Abstract
Hepatocellular carcinoma (HCC) is an aggressive subtype of liver cancer with few effective treatments and the underlying mechanisms that drive HCC pathogenesis remain poorly characterized. Identifying genes and pathways essential for HCC cell growth will aid the development of new targeted therapies for HCC. Using a kinome CRISPR screen in three human HCC cell lines, we identified transformation/transcription domain-associated protein (TRRAP) as an essential gene for HCC cell proliferation. TRRAP has been implicated in oncogenic transformation, but how it functions in cancer cell proliferation is not established. Here, we show that depletion of TRRAP or its co-factor, histone acetyltransferase KAT5, inhibits HCC cell growth via induction of p53- and p21-independent senescence. Integrated cancer genomics analyses using patient data and RNA-sequencing identified mitotic genes as key TRRAP/KAT5 targets in HCC, and subsequent cell cycle analyses revealed that TRRAP- and KAT5-depleted cells are arrested at G2/M phase. Depletion of TOP2A, a mitotic gene and TRRAP/KAT5 target, was sufficient to recapitulate the senescent phenotype of TRRAP/KAT5 knockdown. Conclusion: Our results uncover a role for TRRAP/KAT5 in promoting HCC cell proliferation via activation of mitotic genes. Targeting the TRRAP/KAT5 complex is a potential therapeutic strategy for HCC.
Keywords: Hepatocellular carcinoma, CRISPR screen, histone acetyltransferase, G2/M arrest, senescence
Liver cancer accounts for more than 27,000 deaths in the United States and more than 700,000 deaths worldwide each year (1, 2). The 5-year survival rate for liver cancer patients is 18% and the major subtype of liver cancer is hepatocellular carcinoma (HCC) (3). Current approved advanced HCC treatments include multi-kinase inhibitors regorafenib, lenvatinib and sorafenib, which extend patient survival by several months only, and the immune checkpoint inhibitors Nivolumab and Pembrolizumab, which show ~20% response rates as a second line of therapy (3). The clinical need for more effective HCC treatments remains unmet partially because HCC is genetically heterogeneous and because HCC driver genes amenable to targeted therapy are largely unknown (1, 4). Identifying genes whose depletion can inhibit HCC growth, and determining the mechanisms involved, will aid the development of targeted therapies for HCC patients.
Using a kinome CRISPR screen in three HCC cell lines, we identified transformation/transcription domain-associated protein (TRRAP) as the top-ranking candidate gene required for HCC cell growth. TRRAP, a pseudokinase member of the PI3 kinase-like family, acts as a scaffold protein in histone acetyltransferase (HAT) complexes (5–7) that regulate transcription, DNA repair, and replication (8). TRRAP is also a key regulator of cell cycle progression and stemness in embryonic stem cells and cortical apical neural progenitors (9, 10).
TRRAP is mutated and amplified in different cancer types, including melanomas, gastric, and uterine cancers (11–14), and has been implicated in oncogenic transformation. Indeed, the TRRAP S722F hotspot mutation in melanomas can transform NIH3T3 cells (11). Moreover, wildtype TRRAP is required for transformation by c-myc/Hras and E1A/Hras in rat embryo fibroblasts (15) and MYC-dependent tumor initiation in a breast cancer model (16). By contrast, depletion of TRRAP can induce apoptosis and differentiation in lymphoma and brain-tumor initiating cells (17, 18). Given its role in diverse cellular processes, it is possible that TRRAP and its regulated pathways are altered in cancer. However, the extent of TRRAP alterations and its function in cancer cells, particularly in the context of HCC, is not established.
Here, we show that depletion of TRRAP/KAT5 in HCC cells induces G2/M arrest and senescence in a p53- and p21-independent manner. We found that TRRAP/KAT5 activates a subset of mitotic genes that is highly expressed in HCC and is associated with poor survival in HCC patients. Depletion of TOP2A, a mitotic gene and TRRAP/KAT5 target, is sufficient to recapitulate the senescent phenotype observed in TRRAP- and KAT5-depleted cells. Together, our study uncover a role for TRRAP/KAT5 in HCC cell growth and disruption of TRRAP/KAT5 is a potential therapeutic strategy for HCC.
Materials and Methods
Cell culture
Huh7 and SNU475 cells were provided by Dr. Scott Lowe. Huh7 cells were cultured in DMEM. SNU-475 cells were cultured in RPMI supplemented with 10 mM HEPES, 1 mM sodium pyruvate, and 4500 mg/L glucose. Hep3B and HepG2 cells were provided by Dr. Junwei Shi and cultured in MEM. All cell lines were grown in media supplemented with 10% FBS and 1% penicillin-streptomycin and maintained in a 37°C incubator with 5% CO2. Huh7, HepG2, Hep3B and SNU-475 cells were authenticated using ATCC’s cell authentication service. MG132 and chloroquine were purchased from Millipore Sigma.
Kinome CRISPR screen
The human kinome CRISPR pooled library was a gift from John Doench and David Root (Addgene #1000000083). The library was amplified according to Addgene’s library amplification protocol. Lentivirus containing the kinome library was packaged using 293fs cells. For each cell line, duplicates were performed. For each replicate, 2 × 107 cells were infected with lentivirus. Huh7 and Hep3B cells were selected with 2 μg/mL puromycin for 3 days and HepG2 cells were selected with 4 μg/mL puromycin for 4 days. For each sample, genomic DNA was harvested from at least 6 × 106 cells using the PureLink Genomic DNA mini kit (Thermo Fisher Scientific, Waltham, MA). sgRNA was amplified using Phusion flash high-fidelity PCR master mix (Thermo Fisher Scientific). All sequencing datasets were evaluated using FastQC (version 0.11.2) to ensure high quality. Depleted genes were identified as previously described (19).
Bioinformatics analysis
To analyze gene expression levels between tumor and non-tumor tissues, the GSE14520 (20) and TCGA HCC data sets were used. Gene expression data of tumor (n=225) and non-tumor tissue (n=220) in the GSE14520 data set were analyzed using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/). Briefly, P-values were calculated using moderated t-statistics and adjusted using the Benjamini-Hochberg method. Gene expression data of TCGA HCC tumors and matched non-tumors (n=50) were obtained from the UCSC Xena Browser (http://xena.ucsc.edu/) and analyzed using the Wilcoxon signed-rank test. The list of genes that predict prognosis of HCC patients was obtained from the Human Protein Atlas (version 18, https://www.proteinatlas.org/). For survival analysis, FKPM values and survival data were obtained from the Human Protein Atlas and cBioPortal (http://www.cbioportal.org/) respectively. The LEC data is available on GEO with accession numbers GSE1898 and GSE4024 (21, 22). FKPM values were grouped in tertiles and the highest and lowest tertiles were designated as ‘high expression’ and ‘low expression’ groups. P-values were calculated using the log-rank Mantel-Cox test with GraphPad Prism. TRRAP mutations and copy-number alterations in HCC were obtained from cBioPortal. For gene correlation analyses, correlation data from the TCGA HCC (n=360) and GBM (n=136) data sets were obtained from cBioPortal. Briefly, gene expression data were analyzed using Spearman’s correlation analysis. For GO analyses, the list of genes was analyzed using the PANTHER overrepresentation test and annotated with the GO Ontology database (released 2018-10-08). P-values were calculated using Fisher’s Exact test and corrected by the Benjamini-Hochberg method. To determine KAT5 binding sites, we analyzed ChIP-sequencing data from GSE69671 (23).
sgRNA design and lentivirus infection
sgRNAs were designed using https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design, cloned into the LentiCRISPRv2 backbone (Addgene #52961) and packaged into lentivirus using 293fs cells. HCC cells infected with lentivirus were selected with puromycin for 2–4 days. To generate double sgRNA-infected cells, sgNT and sgp21 were cloned into the LentiCRISPRv2 hygro backbone (Addgene #98291). SNU-475 and Huh7 cells infected with lentivirus were selected with 100 and 500 μg/mL hygromycin for 6 and 10 days respectively, then infected with sgNT, sgTRRAP, sgKAT5, and sgKAT2A, and selected with puromycin. Sequences of sgRNAs are listed in Supplementary Table 1.
Immunoblot Analysis
Cells were washed twice with ice-cold PBS and harvested in RIPA buffer (Boston Bioproducts, Ashland MA) supplemented with protease (Roche, Indianapolis, IN) and phosphatase inhibitor cocktails (Thermo Fisher Scientific). The concentration of protein was measured using the BCA assay (Thermo Fisher Scientific). For each sample, 25 μg of protein was loaded onto an SDS-PAGE gel. The following antibodies were used to probe against: TRRAP (#3967; Cell Signaling Technology, Danvers, MA), p21 (#2947; Cell Signaling Technology), KAT2A (#3305; Cell Signaling Technology), KAT5 (sc-166323; Santa-Cruz Biotechnology, Dallas, TX), NIK (#4994; Cell Signaling Technology), LC3B (#2775; Cell Signaling Technology), TOP2A (#12286; Cell Signaling Technology), p53 (sc-126; Santa-Cruz Biotechnology), GAPDH (MAB374; Millipore Sigma, Burlington, MA), Hsp90 (#610419; BD Bioscience, San Jose, CA), and Flag (#2368; Cell Signaling Technology and #F1804; Millipore Sigma). Bands were visualized with an immunofluorescent secondary antibody (LICOR) using the Odyssey Imaging system.
Cell growth and colony formation assays
To measure cell growth, cells were seeded onto 96-well plates. CellTitre-glo assay (Promega, Madison, WI) was performed according to the manufacturer’s protocol. The growth of cells at day 3 and day 5 was normalized to measurements taken one day after the cells were plated. Each repeat was averaged between 6 wells. For colony formation assays, Huh7 and SNU-475 cells were seeded at 3000 and 2000 cells per well respectively onto 6-well plates. After 11 days, cells were fixed with 4% formalin and stained with 0.5% crystal violet. All experiments were repeated three times.
SA-β-gal assay
Huh7, Hep3B, and SNU-475 cells were seeded onto 12-well plates, senescence-associated β-galactosidase (SA-β-gal) staining was performed at pH = 6.0 and positively stained cells were quantified as previously described (24).
RNA extraction and RT-qPCR
RNA was extracted from cells using the RNeasy mini kit (Qiagen, Germantown, MD) and DNA was removed by on-column DNase digestion (Qiagen) according to the manufacturer’s protocol. One microgram of RNA was used to synthesize cDNA using the high-capacity cDNA reverse transcription kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. RT-qPCR analyses were performed using SsoFast EvaGreen supermix (Bio-Rad, Hercules, CA) according to the manufacturer’s protocol and GAPDH was used as a control. Primer sequences used for qRT-PCR are listed in Supplementary Table 2.
Xenografts in nude mice
Female 6-week-old NCRNu/Nu mice were purchased from Taconic Bioscience (Rensselaer, NY) and injected in the right or left flank with 1×106 TRRAP stable knockdown Huh7 cells. Cells were resuspended in PBS and mixed in a 2:1 ratio with matrigel (Westnet Inc., Canton, MA) to a final volume of 200 μL. Tumor size was measured by calipers and volume was calculated using the formula . All mice were housed in JAG75 (Allentown) ventilated cage systems with 1/4″ Bed-o’Cobs (Andersons Lab Bedding) in facilities accredited by the American Association for Laboratory Animal Care (AALAC) at the University of Massachusetts (UMass) Medical School. Mice were provided with nestlets (Ancare) and Bed-r’Nests (Andersons Lab Bedding) and maintained in a pathogen‐free animal facility at 22°C under the 12‐hour light/dark cycle. Mice were injected during the light cycle and were not fasted prior to injection. Mice were maintained on a standard diet of Prolab Isopro RMH 3000 (LabDiet) throughout the course of the experiment. All animal protocols were approved by the UMass Medical School Institutional Animal Care and Use Committee (IACUC) and comply with all relevant federal guidelines and institutional policies.
RNA-sequencing and bioinformatics analysis
Ribosomal RNA was depleted and RNA-sequencing libraries were prepared as previously described (25). The libraries were quantified using the KAPA Library Quantification Kit (Kapa Biosystems, Wilmington, MA) and sequenced using the Illumina NextSeq 500 system. RNA-seq reads from each sample were aligned using STAR (version 2.5.2b) (26) against the GRCh37/hg19 human reference genome. Gene expression was quantified through RSEM (v1.2.31) (27) with GENCODE V19 gene annotation. DESeq2 (1.22.0) (28) was employed to identify differentially expressed genes using raw read counts quantified by htseq-count (29).
Flow cytometry analyses
For cell cycle analysis, Huh7 and SNU-475 cells were pulsed with 30 μg/mL 5-Bromo-2´-Deoxyuridine (BrdU, Thermo Fisher Scientific) for 1 hour at 37°C. Cells were then trypsinized and fixed with 70% ethanol, permeabilized with 0.3% Triton-X 100 and incubated with a BrdU-FITC conjugated antibody (Thermo Fisher Scientific). Cells were stained with 50 ng/mL propidium iodide (Thermo Fisher Scientific), 10 μg/mL RNase A (Thermo Fisher Scientific), and incubated for 30 minutes at 37°C. For γH2AX analysis, cells were fixed in 70% ethanol, permeabilized with 0.3% Triton-X100 and blocked in 0.8% BSA. Cells were then incubated with an anti-γH2A.X antibody (#9718; Cell Signaling Technology), which was detected using a mouse-FITC IgG. Cells were also stained with propidium iodide as described above to identify 2N and >2N populations. For each sample, at least 40,000 cells were analyzed using the MACSQuant VYB Flow cytometer (MACS Miltenyi Biotec, Auburn, CA). All data were analyzed by FlowJo 10.0 software.
Statistics
Statistical analysis was performed using GraphPad Prism software and data were presented as means ± standard deviation. Student’s t-test was used to determine P-values unless indicated otherwise. P-values of <0.05 were considered to be statistically significant.
Results
A kinome CRISPR screen identifies TRRAP as a regulator of cell growth in HCC
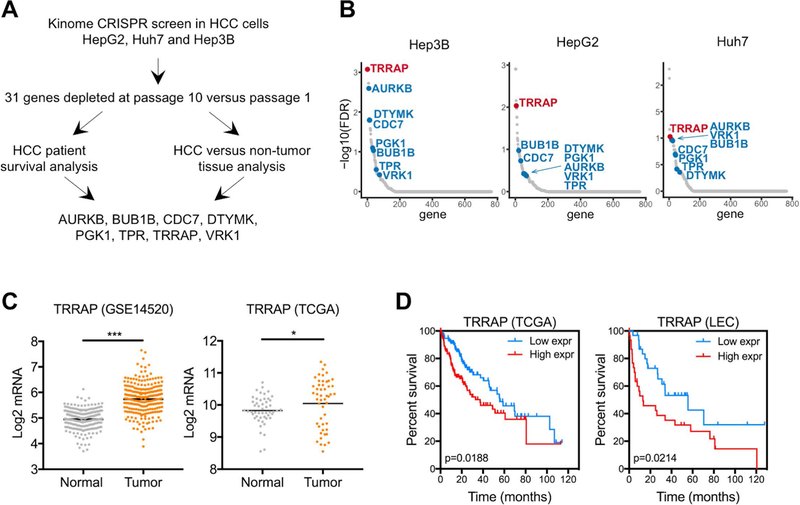
To identify genes that are required for cell growth in HCC, we infected three HCC cell lines (Huh7, HepG2, and Hep3B) with a kinome CRISPR library that targets 763 kinases with 8 single guide RNAs (sgRNAs) per gene. To determine changes in sgRNA representation, we harvested genomic DNA from cells of an early passage and after 10 passages, then amplified and identified sgRNAs using PCR and deep sequencing, respectively. We found that 31 genes were significantly depleted in all three HCC cell lines (Figure 1A). Among these 31 genes, 8 genes (AURKB, BUB1B, CDC7, DTYMK, PGK1, TPR, TRRAP, VRK1) were expressed at least 1.5-fold higher in HCC compared to non-tumor tissue (Figure 1B, 1C, and Supplementary Figure 1A). High expression levels of these genes were associated with poor survival in HCC patients (Figure 1D and Supplementary Figure 1B). Of note, previous studies have shown that AURKB, DTYMK, PGK1, TPR, and VRK1 promote HCC tumorigenesis (30–34).
Figure 1. A kinome CRISPR screen identifies TRRAP as an essential gene for HCC cell growth.
A) Three HCC cell lines were used to identify genes whose depletion inhibit HCC proliferation. Eight candidate genes were identified by integrating CRISPR screen results with HCC patient data. B) Genes from the CRISPR screen were ranked by their false discovery rate, the 8 candidate genes are indicated. C) mRNA levels of TRRAP in non-tumor and tumor samples from the GSE14520 (left) and TCGA (right) data sets, p-values were calculated using moderated t-test and Wilcoxon signed-rank test respectively. Black lines indicate the geometric mean of each group. D) Kaplan Meier curves of HCC patients from the TCGA (left) and NIH Laboratory of Experiment Carcinogenesis (LEC, right) cohorts with high (TCGA n=115 and LEC n=31) or low (TCGA n=118 and LEC n=31) TRRAP expression. P-values were calculated using the log-rank Mantel-Cox test. *p < 0.05, ***p < 0.001.
Out of the 8 genes, TRRAP is the top depleted gene in all three cell lines (Figure 1B). Analysis of the TCGA data set revealed that 12% of HCC patient samples have increased TRRAP mRNA expression, amplification and/or mutations in the TRRAP gene (Supplementary Figure 1C). Using additional published patient data sets, we confirmed that TRRAP expression is higher in tumor compared to matched normal tissue (Figure 1C), and increased TRRAP expression is correlated with poor survival (Figure 1D). We also investigated whether TRRAP expression is correlated with certain gene signatures in HCC. We performed Ingenuity pathway analysis and found that TRRAP expression is positively correlated with expression of genes that are involved in ‘molecular mechanisms of cancer’, ‘role of BRCA1 in DNA damage response’ and growth factor signaling pathways (Supplementary Figure 1D). These results suggest that TRRAP may have an oncogenic role in HCC.
Depletion of TRRAP induces senescence in HCC cells
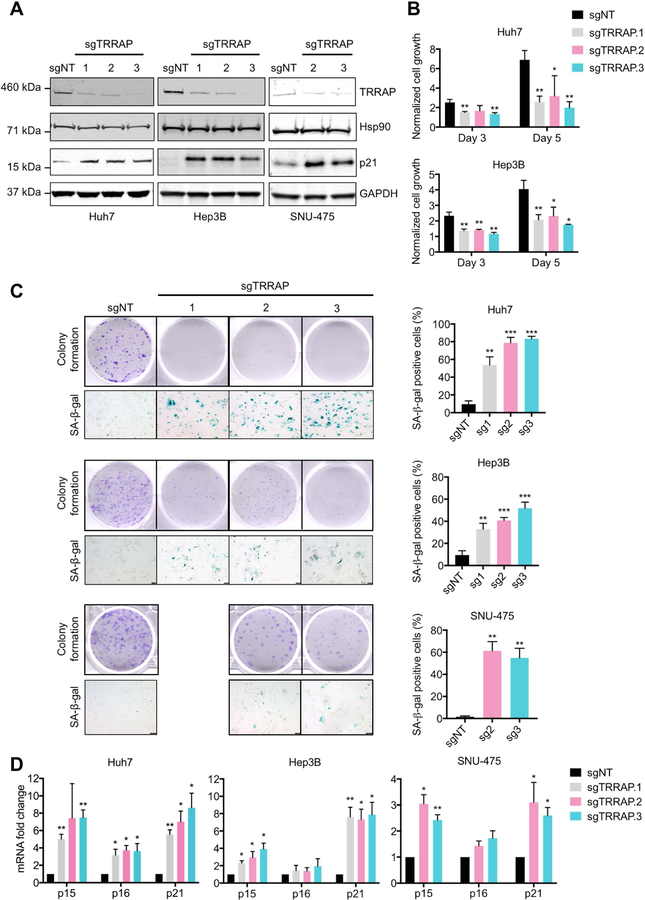
To understand the function of TRRAP in HCC, we depleted TRRAP expression in Huh7 Hep3B and SNU-475 cells using CRISPR (sgTRRAP) (Figure 2A). Loss of TRRAP resulted in decreased cell growth and colony formation in these cell lines (Figure 2B and 2C). Morphologically, the cells became flatter and enlarged after TRRAP depletion, reminiscent of a senescent phenotype (35). Indeed, loss of TRRAP in Huh7 and Hep3B cells resulted in positive senescence-associated-beta-galactosidase (SA-β-gal) staining (Figure 2C) (35, 36). Furthermore, the molecular markers of senescence—p15, p16, and p21 (35, 36)—were increased in sgTRRAP cells compared to non-targeting controls (Figure 2A and 2D). Collectively, these data suggest that loss of TRRAP inhibits cell growth by inducing senescence.
Figure 2. Loss of TRRAP impairs cell growth and induces senescence in HCC cells.
A) TRRAP and p21 protein expression in Huh7, Hep3B, and SNU-475 cells infected with non-target (sgNT) and 3 individual TRRAP sgRNAs (sgTRRAP). B and C) Cell growth, colony formation and induction of senescence of Huh7, Hep3B, and SNU-475 cells infected with sgNT and sgTRRAP. B) Cell growth was measured at 1, 3, and 5 days after plating using the CTG assay and normalized to day 1. C) Cells were stained with crystal violet and senescence was measured by SA-β-gal staining. D) mRNA levels of p15, p16, and p21 in sgTRRAP cells were measured by qRT-PCR and normalized to sgNT cells. Data was presented as mean ± SD; p-values were calculated by comparing to sgNT, *p < 0.05, **p < 0.01, ***p < 0.001 (student’s t test).
Histone acetyltransferase KAT5 is required for cell growth in HCC
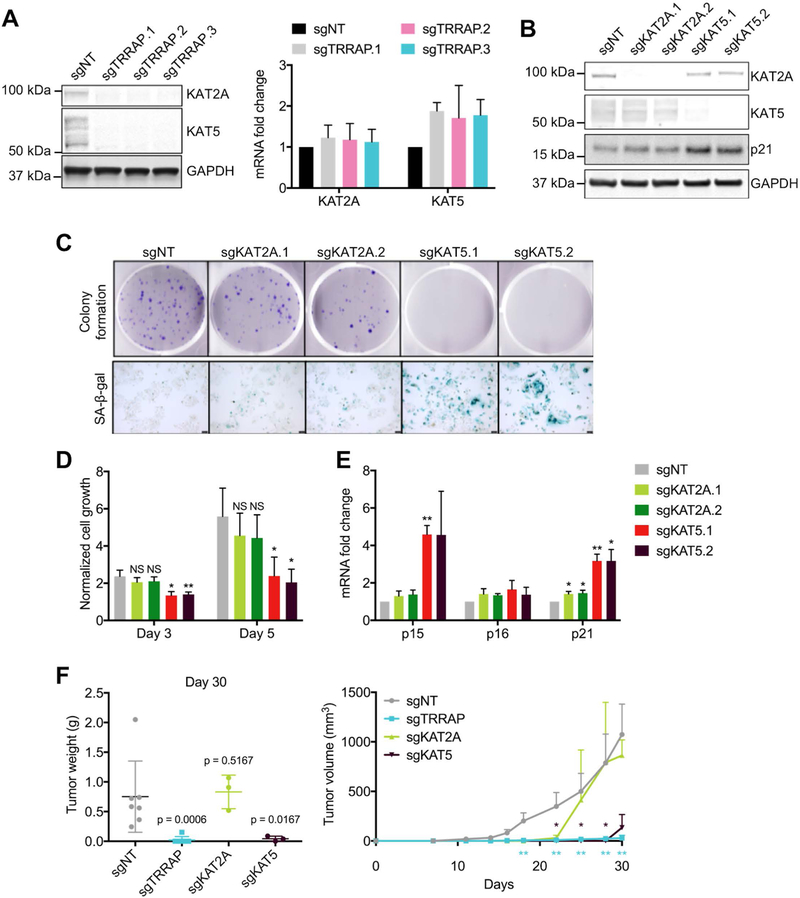
TRRAP is an adaptor protein in several HAT complexes. In mammalian cells, TRRAP predominantly binds to KAT2A and KAT5 (5–7). We found that TRRAP and KAT5 co-localized to the nucleus in Huh7 and SNU-475 cells (Supplementary Figure 2A). We found that TRRAP depletion resulted in decreased KAT5 and KAT2A expression at the protein, but not mRNA level, in Huh7 and SNU-475 cells (Figure 3A and Supplementary Figure 2B). To investigate whether KAT2A and KAT5 protein become unstable due to increased protein degradation in the absence of TRRAP, we inhibited proteasomal and lysosomal protein degradation in sgTRRAP cells with MG132 and chloroquine respectively. However, we found that MG132 and chloroquine could not rescue KAT2A and KAT5 protein expression in sgTRRAP cells (Supplementary Figure 2C), suggesting a degradation-independent mechanism. To determine whether TRRAP depletion induces senescence due to loss of KAT2A or KAT5 expression, we depleted KAT2A and KAT5 using CRISPR (sgKAT2A and sgKAT5) in Huh7 and SNU-475 cells (Figure 3B and Supplementary Figure 2D). We found that depletion of KAT5, but not KAT2A, reduced cell growth and colony formation, and induced senescence similarly to TRRAP-depleted cells (Figure 3C, 3D, and Supplementary Figure 2E). We also found a higher increase in p15 and p21 expression in sgKAT5 cells compared to non-targeting control and sgKAT2A cells (Figure 3E and Supplementary Figure 2F).
Figure 3. Loss of histone acetyltransferase KAT5 impairs growth in HCC and induces senescence.
A) Protein (left) and mRNA (right) levels of KAT2A and KAT5 in Huh7 cells infected with sgNT and sgTRRAP as measured by western blot and qRT-PCR. B) Western blot of KAT2A, KAT5, and p21 levels in Huh7 cells infected with sgNT, sgKAT2A and sgKAT5. C and D) Colony formation, senescence and cell growth of Huh7 cells infected with sgNT, sgKAT2A, and sgKAT5. C) Cells were stained with crystal violet and SA-β-gal. D) Cell growth was measured at 1, 3 and 5 days after plating using the CTG assay and normalized to day 1. E) mRNA levels of p15, p16, and p21 in sgKAT2A and sgKAT5 cells were measured by qRT-PCR and normalized to sgNT cells. F) Tumor weight at day 30 (left) and tumor sizes (right) in nude mice subcutaneously injected with Huh7 cells. P-values were calculated using Mann-Whitney test against the sgNT group. Data was presented as mean ± SD; p-values were calculated by comparing to sgNT, *p < 0.05, **p < 0.01, ***p < 0.001, NS = not significant (student’s t test).
To investigate whether depletion of TRRAP, KAT2A, and KAT5 affect tumor growth in vivo, we injected nude mice subcutaneously with Huh7 cells expressing sgTRRAP, sgKAT2A-, or sgKAT5. TRRAP- and KAT5-depleted tumors were significantly smaller than tumors in non-targeting controls. However, KAT2A depletion did not have any effect on tumor size at the end point of the study (Figure 3F). Together, these results suggest that TRRAP depletion leads to decreased KAT2A and KAT5 expression, but only KAT5 is required for HCC growth.
p53 and p21 are dispensable for senescence induced by loss of TRRAP and KAT5
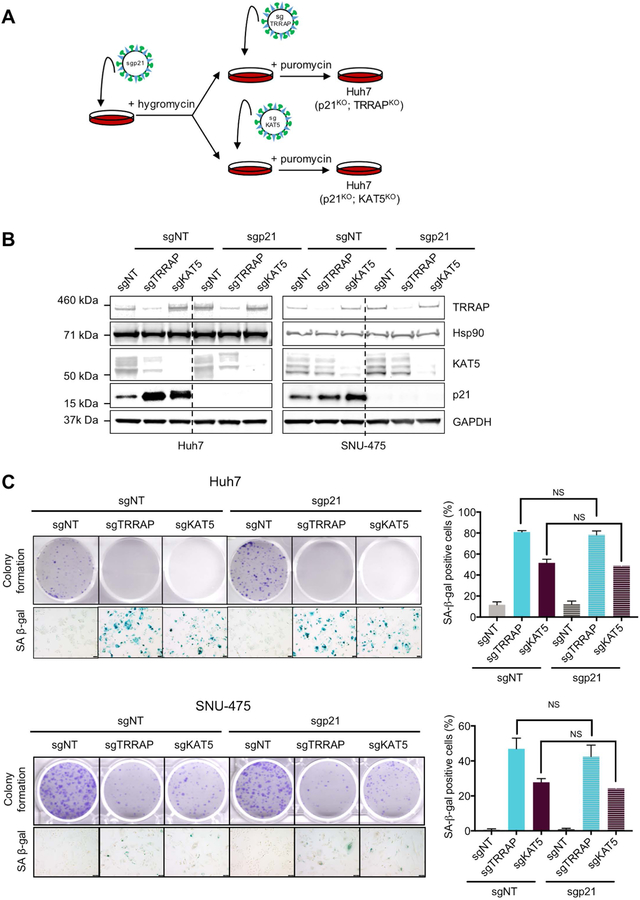
Senescence can be triggered by multiple stimuli, such as DNA damage, replicative stress, activation of oncogenes, and oxidative stress (37). A key molecular event in induction of senescence is p53 activation (38). However, Huh7 and SNU-475 cells are p53-mutant, and Hep3B cells are p53-null, suggesting that the observed induction of senescence due to depleted TRRAP and KAT5 is independent of p53. Since we observed an increase in the expression of p21, a p53 target that is also important in inducing senescence (38), we asked whether p21 is required to induce senescence in TRRAP- and KAT5-depleted cells. To this end, we co-depleted p21 and either TRRAP or KAT5 using CRISPR in Huh7 and SNU-475 cells (Figure 4A). We found that, even in the absence of p21, depletion of TRRAP and KAT5 inhibited colony formation and induced senescence (Figure 4B and 4C), suggesting that the canonical p53/p21 pathway is not required for senescence in this context.
Figure 4. Senescence induced by TRRAP and KAT5 depletion is independent of p21.
A) Schematic illustrating generation of p21 and TRRAP/KAT5 double knockout cells using CRISPR. B) Western blot analysis of TRRAP, KAT5 and p21 levels in Huh7 and SNU-475 cells infected with the indicated sgRNAs. C) Colony formation and SA-β-gal staining of Huh7 cells infected with the indicated sgRNAs. NS = not significant (student’s t test).
TRRAP and KAT5 activate transcription of mitotic genes
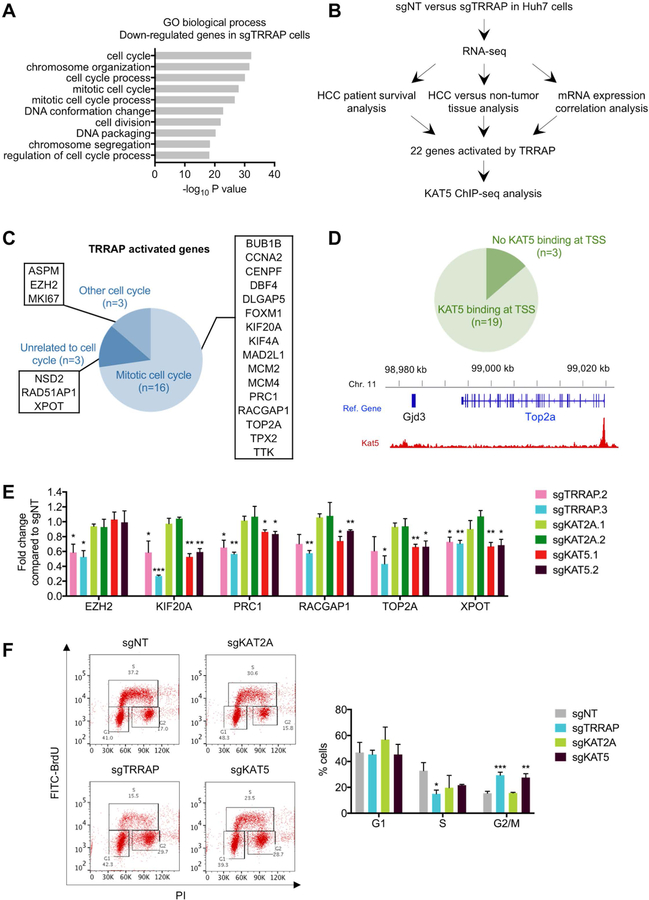
The function of TRRAP in transcriptional regulation has been well described (8). To identify genes that are regulated by TRRAP to promote HCC cell growth, we performed RNA-sequencing in sgTRRAP Huh7 cells to identify differentially-expressed genes. Gene ontology (GO) analysis of genes that were down-regulated in sgTRRAP cells were enriched in cell cycle processes, including mitosis, chromosome segregation, and cell division (Figure 5A). Genes that were up-regulated in sgTRRAP cells were enriched in nucleic acid processing (Supplementary Figure 3A).
Figure 5. TRRAP/KAT5 activates transcription of mitotic genes and depletion of TRRAP/KAT5 leads to G2/M arrest.
A) Gene ontology (GO) analysis of down-regulated genes in sgTRRAP cells compared to non-targeting control. The 10 most significant annotation clusters are shown here. B) Pipeline for identifying HCC-relevant TRRAP target genes. C) GO analysis of the 22 TRRAP-activated genes identified in B. D) KAT5 binding analysis at the transcriptional start sites (TSS) of the 22 TRRAP-activated genes using published ChIP-seq data (top). KAT5 binds to the TSS of TOP2A (bottom). E) mRNA levels of 6 TRRAP-activated genes in Huh7 cells infected with the indicated sgRNAs as measured by qRT-PCR. F) Cell cycle analysis of Huh7 infected with the indicated sgRNAs by BrdU and PI staining. Representative data (left) and quantification (right) are shown here. Data was presented as mean ± SD; p-values were calculated by comparing to non-targeting control, *p < 0.05, **p < 0.01, ***p < 0.001 (student’s t test).
Next, we cross referenced our RNA-sequencing results with published data sets to identify candidate genes that are activated by TRRAP in HCC. We looked for genes that: (1) were down-regulated in sgTRRAP cells from our RNA-sequencing data (fold change<0.5), (2) were positively correlated with TRRAP expression in the TCGA HCC data set (Spearman’s correlation≥0.3), (3) were overexpressed in HCC compared to non-tumor tissue in the GSE14520 data set (fold change≥2), and 4) predict poor survival in HCC patients when highly expressed. We also performed this analysis in the opposite direction to identify genes that were repressed by TRRAP (Figure 5B). Using these criteria, we identified 22 HCC-relevant genes that were activated and 3 that were repressed by TRRAP (Table 1, Supplementary Table 3, Supplementary Figures 3B, 3C, 4 and 5). The 3 TRRAP-repressed genes (BAAT, ITIH1 and RDH16) were liver specific, whereas 19 of the 22 TRRAP-activated genes were involved in either ‘cell cycle’ or ‘mitotic cell cycle’ from the GO analysis (Figure 5C).
Table 1.
List of genes that are down-regulated in the absence of TRRAP and identified from our bioinformatic analyses.
| Gene Symbol | Entrez gene name | RNA-seq sgTRRAP/sgNT | GSE14520 HCC/non-tumor tissue | TCGA HCC co-expressed with TRRAP | |||
|---|---|---|---|---|---|---|---|
| Fold change | P-value | Fold change | P-value | Spearman’s coefficient | P-value | ||
| ASPM | Abnormal spindle microtubule assembly | 0.46 | 1.8E-02 | 8.22 | 8.3E-112 | 0.357 | 1.7E-11 |
| BUB1B | BUB1 mitotic checkpoint serine/threonine kinase B | 0.42 | 2.6E-07 | 3.33 | 1.4E-73 | 0.428 | 2.0E-16 |
| CCNA2 | Cyclin A2 | 0.35 | 2.2E-03 | 2.45 | 1.7E-59 | 0.309 | 9.8E-09 |
| CENPF | Centromere protein F | 0.39 | 5.7E-03 | 5.25 | 3.9E-101 | 0.452 | 2.3E-18 |
| DBF4 | DBF4 zinc finger | 0.46 | 8.4E-06 | 2.41 | 2.6E-71 | 0.48 | 7.2E-21 |
| DLGAP5 | DLG associated protein 5 | 0.42 | 2.9E-06 | 2.49 | 6.9E-60 | 0.316 | 3.9E-09 |
| EZH2 | Enhancer of zeste 2 polycomb repressive complex 2 subunit | 0.38 | 5.5E-07 | 3.41 | 2.4E-78 | 0.408 | 6.1E-15 |
| FOXM1 | Forkhead box M1 | 0.38 | 4.5E-08 | 2.27 | 5.9E-67 | 0.414 | 2.2E-15 |
| KIF20A | Kinesin family member 20A | 0.23 | 1.2E-04 | 3.24 | 3.8E-80 | 0.332 | 5.9E-10 |
| KIF4A | Kinesin family member 4A | 0.41 | 5.5E-06 | 2.70 | 1.4E-69 | 0.314 | 5.3E-09 |
| MAD2L1 | Mitotic arrest deficient 2 like 1 | 0.45 | 1.2E-06 | 2.88 | 4.9E-62 | 0.321 | 2.3E-09 |
| MCM2 | Minichromosome maintenance complex component 2 | 0.38 | 1.9E-07 | 3.13 | 6.3E-69 | 0.342 | 1.5E-10 |
| MCM4 | Minichromosome maintenance complex component 4 | 0.46 | 1.0E-06 | 3.03 | 3.2E-72 | 0.53 | 5.9E-26 |
| MKI67 | Marker of proliferation ki-67 | 0.41 | 4.3E-02 | 2.38 | 9.5E-51 | 0.403 | 1.6E-14 |
| NSD2 | Nuclear receptor binding SET domain protein 2 | 0.46 | 3.5E-06 | 2.10 | 3.0E-61 | 0.605 | 2.6E-35 |
| PRC1 | Protein regulator of cytokinesis 1 | 0.40 | 4.9E-09 | 5.49 | 9.2E-106 | 0.378 | 8.2E-13 |
| RACGAP1 | Rac gtpase activating protein 1 | 0.39 | 1.2E-07 | 4.66 | 2.5E-108 | 0.333 | 5.1E-10 |
| RAD51AP1 | RAD51 associated protein 1 | 0.50 | 3.3E-04 | 2.59 | 2.0E-62 | 0.411 | 4.1E-15 |
| TOP2A | DNA topoisomerase II alpha | 0.34 | 5.2E-03 | 8.27 | 8.5E-98 | 0.406 | 8.6E-15 |
| TPX2 | TPX2, microtubule nucleation factor | 0.37 | 5.6E-08 | 3.22 | 7.3E-71 | 0.317 | 3.6E-09 |
| TTK | TTK protein kinase | 0.47 | 2.0E-05 | 3.22 | 9.7E-69 | 0.306 | 1.4E-08 |
| XPOT | Exportin for trna | 0.47 | 1.5E-08 | 2.44 | 1.9E-62 | 0.485 | 2.5E-21 |
We focused on TRRAP-activated genes for further analysis as they might be potential targets for inhibiting HCC growth. To assess whether TRRAP may regulate the same genes in another cancer type, we analyzed gene expression data from the TCGA glioblastoma multiforme (GBM) data set. TRRAP is required for restricting differentiation of brain tumor-initiating cells that were derived from human GBM samples (10). We found that 14 of the 22 genes activated by TRRAP in HCC were positively correlated with TRRAP expression in GBM (Spearman’s correlation≥0.3, Supplementary Table 4), suggesting that TRRAP regulates a similar set of genes in both cancer types.
Since TRRAP- and KAT5-depleted cells display a similar senescent phenotype, we asked whether TRRAP target genes were also regulated by KAT5. We analyzed previously published ChIP-sequencing data in mouse embryonic stem cells for KAT5 binding sites (23) and found that KAT5 binds to the transcriptional start sites of 19/22 genes activated by TRRAP (Figure 5D and Supplementary Figure 6). After validating our RNA-sequencing data by confirming the downregulation of 6 genes after TRRAP depletion using qRT-PCR (Figure 5E and Supplementary Figure 7A), we investigated the effect of KAT5 and KAT2A depletion on the expression of TRRAP target genes. We found that mRNA expression of TRRAP-activated genes were downregulated in sgKAT5 cells, but not in sgKAT2A cells (Figure 5E and Supplementary Figure 7A). In summary, TRRAP and KAT5 are required for transcriptional activation of mitotic genes. In HCC, these genes are clinically relevant since they are overexpressed and confer poor survival to patients.
TRRAP and KAT5 depletion induces G2/M arrest
Previous studies have found that cells undergoing oncogene-induced senescence are arrested at G1 phase (24), but more recent evidence suggests that G2 arrest can also induce senescence (39). Since we found that the majority of TRRAP target genes regulate mitosis, we reasoned that TRRAP depletion may cause alterations in cell cycle progression. We analyzed cell cycle profiles using BrdU and PI staining and found that sgTRRAP and sgKAT5 cells accumulated at the G2/M phase. As expected, sgKAT2A cells did not accumulate at G2/M phase and displayed a cell cycle profile similar to non-targeting controls (Figure 5F and Supplementary Figure 7B). We also measured DNA damage using the γH2AX marker in sgTRRAP, sgKAT2A, and sgKAT5 cells, and found similar levels compared to non-targeting controls (Supplementary Figures 7C and 7D). This suggests that DNA damage is unlikely the reason for G2/M arrest and senescence upon TRRAP and KAT5 depletion.
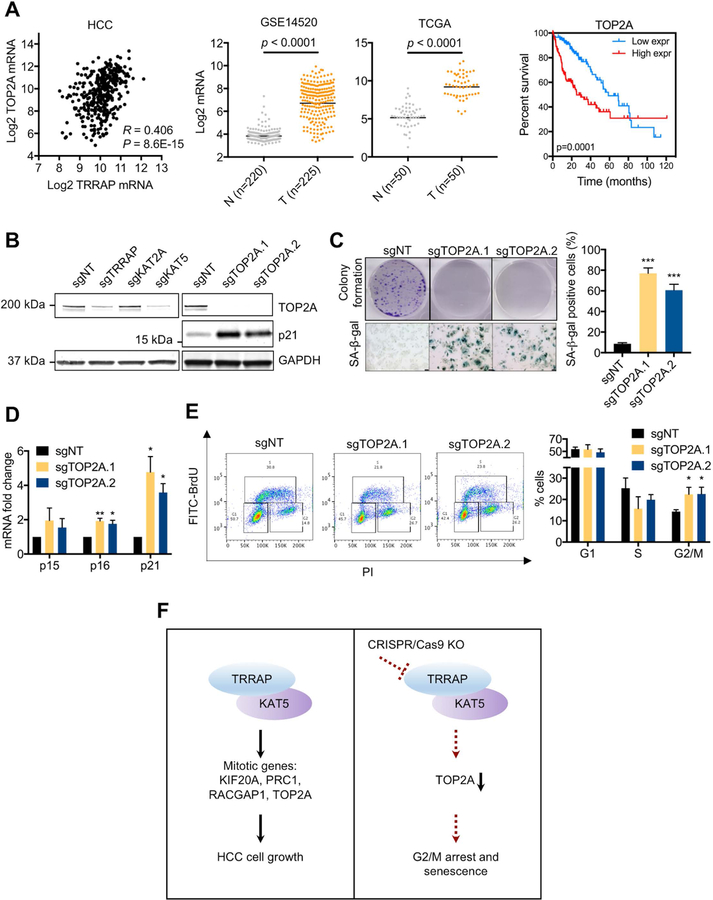
Depletion of TOP2A is sufficient to recapitulate the senescent phenotype of TRRAP-depleted cells
Based on our RNA-sequencing and bioinformatic analyses, we hypothesized that TRRAP depletion induces senescence by down-regulating mitotic genes. To investigate this, we focused on a downstream target of TRRAP and KAT5—Topoisomerase II alpha (TOP2A) (Table 1). TOP2A is highly expressed at G2/M phase and a key regulator of DNA decatenation during mitosis (40). We found that TOP2A expression was positively correlated to TRRAP expression in HCC and GBM patient samples (Figure 6A and Supplementary Figure 8A). Moreover, TOP2A was overexpressed in HCC compared to non-tumor tissue, and its increased expression was associated with poor survival in HCC patients (Figure 6A). We confirmed that TRRAP and KAT5 depletion reduced TOP2A at the mRNA and protein level (Figures 5E, 6B, Supplementary Figures 7A and 9A). Analysis of published ChIP-sequencing data (23) revealed that KAT5 binding to the transcriptional start site of TOP2A (Figure 5D). Using the ChIP-sequencing data, we predicted the KAT5 binding site in the human TOP2A promoter. We cloned a 500 bp (−133/+367) and 1 kb (−383/+617) region spanning the transcriptional start site of TOP2A to a luciferase promoter reporter and found that overexpression of TRRAP and KAT5 induced TOP2A promoter activity (Supplementary Figure 9B). Next, we depleted TOP2A with CRISPR (sgTOP2A) and found reduced colony formation, induction of senescence, and G2/M arrest (Figures 6B–E and Supplementary Figures 9C–E), phenotypes resembling those of TRRAP or KAT5 depletion. Furthermore, we found that γH2AX levels in sgTOP2A cells were similar to non-targeting controls, suggesting that DNA damage is not the cause for the observed G2/M arrest and senescence (Supplementary Figure 9F). Together, our results suggest that TOP2A is a key mitotic target of TRRAP and KAT5 in regulating HCC cell growth.
Figure 6. Loss of TRRAP and KAT5 induce senescence through down-regulation of TOP2A.
A) Correlation between TRRAP and TOP2A mRNA expression in the TCGA HCC data set (n=360) was determined using Spearman’s correlation analysis (left). mRNA expression of TOP2A in non-tumor (N) and HCC (T) samples in the GSE14520 (second left) and TCGA HCC (second right) data sets, p-values were calculated using moderated t-test and the Wilcoxon signed-rank test respectively. Kaplan Meier curves of TCGA HCC patients with high or low TOP2A expression (right), p-value was calculated using the log-rank Mantel-Cox test. B) Protein levels of TOP2A and p21 in Huh7 cells infected with the indicated sgRNAs. C) Colony formation and SA-β-gal staining of Huh7 cells infected with sgTOP2A. D) mRNA levels of senescence markers p15, p16, and p21 in Huh7 cells infected with sgTOP2A. E) Cell cycle analysis of Huh7 cells infected with the indicated sgRNAs. Representative data (left) and quantification (right) are shown here F) Working model. TRRAP and KAT5 induces HCC cell growth by activating expression of mitotic genes. Loss of TRRAP or KAT5 downregulate TOP2A, induce G2/M arrest and senescence. Data was presented as mean ± SD; p-values were calculated by comparing to non-targeting control, *p < 0.05, **p < 0.01, ***p < 0.001 (student’s t test).
Discussion
Here, we demonstrate that TRRAP functions in HCC cell proliferation by promoting G2/M progression. TRRAP depletion led to cell cycle arrest at G2/M phase and induction of senescence (Figure 6F). Previous studies in other cell types have identified several cell cycle genes that are regulated by TRRAP, including cyclins A2, D1, D2 and E, Mad1, Mad2, and MKI67 (17, 41–43). We found that the majority of TRRAP-activated genes in HCC are involved in mitosis, and include genes not previously known to be TRRAP targets. Transcriptional activation of TRRAP-activated genes also required KAT5, indicating a specificity in recruitment of chromatin factors by TRRAP. Other TRRAP-interacting transcription factors, such as Myc and β-catenin, are frequently altered in HCC (8, 44). Investigations into whether these factors are important for TRRAP-dependent cell cycle control in HCC are warranted.
The oncogenic roles of G2/M genes in tumorigenesis have been reported. Over-expression of genes that regulate DNA replication and chromosomal segregation are involved in promoting chromosomal instability, a phenotype of aggressive cancers (45). In liver cancer, YAP and FOXM1 promote chromosomal instability, and predict poor prognosis in patients (46). Of note, our study has identified FOXM1 is one of the TRRAP-activated genes. We also found that 11 of our 22 TRRAP-activated genes overlapped with the chromosomal instability gene signature (45). Thus, TRRAP overexpression may contribute to chromosomal instability in HCC.
The function of TRRAP in other cancer types is unclear. A previous study found that TRRAP depletion induces differentiation of brain tumor-initiating cells derived from GBM patients, partially due to reduced cyclin A2 expression and S/G2 progression (17). We found that cyclin A2 is also a TRRAP-activated gene in HCC, although we did not observe a strong positive correlation between cyclin A2 and TRRAP mRNA expression in TCGA GBM patient samples. However, we found an overlap in TRRAP-activated genes (14 of the 22) between GBM and HCC. Thus, TRRAP may regulate a similar set of genes between different cancer types. However, unlike in HCC, TRRAP mRNA expression is not a prognostic predictor in either GBM or other cancers (Supplementary Figure 8B and based on data from the Human protein atlas). TRRAP mutations are reported in several cancer types, but their effects have not been studied.
Generally, p53 activation is a key event in senescence, but we discovered that TRRAP depletion induces senescence independent of the p53/p21 pathway in HCC cells. This finding has clinical implications for HCC treatment as 30% of HCC patients has p53 mutations (44) and p53 is more frequently mutated in HCC patients with high TRRAP expression (Supplementary Figure 10). A p53- and p21-independent mechanism in immortalized human diploid fibroblasts has been described (47). In this previous study, loss of the histone acetyltransferase p300 induced senescence due to global H3 and H4 hypoacetylation and subsequent alterations in DNA replication timing and fork velocity (47). In the present study, we propose a different mechanism—TRRAP depletion leads to senescence due to downregulation of mitotic genes, such as TOP2A, and G2/M arrest. Our mechanism identifies the TRRAP complex as a potential therapeutic target in HCC. Given that the crystal structure of the TRRAP/KAT5 complex was recently characterized in yeast, it may be possible to target the interaction between TRRAP and KAT5 and disrupt the histone acetyltransferase activity of the complex (48). Moreover, small molecule inhibitors targeting KAT5 and mitotic proteins are currently under development (49, 50). Taken together, targeting the TRRAP/KAT5 complex or its downstream mitotic genes may be a pro-senescent strategy for treating HCC patients.
Our results provide an important discovery for a role for TRRAP and its co-factor KAT5 in activating downstream mitotic genes in HCC cell growth. Future work should determine how TRRAP overexpression and mutations affect its function to understand the extent and significance of TRRAP alterations in other cancers. Moreover, because in vivo models of TRRAP consist of only genetic knockouts, it will be critical to establish constitutively-active TRRAP and mutant TRRAP models to further characterize its role in tumor development.
Supplementary Material
Supplementary Figure 1. A kinome CRISPR screen identifies essential genes for HCC cell growth. A) mRNA levels of candidate genes in non-tumor and tumor samples in the GSE14520 data set. P-values were calculated using moderated t-test. Black lines indicate the geometric mean of each group. B) Kaplan Meier curves of TCGA liver cancer patients with high or low expression of the candidate genes in A. P-values were calculated using the log-rank Mantel-Cox test. C) TRRAP mRNA up-regulation, copy-number alterations and/or mutations in TCGA patient HCC samples. D) Ingenuity Pathway analysis of genes that were positively correlated with TRRAP expression (Spearman’s correlation≥0.3) in TCGA HCC samples.
Supplementary Figure 2. Loss of KAT5 impairs cell growth and induces senescence in HCC cells. A) Immunofluorescence images of Huh7 and SNU-475 cells. B) Protein and mRNA levels of KAT2A and KAT5 in SNU-475 cells infected with sgNT and sgTRRAP. C) Huh7 cells were infected with the indicated sgRNAs, then treated with 2.5 μM MG132 or 50 μM chloroquine for 17 hours. Increased expression of NIK and LC3B were used as positive controls to show inhibition of proteasomal and lysosomal protein degradation. D) Western blot analysis of KAT2A, KAT5, and p21 levels in SNU-475 cells. E) Colony formation and SA-β-gal staining of SNU-475 cells. F) mRNA levels of p15, p16, and p21 in SNU-475 cells. Data was presented as mean ± SD; p-values were calculated by comparing to sgNT, *p < 0.05, **p < 0.01, ***p < 0.001 (student’s t test).
Supplementary Figure 3. Identification of genes repressed by TRRAP. A) GO analysis of up-regulated genes in sgTRRAP cells compared to non-targeting control. The 10 most significant annotation clusters are shown here. B) Kaplan Meier curves of TCGA HCC patients with high or low expression of genes listed in Supplementary Table 3. C) Negative correlation between mRNA expression of TRRAP and TRRAP-inhibited genes in the TCGA HCC data set (n=360). Correlation was determined using Spearman’s correlation analysis.
Supplementary Figure 4. Increased expression of TRRAP-activated genes predicted poor prognosis in HCC patients. Kaplan Meier curves of TCGA HCC patients with high or low expression of TRRAP-activated genes listed in Table 1. P-values were calculated using the log-rank Mantel-Cox test. The Kaplan Meier curve of BUB1B is shown in Supplementary Figure 1B.
Supplementary Figure 5. Positive correlation between mRNA expression of TRRAP and TRRAP-activated genes. mRNA levels were downloaded from the TCGA HCC data set (n=360). Correlation was determined using Spearman’s correlation analysis.
Supplementary Figure 6. KAT5 binds to the transcriptional start sites (TSS) of TRRAP-activated genes. Analysis of published ChIP-sequencing data for KAT5 binding sites in mouse embryonic stem cells at TRRAP-activated genes (blue) identified in Table 1. Of note, no prominent peaks were observed at the TSS of Bub1b, Dlgap5 and Nsd2.
Supplementary Figure 7. Depletion of TRRAP and KAT5 induce G2/M arrest without DNA damage. A) mRNA levels of TRRAP-activated genes in SNU-475 cells infected with the indicated sgRNAs compared to sgNT as measured by qRT-PCR. B) Cell cycle analysis of SNU-475 cells infected with the indicated sgRNAs by BrdU and PI staining. Representative data (left) and quantification (right) are shown here. C) Flow cytometry analysis of DNA damage in Huh7 cells using γH2A.X in the total cell population, 2N and >2N populations. DNA content was determined using PI staining. This experiment was repeated twice. Data was presented as mean ± SD; p-values were calculated by comparing to non-targeting control, *p < 0.05, **p < 0.01, ***p < 0.001 (student’s t test).
Supplementary Figure 8. TRRAP expression is not a predictor for prognosis in GBM. A) Correlation between TRRAP and TOP2A mRNA expression in the TCGA GBM data set (n=136) was determined using Spearman’s correlation analysis. B) Kaplan Meier curve of TCGA GBM patients with high (n=50) or low (n=51) TRRAP expression. P-value was calculated using the log-rank Mantel-Cox test.
Supplementary Figure 9. Depletion of TOP2A induces G2/M arrest without DNA damage. A) Protein levels of TOP2A and p21 in SNU-475 cells infected with the indicated sgRNAs. B) Top2A promoter luciferase assay in 293fs cells transfected with the indicated plasmids. Luciferase activity was normalized to empty pBV-luc control. C) Colony formation and SA-β-gal staining of SNU-475 cells infected with sgTOP2A. D) mRNA levels of p15, p16, and p21 in SNU-475 cells infected with sgTOP2A. E) Cell cycle analysis of SNU-475 cells infected with the indicated sgRNAs by BrdU and PI staining. Representative data (left) and quantification (right) are shown here. F) Flow cytometry analysis of DNA damage in Huh7 cells using γH2A.X in the total cell population, 2N and >2N populations. DNA content was determined using PI staining. This experiment was repeated twice. Data was presented as mean ± SD; p-values were calculated by comparing to non-targeting control, *p < 0.05, **p < 0.01, ***p < 0.001 (student’s t test).
Supplementary Figure 10. p53 is more frequently mutated in high expressing TRRAP patient samples. p53 status in TRRAP high (n=115) and TRRAP low (n=118) TCGA HCC patient samples. P-value was calculated by Fisher’s exact test.
Supplementary Table 1. sgRNA sequences used in this study.
Supplementary Table 2. Primers used for qRT-PCR in this study.
Supplementary Table 3. List of genes that are up-regulated in the absence of TRRAP and identified from our bioinformatics analyses.
Supplementary Table 4. TRRAP regulates a similar set of genes in HCC and GBM. Correlation analysis between TRRAP expression and TRRAP-activated genes identified in Table 1 in TCGA GBM patients (n=136). Genes in bold indicate a positive correlation with TRRAP expression (Spearman’s correlation > 0.3).
Supplementary Table 5. Primers used for cloning the TOP2A promoter.
Acknowldegements
We thank C. Mello, P. Zamore, S. Wolfe, F. Sanchez-Rivera, J. Zuber, J. Shi, T. Fazzio, and E. Sontheimer for discussions. We thank Y. Liu and E. Kittler in the UMass Morphology and Deep Sequencing Cores for support.
Financial Support: W.X. was supported by grants from the National Institutes of Health (DP2HL137167, P01HL131471 and UG3HL147367), American Cancer Society (129056-RSG-16–093), the Lung Cancer Research Foundation, Hyundai Hope on Wheels, and ALS Association. A.S. was supported by F30CA232657 (NCI). H.D. and X.W.W. were supported by grants (Z01-BC 010313, Z01-BC 010876, and Z01-BC 010877) from the intramural research program of the Center for Cancer Research, National Cancer institute. H.Y. was supported by the Young Thousand Talented Program and the startup funding from Wuhan University. X.Z. and Z.W. were partly supported by NIH grant U24-HG009446.
List of abbreviations:
- HCC
Hepatocellular carcinoma
- TRRAP
transformation/transcription domain-associated protein
- HAT
histone acetyltransferase
- GBM
glioblastoma multiforme
- TOP2A
Topoisomerase II alpha
Footnotes
Conflicts of interest: The authors disclose no conflicts.
References
- 1.Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 2015;149:1226–1239. [DOI] [PubMed] [Google Scholar]
- 2.Marquardt JU, Andersen JB, Thorgeirsson SS. Functional and genetic deconstruction of the cellular origin in liver cancer. Nat Rev Cancer 2015;15:653–667. [DOI] [PubMed] [Google Scholar]
- 3.Villanueva A Hepatocellular Carcinoma. N Engl J Med 2019;380:1450–1462. [DOI] [PubMed] [Google Scholar]
- 4.Xue W, Wang XW. The search for precision models clinically relevant to human liver cancer. Hepatic Oncology 2015;2:315–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, et al. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol 2001;21:6782–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, et al. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J 1999;18:5108–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 2000;102:463–473. [DOI] [PubMed] [Google Scholar]
- 8.Murr R, Vaissiere T, Sawan C, Shukla V, Herceg Z. Orchestration of chromatin-based processes: mind the TRRAP. Oncogene 2007;26:5358–5372. [DOI] [PubMed] [Google Scholar]
- 9.Sawan C, Hernandez-Vargas H, Murr R, Lopez F, Vaissiere T, Ghantous AY, Cuenin C, et al. Histone acetyltransferase cofactor Trrap maintains self-renewal and restricts differentiation of embryonic stem cells. Stem Cells 2013;31:979–991. [DOI] [PubMed] [Google Scholar]
- 10.Tapias A, Zhou ZW, Shi Y, Chong Z, Wang P, Groth M, Platzer M, et al. Trrap-dependent histone acetylation specifically regulates cell-cycle gene transcription to control neural progenitor fate decisions. Cell Stem Cell 2014;14:632–643. [DOI] [PubMed] [Google Scholar]
- 11.Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, Davis S, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet 2011;43:442–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, et al. A landscape of driver mutations in melanoma. Cell 2012;150:251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Research N, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 1998;94:363–374. [DOI] [PubMed] [Google Scholar]
- 16.Kalkat M, Resetca D, Lourenco C, Chan PK, Wei Y, Shiah YJ, Vitkin N, et al. MYC Protein Interactome Profiling Reveals Functionally Distinct Regions that Cooperate to Drive Tumorigenesis. Mol Cell 2018. [DOI] [PubMed] [Google Scholar]
- 17.Wurdak H, Zhu S, Romero A, Lorger M, Watson J, Chiang CY, Zhang J, et al. An RNAi screen identifies TRRAP as a regulator of brain tumor-initiating cell differentiation. Cell Stem Cell 2010;6:37–47. [DOI] [PubMed] [Google Scholar]
- 18.Jethwa A, Slabicki M, Hullein J, Jentzsch M, Dalal V, Rabe S, Wagner L, et al. TRRAP is essential for regulating the accumulation of mutant and wild-type p53 in lymphoma. Blood 2018;131:2789–2802. [DOI] [PubMed] [Google Scholar]
- 19.Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 2014;15:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roessler S, Long EL, Budhu A, Chen Y, Zhao X, Ji J, Walker R, et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology 2012;142:957–966 e912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JS, Chu IS, Mikaelyan A, Calvisi DF, Heo J, Reddy JK, Thorgeirsson SS. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat Genet 2004;36:1306–1311. [DOI] [PubMed] [Google Scholar]
- 22.Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, Mikaelyan A, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med 2006;12:410–416. [DOI] [PubMed] [Google Scholar]
- 23.Ravens S, Yu C, Ye T, Stierle M, Tora L. Tip60 complex binds to active Pol II promoters and a subset of enhancers and co-regulates the c-Myc network in mouse embryonic stem cells. Epigenetics Chromatin 2015;8:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997;88:593–602. [DOI] [PubMed] [Google Scholar]
- 25.Fu Y, Wu PH, Beane T, Zamore PD, Weng Z. Elimination of PCR duplicates in RNA-seq and small RNA-seq using unique molecular identifiers. BMC Genomics 2018;19:531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin ZZ, Jeng YM, Hu FC, Pan HW, Tsao HW, Lai PL, Lee PH, et al. Significance of Aurora B overexpression in hepatocellular carcinoma. Aurora B Overexpression in HCC. BMC Cancer 2010;10:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeh HW, Lee SS, Chang CY, Hu CM, Jou YS. Pyrimidine metabolic rate limiting enzymes in poorly-differentiated hepatocellular carcinoma are signature genes of cancer stemness and associated with poor prognosis. Oncotarget 2017;8:77734–77751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu H, Zhu W, Qin J, Chen M, Gong L, Li L, Liu X, et al. Acetylation of PGK1 promotes liver cancer cell proliferation and tumorigenesis. Hepatology 2017;65:515–528. [DOI] [PubMed] [Google Scholar]
- 33.Yang XM, Cao XY, He P, Li J, Feng MX, Zhang YL, Zhang XL, et al. Overexpression of Rac GTPase activating Protein 1 Contributes to Proliferation of Cancer Cells by Reducing Hippo Signaling to Promote Cytokinesis. Gastroenterology 2018. [DOI] [PubMed] [Google Scholar]
- 34.Lee N, Kwon JH, Kim YB, Kim SH, Park SJ, Xu W, Jung HY, et al. Vaccinia-related kinase 1 promotes hepatocellular carcinoma by controlling the levels of cell cycle regulators associated with G1/S transition. Oncotarget 2015;6:30130–30148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 2003;113:703–716. [DOI] [PubMed] [Google Scholar]
- 36.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 2007;445:656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hernandez-Segura A, Nehme J, Demaria M. Hallmarks of Cellular Senescence. Trends Cell Biol 2018;28:436–453. [DOI] [PubMed] [Google Scholar]
- 38.Munoz-Espin D, Serrano M. Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol 2014;15:482–496. [DOI] [PubMed] [Google Scholar]
- 39.Gire V, Dulic V. Senescence from G2 arrest, revisited. Cell Cycle 2015;14:297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen T, Sun Y, Ji P, Kopetz S, Zhang W. Topoisomerase IIalpha in chromosome instability and personalized cancer therapy. Oncogene 2015;34:4019–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shukla V, Cuenin C, Dubey N, Herceg Z. Loss of histone acetyltransferase cofactor transformation/transcription domain-associated protein impairs liver regeneration after toxic injury. Hepatology 2011;53:954–963. [DOI] [PubMed] [Google Scholar]
- 42.Herceg Z, Hulla W, Gell D, Cuenin C, Lleonart M, Jackson S, Wang ZQ. Disruption of Trrap causes early embryonic lethality and defects in cell cycle progression. Nat Genet 2001;29:206–211. [DOI] [PubMed] [Google Scholar]
- 43.Li H, Cuenin C, Murr R, Wang ZQ, Herceg Z. HAT cofactor Trrap regulates the mitotic checkpoint by modulation of Mad1 and Mad2 expression. EMBO J 2004;23:4824–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cancer Genome Atlas Research Network. Electronic address wbe, Cancer Genome Atlas Research N. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017;169:1327–1341 e1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet 2006;38:1043–1048. [DOI] [PubMed] [Google Scholar]
- 46.Weiler SME, Pinna F, Wolf T, Lutz T, Geldiyev A, Sticht C, Knaub M, et al. Induction of Chromosome Instability by Activation of Yes-Associated Protein and Forkhead Box M1 in Liver Cancer. Gastroenterology 2017;152:2037–2051 e2022. [DOI] [PubMed] [Google Scholar]
- 47.Prieur A, Besnard E, Babled A, Lemaitre JM. p53 and p16(INK4A) independent induction of senescence by chromatin-dependent alteration of S-phase progression. Nat Commun 2011;2:473. [DOI] [PubMed] [Google Scholar]
- 48.Wang X, Ahmad S, Zhang Z, Cote J, Cai G. Architecture of the Saccharomyces cerevisiae NuA4/TIP60 complex. Nat Commun 2018;9:1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao C, Bourke E, Scobie M, Famme MA, Koolmeister T, Helleday T, Eriksson LA, et al. Rational design and validation of a Tip60 histone acetyltransferase inhibitor. Sci Rep 2014;4:5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dominguez-Brauer C, Thu KL, Mason JM, Blaser H, Bray MR, Mak TW. Targeting Mitosis in Cancer: Emerging Strategies. Mol Cell 2015;60:524–536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. A kinome CRISPR screen identifies essential genes for HCC cell growth. A) mRNA levels of candidate genes in non-tumor and tumor samples in the GSE14520 data set. P-values were calculated using moderated t-test. Black lines indicate the geometric mean of each group. B) Kaplan Meier curves of TCGA liver cancer patients with high or low expression of the candidate genes in A. P-values were calculated using the log-rank Mantel-Cox test. C) TRRAP mRNA up-regulation, copy-number alterations and/or mutations in TCGA patient HCC samples. D) Ingenuity Pathway analysis of genes that were positively correlated with TRRAP expression (Spearman’s correlation≥0.3) in TCGA HCC samples.
Supplementary Figure 2. Loss of KAT5 impairs cell growth and induces senescence in HCC cells. A) Immunofluorescence images of Huh7 and SNU-475 cells. B) Protein and mRNA levels of KAT2A and KAT5 in SNU-475 cells infected with sgNT and sgTRRAP. C) Huh7 cells were infected with the indicated sgRNAs, then treated with 2.5 μM MG132 or 50 μM chloroquine for 17 hours. Increased expression of NIK and LC3B were used as positive controls to show inhibition of proteasomal and lysosomal protein degradation. D) Western blot analysis of KAT2A, KAT5, and p21 levels in SNU-475 cells. E) Colony formation and SA-β-gal staining of SNU-475 cells. F) mRNA levels of p15, p16, and p21 in SNU-475 cells. Data was presented as mean ± SD; p-values were calculated by comparing to sgNT, *p < 0.05, **p < 0.01, ***p < 0.001 (student’s t test).
Supplementary Figure 3. Identification of genes repressed by TRRAP. A) GO analysis of up-regulated genes in sgTRRAP cells compared to non-targeting control. The 10 most significant annotation clusters are shown here. B) Kaplan Meier curves of TCGA HCC patients with high or low expression of genes listed in Supplementary Table 3. C) Negative correlation between mRNA expression of TRRAP and TRRAP-inhibited genes in the TCGA HCC data set (n=360). Correlation was determined using Spearman’s correlation analysis.
Supplementary Figure 4. Increased expression of TRRAP-activated genes predicted poor prognosis in HCC patients. Kaplan Meier curves of TCGA HCC patients with high or low expression of TRRAP-activated genes listed in Table 1. P-values were calculated using the log-rank Mantel-Cox test. The Kaplan Meier curve of BUB1B is shown in Supplementary Figure 1B.
Supplementary Figure 5. Positive correlation between mRNA expression of TRRAP and TRRAP-activated genes. mRNA levels were downloaded from the TCGA HCC data set (n=360). Correlation was determined using Spearman’s correlation analysis.
Supplementary Figure 6. KAT5 binds to the transcriptional start sites (TSS) of TRRAP-activated genes. Analysis of published ChIP-sequencing data for KAT5 binding sites in mouse embryonic stem cells at TRRAP-activated genes (blue) identified in Table 1. Of note, no prominent peaks were observed at the TSS of Bub1b, Dlgap5 and Nsd2.
Supplementary Figure 7. Depletion of TRRAP and KAT5 induce G2/M arrest without DNA damage. A) mRNA levels of TRRAP-activated genes in SNU-475 cells infected with the indicated sgRNAs compared to sgNT as measured by qRT-PCR. B) Cell cycle analysis of SNU-475 cells infected with the indicated sgRNAs by BrdU and PI staining. Representative data (left) and quantification (right) are shown here. C) Flow cytometry analysis of DNA damage in Huh7 cells using γH2A.X in the total cell population, 2N and >2N populations. DNA content was determined using PI staining. This experiment was repeated twice. Data was presented as mean ± SD; p-values were calculated by comparing to non-targeting control, *p < 0.05, **p < 0.01, ***p < 0.001 (student’s t test).
Supplementary Figure 8. TRRAP expression is not a predictor for prognosis in GBM. A) Correlation between TRRAP and TOP2A mRNA expression in the TCGA GBM data set (n=136) was determined using Spearman’s correlation analysis. B) Kaplan Meier curve of TCGA GBM patients with high (n=50) or low (n=51) TRRAP expression. P-value was calculated using the log-rank Mantel-Cox test.
Supplementary Figure 9. Depletion of TOP2A induces G2/M arrest without DNA damage. A) Protein levels of TOP2A and p21 in SNU-475 cells infected with the indicated sgRNAs. B) Top2A promoter luciferase assay in 293fs cells transfected with the indicated plasmids. Luciferase activity was normalized to empty pBV-luc control. C) Colony formation and SA-β-gal staining of SNU-475 cells infected with sgTOP2A. D) mRNA levels of p15, p16, and p21 in SNU-475 cells infected with sgTOP2A. E) Cell cycle analysis of SNU-475 cells infected with the indicated sgRNAs by BrdU and PI staining. Representative data (left) and quantification (right) are shown here. F) Flow cytometry analysis of DNA damage in Huh7 cells using γH2A.X in the total cell population, 2N and >2N populations. DNA content was determined using PI staining. This experiment was repeated twice. Data was presented as mean ± SD; p-values were calculated by comparing to non-targeting control, *p < 0.05, **p < 0.01, ***p < 0.001 (student’s t test).
Supplementary Figure 10. p53 is more frequently mutated in high expressing TRRAP patient samples. p53 status in TRRAP high (n=115) and TRRAP low (n=118) TCGA HCC patient samples. P-value was calculated by Fisher’s exact test.
Supplementary Table 1. sgRNA sequences used in this study.
Supplementary Table 2. Primers used for qRT-PCR in this study.
Supplementary Table 3. List of genes that are up-regulated in the absence of TRRAP and identified from our bioinformatics analyses.
Supplementary Table 4. TRRAP regulates a similar set of genes in HCC and GBM. Correlation analysis between TRRAP expression and TRRAP-activated genes identified in Table 1 in TCGA GBM patients (n=136). Genes in bold indicate a positive correlation with TRRAP expression (Spearman’s correlation > 0.3).
Supplementary Table 5. Primers used for cloning the TOP2A promoter.