Abstract
BACKGROUND & AIMS:
Inhibitors of MET have not produced satisfactory outcomes in trials of patients with liver cancer. We investigated the mechanisms of liver tumor resistance to MET inhibitors in mice.
METHODS:
We tested the effects of MET inhibitors tivantinib and capmatinib in the mouse hepatocellular carcinoma (HCC) cell line HCA-1 and in immune-competent and immunodeficient mice with subcutaneous tumors grown from this cell line. Tumors were collected from mice and tumor cells were analyzed by time-of-flight mass cytometry. We used short hairpin RNAs to weaken expression of MET in Hep3B, SK-HEP-1, HA59T, and HA22T liver cancer cell lines and analyzed cells by immunoblot, immunofluorescence, and immunoprecipitation assays. Mass spectrometry was used to assess interactions between MET and glycogen synthase kinase 3β (GSK3B), and GSK3B phosphorylation, in liver cancer cell lines. C57/BL6 mice with orthotopic tumors grown from Hep1-6 cells were given combinations of capmatinib or tivantinib and antibodies against programmed cell death 1 (PDCD1; also called PD1); tumors were collected and analyzed by immunofluorescence. We analyzed 268 HCCsamples in a tissue microarray by immunohistochemistry.
RESULTS:
Exposure of liver cancer cell lines to MET inhibitors increased their expression of PD ligand 1 (PDL1) and inactivated cocultured T cells. MET phosphorylated and activated GSK3B at tyrosine 56, which decreased the expression of PDL1 by liver cancer cells. In orthotopic tumors grown in immune-competent mice, MET inhibitors decreased the antitumor activity of T cells. However, addition of anti-PD1 decreased orthotopic tumor growth and prolonged survival of mice compared with anti-PD1 or MET inhibitors alone. Tissue microarray analysis of HCC samples showed an inverse correlation between levels of MET and PDL1 and a positive correlation between levels of MET and phosphorylated GSK3B.
CONCLUSIONS:
In studies of liver cancer cell lines and mice with orthotopic tumors, MET mediated phosphorylation and activated GSK3B, leading to decreased expression of PDL1. Combined with a MET inhibitor, anti-PD1 and anti-PDL1 produced additive effect to slow growth of HCCs in mice.
Keywords: Programmed Cell Death Ligand 1, Hepatocellular Carcinoma, Tumor Necrosis Factor Receptor-Associated Factor 6, Glycogen Synthase Kinase 3
Hepatocellular carcinoma (HCC) is the fourth most common cause of cancer-related mortality world-wide. Current treatments for HCC include surgical resection, liver transplantation, and local ablation for early-stage disease,1 but they are not effective. The small-molecule multikinase inhibitors sorafenib (first-line use),2 regorafenib (second-line use),3 and lenvatinib (first-line use)4 have been approved by the US Food and Drug Administration to treat advanced HCC. Cabozantinib treatment showed positive results in second-line use in patients previously treated for advanced HCC.5 However, those drugs improved the median overall survival duration by only less than 4 months in patients with advanced HCC and showed a low overall response rate.2-5 Anti-programmed cell death protein 1 (PD1), which was recently approved for HCC therapy, achieves only a 20% response rate.6 Although the improved response rate of anti-PD1 is encouraging, the development of more effective therapeutic approaches for HCC is urgently needed.
Patients with HCC and high expression of receptor tyrosine kinase MET (also known as hepatocyte growth factor) were reported to have lower survival rates than those with no or low MET expression.7-9 Thus, MET is considered an excellent therapeutic target for HCC. Indeed, Gherardi et al10 showed that MET inhibitors effectively suppressed tumor growth in xenograft animal models. To date, 6 MET inhibitors have been developed and are under investigation in 10 HCC clinical trials.11 However, those drugs, aimed at directly inhibiting MET activities, have shown limited benefits. For instance, the non-adenosine triphosphate-competitive MET inhibitor, tivantinib (ARQ197), failed to meet its primary end point of improving overall survival in a phase 3 HCC clinical trial.12 It is not yet clear why MET inhibitors failed to show benefits in HCC,12 which is known to overexpress MET.9
Programmed death-ligand 1 (PDL1), also known as B7-H1 and CD274, binds to its receptor, programmed cell death protein 1 (PD1), to suppress tumor-infiltrating lymphocytes in tumors.13,14 Inhibition of the PD1-PDL1 pathway has shown promising results for the treatment of different cancer types including HCC,15-17 and a phase 1–2 trial for HCC indicated that nivolumab treatment produced favorable responses in patients with advanced disease.6 The US Food and Drug Administration recently approved nivolumab for the treatment of advanced HCC refractory to sorafenib, and the objective response rate for nivolumab is 20% in patients with advanced HCC.6 The result is highly encouraging and provides an excellent baseline to develop more effective combination therapies to treat HCC. In addition, recent studies showed that PDL1 expression is positively correlated with the anti-PD1 and anti-PDL1 therapy response rate in certain tumor cells.18 Therefore, understanding the molecular regulation of PDL1 expression could identify a biomarker to predict response to immune checkpoint blockade for HCC treatment.
Glycogen synthase kinase 3b (GSK3B), a serine- and threonine-specific protein kinase and an essential component of the Wnt signaling pathway, plays a significant role in tumorigenesis and embryonic development.19,20 GSK3B induces phosphorylation-dependent proteasome degradation of Snail,21 Mcl-1,22 and EZH2,23 resulting in cancer cell mesenchymal-epithelial transition, apoptosis, and chromosome stability. Our recent studies also showed that GSK3B stringently downregulates PDL1 protein stability by increasing its phosphorylation at T180 and S184 and E3 ligase-mediated degradation.24 Inhibition of epidermal growth factor receptor signaling in cancer cells up-regulates GSK3B activity and inactivates AKT25 and extracellular signal-regulated kinase,26 thereby destabilizing PDL1.24 In contrast, GSK3B activity is down-regulated under MET inhibition.27 Therefore, understanding how MET regulates GSK3B activity could uncover a novel molecular mechanism of PDL1 protein stability.
Methods
Animal Studies
All procedures using C3H, nonobese diabetic, and severe combined immunodeficiency gamma (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ), and C57BL/6 mice (male, 6 weeks old; The Jackson Laboratory, Bar Harbor, ME) were conducted under guidelines approved by the Institutional Animal Care and Use Committee of The University of Texas MD Anderson Cancer Center (Houston, TX). For antibody-based drug intervention, PD1 antibody 100 μg (RMP1-14; Bio X Cell, West Lebanon, NH) or rat immunoglobulin G (control; Bio X Cell) was injected intraperitoneally into mice every 3 days 1 week after tumor-cell inoculation. For drug-based drug intervention, mice were given daily oral doses of capmatinib 10 mg/kg reconstituted in 0.5% methylcellulose and 5% dimethylacetamide28 or tivantinib 100 mg/kg12 formulated in tocopherol polyethylene glycol 1000 succinate (BioXtra, water-soluble vitamin E conjugate). Tumor volumes were calculated using the formula (length × width2)/2. At the experimental end point, mice were killed using CO2 exposure followed by cervical dislocation, and tumors were excised for subsequent histologic analysis or processed immediately for mass cytometric and flow cytometric analyses.
All further materials and methods can be found in the Supplementary Methods.
Results
MET Inhibition Up-regulates PDL1 Expression in HCC Cells and Induces T-Cell Suppression
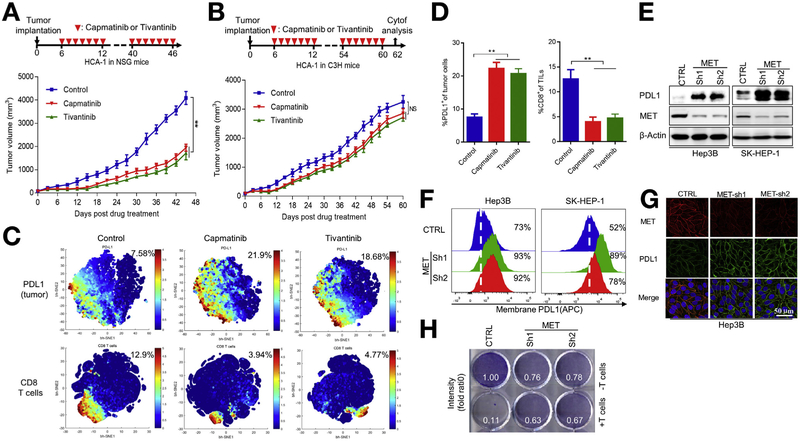
The failure of MET inhibitor tivantinib in phase 3 HCC clinical trials prompted us to refine the therapeutic models currently used in HCC preclinical studies. To improve the therapeutic efficacy of MET inhibitors, we tested the response of the mouse HCC cell line HCA-1 to tivantinib and another MET inhibitor, capmatinib, which is a potent and selective MET kinase inhibitor currently in HCC clinical trial.11,28 As expected, tivantinib and capmatinib decreased cell proliferation of HCA-1 (Supplementary Figure 1A). Next, we subcutaneously inoculated HCA-1 cells into immunocompetent (C3H) and immunodeficient (nonobese diabetic and severe combined immunodeficiency gamma) mice. Drug intervention with capmatinib or tivantinib decreased tumor progression in nonobese diabetic and severe combined immunodeficiency gamma mice (Figure 1A) but failed to inhibit tumor growth in C3H mice (Figure 1B). These results suggested that an intact immune system compromises the therapeutic efficacy of MET inhibitors.
Figure 1.
MET inhibition up-regulates PDL1 expression in HCC cells and induces T-cell suppression. (A) Growth of tumors generated by HCA-1 cells in NSG mice after drug intervention with capmatinib or tivantinib. (Top) Summary of drug intervention protocol. Tumors were measured at the indicated time points. (Bottom) Quantification of tumor-volume changes. (B) HCA-1 tumor growth in C3H mice after drug intervention with capmatinib or tivantinib. (Bottom) Quantification of tumor-volume changes. (C) viSNE map derived from time-of-flight mass spectrometric analysis of HCA-1 tumors in mice after drug intervention with capmatinib or tivantinib. Tumor cell populations were detected using PDL1 and CD8 as markers. Cells in the map are color coded according to the intensity of the expression of the indicated markers. (D) Quantification of PDL1+ tumor cell populations and CD8+ TIL populations in HCA-1 tumors assessed using time-of-flight mass spectrometric analysis and analyzed using viSNE (n = 3). **P < .01 by Student t test. All error bars represent mean ± standard deviation. (E) Immunoblot of MET protein expression and PDL1 expression induced by a vector control and MET knockdown in Hep3B and SK-HEP-1 cells. (F) Flow cytometric analysis of cell-surface PDL1 expression in Hep3B (left) and SK-HEP-1 (right) cells. (G) Confocal microscopic images showing MET and PDL1 protein expression in vector control and MET-knockdown Hep3B cells. Scale bar, 50 γm. (H) T-cell-mediated tumor cell killing assay of Hep3B cells expressing vector control or MET shRNA. Activated T cells and Hep3B cells were cocultured in 12-well plates for 4 days, and the surviving tumor cells were visualized using crystal violet staining. Relative fold ratios of surviving cell intensities are shown. APC, allophycocyanin; CTRL, control; Cytof, time-of-flight mass cytometry; NSG, nonobese diabetic and severe combined immunodeficiency gamma; TIL, tumor-infiltrating lymphocyte.
To determine how immune cells contribute to the failure of MET inhibitors, we used time-of-flight mass cytometry to systematically analyze the immunophenotypes of tumor-infiltrating T cells and tumor cells from HCA-1 tumors with or without capmatinib- or tivantinib-based drug intervention using 2 separate antibody panels (Supplementary Table 1). Unexpectedly, the expression of PDL1 increased substantially in the tumor region with a concomitant decrease in CD8+ T-cell activity when mice received capmatinib or tivantinib (Figure 1C and D). The MET inhibitor-mediated up-regulation of PDL1 was further validated in multiple HCC cell lines, including mouse HCA-1 and human Hep3B, by western blot analysis (Supplementary Figure 1B). Because patients with HCC and high PDL1 expression (>5% of HCC cells) had markedly shorter overall and progression-free survival durations than patients with low PDL1 expression (<5% of HCC cells; Supplementary Figure 1C-E),29,30 we focused on the relation between PDL1 and MET in HCC for all subsequent experiments. Together, our studies in various syngeneic liver cancer models of different hosts enabled us to understand the adverse effect of MET inhibitors against HCC in clinical trials and identify an unexpected MET inhibitor-mediated up-regulation of PDL1.
To determine whether capmatinib- or tivantinib-mediated PDL1 up-regulation is MET dependent, we used 2 independent short hairpin RNAs (shRNAs) to downregulate MET expression in liver cancer cells. In Hep3B and SK-HEP-1 cells, knocking down MET induced PDL1 expression (Figure 1E), cell-surface PDL1 expression (Figure 1F), and PDL1 membrane localization (Figure 1G). Because SK-HEP-1 is of endothelial origin, it might not be a good model for HCC.31,32 However, the data validated our findings in other tumor types and tissues. We also tested the effects of MET inhibition in 6 more HCC cell lines, HA59T, HA22T, PLC, WRL68, Huh-7, and Tong. We found that MET knockdown led to PDL1 stabilization in those 6 HCC cell lines (Supplementary Figure 1F). It is interesting to note that the effects of MET inhibition on PDL1 stability seemed to be more profound in HA59T, which expresses higher levels of MET, than in HA22T, suggesting that PDL1 stability is highly dependent on the MET expression in HCC. In addition, we screened multiple HCC cell lines and showed PDL1 and phosphorylated (p-) MET are inversely correlated (Supplementary Figure 1G). Together, the result supported that inhibition of MET activity increases PDL1 expression in HCC. Similarly, kinase-dead MET also increased PDL1 expression, whereas re-expression of wild-type (WT) MET decreased PDL1 expression (Supplementary Figure 1H). Moreover, MET-knockdown Hep3B cells were more resistant to human T-cell-mediated cytolysis than control cells, likely due to the increased PDL1 expression (Figure 1H). Together, these results indicated that MET inhibition stabilizes PDL1 expression in HCC and allows HCC to escape from T-cell killing.
MET Inhibition Drives PDL1 Expression by Suppressing GSK3B-Mediated PDL1 Degradation
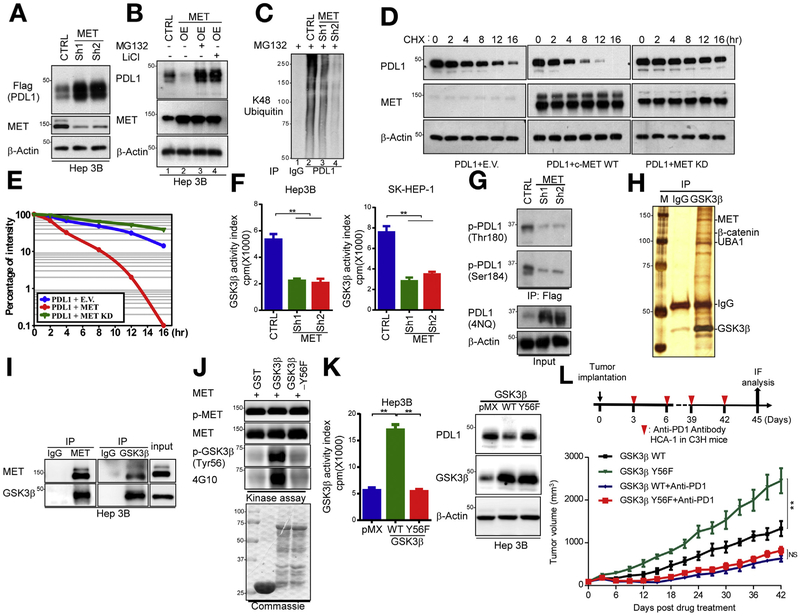
Next, we investigated the mechanisms by which MET regulates PDL1 in HCC cells. Knocking down MET had no effects on PDL1 mRNA expression (Supplementary Figure 1I) in Hep3B or SK-HEP-1 cells, implying that the regulation is not at the transcriptional level and might be at the post-translational level. Indeed, similar to the endogenous PDL1 (Figure 1E), exogenous expression of Flag-tagged PDL1 (Flag-PDL1; cytomegalovirus promoter) was increased in MET-knockdown Hep3B cells (Figure 2A). The addition of proteasome inhibitor MG132 consistently abrogated MET-mediated PDL1 down-regulation in Hep3B cells (Figure 2B, lanes 3 vs 2), suggesting that MET downregulates PDL1 protein expression in a proteasome-dependent manner.
Figure 2.
MET blockade drives PDL1 expression by suppression of GSK3B-mediated PDL1 degradation in HCC cells. (A) Western blot analysis of Flag-PDL1 expression using Flag antibody in MET-knockdown Hep3B cells transfected with Flag-PDL1. (B) Immunoblot analysis of whole cell lysates derived from vector control or MET-overexpressing Hep3B cells given MG132 (5 μmol/L) for 16 hours and LiCl (25 μmol/L) for 5 hours. (C) Ubiquitination assay of PDL1 in MET-knockdown Hep3B cells. Ubiquitinated PDL1 was immuno-precipitated and subjected to western blot analysis with antibody against ubiquitin. Cells were treated with MG132 before ubiquitination analysis. (D) Stability of PDL1 protein in Hep3B cells transfected with HA-PDL1 and WT Flag-MET or MET-KD. Cells were treated with CHX 20 mmol/L at the indicated intervals and subjected to western blot analysis. (E) Quantification of PDL1 half-life in indicated groups. (F) Kinase activity of GSK3B in MET-knockdown Hep3B and SK-HEP-1 cells according to an in vitro kinase assay and phosphorylation analysis. Columns indicate mean activity after subtraction of background phosphorylation. **P < .01. (G) Western blot analysis of PDL1 phosphorylation at T180 and S184 by phospho-Y180 and phospho-S184 PDL1 antibodies in vector control and MET-knockdown Hep3B cells, respectively. (H) Coomassie blue staining of GSK3B-interacting proteins in Hep3B cells. Interacting proteins were isolated and identified by mass spectrometry. (I) Endogenous co-immunoprecipitation of Hep3B cells using MET and GSK3B antibodies. Cell lysates were analyzed by western blotting. (J) In vitro assay of GSK3B phosphorylation by MET at Y56. Purified WT GST-GSK3B and Y56F were incubated with recombinant MET kinase in the presence of adenosine triphosphate at 30°C for 30 minutes. Protein lysates were analyzed by western blotting. (K) (Left) Kinase activity of GSK3B in Hep3B and SK-HEP-1 cells expressing pMX (empty vector), WT GSK3B, or GSK3B Y56F by in vitro kinase assay and phosphorylation analysis. **P < .01. (Right) immunoblot of PDL1 and GSK3B expression in Hep3B cells transiently transfected with pMX (empty vector), WT GSK3B, and/or GSK3B Y56F. (L) (Top) Schematic of drug intervention protocol for PD1 antibody in C3H mice. At the drug intervention end point, tumors were isolated for immunofluorescent analysis. (Bottom) Growth of HCA-1 tumors in C3H mice that were treated with or without the PD1 antibody. Tumors were measured at the indicated time points. CHX, cycloheximide; CTRL, control; E.V., empty vector; GST, glutathione S-transferase; HA-PDL1, hemagglutinin-tagged PDL1; IgG, immunoglobulin G; IP, immuno-precipitated; KD, kinase-dead; OE, overexpression.
Because GSK3B is an essential kinase that downregulates PDL1 protein stability24 and intervention with a MET inhibitor was reported to inhibit GSK3B activity in cancer cells,27 we investigated whether MET destabilizes PDL1 via GSK3B-mediated PDL1 K48 ubiquitination. To this end, we showed that GSK3B was required for MET-mediated PDL1 down-regulation (Figure 2B, lanes 4 vs 2). We observed PDL1 K48 ubiquitination in the presence of MG132 (Figure 2C, lanes 2 vs 1), which was abolished by MET knockdown in Hep3B cells (Figure 2C, lanes 3 and 4 vs 2). Pulse-chase analysis using cycloheximide indicated that overexpression of WT but not kinase-dead MET shortened the PDL1 protein half-life in Hep3B cells (Figure 2D and E), suggesting that MET-mediated PDL1 down-regulation requires the enzyme activity of MET. Next, we immuno-precipitated endogenous GSK3B and measured the kinase activity of GSK3B in MET-knockdown Hep3B and SK-HEP-1 cells using peptides specifically phosphorylated by GSK3B.26 Knocking down MET inhibited the kinase activity of GSK3B (Figure 2F), supporting the notion that MET blockade downregulates GSK3B activity.27 Because phosphorylation of PDL1 at T180 and S184 by GSK3B primes PDL1 for protein ubiquitination and degradation,24 we established that knocking down MET decreased PDL1 phosphorylation at those 2 sites (Figure 2G). Together, these results indicated that MET blockade stabilizes PDL1 by inhibiting GSK3B-mediated PDL1 phosphorylation and degradation.
MET Binds to and Phosphorylates GSK3B at Tyrosine 56 to Activate its Kinase Activity
To determine whether MET binds to and activates GSK3B, we immuno-precipitated endogenous GSK3B complexes from Hep3B cells followed by tandem multi-time-of-flight mass spectrometric analysis to identify GSK3B-interacting proteins (Figure 2H). In addition to β-catenin, a well-known binding partner for GSK3B, we identified MET, ubiquitin-like modifier-activating enzyme 1, and ubiquitin as GSK3B-binding proteins with high sequence coverage (Figure 2H and Supplementary Table 2). The interaction between MET and GSK3B in Hep3B cells was further validated by co-immunoprecipitation (Figure 2I). To determine whether GSK3B is a physiologic substrate of MET, we induced ectopic expression of GSK3B with MET in Hep3B cells followed by immunoprecipitation to examine their phosphorylation status. We discovered strong tyrosine phosphorylation of GSK3B by MET using a phospho-tyrosine antibody (4G10; Supplementary Figure 2A). In contrast, MET failed to phosphorylate other kinases such as IkB kinases or extracellular signal-regulated kinase (Supplementary Figure 2A), suggesting the MET-mediated GSK3B phosphorylation is specific. Mass spectrometric analysis identified phosphorylation of GSK3B at tyrosine 56 (Y56) and Y216 in vivo (Supplementary Figure 2B), whereas GSK3B was phosphorylated directly by MET only at Y56 in vitro (Supplementary Figure 2C). An in vitro kinase assay using purified glutathione S-transferase-GSK3B-kinase-dead protein and full-length recombinant MET kinase confirmed that MET phosphorylates GSK3B at Y56 because the Y56F mutant virtually lost the phosphorylation by MET (Figure 2J, 4G10). To recapitulate this phosphorylation under various experimental conditions, we generated a polyclonal antibody that specifically recognizes p-GSK3B Y56 (anti-phospho-Y56). Mutation of GSK3B Y56 to phenylalanine (Y56F) consistently abrogated its phosphorylation (Figure 2J, p-GSK3B). In a functional assessment, HCC cells expressing GSK3B Y56F mutant, which was no longer phosphorylated by MET, had lower GSK3B kinase activity and higher PDL1 expression than did cells harboring WT GSK3B (Figure 2K). In vivo, implanted tumors expressing GSK3B Y56F grew faster than did tumors expressing WT GSK3B in C3H mice, and the difference in tumor burden between the WT GSK3B and GSK3B Y56F groups was largely decreased after drug intervention with a PD1 antibody (Figure 2L). Notably, tumors expressing GSK3B Y56F had markedly increased PDL1 expression and decreased CD8+ T-cell populations and granzyme B expression (T-cell activity), which were reversed by PD1 antibody (Supplementary Figure 2D). Taken together, these findings demonstrated that MET phosphorylates GSK3B at Y56 in HCC cells, leading to increased GSK3B activity and decreased PDL1 expression, and the enhanced tumorigenic potential of GSK3B Y56F cells is caused by enhanced PDL1 expression, resulting in increased immune evasion.
Y56 Phosphorylation Up-regulates GSK3B Activity by Suppression of Tumor Necrosis Factor Receptor-Associated Factor 6-Mediated GSK3B K63 Ubiquitination
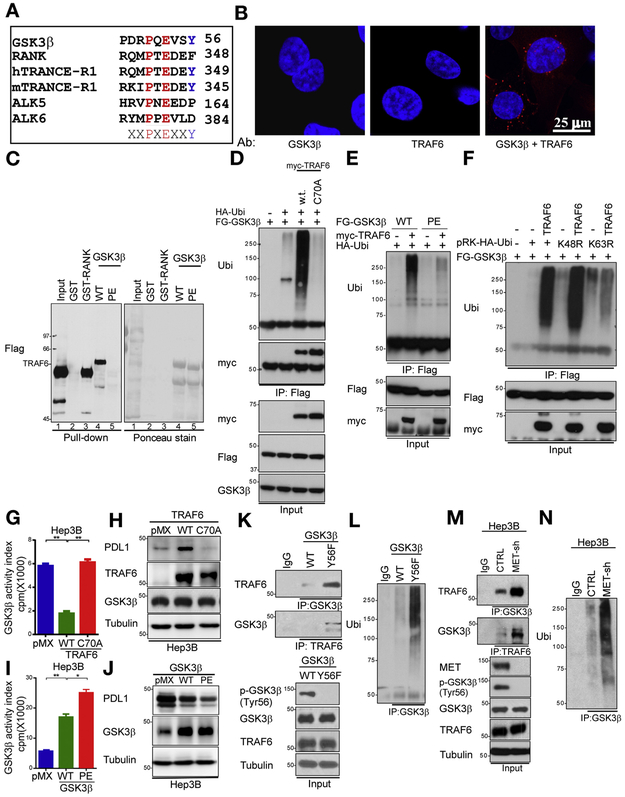
Regarding the underlying molecular mechanism by which Y56 phosphorylation causes functional GSK3B alterations in HCC cells, it has been reported that K63 ubiquitination can regulate protein functions in cancer cells33 and that tumor necrosis factor receptor-associated factor 6 (TRAF6), which plays a role in interleukin-1 receptor-Tolllike receptor signaling, also is a direct E3 ligase for GSK3B K63 ubiquitination.34 Of note, we found that the Y56 of GSK3B is located in a conserved kinase loop adjacent to the PE motif (TRAF6-binding motif [PXEXXAc])35 (Figure 3A) and that endogenous GSK3B and TRAF6 protein interacted strongly in the cytoplasm of Hep3B cells as observed by a Duolink II assay (Sigma-Aldrich, St Louis, MO; Figure 3B). The PE motif was further shown to be required for the interaction between GSK3B and TRAF6 by a yeast 2-hybrid assay (Supplementary Figure 3A). To this end, we spotted a 10-fold serial dilution of yeast co-transformants on tryptophan- and leucine-minus plates with or without 3-aminotrizole to analyze the ability of the yeast to survive in the absence of histidine. The results demonstrated that only yeast cells containing TRAF6 and GSK3B, but not GSK3B-PE mutant (P51A), showed strong interaction, suggesting that the PE motif of GSK3B is critical for GSK3B and TRAF6 interaction (Supplementary Figure 3A, bottom 2 rows). In addition, WT GSK3B exhibited a strong interaction with TRAF6, similar to receptor activator of nuclear factor kB, a known TRAF6-interacting protein (Figure 3C, lanes 4 vs 3), whereas mutation of the PE motif disrupted GSK3B-TRAF6 interaction (Figure 3C, lanes 5 vs 4, and Supplementary Figure 3A). These results suggested that the PE motif of GSK3B is required for its interaction with TRAF6.
Figure 3.
Y56 phosphorylation up-regulates GSK3B activity by suppression of TRAF6-mediated GSK3B K63 ubiquitination. (A) Sequence alignment of TRAF6 consensus motif in GSK3B. The MET phosphorylation site (GSK3B Y56) is located in a consensus TRAF6-binding motif (PXEXXAc). (B) Interaction of endogenous GSK3B and TRAF6 proteins in Hep3B cells. Cells were immunostained with GSK3B and TRAF6 antibodies and assessed by a Duolink II assay. Red foci indicate associations between GSK3B and TRAF6 proteins. Scale bar, 25 μm. (C) Hep3B cells expressing TRAF6 were incubated with bacteria-purified GST-RANK, WT GSK3B, or GSK3B PE. An in vitro binding assay was performed, and the results were analyzed by western blotting. (D) Hep3B cells were transfected with the indicated plasmids and subjected to immunoprecipitation using a Flag antibody. Ubiquitination of transfected cells was analyzed by western blotting. (E) Hep3B cells were transfected with the indicated plasmids and subjected to immunoprecipitation using a Flag antibody. Ubiquitination of transfected cells was analyzed by western blotting. (F) Hep3B cells were transfected with the indicated plasmids. Then, cells were immuno-precipitated with a Flag antibody and subjected to western blotting with a ubiquitin antibody. (G) The kinase activity of GSK3B in Hep3B cells expressing pMX (empty vector), WT TRAF6, or TRAF6 C70A as determined by in vitro kinase assay and phosphorylation analysis. **P < .01. (H) Immunoblot of PDL1 expression in Hep3B cells transiently transfected with pMX (empty vector), WT TRAF6, or TRAF6 C70A. (I) Kinase activity of GSK3B in Hep3B cells expressing pMX (empty vector), WT GSK3B, or GSK3B PE as determined by in vitro kinase assay and phosphorylation analysis. **P < .01. (J) Immunoblot of PDL1 expression in Hep3B cells transiently transfected with pMX (empty vector), WT GSK3B, or GSK3B PE. (K) Co-immunoprecipitation of TRAF6 and GSK3B in WT GSK3B and GSK3B PE Hep3B cells with GSK3B and TRAF6 antibodies. (L) GSK3B was immuno-precipitated from Hep3B cells transfected with WT GSK3B and GSK3B PE. Ubiquitination in the transfected cells was examined by Western blotting. (M) Co-immunoprecipitation of TRAF6 and GSK3B in vector control and MET-knockdown Hep3B cells with GSK3B and TRAF6 antibodies. (N) Endogenous GSK3B was immuno-precipitated from Hep3B cells transfected with a vector control or MET shRNA. A ubiquitination assay in vector control and MET-knockdown Hep3B cells was performed by western blotting. Ab, antibody; GST, glutathione S-transferase; IgG, immunoglobulin G; IP, immunoprecipitation; RANK, receptor activator of nuclear factor κB; Ub, ubiquitination.
Because TRAF6 is a bona fide ubiquitin E3 ligase, we examined whether GSK3B is a potential TRAF6 substrate by transfecting Hep3B cells with plasmids expressing His6-fused ubiquitin together with GSK3B and TRAF6. We pulled down substrates covalently conjugated with His6-ubiquitin in the cells using Ni2+ agarose beads. The results indicated that GSK3B was indeed ubiquitinated in the presence of TRAF6 (Supplementary Figure 3B). In addition, mass spectrometric analysis identified Lys86 and Lys197 as GSK3B ubiquitin-conjugating sites in Hep3B cells (Supplementary Figure 3C). Of note, TRAF6 specifically catalyzed GSK3B ubiquitination, whereas 6 other ubiquitin E3 ligases failed to do so (Supplementary Figure 3D). Moreover, TRAF6 could not catalyze neddylation, SUMOylation, or ISGylation of GSK3B (Supplementary Figure 3B), which is consistent with the enzyme activity of TRAF6. Similarly, disruption of the first Zn2+-chelating structure in the RING domain of TRAF6 (TRAF6 C70A) abrogated TRAF6-mediated GSK3B ubiquitination in Hep3B cells in vivo (Figure 3D) and in vitro (Supplementary Figure 3E). In fact, WT TRAF6 and the C70A mutant demonstrated binding affinity for GSK3B, excluding the possibility that the inability of the C70A mutant to enhance GSK3B ubiquitination was due to the loss of interaction between TRAF6 with GSK3B (Supplementary Figure 3F). Mutation of the GSK3B PE motif consistently resulted in the loss of its ability to be ubiquitinated by TRAF6 (Figure 3E). Collectively, these results demonstrated that the integrity of the RING domain of TRAF6 and PE motif of GSK3B is required for TRAF6-mediated GSK3B ubiquitination.
Because TRAF6 mediates K63-specific ubiquitination, mutation of ubiquitin at Lys63 (K63R), but not Lys48 (K48R), abrogated TRAF6-induced GSK3B ubiquitination in Hep3B cells (Figure 3F). Next, we sought to determine the function of TRAF6-mediated GSK3B ubiquitination and found that the GSK3B kinase activity in TRAF6 C70A cells was 3 times higher than that of endogenous GSK3B in WT TRAF6-expressing Hep3B cells (Figure 3G). Moreover, TRAF6 C70A down-regulated the PDL1 expression compared with WT TRAF6 by enhancing GSK3B activity (Figure 3H). These findings suggested that TRAF6 decreased GSK3B activity by protein K63 ubiquitination, which results in up-regulation of PDL1 expression. Mutation of GSK3B PE motif in Hep3B cells consistently prevented the association of GSK3B with TRAF6, exhibited increased kinase activity (Figure 3I), and had lower PDL1 expression than in those harboring WT GSK3B (Figure 3J). As expected, GSK3B Y56F induced more interaction between GSK3B (Figure 3K) and TRAF6 and more TRAF6-mediated GSK3B K63 ubiquitination than did the WT GSK3B (Figure 3L). MET knockdown consistently decreased GSK3B phosphorylation at Y56 and increased the interaction between GSK3B and TRAF6 (Figure 3M), which induced TRAF6-mediated GSK3B K63 ubiquitination (Figure 3N). Taken together, these results suggested that MET inhibition, which prevents GSK3B phosphorylation at Y56, enhanced the binding between the GSK3B PE motif and TRAF6 and decreased GSK3B activity due to TRAF6-mediated GSK3B K63 ubiquitination, subsequently increasing the expression of PDL1 in HCC cells.
Anti-PD1 Therapy in Combination With MET Blockade Improves Antitumor Activity
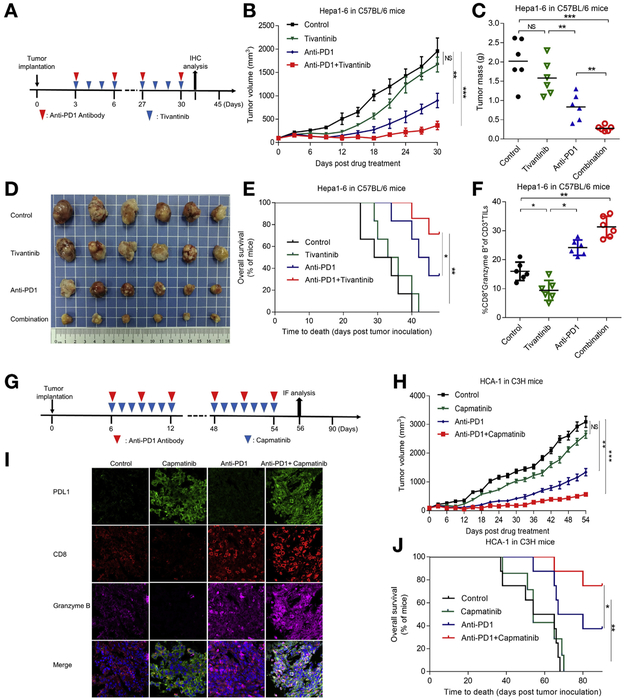
Our data demonstrated that MET blockade stabilizes PDL1 for immune evasion, which could be the underlying factor contributing to the adverse effect of MET blockade against HCC in clinical trials. To determine whether blocking the PDL1 pathway sensitizes HCC to MET inhibition, we administered tivantinib or PD1 antibody alone or in combination to mice bearing orthotopic Hep1-6 tumors and analyzed tumor growth until tumors reached >100 mm3 (Figure 4A) or 300 mm3 (Supplementary Figure 4B) in volume. The combination of tivantinib and anti-PD1 impaired tumor growth and decreased tumor burden more effectively than in control mice or mice receiving tivantinib or anti-PD1 alone (Figure 4B-D and Supplementary Figure 4A-C). Moreover, the combination of tivantinib and anti-PD1 substantially prolonged the overall survival of mice bearing orthotopic Hep1-6 tumors compared with tivantinib alone (Figure 4E and Supplementary Figure 4D). In mice given tivantinib alone or in combination with anti-PD1, the expression of PDL1 was up-regulated in the tumors (Supplementary Figure 4E and F). Importantly, the activated tumor-infiltrating CD8+ T-cell population and expression of granzyme B increased in mice given the tivantinib and anti-PD1 combination (Figure 4F and Supplementary Figure 4E and F), indicating that the combination therapy increased antitumor immunity in mice. Knocking out MET in Hepa1-6 cells abolished the synergistic killing effects of the combination therapy, further validating that MET expression is indispensable for the combination therapy and that the on-target effect of MET inhibitor contributes to the increased antitumor effect of combination therapy (Supplementary Figure 4G).
Figure 4.
Anti-PD1 therapy in combination with MET blockade improves antitumor activity of MET inhibitor in HCC. (A) Schematic of drug intervention protocol for tivantinib and PD1 antibody in C57/BL6 mice. At the drug intervention end point, tumors were obtained for immunohistochemical analysis. (B) Growth of orthotopic Hep1-6 tumors in tivantinib- and/or PD1 antibody-treated C57/BL6 mice. Tumors were measured at the indicated time points. (C) Weights of tumors at drug intervention end point. (D) Representative images of orthotopic C57/BL6 mouse tumors without the liver. (E) Survival of mice bearing Hep1-6 tumors after drug intervention with capmatinib and/or PD1 antibody. Significance was determined using log-rank test. (F) Intracellular cytokine staining of CD8+ and granzyme B+ cells in CD3+ T-cell populations from isolated tumor-infiltrating lymphocytes. Results are presented as mean ± standard deviation from a representative experiment. (G) Schematic of drug intervention protocol for the MET inhibitor capmatinib and PD1 antibody in C3H mice. At the drug intervention end point, tumors were isolated for immunofluorescent analysis. (H) Growth of HCA-1 tumors in C3H mice given capmatinib and/or PD1 antibody. Tumors were measured at indicated time points. (I) Immunofluorescent staining of PDL1, CD8, and granzyme B protein expression patterns in HCA-1 cells. (J) Survival of mice bearing HCA-1 tumors after drug intervention with capmatinib and/or PD1 antibody. *P < .05; **P < .01; ***P < .001. All error bars represent mean ± standard deviation. NS, not significant.
We also compared the combination and single agent therapy in a subcutaneous HCA-1 liver cancer model (Figure 4G). The combination of capmatinib and PD1 antibody also improved tumor-growth inhibition in the subcutaneous model (Figure 4H). Mice given capmatinib plus anti-PD1 exhibited longer survival than those given capmatinib or anti-PD1 monotherapy (Figure 4J). The expression of PDL1 was consistently up-regulated in the tumor tissues of mice given capmatinib alone or in combination with anti-PD1 (Figure 4I). Furthermore, the combination therapy also increased the CD8+ T-cell population and granzyme B expression, which is consistent with the earlier results in the orthotopic model. In mice given capmatinib alone, the expression of p-GSK3B (Y56) was down-regulated and PDL1 was up-regulated in the tumors, which confirmed the correlation between p-GSK3B (Y56) and PDL1 observed in vitro (Supplementary Figure 4H).
Next, we investigated whether the therapy doses used in the experiments were safe. To this end, we compared the body weight and the main biochemistry index, including aspartate transaminase, alanine aminotransferase, creatinine, and serum urea nitrogen levels of mice before and over the course of the drug intervention (Supplementary Figure 4I and J). The results indicated no relevant changes in body weight or the indicators of liver and kidney functions in the experimental and control groups. Taken together, our findings in subcutaneous and orthotopic liver tumor models illustrated the potential therapeutic benefits of combining MET inhibition with PD1 blockade.
p-GSK3B Y56 Expression Associates With Granzyme B Expression and Down-regulation of PDL1 in Patients With HCC
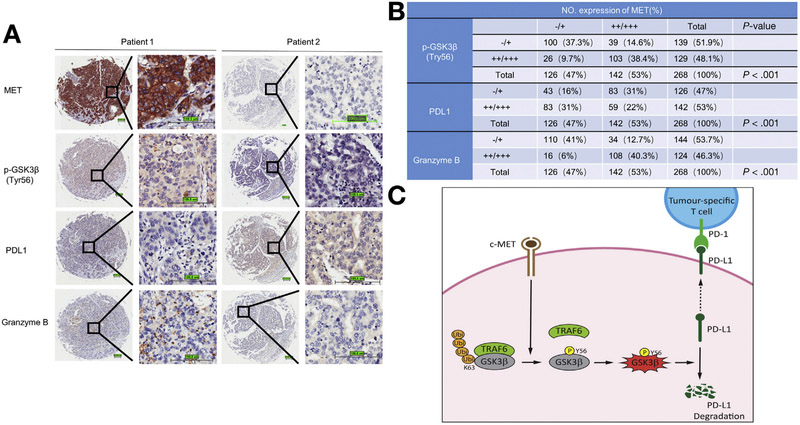
To elucidate the clinical relevance of MET, p-GSK3B (Y56), PDL1, and granzyme B expression in patients with HCC, we measured their expression in 268 HCC samples by immunohistochemical staining (Figure 5A). As expected, the expression level of MET was negatively associated with PDL1 status (P < .001) but positively associated with p-GSK3B (Y56; P < .001) and granzyme B expression level (P < .001; Figure 5B). Specifically, approximately 66% of tumor samples with low MET expression exhibited strong PDL1 staining, and 60% of those with high MET expression exhibited weak or no PDL1 staining. Moreover, compared with tumors with high MET expression, those with low MET expression had lower levels of granzyme B. Collectively, these data indicated that high MET expression is associated with high p-GSK3B (Y56), low PDL1 expression, and high granzyme B levels in clinical HCC samples, further supporting the notion that MET inhibition inactivates GSK3B and in turn stabilizes PDL1 expression in HCC, allowing HCC to escape from T-cell killing. Our findings that higher MET expression accompanied by higher granzyme B secretion suggested that MET inhibitor in combination with anti-PD1 might be more effective in patients with HCC and high MET expression.
Figure 5.
Clinical relevance of MET, p-GSK3B (Y56), PDL1, and granzyme B expression in patients with HCC. (A) Representative immunohistochemical staining of HCC tumors for MET, p-GSK3B (Y56), PDL1, and granzyme B expression. (B) Correlations among MET, p-GSK3B (Y56), PDL1, and granzyme B expression levels in patients with liver cancer. P value by Pearson χ2; −/+, negative or low expression; ++/+++, medium or high expression. Scale bar, 100 μm. (C) Proposed working model. MET directly phosphorylates GSK3B at Y56, activates GSK3B by blocking TRAF6-mediated K63 ubiquitination, and decreases PDL1 expression by K48 ubiquitination in HCC. Ubi, ubiquitination.
Discussion
In the present study, high MET expression was associated with high p-GSK3B (Y56), low PDL1 expression, and high granzyme B levels in clinical HCC samples (Figure 5A and B) with detailed mechanisms (Figure 5C). Although several studies showed inconsistent correlation between MET amplification and PDL1 expression,36,37 it is not yet clear whether and how MET directly regulates PDL1 expression in different cell types. Although MET positively correlates with PDL1 in non-small cell lung cancer,36 the correlation in clear cell renal cell carcinoma seems to be inconsistent.37 To validate this, we compared the correlation between p-MET and PDL1 in several lung cancer and HCC cell lines. p-MET showed a positive correlation with PDL1 in lung cancer (Supplementary Figure 4M), whereas p-MET negatively correlated with PDL1 in HCC (Supplementary Figure 1G). PDL1 is likely regulated by multiple factors, which are differentially activated in different cancer types and/or tissues. Thus, the tissue- and/or cancer-specific role of MET and immune regulation is worthy of further investigation, especially when a combination therapy will be explored. Collectively, our model shown in Figure 5C provides a plausible mechanism for the adverse effect of MET inhibitors in clinical trials, namely inhibition of MET reverses the pathway to inactivate GSK3B, which in turn stabilizes PDL1 and allows tumor cells to escape from immunosurveillance.
A recent study reported that MET deletion in mouse neutrophils enhances tumor growth and metastasis in an immunocompetent mouse model,38 implying that MET blockade in turn induces immune resistance. That study and the present report suggested that the MET oncoprotein might limit antitumor immunity by different mechanisms. Because multiple MET inhibitors have been developed as antitumor drugs and are being tested in clinical trials, it has become critical to address the immunosuppressive capacity of MET inhibitors to develop effective therapy strategies. In this regard, eliminating the immunosuppressive activity of MET inhibitors by combining them with immunotherapy could be a new way to ameliorate the adverse effect of MET inhibitors in patients with HCC in the clinic. In the present report, concurrent blockade of MET and the PD1-PDL1 pathway dramatically improved the efficacy of the MET inhibitor and prolonged mouse survival with enhanced T-cell infiltration and activation in subcutaneous and orthotopic HCC animal models (Figure 4). Recently, Glodde et al39 reported that anti-PD1 immunotherapy consistently led to reactive recruitment of MET+ neutrophils to tumors and regional lymph nodes, which limited the efficacy of immunotherapy and promoted tumor growth, and concomitant MET inhibition enhanced the efficacy of anti-PD1 immunotherapy. Although anti-PD1 increased CD8+ T-cell activity, it did not induce MET+ neutrophils in the HCC model in our studies (Supplementary Figure 4K and L). Thus, the neutrophil-mediated immunosuppression might be specific to cancer type. However, the findings by Glodde et al,39 particularly on the combination therapy strategy, support our data showing that adding anti-PD1 therapy to MET inhibitor can overcome MET inhibitor-induced PDL1 expression and maximize its antitumor activity.
The US Food and Drug Administration approved anti-PD1 therapy for advanced HCC refractory to sorafenib treatment in 2017. In the present study, we identified an intrinsic immune-sensitive mechanism that is important in guiding anti-PD1 therapy for HCC. In subcutaneous and orthotopic models, anti-PD1 therapy ameliorated the adverse effect of MET blockade with minimal cytotoxicity. Specifically, tumors regressed and mouse survival durations increased by up to 80%. It has been reported that the antitumor effect of tivantinib could be attributed in part to the nonspecific killing of proliferating cells independent from MET.40,41 However, in our study, we showed consistent results from multiple experiments, including MET knockout cells (Supplementary Figure 4G) and different MET inhibitors (Figure 4B and H), indicating the conclusion is unlikely coming from the “off-target” effects of tivantinib. The presence of tumor-infiltrating lymphocytes in HCC is relatively rare.42 Thus, the expression of tumor-infiltrating lymphocytes also should be considered when identifying the right patient population for the combination therapy in the future. In our study, we found that high MET expression was accompanied by higher granzyme B secretion, suggesting that the combination therapy might be more effective in patients with MET-high HCC. Collectively, because PDL1 up-regulation is a biomarker for the effectiveness of combined anti-PD1 or anti-PDL1 therapy,43,44 this study provides a safe and potentially effective strategy to overcome the adverse effect of MET inhibitors by combining with anti-PD1 or anti-PDL1 therapy. The encouraging preclinical data warrant the strategy to be tested in future clinical trials for HCC.
Supplementary Material
WHAT YOU NEED TO KNOW.
BACKGROUND AND CONTEXT
MET inhibitor treatment has failed to produce satisfactory outcomes in clinical trials for HCC. Thus, it is critical to understand the underlying mechanisms and develop effective combination strategies.
NEW FINDINGS
MET inhibition leads to PDL1 upregulation and immunosuppression. The combination of MET inhibitor and anti-PD1 therapy potently suppresses tumor growth, prolonging survival.
LIMITATIONS
This study did not measure the PDL1 expression in human HCC samples treated by MET inhibitor.
IMPACT
This study provides a safe and potentially effective strategy to overcome the ineffectiveness of MET inhibitors by combining with anti-PD1/PDL1 therapy.
Acknowledgments
We thank the Department of Scientific Publications at The University of Texas MD Anderson Cancer Center for editing this manuscript.
Funding
This work was funded in part by the National Institutes of Health (grants P30CA016672, and U01 CA201777); the Cancer Prevention & Research Institute of Texas (grants RP150245 and RP160710); The University of Texas MD Anderson Cancer Center-China Medical University and Hospital Sister Institution Fund (to Mien-Chie Hung); the Ministry of Health and Welfare, China Medical University Hospital Cancer Research Center of Excellence (grants MOHW108-TDU-B-212-124024 and MOHW108-TDU-B-212-122015); the Program of Shanghai Subject Chief Scientist (16XD1400800 to Qinghai Ye); the National Natural Science Foundation of China (81572301 to Qinghai Ye, 81802893 to Shuang Liu, and 81502487 to Lei Guo); the Natural Science Foundation of Guangdong Province of China (2017A030310641 to Xiaoqiang Li); the Medical Scientific Research Foundation of Guangdong Province of China (A2017327 to Xiaoqiang Li); and the State Scholarship Fund of China (201606100205 to Hui Li).
Abbreviations used in this paper:
- GSK3B
glycogen synthase kinase 3β
- HCC
hepatocellular carcinoma
- p
phosphorylated
- PD1
programmed cell death protein 1
- PDL1
programmed death-ligand 1
- shRNA
short hairpin RNA
- TRAF6
tumor necrosis factor receptor-associated factor 6
- WT
wild-type
- Y56F
mutation of GSK3B tyrosine 56 to phenylalanine
- Y56
tyrosine 56
Footnotes
Supplementary Material
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at https://doi.org/10.1053/j.gastro.2019.01.252
Conflicts of interest
The authors disclose no competing financial interests (financial, professional, and personal).
References
- 1.El-Serag HB. Hepatocellular carcinoma. N Engl J Med 2011;365:1118–1127. [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008; 359:378–390. [DOI] [PubMed] [Google Scholar]
- 3.Bruix J, Qin S, Merle P, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017; 389:56–66. [DOI] [PubMed] [Google Scholar]
- 4.Kudo M, Finn RS, Qin S, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet 2018; 391:1163–1173. [DOI] [PubMed] [Google Scholar]
- 5.Abou-Alfa GK, Meyer T, Cheng AL, et al. Cabozantinib in patients with advanced and progressing hepatocellular carcinoma. N Engl J Med 2018;379:54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Khoueiry AB, Sangro B, Yau T, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 2017;389:2492–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boccaccio C, Comoglio PM. Invasive growth: a MET-driven genetic programme for cancer and stem cells. Nat Rev Cancer 2006;6:637–645. [DOI] [PubMed] [Google Scholar]
- 8.Birchmeier C, Birchmeier W, Gherardi E, et al. Met, metastasis, motility and more. Nat Rev Mol Cell Biol 2003;4:915–925. [DOI] [PubMed] [Google Scholar]
- 9.Ueki T, Fujimoto J, Suzuki T, et al. Expression of hepatocyte growth factor and its receptor, the c-met proto-oncogene, in hepatocellular carcinoma. Hepatology 1997;25:619–623. [DOI] [PubMed] [Google Scholar]
- 10.Gherardi E, Birchmeier W, Birchmeier C, et al. Targeting MET in cancer: rationale and progress. Nat Rev Cancer 2012;12:89–103. [DOI] [PubMed] [Google Scholar]
- 11.Bouattour M, Raymond E, Qin S, et al. Recent developments of c-Met as a therapeutic target in hepatocellular carcinoma. Hepatology 2018;67:1132–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rimassa L, Assenat E, Peck-Radosavljevic M, et al. Second-line tivantinib (ARQ 197) vs placebo in patients (Pts) with MET-high hepatocellular carcinoma (HCC): results of the METIV-HCC phase III trial. J Clin Oncol 2017;35:4000. [Google Scholar]
- 13.Baumeister SH, Freeman GJ, Dranoff G, et al. Coinhibitory pathways in immunotherapy for cancer. Annu Rev Immunol 2016;34:539–573. [DOI] [PubMed] [Google Scholar]
- 14.Butte MJ, Keir ME, Phamduy TB, et al. Programmed death-1 ligand 1 interacts specifically with the B7-1 costimulatory molecule to inhibit T cell responses. Immunity 2007;27:111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Postow MA, Callahan MK, Wolchok JD. Immune checkpoint blockade in cancer therapy. J Clin Oncol 2015;33:1974–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Li X, Liu S, et al. PD1 checkpoint blockade in combination with an mTOR inhibitor restrains hepatocellular carcinoma growth induced by hepatoma cell-intrinsic PD1. Hepatology 2017;66:1920–1933. [DOI] [PubMed] [Google Scholar]
- 17.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD1 antibody in cancer. N Engl J Med 2012;366:2443–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herbst RS, Soria JC, Kowanetz M, et al. Predictive correlates of response to the anti-PDL1 antibody MPDL3280A in cancer patients. Nature 2014;515:563–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farago M, Dominguez I, Landesman-Bollag E, et al. Kinase-inactive glycogen synthase kinase 3beta promotes Wnt signaling and mammary tumorigenesis. Cancer Res 2005;65:5792–5801. [DOI] [PubMed] [Google Scholar]
- 20.Hoeflich KP, Luo J, Rubie EA, et al. Requirement for glycogen synthase kinase-3beta in cell survival and NF-kappaB activation. Nature 2000;406:86–90. [DOI] [PubMed] [Google Scholar]
- 21.Zhou BP, Deng J, Xia W, et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol 2004; 6:931–940. [DOI] [PubMed] [Google Scholar]
- 22.Ding Q, He X, Hsu JM, et al. Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol 2007;27:4006–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ko HW, Lee HH, Huo L, et al. GSK3beta inactivation promotes the oncogenic functions of EZH2 and enhances methylation of H3K27 in human breast cancers. Oncotarget 2016;7:57131–57144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li CW, Lim SO, Xia W, et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun 2016;7:12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horn S, Endl E, Fehse B, et al. Restoration of SHIP activity in a human leukemia cell line downregulates constitutively activated phosphatidylinositol 3-kinase/Akt/GSK-3beta signaling and leads to an increased transit time through the G1 phase of the cell cycle. Leukemia 2004;18:1839–1849. [DOI] [PubMed] [Google Scholar]
- 26.Ding Q, Xia W, Liu JC, et al. Erk associates with and primes GSK-3beta for its inactivation resulting in up-regulation of beta-catenin. Mol Cell 2005;19:159–170. [DOI] [PubMed] [Google Scholar]
- 27.Remsing Rix LL, Kuenzi BM, Luo Y, et al. GSK3 alpha and beta are new functionally relevant targets of tivantinib in lung cancer cells. ACS Chem Biol 2014;9:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Wang Q, Yang G, et al. A novel kinase inhibitor, INCB28060, blocks c-MET-dependent signaling, neoplastic activities, and cross-talk with EGFR and HER-3. Clin Cancer Res 2011;17:7127–7138. [DOI] [PubMed] [Google Scholar]
- 29.Gao Q, Wang XY, Qiu SJ, et al. Overexpression of PDL1 significantly associates with tumor aggressiveness and postoperative recurrence in human hepatocellular carcinoma. Clin Cancer Res 2009;15:971–979. [DOI] [PubMed] [Google Scholar]
- 30.Calderaro J, Rousseau B, Amaddeo G, et al. Programmed death ligand 1 expression in hepatocellular carcinoma: relationship with clinical and pathological features. Hepatology 2016;64:2038–2046. [DOI] [PubMed] [Google Scholar]
- 31.Rebouissou S, Zucman-Rossi J, Moreau R, et al. Note of caution: contaminations of hepatocellular cell lines. J Hepatol 2017;67:896–897. [DOI] [PubMed] [Google Scholar]
- 32.Heffelfinger SC, Hawkins HH, Barrish J, et al. HEP-1: a human cell line of endothelial origin. In Vitro Cell Dev Biol 1992;28A:136–142. [DOI] [PubMed] [Google Scholar]
- 33.Yang WL, Wang J, Chan CH, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science 2009;325:1134–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ko R, Park JH, Ha H, et al. Glycogen synthase kinase 3beta ubiquitination by TRAF6 regulates TLR3-mediated pro-inflammatory cytokine production. Nat Commun 2015;6:6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bidere N, Snow AL, Sakai K, et al. Caspase-8 regulation by direct interaction with TRAF6 in T cell receptor-induced NF-kappaB activation. Curr Biol 2006;16:1666–1671. [DOI] [PubMed] [Google Scholar]
- 36.Albitar M, Sudarsanam S, Ma W, et al. Correlation of MET gene amplification and TP53 mutation with PDL1 expression in non-small cell lung cancer. Oncotarget 2018;9:13682–13693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lalani AA, Gray KP, Albiges L, et al. Differential expression of c-Met between primary and metastatic sites in clear-cell renal cell carcinoma and its association with PDL1 expression. Oncotarget 2017;8:103428–103436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finisguerra V, Di Conza G, Di Matteo M, et al. MET is required for the recruitment of anti-tumoural neutrophils. Nature 2015;522:349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glodde N, Bald T, van den Boorn-Konijnenberg D, et al. Reactive neutrophil responses dependent on the receptor tyrosine kinase c-MET limit cancer immunotherapy. Immunity 2017;47:789–802 e9. [DOI] [PubMed] [Google Scholar]
- 40.Rebouissou S, La Bella T, Rekik S, et al. Proliferation markers are associated with MET expression in hepatocellular carcinoma and predict tivantinib sensitivity in vitro. Clin Cancer Res 2017;23:4364–4375. [DOI] [PubMed] [Google Scholar]
- 41.Michieli P, Di Nicolantonio F. Targeted therapies: tivantinib—a cytotoxic drug in MET inhibitor’s clothes? Nat Rev Clin Oncol 2013;10:372–374. [DOI] [PubMed] [Google Scholar]
- 42.Shirabe K, Motomura T, Muto J, et al. Tumor-infiltrating lymphocytes and hepatocellular carcinoma: pathology and clinical management. Int J Clin Oncol 2010;15:552–558. [DOI] [PubMed] [Google Scholar]
- 43.Jiao S, Xia W, Yamaguchi H, et al. PARP inhibitor up-regulates PDL1 expression and enhances cancer-associated immunosuppression. Clin Cancer Res 2017; 23:3711–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Bu X, Wang H, et al. Cyclin D-CDK4 kinase destabilizes PDL1 via cullin 3-SPOP to control cancer immune surveillance. Nature 2018;553:91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.