Significance
Forested ecosystems provide many ecological, economic, and societal benefits, but those benefits are threatened by climate change. Conservation strategies often assume that plants are currently growing in conditions well-suited to their growth, survival, and reproduction, regardless of whether this assumption is valid. We show that an ecosystem-foundational species in California, valley oak (Quercus lobata), is already mismatched to current temperature and will likely experience further declines in growth rates as temperatures rise over the next century. Given this mismatch, new approaches to climate change management are needed. By using genomic information and identifying genotypes with faster growth rates under warmer temperatures, we present an approach to mitigate negative consequences of rising temperatures for species that may already be experiencing maladaptation.
Keywords: adaptational lag, climate change, ecological genomics, local adaptation, temperature
Abstract
Climate change over the next century is predicted to cause widespread maladaptation in natural systems. This prediction, as well as many sustainable management and conservation practices, assumes that species are adapted to their current climate. However, this assumption is rarely tested. Using a large-scale common garden experiment combined with genome-wide sequencing, we found that valley oak (Quercus lobata), a foundational tree species in California ecosystems, showed a signature of adaptational lag to temperature, with fastest growth rates occurring at cooler temperatures than populations are currently experiencing. Future warming under realistic emissions scenarios was predicted to lead to further maladaptation to temperature and reduction in growth rates for valley oak. We then identified genotypes predicted to grow relatively fast under warmer temperatures and demonstrated that selecting seed sources based on their genotype has the potential to mitigate predicted negative consequences of future climate warming on growth rates in valley oak. These results illustrate that the belief of local adaptation underlying many management and conservation practices, such as using local seed sources for restoration, may not hold for some species. If contemporary adaptational lag is commonplace, we will need new approaches to help alleviate predicted negative consequences of climate warming on natural systems. We present one such approach, “genome-informed assisted gene flow,” which optimally matches individuals to future climates based on genotype–phenotype–environment associations.
Maladaptation due to rapidly changing climate conditions is one of the greatest modern threats to biodiversity (1). Underlying many predictions of the severity of climate change on natural systems is the assumption that populations are locally adapted or well-suited to their current climate conditions. Additionally, many conservation and management strategies, such as using local seed sources in habitat restoration, assume that populations are locally adapted (2, 3). However, local adaption may be less common than frequently assumed. For example, only 45% of plant populations in a recent meta-analysis showed patterns of local adaptation (4, 5). Such deviations from local adaptation could fundamentally alter currently used practices to manage the impacts of climate change on natural systems.
Long-lived organisms like trees are particularly vulnerable to maladaptation because of their long generation times (6–8). Adaptation to climate in trees is typically tested through common garden experiments, which test the expectation that if trees are locally adapted, optimal growth or survival will occur when trees are planted in sites with climates similar to where they originate (7–12). This approach, also referred to as a provenance trial, has generated several classic studies that show that in many cases trees grow best when they are planted in locations similar to their climate of origin (10, 11), much like studies on other plant taxa (4, 5). However, a number of studies have documented tree species maladapted to their current environment (13–15). For example, populations of lodgepole pine (Pinus contorta) have been shown to occupy suboptimal climates, with moderate warming predicted to lead up to a 7% increase in growth (15, 16), while other tree species are predicted to have higher growth rates in cooler climates than they are currently experiencing (13). A variety of mechanisms could lead to these patterns of maladaptation, which include but are not limited to inbreeding, genetic drift, selection on other traits, and the inability to keep up with changing environments via dispersal (17). Despite the potential for trees not to be locally adapted, relatively few studies have addressed contemporary patterns of adaptational lag caused by either past (18) or present-day climate change (19), and instead focus on how future climate change may lead to maladaptation as populations shift away from their assumed climate optimum.
A key challenge facing strategies to mitigate the negative effects of climate change, such as assisted gene flow where individuals are intentionally moved to mitigate maladaptation (20), is identifying seed sources that are “preadapted” to future climate conditions (21, 22). Most commonly, seed selection is done by choosing seed sources that will match future predicted climates at a given site, often under the assumption that plants are locally adapted to their current climate conditions (21). However, because many species may not demonstrate local adaptation (4), alternative approaches are needed to identify seed sources preadapted to future climates without assuming local adaptation. These alternatives are especially relevant for the management and conservation of forests to maintain the numerous ecological, cultural, and ecosystem services they provide, given that forested ecosystems cover ∼30% of the Earth’s land surface (23) and are a major terrestrial carbon sink (24).
Integrating genomic sequencing data with phenotypic data measured in common garden experiments may be a particularly powerful approach to identify genotypes preadapted to future climates (8, 25, 26). Advances in modeling the genomic basis of complex traits through genomic-estimated breeding values (GEBVs: the sum of the effects of genome-wide markers capturing variation in a target trait, also known as polygenic scores) enable the prediction and selection of desired phenotypes based on a panel of genome-wide markers, a process that has transformed animal and plant breeding programs but has only rarely been applied to natural systems (25–29). Together, linking genotype to phenotype with common garden experiments and genome-wide sequencing could form the foundation of a “genome-informed” assisted gene flow strategy, where seed sources are chosen based on the observed relationship between genotype and phenotype, in order to assist adaptation under future climate scenarios.
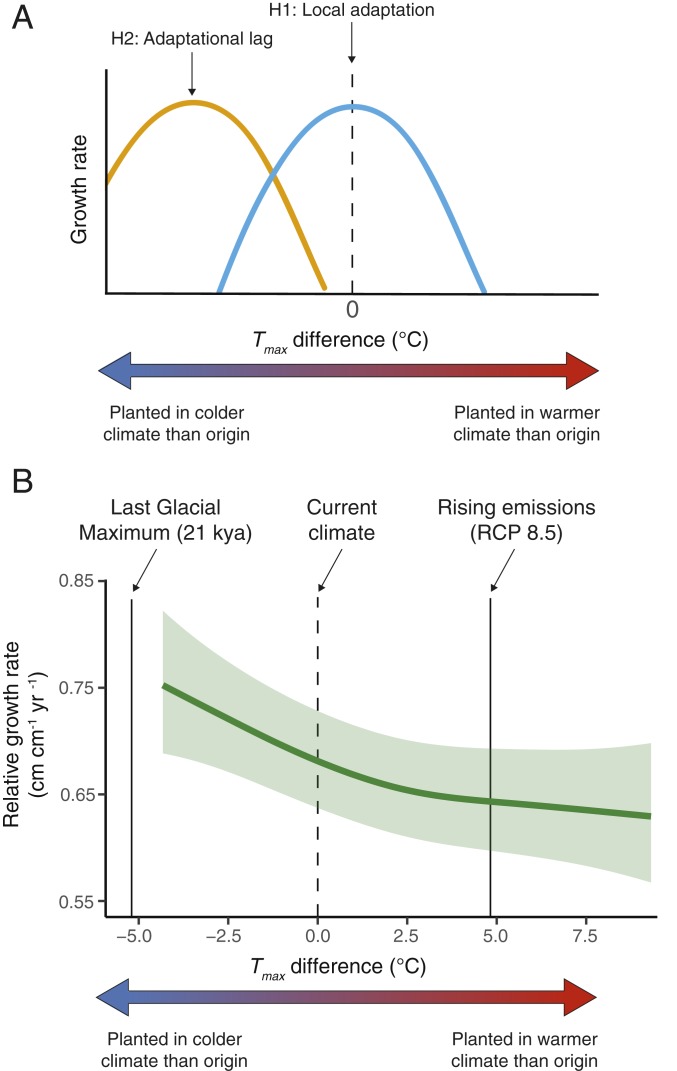
In this study, our first objective was to test the hypothesis of local adaptation to temperature for a long-lived foundational tree species, valley oak (Quercus lobata Née), using data from a large-scale common garden experiment (30). We estimate how early-life stage growth in this species is related to the difference in temperature between where a tree is planted and where the tree originated, which provides a space-for-time substitution for climate change because some individuals are planted into warmer environments than where they originated (31). Here, we assume growth is a component of fitness (9), because faster growth leads to larger trees, which are more likely to produce more offspring over their lifetime due to larger crown size and to avoid mortality from herbivory; however, we recognize the potential trade-off between growth and response to biotic or abiotic stress (32). We focus on the effects of Tmax, the average maximum temperature of the hottest months from June through August, which has been shown to be important in shaping the genetic variation and geographic distribution of valley oak (33, 34). If valley oaks are locally adapted to temperature, we expect to see a characteristic pattern of local adaptation where maximum growth rate occurs when there is no difference in temperature between planting site and origin site (i.e., 0 °C Tmax difference; H1, Fig. 1A). Alternatively, if valley oaks are currently in a state of adaptational lag to temperature, we expect to see the highest growth rates away from 0 °C Tmax difference (H2, Fig. 1A). Our second objective was to evaluate the potential for a “genome-informed” assisted gene flow strategy to mitigate the predicted negative consequences of rising temperatures on growth rates in valley oak. We develop this framework by estimating genotype–phenotype–environment associations in our common garden experiment to identify genomic variation associated with increased growth rates under warmer temperatures.
Fig. 1.
Effects of Tmax difference on valley oak (Q. lobata) growth rate. (A) Conceptual diagram illustrating hypotheses of potential growth rate responses to Tmax difference (i.e., temperature difference between site of planting and site of origin). (B) Predicted relative growth rates (i.e., growth relative to size) and approximate 95% confidence interval across Tmax difference estimated from 2 common gardens. Dashed vertical line shows where planting site matches climate of origin (Tmax difference = 0) and solid lines show Tmax difference between Last Glacial Maximum 21,000 y ago and current climate (5.2° cooler) and predicted increase in temperature of 4.8 °C for rising emissions scenarios by 2100 (RCP 8.5).
Results
Evidence of Adaptational Lag to Temperature in Valley Oak.
Using growth data from 5,051 valley oaks planted across 2 common gardens (SI Appendix, Fig. S1), we found that growth rates on average decreased as valley oaks were planted in climates warmer than where they originated, after controlling for block effects, site, and climate of origin, family, and initial height (Fig. 1B and SI Appendix, Table S1). Further warming under a standard emissions scenario (average 4.8 °C increase in temperature across the valley oak range by 2070 to 2099 under RCP-8.5) would lead to a predicted 5.6% reduction in annualized relative growth rates (Fig. 1B). This 5.6% reduction in growth rates would result in a 10-cm difference in absolute height over a 3-y period for a hypothetical tree starting at a height of 50 cm and with relative growth rates decreasing with size, which may have long-standing consequences for tree size and other fitness components of individual trees, as well as the viability of valley oak populations.
Growth rates were highest on average for valley oaks planted into cooler climates than where they originated as acorns (Fig. 1B and SI Appendix, Table S1). The lack of a peak in growth rates near 0 °C Tmax difference expected under local adaptation to current conditions (H1, Fig. 1A) indicates that valley oaks may be in a state of adaptational lag to contemporary temperature (Fig. 1B) and seem more adapted to cooler temperatures approaching levels that were last experienced during the Last Glacial Maximum 21,000 y ago. We found similar patterns when the 2 common gardens were analyzed separately (SI Appendix, Fig. S2). Because the common garden sites in this study were irrigated to maximize the probability of seedling establishment, we were not able to provide an analysis of how precipitation differences impact valley oak growth. Overall, the full model explained 72% of the variation in relative growth rates, with initial height, block effects, Tmax difference, and locality showing the strongest effects (SI Appendix, Table S1 and Fig. S3).
Genome-Informed Assisted Gene Flow Mitigates Adaptational Lag to Temperature.
Our second objective was to assess whether genetic information could be used in management strategies to mitigate adaptational lag to temperature for valley oak. Here, we estimated GEBVs (25, 27–29) for high growth rates under future climate scenarios, which could form the basis for genome-informed assisted gene flow, and validated our analysis approach using data simulations (SI Appendix). We first used a genome-wide association study (GWAS) framework to estimate genotype-by-environment interactions of relative growth rates after adjusting for covariates not of primary interest (e.g., block effects and initial height) with Tmax difference across 12,357 single-nucleotide polymorphisms (SNPs) (Fig. 2A) and used these estimates to calculate GEBVs. We calculated GEBVs based on the maternal genotype of each seedling, and as a result we interpret the GEBVs to indicate the genetic value of each maternal tree in relation to their average progeny performance when progeny are planted into warmer temperatures. The explanatory power of GEBVs calculated with 600 SNPs (i.e., the ability of GEBVs to explain variation in adjusted relative growth rates in the same dataset the GEBVs were estimated from) was 9.5% (R2adj) for the full dataset (n = 2,295 seedlings). The predictive power of the GEBVs (i.e., the ability of the model to explain variation in observations not used in fitting the model) estimated via 10-fold cross-validation ranged from 3.5 to 4.0% (R2adj) depending on whether individuals from the same family were included in the training and testing sets and was higher than expected if there was no association between GEBVs and adjusted relative growth rates (SI Appendix, Table S2). The predictive power of GEBVs was lower than the explanatory power, indicating that GEBVs are less effective at predicting growth responses for individuals that are not included in the GEBV estimation, as is common in many studies using breeding values or polygenic scores (35).
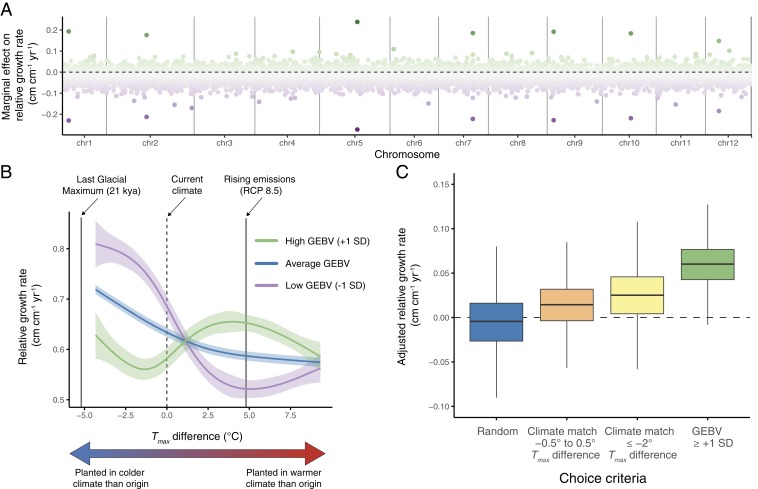
Fig. 2.
Genotype-by-temperature interactions in valley oak (Q. lobata). (A) Predicted marginal effects of genotypes on relative growth rates of progeny planted into warmer temperatures estimated by genotype by Tmax difference interactions of 12,357 SNPs across 12 chromosomes of valley oak. (B) Contrasting progeny growth responses for Tmax difference and approximate 95% confidence intervals for maternal trees with GEBVs for optimal growth rate under warmer temperatures at the average value, +1 SD above average, and −1 SD below average. (C) Box plots showing observed adjusted relative growth rates for hypothetical sets of seedlings chosen randomly, or based on matching individuals to their future climate, matching individuals to future climate and accounting for adaptational lag in valley oak, or selecting maternal trees with high GEBVs.
As expected, progeny of maternal trees with high GEBVs showed a pattern of “preadaptation” to warmer temperatures. The overall shape of the growth response to Tmax difference for progeny of maternal trees with high GEBVs (+1 SD away from average) peaked in warmer temperatures, which contrasts with the average valley oak response of decreasing growth rates in warmer temperatures (Fig. 2B). Specifically, at a 4.8 °C increase in temperature (predicted under RCP-8.5 by 2070 to 2099), progeny of maternal trees with GEBVs +1 SD above average estimated from the full dataset are predicted to have 11% and 25% higher relative growth rates than the progeny of maternal trees with average GEBVs or GEBVs −1 SD below average, respectively (Fig. 2B). Further analyses quantified the benefits of selecting source trees based on GEBVs under different climate scenarios. We used bootstrapping to simulate 10,000 hypothetical sets of 50 progeny selected based on the following criteria: 1) matching progeny to their future climate, which would maintain a Tmax difference of −0.5° to 0.5 °C between the source and planting site under future climate conditions; 2) optimizing growth by maintaining a Tmax difference ≤−2.0 °C between the source and planting site in future climate conditions to account for the species-level preference for cooler temperatures; 3) selecting progeny based on maternal trees with high GEBVs (+1 SD higher than the mean) under a scenario of warming temperatures (Tmax difference ≥ 0 °C between the source and planting site in future climate conditions); and 4) selecting progeny randomly without regard to GEBVs or Tmax difference. For each of these groups, we compared observed relative growth rates after adjusting for block effects, initial height, locality, family identification, genetic kinship, and climate of origin. Average observed growth rates were highest for progeny selected based on maternal GEBVs +1 SD above average (Fig. 2C), leading to a growth difference of 3 to 5 cm of height per year (assuming a 100-cm starting height) between individuals selected based on high maternal GEBVs compared to those selected based on climate matching alone.
Our approach could be extended to identify regions where trees may be especially vulnerable to growth rate declines or resilient to warming temperatures by mapping the current distribution of GEBVs across sampled populations of valley oak based on climate and spatial associations (SI Appendix, Fig. S5 and Table S3). The northern edge of the valley oak range, along the Sierra Nevada foothills in the eastern edge of the range, and the southeastern edge of the range all showed relatively high GEBVs and correspondingly high predicted progeny growth rates under warmer temperatures (RCP-8.5, 2070 to 2099) (Fig. 3A) and thus may be of lesser concern from a conservation and management perspective. In contrast, trees from the western, southwestern, and central parts of the valley oak range had relatively lower GEBVs and predicted progeny growth rates under warmer temperatures and, consequently, may be especially vulnerable to increasing temperatures from climate change (Fig. 3A). These results are in agreement with Kueppers et al. (33), who predicted persistence and expansion of valley oak in the northwestern, eastern, and southeastern parts of the range based on bioclimatic modeling, and with Sork et al. (34), who identified western valley oak populations as especially vulnerable to climate change based on neutral genetic variation. Together, these studies provide supporting evidence that different regions of valley oak vary in their vulnerability to climate change.
Fig. 3.
Landscape distribution of predicted changes in relative growth rates for valley oak based on GEBVs. (A) Predicted changes in progeny relative growth rates by 2070 to 2099 under a business-as-usual emissions scenario (RCP 8.5) based on current distribution of maternal GEBVs across valley oak populations. Black circles indicate sampled localities. Black outlines indicate contemporary valley oak range. (B) Predicted changes in progeny relative growth rates by 2070 to 2099 for a scenario where maternal trees with the highest GEBV within 50 km of each planting site are used as a seed source.
Our last analysis illustrates how growth rates across the range of valley oak would change if trees with the highest GEBVs were chosen as potential seed sources, as would be done under a genome-informed assisted gene flow program. Using the highest predicted GEBVs within a 50-km radius (GEBV distribution shown in SI Appendix, Fig. S5) for each potential planting site across our study area, we predicted progeny growth rates under warmer temperatures (RCP-8.5, 2070 to 2099) for those highest GEBVs. We found that choosing individuals with the highest GEBVs as seed sources leads to substantial increases in predicted progeny growth rates under warmer temperatures compared to selecting seeds regardless of GEBVs and in many locations offset the predicted declines in growth rates under future climate scenarios (Fig. 3B). In other words, these results demonstrate that using assisted gene flow informed by genomic data and how it relates to environmental conditions has the potential to mitigate the negative effects of rising temperatures from climate change in this species.
Discussion
Maladaptation is widely expected to be a consequence of future climate change (36), but this study demonstrates that contemporary populations of an ecosystem-foundational tree species, valley oak, are already maladapted to temperature under current climate conditions. This finding brings into question the assumption of local adaptation that forms the basis for typical management and conservation strategies (2). Using genome-wide sequence data, we showed the potential of genome-informed assisted gene flow to mitigate the negative effects of future increases in temperature by selecting for genotypes predicted to have relatively high productivity in future climate scenarios. The challenges associated with mitigating the effects of climate change through assisted gene flow are multifaceted and complex (9, 22, 37), but genomic-based approaches like the one outlined in this study are likely to play a key role in informing effective mitigation strategies.
For long-lived species, such as trees, that have occupied dynamic environments across geologic time, maladaptation to contemporary climate may be relatively common. Evidently, many populations of forest tree species seem to grow well when planted at their current latitude or climate niche (e.g., refs. 10 and 38), but maladaptation has been found in a variety of tree taxa such as poplar (Populus, ref. 39), eucalyptus (Eucalyptus, ref. 6), pine (Pinus, ref. 14), spruce (Picea, ref. 40), and oak (Quercus, ref. 41), with each taxon showing varying directions and degrees of maladaptation. For example, in Pinus sylvestris and P. contorta, populations tended to grow optimally in climates warmer than the climates they inhabit (14, 15), while 8 out of 10 species in the eastern United States would grow optimally in cooler climates (13). Across 15 species in western North America, bioclimatic envelope modeling suggests that populations lag behind their optimal climate by ∼130 km in latitude or 60 m in elevation (42). In our study, we found that species-wide optimal growth in valley oak occurs at cooler temperatures than populations currently experience, which could potentially be explained by past adaptation to colder, historical climates that has resulted in the current state of maladaptation. Supporting this idea, a previous study using putatively neutral genetic markers has found that climate 21,000 y ago from the Last Glacial Maximum explains a similar proportion of genetic variation in valley oak as current climate conditions, even after controlling for geographic effects, likely due to the avoidance of extreme bottlenecks from glaciation and the relative stability of valley oak populations across time (43), although we cannot rule out that the correlation between allele frequencies and climate has stayed proportional through time. Our findings that species-wide growth rates in valley oak were highest at cooler temperatures are not unprecedented and may indicate that many tree species may be challenged by climate warming (13).
Maladaptation to climate does not mean that tree populations have not responded to other selective pressures, such as soil properties and pathogens, or that their genetic composition is not shaped by genetic correlations and trade-offs among traits. Alternatively, maladaptation could also arise from the joint or combined effects of inbreeding, dispersal lag, gene flow, deleterious mutations, and genetic drift that are dependent on a species’ demographic and evolutionary history (17, 32, 44). Because climate is a complex selection pressure that involves many distinct and correlated factors (e.g., temperature, precipitation, and seasonality), it is important to consider how different aspects of the climate contribute to patterns of both local adaptation and maladaptation. While we show that changes in temperature are predicted to have substantial effects on growth rates, it is unlikely that temperature alone will solely determine the trajectory of valley oak populations over the next century. Rather, the independent and interacting influences of other aspects of climate (i.e., water availability) along with changes in biotic factors (i.e., pathogens and mutualists and competition) are likely to also have important influences on valley oak response to climate change. Additionally, selection for high growth rates may involve a trade-off for cold hardiness, drought, and other forms of abiotic stress (9, 32), so care must be taken not to maximize growth rates at the expense of increasing vulnerability to other stresses. Future studies should explicitly test these potential trade-offs to provide a more holistic perspective on how changing climate will affect valley oak fitness. Growth in early life stages is likely an important component of fitness for valley oak (45), but further analyses should examine whether the growth consequences of adaptational lag in early-life growth will persist into adulthood, which is a question that will be answered as the valley oak provenance trial continues. In sum, many evolutionary processes could influence climate-associated maladaptation, which calls for an assessment of the underlying mechanisms driving these patterns. Nonetheless, the possibility of contemporary maladaptation to climate supports a reevaluation of management strategies that assume local adaptation and provides an important factor for incorporation into future practices.
Assisted gene flow remains a controversial, yet promising, approach to alleviate the potential negative effects of climate change. An essential component of designing assisted gene flow programs is to understand contemporary patterns of adaptation to climate and other selection pressures (e.g., biotic interactions and edaphic factors) (20). In light of the results of this study, we caution against strategies that assume that local seed sources are preferable unless supported by empirical data. Although logistically challenging and currently rarely done, some species may be sufficiently important to use an approach that integrates common garden experiments with genome-wide sequencing to develop assisted gene flow programs by identifying genotypes preadapted to future climates based on observed genotype-by-climate interactions.
A key benefit of this genome-assisted approach is the ability to sequence adult trees in the field and choose seed sources based on GEBVs for a planting site, given the local current and predicted climate. With the increasing availability and decreasing cost of genotyping technologies, this approach is likely to become more feasible over time. We have focused here on a business-as-usual climate scenario where emissions continue to rise over the next century (RCP-8.5), but the high levels of uncertainty both within and across different climate scenarios warrants thought about what time point in the future should be chosen as the reference point for preadaptation. The flexibility of the breeding value approach presented in this study allows the possibility of calculating breeding values for different time points and different climate scenarios to account for this uncertainty. To help address these challenges and uncertainties, composite and admixture seed selection strategies that utilize multiple source populations for assisted gene flow may increase the probability of success by ensuring that adequate levels of genetic diversity are present to hedge against uncertainty in future climate scenarios and nonclimate-related factors (9, 37). For species with demonstrated maladaptation, we conclude that genome-informed assisted gene flow that utilizes genome-wide sequencing data to identify genotypes preadapted to future climate conditions may be effective in mitigating the negative impacts of climate change on natural ecosystems.
Materials and Methods
Study System: Valley Oak Provenance Trial.
Valley oak (Q. lobata Née) is endemic to California woodlands and savannas, with populations ranging from 0- to 1,700-m elevation along the central valley and foothills of the Sierra Nevada and Coastal Ranges. Populations of valley oak have declined dramatically over the past few centuries due to increasing development and conversion of habitat and is further threatened by a combination of recruitment limitation (45) and climate change (33, 34). The valley oak provenance trial was initiated in 2012 when >11,000 open-pollinated acorns were collected from 674 adults distributed across over 95 localities within the range of valley oak (for design and early data see ref. 30). Then, in 2014, 6,945 of those seedlings were out-planted into 2 common gardens sites (SI Appendix, Fig. S1).
Climate Data.
We obtained historical and projected climate data from the Basin Characterization Model dataset developed for California (46). To estimate Tmax difference (i.e., the difference in Tmax between where a seedling was planted and its origin in units of degrees Celsius), we calculated the difference between the 2014 to 2016 average Tmax of each planting site and the 1951 to 1980 30-y average Tmax of the climate of origin (SI Appendix). Similar patterns were found using different reference points of Tmax (e.g., 1921 to 1950). Note that Tmax difference estimated in this way would include any recent (i.e., 1951 to 2016) changes in Tmax between source and planting sites due to anthropogenic climate change. Positive values of Tmax indicate a seedling being planted into a warmer climate than where it originated, and negative values indicate cooler climates, with 0 °C Tmax difference indicating no change in temperature.
Modeling Growth Based on Tmax Difference.
We used a generalized additive model (GAM) framework (47) to model the effect of Tmax difference on the growth of valley oak in the provenance trials. GAMs are a suitable framework for this analysis because they easily and flexibly accommodate nonlinear responses in growth (11), without imposing limits on the shape that responses can take (e.g., parabola in a quadratic regression). Our response variable was relative growth rate, measured as , where H2014 and H2017 are the height of the tallest stem as measured during the 2014 and 2017 census, respectively. Timedif is the difference in years between the 2014 and 2017 censuses. Our model controlled for block effects, site and climate of origin, family, and initial height. See SI Appendix for full model specification.
Genotyping by Sequencing.
To relate the genotypes of adults with progeny performance in the provenance trial, we used genotyping by sequencing to obtain information on SNPs for 451 adults across the valley oak range, with 304 of the 451 adults having progeny represented in the provenance trial (SI Appendix, Fig. S1). Further details on sampling and sequencing are available in SI Appendix. After filtering (SI Appendix), we retained 12,357 SNPs from 421 valley oak adults that passed quality and missing data filters.
Genome-Wide Association Analysis with Tmax Difference.
To estimate how genetic variation was associated with differential growth response to Tmax difference, we followed a GWAS framework, where each SNP is modeled independently using a GAM framework similar to the one described above. We used a 2-stage residual-outcome approach to aid in computational efficiency, where in stage 1 our dependent variable relative growth rate was regressed against covariates that are not of primary interest (e.g., block effects and initial height of each individual; SI Appendix). In stage 2, the residuals from the model in stage 1 (i.e., adjusted relative growth rate) were then used as the dependent variable in the GWAS to estimate allelic effects on relative growth rates after controlling for potential confounding factors (SI Appendix).
GEBVs.
To evaluate the genomic basis of growth responses to Tmax difference, we calculated GEBVs. To estimate GEBVs, we summed the predicted adjusted relative growth rates in warmer temperatures (Tmax difference > 0 °C) for each genotype and evaluated the predictive ability of GEBVs using 10-fold cross-validation (28) (SI Appendix). We further validated our approach using data simulations to assess how well the GEBVs estimated from the approach outline above were correlated with “true” breeding values known from simulated data (SI Appendix). Estimating GEBVs using best linear unbiased predictors produced similar results (SI Appendix).
Data Availability.
Phenotypic and genotypic data from the valley oak provenance trial along with scripts used for statistical analyses are publicly available on figshare (https://figshare.com/articles/Data_and_analysis_scripts_for_Adaptational_lag_to_temperature_in_valley_oak_Quercus_lobata_can_be_mitigated_by_genome-informed_assisted_gene_flow_/9999629/1). Climate data used in this study are publicly available through the Basin Characterization Model website (https://ca.water.usgs.gov/projects/reg_hydro/basin-characterization-model.html).
Supplementary Material
Acknowledgments
We acknowledge the native peoples of California as the traditional caretakers of oak ecosystems sampled for this project. We thank Annette Delfino-Mix and the many data collectors and volunteers who have worked to help establish and maintain the valley oak provenance trial. We thank D. Ackerly, G. DiRenzo, R. Harrigan, J. Lloyd-Smith, K. Lohmueller, members of the V.L.S. laboratory, and 2 anonymous reviewers for valuable feedback. This project was supported in part by a research award to V.L.S. and colleagues from the NSF Plant Genome Research Program (NSF IOS-1444661). Funding for L.B. was provided by the La Kretz Center for California Conservation Science at the University of California, Los Angeles, and the NSF award. Provenance trials were supported by the US Department of Agriculture Forest Service Pacific Southwest Research Station. Any use of product names is for informational purposes only and does not imply endorsement by the US Government.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Raw sequence reads reported in this paper have been deposited in the NCBI GenBank BioProject database (accession no. PRJNA587528).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908771116/-/DCSupplemental.
References
- 1.Urban M. C., Accelerating extinction risk from climate change. Science 348, 571–573 (2015). [DOI] [PubMed] [Google Scholar]
- 2.McKay J. K., Christian C. E., Harrison S., Rice K. J., “How local is local?” A review of practical and conceptual issues in the genetics of restoration. Restor. Ecol. 13, 432–440 (2005). [Google Scholar]
- 3.Broadhurst L. M., et al. , Seed supply for broadscale restoration: Maximizing evolutionary potential. Evol. Appl. 1, 587–597 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leimu R., Fischer M., A meta-analysis of local adaptation in plants. PLoS One 3, e4010 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hereford J., A quantitative survey of local adaptation and fitness trade-offs. Am. Nat. 173, 579–588 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Gellie N. J. C., Breed M. F., Thurgate N., Kennedy S. A., Lowe A. J., Local maladaptation in a foundation tree species: Implications for restoration. Biol. Conserv. 203, 226–232 (2016). [Google Scholar]
- 7.Aitken S. N., Yeaman S., Holliday J. A., Wang T., Curtis-McLane S., Adaptation, migration or extirpation: Climate change outcomes for tree populations. Evol. Appl. 1, 95–111 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sork V. L., et al. , Putting the landscape into the genomics of trees: Approaches for understanding local adaptation and population responses to changing climate. Tree Genet. Genomes 9, 901–911 (2013). [Google Scholar]
- 9.Aitken S. N., Bemmels J. B., Time to get moving: Assisted gene flow of forest trees. Evol. Appl. 9, 271–290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matyas C., Climatic adaptation of trees: Rediscovering provenance tests. Euphytica 92, 45–54 (1996). [Google Scholar]
- 11.Schmidtling R. C., Use of provenance tests to predict response to climate change: Loblolly pine and Norway spruce. Tree Physiol. 14, 805–817 (1994). [DOI] [PubMed] [Google Scholar]
- 12.Wang T., O’Neill G. A., Aitken S. N., Integrating environmental and genetic effects to predict responses of tree populations to climate. Ecol. Appl. 20, 153–163 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Carter K. K., Provenance tests as indicators of growth response to climate change in 10 north temperate tree species. Can. J. For. Res. 26, 1089–1095 (1996). [Google Scholar]
- 14.Rehfeldt G. E., et al. , Intraspecific responses to climate in Pinus sylvestris. Glob. Change Biol. 8, 912–929 (2002). [Google Scholar]
- 15.Rehfeldt G. E., Ying C. C., Spittlehouse D. L., Hamilton D. A. Jr, Genetic responses to climate in Pinus contorta: Niche breadth, climate change, and reforestation. Ecol. Monogr. 69, 375–407 (1999). [Google Scholar]
- 16.Wang T., Hamann A., Yanchuk A., O’Neill G. A., Aitken S. N., Use of response functions in selecting lodgepole pine populations for future climates. Glob. Change Biol. 12, 2404–2416 (2006). [Google Scholar]
- 17.Crespi B. J., The evolution of maladaptation. Heredity 84, 623–629 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Davis M. B., Shaw R. G., Etterson J. R., Evolutionary responses to changing climate. Ecology 86, 1704–1714 (2005). [Google Scholar]
- 19.Wilczek A. M., Cooper M. D., Korves T. M., Schmitt J., Lagging adaptation to warming climate in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 111, 7906–7913 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aitken S. N., Whitlock M. C., Assisted gene flow to facilitate local adaptation to climate change. Annu. Rev. Ecol. Evol. Syst. 44, 367–388 (2013). [Google Scholar]
- 21.Hoegh-Guldberg O., et al. , Ecology. Assisted colonization and rapid climate change. Science 321, 345–346 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Schwartz M. W., et al. , Managed relocation: Integrating the scientific, regulatory, and ethical challenges. Bioscience 62, 732–743 (2012). [Google Scholar]
- 23.Bonan G. B., Forests and climate change: Forcings, feedbacks, and the climate benefits of forests. Science 320, 1444–1449 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Pan Y., et al. , A large and persistent carbon sink in the world’s forests. Science 333, 988–993 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Gienapp P., et al. , Genomic quantitative genetics to study evolution in the wild. Trends Ecol. Evol. 32, 897–908 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Holliday J. A., et al. , Advances in ecological genomics in forest trees and applications to genetic resources conservation and breeding. Mol. Ecol. 26, 706–717 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Meuwissen T. H., Hayes B. J., Goddard M. E., Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desta Z. A., Ortiz R., Genomic selection: Genome-wide prediction in plant improvement. Trends Plant Sci. 19, 592–601 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Lasky J. R., et al. , Genome-environment associations in sorghum landraces predict adaptive traits. Sci. Adv. 1, e1400218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delfino-Mix A., Wright J. W., Gugger P. F., Liang C., Sork V. L., “Establishing a range-wide provenance test in valley oak (Quercus lobata Née) at two California sites” in Proceedings of the Seventh Oak Symposium: Managing Oak Woodlands in a Dynamic World (US Forest Service, US Department of Agriculture, 2015), pp. 413–424. [Google Scholar]
- 31.Blois J. L., Williams J. W., Fitzpatrick M. C., Jackson S. T., Ferrier S., Space can substitute for time in predicting climate-change effects on biodiversity. Proc. Natl. Acad. Sci. U.S.A. 110, 9374–9379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leites L. P., Rehfeldt G. E., Steiner K. C., Adaptation to climate in five eastern North America broadleaf deciduous species: Growth clines and evidence of the growth-cold tolerance trade-off. Perspect. Plant Ecol. Evol. Syst. 37, 64–72 (2019). [Google Scholar]
- 33.Kueppers L. M., Snyder M. A., Sloan L. C., Zavaleta E. S., Fulfrost B., Modeled regional climate change and California endemic oak ranges. Proc. Natl. Acad. Sci. U.S.A. 102, 16281–16286 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sork V. L., et al. , Gene movement and genetic association with regional climate gradients in California valley oak (Quercus lobata Née) in the face of climate change. Mol. Ecol. 19, 3806–3823 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Wray N. R., et al. , Pitfalls of predicting complex traits from SNPs. Nat. Rev. Genet. 14, 507–515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw R. G., Etterson J. R., Rapid climate change and the rate of adaptation: Insight from experimental quantitative genetics. New Phytol. 195, 752–765 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Breed M. F., Stead M. G., Ottewell M., Gardner M. G., Lowe A. J., Which provenance and where? Seed sourcing strategies for revegetation in a changing environment. Conserv. Genet. 14, 1–10 (2013). [Google Scholar]
- 38.O’Neill G. A., Nigh G., Linking population genetics and tree height growth models to predict impacts of climate change on forest production. Glob. Change Biol. 17, 3208–3217 (2011). [Google Scholar]
- 39.Gray L. K., Gylander T., Mbogga M. S., Chen P. Y., Hamann A., Assisted migration to address climate change: Recommendations for aspen reforestation in western Canada. Ecol. Appl. 21, 1591–1603 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Andalo C., Beaulieu J., Bousquet J., The impact of climate change on growth of local white spruce populations in Quebec, Canada. For. Ecol. Manage. 205, 169–182 (2005). [Google Scholar]
- 41.Sáenz-Romero C., et al. , Adaptive and plastic responses of Quercus petraea populations to climate across Europe. Glob. Change Biol. 23, 2831–2847 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gray L. K., Hamann A., Tracking suitable habitat for tree populations under climate change in western North America. Clim. Change 117, 289–303 (2013). [Google Scholar]
- 43.Gugger P. F., Ikegami M., Sork V. L., Influence of late Quaternary climate change on present patterns of genetic variation in valley oak, Quercus lobata Née. Mol. Ecol. 22, 3598–3612 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Hellmann J. J., Pineda-Krch M., Constraints and reinforcement on adaptation under climate change: Selection of genetically correlated traits. Biol. Conserv. 137, 599–609 (2007). [Google Scholar]
- 45.Tyler C. M., Kuhn B., Davis F. W., Demography and recruitment limitations of three oak species in California. Q. Rev. Biol. 81, 127–152 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Flint L. E., Flint A. L., Thorne J. H., Boynton R., Fine-scale hydrologic modeling for regional landscape applications: The California Basin characterization model development and performance. Ecol. Process. 2, 25 (2013). [Google Scholar]
- 47.Wood S. N., mgcv: GAMs and generalized ridge regression for R. R. News 1, 20–25 (2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Phenotypic and genotypic data from the valley oak provenance trial along with scripts used for statistical analyses are publicly available on figshare (https://figshare.com/articles/Data_and_analysis_scripts_for_Adaptational_lag_to_temperature_in_valley_oak_Quercus_lobata_can_be_mitigated_by_genome-informed_assisted_gene_flow_/9999629/1). Climate data used in this study are publicly available through the Basin Characterization Model website (https://ca.water.usgs.gov/projects/reg_hydro/basin-characterization-model.html).