Abstract
An in vitro model of intestinal epithelium with an immune component was bioengineered to mimic immunologic responses seen in inflammatory bowel disease. While intestinal immune phenomena can be modeled in transwells and 2D culture systems, 3D tissue models improve physiological relevance by providing a 3D substrate which enable migration of macrophages towards the epithelium. An intestinal epithelial layer comprised of non-transformed human colon organoid cells and a subepithelial layer laden with monocyte-derived macrophages was bioengineered to mimic native intestinal mucosa cell organization using spongy biomaterial scaffolds. Confluent monolayers with microvilli, a mucus layer, and infiltration of macrophages to the basal side of the epithelium were observed. Inflammation, induced by E. coli O111:B4 lipopolysaccharide and interferon γ resulted in morphological changes to the epithelium, resulting in ball-like structures, decreased epithelial coverage, and increased migration of macrophages to the epithelium. Analysis of cytokines present in the inflamed tissue model demonstrated significantly upregulated secretion of pro-inflammatory cytokines that are often associated with active inflammatory bowel disease, including CXCL10, IL-1β, IL-6, MCP-2, and MIP-1β. The macrophage layer enhanced epithelial and biochemical responses to inflammatory insult, and this new tissue system may be useful to study and develop potential therapies for inflammatory bowel disease.
Keywords: Tissue Engineering, Intestine tissue, Inflammatory bowel disease, Intestinal immune system, Colonoid, Monocyte-derived macrophage
Introduction
The intestine presents a surface area of 250 m3 – 400 m3 to an external environment which comprises of digesting food and a population of 1013 – 1014 bacteria known as the gut microbiota [1, 2]. A single layer of the intestinal epithelium ensures that these microorganisms do not invade into the host with coordination from the intestinal immune system which both directly eliminates pathogens and secretes factors contributing to epithelial renewal and remodeling [3–5]. Dysregulation of the intestinal immune system can result in inflammatory bowel disease (IBD) in which the intestinal immune system becomes hyperactive and causes unnecessary inflammation in the intestinal mucosa [5]. Intestinal macrophages are one of the largest populations of macrophages in the body and are primarily concentrated directly underneath the single layer of epithelium, acting as the first line of defense [6–8]. This strategic subepithelial localization allows for recognition and phagocytosis of invading pathogens within minutes and modulation of bacterial populations in the microbiota [7, 9]. Moreover, intestinal macrophages secrete factors promoting epithelial cell renewal including Wnt signaling molecules and hepatocyte growth factor [10, 11].
Investigations of intestinal immune phenomena are often conducted using in vitro cell or tissue models or in vivo mice models. Though in vitro 2D models include immune cell co-cultures with epithelial cells, many use cell-line based epithelial modeling [12–14]. A recently published in vitro 3D model of inflammatory bowel disease used Caco-2 and HT29-MTX intestinal epithelial cell lines, but does not include contributions of the immune system in epithelial barrier pathologies [15]. Mice models present an alternative for intestinal epithelial-immune studies. For example, macrophage ablation in mice reduced levels of Leucine Rich Repeat Containing G Protein-Coupled Receptor 5 positive (LGR5+) intestinal stem cells, disrupted development of M cells, and enhanced goblet cell density [16]. This work demonstrates important functions of macrophages in the intestinal epithelium; however, the study was carried out in mice and the relevance to human physiology is unclear. In the present study, we utilized an in vitro 3D tissue engineered system which models colonic epithelium-immune responses using human LGR5+ colonic organoids (colonoids) to better mimic physiologic responses of the human intestinal epithelium. Furthermore, we utilized co-cultures with human primary monocyte-derived macrophages layered underneath the epithelial layer, mimicking intestinal tissue organization. Non-transformed human cells were used to improve relevance vs the more commonly used cell-line-based cultures.
To ensure physiologic responses of the intestinal epithelium, human colonoids were cultivated. Colonoids are untransformed LGR5+ epithelial stem cells derived from colonic crypts, are capable of indefinite propagation, and can differentiate towards all epithelial lineages [17]. Under inflammatory conditions, intestinal epithelial cells direct immune cell responses against pathogens and experience impaired proliferation resulting in a loss of intestinal epithelial coverage [18–20]. In a previous study examining the expression of inflammation-associated genes between intestinal epithelia derived from cell-lines, human primary cells, and intestinal organoids in a 3D system, organoids were shown to be the most sensitive to inflammation cues [21]. Moreover, colonoids were used to model epithelial-immune interactions as opposed to small intestine organoids, due to the colonic lamina propria containing a relatively higher population of macrophages than in the small intestine lamina propria [3].
A cylindrical scaffold shape was used due to previous intestinal modeling showing that intestinal epithelia arranged in a tubular configuration results in low oxygen tensions within the lumen, enabling the growth of anaerobic bacteria [21, 22]. Moreover, the physiology of the intestine is tubular and the cylindrical design of the model mimics intestine architecture. To achieve the 3D intestinal tissue organization, we modified our previous small intestine epithelium tissue model cultured on lyophilized silk protein sponge matrices by introducing the modular secondary macrophage laden 3D tissue layer [21–23]. The double cylindrical scaffold is a critical feature, enabling replacement of the macrophage-laden outer layer without disruption of the inner scaffold layer containing the epithelium and the observation of macrophage migration towards the epithelial layer. While intestinal macrophages are constantly replenished by bone marrow derived monocytes from peripheral blood, 3D tissue engineered constructs with immune cells are typically cultured to a terminal timepoint and are not able to replenish immune cell populations [24–27]. The current tissue model is a significant advance in both biomaterial and biology over the previously published system.
During active IBD, monocytes massively infiltrate the epithelium and elevated levels of proinflammatory cytokines including interleukins 1β and 6 (IL-1β & IL-6), monocyte chemoattractant proteins 1 and 2 (MCP-1 & MCP-2), and macrophage inflammatory protein 1β (MIP-1β/CCL4) are detected [28–31]. This 3D silk scaffold system presents substrates for cultivation and movement of macrophages towards the colonic epithelial layer allowing for evaluation of inflammation in vitro. The silk sponges show formation of a porous film for the cultivation of an epithelial layer with appropriate material properties for intestinal tissue engineering. Analysis of the epithelium of inflamed 3D colonoid-epithelial co-cultures showed significant decreases in epithelial coverage and morphology in addition to the infiltration of macrophages towards the basal side of the epithelium. Cytokine analysis showed enhanced secretion of inflammatory cytokines mimicking cytokine profiles found in vivo in response to inflammation. This paper presents a novel system using human colonoids and human primary monocyte-derived macrophages cultured in a 3D sponge format with epithelial-immune interactions reflective of IBD.
Materials
Cell Culture
Human colonoid culture –
Human colonoids isolated from the large intestine epithelium were provided by Dr. Mary Estes from Baylor College of Medicine under approved protocols. Colonoids were cultured in 24 well tissue culture plates in Matrigel (Corning Inc, Corning, NY) droplet cultures based on previously described protocols [32]. Frozen vials of colonoids were thawed and suspended in Matrigel and plated in 10 μL droplets with 3 droplets in each well. Each well of colonoids received 500 μL of high Wingless/Integrated (Wnt) media and was replaced every other day. High Wnt media consists of 75% Wnt-3A conditioned media harvested from L Wnt-3A cells (American Type Culture Collection, Manassas, VA), 10% R-spondin conditioned media harvested from R-spondin generating cells (courtesy of Dr. Calvin Kuo, Palo Alto, CA), 5% Noggin conditioned media harvested from Noggin generating cells (courtesy of Dr. Gijs van den Brink, Amsterdam, Netherlands), and 10% complete media without growth factors (CMGF- media) and growth factors. CMGF- media consists of Advanced Dulbecco’s Modified Eagle Medium (DMEM)/Ham’s F-12 (ThermoFisher Scientific, Waltham, MA) supplemented with 1x GlutaMAX (ThermoFisher), 1x Penicillin-Streptomycin (ThermoFisher), and 10 mM HEPES buffer (ThermoFisher). Final concentrations of growth factors in the high Wnt media are 0.5x B-27 supplement (ThermoFisher), 0.5x N-2 supplement (ThermoFisher), 0.5 mM N-Acetyl-L-cysteine (Sigma-Aldrich, St. Louis, MO), 25 ng/mL mouse recombinant epidermal growth factor (ThermoFisher), 250 nM A-83–01 (Tocris Bioscience, Bristol, United Kingdom), 5 mM SB 202190 (Sigma-Aldrich), 5 mM nicotinamide (Sigma-Aldrich), 5 nM [15-Leucine]-Gastrin I (Sigma-Aldrich), and 100 ug/mL Primocin (Invivogen, San Diego, CA).
Human monocyte-derived macrophage culture –
Monocytes were isolated using the Pan Monocyte Isolation Kit (Miltenyi Biotec Inc, Bergisch Gladbach, Germany) per manufacturer’s instructions from unpurified buffy coats (Research Blood Components LLC, Boston, MA). Isolated monocytes were frozen in 10% DMSO (Sigma-Aldrich) and 90% in macrophage media consisting of RPMI 1640 medium with 10% heat inactivated FBS (ThermoFisher), 10 mM HEPES (ThermoFisher), 1mM sodium pyruvate (ThermoFisher), 0.05 mM 2-mercaptoethanol (ThermoFisher), and 1X Penicillin-Streptomycin (ThermoFisher). Monocytes were thawed as needed and differentiated into macrophages by culturing in 50 ng/mL macrophage colony-stimulating factor (ThermoFisher) supplemented macrophage media for 6 days with media changes every other day. For M1 polarization, monocyte-derived macrophages were then cultured in macrophage media with 100 ng/mL lipopolysaccharides (LPS) from E. coli 0111:B4 (Sigma-Aldrich) and recombinant human interferon γ (IFNγ) (PeproTech, Rocky Hill, NJ).
Silk Scaffolds
Silk fibroin (hereafter referred to as silk) extracted from Bombyx mori silkworm cocoons was processed to a 5–7% (wt/vol) silk solution following established protocols [33]. Hollow channels were formed in silk scaffolds using a cylindrical mold cast comprised of polydimethylsiloxane (Dow Corning, Midland, MI) following previous protocols [22]. A 2 mm diameter Teflon-coated stainless-steel wire (McMaster-Carr, Elmhurst, IL) was inserted through the cross section of the cylindrical mold to form the channel. The silk solution was then poured into the molds and frozen overnight at −20°C. The frozen silk was then placed in a lyophilizer for freeze-drying. Once fully dried, the silk scaffolds were autoclaved to induce β-sheet conformation, stabilizing the scaffold structure and preventing solubilization. The scaffolds were then soaked in distilled water overnight and cut into silk sections with 8 mm long hollow channels. A 6 mm biopsy punch (Acuderm Inc, Fort Lauderdale, FL) was used to punch out an inner silk scaffolds with a diameter of 6 mm, a 2 mm diameter hollow center and length of 8 mm. A 10 mm biopsy punch (Acuderm Inc) was then used to punch out a 10 mm diameter outer scaffold with a 6 mm diameter hollow center and length of 8 mm.
Cell seeding
Inner scaffolds –
Human colonoids were seeded on the inner surface of the 2 mm diameter hollow channels following previously described protocols for human intestinal organoids [21]. For each scaffold, 500 μl of collagen gel was added into the spongy silk scaffold and gelled at 37°C for 45 minutes. The collagen gel was 80% 2.01 mg/ml rat tail collagen type I (First Link Ltd, Wolverhampton, United Kingdom), 10% 10X DMEM (Sigma-Aldrich), and 10% CMGF- media. 5 M NaOH was used to neutralize the collagen gel. Two wells of confluent colonoids were dissociated and seeded following a previously established protocol e. In short, Matrigel droplets containing confluent colonoids were suspended in 500 μL/well of 4°C 0.5 mM ethylenediamine tetraacetic acid (ThermoFisher) and centrifuged at 200 g at 4°C for 5 minutes. The resultant cell pellet was then digested with 0.05% Trypsin-EDTA (ThermoFisher) for 4 minutes at 37°C. The trypsin was inhibited with CMGF- media containing 10% fetal bovine serum (ThermoFisher) and the colonoid pellet was further dissociated via pipetting. The resulting cell suspension was passed through a 40 μm cell strainer (Corning) and centrifuged again at 1,500 rpm for 5 minutes to obtain a cell pellet. The pellet was resuspended in 30 μL of high Wnt media supplemented with 10 μM Y-27632 (Sigma-Aldrich) and pipetted onto one side of the hollow channel of the scaffold. The scaffolds were transferred to 37°C fo r 45 minutes. The scaffold was then flipped 180°and another 30 μL of colonoid suspension made from another 2 wells of confluent colonoids was pipetted into the hollow channel to completely coat the inner surface of the channel. The inner scaffolds were then cultured in high Wnt media with 10 MM Y-27632 for 1 day before being cultured in high Wnt media for a total of 7 days. Media changes occurred every other day. Seeded colonoids were differentiated by culturing in differentiation medium comprised of 90% CMGF- supplemented with 10% R-spondin conditioned media, 1x B-27 supplement, 1x N-2 supplement, 1 mM N-Acetyl-L-cysteine, 50 ng/mL mouse recombinant epidermal growth factor, 500 nM A-83–01, 10 nM [15-Leucine]-Gastrin I, and 100 ug/mL Primocin.
Outer scaffolds –
Monocytes suspended in collagen gel (80% 2.01 mg/ml rat tail collagen type I (First Link Ltd), 10% 10X DMEM (Sigma-Aldrich), and 10% macrophage media) were seeded throughout the bulk of the outer scaffold at 3×106 cells/scaffold. Seeded monocytes were differentiated to uncommitted macrophages (M0) by 6 days of culture in macrophage media supplemented with 50 ng/mL MCSF. The outer scaffold was then either cultured for 1 day in macrophage media with 100 ng/mL LPS and 20 ng/mL IFNγ for M1 polarization or in plain macrophage media. The outer scaffold was combined with the inner scaffold on day 8 as shown in Figure 2D. Once combined M1 differentiation was maintained by 100 ng/mL LPS and 20 ng/mL IFNγ in the co-culture media.
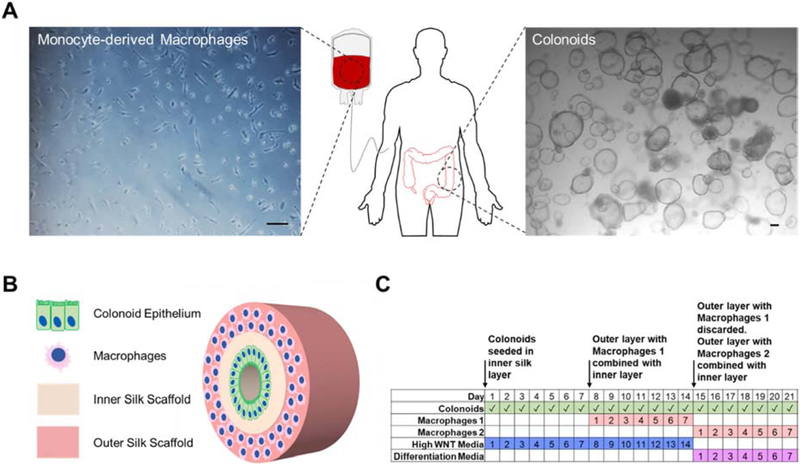
Figure 2. Colonoid and macrophage cultivation scheme in the 3D bilayer system.
(A) Human monocytes were isolated from whole blood and human colonoids from large intestine biopsies were cultured according to established protocols. (B) Cell suspensions of colonoids were seeded on the film surface on the inner silk scaffold and monocyte-derived macrophages were seeded throughout the porous outer silk scaffold. (C) The model is cultured for 3 weeks total with 2 weeks in High WNT media and 1 week in differentiation media. Colonoids are present in the model throughout the 3 week culture time. 2 sets of macrophages are added with the first set added after the first week of culture and the second set replacing the first set after the second week.
Scanning Electron Microscopy
Samples were fixed in 1% glutaraldehyde (Sigma-Aldrich) for 1 hour and then rinsed in PBS and then deionized water. After the rinses, the sample was dehydrated in a graded series of ethanol (30%, 50%, 70%, 90%, 100%) before being dehydrated overnight in 100% ethanol. The samples were dried via critical point drying using a liquid CO2 dryer (AutoSamdri-815, Tousimis Research Corp.). Samples were then coated with a 10 nm thick layer of Pt/Pd using a sputter coater (208HR, Cressington Scientific Instruments Inc.). Samples were imaged on a scanning electron microscope (Zeiss UltraPlus SEM or Zeiss Supra 55 VP SEM, Carl Zeiss SMT Inc.) at 2–3 kV.
Mechanical Characterization of Scaffolds
The mechanical properties of silk sponges were characterized using an Instron 3366 (Instron Inc.). The outer sponges have an outer dimeter of 10 mm, an inner diameter of 6 mm, and a length of 8 mm. The inner sponges have an outer diameter of 6 mm, an inner diameter of 2 mm, and a length of 8 mm. The compression tests were done on individual inner and outer sponges along the longitudinal direction. In each test, the hydrated samples were loaded onto the testing frame at room temperature and then were compressed with a rate of 2.0 mm/min. Compressive modulus were calculated at 5% strains for comparison. At least n = 3 samples were used to calculate average modulus with standard deviation.
Immunofluorescence and Epithelium Measurements
Seeded 3D silk scaffolds and transwell membranes were fixed in 4% paraformaldehyde (Santa Cruz Biotechnology Inc, Dallas, TX). For luminal surface imaging, inner scaffolds were cut along the hollow channel to expose the colonoid seeded lumen surface to blocking solutions and antibodies. To obtain transverse sections of the model, fixed samples were suspended in an increasing gradient of sucrose (Sigma) and optimal cutting temperature (OCT) compound (VWR, Radnor, PA) over 4 days. Scaffolds were then embedded in OCT and sectioned at 20 μm thickness. All samples were permeabilized and blocked with 5% bovine serum albumin (Sigma Aldrich) and 0.1% Triton X-100 (Fisher Scientific, Hampton, NH) in phosphate-buffered saline (PBS, Thermofisher) for 30 minutes. Samples were incubated overnight at 4°C in PBS with human e-cadherin antibody (1:250, ab1416) (Abcam, Cambridge, United Kingdom), human mucin-2 antibody (1:50, H-300) (Santa Cruz Biotechnology Inc), human CD-68 antibody (1:200, ab955), and human iNOS antibody (1:150, ab15327). The samples were rinsed with PBS then incubated in goat anti-mouse Alexa Fluor 488 (1:250, A28175) (Thermofisher) or goat anti-rabbit Alexa Fluor 546 (1:1000, A-11010) (Thermofisher). Scaffolds were counterstained with 4’,6-diamidino-2-phenylindole, dihydrochloride (DAPI) (Thermofisher). A Leica SP8 confocal microscope (Leica Microsystems, Wetzlar, Germany) with z-series capability was used to image samples. Scaffolds were observed under preset filters for DAPI (Ex/Em: 359/461 nm), AF488 (Ex/Em: 499/520) and AF546 (556/573). Leica Application Suite X (Leica Microsystems) ver 3.5.2.18963 was used to assemble confocal maximum projection images. ImageJ with the Fiji plugin was used to measure epithelial coverage and epithelial heights on histologic sections of the models. Epithelial height was measured at every 5 Mm where epithelium was undisrupted.
Cytokine Array
Cytokines were measured by chemiluminescence using commercial arrays (RayBiotech Inc, Norcross, GA) following manufacturer’s instructions. Media was collected at days 3 of differentiation media culture. Blot intensities were measured using BD ChemiDoc Imaging System (Becton, Dickinson and Company, Franklin Lakes, NJ) and blot intensities were all normalized to each other using ImageJ. Principle component analysis and biplots were calculated using Matlab R2018b.
Mucus Thickness Measurements
Transverse sections of the inner scaffold were obtained using the sucrose-OCT gradient equilibration and cryo-embedding procedure outlined for immunofluorescence. 20 Mm slices were obtained and rinsed with tap water for 2 minutes. The slides were then dried for 20 minutes and rinsed with 3% acetic acid (pH 2.5) for 2 minutes and stained in Alcian blue (Sigma) for 15 minutes. The samples were then rinsed in running tap water for 5 minutes and dipped in deionized water once. The slides were dehydrated through an alcohol series, cleared in xylene and coverslipped using DPX Mountant (Sigma). A light microscope was used to evaluate stained samples. Mucus thickness measurements were taken every 5 μm where the epithelium remained intact using ImageJ with the Fiji plugin.
Statistical Analysis
Statistical significances were calculated using Analysis of Variance (ANOVA) paired with Tukey’s post-hoc test using Minitab 18 (Minitab LLC, State College, PA). Data were reported as mean ± standard deviation. P-values < 0.05 were considered significant.
Results
Characterization of 3D silk sponges with a porous film for intestinal tissue engineering
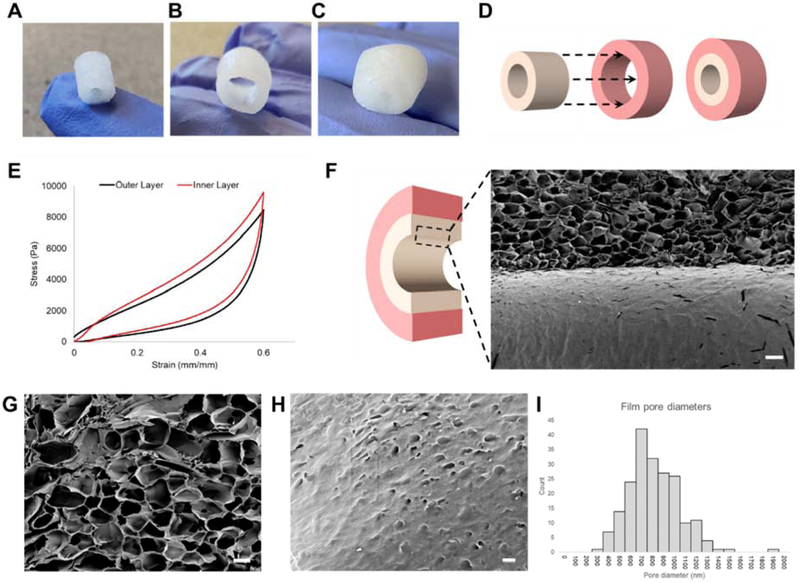
The bilayer components were generated using silk fibroin isolated from Bombyx mori cocoons and processed following established protocols into lyophilized silk sponges with a hollow lumen space (diameter: 2mm) [21, 22, 33]. A cylindrical inner scaffold was shaped using a 6 mm biopsy punch (inner diameter: 2mm, outer diameter: 6mm, length: 8mm) and a hollow outer scaffold with a 10 mm biopsy punch (inner diameter: 6mm, outer diameter: 10mm, length: 8mm) (Figure 1A–C). The scaffold dimensions were designed to enable simple assembly of a bilayered silk sponge format in which the colonoid epithelial monolayer is cultured on the filmy surface of the inner layer and the macrophages are cultured throughout the outer layer (Figure 1C and D). Compression testing of silk scaffolds yielded Young’s moduli averaged at 18.16 kPa, within soft tissue moduli of 0.2–190 kPa [34] (Figure 1E). Bulk properties of the lyophilized sponge have been previously characterized for pore size and degradation properties [35]. SEM imaging of the sponge microstructure showed a lumen surface with a film and a porous spongy bulk matrix directly underneath (Figure 2F–H). The film consistently formed on the inner surface of silk scaffolds though the mechanism for its formation is unclear. We postulate that the interaction between the Teflon coated stainless-steel wire and the silk solution during the freezing process contributed to the film formation. Quantification of pore diameters on the film showed an average diameter of 767 nm (Figure 1I).
Figure 1. Dimensions, stiffness, and porous film characterization of lyophilized silk matrices.
3D scaffold systems for intestinal tissue engineering were fabricated used silk fibroin. (A) Inner scaffolds are cylindrical constructs with an outer Ø of 6 mm, inner Ø of 2 mm, and length of 8 mm. (B) Outer scaffolds are wider cylindrical constructs capable of enclosing the inner scaffold with an outer Ø of 10 mm, inner Ø of 6 mm, and length of 8 mm. (C) Silk scaffold system with inner and outer scaffold combined (D) Dimensions of inner and outer scaffolds allow sliding of the inner scaffold into the hollow center of the outer scaffold. (E) Representative tress-strain curves of both outer and inner scaffolds (F) SEM cutaway of the interface between the film and sponge (scalebar: 200 μm) (G) Interconnected pores in the spongey bulk of scaffolds (scalebar: 100 μm) (H) Nanopores in the film (scalebar: 2 μm). (I) Aggregate diameters of nanopores (n=201) averaged at 767 nm (s.d. = 233 nm).
Macrophage, colonoid, and tissue model cultivation
Human colonoids were cultured in Matrigel using established protocols prior to seeding in the tissue models [21, 32, 36] (Figure 2A). Human monocytes were isolated from unpurified buffy coats using magnetic activated cell sorting and cryopreserved until needed (Figure 2A). The colonoid epithelium of the tissue model was seeded by creating a single cells suspension of the colonoids and directly pipetting the suspension into the lumen space. Colonoid cells attached to the inner surface, forming a monolayer throughout the lumen surface (Figure 2B). Monocytes were seeded in the bulk throughout the sponge of the outer layer and differentiated into a macrophage lineage prior to combining with the inner layer (Figure 2B). The tissue model culture schedule consisted of 2 weeks of culture in High WNT media to ensure formation of a confluent colonoid epithelial layer within the tissue model, while preventing the colonoid monolayer from differentiating (Figure 2C). The tissue model was then cultured in differentiation media for up to a week (Figure 2C). A week was chosen as the terminal timepoint since colonoid monolayers seeded on transwell membranes remain functional for up to 7 days of differentiation (Supplementary Figure S1). The outer layer with monocytes was differentiated independently into a macrophage lineage and added to the model 7 days prior to differentiation and again at the start of differentiation (Figure 2C). The outer layer was replaced every 7 days due to up to 50% decreased metabolic activity of monocyte-derived macrophages in either high WNT or differentiation media by day 7 (Supplementary Figure S2A).
Establishment of the tissue epithelial layer and macrophage outer layer
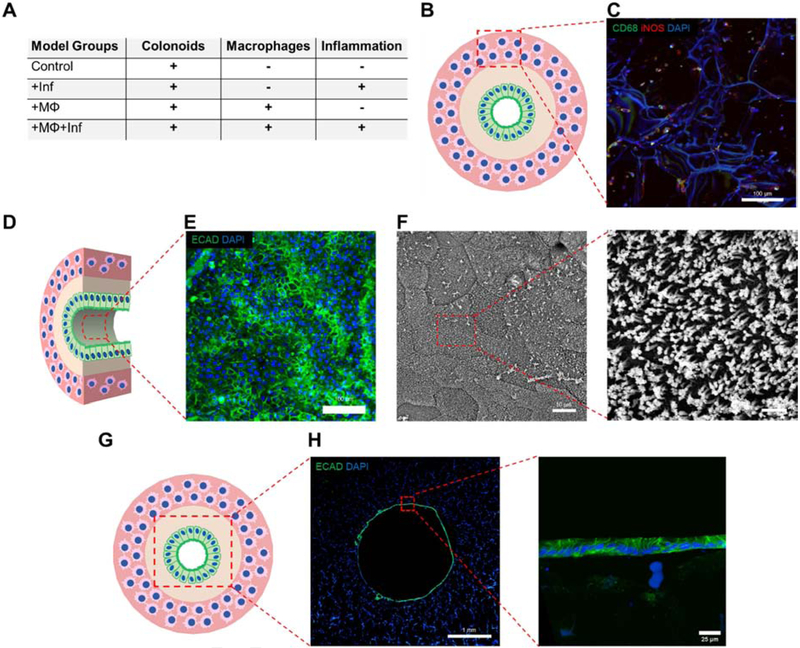
Figure 3A outlines the tissue models constructed and analyzed to investigate the effects of macrophages and inflammatory cues on the colonoid epithelial layer. Staining for CD68, a panmacrophage marker, and iNOS, an M1 polarized macrophage marker showed the presence of macrophages dispersed through the outer silk scaffold (Figure 3B, C). iNOS expression was seen in macrophages regardless of culture in high WNT or differentiation media (Supplementary Figure S2D). Control models were analyzed at day 3 and day 7 of differentiation to ensure lumen surface coverage by a single cell layer of polarized epithelial cells as seen in vivo (Figure 3D). E-cadherin, a cell adhesion molecule selective for colonoid cells but not macrophages was used as an immunocytochemical marker for colonoids. At day 3, seeding of colonoids in the lumen resulted in the formation of an epithelial layer expressing E-cadherin (Figure 3E). SEM imaging of the epithelial surface at day 7 confirmed a polarized epithelial surface with microvilli on the apical surface with defined borders between individual cells, indicating a functional and properly differentiated intestinal epithelium (Figure 3F). Transverse sections of controls showed a fully confluent monolayer at day 3, replicating the intestinal monolayer found in vivo (Figure 3G, H). A closer look at the epithelium showed individual cells arranged in a monolayer with e-cadherin expression between cells.
Figure 3. Colonoid epithelial monolayer forMs a polarized and confluent monolayer.
(A) Table showing different group conditions for analyzing inflammation and macrophage contributions to model. All groups are seeded with colonoids. (B, C) The macrophage laden outer layer of the +MΦ+Inf group was imaged for CD68 and iNOS to show even distribution of macrophages throughout outer scaffold (scalebar: 100 μm). (D) The surface of the epithelial monolayer was imaged using immunofluorescence and SEM. (E) Colonoids seeded on inner scaffold lumen surface show consistent expression of e-cadherin at day 3 (scalebar: 100 μm) (F) SEM shows microvilli on the apical side of colonoids (scalebar: 10 μm and 1 μm respectively). (G) Transverse histological sections (thickness: 20 μm) of the 3D models were immunostained with e-cadherin to analyze epithelial coverage. (H) 3 days differentiated control models show a single cell layer of epithelium on the lumen surface. (scalebar: 1 mm and 25 μm).
Macrophage infiltration into epithelium
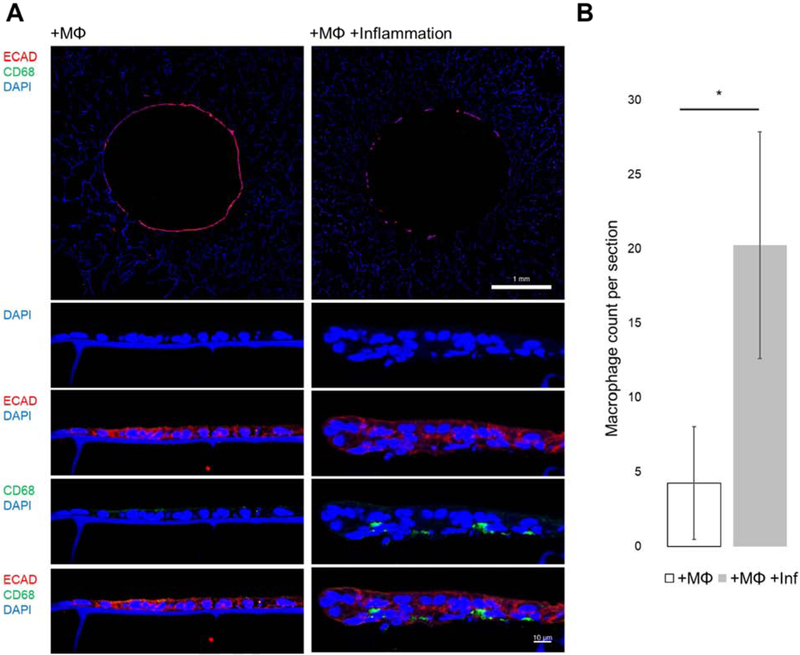
Transverse histological sections of macrophage laden models at day 3 of differentiation (+MΦ,+MΦ+Inf) were examined for the presence of macrophage along the epithelium. Macrophages were identified by CD68 positive staining which does not stain intestinal epithelium. At 10X, CD68+ staining was not readily observed (Figure 4A), however 40X imaging of the epithelium showed CD68+ cells along the basal side of the epithelium (Figure 4A). Quantification of CD68+ cells on histological sections showed a statistically significant difference between +MΦ and +MΦ+Inf Models (Figure 4B). Observation of CD68+ cells along the epitheliuM suggests that Macrophages in the outer layer of the model system were capable of movement through the silk-collagen matrix. The increased macrophage count in the +MΦ+Inf Models over +MΦ was explained by increased levels chemotactic cues released by the inflamed epithelium (Figure 6A).
Figure 4. Monocyte-derived macrophages infiltrating the epithelial monolayer under inflammation.
(A) Transverse histological sections of models at day 3 show e-cadherin positive epithelial cells and little to no CD68+ cells at 10X (top scalebar 1 mm). Higher magnification images of the epithelium show CD68+ macrophages on the basal side of the epithelium of inflamed conditions (bottom scalebar 10 μm). (B) Quantification of CD68+ macrophage cells on transverse sections between +MO and +MO+Inflammation show statistical significance (n=4) (p<0.05).
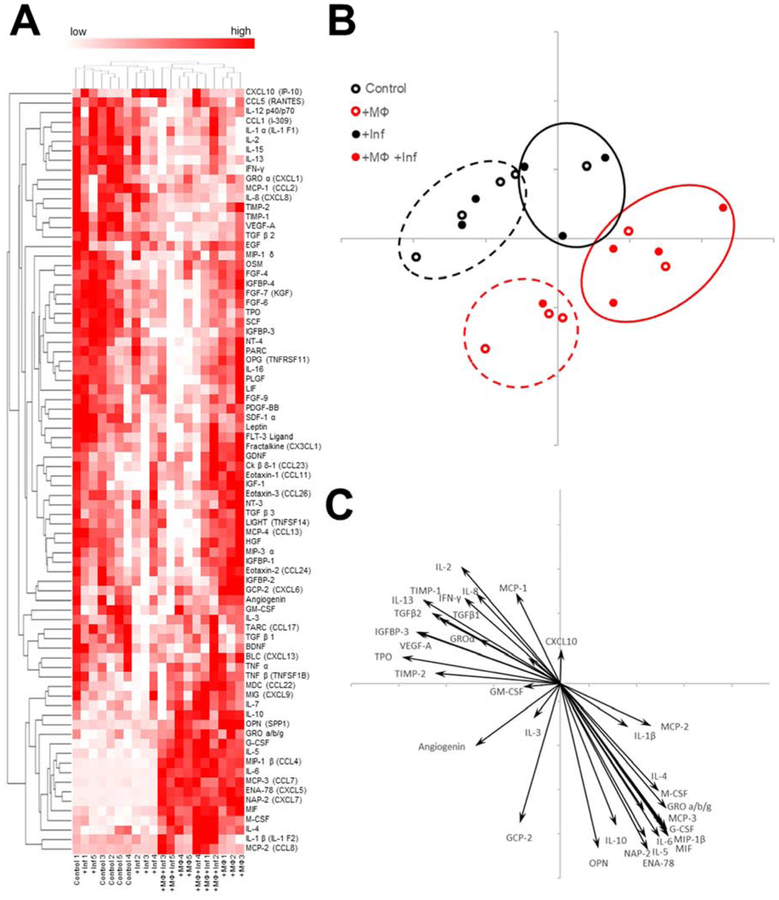
Figure 6. Addition of macrophages and inflammatory cues each contribute to unique cytokine profiles.
(A) 80 cytokines were assayed in the model media at day 3 of differentiation. Cytokine blot intensities were standardized by z-score and reported in a heat Map. Heat scores for each individual sample were represented with red denoting higher relative concentration. (n=5). (B) Principal component analysis was used to interpret the cytokine profiles of each sample. Hierarchical clustering of the cytokine data was used to group each cytokine profile. (C) Biplot of statistically significant cytokine z-scores based on ANOVA shows contributions of cytokines to principal component analysis placements (p<0.05).
Effects of Macrophage co-culture and inflammatory cues on epithelial Morphology and coverage
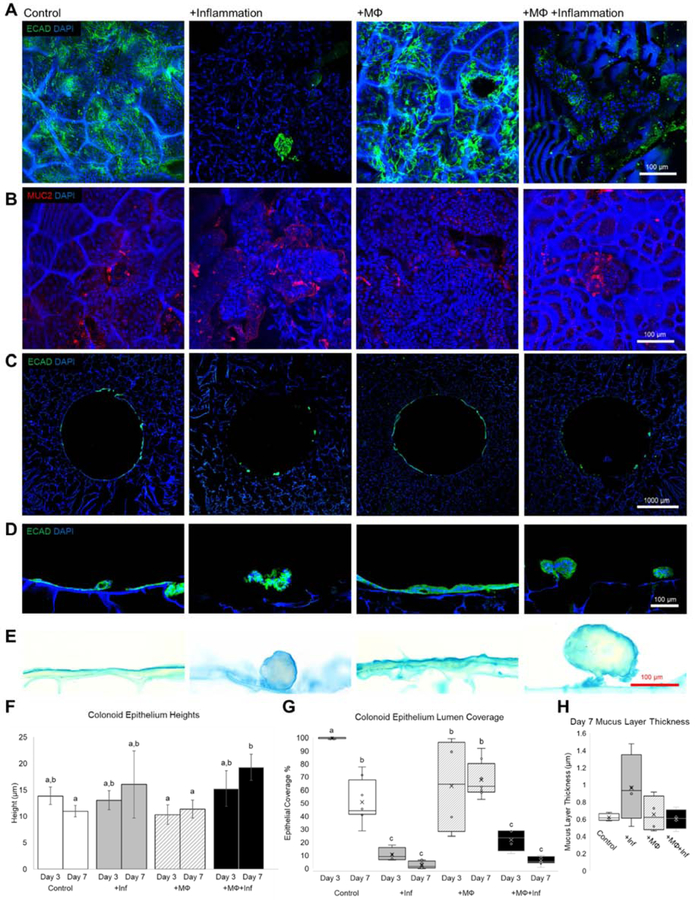
Lumen surfaces of each Model group were stained with E-cadherin and MUC2 (Figure 5A, B). E-cadherin staining showed clear differences in epithelial coverage at day 7 between inflamed and uninflamed groups (Figure 5A). MUC2 staining which is indicative of goblet cells showed the presence of Mucus producing goblet cells (Figure 5B). To quantify epithelial coverage, transverse histological sections of the monolayer on the whole lumen were stained with E-cadherin (Figure 5C). Epithelial coverage at day 7 evaluated via transverse sections reflect the epithelial coverage seen via lumen surface staining (Figures 5A, C). Close examination of the epithelial layer showed differences in morphology between inflamed and uninflamed groups with a monolayer on uninflamed groups and a ball-like epithelial structure forming for inflamed groups (Figure 5D). Alcian blue staining showed a thin layer of mucin on the epithelial cells in all groups (Figure 5E). Epithelial height measurements reflected the morphological differences between groups observed in Figure 5D with +MΦ+Inf displaying a higher epithelial height at day 7 compared to +MΦ at day 3 or 7 and control at Day 7 (Figure 5F). This was explained by the overall higher epithelial heights seen in the ball-like structures in the inflamed groups. Lumen coverage was quantified based on the transverse sections at day 3 and day 7 (Figure 5G). Epithelial coverage was highest for day 3 control tissue models followed by day 3 +MΦ models, though day 7 coverages between control and +MΦ were not statistically significant. Overall, the +MΦ group displayed a higher average coverage. +MΦ+Inf and +lnf groups at both days 3 and 7 had statistically significant coverage compared to the uninflamed groups. Though inflammation is known to play a role in goblet cell differentiation, the thickness of the mucus layers did not change between groups (Figure 5H).
Figure 5. Inflammatory cues and macrophages influence in vitro colonic epithelial coverage and morphology but not mucus thickness.
(A) Lumen surface immunostaining for e-cadherin show differences in epithelial coverages between inflamed versus uninflamed groups at day 7 of differentiation (scalebar: 100 μm). (B) Luminal MUC2 staining shows positive staining on all groups indicating presence of goblet cells (scalebar: 100 μm). (C) Transverse histological sections of models at day 7 show variable epithelial coverages as seen by the e-cadherin immunostaining (scalebar 1 mm). (D) A closer look at the epithelia shows monolayers for uninflamed groups and ball like structures in inflamed groups (scalebar: 100 μm). (E) Alcian blue staining of transverse sections stains mucus layers (scalebar: 100 μm). (F) Quantification of epithelial heights between Day 3 and Day 7 for all groups show that the combination of macrophages and inflammatory cues results in morphologic differences (n=4–8) (p<0.05). (G) Epithelial coverage was quantified showing a decreased coverage in inflamed groups and over time (n=4–8) (p<0.05). (H) Mucus layer thickness at day 7 showed no significant changes between groups (n=4–8) (p<0.05).
Effects of macrophages and inflammatory cues on secreted cytokines
The cytokine profiles of the tissue models were assayed using a cytokine array capable of measuring the presence of 80 cytokines and standardized by z-score to highlight relative differences of secreted cytokines between groups (Figure 6A). To simplify cytokine array data, principle component analysis (PCA) was used to visualize compare overall cytokine profiles.
The placement of PCA points in figure 6B showed that the cytokine profiles of each sample generally correspond to its treatment: control PCA placements were largely closest to each other as was the case with +MΦ, +lnf, and +MΦ+Inf samples (Figure 6B). Hierarchical clusters were formed based on cytokine z-scores and used on the PCA plot to highlight distinct groups of samples (Figure 6B). Four clusters were formed, mirroring the treatment groups in this study (Figure 6B). To visualize how each individual cytokine contributed to PCA placements, a biplot showing the magnitude and direction of each cytokine’s contribution to PCA placements was shown (Figure 6C). To simplify the visualization, the 34 cytokines that were statistically significant between groups based on ANOVA were shown (Figure 6C). Comparisons between groups for levels of cytokine secretions were tested for statistical significance via Tukey’s Post Hoc test and all instances of reported higher secretion reflect statistical significance. Both macrophage groups +MΦ and +MΦ+Inf resulted in the higher secretions of ENA-78, G-CSF, GRO a/b/g, IL-10, IL-4, IL-5, MCP-3, MIF, and NAP-2 compared to control and +lnf. Proinflammatory cytokines IL-1β, IL-6, MCP-2, M-CSF, and MIP-1β were the highest secreted in +MΦ+Inf samples while +lnf showed similar levels of these proinflammatory cytokines as control. GCP-2 was the most highly secreted in the +MΦ group. Interestingly, control and +lnf which did not contain macrophages both had higher levels of IFN-γ, IL-13, IL-2, IL-8, TIMP-1, and VEGF-A than +MΦ or +MΦ+Inf groups. The control group had the highest secretion of GROa and MCP-1. All cytokines that were secreted in +lnf groups were secreted similar levels as control except for CXCL10. Both +lnf and +MΦ+Inf samples had similarly higher secretions of CXCL10 compared to control and +MΦ.
Discussion
The secondary layer of macrophages significantly contributed towards inflammatory responses in a 3D tissue model of the large intestine epithelium by demonstrating leukocyte infiltration and pro-inflammatory cytokine secretion. These features have been reported in inflamed intestinal epithelial tissue from IBD patients and in serum samples of IBD patients with active inflammation [12, 37–39], This 3D tissue system, comprised of non-transformed human intestinal epithelia and macrophage immune compartments in a modular scaffold provides a useful 3D epithelial barrier model with utility in IBD and intestinal inflammation studies.
Biomaterial characterization and cell behavior suggested that the lyophilized silk sponge formt was suitable as scaffolding material for engineering an intestinal epithelium with a 3D subepithelial space. Bombyx mori derived silk fibroin is a biocopatible scaffolding material which has been successfully used in intestinal epithelium systems and immune cell co-cultures [21–23, 40], Furthermore, silk is a well characterized bio aterial used in in vivo implants and tissue engineered systems due to its bio-inertness and versatility of material formats including films, hydrogels, and sponges [33], In this study, we used lyophilized silk sponge matrices capable of forming a porous film layer for the cultivation of epithelial surfaces. SEM imaging of silk scaffolds used in this study showed a porous film capable of supporting a monolayer of epithelium for up to 21 days of culture (Figure 1H, I 5A). The formation of monolayers supported the physiological relevance of colonoids as used in this study since the in vivo intestinal epithelium is also a single cell thick layer [18]. Moreover, SEM imaging of the epithelial monolayer at day 3 of differentiation showed dense microvilli formation on the apical side of the colonoid epithelium which is indicative of a functioning and differentiated epithelium [41]. Directly beneath the film structure was the spongey bulk, which presented a porous 3D matrix with interconnected pores which mimicked soft tissue stiffness (Figure 1G,E) [34].
The presence of macrophages on the basal side of the epithelium suggested that the spongey silk construct was capable of supporting macrophage infiltration from the outer layer through the inner layer to the epithelium (Figure 4A,B). Infiltration of macrophages towards an epithelium was a unique feature of this in vitro tissue model which is reflected in intestine tissue samples of IBD patients with active inflammation [12]. In Lissner et al., counts of CD68 positive cells in the tissue layer directly underneath the intestinal epithelium was significantly higher in samples of Crohn’s disease than in normal intestine samples. Intestinal epithelial cells are key regulators of the intestinal immune system, releasing cytokines in response to inflammatory stimuli as well as under normal conditions [18, 42], The release of pro-inflammatory cytokines provides chemotactic cues which cause macrophages to migrate towards the inflamed epithelium.
Evaluation of the epithelial barrier in our tissue model demonstrated physiological responses to inflammation cues and macrophage co-culture. In vitro epithelial coverage was identified as a key metric based on increased epithelial cell death in inflamed IBD tissue compared to uninflamed tissue [43], At both day 3 and 7, +lnf and MΦ+lnf tissue models presented lower epithelial coverage compared to uninflamed models, suggesting increased cell death, recapitulating inflamed tissue characteristics (Figure 5G). Uninflamed models showed improved cell viability when compared to inflamed models (Figure 5G). Furthermore, no significant differences in coverage were detected between control and +MΦ at day 7, suggesting cell death was unaffected by differences in co-culture conditions. Epithelial height was also measured since morphological changes can be observed in macrophage co-cultures and with inflammatory stimuli can be observed in transwell models [13]. Epithelial height was most pronounced in the +MΦ+Inf group, suggesting macrophages and inflammation contributed to differences in epithelial morphology (Figure 5F). Histological sections of epithelia at day 7 reflected morphological changes with ball-like structures present in +MΦ+Inf groups instead of a monolayer as seen in the control or +MΦ groups (Figure 5D). Mucus thickness was measured since cytokines such as IL-6, 10, and 33 are known to influence the secretion of mucin or the proliferation of intestinal goblet cells [44–46], While the presence of a mucus layer was confirmed and its thickness measured, no significant differences were detected between the study groups, suggesting effects on mucus thickness may require a longer time to reflect in vivo mucus layer thickness changes observed in vivo (Figure 5H).
Cytokines mediate inflammatory responses as well as immune functions under homeostasis, as a result systemic levels of specific cytokines differ between patients with IBD and healthy patients [38, 48]. Our tissue models also reflected differences in secreted cytokines with each study group presenting unique cytokine profiles, however, the addition of macrophages contributed to a more physiologically relevant cytokine profile in both the uninflamed and inflamed cases (Figure 5B, C). Statistical significance of cytokine levels between groups was tested via ANOVA followed by Tukey’s Post Hoc test and all instances of reported higher secretion reflect statistical significance. Control and +lnf groups which did not have macrophages had higher levels (compared to +MΦ and +MΦ+Inf) of pleiotropic cytokine IL-2 [49], extracellular remodeling cytokine TIMP-1 [50], and angiogenic cytokines IL-8 and VEGF-A [51, 52]. Interestingly, the control group also expressed the highest levels of proinflammatory protein MCP-1 in addition to angiogenic cytokine GROa, a CXC chemokine which binds to IL8RB and mediates the same angiogenic activity as IL-8 [51, 53], The effects of inflammatory cues on the colonoid epithelium without macrophages showed elevated secretion of only CXCL10, a pro-inflammatory cytokine that is elevated in IBD patient sera [38], However, levels of CXCL10 were statistically significant and similarly high in the +MΦ+Inf group compared to control (Figure 6A, C). Overall, the groups without macrophages (control and +Inf) displayed statistically significant higher expression of cytokines mediating angiogenesis which was not reflected in the groups with macrophages (+MΦ and +MΦ+Inf). Elevated cytokines as confirmed by statistical significance via Tukey’s Post Hoc test in both +MΦ and +MΦ+Inf Models included pro-inflammatory cytokine MIF [54], anti-inflammatory cytokine IL-10 and IL-4 [55, 56], monocyte chemotactic cytokines MCP-3 and GRO a/b/g [57–59], neutrophil chemotactic cytokine ENA-78/CXCL5 and NAP-2 [57, 60], and granulocyte and neutrophil mediating cytokine G-CSF [61]. Pro-inflammatory cytokines with higher expression in patients biopsy samples or sera with active IBD compared to healthy patients including IL-1β [38], IL-6 [48, 62], MCP-2 [63], and MIP-1β [63] were statistically significant and highly expressed in the +MΦ+Inf samples compared to all other groups. Elevated secretion of multiple pro-inflammatory cytokines associated with active IBD in +MΦ+Inf samples but not in +Inf samples suggested that the macrophage laden outer layer enhanced the biochemical response and as a result, the physiologic relevance of the 3D colonoid model.
Conclusions
Our in vitro model displays phenotypic responses to inflammation through epithelial barrier disruption, macrophage migration, and the secretion of cytokines associated with IBD. Culture of the colonoid monolayers on the silk scaffolds resulted in a polarized epithelium displaying microvilli on the apical surface with infiltration of macrophages from the outer layer to the basal surface of the epithelium. Our results confirmed a need for an immune cell component in intestinal epithelial models and demonstrates the utility of this tissue system consisting of colonoids and macrophages, versus an epithelium only model, in evaluating the effects of inflammatory cues on epithelial morphological features and biochemical composition. Further development of the in vitro tissue model system to enable longer-term cultures for the study of chronic effects of inflammation are suggested based on the responses reported here. As a tissue platforM for studying epithelial-immune reactions against inflammatory cues, this in vitro model may be used for studying the response of a human intestinal epithelium and immune system against acute pathogenic insults and modulation of active inflammation in IBD.
Supplementary Material
Funding Sources:
Support from the NIH (P41EB002520, U19-AI131126) and the Air Force Office of Scientific Research, National Defense Science and Engineering Graduate (NDSEG) Fellowship is greatly appreciated. This work was also supported by the NIH Research Infrastructure grant NIH S10 OD021624. Scanning electron microscopy was performed at the Center for Nanoscale Systems (CNS), a member of the National Nanotechnology Infrastructure Network (NNIN). CNS is part of the Faculty of Arts and Sciences at Harvard University.
Footnotes
Declarations of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Sender R, Fuchs S, Milo R, Revised Estimates for the Number of Human and Bacteria Cells in the Body, PLOS Biology 14(8) (2016) e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Thursby E, Juge N, Introduction to the human gut microbiota, Biochem J 474(11) (2017) 1823–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mowat AM, Agace WW, Regional specialization within the intestinal immune system, Nature Reviews Immunology 14 (2014) 667. [DOI] [PubMed] [Google Scholar]
- [4].Davies LC, Jenkins SJ, Allen JE, Taylor PR, Tissue-resident macrophages, Nature immunology 14(10) (2013) 986–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kühl AA, Erben U, Kredel LI, Siegmund B, Diversity of Intestinal Macrophages in Inflammatory Bowel Diseases, Frontiers in immunology 6 (2015) 613–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mowat AM, Scott CL, Bain CC, Barrier-tissue macrophages: functional adaptation to environmental challenges, Nature Medicine 23 (2017) 1258. [DOI] [PubMed] [Google Scholar]
- [7].Smith PD, Smythies LE, Shen R, Greenwell-Wild T, Gliozzi M, Wahl SM, Intestinal macrophages and response to microbial encroachment, Mucosal Immunology 4 (2010) 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lee SH, Starkey PM, Gordon S, Quantitative analysis of total macrophage content in adult mouse tissues. ImmunocheMical studies with monoclonal antibody F4/80, The Journal of experiMental medicine 161(3) (1985) 475–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lahiri A, Abraham C, Activation of pattern recognition receptors up-regulates metallothioneins, thereby increasing intracellular accumulation of zinc, autophagy, and bacterial clearance by macrophages, Gastroenterology 147(4) (2014) 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].D’Angelo F, Bernasconi E, Schäfer M, Moyat M, Michetti P, Maillard MH, Velin D, Macrophages promote epithelial repair through hepatocyte growth factor secretion, Clinical & Experimental Immunology 174(1) (2013) 60–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ortiz-Masiá D, Cosin-Roger J, Calatayud S, Hernández C, Alós R, Hinojosa J, Apostolova N, Alvarez A, Barrachina MD, Hypoxic macrophages impair autophagy in epithelial cells through Wnt1: relevance in IBD, Mucosal Immunology 7 (2013) 929. [DOI] [PubMed] [Google Scholar]
- [12].Lissner D, Schumann M, Batra A, Kredel L-I, Kühl AA, Erben U, May C, Schulzke J-D, Siegmund B, Monocyte and M1 Macrophage-induced Barrier Defect Contributes to Chronic Intestinal Inflammation in IBD, Inflammatory Bowel Diseases 21(6) (2015) 1297–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Noel G, Baetz NW, Staab JF, Donowitz M, Kovbasnjuk O, Pasetti MF, Zachos NC, A priMary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions, Scientific Reports 7 (2017) 45270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kim HJ, Li H, Collins JJ, Ingber DE, Contributions of microbiome and mechanical deforMation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip, Proceedings of the National Academy of Sciences 113(1) (2016) E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dosh RH, Jordan-Mahy N, Sammon C, Le Maitre CL, Long-term in vitro 3D hydrogel co-culture model of inflammatory bowel disease, Scientific Reports 9(1) (2019) 1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sehgal A, Donaldson DS, Pridans C, Sauter KA, Hume DA, Mabbott NA, The role of CSF1R-dependent macrophages in control of the intestinal stem-cell niche, Nature Communications 9(1) (2018) 1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sato T, Stange DE, Ferrante M, Vries RGJ, van Es JH, van den Brink S, van Houdt WJ, Pronk A, van Gorp J, Siersema PD, Clevers H, Long-term Expansion of Epithelial Organoids From Human Colon, Adenoma, Adenocarcinoma, and Barrett’s Epithelium, Gastroenterology 141(5) (2011) 1762–1772. [DOI] [PubMed] [Google Scholar]
- [18].Peterson LW, Artis D, Intestinal epithelial cells: regulators of barrier function and immune homeostasis, Nature Reviews Immunology 14 (2014) 141. [DOI] [PubMed] [Google Scholar]
- [19].Weber CR, Turner JR, Inflammatory bowel disease: is it really just another break in the wall?, Gut 56(1) (2007) 6–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Andrews C, McLean MH, Durum SK, Cytokine Tuning of Intestinal Epithelial Function, Frontiers in immunology 9 (2018) 1270–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chen Y, Zhou W, Roh T, Estes MK, Kaplan DL, In vitro enteroid-derived three-dimensional tissue model of human small intestinal epithelium with innate immune responses, PLoS ONE 12(11) (2017) e0187880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen Y, Lin Y, Wang Q, Rnjak-Kovacina J, Li C, Isberg RR, Kumamoto CA, Mecsas J, Kaplan D, Robust bioengineered 3D functional human intestinal epithelium, 2015. [DOI] [PMC free article] [PubMed]
- [23].Zhou W, Chen Y, Roh T, Lin Y, Ling S, Zhao S, Lin JD, Khalil N, Cairns DM, Manousiouthakis E, Tse M, Kaplan DL, Multifunctional Bioreactor SysteM for Human Intestine Tissues, ACS Biomaterials Science & Engineering 4(1) (2018) 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Barthes J, Dollinger C, Muller CB, Liivas U, Dupret-Bories A, Knopf-Marques H, Vrana NE, Immune Assisted Tissue Engineering via Incorporation of Macrophages in Cell-Laden Hydrogels Under Cytokine Stimulation, Frontiers in bioengineering and biotechnology 6 (2018) 108–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dohle E, Bischoff I, Bose T, Marsano A, Banfi A, Unger RE, Kirkpatrick CJ, Macrophage-mediated angiogenic activation of outgrowth endothelial cells in co-culture with primary osteoblasts, European cells & materials 27 (2014) 149–64; discussion 164–5. [DOI] [PubMed] [Google Scholar]
- [26].Dollinger C, Ciftci S, Knopf-Marques H, Guner R, GhaemmaghaMi AM, Debry C, Barthes J, Vrana NE, Incorporation of resident macrophages in engineered tissues: Multiple cell type response to microenvironment controlled macrophage-laden gelatine hydrogels, Journal of Tissue Engineering and Regenerative Medicine 12(2) (2018) 330–340. [DOI] [PubMed] [Google Scholar]
- [27].Bain CC, Bravo-Blas A, Scott CL, Gomez Perdiguero E, Geissmann F, Henri S, Malissen B, Osborne LC, Artis D, Mowat AM, Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice, Nature Immunology 15 (2014)929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Thiesen S, Janciauskiene S, Uronen-Hansson H, Agace W, Högerkorp C-M, Spee P, Håkansson K, Grip O, CD14hiHLA-DRdim macrophages, with a resemblance to classical blood monocytes, dominate inflamed mucosa in Crohn’s disease, Journal of Leukocyte Biology 95(3) (2014)531–541. [DOI] [PubMed] [Google Scholar]
- [29].Grainger JR, Wohlfert EA, Fuss IJ, Bouladoux N, Askenase MH, Legrand F, Koo LY, Brenchley JM, Fraser IDC, Belkaid Y, Inflammatory monocytes regulate pathologic responses to commensals during acute gastrointestinal infection, Nature medicine 19(6) (2013) 713–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Asano K, Takahashi N, Ushiki M, Monya M, Aihara F, Kuboki E, Moriyama S, Iida M, Kitamura H, Qiu C-H, Watanabe T, Tanaka M, Intestinal CD169(+) macrophages initiate mucosal inflammation by secreting CCL8 that recruits inflammatory monocytes, Nature communications 6 (2015) 7802–7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bain CC, Mowat AM, Macrophages in intestinal homeostasis and inflammation, Immunol Rev 260(1) (2014) 102–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].In J, Foulke-Abel J, Zachos NC, Hansen A-M, Kaper JB, Bernstein HD, Halushka M, Blutt S, Estes MK, Donowitz M, Kovbasnjuk O, Enterohemorrhagic Escherichia coli reduce mucus and intermicrovillar bridges in human stem cell-derived colonoids, Cellular and molecular gastroenterology and hepatology 2(1) (2015) 48–62.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rockwood DN, Preda RC, Yücel T, Wang X, Lovett ML, Kaplan DL, Materials fabrication from Bombyx mori silk fibroin, Nature Protocols 6(10) (2011) 1612–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Feiner R, Dvir T, Tissue-electronics interfaces: from implantable devices to engineered tissues, Nature Reviews Materials 3 (2017) 17076. [Google Scholar]
- [35].Rnjak-Kovacina J, Wray LS, Burke KA, Torregrosa T, Golinski JM, Huang W, Kaplan DL, Lyophilized Silk Sponges: A Versatile Biomaterial Platform for Soft Tissue Engineering, ACS biomaterials science & engineering 1(4) (2015) 260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H, Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche, Nature 459 (2009) 262. [DOI] [PubMed] [Google Scholar]
- [37].Blander JM, Death in the intestinal epithelium-basic biology and implications for inflammatory bowel disease, The FEBS journal 283(14) (2016) 2720–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Singh UP, Singh NP, Murphy EA, Price RL, Fayad R, Nagarkatti M, Nagarkatti PS, Chemokine and cytokine levels in inflammatory bowel disease patients, Cytokine 77 (2016) 44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Neurath MF, Cytokines in inflammatory bowel disease, Nature Reviews Immunology 14 (2014)329. [DOI] [PubMed] [Google Scholar]
- [40].Vidal SEL, Tamamoto KA, Nguyen H, Abbott RD, Cairns DM, Kaplan DL, 3D biomaterial matrix to support long terM, full thickness, immuno-competent human skin equivalents with nervous system components, Biomaterials (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Crawley SW, Mooseker MS, Tyska MJ, Shaping the intestinal brush border, The Journal of Cell Biology 207(4) (2014) 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Onyiah JC, Colgan SP, Cytokine responses and epithelial function in the intestinal mucosa, Cell mol Life Sci 73(22) (2016) 4203–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Di Sabatino A, Ciccocioppo R, Luinetti O, Ricevuti L, Morera R, Cifone MG, Solcia E, Corazza GR, Increased Enterocyte Apoptosis in Inflamed Areas of Crohn’s Disease, Diseases of the Colon & Rectum 46(11) (2003) 1498–1507. [DOI] [PubMed] [Google Scholar]
- [44].Hasnain SZ, Tauro S, Das I, Tong H, Chen ACH, Jeffery PL, McDonald V, Florin TH, McGuckin MA, IL-10 Promotes Production of Intestinal Mucus by Suppressing Protein Misfolding and Endoplasmic Reticulum Stress in Goblet Cells, Gastroenterology 144(2) (2013) 357–368.e9. [DOI] [PubMed] [Google Scholar]
- [45].Kuhn KA, Schulz HM, Regner EH, Severs EL, Hendrickson JD, Mehta G, Whitney AK, Ir D, Ohri N, Robertson CE, Frank DN, Campbell EL, Colgan SP, Bacteroidales recruit IL-6-producing intraepithelial lymphocytes in the colon to promote barrier integrity, Mucosal immunology 11(2) (2018) 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Waddell A, Vallance JE, Moore PD, Hummel AT, Wu D, Shanmukhappa SK, Fei L, Washington MK, Minar P, Coburn LA, Nakae S, Wilson KT, Denson LA, Hogan SP, Rosen MJ, IL-33 Signaling Protects from murine Oxazolone Colitis by Supporting Intestinal Epithelial Function, Inflammatory bowel diseases 21(12) (2015) 2737–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kämpfer AAM, Urbán P, Gioria S, Kanase N, Stone V, Kinsner-Ovaskainen A, Development of an in vitro co-culture model to, mimic the human intestine in healthy and diseased state, Toxicol In Vitro 45(Pt 1) (2017) 31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mudter J, Neurath MF, Il-6 signaling in inflammatory bowel disease: Pathophysiological role and clinical relevance, Inflammatory Bowel Diseases 13(8) (2007) 1016–1023. [DOI] [PubMed] [Google Scholar]
- [49].Arenas-Ramirez N, Woytschak J, Boyman O, Interleukin-2: Biology, Design and Application, Trends in Immunology 36(12) (2015) 763–777. [DOI] [PubMed] [Google Scholar]
- [50].Brew K, Dinakarpandian D, Nagase H, Tissue inhibitors of metalloproteinases: evolution, structure and function 11 Dedicated to Professor H. Neurath on the occasion of his 90th birthday, Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology 1477(1) (2000) 267–283. [DOI] [PubMed] [Google Scholar]
- [51].Inoue K, Slaton JW, Eve BY, Kim SJ, Perrotte P, Balbay MD, Yano S, Bar-Eli M, Radinsky R, Pettaway CA, Dinney CPN, Interleukin 8 Expression Regulates Tumorigenicity and Metastases in Androgen-independent Prostate Cancer, Clinical Cancer Research 6(5) (2000)2104. [PubMed] [Google Scholar]
- [52].Hoeben A, Landuyt B, Highley MS, Wildiers H, Van Oosterom AT, De Bruijn EA, Vascular Endothelial Growth Factor and Angiogenesis, Pharmacological Reviews 56(4) (2004) 549. [DOI] [PubMed] [Google Scholar]
- [53].Miyake M, Goodison S, Urquidi V, GoMes Giacoia E, Rosser CJ, Expression of CXCL1 in human endothelial cells induces angiogenesis through the CXCR2 receptor and the ERK1/2 and EGF pathways, Laboratory Investigation 93 (2013) 768. [DOI] [PubMed] [Google Scholar]
- [54].Calandra T, Roger T, Macrophage migration inhibitory factor: a regulator of innate immunity, Nature Reviews Immunology 3(10) (2003) 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Krause P, Morris V, Greenbaum JA, Park Y, Bjoerheden U, Mikulski Z, Muffley T, Shui J-W, Kim G, Cheroutre H, Liu Y-C, Peters B, Kronenberg M, Murai M, IL-10-producing intestinal macrophages prevent excessive antibacterial innate immunity by limiting IL-23 synthesis, Nature communications 6 (2015) 7055–7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hart PH, Vitti GF, Burgess DR, Whitty GA, Piccoli DS, Hamilton JA, Potential antiinflammatory effects of interleukin 4: suppression of human Monocyte tumor necrosis factor alpha, interleukin 1, and prostaglandin E2, Proceedings of the National Academy of Sciences of the United States of America 86(10) (1989) 3803–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ahuja SK, Murphy PM, The CXC Chemokines Growth-regulated Oncogene (GRO) α, GROβ, GROγ, Neutrophil-activating Peptide-2, and Epithelial Cell-derived Neutrophil-activating Peptide-78 Are Potent Agonists for the Type B, but Not the Type A, Human Interleukin-8 Receptor, Journal of Biological Chemistry 271(34) (1996) 20545–20550. [DOI] [PubMed] [Google Scholar]
- [58].Persson T, Monsef N, Andersson P, Bjartell A, Malm J, Calafat J, Egesten A, Expression of the neutrophil-activating CXC chemokine ENA-78/CXCL5 by human eosinophils, Clinical & Experimental Allergy 33(4) (2003) 531–537. [DOI] [PubMed] [Google Scholar]
- [59].Xuan W, Qu Q, Zheng B, Xiong S, Fan G-H, The Chemotaxis of M1 and M2 macrophages is regulated by different chemokines, Journal of Leukocyte Biology 97(1) (2015) 61–69. [DOI] [PubMed] [Google Scholar]
- [60].Walz A, Dewald B, von Tscharner V, Baggiolini M, Effects of the neutrophil-activating peptide NAP-2, platelet basic protein, connective tissue-activating peptide III and platelet factor 4 on human neutrophils, The Journal of experimental medicine 170(5) (1989) 1745–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Roberts AW, G-CSF: A key regulator of neutrophil production, but that’s not all!, Growth Factors 23(1) (2005) 33–41. [DOI] [PubMed] [Google Scholar]
- [62].Suzuki Y, Saito H, Kasanuki J, Kishimoto T, Tamura Y, Yoshida S, Significant increase of interleukin 6 production in blood mononuclear leukocytes obtained from patients with active imflammatory bowel disease, Life Sciences 47(24) (1990) 2193–2197. [DOI] [PubMed] [Google Scholar]
- [63].Banks C, Bateman A, Payne R, Johnson P, Sheron N, Chemokine expression in IBD. Mucosal chemokine expression is unselectively increased in both ulcerative colitis and Crohn’s disease, The Jousrnal of Pathology 199(1) (2003) 28–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.