BACKGROUND
Multiple pathogenic mechanisms have been implicated in autoimmune hepatitis, but they have not fully explained susceptibility, triggering events, and maintenance or escalation of the disease. Furthermore, they have not identified a critical defect that can be targeted. The goals of this review are to examine the diverse pathogenic mechanisms that have been considered in autoimmune hepatitis, indicate investigational opportunities to validate their contribution, and suggest interventions that might evolve to modify their impact. English abstracts were identified in PubMed by multiple search terms. Full length articles were selected for review, and secondary and tertiary bibliographies were developed. Genetic and epigenetic factors can affect susceptibility by influencing the expression of immune regulatory genes. Thymic dysfunction, possibly related to deficient production of programmed cell death protein-1, can allow autoreactive T cells to escape deletion, and alterations in the intestinal microbiome may help overcome immune tolerance and affect gender bias. Environmental factors may trigger the disease or induce epigenetic changes in gene function. Molecular mimicry, epitope spread, bystander activation, neo-antigen production, lymphocytic polyspecificity, and disturbances in immune inhibitory mechanisms may maintain or escalate the disease. Interventions that modify epigenetic effects on gene expression, alter intestinal dysbiosis, eliminate deleterious environmental factors, and target critical pathogenic mechanisms are therapeutic possibilities that might reduce risk, individualize management, and improve outcome. In conclusion, diverse pathogenic mechanisms have been implicated in autoimmune hepatitis, and they may identify a critical factor or sequence that can be validated and used to direct future management and preventive strategies.
Keywords: Autoimmune hepatitis, Pathogenesis, Epigenetics, Molecular mimicry, Epitope spread, Intestinal microbiome
Core tip: The next generation of management of autoimmune hepatitis will depend on clarification of the pathogenic sequence from susceptibility to triggering events to disease maintenance or escalation. Studies that move the pathogenesis of autoimmune hepatitis closer to its cause have the potential to replace blanket immunosuppressive regimens and even suggest strategies for prevention. Investigations that expand concepts of acquired epigenetic change in gene expression, pathogenic molecular mimicries, epitope spread, bystander activation, and intestinal dysbiosis may narrow the knowledge gap between environmental factors and disease occurrence and behavior. Protective as well as site-specific corrective interventions may emerge.
INTRODUCTION
Autoimmune hepatitis is an immune-mediated chronic liver disease that lacks a demonstrable etiologic agent[1]. The diagnosis requires histological features of interface hepatitis, increased serum immunoglobulin G level, the presence of autoantibodies, and the exclusion of virus-related, drug-induced, metabolic, and hereditary diseases[1,2]. Type 1 autoimmune hepatitis is characterized by the presence of antinuclear antibodies and/or smooth muscle antibodies, and type 2 autoimmune hepatitis is characterized by the presence of antibodies to liver kidney microsome type 1 (anti-LKM1)[3,4]. Other autoantibodies that may be useful in the diagnosis of type 1 disease are antibodies to actin (anti-actin)[5-8], antibodies to soluble liver antigen[9,10], and atypical perinuclear anti-neutrophil cytoplasm antibodies[11,12]. Antibodies to liver cytosol type 1 may be useful in the diagnosis of type 2 disease[13-15].
Pathogenic concepts have indicated a genetic susceptibility for autoimmune hepatitis[16-18], and deficiencies or disruptions in homeostatic mechanisms have been described that can overcome self-tolerance[19-22]. These elements have not been formulated into a validated sequence that fully explains susceptibility, disease onset, and course. Furthermore, a critical pathogenic defect has not been identified that can be selectively targeted by a designed intervention.
A pathogenic hypothesis that can accommodate the clinical experiences and science of autoimmune hepatitis cannot exclude genetic makeup as a predisposing factor, but it must also integrate other factors to help explain the risk burden[19,23,24]. Epigenetic changes could translate environmental cues into alterations of gene expression that disrupt pathways modulating the immune response[25,26], and thymic failure may allow autoreactive T cells to escape negative selection[27-29] and help explain susceptibility to the disease.
Molecular mimicry could be a mechanism for foreign antigens (infectious agents, drugs, xenobiotic chemicals, and gut-derived bacteria) to overcome self-tolerance[30-33]. Bystander activation could release pro-inflammatory cytokines, generate autoreactive T cells, and intensify the immune response[34-36]. Epitope spread could extend immune reactivity to less dominant epitopes within the same or other molecules[37-41]; neo-antigens generated by tissue injury could expand the number of antigenic targets[34,42]; and polyspecificity of the T cell antigen receptors (TCRs) could facilitate cross-reactivity between foreign and self-antigens[43-45]. These mechanisms could help explain how environmental triggers can overcome immune tolerance and induce the disease. Furthermore, variations in the composition of the intestinal microbiome could allow microbial antigens and gut-driven immune cells to individualize the risk, clinical phenotype, and consequences of autoimmune hepatitis[46].
The goals of this review are to examine the diverse mechanisms that have been implicated in the susceptibility, onset, and maintenance of autoimmune hepatitis, indicate investigational opportunities to validate their pathogenic roles, and suggest future interventions that might modify their impact.
METHODS
English abstracts were identified in PubMed using the primary search words, “pathogenesis of autoimmune hepatitis”, “genetics of autoimmune hepatitis”, “epigenetics and autoimmunity”, “pathogens and autoimmune hepatitis”, “drug-induced autoimmune hepatitis”, “molecular mimicry and autoimmune disease,” “neo-antigens”, “bystander activation”, “epitope spread”, “intestinal microbiome and autoimmune disease”, and “environment and autoimmunity”. Abstracts judged pertinent to the review were identified; key aspects were recorded; and full-length articles were selected from relevant abstracts. A secondary bibliography was developed from the references cited in the selected full-length articles, and additional PubMed searches were performed to expand the concepts developed in these articles. The discovery process was repeated, and a tertiary bibliography was developed after reviewing selected articles from the secondary bibliography. Over 1500 abstracts and 100 full length articles were reviewed.
FACTORS AFFECTING SUSCEPTIBILITY
Genetic predisposition
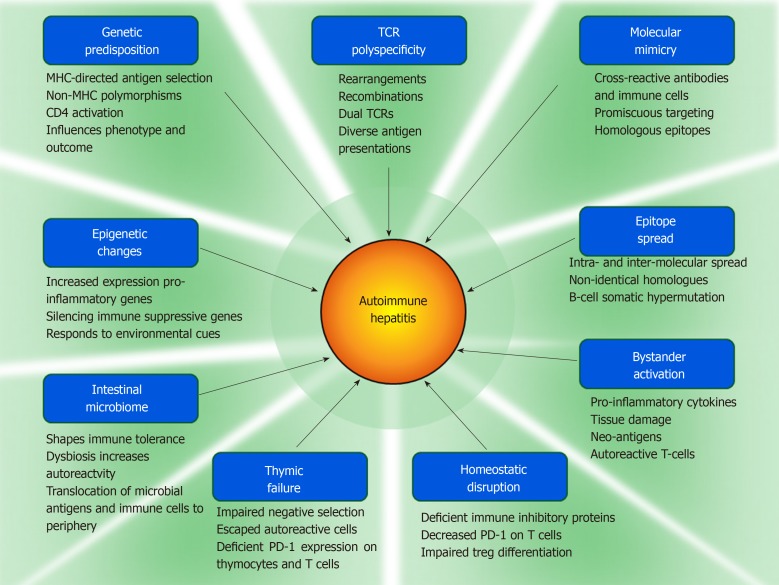
Genetic associations inside and outside the major histocompatibility complex (MHC) have been associated with the occurrence, serological phenotype, and severity of autoimmune hepatitis[18,47,48], and these associations have varied between ethnic groups[16,49-51], geographical regions[52-56], and age ranges[57,58] (Table 1). Genetic factors within the MHC may predispose to autoimmune hepatitis by encoding antigen binding grooves on class II MHC molecules that select and present antigens with certain structural and conformational properties[17,59], and genetic factors outside the MHC (polymorphisms of key immune regulatory and cytokine-producing genes) may promote immune reactivity and inflammatory responses that affect clinical phenotype and disease severity[48,60-62] (Figure 1).
Table 1.
Factors affecting susceptibility to autoimmune hepatitis
| Factors | Features | Pathogenic Implications in AIH |
| Genetic predispositions | DRB1*03:01 and DRB1*04:01 in white European and North American patients[16,18,47] | DRB1*03:01 associated with young age, severity, cirrhosis, and poor outcome[18,47] |
| DRB1*04:01 associated with elderly, concurrent immune diseases, treatment response[16,18,373] | ||
| DRB1*04:04 and DRB1*04:05 in Asian and Mexican patients[49,51,52,55,367] | ||
| DRB1*13:01 in South American children in and DRB1*04:05 in adults[50,53,58,368] | ||
| DRB1*13:01 distinguishes South American children from adults[58,368] | ||
| DQB1*0201, DRB1*07 and DRB1*03 in patients (mainly children) with anti-LKM1[369] | ||
| DQB1*02:01 and DRB1*07 associated with type 2 (anti-LKM1-positive) AIH[369,374,375] Genetics explain 51%-55% of risk-burden[23,25,47,65] | ||
| Polymorphisms of TNFα, Fas, CTLA4, and SH2B3 variably involved[48,56,60-62,65,370-372] | ||
| Polymorphisms may be discovered by GWAS[23] | ||
| Epigenetic changes | Alter structure of nucleosomes[25,26] | miR-21 and miR-122 increased in AIH[103] |
| Affect transcriptional activity of genes[67,69] | Hypomethylation of gene promoters in SLE and PBC may promote autoimmunity[82-85] | |
| Responsive to environmental cues[26,67] | ||
| Changes may be inherited[26,67] | Histone acetylation can increase Tregs or expression of pro-inflammatory genes[88,89] | |
| DNA methylation represses gene activity[70] DNA hypomethylation activates gene[77-81] | ||
| Histone changes can weaken self-tolerance[71,93] | ||
| Histone acetylation, phosphorylation, methylation, and ubiquitination can activate or repress gene activity[72,86,87,92] | ||
| May explain population risk differences[25,26] | ||
| Contributes to risk burden of AIH[23,25] | ||
| MiRNAs silence genes[73,94-96] | Epigenetics in AIH under-evaluated[25] | |
| Escaped autoreactive lymphocytes | Self-reactive thymocytes normally eliminated (negative selection)[27-29] | Escaped self-reactive CD4+ T cells may promote autoimmunity[112-114] |
| Thymocytes recognizing foreign antigens normally retained (positive selection)[27-29] | ||
| PD-1 expression on thymocytes and lymphocytes may be impaired[109,112,116] | ||
| Escaped self-reactive CD4+ T cells become self-tolerant, autoreactive, or Tregs depending on PD-1 and FoxP3 expression[112-114] | ||
| PD-1 expression in AIH unassessed[22] | ||
| Regulatory role of sPD-1 unknown in AIH[22] |
Superscripted numbers are references. AIH: Autoimmune hepatitis; anti-LKM1: Antibodies to liver kidney microsome type 1; CTLA4: Cytotoxic T lymphocyte antigen 4 gene; DNA: Deoxyribonucleic acid; Fas: Tumor necrosis factor receptor superfamily member 6 gene; FoxP3: Forkhead box P3; GWAS: Genome-wide association studies; HLA: Human leukocyte antigen; PBC: Primary biliary cholangitis; PD-1: Programmed cell death antigen-1; SH2B3: Src homology 2-B adaptor protein 3 gene; miRNAs: Micro-ribonucleic acids; TNF-α: Tumor necrosis factor-alpha gene; sPD-1: Soluble programmed cell death antigen-1; Tregs: Regulatory T cells.
Figure 1.
Pathogenic mechanisms implicated in autoimmune hepatitis. The components of each putative mechanism are shown under each panel. MHC: Major histocompatibility complex; TCR: T cell antigen receptor; PD-1: Programmed cell death protein-1.
The triggering peptides may be common in particular sub-populations, such as the young (viral infections) or the elderly (polypharmacy), and they may reflect the genetic composition and environment of the population at risk. HLA DRB1*03 has a low frequency in the normal Japanese population, and autoimmune hepatitis in Japan is associated mainly with HLA DRB1*04[49]. In contrast, HLA DRB1*04 is less frequent in the normal Italian population than in healthy North American adults (16% vs 34%, P = 0.0003)[63]. These differences in the genetic composition of the populations at risk could affect antigen selection, disease occurrence, and clinical phenotype.
Future studies of genetic and environmental factors associated with autoimmune hepatitis should be population-based and correlate genetic determinants with age, gender, ethnicity, and exposure to possible antigenic triggers[64]. Importantly, the risk-burden for autoimmune hepatitis cannot be fully explained by genetic factors[23]. The key susceptibility alleles (DRB1*03:01 and DRB1*04:01) occur in only 51%-55% of white North American and northern European patients[16], and polymorphisms of genes outside the MHC, which have yet to be ascribed biological significance, have similar frequencies of occurrence[23,25,60,65,66]. Population-based genome-wide association studies may define the genetic fabric outside the MHC that is still undiscovered in autoimmune hepatitis, and epigenetic changes, cued by the environment, may also explain the risk.
Epigenetic changes
Epigenetic changes influence the function of genes without altering the sequence of deoxyribonucleic acid (DNA), and they may contribute to the occurrence and phenotype of autoimmune hepatitis[25,26] (Table 1). They may also explain the risk-burden for autoimmune hepatitis that cannot be linked to classical genetic associations. Epigenetic changes can occur at sites throughout the genome and respond to pressures that reflect diverse environmental factors[26,67] (Figure 1). The structural adaptations occur within the nucleosomes of chromatin, and they affect the packaging of DNA, the activity of ribonucleic acid polymerase (RNAP), the ability of RNAP to open double-stranded DNA, and the accessibility of transcription factors to DNA binding sites[67-69]. DNA methylation[70] and histone modification[71,72] are the principal mechanisms that modify chromatin structure, and noncoding micro-ribonucleic acids (miRNAs)[73-76] are the main agents that silence gene activity.
DNA methylation, histone modifications, and miRNAs
DNA methylation typically represses gene activity by inhibiting the binding of transcription factors to DNA, and it can be reversed by oxidation of the methylated site[26,77,78] (Table 1). The ten eleven translocation oxygenases catalyze this conversion, increase gene translational activity, and counterbalance the repressive effects of DNA methylation[79-81]. Hypomethylation of gene promoters has been described in systemic lupus erythematosus (SLE)[82,83] and primary biliary cholangitis (PBC)[84], and epigenetic alterations in the expression of immune regulatory genes may promote loss of self-tolerance[85].
Histone modifications involve enzymatic alterations of amino acids in the tail of the histone proteins comprising the nucleosomes, and these modifications (phosphorylation, methylation, acetylation, and ubiquitination) can in turn loosen the wrap of DNA and increase the transcriptional activity of the gene[25,72,86,87] (Table 1). Histone acetylation by histone acetyltransferase characterizes activated genes which may have stimulatory or repressive effects on disease activity by increasing the number and function of regulatory T cells (Tregs)[88] or increasing the expression of pro-inflammatory genes[89]. Histone deacetylase can modulate transcription activity by removing acetyl groups and repressing gene activity[90-92]. The number and location of possible histone modifications within the nucleosome contribute to genomic instability, and histone modifications have been associated with the loss of self-tolerance[71,92,93].
Noncoding miRNAs are small double-stranded molecules that can pair with similarly sequenced messenger RNAs (mRNAs) within the cytosol. The complex is destined for degradation by a RNA-induced silencing complex[73,94-96] (Table 1). Gene silencing can have organ specificity, and it can occur by degradation or repression of the mRNAs[97]. A single miRNA can affect the expression of multiple genes, and many different miRNAs can target the same mRNA. MiRNAs can thereby modulate disease activity by affecting immune (gene product production), inflammatory (cytokine production), and proliferative (lymphocyte differentiation) pathways[98-100]. The investigational challenge is to identify within the body of described miRNAs (> 1400 types) the key gene products that are targeted in autoimmune hepatitis[101,102].
Epigenetic changes and autoimmune hepatitis
Serum levels of micro-RNA 21 (miR-21) and micro-RNA 122 (miR-122) are increased in autoimmune hepatitis[103] (Table 1). MiR-21 is expressed by T lymphocytes, and the programmed cell death 4 (PDCD4) gene is one of its gene targets[103,104]. The gene product of PDCD4 is the immune inhibitory protein, PDCD-4, and the down-regulation of PDCD4 by miR-21 could promote immune reactivity. PDCD-4 increases the apoptosis of activated T lymphocytes and decreases the production of pro-inflammatory cytokines[105], and serum levels of miR-21 have correlated with the histological grade of liver inflammation[103].
Similarly, miR-122 has been associated with inflammatory activity (serum alanine aminotransferase levels) in autoimmune hepatitis[103], and miR-122 has increased the production of the pro-inflammatory cytokine, interferon type 1, by de-repressing cytokine signaling in a murine model of PBC[105]. Both miR-21 and miR-122 have been proposed as biomarkers of inflammatory activity in autoimmune hepatitis, but the nature and scope of their actions, disease-specificity, and value as therapeutic targets remain uncertain[103].
Thymic failure and escaped autoreactive T cells
Susceptibility to autoimmune disease may also relate to the escape of autoreactive immune cells from negative selection by the thymus and their persistence in the circulation (Figure 1). The existence of circulating autoreactive T cells implies a central defect in their thymic elimination or a peripheral failure to suppress their activity. Disruptions in the homeostatic molecular pathways that modulate negative selection within the thymus and lymphocyte differentiation in the lymphatic tissue have been proposed, and the expression of programmed cell death antigen-1 (PD-1) on thymocytes and maturing lymphocytes may be a key factor in limiting the escape and activity of autoreactive T cells[22].
Failures in thymic selection and peripheral protection
Immature CD4-CD8- thymocytes differentiate into CD4+CD8+ thymocytes that are characterized by TCR beta (β)-chains (β-selection)[106-109] (Table 1). The CD4+CD8+ thymocytes with β-chains develop TCRs consisting of alpha (α)- and β-chains, and only the thymocytes that survive selection by demonstrating TCR specificity can mature into CD4+ or CD8+ T lymphocytes (repertoire selection)[110,111]. Thymocytes with self-reactive antigen receptors are eliminated by apoptosis (negative selection), and thymocytes that recognize foreign peptides presented by MHC molecules are selected to mature (positive selection)[27-29].
Self-reactive CD4+ T cells that escape thymic deletion can autonomously express the immune inhibitory protein, PD-1, to induce self-tolerance[112] (Table 1). Otherwise, the majority will persist as autoreactive T cells unless they express forkhead box P3 (FoxP3)[112-114]. Self-reactive CD4+ T cells expressing FoxP3 can differentiate into Tregs and counteract the autoreactive response. This transformation into Tregs occurs in only a minority of the self-reactive T cells that have evaded thymic elimination, and its dampening effect on the autoreactive response may be incomplete or negligible[112,113,115].
The expression of PD-1 on thymocytes[109,116] and maturing lymphocytes[112] is critical in limiting the emergence of autoreactive lymphocytes in the periphery. Clarification of the factors that regulate production and function of PD-1 should be an investigational objective in autoimmune hepatitis[22]. Soluble PD-1 is an alternatively spliced transcript encoded by the PD-1 gene which lacks a membrane-spanning portion and is rendered soluble[117]. The shortened soluble molecule is a variant of PD-1 that can competitively inhibit ligation of the full length molecule and prevent generation of an immune inhibitory signal[117-119]. Its counter effects on the immune inhibitory actions of full-length PD-1 may contribute to the escape of autoreactive T cells, and soluble PD-1 should be evaluated as a potential biomarker of disease activity and therapeutic target in autoimmune hepatitis.
TRIGGERING EVENTS
Microbial infection
Clinical experiences have described temporal associations between viral infections and the onset of autoimmune hepatitis in isolated cases. Infections with hepatitis A virus[120-125], hepatitis B virus (HBV)[126], hepatitis C virus (HCV)[127], Epstein-Barr virus (EBV)[128-132], varicella zoster[133], and human immunodeficiency virus[134] have been proposed as triggering events (Table 2). Other studies have described evidence of HCV[135-139], measles virus[140-142], cytomegalovirus (CMV)[143], and EBV[144] in the serum or liver tissue of patients with autoimmune hepatitis. These findings have supported an association between viral infection and autoimmune hepatitis, but the nature of the relationship (coincidental vs etiologic) has been uncertain[145,146].
Table 2.
Possible triggering events for autoimmune hepatitis
| Factors | Features | Pathogenic Implications in AIH |
| Microbial triggers | Infections with multiple viruses temporally associated with onset of AIH[120,126-128,133] | Low frequency of viral markers in AIH[136,147] |
| Undiscovered viral agents possible[151] | ||
| Rarity of AIH contrasts with ubiquity of viruses and supports host-related predisposing factors[147] | ||
| Multiple viral antigens discovered in serum and liver tissue of patients with AIH[139,142-144] | ||
| Microbial infection as direct cause unlikely[147] | ||
| Superantigens | Induced by viruses and bacteria[158,159] | Superantigens implicated in model of MS and patients with RA[165-167] |
| No MHC-restricted antigen presentation[158] | ||
| Associated with nearly monoclonal single type Vβ T cells in RA[166] | ||
| Bind to class II MHC molecule on APC and Vβ region of TCR[160] | ||
| Microbial basis inferred in RA[159,166,167] | ||
| Generate polyclonal T cell response[161] | ||
| Can induce T cell exhaustion[376,377] | ||
| Superantigens unassessed in AIH[158,159] | ||
| Drug exposure | Metabolites can interact with self-proteins to promote loss of tolerance[176,178,179] | Idiosyncratic drug-induced liver injury can resemble AIH[168,169] |
| Immune checkpoint inhibitors enhance reactivity against neo-antigens[182,184] | Immune-mediated hepatitis associated with blockade of immune inhibitors[190-194] | |
| Immune checkpoint inhibitors induce diverse autoimmune diseases[186,187] | Hepatitis may occur months after cessation of immune checkpoint inhibitor[189] | |
| Drugs can cause DNA demethylation[199-203] | DNA demethylating drugs induce lupus-like reactions in animal models[202] | |
| Environmental pressures | Diet, drug or alcohol abuse, pollutants, sanitation, polypharmacy, and socioeconomic status are potential but unevaluated risk factors for AIH[25] | Vitamin D deficiency in refractory AIH[209,210] |
| Vitamin D response element in genes[378,379] | ||
| Gene expressions affected by vitamin D deficiency[35,85,207,208,211] | ||
| Environment can affect antigen exposures, gene expression, and immune responses[25] | ||
| Polymorphisms of VDR associated with occurrence of AIH[212,213] | ||
| Other environmental factors unexplored in AIH |
Superscripted numbers are references. AIH: Autoimmune hepatitis; APC: Antigen-presenting cell; DNA: Deoxyribonucleic acid; MHC: Major histocompatibility complex; MS: Multiple sclerosis; RA: Rheumatoid arthritis; TCR: T cell antigen receptor; VDR: Vitamin D receptor; Vβ region: Variable region of the β chain.
The frequency of viral markers in patients with characteristic features of autoimmune hepatitis has been low (2%-11% for HCV and HBV)[136,147,148], often associated with false positive first generation assays confounded by hypergammag-lobulinemia[149,150], and commonly unconfirmed by second generation assays[147] (Table 2). The possibility of an undiscovered viral cause of autoimmune hepatitis cannot be excluded, but assessments for unusual viruses have been unrewarding (albeit few in number)[151,152]. After decades of observation and pursuit, a particular viral trigger for autoimmune hepatitis has not been established. Instead, viruses may constitute one of several categories of environmental agents that expose individuals to antigens that challenge immune tolerance in susceptible individuals.
The ubiquity and diversity of the presumed viral agents and the rarity of autoimmune hepatitis suggest that infection must be accompanied by host specific factors to trigger autoimmunity[147]. Genetic makeup[17,153], dysregulated critical homeostatic pathways[34], pre-conditioning by previous viral infections[154], molecular mimicries between microbial and self-antigens[30,31,155,156], and unmasked or newly created antigens (neo-antigens[34,146] or superantigens[157-159]) may constitute the host-specific factors that translate microbial infection into an autoimmune disease.
Superantigens
Viruses and bacteria can induce the formation of superantigens which can activate autoreactive T cells without antigen presentation by MHC molecules[158,159] (Table 2). The superantigens are a family of proteins that can bind to class II molecules of the MHC on antigen presenting cells (APCs) at a site distant from the peptide binding groove[158]. Simultaneously, they can bind to the TCR of T lymphocytes and activate them[158]. Superantigens bind to the variable region of the β chain within the TCR[160], and they frequently bind to several TCRs, thereby generating a polyclonal response[161]. Virtually all T cells bearing a particular TCR Vβ are activated by a superantigen[160], and the net effect is to trigger a T cell response manifested by the release of pro-inflammatory cytokines[162] and the proliferation of T lymphocytes[163,164].
Superantigens have been implicated as a reactivating factor in an experimental model of multiple sclerosis[165] and a triggering factor in patients with rheumatoid arthritis[166,167] (Table 2). The presence of a superantigen in patients with rheumatoid arthritis has been suspected because of the high percentage of Vβ14+ T cells in the synovial fluid and the oligoclonality of the Vβ14+ population[166]. Similar studies assessing the Vβ repertoire of T cells in the liver tissue and peripheral blood in autoimmune hepatitis might also implicate a superantigen and extend the search for microbial triggers.
Drug exposure
Multiple drugs, especially minocycline, nitrofurantoin, and infliximab, can induce an idiosyncratic liver injury indistinguishable from autoimmune hepatitis[168-170], and herbal medicines[171-174] and environmental pollutants[175] may also be initiating factors (Table 2). Drug-induced liver injury resembling autoimmune hepatitis is typically self-limited after drug withdrawal[168,169], and the rare instances of chronic self-perpetuating liver injury may reflect host-specific deficiencies in immune regulation and antigens generated by interactions between reactive drug metabolites and self-proteins that promote loss of self-tolerance[176-179].
The immune checkpoint inhibitors constitute an emerging category of biological agents that have been designed to block key immune inhibitory proteins [PD-1, programmed cell death antigen ligand-1, and cytotoxic T lymphocyte antigen-4 (CTLA-4)][22,180,181] (Table 2). The immune checkpoint inhibitors have enhanced the adaptive immune response to neo-antigens expressed by tumor cells in animal models and patients[182-185], and they have been associated with diverse immune-related adverse events[186-189], including the development of an immune-mediated hepatitis[190-194]. The clinical phenotype of liver injury induced by the immune checkpoint inhibitors has lacked the laboratory and histological features characteristic of autoimmune hepatitis[191,195-198], but the delayed development of hepatitis 8 mo after discontinuation of an anti-PD-1 preparation suggests that induced disturbances in the immune inhibitory axis may endure and pose a long-term risk for immune-mediated liver disease[189].
Drugs may also induce epigenetic changes that alter the expression of immune regulatory genes, and in turn this alteration that facilitate the loss of self-tolerance (Table 2). DNA demethylation has been associated with multiple drugs that can induce lupus-like reactions (procainamide, phenytoin, isoniazid, chlorpromazine, hydralazine, and 5-azacytidine)[199-203], and demethylating drugs as initiating agents in autoimmune hepatitis have been under-evaluated[25,169]. Aging may compound the effects of the environmental pressures on gene expression by promoting genomic instabilities through repeated cycles of DNA replication[204-206].
Most cases of autoimmune hepatitis lack a definable etiological trigger, and the possibilities that an antecedent microbial infection or drug exposure has been overlooked or that multiple different microbial and drug exposures have had a cumulative deleterious effect on the maintenance of self-tolerance cannot be proven or denied[146].
Environmental pressures
Epigenetic modifications can be induced by environmental pressures, and environmental cues that alter gene expression may help explain the occurrence of autoimmune hepatitis in diverse populations and geographical regions[35,85,207,208] (Table 2). Vitamin D deficiency has been associated with histological severity, advanced hepatic fibrosis, and non-response to conventional glucocorticoid therapy in autoimmune hepatitis[209-211]. 1, 25 dihydroxyvitamin D binds to the vitamin D receptor (VDR), and this complex in turn activates the vitamin D response element in certain genes. This epigenetic change may enhance the transcription of genes that affect the inflammatory and immune responses[211], and genetic polymorphisms affecting the structure of the VDR have been associated with increased susceptibility to autoimmune hepatitis[212,213]. Other environmental pressures related to diet, socioeconomic status, drug or alcohol abuse, sanitation, and polypharmacy have an uncertain effect on the occurrence and outcome of autoimmune hepatitis, and they warrant further assessment in population-based studies.
FACTORS AFFECTING MAINTENANCE OR ESCALATION
Molecular mimicry
Molecular mimicry between foreign and self-antigens has been proposed as a mechanism by which the adaptive immune response can be sensitized to epitopes that are similar but not identical to self[30-33,214,215] (Table 3). Invading pathogens[216-218], naturally occurring (environmental) antigens[35], synthetic peptides (chemical agents, halothane)[219-223], and vaccines[224,225] have been implicated as xenobiotic triggers that overcome self-tolerance by generating cross-reacting antibodies and immune cells (Figure 1). The molecular mimics have structural or conformational similarities with self-antigens, and they are introduced by infection, environmental exposure, or xenobiotic modification of proteins in situ[31,222,223].
Table 3.
Factors affecting maintenance or escalation of autoimmune hepatitis
| Factors | Features | Pathogenic implications in AIH |
| Molecular mimicry | Structural or conformational similarity between foreign and self-antigen[30-33,215] | Mimicries between CYP 2D6 of AIH and HCV, herpes simplex, CMV[244-246] |
| Introduced by infection, environment, or xenobiotic modification of self-antigen[215,222] | Structural mimicry with bacteria in PBC[247-251] | |
| Generates cross-reacting antibodies and immune cells[31,216] | Virus expressing human CYP 2D6 induces experimental AIH[226,252] | |
| More mimicries between bacterial motifs and self-antigens than AIH occurrence suggest low impact or other factors involved[64,253,257] | ||
| Requires similarity not identity to self-epitope[31] | ||
| Must mimic biologically active homologue[31] | ||
| Epitope spread | Antibodies or immune cells target multiple epitopes on same or other molecules[37,38,264,268] | Autoantibody-response in murine AIH model spreads from immune-dominant epitope to neighboring and remote regions[40] |
| Initiating immune-dominant epitope may be lost as range of immune reactivity increases[40,268] | ||
| Patients with AIH show similar response[40] | ||
| Enhanced by endocytic processing and variability of peptide fragments presented by class II MHC molecules[271,380,381] | ||
| Somatic hypermutation diversifies B cell receptors and their reactivity to wider spectrum of antigens[41,266,272,273] | ||
| Neo-antigens | Antigens released from injured tissue or formed during inflammatory activity[42,264] | Can increase epitope spreading[264,265] |
| May re-enforce immune response[42] | ||
| Expressed only under certain conditions[42] | Unassessed in AIH | |
| Can be variable between individuals[42,264] | ||
| Bystander activation | Induced by viral infection, bacterial products, and virus-mimetics (vaccines)[274-278] | Can intensify collateral tissue injury[274,282] |
| Activate APCs (dendritic cells)[274] | ||
| Pro-inflammatory cytokines released from T cells and macrophages activate pre-primed polyclonal memory T cells[274] | ||
| Mobilize autoreactive T cells[274] | ||
| Unassessed in AIH | ||
| Memory CD8+ T cells mainly involved[275,276] | ||
| Memory CD4+ T cells also activated[279,280] |
Superscripted numbers are references. AIH: Autoimmune hepatitis; APCs: Antigen presenting cells; CMV: Cytomegalovirus; CYP 2D6: Cytochrome P450 2D6; HCV: Hepatitis C virus; MHC: Major histocompatibility complex; PBC: Primary biliary cholangitis.
Molecular identity rather than mimicry induces immune tolerance[226], and it is a mechanism by which invading pathogens may avoid recognition and elimination by immune defenses[227]. Molecular mimicry implies that the antigenic homologue is sufficiently different from the self-antigen to generate cross-reactive immune responses[31]. The molecular mimic must also resemble a self-antigen with biological activity that could promote autoimmune disease[31], and the structural or conformational similarities to the self-antigen must be sufficient to ensure binding to the same MHC class II molecules and activation of the same lymphocyte populations[228].
Pathogenic molecular mimicry has been incriminated in diverse autoimmune diseases that have been associated with bacterial and viral infections, including rheumatic fever[229-234], Guillain Barre syndrome[235,236], Lyme disease[237,238], Reiter’ syndrome[239], ankylosing spondylitis[240,241], rheumatoid arthritis[242], multiple sclerosis[217], and SLE[243].
Molecular mimicry and autoimmune hepatitis
Molecular mimicries have been demonstrated between an autoantigen of autoimmune hepatitis (cytochrome P450 2D6) and infectious agents (Table 3). The anti-LKM1 associated with type 2 autoimmune hepatitis recognize a short-linear amino acid sequence on cytochrome P450 2D6 as its principal epitope, and antibodies to herpes simplex type 1 virus, HCV, and CMV have demonstrated cross-recognition of this epitope[244-246]. Structural similarities between bacterial agents (Escherichia coli and Novoshingobium aromaticivorans) and pyruvate dehydrogenase-E2 complex (PDC-E2) have also suggested a pathogenic role for molecular mimicry in PBC[247-251], and infection with a virus expressing a human P450 2D6 that is homologous to mouse P450 2D6 has induced severe liver damage in a murine model of autoimmune hepatitis[226,252]. The main investigational challenge is to associate these molecular mimicries with pertinent pathogenic mechanisms of the disease.
Pathogenic uncertainties and future investigations
Studies analyzing exact peptide matches at penta-, hexa-, hepta-, and octapeptide levels have demonstrated that virtually all human proteins include a bacterial pentapeptide or hexapeptide motif[253] (Table 3). Furthermore, the amino acid sequences necessary for peptide binding by MHC I and MHC II molecules are short (8-18 amino acids), and only a small portion of the antigen is required for recognition by a TCR[254-257]. The relative rarity of autoimmune diseases in the general population (5%-8%)[32] and low annual incidence of autoimmune hepatitis (0.85-1.9 cases per 100000 persons)[64] suggest that molecular mimicry is either an unimportant cause of autoimmune hepatitis or that its impact is mitigated by mechanisms that are still undiscovered.
Molecular mimicry induced by a non-infectious environmental agent has been uncommonly described in autoimmune disease, but it has been demonstrated in PBC. Immunization of a murine model with 2-octynoic acid, a chemical used widely in cosmetic products and food flavorings[221,258,259], has induced autoimmune cholangitis and antimitochondrial antibodies. The mechanism has been ascribed to a molecular mimicry in which the xenobiotic chemical has modified the lipoic binding region of PDC-E2 in situ and compromised immune tolerance of the self-antigen[222,223].
Future investigations in autoimmune hepatitis should explore molecular mimicries between non-infectious environmental agents and key antigenic targets in autoimmune hepatitis, such as cytochrome P450 2D6[244], formiminotransferase cyclodeaminase[14,15], and Sep [O-phosphoserine] tRNA: Sec [selenocysteine] tRNA synthase[260-263].
Epitope spread and neo-antigens
Autoantibodies and activated immune cells that target epitopes different from the immune-dominant epitope that initiated the immune response constitute epitope spread[37,38,40,41,264-266] (Table 3). The reactivity may be against epitopes on the same molecule or homologous sequences on other molecules. Infections and environmental agents (chemicals, toxins, and drugs) may generate, amplify or sustain the autoimmune response by releasing neo-antigens directly from the injured tissue[37,38,264,267] (Figure 1), and the neo-antigens may vary between individuals[42]. The autoantibodies or immune cells activated by the neo-antigens can broaden the range of immune reactivity and promote the spread of antigenic targets[37,41,268,269].
The initiating immune-dominant epitope may be lost during this process as the targeted homologous sequences in neighboring and remote regions become increasingly different from the original epitope[40]. In this fashion, the relationship to the initial immune-dominant epitope may be lost before clinical onset of the disease, and the original invading pathogen, environmental agent, or triggering antigen may be undetectable[270]. Epitope spread has been demonstrated in patients and animal models of autoimmune hepatitis characterized by antibodies to cytochrome P450 2D6[40].
Epitope spread can be enhanced by endocytic processing, variability in the peptide fragments that are loaded into the class II MHC molecules, and somatic hypermutation[35,41] (Table 3). The peptide segments loaded into the class II MHC complexes after endocytic processing can vary in length and affinity[271]. Similar molecules can generate homologous peptide fragments that vary sufficiently to favor cross-reactive immune responses and thereby induce intermolecular epitope spread. Somatic hypermutation can also contribute to epitope spread by diversifying B cell receptors after antigen recognition in order to recognize a wider spectrum of foreign antigens[41,266,272] (Table 3). Somatic hypermutation is an adaptive mutation involving single base substitutions mainly in the hypervariable regions of the immunoglobulin genes[272,273]. The roles of neo-antigens and somatic hypermutation in the severity of autoimmune hepatitis have not been assessed.
Bystander activation
Bystander activation can increase the immune response and tissue damage by activating pre-primed T cells and spreading the damage to uninvolved neighboring cells[274] (Table 3). Viral infections[275], bacterial products (lipopolysaccharide)[276], and virus-mimetics (vaccines)[277,278] can induce the secretion of cytokines which in turn can stimulate the proliferation of polyclonal memory T cells. Memory CD8+ T cells are mainly affected by bystander activation, and their proliferation is mediated by interferon-gamma and interleukin (IL)-12, IL-15, and IL-18[275,276,279,280]. Memory CD4+ T cells can also undergo bystander activation (to a lesser degree), and their proliferation is mediated mainly by IL-2[279,280].
The bystander activation of pre-primed autoreactive T cells does not require stimulation of the TCR by a specific antigen[279,281]. Bystander activation with the release of pro-inflammatory cytokines can intensify the collateral damage associated with microbial infection[274,281-283], and it may be a mechanism that contributes to the emergence and progression of autoimmune disease by activating APCs and mobilizing autoreactive T cells[274,279] (Figure 1). Bystander activation has not been assessed in autoimmune hepatitis, and the role of gut-derived microbial products in inducing a bystander effect has not been determined.
TCR polyspecificity
TCRs can undergo continuous rearrangements and re-combinations that broaden the range of antigens that they recognize, and this plasticity can contribute to an autoreactive response[284,285] (Figure 1). TCRs recognize only a small portion of the peptide presented by class II MHC molecules[254,256,257], and the same TCR can respond to multiple antigens[45,286]. Polyspecificity of the TCR can generate cross-reactivity between T cells and facilitate promiscuous targeting of self-antigens.
T cell targeting can also be influenced by the surface expression of dual TCRs[287,288] (Table 4). Thirty percent of human T cells express two functional α-chains. They may also express two β-chains at a lower frequency[43,289]. T cells expressing dual TCRs are more difficult to stimulate[44], and they may escape negative selection in the thymus by failing to demonstrate strong reactivity to self-antigens[290]. In the peripheral circulation, the T cells with dual TCRs may be stimulated by foreign antigen and self-peptide, and the dual stimulation may overcome self-tolerance[44,288]. Future investigations in autoimmune hepatitis that determine the number and nature of circulating T lymphocytes with dual TCRs may reveal a means by which to measure or monitor the autoreactive propensity.
Table 4.
Pathogenic implications of T cell antigen receptor polyspecificity and intestinal dysbiosis in autoimmune hepatitis
| Factors | Features | Pathogenic implications in AIH |
| TCR polyspecificity | TCRs have plasticity that increase cross-reactivity and polyspecificity[45,284-286] | Increased cross-reactivity, promiscuous targeting, and less self-tolerance[45] |
| Dual TCRs escape thymic negative selection[290] | ||
| Unassessed in autoimmune hepatitis | ||
| Dual TCRs may recognize both foreign and self-antigens[44,288] | ||
| Intestinal dysbiosis | Intestinal dysbiosis associated with activation of TLRs, inflammasomes, and stimulation of immune response[296,303,304,306,307,309] | Present in diverse liver and non-liver autoimmune diseases[249,302,303,318,323,325] |
| Deficient structural proteins of mucosal barrier in AIH[325] | ||
| Gut-derived activated immune cells migrate to peripheral lymph nodes[310,311] | ||
| Circulating gut-derived bacterial lipopolysaccharide in AIH[325] | ||
| Transfer experiments using intestinal microbiota affect female bias for diabetes[300,332,333] | ||
| Decreased intestinal anaerobes in AIH[325] | ||
| Exposure to gut-derived microbial products at young age may protect against intolerance to self-antigens (“hygiene hypothesis”)[334-337] | ||
| Dysbiosis associated with flares in experimental AIH[326] | ||
| May influence female gender bias in autoimmune disease[300,332,333] |
AIH: Autoimmune hepatitis; TCR: T cell antigen receptor; TLRs: Toll-like receptors; Tregs: Regulatory T cells.
Intestinal dysbiosis
The intestinal microbiome shapes the intestinal and systemic immune responses[291-296] (Figure 1). Its composition varies by gender, ethnicity, age, long-term diet, and socioeconomic status[297-300], and it constitutes an environmental variable that may impact on the predisposition, occurrence, and clinical phenotype of autoimmune disease. Bacterial components of the intestinal microbiome can activate Toll-like receptors (TLRs)[296,301-303], contribute to the formation of inflammasomes[304-307], stimulate the systemic immune response[293,296,308,309], and activate immune cells within the intestine that migrate to peripheral lymphoid tissue[310,311]. Changes in the microbial composition of the intestine (dysbiosis) have already been implicated in type 1 diabetes[312-314], rheumatoid arthritis[303,315-317], multiple sclerosis[318], inflammatory bowel disease[319-321], and diverse liver diseases, including NAFLD[306], PBC[249,322], PSC[323,324], and autoimmune hepatitis[325,326].
Intestinal microbiome and autoimmune hepatitis
Patients with autoimmune hepatitis have been distinguished from healthy individuals by deficiencies in the structural proteins (zona occludens 1 and occludin) that maintain integrity of the gastrointestinal mucosal barrier[325] (Table 4). They also have had increased plasma levels of gut-derived bacterial lipopolysaccharide and decreased numbers of intestinal anaerobes (dysbiosis)[325]. Reduced diversity and total load of gut bacteria have also been associated with exacerbations of experimental autoimmune hepatitis in HLA DRB1*03-positive transgenic mice[326]. The findings suggest that autoimmune hepatitis is associated with dysbiosis, weakened gastrointestinal mucosal barrier, and translocation of gut-derived microbial products into the systemic circulation. The challenge is to establish the sequence of pathogenic events and distinguish causes from consequences of the liver disease.
Intestinal microbiome and gender bias
Autoimmune hepatitis occurs predominately in females, and this strong gender bias is evident in both children (where girls constitute 60%-76% of patients with autoimmune hepatitis) and adults (where women constitute 71%-95% of patients with autoimmune hepatitis)[64]. Explanations for the female propensity for autoimmune diseases have included estrogen effects on cytokine pathways[327] and gene expression[328], fetal microchimerism[329,330], and variable X-chromosomal inactivation[331]. The female bias for autoimmune disease may also be explained in part by gender-specific differences in the composition of the intestinal microbiome[300,332,333] (Table 4).
Commensal microbes colonizing the intestine of male non-obese diabetic mice (NOD) can raise serum testosterone levels and protect them against the development of type 1 diabetes[300]. Transfer of the intestinal microbiota from mature male NOD mice to immature female NOD mice can protect the females from developing type 1 diabetes. Blockade of the androgen receptor can have a similar protective effect[300]. Investigations of gender differences in the intestinal microbiome of patients with autoimmune hepatitis are needed to understand and possibly manipulate this pathogenic aspect.
Intestinal microbiome and the “hygiene hypothesis”
The “hygiene hypothesis” speculates that the rising risk of allergy (asthma, hay fever) and autoimmunity (type 1 diabetes, multiple sclerosis) in western countries relates to the decreasing incidence of infection and microbial exposure in childhood which in turn impairs development of the immune system[334-337]. Microbial exposure can promote maturation of the immune system by stimulating TLRs, inducing Tregs, increasing production of anti-inflammatory cytokines (e.g., IL-10), and generating an immune response that diminishes immune reactivity against self-antigens[334,338,339].
The intestinal microbiome has emerged as a key factor in maintaining immune tolerance by allowing systemic exposure to microbial products and activated immune cells during childhood and protecting against intestinal colonization by non-commensal micro-organisms[33,340] (Table 4). Management strategies using extracts of bacterial and parasitic components have reduced the risk of type 1 diabetes in NOD mice, and the findings suggest that manipulations of the intestinal microbiome at a young age may protect against autoimmune disease[341,342]. The possibility that the intestinal microbiome can influence the risk, gender bias, severity, and outcome of autoimmune hepatitis is a compelling reason to investigate it further as a protective and pathogenic factor in this disease.
TRANSITIONING FROM PATHOGENIC CONCEPTS TO NEXT GENERATION MANAGEMENT
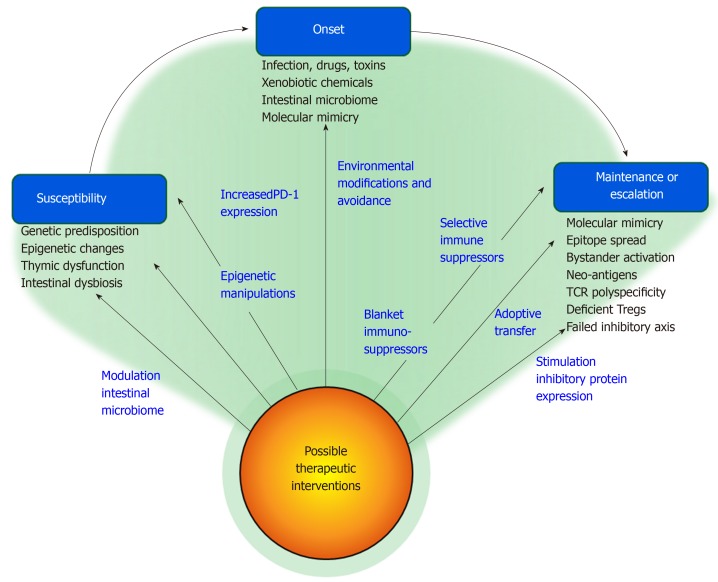
Translation of the pathogenic concepts into next generation management requires an understanding of each phase of autoimmune hepatitis from susceptibility to triggering events to mechanisms that sustain or intensify the inflammatory activity (Figure 2). The science of autoimmune hepatitis has not yet achieved this level of comprehension, but the opportunities to enlarge the knowledge base and impact on future management are plentiful.
Figure 2.
Pathogenic mechanisms and factors associated with susceptibility, onset, and maintenance of autoimmune hepatitis and possible therapeutic interventions. Pathogenic mechanisms and factors are shown, and possible therapeutic interventions for each aspect of the disease are indicated alongside the arrows. PD-1: Programmed cell death protein-1.
Increased host susceptibility to an environmental factor is supported by genetic and epigenetic findings[16,18,25], and this susceptibility may be enhanced by defects in the negative selection of autoreactive immune cells within the thymus and alterations in the intestinal microbiome[27,46,112] (Figure 2). Host susceptibility can be difficult to modify, but future investigations might validate mechanisms that ensure the expression of PD-1 on thymocytes and maturing lymphocytes[22], prevent or reverse DNA demethylation[25,343], or prime the intestinal microbiome to protect against autoreactive responses[341,342].
Progress has already been made in demonstrating that epigenetic manipulation is possible in the laboratory[344,345]. The methyl residue on S-adenosylmethionine can silence aberrant gene expression by inhibiting DNA demethylation directly or indirectly in cell culture[344]. The methyl groups donated by S-adenosylmethionine have a direct inhibitory effect on DNA demethylation, and they also impair the activity of the nuclear protein, methyl-DNA-binding domain protein 2 (MBD2). MBD2 binds specifically to methylated DNA, has demethylase activity[89,346,347], and is inversely associated with DNA hypo-methylation in patients with SLE[348]. Investigational interventions that alter DNA demethylation directly or indirectly by methyl group donation[344] or the administration of anti-sense oligonucleotides that inhibit MBD2[345] have the prospect of dampening transcriptional activity of pro-inflammatory genes and reducing susceptibility for autoimmune hepatitis.
The identification of the triggering factors for autoimmune hepatitis is a key requirement for the development of protective strategies that might include environmental modifications or specific avoidance behaviors (Figure 2). Population-based epidemiological studies that are designed to evaluate the association of infection, drugs, toxins, and xenobiotic chemicals with autoimmune hepatitis would help in identifying these triggers, and clarification of the role of the intestinal microbiome in affecting susceptibility, gender bias, and outcome could identify a modifiable source of microbial products and activated immune cells that might affect the occurrence and course of autoimmune hepatitis.
Multiple immune-mediated liver (NAFLD[306], PSC[323,324], and autoimmune hepatitis[325,326]) and non-liver (type 1 diabetes[313,314], rheumatoid arthritis[303,315,316], multiple sclerosis[318], and inflammatory bowel disease[319-321]) diseases have already been linked to intestinal dysbiosis, and modifications of the intestinal microbiome by diet, probiotics, antibiotics, or re-colonization are possible[46]. A preliminary randomized clinical trial of antibiotic therapy in PSC has suggested that therapy with vancomycin or metronidazole has the potential to improve liver tests and Mayo PSC risk score[349], presumably by favorably altering the intestinal microbiome[350].
Molecular mimicry, epitope spread, bystander activation, neo-antigen formation, TCR polyspecificity, and deficient immune inhibitory mechanisms (Tregs[351] and immune inhibitory proteins[22]) may extend an inciting event into a self-perpetuating autoimmune process (Figure 2). Multiple disturbances in the homeostatic pathways affecting activation[352], migration[353], and survival[354] of T lymphocytes have already been described that may be contributory. Deficiencies in the number and function of Tregs have been implicated[355,356] (and challenged[357]), and failure of the immune inhibitory proteins, especially PD-1, to dampen the differentiation and proliferation of autoreactive cells has been an evolving area of investigational interest[22,358]. These disturbances may in turn relate to epigenetic changes that affect the transcription of immune regulatory genes, and these genes are already being manipulated experimentally[89,343,346].
Management strategies when the liver disease is established will depend on whether multiple factors must participate concurrently to sustain or escalate the disease or a predominant critical defect can be identified and targeted. Blanket immunosuppression may be necessary to suppress multiple concurrent pathogenic pathways, whereas highly specific interventions would require demonstration of a correctable pathogenic linchpin that supports the disease (Figure 2). Monoclonal antibodies against pro-inflammatory cytokines or B cells[359,360] and recombinant molecules that dampen T cell activation by increasing expression of the immune inhibitory proteins (recombinant CTLA-4[361-363] or PD-1[364]) are possible supplemental therapies. The adoptive transfer of antigen-specific, induced Tregs, mesenchymal stromal cells, or myeloid-derived suppressor cells exemplifies a next-generation intervention that might replace blanket immunosuppression[365,366].
CONCLUSION
Multiple pathogenic concepts have emerged to explain the susceptibility, onset, and maintenance of autoimmune hepatitis (Figure 1). Each concept is insufficient or incomplete. The risk burden for autoimmune hepatitis cannot be explained by genetic susceptibility; the triggering events of autoimmune hepatitis remain unknown; the factors that sustain or escalate the disease are unclear; and the dominant defect or particular aggregate of defects in immune homeostasis that could shape next generation therapy is unresolved.
Pathogenic concepts that are insufficient or incomplete are not wrong, and their deficiencies should drive investigations that define their impact more fully. The knowledge base of autoimmune hepatitis now includes evolving concepts of susceptibility, onset, and maintenance that should encourage investigations of acquired epigenetic change in gene expression, thymic dysfunction, molecular mimicry, epitope spread, and intestinal dysbiosis (Figure 2).
The next generation of management for autoimmune hepatitis will depend on clarification of the pathogenic sequence from susceptibility to established clinical phenotype, and it could include a strategy for prevention. Certain mechanisms may be evident throughout the course of the disease, and they may indicate a pathogenic linchpin that can be targeted selectively. Alternatively, certain mechanisms may require the alignment of several concurrent disruptions in immune homeostasis, and they may require blanket immunosuppression, possibly supplemented by site-specific interventions that target prominent contributory mechanisms. Studies that move the pathogenesis of autoimmune hepatitis closer to its cause may result in management strategies with a greater potential to replace blanket immunosuppressive regimens than interventions directed at downstream defects in the homeostatic network which are likely to yield supplemental rather than replacement interventions.
Footnotes
Conflict-of-interest statement: Albert J Czaja has no conflict of interests to declare.
Manuscript source: Invited Manuscript
Peer-review started: November 1, 2019
First decision: November 22, 2019
Article in press: November 29, 2019
Specialty type: Gastroenterology and hepatology
Country of origin: United States
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Aguilera I, Muratori P, Rajcani J S-Editor: Tang JZ L-Editor: A E-Editor: Ma YJ
References
- 1.Alvarez F, Berg PA, Bianchi FB, Bianchi L, Burroughs AK, Cancado EL, Chapman RW, Cooksley WG, Czaja AJ, Desmet VJ, Donaldson PT, Eddleston AL, Fainboim L, Heathcote J, Homberg JC, Hoofnagle JH, Kakumu S, Krawitt EL, Mackay IR, MacSween RN, Maddrey WC, Manns MP, McFarlane IG, Meyer zum Büschenfelde KH, Zeniya M. International Autoimmune Hepatitis Group Report: review of criteria for diagnosis of autoimmune hepatitis. J Hepatol. 1999;31:929–938. doi: 10.1016/s0168-8278(99)80297-9. [DOI] [PubMed] [Google Scholar]
- 2.Manns MP, Czaja AJ, Gorham JD, Krawitt EL, Mieli-Vergani G, Vergani D, Vierling JM American Association for the Study of Liver Diseases. Diagnosis and management of autoimmune hepatitis. Hepatology. 2010;51:2193–2213. doi: 10.1002/hep.23584. [DOI] [PubMed] [Google Scholar]
- 3.Czaja AJ, Manns MP. The validity and importance of subtypes in autoimmune hepatitis: a point of view. Am J Gastroenterol. 1995;90:1206–1211. [PubMed] [Google Scholar]
- 4.Czaja AJ. Performance parameters of the conventional serological markers for autoimmune hepatitis. Dig Dis Sci. 2011;56:545–554. doi: 10.1007/s10620-010-1501-1. [DOI] [PubMed] [Google Scholar]
- 5.Czaja AJ, Cassani F, Cataleta M, Valentini P, Bianchi FB. Frequency and significance of antibodies to actin in type 1 autoimmune hepatitis. Hepatology. 1996;24:1068–1073. doi: 10.1002/hep.510240515. [DOI] [PubMed] [Google Scholar]
- 6.Chretien-Leprince P, Ballot E, Andre C, Olsson NO, Fabien N, Escande A, Oksman F, Dubuquoi S, Jego S, Goetz J, Chevailler A, Sanmarco M, Humbel RL, Johanet C. Diagnostic value of anti-F-actin antibodies in a French multicenter study. Ann N Y Acad Sci. 2005;1050:266–273. doi: 10.1196/annals.1313.028. [DOI] [PubMed] [Google Scholar]
- 7.Frenzel C, Herkel J, Lüth S, Galle PR, Schramm C, Lohse AW. Evaluation of F-actin ELISA for the diagnosis of autoimmune hepatitis. Am J Gastroenterol. 2006;101:2731–2736. doi: 10.1111/j.1572-0241.2006.00830.x. [DOI] [PubMed] [Google Scholar]
- 8.Couto CA, Bittencourt PL, Porta G, Abrantes-Lemos CP, Carrilho FJ, Guardia BD, Cançado EL. Antismooth muscle and antiactin antibodies are indirect markers of histological and biochemical activity of autoimmune hepatitis. Hepatology. 2014;59:592–600. doi: 10.1002/hep.26666. [DOI] [PubMed] [Google Scholar]
- 9.Manns M, Gerken G, Kyriatsoulis A, Staritz M, Meyer zum Büschenfelde KH. Characterisation of a new subgroup of autoimmune chronic active hepatitis by autoantibodies against a soluble liver antigen. Lancet. 1987;1:292–294. doi: 10.1016/s0140-6736(87)92024-1. [DOI] [PubMed] [Google Scholar]
- 10.Montano-Loza AJ, Shums Z, Norman GL, Czaja AJ. Prognostic implications of antibodies to Ro/SSA and soluble liver antigen in type 1 autoimmune hepatitis. Liver Int. 2012;32:85–92. doi: 10.1111/j.1478-3231.2011.02502.x. [DOI] [PubMed] [Google Scholar]
- 11.Targan SR, Landers C, Vidrich A, Czaja AJ. High-titer antineutrophil cytoplasmic antibodies in type-1 autoimmune hepatitis. Gastroenterology. 1995;108:1159–1166. doi: 10.1016/0016-5085(95)90215-5. [DOI] [PubMed] [Google Scholar]
- 12.Zauli D, Ghetti S, Grassi A, Descovich C, Cassani F, Ballardini G, Muratori L, Bianchi FB. Anti-neutrophil cytoplasmic antibodies in type 1 and 2 autoimmune hepatitis. Hepatology. 1997;25:1105–1107. doi: 10.1002/hep.510250510. [DOI] [PubMed] [Google Scholar]
- 13.Abuaf N, Johanet C, Chretien P, Martini E, Soulier E, Laperche S, Homberg JC. Characterization of the liver cytosol antigen type 1 reacting with autoantibodies in chronic active hepatitis. Hepatology. 1992;16:892–898. doi: 10.1002/hep.1840160407. [DOI] [PubMed] [Google Scholar]
- 14.Lapierre P, Hajoui O, Homberg JC, Alvarez F. Formiminotransferase cyclodeaminase is an organ-specific autoantigen recognized by sera of patients with autoimmune hepatitis. Gastroenterology. 1999;116:643–649. doi: 10.1016/s0016-5085(99)70186-1. [DOI] [PubMed] [Google Scholar]
- 15.Muratori L, Sztul E, Muratori P, Gao Y, Ripalti A, Ponti C, Lenzi M, Landini MP, Bianchi FB. Distinct epitopes on formiminotransferase cyclodeaminase induce autoimmune liver cytosol antibody type 1. Hepatology. 2001;34:494–501. doi: 10.1053/jhep.2001.27179. [DOI] [PubMed] [Google Scholar]
- 16.Strettell MD, Donaldson PT, Thomson LJ, Santrach PJ, Moore SB, Czaja AJ, Williams R. Allelic basis for HLA-encoded susceptibility to type 1 autoimmune hepatitis. Gastroenterology. 1997;112:2028–2035. doi: 10.1053/gast.1997.v112.pm9178696. [DOI] [PubMed] [Google Scholar]
- 17.Czaja AJ. Genetic factors affecting the occurrence, clinical phenotype, and outcome of autoimmune hepatitis. Clin Gastroenterol Hepatol. 2008;6:379–388. doi: 10.1016/j.cgh.2007.12.048. [DOI] [PubMed] [Google Scholar]
- 18.van Gerven NM, de Boer YS, Zwiers A, Verwer BJ, Drenth JP, van Hoek B, van Erpecum KJ, Beuers U, van Buuren HR, den Ouden JW, Verdonk RC, Koek GH, Brouwer JT, Guichelaar MM, Vrolijk JM, Coenraad MJ, Kraal G, Mulder CJ, van Nieuwkerk CM, Bloemena E, Verspaget HW, Kumar V, Zhernakova A, Wijmenga C, Franke L, Bouma G Dutch Autoimmune Hepatitis Study Group. HLA-DRB1*03:01 and HLA-DRB1*04:01 modify the presentation and outcome in autoimmune hepatitis type-1. Genes Immun. 2015;16:247–252. doi: 10.1038/gene.2014.82. [DOI] [PubMed] [Google Scholar]
- 19.Czaja AJ. Transitioning from Idiopathic to Explainable Autoimmune Hepatitis. Dig Dis Sci. 2015;60:2881–2900. doi: 10.1007/s10620-015-3708-7. [DOI] [PubMed] [Google Scholar]
- 20.Liberal R, Mieli-Vergani G, Vergani D. Autoimmune hepatitis: From mechanisms to therapy. Rev Clin Esp. 2016;216:372–383. doi: 10.1016/j.rce.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Mieli-Vergani G, Vergani D, Czaja AJ, Manns MP, Krawitt EL, Vierling JM, Lohse AW, Montano-Loza AJ. Autoimmune hepatitis. Nat Rev Dis Primers. 2018;4:18017. doi: 10.1038/nrdp.2018.17. [DOI] [PubMed] [Google Scholar]
- 22.Czaja AJ. Immune inhibitory proteins and their pathogenic and therapeutic implications in autoimmunity and autoimmune hepatitis. Autoimmunity. 2019;52:144–160. doi: 10.1080/08916934.2019.1641200. [DOI] [PubMed] [Google Scholar]
- 23.Webb GJ, Hirschfield GM. Using GWAS to identify genetic predisposition in hepatic autoimmunity. J Autoimmun. 2016;66:25–39. doi: 10.1016/j.jaut.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 24.Czaja AJ. Under-Evaluated or Unassessed Pathogenic Pathways in Autoimmune Hepatitis and Implications for Future Management. Dig Dis Sci. 2018;63:1706–1725. doi: 10.1007/s10620-018-5072-x. [DOI] [PubMed] [Google Scholar]
- 25.Czaja AJ. Epigenetic changes and their implications in autoimmune hepatitis. Eur J Clin Invest. 2018;48 doi: 10.1111/eci.12899. [DOI] [PubMed] [Google Scholar]
- 26.Mann DA. Epigenetics in liver disease. Hepatology. 2014;60:1418–1425. doi: 10.1002/hep.27131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robey E, Fowlkes BJ. Selective events in T cell development. Annu Rev Immunol. 1994;12:675–705. doi: 10.1146/annurev.iy.12.040194.003331. [DOI] [PubMed] [Google Scholar]
- 28.Nossal GJ. Negative selection of lymphocytes. Cell. 1994;76:229–239. doi: 10.1016/0092-8674(94)90331-x. [DOI] [PubMed] [Google Scholar]
- 29.Eichmann K. A signal strength hypothesis of thymic selection: preliminary considerations. Immunol Lett. 1995;44:87–90. doi: 10.1016/0165-2478(94)00197-y. [DOI] [PubMed] [Google Scholar]
- 30.Cusick MF, Libbey JE, Fujinami RS. Molecular mimicry as a mechanism of autoimmune disease. Clin Rev Allergy Immunol. 2012;42:102–111. doi: 10.1007/s12016-011-8294-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oldstone MB. Molecular mimicry: its evolution from concept to mechanism as a cause of autoimmune diseases. Monoclon Antib Immunodiagn Immunother. 2014;33:158–165. doi: 10.1089/mab.2013.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose NR. Molecular mimicry and clonal deletion: A fresh look. J Theor Biol. 2015;375:71–76. doi: 10.1016/j.jtbi.2014.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rojas M, Restrepo-Jiménez P, Monsalve DM, Pacheco Y, Acosta-Ampudia Y, Ramírez-Santana C, Leung PSC, Ansari AA, Gershwin ME, Anaya JM. Molecular mimicry and autoimmunity. J Autoimmun. 2018;95:100–123. doi: 10.1016/j.jaut.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 34.Getts DR, Chastain EM, Terry RL, Miller SD. Virus infection, antiviral immunity, and autoimmunity. Immunol Rev. 2013;255:197–209. doi: 10.1111/imr.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Floreani A, Leung PS, Gershwin ME. Environmental Basis of Autoimmunity. Clin Rev Allergy Immunol. 2016;50:287–300. doi: 10.1007/s12016-015-8493-8. [DOI] [PubMed] [Google Scholar]
- 36.Vogel A, Manns MP, Strassburg CP. Autoimmunity and viruses. Clin Liver Dis. 2002;6:739–753. doi: 10.1016/s1089-3261(02)00024-7. [DOI] [PubMed] [Google Scholar]
- 37.Vanderlugt CJ, Miller SD. Epitope spreading. Curr Opin Immunol. 1996;8:831–836. doi: 10.1016/S0952-7915(96)80012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanderlugt CL, Begolka WS, Neville KL, Katz-Levy Y, Howard LM, Eagar TN, Bluestone JA, Miller SD. The functional significance of epitope spreading and its regulation by co-stimulatory molecules. Immunol Rev. 1998;164:63–72. doi: 10.1111/j.1600-065x.1998.tb01208.x. [DOI] [PubMed] [Google Scholar]
- 39.Tsunoda I, Libbey JE, Fujinami RS. Sequential polymicrobial infections lead to CNS inflammatory disease: possible involvement of bystander activation in heterologous immunity. J Neuroimmunol. 2007;188:22–33. doi: 10.1016/j.jneuroim.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hintermann E, Holdener M, Bayer M, Loges S, Pfeilschifter JM, Granier C, Manns MP, Christen U. Epitope spreading of the anti-CYP2D6 antibody response in patients with autoimmune hepatitis and in the CYP2D6 mouse model. J Autoimmun. 2011;37:242–253. doi: 10.1016/j.jaut.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Cornaby C, Gibbons L, Mayhew V, Sloan CS, Welling A, Poole BD. B cell epitope spreading: mechanisms and contribution to autoimmune diseases. Immunol Lett. 2015;163:56–68. doi: 10.1016/j.imlet.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Rosen A, Casciola-Rosen L. Autoantigens in systemic autoimmunity: critical partner in pathogenesis. J Intern Med. 2009;265:625–631. doi: 10.1111/j.1365-2796.2009.02102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Padovan E, Casorati G, Dellabona P, Meyer S, Brockhaus M, Lanzavecchia A. Expression of two T cell receptor alpha chains: dual receptor T cells. Science. 1993;262:422–424. doi: 10.1126/science.8211163. [DOI] [PubMed] [Google Scholar]
- 44.Blichfeldt E, Munthe LA, Røtnes JS, Bogen B. Dual T cell receptor T cells have a decreased sensitivity to physiological ligands due to reduced density of each T cell receptor. Eur J Immunol. 1996;26:2876–2884. doi: 10.1002/eji.1830261211. [DOI] [PubMed] [Google Scholar]
- 45.Wucherpfennig KW, Allen PM, Celada F, Cohen IR, De Boer R, Garcia KC, Goldstein B, Greenspan R, Hafler D, Hodgkin P, Huseby ES, Krakauer DC, Nemazee D, Perelson AS, Pinilla C, Strong RK, Sercarz EE. Polyspecificity of T cell and B cell receptor recognition. Semin Immunol. 2007;19:216–224. doi: 10.1016/j.smim.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Czaja AJ. Factoring the intestinal microbiome into the pathogenesis of autoimmune hepatitis. World J Gastroenterol. 2016;22:9257–9278. doi: 10.3748/wjg.v22.i42.9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Czaja AJ, Strettell MD, Thomson LJ, Santrach PJ, Moore SB, Donaldson PT, Williams R. Associations between alleles of the major histocompatibility complex and type 1 autoimmune hepatitis. Hepatology. 1997;25:317–323. doi: 10.1002/hep.510250211. [DOI] [PubMed] [Google Scholar]
- 48.Czaja AJ, Cookson S, Constantini PK, Clare M, Underhill JA, Donaldson PT. Cytokine polymorphisms associated with clinical features and treatment outcome in type 1 autoimmune hepatitis. Gastroenterology. 1999;117:645–652. doi: 10.1016/s0016-5085(99)70458-0. [DOI] [PubMed] [Google Scholar]
- 49.Seki T, Kiyosawa K, Inoko H, Ota M. Association of autoimmune hepatitis with HLA-Bw54 and DR4 in Japanese patients. Hepatology. 1990;12:1300–1304. doi: 10.1002/hep.1840120609. [DOI] [PubMed] [Google Scholar]
- 50.Goldberg AC, Bittencourt PL, Mougin B, Cançado EL, Porta G, Carrilho F, Kalil J. Analysis of HLA haplotypes in autoimmune hepatitis type 1: identifying the major susceptibility locus. Hum Immunol. 2001;62:165–169. doi: 10.1016/s0198-8859(00)00234-2. [DOI] [PubMed] [Google Scholar]
- 51.Qiu DK, Ma X. Relationship between human leukocyte antigen-DRB1 and autoimmune hepatitis type I in Chinese patients. J Gastroenterol Hepatol. 2003;18:63–67. doi: 10.1046/j.1440-1746.2003.02918.x. [DOI] [PubMed] [Google Scholar]
- 52.Vázquez-García MN, Aláez C, Olivo A, Debaz H, Pérez-Luque E, Burguete A, Cano S, de la Rosa G, Bautista N, Hernández A, Bandera J, Torres LF, Kershenobich D, Alvarez F, Gorodezky C. MHC class II sequences of susceptibility and protection in Mexicans with autoimmune hepatitis. J Hepatol. 1998;28:985–990. doi: 10.1016/s0168-8278(98)80347-4. [DOI] [PubMed] [Google Scholar]
- 53.Czaja AJ, Souto EO, Bittencourt PL, Cancado EL, Porta G, Goldberg AC, Donaldson PT. Clinical distinctions and pathogenic implications of type 1 autoimmune hepatitis in Brazil and the United States. J Hepatol. 2002;37:302–308. doi: 10.1016/s0168-8278(02)00182-4. [DOI] [PubMed] [Google Scholar]
- 54.Fortes Mdel P, Machado IV, Gil G, Fernández-Mestre M, Dagher L, León RV, Bianco NE, Tassinari P. Genetic contribution of major histocompatibility complex class II region to type 1 autoimmune hepatitis susceptibility in Venezuela. Liver Int. 2007;27:1409–1416. doi: 10.1111/j.1478-3231.2007.01581.x. [DOI] [PubMed] [Google Scholar]
- 55.Lim YS, Oh HB, Choi SE, Kwon OJ, Heo YS, Lee HC, Suh DJ. Susceptibility to type 1 autoimmune hepatitis is associated with shared amino acid sequences at positions 70-74 of the HLA-DRB1 molecule. J Hepatol. 2008;48:133–139. doi: 10.1016/j.jhep.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 56.Chaouali M, Carvalho A, Tezeghdenti A, Ben Azaiez M, Cunha C, Ghazouani E, Kochkar R. Cytotoxic T lymphocyte antigen-4 gene polymorphisms and susceptibility to type 1 autoimmune hepatitis in the Tunisian population. Genes Dis. 2018;5:256–262. doi: 10.1016/j.gendis.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Czaja AJ, Carpenter HA. Distinctive clinical phenotype and treatment outcome of type 1 autoimmune hepatitis in the elderly. Hepatology. 2006;43:532–538. doi: 10.1002/hep.21074. [DOI] [PubMed] [Google Scholar]
- 58.Pando M, Larriba J, Fernandez GC, Fainboim H, Ciocca M, Ramonet M, Badia I, Daruich J, Findor J, Tanno H, Cañero-Velasco C, Fainboim L. Pediatric and adult forms of type I autoimmune hepatitis in Argentina: evidence for differential genetic predisposition. Hepatology. 1999;30:1374–1380. doi: 10.1002/hep.510300611. [DOI] [PubMed] [Google Scholar]
- 59.Czaja AJ, Donaldson PT. Genetic susceptibilities for immune expression and liver cell injury in autoimmune hepatitis. Immunol Rev. 2000;174:250–259. doi: 10.1034/j.1600-0528.2002.017401.x. [DOI] [PubMed] [Google Scholar]
- 60.Agarwal K, Czaja AJ, Jones DE, Donaldson PT. Cytotoxic T lymphocyte antigen-4 (CTLA-4) gene polymorphisms and susceptibility to type 1 autoimmune hepatitis. Hepatology. 2000;31:49–53. doi: 10.1002/hep.510310110. [DOI] [PubMed] [Google Scholar]
- 61.Agarwal K, Czaja AJ, Donaldson PT. A functional Fas promoter polymorphism is associated with a severe phenotype in type 1 autoimmune hepatitis characterized by early development of cirrhosis. Tissue Antigens. 2007;69:227–235. doi: 10.1111/j.1399-0039.2006.00794.x. [DOI] [PubMed] [Google Scholar]
- 62.Cookson S, Constantini PK, Clare M, Underhill JA, Bernal W, Czaja AJ, Donaldson PT. Frequency and nature of cytokine gene polymorphisms in type 1 autoimmune hepatitis. Hepatology. 1999;30:851–856. doi: 10.1002/hep.510300412. [DOI] [PubMed] [Google Scholar]
- 63.Muratori P, Czaja AJ, Muratori L, Pappas G, Maccariello S, Cassani F, Granito A, Ferrari R, Mantovani V, Lenzi M, Bianchi FB. Genetic distinctions between autoimmune hepatitis in Italy and North America. World J Gastroenterol. 2005;11:1862–1866. doi: 10.3748/wjg.v11.i12.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Czaja AJ. Global Disparities and Their Implications in the Occurrence and Outcome of Autoimmune Hepatitis. Dig Dis Sci. 2017;62:2277–2292. doi: 10.1007/s10620-017-4675-y. [DOI] [PubMed] [Google Scholar]
- 65.de Boer YS, van Gerven NM, Zwiers A, Verwer BJ, van Hoek B, van Erpecum KJ, Beuers U, van Buuren HR, Drenth JP, den Ouden JW, Verdonk RC, Koek GH, Brouwer JT, Guichelaar MM, Vrolijk JM, Kraal G, Mulder CJ, van Nieuwkerk CM, Fischer J, Berg T, Stickel F, Sarrazin C, Schramm C, Lohse AW, Weiler-Normann C, Lerch MM, Nauck M, Völzke H, Homuth G, Bloemena E, Verspaget HW, Kumar V, Zhernakova A, Wijmenga C, Franke L, Bouma G Dutch Autoimmune Hepatitis Study Group; LifeLines Cohort Study; Study of Health in Pomerania. Genome-wide association study identifies variants associated with autoimmune hepatitis type 1. Gastroenterology. 2014;147:443–52.e5. doi: 10.1053/j.gastro.2014.04.022. [DOI] [PubMed] [Google Scholar]
- 66.Agarwal K, Jones DE, Daly AK, James OF, Vaidya B, Pearce S, Bassendine MF. CTLA-4 gene polymorphism confers susceptibility to primary biliary cirrhosis. J Hepatol. 2000;32:538–541. doi: 10.1016/s0168-8278(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 67.Jeffries MA, Sawalha AH. Autoimmune disease in the epigenetic era: how has epigenetics changed our understanding of disease and how can we expect the field to evolve? Expert Rev Clin Immunol. 2015;11:45–58. doi: 10.1586/1744666X.2015.994507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sydow JF, Cramer P. RNA polymerase fidelity and transcriptional proofreading. Curr Opin Struct Biol. 2009;19:732–739. doi: 10.1016/j.sbi.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 69.Tough DF, Prinjha RK. Immune disease-associated variants in gene enhancers point to BET epigenetic mechanisms for therapeutic intervention. Epigenomics. 2017;9:573–584. doi: 10.2217/epi-2016-0144. [DOI] [PubMed] [Google Scholar]
- 70.Campanero MR, Armstrong MI, Flemington EK. CpG methylation as a mechanism for the regulation of E2F activity. Proc Natl Acad Sci USA. 2000;97:6481–6486. doi: 10.1073/pnas.100340697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 72.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 74.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 75.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 76.Iborra M, Bernuzzi F, Invernizzi P, Danese S. MicroRNAs in autoimmunity and inflammatory bowel disease: crucial regulators in immune response. Autoimmun Rev. 2012;11:305–314. doi: 10.1016/j.autrev.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 77.Cedar H, Bergman Y. Programming of DNA methylation patterns. Annu Rev Biochem. 2012;81:97–117. doi: 10.1146/annurev-biochem-052610-091920. [DOI] [PubMed] [Google Scholar]
- 78.Schomacher L. Mammalian DNA demethylation: multiple faces and upstream regulation. Epigenetics. 2013;8:679–684. doi: 10.4161/epi.24977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guo JU, Su Y, Zhong C, Ming GL, Song H. Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle. 2011;10:2662–2668. doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, Berdasco M, Fraga MF, O'Hanlon TP, Rider LG, Jacinto FV, Lopez-Longo FJ, Dopazo J, Forn M, Peinado MA, Carreño L, Sawalha AH, Harley JB, Siebert R, Esteller M, Miller FW, Ballestar E. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Coit P, Jeffries M, Altorok N, Dozmorov MG, Koelsch KA, Wren JD, Merrill JT, McCune WJ, Sawalha AH. Genome-wide DNA methylation study suggests epigenetic accessibility and transcriptional poising of interferon-regulated genes in naïve CD4+ T cells from lupus patients. J Autoimmun. 2013;43:78–84. doi: 10.1016/j.jaut.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lleo A, Zhang W, Zhao M, Tan Y, Bernuzzi F, Zhu B, Liu Q, Tan Q, Malinverno F, Valenti L, Jiang T, Tan L, Liao W, Coppel R, Invernizzi P, Lu Q, Adams DH, Gershwin ME PBC Epigenetic Study Group. DNA methylation profiling of the X chromosome reveals an aberrant demethylation on CXCR3 promoter in primary biliary cirrhosis. Clin Epigenetics. 2015;7:61. doi: 10.1186/s13148-015-0098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Javierre BM, Hernando H, Ballestar E. Environmental triggers and epigenetic deregulation in autoimmune disease. Discov Med. 2011;12:535–545. [PubMed] [Google Scholar]
- 86.Osley MA, Fleming AB, Kao CF. Histone ubiquitylation and the regulation of transcription. Results Probl Cell Differ. 2006;41:47–75. doi: 10.1007/400_006. [DOI] [PubMed] [Google Scholar]
- 87.Wysocka J, Allis CD, Coonrod S. Histone arginine methylation and its dynamic regulation. Front Biosci. 2006;11:344–355. doi: 10.2741/1802. [DOI] [PubMed] [Google Scholar]
- 88.Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, Li B, Turka LA, Olson EN, Greene MI, Wells AD, Hancock WW. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13:1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- 89.Szyf M. Epigenetic therapeutics in autoimmune disease. Clin Rev Allergy Immunol. 2010;39:62–77. doi: 10.1007/s12016-009-8172-8. [DOI] [PubMed] [Google Scholar]
- 90.Gregory PD, Wagner K, Hörz W. Histone acetylation and chromatin remodeling. Exp Cell Res. 2001;265:195–202. doi: 10.1006/excr.2001.5187. [DOI] [PubMed] [Google Scholar]
- 91.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thiagalingam S, Cheng KH, Lee HJ, Mineva N, Thiagalingam A, Ponte JF. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci. 2003;983:84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- 93.Hu N, Qiu X, Luo Y, Yuan J, Li Y, Lei W, Zhang G, Zhou Y, Su Y, Lu Q. Abnormal histone modification patterns in lupus CD4+ T cells. J Rheumatol. 2008;35:804–810. [PubMed] [Google Scholar]
- 94.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 95.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 96.Pratt AJ, MacRae IJ. The RNA-induced silencing complex: a versatile gene-silencing machine. J Biol Chem. 2009;284:17897–17901. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 98.Hoefig KP, Heissmeyer V. MicroRNAs grow up in the immune system. Curr Opin Immunol. 2008;20:281–287. doi: 10.1016/j.coi.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 99.Bi Y, Liu G, Yang R. MicroRNAs: novel regulators during the immune response. J Cell Physiol. 2009;218:467–472. doi: 10.1002/jcp.21639. [DOI] [PubMed] [Google Scholar]
- 100.Harris A, Krams SM, Martinez OM. MicroRNAs as immune regulators: implications for transplantation. Am J Transplant. 2010;10:713–719. doi: 10.1111/j.1600-6143.2010.03032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tomankova T, Petrek M, Gallo J, Kriegova E. MicroRNAs: Emerging Regulators of Immune-Mediated Diseases. Scand J Immunol. 2012;75:129–141. doi: 10.1111/j.1365-3083.2011.02650.x. [DOI] [PubMed] [Google Scholar]
- 102.Haider BA, Baras AS, McCall MN, Hertel JA, Cornish TC, Halushka MK. A critical evaluation of microRNA biomarkers in non-neoplastic disease. PLoS One. 2014;9:e89565. doi: 10.1371/journal.pone.0089565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Migita K, Komori A, Kozuru H, Jiuchi Y, Nakamura M, Yasunami M, Furukawa H, Abiru S, Yamasaki K, Nagaoka S, Hashimoto S, Bekki S, Kamitsukasa H, Nakamura Y, Ohta H, Shimada M, Takahashi H, Mita E, Hijioka T, Yamashita H, Kouno H, Nakamuta M, Ario K, Muro T, Sakai H, Sugi K, Nishimura H, Yoshizawa K, Sato T, Naganuma A, Komatsu T, Oohara Y, Makita F, Tomizawa M, Yatsuhashi H. Circulating microRNA Profiles in Patients with Type-1 Autoimmune Hepatitis. PLoS One. 2015;10:e0136908. doi: 10.1371/journal.pone.0136908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 105.Ando Y, Yang GX, Kenny TP, Kawata K, Zhang W, Huang W, Leung PS, Lian ZX, Okazaki K, Ansari AA, He XS, Invernizzi P, Ridgway WM, Lu Q, Gershwin ME. Overexpression of microRNA-21 is associated with elevated pro-inflammatory cytokines in dominant-negative TGF-β receptor type II mouse. J Autoimmun. 2013;41:111–119. doi: 10.1016/j.jaut.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Adkins B, Mueller C, Okada CY, Reichert RA, Weissman IL, Spangrude GJ. Early events in T-cell maturation. Annu Rev Immunol. 1987;5:325–365. doi: 10.1146/annurev.iy.05.040187.001545. [DOI] [PubMed] [Google Scholar]
- 107.Godfrey DI, Zlotnik A. Control points in early T-cell development. Immunol Today. 1993;14:547–553. doi: 10.1016/0167-5699(93)90186-O. [DOI] [PubMed] [Google Scholar]
- 108.von Boehmer H, Aifantis I, Azogui O, Feinberg J, Saint-Ruf C, Zober C, Garcia C, Buer J. Crucial function of the pre-T-cell receptor (TCR) in TCR beta selection, TCR beta allelic exclusion and alpha beta versus gamma delta lineage commitment. Immunol Rev. 1998;165:111–119. doi: 10.1111/j.1600-065x.1998.tb01234.x. [DOI] [PubMed] [Google Scholar]
- 109.Nishimura H, Honjo T, Minato N. Facilitation of beta selection and modification of positive selection in the thymus of PD-1-deficient mice. J Exp Med. 2000;191:891–898. doi: 10.1084/jem.191.5.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.von Boehmer H. Developmental biology of T cells in T cell-receptor transgenic mice. Annu Rev Immunol. 1990;8:531–556. doi: 10.1146/annurev.iy.08.040190.002531. [DOI] [PubMed] [Google Scholar]
- 111.von Boehmer H. Positive selection of lymphocytes. Cell. 1994;76:219–228. doi: 10.1016/0092-8674(94)90330-1. [DOI] [PubMed] [Google Scholar]
- 112.Jiang TT, Martinov T, Xin L, Kinder JM, Spanier JA, Fife BT, Way SS. Programmed Death-1 Culls Peripheral Accumulation of High-Affinity Autoreactive CD4 T Cells to Protect against Autoimmunity. Cell Rep. 2016;17:1783–1794. doi: 10.1016/j.celrep.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hsieh CS, Zheng Y, Liang Y, Fontenot JD, Rudensky AY. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat Immunol. 2006;7:401–410. doi: 10.1038/ni1318. [DOI] [PubMed] [Google Scholar]
- 114.Moon JJ, Dash P, Oguin TH, 3rd, McClaren JL, Chu HH, Thomas PG, Jenkins MK. Quantitative impact of thymic selection on Foxp3+ and Foxp3- subsets of self-peptide/MHC class II-specific CD4+ T cells. Proc Natl Acad Sci USA. 2011;108:14602–14607. doi: 10.1073/pnas.1109806108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bautista JL, Lio CW, Lathrop SK, Forbush K, Liang Y, Luo J, Rudensky AY, Hsieh CS. Intraclonal competition limits the fate determination of regulatory T cells in the thymus. Nat Immunol. 2009;10:610–617. doi: 10.1038/ni.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Blank C, Brown I, Marks R, Nishimura H, Honjo T, Gajewski TF. Absence of programmed death receptor 1 alters thymic development and enhances generation of CD4/CD8 double-negative TCR-transgenic T cells. J Immunol. 2003;171:4574–4581. doi: 10.4049/jimmunol.171.9.4574. [DOI] [PubMed] [Google Scholar]
- 117.Nielsen C, Ohm-Laursen L, Barington T, Husby S, Lillevang ST. Alternative splice variants of the human PD-1 gene. Cell Immunol. 2005;235:109–116. doi: 10.1016/j.cellimm.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 118.Pen JJ, Keersmaecker BD, Heirman C, Corthals J, Liechtenstein T, Escors D, Thielemans K, Breckpot K. Interference with PD-L1/PD-1 co-stimulation during antigen presentation enhances the multifunctionality of antigen-specific T cells. Gene Ther. 2014;21:262–271. doi: 10.1038/gt.2013.80. [DOI] [PubMed] [Google Scholar]
- 119.Guo Y, Walsh AM, Canavan M, Wechalekar MD, Cole S, Yin X, Scott B, Loza M, Orr C, McGarry T, Bombardieri M, Humby F, Proudman SM, Pitzalis C, Smith MD, Friedman JR, Anderson I, Madakamutil L, Veale DJ, Fearon U, Nagpal S. Immune checkpoint inhibitor PD-1 pathway is down-regulated in synovium at various stages of rheumatoid arthritis disease progression. PLoS One. 2018;13:e0192704. doi: 10.1371/journal.pone.0192704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vento S, Garofano T, Di Perri G, Dolci L, Concia E, Bassetti D. Identification of hepatitis A virus as a trigger for autoimmune chronic hepatitis type 1 in susceptible individuals. Lancet. 1991;337:1183–1187. doi: 10.1016/0140-6736(91)92858-y. [DOI] [PubMed] [Google Scholar]
- 121.Huppertz HI, Treichel U, Gassel AM, Jeschke R, Meyer zum Büschenfelde KH. Autoimmune hepatitis following hepatitis A virus infection. J Hepatol. 1995;23:204–208. doi: 10.1016/0168-8278(95)80336-x. [DOI] [PubMed] [Google Scholar]
- 122.Hilzenrat N, Zilberman D, Klein T, Zur B, Sikuler E. Autoimmune hepatitis in a genetically susceptible patient: is it triggered by acute viral hepatitis A? Dig Dis Sci. 1999;44:1950–1952. doi: 10.1023/a:1026645629103. [DOI] [PubMed] [Google Scholar]
- 123.Grünhage F, Spengler U, Fischer HP, Sauerbruch T. Autoimmune hepatitis--sequel of a relapsing hepatitis A in a 75-year-old woman. Digestion. 2004;70:187–191. doi: 10.1159/000082253. [DOI] [PubMed] [Google Scholar]
- 124.Tabak F, Ozdemir F, Tabak O, Erer B, Tahan V, Ozaras R. Autoimmune hepatitis induced by the prolonged hepatitis A virus infection. Ann Hepatol. 2008;7:177–179. [PubMed] [Google Scholar]
- 125.Kim YD, Kim KA, Rou WS, Lee JS, Song TJ, Bae WK, Kim NH. [A case of autoimmune hepatitis following acute hepatitis A] Korean J Gastroenterol. 2011;57:315–318. doi: 10.4166/kjg.2011.57.5.315. [DOI] [PubMed] [Google Scholar]
- 126.Laskus T, Slusarczyk J. Autoimmune chronic active hepatitis developing after acute type B hepatitis. Dig Dis Sci. 1989;34:1294–1297. doi: 10.1007/BF01537282. [DOI] [PubMed] [Google Scholar]
- 127.Vento S, Cainelli F, Renzini C, Concia E. Autoimmune hepatitis type 2 induced by HCV and persisting after viral clearance. Lancet. 1997;350:1298–1299. doi: 10.1016/S0140-6736(05)62476-2. [DOI] [PubMed] [Google Scholar]
- 128.Vento S, Guella L, Mirandola F, Cainelli F, Di Perri G, Solbiati M, Ferraro T, Concia E. Epstein-Barr virus as a trigger for autoimmune hepatitis in susceptible individuals. Lancet. 1995;346:608–609. doi: 10.1016/s0140-6736(95)91438-2. [DOI] [PubMed] [Google Scholar]
- 129.Kojima K, Nagayama R, Hirama S, Maeda T, Takikawa H, Miyake K, Yamanaka M, Shiga J. Epstein-Barr virus infection resembling autoimmune hepatitis with lactate dehydrogenase and alkaline phosphatase anomaly. J Gastroenterol. 1999;34:706–712. doi: 10.1007/s005350050324. [DOI] [PubMed] [Google Scholar]
- 130.Nobili V, Comparcola D, Sartorelli MR, Devito R, Marcellini M. Autoimmune hepatitis type 1 after Epstein-Barr virus infection. Pediatr Infect Dis J. 2003;22:387. doi: 10.1097/01.inf.0000060825.68086.9c. [DOI] [PubMed] [Google Scholar]
- 131.Cabibi D. Autoimmune hepatitis following Epstein-Barr virus infection. BMJ Case Rep. 2008;2008:bcr0620080071. doi: 10.1136/bcr.06.2008.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zellos A, Spoulou V, Roma-Giannikou E, Karentzou O, Dalekos GN, Theodoridou M. Autoimmune hepatitis type-2 and Epstein-Barr virus infection in a toddler: art of facts or an artifact? Ann Hepatol. 2013;12:147–151. [PubMed] [Google Scholar]
- 133.Al-Hamoudi WK. Severe autoimmune hepatitis triggered by varicella zoster infection. World J Gastroenterol. 2009;15:1004–1006. doi: 10.3748/wjg.15.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hagel S, Bruns T, Herrmann A, Tannapfel A, Stallmach A. Autoimmune hepatitis in an HIV-infected patient: an intriguing association. Int J STD AIDS. 2012;23:448–450. doi: 10.1258/ijsa.2009.009337. [DOI] [PubMed] [Google Scholar]
- 135.Lenzi M, Ballardini G, Fusconi M, Cassani F, Selleri L, Volta U, Zauli D, Bianchi FB. Type 2 autoimmune hepatitis and hepatitis C virus infection. Lancet. 1990;335:258–259. doi: 10.1016/0140-6736(90)90070-l. [DOI] [PubMed] [Google Scholar]
- 136.Czaja AJ, Taswell HF, Rakela J, Schimek CM. Frequency and significance of antibody to hepatitis C virus in severe corticosteroid-treated autoimmune chronic active hepatitis. Mayo Clin Proc. 1991;66:572–582. doi: 10.1016/s0025-6196(12)60515-1. [DOI] [PubMed] [Google Scholar]
- 137.Lunel F, Abuaf N, Frangeul L, Grippon P, Perrin M, Le Coz Y, Valla D, Borotto E, Yamamoto AM, Huraux JM. Liver/kidney microsome antibody type 1 and hepatitis C virus infection. Hepatology. 1992;16:630–636. doi: 10.1002/hep.1840160304. [DOI] [PubMed] [Google Scholar]
- 138.Zeniya M, Aizawa Y, Watanabe F, Kawabe T, Hara M, Sakaguchi M, Toda G. HCV-marker-positive autoimmune-type chronic active hepatitis: a possible relation between HCV infection and liver autoreaction. Liver. 1994;14:206–212. doi: 10.1111/j.1600-0676.1994.tb00075.x. [DOI] [PubMed] [Google Scholar]
- 139.Savage K, Dhillon AP, Schmilovitz-Weiss H, el-Batanony M, Brown D, Dusheiko G, Scheuer PJ. Detection of HCV-RNA in paraffin-embedded liver biopsies from patients with autoimmune hepatitis. J Hepatol. 1995;22:27–34. doi: 10.1016/0168-8278(95)80256-8. [DOI] [PubMed] [Google Scholar]
- 140.Christie KE, Haukenes G. Measles virus-specific IgM antibodies in sera from patients with chronic active hepatitis. J Med Virol. 1983;12:267–272. doi: 10.1002/jmv.1890120406. [DOI] [PubMed] [Google Scholar]
- 141.Robertson DA, Zhang SL, Guy EC, Wright R. Persistent measles virus genome in autoimmune chronic active hepatitis. Lancet. 1987;2:9–11. doi: 10.1016/s0140-6736(87)93051-0. [DOI] [PubMed] [Google Scholar]
- 142.Vento S, Cainelli F, Ferraro T, Concia E. Autoimmune hepatitis type 1 after measles. Am J Gastroenterol. 1996;91:2618–2620. [PubMed] [Google Scholar]
- 143.Toyoda-Akui M, Yokomori H, Kaneko F, Shimizu Y, Takeuchi H, Tahara K, Yoshida H, Kondo H, Motoori T, Ohbu M, Oda M, Hibi T. Association of an overlap syndrome of autoimmune hepatitis and primary biliary cirrhosis with cytomegalovirus infection. Int J Gen Med. 2011;4:397–402. doi: 10.2147/IJGM.S19245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chiba T, Goto S, Yokosuka O, Imazeki F, Tanaka M, Fukai K, Takahashi Y, Tsujimura H, Saisho H. Fatal chronic active Epstein-Barr virus infection mimicking autoimmune hepatitis. Eur J Gastroenterol Hepatol. 2004;16:225–228. doi: 10.1097/00042737-200402000-00017. [DOI] [PubMed] [Google Scholar]
- 145.Christen U, Hintermann E. Pathogen infection as a possible cause for autoimmune hepatitis. Int Rev Immunol. 2014;33:296–313. doi: 10.3109/08830185.2014.921162. [DOI] [PubMed] [Google Scholar]
- 146.Christen U, Hintermann E. Pathogens and autoimmune hepatitis. Clin Exp Immunol. 2019;195:35–51. doi: 10.1111/cei.13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Czaja AJ, Carpenter HA, Santrach PJ, Moore SB, Taswell HF, Homburger HA. Evidence against hepatitis viruses as important causes of severe autoimmune hepatitis in the United States. J Hepatol. 1993;18:342–352. doi: 10.1016/s0168-8278(05)80279-x. [DOI] [PubMed] [Google Scholar]
- 148.Czaja AJ, Magrin S, Fabiano C, Fiorentino G, Diquattro O, Craxi A, Pagliaro L. Hepatitis C virus infection as a determinant of behavior in type 1 autoimmune hepatitis. Dig Dis Sci. 1995;40:33–40. doi: 10.1007/BF02063938. [DOI] [PubMed] [Google Scholar]
- 149.McFarlane IG, Smith HM, Johnson PJ, Bray GP, Vergani D, Williams R. Hepatitis C virus antibodies in chronic active hepatitis: pathogenetic factor or false-positive result? Lancet. 1990;335:754–757. doi: 10.1016/0140-6736(90)90870-b. [DOI] [PubMed] [Google Scholar]
- 150.Czaja AJ, Taswell HF, Rakela J, Rabe D. Duration and specificity of antibodies to hepatitis C virus in chronic active hepatitis. Gastroenterology. 1992;102:1675–1679. doi: 10.1016/0016-5085(92)91729-n. [DOI] [PubMed] [Google Scholar]
- 151.Czaja AJ, Abdulkarim AS, Carpenter HA, Perez RG, Persing DH, Zein NN. GB virus-C infection in type 1 autoimmune hepatitis. Mayo Clin Proc. 1998;73:412–418. doi: 10.1016/S0025-6196(11)63722-1. [DOI] [PubMed] [Google Scholar]
- 152.Charlton M, Adjei P, Poterucha J, Zein N, Moore B, Therneau T, Krom R, Wiesner R. TT-virus infection in North American blood donors, patients with fulminant hepatic failure, and cryptogenic cirrhosis. Hepatology. 1998;28:839–842. doi: 10.1002/hep.510280335. [DOI] [PubMed] [Google Scholar]
- 153.Fainboim L, Cañero Velasco MC, Marcos CY, Ciocca M, Roy A, Theiler G, Capucchio M, Nuncifora S, Sala L, Zelazko M. Protracted, but not acute, hepatitis A virus infection is strongly associated with HLA-DRB*1301, a marker for pediatric autoimmune hepatitis. Hepatology. 2001;33:1512–1517. doi: 10.1053/jhep.2001.24562. [DOI] [PubMed] [Google Scholar]
- 154.Christen U, von Herrath MG. Do viral infections protect from or enhance type 1 diabetes and how can we tell the difference? Cell Mol Immunol. 2011;8:193–198. doi: 10.1038/cmi.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Albert LJ, Inman RD. Molecular mimicry and autoimmunity. N Engl J Med. 1999;341:2068–2074. doi: 10.1056/NEJM199912303412707. [DOI] [PubMed] [Google Scholar]
- 156.Oldstone MB. Molecular mimicry, microbial infection, and autoimmune disease: evolution of the concept. Curr Top Microbiol Immunol. 2005;296:1–17. doi: 10.1007/3-540-30791-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Scherer MT, Ignatowicz L, Winslow GM, Kappler JW, Marrack P. Superantigens: bacterial and viral proteins that manipulate the immune system. Annu Rev Cell Biol. 1993;9:101–128. doi: 10.1146/annurev.cb.09.110193.000533. [DOI] [PubMed] [Google Scholar]
- 158.Llewelyn M, Cohen J. Superantigens: microbial agents that corrupt immunity. Lancet Infect Dis. 2002;2:156–162. doi: 10.1016/s1473-3099(02)00222-0. [DOI] [PubMed] [Google Scholar]
- 159.Schiffenbauer J. Superantigens and their role in autoimmune disorders. Arch Immunol Ther Exp (Warsz) 1999;47:17–24. [PubMed] [Google Scholar]
- 160.Marrack P, Kappler J. The staphylococcal enterotoxins and their relatives. Science. 1990;248:705–711. doi: 10.1126/science.2185544. [DOI] [PubMed] [Google Scholar]
- 161.Li H, Llera A, Malchiodi EL, Mariuzza RA. The structural basis of T cell activation by superantigens. Annu Rev Immunol. 1999;17:435–466. doi: 10.1146/annurev.immunol.17.1.435. [DOI] [PubMed] [Google Scholar]
- 162.Miethke T, Wahl C, Heeg K, Echtenacher B, Krammer PH, Wagner H. T cell-mediated lethal shock triggered in mice by the superantigen staphylococcal enterotoxin B: critical role of tumor necrosis factor. J Exp Med. 1992;175:91–98. doi: 10.1084/jem.175.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Norrby-Teglund A, Norgren M, Holm SE, Andersson U, Andersson J. Similar cytokine induction profiles of a novel streptococcal exotoxin, MF, and pyrogenic exotoxins A and B. Infect Immun. 1994;62:3731–3738. doi: 10.1128/iai.62.9.3731-3738.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Choi Y, Lafferty JA, Clements JR, Todd JK, Gelfand EW, Kappler J, Marrack P, Kotzin BL. Selective expansion of T cells expressing V beta 2 in toxic shock syndrome. J Exp Med. 1990;172:981–984. doi: 10.1084/jem.172.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Schiffenbauer J, Johnson HM, Butfiloski EJ, Wegrzyn L, Soos JM. Staphylococcal enterotoxins can reactivate experimental allergic encephalomyelitis. Proc Natl Acad Sci U S A. 1993;90:8543–8546. doi: 10.1073/pnas.90.18.8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Paliard X, West SG, Lafferty JA, Clements JR, Kappler JW, Marrack P, Kotzin BL. Evidence for the effects of a superantigen in rheumatoid arthritis. Science. 1991;253:325–329. doi: 10.1126/science.1857971. [DOI] [PubMed] [Google Scholar]
- 167.Howell MD, Diveley JP, Lundeen KA, Esty A, Winters ST, Carlo DJ, Brostoff SW. Limited T-cell receptor beta-chain heterogeneity among interleukin 2 receptor-positive synovial T cells suggests a role for superantigen in rheumatoid arthritis. Proc Natl Acad Sci U S A. 1991;88:10921–10925. doi: 10.1073/pnas.88.23.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Björnsson E, Talwalkar J, Treeprasertsuk S, Kamath PS, Takahashi N, Sanderson S, Neuhauser M, Lindor K. Drug-induced autoimmune hepatitis: clinical characteristics and prognosis. Hepatology. 2010;51:2040–2048. doi: 10.1002/hep.23588. [DOI] [PubMed] [Google Scholar]
- 169.Czaja AJ. Drug-induced autoimmune-like hepatitis. Dig Dis Sci. 2011;56:958–976. doi: 10.1007/s10620-011-1611-4. [DOI] [PubMed] [Google Scholar]
- 170.Björnsson ES, Gunnarsson BI, Gröndal G, Jonasson JG, Einarsdottir R, Ludviksson BR, Gudbjörnsson B, Olafsson S. Risk of drug-induced liver injury from tumor necrosis factor antagonists. Clin Gastroenterol Hepatol. 2015;13:602–608. doi: 10.1016/j.cgh.2014.07.062. [DOI] [PubMed] [Google Scholar]
- 171.Kamiyama T, Nouchi T, Kojima S, Murata N, Ikeda T, Sato C. Autoimmune hepatitis triggered by administration of an herbal medicine. Am J Gastroenterol. 1997;92:703–704. [PubMed] [Google Scholar]
- 172.Cohen SM, O'Connor AM, Hart J, Merel NH, Te HS. Autoimmune hepatitis associated with the use of black cohosh: a case study. Menopause. 2004;11:575–577. doi: 10.1097/01.gme.0000142914.55849.6a. [DOI] [PubMed] [Google Scholar]
- 173.Guzman G, Kallwitz ER, Wojewoda C, Chennuri R, Berkes J, Layden TJ, Cotler SJ. Liver Injury with Features Mimicking Autoimmune Hepatitis following the Use of Black Cohosh. Case Rep Med. 2009;2009:918156. doi: 10.1155/2009/918156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Borum ML. Fulminant exacerbation of autoimmune hepatitis after the use of ma huang. Am J Gastroenterol. 2001;96:1654–1655. doi: 10.1111/j.1572-0241.2001.03827.x. [DOI] [PubMed] [Google Scholar]
- 175.Gilbert KM, Przybyla B, Pumford NR, Han T, Fuscoe J, Schnackenberg LK, Holland RD, Doss JC, Macmillan-Crow LA, Blossom SJ. Delineating liver events in trichloroethylene-induced autoimmune hepatitis. Chem Res Toxicol. 2009;22:626–632. doi: 10.1021/tx800409r. [DOI] [PubMed] [Google Scholar]
- 176.Pohl LR, Kenna JG, Satoh H, Christ D, Martin JL. Neoantigens associated with halothane hepatitis. Drug Metab Rev. 1989;20:203–217. doi: 10.3109/03602538909103537. [DOI] [PubMed] [Google Scholar]
- 177.Zimmerman HJ. Drug-induced liver disease. Clin Liver Dis. 2000;4:73–96, vi. doi: 10.1016/s1089-3261(05)70097-0. [DOI] [PubMed] [Google Scholar]
- 178.Robin MA, Le Roy M, Descatoire V, Pessayre D. Plasma membrane cytochromes P450 as neoantigens and autoimmune targets in drug-induced hepatitis. J Hepatol. 1997;26:23–30. doi: 10.1016/s0168-8278(97)82329-x. [DOI] [PubMed] [Google Scholar]
- 179.Eliasson E, Gardner I, Hume-Smith H, de Waziers I, Beaune P, Kenna JG. Interindividual variability in P450-dependent generation of neoantigens in halothane hepatitis. Chem Biol Interact. 1998;116:123–141. doi: 10.1016/s0009-2797(98)00081-7. [DOI] [PubMed] [Google Scholar]
- 180.Hokland P, Hokland M, Cotter F. The Nobel Prize for Medicine awarded for cancer therapy by inhibition of negative immune regulation. Br J Haematol. 2018;183:698–700. doi: 10.1111/bjh.15694. [DOI] [PubMed] [Google Scholar]
- 181.Wei SC, Duffy CR, Allison JP. Fundamental Mechanisms of Immune Checkpoint Blockade Therapy. Cancer Discov. 2018;8:1069–1086. doi: 10.1158/2159-8290.CD-18-0367. [DOI] [PubMed] [Google Scholar]
- 182.Peggs KS, Quezada SA, Chambers CA, Korman AJ, Allison JP. Blockade of CTLA-4 on both effector and regulatory T cell compartments contributes to the antitumor activity of anti-CTLA-4 antibodies. J Exp Med. 2009;206:1717–1725. doi: 10.1084/jem.20082492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Topalian SL, Drake CG, Pardoll DM. Targeting the PD-1/B7-H1(PD-L1) pathway to activate anti-tumor immunity. Curr Opin Immunol. 2012;24:207–212. doi: 10.1016/j.coi.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Quezada SA, Peggs KS. Exploiting CTLA-4, PD-1 and PD-L1 to reactivate the host immune response against cancer. Br J Cancer. 2013;108:1560–1565. doi: 10.1038/bjc.2013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Riaz N, Morris L, Havel JJ, Makarov V, Desrichard A, Chan TA. The role of neoantigens in response to immune checkpoint blockade. Int Immunol. 2016;28:411–419. doi: 10.1093/intimm/dxw019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Tocut M, Brenner R, Zandman-Goddard G. Autoimmune phenomena and disease in cancer patients treated with immune checkpoint inhibitors. Autoimmun Rev. 2018;17:610–616. doi: 10.1016/j.autrev.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 187.King GT, Sharma P, Davis SL, Jimeno A. Immune and autoimmune-related adverse events associated with immune checkpoint inhibitors in cancer therapy. Drugs Today (Barc) 2018;54:103–122. doi: 10.1358/dot.2018.54.2.2776626. [DOI] [PubMed] [Google Scholar]
- 188.Akturk HK, Alkanani A, Zhao Z, Yu L, Michels AW. PD-1 Inhibitor Immune-Related Adverse Events in Patients With Preexisting Endocrine Autoimmunity. J Clin Endocrinol Metab. 2018;103:3589–3592. doi: 10.1210/jc.2018-01430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Parakh S, Cebon J, Klein O. Delayed Autoimmune Toxicity Occurring Several Months After Cessation of Anti-PD-1 Therapy. Oncologist. 2018;23:849–851. doi: 10.1634/theoncologist.2017-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Suzman DL, Pelosof L, Rosenberg A, Avigan MI. Hepatotoxicity of immune checkpoint inhibitors: An evolving picture of risk associated with a vital class of immunotherapy agents. Liver Int. 2018;38:976–987. doi: 10.1111/liv.13746. [DOI] [PubMed] [Google Scholar]
- 191.Zen Y, Yeh MM. Hepatotoxicity of immune checkpoint inhibitors: a histology study of seven cases in comparison with autoimmune hepatitis and idiosyncratic drug-induced liver injury. Mod Pathol. 2018;31:965–973. doi: 10.1038/s41379-018-0013-y. [DOI] [PubMed] [Google Scholar]
- 192.De Martin E, Michot JM, Papouin B, Champiat S, Mateus C, Lambotte O, Roche B, Antonini TM, Coilly A, Laghouati S, Robert C, Marabelle A, Guettier C, Samuel D. Characterization of liver injury induced by cancer immunotherapy using immune checkpoint inhibitors. J Hepatol. 2018;68:1181–1190. doi: 10.1016/j.jhep.2018.01.033. [DOI] [PubMed] [Google Scholar]
- 193.Jennings JJ, Mandaliya R, Nakshabandi A, Lewis JH. Hepatotoxicity induced by immune checkpoint inhibitors: a comprehensive review including current and alternative management strategies. Expert Opin Drug Metab Toxicol. 2019;15:231–244. doi: 10.1080/17425255.2019.1574744. [DOI] [PubMed] [Google Scholar]
- 194.Nishida N, Kudo M. Liver damage related to immune checkpoint inhibitors. Hepatol Int. 2019;13:248–252. doi: 10.1007/s12072-018-9921-7. [DOI] [PubMed] [Google Scholar]
- 195.Reddy HG, Schneider BJ, Tai AW. Immune Checkpoint Inhibitor-Associated Colitis and Hepatitis. Clin Transl Gastroenterol. 2018;9:180. doi: 10.1038/s41424-018-0049-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kim KW, Ramaiya NH, Krajewski KM, Jagannathan JP, Tirumani SH, Srivastava A, Ibrahim N. Ipilimumab associated hepatitis: imaging and clinicopathologic findings. Invest New Drugs. 2013;31:1071–1077. doi: 10.1007/s10637-013-9939-6. [DOI] [PubMed] [Google Scholar]
- 197.Johncilla M, Misdraji J, Pratt DS, Agoston AT, Lauwers GY, Srivastava A, Doyle LA. Ipilimumab-associated Hepatitis: Clinicopathologic Characterization in a Series of 11 Cases. Am J Surg Pathol. 2015;39:1075–1084. doi: 10.1097/PAS.0000000000000453. [DOI] [PubMed] [Google Scholar]
- 198.Doherty GJ, Duckworth AM, Davies SE, Mells GF, Brais R, Harden SV, Parkinson CA, Corrie PG. Severe steroid-resistant anti-PD1 T-cell checkpoint inhibitor-induced hepatotoxicity driven by biliary injury. ESMO Open. 2017;2:e000268. doi: 10.1136/esmoopen-2017-000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Richardson B. Effect of an inhibitor of DNA methylation on T cells. II. 5-Azacytidine induces self-reactivity in antigen-specific T4+ cells. Hum Immunol. 1986;17:456–470. doi: 10.1016/0198-8859(86)90304-6. [DOI] [PubMed] [Google Scholar]
- 200.Cornacchia E, Golbus J, Maybaum J, Strahler J, Hanash S, Richardson B. Hydralazine and procainamide inhibit T cell DNA methylation and induce autoreactivity. J Immunol. 1988;140:2197–2200. [PubMed] [Google Scholar]
- 201.Scheinbart LS, Johnson MA, Gross LA, Edelstein SR, Richardson BC. Procainamide inhibits DNA methyltransferase in a human T cell line. J Rheumatol. 1991;18:530–534. [PubMed] [Google Scholar]
- 202.Quddus J, Johnson KJ, Gavalchin J, Amento EP, Chrisp CE, Yung RL, Richardson BC. Treating activated CD4+ T cells with either of two distinct DNA methyltransferase inhibitors, 5-azacytidine or procainamide, is sufficient to cause a lupus-like disease in syngeneic mice. J Clin Invest. 1993;92:38–53. doi: 10.1172/JCI116576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Christman JK. 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 204.Issa JP. Aging and epigenetic drift: a vicious cycle. J Clin Invest. 2014;124:24–29. doi: 10.1172/JCI69735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Maegawa S, Gough SM, Watanabe-Okochi N, Lu Y, Zhang N, Castoro RJ, Estecio MR, Jelinek J, Liang S, Kitamura T, Aplan PD, Issa JP. Age-related epigenetic drift in the pathogenesis of MDS and AML. Genome Res. 2014;24:580–591. doi: 10.1101/gr.157529.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Dozmorov MG, Coit P, Maksimowicz-McKinnon K, Sawalha AH. Age-associated DNA methylation changes in naive CD4+ T cells suggest an evolving autoimmune epigenotype in aging T cells. Epigenomics. 2017;9:429–445. doi: 10.2217/epi-2016-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Somers EC, Richardson BC. Environmental exposures, epigenetic changes and the risk of lupus. Lupus. 2014;23:568–576. doi: 10.1177/0961203313499419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Cañas CA, Cañas F, Bonilla-Abadía F, Ospina FE, Tobón GJ. Epigenetics changes associated to environmental triggers in autoimmunity. Autoimmunity. 2016;49:1–11. doi: 10.3109/08916934.2015.1086996. [DOI] [PubMed] [Google Scholar]
- 209.Efe C, Kav T, Aydin C, Cengiz M, Imga NN, Purnak T, Smyk DS, Torgutalp M, Turhan T, Ozenirler S, Ozaslan E, Bogdanos DP. Low serum vitamin D levels are associated with severe histological features and poor response to therapy in patients with autoimmune hepatitis. Dig Dis Sci. 2014;59:3035–3042. doi: 10.1007/s10620-014-3267-3. [DOI] [PubMed] [Google Scholar]
- 210.Ebadi M, Bhanji RA, Mazurak VC, Lytvyak E, Mason A, Czaja AJ, Montano-Loza AJ. Severe vitamin D deficiency is a prognostic biomarker in autoimmune hepatitis. Aliment Pharmacol Ther. 2019;49:173–182. doi: 10.1111/apt.15029. [DOI] [PubMed] [Google Scholar]
- 211.Czaja AJ, Montano-Loza AJ. Evolving Role of Vitamin D in Immune-Mediated Disease and Its Implications in Autoimmune Hepatitis. Dig Dis Sci. 2019;64:324–344. doi: 10.1007/s10620-018-5351-6. [DOI] [PubMed] [Google Scholar]
- 212.Vogel A, Strassburg CP, Manns MP. Genetic association of vitamin D receptor polymorphisms with primary biliary cirrhosis and autoimmune hepatitis. Hepatology. 2002;35:126–131. doi: 10.1053/jhep.2002.30084. [DOI] [PubMed] [Google Scholar]
- 213.Fan L, Tu X, Zhu Y, Zhou L, Pfeiffer T, Feltens R, Stoecker W, Zhong R. Genetic association of vitamin D receptor polymorphisms with autoimmune hepatitis and primary biliary cirrhosis in the Chinese. J Gastroenterol Hepatol. 2005;20:249–255. doi: 10.1111/j.1440-1746.2005.03532.x. [DOI] [PubMed] [Google Scholar]
- 214.Christen U, Hintermann E, Holdener M, von Herrath MG. Viral triggers for autoimmunity: is the 'glass of molecular mimicry' half full or half empty? J Autoimmun. 2010;34:38–44. doi: 10.1016/j.jaut.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Alam J, Kim YC, Choi Y. Potential role of bacterial infection in autoimmune diseases: a new aspect of molecular mimicry. Immune Netw. 2014;14:7–13. doi: 10.4110/in.2014.14.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Fujinami RS, Oldstone MB, Wroblewska Z, Frankel ME, Koprowski H. Molecular mimicry in virus infection: crossreaction of measles virus phosphoprotein or of herpes simplex virus protein with human intermediate filaments. Proc Natl Acad Sci USA. 1983;80:2346–2350. doi: 10.1073/pnas.80.8.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Fujinami RS, Oldstone MB. Amino acid homology between the encephalitogenic site of myelin basic protein and virus: mechanism for autoimmunity. Science. 1985;230:1043–1045. doi: 10.1126/science.2414848. [DOI] [PubMed] [Google Scholar]
- 218.Wucherpfennig KW. Mechanisms for the induction of autoimmunity by infectious agents. J Clin Invest. 2001;108:1097–1104. doi: 10.1172/JCI14235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Gut J, Christen U, Huwyler J. Mechanisms of halothane toxicity: novel insights. Pharmacol Ther. 1993;58:133–155. doi: 10.1016/0163-7258(93)90047-h. [DOI] [PubMed] [Google Scholar]
- 220.Christen U, Quinn J, Yeaman SJ, Kenna JG, Clarke JB, Gandolfi AJ, Gut J. Identification of the dihydrolipoamide acetyltransferase subunit of the human pyruvate dehydrogenase complex as an autoantigen in halothane hepatitis. Molecular mimicry of trifluoroacetyl-lysine by lipoic acid. Eur J Biochem. 1994;223:1035–1047. doi: 10.1111/j.1432-1033.1994.tb19082.x. [DOI] [PubMed] [Google Scholar]
- 221.Long SA, Quan C, Van de Water J, Nantz MH, Kurth MJ, Barsky D, Colvin ME, Lam KS, Coppel RL, Ansari A, Gershwin ME. Immunoreactivity of organic mimeotopes of the E2 component of pyruvate dehydrogenase: connecting xenobiotics with primary biliary cirrhosis. J Immunol. 2001;167:2956–2963. doi: 10.4049/jimmunol.167.5.2956. [DOI] [PubMed] [Google Scholar]
- 222.Wakabayashi K, Lian ZX, Leung PS, Moritoki Y, Tsuneyama K, Kurth MJ, Lam KS, Yoshida K, Yang GX, Hibi T, Ansari AA, Ridgway WM, Coppel RL, Mackay IR, Gershwin ME. Loss of tolerance in C57BL/6 mice to the autoantigen E2 subunit of pyruvate dehydrogenase by a xenobiotic with ensuing biliary ductular disease. Hepatology. 2008;48:531–540. doi: 10.1002/hep.22390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wakabayashi K, Yoshida K, Leung PS, Moritoki Y, Yang GX, Tsuneyama K, Lian ZX, Hibi T, Ansari AA, Wicker LS, Ridgway WM, Coppel RL, Mackay IR, Gershwin ME. Induction of autoimmune cholangitis in non-obese diabetic (NOD).1101 mice following a chemical xenobiotic immunization. Clin Exp Immunol. 2009;155:577–586. doi: 10.1111/j.1365-2249.2008.03837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Agmon-Levin N, Paz Z, Israeli E, Shoenfeld Y. Vaccines and autoimmunity. Nat Rev Rheumatol. 2009;5:648–652. doi: 10.1038/nrrheum.2009.196. [DOI] [PubMed] [Google Scholar]
- 225.Kanduc D. Peptide cross-reactivity: the original sin of vaccines. Front Biosci (Schol Ed) 2012;4:1393–1401. doi: 10.2741/s341. [DOI] [PubMed] [Google Scholar]
- 226.Ehser J, Holdener M, Christen S, Bayer M, Pfeilschifter JM, Hintermann E, Bogdanos D, Christen U. Molecular mimicry rather than identity breaks T-cell tolerance in the CYP2D6 mouse model for human autoimmune hepatitis. J Autoimmun. 2013;42:39–49. doi: 10.1016/j.jaut.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 227.Damian RT. Molecular Mimicry in Biological Adaptation. Science. 1965;147:824. doi: 10.1126/science.147.3660.824-b. [DOI] [PubMed] [Google Scholar]
- 228.Pahari S, Chatterjee D, Negi S, Kaur J, Singh B, Agrewala JN. Morbid Sequences Suggest Molecular Mimicry between Microbial Peptides and Self-Antigens: A Possibility of Inciting Autoimmunity. Front Microbiol. 2017;8:1938. doi: 10.3389/fmicb.2017.01938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Krisher K, Cunningham MW. Myosin: a link between streptococci and heart. Science. 1985;227:413–415. doi: 10.1126/science.2578225. [DOI] [PubMed] [Google Scholar]
- 230.Ellis NM, Li Y, Hildebrand W, Fischetti VA, Cunningham MW. T cell mimicry and epitope specificity of cross-reactive T cell clones from rheumatic heart disease. J Immunol. 2005;175:5448–5456. doi: 10.4049/jimmunol.175.8.5448. [DOI] [PubMed] [Google Scholar]
- 231.Guilherme L, Kalil J, Cunningham M. Molecular mimicry in the autoimmune pathogenesis of rheumatic heart disease. Autoimmunity. 2006;39:31–39. doi: 10.1080/08916930500484674. [DOI] [PubMed] [Google Scholar]
- 232.Faé KC, da Silva DD, Oshiro SE, Tanaka AC, Pomerantzeff PM, Douay C, Charron D, Toubert A, Cunningham MW, Kalil J, Guilherme L. Mimicry in recognition of cardiac myosin peptides by heart-intralesional T cell clones from rheumatic heart disease. J Immunol. 2006;176:5662–5670. doi: 10.4049/jimmunol.176.9.5662. [DOI] [PubMed] [Google Scholar]
- 233.Cunningham MW. Streptococcus and rheumatic fever. Curr Opin Rheumatol. 2012;24:408–416. doi: 10.1097/BOR.0b013e32835461d3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Cunningham MW. Rheumatic fever, autoimmunity, and molecular mimicry: the streptococcal connection. Int Rev Immunol. 2014;33:314–329. doi: 10.3109/08830185.2014.917411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Sheikh KA, Zhang G, Gong Y, Schnaar RL, Griffin JW. An anti-ganglioside antibody-secreting hybridoma induces neuropathy in mice. Ann Neurol. 2004;56:228–239. doi: 10.1002/ana.20173. [DOI] [PubMed] [Google Scholar]
- 236.Ang CW, Jacobs BC, Laman JD. The Guillain-Barré syndrome: a true case of molecular mimicry. Trends Immunol. 2004;25:61–66. doi: 10.1016/j.it.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 237.Gross DM, Forsthuber T, Tary-Lehmann M, Etling C, Ito K, Nagy ZA, Field JA, Steere AC, Huber BT. Identification of LFA-1 as a candidate autoantigen in treatment-resistant Lyme arthritis. Science. 1998;281:703–706. doi: 10.1126/science.281.5377.703. [DOI] [PubMed] [Google Scholar]
- 238.Benoist C, Mathis D. Autoimmunity provoked by infection: how good is the case for T cell epitope mimicry? Nat Immunol. 2001;2:797–801. doi: 10.1038/ni0901-797. [DOI] [PubMed] [Google Scholar]
- 239.Schwimmbeck PL, Yu DT, Oldstone MB. Autoantibodies to HLA B27 in the sera of HLA B27 patients with ankylosing spondylitis and Reiter's syndrome. Molecular mimicry with Klebsiella pneumoniae as potential mechanism of autoimmune disease. J Exp Med. 1987;166:173–181. doi: 10.1084/jem.166.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Schwimmbeck PL, Oldstone MB. Klebsiella pneumoniae and HLA B27-associated diseases of Reiter's syndrome and ankylosing spondylitis. Curr Top Microbiol Immunol. 1989;145:45–56. doi: 10.1007/978-3-642-74594-2_4. [DOI] [PubMed] [Google Scholar]
- 241.Fielder M, Pirt SJ, Tarpey I, Wilson C, Cunningham P, Ettelaie C, Binder A, Bansal S, Ebringer A. Molecular mimicry and ankylosing spondylitis: possible role of a novel sequence in pullulanase of Klebsiella pneumoniae. FEBS Lett. 1995;369:243–248. doi: 10.1016/0014-5793(95)00760-7. [DOI] [PubMed] [Google Scholar]
- 242.Albani S, Keystone EC, Nelson JL, Ollier WE, La Cava A, Montemayor AC, Weber DA, Montecucco C, Martini A, Carson DA. Positive selection in autoimmunity: abnormal immune responses to a bacterial dnaJ antigenic determinant in patients with early rheumatoid arthritis. Nat Med. 1995;1:448–452. doi: 10.1038/nm0595-448. [DOI] [PubMed] [Google Scholar]
- 243.McClain MT, Heinlen LD, Dennis GJ, Roebuck J, Harley JB, James JA. Early events in lupus humoral autoimmunity suggest initiation through molecular mimicry. Nat Med. 2005;11:85–89. doi: 10.1038/nm1167. [DOI] [PubMed] [Google Scholar]
- 244.Manns MP, Griffin KJ, Sullivan KF, Johnson EF. LKM-1 autoantibodies recognize a short linear sequence in P450IID6, a cytochrome P-450 monooxygenase. J Clin Invest. 1991;88:1370–1378. doi: 10.1172/JCI115443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ma Y, Thomas MG, Okamoto M, Bogdanos DP, Nagl S, Kerkar N, Lopes AR, Muratori L, Lenzi M, Bianchi FB, Mieli-Vergani G, Vergani D. Key residues of a major cytochrome P4502D6 epitope are located on the surface of the molecule. J Immunol. 2002;169:277–285. doi: 10.4049/jimmunol.169.1.277. [DOI] [PubMed] [Google Scholar]
- 246.Kerkar N, Choudhuri K, Ma Y, Mahmoud A, Bogdanos DP, Muratori L, Bianchi F, Williams R, Mieli-Vergani G, Vergani D. Cytochrome P4502D6(193-212): a new immunodominant epitope and target of virus/self cross-reactivity in liver kidney microsomal autoantibody type 1-positive liver disease. J Immunol. 2003;170:1481–1489. doi: 10.4049/jimmunol.170.3.1481. [DOI] [PubMed] [Google Scholar]
- 247.Selmi C, Balkwill DL, Invernizzi P, Ansari AA, Coppel RL, Podda M, Leung PS, Kenny TP, Van De Water J, Nantz MH, Kurth MJ, Gershwin ME. Patients with primary biliary cirrhosis react against a ubiquitous xenobiotic-metabolizing bacterium. Hepatology. 2003;38:1250–1257. doi: 10.1053/jhep.2003.50446. [DOI] [PubMed] [Google Scholar]
- 248.Olafsson S, Gudjonsson H, Selmi C, Amano K, Invernizzi P, Podda M, Gershwin ME. Antimitochondrial antibodies and reactivity to N. aromaticivorans proteins in Icelandic patients with primary biliary cirrhosis and their relatives. Am J Gastroenterol. 2004;99:2143–2146. doi: 10.1111/j.1572-0241.2004.40397.x. [DOI] [PubMed] [Google Scholar]
- 249.Bogdanos DP, Baum H, Grasso A, Okamoto M, Butler P, Ma Y, Rigopoulou E, Montalto P, Davies ET, Burroughs AK, Vergani D. Microbial mimics are major targets of crossreactivity with human pyruvate dehydrogenase in primary biliary cirrhosis. J Hepatol. 2004;40:31–39. doi: 10.1016/s0168-8278(03)00501-4. [DOI] [PubMed] [Google Scholar]
- 250.Padgett KA, Selmi C, Kenny TP, Leung PS, Balkwill DL, Ansari AA, Coppel RL, Gershwin ME. Phylogenetic and immunological definition of four lipoylated proteins from Novosphingobium aromaticivorans, implications for primary biliary cirrhosis. J Autoimmun. 2005;24:209–219. doi: 10.1016/j.jaut.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 251.Ortega-Hernandez OD, Levin NA, Altman A, Shoenfeld Y. Infectious agents in the pathogenesis of primary biliary cirrhosis. Dis Markers. 2010;29:277–286. doi: 10.3233/DMA-2010-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Holdener M, Hintermann E, Bayer M, Rhode A, Rodrigo E, Hintereder G, Johnson EF, Gonzalez FJ, Pfeilschifter J, Manns MP, Herrath Mv, Christen U. Breaking tolerance to the natural human liver autoantigen cytochrome P450 2D6 by virus infection. J Exp Med. 2008;205:1409–1422. doi: 10.1084/jem.20071859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Trost B, Lucchese G, Stufano A, Bickis M, Kusalik A, Kanduc D. No human protein is exempt from bacterial motifs, not even one. Self Nonself. 2010;1:328–334. doi: 10.4161/self.1.4.13315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Reay PA, Kantor RM, Davis MM. Use of global amino acid replacements to define the requirements for MHC binding and T cell recognition of moth cytochrome c (93-103) J Immunol. 1994;152:3946–3957. [PubMed] [Google Scholar]
- 255.Sinigaglia F, Hammer J. Defining rules for the peptide-MHC class II interaction. Curr Opin Immunol. 1994;6:52–56. doi: 10.1016/0952-7915(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 256.Sinigaglia F, Hammer J. Rules for peptide binding to MHC class II molecules. APMIS. 1994;102:241–248. doi: 10.1111/j.1699-0463.1994.tb04871.x. [DOI] [PubMed] [Google Scholar]
- 257.Wucherpfennig KW, Sette A, Southwood S, Oseroff C, Matsui M, Strominger JL, Hafler DA. Structural requirements for binding of an immunodominant myelin basic protein peptide to DR2 isotypes and for its recognition by human T cell clones. J Exp Med. 1994;179:279–290. doi: 10.1084/jem.179.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Amano K, Leung PS, Rieger R, Quan C, Wang X, Marik J, Suen YF, Kurth MJ, Nantz MH, Ansari AA, Lam KS, Zeniya M, Matsuura E, Coppel RL, Gershwin ME. Chemical xenobiotics and mitochondrial autoantigens in primary biliary cirrhosis: identification of antibodies against a common environmental, cosmetic, and food additive, 2-octynoic acid. J Immunol. 2005;174:5874–5883. doi: 10.4049/jimmunol.174.9.5874. [DOI] [PubMed] [Google Scholar]
- 259.Rieger R, Leung PS, Jeddeloh MR, Kurth MJ, Nantz MH, Lam KS, Barsky D, Ansari AA, Coppel RL, Mackay IR, Gershwin ME. Identification of 2-nonynoic acid, a cosmetic component, as a potential trigger of primary biliary cirrhosis. J Autoimmun. 2006;27:7–16. doi: 10.1016/j.jaut.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 260.Gelpi C, Sontheimer EJ, Rodriguez-Sanchez JL. Autoantibodies against a serine tRNA-protein complex implicated in cotranslational selenocysteine insertion. Proc Natl Acad Sci U S A. 1992;89:9739–9743. doi: 10.1073/pnas.89.20.9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Costa M, Rodríguez-Sánchez JL, Czaja AJ, Gelpí C. Isolation and characterization of cDNA encoding the antigenic protein of the human tRNP(Ser)Sec complex recognized by autoantibodies from patients withtype-1 autoimmune hepatitis. Clin Exp Immunol. 2000;121:364–374. doi: 10.1046/j.1365-2249.2000.01280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Wies I, Brunner S, Henninger J, Herkel J, Kanzler S, Meyer zum Büschenfelde KH, Lohse AW. Identification of target antigen for SLA/LP autoantibodies in autoimmune hepatitis. Lancet. 2000;355:1510–1515. doi: 10.1016/s0140-6736(00)02166-8. [DOI] [PubMed] [Google Scholar]
- 263.Volkmann M, Luithle D, Zentgraf H, Schnölzer M, Fiedler S, Heid H, Schulze-Bergkamen A, Strassburg CP, Gehrke SG, Manns MP. SLA/LP/tRNP((Ser)Sec) antigen in autoimmune hepatitis: identification of the native protein in human hepatic cell extract. J Autoimmun. 2010;34:59–65. doi: 10.1016/j.jaut.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 264.McRae BL, Vanderlugt CL, Dal Canto MC, Miller SD. Functional evidence for epitope spreading in the relapsing pathology of experimental autoimmune encephalomyelitis. J Exp Med. 1995;182:75–85. doi: 10.1084/jem.182.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Vanderlugt CL, Miller SD. Epitope spreading in immune-mediated diseases: implications for immunotherapy. Nat Rev Immunol. 2002;2:85–95. doi: 10.1038/nri724. [DOI] [PubMed] [Google Scholar]
- 266.Deshmukh US, Bagavant H, Lewis J, Gaskin F, Fu SM. Epitope spreading within lupus-associated ribonucleoprotein antigens. Clin Immunol. 2005;117:112–120. doi: 10.1016/j.clim.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 267.Horwitz MS, Bradley LM, Harbertson J, Krahl T, Lee J, Sarvetnick N. Diabetes induced by Coxsackie virus: initiation by bystander damage and not molecular mimicry. Nat Med. 1998;4:781–785. doi: 10.1038/nm0798-781. [DOI] [PubMed] [Google Scholar]
- 268.Lehmann PV, Forsthuber T, Miller A, Sercarz EE. Spreading of T-cell autoimmunity to cryptic determinants of an autoantigen. Nature. 1992;358:155–157. doi: 10.1038/358155a0. [DOI] [PubMed] [Google Scholar]
- 269.Christen U, Hintermann E. Autoantibodies in Autoimmune Hepatitis: Can Epitopes Tell Us about the Etiology of the Disease? Front Immunol. 2018;9:163. doi: 10.3389/fimmu.2018.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Ma WT, Chang C, Gershwin ME, Lian ZX. Development of autoantibodies precedes clinical manifestations of autoimmune diseases: A comprehensive review. J Autoimmun. 2017;83:95–112. doi: 10.1016/j.jaut.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 271.Rammensee HG. Chemistry of peptides associated with MHC class I and class II molecules. Curr Opin Immunol. 1995;7:85–96. doi: 10.1016/0952-7915(95)80033-6. [DOI] [PubMed] [Google Scholar]
- 272.Li Z, Woo CJ, Iglesias-Ussel MD, Ronai D, Scharff MD. The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev. 2004;18:1–11. doi: 10.1101/gad.1161904. [DOI] [PubMed] [Google Scholar]
- 273.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 274.Fujinami RS, von Herrath MG, Christen U, Whitton JL. Molecular mimicry, bystander activation, or viral persistence: infections and autoimmune disease. Clin Microbiol Rev. 2006;19:80–94. doi: 10.1128/CMR.19.1.80-94.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Tough DF, Borrow P, Sprent J. Induction of bystander T cell proliferation by viruses and type I interferon in vivo. Science. 1996;272:1947–1950. doi: 10.1126/science.272.5270.1947. [DOI] [PubMed] [Google Scholar]
- 276.Tough DF, Sun S, Sprent J. T cell stimulation in vivo by lipopolysaccharide (LPS) J Exp Med. 1997;185:2089–2094. doi: 10.1084/jem.185.12.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Di Genova G, Roddick J, McNicholl F, Stevenson FK. Vaccination of human subjects expands both specific and bystander memory T cells but antibody production remains vaccine specific. Blood. 2006;107:2806–2813. doi: 10.1182/blood-2005-08-3255. [DOI] [PubMed] [Google Scholar]
- 278.Di Genova G, Savelyeva N, Suchacki A, Thirdborough SM, Stevenson FK. Bystander stimulation of activated CD4+ T cells of unrelated specificity following a booster vaccination with tetanus toxoid. Eur J Immunol. 2010;40:976–985. doi: 10.1002/eji.200940017. [DOI] [PubMed] [Google Scholar]
- 279.Boyman O. Bystander activation of CD4+ T cells. Eur J Immunol. 2010;40:936–939. doi: 10.1002/eji.201040466. [DOI] [PubMed] [Google Scholar]
- 280.Eberl G, Brawand P, MacDonald HR. Selective bystander proliferation of memory CD4+ and CD8+ T cells upon NK T or T cell activation. J Immunol. 2000;165:4305–4311. doi: 10.4049/jimmunol.165.8.4305. [DOI] [PubMed] [Google Scholar]
- 281.Duke RC. Self recognition by T cells. I. Bystander killing of target cells bearing syngeneic MHC antigens. J Exp Med. 1989;170:59–71. doi: 10.1084/jem.170.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Smyth MJ, Sedgwick JD. Delayed kinetics of tumor necrosis factor-mediated bystander lysis by peptide-specific CD8+ cytotoxic T lymphocytes. Eur J Immunol. 1998;28:4162–4169. doi: 10.1002/(SICI)1521-4141(199812)28:12<4162::AID-IMMU4162>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 283.Nakamoto N, Kanai T. Role of toll-like receptors in immune activation and tolerance in the liver. Front Immunol. 2014;5:221. doi: 10.3389/fimmu.2014.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Nikolich-Zugich J, Slifka MK, Messaoudi I. The many important facets of T-cell repertoire diversity. Nat Rev Immunol. 2004;4:123–132. doi: 10.1038/nri1292. [DOI] [PubMed] [Google Scholar]
- 285.Warren RL, Freeman JD, Zeng T, Choe G, Munro S, Moore R, Webb JR, Holt RA. Exhaustive T-cell repertoire sequencing of human peripheral blood samples reveals signatures of antigen selection and a directly measured repertoire size of at least 1 million clonotypes. Genome Res. 2011;21:790–797. doi: 10.1101/gr.115428.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Harbige J, Eichmann M, Peakman M. New insights into non-conventional epitopes as T cell targets: The missing link for breaking immune tolerance in autoimmune disease? J Autoimmun. 2017;84:12–20. doi: 10.1016/j.jaut.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 287.Libbey JE, Cusick MF, Tsunoda I, Fujinami RS. Antiviral CD8⁺ T cells cause an experimental autoimmune encephalomyelitis-like disease in naive mice. J Neurovirol. 2012;18:45–54. doi: 10.1007/s13365-012-0077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Cusick MF, Libbey JE, Fujinami RS. Multiple sclerosis: autoimmunity and viruses. Curr Opin Rheumatol. 2013;25:496–501. doi: 10.1097/BOR.0b013e328362004d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Padovan E, Giachino C, Cella M, Valitutti S, Acuto O, Lanzavecchia A. Normal T lymphocytes can express two different T cell receptor beta chains: implications for the mechanism of allelic exclusion. J Exp Med. 1995;181:1587–1591. doi: 10.1084/jem.181.4.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Sarukhan A, Garcia C, Lanoue A, von Boehmer H. Allelic inclusion of T cell receptor alpha genes poses an autoimmune hazard due to low-level expression of autospecific receptors. Immunity. 1998;8:563–570. doi: 10.1016/s1074-7613(00)80561-0. [DOI] [PubMed] [Google Scholar]
- 291.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 292.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Geuking MB, Köller Y, Rupp S, McCoy KD. The interplay between the gut microbiota and the immune system. Gut Microbes. 2014;5:411–418. doi: 10.4161/gmic.29330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Van Praet JT, Donovan E, Vanassche I, Drennan MB, Windels F, Dendooven A, Allais L, Cuvelier CA, van de Loo F, Norris PS, Kruglov AA, Nedospasov SA, Rabot S, Tito R, Raes J, Gaboriau-Routhiau V, Cerf-Bensussan N, Van de Wiele T, Eberl G, Ware CF, Elewaut D. Commensal microbiota influence systemic autoimmune responses. EMBO J. 2015;34:466–474. doi: 10.15252/embj.201489966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Sánchez B, Hevia A, González S, Margolles A. Interaction of Intestinal Microorganisms with the Human Host in the Framework of Autoimmune Diseases. Front Immunol. 2015;6:594. doi: 10.3389/fimmu.2015.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Ignacio A, Morales CI, Câmara NO, Almeida RR. Innate Sensing of the Gut Microbiota: Modulation of Inflammatory and Autoimmune Diseases. Front Immunol. 2016;7:54. doi: 10.3389/fimmu.2016.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD, Macpherson AJ, Danska JS. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 301.Miyake Y, Yamamoto K. Role of gut microbiota in liver diseases. Hepatol Res. 2013;43:139–146. doi: 10.1111/j.1872-034X.2012.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Henao-Mejia J, Elinav E, Thaiss CA, Licona-Limon P, Flavell RA. Role of the intestinal microbiome in liver disease. J Autoimmun. 2013;46:66–73. doi: 10.1016/j.jaut.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 303.Rogier R, Koenders MI, Abdollahi-Roodsaz S. Toll-like receptor mediated modulation of T cell response by commensal intestinal microbiota as a trigger for autoimmune arthritis. J Immunol Res. 2015;2015:527696. doi: 10.1155/2015/527696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Watanabe A, Sohail MA, Gomes DA, Hashmi A, Nagata J, Sutterwala FS, Mahmood S, Jhandier MN, Shi Y, Flavell RA, Mehal WZ. Inflammasome-mediated regulation of hepatic stellate cells. Am J Physiol Gastrointest Liver Physiol. 2009;296:G1248–G1257. doi: 10.1152/ajpgi.90223.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 306.Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, Camporez JP, Shulman GI, Gordon JI, Hoffman HM, Flavell RA. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Boaru SG, Borkham-Kamphorst E, Tihaa L, Haas U, Weiskirchen R. Expression analysis of inflammasomes in experimental models of inflammatory and fibrotic liver disease. J Inflamm (Lond) 2012;9:49. doi: 10.1186/1476-9255-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Wu HJ, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. 2012;3:4–14. doi: 10.4161/gmic.19320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, McCoy KD, Macpherson AJ. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 310.Hänninen A, Salmi M, Simell O, Jalkanen S. Mucosa-associated (beta 7-integrinhigh) lymphocytes accumulate early in the pancreas of NOD mice and show aberrant recirculation behavior. Diabetes. 1996;45:1173–1180. doi: 10.2337/diab.45.9.1173. [DOI] [PubMed] [Google Scholar]
- 311.Paronen J, Klemetti P, Kantele JM, Savilahti E, Perheentupa J, Akerblom HK, Vaarala O. Glutamate decarboxylase-reactive peripheral blood lymphocytes from patients with IDDM express gut-specific homing receptor alpha4beta7-integrin. Diabetes. 1997;46:583–588. doi: 10.2337/diab.46.4.583. [DOI] [PubMed] [Google Scholar]
- 312.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, Gordon JI, Chervonsky AV. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Li Y, Liu Y, Chu CQ. Th17 Cells in Type 1 Diabetes: Role in the Pathogenesis and Regulation by Gut Microbiome. Mediators Inflamm. 2015;2015:638470. doi: 10.1155/2015/638470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Mejía-León ME, Barca AM. Diet, Microbiota and Immune System in Type 1 Diabetes Development and Evolution. Nutrients. 2015;7:9171–9184. doi: 10.3390/nu7115461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Abdollahi-Roodsaz S, Joosten LA, Koenders MI, Devesa I, Roelofs MF, Radstake TR, Heuvelmans-Jacobs M, Akira S, Nicklin MJ, Ribeiro-Dias F, van den Berg WB. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J Clin Invest. 2008;118:205–216. doi: 10.1172/JCI32639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Abdollahi-Roodsaz S, Koenders MI, Walgreen B, Bolscher J, Helsen MM, van den Bersselaar LA, van Lent PL, van de Loo FA, van den Berg WB. Toll-like receptor 2 controls acute immune complex-driven arthritis in mice by regulating the inhibitory Fcγ receptor IIB. Arthritis Rheum. 2013;65:2583–2593. doi: 10.1002/art.38087. [DOI] [PubMed] [Google Scholar]
- 318.Miyake S, Kim S, Suda W, Oshima K, Nakamura M, Matsuoka T, Chihara N, Tomita A, Sato W, Kim SW, Morita H, Hattori M, Yamamura T. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS One. 2015;10:e0137429. doi: 10.1371/journal.pone.0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Sokol H, Seksik P, Rigottier-Gois L, Lay C, Lepage P, Podglajen I, Marteau P, Doré J. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm Bowel Dis. 2006;12:106–111. doi: 10.1097/01.MIB.0000200323.38139.c6. [DOI] [PubMed] [Google Scholar]
- 320.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, Gordon JI, Onderdonk AB, Glimcher LH. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Hopf U, Möller B, Stemerowicz R, Lobeck H, Rodloff A, Freudenberg M, Galanos C, Huhn D. Relation between Escherichia coli R(rough)-forms in gut, lipid A in liver, and primary biliary cirrhosis. Lancet. 1989;2:1419–1422. doi: 10.1016/s0140-6736(89)92034-5. [DOI] [PubMed] [Google Scholar]
- 323.Tabibian JH, O'Hara SP, Trussoni CE, Tietz PS, Splinter PL, Mounajjed T, Hagey LR, LaRusso NF. Absence of the intestinal microbiota exacerbates hepatobiliary disease in a murine model of primary sclerosing cholangitis. Hepatology. 2016;63:185–196. doi: 10.1002/hep.27927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Mueller T, Beutler C, Picó AH, Shibolet O, Pratt DS, Pascher A, Neuhaus P, Wiedenmann B, Berg T, Podolsky DK. Enhanced innate immune responsiveness and intolerance to intestinal endotoxins in human biliary epithelial cells contributes to chronic cholangitis. Liver Int. 2011;31:1574–1588. doi: 10.1111/j.1478-3231.2011.02635.x. [DOI] [PubMed] [Google Scholar]
- 325.Lin R, Zhou L, Zhang J, Wang B. Abnormal intestinal permeability and microbiota in patients with autoimmune hepatitis. Int J Clin Exp Pathol. 2015;8:5153–5160. [PMC free article] [PubMed] [Google Scholar]
- 326.Yuksel M, Wang Y, Tai N, Peng J, Guo J, Beland K, Lapierre P, David C, Alvarez F, Colle I, Yan H, Mieli-Vergani G, Vergani D, Ma Y, Wen L. A novel "humanized mouse" model for autoimmune hepatitis and the association of gut microbiota with liver inflammation. Hepatology. 2015;62:1536–1550. doi: 10.1002/hep.27998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Whitacre CC, Reingold SC, O'Looney PA. A gender gap in autoimmunity. Science. 1999;283:1277–1278. doi: 10.1126/science.283.5406.1277. [DOI] [PubMed] [Google Scholar]
- 328.Young NA, Wu LC, Burd CJ, Friedman AK, Kaffenberger BH, Rajaram MV, Schlesinger LS, James H, Shupnik MA, Jarjour WN. Estrogen modulation of endosome-associated toll-like receptor 8: an IFNα-independent mechanism of sex-bias in systemic lupus erythematosus. Clin Immunol. 2014;151:66–77. doi: 10.1016/j.clim.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Tanaka A, Lindor K, Ansari A, Gershwin ME. Fetal microchimerisms in the mother: immunologic implications. Liver Transpl. 2000;6:138–143. doi: 10.1002/lt.500060225. [DOI] [PubMed] [Google Scholar]
- 330.Lambert NC, Evans PC, Hashizumi TL, Maloney S, Gooley T, Furst DE, Nelson JL. Cutting edge: persistent fetal microchimerism in T lymphocytes is associated with HLA-DQA1*0501: implications in autoimmunity. J Immunol. 2000;164:5545–5548. doi: 10.4049/jimmunol.164.11.5545. [DOI] [PubMed] [Google Scholar]
- 331.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 332.Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, Antonopoulos D, Umesaki Y, Chervonsky AV. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Markle JG, Frank DN, Adeli K, von Bergen M, Danska JS. Microbiome manipulation modifies sex-specific risk for autoimmunity. Gut Microbes. 2014;5:485–493. doi: 10.4161/gmic.29795. [DOI] [PubMed] [Google Scholar]
- 334.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 335.Okada H, Kuhn C, Feillet H, Bach JF. The 'hygiene hypothesis' for autoimmune and allergic diseases: an update. Clin Exp Immunol. 2010;160:1–9. doi: 10.1111/j.1365-2249.2010.04139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Rook GA. Hygiene hypothesis and autoimmune diseases. Clin Rev Allergy Immunol. 2012;42:5–15. doi: 10.1007/s12016-011-8285-8. [DOI] [PubMed] [Google Scholar]
- 337.Daley D. The evolution of the hygiene hypothesis: the role of early-life exposures to viruses and microbes and their relationship to asthma and allergic diseases. Curr Opin Allergy Clin Immunol. 2014;14:390–396. doi: 10.1097/ACI.0000000000000101. [DOI] [PubMed] [Google Scholar]
- 338.Pross HF, Eidinger D. Antigenic competition: a review of nonspecific antigen-induced suppression. Adv Immunol. 1974;18:133–168. doi: 10.1016/s0065-2776(08)60309-0. [DOI] [PubMed] [Google Scholar]
- 339.Liacopoulos P, Ben-Efraim S. Antigenic competition. Prog Allergy. 1975;18:97–204. doi: 10.1159/000395257. [DOI] [PubMed] [Google Scholar]
- 340.Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, Hickman HD, McCulloch JA, Badger JH, Ajami NJ, Trinchieri G, Pardo-Manuel de Villena F, Yewdell JW, Rehermann B. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell. 2017;171:1015–1028.e13. doi: 10.1016/j.cell.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Alyanakian MA, Grela F, Aumeunier A, Chiavaroli C, Gouarin C, Bardel E, Normier G, Chatenoud L, Thieblemont N, Bach JF. Transforming growth factor-beta and natural killer T-cells are involved in the protective effect of a bacterial extract on type 1 diabetes. Diabetes. 2006;55:179–185. [PubMed] [Google Scholar]
- 342.Zaccone P, Burton O, Miller N, Jones FM, Dunne DW, Cooke A. Schistosoma mansoni egg antigens induce Treg that participate in diabetes prevention in NOD mice. Eur J Immunol. 2009;39:1098–1107. doi: 10.1002/eji.200838871. [DOI] [PubMed] [Google Scholar]
- 343.Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010;28:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Detich N, Hamm S, Just G, Knox JD, Szyf M. The methyl donor S-Adenosylmethionine inhibits active demethylation of DNA: a candidate novel mechanism for the pharmacological effects of S-Adenosylmethionine. J Biol Chem. 2003;278:20812–20820. doi: 10.1074/jbc.M211813200. [DOI] [PubMed] [Google Scholar]
- 345.Campbell PM, Bovenzi V, Szyf M. Methylated DNA-binding protein 2 antisense inhibitors suppress tumourigenesis of human cancer cell lines in vitro and in vivo. Carcinogenesis. 2004;25:499–507. doi: 10.1093/carcin/bgh045. [DOI] [PubMed] [Google Scholar]
- 346.Radic M, Muller S. Epigenetics of autoantigens: new opportunities for therapy of autoimmune diseases. Genet Epigenet. 2013;5:63–70. doi: 10.4137/GEG.S12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Hamm S, Just G, Lacoste N, Moitessier N, Szyf M, Mamer O. On the mechanism of demethylation of 5-methylcytosine in DNA. Bioorg Med Chem Lett. 2008;18:1046–1049. doi: 10.1016/j.bmcl.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 348.Balada E, Ordi-Ros J, Serrano-Acedo S, Martinez-Lostao L, Vilardell-Tarrés M. Transcript overexpression of the MBD2 and MBD4 genes in CD4+ T cells from systemic lupus erythematosus patients. J Leukoc Biol. 2007;81:1609–1616. doi: 10.1189/jlb.0107064. [DOI] [PubMed] [Google Scholar]
- 349.Tabibian JH, Weeding E, Jorgensen RA, Petz JL, Keach JC, Talwalkar JA, Lindor KD. Randomised clinical trial: vancomycin or metronidazole in patients with primary sclerosing cholangitis - a pilot study. Aliment Pharmacol Ther. 2013;37:604–612. doi: 10.1111/apt.12232. [DOI] [PubMed] [Google Scholar]
- 350.Tabibian JH, Talwalkar JA, Lindor KD. Role of the microbiota and antibiotics in primary sclerosing cholangitis. Biomed Res Int. 2013;2013:389537. doi: 10.1155/2013/389537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Liberal R, Grant CR, Longhi MS, Mieli-Vergani G, Vergani D. Regulatory T cells: Mechanisms of suppression and impairment in autoimmune liver disease. IUBMB Life. 2015;67:88–97. doi: 10.1002/iub.1349. [DOI] [PubMed] [Google Scholar]
- 352.Montano-Loza AJ, Czaja AJ. Cell mediators of autoimmune hepatitis and their therapeutic implications. Dig Dis Sci. 2015;60:1528–1542. doi: 10.1007/s10620-014-3473-z. [DOI] [PubMed] [Google Scholar]
- 353.Czaja AJ. Review article: chemokines as orchestrators of autoimmune hepatitis and potential therapeutic targets. Aliment Pharmacol Ther. 2014;40:261–279. doi: 10.1111/apt.12825. [DOI] [PubMed] [Google Scholar]
- 354.Czaja AJ. Targeting apoptosis in autoimmune hepatitis. Dig Dis Sci. 2014;59:2890–2904. doi: 10.1007/s10620-014-3284-2. [DOI] [PubMed] [Google Scholar]
- 355.Longhi MS, Ma Y, Bogdanos DP, Cheeseman P, Mieli-Vergani G, Vergani D. Impairment of CD4(+)CD25(+) regulatory T-cells in autoimmune liver disease. J Hepatol. 2004;41:31–37. doi: 10.1016/j.jhep.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 356.Longhi MS, Hussain MJ, Mitry RR, Arora SK, Mieli-Vergani G, Vergani D, Ma Y. Functional study of CD4+CD25+ regulatory T cells in health and autoimmune hepatitis. J Immunol. 2006;176:4484–4491. doi: 10.4049/jimmunol.176.7.4484. [DOI] [PubMed] [Google Scholar]
- 357.Peiseler M, Sebode M, Franke B, Wortmann F, Schwinge D, Quaas A, Baron U, Olek S, Wiegard C, Lohse AW, Weiler-Normann C, Schramm C, Herkel J. FOXP3+ regulatory T cells in autoimmune hepatitis are fully functional and not reduced in frequency. J Hepatol. 2012;57:125–132. doi: 10.1016/j.jhep.2012.02.029. [DOI] [PubMed] [Google Scholar]
- 358.McKinney EF, Smith KG. T cell exhaustion and immune-mediated disease-the potential for therapeutic exhaustion. Curr Opin Immunol. 2016;43:74–80. doi: 10.1016/j.coi.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 359.Weiler-Normann C, Schramm C, Quaas A, Wiegard C, Glaubke C, Pannicke N, Möller S, Lohse AW. Infliximab as a rescue treatment in difficult-to-treat autoimmune hepatitis. J Hepatol. 2013;58:529–534. doi: 10.1016/j.jhep.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 360.Burak KW, Swain MG, Santodomingo-Garzon T, Lee SS, Urbanski SJ, Aspinall AI, Coffin CS, Myers RP. Rituximab for the treatment of patients with autoimmune hepatitis who are refractory or intolerant to standard therapy. Can J Gastroenterol. 2013;27:273–280. doi: 10.1155/2013/512624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Kremer JM, Westhovens R, Leon M, Di Giorgio E, Alten R, Steinfeld S, Russell A, Dougados M, Emery P, Nuamah IF, Williams GR, Becker JC, Hagerty DT, Moreland LW. Treatment of rheumatoid arthritis by selective inhibition of T-cell activation with fusion protein CTLA4Ig. N Engl J Med. 2003;349:1907–1915. doi: 10.1056/NEJMoa035075. [DOI] [PubMed] [Google Scholar]
- 362.Kremer JM, Dougados M, Emery P, Durez P, Sibilia J, Shergy W, Steinfeld S, Tindall E, Becker JC, Li T, Nuamah IF, Aranda R, Moreland LW. Treatment of rheumatoid arthritis with the selective costimulation modulator abatacept: twelve-month results of a phase iib, double-blind, randomized, placebo-controlled trial. Arthritis Rheum. 2005;52:2263–2271. doi: 10.1002/art.21201. [DOI] [PubMed] [Google Scholar]
- 363.Larsen CP, Pearson TC, Adams AB, Tso P, Shirasugi N, Strobert E, Anderson D, Cowan S, Price K, Naemura J, Emswiler J, Greene J, Turk LA, Bajorath J, Townsend R, Hagerty D, Linsley PS, Peach RJ. Rational development of LEA29Y (belatacept), a high-affinity variant of CTLA4-Ig with potent immunosuppressive properties. Am J Transplant. 2005;5:443–453. doi: 10.1111/j.1600-6143.2005.00749.x. [DOI] [PubMed] [Google Scholar]
- 364.Rigby MR, Harris KM, Pinckney A, DiMeglio LA, Rendell MS, Felner EI, Dostou JM, Gitelman SE, Griffin KJ, Tsalikian E, Gottlieb PA, Greenbaum CJ, Sherry NA, Moore WV, Monzavi R, Willi SM, Raskin P, Keyes-Elstein L, Long SA, Kanaparthi S, Lim N, Phippard D, Soppe CL, Fitzgibbon ML, McNamara J, Nepom GT, Ehlers MR. Alefacept provides sustained clinical and immunological effects in new-onset type 1 diabetes patients. J Clin Invest. 2015;125:3285–3296. doi: 10.1172/JCI81722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Czaja AJ. Adoptive cell transfer in autoimmune hepatitis. Expert Rev Gastroenterol Hepatol. 2015;9:821–836. doi: 10.1586/17474124.2015.1019470. [DOI] [PubMed] [Google Scholar]
- 366.Lapierre P, Béland K, Yang R, Alvarez F. Adoptive transfer of ex vivo expanded regulatory T cells in an autoimmune hepatitis murine model restores peripheral tolerance. Hepatology. 2013;57:217–227. doi: 10.1002/hep.26023. [DOI] [PubMed] [Google Scholar]
- 367.Seki T, Ota M, Furuta S, Fukushima H, Kondo T, Hino K, Mizuki N, Ando A, Tsuji K, Inoko H. HLA class II molecules and autoimmune hepatitis susceptibility in Japanese patients. Gastroenterology. 1992;103:1041–1047. doi: 10.1016/0016-5085(92)90041-v. [DOI] [PubMed] [Google Scholar]
- 368.Fainboim L, Marcos Y, Pando M, Capucchio M, Reyes GB, Galoppo C, Badía I, Remondino G, Ciocca M, Ramonet M. Chronic active autoimmune hepatitis in children. Strong association with a particular HLA-DR6 (DRB1*1301) haplotype. Hum Immunol. 1994;41:146–150. doi: 10.1016/0198-8859(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 369.Djilali-Saiah I, Fakhfakh A, Louafi H, Caillat-Zucman S, Debray D, Alvarez F. HLA class II influences humoral autoimmunity in patients with type 2 autoimmune hepatitis. J Hepatol. 2006;45:844–850. doi: 10.1016/j.jhep.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 370.Hiraide A, Imazeki F, Yokosuka O, Kanda T, Kojima H, Fukai K, Suzuki Y, Hata A, Saisho H. Fas polymorphisms influence susceptibility to autoimmune hepatitis. Am J Gastroenterol. 2005;100:1322–1329. doi: 10.1111/j.1572-0241.2005.41053.x. [DOI] [PubMed] [Google Scholar]
- 371.Tang J, Zhou C, Zhang ZJ, Zheng SS. Association of polymorphisms in non-classic MHC genes with susceptibility to autoimmune hepatitis. Hepatobiliary Pancreat Dis Int. 2012;11:125–131. doi: 10.1016/s1499-3872(12)60136-2. [DOI] [PubMed] [Google Scholar]
- 372.Li S, Huang X, Zhong H, Chen Z, Peng Q, Deng Y, Qin X. Tumour necrosis factor alpha (TNF-α) genetic polymorphisms and the risk of autoimmune liver disease: a meta-analysis. J Genet. 2013;92:617–628. [PubMed] [Google Scholar]
- 373.Czaja AJ, Carpenter HA, Santrach PJ, Moore SB. Significance of HLA DR4 in type 1 autoimmune hepatitis. Gastroenterology. 1993;105:1502–1507. doi: 10.1016/0016-5085(93)90157-8. [DOI] [PubMed] [Google Scholar]
- 374.Bittencourt PL, Goldberg AC, Cançado EL, Porta G, Carrilho FJ, Farias AQ, Palacios SA, Chiarella JM, Abrantes-Lemos CP, Baggio VL, Laudanna AA, Kalil J. Genetic heterogeneity in susceptibility to autoimmune hepatitis types 1 and 2. Am J Gastroenterol. 1999;94:1906–1913. doi: 10.1111/j.1572-0241.1999.01229.x. [DOI] [PubMed] [Google Scholar]
- 375.Czaja AJ, Kruger M, Santrach PJ, Moore SB, Manns MP. Genetic distinctions between types 1 and 2 autoimmune hepatitis. Am J Gastroenterol. 1997;92:2197–2200. [PubMed] [Google Scholar]
- 376.O'Hehir RE, Lamb JR. Induction of specific clonal anergy in human T lymphocytes by Staphylococcus aureus enterotoxins. Proc Natl Acad Sci U S A. 1990;87:8884–8888. doi: 10.1073/pnas.87.22.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Miethke T, Wahl C, Heeg K, Wagner H. Acquired resistance to superantigen-induced T cell shock. V beta selective T cell unresponsiveness unfolds directly from a transient state of hyperreactivity. J Immunol. 1993;150:3776–3784. [PubMed] [Google Scholar]
- 378.Carlberg C. The vitamin D(3) receptor in the context of the nuclear receptor superfamily : The central role of the retinoid X receptor. Endocrine. 1996;4:91–105. doi: 10.1007/BF02782754. [DOI] [PubMed] [Google Scholar]
- 379.Pike JW, Meyer MB. The vitamin D receptor: new paradigms for the regulation of gene expression by 1,25-dihydroxyvitamin D(3) Endocrinol Metab Clin North Am. 2010;39:255–269, table of contents. doi: 10.1016/j.ecl.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Lennon-Duménil AM, Bakker AH, Wolf-Bryant P, Ploegh HL, Lagaudrière-Gesbert C. A closer look at proteolysis and MHC-class-II-restricted antigen presentation. Curr Opin Immunol. 2002;14:15–21. doi: 10.1016/s0952-7915(01)00293-x. [DOI] [PubMed] [Google Scholar]
- 381.Fiebiger E, Meraner P, Weber E, Fang IF, Stingl G, Ploegh H, Maurer D. Cytokines regulate proteolysis in major histocompatibility complex class II-dependent antigen presentation by dendritic cells. J Exp Med. 2001;193:881–892. doi: 10.1084/jem.193.8.881. [DOI] [PMC free article] [PubMed] [Google Scholar]