compositions.
Critical consequences of clumping: 4. Enhanced colonization?
Interestingly, although the previously described consequences of sIgA binding and clumping were proposed as negative selective pressures, it has been suggested that sIgA can also promote colonization of mucosal surfaces. Humans with IgA deficiency present an alteration in their microbiota composition, which includes absence of some species that are normally sIgA‐coated.42 This is not consistent with sIgA exerting a strong selective pressure on all bound bacteria.
One hypothesis that has been put forward suggests a role for sIgA as a factor that increases adhesion and thereby helps establish a niche at the mucosal surface. In fact, sIgA‐driven clumping of B. fragilis has been observed in the descending colonic mucus.14 Although not directly tested, this could suggest that at least in this niche of the intestine, planktonic bacteria could use sIgA binding to invade the mucus and subsequently begin to grow; generating embedded enchained clumps that might be shed more slowly than planktonic bacteria. As discussed above with microbial physiology, the relevance of these clumps for the total luminal B. fragilis population remains to be determined, and alternative explanations may be required to explain total population effects.
However, there are at least two alternative explanations of the pro‐colonization effects of sIgA. First, bacteria can interact with sIgA in a non‐cognate way by binding to the extensive O‐glycans at the hinge regions and N‐glycans attached to the secretory component.58 Indeed many Bacteroides strains carry the complete machinery required to degrade mammalian N‐glycans59 and may reasonably use these as a carbon source in the gut lumen, similar to the accepted role of mucins as a bacterial carbon source. Non‐cognate binding of sIgA to B. thetaiotaomicron mediated an effect on the expression of polysaccharide utilization loci and facilitated symbiosis with other phyla such Firmicutes.60 A simple nutritional explanation for pro‐colonization effects has therefore not yet been excluded.
A second alternative relies on the indirect effects of sIgA‐driven clumping/immune exclusion on the total bacterial ecosystem in the gut. As discussed above, any inflammatory signals, which may be inhibited when sIgA is present, are expected to have major consequences ranging from altering intestinal motility and concentrations of electron acceptors61 to changing the rate of reactivation of temperate phage,56 and to changing intestinal transit time (potentially affecting clearance rates of all species present in the gut). Moreover, generating or altering community structure through sIgA‐mediated clumping may influence cross‐feeding and therefore growth rates in both targeted and untargeted species, as has been observed in biofilms.62 Finally, highly cooperative species may actually grow faster when kin‐association is enforced63 by enchained growth.
Therefore, colonization‐promoting effects of sIgA can be envisaged as both direct and indirect effects of bacterial clumping. However, further effects such as metabolism of sIgA N‐glycans will probably be required to fully explain these phenomena.
Outlook and conclusions
In conclusion, it is possible to explain a considerable amount of sIgA‐associated phenotypes in the gut through a simple but elegant mechanism: i.e. the most basic property of antibodies (target binding and clumping) enhancing the normal physiological processes of bacterial clearance under flow. This can generate apparently complex consequences (such as clonal extinction, growth rate selection, altered ability to evolve of bacteria, and changes in host inflammatory mediators), that shape the gut microbiota community. Superimposing immunological knowledge onto an understanding of intestinal and microbial physiology is therefore a powerful technique to discern functional mechanisms. Nevertheless, much remains to be uncovered: the apparent promotion of colonization by sIgA remains incompletely explained in most cases and the full consequences of sIgA for bacterial evolution in the intestine and transmission between hosts remain under‐explored. Given the potential for sIgA to ‘peacefully’ manage intestinal microbiota composition, there is an urgent need to understand the remaining mechanistic problems and to hone strategies for inducing specifically targeted sIgA responses in humans and relevant veterinary contexts.
Disclosures
The authors declare not conflict of interest.
Author contributions
DH planned and wrote the manuscript and made the figures. MA, CL, MD and ES wrote short sections and extensively edited the text.
Acknowledgements
The authors would like to thank Daniela Latorre and Verena Lentsch for their critical reading of the manuscript.
References
- 1. Jahnsen FL, Bækkevold ES, Hov JR, Landsverk OJ, Baekkevold ES, Hov JR et al Do long‐lived plasma cells maintain a healthy microbiota in the gut? Trends Immunol 2018; 39:196–208. [DOI] [PubMed] [Google Scholar]
- 2. Brandtzaeg P. Secretory IgA: designed for anti‐microbial defense. Front Immunol 2013; 4:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kadaoui KA, Corthésy B. Secretory IgA mediates bacterial translocation to dendritic cells in mouse Peyer's patches with restriction to mucosal compartment. J Immunol 2007; 179:7751–7. [DOI] [PubMed] [Google Scholar]
- 4. Slack E, Balmer ML, Fritz JH, Hapfelmeier S. Functional flexibility of intestinal IgA – broadening the fine line. Front Immunol 2012; 3:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pabst O, Cerovic V, Hornef M. Secretory IgA in the coordination of establishment and maintenance of the microbiota. Trends Immunol 2016; 37:287–96. [DOI] [PubMed] [Google Scholar]
- 6. Macpherson AJ, Yilmaz B, Limenitakis JP, Ganal‐Vonarburg SC. IgA function in relation to the intestinal microbiota. Annu Rev Immunol 2018; 36:359–81. [DOI] [PubMed] [Google Scholar]
- 7. Bunker JJ, Bendelac A. IgA responses to microbiota. Immunity 2018; 49:211–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sterlin D, Fadlallah J, Slack E, Gorochov G. The antibody/microbiota interface in health and disease. Mucosal Immunol 2019; 14:1–9. [DOI] [PubMed] [Google Scholar]
- 9. Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA Response to Symbiotic Bacteria as a Mediator of Gut Homeostasis. Cell Host Microbe 2007; 2:328–39. [DOI] [PubMed] [Google Scholar]
- 10. Moor K, Wotzka SY, Toska A, Diard M, Hapfelmeier S, Slack E. Peracetic acid treatment generates potent inactivated oral vaccines from a broad range of culturable bacterial species. Front Immunol 2016; 7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moor K, Diard M, Sellin ME, Felmy B, Wotzka SY, Toska A et al High‐avidity IgA protects the intestine by enchaining growing bacteria. Nature 2017; 544:498–502. [DOI] [PubMed] [Google Scholar]
- 12. Endt K, Stecher B, Chaffron S, Slack E, Tchitchek N, Benecke A et al The microbiota mediates pathogen clearance from the gut lumen after non‐typhoidal Salmonella diarrhea. Stebbins CE, editor. PLoS Pathog 2010;6:e1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Uchimura Y, Fuhrer T, Li H, Lawson MA, Zimmermann M, Yilmaz B et al Antibodies set boundaries limiting microbial metabolite penetration and the resultant mammalian host response. Immunity 2018; 49:545–59.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou W‐C et al Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 2018; 360:795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cremer J, Arnoldini M, Hwa T. Effect of water flow and chemical environment on microbiota growth and composition in the human colon. Proc Natl Acad Sci U S A. 2017; 114:6438–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tropini C, Moss EL, Merrill BD, Ng KM, Higginbottom SK, Casavant EP et al Transient osmotic perturbation causes long‐term alteration to the gut microbiota. Cell 2018; 173:1742–54.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Korem T, Zeevi D, Suez J, Weinberger A, Avnit‐Sagi T, Pompan‐Lotan M et al Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science 2015;349:1101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao S, Lieberman TD, Poyet M, Kauffman KM, Gibbons SM, Groussin M et al Adaptive evolution within gut microbiomes of healthy people. Cell Host Microbe 2019; 25:656–67.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brown CT, Olm MR, Thomas BC, Banfield JF. Measurement of bacterial replication rates in microbial communities. Nat Biotechnol 2016; 34:1256–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 2013; 152:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE et al Extensive impact of non‐antibiotic drugs on human gut bacteria. Nature 2018; 555:623–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reyes A, Wu M, McNulty NP, Rohwer FL, Gordon JI. Gnotobiotic mouse model of phage–bacterial host dynamics in the human gut. Proc Natl Acad Sci U S A 2013; 110:20236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shkoporov AN, Hill C. Bacteriophages of the human gut: the “Known Unknown” of the microbiome. Cell Host Microbe 2019; 25:195–209. [DOI] [PubMed] [Google Scholar]
- 24. Maier L, Diard M, Sellin ME, Chouffane E‐S, Trautwein‐Weidner K, Periaswamy B et al Granulocytes impose a tight bottleneck upon the gut luminal pathogen population during Salmonella typhimurium colitis. Luo Z‐Q, editor. PLoS Pathog 2014; 10:e1004557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee H, Popodi E, Tang H, Foster PL. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole‐genome sequencing. Proc Natl Acad Sci U S A 2012; 109:E2774–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heaton KW, Radvan J, Cripps H, Mountford RA, Braddon FE, Hughes AO. Defecation frequency and timing, and stool form in the general population: a prospective study. Gut 1992; 33:818–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hapfelmeier S, Lawson MAE, Slack E, Kirundi JK, Stoel M, Heikenwalder M et al Reversible microbial colonization of germ‐free mice reveals the dynamics of IgA immune responses. Science 2010; 328:1705–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barrett KE, editor. Water and electrolyte absorption and secretion In: Gastroinstestinal Physiology, 2nd edn Columbus: McGraw‐Hill, 2014:132–7. [Google Scholar]
- 29. Phillips SF, Giller J. The contribution of the colon to electrolyte and water conservation in man. J Lab Clin Med. 1973; 81:733–46. [PubMed] [Google Scholar]
- 30. Turner L, Ping L, Neubauer M, Berg HC. Visualizing flagella while tracking bacteria. Biophys J 2016; 111:630–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zoetendal EG, von Wright A, Vilpponen‐Salmela T, Ben‐Amor K, Akkermans ADL, de Vos WM. Mucosa‐associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol 2002; 68:3401–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gorbach SL, Plaut AG, Nahas L, Weinstein L, Spanknebel G, Levitan R. Studies of intestinal microflora. II. Microorganisms of the small intestine and their relations to oral and fecal flora. Gastroenterology 1967; 53:856–67. [PubMed] [Google Scholar]
- 33. Marteau P, Pochart P, Doré J, Béra‐Maillet C, Bernalier A, Corthier G. Comparative study of bacterial groups within the human cecal and fecal microbiota. Appl Environ Microbiol 2001; 67:4939–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. 2008; 6:121–31. [DOI] [PubMed] [Google Scholar]
- 35. Hammer J, Phillips SF. Fluid loading of the human colon: Effects on segmental transit and stool composition. Gastroenterology 1993; 105:988–98. [DOI] [PubMed] [Google Scholar]
- 36. Mantis NJ, Rol N, Corthésy B. Secretory IgA's complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol 2011; 4:603–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Forbes SJ, Eschmann M, Mantis NJ. Inhibition of Salmonella enterica serovar typhimurium motility and entry into epithelial cells by a protective antilipopolysaccharide monoclonal immunoglobulin A antibody. Infect Immun 2008; 76:4137–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wotzka SY, Nguyen BD, Hardt WD. Salmonella typhimurium diarrhea reveals basic principles of enteropathogen infection and disease‐promoted DNA exchange. Cell Host Microbe 2017; 21:443–54. [DOI] [PubMed] [Google Scholar]
- 39. Bunker JJ, Erickson SA, Flynn TM, Henry C, Koval JC, Meisel M et al Natural polyreactive IgA antibodies coat the intestinal microbiota. Science 2017; 358:eaan6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bunker JJ, Drees C, Watson AR, Plunkett CH, Nagler CR, Schneewind O et al B cell superantigens in the human intestinal microbiota. Sci Transl Med 2019; 11:eaau9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Palm NW, De Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L et al Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014; 158:1000–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fadlallah J, El Kafsi H, Sterlin D, Juste C, Parizot C, Dorgham K et al Microbial ecology perturbation in human IgA deficiency. Sci Transl Med 2018; 10:eaan1217. [DOI] [PubMed] [Google Scholar]
- 43. Catanzaro JR, Strauss JD, Bielecka A, Porto AF, Lobo FM, Urban A et al IgA‐deficient humans exhibit gut microbiota dysbiosis despite secretion of compensatory IgM. Sci Rep 2019; 9:13574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kuhn HM, Meier‐Dieter U, Mayer H. ECA, the enterobacterial common antigen. FEMS Microbiol Rev 1988; 4:195–222. [DOI] [PubMed] [Google Scholar]
- 45. Avci FY, Kasper DL. How bacterial carbohydrates influence the adaptive immune system. Annu Rev Immunol 2010; 28:107–30. [DOI] [PubMed] [Google Scholar]
- 46. Amarasinghe JJ, D'Hondt RE, Waters CM, Mantis NJ. Exposure of Salmonella enterica Serovar typhimurium to a protective monoclonal IgA triggers exopolysaccharide production via a diguanylate cyclase‐dependent pathway. Infect Immun. 2013; 81:653–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McLoughlin K, Schluter J, Rakoff‐Nahoum S, Smith AL, Foster KR. Host selection of microbiota via differential adhesion. Cell Host Microbe 2016; 19:550–9. [DOI] [PubMed] [Google Scholar]
- 48. van der Woude MW. Phase variation: how to create and coordinate population diversity. Curr Opin Microbiol 2011; 14:205–11. [DOI] [PubMed] [Google Scholar]
- 49. Mostowy RJ, Holt KE. Diversity‐generating machines: genetics of bacterial sugar‐coating. Trends Microbiol 2018; 26:1008–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Porter NT, Canales P, Peterson DA, Martens EC. A subset of polysaccharide capsules in the human symbiont Bacteroides thetaiotaomicron promote increased competitive fitness in the mouse gut. Cell Host Microbe 2017; 22:494–506.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gencay YE, Sørensen MCH, Wenzel CQ, Szymanski CM, Brøndsted L. Phase variable expression of a single phage receptor in Campylobacter jejuni NCTC12662 influences sensitivity toward several diverse CPS‐dependent phages. Front Microbiol 2018; 9:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Porter NT, Hryckowian AJ, Merrill BD, Gardner JO, Singh S, Sonnenburg JL et al Multiple phase‐variable mechanisms, including capsular polysaccharides, modify bacteriophage susceptibility in Bacteroides thetaiotaomicron . bioRxiv. 2019; 521070 10.1101/521070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bansept F, Schumann‐Moor K, Diard M, Hardt W‐D, Slack E, Loverdo C. Enchained growth and cluster dislocation: a possible mechanism for microbiota homeostasis. PLOS Comput Biol 2019; 15:e1006986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gopalakrishna KP, Macadangdang BR, Rogers MB, Tometich JT, Firek BA, Baker R et al Maternal IgA protects against the development of necrotizing enterocolitis in preterm infants. Nat Med. 2019; 25:1110–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mirpuri J, Raetz M, Sturge CR, Wilhelm CL, Benson A, Savani RC et al Proteobacteria‐specific IgA regulates maturation of the intestinal microbiota. Gut Microbes 2014; 5:28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Diard M, Bakkeren E, Cornuault JK, Moor K, Hausmann A, Sellin ME et al Inflammation boosts bacteriophage transfer between Salmonella spp. Science 2017;355:1211–5. [DOI] [PubMed] [Google Scholar]
- 57. Van Belleghem JD, Dąbrowska K, Vaneechoutte M, Barr JJ, Bollyky PL. Interactions between bacteriophage, bacteria, and the mammalian immune system.Viruses 2018; 11:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mestecky J, Russell MW. Specific antibody activity, glycan heterogeneity and polyreactivity contribute to the protective activity of S‐IgA at mucosal surfaces. Immunol Lett 2009; 124:57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Briliūtė J, Urbanowicz PA, Luis AS, Baslé A, Paterson N, Rebello O et al Complex N‐glycan breakdown by gut Bacteroides involves an extensive enzymatic apparatus encoded by multiple co‐regulated genetic loci. Nat Microbiol 2019; 4:1571–81. [DOI] [PubMed] [Google Scholar]
- 60. Nakajima A, Vogelzang A, Maruya M, Miyajima M, Murata M, Son A et al IgA regulates the composition and metabolic function of gut microbiota by promoting symbiosis between bacteria. J Exp Med 2018; 215:2019–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Litvak Y, Byndloss MX, Bäumler AJ. Colonocyte metabolism shapes the gut microbiota. Science 2018; 362:eaat9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pande S, Kaftan F, Lang S, Svatoš A, Germerodt S, Kost C. Privatization of cooperative benefits stabilizes mutualistic cross‐feeding interactions in spatially structured environments. ISME J 2016; 10:1413–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dal Co A, Ackermann M, van Vliet S. Metabolic activity affects the response of single cells to a nutrient switch in structured populations. J R Soc Interface 2019; 16:20190182. [DOI] [PMC free article] [PubMed] [Google Scholar]


