Abstract
Ewing sarcoma (EwS) is an aggressive pediatric tumor treated with intensive cytotoxic chemotherapies. Overall survival for metastatic or relapsed disease is only 20-30%. Metformin has long been an attractive therapeutic option for EwS, but hypoxia limits its efficacy. Through a systematic integration of drug combination screening, bioinformatics analyses, functional and in vivo studies, and correlation with clinical outcome, we identified another known drug, imatinib that could augment the in vivo anti-tumor capacity of metformin by attenuating tumor hypoxic response. This drug combination regimen widely suppressed multiple dominant mechanisms in EwS genesis, growth, and metastasis, including key EWS-FLI1 downstream targets that converge into the PI3K/AKT/mTOR signaling pathway. In addition, the combination significantly enhanced inhibition on tumor cell proliferation by standard EwS chemotherapy drugs, including cyclophosphamide and ifosfamide. This suggests a potential clinical benefit of the metformin/imatinib combination by allowing the reduction in dose intensity of standard chemotherapy without compromising survival outcome and represents a potential faster track application for EwS patients.
Keywords: Ewing sarcoma, metformin, imatinib, drug combinations, drug repositioning
Introduction
Ewing sarcoma (EwS) is an aggressive bone or soft-tissue tumor that mainly affects children, adolescents, and young adults. With the strategy of dose intensification of multi-agent cytotoxic chemotherapies, the 5-year overall survival rate for localized disease has increased to 70-75% but not for metastatic or relapsed disease which remains at only 20-30% [1]. Severe toxicity and high rates of life-threatening events such as secondary malignancies are associated with high dose multi-drug chemotherapy [1]. Even in the best situation of localized tumors, there is a need for complementary approaches to help minimize treatment-related toxicity. Unfortunately, the development of new treatment for EwS proves to be challenging. Only a limited number of new treatments for EwS have been explored in the past decade: less than 100 studies for EwS, in comparison with over 4,000 studies for breast cancer and 3,000 studies for lung cancer registered in ClinicalTrials.gov.
The insulin-like growth factor (IGF) system is recognized to be a critical mechanism for EwS cell survival and malignant behavior [1]. EWS-FLI1, the genetic hallmark and primary oncogenic driver of the majority of EwS, is only capable of driving transformation in the presence of IGF1 receptor (IGF-1R) [2] and is known to interfere with the IGF system either by blocking IGFBP-3 expression [3], a molecule that inhibits IGF signaling by sequestering IGF1 and preventing its interaction with IGF-1R, or by inducing IGF1 expression. Excellent clinical responses to anti-IGF-1R monoclonal antibodies were achieved in 10-14% of EwS patients [1]. However, IGF-1R and insulin receptor (IR) are expressed in virtually all EwS tumors [4]. IGFs that are constantly produced by EwS cells can also bind to IR, albeit with a lower affinity than IGF-1R. Cancer cells resistant to the anti-IGF-1R therapies overexpress IR to sustain their growth by autocrine loops mediated by IGFs and/or insulin [5, 6]. Therefore, an effective treatment strategy for EwS would need to target both the IGF system and IR signaling. Metformin, an anti-hyperglycemic agent, has been reported to down-regulate both the IGF system and IR signaling pathways in vivo [7, 8]. It has been widely used clinically since 1958 and gained much attention recently in cancer research. For EwS, metformin has a remarkable anti-proliferative effect on both chemo-sensitive and chemo-resistant tumor cells, and even cells resistant to anti-IGF-1R therapies [9]. Metformin was also shown to cooperate with different chemotherapeutic agents to increase their anti-cancer activity [10]. Unfortunately, its therapeutic efficacy is impeded due to hypoxia [9], a major factor found in the EwS tumor microenvironment [11–13]. EwS cells are reported to adapt to hypoxia by redefining their transcriptome and acquiring a distinct hypoxic phenotype [11, 14]. We hypothesize that other known drugs could be combined with metformin to kill EwS cells under hypoxic condition and restore the in vivo efficacy of metformin. We therefore performed a drug combination screen under hypoxic condition (1% O2) to identify combinations of existing EwS-sensitive drugs with metformin that could overcome treatment resistance caused by hypoxia.
We assembled a pool of 57 previously reported anti-EwS drugs and compiled a customized drug library. Then, we performed EwS cell viability assay with 96 well-plate combining metformin with the 57 anti-EwS drugs individually. The anti-proliferation effect of metformin was significantly enhanced by seven of the 57 drugs. In particular, imatinib stood out as the most potent and showed a strong synergy with metformin in inhibiting EwS cell growth on multiple EwS cell lines. Importantly, the in vitro synergistic effect of the combination of metformin and imatinib remained evident when clinically relevant plasma concentrations of each drug were applied. Furthermore, the combination significantly enhanced in vitro cell-killing efficacy of two standard EwS chemotherapeutic drugs, cyclophosphamide and ifosfamide, suggesting a potential benefit of lower dosage and better outcomes. In vivo studies on two EwS xenograft mouse models confirmed the superior anti-tumor efficacy of the combination.
Mechanistic exploration revealed that imatinib strikingly reversed the hypoxia-specific transcriptional signature in EwS cells, and the metformin-imatinib drug combination significantly induced cell cycle arrest and apoptosis through regulating a network of sixty-one signaling molecules that inhibited the activity of PI3K-AKT-mTOR signaling pathway. The genes regulated by the drug combination highly correlate with poor clinical outcome of clinical EwS cohorts, thus supporting the potential efficacy of this combination regimen for EwS.
Materials and Methods
Cell lines, cell culture and reagents
EwS cell lines TC71, TC32, and A673 used in this study have been previously described[15]. Stable luciferase reporter cell lines for TC71-Luc and TC32-Luc were created using a modified pGL4.32 reporter (Promega, Madison, WI), which contains the luc2P reporter and hygromycin resistance. Successful cloning was verified by complete sequencing and cells were selected in 125μg/mL hygromycin B for 4 weeks. Non-malignant human bone cell line, hFOB, was purchased from ATCC. Cell line authentication was performed with short-tandem repeat profiling and passaged in our laboratory for less than six months after receipt. All cell lines were tested negative for mycoplasma. Metformin was purchased from Sigma Aldrich (St. Louis, MO), dissolved as a 100 mM stock solution in PBS. Imatinib was purchased from Selleckchem (Houston, TX, US), dissolved as a 10 mM stock solution in DMSO. The stock solutions were frozen in aliquots for use in the in vitro and in vivo experiments.
Drug library
A customized library consisting of 57 clinical drugs (passed phase I trials or above) was used for the screening (Selleckchem, Houston, TX) (Table S1). Seventeen drugs are previously defined as “sensitive” on EwS or osteosarcoma cell lines in the Genomics of Drug Sensitivity in Cancer database [16]. Twenty-seven drugs showed significant anti-EwS effects from the MIPE4.0 small molecule library screening on multiple EwS cell lines [17], 13 drugs have published evidence for EwS treatment in PubMed. The library drugs are all placed in one 96-well plate with 10mM stock concentration in DMSO for each drug, 3 wells as negative controls (DMSO), and no drugs in all edge wells.
In vitro cell-based screening and combination index
TC71 cell line was used for drug screening. It was derived from a tumor recurrent after chemotherapy in a 22-year-old EwS patient, and was previously characterized as one of the most chemo-resistant EwS cell lines [18]. When 5 × 103 cells were grown to 80% confluence in 96-well plates, they were gently washed with PBS twice. Then medium was replaced with DMEM containing 2.5mM library drugs and 5mM of metformin, or the library drugs only. 5mM is about the IC25 of metformin on TC71 cell line under hypoxia condition (Fig. S1A). The cells were then subjected to hypoxia in a hypoxia incubator (Sanyo, Japan) containing a gas mixture of 94% N2, 5% CO2, and 1% O2. Cell viability was measured at 48h by the highly sensitive Cell Counting kit-8 (CCK-8; Dojindo Molecular Technology Inc.). Three rounds of independent screening were performed using the same generation and same passage number of TC71 cells. For drug combination effect tested under non-constant ratios on multiple EwS cell lines, as suggested [19], we kept imatinib as 2.5uM and varying the metformin dosage, to test for CI values under various ratios. The Log(CI) scatter plot was calculated by ComboSyn software version 2.1 (ComboSyn Incorporated.) according to its user guide.
RNA isolation, RNA-Seq library preparation and sequencing
TC71 cells treated for 48 hours with 5mM metformin alone, or 2.5μM imatinib alone or the combination were harvested for RNA-seq analysis at BGI-Shenzhen (Guangdong, China), as described previously [20]. For RNAseq data preprocessing and normalization, raw reads in fastq files were mapped to human reference genome (hg19) using Tophat2 [21–23]. Gene FPKM values were generated using cufflinks[23]. Differential analysis under different treatments were done by cuffdiff [23].
Reverse phase protein array (RPPA) analysis
Cell lysates from treated TC71 cells were used for RPPA analysis at the University of Texas MD Anderson Cancer Center’s RPPA Core. The assay procedure and data normalization are described in the core website (https://www.mdanderson.org/research/research-resources/core-facilities/functional-proteomics-rppa-core.html).
Pathway analysis
The ConsensusPathDB database (http://ConsensusPathDB.org)[24] and Ingenuity Pathway Analysis (IPA) (Qiagen, US) were used to define molecular functional categories and pathways of deregulated genes/proteins (for both RNA-seq and RPPA data) under metformin and imatinib combination treatment.
Western blot
Cell or tissue lysates and immunoblotting analysis were performed as described [25]. The antibodies are described in Supplementary Materials. Densitometry analysis was performed using ImageJ software (version 1.8.0, NIH).
In vivo animal experiments
Animal procedures were conducted under the approval of Institutional Animal Care and Use Committee (IACUC) of Houston Methodist Research Institute. Both male and female NOD-SCID mice (6-8 weeks-of-age; Charles River Laboratories, Boston, MA) were anesthetized with isoflurane/O2 and 2×106 viable TC71-Luc or TC32-Luc single cells were injected into the gastrocnemius muscle. Tumor growth was monitored by two independent methods; one is caliper measurement on tumor diameter, and the other is repeated non-invasive bioluminescence imaging (BLI) twice per week using an IVIS 200 system (Xenogen). Animals were randomly assigned into four treatment groups of 10 mice per group when tumors reached a diameter of ~8mm (day 8 after tumor cell injection in TC71 model and day 12 after tumor cell injection in TC32 model): metformin only group (150 mg/kg, twice a day, by oral gavage), imatinib only group (50 mg/kg, twice daily, by oral gavage), metformin plus imatinib combination group (same regimen as single treatment), and vehicle control (animals receiving an equivalent volume of sterile normal saline by oral gavage). The maximum treatment duration is 22 days. Tumor growth for mice that had already reached diameter of 2cm were carried forward until all mice in the group reached endpoint or the experiment was terminated at day 29. Lung, contralateral tibia, and spine were dissected for ex vivo bioluminescence imaging to examine metastasis, and primary tumors were harvested for histology examination at the time the mice were sacrificed.
Microarray data analysis
GSE19197 dataset was used to analyze the hypoxia gene expression of EwS cells [14]. Six independent clinical datasets of EwS or sarcoma cohorts: Savola Ewing sarcoma (GSE17679, n=88 EwSs and n=18 tumor-adjacent normal skeletal muscle) [26], Ohali Ewing sarcoma (n=21 EwSs) [27], Ferreira Ewing sarcoma (GSE8303, n=27 EwSs) [28], Chibon sarcomas (GSE21050, n=310), and TCGA sarcoma (n=245), for which publicly available microarray and clinical data (overall survival time, metastatic-free survival time, event-free survival time, or metastatic status) were used for gene expression correlation analysis. Postel-Vinay EwS clinical cohort (GSE34620, n=117 EwSs) [29] was used to compare the expression of drug combination regulated-gene set with the published EwS prognosis gene signature [27].
Statistical analysis
All experimental data presented are representative of at least two independent experiments performed in triplicate. Data are presented as mean±s.e.m. Statistical significance between two groups was analyzed by two-tailed Student’s t-test. Experiments with more than two groups were analyzed with one-way analysis of variance (ANOVA) and Bonferroni’s post-hoc test. Tumor burden was compared between groups using a Wilcoxon rank-sum test at serial time points. Survival proportions were assessed by Kaplan-Meier method with log-rank test. A p value of less than 0.05 was considered significant. All data were analyzed by using GraphPad Prism (GraphPad Software, USA).
Result
Combination of metformin and imatinib exhibits potent and synergistic effect against EwS cell growth under hypoxic condition
Our goal was to screen for combinatory drug hits that can sensitize EwS cells to metformin under hypoxic condition (Fig. 1A). The drug screening was conducted under 1% oxygen condition, a generally accepted oxygen level in hypoxic tumor tissues [30]. Under this condition, metformin was much less effective in inhibiting proliferation of the EwS cells. Specifically, in TC71 cell line, the IC50 increased to 13.74 mM under hypoxia from 7.78 mM under normoxic condition (Fig. S1A). Under the screening dosage, metformin (5mM), could only inhibit ~25% of TC71 and TC32 cells under hypoxia, while these cells could be killed more than 50% under normoxia (Fig. S1B). The screening results were consistent and reproducible from three rounds of independent experiments, which yielded a high correlation coefficient (median R2 = 0.86; range = 0.80 – 0.90) (Fig. 1B).
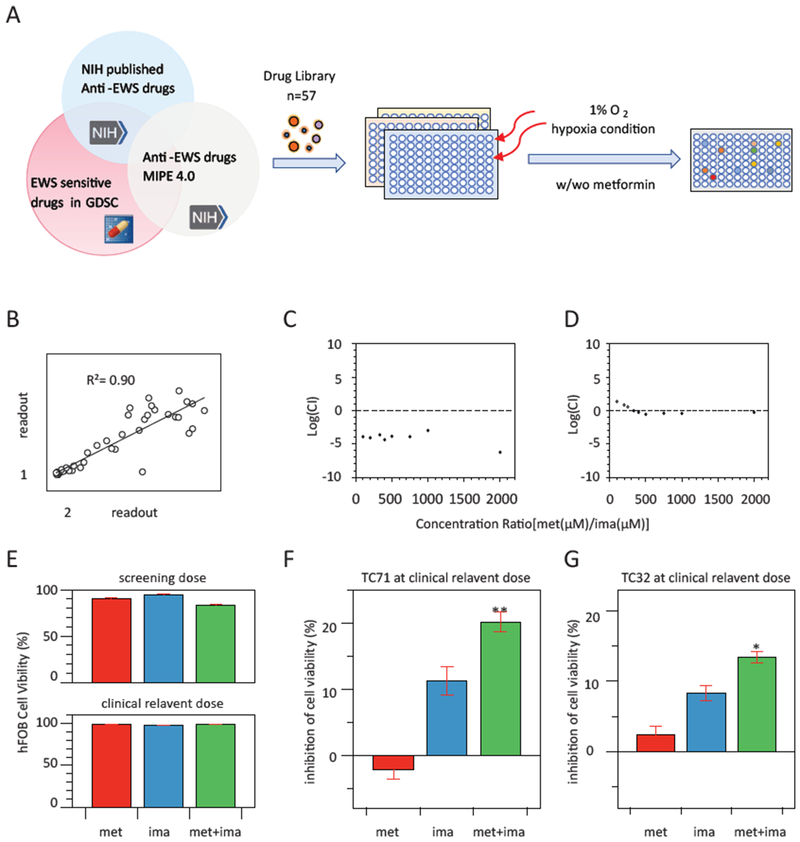
Figure 1. Combination of metformin and imatinib exhibits potent and synergistic effect against EwS cell growth under hypoxia condition.
A. Workflow of the drug combination screening on Ewing sarcoma cell line TC71 under hypoxia condition for 48 hours. B. Good correlation between two out of three rounds of independent screening experiments. C-D. Log(CI) of various metformin and imatinib combination (met/ima) dose ratios on the viability of TC71 (C) and TC32 (D), Log(CI) < 0 indicates synergy. E. Combination of metformin (10mM) and imatinib (5μM) had no oblivious cell toxicity on non-malignant human bone cell line hFOB. F and G. Combination of clinical relevant doses of metformin (10μM) and imatnib (1μM) significantly inhibited the viability of TC71 and TC32 cells at 72 hours. ** p<0.01, *p<0.05, vs metformin alone (met) and imatinib alone (ima). Experiments were triplicated, and mean±s.e.m. was presented.
As shown in Table 1, seven out of the 57 drugs were able to significantly enhance the anti-proliferation activity of metformin (p<0.05 vs. single drug treatment), with 1.16 – 2.49 fold decrease of the cell viability. Combination of imatinib with metformin was most potent in reducing cell viability in all three screenings: 34.33±1.14 % viable cells in the combination group vs. 82.91±5.06 % in the imatinib only group. Imatinib was previously reported as being partially active at high concentrations (>23.4 μM) against TC71 cell growth (https://tripod.nih.gov/matrix-client/). Thus, our screening result that imatinib at 2.5μM synergizes with metformin at an IC25 dose achieved a >65% inhibition on TC71 cell growth is striking, indicating a unique mechanism of the drug combination.
Table 1.
Seven out of the 57 drugs significantly boost the anti-proliferation action of metformin in TC-71 EwS cells
| Name | Targets | Target pathway | log2(FC) | p value |
|---|---|---|---|---|
| Imatinib | ABL, KIT, PDGFR | Protein Tyrosine Kinase/RTK | 1.320 | 0.003 |
| Dacarbazine | POLA2, MMP9, DNA/RNA synthesis | Cell Cycle/DNA Damage | 0.711 | 0.039 |
| Sunitinib | PDGFR, KIT, VEGFR, FLT3, RET, CSF1R | Protein Tyrosine Kinase/RTK | 0.450 | 0.001 |
| Isotretinoin | RAR/RXR | Metabolism | 0.352 | 0.029 |
| Hydroxyurea | RRM2, RRM1, NFKB1 | DNA Damage/DNA Repair | 0.236 | 0.010 |
| Lenalidomide | CRBN | others | 0.227 | 0.009 |
| Clafen | DNA alkylator/crosslinker | Cell Cycle/DNA Damage | 0.215 | 0.005 |
Synergy between metformin and imatinib on inhibiting the growth of TC71 cell line was confirmed by the combination index (CI) method that provides a quantitative definition for additive effect (CI=1, Log(CI)=0), synergism (CI<1, Log(CI)<0), and antagonism (CI>1, Log(CI)>0) in drug combination. As shown in Fig. 1C, metformin and imatinib performed synergistically under all tested combination ratios, with the log(CI)<0. Additionally, the synergistic effects between these two drugs were also examined on two other EwS cell lines, i.e., A673 and TC32. In TC32 cells, single treatment with 5mM metformin or 2.5μM imatinib resulted in 31.47±3.02% and 2.78±0.12% inhibition on cell viability, respectively but increased to 47.64±5.11% in the combination treatment, CI is 0.9 (log CI= −0.05) (Fig. 1D). In A673 cells, treatment with either 5mM metformin or 2.5μM imatinib alone resulted in 18.25±1.02% and 8.57±0.62% inhibition on cell viability, but when metformin and imatinib were used together, the combination index is 0.7 (log CI= −0.15), suggesting a strong synergism. On the other hand, in the non-malignant human bone cell line hFOB, metformin alone or in imatinib or combination showed no significant cytotoxicity under the tested dosage or clinically relevant dose (Fig. 1F). All these findings indicate that the synergistic anti-tumor cell growth effect of the combined metformin and imatinib treatment is tumor cell-specific.
The systemic plasma concentration of metformin is 10–40μM in human under the maximal approved total daily dose for treatment of diabetes mellitus at 2.5g (35mg/kg body weight) [31]. For imatinib, it is 1.7μM in plasma with standard-dose imatinib in chronic myeloid leukemia patients [32]. When applying the combination of 10μM metformin and 1μM imatinib, synergistic effects were still observed in inhibiting the viability of TC71 and TC32 cells (Fig. 1G–H), and no toxicity in hFOB as mentioned above.
Imatinib suppresses metformin-induced HIFl-α stabilization and transcriptional activation of hypoxia-responsive genes in EwS cells
We noticed that under the drug screening condition, metformin alone down-regulated the insulin and IGF1 signaling pathways in the TC71 cells (Table S2), however, concurrent imatinib treatment did not further potentiate the effects, suggesting other mechanisms underlying the synergy.
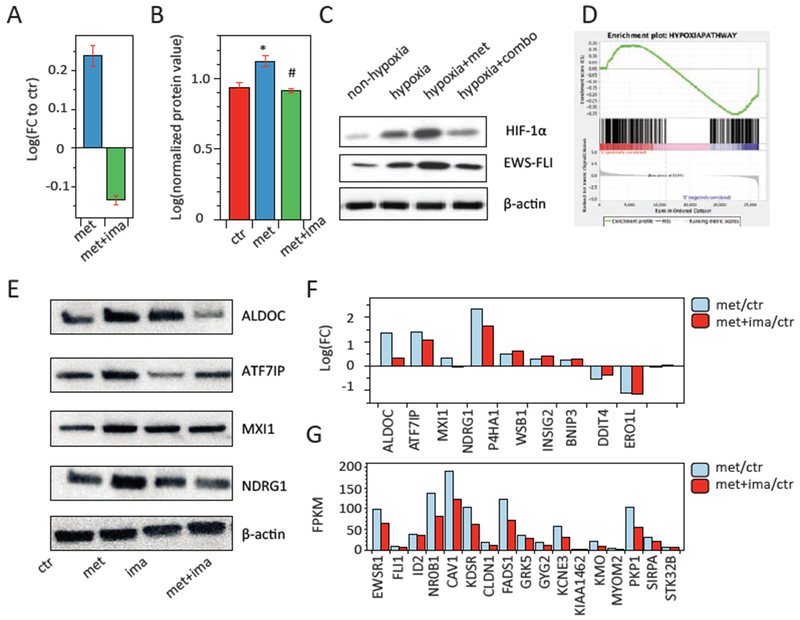
As a central adaptive response to tumor hypoxia, the transcriptional activity of hypoxia-inducible factor 1-α (HIF1-α) is stabilized and dramatically increased at the protein level, and HIF1-α plays key roles in EwS metastasis and chemo-resistance [33]. Upon metformin treatment, we examined a further increase of HIFl-α expression at both transcriptional and protein levels under hypoxia, and the elevation was completely suppressed by concurrent imatinib treatment (Fig. 2A–C). Furthermore, among the ten most well conserved and universal hypoxia targets [34], 7 of them were up-regulated by metformin in TC7l cells, and 4 out of the 7 were variably attenuated by imatinib (Fig. 2E, F). In addition, approximately 1-1.5% of the genome in tumor cells is transcriptionally responsive to hypoxia, and previous studies have identified a EwS-specific hypoxia gene signature consisting of 352 genes [14]. By performing a gene set enrichment analysis (GSEA), we found that TC71 cells treated with imatinib showed an inverse correlation with the hypoxia gene profile (p<0.01) (Fig. 2D) and the reversal of key genes were even verified at protein level (Fig. S1C).
Figure 2. Imatinib suppresses metformin-induced hypoxia response in EwS cells.
A and B. Metformin alone (met) up-regulated the expressions of Hif-1α mRNA (A) by RNA-seq analysis and HIF-1α protein (B) by RPPA analysis on TC71 cells; cocurrent metformin and imatinib treatment (met+ima) abrogated the effects. * p<0.05, vs. control; # p<0.05, vs. met. C. Representative western blot analysis of HIF-1α and EWS-FLI under hypoxia and metformin alone or combination treatment. Experiments were triplicated. D. Gene set enrichment analysis (GSEA) of the EwS-specific hypoxia gene signature consisting of 352 genes (12) was reversed by imatinib treatment (p=0.015). E-F. Expression of important hypoxia related genes upon metformin alone or combination treatment by western blot analysis (E) and RNA-seq analysis (F). G. RNA-seq FPKM values of the EWS-FLI downstream targets upon metformin alone or combination treatment. In panels C and E, western blots images are cropped to show the protein of interest, and all blots were performed under the same experimental conditions.
HIF1-α knockout was conducted in TC71 cells using a lentivirus-mediated CRISPR/Cas system with small guide RNA-721 as previously reported [35]. The growth characteristics of the TC71-HIF1α-KO cells under the hypoxic condition was analyzed, cell proliferation was completely suppressed at 48hr post infection in comparing with the control group. When treated with metformin alone at 5mM or metformin-imatinib combination, nearly all cells were dead at 48 hr and no difference between the two groups was observed (data not shown). These results indicate the importance of HIF1-α in EwS tumorigenesis and treatment response to metformin that is concordance with the previous publication [9].
The EWS-FLI1 protein expression that characterizes EwS is up-regulated by hypoxia in a HIF-1α-dependent manner [14](Fig. 2C). The concurrent treatment significantly inhibited the EWS-FLI1 expression (Fig. 2C) and consistently down-regulated the transcription of twenty-one out of the 28 EWS-FLI1 downstream target genes [1] (Fig. 2G), including a 32.8% inhibition on NROB1 and 28.9% inhibition on CAV1. Taken together, our experimental data demonstrate that metformin augments the cellular hypoxic response in EwS cells, limiting its therapeutic effects, and this situation can be attenuated by concurrent imatinib treatment.
Combination of metformin and imatinib induces G1/S and G2/M cell cycle arrest through modulating a signaling network of 61 genes/proteins
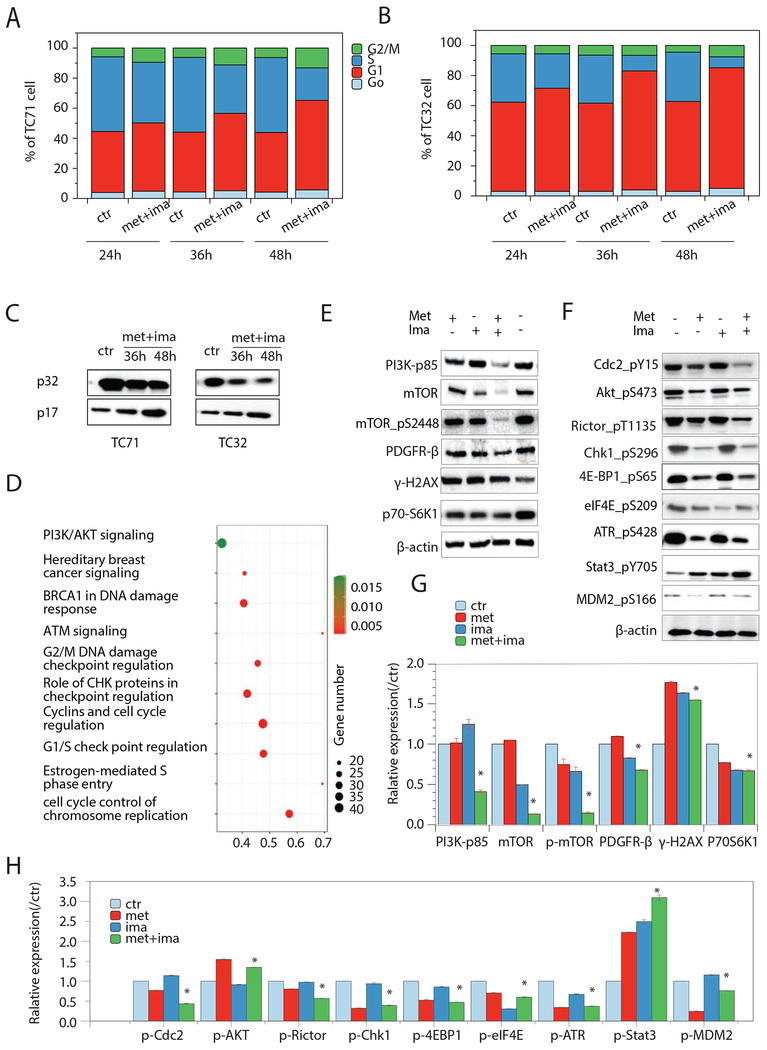
To reveal whether the cell inhibition effect of the combination regimen was cytotoxic or cytostatic, we performed flow cytometry-based apoptosis and cell cycle analyses. As shown in Fig. 3A, a significant increase of cell cycle arrest at G1/S and G2/M (p<0.05) was observed starting 24 hr post-combination treatment in TC71 cells, and further increased until 48 hr. As single agent, metformin at 5μM only moderately induced G1/S arrest on TC71 cells at 48 hours (34.7±4% to 39.2±4%, p>0.05). Imatinib at concentrations greater than 10μM was able to induce significant apoptosis between 36 and 72 hours after treatment, however, at 2.5μM the cell cycle progression was not significantly affected (data not shown). Similarly, the combination treatment induced G1/S and G2/M cell cycle arrest and a late onset apoptosis on TC32 cells (p<0.05) (Fig. 3B). In both TC71 and TC32 cells, we were able to demonstrate caspase activation concomitant with the onset of apoptosis. In controls, caspase-3 was expressed predominantly in its inactive 32-kDa procaspase state (p32). Conversion of p32 to the active 17-kDa form of caspase-3 (p17) was observed after treatment with the drug combination for 36 and 48 hr (Fig. 3C). These results suggested that cell apoptosis was a consequence following their G1/S and G2/M cell cycle arrest.
Figure 3. Combination of metformin and imatinib induces G1/S and G2/M cell cycle arrest and apoptosis in EwS cells.
A and B. Cell cycle analysis of 24h-48h combination treatment on TC71 and TC32 cells. C. Representative immunoblot of p32 and p17 expression in TC71 and TC32 cells upon cimbination treatment for 36h and 48h. D. IPA enriched pathways of the differentially expressed genes in the combination regimen treated TC71 cells. E. Representative immunoblots of RPPA analysis identified proteins in TC71 cells. Experiments were triplicated. In panels C and E, western blots images are cropped to show the protein of interest, and all blots were performed under the same experimental conditions.
Since the combination treatment mainly displays a cytostatic effect, we further added the clinical standard chemotherapeutic drugs for EwS treatment into the regimen. With less than 5% inhibition rate of cyclophosphamide (cyc) or ifosfamide (ifo) alone at 10μM, which is 11 or 470 fold lower than their IC50 doses on TC71 cell line (the IC50 for CTX is 114.8μM [16] and IFOS is 4.7mM), the metformin-imatinib combination at clinically relevance doses synergistically increased their cell inhibition to 30-50% on both TC71 and TC32 EwS cells (Fig. S1D). These results indicate the potential benefit of the metformin-imatinib combination in lowering the dosage of toxic chemotherapeutics in EwS patients.
RNA-seq analysis of the TC71 cells identified ~4,000 differentially regulated genes (log2 fold change>|1.5| and p<0.05) by the combination treatment for 48 hr, and cell cycle regulation dominated the top ten enriched pathways (Fig. 3D), i.e., cell cycle control of chromosome replication (p<0.001), estrogen-mediated s-phase entry (p<0.001), G1/S check point regulation (p<0.001), cyclins and cell cycle regulation (p<0.001), role of CHK proteins in checkpoint regulation (p<0.001), G2/M DNA damage checkpoint regulation (p<0.001), ATM signaling (p<0.001), BRCA1 in DNA damage response (p<0.001), hereditary breast cancer signaling (p<0.001), and PI3K/AKT signaling (p=0.02).
RPPA analysis for the 92 significantly modulated proteins (p<0.05) of the treated TC71 cells also indicate cell cycle (p=0.004) and G1 to S cell cycle control (p=0.007) as the top enriched pathways and cell cycle process (p=0.002) as the enriched top gene ontology (GO). Others include mTOR signaling pathway (p=0.007) and skeletal muscle hypertrophy regulated via AKT/mTOR pathway (p=0.009), GOs: 0044237/cellular metabolic process (p=0.004) and 0009058/biosynthetic process (p=0.005). Fifteen key proteins involved in these pathways are validated by immunoblotting as represented and quantified in Fig. 3E, G and Fig. 3F, H. From the RPPA analysis, twenty-seven of the 92 significantly changed proteins were examined at the phosphorylation level. Functional annotation of this protein set indicated PI3K-AKT signaling pathways (p=0.002), mTOR signaling pathways (p=0.0009), VEGFA-VEGFR2 signaling pathways (p=0.0001), JAK-STAT signaling pathways (p=0.0006), insulin signaling (p=0.001), and KIT receptor signaling (p=0.008).
Overall, a list of sixty-one genes and proteins were derived to represent the molecular effects of the drug combination on EwS cells, from which 48 genes/proteins showed consistent changes on both transcription and protein expression level and 13 proteins showed significant changes on the protein phosphorylation level (Table S3). These data not only provided molecular evidence supporting the effects of the combination on impairing cell cycle regulation and subsequently leading to tumor cell apoptosis, but also pointed out possible signaling pathways underlying the effects.
Combination of metformin and imatinib inhibits tumor metastasis and growth in EwS xenograft models
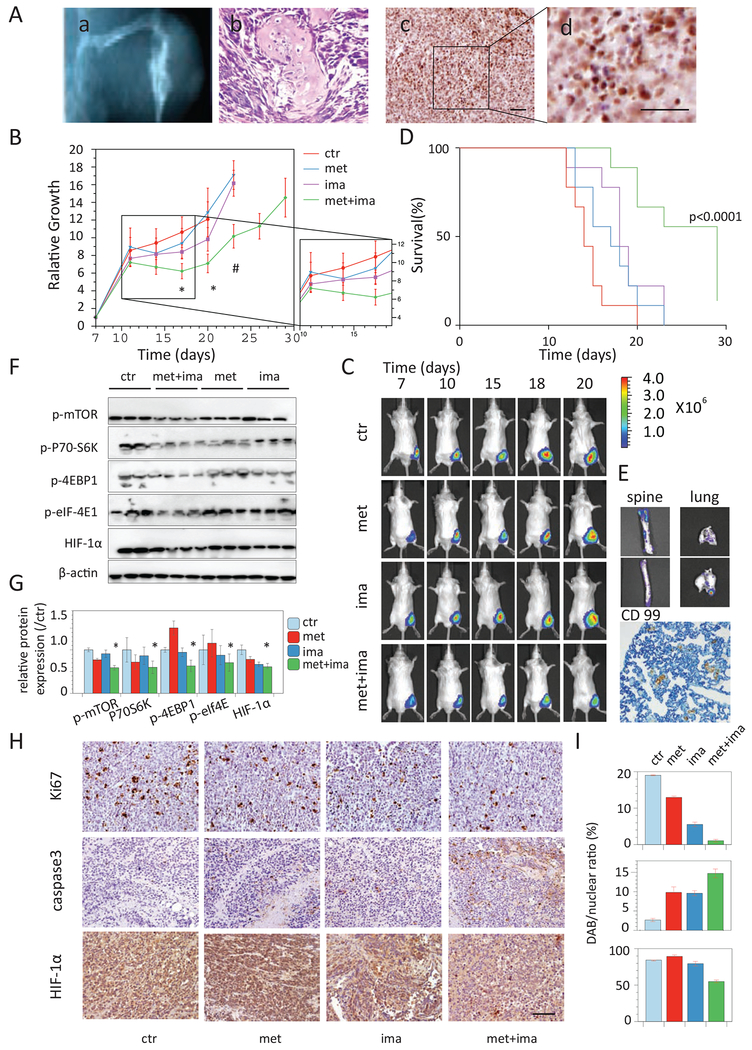
Based on a previously published EwS xenograft model that allows for tumor growth in two natural EwS environments, muscles and bones and resulting in distant metastases typically affected in humans [36], we injected EwS cells into the gastrocnemius muscle of NOD-SCID mice (Fig. 4A). With the TC71 xenograft model, rapid tumor growth was observed. Almost all tumors reached 2 cm diameter by day 29, thus we chose to administer the treatment for 21 days starting day 8-post injection. From the middle stage of tumor growth around day 15, quantitative BLI imaging started to detect a significant abrupt signal decline in ~30% mice, especially in the metformin group, while caliper measurement showed continuous tumor growth. Histological examination of these tumors showed necrosis in tumor vessels, which may lead to the lack of luciferin substrate in local tissue, suggesting tumor hypoxia and was further confirmed by extensive HIF-1α-positive staining and the signs of hypoxia-induced cell shrinkage and loose structure (Fig. 4A). As shown in Fig. 4B–C, the combination treatment suppressed tumor growth during the treatment period. Survival to endpoint was superior in the combination group (p<0.001, by log-rank test) (Fig. 4D). Most strikingly, five out of the 10 mice in the vehicle group had metastasis in lung and spine, but none was found with metastasis in the combination treatment group (0/10, p=0.03, by fisher exact test) (Fig. 4E). Importantly, in vivo cellular response to hypoxia was attenuated remarkably in the combination-treated tumors (Fig. 4F).
Figure 4. Combination of metformin and imatinib inhibits tumor growth and metastasis in TC71 EwS xenograft model.
A. A computed tomography (CT) image (a) and H.E. staining image (b) showing the tumor growth in muscles and bones in TC71 xenograft model, with extensive HIF-1α immuno-reactivity (c) and hypoxia-induced tumor cell shrinkage and loose structure (d). B. Tumor growth of TC71 xenograft mice treated with vehicle (ctr), metformin, imatinib, or the combination. N=10. *p<0.05, vs vehicle, metformin and imatinib; #p<0.05, vs metformin and imatinib. C. Representative BLI images showing tumor growth in each treatment group. D. Kaplan–Meier survival analysis of the TC71 xenograft mice in each treatment group. p<0.0001, log-rank test. E. Representative BLI images and a immunohistochemistry image showing tumor metasis to spine and lung in the TC71 xenograft mice. F. Western blot anaylais of key mechanism molecules in in vivo tumors. Three tumor samples in each treatment group were shown. G. Quantification of the in vivo tumor expression of the key mechanism molecules. *p<0.05, vs ctr. H. Representative immunohistochemistry images showing the tumor cell proliferation (ki67) apoptosis (caspase-3) and HIF-1α in TC71 tumors in each group. I. Quantification of the proliferation index (upper panel),% of apoptic cells (middle panel) and H score of HIF-1α (lower panel) in the tumors. n=5 mice per group. *p<0.05, vs ctr, met and ima. Scale bar 20 μm.
Similarly, the combination treatment steadily suppressed tumor growth in the TC32 animals during the treatment period (p<0.05) and significantly improved overall survival of the mice (p=0.01) (Fig. S1D–E). Tumor tissue was assessed for tumor cell proliferation (anti-Ki67), apoptosis (anti-cleaved caspase 3), and expression of HIF-1α. Consistent with the in vitro finding, tumors in both xenograft models showed decreased proliferation index, increased apoptotic rate, and attenuated HIF-1α expression with the combination treatment (Fig. 4H–I).
Clinical correlation studies of the metformin and imatinib combination with EwS and sarcoma patient cohorts
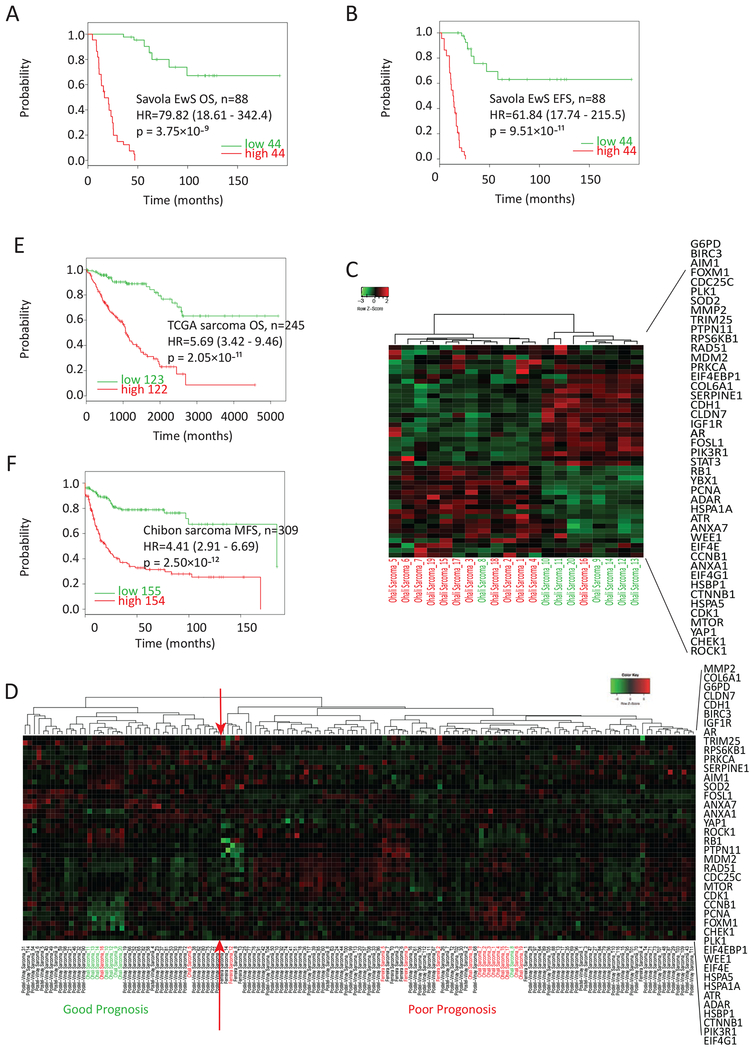
We conducted a series of correlative studies using the drug combination-associated 48 genes/proteins and several clinically annotated EwS microarray cohorts [37]. In the Savola Ewing sarcoma cohort, forty out of 48 genes showed significantly differential expression between the 88 patient tumor samples and 18 tumor-adjacent normal skeletal muscle tissues, and the genes that were down-regulated by the drug combination treatment in EwS cells always showed elevated expression in the EwS tumors, and vice versa (Table S4). Furthermore, the high expression of the forty-eight genes was correlated with poor overall survival (p=3.75×10-9) and event-free survival (p=9.51×10-11) in the Savola EwS patients (Fig. 5A–B), and poor prognosis in the Ohali EwS patients (Fig. 5C, Table S5). The Postel-Vinay EwS cohort does not have clinical information but has a relatively large sample size (n=117 EwS). Thus, we did an unsupervised hierarchical clustering using our 48 genes on the combined Oncomine standardized datasets consisted of 117 cases from the Postel-Vinay cohort together with 20 cases from the Ohali cohort and 4 cases from the Ferreira cohort that have matched clinical data for a total of 141. The patients were stratified into two clusters, each containing 42 and 99 patients, 14 out of 16 patients with known good prognosis were clustered together, as were the 7 out of 8 poor prognosis patients (Fig. 5D). In comparison, the published EwS prognosis gene signature [27] failed to stratify the same 141 patients into distinct clusters (Fig. S2).
Figure 5. The gene expression of 48 drug combination-associated genes/proteins correlates with poor prognosis in clinical EwS and sarcoma cohorts.
A and B. Kaplan–Meier survival analysis of overall survival (OS) and event-free survival (EFS) according to the expression of the 48 genes in the Savola EwS cohort (n=88). C. Heatmap of the 48-gene expression in the Ohali EwS cohort by unsupervised hierarchical clustering. Green patient label: good prognosis; red patient label: poor prognosis. D. Heatmap of the 48-gene expression in the combined dataset of Postel-Vinay (n=117) with Ohali (n=20) and 4 data from the Ferreira cohort with confirmed clinical prognosis status (n=141) by unsupervised hierarchical clustering. Patients at the left to the red arrows are enriched with good prognosis cases while at the right are enriched with poor prognosis cases. E and F. Kaplan–Meier survival analysis of patient OS and metastasis-free survival (MFS) according to the expression of the 48 genes in TCGA sarcoma cohort (E) and Chibon sarcoma cohort (F).
In addition to EwS, the expression of the forty-eight genes was correlated with poor overall survival in two sarcoma cohorts (p=2.05×10−11, in the TCGA dataset, n=245) and metastasis-free survival (p=2.49×10−12, in the Chibon dataset, n=310) (Fig. 5E–F). These results indicate that the metformin-imatinib combination could be potentially applied to EwS and sarcoma patients with poor prognosis.
Discussion
The novel drug combination identified in this work revives the potential for metformin in treating EwS. This combination regimen includes oral administration of standard clinical dosages of metformin and imatinib. Using a multi-omics analytic approach, we discovered that this combination regimen suppressed multiple dominant mechanisms in EwS genesis, growth, and metastasis, including the IGF system, key EWS-FLI1 downstream targets, and the PDGFR-β/PI3K/AKT/mTOR signaling pathway. More importantly, we demonstrated that imatinib could attenuate tumor hypoxic response to augment the in vivo anti-tumor capacity of metformin. Metformin is actively under clinical evaluation as a safe anti-cancer drug for almost all types of cancer, but hypoxia persistently impacts negatively to its efficacy. Our findings indicate a promising potential for combining it with imatinib to increase metformin’s in vivo efficacy in cancer treatment.
Repositioning drugs for the treatment of rare tumors represents an alternative and cost-effective strategy to accelerate the discovery of new therapeutics [38]. Metformin represents such a candidate, particularly in EwS, for which the IGF system is important for pathogenesis and progression. Metformin could inhibit EwS cell growth even in the presence of high concentrations of IGFs [9]. This drug is relatively safe for long-term use and inexpensive. Although the in vitro dose of metformin in our screening was above the feasible therapeutic plasma levels in humans, we fully demonstrated that the therapeutic plasma level of metformin in combination with imatinib is effective in inhibiting EwS tumor growth in vitro and in vivo. Furthermore, 57 clinical drugs that all have evidence as useful against EwS as individual agents were tested in combination with metformin in our in vitro screening study, and metformin with imatinib was the most potent combination in reducing EwS cell viability. This combination can also enhance the inhibitory effect of cyclophosphamide and ifosfamide in EwS cells, showing a potential benefit to patients of less dosage. Imatinib is also a relatively safe and inexpressive generic drug. While neither metformin [9] nor imatinib [39] alone was effective in EwS animal models or patients, their combination manifested profound anti-EwS effects. In addition, there is no clinical contraindication for this combination reported in public databases (e.g., Drug Interactions Checker and Drug Interaction Lookup), indicating a potential faster track to clinical trials for both drugs in combating EwS.
Both metformin and imatinib exhibit polypharmacology – they interact with more than one molecular target, and this promiscuous multi-targeting nature leads to their potential uses in a variety of other diseases [40, 41]. In our study, we demonstrated that the metformin-imatinib combination possesses a synergistic EwS tumor inhibition through modulating a network of targets. Extensive data demonstrate that molecular signals within a tumor are transmitted through a network of proteins rather than hierarchical signaling pathways [42], providing the rationale that targeting a single component in a canonical pathway instead of simultaneously inhibiting network proteins is insufficient for cancer treatment. Specifically in EwS, the HIF1-α mediated tumor hypoxic response in limiting the in vivo efficacy of metformin was significantly attenuated by concurrent imatinib treatment; this lays the foundation for the combination efficacy. Suppressing the convergent tumor-promoting signaling (mainly the PI3K/AKT/mTOR signals) by targeting different receptors using the two drugs (imatinib inhibits PDGFR-β and metformin inhibits IGF1-R and IR) further contributes to the synergistic anti-tumor effects. Imatinib has been previously shown to inhibit HIF1-α activity and intercept tumor hypoxic response in chronic myeloid leukemia [43] and lung cancer [44]. Our data provide new evidence that this effect can be extended not only to another tumor type – EwS, but also to rescue drug resistance due to hypoxia (metformin as an example). It is noted that the convergent therapeutic strategy by drug combinations could better tackle tumor heterogeneity and overcome acquired resistance [45]. Although the primary EwS tumors, which are defined and diagnosed by the presence of the EWS-FLI1 fusion oncogene, are not as heterogeneous as many other tumor types, the development to metastatic EwS disease is [46]. In the TC71 xenograft treatment study, a remarkable anti-metastasis effect was observed in the combination treatment group, and animals showed much improved overall survival, suggesting potential benefits of the combination, particularly for patients with advanced stage disease.
In addition to EwS and other sarcomas, the IGF system, PI3K/AKT/mTOR tumor-promoting signaling, and HIF1-α/hypoxia factors have been implicated in tumor growth and metastasis in many other cancers. There are more than 300 clinical trials evaluating metformin against various types of cancer (clinicatrials.gov). Our analysis on clinical cohorts of glioblastoma (GBM), stage III-IV ovarian cancer, and early stage breast cancer for which the metformin clinical trials are actively ongoing, indicated that the reverted expression pattern of the forty-eight genes correlated with poor patient outcomes (Fig. S3), which means that if the tumors were treated with the drug combination, they might have better outcome. However, much more investigation than a correlational analysis of patient data is needed to explore potential drug success.
In conclusion, our study demonstrates that imatinib could potentiate the anti-tumor effect of metformin, especially at the clinically relevant dose, through attenuating tumor hypoxic response and inhibiting a convergent tumor-promoting signaling pathway. The expression and activity of genes perturbed by the metformin/imatinib combination are found to be associated with tumors of poor prognosis, providing more evidence that the combination could be valuable addition to the current treatment regimen of metastatic or resistant tumors. In addition, the combination significantly enhances in vitro cell inhibition efficacy of chemotherapy drugs, cyclophosphamide and ifosfamide, suggesting that it might allow the reduction of dose intensity of the chemotherapy for EwS which could be applicable to localized disease by reducing adverse side effects of intensive cytotoxic chemotherapy. Finally, because of the ability of the metformin/imatinib combination to reverse the drug resistant effects of hypoxia which are quite prevalent in many cancers, there could be potential applications for other types of sarcoma that need to be explored.
Supplementary Material
Highlights.
A drug repositioning and combination screening identified seven synergistic drug combinations in inhibiting the viability of Ewing sarcoma cell lines under hypoxia.
The imatinib and metformin combination regimen has potent synergistic anti-tumor effect in Ewing sarcoma in vitro and in vivo models.
This combination suppresses dominant mechanisms in Ewing sarcoma including the key EWS-FLI1 downstream targets that are convergent into the PI3K/AKT/mTOR signaling pathway.
The drug combination-related 48 gene signature showed significant correlation with clinical outcomes in Ewing sarcoma, sarcoma, GBM, stage III-IV ovarian cancer and early stage breast cancer cohorts.
Acknowledgements
This research was funded by NIH U54 CA149196, NIH U01CA188388, TT and WF Chao Foundation, John S. Dunn Research Foundation, and Cancer Fighter of Houston to S.T.C. Wong, and China Scholarship Council to X. Nan. The authors would like to thank Rebecca Danforth for proofreading the manuscript and Lei Huang for valuable discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
Reference
- [1].Grunewald TGP, Cidre-Aranaz F, Surdez D, Tomazou EM, de Alava E, Kovar H, Sorensen PH, Delattre O, Dirksen U, Ewing sarcoma, Nat Rev Dis Primers, 4 (2018) 5. [DOI] [PubMed] [Google Scholar]
- [2].Toretsky JA, Kalebic T, Blakesley V, LeRoith D, Helman LJ, The insulin-like growth factor-I receptor is required for EWS/FLI-1 transformation of fibroblasts, J Biol Chem, 272 (1997) 30822–30827. [DOI] [PubMed] [Google Scholar]
- [3].Prieur A, Tirode F, Cohen P, Delattre O, EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3, Mol Cell Biol, 24 (2004) 7275–7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Scotlandi K, Manara MC, Serra M, Marino MT, Ventura S, Garofalo C, Alberghini M, Magagnoli G, Ferrari S, Lopez-Guerrero JA, Llombard-Bosch A, Picci P, Expression of insulin-like growth factor system components in Ewing’s sarcoma and their association with survival, Eur J Cancer, 47 (2011) 1258–1266. [DOI] [PubMed] [Google Scholar]
- [5].Ulanet DB, Ludwig DL, Kahn CR, Hanahan D, Insulin receptor functionally enhances multistage tumor progression and conveys intrinsic resistance to IGF-1R targeted therapy, Proc Natl Acad Sci U S A, 107 (2010) 10791–10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Buck E, Gokhale PC, Koujak S, Brown E, Eyzaguirre A, Tao N, Rosenfeld-Franklin M, Lerner L, Chiu MI, Wild R, Epstein D, Pachter JA, Miglarese MR, Compensatory insulin receptor (IR) activation on inhibition of insulin-like growth factor-1 receptor (IGF-1R): rationale for cotargeting IGF-1R and IR in cancer, Mol Cancer Ther, 9 (2010) 2652–2664. [DOI] [PubMed] [Google Scholar]
- [7].Karnevi E, Said K, Andersson R, Rosendahl AH, Metformin-mediated growth inhibition involves suppression of the IGF-I receptor signalling pathway in human pancreatic cancer cells, BMC Cancer, 13 (2013) 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sarfstein R, Friedman Y, Attias-Geva Z, Fishman A, Bruchim I, Werner H, Metformin downregulates the insulin/IGF-I signaling pathway and inhibits different uterine serous carcinoma (USC) cells proliferation and migration in p53-dependent or -independent manners, PLoS One, 8 (2013) e61537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Garofalo C, Capristo M, Manara MC, Mancarella C, Landuzzi L, Belfiore A, Lollini PL, Picci P, Scotlandi K, Metformin as an adjuvant drug against pediatric sarcomas: hypoxia limits therapeutic effects of the drug, PLoS One, 8 (2013) e83832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rocha GZ, Dias MM, Ropelle ER, Osorio-Costa F, Rossato FA, Vercesi AE, Saad MJ, Carvalheira JB, Metformin amplifies chemotherapy-induced AMPK activation and antitumoral growth, Clin Cancer Res, 17 (2011) 3993–4005. [DOI] [PubMed] [Google Scholar]
- [11].Knowles HJ, Schaefer KL, Dirksen U, Athanasou NA, Hypoxia and hypoglycaemia in Ewing’s sarcoma and osteosarcoma: regulation and phenotypic effects of Hypoxia-Inducible Factor, BMC Cancer, 10 (2010) 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kilic M, Kasperczyk H, Fulda S, Debatin K-MJO, Role of hypoxia inducible factor-1 alpha in modulation of apoptosis resistance, 26 (2007) 2027. [DOI] [PubMed] [Google Scholar]
- [13].Li Y, Zhang W, Li S, Tu CJS, Prognosis value of Hypoxia-inducible factor-1α expression in patients with bone and soft tissue sarcoma: a meta-analysis, 5 (2016) 1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Aryee DN, Niedan S, Kauer M, Schwentner R, Bennani-Baiti IM, Ban J, Muehlbacher K, Kreppel M, Walker RL, Meltzer P, Poremba C, Kofler R, Kovar H, Hypoxia modulates EWS-FLI1 transcriptional signature and enhances the malignant properties of Ewing’s sarcoma cells in vitro, Cancer Res, 70 (2010) 4015–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kovar H, Context matters: the hen or egg problem in Ewing’s sarcoma, Semin Cancer Biol, 15 (2005) 189–196. [DOI] [PubMed] [Google Scholar]
- [16].Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, Bindal N, Beare D, Smith JA, Thompson IR, Ramaswamy S, Futreal PA, Haber DA, Stratton MR, Benes C, McDermott U, Garnett MJ, Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells, Nucleic Acids Res, 41 (2013) D955–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Heske CM, Davis MI, Baumgart JT, Wilson K, Gormally MV, Chen L, Zhang X, Ceribelli M, Duveau DY, Guha R, Ferrer M, Arnaldez FI, Ji J, Tran HL, Zhang Y, Mendoza A, Helman LJ, Thomas CJ, Matrix Screen Identifies Synergistic Combination of PARP Inhibitors and Nicotinamide Phosphoribosyltransferase (NAMPT) Inhibitors in Ewing Sarcoma, Clin Cancer Res, 23 (2017) 7301–7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].May WA, Grigoryan RS, Keshelava N, Cabral DJ, Christensen LL, Jenabi J, Ji L, Triche TJ, Lawlor ER, Reynolds CP, Characterization and drug resistance patterns of Ewing’s sarcoma family tumor cell lines, PLoS One, 8 (2013) e80060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chou TC, Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies, Pharmacol Rev, 58 (2006) 621–681. [DOI] [PubMed] [Google Scholar]
- [20].Ren D, Zhu X, Kong R, Zhao Z, Sheng J, Wang J, Xu X, Liu J, Cui K, Zhang XH, Zhao H, Wong STC, Targeting Brain-Adaptive Cancer Stem Cells Prohibits Brain Metastatic Colonization of Triple-Negative Breast Cancer, Cancer Res, 78 (2018) 2052–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL, TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions, Genome Biol, 14 (2013) R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Trapnell C, Pachter L, Salzberg SL, TopHat: discovering splice junctions with RNA-Seq, Bioinformatics, 25 (2009) 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L, Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks, Nat Protoc, 7 (2012) 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Herwig R, Hardt C, Lienhard M, Kamburov A, Analyzing and interpreting genome data at the network level with ConsensusPathDB, Nat Protoc, 11 (2016) 1889–1907. [DOI] [PubMed] [Google Scholar]
- [25].Zhao H, Jin G, Cui K, Ren D, Liu T, Chen P, Wong S, Li F, Fan Y, Rodriguez A, Chang J, Wong ST, Novel modeling of cancer cell signaling pathways enables systematic drug repositioning for distinct breast cancer metastases, Cancer research, 73 (2013) 6149–6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Savola S, Klami A, Myllykangas S, Manara C, Scotlandi K, Picci P, Knuutila S, Vakkila J, High Expression of Complement Component 5 (C5) at Tumor Site Associates with Superior Survival in Ewing’s Sarcoma Family of Tumour Patients, ISRN Oncol, 2011 (2011) 168712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ohali A, Avigad S, Zaizov R, Ophir R, Horn-Saban S, Cohen IJ, Meller I, Kollender Y, Issakov J, Yaniv I, Prediction of high risk Ewing’s sarcoma by gene expression profiling, Oncogene, 23 (2004) 8997–9006. [DOI] [PubMed] [Google Scholar]
- [28].Ferreira BI, Alonso J, Carrillo J, Acquadro F, Largo C, Suela J, Teixeira MR, Cerveira N, Molares A, Gomez-Lopez G, Pestana A, Sastre A, Garcia-Miguel P, Cigudosa JC, Array CGH and gene-expression profiling reveals distinct genomic instability patterns associated with DNA repair and cell-cycle checkpoint pathways in Ewing’s sarcoma, Oncogene, 27 (2008) 2084–2090. [DOI] [PubMed] [Google Scholar]
- [29].Postel-Vinay S, Veron AS, Tirode F, Pierron G, Reynaud S, Kovar H, Oberlin O, Lapouble E, Ballet S, Lucchesi C, Kontny U, Gonzalez-Neira A, Picci P, Alonso J, Patino-Garcia A, de Paillerets BB, Laud K, Dina C, Froguel P, Clavel-Chapelon F, Doz F, Michon J, Chanock SJ, Thomas G, Cox DG, Delattre O, Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma, Nat Genet, 44 (2012) 323–327. [DOI] [PubMed] [Google Scholar]
- [30].Muz B, de la Puente P, Azab F, Azab AK, The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy, Hypoxia (Auckl), 3 (2015) 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].He L, Wondisford FE, Metformin action: concentrations matter, Cell Metab, 21 (2015) 159–162. [DOI] [PubMed] [Google Scholar]
- [32].Picard S, Titier K, Etienne G, Teilhet E, Ducint D, Bernard MA, Lassalle R, Marit G, Reiffers J, Begaud B, Moore N, Molimard M, Mahon FX, Trough imatinib plasma levels are associated with both cytogenetic and molecular responses to standard-dose imatinib in chronic myeloid leukemia, Blood, 109 (2007) 3496–3499. [DOI] [PubMed] [Google Scholar]
- [33].El-Naggar AM, Veinotte CJ, Cheng H, Grunewald TG, Negri GL, Somasekharan SP, Corkery DP, Tirode F, Mathers J, Khan D, Kyle AH, Baker JH, LePard NE, McKinney S, Hajee S, Bosiljcic M, Leprivier G, Tognon CE, Minchinton AI, Bennewith KL, Delattre O, Wang Y, Dellaire G, Berman JN, Sorensen PH, Translational Activation of HIF1alpha by YB-1 Promotes Sarcoma Metastasis, Cancer Cell, 27 (2015) 682–697. [DOI] [PubMed] [Google Scholar]
- [34].Lendahl U, Lee KL, Yang H, Poellinger L, Generating specificity and diversity in the transcriptional response to hypoxia, Nat Rev Genet, 10 (2009) 821–832. [DOI] [PubMed] [Google Scholar]
- [35].Liu Q, Fan D, Adah D, Wu Z, Liu R, Yan QT, Zhang Y, Du ZY, Wang D, Li Y, Bao SY, Liu LP, CRISPR/Cas9mediated hypoxia inducible factor1alpha knockout enhances the antitumor effect of transarterial embolization in hepatocellular carcinoma, Oncol Rep, 40 (2018) 2547–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hong SH, Tilan JU, Galli S, Izycka-Swieszewska E, Polk T, Horton M, Mahajan A, Christian D, Jenkins S, Acree R, Connors K, Ledo P, Lu C, Lee YC, Rodriguez O, Toretsky JA, Albanese C, Kitlinska J, High neuropeptide Y release associates with Ewing sarcoma bone dissemination - in vivo model of site-specific metastases, Oncotarget, 6 (2015) 7151–7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Aguirre-Gamboa R, Gomez-Rueda H, Martínez-Ledesma E, Martínez-Torteya A, Chacolla-Huaringa R, Rodriguez-Barrientos A, Tamez-Pena JG, Trevino V.J.P.o., SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis, 8 (2013) e74250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sardana D, Zhu C, Zhang M, Gudivada RC, Yang L, Jegga AG, Drug repositioning for orphan diseases, Brief Bioinform, 12 (2011) 346–356. [DOI] [PubMed] [Google Scholar]
- [39].Chao J, Budd GT, Chu P, Frankel P, Garcia D, Junqueira M, Loera S, Somlo G, Sato J, Chow WA, Phase II clinical trial of imatinib mesylate in therapy of KIT and/or PDGFRalpha-expressing Ewing sarcoma family of tumors and desmoplastic small round cell tumors, Anticancer Res, 30 (2010) 547–552. [PubMed] [Google Scholar]
- [40].Lee SJ, Wang JY, Exploiting the promiscuity of imatinib, J Biol, 8 (2009) 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pryor R, Cabreiro F, Repurposing metformin: an old drug with new tricks in its binding pockets, Biochem J, 471 (2015) 307–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Smock RG, Gierasch LM, Sending signals dynamically, Science, 324 (2009) 198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ng KP, Manjeri A, Lee KL, Huang W, Tan SY, Chuah CT, Poellinger L, Ong ST, Physiologic hypoxia promotes maintenance of CML stem cells despite effective BCR-ABL1 inhibition, Blood, 123 (2014) 3316–3326. [DOI] [PubMed] [Google Scholar]
- [44].Litz J, Krystal GW, Imatinib inhibits c-Kit-induced hypoxia-inducible factor-1alpha activity and vascular endothelial growth factor expression in small cell lung cancer cells, Mol Cancer Ther, 5 (2006) 1415–1422. [DOI] [PubMed] [Google Scholar]
- [45].Hazar-Rethinam M, Kleyman M, Han GC, Liu D, Ahronian LG, Shahzade HA, Chen L, Parikh AR, Allen JN, Clark JW, Kwak EL, Faris JE, Murphy JE, Hong TS, Van Seventer EE, Nadres B, Hong CB, Gurski JM Jr., Jessop NA, Dias-Santagata D, Iafrate AJ, Van Allen EM, Corcoran RB, Convergent Therapeutic Strategies to Overcome the Heterogeneity of Acquired Resistance in BRAF(V600E) Colorectal Cancer, Cancer Discov, 8 (2018) 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sheffield NC, Pierron G, Klughammer J, Datlinger P, Schonegger A, Schuster M, Hadler J, Surdez D, Guillemot D, Lapouble E, Freneaux P, Champigneulle J, Bouvier R, Walder D, Ambros IM, Hutter C, Sorz E, Amaral AT, de Alava E, Schallmoser K, Strunk D, Rinner B, Liegl-Atzwanger B, Huppertz B, Leithner A, de Pinieux G, Terrier P, Laurence V, Michon J, Ladenstein R, Holter W, Windhager R, Dirksen U, Ambros PF, Delattre O, Kovar H, Bock C, Tomazou EM, DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma, Nat Med, 23 (2017) 386–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.