Abstract
Aims: Members of the thioredoxin (Trx) protein family participate mainly in redox pathways and have not been associated with Fe/S binding, in contrast to some closely related glutaredoxins (Grxs). Cestode parasites possess an unusual diversity of Trxs and Trx-related proteins with unexplored functions. In this study, we addressed the biochemical characterization of a new class of Trx-related protein (IsTRP) and a classical monothiol Grx (EgGrx5) from the human pathogen Echinococcus granulosus. Results: The dimeric form of IsTRP coordinates Fe2S2 in a glutathione-independent manner; instead, Fe/S binding relies on the CXXC motif conserved among Trxs. This novel binding mechanism allows holo-IsTRP to be highly resistant to oxidation. IsTRP lacks canonical reductase activities. Mitochondrially targeted IsTRP aids growth of a Grx5 null yeast strain. Similar complementation assays performed with EgGrx5 revealed functional conservation for class II Grxs, despite the presence of nonconserved structural elements. IsTRP is a cestode lineage-specific protein highly expressed in the gravid adult worm, which releases the infective stage critical for dissemination. Innovation: IsTRP is the first member from the Trx family to be reported to bind Fe/S. We disclose a novel mechanism of Fe/S coordination within the Trx folding unit, which renders the cluster highly resistant to oxidation-mediated disassembly. Conclusion: We demonstrate that IsTRP defines a new protein family within the Trx superfamily, confirm the conservation of function for class II Grx from nonphylogenetically related species, and highlight the versatility of the Trx folding unit to acquire Fe/S binding as a recurrent emergent function. Antioxid. Redox Signal. 24, 205–216.
Introduction
The thioredoxin (Trx) superfamily includes a wide range of proteins with diverse functions that share a common structural pattern, the Trx fold unit, comprising a β-sheet consisting of four beta strands, surrounded by three α-helices (35). The prototypical Trx is a thiol-dependent oxidoreductase with a characteristic WCGPC dithiol active site motif that functions mainly as protein disulfide reductase (16). Thioredoxin reductase (TR) maintains Trxs in their reduced form (19, 33) at the expense of NADPH oxidation. Trx-related proteins (TRPs) or Trx-like proteins also possess a Trx fold with a canonical Trx active site and are involved in redox homeostasis (34). Glutaredoxins (Grxs), which utilize glutathione (GSH) as a cofactor for their functions, also belong to the Trx superfamily (26). Grxs are clustered into three classes (44), with classes I and II being widely distributed.
Innovation.
IsTRP defines a new class of a thioredoxin (Trx)-related protein able to bind Fe/S. We disclose a novel mechanism of Fe/S coordination within the Trx folding unit, which renders the cluster highly resistant to oxidation-mediated disassembly. IsTRP is present exclusively in some flatworm parasites and highly expressed in the gravid adult worm. In addition, we confirm the function of Echinococcus granulosus Grx5 in mitochondrial Fe/S biosynthesis, despite possessing distinctive parasite-specific features. Our results uncover lineage-specific adaptations of the Trx fold and provide a putative pharmacological target for neglected diseases caused by flatworm infections.
Class I Grxs possess a canonical CPYC active site and their main functions rely on their redox activity, particularly as protein disulfide and GSH-protein disulfide reductases, involved in an oxidoreduction cascade with GSH, glutathione reductase (GR), and NADPH (33). On the other hand, class II Grxs possess a typical CGFS active site and are functionally distinct: they exhibit low or no thiol–disulfide oxidoreductase activity (14, 21, 47, 53) and their main role relates to iron homeostasis (24). Mitochondrial class II Grxs, such as Saccharomyces cerevisiae Grx5 (ScGrx5), are usually indispensable components of the mitochondrial iron–sulfur cluster assembly machinery (ISC), participating in Fe/S trafficking and assembly into apoproteins (24, 37). Fe/S are essential cofactors for oxidative phosphorylation, some Krebs cycle enzymes, and protein synthesis, among other important pathways. These requirements provide the basis for assessing the function of Grx5 (23).
The ISC includes several proteins and is essential for Fe/S synthesis in the mitochondria and, most likely, in the cytosol (24). The dependence to the mitochondria for de novo synthesis in the cytosol is controversial (38), but the most accepted view emphasizes that Fe/S synthesis starts in the mitochondria from which a sulfur-containing moiety is exported to the cytosol, in a process dependent of an ABC transporter and GSH, but details are not fully understood (43). In mammals, the presence of cytosolic and nuclear forms of some components of the ISC has raised the possibility that Fe/S could be synthesized de novo in these compartments (38). In any case, the cytosolic Fe/S protein assembly machinery (CIA) is necessary for further processing the Fe/S in the cytosol (23).
Finally, class III Grxs are restricted to angiosperms, possess a CC(M/L)(C/S) active site sequence, and represent the large majority of the Grxs encoded in higher plant genomes. These proteins seem to be involved in the resistance of plants to a variety of different environmental conditions (44).
Generally speaking, Trx-fold proteins mainly serve as thiol/disulfide oxidoreductases and only a small subset binds metal ions (46). Class II and some class I Grxs bind Fe/S in a dimeric manner, where the iron atoms of the cluster are coordinated by the N-terminal active site cysteine residue (Cys) of each subunit and by two noncovalently bound GSH molecules (4). Different types of Fe/S have been shown to be bound to Grxs, including rhombic [Fe2S2]2+, linear [Fe3S4]+, and [Fe4S4]2+ (54). In addition to this canonical binding through GSH, an unusual mode of Fe/S coordination was reported in Grx2 from the teleost fish (8). The monomeric form of this protein can bind Fe/S using four Cys residues different from the CxxC active site. Trx-like ferredoxins (52) are also able to bind Fe/S. This type of ferredoxin forms homodimers bound by protein/protein interactions, in which each subunit coordinates one [Fe2S2] through four Cys residues (29, 52). Although the function of these proteins is still unclear, they might be involved in nitrogen fixation since it has been shown that they interact with a molybdenum-containing nitrogenase (15, 29). Finally, TrxA from Escherichia coli (27) and human Trx1 (46) are able to coordinate Fe/S when specific non-natural mutations are introduced in their sequences.
Parasitic tapeworm genomes have revealed an unexpected diversity of Trxs and Grxs (5, 49), but the functions of most of these proteins have not been examined. This diversity contrasts with the minimalistic arrangement of the upstream pathway, where GR and TR have been replaced by a single enzyme, the thioredoxin glutathione reductase (TGR), which provides electrons to Trxs and Grxs at the expense of NADPH (51).
In this study, we characterized an ortholog of yeast Grx5 in the tapeworm Echinococcus granulosus (Grx5) and a new Trx-fold protein, which we designate as iron–sulfur Trx-related protein (IsTRP) that is the first wild-type Trx-like protein with Fe/S coordination capacity to be reported. We show that its Fe/S binding is GSH independent, involves dimerization, and uses two Cys residues from each monomer as ligands for iron coordination of the cluster. A model for the relevance of IsTRP for Fe/S supply during infection of the intermediate host is proposed.
Results
Common and unique features of cestode IsTRP and Grx5
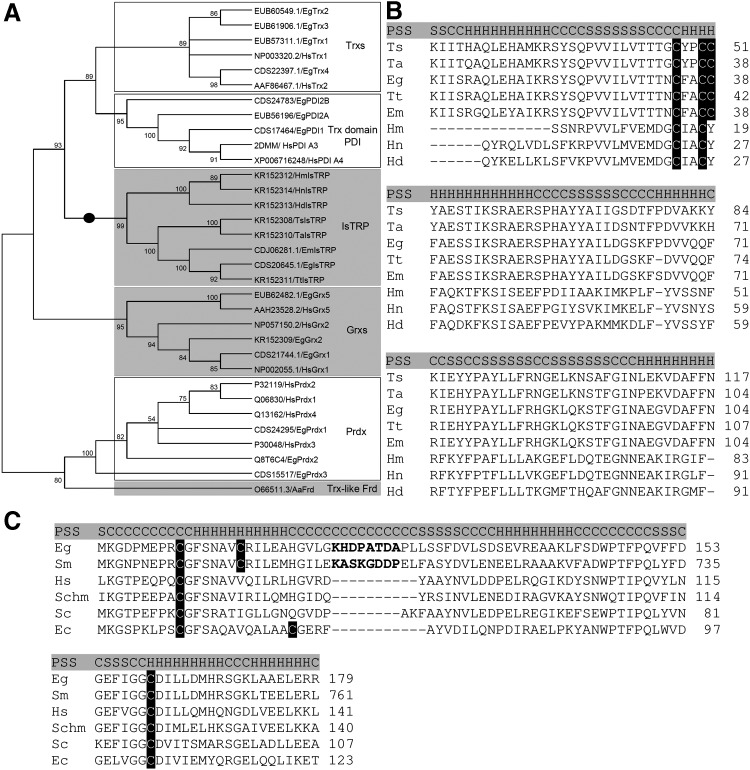
IsTRP possesses a Trx-like domain, which unequivocally clusters with Trxs and Trx-related proteins in the phylogenetic analysis, and is distant to all other Trx-fold proteins (Fig. 1A). A conspicuous feature of IsTRP is the presence of an additional Cys residue immediately adjacent to the canonical active site CxxC motif, resulting in the CxxCC motif. Data mining of orthologous genes showed that IsTRP is encoded exclusively by the Cyclophyllidea order within the class Cestoda and is absent in any other sequenced lineage. Interestingly, the third Cys is conserved in most, but not all, IsTRPs (Fig. 1B). None of the cestode IsTRPs has a sorting signal peptide. Since there are no RNASeq data of IsTRP in E. granulosus, we examined the expression data from Echinococcus multilocularis kindly provided by Dr. Matt Berriman. This data mining indicated that IsTRP is highly expressed in the gravid adult (the stage that releases embryonated eggs to the environment), while very low expression is observed in other stages of the parasite (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/ars).
FIG. 1.
IsTRP and Grx5 alignment and phylogenetic analysis. (A) Simplified unrooted neighbor-joining phylogeny of some Trx superfamily members (Trxs, thioredoxins; Trx domain of PDI, thioredoxin domain of protein disulfide isomerases; IsTRP, Iron–sulfur thioredoxin-related protein; Grxs, glutaredoxins; Prdxs, peroxiredoxins; Trx-like Frd, thioredoxin-like ferredoxin). Bootstrap values were calculated using 500 iterations. The members of the Trx fold that can coordinate Fe/S are shown in gray boxes (for IsTRP, the evidence stems from this work). Sequences specific to the Cyclophyllidea order within the class Cestoda are pointed with a black circle on the branches. (B, C) Protein sequence alignment of IsTRP and Grx5 ortholog proteins, respectively. Cysteine residues are highlighted in black. The residues of the insertion of tapeworm Grx5 are shown in bold. PSS elements (highlighted in gray) are represented with (H) when it is α-helix, (S) when it is β-sheet, and (C) when it is coiled. Ts. Taenia solium (IsTRP KR152308). Ta. Taenia asiatica (IsTRP KR152310). Eg. Echinococcus granulosus (IsTRP CDS20645, Grx5 EUB62482). Tt. Taenia taeniaeformis (IsTRP KR152311). Em. Echinococcus multilocularis (IsTRP CDJ06281). Hm. Hymenolepis microstoma (IsTRP KR152312). Hn. Hymenolepis nana (IsTRP KR152314). Hd. Hymenolepis diminuta (IsTRP KR152313). Sm. Schistosoma mansoni (Grx5 CCD60523). Hs. Homo sapiens (Grx5 AAH23528). Schm. Schmidtea mediterranea. Sc. Saccharomyces cerevisiae (Grx5 3GX8). Ec. Escherichia coli (Grx5 2WCI). Fe/S, iron–sulfur cluster; PSS, predicted secondary structure.
In contrast, Grx5 is a highly conserved mitochondrial class II Grx, with orthologous proteins present in a wide range of eukaryotic lineages. A remarkable feature of parasitic flatworm Grx5s is that they possess a unique insertion of eight amino acids in an infrequent insertion site in the Trx superfamily, located between the active site containing α-helix and the following β-strand (Fig. 1C). Analysis of a homology-based model of E. granulosus Grx5 indicates that this element does not impose any significant constrain to the Trx domain folding core (Supplementary Fig. S2). Interestingly, this insertion defines an extended loop located near the class II Grx acidic surface that is proposed to be important for the interaction with its partners (21). Grx5s from parasitic flatworms also harbor a nonconserved cysteine residue at the C-terminus of the α-helix containing the active site.
Dimeric IsTRP and Grx5 coordinate Fe/S
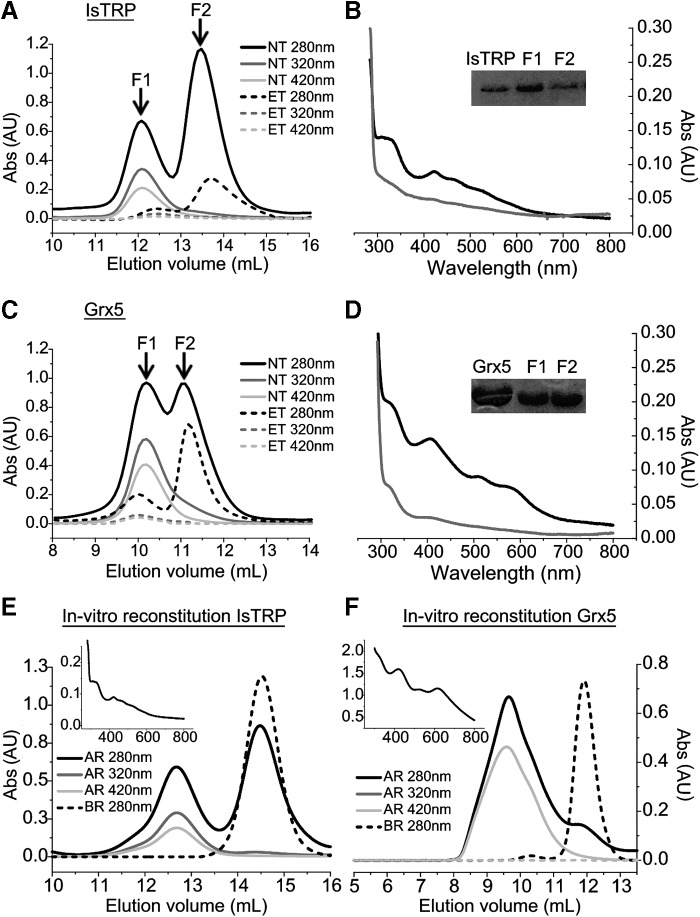
IsTRP and Grx5 were expressed as His-tagged recombinant proteins in E. coli and affinity purified as a brownish solution with UV-visible spectra compatible with the presence of bound Fe/S. Size exclusion chromatography (SEC) of purified proteins revealed a mixture of monomeric and dimeric forms for both proteins (Fig. 2A, C). IsTRP eluted with an apparent molecular mass of 22 and 11 kDa, corresponding to the dimeric and monomeric forms, respectively (theoretical mass of the monomer 15.6 kDa). In the case of Grx5, the protein eluted with an apparent molecular mass of 54 and 35 kDa, likely corresponding to the dimer and monomer even though the theoretical mass of the monomer is 20.2 kDa. This shift in the apparent molecular mass of Grx5 is probably the consequence of an increased radius due to a flexible N-terminal extension present in the recombinant protein (see full sequence in Supplementary Fig. S3). The fractions containing the dimers for IsTRP and Grx5, but not those containing the monomers, displayed absorbance at 320 and 420 nm (Fig. 2A, C) and UV-visible spectra (Fig. 2B, D) similar to Fe/S-binding proteins (4, 54), suggesting Fe/S binding. Furthermore, the formation of dimers was dependent on cluster binding since EDTA treatment (Fig. 2A, B) decreased the dimer/monomer ratio. Moreover, in vitro reconstitution of Fe/S (Fig. 2E, F) increased the dimer/monomer ratio.
FIG. 2.
SEC and spectroscopic analysis of E. granulosus IsTRP and Grx5. Size exclusion chromatography analysis of IsTRP and Grx5 is shown in (A, C), respectively, and was performed on a Superdex 75 10/300 GL column. Absorbance at 280, 320, and 420 nm is shown for EDTA-treated (ET) and nontreated (NT) samples. The UV-visible spectra and SDS-PAGE of fractions F1 (black) and F2 (gray) of the untreated samples are shown for IsTRP and Grx5 in (B, D), respectively. SDS-PAGE of recombinant proteins before (left lane) and after (right lane) gel filtration is shown as an inset. (E, F) Size exclusion chromatography of Fe/S reconstitution mixture for tag-free IsTRP and Grx5, respectively. Absorbance of the proteins AR and BR is shown. The spectra of the reconstituted proteins are shown in an inset. AR, after reconstitution; BR, before reconstitution; ET, EDTA-treated; NT, nontreated; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
IsTRP coordinates 2Fe2S clusters
The UV-visible spectrum of holo-IsTRP suggested the presence of a 2Fe2S in the cluster bound by this protein. To confirm the nature and the stoichiometry of the cluster in holo-IsTRP obtained in vivo from E. coli, the iron and sulfur content was assessed using analytical techniques (Table 1). Iron was measured with 4,7-diphenyl-1,10-phenanthroline and by atomic absorption, and sulfide was measured by the methylene blue technique. The results were consistent with the presence of one 2Fe2S center per dimer.
Table 1.
Concentration Ratio of Iron and Sulfur to Holo-IsTRP
| [Fe]/[HoloProt] | [S2-]/[HoloProt] | |
|---|---|---|
| IsTRP | 0.9 ± 0.1 | 0.89 ± 0.07 |
Iron was measured using 4,7-diphenyl-1,10-phenanthroline, and acid-labile sulfur was measured using the methylene blue method (see the Materials and Methods section). Concentration of holoprotein in the analyzed fraction was estimated by analytical SEC.
IsTRP, iron–sulfur thioredoxin-related protein; SEC, size exclusion chromatography.
Fe/S coordination by IsTRP is stable, dependent on two Cys residues of the polypeptide chain, and GSH independent
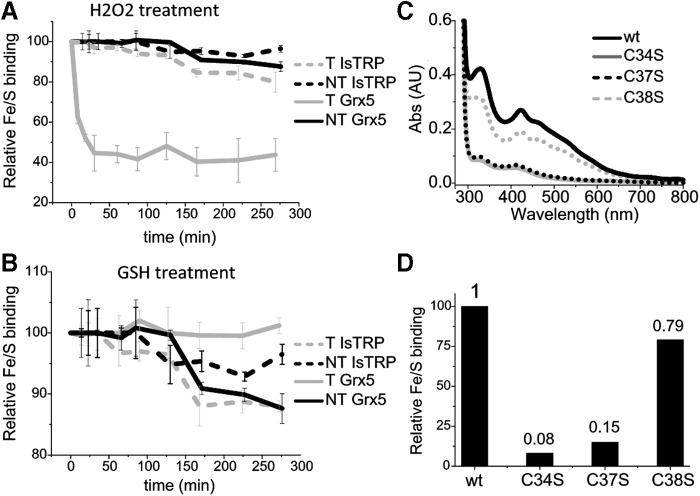
To characterize Fe/S binding by IsTRP, absorbance of the holocomplex was followed over time in the presence or absence of GSH and/or H2O2 (Fig. 3A). Experiments with Grx5 were carried out in parallel for comparison (Fig. 3B). Interestingly, GSH did not increase stability of the Fe/S bound to IsTRP, in contrast to Fe/S coordinated by Grx5. In addition, the IsTRP-bound Fe/S was far more stable (50-fold) to H2O2 treatment than that bound by Grx5, even when GSH was present in the Grx5 assay mixture (data not shown). We next examined the importance of IsTRP Cys residues in Fe/S binding using Cys→Ser mutants of each Cys residue (i.e., SxxCC, CxxSC, and CxxCS). Fe/S coordination by wild-type and mutant proteins was further assessed by a spectrophotometric analysis of recombinant proteins (Fig. 3C, D). Two cysteine residues, Cys 34 and Cys 37, corresponding to nucleophilic and resolving positions of catalytic cysteines in Trxs, respectively, were found to be essential for cluster binding by IsTRP since the 320/280 nm absorbance ratios of these mutants were 15% and 8% of the wild-type protein ratio. In addition, these mutants displayed mostly a monomeric behavior when analyzed by size exclusion chromatography (data not shown). In contrast, the contiguous Cys 38 was not required for Fe/S binding by IsTRP.
FIG. 3.
IsTRP and Grx5 Fe/S-binding properties. (A, B) Kinetics for the disassembly of holo-IsTRP and holo-Grx5 (NT) and upon treatment (T) with 10 mM of H2O2 or 3 mM of GSH. The loss of the Fe/S was recorded by following the decrease in absorbance at 320 nm. Error bars correspond to standard deviations of three replicates. (C, D) The Fe/S coordination ability of wild-type, Cys38Ser, Cys37Ser, and Cys34Ser mutants was analyzed by measuring the absorbance at 320 nm of the copurified holoproteins. Protein concentration was between 300 and 200 μM. A representative UV-visible spectrum (normalized to protein concentration) of each protein species is shown (C) and the corresponding 320/280 nm ratio relative to the binding of the wild-type protein is shown (D). GSH, glutathione.
These data, the lack of a stabilizing effect of GSH on holo-IsTRP, and the absence of the characteristic GSH-binding site residues in the sequence, strongly support that IsTRP binds Fe/S by a GSH-independent mechanism. To further test this model, the holo and apo protein fractions of IsTRP and Grx5 obtained from E. coli were acid precipitated and total low-molecular-weight thiols and GSH were measured (Table 2). The results indicated that neither GSH nor any other low-molecular-weight thiol was an integral component of the holo-IsTRP, while, as previously reported for canonical Fe/S Grxs (4), GSH was found as a cofactor for the holo-Grx5.
Table 2.
Concentration Ratio Between Low-Molecular-Weight Thiols and GSH and Apo or Holo Form of Grx5 and IsTRP Purified from Escherichia coli
| LMWT/Prot. | GSH/Prot. | |||
|---|---|---|---|---|
| Grx5 | IsTRP | Grx5 | IsTRP | |
| Holoprotein | 0.51 | 0.01 | 0.43 | <0.01 |
| Apoprotein | 0.11 | 0.01 | 0.09 | <0.01 |
GSH was added to all buffers during IMAC purification of both proteins, and free GSH was removed afterward by SEC.
GSH, glutathione; LMWT, low-molecular-weight thiols; IMAC, immobilized metal affinity chromatograph.
IsTRP did not exhibit canonical reductase activities
The disulfide redox activity of the recombinant IsTRP and Grx5 was tested using the insulin (17) and HED (18) reduction assays; a canonical E. granulosus Trx (EUB56960.1) (7) and Grx (CDS21744.1) were used as positive controls. The apo and holo forms of IsTRP and apo-Grx5 were unable to efficiently catalyze the reduction of these substrates under the reaction conditions tested (Fig. 4A, B). A marginal catalytic activity in the HED assay by Grx5 was observed. In addition, it is worth mentioning that the oxidized apo-IsTRP was reduced by TGR (AAN63052) in an NADPH-dependent assay, but only at high TGR concentration (Fig. 4C). The Cys 34 and Cys 37 residues contributed to disulfide formation since the corresponding Ser mutants showed a significant reduction in NADPH consumption by TGR. In particular, mutation of Cys 34, corresponding to the nucleophilic Cys in Trxs, led to a complete loss of TGR reductase activity with this substrate, indicating that this residue is involved in the disulfide formation.
FIG. 4.
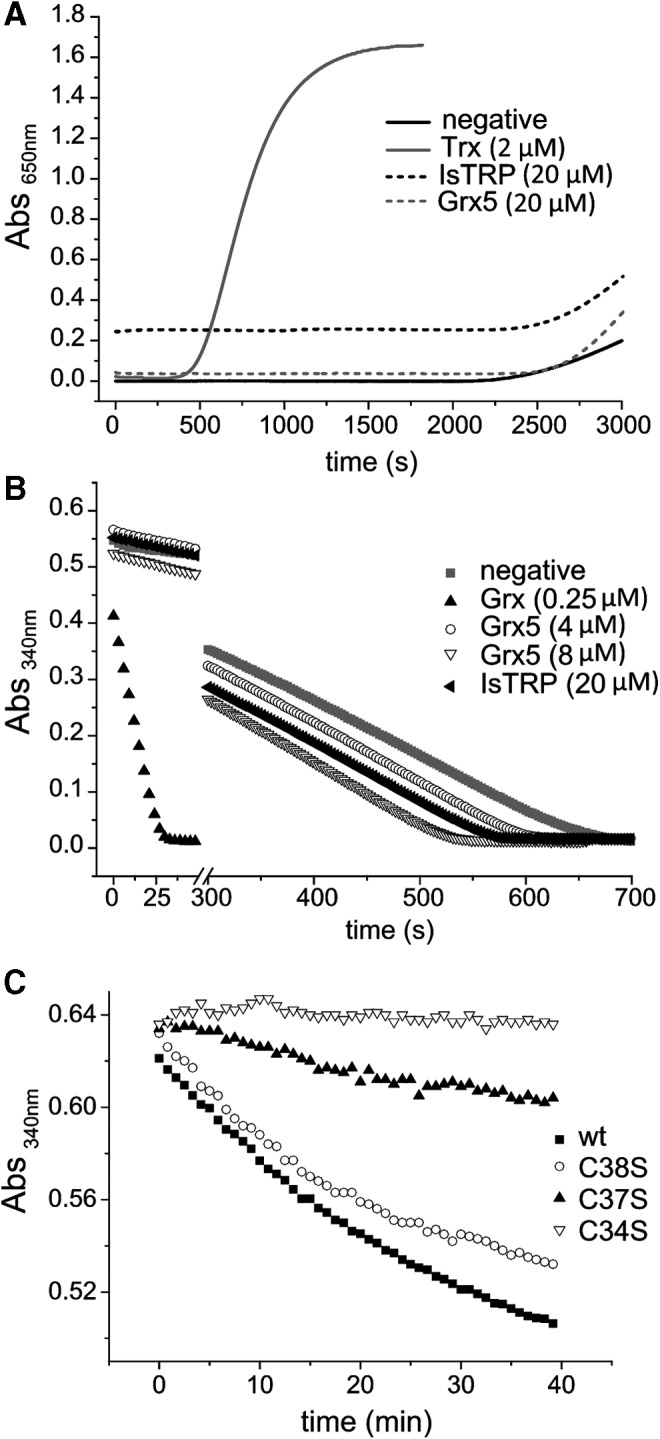
Activities of IsTRP and Grx5. (A) Insulin reduction by dithiothreitol. Twenty micromolars of IsTRP and Grx5; and 2 μM of a Trx from E. granulosus. The absorbance at 650 nm is plotted against time. (B) HED activity assay. The capacity of IsTRP, Grx5, and a Grx from E. granulosus to deglutathionylate was evaluated. NADPH oxidation was followed by measuring absorbance at 340 nm. The full time courses obtained are shown. (C) Reduction of 20 μM wild-type IsTRP (black) and mutants Cys to Ser by the TGRC31S (a TGR mutant that conserves full TR activity, but lacks gutathione reductase activity). One hundred twenty-five nanomolars of TGRC31S was used for the assay. Representative full time courses of NADPH oxidation are shown. TGR, thioredoxin glutathione reductase; TR, Thioredoxin reductase.
To test if IsTRP could catalyze the reduction of specific targets, pull-down experiments were performed using the CxxS mutant. This strategy has proved to be very efficient, particularly for identifying targets of Trxs and Grxs in different organisms [e.g., (45)]. We examined protein extracts from the larval stage of E. granulosus (protoscolex) and gravid adult worms (the stage where IsTRP is highly expressed) from the closely related parasite Hymenolepis microstoma [an experimental model noninfective to humans (12)]. No specific targets were trapped: the same proteins were identified in the CxxS mutant, wild-type, and buffer control samples. Finally, we tested whether IsTRP possesses ferredoxin activity in the cytochrome c reduction assay in vitro. No ferredoxin activity was detected for IsTRP, a spinach ferredoxin was used as a positive control (data not shown).
Grx5 and IsTRP can rescue the S. cerevisiae Grx5-deficient yeast phenotype
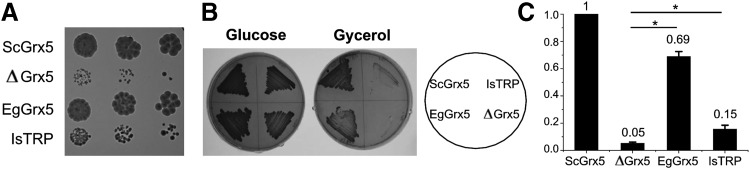
We tested whether the E. granulosus Grx5 replaces the canonical yeast mitochondrial class II Grx (Grx5). As IsTRP belongs to the same protein superfamily as mitochondrial class II Grxs and both are able to coordinate Fe/S, we also examined whether IsTRP can replace S. cerevisiae Grx5. We expressed the E. granulosus IsTRP and Grx5 in the mitochondria of the S. cerevisiae Grx5 null mutant strain (30). This null strain grows slowly in glucose and is unable to grow using glycerol (nonfermentable substrate) as the sole carbon source (30). As shown in Figure 5A and B, the mitochondrial targeted E. granulosus Grx5 rescued the growth defects of the mutant lacking Grx5. To a lower extent, the expression of IsTRP also allows growth on glycerol of the Grx5 null yeast strain (Fig. 5A, B). The aconitase to malate dehydrogenase activity ratios were also measured to assess the efficiency of Fe/S assembly (Fig. 5C), confirming the magnitude of the yeast rescued phenotype for both strains.
FIG. 5.
Rescue of the ΔGrx5 mutant defects by Grx5 and IsTRP. (A) Growth plates after a 3-day incubation at 30°C on YPD. From left to right, 1:5 serial dilutions of exponential cultures were spotted. (B) Growth on YPD or YPG plates after 3 days at 30°C. S. cerevisiae Grx5 (ScGrx5), E. granulosus (EgGrx5), E. granulosus IsTRP (IsTRP), and null mutant (ΔGrx5) are shown. (C) Ratio between the Fe/S-containing enzyme aconitase and the non-Fe/S-containing enzyme malate dehydrogenase activities in exponential cultures at 30°C in YPGal medium. Values were normalized with respect to strain MML240 that expresses yeast Grx5-HA. The values are the mean of three experiments with standard deviations. Statistical analysis was performed using one-way ANOVA (*p < 0.05).
The Fe/S synthesis pathway genes are encoded in the genome of E. granulosus and are controlled at the transcriptional level
Since Fe/S synthesis in platyhelminthes has not been characterized yet, we examined the presence of the known key genes involved in mitochondrial and cytosolic iron–sulfur protein assembly machineries (ISC and CIA, respectively) in E. granulosus and E. multilocularis (a closely related species with a similar life cycle) genomes. This survey indicated that complete ISC and CIA machineries are present in these organisms. Furthermore, the RNAseq data from E. granulosus (55) indicated that the genes are actively transcribed during most of the parasite life cycle, with the exception of activated oncospheres (the newly released embryo in the intermediate host after hatching of the ingested egg), where almost all genes of the CIA Fe/S synthesis pathway and some of the ISC machinery (including Grx5) are completely silenced (Table 3).
Table 3.
Transcription Profiles of Genes Involved in the ISC and CIA Assembly Machinery from Adult, Oncosphere, Protoscolex, and Hydatid Cyst Membrane of Echinococcus granulosus
| RPKMa | |||||
|---|---|---|---|---|---|
| Gene ID | Adult | Oncb | PSCb | Cyst | |
| Components of the ISC assembly machinery | |||||
| NFS1 | CDS19765 | 56 | 33 | 100 | 52 |
| ISD11 | EUB55154 | 695 | 964 | 131 | 306 |
| FDXR | CDS18240 | 13 | 0 | 36 | 20 |
| FDX2 | EUB56835 | 16 | 105 | 52 | 0 |
| FXN | CDS24551 | 74 | 122 | 61 | 85 |
| ISCU | EUB64018 | 444 | 455 | 91 | 159 |
| GRP75c | CDS17875 | 280 | 2170 | 372 | 610 |
| HSC20d | CDS23455 | 62 | 0 | 77 | 18 |
| GRPE-L1/L2 | EUB64601 | 82 | 77 | 96 | 81 |
| Grx5 | EUB62482 | 114 | 0 | 19 | 65 |
| Components of the CIA assembly machinery | |||||
| NUBP1 | CDS22073 | 64 | 0 | 48 | 157 |
| NUBP2 | CDS21766 | 118 | 0 | 44 | 55 |
| CIAPIN1 | EUB62290 | 78 | 0 | 369 | 125 |
| NDOR1 | EUB64339 | 0 | 0 | 31 | 26 |
| CIAO1 | CDS16778 | 24 | 78 | 19 | 24 |
| MIP18 | CDS20555 | 50 | 0 | 11 | 0 |
| Mms19 | CDS16337 | 32 | 13 | 59 | 18 |
| NARFL | CDS21645 | 30 | 0 | 19 | 39 |
Transcriptomic data were obtained from (55). An adapted figure from (25) illustrating Fe/S synthesis is shown in Supplementary Figure S5.
Reads per kilobase per million mapped reads.
Onc and PSC were treated for activation before transcriptomic analysis.
Generic chaperone.
Specific cochaperone.
Cyst, hydatid cyst membrane; Fe/S, iron–sulfur cluster; ISC, mitochondrial iron–sulfur cluster assembly machinery; ONC, oncosphere; PSC, protoscolex.
Discussion
Parasitic flatworms have been previously shown to possess specific adjustments in their Trx and GSH pathways. Some of the striking features of these pathways were the replacement of conventional TR and GR by TGR and the GSH-independent reduction of glutathionylated proteins by TGR (6). Since Trx and GSH pathways are linked and streamlined in these organisms, the presence of several Trx and Grx genes in tapeworm genomes is also intriguing (49). In the current study, we identified and characterized two Trx-fold proteins of the tapeworm E. granulosus: a mitochondrial Fe/S-binding Grx5 that possesses an unusual extended loop and IsTRP, a novel class of Trx-related protein that bound Fe/S independently of GSH and lacked classical thiol–disulfide oxidoreductase activity.
Metal coordination by Trx-fold proteins is not a random event since the majority of oxidoreductases within this superfamily undergo sequence and structural modifications to avoid metal binding (46). As shown in this study, IsTRP coordinates Fe/S through the Cys 34 and Cys 37 residues, which correspond to the homologous positions of nucleophilic and resolving Cys of Trxs, respectively. These highly conserved residues in Trxs do not coordinate Fe/S and, in fact, this binding would be highly disadvantageous for the oxidoreductase activity. In the reduced state, the resolving Cys of Trxs is partially occluded, avoiding Fe/S binding, and therefore there must be some adaptations (permanent or induced) on the structure of IsTRP to locate the resolving Cys in the correct spatial location to coordinate the Fe/S. The homology model for IsTRP revealed that Cys 37 would be also occluded (Supplementary Fig. S4). If the model is correct, the assembly of Fe/S in the apo-IsTRP must induce a conformational change to allow Cys 37 to be available for binding. This conformational change could be similar to what has been proposed for the mechanism of E. coli TrxA mutant, with a CACA active site that binds Fe/S (27). Upon Fe/S coordination, this protein would form dimers and one of the subunits would display a partial unfolding at the N-terminus of the active site α-helix (11). A similar conformational change may expose Cys 37 and allows this residue to coordinate Fe/S. Future efforts will be directed at gaining further insights into the structural features of IsTRP that allow this protein to replace the canonical oxidoreduction activity of TRPs for Fe/S binding.
Thus, if Fe/S is not a random event and requires structural adjustments of the fold, the coordination capacity of IsTRP is most likely physiologically relevant. A possible function for the Fe/S might be the regulation of the redox activity of the apoprotein as proposed for class I Grxs (4). However, oxidoreductase activity was not detected in the canonical Trx and Grx assays, and we did not succeed in identifying IsTRP targets using the CxxS mutant in pull-down experiments. In any case, the high stability of the holocomplex implies that cluster disassembly to render the active form of IsTRP would demand harsh oxidizing conditions or the action of specific stimuli (e.g., protein partners). Alternatively, IsTRP may function as storage for Fe/S. The ability of IsTRP to slightly suppress the phenotype of the Grx5 null mutant yeast would suggest that this protein might be able to incorporate Fe/S in a eukaryotic system and collaborate in Fe/S transfer to other proteins. It should be noted that overexpression of other members of ISC, such as ferredoxin, can partially rescue the Grx5 knockout phenotype (37). The lack of ferredoxin activity of IsTRP ruled out this possibility. Additional experiments need to be performed to obtain conclusive evidence regarding IsTRP-specific function(s).
On the other hand, we demonstrated here that E. granulosus Grx5 is able to replace Grx5 function in yeasts, despite having an extended loop in a noncanonical insertion site of the Trx fold. This provides further evidence of the functional conservation of mitochondrial class II Grxs among different lineages of life (31). Thus, the structural singularities of E. granulosus Grx5 may play a regulatory role or be important for parasite-specific functions (i.e., interaction with parasite-specific partners).
Although Fe/S biosynthesis has not been studied in platyhelminth parasites, all genes involved in this metabolic process are present in these organisms, suggesting that Fe/S are synthesized de novo. Interestingly, the RNASeq data indicate that most of the genes of the Fe/S biosynthesis are silenced in activated oncospheres. The oncosphere is activated by gastrointestinal cues (e.g., pH), penetrates the intestinal wall, and migrates through the circulatory system to internal organs (preferentially the liver and lung), where it develops as the metacestode larval stage. The decrease in the synthesis of weakly bound Fe/S and the concomitant decrease of the source of free iron and sulfide are likely important during migration and establishment, when the parasite is particularly exposed to the hostile environment imposed by the host. This raises the question of how are Fe/S supplied to newly synthesized essential enzymes (3, 23) during early infection in the intermediate host. We hypothesize that IsTRP might be relevant at this stage as ready-to-use storage of preformed Fe/S, synthesized during egg generation. Indeed, IsTRP is highly expressed in the gravid adult (while Fe/S are still being synthesized) and the Fe/S bound to IsTRP are resistant to oxidation.
To sum up, this study describes an entirely new class of proteins of the Trx fold unit: a Trx-related protein that lacks classical oxidoreductase activity, but can bind Fe/S by a unique mechanism, involving both Cys from the active site and no GSH. Overall, our results recapitulate some laboratory evolution experiments (27, 45), highlighting the versatility of the Trx folding unit that (in addition to the different adaptations for redox functions) also displays Fe/S binding as a recurrent emergent function.
Materials and Methods
Bioinformatic analysis
Genomic data were obtained from the Sanger Institute or generic databases. Multiple protein sequence alignments were generated with ClustalW (22). Maximum likelihood and neighbor-joining tree bootstrap values (500 replicates) were calculated using the MEGA 6 package/software (48).
Cloning of E. granulosus proteins for bacterial and yeast expression
The open reading frame of the IsTRP and Grx5 was amplified from cDNA of E. granulosus protoscolex total mRNA and cloned into pET28a (Novagen) using standard molecular cloning techniques, thereby introducing an N-terminal His-tag. Mutants of IsTRP were generated by site-directed mutagenesis using the overlap extension method (39). IsTRP and Grx5 constructs for yeast expression were generated by polymerase chain reaction amplification using pET28a constructs as DNA templates and cloned into the pMM221 plasmid (30). All primers and vectors generated are shown in Supplementary Table S1. Constructs were sequenced in all cases.
Expression and purification of recombinant proteins
The recombinant protein expression was carried out in E. coli BL21(DE3) cells following standard protocols. Essentially, recombinant clones were grown on Luria broth in the presence of kanamycin, and induction of recombinant proteins was carried out with 100 μM isopropyl-1-thio-β-D-galactopyranoside at the early exponential phase (Abs600nm = 0.6) overnight at 25°C. The bacterial cultures were centrifuged, and the pellets were resuspended in lysis buffer (300 mM NaCl, 50 mM sodium phosphate, 10 mM imidazole, pH 7.2) containing 1 mM phenylmethylsulfonyl fluoride, and sonicated. The lysates were centrifuged for 30 min at 20,000 g, and supernatants were applied to a HisPur™ Cobalt Superflow Agarose column (Thermo), washed with 300 mM NaCl, 50 mM sodium phosphate, and 20 mM imidazole, pH 7.2, and eluted with 250 mM imidazole. When it is specified, 1 mM GSH(Sigma) was added during all steps of purification. Protein concentrations were determined spectrophotometrically at 280 nm (ɛ280 = 7575 and 5625 M−1 cm−1 for IsTRP and Grx5, respectively) and purity was assessed on 15% sodium dodecyl sulfate–polyacrylamide gels under reducing conditions. TGRC31S was produced and purified as previously described (7).
Recombinant protein analysis
Protein fractions were subjected to gel chromatography on Superdex 75 10/300 GL column (GE Healthcare), pre-equilibrated in phosphate-buffered saline (pH 7.0), coupled to the ÄKTA-FPLC system (GE Healthcare) with online UV-visible detection. Standard globular proteins (6.5–75 kDa; GE Healthcare kit) were used for the calibration of the columns. Absorbance at 280, 320, and 420 nm was recorded. UV-visible spectra and reducing sodium dodecyl sulfate–polyacrylamide gel electrophoresis were carried out for the purified fractions. For low-molecular-weight thiol-containing compound determinations, size exclusion chromatography fractions were subjected to 5% trichloroacetic acid treatment during 30 min on ice, for complete Fe/S disassembly, and 20,000 g centrifugation, for protein removal. Samples were neutralized in all cases. DTNB recycling assay was performed, as previously described (32), for measuring total GSH and the Ellman's reaction was used to measure total free thiols, following the manufacturer's specifications (Thermo). Protein concentration was measured before Fe/S disassembly. Iron determination was done using 4,7-diphenyl-1,10-phenanthroline (Sigma) as previously described (28). Ferric iron was reduced using ascorbic acid (Fisher) and standard curves were performed using ammonium iron (II) sulfate (Aldrich). Sulfur determination was performed as described (9, 41). Briefly, 150 μl of 20 mM N.N-dimethyl-p-phenylenediamine dissolved in 7.2 N hydrochloric acid and 150 μl of 30 mM ferric chloride in 1.2 N hydrochloric acid were added to a final volume of 1 mL; 1.5 mM zinc acetate was added to the reaction to avoid sulfide gas loss. The absorbance at 670 nm was determined after 20 min of incubation. Standard curves were performed using sodium sulfide. The Fe content of IsTRP was determined by atomic absorption using a Plasma Emission Spectrometer (Jarrell-Ash 965 ICP) in the Chemical Analysis Laboratory, University of Georgia.
Fe/S reconstitution assay
Purified IsTRP was treated with thrombin to obtain the tag-free protein and afterward thrombin was removed by a p-aminobenzamidine–agarose prepacked column (Sigma). In vitro Fe/S reconstitution of IsTRP was performed as described (4). One hundred to 200 μM of IsTRP was incubated under argon atmosphere at room temperature with 10 mM equivalents of E. coli cysteine desulfurase IscS, 2.5–5 equivalents of cysteine, 2–4 equivalents of Fe(NH4)2(SO4)2, 1 mM GSH, 5 mM DTT, and 10 μM pyridoxal phosphate in 50 mM sodium phosphate buffer, pH 8.0, containing 200 mM NaCl.
Expression of IsTRP and Grx5 in Grx5-deficient S. cerevisiae
All employed yeast strains belong to the W303 genetic background and are summarized in Supplementary Table S2. The plasmids were linearized by ClaI digestion and then used for transformation by the standard lithium salts method (1). Crosses between yeast strains, sporulation, and tetrad analyses were carried out by standard genetic techniques. S. cerevisiae cultures were grown at 30°C in YPD, YPG (as YPD, but with 3% glycerol instead of dextrose), or YPGal (as YPD, but with 2% galactose instead of dextrose), as previously described (30).
Enzymatic assays
The insulin interchain disulfide reduction was performed as described (17). The reduction of two interchain disulfides of insulin catalyzed by IsTRP in the presence of DTT was used as a measure of Trx activity. The reaction was followed by the increase in absorbance at 650 nm due to the precipitation of free insulin β-chain. The 0.8 ml reaction mixtures contained 0.33 mM DTT, 130 μM insulin, and 2 mM non-EDTA in 100 mM potassium phosphate buffer, pH 7.0. Time courses with DTT alone and the reaction catalyzed by an E. granulosus Trx (2 μM, EUB56960.1) were performed as controls. The Grx assay was performed as described previously (18). A reaction mixture containing 1 mM GSH (Sigma), 2 mM non-EDTA, 0.1 mg ml−1 bovine serum albumin (BSA), 0.7 mM β-hydroxyethyl disulfide (Acros Organics), and 0.4 units of yeast (GR; Sigma) was preincubated for 2 min. Afterward, the protein was added, and then the reduction of the mixed glutathione-hydroxyethyl disulfide, followed by the oxidation of NADPH at 340 nm, was performed. Reduction of spontaneously oxidized IsTRP by the TGRC31S mutant (lacking the Grx and GR activities) (6) was performed as described (20). The mixture contained 100 mM potassium phosphate buffer, pH 7.0, 1 mM EDTA, 0.1 mg/ml BSA, 100 μM NADPH, and 20 μM IsTRP. The reaction was started by addition of 125 nM of TGRC31S. Aconitase and malate dehydrogenase activities were assayed following the methods described (36). Ferredoxin activity was determined following the generation of reduced cytochrome C at 550 nm, 0.3 mM NADPH, 50 mM Tris pH 8.5, 50 μM cytochrome c (Sigma), and 47 nM of pea ferredoxin reductase (40). Up to 50 μM IsTRP was added to the assay and 2.5 μM spinach ferredoxin (Sigma) was used as positive control.
Pull-down experiments
One milligram of IsTRPwt, IsTRPCxxS mutant, or only buffer samples was incubated for 30 min at 4°C under gentle agitation with Ni-NTA Magnetic Agarose Beads (Qiagen). The immobilized bait was then incubated with aqueous extracts obtained from 100 μL of packed E. granulosus protoscolex or H. microstoma adult worms homogenized using a mortar and pestle under liquid nitrogen and sonicated. After washing 10 times with 300 mM NaCl, 50 mM sodium phosphate, and 30 mM imidazole, pH 7.2, elution was performed with 10 mM DTT and 8 M urea. In-gel trypsin digestion was performed and the samples extracted from the gel were analyzed in a reverse phase EASY-Spray column, 50 cm ×75 μm ID, PepMap RSLC C18, 2 μm, (Thermo Scientific), and an LTQ VELOS nano-ESI (Thermo Scientific). PatternLab for Proteomics (version 3.2.0.3) was used for spectral analysis and protein identification (10).
Homology model generation
Crystal structures were retrieved from the PDB using the HHpred suite for distant homology detection (42). A multiple-template approach was taken using Modeller version 9v11 (50) to build models from the multiple structural alignments gathered by the HHsuite. 3ipzA, 2wulA, 2yanA, and 3zywA were used as templates for Grx5. For TRP, the templates were 3m9jA, 2vimA, 4aj8A, 2yoiA, and 4i8bA. The best models obtained from 50 iterations were determined with the Discrete Optimized Protein Energy method, included in the Modeller suite. Overall quality assessment of the final models was done with Coot (13). Electrostatic calculations were made with the Adaptive Poisson–Boltzmann Solver (2). All figures illustrating the protein structure were prepared using open-source PyMOL (http://www.pymol.org/).
Supplementary Material
Abbreviations Used
- CIA
cytosolic iron–sulfur protein assembly machinery
- DTNB
dithio-nitrobenzoic acid
- Fe/S
iron–sulfur cluster
- GR
glutathione reductase
- Grx
glutaredoxin
- GSH
glutathione
- ISC
mitochondrial iron–sulfur cluster assembly machinery
- IsTRP
iron–sulfur thioredoxin-related protein
- SEC
size exclusion chromatography
- TGR
thioredoxin glutathione reductase
- TR
thioredoxin reductase
- TRP
thioredoxin-related protein
- Trx
thioredoxin
Acknowledgments
The authors thank Dr. Enrique Herrero (Universitat de Lleida, Spain) for provision of yeast strains and helpful experimental assistance, Dr. Matt Berriman (Parasite Genomics, Wellcome Trust Sanger Institute) for provision of data from E. multilocularis transcriptome, Dr. Rosario Durán (Analytical and Biochemical Proteomic Unit, Institut Pasteur Montevideo) for technical assistance with proteomics, Dr. Néstor Carrillo for provision of purified pea ferredoxin and assistance with the ferredoxin assay, Dr. Uriel Koziol (Facultad de Ciencias, Udelar, Uruguay) for provision of H. microstoma worms, and Dr. Massimo Bellanda (University of Padova) for helpful discussions. This work was supported by Universidad de la República, Uruguay (Grant Number I + D 625 to G.S. and INI 915 to H.B.), Agencia Nacional de Innovación e Investigación (POS_NAC_2012_1_8789), ICGEB Research Grant URU14-01 to G.S. and M.C, FOCEM (MERCOSUR Structural Convergence Fund, [COF 03/11]), and Seeding Labs.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Agatep R. Transformation of Saccharomyces cerevisiae by the lithium acetate/single-stranded carrier DNA/polyethylene glycol protocol. Technical Tips Online 3: 133–137, 1998 [Google Scholar]
- 2.Baker NA, Sept D, Joseph S, Holst MJ, and McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 98: 10037–10041, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balk J. and Pilon M. Ancient and essential: the assembly of iron-sulfur clusters in plants. Trends Plant Sci 16: 218–226, 2011 [DOI] [PubMed] [Google Scholar]
- 4.Berndt C, Hudemann C, Hanschmann EM, Axelsson R, Holmgren A, and Lillig CH. How does iron-sulfur cluster coordination regulate the activity of human glutaredoxin 2? Antioxid Redox Signal 9: 151–157, 2007 [DOI] [PubMed] [Google Scholar]
- 5.Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, Cerqueira GC, Mashiyama ST, Al-Lazikani B, Andrade LF, Ashton PD, Aslett MA, Bartholomeu DC, Blandin G, Caffrey CR, Coghlan A, Coulson R, Day TA, Delcher A, DeMarco R, Djikeng A, Eyre T, Gamble JA, Ghedin E, Gu Y, Hertz-Fowler C, Hirai H, Hirai Y, Houston R, Ivens A, Johnston DA, Lacerda D, Macedo CD, McVeigh P, Ning Z, Oliveira G, Overington JP, Parkhill J, Pertea M, Pierce RJ, Protasio AV, Quail MA, Rajandream MA, Rogers J, Sajid M, Salzberg SL, Stanke M, Tivey AR, White O, Williams DL, Wortman J, Wu W, Zamanian M, Zerlotini A, Fraser-Liggett CM, Barrell BG, and El-Sayed NM. The genome of the blood fluke Schistosoma mansoni. Nature 460: 352–358, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonilla M, Denicola A, Marino SM, Gladyshev VN, and Salinas G. Linked thioredoxin-glutathione systems in platyhelminth parasites: alternative pathways for glutathione reduction and deglutathionylation. J Biol Chem 286: 4959–4967, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonilla M, Denicola A, Novoselov SV, Turanov AA, Protasio A, Izmendi D, Gladyshev VN, and Salinas G. Platyhelminth mitochondrial and cytosolic redox homeostasis is controlled by a single thioredoxin glutathione reductase and dependent on selenium and glutathione. J Biol Chem 283: 17898–17907, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brautigam L, Johansson C, Kubsch B, McDonough MA, Bill E, Holmgren A, and Berndt C. An unusual mode of iron-sulfur-cluster coordination in a teleost glutaredoxin. Biochem Biophys Res Commun 436: 491–496, 2013 [DOI] [PubMed] [Google Scholar]
- 9.Carballal S, Trujillo M, Cuevasanta E, Bartesaghi S, Moller MN, Folkes LK, Garcia-Bereguiain MA, Gutierrez-Merino C, Wardman P, Denicola A, Radi R, and Alvarez B. Reactivity of hydrogen sulfide with peroxynitrite and other oxidants of biological interest. Free Radic Biol Med 50: 196–205, 2011 [DOI] [PubMed] [Google Scholar]
- 10.Carvalho PC, Fischer JS, Xu T, Yates JR, 3rd, and Barbosa VC. PatternLab: from mass spectra to label-free differential shotgun proteomics. Curr Protoc Bioinformatics Chapter 13: Unit13.19, 2012 [DOI] [PubMed] [Google Scholar]
- 11.Collet JF, Peisach D, Bardwell JC, and Xu Z. The crystal structure of TrxA(CACA): insights into the formation of a [2Fe-2S] iron-sulfur cluster in an Escherichia coli thioredoxin mutant. Protein Sci 14: 1863–1869, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunningham LJ. and Olson PD. Description of Hymenolepis microstoma (Nottingham strain): a classical tapeworm model for research in the genomic era. Parasit Vectors 3: 123–131, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emsley P, Lohkamp B, Scott WG, and Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandes AP, Fladvad M, Berndt C, Andresen C, Lillig CH, Neubauer P, Sunnerhagen M, Holmgren A, and Vlamis-Gardikas A. A novel monothiol glutaredoxin (Grx4) from Escherichia coli can serve as a substrate for thioredoxin reductase. J Biol Chem 280: 24544–24552, 2005 [DOI] [PubMed] [Google Scholar]
- 15.Golinelli MP, Gagnon J, and Meyer J. Specific interaction of the [2Fe-2S] ferredoxin from Clostridium pasteurianum with the nitrogenase MoFe protein. Biochemistry 36: 11797–11803, 1997 [DOI] [PubMed] [Google Scholar]
- 16.Hanschmann EM, Godoy JR, Berndt C, Hudemann C, and Lillig CH. Thioredoxins, glutaredoxins, and peroxiredoxins—molecular mechanisms and health significance: from cofactors to antioxidants to redox signaling. Antioxid Redox Signal 19: 1539–1605, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem 254: 9627–9632, 1979 [PubMed] [Google Scholar]
- 18.Holmgren A. and Aslund F. Glutaredoxin. Methods Enzymol 252: 283–292, 1995 [DOI] [PubMed] [Google Scholar]
- 19.Holmgren A, Johansson C, Berndt C, Lonn ME, Hudemann C, and Lillig CH. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem Soc Trans 33: 1375–1377, 2005 [DOI] [PubMed] [Google Scholar]
- 20.Johansson C, Lillig CH, and Holmgren A. Human mitochondrial glutaredoxin reduces S-glutathionylated proteins with high affinity accepting electrons from either glutathione or thioredoxin reductase. J Biol Chem 279: 7537–7543, 2004 [DOI] [PubMed] [Google Scholar]
- 21.Johansson C, Roos AK, Montano SJ, Sengupta R, Filippakopoulos P, Guo K, von Delft F, Holmgren A, Oppermann U, and Kavanagh KL. The crystal structure of human GLRX5: iron-sulfur cluster co-ordination, tetrameric assembly and monomer activity. Biochem J 433: 303–311, 2011 [DOI] [PubMed] [Google Scholar]
- 22.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, and Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948, 2007 [DOI] [PubMed] [Google Scholar]
- 23.Lill R. Function and biogenesis of iron-sulphur proteins. Nature 460: 831–838, 2009 [DOI] [PubMed] [Google Scholar]
- 24.Lill R, Hoffmann B, Molik S, Pierik AJ, Rietzschel N, Stehling O, Uzarska MA, Webert H, Wilbrecht C, and Muhlenhoff U. The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim Biophys Acta 1823: 1491–1508, 2012 [DOI] [PubMed] [Google Scholar]
- 25.Lill R, Srinivasan V, and Muhlenhoff U. The role of mitochondria in cytosolic-nuclear iron-sulfur protein biogenesis and in cellular iron regulation. Curr Opin Microbiol 22: 111–119, 2014 [DOI] [PubMed] [Google Scholar]
- 26.Lillig CH, Berndt C, and Holmgren A. Glutaredoxin systems. Biochim Biophys Acta 1780: 1304–1317, 2008 [DOI] [PubMed] [Google Scholar]
- 27.Masip L, Pan JL, Haldar S, Penner-Hahn JE, DeLisa MP, Georgiou G, Bardwell JC, and Collet JF. An engineered pathway for the formation of protein disulfide bonds. Science 303: 1185–1189, 2004 [DOI] [PubMed] [Google Scholar]
- 28.Massey V. Studies on succinic dehydrogenase. VII. Valency state of the iron in beef heart succinic dehydrogenase. J Biol Chem 229: 763–770, 1957 [PubMed] [Google Scholar]
- 29.Meyer J. Ferredoxins of the third kind. FEBS Lett 509: 1–5, 2001 [DOI] [PubMed] [Google Scholar]
- 30.Molina MM, Belli G, de la Torre MA, Rodriguez-Manzaneque MT, and Herrero E. Nuclear monothiol glutaredoxins of Saccharomyces cerevisiae can function as mitochondrial glutaredoxins. J Biol Chem 279: 51923–51930, 2004 [DOI] [PubMed] [Google Scholar]
- 31.Molina-Navarro MM, Casas C, Piedrafita L, Belli G, and Herrero E. Prokaryotic and eukaryotic monothiol glutaredoxins are able to perform the functions of Grx5 in the biogenesis of Fe/S clusters in yeast mitochondria. FEBS Lett 580: 2273–2280, 2006 [DOI] [PubMed] [Google Scholar]
- 32.Morgan B, Ezerina D, Amoako TN, Riemer J, Seedorf M, and Dick TP. Multiple glutathione disulfide removal pathways mediate cytosolic redox homeostasis. Nat Chem Biol 9: 119–125, 2013 [DOI] [PubMed] [Google Scholar]
- 33.Nordberg J. and Arner ES. Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic Biol Med 31: 1287–1312, 2001 [DOI] [PubMed] [Google Scholar]
- 34.Pader I, Sengupta R, Cebula M, Xu J, Lundberg JO, Holmgren A, Johansson K, and Arner ES. Thioredoxin-related protein of 14 kDa is an efficient L-cystine reductase and S-denitrosylase. Proc Natl Acad Sci U S A 111: 6964–6969, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan JL. and Bardwell JC. The origami of thioredoxin-like folds. Protein Sci 15: 2217–2227, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson JB, Jr., Brent LG, Sumegi B, and Srere PA. An enzymatic approach to the study of the Krebs tricarboxylic acid cycle. In: Mitochondria: A Practical Approach, edited by Darley-Usmar VM, Rickwood D, and Wilson MT. Washington, DC: IRL Press, 1987, pp. 153–169 [Google Scholar]
- 37.Rodriguez-Manzaneque MT, Tamarit J, Belli G, Ros J, and Herrero E. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol Biol Cell 13: 1109–1121, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouault TA. Biogenesis of iron-sulfur clusters in mammalian cells: new insights and relevance to human disease. Dis Model Mech 5: 155–164, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch EF, and Maniatis T. (Eds). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory, 1989, p. 1659 [Google Scholar]
- 40.Shin M. Ferredoxin-NADP reductase from spinach. Methods Enzymol 23: 440–447, 1971 [Google Scholar]
- 41.Siegel LM. A direct microdetermination for sulfide. Anal Biochem 11: 126–132, 1965 [DOI] [PubMed] [Google Scholar]
- 42.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics 21: 951–960, 2005 [DOI] [PubMed] [Google Scholar]
- 43.Srinivasan V, Pierik AJ, and Lill R. Crystal structures of nucleotide-free and glutathione-bound mitochondrial ABC transporter Atm1. Science 343: 1137–1140, 2014 [DOI] [PubMed] [Google Scholar]
- 44.Stroher E. and Millar AH. The biological roles of glutaredoxins. Biochem J 446: 333–348, 2012 [DOI] [PubMed] [Google Scholar]
- 45.Sturm N, Jortzik E, Mailu BM, Koncarevic S, Deponte M, Forchhammer K, Rahlfs S, and Becker K. Identification of proteins targeted by the thioredoxin superfamily in Plasmodium falciparum. PLoS Pathog 5: e1000383, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su D, Berndt C, Fomenko DE, Holmgren A, and Gladyshev VN. A conserved cis-proline precludes metal binding by the active site thiolates in members of the thioredoxin family of proteins. Biochemistry 46: 6903–6910, 2007 [DOI] [PubMed] [Google Scholar]
- 47.Tamarit J, Belli G, Cabiscol E, Herrero E, and Ros J. Biochemical characterization of yeast mitochondrial Grx5 monothiol glutaredoxin. J Biol Chem 278: 25745–25751, 2003 [DOI] [PubMed] [Google Scholar]
- 48.Tamura K, Stecher G, Peterson D, Filipski A, and Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsai IJ, Zarowiecki M, Holroyd N, Garciarrubio A, Sanchez-Flores A, Brooks KL, Tracey A, Bobes RJ, Fragoso G, Sciutto E, Aslett M, Beasley H, Bennett HM, Cai J, Camicia F, Clark R, Cucher M, De Silva N, Day TA, Deplazes P, Estrada K, Fernandez C, Holland PW, Hou J, Hu S, Huckvale T, Hung SS, Kamenetzky L, Keane JA, Kiss F, Koziol U, Lambert O, Liu K, Luo X, Luo Y, Macchiaroli N, Nichol S, Paps J, Parkinson J, Pouchkina-Stantcheva N, Riddiford N, Rosenzvit M, Salinas G, Wasmuth JD, Zamanian M, Zheng Y, Cai X, Soberon X, Olson PD, Laclette JP, Brehm K, and Berriman M. The genomes of four tapeworm species reveal adaptations to parasitism. Nature 496: 57–63, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webb B. and Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics 47: 5.6.1–5.6.32, 2014 [DOI] [PubMed] [Google Scholar]
- 51.Williams DL, Bonilla M, Gladyshev VN, and Salinas G. Thioredoxin glutathione reductase-dependent redox networks in platyhelminth parasites. Antioxid Redox Signal 19: 735–745, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yeh AP, Chatelet C, Soltis SM, Kuhn P, Meyer J, and Rees DC. Structure of a thioredoxin-like [2Fe-2S] ferredoxin from Aquifex aeolicus. J Mol Biol 300: 587–595, 2000 [DOI] [PubMed] [Google Scholar]
- 53.Zaffagnini M, Michelet L, Massot V, Trost P, and Lemaire SD. Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii reveals the unique properties of a chloroplastic CGFS-type glutaredoxin. J Biol Chem 283: 8868–8876, 2008 [DOI] [PubMed] [Google Scholar]
- 54.Zhang B, Bandyopadhyay S, Shakamuri P, Naik SG, Huynh BH, Couturier J, Rouhier N, and Johnson MK. Monothiol glutaredoxins can bind linear [Fe3S4]+ and [Fe4S4]2+ clusters in addition to [Fe2S2]2+ clusters: spectroscopic characterization and functional implications. J Am Chem Soc 135: 15153–15164, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng H, Zhang W, Zhang L, Zhang Z, Li J, Lu G, Zhu Y, Wang Y, Huang Y, Liu J, Kang H, Chen J, Wang L, Chen A, Yu S, Gao Z, Jin L, Gu W, Wang Z, Zhao L, Shi B, Wen H, Lin R, Jones MK, Brejova B, Vinar T, Zhao G, McManus DP, Chen Z, Zhou Y, and Wang S. The genome of the hydatid tapeworm Echinococcus granulosus. Nat Genet 45: 1168–1175, 2013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.