Abstract
Environmental adaptation of organisms relies on fast perception and response to external signals, which lead to developmental changes. Plant cell growth is strongly dependent on cell wall remodeling. However, little is known about cell wall‐related sensing of biotic stimuli and the downstream mechanisms that coordinate growth and defense responses. We generated genetically encoded pH sensors to determine absolute pH changes across the plasma membrane in response to biotic stress. A rapid apoplastic acidification by phosphorylation‐based proton pump activation in response to the fungus Fusarium oxysporum immediately reduced cellulose synthesis and cell growth and, furthermore, had a direct influence on the pathogenicity of the fungus. In addition, pH seems to influence cellulose structure. All these effects were dependent on the COMPANION OF CELLULOSE SYNTHASE proteins that are thus at the nexus of plant growth and defense. Hence, our discoveries show a remarkable connection between plant biomass production, immunity, and pH control, and advance our ability to investigate the plant growth‐defense balance.
Keywords: cellulose, defense, growth, pH
Subject Categories: Development & Differentiation; Membrane & Intracellular Transport; Microbiology, Virology & Host Pathogen Interaction
Induction of apoplast acidification and modification of cell wall properties to control fungal infection are regulated by the COMPANION OF CELLULOSE SYNTHASE proteins at the nexus of plant growth and defense.
Introduction
Developmental adaptation to the environment, such as growth and differentiation, is the driving force of evolution and key for the survival of organisms. Therefore, unraveling the mechanisms by which signal perception and response are coupled to growth at the cellular level is one of the central challenges in biology. Plants adapt their growth to the environment through precisely controlled changes in cell expansion and division, which rely on the accurate remodeling of their cell walls. This strong, yet extensible, polysaccharide‐based structure is the most external cell compartment that surrounds the plasma membrane, while being connected with the cytosol (Liu et al, 2015; Cosgrove, 2018). The cell wall, as the structure occupying most of the apoplastic space, is the first cellular compartment, which encounters environmental signals and is directly involved in perception, transduction, and response to these stimuli (Kesten et al, 2017; Wolf, 2017). Cellulose is an essential constituent of plant cell walls, composed of β‐1,4‐linked D‐glucose chains that are synthesized and extruded to the apoplast by plasma membrane localized cellulose synthase (CesA) complexes (CSCs; McFarlane et al, 2014). The catalytically driven CSCs traverse the plasma membrane, a process that is guided by cortical microtubules (Paredez et al, 2006; Watanabe et al, 2015). CSCs and cortical microtubules are physically connected by at least two protein families, the CELLULOSE SYNTHASE‐INTERACTIVE PROTEIN 1/POM2 (CSI1/POM2; Bringmann et al, 2012; Li et al, 2012) and COMPANION OF CELLULOSE SYNTHASE proteins (CCs) (Endler et al, 2015). Importantly, the CSCs and cortical microtubules are interdependent, as alterations in one affect the activity of the other. This mutual support has been reported by several studies, especially under abiotic stress conditions such as drought and salt (Gutierrez et al, 2009; Nick, 2013; Endler et al, 2015; Wang et al, 2016). Recently, CCs were shown to be indispensable for the recovery of the cortical microtubule array and CSC activity under salt stress (Endler et al, 2015), and the microtubule‐interacting N‐terminus of CC1 to be key for this function (Kesten et al, 2019).
The alteration of cellulose synthase activity in response to extra‐ and intracellular signals is vital for growth modification under dynamic environmental conditions. Changes in pH within distinct cell compartments, including the apoplast/cell wall, act as messengers to transfer such signals (Felle, 2001; Sze & Chanroj, 2018). Plants and fungi primarily regulate pH across the plasma membrane via the activity of proton pumps, such as plasma membrane H+‐ATPases. The active translocation of protons to the apoplast by these pumps provides the chemical and electrical potential needed for solute transport (Haruta et al, 2015). The activity of Arabidopsis H+‐ATPases (AHAs) is essential for plant growth and development under different environmental conditions. Dynamic phosphorylation of specific AHA amino acid residues rapidly regulates proton pump activity in response to external and internal stimuli, such as abiotic and biotic stresses or hormones (Olsson et al, 1998; Gao et al, 2004; Jeworutzki et al, 2010; Stecker et al, 2014; Haruta et al, 2015; Falhof et al, 2016; Geilfus, 2017). The activation of AHAs, for example, is known to require the phosphorylation of their penultimate C‐terminal threonine residue (Inoue & Kinoshita, 2017). Moreover, rapid adjustment of cell wall/apoplastic pH can influence cell wall loosening and stiffening, and thus the connection of the cell wall to the underlying cell, although the exact mechanism is not yet fully understood (Fendrych et al, 2016; Barbez et al, 2017; Mangano et al, 2018). Plant microbes were shown to secrete various cell wall‐degrading enzymes depending on the ambient pH (Li et al, 2012). In planta, the apoplastic pH (pHapo) plays an essential role in the infection process of some pathogens, such as the root vascular fungus Fusarium oxysporum (Masachis et al, 2016; López‐Díaz et al, 2018). Fusarium oxysporum hyphae attach to the root surface, penetrate through natural openings, and then grow intercellularly toward xylem vessels where they can proliferate throughout the plant. Once in the xylem, it secretes a complex collection of proteins named Secreted In Xylem (SIX), which serve to evade plant defense responses (Pietro et al, 2003; Houterman et al, 2007). Over a hundred crop species can be infected by F. oxysporum, with single strains infecting a narrow host‐range. Among them, F. oxysporum 5176 (Fo5176) is one of the genetic strains that can infect Arabidopsis (Thatcher et al, 2012).
The regulation of cellulose synthesis is crucial for plant development and environmental adaptation. Yet the mechanisms underlying stress‐induced changes in microtubule and CSC organization, their crosstalk with cellular signaling messengers, and broad cellular implications remain elusive, especially upon biotic stress (Kesten et al, 2017). This knowledge would be particularly relevant at the root level considering the essential role of this organ in nutrient uptake and response to abiotic and biotic stresses (Zamioudis et al, 2015; Feng et al, 2016; Smakowska et al, 2016; Morris et al, 2017). In this study, we use the Arabidopsis‐Fo5176 pathosystem to characterize plant responses to microbes at a cellular level and how these influence plant adaptation to biotic stress.
Results
Rapid alteration of the cellulose synthesis machinery and cell elongation upon fungal contact is pH dependent
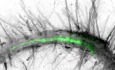
Plant cell wall reorganization in response to stress is essential for proper environmental adaptation. To determine the influence of biotic stress on the regulation of plant cellulose synthesis, we visualized CesAs and cortical microtubules in the elongation zone of A. thaliana roots exposed to Fo5176 hyphae. We aimed to mimic early contact events at the plant–microbe interface and therefore used young hyphae obtained directly after spores germinated overnight. We observed a depletion of CesA foci at the plasma membrane (GFP‐CesA3; Desprez et al, 2007) simultaneously to depolymerization of cortical microtubules (mCherry‐TUA5; Gutierrez et al, 2009) within five min of hyphae contact, which were not observed in control roots (Fig 1A–C; Movie EV1). Additionally, the speed of the remaining CesA foci at the plasma membrane was reduced in comparison with mock conditions (Fig EV1A). In agreement with the observed molecular changes, roots pre‐exposed to fungal hyphae for five min had a reduced root growth rate as compared to control plants (Fig 1D and E). Next, we exposed roots to a Fo5176 elicitor mix containing solubilized molecules from a fungal culture (Baldrich et al, 2014) to exclude that the observed response was solely based on pressure applied by the hyphae. The fungal elicitor mix induced similar, rapid changes on the cellulose synthase machinery, as well as on cortical microtubules, to those observed in response to live hyphae, i.e., cortical microtubules depolymerized and plasma membrane localized CesA density and speed was reduced as compared to mock‐treated roots (Figs 1F–H and EV1B; Movie EV2). Consequently, plants reacted with an immediate root growth reduction once the elicitors were added to the imaging media (Fig 1I and J, Movie EV3), suggesting that a fungal molecule induced the rapid changes in the CSC‐microtubule machinery and root growth.
Figure 1. Fo5176 hyphae and elicitors cause simultaneous depletion of the cellulose synthase machinery at the plasma membrane and root growth rate reduction.
- Representative image of a 5‐day‐old WT (Col‐0) GFP‐CesA3 and mCh‐TUA5 dual‐labeled root epidermal cell under mock conditions (left panel) or upon 5 min of Fo5176 hyphae contact (right panel). A green dashed line in the brightfield (BF) channel highlights Fo5176 hypha. Scale bar = 5 μm.
- GFP‐CesA3 density at the plasma membrane after Fo5176 hyphae contact as depicted in (A). Box plots: centerlines show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend to the minimum and maximum. N ≥ 25 cells from 14 roots and three independent experiments. Welch's unpaired t‐test; ***P‐value ≤ 0.001.
- Microtubule density at the cell cortex after Fo5176 hyphae contact as depicted in (A). Box plots as described in (B). N ≥ 20 cells from 8 roots and three independent experiments. Welch's unpaired t‐test; ***P‐value ≤ 0.001.
- Growth progression of roots grown in half MS or half MS + Fo5176 hyphae. Left panels: representative images of roots at different times after the corresponding treatment. Images of hyphae treatment were modified with an unsharp mask to enhance visibility of hyphae. Scale bar = 100 μm. Right panel: kymographs depicting growth of roots in the left panel.
- Growth rate of roots in half MS or half MS + Fo5176 hyphae, analyzed from images as in (D). Roots were exposed to hyphae for 5 min before imaging started. Average growth rate in half MS: 0.0047 ± 0.0005 μm/s; average growth rate in half MS + Fo5176 hyphae: 0.0031 ± 0.0005 μm/s. Values are mean ± SEM, N ≥ 11 seedlings from three independent experiments. Welch's unpaired t‐test; P‐value ≤ 0.05.
- Representative image of a 5‐day‐old WT (Col‐0) GFP‐CesA3 and mCh‐TUA5 dual‐labeled root epidermal cell under mock conditions (left panel) or upon 5‐min elicitor treatment (right panel). Scale bar = 5 μm.
- GFP‐CesA3 density at the plasma membrane after elicitor treatment as depicted in (F). Box plots as described in (B). N ≥ 35 cells from 16 roots and three independent experiments. Welch's unpaired t‐test; **P‐value ≤ 0.01.
- Microtubule density at the cell cortex, after elicitor treatment as depicted in (F). Box plots as described in (B). N ≥ 37 cells from 16 roots and three independent experiments. Welch's unpaired t‐test; ***P‐value ≤ 0.001.
- Growth progression of roots exposed to fungal elicitor mix. Upper panel: representative images of roots grown in half MS (−17 to 0 min) and after being exposed to the fungal elicitor mix added at 0 min. Scale bar = 100 μm. Lower panel: kymograph depicting growth of roots in the upper panel.
- Growth rate of roots exposed to fungal elicitors, analyzed from images as in (I). After 17 min of growth in half MS, H2O or elicitors were added and the growth rate was measured for additional 17 min. Average growth rate before treatment (−17 to 0 min): H2O: 0.0050 ± 0.0009 μm/s; elicitor mix: 0.0053 ± 0.0005 μm/s. Average growth rate after treatment (0–17 min): H2O: 0.0048 ± 0.0009 μm/s; elicitor mix: 0.0028 ± 0.0004 μm/s. Values are mean ± SEM, N ≥ 10 seedlings from three independent experiments. Welch's unpaired t‐test for roots before and after elicitor treatment; P‐value ≤ 0.01.
Source data are available online for this figure.
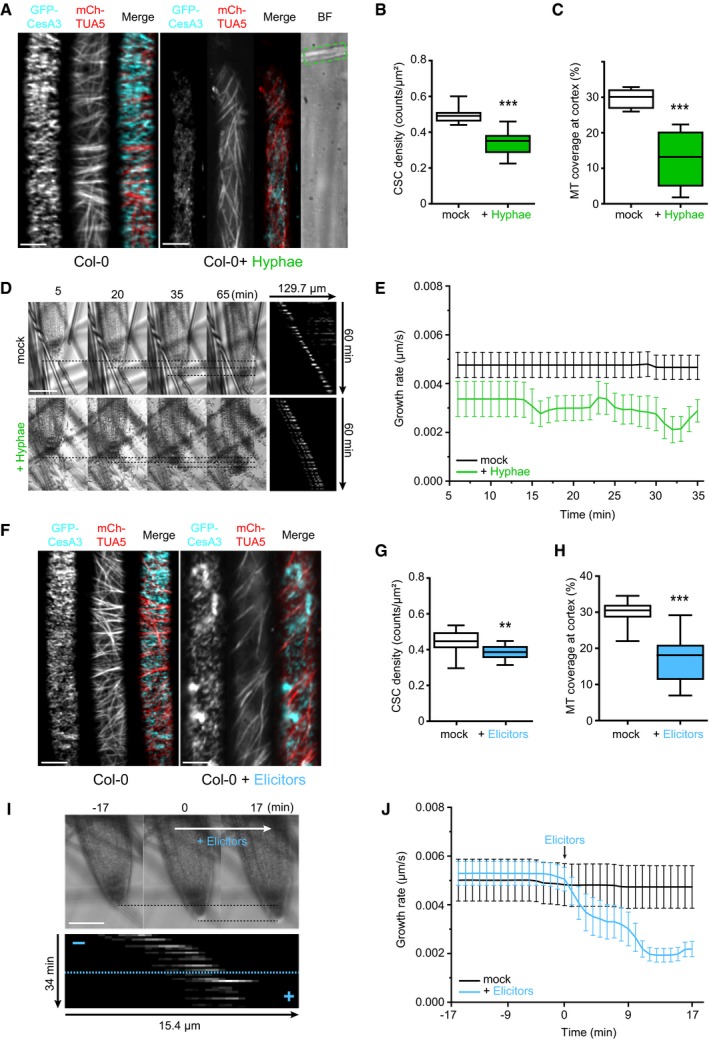
Figure EV1. Fo5176 hyphae and elicitors cause an immediate speed decrease of plasma membrane localized cellulose synthase complexes.
- GFP‐CesA3 speed at the plasma membrane of WT (Col‐0) root cells 5 min after half MS treatment or Fo5176 contact (as depicted in Fig 1A). Box plots: centerlines show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend to the minimum and maximum. N ≥ 369 particles from 6 cells and 6 roots and three independent experiments; Welch's unpaired t‐test; ***P‐value ≤ 0.001.
- GFP‐CesA3 speed at the plasma membrane of WT (Col‐0) root cells 5 min after half MS or half MS + elicitor treatment (as depicted in Fig 1F). Box plots as described in (A). N ≥ 647 particles from 9 cells and 6 roots and three independent experiments; Welch's unpaired t‐test; ***P‐value ≤ 0.001.
- GFP‐CesA3 speed at the plasma membrane of WT (Col‐0) root cells 5 min after being exposed to half MS + 5 mM MES and half MS + 5 mM MES + elicitor treatment (as depicted in Fig 2A). Box plots as described in (A). N ≥ 786 particles from 13 cells and 11 roots and three independent experiments.
- Growth rate of WT (Col‐0) roots exposed to fungal elicitors and 5 mM MES. After 17 min of growth in half MS + 5 mM MES, H2O or elicitors were added and the growth rate was measured for an additional 17 min. Average growth rate before treatment (−17 to 0 min): 5 mM MES: 0.0074 ± 0.0009 μm/s; elicitors + 5 mM MES: 0.0068 ± 0.0010 μm/s. Average growth rate after treatment (0–17 min): 5 mM MES: 0.0077 ± 0.0024 μm/s; elicitors + 5 mM MES: 0.0069 ± 0.0020 μm/s. Values are mean ± SEM, N ≥ 11 seedlings from three independent experiments. Welch's unpaired t‐test for roots before and after elicitor treatment; P‐value = 0.33.
Source data are available online for this figure.
Alteration of pH transients in distinct cell compartments is a fast cellular signaling event reported in response to stress (Behera et al, 2018). Therefore, we investigated the influence of pH on the observed Fo5176 effect on roots. Reduction of CesA density and speed, cortical microtubule depolymerization, as well as root growth reduction were completely reversible by buffering the imaging media with 5 mM 2‐(N‐morpholino)ethanesulfonic acid‐KOH (MES) (Figs 2A–C and EV1C and D; Movie EV4), a common buffer in plant growth media (Good et al, 1966; Bugbee & Salisbury, 1985; Stahl et al, 1999). This implies the direct involvement of pH changes in the observed responses of plant roots to Fo5176.
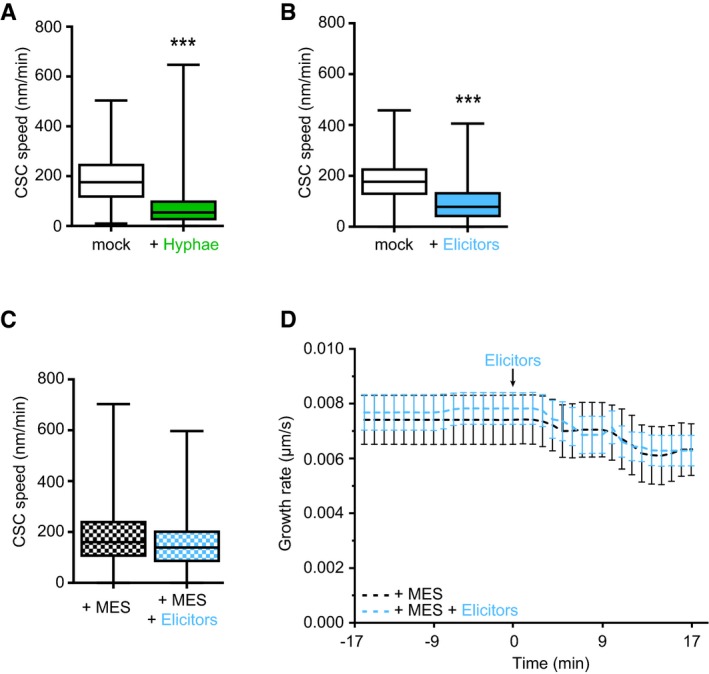
Figure 2. Fo5176‐induced depletion of the cellulose synthase machinery and acidification of the plasma membrane interface can be buffered and are dependent on upregulation of AHA activity.
- Representative image of a 5‐day‐old WT (Col‐0) GFP‐CesA3 and mCh‐TUA5 dual‐labeled root epidermal cell in half MS + 5 mM MES (left panel) or upon 5‐min elicitor treatment in half MS + 5 mM MES (right panel). Scale bar = 5 μm.
- GFP‐CesA3 density at the plasma membrane after elicitor treatment in half MS + 5 mM MES as depicted in (A). Box plots: centerlines show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend to the minimum and maximum. N ≥ 35 cells from 10 roots and three independent experiments.
- Microtubule density at the cell cortex after elicitor treatment in half MS + 5 mM MES as depicted in (A). Box plots as described in (B). N ≥ 29 cells from 10 roots and three independent experiments.
- Representative surface plot of a WT root expressing the pHapo sensor SYP122‐pHusion grown in half MS (−5 to 0 min). At 0 min, either H2O (upper panel) or an elicitor mix (lower panel) was added. Upon elicitor treatment, the signal intensity in the depicted 488 nm channel drastically decreases (highlighted with a blue square).
- Apoplastic pH in WT roots expressing the pHapo sensor SYP122‐pHusion over time, either in half MS or in half MS + 5 mM MES. Imaging started 5 min before either H2O or a fungal elicitor mix was added (0 min). Values are mean ± SEM, N ≥ 15 seedlings from three independent experiments. RM two‐way ANOVA on half MS + H2O versus half MS + elicitors: P ≤ 0.05 (treatment), P ≤ 0.001 (time), P ≤ 0.001 (treatment × time).
- Cortical pH of WT roots expressing the pHcortical sensor pHGFP‐Lti6b over time, either in half MS or in half MS + 5 mM MES. Imaging started 5 min before either H2O or a fungal elicitor mix was added (0 min). Values are mean ± SEM, N = 16 seedlings from three independent experiments. Mixed‐effects model on half MS + H2O versus half MS + elicitors: P = 0.80 (treatment), P ≤ 0.001 (time), P ≤ 0.001 (treatment × time).
- Representative surface plot of WT root expressing the pHapo sensor SYP122‐pHusion grown 30 min in half MS (left panel) or half MS + Fo5176 hyphae (right panel). The hyphae treated root shows drastically reduced signal intensity in the depicted 488 nm channel (highlighted with a green square).
- Apoplastic pH of WT roots expressing the pHapo sensor SYP122‐pHusion over time, either in half MS or half MS + Fo5176 hyphae. Roots were exposed to hyphae for 5 min before imaging started. Values are mean ± SEM, N ≥ 12 seedlings from three independent experiments. RM two‐way ANOVA on half MS versus half MS + elicitors: P ≤ 0.01 (treatment), P ≤ 0.001 (time), P ≤ 0.001 (treatment × time).
- Cortical pH variation of WT roots expressing the pHcortical sensor pHGFP‐Lti6b over time, either in half MS or in half MS + Fo5176 hyphae. Roots were exposed to hyphae for 5 min before imaging started. Values are mean ± SEM, N ≥ 13 seedlings from three independent experiments. RM two‐way ANOVA on half MS versus half MS + elicitors: P ≤ 0.01 (treatment), P = 0.23 (time), P ≤ 0.001 (treatment × time).
- Western blots showing chemiluminescent signals of an anti‐pThr or anti‐AHA incubated membrane loaded with Arabidopsis root samples treated for 8 min with either half MS or half MS + Fo5176 hyphae. The Ponceau S panel shows total protein content. The AHA band used for quantification is indicated with an arrowhead. Dashed line separates different treatments of the same membrane.
- Relative AHA phosphorylation status from Western blots as shown in (J). Normalized signal intensity ratio of anti‐pThr in respect to anti‐AHA is shown. Box plots as described in (B). N = 5 independent experiments. Welch's unpaired t‐test; *P‐value ≤ 0.05.
Source data are available online for this figure.
New sensors to measure the pH at both sides of the plant plasma membrane
To test this hypothesis, we sought to measure the pH of plant root cells in response to live Fo5176 hyphae and elicitors. As previously reported, the apoplastic pH sensor apo‐pHusion suffers from high background signal in the endoplasmic reticulum (ER), which compromises precise pH measurements in the apoplast (Gjetting et al, 2012; Martinière et al, 2018). Therefore, we generated a pHapo sensor with as low as possible intracellular background signal by fusing the ratiometric probe pHusion (Gjetting et al, 2012) to the C‐terminus of the plasma membrane localized SNARE (soluble N‐ethyl‐maleimide sensitive factor attachment protein receptor) protein Syntaxin of Plants 122 (SYP122; Assaad et al, 2004; Uemura et al, 2004). Confocal microscopy of stable transgenic lines suffered considerably less from intracellular signal for pUB10::SYP122‐pHusion than for apo‐pHusion (Fig EV2A). In vivo calibration revealed a sigmoidal correlation between pH and the EGFP/RFP ratio of the sensor with a linear range between pH 5.2 and 6.8 (Fig EV2B). pH measurements in control epidermal/cortex cells of the root elongation zone indicated an average pHapo over five min of 5.30 ± 0.45 (Fig 2D and E), which is in good agreement with previously reported values obtained with fluorescent dyes or surface electrode measurements in the root elongation zone of Arabidopsis (Staal et al, 2011; Barbez et al, 2017). To measure pH on both sides of the plasma membrane, we targeted the cortical side by fusing pHGFP (Moseyko & Feldman, 2001) to the N‐terminus of the plasma membrane localized low temperature induced protein 6b (Lti6b; Cutler et al, 2000), also driven by pUB10. Counterstaining of cell walls with propidium iodide indicated plasma membrane localization of pHGFP‐Lti6b in root cells (Fig EV2C). In vivo calibration of the cortical pH (pHcortical) sensor pHGFP‐Lti6b revealed an almost linear relationship of emission ratio and pH in the range from pH 5.6–8.0 (Fig EV2D). Cortical pH measurements in control epidermal/cortex cells of the root elongation zone indicated an average pHcortical over 5 min of 7.03 ± 0.36 (Fig 2F), which is in agreement with previous reports (Moseyko & Feldman, 2001; Gao et al, 2004; Schulte et al, 2006). Cytoplasmic pH (pHcyto) measurements were approached by expressing free pHGFP in the cytosol (Moseyko & Feldman, 2001; Fendrych et al, 2014). In control epidermal/cortex cells of the root elongation zone, we measured an average pHcyto over 5 min of 6.62 ± 0.92 (Fig EV2E), which is also in agreement with previous reports using fluorescent methods (Moseyko & Feldman, 2001; Gao et al, 2004; Schulte et al, 2006).
Figure EV2. Fo5176‐induced depletion of the cellulose synthase machinery and acidification of the plasma membrane interface, measured with newly designed sensors, can be buffered.
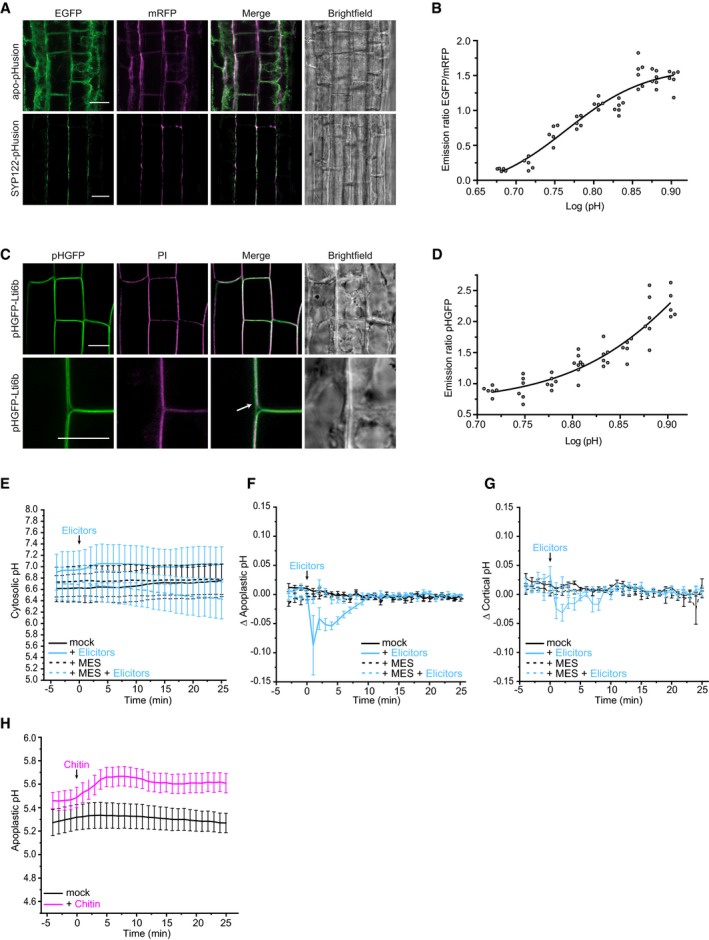
- Representative images of 6‐day‐old WT (Col‐0) root epidermal and cortex cells expressing apo‐pHusion or SYP122‐phusion. Much less intracellular signal of pHusion is observed for SYP122‐pHusion than for apo‐pHusion. Scale bar = 20 μm.
- In vivo calibration of SYP122‐pHusion in 6‐day‐old roots. Dots represent individual samples with N ≥ 5 seedlings per standard buffer. Data points were fitted using sigmoidal regression.
- Representative images of 6‐day‐old WT (Col‐0) root epidermal and cortex cells expressing pHGFP‐Lti6b. Cell walls of seedlings were counterstained with 3 μg/ml propidium iodide to illustrate plasma membrane localization of pHGFP‐Lti6b of two adjacent cells (white arrow). Scale bars = 10 μm.
- In vivo calibration of pHGFP‐Lti6b in 6‐day‐old roots. Dots represent individual samples with N ≥ 5 seedlings per standard buffer. Data points were fitted using sigmoidal regression.
- Cytoplasmic pH variation of WT roots expressing the pHcyto free pHGFP sensor over time, either in half MS or in half MS + 5 mM MES. Imaging started 5 min before either H2O or a fungal elicitor mix was added (0 min). Values are mean ± SEM, N ≥ 12 seedlings from three independent experiments. RM two‐way ANOVA on half MS + H2O versus half MS + elicitors: P = 0.42 (treatment), P = 0.06 (time), P = 0.08 (treatment × time).
- Apoplastic ∆pH of WT roots expressing the pHapo SYP122‐pHusion sensor over time, either in half MS or in half MS + 5 mM MES. Imaging started 5 min before either H2O or a fungal elicitors were added (0 min). Values are mean ± SEM, N ≥ 16 seedlings from three independent experiments. RM two‐way ANOVA on half MS + H2O versus half MS + elicitors: P ≤ 0.001 (treatment), P ≤ 0.05 (time), P ≤ 0.001 (treatment × time).
- Cortical ∆pH of WT roots expressing the pHcortical pHGFP‐Lti6b sensor over time, either in half MS or in half MS + 5 mM MES. Imaging started 5 min before either H2O or a fungal elicitor mix was added (0 min). Mixed‐effects model on half MS + H2O versus half MS + elicitors: P ≤ 0.01 (treatment), P ≤ 0.001 (time), P ≤ 0.001 (treatment × time).
- Apoplastic pH of WT roots expressing the pHapo SYP122‐pHusion sensor over time in half MS. Imaging started 5 min before either H2O (mock) or 1 mM chitin were added (0 min). Values are mean ± SEM, N = 15 seedlings from three independent experiments. RM two‐way ANOVA on half MS + H2O versus half MS + chitin: P ≤ 0.05 (treatment), P ≤ 0.001 (time), P ≤ 0.001 (treatment × time). This chitin assay was done simultaneously to the elicitor treatments (Fig 2E); therefore, they share the same mock (H2O) curve.
Source data are available online for this figure.
Fungal‐induced pH changes across the plasma membrane are generated by the activation of proton pumps
Upon elicitor treatment, pHapo dropped below 5.0 within five min (Figs 2D and E and EV2F; Movie EV5), simultaneously with the observed reduction of root growth (Fig 1I and J). It is important to note that the pH of the added elicitor mix ranged between 5.5 and 5.8 in individual preparations. The cortical side of the plasma membrane also acidified (Figs 2F and EV2G), but the response was delayed by 1 min in comparison with the apoplastic side. In this first minute, the pHcortical rose as an immediate response to acidification of the apoplast (Figs 2E and EV2F), pointing toward a translocation of protons from the cortical to the apoplastic side of the plasma membrane. The pHcortical recovered to control levels approx. 7 min after elicitor application. We could not detect any apparent changes in the pHcyto in response to fungal elicitors (Fig EV2E), highlighting that the immediate pH response was constraint to the plasma membrane environment. When we buffered the media with MES before adding the elicitor mix, no significant change of pH could be observed on any side of the plasma membrane (Figs 2E and F, and EV2F and G). This suggests that the depletion of CSCs and cortical microtubules, as well as the reduction of root growth upon elicitor treatment, is linked to plasma membrane pH changes.
Next, we explored whether the cellular pH fluctuations were also detectable upon fungal contact by imaging plants on an agarose cushion that was coated with live Fo5176 hyphae. Under these conditions, the pHapo and pHcortical were comparable to the elicitor mix experiments under mock conditions (5.44 ± 0.33 and 6.99 ± 0.73, respectively). In line with the elicitor mix treated roots, five min of contact with Fo5176 hyphae also induced a drastic acidification of both the apoplast and the cortical side of the plasma membrane (Fig 2G–I). This was unexpected since the main component of the fungal cell wall, chitin, was previously reported to cause a rapid alkalinization of the leaf apoplast (Felle et al, 2009). We could confirm the alkalinization of the apoplast in roots with the pHapo sensor (Fig EV2H), indicating that the rapid acidification in response to live Fo5176 hyphae was not caused by chitin. The fast apoplastic acidification in response to Fo5176 points toward an increase in plant plasma membrane proton pump activity upon hyphae and elicitor mix contact. We approached this hypothesis by Western blotting with well‐characterized antibodies against the catalytic domain of the AHAs and their upregulated form that is phosphorylated on the penultimate threonine residue (Hayashi et al, 2010). Indeed, Western blotting revealed increased phosphorylation of AHAs upon Fo5176 hyphae contact (Fig 2J and K), supporting the observed pHapo drop upon Fo5176 exposure (Fig 2G and H). These results suggest that a ∆pH change across the plasma membrane induced the detected CSC depletion, cortical microtubule depolymerization, and root growth inhibition upon hyphae and elicitor treatment.
CSC and cortical microtubule regulation upon biotic stress requires the CC proteins
Our data show a simultaneous response of CSCs and cortical microtubules to biotic stress, highlighting their reported interdependence. Considering their physical interaction, we evaluated the role of CSC‐microtubule connecting proteins in response to Fo5176. The first obvious candidates were the CC proteins, as major players in regulating cellulose synthesis and microtubule dynamics under abiotic stress conditions, such as salt (Endler et al, 2015). We exposed a cc1cc2 YFP‐CesA6 mChTUA dual‐label line (Endler et al, 2015) to Fo5176 hyphae, which lacks the two most important CC proteins and keeps a wild‐type (WT)‐like CSC‐microtubule pattern under mock conditions (Fig 3A–D). In contrast to WT plants, in cc1cc2 roots CSC density and speed were partially maintained and cortical microtubule density was indistinguishable from mock conditions when treated with Fo5176 hyphae (Figs 3A–D and EV3A). These observations indicated that the fast CSC depletion from the plasma membrane and depolymerization of microtubules in WT roots upon Fo5176 contact is dependent on the CC proteins. Further supporting these observations, no immediate growth rate reduction was detected for cc1cc2 when exposed to Fo5176 hyphae (Fig 3E). We next tested the function of the CC1 microtubule‐interacting domain, recently reported to be essential for plants to regulate microtubule bundling and dynamics under salt stress (Endler et al, 2015; Kesten et al, 2019). Therefore, we used cc1cc2 mutant lines complemented with a fully functional, GFP tagged CC1 (GFP‐CC1), or a truncated CC1 version, in which the microtubule‐interacting domain is exchanged to GFP (GFP‐CC1ΔN120) ((Endler et al, 2015). We employed GFP‐CC1ΔN120 or GFP‐CC1 as proxy for CSCs as they are part of the same complex and co‐localize at the plasma membrane (Endler et al, 2015). No alteration of CSC and microtubule density as well as no CSC speed reduction could be observed in cc1cc2 GFP‐CC1ΔN120, rendering the plants indistinguishable from mock‐treated plants (Figs 3F–I and EV3B). These results demonstrate that the CC protein family, via its microtubule‐interacting domain, does participate in plant responses to abiotic and biotic stress, but with contrary roles.
Figure 3. Fo5176 hyphae do not affect the cellulose synthase machinery and growth rate of cc1cc2 mutant roots.
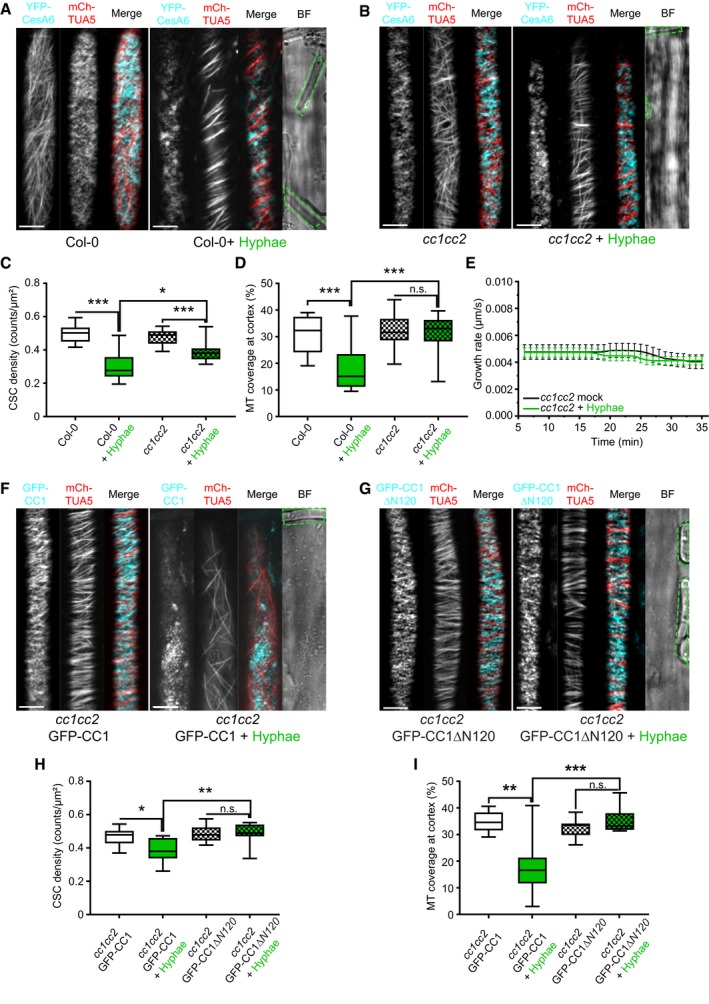
- Representative image of a 5‐day‐old WT (Col‐0) YFP‐CesA6 and mCh‐TUA5 dual‐labeled root epidermal cell under mock conditions (left panel) or upon 5 min of Fo5176 hyphae contact (right panel). A green dashed line in the bright field (BF) channel highlights Fo5176 hyphae. Scale bar = 5 μm.
- Representative image of a 5‐day‐old cc1cc2 YFP‐CesA6 and mCh‐TUA5 dual‐labeled root epidermal cell under mock conditions (left panel) or upon 5 min of Fo5176 hyphae contact (right panel). A green dashed line in the BF channel highlights Fo5176 hyphae. Scale bar = 5 μm.
- YFP‐CesA6 density at the plasma membrane of WT (Col‐0) or cc1cc2 root cells after Fo5176 hyphae contact as depicted in (A and B). Box plots: centerlines show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend to the minimum and maximum. N ≥ 21 cells from 10 roots and three independent experiments; Welch's unpaired t‐test; *P‐value ≤ 0.05, ***P‐value ≤ 0.001.
- Microtubule density at the cell cortex of WT (Col‐0) or cc1cc2 root cells after Fo5176 hyphae contact as depicted in (A and B). Box plots as described in (C). N ≥ 24 cells from 9 roots and three independent experiments; Welch's unpaired t‐test; ***P‐value ≤ 0.001.
- Growth rate of cc1cc2 roots grown in half MS or half MS + Fo5176 hyphae. Average growth rate in half MS: 0.0046 ± 0.0005 μm/s; average growth rate in half MS + Fo5176 hyphae: 0.0045 ± 0.0003 μm/s. Values are mean ± SEM, N = 17 seedlings from three independent experiments. Welch's unpaired t‐test; P‐value = 0.78.
- Representative image of a 5‐day‐old cc1cc2 GFP‐CC1 and mCh‐TUA5 dual‐labeled root epidermal cell under mock conditions (left panel) or upon 5 min of Fo5176 hyphae contact (right panel). A green dashed line in the BF channel highlights Fo5176 hypha. Scale bar = 5 μm.
- Representative image of a 5‐day‐old cc1cc2 GFP‐CC1ΔN120 and mCh‐TUA5 dual‐labeled root epidermal cell under mock conditions (left panel) or upon 5 min of Fo5176 hyphae contact (right panel). A green dashed line in the BF channel highlights Fo5176 hypha. Scale bar = 5 μm.
- GFP‐CC1 or GFP‐CC1ΔN120 density (as proxy for the CSC) at the plasma membrane of cc1cc2 root cells after Fo5176 hyphae contact as depicted in (F and G). Box plots as described in (C). N ≥ 22 cells from 8 roots and three independent experiments; Welch's unpaired t‐test; *P‐value ≤ 0.05, **P‐value ≤ 0.01.
- Microtubule density at the cell cortex of cc1cc2 GFP‐CC1 or cc1cc2 GFP‐CC1ΔN120 root cells after Fo5176 hyphae contact as depicted in (F and G). Box plots as described in (C). N ≥ 24 cells from 7 roots and three independent experiments; Welch's unpaired t‐test; **P‐value ≤ 0.01, ***P‐value ≤ 0.001, n.s. = not significant.
Source data are available online for this figure.
Figure EV3. The speed of plasma membrane localized cellulose synthase complexes is differentially affected by Fo5176 hyphae contact in cc1cc2 mutants compared to wild‐type.
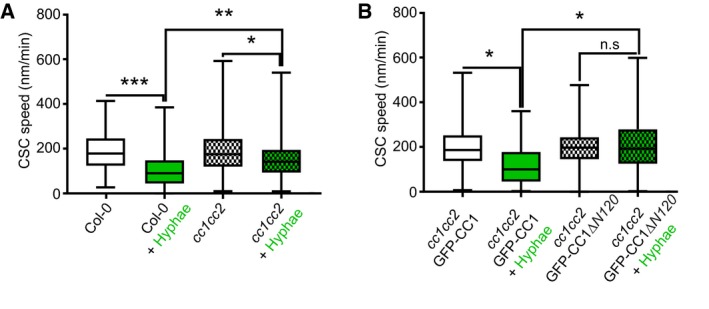
- CSC speed at the plasma membrane of WT (Col‐0) and cc1cc2 root cells, as depicted in Fig 3A and B, upon Fo5176 hyphae contact. Box plots: centerlines show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend to the minimum and maximum. N ≥ 270 particles from 8 cells and 7 roots and three independent experiments; Welch's unpaired t‐test; *P‐value ≤ 0.05, **P‐value ≤ 0.01, ***P‐value ≤ 0.001.
- GFP‐CC1 and GFP‐CC1ΔN120 speed (as proxy for the CSC) at the plasma membrane of cc1cc2 root cells upon Fo5176 hyphae contact, as depicted in Fig 3F and G. Box plots as described in (A). N ≥ 288 particles from 12 cells and 7 roots and three independent experiments; Welch's unpaired t‐test; *P‐value ≤ 0.05, n.s. = not significant.
Source data are available online for this figure.
cc1cc2 is less susceptible to Fo5176 vasculature colonization than WT plants
Plant growth rate and cellulose synthase machinery were obviously affected by Fo5176 contact in a CC‐ and pH‐dependent manner. Hence, we questioned whether this response was maintained during Fo5176 root colonization and designed an in‐plate assay that allows for monitoring both root growth and fungal vascular colonization at the same time. We generated a Fo5176 line harboring a GFP expression cassette under the control of the Fo5176 SIX1 effector promoter (for Secreted in Xylem 1; Fo5176 pSIX1::GFP), whose activation is needed for F. oxysporum virulence in different hosts (Rep et al, 2004; van der Does et al, 2008a,b). In our plate infection assay, we observed GFP signal in Fo5176 pSIX1::GFP specifically when it reached the xylem of the host vasculature (Fig 4A). Consequently, the sensor allowed us to quantify fungal vasculature penetration events over the progression of the infection, while root growth inhibition could be quantified as a fungal disease symptom. In the course of the experiment, cc1cc2 roots outgrew the WT when treated with Fo5176 pSIX1::GFP, while both genotypes were indistinguishable under mock conditions (Fig 4B–E). The same was observed for plants inoculated with WT Fo5176, which confirmed that the Fo5176 virulence is not affected by the pSIX1::GFP construct (Fig EV4A). Going along with root growth reduction, WT roots exhibited the first events of fungal entry into the xylem at 3 days post‐fungal inoculation (dpi), and the vascular penetration rate increased over the course of the experiment (Fig 4E). The vasculature of cc1cc2 mutants was in contrast barely colonized and root growth less affected than in WT (Fig 4E). Likewise to inoculation with pSIX1::GFP, we detected an increased SIX1 expression in WT plants inoculated with WT Fo5176 in comparison with cc1cc2, both relative to a fungal and to a plant reference gene (FoβTUB and AtGAPDH, respectively) (Fig 4F and G). To determine whether the observed reduced Fo5176 vascular colonization in cc1cc2 was a consequence of a reduced total colonization of the root, we quantified fungal biomass inside the roots following two complementary methods. We measured the amount of N‐acetylglucosamine within infected roots that is proportional to the amount of fungal cell wall (i.e., chitin; Fig EV4B and C) and quantified the amount of live fungus by counting fungal colonies that developed after grinding and plating surface‐sterilized roots (Durán et al, 2018) (Fig EV4D). Both analyses showed no differences between total fungal biomass in WT and cc1cc2 roots, indicating that the cc1cc2 mutation hinders Fo5176 from colonizing the vasculature but not previous root cell layers. To gain further insights into cc1cc2 resistance to Fo5176 vascular colonization, we measured the expression of At1g51890, WRKY45, and WRKY53, genes known to be upregulated in response to stress (Roux et al, 2011; Masachis et al, 2016; de Souza et al, 2017; Wang et al, 2018). WRKY45 and WRKY53 showed significantly upregulated expression in cc1cc2 under mock conditions (Fig 4H).
Figure 4. cc1cc2 mutants are less susceptible than wild‐type to Fo5176 vasculature colonization.
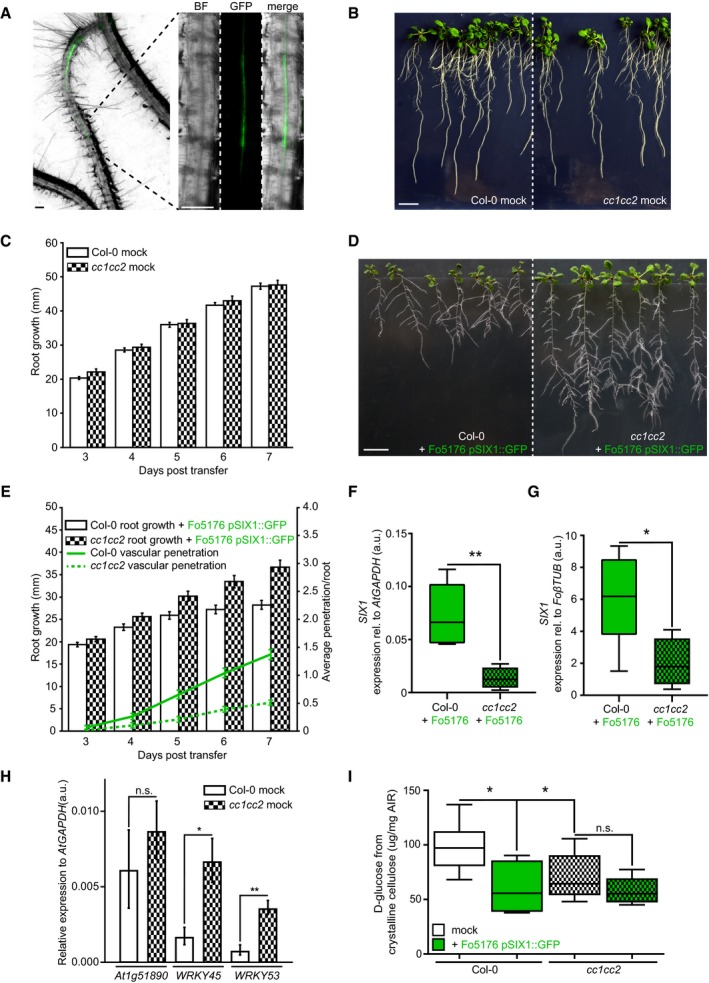
- Arabidopsis root vasculature colonized by Fo5176 pSIX1::GFP. Right panel shows a magnification separated into brightfield (BF) and GFP channel. GFP fluorescence is apparent when Fo5176 colonized the vasculature (dark gray, root central line in BF channel). Scale bars = 125 μm.
- Representative image of WT (Col‐0) and cc1cc2 plants 7 days post‐transfer to half MS mock plates. Scale bar = 10 mm.
- Root elongation of WT (Col‐0) and cc1cc2 plants at various days post‐transfer to mock plates, as depicted in (B). Values are mean ± SEM, N ≥ 103 plants from three independent experiments. RM two‐way ANOVA: P = 0.65 (genotype), P ≤ 0.001 (time), P = 0.33 (genotype × time).
- Representative image of WT (Col‐0) and cc1cc2 plants 7 days post‐transfer to Fo5176 pSIX1::GFP spore containing plates. Scale bar = 10 mm.
- Root elongation and vascular penetration of WT (Col‐0) and cc1cc2 mutant plants at various days post‐transfer to Fo5176 pSIX1::GFP spore containing plates, as depicted in (D). Values are mean ± SEM, N ≥ 103 plants from three independent experiments. RM two‐way ANOVA on root growth: P ≤ 0.001 (genotype), P ≤ 0.001 (time), P ≤ 0.01 (genotype × time). RM two‐way ANOVA on vascular penetration rate: P ≤ 0.01 (genotype), P ≤ 0.001 (time), P ≤ 0.001 (genotype × time).
- SIX1 expression relative to AtGAPDH in WT (Col‐0) or cc1cc2 roots 7 days post‐inoculation with Fo5176 spores. Box plots: centerlines show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend to the minimum and maximum. N ≥ 4 biological replicates; Welch's unpaired t‐test; **P‐value ≤ 0.01.
- SIX1 expression relative to FoβTUB in WT (Col‐0) or cc1cc2 roots 7 days post‐inoculation with Fo5176. Box plots as described in (F). N ≥ 4 biological replicates; Welch's unpaired t‐test; *P‐value ≤ 0.05.
- Expression of At1g51890, WRKY45, and WRKY53 relative to AtGAPDH in WT (Col‐0) and cc1cc2 roots under mock conditions. Values are mean ± SEM, N ≥ 4 biological replicates; Welch's unpaired t‐test; *P‐value ≤ 0.05. **P value ≤ 0.01.
- Cellulose content of roots grown as depicted in (B and D), represented as μg of D‐glucose derived from crystalline cellulose per mg of dried alcohol‐insoluble residue (AIR). Box plots as described in (F). N ≥ 5 biological replicates; 2 technical replicates per biological replicate. Welch's unpaired t‐test; *P‐value ≤ 0.05, n.s. = not significant.
Source data are available online for this figure.
Figure EV4. cc1cc2 resistance to Fo5176 root colonization can be mimicked in wild‐type plants by buffering the growth media.
- Root elongation at various days post‐transfer to Fo5176 spore containing plates. Values are mean ± SEM, N ≥ 33 plants from three independent experiments. RM two‐way ANOVA: P ≤ 0.01 (genotype), P ≤ 0.001 (time), P ≤ 0.05 (genotype × time).
- Left panel: Monosaccharide elution profile of a 1 μg/ml standard (black) compared to 1:10 dilutions of hydrolyzed samples (Saeman hydrolysis, as indicated in methods section) derived from roots after 7 days of either mock (pink) or Fo5176 treatment (blue). The N‐acetylglucosamine (GlcNAc) peak (4) is only identifiable in hydrolyzed infected samples (blue). Right panel: Strong and weak hydrolyses of fungal‐derived cell wall material indicate negligible presence of glucose non‐extractable with the matrix hydrolysis method, as the glucose peak is similar in either hydrolysis method. In plants, there is a clear difference between the glucose peaks in either hydrolysis method.
- Quantification of the N‐acetylglucosamine peak of Fo5176 pSix1::GFP‐infected roots derived from the fungal cell wall as shown in (B). Box plots: centerlines show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend to the minimum and maximum. N ≥ 5 biological replicates; Welch's unpaired t‐test; P = 0.34.
- Quantification of Fo5176 colonies developing after surface sterilization of infected roots. Box plots as described in (C). N = 5 biological replicates; Welch's unpaired t‐test; P = 0.75.
- Vascular penetration of WT (Col‐0) and cc1cc2 plants at various days post‐inoculation with Fo5176 pSIX1::GFP spores with or without 5 mM MES. Values are mean ± SEM, N ≥ 33 plants from three independent experiments. RM two‐way ANOVA (Col‐0 half MS versus Col‐0 half ME + MES): P ≤ 0.001 (genotype), P ≤ 0.001 (time), P ≤ 0.001 (genotype × time).
- Root elongation of WT (Col‐0) and cc1cc2 plants at various days post‐inoculation with Fo5176 pSIX1::GFP spores with or without 5 mM MES. Values are mean ± SEM, N ≥ 33 plants from three independent experiments. RM two‐way ANOVA (Col‐0 half MS versus Col‐0 half MS + MES): P ≤ 0.001 (genotype), P ≤ 0.001 (time), P ≤ 0.001 (genotype × time).
- Cellulose content of roots 7 days post‐transfer to Fo5176 pSIX1::GFP spore containing half MS + 5 mM MES plates or post‐transfer to control (half MS +5 mM MES) plates, represented as μg of D‐glucose derived from crystalline cellulose per mg of dried alcohol‐insoluble residue (AIR). Box plots as described in (C). N ≥ 5 biological replicates; 2 technical replicates per biological replicate. Welch's unpaired t‐test; **P‐value ≤ 0.01.
- CSC (N ≥ 27 cells from 11 roots) and microtubule (N ≥ 17 cells from 7 roots) density and CSC speed (N ≥ 366 particles from 17 cells and 9 roots) at the plasma membrane of WT (Col‐0) or cc1cc2 root cells expressing YFP‐CesA6 and mCh‐TUA5 grown on half MS + 5 mM MES. Data are from three independent experiments. Box plots as described in (C). Welch's unpaired t‐test indicates no significant difference for all measurements.
- Quantification of the N‐acetylglucosamine peak of Fo5176 infected plants grown on half MS + 5 mM MES derived from the fungal cell wall as shown in (B). Box plots as described in (C). N = 3 biological replicates; Welch's unpaired t‐test; P = 0.41.
Source data are available online for this figure.
Since the short‐term effect of hyphae contact on the cellulose synthase machinery could be inhibited by buffering the media with MES, we also aimed to confirm the influence of the plasma membrane ∆pH during progression of Fo5176 root colonization. Both the average vasculature penetration rate of Fo5176 pSIX1::GFP and root growth inhibition were significantly reduced when WT plants were grown on buffered media but did not further change in cc1cc2 (Fig EV4E and F), i.e., we could partially mimic the cc1cc2 phenotype by growing WT plants on buffered media. Finally, we measured the influence of fungal root colonization on the cellulose content of the host. We adapted a protocol from Yeats & Vellosillo et al to identify all cell wall sugars (Yeats et al, 2016a,b) and were able to reliably distinguish the N‐acetylglucosamine of the fungal, chitin‐based cell wall from the plant cell wall, derived from crystalline cellulose as glucose (Figs 4I and EV4B). Furthermore, the presence of the fungus in cell wall samples of infected plants did not affect crystalline cellulose analyses (Fig EV4B right panel). We observed a significant reduction of cellulose content in WT roots upon Fo5176 pSIX1::GFP colonization, while the fungal infection did not alter the cellulose amount in cc1cc2 mutants, which was already reduced under mock conditions (Fig 4I). This observation was conflicting with other publications, reporting no changes in cellulose levels in these mutants under mock conditions when using buffered media (Endler et al, 2015; Kesten et al, 2019). Hence, we repeated the assay under buffered conditions, which fully recovered cellulose levels in cc1cc2 when compared to WT without changing CesA density and speed or microtubule density in any genotype (Fig EV4G and H; compare to Figs 3C and D, and EV3A), confirming previous results (Endler et al, 2015; Kesten et al, 2019). Buffering the media furthermore led to a reduction of cellulose levels in infected cc1cc2 roots, while it did not affect the cellulose decrease in WT roots caused by Fo5176 pSIX1::GFP (Fig EV4G; compare to Fig 4I). In addition, buffering the media did not alter total fungal biomass in any genotype (Fig EV4I).
Based on these results, we conclude that the observed short‐term fungal effects, i.e., depletion of the cellulose synthase machinery and growth rate reduction, also affect the overall infection process. Furthermore, these effects are repressed in the cc1cc2 mutant and under buffered media conditions independently of cellulose levels in these plants, indicating that the plant response to the fungus is pH but not cellulose dependent.
cc1cc2 exhibits an elevated ΔpH across the plasma membrane
The cc1cc2 mutant did exhibit less sensitivity to Fo5176 than WT plants, both during short contact and infection (Figs 3 and 4). As we observed a clear influence of plant plasma membrane ∆pH changes on the root response to the fungus (Fig 2) and could partially mimic the cc1cc2 phenotype by growing WT plants on buffered media (Fig EV4E and F), we speculated that the pH at the plasma membrane interface of cc1cc2 might be altered. Therefore, we introgressed both pHapo and pHcortical sensors into the cc1cc2 knockout line. cc1cc2 plants already showed significantly different pHapo and pHcortical when compared to WT plants under mock conditions. We measured an average pHapo over 5 min of 5.21 ± 0.20 (Fig 5A; compare to WT pHapo 5.42 ± 0.32 in Fig 2H; values are mean ± SD, N ≥ 12 seedlings from three independent experiments; Welch's unpaired t‐test; P‐value ≤ 0.05) and an average pHcortical over 5 min of 7.67 ± 0.38 (Fig 5B; compare to WT pHcortical 7.07 ± 0.64 in Fig 2I; values are mean ± SD, N ≥ 11 seedlings from three independent experiments; Welch's unpaired t‐test; P‐value ≤ 0.05). This implicates an enhanced proton chemical gradient across the plasma membrane in the cc1cc2 mutant background (WT ΔpH = 1.55, cc1cc2 ΔpH = 2.45). The altered plasma membrane ∆pH of cc1cc2 roots points toward an elevated plasma membrane proton pump activity. Hyperactivation of AHAs in both cc1cc2 and cc1cc2 GFP‐CC1ΔN120 mutants was confirmed by measuring the acidification process of an alkaline growth media, which both lines acidified significantly faster than WT plants (Fig 5C). To exclude that the enhanced proton efflux was induced by the cellulose deficiency of cc1cc2 in unbuffered conditions, we repeated the assay with two other cellulose‐deficient mutants, prc1‐1 and pom2‐4 (Fagard et al, 2000; Bringmann et al, 2012). Both lines did not acidify the growth media faster than WT, with prc1‐1 being indistinguishable from WT and pom2‐4 showing less proton efflux than the other genotypes, suggesting that the observed ∆pH alteration in cc1cc2 is not purely based on its cellulose deficiency (Fig EV5A). Western blotting corroborated an increased phosphorylation of the penultimate AHA threonine residue in cc1cc2 mutants as compared to WT (Fig 5D and E). These results confirm the observed lower pHapo and higher pHcortical measured in mock‐treated cc1cc2 root cells (Fig 5A and B). We detected the same hyperactivation in the cc1cc2 GFP‐CC1ΔN120 line (Fig 5F and G), pointing toward an important role of the CC1 microtubule‐ interacting domain for pH regulation at the plasma membrane. Confirming the higher plasma membrane ΔpH of cc1cc2, both CC‐impaired lines were more sensitive to low amounts of hygromycin B as compared to WT (Fig EV5B–E), which is primarily internalized through the energy generated by the proton chemical gradient (Haruta et al, 2010).
Figure 5. cc1cc2 mutants exhibit an acidic apoplast, alkaline cortex and high basal AHA phosphorylation state, which are not affected by Fo5176 contact.
- Apoplastic pH variation of cc1cc2 roots expressing the pHapo sensor SYP122‐pHusion over time, either in half MS or in half MS + Fo5176 hyphae. Values are mean ± SEM, N = 17 seedlings from three independent experiments. Welch's unpaired t‐test on total average pH; P‐value = 0.84.
- Cortical pH variation of cc1cc2 roots expressing the pHcortical sensor pHGFP‐Lti6b over time, either in half MS or in half MS + Fo5176 hyphae. Values are mean ± SEM, N ≥ 12 seedlings from three independent experiments. Welch's unpaired t‐test on total average pH; P‐value = 0.67.
- Media pH development over time when 10‐day‐old WT (Col‐0), cc1cc2, and cc1cc2 GFP‐CC1ΔN120 plants were transferred to a liquid, alkaline media (pH 6.45). Values are mean ± SEM, N ≥ 23 plants from three independent experiments. Welch's unpaired t‐test after 6 h; *P‐value ≤ 0.05.
- Western blots showing chemiluminescent signals of anti‐pThr or anti‐AHA incubated membranes loaded with Arabidopsis WT (Col‐0) or cc1cc2 root samples. Roots were treated for 8 min with either half MS or half MS + Fo5176 pSIX1::GFP hyphae. The Ponceau S panel shows total protein content. The AHA band used for quantification is highlighted with an arrowhead. Dashed line separates different treatments of the same membrane.
- Relative AHA phosphorylation status from Western blots as shown in (D). Normalized signal intensity ratio of anti‐pThr in respect to anti‐AHA is shown. Box plots: centerlines show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend to the minimum and maximum. N = 5 independent experiments. Welch's unpaired t‐test in comparison with Col‐0 mock conditions; *P‐value ≤ 0.05, **P‐value ≤ 0.01.
- Western blots showing chemiluminescent signals of anti‐pThr or anti‐AHA incubated membranes loaded with Arabidopsis WT (Col‐0) or cc1cc2 GFP‐CC1CC1ΔN120 root samples. Roots were treated for 8 min with either half MS or half MS + Fo5176 pSIX1::GFP hyphae. The Ponceau S panel shows total protein content. The AHA band used for quantification is highlighted with an arrowhead. Dashed line separates different treatments of the same membrane.
- Relative AHA phosphorylation status from Western blots as shown in (F). Normalized signal intensity ratio of anti‐pThr in respect to anti‐AHA is shown. Box plots as described in (E). N = 6 independent experiments. Welch's unpaired t‐test in comparison with Col‐0 mock conditions; *P‐value ≤ 0.05, **P‐value ≤ 0.01.
Source data are available online for this figure.
Figure EV5. cc1cc2 mutants rapidly acidify the media, are hypersensitive to hygromycin B, and show a delay of Fo5176‐induced media alkalinization.
- Media pH development over time when 10‐day‐old WT (Col‐0), cc1cc2, pom2‐4, and prc1‐1 plants were transferred to a liquid, alkaline media (pH 6.45). Values are mean ± SEM, N ≥ 17 plants from three independent experiments. Welch's unpaired t‐test versus Col‐0 at 6 h: cc1cc2 P‐value ≤ 0.001; pom2‐4 P‐value ≤ 0.01; prc1‐1 P‐value = 0.29.
- Representative image of WT (Col‐0) and cc1cc2 plants 7 days post‐transfer to half MS + 5 μg/ml hygromycin B plates. Scale bar = 10 mm.
- Root growth of plants grown as in (B). Values are mean ± SEM, N ≥ 15 plants from three independent experiments. Welch's unpaired t‐test; **P‐value ≤ 0.01.
- Representative image of WT (Col‐0) and cc1cc2 GFP‐CC1ΔN120 plants 7 days post‐transfer to half MS + 5 μg/ml hygromycin B plates. Scale bar = 10 mm.
- Root growth of plants grown as in (D). Values are mean ± SEM, N ≥ 16 plants from three independent experiments. Welch's unpaired t‐test; ***P‐value ≤ 0.001.
- Representative image of WT (Col‐0) and cc1cc2 plants 7 days post‐inoculation with Fo5176 pSIX1::GFP spores in liquid half MS media. Scale bar = 10 mm.
- Media pH development of an experiment as depicted in (F). Values are mean ± SEM, N = 36 plants from three independent experiments. RM two‐way ANOVA: P = 0.069 (genotype), P ≤ 0.001 (time), P ≤ 0.05 (genotype × time).
Source data are available online for this figure.
Unlike WT plants, cc1cc2 did not show any significant change of pH on both plasma membrane sides upon Fo5176 hyphae contact (Fig 5A and B). While the penultimate AHA threonine residue in WT roots was significantly more phosphorylated upon hyphae contact, this was not observed in cc1cc2 and cc1cc2 GFP‐CC1ΔN120 mutant roots (Fig 5D–G), confirming the pH measurements. To assess whether the changed plasma membrane ∆pH in cc1cc2, a consequence of hyperactive AHAs, plays a role in the long‐term response to the Fo5176 infection, we measured the media pH of liquid Fo5176 pSIX1::GFP‐infected plants at various dpi. We observed an alkalinization of the media during the infection process (Fig EV5F and G), confirming previous data reported in the pathosystem tomato‐Fusarium oxysporum f. sp. lycopersici (Masachis et al, 2016). The process was significantly delayed for media containing cc1cc2 plants and cc1cc2 plants were more vital than WT (Fig EV5F and G), implicating that the changed plasma membrane ∆pH and hyperactivation of AHAs lead to a delay in long‐term media alkalinization during Fo5176 infection.
Discussion
Under natural conditions, plants are constantly exposed to abiotic and biotic stresses. Studying how plants cope with these factors is therefore of utmost importance in times of uncertain environmental and climate changes. Here, we outline a detailed analysis of early and long‐term plant cellular responses to pathogen‐induced stress at the host plasma membrane interface. Our work demonstrates that microbe contact can induce a rapid plasma membrane AHA activation, leading to an apoplastic acidification and a concomitant change in the ΔpH at the plasma membrane (Fig 2). This fast and local pH alteration negatively affects cell elongation as well as cellulose synthesis and could be blocked by MES buffer (Figs 1 and 2, and EV2). The cortical microtubule array has been reported to concentrate on the contact site of plant cells with hyphae or a microneedle (Hardham et al, 2008), while a low degree of depolymerization was observed 75 min after treatment with VD toxin from the fungus Verticillium dahliae (Yao et al, 2011). Here, we show a rapid depolymerization of microtubules within 5 min upon contact with Fo5176 hyphae and Fo5176‐derived molecules (Fig 1). The influence of intracellular pH changes on the cortical microtubule array has recently been reported in Chlamydomonas (Liu et al, 2017). These and our observations point toward a conserved mechanism in algae and land plants, in which stress‐induced cellular pH changes alter the microtubule network. Moreover, our assays showed a simultaneous decrease of density and speed of plasma membrane located CSCs (Fig 1) that likely does not stem from microtubule depolymerization, as the CSC status is not per se affected by chemically induced microtubule depolymerization (Gutierrez et al, 2009; Endler et al, 2015). Extracellular alkalization is considered as one of the first plant responses to biotic stress as a consequence of negative AHA regulation mediated by phosphorylation. This posttranscriptional modification has been reported in Arabidopsis cell cultures exposed to the bacterial elicitor flg22 (Benschop et al, 2007; Nühse et al, 2007) and the fungal elicitor chitin seems to alkalize the apoplast following a similar molecular mechanism (Felix et al, 1993; Liu et al, 2009). In contrast, our data show a fast AHA activation, hence apoplastic acidification, in intact Arabidopsis roots that we could directly link to changes of pH at both sides of the plasma membrane with newly developed pH sensors (Fig 2). Our results add a detailed layer of spatial resolution to the analysis of subcellular pH and demonstrate a high level of cell compartmentalization and the ability of plant cells to act as a rheostat, as ΔpH changes were restricted to the plasma membrane interface (Figs 2 and EV2E), probably due to a buffering effect of the tonoplast (Pittman, 2012). Our data are in line with previously reported root surface acidification in response to the mycorrhizal fungus Piriformospora indica (Felle et al, 2009). Furthermore, we confirmed a chitin‐induced rapid apoplastic alkalinization in roots (Fig EV2H), as previously reported in leaves (Felle et al, 2009). Our work therefore broadens the “apoplastic alkalization” paradigm in response to microbes and we would rather suggest “apoplastic pH alterations” as a more accurate option. In addition, our data indicate that Fo5176 exposes or secretes a molecule with a rapid and strong capacity to acidify the host apoplast, counteracting the activity of chitin that is present in the fungal cell wall and elicitor mix. The nature of that molecule and its conservation in other fungi need to be studied further.
The fast plant cell responses to Fo5176 contact required active CC proteins (Fig 3). CC proteins and more specifically the CC1 microtubule‐binding domain are involved in plasma membrane pH regulation by altering AHA phosphorylation state, as the proton pumps showed a permanently elevated basal activity in cc1cc2 and cc1cc2 GFP‐CC1ΔN120, which did not get further activated in response to the fungus (Fig 5). Consequently, cc1cc2 mutants exhibited a more acidic apoplast and basic cortical side of the plasma membrane, could acidify a basic growth media faster, and were more sensitive to hygromycin than WT plants (Figs 5 and EV5). Salt stress was reported to alkalinize the apoplast and the cytosol (Gao et al, 2004; Geilfus, 2017). This highlights the inverse effect of biotic and abiotic stress on plant cellular pH modulation at the plasma membrane and might explain the opposite phenotypes of cc1cc2 on abiotic and biotic stress. We therefore postulate that the higher plasma membrane ΔpH of cc1cc2 is the molecular reason for their insensitivity to fungal contact (Figs 3 and 4), their inability to rearrange the CSC‐microtubule machinery in response to salt stress, and their stunted growth on salt containing media (Endler et al, 2015; Kesten et al, 2019). The acidic apoplast of cc1cc2 that does not change upon fungal contact might also explain its reduced susceptibility to Fo5176, measured as the ability of the fungus to express SIX1, reach the xylem, and inhibit root growth (Fig 4). Actually, buffered media (half MS, 5 mM MES, pH 5.7) reduced Fo5176 virulence in WT plants to cc1cc2 levels (Fig EV4), supporting previous data that reported the need of an active apoplastic pH modulation by F. oxysporum to increase infection (Masachis et al, 2016). Our plate infection method allows for visualizing and tracking pathogenic colonization of the plant vasculature and enabled us to distinguish microbe growth in the xylem from growth in other root cell layers (Fig 4). We quantified less Fo5176 xylem colonization in cc1cc2 roots, correlated with lower expression of SIX1 (Fig 4), a gene encoding a protein that contributes to the virulence of different F. oxysporum f.sp (van der Does et al, 2008a). However, the total amount of fungus was the same in cc1cc2 and WT roots (Fig 4), indicating that the cc1cc2 mutations specifically hinder Fo5176 to enter the vasculature. Since cc1cc2 has a more acidic apoplastic milieu that alkalinizes slower in response to Fo5176 (Figs 5 and EV5), this is in line with previous reports showing the need of an alkaline environment for Fo5176 to produce invasive hyphae (Masachis et al, 2016). Vascular microbes establish a compatible pathogenic interaction with the host only if they can reach xylem vessels. Indeed, non‐pathogenic endophytes, like several F. oxysporum f.sp, are unable to colonize the xylem and some of them confer protection to vascular pathogens (Brader et al, 2017). The fungal growth in cc1cc2 therefore resembles that of non‐pathogenic, endophytic F. oxysporum strains, which colonize the root cell layers, but cannot reach the xylem. The resistance to Fo5176 conferred by cc1cc2 should not interfere with the growth of beneficial endophytes or endosymbionts and might render this mutation an interesting option for agronomical applications in conditions of no salinity stress.
A second consequence of the AHA hyperactivation in cc1cc2 is a reduction of the amount of cellulose in its cell wall (Fig 4). This deficiency was restored by adding MES to the media, while the buffer did not alter the cellulose content of WT roots (Fig EV4). This confirms previous studies reporting no cellulose deficiencies in cc1cc2 etiolated hypocotyls grown on media supplemented with sucrose and MES (Endler et al, 2015; Kesten et al, 2019). Our results indicate that the pH at the plasma membrane influences cellulose content of plants. Cellulose is measured as glucose contained in the crystalline fraction of the cell wall (Yeats et al, 2016b) but the polymer has a paracrystalline structure that is influenced by hydrogen bonds between glucan chains and interactions with other polysaccharides (Kulasinski et al, 2014). We therefore hypothesize that the apoplastic pH impacts glucan chain assembly into crystalline structures, since polysaccharide interactions and protein activity in the cell wall are regulated by pH (Cosgrove, 2015). Our theory would explain why CSC density and speed was not affected by media with or without a buffering component in cc1cc2 mutants, while we observed less (crystalline) cellulose in comparison with WT plants under unbuffered conditions (Figs 3 and 4I, and EV3 and EV4). Our data could also be explained by considering that the speed of the CSCs at the plasma membrane is not only dependent on their catalytic activity since CSC speed and density is comparable between WT and cc1cc2 independently of pH conditions (i.e., buffered or unbuffered; Figs 3 and EV3). Based on the reduced cellulose content of cc1cc2 under unbuffered conditions, we would have expected a reduction of CSC density and/or speed in cc1cc2 cells, which we did not (Figs 3 and 4). The increased plasma membrane ΔpH of cc1cc2 might have an impact on the plasma membrane ionic state, CSC enzymatic activity and shape as well as on the charge state of its substrate UDP‐glucose, which in consequence might alter the activity of the complex (Colombani et al, 2004; Cho et al, 2017). In addition, the upregulated ΔpH might influence the speed of the CSCs by changing the plasma membrane lipid composition. Further research is required to fully understand the mechanism of cellulose synthesis and assembly and to elucidate the influence of apoplastic ionic state on cell wall architecture. Detailed information about the signaling cascade downstream of cellulose deficiencies still remains scarce. Low cellulose content is known to induce the expression of genes involved in plant defense (Engelsdorf et al, 2018), which we also observed in cc1cc2 grown on unbuffered media (Fig 4). In any case, the cc1cc2 cellulose deficiencies are not the reason for its low susceptibility to Fo5176, as the mutant maintained its resistance to fungal vascular colonization and still outgrew WT plants when grown on MES, while its cellulose content was restored to WT levels (Fig EV4). Moreover, the (crystalline) cellulose reduction upon fungal infection in WT plants was not altered by buffering the media and is similar to that observed in cc1cc2 plants grown on MES, despite a reduction in vascular colonization rate under these conditions (Fig EV4). This observation clearly highlights that pH regulation rather than cellulose‐based effects causes the cc1cc2 resistance to Fo5176 vascular colonization.
Our work supports and expands previous data showing the essential role of pH in plant infection by different Fusarium sps. We show that Fo5176 induces an apoplastic acidification at the beginning of the interaction (Figs 1 and 2) that might be needed for mycotoxin production (Gardiner et al, 2009). Furthermore, the acidification might enable host cell wall loosening and degradation, as cell wall loosening enzymes are activated by low pH (Cosgrove, 2015) and might reduce cellulose crystallinity (Fig EV4G). Rapid, chitin‐mediated apoplastic alkalinization (Fig EV2H, Felle et al, 2009) might therefore be a plant evolutionary response to counteract the initial pHapo drop caused by the fungus. Supporting previous data, we showed that F. oxysporum promotes virulence during plant infection (Fig EV5G), by stimulating (F)‐RALF (rapid alkalinization factor)‐dependent long‐term apoplastic alkalization (Masachis et al, 2016). Blocking this alkalinization promoted plant survival, either by buffering the media or by introducing plant mutations that induce a natural unresponsive pH at the plasma membrane interface (Figs 4E–I and EV4C–F and I; Masachis et al, 2016).
In conclusion, our data extend the current view of plant adaptation to the environment and indicate that the plasma membrane proton chemical gradient is modulated in a CC‐dependent manner. The gradient therefore might act as a decisive starting point for plant adaptation to both biotic and abiotic stress and a definitive factor in xylem colonization by F. oxysporum. We also suggest a direct influence of pH on cellulose structure that might open up potential applications for biomaterial and biofuel industries. Furthermore, we revealed a set of proteins that connects growth and defense with opposite functions upon biotic and abiotic stress, which should be kept in mind in the attempt to create stress‐tolerant plants.
Materials and Methods
Plant material and growth
Arabidopsis thaliana (Col‐0) lines expressing pUB10::SYP122‐pHusion and pUB10::pHGFP‐Lti6b (see below under “Constructs”) were transformed according to standard procedures (Hellens et al, 2000). Transgenic lines were isolated on plates containing 0.5× MS (Duchefa), 0.5% sucrose, pH 5.8 (KOH), and 0.55% phytoagar (Duchefa) supplemented with either 11.25 μg/ml sulfadiazine for pUB10::SYP122‐pHusion or 50 μg/ml hygromycin B for pUB10::pHGFP‐Lti6b. Transgenic lines were screened for 3:1 segregation of the resistance marker and fluorescence intensities of the respective sensors. The pHcyto sensor line, pUB10::pHGFP, and the cc1cc2, cc1cc2 GFP‐CC1 and cc1cc2 GFP‐CC1ΔN120 lines were published previously (Fendrych et al, 2014; Endler et al, 2015). If not stated otherwise, seedlings were grown upright on non‐buffered, solid half MS media (pH 5.7) in a 16‐h light/8‐h dark cycle at 21°C for the timeframes as indicated below.
Fungal strains, culture conditions, and elicitor mix preparation
Fusarium oxysporum Fo5176 was used throughout this study. Strain culture and storage were performed as described earlier (Di Pietro et al, 2001). Fungal elicitor mix was prepared based on a method published previously (Baldrich et al, 2014) with the following modifications: Fo5176 was grown in liquid potato dextrose broth (PDB) at 27°C in the dark for 5 days. The culture was filtered through miracloth and washed thoroughly with water to remove excess PDB. Mycelia were harvested, frozen in liquid nitrogen, and lyophilized until dry. Dried mycelia were ground to powder in a Geno/Grinder (SPEX SamplePrep, USA). The powder was diluted with distilled water to obtain a concentration of 30 mg/ml. The slurry was autoclaved for 15 min at 121°C, aliquoted, and stored at −20°C.
Generation of Fo5176 hyphae
1 ml half MS (pH 5.7) + 1% sucrose containing 107 Fo5176 or Fo5176 pSIX1::GFP spores was shaken horizontally overnight at 150 rpm in a 2‐ml tube on a rotating device. Germinated spores were spun down at 2,000 g for 5 min. Supernatant was discarded, and hyphae were washed three times with half MS to remove excess sucrose. Hyphae were used for short‐term root treatments (5–8 min) and measurement of pH changes in the media in response to Fo5176 (see below).
Constructs
pUB10::SYP122‐pHusion and pUB10::pHGFP‐Lti6b constructs were generated and assembled using GreenGate (GG) cloning (Lampropoulos et al, 2013).
For pUB10::SYP122‐pHusion, the full‐length coding sequence (CDS) of SYP122 (Syntaxin of Plants 122; At3g52400) was amplified from Arabidopsis thaliana Col‐0 cDNA with primers 1 and 2 listed in Table EV1, attaching Eco31I recognition sites and specific GG‐overhangs. The Stop codon was removed from the SYP122 CDS and the resulting 1056‐bp fragment was subcloned into pGGC000 (Lampropoulos et al, 2013). The CDS of pHusion was amplified from p16‐SYP61‐pHusion (Luo et al, 2015) with primers 3 and 4 listed in Table EV1 and the resulting 1,439‐bp fragment was subcloned into pGGD000 (Lampropoulos et al, 2013). The final construct was assembled in a GG reaction from modules listed below.
pGGA006 (UBIQUITIN10 promoter; Lampropoulos et al, 2013).
pGGB003 (B‐dummy; Lampropoulos et al, 2013).
pGGC‐SYP122 (SYP122 CDS).
pGGD‐pHusion (pHusion CDS).
pGGE001 (RBCS terminator from pea; Lampropoulos et al, 2013).
pGGF012 (pMAS:SulfRm:t35S; Lampropoulos et al, 2013).
pGGZ001 (Vector backbone with plant resistance at RB; Lampropoulos et al, 2013).
To generate pUB10::pHGFP‐Lti6b, the CDS of pHGFP was amplified from proUBQ10:pHGFP (Fendrych et al, 2014) in two parts, to remove an internal Eco31I site by introducing a silent point mutation. Eco31I recognition sites and specific GG‐overhangs were attached to amplicons with primers 5–8 listed in Table EV1. The internal Eco31I site and the Stop codon were removed from pHGFP and the resulting 676‐bp and 99‐bp fragments were subcloned into pGGC000 (Lampropoulos et al, 2013). The CDS of Lti6b was amplified from PM‐YC3.6‐Lti6b (Krebs et al, 2012) with primers 9 and 10 listed in Table EV1, and the resulting 195‐bp fragment was subcloned into pGGD000 (Lampropoulos et al, 2013). The final construct was assembled in a GG reaction from modules listed below.
pGGA006 (UBIQUITIN10 promoter; Lampropoulos et al, 2013).
pGGB003 (B‐dummy; Lampropoulos et al, 2013).
pGGC‐pHGFP (pHGFP CDS).
pGGD‐Lti6b (Lti6b CDS).
pGGE001 (RBCS terminator from pea; Lampropoulos et al, 2013).
pGGF005 (pUBQ10:HygrR:tOCS; Lampropoulos et al, 2013).
pGGZ003 (Vector backbone with plant resistance at LB; Lampropoulos et al, 2013).
For plant transformation, pUB10::SYP122‐pHusion and pUB10::pHGFP‐Lti6b were transformed into Agrobacterium tumefaciens ASE strain harboring the pSOUP plasmid.
To obtain the Fo5176 pSIX1::GFP line, the pRW2h binary vector (Houterman et al, 2008) was digested (XbaI/HindIII) and a three point ligation was performed using two inserts, which were amplified with primers 11–18 listed in Table EV1: (i) the 1 kb region downstream of the Fo5176‐SIX1 ORF, digested with XbaI/EcoRI, and (ii) the herpes simplex virus thymidine kinase (HSVtk) gene under the control of the Cochliobolus heterostrophus glyceraldehyde‐3‐phosphate dehydrogenase (ChGPD) gene promoter and the Neurospora crassa β‐tubulin gene terminator (EcoRI/HindIII). Since this vector was meant for use in a knockout experiment of SIX1, HSVtk was inserted as a conditional negative selection marker against ectopic transformants (Khang et al, 2005). Subsequently, the 1 kb region upstream of Fo5176‐SIX1 was inserted into the vector using PacI/ClaI, and the GFP coding sequence was inserted next to this promoter region using primers with ClaI and KpnI linkers. The plasmid was inserted into F. oxysporum Fo5176 by Agrobacterium‐mediated transformation as described previously (Takken et al, 2004). One ectopic transformant was selected for having low in vitro and higher in planta GFP expression.
Spinning disk live cell imaging and data processing
Five‐day‐old plant roots were covered with a 1% agarose cushion as described previously (Gutierrez et al, 2009). Treatments were added to coverslips before plant transfer and roots were subsequently placed directly into the treatment solution. 10 μl of a Fo5176 hyphae suspension containing 106 hyphae/ml in half MS (originated from overnight germination of Fo5176 spores, see above) or an elicitor mix in a 1:3.66 dilution with half MS were used as treatment. The whole process (transferring to the microscope and adjustments) took approximately 5 min.
XFP‐tagged proteins were imaged with a CSU‐W1 Yokogawa spinning disk head fitted to a Nikon Eclipse Ti‐E‐inverted microscope with a CFI PlanApo × 100 N.A. 1.40 oil immersion objective, an EM‐CCD ImageEM 1K (C9100‐14) (Hamamatsu Photonics, Japan), and a ×1.2 lens between the spinning disk and camera. GFP was imaged using a 488 nm solid‐state diode laser and a 525/50 nm emission filter; RFP was detected with a 561 nm solid‐state diode laser and a 609/54 nm emission filter. Alternatively, a CSU‐W1 Yokogawa spinning disk head fitted to a Nikon Eclipse Ti‐E‐inverted microscope with a CFI PlanApo × 100 N.A. 1.40 oil immersion objective, two iXon Ultra EM‐CCD cameras (Andor, GB), and a ×1.2 lens between the spinning disk and camera was used. For this system, GFP was imaged using a 488 nm solid‐state diode laser and a 525/50 nm emission filter; RFP was detected with a 561 nm solid‐state diode laser and a 630/75 nm emission filter and YFP was detected with a 515 nm solid‐state diode laser and a 535/30 nm emission filter. Time lapse images were processed and analyzed with Fiji (Schindelin et al, 2012). Drifts were corrected by using the plugin StackReg or MultiStackReg in cases where two channels were imaged (Thevenaz et al, 1998). Backgrounds were subtracted by the “Subtract Background” tool (rolling ball radius, 30–50 pixels). To quantify CesA velocities, three frames were averaged by “WalkingAverage” and kymograph analysis was performed with the kymograph tool of FIESTA (Ruhnow et al, 2011).
CesA and microtubule density measurements
Both methods were described earlier (Endler et al, 2015). Microtubule density measurements were done following the basic principle of the method described above but it was transferred to Fiji. Cell boundaries were detected by applying a Gaussian kernel (sigma = 1.33 μm) into each image and applying various thresholds with the Otsu algorithm (Otsu, 1979). Instead of using the mentioned Sobel edge‐detection algorithm, a Laplacian image was generated (Gaussian kernel with sigma = 1.5 μm) using FeatureJ (Erik Meijering, Biomedical Imaging Group, EPFL Lausanne). Various thresholds, based on signal to noise ratio of each individual image series, were applied to detect most microtubules and least noise pixel. The total area of microtubules was then set in relation to the cell area, which resulted in microtubule density.
Live cell ratiometric pH sensor imaging including flat and dark field correction and data processing
Five‐day‐old A. thaliana seedlings were transferred to imaging chambers as described earlier (Krebs & Schumacher, 2013). 400 μl of half MS (pH 5.7) or half MS containing 5 mM MES (pH 5.7) was used as imaging media. 150 μl fungal elicitor mix was added at the indicated time points through the opening in the lid of the imaging chambers. For hyphae treatment and growth measurements, as a minor modification, the seedlings were not glued to the surface of a coverslip but placed on top of a 1% agarose cushion as described previously (Gutierrez et al, 2009). 10 μl of a Fo5176 hyphae suspension containing 107 hyphae/ml in half MS (originated from overnight germination of Fo5176 spores, see above) was spread across the agarose sandwich and air‐dried for 5 min before seedlings were placed on top. Imaging was started immediately afterward, and the whole process (transferring the chamber to the microscope and adjustments) took approximately 5 min.
Imaging of plant roots was performed on a Leica TCS SP8‐AOBS (Leica Microsystems, Germany) confocal laser scanning microscope equipped with a Leica 10× 0.3NA HC PL Fluotar Ph1 objective. pHusion was excited simultaneously, where GFP was excited with 488 nm and detected between 500 and 545 nm and mRFP was excited with 561 nm and detected between 600 and 640 nm. pHGFP‐Lti6b and free pHGFP were excited sequentially, where GFP was first excited at 405 nm and detected between 500 and 545 nm and then excited with 488 nm and detected between 500 and 545 nm. HyD detectors were used for all image acquisitions. Image settings were kept identical for calibration and pH measurements with offsets being deactivated. Except for the standards, all images were collected as an XYt series for 30 min with a time frame of 1 min.
To collect standard curves for the pH sensors, 6–12 seedlings each were incubated for 15 min in a buffer series with pH values between 4.8 and 8.0. The buffers contained 50 mM 4‐(2‐hydroxyethyl)‐1‐piperazineethanesulfonic acid (HEPES) (pH 6.8–8.0) or 50 mM 2‐(N‐morpholino)ethanesulfonic acid (MES) (pH 5.2–6.4) and 50 mM ammonium acetate. The pH was adjusted to the desired value by adding Bis–tris propane (BTP). If necessary, the pH 5.2 buffer was additionally adjusted by adding a small amount of diluted HCl. Buffer pH 4.8 was composed of 22 mM citric acid, 27 mM trisodium citrate, and 50 mM ammonium acetate. pH was adjusted with small amounts of KOH and HCl. After incubation, seedlings were placed between a microscope slide and coverslip and imaged as described above.
Flat field images were collected as described previously (Model & Burkhardt, 2001; Model, 2006). The procedure was done for each detector‐objective combination. Dark images were acquired by setting the lightpath of the microscope to the eyepiece and therefore blocking all light to reach the detector. Subsequently, a time series of 50 images was acquired and averaged using the Z‐projection tool of Fiji in average intensity mode. Flat field images were acquired by using the following dyes and concentrations to prepare imaging slides:
7‐Diethylamino‐4‐methylcoumarin (Sigma D87759‐5G, 50 mg/ml in DMSO).
Fluorescein sodium salt (Sigma 46960‐25G‐F, 100 mg/ml in H2O).
Rose bengal (Sigma 198250‐5G, 100 mg/ml in H2O).
Brilliant Blue FCD (Sigma 80717‐100MG, 100 mg/ml in H2O).
A drop of the specific dye solution was placed between slide and coverslip. A uniform dye area was selected and the focus was set to the area just below the coverslip. Subsequently, 30 different spots were imaged with settings that used the whole range of the detector (i.e., no over‐ or underexposure). These images were averaged using the Z‐projection tool of Fiji in median intensity mode. The image was converted back to 8 bit with disabled scaling. All acquired XYt image series and standards were finally automatically corrected with a self‐written Fiji macro, which did the following modifications:
Dark images were subtracted from each single image of a series by using the Fiji plugin Calculator Plus.
Each single image of the resulting dark corrected series was divided by the median flat field image with Calculator Plus.
Each single image of the resulting dark and flat field‐corrected image series was multiplied with the average intensity of the corresponding flat field image using Calculator Plus to raise intensity back to appropriate values.
Subsequently, ratio calculations (488 nm/561 nm for pHusion, 405 nm/488 nm for pHGFP) were performed on the corrected image series. The calculations were performed automatically with a self‐written Fiji macro, which did the following operations:
A maximum projection over the whole image series was generated to estimate an average root position over time and correct for root growth and (slight) drifting of the root.
Root boundaries were detected on this maximum projection by applying a Gaussian kernel (sigma = 1.33 μm) into each image and applying various thresholds with the Otsu algorithm (Otsu, 1979). All following operations were performed in these boundaries.
For each channel, a specific LUT was applied with the set Min and Max function, which was kept constant for all corresponding image series in this manuscript. At a minimum, the range was set to 1–254 to remove all black (0) and oversaturated (255) pixel.
The macro then iterated through the slices and channels of the image series and automatically divided the mean gray value of the first channel by the corresponding second channel (e.g., 488 nm/561 nm) in each slice and exported the ratio over time.
Standard curves were calculated using sigmoidal regression (4PL fitting algorithm) with Prism (version 8, GraphPad, USA). The pH over time of the image series was subsequently calculated based on the equations of the standard curve. The replicates of individual time series were averaged and visualized with the multiple curve averaging function of OriginPro 2019 (OriginLab, USA).
Root growth analysis
Five‐day‐old seedlings were mounted into an imaging chamber and treated as described above in the “live cell ratiometric pH sensor imaging” section, and experiments were undertaken in the same manner. Root growth was measured by performing a time‐phased image subtraction with a phase shift of one frame. The detailed method was described earlier (Schneider et al, 2019). The resulting “root growth fronts” were transformed into kymographs by drawing a segmented line (line width = 2 pixel) through the tip of the root over time, followed by the kymograph plugin of Fiji. Positions of the root tip over time were extracted from the kymographs by following the kymographs with a segmented line and finally exporting the coordinates with the “save as XY Coordinates” function of Fiji.
Plate infection assay
1 × 10 cm Whatman paper strips were heat‐sterilized, and two strips placed on 12 × 12 cm square plate containing 50 ml solid half MS media. 6–10 sterilized and stratified A. thaliana seeds were placed on each strip and grown under the conditions indicated in the “Plant material and growth” section. After 8 days, the paper strip was transferred to a mock or infection plate. The infection plate was generated by equally spreading 100 μl of a Fo5176 or Fo5176 pSIX1::GFP spore suspension containing 107 spores/ml in water on a 12 × 12 cm square plate containing 50 ml solid half MS media. Each plate was scanned daily to assess root growth. Root length was measured with Fiji with a macro employing the above described method on whole plates (see “Root growth analysis”). Fo5176 pSIX1::GFP penetration of the root vasculature was assessed with a Leica M205 FCA fluorescent stereo microscope, equipped with a long pass GFP filter (ET GFP LP; Excitation nm: ET480/40x; Emission nm: ET510 LP). Vascular penetration/infection was counted when clear GFP signal was observed. In brief, we scanned each plate manually with the stereo microscope for GFP signal that showed a clear, linear, root central pattern (see Fig 4A), which is typical for the penetration of the xylem. Then, the number of xylem penetrations per root was calculated by dividing the total, cumulative number of penetrations per day by the total number of plants.
Live fungus quantification
Plants were grown and treated as described under “Plate infection assay”. Subsequently, roots of infected plants were harvested and weighed. The roots were surface‐sterilized for 1 min in 80% EtOH, followed by a second sterilization step for 1 min in 0.25% NaClO as recently described (Durán et al, 2018) to remove and kill the fungus growing at the root surface. Roots were washed three times with sterile H2O and finally taken up in 1 ml H2O/g fresh weight. The tissue was ground using the TissueLyser II (Qiagen) and glass beads for 1 min. The ground material was spread on PDB plates and incubated at 25°C in the dark for 3 days. The third washing fraction was treated equally to access if the surface sterilization was successful. Developing, fungal colonies were finally counted and set in relation to the initial measured fresh weight.
Plant cell wall and fungal chitin analysis
Plants were grown and treated as described under “Plate infection assay”, harvested, flash‐frozen, and subsequently lyophilized using a freeze‐dryer (Christ, Alpha 2–4). Lyophilized roots were ground with glass beads (2.85–3.45 mm beads, Roth, Article number A557.1) using a tissue homogenizer (Retsch MM301). Starch degradation was performed as previously described (Hostettler et al, 2011). Alcohol‐insoluble residue (AIR) production, subsequent sample preparation, hydrolysis, and data analysis for monosaccharide quantification were performed as previously described (Yeats et al, 2016a,b) with some modifications. Briefly, crystalline cellulose was calculated by subtracting glucose released by weak “matrix hydrolysis” (4% sulfuric acid at 121°C for 1 h) from glucose released by strong “Saeman hydrolysis” (72% sulfuric acid at room temperature 1 h), followed by “matrix hydrolysis”. Prior to hydrolysis, 150 μg of sedoheptulose (CarboSynth, Item Number MS139006) was added to each sample as an internal standard. 10 μl of 1:10 dilutions of hydrolyzed samples was measured on a Dionex ICS‐5000 using a ThermoFischer Scientific CarboPac PA20 column (3 × 150 mm, Product Number 060142) and accompanying CarboPac PA20 guard column (3 × 30 mm, Product Number 060144). Eluents for HPLC analysis were prepared as follows: Eluent A: water, Eluent B: 50 mM NaOH, Eluent C: 100 mM Na‐Acetate, 100 mM NaOH, Eluent D: 200 mM NaOH. All eluents were purged with and maintained under helium gas as described by the manufacturer. Column temperature was maintained at 36°C as well as a constant eluent flow of 0.4 ml/min of the following gradient to elute monosaccharides: 0–18 min 4.8% B, 95.2% A; 18–20 min linear increase to next condition; 20–30 min 50% D, 50% A; 30–40 min linear increase to next condition; 40–56 min 100% C; 56–56.1 min linear decrease to 50% D; 56.1–60 min 50% D; 60–60.1 min linear decrease to next condition; 60.1–80 min 4.8% B, 95.2% A to equilibrate column back to starting conditions. All standard curve and hydrolysis sample peaks were integrated using Chromeleon 8.0 software.
Quantification of AHA phosphorylation levels
Ten‐day‐old plants were transferred to solid half MS media plates. Their roots were treated for 8 min with 50 μl half MS or with 50 μl of a hyphae suspension containing 106 hyphae/ml in half MS (originated from overnight germination of Fo5176 spores, see above). Roots were collected and ground with a pestle in 30 μl pre‐heated (65°C) Laemmli sample buffer (Laemmli, 1970) and solubilized for 4 min. The homogenates were centrifuged at room temperature (10,000× g for 5 min), and 20 μl of the supernatant were loaded onto 4–12% (w/v) acrylamide gradient gels (Expedeon, GB). SDS–PAGE and protein transfer to nitrocellulose membranes were performed with a Trans‐Blot Turbo Transfer System (Bio‐Rad, USA) according to the manufacturer's protocol. The amount of transferred protein was analyzed by staining the membrane with Ponceau S Staining Solution (0.1% (w/v) Ponceau S in 5% (v/v) acetic acid) for 10 min and washing with distilled water to remove the background. Two polyclonal AHA antibodies were subsequently used: first, one against the region including the phosphorylated penultimate threonine residue (anti‐pThr), then one against the conserved catalytic domain of the AHA (anti‐AHA) (Hayashi et al, 2010). A goat anti‐rabbit IgG conjugated to horseradish peroxidase (AC2114, Azure Biosystems, USA) was utilized as a secondary antibody and the chemiluminescence from the horseradish peroxidase reaction with a chemiluminescence substrate (WesternBright ECL, Advansta, USA) was detected using the Chemidoc Touch Image System (Bio‐Rad, USA). The membrane was stripped with 0.5M glycine, 0.1% SDS, and 0.01% Tween‐20 (pH 2.2) and reused with the anti‐AHA antibody. The signal intensity of bands was analyzed using the “Gel function” of Fiji and processed as described previously (Kesten et al, 2016). The signal intensity of the anti‐phThr antibody was normalized with respect to the intensity of the anti‐AHA signal. Data were finally normalized to signal intensity ratios of Col‐0 in mock conditions.
Measurement of media pH
Ten‐day‐old seedlings grown as described under “Plant material and growth” were transferred into liquid half MS (pH 5.7) media. Media pH was monitored with a pH Microsensor from Mettler Toledo Inlab (USA). The proton secretion assay was performed by transferring one plant into 250 μl half MS (pH 6.45), and the change of media pH was measured over 6 h. The pH changes in the media in response to Fo5176 were assessed by transferring one plant into 1 ml liquid half MS (pH 5.7), inoculated with 50 μl hyphae suspension containing 106 hyphae/ml in half MS (originated from overnight germination of Fo5176 spores, see above), and the pH of the media was monitored until the death of the plants (6 dpi).
Hygromycin B sensitivity assay
Seven‐day‐old seedlings were transferred to plates containing solid half MS media supplemented with or without 5 μg/ml of Hygromycin (Roth, Germany). After 7 days, the plates were scanned and root growth was quantified by employing the Fiji software.
qRT–PCR
For the quantification of stress‐related genes, 10‐day‐old roots were harvested, immediately frozen in liquid nitrogen, and subsequently ground using a TissueLyser II (Qiagen) and glass beads. To quantify the expression of SIX1, plants were treated as described under “Plate infection assay”, harvested 7 days post‐infection, and then ground as described above. Total RNA was extracted using isol‐RNA lysis reagent (5 PRIME) following the manufacturer's protocol. 1 μg of total RNA per 20‐μl reaction was used to generate first‐strand cDNA using Thermo Scientific Maxima™ H Minus cDNA Synthesis Master Mix with dsDNase (Thermofisher) following the manufacturer's protocol. Primers (Table EV1, primers 19–30) (Roux et al, 2011; Masachis et al, 2016; de Souza et al, 2017) were used in the qRT–PCR to amplify corresponding cDNA sequences under the following PCR conditions: 95°C for 3 min, 40 cycles of 94°C for 10 s, 58°C for 15 s, and 72°C for 10 s, using Fast SYBR Green Master Mix (Thermofisher) in a 10‐μl reaction. The reference gene GAPDH 600b (Czechowski et al, 2005) or FoβTUB (FOXG_06228) was amplified in parallel on each plate for normalization. No‐template controls and melting curves were examined to ensure no contamination and primer–dimer formation was present. The 2ΔCT method was used to quantify the relative expression of each gene (Schmittgen & Livak, 2008).
Statistical analysis and experimental design
For statistical analyses, Welch's unpaired t‐test, repeated‐measures two‐way ANOVA, or mixed‐effects model analyses were performed using GraphPad Prism 8. For the two latter methods, we did not assume sphericity and used Greenhouse–Geisser correction. A P‐value of < 0.05 was considered as statistically significant. Statistical methods and the resulting P‐values are defined in the corresponding figure legends. For CSC speed measurements, box plots of the particle speed were plotted to give information about the total distribution (from min to max), while statistical analysis was performed on the average CSC speeds of individual roots. If the fluorescent ratios of the pH sensors (pHusion, pHGFP) were measured to be outside of the standard curve range, they were excluded from the analysis. If samples drifted during image acquisition and this could not be corrected as described above, they were excluded from the analysis.
Author contributions
CK, FMG‐A, AM, AIH, MK, KS, and CS‐R designed the research. CK, FMG‐A, AM, SD, AIH, H‐YH, and MK performed the research. CK, FMG‐A, AM, SD, and CS‐R analyzed data. SS, NT, TK, MR, MK, and KS provided essential material. CK, FMG‐A, and CS‐R wrote the article. CK, FMG‐A, MK, KS, and CS‐R revised the article.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Acknowledgements
We are grateful to G. Sancho‐Andrés, M. Fischer‐Stettler, B. Pfister, and P. van Dam for technical support. We thank S. Persson for the cc1cc2, cc1cc2 GFP‐CC1, and cc1cc2 GFP‐CC1ΔN120 lines and J.M. Pardo, A. Molina, G. Bissoli, and E. López‐Solanilla for constructive discussions. Live cell imaging was performed with equipment maintained by the Center for Microscopy and Image Analysis (University of Zurich) and Scientific Center for Optical and Electron Microscopy (ScopeM, ETH Zurich). The authors would like to thank Lotte Bald and Rainer Waadt (COS Heidelberg) for cloning pUB10:SYP122‐pHusion and pGGC‐pHGFP, respectively. We thank David Vukovic (Plückthun group/University of Zurich) for advice on imaging and data analysis. The research leading to these results has received funding from the Peter und Traudl Engelhorn‐Stiftung to C.K., Swiss National Foundation to C.S.‐R. (SNF 31003A_163065/1; A.M.), ETHZ Foundation to C.S.‐R. (0‐20172‐16; H‐Y.H.), Heinz Imhof Foundation to C.S.‐R. (2‐72160‐16; A.I.H.), Grants‐in‐Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, to T.K. (15H05956), Innovational Research Incentives Scheme Vici of The Netherlands Organisation for Scientific Research (NWO) and the Horizon program of the Netherlands Genomics Initiative through grants to M.R., German Research Foundation (Deutsche Forschungsgemeinschaft, DFG) to K.S. (SFB 1101‐TPA02).
The EMBO Journal (2019) 38: e101822
References
- Assaad FF, Qiu J‐L, Youngs H, Ehrhardt D, Zimmerli L, Kalde M, Wanner G, Peck SC, Edwards H, Ramonell K et al (2004) The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol Biol Cell 15: 5118–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldrich P, Kakar K, Siré C, Moreno AB, Berger A, García‐Chapa M, López‐Moya JJ, Riechmann JL, San Segundo B (2014) Small RNA profiling reveals regulation of Arabidopsis miR168 and heterochromatic siRNA415 in response to fungal elicitors. BMC Genom 15: 1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbez E, Dünser K, Gaidora A, Lendl T, Busch W (2017) Auxin steers root cell expansion via apoplastic pH regulation in Arabidopsis thaliana . Proc Natl Acad Sci USA 114: E4884–E4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera S, Xu Z, Luoni L, Bonza C, Doccula FG, DeMichelis MI, Morris RJ, Schwarzländer M, Costa A (2018) Cellular Ca2+ signals generate defined pH signatures in plants. Plant Cell 30: 2704–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop JJ, Mohammed S, O'Flaherty M, Heck AJR, Slijper M, Menke FLH (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis . Mol Cell Proteomics 6: 1198–1214 [DOI] [PubMed] [Google Scholar]
- Brader G, Compant S, Vescio K, Mitter B, Trognitz F, Ma L‐J, Sessitsch A (2017) Ecology and Genomic Insights into Plant‐Pathogenic and Plant‐Nonpathogenic Endophytes. Annu Rev Phytopathol 55: 61–83 [DOI] [PubMed] [Google Scholar]
- Bringmann M, Li E, Sampathkumar A, Kocabek T, Hauser M‐T, Persson S (2012) POM‐POM2/CELLULOSE SYNTHASE INTERACTING1 Is Essential for the Functional Association of Cellulose Synthase and Microtubules in Arabidopsis . Plant Cell 24: 163–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugbee BG, Salisbury FB (1985) An evaluation of MES (2(N‐Morpholino)ethanesulfonic acid) and amberlite IRC‐50 as pH buffers for nutrient solution studies. J Plant Nutr 8: 567–583 [DOI] [PubMed] [Google Scholar]
- Cho SH, Purushotham P, Fang C, Maranas C, Díaz‐Moreno SM, Bulone V, Zimmer J, Kumar M, Nixon BT (2017) Synthesis and Self‐Assembly of Cellulose Microfibrils from Reconstituted Cellulose Synthase. Plant Physiol 175: 146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombani A, Djerbi S, Bessueille L, Blomqvist K, Ohlsson A, Berglund T, Teeri TT, Bulone V (2004) In vitro synthesis of (1→3)‐β‐D‐glucan (callose) and cellulose by detergent extracts of membranes from cell suspension cultures of hybrid aspen. Cellulose 11: 313–327 [Google Scholar]
- Cosgrove DJ (2015) Plant expansins: diversity and interactions with plant cell walls. Curr Opin Plant Biol 25: 162–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ (2018) Nanoscale structure, mechanics and growth of epidermal cell walls. Curr Opin Plant Biol 46: 77–86 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR (2000) Random GFP∷cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W‐R (2005) Genome‐wide identification and testing of superior reference genes for transcript normalization in Arabidopsis . Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana . Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro A, Garcia‐Maceira FI, Meglecz E, Roncero MIG (2001) A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol Microbiol 39: 1140–1152 [PubMed] [Google Scholar]
- van der Does HC, Duyvesteijn RGE, Goltstein PM, van Schie CCN, Manders EMM, Cornelissen BJC, Rep M (2008a) Expression of effector gene SIX1 of Fusarium oxysporum requires living plant cells. Fungal Genet Biol 45: 1257–1264 [DOI] [PubMed] [Google Scholar]
- van der Does HC, Lievens B, Claes L, Houterman PM, Cornelissen BJC, Rep M (2008b) The presence of a virulence locus discriminates Fusarium oxysporum isolates causing tomato wilt from other isolates. Environ Microbiol 10: 1475–1485 [DOI] [PubMed] [Google Scholar]
- Durán P, Thiergart T, Garrido‐Oter R, Agler M, Kemen E, Schulze‐Lefert P, Hacquard S (2018) Microbial Interkingdom Interactions in Roots Promote Arabidopsis Survival. Cell 175: 973–983.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler A, Kesten C, Schneider R, Zhang Y, Ivakov A, Froehlich A, Funke N, Persson S (2015) A Mechanism for Sustained Cellulose Synthesis during Salt Stress. Cell 162: 1353–1364 [DOI] [PubMed] [Google Scholar]
- Engelsdorf T, Gigli‐Bisceglia N, Veerabagu M, McKenna JF, Vaahtera L, Augstein F, Van der Does D, Zipfel C, Hamann T (2018) The plant cell wall integrity maintenance and immune signaling systems cooperate to control stress responses in Arabidopsis thaliana . Sci Signal 11: eaao3070 [DOI] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Höfte H (2000) PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark‐grown hypocotyls of Arabidopsis . Plant Cell 12: 2409–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falhof J, Pedersen JT, Fuglsang AT, Palmgren M (2016) Plasma Membrane H(+)‐ATPase Regulation in the Center of Plant Physiology. Mol. Plant 9: 323–337 [DOI] [PubMed] [Google Scholar]
- Felix G, Regenass M, Boller T (1993) Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J. 4: 307–316 [Google Scholar]
- Felle HH (2001) pH: signal and messenger in plant cells. Plant Biol. 3: 577–591 [Google Scholar]
- Felle HH, Waller F, Molitor A, Kogel K‐H (2009) The Mycorrhiza Fungus Piriformospora indica Induces Fast Root‐Surface pH Signaling and Primes Systemic Alkalinization of the Leaf Apoplast Upon Powdery Mildew Infection. Mol Plant Microbe Interact 22: 1179–1185 [DOI] [PubMed] [Google Scholar]
- Fendrych M, Van Hautegem T, Van Durme M, Olvera‐Carrillo Y, Huysmans M, Karimi M, Lippens S, Guérin CJ, Krebs M, Schumacher K et al (2014) Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis . Curr Biol 24: 931–940 [DOI] [PubMed] [Google Scholar]
- Fendrych M, Leung J, Friml J (2016) TIR1/AFB‐Aux/IAA auxin perception mediates rapid cell wall acidification and growth of Arabidopsis hypocotyls. Elife 5: e19048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Lindner H, Robbins NE II, Dinneny JR (2016) Growing Out of Stress: the Role of Cell‐ and Organ‐Scale Growth Control in Plant Water‐Stress Responses. Plant Cell 28: 1769–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Knight MR, Trewavas AJ, Sattelmacher B, Plieth C (2004) Self‐Reporting Arabidopsis Expressing pH and [Ca2+] Indicators Unveil Ion Dynamics in the Cytoplasm and in the Apoplast under Abiotic Stress. Plant Physiol 134: 898–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner DM, Osborne S, Kazan K, Manners JM (2009) Low pH regulates the production of deoxynivalenol by Fusarium graminearum. Microbiology 155: 3149–3156 [DOI] [PubMed] [Google Scholar]
- Geilfus C‐M (2017) The pH of the Apoplast: dynamic Factor with Functional Impact Under Stress. Mol. Plant 10: 1371–1386 [DOI] [PubMed] [Google Scholar]
- Gjetting KSK, Ytting CK, Schulz A, Fuglsang AT (2012) Live imaging of intra‐ and extracellular pH in plants using pHusion, a novel genetically encoded biosensor. J Exp Bot 63: 3207–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good NE, Winget GD, Winter W, Connolly TN, Izawa S, Singh RMM (1966) Hydrogen Ion Buffers for Biological Research. Biochemistry 5: 467–477 [DOI] [PubMed] [Google Scholar]
- Gutierrez R, Lindeboom JJ, Paredez AR, Emons AMC, Ehrhardt DW (2009) Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol 11: 797–806 [DOI] [PubMed] [Google Scholar]
- Hardham AR, Takemoto D, White RG (2008) Rapid and dynamic subcellular reorganization following mechanical stimulation of Arabidopsis epidermal cells mimics responses to fungal and oomycete attack. BMC Plant Biol 8: 63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Burch HL, Nelson RB, Barrett‐Wilt G, Kline KG, Mohsin SB, Young JC, Otegui MS, Sussman MR (2010) Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J Biol Chem 285: 17918–17929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Gray WM, Sussman MR (2015) Regulation of the plasma membrane proton pump (H(+)‐ATPase) by phosphorylation. Curr Opin Plant Biol 28: 68–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Nakamura S, Takemiya A, Takahashi Y, Shimazaki K‐I, Kinoshita T (2010) Biochemical Characterization of In Vitro Phosphorylation and Dephosphorylation of the Plasma Membrane H ‐ATPase. Plant Cell Physiol 51: 1186–1196 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium‐mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hostettler C, Kölling K, Santelia D, Streb S, Kötting O, Zeeman SC (2011) Analysis of starch metabolism in chloroplasts. Methods Mol Biol 775: 387–410 [DOI] [PubMed] [Google Scholar]
- Houterman PM, Speijer D, Dekker HL, De Koster CG, Cornelissen BJC, Rep M (2007) The mixed xylem sap proteome of Fusarium oxysporum‐infected tomato plants. Mol. Plant Pathol. 8: 215–221 [DOI] [PubMed] [Google Scholar]
- Houterman PM, Cornelissen BJC, Rep M (2008) Suppression of plant resistance gene‐based immunity by a fungal effector. PLoS Pathog 4: e1000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S‐I, Kinoshita T (2017) Blue Light Regulation of Stomatal Opening and the Plasma Membrane H+‐ATPase. Plant Physiol 174: 531–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeworutzki E, Roelfsema MRG, Anschütz U, Krol E, Elzenga JTM, Felix G, Boller T, Hedrich R, Becker D (2010) Early signaling through the Arabidopsis pattern recognition receptors FLS2 and EFR involves Ca‐associated opening of plasma membrane anion channels. Plant J. 62: 367–378 [DOI] [PubMed] [Google Scholar]
- Kesten C, Schneider R, Persson S (2016) In vitro Microtubule Binding Assay and Dissociation Constant Estimation. BIO‐PROTOCOL 6: e1759 [Google Scholar]
- Kesten C, Menna A, Sánchez‐Rodríguez C (2017) Regulation of cellulose synthesis in response to stress. Curr Opin Plant Biol 40: 106–113 [DOI] [PubMed] [Google Scholar]
- Kesten C, Wallmann A, Schneider R, McFarlane HE, Diehl A, Khan GA, van Rossum B‐J, Lampugnani ER, Szymanski WG, Cremer N et al (2019) The companion of cellulose synthase 1 confers salt tolerance through a Tau‐like mechanism in plants. Nat Commun 10: 857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khang CH, Park S‐Y, Lee Y‐H, Kang S (2005) A dual selection based, targeted gene replacement tool for Magnaporthe grisea and Fusarium oxysporum. Fungal Genet Biol 42: 483–492 [DOI] [PubMed] [Google Scholar]
- Krebs M, Held K, Binder A, Hashimoto K, Den Herder G, Parniske M, Kudla J, Schumacher K (2012) FRET‐based genetically encoded sensors allow high‐resolution live cell imaging of Ca2+ dynamics. Plant J. 69: 181–192 [DOI] [PubMed] [Google Scholar]
- Krebs M, Schumacher K (2013) Live cell imaging of cytoplasmic and nuclear Ca2+ dynamics in Arabidopsis roots. Cold Spring Harb. Protoc. 2013: 776–780 [DOI] [PubMed] [Google Scholar]
- Kulasinski K, Keten S, Churakov SV, Derome D, Carmeliet J (2014) A comparative molecular dynamics study of crystalline, paracrystalline and amorphous states of cellulose. Cellulose 21: 1103–1116 [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lampropoulos A, Sutikovic Z, Wenzl C, Maegele I, Lohmann JU, Forner J (2013) GreenGate—a novel, versatile, and efficient cloning system for plant transgenesis. PLoS ONE 8: e83043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Wang W, Zong Y, Qin G, Tian S (2012) Exploring pathogenic mechanisms of Botrytis cinerea secretome under different ambient pH based on comparative proteomic analysis. J Proteome Res 11: 4249–4260 [DOI] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Fuglsang AT, Palmgren MG, Staskawicz BJ, Coaker G (2009) RIN4 Functions with Plasma Membrane H+‐ATPases to Regulate Stomatal Apertures during Pathogen Attack. PLoS Biol 7: e1000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Persson S, Sánchez‐Rodríguez C (2015) At the border: the plasma membrane‐cell wall continuum. J Exp Bot 66: 1553–1563 [DOI] [PubMed] [Google Scholar]
- Liu Y, Visetsouk M, Mynlieff M, Qin H, Lechtreck KF, Yang P (2017) H+‐ and Na+‐ elicited rapid changes of the microtubule cytoskeleton in the biflagellated green alga Chlamydomonas. Elife 6: e26002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Díaz C, Rahjoo V, Sulyok M, Ghionna V, Martín‐Vicente A, Capilla J, Di Pietro A, López‐Berges MS (2018) Fusaric acid contributes to virulence of Fusarium oxysporum on plant and mammalian hosts. Mol. Plant Pathol. 19: 440–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Scholl S, Doering A, Zhang Y, Irani NG, Di Rubbo S, Neumetzler L, Krishnamoorthy P, Van Houtte I, Mylle E et al (2015) V‐ATPase activity in the TGN/EE is required for exocytosis and recycling in Arabidopsis . Nature Plants 1: 15094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangano S, Martínez Pacheco J, Marino‐Buslje C, Estevez JM (2018) How does pH fit in with oscillating polar growth? Trends Plant Sci 23: 479–489 [DOI] [PubMed] [Google Scholar]
- Martinière A, Gibrat R, Sentenac H, Dumont X, Gaillard I, Paris N (2018) Uncovering pH at both sides of the root plasma membrane interface using noninvasive imaging. Proc Natl Acad Sci USA 115: 6488–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masachis S, Segorbe D, Turrà D, Leon‐Ruiz M, Fürst U, El Ghalid M, Leonard G, López‐Berges MS, Richards TA, Felix G et al (2016) A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nature Microbiology 1: 16043 [DOI] [PubMed] [Google Scholar]
- McFarlane HE, Döring A, Persson S (2014) The cell biology of cellulose synthesis. Annu Rev Plant Biol 65: 69–94 [DOI] [PubMed] [Google Scholar]
- Model MA, Burkhardt JK (2001) A standard for calibration and shading correction of a fluorescence microscope. Cytometry 44: 309–316 [DOI] [PubMed] [Google Scholar]
- Model MA (2006) Intensity calibration and shading correction for fluorescence microscopes. Curr. Protoc. Cytom. Chapter 10: Unit10.14 [DOI] [PubMed] [Google Scholar]
- Morris EC, Griffiths M, Golebiowska A, Mairhofer S, Burr‐Hersey J, Goh T, von Wangenheim D, Atkinson B, Sturrock CJ, Lynch JP et al (2017) Shaping 3D Root System Architecture. Curr Biol 27: R919–R930 [DOI] [PubMed] [Google Scholar]
- Moseyko N, Feldman LJ (2001) Expression of pH‐sensitive green fluorescent protein in Arabidopsis thaliana . Plant, Cell Environ 24: 557–563 [DOI] [PubMed] [Google Scholar]
- Nick P (2013) Microtubules, signalling and abiotic stress. Plant J. 75: 309–323 [DOI] [PubMed] [Google Scholar]
- Nühse TS, Bottrill AR, Jones AME, Peck SC (2007) Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 51: 931–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A, Svennelid F, Ek B, Sommarin M, Larsson C (1998) A phosphothreonine residue at the C‐terminal end of the plasma membrane H+‐ATPase is protected by fusicoccin‐induced 14‐3‐3 binding. Plant Physiol 118: 551–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu N (1979) A threshold selection method from gray‐level histograms. IEEE Trans. Syst. Man Cybern. 9: 62–66 [Google Scholar]
- Paredez AR, Somerville C, Ehrhardt D (2006) Visualization of Cellulose Synthase with Microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Pietro AD, Madrid MP, Caracuel Z, Delgado‐Jarana J, Roncero MIG (2003) Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 4: 315–325 [DOI] [PubMed] [Google Scholar]
- Pittman JK (2012) Multiple Transport Pathways for Mediating Intracellular pH Homeostasis: the Contribution of H(+)/ion Exchangers. Front. Plant Sci. 3: 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, Van Der Does HC, Meijer M, Van Wijk R, Houterman PM, Dekker HL, De Koster CG, Cornelissen BJC (2004) A small, cysteine‐rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I‐3‐mediated resistance in tomato. Mol Microbiol 53: 1373–1383 [DOI] [PubMed] [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tör M, de Vries S, Zipfel C (2011) The Arabidopsis leucine‐rich repeat receptor–like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhnow F, Zwicker D, Diez S (2011) Tracking single particles and elongated filaments with nanometer precision. Biophys J 100: 2820–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real‐time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schneider R, Sampathkumar A, Persson S (2019) Quantification of Cytoskeletal Dynamics in Time‐Lapse Recordings. Current Protocols in Plant Biology 2: e20091 [DOI] [PubMed] [Google Scholar]
- Schulte A, Lorenzen I, Böttcher M, Plieth C (2006) A novel fluorescent pH probe for expression in plants. Plant Methods 2: 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smakowska E, Kong J, Busch W, Belkhadir Y (2016) Organ‐specific regulation of growth‐defense tradeoffs by plants. Curr Opin Plant Biol 29: 129–137 [DOI] [PubMed] [Google Scholar]
- de Souza CA, Li S, Lin AZ, Boutrot F, Grossmann G, Zipfel C, Somerville SC (2017) Cellulose‐derived oligomers act as damage‐associated molecular patterns and trigger defense‐like responses. Plant Physiol 173: 2383–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal M, De Cnodder T, Simon D, Vandenbussche F, Van der Straeten D, Verbelen J‐P, Elzenga T, Vissenberg K (2011) Apoplastic alkalinization is instrumental for the inhibition of cell elongation in the Arabidopsis root by the ethylene precursor 1‐aminocyclopropane‐1‐carboxylic acid. Plant Physiol 155: 2049–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl RS, Grossl P, Bugbee B (1999) Effect of 2(N‐morpholino)ethane)sulfonic acid (MES) on the growth and tissue composition of cucumber. J Plant Nutr 22: 315–330 [Google Scholar]
- Stecker KE, Minkoff BB, Sussman MR (2014) Phosphoproteomic Analyses Reveal Early Signaling Events in the Osmotic Stress Response. Plant Physiol 165: 1171–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze H, Chanroj S (2018) Plant Endomembrane Dynamics: studies of K+/H+ Antiporters Provide Insights on the Effects of pH and Ion Homeostasis. Plant Physiol 177: 875–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken FLW, Van Wijk R, Michielse CB, Houterman PM, Ram AFJ, Cornelissen BJC (2004) A one‐step method to convert vectors into binary vectors suited for Agrobacterium‐mediated transformation. Curr Genet 45: 242–248 [DOI] [PubMed] [Google Scholar]
- Thatcher LF, Gardiner DM, Kazan K, Manners JM (2012) A highly conserved effector in Fusarium oxysporum is required for full virulence on Arabidopsis . Mol Plant Microbe Interact 25: 180–190 [DOI] [PubMed] [Google Scholar]
- Thevenaz P, Ruttimann UE, Unser M (1998) A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process 7: 27–41 [DOI] [PubMed] [Google Scholar]
- Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH (2004) Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post‐Golgi network in plant cells. Cell Struct Funct 29: 49–65 [DOI] [PubMed] [Google Scholar]
- Wang T, McFarlane HE, Persson S (2016) The impact of abiotic factors on cellulose synthesis. J Exp Bot 67: 543–552 [DOI] [PubMed] [Google Scholar]
- Wang J, Zhou L, Shi H, Chern M, Yu H, Yi H, He M, Yin J, Zhu X, Li Y et al (2018) A single transcription factor promotes both yield and immunity in rice. Science 361: 1026–1028 [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Meents MJ, McDonnell LM, Barkwill S, Sampathkumar A, Cartwright HN, Demura T, Ehrhardt DW, Samuels AL, Mansfield SD (2015) Visualization of cellulose synthases in Arabidopsis secondary cell walls. Science 350: 198–203 [DOI] [PubMed] [Google Scholar]
- Wolf S (2017) Plant cell wall signalling and receptor‐like kinases. Biochem. J 474: 471–492 [DOI] [PubMed] [Google Scholar]
- Yao L‐L, Zhou Q, Pei B‐L, Li Y‐Z (2011) Hydrogen peroxide modulates the dynamic microtubule cytoskeleton during the defence responses to Verticillium dahliae toxins in Arabidopsis . Plant, Cell Environ 34: 1586–1598 [DOI] [PubMed] [Google Scholar]
- Yeats TH, Sorek H, Wemmer DE, Somerville CR (2016a) Cellulose Deficiency Is Enhanced on Hyper Accumulation of Sucrose by a H+‐Coupled Sucrose Symporter. Plant Physiol 171: 110–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats T, Vellosillo T, Sorek N, Ibáñez AB, Bauer S (2016b) Rapid determination of cellulose, neutral sugars, and uronic acids from plant cell walls by one‐step two‐step hydrolysis and HPAEC‐PAD. Bio‐Protocol 6: e1978 [Google Scholar]
- Zamioudis C, Korteland J, Van Pelt JA, van Hamersveld M, Dombrowski N, Bai Y, Hanson J, Van Verk MC, Ling H‐Q, Schulze‐Lefert P et al (2015) Rhizobacterial volatiles and photosynthesis‐related signals coordinate MYB72 expression in Arabidopsis roots during onset of induced systemic resistance and iron‐deficiency responses. Plant J. 84: 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5


