SUMMARY
Influenza virus neuraminidase is a major target for small molecule antiviral drugs. Antibodies targeting the neuraminidase surface antigen could also inhibit virus entry and egress to provide host protection. However, our understanding of the nature and range of target epitopes is limited due to a lack of human antibody structures with influenza neuraminidase. Here, we describe crystal and cryo-EM structures of neuraminidases from human-infecting avian H7N9 viruses in complex with five human anti-N9 antibodies, systematically defining several antigenic sites and antibody epitope footprints. These antibodies fully or partially block the neuraminidase active site or bind to epitopes distant from the active site while still showing neuraminidase inhibition. The inhibition of antibodies to neuraminidases was further analyzed by glycan array and solution-based neuraminidase activity assays. Together, our structural studies provide insights into protection by anti-neuraminidase antibodies and templates for the development of NA-based influenza virus vaccines and therapeutics.
Keywords: H7N9 influenza virus, neuraminidase, antibody, neuraminidase-Fab structures, X-ray crystallography, cryo-electron microscopy, inhibition mechanism, glycan array
eTOC Blurb (In Brief)
Antibodies to influenza neuraminidase can provide host protection against influenza virus. Zhu et al. report crystal and cryo-EM structures of N9 neuraminidases from human-infecting H7N9 viruses in complex with five human anti-N9 antibodies, thereby systematically defining neuraminidase antibody epitopes, including the active site.
Graphical Abstract
INTRODUCTION
The two surface glycoproteins on influenza A and B viruses, hemagglutinin (HA) and neuraminidase (NA), have essential functions in the influenza life cycle. HA mediates binding of the virus to the host cell and then triggers the fusion of viral and host membranes in the endosome. NA cleaves off terminal sialic acid from glycans on the host cell and on the emerging virions, thereby enabling release of progeny viruses from the host cell. Current licensed influenza vaccines are selected based on their abilities to induce antibodies against HA, which is also the focus for development of more broadly protective vaccines (Berlanda Scorza et al., 2016). It has been recently appreciated that NA is also highly immunogenic and NA-specific antibodies likely contribute to immunity by limiting viral replication and disease severity (Eichelberger et al., 2018). However, NA has largely been ignored in the vaccine development process. NA might be contained in current influenza vaccines, but the NA quantity and quality vary and are not standardized (Chen et al., 2018; Krammer et al., 2018). There are therefore substantial knowledge gaps about NA immunity and its protective effects, such as the nature of the NA antigenic sites or epitopes, which are also important for understanding NA evolution and antigenic drift (Krammer et al., 2018).
To date, atomic structures of only four mouse antibodies in complex with influenza NAs have been reported for N9 (Malby et al., 1994; Tulip et al., 1992), N2 (Venkatramani et al., 2006) or N1 (Wan et al., 2015) by X-ray crystallography, and two mouse antibodies with influenza B virus NA by negative-stain electron microscopy (Wohlbold et al., 2017). Thus, there is only very limited knowledge on the antigenic sites on NA, especially for human antibodies.
Avian H7N9 viruses that emerged in China in 2013 have caused severe human infections. Since then, infection with the H7N9 viruses has resulted in a high mortality rate (at least 615 deaths, ~ 40%) of 1,567 laboratory-confirmed human cases (https://www.who.int/csr/don/05-september-2018-ah7n9-china/en/). H7N9 infection is generally considered an avian disease, as these viruses spread extensively among poultry and wild birds, but only occasionally infect humans (Gao et al., 2013). Rapid evolution of H7N9 viruses from low pathogenic influenza viruses to highly pathogenic strains also occurred in 2017 (Shi et al., 2018). Great efforts have therefore been made to develop vaccines against H7N9 viruses and have successfully reduced the H7N9 prevalence in poultry (Zeng et al., 2018).
Here, we report crystal and cryo-EM structures of NAs from H7N9 viruses in complex with five human anti-N9 antibodies, NA-22, NA-45, NA-63, NA-73 and NA-80, that were identified in one healthy subject experimentally vaccinated with a monovalent inactivated A/Shanghai/02/2013 (Sh2) H7N9 vaccine (Gilchuk et al., co-submitted). These antibodies inhibited NA activity in an enzyme-linked lectin assay (ELLA) with the glycoprotein fetuin as substrate and protected mice from H7N9 influenza virus infection (NA-63, which shared almost the same epitope as NA-80, was not tested for mouse protection) (Gilchuk et al., co-submitted). We further analyzed antibody inhibition of NA activity in a glycan array based assay using diversified sialosides and in a solution based assay using a fluorescent substrate 2’-(4-methylumbelliferyl)-α-D-N-acetylneuraminic acid (4-MU-NANA). These data define the epitopes of five human NA antibodies and elucidate the antibody inhibition mechanism, providing important insights for NA-based development of vaccines and therapeutics.
RESULTS
Ultra-high resolution crystal structure of a stabilized NA ectodomain from A/Shanghai/2/2013 (H7N9) using a single amino-acid mutation Y169aH
The NA ectodomain (residues 82 to 468, in N2 numbering) from a human-infecting H7N9 virus from A/Shanghai/2/2013 (Sh2) was overexpressed in a baculovirus expression system as previously described (Xu et al., 2008) (see STAR Methods). After thrombin cleavage to remove the tetramerization domain and His-tag, the purified Sh2 N9 NA protein was recovered in mostly monomeric form in our experiments. To stabilize the tetramer, we considered residues that might affect quaternary formation (Colacino et al., 1997) and searched for amino-acid differences between Hunan N9 NA that forms a stable tetramer and Sh2 N9 NA that is largely monomeric. Four differences were found (see below) and mutation of Y169aH in the center of the Sh2 N9 NA tetramer (Figure 1A) resulted in recovery of tetramers, even after thrombin cleavage.
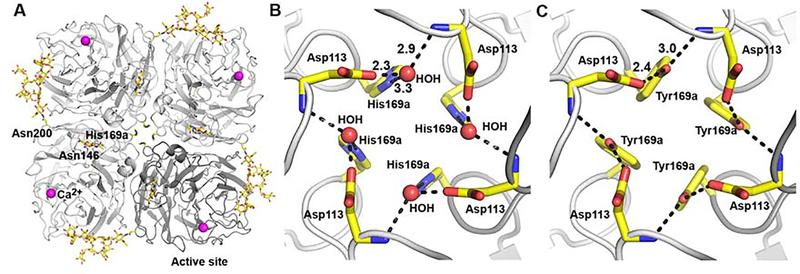
Figure 1. Crystal structure of Sh2 N9 stabilized ectodomain with single mutation Y169aH.
(A) The N9 tetramer as viewed from above the viral surface consists of four identical monomers in C4 symmetry. One monomer is colored in dark grey while other three monomers are colored in light grey. The active site is located on the membrane-distal surface (on top of the molecule). The two N-linked glycosylation sites in each monomer are shown with attached carbohydrates. Single calcium ions are shown as magenta spheres.
(B) A close-up view of the tetramer center of Sh2 N9 Y169aH showing the Y169aH mutation. The side-chain of His169a makes a hydrogen bond to a water molecule, which then hydrogen bonds to the side-chain of Asp113 and the main-chain nitrogen of Asp113 from an adjacent monomer. The hydrogen bonds are depicted with dotted lines with distances (in A) shown for one monomer, as also in (C).
(C) A close-up view of the tetramer center of wild-type Sh2 N9 NA (PDB code 5L14). The side-chain of Y169a makes hydrogen bonds directly to the side-chain of Asp113 and the main-chain nitrogen of Asp113 from adjacent monomer.
To elucidate the structural differences between mutant and wild-type Sh2 N9 structures, we determined the crystal structure of Sh2 N9 Y169aH ectodomain at 1.12 Å resolution (Figure 1A, Table S1). The mutant Sh2 N9 NA structure is nearly identical to that of wild-type Sh2 N9 NA (PDB code 5L14, 1.9 Å resolution) with a root-mean-square deviation (RMSD) of 0.12 Å for all Cα atoms. Electron density for oligosaccharides was observed for two out of three potential N-linked glycosylation sites Asn146 and Asn200 (but not Asn86) at which one and ten sugar residues could be modeled, respectively (Figures 1A). Our structure confirmed that the mutated His169a residues clustered near the four-fold axis (Figure 1B) similar to Tyr169a in the wild-type NA (Figure 1C). In the Y169aH mutant, the H169a side-chain hydrogen bonds to a water molecule, which in turn hydrogen bonds to the Asp113 side-chain in the same protomer and the Asp113 main-chain nitrogen atom from the neighboring protomer (Figure 1B). In contrast, in the wild-type Sh2 N9 NA, the Tyr169a side-chain makes a direct hydrogen bond with the Asp113 side-chain from the same protomer as well as the Asp113 main-chain nitrogen atom from the neighboring protomer to form a similar hydrogen bond network between the four protomers in center of the N9 tetramer (Figure 1C). This small change was sufficient to stabilize the N9 NA as a tetramer. We then used this stabilized NA tetramer in the crystallographic analyses with anti-N9 antibody.
Structures of N9 NA with human antibodies
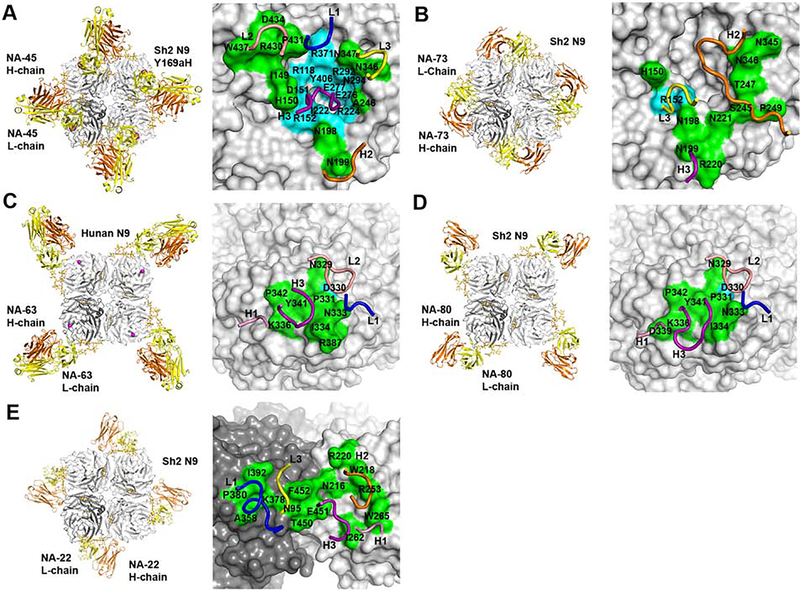
To elucidate the NA antigenic sites and how anti-NA antibodies protect against influenza virus infection, we determined structures of five human anti-N9 antibodies, NA-45, NA-73, NA-63, NA-80 and NA-22 in complexes with N9 NAs by X-ray crystallography and cryo-EM (Figure 2, Tables S1 and S2).
Figure 2. Structures of five anti-N9 antibody Fabs in complex with N9 NAs.
(A) Crystal structure of antibody NA-45 Fab in complex with the NA ectodomain tetramer of Sh2 N9 Y169aH at 2.3 Å resolution (left). One NA-Fab protomer is colored with NA in dark grey, Fab light chain (L-chain) in yellow and Fab heavy chain (H-chain) in orange. The other NA and Fab protomers are in light grey. N-linked glycans are shown in stick representation with yellow carbon atoms. The epitope of NA-45, with conserved NA residues in cyan and non-conserved NA residues in green, is mapped onto the N9 NA with the CDR loops involved in the interaction superimposed (right). CDR L1 is colored in blue, CDR L2 in salmon, CDR L3 in yellow, CDR H1 in pink, CDR H2 in orange and CDR H3 in purple. The same coloring scheme is used in (B), (C), (D), and (E).
(B) Cryo-EM structure of antibody NA-73 Fab in complex with ectodomain plus stalk tetramer of Sh2 N9 NA at 3.2 Å resolution (left). Only the Fv domain of the Fab was modeled. The epitope of NA-73 is mapped onto the N9 NA with the CDR loops involved in the interaction illustrated (right).
(C) Crystal structure of antibody NA-63 Fab in complex with ectodomain tetramer of Hunan N9 at 2.80 Å resolution (left). The epitope of NA-63 is mapped onto the N9 NA with the CDR loops involved in the interaction (right).
(D) Cryo-EM structure of antibody NA-80 Fab in complex with ectodomain plus stalk tetramer of Sh2 N9 at 3.6 Å resolution (left). Only the Fv domain of the Fab was modeled. The epitope of NA-80 is mapped onto the N9 NA with the CDR loops involved in the interaction (right).
(E) Cryo-EM structure of antibody NA-22 Fab in complex with ectodomain plus stalk of Sh2 N9 tetramer at 3.0 Å resolution (left). Only the Fv domain of the Fab was modeled. NA-22 interacts with two adjacent protomers of the N9 tetramer. The epitope of NA-22 is mapped onto the N9 NA with the CDR loops involved in the interaction (right).
See also Figures S1, S2, S3, S4, S5 and S6, and Tables S1, S2, S3 and S4.
Antibody NA-45 fully blocks the N9 active site with partial substrate mimicry
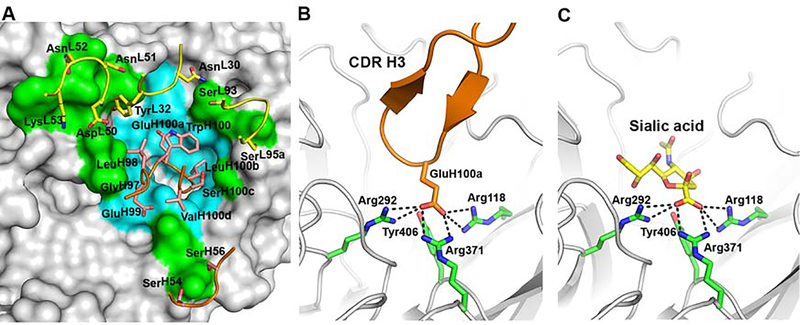
The crystal structure of NA-45 Fab with the Sh2 N9 Y169aH mutant was determined to 2.3 Å resolution (Table S1) with one NA-45 Fab per N9 NA protomer (Figure 2A). The Sh2 N9 NA mutant is nearly identical to its unliganded structure, with an RMSD of 0.18 Å for all Cα atoms. NA-45 Fab fully blocks the NA active site (Figures 2A and 3), which is unique among the structurally characterized NA antibodies (Malby et al., 1994; Tulip et al., 1992; Venkatramani et al., 2006; Wan et al., 2015). The conformational epitope consists of two main components: active site residues and residues surrounding the active site (Figures 2A and 3). NA-45 recognizes its epitope using both heavy and light chains. A total surface area of 810 Å2 is buried, of which 59% arises from the heavy chain and 41% from the light chain. NA is specifically recognized by light chain complementarity determining regions (CDR) L1, L2 and L3, and heavy chain CDRs H2 and H3.
Figure 3. Crystal structure of NA-45 Fab with Sh2 N9 Y169aH NA mutant.
(A) Zoomed-in molecular surface depicting the epitope on the NA with cyan for conserved NA active site residues and green for other non-conserved NA residues. The key interacting residues on the antibody are indicated on the top of the surface with labels. NA-45 binds to the NA active site using substrate mimicry.
(B) The carboxylate of GluH100a makes salt bridge interactions (black dashed lines) with key residues in the NA catalytic site.
(C) Sialic acid uses identical salt bridge interactions as the NA-45 Fab HCDR3 as shown in (B) consistent with a substrate mimicry mechanism.
See also Figures S1 and S2, and Tables S1 and S3.
Antibody NA-45 was shown to be an N9-specific antibody (Gilchuk et al., co-submitted). We therefore inspected the NA-45 epitope conservation across influenza A virus subtypes N1 to N9 and influenza B virus using nonredundant NA sequences in the National Center for Biotechnology Information Flu database (Bao et al., 2008) (Table S3). N10 and N11 were not included into the analysis in this study as they are of bat origin and have no known sialic acid cleavage activity (Karakus et al., 2019; Tong et al., 2013; Zhu et al., 2012). Within the NA-45 epitope (Figure 2A), 11 residues in the active site, which account about half of the total buried surface area, are >99% identical across all 10 NA subtypes, while the other 11 epitope residues on the rim of the NA active site are not conserved across different NA subtypes (Table S3). In NA-45, its 17-residue CDR H3 interacts with the conserved residues, while CDRs L1, L2, L3 and H2 largely make contacts with non-active site residues (Figures 2A, 3A and S1A).
NA-45 CDR H3 adopts a protruding conformation with a β-turn at its tip that inserts into the NA active site (Figure 3). The GluH100a carboxylate closely aligns with the carboxylate of sialic acid and uses the same salt-bridge interaction network with NA Arg118, Arg292, and Arg371, which are critical for NA enzymatic activity (Xu et al., 2008). To our knowledge, the NA-45 substrate mimicry is unique among all structurally characterized anti-NA antibodies. Very similar sialic acid receptor mimicry also has been reported in influenza anti-HA antibodies, such as 5J8 (Hong et al., 2013), F045–092 (Lee et al., 2014), CH65 (Whittle et al., 2011) and 641 I-9 among others (Schmidt et al., 2015). In these antibodies, the carboxylate side-chain of an Asp aligns with the carboxylate side-chain of sialic acid, and, in addition, a Pro/Leu/Val/Val hydrophobic side-chain prior to the Asp residue can be aligned with the acetamide of sialic acid, which is not the case for NA-45. This hydrophobic interaction is a general feature of all broad neutralizing antibodies to the HA receptor binding site (RBS) (Lee et al., 2014; Schmidt et al., 2015).
Antibody NA-73 partially blocks the NA active site
The cryo-EM structure of NA-73 Fab with Sh2 N9 NA stalk construct that included the ectodomain and stalk (residues 37 to 468) was determined to 3.2 Å resolution (Table S2). In this and the following cryo-EM structures, the N9 ectodomain and variable domain of the Fab were modeled, while the more flexible NA stalk and Fab constant domains were not modeled due to weak cryo-EM density in these regions.
One NA-73 Fab binds per NA protomer (Figure 2B), burying 490 Å2 of NA surface where 71% arises from interaction with the heavy chain. No large conformational changes are observed for the Sh2 N9 NA ectodomain in the NA-73 Fab complex as compared to its unliganded structure, with an RMSD of 0.47 Å for all Cα atoms. NA-73 binds to the NA active site, blocking approximately half of the entrance to the active site (Figure 2B). Three NA-73 CDR loops L3, H2 and H3 are involved in NA binding (Figures 2B and S1B), and its epitope includes one active site residue Arg152, which is conserved in influenza A and B NAs, and ten active site rim residues, which are not conserved (Table S4A). CDR L3 interacts with the active site Arg152 and active site rim His150, Asn198 and Asn221, CDR H2 with the rim Ser245, Thr247, Pro249, Asn345 and Asn346, while CDR H3 only contacts the rim Asn199 and Arg220 (Figure S1B).
Antibodies NA-63 and NA-80 bind to a similar linear epitope outside the active site but utilize heavy chains encoded by different germline genes
The crystal structure of NA-63 Fab with N9 NA from A/Hunan/02650/2016 (H7N9) (Hunan N9) was determined to 2.8 Å resolution (Table S1). This Hunan H7N9 strain was from wave 4 of H7N9 infections and is an H7N9 vaccine strain recommended by the World Health Organization (https://www.who.int/influenza/vaccines/virus/candidates_reagents/summary_a_h7n9_cvv_20170928.pdf). Four residues differ in the ectodomains between Sh2 N9 NA and Hunan N9 NA; Y169aH, S245P, I303V and N325S (Figure S2). The overall structure of the Hunan N9 NA ectodomain is nearly identical to Sh2 N9 NA in its apo form (no ligand bound) (PDB code 5L14) with an RMSD of 0.20 Å for all Cα atoms (Figure S2).
NA-63 binds Hunan N9 NA with one Fab for per NA protomer. Interestingly, the NA-63 epitope is focused mainly on a linear epitope from Asn329 to Pro342 with the only other interaction to Arg387 (Figure 2C), all of which are distant from the NA active site. The NA-63 footprint includes one conserved residue D330 and eight non-conserved NA residues (Table S4B). NA-63 recognizes its epitope using both heavy and light chains where 51% of the total surface buried surface (460 Å2) arises from heavy chain. The N9 NA interacts with CDRs L1, L2, H1, and H3 (Figures 2C and S3A).
We also determined a cryo-EM structure of NA-63 Fab with Sh2 N9 NA ectodomain plus stalk to 3.1 Å resolution (Table S2). No large conformational changes were found for Sh2 N9 NA ectodomain upon binding to NA-63 Fab as compared to its unliganded structure, with an RMSD of 0.37 Å for all Cα atoms. As expected, the NA-63 recognizes the same epitope in Sh2 N9 NA as in Hunan N9 NA (Figure S3B), since the four-residue differences between these strains are distant from the NA-63 binding site.
Antibody NA-80 binds a similar N9 epitope to that of NA-63 as seen from the cryo-EM structure of NA-80 Fab with Sh2 N9 NA ectodomain plus stalk at 3.6 Å resolution (Table S2, Figure 2D). One NA-80 Fab binds per NA protomer, burying 470 Å2 of NA surface where 54% arises from the heavy chain. The Sh2 N9 NA ectodomain is nearly identical to its unliganded structure, with an RMSD of 0.28 Å for all Cα atoms. NA-80 recognizes a linear epitope from Asn329 to Pro342, which is almost the same as that of NA-63 except for no interaction with Asp339 for NA-63 and with Arg387 for NA-80. These NA-80 footprint residues are not conserved except for Asp330 (Table S4C). The similar epitopes between these two antibodies were unexpected as the two antibodies are encoded by different heavy chain germline V-genes, with IGHV3–7 for NA-63 and IGVH1–18 for NA-80, although they share the same light chain germline V-gene IGK1–39 (Figure S4A). NA-80 also recognizes N9 NA with a similar angle of approach versus NA-63 (Figure S4B) using CDRs L1. L2, H1, and H3 (Figure 2D). The conformation of CDR L1, L2 and H1 are similar, with the same residues involved into binding to N9 NA: SerL31/ThrL31, TyrL32, TyrL49, AlaL50, SerL53, LeuL54, SerH31, TyrH32 (Figures S4C and S3A). However, the CDR H3 conformation differs with NA-63 using ThrH96, AlaH97, GluH99, and PheH100 (Figure S3A), while NA-80 interacts with IleH96, ProH97, Ala100 and AspH100b (Figure S4C).
The crystal structure of apo NA-80 Fab also was determined to 1.59 Å resolution (Table S1) and was used as the starting model for refinement into the cryo-EM density map of NA-80 with Sh2 N9 NA. The RMSD for the Cα atoms of NA-80 Fv domain in its apo and bound forms is 1.3 Å, and a significant conformational movement was observed for CDR H3 (residues 98–100a), where the Cα atoms shifted 6.0 to 8.7 Å upon N9 NA binding (Figure S4D).
Antibody NA-22 binds an epitope at the NA protomer interface
The cryo-EM structure of NA-22 Fab with Sh2 N9 NA ectodomain plus stalk was determined to 3.0 Å resolution (Table S2) with one NA-22 Fab binding to two adjacent protomers in the NA tetramer (four Fabs per NA tetramer) (Figure 2E). 660 Å2 of buried NA surface includes 380 Å2 from one monomer and 280 Å2 from the adjacent monomer, with 57% of the binding surface coming from the heavy chain. The first NA monomer accounts for all buried surface of the light chain and 29% buried surface of the heavy chain, while second NA monomer accounts for 71% buried surface of the heavy chain only. The Sh2 N9 NA ectodomain did not exhibit any major conformational changes upon binding to NA-22 Fab as compared to its unliganded structure, with an RMSD of 0.40 Å for all Cα atoms. A total of 14 non-conserved NA residues from different regions of two NA protomers outside of the active site participate in the NA-Fab contacts (Figure 2E, Table S4D). NA-22 CDRs L1 and L3 interact with one NA protomer, whereas CDRs H1 and H2 interact with the adjacent NA protomer; CDR H3 interacts with both NA protomers (Figures 2E and S5A). NA-22 CDR L1 interacts with Asn95, Thr93, Ala358, Lys378, Pro380 and Ile392, and CDR L3 with Ile392 and Phe452 of the same NA protomer (Figure S5A). Thr450, Glu451 and Phe452 from this NA protomer, and Asn216, Ile262, Leu263 and Trp265 from the adjacent protomer interact with the CDR H3 residues. NA-22 CDR H1 contacts Trp265 and CDR H2 interacts with Trp218, Arg220, Arg253, Tyr255 and Trp265, all from the second protomer. Through these antibody-NA interactions, NA-22 bridges between adjacent NA protomers. Interestingly, NA-22 also interacts with N-linked glycans from Asn200 of one Sh2 N9 NA protomer (Figure S5B). The glycan residues attached to Asn200 are displaced compared to the unliganded structure to avoid a clash with the NA-22 light chain.
The crystal structure of apo NA-22 Fab also was determined to 2.3 Å resolution (Table S1) and used as the starting model for refinement into the cryo-EM density map of NA-22 with Sh2 N9 NA. The RMSD for Cα atoms of NA-22 Fv domain in apo and bound forms is 0.4 Å, with no major conformational changes observed upon N9 NA binding (Figure S5C).
Antibody inhibition of NA activity from glycan array analysis and a solution-based assay with substrate 4-MU-NANA
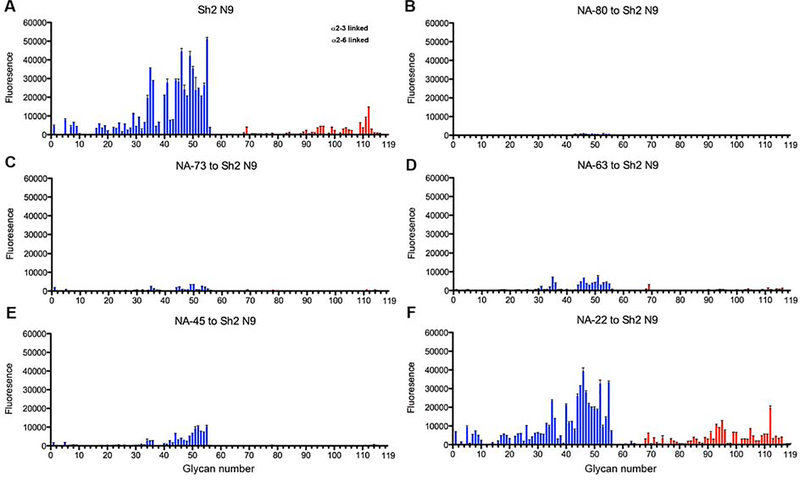
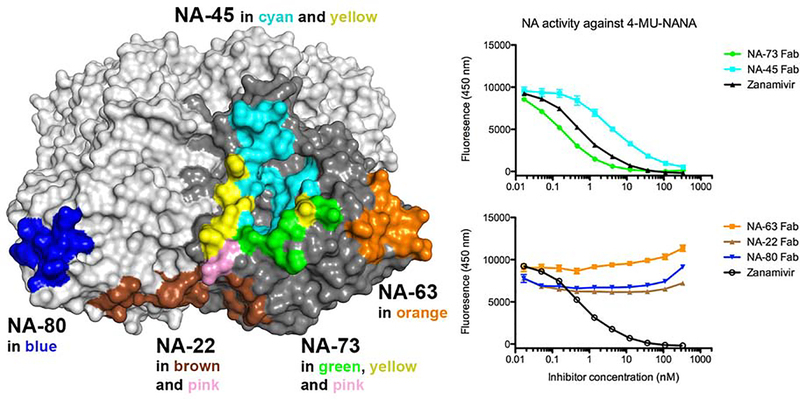
In the fetuin-based ELLA NA activity assay, NA-80, NA-73, NA-63, NA-45 and NA-22, all exhibited strong inhibition to the NA activity (Gilchuk et al., co-submitted). To assess substrate specificity, we previously developed a method to screen NA antibodies for inhibition of NA activity against diverse sialoside sequences on a glycan microarray NA cleavage analysis (Peng et al., 2017; Xu et al., 2012). Briefly, we incubated the Sh2 N9 NA ectodomain plus stalk with antibody IgG using a molar ratio of 1:4 for Sh2 N9 protomer to antibody IgG for one hour at room temperature. The antibody-NA mixture was then incubated with the sialoside glycan array (Data S1). NA digestion of the terminal sialic acid from the imprinted glycans was observed using a lectin to detect the newly exposed terminal galactose (Figure 4). Sh2 N9 NA exhibited a preference to cleave α2–3-linked sialosides over α2–6 sialosides, as observed for other NAs, such as the 1918 or 2009 pandemic H1N1 NAs (Xu et al., 2012). Antibodies NA-80 and NA-73 almost completely inhibited NA activity against α2–3 and α2–6-linked sialosides, while NA-63 and NA-45 showed substantial but not complete inhibition. Interestingly, NA-22 showed little or no inhibition for hydrolysis of either α2–3 or α2–6-linked sialosides. The finding that NA-22 showed strong inhibition in the fetuin-based ELLA assay suggests that it causes steric hindrance to substrate glycans on fetuin, a large glycoprotein with a molecular weight of 49 kDa, but not to printed glycans on the array.
Figure 4. Antibody inhibition of NA enzymatic activity on a glycan microarray for Sh2 N9 NA.
Different categories of glycans on the array are highlighted in colors; the bars denote the fluorescence signal intensity, and the error bars indicate the standard errors for four replicates. The control (A) and inhibition of Sh2 N9 NA cleavage by antibody IgGs (B-F).
(A) Sh2 N9 NA positive control.
(B) NA-80
(C) NA-73
(D) NA-63
(E) NA-45
(F) NA-22.
Antibody inhibition of NA activity was further assessed by cleavage of a small substrate 4-MU-NANA (Figure 5). Antibody NA-45 and NA-73 exhibited strong inhibition to NA cleaving activity toward 4-MU-NANA similar to zanamivir, whereas NA-63, NA-22 and NA-80 did not show any inhibition. These data are consistent with NA-45 and NA-73 fully blocking the active site entrance (Figure 5). The other three antibodies NA-63, NA-22 and NA-80 bind to the epitope distant from the active site (Figure 5); although they can sterically prevent access to glycan substrates on fetuin in the ELLA assay and/or the glycan array (Figure 4), they do not inhibit NA from cleaving the small 4-MU-NANA substrate. This finding also confirms that binding by these three antibodies do not have an allosteric effect on the NA active site.
Figure 5. Antibody inhibition of NA enzymatic activity using a 4-MU-NANA based assay for Sh2 N9 NA.
Antibodies inhibit the NA activity of Sh2 N9 by blocking the access of 4-MU-NANA to the active site. The epitopes of five antibodies are mapped onto the N9 NA tetramer (with one protomer in dark grey) to show the relative location of the epitopes (left). NA-45 binds to the NA active site, and NA-73 binds to part of the active site, while NA-63, NA-80 and NA-22 bind to residues outside of the active site. Because NA-63 and NA-80 epitopes are nearly identical, they are shown on different protomers. Antibodies NA-45 and NA-73 inhibit the NA cleavage activity against 4-MU-NANA, while NA-63, NA-80 and NA-22 do not show detectable inhibition of the NA activity against 4-MU-NANA (right).
DISCUSSION
Influenza vaccines are the first line of defense for influenza prevention. To develop a more effective and broad vaccine, it is necessary to fill in the current knowledge gaps of the NA-based immunity for influenza virus protection (Krammer et al., 2018). Our understanding of NA epitopes targeted by human antibodies is surprisingly limited. In this study, we structurally characterized five human anti-N9 antibodies in complex with influenza NAs to atomic resolution, providing the structural basis for influenza vaccine development and immunotherapy. These antibodies target diversified N9 NA epitopes: active site (NA-45), active site rim (NA-73), linear epitope on NA tip (NA-63 and NA-80), and interface between adjacent NA monomers also outside of the active site (NA-22) (Figure 5).
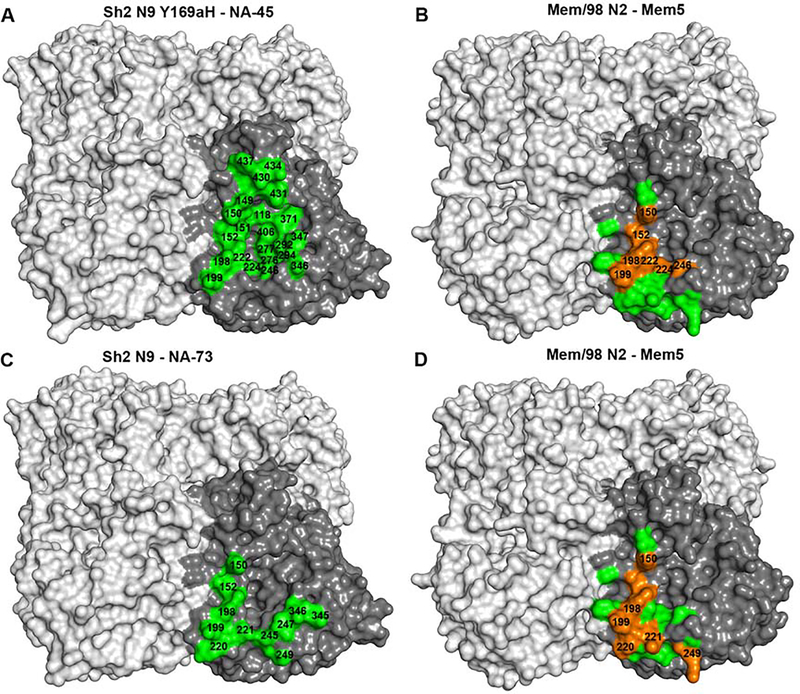
Thus far, only four atomic structures of mouse antibodies in complex with NAs have been elucidated: NC10, NC41, Mem5 and CD6 N9 (Malby et al., 1994; Tulip et al., 1992; Venkatramani et al., 2006; Wan et al., 2015). In comparison to these mouse antibodies, human NA-45 is unique in that it encompasses the whole NA active site, although its epitope does partially overlap with the Mem5 epitope on N2 NA on the active site rim (Figure 6). It is a significant finding that the recessed NA active site is also an antigenic site as it is highly conserved among influenza A and B viruses. It remains to be seen if it is possible to discover broadly reactive antibodies against the NA active site. The NA-73 epitope also partially overlaps with the Mem5 epitope (Figure 6). The NA-63 and NA-80 epitopes are similar, and their linear epitopes have not been previously described but partially overlap with NC-10 and NC-41 N9 epitopes, which are discontinuous (Figure S6). Interestingly, the NA-22 epitope on N9 NA corresponds well to the CD6 epitope on N1 NA (Figure S6).
Figure 6. Epitope comparison of human antibodies NA-45 and NA-73 with N9 NA versus mouse antibody Mem5 with N2 NA.
(A) The epitope of NA-45 is mapped onto Sh2 N9 Y169aH NA mutant (green).
(B) The epitope of Mem5 is mapped onto Mem/98 N2 NA (green and orange, PDB code 2AEP) with overlapping epitope residues with NA-45 shown in orange.
(C) The epitope of NA-73 is mapped onto Sh2 N9 NA (green).
(D) The epitope of Mem5 is mapped onto Mem/98 N2 NA (green and orange, PDB code 2AEP) with the overlapped epitope residues with NA-45 shown in orange.
Anti-NA antibodies function by inhibiting NA cleavage activity. Our results show that some antibodies bind to residues outside of the NA active site, such as NA-63, NA-80 and NA-22, but can inhibit NA activity against the large fetuin molecule but not against the small substrate 4-MA-NANA. This activity is associated with the capacity to block virus egress from infected cells (Gilchuk et al., co-submitted). These findings support a mechanism of blocking viral egress by steric hindrance of the NA enzyme site, which was also proposed with antibody CD6 (Wan et al., 2015). It is interesting that NA-22 showed weak or no obvious inhibition to N9 NA cleavage of sialic acid from glycans attached to flexible linkers on the microarray, while NA-63 and NA-80 strongly inhibited N9 NA activity in the same experiment, which may be due to their different epitopes and angles of approach. Antibodies NA-45 and NA-73 that fully or partially block the entrance of the NA active site pocket inhibit NA cleavage of both fetuin and glycans on the microarray or 4-MU-NANA cleavage.
Vaccines and antiviral drugs are key to control and eradicate viral pathogens, and the same is true for influenza virus. Here, the structures of human antibodies with NA provides insights to advance vaccine strategies, and epitope-based vaccine design. In addition, NA inhibitors, oseltamivir, zanamivir, peramivir, and laninamivir, and a recently approved polymerase acidic (PA) inhibitor baloxavir marboxil, are currently used to treat influenza virus infections. However, the emergence of antiviral drug resistance is a major concern for these NA inhibitors (Hussain et al., 2017) and even for the more recent PA inhibitor (Jones et al., 2018). Our results reveal sites of vulnerability on NA that may inform design of new drugs, such as therapeutic antibodies, small proteins, peptides or small molecules [as for the HA (Chevalier et al., 2017; Fleishman et al., 2011; Kadam et al., 2017; Laursen et al., 2018; Strauch et al., 2017; van Dongen et al., 2019)], to deal with NA-resistant viruses. Finally, the structures presented almost certainly represent only the tip of the iceberg in terms of the types of antibodies that target NA. We predict that just as has borne out with antibodies that recognize HA, there will be an explosion of discovery of NA antibodies in the near future, with a focused pursuit of the most potent and cross-reactive examples.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ian A. Wilson (wilson@scripps.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS Cell cultures
Sf9 cells (Spodoptera frugiperda ovarian cells, female) were maintained in HyClone insect cell culture medium. ExpiCHO cells (Chinese hamster ovary cells, female) were maintained according to the manufacturer’s instructions (Thermo Fisher Scientific).
METHOD DETAILS
Cloning, baculovirus expression and purification of neuraminidases for x-ray, cryo-EM and activity studies
The ectodomains of influenza Sh2 N9 and Hunan N9 NAs were expressed essentially as previously described (Xu et al., 2008). Briefly, the ectodomain (residues 83–470, 82–468 in N2 numbering) and ectodomain plus stalk region (residues 37–470, 37–468 in N2 numbering) of N9 NA from A/Shanghai/02/2013 (H7N9) (Sh2 N9, GenBank accession number KF021599) and from A/Hunan/02650/2016 (H7N9) (Hunan N9, GISAID accession number EPI961189) were expressed in a baculovirus system for structural and functional analyses. The cDNAs corresponding to the N9 ectodomain and the ectodomain plus stalk region of Sh2 N9 and Hunan N9 NAs were incorporated into a baculovirus transfer vector, pFastbacHT-A (Invitrogen) with an N-terminal gp67 signal peptide, thrombin cleavage site, and His6-tag (Xu et al., 2008). Once sequence verified, the plasmid was used for mutagenesis to engineer an Sh2 N9 NA construct with a single mutation Y169aH. The constructed plasmids were used to transform DH10bac competent bacterial cells by site-specific transposition (Tn-7 mediated) to form a recombinant Bacmid with beta-galactosidase blue-white receptor selection. The purified recombinant bacmids were used to transfect Sf9 insect cells for overexpression. NA protein was produced in suspension cultures of Sf9 cells with recombinant baculovirus at an MOI of 5–10 and incubated at 28°C shaking at 110 RPM. After 72 hours, Sf9 cells were removed by centrifugation and supernatants containing secreted, soluble NA protein was concentrated and buffer-exchanged into 20 mM Tris pH 8.0, 150 mM NaCl, and then further purified by metal affinity chromatography using Ni-nitrilotriacetic acid (NTA) resin (Qiagen). For crystal structure determination, the His6-tag was cleaved from the N9 NA ectodomain with thrombin and purified further by size exclusion chromatography on a Hiload 16/90 Superdex 200 column (GE healthcare) in 20 mM Tris pH 8.0, 150 mM NaCl and 0.02% NaN3. For cryo-EM and for NA glycan array-based or solution-based activity assays, the ectodomain plus stalk region of the Sh2 N9 NA with its His6-tag attached was concentrated after Ni-NTA purification, in 20 mM Tris pH 8.0, 150 mM NaCl and 0.02% NaN3. The purified NAs were quantified by optical absorbance at 280 nm, and purity and integrity were analyzed by reducing and nonreducing SDS-PAGE.
Expression of recombinant antibody Fabs in insect and mammalian cells
The light and heavy chain variable regions of human antibodies NA-73 were cloned into the vector pFastbac-dual containing the corresponding lambda CL region, and the CH1 region of human IgG1 appended to His6-tags, respectively. The NA-73 Fab was expressed in Sf9 insect cells using the same method as above for neuraminidase expression.
The light and heavy chain variable regions of human antibodies NA-45, NA-63, NA-80 and NA-22 were cloned into the vector phCMV containing the corresponding lambda or kappa CL region, and the CH1 region of human IgG1 appended to His6-tags, respectively. The Fabs were expressed by transient co-transfection of the expression vectors containing heavy chain and light chain into ExpiCHO cells (Thermo Fisher Scientific) at 37 °C with 8% CO2. Recombinant Fabs were purified from culture supernatant using metal affinity chromatography using Ni-nitrilotriacetic acid (NTA) resin (Qiagen) followed by size exclusion chromatography using a Superdex 200 column (GE Healthcare). Purified Fabs in 20 mM Tris pH 8.0, 150 mM NaCl and 0.02% NaN3 were quantified by optical absorbance at 280 nm, and purity and integrity were analyzed by reducing and nonreducing SDS-PAGE.
Crystal structure determination
Crystallization experiments were set up using the sitting drop vapor diffusion method using automated robotic crystal screening on our custom-designed CrystalMation system (Rigaku). Diffraction quality crystals for the Sh2 N9 Y169aH NA ectodomain tetramer were obtained by mixing 0.1 μL of the NA protein at 6.9 mg/ml in 20 mM Tris pH 8.0, 150 mM NaCI and 0.02% (v/v) NaN3 with 0.1 μL of the well solution in 0.1 M MES pH 6.5 and 12% (w/v) polyethylene glycol 20,000 at 20°C. The Sh2 N9 Y169aH NA crystals were cryoprotected in mother liquor with addition of 15% (w/v) ethylene glycol before being flash-cooled at 100 K. The complex of Sh2 N9 Y169aH NA and NA-45 Fab at 6.0 mg/mL was crystallized in 0.1 M Hepes, pH 7.0 and 10% (w/v) polyethylene glycol 6000 at 20°C. The complex crystals were cryoprotected in mother liquor with addition of 15% (v/v) ethylene glycol before being flash-cooled at 100 K. The complex of Hunan N9 NA and NA-63 Fab at 5.7 mg/mL was crystallized in 0.1 M Tris, pH 9.0, 10% (v/v) glycerol, and 10% (w/v) polyethylene glycol 6000 at 20°C. The complex crystals were flashed-cooled at 100 K without adding additional cryo-protectant. NA-80 Fab at 10.0 mg/ml was crystallized in 0.1 M sodium citrate, pH 5.6, 40% (w/v) polyethylene glycol 600 at 20°C. The NA-80 Fab crystals were flashed-cooled at 100 K without additional cryo-protectant. NA-22 Fab at 12.0 mg/mL was crystallized in 0.1 M Tris, pH 8.5, 40% (v/v) ethanol and 0.02% (w/v) MgCl2 at 4°C. The NA-22 Fab crystals were flashed-cooled at 100 K without additional cryo-protectant.
Diffraction data were collected at synchrotron beamlines (Table S1). Data for all crystals were integrated and scaled with HKL2000 (Otwinowski and Minor, 1997). Data collection statistics are summarized in Table S1.
All of the crystal structures were determined by molecular replacement (MR) using the program Phaser (McCoy et al., 2005). The Sh2 N9 Y169aH NA structure was determined using wild-type Sh2 N9 NA (PDB code 5L14) as an MR model. The Sh2 N9 Y169aH NA and NA-45 complex structure was subsequently determined using the refined Sh2 N9 Y169aH NA and individual light and heavy chains (PDB codes 5T93 and 5BV7, respectively) as input MR models. The complex structure of Hunan N9 NA with NA-63 was determined using wild-type Sh2 N9 NA (PDN code 5L14) and Fab light and heavy chains (PDB codes 5IT2 and 5V7R, respectively) as input MR models. Fab light and heavy chains from two Fabs (PDB codes 4KMT and 5AZE, respectively) were used to determine the structure of NA-80, while light and heavy chains from another two Fabs (PDB codes 5C2B and 4V1D, respectively) were used to determine the NA-22 structure. Initial rigid body refinement was performed in REFMAC5 (Murshudov et al., 1997), and simulated annealing and restrained refinement (including TLS refinement) were carried out in Phenix (Adams et al., 2010). Between rounds of refinements, model building was carried out with the program Coot (Emsley et al., 2010). Final statistics for these structures are summarized in Table S1. The quality of the structures was analyzed using the JCSG validation suite (www.jcsg.org) and MolProbity (Chen et al., 2010). All figures were generated with PyMol (www.pymol.org).
Cryo-EM structure determination
For each complex with NA anti-N9 Fabs NA-73, NA-63, NA-80 or NA-22 were added at 5 times molar excess to the Sh2 N9 NA ectodomain plus stalk and His6-tag and incubated on ice for 5 hours before cryo-grid preparation. 0.01 mM LMNG detergent was added to the complex to aid in particle tumbling. The final concentration of sample was 0.12 mg/mL on the grid. 3 μL of this mixture was added to glow discharged 1.2/1.3 copper Quantifoils 400 mesh grids and then plunged into liquid ethane using a Vitrobot.
Cryo grids were imaged on a Titan Krios at 300 kV using a Gatan K2 Summit camera. Data were collected using Leginon (Suloway et al., 2005) and transferred to Appion (Lander et al., 2009) for initial image processing. Movies were motion corrected with MotionCor2 (Zheng et al., 2017) and particles were picked using DoG Picker (Voss et al., 2009) before being added to CryoSparc1 (Punjani et al., 2017). Final maps were generated with C4 symmetry with a global resolution between 3.0 and 3.6 Å (Table S2).
Crystal structures of NA-80 Fab and NA-22 Fab, and Sh2 N9 (PDB codes 5L14 or 4MWJ) were docked into the cryo-EM density and C4 symmetry was applied. The models were relaxed into cryoEM maps using Rosetta (DiMaio et al., 2015) and manually adjusted with Coot (Emsley et al., 2010).
Glycan array neuraminidase inhibition assay
The Sh2 N9 NA tetramer with ectodomain plus stalk and His6-tag was diluted to 20 pg/mL and incubated with antibody IgG at a molar ratio of one NA protomer to four antibody IgGs for 1 hour at room temperature (~ 22 °C) in 100 mM imidazole-malate pH 6.15, 150 mM NaCl, 10 mM CaCl2, and 0.02% NaN3. The mixtures were then applied to glycan arrays (see Data S1 for glycan list) (Peng et al., 2017) for 1 hour. Then each array was washed to remove the NA solution by dipping 3 times with 1× PBS + 0.05% Tween, pH 7.4 at room temperature. Following washing, 250 μL of a pre-complexed solution of biotinylated Erythrina cristagalli lectin (ECA) (10 μg/mL, VectorLabs) + Streptavidin-AlexaFluor555 (2 μg/mL, Thermo Fisher Scientific) was applied directly to the array surface and incubated for 2 hours. Following incubation, ECA-Streptavidin solution was removed by dipping 3 times with 1× PBS + 0.05% Tween and, subsequently, by dipping 3 times in 1× PBS and then 3 times in distilled H2O. Washed slides were dried by centrifugation and scanned on an Innoscan1100AL (Innopsys) confocal slide scanner for 532 emission. Image data were stored as a TIFF image and signal data were collected using Mapix (Innopsys) imaging software. Collected signal data were processed to determine the averaged (mean signal minus mean background) values of 4 replicate spots on the array for each unique printed glycan.
Neuraminidase inhibition assay with substrate 4-MU-NANA
We measured the inhibition of NA enzymatic activity in 33.3 mM MES pH 6. 5, 4 mM CaCl2 0.02% NaN3 buffer by using fluorescent substrate 2’-(4-methylumbelliferyl)-α-D-N-acetylneuraminic acid (4-MU-NANA) (Potier et al., 1979) with excitation and emission wavelengths of 365 nm and 450 nm, respectively. The mixtures of Sh2 N9 NA at 2.5 μg/mL and antibody or control inhibitor zanamivir at 10 different concentrations (starting solutions at 333 nM were serially diluted 1:3) were incubated 1 hour at 37°C before adding 0.05 Mm substrate 4-MU-NANA. The reaction was conducted for 1 hour at 37°C in a total volume of 100 μL for the N9 NA protein with ectodomain plus stalk that was expressed as a tetramer. The reactions were all performed in duplicate and were stopped by adding 100 μL of 0.2 M Na2CO3.
QUANTIFICATION AND STATISTICAL ANALYSIS
The statistical details of experiments can be found above in the Methods Details section for each type of analysis.
DATA AND SOFTWARE AVAILABILITY
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession codes 6PZD and 6PZE for Sh2 N9 Y169aH in apo form and in complex with NA-45 Fab, 6PZF for Hunan N9 in complex with NA-63, and 6PZG and 6PZH for NA-80 Fab and NA-22 Fab in apo form. The cryo-EM maps and fitted coordinates of Sh2 N9 NA in complex with NA-22, NA-63, NA-73 and NA-80 Fabs are deposited in the EMDataResource and PDB with accession codes EMD-20538/PDB 6PZW, EMD-20594/PDB 6U02, EMD-20540/PDB 6PZY and EMD-20541/PDB 6PZZ, respectively.
Supplementary Material
Data S1. Glycan array compound list. Related to STAR Methods.
Highlights.
H7N9 neuraminidase (NA) stabilization mutant enables formation of ectodomain tetramer
Two human anti-N9 antibody Fabs are co-crystallized in complex with N9 NAs
Four human anti-N9 antibody Fab cryo-EM structures are elucidated with an N9 NA
Isolated anti-NA antibodies target diverse epitopes to neutralize NA activity
ACKNOWLEDGEMENTS
We thank Henry Tien for automated robotic crystal screening. The work was supported by National Institutes of Health grants U19 AI117905 (J.E.C., Jr., A.B.W., and I.A.W.), and AI114730 (J.C.P.), National Institutes of Health contract HHSN272201400024C (J.E.C., Jr., A.B.W., and I.A.W.), and the Skaggs Institute for Chemical Biology (I.A.W.). X-ray diffraction data were collected at the Advanced Photon Source (APS) beamline 23ID-D (GM/CA CAT) and the Stanford Synchrotron Radiation Lightsource (SSRL) beamlines 9-2 and 12-2. GM/CA CAT is funded in whole or in part with federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104). Use of the APS was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357. The SSRL is a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393) and the National Center for Research Resources (P41RR001209). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIAID, NIGMS, NCRR or NIH. This is publication MS29847 from The Scripps Research Institute.
Footnotes
DECLARATION OF INTERESTS
J.E.C. has served as a consultant for Takeda Vaccines, Sanofi Pasteur, Pfizer, and Novavax, is on the Scientific Advisory Boards of CompuVax and Meissa Vaccines, and is Founder of IDBiologics, Inc. All other authors declare no conflict of interest. Vanderbilt University has applied for a patent related to the NA antibodies.
SUPPLEMENTAL INFORMATION
Figures S1-S6 and Tables S1-S5.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, and Lipman D (2008). The influenza virus resource at the National Center for Biotechnology Information. J. Virol. 82, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barad BA, Echols N, Wang RY, Cheng Y, DiMaio F, Adams PD, and Fraser JS (2015). EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlanda Scorza F, Tsvetnitsky V, and Donnelly JJ (2016). Universal influenza vaccines: Shifting to better vaccines. Vaccine 34, 2926–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YQ, Wohlbold TJ, Zheng NY, Huang M, Huang Y, Neu KE, Lee J, Wan H, Rojas KT, Kirkpatrick E, et al. (2018). Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell 173, 417–429 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier A, Silva DA, Rocklin GJ, Hicks DR, Vergara R, Murapa P, Bernard SM, Zhang L, Lam KH, Yao G, et al. (2017). Massively parallel de novo protein design for targeted therapeutics. Nature 550, 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colacino JM, Chirgadze NY, Garman E, Murti KG, Loncharich RJ, Baxter AJ, Staschke KA, and Laver WG (1997). A single sequence change destabilizes the influenza virus neuraminidase tetramer. Virology 236, 66–75. [DOI] [PubMed] [Google Scholar]
- DiMaio F, Song Y, Li X, Brunner MJ, Xu C, Conticello V, Egelman E, Marlovits T, Cheng Y, and Baker D (2015). Atomic-accuracy models from 4.5 Å cryo-electron microscopy data with density-guided iterative local refinement. Nat. Methods 12, 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichelberger MC, Morens DM, and Taubenberger JK (2018). Neuraminidase as an influenza vaccine antigen: a low hanging fruit, ready for picking to improve vaccine effectiveness. Curr. Opin. Immunol. 53, 38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleishman SJ, Whitehead TA, Ekiert DC, Dreyfus C, Corn JE, Strauch EM, Wilson IA, and Baker D (2011). Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science 332, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, et al. (2013). Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 368, 1888–1897. [DOI] [PubMed] [Google Scholar]
- Gilchuk IM, Bangaru S, Gilchuk P, Irving RP, Kose N, Bombardi RG, Thornburg NJ, Creech B, Edwards KM, Li S, et al. Influenza H7N9 virus neuraminidase-specific human monoclonal antibodies inhibit viral egress and protect from influenza virus lethal infection in mice. Cell Host Microbe co-submission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Lee PS, Hoffman RM, Zhu X, Krause JC, Laursen NS, Yoon SI, Song L, Tussey L, Crowe JE Jr., et al. (2013). Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J. Virol. 87, 12471–12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M, Galvin HD, Haw TY, Nutsford AN, and Husain M (2017). Drug resistance in influenza A virus: the epidemiology and management. Infect. Drug Resist. 10, 121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Kumar G, Barman S, Najera I, White SW, Webby RJ, and Govorkova EA (2018). Identification of the I38T PA substitution as a resistance marker for next-generation influenza virus endonuclease inhibitors. mBio 9, e00430–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam RU, Juraszek J, Brandenburg B, Buyck C, Schepens WBG, Kesteleyn B, Stoops B, Vreeken RJ, Vermond J, Goutier W, et al. (2017). Potent peptidic fusion inhibitors of influenza virus. Science 358, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakus U, Thamamongood T, Ciminski K, Ran W, Gunther SC, Pohl MO, Eletto D, Jeney C, Hoffmann D, Reiche S, et al. (2019). MHC class II proteins mediate cross-species entry of bat influenza viruses. Nature 567, 109–112. [DOI] [PubMed] [Google Scholar]
- Krammer F, Fouchier RAM, Eichelberger MC, Webby RJ, Shaw-Saliba K, Wan H, Wilson PC, Compans RW, Skountzou I, and Monto AS (2018). NAction! How can neuraminidase-based immunity contribute to better influenza virus vaccines? mBio 9, e02332–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. (2009). Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen NS, Friesen RHE, Zhu X, Jongeneelen M, Blokland S, Vermond J, van Eijgen A, Tang C, van Diepen H, Obmolova G, et al. (2018). Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin. Science 362, 598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, Kurosawa Y, and Wilson IA (2014). Receptor mimicry by antibody F045–092 facilitates universal binding to the H3 subtype of influenza virus. Nat. Commun. 5, 3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malby RL, Tulip WR, Harley VR, McKimm-Breschkin JL, Laver WG, Webster RG, and Colman PM (1994). The structure of a complex between the NC10 antibody and influenza virus neuraminidase and comparison with the overlapping binding site of the NC41 antibody. Structure 2, 733–746. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, and Read RJ (2005). Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61, 458–464. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, and Dodson EJ (1997). Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, and Minor W (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Peng W, de Vries RP, Grant OC, Thompson AJ, McBride R, Tsogtbaatar B, Lee PS, Razi N, Wilson IA, Woods RJ, et al. (2017). Recent H3N2 viruses have evolved specificity for extended, branched human-type receptors, conferring potential for increased avidity. Cell Host Microbe 21, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Potier M, Mameli L, Belisle M, Dallaire L, and Melancon SB (1979). Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-α-D-N-acetylneuraminate) substrate. Anal. Biochem. 94, 287–296. [DOI] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Schmidt AG, Therkelsen MD, Stewart S, Kepler TB, Liao HX, Moody MA, Haynes BF, and Harrison SC (2015). Viral receptor-binding site antibodies with diverse germline origins. Cell 161, 1026–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Deng G, Ma S, Zeng X, Yin X, Li M, Zhang B, Cui P, Chen Y, Yang H, et al. (2018). Rapid evolution of H7N9 highly pathogenic viruses that emerged in China in 2017. Cell Host Microbe 24, 558–568.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch EM, Bernard SM, La D, Bohn AJ, Lee PS, Anderson CE, Nieusma T, Holstein CA, Garcia NK, Hooper KA, et al. (2017). Computational design of trimeric influenza-neutralizing proteins targeting the hemagglutinin receptor binding site. Nat. Biotechnol. 35, 667–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, and Carragher B (2005). Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60. [DOI] [PubMed] [Google Scholar]
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, et al. (2013). New world bats harbor diverse influenza A viruses. PLoS Pathog. 9, e1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulip WR, Varghese JN, Laver WG, Webster RG, and Colman PM (1992). Refined crystal structure of the influenza virus N9 neuraminidase-NC41 Fab complex. J. Mol. Biol. 227, 122–148. [DOI] [PubMed] [Google Scholar]
- van Dongen MJP, Kadam RU, Juraszek J, Lawson E, Brandenburg B, Schmitz F, Schepens WBG, Stoops B, van Diepen HA, Jongeneelen M, et al. (2019). A small-molecule fusion inhibitor of influenza virus is orally active in mice. Science 363, eaar6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatramani L, Bochkareva E, Lee JT, Gulati U, Graeme Laver W, Bochkarev A, and Air GM (2006). An epidemiologically significant epitope of a 1998 human influenza virus neuraminidase forms a highly hydrated interface in the NA-antibody complex. J. Mol. Biol. 356, 651–663. [DOI] [PubMed] [Google Scholar]
- Voss NR, Yoshioka CK, Radermacher M, Potter CS, and Carragher B (2009). DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Yang H, Shore DA, Garten RJ, Couzens L, Gao J, Jiang L, Carney PJ, Villanueva J, Stevens J, et al. (2015). Structural characterization of a protective epitope spanning A(H1N1)pdm09 influenza virus neuraminidase monomers. Nat. Commun. 6, 6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, et al. (2011). Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. U S A 108, 14216–14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlbold TJ, Podolsky KA, Chromikova V, Kirkpatrick E, Falconieri V, Meade P, Amanat F, Tan J, tenOever BR, Tan GS, et al. (2017). Broadly protective murine monoclonal antibodies against influenza B virus target highly conserved neuraminidase epitopes. Nat. Microbiol. 2, 1415–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Zhu X, McBride R, Nycholat CM, Yu W, Paulson JC, and Wilson IA (2012). Functional balance of the hemagglutinin and neuraminidase activities accompanies the emergence of the 2009 H1N1 influenza pandemic. J. Virol. 86, 9221–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Zhu X, Dwek RA, Stevens J, and Wilson IA (2008). Structural characterization of the 1918 influenza virus H1N1 neuraminidase. J. Virol. 82, 10493–10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Tian G, Shi J, Deng G, Li C, and Chen H (2018). Vaccination of poultry successfully eliminated human infection with H7N9 virus in China. Sci. China Life Sci. 61, 1465–1473. [DOI] [PubMed] [Google Scholar]
- Zhang K (2016). Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Yang H, Guo Z, Yu W, Carney PJ, Li Y, Chen LM, Paulson JC, Donis RO, Tong S, et al. (2012). Crystal structures of two subtype N10 neuraminidase-like proteins from bat influenza A viruses reveal a diverged putative active site. Proc. Natl. Acad. Sci. U S A 109, 18903–18908. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Glycan array compound list. Related to STAR Methods.
Data Availability Statement
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession codes 6PZD and 6PZE for Sh2 N9 Y169aH in apo form and in complex with NA-45 Fab, 6PZF for Hunan N9 in complex with NA-63, and 6PZG and 6PZH for NA-80 Fab and NA-22 Fab in apo form. The cryo-EM maps and fitted coordinates of Sh2 N9 NA in complex with NA-22, NA-63, NA-73 and NA-80 Fabs are deposited in the EMDataResource and PDB with accession codes EMD-20538/PDB 6PZW, EMD-20594/PDB 6U02, EMD-20540/PDB 6PZY and EMD-20541/PDB 6PZZ, respectively.