Abstract
Staphylococcus aureus (S. aureus) causes persistent biofilm-related infections. Biofilm formation by S. aureus is affected by the culture conditions and is associated with certain genotypic characteristics. Here, we show that glucose and sodium chloride (NaCl) supplementation of culture media, a common practice in studies of biofilms in vitro, influences both biofilm formation by 40 S. aureus clinical isolates (methicillin-resistant and methicillin-sensitive S. aureus) and causes variations in biofilm quantification. Methicillin-resistant strains formed more robust biofilms than methicillin-sensitive strains in tryptic soy broth (TSB). However, glucose supplementation in TSB greatly promoted and stabilized biofilm formation of all strains, while additional NaCl was less efficient in this respect and resulted in significant variation in biofilm measurements. In addition, we observed that the ST239-SCCmec (Staphylococcal Cassette Chromosome mec) type III lineage formed strong biofilms in TSB supplemented with glucose and NaCl. Links between biofilm formation and accessory gene regulator (agr) status, as assessed by δ-toxin production, and with mannitol fermentation were not found. Our results show that TSB supplemented with 1.0% glucose supports robust biofilm production and reproducible quantification of S. aureus biofilm formation in vitro, whereas additional NaCl results in major variations in measurements of biofilm formation.
Keywords: S. aureus, methicillin-resistant Staphylococcus aureus (MRSA), biofilm, biofilm formation assay, glucose supplementation, SCCmec type, delta-toxin
1. Introduction
Staphylococcus aureus is one of the most common bacterial pathogens that colonize mammalian skin and/or mucosal membranes. S. aureus has major clinical importance both as a commensal bacterium and an opportunistic pathogen, causing a wide variety of infections, including simple soft skin infections, endocarditis, bacteremia, and severe pneumonia [1,2,3]. Biofilms are highly organized multicellular bacterial communities embedded within a complex matrix composed of polysaccharide, proteins, and/or extracellular DNA (eDNA), and contribute to reduced susceptibility to antimicrobials and persistence of biofilm-associated infections [4]. Biofilm formation by bacteria significantly contributes their survival in the host and has been considered as a key virulence factor responsible for serious chronic infections [5,6]. In clinical scenarios, such as indwelling medical devices or catheter-associated infections, the ability to form biofilms is crucial for S. aureus pathogenicity [7] and biofilm-associated S. aureus infections resist antimicrobial therapy and innate host defense mechanisms [8]. Furthermore, biofilm formation by S. aureus and antimicrobial resistance are functionally linked, as the biofilm phenotype expressed can be influenced by the acquisition of antimicrobial resistance [4,9]. Many foodborne diseases are also associated with biofilms and considered as an emergent public health concern [10].
The S. aureus responsible for biofilm-associated infections can have different genetic backgrounds and, therefore, express a different spectrum of virulence factors during infection [11]. For example, biofilm formation appears to be associated with several regulatory factors, including the agr quorum sensing system [12]. Agr upregulates expression of various toxins, including δ-toxin (a molecule with surfactant-like properties) [13], which contributes to S. aureus adhesion and biofilm development [14]. In addition, biofilm formation requires polysaccharide intercellular adhesin (PIA), which is produced and regulated by the intercellular adhesion (ica) ADCB operon [15]. The icaADCB operon includes an N-acetylglucosamine transferase (icaA and icaB) [16], a deacetylase (icaD) [17], and a predicted exporter (icaC) [16]. Furthermore, some surface components, such as Staphylococcal protein A (spa), contribute to adhesion within biofilms [18]. Genotypic variation among S. aureus strains may additionally influence biofilm formation [19], but these associations are not consistently reported. In addition, the molecular epidemiology of methicillin-resistant S. aureus (MRSA) strains, as identified by staphylococcal cassette chromosome mec (SCCmec) typing, are also associated with biofilm formation [20].
A 96-well microtiter plate assay has been considered as the standard method for quantification of S. aureus biofilm formation in vitro [21]. This assay allows rapid, high-throughput screening of antibiofilm compounds and typically serves as a precursor to in vivo studies. Numerous studies have examined the ability of S. aureus clinical strains to form biofilms by this method [19,22,23,24,25]. These studies vary widely in assay conditions, the culture media used, supplementation of sugar or salt in culture media, and categorization of biofilm formation; however, several did not fully evaluate the relationship between biofilm formation and the molecular characteristics of strain relatedness and genetic heterogeneity.
Tryptic soy broth (TSB) is the most commonly used laboratory culture media for biofilm formation assay [26,27,28]. Furthermore, glucose is normally added in the TSB to promote S. aureus biofilm formation in vitro [29]; however, various glucose concentrations have been used in different studies. For example, TSB (which already contains 2.5 g/L glucose and 5.0 g/L NaCl) is often supplemented with additional glucose (0.25%, 0.4%, 0.5%, and 1.0%) [22,30,31,32,33,34]. In addition, brain heart infusion (BHI) (which already contains 2.0 g/L glucose) is also used as a culture media for S. aureus biofilm formation in vitro [35], with additional glucose supplementation (0.5%, 1.0%, and 1.5%) [36,37]. Lennox broth supplemented with different concentration of glucose (0 to 320 mg/dL in 20 mg/dL intervals) was also used to study the enhanced effect on biofilm formation by S. aureus [38]. Moreover, Lim et al. carried out a comprehensive study to elucidate the effect of NaCl (0.1% to 5.6%) on biofilm formation by S. aureus in TSB [39]. It is interesting to note that strain categorization, when based on biofilm production capacity, may not ensure reproducibility in a laboratory setting. Thus, there is likely to be considerable variability in biofilm formation data generated by different studies.
In medical biofilm study, sensitivity and assay reproducibility are crucially important for diagnosis and determining the efficacy of drugs given to prevent or reduce biofilm formation on indwelling medical devices [40]. The reproducible assay method provides researchers with evidence that data generated are objective and reliable and not influenced by bias. Therefore, even with complex systems simulating real medical devices, the culture media should be standardized to minimize variability in the biofilm formation assay. Here, we hypothesized that the culture media is the most influential parameter that results in data variability of the 96-well microtiter plate biofilm assay. Thus, we have (i) investigated the effects of glucose and NaCl on S. aureus biofilm formation, and (ii) undertaken genotypic and phenotypic characterization of S. aureus clinical isolates to determine any association with biofilm formation capacity.
2. Methods
2.1. Bacterial Strains and Culture Conditions
This study used 40 randomly selected S. aureus isolates (21 MRSA and 19 methicillin-sensitive S. aureus, MSSA) from a total of 360 strains that were recovered from blood samples between 2005 and 2014 at a university-affiliated hospital in Korea. Bacterial identification was performed using matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) (Bruker Microflex LT, Bruker Daltonik GmbH, Bremen, Germany). Antimicrobial susceptibility of the strains was tested using the MicroScan WalkAway 96 plus system (Beckman Coulter, Atlanta, GA, USA). S. aureus strains were grown on mannitol salt agar (MSA) and/or mannitol salt broth (MSB) (KisanBio, Seoul, Korea). Stock cultures were stored in skimmed milk at −70 °C and revived onto blood agar plates (BAP) (Shinyang Diagnostics, Seoul, Korea) for 16–18 h at 37 °C under 5% CO2. For the biofilm formation assay, cultures were grown in TSB (Becton Dickinson, Franklin Lakes, NJ, USA) (TSB composition: tryptone (pancreatic digest of casein) 17.0 g/L, soytone (peptic digest of soybean) 3.0 g/L, glucose (= dextrose) 2.5 g/L, sodium chloride 5.0 g/L, and dipotassium phosphate 2.5 g/L).
2.2. Biofilm Formation Assay
Quantification of S. aureus biofilm formation was performed by 96-well microtiter plate assay as previously described, with the following modifications [21]. Briefly, S. aureus isolates were grown overnight at 37 °C on BAP under 5% CO2. A single colony from BAP was then cultured in TSB at 37 °C overnight and used to prepare a bacterial suspension equivalent of a 0.5 McFarland turbidity standard (MicroScan turbidity meter, Beckman Coulter, Inc., Atlanta, Georgia). The assay media was either TSB, or TSB supplemented with 0.5% or 1.0% d-(+)-glucose, 1.0% or 2.0% NaCl, or with both 1.0% glucose and 1.0% NaCl. Therefore, the total sugar or salt concentrations in TSB were either 7.5 g/L or 12.5 g/L d-(+)-glucose, 15.0 g/L or 25.0 g/L NaCl, or both 12.5 g/L glucose and 15.0 g/L NaCl. The culture media were inoculated to give a final bacterial concentration of 1 × 106 CFU/mL and dispensed (200 μL/well) into wells of microtiter plates (Falcon, Corning Inc., Corning, NJ, USA) [22,41]. The uninoculated media served as a negative control. Biofilms were grown under stationary conditions for 24 h at 37 °C with 5% CO2. After incubation, the bacterial culture from each microtiter plate well was gently aspirated, the wells washed twice with 200 μL of phosphate-buffered saline (PBS, pH 7.4) to remove nonadherent bacteria, the adherent bacteria fixed by heating at 65 °C for 1 h and then stained with 150 μL of 0.1% (w/v) crystal violet (Sigma-Aldrich, St. Louis, MO, USA) for 5 min. Excess crystal violet stain was then discarded and the plates were washed twice with PBS (200 μL) to remove residual dye and then allowed to dry for 30 min at room temperature. The stain adherent biofilm was dissolved in 150 μL of 33% glacial acetic acid (v/v) per well for 30 min. Resulting biofilm formation was quantified by measuring the absorbance at 595 nm with a 96-well ELISA reader (Multiskan FC, Thermo Fisher Scientific, Waltham, MA, USA). Each assay consisted of eight replicates for each condition and performed on at least two occasions. For each assay, the outlier values were identified by Z-score method and the nonoutlier data used to calculate average biofilm formation. The negative control value was subtracted, and the data displayed as the mean absorbance ± standard deviation (SD). S. aureus strains were considered to form biofilms when the ABS595 value was three times the SD of mean absorbance of the negative control [42].
The S. aureus RN4220 strain, a well-characterized biofilm-producing strain, was included in the assay as a positive control for biofilm formation [22]. Biofilm formation was categorized into three groups using an arbitrarily established baseline (the biofilm formation by the positive control strain grown for 24 h in TSB). The S. aureus isolates that formed (i) biofilms ≥75% of the positive control were designated as strong biofilm producers, (ii) 25–75% biofilms of the positive control as moderate biofilm producers, and (iii) <25% biofilms of the positive control as weak biofilm producers [23].
2.3. SCCmec Typing and Detection of ica Genes
DNA for PCR amplification was extracted using the HiYield™ Genomic DNA Mini kit (Real Biotech Corp., Taipei, Taiwan) as per the manufacturer’s instructions. MRSA strains were subjected to SCCmec typing by discriminating the mec gene complex and the cassette chromosome recombinases (ccr) gene complex types, as previously described [43]. The presence of icaA, icaD, icaB, and icaC genes involved in biofilm formation of S. aureus was identified by PCR assay. Primers for typing ica gens were as described previously [44,45].
2.4. Multilocus Sequence Typing (MLST) and spa Analysis
All strains were screened for MLST based on seven housekeeping genes (arcC, aroE, glpF, gmK, pta, tpi, and yqiL) as described previously [46]. The spa-typing was performed using the protocol and primers published previously [47]. Sequence types (STs) and spa types were assigned using the BioNumerics software v.7.5 (Applied Math, Sint-Martens-Latem, Belgium).
2.5. Delta-Toxin Production
Agr functionality was assessed by the detection of δ-toxin production by MALDI-TOF MS [48]. The presence of the δ-toxin peak at m/z 3004 or its allelic variant at m/z 3034 was manually searched in the MALDI-TOF spectrum. The dysfunction of agr was defined as the absence of δ-toxin production [48].
2.6. Statistical Analysis
Statistical analyses were performed using SPSS software ver. 24.0 (SPSS Inc., IBM, Chicago, IL, USA) and Microsoft Excel 2016 (Microsoft, Redmond, WA, USA). Student’s t-test was used to compare the biofilm formation between the MRSA and MSSA strains. The level of significance was determined using one-way ANOVA with Tukey′s multiple comparison test. P ≤ 0.05 was considered significant.
3. Results
3.1. Effect of Glucose and NaCl Supplementation on Biofilm Formation
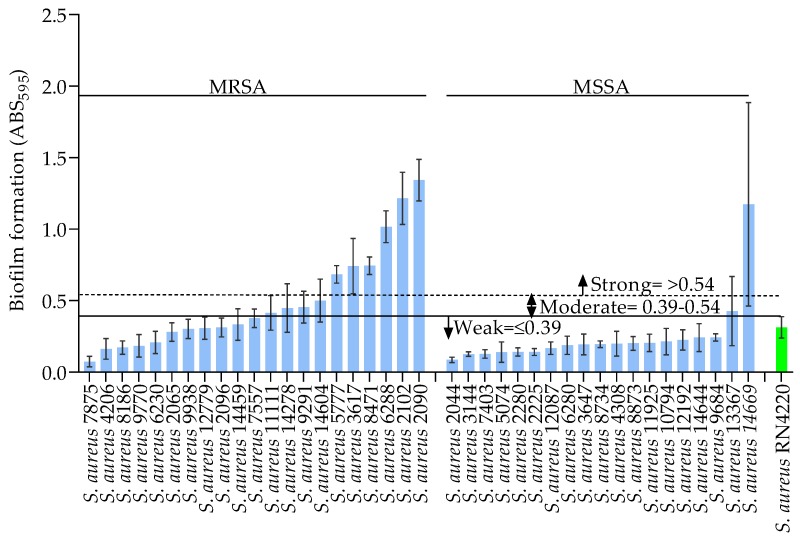
Prior to biofilm quantification, the cumulative bacterial growth consisting of the biofilm and planktonic cells in a 96-well microtiter plate was determined by measuring the absorbance at 595 nm (ABS595). This revealed robust growth for all 40 of the S. aureus clinical isolates when cultured in either TSB or TSB supplemented with different concentrations of glucose and NaCl (data not shown). Nevertheless, some differences in the level of growth were observed between strains and under the different culture media compositions. The biofilm formed on the bottom surfaces of microtiter plate wells was then assessed by crystal violet staining. Firstly, we established an arbitrary baseline for the categorization of biofilm formation by the S. aureus clinical isolates using the biofilm formed by a positive control strain, S. aureus RN4220, when grown in TSB alone. TSB supported growth and biofilm formation by all of the S. aureus strains when compared with that of the positive control strain (RN4220, ABS595 0.31). On this basis, the strains were categorized as strong (ABS595 ≥ 0.54), moderate (ABS595 0.39–0.54), or weak (ABS595 < 0.39) biofilm producers, as seen in Figure 1. As illustrated in Figure 1, distinct biofilm formation was observed between the S. aureus strains when grown in TSB. Notably, several MRSA strains showed significantly greater biofilm formation than the MSSA strains (p < 0.05). Among the 21 MRSA strains, 28.5% (n = 6/21) showed strong biofilm formation compared with only 5.2% (n = 1/19) of the MSSA strains. Furthermore, 19.0% (n = 4/21) of the MRSA and 5.2% (n = 1/19) of the MSSA strains formed moderate biofilms in TSB, while weak biofilms were formed by 52.3% (n = 11/21) and 89.4% (n = 17/19) of the MRSA and MSSA strains, respectively. Interestingly, although most of the MSSA strains formed weak biofilms in TSB, the S. aureus 14,669 strain formed a strong biofilm (ABS595 1.17).
Figure 1.
Biofilm formation (ABS595) of MRSA and MSSA strains grown in TSB. The green bar shows results for the positive control strain RN4220. The data show the mean, and error bars represent the standard deviation within the replicates, for at least two independent experiments.
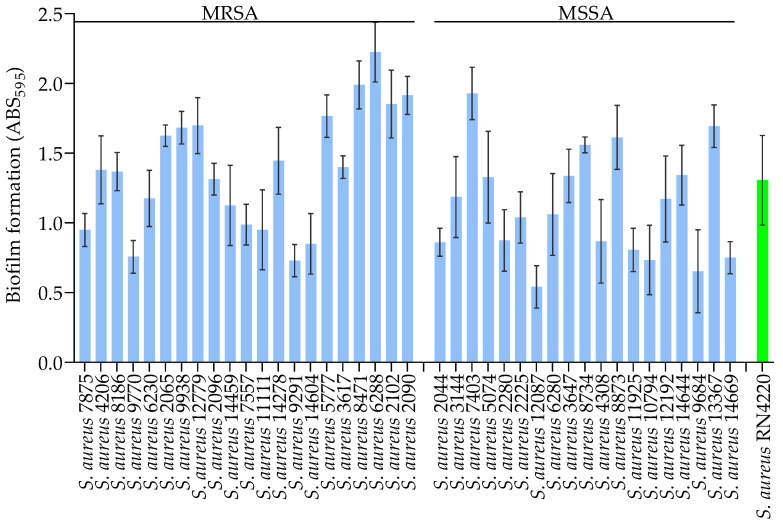
The addition of glucose to the culture media (TSB) strongly promoted biofilm formation by both the MRSA and MSSA strains. All 21 MRSA strains showed increased biofilm formation in TSB supplemented with 0.5% and 1.0% glucose, as seen in Figure S1 and Figure 2, when compared with TSB alone. Additionally, glucose supplementation strongly promoted biofilm formation by most of the MSSA strains (0.5% glucose; 84.2%, n = 16/19 and 1.0% glucose; 94.7%, n = 18/19), whereas the other strains showed moderate biofilm formation (0.5% glucose; 15.7%, n = 3/19 and 1.0% glucose; 5.2%, n = 1/19). However, there was no statistically significant difference in biofilm formation between the MRSA and MSSA strains when they were grown in TSB supplemented with 0.5 or 1.0% glucose. These results show that glucose supplementation promotes biofilm formation of all of the strains tested, and suggest that the rate of biofilm formation is distinct for MRSA and MSSA strains.
Figure 2.
Biofilm formation (ABS595) of MRSA and MSSA strains grown in TSB supplemented with 1.0% glucose. The green bar shows results for the positive control strain RN4220. The data show the mean, and error bars represent the standard deviation within the replicates, for at least two independent experiments.
Addition of 1.0% NaCl to TSB resulted in different levels of biofilm formation for both the MRSA and MSSA strains, as seen in Figure S2. For the MRSA strains, 57.1% (n = 12/21) showed robust biofilm formation, whereas robust biofilm was only seen for 15.7% (n = 3/19) of MSSA strains. When NaCl was increased to 2.0%, 80.9% (n = 17/21) of the MRSA and 52.6% (n = 10/19) of the MSSA strains formed strong biofilms, as seen in Figure S3. However, weak biofilm formation was also seen for 2.8% (n = 9/21) of the MRSA and 73.6% (n = 14/19) of the MSSA strains in 1.0% NaCl, and for 9.2% (n = 2/21) of the MRSA and 31.5% (n = 6/19) of the MSSA strains in 2.0% NaCl. Interestingly, the MSSA strain 14669 formed a strong biofilm in TSB supplemented with either 1.0% or 2.0% NaCl (ABS595 1.74 and 3.43, respectively). Additionally, the MSSA strain 7403 also formed considerably strong biofilm in TSB supplemented with 2.0% NaCl (ABS595 2.38), as seen in Figure S3. Notably, when compared with biofilm formation in TSB alone, the addition of 2.0% NaCl significantly promoted biofilm formation for the MRSA and MSSA strains; however, considerable variation in biofilms was observed. Our results show that average MRSA strains generated significantly stronger biofilms than MSSA strains when they were grown in TSB supplemented with 1.0% NaCl. The coefficient of variation (CV) was used to assess the variability of the data from the biofilm formation assay. This showed that the CV for the assay in TSB supplemented with NaCl ranged from 2.4% to 50% for 1.0% NaCl, and between 2.0% and 53.7% for 2.0% NaCl (data not shown), indicating poor reproducibility of biofilm formation in the presence of additional NaCl.
We also investigated the effects on biofilms of a combination of 1.0% glucose and 1.0% NaCl. The results show that 85.7% (n = 18/21) of the MRSA and 68.4% (n = 13/19) of the MSSA strains formed strong biofilms under these conditions, as seen in Figure S4. Moreover, 14.2% (n = 3/21) of the MRSA and 21.0% (n = 4/19) of the MSSA strains formed moderate biofilms when additional glucose and NaCl were both present, while two MSSA strains (S. aureus 10794 and S. aureus 9684) produced weak biofilms. However, the effect of adding both glucose and NaCl to TSB on biofilm formation by the MRSA and MSSA strains was not significantly different.
3.2. SCCmec Types and Biofilm Formation
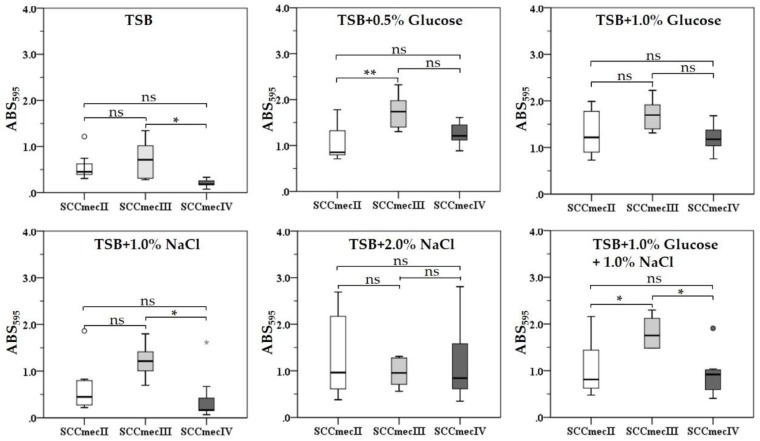
Next, we investigated if different SCCmec types influenced biofilm formation by the MRSA strains when grown in different culture media compositions. Molecular epidemiology analysis of MRSA strains revealed the following SCCmec types: 38.0% were SCCmec type II (n = 8), 33.3% (n = 7) SCCmec type IV, and 28.5%, (n = 6) SCCmec type III, as seen in Table 1. Notably, only 66.6% (n = 4) of the SCCmec type III strains formed strong biofilms in TSB, while two strains (S. aureus 2065 and S. aureus 2096) showed weak biofilm formation. All SCCmec type III strains formed strong biofilms in glucose (0.5% and 1.0%), NaCl (1.0% and 2.0%), and glucose plus NaCl (1.0% each) supplemented TSB, as seen in Figure 3. Similarly, although the SCCmec type IV strains showed weak biofilm formation in TSB, they showed strong biofilm formation in TSB supplemented with glucose, while their biofilm formation differed when they were grown in TSB supplemented with NaCl and TSB supplemented with glucose plus NaCl. No statistically significant difference in biofilm formation by different SCCmec type strains was observed when they were cultured in TSB supplemented with 1.0% glucose or 2.0% NaCl, although supplementation with 2.0% NaCl resulted in wide differences in biofilm formation between strains.
Table 1.
Phenotypic and genotypic characteristics of the S. aureus strains used in this study.
| S. aureus Strains | SCCmec Type (Only for MRSA) |
MLST ST |
spa Types | Mannitol Fermentation |
δ-toxin (m/z 3004 and 3034*) |
|---|---|---|---|---|---|
| MRSA | |||||
| 7875 | IV | 72 | t664 | N | + |
| 4206 | IV | 1 | t286 | P | +* |
| 8186 | IV | 72 | t324 | P | + |
| 9770 | IV | 72 | t148 | N | + |
| 6230 | IV | 72 | t324 | P | + |
| 2065 | III | 239 | t037 | P | + |
| 9938 | IV | 1 | t286 | P | +* |
| 12779 | II | 5 | t2460 | P | − |
| 2096 | III | 239 | t037 | N | + |
| 14459 | IV | 72 | t324 | P | + |
| 7557 | II | 5 | t9353 | P | + |
| 11111 | II | 5 | t601 | P | − |
| 14278 | II | 5 | t9353 | P | + |
| 9291 | II | 5 | t601 | P | − |
| 14604 | II | 5 | t9353 | P | + |
| 5777 | III | 239 | t037 | P | + |
| 3617 | III | 239 | t037 | P | + |
| 8471 | II | 5 | t9353 | P | + |
| 6288 | III | 239 | t037 | P | + |
| 2102 | II | 686 | t5607 | P | + |
| 2090 | III | 239 | t037 | P | + |
| MSSA | |||||
| 2044 | O | 97 | t267 | P | + |
| 3144 | O | 513 | t164 | P | + |
| 7403 | O | 97 | t9353 | P | + |
| 5074 | O | 45 | t1460 | P | + |
| 2280 | O | 188 | t189 | P | + |
| 2225 | O | 1 | t127 | P | +* |
| 12087 | O | 188 | t189 | P | + |
| 6280 | O | 1 | t127 | N | − |
| 3647 | O | 30 | t318 | P | + |
| 8734 | O | 188 | t189 | P | + |
| 4308 | O | 59 | t1950 | N | +* |
| 8873 | O | 72 | t126 | P | + |
| 11925 | O | 6 | t701 | P | + |
| 10794 | O | 1970 | t065 | P | + |
| 12192 | O | 8 | t008 | P | + |
| 14644 | O | 72 | t126 | P | + |
| 9684 | O | 59 | t1151 | P | +* |
| 13367 | O | 8 | t008 | P | + |
| 14669 | O | 1821 | t5554 | P | + |
MLST: Multilocus sequence typing, CC: Clonal complexes, ST: Sequence type, spa: Staphylococcal protein A, O: Nontypable, P: Positive, N: Negative, +: Present, −: Absent, m/z 3034*: Allelic variant of δ-toxin.
Figure 3.
Biofilm formation by MRSA strains with distinct SCCmec types (SCCmec II, n = 8; SCCmec III, n = 6; and SCCmec IV, n = 7) grown in different culture media compositions. Box plot shows the median (horizontal thick blank line) of biofilms formed by MRSA strains. The lower and higher edges of the box represent the 25th and 75th percentiles, respectively. Box plot whiskers represent the 90th and 10th percentiles. P values were determined using Student′s t-test and asterisks indicate statistical significance (* = p ≤ 0.05, ** = p < 0.01). (ns = p > 0.05, not statistically significant).
3.3. Multilocus Sequence Typing (MLST) and spa Analysis
The evolutionary and genetic diversity of the S. aureus strains was analyzed by MLST and revealed a total of 15 distinct STs, as seen in Table 1. The MSSA strains were more diverse than MRSA strains with a higher number (n = 12) of STs (ST1, ST6, ST8, ST30, ST45, ST59, ST72, ST97, ST188, ST513, ST1821, and ST1970). Of these, the most common were ST188 (15.7%, n = 3), ST1, ST8, ST59, ST72, and ST97 (each 10.5%, n = 2). Among the 21 MRSA strains, MLST analysis identified five different STs (ST1, ST5, ST72, ST239, and ST686), with the most common being ST5 (33.3%, n = 7), ST72 (23.8%, n = 5), and ST239 (28.5%, n = 6). A new ST was identified for MRSA strain 2102 and designated ST686 and deposited in the online database (https://pubmlst.org/saureus/). The genetic variation analysis among the S. aureus strains showed a total of 22 spa types, as seen in Table 1. Among the MRSA strains, spa t037 (28.5%, n = 6) was the most dominant, followed by t9353 (19.0%, n = 4), t324 (14.2%, n = 3), and t286 and t601 (each 9.5%, n = 2). The following spa types were seen only once: t148, t2460, t5607, and t664 (each 4.7%, n = 1). Of the MSSA strains, 14 spa types were identified, among which types t008, t126, and t127 (10.5%, n = 2), and t189 (15.7%, n = 3) were most frequently seen. Each of the remaining spa types were identified in 10 MSSA strains.
3.4. The icaADCB Genes and Mannitol Fermentation
Our results showed that all of the genes encoded by the icaADCB operon were present in all 40 S. aureus strains. Therefore, an association between S. aureus biofilm formation and the prevalence of ica genes could not be evaluated. Results of growth on mannitol salt agar identified three MRSA (S. aureus 7875, S. aureus 9770, and S. aureus 2096) and two MSSA (S. aureus 6280 and S. aureus 4308) strains as mannitol-negative, as seen in Table 1. All of the mannitol-negative MRSA and MSSA strains formed weak biofilms in TSB. Additionally, some of the mannitol-positive MRSA (44.4%, n = 8/18) and MSSA (88.2%, n = 15/17) strains also showed weak biofilm formation in TSB. All of the mannitol-negative and mannitol-positive MRSA strains (n = 21) showed strong biofilm formation when the media was supplemented with glucose (0.5% and 1.0%).
3.5. Agr Status and Biofilm Formation
MALDI-TOF MS confirmed the presence of δ-toxin (m/z 3004 and/or its allelic variant at m/z 3034) for 36 isolates; MRSA strains (n = 18) and MSSA strains (n = 18), as seen in Table 1. Two of the agr dysfunctional strains (MRSA, S. aureus 12779 and MSSA, S. aureus 6280), identified by the lack of δ-toxin, formed weak biofilms, while two other MRSA (S. aureus 11111 and S. aureus 9291) strains formed moderate biofilms in TSB, as seen in Figure 1. All strains with a dysfunctional agr showed strong biofilm formation in TSB supplemented with 0.5% and 1.0% glucose, as seen in Figure S1 and Figure 2, weak biofilm formation in TSB supplemented with 1.0% NaCl, as seen in Figure S2, different levels of biofilm formation in TSB supplemented 2.0% NaCl, as seen in Figure S3, and strong to moderate biofilm formation in TSB supplemented with 1.0% glucose and 1.0% NaCl, as seen in Figure S4. Similarly, agr functional strains also formed strong to moderate biofilms in glucose supplemented TSB, distinct biofilms in NaCl supplemented TSB, and strong to moderate biofilms in TSB supplemented with both glucose and NaCl. Collectively, these data show that there was no significant difference in biofilm formation by strains with functional or dysfunctional agr under the tested media compositions.
4. Discussion
TSB, a less nutrient-rich laboratory medium which is commonly used for biofilm formation studies [26,27,28] was shown to support the growth of S. aureus, but with insufficient biofilm formation [22]. However, supplementing TSB with 1.0% glucose resulted in promoted biofilm formation of 40 S. aureus clinical strains with different genetic backgrounds in vitro, as seen in Figure 2. We also found that the addition of glucose resulted in more consistent biofilm quantification when using a 96-well microtiter plate assay, as less variation (i.e., lower SD) was seen between replicates and between assays. The evidence of glucose-induced biofilm formation for S. aureus has been observed [10], and subsequently confirmed in several studies [14,19,22,49]. We did not determine pH changes in the culture media, but glucose represses the agr quorum sensing system [29] due to the excretion of short-chain fatty acids that result from glucose metabolism, which lowers the pH of the surrounding medium [50]. An acidic pH represses extracellular protease production [51], stimulates the association of biofilm matrix proteins on cell surfaces [52,53], and instigates functional amyloid assembly [54], which promotes biofilm formation.
Addition of NaCl to TSB also enhanced the biofilm formation of several S. aureus strains tested; however, considerable differences in biofilm formation and experimental data was observed. This is likely due to the loose attachment of S. aureus biofilms to the well surface when excess NaCl is present. We also observed that NaCl (1.0% and 2.0%) induced biofilm formation more effectively for MRSA (57.1% and 80.9%) than MSSA strains (15.7% and 52.6%). These results are contrary to previously published work, where biofilm formation in the presence of additional NaCl was more efficient for MSSA strains over MRSA strains [22]. However, this discrepancy could be explained by the use of different NaCl concentrations between the different studies, as the study by Sugimoto et al. used 4.0% NaCl [22], which is twice the salt concentration we tested. As described, NaCl activates expression of the icaADBC operon in S. aureus, resulting in PIA production [14], which is required for biofilm formation and stability. However, although NaCl-induced activation of transcription from the icaADCB operon is stronger for MRSA than MSSA strains, it does not result in PIA production in MRSA strains [14,18]. Our results showing poor reproducibility and large differences in CV (2.0–53.7%) between replicates and between assays, when biofilm formation is determined by the 96-well microtiter plate assay in the presence of additional NaCl, will be significant for the development of a consistent assay method. In addition, TSB supplemented with 1.0% glucose produced robust and consistent biofilms, leading to greater assay reproducibility, and thus should result in improved quantification of S. aureus biofilms in vitro.
We also investigated the association between genotypic and phenotypic characteristics of S. aureus strains with biofilm formation when varying concentrations of glucose and NaCl were added to the culture media. Our results suggest that MRSA strains with SCCmec type III form strong biofilm in glucose and NaCl supplemented TSB, whereas SCCmec type I and II strains formed strong biofilms in the presence of glucose and differed when grown in NaCl supplemented TSB. The most prevalent hospital associated (HA) MRSA lineage that carries the SCCmec type III is MRSA-ST239 [55] and recognized as multidrug resistance across the globe [56]. In the present study, we found that 28.5% (n = 6/21) of the MRSA strains tested were of the ST239 lineage (typically referred as HA strains), and of these, the ST5-SCCmec II and ST72 lineages predominated. These finding are consistent with a previous report that showed the prevalence of ST239 (53.0%, n = 100/188) and ST5 (34.0%, n = 63/188) MRSA clones in Korean hospitals during 2001–2004 [57]. In addition, the ST5 [58] and ST239 [59] lineages have been identified as the dominant clones in clinical specimens from other geographical regions. We detected icaADCB genes in all the 40 S. aureus strains tested. However, previous studies on biofilm formation by S. aureus strains did not identify all of the ica genes, and the icaA and icaD genes were more frequently observed [24,60,61]. Regardless of mannitol fermentation, different levels of biofilm formation were observed among strains grown in TSB supplemented with 1.0% and 2.0% NaCl as well as in TSB supplemented with combination of 1.0% glucose and 1.0% NaCl. In our study, an association between mannitol fermentation and biofilm formation was not observed in the S. aureus strains tested.
We found no association between agr status (as assessed by δ-toxin production) and biofilm formation under any of the assay conditions. However, previous studies suggest that agr dysfunction is associated with increased biofilm formation in S. aureus [17,62]. A limited number of δ-toxin negative strains may have impaired the ability to establish association between robust biofilm formation and agr dysfunction. Here, to minimize the sample size bias, the S. aureus clinical isolates were randomly selected from a total of 360 strains recovered between 2005 and 2014 and were blind to the biofilm formation ability. Additionally, we used S. aureus RN4220 as a positive-control biofilm-forming strain to ensure comparability between results.
In conclusion, our findings show that biofilm phenotype of S. aureus clinical strains can vary considerably depending on the composition of the culture media. In particular, glucose added to TSB promoted robust biofilm formation and resulted in improved assay results, while the addition of NaCl resulted in less biofilm formation and significant variation in biofilm quantification when using a 96-well microtiter plate assay. Associations between mannitol fermentation and agr functionality with biofilm formation were not observed under all of the tested culture media compositions. This study shows that TSB supplemented with 1.0% glucose is the most appropriate culture medium for promoting biofilm formation and results in higher assay reproducibility when quantifying S. aureus biofilm formation in vitro.
Supplementary Materials
The following are available online at https://www.mdpi.com/2077-0383/8/11/1853/s1, Figure S1: Biofilm formation (ABS595) of MRSA and MSSA strains grown in TSB supplemented with 0.5% glucose, Figure S2: Biofilm formation (ABS595) of MRSA and MSSA strains grown in TSB supplemented with 1.0% NaCl, Figure S3: Biofilm formation (ABS595) of MRSA and MSSA strains grown in TSB supplemented with 2.0% NaCl, Figure S4. Biofilm formation (ABS595) of MRSA and MSSA strains grown in TSB supplemented with 1.0% glucose and 1.0% NaCl.
Author Contributions
Conceived and designed the experiment, H.L., H.-S.J. and J.-S.K.; performed the experiments, H.L., J.H.P., S.H.C., I.H.K. and J.-M.K.; analyzed the data, H.L., H.-S.J., J.-S.K.; wrote the paper, H.L., H.-S.J. and J.-S.K.; edited the paper, H.L., J.-M.K., H.-S.J. and J.-S.K.
Funding
This study was supported by the Bio and Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government, Ministry of Science and ICT (MSIT) 2017M3A9E4077232. This study was also supported by the Hallym university research fund.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Gillet Y., Issartel B., Vanhems P., Fournet J.C., Lina G., Bes M., Vandenesch F., Piémont Y., Brousse N., Floret D., et al. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet. 2002;359:753–759. doi: 10.1016/S0140-6736(02)07877-7. [DOI] [PubMed] [Google Scholar]
- 2.Røder B.L., Wandall D.A., Frimodt-Møller N., Espersen F., Skinhøj P., Rosdahl V.T. Clinical features of staphylococcus aureus endocarditis. Arch. Intern. Med. 1999;159:462–469. doi: 10.1001/archinte.159.5.462. [DOI] [PubMed] [Google Scholar]
- 3.Wyllie D.H., Crook D.W., Peto T.E.A. Mortality after Staphylococcus aureus bacteraemia in two hospitals in Oxfordshire, 1997–2003: Cohort study. BMJ. 2006;333:281. doi: 10.1136/bmj.38834.421713.2F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stewart P.S. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 2002;292:107–113. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- 5.Parrino B., Schillaci D., Carnevale I., Giovannetti E., Diana P., Cirrincione G., Cascioferro S. Synthetic small molecules as anti-biofilm agents in the struggle against antibiotic resistance. Eur. J. Med. Chem. 2019;161:154–178. doi: 10.1016/j.ejmech.2018.10.036. [DOI] [PubMed] [Google Scholar]
- 6.Parrino B., Diana P., Cirrincione G., Cascioferro S. Bacterial biofilm inhibition in the development of effective anti-virulence strategy. Open Med. Chem. J. 2018;12:84–87. doi: 10.2174/1874104501812010084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suresh M.K., Biswas R., Biswas L. An update on recent developments in the prevention and treatment of Staphylococcus aureus biofilms. Int. J. Med. Microbiol. 2019;309:1–12. doi: 10.1016/j.ijmm.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Otto M. Current Topics in Microbiology and Immunology. Volume 322. Springer; Berlin/Heidelberg, Germany: 2008. Staphylococcal biofilms; pp. 207–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarthy H., Rudkin J.K., Black N.S., Gallagher L., O’Neill E., O’Gara J.P. Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2015;5:1. doi: 10.3389/fcimb.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srey S., Jahid I.K., Ha S.-D. Biofilm formation in food industries: A food safety concern. Food Control. 2013;31:572–585. doi: 10.1016/j.foodcont.2012.12.001. [DOI] [Google Scholar]
- 11.Jarraud S., Mougel C., Thioulouse J., Lina G., Meugnier H., Forey F., Nesme X., Etienne J., Vandenesch F. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 2002;70:631–641. doi: 10.1128/IAI.70.2.631-641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vuong C., Kocianova S., Voyich J.M., Yao Y., Fischer E.R., DeLeo F.R., Otto M. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J. Biol. Chem. 2004;279:54881–54886. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- 13.Jiang Q., Jin Z., Sun B. MgrA negatively regulates biofilm formation and detachment by repressing the expression of psm operons in Staphylococcus aureus. Appl. Environ. Microbiol. 2018;84:e01008–e01018. doi: 10.1128/AEM.01008-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Neill E., Pozzi C., Houston P., Smyth D., Humphreys H., Robinson D.A., O’Gara J.P. Association between Methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J. Clin. Microbiol. 2007;45:1379–1388. doi: 10.1128/JCM.02280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cramton S.E., Gerke C., Schnell N.F., Nichols W.W., Götz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 1999;67:5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerke C., Kraft A., Süßmuth R., Schweitzer O., Götz F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 1998;273:18586–18593. doi: 10.1074/jbc.273.29.18586. [DOI] [PubMed] [Google Scholar]
- 17.Vuong C., Saenz H.L., Götz F., Otto M. Impact of the agr quorum-sensing system on adherence to Polystyrene in Staphylococcus aureus. J. Infect. Dis. 2000;182:1688–1693. doi: 10.1086/317606. [DOI] [PubMed] [Google Scholar]
- 18.Lasa I., Penadés J.R. Bap: A family of surface proteins involved in biofilm formation. Res. Microbiol. 2006;157:99–107. doi: 10.1016/j.resmic.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Croes S., Deurenberg R.H., Boumans M.-L.L., Beisser P.S., Neef C., Stobberingh E.E. Staphylococcus aureus biofilm formation at the physiologic glucose concentration depends on the S. aureus lineage. BMC Microbiol. 2009;9:229. doi: 10.1186/1471-2180-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim Y., Shin H.J., Kwon A.S., Reu J.H., Park G., Kim J. Predictive genetic risk markers for strong biofilm-forming Staphylococcus aureus: fnbB gene and SCCmec type III. Diagn. Microbiol. Infect. Dis. 2013;76:539–541. doi: 10.1016/j.diagmicrobio.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 21.Christensen G.D., Simpson W.A., Younger J.J., Baddour L.M., Barrett F.F., Melton D.M., Beachey E.H. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: A quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985;22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugimoto S., Sato F., Miyakawa R., Chiba A., Onodera S., Hori S., Mizunoe Y. Broad impact of extracellular DNA on biofilm formation by clinically isolated Methicillin-resistant and -sensitive strains of Staphylococcus aureus. Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-018-20485-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanhommerig E., Moons P., Pirici D., Lammens C., Hernalsteens J.-P., De Greve H., Kumar-Singh S., Goossens H., Malhotra-Kumar S. Comparison of biofilm formation between major clonal lineages of Methicillin resistant Staphylococcus aureus. PLoS ONE. 2014;9:e104561. doi: 10.1371/journal.pone.0104561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khoramian B., Jabalameli F., Niasari-Naslaji A., Taherikalani M., Emaneini M. Comparison of virulence factors and biofilm formation among Staphylococcus aureus strains isolated from human and bovine infections. Microb. Pathog. 2015;88:73–77. doi: 10.1016/j.micpath.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 25.Smith K., Perez A., Ramage G., Lappin D., Gemmell C.G., Lang S. Biofilm formation by Scottish clinical isolates of Staphylococcus aureus. J. Med. Microbiol. 2008;57:1018–1023. doi: 10.1099/jmm.0.2008/000968-0. [DOI] [PubMed] [Google Scholar]
- 26.Mashruwala A.A., van de Guchte A., Boyd J.M. Impaired respiration elicits SrrAB-dependent programmed cell lysis and biofilm formation in Staphylococcus aureus. Elife. 2017;6:1–29. doi: 10.7554/eLife.23845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manner S., Goeres D.M., Skogman M., Vuorela P., Fallarero A. Prevention of Staphylococcus aureus biofilm formation by antibiotics in 96-Microtiter Well Plates and Drip Flow Reactors: Critical factors influencing outcomes. Sci. Rep. 2017;7:43854. doi: 10.1038/srep43854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong C., Chee C.-F., Richter K., Thomas N., Abd. Rahman N., Nathan S. Suppression of Staphylococcus aureus biofilm formation and virulence by a benzimidazole derivative, UM-C162. Sci. Rep. 2018;8:2758. doi: 10.1038/s41598-018-21141-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Regassa L.B., Novick R.P., Betley M.J. Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infect. Immun. 1992;60:3381–3388. doi: 10.1128/iai.60.8.3381-3388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernández-Barat L., Ben-Aicha S., Motos A., Vila J., Marco F., Rigol M., Muñoz L., Li Bassi G., Ferrer M., Torres A. Assessment of in vivo versus in vitro biofilm formation of clinical methicillin-resistant Staphylococcus aureus isolates from endotracheal tubes. Sci. Rep. 2018;8:11906. doi: 10.1038/s41598-018-30494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goncheva M.I., Flannagan R.S., Sterling B.E., Laakso H.A., Friedrich N.C., Kaiser J.C., Watson D.W., Wilson C.H., Sheldon J.R., McGavin M.J., et al. Stress-induced inactivation of the Staphylococcus aureus purine biosynthesis repressor leads to hypervirulence. Nat. Commun. 2019;10:775. doi: 10.1038/s41467-019-08724-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He L., Le K.Y., Khan B.A., Nguyen T.H., Hunt R.L., Bae J.S., Kabat J., Zheng Y., Cheung G.Y.C., Li M., et al. Resistance to leukocytes ties benefits of quorum sensing dysfunctionality to biofilm infection. Nat. Microbiol. 2019;4:1114–1119. doi: 10.1038/s41564-019-0413-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeFrancesco A.S., Masloboeva N., Syed A.K., DeLoughery A., Bradshaw N., Li G.-W., Gilmore M.S., Walker S., Losick R. Genome-wide screen for genes involved in eDNA release during biofilm formation by Staphylococcus aureus. Proc. Natl. Acad. Sci. USA. 2017;114:E5969–E5978. doi: 10.1073/pnas.1704544114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu L., Hisatsune J., Hayashi I., Tatsukawa N., Sato’o Y., Mizumachi E., Kato F., Hirakawa H., Pier G.B., Sugai M. A novel repressor of the ica locus discovered in clinically isolated super-biofilm-elaborating Staphylococcus aureus. MBio. 2017;8:1–17. doi: 10.1128/mBio.02282-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oyama L.B., Girdwood S.E., Cookson A.R., Fernandez-Fuentes N., Privé F., Vallin H.E., Wilkinson T.J., Golyshin P.N., Golyshina O.V., Mikut R., et al. The rumen microbiome: An underexplored resource for novel antimicrobial discovery. NPJ Biofilms Microbiomes. 2017;3:33. doi: 10.1038/s41522-017-0042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu C.-Y., Shu J.-C., Lin M.-H., Chong K.-Y., Chen C.-C., Wen S.-M., Hsieh Y.-T., Liao W.-T. High glucose concentration promotes vancomycin-enhanced biofilm formation of vancomycin-non-susceptible Staphylococcus aureus in diabetic mice. PLoS ONE. 2015;10:e0134852. doi: 10.1371/journal.pone.0134852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshii Y., Okuda K., Yamada S., Nagakura M., Sugimoto S., Nagano T., Okabe T., Kojima H., Iwamoto T., Kuwano K., et al. Norgestimate inhibits staphylococcal biofilm formation and resensitizes methicillin-resistant Staphylococcus aureus to β-lactam antibiotics. NPJ Biofilms Microbiomes. 2017;3:18. doi: 10.1038/s41522-017-0026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waldrop R., McLaren A., Calara F., McLemore R. Biofilm growth has a threshold response to glucose in vitro. Clin. Orthop. Relat. Res. 2014;472:3305–3310. doi: 10.1007/s11999-014-3538-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim Y., Jana M., Luong T.T., Lee C.Y. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 2004;186:722–729. doi: 10.1128/JB.186.3.722-729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malone M., Goeres D.M., Gosbell I., Vickery K., Jensen S., Stoodley P. Approaches to biofilm-associated infections: The need for standardized and relevant biofilm methods for clinical applications. Expert Rev. Anti. Infect. Ther. 2017;15:147–156. doi: 10.1080/14787210.2017.1262257. [DOI] [PubMed] [Google Scholar]
- 41.Fernández L., González S., Campelo A.B., Martínez B., Rodríguez A., García P. Low-level predation by lytic phage phiIPLA-RODI promotes biofilm formation and triggers the stringent response in Staphylococcus aureus. Sci. Rep. 2017;7:40965. doi: 10.1038/srep40965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stepanović S., Vuković D., Ježek P., Pavlović M., Švabic-Vlahović M. Influence of dynamic conditions on biofilm formation by Staphylococci. Eur. J. Clin. Microbiol. Infect. Dis. 2001;20:0502–0504. doi: 10.1007/s100960100534. [DOI] [PubMed] [Google Scholar]
- 43.Kondo Y., Ito T., Ma X.X., Watanabe S., Kreiswirth B.N., Etienne J., Hiramatsu K. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: Rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 2007;51:264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rohde H., Knobloch J.K., Horstkotte M.A., Mack D. Correlation of Staphylococcus aureus icaADBC genotype and biofilm expression phenotype. J. Clin. Microbiol. 2001;39:4595–4596. doi: 10.1128/JCM.39.12.4595-4596.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiem S., Oh W.S., Peck K.R., Lee N.Y., Lee J.-Y., Song J.-H., Hwang E.S., Kim E.-C., Cha C.Y., Choe K.-W. Phase variation of biofilm formation in Staphylococcus aureus by IS 256 insertion and its impact on the capacity adhering to polyurethane surface. J. Korean Med. Sci. 2004;19:779–782. doi: 10.3346/jkms.2004.19.6.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Enright M.C., Day N.P., Davies C.E., Peacock S.J., Spratt B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harmsen D., Claus H., Witte W., Rothgänger J., Claus H., Turnwald D., Vogel U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 2003;41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gagnaire J., Dauwalder O., Boisset S., Khau D., Freydière A.-M., Ader F., Bes M., Lina G., Tristan A., Reverdy M.-E., et al. Detection of Staphylococcus aureus delta-toxin production by whole-cell MALDI-TOF mass spectrometry. PLoS ONE. 2012;7:e40660. doi: 10.1371/journal.pone.0040660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tasse J., Trouillet-Assant S., Josse J., Martins-Simões P., Valour F., Langlois-Jacques C., Badel-Berchoux S., Provot C., Bernardi T., Ferry T., et al. Association between biofilm formation phenotype and clonal lineage in Staphylococcus aureus strains from bone and joint infections. PLoS ONE. 2018;13:e0200064. doi: 10.1371/journal.pone.0200064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olson M.E., King J.M., Yahr T.L., Horswill A.R. Sialic acid catabolism in Staphylococcus aureus. J. Bacteriol. 2013;195:1779–1788. doi: 10.1128/JB.02294-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boles B.R., Horswill A.R. Staphylococcal biofilm disassembly. Trends Microbiol. 2011;19:449–455. doi: 10.1016/j.tim.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Foulston L., Elsholz A.K.W., DeFrancesco A.S., Losick R. The extracellular matrix of Staphylococcus aureus biofilms comprises cytoplasmic proteins that associate with the cell surface in response to decreasing pH. MBio. 2014;5:e01667-14. doi: 10.1128/mBio.01667-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dengler V., Foulston L., DeFrancesco A.S., Losick R. An electrostatic net model for the role of extracellular DNA in biofilm formation by Staphylococcus aureus. J. Bacteriol. 2015;197:3779–3787. doi: 10.1128/JB.00726-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taglialegna A., Navarro S., Ventura S., Garnett J.A., Matthews S., Penades J.R., Lasa I., Valle J. Staphylococcal bap proteins build amyloid scaffold biofilm matrices in response to environmental signals. PLoS Pathog. 2016;12:e1005711. doi: 10.1371/journal.ppat.1005711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson D.A., Enright M.C. Evolution of Staphylococcus aureus by large chromosomal replacements. J. Bacteriol. 2004;186:1060–1064. doi: 10.1128/JB.186.4.1060-1064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Botelho A.M.N., Cerqueira e Costa M.O., Moustafa A.M., Beltrame C.O., Ferreira F.A., Côrtes M.F., Costa B.S.S., Silva D.N.S., Bandeira P.T., Lima N.C.B., et al. Local diversification of Methicillin- resistant Staphylococcus aureus ST239 in South America after its rapid worldwide dissemination. Front. Microbiol. 2019;10:82. doi: 10.3389/fmicb.2019.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cha H.Y., Moon D.C., Choi C.H., Oh J.Y., Jeong Y.S., Lee Y.C., Seol S.Y., Cho D.T., Chang H.-H., Kim S.-W., et al. Prevalence of the ST239 clone of methicillin-resistant Staphylococcus aureus and differences in antimicrobial susceptibilities of ST239 and ST5 clones identified in a Korean hospital. J. Clin. Microbiol. 2005;43:3610–3614. doi: 10.1128/JCM.43.8.3610-3614.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song Y., Du X., Li T., Zhu Y., Li M. Phenotypic and molecular characterization of Staphylococcus aureus recovered from different clinical specimens of inpatients at a teaching hospital in Shanghai between 2005 and 2010. J. Med. Microbiol. 2013;62:274–282. doi: 10.1099/jmm.0.050971-0. [DOI] [PubMed] [Google Scholar]
- 59.Chen H., Liu Y., Jiang X., Chen M., Wang H. Rapid change of methicillin-resistant Staphylococcus aureus clones in a Chinese tertiary care hospital over a 15-year period. Antimicrob. Agents Chemother. 2010;54:1842–1847. doi: 10.1128/AAC.01563-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arciola C.R., Baldassarri L., Montanaro L. Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J. Clin. Microbiol. 2001;39:2151–2156. doi: 10.1128/JCM.39.6.2151-2156.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tekeli A., Ocal D.N., Ozmen B.B., Karahan Z.C., Dolapci I. Molecular characterization of Methicillin-resistant Staphylococcus aureus bloodstream isolates in a Turkish university hospital between 2002 and 2012. Microb. Drug Resist. 2016;22:564–569. doi: 10.1089/mdr.2015.0116. [DOI] [PubMed] [Google Scholar]
- 62.Sakoulas G., Moise P.A., Rybak M.J. Accessory gene regulator dysfunction: An advantage for Staphylococcus aureus in health-care settings? J. Infect. Dis. 2009;199:1558–1559. doi: 10.1086/598607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.