Abstract
Human modification of the environment, particularly through land-use change, often reduces animal species diversity. However, the effect of land-use change on the gut microbiome of wildlife in human-dominated landscapes is not well understood despite its potential consequences for host health. We sought to quantify the effect of land-use change on wild bird gut microbiomes in a countryside landscape in Costa Rica, comprising a range of habitat types, ranging from primary and secondary forests to diversified and monoculture farms. We collected 280 fresh fecal samples from individuals belonging to six common species of saltator, thrushes, and warblers at 24 sites across this land-use gradient. Through 16S rRNA community profiling, we found that bacterial species composition responded to host species identity more strongly than to habitat type. In addition, we found evidence that habitat type affected microbial composition only for two of the six bird species. Our findings indicate that some host species and their microbiota may be more vulnerable to human disturbances than others.
Subject terms: Biodiversity, Microbial ecology
Introduction
To understand the structure and function of the gut microbiome, it is necessary to consider factors beyond the host to include environmental conditions [1–4]. Consideration of the host’s habitat may be especially informative in environments undergoing major anthropogenic changes. Human influences, such as land-use change, alter food sources, and environmental microbiota, a potential source of gut microbial species [5, 6]. Recent studies on this topic, on the black howler monkey (Alouatta pigra) and the rice frog (Fejervarya limnocharis), suggest that land-use change does alter the structure of gut microbial communities through changes in food sources [7, 8]. However, it remains unknown if this effect is widespread across host species or specific to certain hosts. To address this question, multiple host species should be examined simultaneously in a comparable fashion within the same landscape, but few studies have taken this approach.
We sought to quantify changes in the gut microbiomes of multiple bird species across a land-use gradient in Costa Rica. We characterized the effects of different habitat types, ranging from primary forests to coffee monocultures, on the microbial communities of six bird species: Saltator maximus (Buff-throated Saltator), Turdus grayi (Clay-colored Thrush), Catharus aurantiirostris (Orange-billed Nightingale-Thrush), Catharus ustulatus (Swainson’s Thrush), Basileuterus rufifrons (Rufous-capped Warbler), and Dendroica petechia (Yellow Warbler). We focused on these species because all were abundant with high capture rates and present in more than one habitat type based on the long-term bird banding data in this study system. We tested the hypotheses that host species identity and habitat type affected gut microbial communities and that the effect of habitat type on microbiota varied among host species.
Methods and results
To quantify changes in the gut microbiome, we sequenced the V4 region of bacterial 16S rRNA gene of 280 fecal samples (Supplementary Table 1). Samples were collected in January–April 2016 across six habitat types with different degrees of land-use intensity, varying in tree cover and crop diversity within a long-term study system that spans ~43 km2 [9] (Supplementary Table 2). These land cover types include: (1) primary forests, (2) secondary forests, (3) riparian corridors, (4) forest fragments, (5) diversified agriculture, and (6) monoculture (see Supplementary Information).
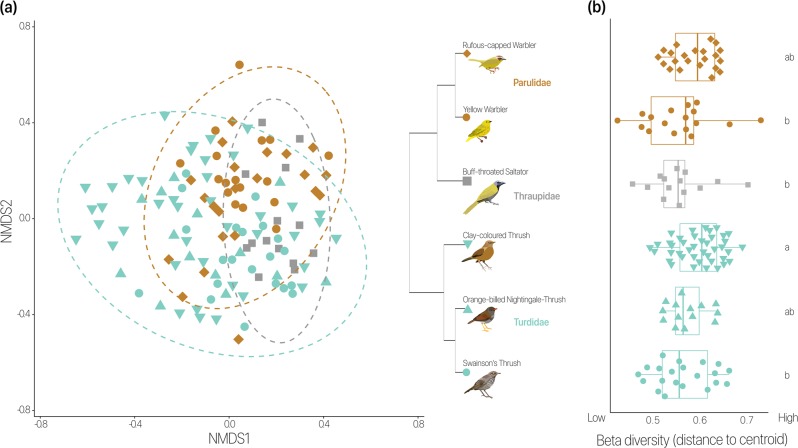
To determine whether host species identity, habitat type, and their interaction explained microbiome composition, we used PERMANOVA, which revealed that host species identity was a significant predictor (r2 = 0.09, p = 0.001), whereas habitat type was a weaker predictor (r2 = 0.04, p = 0.09) (Supplementary Table 3). The interaction of host species identity and habitat had a stronger relationship with gut microbial composition than either host species or habitat alone (r2 = 0.12, p = 0.049). In NMDS analysis, gut microbial assemblages clustered by host family and species (Fig. 1a). Microbial composition among individuals overlapped across most host species at the phylum level (Fig. S1), but these microbial communities still clustered by host species (Fig. S2).
Fig. 1.
Microbial composition shows some differences among host species despite high conspecific variation. a NMDS patterns show host specificity. There is overlap between individuals from different bird species, yet some significant clustering at bird family and species levels. Color indicates bird family, while shape represents species. Ellipses denote 95% confidence level. b Beta diversity varies between bird species. Distance to centroid was calculated using a multivariate version of the Levene’s test. Lower values indicate more shared bacterial taxa among individuals of the same bird species. Higher values show higher bacterial taxa variability among individuals of the same host species. Color and shape are as in a. Labels indicate significant differences between bird species
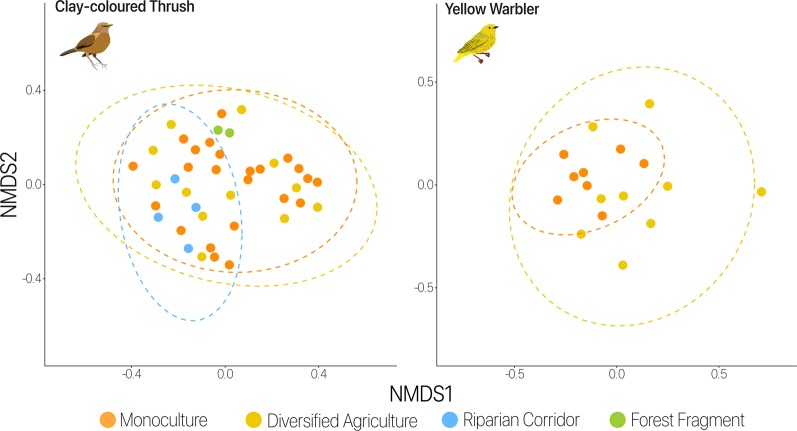
Habitat was a significant predictor of microbial communities in just two species, the Clay-colored Thrush and Yellow Warbler (Fig. 2 and Supplementary Table 4). Among the clay-colored thrush individuals, forest fragment samples clustered together, as well as riparian corridor samples, though with a small sample size in each habitat type (r2 = 0.10, p = 0.023). Among the Yellow Warbler individuals, monoculture samples formed a significant cluster (r2 = 0.11, p = 0.003), distinguishing that habitat type from diversified agriculture.
Fig. 2.
Habitat type influences microbial composition. PERMANOVA results for individual bird species show habitat as a predictor of microbial variance for the Clay-colored Thrush (r2 = 0.1, p = 0.023) and Yellow Warbler (r2 = 0.11, p = 0.003), as illustrated in the NMDS plot. Each color corresponds to a particular habitat type. Ellipses are at 95% confidence level
To describe the variation in microbial composition within and among conspecific individuals and determine whether certain bird species have higher variance than others, we calculated alpha and beta diversity. We found no significant effect of host species identity (Fig. S3, p = 0.802) or habitat type (Fig. S4, p = 0.498) on alpha diversity. Reporting the distance to centroid as an index of beta diversity, we found that conspecific differences in gut microbial community composition were lowest in the Buff-throated Saltator and highest in the Clay-colored Thrush (Fig. 1b, p = 0.042). Within-species variation in gut microbial composition in the Clay-colored Thrush was significantly higher than in the Yellow Warbler (p = 0.015), Buff-throated Saltator (p = 0.017), and Swainson’s Thrush (p = 0.026) (Fig. 1b).
Discussion
Our results indicate that the effect of land-use change on avian gut microbiomes can depend on both host species identity and habitat type, although there was considerable overlap across host species and habitat type (Fig. 1). Causes of these significant patterns remain unclear, but one speculation is that host genetics have selected for certain communities [10, 11]. Similarities across bird species might reflect commonalities in diet and habitat use, which would expose them to similar microbes. All six species consume invertebrates and fruits. The Orange-billed Nightingale-Thrush and Swainson’s Thrush also eat seeds, whereas the Clay-colored Thrush includes reptiles in its diet (Supplementary Table 1). Another trait to consider is migratory status. The Swainson’s Thrush and Yellow Warbler are latitudinal migrants, which may expose them to different microbes during stopover events [12, 13]. Moreover, there are resident and migratory individuals of the Swainson’s Thrush with similar microbiomes, suggesting that bacterial composition is conserved within host species or that their compositions are converging to look like their resident counterparts. Alternatively, the gut microbiomes of these species may be resistant to external influences, like habitat type, shown in the case of the New Zealand kākāpō (Strigops habroptilus) [14].
We found a marginally significant effect of habitat type on microbial composition. We suspected that bird species found in more than one habitat type might modify their diets due to changes in plant and animal composition associated with land-use change [7, 8]. It remains uncertain to what extent individual birds were using multiple habitat types. Regardless of the actual extent of individual movements across habitat types, our data indicated high beta diversity in the Clay-colored Thrush, meaning there were fewer shared microbial taxa across individuals of this species. It is a generalist that is estimated to consume, on average, 60% fruits, 30% invertebrates, and 10% reptiles [15, 16], but this ratio may change spatially, from one habitat to another, or seasonally. Although not studied in birds, seasonal diet shifts seem to be accompanied by microbial compositional changes in other vertebrates, including wild wood mice (Apodemus sylvaticus), black howler monkeys (Alouatta pigra), and the American bison (Bison bison) [17–19].
The focal birds co-occur in some habitat types, which may explain why their microbiomes are similar, shaped via exposure to similar microbes in diet, environment, and their interactions with other bird species. In addition to being in the same family, the Rufous-capped Warbler and Yellow Warbler occupy similar habitats, such as secondary forests and agricultural areas [20]. They also belong to a similar feeding guild where the former eats 80% invertebrates and 20% fruit and the latter consumes mostly invertebrates and some fruit [15, 16]. Despite being a latitudinal migrant, the Yellow Warbler shares similar bacterial taxa with the non-migratory Rufous-capped Warbler (Fig. 1a), suggesting that more closely related species are more likely to have similar microbiomes.
The significant interactive effect of host species and habitat type indicates that some species may be affected more by land-use change. Both the Clay-colored Thrush and Yellow Warbler gut microbiomes appear to have shifted with habitat. Consequences of these patterns for host health remain largely unknown. To understand the relationship between land-use change and avian gut microbiota further, additional data on diet, environmental microbiota, and microbial function across habitats could be informative. For example, diet information derived from the same fecal samples may explain patterns observed in microbial composition. Furthermore, functional profiling of the gut microbial communities through metagenomic or metatranscriptomic approaches would allow us to test whether the changes in species composition across hosts and habitat types had functional consequences [21]. Functional profiles, coupled with animal fitness data, could link these changes in gut microbial communities to their impact on host health.
Supplementary information
Acknowledgements
We thank Manpreet Dhami for molecular techniques, Po-Ju Ke for bioinformatics help, Andrew Letten for statistical advice, three anonymous reviewers for comments, and Henry Sandi Amador, Erick Barrantes, Jeisson Figueroa, Esteban Calderón Porras, and Dunia Villalobos for handling birds and collecting samples. This work was funded by the Moore Family Foundation and the Winslow Foundation through the Stanford Center for Conservation Biology, the Stanford University Terman Fellowship, and the Organization of Tropical Studies Thesis Research Fellowship.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-019-0535-4) contains supplementary material, which is available to authorized users.
References
- 1.Miller ET, Svanbäck R, Bohannan BJM. Microbiomes as metacommunities: understanding host-associated microbes through metacommunity ecology. Trends Ecol Evol. 2018;33:926–35. doi: 10.1016/j.tree.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Rothschild Daphna, Weissbrod Omer, Barkan Elad, Kurilshikov Alexander, Korem Tal, Zeevi David, Costea Paul I., Godneva Anastasia, Kalka Iris N., Bar Noam, Shilo Smadar, Lador Dar, Vila Arnau Vich, Zmora Niv, Pevsner-Fischer Meirav, Israeli David, Kosower Noa, Malka Gal, Wolf Bat Chen, Avnit-Sagi Tali, Lotan-Pompan Maya, Weinberger Adina, Halpern Zamir, Carmi Shai, Fu Jingyuan, Wijmenga Cisca, Zhernakova Alexandra, Elinav Eran, Segal Eran. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555(7695):210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 3.Ezenwa VO, Gerardo NM, Inouye DW, Medina M, Xavier JB. Animal behavior and the microbiome. Science. 2012;338:198–9. doi: 10.1126/science.1227412. [DOI] [PubMed] [Google Scholar]
- 4.Moeller AH, Foerster S, Wilson ML, Pusey AE, Hahn BH, Ochman H. Social behavior shapes the chimpanzee pan-microbiome. Sci Adv. 2016;2:e1500997. doi: 10.1126/sciadv.1500997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith CC, Snowberg LK, Gregory Caporaso J, Knight R, Bolnick DI. Dietary input of microbes and host genetic variation shape among-population differences in stickleback gut microbiota. ISME J. 2015;9:2515–26. doi: 10.1038/ismej.2015.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnes AD, Allen K, Kreft H, Corre MD, Jochum M, Veldkamp E, et al. Direct and cascading impacts of tropical land-use change on multi-trophic biodiversity. Nat Ecol Evol. 2017;1:1511–19. doi: 10.1038/s41559-017-0275-7. [DOI] [PubMed] [Google Scholar]
- 7.Amato KR, Yeoman CJ, Kent A, Righini N, Carbonero F, Estrada A, et al. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 2013;716:1344–53. doi: 10.1038/ismej.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang C-W, Huang B-H, Lin S-M, Huang C-L, Liao P-C. Changes of diet and dominant intestinal microbes in farmland frogs. BMC Microbiol. 2016;16:1–13. doi: 10.1186/s12866-015-0617-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belisle M, Mendenhall CD, Oviedo Brenes F, Fukami T. Temporal variation in fungal communities associated with tropical hummingbirds and nectarivorous bats. Fungal Ecol. 2014;12:44–51. doi: 10.1016/j.funeco.2014.02.007. [DOI] [Google Scholar]
- 10.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–90. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 11.Goodrich Julia K., Davenport Emily R., Clark Andrew G., Ley Ruth E. The Relationship Between the Human Genome and Microbiome Comes into View. Annual Review of Genetics. 2017;51(1):413–433. doi: 10.1146/annurev-genet-110711-155532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis WB, Moore FR, Wang S. Characterization of the gut microbiota of migratory passerines during stopover along the northern coast of the Gulf of Mexico. J Avian Biol. 2016;47:659–68. doi: 10.1111/jav.00954. [DOI] [Google Scholar]
- 13.Wu Y, Yang Y, Cao L, Yin H, Xu M, Wang Z, et al. Habitat environments impacted the gut microbiome of long-distance migratory swan geese but central species conserved. Sci Rep. 2018;8:13314. doi: 10.1038/s41598-018-31731-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perry EK, Digby A, Taylor MW. The low-diversity fecal microbiota of the critically endangered kakapo is robust to anthropogenic dietary and geographic influences. Front Microbiol. 2017;8:1–13. [DOI] [PMC free article] [PubMed]
- 15.Wilman H, Belmaker J, Simpson J, de la Rosa C, Rivadeneira MM, Jetz W. Elton traits 1.0: species-level foraging attributes of the world’s birds and mammals. Ecology. 2014;95:2027. doi: 10.1890/13-1917.1. [DOI] [Google Scholar]
- 16.Foster MS. The potential of fruit trees to enhance converted habitats for migrating birds in southern Mexico. Bird Conserv Int. 2007;17:45–61. doi: 10.1017/S0959270906000554. [DOI] [Google Scholar]
- 17.Maurice CF, CL Knowles S, Ladau J, Pollard KS, Fenton A, Pedersen AB, et al. Marked seasonal variation in the wild mouse gut microbiota. ISME J. 2015;9:2423–34. doi: 10.1038/ismej.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amato KR, Leigh SR, Kent A, Mackie RI, Yeoman CJ, Stumpf RM, et al. The gut microbiota appears to compensate for seasonal diet variation in the wild black howler monkey (Alouatta pigra) Micro Ecol. 2014;69:434–43. doi: 10.1007/s00248-014-0554-7. [DOI] [PubMed] [Google Scholar]
- 19.Bergmann GT, Craine JM, Robeson MS, Fierer N. Seasonal shifts in diet and gut microbiota of the American bison (Bison bison) PLoS One. 2015;10:1–14. doi: 10.1371/journal.pone.0142409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stiles FG, Skutch AF. A guide to the birds of Costa Rica. Ithaca: Cornell University Press; 1989. [Google Scholar]
- 21.The Human Microbiome Project Consortium. Structure, function, and diversity of the healthy human microbiome. Nature. 2014;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.