We supplemented knowledge of the broad metabolic diversity of the Beijerinckiaceae by characterizing new members of this family that rely on lanthanides for methanol oxidation and that possess additional lanthanide-dependent enzymes. Considering that lanthanides are critical resources for many modern applications and that recovering them is expensive and puts a heavy burden on the environment, lanthanide-dependent metabolism in microorganisms is an exploding field of research. Further research into how isolated Beijerinckiaceae and other microbes utilize lanthanides is needed to increase our understanding of lanthanide-dependent metabolism. The diversity and widespread occurrence of lanthanide-dependent enzymes make it likely that lanthanide utilization varies in different taxonomic groups and is dependent on the habitat of the microbes.
KEYWORDS: Beijerinckiaceae, methylotrophy, methanol dehydrogenases, lanthanides
ABSTRACT
Methylotrophic bacteria use methanol and related C1 compounds as carbon and energy sources. Methanol dehydrogenases are essential for methanol oxidation, while lanthanides are important cofactors of many pyrroloquinoline quinone-dependent methanol dehydrogenases and related alcohol dehydrogenases. We describe here the physiological and genomic characterization of newly isolated Beijerinckiaceae bacteria that rely on lanthanides for methanol oxidation. A broad physiological diversity was indicated by the ability to metabolize a wide range of multicarbon substrates, including various sugars, and organic acids, as well as diverse C1 substrates such as methylated amines and methylated sulfur compounds. Methanol oxidation was possible only in the presence of low-mass lanthanides (La, Ce, and Nd) at submicromolar concentrations (>100 nM). In a comparison with other Beijerinckiaceae, genomic and transcriptomic analyses revealed the usage of a glutathione- and tetrahydrofolate-dependent pathway for formaldehyde oxidation and channeling methyl groups into the serine cycle for carbon assimilation. Besides a single xoxF gene, we identified two additional genes for lanthanide-dependent alcohol dehydrogenases, including one coding for an ExaF-type alcohol dehydrogenase, which was so far not known in Beijerinckiaceae. Homologs for most of the gene products of the recently postulated gene cluster linked to lanthanide utilization and transport could be detected, but for now it remains unanswered how lanthanides are sensed and taken up by our strains. Studying physiological responses to lanthanides under nonmethylotrophic conditions in these isolates as well as other organisms is necessary to gain a more complete understanding of lanthanide-dependent metabolism as a whole.
IMPORTANCE We supplemented knowledge of the broad metabolic diversity of the Beijerinckiaceae by characterizing new members of this family that rely on lanthanides for methanol oxidation and that possess additional lanthanide-dependent enzymes. Considering that lanthanides are critical resources for many modern applications and that recovering them is expensive and puts a heavy burden on the environment, lanthanide-dependent metabolism in microorganisms is an exploding field of research. Further research into how isolated Beijerinckiaceae and other microbes utilize lanthanides is needed to increase our understanding of lanthanide-dependent metabolism. The diversity and widespread occurrence of lanthanide-dependent enzymes make it likely that lanthanide utilization varies in different taxonomic groups and is dependent on the habitat of the microbes.
INTRODUCTION
Methylotrophic bacteria utilize reduced carbon substrates without carbon-carbon bonds (1) and are widespread in nature, where they participate in bottom-up carbon cycling. While methanotrophic methylotrophs are an important subgroup as they capture the majority of methane produced on Earth (119), nonmethanotrophic methylotrophs are ecologically relevant as sinks for diverse C1 compounds released during carbon turnover, including methanol released from plants and lignin cleavage (2), methylamines released during biomass breakdown in aquatic systems (3), and naturally occurring and man-made methylated sulfur species and halogenated methanes (4, 5). Previously considered to be restricted to few taxonomic groups, methylotrophic bacteria from Alpha-, Beta-, and Gammaproteobacteria, high- and low-G+C Gram-positive bacteria, and Verrucomicrobia and the NC10 phylum (6) are now known. Recent work focusing on Xox-type methanol dehydrogenases (MDHs) suggests methylotrophy or at least methylovory (that is, the supplemental use of C1 compounds as energy sources [7, 8]) in many more taxonomic groups (8), including less-well-characterized taxa such as Rokubacteria (9) and Tectomicrobia (10). In comparison to this broad taxonomic distribution of methylotrophy, the family Beijerinckiaceae within the Alphaproteobacteria represents an example of high metabolic diversity over a limited evolutionary distance (approximately 4% dissimilarity of 16S rRNA gene sequences). Members of this family include heterotrophs (Beijerinckia), facultative methylotrophs (Methylovirgula), facultative methanotrophs (Methylocapsa and Methylocella), and the USC-α clade of atmospheric methane oxidizers (11–16). Most methylotrophic Beijerinckiaceae have in common that they possess genes for Mxa- and Xox-type MDHs, which are central to methanol oxidation.
Since its discovery in the 1960s (17), the pyrroloquinoline quinone (PQQ)-containing, calcium-dependent, heterotetrameric Mxa-type MDH was considered the exclusive enzyme for methanol oxidation. The methanol oxidation activity of Xox-type MDHs was questioned, given the distant relatedness to Mxa-type MDHs (<50% amino acid sequence identity), the lack of small subunits characteristic of Mxa-type MDHs (18), and their low expression level under laboratory conditions (19). Only the observation of their dependence on rare earth elements (REEs) (lanthanides) proved their activity in nature. Supplementation with lanthanides restored methylotrophy in an mxaF mutant of Methylorubrum extorquens AM1 by stimulating the activity of its Xox-type MDH (20). REE addition promoted growth and induced the activity of XoxF in Methylobacterium radiotolerans and Bradyrhizobium (21, 22). Similar evidence was found for the Verrucomicrobia Methylacidiphilum fumariolicum SolV (23). The widespread occurrence of xoxF genes across diverse environments (18) and the finding that mxaF expression was suppressed by natural lanthanide concentrations suggest that Xox-type MDHs are ecologically highly relevant (24). A recent genome screening confirmed that Xox-type MDHs are more widespread among Proteobacteria, including nonmethylotrophic taxa, than Mxa-type MDHs (25).
While lanthanides were hypothesized to be superior to calcium in enzymes due to their stronger Lewis acidity, the evolution of lanthanide-dependent enzymes was considered unlikely given their low bioavailability (26). Lanthanides make up on average 0.015% of the Earth’s crust (27, 28), and an elevated level of lanthanides can be a consequence of volcanic activity (23, 29), coal and ore mining, or acid mine drainage (27). The overall low bioavailability of lanthanides, due to sequestration into carbonates (30) or phosphate minerals (31–33) or due to being bound to organic substances (34–36), makes dedicated uptake mechanisms in microorganisms using lanthanides necessary. Recent work with Methylorubrum extorquens PA1 and M. extorquens AM1 suggested a mechanism relying on TonB-dependent receptors for the uptake of lanthanides into the periplasm, ABC transporters for uptake into the cytoplasm, and lanthanide-binding proteins such as lanmodulin for shuttling lanthanides to XoxF and other potential lanthanide-dependent enzymes (37–40). The involvement of a TonB-dependent receptor hints toward a chelator-assisted uptake of lanthanides, similar to the case for iron and copper (41).
In the present work, we used soft coal slags, which are residuals of early-industrial mineral leaching (42) and feature an increased load of mobilizable lanthanides, as starting material for the isolation of microorganisms. We assumed that the increased availability of lanthanides might favor microbes that are able to utilize them. Our efforts led to the isolation of three facultative methylotrophic Beijerinckiaceae. The isolates share common features, such as morphology, with other members of the Beijerinckiaceae; however, phenotypic and genomic characteristics, including their metabolic diversity relating to utilizable C1 and multicarbon substrates and their dependency on lanthanides for methanol oxidation, set them apart.
RESULTS AND DISCUSSION
Phylogenetic and phylogenomic analysis.
Early-industrial soft coal slags, resulting from the extraction of aluminum sulfur minerals, were recently described as an unusual habitat for microbial life (42). We isolated three pure cultures of methylotrophic bacteria using these aluminum-rich soft coal slags as an inoculum. The usage of media containing aluminum at acidic pH to mimic the conditions in the slags had a strong selective effect, and we obtained only sporadic colonies after 6 to 8 weeks of incubation, presumably due to aluminum toxicity (43). Most of the resulting colonies featured a distinct red color (see Fig. S1 in the supplemental material), reminiscent of pink-pigmented facultative methylotrophs known from phyllosphere and plant root environments (44, 45). After repeated transfers, with methanol as a carbon source, three methylotrophic isolates (here designated CH11, RH AL1, and RH AL8) could be stably maintained on solid medium.
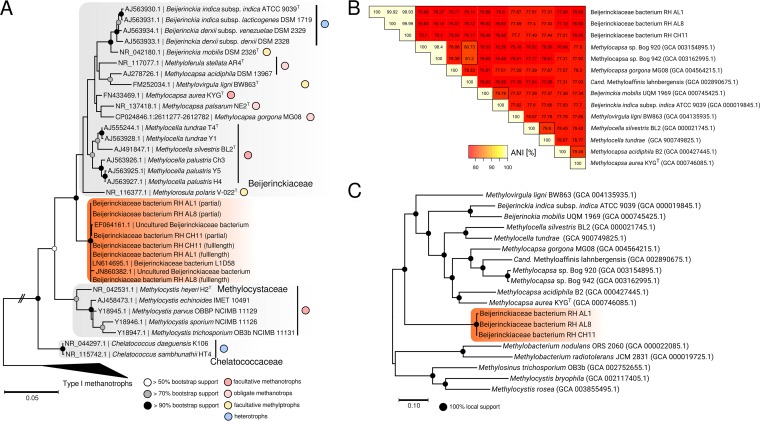
Phylogenetic analysis of 16S rRNA gene sequences obtained from colony PCR followed by Sanger sequencing placed the three isolates within the Beijerinckiaceae (Fig. 1A). The genome sequences of the three isolates were determined by high-throughput (Illumina) and long-read (Pacific Biosciences) sequencing. The obtained full-length 16S rRNA gene sequences confirmed the placement of the isolates (Fig. 1A). The closest neighbors were sequences of uncultured (GenBank accession numbers EF064161, HM843775, and JN860382) or uncharacterized (LN614695) Beijerinckiaceae (Fig. 1A), with sequence identities between 98.0 and 99.9% (see Table S1 in the supplemental material). The sources of these sequences are diverse, including acid mine drainage (EF064161), hydrothermal systems (JN860832), diabetic skin microbiomes (HM843775), and biological soil crusts in Canyonlands National Park (Utah, USA) (LN614695). The closest cultured representatives were Beijerinckia indica, Beijerinckia mobilis, Methylocapsa aurea, and Methylocapsa palsarum (95.2 to 96.2%) (Table S1). Additional blastn searches against the NCBI nonredundant nucleotide collection (see Table S2 in the supplemental material) were consistent with the results from phylogenetic tree construction and DNA distance matrix calculations (Table S1). The dissimilarity between the obtained isolates and cultured representatives of the Beijerinckiaceae suggested that our isolates form a new genus. After sequencing the genomes of the isolates, we were able to calculate average nucleotide identities (ANI) (Fig. 1B), which showed that the isolates form one species according to an ANI criterion of >95% for species delineation (46). Phylogenomics based on a concatenated alignment of 270 concatenated single-copy core genes showed a distinct grouping, supporting that the isolates constitute a new genus within the Beijerinckiaceae (Fig. 1C). We obtained additional support for this claim by determining the proportion of conserved proteins (POCP) (47) by comparing our genomes to available Beijerinckiaceae genomes. A POCP of 50% was previously introduced as a threshold for genus-level delineation (47). Our results indicated that this threshold is too low for Beijerinckiaceae (see Table S3 and Fig. S2 in the supplemental material). Instead, we found a POCP of 65% to be suitable for genus-level delineation in Beijerinckiaceae. Based on this threshold, our isolates form a new genus-level group.
FIG 1.
Phylogenetic relationship, average nucleotide identities (ANI), and phylogenomic analysis of soft coal slag methylotrophs and related Beijerinckiaceae. (A) An unrooted maximum-likelihood tree of 16S rRNA gene sequences of related Beijerinckiaceae was calculated using the Tamura-Nei model (114) and bootstrapping (n = 100) (105). Bootstrap support is indicated by circles placed on the respective nodes. Colored circles show the metabolic grouping of the corresponding organisms. The grouping is based on available literature information for M. palustris (55), M. silvestris (49), M. tundrae (50), M. acidiphila (115), M. aurea (11), M. palsarum (12), M. gorgona (13), M. stellata (116), M. polaris (59), M. ligni (14), and Beijerinckia spp. (117, 118). The scale bar refers to nucleotide substitutions. (B) A triangular matrix of pairwise ANI between soft coal slag methylotrophs and genome-sequenced relatives (assembly levels: complete, chromosome, scaffold). NCBI assembly accession numbers are given in parentheses. The color code refers to ANI in percentages. (C) A phylogenomic tree was calculated using a set of 270 shared protein sequences encoded by single-copy core genes. Alignments for individual gene products were concatenated, and the concatenated alignment was subjected to treeing using fasttree (version 2.1.8) (74) as described in Materials and Methods. Local support values are indicated by circles placed on nodes. The scale bar indicates amino acid substitutions.
C1 metabolism gene screening.
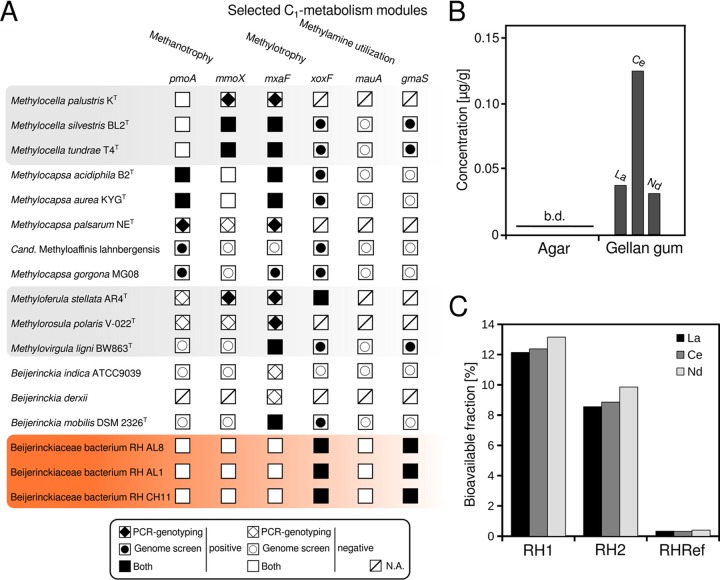
A PCR-based screening showed that the yielded isolates possess genes for only an Xox-type MDH (Fig. 2A; see Fig. S3 in the supplemental material) and not for an Mxa-type MDH. We confirmed these results by screening the sequenced genomes using custom profile hidden Markov models (pHMMs). A lack of genes for Mxa-type MDHs in methylotrophic Beijerinckiaceae is so far known only for the genome of “Candidatus Methyloaffinis lahnbergensis” (48) and two other metagenome assembled genomes of the USC-α group (13) of atmospheric methane oxidizers. Considering that these three genomes are incomplete (completeness between 72 and 88% based on the presence of single-copy core genes) (13, 48) and that the xoxF and mxaF genes were identified in the first cultivated member of the USC-α group, Methylocapsa gorgona MG08 (13), it is currently unclear whether the presence or absence of mxaF is a common feature of USC-α group members.
FIG 2.
Screening of C1 metabolism modules in Beijerinckiaceae and lanthanide availability in solidifying agents and sampled slag material. (A) The presence of selected key genes of C1 metabolism modules (methane oxidation, methanol oxidation, and methylamine utilization) in isolated soft coal slag methylotrophs was assessed by PCR and screening of sequenced genomes by using profile hidden Markov models (pHMMs) (pmoA [particulate methane monooxygenase], mmoX [soluble methane monooxygenase], mxaF [MxaF methanol dehydrogenase], xoxF [XoxF methanol dehydrogenase], mauA [methylamine dehydrogenase], and gmaS [gamma-glutamylmethylamide synthetase]). Information for related Beijerinckiaceae was deduced from available literature for M. palustris (55), M. silvestris (49), M. tundrae (50), M. acidiphila (115), M. aurea (11), M. palsarum (12), M. gorgona (13), M. stellata (116), M. polaris (59), M. ligni (14), and Beijerinckia spp. (117, 118). Whenever possible, available genome sequences of Beijerinckiaceae (assembly levels: chromosome, complete, scaffold) were used to complement literature information by screening genomes using the aforementioned set of pHMMs. Filled and empty symbols indicate the presence of the respective gene based on PCR genotyping as described in the respective references and genome screening (this study). N.A., not available (the presence of genes was not tested by PCR previously, nor is a representative genome for a pHMM-based screening yet available. (B) Lanthanide availability in solidifying agents was determined by microwave digestion followed by ICP-MS, b.d., below detection limit. (C) The bioavailable fraction of lanthanides present in the sampled slag material was assessed by total digestion plus ICP-MS to determine the absolute quantities of lanthanides present (see Table S4 in the supplemental material) and by sequential digestion plus ICP-MS. We define bioavailable here as fractions I (mobile) and II (exchangeable) according to the sequential digestion method of Zeien and Brümmer (53, 89). The nomenclature for the sampling sites is according to Wegner and Liesack (42). RH1 and RH2 refer to slag deposition sites from which the slag material that was used for isolation was collected. RHRef refers to undisturbed forest soil collected from the surroundings of sites RH1 and RH2.
We could not identify genes for particulate or soluble methane monooxygenase but found gmaS genes, encoding N-glutamylmethylamide synthetases as part of the N-methylglutamate pathway for methylamine utilization, in RH AL1, RH AL8, and RH CH11. Genes coding for the light chain of methylamine dehydrogenase (mauA) could not be identified. Methylamine utilization is not a common trait in Beijerinckiaceae, and we found gmaS genes in addition only in the genomes of Methylocella silvestris, Methylocella tundrae, and Methylovirgula ligni (Fig. 2A). Growth on methylamine was previously described for Methylocella silvestris and M. tundrae but not for Methylovirgula ligni (14, 49, 50).
Lanthanide analysis.
The lack of mxaF genes in the methylotrophic isolates hinted toward a dependency on lanthanides for methanol oxidation, as lanthanides are needed as cofactors by Xox-type MDH (18). This finding met our assumption that the soft coal material could favor lanthanide-utilizing microbes. We could stably maintain RHAL1, RHAL8, and RHCH11 on methanol-containing solid MM2 medium (1.5% gellan gum) (51), but our first attempts to grow the isolates in liquid culture using the same medium failed. Taking into account that we did not supplement media with any additional lanthanides, this observation suggested a concealed lanthanide supply in the solid medium.
We used microwave-assisted extraction combined with inductively coupled plasma mass spectrometry (ICP-MS) to determine the lanthanide content of the gellan gum and also of agar as a commonly used solidifying agent and reference (Fig. 2B). We could not detect any lanthanides in agar but found concentrations between 0.03 and 0.12 μg/g for lighter lanthanides (La, Ce, and Nd) in gellan gum, and these concentrations were high enough to allow growth without any additional lanthanides added to the medium. This finding was surprising, as gellan gum is considered a very pure solidifying agent (52). We similarly assessed the lanthanide content of the slag material that we used for isolation. Undisturbed forest soil from the surroundings of our sampling sites was used for comparison. The absolute and mobilizable contents of lanthanides were determined by total and sequential digestion followed by ICP-MS. La, Ce, and Nd were generally the most abundant lanthanides, and we saw no difference between undisturbed forest soil and slag material in terms of absolute quantities. Determined concentrations for La, Ce, and Nd ranged from 30.7 to 31.1 μg/g, 64.4 to 66.3 μg/g, and 27.6 to 30.4 μg/g, respectively (see Table S4 in the supplemental material). The bioavailable (53, 54) fraction of lanthanides was, however, 24 to 37 times higher for slag material than for undisturbed forest soil (Fig. 2C; Table S4). The slag deposits provide enough lanthanides to promote lanthanide-dependent methylotrophy, given a sufficient supply of C1 substrates.
Morphological characterization.
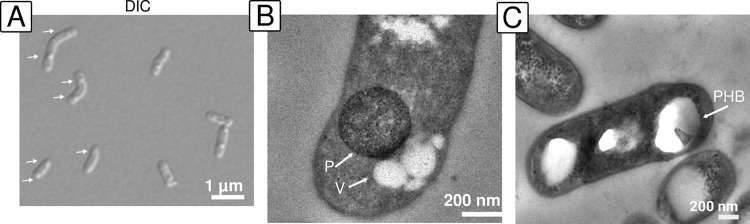
Beijerinckiaceae feature a distinct bipolar morphology (15) that is characterized by vacuoles associated with the storage of polyhydroxybutyrate (PHB). These can even be seen by light microscopy, where they appear as refractile cytoplasmic inclusions (55). Accumulating PHB is a strategy widely used by bacteria in response to nutrient limitation and other physiological stresses (56). Light microscopic examinations of the soft coal slag methylotrophs revealed a similar morphology (Fig. 3A). Transmission electron microscopy (TEM) of cell material grown on solid medium (for more than 2 weeks) with methanol but without lanthanide supplementation revealed contrast-rich, polar bodies featuring a regular, round structure. These bodies were found to commonly cooccur with smaller vesicles (Fig. 3B). PHB usually shows low contrast in TEM images due to its hydrophobicity, which prevents the binding of common stains, such as uranyl acetate or lead citrate, that are often used sequentially for enhancing contrast during TEM sample preparation. We hypothesize that these contrast-rich polar bodies might reflect a stage of starvation. When grown in liquid medium until exponential phase and with additional lanthanide supplementation (1 μM lanthanum), our strains did not show these contrast-rich polar bodies. Instead, we noticed bright polar bodies indicating the presence of accumulated PHB (Fig. 3C). This difference might be a consequence of cells being in different growth phases. Recent analysis based on energy-dispersive X-ray spectroscopy revealed that M. extorquens AM1 can store lanthanides in its cytoplasm (57).
FIG 3.
Morphology of methylotrophic isolates from soft coal slag. (A) Light microscopy of Beijerinckiaceae bacterium RH CH11 was done using differential interference contrast (DIC). Arrows point to refractile cytoplasmic inclusions. (B) Transmission electron microscopy (TEM) of Beijerinckiaceae bacterium RH AL1 grown on solid medium with methanol as a carbon source and without lanthanide supplementation for 2 weeks revealed contrast-rich polar bodies (P) partially surrounded by vesicle-like structures (V). (C) TEM analysis of Beijerinckiaceae bacterium RH AL1 during exponential growth in liquid culture with methanol as a carbon source and with additional lanthanide supplementation (1 μM lanthanum) revealed bright polar bodies, which are characteristic of PHB-storing vacuoles (PHB).
Carbon utilization.
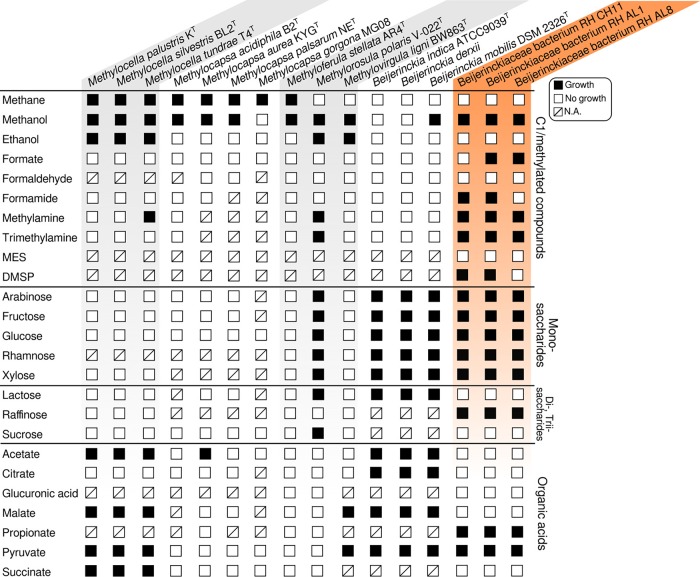
Beijerinckiaceae comprise a broad physiological diversity over a small evolutionary distance (approximately 4% dissimilarity of 16S rRNA gene sequences) (16). Incubation experiments testing various classes of carbon substrates revealed a broad substrate range, including various sugars (Fig. 4). The physiological capacity of the yielded isolates expanded well beyond methylotrophy.
FIG 4.
Carbon utilization by soft coal slag methylotrophs. The carbon utilization range was assessed by growing isolates in triplicates in MM2 medium with different carbon sources (see the text in the supplemental material). Growth was assessed against negative controls not supplemented with any carbon source. Information about other Beijerinckiaceae was taken from the literature for M. palustris (55), M. silvestris (49), M. tundrae (50), M. acidiphila (115), M. aurea (11), M. palsarum (12), M. gorgona (13), M. stellata (116), M. polaris (59), M. ligni (14), and Beijerinckia spp. (117, 118). N.A., data not available/not tested.
Utilizable C1/methylated substrates include methanol, monomethylamine for isolates RH AL1 and AL8, and trimethylamine for RH CH11 and AL8 (Fig. 4). Growth was weaker for formate, formamide, and dimethylsulfoniopropionate (DMSP) than for methanol. No growth was observed for formaldehyde. Sources of methanol in soft coal slags include demethoxylation during the breakdown of lignin components that are present. Methylamine and trimethylamine are common breakdown products of proteins and amino acids, as well as osmolytes (58). Coal tars and slags may contain anilines and related methylanilines as potential parent material for methylamines. The positive growth responses for methylamines were in line with the detection of gmaS (Fig. 3A). Weak responses to DMSP and 2-(N-morpholino)ethanesulfonic acid (MES) were unexpected, as the slag material presumably contains high quantities of lignin-derived compounds containing sulfur and methylated sulfur compounds. Considering the utilization of monosaccharides and di-/trisaccharides, the isolated strains showed a broad range of potential substrates, matched only by Methylorosula polaris (59) and the heterotroph Beijerinckia (15). Regarding organic acid utilization, we observed growth for propionate as well as pyruvate (Fig. 4).
Lanthanide-dependent methylotrophy.
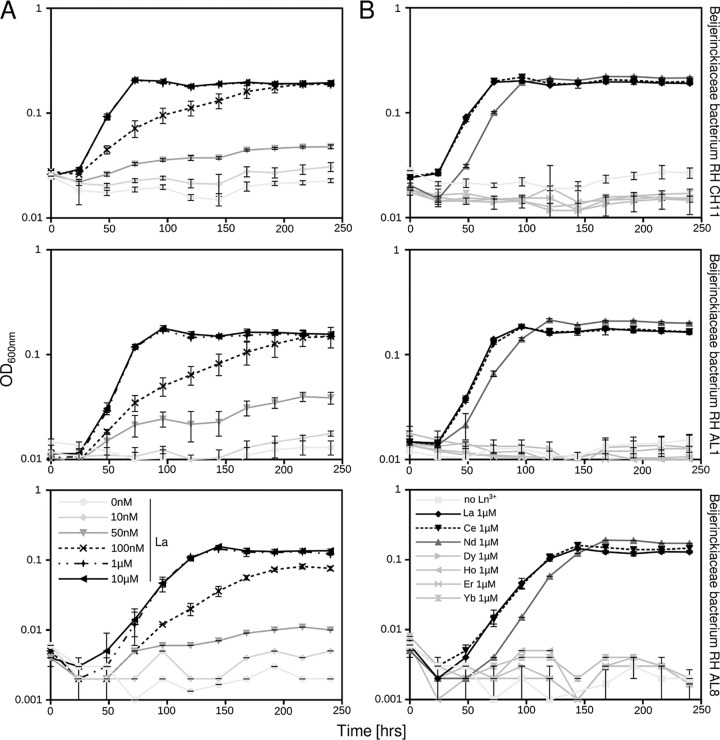
Growing cultures with different concentrations of La3+ revealed growth at concentrations higher than 100 nM (Fig. 5A). Similar to previous findings for M. extorquens AM1 (24, 60), optimal growth was observed at concentrations higher than 1 μM. Growth rates (μ) ranged between 0.021 and 0.026 h−1 with 100 nM La and between 0.046 and 0.050 h−1 with 1 μM La. The minimum concentration of lanthanides to promote (limited) growth for M. extorquens AM1 was 2.5 nM (24), which is significantly lower than what we have observed for our isolates.
FIG 5.
Methylotrophic growth dependent on lanthanide concentration (0 nM to 10 μM) (A) and species (La, Ce, Nd, Dy, Ho, Er, and Yb) (B). Soft coal slag methylotrophs were grown in MM2 medium (pH 5 to 5.5) supplemented with methanol (1% [vol/vol]) as a carbon source. Cultures were grown in triplicates (n = 3); error bars represent standard deviation. Ln3+, lanthanides; La, lanthanum; Ce, cerium; Nd, neodymium; Dy, dysprosium; Ho, holmium; Er, erbium; Yb, ytterbium.
Replacing La3+ with lanthanides of increasing atomic mass (Ce [cerium], Nd [neodymium], Dy [dysprosium], Ho [holmium], Er [erbium], and Yb [ytterbium]) showed that growth was supported only by lower-mass lanthanides (La, Ce, and Nd) (Fig. 5B). Poor growth on higher-mass lanthanides was first described by Pol et al. (23), who were among the first to describe the positive REE-mediated effect on methylotrophy in M. fumariolicum SolV. More recently, similar observations were made for Methylobacterium aquaticum (61) and for the nonmethylotrophic organism Pseudomonas putida KT2440, which utilizes an REE-dependent PQQ alcohol dehydrogenase (ADH), PedH (62). Early on, it was suggested that Xox-type MDHs are promiscuous in their use of REEs (18). Detailed structural and kinetic analyses of XoxF from M. fumariolicum SolV confirmed that although REEs are similar in properties, differences in atomic mass and ionic radii alter the coordination of the REE in the active site, thus affecting the activity and catalytic efficiency of XoxF (63).
PQQ-dependent ADHs.
Xox-type MDHs belong to the broad group of eight-bladed propeller quinoproteins and can be subdivided into at least five different clades (XoxF1 to -5) based on amino acid sequence divergence (18, 64). We observed positive PCR amplification for our isolates when using xoxF4- and xoxF5-specific primers (65). XoxF4 is until now known exclusively from Methylophilales (66). We assume that the positive amplification with xoxF4 primers was due to the inherent cross-specificity of the xoxF4 primer set for xoxF5 (65). We could confirm this using a collection of pHMMs for the different XoxF and PQQ-dependent alcohol dehydrogenase (PQQ ADH) clades to screen the isolates’ genomes (see Table S5 in the supplemental material). Assessing xoxF5 gene expression by reverse transcription-PCR, we showed expression of xoxF5 in all three strains (see Fig. S4 in the supplemental material) when grown with methanol and lanthanum. The constructed pHMMs allowed us to identify two additional PQQ ADH genes in all three isolates, one encoding a PQQ ADH type 9 and one encoding an ExaF/PedH-type ADH (Table S5). The affiliation of the three PQQ ADHs was confirmed by phylogenetic tree construction (see Fig. S5 in the supplemental material). The XoxF5 sequences of RH CH11, AL1, and AL8 were grouped together with Methylobacterium sequences, while ExaF and PQQ ADH 9 were related to Methylobacterium and Methylophaga/Methylotenera, respectively.
PedH/ExaF and PQQ ADH type 9 are both clades that rely on lanthanides as cofactors (18, 25). While there is little known about the putative function of PQQ ADH 9, ExaF was the first non-Xox-type alcohol oxidation system shown to require lanthanides for activity and to act on multicarbon substrates (67). PedH represented the first characterized lanthanide-dependent enzyme in a nonmethylotrophic organism (62).
Screening available Beijerinckiaceae genomes using the constructed pHMMs revealed a high heterogeneity with respect to encoded PQQ ADHs (see Fig. S6 in the supplemental material). Methylocella and Methylovirgula possess up to five genes encoding Mxa- and Xox-type MDHs. Beijerinckia mobilis carries five genes coding for poorly known type 4 and 9 PQQ ADHs, while RH CH11, AL1, AL8, and Rhodoblastus (68) are the only Beijerinckiaceae possessing genes for ExaF/PedH. The presence of diverse sets of genes encoding lanthanide-dependent PQQ AHDs suggests that their physiological role is not limited to methylotrophy in Beijerinckiaceae.
Genomic analysis and transcriptomics.
Genome sequencing and assembly led to draft genomes for RH AL8 and RH CH11. In the case of RH AL1, we obtained a closed, circular genome (4.23 Mb), including a plasmid (119.5 kb) (see Table S6 in the supplemental material). Genomic and transcriptome analyses of RH AL1 under methylotrophic growth conditions confirmed the physiological data and provided additional insights into C1 metabolism.
Of the three identified genes encoding lanthanide-dependent PQQ ADHs, xoxF5 showed the highest average gene expression, with 10.5 log2 reads per kilobase per million (RPKM). Values for the genes encoding PQQ ADH 9 and ExaF were lower, with 5.7 and 6.3 log2 RPKM, respectively (see Tables S7 and S8 in the supplemental material). Although we cannot rule out that ExaF and PQQ ADH 9 participate in methanol oxidation, XoxF is the dominant MDH based on gene expression data. ExaF has a substrate preference for ethanol but was shown to oxidize methanol and ethanol at similar rates and was found to function as an efficient formaldehyde dehydrogenase as well (67, 69). The latter suggests that ExaF might contribute to methanol oxidation in Beijerinckiaceae bacterium RHAL1 during formaldehyde oxidation. This could limit the accumulation of toxic formaldehyde, but without additional data, this question remains unanswered for now.
Screening the genome of RHAL1 confirmed the absence of genes linked to the tetrahydromethanopterin (H4MPT) pathway for formaldehyde oxidation. This is in contrast to most methylotrophs/methanotrophs within the Beijerinckiaceae (16), including members of the USC-α clade (13, 48). Instead, we identified fhs (formate-tetrahydrofolate ligase) and folD (bifunctional 5,10-methylene-tetrahydrofolate dehydrogenase and 5,10-methylene-tetrahydrofolate cyclohydrolase), which are part of the tetrahydrofolate pathway and are used for channeling methyl groups from formate into the serine cycle for carbon assimilation. Together with gfa (glutathione-dependent formaldehyde-activating enzyme) and frmAB (glutathione-dependent formaldehyde dehydrogenase, S-formylglutathione hydrolase), these genes encode a glutathione-dependent pathway of formaldehyde oxidation (Fig. 6A). We detected gene expression for all of these genes, and expression ranged between 5.9 (fhs) and 8.5 (frmA) log2 RPKM (Table S7). Similarly, we identified all genes of the serine cycle for carbon assimilation, and all of these genes were expressed (Tables S7 and S8). With respect to formate oxidation, we found genes coding for a formate dehydrogenase (Fdh).
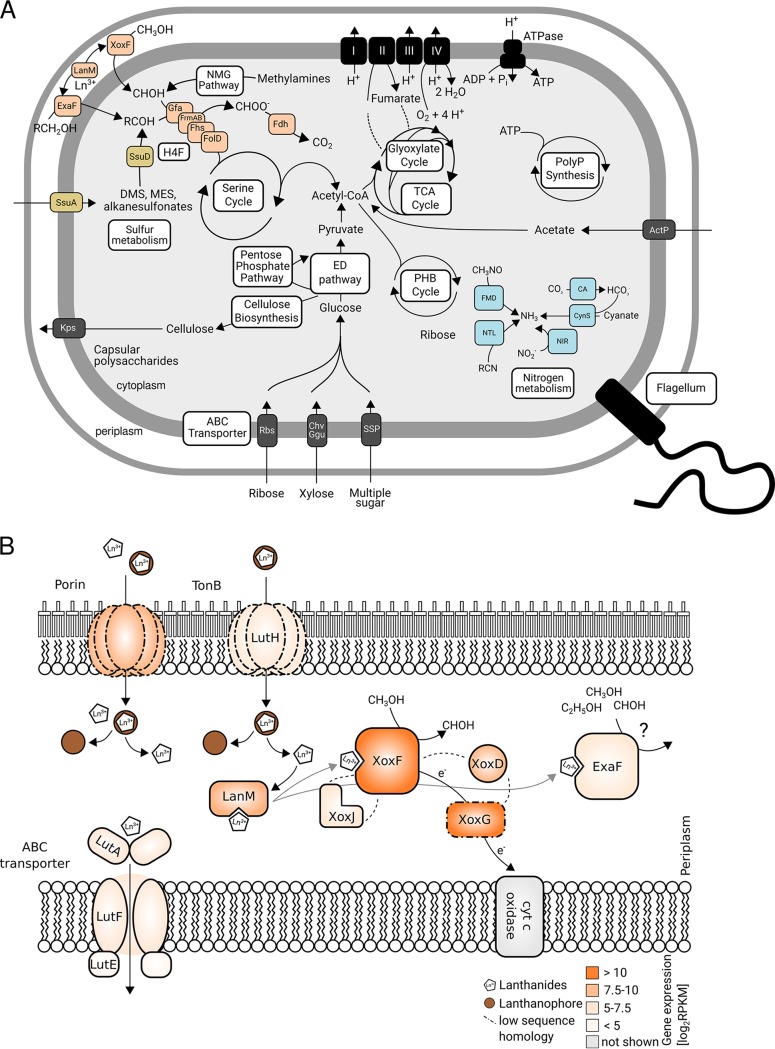
FIG 6.
Graphical representation of the genomic potential of Beijerinckiaceae bacterium RHAL1 (A) and gene expression of genes encoding products that might be involved in lanthanide-dependent metabolism (B). Methanol oxidation-related gene products are highlighted in orange, nitrogen metabolism-related gene products in blue, and sulfur metabolism-related gene products in yellow. Abbreviations: XoxF, XoxF-MDH; LanM, lanmodulin; ExaF, ExaF PQQ ADH; Gfa, glutathione-dependent formaldehyde-activating enzyme; FrmAB, glutathione-dependent formaldehyde dehydrogenase, S-formylglutathione hydrolase; Fhs, formate-tetrahydrofolate ligase; FolD, bifunctional 5,10-methylene-tetrahydrofolate dehydrogenase and 5,10-methylene-tetrahydrofolate cyclohydrolase; Fdh, formate dehydrogenase; NMG, N-methylglutamate pathway; SsuD, alkanesulfonate monooxygenase; SsuA, alkanesulfonate transporter; H4F, tetrahydrofolate; ED pathway, Entner-Doudoroff pathway; PHB, polyhydroxybutyrate; PolyP, polyphosphate; FMD, formamidase; NTL, nitrilase; CynS, cyanase; CA, carbonic anhydrase; NIR, nitrite reductase; SSP, simple sugar permease; Chv, multiple sugar-binding periplasmic receptor Chv; Ggu, multiple sugar transport system; Rbs, ribose transporter; Kps, capsular polysaccharide transporter. A list of genes, gene products, and their gene expression is given in Table S7 in the supplemental material. Gene expression data for all genes in the genome can be found in Table S8 in the supplemental material. Homologs of gene products linked to lanthanide-dependent metabolism in Beijerinckiaceae were identified by blastp (79) queries of the amino acid sequences of the corresponding gene products in M. extorquens AM1 against the amino acid sequences of all gene products of the genome of RH AL1. Detailed blast results are given in Table S9 in the supplemental material. RPKM, reads per kilobase million; LutAEF, ABC transporter linked to lanthanide uptake into the cytoplasm (LutA, ABC transporter cytoplasmic binding component; LutE, ABC transporter ATP binding component; LutF, ABC transporter membrane component); LutH, TonB-dependent receptor involved in lanthanide uptake into the periplasm; LanM, lanthanide-binding protein lanmodulin; XoxJ, periplasmic binding protein potentially involved in XoxF activation; XoxG, cL-type cytochrome accepting electrons from XoxF; XoxD, MxaD homolog interacting with XoxF and XoxG. The color code indicates the expression of genes encoding gene products linked to lanthanide-dependent metabolism. Dotted lines indicate a low degree of homology (≤40%) based on amino acid sequence identity. The presented model for lanthanide uptake and utilization was adapted based on work from the laboratories of Cotruvo (37, 39, 71), Martinez-Gomez and Skovran (57), and Vorholt (40).
We identified genes coding for respiratory complexes I to IV. With respect to central carbon metabolism, we detected full gene sets for the Entner-Doudoroff pathway, the oxidative tricarboxylic acid cycle, the glyoxylate cycle, and the pentose phosphate pathway (Fig. 6A; Table S7). These pathways, together with genes encoding multiple sugar transporters (Rbs, Chv/Ggu, and SSP), enable RHAL1 to metabolize diverse carbon sources, which confirmed our results with respect to utilizable carbon sources (Fig. 4). We also found an acetate permease gene, actP. This gene is absent in Methylocapsa gorgona MG08 and Methylocapsa acidiphila B2T (13), although both carry the necessary genes for metabolizing acetate. It was previously hypothesized that the substrate limitation of methylotrophic/methanotrophic Beijerinckiaceae is due to a lack of specific membrane transporters, and our results support this idea. The Beijerinckiaceae bacterium RHAL1 genome features an incomplete Calvin-Benson-Bassham (CBB) cycle and does not include any genes linked to nitrogen fixation. The latter is a common feature for most Beijerinckiaceae (15), including Methylocapsa gorgona MG08 of the USC-α clade of atmospheric methane oxidizers (13). The presence of complete pathways for polyphosphate and polyhydroxybutyrate (PHB) metabolism suggested that these compounds can be used for energy and carbon storage, and genes linked to both pathways were expressed under methanol oxidation conditions (Tables S7 and S8).
Beijerinckiaceae bacterium RHAL1 carries genes for nitrilases (NTLs), formidases (FMDs) and cyanases (CynS) (Fig. 6A). This indicated that nitrogen assimilation might benefit from tapping additional sources. The presence of multiple genes for dimethylsulfone and alkanesulfonate monooxygenases (SsuD) is likely an adaptation to an environment enriched in organosulfur compounds. RHAL1 possesses genes for utilizing mono-, di-, and trimethylated amines via the N-methylglutamate pathway, thus confirming our growth experiments with mono- and trimethylamine (Fig. 4). Similar to “Ca. Methyloaffinis lahnbergensis” (48), RHAL1 encodes multiple proteins linked to (exo)capsular polysaccharide production and export (Kps), implying that our isolates might be able to form biofilms or aggregates. The presence of genes coding for flagellar components (hook, filament, and motor/switch) indicated the potential motility of RHAL1.
Lanthanide utilization.
Lanthanide sensing and uptake are poorly understood, but uptake is suspected to be similar to siderophore- and chalcophore-mediated iron and copper uptake (41), involving chelators termed lanthanophores (70). In Methylorubrum extorquens, a cluster of 10 genes was identified to be involved in lanthanide utilization and transport (37, 39, 40, 57). These genes encode a TonB-dependent receptor for taking up lanthanides into the periplasm (LutH), an ABC transporter for uptake into the cytoplasm (LutAEF), multiple exported hypothetical proteins, and lanthanide-binding proteins such as lanmodulin (LanM) for shuttling lanthanides to XoxF and other potential lanthanide-dependent enzymes. A recent mutational analysis of M. extorquens AM1 identified more gene products potentially linked to lanthanide-dependent metabolism (57), including a LysR-type transcriptional regulator, XoxG, XoxJ, XoxD, a homospermidine synthase, and a porin protein. XoxG is a cL-type cytochrome functioning as an electron acceptor from XoxF. XoxJ is a poorly characterized periplasmic binding protein; however, it was recently postulated that XoxJ participates in XoxF activation (71). The genes xoxFGJ are colocalized in the genome of RH AL1. XoxD is a homolog of MxaD and presumably supports interactions between XoxF and XoxG (72).
We were able to detect homologs for most of these genes in the genome of RHAL1 (see Table S9 in the supplemental material). Transcriptome analysis showed that xoxF, xoxG, xoxD, and lanM are among the most highly expressed genes (top 10% of expressed genes; 9.0 log2 RPKM) under methanol oxidation conditions (Fig. 6B; Table S9). A biochemical characterization of XoxG in M. extorquens AM1 (71) suggested that the reduction potential of XoxG determines the range of lanthanides utilizable by the associated XoxF. Similar to M. extorquens AM1, our strains are able to use only lighter lanthanides. The homology between the XoxG proteins of M. extorquens AM1 and RH AL1 is rather low (29.5% sequence identity) (Fig. 6B; Table S9). It was shown that the XoxG phylogeny is diverse, with multiple subgroups that are partially specific for certain taxa (73), thus providing an explanation for such low homology; additionally, this diversity might be a consequence of organisms differing with respect to their range of utilizable lanthanides, likely due to the environmental conditions in their habitat. Differences in pH affect, for example, the solubility of lanthanides.
The high expression of xoxD (9.0 log2 RPKM) underscores its importance for methanol oxidation in RHAL1. Lanmodulin was the first characterized lanthanide-binding protein (37) and plays an important, but nonessential (40, 57), role in lanthanide-dependent metabolism in M. extorquens. Our gene expression data (9.6 log2 RPKM) suggest a relevance for shuttling lanthanides in RHAL1 as well (Fig. 6B). Comparing the lanmodulin of RHAL1 with that of M. extorquens AM1 by sequence analysis revealed differences. Similar to calmodulin, a ubiquitous eukaryotic calcium-binding protein, lanmodulin is an EF hand-containing protein. These motifs include a metal-binding loop, flanked by alpha helices. Lanmodulin features proline and aspartate residues in the metal-binding loop; these residues are not found in calmodulin, and they contribute to lanthanide selectivity. Like lanmodulin, the homolog in RH AL1 features four EF hand motifs, but it lacks one proline and one aspartate residue in one EF hand motif (see Fig. S7 in the supplemental material). These differences might reflect differences in the lanthanide-binding capability.
We expected that the expression of the genes encoding homologs of the assumed core components of the proposed lanthanide uptake machinery, a TonB-dependent receptor and an ABC-type transporter, would be similar to the expression of genes encoding components of the methanol oxidation machinery (xoxDFG). Instead, their gene expression was lower, not exceeding 6.6 log2 RPKM (Table S9). In case of LutH, the sequence identity of the RHAL1 homolog was comparably low (36.8%) (Fig. 6B; Table S9). With the data at hand, it is not possible to say whether the detected homologs are essential for lanthanide utilization in RHAL1. For M. extorquens AM1, it was recently shown that exogenous lanthanides repress lutH expression (57). The mutagenesis screen carried out in the same study showed that the loss of a porin gene resulted in a reduced growth rate and extended lag phase in M. extorquens AM1. Based on the phenotype of a mutant lacking the TonB-dependent receptor, it was postulated that non-chelator-bound lanthanides might enter cells through porins. Intriguingly, the RHAL1 homolog of the M. extorquens porin protein was highly expressed (9.3 log2 RPKM). Screening the transcriptome data showed that multiple genes encoding porins were expressed at a similar level (Table S8), which might indicate the passive uptake of lanthanides through porins in RHAL1. Additional experiments are necessary to answer these intriguing questions.
Concluding remarks.
In summary, we have characterized three newly isolated Beijerinckiaceae that presumably form a new genus, and we could show that they rely on lanthanides for methanol oxidation. This study presents Beijerinckiaceae isolates that possess only an Xox-type and not an Mxa-type MDH. Recent work revealed a wide taxonomic distribution of ExaF/PedH enzymes (25). Beijerinckiaceae bacterium RHAL1, AL8, CH11, and the purple nonsulfur bacterium Rhodoblastus (68) are the only members of the Beijerinckiaceae that encode ExaF/PedH PQQ ADHs. Although we detected homologs for most gene products of the postulated lanthanide utilization and transport gene cluster, the diversity and wide distribution of lanthanide-dependent enzymes suggest that multiple mechanisms for lanthanide acquisition and utilization do exist in different taxa. The metabolic diversity of our isolates allows the study of physiological responses to lanthanides under different, nonmethylotrophic conditions. Examining how lanthanides are sensed and utilized by different organisms in cellular metabolism and how far this expands beyond C1 metabolism will deepen our understanding of this uncharted territory of microbial physiology.
MATERIALS AND METHODS
Sampling and sampling site description.
Sampling was carried out as described previously (74). In brief, two different sites were sampled at the slag deposit Red Hill (RH1 [50°44′21.45′′N, 7°10′27.87′′E] and RH2 [50°44′21.85′′N, 7°10′27.78′′E]) in September 2014. The top layer, soil covering the slag material (approximately 5 cm), was removed and underlying slag material sampled in plastic containers. In addition, undisturbed nearby forest soil (Ref [50°43′49.63′′N, 7°10′30.44′′E) was sampled as a reference. Sampled slags showed, in general, a pH of 3.2 to 3.6. The pH of the reference forest soil is near neutral (pH 6.8). The slag material featured elevated levels of aluminum, iron, and sulfur (42).
Isolation.
Collected soil/slag material was used to set up slurries in 200-ml Schott bottles by mixing 10 g of sample material in 100 ml double-distilled water. Slurries were horizontally agitated on a shaker at 200 rpm and room temperature for 2 h. Agitated slurry solutions were serially diluted in 10-fold steps in basal VL55 medium (75) and used for inoculating a selection of media with different carbon sources as described in the supplemental material. After 16S rRNA-gene based identification, obtained colonies were repeatedly restreaked for purification and subsequently maintained on MM2 plates (51).
16S rRNA gene sequencing and sequence analysis.
Partial 16S rRNA gene sequences were obtained by colony PCR followed by Sanger sequencing (see the supplemental material) and subsequently analyzed in mega (version X) (76) using related representative sequences taken from SILVA (release 132) (77) as a reference. Phylogenetic tree construction for representative sequences was done by the maximum-likelihood (ML) method with the following settings: bootstrapping, 100 replicates; substitution type, nucleotide; model, Tamura-Nei; ML heuristics, nearest neighborhood-interchange; and initial tree, NJ/BioNJ. Obtained sequences were queried against SILVA (release 132) (77) with SINA (version 1.2.11) (78) and against the NCBI nonredundant nucleotide collection and the NCBI 16S rRNA sequence collection with blastn (version 2.9.0) (79). DNA distance matrices based on partial 16S rRNA gene sequence alignments were done with dnadist (version 3.697) (80).
Assaying the C1 metabolism-related functional gene repertoire.
The presence of marker genes (pmoA [particulate methane monooxygenase], mmoX [soluble methane monooxygenase], mxaF [MxaF methanol dehydrogenase], xoxF [XoxF methanol dehydrogenase], mauA [methylamine dehydrogenase], and gmaS [gamma-glutamylmethylamide synthetase]) linked to distinct functional modules of C1 metabolism (methanotrophy [pmoA and mmoX], methylotrophy [mxaF and xoxF], and methylamine utilization [mauA and gmaS]) was checked by functional gene PCR as outlined in the supplemental material and previously described (3, 11, 65, 81–86). In addition, we screened sequenced genomes and the genomes of related Beijerinckiaceae using a collection of constructed profile hidden Markov models. For these we collected available full-length amino acid sequences of gene products of interest (e.g., GmaS) from the NCBI protein database. Sequences were subsequently aligned using muscle (version 3.8.31) (87). The alignments were inspected and used for generating pHMMs with hmmbuild (version 3.1b2). These profiles were then merged and used to query the amino acid sequences of coding sequences in our sequenced genomes and related Beijerinckiaceae genomes with hmmsearch (version 3.1b2) (88).
Cultivation and strain maintenance.
Various commonly used media for methanotrophs and methylotrophs were tested for cultivating the yielded methylotrophs (51). All tested media supported growth on solid media using gellan gum as solidification agent. Liquid cultivation was possible only after supplementation with lanthanides. Isolated strains were maintained on MM2 plates and physiologically characterized in liquid MM2 medium, which contained, per liter (milligrams), KH2PO4 (100), (NH4)2SO4 (100), MgSO4·7 H2O (50), CaCl2·2H2O (20), and 1 ml trace element solution no. 1 (51) (pH 5 to 5.5). Methanol (1%, vol/vol) was added as carbon source and 1 μM Ln3+ as a necessary growth supplement. Culture purity was checked by streaking isolates regularly on diluted nutrient broth plates (no. 70122; Sigma-Aldrich, Taufkirchen, Germany) to check for the presence of contaminating satellite bacteria and by microscopic inspection before transfers.
Elemental analysis.
The total and mobilizable fractions of lanthanides in slag material (RH1 and RH2), undisturbed forest soil (RHRef [50°43′49.63′′N, 7°10′30.44′′E]), and solidifying agents (Gelrite [Carl Roth, no. 0039.1] and agar-agar [Carl Roth, no. 2266.1]) was determined by total and sequential digestion as outlined before (89, 90) in combination with ICP-MS.
Physiological characterization.
Growth was monitored by measuring the optical density at 600 nm (OD600) using a DR3900 photometer (Hach, Düsseldorf, Germany) or a Synergy H4 plate reader (Biotek, Bad Friedrichshall, Germany). Physiological characteristics, including growth promotion by various lanthanides, metabolizable C1 compounds, utilizable N sources, and usable carbon substrates, were assayed with triplicate incubation experiments. The dependence on REEs for methanol oxidation was investigated by growing isolated strains in MM2 medium with 1% (vol/vol) methanol as a carbon source plus different concentrations of La (0, 10, 50, and 100 nM and 1 and 10 μM). Various REEs (La, Ce, Dy, Nd, Ho, Er, and Yb) were tested to determine whether they promote the growth of the isolated strains by growing cultures in MM2 with 1% (vol/vol) methanol as a carbon source and the respective lanthanide (final concentration, 1 μM). Cultures were generally inoculated using precultures that were washed twice with basal medium to minimize the risk of carrying over REE. Growth was monitored spectrophotometrically at 600 nm. The range of utilizable C1 substrates was determined by replacing methanol with formaldehyde, formamide, formate, methylamine, trimethylamine, ethanol, methanesulfonic acid, or dimethylsulfoniopropionate (0.05% [vol/vol]). Metabolizable N sources were identified by replacing (NH4)2SO4 with either NaNO2, KNO3, urea, yeast extract, peptone, asparagine, glutamine, or cysteine (0.05% [wt/vol]). We observed growth for all tested N sources except glutamine. Tested multicarbon substrates (each at 0.05% [wt/vol]) included glucose, arabinose, xylose, lactose, rhamnose, raffinose, sucrose, fructose, acetate, malate, pyruvate, propionate, succinate, citrate, and glucuronic acid. Growth responses to the carbon sources were tested in triplicates in the absence of lanthanides. We define positive growth responses as a significant (paired t test, P ≤ 0.05) increase in OD600 over a time period of 2 weeks in comparison to that of a negative growth control without an added carbon source. Growth was considered weak if the increase in OD600 over 2 weeks was significantly lower (paired t test, P ≤ 0.05) than that of positive growth controls grown with 1% (vol/vol) methanol.
Reverse transcription-PCR.
Total RNA was extracted from cultures grown to the mid-exponential phase using the SV total RNA isolation system (Promega, Mannheim, Germany) according to the manufacturer’s instructions. RNA extracts were column purified, using the RNA clean and concentrator kit (Zymo Research, Freiburg, Germany), before cDNA synthesis with the GoScript reverse transcription system (Promega) using random priming. The cDNA obtained was used for xoxF-targeting PCR.
Light and electron microscopy.
Morphological characteristics were determined by light microscopy using a Zeiss Axioplan microscope (Carl Zeiss AG, Oberkochen, Germany) and agar-coated slides (91), as well as electron microscopy. For electron microscopy, cell material was fixed in 2.5% (vol/vol) glutaraldehyde in cacodylate buffer (100 mM, pH 7.4) for 2 h at 20°C and processed as described in the supplemental material.
Genome sequencing, assembly, and annotation.
Genome sequencing, assembly, and annotation were carried out as described in the supplemental material. In short, high-molecular-weight DNA from strains RH AL1, AL8, and CH11 was extracted by a column-based procedure and subsequently subjected to PacBio long-read sequencing (RH AL1) and Illumina high-throughput sequencing (RH AL8, and CH11). Genome assembly was done using canu (version 1.5) (92) and spades (version 3.13) (93). The preliminary assembly of RH AL1 with canu was refined with racon (version 1.3.3) (94) and pilon (version 1.23) (95) and indel corrected with pacbio-utilities (https://github.com/douglasgscofield/PacBio-utilities ). RH CH11 and RH AL8 were assembled with spades, and the assembly of RH AL1 was used to improve the assembly of the other two genomes with ragout (version 2.2) (96). Detailed annotations were generated with MaGe (version 3.12) (97) and prokka (version 1.13.7) (98). Gene products that might be linked to lanthanide-dependent metabolism were identified by querying the genome of RH AL1 with blastp (79) using gene products recently found to be linked to lanthanide-dependent metabolism in Methylorubrum extorquens AM1 as queries (57). Lanmodulin amino acid sequences were aligned and inspected for differences in JalView (version 2.11) (99).
Phylogenomics.
Average nucleotide identity (ANI) values were calculated using fastANI (46). The genomes of related Beijerinckiaceae were retrieved in the form of nucleotide fasta files with ncbi-genome-download (https://github.com/kblin/ncbi-genome-download) (assembly level: complete, chromosome, scaffold). Rhodoblastus genomes were not considered for ANI calculations. A phylogenomic tree using 270 single-copy core genes was calculated using fastTree (version 2.1.8) (74) with the WAG model (100), nearest-neighbor interchange (NNI) for optimizing tree topology, and the CAT approximation to account for evolutionary rate heterogeneity (101). The 270 single-copy core genes were identified by pangenomic analysis in anvio (version 5.5) (102), based on a pangenome analysis workflow described before (103). The percentage of conserved proteins (POCP) was calculated as previously described (47). Additional details about the phylogenomic analysis can be found in the supplemental material.
Analysis of PQQ ADH sequences.
We reconstructed the PQQ ADH database described by Keltjens et al. (18) by collecting corresponding sequences using the NCBI Batch Entrez tool. Sequences were subsequently aligned using muscle (version 3.8.31) (87). Phylogenetic tree construction was done with mega (version X) using the neighbor-joining method (104) and validated by bootstrapping (105). PQQ ADH/XoxF clade-specific sequence subsets were extracted from the compiled database and used for generating profile hidden Markov models as outlined above. Amino acid sequences of coding sequences in the assembled genomes and related Beijerinckiaceae genomes were queried with hmmsearch (version 3.1b2) (88) using the merged profiles. Potential hits were validated by phylogenetic tree construction based on the previously assembled PQQ ADH database.
Transcriptomics.
Beijerinckiaceae bacterium RH AL1 was grown in triplicates in MM2 medium supplemented with methanol (1% [vol/vol]) and lanthanum (1 μM) to an OD600 of 0.15. Biomass was harvested by centrifugation and subjected to RNA extraction using a protocol based on that described by Wegner et al. (106). Total RNA was enriched for mRNA by rRNA depletion using the MICROBExpress bacterial mRNA enrichment kit (Thermo Fisher Scientific, Schwerte, Germany). Transcriptome sequencing (RNA-Seq) libraries were prepared using the NEBNext Ultra II directional RNA library prep kit for Illumina (New England Biolabs, Frankfurt, Germany). Libraries were sequenced on an Illumina HiSeq 2500 platform in rapid-run mode (2 × 150 bp) by CeGaT GmbH (Tübingen, Germany). The quality of raw, demultiplexed RNA-Seq data sets was inspected using FastQC (version 0.11.7) (107). Quality filtering (settings: minlen = 75, qtrim = rl, ktrim = rl, k = 25, mink = 11, trimq = 20, and qtrim = rl) and the removal of eventually still-present adapter sequences were done with bbduk (version 38.26) (108) using the included set of common sequence contaminants and adapters. rRNA-derived sequences as well as noncoding RNA sequences were filtered out with sortmerna (version 2.1) (109) and its precompiled databases of SILVA (77) and Rfam (110). The remaining, putatively mRNA-derived sequences were mapped onto the genome of Beijerinckiaceae bacterium RH AL1 using bbmap (version 38.26) (108) (settings: slow, k = 11). The resulting bam files were sorted and indexed with samtools (version 1.3.1) (120). Read counts, meaning the number of mapped reads per coding gene, were deduced from the generated bam files using the program featureCounts implemented in subread (version 1.6.3) (111, 112). Reads per kilobase per million (RPKM) values were calculated using edgeR (version 3.24.3) (113).
Data availability.
Genome assemblies, raw genome sequencing data, and transcriptome data sets have been deposited at EBI/ENA and are available under BioProject numbers PRJEB32205 (https://www.ebi.ac.uk/ena/data/view/PRJEB32205) and PRJNA558391 (https://www.ebi.ac.uk/ena/data/view/PRJNA558391) and ArrayExpress submission E-MTAB-8184 (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-8184/), respectively. Genome assemblies can be directly accessed via the following EBI accession numbers: LR590083 (https://www.ebi.ac.uk/ena/data/view/LR590083) and LR699074 (https://www.ebi.ac.uk/ena/data/view/LR699074) (RH AL1, genome and plasmid), CABDXJ020000000 (https://www.ebi.ac.uk/ena/data/view/CABDXJ020000000) (RH AL8, assembly contigs), and CABDXI020000000 (https://www.ebi.ac.uk/ena/data/view/CABDXI020000000) (RH CH11, assembly contigs).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dirk Merten (Institute of Geosciences, Friedrich Schiller University Jena) and Monika Ballmann (Philipps University Marburg) for excellent technical assistance. Carl-Eric Wegner thanks Svetlana N. Dedysh (Russian Academy of Sciences), Rehab Z. Abdallah (MPI Marburg), and Rebecca E. Cooper (Friedrich Schiller University Jena) for helpful discussions.
We acknowledge the LABGeM (CEA/Genoscope and CNRS UMR8030) and the France Génomique and French Bioinformatics Institute national infrastructures (funded as part of the Investissement d’Avenir program managed by the Agence Nationale pour la Recherche, contracts ANR-10-INBS-09 and ANR-11-INBS-0013) for support within the MicroScope annotation platform. This research was partially supported by funds from the Collaborative Research Centre AquaDiva (CRC 1076 AquaDiva) of the Friedrich Schiller University Jena, funded by the Deutsche Forschungsgemeinschaft.
We declare no conflicting interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01830-19.
REFERENCES
- 1.Chistoserdova L, Kalyuzhnaya MG, Lidstrom ME. 2009. The expanding world of methylotrophic metabolism. Annu Rev Microbiol 63:477–499. doi: 10.1146/annurev.micro.091208.073600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warneke C, Karl T, Judmaier H, Hansel A, Jordan A, Lindinger W, Crutzen PJ. 1999. Acetone, methanol, and other partially oxidized volatile organic emissions from dead plant matter by abiological processes: significance for atmospheric HOx chemistry. Global Biogeochem Cycles 13:9–17. doi: 10.1029/98GB02428. [DOI] [Google Scholar]
- 3.Neufeld JD, Schäfer H, Cox MJ, Boden R, McDonald IR, Murrell JC. 2007. Stable-isotope probing implicates Methylophaga spp. and novel Gammaproteobacteria in marine methanol and methylamine metabolism. ISME J 1:480–491. doi: 10.1038/ismej.2007.65. [DOI] [PubMed] [Google Scholar]
- 4.Boden R, Kelly DP, Murrell JC, Schäfer H. 2010. Oxidation of dimethylsulfide to tetrathionate by Methylophaga thiooxidans sp. nov.: a new link in the sulfur cycle. Environ Microbiol 12:2688–2699. doi: 10.1111/j.1462-2920.2010.02238.x. [DOI] [PubMed] [Google Scholar]
- 5.Leisinger T, Braus-Stromeyer SA. 1995. Bacterial growth with chlorinated methanes. Environ Health Perspect 5:33–36. doi: 10.1289/ehp.95103s433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chistoserdova L, Lidstrom ME. 2013. Aerobic methylotrophic prokaryotes, p 267–285. In Rosenberg E. (ed), The prokaryotes. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 7.Sun J, Steindler L, Thrash JC, Halsey KH, Smith DP, Carter AE, Landry ZC, Giovannoni SJ. 2011. One carbon metabolism in SAR11 pelagic marine bacteria. PLoS One 6:e23973. doi: 10.1371/journal.pone.0023973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chistoserdova L, Kalyuzhnaya MG. 2018. Current trends in methylotrophy. Trends Microbiol 26:703–714. doi: 10.1016/j.tim.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Butterfield CN, Li Z, Andeer PF, Spaulding S, Thomas BC, Singh A, Hettich RL, Suttle KB, Probst AJ, Tringe SG, Northen T, Pan C, Banfield JF. 2016. Proteogenomic analyses indicate bacterial methylotrophy and archaeal heterotrophy are prevalent below the grass root zone. PeerJ 4:e2687. doi: 10.7717/peerj.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson MC, Mori T, Rückert C, Uria AR, Helf MJ, Takada K, Gernert C, Steffens UAE, Heycke N, Schmitt S, Rinke C, Helfrich EJN, Brachmann AO, Gurgui C, Wakimoto T, Kracht M, Crüsemann M, Hentschel U, Abe I, Matsunaga S, Kalinowski J, Takeyama H, Piel J. 2014. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506:58–62. doi: 10.1038/nature12959. [DOI] [PubMed] [Google Scholar]
- 11.Dunfield PF, Belova SE, Vorob'ev AV, Cornish SL, Dedysh SN. 2010. Methylocapsa aurea sp. nov., a facultative methanotroph possessing a particulate methane monooxygenase, and emended description of the genus Methylocapsa. Int J Syst Evol Microbiol 60:2659–2664. doi: 10.1099/ijs.0.020149-0. [DOI] [PubMed] [Google Scholar]
- 12.Dedysh SN, Didriksen A, Danilova OV, Belova SE, Liebner S, Svenning MM. 2015. Methylocapsa palsarum sp. nov., a methanotroph isolated from a subarctic discontinuous permafrost ecosystem. Int J Syst Evol Microbiol 65:3618–3624. doi: 10.1099/ijsem.0.000465. [DOI] [PubMed] [Google Scholar]
- 13.Tveit AT, Hestnes AG, Robinson SL, Schintlmeister A, Dedysh SN, Jehmlich N, von Bergen M, Herbold C, Wagner M, Richter A, Svenning MM. 2019. Widespread soil bacterium that oxidizes atmospheric methane. Proc Natl Acad Sci U S A 116:8515–8524. doi: 10.1073/pnas.1817812116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vorob'ev AV, de Boer W, Folman LB, Bodelier PLE, Doronina NV, Suzina NE, Trotsenko YA, Dedysh SN. 2009. Methylovirgula ligni gen. nov., sp. nov., an obligately acidophilic, facultatively methylotrophic bacterium with a highly divergent mxaF gene. Int J Syst Evol Microbiol 59:2538–2545. doi: 10.1099/ijs.0.010074-0. [DOI] [PubMed] [Google Scholar]
- 15.Haupt ES, Dedysh SN, Dunfield PF. 2016. Emended description of the family Beijerinckiaceae and transfer of the genera Chelatococcus and Camelimonas to the family Chelatococcaceae fam. nov. Int J Syst Evol Microbiol 66:3177–3182. doi: 10.1099/ijsem.0.001167. [DOI] [PubMed] [Google Scholar]
- 16.Tamas I, Smirnova AV, He Z, Dunfield PF. 2014. The (d)evolution of methanotrophy in the Beijerinckiaceae—a comparative genomics analysis. ISME J 8:369–382. doi: 10.1038/ismej.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anthony C, Zatman LJ. 1967. The microbial oxidation of methanol. The prosthetic group of the alcohol dehydrogenase of Pseudomonas sp. M27: a new oxidoreductase prosthetic group. Biochem J 104:960–969. doi: 10.1042/bj1040960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keltjens JT, Pol A, Reimann J, Op Den Camp H. 2014. PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl Microbiol Biotechnol 98:6163–6183. doi: 10.1007/s00253-014-5766-8. [DOI] [PubMed] [Google Scholar]
- 19.Chistoserdova L, Lidstrom ME. 1997. Molecular and mutational analysis of a DNA region separating two methylotrophy gene clusters in Methylobacterium extorquens AM1. Microbiology 143:1729–1736. doi: 10.1099/00221287-143-5-1729. [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa T, Mitsui R, Tani A, Sasa K, Tashiro S, Iwama T, Hayakawa T, Kawai K. 2012. A catalytic role of XoxF1 as La3+-dependent methanol dehydrogenase in Methylobacterium extorquens strain AM1. PLoS One 7:e50480–7. doi: 10.1371/journal.pone.0050480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hibi Y, Asai K, Arafuka H, Hamajima M, Iwama T, Kawai K. 2011. Molecular structure of La3+-induced methanol dehydrogenase-like protein in Methylobacterium radiotolerans. J Biosci Bioeng 111:547–549. doi: 10.1016/j.jbiosc.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 22.Fitriyanto NA, Fushimi M, Matsunaga M, Pertiwiningrum A, Iwama T, Kawai K. 2011. Molecular structure and gene analysis of Ce3+-induced methanol dehydrogenase of Bradyrhizobium sp. MAFF211645. J Biosci Bioeng 111:613–617. doi: 10.1016/j.jbiosc.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 23.Pol A, Barends TRM, Dietl A, Khadem AF, Eygensteyn J, Jetten MSM, Op den Camp H. 2014. Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ Microbiol 16:255–264. doi: 10.1111/1462-2920.12249. [DOI] [PubMed] [Google Scholar]
- 24.Vu HN, Subuyuj GA, Vijayakumar S, Good NM, Martinez-Gomez NC, Skovran E. 2016. Lanthanide-dependent regulation of methanol oxidation systems in Methylobacterium extorquens AM1 and their contribution to methanol growth. J Bacteriol 198:1250–1259. doi: 10.1128/JB.00937-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang J, Yu Z, Groom J, Cheng JF, Tarver A, Yoshikuni Y, Chistoserdova L. 2019. Rare earth element alcohol dehydrogenases widely occur among globally distributed, numerically abundant and environmentally important microbes. ISME J 13:2005–2017. doi: 10.1038/s41396-019-0414-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim S, Franklin SJ. 2004. Lanthanide-binding peptides and the enzymes that might have been. Cell Mol Life Sci 61:2184–2188. doi: 10.1007/s00018-004-4156-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Migaszewski ZM, Gałuszka A. 2015. The characteristics, occurrence, and geochemical behavior of rare earth elements in the environment: a review. Crit Rev Environ Sci Technol 45:429–471. doi: 10.1080/10643389.2013.866622. [DOI] [Google Scholar]
- 28.Picone N, Op den Camp HJ. 2019. Role of rare earth elements in methanol oxidation. Curr Opin Chem Biol 49:39–44. doi: 10.1016/j.cbpa.2018.09.019. [DOI] [PubMed] [Google Scholar]
- 29.Gammons CH, Wood SA, Pedrozo F, Varekamp JC, Nelson BJ, Shope CL, Baffico G. 2005. Hydrogeochemistry and rare earth element behavior in a volcanically acidified watershed in Patagonia, Argentina. Chem Geol 222:249–267. doi: 10.1016/j.chemgeo.2005.06.002. [DOI] [Google Scholar]
- 30.Zaharescu DG, Burghelea CI, Dontsova K, Presler JK, Maier RM, Huxman T, Domanik KJ, Hunt EA, Amistadi MK, Gaddis EE, Palacios-Menendez MA, Vaquera-Ibarra MO, Chorover J. 2017. Ecosystem composition controls the fate of rare earth elements during incipient soil genesis. Sci Rep 7:43208. doi: 10.1038/srep43208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banfield JF, Eggleton RA. 1989. Apatite replacement and rare earth mobilization, fractionation, and fixation during weathering. Clays Clay Min 37:113–127. [Google Scholar]
- 32.Firsching FH, Brune SN. 1991. Solubility products of the trivalent rare-earth phosphates. J Chem Eng Data 36:93–95. doi: 10.1021/je00001a028. [DOI] [Google Scholar]
- 33.Taunton AE, Welch SA, Banfield JF. 2000. Microbial controls on phosphate and lanthanide distributions during granite weathering and soil formation. Chem Geol 169:371–382. doi: 10.1016/S0009-2541(00)00215-1. [DOI] [Google Scholar]
- 34.Tang J, Johannesson KH. 2010. Ligand extraction of rare earth elements from aquifer sediments: implications for rare earth element complexation with organic matter in natural waters. Geochim Cosmochim Acta 74:6690–6705. doi: 10.1016/j.gca.2010.08.028. [DOI] [Google Scholar]
- 35.Marsac R, Davranche M, Gruau G, Dia A, Pédrot M, Le Coz-Bouhnik M, Briant N. 2013. Effects of Fe competition on REE binding to humic acid: origin of REE pattern variability in organic waters. Chem Geol 342:119–127. doi: 10.1016/j.chemgeo.2013.01.020. [DOI] [Google Scholar]
- 36.Vázquez-Ortega A, Huckle D, Perdrial J, Amistadi MK, Durcik M, Rasmussen C, McIntosh J, Chorover J. 2016. Solid-phase redistribution of rare earth elements in hillslope pedons subjected to different hydrologic fluxes. Chem Geol 426:1–18. doi: 10.1016/j.chemgeo.2016.01.001. [DOI] [Google Scholar]
- 37.Cotruvo JA Jr, Featherston ER, Mattocks JA, Ho JV, Laremore TN. 2018. Lanmodulin: a highly selective lanthanide-binding Protein from a lanthanide-utilizing bacterium. J Am Chem Soc 140:15056–15061. doi: 10.1021/jacs.8b09842. [DOI] [PubMed] [Google Scholar]
- 38.Cook EC, Featherston ER, Showalter SA, Cotruvo JA Jr. 2019. Structural basis for rare earth element recognition by Methylobacterium extorquens lanmodulin. Biochemistry 58:120–125. doi: 10.1021/acs.biochem.8b01019. [DOI] [PubMed] [Google Scholar]
- 39.Mattocks JA, Ho JV, Cotruvo JA Jr. 2019. A selective, protein-based fluorescent sensor with picomolar affinity for rare earth elements. J Am Chem Soc 141:2857–2861. doi: 10.1021/jacs.8b12155. [DOI] [PubMed] [Google Scholar]
- 40.Ochsner AM, Hemmerle L, Vonderach T, Nüssli R, Bortfeld-Miller M, Hattendorf B, Vorholt JA. 2019. Use of rare-earth elements in the phyllosphere colonizer Methylobacterium extorquens PA1. Mol Microbiol 111:1152–1166. doi: 10.1111/mmi.14208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Semrau JD, DiSpirito AA, Gu W, Yoon S. 2018. Metals and methanotrophy. Appl Environ Microbiol 84:e02289-17. doi: 10.1128/AEM.02289-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wegner CE, Liesack W. 2017. Unexpected dominance of elusive acidobacteria in early industrial soft coal slags. Front Microbiol 8:1023. doi: 10.3389/fmicb.2017.01023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piña RG, Cervantes C. 1996. Microbial interactions with aluminium. Biometals 9:311–316. doi: 10.1007/BF00817932. [DOI] [PubMed] [Google Scholar]
- 44.Corpe WA, Rheem S. 1989. Ecology of the methylotrophic bacteria on living leaf surfaces. FEMS Microbiol Lett 62:243–249. doi: 10.1111/j.1574-6968.1989.tb03698.x. [DOI] [Google Scholar]
- 45.Omer ZS, Tombolini R, Gerhardson B. 2004. Plant colonization by pink-pigmented facultative methylotrophic bacteria (PPFMs). FEMS Microbiol Ecol 47:319–326. doi: 10.1016/S0168-6496(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 46.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin QL, Xie BB, Zhang XY, Chen XL, Zhou BC, Zhou J, Oren A, Zhang YZ. 2014. A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol 196:2210–2215. doi: 10.1128/JB.01688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pratscher J, Vollmers J, Wiegand S, Dumont MG, Kaster AK. 2018. Unravelling the identity, metabolic potential and global biogeography of the atmospheric methane-oxidizing upland soil cluster α. Environ Microbiol 20:1016–1029. doi: 10.1111/1462-2920.14036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dunfield PF, Khmelenina VN, Suzina NE, Trotsenko YA, Dedysh SN. 2003. Methylocella silvestris sp. nov., a novel methanotroph isolated from an acidic forest cambisol. Int J Syst Evol Microbiol 53:1231–1239. doi: 10.1099/ijs.0.02481-0. [DOI] [PubMed] [Google Scholar]
- 50.Dedysh SN, Berestovskaya YY, Vasylieva LV, Belova SE, Khmelenina VN, Suzina NE, Trotsenko YA, Liesack W, Zavarzin GA. 2004. Methylocella tundrae sp. nov., a novel methanotrophic bacterium from acidic tundra peatlands. Int J Syst Evol Microbiol 54:151–156. doi: 10.1099/ijs.0.02805-0. [DOI] [PubMed] [Google Scholar]
- 51.Dedysh SN, Dunfield PF. 2014. Cultivation of methanotrophs, p 231–247. In McGenity T, Timmis K, Nogales B (ed), Hydrocarbon and lipid microbiology protocols. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 52.Dedysh SN. 2011. Cultivating uncultured bacteria from northern wetlands: knowledge gained and remaining gaps. Front Microbiol 2:184. doi: 10.3389/fmicb.2011.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weber J, Dradrach A, Karczewska A, Kocowicz A. 2018. The distribution of sequentially extracted Cu, Pb, and Zn fractions in Podzol profiles under dwarf pine of different stages of degradation in subalpine zone of Karkonosze Mts (central Europe). J Soils Sediments 18:2387–2398. doi: 10.1007/s11368-017-1715-3. [DOI] [Google Scholar]
- 54.Schmidt A, Haferburg G, Schmidt A, Lischke U, Merten D, Ghergel F, Büchel G, Kothe E. 2009. Heavy metal resistance to the extreme: Streptomyces strains from a former uranium mining area. GeoChem Explor Environ Anal 69:35–44. doi: 10.1016/j.chemer.2007.11.002. [DOI] [Google Scholar]
- 55.Dedysh SN, Liesack W, Khmelenina VN, Suzina NE, Trotsenko YA, Semrau JD, Bares AM, Panikov NS, Tiedje JM. 2000. Methylocella palustris gen. nov., sp nov., a new methane-oxidizing acidophilic bacterium from peat bags, representing a novel subtype of serine-pathway methanotrophs. Int J Syst Evol Microbiol 50:955–969. doi: 10.1099/00207713-50-3-955. [DOI] [PubMed] [Google Scholar]
- 56.Jendrossek D, Handrick R. 2002. Microbial degradation of polyhydroxyalkanoates. Annu Rev Microbiol 56:403–432. doi: 10.1146/annurev.micro.56.012302.160838. [DOI] [PubMed] [Google Scholar]
- 57.Roszczenko-Jasińska P, Vu HN, Subuyuj GA, Crisostomo RV, Cai J, Raghuraman C, Ayala EM, Clippard EJ, Lien NF, Ngo RT, Yarza F, Hoeber CA, Martinez-Gomez NC, Skovran E. 2019. Lanthanide transport, storage, and beyond: genes and processes contributing to XoxF function in Methylorubrum extorquens AM1. bioRxiv https://www.biorxiv.org/content/10.1101/647677v1.
- 58.Burg MB, Ferraris JD. 2008. Intracellular organic osmolytes: function and regulation. J Biol Chem 283:7309–7313. doi: 10.1074/jbc.R700042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berestovskaya JJ, Kotsyurbenko OR, Tourova TP, Kolganova TV, Doronina NV, Golyshin PN, Vasilyeva LV. 2012. Methylorosula polaris gen. nov., sp. nov., an aerobic, facultatively methylotrophic psychrotolerant bacterium from tundra wetland soil. Int J Syst Evol Microbiol 62:638–646. doi: 10.1099/ijs.0.007005-0. [DOI] [PubMed] [Google Scholar]
- 60.Zheng Y, Huang J, Zhao F, Chistoserdova L. 2018. Physiological effect of XoxG(4) on lanthanide-dependent methanotrophy. mBio 9:e02430-17. doi: 10.1128/mBio.02430-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masuda S, Suzuki Y, Fujitani Y, Mitsui R, Nakagawa T, Shintani M, Tani A. 2018. Lanthanide-dependent regulation of methylotrophy in Methylobacterium aquaticum strain 22A. mSphere 3:e00462-17. doi: 10.1128/mSphere.00462-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wehrmann M, Billard P, Martin-Meriadec A, Zegeye A, Klebensberger J. 2017. Functional role of lanthanides in enzymatic activity and transcriptional regulation of pyrroloquinoline quinone-dependent alcohol dehydrogenases in Pseudomonas putida KT2440. mBio 8:e00570-17. doi: 10.1128/mBio.00570-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jahn B, Pol A, Lumpe H, Barends TRM, Dietl A, Hogendoorn C, Op den Camp HJM, Daumann LJ. 2018. Similar but not the same: first kinetic and structural analyses of a methanol dehydrogenase containing a europium ion in the active site. Chembiochem 19:1147–1153. doi: 10.1002/cbic.201800130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chistoserdova L. 2011. Modularity of methylotrophy, revisited. Environ Microbiol 13:2603–2622. doi: 10.1111/j.1462-2920.2011.02464.x. [DOI] [PubMed] [Google Scholar]
- 65.Taubert M, Grob C, Howat AM, Burns OJ, Dixon JL, Chen Y, Murrell JC. 2015. XoxF encoding an alternative methanol dehydrogenase is widespread in coastal marine environments. Environ Microbiol 17:3937–3948. doi: 10.1111/1462-2920.12896. [DOI] [PubMed] [Google Scholar]
- 66.Huang J, Yu Z, Chistoserdova L. 2018. Lanthanide-dependent methanol dehydrogenases of XoxF4 and XoxF5 clades are differentially distributed among methylotrophic bacteria and they reveal different biochemical properties. Front Microbiol 9:1366. doi: 10.3389/fmicb.2018.01366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Good NM, Vu HN, Suriano CJ, Subuyuj GA, Skovran E, Martinez-Gomez NC. 2016. Pyrroloquinoline quinone ethanol dehydrogenase in Methylobacterium extorquens AM1 extends lanthanide-dependent metabolism to multicarbon substrates. J Bacteriol 198:3109–3118. doi: 10.1128/JB.00478-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kulichevskaya IS, Guzev VS, Gorlenko VM, Liesack W, Dedysh SN. 2006. Rhodoblastus sphagnicola sp. nov., a novel acidophilic purple non-sulfur bacterium from Sphagnum peat bog. Int J Syst Evol Microbiol 56:1397–1402. doi: 10.1099/ijs.0.63962-0. [DOI] [PubMed] [Google Scholar]
- 69.Good NM, Walser ON, Moore RS, Suriano C, Huff AF, Martinez-Gomez NC. 2018. Investigation of lanthanide-dependent methylotrophy uncovers complementary roles for alcohol dehydrogenase enzymes. bioRxiv https://www.biorxiv.org/content/biorxiv/early/2018/05/23/329011.
- 70.Daumann L. 2019. Essential and ubiquitous: the emergence of lanthanide metallobiochemistry. Angew Chem Int Ed Engl 58:12795–12802. doi: 10.1002/anie.201904090. [DOI] [PubMed] [Google Scholar]
- 71.Featherston ER, Rose HR, McBride MJ, Taylor E, Boal AK, Cotruvo J Jr. 2019. Biochemical and structural characterization of XoxG and XoxJ and their roles in activity of the lanthanide-dependent methanl dehydrogenase, XoxF. Chembiochem 20:2360–2372. doi: 10.1002/cbic.201900184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Toyama H, Inagaki H, Matsushita K, Anthony C, Adachi O. 2003. The role of the MxaD protein in the respiratory chain of Methylobacterium extorquens during growth on methanol. Biochim Biophys Acta 1647:372–375. doi: 10.1016/s1570-9639(03)00097-9. [DOI] [PubMed] [Google Scholar]
- 73.Yu Z, Beck DAC, Chistoserdova L. 2017. Natural selection in synthetic communities highlights the roles of Methylococcaceae and Methylophilaceae and suggests differential roles for alternative methanol dehydrogenases in methane consumption. Front Microbiol 8:2392. doi: 10.3389/fmicb.2017.02392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sait M, Hugenholtz P, Janssen PH. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ Microbiol 4:654–666. doi: 10.1046/j.1462-2920.2002.00352.x. [DOI] [PubMed] [Google Scholar]
- 76.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pruesse E, Peplies J, Glöckner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Felsenstein J. 1989. PHYLIP–phylogeny inference package (version 3.2). Cladistics 5:164–166. [Google Scholar]
- 81.Bourne DG, McDonald IR, Murrell JC. 2001. Comparison of pmoA PCR primer sets as tools for investigating methanotroph diversity in three danish soils. Appl Environ Microbiol 67:3802–3809. doi: 10.1128/aem.67.9.3802-3809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Holmes AJ, Costello A, Lidstrom ME, Murrell JC. 1995. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett 132:203–208. doi: 10.1016/0378-1097(95)00311-r. [DOI] [PubMed] [Google Scholar]
- 83.Costello AM, Lidstrom ME. 1999. Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl Environ Microbiol 65:5066–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McDonald IR, Murrell JC. 1997. The methanol dehydrogenase structural gene mxaF and its use as a functional gene probe for methanotrophs and methylotrophs. Appl Environ Microbiol 63:3218–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lau E, Fisher MC, Steudler PA, Cavanaugh CM. 2013. The methanol dehydrogenase gene, mxaF, as a functional and phylogenetic marker for proteobacterial methanotrophs in natural environments. PLoS One 8:e56993. doi: 10.1371/journal.pone.0056993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wischer D, Kumaresan D, Johnston A, Khawand ME, Stephenson J, Hillebrand-Voiculescu AM, Chen Y, Murrell JC. 2015. Bacterial metabolism of methylated amines and identification of novel methylotrophs in movile cave. ISME J 9:195–112. doi: 10.1038/ismej.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zeien H, Brümmer GW. 1989. Chemische Extraktion zur Bestimmung der Schwermetallbindungsformen in Böden. Mittlgn Dtsch Bodenkundl Gesellsch 39:505–510. [Google Scholar]
- 90.Grawunder A, Merten D, Büchel G. 2014. Origin of middle rare earth element enrichment in acid mine drainage-impacted areas. Environ Sci Pollut Res Int 21:6812–6823. doi: 10.1007/s11356-013-2107-x. [DOI] [PubMed] [Google Scholar]
- 91.Pfennig N, Wagener S. 1986. An improved method of preparing wet mounts for photomicrographs of microorganisms. J Microbiol Methods 4:303–306. doi: 10.1016/0167-7012(86)90043-6. [DOI] [Google Scholar]
- 92.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev M, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vaser R, Sović I, Nagarajan N, Šikić M. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 27:737–746. doi: 10.1101/gr.214270.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kolmogorov M, Raney B, Paten B, Pham S. 2014. Ragout—a reference-assisted assembly tool for bacterial genomes. Bioinformatics 30:I302–I309. doi: 10.1093/bioinformatics/btu280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vallenet D, Engelen S, Mornico D, Cruveiller S, Fleury L, Lajus A, Rouy Z, Roche D, Salvignol G, Scarpelli C, Médigue C. 2009. MicroScope: a platform for microbial genome annotation and comparative genomics. Database (Oxford) 2009:bap021. doi: 10.1093/database/bap021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 99.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Whelan S, Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 101.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 102.Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO. 2015. Anvi’o: an advanced analysis and visualization platform for ’omics data. PeerJ 3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Delmont TO, Eren AM. 2018. Linking pangenomes and metagenomes: the Prochlorococcus metapangenome. PeerJ 6:e4320. doi: 10.7717/peerj.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 105.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 106.Wegner CE, Richter-Heitmann T, Klindworth A, Klockow C, Richter M, Achstetter T, Glöckner FO, Harder J. 2013. Expression of sulfatases in Rhodopirellula baltica and the diversity of sulfatases in the genus Rhodopirellula. Mar Genomics 9:51–61. doi: 10.1016/j.margen.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 107.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 108.Bushnell B. 2016. BBMap short read aligner. https://www.sourceforge.net/projects/bbmap/.
- 109.Kopylova E, Noé L, Touzet H, Noe L, Touzet H. 2012. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 110.Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, Eddy SR, Gardner PP, Bateman A. 2013. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res 41:D226–D232. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liao Y, Smyth GK, Shi W. 2013. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 113.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 115.Dedysh SN, Khmelenina VN, Suzina NE, Trotsenko YA, Semrau JD, Liesack W, Tiedje JM. 2002. Methylocapsa acidiphila gen. nov., sp. nov., a novel methane-oxidizing and dinitrogen-fixing acidophilic bacterium from Sphagnum bog. Int J Syst Evol Microbiol 52:251–261. doi: 10.1099/00207713-52-1-251. [DOI] [PubMed] [Google Scholar]
- 116.Vorobev AV, Baani M, Doronina NV, Brady AL, Liesack W, Dunfield PF, Dedysh SN. 2011. Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium that possesses only a soluble methane monooxygenase. Int J Syst Evol Microbiol 61:2456–2463. doi: 10.1099/ijs.0.028118-0. [DOI] [PubMed] [Google Scholar]
- 117.Kennedy C. 2005. Genus I, Beijerinckia Derx 1950a, 145, p 423–433. In Garrity G, Brenner DJ, Krieg NR, Staley JT (ed), Bergey’s manual of systematic bacteriology, vol 2. The Proteobacteria (Part C). Springer, New York, NY. [Google Scholar]
- 118.Dedysh SN, Smirnova KV, Khmelenina VN, Suzina NE, Liesack W, Trotsenko YA. 2005. Methylotrophic autotrophy in Beijerinckia mobilis. J Bacteriol 187:3884–3888. doi: 10.1128/JB.187.11.3884-3888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Trotsenko YA, Murrell JC. 2008. Metabolic aspects of aerobic obligate methanotrophy. Adv Appl Microbiol 63:183–229. doi: 10.1016/S0065-2164(07)00005-6. [DOI] [PubMed] [Google Scholar]
- 120.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome assemblies, raw genome sequencing data, and transcriptome data sets have been deposited at EBI/ENA and are available under BioProject numbers PRJEB32205 (https://www.ebi.ac.uk/ena/data/view/PRJEB32205) and PRJNA558391 (https://www.ebi.ac.uk/ena/data/view/PRJNA558391) and ArrayExpress submission E-MTAB-8184 (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-8184/), respectively. Genome assemblies can be directly accessed via the following EBI accession numbers: LR590083 (https://www.ebi.ac.uk/ena/data/view/LR590083) and LR699074 (https://www.ebi.ac.uk/ena/data/view/LR699074) (RH AL1, genome and plasmid), CABDXJ020000000 (https://www.ebi.ac.uk/ena/data/view/CABDXJ020000000) (RH AL8, assembly contigs), and CABDXI020000000 (https://www.ebi.ac.uk/ena/data/view/CABDXI020000000) (RH CH11, assembly contigs).