Abstract
Nonviral gene delivery has seen major progress in the last two decades owing to facile synthesis, low toxicity, and ease of modification of nanocarriers that take nucleic acids to cells and tissues. Gene delivery nanocomplexes need to reach the target locations in significant amounts by overcoming multiple barriers. While the importance of nanocomplex stability, cellular uptake, intracellular trafficking, and nuclear localization has been studied extensively, the role of cellular retention and recycling of these nanocomplexes is less understood in the context of gene delivery. In this study, we used different DNA carriers and made efforts to understand the role played by cellular retention in determining their gene delivery efficiency across multiple cell lines. In addition, we also analyzed whether state of complexation and localization of the nanocomplexes play a role in conjunction with cellular retention. We observed higher transfection efficiencies for nanocomplexes showing better retention, lower unpackaging, and low recycling. Our data also suggests that nanocomplexes made of peptides with terminal cysteine modification show enhanced retention and transfection efficiency compared to their counterparts with no terminal cysteine. Overall, the work highlights myriad of factors to be considered for improving gene delivery efficiency of nanocomplexes.
Introduction
Over the years, research in gene therapy has expanded its scope to encompass the entire gamut from replacing an absent or defective genetic material with its functional form up to the realm of alteration of expression (increase or decrease) of a particular gene by delivering appropriate nucleic acids.1,2 Although gene therapy holds great potential as a powerful tool for altering gene expression in patients, the challenges of safety and efficiency of delivery still persist. One of the basic requirements of efficient gene therapy is appropriate gene delivery vector systems used for introducing the therapeutic nucleic acids. Gene delivery vector systems can be broadly classified as viral (adenoviral, retroviral, helper-dependent adenoviral systems, hybrid adenoviral systems, lentiviral, herpes simplex, pox virus, Epstein–Barr virus-associated, etc.) and nonviral (cationic lipids, different cationic polymers, lipid polymers, peptides, microinjection, DNA bombardment, etc.) methods, with each class having its advantages and limitations. Viral methods are considered more promising owing to their better transduction efficiency and long-term expression; however, they suffer from undesired effects of random integration and associated problems.3,4 On the other hand, nonviral vectors, where the nucleic acid is complexed with the carrier to form nanometer-sized complexes, could be advantageous because of their lower immunogenicity, ease of synthesis, and prospects of modifications for better organ targeting.5−7 Despite the efforts toward the development of a number of nonviral methods of gene delivery with the ultimate goal of single administration replacement of nonfunctional gene, it is hard to design delivery methods with high efficiency in vitro and in vivo, effective targeting, and absence of any side effects.
In the case of nonviral gene delivery, the vector–nucleic acid nanocomplex has to overcome different in vivo barriers like interaction with serum proteins and destabilization by nucleases.8,9 Even if the complex reaches the desired cells, there are a number of intracellular barriers like endosomal entrapment, balance of tight packaging and intracellular cargo release, cytoplasmic stability, and barriers for nuclear entry.1,10−12 Most of the current studies focus on overcoming these barriers by chemical modifications on the carrier, design of multifunctional carriers, application of different surface coatings, and so on.13−16 However, although significant advances have been made in these directions, nonviral vectors which are as efficient as the viral ones are yet to be developed. This indicates that the different barriers for nonviral gene delivery are not completely understood.17
One barrier that has often been overlooked in the literature is the problem of cellular retention of nanocomplexes. After entering the cells, the nanocomplexes are recycled through exocytosis, which can result in lower cellular retention with a possible effect on gene delivery.18 Low cellular retention is likely to cause removal of the nanocomplexes from the cell, which will eventually allow less nucleic acid to reach the desired cellular locale. Unlike in the case of gene delivery nanocomplexes, exocytosis of inorganic nanoparticles has been studied extensively in the literature.19−22 There is a large volume of literature which analyzes the role of shape, size, surface modification, uptake, and egress routes of these nanoparticles to derive some parameters that can be controlled to improve cellular retention.23−25 On the other hand, there are only few studies in the literature on the role of exocytosis in gene delivery.18,26,27 Isolated reports discuss exocytosis of both uptaken siRNA and plasmid DNA from cells and its effect on the functional activity.18,28 For example, electron microscopy has been employed to demonstrate that polyethyleneimine (PEI)–DNA nanocomplexes and poly-l-lysine–DNA complexes are recycled from airway epithelial cells through exosome-like pathways.15 In an extensive study, Sahay et al. have shown using fluorescent probes and flow cytometry that there is significant endocytic recycling of si-RNA lipid nanocomplexes in HeLa cells.28 Up to 70% of the internalized si-RNA is recycled back and multivesicular bodies (MVB) are involved in the cellular egress. When HeLa cells were treated with radiolabeled poly(ethyleneimine)–DNA complexes, there was exocytosis of the complexes as indicated by decrease in intracellular radioactivity over time and concomitant increase of the same in the supernatant fraction.27 Further, quantitative-polymerase chain reaction (q-PCR)-based estimation of amounts of exocytosed and retained plasmid DNA inside cells demonstrated the highest retention profile in the case of poly(ethyleneimine)-mediated delivery among the vectors studied.18 However, there was a poor correlation between transgene expression and retention of plasmid DNA in this case. On the other hand, transgene expression of plasmid DNA delivered using PEI (polyethyleneimine) in sponges was reported to show a significant correlation with plasmid retention.26
While such isolated efforts aim to understand correlations among exocytosis, cellular retention, and gene delivery, as described above, there are also few efforts to characterize recycling pathways of the nanocomplexes.29 Available literature suggests involvement of multiple recycling pathways for different nanocomplexes, for example, early recycling of poly(lactic-co-glycolic acid) nanoparticle30 and MVB-mediated recycling of lipid-siRNA complexes.28 There are also limited efforts to analyze whether strategies to prevent exocytosis and increase cellular retention can help in drug/gene delivery. Silencing the NPC-1 gene which is associated with the surface of multivesicular late endosome has been found to enhance cellular retention of lipid nanoparticle–siRNA complexes.28 There are also studies on whether small molecules which can impair cholesterol metabolism can help in cellular accumulation of nanocomplexes.31 However, these studies are few and far between. A comprehensive picture of the correlation between cellular retention and transfection efficiency and whether it depends on factors like nature of the cationic carrier, nanocomplex size and charge, localization of the nanocomplex, and other factors has not yet emerged.
In this study, we have taken a number of cationic vectors, which are either developed in our laboratory or commercially available. We have analyzed the cellular uptake, retention, and exocytosis of nanocomplexes prepared using these agents in four different cell lines using q-PCR studies and confocal microscopy. The intracellular state of the nanocomplexes and localization have also been analyzed. Comparisons in the retention of the DNA and its state and localization were made across different agents and different cell lines and analyzed with respect to the transfection efficiencies.
Results
Choice of Carriers and Characterization of the Nanocomplexes
Four amphipathic peptides previously designed in our lab M1, M3, M4, and M932,33 were selected along with poly(ethyleneimine) (PEI) (cationic polymer) and Lipofectamine (lipid-based reagent) for making the nanocomplexes. M1 was derived from human protein phosphatase E1; M3 represents a secondary amphipathic peptide derived from M1, which was designed to improve the transfection efficiency of M1 by increasing the number of positive charges;32 M4 is the primary amphipathic variant of M3; and M9 represents terminal cysteine-modified M3.33 While M1 has been shown to have low transfection efficiency, all of the derived sequences have been shown to have improved transfection efficiency.32 PEI and Lipofectamine are efficient commercial transfection agents. The details of all of the carriers used are listed in Table 1.
Table 1. Cationic Nanocomplexes (NC) Used for the Study and Their Respective Size and Surface Charge Measurements Using Dynamic Light Scattering (DLS)a.
| cationic NCs | sequence/nature | size (d nm) | PDI | ζ-potential (mV) |
|---|---|---|---|---|
| Lipofectamine NC | lipid-based | 152.9 ± 8.80 | 0.17 ± 0.02 | –26 ± 1.90 |
| M3 NC | RRLRHLRHHYRRRWHRFR | 240.55 ± 6.05 | 0.13 ± 0.01 | 14.85 ± 0.85 |
| M4 NC | LLYWFRRRHRHHRRRHRR | 215.5 ± 1.70 | 0.15 ± 0.01 | 28.8 ± 1.90 |
| M9 NC | CRRLRHLRHHYRRRWHRFRC | 89.13 ± 1.98 | 0.29 ± 0.04 | 32.75 ± 2.45 |
| M1 NC | SRLSHLRHHYSKKWHRFR | 411.65 ± 0.55 | 0.26 ± 0.00 | 19.65 ± 3.65 |
| PEI NC | (C2H5N)n, polymer-based | 187.85 ± 7.85 | 0.15 ± 0.07 | 40 ± 3.20 |
Size and ζ-potential were measured in two separate experiments each comprising three measurements of 12 runs.
Nanocomplexes were prepared at a charge ratio of 5 between the pMIR plasmid and the different carriers, as described in Materials and Methods. The nanocomplexes were characterized for their shape, size, and surface charge using Atomic Force Microscopy (AFM) (Figure S1) and Dynamic Light Scattering (Table 1). All of the nanocomplexes appeared to be more or less spherical in shape, as indicated by the AFM images, and homogeneous in nature as indicated by the low PDI values (Table 1). The size of M1 nanocomplexes was found to be around 400 nm, indicating relatively loose complexation. Nanocomplexes formed with M3, M4, PEI, and Lipofectamine showed sizes larger than 100 nm at this charge ratio, while M9 peptide showed the highest complexation with hydrodynamic radii of about 80 nm and positive surface charge (Table 1). These are along expected lines and comparable to the values obtained with the nanocomplexes in our earlier studies carried out at different preparative conditions.32,33 All of the nanocomplexes except those formed with Lipofectamine were found to have positive surface charge (Table 1).
Uptake of the Nanocomplexes
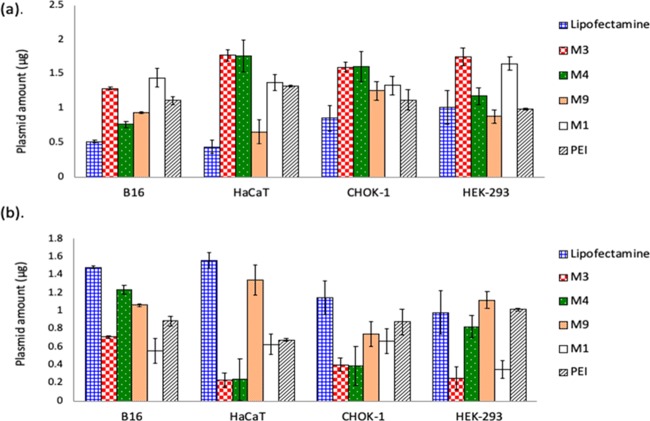
Cellular uptake of all of the nanocomplexes in different cell lines was measured using the protocol described in Materials and Methods. Four different cell lines were chosen for the study to cover wide spectra of cellular origin. The uptake was monitored for over a time period of 4 h to ensure saturation conditions (data not shown). After the incubation, the amount of DNA left over in the media was measured using q-PCR. Since the leftover DNA in the media is a measure of the DNA in the nanocomplexes that was not taken up by the cells, it can be used to calculate the cellular uptake efficiency of the different carriers. Figure 1a depicts the DNA content in the leftover fraction after a period of 4 h. Figure 1b shows the corresponding intracellular DNA calculated by subtracting the amount of DNA in the media from the total amount of DNA delivered.
Figure 1.
(a) q-PCR-based estimation of pMIR plasmid in leftover media: B16-F10, HaCaT, CHOK-1, and HEK-293 cells were incubated with different nanocomplexes for 4 h, followed by harvesting the leftover treatment media and q-PCR-based estimation of plasmid DNA. (b) Amounts of plasmid DNA inside the cells after 4 h incubation (DNA estimated in media fraction was subtracted from the total amount in each case, for estimating respective cellular uptake).
Nanocomplexes formed by M1, M3, and M4 peptides showed lower uptake in almost all of the cases compared to the other carriers. PEI nanocomplexes showed intermediate uptake, while efficient uptake was observed for M9 nanocomplexes and Lipofectamine nanocomplexes. Overall, B16-F10 cells showed the highest uptake of nanocomplexes, whereas the least uptake was observed in HaCaT cells.
Cellular Retention of Plasmid DNA with Time
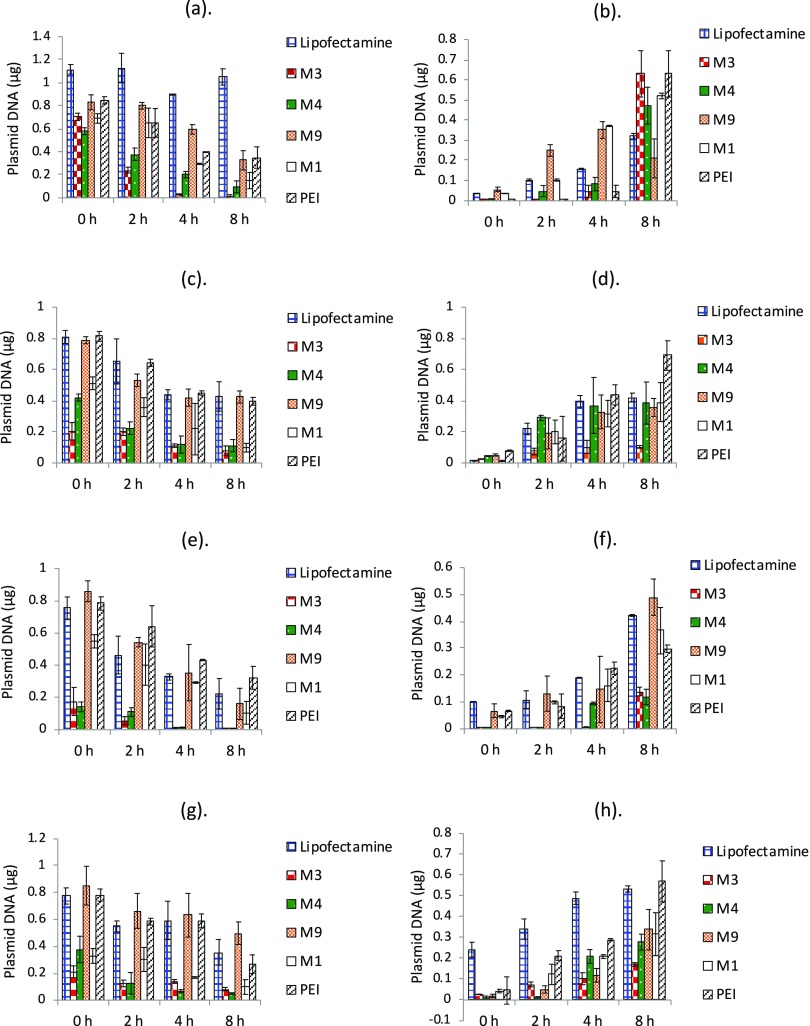
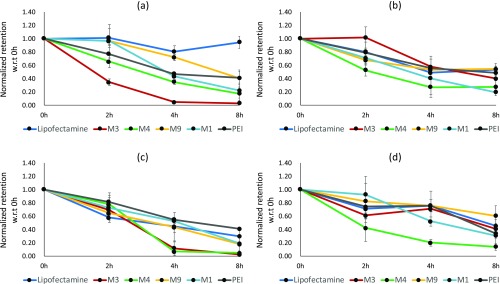
We monitored the retention of DNA delivered through the nanocomplexes over a period of 8 h through q-PCR-based estimation using the protocol described in the Materials and Methods section. Figure 2a–d depicts the trends in cellular retention of all of the samples in the four different cell lines. We have in parallel measured the presence of DNA at the same time points in the media fraction to get an estimate of the amount of DNA that is thrown out of the cells during this time period. The corresponding amounts in the media fraction are shown in Figure 2e–h. The 0 h reading in all of the cases denotes the DNA present in the cells when the uptake is saturated. The media is replaced, and the time course of cellular retention and exocytosis is followed for 8 h from this time point. In selected cases, we also monitored the retention till 16 h (data not shown). We did not observe any significant change in retention beyond 8 h; hence, we present the data till this time point in all of the cases. Since the uptake of the nanocomplexes is different in different cell lines and with different carriers, as shown in Figure 1, we have also normalized the retention amount with respect to the uptake of nanocomplexes in each case (see Figure 3).
Figure 2.
Amounts of plasmid DNA quantified in cellular fraction and media fraction in different cell lines. (a) B16-F10 cellular fraction, (b) B16-F10 media fraction, (c) CHOK-1 cellular fraction, (d) CHOK-1 media fraction, (e) HaCaT cellular fraction, (f) HaCaT media fraction, (g) HEK-293 cellular fraction, and (h) HEK-293 media fraction. Cells were incubated with different nanocomplexes for 4 h, followed by q-PCR-based estimation of pMIR plasmid at 0, 2, 4, and 8 h in both media and cellular fractions. Data are presented as mean ± standard deviation of at least three independent experiments.
Figure 3.
Normalized cellular fraction retention trends in different cell lines. (a) B16-F10, (b) CHOK-1, (c) HaCaT, and (d) HEK-293 cells were incubated with different nanocomplexes for 4 h, followed by q-PCR-based estimation of pMIR plasmid at 0, 2, 4, and 8 h. Normalization was performed by dividing the estimated cellular fraction amount at any time points with estimated amounts of that nanocomplex at 0 h after media change. Data are presented as mean ± standard deviation of at least three independent experiments.
For all nanocomplexes, there was reduction in amount of plasmid DNA present inside the cells over time (Figure 2). Nanocomplexes formed with Lipofectamine, M9, and PEI showed comparatively higher retention and slower cellular egress, while a higher loss of DNA from the cells was observed in M1, M3, and M4 nanocomplexes over time. M9 (cysteine-modified peptide nanocomplex) exhibited a good retention in HaCaT cells as well, which otherwise shows less retention. B16-F10 melanoma cells and CHO-K1 cells exhibited comparatively higher retention of nanocomplexes in general. When the presence of DNA in the media was analyzed, it was observed that for all nanocomplexes, the amount of estimated plasmid DNA increased with time in the media, indicating cellular egress. The trends matched with that observed in the case of cellular retention. Since the uptake of the nanocomplexes was different, we analyzed the data for cellular retention by normalizing against the cellular uptake (Figure 3) as well. While M1, M3, and M4 showed the maximum cellular egress by 8 h, the best retention was observed in Lipofectamine in some cell lines and by M9 or PEI in some others. This indicates that the overall amounts retained in the cells (irrespective of the uptake) are higher for M9 and the two commercial agents compared to M1, M3, and M4. Moreover, in most cases, the loss of DNA was quite sharp in the early time points for M3, M4, and M1, indicating considerable early recycling.
We further validated this by carrying out fixed cell confocal microscopy using fluorescein isothiocyanate (FITC)-labeled nanocomplexes (as described in Materials and Methods) for M9 and Lipofectamine nanocomplexes in B16-F10 cells. Hoechst was used for staining the nucleus, and Cell Mask Orange was used for staining plasma membrane. Nanocomplexes could be visualized as punctate green signal, localized both in cytoplasm and nucleus. As expected, the nanocomplex signal decreased gradually with time, corroborating the data from q-PCR experiments (Figure S2). In the case of HaCaT cells, overall nanocomplexes visualized were less than those seen in B16-F10 cells, which could be because of the lower uptake in the former.
State of the Nanocomplex Inside the Cells
We analyzed the state of the different nanocomplexes in the cellular milieu by detecting the amount of uncomplexed plasmid DNA present at different time points (0–8 h) after the initial 4 h of incubation. We chose Lipofectamine and M9 nanocomplexes for this study since these appeared to be showing best retention across cell lines. We also included M3 nanocomplexes to compare with M9 nanocomplexes and observe if terminal cysteines in the peptide have any effect.
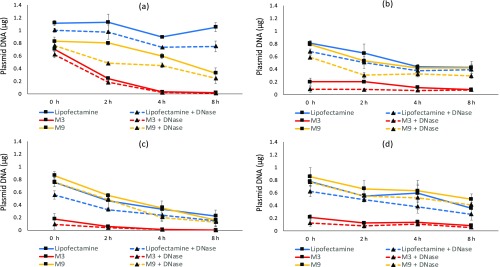
For estimating the unbound plasmid amounts, cellular lysate of each sample was divided in two halves, where DNase treatment was given to chop off unbound plasmid DNA in one half and no treatment was given to the other (detailed protocol mentioned in Materials and Methods). The difference between the measured values of untreated and DNase-treated samples would give an estimate of unbound DNA present in each sample. Figure 4 shows that there was a reduction in the estimated amount of plasmid DNA after DNase treatment in almost all samples studied, indicating that there is decomplexation in all of the cases over time. Apart from the CHO-K1 cell line, a gradual decrease in the percentage of bound plasmid DNA was observed in B16-F10, HEK-293, and HaCaT cells for all nanocomplexes. Figure S3 depicts the bound DNA as a percentage of total DNA (derived from the measurements shown in Figure 4). On normalization against total DNA, it was observed that for M3 nanocomplexes, the plasmid release at lower time points was comparatively higher, suggesting that the nanocomplexes were more in uncomplexed state. In the case of M9 and Lipofectamine, in most cases, at low time points of 0 and 2 h, more DNA was present in the complexed state than the uncomplexed state. However, the uncomplexation appears to increase at 4 and 8 h time points.
Figure 4.
Estimating the amount of uncomplexed plasmid DNA in cellular factions: (a) B16-F10, (b) CHOK-1, (c) HaCaT, and (d) HEK-293 cells were incubated with different nanocomplexes for 4 h, followed by cell harvesting and lysis at 0, 2, 4, and 8 h. Cellular lysate of each sample was divided in two halves and was either subjected to DNase or given no treatment, prior to q-PCR estimation of plasmid DNA. The solid lines represent the total DNA, while the corresponding dotted line represents the DNA remaining after DNase treatment. Data are presented as mean ± standard deviation of at least three independent experiments.
Lysosomal Colocalization of Nanocomplexes
In addition, we also analyzed whether the localization of the nanocomplexes is different in the different cases. Evolving lysosomal colocalization of M3, M9, and Lipofectamine nanocomplexes with time was observed at 0, 2, 4, and 8 h (Figure S4) in the B16-F10 cell line. M3 nanocomplexes displayed the highest average colocalization over time, followed by Lipofectamine and M9, as indicated by the Pearson’s correlation coefficient in Supporting Information Table S2. Temporal changes in the lysosomal colocalization of the nanocomplexes were found to be completely different for M3, higher colocalization was observed at early hours, while for M9 and Lipofectamine, colocalization increased with time. This indicates that for M3, a larger fraction of the nanocomplexes becomes unavailable for further downstream processing in early time points (0–2 h). In the case of M9 and Lipofectamine, colocalization remains similar over time or drops slightly. M9 showed overall lower lysosomal colocalization than Lipofectamine.
Nuclear colocalization of the M9 and Lipofectamine nanocomplexes was studied using confocal live cell microscopy for 8 h in B16-F10 cells. Images were also captured at 0, 2, 4, and 8 h (Figure S5). For both nanocomplexes, colocalization with nucleus was found to increase between 2 and 6 h, after which it stabilized. A higher nuclear colocalization was observed in the case of Lipofectamine NC than M9 NC.
Transfection Efficiency of the Nanocomplexes
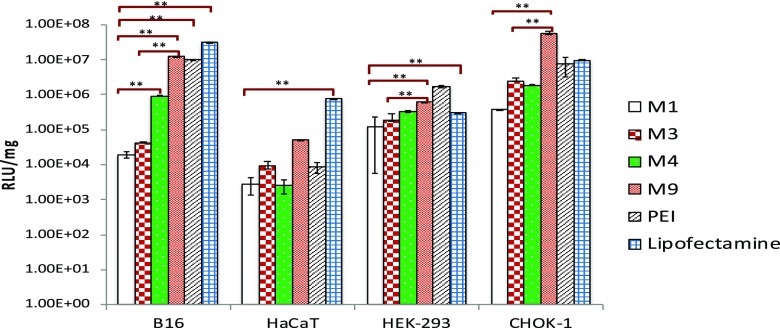
Luciferase expression after 24 h in different cell lines was found to be differing for different vector system and cell line (Figure 5). For all of the cell lines studied, HaCaT showed the least transgene expression with all nanocomplexes, while B16-F10 and HEK-293 showed highest expression levels. Lipofectamine nanocomplexes exhibited the highest transfection efficiency in majority of the cell lines, followed by M9- and PEI-based nanocomplexes, while M3, M4, and M1 were lower in transfection efficiency in most of the cases.
Figure 5.
Cellular transfection of nanocomplexes was estimated at 24 h post media by measuring luciferase gene expression. Data are presented as mean ± standard deviation of at least three independent experiments. (Few of the transfection efficiencies were reported earlier for M3, M4, and M9. However, for the sake of comparison, and since experimental conditions were not identical, we had to carry out fresh experiments and plotted all data carried out under similar conditions together). (**p<0.05).
Discussion
In this work, we are trying to understand the role of cellular retention and exocytosis in controlling the gene delivery efficiency of a number of peptide cationic vectors in comparison to two commercial gene delivery agents. We observed that while the commercial agents showed good transfection, only the cysteine-modified peptide showed high transfection efficiency while the others were not efficient in transfection. To check for the possible role of cellular retention in governing transfection efficiency, we measured the retained and exocytosed nanocomplexes in all of the cases. We devised an experimental methodology in which cells were initially incubated with a nanocomplex for 4 h, and then media was changed, followed by the q-PCR-based estimation of plasmid DNA at different time points. Although this does not give us the information whether the DNA is in a complexed state or existing as free DNA, the total amount of DNA from the plasmid present inside the cells and in the media can be estimated. Moreover, we used heparin to release the DNA in all of the cases to avoid any measurement artifact between DNA in compacted form and free/partially complexed DNA. It also needs to be noted here that there is small loss during the process of measurement because of which the sum of the total amount retained in the cells and the total amount in the media fraction is always slightly less than the total amount added initially. This is however a small experimental artifact across all of the samples and is unlikely to change the trends we observe. Also, small errors could arise in the case of measurements of very low concentrations since regression line obtained from standard curve (Ct value vs concentration) was exponential. However, the overall trends are largely not affected. Another possible anomaly can arise because of the presence of some nanocomplexes on the cell surface itself, the information from which might not get incorporated in the measurements. To minimize any effects of this phenomenon, we have used multiple washing steps with 1× phosphate-buffered saline (PBS) to remove the cell-surface-bound nanocomplexes.
We observed a drop in the amount of DNA retained in the cells over a period of time along with a concomitant increase in DNA in the media in all of the cases. The pattern of cellular egress, however, varied depending upon the nature of the carrier, as described in the Results section. The size of nanocomplexes showed a clear bearing on the retention, with smaller-sized particles showing more retention (M9, Lipofectamine, and PEI) and low retention in the case of comparatively larger nanocomplexes (M1, M3, and M4). Biophysical properties like nanoparticle size can be an important parameter that controls cellular uptake.24 In this case, even when the retention was normalized against the uptake, better retention was seen in the case of smaller nanocomplexes. Normalized retention trends showed fast recycling during initial hours (0–2 h) for M1, M3, and M4, suggesting a significant egress of these nanocomplexes. Both highly positively charged nanocomplexes like those prepared with M9 and highly negatively charged nanocomplexes like those with Lipofectamine showed high retention. Overall, the nanocomplexes showing high retention also showed high transfection efficiency.
The nature of the carrier also appeared to play a role in the retention and egress. Peptide M9 (which has two terminal cysteines added in the M3 sequence) not only showed higher uptake in almost all of the cell lines but also far slower recycling, as indicated by larger amounts of DNA retained beyond 2 h in most cases. Thus, it appears that more DNA is uptaken as well as retained in the cells in this case over time. Cysteine modification of peptide has been reported to improve not only stability of nanocomplexes34 but also transfection efficiencies32,35 by facilitating disulfide cross-linkages. While it is not clear how this could help in increased retention, one of the contributing factors that could be important is enhanced membrane-anchoring activity rendered by terminal cysteines.33,34 Since the peptide carrying the corresponding non-cysteine-modified sequence (M3) did not show high retention, the mechanisms and kinetics of cellular egress might be controlled by this subtle change in the peptide sequence.
Another observation was that the trends were observed to be mostly similar in all of the cell lines and no drastic variation in retention trends was seen. Multiple reports in the literature suggest higher recycling of drugs and nanomedicine in cancer cells;36 thus, for our study, we chose three immortalized cell lines of different tissue origin and one cancerous cell line (B16-F10). However, although the uptakes varied from cell line to cell line, our results on normalized cellular retention trends do not show much difference between the two cell types, suggesting a higher dependence on the type of vector used rather than the cell line. The available literature also suggests that all of these nanocomplexes employ multiple endocytotic pathways for nanocomplex uptake. The lack of the effect of cell line on retention might indicate a higher importance of the physicochemical nature of the nanocomplexes rather than the endocytotic pathways of entry.
We additionally also looked at the state of the retained nanocomplex and its localization to check whether these factors additionally influence transfection. Nanocomplex unpackaging to release transgene has been described as one of the major hurdles in dictating the gene delivery efficiency.37−39 However, early unpackaging makes the DNA prone to cytoplasmic degradation, while low unpackaging or strong complexation will result in less amounts being available for expression. In view of this, late unpackaging of nanocomplexes preferably near nucleus would be the ideal scenario for DNA delivery and subsequent expression of transgene. As described earlier, for estimating this, nanocomplexes were subjected to DNase treatment, followed by q-PCR-based measurement. It may be noted that one possible source of error in such measurement could be amplification through fragmented plasmid DNA. Even though we confirmed that DNase treatment was able to completely degrade the plasmid DNA through agarose gel electrophoresis (data not shown), this experimental limitation might have resulted in slightly lower estimation of free DNA. However, this will be across all of the cases, and therefore, the trends of the result would not be affected. We observed a higher unpackaging in M3 nanocomplexes compared to M9 nanocomplexes. This could be because the M3 nanocomplexes are comparatively less packaged as seen by the higher size. This could also be attributed to the absence of terminal cysteine modification in M3. Moreover, more uncomplexed DNA was observed at earlier time point (Figure S3) in M3, while unpackaging for M9 was delayed and was observed at higher time points, which might facilitate gene delivery to nucleus and thus transgene expression. However, it was intriguing to note that in the case of Lipofectamine, although the nanocomplex size was large, the unpackaging was less. The nature of peptide–DNA and lipid–DNA interactions for nanocomplex formation might be different in the two cases and might be responsible for this effect.
Lysosome-based recycling of nanocomplexes and nanoparticles has been identified as a significant metabolic pathway in the literature.40 Thus, nanocomplexes colocalizing with lysosomes have higher chances of being recycled. A significantly higher lysosomal colocalization of M3 NCs at 0 h (just after media change) indicates that a large amount of the nanocomplexes are going to get recycled through the lysosomal pathway or will undergo lysosomal degradation. With time, lysosomal colocalization of M3 decreased; however, cellular retention is poor at these time points and the retained DNA is largely uncomplexed. Thus, even if the lysosomal degradation is avoided, the uncomplexed DNA may not be suitably stable to reach the nucleus for gene expression. Lipofectamine nanocomplexes exhibited a higher lysosomal colocalization than M9 nanocomplexes; however, cellular retention of Lipofectamine nanocomplexes was slightly higher than that of the M9 nanocomplex, indicating that the fraction of DNA available for downstream processing may not be significantly different between the two. In both these cases, the DNA release from the nanocomplexes occurs at higher time points, which might mean that the nanocomplexes which avoided the recycling would be more stable for the cytoplasmic journey and eventual nuclear entry. Since nuclear entry is an identified prerequisite for successful gene delivery,41 we also studied colocalization of M9 and Lipofectamine nanocomplexes with nucleus. We observed a significant nuclear colocalization for both M9 and Lipofectamine nanocomplexes, starting from 3 h post media change (Figure S5). This further reiterates the earlier observations. All of the nanocomplexes used for studying the effect of retention were screened for toxicity using the MTT assay and showed nonsignificant cellular killing (Figure S6).
Conclusions
The results suggest that cellular retention and exocytosis are important factors that can have an effect on transfection efficiencies of peptide, lipid, and polymer-based vectors. In addition, subcellular distribution/localization along with unpackaging dynamics of DNA can further have an effect on the DNA retained in the cells. The chemical nature of the carrier and size of the nanocomplexes are important determinants of cellular retention, and the cell line used does not appear to have any effect. High transfection efficiencies for Lipofectamine and a cysteine-modified peptide M9 in comparison to other nanocomplexes can be rationalized as a combinatorial effect of high uptake, enhanced retention, lower unpackaging, and poor colocalization with lysosomes at an early stage after entering the cells. Further, the role of terminal cysteine modification stands out in enhancing the retention. Further work may involve modification of the nanocomplexes through different moieties/signal sequences to alter the retention and trafficking of these moieties and thereby used as a strategy to enhance the transfection efficiency.
Materials and Methods
Chemicals, Reagents, and Kits
All of the peptides were custom-synthesized with >95% purity grade from G.L. Biochem (Shanghai) Ltd. The plasmid used for this study, pMIR-REPORT Luciferase (pDNA), was maintained in E. coli DH5α cells and purified using GenElute HP Endotoxin-Free Plasmid MaxiPrep Kit (Sigma). Polyethyleneimine (PEI, MW ∼ 25 kDa, branched) was purchased from Sigma-Aldrich. Lipofectamine 2000, heparin salt was purchased from Invitrogen. Primers for q-PCR were designed using Primer3 Input version 0.4.0 and synthesized through (Integrated DNA Technologies) IDT (Supporting Information Table S1). Fluorescein DNA labeling kits were purchased from Mirus Bio Corporation. Cell viability assay kit CellTiter Glow was obtained from Promega, and 35 mm glass-bottom imaging dishes for confocal microscopy were purchased from ibidi cells in focus. KAPA SYBR fast 5× was purchased from Sigma-Aldrich. Dulbecco’s modified Eagle’s medium (DMEM) culture medium, phosphate-buffered saline (PBS), calf fetal serum, and penicillin/streptomycin antibiotics mixture were purchased from Invitrogen.
Nanocomplex Formation
Nanocomplexes were formed with different cationic agents and plasmid DNA. The charge ratio or ratio of the amount of cationic agent to DNA was chosen in such a way as to ensure complete condensation in all of the cases. The cationic agents used are listed in Table 1. Peptide–DNA nanocomplexes were prepared at a charge ratio Z (±) of 5 (charge ratio = total positive charge of peptide/total negative charge of DNA). This charge ratio was chosen since we have earlier observed a complete complexation of plasmid DNA with these peptides under this condition.33 For this, plasmid DNA dilutions of 40 ng/μL were added dropwise to equal volumes of appropriate peptide dilution while vortexing at a steady speed. Lipofectamine nanocomplexes were prepared according to the manufacturer’s protocol. For preparation of PEI nanocomplexes, a 10 mM solution of branched PEI (25 kDa) was added dropwise to 40 ng/μL DNA dilution.18 The complexes so formed were incubated for 30–45 min at room temperature before performing any experiment.
Characterization of Nanocomplexes
Dynamic Light Scattering (DLS)
The size of the nanocomplexes and their surface charge (ζ-potential) were measured using Zetasizer ZS90 (Malvern Instrument, U.K.) at a fixed angle of 90°. Nanocomplexes were prepared with 40 ng/μL plasmid DNA at a charge ratio Z (±) of 5. Data were represented as mean ± standard deviation. The sizes and charge of the nanocomplexes are listed in Table 1.
Atomic Force Microscopy (AFM)
To observe the morphology of the nanocomplexes, 10 μL of different samples were prepared at a charge ratio of 5 (as mentioned in the previous section) and deposited on mica followed by drying. The imaging was performed using a 5500 Scanning Probe Microscope (Agilent Technologies, Inc., AZ) using Picoview software 1.4.4. Images were taken in the AAC mode in air with silicon cantilever at 75 kHz of resonance frequency and 2.8 N/m constant force. The scanning speed was set at 1 line/s. Picoview software was used for minimum image processing and analysis.
Cell Culture
HEK-293 (human embryonic kidney cells) and CHO-K1 (epithelial cells from Chinese hamster ovary) cells lines were obtained from American Type Culture Collection (ATCC). B16-F10 (mouse melanoma) and HaCaT (human keratinocytes) cell lines were a kind gift from Dr T.N. Vivek (CSIR-IGIB). The HEK-293 cells were maintained in high-glucose DMEM; CHO-K1 in Hem’s F-12K; and B16-F10 and HaCaT cells were maintained in DMEM-F12 media, supplemented with heat-inactivated 10% (v/v) fetal bovine serum (Life Technologies) at 37 °C and 5% CO2 in a humidified incubator. Before proceeding for any treatment, all cells were allowed to reach a confluency level of 70–80%. The cell lines were selected to cover wide tissue origin and included both cancerous and noncancerous cell line, and the rate of exocytosis is found to be higher in cancerous cell lines.36
Cellular Uptake Assay
Cells were seeded in a 24-well plate at different densities (55 000 cells/well for B16-F10, 75 000/well for HaCaT, 50 000/well for CHO-K1 and HEK-293) 1 day prior to treatment with nanocomplexes. Nanocomplexes were formed using pMIR plasmid DNA nanocomplexes containing 2 μg of DNA added to each well in Opti-MEM. After 4 h, the leftover media was harvested from the cells and stored at −20 °C before setting up q-PCR reactions. For estimating the amount of plasmid DNA present in the leftover fraction, 2 μL of diluted media fraction samples were used for setting up 15 μL of q-PCR reactions. The uptaken amounts of plasmid DNA were estimated by subtracting the measured amounts from 2 μg (initial DNA amount). In the uptake experiment, multiple washing steps with 1× PBS was carried out so as to minimize the amount of nanocomplex attached to the cell surface.
q-PCR-Based Cellular Retention Study
Cells were seeded at different densities (55 000 cells/well for B16-F10, 75 000/well for HaCaT, 50 000/well for CHO-K1 and HEK-293) in a 24-well plate 1 day prior to treatment. Once subconfluency was achieved, the cells were treated with nanocomplexes containing 2 μg of plasmid DNA per well in 550 μL of reduced serum medium (Gibco Opti-MEM) for 4 h. After 4 h, the leftover media was collected and labeled as leftover fraction. The cells were washed with DPBS twice and 550 μL of respective complete media was added to each well. From this time onward, sampling was done at 0, 2, 4, and 8 h for both cellular fraction and media fraction. For harvesting the cellular fraction, the cells were washed twice with 1× DPBS and then trypsinized using 150 μL of trypsin. The cells were harvested by centrifugation at 5000 rpm for 5 min and then finally suspended in 500 μL of MQ for lysis. All of the cellular and media fractions were stored at −20 °C, and q-PCR was carried out subsequently for estimating the amount of plasmid DNA in different samples (cellular and media fractions in all of the samples). Prior to setting up q-PCR reactions, all of the samples were appropriately diluted and subjected to 9 mM heparin challenge for uncomplexing the bound plasmid DNA. Primers were designed against the backbone of pMIR-REPORT Luciferase plasmid with amplicon size of ∼110 nm. The primers are listed in Supporting Information Table S1. Designed PCR primers were checked for nonspecificity by NCBI blast tool; 15 μL PCR reactions were set up in Roche 384-well white plates (7.5 μL of SYBR mix, 1.5 μL of primer mix, 4 μL of nuclease free water, and 2 μL of samples). With each q-PCR assay, reactions were setup to obtain a standard curve for the Ct value versus known plasmid dilution. The amount of plasmid DNA was estimated for each sample by using linear equation of standard curve and the obtained Ct value. To minimize the effect of cellular lysate on q-PCR-based amplification, optimization for cellular fraction dilution was performed. A 100× dilution of cellular fraction was first prepared, and then, 2 μL of this was used for setting up q-PCR reaction. Further, the standard curves for cellular fraction were prepared by spiking plasmid DNA in similarly diluted cell lysate to take care of any effect of cell lysate.
Assay for Uncomplexation of Plasmid DNA
After cellular uptake, the amount of uncomplexed DNA present inside the cell at different time points was estimated using a q-PCR-based strategy in all of the cell lines. The cells were treated with nanocomplexes for 4 h followed by media change, as described above. After media replacement, cellular fractions were harvested at 0, 2, 4, and 8 h and lysed in Milli-Q water. Each lysed cellular fraction was divided into two aliquots of 200 μL each. One of the aliquots of each sample was treated with 20 μL of 1 mg/mL DNase solution for 15 min at room temperature, while the other aliquot remained untreated. Post DNase treatment, both aliquots of each sample were subjected to 9 mM of Heparin challenge for 30 min. The samples were then diluted 100×, and 2 μL solutions were used for setting up q-PCR reaction to estimate the complexed and uncomplexed DNA present in each sample.
Cellular Retention and Subcellular Localization of Nanocomplexes Using Confocal Imaging
Cells were grown on a coverslip in a six-well plate by seeding B16-F10 cells at a density of 80 000 cells/well and incubating for 24 h. FITC-labeled pMIR plasmid was used for preparing nanocomplexes at a Z (±) of 5 and added to the cells (2 μg pDNA per well) in 500 μL of Opti-MEM. DNA labeling was performed using label IT tracker FITC kit (Mirus Bio LLC), according to manufacturer’s instruction. After 4 h of incubation, the treatment media was replaced by complete media and cells were again incubated. At time points of 0, 2, 4, and 8 h after this, respective coverslips were treated with 1000× Cell Mask Orange and 330 ng/μL of Hoechst 33342 (Invitrogen) followed by 30 min incubation and then washing. The cells were fixed using 4% paraformaldehyde (Sigma) and mounted on slides using DPX mountant (Sigma).
For studying colocalization of nanocomplexes with lysosomes, 106 B16-F10 cells were seeded in ibidi glass-bottom (35 mm) dishes 24 h prior to treatment. The cells were incubated for 4 h in Opti-MEM with nanocomplexes (prepared using FITC-labeled pMIR at a Z (±) of 5). After 4 h of incubation, the treatment media was replaced with complete media after washing the cells with 1× PBS. Lysosomal staining was performed by incubating the cells with Lysotracker RED for 1 h prior to imaging. The cells were washed with 0.04% trypan blue solution in 1× PBS, followed by washing with 1× PBS. Finally, 1000 μL of Opti-MEM was added to the dishes and live cell imaging was performed in a Leica SP8 confocal microscope. Analysis of lysosomal colocalization with nanocomplexes was performed using Volocity software, for which 50 cells were selected from three different fields and Pearson’s correlation coefficients were estimated. Average Pearson’s coefficient values from two independent sets of experiments were calculated with standard deviation.
Nuclear colocalization of nanocomplexes was studied by live cell continuous microscopy over a period of 8 h after media change. For this, B16-F10 cells were seeded in ibidi microchambered slides, at a density of 20 000 cells per well 24 h prior to treatment. The cells were treated with FITC-labeled nanocomplexes in Opti-MEM for 4 h, followed by media change. Hoechst staining of cells was performed by incubating with 1000× diluted working solution of Hoechst (5 mg/mL) in 1× PBS for 10 min. The same cells were continuously imaged in a CO2-maintained chamber for 8 h.
Transfection Efficiency through Luciferase Expression
For measurement of transfection in all of the cell lines involved in this study, the cells were seeded in 24-well plates 1 day prior to treatment (when cellular confluency reached ∼70%). Nanocomplexes were prepared fresh at a Z (±) of 5 and added to cells after 1 h incubation at room temperature, and 100 μL of nanocomplex was added to each well in Opti-MEM (serum-free media). After 4 h of incubation in a humidified incubator at 37 °C and 5% CO2, media was discarded and cells were washed with phosphate-buffered saline (PBS, pH 7.4), followed by addition of 500 μL of complete media for respective cell type. For measurement of luciferase expression, the cells were washed with PBS after 24 h and lysed using 100 μL of 1× cell culture lysis buffer (Promega). The measurement of expression in 50 μL of lysate using luciferase assay substrate (Promega) was taken as light emission by integration over 10 s in Orion microplate luminometer (Berthold Detection System, Germany). Luciferase activity was normalized with respect to the total protein content of the cell (measured using BCA).
Cellular Viability Assay
Cellular toxicity of all of the nanocomplexes used in this study was assessed after 24 h of treatment using the MTT assay. Briefly, the cells were seeded in a 96-well plate 1 day prior to incubation with nanocomplexes. At subconfluency, the cells were incubated with 30 μL of different nanocomplexes for 4 h in serum-free media, after which the media was aspirated and cells were washed with 1× PBS and then supplemented with 100 μL complete media. After 24 h, the cells were incubated for another 2 h after adding the MTT reagent (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) in each well. The media was then aspirated and 100 μL of dimethyl sulfoxide was added followed by absorbance recording.
Acknowledgments
The authors acknowledge the valuable scientific discussions and suggestions by Dr Sheetal Gandotra (CSIR-IGIB). They are also thankful to Manish Kumar (CSIR-IGIB) for helping with the confocal microscopy. The funding support from Department of Biotechnology (DBT), India, is acknowledged. U.R.D., S.M., and A.K. acknowledge University Grants Commission (UGC) for providing fellowships.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b02401.
AFM-based shape characterization of nanocomplexes, primer sequence, visualization of nanocomplex retention using confocal imaging, plasmid DNA release profile of different nanocomplexes, lysosomal colocalization of nanocomplexes, nuclear colocalization of nanocomplexes, and cellular viability assay (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Zhang Y.; Satterlee A.; Huang L. In Vivo Gene Delivery by Nonviral Vectors: Overcoming Hurdles. Mol. Ther. 2012, 20, 1298–1304. 10.1038/mt.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris E.; Heck K.; Lampe A. T.; Brown D. M.; Ramer-Tait A. E.; Pannier A. K. Oral Non-Viral Gene Delivery for Applications in DNA Vaccination and Gene Therapy. Curr. Opin. Biomed. Eng. 2018, 7, 51–57. 10.1016/j.cobme.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther W.; Stein U. Viral Vectors for Gene Transfer. Drugs 2000, 60, 249–271. 10.2165/00003495-200060020-00002. [DOI] [PubMed] [Google Scholar]
- Al-Dosari M. S.; Gao X. Nonviral Gene Delivery: Principle, Limitations, and Recent Progress. AAPS J. 2009, 11, 671–681. 10.1208/s12248-009-9143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemejane J.; Mir L. M. Physical Methods of Nucleic Acid Transfer: General Concepts and Applications. Br. J. Pharmacol. 2009, 157, 207–219. 10.1111/j.1476-5381.2009.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombu C. Y.; Kroubi M.; Zibouche R.; Matran R.; Betbeder D. Characterization of Endocytosis and Exocytosis of Cationic Nanoparticles in Airway Epithelium Cells. Nanotechnology 2010, 21, 355102 10.1088/0957-4484/21/35/355102. [DOI] [PubMed] [Google Scholar]
- Hama S.; Akita H.; Iida S.; Mizuguchi H.; Harashima H. Quantitative and Mechanism-Based Investigation of Post-Nuclear Delivery Events between Adenovirus and Lipoplex. Nucleic Acids Res. 2007, 35, 1533–1543. 10.1093/nar/gkl1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Satterlee A.; Huang L. In Vivo Gene Delivery by Nonviral Vectors: Overcoming Hurdles. Mol. Ther. 2012, 20, 1298–1304. 10.1038/mt.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon C.; Billiet L.; Midoux P. Chemical Vectors for Gene Delivery: Uptake and Intracellular Trafficking. Curr. Opin. Biotechnol. 2010, 21, 640–645. 10.1016/j.copbio.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Suh J.; Wirtz D.; Hanes J. Efficient Active Transport of Gene Nanocarriers to the Cell Nucleus. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 3878–3882. 10.1073/pnas.0636277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganta S.; Devalapally H.; Shahiwala A.; Amiji M. A Review of Stimuli-Responsive Nanocarriers for Drug and Gene Delivery. J. Controlled Release 2008, 126, 187–204. 10.1016/j.jconrel.2007.12.017. [DOI] [PubMed] [Google Scholar]
- Oh N.; Park J. H. Surface Chemistry of Gold Nanoparticles Mediates Their Exocytosis in Macrophages. ACS Nano 2014, 8, 6232–6241. 10.1021/nn501668a. [DOI] [PubMed] [Google Scholar]
- Wiley D. T.; Webster P.; Gale A.; Davis M. E. Transcytosis and Brain Uptake of Transferrin-Containing Nanoparticles by Tuning Avidity to Transferrin Receptor. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 8662–8667. 10.1073/pnas.1307152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T.; Kosuge M.; Tadokoro A.; Sugiura Y.; Nishi M.; Kawata M.; Sakai N.; Matile S.; Futaki S. Direct and Rapid Cytosolic Delivery Using Cell-Penetrating Peptides Mediated by Pyrenebutyrate. ACS Chem. Biol. 2006, 1, 299–303. 10.1021/cb600127m. [DOI] [PubMed] [Google Scholar]
- Grosse S.; Aron Y.; Thévenot G.; François D.; Monsigny M.; Fajac I. Potocytosis and Cellular Exit of Complexes as Cellular Pathways for Gene Delivery by Polycations. J. Gene Med. 2005, 7, 1275–1286. 10.1002/jgm.772. [DOI] [PubMed] [Google Scholar]
- Serda R. E.; MacK A.; Van De Ven A. L.; Ferrati S.; Dunner K.; Godin B.; Chiappini C.; Landry M.; Brousseau L.; Liu X.; et al. Logic-Embedded Vectors for Intracellular Partitioning, Endosomal Escape, and Exocytosis of Nanoparticles. Small 2010, 6, 2691–2700. 10.1002/smll.201000727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A.; Lopes C.; Sousa Lobo J.; Amaral M. Nucleic Acids Delivery Systems: A Challenge for Pharmaceutical Technologists. Curr. Drug Metab. 2015, 16, 3–16. 10.2174/1389200216666150401110211. [DOI] [PubMed] [Google Scholar]
- Ruponen M.; Arkko S.; Urtti A.; Reinisalo M.; Ranta V. P. Intracellular DNA Release and Elimination Correlate Poorly with Transgene Expression after Non-Viral Transfection. J. Controlled Release 2009, 136, 226–231. 10.1016/j.jconrel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Bae Y. M.; Park Y. Il; Nam S. H.; Kim J. H.; Lee K.; Kim H. M.; Yoo B.; Choi J. S.; Lee K. T.; Hyeon T.; et al. Endocytosis, Intracellular Transport, and Exocytosis of Lanthanide-Doped Upconverting Nanoparticles in Single Living Cells. Biomaterials 2012, 33, 9080–9086. 10.1016/j.biomaterials.2012.08.039. [DOI] [PubMed] [Google Scholar]
- Jiang X.; Röcker C.; Hafner M.; Brandholt S.; Dörlich R. M.; Nienhaus G. U. Endo- and Exocytosis of Zwitterionic Quantum Dot Nanoparticles by Live Hela Cells. ACS Nano 2010, 4, 6787–6797. 10.1021/nn101277w. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Wu Q.; Sui K.; Chen X.-X.; Fang J. H. A Quantitative Study of Exocytosis of Titanium Dioxide Nanoparticles from Neural St.Pdf. Nanoscale 2013, 5, 4737–4743. 10.1039/c3nr00796k. [DOI] [PubMed] [Google Scholar]
- Sha L.; Zhao Q.; Wang D.; Li X.; Wang X.; Guan X.; Wang S. “Gate” Engineered Mesoporous Silica Nanoparticles for a Double Inhibition of Drug Efflux and Particle Exocytosis to Enhance Antitumor Activity. J. Colloid Interface Sci. 2019, 535, 380–391. 10.1016/j.jcis.2018.09.089. [DOI] [PubMed] [Google Scholar]
- Chu Z.; Huang Y.; Tao Q.; Li Q. Cellular Uptake, Evolution, and Excretion of Silica Nanoparticles in Human Cells. Nanoscale 2011, 3, 3291–3299. 10.1039/c1nr10499c. [DOI] [PubMed] [Google Scholar]
- Foroozandeh P.; Aziz A. A. Insight into Cellular Uptake and Intracellular Trafficking of Nanoparticles. Nanoscale Res. Lett. 2018, 13, 339 10.1186/s11671-018-2728-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh N.; Park J.-H. Surface Chemistry of Gold Nanoparticles Mediates Their Exocytosis in Macrophages. ACS Nano 2014, 8, 6232–6241. 10.1021/nn501668a. [DOI] [PubMed] [Google Scholar]
- Rose L.; Mahdipoor P.; Kucharski C.; Uludağ H. Pharmacokinetics and Transgene Expression of Implanted Polyethylenimine- Based PDNA Complexes. Biomater. Sci. 2014, 2, 833–842. 10.1039/C3BM60200A. [DOI] [PubMed] [Google Scholar]
- Shi J.; Chou B.; Choi J. L.; Ta A. L.; Pun S. H. Investigation of Polyethylenimine/DNA Polyplex Transfection to Cultured Cells Using Radiolabeling and Subcellular Fractionation Methods. Mol. Pharm. 2013, 10, 2145–2156. 10.1021/mp300651q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahay G.; Querbes W.; Alabi C.; Eltoukhy A.; Sarkar S.; Zurenko C.; Karagiannis E.; Love K.; Chen D.; Zoncu R.; et al. Efficiency of SiRNA Delivery by Lipid Nanoparticles Is Limited by Endocytic Recycling. Nat. Biotechnol. 2013, 31, 653–658. 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya U. R.; Ganguli M. Exocytosis—a Putative Road-Block in Nanoparticle and Nanocomplex Mediated Gene Delivery. J. Controlled Release 2019, 12 10.1016/j.jconrel.2019.04.012. [DOI] [PubMed] [Google Scholar]
- Cartiera M. S.; Johnson K. M.; Rajendran V.; Caplan M. J.; Saltzman W. M. The Uptake and Intracellular Fate of PLGA Nanoparticles in Epithelial Cells. Biomaterials 2009, 30, 2790–2798. 10.1016/j.biomaterials.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombu C. Y.; Kroubi M.; Zibouche R.; Matran R.; Betbeder D. Characterization of Endocytosis and Exocytosis of Cationic Nanoparticles in Airway Epithelium Cells. Nanotechnology 2010, 21, 355102 10.1088/0957-4484/21/35/355102. [DOI] [PubMed] [Google Scholar]
- Sharma R.; Shivpuri S.; Anand A.; Kulshreshtha A.; Ganguli M. Insight into the Role of Physicochemical Parameters in a Novel Series of Amphipathic Peptides for Efficient DNA Delivery. Mol. Biopharm. 2013, 10, 2588–2600. 10.1021/mp400032q. [DOI] [PubMed] [Google Scholar]
- Sharma R.; Nisakar D.; Shivpuri S.; Ganguli M. Contrasting Effects of Cysteine Modification on the Transfection Efficiency of Amphipathic Peptides. Biomaterials 2014, 35, 6563–6575. 10.1016/j.biomaterials.2014.04.046. [DOI] [PubMed] [Google Scholar]
- Åmand H. L.; Nordén B.; Fant K. Functionalization with C-Terminal Cysteine Enhances Transfection Efficiency of Cell-Penetrating Peptides through Dimer Formation. Biochem. Biophys. Res. Commun. 2012, 418, 469–474. 10.1016/j.bbrc.2012.01.041. [DOI] [PubMed] [Google Scholar]
- Wang S.; Yan C.; Zhang X.; Shi D.; Chi L.; Luo G.; Deng J. Antimicrobial Peptide Modification Enhances the Gene Delivery and Bactericidal Efficiency of Gold Nanoparticles for Accelerating Diabetic Wound Healing. Biomater. Sci. 2018, 6, 2757–2772. 10.1039/C8BM00807H. [DOI] [PubMed] [Google Scholar]
- Brownlee W. J.; Seib F. P. Impact of the Hypoxic Phenotype on the Uptake and Efflux of Nanoparticles by Human Breast Cancer Cells. Sci. Rep. 2018, 8, 12318 10.1038/s41598-018-30517-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong K. W.; Grigsby C. L. Balancing Protection and Release of DNA: Tools to Address a Bottleneck of Non-Viral Gene Delivery. J. R. Soc. Interface 2010, 7, S67–S82. 10.1098/rsif.2009.0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielson N. P.; Pack D. W. Acetylation of Polyethylenimine Enhances Gene Delivery via Weakened Polymer/DNA Interactions. Biomacromolecules 2006, 7, 2427–2435. 10.1021/bm060300u. [DOI] [PubMed] [Google Scholar]
- Schaffer D. V.; Fidelman N. A.; Dan N.; Lauffenburger D. A. Vector Unpacking as a Potential Barrier for Receptor-Mediated Polyplex Gene Delivery. Biotechnol. Bioeng. 2000, 67, 598–606. . [DOI] [PubMed] [Google Scholar]
- Yanes R. E.; Tarn D.; Hwang A. A.; Ferris D. P.; Sherman S. P.; Thomas C. R.; Lu J.; Pyle A. D.; Zink J. I.; Tamanoi F. Involvement of Lysosomal Exocytosis in the Excretion of Mesoporous Silica Nanoparticles and Enhancement of the Drug Delivery Effect by Exocytosis Inhibition. Small 2013, 9, 697–704. 10.1002/smll.201201811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean D. A.; Strong D. D.; Zimmer W. E. Nuclear Entry of Nonviral Vectors. Gene Ther. 2005, 12, 881–890. 10.1038/sj.gt.3302534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.