Abstract
Background
Tetraploid Thinopyrum elongatum (2n = 4x = 28) is a promising source of useful genes, including those related to adaptability and resistance to diverse biotic (Fusarium head blight, rust, powdery mildew, and yellow dwarf virus) and abiotic (cold, drought, and salt) stresses. However, gene transfer rates are low for this species and relatively few species-specific molecular markers are available.
Results
The wheat-tetraploid Th. elongatum line K17–841-1 derived from a cross between a hexaploid Trititrigia and Sichuan wheat cultivars was characterized based on sequential genomic and fluorescence in situ hybridizations and simple sequence repeat markers. We revealed that K17–841-1 is a 1E (1D) chromosomal substitution line that is highly resistant to stripe rust pathogen strains prevalent in China. By comparing the sequences generated during genotyping-by-sequencing (GBS), we obtained 597 specific fragments on the 1E chromosome of tetraploid Th. elongatum. A total of 235 primers were designed and 165 new Th. elongatum-specific markers were developed, with an efficiency of up to 70%. Marker validation analyses indicated that 25 specific markers can discriminate between the tetraploid Th. elongatum chromosomes and the chromosomes of other wheat-related species. An evaluation of the utility of these markers in a F2 breeding population suggested these markers are linked to the stripe rust resistance gene on chromosome 1E. Furthermore, 28 markers are unique to diploid Th. elongatum, tetraploid Th. elongatum, or decaploid Thinopyrum ponticum, which carry the E genome. Finally, 48 and 74 markers revealed polymorphisms between Thinopyrum E-genome- containing species and Thinopyrum bessarabicum (Eb) and Pseudoroegneria libanotica (St), respectively.
Conclusions
This new substitution line provide appropriate bridge–breeding–materials for alien gene introgression to improve wheat stripe rust resistance. The markers developed using GBS technology in this study may be useful for the high-throughput and accurate detection of tetraploid Th. elongatum DNA in diverse materials. They may also be relevant for investigating the genetic differences and phylogenetic relationships among E, Eb, St, and other closely-related genomes and for further characterizing these complex species.
Keywords: Tetraploid Thinopyrum elongatum, Chromosome substitution line, Stripe rust, Species- specific molecular markers, GBS
Background
Common wheat (Triticum aestivum L., 2n = 6x = 42, AABBDD) is a staple cereal cultivated worldwide, with a predicted global grain yield of 757.4 million tons in 2019 [1]. However, the domestication of wheat decreased its genetic diversity as well as tolerance to biotic and abiotic stresses, which has restricted further improvements to wheat productivity and quality [2]. Stripe rust caused by Puccinia striiformis f. sp. tritici (Pst) is a serious wheat disease that threatens global wheat production [3, 4]. The identification and application of new disease-resistance genes and the development of disease-resistant cultivars represent the most effective means of decreasing the reliance on fungicides to control stripe rust in large-scale commercial production systems [5]. Wild relatives are a largely unexploited source of genes for agronomically important traits that can be transferred to common wheat via wide hybridizations to enrich wheat genetic diversity [6, 7].
Thinopyrum elongatum (syn. Agropyron elongatum or Lophopyrum elongatum) is a distant wild relative of common wheat and has long been the focus of wheat breeders. The taxon comprises the following three ploidy levels involving the E-genome: diploid (2n = 2x = 14, EE), tetraploid (2n = 4x = 28, EEEE), and decaploid (2n = 10x = 70, EEEEEEStStStSt) [8]. This species possesses many desirable traits, including strong adaptability, high tolerance to cold, drought, and salt stresses, and resistance to Fusarium head blight, rust, powdery mildew, and the yellow dwarf virus [9, 10]. To transfer desirable traits from Th. elongatum into wheat, wide hybridizations between Th. elongatum and common wheat began in the 1980s [8]. Progeny lines harboring Th. elongatum chromosomes (segments) incorporated into the wheat genome were obtained as lines with chromosomal additions, substitutions, or translocations [7, 11–14]. However, these introgressions mainly involved the diploid Th. elongatum and decaploid Thinopyrum ponticum. There are relatively few reports describing attempts to transfer tetraploid Th. elongatum genes into wheat [15–18]. Thus, identifying new elite alien genes and incorporating them into common wheat are critical for increasing wheat genetic diversity through the development of wheat–tetraploid Th. elongatum introgression lines.
Marker-assisted selection is useful for detecting genes associated with a trait of interest based on a linked marker, with implications for breeding involving multiple traits [11]. Developing species- specific molecular markers that facilitate the identification of alien chromosomes or segments is very important for wheat breeding programs [19, 20]. Diverse molecular markers specific to diploid Th. elongatum and Th. ponticum have been reported, including random-amplified polymorphic DNAs (RAPDs), simple sequence repeats (SSRs), expressed sequence tags, cleaved amplified polymorphic sequences, sequence-tagged sites, sequence-characterized amplified regions, amplified fragment length polymorphisms, single nucleotide polymorphisms, and specific-locus amplified fragments (SLAFs) [11, 12, 19, 21–25]. However, there are still relatively few of these markers, and they do not cover the whole Thinopyrum genome. Consequently, there is an urgent need to develop more molecular markers, especially those with a wide genomic distribution and that are amenable to high-throughput genotyping [12]. Genotyping-by-sequencing (GBS) is a high-throughput, highly accurate, inexpensive, and relatively simple method for developing several markers in Triticale species [26]. However, developing specific molecular markers for tetraploid Th. elongatum remains difficult. Therefore, generating chromosome-specific molecular markers is critical for detecting and tracing alien chromosomes in wheat–tetraploid Th. elongatum hybrid derivatives.
Tetraploid Th. elongatum is an important genetic resource for improving wheat, and some wheat–tetraploid Th. elongatum derivative lines have been developed by crossing hexaploid Trititrigia with Sichuan wheat cultivars [17]. Li et al. [18] produced a fluorescence in situ hybridization (FISH) karyotype of the E-genome chromosomes of tetraploid Th. elongatum based on various repetitive sequence probes, which may enhance the utility of tetraploid Th. elongatum for the introgression of alien genes into wheat. The main objectives of the current study were to: (1) characterize the chromosomal constitution of a wheat–tetraploid Th. elongatum substitution line and evaluate its effects on stripe rust resistance and agronomic traits and (2) develop and validate specific and easy-to-use molecular markers based on GBS, which may be useful for the efficient and reliable identification of tetraploid Th. elongatum chromatin in several species, including common wheat.
Results
Chromosomal constitution of K17–841-1
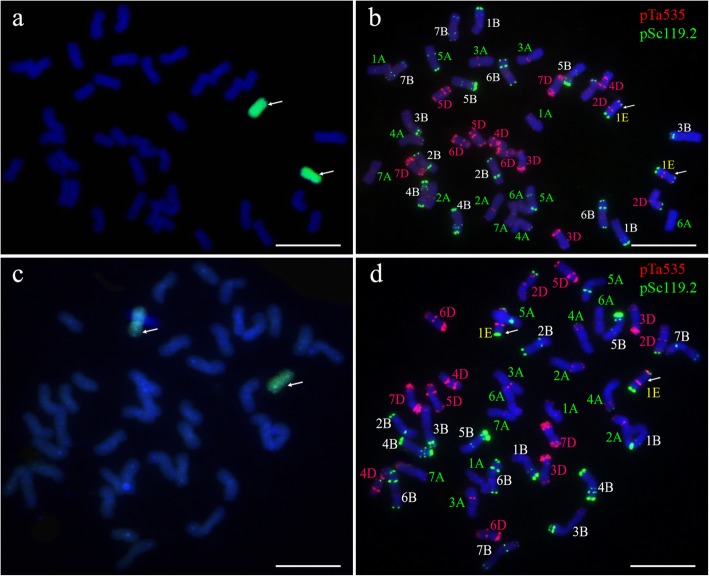
Genomic in situ hybridization (GISH), FISH, and SSR marker analyses were performed to determine the chromosomal constitution of wheat–tetraploid Th. elongatum line K17–841-1. When tetraploid Th. elongatum total genomic DNA and the J-11 genomic DNA were used as the probe and the blocking DNA, respectively, we observed that line K17–841-1 carried 40 wheat chromosomes and two E chromosomes (Fig. 1a). The GISH signals were sequentially removed and the slides were used in a FISH analysis involving pSc119.2 and pTa535 probes. A pair of E chromosomes produced strong terminal pSc119.2 signals on both arms as well as strong pTa535 signals on the subterminal regions of the short arm and near the centromeric region of the long arm (Fig. 1b). These results are consistent with the previously reported FISH pattern for the 1E chromosome of tetraploid Th. elongatum [18]. Thus, the FISH karyotype of the E-genome chromosomes of tetraploid Th. elongatum and common wheat based on probes pSc119.2 and pTa535 suggested that K17–841-1 is a 1E (1D) chromosomal substitution line. To determine the cytological stability of K17–841-1, we used GISH and FISH to analyze 20 randomly selected seeds of K17–841-1 self-progeny. We revealed that all seeds contained 14 A- (1A–7A), 14 B- (1B–7B), and 12 D- (2D–7D) genomes, as well as a pair of 1E chromosomes (Fig. 1c, d).
Fig. 1.
GISH and FISH identification of the wheat–tetraploid Th. elongatum substitution line K17–841-1. The probes used for in situ hybridization were tetraploid Th. elongatum genomic DNA (a, c); pSc119.2 and pTa535 (b, d). Arrows indicate 1E-genome chromosomes. Scale bar: 10 μm
We completed a PCR analysis involving the wheat chromosome 1D-specific SSR markers to confirm the chromosomal constitution of K17–841-1. As expected, amplified products for the chromosome 1D SSR markers (i.e., wmc147, wmc222, gwm337, and Xcfd63) were detected for Chinese Spring (CS), Shumai482 (SM482), and Shumai921 (SM921). In contrast, amplicons were not genetated for 8801 and K17–841-1 (Fig. 2). Our results verified that in line K17–841-1, wheat chromosome 1D had been replaced by chromosome 1E of tetraploid Th. elongatum.
Fig. 2.
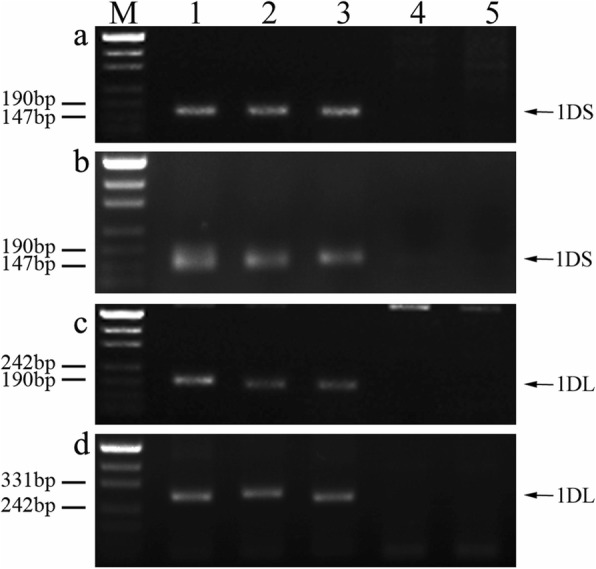
PCR amplification of SSR markers wmc147 (a), wmc222 (b), gwm337 (c), and Xcfd63 (d). Lanes M marker, 1 CS, 2 wheat cultivar Shumai482, 3 wheat cultivar Shumai921, 4 8801 (T. durum-tetraploid Th. elongatum amphidiploid), 5 K17–841-1(wheat–tetraploid Th. elongatum substitution line). Arrows show the diagnostic amplification products of SSR markers
Morphology of K17–841-1
We analyzed the agronomic traits of K17–841-1 and the donor parents in two growing seasons. Line K17–841-1 displayed stable morphological traits, which were similar to those of the wheat parents SM482 and SM921 (Table 1; Fig. 3). The average plant height and spike length of line K17–841-1 were significantly lower than those of the Triticum durum–tetraploid Th. elongatum partial amphidiploid 8801, but were greater than those of SM482 and similar to those of SM921 (Fig. 3a, b). The number of spikelets per spike was lower than that of SM482 or SM921, but was similar to that of 8801. The grain number per spike was greater than that of 8801, but was lower than that of SM921 and similar to that of SM482 (Fig. 3c). There were no significant differences between K17–841-1 and either SM482 or SM921 regarding the tiller number, thousand-kernel weight, and seed setting rate (Fig. 3d).
Table 1.
Agronomic traits of K17–841-1 and its parental lines
| Lines | Year | Plant height (cm) | Tiller number | Spike length (cm) | Spikelet per spike | Grains per spike | Thousand-grain weight (g) | Seed setting rate (%) |
|---|---|---|---|---|---|---|---|---|
| 8801 | 2017–2018 | 143.6 ± 6.0a | 8.2 ± 2.3a | 15.5 ± 2.1a | 17.5 ± 1.2c | 39.7 ± 2.9c | 24.5 ± 0.5b | 84.0 ± 8.4b |
| 2018–2019 | 132.4 ± 4.5a | 8.5 ± 0.6a | 17.3 ± 2.1a | 17.5 ± 1.3b | 31.3 ± 5.0c | 29.3 ± 0.2b | 68.5 ± 12.8b | |
| SM482 | 2017–2018 | 82.5 ± 4.3c | 9.2 ± 1.6a | 13.4 ± 1.3b | 22.0 ± 2.1b | 63.3 ± 9.4b | 44.7 ± 0.2a | 99.6 ± 0.9a |
| 2018–2019 | 79.0 ± 5.2c | 8.8 ± 0.4a | 12.5 ± 0.9b | 18.6 ± 1.7ab | 58.2 ± 7.9b | 43.2 ± 0.8a | 97.9 ± 2.3a | |
| SM921 | 2017–2018 | 86.1 ± 3.1bc | 9.3 ± 2.3a | 13.9 ± 1.4ab | 24.7 ± 2.9a | 84.3 ± 12.3a | 44.5 ± 2.0a | 99.4 ± 1.6a |
| 2018–2019 | 87.7 ± 2.6b | 4.8 ± 1.3b | 13.2 ± 0.8b | 20.6 ± 0.9a | 72.8 ± 8.6a | 38.6 ± 0.8b | 92.7 ± 3.2a | |
| K17–841-1 | 2017–2018 | 89.9 ± 4.2b | 8.2 ± 1.3a | 13.5 ± 0.8b | 19.5 ± 1.0c | 63.0 ± 8.2b | 44.6 ± 1.7a | 95.7 ± 1.3a |
| 2018–2019 | 94.1 ± 4.8b | 9.3 ± 1.8a | 14.0 ± 0.6b | 17.8 ± 1.0b | 51.8 ± 11.0b | 44.3 ± 1.2a | 93.8 ± 5.4a |
Data in the columns indicate means ± standard errors
Lowercase letters following the means indicate significant differences at P < 0.05 as determined by the least significant differences
Fig. 3.
Plant morphology of the wheat–tetraploid Th. elongatum substitution line K17–841-1 and its parents. Adult plants (a), spikes (b), spikelets(c), and grains(d). Numbers 1–4 represent 8801 (T. durum-tetraploid Th. elongatum amphidiploid), wheat cultivar Shumai482, wheat cultivar Shumai 921, and K17–841-1, respectively
Stripe rust resistance evaluation
We evaluated the stripe rust resistance of K17–841-1, 8801, SM482, SM921, and SY95–71 plants inoculated with a mixture of Pst races (CYR-32, CYR-33, CYR-34, and V26/Gui22–14) at Chengdu, Sichuan, China. An analysis of three replicates revealed that the adult SM482, SM921, and SY95–71 plants were susceptible to these Pst races, with infection types (ITs) of 4, 3, and 4, respectively. In contrast, 8801 and K17–841-1 plants were highly resistant to these races (i.e., IT of 0) (Fig. 4).
Fig. 4.
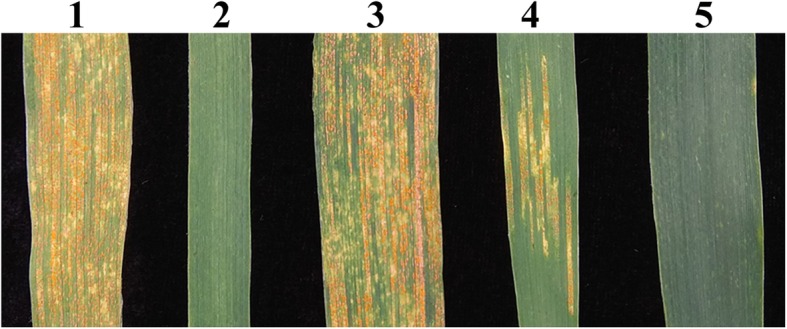
Stripe rust resistance of the wheat–tetraploid Th. elongatum substitution line K17–841-1 and the controls. 1 wheat line SY95–71, 2 8801 (T. durum-tetraploid Th. elongatum amphidiploid), 3 wheat cultivar Shumai482, 4 wheat cultivar Shumai921, 5 K17–841-1
Development and validation of specific molecular markers for the 1E chromosome of tetraploid Th. elongatum
The GBS approach was applied to identify tetraploid Th. elongatum 1E chromosome-specific sequences. Details regarding the sequencing, including raw reads, clean reads, effective rate, error rate, Q20, Q30, and GC content, are summarized in Table 2. The results revealed a high sequencing quality (Q20 ≥ 94% and Q30 ≥ 86%) and a normal GC content distribution. A total of 45,115,502, 7,389,702, 8,710,288, and 8,831,260 effective GBS sequences were obtained for PI531718, PI531750, 8801, and K17–841-1, respectively (Table 3). The sequencing depth was more than 10.32×. A sequence comparison revealed 73,744 K17–841-1 clean reads that were less than 23% homologous with CS sequences. Additional sequence alignments uncovered 2952 K17–841-1 reads that were more than 23% homologous with 8801 and PI531750 fragments obtained by GBS. Finally, 1194 K17–841-1 unique reads (597 specific fragments) that were less than 10% homologous with sequences from PI531718 and the other six substitution lines were obtained and were considered the tetraploid Th. elongatum 1E chromosome-specific fragments.
Table 2.
Quality of GBS data
| Genotype | Raw base (bp) |
Clean base (bp) |
Effective rate (%) |
Error rate (%) |
Q20 (%) |
Q30 (%) |
GC (%) |
|---|---|---|---|---|---|---|---|
| PI531718 | 6,496,632,288 | 6,496,632,288 | 100 | 0.03 | 95.97 | 93.68 | 43.95 |
| PI531750 | 1,064,120,832 | 1,064,117,088 | 100 | 0.04 | 95.96 | 89.20 | 42.99 |
| 8801 | 1,254,284,064 | 1,254,281,472 | 100 | 0.05 | 95.00 | 87.17 | 42.13 |
| K17–841-1 | 1,271,704,608 | 1,271,701,440 | 100 | 0.05 | 94.71 | 86.58 | 43.17 |
Table 3.
Sequence alignment between K17–841-1 and its parental lines
| Genotype | Total reads | Unmapped reads | Map2 parent reads | uniqReads |
|---|---|---|---|---|
| PI531718 | 45,115,502 | 5,309,332 | ||
| PI531750 | 7,389,702 | 776,849 | ||
| 8801 | 8,710,288 | 300,048 | ||
| K17–841-1 | 8,831,260 | 73,744 | 2952 | 1194 |
Unmapped reads: reads unmapped on CS
Map 2 parent reads: reads mapped on PI531750 and 8801
UniqReads: unique reads of K17–841-1
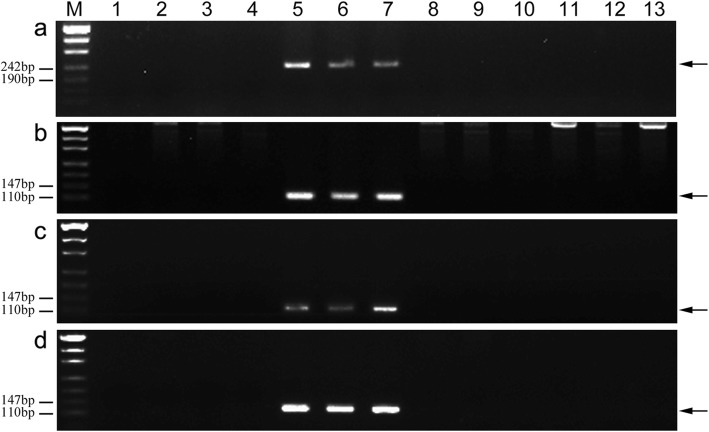
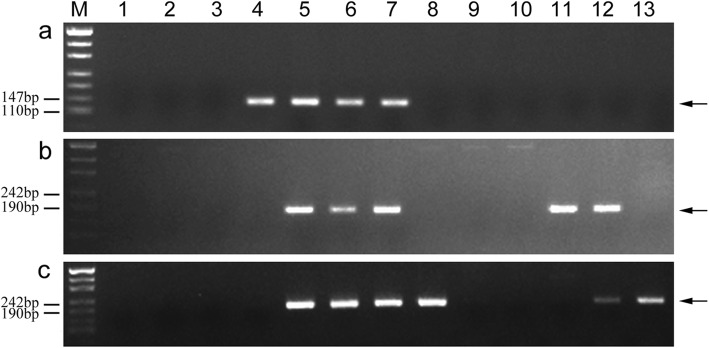
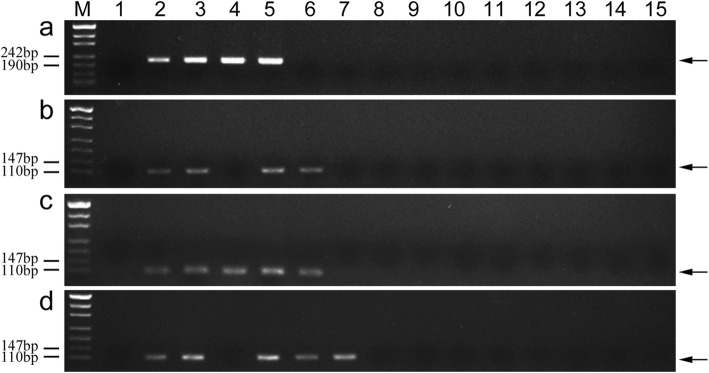
To develop tetraploid Th. elongatum 1E chromosome-specific markers, 235 PCR primers pairs were designed based on these specific fragments and used to amplify sequences from CS, SM482, SM921, PI531718, PI531750, 8801, K17–841-1, and six wheat-tetraploid Th. elongatum disomic substitution (TDS) lines (2E-7E). A total of 165 (70%) Th. elongatum-specific molecular markers were successfully developed (see Additional file 1: Table S1), of which 132 markers amplified specific sequences only from PI531750, 8801, and K17–841-1 (Type I) (Fig. 5a-d). Therefore, these markers were regarded as tetraploid Th. elongatum 1E chromosome-specific molecular markers, with a success rate of up to 56.2%. Additionally, 21 markers amplified specific sequences only from PI531718, PI531750, 8801, and K17–841-1 (Type II) (Fig. 6a). Moreover, 12 markers were also detected on the other E chromosomes of tetraploid Th. elongatum (Fig. 6b, c).
Fig. 5.
PCR amplification of markers TTE1E-12 (a), TTE1E-140 (b), TTE1E-189 (c), and TTE1E-193 (d). Lanes M marker, 1 CS, 2 wheat cultivar Shumai482, 3 wheat cultivar Shumai921, 4 diploid Thinopyrum elongatum, 5 tetraploid Thinopyrum elongatum, 6 8801 (T. durum-tetraploid Th. elongatum amphidiploid), 7 K17–841-1 (wheat–tetraploid Th. elongatum substitution line), 8 TDS2E(2A), 9 TDS3E(3D), 10 TDS4E(4D), 11 TDS5E(5D), 12 TDS6E(6D), 13 TDS7E(7D). Arrows show the diagnostic amplification products of tetraploid Th. elongatum 1E chromosome
Fig. 6.
PCR amplification of markers TTE1E-214 (a), TTE1E-3 (b), and TTE1E-58 (c). Lanes M marker, 1 CS, 2 wheat cultivar Shumai482, 3 wheat cultivar Shumai921, 4 diploid Thinopyrum elongatum, 5 tetraploid Thinopyrum elongatum, 6 8801 (T. durum-tetraploid Th. elongatum amphidiploid), 7 K17–841-1 (wheat–tetraploid Th. elongatum substitution line), 8 TDS2E(2A), 9 TDS3E(3D), 10 TDS4E(4D), 11 TDS5E(5D), 12 TDS6E(6D), 13 TDS7E(7D). Arrows show the diagnostic amplification products of tetraploid Th. elongatum 1E chromosome
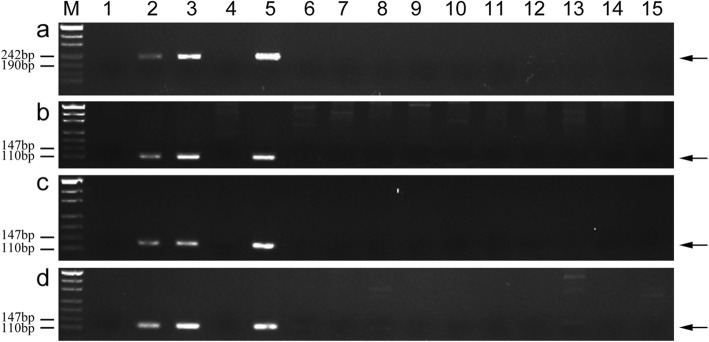
To confirm marker specificity and stability, 153 markers (Types I and II) were used to further analyze 11 wheat-related species. The PCR results are presented in Additional file 2: Table S2. Among the 132 tetraploid Th. elongatum 1E chromosome-specific markers, 25 amplified sequences from tetraploid Th. elongatum, but not from the analyzed related species (Table 4; Fig. 7a-d). In contrast, 21 and 106 markers amplified specific sequences from not only tetraploid Th. elongatum, but also from diploid Th. elongatum and decaploid Th. ponticum, respectively (Fig. 8a, b). Additionally, 28 markers amplified a common sequence from tetraploid Th. elongatum, diploid Th. elongatum, and Th. ponticum, but did not amplify any sequences from the other related species (Fig. 8c). Five markers amplified specific sequences not only from tetraploid Th. elongatum and Th. ponticum, but also from Thinopyrum bessarabicum (Fig. 8d). Four markers amplified a common sequence from diploid Th. elongatum, tetraploid Th. elongatum, Th. ponticum, Th. bessarabicum, Pseudoroegneria libanotica, Trichopyrum caespitosum, and Psammopyrum athericum. Furthermore, 58, 32, 5, 8, 28, 8, 13, 50, and 73 markers amplified sequences from Th. bessarabicum, Pse. libanotica, Dasypyrum villosum, Hordeum bogdanii, Agropyron cristatum, Secale cereale, Psathyrostachys huashanica, Tr. caespitosum, and Psa. athericum, respectively.
Table 4.
Specific amplification results of 1E-chromosomal markers in wheat-related species
| Number of markers | Amplification result | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PI531718 (E) |
PI531750 (EE) |
PI531737 (EEEEE) |
W6–10232 (Eb) |
PI228391 (St) |
PI251477 (V) |
Y1819 (H) |
PI610892 (P) |
QL (R) |
ZY3156 (Ns) |
PI634264 (StE) |
PI531744 (StEP) |
|
| 25 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 24 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| 4 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| 2 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| 5 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| 11 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 4 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
“1” or “0” indicates the presence or absence of the specific band, respectively
Fig. 7.
Stability and specificity of markers TTE1E-12 (a), TTE1E-140 (b), TTE1E-189 (c), and TTE1E-193 (d) in other wheat-related species. Lanes M marker, 1 CS, 2 8801 (T. durum-tetraploid Th. elongatum amphidiploid), 3 K17–841-1 (wheat–tetraploid Th. elongatum substitution line), 4 diploid Thinopyrum elongatum, 5 tetraploid Thinopyrum elongatum, 6 Thinopyrum ponticum, 7 Thinopyrum bessarabicum, 8 Pseudoroegneria libanotica, 9 Dasypyrum villosum, 10 Hordeum bogdanii, 11 Agropyron cristatum, 12 Secale cereale, 13 Psathyrostachys huashanica, 14 Trichopyrum caespitosum, 15 Psammopyrum athericum. Arrows show the diagnostic amplification products of tetraploid Th. elongatum 1E chromosome
Fig. 8.
Stability and specificity of markers TTE1E-59 (a), TTE1E-184 (b), TTE1E-222 (c), and TTE1E-192 (d) in other wheat-related species. Lanes M marker, 1 CS, 2 8801 (T. durum-tetraploid Th. elongatum amphidiploid), 3 K17–841-1 (wheat–tetraploid Th. elongatum substitution line), 4 diploid Thinopyrum elongatum, 5 tetraploid Thinopyrum elongatum, 6 Thinopyrum ponticum, 7 Thinopyrum bessarabicum, 8 Pseudoroegneria libanotica, 9 Dasypyrum villosum, 10 Hordeum bogdanii, 11 Agropyron cristatum, 12 Secale cereale, 13 Psathyrostachys huashanica, 14 Trichopyrum caespitosum, 15 Psammopyrum athericum. Arrows show the diagnostic amplification products of tetraploid Th. elongatum 1E chromosome
Utility of the tetraploid Th. elongatum-specific markers in the F2 population
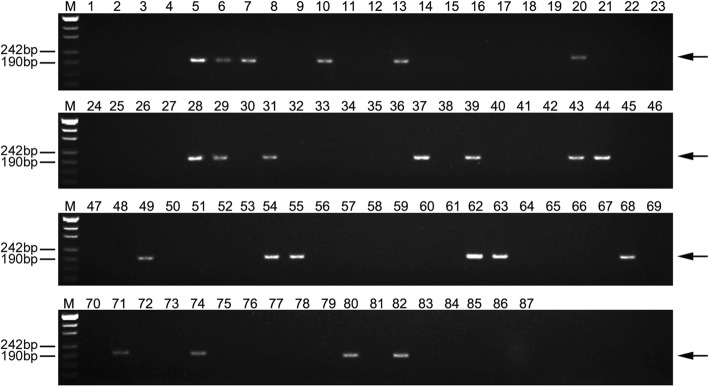
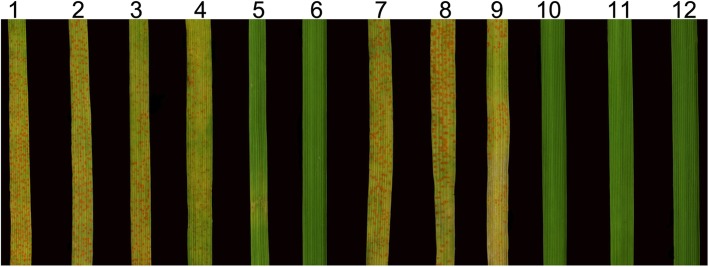
To verify the utility of the newly developed molecular markers, 80 F2 individuals of K17–841-1 and wheat cultivar Shumai969 (SM969) were analyzed by PCR. We detected specific amplicons for only 20 individuals (Fig. 9, Additional file 3: Table S3). The results of a GISH indicated these 20 plants with specific amplicons carried 1E (1D) chromosomal substitutions, while the 60 plants without specific amplicons had no GISH signal in their chromosome preparation. More importantly, an evaluation of stripe rust resistance at the seedling stage revealed that 8801, K17–841-1, and the 20 positive F2 individuals carrying 1E chromosomal markers were highly resistant to Pst race CYR-34. In contrast, the 60 F2 plants lacking amplicons as well as SY95–71 and the parental lines SM482, SM921, and SM969 were highly susceptible (Fig. 10, Additional file 3: Table S3). These observations implied that the specific markers developed in this study may be useful for detecting the stripe rust resistance gene on the 1E chromosome of tetraploid Th. elongatum during the breeding of new disease-resistant wheat varieties.
Fig. 9.
Utility of marker TTE1E-21 in 80 F2 individuals of K17–841-1 and SM969. 1 CS, 2 wheat cultivar Shumai482, 3 wheat cultivar Shumai921, 4 wheat cultivar Shumai969, 5 tetraploid Thinopyrum elongatum, 6 8801 (T. durum-tetraploid Th. elongatum amphidiploid), 7 K17–841-1 (wheat–tetraploid Th. elongatum substitution line), 8–87 80 F2 individuals. Arrows show the diagnostic amplification products of tetraploid Th. elongatum 1E chromosome
Fig. 10.
Evaluation for seedling stage reacctions to Pst race CYR-34 in F2 individuals of K17–841-1 and SM969 and the controls. 1 wheat line SY95–71, 2 wheat cultivar Shumai482, 3 wheat cultivar Shumai921, 4 wheat cultivar Shumai969, 5 8801 (T. durum-tetraploid Th. elongatum amphidiploid), 6 K17–841-1 (wheat–tetraploid Th. elongatum substitution line), 7–9 susceptible F2 individuals, 10–12 resistant F2 individuals
Discussion
Distant hybridizations may be useful for transferring agronomically valuable genes from wild relatives to common wheat varieties. Creating intermediate lines carrying alien chromosomes of wheat relatives provides the basis for using these germplasm resources to improve domesticated wheat [27]. Wheat–alien chromosomal substitution lines are important bridge materials for transferring alien genes to common wheat. In recent decades, there have been many attempts by wheat breeders to generate wheat–Th. elongatum substitution lines [8]. However, most of the introgressions from Th. elongatum into wheat involved the diploid Th. elongatum and decaploid Th. ponticum. For example, CS–diploid Th. elongatum addition and substitution lines have been produced [28]. Additionally, Zheng et al. [29] generated the wheat–Th. ponticum 4Ag (4D) disomic substitution line Blue 58 from a hybridization between Th. ponticum and common wheat. Fu et al. [30] developed a wheat–diploid Th. elongatum 7E (7D) substitution line resistant to Fusarium head blight. Wang et al. [31] produced a wheat–Th. ponticum St (6A) disomic substitution line exhibiting powdery mildew resistance. The 7JSt (7B) substitution line CH1113-B13, which is resistant to stripe rust, was identified from among the progeny of a cross between common wheat and Th. ponticum [32]. The tetraploid Th. elongatum harbors many beneficial genes that provide protection from diseases as well as cold, drought, and salinity stresses. Thus, it is a valuable genetic resource for improving the tolerance of common wheat to biotic and abiotic stresses [9, 10, 17]. To date, there are few reports regarding wheat–tetraploid Th. elongatum substitution lines. Li et al. [17] developed 50 wheat–tetraploid Th. elongatum introgression lines by crossing the T. durum–tetraploid Th. elongatum partial amphidiploid line 8801 with wheat varieties native to the Sichuan Basin, China. These lines comprise 40–47 chromosomes and most of them are cytologically unstable. In the current study, we further characterized the wheat–tetraploid Th. elongatum 1E (1D) disomic substitution line K17–841-1 derived from the 8801/SM482/SM921 F5 progeny. The FISH and SSR analyses confirmed that common wheat chromosome 1D was substituted with the homologous chromosome 1E from tetraploid Th. elongatum, providing further evidence that the E genome of Th. elongatum is closely related to the D genome of common wheat [33, 34]. Interestingly, an evaluation of the stripe rust resistance indicated that the wheat parents SM482 and SM921 are susceptible and that the resistance of K17–841-1 was derived from tetraploid Th. elongatum. Some stripe rust resistance genes have been identified in Thinopyrum species and have been transferred to common wheat, including Yr50 from Th. intermedium, YrE from diploid Th. elongatum, and Yr69, YrTP1, YrTP2, and YrCH7056 from Th. ponticum [35, 36]. To the best of our knowledge, this study is the first to demonstrate the successful transfer of a new and high-level stripe rust resistance gene from tetraploid Th. elongatum. Furthermore, the grain yield of line K17–841-1 is similar to that of the wheat parents SM482 and SM921. Accordingly, K17–841-1 represents an appropriate bridge breeding material for the introgression of alien genes to enhance wheat disease resistance.
Molecular markers are widely used to detect and track alien chromosomes and/or chromosomal segments during crossing and selection [10]. Conventional methods for developing markers, including SSRs, RAPDs, and amplified fragment length polymorphisms, have low success rates because of the high genomic sequence homology between Th. elongatum and common wheat. For example, using 40 SSR primers, You et al. [37] amplified 108 diploid Th. elongatum-specific fragments, but only one genome-specific molecular marker was developed. In another previous study, 94 diploid Th. elongatum-specific fragments were generated with 26 pairs of RAPD primers, but only three 1E or 3E chromosome-specific markers were developed [38]. Additionally, Ge et al. [39] developed one diploid Th. elongatum-specific marker from 65 specific fragments based on suppression subtractive hybridization. Furthermore, inter-SSR, inter-retrotransposon amplified polymorphism, and retrotransposon microsatellite amplified polymorphism techniques were applied to develop 13 specific sequence-characterized amplified region markers based on 34 specific diploid Th. elongatum sequences [40]. In a recent study, 135 randomly selected fragments of diploid Th. elongatum generated by SLAF-sequencing (SLAF-seq) were used to develop 89 specific molecular markers, including 61 7E chromosome-specific markers [11]. Chen et al. [25] developed 20 diploid Th. elongatum 1E chromosome-specific and two E genome-specific molecular markers based on SLAF-seq data, with an efficiency of up to 60%. Approximately 89% of the mined SNPs of diploid Th. elongatum may be authentic with respect to their polymorphisms and chromosomal locations as determined by a high-resolution melting curve assay [12]. Moreover, SLAF-seq data were used to develop 67 Th. ponticum-specific molecular markers from a wheat–Th. ponticum translocation line, with a success rate of up to 39% [19]. Furthermore, GBS, which is a robust, simple, and high-throughput next-generation sequencing approach that can decrease the complexity of large genomes, has been applied to develop many molecular markers in Triticale species [26, 41]. This approach can generate considerable sequence information and is appropriate for all whole-genome density distributions. The flexibility and low cost of GBS makes it ideal for genomics-assisted breeding [42]. To date, there have been no reports describing the development of tetraploid Th. elongatum-specific molecular markers. In this study, the GBS method was applied to develop 165 specific molecular markers, including 132 tetraploid Th. elongatum chromosome 1E-specific PCR markers, with a success rate of up to 70%. The greater success rate for our study relative to those of earlier investigations indicates that GBS technology may be used to efficiently develop alien-specific molecular markers in a wheat background.. The markers developed in this study may be useful for the high-throughput and accurate detection of tetraploid Th. elongatum DNA in diverse materials. Most importantly, the verification of their utility for analyzing a F2 breeding population implies that these specific markers are appropriate for tracing the stripe rust resistance gene carried on chromosome 1E of tetraploid Th. elongatum for the breeding of enhanced materials.
Even after removing K17–841-1 sequences that were more than 90% homologous with sequences from six other substitution lines (2E–7E), 12 markers still amplified specific sequences in the other substitution lines, confirming that the Th. elongatum E chromosomes sequences are highly homologous, likely because of chromosomal rearrangements. Earlier investigations involving biochemical and molecular markers as well as SLAF-seq revealed similar sequence identity levels for the diploid Th. elongatum [11, 43]. After analyzing the hybrids from crosses between tetraploid Th. elongatum and diploid Th. elongatum and Th. bessarabicum, Dvořák [44] concluded that tetraploid Th. elongatum is a segmental allotetraploid with two closely related E genomes. On the basis of examinations of chromosomal pairs, karyotypes, and C-banding patterns,Liu and Wang [45] also determined that tetraploid Th. elongatum is a segmental allotetraploid carrying two E genomes with slight differences. In a previous study involving a FISH with repetitive clones, two tetraploid Th. elongatum accessions were analyzed to uncover the significant chromosomal polymorphisms between the E genome of the putative diploid progenitor Th. elongatum and the other genome [18]. The verification of 153 markers specific to tetraploid Th. elongatum confirmed that 21, 106, 50, and 73 markers can detect E genome chromosomes in diploid Th. elongatum, Th. ponticum, Tr. caespitosum (StE), and Psa. athericum (StEP), respectively (see Additional file 2: Table S2). Our results provide further evidence that tetraploid Th. elongatum is an allotetraploid harboring two different E genomes, and Thinopyrum E genomes were differentiated during polyploidization events. Two specific markers, TTE1E-212 and TTE1E-222, were unique to diploid Th. elongatum, tetraploid Th. elongatum, and Th. ponticum, suggesting they can detect the E chromosomal DNA in all materials carrying the E genome. Interestingly, 58, 32, 5, 8, 28, 8, and 13 markers based on specific tetraploid Th. elongatum sequences successfully amplified products from Th. bessarabicum (Eb), Pse. libanotica (St), D. villosum (V), H. bogdanii (H), Ag. cristatum (P), S. cereale (R), and P. huashanica (Ns), respectively. Our specificity analysis indicated that the marker amplification frequencies are considerably higher in E, Eb, and St genomic species than in other wheat-related species, which is consistent with the findings of previous studies, in which the E genomes of Th. elongatum and Th. ponticum were more closely related to the Eb genome of Th. bessarabicum and the St genome of Pseudoroegneria species than to the other genomes [33, 46]. Additionally, 48 and 74 markers uncovered polymorphisms between Thinopyrum E-genome-containing species and Th. bessarabicum and Pse. libanotica, respectively, indicating that they will likely be applicable for investigating the genetic differences between E, Eb, and St genomes and for revealing the genetic diversity among species (populations) carrying E, Eb, and/or St genomes. The resulting information will further characterize these complex species and may be relevant for the genetic improvement of these important forage crops.
Conclusions
We characterized a wheat-tetraploid Th. elongatum 1E (1D) disomic substitution line based on GISH, FISH, and SSR analyses. This line is highly resistant to the stripe rust pathogen strains prevalent in China. Moreover, it represents an appropriate bridge breeding material for the introgression of alien genes to improve wheat disease resistance. Additionally, we analyzed GBS data to generate and validate a new 1E chromosome-specific, easy-to-use marker set that may be applicable for identifying and characterizing the tetraploid Th. elongatum chromosomes along with the chromosomes of all other wheat-related species and for investigating the genetic differences and phylogenetic relationships among E, Eb, St, and other closely-related genomes. The use of this marker set will further increase our understanding of these complex species.
Materials and methods
Plant materials
The plant materials used in the current study are listed in Table 5. The hexaploid Trititrigia line 8801 (2n = 6x = 42, AABBEE), which is tolerant to cold, drought, and salt stresses and resistant to Fusarium head blight, rust, and powdery mildew, was originally produced and identified at the Eastern Cereal and Oilseed Research Center, Ottawa, Canada [17]. The native wheat cultivars SM482, SM921, and SM969 exhibit superior agronomic traits, but are susceptible to the stripe rust pathogens prevalent in southwestern China. To produce wheat–tetraploid Th. elongatum derivative line, we first crossed 8801 with SM482, after which the resulting F1 plants were further crossed with SM921 to obtain the BC1F1 population. Seeds from the BC1F1 plants were bulked and advanced to the BC1F5 generation by single seed descent, ultimately resulting in the isolation of line K17–841-1 (Fig. 11). The F2 population comprising 80 individuals was derived from a cross between K17–841-1 and SM969. We developed six wheat–tetraploid Th. elongatum disomic substitution lines, TDS2E (2A), TDS3E (3D), TDS4E (4D), TDS5E (5D), TDS6E (6D), and TDS7E (7D), via the hybridization between native wheat cultivars and Trititrigia line 8801 (data not published). Wheat cultivar Chinese Spring (CS) was used as a positive control for the molecular marker analysis. Wheat line SY95–71 was used as a susceptible control for the evaluations of stripe rust responses. Regarding the GISH analysis, wheat cultivar J-11 was used as a source of blocking DNA, whereas tetraploid Th. elongatum accession PI531750 (2n = 4x = 28, EEEE) was used as a source of probe DNA. The molecular markers were validated with the following wheat-related species: Thinopyrum elongatum (2n = 2x = 14, EE), Thinopyrum ponticum (2n = 10x = 70, EEEEEEEEEE/StStStStEEEEEE), Thinopyrum bessarabicum (2n = 2x = 14, EbEb), Pseudoroegneria libanotica (2n = 2x = 14, StSt), Dasypyrum villosum (2n = 2x = 14, VV), Hordeum bogdanii (2n = 2x = 14, HH), Agropyron cristatum (2n = 2x = 14, PP), Secale cereale (2n = 2x = 14, RR), Psathyrostachys huashanica (2n = 2x = 14, NsNs), Trichopyrum caespitosum (2n = 4x = 28, StStEE), and Psammopyrum athericum (2n = 6x = 42, StStEEPP). Voucher specimens have been deposited in the herbarium of the Triticeae Research Institute, Sichuan Agricultural University, China (SAUTI).
Table 5.
Plant materials used in this study
| Accession | Species/Materials | Chromosome numbers | Genome |
|---|---|---|---|
| PI531718 | Thinopyrum elongatum | 14 | E |
| PI531750 | Thinopyrum elongatum | 28 | EE |
| PI531737 | Thinopyrum ponticum | 70 | EEEEE/StStEEE |
| W6–10232 | Thinopyrum bessarabicum | 14 | Eb |
| PI228391 | Pseudoroegneria libanotica | 14 | St |
| PI251477 | Dasypyrum villosum | 14 | V |
| Y1819 | Hordeum bogdanii | 14 | H |
| PI610892 | Agropyron cristatum | 14 | P |
| QL | Secale cereale | 14 | R |
| ZY3156 | Psathyrostachys huashanica | 14 | Ns |
| PI634264 | Trichopyrum caespitosum | 28 | StE |
| PI531744 | Psammopyrum athericum | 42 | StEP |
| 8801 | Triticum durum-tetraploid Thinopyrum elongatum amphidiploid | 42 | ABE |
| SM482 | Shumai482 | 42 | ABD |
| SM921 | Shumai921 | 42 | ABD |
| SM969 | Shumai969 | 42 | ABD |
| CS | Chinese Spring | 42 | ABD |
| J-11 | wheat cultivar J-11 | 42 | ABD |
| SY95–71 | wheat line SY95–71 | 42 | ABD |
| K17–841-1 | Wheat-tetraploid Thinopyrum elongatum derivative line | 42 | ABDE |
| TDS2E(2A),TDS3E(3D), TDS4E(4D),TDS5E(5D), TDS6E(6D),TDS7E(7D) | Wheat-tetraploid Thinopyrum elongatum disomic substitution lines | 42 | ABDE |
| F2 population of K17–841-1/Shumai969 | 42 |
Fig. 11.
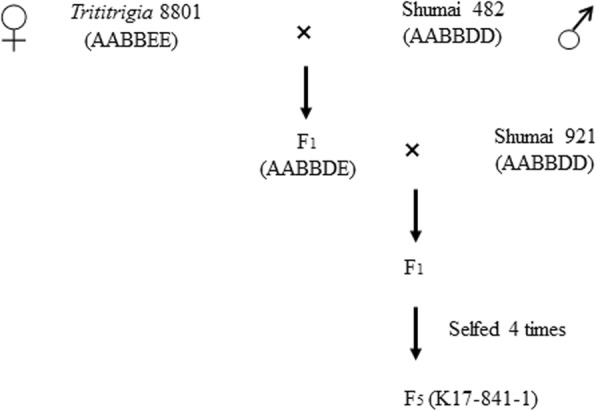
Pedigree details of K17–841-1
GISH and FISH analyses
Root tips from germinating seeds were treated with nitrous oxide for 2 h and 90% acetic acid for 10 min, after which they were digested with pectinase and cellulase (Yakult Pharmaceutical, Tokyo, Japan) [47]. Slides were then prepared for the GISH analysis as described by Han et al. [48]. The cetyltrimethyl ammonium bromide method was used to extract genomic DNA from the freshly collected leaves of tetraploid Th. elongatum PI531750 and wheat cultivar J-11 [49]. The PI531750 DNA was labeled with fluorescein-12-dUTP according to the nick translation method (Thermo Fisher Scientific, Eugene, OR, USA) and was used as the hybridization probe. The GISH analysis was completed according to the method described by Han et al. [50], with a 1:150 probe DNA:blocking DNA ratio. Specifically, 10 μL hybridization solution containing 2 × saline sodium citrate (SSC), 10% dextran sulfate, and 10 ng/μL labeled probe DNA together with blocking DNA was added to each slide. Samples were denatured by heating at 85 °C for 5 min, incubated at 37 °C for 8 h, and washed with 2× SSC. Finally, the chromosomes were counterstained with 4,6-diamino-2- phenylindole solution (Vector Laboratories, Burlingame, CA, USA). The GISH slides were examined with the BX-63 microscope (Olympus, Tokyo, Japan) and images were captured with a DP-70 CCD camera.
The GISH slides were washed sequentially with 70% (v/v) ethanol for 5 min, 2× SSC at 60 °C for 30 min, ddH2O for 10 min, and 100% (v/v) ethanol for 5 min. A FISH analysis was subsequently completed to identify the chromosomal constitution of line K17–841-1, with pSc119.2 and pTa535 as probes [47, 51]. The FISH was performed as described by Han et al. [48], with minor modifications. The probe mixture (0.35 μL each probe in 2× SSC and 1× TE buffer, pH 7.0; total volume = 10 μL) was added to a slide, covered with a coverslip, incubated in a moist box at 37 °C for 2 h, and then washed with 2× SSC at room temperature. The FISH signals were observed with the BX63 microscope (Olympus). Images were captured with the DP-70 CCD camera and analyzed with Adobe Photoshop software.
SSR marker analysis
Primer pairs for four SSR markers (wmc147, wmc222, gwm337, and Xcfd63) on wheat chromosomes 1DS and 1DL were used to characterize the 1D chromatin in the wheat–tetraploid Th. elongatum line K17–841-1. Details regarding all primers are listed in Table 6. Additionally, CS, SM482, and SM921 were used as positive controls, whereas 8801 was used as a negative control. The PCR amplification was completed as described by Somers et al. [52], with minor modifications.
Table 6.
Sequences of wheat 1DS and 1DL SSR markers
| Marker | Primer sequence (5′-3′) | Annealing temperature (°C) | Arm location | Amplification size (bp) |
|---|---|---|---|---|
| wmc147 |
F: AGAACGAAAGAAGCGCGCTGAG R: ATGTGTTTCTTATCCTGCGGGC |
58 | 1DS | 154 |
| wmc222 |
F: AAAGGTGCGTTCATAGAAAATTAGA R:AGAGGTGTTTGAGACTAATTTGGTA |
54 | 1DS | 184 |
| gwm337 |
F: CCTCTTCCTCCCTCACTTAGC R: TGCTAACTGGCCTTTGCC |
59 | 1DL | 191 |
| Xcfd63 |
F: TCCTGAGGATGTTGAGGACC R: GAGAGAGGCGAAACATGGAC |
58 | 1DL | 282 |
F forward primer, R reverse primer
Agronomic trait evaluation
The morphological traits of K17–841-1 and its parents were evaluated in a field trial in Wenjiang, Sichuan province, China, with three replicates in the 2017–2018 and 2018–2019 growing seasons. For each replicate, 15 grains of each line were evenly planted in 1.5-m rows separated by 0.3 m. The plant height, tiller number, spike length, spikelet per spike, grains per spike, thousand-grain weight, and seed setting rate were evaluated for 10 samples per replicate. Significant differences in traits were determined with the SAS 8.2 program (SAS Institute Inc., Cary, NC, USA).
Stripe rust resistance screening
The smear method [53] was used to evaluate the responses of adult K17–841-1, 8801, SM482, SM921, and SY95–71 plants to a mixture of Pst races (CYR-32, CYR-33, CYR-34, and V26/Gui22–14) in a field trial in Chengdu, Sichuan, China during the 2017–2018 growing season. To assess the utility of the developed tetraploid Th. elongatum 1E chromosome-specific markers, the K17–841-1/SM969 F2 individuals as well as 8801, SM482, SM921, SM969, and SY95–71 seedling were evaluated for their reactions to Pst race CYR-34 in a growth chamber. The plants were inoculated at the two-leaf stage and the stripe rust reaction of the first leaf of each plant was evaluated at 14 days after inoculation. Wheat line SY95–71 was used as the susceptible control. The stripe rust responses were evaluated with three replicates as described by Li et al. [17]. The stripe rust IT was based on the following scale: 0, 0;, 1, 2, 3, and 4, in which 0 = immunity, 0; = necrotic flecks, and 1–4 = increasing sporulation and decreasing necrosis or chlorosis. Plants with an IT of 2 or lower were considered resistant, whereas plants with an IT of 3 or 4 were considered susceptible.
Genotyping-by-sequencing, sequence alignment, and tetraploid Th. elongatum 1E chromosome– specific fragment acquisition
Total genomic DNA was extracted from fresh young leaves with the cetyltrimethyl ammonium bromide method, after which the DNA concentration was adjusted to 150 ng/μL. Genomic DNA samples of diploid Th. elongatum PI531718, PI531750, 8801, K17–841-1, and six wheat–tetraploid Th. elongatum (2E-7E) chromosomal substitution lines were subjected to GBS, which was completed by Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). High-Quality DNA libraries were constructed and sequenced with the Illumina HiSeq™ system. The raw reads in FASTQ format were filtered by removing low-quality reads and reads with adapter and/or poly-N sequences to obtain clean reads (140 bp). Tetraploid Th. elongatum 1E chromosome-specific sequences were obtained as follows. First, the high-quality K17–841-1 sequences were compared with the CS reference genome sequences (https://urgi.versailles.inra.fr/download/iwgsc/IWGSC_RefSeq_Assemblies/v1.0/), after which the sequences with identities greater than 23% were eliminated [54]. Second, the sequences were compared with those of 8801 and PI531750 acquired by GBS in this study, and the sequences with identities greater than 23% were selected. Finally, the retained K17–841-1 sequences were compared with the sequences of PI531718 and the six chromosomal substitution lines. Sequences with identities exceeding 90% were removed. The remaining sequences were considered to be specific to chromosome 1E of tetraploid Th. elongatum.
Development and validation of the tetraploid Th. elongatum-specific molecular markers
On the basis of these specific sequences, PCR primers were designed with the Primer3 Plus online tool (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) and then synthesized by TSINGKE (Chengdu, China). Details regarding the PCR primers are presented in Additional file 1: Table S1. The amplified products were examined by 3% agarose electrophoresis. The markers that amplified specific sequences in PI531750, 8801, and K17–841-1, but not in SM482, SM921, CS, PI531718, and the 2E-7E substitution lines, were identified as tetraploid Th. elongatum 1E chromosome-specific molecular markers. The stability, repeatability, and specificity of these markers were validated in the CS, PI531750, 8801, and K17–841-1 lines as well as in 11 wheat-related species.
The PCR amplifications were completed in a reaction volume of 25 μL, which included 1.0 μL template DNA (100 ng/μL), 12.5 μL 2× Taq Master Mix for PAGE (Dye plus), 1.0 μL each primer (10 μM), and 9.5 μL ddH2O. The PCR program was as follows: 94 °C for 5 min; 35 cycles of 94 °C for 30 s, appropriate annealing temperature of 50–60 °C for 30 s, and 72 °C for 1 min; 72 °C for 10 min.
Supplementary information
Additional file 1: Table S1. PCR amplification results of tetraploid Th. elongatum specific molecular markers.
Additional file 2: Table S2. Specific amplification of 1E chromosome markers in wheat-related species.
Additional file 3: Table S3. Stripe rust response at seedling stage and specific amplification of 1E chromosome markers in F2 population.
Acknowledgments
We thank Dr. L Huang, Sichuan Agricultural University, Sichuan, China, for technical guidance in stripe rust resistance screening at seedling stage. We thank Dr. George Fedak, Eastern Cereal and Oilseed Research Center, Ottawa, Canada, for kindly supplying the Trititrigia 8801 material used in this study. We also thank Prof. QZ Jia, Plant Protection Institute of Gansu Academy of Agricultural Sciences, Gansu, China, for kindly supplying the urediniospores of the Pst races.
Abbreviations
- CS
Chinese Spring
- FISH
Fluorescence in situ hybridization
- GBS
Genotyping-by-sequencing
- GISH
Genome in situ hybridization
- MAS
Marker-assisted selection
- PCR
Polymerase chain reaction
- Pst
Puccinia striiformis f. sp. tritici
- SM482
Shumai482
- SM921
Shumai921
- SM969
Shumai969
- SSR
Simple sequence repeats
Author contributions
HYK and YHZ conceived and designed the research. DYL, JWZ, HJL, BWT, and WZ conducted the experiments. LLX, YW, JZ, XF, and LNS participated in the preparation of the reagents and materials. HQZ, JM, and GYC analyzed the data. DYL and HYK wrote the manuscript. All the authors read and approved the manuscript.
Funding
This work was funded by the National Key Research and Development Program of China (2016YFD0102000 and 2017YFD0100905), the National Natural Science Foundation of China (No. 31771781, 31971883), and the Science and Technology Bureau of Sichuan Province. The funding bodies were not involved in the design of the study, data collection, interpretation of data, or in writing the manuscript.
Availability of data and materials
The datasets generated and analyzed during the present study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Daiyan Li and Juwei Zhang contributed equally to this work.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-019-6359-9.
References
- 1.Food and Agriculture Organisation of the United Nations (FAO) Crop prospects and food situation. 2019. [Google Scholar]
- 2.Dubcovsky J, Dvorak J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science. 2007;316:1862–1866. doi: 10.1126/science.1143986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wellings CR. Global status of stripe rust: a review of historical and current threats. Euphytica. 2011;179:129–141. doi: 10.1007/s10681-011-0360-y. [DOI] [Google Scholar]
- 4.Nsabiyera V, Bariana HS, Qureshi N, Wong D, Hayden MJ, Bansal UK. Characterisation and mapping of adult plant stripe rust resistance in wheat accession Aus27284. Theor Appl Genet. 2018;131:1459–1467. doi: 10.1007/s00122-018-3090-x. [DOI] [PubMed] [Google Scholar]
- 5.Chen XM. Epidemiology and control of stripe rust [Puccinia striiformis f. sp. tritici] on wheat. Can J Plant Pathol. 2005;27:314–337. doi: 10.1080/07060660509507230. [DOI] [Google Scholar]
- 6.Mujeeb-Kazi A, Kazi AG, Dundas I, Rasheed A, Ogbonnaya F, Kishii M, Bonnett D, Wang RRC, Xu S, Chen P, Mahmood T, Bux H, Farrakh S. Genetic diversity for wheat improvement as a conduit to food security. Adv Agron. 2013;122:179–257. doi: 10.1016/B978-0-12-417187-9.00004-8. [DOI] [Google Scholar]
- 7.Ceoloni C, Forte P, Kuzmanović L, Tundo S, Moscetti I, De Vita P, Virili ME, D’Ovidio R. Cytogenetic mapping of a major locus for resistance to Fusarium head blight and crown rot of wheat on Thinopyrum elongatum 7EL and its pyramiding with valuable genes from a Th. ponticum homoeologous arm onto bread wheat 7DL. Theor Appl Genet. 2017;130:2005–2024. doi: 10.1007/s00122-017-2939-8. [DOI] [PubMed] [Google Scholar]
- 8.Dvořák J, Chen KC. Phylogenetic relationships between chromosomes of wheat and chromosome 2E of Elytrigia elongata. Can J Genet Cytol. 1984;26:128–132. doi: 10.1139/g84-021. [DOI] [Google Scholar]
- 9.Dvořák J, Edge M, Ross K. On the evolution of the adaptation of Lophopyrum elongatum to growth in saline environments. PNAS. 1988;85:3805–3809. doi: 10.1073/pnas.85.11.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fedak G. Molecular aids for integration of alien chromatin through wide crosses. Genome. 1999;42:584–591. doi: 10.1139/g99-046. [DOI] [Google Scholar]
- 11.Chen SQ, Huang ZF, Dai Y, Qin SW, Gao YY, Zhang LL, Gao Y, Chen JM. The development of 7E chromosome-specific molecular markers for Thinopyrum elongatum based on SLAF-seq technology. PLoS One. 2013;8:e65122. doi: 10.1371/journal.pone.0065122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lou H, Dong L, Zhang K, Wang DW, Zhao M, Li Y, Rong C, Qin H, Zhang A, Dong Z, Wang D. High-throughput mining of E-genome-specific SNPs for characterizing Thinopyrum elongatum introgressions in common wheat. Mol Ecol Resour. 2017;17:1318–1329. doi: 10.1111/1755-0998.12659. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka H, Nabeuchi C, Kurogaki M, Garg M, Saito M, Ishikawa G, Nakamura T, Tsujimoto H. A novel compensating wheat–Thinopyrum elongatum Robertsonian translocation line with a positive effect on flour quality. Breed Sci. 2017;67:509–517. doi: 10.1270/jsbbs.17058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar A, Garg M, Kaur N, Chunduri V, Sharma S, Misser S, Kumar A, Tsujimoto H, Dou QW, Gupta RK. Rapid development and characterization of chromosome specific translocation line of Thinopyrum elongatum with improved dough strength. Front Plant Sci. 2017;8:1593. doi: 10.3389/fpls.2017.01593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai Y, Duan Y, Chi D, Liu H, Huang S, Cao W, Gao Y, Fedak G, Chen J. Chromosome identification by new molecular markers and genomic in situ hybridization in the Triticum–Secale–Thinopyrum trigeneric hybrids. Genome. 2017;60:687–694. doi: 10.1139/gen-2017-0025. [DOI] [PubMed] [Google Scholar]
- 16.Dai Y, Duan Y, Liu H, Chi D, Cao W, Xue A, Gao Y, Fedak G, Chen J. Molecular cytogenetic characterization of two Triticum-Secale-Thinopyrum trigeneric hybrids exhibiting superior resistance to Fusarium head blight, leaf rust, and stem rust race Ug99. Front Plant Sci. 2017;8:797. doi: 10.3389/fpls.2017.00797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li DY, Long D, Li TH, Wu YL, Wang Y, Zeng J, Xu LL, Fan X, Sha LN, Zhang HQ, Zhou YH, Kang HY. Cytogenetics and stripe rust resistance of wheat–thinopyrum elongatum hybrid derivatives. Mol Cytogenet. 2018;11:16. doi: 10.1186/s13039-018-0366-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li DY, Li TH, Wu YL, Zhang XH, Zhu W, Wang Y, Zeng J, Xu LL, Fan X, Sha LN, Zhang HQ, Zhou YH, Kang HY. FISH-based markers enable identification of chromosomes derived from tetraploid Thinopyrum elongatum in hybrid lines. Front Plant Sci. 2018;9:526. doi: 10.3389/fpls.2018.00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu LQ, Luo QL, Teng W, Li B, Li HW, Li YW, Li ZS, Zheng Q. Development of Thinopyrum ponticum- specific molecular markers and FISH probes based on SLAF-seq technology. Planta. 2018;247:1099–1108. doi: 10.1007/s00425-018-2845-6. [DOI] [PubMed] [Google Scholar]
- 20.Zhang JP, Liu WH, Lu YQ, Liu QX, Yang XM, Li XQ, Li LH. A resource of large-scale molecular markers for monitoring Agropyron cristatum chromatin introgression in wheat background based on transcriptome sequences. Sci Rep. 2017;7:11942. doi: 10.1038/s41598-017-12219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li XM, Lee BS, Mammadov AC, Koo BC, Mott IW, Wang RR. CAPS markers specific to Eb, Ee, and R genomes in the tribe Triticeae. Genome. 2007;50:400–411. doi: 10.1139/G07-025. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, Yan ZH, Zheng YL, Liu DC, Dai SF, Zhang LQ, Wei YM. Development of Ee-chromosome specific AFLP and STS molecular marker for Lophopyrum elongatum in Chinese spring wheat background. J Agric Biotechnol. 2008;16:465–473. [Google Scholar]
- 23.Wang RRC, Larson SR, Jensen KB. Analyses of Thinopyrum bessarabicum, T. elongatum, and T. junceum chromosomes using EST-SSR markers. Genome. 2010;53:1083. doi: 10.1139/G10-088. [DOI] [PubMed] [Google Scholar]
- 24.Xu GH, Su WY, Shu YJ, Cong WW, Wu L, Guo CH. RAPD and ISSR-assisted identification and development of three new SCAR markers specific for the Thinopyrum elongatum E (Poaceae) genome. Genet Mol Res. 2012;11:1741. doi: 10.4238/2012.June.29.7. [DOI] [PubMed] [Google Scholar]
- 25.Chen SQ, Qin SW, Huang ZF, Yi D, Zhang LL, Gao YY, Yong G. Development of specific molecular markers for Thinopyrum elongatum chromosome using SLAF-seq technique. Acta Agron Sin. 2013;39:727. doi: 10.3724/SP.J.1006.2013.00727. [DOI] [Google Scholar]
- 26.Kantarski T, Larson S, Zhang X, DeHaan L, Borevitz J, Anderson J, Poland J. Development of the first consensus genetic map of intermediate wheatgrass (Thinopyrum intermedium) using genotyping-by-sequencing. Theor Appl Genet. 2017;130:137–150. doi: 10.1007/s00122-016-2799-7. [DOI] [PubMed] [Google Scholar]
- 27.Kwiatek M, Belter J, Majka M, Wiśniewska H. Allocation of the S-genome chromosomes of Aegilops variabilis Eig. Carrying powdery mildew resistance in triticale (× Triticosecale Wittmack) Protoplasma. 2016;253:329–343. doi: 10.1007/s00709-015-0813-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dvořák J, Sosulski FW. Effects of additions and substitutions of Agropyron elongatum chromosomes on quantitative characters in wheat. Can J Genet Cytol. 1974;16:627–637. doi: 10.1139/g74-069. [DOI] [Google Scholar]
- 29.Zheng Q, Li B, Mu SM, Zhou HP, Li ZS. Physical mapping of the blue-grained gene(s) from Thinopyrum ponticum by GISH and FISH in a set of translocation lines with different seed colors in wheat. Genome. 2006;49:1109–1114. doi: 10.1139/g06-073. [DOI] [PubMed] [Google Scholar]
- 30.Fu SL, Lv ZL, Qi B, Guo X, Li J, Liu B, Han FP. Molecular cytogenetic characterization of wheat--Thinopyrum elongatum addition, substitution and translocation lines with a novel source of resistance to wheat Fusarium head blight. J Genet Genomics. 2012;39:103–110. doi: 10.1016/j.jgg.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 31.Wang XH, Chen CH, Wang CY, Mo QB, Li H, Ji WQ. Molecular cytogenetic identification of a wheat– Thinopyrum ponticum substitution line with powdery mildew resistance. J Triticeae Crops. 2015;35:596–602. [Google Scholar]
- 32.Zhu MC, Wang MY, Chen MC, Wang C, Zhang MA, Peng MN, Wang Y, Zhang H, Liu X, Ji W. Molecular cytogenetic identification of wheat-Thinopyrum ponticum substitution line with stripe rust resistance. Genome. 2017;60:860–867. doi: 10.1139/gen-2017-0099. [DOI] [PubMed] [Google Scholar]
- 33.Liu Z, Li D, Zhang X. Genetic relationships among five basic genomes St, E, a, B and D in Triticeae revealed by genomic southern and in situ hybridization. J Integr Plant Biol. 2007;49:1080–1086. doi: 10.1111/j.1672-9072.2007.00462.x. [DOI] [Google Scholar]
- 34.Hu LJ, Liu C, Zeng ZX, Li GR, Song XJ, Yang ZJ. Genomic rearrangement between wheat and Thinopyrum elongatum revealed by mapped functional molecular markers. Genes Genomics. 2012;34:67–75. doi: 10.1007/s13258-011-0153-7. [DOI] [Google Scholar]
- 35.Liu J, Chang Z, Zhang X, Yang Z, Li X, Jia J, Zhan H, Guo H, Wang J. Putative Thinopyrum intermedium-derived stripe rust resistance gene Yr50 maps on wheat chromosome arm 4BL. Theor Appl Genet. 2013;126:265–274. doi: 10.1007/s00122-012-1979-3. [DOI] [PubMed] [Google Scholar]
- 36.Hou LY, Jia JQ, Zhang XJ, Li X, Yang ZJ, Ma J, Guo HJ, Zhan HX, Qiao LY, Chang ZJ. Molecular mapping of the stripe rust resistance gene Yr69 on wheat chromosome 2AS. Plant Dis. 2016;100:1717–1724. doi: 10.1094/PDIS-05-15-0555-RE. [DOI] [PubMed] [Google Scholar]
- 37.You MS, Li BY, Tian ZH, Tang ZH, Liu SB, Liu GT. Development of specific SSR marker for E-genome of Thinopyrum spp. using wheat microsatellites. J Agric Biotechnol. 2003;1:143–148. [Google Scholar]
- 38.Liu SB, Jia XZ, Wang HG, Kong LR, Zhou RH. Special chromosome markers for e genome and DNA polymorphism between Agropyron elongatum (2n=14) and common wheat detected by RAPD markers. Acta Agron Sin. 1998;24:687–690. [Google Scholar]
- 39.Ge JY, Chen SQ, Gao YY, Gao Y, Huang ZF, Chen JM. Development of genome- specific molecular markers for Lophopyrum elongatum based on suppression subtractive hybridization. Acta Agron Sin. 2012;38:1818–1826. doi: 10.3724/SP.J.1006.2012.01818. [DOI] [Google Scholar]
- 40.Chen SQ, Gao Y, Zhu X, Zhang C, Cao WG, Fedak G, He ZT, Chen XL, Chen JM. Development of E-chromosome specific molecular markers for Thinopyrum elongatum in a wheat background. Crop Sci. 2015;55:2777–2785. doi: 10.2135/cropsci2014.08.0539. [DOI] [Google Scholar]
- 41.Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One. 2011;6:e19379. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poland JA, Brown PJ, Sorrells ME, Jannink JL. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One. 2012;7:e32253. doi: 10.1371/journal.pone.0032253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu SB, Jia JZ, Wang HG, Kong LR, Zhou RH. Identification of homoeology between the Elytrigia elongatum (2n=14, EE) and wheat chromosomes using biochemical and molecular markers. Acta Agron Sin. 1999;26:37–4227. [Google Scholar]
- 44.Dvořák J. Genome relationships among Elytrigia (=Agropyron) elongata, E. stipifolia, “ E. elongata 4x,” E. caespitosa, E. intermedia, and “E. elongata 10x”. Can J Genet Cytol. 1981;23:481–492. doi: 10.1139/g81-053. [DOI] [Google Scholar]
- 45.Liu ZW, Wang RRC. Genome constitutions of Thinopyrum curvifolium, T. scirpeum, T. distichum, and T. junceum (Triticeae: Gramineae) Genome. 1993;36:641–651. doi: 10.1139/g93-085. [DOI] [PubMed] [Google Scholar]
- 46.Wang RRC, Bothmer RV, Dvorak J, Fedak G, Linde-Laursen I, Muramatsu M. Genome symbols in the Triticeae (Poaceae). In: Wang RRC, Jensen KB, Jaussi C, editors. Proc 2nd Intern Triticeae Symp. Logan: The Utah State University Press; 1994. pp. 29–34.
- 47.Komuro S, Endo R, Shikata K, Kato A. Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome. 2013;56:131–137. doi: 10.1139/gen-2013-0003. [DOI] [PubMed] [Google Scholar]
- 48.Han FP, Lamb JC, Birchler JA. High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. PNAS. 2006;103:3238–3243. doi: 10.1073/pnas.0509650103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 2006;1:2320. doi: 10.1038/nprot.2006.384. [DOI] [PubMed] [Google Scholar]
- 50.Han FP, Gao Z, Birchler JA. Reactivation of an inactive centromere reveals epigenetic and structural components for centromere specification in maize. Plant Cell. 2009;21:1929–1939. doi: 10.1105/tpc.109.066662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McIntyre C, Pereira S, Moran L, Appels R. New Secale cereale (rye) DNA derivatives for the detection of rye chromosome segments in wheat. Genome. 1990;33:635–640. doi: 10.1139/g90-094. [DOI] [PubMed] [Google Scholar]
- 52.Somers DJ, Isaac P, Edwards K. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.) Theor Appl Genet. 2004;109(6):1105–1114. doi: 10.1007/s00122-004-1740-7. [DOI] [PubMed] [Google Scholar]
- 53.Carter AH, Chen XM, Garland-Campbell K, Kidwell KK. Identifying QTL for high-temperature adult-plant resistance to stripe rust (Puccinia striiformis f. sp. tritici) in the spring wheat (Triticum aestivum L.) cultivar ‘Louise’. Theor Appl Genet. 2009;119:1119–1128. doi: 10.1007/s00122-009-1114-2. [DOI] [PubMed] [Google Scholar]
- 54.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. PCR amplification results of tetraploid Th. elongatum specific molecular markers.
Additional file 2: Table S2. Specific amplification of 1E chromosome markers in wheat-related species.
Additional file 3: Table S3. Stripe rust response at seedling stage and specific amplification of 1E chromosome markers in F2 population.
Data Availability Statement
The datasets generated and analyzed during the present study are available from the corresponding author on reasonable request.