Abstract
Molecular characterization of lung squamous cell carcinoma (LUSC), one of the major subtypes of lung cancer, has not sufficiently improved its non-stratified treatment strategies over decades. Accumulating evidence suggests that lineage-specific transcriptional regulators control differentiation states during cancer evolution, and underlie their distinct biological behaviors. In this study, by investigating the super-enhancer landscape of LUSC, we identified a previously undescribed ‘neural’ subtype defined by Sox2 and a neural lineage factor Brn2, as well as the classical LUSC subtype defined by Sox2 and its classical squamous partner p63. Robust protein-protein interaction and genomic co-occupancy of Sox2 and Brn2, in place for p63 in the classical LUSC, indicated their transcriptional cooperation imparting this unique lineage state in the ‘neural’ LUSC. Forced expression of p63 downregulated Brn2 in the ‘neural’ LUSC cells and invoked the classical LUSC lineage with more squamous/epithelial features, which were accompanied by increased activities of ErbB/Akt and MAPK-ERK pathways suggesting differential dependency. Collectively, our data demonstrate heterogeneous cell lineage states of LUSC featured by Sox2 cooperation with Brn2 or p63, for which distinct therapeutic approaches may be warranted.
Keywords: Sox2, Brn2, p63, lung squamous cell carcinoma, super-enhancer
Introduction
Lung cancer is the leading cause of cancer-related death worldwide (1). Despite recent progress in diagnosis and treatment including molecular-targeted therapeutics and immunotherapy which provide a considerable survival benefit for lung cancer patients, the overall 5-year survival remains less than adequate at 19% (2). Lung squamous cell carcinoma (LUSC) is the second most common histological subtype of lung cancer and is strongly associated with cigarette smoking. Although achievements in tobacco control in developed countries have contributed to a decline in mortality rate of lung cancer, LUSC still remains a major cause of death worldwide and shows even higher incidence than lung adenocarcinoma in several countries (3,4). Comprehensive genomic characterization of LUSC conducted by The Cancer Genome Atlas (TCGA) project revealed its heterogeneous features with complex genomic alterations (5). Unfortunately, these findings have not resulted in the successful development of clinically approved targeted drugs for LUSC. Compared to patients with lung adenocarcinoma, where multiple genome-based targeted therapeutics have been approved, LUSC patients have very limited therapeutic options. More recently, immune checkpoint inhibitors have emerged as key therapeutic options for solid tumors including LUSC (6,7); however, the response rate for unselected LUSC population remains only approximately 20% and initially-responded tumors eventually progress in most cases. Therefore, innovative strategies to better characterize and classify LUSC for a better patient stratification for current and future therapeutic options are desperately needed.
We previously identified SOX2 as the most commonly amplified oncogene in LUSC (8). We proposed SOX2 as a lineage-survival oncogene in squamous cell cancers for its essential role during the development in the specification of the squamous cell lineages by opposing the role of Nkx2-1 in the dividing foregut and its essentiality for LUSC cell survival (9). In a following study, we identified another squamous lineage factor, p63 as an important cooperative partner of Sox2 in LUSC (10). SOX2 amplification on chromosome 3q in LUSC often extends to its telomeric side to include the locus of TP63, which encodes p63. While focal amplification and/or overexpression of both SOX2 and TP63 genes is found in only 7 % of LUSC tumors, broader copy number gains on 3q telomeric ends are observed in the vast majority of LUSCs (5). Studies on expression profiles classified LUSCs into four expression subtypes (primitive, classical, secretory, and basal), suggesting the heterogeneity of transcriptional programs within LUSCs (5,11,12). Based on this classification, co-amplification of SOX2 and TP63 is enriched in the classical subtype of LUSC whereas TP63 expression is relatively low in the primitive and secretory subtypes. However, it remains largely unknown what mechanisms are involved in controlling transcriptional programs in the heterogeneous group of LUSCs.
A series of recent genome-wide histone modification analyses have demonstrated the presence of large clusters of putative enhancers in close genomic proximity, coined super-enhancers (13,14). Since these regions typically exhibit cell-lineage-specific patterns in health and in disease, super-enhancer profiling is becoming a powerful tool to identify novel cancer cell lineages governed by specific transcriptional regulators. For example, analysis of the super-enhancer landscape of neuroblastoma has demonstrated heterogeneity of lineage states governed by specific transcriptional programs that may underlie the cause of relapse after chemotherapy (15). Similar analysis of the super-enhancer landscape in acute myeloid leukemia identified a novel epigenomic subtype, which have therapeutic implication for differentiation therapy (16). In the present study, we investigated super-enhancer profiles in LUSC and identified a novel subtype in which Sox2 and a neural transcription factor Brn2 have key roles in determining its distinctive differentiation state. In addition, we show that Brn2 serves as an interacting partner for Sox2 in this novel subtype, instead of p63 in the classical subtype of LUSC and that forced expression of p63 leads to classical squamous cell differentiation and suppression of Brn2 in this novel LUSC subtype.
Materials and Methods
Cell lines
Lung cancer cell lines (HCC95, KNS62, HCC2814, LK2, NCI-H520, SQ-1, EBC-1, HCC2279, NCI-H2887, HCC2450, Calu-1, HARA, LC-1/SQSF) were maintained in RPMI-1640 (Gibco) with 10% FBS and 1% penicillin-streptomycin (Gibco). HEK293T cells were maintained in DMEM with 10% FBS and 1% penicillin-streptomycin (Gibco). Original sources for these cell lines are shown in Supplementary Table S1. HCC95, HCC2814, HCC2279, NCI-H2887 and HCC2450 cells were a gift from Dr. John Minna. Cells were authenticated by copy number variation analyses and regularly tested for mycoplasma using the mycoAlert Detection Kit (Lonza).
Chromatin immunoprecipitation sequencing (ChIP-seq)
For H3K27ac, Sox2 and p63 ChIP, cells were performed as previously described with modifications (10). Cells were crosslinked with 1% formaldehyde in PBS for 10 min at room temperature, washed in 5 mg/ml bovine serum albumin (BSA) in PBS and then in just cold PBS, re-suspended in lysis buffer (50 mM Tris-HCl pH 8.1, 10 mM EDTA, 1% SDS, 1× protease inhibitor cocktail (Thermo Fisher Scientific)) and sonicated with the Covaris M220 sonicator or the Diagenode Bioruptor sonicator to obtain chromatin fragment lengths of 100-to-1,000 bp judged by Bioanalyzer DNA High sensitivity kit (Agilent). Fragmented chromatin was diluted in IP buffer (20 mM Tris-HCl pH 8.1, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100) and incubated overnight at 4°C with Protein G magnetic beads (Dynabeads: ThemoFisher) that had been pre-incubated with anti-H3K27ac (Abcam, ab4729) or anti-Sox2 (R&D, AF2018) or anti-p63 (Santa Cruz, sc-8431) antibodies.
For Brn2 ChIP, cells were crosslinked with 2 mM disuccinimidyl glutarate in PBS with 1 mM MgCl2 for 45 min at room temperature and 1% formaldehyde in PBS for 11 min at room temperature, washed in 5 mg/ml BSA in PBS and then in just cold PBS, re-suspended in nuclear extraction buffer (10 mM Tris-HCl pH 7.5, 10 mM NaCl, 3mM MgCl2, 0.1 % IGEPAL CA-630, 1× protease inhibitor cocktail) and incubated on ice for 10 min. Extracted nuclei were then washed and resuspended in micrococcal nuclease digestion buffer (0.3 M sucrose, 20 mM Tris-HCl pH 7.5, 3 mM CaCl2, 1× protease inhibitor cocktail) and incubated with 12,000 gel units/ml micrococcal nuclease (NEB, M0247S) at 37°C with frequently mixing to digest chromatin to lengths of approximately 100-to-1,000 bp. Digestion was stopped with 25 mM EDTA pH 8.0 and nuclei was re-suspended in lysis buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% SDS, 1× protease inhibitor cocktail) and sonicated with the Diagenode Bioruptor sonicator (low intensity, 30 sec on and 30 sec off, 2 cycles). Fragmented chromatin was diluted in IP buffer (20 mM Tris-HCl pH 8.1, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100) and incubated with anti-Brn2 (Cell Signaling Technology, #12137) antibody overnight at 4°C. Protein G magnetic beads (Dynabeads: ThemoFisher) were then added and the IP reaction was incubated 2 hours at 4°C.
Immunoprecipitates were washed six times with wash buffer (50 mM HEPES pH 7.6, 0.5 M LiCl, 1 mM EDTA, 0.7% Na deoxycholate, 1% IGEPAL CA-630) and twice with TE buffer. Immunoprecipitated (or no IP input) DNA was treated with RNase A and Proteinase K on the beads, recovered in 1% SDS and 0.1 M NaHCO3 over a period of 7 h at 65°C, and purified with DNA clean and concentrator-25 (Zymo Research). Up to 10 ng of DNA was used for the library construction using NEBNext Ultra II DNA Library Prep Kit (NEB, E7645). Sequencing was performed on NextSeq500 (Illumina) for 38 nucleotides each from paired ends according to the manufacturer’s instructions.
ChIP-seq data analysis
For H3K27ac ChIP, ChIP-enriched regions (peaks) were identified by MACS (17) after aligning to the human reference genome hg19/GRCh37. To define ‘super-enhancers’, we used the ROSE2 pipeline (18). H3K27ac enriched peaks were stitched together, using an optimal distance determined by the algorithm per sample up to 12.5 kb. The algorithm generates plots for all stitched enhancers per each sample and defines super-enhancers on the basis of top-ranked enhancers with highest read counts at the tangential cut-off. Unsupervised hierarchical clustering was performed using signals (read counts on defined super-enhancer regions) near transcriptional regulators. A list of the transcriptional regulators was obtained from the genes annotated with gene ontology term, DNA-binding (GO: 0003677). Principle component analysis was performed based on the same signals per region, and top 3 components were depicted in three dimensional plots.
ChIP signals for each sample were visualized on integrative genome viewer (IGV) genome browser (19) using wiggle files with a 10 bp resolution for H3K27ac modification generated by MACS with tag shift that was rescaled to normalize to a total number of uniquely alignable sequences by WigMath function of Java-Genomic Toolkit. Differentially enriched super-enhancers were determined by comparing the ‘neural’ subgroup against the ‘classical’ subgroup using ‘samr’ R package (20), based on cutoffs of fold change>2 and SAM sore>4.
For Sox2, Brn2 and p63 ChIP, peaks were identified by MACS (17) after aligning to the human reference genome hg19/GRCh37. ChIP signals on Sox2, Brn2 and p63 peaks detected on any samples were visualized as a heatmap generated by ‘heatmap’ function in Cistrome analysis pipeline (http://cistrome.org/) or plotHeatmap function of deepTools (21) using wiggle files with a 10 bp resolution generated by MACS with tag shift that was rescaled to normalize to a total number of uniquely alignable sequences by WigMath function of Java-Genomic Toolkit.
RNA-seq
Total RNAs from engineered LK2 cell lines were extracted using the Qiagen RNeasy kit. Poly-adenylated RNA was enriched from 1ug of RNA for each sample with the NEBNext PolyA mRNA Magnetic Isolation Module (NEB, E7490), incubated at 94°C for 15 min and double-strand cDNA was synthesized using SuperScript III reverse transcriptase (ThermoFisher) and NEBNext® Ultra™ II Directional RNA Second Strand Synthesis Module (NEB). Up to 10 ng of cDNA was used for the Illumina sequencing library construction using NEBNext Ultra DNA Library Prep Kit (NEB, E7645). Paired ends sequencing was performed on NextSeq 500 (Illumina) for 38 nucleotides from each end according to the manufacturer’s instructions.
RNA-seq analyses
RNA-seq gene expression data as well as SNP-array copy number data for SOX2 were obtained from Cancer Cell Line Encyclopedia (CCLE) (http://www.broadinstitute.org/ccle/home). RNA-seq data of 501 tumor tissues were obtained from TCGA-LUSC dataset (5). We downloaded htseq-counts as read counts for each gene in May 2018 (https://portal.gdc.cancer.gov/). The TCGA read counts originally aligned for Ensemble transcripts were converted to corresponding RefGene symbol based on the USCS database. mRNA abundance was estimated from read counts in Transcripts Per Million (TPM) as described in Wagner et el. (22). GTEx TPM matrix (https://gtexportal.org/home/datasets) was downloaded from GTEx data portal in July 2018 (23). Data from the brain hypothalamus region were used for the analysis. Log2-transformed TPM values were used as log2(TPM+1) for the following analysis. Two RNA-seq data (EGAD00001001244 and GSE60052) were downloaded for Small Cell Lung Cancer (SCLC) (24,25). We downloaded raw fastq files, aligned to the human reference genome hg19, and estimated mRNA abundance in terms of TPM.
For the TCGA-LUSC RNA-seq data, the 501 LUSC tumor tissues were classified based on the expression level of TP63, SOX2 and POU3F2. We used TPM cutoffs using (mean – standard deviation) for TP63 and SOX2 as their expressions are normally distributed (Fig. 2D and Supplementary Fig. S2D and S2E) and TPM>1 for POU3F2 since most of the tumors have little expression of the gene (Fig. 2A). Hierarchical clustering of the LUSC tumors (n=416) was performed using the differentially expressed genes between POU3F2-high/ TP63-low and POU3F2-low/ TP63-high tumors with complete-linkage clustering (Fig. 3A). Kaplan-Meier curves were plotted to compare survival for patients with POU3F2-high/ TP63-low tumors versus patients with POU3F2-low/ TP63-high tumors (Fig. 3C). To compare the POU3F2-high/ TP63-low tumors with previously reported LUSC expression subtypes, the TCGA tumors were classified based on expression of the 16 subtype exemplar genes for each subtype (5). Using the k-mean clustering with k=4, these genes classified 497 tumors into 4 clusters with 106 primitive, 158 classical, 127 secretory, and 106 basal subtype tumors (Supplementary Fig. S2G).
To perform RNA-seq analyses for engineered LK2 cell lines, sequencing reads were aligned to the human reference genome hg19/GRCh37 using Tophat (v2.1) and read counts per RefGene symbol on the USCS database was estimated using the htseq-count function in SAMtools (26). Then DEseq2 (27) was used to identify the differentially expressed genes between control LK2 cells (two stable GFP-overexpressed LK2 cells and duplicates of inducible DNp63-expressing LK2 cells without doxycycline) and DNp63-overexpressed LK2 cells (two stable DNp63-overexpressed cells and duplicates of inducible DNp63-expressing LK2 cells after treatment with 2 μg/ml doxycycline for 6 days) and to identify genes significantly associated with DNp63 overexpression levels (control -> inducible -> stable). To compare the transcriptomic profiles of the engineered LK2 cells with those of the LUSC cell lines from CCLE, we merged the TPM matrix from the engineered cells with that from the CCLE cell lines based on common genes and performed quantile normalization followed by ComBat to remove potential batch effects.
To identify potentially enriched functions of selected gene sets of interest, we compared these gene sets with the genes annotated by the same Gene Ontology (GO) terms curated in the Molecular Signature Database (MSigDB) (28). Each of 5917 GO terms included in “C5” collection (version 6.0 downloaded in 2017) was compared with query gene sets. Any GO terms consisting of more than 2000 genes were considered non-specific and removed from the analysis.
Immunofluorescence
Cells were grown on glass coverslips coated with poly D-lysine (Neuvitro), fixed in 4% paraformaldehyde for 10 min and permeabilized with 1× PBS containing 0.25% Triton X-100 for 10 min. Primary antibodies were used at the following dilutions: anti-Brn2 at 1:200 (Cell Signaling, #12137), and anti-Sox2 at 1:400 (Abcam, ab171380). Fluorescent signal was detected with secondary antibodies conjugated with Alexa Fluor (ThermoFisher #A-11034, #A-21236) diluted at 1:2000, and coverslips were mounted (ProLong Diamond Antifade Mountant with DAPI: ThermoFisher). Images were obtained with a Leica DM5500 B fluorescence microscope at 20× objective magnification. Images were processed using the Fiji distribution of ImageJ (https://fiji.sc/).
Western blotting and co-immunoprecipitation
To determine protein expression, cell lysates were prepared by incubating cells in lysis buffer A (150 mM NaCl, 50 mM Tris-HCl at pH 8.0, 1% IGEPAL CA-630, 0.5% Na deoxycholate, 0.1% SDS, protease inhibitors) or lysis buffer B (150 mM NaCl, 40 mM Tris-HCl at pH 8.0, 1% IGEPAL CA-630, 0.5% Na deoxycholate, 2mM EDTA at pH 8.0, protease/phosphatase inhibitors) for 30 min at 4°C. After centrifugation to remove insoluble debris, lysates were immunoblotted with use of an anti-Brn2 antibody (Santa Cruz Biotechnology, sc-393324 or Cell Signaling Technology, #12137), anti-p63 antibody (Santa Cruz Biotechnology, sc-8344, sc-8431 or Cell Signaling Technology, #13109), anti-Sox2 antibody (R&D, AF2018 or Cell Signaling Technology, #3579), anti-ErbB4 antibody (Cell Signaling Technology, #4795), anti-phospho-ErbB4 antibody (Cell Signaling Technology, #4757), anti-ErbB3 antibody (Cell Signaling Technology, #12708), anti-phospho-ErbB3 antibody (Cell Signaling Technology, #4791), anti-EGFR antibody (Millipore, #06-847), anti-phospho-EGFR antibody (Abcam, ab40815) anti-phospho-Akt antibody (Cell Signaling Technology, #4060) and anti-phospho-Erk1/2 antibody (Cell Signaling Technology, #4370), anti-Ascl1 antibody (Santa Cruz, sc-374104), anti-SYP antibody (Santa Cruz, sc-17750), anti-β-Actin antibody (Sigma), or anti-vinculin (Sigma). For western blotting to profile ErbB family signaling, engineered LK2 cells were serum-starved for four hours, and then incubated with anti-ErbB4 monoclonal antibody P6-1 (a kind gift from the Hideyuki Saya laboratory, Keio University) for 15 minutes, followed by stimulation with recombinant human NRG1-β1 EGF domain (R&D) for 30 minutes.
For co-immunoprecipitation, whole cell lysate of cells was prepared in IP lysis buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% IGEPAL CA-630, 5% glycerol, protease inhibitors) followed by incubation with anti-Brn2 antibody, anti-Sox2 antibody, anti-p63 antibody or normal IgG antibody and Dynabeads Protein G (ThemoFisher) on a rotator overnight at 4°C. After washing with PBS + 0.05% Tween 20, the beads were boiled for 10 min in 1× SDS sample buffer. The immunoprecipitates were then processed for immunoblotting.
Lentiviral introduction of genes
POU3F2, DNp63α or GFP open reading frame (ORF) was cloned into pLEX_306 (a gift from Dr. David Root, Addgene #41391), pLEX_307 (a gift from Dr. David Root, Addgene #41392) or pLIX_403 (a gift from Dr. David Root, Addgene #41395) using Gateway® cloning methods according to manufacturer’s recommendations. For lentiviral vectors production, HEK293T cells were seeded in 10-cm tissue culture dish and incubated at 37°C and 5% CO2. Cells at 80% confluency were co-transfected with 10 μg of lentiviral expression constructs, 7.5 μg of psPAX2 and 2.5 μg pMD2.G vectors using TransIT-Lenti (Mirus) following manufacturer’s recommendations. At 48 h post transfection, supernatants were collected, filtered (0.45 μm) and stored at −80°C. Cells were infected with supernatant containing lentivirus supplemented with polybrene at a final concentration of 8 μg/mL and then selected with puromycin (2-3 μg/mL for 4-6 days). Ectopic protein expression was confirmed via immunoblotting and compared with physiological expression levels in the LUSC cells with native expression of the transgenes.
CRISPR-Cas9 genome editing
Cells stably expressing human codon-optimized S. pyogenes Cas9 were generated by infection with the lentiCas9-Blast plasmid (a gift from Dr. Feng Zhang, Addgene, # 52962). sgRNAs were cloned at BbsI site downstream of the human U6 promoter in a lentiviral vector containing eGFP downstream of the human PGK promoter (a kind gift from the Brian Brown laboratory, Icahn School of Medicine at Mount Sinai). Lentivirus was produced as above. Cells were first infected with the lentiCas9-Blast lentivirus, and then selected with blasticidin (5 μg/mL for 10 days) for cells expressing the Cas9 nuclease. Cells were then infected with pLenti-GFP-sgRNA. Polyclonal populations of infected cells were used to avoid the effects of selecting single/unique knockout clones. The knockout efficiency was confirmed in the bulk populations via immunoblotting. sgRNA target sequences were selected from Brunello library(29) and Orzol et al. (30). Non-target sgRNAs from the Gecko library v2 (31) were used as scramble sgRNAs. sgRNA target sequences are listed in Supplementary Table S2.
Immunohistochemistry
Immunohistochemical analyses were performed for xenograft tumor specimens and human primary LUSC tumor specimens. Total 23 formalin-fixed, paraffin-embedded (FFPE) specimens of human primary LUSCs resected from patients at Icahn School of Medicine at Mount Sinai (ISMMS, USA) between 2011 and 2013 were obtained and cut into 5-μm-thick sections. Characteristics of the human LUSC specimens are shown in Supplementary Table S3.
Immunohistochemical staining for Brn2, after being deparaffinized and rehydrated, FFPE tissue slides (5 μm thick) were heated at 95°C in 10 mM citrate buffer (pH 6.0) for antigen retrieval for 30 minutes. The sections were incubated with 0.3% H2O2 in TBS for 15 minutes to block endogenous peroxidase activity and were incubated with 10% normal goat serum (Jackson ImmunoResearch) in TBS for 30 minutes to block non-specific staining. After rinsing with TBS + 0.025% Triton X-100, the sections were incubated with anti-Brn2 antibody (1:100; Cell Signaling #12137) at 4°C overnight, followed by incubation with biotin-conjugated Goat anti-Rabbit secondary antibody (Vector Laboratories) at room temperature for 1 hour. Then, the sections were incubated with the ABC reagent (Vector Laboratories) and visualized with ImmPACT-DAB Peroxidase Substrate (Vector Laboratories). Immunohistochemical staining for Sox2 and p63 was conducted using a Leica Bond-III automated slide stainer. The sections were deparaffinized and subjected to heat induced antigen retrieval with high pH EDTA buffer for 20 minutes and 10 minutes, respectively. The sections were incubated with anti-Sox2 antibody (prediluted; Cellmarque clone SP76) or anti-p63 antibody (prediluted; Biocare Medical, clone 4A4) as per manufacturer’s directions. All slides were counterstained with hematoxylin before being mounting.
Cell proliferation assay
Cells were plated onto sextuplicate wells of a 96-well plate; three identical plates were prepared. Cell proliferation was assayed at 24, 72 and 120 h after plating with alamarBlue Cell Viability Reagent (ThermoFisher) and fluorescence at 585 nm was measured on a Spectra Max3 plate reader (Molecular Device) according to the manufacturer’s protocol at excitation of 555 nm. Cell viability at 72 and 120 h were corrected for the ratio to control cells from the 24-h reading to account for plating unevenness.
Cell cycle analysis
Cells were fixed with 70 % ethanol at least overnight at 4°C. After washing with PBS, cells were incubated in triplicate in PBS containing 100 μg/ml RNase and 50 μg/ml propidium iodide overnight at 4°C overnight. DNA content was analyzed by FACS Canto II (BD Bioscience), and quantitative analyses for the proportions of cells in cell cycle were performed using FlowJo software.
Xenograft model
Engineered LK2 cells (4 × 106 cells) were injected with a 1:1 mixture of 50 μl cell suspension and 50 μl Matrigel (Corning) subcutaneously into both flank regions of 4- to 6-week-old male NOD-scid or NOD-scid gamma mice (Jackson Laboratory). Tumor volume (length × width2 /2) was measured twice a week. Before tumor size reached at 1000 mm3, mice were sacrificed and tumors were immersed in formalin for immunohistological analysis. All animal procedures and studies were approved by the Mount Sinai Institutional Animal Care and Use Committee (IACUC) (protocol number, IACUC-2018-0021).
Phospho-RTK array analysis
The Human Phospho-RTK Array Kit (R&D Systems) was used to determine the relative levels of tyrosine phosphorylation of 42 distinct RTKs, according to the manufacturer’s protocol. 300 μg of total protein was used for each membrane. Chemiluminescent signals were captured with a Chemidoc MP Imaging System (Bio-Rad laboratories) and images were processed using Image Lab software (Bio-Rad laboratories).
Quantitative reverse transcription PCR
For expression analysis, RNA was extracted from cells using the Qiagen RNeasy Mini Kit and cDNA was prepared with the Qiagen QuantiTect Reverse Transcription Kit. Real-time quantitative PCRs were performed in triplicate (Applied Biosystems) on a 7500 Real-Time PCR System (Applied Biosystems) with results normalized to GAPDH expression. Primers are listed in Supplementary Table S2.
Statistical analyses
Differentially enriched super-enhancers were determined using ‘samr’ R package (20), based on cutoffs of fold change>2 and SAM sore>4. For the TCGA-LUSC RNA-seq data, Pearson’s correlation coefficient between gene expression levels was calculated and associated P-value was determined. T-test was performed to identify differentially expressed genes between POU3F2-high/ TP63-low and POU3F2-low/ TP63-high tumors based on cutoffs of fold change>2 and FDR<0.01. Significance of the overlap between gene sets was determined using Fisher’s exact test. To compare the overall survival, Kaplan-Meier curves were plotted and log-rank test was performed. DEseq2 (27) was used to identify the differentially expressed genes between control and DNp63-overexpressed LK2 cells based on cutoffs of fold change>1.5 and FDR<0.05. In addition, likelihood ratio test model was used to identify genes significantly associated with conditions based on cutoffs of fold change>2 and FDR<0.01. For cell proliferation assay, xenograft tumor growth measurement and cell cycle analysis, differences between two groups were examined using two-tailed t-test. Bonferroni corrections were performed in case multiple comparisons were conducted.
Data availability
ChIP-seq and RNA-seq data were deposited in the Gene Expression Omnibus under the accession number GSE137461.
Results
Super-enhancer profiling identifies a novel subtype of lung squamous cell carcinoma
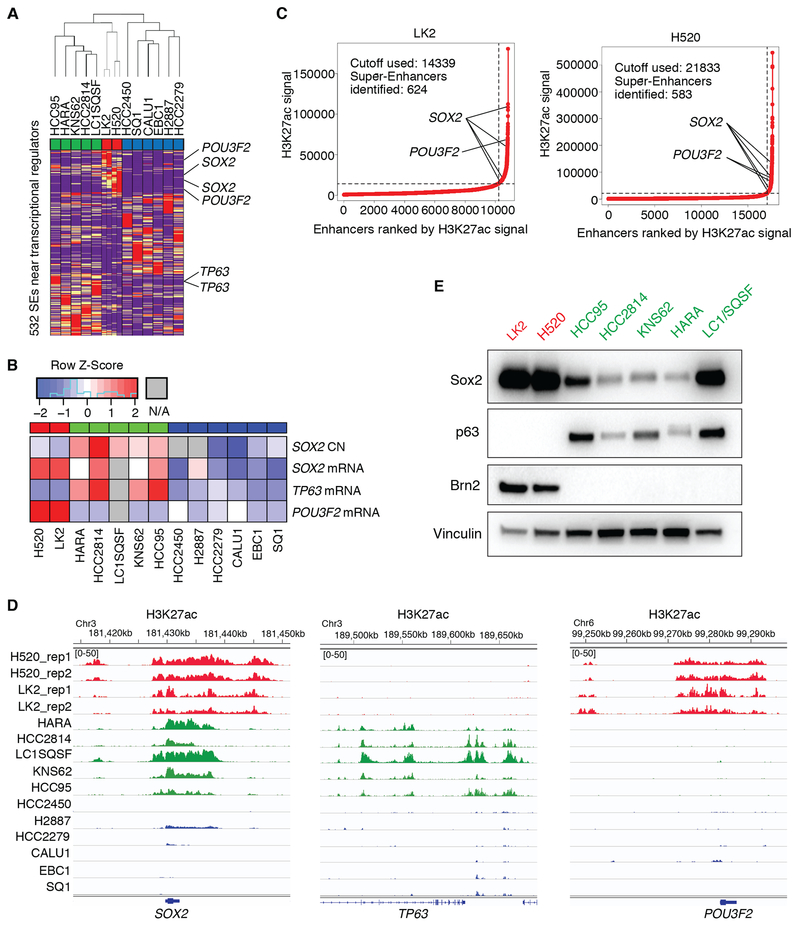
To understand the inter-tumor heterogeneity of cell lineages in LUSC, we examined super-enhancer landscape in a panel of 13 LUSC cell lines. Unsupervised hierarchical clustering of these cell lines using H3K27 acetylation signals over super-enhancer regions near transcriptional regulators identified three subgroups of LUSC (Fig. 1A). Principal component analysis (PCA) using those signals supported this classification (Supplementary Fig. S1A). One of the three subgroups consists of five LUSC cell lines, in which common super-enhancers are observed at genetic loci of 9 transcriptional regulators including SOX2 and TP63 loci (Supplementary Fig. S1B; Supplementary Table S4), consistent with our previous findings that these genes play essential roles as lineage oncogenes in typical LUSCs (8,10). This subgroup is enriched in SOX2-amplified LUSCs with high expression of both SOX2 and TP63 (Fig. 1B). In contrast, LK2 and NCI-H520 cells formed a small subgroup (Fig. 1A; Supplementary Fig. S1A), in which the common super-enhancers with highest signals lie on only two loci at the SOX2 gene and the POU3F2 gene, which encodes Brn2 (Fig. 1C and D; Supplementary Table S5). Brn2 is a well-known lineage factor in neural progenitor cells, particularly in the hypothalamus, where it partners with Sox2 to exert its transcriptional functions (32–34). We also noted that the third subgroup which consists of 5 cell lines shows relatively low expression of both SOX2 and TP63 (Fig. 1B) although human LUSC tumors with low expression (< mean−SD) of both genes accounts for only 6.6 % in the Cancer Genome Atlas Lung Squamous Cell Carcinoma (TCGA-LUSC) dataset. While the SOX2 super-enhancers were shared by the former two subgroups (Fig. 1C and D; Supplementary Fig. S1B) and the TP63 super-enhancers were found only in the first subgroup (Fig. 1D; Supplementary Fig. S1B), POU3F2 super-enhancers were present only in the small subset of LK2 and NCI-H520 (Fig. 1D). We also found the POU3F2 super-enhancers to be among the top differential super-enhancers in this subset compared to the ‘classical’ SOX2/TP63 subgroup (Supplementary Fig. S1C). Analyses using RNA-seq data on these cell lines obtained through Cancer Cell Line Encyclopedia (CCLE) dataset revealed that expression of POU3F2 was indeed significantly higher in this small subset while the squamous lineage factor TP63 was expressed significantly higher in the ‘classical’ SOX2/TP63 subgroup of LUSC (Fig. 1B). By contrast, SOX2 expression was found in both subgroups and highest in the SOX2/POU3F2 subset (Fig. 1B). Immunoblotting showed the expression pattern to be consistent with protein level; the LUSC cell lines that harbor POU3F2 super-enhancers do not express p63 at a detectable level but have high expression of Sox2 and Brn2 (Fig. 1E). These findings suggest that, while SOX2 is highly expressed and likely serves as a lineage factor for both of these two subgroups, this small subgroup represents a unique subset of LUSCs signified by the neural lineage factor Brn2.
Figure 1.
Super-enhancer profiling identifies a novel subtype of LUSC. A, Unsupervised hierarchical clustering of 13 LUSC cell lines using super-enhancer (SE) signals near transcriptional regulator genes. SE signals were normalized to linear scores using the sum of squares of the values with GenePatten. Duplicate data are shown for LK2 and NCI-H520 cells. B, Copy number of SOX2 and mRNA expression of SOX2, TP63 and POU3F2 in LUSC cell lines from CCLE. C, Super-enhancer plots using H3K27ac scores in the small subset of LUSC cell lines; LK2 cells (left) and NCI-H520 cells (right). D, Genome view tracks of H3K27ac signal at loci of SOX2 (left), TP63 (middle) and POU3F2 (right) in LUSC cell lines. Normalized signals are shown in a range from 0 to 50 for all the tracks. E, Protein expression of Brn2, p63, Sox2 and vinculin as a loading control in LUSC cell lines.
BRN2 signifies the ‘neural’ subtype in human LUSCs
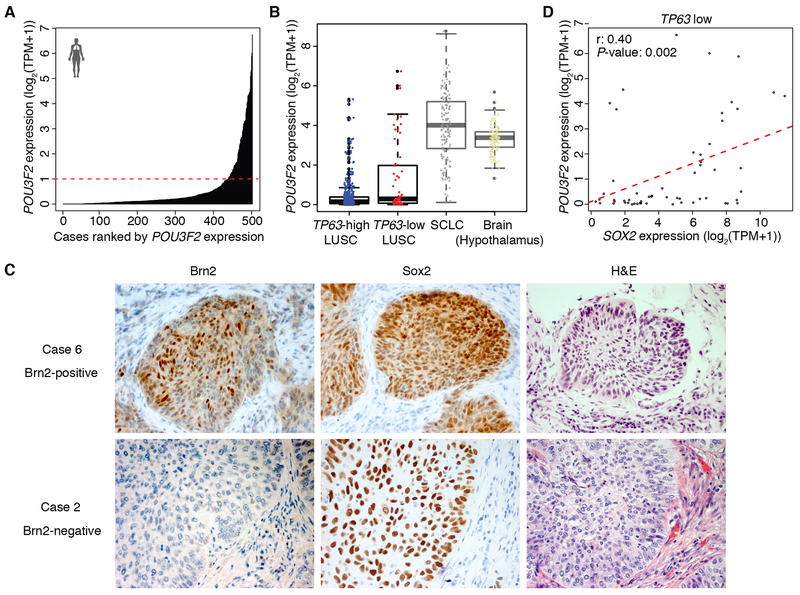
Brn2 has been described to have an oncogenic role in neuroendocrine tumors such as small cell lung cancer (SCLC), neuroendocrine prostate cancer and glioblastoma (35–39). To determine whether this unique ‘neural’ subset exists in human LUSC data and POU3F2 (Brn2) signifies this subgroup, we examined TCGA-LUSC dataset and identified a subset of LUSC that expresses POU3F2 (Fig. 2A) and their expression levels were comparable to that in SCLC (24) and hypothalamus in GTEx data (23) (Fig. 2B), suggesting functional relevancy of Brn2 in this unique subset of LUSC. Of note, this ‘neural’ subset of LUSC has very low expression of neuroendocrine markers (Supplementary Fig. S2A and S2B), suggesting this subtype is not necessarily closer to SCLC or neuroendocrine tumors. In order to investigate how these tumors with Brn2 expression are represented in clinical specimens, we examined Brn2 expression pattern at protein level in human LUSC tissues by immunohistochemical staining. Brn2 expression was found in 6 out of 23 specimens and all of them were positive for Sox2 (Fig. 2C; Supplementary Fig. S2C). Of note, the Brn2 expression was relatively confined to subpopulations of tumor cells in these LUSC tissues. This suggests that some of the tumors with intermediate POU3F2 expression level observed in bulk mRNA in the TCGA-LUSC dataset may reflect heterogeneous tumor populations with partial ‘neural’ differentiation and that, perhaps unlike cell lines, distinct lineage states can coexist within the same LUSC tumor. This further implies trans-differentiation during the evolution of LUSCs. We next examined the correlation of expression of POU3F2 with SOX2 and TP63 expression in the TCGA-LUSC dataset. We found that expression of POU3F2 was anti-correlated with that of TP63 (Fig. 2B; Supplementary Fig. S2D), consistent with the data from LUSC cell lines in which high expression of POU3F2 and TP63 were mutually exclusive. We did not find significant correlation between POU3F2 and SOX2 expression (Supplementary Fig. S2E) in the entire dataset; however, when we focused on tumors with low TP63 expression, we found a significant correlation of POU3F2 with SOX2 (Fig. 2D) whereas there was a significant correlation of TP63 with SOX2 among LUSCs with high TP63 expression (Supplementary Fig. S2F). These findings suggest that Brn2 and p63 have contrasting roles representing distinct lineages within LUSC by partnering with Sox2 to elicit distinct transcriptional outputs.
Figure 2.
Brn2 is expressed in a subset of human primary LUSC tumors. A, Expression of POU3F2 in 501 TCGA LUSC tumor tissues. The red dashed line (TPM=1) shows the cutoff to separate samples into POU3F2 high and low samples. B, Box plots of POU3F2 expression in TP63 high and low LUSC tumors from TCGA, SCLC tumors from publicly available datasets and normal brain tissues (hypothalamus) from GTEx. C, Immunohistochemical staining of Brn2 and Sox2 and H&E staining in Brn2-positive and negative human LUSC tumors. Representative images are shown (original images, ×400). D, Scatter plots of expression of SOX2 and POU3F2 in TP63-low LUSC tumors from TCGA.
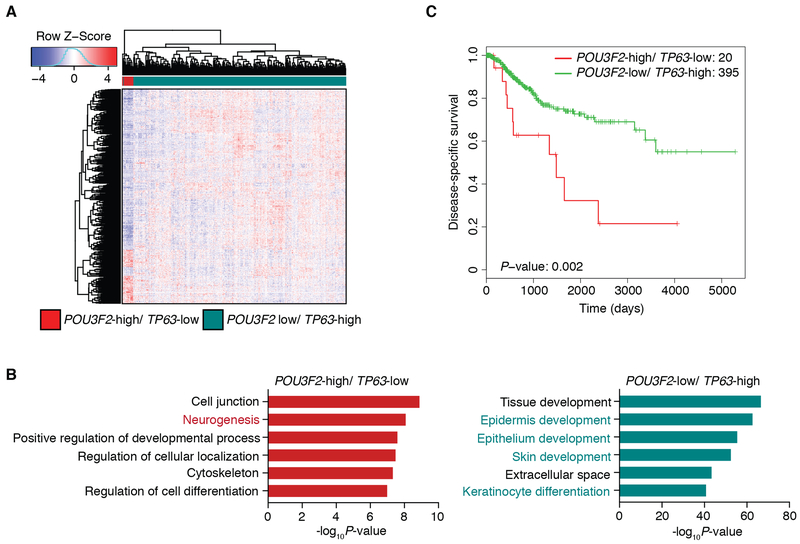
To investigate the difference in gene signatures between POU3F2-expressing LUSC tumors and TP63-expressing LUSC tumors, we next performed differential expression analysis in the TCGA-LUSC dataset comparing POU3F2-high/ TP63-low tumors vs. TP63-high/ POU3F2-low tumors (Fig. 3A). A functional enrichment analysis of differentially expressed genes identified genes which play roles in neural cell differentiation significantly enriched in POU3F2-high/ TP63-low tumors while those involved in epithelial/ epidermal cell differentiation were significantly enriched in TP63-high/ POU3F2-low tumors (Fig. 3B). Further, we evaluated the prognostic significance of TP63/ POU3F2 expression status in the TCGA-LUSC dataset and found that patients with POU3F2-high/ TP63-low tumors showed significantly shorter survival compared to those with TP63-high/ POU3F2-low tumors, suggesting clinically distinct, aggressive features of this ‘neural’ subtype (Fig. 3C). When we examined the relationship between these POU3F2-high/ TP63-low tumors and the LUSC expression subtypes (11), consistent with the report in which the ‘neural’ LK2 and NCI-H520 cells were predicted as primitive (12), all of the POU3F2-high/ TP63-low tumors were classified as the primitive subtype (Supplementary Fig. S2G), which was reported to show worse prognosis among LUSC (11). These findings indicate the presence of a distinct LUSC subset with neural differentiation that has not been described previously.
Figure 3.
Brn2 signifies the ‘neural’ subtype of LUSC in human primary LUSC tumors. A, Heatmap showing hierarchical clustering of LUSC tumors from TCGA using differentially expressed genes between POU3F2-high/ TP63-low (n=20) and POU3F2-low/ TP63-high tumors (n=396). With cutoffs of fold change>2 and FDR<0.01, 196 genes are differentially up-regulated in the POU3F2-high/ TP63-low tumors and 735 genes are differentially up-regulated in the POU3F2-low/ TP63-high tumors. Color scheme represents Z-score distribution. B, Gene ontology analyses for the differentially up-regulated genes in POU3F2-high/ TP63-low tumors (left) and for those in POU3F2-low/ TP63-high tumors (right). Enriched functions for these genes are identified based on Fisher’s exact test against GO terms curated in MSigDB. C, Kaplan-Meier curves plotted to compare the disease-specific survival of POU3F2-high/ TP63-low (n=20) versus POU3F2-low/ TP63-high (n=395) LUSC tumors from TCGA. Log-rank test P-value=0.002.
BRN2 and SOX2 interact and co-localize at genetic loci in the ‘neural’ LUSC cells
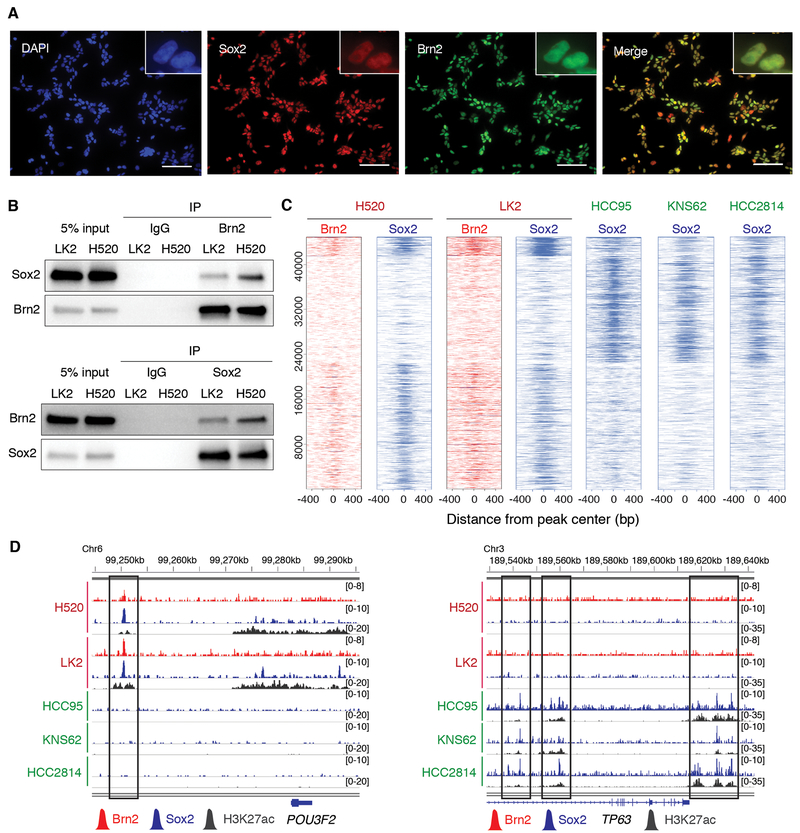
Given the positive correlation of POU3F2 and SOX2 expressions in LUSC tumors with low TP63 expression, and previously reported interaction of Brn2 and Sox2 in neural progenitor cells (33,34), we further investigated the relationship between Brn2 and Sox2 in LUSC. First, using immunofluorescence analysis we found that Sox2 and Brn2 co-localize in the nuclei of the Brn2-positive LUSC cell lines (Fig. 4A; Supplementary Fig. S3A). In contrast, Sox2 but not Brn2 was detected in nuclei of the ‘classical’ Sox2/p63-positive LUSC cells confirming the specificity of the antibody (Supplementary Fig. S3B). Second, we observed their robust protein-protein interaction by endogenous and reciprocal co-immunoprecipitations in the two ‘neural’ LUSC cell lines (Fig. 4B). Given their physical interaction, we hypothesized that Brn2 and Sox2 co-occupy genomic loci to coordinately regulate gene expression in these Brn2-positive LUSC cells. To test for genomic co-occupancy of Brn2 and Sox2, we explored genome-wide binding profiles of Brn2 and Sox2 in the ‘neural’ LUSC cells by ChIP-seq and found that the Brn2 binding peaks substantially overlap with the peaks for Sox2 in these cells (Fig. 4C and D). In contrast, the Sox2 binding peaks from ‘classical’ LUSC cells showed distinct profiles with less overlaps with those for Brn2 in ‘neural’ LUSC cells (Fig. 4C and D), suggesting that in the absence of p63, Brn2 engages Sox2 to localize at genetic loci thereby imparting ‘neural’ features on LUSC cells. To investigate which other factors are involved in collaboration of Sox2 and Brn2 at genetic loci in the ‘neural’ cells, we performed motif analysis for the Sox2/ Brn2 peak-enriched cluster (Supplementary Fig. S3C). As well as SOX family and POU family motifs as expected, we found TCF family, RUNX family, FOX family and AP-1 motifs. Of note, TCF4 super-enhancer was called specifically in the ‘neural’ LUSC subset. Substantial investigation is required to understand if any of these factors contribute to establishing their ‘neural’ differentiation state.
Figure 4.
Brn2 and Sox2 interact and co-localize at genetic loci in the ‘neural’ subset of LUSC. A, Expression of endogenous Brn2 and Sox2 in LK2 cells, determined by immunofluorescence with anti-Sox2 (green) and anti-Brn2 (red) antibodies, respectively. DAPI staining (nuclei; blue) and merged images are also shown. Original magnification, ×200. Scale bar, 100 μm. B, Sox2-Brn2 interaction, shown by co-immunoprecipitation of Sox2 using an antibody against endogenous Brn2 (top) and co-immunoprecipitation of Brn2 using an antibody against endogenous Sox2 (bottom) in LK2 and NCI-H520 cells. C, Heatmap depicting global analysis of ChIP-seq signals for Brn2 and Sox2 in ‘neural’ NCI-H520 and LK2 cells and those for Sox2 in ‘classical’ HCC95, KNS62 and HCC2814 cells at all the peak loci. ChIP-seq signal intensity is shown by color shading. D, Genome view tracks of Brn2, Sox2 and H3K27ac ChIP-seq signals in NCI-H520 and LK2 cells, and Sox2 and H3K27ac ChIP signals in HCC95, KNS62 and HCC2814 cells at loci of POU3F2 (left) and TP63 (right).
p63 suppresses Brn2 expression and induces a classical-squamous tumor state
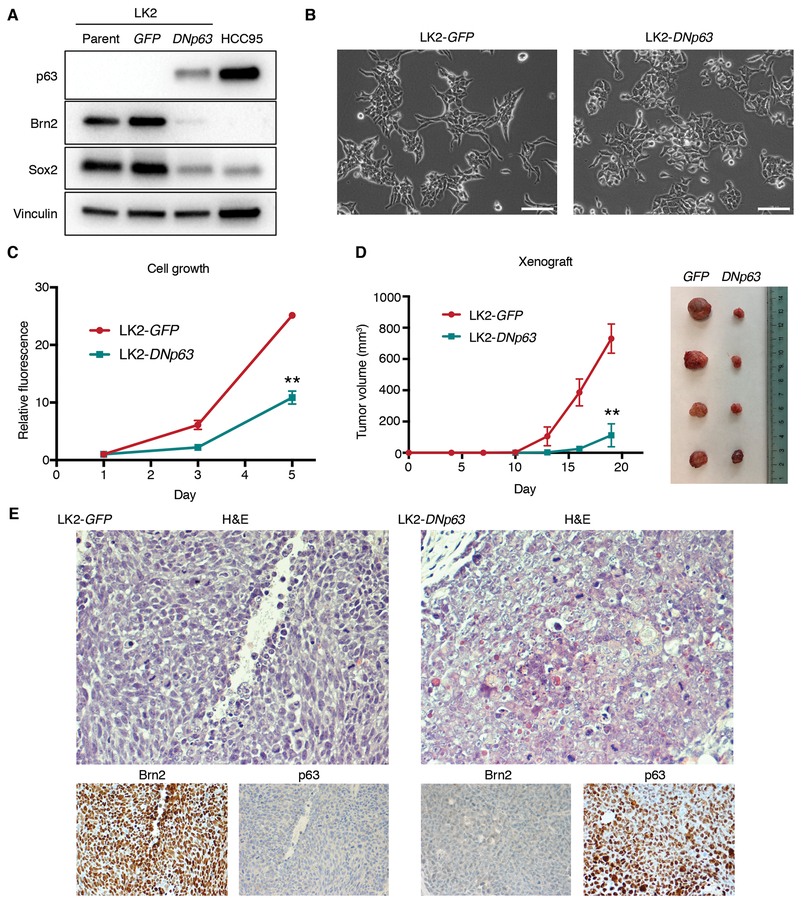
We previously reported that p63 and Sox2 physically interact and exhibit overlapping genomic occupancy, which suggest their cooperative transcriptional programs in ‘classical’ LUSCs (10). The interaction of Brn2, in place of p63, with Sox2 that we observed in this ‘neural’ subset implies counteractive functions of Brn2 and p63 in lineage determination. To date, however, the role of the balance between these two factors in differentiation of squamous tissue, or in any lineages, has not been described. Therefore, to determine whether p63 has any effects on Brn2 or vice versa, we first overexpressed deltaNp63-alpha (DNp63), the predominant isoform in squamous lineages including the normal squamous basal layers (40) and squamous cell cancers (10) (Supplementary Fig. S4A), in the ‘neural’ LUSC cells (Fig. 5A; Supplementary Fig. S4B and S4C). Introduction of DNp63 substantially suppressed Brn2 expression, while it also suppressed expression of Sox2 to a lesser extent in these cells (Fig. 5A; Supplementary Fig. S4B and S4C). Notably, DNp63 overexpression led to morphological changes in these ‘neural’ LUSC cells from small and/or elongated morphology to larger and cuboidal morphology in conventional 2-dimensional culture (Fig. 5B; Supplementary Fig. S4D) and decreased cell proliferation (Fig. 5C; Supplementary Fig. S4E). To confirm whether these observed changes in vitro reflect a shift in differentiation status, we next investigated the effects of DNp63 overexpression on the ‘neural’ LUSC in vivo. In the xenograft model, the tumor growth rates were significantly decreased in DNp63-overexpressed LUSC xenografts compared to control ‘neural’ LUSC (Fig. 5D). In addition, Brn2 expression was suppressed in p63-positive tumor cells (Fig. 5E; Supplementary Fig. S4F and S4G). Consistent with our finding in vitro, DNp63 expression downregulated expression of Sox2 (Supplementary Fig. S4F, S4G and S4H) presumably reflecting the squamous epithelial differentiation induced by p63 (41,42). Notably, patches of p63-negative tumor cells in DNp63-overexpressed xenograft tumor shows reduced expression of Brn2 and Sox2 (Supplementary Fig. S4F and S4G), likely due to imperfect selection or heterogeneous ectopic expression of DNp63, exhibiting a clear effect of DNp63 on suppressing Brn2. Morphologically, p63-positive tumor cells showed open chromatin, larger nucleoli, and on average, had a polygonal shape and more cytoplasm indicative of more classical squamous cell carcinoma histology while p63-negative/ Brn2-positive cells have a slightly oval shape, less cytoplasm, denser chromatin and less frequent/smaller nucleoli (Fig. 5E; Supplementary Fig. S4G). These results suggest that DNp63 counteracts with Brn2 and induces a different tumor state characterized by more squamous/epithelial features of the cells.
Figure 5.
DNp63 overexpression in the ‘neural’ LUSC cells suppresses Brn2 expression and induces phenotypic changes. A, Protein expression of p63, Brn2, Sox2 and vinculin as a loading control in parental, GFP-overexpressed and DNp63-overexpressed LK2 cells. B, Phase-contrast microphotographs of GFP-overexpressed and DNp63-overexpressed LK2 cells. Bar = 100 μm. C, Cell growth of GFP-overexpressed and DNp63-overexpressed LK2 cells. Mean ± SD of sextuplicates are shown. **, P<0.001 vs. GFP-overexpressed LK2 cells, t-test. D, Tumor growth of GFP-overexpressed and DNp63-overexpressed LK2 cells in vivo. Mean ± SD of tetraplicates are shown. **, P<0.001 vs. GFP-overexpressed xenografts, t-test. Xenograft tumors resected 19 days after inoculation are shown in the right picture. E, H&E staining and immunohistochemical staining of Brn2 and p63 in GFP-overexpressed or DNp63-overexpressed LK2 xenograft. Original Images, ×400.
On the other hand, overexpression of Brn2 in p63-positive ‘classical’ LUSC cells did not lead to suppression of p63 while modestly increased expression of Sox2, indicating non-reciprocal regulatory roles of Brn2 and p63, and regulation of Sox2 by Brn2 (Supplementary Fig. S5A and S5B). In addition, by reciprocal co-immunoprecipitations of Sox2 and p63, Brn2 overexpression in the ‘classical’ LUSC cells did not decrease the amount of Sox2 or p63 as a co-immunoprecipitated protein (Supplementary Fig. S5C), suggesting p63 might be a dominant interactor of Sox2 in those cells. To further investigate the role of Brn2 in the ‘neural’ LUSC cells, we ablated expression of Brn2 in these cells by CRISPR-Cas9 mediated deletion of the POU3F2 gene (Supplementary Fig. S5D). Brn2 ablation led to decreased cell proliferation in ‘neural’ LK2 cells (Supplementary Fig. S5E), suggesting its role in maintaining the lineage state; however, it did not lead to changes in p63 expression (Supplementary Fig. S5D), again suggesting that p63 expression is not under the regulation of Brn2. Although Brn2 overexpression led to increased Sox2 expression, Brn2 ablation did not result in suppression of Sox2 (Supplementary Fig. S5D). Instead, p63 ablation increased expression of Sox2 (Supplementary Fig. S5F), suggesting negative regulation of Sox2 by p63, consistent with the findings that p63-high LUSC cells have lower expression of Sox2 than the Brn2-positive subset (Fig. 1E) and DNp63 induction suppressed SOX2 expression in the same subset (Fig. 5A; Supplementary Fig. S4B and S4C).
DNp63 changes the transcriptional program of the ‘neural’ LUSC cells
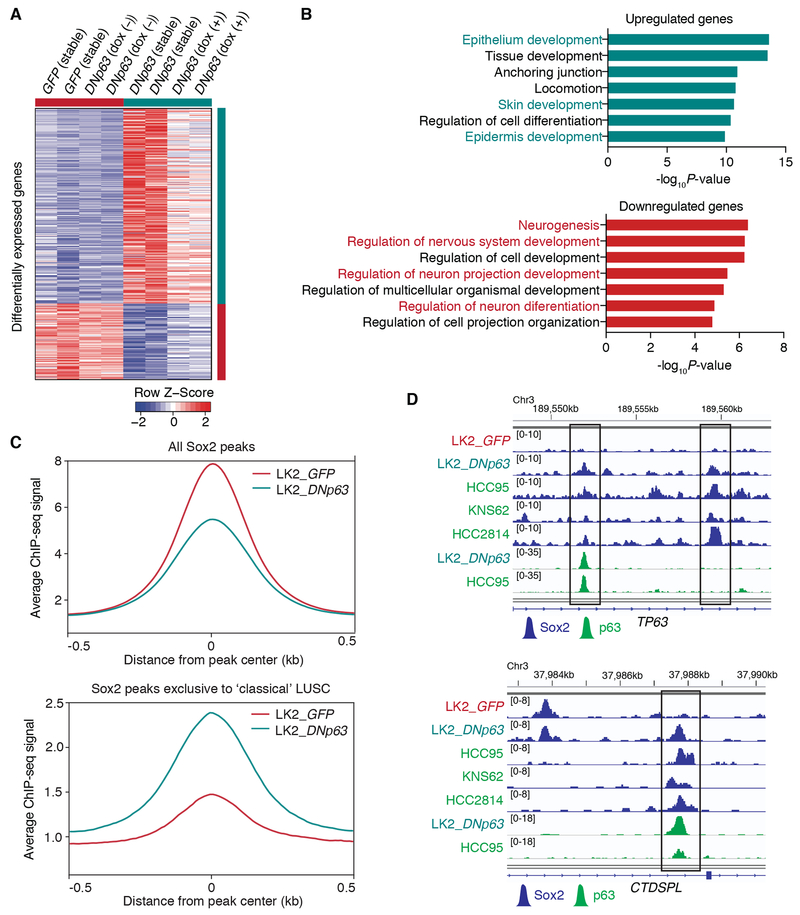
To investigate how DNp63 changed transcriptional programs leading to the phenotypical changes in ‘neural’ LK2 cells, we performed transcriptomic profiling on these engineered cells (Fig. 6A). We found that genes which play roles in neuronal functions were enriched in down-regulated genes while genes involved in epithelial/ epidermal cell development were enriched in up-regulated genes (Fig. 6B). To see whether introduction of DNp63 results in transcriptomic remodeling toward classical LUSC, we compared the transcriptomic profiles of the engineered LK2 cells with those of classical LUSC cells. Principal component analysis among these cells (Supplementary Fig. S6A) showed that DNp63-overexpressed LK2 cells were indeed closer to the classical LUSC cells compared to the control LK2 cells, or located between the control LK2 cells and the classical LUSC cells. When we considered the levels of DNp63 expression depending on the experimental models for the gene ontology analyses, in addition to neuronal genes, those involved in cell cycle/ DNA replication were also enriched in down-regulated genes (Supplementary Fig. S6B). Consistent with this finding, higher proportion of cells in the S phase of cell cycle was found in control LK2 cells compared to the DNp63-overexpressed cells (Supplementary Fig. S6C and S6D). Altered cell-cycle dynamics might be a simply reflection of faster cell proliferation but it is also plausible that it is associated with neuronal differentiation in these ‘neural’ LUSC cells, which needs further investigation (43–45). Of note, we found strong associations (OR=9.81 and 6.17 for up- and down-regulated genes, respectively) between gene signatures in this model and those from human TCGA-LUSC tumors based on POU3F2/ TP63 expression (Fig. 3A; Supplementary Table S6), suggesting biological relevancy of our model in the context of human LUSCs.
Figure 6.
DNp63 induces a classical squamous-cell transcriptional program in the ‘neural’ LUSC cells. A, Heatmap showing 399 differentially expressed genes (287 up-regulated and 112 down-regulated genes) between control LK2 cells (stable GFP-overexpressed cells and doxycycline-inducible DNp63-overexpressing cells without doxycycline in duplicate) and DNp63-overexpressed LK2 cells (stable DNp63-overexpressed cells and doxycycline-inducible DNp63-overexpressing cells with 2 μg/ml doxycycline in duplicate), sorted by fold change. Color scheme represents Z-score distribution. B, Gene ontology analyses for the differentially up-regulated (top) and down-regulated (bottom) genes upon DNp63 overexpression in LK2 cells. Enriched functions for these genes are identified based on Fisher’s exact test against GO terms curated in MSigDB. C, Average ChIP-seq signals for all Sox2 peaks (top) and Sox2 peaks exclusively found in ‘classical’ HCC95, KNS62 and HCC2814 cells (bottom) in GFP-overexpressed and DNp63-overexpressed LK2 cells. D, Genome view tracks of Sox2 ChIP-seq signals in GFP-overexpressed and DNp63-overexpressed LK2 cells, HCC95, KNS62 and HCC2814 cells and p63 ChIP signals in DNp63-overexpressed LK2 cells and HCC95 cells at loci of TP63 (top) and CTDSPL (bottom).
When we examined whether DNp63 introduction resulted in reduced super-enhancer activities specific to the ‘neural’ LUSC cells, we found H3K27ac signals indeed decreased in 5 out of 7 common super-enhancers exclusive to the ‘neural’ LUSC lines including POU3F2 and SOX2 super-enhancers (Supplementary Fig. S7A). We also confirmed expression levels of the genes at these loci significantly decreased upon it (Supplementary Fig. S7B), suggesting DNp63 contributes to suppress the original lineage program of the ‘neural’ LUSC cells.
Next, to investigate whether DNp63 reconstitutes Sox2-binding profile toward classical LUSC, we profiled genomic occupancy of Sox2 in the DNp63-overexpressed cells and compared to the control ‘neural’ LK2 cells. We found that while average ChIP-seq signals of all the Sox2 peaks decreased in the DNp63-overexpressed cells compared to the control LK2 cells, the DNp63-overexpressed cells showed increased signals near the Sox2 peaks exclusive to the ‘classical’ LUSC lines (Fig. 6C), suggesting that presence of DNp63 re-engages Sox2 cistrome in the ‘classical’ LUSC cells specific Sox2-binding regions (Fig. 6D). Furthermore, we found p63 binding profile formed in the DNp63-overexpressed cells similar to that in ‘classical’ HCC95 cells including overlaps with Sox2 binding peaks from these cells (Fig. 6D; Supplementary Fig. S7C). These findings suggest that DNp63 overrides Sox2’s transcriptional programs in the ‘neural’ state to redefine it into ‘classical’ squamous-cell state via superseding Brn2 as a Sox2 partner.
DNp63 changes activities of ErbB family signaling in the ‘neural’ LUSC cells
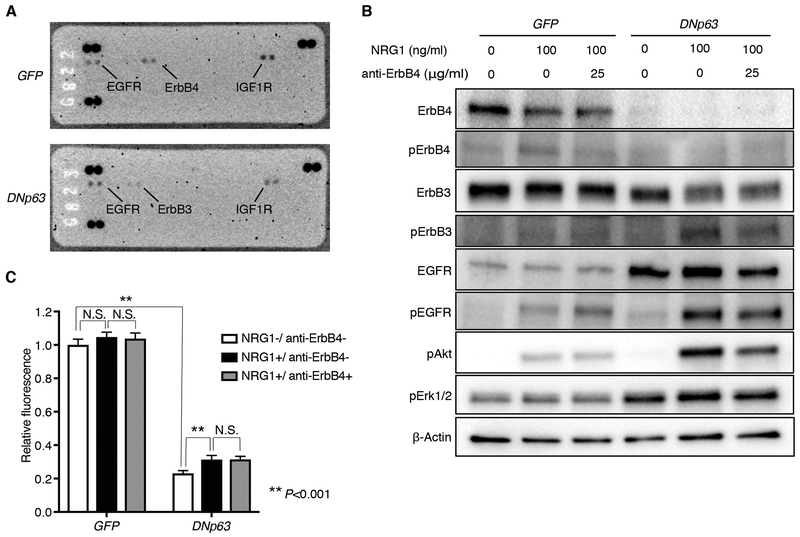
Distinct patterns of cell signaling landscape including receptor tyrosine kinase (RTK) activity are associated with cell differentiation states. To investigate whether DNp63-overexpression induces a differential signaling activity profile in the ‘neural’ LUSC cells, we assayed for human phospho-RTK array on these cells (Fig. 7A). At the original state in the control cells, we detected ErbB4 phosphorylation along with activation of EGFR and IGF1R. In contrast, ErbB4 phosphorylation was diminished and instead higher signals of ErbB3 phosphorylation emerged in the DNp63-overexpressed cells.
Figure 7.
DNp63 alters ErbB family signaling profile in the ‘neural’ LK2 cells. A, Human phospho-receptor tyrosine kinase array was performed for GFP-overexpressed and DNp63-overexpressed LK2 cells. B, Protein expression of ErbB4, phospho-ErbB4, ErbB3, phospho-ErbB3, EGFR, phospho-EGFR, phospho-Akt and phospho-Erk1/2 and β-Actin as a loading control in GFP-overexpressed and DNp63-overexpressed LK2 cells. Cells were serum-starved for 4 hours, and then incubated with anti-ErbB4 monoclonal antibody at the indicated concentrations for 15 minutes, followed by stimulation with NRG1 for 30 minutes at the indicated concentrations. C, Cell proliferation of GFP-overexpressed and DNp63-overexpressed LK2 cells. Cells were cultured with 5% FBS and with or without NRG-1 (100 ng/ml) and anti-ErbB4 monoclonal antibody (25 μg/ml) for 4 days. Mean ± SD of sextuplicates are shown. **, P<0.001 and N.S., not significant (P>0.05), t-test with Bonferroni correction.
We confirmed this switch in activity between ErbB family members by immunoblotting and further examined the difference in ErbB family signaling with the use of recombinant Neuregulin-1 (NRG1), a ligand for ErbB3 and ErbB4, as well as a neutralizing antibody directed against ErbB4 (Fig. 7B; Supplementary Fig. S8A). This line of experiments revealed that diminished phosphorylation of ErbB4 upon DNp63 overexpression was a consequence of suppressed ErbB4 protein level. In contrast, expression levels of ErbB3 are similar between DNp63 overexpressed and control cell lines while its activity was significantly increased upon DNp63 overexpression under the same growth condition (10% FBS) (Supplementary Fig. S8A), suggesting that DNp63 expression led to altered expression levels of its ligands or expression/activity of its heterodimeric partners and/or interacting signaling molecules. Indeed, we observed increased protein level of EGFR upon DNp63 overexpression, and in response to NRG1 stimulation, phosphorylation of ErbB3 and EGFR increased to even higher level in the DNp63-overexpressed cells (Fig. 7B; Supplementary Fig. S8A), suggesting that increased EGFR protein level and their homo/hetero-dimerization led to increased phosphorylation of ErbB3 and EGFR. We also found that p63 binds to the EGFR locus in the DNp63-overexpressed cells suggesting a direct regulation of EGFR gene by p63 at the transcriptional level (Supplementary Fig. S8B).
To determine which major downstream signal pathway mediates ErbB3/EGFR activation, we examined phosphorylation levels of Akt and Erk1/2 in these cells. We observed increased Akt phosphorylation levels corresponding to phosphorylation levels of ErbB3 and EGFR in response to NRG1 stimulation (Fig. 7B; Supplementary Fig. S8A). In contrast, Erk1/2 phosphorylation levels were higher in the DNp63-overexpressed cells, but did not change after NRG1 stimulation, suggesting that the activation of MAPK-ERK pathway by DNp63 overexpression was independent from the phosphorylation of ErbB3 and EGFR. We next examined response to NRG1 stimulation on cell growth in these cells and found significantly higher response in the DNp63-overexpressed cells compared to the control cells (Fig. 7C). These data suggest that the ‘neural’ LUSC cells are less dependent on ErbB/Akt and MAPK-ERK pathways compared to the more epithelial cell differentiation state induced by DNp63, and further imply different therapeutic strategies could be applied by targeting different molecules depending on the cell differentiation status.
Discussion
In this report, we identified a novel subtype of LUSC characterized by the neural transcription factor Brn2. We examined super-enhancer landscape of LUSC cell lines to reveal the heterogeneity of transcriptional programs in LUSC and identified three subgroups of LUSC based on the super-enhancer profiles. Notably, super-enhancers on the locus of the SOX2 gene are shared by two subgroups. We previously reported that SOX2 is the most commonly amplified gene in LUSC and its expression is essential for cell survival (8). Following our study, it has been confirmed that Sox2 promotes lung squamous cancer lineage in mice model (46,47), supporting its important role as a lineage oncogene. However, in one subgroup of LUSC, SOX2 super-enhancers were not found and its expression was low, suggesting that Sox2 does not necessarily govern transcriptional programs among all LUSCs. While this subset is not represented as a significant proportion of primary human LUSC in TCGA cohort, it remains to be seen which lineage state this low Sox2 LUSC subset represents and which transcriptional program may be active. In addition, super-enhancers on the locus of the TP63 gene, which we also identified as a collaborating lineage factor with Sox2 in LUSC, were only found in one ‘classical’ subgroup of LUSC, suggesting that in the other subgroups, other factors play roles in controlling their lineage states. In fact, our super-enhancer profiling found that super-enhancers on the loci of SOX2 and POU3F2 instead of TP63 were commonly shared in a small subset of LUSC. Brn2, encoded by POU3F2, is a Class III POU transcription factor which play an essential role in neural cell differentiation (48–51) and is highly expressed in glioblastoma, neuroendocrine SCLC and neuroendocrine prostate cancer (36–38). This subset exhibited gene expression patterns associated with neural development compared to the ‘classical’ subtype, suggesting the differentiation states of the subset is closer to neural lineages. This subset is not categorized as a neuroendocrine lung cancer which also commonly expresses Brn2 and is close to neural lineages. We speculate that this unique LUSC subset can have a potential to show neuroendocrine phenotypes possibly with additional factors implicated for neuroendocrine lineages such as Rb1 and Ascl1, which warrants further investigation.
In this study, we highlighted the partnership of Sox2 and Brn2 in this unique ‘neural’ subset of LUSC for regulating their transcriptional programs. Sox2 has been described to have a role in determining cell differentiation states by cooperating with other lineage factors such as Oct4 in embryonic stem cells (52), Pax6 in eye lens development (53), p63 in squamous lineages (10) and Brn2 in neural progenitor cells (33,34). Our data support distinct contributions of Sox2 across multiple cell types and confirmed that cooperation of Sox2 and Brn2 in neural progenitor cells is conserved in the ‘neural’ LUSC cells, which has not been described previously.
Prior studies including ours have reported a role of p63 as a lineage oncogene in defining squamous lineages (10,54,55). To support and extend those findings, our study highlighted the critical role of p63 antagonizing Brn2 in defining more classical squamous cell states within the context of LUSC. Our findings suggest that switching the Sox2 collaborating partner from Brn2 to p63 contributes a shift in cell differentiation states in LUSC via dramatically changing Sox2 cistrome. This phenomenon also supports a role of Sox2 in determining cell differentiation states by having specific partners is maintained in malignant cells. As this switch has not been described in normal differentiation or regeneration of lung cells, whether this transition from Brn2 to p63 is unique to LUSC remains to be investigated. Of note, p63 overexpression in the Brn2-positive ‘neural’ LUSC cells decreased cell growth both in vitro and in vivo. It suggests that rather than playing a role as a general oncogene, p63 defines more squamous differentiation states and the lineage program of this Brn2-positive ‘neural’ LUSC could lead to more aggressive phenotypes as cancer cells compared to that of the classical LUSC defined by p63. Our study raised a hypothesis that a loss of p63 in the classical LUSC can unveil a latent neural state that is represented by the novel Brn2-high subtype. However, our classical LUSC cell lines were dependent on p63 for their survival and p63 knockout did not induce Brn2 expression in a short period of time. Further studies are needed to explore this hypothesis.
We also observed a change of RTK signaling activity profile through a shift in cell differentiation states induced by p63. It has been reported that p63 regulates a various signaling pathway molecules depending on cell contexts such as β-catenin, EGFR and Jagged1 in human airway epithelial basal cells (56), FGFR2 in murine squamous cell carcinoma model (57) and NRG1 in mammary basal cells (58). In our study, we identified several RTKs whose expression and phosphorylation level are altered after p63 overexpression. While functions of positive ErbB4 signaling in the ‘neural’ LUSC cells are unclear, the p63-overexpressed LUSC cells showed more responses to ErbB3/EGFR pathway stimulation and high ERK activity even without stimulation. It is possible that the ‘neural’ LUSC cells are less dependent on AKT and MAPK pathways similarly to SCLC. Future studies are needed to investigate the relationships between lineage programs and networks of signaling pathways and this would help us making therapeutic strategies for each lineage cancer program.
Taken together, we identified a novel ‘neural’ lineage signified by Sox2 and Brn2 in LUSC by investigating its super-enhancer landscape. Brn2 cooperates with Sox2 in determining its transcriptional program, which is overwhelmed and reprogrammed by p63, the classical squamous partner of Sox2. Characterization of each cancer lineage to identify its unique vulnerabilities could lead to a novel approach to make therapeutic strategies for this heterogeneous disease.
Supplementary Material
Statement of Significance.
Epigenomic profiling reveals a novel subtype of lung squamous cell carcinoma with neural differentiation.
Acknowledgements
We thank Chiara Vardabasso, Dan Hasson, Katsutoshi Sato, Aleksandra Wroblewska, Almuneda Bosch and Iman Tavassoly for helpful discussions; Aleksandra Wroblewska and Brian D. Brown for providing a lentiviral vector for cloning sgRNAs; NextSeq Sequencing Facility of the Department of Oncological Sciences at Icahn School of Medicine at Mount Sinai (ISMMS), Saboor Hekmaty, Irene Salib, Gayatri Panda and Ravi Sachidanandam for sequencing assistance; and Stephanie Tuminello and Michael William for technical assistance. The authors also thank the Flow Cytometry Core facility and the Biorepository and Pathology Core Facility at ISMMS. This work was supported in part through Tisch Cancer Institute at ISMMS and the computational resources and staff expertise provided by Scientific Computing at ISMMS.
Grant Support: This study was supported by the 2017 ATS Foundation Unrestricted Grant: Pulmonary (H. Watanabe), the American Lung Association of the Northeast (H. Watanabe) and NIH R01CA240342 (H. Watanabe). T. Sato is supported by the Japanese Respiratory Society the 6th Lilly Oncology Fellowship Program and the Uehara Memorial Foundation. R. Kong is supported by Shaanxi Provincial Natural Science Foundation, China (No. 2017JM8046). C.A. Powell and H. Watanabe are supported by NIH (R01CA163772).
Footnotes
Disclosure of Potential Conflicts of Interests
The authors declare no potential conflicts of interest.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- 3.Cheng TY, Cramb SM, Baade PD, Youlden DR, Nwogu C, Reid ME. The International Epidemiology of Lung Cancer: Latest Trends, Disparities, and Tumor Characteristics. J Thorac Oncol 2016;11(10):1653–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Islami F, Torre LA, Jemal A. Global trends of lung cancer mortality and smoking prevalence. Transl Lung Cancer Res 2015;4(4):327–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489(7417):519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brahmer J, Reckamp KL, Baas P, Crino L, Eberhardt WE, Poddubskaya E, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 2015;373(2):123–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herbst RS, Baas P, Kim DW, Felip E, Perez-Gracia JL, Han JY, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387(10027):1540–50. [DOI] [PubMed] [Google Scholar]
- 8.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 2009;41(11):1238–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Que J, Okubo T, Goldenring JR, Nam KT, Kurotani R, Morrisey EE, et al. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development 2007;134(13):2521–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe H, Ma Q, Peng S, Adelmant G, Swain D, Song W, et al. SOX2 and p63 colocalize at genetic loci in squamous cell carcinomas. J Clin Invest 2014;124(4):1636–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilkerson MD, Yin X, Hoadley KA, Liu Y, Hayward MC, Cabanski CR, et al. Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin Cancer Res 2010;16(19):4864–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu D, Pang Y, Wilkerson MD, Wang D, Hammerman PS, Liu JS. Gene-expression data integration to squamous cell lung cancer subtypes reveals drug sensitivity. Br J Cancer 2013;109(6):1599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell 2013;155(4):934–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pott S, Lieb JD. What are super-enhancers? Nat Genet 2015;47(1):8–12. [DOI] [PubMed] [Google Scholar]
- 15.Boeva V, Louis-Brennetot C, Peltier A, Durand S, Pierre-Eugene C, Raynal V, et al. Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat Genet 2017;49(9):1408–13. [DOI] [PubMed] [Google Scholar]
- 16.McKeown MR, Corces MR, Eaton ML, Fiore C, Lee E, Lopez JT, et al. Superenhancer Analysis Defines Novel Epigenomic Subtypes of Non-APL AML, Including an RARalpha Dependency Targetable by SY-1425, a Potent and Selective RARalpha Agonist. Cancer Discov 2017;7(10):1136–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008;9(9):R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin CY, Erkek S, Tong Y, Yin L, Federation AJ, Zapatka M, et al. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature 2016;530(7588):57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol 2011;29(1):24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 2001;98(9):5116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol 2011;12(8):R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci 2012;131(4):281–5. [DOI] [PubMed] [Google Scholar]
- 23.Consortium GTEx. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45(6):580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524(7563):47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang L, Huang J, Higgs BW, Hu Z, Xiao Z, Yao X, et al. Genomic Landscape Survey Identifies SRSF1 as a Key Oncodriver in Small Cell Lung Cancer. PLoS Genet 2016;12(4):e1005895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25(16):2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 2016;34(2):184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orzol P, Nekulova M, Holcakova J, Muller P, Votesek B, Coates PJ. DeltaNp63 regulates cell proliferation, differentiation, adhesion, and migration in the BL2 subtype of basal-like breast cancer. Tumour Biol 2016;37(8):10133–40. [DOI] [PubMed] [Google Scholar]
- 31.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014;11(8):783–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schonemann MD, Ryan AK, McEvilly RJ, O’Connell SM, Arias CA, Kalla KA, et al. Development and survival of the endocrine hypothalamus and posterior pituitary gland requires the neuronal POU domain factor Brn-2. Genes Dev 1995;9(24):3122–35. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka S, Kamachi Y, Tanouchi A, Hamada H, Jing N, Kondoh H. Interplay of SOX and POU factors in regulation of the Nestin gene in neural primordial cells. Mol Cell Biol 2004;24(20):8834–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lodato MA, Ng CW, Wamstad JA, Cheng AW, Thai KK, Fraenkel E, et al. SOX2 co-occupies distal enhancer elements with distinct POU factors in ESCs and NPCs to specify cell state. PLoS Genet 2013;9(2):e1003288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishii J, Sato H, Sakaeda M, Shishido-Hara Y, Hiramatsu C, Kamma H, et al. POU domain transcription factor BRN2 is crucial for expression of ASCL1, ND1 and neuroendocrine marker molecules and cell growth in small cell lung cancer. Pathol Int 2013;63(3):158–68. [DOI] [PubMed] [Google Scholar]
- 36.Ishii J, Sato H, Yazawa T, Shishido-Hara Y, Hiramatsu C, Nakatani Y, et al. Class III/IV POU transcription factors expressed in small cell lung cancer cells are involved in proneural/neuroendocrine differentiation. Pathol Int 2014;64(9):415–22. [DOI] [PubMed] [Google Scholar]
- 37.Bishop JL, Thaper D, Vahid S, Davies A, Ketola K, Kuruma H, et al. The Master Neural Transcription Factor BRN2 Is an Androgen Receptor-Suppressed Driver of Neuroendocrine Differentiation in Prostate Cancer. Cancer Discov 2017;7(1):54–71. [DOI] [PubMed] [Google Scholar]
- 38.Schreiber E, Harshman K, Kemler I, Malipiero U, Schaffner W, Fontana A. Astrocytes and glioblastoma cells express novel octamer-DNA binding proteins distinct from the ubiquitous Oct-1 and B cell type Oct-2 proteins. Nucleic Acids Res 1990;18(18):5495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suva ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 2014;157(3):580–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 1998;2(3):305–16. [DOI] [PubMed] [Google Scholar]
- 41.Truong AB, Khavari PA. Control of keratinocyte proliferation and differentiation by p63. Cell Cycle 2007;6(3):295–9. [DOI] [PubMed] [Google Scholar]
- 42.Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, et al. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 2011;9(4):317–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferguson KL, Slack RS. The Rb pathway in neurogenesis. Neuroreport 2001;12(9):A55–62. [DOI] [PubMed] [Google Scholar]
- 44.Gobert RP, Joubert L, Curchod ML, Salvat C, Foucault I, Jorand-Lebrun C, et al. Convergent functional genomics of oligodendrocyte differentiation identifies multiple autoinhibitory signaling circuits. Mol Cell Biol 2009;29(6):1538–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magri L, Swiss VA, Jablonska B, Lei L, Pedre X, Walsh M, et al. E2F1 coregulates cell cycle genes and chromatin components during the transition of oligodendrocyte progenitors from proliferation to differentiation. J Neurosci 2014;34(4):1481–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferone G, Song JY, Sutherland KD, Bhaskaran R, Monkhorst K, Lambooij JP, et al. SOX2 Is the Determining Oncogenic Switch in Promoting Lung Squamous Cell Carcinoma from Different Cells of Origin. Cancer Cell 2016;30(4):519–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tata PR, Chow RD, Saladi SV, Tata A, Konkimalla A, Bara A, et al. Developmental History Provides a Roadmap for the Emergence of Tumor Plasticity. Dev Cell 2018;44(6):679–93 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugitani Y, Nakai S, Minowa O, Nishi M, Jishage K, Kawano H, et al. Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev 2002;16(14):1760–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin Z, Liu L, Bian W, Chen Y, Xu G, Cheng L, et al. Different transcription factors regulate nestin gene expression during P19 cell neural differentiation and central nervous system development. J Biol Chem 2009;284(12):8160–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iwafuchi-Doi M, Yoshida Y, Onichtchouk D, Leichsenring M, Driever W, Takemoto T, et al. The Pou5f1/Pou3f-dependent but SoxB-independent regulation of conserved enhancer N2 initiates Sox2 expression during epiblast to neural plate stages in vertebrates. Dev Biol 2011;352(2):354–66. [DOI] [PubMed] [Google Scholar]
- 51.Dominguez MH, Ayoub AE, Rakic P. POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cereb Cortex 2013;23(11):2632–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 2005;122(6):947–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kamachi Y, Uchikawa M, Tanouchi A, Sekido R, Kondoh H. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev 2001;15(10):1272–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Devos M, Gilbert B, Denecker G, Leurs K, Mc Guire C, Lemeire K, et al. Elevated DeltaNp63alpha Levels Facilitate Epidermal and Biliary Oncogenic Transformation. J Invest Dermatol 2017;137(2):494–505. [DOI] [PubMed] [Google Scholar]
- 55.Somerville TDD, Xu Y, Miyabayashi K, Tiriac H, Cleary CR, Maia-Silva D, et al. TP63-Mediated Enhancer Reprogramming Drives the Squamous Subtype of Pancreatic Ductal Adenocarcinoma. Cell Rep 2018;25(7):1741–55 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Warner SM, Hackett TL, Shaheen F, Hallstrand TS, Kicic A, Stick SM, et al. Transcription factor p63 regulates key genes and wound repair in human airway epithelial basal cells. Am J Respir Cell Mol Biol 2013;49(6):978–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramsey MR, Wilson C, Ory B, Rothenberg SM, Faquin W, Mills AA, et al. FGFR2 signaling underlies p63 oncogenic function in squamous cell carcinoma. J Clin Invest 2013;123(8):3525–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forster N, Saladi SV, van Bragt M, Sfondouris ME, Jones FE, Li Z, et al. Basal cell signaling by p63 controls luminal progenitor function and lactation via NRG1. Dev Cell 2014;28(2):147–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq and RNA-seq data were deposited in the Gene Expression Omnibus under the accession number GSE137461.