Significance
A vectorial flow of electrons in the membrane generates proton-motive force, which is central to cellular respiration. Organisms, including the last universal common ancestor of living organisms (LUCA), have multiple electron transport systems to use diverse electron donor–acceptor pairs. Such respiratory flexibility enables survival in varying environments. The appearance of oxygen in Earth’s environment due to the Great Oxidation Event (GOE) caused a major transformation in microbial bioenergetics. Here we performed a systems-level analysis to examine the suitability of pre-GOE era respiratory quinone, naphthoquinone, in oxic environments and resource constraint requiring the advent of the high-redox-potential quinone.
Keywords: respiration, naphthoquinone, oxidative stress, genome-scale model
Abstract
Evolution fine-tunes biological pathways to achieve a robust cellular physiology. Two and a half billion years ago, rapidly rising levels of oxygen as a byproduct of blooming cyanobacterial photosynthesis resulted in a redox upshift in microbial energetics. The appearance of higher-redox-potential respiratory quinone, ubiquinone (UQ), is believed to be an adaptive response to this environmental transition. However, the majority of bacterial species are still dependent on the ancient respiratory quinone, naphthoquinone (NQ). Gammaproteobacteria can biosynthesize both of these respiratory quinones, where UQ has been associated with aerobic lifestyle and NQ with anaerobic lifestyle. We engineered an obligate NQ-dependent γ-proteobacterium, Escherichia coli ΔubiC, and performed adaptive laboratory evolution to understand the selection against the use of NQ in an oxic environment and also the adaptation required to support the NQ-driven aerobic electron transport chain. A comparative systems-level analysis of pre- and postevolved NQ-dependent strains revealed a clear shift from fermentative to oxidative metabolism enabled by higher periplasmic superoxide defense. This metabolic shift was driven by the concerted activity of 3 transcriptional regulators (PdhR, RpoS, and Fur). Analysis of these findings using a genome-scale model suggested that resource allocation to reactive oxygen species (ROS) mitigation results in lower growth rates. These results provide a direct elucidation of a resource allocation tradeoff between growth rate and ROS mitigation costs associated with NQ usage under oxygen-replete condition.
Naphthoquinone (NQ) and ubiquinone (UQ) are the 2 major classes of quinones that play a key role in cellular respiration. These redox-active respiratory quinones differ in their reduction potential (−70 to 40 mV for methylated and demethylated NQ, whereas ∼100 mV for UQ) and function as electron carriers between various membrane-bound dehydrogenases (electron donor) and oxidases (electron acceptor) (1, 2). NQ occurred early in evolutionary history, and UQ appeared later, alongside the Great Oxidation Event (GOE) (3, 4). Since then, the microbial respiratory electron transport chain diverged into an NQ-based “low-potential (LP) chain” and a UQ-based “high-potential (HP) chain”. LP-to-HP bioenergetic transition required an overall redox potential up-shift, and, therefore, prokaryotes either have an LP or HP chain. Gammaproteobacteria, like Escherichia coli, contain both NQ and UQ biosynthetic abilities and are reported to switch the quinone type in response to ambient conditions (5). An E. coli strain defective in UQ biosynthesis showed less than 30% growth rate as compared to a UQ-dependent strain and assumed a fermentative behavior (6, 7). Obligate NQ-dependent microbes can efficiently respire aerobically, and this motivated us to examine metabolic limitations of aerobic NQ usage in E. coli.
To determine the consequences of substituting NQ for UQ, we deployed adaptive laboratory evolution (ALE) to evolve a genome-edited strain of E. coli, in which UQ biosynthesis was disabled, in an aerobic environment. To utilize oxygen, the evolved strains had to adapt to use NQ to carry out aerobic respiration. We found that: 1) NQ can support aerobic respiration, albeit at a lower growth rate than UQ; 2) key mutations that enabled the adaptation were associated with the pyruvate dehydrogenase complex transcriptional regulator (PdhR); 3) an adjustment in the activity of 3 transcriptional regulators (PdhR, RpoS, and Fur) supported efficient NQ use in the electron transport system (ETS); 4) the adaptation to NQ use comes with an elevated expression of RpoS-regulated periplasmic superoxide-quenching enzymes; and 5) genome-scale computational models of proteome allocation suggest that the use of NQ requires increased proteome allocation to reactive oxygen species (ROS) mitigation, resulting in lower growth rates. Taken together, these results provide a deep understanding of the adaptive rewiring, proteome cost for the NQ-dependent ETS and empirically establishes that the evolution of UQ conferred a growth advantage by resource conservation in the post-GOE environment.
Results and Discussion
Adaptive Laboratory Evolution of NQ-Dependent E. coli.
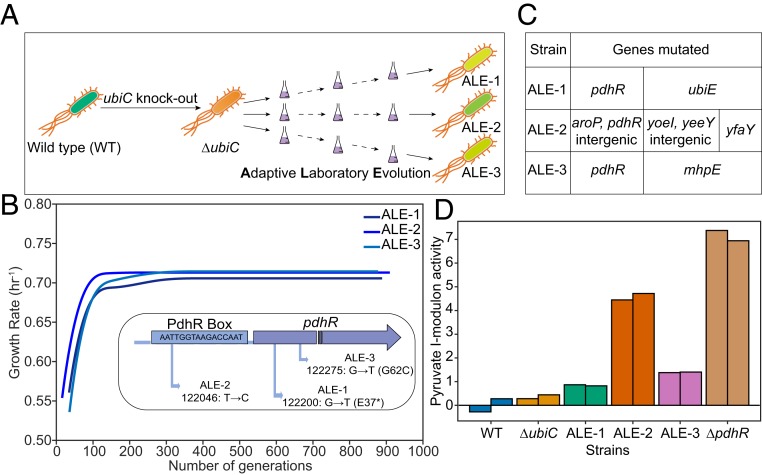
The biosynthetic pathways of respiratory quinones diverge at the chorismate node. Chorismate lyase (UbiC) and isochorismate synthase (MenF) drive chorismate toward UQ and NQ biosynthesis, respectively (SI Appendix, Fig. S1A). We generated a UQ-deficient strain by knocking out the chorismate lyase (ΔubiC) to prevent chorismate flux toward UQ biosynthesis. The initial knockout strain was characterized, and ALE was subsequently used to probe the adaptive capabilities of this strain in an oxygen-rich environment (Fig. 1A). As expected, the deletion of ubiC resulted in diminished UQ levels, and an increased biosynthesis of NQ was observed (SI Appendix, Fig. S1B). Interestingly, all 3 replicates of the ΔubiC strains evolved to grow at a higher growth rate (∼0.70 h−1) (Fig. 1B and SI Appendix, Fig. S1C). However, none of the evolved strains could achieve a growth rate of the evolved WT strain (∼0.95–1.0 h−1) (8). Thus, despite the fitness gain of the ΔubiC strain during ALE, it was less than that of the wild type. We refer to all of the ΔubiC strains together as NQ-dependent strains, preevolved strain as ΔubiC, and evolved strains individually as ALE-1, ALE-2, and ALE-3 (Fig. 1A).
Fig. 1.
Experimental evolution of E. coli ΔubiC strain. (A) The schematic for generation of strains used in the study. (B) The growth rate evolution trajectories (smoothed data) and the convergent mutations (Inset) observed in ΔubiC replicates during laboratory evolution. SI Appendix, Fig. S1C shows the extended axis plot of these trajectories. (C) List of genes mutated during the evolution of the ΔubiC replicates. (D) Activity of the pyruvate i-modulon estimated by independent component analysis (ICA). ΔpdhR has been used as the control strain. The bars with identical colors represent biological replicates of the corresponding strain.
Fitness gains result from the acquisition of beneficial mutations. We performed whole-genome sequencing of the NQ-dependent strains to identify the genetic basis of the increase in growth rate. All of the evolved NQ-dependent strains acquired only 1 common gene mutation (Fig. 1 B and C). ALE-1 and ALE-3 showed a single-base substitution in the gene pdhR (pyruvate dehydrogenase complex regulator), whereas ALE-2 had a base substitution upstream of the pdhR gene (Fig. 1 B, Inset and SI Appendix, Table S1). PdhR is negatively autoregulated, and the base substitution in ALE-2 was found to be located in the PdhR-binding region, also known as PdhR box, upstream of pdhR gene (9). Remarkably, the base substitution in ALE-1 brought in a premature termination codon in the pdhR ORF. This suggested an altered activity of PdhR responsible for the fitness changes of the evolved NQ-dependent strains and motivated us to examine the effect of the absence of this transcriptional regulator on the transcriptome of these strains. We thus generated a pdhR knockout strain (ΔpdhR) and performed transcriptome analysis of ΔpdhR and the NQ-dependent strains under the same conditions.
We applied an effective signal deconvolution algorithm, independent component analysis (ICA), to perform an unbiased analysis of the transcriptional responses (Materials and Methods) (10). This analysis results in cohesive sets of independently modulated genes (called i-modulons), each of which often exhibits a high overlap with the gene targets of a specific transcriptional regulator (called regulons). In addition, ICA estimates an activity for each i-modulon in each expression profile, serving as a proxy for the activity of the linked regulator. We observed a differential activity of the pyruvate-related independently modulated set of genes (Fig. 1D). The activity of this i-modulon was high in the ΔpdhR strain, suggesting a derepression of PdhR i-modulon. The activity of this component was higher in the evolved NQ-dependent strains than in preevolved ΔubiC strain, indicating a partial derepression of PdhR i-modulon due to the mutations in pdhR.
Respiratory Rewiring during NQ-Dependent ETS.
To understand the respiratory changes in the NQ-dependent strains, we estimated the exchange rates of the respiratory metabolites in the WT, ΔubiC, and the 3 evolved ΔubiC strains. We observed a clear tradeoff between fermentative and oxidative respiration, shown by the change in oxygen uptake rate and lactate secretion rate (Table 1). As expected, the loss of UQ in the ΔubiC strain resulted in a significant lactate production (7) and a decreased oxygen uptake rate. However, after evolution, the oxygen consumption of the NQ-dependent strains improved by more than 60%, and lactate secretion was reduced by 80% on average.
Table 1.
Exchange rates of wild-type, ΔubiC, and evolved ΔubiC E. coli K12 MG1655 strains
| Strain | Oxygen uptake rate | Glucose uptake rate | Acetate secretion rate | Lactate secretion rate |
| WT | 14.27 ± 1.01 | 11.23 ± 0.55 | 7.83 ± 0.61 | ND |
| ∆ubiC | 7.41 ± 0.72 | 9.06 ± 0.24 | 6.45 ± 0.13 | 3.41 ± 0.80 |
| ALE-1 | 12.16 ± 0.62 | 9.40 ± 0.36 | 7.38 ± 0.20 | 0.34 ± 0.05 |
| ALE-2 | 11.78 ± 0.38 | 8.86 ± 0.40 | 6.74 ± 0.25 | 0.66 ± 0.19 |
| ALE-3 | 12.17 ± 0.46 | 8.38 ± 0.07 | 6.32 ± 0.08 | 0.70 ± 0.02 |
The unit of metabolite uptake/secretion rate is mmol/gDCW/h. gDCW, gram dry cell weight; ND, not detected.
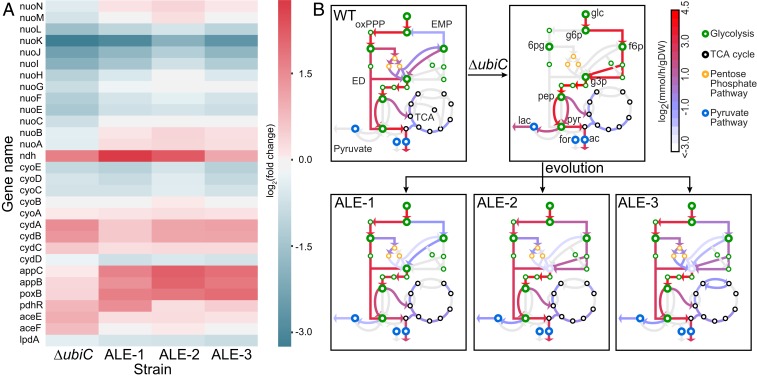
We then explored the transcriptome shift underlying this altered respiratory behavior of the NQ-dependent strains. Since PdhR has been reported to have an impact on the ETS of E. coli (9), we examined expression changes in the ETS enzymes (Fig. 2A). The E. coli genome encodes 2 NADH dehydrogenases; the proton-translocating type I NADH dehydrogenase and nontranslocating type II NADH dehydrogenase (Ndh). In the NQ-dependent strains, there was an up-regulation in the expression of ndh. Interestingly, there appears to be an operational synergy between NQ and Ndh (11). Earlier, an attempt to generate an Ndh-deficient strain of NQ-producing bacterium was unsuccessful (12). ALE-1 strains with the frame-disruptive mutation in pdhR showed the highest up-regulation of ndh. There was also an increase in the expression of type I NADH dehydrogenase; however, deactivation of PdhR resulted in a constitutive expression of the type II NADH dehydrogenase in the evolved strains. Interestingly, there appears to be a subunit-level calibration in the expression of the proton-translocating units of the type I NADH dehydrogenase for the optimization of NQ-based ETS (13, 14).
Fig. 2.
Evolutionary optimization of the respiratory system. (A) Heatmap showing the expression changes of enzymes involved in ETS and pyruvate metabolism. Fold-changes have been calculated with respect to the wild-type strain. (B) Computed metabolic flux maps depict the differences in central metabolism between the WT, the preevolved, and evolved strains (ALE-1, 2, and 3). Major pathways represented in the figure include glycolysis (metabolites colored in green), oxidative pentose phosphate pathway (oxPPP, yellow), the TCA cycle (black), and the pyruvate pathway (blue). Key metabolites are indicated in the figure as follows: glc, glucose; g6p, d-glucose-6-phosphate; g3p, glyceraldehyde-3-phosphate; f6p, d-fructose-6-phosphate; pyr, pyruvate; 6pgc, 6-phospho-d-gluconate; pep, phosphoenolpyruvate; lac, lactate; for, formate. Calculated fluxes of each strain are colored on a log scale.
There are 3 cytochrome oxidases in the E. coli genome that can transfer electrons to oxygen. There was no significant differential regulation of the cytochrome bo3 oxidase (cyoABCDE). Cytochrome bd-I oxidase (cydBD) expression was higher in the NQ-based strains, specifically in the preevolved strain. The evolved strains showed an increased expression of cytochrome bd-II oxidase (appBC). Notably, the specific activity of cytochrome bd-II oxidase is reported to be higher for naphthoquinone than benzoquinone (15), and these alternate oxidases are more ROS-protective as well (16). The ETS of evolved strains thus appeared to be tailored to a more efficient route of electron flow for NQ utilization. A similar transcriptional rewiring was observed in the glycolytic cycle where the evolved strains, despite retaining high activity of pyruvate dehydrogenase complex, up-regulated the expression of pyruvate oxidase (poxB), which is reported to relieve oxidative stress (17).
In addition to the changes in the ETS enzyme usage, the shift in cellular respiration should also be apparent by comparing fluxes through biochemical reactions in central metabolism. Accordingly, we incorporated the experimentally measured metabolite exchange rates and transcriptomics data into a genome-scale model of metabolism and protein expression enhanced by a protein-folding network, FoldME (18) and simulated the flux map for each experimental strain at their specific growth rate (Materials and Methods). The resulting metabolic flux map shows a clear alteration in use of the glycolysis pathway, mixed fermentation pathway, and the tricarboxylic acid (TCA) cycle (Fig. 2B). The preevolved ΔubiC strain, which exhibits a fermentative phenotype determined by experimental measurement, is characterized by heavy lactate secretion and glycolysis flux supported solely by the Embden–Meyerhof–Parnas pathway. After evolution, glucose metabolism becomes heavily dependent on the Entner–Doudoroff pathway and further diversifies to different levels of complexity of using the oxidative pentose phosphate pathway. Our simulations show that the evolved strains reduce the high lactate production level by rebalancing their glycolytic proteome to mimic the WT pathway usage and increase aerobic respiration through the TCA cycle.
Oxidative Stress Mitigation.
Increase in aerobic respiration results in higher oxidative stress. The respiratory electron carriers are a major source of periplasmic superoxide production and, in the absence of UQ, the respiratory chain becomes more autooxidizable (19). Therefore, an increased NQ-based electron flow will require an extensive oxidative defense system.
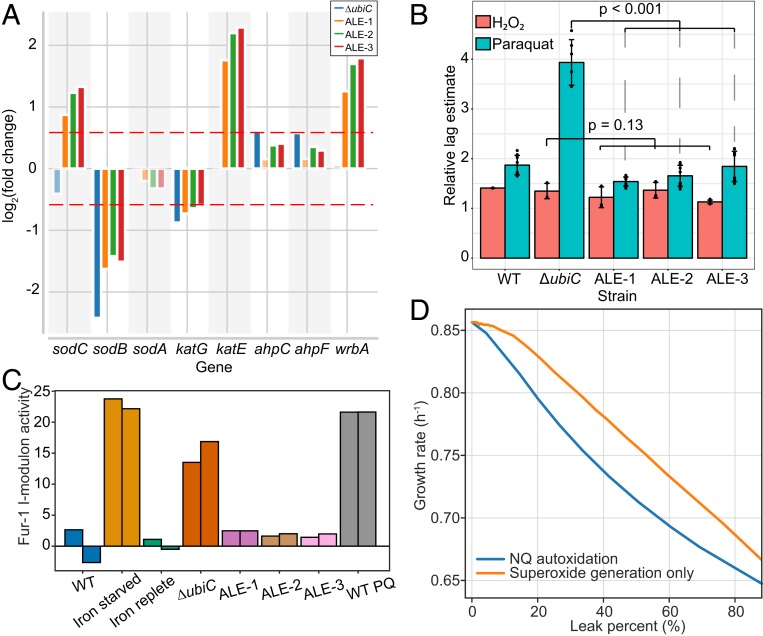
The E. coli genome has elaborate oxidative stress response machinery that is mostly regulated by 3 transcriptional regulators: OxyR, SoxS, and RpoS. We examined the activity of the 3 i-modulons regulated by these transcriptional regulators. There was no significant change in the activity of the canonical peroxide and superoxide stress regulons, OxyR and SoxS, respectively (SI Appendix, Fig. S2). However, we observed a high activity of the alternative sigma factor RpoS i-modulon in the evolved strains (SI Appendix, Fig. S2). There are 3 superoxide dismutases responsible for the conversion of superoxide to less damaging peroxide (SodA, SodB, and SodC). SodA and SodB are the cytoplasmic dismutases, whereas SodC catalyzes the dismutation reaction in the periplasm. Although there was no significant up-regulation in the expression of cytoplasmic superoxide dismutases, the periplasmic superoxide was invariably up-regulated in all 3 of the evolved strains (Fig. 3A).
Fig. 3.
Oxidative stress mitigation. (A) Expression changes in ROS-quenching enzymes. Fold-changes have been calculated with respect to the wild-type strain [periplasmic superoxide dismutase, sodC; cytosolic superoxide dismutase, sodB and sodA; bifunctional hydroperoxidase, katG; monofunctional catalase, katE; alkyl hydroperoxide reductase, ahpC and ahpF; NAD(P)H:quinone oxidoreductase, wrbA]. (B) Estimation of the growth retardation caused due to addition of peroxide (1 mM H2O2) and superoxide (1 µM paraquat) generating chemicals by calculating the relative increase in the lag phase. For each strain, the mean of the estimated lag phase across untreated replicates (control) was calculated. The lag-phase estimates of the conditions with treatment was then divided by the corresponding control condition (yielding the relative lag phase). The error bars represent the SD for the lag estimates across replicates (n ≥ 3). The effect of adaptive evolution was significant in the paraquat treatment (P value < 0.001) but not in peroxide treatment (P value = 0.13) (Materials and Methods). (C) Adaptive changes in the activity of Fur i-modulon in the evolved strains. Iron-starved, iron-replete, and paraquat-treated WT conditions serve as controls. The bars with identical colors represent biological replicates of the corresponding strain. (D) Genome-scale model-based calculation of the impact of periplasmic nonproductive electron leak on the growth rate of E. coli. SI Appendix, Fig. S5 shows the extended axis plot of these calculations.
The peroxide is further converted to water and oxygen by 2 catalases, KatG and KatE. The expression of katE showed a pattern similar to that of the superoxide dismutase sodC (Fig. 3A). Notably, KatE has a higher turnover rate than KatG (20). WrbA, NAD(P)H:Quinone oxidoreductase, is responsible for the reduction of quinones to hydroquinones which prevent interaction of semiquinone to oxygen, thereby preventing the generation of superoxide. This enzyme is active during aerobic growth of E. coli and is proposed to function in a highly oxidizing environment (21). The expression of wrbA was similar to superoxide dismutase and catalase (Fig. 3A). All these up-regulated ROS-mitigating enzymes belong to RpoS regulon. A comprehensive examination of all stress-related i-modulons showed a response similar to RpoS i-modulon in case of EvgA and GadWX/EWX (SI Appendix, Fig. S3). These i-modulons are related to acid stress, and their induction could be either adaptive or an indirect consequence of RpoS i-modulon activation (22). Taken together, these results show that a subset of the E. coli ROS defense system, regulated by RpoS, was up-regulated to quench the ROS generated due to the electron leakage from ETS.
The effect of changes in the ROS mitigation ability was validated by challenging the cells to higher oxidative stress. We examined the response of these strains toward 2 distinct ROS-generating agents: hydrogen peroxide and paraquat (23) (Fig. 3B and SI Appendix, Fig. S4). There was no significant difference in the peroxide tolerance between the preevolved and evolved strains. However, when the strains were challenged by a superoxide-generating molecule, paraquat, a significant growth retardation was observed in the preevolved strain as compared to evolved strains. This suggests that an elevated superoxide stress response state is needed to enable an efficient utilization of NQ-based ETS.
The ICA also revealed an activity change in the Fur-1 i-modulon. This i-modulon is associated with the Fur (ferric uptake regulator) regulon, which is responsible for cellular iron homeostasis (24). The Fur-1 i-modulon was found to be highly active in the ΔubiC strain (Fig. 3C), suggesting a decreased repressor activity of Fur. Superoxide stress is reported to negatively impact Fur functioning, as it is a mononuclear iron protein (25). We observed a similar increase in the activity of this i-modulon in paraquat-treated cells. Thus, increased oxidative stress could be responsible for the activity change in the ΔubiC strain. An inverse relation between the iron uptake system and NQ biosynthesis has been reported to reduce superoxide toxicity in Staphylococcus aureus (26). Along the same lines, the improved ROS defense system in the evolved strains brought down the activity of the Fur-1 i-modulon, which allowed efficient use of the NQ-based ETS without potentiating the ROS toxicity.
Genome-Scale Calculation of Tradeoff between Growth and ROS Defense.
The knowledge of growth rate limitation and ROS defense rewiring encouraged us to compute the cost of respiratory electron dissipation. We used the genome-scale model of metabolism, macromolecule expression, and oxidative stress, OxidizeME (27), to estimate the impact of periplasmic superoxide generation and spontaneous autooxidation of NQ on the growth rate. First, we found that the cost (metabolic and protein expression) of detoxifying superoxide had a strong negative impact on the growth rate (Fig. 3D and SI Appendix, Fig. S5). The chemistry of NQ makes it more prone to autooxidation (28). Upon accounting for the decrease in ETS efficiency by diverting electrons from NQ toward superoxide generation, we computed a further decrease in the growth rate (Fig. 3D). Thus, despite an elevated ROS defense capability, the growth capability of NQ-dependent strains is certainly restricted. The need for ROS mitigation thus explains the limited fitness improvement of the NQ-dependent strains through the need to reallocate the proteome from growth to stress response functions.
The biosynthetic pathways for both respiratory quinones, UQ and NQ, are very similar. Both are derived from the shikimate pathway and undergo prenylation. However, there are some distinct metabolic requirements for the synthesis of these quinones. NQ biosynthesis requires 2-ketoglutarate, thiamine PPi, CoA, and ATP as cofactors, whereas UQ biosynthesis requires oxygen, flavoprotein, and NADH (29). We performed a comparative cost analysis of the 2 biosynthetic pathways using the genome-scale reconstruction of the metabolic network in E. coli K-12 MG1655 (iML1515) (30). The computation showed UQ biosynthesis to be more costly than NQ; however, the major contributor to this difference was a higher oxygen requirement (SI Appendix, Table S2).
Conclusions
Except for aceto- and methanogens (except Methanosarcinales), the bioenergetic chains of all organisms exploit redox-active electron carriers for optimal use of higher-redox-span substrate couples (2). These electron carriers, with the exception of phenazines, contain a quinone scaffold with 2 carbonyl groups at para positions and undergo a keto-hydroxy cycling by a 2-electron transfer to enable the electronic circuit of the ETS. The 2-electron reduction of these quinones proceeds via a single-electron reduction to the semiquinone radical, the autooxidation of which is believed to produce superoxide from ETS (31). The modern-age microbial respiratory systems consist of diverse respiratory quinones. This quinone diversification is believed to be an adaptive consequence of the oxygenation of Earth’s environment. Interestingly, quinone evolution has resulted in the emergence of both high- and low-redox-potential molecules from the ancestral NQ (2). While high-redox-potential quinone, UQ, is believed to be a crucial adaptive feature in post-GOE bioenergetics, owing to its lesser propensity to noncatalyzed oxidation, low-potential quinone, rhodoquinone (RQ), appears to have emerged to optimize the biosynthetic pathways (2, 3, 32). A comprehensive profiling of quinone distribution in the bacterial world indicates an association between quinone composition and respiratory behavior in the Gram-negative species, with obligate anaerobes and aerobes exclusively consisting of NQ and UQ, respectively, whereas facultative aerobes have a mix of the 2 quinone types (33). However, NQ is the only quinone identified in Gram-positive bacterial species (33). The majority of bacterial species, including the most prevalent phyla (actinobacteria and proteobacteria), retained NQ as either exclusive or substitutable respiratory quinone (2). In facultative NQ-dependent genera like Escherichia, Klebsiella, and Proteus, UQ and NQ are associated with aerobic and anaerobic lifestyles, respectively (33–35). The maintenance of efficient aerobic respiratory chains in obligate NQ-dependent species inspired us to examine the cost of oxidative respiration in UQ-deficient E. coli.
We addressed the paradox of the different nature of NQ-based respiration in obligate and facultative NQ-dependent species using adaptive laboratory evolution of engineered NQ-dependent strains. Systems-level examination of the evolved NQ-dependent strains showed an extensive physiological rewiring orchestrated by the activity of 3 transcriptional regulators resulting in proteome reallocation from growth to ROS stress-mitigating functions. Notably, the evolved NQ-dependent strains shifted from fermentative to oxidative energetics. However, the resource partitioning to periplasmic ROS defense system prevented the strain from achieving full growth capacity. Interestingly, a relatively smaller molar ratio of NQ is reported to be required for the functioning of ETS as compared to UQ, which may be due to the need for efficient redox recycling to reduce nonproductive electron dissipation (36–38). In an oxygen-replete condition, the additional cost of UQ biosynthesis may not have a substantial impact, and the resource conservation due to lesser ROS production might have a growth-promoting effect. This reverse tracing of the natural course of quinone evolution thus provided a fundamental understanding of the environment-specific cost of the electron carriers. Extrapolation of this study, especially in pathogenic bacteria with NQ as the sole electron carrier, may enable therapeutic targeting of microbial energetics. RQ, a respiratory quinone with redox potential similar to NQ, enables hypoxic survival of several eukaryotes including parasitic helminths (39). Thus, understanding the physiological properties and vulnerabilities associated with the functioning of low-redox-potential quinone holds translatable potential across life forms.
Materials and Methods
Genome-edited E. coli strain was evolved in the laboratory using an automated system. Detailed materials and methods used for strain generation, adaptive laboratory evolution, DNA sequencing, RNA sequencing, i-modulon decomposition, phenotype characterization, respiratory quinone quantitation, proteome-constrained simulation, and genome-scale computations are provided as SI Appendix.
Data Availability.
Resequencing and expression profiling data that support the findings of this study have been deposited to NCBI Sequence Read Archive (SRA accession: PRJNA560068) and Gene Expression Omnibus (GSE135867), respectively.
Supplementary Material
Acknowledgments
This work was funded by Novo Nordisk Foundation Grant NNF10CC1016517 and NIH Grants R01GM057089 and U01AI124316. We thank Marc Abrams (Systems Biology Research Group, University of California San Diego) for assistance with manuscript editing. The support of University of California San Diego, Chemistry and Biochemistry Molecular Mass Spectrometry Facility is duly acknowledged.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Resequencing and expression profiling data that support the findings of this study have been deposited to National Center for Biotechnology Information Sequence Read Archive (SRA accession no. PRJNA560068) and Gene Expression Omnibus (accession no. GSE135867), respectively.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909987116/-/DCSupplemental.
References
- 1.Unden G., Bongaerts J., Alternative respiratory pathways of Escherichia coli: Energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320, 217–234 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Schoepp-Cothenet B., et al. , On the universal core of bioenergetics. Biochim. Biophys. Acta 1827, 79–93 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Schoepp-Cothenet B., et al. , Menaquinone as pool quinone in a purple bacterium. Proc. Natl. Acad. Sci. U.S.A. 106, 8549–8554 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Degli Esposti M., A journey across genomes uncovers the origin of ubiquinone in cyanobacteria. Genome Biol. Evol. 9, 3039–3053 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones R. W., Garland P. B., “The function of ubiquinone and menaquinone in the respiratory chain of Escherichia coli” in Function of Quinones in Energy Conserving Systems (Elsevier, 1982), pp. 465–476. [Google Scholar]
- 6.Nitzschke A., Bettenbrock K., All three quinone species play distinct roles in ensuring optimal growth under aerobic and fermentative conditions in E. coli K12. PLoS One 13, e0194699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wallace B. J., Young I. G., Role of quinones in electron transport to oxygen and nitrate in Escherichia coli. Studies with a ubiA- menA- double quinone mutant. Biochim. Biophys. Acta 461, 84–100 (1977). [DOI] [PubMed] [Google Scholar]
- 8.LaCroix R. A., et al. , Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl. Environ. Microbiol. 81, 17–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogasawara H., Ishida Y., Yamada K., Yamamoto K., Ishihama A., PdhR (pyruvate dehydrogenase complex regulator) controls the respiratory electron transport system in Escherichia coli. J. Bacteriol. 189, 5534–5541 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sastry A. V, et al. , The Escherichia coli transcriptome consists of independently regulated modules. 10.1101/620799 (29 April 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boersch M., Rudrawar S., Grant G., Zunk M., Menaquinone biosynthesis inhibition: A review of advancements toward a new antibiotic mechanism. RSC Adv. 8, 5099–5105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vilchèze C., Weinrick B., Leung L. W., Jacobs W. R. Jr, Plasticity of Mycobacterium tuberculosis NADH dehydrogenases and their role in virulence. Proc. Natl. Acad. Sci. U.S.A. 115, 1599–1604 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torres-Bacete J., et al. , Roles of subunit NuoK (ND4L) in the energy-transducing mechanism of Escherichia coli NDH-1 (NADH:quinone oxidoreductase). J. Biol. Chem. 287, 42763–42772 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Efremov R. G., Baradaran R., Sazanov L. A., The architecture of respiratory complex I. Nature 465, 441–445 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Sturr M. G., Krulwich T. A., Hicks D. B., Purification of a cytochrome bd terminal oxidase encoded by the Escherichia coli app locus from a delta cyo delta cyd strain complemented by genes from Bacillus firmus OF4. J. Bacteriol. 178, 1742–1749 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giuffrè A., Borisov V. B., Arese M., Sarti P., Forte E., Cytochrome bd oxidase and bacterial tolerance to oxidative and nitrosative stress. Biochim. Biophys. Acta 1837, 1178–1187 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Moreau P. L., Diversion of the metabolic flux from pyruvate dehydrogenase to pyruvate oxidase decreases oxidative stress during glucose metabolism in nongrowing Escherichia coli cells incubated under aerobic, phosphate starvation conditions. J. Bacteriol. 186, 7364–7368 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen K., et al. , Thermosensitivity of growth is determined by chaperone-mediated proteome reallocation. Proc. Natl. Acad. Sci. U.S.A. 114, 11548–11553 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korshunov S., Imlay J. A., Detection and quantification of superoxide formed within the periplasm of Escherichia coli. J. Bacteriol. 188, 6326–6334 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mishra S., Imlay J., Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch. Biochem. Biophys. 525, 145–160 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patridge E. V., Ferry J. G., WrbA from Escherichia coli and Archaeoglobus fulgidus is an NAD(P)H:quinone oxidoreductase. J. Bacteriol. 188, 3498–3506 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanjee U., Houry W. A., Mechanisms of acid resistance in Escherichia coli. Annu. Rev. Microbiol. 67, 65–81 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Abrashev R., et al. , Differential effect of paraquat and hydrogen peroxide on the oxidative stress response in Vibrio cholerae Non {O1} 26/06. Biotechnol. Biotechnol. Equip. 25, 72–76 (2011). [Google Scholar]
- 24.Seo S. W., et al. , Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat. Commun. 5, 4910 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imlay J. A., The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol. 11, 443–454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wakeman C. A., et al. , Menaquinone biosynthesis potentiates haem toxicity in Staphylococcus aureus. Mol. Microbiol. 86, 1376–1392 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L., et al. , Cellular responses to reactive oxygen species are predicted from molecular mechanisms. Proc. Natl. Acad. Sci. U.S.A. 116, 14368–14373 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anand A., et al. , Polyketide quinones are alternate intermediate electron carriers during mycobacterial respiration in oxygen-deficient niches. Mol. Cell 60, 637–650 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meganathan R., Kwon O., Biosynthesis of menaquinone (vitamin K2) and ubiquinone (coenzyme Q). EcoSal Plus, 10.1128/ecosalplus.3.6.3.3 (23 December 2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monk J. M., et al. , iML1515, a knowledgebase that computes Escherichia coli traits. Nat. Biotechnol. 35, 904–908 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dykens J. A., “Redox enzymes” in Comprehensive Medicinal Chemistry II, Taylor J. B. and Triggle D. J., Eds. (Elsevier Science, 2007), vol. 2, pp. 1053–1087. [Google Scholar]
- 32.Brajcich B. C., et al. , Evidence that ubiquinone is a required intermediate for rhodoquinone biosynthesis in Rhodospirillum rubrum. J. Bacteriol. 192, 436–445 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collins M. D., Jones D., Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol. Rev. 45, 316–354 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Beilen J. W. A., Hellingwerf K. J., All three endogenous quinone species of Escherichia coli are involved in controlling the activity of the aerobic/anaerobic response regulator ArcA. Front. Microbiol. 7, 1339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kröger A., Dadák V., Klingenberg M., Diemer F., On the role of quinones in bacterial electron transport. Differential roles of ubiquinone and menaquinone in Proteus rettgeri. Eur. J. Biochem. 21, 322–333 (1971). [DOI] [PubMed] [Google Scholar]
- 36.Kröger A., Klingenberg M., “On the role of ubiquinone” in Current Topics in Bioenergetics (Elsevier, 1967), pp. 151–193. [Google Scholar]
- 37.Newton N. A., Cox G. B., Gibson F., Function of ubiquinone in Escherichia coli: A mutant strain forming a low level of ubiquinone. J. Bacteriol. 109, 69–73 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kröger A., Dadák V., On the role of quinones in bacterial electron transport. The respiratory system of Bacillus megaterium. Eur. J. Biochem. 11, 328–340 (1969). [DOI] [PubMed] [Google Scholar]
- 39.Del Borrello S., et al. , Rhodoquinone biosynthesis in C. elegans requires precursors generated by the kynurenine pathway. eLife 8, e48165 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Resequencing and expression profiling data that support the findings of this study have been deposited to NCBI Sequence Read Archive (SRA accession: PRJNA560068) and Gene Expression Omnibus (GSE135867), respectively.