Highlights
-
•
Automatic intracranial volume segmentation.
-
•
Fetal and neonatal MRI.
-
•
Deep learning.
Keywords: Brain extraction, Neonatal MRI, Fetal MRI, Skull stripping, Brain segmentation, Deep learning, Intracranial volume segmentation
Abstract
MR images of infants and fetuses allow non-invasive analysis of the brain. Quantitative analysis of brain development requires automatic brain tissue segmentation that is typically preceded by segmentation of the intracranial volume (ICV). Fast changes in the size and morphology of the developing brain, motion artifacts, and large variation in the field of view make ICV segmentation a challenging task.
We propose an automatic method for segmentation of the ICV in fetal and neonatal MRI scans. The method was developed and tested with a diverse set of scans regarding image acquisition parameters (i.e. field strength, image acquisition plane, image resolution), infant age (23–45 weeks post menstrual age), and pathology (posthaemorrhagic ventricular dilatation, stroke, asphyxia, and Down syndrome). The results demonstrate that the method achieves accurate segmentation with a Dice coefficient (DC) ranging from 0.98 to 0.99 in neonatal and fetal scans regardless of image acquisition parameters or patient characteristics. Hence, the algorithm provides a generic tool for segmentation of the ICV that may be used as a preprocessing step for brain tissue segmentation in fetal and neonatal brain MR scans.
1. Introduction
Magnetic resonance imaging (MRI) is a clinically used non-invasive tool for monitoring brain development in fetuses and neonates. The analysis usually comprises of quantification of brain tissue volumes and cortical morphology to extract meaningful information for diagnosis or prognosis (Claessens et al., 2016, Drost et al., 2018, Dubois, Benders, Cachia, Lazeyras, Ha-Vinh Leuchter, Sizonenko, Borradori-Tolsa, Mangin, Hüppi, 2007, Inder, Huppi, Warfield, Kikinis, Zientara, Barnes, Jolesz, Volpe, 1999, Kersbergen, Leroy, Išgum, Groenendaal, de Vries, Claessens, van Haastert, Moeskops, Fischer, Mangin, et al., 2016, Moeskops, Benders, Kersbergen, Groenendaal, de Vries, Viergever, Išgum, 2015, Moeskops, Išgum, Keunen, Claessens, Haastert, Groenendaal, Vries, Viergever, Benders, 2017). Automatic quantification of these indices requires segmentation of brain tissue classes. To allow dedicated analysis within the brain, automatic methods typically perform extraction of the intracranial volume (ICV) prior to further analysis (Išgum, Benders, Avants, Cardoso, Counsell, Gomez, Gui, Hűppi, Kersbergen, Makropoulos, et al., 2015, Moeskops, Viergever, Mendrik, de Vries, Benders, Išgum, 2016).
A number of methods for segmentation of ICV in adult MR scans have been applied to analysis of T1- and T2-weighted neonatal MR images (Eskildsen, Coupé, Fonov, Manjón, Leung, Guizard, Wassef, Østergaard, Collins, The Alzheimer’s Disease Neuroimaging Initiative, 2012, Iglesias, Liu, Thompson, Tu, 2011, Ségonne, Dale, Busa, Glessner, Salat, Hahn, Fischl, 2004, Smith, 2002). Brain Extraction Tool (BET) Smith (2002) is a publicly available tool used as a preprocessing step by many automatic brain segmentation methods (Išgum et al., 2015). BET iteratively deforms a sphere to fit it on the brain surface using a geometric algorithm. Robust Brain Extraction tool (ROBEX) is another commonly used and publicly available tool for segmentation of the ICV in adult MR images (Iglesias et al., 2011). ROBEX first employs a Random Forest classifier to detect the brain boundary and thereafter uses a point distribution model that ensures a plausible result. Furthermore, Brain Extraction based on non-local Segmentation Technique (BEaST) is a publicly available tool for ICV segmentation (Eskildsen et al., 2012). BEaST is a patch-based segmentation method exploiting the similarity between the patches in the region of interest and predefined patches in a library.
Because of the lack of publicly available tools developed for ICV segmentation of neonatal brain MRI, these methods designed to analyze brain MR scans of adults are frequently used to segment the ICV in neonatal scans. Consequently, they generally do not produce highly accurate results when applied to neonatal brain MR scans (Yamaguchi et al., 2010). Moreover, these methods typically fail when applied to fetal MR scans. Hence, several methods specifically designed to extract the ICV in MR scans of neonates have been proposed. Serag et al. (2016); Yamaguchi et al. (2010) proposed a method for segmentation of the ICV in brain MRI of neonates and children aged between 36 weeks post menstrual age (PMA) and 4 years. The method uses fuzzy logic and it is applicable to images without severe pathology acquired sagittally. In the first step the intensity distributions of white matter (WM), gray matter (GM), cerebrospinal fluid (CSF), fat, and other tissues visible in the scan are estimated using Bayesian classification and a Gaussian mixture model. Segmentation of brain tissue classes is thereafter performed by means of a fuzzy active surface model using distributions of WM, GM and CSF from the previous step. The qualitative evaluation of this method demonstrated improved performance over BET. Later, Mahapatra (2012) proposed a shape model with graph cuts for segmentation of the ICV in neonatal MRI. The shape model is generated by averaging manually labeled images which is afterwards used with graph cut for segmentation. This method was applied to term-born infants imaged at about three weeks of age. Serag et al. (2016) proposed an atlas-based segmentation of the ICV. To eliminate the need for representative training data i.e. data coming from the same distribution, atlases that are uniformly distributed were selected. The algorithm was applied to T1-weighted and T2-weighted MR scans without visible pathology of preterm infants scanned at term equivalent age. The method showed high segmentation accuracy and it outperformed publicly available tools such as BET and ROBEX.
Similar to methods dedicated to segmentation of the ICV in neonatal MR scans, a number of studies proposed segmentation of the ICV in fetal MRI. Anquez et al. (2009) proposed a method that first localizes the eyes and exploits this information to segment the ICV using a graph cut approach guided by shape, contrast, and biometrical priors. The method was applied to scans with unknown fetal orientation and the results demonstrated high segmentation accuracy.
In recent years, convolutional neural networks (CNNs) have become the most popular method for automatic image segmentation in medical images (Litjens et al., 2017). Several studies investigated different CNN architectures for brain tissue segmentation (Akkus, Galimzianova, Hoogi, Rubin, Erickson, 2017, Chen, Dou, Yu, Qin, Heng, 2018, Dolz, Desrosiers, Ayed, 2018, Makropoulos, Counsell, Rueckert, 2018, Tu, Bai, 2010) and brain extraction (Dey, Hong, Dolz, Desrosiers, Wang, Yuan, Shen, Ayed, Kleesiek, Urban, Hubert, Schwarz, Maier-Hein, Bendszus, Biller, 2016) in adult MRI. Wachinger et al. (2018) proposed a network that combines brain extraction and brain tissue segmentation.
A few studies used CNNs to segment ICV from fetal or neonatal MRI. Rajchl et al. (2017) proposed a weakly supervised deep learning approach for ICV segmentation in fetal MRI that combines a convolutional neural network and iterative graph optimization. The network was trained with bounding boxes around the brain as weak labels. The method was applied to fetal MR scans and achieved high segmentation accuracy. In another study, Rajchl et al. (2016) investigated the use of crowd sourcing platform for ICV segmentation of fetal MRI using convolutional neural network. Salehi et al. (2017) proposed an iterative deep learning segmentation method that uses U-net-like convolutional neural network (Auto-net). In this approach, the fetal brain is segmented from a localized bounding box which was defined manually using ITKSNAP (Yushkevich et al., 2006). In a subsequent study, Salehi et al. (2018) evaluated Auto-net on fetal MRI without any preprocessing steps such as defining a bounding box. The method was trained on a very large number of manually annotated fetal MRI and demonstrated accurate segmentation results in fetal scans. Recently, Khalili et al. (2017) proposed multi-scale convolutional neural network for ICV segmentation of fetal MRI.
Unlike methods performing ICV segmentation directly, several methods perform brain localization as a step prior to fetal ICV segmentation (Ison, Donner, Dittrich, Kasprian, Prayer, Langs, 2012, Keraudren, Kyriakopoulou, Rutherford, Hajnal, Rueckert, 2013, Keraudren, Kuklisova-Murgasova, Kyriakopoulou, Malamateniou, Rutherford, Kainz, Hajnal, Rueckert, 2014, Taimouri et al., 2015). Recently, Tourbier et al. (2017) proposed a pipeline that sequentially performs ICV localization, ICV segmentation and super-resolution reconstruction in fetal MR scans. In this method a template-matching approach, with age as prior knowledge, is used to segment the ICV in fetal MRI. A limitation of template based techniques is that they are typically computationally more expensive than machine learning algorithms. In addition, they have a high chance of failure if representative age-matched templates are not available. Moreover, to segment brain tissue classes, methods employing brain localization require subsequent segmentation of the ICV.
All aforementioned methods were evaluated either on neonatal or fetal MR scans, without visible pathology. To the best of our knowledge, thus far no study proposed a generic method that performs segmentation of the ICV in neonatal and fetal MRI. In this study, we propose a method for automatic segmentation of the ICV in neonatal and fetal T2-weighted MR scans that is robust to imaging parameters (field strength, image acquisition plane, image resolution), and pathology and patient characteristics (posthaemorrhagic ventricular dilatation (PHVD), stroke, asphyxia, Down syndrome). The method employs a convolutional neural network with a U-net architecture (Ronneberger et al., 2015). The network was trained with a combination of fetal and preterm born neonatal scans acquired in axial, coronal and sagittal orientation. The age of patients at the time of scanning in the training set ranged from 23 to 35 weeks PMA. The method was evaluated using images of fetuses and infants between 23 weeks PMA and 3 months of age at the time of scanning, ranging from absence of visible pathology to presence of severe pathology such as stroke or PHVD. This work builds upon our preliminary study that described segmentation of the ICV in fetal MRI using a multi-scale convolutional neural network (Khalili et al., 2017).
2. Data
In this study a diverse set of fetal and neonatal T2-weighted MR scans was used. Fetal scans were acquired in axial, sagittal and coronal image planes and did not contain visible pathology. Neonatal images include scans of preterm and term-born infants. The scans were acquired in axial or coronal image planes, and include images without and with pathology. Examples of fetal and neonatal images included in the study are illustrated in Fig. 1. As shown in the figure, fetal MRIs have a larger field of view that visualizes the entire fetus as well as parts of the maternal body. Moreover, we include scans which were acquired with different scanner-vendors (Philips, Siemens) and field strength (1,5T and 3T). The neuroimaging data were obtained as part of the clinical protocol, written informed consent for use of the clinically acquired data and approval of the experiments and methodology was waived by the institutional review board of the University Medical Center Utrecht, The Netherlands.
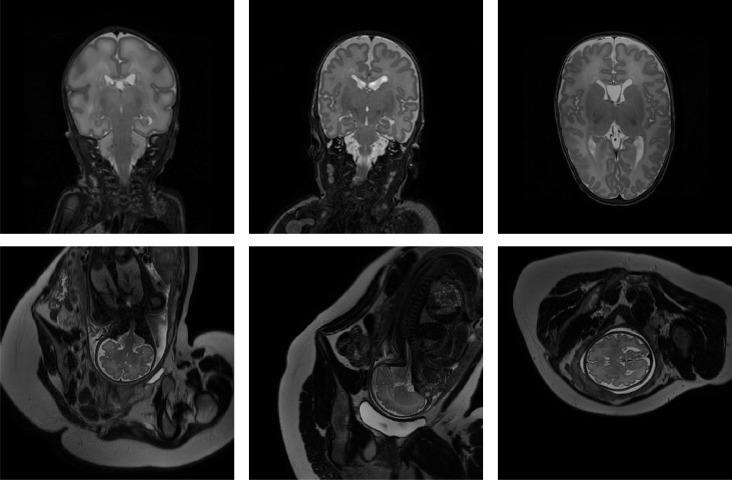
Fig. 1.
Examples of preterm neonatal and fetal MR scans included in the study. Top: coronal MRI acquired at 30 weeks PMA (left), coronal MRI acquired at 40 weeks PMA (middle), axial MRI acquired at 40 weeks PMA (right). Bottom: fetal MRI acquired in coronal (left), sagittal (middle) and axial (right) directions.
2.1. Fetal MRI
Two sets of fetal MR scans were used. The first set (Set 1) includes T2-weighted MR scans of fetuses (age: 23–35 weeks PMA). Images were acquired on a Philips Achieva 3T scanner at the University Medical Center (UMC) Utrecht, Utrecht, the Netherlands using a turbo fast spin-echo sequence. The dataset contains 45 scans in total: 17 scans acquired in axial direction, 15 scans in coronal direction and 13 scans in sagittal direction. The images were acquired with voxel sizes of 1.25 × 1.25 × 2.5 mm3 and reconstructed to 0.7 × 0.7 × 1.25 mm3 with reconstruction matrix of 512 × 512 × 80. The scans were reconstructed by the scanners algorithm and no further reconstruction (e.g. super-resolution processing) of the acquired images was performed. Furthermore, the proposed approach was applied to the 2D MRI slices without any prepossessing steps such as intensity inhomogeneity or motion correction.
The second set (Set 2) contains publicly available T2-weighted MR scans of 17 fetuses (age range: 29 ± 5 weeks PMA) which present a subset of scans described by Salehi et al. (2018). Scans were acquired on a 3T Siemens Skyra scanner at Boston Childeren’s Hospital, Boston, US in axial, sagittal and coronal direction. The scans were acquired with voxel sizes of 1 × 1 × 2 mm3 with a reconstruction matrix of 256 × 256; the number of slices varied from 48 to 54.
The third set (Set 3) includes fetal T2-weighted MR scans acquired on Philips Achieva 1.5T scanner at the UMC Utrecht, Utrecht, the Netherlands. The dataset contains 18 scans: 6 scans were acquired in axial direction, 6 in coronal and 6 in sagittal direction. The scans were reconstructed to a voxel size of 1.18 × 1.18 × 1.25 mm3 and reconstruction matrix of 288 × 288 × 80.
2.2. Neonatal MRI
All neonatal scans were acquired on a Philips Achieva 3T scanner at the University Medical Center Utrecht, Utrecht, the Netherlands. We divided the data according to age of the infants at the time of acquisition, image acquisition plane, and presence and type of visible pathology. As shown in Fig. 1, there are variations in the neonatal scans, especially between 30 and 40 weeks PMA, when the brains exhibit important structural development, including cortical folding, and changes in shape and volume.
2.2.1. Preterm born infants without visible pathology
This set consists of three different subsets. The first one - 30-weeks coronal MRI - comprises 20 scans of preterm born infants imaged at 30 weeks PMA. The second set - 40-weeks coronal MRI - contains 17 scans of preterm born infants imaged at term equivalent age. The third set - 40-weeks axial MRI - contains 15 scans of preterm born infants imaged at term equivalent age. This set includes all 22 scans from the NeoBrainS12 challenge. Detailed data description is provided in a former study (Išgum et al., 2015).
2.2.2. Cross-sectional cohort
A set of 10 T2-weighted MRI scans were taken from a study investigating neonatal brain development that were made shortly after birth (29–43 weeks PMA) Keunen (2017). The scans were selected to include images of 10 neonates covering the complete available infant age range. Hence, this set includes preterm and full-term born infants.
2.2.3. Infants with congenital heart disease (CHD)
The set consists of 10 T2-weighted MRI scans of 10 patients with critical congenital heart disease (CHD). These infants were scanned before and after univentricular or biventricular cardiac repair using cardiopulmonary bypass within the first 30 days of life (Claessens et al., 2018). We selected 5 scans made before and 5 scans after surgery, of different patients. The images visualized WM lesions indicating mild to moderate brain injuries. However, the brain morphology was not significantly altered.
2.2.4. Infants with PHVD
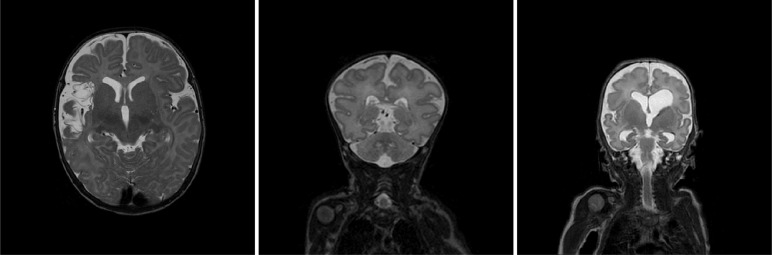
A set of 10 T2-weigted MRI scans of 10 infants with germinal matrix-intraventricular hemorrhage (GMH-IVH) and subsequent PHVD requiring intervention were selected randomly from a clinical study on PHVD infants (Brouwer et al., 2016). The infants included in this study received a ventricular shunt that is next to the substantial ventricular dilatation visible in MR images. An example of this is illustrated in Fig. 2. Note that the ventricles are substantially enlarged typically resulting in a deformed brain shape. Moreover, these patients often have a temporary ventricular shunt which is visible in a number of scan slices.
Fig. 2.
Examples of T2-weighted MR scans of preterm born neonates with ischemic stroke (left), Down syndrome (middle), and PHVD (right).
2.2.5. Infants with stroke
This set consists of 10 T2-weighted MRI scans of 10 infants with arterial ischemic stroke (Benders et al., 2014). These neonates were treated with 1000 IU/kg rhEPO immediately after diagnosis. A secondary MRI was performed when the patients were 3 months of age. We included 5 primary and 5 secondary scans showing WM degradation. Primary and secondary scans were not showing the same patients. Fig. 2 illustrates an example of a secondary scan when the stroke-affected area is filled with CSF.
2.2.6. Infants with asphyxia
This set consists of 9 T2-weighted MRI scans of 9 patients with perinatal asphyxia (Alderliesten et al., 2017). These scans present diffuse hypoxic-ischemic injury demonstrated as hypointensities in the images that can be present throughout the brain tissue.
2.2.7. Infants with down syndrome (DS)
This set consist of 10 T2-weighted MRI scans imaging 10 infants with Down syndrome. In these patients, the brain volume is smaller because of delayed brain growth and gyrification compared with healthy infants (Coyle et al., 1986). Fig. 2 illustrates a typical example of a Down syndrome infant demonstrating abnormal shape of the head, the brain and delayed gyrifcation.
2.2.8. Infant scans with artifacts
This set consist of 10 T2-weighted MRI scans acquired in coronal orientation from preterm born infants imaged at term equivalent age (40 weeks of post menstrual age). 5 scans contain intensity inhomogeneity artifacts and 5 scans show motion artifacts.
Details on image acquisition parameters for all sets are listed in Table 1.
Table 1.
Parameters of neonatal MRI scans. For each set the table lists total number of scans (Nr) in a set, average and standard deviation of the infant age at the time of scanning expressed in weeks of PMA (Age), image acquisition (Orientation) as axial (Ax) or coronal (Cor), reconstruction matrix (Matrix), reconstructed voxel sizes expressed in mm and the number of scans used in training and test set (Training/Test).
| Nr | Age | Orientation | Matrix | Voxel size | Training / test | |
|---|---|---|---|---|---|---|
| 30-weeks coronal | 20 | 30.7 ± 1.0 | Cor | 384 × 384 × 50 | 0.34 × 0.34 × 2.0 | 3 / 17 |
| 40-weeks coronal | 15 | 41.2 ± 0.9 | Cor | 512 × 512 × 110 | 0.35 × 0.35 × 1.2 | 3 / 12 |
| 40-weeks axial | 17 | 41.3 ± 0.8 | Ax | 512 × 512 × 50 | 0.35 × 0.35 × 2.0 | 3 / 14 |
| Cross-sectional cohort | 10 | 36.9 ± 5.0 | Cor | 512 × 512 × 110 | 0.35 × 0.35 × 1.2 | 0 / 10 |
| Infants with CHD | 10 | 41.0 ± 1.7 | Cor | 512 × 512 × 110 | 0.35 × 0.35 × 1.2 | 0 / 10 |
| Infants with PHVD | 10 | 41.0 ± 0.7 | Cor | 512 × 512 × 110 | 0.35 × 0.35 × 1.2 | 0 / 10 |
| Infants with stroke | 10 | 44.1 ± 6.2 | Ax | 512 × 512 × 50 | 0.35 × 0.35 × 2.0 | 0 / 10 |
| Infants with asphyxia | 9 | 39.2 ± 1.7 | Ax | 512 × 512 × 50 | 0.35 × 0.35 × 2.0 | 0 / 10 |
| Infants with DS | 10 | 37.9 ± 5.9 | Cor and Ax | 512 × 512 × 110 | 0.35 × 0.35 × 1.2 | 0 / 10 |
| Scans with artifacts | 10 | 41.12 ± 0.7 | Cor | 512 × 512 × 110 | 0.35 × 0.35 × 1.2 | 0/10 |
2.3. Reference standard
To establish the reference standard, manual segmentation of the ICV was performed by a trained medical student. Manual annotation was accomplished using in-house developed software by painting ICV voxels in each image slice. ICV included brain, cerebellum and extracranial cerebrospinal fluid. Skull and skin were excluded from the segmentation. We followed the definition of the eight tissue types provided by the NeoBrainS12 challenge for ending point of the brain stem (Išgum et al., 2015). Note that the reference standard for Set 3 of fetal MRI and infants scans with artifacts was not available, hence the segmentation performance on these two sets was evaluated visually.
To estimate inter-observer variability, three slices of 7 scans were segmented by different observers. Two scans from 30 weeks coronal MRI, three scans from 40 weeks coronal MRI and two scans from 40 weeks axial MRI were selected. Furthermore, the slice representing the middle of the brain and subsequently, the first and last slice on which each tissue was visible were identified.
3. Method
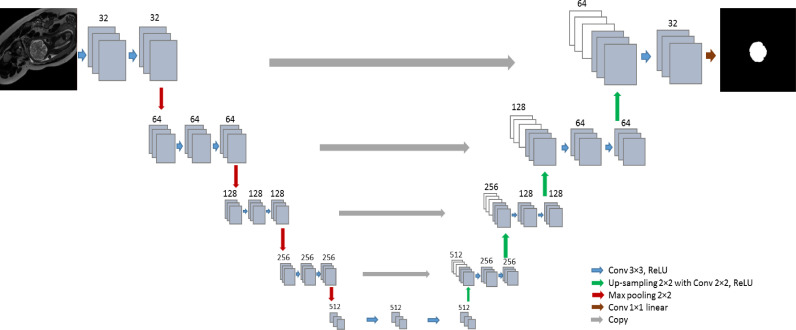
Our aim is to train a single network that is able to perform segmentation of the ICV in a diverse set of scans where the diversity comprises differences in field of view, age of the scanned subjects, orientation of image acquisition, image resolution and presence of pathology1 Our method employs a fully convolutional network (FCN) with U-net like architecture (Ronneberger et al., 2015) since such networks have demonstrated accurate segmentation performance in a number of different segmentation tasks (Litjens et al., 2017). We have used a smaller version of U-net to avoid over-fitting. The network has a contracting path and an expanding path. The contracting path consists of repeated 3 × 3 zero padded convolutions where each convolution is followed by a rectified linear unit (ReLU). 2 × 2 max pooling layers with stride 2 downsample the feature maps. The number of the feature maps doubles after every two convolutional layers. In the expanding path, up-sampling with stride 2 is followed by a 2 × 2 transposed convolution which halves the number of feature channels. The resulting feature maps were concatenated with the corresponding feature map of the contracting path and convolved by two 3 × 3 convolutional layers followed by ReLU. At the final layer, a 1 × 1 convolutions map each component of feature vector to the desired number of classes (Fig. 3). A softmax function is applied in the last layer to classify ICV and background. As a loss function, cross-entropy between the output layer and the manual segmentation reference is used. For optimization, Nesterov Adam optimizer is applied (Dozat, 2016, Kingma, Adam, 2015). In order to increase the mean learning rate, batch normalization (Ioffe and Szegedy, 2015) is used after each convolutional layer (Convolution, Batch Normalization, ReLU) Ioffe and Szegedy (2015). The learning rate of Adam optimization is set to 0.0001. The hyper-parameters were tuned using cross-validation on the training set. The training was stopped after 300 epochs when the loss function became stable. The network is trained with 2D slices and batch size is 30 for each iteration. The image intensity were normalized to the range [0, 1023] before feeding them to the network. Data augmentation was applied during the training by random flipping and rotation of 2D slices. The rotations ranged between 0 to 360 degrees to mimic fetal brain angle variations. As all image intensities were normalized between [0, 1023], we did not vary image intensities nor the contrast as an augmentation. We have implemented the network in Keras, an open-source neural-network library written in Python (Chollet et al., 2015).
Fig. 3.
Network architecture: The network consists of a contracting path and an expanding path. The contracting path consists of repeated convolution layers followed by max pooling, and the expansion path consists of convolution layers followed by up-sampling.
Given that the network performs voxel classification, ICV segmentation may result in small isolated clusters of voxels outside the ICV. To prevent this, 3D connected components smaller than 3 cm3 are discarded. Similarly, possibly remaining holes in the binary mask automatically are filled.
4. Evaluation
The automatic ICV segmentations were evaluated in 3D by means of the Dice coefficient, the mean surface distance and the Hausdorff distance (Taha and Hanbury, 2015) between the manual and automatic segmentations per image per set.
5. Experiments and results
5.1. Training with joint neonatal and fetal scans
We performed segmentation in fetal and neonatal MRI scans using a single trained network. The training set consisted of 21 fetal and 9 neonatal scans. Fetal scans in the training contained 7 scans acquired in axial, 7 scans acquired in coronal and 7 scans acquired in sagittal imaging orientations (21 scans) of 7 patients from Set 1. Neonatal scans were from preterm born infants without visible pathology. Neonatal scans included in the training consisted of 3 coronal scans acquired at 30 weeks PMA, 3 coronal scans acquired at 40 weeks PMA, and 3 axial scans acquired at 40 weeks PMA. Note that the training and test set were separated per subject. During the training, only in joint training scenario, each batch was balanced between fetal scans, 30 weeks coronal neonatal, 40 weeks axial neonatal and 40 weeks coronal neonatal scans.
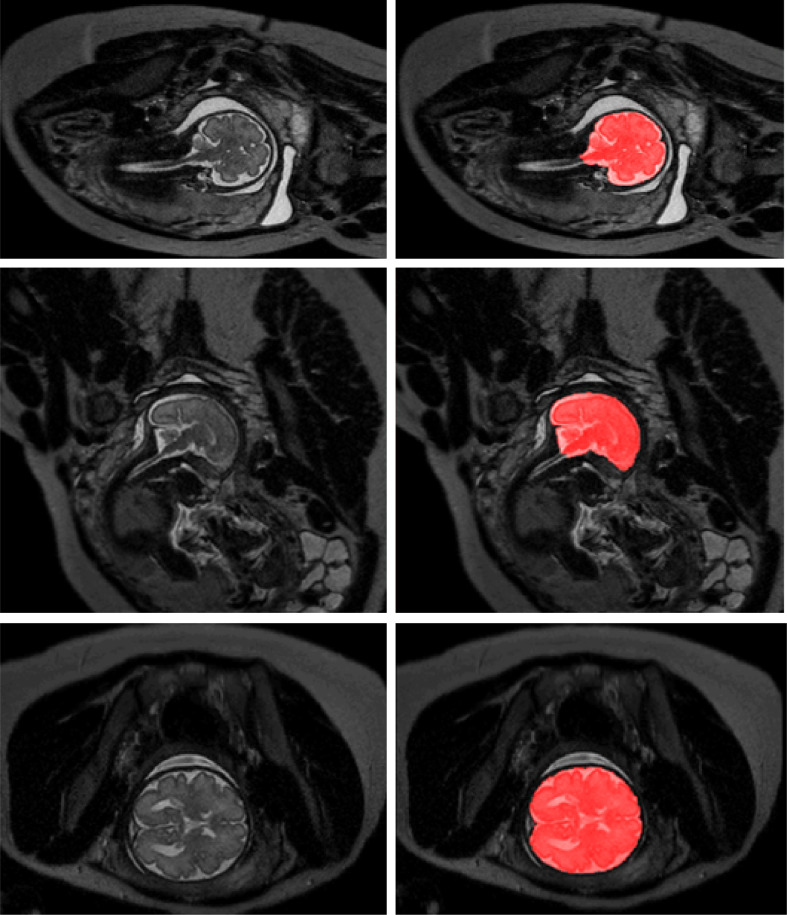
The method was tested with the remaining 24 fetal scans from Set 1 that were acquired in axial, coronal and sagittal orientation, and neonatal scans of the remaining 110 patients. The obtained quantitative results are listed in Table 2 (first three columns). Fig. 4 illustrates examples of the obtained ICV segmentations in images acquired in axial, coronal and sagittal image planes. Fig. 5 illustrates ICV segmentation results in one scan acquired in sagittal imaging plane. The segmentation results are shown in the acquisition plane as well as in planes perpendicular to the acquisition plane. Furthermore, Fig. 6 illustrates examples of the ICV segmentations in slices with intensity inhomogeneity.
Table 2.
Performance of the automatic segmentation expressed by the average Dice coefficient (DC), mean surface distance (MSD) in mm, and Hausdorff distance (HD) in mm. Columns show experiments where the network was trained with: 1) a combination of fetal and neonatal MRI (Fetal and Neonatal) 2) fetal MRI when the test images were from fetuses (Fetal) or neonatal MRI when test images were from neonates (Neonatal) 3) Only a representative set of images. For each test set best results among the three experiments are indicated in bold.
| Neonatal and Fetal |
Fetal |
Representative |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| DC | MSD | HD | DC | MSD | HD | DC | MSD | HD | |
| Set 1 | 0.976 | 0.34 | 10.58 | 0.980 | 0.32 | 16.89 | 0.978 | 0.36 | 15.45 |
| Neonatal and Fetal |
Neonatal |
Representative |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| DC | MSD | HD | DC | MSD | HD | DC | MSD | HD | |
| DC | MSD | HD | DC | MSD | HD | DC | MSD | HD | |
| 30-weeks coronal | 0.993 | 0.11 | 6.19 | 0.988 | 0.18 | 7.87 | 0.992 | 0.12 | 10.90 |
| 40-weeks coronal | 0.993 | 0.18 | 7.98 | 0.994 | 0.14 | 8.32 | 0.993 | 0.16 | 9.64 |
| 40-weeks axial | 0.988 | 0.22 | 7.60 | 0.988 | 0.24 | 7.67 | 0.987 | 0.44 | 25.89 |
| Cross-sectional cohort | 0.987 | 0.35 | 11.13 | 0.990 | 0.19 | 8.88 | 0.991 | 0.22 | 13.94 |
| Infants with CHD | 0.987 | 0.53 | 17.19 | 0.990 | 0.19 | 8.57 | 0.985 | 0.64 | 30.26 |
| Infants with PHVD | 0.987 | 0.29 | 14.67 | 0.988 | 0.31 | 15.89 | 0.986 | 0.35 | 17.34 |
| Infants with stroke | 0.987 | 0.30 | 14.68 | 0.988 | 0.46 | 11.09 | 0.984 | 0.58 | 19.05 |
| Infants with asphyxia | 0.980 | 0.34 | 10.58 | 0.970 | 0.62 | 15.54 | 0.963 | 0.80 | 16.41 |
| Infants with DS | 0.982 | 0.46 | 12.52 | 0.983 | 0.38 | 14.36 | 0.983 | 0.58 | 25.87 |
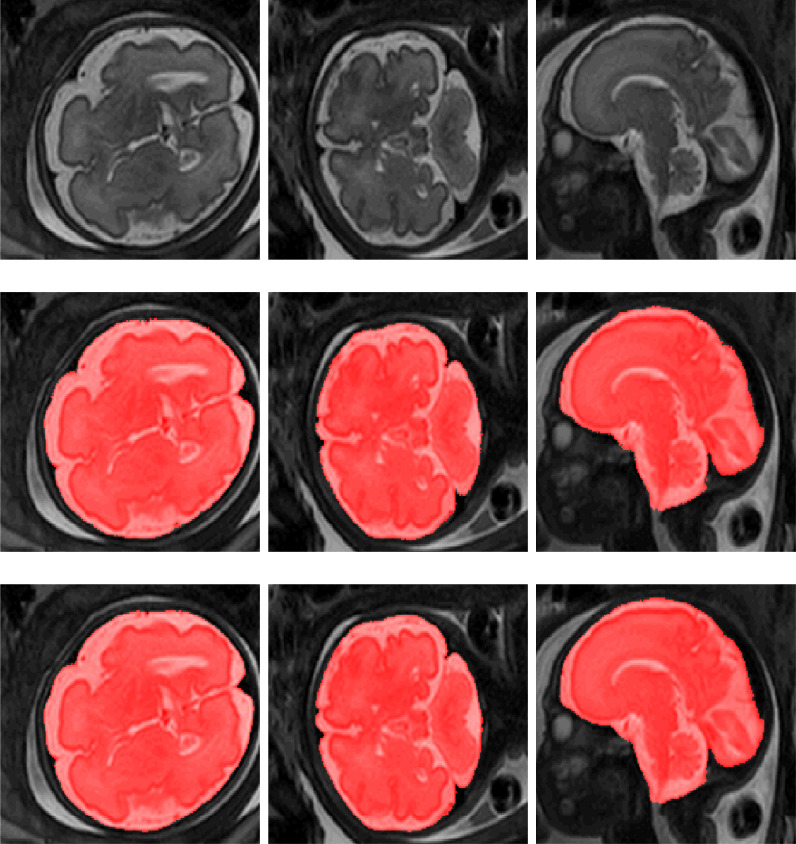
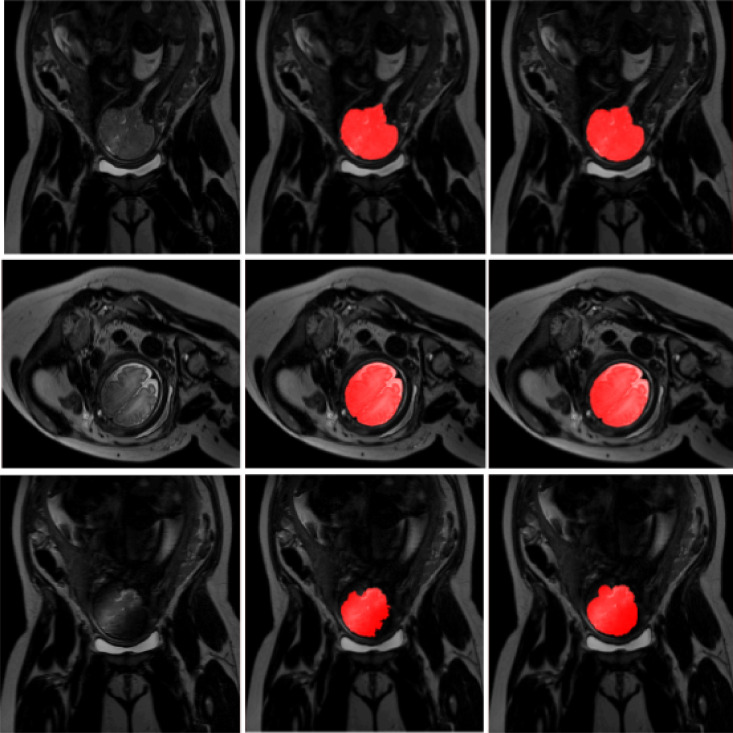
Fig. 4.
Examples of ICV segmentation in slices from fetal scans acquired in axial (left), coronal (middle) and sagittal (right) image planes. The images are selected from the test set. A slice from T2-weighted image (top); segmentation achieved by the proposed method trained with a combination of neonatal and fetal MRI (middle); manual segmentation (bottom).
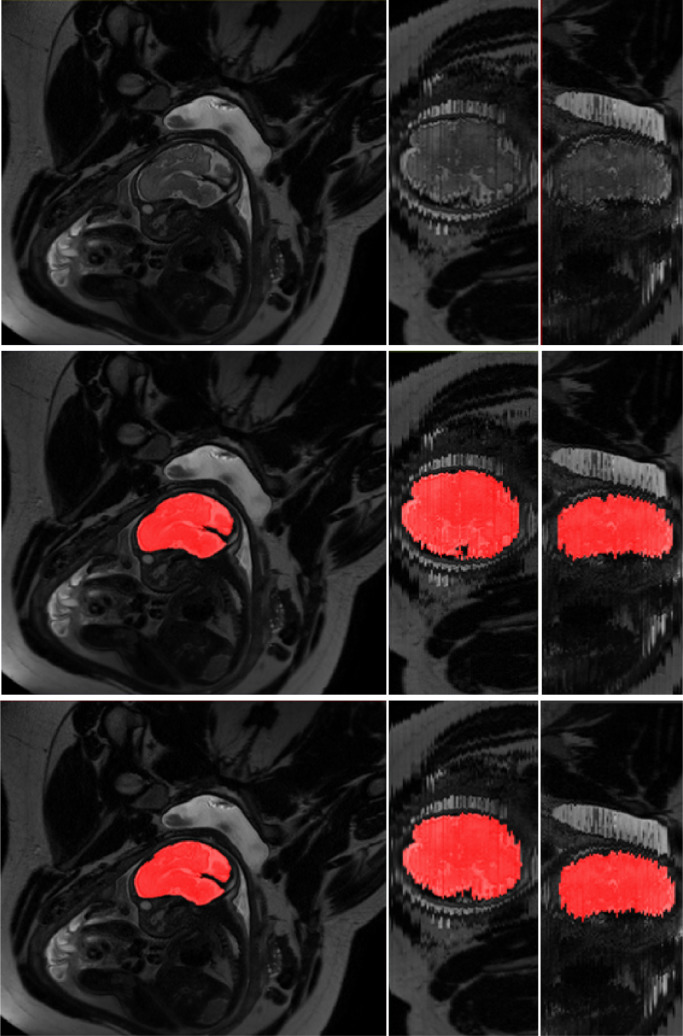
Fig. 5.
Examples of ICV segmentation in a scan acquired in sagittal plane. A slice from T2-weighted fetal MRI scan (top row), segmentation obtained with joint training (middle row) and manual segmentation (bottom row). The first column illustrates the segmentation in the in-plane view. Second and third columns illustrated out-of-plane views. The slices were selected from the test set.
Fig. 6.
Examples of ICV segmentation in slices from fetal scans that visualized intensity inhomogeneity. A slice from T2-weighted image (left); segmentation achieved by the proposed method trained with a combination of neonatal and fetal MRI (middle) and manual segmentation (right). The images were selected from the test set.
Moreover, Fig. 7 shows an example of ICV segmentation in a neonate with PHVD. The automatic segmentation excluded the inserted shunt from the brain mask even though PHVD scans were not included in the training data. Fig. 8 illustrates another example of ICV segmentation in a neonate with PHVD where the cerebellum was undersegmented. It may be observed that in this case cerebellum has voxels of lower intensity than images without visible pathology.
Fig. 7.
Example of ICV segmentation in one test neonate with PHVD on the left compared with manual segmentation on the right.The infants received a temporary ventricular shunt that is visible in some slices. The images were selected from the test set.
Fig. 8.
A slice from a scan of infant with PHVD (left) where the joint training undersegmented cerebellum (middle) compared with reference annotation (right). The cerebellar volume, shape and image intensity are typically different in infants with PHVD from infants without visible pathology. The images were selected from the test set.
5.2. Training with neonatal or fetal scans
Manual annotation in a large set of scans is time-consuming and expensive. Hence, to estimate whether the method performs better on fetal images when trained with fetal images only and whether it performs better on neonatal images when trained with neonatal images only, additional experiments were performed. For this, two separate networks were trained. The first network was trained using only fetal images. This set included scans of 7 fetuses with images acquired in axial, coronal and sagittal directions. The second network was trained using only neonatal images. This training set included images of infants scanned at 30- and 40- weeks PMA acquired in axial and coronal directions. In both experiments, the training images were the same training images that were used in the experiment described in Section 4 A when the fetal and neonatal training images were used together in the training. No other changes in the network architecture or training procedure were applied. The obtained results are listed in Table 2 (middle three columns).
5.3. Training with representative scans
To evaluate whether it might be advantageous to train the network using representative data only, three instances of the original network were trained. One instance was trained and tested with scans of neonates acquired at 30 weeks PMA coronal, another instance was trained and tested with scans of neonates acquired at 40 weeks PMA coronal, and last instance was trained and tested with scan of neonates acquired at 40 weeks PMA axial. Training images represent subsets of scans used in the experiment where all training data was mixed. The obtained results are listed in Table 2 (last three columns).
5.4. Second observer evaluation
To evaluate inter-observer variability, we obtained second observer manual annotations for small subset of neonatal data. The evaluation was performed on 3 slices of 7 scans, i.e 21 slices in total and the results were compared to corresponding slices annotated by the first observer. The results are listed in Table 6.
Table 6.
Evaluating joint training segmentation performance with manual segmentation obtained by two different observers. The results are expressed in terms of Dice coefficient (DC), Hausdorff distance (HD) and mean surface distance (MSD). The HD and MSD are expressed in mm. Note that the evaluation was performed in 3 slices in 7 scans, totalling to 21 slices. The segmentations are compared with the segmentations of the first observer in the same slices.
| First Observer |
Second Observer |
|||||
|---|---|---|---|---|---|---|
| DC | MSD | HD | DC | MSD | HD | |
| 30-weeks coronal | 0.993 | 0.656 | 11.52 | 0.983 | 1.660 | 16.92 |
| 40-weeks coronal | 0.992 | 0.891 | 9.231 | 0.994 | 0.474 | 6.332 |
| 40-weeks axial | 0.993 | 0.828 | 8.752 | 0.992 | 0.979 | 9.620 |
5.5. Comparison with state-of-the art methods
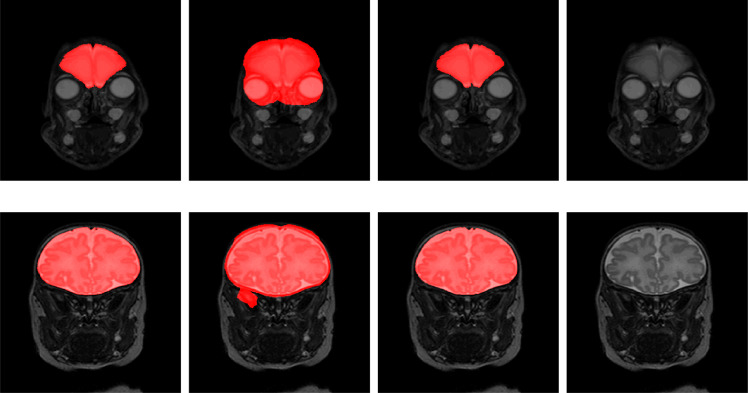
The performance of the proposed method was compared with publicly available ICV segmentation tools. Given that BET is frequently used to segment the ICV in premature neonatal images (Moeskops et al., 2016), we have applied it to segment images in our test set. The fractional intensity threshold (-f) is empirically set to 0.3. The obtained results are presented in Table 5. They demonstrate that BET achieved better performance in neonatal MRI acquired at 40 weeks PMA than in neonatal MRI acquired at 30 weeks PMA. Fig. 10 shows segmentations obtained with BET and joint training in a slice from a scan acquired at 30 weeks PMA and one acquired at 40 weeks PMA.
Table 5.
Performance of the joint training using fetal and neonatal scans for ICV segmentation compared with BET. The results are expressed using the average Dice coefficient (DC), mean surface distance (MSD) in mm, and Hausdorff distance (HD) in mm.
| Joint training |
BET |
|||||
|---|---|---|---|---|---|---|
| DC | MSD | HD | DC | MSD | HD | |
| 30-weeks coronal | 0.99 | 0.11 | 6.19 | 0.91 | 2.05 | 24.92 |
| 40-weeks coronal | 0.99 | 0.18 | 7.98 | 0.94 | 1.92 | 27.91 |
| 40-weeks axial | 0.99 | 0.22 | 7.60 | 0.94 | 1.39 | 34.63 |
| Cross-sectional cohort | 0.99 | 0.35 | 11.13 | 0.93 | 1.83 | 26.76 |
| Infants with CHD | 0.99 | 0.53 | 17.19 | 0.95 | 2.36 | 36.46 |
| Infants with PHVD | 0.99 | 0.29 | 14.67 | 0.94 | 1.72 | 24.16 |
| Infants with stroke | 0.99 | 0.30 | 14.68 | 0.95 | 1.33 | 30.80 |
| Infants with asphyxia | 0.98 | 0.34 | 10.58 | 0.95 | 1.33 | 32.88 |
| Infants with DS | 0.98 | 0.46 | 12.52 | 0.95 | 1.43 | 14.69 |
Fig. 10.
Examples of ICV segmentation in neonates acquired at 30 weeks PMA (top row) and 40 weeks PMA (bottom row). Results of the joint training (first column), the result obtained with BET (second column), manual annotation (third column) and the original T2-weighted MRI (last column).The images were selected from the test set.
Both slices illustrate oversegmentation of the ICV along the whole boundary, which is a frequent error of the BET tool visible in our test set. Quantitative results listed in Table 5 show that joint training consistently achieved higher DC and lower HD and MSD than BET.
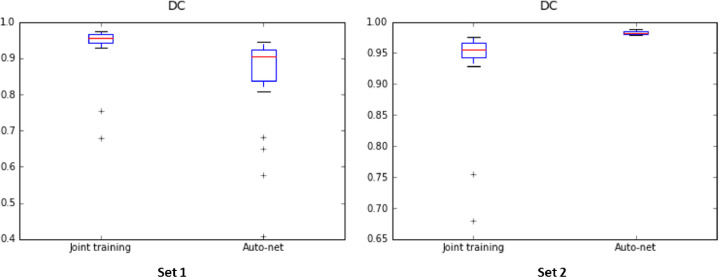
To investigate robustness of our method to variation in scanner characteristics and patient population, the joint training model was evaluated using publicly available fetal MRI scans from another hospital (Set 2). The results were compared with a publicly available Auto-net (Salehi et al., 2018)2 model trained on a much larger set of representative fetal scans from the same hospital. Even though, U-net or any fully convolutional neural network can take any arbitrary image size but the segmentation performance will likely drop if the images in the training and test set do not have the same resolution. Given that scans in Set 2 have different voxel sizes than our fetal images (Set 1) used in the training, prior to analysis scans from Set 2 were resampled to the resolution of our training images. Furthermore, the images were normalized between [0, 1023]. All obtained results are listed in Table 3 and shown in Fig. 9. Note that this model was only trained on fetal MRI and training data did not include any neonatal MRI data. Therefore, the evaluation was performed on fetal MRI only.
Table 3.
Performance of the proposed method using joint training with 21 fetal and 9 neonatal MRI scans, and performance of the publicly provided Auto-net trained with 260 fetal MRI scans. Both methods were tested on publicly available fetal scans from Set 2. The results are expressed by the average Dice coefficient (DC), mean surface distance (MSD) in mm, and Hausdorff distance (HD) in mm.
| DC | MSD | HD | |
|---|---|---|---|
| Joint training | 0.94 ± 0.02 | 1.7 ± 0.72 | 34.5 ± 16.11 |
| Auto-net | 0.98 ± 0.01 | 0.2 ± 0.04 | 10.1 ± 5.45 |
Fig. 9.
Dice coefficients achieved by the proposed method using joint training with 21 fetal and 9 neonatal MRI scans, and by the publicly provided Auto-net trained with 260 fetal MRI scans. Both methods were tested on Set 1 (left) and Set 2 (right).
In addition, Auto-net was evaluated on fetal images from our hospital (Set 1). Quantitative results are listed in Table 4. As in the previous experiment scans from Set 1 were resampled to the same resolution of the images used to train Auto-net (Set 2). Furthermore, the scans were normalized between [0, 1023]. Fig. 9 illustrates the segmentation performance in a box plot. Note that even though the scans were resampled to the same resolution in both experiments, the images had different field of view.
Table 4.
Performance of the proposed method using joint training with 21 fetal and 9 neonatal MRI scans and performance of the publicly available Auto-net trained with 260 MRI scans. Both methods were tested with fetal images from Set 1. The results are expressed by average Dice coefficient (DC), Mean surface distance (MSD) in mm, and Hausdorff distance (HD) in mm.
| DC | MSD | HD | |
|---|---|---|---|
| Joint training | 0.98 ± 0.02 | 0.3 ± 0.36 | 10.6 ± 5.73 |
| Auto-net | 0.87 ± 0.10 | 3.2 ± 1.84 | 72.41 ± 50.34 |
5.6. Evaluation on scans acquired with 1.5 Tesla scanner
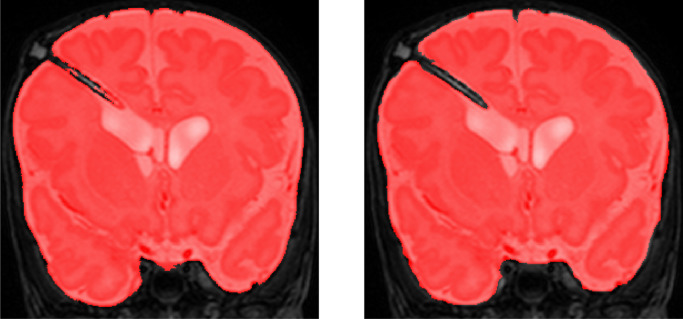
To demonstrate the performance of the proposed method on images acquired with a scanner exploiting a different field strength, the joint training model was evaluated on fetal MRI scans (Set 3) acquired with 1.5 Tesla scanner. We illustrate segmentation results in the three scans without reference standard in Fig. 11. Visual inspection of the results in these scans reveals that the joint training model produced accurate ICV segmentations in scans with different field strength, although the model was not trained with such scans.
Fig. 11.
Examples of ICV segmentation in slices from fetal scans acquired with 1.5 Tesla scanner in coronal (top), sagittal (middle) and axial (bottom) image planes. A slice from T2-weighted image (left) and segmentation achieved by the proposed method trained with a combination of neonatal and fetal MRIs (right).
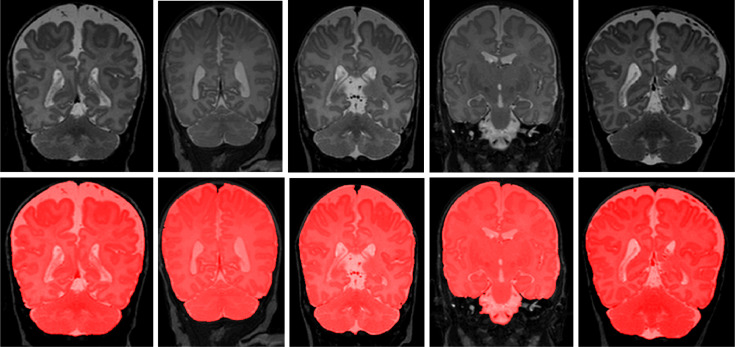
5.7. Evaluation on scans with artifacts
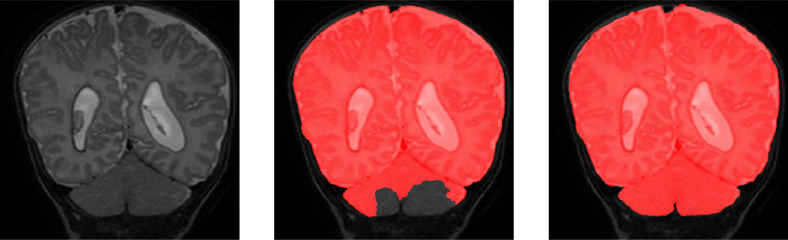
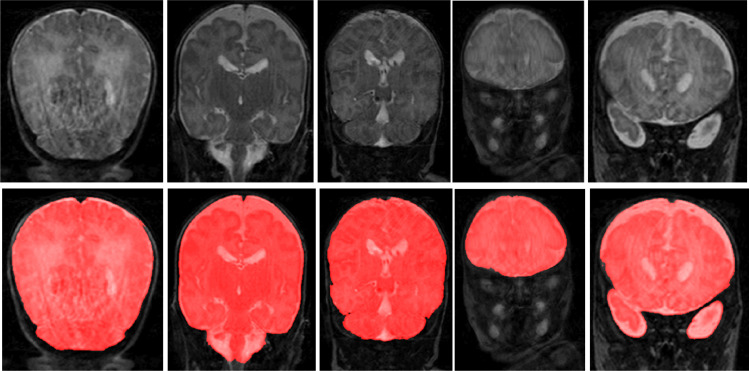
To demonstrate the performance of proposed method on scans with intensity inhomogeneity and motion artifacts, the joint training model was evaluated on 5 neonatal scans with intensity inhomogeneity and 5 neonatal scans with motion artifacts. We illustrate segmentation results in the five scans with intensity inhomogeneity in Fig. 12 and five scans with motion artifacts in Fig. 13. Visual inspection of the results in these scans reveals that the joint training model produced accurate ICV segmentation in scans with motion artifacts and intensity inhomogeneity.
Fig. 12.
Examples of ICV segmentation in 5 neonatal MR scans with intensity inhomogeneity artifacts. A slice from T2-weighted fetal MRI scan (first row); segmentation obtained with joint training (second row).
Fig. 13.
Examples of ICV segmentation in 5 neonatal MR scans with motion artifacts. A slice from T2-weighted fetal MRI scan (first row); segmentation obtained with joint training (second row).
5.8. Second observer evaluation
Manual annotation in a large set of scans is time-consuming and expensive. Hence, to estimate inter-observer variability, second observer performed manual annotations in a small subset of neonatal data. The evaluation was performed on 3 slices of 7 scans, totally in 21 slices. These segmentations were compared with the segmentations of the first observer in the same slices. The results are listed in Table 6.
6. Discussion and conclusion
An automatic method for segmentation of the ICV in fetal and neonatal brain MR scans was presented. The proposed method employs a fully convolutional network with U-net architecture. It was trained by using a combination of neonatal and fetal MRI and the results demonstrate accurate segmentation of ICV in fetal and neonatal MR scans regardless the orientation of the image acquisition, the age of infants at the time of scanning or the presence of pathology. Unlike previous ICV segmentation methods developed for fetal or neonatal MRI (Anquez, Angelini, Bloch, 2009, Keraudren, Kyriakopoulou, Rutherford, Hajnal, Rueckert, 2013, Taimouri et al., 2015), the proposed method does not require brain localization or prior information about the patient age or expected anatomy.
In this study, 2D analysis was applied. Even though 3D analysis regularly allows better exploitation of the available information compared to 2D analysis, 2D analysis was advantageous as it minimized the risk of overfitting and allowed analysis of scans with large slice thickness that led to substantial changes in the anatomy (Havaei, Davy, Warde-Farley, Biard, Courville, Bengio, Pal, Jodoin, Larochelle, 2017, Moeskops, Viergever, Mendrik, de Vries, Benders, Išgum, 2016, Wolterink, Leiner, de Vos, van Hamersvelt, Viergever, Išgum, 2016). Moreover, 2D analysis was less influenced by missing and corrupted slices resulting from continuous fetal motion.
Generating manual segmentation is a cumbersome and extremely time-consuming task. The results illustrate that with a small number of available manual segmentation used for training, the network achieves a competitive and robust results in a large test set. Using semi-automatic segmentation comprised of automatic presegmentation and subsequent manual correction to gnerate refrence standard for training purposes could make the process faster. Availability of a large training set could offer possibility to investigate the impact of the size of the training set on the method performance as well as research towards the requirements regarding characteristics of the training set for employment in the MRIs presenting pathology or artifacts. This may be interesting direction for future research.
Although the network was trained with images containing no visible pathology, the evaluation was performed on a large and diverse set of scans, which includes scans with pathology. The segmentation results in neonatal scans with or without lesions are comparable. Note that large lesions in the brain strongly affecting tissue appearance (infants with stroke), morphological changes (infants with Down syndrome and PHVD), and presence of implants (shunts) that were mostly excluded (PHVD) (see Fig. 7).
We evaluated the proposed method on scans with artifacts such as intensity inhomogeneity and motion artifacts. The visual inspection demonstrate that even thought scans with artifacts were not in the training, the proposed approach is able to segment ICV.
Furthermore, we investigated whether it is feasible to train a single instance of the network applicable to both fetal and neonatal scans, or whether better performance can be achieved by training a separate network using only fetal or only neonatal scans. The results show that in both cases DC ranges from 0.98 to 0.99.
Moreover, we compared performance using joint training with fetal and neonatal scans against training using representative scans only. The results demonstrate that in both cases accurate segmentation was achieved when evaluating the overlap between automatic and reference segmentations (0.98 to 0.99 Dice coefficient). The results also demonstrate that training with diverse images using fetal and neonatal scans reduced false positive voxels far from the intracranial volume surface leading to lower Hausdorff distances and mean surface distances in all sets. Training with both fetal and neonatal scans indicated the most noticeable improvement in infants with asphyxia. Despite the differences in image acquisition, image orientation, and brain morphology, fetal and neonatal scans share common features that improve the ability of the network to generalize, making it more robust and compensating for the lack of representative data.
To investigate robustness of the proposed method to variations in scanner characteristics and patient population, the method was evaluated using publicly available fetal MRI scans from another hospital (Set 2). The results were compared with a publicly available Auto-net (Salehi et al., 2018) model trained on a much larger set of representative fetal scans from the same hospital. The results show that our model did not outperform the dedicated data-specific approach. Nevertheless, it achieved DC, MSD and HD of 0.94, 1.7 and 34.5 respectively. Similarly, we evaluated Auto-net on fetal scans from our hospital (Set 1). The results demonstrate that the proposed method trained on representative fetal scans from our hospital outperformed Auto-net trained on different data. The two experiments indicate that reasonable performance can be achieved using different scans but also underline the importance of training with representative data. In future research, investigating interpretability of model using saliency map (Simonyan et al., 2013) can demonstrate a better understanding of limitations in network performance.
In addition, the proposed method was compared with the publicly available and widely used BET for the segmentation of neonatal MRIs. Although BET is known to achieve accurate segmentation of ICV in adults, our results demonstrate that it is less suited for neonatal brain. Our dedicated method clearly outperformed BET.
To conclude, this study presented a method for automatic ICV segmentation in neonatal and fetal MRI. Despite the variability among the evaluated scans, the method obtained accurate segmentation results in both fetal and neonatal MR scans. Hence, the algorithm provides a generic tool for segmentation of the ICV that may be used as a preprocessing step for brain tissue segmentation in fetal and neonatal brain MR scans.
Declaration of Competing Interest
There is no conflict between authors.
Acknowledgments
This study was sponsored by the Research Program Specialized Nutrition of the Utrecht Center for Food and Health, through a subsidy from the Dutch Ministry of Economic Affairs, the Utrecht Province and the Municipality of Utrecht.
Footnotes
We are aiming to make the code publicly available upon the paper acceptance.
References
- Akkus Z., Galimzianova A., Hoogi A., Rubin D.L., Erickson B.J. Deep learning for brain MRI segmentation: state of the art and future directions. J. Digit. Imaging. 2017;30(4):449–459. doi: 10.1007/s10278-017-9983-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alderliesten T., de Vries L.S., Staats L., van Haastert I.C., Weeke L., Benders M.J., Koopman-Esseboom C., Groenendaal F. MRI and spectroscopy in (near) term neonates with perinatal asphyxia and therapeutic hypothermia. Arch. Dis. Child. Fetal Neonatal Edition. 2017;102(2):F147–F152. doi: 10.1136/archdischild-2016-310514. [DOI] [PubMed] [Google Scholar]
- Anquez J., Angelini E.D., Bloch I. The IEEE International Symposium on Biomedical Imaging (ISBI) IEEE; 2009. Automatic segmentation of head structures on fetal MRI; pp. 109–112. [Google Scholar]
- Benders M.J., van der Aa N.E., Roks M., van Straaten H.L., Isgum I., Viergever M.A., Groenendaal F., de Vries L.S., van Bel F. Feasibility and safety of erythropoietin for neuroprotection after perinatal arterial ischemic stroke. J. Pediatr. 2014;164(3):481–486. doi: 10.1016/j.jpeds.2013.10.084. [DOI] [PubMed] [Google Scholar]
- Brouwer M.J., De Vries L.S., Kersbergen K.J., Van Der Aa N.E., Brouwer A.J., Viergever M.A., Išgum I., Han K.S., Groenendaal F., Benders M.J. Effects of posthemorrhagic ventricular dilatation in the preterm infant on brain volumes and white matter diffusion variables at term-equivalent age. J. Pediatr. 2016;168:41–49. doi: 10.1016/j.jpeds.2015.09.083. [DOI] [PubMed] [Google Scholar]
- Chen H., Dou Q., Yu L., Qin J., Heng P.-A. Voxresnet: deep voxelwise residual networks for brain segmentation from 3D MR images. Neuroimage. 2018;170:446–455. doi: 10.1016/j.neuroimage.2017.04.041. [DOI] [PubMed] [Google Scholar]
- Chollet, F., et al., 2015. Keras. https://keras.io.
- Claessens N.H., Algra S.O., Jansen N.J., Groenendaal F., de Wit E., Wilbrink A.A., Haas F., Schouten A.N., Nievelstein R.A., Benders M.J. Clinical and neuroimaging characteristics of cerebral sinovenous thrombosis in neonates undergoing cardiac surgery. J. Thorac. Cardiovasc. Surg. 2018:1150–1158. doi: 10.1016/j.jtcvs.2017.10.083. [DOI] [PubMed] [Google Scholar]
- Claessens N.H., Moeskops P., Buchmann A., Latal B., Knirsch W., Scheer I., Išgum I., De Vries L.S., Benders M.J., Von Rhein M. Delayed cortical gray matter development in neonates with severe congenital heart disease. Pediatr. Res. 2016;80(5):668–674. doi: 10.1038/pr.2016.145. [DOI] [PubMed] [Google Scholar]
- Coyle J.T., Oster-Granite M., Gearhart J.D. The neurobiologie consequences of down syndrome. Brain Res. Bull. 1986;16(6):773–787. doi: 10.1016/0361-9230(86)90074-2. [DOI] [PubMed] [Google Scholar]
- Dey, R., Hong, Y., 2018. Compnet: complementary segmentation network for brain MRI extraction. arXiv preprint arXiv:1804.00521.
- Dolz J., Desrosiers C., Ayed I.B. 3D fully convolutional networks for subcortical segmentation in mri: a large-scale study. Neuroimage. 2018;170:456–470. doi: 10.1016/j.neuroimage.2017.04.039. [DOI] [PubMed] [Google Scholar]
- Dolz, J., Desrosiers, C., Wang, L., Yuan, J., Shen, D., Ayed, I. B., 2017. Deep CNN ensembles and suggestive annotations for infant brain MRI segmentation. arXiv preprint arXiv:1712.05319. [DOI] [PubMed]
- Dozat T. 4th International Conference on Learning Representations, Workshop. 2016. Incorporating nesterov momentum into Adam. [Google Scholar]
- Drost F.J., Keunen K., Moeskops P., Claessens N.H., van Kalken F., Išgum I., Voskuil-Kerkhof E.S., Groenendaal F., de Vries L.S., Benders M.J. Severe retinopathy of prematurity is associated with reduced cerebellar and brainstem volumes at term and neurodevelopmental deficits at two years. Pediatr. Res. 2018:818–824. doi: 10.1038/pr.2018.2. [DOI] [PubMed] [Google Scholar]
- Dubois J., Benders M., Cachia A., Lazeyras F., Ha-Vinh Leuchter R., Sizonenko S., Borradori-Tolsa C., Mangin J., Hüppi P.S. Mapping the early cortical folding process in the preterm newborn brain. Cereb. Cortex. 2007;18(6):1444–1454. doi: 10.1093/cercor/bhm180. [DOI] [PubMed] [Google Scholar]
- Eskildsen S.F., Coupé P., Fonov V., Manjón J.V., Leung K.K., Guizard N., Wassef S.N., Østergaard L.R., Collins D.L., The Alzheimer’s Disease Neuroimaging Initiative Beast: brain extraction based on nonlocal segmentation technique. Neuroimage. 2012;59(3):2362–2373. doi: 10.1016/j.neuroimage.2011.09.012. [DOI] [PubMed] [Google Scholar]
- Havaei M., Davy A., Warde-Farley D., Biard A., Courville A., Bengio Y., Pal C., Jodoin P.-M., Larochelle H. Brain tumor segmentation with deep neural networks. Med. Image Anal. 2017;35:18–31. doi: 10.1016/j.media.2016.05.004. [DOI] [PubMed] [Google Scholar]
- Iglesias J.E., Liu C.-Y., Thompson P.M., Tu Z. Robust brain extraction across datasets and comparison with publicly available methods. IEEE Trans. Med. Imaging. 2011;30(9):1617–1634. doi: 10.1109/TMI.2011.2138152. [DOI] [PubMed] [Google Scholar]
- Inder T.E., Huppi P.S., Warfield S., Kikinis R., Zientara G.P., Barnes P.D., Jolesz F., Volpe J.J. Periventricular white matter injury in the premature infant is followed by reduced cerebral cortical gray matter volume at term. Ann. Neurol. 1999;46(5):755–760. doi: 10.1002/1531-8249(199911)46:5<755::aid-ana11>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Ioffe S., Szegedy C. The 32nd International Conference on Machine Learning (ICML-15) 2015. Batch normalization: accelerating deep network training by reducing internal covariate shift; pp. 448–456. [Google Scholar]
- Išgum I., Benders M.J., Avants B., Cardoso M.J., Counsell S.J., Gomez E.F., Gui L., Hűppi P.S., Kersbergen K.J., Makropoulos A. Evaluation of automatic neonatal brain segmentation algorithms: the NeoBrainS12 challenge. Med. Image Anal. 2015;20(1):135–151. doi: 10.1016/j.media.2014.11.001. [DOI] [PubMed] [Google Scholar]
- Ison M., Donner R., Dittrich E., Kasprian G., Prayer D., Langs G. Workshop on Paediatric and Perinatal Imaging, MICCAI. 2012. Fully automated brain extraction and orientation in raw fetal MRI; pp. 17–24. [Google Scholar]
- Keraudren K., Kuklisova-Murgasova M., Kyriakopoulou V., Malamateniou C., Rutherford M.A., Kainz B., Hajnal J.V., Rueckert D. Automated fetal brain segmentation from 2D MRI slices for motion correction. Neuroimage. 2014;101:633–643. doi: 10.1016/j.neuroimage.2014.07.023. [DOI] [PubMed] [Google Scholar]
- Keraudren K., Kyriakopoulou V., Rutherford M., Hajnal J.V., Rueckert D. MICCAI, LNCS. Springer; 2013. Localisation of the brain in fetal MRI using bundled sift features; pp. 582–589. [DOI] [PubMed] [Google Scholar]
- Kersbergen K.J., Leroy F., Išgum I., Groenendaal F., de Vries L.S., Claessens N.H., van Haastert I.C., Moeskops P., Fischer C., Mangin J.-F. Relation between clinical risk factors, early cortical changes, and neurodevelopmental outcome in preterm infants. Neuroimage. 2016;142:301–310. doi: 10.1016/j.neuroimage.2016.07.010. [DOI] [PubMed] [Google Scholar]
- Keunen K. The Neonatal Brain: Early Connectome Development and Childhood Cognition. Utrecht University; 2017. Ph.D. thesis. [Google Scholar]
- Khalili N., Moeskops P., Claessens N., Scherpenzeel S., Turk E., de Heus R., Benders M., Viergever M., Pluim J., Išgum I. Fetal, Infant and Ophthalmic Medical Image Analysis. 2017. Automatic segmentation of the intracranial volume in fetal MR images; pp. 42–51. [Google Scholar]
- Kingma D., Adam J.B. International Conference on Learning Representations. 2015. A method for stochastic optimisation. [Google Scholar]
- Kleesiek J., Urban G., Hubert A., Schwarz D., Maier-Hein K., Bendszus M., Biller A. Deep MRI brain extraction: a 3D convolutional neural network for skull stripping. Neuroimage. 2016;129:460–469. doi: 10.1016/j.neuroimage.2016.01.024. [DOI] [PubMed] [Google Scholar]
- Litjens G., Kooi T., Bejnordi B.E., Setio A.A.A., Ciompi F., Ghafoorian M., van der Laak J.A., van Ginneken B., Sánchez C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- Mahapatra D. Skull stripping of neonatal brain MRI: using prior shape information with graph cuts. J. Digit. Imaging. 2012;25(6):802–814. doi: 10.1007/s10278-012-9460-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makropoulos A., Counsell S.J., Rueckert D. A review on automatic fetal and neonatal brain MRI segmentation. Neuroimage. 2018;170:231–248. doi: 10.1016/j.neuroimage.2017.06.074. [DOI] [PubMed] [Google Scholar]
- Moeskops P., Benders M.J., Kersbergen K.J., Groenendaal F., de Vries L.S., Viergever M.A., Išgum I. Development of cortical morphology evaluated with longitudinal MR brain images of preterm infants. PLoS One. 2015;10(7):e0131552. doi: 10.1371/journal.pone.0131552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeskops P., Išgum I., Keunen K., Claessens N.H., Haastert I.C., Groenendaal F., Vries L.S., Viergever M.A., Benders M.J. Prediction of cognitive and motor outcome of preterm infants based on automatic quantitative descriptors from neonatal brain images. Sci. Rep. 2017;7(1):2163. doi: 10.1038/s41598-017-02307-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeskops P., Viergever M.A., Mendrik A.M., de Vries L.S., Benders M.J., Išgum I. Automatic segmentation of MR brain images with a convolutional neural network. IEEE Trans. Med. Imaging. 2016;35(5):1252–1261. doi: 10.1109/TMI.2016.2548501. [DOI] [PubMed] [Google Scholar]
- Rajchl M., Lee M.C., Oktay O., Kamnitsas K., Passerat-Palmbach J., Bai W., Damodaram M., Rutherford M.A., Hajnal J.V., Kainz B. Deepcut: object segmentation from bounding box annotations using convolutional neural networks. IEEE Trans. Med. Imaging. 2017;36(2):674–683. doi: 10.1109/TMI.2016.2621185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajchl, M., Lee, M. C., Schrans, F., Davidson, A., Passerat-Palmbach, J., Tarroni, G., Alansary, A., Oktay, O., Kainz, B., Rueckert, D., 2016. Learning under distributed weak supervision. arXiv preprint arXiv:1606.01100.
- Ronneberger O., Fischer P., Brox T. MICCAI, LNCS. Springer; 2015. U-net: Convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- Salehi S.S.M., Erdogmus D., Gholipour A. Auto-context convolutional neural network (auto-net) for brain extraction in magnetic resonance imaging. IEEE Trans. Med. Imaging. 2017;36(11):2319–2330. doi: 10.1109/TMI.2017.2721362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi S.S.M., Hashemi S.R., Velasco-Annis C., Ouaalam A., Estroff J.A., Erdogmus D., Warfield S.K., Gholipour A. Biomedical Imaging (ISBI 2018), 2018 IEEE 15th International Symposium on. IEEE; 2018. Real-time automatic fetal brain extraction in fetal MRI by deep learning; pp. 720–724. [Google Scholar]
- Ségonne F., Dale A.M., Busa E., Glessner M., Salat D., Hahn H.K., Fischl B. A hybrid approach to the skull stripping problem in MRI. Neuroimage. 2004;22(3):1060–1075. doi: 10.1016/j.neuroimage.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Serag A., Blesa M., Moore E.J., Pataky R., Sparrow S.A., Wilkinson A., Macnaught G., Semple S.I., Boardman J.P. Accurate learning with few atlases (alfa): an algorithm for MRI neonatal brain extraction and comparison with 11 publicly available methods. Sci. Rep. 2016;6:23470. doi: 10.1038/srep23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan, K., Vedaldi, A., Zisserman, A., 2013. Deep inside convolutional networks: visualising image classification models and saliency maps. arXiv preprint arXiv:1312.6034.
- Smith S.M. Fast robust automated brain extraction. Hum. Brain Mapp. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha A.A., Hanbury A. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med. Imaging. 2015;15(1):29. doi: 10.1186/s12880-015-0068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taimouri, V., Gholipour, A., Velasco-Annis, C., Estroff, J.A., Warfield, S.K., 2015. A template-to-slice block matching approach for automatic localization of brain in fetal MRI, Taimouri, V., Gholipour, A., Velasco-Annis, C., Estroff, J. A., & Warfield, S. K. (2015, April). A template-to-slice block matching approach for automatic localization of brain in fetal MRI. In 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), 144–147.
- Tourbier S., Velasco-Annis C., Taimouri V., Hagmann P., Meuli R., Warfield S.K., Cuadra M.B., Gholipour A. Automated template-based brain localization and extraction for fetal brain MRI reconstruction. Neuroimage. 2017;155:460–472. doi: 10.1016/j.neuroimage.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z., Bai X. Auto-context and its application to high-level vision tasks and 3d brain image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2010;32(10):1744–1757. doi: 10.1109/TPAMI.2009.186. [DOI] [PubMed] [Google Scholar]
- Wachinger C., Reuter M., Klein T. Deepnat: deep convolutional neural network for segmenting neuroanatomy. Neuroimage. 2018;170:434–445. doi: 10.1016/j.neuroimage.2017.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolterink J.M., Leiner T., de Vos B.D., van Hamersvelt R.W., Viergever M.A., Išgum I. Automatic coronary artery calcium scoring in cardiac ct angiography using paired convolutional neural networks. Med. Image Anal. 2016;34:123–136. doi: 10.1016/j.media.2016.04.004. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Fujimoto Y., Kobashi S., Wakata Y., Ishikura R., Kuramoto K., Imawaki S., Hirota S., Hata Y. Fuzzy Systems (FUZZ), 2010 IEEE International Conference. 2010. Automated fuzzy logic based skull stripping in neonatal and infantile MR images; pp. 1–7. [Google Scholar]
- Yushkevich P.A., Piven J., Hazlett H.C., Smith R.G., Ho S., Gee J.C., Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]