Abstract
Background
Asthma is a common respiratory disorder with a highly heterogeneous nature that remains poorly understood. The objective was to use whole genome sequencing (WGS) data to identify regions of common genetic variation contributing to lung function in individuals with a diagnosis of asthma.
Methods
WGS data were generated for 1,053 individuals from trios and extended pedigrees participating in the family-based Genetic Epidemiology of Asthma in Costa Rica study. Asthma affection status was defined through a physician’s diagnosis of asthma, and most participants with asthma also had airway hyperresponsiveness (AHR) to methacholine. Family-based association tests for single variants were performed to assess the associations with lung function phenotypes.
Results
A genome-wide significant association was identified between baseline FEV1/FVC ratio and a single-nucleotide polymorphism in the top hit cysteine-rich secretory protein LCCL domain-containing 2 (CRISPLD2) (rs12051168; P = 3.6 × 10−8 in the unadjusted model) that retained suggestive significance in the covariate-adjusted model (P = 5.6 × 10−6). Rs12051168 was also nominally associated with other related phenotypes: baseline FEV1 (P = 3.3 × 10−3), postbronchodilator (PB) FEV1 (7.3 × 10−3), and PB FEV1/FVC ratio (P = 2.7 × 10−3). The identified baseline FEV1/FVC ratio and rs12051168 association was meta-analyzed and replicated in three independent cohorts in which most participants with asthma also had confirmed AHR (combined weighted z-score P = .015) but not in cohorts without information about AHR.
Conclusions
These findings suggest that using specific asthma characteristics, such as AHR, can help identify more genetically homogeneous asthma subgroups with genotype-phenotype associations that may not be observed in all children with asthma. CRISPLD2 also may be important for baseline lung function in individuals with asthma who also may have AHR.
Key Words: airway hyperresponsiveness, asthma, lung function, whole genome sequencing
Abbreviations: AHR, airway hyperresponsiveness; CAMP, Childhood Asthma Management Program; COPSAC2000, Copenhagen Prospective Study on Asthma in Childhood 2000; CRISPLD2, cysteine-rich secretory protein LCCL domain-containing 2; FBAT, family-based association test; GACRS, Genetic Epidemiology of Asthma in Costa Rica study; GALA II, Genes-Environments and Admixture in Latino Americans; GEE, generalized estimating equation; HPR, Hartford-Puerto Rico; LGL1, late gestation lung 1; MAF, minor allele frequency; NHLBI, National Heart, Lung, and Blood Institute; PB, postbronchodilator; SAGE, Study of African Americans, Asthma, Genes and Environments; SAPPHIRE, Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-Ethnicity; SNP, single-nucleotide polymorphism; TOPMed, Trans-Omics for Precision Medicine; WGS, whole genome sequencing
Asthma is a disease with a strong genetic basis and substantial heterogeneity1, 2, 3 combined with early life and environmental factors4 that is characterized in three general domains based on most traditional approaches: (1) reversible airway obstruction, (2) airway hyperresponsiveness (AHR), and (3) airway inflammation.3,5 Despite these features, patients differ across a wide range of clinical manifestations, inflammatory characteristics, severity, and outcomes,3 stemming from differing underlying endotypes and causes.2,6 Asthma outcome has benefited from personalized care programs; however, despite the current national asthma guidelines,7,8 its prevalence and economic burden continues to increase worldwide.3,5 Common clinical symptoms shared across different asthma endotypes, along with variables including age and time of onset, sex, and lung function, make it extremely complicated to define appropriate treatment regimens and strategies for disease control. Although targeted therapies based on disease stratification and severity are available,9,10 precise identification and classification of phenotypes and endotypes has become imperative and will benefit from conducting data-driven large multicenter genome-wide studies.
Albeit genome-wide association studies of asthma have identified and replicated associations of several single-nucleotide polymorphisms (SNPs) with asthma,11, 12, 13, 14 these variants explain only a fraction of the total of disease variants.1 The use of broad categorizations tailored toward clinical classifications further complicate gene discovery through misclassification of case and control subjects that leads to reduced statistical power from misclassification. This explanation has become an impetus for creating more genetically homogeneous disease subgroups.
Whole genome sequencing (WGS) enables a more focused assessment of low-frequency and rare genetic variants; however, this assessment is not feasible until WGS data are available in a large enough number of individuals for sufficient statistical power to evaluate these low-frequency variants. To date, to our knowledge, only two WGS studies of asthma have been published.15,16 A recently published WGS study identified several viral and bacterial species in children with asthma and pneumonia17; however, to our knowledge, none have focused on the genetic basis of lung function among individuals with asthma.15,16 In this study, we present, to our knowledge, the first WGS analysis of lung function phenotypes by using family-based association tests (FBATs)18,19 for extended pedigrees of 1,053 individuals who participated in the Genetic Epidemiology of Asthma in Costa Rica study (GACRS) cohort.20
Materials and Methods
Study Population
The study included children with asthma who were aged 6 to 14 years from the GACRS family-based trios and probands from extended pedigrees who were recruited from a genetically homogeneous Hispanic population isolate in the Central Valley of Costa Rica with one of the highest prevalences of asthma worldwide (24% in children).21 Enrollment and baseline characteristics of the Costa Rican trios have been described previously in detail.20 In brief, unrelated children were eligible if they had a high probability of having at least six great-grandparents born in the Central Valley of Costa Rica and they had asthma, defined as physician-diagnosed asthma plus a history of at least two episodes of troublesome lung symptoms or asthma attacks in the prior year.22 Children with an asthma diagnosis in this cohort also were evaluated for AHR by using a methacholine challenge. Additional probands were included from the extended pedigrees in which asthma was diagnosed by using the same diagnostic criteria. Further details can be found in e-Appendix 1.
Ethics
Oral and written parental consent and participating child’s assent were obtained. The study was approved by the Partners Human Research Committee at Brigham and Women’s Hospital (Boston, MA; Protocol No. 2000-P-001130/55).
Whole Genome Sequencing
WGS was performed as a part of the TOPMed program offered through the NHLBI. Details about the TOPMed sequencing and variant calling are available in e-Appendix 1.
Statistical Analysis
Single-Variant Family-Based Association Scans
Single-variant association analyses across four quantitative lung function phenotypes were performed using FBATs18,19 that use the within-family information and are therefore robust to population stratification.23 The FBAT approach tests for association between the offspring phenotype and the Mendelian residual. The Mendelian residual is described by the offspring genotype minus the expectation under Mendel’s law, computed using the parental genotypes. If parental genotypes are missing, FBAT uses the sufficient statistic approach to compute the expectation under the null hypothesis.24 Treating the parental genetic data and the offspring phenotype in this manner means that this approach is robust against population stratification and phenotype misspecification. For the analysis, we used the publicly available software implementation FBAT Toolkit (https://sites.google.com/view/fbat-web-page).
The association analyses for the lung function phenotypes (prebronchodilator FEV1, prebronchodilator FEV1/FVC ratio, postbronchodilator [PB] FEV1 , PB FEV1/FVC ratio) were performed using two primary statistical models: (1) an unadjusted model in which the phenotype was the mean-centered lung function variable and (2) an adjusted model in which the phenotype was the lung function variable residual of a linear regression model that adjusted for sex, height, and dichotomized age (< or ≥18 years). We performed these analyses by using (1) all probands with available phenotypic information (531 probands of 1,053 individuals from the extended pedigrees and trios), including individuals with and those without an asthma diagnosis, and (2) only probands with an asthma diagnosis (n = 302). A minimum of five informative families was required for all of these analyses, which corresponds approximately to a minor allele frequency (MAF) cutoff of 0.3%.
Multivariate Family-Based Association Scan
Given the correlation between lung function phenotypes, we also used a multivariate generalized estimating equation (GEE) FBAT-GEE model.25 FBAT-GEE accounts for the correlation between phenotypes and therefore minimizes multiple comparisons by analyzing all phenotypes simultaneously, resulting in one joint P value to evaluate whether the genetic variant is associated with any of the lung function phenotypes.
Rare Variant Analysis
Because of the limited sample size with WGS in GACRS, rare variant analyses were underpowered. Therefore, we did not present them here.
Replication Cohorts
We identified six validation cohorts of children with an asthma diagnosis including four from the National Heart, Lung, and Blood Institute (NHLBI) Trans-Omics for Precision Medicine (TOPMed) consortium (CAMP26: Childhood Asthma Management Program, GALAII27: Gene-Environments and Admixture in Latino Americans, SAGE27: Study of African Americans, Asthma, Genes and the Environment and SAPPHIRE28: Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity) and two independent cohorts (HPR29: Hartford-Puerto Rico (HPR) and COPSAC2000: Copenhagen Prospective Study on Asthma in Childhood30, 31). Among these, three cohorts also identified that most of these children with asthma also had confirmed AHR (CAMP,26 HPR,29 COPSAC30). Rest of the three cohorts did not have information about AHR (GALAII, SAGE,27 SAPPHIRE28). Details about the replication study populations can be found in e-Appendix 1.
Replication Analyses
Children with a diagnosis of asthma were included from each cohort for the replication. A total of 5,451 subjects with both genotype and phenotype data were included from CAMP (n = 769), HPR (n = 490), SAPPHIRE (n = 802), GALA II (n = 2,203), SAGE (n = 1,124), and COPSAC2000 (n = 63) as described in Table 1. Linear models with lung function variables, prebronchodilator and PB FEV1 and FEV1/FVC ratios, as outcome were used in both adjusted and unadjusted models in which the adjusted model included age, sex, and height. For CAMP, the models were additionally adjusted for the first two principal components, race and the interaction between genotype and race because this was the only replication population with multiple ethnicities. In SAPPHIRE, the models were adjusted additionally for genome-wide percentage of African ancestry. Combined P values were calculated across the replication cohorts by means of ascertainment schemas. P values were combined using the weighted z-score method, a widely used and robust method for combining P values in meta-analysis,32 from the Bioconductor survcomp33 package in R because of variable sample sizes across cohorts.
Table 1.
Baseline Characteristics and Lung Function Measures of the Whole Genome Sequencing Population
| Characteristic | Cohort |
||||||
|---|---|---|---|---|---|---|---|
| GACRS | CAMP | COPSAC2000 | HPR | SAPPHIRE | GALA II | SAGE | |
| No. of probands | Total: 531 With Asthma: 302 |
769 | 63 | 490 | 802 | 2,203 | 1,124 |
| Age, mean (SD), y | 19.23 (16.49) | 8.92 (2.14) | 7.09 (0.32) | 10.1 (2.67) | 31.72 (14.72) | 12.7 (3.3) | 14.9 (5.3) |
| Male, No. (%) | 271 (51) | 469 (61) | 33 (52) | 267 (54) | 304 (38) | 1,199 (54) | 551 (49) |
| FEV1, mean (SD), L | 2.25 (0.93) | 1.66 (0.48) | 1.39 (0.23) | 1.95 (0.70) | 2.58 (0.77) | 2.46 (0.83) | 2.56 (0.76) |
| FEV1/FVC ratio, mean (SD) | 0.84 (0.08) | 0.79 (0.08) | 0.91 (0.06) | 0.83 (0.09) | 0.76 (9.56) | 0.84 (0.08) | 0.83 (0.09) |
| PB FEV1, mean (SD), L | 2.35 (0.93) | 1.82 (0.51) | 1.47 (0.23) | 2.06 (0.73) | 2.81 (0.76) | 2.71 (0.87) | 2.79 (0.81) |
| PB FEV1/FVC ratio, mean (SD) | 0.87 (0.07) | 0.85 (0.07) | 0.96 (0.04) | 0.85 (0.09) | 0.80 (8.96) | 0.89 (0.06) | 0.87 (0.07) |
No. of children indicates those with asthma diagnosed. GACRS corresponds to the discovery cohort; CAMP, COPSAC2000, and HPR correspond to the replication cohorts in which individuals had a physician’s diagnosis of asthma that was confirmed with airway hyperresponsiveness in most participants; SAPPHIRE, GALA II, and SAGE correspond to the replication cohorts that used a physician’s diagnosis of asthma with no information about airway hyperresponsiveness.
CAMP = Childhood Asthma Management Program; COPSAC2000 = Copenhagen Prospective Study on Asthma in Childhood 2000; GACRS = Genetic Epidemiology of Asthma in Costa Rica study; GALA II = Genes-Environments and Admixture in Latino Americans; HPR = Hartford-Puerto Rico; PB = postbronchodilator; SAGE = Study of African Americans, Asthma, Genes and Environments; SAPPHIRE = Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-Ethnicity.
Results
Baseline Characteristics
On the basis of 1,053 individuals from extended pedigrees, FBAT identified 317 nonsingleton nuclear families and 248 trios for the analysis. Baseline characteristics for the 531 GACRS samples that were used for phenotype construction and the replication cohorts are listed in Table 1.
Single-Variant FBAT
After the quality control and data cleaning procedure, 25.8 million autosomal SNPs remained for single-variant analysis. Additional details are available in e-Appendix 1.
Lung Function Phenotypes
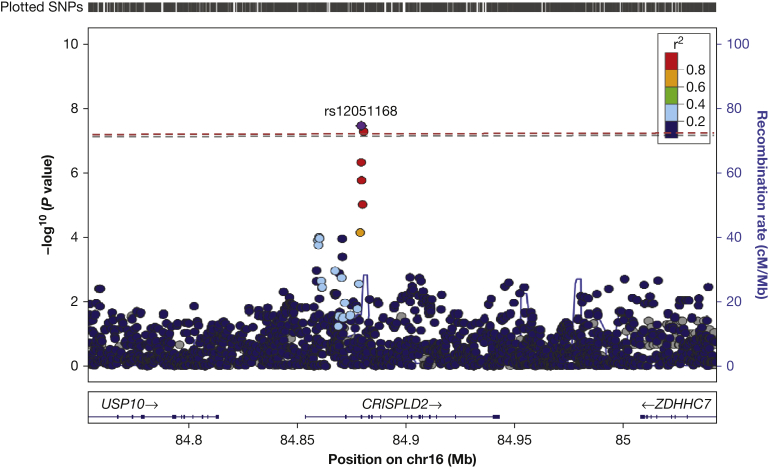
Single-variant association analyses for the four lung function phenotypes led to association P values for 12.2 million SNPs, with λ inflation factors that ranged, across variants with more than 10 informative families, between 1.023 and 1.067. e-Tables 1 through 4 provide lists of all SNPs with association P values lower than 10−5 for each lung function phenotype analyzed in all children in the GACRS. In the evaluation of the covariate-unadjusted analysis, rs12051168 (Fig 1) achieved genome-wide significance for unadjusted baseline FEV1/FVC ratio (P = 3.6 × 10−8) (e-Table 1). SNP rs12051168 is on chromosome 16 (16: 84879324) in the intronic region of the cysteine-rich secretory protein LCCL domain-containing 2 (CRISPLD2) gene with an MAF of 0.39 in Hispanics (Table 2). Another SNP, rs12919905, also in the intronic region of CRISPLD2 gene (chromosome 16: 84880424) suggestively achieved the genome-wide significance threshold (P = 5.34 × 10−8) (e-Table 1). The corresponding P value for rs12051168 in the covariate-adjusted model for FEV1/FVC ratio was 5.61 × 10−6 (e-Table 1, Table 3). The adjusted models also showed similar associations for the other lung function phenotypes: baseline FEV1 (P = 3.3 × 10−3), PB FEV1 (P = 7.3 × 10−3), and PB FEV1/FVC ratio (P = 2.7 × 10−3)(Table 3). The associations with lung function phenotypes were slightly attenuated in the covariate-adjusted model including those with asthma only (Table 3).
Figure 1.
LocusZoom plot highlighting the top hit (rs12051168) of the single-variant whole genome sequencing family-based association test analysis. The plot shows the covariate-unadjusted results with −log10 P values (y-axis 1), recombination rates (y-axis 2), and genome-wide significance threshold (dashed line) for SNPs (rs12051168 as highlighted by the red dashed line, rs12919905 adjacent to rs12051168 as highlighted by the gray dashed line) in the CRISPLD2 gene on chromosome 16. cM = centimorgan; Mb = Megabase; SNP = single-nucleotide polymorphism.
Table 2.
Replication SNPs, Their MAFs, and Ethnicity Across Cohorts
| Cohort | |||
|---|---|---|---|
| Discovery | SNP | MAF | Ethnicity |
| GACRS | rs12051168 | 0.392 | Costa Rican |
| rs12051468 | 0.375 | ||
| Replication | |||
| CAMP | rs12051168 | 0.37 | White |
| rs12051468 | 0.38 | ||
| CAMP | rs12051168 | 0.14 | Black |
| rs12051468 | 0.38 | ||
| HPR | rs12051168 | 0.32 | Puerto Rican |
| rs12051468 | 0.39 | ||
| COPSAC2000 | rs12051168 | 0.46 | European, Danish |
| rs12051468 | 0.43 | ||
| SAPPHIRE | rs12051168 | 0.15 | Black |
| rs12051468 | 0.40 | ||
| GALA II | rs12051168 | 0.32 | Hispanic |
| rs12051468 | 0.34 | ||
| SAGE | rs12051168 | 0.15 | Black |
| rs12051468 | 0.39 | ||
MAF = minor allele frequency; SNP = single-nucleotide polymorphism. See Table 1 legend for expansion of other abbreviations.
Table 3.
Phenotypes Across Discovery and All Replication Cohorts for SNP rs12051168
| Discovery Cohort | |||||
|---|---|---|---|---|---|
| Phenotype | Cohort | z Score | P Valuea (All Samples) | P Valuea (With Asthma) | FBAT-GEE (Joint Phenotypes) |
| FEV1 | GACRS | −2.940 | 3.3 × 10−3 | 1.3 × 10−2 | 4.5 × 10−5 |
| FEV1/FVC ratio | −2.685 | 5.6 × 10−6 | 9.1 × 10−6 | … | |
| PB FEV1 | −4.541 | 7.3 × 10−3 | 1.6 × 10−2 | … | |
| PB FEV1/FVC ratio | 0.003 | 2.7 × 10−3 | 3.5 × 10−3 | … | |
| Replication Cohorts With Asthma Diagnosis and Confirmed AHR in Most Participants | |||||
|---|---|---|---|---|---|
| Phenotype | Cohort | P Valuea | Meta-P Weighted z Score | ||
| Baseline FEV1/FVC ratio | CAMP | .055 | .015 | ||
| HPR | .076 | … | |||
| COPSAC2000 | .11 | … | |||
| Replication in All Cohorts | |||||
|---|---|---|---|---|---|
| Phenotype | Cohort | P Valuea | Meta-P Weighted z Score | ||
| Baseline FEV1/FVC ratio | CAMP | .055 | .35 | ||
| HPR | .076 | … | |||
| COPSAC2000 | .11 | … | |||
| SAPPHIRE | .71 | … | |||
| GALA II | .59 | … | |||
| SAGE | .58 | … | |||
Boldface indicates significant or suggestive P values. The CAMP cohort was adjusted additionally for race and interaction between genotype and race, which was either significant or suggestive in the models for baseline FEV1/FVC ratio (P = .051) and PB FEV1 (P = .054), and PB FEV1/FVC ratio (P = .023); however, race was not significant in any model.
AHR = airway hyperresponsiveness; FBAT = family-based association test; GEE = generalized estimating equation. See Tables 1 and 2 legends for expansion of other abbreviations.
Adjusted for sex, age, and height.
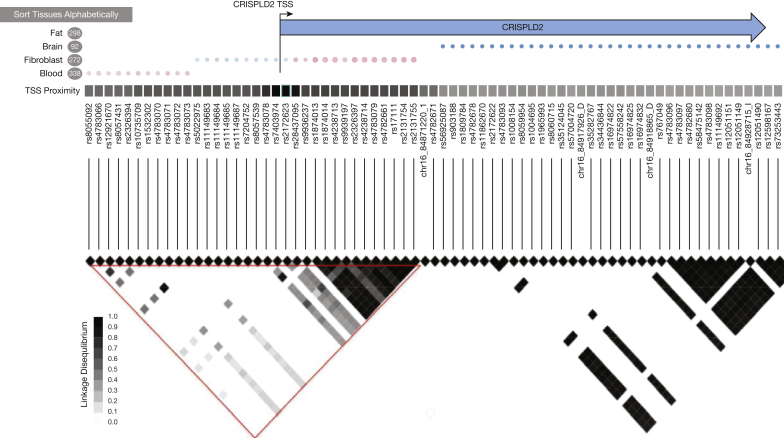
The closest functional variant to the rs12051168 SNP in the CRISPLD2 gene was a splice donor at another SNP, rs185968132, which is illustrated in Figure 2. Furthermore, the SNPs within the 17q21 region, the most replicated childhood asthma locus, which encompasses the ORMDL3 (ORMDL Sphingolipid Biosynthesis Regulator 3) gene12, 13, 34, had nominally significant (P <.05) associations with lung function. In the unadjusted analysis, the functional SNP, rs12936231,35 yielded nominal associations with the baseline FEV1/FVC ratio (P = .044), FEV1 (P = 7.6 × 10−3) and PB FEV1 (P = 8.9 × 10−3).
Figure 2.
The figure provides a haplotype overview of the SNPs in the CRISPLD2 gene on chromosome 16 showing linkage disequilibrium (LD) between the SNPs and corresponding haplotype blocks. TSS = transcription start site.
Multivariate FBAT and Haplotype Analyses
Using FBAT-GEE, we observed an overall significant association between the rs12051168 SNP and the lung function phenotypes (P = 4.5 × 10−5) (Table 3). To investigate further, we used the FBAT haplotype test18 for rs12051168, and the closest coding variant in CRISPLD2, rs12051468, was in linkage disequilibrium with rs12051168 (Table 4). This finding indicated that these two associated SNPs do not show an independent signal and that minor alleles are associated with decreased lung function. Haplotype TA has a frequency of 0.58 and is associated with increased lung function, whereas haplotype CG has a frequency of 0.35 and is associated with decreased lung function.
Table 4.
Haplotypes Showing Significant Associations for rs12051168 and for rs12051468
| Haplotype | Allele Frequency | z Score | P Value |
|---|---|---|---|
| 2-2 (TA) | 0.58 | 4.38 | 1.2 × 10−5 |
| 1-1 (CG) | 0.35 | −4.30 | 1.8 × 10−5 |
| 1-2 | 0.04 | −1.14 | .25 |
| 2-1 | 0.02 | 0.29 | .77 |
Replication in Independent Asthma Cohorts
Table 2 describes the MAFs of the two lung function SNPs of interest, rs12051168 and its coding variant rs12051468, in discovery and in replication cohorts. Notably, although white and Hispanic cohorts had a similar MAF for rs12051168, the black cohorts had a much lower MAF (∼0.15). Table 3 summarizes the replication findings in all cohorts. Although rs12051168 was not significant overall in all of the cohorts, this finding was replicated across the three independent cohorts (HPR, CAMP, and COPSAC2000) in which most children with asthma also had confirmed AHR (combined P value for baseline FEV1/FVC ratio = .015) (Table 3).
Discussion
In this WGS family-based analysis of lung function phenotypes among children with asthma, we identified a plausible suggestive association between baseline FEV1/FVC ratio and the rs12051168 SNP in the CRISPLD2 gene, a gene that has been implicated in pharmacogenetics mechanisms of asthma previously.36 Nominally significant associations also were detected between rs12051168 and the baseline FEV1 and PB FEV1/FVC ratio. A multivariate GEE model detected a significant joint association between rs12051168 and the lung function phenotypes. This finding was replicated and showed a trend for all lung function phenotypes when meta-analyzed in three cohorts with a parental report of physician-diagnosed asthma with AHR confirmed by means of a methacholine challenge in most subjects, suggesting that this lung function association at rs12051168 may be specific to individuals with AHR. We were not able to replicate this finding in other cohorts because AHR was not measured in the other cohorts, so we could not evaluate the subjects on the basis of this criterion. Therefore, our results suggest that we can achieve better accuracy with a more stringent asthma definition and ascertainment. Together, these findings further help confirm those of previous studies36, 37, 38 that suggest CRISPLD2 is a lung function candidate gene for children with asthma that may be specific to those who also have confirmed AHR.
There is significant prior evidence that CRISPLD2, a glucocorticoid-responsive gene and regulator of immune response, could be a potential candidate for pharmacologic targeting to treat asthma. We previously reported that the CRISPLD2 gene on chromosome 16 may play a role in modulating two important asthma pharmacogenetic phenotypes in response to inhaled corticosteroid use.36 The study highlighted several SNPs within CRISPLD2, or spanning 50 kilobase pairs on either side, that were nominally associated with inhaled corticosteroid resistance and bronchodilator response.36 This prior work also identified nominally significant associations between SNPs in the CRISPLD2 gene and changes in lung function after investigating bronchodilator response to an inhaled short-acting β2-agonist36 in a discovery and replication study in more than 2,000 people with asthma.37
Previous functional RNA-sequencing studies of human airway smooth muscle cell lines found that treatment with dexamethasone, a glucocorticoid,36 was associated with increased CRISPLD2 messenger RNA and protein expression and that CRISPLD2 expression was induced by the proinflammatory cytokine IL-1β, suggesting that CRISPLD2 regulates antiinflammatory effects of glucocorticoids in the airways.36 These findings were replicated in other studies by analyzing publicly available expression data from human airway smooth muscle cells treated with dexamethasone38 and fluticasone.39 An immune regulatory role of the CRISPLD2 protein is further supported by studies showing its potential for regulating endotoxin function.40 However, it has been reported that even mutation or overexpression of late gestation lung 1 (LGL1) can lead to cell proliferation and metastasis via binding and upregulation of miR-652-3p in patients with non-small cell lung cancer.41
Although the regulatory role of CRISPLD2 in maintaining proper lung function is accumulating and being realized in humans, its role in crucial developmental processes leading to lung formation are widely reported in mouse studies. CRISPLD2, also known as LGL1, has been shown to regulate fetal lung development in experimental studies of rats.42 Several studies of rat lung models have shown that the glycoprotein-inducible protein encoded by CRISPLD2 is implicated in both branching morphogenesis and alveologenesis and that the absence of LGL1 is lethal to embryos.42, 43, 44 One of the studies highlights that postnatal LGL1 deficiency and knockout leads to production of inflammatory cytokines, altered pulmonary lung function, lung injury, and possible predisposition to an early onset of adult lung disease.43 A 2015 human fetal airway fibroblast study45 confirmed the importance of CRISPLD2 for fetal lung development, which may affect lung function phenotypes in childhood.
The association between CRISPLD2 and baseline FEV1/FVC ratio in this study suggests that CRISPLD2 not only regulates treatment response to inhaled β2-agonists and glucocorticoids but also may play a larger role in asthma pathogenesis by altering baseline lung function. These findings expand on our previous work and suggest that variants within CRISPLD2 may (1) affect altered baseline lung function, a key characteristic of asthma, and (2) be specific to children with confirmed AHR.
The top hit for the rs12051168 SNP within the CRISPLD2 gene was for baseline FEV1/FVC ratio but not FEV1. Children with asthma can have an abnormal FEV1/FVC ratio despite a normal FEV1 or FVC, a condition termed dysanapsis, which is believed to occur when early life lung volume and airway length growth outpace the increase in airway caliber.46 We also previously have shown that airway dysanapsis was associated with worse lung function outcomes47 in children with asthma who were obese,46 highlighting further the importance of reevaluating asthma phenotypes and definitions. Therefore, to avoid potential observation bias, we chose to focus on lung function, one of the common attributes and features of asthma phenotypes in general.
The major strength of our WGS data is the broad coverage of the genome, providing in-depth information about copy number variations; noncoding and intergenic regions; and ability to access rare frequencies, particularly rare variants with MAF lower than 1%. However, we were not able to evaluate rare variants because of limited statistical power. Notably, we still identified a compelling common variant in our WGS analysis: the rs12051168 SNP with an MAF of 0.39 in the population in our study and 0.30 in the 1000 Genomes Project population.48 The family-based single-variant FBAT approach,49 with analysis of trios of children, parents, and extended pedigrees used in our study, is a significant advantage compared with population-based designs because it is robust against population admixture and stratification and allows both linkage and association tests,19 which could be used in future studies.
Despite these strengths, our study was subject to several limitations. Most notably, the small sample size makes the analysis underpowered and prohibits the evaluation of rare genetic variants. Another limitation may be the lack of consistency of phenotypes between the discovery and replication populations; most of the children had mild to moderate asthma with AHR in the discovery population, while the asthma phenotypes in the replication populations were heterogeneous.
Conclusions
In conclusion, we present the first, to our knowledge, WGS analysis of lung function phenotypes among children with asthma, identifying a variant in CRISPLD2 as associated with the baseline FEV1/FVC ratio. Although this finding did not replicate in all six cohorts, there was nominal replication in three cohorts of children with a diagnosis of asthma among whom most also had confirmed AHR. Our findings suggest that more refined evaluation of asthma phenotypes focused on the physiologic characteristics of asthma is useful for identifying genetic variants for specific asthma domains. With CRISPLD2 known to regulate the antiinflammatory effects of glucocorticoids in airway smooth muscle cells, our WGS data extend those findings and suggest that CRISPLD2 may play a larger role in asthma pathogenesis, which is biologically plausible because mechanistic studies have proposed a role of CRISPLD2 in lung organogenesis.
Acknowledgments
Author contributions: P. K. takes full responsibility for this manuscript and had the final responsibility to submit it for publication. P. K. and J. H. take responsibility for the integrity and accuracy of the data analysis and had full access to the data. J. C.C. and S. T. W. conceived and designed the initial Costa Rican study. P. K., J. H., B. L. C., M. H. C., D. Q. analyzed the data. P. K., T. S. A., K. C. B., E. G. B., C. E., D.H., J. C. C., M. D., A. M. L., H. G., L. K. W., E.F., A. C. Y. M., L. A., M. E. S. Q., M. M. C., E.A. P., G. C., K. B., and H. B. performed replication in their cohorts and assisted understanding of the data. C. L. advised and provided the statistical supervision. P. K., J.H., B. L. C., and J. A. L. S. wrote the manuscript. R. S. K., S. H. C., Y. V. V., M. H. and B. A. R. contributed to writing and critical reviewing of the manuscript. All authors reviewed and approved of the final manuscript. We thank all the participants, investigators and staff without whom this work could not have been accomplished.
Financial/nonfinancial disclosures: The authors have reported to CHEST the following: M. H. C. has received consulting fees from Genentech and grant funding from GlaxoSmithKline. J. C. C. has received research materials from GlaxoSmithKline and Merck (inhaled steroids) and Pharmavite (vitamin D and placebo capsules) to provide medications free of cost to participants in studies funded by the National Institutes of Health, unrelated to the current work. L. A. has participated in speaking activities and symposia for Abbott Laboratories, AstraZeneca, GlaxoSmithKline, Sandoz, and Sanofi Pasteur. M. E. S. Q. has participated in speaking activities for AstraZeneca. B. A. R. has received royalties from UpToDate, Inc., as a section editor for Genetics, and has received honoraria for serving on the advisory boards for Sanofi Genzyme and Teva Pharmaceuticals but has no conflicts of interest directly related to this work. J. A. L. S. has been a consultant to Metabolon, Inc. None declared (P. K., J. H., B. L. C., T. S. A., D. Q., R. S. K., S. H. C., Y. V. V., M. H., K. C. B., E. G. B., C. E., D. H., M. D., A. M. L., H. G., L. K. W., E. F., A. C. Y. M., M. M. C., E. A. P., G. C., K. B., H. B., C. L., S. T. W.).
Role of sponsors: The sponsor had no role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript.
*National Heart, Lung, and Blood Institute Trans-Omics for Precision Medicine (TOPMed) Consortium Collaborators: Namiko Abe, New York Genome Center; Goncalo Abecasis, University of Michigan; Christine Albert, Massachusetts General Hospital; Nicholette (Nichole) Palmer Allred, Wake Forest Baptist Health; Laura Almasy, Children's Hospital of Philadelphia, University of Pennsylvania; Alvaro Alonso, Emory University; Seth Ament, University of Maryland; Peter Anderson, University of Washington; Pramod Anugu, University of Mississippi; Deborah Applebaum-Bowden, National Institutes of Health; Dan Arking, Johns Hopkins University; Donna K. Arnett, University of Kentucky; Allison Ashley-Koch, Duke University; Stella Aslibekyan, University of Alabama; Tim Assimes, Stanford University; Paul Auer, University of Wisconsin Milwaukee; Dimitrios Avramopoulos, Johns Hopkins University; John Barnard, Cleveland Clinic; Kathleen Barnes, University of Colorado at Denver; R. Graham Barr, Columbia University; Emily Barron-Casella, Johns Hopkins University; Terri Beaty, Johns Hopkins University; Diane Becker, Johns Hopkins University; Lewis Becker, Johns Hopkins University; Rebecca Beer, National Heart, Lung, and Blood Institute, National Institutes of Health; Ferdouse Begum, Johns Hopkins University; Amber Beitelshees, University of Maryland; Emelia Benjamin, Boston University, Massachusetts General Hospital; Marcos Bezerra, Fundação de Hematologia e Hemoterapia de Pernambuco—Hemope; Larry Bielak, University of Michigan; Joshua Bis, University of Washington; Thomas Blackwell, University of Michigan; John Blangero, University of Texas Rio Grande Valley School of Medicine; Eric Boerwinkle, University of Texas Health at Houston; Ingrid Borecki, University of Washington; Russell Bowler, National Jewish Health; Jennifer Brody, University of Washington; Ulrich Broeckel, Medical College of Wisconsin; Jai Broome, University of Washington; Karen Bunting, New York Genome Center; Esteban Burchard, University of California, San Francisco; Jonathan Cardwell, University of Colorado at Denver; Cara Carty, Women's Health Initiative; Richard Casaburi, University of California, Los Angeles; James Casella, Johns Hopkins University; Mark Chaffin, The Broad Institute; Christy Chang, University of Maryland; Daniel Chasman, Brigham and Women's Hospital; Sameer Chavan, University of Colorado at Denver; Bo-Juen Chen, New York Genome Center; Wei-Min Chen, University of Virginia; Yii-Der Ida Chen, Los Angeles Biomedical Research Institute; Michael H. Cho, Brigham and Women's Hospital; Seung Hoan Choi, The Broad Institute; Lee-Ming Chuang, National Taiwan University; Mina Chung, Cleveland Clinic; Elaine Cornell, University of Vermont; Adolfo Correa, University of Mississippi; Carolyn Crandall, University of California, Los Angeles; James Crapo, National Jewish Health; L. Adrienne Cupples, Boston University; Joanne Curran, University of Texas Rio Grande Valley School of Medicine; Jeffrey Curtis, University of Michigan; Brian Custer, Blood Systems Research Institute UCSF; Coleen Damcott, University of Maryland; Dawood Darbar, University of Illinois at Chicago; Sayantan Das, University of Michigan; Sean David, Stanford University; Colleen Davis, University of Washington; Michelle Daya, University of Colorado at Denver; Mariza de Andrade, Mayo Clinic; Michael DeBaun, Vanderbilt University; Ranjan Deka, University of Cincinnati; Dawn DeMeo, Brigham and Women's Hospital; Scott Devine, University of Maryland; Ron Do, Icahn School of Medicine at Mount Sinai; Qing Duan, University of North Carolina; Ravi Duggirala, University of Texas Rio Grande Valley School of Medicine; Peter Durda, University of Vermont; Susan Dutcher, Washington University in St. Louis; Charles Eaton, Brown University; Lynette Ekunwe, University of Mississippi; Patrick Ellinor, Massachusetts General Hospital; Leslie Emery, University of Washington; Charles Farber, University of Virginia; Leanna Farnam, Brigham and Women's Hospital; Tasha Fingerlin, National Jewish Health; Matthew Flickinger, University of Michigan; Myriam Fornage, University of Texas Health at Houston; Nora Franceschini, University of North Carolina; Mao Fu, University of Maryland; Stephanie M. Fullerton, University of Washington; Lucinda Fulton, Washington University in St. Louis; Stacey Gabriel, The Broad Institute; Weiniu Gan, National Heart, Lung, and Blood Institute, National Institutes of Health; Yan Gao, University of Mississippi; Margery Gass, Fred Hutchinson Cancer Research Center; Bruce Gelb, Icahn School of Medicine at Mount Sinai; Xiaoqi (Priscilla) Geng, University of Michigan; Soren Germer, New York Genome Center; Chris Gignoux, Stanford University; Mark Gladwin, University of Pittsburgh; David Glahn, Yale University; Stephanie Gogarten, University of Washington; Da-Wei Gong, University of Maryland; Harald Goring, University of Texas Rio Grande Valley School of Medicine; C. Charles Gu, Washington University in St. Louis; Yue Guan, University of Maryland; Xiuqing Guo, Los Angeles Biomedical Research Institute; Jeff Haessler, Fred Hutchinson Cancer Research Center, Women's Health Initiative; Michael Hall, University of Mississippi; Daniel Harris, University of Maryland; Nicola Hawley, Yale University; Jiang He, Tulane University; Ben Heavner, University of Washington; Susan Heckbert, University of Washington; Ryan Hernandez, University of California, San Francisco; David Herrington, Wake Forest Baptist Health; Craig Hersh, Brigham and Women's Hospital; Bertha Hidalgo, University of Alabama; James Hixson, University of Texas Health at Houston; John Hokanson, University of Colorado at Denver; Kramer Holly, Loyola University; Elliott Hong, University of Maryland; Karin Hoth, University of Iowa; Chao (Agnes) Hsiung, National Health Research Institute Taiwan; Haley Huston, Blood Works Northwest; Chii Min Hwu, Taichung Veterans General Hospital Taiwan; Marguerite Ryan Irvin, University of Alabama; Rebecca Jackson, Ohio State University Wexner Medical Center; Deepti Jain, University of Washington; Cashell Jaquish, National Heart, Lung, and Blood Institute, National Institutes of Health; Min A. Jhun, University of Michigan; Jill Johnsen, Blood Works Northwest, University of Washington; Andrew Johnson, National Heart, Lung, and Blood Institute, National Institutes of Health; Craig Johnson, University of Washington; Rich Johnston, Emory University; Kimberly Jones, Johns Hopkins University; Priyadarshini Kachroo, Brigham and Women's Hospital; Hyun Min Kang, University of Michigan; Robert Kaplan, Albert Einstein College of Medicine; Sharon Kardia, University of Michigan; Sekar Kathiresan, The Broad Institute; Laura Kaufman, Brigham and Women's Hospital; Shannon Kelly, Vitalant; Eimear Kenny, Icahn School of Medicine at Mount Sinai; Michael Kessler, University of Maryland; Alyna Khan, University of Washington; Greg Kinney, University of Colorado at Denver; Barbara Konkle, Blood Works Northwest; Charles Kooperberg, Fred Hutchinson Cancer Research Center; Stephanie Krauter, University of Washington; Christoph Lange, Harvard School of Public Health; Ethan Lange, University of Colorado at Denver; Leslie Lange, University of Colorado at Denver; Cathy Laurie, University of Washington; Cecelia Laurie, University of Washington; Meryl LeBoff, Brigham and Women's Hospital; Seunggeun Shawn Lee, University of Michigan; Wen-Jane Lee, Taichung Veterans General Hospital Taiwan; Jonathon LeFaive, University of Michigan; David Levine, University of Washington; Dan Levy, National Heart, Lung, and Blood Institute, National Institutes of Health; Joshua Lewis, University of Maryland; Yun Li, University of North Carolina; Honghuang Lin, Boston University; Keng Han Lin, University of Michigan; Simin Liu, Brown University, Women's Health Initiative; Yongmei Liu, Wake Forest Baptist Health; Ruth Loos, Icahn School of Medicine at Mount Sinai; Steven Lubitz, Massachusetts General Hospital; Kathryn Lunetta, Boston University; James Luo, National Heart, Lung, and Blood Institute, National Institutes of Health; Michael Mahaney, University of Texas Rio Grande Valley School of Medicine; Barry Make, Johns Hopkins University; Ani Manichaikul, University of Virginia; JoAnn Manson, Brigham and Women's Hospital; Lauren Margolin, The Broad Institute; Lisa Martin, George Washington University; Susan Mathai, University of Colorado at Denver; Rasika Mathias, Johns Hopkins University; Patrick McArdle, University of Maryland; Merry-Lynn McDonald, University of Alabama; Sean McFarland, Harvard University; Stephen McGarvey, Brown University; Hao Mei, University of Mississippi; Deborah A. Meyers, University of Arizona; Julie Mikulla, National Heart, Lung, and Blood Institute, National Institutes of Health; Nancy Min, University of Mississippi; Mollie Minear, National Heart, Lung, and Blood Institute, National Institutes of Health; Ryan L. Minster, University of Pittsburgh; Braxton Mitchell, University of Maryland; May E. Montasser, University of Maryland; Solomon Musani, University of Mississippi; Stanford Mwasongwe, University of Mississippi; Josyf C. Mychaleckyj, University of Virginia; Girish Nadkarni, Icahn School of Medicine at Mount Sinai; Rakhi Naik, Johns Hopkins University; Pradeep Natarajan, The Broad Institute, Harvard University, Massachusetts General Hospital; Sergei Nekhai, Howard University; Deborah Nickerson, University of Washington; Kari North, University of North Carolina; Jeff O'Connell, University of Maryland; Tim O'Connor, University of Maryland; Heather Ochs-Balcom, University at Buffalo; James Pankow, University of Minnesota; George Papanicolaou, National Heart, Lung, and Blood Institute, National Institutes of Health; Margaret Parker, Brigham and Women's Hospital; Afshin Parsa, University of Maryland; Sara Penchev, National Jewish Health; Juan Manuel Peralta, University of Texas Rio Grande Valley School of Medicine; Marco Perez, Stanford University; James Perry, University of Maryland; Ulrike Peters, Fred Hutchinson Cancer Research Center, University of Washington; Patricia Peyser, University of Michigan; Lawrence S. Phillips, Emory University; Sam Phillips, University of Washington; Toni Pollin, University of Maryland; Wendy Post, Johns Hopkins University; Julia Powers Becker, University of Colorado at Denver; Meher Preethi Boorgula, University of Colorado at Denver; Michael Preuss, Icahn School of Medicine at Mount Sinai; Dmitry Prokopenko, Harvard University; Bruce Psaty, University of Washington; Pankaj Qasba, National Heart, Lung, and Blood Institute, National Institutes of Health; Dandi Qiao, Brigham and Women's Hospital; Zhaohui Qin, Emory University; Nicholas Rafaels, University of Colorado at Denver; Laura Raffield, University of North Carolina; Vasan Ramachandran, Boston University; D. C. Rao, Washington University in St. Louis; Laura Rasmussen-Torvik, Northwestern University; Aakrosh Ratan, University of Virginia; Susan Redline, Brigham and Women's Hospital; Robert Reed, University of Maryland; Elizabeth Regan, National Jewish Health; Alex Reiner, Fred Hutchinson Cancer Research Center, University of Washington; Ken Rice, University of Washington; Stephen Rich, University of Virginia; Dan Roden, Vanderbilt University; Carolina Roselli, The Broad Institute; Jerome Rotter, Los Angeles Biomedical Research Institute; Ingo Ruczinski, Johns Hopkins University; Pamela Russell, University of Colorado at Denver; Sarah Ruuska, Blood Works Northwest; Kathleen Ryan, University of Maryland; Phuwanat Sakornsakolpat, Brigham and Women's Hospital; Shabnam Salimi, University of Maryland; Steven Salzberg, Johns Hopkins University; Kevin Sandow, Los Angeles Biomedical Research Institute; Vijay Sankaran, Harvard University; Christopher Scheller, University of Michigan; Ellen Schmidt, University of Michigan; Karen Schwander, Washington University in St. Louis; David Schwartz, University of Colorado at Denver; Frank Sciurba, University of Pittsburgh; Christine Seidman, Harvard Medical School; Jonathan Seidman, Harvard Medical School; Vivien Sheehan, Baylor College of Medicine; Amol Shetty, University of Maryland; Aniket Shetty, University of Colorado at Denver; Wayne Hui-Heng Sheu, Taichung Veterans General Hospital Taiwan; M. Benjamin Shoemaker, Vanderbilt University; Brian Silver, UMass Memorial Medical Center; Edwin Silverman, Brigham and Women's Hospital; Jennifer Smith, University of Michigan; Josh Smith, University of Washington; Nicholas Smith, University of Washington; Tanja Smith, New York Genome Center; Sylvia Smoller, Albert Einstein College of Medicine; Beverly Snively, Wake Forest Baptist Health; Tamar Sofer, Brigham and Women's Hospital; Nona Sotoodehnia, University of Washington; Adrienne Stilp, University of Washington; Elizabeth Streeten, University of Maryland; Yun Ju Sung, Washington University in St. Louis; Jessica Su-Lasky, Brigham and Women's Hospital; Jody Sylvia, Brigham and Women's Hospital; Adam Szpiro, University of Washington; Carole Sztalryd, University of Maryland; Daniel Taliun, University of Michigan; Hua Tang, Stanford University; Margaret Taub, Johns Hopkins University; Kent Taylor, Los Angeles Biomedical Research Institute; Simeon Taylor, University of Maryland; Marilyn Telen, Duke University; Timothy A. Thornton, University of Washington; Lesley Tinker, Women's Health Initiative; David Tirschwell, University of Washington; Hemant Tiwari, University of Alabama; Russell Tracy, University of Vermont; Michael Tsai, University of Minnesota; Dhananjay Vaidya, Johns Hopkins University; Peter VandeHaar, University of Michigan; Scott Vrieze, University of Colorado at Boulder, University of Minnesota; Tarik Walker, University of Colorado at Denver; Robert Wallace, University of Iowa; Avram Walts, University of Colorado at Denver; Emily Wan, Brigham and Women's Hospital; Fei Fei Wang, University of Washington; Karol Watson, University of California, Los Angeles; Daniel E. Weeks, University of Pittsburgh; Bruce Weir, University of Washington; Scott Weiss, Brigham and Women's Hospital; Lu-Chen Weng, Massachusetts General Hospital; Cristen Willer, University of Michigan; Kayleen Williams, University of Washington; L. Keoki Williams, Henry Ford Health System; Carla Wilson, Brigham and Women's Hospital; James Wilson, University of Mississippi; Quenna Wong, University of Washington; Huichun Xu, University of Maryland; Lisa Yanek, Johns Hopkins University; Ivana Yang, University of Colorado at Denver; Rongze Yang, University of Maryland; Norann Zaghloul, University of Maryland; Maryam Zekavat, The Broad Institute; Yingze Zhang, University of Pittsburgh; Snow Xueyan Zhao, National Jewish Health; Wei Zhao, University of Michigan; Xiuwen Zheng, University of Washington; Degui Zhi, University of Texas Health at Houston; Xiang Zhou, University of Michigan; Michael Zody, New York Genome Center; Sebastian Zoellner, University of Michigan
Other contributions: The National Heart, Lung, and Blood Institute (NHLBI) supported whole genome sequencing (WGS) for the Trans-Omics for Precision Medicine (TOPMed) program. WGS for NHLBI TOPMed: Genes-Environments and Admixture in Latino Americans (GALA II) study (phs#000920) was performed at the New York Genome Center (NYGC) (3R01HL117004-02S3). WGS for NHLBI TOPMed: Study of African Americans, Asthma, Genes and Environments (SAGE) (phs#000921) was performed at NYGC (3R01HL117004-02S3). WGS for NHLBI TOPMed: Genetic Epidemiology of Asthma in Costa Rica study (GACRS) (phs#000988) was performed at the University of Washington Northwest Genomics Center (3R37HL066289-13S1).
Centralized read mapping and genotype calling, along with variant quality metrics and filtering were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1). Phenotype harmonization, data management, sample-identity quality control, and general study coordination were provided by the TOPMed Data Coordinating Center (3R01HL-120393-02S1).
We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed. The authors wish to acknowledge the following GALA II coinvestigators for subject recruitment, sample processing, and quality control: Sandra Salazar, Scott Huntsman, MSc; Lisa Caine; Shannon Thyne, MD; Harold J. Farber, MD, MSPH; Pedro C. Avila, MD; Denise Serebrisky, MD; William Rodriguez-Cintron, MD; Jose R. Rodriguez-Santana, MD; Rajesh Kumar, MD; Luisa N. Borrell, DDS, PhD; Emerita Brigino-Buenaventura, MD; Adam Davis, MA, MPH; Michael A. LeNoir, MD; Kelley Meade, MD; Saunak Sen, PhD; Fred Lurmann, MS; and Harold J. Farber, MD, MSPH. The authors also wish to thank the staff and participants who contributed to GALA II and SAGE.
Additional information: The e-Appendix and e-Tables can be found in the Supplemental Materials section of the online article.
Footnotes
Drs Kachroo, Hecker, and Chawes contributed equally to this manuscript.
FUNDING/SUPPORT: The Genetic Epidemiology of Asthma in Costa Rica study was supported by the National Institutes of Health [Grants HL066289, HL04370]. P. K., J. H., M. H. C., D. Q., S. T. W., and J. A. L. S. were supported by the National Institutes of Health/National Heart Lung and Blood Institute [Grant P01HL132825]. R. S. K. and J. A. L. S. were supported by the National Heart Lung and Blood Institute and the Department of Defense [Grants R01HL123915, W81XWH-17-1-0533]. S.H.C., M. H. and J. A. L. S. were supported by the National Heart Lung and Blood Institute [Grant R01HL141826]. The Hartford-Puerto Rico study was supported by the National Institutes of Health [Grants HL079966, HL117191]. Y. V. V. was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health [Grant K23AI130408]. L. K. W. received support from the Fund for Henry Ford Hospital and grants from the American Asthma Foundation and the National Institutes of Health [R01HL118267, R01AI079139, R01DK113003]. The Copenhagen Prospective Studies on Asthma in Childhood authors, B. L. C., T. S. A., K. B., and H. B., were funded by private and public research funds all listed on http://copsac.com/home/about/funding/. The Lundbeck Foundation, Danish State Budget, Danish Council for Strategic Research, Danish Council for Independent Research, and Capital Region Research Foundation provided core support for the Copenhagen Prospective Studies on Asthma in Childhood. The Genes-Environments and Admixture in Latino Americans study; the Study of African Americans, Asthma, Genes and Environments; E. G. B.; and A. C. Y. M. were supported by the Sandler Family Foundation; American Asthma Foundation; Robert Wood Johnson Foundation Harold Amos Medical Faculty Development Program; Harry Wm. and Diana V. Hind Distinguished Professorship in Pharmaceutical Sciences II; and National Heart, Lung, and Blood Institute [Grants R01HL117004, R01HL128439, R01HL135156, X01HL134589]; the National Institute of Environmental Health Sciences [Grants R01ES015794, R21ES24844]; the National Institute on Minority Health and Health Disparities [Grants P60MD006902, R01MD010443, RL5GM118984]; and the Tobacco-Related Disease Research Program [Grant 24RT-0025].
Contributor Information
Jessica A. Lasky-Su, Email: rejas@channing.harvard.edu.
National Heart, Lung, and Blood Institute Trans-Omics for Precision Medicine (TOPMed) Consortium:
Namiko Abe, Goncalo Abecasis, Christine Albert, Nicholette (Nichole) Palmer Allred, Laura Almasy, Alvaro Alonso, Seth Ament, Peter Anderson, Pramod Anugu, Deborah Applebaum-Bowden, Dan Arking, Donna K. Arnett, Allison Ashley-Koch, Stella Aslibekyan, Tim Assimes, Paul Auer, Dimitrios Avramopoulos, John Barnard, Kathleen Barnes, R. Graham Barr, Emily Barron-Casella, Terri Beaty, Diane Becker, Lewis Becker, Rebecca Beer, Ferdouse Begum, Amber Beitelshees, Emelia Benjamin, Marcos Bezerra, Larry Bielak, Joshua Bis, Thomas Blackwell, John Blangero, Eric Boerwinkle, Ingrid Borecki, Russell Bowler, Jennifer Brody, Ulrich Broeckel, Jai Broome, Karen Bunting, Esteban Burchard, Jonathan Cardwell, Cara Carty, Richard Casaburi, James Casella, Mark Chaffin, Christy Chang, Daniel Chasman, Sameer Chavan, Bo-Juen Chen, Wei-Min Chen, Yii-Der Ida Chen, Michael H. Cho, Seung Hoan Choi, Lee-Ming Chuang, Mina Chung, Elaine Cornell, Adolfo Correa, Carolyn Crandall, James Crapo, L. Adrienne Cupples, Joanne Curran, Jeffrey Curtis, Brian Custer, Coleen Damcott, Dawood Darbar, Sayantan Das, Sean David, Colleen Davis, Michelle Daya, Mariza de Andrade, Michael DeBaun, Ranjan Deka, Dawn DeMeo, Scott Devine, Ron Do, Qing Duan, Ravi Duggirala, Peter Durda, Susan Dutcher, Charles Eaton, Lynette Ekunwe, Patrick Ellinor, Leslie Emery, Charles Farber, Leanna Farnam, Tasha Fingerlin, Matthew Flickinger, Myriam Fornage, Nora Franceschini, Mao Fu, Stephanie M. Fullerton, Lucinda Fulton, Stacey Gabriel, Weiniu Gan, Yan Gao, Margery Gass, Bruce Gelb, Xiaoqi (Priscilla) Geng, Soren Germer, Chris Gignoux, Mark Gladwin, David Glahn, Stephanie Gogarten, Da-Wei Gong, Harald Goring, C. Charles Gu, Yue Guan, Xiuqing Guo, Jeff Haessler, Michael Hall, Daniel Harris, Nicola Hawley, Jiang He, Ben Heavner, Susan Heckbert, Ryan Hernandez, David Herrington, Craig Hersh, Bertha Hidalgo, James Hixson, John Hokanson, Kramer Holly, Elliott Hong, Karin Hoth, Chao (Agnes) Hsiung, Haley Huston, Chii Min Hwu, Marguerite Ryan Irvin, Rebecca Jackson, Deepti Jain, Cashell Jaquish, Min A. Jhun, Jill Johnsen, Andrew Johnson, Craig Johnson, Rich Johnston, Kimberly Jones, Priyadarshini Kachroo, Hyun Min Kang, Robert Kaplan, Sharon Kardia, Sekar Kathiresan, Laura Kaufman, Shannon Kelly, Eimear Kenny, Michael Kessler, Alyna Khan, Greg Kinney, Barbara Konkle, Charles Kooperberg, Stephanie Krauter, Christoph Lange, Ethan Lange, Leslie Lange, Cathy Laurie, Cecelia Laurie, Meryl LeBoff, Seunggeun Shawn Lee, Wen-Jane Lee, Jonathon LeFaive, David Levine, Dan Levy, Joshua Lewis, Yun Li, Honghuang Lin, Keng Han Lin, Simin Liu, Yongmei Liu, Ruth Loos, Steven Lubitz, Kathryn Lunetta, James Luo, Michael Mahaney, Barry Make, Ani Manichaikul, JoAnn Manson, Lauren Margolin, Lisa Martin, Susan Mathai, Rasika Mathias, Patrick McArdle, Merry-Lynn McDonald, Sean McFarland, Stephen McGarvey, Hao Mei, Deborah A. Meyers, Julie Mikulla, Nancy Min, Mollie Minear, Ryan L. Minster, Braxton Mitchell, May E. Montasser, Solomon Musani, Stanford Mwasongwe, Josyf C. Mychaleckyj, Girish Nadkarni, Rakhi Naik, Pradeep Natarajan, Sergei Nekhai, Deborah Nickerson, Kari North, Jeff O'Connell, Tim O'Connor, Heather Ochs-Balcom, James Pankow, George Papanicolaou, Margaret Parker, Afshin Parsa, Sara Penchev, Juan Manuel Peralta, Marco Perez, James Perry, Ulrike Peters, Patricia Peyser, Lawrence S. Phillips, Sam Phillips, Toni Pollin, Wendy Post, Julia Powers Becker, Meher Preethi Boorgula, Michael Preuss, Dmitry Prokopenko, Bruce Psaty, Pankaj Qasba, Dandi Qiao, Zhaohui Qin, Nicholas Rafaels, Laura Raffield, Vasan Ramachandran, D.C. Rao, Laura Rasmussen-Torvik, Aakrosh Ratan, Susan Redline, Robert Reed, Elizabeth Regan, Alex Reiner, Ken Rice, Stephen Rich, Dan Roden, Carolina Roselli, Jerome Rotter, Ingo Ruczinski, Pamela Russell, Sarah Ruuska, Kathleen Ryan, Phuwanat Sakornsakolpat, Shabnam Salimi, Steven Salzberg, Kevin Sandow, Vijay Sankaran, Christopher Scheller, Ellen Schmidt, Karen Schwander, David Schwartz, Frank Sciurba, Christine Seidman, Jonathan Seidman, Vivien Sheehan, Amol Shetty, Aniket Shetty, Wayne Hui-Heng Sheu, M. Benjamin Shoemaker, Brian Silver, Edwin Silverman, Jennifer Smith, Josh Smith, Nicholas Smith, Tanja Smith, Sylvia Smoller, Beverly Snively, Tamar Sofer, Nona Sotoodehnia, Adrienne Stilp, Elizabeth Streeten, Yun Ju Sung, Jessica Su-Lasky, Jody Sylvia, Adam Szpiro, Carole Sztalryd, Daniel Taliun, Hua Tang, Margaret Taub, Kent Taylor, Simeon Taylor, Marilyn Telen, Timothy A. Thornton, Lesley Tinker, David Tirschwell, Hemant Tiwari, Russell Tracy, Michael Tsai, Dhananjay Vaidya, Peter VandeHaar, Scott Vrieze, Tarik Walker, Robert Wallace, Avram Walts, Emily Wan, Fei Fei Wang, Karol Watson, Daniel E. Weeks, Bruce Weir, Scott Weiss, Lu-Chen Weng, Cristen Willer, Kayleen Williams, L. Keoki Williams, Carla Wilson, James Wilson, Quenna Wong, Huichun Xu, Lisa Yanek, Ivana Yang, Rongze Yang, Norann Zaghloul, Maryam Zekavat, Yingze Zhang, Snow Xueyan Zhao, Wei Zhao, Xiuwen Zheng, Degui Zhi, Xiang Zhou, Michael Zody, and Sebastian Zoellner
Supplementary Data
References
- 1.Gibson G. Hints of hidden heritability in GWAS. Nat Genet. 2010;42:558–560. doi: 10.1038/ng0710-558. [DOI] [PubMed] [Google Scholar]
- 2.Barnes K.C. Genetic studies of the etiology of asthma. Proc Am Thorac Soc. 2011;8:143–148. doi: 10.1513/pats.201103-030MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carr T.F., Bleecker E. Asthma heterogeneity and severity. World Allergy Organ J. 2016;9:41. doi: 10.1186/s40413-016-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnelykke K., Ober C. Leveraging gene-environment interactions and endotypes for asthma gene discovery. J Allergy Clin Immunol. 2016;137:667–679. doi: 10.1016/j.jaci.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinberger M. Why clinical practice guidelines hinder rather than help. Paediatr Respir Rev. 2018;25:85–87. doi: 10.1016/j.prrv.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Fanta C.H. Asthma. N Engl J Med. 2009;360:1002–1014. doi: 10.1056/NEJMra0804579. [DOI] [PubMed] [Google Scholar]
- 7.Becker A.B., Abrams E.M. Asthma guidelines: the Global Initiative for Asthma in relation to national guidelines. Curr Opin Allergy Clin Immunol. 2017;17:99–103. doi: 10.1097/ACI.0000000000000346. [DOI] [PubMed] [Google Scholar]
- 8.Global Initiative for Asthma Global strategy for asthma management and prevention. www.ginasthma.org
- 9.Kaur R., Chupp G. Phenotypes and endotypes of adult asthma: moving toward precision medicine. J Allergy Clin Immunol. 2019;144:1–12. doi: 10.1016/j.jaci.2019.05.031. [DOI] [PubMed] [Google Scholar]
- 10.Santus P., Saad M., Damiani G., Patella V., Radovanovic D. Current and future targeted therapies for severe asthma: managing treatment with biologics based on phenotypes and biomarkers. Pharmacol Res. 2019;146:104296. doi: 10.1016/j.phrs.2019.104296. [DOI] [PubMed] [Google Scholar]
- 11.Bønnelykke K., Sleiman P., Nielsen K. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. 2014;46:51–55. doi: 10.1038/ng.2830. [DOI] [PubMed] [Google Scholar]
- 12.Moffatt M.F., Gut I.G., Demenais F. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torgerson D.G., Ampleford E.J., Chiu G.Y. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Himes B.E., Hunninghake G.M., Baurley J.W. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009;84:581–593. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell C.D., Mohajeri K., Malig M. Whole-genome sequencing of individuals from a founder population identifies candidate genes for asthma. PLoS One. 2014;9 doi: 10.1371/journal.pone.0104396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith D., Helgason H., Sulem P. A rare IL33 loss-of-function mutation reduces blood eosinophil counts and protects from asthma. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romero-Espinoza J.A., Moreno-Valencia Y., Coronel-Tellez R.H. Virome and bacteriome characterization of children with pneumonia and asthma in Mexico City during winter seasons 2014 and 2015. PLoS One. 2018;13 doi: 10.1371/journal.pone.0192878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horvath S., Xu X., Lake S.L., Silverman E.K., Weiss S.T., Laird N.M. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol. 2004;26:61–69. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- 19.Laird N.M., Lange C. Family-based designs in the age of large-scale gene-association studies. Nat Rev Genet. 2006;7:385–394. doi: 10.1038/nrg1839. [DOI] [PubMed] [Google Scholar]
- 20.Hunninghake G.M., Soto-Quiros M.E., Avila L. Sensitization to Ascaris lumbricoides and severity of childhood asthma in Costa Rica. J Allergy Clin Immunol. 2007;119:654–661. doi: 10.1016/j.jaci.2006.12.609. [DOI] [PubMed] [Google Scholar]
- 21.Pearce N., Aït-Khaled N., Beasley R. Worldwide trends in the prevalence of asthma symptoms: phase III of the International Study of Asthma and Allergies in Childhood (ISAAC) Thorax. 2007;62:758–766. doi: 10.1136/thx.2006.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Escamilla M.A., Spesny M., Reus V.I. Use of linkage disequilibrium approaches to map genes for bipolar disorder in the Costa Rican population. Am J Med Genet. 1996;67:244–253. doi: 10.1002/(SICI)1096-8628(19960531)67:3<244::AID-AJMG2>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 23.Price A.L., Zaitlen N.A., Reich D., Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rabinowitz D., Laird N. A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered. 2000;50 doi: 10.1159/000022918. 211–23. [DOI] [PubMed] [Google Scholar]
- 25.Lange C., Silverman E.K., Xu X., Weiss S.T., Laird N.M. A multivariate family-based association test using generalized estimating equations: FBAT-GEE. Biostatistics. 2003;4:195–206. doi: 10.1093/biostatistics/4.2.195. [DOI] [PubMed] [Google Scholar]
- 26.The Childhood Asthma Management Program (CAMP): design, rationale, and methods—Childhood Asthma Management Program Research Group. Control Clin Trials. 1999;20(1):91–120. [PubMed] [Google Scholar]
- 27.Nishimura K.K., Galanter J.M., Roth L.A. Early-life air pollution and asthma risk in minority children: the GALA II and SAGE II studies. Am J Respir Crit Care Med. 2013;188:309–318. doi: 10.1164/rccm.201302-0264OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wells K.E., Cajigal S., Peterson E.L. Assessing differences in inhaled corticosteroid response by self-reported race-ethnicity and genetic ancestry among asthmatic subjects. J Allergy Clin Immunol. 2016;137 doi: 10.1016/j.jaci.2015.12.1334. 1364.e2-1369.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brehm J.M., Acosta-Perez E., Klei L. African ancestry and lung function in Puerto Rican children. J Allergy Clin Immunol. 2012;129 doi: 10.1016/j.jaci.2012.03.035. 1484.e6-1490.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bisgaard H. The Copenhagen Prospective Study on Asthma in Childhood (COPSAC): design, rationale, and baseline data from a longitudinal birth cohort study. Ann Allergy Asthma Immunol. 2004;93:381–389. doi: 10.1016/S1081-1206(10)61398-1. [DOI] [PubMed] [Google Scholar]
- 31.Bisgaard H., Jensen S.M., Bonnelykke K. Interaction between asthma and lung function growth in early life. Am J Respir Crit Care Med. 2012;185:1183–1189. doi: 10.1164/rccm.201110-1922OC. [DOI] [PubMed] [Google Scholar]
- 32.Zaykin D.V. Optimally weighted Z-test is a powerful method for combining probabilities in meta-analysis. J Evol Biol. 2011;24(8):1836–1841. doi: 10.1111/j.1420-9101.2011.02297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schroder M.S., Culhane A.C., Quackenbush J., Haibe-Kains B. survcomp: an R/Bioconductor package for performance assessment and comparison of survival models. Bioinformatics. 2011;27:3206–3208. doi: 10.1093/bioinformatics/btr511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galanter J., Choudhry S., Eng C. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am J Respir Crit Care Med. 2008;177:1194–1200. doi: 10.1164/rccm.200711-1644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verlaan D.J., Berlivet S., Hunninghake G.M. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85:377–393. doi: 10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Himes B.E., Jiang X., Wagner P. RNA-Seq transcriptome profiling identifies CRISPLD2 as a glucocorticoid responsive gene that modulates cytokine function in airway smooth muscle cells. PLoS One. 2014;9 doi: 10.1371/journal.pone.0099625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Himes B.E., Jiang X., Hu R. Genome-wide association analysis in asthma subjects identifies SPATS2L as a novel bronchodilator response gene. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masuno K., Haldar S.M., Jeyaraj D. Expression profiling identifies Klf15 as a glucocorticoid target that regulates airway hyperresponsiveness. Am J Respir Cell Mol Biol. 2011;45:642–649. doi: 10.1165/rcmb.2010-0369OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Misior A.M., Deshpande D.A., Loza M.J., Pascual R.M., Hipp J.D., Penn R.B. Glucocorticoid- and protein kinase A-dependent transcriptome regulation in airway smooth muscle. Am J Respir Cell Mol Biol. 2009;41:24–39. doi: 10.1165/rcmb.2008-0266OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z.Q., Xing W.M., Fan H.H. The novel lipopolysaccharide-binding protein CRISPLD2 is a critical serum protein to regulate endotoxin function. J Immunol. 2009;183:6646–6656. doi: 10.4049/jimmunol.0802348. [DOI] [PubMed] [Google Scholar]
- 41.Yang W., Zhou C., Luo M. MiR-652-3p is upregulated in non-small cell lung cancer and promotes proliferation and metastasis by directly targeting Lgl1. Oncotarget. 2016;7:16703–16715. doi: 10.18632/oncotarget.7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oyewumi L., Kaplan F., Gagnon S., Sweezey N.B. Antisense oligodeoxynucleotides decrease LGL1 mRNA and protein levels and inhibit branching morphogenesis in fetal rat lung. Am J Respir Cell Mol Biol. 2003;28(2):232–240. doi: 10.1165/rcmb.4877. [DOI] [PubMed] [Google Scholar]
- 43.Lan J., Ribeiro L., Mandeville I. Inflammatory cytokines, goblet cell hyperplasia and altered lung mechanics in Lgl1+/- mice. Respir Res. 2009;10:83. doi: 10.1186/1465-9921-10-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nadeau K., Montermini L., Mandeville I. Modulation of Lgl1 by steroid, retinoic acid, and vitamin D models complex transcriptional regulation during alveolarization. Pediatr Res. 2010;67(4):375–381. doi: 10.1203/PDR.0b013e3181d23656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H., Sweezey N.B., Kaplan F. LGL1 modulates proliferation, apoptosis, and migration of human fetal lung fibroblasts. Am J Physiol Lung Cell Mol Physiol. 2015;308:L391–L402. doi: 10.1152/ajplung.00119.2014. [DOI] [PubMed] [Google Scholar]
- 46.Forno E., Weiner D.J., Mullen J. Obesity and airway dysanapsis in children with and without asthma. Am J Respir Crit Care Med. 2017;195:314–323. doi: 10.1164/rccm.201605-1039OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones M.H., Roncada C., Fernandes M.T.C. Asthma and obesity in children are independently associated with airway dysanapsis. Front Pediatr. 2017;5:270. doi: 10.3389/fped.2017.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abecasis G.R., Altshuler D., Auton A. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loehlein Fier H., Prokopenko D., Hecker J. On the association analysis of genome-sequencing data: a spatial clustering approach for partitioning the entire genome into nonoverlapping windows. Genet Epidemiol. 2017;41(4):332–340. doi: 10.1002/gepi.22040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.