Abstract
The human gastrointestinal tract is home to a thriving community of microbes including the fungal ‘mycobiota’. Although sequencing methodology has enumerated diverse fungal genera within this niche, discerning persistent symbiotic residents from contaminants and purely environmental transients remains a challenge. Recent advances in culturomics and sequencing employing metagenomics, metatranscriptomics and longitudinal studies have begun to reveal a human symbiont ‘core mycobiome’ that may contribute to human health and disease. Trans-kingdom interactions between the bacterial microbiota and evolution within the niche have defined C. albicans as a true symbiont, setting a bar for defining other fungi. Additionally, elegant investigations of mammalian antifungal immunity have examined mononuclear phagocytes, neutrophils, antigen-specific recognition by T cells and other mechanisms important for local and systemic effects on the host, providing further evidence supporting gut persistence. In this review we discuss current research aimed at investigating the symbiotic mycobiota and propose four criteria aiding in the differentiation of fungal symbionts from environmental transients.
Introduction
The barrier surfaces of the human body serve as habitats to a variety of fungal organisms collectively known as the human ‘mycobiota’. The intestinal mycobiota has been linked to maintenance of healthy homeostasis and prevention of systemic infection [1–4]. Dysbiosis of the human mycobiota and expansion of Candida spp., fungal species highly represented in the healthy gastrointestinal tract, have been identified as signatures of inflammatory bowel disease (IBD) and several other inflammatory and non-inflammatory diseases [5–9]. Patients with genetic polymorphisms in receptors involved in fungal recognition[9] display increased intestinal disease severity, while a deficiency in CX3CR1, a fractalkine receptor flagging a population of mononuclear phagocytes, is associated with decreased IgG responses to fungi in patients with Crohn’s Disease[10]. Moreover, overgrowth of opportunistic pathogens, such as C. albicans or C. parapsilosis, has been linked to an increased risk of translocation and systemic infection in patients with compromised immune function [11]. The growing consensus on the importance of the intestinal mycobiota has prompted the investigation of which fungi are capable of surviving, residing and replicating in the gastrointestinal tract and thereby maximally able to influence the host over a prolonged period.
The mycobiota poses unique challenges in terms of identifying and validating constituents of this intestinal community. In contrast to bacteria, fungi are found in relatively low abundance in the gut, but are common in various food sources and the environment (cheese, beer, bread, wine, airborne spores, etc.). While the development of deep sequencing approaches have provided researchers with utmost opportunities to investigate the fungal variety making up the gastrointestinal (GI) “mycobiome”, the combination of low fungal DNA abundance in the gut and high dietary/environmental abundance provides data sets susceptible to serendipitous contamination from sources outside of the body and from other bodily surfaces (i.e. face, hands, sweat etc.). In addition to outnumbering fungal organisms, many members of the bacterial community are dependent on the anaerobic environment provided by the gut, greatly increasing their probability of persisting within the intestines. Fungi which are typically either facultative anaerobes or able to withstand hypoxic environments lack this certainty. These discrepancies between bacterial and fungal communities highlight the urgent need for more specific mycobiota-tailored criteria to differentiate resident fungi, referred here as fungal symbionts (Box 1) from transient passengers and contaminants.
Box 1.
Symbiont refers to any organism persistently living in contact with another. In the context of this review, this term specifies fungi living and replicating within the gastrointestinal tract. Symbionts include all manner of non-transient relationships including: mutualism, parasitism and commensalism, among others. In delineating symbionts from transients, or passing environmental/dietary organisms, specific relationships may act as qualifiers justifying potential symbiont classification. The precise type of symbiosis may be altered between fungi and the host depending on body site, extent of colonization and immune integrity, as is the case for pathobionts and opportunistic pathogens which transition from neutral or beneficial relationship to one of a pathogenic nature.
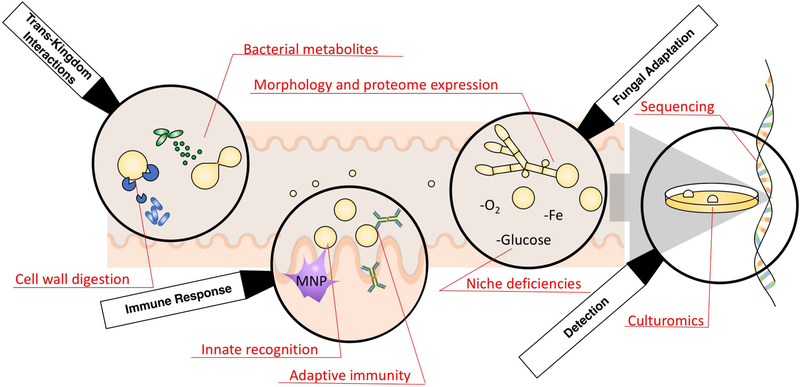
We propose the use of four primary lenses for examining suspect fungal symbionts : 1) Detection by both, sequencing and culturomics, 2) Activation of host immune responses, 3) Ability to populate and evolve in the gastrointestinal niche through fungal adaptation, and 4) Evidence of trans-kingdom interactions between bacteria and fungi. (Figure 1) Herein we review these strategies for studying distinct members of the mycobiota, focusing on the last two years of efforts to define the fungal symbiotic relationships in the mammalian gut.
Figure 1.
Maturing perspectives for the investigations of symbiotic members of the mycobiota. Both bacterial-fungal and host immunity interactions supplement the direct observations of species evolution, sequencing and culturomics observed in gut fungi. Together these tools have been used to validate C. albicans as a model intestinal symbiont.
Detection-Sequencing and Culturomics
Just as detectives wear gloves to protect the crime scene, mycobiome researchers also pay close attention to their choice of methods, aiming to accurately elucidate a microscopic reality in the fungal world hidden inside the mammalian GI tract. High throughput sequencing (HTS) for the gastrointestinal mycobiome has become a mainstay and the first step toward studying fungal symbiosis in the gut. The most common HTS method is amplicon sequencing of the Internal Transcribed Spacer region 1 or 2 (ITS1/2)(Table 1). The convenience of amplicon sequencing results from the amplification of fungal DNA the identification of fungi in microbial communities residing the GI tract that are naturally dominated by bacteria. However, several caveats of this approach have also been demonstrated, such as primer bias, amplification preference and choice of optimally curated database for fungal identification [12–14]. Reverse transcriptomic sequencing of rRNA has been proposed to decrease the primer bias but has thus far been shown more suitable for bacterial identification [15]. In contrast, shotgun metagenomic sequencing omits targeted PCR amplification, thus improving result fidelity [16]. An intrinsic disadvantage to this approach is that fungal sequences can be buried by the overwhelming abundance of fecal bacterial DNA [17]. The present hope is that this issue is partially resolved as the cost-to-sequencing depth ratio decreases over time, and databases record more complete fungal genomes. Alternatively, metatranscriptomic analysis has been performed on environmental mycobiomes and has the advantage of identifying living, actively-transcribing organisms. However, the current classification of fungal metatranscriptomic contigs is chiefly reliable only at the phylum level [18].
Table 1.
| Method | Technology | Target | Advantages | Disadvantages |
|---|---|---|---|---|
| High throughput sequencing (HTS) | Amplicon sequencing | Internal Transcribed Spacer 1 and 2 (ITS1/2) | Database availability; amplification of target sequences | Primary bias; amplification preference |
| 18S/26S rRNA genes | ||||
| Reverse transcribed rRNA | Decreased primer bias | Amplification preference | ||
| Shotgun sequencing | Metagenome | No amplification preference | Low cost efficiency due to fungal DNA exiguity | |
| Meta-transcriptome | Identification of live organisms | Low cost efficiency; classification mostly reliable at phylum level | ||
| Culturomics | Culturing in selective media | Live organisms | Identification of live organisms | Potential to miss anaerobic or fastidious fungi (ex. anaerobic fungi from ruminant gut) |
| Mass spectrometry (MS) | Matrix Assisted Laser Desorption/ionization Time-Of-Flight (MALDI-TOF) | Protein | Fast process; strain-level identification | Limited and often proprietary databases |
To eliminate the possibility of identifying fungal nucleic acids from dead or non-proliferating cells, culturomics is still used as the gold standard for identifying live fungi when they are culturable [19]. Fungal isolation is traditionally coupled with morphological analysis and whole genome sequencing but recently these methodologies have been supplemented with mass spectrometry. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) has been applied for yeast identification in both academic settings as well as for clinical isolates [20]. Better curated databases in the future will improve the power and accuracy of this rapid identification method, allowing for further expansion of fidelity to strain-level identification [21]. Further development of culturomic approaches that capture a vast majority of fungal species found in GI tract will further advance the field of gut mycology, mirroring success recently achieved for intestinal bacteria [22].
Recent efforts are aiming to broadly define the human intestinal fungal communities with established sequencing platforms. Nash and colleagues studied the fecal mycobiome of 317 healthy donors from the Human Microbiome Project and characterized a high prevalence of the genera Saccharomyces, Malassezia and Candida, with the highest abundance represented by the species S. cerevisiae, M. restricta and C. albicans, respectively [17]. The authors applied amplicon sequencing of the ITS2 region and verified their results via sequencing 18S rRNA gene and shotgun metagenomics. Despite the generally large variability in mycobiomes, these taxa were commonly identified both across individuals and longitudinally within individuals. Previously published work by Hoffman and colleagues sequenced ITS1 of 98 individuals. They identified many fungal taxa in common, with the top three being Saccharomyces, Candida, and Cladosporium [23]. In addition to these identified most prevalent taxa, both studies respectively reported another 177 and 63 fungal genera. These findings suggest a ‘core mycobiome’ across populations of healthy humans, with a highly diverse remainder of less represented fungi. The latter diversity arises either from environmental/dietary transient variability or from rare fungal symbionts (a less plausible possibility).
Candida species, including C. albicans, are generally accepted as true gut symbiotic fungi. C. albicans is frequently sequenced and isolated from feces of healthy humans. However, the symbiotic status of S. cerevisiae and M. restricta are still debated, mainly due to their abundance in food sources and skin mycobiota, respectively [24,25]. Recent studies have gone as far as to challenge the origin of fecal C. albicans, suggesting that it arises from the oral mycobiota or food sources [26,27]. As these questions remain unanswered, another direction of evaluating symbiosis is to study the sources of fungi found within the GI tract. Boix-Amorós and colleagues have conducted two studies on breast milk mycobiomes [28,29]. By combining 28S rRNA sequencing with culturomics, they first identified Candida, Saccharomyces and Malassezia as the most prevalent genera. They further expanded the study to healthy volunteers spanning across four countries. Despite different environmental and dietary factors, Malassezia maintained an average of 40% abundance across four cohorts. While these studies do not answer the question whether Malassezia is a true gut symbiont, they do reveal a feasible route for transient early-life inoculation of the infant gut through mother breast skin-derived Malassezia during breastfeeding [30]. Indeed, transient Malassezia presence in infant fecal samples have been detected by ITS sequencing until 4 months of age whereupon it is overtaken by mycobiota members belonging to the Saccharomycetales order with the development of stable intestinal microbiota later in life [30].
In summary, mycobiome studies shortlist core fungal taxa that are strongly suspected gut residents. However, mycobiome detection does not readily prove or disprove their symbiotic nature. Further studies on fungal interactions with the host and bacteria are needed to add confidence in making this distinction (Fig 1).
Immune Response- Antifungal Immunity
For symbiotic fungi to persist and proliferate in the gut, they face the challenge of interacting with the host immune system. Hence, discovery of bidirectional interactions between fungi and the host immune system can serve as supportive evidence for symbiosis. On one hand, the host immune system monitors fungal expansion and penetration. On the other hand, symbiotic fungi may modulate the tone of immune responses locally or even distally [1,31]. In instances when direct exposure would endanger fungal persistence, fungal cell wall rearrangement strategies have been reported, although “in situ” evidence is still scarce [32]. Most studies on intestinal immunity to mycobiota have been carried out on C. albicans, the most prevalent fungal pathobiont in the human gut. The specific immune system responses to C. albicans and some other fungi involve innate pattern recognition receptors (PRRs), CARD9/Syk signaling pathways and Th17 adaptive immunity, which have been previously reviewed [33,34]. In recent years, new discoveries on C. albicans and other potential fungal gut symbionts have added to our understanding of this dynamic, two-way relationship.
It was recently described that CX3CR1+ mononuclear phagocytes (MNPs) express several fungal-recognizing C-type lectin receptors, including dectin-1, dectin-2 and mincle. These phagocytes mediate fungal recognition and antigen-specific Th17 response to C. albicans in the mouse colon during the steady state and are crucial for control of C. albicans during intestinal disease [10]. A further study showed the importance of CX3CR1+ MNPs in mediating the effect of gut fungal dysbiosis on house dust mite (HDM)-induced allergic airway disease (AAD), providing an explanation as to how intestinal fungal dysbiosis aggravates lung allergy [1]. Intestinal fungal colonization of mycobiota-free altered Schaedler flora-colonized mice aggravated AAD without altering bacterial composition, suggesting the enhanced allergic responses are dependent on direct interaction with fungal constituents of the gut [1].
Focusing on adaptive antifungal immunity, Bacher and colleagues [31] found that among 30 mycobiota members, C. albicans was the strongest inducer of CD4+ memory Th17 cells in humans, suggesting an “immunological imprint” of this fungal symbiont. C .albicans induces Th17 responses in the mouse gut upon intestinal colonization independently of the preexisting gut mycobiome composition [10,35,36]. Th17 cells induced by intestinal C. albicans colonization exert also a systemic effect by protecting mice against systemic C. albicans and Staphylococcus aureus infections[37].
Researchers have expanded similar studies to minor components of gastrointestinal mycobiota identified by deep sequencing. Intestinal delivery of three fugal species, A. amstelorami, E. nigrum and W. mellicola, exacerbate HDM-induced allergic airway disease [38]. Oral gavage of M. restricta resulted in exacerbated DSS-induced colitis in mice in a CARD9-dependent manner[39]. M. restricta also augmented Th1 and Th17 responses induced by acute DSS exposure in the mouse colon. However whether these minor components of the mycobiota can naturally colonize the gut and whether can they can prime intestinal immunity in the absence of inflammation remains to be determined.
The extensive new studies on mucosal immunity to gut-residing C. albicans, together with other characteristics discussed later in this review, have begun to provide strong evidence supporting this organism’s true symbiotic nature. Increasing findings pertaining to immune responses against S. cerevisiae, M. restricta, W. mellicola, A. amstelodami, E. nigrum and other Candida and non-Candida species do not by themselves assure their symbiotic status (Figure 1), but at least demonstrate scenarios where they promote persistent influences upon the host.
Fungal Adaption- Evolution towards Persistence
The mammalian gastrointestinal tract presents a balance of challenges and benefits for potential fungal residents. This intestinal niche, from proximal duodenum to distal colon, represents an increasingly hypoxic, hypercarbic, anemic and hypoglycemic residence. Additional stressors include the innate and adaptive immune systems (vide supra) and a diverse bacterial clientele (vide infra). Occupants able to navigate these challenges are rewarded with a warm, moist and dark environment equipped with a natural defense from eukaryote-targeting pathogens (i.e. the host immune system). As such, potential long-term clientele of the gut would be anticipated to adapt to residence, providing another clue useful in identifying true symbiotic fungi.
Many fungi are opportunistic pathogens, intrinsically harboring the cellular machinery necessary for host invasion and infection which poses a liability to gut symbiosis. Elucidating the mechanisms by which these fungi achieve an evolutionary equilibrium conducive to symbiosis is a recently burgeoning field of mycobiota research. In this regard, Bennett and coworkers [4] have revealed a novel mechanism by which C. albicans evolves to compete in the GI tract through utilization of hyphal-regulating transcription factors EFG1 and WOR1 via genetic mutation and epigenetic regulation, respectively. Authors reported human isolates displayed a high percentage (11%) of disruptive efg1 mutations resulting in efg1+/− and efg1−/− genotypes uniquely able to switch to a gray colony state in vitro and exhibiting a competitive advantage in the mouse GI tract. This work aligns well with several observations of loss of efg1 functionality often linked with hypercompetitive phenotypes in systemic and gastrointestinal candidiasis [40–42], although conclusions as to regulation of WOR1 must be viewed in the context of gut-focused research showing that expression of this transcription factor can bypass the heterozygosity in the mating locus in vivo [42].
A more systematic study relying on a C. albicans gene disruption library published by the Noble lab [32] revealed a series of transcription factors (EFG1, BRG1, TEC1) targeting morphogenesis and virulence whose gene disruption increased gastrointestinal colonization competitive fitness in vivo. Intriguingly, these transcription factors all influence UME6, a master regulator of hyphal programing. Functional deletion of UME6 resulted in reduced filamentation in vitro, restoration of competitive fitness and, surprisingly, no change to in vivo morphology by FISH-assisted microscopy analysis of gastrointestinal contents. Further nanostring, RNAseq and mutational analyses identified two UME6-regulated proteins, SAP6 (secreted aspartyl protease 6) and, to a lesser extent, HYR1 (hyphally regulated gene 1) as culpable in reducing gastrointestinal competition of C. albicans strains independently of gross morphology. SAP6 was previously shown [43] to be adept at activating the inflammasome in dendritic cells and macrophages, possibly targeting the fungus for elimination by the immune system. Additionally, SAP6 was differentially expressed in an independent experimental approach [3] which identified hyper-competition in FLO8 mutants in a murine model of gastrointestinal colonization. Combined these studies highlight the intriguing evolution of one fungi as it adapts to the challenges of the intestinal niche through diminishing its own proclivities towards a more destructive, pathogenic character. Finally, this emerging evolutionary adaption approach may prove invaluable when expanded to other fungal members of the mycobiota, providing a new lens by which to judge suspected symbionts (Fig. 1).
Trans-Kingdom Interactions- Bacterial Modulation of Fungi
The environmental and host immunity-mediated challenges present in the mammalian intestinal tract are compounded by the abundance of the facultative and obligate anaerobic bacteria that grossly outnumber other eukaryotic microorganisms. While transient microorganisms might transition through the gut without notice, true symbiotic fungi, persisting in the gut and competing for resources, would be expected to induce a retaliatory or mutalistic response from the bacterial microbiota (Fig. 1). In light of this constant recognition, specific trans-kingdom interactions serve as an additional means of qualifying possible fungal symbionts in the gastrointestinal tract.
Bacteria depend on chemical interactions with other organisms both through a suite of small-molecule metabolites as well as protein gene products. Their diffusible metabolites arise from both primary (such as fatty acids or polysaccharides) and secondary (such as polyketides, non-ribosomal peptides, terpenes) metabolic processes. Bacterial pathobionts of the GI tract have recently been shown to produce both fungal modifying compounds as well as secreted enzymes. Coulthurst and coworkers [44] have recently elucidated and interrogated two new type VI secretion system proteins, Tfe1 and Tfe2, with antifungal properties against Candida albicans, C. glabrata and S. cerevisiae from Serratia marcescens. Interestingly, this relationship appears to run contrary to recent sequencing data in Crohn’s disease showing a strong positive correlation between expansion of Serratia and Candida, possibly implicating an underlying symbiotic relationship in vivo, an altered dynamic during disease-induced inflammation, or spatially distinct niches for these organisms within the gastrointestinal tract [45].
Obligate anaerobic bacteria predominate the mammalian gastrointestinal tract and offer a snapshot into the neighboring organisms fungi must tolerate to maintain their symbiotic status in the unique environment of the gut. From the phylum Firmicutes, the opportunistic pathogen C. difficile maintains its hold over the intestinal niche through its distinctive production of p-cresol, an antibacterial agent derived from tyrosine metabolism [46]. This phenol metabolite was shown [47] to inhibit the yeast-hyphae transition and biofilm formation in C. albicans. Moreover, in co-culture with C. albicans, C. difficile was able to tolerate aerobic growth conditions independently of adhesion or bi-species biofilm formation [47] as previously shown for the bacterium C. perfringens [48]. Interestingly, these studies also align with an examination of both fecal material transplants (FMT) in patients and mouse models of C. difficile infection correlating reduced mycobiota diversity and increased C. albicans abundance to poor FMT recipient outcomes [49], further implicating the physiological relevancy of this cross-kingdom interaction. Sellam and colleagues [50] have confirmed the relevance of the Firmicutes-Candida interaction as well as implicated a member of the phylum Bacteroidetes with their discovery of morphology-modifying and fungistatic properties from both Roseburia and Bacteroides spp., although the exact molecules at play remain unknown.
Bacteria of the gastrointestinal tract serve a vital role in processing dietary fiber to extract carbon and nitrogen from sources otherwise inaccessible to the host. Fungal cell walls contain a complex polysaccharide coating consisting of mannan, β-glucan and chitin, providing an alternative source of fiber in the gastrointestinal environment. Both β-glucan and chitin have been found to be modified by bacteria of the microbiota through small molecule signaling [51] and enzymatic degradation [52], respectively. Specifically, Bacillus spp. were found to harbor chitinases capable of altering C. albicans morphology, inhibiting biofilm formation and antagonizing growth [52]. Lactic acid, a metabolite produced by lactobacilli in the GI tract, was also shown to promote restructuring of C. albicans cell wall architecture, masking β-glucan exposure in the cell wall [51]. A recent surge of interest in polysaccharide utilizing loci (PUL) chiefly from Bacteroidetes spp. within the mammalian gut has revealed discrete PUL responsible for fungal β−1,6-glucan [53] and S. cerevisiae mannan [54] catabolism. Although all members of the fungal kingdom contain β-(1,6 or 1,3)glucan and chitin (polymeric 1,4-N-acetylglucosamine) within their cell wall, mannan serves as a means of concealing potential pathogen associated molecular pattern (PAMP) ligands from the host [55] and varies significantly in linkages and gross structure across species [56]. Thus, the further identification and evaluation of bacterial enzymes dedicated to the cleavage of genus/species-specific mannosyl linkages could serve to validate stable predator-prey interactions conserved in the native microbiota.
Conclusions
As mycobiome sequencing technology continues to mature, we are ultimately faced with the question: How do we best categorize the diverse members of gastrointestinal mycobiota in order to separate true residents from transient passersby? Sequencing and culturomics have profiled the possible suspect fungi, but there is a need for additional longitudinal work across a variety of geographic regions and dietary conditions to delineate environmental and nutritional constituents from less variant members of the mycobiota. Specific interactions with both the host (immunity) and bacteria (microbiota) may serve as traits to gauge the role of fungi in the gut, but the scope of study has thus far been limited to chiefly C. albicans and S. cerevisiae. Expanding these investigations as well as interrogating the propensity of each fungal species and even of specific strains to evolve within the gastrointestinal niche offer the most promising strategies forward in defining a symbiotic mycobiota. In this regard, Candida species and C. albicans specifically may serve vital roles as not only a prototypical symbiont, but also as a future road map for studying important fungal constituents and distinguishing true fungal symbionts of the human gut.
Highlights.
Deep-sequencing and culturomics data suggest a core fungal community within the human gut.
We propose four criteria: detection, fungal adaptation, immune response and trans-kingdom interactions to define symbionts.
C. albicans is a model symbiont capable of adaptation in the gastrointestinal niche.
Acknowledgements
The authors are supported by the National Institutes of Health (DK113136, AI146957 and F32DK120228), Irma Hirschl Research Scientist and Crohn’s and Colitis Foundation awards.
References
*Denote special interest, **Denotes outstanding interest
- [1].Li X, Leonardi I, Semon A, Doron I, Gao IH, Putzel GG, Kim Y, Kabata H, Artis D, Fiers WD, Ramer-Tait AE, Iliev ID, Response to Fungal Dysbiosis by Gut-Resident CX3CR1+ Mononuclear Phagocytes Aggravates Allergic Airway Disease, Cell Host Microbe. 24 (2018) 847–856.e4. doi: 10.1016/j.chom.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; *The study reports an expansion of Th2 cells in the lung upon gut fungal dysbiosis. The observed systemic responses to fungi in the gut are mediated by gut-resident CX3CR1+ mononuclear phagocytes.
- [2].Jiang TT, Shao T-Y, Ang WXG, Kinder JM, Turner LH, Pham G, Whitt J, Alenghat T, Way SS, Commensal Fungi Recapitulate the Protective Benefits of Intestinal Bacteria, Cell Host Microbe. 22 (2017) 809–816.e4. doi: 10.1016/j.chom.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tso GHW, Reales-Calderon JA, Tan ASM, Sem X, Le GTT, Tan TG, Lai GC, Srinivasan KG, Yurieva M, Liao W, Poidinger M, Zolezzi F, Rancati G, Pavelka N, Experimental evolution of a fungal pathogen into a gut symbiont, Science. 362 (2018) 589–595. doi: 10.1126/science.aat0537. [DOI] [PubMed] [Google Scholar]; *Authors were able to evolve C. albicans through serial gastrointestinal passage experiments into hyper-competitive strains often resulting from Flo8 transcriptional factor mutations. Interesting, a few strains bestowed protective benefits to the host from a range of pathogens including A. fumigatus, P. aeruginosa, S. aureus, and C. albicans, in a T and B cell independent manner.
- [4].Liang S-H, Anderson MZ, Hirakawa MP, Wang JM, Frazer C, Alaalm LM, Thomson GJ, Ene IV, Bennett RJ, Hemizygosity Enables a Mutational Transition Governing Fungal Virulence and Commensalism, Cell Host Microbe. 25 (2019) 418–431.e6. doi: 10.1016/j.chom.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Authors investigated phenotypic switching in C. albicans and revealed that the genetic basis for metabolic states is dependent on zygosity of the transcription factor EFG1 in combination with the mating locus a1/a2. Oral C. albicans strains containing hemizygous EFG1 were isolated from patients and this genetic state was found to confer increased fitness in the gastrointestinal tract and increased mortality in a mouse model of systemic candidiasis.
- [5].Iliev ID, Leonardi I, Fungal dysbiosis: immunity and interactions at mucosal barriers, Nat. Rev. Immunol 17 (2017) 635–646. doi: 10.1038/nri.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sokol H, Leducq V, Aschard H, Pham H-P, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, Cosnes J, Seksik P, Langella P, Skurnik D, Richard ML, Beaugerie L, Fungal microbiota dysbiosis in IBD, Gut. 66 (2017) 1039–1048. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]; *A comprehensive study of the human mycobiota in IBD patients revealed an increase in C. albicans IBD (especially during inflammatory flare). Additionally, the microbiota in IBD patients displayed distinctly unbalanced correlations with the mycobiota in comparison to healthy controls, suggesting a dysbiotic state conducive to fungal expansion.
- [7].Lewis JD, Chen EZ, Baldassano RN, Otley AR, Griffiths AM, Lee D, Bittinger K, Bailey A, Friedman ES, Hoffmann C, Albenberg L, Sinha R, Compher C, Gilroy E, Nessel L, Grant A, Chehoud C, Li H, Wu GD, Bushman FD, Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn’s Disease, Cell Host Microbe. 18 (2015) 489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mar JS, LaMere BJ, Lin DL, Levan S, Nazareth M, Mahadevan U, Lynch SV, Disease Severity and Immune Activity Relate to Distinct Interkingdom Gut Microbiome States in Ethnically Distinct Ulcerative Colitis Patients, MBio. 7 (2016) e01072–16. doi: 10.1128/mBio.01072-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li XV, Leonardi I, Iliev ID, Gut Mycobiota in Immunity and Inflammatory Disease, Immunity. 50 (2019) 1365–1379. doi: 10.1016/j.immuni.2019.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Leonardi I, Li X, Semon A, Li D, Doron I, Putzel G, Bar A, Prieto D, Rescigno M, McGovern DPB, Pla J, Iliev ID, CX3CR1+ mononuclear phagocytes control immunity to intestinal fungi, Science. 359 (2018) 232–236. doi: 10.1126/science.aao1503. [DOI] [PMC free article] [PubMed] [Google Scholar]; **CX3CR1+ macrophages, a cell population of inherently expressing high amounts of antifungal C-leptin receptors, were recently identified as key cell types in sensing and sampling intestinal fungi. Genetic ablation of CX3CR1+ mononuclear phagocytes resulted in aggravated chemically-induced colitis in mice. Furthermore, patients with polymorphisms in the CX3CR1 gene presented with attenuated serum IgG response to fungi.
- [11].Nucci M, Anaissie E, Revisiting the Source of Candidemia: Skin or Gut?, Clin. Infect. Dis 33 (2001) 1959–1967. doi: 10.1086/323759. [DOI] [PubMed] [Google Scholar]
- [12].Prakash PY, Irinyi L, Halliday C, Chen S, Robert V, Meyer W, Online Databases for Taxonomy and Identification of Pathogenic Fungi and Proposal for a Cloud-Based Dynamic Data Network Platform, J. Clin. Microbiol 55 (2017) 1011–1024. doi: 10.1128/JCM.02084-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Filippis FD, Laiola M, Blaiotta G, Ercolini D, Different Amplicon Targets for Sequencing-Based Studies of Fungal Diversity, Appl. Environ. Microbiol 83 (2017) e00905–17. doi: 10.1128/AEM.00905-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tang J, Iliev ID, Brown J, Underhill DM, Funari VA, Mycobiome: Approaches to analysis of intestinal fungi, J. Immunol. Methods 421 (2015) 112–121. doi: 10.1016/j.jim.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Karst SM, Dueholm MS, McIlroy SJ, Kirkegaard RH, Nielsen PH, Albertsen M, Retrieval of a million high-quality, full-length microbial 16S and 18S rRNA gene sequences without primer bias, Nat. Biotechnol 36 (2018) 190–195. doi: 10.1038/nbt.4045. [DOI] [PubMed] [Google Scholar]
- [16].Costea PI, Zeller G, Sunagawa S, Pelletier E, Alberti A, Levenez F, Tramontano M, Driessen M, Hercog R, Jung F-E, Kultima JR, Hayward MR, Coelho LP, Allen-Vercoe E, Bertrand L, Blaut M, Brown JRM, Carton T, Cools-Portier S, Daigneault M, Derrien M, Druesne A, de Vos WM, Finlay BB, Flint HJ, Guarner F, Hattori M, Heilig H, Luna RA, van Hylckama Vlieg J, Junick J, Klymiuk I, Langella P, Le Chatelier E, Mai V, Manichanh C, Martin JC, Mery C, Morita H, O’Toole PW, Orvain C, Patil KR, Penders J, Persson S, Pons N, Popova M, Salonen A, Saulnier D, Scott KP, Singh B, Slezak K, Veiga P, Versalovic J, Zhao L, Zoetendal EG, Ehrlich SD, Dore J, Bork P, Towards standards for human fecal sample processing in metagenomic studies, Nat. Biotechnol 35 (2017) 1069–1076. doi: 10.1038/nbt.3960. [DOI] [PubMed] [Google Scholar]
- [17].Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, Stewart CJ, Metcalf GA, Muzny DM, Gibbs RA, Ajami NJ, Petrosino JF, The gut mycobiome of the Human Microbiome Project healthy cohort, Microbiome. 5 (2017) 153. doi: 10.1186/s40168-017-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Authors analyzed fungal ITS1/ITS2 sequences within the Human Microbiome Project fecal samples, revealing domination and persistence by the yeast genera Saccharomyces, Malassezia, and Candida. In general, fungal populations displayed lower alpha diversity but longitudinal studies revealed that outside the ‘core mycobiota’ other genera are extremely variable.
- [18].Žifčáková L, Větrovský T, Lombard V, Henrissat B, Howe A, Baldrian P, Feed in summer, rest in winter: microbial carbon utilization in forest topsoil, Microbiome. 5 (2017) 122. doi: 10.1186/s40168-017-0340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gouba N, Raoult D, Drancourt M, Eukaryote Culturomics of the Gut Reveals New Species, PLOS ONE. 9 (2014) e106994. doi: 10.1371/journal.pone.0106994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hamad I, Ranque S, Azhar EI, Yasir M, Jiman-Fatani AA, Tissot-Dupont H, Raoult D, Bittar F, Culturomics and Amplicon-based Metagenomic Approaches for the Study of Fungal Population in Human Gut Microbiota, Sci. Rep 7 (2017) 16788. doi: 10.1038/s41598-017-17132-4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [21].Borges FM, de Paula TO, Sarmiento MRA, de Oliveira MG, Pereira MLM, Toledo IV, Nascimento TC, Ferreira-Machado AB, Silva VL, Diniz CG, Fungal Diversity of Human Gut Microbiota Among Eutrophic, Overweight, and Obese Individuals Based on Aerobic Culture-Dependent Approach, Curr. Microbiol 75 (2018) 726–735. doi: 10.1007/s00284-018-1438-8. [DOI] [PubMed] [Google Scholar]
- [22].Poyet M, Groussin M, Gibbons SM, Avila-Pacheco J, Jiang X, Kearney SM, Perrotta AR, Berdy B, Zhao S, Lieberman TD, Swanson PK, Smith M, Roesemann S, Alexander JE, Rich SA, Livny J, Vlamakis H, Clish C, Bullock K, Deik A, Scott J, Pierce KA, Xavier RJ, Alm EJ, A library of human gut bacterial isolates paired with longitudinal multiomics data enables mechanistic microbiome research, Nat. Med 25 (2019) 1442–1452. doi: 10.1038/s41591-019-0559-3. [DOI] [PubMed] [Google Scholar]
- [23].Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD, Lewis JD, Bushman FD, Archaea and Fungi of the Human Gut Microbiome: Correlations with Diet and Bacterial Residents, PLOS ONE. 8 (2013) e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Richard ML, Sokol H, The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases, Nat. Rev. Gastroenterol. Hepatol 16 (2019) 331. doi: 10.1038/s41575-019-0121-2. [DOI] [PubMed] [Google Scholar]
- [25].Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, NIH Intramural Sequencing Center Comparative Sequencing Program, Becker J, Benjamin B, Blakesley R, Bouffard G, Brooks S, Coleman H, Dekhtyar M, Gregory M, Guan X, Gupta J, Han J, Hargrove A, Ho S, Johnson T, Legaspi R, Lovett S, Maduro Q, Masiello C, Maskeri B, McDowell J, Montemayor C, Mullikin J, Park M, Riebow N, Schandler K, Schmidt B, Sison C, Stantripop M, Thomas J, Thomas P, Vemulapalli M, Young A, Kong HH, Segre JA, Topographic diversity of fungal and bacterial communities in human skin, Nature. 498 (2013) 367–370. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Auchtung TA, Fofanova TY, Stewart CJ, Nash AK, Wong MC, Gesell JR, Auchtung JM, Ajami NJ, Petrosino JF, Investigating Colonization of the Healthy Adult Gastrointestinal Tract by Fungi, MSphere. 3 (2018) e00092–18. doi: 10.1128/mSphere.00092-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ, Diet rapidly and reproducibly alters the human gut microbiome, Nature. 505 (2014) 559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Boix-Amorós A, Martinez-Costa C, Querol A, Collado MC, Mira A, Multiple Approaches Detect the Presence of Fungi in Human Breastmilk Samples from Healthy Mothers, Sci. Rep 7 (2017) 13016. doi: 10.1038/s41598-017-13270-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Boix-Amorós A, Puente-Sánchez F, du Toit E, Linderborg KM, Zhang Y, Yang B, Salminen S, Isolauri E, Tamames J, Mira A, Collado MC, Mycobiome Profiles in Breast Milk from Healthy Women Depend on Mode of Delivery, Geographic Location, and Interaction with Bacteria, Appl. Environ. Microbiol 85 (2019) e02994–18. doi: 10.1128/AEM.02994-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, Panzer AR, LaMere B, Rackaityte E, Lukacs NW, Wegienka G, Boushey HA, Ownby DR, Zoratti EM, Levin AM, Johnson CC, Lynch SV, Neonatal gut microbiota associates with childhood multi–sensitized atopy and T–cell differentiation, Nat. Med 22 (2016) 1187–1191. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bacher P, Hohnstein T, Beerbaum E, Röcker M, Blango MG, Kaufmann S, Röhmel J, Eschenhagen P, Grehn C, Seidel K, Rickerts V, Lozza L, Stervbo U, Nienen M, Babel N, Milleck J, Assenmacher M, Cornely OA, Ziegler M, Wisplinghoff H, Heine G, Worm M, Siegmund B, Maul J, Creutz P, Tabeling C, Ruwwe-Glösenkamp C, Sander LE, Knosalla C, Brunke S, Hube B, Kniemeyer O, Brakhage AA, Schwarz C, Scheffold A, Human Anti-fungal Th17 Immunity and Pathology Rely on Cross-Reactivity against Candida albicans, Cell. 176 (2019) 1340–1355.e15. doi: 10.1016/j.cell.2019.01.041. [DOI] [PubMed] [Google Scholar]; **Authors used in vitro priming of human T cells with fungal lysates to show a novel cross-reactivity between the sporulating fungus A. fumigatus and the common intestinal opportunistic pathogen C. albicans. Cross reactive cells for C. albicans uniquely expressed high amounts of IL-17A in Crohn’s disease and asthmatic patients and these Th17 cells were selectively expanded in patients with aspergillosis.
- [32].Witchley JN, Penumetcha P, Abon NV, Woolford CA, Mitchell AP, Noble SM, Candida albicans Morphogenesis Programs Control the Balance between Gut Commensalism and Invasive Infection, Cell Host Microbe. 25 (2019) 432–443.e6. doi: 10.1016/j.chom.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Researchers utilized a homozygous gene deletion library of C. albicans to identify several transcription factors that attenuate competitive fitness in the gastrointestinal tract. Genetic ablation of a downstream hyphal programming regulator, UME6, reproduced this competitive edge through reduction of the proteins SAP6 and HYR1.
- [33].Plato A, Hardison SE, Brown GD, Pattern recognition receptors in antifungal immunity, Semin. Immunopathol 37 (2015) 97–106. doi: 10.1007/s00281-014-0462-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hernández-Santos N, Gaffen SL, Th17 Cells in Immunity to Candida albicans, Cell Host Microbe. 11 (2012) 425–435. doi: 10.1016/j.chom.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, Ishikawa E, Shima T, Hara T, Kado S, Jinnohara T, Ohno H, Kondo T, Toyooka K, Watanabe E, Yokoyama S, Tokoro S, Mori H, Noguchi Y, Morita H, Ivanov II, Sugiyama T, Nuñez G, Camp JG, Hattori M, Umesaki Y, Honda K, Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells, Cell. 163 (2015) 367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Doron I, Leonardi I, Iliev ID, Profound mycobiome differences between segregated mouse colonies do not influence Th17 responses to a newly introduced gut fungal commensal, Fungal Genet. Biol. FG B 127 (2019) 45–49. doi: 10.1016/j.fgb.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Shao T-Y, Ang WXG, Jiang TT, Huang FS, Andersen H, Kinder JM, Pham G, Burg AR, Ruff B, Gonzalez T, Hershey GKK, Haslam DB, Way SS, Commensal Candida albicans Positively Calibrates Systemic Th17 Immunological Responses, Cell Host Microbe. 25 (2019) 404–417.e6. doi: 10.1016/j.chom.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This study showed that Th17 cells and neutrophils induced by Candida colonization in the gut can be found systemically and offer protection against fungal and bacterial infection while aggravating house dust mite-induced lung inflammation.
- [38].Wheeler ML, Limon JJ, Bar AS, Leal CA, Gargus M, Tang J, Brown J, Funari VA, Wang HL, Crother TR, Arditi M, Underhill DM, Iliev ID, Immunological Consequences of Intestinal Fungal Dysbiosis, Cell Host Microbe. 19 (2016) 865–873. doi: 10.1016/j.chom.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Limon JJ, Tang J, Li D, Wolf AJ, Michelsen KS, Funari V, Gargus M, Nguyen C, Sharma P, Maymi VI, Iliev ID, Skalski JH, Brown J, Landers C, Borneman J, Braun J, Targan SR, McGovern DPB, Underhill DM, Malassezia Is Associated with Crohn’s Disease and Exacerbates Colitis in Mouse Models, Cell Host Microbe. 25 (2019) 377–388.e6. doi: 10.1016/j.chom.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Authors reported increased Malassezia DNA in the gut of a cohert of CD patients. M. restricta was further shown to exacerbate chemical colitis in mice in a CARD9-dependent fashion.
- [40].Pierce JV, Kumamoto CA, Variation in Candida albicans EFG1 Expression Enables Host-Dependent Changes in Colonizing Fungal Populations, MBio. 3 (2012) e00117–12. doi: 10.1128/mBio.00117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pierce JV, Dignard D, Whiteway M, Kumamoto CA, Normal Adaptation of Candida albicans to the Murine Gastrointestinal Tract Requires Efg1p-Dependent Regulation of Metabolic and Host Defense Genes, Eukaryot. Cell 12 (2013) 37–49. doi: 10.1128/EC.00236-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Pande K, Chen C, Noble SM, Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism, Nat. Genet 45 (2013) 1088–1091. doi: 10.1038/ng.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pietrella D, Pandey N, Gabrielli E, Pericolini E, Perito S, Kasper L, Bistoni F, Cassone A, Hube B, Vecchiarelli A, Secreted aspartic proteases of Candida albicans activate the NLRP3 inflammasome, Eur. J. Immunol 43 (2013) 679–692. doi: 10.1002/eji.201242691. [DOI] [PubMed] [Google Scholar]
- [44].Trunk K, Peltier J, Liu Y-C, Dill BD, Walker L, Gow NAR, Stark MJR, Quinn J, Strahl H, Trost M, Coulthurst SJ, The type VI secretion system deploys antifungal effectors against microbial competitors, Nat. Microbiol 3 (2018) 920. doi: 10.1038/s41564-018-0191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Authors report unique antifungal effector proteins secreted by Serratia marcescens with activity against various candida species and S. cerevisiae. Although two of the secreted proteins both result in fungal cell death, they act through disparate mechanisms and pathogenic morphologies.
- [45].Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, Neut C, Vermeire S, Clemente J, Colombel JF, Fujioka H, Poulain D, Sendid B, Ghannoum MA, Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn’s Disease, MBio. 7 (2016) e01250–16. doi: 10.1128/mBio.01250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Passmore IJ, Letertre MPM, Preston MD, Bianconi I, Harrison MA, Nasher F, Kaur H, Hong HA, Baines SD, Cutting SM, Swann JR, Wren BW, Dawson LF, Para-cresol production by Clostridium difficile affects microbial diversity and membrane integrity of Gram-negative bacteria, PLOS Pathog. 14 (2018) e1007191. doi: 10.1371/journal.ppat.1007191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].van Leeuwen PT, van der Peet JM, Bikker FJ, Hoogenkamp MA, Paiva AMO, Kostidis S, Mayboroda OA, Smits WK, Krom BP, Interspecies Interactions between Clostridium difficile and Candida albicans, MSphere. 1 (2016) e00187–16. doi: 10.1128/mSphere.00187-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Fox EP, Cowley ES, Nobile CJ, Hartooni N, Newman DK, Johnson AD, Anaerobic Bacteria Grow within Candida albicans Biofilms and Induce Biofilm Formation in Suspension Cultures, Curr. Biol 24 (2014) 2411–2416. doi: 10.1016/j.cub.2014.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zuo T, Wong SH, Cheung CP, Lam K, Lui R, Cheung K, Zhang F, Tang W, Ching JYL, Wu JCY, Chan PKS, Sung JJY, Yu J, Chan FKL, Ng SC, Gut fungal dysbiosis correlates with reduced efficacy of fecal microbiota transplantation in Clostridium difficile infection, Nat. Commun 9 (2018) 3663. doi: 10.1038/s41467-018-06103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Authors reveal a loss in fungal diversity and an increase in C. albicans abundance associated with C. difficile infection. FMT treatment responders resulted in increased fungal diversity and a decrease C. albicans abundance with a corresponding increase in Saccharomyces spp. Furthermore, a mouse model of CDI and FMT treatment implicated C. albicans as a causative factor in treatment response failure, reducing FMT efficacy and increasing C. difficile abundance.
- [50].García C, Tebbji F, Daigneault M, Liu N-N, Köhler JR, Allen-Vercoe E, Sellam A, The Human Gut Microbial Metabolome Modulates Fungal Growth via the TOR Signaling Pathway, MSphere. 2 (2017) e00555–17. doi: 10.1128/mSphere.00555-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ballou ER, Avelar GM, Childers DS, Mackie J, Bain JM, Wagener J, Kastora SL, Panea MD, Hardison SE, Walker LA, Erwig LP, Munro CA, Gow NAR, Brown GD, MacCallum DM, Brown AJP, Lactate signalling regulates fungal β-glucan masking and immune evasion, Nat. Microbiol 2 (2017) 16238. doi: 10.1038/nmicrobiol.2016.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Mayer FL, Kronstad JW, Disarming Fungal Pathogens: Bacillus safensis Inhibits Virulence Factor Production and Biofilm Formation by Cryptococcus neoformans and Candida albicans, MBio. 8 (2017) e01537–17. doi: 10.1128/mBio.01537-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Temple MJ, Cuskin F, Baslé A, Hickey N, Speciale G, Williams SJ, Gilbert HJ, Lowe EC, A Bacteroidetes locus dedicated to fungal 1,6-β-glucan degradation: Unique substrate conformation drives specificity of the key endo-1,6-β-glucanase, J. Biol. Chem 292 (2017) 10639–10650. doi: 10.1074/jbc.M117.787606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron EA, Pudlo NA, Porter NT, Urs K, Thompson AJ, Cartmell A, Rogowski A, Hamilton BS, Chen R, Tolbert TJ, Piens K, Bracke D, Vervecken W, Hakki Z, Speciale G, Munōz-Munōz JL, Day A, Peña MJ, McLean R, Suits MD, Boraston AB, Atherly T, Ziemer CJ, Williams SJ, Davies GJ, Abbott DW, Martens EC, Gilbert HJ, Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism, Nature. 517 (2015) 165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Graus MS, Wester MJ, Lowman DW, Williams DL, Kruppa MD, Martinez CM, Young JM, Pappas HC, Lidke KA, Neumann AK, Mannan Molecular Substructures Control Nanoscale Glucan Exposure in Candida, Cell Rep. 24 (2018) 2432–2442.e5. doi: 10.1016/j.celrep.2018.07.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Shibata N, Ikuta K, Imai T, Satoh Y, Satoh R, Suzuki A, Kojima C, Kobayashi H, Hisamichi K, Suzuki S, Existence of Branched Side Chains in the Cell Wall Mannan of Pathogenic Yeast, Candida albicans Structure- antigenicity relationship between the cell wall mannans of Candida albicans and Candida parapsilosis, J. Biol. Chem 270 (1995) 1113–1122. doi: 10.1074/jbc.270.3.1113. [DOI] [PubMed] [Google Scholar]