SUMMARY
Brown adipose tissue harbors UCP1 to dissipate chemical energy as heat. However, the transcriptional network that governs the thermogenic gene program is incompletely understood. Zc3h10, a CCCH-type zinc finger protein, has recently been reported to bind RNA. However, we report here that Zc3h10 functions as a transcription factor to activate UCP1 not through the enhancer region, but by binding to a far upstream region of the UCP1 promoter. Upon sympathetic stimulation, Zc3h10 is phosphorylated at S126 by p38 mitogen-activated protein kinase (MAPK) to increase binding to the distal region of the UCP1 promoter. Zc3h10, as well as mutant Zc3h10, which cannot bind RNA, enhances thermogenic capacity and energy expenditure, protecting mice from diet-induced obesity. Conversely, Zc3h10 ablation in UCP1+ cells in mice impairs thermogenic capacity and lowers oxygen consumption, leading to weight gain. Hence, Zc3h10 plays a critical role in the thermogenic gene program and may present future targets for obesity therapeutics.
Graphical Abstract

In Brief
Zc3h10 is a RNA-binding protein. Here, Yi et al. report Zc3h10 is a transcription factor that activates UCP1 and other BAT genes. Cold/β3 stimulation causes phosphorylation of Zc3h10 at S126 by p38 MAPK to increase its binding to targets genes and, thus, promotes thermogenic capacity and energy expenditure.
INTRODUCTION
White adipose tissue (WAT) primarily functions as the storage site of excess energy, whereas brown adipose tissue (BAT) dissipates energy as heat to maintain body temperature. BAT is enriched with a high number of mitochondria that possess a specialized inner mitochondrial H+/fatty acid symporter, Uncoupling Protein 1 (UCP1), for thermogenesis (Fedorenko et al., 2012). In mice, the expression of mouse UCP1 is restricted to BAT only in an unstimulated condition. However, upon cold exposure, UCP1+ thermogenic adipocytes, termed “beige” or “brite” cells, can arise in WAT depots, especially subcutaneous WAT, although whether this is from recruitment and/or transdifferentiation of white adipocytes is currently not well understood (Long et al., 2014; Kalinovich et al., 2017). In adult humans, the expression pattern of thermogenic cells show similarity to either mouse beige or brown adipocytes depending on the depot surveyed (Jespersen et al., 2013; Cypess et al., 2013; Lidell et al., 2013). Therefore, stimulating BAT or inducing browning in WAT in humans seems to be a promising avenue to combat obesity and type 2 diabetes. Thus, unraveling the mechanisms underlying the thermogenic gene program has drawn growing attention in obesity research.
Transcription of UCP1 and the thermogenic gene program is regulated by a web of transcription factors and cofactors. Thus, a multitude of transcriptional regulators have been implicated in the transcription of UCP1, including transcription factors Zfp516, ATF2, and EBF2 and transcriptional cofactors PRDM16 and PGC1α (Cao et al., 2004; Seale et al., 2008; Kleiner et al., 2012; Rajakumari et al., 2013; Dempersmier et al., 2015; Puigserver et al., 1998). Expressions of a number of these factors, such as PGC1α, ATF2, IRF4, and Zfp516, are increased also by cold exposure or β3-adrenergic stimulation (Puigserver et al., 1998; Kong et al., 2014). Moreover, the α3-adrenergic receptor-cyclic AMP (cAMP)-PKA pathway for cold-induced thermogenesis, with p38 MAPK as its downstream effector, has been shown to phosphorylate ATF2 and PGC1α to increase their activation and/or stability for UCP1 transcription (Knutti et al., 2001; Puigserver et al., 2001; Watson et al., 2017; Cao et al., 2004).
Zc3h10 belongs to the CCCH-type zinc finger family of proteins that has been found to be involved in RNA metabolism, including processing, splicing, and stability (Ray et al., 2013; Fu and Blackshear, 2017). Treiber et al. (2017) recently reported that Zc3h10 is a RNA-binding protein functioning in specific pri-miRNA processing. However, here, we identify Zc3h10 as a DNA-binding transcription factor that binds 5′-TYCCNG-3′. We demonstrate that Zc3h10 binds to an upstream UCP1 promoter region through its bZIP domain, and this function does not require RNA-binding zinc fingers, revealing its distinct function. Moreover, we show that Zc3h10 is phosphorylated by p38 MAPK in response to cold, resulting in an increase in DNA binding for transcriptional activation of UCP1. Thus, ablation of Zc3h10 in UCP1+ cells in vitro and in vivo impairs thermogenic program, whereas ectopic Zc3h10 expression enhances it, protecting mice from diet-induced obesity.
RESULTS
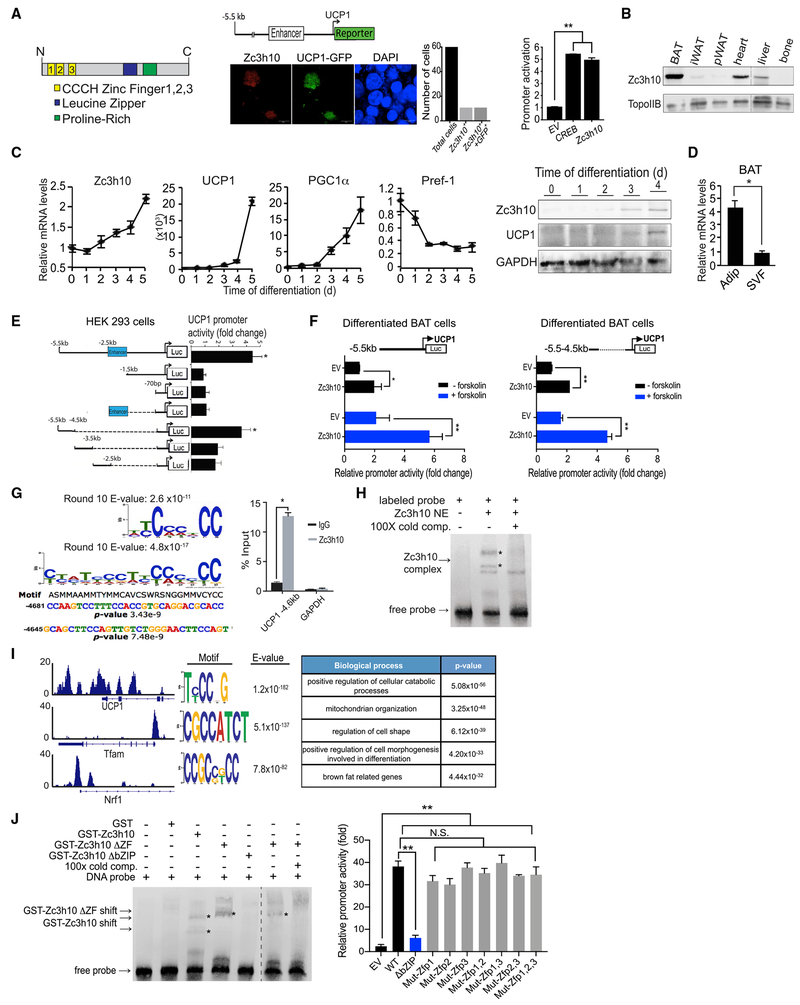
Zc3h10 Activates UCP1 Promoter by Binding to a Distal Upstream Region
In an effort to identify transcription factors that activate the UCP1 promoter, we co-transfected known and putative transcription factors individually along with −5.5 kb UCP1 promoter-GFP reporter (Dempersmier et al., 2015). Zc3h10 was one such transcription factor that was identified to confer strong GFP signal (Figure 1A, middle). Quantification of the reporter activation by using the −5.5 kb UCP1 promoter-luciferase construct showed that, similar to CREB, Zc3h10 could activate the UCP1 promoter by approximately 5-fold (Figure 1A, right; Figure S1A). Zc3h10 contains an N-terminal domain of 3 CCCH-type zinc fingers (ZF1, ZF2, and ZF3) and a proline-rich domain. We also identified a basic leucine-zipper domain (bZIP) positioned between the zinc finger domain and the proline-rich domain (Figure 1A, left). Tissue expression profiling by immunoblotting indicated that Zc3h10 was enriched in brown adipose tissue (BAT), detected at the highest level among the tissues examined. Zc3h10 was found at a significant level in heart and liver but barely detectable in bone and in white adipose tissue depots, such as inguinal (iWAT) and perigonadal (pWAT) (Figure 1B). We next examined Zc3h10 expression during brown adipocyte differentiation of BAT cells in vitro. As expected, expression of BAT-enriched genes, such as UCP1 and PGC1α, increased during the course of brown adipocyte differentiation, whereas a preadipocyte gene, Pref-1, mRNA level was decreased during the differentiation (Figure 1C, left). More importantly, Zc3h10 mRNA and protein levels were increased over the course of brown adipocyte differentiation, confirming the concept that Zc3h10 is enriched in mature brown adipocytes (Figure 1C, left and right). Fractionation of BAT showed that the Zc3h10 mRNA level was 4-fold higher in adipocyte fraction than in stromal vascular fraction (SVF) that contains preadipocytes as well as other cell types (Figure 1D). These data show that Zc3h10 is enriched in BAT and can activate the UCP1 promoter.
Figure 1. Zc3h10 Activates the UCP1 Promoter by Binding at a Distal Region.
(A) (Left) Diagram of Zc3h10 structure. (Middle) Immunofluorescence of HEK293FT cells transfected with Zc3h10 (red) and −5.5 kb UCP1-GFP (green). (Right) Luciferase activity in HEK293FT cells cotransfected with the −5.5 kb UCP1-luc and indicated vector.
(B) Western blot analysis of Zc3h10 protein in various tissues from 10-week-old C57BL/6 mice.
(C) (Left) qRT-PCR for indicated genes in BAT cells at day 0–5 of differentiation. (Right) Immunoblotting for indicated proteins during BAT cell differentiation.
(D) Zc3h10 mRNA levels in the adipocyte fraction and SVF from BAT.
(E) Schematic representation of 5′ deletion constructs of the UCP1 promoter-Luc. Luciferase activity in HEK293FT cells, co-transfected with Zc3h10.
(F) Luciferase activity in differentiated BAT cells −/+ forskolin, cotransfected with indicated plasmids.
(G) (Top left) Zc3h10-binding sequences after 10 rounds of SELEX enrichment followed by MEME using 8-mer and 30-mer analysis. (Bottom left) Analysis of the UCP1 promoter using the IUPAC nucleotide for the Zc3h10 binding sites identified by SELEX. (Right) ChIP-qPCR of Zc3h10 at the binding region identified by SELEX using BAT cells.
(H) Gel shift assay performed with nuclear extracts of HEK293FT cells transfected with FLAG-Zc3h10 and oligos corresponding to the −4.6 kb UCP1 binding site.
(I) (Left) ChIP-seq of Zc3h10 occupancy at the promoters of UCP1, Tfam and Nrf1 in BAT of C57BL/6 mice. (Middle) De novo binding motif from Zc3h10ChIP-seq in BAT of WT mice. (Right) Gene Ontology of biological processes.
(J) (Left) Gel shift assay using GST-Zc3h10 fusion proteins/various GST-Zc3h10 domain deletions and oligos corresponding to the −4.6 kb UCP1 binding site. (Right) Luciferase activity in HEK293FT cells cotransfected with the −5.5 kb UCP1 promoter and Zc3h10 mutants (n = 4).
Data presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. See also Figure S1.
To examine how Zc3h10 activates the UCP1 promoter, we performed UCP1 promoter-reporter assays by using various UCP1 promoter deletion constructs in HEK293 cells. First, as above, Zc3h10 activated the −5.5 kb UCP1 promoter-luciferase (Luc) by 4.5-fold (Figure 1A, right); however, Zc3h10 could not activate the shorter −1.5 kb or −70 bp UCP1 promoter-Luc (Figure 1E). We next generated a series of reporter constructs containing a 1 kb deletion of the UCP1 promoter region starting from the upstream 5′ end, linked to −70 bp UCP1 proximal promoter. Co-transfection of Zc3h10 with the −5.5 to −4.5 kb linked to the 70 bp UCP1 promoter resulted in a robust promoter activation of over 4-fold. However, the −4.5 to −3.5, −3.5 to −2.5, and −2.5 kb sequences linked to the −70 bp UCP1 promoter showed a significantly lower promoter activation (Figure 1E). These data indicate Zc3h10 activates the UCP1 promoter mainly by a −5.5 to −4.5 kb upstream region. We also performed luciferase assays in differentiated BAT cells (Figure 1F, left). In the basal condition, Zc3h10 activated the −5.5 kb UCP1 promoter by only 2-fold compared to 4-fold observed in HEK293 cells (Figure 1E), probably due to the higher UCP1 promoter activity from endogenous Zc3h10 in differentiated BAT cells in the absence of transfected Zc3h10. After 6 h of forskolin treatment, Zc3h10 overexpression activated the −5.5 kb UCP1 promoter to nearly 6-fold compared to the EV transfection. When we used the −5.5 to −4.5 kb fragment linked to −70 bp UCP1-Luc, Zc3h10 activated the −5.5 to −4.5kb UCP1-Luc similarly by 2-fold (Figure 1F, right). In the forskolin-treated condition, Zc3h10 overexpression increased the −5.5 to −4.5 kb UCP1 promoter activity by 4.5-fold compared to the EV transfection. Importantly, the degree of the UCP1 promoter activation by Zc3h10 in the stimulated condition was 2.5-fold, similar to that of the −5.5 kb UCP1 promoter, confirming that Zc3h10 binds to the UCP1 promoter by the −5.5 to −4.5 kb region.
Identification of the Zc3h10-Binding Site and Its Target Genes
Next, to further define the binding sequence for Zc3h10, we performed systematic evolution of ligands by exponential enrichment (SELEX) for cyclic amplification and sequencing. FLAG-tagged Zc3h10 was used to select the DNA-binding sequence from a pool of oligonucleotide libraries. After 10 rounds of binding and elution, the eluted DNAs were sequenced and analyzed by de novo motif analysis (MEME). The top-scoring motifs for Zc3h10 binding were GC-rich sequences with repeated CC doublets, achieved by 8-mer analysis. Similar sequences were also identified by 30-mer analysis (Figure 1G, top left). An analysis of the proximal UCP1 promoter for the Zc3h10-binding site identified by SELEX revealed the 2 tandem Zc3h10-binding sites at −4.6 kb upstream of the UCP1 transcription start site (Figure 1G, bottom left). This region contains the sequence responsive to Zc3h10 by the above UCP1 promoter-luciferase reporter assay (Figures 1E and 1F). Indeed, chromatin immunoprecipitation (ChIP) showed strong enrichment of Zc3h10 at the −4.6 kb UCP1 locus (Figure 1F, right), confirming this site as a bona fide Zc3h10-binding site. We also examined whether Zc3h10 directly binds to DNA by electrophoresis mobility shift assay (EMSA) by using nuclear extracts from HEK293FT cells overexpressing Zc3h10 and oligonucleotides corresponding to the −4.6 kb UCP1 binding site. This EMSA showed a complex formation of Zc3h10 with the labeled probe, and the addition of cold competitor prevented the binding, demonstrating that Zc3h10 directly binds to the −4.6 kb UCP1 region (Figure 1H). Interestingly, Zc3h10 has recently been reported to be a RNA-binding protein (Treiber et al., 2017). However, our present study demonstrates that Zc3h10 can function as a DNA-binding transcription factor that activates the UCP1 promoter. In order to examine genome-wide Zc3h10 targets in BAT, we performed chromatin immunoprecipitation followed by next-generation sequencing (ChIP-seq) by using BAT from C57BL/6 mice (Figure 1I). We found a Zc3h10 binding peak at the −4.6 kb of the UCP1 promoter region, in agreement with the ChIP-qPCR, although there were multiple peaks of Zc3h10 binding in the UCP1 enhancer, proximal promoter, as well as intronic regions. We also detected Zc3h10 binding at the promoter regions of Tfam and Nrf1, which are transcription factors for mitochondrial biogenesis (Figure 1I). Gene Ontology (GO) analysis of the genes associated with Zc3h10-binding peaks confirmed a role in mitochondrial biogenesis (termed mitochondrion organization), and those genes linked to BAT-related genes, such as C/EBPβ (Figure 1H, right). Using biotinylated oligonucleotides corresponding to +50 bp Tfam and −200 bp Nrf1 regions in EMSA, binding of Zc3h10 to the Tfam and Nrf1 promoters, respectively, was detected (Figure S1B). Activation of the Tfam and Nrf1 promoters by Zc3h10 was also revealed in promoter-luciferase assays in differentiated BAT cells (Figure S1C). Together, these results show that, as a BAT-enriched transcription factor, Zc3h10 binds to UCP1 to transcriptionally activate the UCP1 and other genes, including those for mitochondrial biogenesis.
Zc3h10 Binds DNA by Its bZIP Domain
Next, we sought to identify the domain of Zc3h10 responsible for the DNA-binding property. We generated full-length Zc3h10-GST fusion proteins as well as truncations lacking either the N-terminal zinc finger domain or the C-terminal basic leucine zipper (bZIP) domain. Various purified Zc3h10-GST fusion proteins were used in EMSA as described above. Full-length Zc3h10 and Zc3h10 lacking the CCCH zinc finger domains had distinct shifted bands that were competed out with the addition of cold oligos. In contrast, Zc3h10 lacking the ΔbZIP domain did not result in a Zc3h10 complex formation (Figure 1J, left). These EMSA results show that the bZIP domain is required for the DNA-binding function. We conclude that Zc3h10 binds to the distal UCP1 promoter through the bZIP domain and not the zinc finger domain (Figure 1J, left). Treiber et al. (2017) reported that, as an RNA-binding protein, Zc3h10 regulates pre-miRNA processing and that the first two CCCH zinc fingers ZF1 and ZF2 serve as RNA-binding motifs. Therefore, we tested the potential role of the zinc fingers of Zc3h10 in activating the UCP1 promoter. HEK293FT cells were transfected with the zinc finger mutants of Zc3h10 (second and third cysteines, which are Zn2+-chelating residues, mutated to serine, provided by Treiber et al., 2017) along with the −5.5 kb UCP1 promoter-Luc reporter. All of the zinc finger mutant constructs could still activate the UCP1 promoter to a similar degree as that by the wild-type-Zc3h10, whereas the Zc3h10 DbZIP construct could activate UCP1 promoter to a significantly lower degree (Figure 1J, right). These results demonstrate that the zinc fingers for RNA-binding function are not required for activation of the UCP1 promoter, confirming that Zc3h10 activates the UCP1 promoter as a DNA-binding transcription factor.
Zc3h10 Promotes Thermogenic Gene Program In Vitro
We next performed a loss-of-function study for Zc3h10 in BAT gene program by using cultured BAT cells. BAT cells at day 2 of differentiation were transduced with adenoviruses expressing short hairpin RNAs targeting Zc3h10 (shZc3h10) for knock down (KD), and the cells were stimulated with forskolin on day 6 of differentiation. Transduction of shZc3h10 adenovirus in BAT cells decreased Zc3h10 mRNA and protein levels by approximately 50% (Figure 2A, left). There were no apparent changes in brown adipocyte differentiation of Zc3h10 KD cells, as observed by oil red O staining for lipid accumulation, as well as by expression levels of general differentiation markers, such as PPARγ, AdipoQ, and FABP4 (Figure 2B, left and right). However, the UCP1 mRNA level was reduced by 50%, and UCP1 protein levels in both basal and stimulated conditions were severely abrogated by Zc3h10 KD (Figure 2B). Moreover, Tfam and Nrf1 mRNA levels were also significantly lower (Figure 2B). Indeed, Zc3h10 KD cells showed decreased oxygen consumption under oligomycin-, FCCP-, and rotenone/antimycin A-treated conditions, and the uncoupled respiration in these cells was significantly lower (Figure 2C). In accordance of decreased Tfam and Nrf1 expression levels, Zc3h10 KD BAT cells had a significantly lower mitochondria DNA/genomic DNA ratio, a standard measurement of mitochondrial number. (Figure 2D, left). We also observed markedly decreased mitochondria in Zc3h10 KD cells by using MitoTracker Green (Figure 2D, middle). Quantification of the MitoTracker signal showed that Zc3h10-KD BAT cells had nearly 40% reduced mitochondria compared to control cells (Figure 2D, right). Similar results were obtained by MitoTracker Red that detects membrane potentials (data not shown). Together, these data demonstrate that Zc3h10 is critical for the activation of BAT genes, including UCP1 and those for mitochondriogenesis, and thus for thermogenic function in BAT cells.
Figure 2. Zc3h10 Promotes Thermogenic Gene Program In Vitro.
(A) (Left) qRTqPCR and immunoblotting for Zc3h10KD in BAT cells.
(B) (Left) qRT-PCR for indicated genes in forskolin-treated differentiated Zc3h10KD BAT cells (Middle) Immunoblotting for indicated proteins. (Right) ORO staining of BAT cells (20× magnification) at day 5 of differentiation.
(C) OCR (pMol/min) measured using Seahorse XF24 analyzer in Zc3h10 KD BAT cells.
(D) (Left) Quantification of mitochondria number in Zc3h10KD BAT cells. (Middle) Mitochondria staining and quantification of MitoTraker Green signals in shZc3h10-infected cells (shown in red, n = 30).
(E) qRT-PCR/western blot for Zc3h10 in 3T3-L1 cells infected with Zc3h10 adenovirus.
(F) qRT-PCR/western blot for indicated genes in 3T3-L1 cells after 6 h of forskolin (10 μM) treatment.
(G) OCR (pMol/min) measured in Zc3h10-overexpressing 3T3-L1 cells.
(H) qRT-PCR for Zc3h10 in primary white adipocytes differentiated from SVF of WT and AQ-Zc3h10 TG.
(I) qRT-PCR/western blot for indicated genes in primary white adipocytes after 6 h of forskolin (10 μM) treatment.
(J) OCR measured by Seahorse assay.
Data presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
Next, to test the ability of Zc3h10 to induce UCP1 and other thermogenic genes by a gain-of-function study, we overexpressed Zc3h10 in 3T3-L1 cells. Ectopic overexpression of Zc3h10 was verified by qRT-PCR and western blot analysis (Figure 2E, left). Overexpression of Zc3h10 in 3T3-L1 cells (Zc3h10 OE) increased UCP1 mRNA levels significantly, with the average cycle threshold (Ct) value for the control 27.91 and for Zc3h10 overexpression 25.78, (Figure 2F, right) The increase in UCP1 expression was also detected at the protein level. Expression of other genes, including Tfam, Dio2, and CideA, was significantly increased as well (Figure 2F, right). The OCR of Zc3h10 OE cells was higher than the control in all conditions, and the uncoupled respiration measured under oligomycin was significantly increased (Figure 2G). We also used primary adipocytes differentiated from SVF from WAT of wild-type (WT) and AQ-Zc3h10 TG mice. As expected, the primary adipocytes from AQ-Zc3h10 TG mice compared to those from WT mice had significantly higher UCP1 and Tfam at the mRNA and protein levels. Adipocytes of AQ-Zc3h10 TG compared to WT mice also showed higher OCR at all time points, including basal, uncoupled, maximal OCR (Figures 2J and 2I). These data further demonstrate the significance of Zc3h10 in browning of white adipocytes in vitro, by activation of BAT gene program.
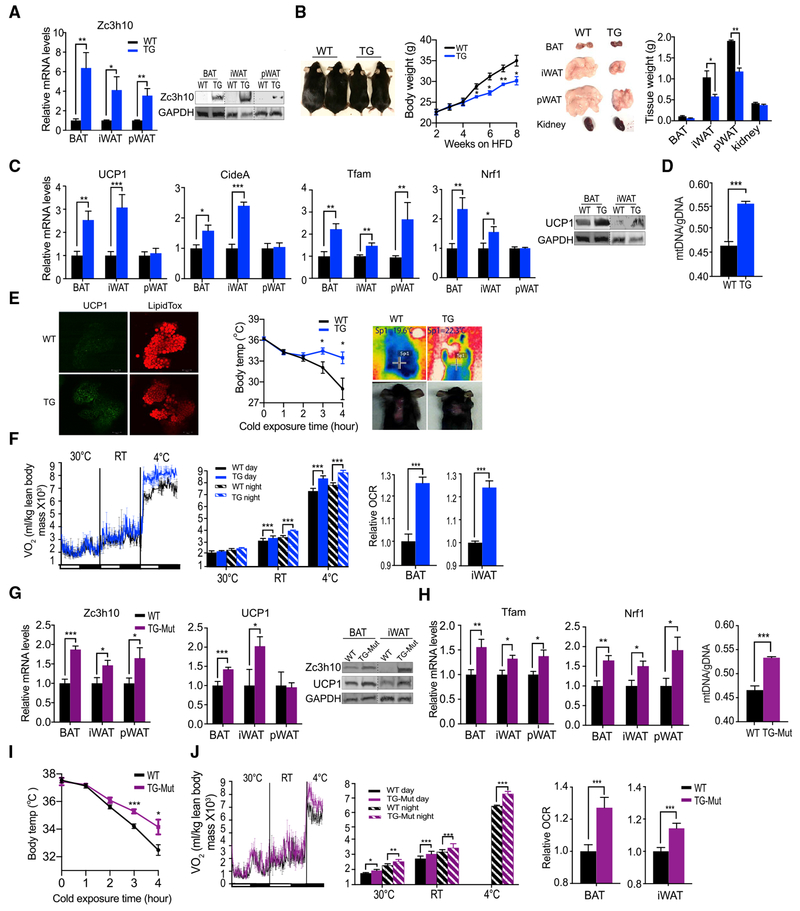
Overexpression of Zc3h10 Promotes BAT Gene Program to Enhance Energy Expenditure, Preventing Diet-Induced Obesity
To examine the effect of Zc3h10 in vivo, we next performed gain-of-function analyses by generating transgenic mouse lines overexpressing Zc3h10, driven by the −5.2 kb adiponectin promoter (AQ-Zc3h10). This allowed us to study the role of Zc3h10 in BAT, as well as its role in browning of subcutaneous WAT. In these AQ-Zc3h10 mice, Zc3h10 mRNA levels were higher by 6- and 4-fold in BAT and iWAT/pWAT, respectively. Zc3h10 protein levels were also significantly higher in BAT and iWAT/pWAT of AQ-Zc3h10 transgenic mice (Figure 3A). When we measured gene expression, UCP1 mRNA levels were increased by 2.5- and 3-fold in BAT and iWAT of transgenic mice, respectively (Figure 3C, left). Similarly, UCP1 protein levels were increased in both BAT and iWAT (Figure 3C, right), corroborated by whole-mount immunostaining of BAT (Figure 3E, left). Interestingly, even at room temperature, an increase in UCP1 at both mRNA and protein levels in iWAT was higher than those in BAT, suggesting its role in browning of iWAT. The AQ-Zc3h10 transgenic mice also had higher mRNA levels of CideA, Tfam, and Nrf1 (Figure 3C), accompanied with an increase of mitochondria number in BAT of TG mice (Figure 3D).
Figure 3. Overexpression Zc3h10 Promotes BAT Gene Program and Enhances Energy Expenditure, Preventing Diet-Induced Obesity.
(A) qRT-PCR/immunoblotting for Zc3h10 in adipose depots of WT and AQ-Zc3h10 mice (TG).
(B) Representative photograph and body weight of 16-week-old WT and TG mice fed high-fat diet (HFD) for 8 weeks (n = 5–6 mice per group).
(C) (Left) qRT-PCR for indicated genes in adipose depots of WT and TG mice housed at RT (n = 5-6 mice per group) (Right) Immunoblotting for UCP1 in BAT and iWAT.
(D) Quantification of mitochondria number in BAT (n = 12).
(E) (Left) Whole-mount immunostaining for UCP1 (green) and LipidTox staining (red) in BAT of 10-week-old mice. (Middle) Rectal temperature measured in 15-week-old mice at 4°C (n = 6 mice per group). (Right) Infrared thermography of 4-h cold-induced BAT activity; Sp1, temperature of specific area of BAT.
(F) (Left, middle) VO2 assayed in mice, housed at indicated ambient temperatures by indirect calorimetry using CLAMS (n = 5 per group). (Right) OCR measured in BAT and iWAT of mice using Seahorse XF24 Analyzer (n = 5).
(G) qRT-PCR/immunoblotting for Zc3h10 and UCP1 in various adipose depots of WT and TG-Mut, overexpressing Zc3h10-mutated RNA-binding domains, ZF1 and ZF2.
(H) (Left) qRT-PCR for indicated genes in adipose depots of WT and TG-Mut mice. (Right) Quantification of mitochondria number in BAT of WT and TG-Mut mice.
(I) Rectal temperature measured in 15-week-old mutant mice at 4°C at indicated time points (h) (n = 5–6 mice per group).
(J) VO2 assayed in WT and TG-Mut mice on CD, housed at indicated ambient temperature (n = 5–6 per group), measured by indirect calorimetry using CLAMS.
Data presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. See also Figure S2.
We next performed an acute cold exposure at 4°C. Upon 3 h of cold challenge, the TG had 2°C higher core body temperature than WT littermates (Figure 3E, right). The difference in core body temperature was even greater after 4 h, and the body temperature of transgenic mice was higher by 5°C than that of WT littermates. We also measured thermal changes by infrared thermography as a means to access BAT-derived thermogenesis. Indeed, upon 4 h of cold exposure, the BAT of AQ-Zc3h10 transgenic mice displayed higher temperature than that of WT littermates detected by infrared thermography (method reviewed in Crane et al., 2014) (Figure 3E, right). Considering an increased thermogenic capacity in AQ-Zc3h10 transgenic mice, we explored the role of Zc3h10 on energy balance by indirect calorimetry. The TG mice had significantly higher OCR at both room (23°C) and cold ambient temperature (4°C), including during days and nights, without changes in locomotor activity or food intake (Figure 3F, left and middle; Figure S2A). We next tested whether changes in respiratory activity specifically from BAT and iWAT were responsible for changes observed in whole-body OCR. Indeed, we detected increased OCR in BAT and iWAT isolated from AQ-Zc3h10 transgenic mice, as measured by a Seahorse assay, which is evidence of the contribution of BAT and iWAT to the enhanced whole-body metabolic activity (Figure 3F, right).
Observing enhanced energy expenditure measured by a striking increase in OCR, we tested whether AQ-Zc3h10 TG mice were protected against diet-induced obesity. AQ-Zc3h10 mice were placed on a high-fat diet (HFD) for 8 weeks. Starting at week 5, TG mice were significantly leaner than WT. And by week 8, TG mice were 6 g leaner than WT littermates. Although organ weights of BAT and kidney were comparable in WT and TG mice, TG mice had significantly smaller WAT depots, including iWAT and pWAT, (Figure 3B, right). In line with these results, the TG mice showed improved glucose tolerance and insulin sensitivity compared to WT littermates, especially those on HFD (Figures S2B and S2C). We also generated an additional transgenic mouse model overexpressing Zc3h10, driven by −5.4 kb aP2 (FABP4) promoter and detected similar alterations (Figures S2E-S2I). Thus, the aP2-Zc3h10 transgenic mice had increased UCP1 mRNA and protein levels, and they were more cold tolerant and lean (Figures S2G-S2I). Overall, these results demonstrate that Zc3h10 increases UCP1 and other BAT gene programs to increase cold tolerance and to increase energy expenditure, leading to protection against diet-induced obesity.
Zc3h10 RNA-Binding Activity Is Not Required for Activation of BAT Gene Program
To further address the function of Zc3h10 as an RNA-binding protein in thermogenic gene activation, we generated transgenic mice overexpressing a Zc3h10 mutant lacking RNA-binding ability, driven by the −5.2 kb adiponectin promoter (AQ-Zc3h10-mutZF1/2). This allowed us to examine Zc3h10 function only as a transcription factor, not as a RNA-binding protein. The Zc3h10-mutZF1/2 overexpression in AQ-Zc3h10-mutZF1/2 transgenic mice detected by qRT-PCR was 2- and 1.5-fold in BAT and iWAT, respectively. Similar to the results we obtained using AQ-Zc3h10 mice, the AQ-Zc3h10-mutZF1/2 mice showed increased mRNA and protein levels of UCP1 in BAT and iWAT compared to their WT littermates (Figure 3G). The AQ-Zc3h10-mutZF1/2 mice also had higher mRNA levels of CideA, Tfam, and Nrf1 (Figure 3H, left), accompanied with an increase of mitochondria number in BAT of mutant mice compared to BAT of WT mice (Figure 3H, right). As a result of an increase in thermogenic gene expression, the AQ-Zc3h10-mutZF1/2 transgenic mice had higher thermogenic capacity at 4°C so that their core body temperatures were significantly higher than those of WT littermates, especially after 3 h of cold exposure (Figure 3I). Moreover, AQ-Zc3h10-mutZF1/2 transgenic mice had higher VO2 at all temperatures (Figure 3J), with no differences in food intake or physical activity (Figure S2D). The enhanced energy expenditure measured by all tissues was contributed by significantly increased OCR, as measured by a Seahorse assay using BAT and iWAT from AQ-Zc3h10-mutZF1/2 transgenic mice (Figure 3J, right). The degree of changes observed was somewhat lower than those of the above AQ-Zc3h10 transgenic mice, probably due to the lower degree of Zc3h10-mutZF1/2 overexpression (2- and 1.5-fold in BAT and iWAT) than Zc3h10-WT (6- and 4-fold in BAT and iWAT). Regardless, Zc3h10-mutZF1/2 overexpression in transgenic mice clearly showed promotion of thermogenesis. Collectively, these results support the notion that Zc3h10 is a critical transcription factor that upregulates thermogenic gene program by binding to the UCP1 and other target promoter regions and that the RNA binding of Zc3h10 does not play a role in this process.
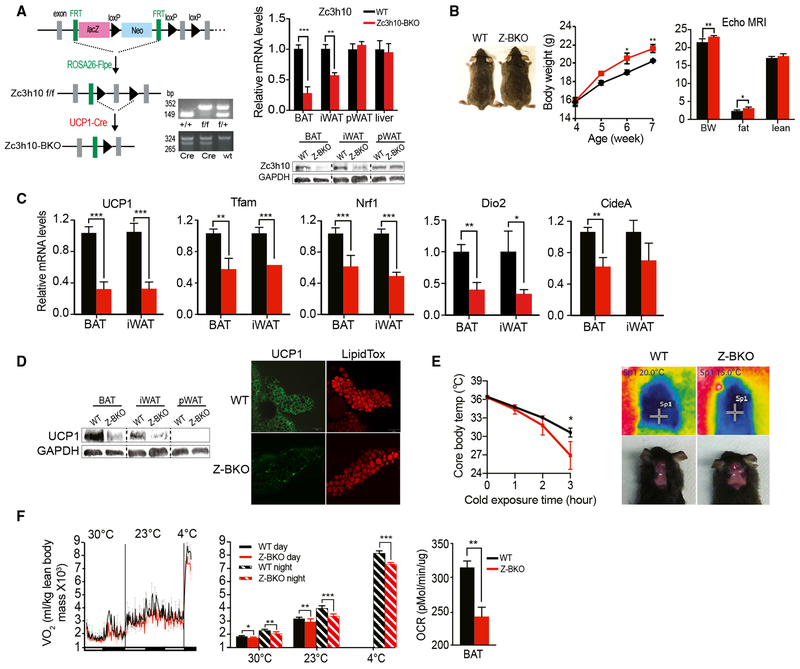
Zc3h10 Is Required for Activation of BAT Gene Program In Vivo
Next, to further examine the role of Zc3h10 in vivo, we performed loss-of-function studies by ablating Zc3h10 in UCP1+ cells in mice. We first generated Zc3h10 floxed mice by using embryonic stem cells (ESCs) obtained from EUCOMM. Zc3h10 floxed ESCs contained two FRT sites, flanking lacZ and neo genes between exons 2 and 3, followed by the two loxP sites flanking exon 3. Germline-transmitted mice were mated with ROSA26-Flp recombinase mice for LacZ-neo cassette excision. Flox/flox mice were bred to UCP1-Cre mice to ablate Zc3h10 in UCP1+ cells (Figure 4A, left). These Zc3h10 UCP1+-cell-specific KO (Zc3h10-BKO) mice were compared to control, flox/flox littermates for all experiments. In addition to genotyping using DNA, we also confirmed Zc3h10 deficiency by measuring Zc3h10 mRNA levels. Indeed, Zc3h10 mRNA levels were lower by 70% and 50% in BAT and iWAT, respectively (Figure 4A, right). Because these mice were kept at room temperature at 23°C, which is considered to be mildly cold, the Zc3h10 level was also lower in iWAT, probably due to the emergence of UCP1+ beige adipocytes in this WAT depot. Similarly, Zc3h10 protein levels were lower significantly in both BAT and iWAT, but not in pWAT, of Zc3h10-BKO mice. Zc3h10 deficiency was accompanied by a substantial reduction in the expression of genes associated with thermogenesis. We detected a 70% reduction in UCP1 mRNA levels in BAT (Figure 4C, left). Expressions of other BAT-enriched genes, such as Dio2 and CideA, as well as other Zc3h10 target genes, Tfam and Nrf1, were also lower in BAT and iWAT of Zc3h10-BKO mice (Figure 4C). The immunostaining for UCP1 showed drastically lower protein levels in BAT and iWAT of Zc3h10-BKO than in wild-type (WT) mice (Figure 4D, lower left). The decrease in UCP1 protein levels was corroborated by a reduced UCP1 staining in the whole-mount immunostaining of BAT tissue (Figure 4D, bottom right).
Figure 4. Zc3h10 Is Required for Full Activation of BAT Gene Program.
(A) (Left) Schematic diagram of the strategy used to generate BAT-specific Zc3h10 conditional knockout mice. PCR genotyping of the mice: (top) Zc3h10 allele; (bottom) Cre. (Right) qRT-PCR/immunoblotting for Zc3h10 with GAPDH as the loading control in tissues from Zc3h10 f/f (WT) and Zc3h10-BKO mice.
(B) Representative photograph of 8-week-old mice and their body weights and body composition.
(C) qRT-PCR for indicated genes in BAT/iWAT of mice.
(D) (Left) Immunoblotting for UCP1 and the same loading control GAPDH blot in various tissues (n = 5) as Zc3h10 and UCP1 were blotted using the same amount of lysate. (Right) Whole-mount immunostaining of UCP1 (shown in green) and LipidTox (shown in red) staining in BAT of 10-week-old mice (n = 5).
(E) (Left) Rectal temperature of 13-week-old mice maintained at 4°C. (Right) Infrared thermography of 4-h cold-induced BAT activity.
(F) (Left) VO2 assayed in WT and Zc3h10-BKO mice on CD using CLAMS. (Right) OCR measured in BAT from mice using Seahorse XF24 (n = 5).
Data presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. See also Figure S3.
Next, to study the physiological outcome from decreased expression of thermogenic genes, we subjected the Zc3h10-BKO mice to acute cold exposure at 4°C. Both WT and Zc3h10-BKO mice had a body temperature of 36°C prior to the cold exposure. After 3 h of cold exposure, the Zc3h10-BKO mice were cold intolerant compared to WT mice (Figure 4E, left), as the rectal temperature significantly dropped to 26°C, whereas WT littermates could maintain their body temperature of 31°C. Moreover, the BAT of Zc3h10-BKO mice displayed markedly lower temperature than that of WT littermates (Figure 4E, left) upon 4 h of cold exposure, as assessed by infrared thermography (Figure 4E, right). These data clearly show that Zc3h10-BKO mice had impaired thermogenic capacity with lower UCP1 expression in BAT and iWAT. We further assessed the functional consequences of the loss of Zc3h10 in UCP1+ cells on energy balance. We then examined the energy expenditure by measuring whole-body O2 consumption by CLAMS. Indeed, the Zc3h10-BKO mice showed a significantly lower VO2 than WT littermates at all temperatures and showed an even greater VO2 reduction at 4°C, whereas food intake and locomotive activity were similar (Figure 4F, left; Figure S3B). By using a Seahorse XF24 analyzer, we found a 30% decrease in OCR in BAT isolated from Z-BKO mice, showing BAT contribution to changes in whole-body energy expenditure (Figure 4F, right). Reflecting the impairment of thermogenesis and energy expenditure, Zc3h10-BKO mice displayed significantly larger body weight than control floxed mice on a chow diet. The body composition analysis by EchoMRI showed that Z-BKO mice had increased fat mass without changes in lean mass compared to WT, and the weight gain was primarily accounted for by adipose accumulation (Figure 4B; Figure S3A). In addition, blood glucose levels of Zc3h10-BKO mice were significantly higher during the course of glucose tolerance test (GTT) especially when on HFD, and they had impaired insulin sensitivity when subjected to an insulin tolerance test (Figure S3C and S3D). Together, we conclude that Zc3h10 is essential for thermogenesis by inducing UCP1 and other target genes. Thus, Zc3h10 can increase energy expenditure to prevent diet-induced adiposity.
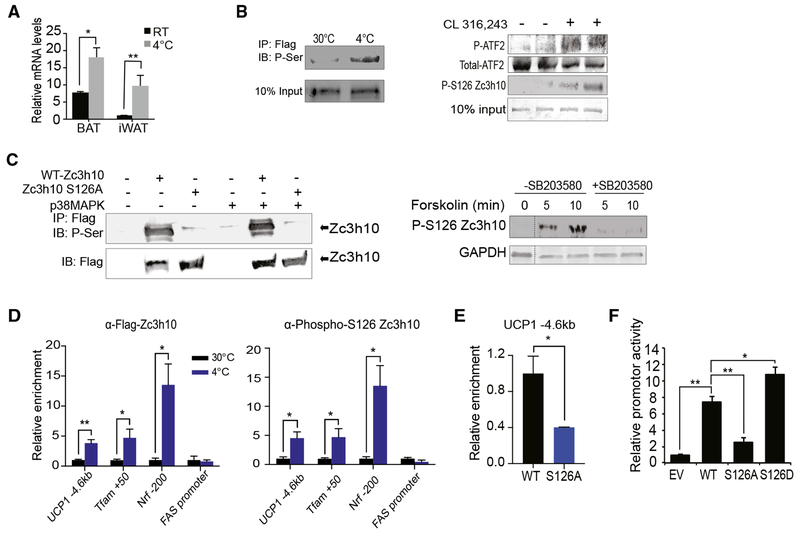
Phosphorylation of Zc3h10 at S126 by p38 MAPK in Response to Cold in Activating UCP1 Transcription
We found that Zc3h10 expression at mRNA and protein levels is induced upon cold exposure (Figure 5A). However, Zc3h10 may also be regulated by post-translational modification in promoting the BAT gene program in conditions when thermogenesis is activated through β3-adrenergic stimulation. Indeed, Phosphosite indicated multiple phosphorylation sites of Zc3h10. We also tested and compared the phosphorylation status of Zc3h10 in BAT of mice under thermoneutral conditions and cold exposure. Indeed, cold-exposed mice showed a strong phosphorylation of Zc3h10 by immunoblotting using a pan-phosphoserine antibody (Figure 5B, left), demonstrating that Zc3h10 is phosphorylated upon cold exposure. We examined the role of S126 phosphorylation due to its high-frequency detection among serine/threonine residues. To test whether the S126 is indeed specifically phosphorylated during thermogenesis, we generated an anti-peptide antibody specific to the Zc3h10 peptide sequence corresponding to phosphorylated S126. Next, we performed immunoblotting of BAT from mice intraperitoneally administered with CL-316,243 by using anti-phospho-S126 antibody. Indeed, we detected S126 phosphorylation of Zc3h10 in BAT from mice when treated with CL-316,243, in parallel to an increase in p38 MAPK activity, as detected by ATF2 phosphorylation (Figure 5B, right). It has been reported that p38 MAPK is an essential component of the cAMP signaling cascade that activates thermogenic gene program in response to β3-adrenergic activation. Examination of the amino acid sequence near S126 suggested it to be a potential p38 MAPK site. To study Zc3h10 as a downstream substrate of p38 MAPK for the promotion of UCP1 transcription, we generated a Zc3h10 expression construct mutated of the putative phosphorylation site of S126 to non-phosphorylatable S126A. HEK293FT cells were transfected with either Zc3h10 or Zc3h10 with S126A mutation. Using a phosphoserine antibody, we detected strong phosphorylation of Zc3h10 but not the S126A mutant, (Figure 5C, left), which demonstrated that S126 is the major site of Zc3h10 phosphorylation. We next tested whether S126 is the target of p38 MAPK. HEK293FT cells were co-transfected with p38 MAPK and Zc3h10 or S126A mutant. We found that phosphorylation of Zc3h10 was higher, whereas the S126A mutant showed the same weak signal as in control cells without transfection of p38 MAPK. The weak signaling probably was from presence of endogenous p38 MAPK in HEK293 cells. These results support the idea that phosphorylation of Zc3h10 at S126 is by p38 MAPK (Figure 5C, left). To further address the role of p38 MAPK, we pretreated the cells with a p38 MAPK inhibitor, SB203580, for 30 min before the forskolin treatment. Indeed, phosphorylation of S126 was greatly increased by 5 and 10 min of forskolin treatment, but this increase was totally abolished by SB203580 pretreatment (Figure 5C, right), revealing that p38 MAPK phosphorylates Zc3h10 at S126.
Figure 5. Phosphorylation of Zc3h10-S126 by p38 MAPK for UCP1 Transcription during Cold Exposure.
(A) qRT-PCR for Zc3h10 in BAT/iWAT 10-week-old C57BL76 mice housed at RT or at 4°C (n = 4).
(B) (Left) Immunoblotting of BAT from AQ-Zc3h10 TG mice housed at indicated ambient temperatures using a P-Serine antibody. (Right) Immunoblotting of BAT from AQ-Zc3h10 TG mice that are injected with saline or CL 316,243 using a P-S126 Zc3h10 specific antibody. p38MAPK activity was detected by P-ATF2.
(C) (Left) Immublotting for P-Serine of lysates of HEK293FT cells. (Right) HEK293FT cells were treated with p38 MAPK inhibitor SB203580 (10 μM) for 30 min before the forskolin (1 μM) treatment for the indicated duration.
(D) ChIP-qPCR for Zc3h10 and P-S126 Zc3h10 occupancy to Zc3h10 target genes using BAT chromatin from AQ-Zc3h10 TG.
(E) ChIP-qPCR for Zc3h10 binding at the −4.6 kb region of UCP1 promoter using FLAG antibody.
(F) UCP1 promoter-Luc activity using HEK293FT cells (n = 4).
Data presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. See also Figure S4.
Having determined that Zc3h10 is phosphorylated at S126 by p38 MAPK, we next examined the effect of phosphorylation on enrichment of Zc3h10 at the UCP1 promoter region. ChIP-qPCR was performed using BAT from AQ-Zc3h10 transgenic mice housed either at thermoneutrality (30°C) or at 4°C. In effect, upon cold exposure, the enrichment of Zc3h10 at the −4.6 kb region of the UCP1 promoter was increased by 4-fold (Figure 5D, left). Use of our phospho-S126 Zc3h10 antibody also showed a similar fold increase in Zc3h10 binding at the −4.6 kb UCP1 (Figure 5D, right). The enrichments of Zc3h10 at other Zc3h10 target genes, namely, Tfam and Nrf1, were also increased significantly. These data support that phosphorylation of Zc3h10 at S126 enhances binding to its target genes. However, because Zc3h10 expression is increased upon cold exposure, we could not preclude that this was due to an increase in total Zc3h10 protein in response to cold. To this end, we overexpressed S126A and WT-Zc3h10 in HEK293FT cells and performed ChIP-qPCR for Zc3h10 binding at the −4.6 kb region of UCP1 promoter. The enrichment of S126A Zc3h10 at the −4.6 kb region of the UCP1 promoter was significantly lower by 60% than that of WT-Zc3h10 (Figure 5E). We next examined whether Zc3h10 phosphorylation results in an increase in promoter activity as a consequence of affecting DNA binding. In addition to the S126A mutant, we generated a Zc3h10 expression construct mutated of S126 to hyperphosphorylation mimicking S126D. WT-Zc3h10 as well as S126A and S126D mutants were co-transfected each with the −5.5 kb UCP1-Luc reporter into HEK293FT cells. WT-Zc3h10 activated the UCP1 promoter by over 7-fold relative to the vector control, and the S126D resulted in an even higher UCP1 promoter activation by 10-fold (Figure 5F). In contrast, the co-transfection of the S126A mutant resulted in a 60% lower UCP1 promoter activity than WT, signifying that phosphorylation of S126 of Zc3h10 is essential for Zc3h10-mediated activation of the UCP1 promoter (Figure 5F). Collectively, these results demonstrate that Zc3h10 is phosphorylated by p38 MAPK at S126 upon cold exposure, and this phosphorylation is essential for its binding to the UCP1 promoter, thereby activating transcription.
DISCUSSION
It is well established that thermogenic gene program is regulated by a network of DNA-binding proteins as well as a large number of cofactors and epigenetic factors that are recruited to activate or repress transcription. Thus, it is imperative to understand how transcription factors and cofactors interplay to orchestrate transcription of UCP1 and other thermogenic genes. In the present study, we identified Zc3h10 as a transcriptional activator of UCP1 by using an unbiased transcription factor library screening. Zc3h10 belongs to the family of CCCH-type zinc finger proteins. Zc3h10 is brown fat enriched and cold inducible and transcriptionally activates UCP1. Overexpression of Zc3h10 increases thermogenic gene expressions in BAT and iWAT in vivo. Conversely, Zc3h10 ablation in UCP1+ cells in mice drives to lower thermogenic gene expression, impairing thermogenic capacity, lowering OCR in BAT, and leading to weight gain. Importantly, we show that upon sympathetic signaling, Zc3h10 is phosphorylated by p38 MAPK to increase its binding to the target genes.
By using several approaches, such as EMSA and SELEX, we demonstrate that Zc3h10 is a DNA-binding transcription factor. In this regard, as a member of CCCH zinc finger proteins that make up the second most common group of RNA-binding proteins (RBPs), Zc3h10 has been reported as a RBP, specifically binding to pri-miRNA to prevent processing to pre-miRNA through binding of ZF1 and ZF2 of Zc3h10 (Ray et al., 2013; Treiber et al., 2017). Also, the mutation of either of these zinc fingers was sufficient to block Zc3h10 binding to target miRNAs. However, we show that the DNA-binding activity of Zc3h10 does not require the RNA-binding zinc fingers because the mutation of the RNA-binding domain did not affect Zc3h10 activation of the UCP1 promoter. Furthermore, in EMSA, Zc3h10 lacking the CCCH zinc finger domains still can form a Zc3h10-DNA complex, whereas the complex formation was completely hindered when the bZIP domain of Zc3h10 was deleted. This demonstrates that the bZIP domain is required for the DNA-binding function. Moreover, overexpression of mutated Zc3h10, which lacks the RNA-binding ability in mice, still results in an increase in thermogenic gene expression and thermogenic capacity. Through both in vitro and in vivo studies, we unequivocally demonstrate that the zinc fingers for the RNA-binding property are not required for activation of the UCP1 promoter, revealing that Zc3h10 has a dual role as a DNA-binding transcription factor and a RNA-binding protein.
Interestingly, the majority of known transcription factors, such as ATF2, TR, and PPARγ, all act through the −2.5 kb enhancer region of the UCP1 gene. In contrast, Zc3h10 binds at the far upstream −4.6 kb region (Villarroya et al., 2017). Recently, there have been an increasing number of studies supporting that larger enhancer domains, so-called super enhancers (SEs), that are occupied by master transcription factors and are associated with Mediator to facilitate the ability of enhancer-bound transcription factors to recruit RNA polymerase II (Pol II) or writers of activation marks to the target genes (Whyte et al., 2013; Harms et al., 2015). Importantly, these SEs appear to span from the distal −4 kb region to −5.5 kb upstream of UCP1 promoter. These findings strongly suggest that yet-to-be-identified transcription factors may bind at the distal region to play an important role in tissue-specific and regulated UCP1 transcription (Brunmeir et al., 2016; Lai et al., 2017). We propose that Zc3h10 is one such transcription factor that binds at the distal UCP1 promoter region to be critical for transcriptional activation.
We report here that, in addition to UCP1, Zc3h10 binds to Tfam and Nrf1 promoter regions for mitochondrial biogenesis required for thermogenic BAT program. Nrf1 is an important transcription factor that increases transcription of key mitochondrial enzymes and upregulates multiple mitochondrial genes, including Tfam that promotes transcription and replication of mtDNA (Jornayvaz and Shulman, 2010). During the preparation of our manuscript, Audano et al. (2018) reported that Zc3h10 regulates mitochondria function by finding that knock down of Zc3h10 impairs differentiation of C2C12 into myotubes. These authors proposed that Zc3h10 may regulate mitochondrial function through its RNA binding activity by an unknown mechanism, even when they identified Zc3h10 by using Tfam promoter (Audano et al., 2018). In fact, BAT develops from Myf5+ precursors that can differentiate into both muscle cells and brown adipocytes (Seale et al., 2008); it is intriguing that Zc3h10 has similar effects in both of tissues through Tfam. In the present study, however, we clearly show by EMSA and luciferase assays that Zc3h10 binds and activates the promoter of Tfam and Nrf1 known for mitochondrial biogenesis, which also supports the functional significance of Zc3h10 as a DNA-binding protein. Moreover, by generating mice overexpressing Zc3h10 containing zinc finger mutations, we show that the Zc3h10, which lacks the RNA-binding property, can increase transcription of Tfam and Nrf1 and results in an increase in mitochondria number and, thus, further supporting Zc3h10 function as a DNA-binding transcription factor to promote mitochondrial biogenesis.
Adaptive thermogenesis relies on a coordinated signaling cascade in response to cold. It is well established that p38 MAPK is an essential component of signaling cascade mediating β3-adrenergic regulation of UCP1 expression in BAT for cold acclimation. In particular, increased UCP1 transcription has been reported to be dependent on β3-adrenergic receptor-PKA-p38 MAPK pathway by phosphorylation of ATF2 and PGC1α (Cao et al., 2001, 2004). p38 MAPK has been reported to protect ATF2 from protein degradation upon phosphorylation at T69 and T71, resulting in an increase in transcriptional activity (Watson et al., 2017). Furthermore, although p38 MAPK enhances PGC1α stability and transcription (Knutti et al., 2001; Puigserver et al., 2001), phosphorylation has also been reported to remove the binding and repression by p160 Myb binding protein (Fan et al., 2004), resulting in increased PGC1a transcriptional capacity. Here, we show that p38 MAPK phosphorylates Zc3h10 at S126 upon cold exposure or β3-adrenergic agonist. By a combination of ChIP-qPCR and luciferase assay, we demonstrate that phosphorylation of Zc3h10 at S126 by p38 MAPK is required to increase Zc3h10 binding to the distal UCP1 promoter region for transcriptional activation of the UCP1 promoter, as S126A-Zc3h10 results in significantly lower binding at its target sites (Figures 5E and 5F). It is possible that upon cold exposure, Zc3h10 that binds the −4.6 kb region of UCP1 and ATF2 and PGC1α that bind at the −2.5 kb enhancer region of UCP1 are all phosphorylated by p38 MAPK to work together in a cooperative manner to activate the UCP1 promoter.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hei Sook Sul (hsul@berkeley.edu). All plasmids generated in this study will be made available on request but we may require a payment and/or a completed Materials Transfer Agreement if there is potential for commercial application.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Adiponectin-Zc3h10 mice were generated by cloning Zc3h10-3xFLAG into −5.2 kb Adiponectin promoter-Cre in pbetaGal vector provided by Dr. P. Scherer. Cre insert and small T intron were removed and replaced with Beta globin intron and poly(A) signal. Phusion polymerase was used to amplify and add restriction sites to Zc3h10 using the following primers: 5′-AATAGTCGACACCATGTGGAGGTGGAGACTC-3′ and 5′-CTCCATCGATCTATTTATCGTCATCATCTTTGTAGTCC-3′. SalI and ClaI sites were used to ligate Zc3h10 to the vector. Generation of mice was performed by the UC Berkeley Gene Targeting Facility. ES cells containing the KO first, conditional Zc3h10 allele knock-in (Zc3h10tm1a(EUCOMM)Wtsi) were obtained from the International Mouse Phenotyping Consortium and injected at the Gladstone Institutes Transgenic Gene-Targeting Core. Zc3h10 conditional knockout mice were generated by first mating with FLPe mice (ROSA26::FLPe knock in) from Jackson Lab. FLP mediated recombination was confirmed by PCR. Resultant progenies were mated with UCP1-cre mice (B6.FVB-Tg(Ucp1-cre)1Evdr/J) also from Jackson Lab. Unless otherwise stated, mice used in experiments were male and between 8-12 weeks of age. Mice were fed a chow diet or a palatable high fat diet (45% fat derived calories-Dyets) ad libitum. All protocols for mice studies were approved from the University of California at Berkeley Animal Care and Use Committee.
Cell lines
HEK293FT and 3T3-L1 cell lines were obtained from UCB Cell Culture Facility which is supported by The University of California Berkeley. The immortalized BAT cell line was from Dr. Shingo Kajimura (UCSF) (Ohno et al., 2013). Cells were grown in standard condition with 5% CO2, at 37°C. Primary SVF cells were isolated from 12 wk-old male mice of indicated genotype.
METHOD DETAILS
Cold-induced thermogenesis
Cold exposed mice were housed for 10 days at thermoneutrality before cold exposure at 4°C. Core body temperature was determined using a Physitemp BAT-12 probe. CL-316,243 (Sigma) was injected intraperitoneally into mice at 1mg/kg. Infrared thermography of BAT was taken by FLIR E9-XT thermal camera.
Indirect calorimetry
Oxygen Consumption was measured using the Comprehensive Laboratory Animal Monitoring System (CLAMS). Data were normalized to lean body mass determined by EchoMRI. Mice were individually caged and maintained under a 12 hr light/12 hr dark cycle. Food consumption and locomotor activity were tracked.
Whole mount staining
1mm piece of tissue was excised, fixed with 1% PFA, incubated with UCP1 antibody, LipidTox Red Reagent (Thermo Fisher) and DAPI. Tissues were immobilized on a slide with mounting medium and visualized using a confocal microscope.
Cell culture
BAT cells, 3T3-L1 and HEK293FT cells were maintained in DMEM containing 10% FBS and 1% pen/strep prior to differentiation/transfection. Brown adipocyte differentiation was induced by treating confluent cells in DMEM containing 10% FBS, 0.5 mM isobutyl-methylxanthine, 125 nM indomethacin, 5 μM dexamethasone, 850 nM insulin, 1 nM T3, and 1 μM rosiglitazone. Two days after induction, cells were switched to the maintenance medium containing 10% FBS, 850 nM insulin, 1 nM T3, 1 μM rosiglitazone. shZc3h10 adenoviruses were purchased from Vector Biolabs while Ad-Zc3h10-flag adenovirus was purchased from Viraquest. Knockdown of Zc3h10 accomplished using adenovirus expressing short hairpin targeting Zc3h10 (shZc3h10) of MOI of 50 at D2 of adipogenic differentiation. 3T3-L1 cells were induced to differentiate by treatment of confluent cells with DMEM containing 10% FBS, 850 nM insulin, 0.5 mM isobutyl-methylxanthine, 1 μM dexamethasone, 1 nM T3, 125 nM indomethacin. After 48 h of induction, cells were switched to a maintenance medium containing 10% FBS, 850 nM insulin, 1nM T3. Cells were infected with GFP or Zc3h10 adenovirus at Day 4. The following day, medium was replaced to a maintenance medium. To stimulate browning/beiging, cells at Day 6 were treated with 10 μM forskolin for 6 hr. Total RNA was extracted using TRIzol reagent (Invitrogen) on the days indicated. Wells used for ORO staining were washed once with phosphate-buffered saline and subsequently fixed for 30 minutes in 4% paraformaldehyde. Cells were incubated in 60% isopropanol then stained with Oil Red O working solution for 1hour. Cells were washed twice with distilled water prior to visualization.
Mitochondrial analysis
MitoTracker Green was added into the culture media at final concentrations of 300nM. The cells were incubated under normal culture conditions for 30 min, mounted with Duolink Mounting medium with DAPI, and then visualized by fluorescence microscopy. shZc3h10-transfected cells were marked using ImageJ software, and MitoTracker Green intensity within the cells was measured.
Seahorse assay
BAT cell line cells were differentiated in 12-well plates, trysinized, and reseeded in XF24 plates at 50K cells per well at day 4 of differentiation and assayed on day 5 of differentiation. On the day of experiments, the cells were washed three times and maintained in XF-DMEM (Sigma-Aldrich) supplemented with 1 mM sodium pyruvate and 17.5 mM glucose. Oxygen consumption was blocked by 1 μM oligomycin. Maximal respiratory capacity was assayed by the addition of 1 μM FCCP. Tissues were incubated for 1 hour at 37°C without CO2 prior to analysis on the XF24 Analyzer.
RT-PCR Analysis and western blotting
Reverse transcription was performed with 500ng of total RNA using SuperScript III (Invitrogen). RT-qPCR was performed in triplicate with an ABI PRISM 7500 sequence detection system (Applied Biosystems) to quantify the relative mRNA levels for various genes. Statistical analysis of the qPCR was obtained using the ΔΔCt method with 18 s as the control. Primer sets used are listed in Table S1. For western blot analysis, total cell lysates were prepared using RIPA buffer, and nuclear extracts were isolated using the NE-PER Nuclear and Cytoplasmic Extraction kit (Thermo). Proteins were separated by SDS-PAGE, transferred to nitrocellulose membrane and probed with the indicated antibodies.
Luciferase Assays
293FT cells were transfected with 300ng Zc3h10 (WT or Mutants) or empty vector with 100 ng of indicated luciferase reporter construct and 0.5ng pRL-CMV in 24-well plates with Lipofectamine2000 reagent. Cells were lysed 48h post-transfection and assayed for luciferase activity using the Dual-Luciferase Kit (Promega) according to manufacturer recommended protocol. BAT cells were transfected with 2ug Zc3h10 or empty vector along with 1ug promoter-reporter constructs using Lipofectamine3000 reagent. Cells were differentiated first and, at Day 4, cells were treated with forskolin(10 uM) for 6 hr before lysing and assaying for luciferase activity.
Plasmid Constructs
The Zc3h10 sequence was subcloned by PCR amplifying and inserting into 3Tag-3a from Agilent as well as pGEX-4T-3 from GE. Zc3h10 deletion constructs were created by PCR amplifying the non-deleted part of the vector and inserting a HindIII site. Zc3h10 phosphomimetic mutants were created by site directed mutagenesis using the following primers: S126A F: TGGGTCTCGCACCCGCCGACCTACCCA R: TCGGCGGGTGCGAGACCCAGCCCAGCG S126D F: TGGGTCTCGATCCCGCCGACCTACCCA
R: TCGGCGGGATCGAGACCCAGCCCAGCG.
ChIP and ChIPseq
For ChIP experiments in BAT, tissues from 3 BATs were combined and minced on ice. Crosslinked was performed using 1% methanol-free formaldehyde in crosslinking buffer from the truChIP chromatin shearing kit (Covaris) for 5 min. The manufacturer recommended protocol was followed with the exception of adding 2 dounce steps following the neutralization of the crosslinking with the loose pestle and after addition of the lysis buffer with the tight pestle. Samples were filtered through a 100uM mesh following resuspension in wash buffer. Nuclei were sonicated on an M220 for 8mins at the manufacturer recommended power settings. Soluble chromatin was quantified by absorbance at 260 nm, and 30ug of chromatin was immunoprecipitated using 3 μg of the indicated antibodies or normal mouse IgG control using buffers and protocol from the Simple ChIP kit (cell signaling). Samples were analyzed by qPCR using the primer sets in Table S1 for enrichment in target areas. ChIP-seq samples were then quantified by PicoGreen. Library construction and sequencing were performed by the UC Berkeley Functional Genomics laboratory (FGL) and the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley on an Illumina HiSeq 4000 using 75-nt reads. Data was mapped to mm10 using BWA and peak calling was performed using MACS with the minimum FDR (q-value) set to p0 < 0.005. Binding motifs were identified by MEME-ChIP and ontological data was analyzed by GREAT (McLean et al., 2010).
SELEX
Zc3h10-flag was transfected into 293FT cells and 20ug of the resultant nuclear extract (NE-PER kit-Thermo) was immunoprecipitated using 15ul of magnetic M2-FLAG beads (Sigma) for 1 hour. Immobilized Zc3h10 was washed once with FLAG-wash buffer, and once with SELEX buffer before incubation of 40N aptamer complexes. SELEX was performed using the XELEX kit (Roboklon) according the manufacturer suggested protocol with aptamer incubation at 25°C. DNA aptamer eluates after 5 and 10 rounds of selection were subcloned into pGEM T-Easy (Promega). 100 colonies from each ligation were grown and the resultant plasmids were sequenced. Adaptor sequences were manually excised and Zc3h10 binding motifs were identified by MEME-ChIP.
CoIP
For Co-IP experiments using tagged constructs, 293FT cells were transfected using Lipofectamine2000 to express Flag-tagged Zc3h10. Cells were lysed in IP buffer containing 20 mM Tris, pH 7.4, 150 mM NaCl, 1mM EDTA, 10% glycerol, 1% NP-40 supplemented with proteases inhibitors. Total cell lysates were incubated 2 hours at 4°C with either anti-FLAG M2. For Co-IP in brown adipose tissue of wild-type mice, nuclear extraction was carried out using the NE-PER Nuclear and Cytoplasmic Extraction kit (Thermo). Equal amounts of nuclear extracts were incubated with specific antibodies and protein A/G agarose beads overnight at 4° C. Agarose beads were washed 3 times and bound proteins were eluted by boiling in Laemmli sample buffer and analyzed by immunoblotting using the indicated antibodies.
Gel Shift
GST and GST-Zc3h10 full length and deletion plasmids were transformed into BL21(DE3) E. coli (NEB) and production was induced by induction by 0.1M IPTG. Resultant proteins were purified using Glutatione S-Sepharose 4B (GE) according to the manufacturer recommended protocol. Oligos corresponding to the −4.6 kb UCP1 binding site F: 5′-TTTACCAAGTCCTTTCCACCGTGCAGGACGCACCATTCGGCAGCTTCCAGTTGTCTGGGAACTTCCAGTTGAAGGGATGA-3′ and R: 5′-TCATCCCTTCAACTGGAAGTTCCCAGACAACTGGAAGCTGCCGAATGGTGCGTCCTGCACGGTGGAAAGGACTTGGTAAA-3′ were biotinylated using the Pierce Biotin 3′ End DNA Labeling Kit according to the manufacturer protocol. Gel shift assays were performed using the 10nM of annealed labeled oligos and 5ug of purified protein using the LightShift Chemiluminescent EMSA Kit (Thermo) according the manufacturer recommended protocol.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed using two tailed t test. The error bars represent standard error mean (SEM). Data are expressed as mean ± SEM, p value < 0.05 was considered statistically significant, *p < 0.05, ** p < 0.01, *** p < 0.001. Number of mice or replicates used in each experiment is indicated in figure legends. Experiments were repeated at least three times; representative data are shown. Cells and mice were randomly assigned into experimental groups. Subjects were excluded from analysis only due to health conditions.
DATA AND CODE AVAILABILITY
ChIP-sequencing data have been deposited in NCBI’s GEO (GSE139029).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Topo IIB | Santa Cruz | Cat#SC-48429; RRID:AB_628383 |
| GAPDH | Santa Cruz | Cat#SC-25778; RRID:AB_10167668 |
| UCP1 | Abcam | Cat#ab10983; RRID:AB_2241462 |
| FLAG | Cell signaling | Cat#14793S; RRID:AB_2572291 |
| Zc3h10 (Immunoblotting) | ThermoFisher | Cat#PA5-31814; RRID:AB_2549287 |
| Zc3h10 (ChIP) | Abnova | Cat#H00084872-B01P; RRID:AB_1643833 |
| P-S126 Zc3h10 | Genemed Synthesis Inc. | N/A |
| ATF2 | Cell signaling | Cat#35031; RRID:AB_2799069 |
| P-ATF2 | Abcam | Cat#ab32019; RRID:AB_725567 |
| Phosphoserine | Millipore | Cat#ab1603; RRID:AB_390205 |
| Bacterial and Virus Strains | ||
| Ad-Zc3h10-flag | ViraQuest | N/A |
| Ad-Zc3h10-shRNA | Vector Biolabs | Cat#shADV-276549 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MitoTracker Green | Cell signaling | Cat#9074 |
| LipidTox Red Reagent | ThermoFisher | Cat#H34476 |
| Duolink Mounting medium with DAPI | Sigma-Aldrich | Cat#DUO82040 |
| CL-316,243 | Sigma-Aldrich | Cat#C5976 |
| SB203580 | Sigma-Aldrich | Cat# S8307 |
| Critical Commercial Assays | ||
| LightShift Chemiluminescent EMSA Kit | Thermofisher | Cat#20148 |
| SimpleChIP kit | Cell Signaling | Cat##56383 |
| Mitochondria Isolation Kit for Tissue | Thermofisher | Cat#89801 |
| NE-PER Nuclear and Cytoplasmic Extraction kit | ThermoFisher | Cat#78833 |
| Dual-Luciferase Kit | Promega | Cat#E1980 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE139029 |
| Experimental Models: Cell Lines | ||
| 3T3-L1 | UCB Cell Culture Facility | N/A |
| HEK293FT | UCB Cell Culture Facility | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: AQ-Zc3h10 | This Laboratory | N/A |
| Mouse: AQ-Zc3h10-mutZfp1/2 | This Laboratory | N/A |
| Mouse: UCP1-Cre B6.FVB-Tg(Ucp1-cre)1Evdr/J | The Jackson Laboratory | 024670 |
| Oligonucleotides | ||
| See Table S1 for RT-qPCR and ChIP primers | N/A | |
| See Table S2 for gel shift probes | N/A | |
| Recombinant DNA | ||
| pCMV-3Tag3A | Agilent | Cat#240195 |
| pGEX-4T-3 | GE Healthcare | Cat#28-9545-52 |
| Software and Algorithms | ||
| MACS | Zhang et al., 2008 | https://github.com/taoliu/MACS/ |
| GREAT | McLean et al., 2010 | http://great.stanford.edu/public/html/ |
| MEME | Bailey and Elkan, 1994 | http://meme-suite.org/tools/meme |
Highlights.
Zc3h10 directly binds and activates the UCP1 and other BAT gene promoter regions
Zc3h10 promotes BAT gene expression and protects mice from diet-induced obesity
p38 MAPK phosphorylates Zc3h10 to allow binding to target sites for transcription
ACKNOWLEDGMENTS
We thank Dr. Meister for Zc3h10 zinc finger mutant constructs. We thank T. Lou and M. Zhu for technical assistance. The work was supported by NIH grant DK120075-01 to H.S.S.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.10.099.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Audano M, Pedretti S, Cermenati G, Brioschi E, Diaferia GR, Ghisletti S, Cuomo A, Bonaldi T, Salerno F, Mora M, et al. (2018). Zc3h10 is a novel mitochondrial regulator. EMBO Rep. 19, e45531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, and Elkan C (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36. [PubMed] [Google Scholar]
- Brunmeir R, Wu J, Peng X, Kim SY, Julien SG, Zhang Q, Xie W, and Xu F (2016). Comparative Transcriptomic and Epigenomic Analyses Reveal New Regulators of Murine Brown Adipogenesis. PLoS Genet. 12, e1006474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Medvedev AV, Daniel KW, and Collins S (2001). beta-Adrenergic activation of p38 MAP kinase in adipocytes: cAMP induction of the uncoupling protein 1 (UCP1) gene requires p38 MAP kinase. J. Biol. Chem. 276, 27077–27082. [DOI] [PubMed] [Google Scholar]
- Cao W, Daniel KW, Robidoux J, Puigserver P, Medvedev AV, Bai X, Floering LM, Spiegelman BM, and Collins S (2004). p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol. Cell. Biol. 24, 3057–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane JD, Mottillo EP, Farncombe TH, Morrison KM, and Steinberg GR (2014). A standardized infrared imaging technique that specifically detects UCP1-mediated thermogenesis in vivo. Mol. Metab. 3, 490–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess AM, White AP, Vernochet C, Schulz TJ, Xue R, Sass CA, Huang TL, Roberts-Toler C, Weiner LS, Sze C, et al. (2013). Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat. Med. 19, 635–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempersmier J, Sambeat A, Gulyaeva O, Paul SM, Hudak CS, Raposo HF, Kwan HY, Kang C, Wong RH, and Sul HS (2015). Cold-inducible Zfp516 activates UCP1 transcription to promote browning of white fat and development of brown fat. Mol. Cell 57, 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Rhee J, St-Pierre J, Handschin C, Puigserver P, Lin J, Jäeger S, Erdjument-Bromage H, Tempst P, and Spiegelman BM (2004). Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1alpha: modulation by p38 MAPK. Genes Dev. 18, 278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorenko A, Lishko PV, and Kirichok Y (2012). Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell 151, 400–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, and Blackshear PJ (2017). RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat. Rev. Immunol. 17, 130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MJ, Lim HW, Ho Y, Shapira SN, Ishibashi J, Rajakumari S, Steger DJ, Lazar MA, Won KJ, and Seale P (2015). PRDM16 binds MED1 and controls chromatin architecture to determine a brown fat transcriptional program. Genes Dev. 29, 298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen NZ, Larsen TJ, Peijs L, Daugaard S, Homøe P, Loft A, de Jong J, Mathur N, Cannon B, Nedergaard J, et al. (2013). A classical brown adipose tissue mRNA signature partly overlaps with brite in the supra-clavicular region of adult humans. Cell Metab. 17, 798–805. [DOI] [PubMed] [Google Scholar]
- Jornayvaz FR, and Shulman GI (2010). Regulation of mitochondrial biogenesis. Essays Biochem. 47, 69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinovich AV, de Jong JMA, Cannon B, and Nedergaard J (2017). UCP1 in adipose tissues: two steps to full browning. Biochimie 134, 127–137. [DOI] [PubMed] [Google Scholar]
- Kleiner S, Mepani RJ, Laznik D, Ye L, Jurczak MJ, Jornayvaz FR, Estall JL, Chatterjee Bhowmick D, Shulman GI, and Spiegelman BM (2012). Development of insulin resistance in mice lacking PGC-1α in adipose tissues. Proc. Natl. Acad. Sci. USA 109, 9635–9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutti D, Kressler D, and Kralli A (2001). Regulation of the transcriptional coactivator PGC-1 via MAPK-sensitive interaction with a repressor. Proc. Natl. Acad. Sci. USA 98, 9713–9718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Banks A, Liu T, Kazak L, Rao RR, Cohen P, Wang X, Yu S, Lo JC, Tseng YH, et al. (2014). IRF4 is a key thermogenic transcriptional partner of PGC-1 a. Cell 158, 69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai B, Lee J-E, Jang Y, Wang L, Peng W, and Ge K (2017). MLL3/MLL4 are required for CBP/p300 binding on enhancers and super-enhancer formation in brown adipogenesis. Nucleic Acids Res. 45, 6388–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidell ME, Betz MJ, Dahlqvist Leinhard O, Heglind M, Elander L, Slawik M, Mussack T, Nilsson D, Romu T, Nuutila P, et al. (2013). Evidence for two types of brown adipose tissue in humans. Nat. Med. 19, 631–634. [DOI] [PubMed] [Google Scholar]
- Long JZ, Svensson KJ, Tsai L, Zeng X, Roh HC, Kong X, Rao RR, Lou J, Lokurkar I, Baur W, et al. (2014). A smooth muscle-like origin for beige adipocytes. Cell Metab. 19, 810–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, and Bejerano G (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H, Shinoda K, Ohyama K, Sharp LZ, and Kajimura S (2013). EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature 504, 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, and Spiegelman BM (1998). A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92, 829–839. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Lin J, Wu Z, Yoon JC, Zhang CY, Krauss S, Mootha VK, Lowell BB, and Spiegelman BM (2001). Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma co-activator-1. Mol. Cell 8, 971–982. [DOI] [PubMed] [Google Scholar]
- Rajakumari S, Wu J, Ishibashi J, Lim HW, Giang AH, Won KJ, Reed RR, and Seale P (2013). EBF2 determines and maintains brown adipocyte identity. Cell Metab. 17, 562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. (2013).A compendium of RNA-binding motifs for decoding gene regulation. Nature 499, 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scimè A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. (2008). PRDM16 controls a brown fat/skeletal muscle switch. Nature 454, 961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber T, Treiber N, Plessmann U, Harlander S, Daiß J-L, Eichner N, Lehmann G, Schall K, Urlaub H, and Meister G (2017). A Compendium of RNA-Binding Proteins that Regulate MicroRNA Biogenesis. Mol. Cell 66, 270–284. e13. [DOI] [PubMed] [Google Scholar]
- Villarroya F, Peyrou M, and Giralt M(2017).Transcriptional regulation of the uncoupling protein-1 gene. Biochimie 134, 86–92. [DOI] [PubMed] [Google Scholar]
- Watson G, Ronai ZA, and Lau E (2017). ATF2, a paradigm of the multifaceted regulation of transcription factors in biology and disease. Pharmacol. Res. 119, 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, and Young RA (2013). Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-sequencing data have been deposited in NCBI’s GEO (GSE139029).