Activation of the kiwifruit suberin biosynthesis gene AchnFHT is coordinately controlled via repression by AchnMYB4 and promotion by AchnABF2, AchnMYB41, and AchnMYB107.
Keywords: Abscisic acid, Actinidia chinensis, kiwifruit, Nicotiana benthamiana, suberization, transcription factor, transcriptional regulation, wound healing
Abstract
Suberin is a cell-wall biopolymer with aliphatic and aromatic domains that is synthesized in the wound tissues of plants in order to restrict water loss and pathogen infection. ω-hydroxyacid/fatty alcohol hydroxycinnamoyl transferase (FHT) is required for cross-linking of the aliphatic and aromatic domains. ABA is known to play a positive role in suberin biosynthesis but it is not known how it interacts with FHT. In this study, the kiwifruit (Actinidia chinensis) AchnFHT gene was isolated and was found to be localized in the cytosol. Transient overexpression of AchnFHT in leaves of Nicotiana benthamiana induced massive production of ferulate, ω-hydroxyacids, and primary alcohols, consistent with the in vitro ability of AchnFHT to catalyse acyl-transfer from feruloyl-CoA to ω-hydroxypalmitic acid and 1-tetradecanol. A regulatory function of four TFs (AchnABF2, AchnMYB4, AchnMYB41, and AchnMYB107) on AchnFHT was identified. These TFs localized in the nucleus and directly interacted with the AchnFHT promoter in yeast one-hybrid assays. Dual-luciferase analysis indicated that AchnABF2, AchnMYB41, and AchnMYB107 activated the AchnFHT promoter while AchnMYB4 repressed it. These findings were supported by the results of transient overexpression in N. benthamiana, in which AchnABF2, AchnMYB41, and AchnMYB107 induced expression of suberin biosynthesis genes (including FHT) and accumulation of suberin monomers, whilst AchnMYB4 had the opposite effect. Exogenous ABA induced the expression of AchnABF2, AchnMYB41, AchnMYB107, and AchnFHT and induced suberin monomer formation, but it inhibited AchnMYB4 expression. In addition, fluridone (an inhibitor of ABA biosynthesis) was found to counter the inductive effects of ABA. Activation of suberin monomer biosynthesis by AchnFHT was therefore controlled in a coordinated way by both repression of AchnMYB4 and promotion of AchnABF2, AchnMYB41, and AchnMYB107.
Introduction
Suberin forms a lipophilic extracellular barrier and is usually deposited on the inner side of the primary cell wall in plants (Ranathunge et al., 2011). It is a complex biopolymer composed of cross-linked aliphatic and aromatic domains (Graça et al., 2015) and is constitutively synthesized in a variety of both internal and exposed plant tissues (Vishwanath et al., 2015) to protect wound tissue from water loss and from bacterial and fungal attack (Leide et al., 2012). The aliphatic domain consists of a glycerol-based fatty acid-derived polyester comprised primarily of ω-hydroxyacids, α, ω-dicarboxylic acids, fatty alcohols, and small amounts of p-hydroxycinnamic acids (mainly ferulate) (Pollard et al., 2008; Graça et al., 2015), while the aromatic domain is principally composed of p-hydroxycinnamates (ferulate, p-coumarate, and sinapate) and their derivatives (Bernards and Razem, 2001). Fatty ω-hydroxyacid/fatty alcohol hydroxycinnamoyl transferase (FHT) is a member of the BAHD family of HxxxD-type acyltransferases, and transfers feruloyl from feruloyl-CoA to fatty ω-hydroxyacids and fatty alcohols to form suberin monomers (Serra et al., 2010). Genes encoding FHT required for suberin biosynthesis have been identified in Arabidopsis roots (ASFT/HHT) and potato tubers (FHT) (Gou et al., 2009; Molina et al., 2009; Boher et al., 2013).
Wound damage commonly occurs in fresh fruit and vegetables during harvest and post-harvest processing, and rapid wound healing including suberin deposition at the wound site can extend the subsequent storage life (Leide et al., 2012; Fugate et al., 2016). ABA is a broad-spectrum phytohormone involved in a host of biological processes, including responses to biotic and abiotic factors (Yoshida et al., 2010, 2015). There is accumulating evidence to indicate that ABA plays a positive role in suberin biosynthesis in a variety of different species and tissues, including Arabidopsis roots (Efetova et al., 2007), potato tubers (Boher et al., 2013), tomato fruit (Leide et al., 2012), and kiwifruit (Han et al., 2017). Our previous studies have indicated that ABA can increase wound-associated deposition of suberin, with a concomitant increase in the expression of suberin biosynthetic genes in kiwifruit (Han et al., 2017, 2018) and tomato fruit (Tao et al., 2016). The blocking of ABA biosynthesis by fluridone (FLD) provides a reliable means of determining the role of ABA in wound-induced suberization in potato tubers (Lulai et al., 2008) and tomato fruit (Tao et al., 2016).
Transcriptional regulation plays a crucial role in the ABA signaling pathway. Many transcription factors (TFs) have been identified that mediate ABA regulation through the cis-acting regulatory elements of ABA/stress-inducible genes (Agarwal and Jha, 2010). MYB genes constitute a large TF family with diverse functions including suberin monomer biosynthesis (Kosma et al., 2015; Lashbrooke et al., 2016; Legay et al., 2016). Arabidopsis mutants of MYB107 display significant reductions in ASFT expression and in ferulate accumulation in seeds (Lashbrooke et al., 2016), while overexpression of AtMYB41 in Arabidopsis and Nicotiana benthamiana induces ASFT expression and ferulate accumulation (Kosma et al., 2015). ABF2 is one of the master TFs of ABA signaling in response to abiotic stress (Gao et al., 2016). MYB4 functions as a repressor of the cinnamate 4-hydroxylase (C4H) and 4-coumarate (4CL; CoA Ligase) genes to control the formation of the sinapate ester (hydroxycinnamic acid derivatives) (Jin et al., 2000). However, the identity of the TFs that directly control the ABA-mediated suberin biosynthetic genes has not been determined.
In this study, we isolated the kiwifruit AchnFHT and four TF genes, AchnABF2, AchnMYB4, AchnMYB41 and AchnMYB107. The functional characterization of AchnFHT was determined by the in vitro activity of the purified protein and by transient overexpression in N. benthamiana leaves. The regulatory functions of AchnABF2, AchnMYB4, AchnMYB41, and AchnMYB107 on AchnFHT were investigated using yeast one-hybrid and dual-luciferase assays, and by transient overexpression in N. benthamiana leaves. In addition, the responses of gene expression and accumulation of suberin monomers to exogenous ABA and FLD were determined.
Materials and methods
Plant material and treatments
Kiwifruit (Actinidia chinensis Planch, cv. Xuxiang) free from physical injuries and infections, and of similar size and shape were harvested at commercial maturity in Hangzhou, Zhejiang Provence, China. The harvested fruit were surface-sterilized with 0.5% (v/v) sodium hypochlorite solution for 3 min, washed with sterile deionized water, surface-dried, and then cut longitudinally in half using a sterilized blade on a sterile bench (SW-CJ-1D, Suzhou Purification Equipment Co., China) at room temperature. The fruit halves were treated with either deionized water, 0.1 mmol l–1 FLD, or 0.5 mmol l–1 ABA using vacuum-infiltration as described previously (Tao et al., 2016; Han et al., 2018). A total of 270 fruit were harvested from different plants and randomly divided into three groups of 90 for each treatment with three replicates. After treatment, the fruit halves were placed into a sterile incubator (HWS, Ningbo Southeast Instrument Co., China) in darkness for wound healing at 20 °C and 85% RH (relative humidity). Wound tissue samples consisted of slices of the outer pericarp region of 1.0−1.5 mm thickness below the cut surface of the fruit halves (Wei et al. (2018). The collected samples were frozen in liquid nitrogen and stored at –80 °C until further analysis.
RNA extraction and cDNA synthesis
Total RNA was extracted from the wound tissues of kiwifruit and from leaves of Nicotiana benthamiana using the cetyltrimethylammonium bromide (CTAB) method (Reid et al., 2006). The extraction buffer contained 0.3 M Tris HCl (pH 8.0), 25 mM EDTA, 2 M NaCl, 2% CTAB, 2% PVPP, and 0.05% spermidine trihydrochloride. Samples were ground to a fine powder in liquid nitrogen using a mortar and pestle. The powder was added to pre-warmed extraction buffer with 2% β-mercaptoethanol, incubated at 65 °C for 10 min, and centrifuged at 10 000 g at 4 °C for 10 min. The supernatant was extracted twice with an equal volume of chloroform and then centrifuged, followed by mixing with 0.2 volumes of LiCl (12 M), and then stored at 4 °C for 12 h. The supernatant was centrifuged at 10 000 g at 4 °C for 20 min, and the RNA was dissolved in SSTE buffer. RNA was precipitated by mixing the solution with two volumes of pre-chilled absolute ethanol at –80 °C for 30 min and pelleted by centrifugation. The RNA pellet was washed with 75% ethanol (–20 °C), air-dried, and dissolved in RNase-free water. RNA concentrations and 260/280 nm ratios were determined using a NanoDrop 2000 (NanoDrop Technologies, Inc.). Contaminating gDNA was removed using a TURBO DNA-free kit (ThermoFisher Scientific). A 1-μg aliquot of RNA was used for cDNA synthesis using an iScriptTM cDNA Synthesis kit (Bio-Rad).
Real-time PCR analysis
The cDNA of kiwifruit and N. benthamiana genes for RT-qPCR were obtained by tblastx analysis against the kiwifruit genome database (http://bioinfo.bti.cornell.edu/cgi-bin/kiwi/home.cgi) and the SOL Genomics Network database (https://solgenomics.net/), respectively, using Arabidopsis suberin biosynthetic genes as the queries (see Supplementary Table S1 at JXB online). Gene-specific primers were designed using Primer 5.0 and are listed in Supplementary Table S1. Real-time PCR was performed in 96-well plates using SYBR® Premix Ex TaqTM (Takara Biomedical Technology Co., China) with an Applied Biosystems 7500 real-time PCR system (ThermoFisher Scientific). The 2−△△ CT method was used to determine the relative fold-differences in template abundance for each sample. The gene-specific primers, SYBR® Premix Ex TaqTM II (Tli RNaseH Plus), and the cDNA template were mixed to obtain the 25-μl PCR reaction mix. Reactions were run using the recommended cycling parameters of 95 °C for 2 min followed by 95 °C for 5 s, 60 °C for 30 s, for 40 cycles. Controls without the cDNA template were included for each primer pair, and each PCR reaction was conducted in triplicate.
AchnFHT heterologous expression and enzyme activity assays
The AchnFHT coding sequence was amplified using forward and reverse primers with the recognition sites SacI-SalI to subclone into the pET-28a vector (Supplementary Table S2). A His tag was fused in-frame at the N terminus. This construct was transferred to Escherichia coli BL21 (DE3) cells by heat-shock and they were grown in Luria-Bertani (LB) plates containing 50 µg ml−1 carbenicillin to express the fused proteins. A single bacterial colony was used to inoculate a liquid LB medium with 50 µg ml−1 carbenicillin and was grown overnight at 37 °C with shaking. The pre-culture (5 ml) was used to inoculate 500 ml of fresh medium until OD600=0.5, and expression of AchnFHT was induced for 15 h at 18 °C with 0.1 mM isopropyl-β-thiogalactopyranoside (IPTG). The procedures for protein extraction and subsequent purification were as described by Beuerle and Pichersky (2002). The soluble recombinant protein was purified by Ni2+-chelating chromatography, and the presence of the fusion protein was confirmed by SDS-PAGE.
Feruloyl transferase assays were performed according to Serra et al. (2010). The reaction mixture (1 ml) contained 0.2 mM feruloyl-CoA, 1 mM dithiothreitol, 50 mM Tris-HCl, 0.2 mM ω-hydroxypalmitic acid or 1-tetradecanol, and 20 µl of recombinant AchnFHT protein. The reaction mixture was incubated for 20 min at 30 °C, stopped with the same volume of hot acetonitrile (60 °C), heated at 96 °C for 1 min, and then the mixture was filtered through 0.22-μm membranes before HPLC/LC-MS analysis. Reactions were performed with three replicates. The reaction products were analysed using a Waters e2695 HPLC equipped with a C18 column (Intertsil ODS-3, 250×4.6 mm, 5 μm; Shimadzu) and a detector set at 320 nm. of the detailed chromatographic conditions were as described by Lotfy et al. (1994). For LC-MS analysis, the reaction products were resolved with a reverse-phase C18 column (XDB-18, 250×4.6 mm, 5 μm; Agilent) at a constant flow rate of 0.3 ml min−1 with the gradient mobile phase of 50% acetonitrile (solvent B) in 0.1% acetic acid in water (solvent A) for 3 min, then increased to 100% B within 12 min and maintained for another 3 min. The eluent was injected into a mass spectrometer equipped with an AJS ESI source (Agilent). The MS spectrum of ω-feruloyloxypalmitic acid was collected in positive and negative modes at 3.0 kV and 4.0 kV spray voltage, respectively, 45 psi nebulizer, 5 L min−1 dry gas at 325 °C, and 350 °C vaporizer temperature.
Yeast one-hybrid assays
The full-length sequences of kiwifruit AchnFHT, AchnABF2, AchnMYB4, AchnMYB41, and AchnMYB107 were obtained by tblastx analysis against the kiwifruit genome database using Arabidopsis AtASFT, AtABF2, AtMYB4, AtMYB41, and AtMYB107 as the queries, respectively. Yeast one-hybrid (Y1H) screening was carried out using the MatchmakerTM Gold Yeast One-Hybrid Library Screening System (Clontech) (Zeng et al., 2015). The bait fragment (the 1475-bp fragment of the AchnFHT promoter) was cloned into the pAbAi vector. AchnFHT-AbAi and p53-AbAi were linearized and transformed into Y1HGold to make individual bait-reporter strains. Transformants were initially screened on plates containing SD medium without Ura (SD/–Ura) supplemented with 0–1000 ng ml−1 aureobasidin A (AbA) for auto-activation analysis. Full-length coding sequences of AchnABF2, AchnMYB4, AchnMYB41, and AchnMYB107 were cloned into the pGADT7 (AD) prey vector and transferred into individual bait-reporter yeast strains. The transformed Y1HGold were cultured on SD medium with 50 ng ml−1 AbA and without leucine (SD/–Leu+AbA50) at 28 °C for 3 d to test the interactions. pGADT7-Rec (AD-Rec-P53) was co-transformed with the p53-promoter fragment to Y1HGold as positive control, while AD-empty and AchnFHT-AbAi were used as negative controls. The primers used for the Y1H assays are listed in Supplementary Table S2.
Dual luciferase assays
Dual luciferase assays were performed according to Min et al. (2012). Full-length coding sequences of AchnABF2, AchnMYB4, AchnMYB41, and AchnMYB107 were individually inserted into the pGreen II 0029 62-SK vector (SK), and the fragment of AchnFHT promoter was inserted into the pGreen II 0800-LUC vector. All the constructs were transformed into Agrobacterium tumefaciens GV3101 using the freeze–thaw method (Holsters et al., 1978). The dual luciferase assays were performed with N. benthamiana that had been grown in controlled environment chambers for 5 weeks. Agrobacterium cultures were prepared with infiltration buffer (10 mM MgCl2, 10 mM MES, and 150 mM acetosyringone) to OD600=0.8. Agrobacterium culture mixtures of TF genes (1 ml) and promoter (100 μl) were infiltrated into the abaxial side of the leaves using needleless syringes. Leaves were sampled 3 d after infiltration for analyses of FLUC (Firefly luciferase) and RLUC (Renilla luciferase) activities using a Dual-Luciferase Reporter Assay System (Promega) equipped with Modulus Luminometers (Promega). Luciferase activity was analysed in three independent experiments with six replications for each assay. The primers used are listed in Supplementary Table S2.
Plasmid construction for transient overexpression
The full-length coding sequences of AchnFHT, AchnABF2, AchnMYB4, AchnMYB41, and AchnMYB107 were individually cloned into the plant binary vector pBI121, replacing the GUS gene, behind the CaMV 35S promoter. The recombinant plasmids were transformed into Agrobacterium cells (EHA105) by the freeze–thaw method (Holsters et al., 1978) and grown in Yeast Extract Peptone (YEP) plates containing 100 µg ml−1 ampicillin and 20 µg ml−1 rifampin. A single bacterial colony was inoculated into 50 ml liquid YEP medium and was grown to saturation. After centrifugation, the pellet was re-suspended in the infection solution (10 mM MES, 10 mM MgCl2, and 150 mM acetosyringone). Leaves of 5-week-old N. benthamiana were used for infiltration, and the pBI121 empty vector was used as the control. The experiments were independently repeated at least three times. At 6 d after infiltration, leaves were harvested for qRT-PCR and suberin monomer analysis. The primers are listed in Supplementary Table S2.
Subcellular localization analysis
Transient expression was conducted to examine the subcellular localization of AchnFHT, AchnABF2, AchnMYB4, AchnMYB41, and AchnMYB107 according to Voinnet et al. (2003). The coding sequences of AchnFHT, AchnABF2, and the three AchnMYB genes without termination codons were inserted into the pBI221-EGFP vector and transiently expressed under the control of the CaMV 35S promoter. The fusion constructs 35S::FHT-GFP, 35S::ABF2-GFP, 35S::MYB4-GFP, 35S::MYB41-GFP, 35S::MYB107-GFP, and control vector pBI221-EGFP (35S::GFP) were transformed into Agrobacterium cells (EHA105) and grown in YEP plates containing 100 µg ml−1 ampicillin and 20 µg ml−1 rifampin. The bacterium was grown to saturation in the liquid YEP medium. After centrifugation, the pellet was re-suspended in the infection solution (10 mM MES, 10 mM MgCl2, and 150 mM acetosyringone), and was injected into leaves of 4-week-old of N. benthamiana. The green was observed 3 d after infiltration under a confocal microscope (TCS SP8, Leica) with excitation at 488 nm and detection at 495−531 nm. All transient expression assays were repeated at least three times. The primers used for subcellular localization are listed in Supplementary Table S2.
Suberin depolymerization and monomer analysis
The soluble lipid fraction was extracted from wound tissue samples as described by Legay et al. (2016). The dry residue was depolymerized using acid-catalysed methanolysis following the method described by DeBolt et al. (2009). Briefly, the samples (1 g) were placed in glass vials, 4 ml of sulfuric acid/methanol (1:20, v/v) was added, and the vials were immediately incubated at 85 °C for 3 h. Methyl heptadecanoate and ω-pentadecalactone were used as internal standards. The suberin monomers were extracted by adding two volumes of dichloromethane and one volume of 0.9% (w/v) NaCl. After washing with distilled water, the organic phase was dried over anhydrous sodium sulfate and evaporated under nitrogen gas. The suberin depolymerization residue was derivatized by adding 100 μl of pyridine and 100 μl of N, O-bis(trimethylsilyl)-trifluoroacetamide (BSTFA) at 70 °C for 40 min. After derivatization, the samples were dried under nitrogen gas and re-dissolved in 500 μl dichloromethane. A 20-µl aliquot was then analysed using an Agilent Technologies 7890B-5977A Gas Chromatograph–Mass Spectrometer Detector (GC–MSD) system. Details of the chromatographic conditions are given in Han et al. (2017). The analyses were repeated at three times.
Accession numbers
Sequence data from this study can be found in the Kiwifruit Genome Database, SOL Genomics Network Database, TAIR, and NCBI under the following gene ID and accession numbers: AchnFHT (Achn348111), StFHT (FJ825138), NbFHT (Niben101Scf08936g06001.1), QsFHT (XM024033912), AtASFT (At5g41040), AchnABF2 (315671), AtABF1 (At1g49720), AtABF2 (At1g45249), AtABF3 (At4g34000), AtABF4 (At3g19290), AchnMYB4 (Achn020361), AchnMYB41 (Achn345001), AchnMYB107 (Achn267491), AtMYB3 (At1g22640), AtMYB4 (At4g38620), AtMYB7 (At2g16720), AtMYB9 (At5g16770), AtMYB32 (At4g34990), AtMYB41 (At4g28110), AtMYB74 (At4g05100), AtMYB102 (At4g21440), and AtMYB107 (At3g02940).
Results
Gene isolation and analysis
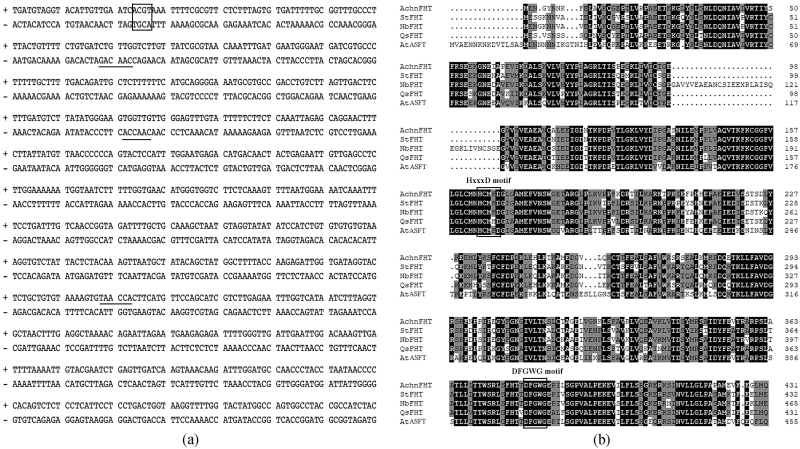
A fragment (1475 bp) of the AchnFHT promoter was obtained from kiwifruit DNA and cis-acting regulatory elements were analysed using the PLACE database (https://www.dna.affrc.go.jp/PLACE/). The AchnFHT promoter contained an ABA-responsive element (ABRE; Fig. 1a, box) and three MYB recognition elements (MYBREs, underlined), which could be bound with bZIP and MYB proteins, respectively.
Fig. 1.
Sequence analysis of AchnFHT. (a) The cis-acting regulatory elements of the AchnFHT promoter. The ABRE is indicated by the box and the MYB recognition elements are underlined in the part fragment of the promoter. (b) Amino acid sequences of Actinidia chinensis AchnFHT, Solanum tuberosum StFHT, Nicotiana benthamiana NbFHT, Quercus suber QsFHT, and Arabidopsis thaliana AtFHT aligned using ClustalX. The HxxxD and DFGWG motifs are indicated.
The full-length coding sequence of AchnFHT (1299 bp) was obtained from kiwifruit cDNA, and encoded a predicated protein of 432 amino acids of deduced molecular mass 47.9 kDa with a theoretical PI of 5.3. AchnFHT displayed high similarity with potato StFHT (81%), tobacco NbFHT (76.2%), and Arabidopsis AtASFT (70%). Multiple alignments of the proteins highlighted two conserved motifs that were common to the plant BAHD acyltransferase (Fig. 1b). The HxxxD motif (His164–Asp168) is involved in the catalytic activity of acyl transfer from acyl-CoA to fatty ω-hydroxyacids and fatty alcohols (El-Sharkawy et al., 2005), whilst the DFGWG motif (Asp380–Gly384) was located away from active sites and plays a structural role (Ma et al., 2005).
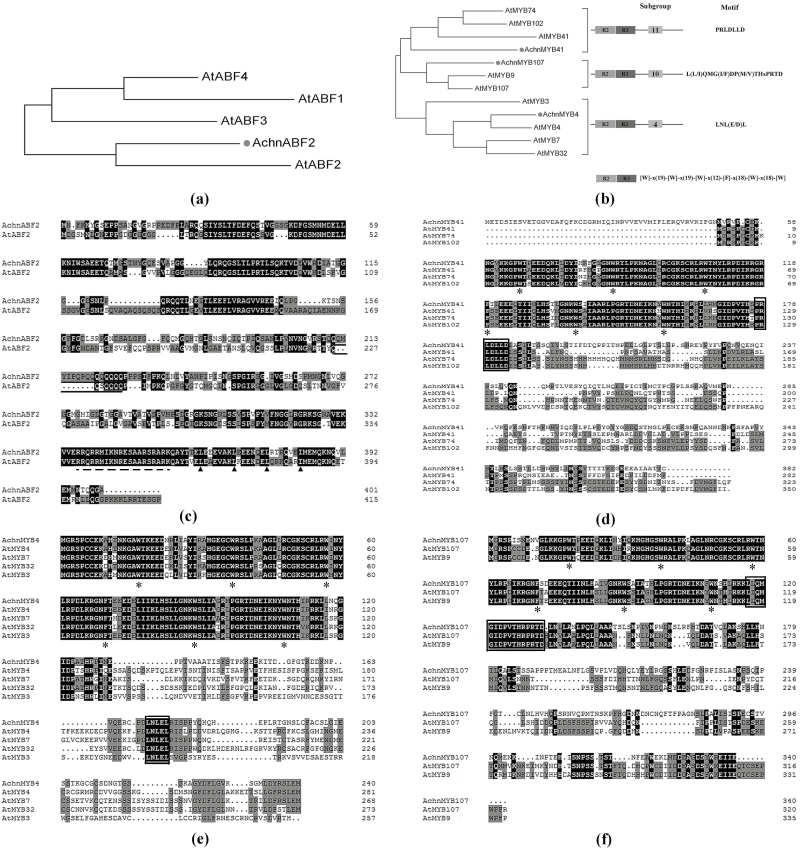
The full-length coding sequence of AchnABF2 was isolated from kiwifruit cDNA. Based on a phylogenetic tree constructed using FigTree (Fig. 2a), AchnABF2 clustered with Arabidopsis AtABF2, which has was identified as a transcriptional activator of ABA signaling in responses to abiotic stresses (Yoshida et al., 2010). Examination of the amino acid sequences indicated that they shared a basic region near the C terminus (Fig. 2c, dashed line), and immediately downstream they contained four heptad repeats of leucine (arrowheads), indicating that AchnABF2 encoded a bZIP protein (Landschulz et al., 1988). In addition, AchnABF2 also possessed a glutamine-rich domain (Fig. 2c, solid lines), which is a transcriptional activation domain of bZIP proteins (Meshi and Iwabuchi, 1995). Three MYB genes, designated as AchnMYB4, AchnMYB41, and AchnMYB107, were isolated from kiwifruit. Phylogenetic analysis showed that AchnMYB107, AchnMYB41, and AchnMYB4 clustered with Arabidopsis AtMYB107, AtMYB41, and AtMYB4, respectively (Fig. 2b). AchnMYB41 and AchnMYB107 belonged to R2R3 subgroups 11 and 10, respectively, and shared their conserved motifs (Fig. 2d, f, black box) (Stracke et al., 2001). AchnMYB4 belonged to subgroup 4 proteins, which is the repressor-type MYB group in regulating hydroxycinnamic acid metabolism (Jin et al., 2000). Alignment analysis showed that AchnMYB4 shared a conserved motif of subgroup 4 (Fig. 2e, black box).
Fig. 2.
Alignment and phylogenetic analyses of ABF2 and MYB transcription factors from kiwifruit and Arabidopsis. (a, b) Phylogenetic trees of AchnABF2 and AchnMYB transcription factors with homologs from Arabidopsis. (c–f) The amino acid sequence alignments of AchnABF2, AchnMYB41, AchnMYB4, and AchnMYB107 with homologs from Arabidopsis. The basic region and leucine repeats of AchnABF2 are indicated by the dashed line and arrowheads, respectively, and glutamine-rich regions are indicated by solid lines. The primary structures of R2R3-MYB and the conserved motifs of MYB41, MYB4, and MYB107 are indicated by asterisks and boxes, respectively. The phylogenetic trees were constructed using FigTree (http://tree.bio.ed.ac.uk/software/figtree/), and the sequences were aligned using ClustalX.
Recombinant AchnFHT functions as a feruloyl transferase
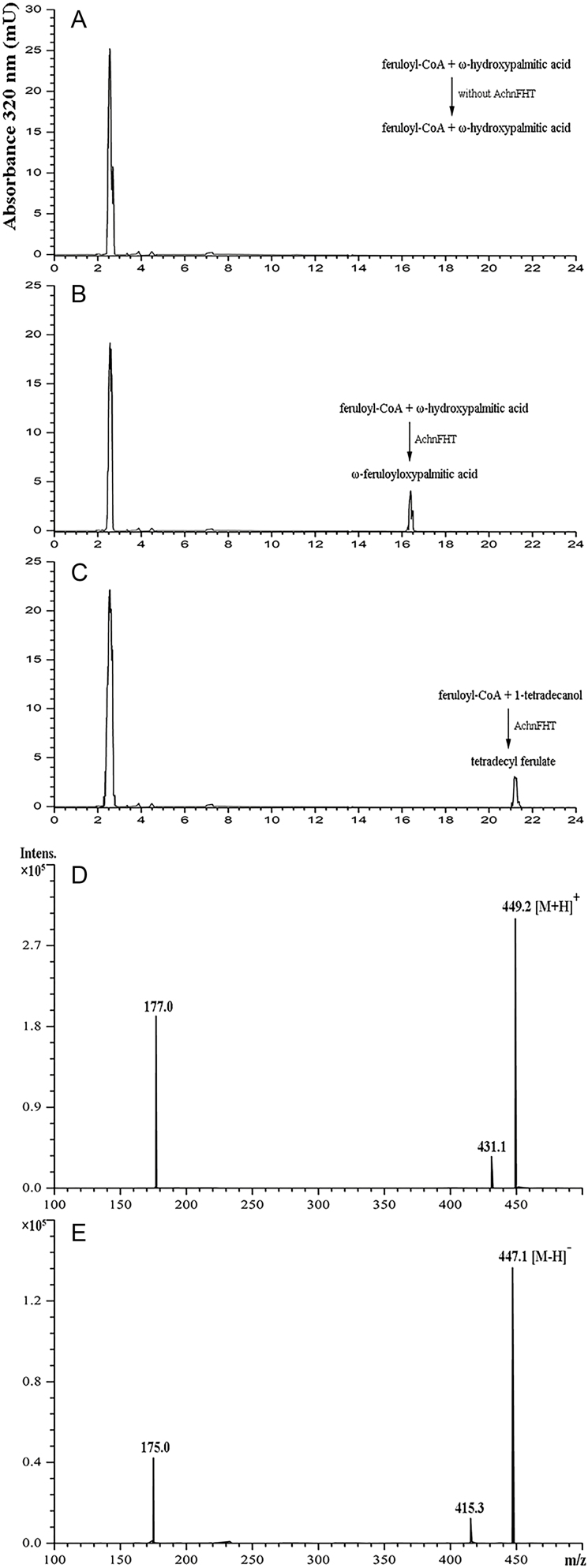
The recombinant AchnFHT protein was used to test for enzyme activity in vitro. SDS-PAGE analysis showed confirmed the purity of the AchnFHT protein as it appeared as a single band (Supplementary Fig. S1). Feruloyl-CoA was used as the acyl donor, and a primary alcohol (1-tetradecanol) and a ω-hydroxyacid (ω-hydroxypalmitic acid) were used as acyl acceptors. Only feruloyl-CoA was detected in reactions without the AchnFHT protein, at a retention time of 2.55 min (Fig. 3a). New and unique peaks corresponding to ω-feruloyloxypalmitic acid (Fig. 3b) and tetradecyl ferulate (Fig. 3c) appeared in the reactions at retention times of 16.39 min and 21.16 min, respectively. The reaction system of ω-hydroxypalmitic acid was further confirmed by LC-MS. The mass of the main molecular ion [(M-H)+=449 m/z and (M-H)−=447 m/z] indicated that ferulic acid was conjugated with ω-hydroxypalmitic acid to form ω-feruloyloxypalmitic acid (Fig. 3d, e), which was consistent with previous studies (Gou et al., 2009; Serra et al., 2010).
Fig. 3.
In vitro activity of AchnFHT. (a–c) HPLC chromatograms monitored at 320 nm showing the reaction products after incubating feruloyl-CoA and ω-hydroxypalmitic acid without the AchnFHT protein (a), feruloyl-CoA and ω-hydroxypalmitic acid with the AchnFHT protein (b), and feruloyl-CoA and 1-tetradecanol with the AchnFHT protein (c). (d, e) The mass spectra obtained in the AchnFHT enzymatic assay using feruloyl-CoA and ω-hydroxypalmitic acid as substrates. The mass of the main molecular ions at 449.2 m/z in positive mode (d) and at 447.1 m/z in negative mode (e), corresponding to ω-feruloyloxypalmitic acid.
Catalysation of suberin monomer biosynthesis by AchnFHT
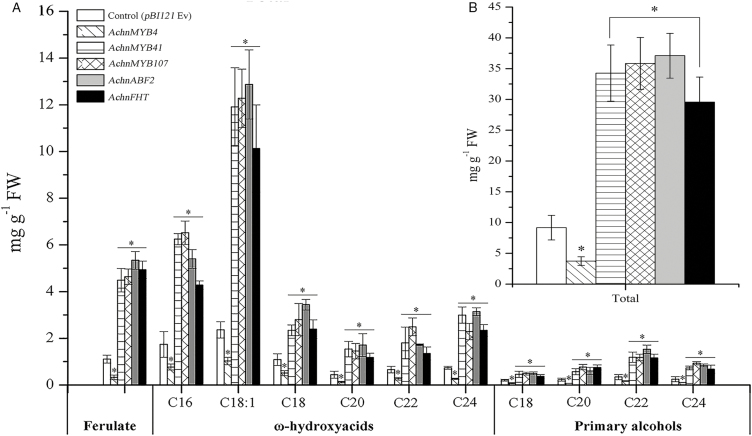
To investigate the in vivo function of AchnFHT, it was transiently overexpressed in N. benthamiana leaves, and the suberin monomers ferulate, ω-hydroxyacids, and primary alcohols were examined. Overexpression of AchnFHT resulted in significant production of ferulate, ω-hydroxyacids, and primary alcohols (Fig. 4a). The ferulate content in N. benthamiana leaves after 6 d of AchnFHT expression was 4.5-fold higher than that of the empty vector control. Likewise, significant induction of ω-hydroxyacids with chain lengths C16–C24 was detected in plants overexpressing AchnFHT, in particular the C18:1 ω-hydroxyacid, which was 4.3-fold higher than in the control. In addition, pronounced increases were also detected in primary alcohols of chain lengths C18–C24. In total, the suberin monomers in leaves overexpressing AchnFHT amounted to 29.6 mg g−1, an increase of 3.2-fold compared to the control (Fig. 4b).
Fig. 4.
(a) Suberin monomers from leaves of N. benthamiana infiltrated with AchnFHT, AchnABF2, AchnMYB4, AchnMYB41, and AchnMYB107 compared with the empty vector control (pBI121 Ev). (b) Total content of suberin monomers. Data are means (±SD), n=3. Significant differences were determined using Student’s t-test: *P<0.05.
AchnABF2, AchnMYB41, and AchnMYB107 activate the AchnFHT promoter and AchnMYB4 represses it
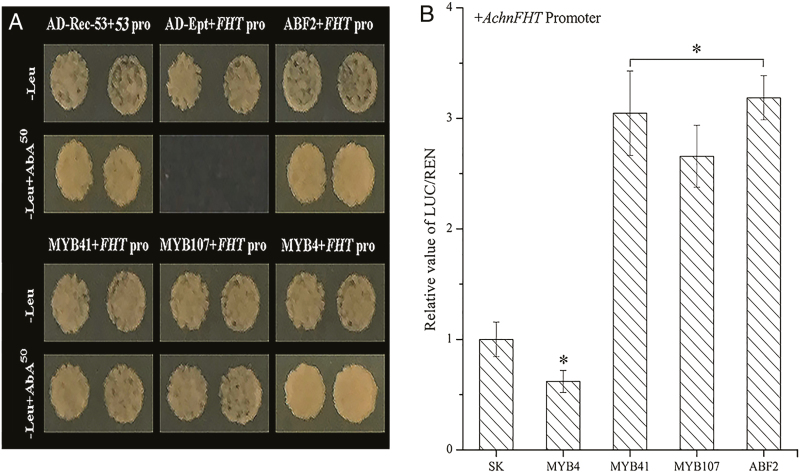
Y1H assays were used to determine whether AchnABF2, AchnMYB41, AchnMYB107, and AchnMYB4 were capable of physically interacting with the AchnFHT promoter. Linearized AchnFHT-AbAi was transformed into Y1HGold and grown on SD/–Ura medium with aureobasidin A (AbA) from 0–1000 ng ml−1, and we found that the AchnFHT promoter was suppressed by 50 ng ml−1 AbA. In the interaction tests, expression of AchnABF2, AchnMYB41, AchnMYB107, and AchnMYB4 separately induced the expression of the AbA-resistance reporter gene driven by the AchnFHT promoter (Fig. 5a), indicating that all the four TFs could directly interact with the AchnFHT promoter.
Fig. 5.
AchnABF2, AchnMYB41, AchnMYB107, and AchnMYB4 individually activate or repress the AchnFHT promoter. (a) Yeast one-hybrid assays. AD-Rec-p53 with p53-AbAi was used as the positive control and AD-empty with AchnFHT-AbAi was used as the negative control. (b) Dual luciferase assays demonstrating that AchnABF2, AchnMYB41, and AchnMYB107 activate the AchnFHT promoter, and AchnMYB4 repress the promoter. The ratio of LUC/REN of the empty vector (SK) plus the promoter was used as the reference (set as 1). Pro, promoter; Ept, empty. Data are means (±SD) of three independent experiments each with six replicates (i.e. n=18). Significant differences were determined using Student’s t-test: *P<0.05. (This figure is available in colour at JXB online.)
To verify the regulatory role of the four TFs on AchnFHT, dual luciferase assays were performed in N. benthamiana leaves (Fig. 5b). AchnMYB41, AchnMYB107, and AchnABF2 could significantly activate the promoter of AchnFHT, which reached levels that were 3.0-, 2.7-, and 3.2-fold higher than that of the control, respectively. In contrast, AchnMYB4 repressed AchnFHT promoter, resulting in only 0.6-fold expression of the control.
Regulation of expression of suberin biosynthetic genes and accumulation of suberin monomers by AchnABF2 and AchnMYBs
To validate the regulatory role of kiwifruit AchnABF2, AchnMYB4, AchnMYB41, and AchnMYB107 on AchnFHT, the four TF genes were separately transiently overexpressed in N. benthamiana leaves and expression of suberin biosynthetic genes (Table 1) and accumulation of monomers (Fig. 4) were determined. Genes implicated in the aliphatic and phenolic pathways were largely altered by overexpression of AchnABF2 and AchnMYB. Two N. benthamiana homologs of AchnFHT, namely NbFHT and NbFHT2, were significantly induced by overexpression of AchnMYB41, AchnMYB107, and AchnABF2, particularly NbFHT that shares high homology with AchnFHT (Fig. 1b), which displayed increases of 7.1-, 8.0-, and 8.2-fold relative to the control, respectively. In addition, expression of the 3-ketoacyl-CoA synthase (KCS) genes, NbKCS2, NbKCS4, and NbKCS11, were significantly elevated. The KCS proteins are part of the fatty acid elongation complex that generates very long-chain fatty acids in suberin biosynthesis (Vishwanath et al., 2015). Genes encoding fatty acid ω-hydroxylases (NbCYP86A1, NbCYP86B1) and fatty acyl-reductases (NbFAR2, NbFAR3), which are involved in suberin monomer biosynthesis (Höfer et al., 2008; Compagnon et al., 2009; Domergue et al., 2010), were also strongly up-regulated. In the phenolic pathway, phenylalanine ammonia lyase (NbPAL2) and 4-coumarate CoA ligase (Nb4CL1) are tightly associated with suberin biosynthesis (Kosma et al., 2015; Lashbrooke et al., 2016), and were also elevated by overexpression of AchnMYB41, AchnMYB107, or AchnABF2. Conversely, significant repression of the suberin biosynthetic genes was detected in leaves overexpressing AchnMYB4, specifically NbFHT and NbFHT2, which were 10.9- and 8.2-fold lower than that of the control, respectively.
Table 1.
Relative change in expression of kiwifruit suberin biosynthetic genes expressed in leaves of N. benthamiana at 6 d after infiltration
| Suberin biosynthetic gene | Description | Overexpressed gene | |||
|---|---|---|---|---|---|
| AchnMYB4 | AchnMYB41 | AchnMYB107 | AchnABF2 | ||
| NbFHT | Fatty ω-hydroxyacid/alcohol hydroxycinnamoyl transferase 1 | –10.9±1.2 | 7.1±0.6 | 8.0±0.9 | 8.2±1.1 |
| NbFHT2 | Fatty ω-hydroxyacid/alcohol hydroxycinnamoyl transferase 2 | –8.2±1.4 | 5.1±0.7 | 4.8±0.6 | 7.1±0.8 |
| NbKCS2 | 3-ketoacyl-synthase 2 | –10.6±2.0 | 6.2±0.6 | 3.4±0.2 | 3.2±0.5 |
| NbKCS4 | 3-ketoacyl-synthase 4 | –5.7±0.6 | 3.1±0.5 | 3.7±0.5 | 8.0±1.0 |
| NbKCS11 | 3-ketoacyl-synthase 11 | –8.7±0.7 | 2.5±0.4 | 1.8±0.6 | 2.8±0.1 |
| NbCYP86 A1 | Cytochrome p450 86 A1 | –6.6±0.9 | 8.0±0.8 | 6.8±0.7 | 11.4±0.9 |
| NbCYP86 B1 | Cytochrome p450 86 B1 | –2.7±0.4 | 2.3±0.3 | 4.2±0.2 | 6.7±0.9 |
| NbFAR2 | Fatty acyl-reductase 2 | –5.0±0.5 | 4.1±0.2 | 4.7±0.4 | 6.4±0.5 |
| NbFAR3 | Fatty acyl-reductase 3 | –6.5±0.7 | 5.1±0.6 | 5.4±0.6 | 7.4±0.4 |
| Nb4CL1 | 4-coumarate-CoA ligase 1 | –10.8±1.3 | 3.7±0.5 | 4.6±0.4 | 3.1±0.6 |
| NbPAL2 | Phenylalanine ammonia lyase 2 | –2.9±0.3 | 1.8±0.4 | 3.4±0.2 | 1.7±0.3 |
Data are presented as the mean (±SE) fold-change (log2) relative to the empty vector control from three biological replicates. Full details of the genes are given in Supplementary Table S1.
We examined the suberin monomers ferulate, ω-hydroxyacids, and primary alcohols in N. benthamiana leaves (Fig. 4). Ferulate content was significantly increased by overexpression of AchnMYB41, AchnMYB107, and AchnABF2, by 4.0-, 4.2-, and 4.8-fold relative to the control, respectively. Similar results were also found for ω-hydroxyacids and primary alcohols, which were mainly produced by CYP86A1 and CYP86B1, and by FARs, respectively. Pronounced increases in C16–C24 ω-hydroxyacids and C18–C24 primary alcohols were detected in leaves overexpressing AchnMYB41, AchnMYB107, and AchnABF2. However, overexpression of AchnMYB4 significantly reduced the production of ferulate, ω-hydroxyacids, and primary alcohols. In total, the contents of suberin monomers in leaves overexpressing AchnMYB4, AchnMYB41, AchnMYB107, and AchnABF2 were 0.4-, 3.7-, 3.9-, and 4.0-fold that of the control (Fig. 4b).
Subcellular localization
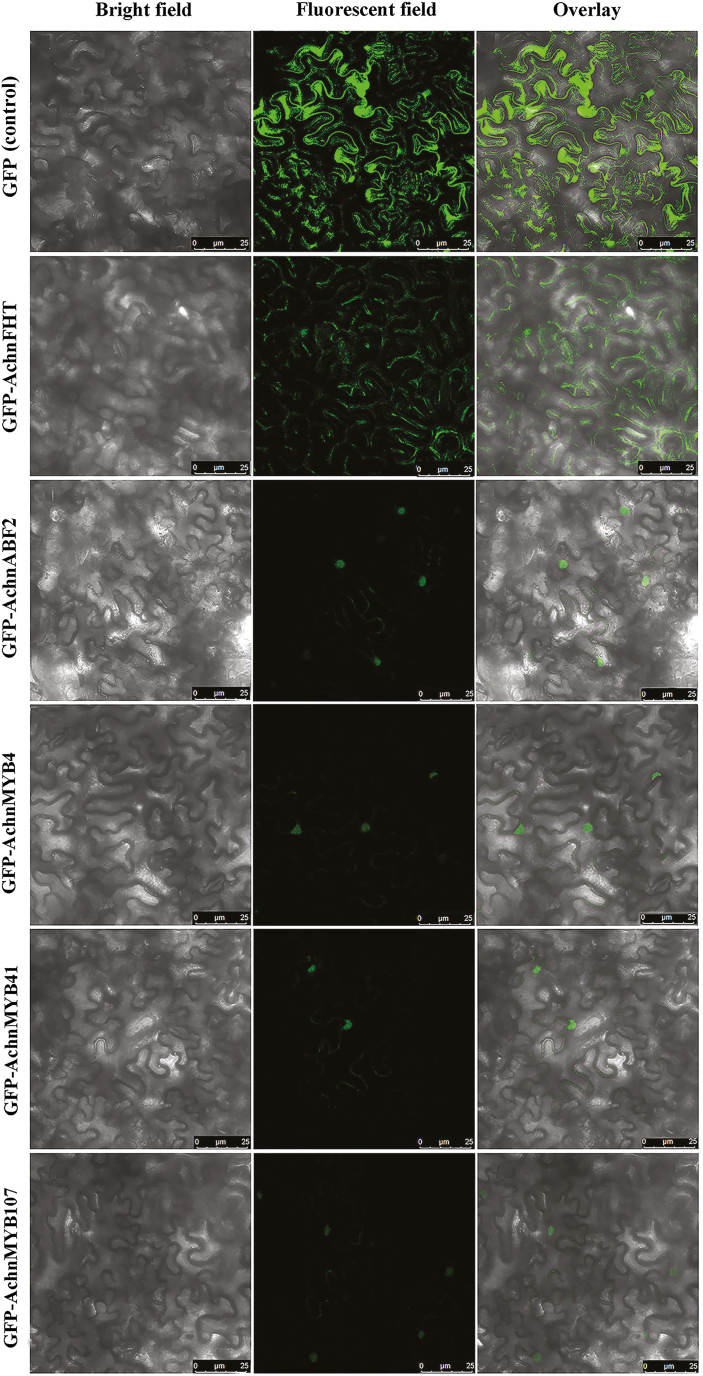
The subcellular localization of AchnABF2, AchnMYB4, AchnMYB41, AchnMYB107, and AchnFHT were examined in N. benthamiana leaves by GFP tagging, with the GFP-only vector used as the control (Fig. 6). As the GFP plasmid was transformed, green fluorescent signals were observed in the whole cell. The fluorescent signals of AchnABF2, AchnMYB4, AchnMYB41, and AchnMYB107 were observed in the nucleus, while the signal of AchnFHT was detected in the cytosol, thus indicating their respective localizations.
Fig. 6.
Subcellular localization of AchnFHT, AchnABF2, AchnMYB4, AchnMYB41. and AchnMYB107 agro-infiltrated into N. benthamiana leaves. The control was the vector containing only GFP. Scale bars are 25 μm. (This figure is available in colour at JXB online.)
Induction of gene expression and suberin monomer accumulation by exogenous ABA
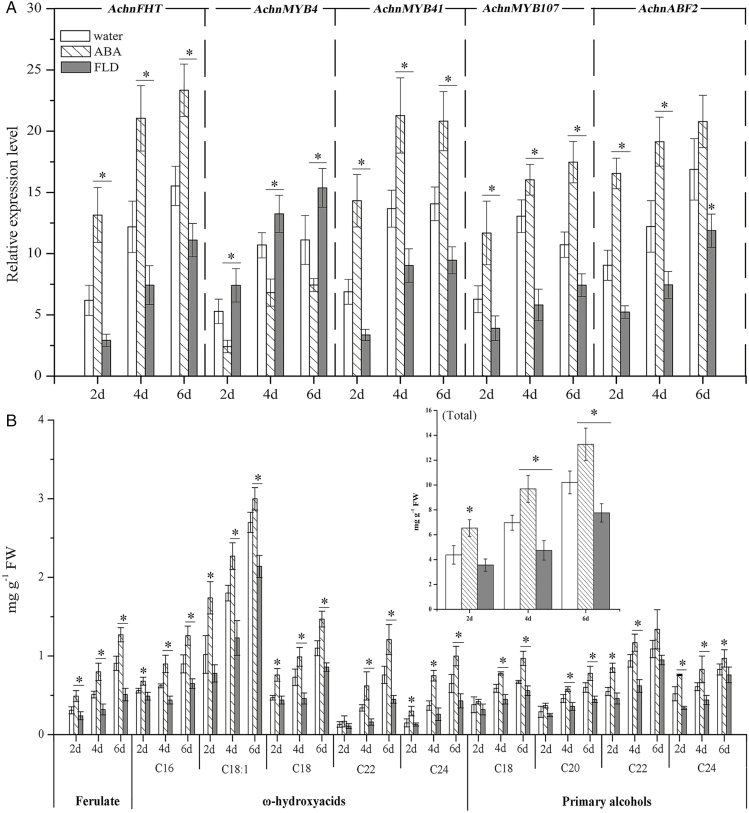
The expression levels of AchnFHT, AchnMYB4, AchnMYB41, AchnMYB107, and AchnABF2 in wound tissues in response to ABA and FLD were examined in kiwifruit (Fig. 7a). AchnFHT, AchnMYB41, AchnMYB107, and AchnABF2 were induced by wounding and by ABA treatment over a 6-d healing period. These genes were also induced under FLD treatment, but to a lower extent. By day 6, AchnFHT expression was 1.5-fold that of the water treatment in ABA-treated tissues but only 0.7-fold of the water treatment in FLD-treated tissues. Similar results were seen for AchnMYB41, AchnMYB107, and AchnABF2. In contrast, expression of AchnMYB4 was significantly increased in FLD-treated tissues compared with the water treatment, but it was inhibited by ABA.
Fig. 7.
Gene expression and suberin monomers in kiwifruit wound tissues treated with water (control), abscisic acid (ABA), or fluridone (FLD). (a) Expression levels of AchnFHT, AchnMYB4, AchnMYB41, AchnMYB107, and AchnABF2. Relative mRNA abundance was evaluated using real-time PCR and is expressed as fold-change relative to the initial value upon wounding. (b) Accumulation of ferulate, ω-hydroxyacids, and primary alcohols. The inset graph shows the total content of suberin monomers. Data are means (±SD) of three biological replicates. Significant differences were determined using Student’s t-test: *P<0.05.
Compared with water-treated fruit, ferulate, ω-hydroxyacids, and primary alcohols were increased and reduced by the ABA and FLD treatments, respectively, over 6 d of wound healing (Fig. 7b). By day 6, the ferulate content in ABA-treated tissues was up to 1.3 mg g−1, which was 1.4-fold and 2.5-fold that of the water and FLD treatments, respectively. Significant increases in C16–C24 ω-hydroxyacids and C18–C24 primary alcohols were also detected in ABA-treated tissues. In contrast, the FLD treatment significantly reduced the contents of the three types of monomers. By day 6 of wound healing, the total content of the suberin monomers in the ABA treatment was 13.3 mg g−1, with increases of 1.3- and 1.7-fold relative to the water and FLD treatments, respectively (Fig. 7b).
Discussion
FHT plays a key role in the suberization process that is one of the characteristic features of wound damage and it is also known to occur in cases of abiotic stress (Jin et al., 2018; Wei et al., 2018). Suberin phenolics are presumed to cross-link to form an aromatic domain that is anchored to the cell wall through covalent bonds that are formed with polysaccharides (Yan and Stark, 2000). The phenolics in the aromatic domain (mainly ferulic acid) are esterified with fatty ω-hydroxyacids and with fatty alcohols that are catalysed by FHT to link with the aliphatic domain (Pollard et al., 2008; Ranathunge et al., 2011). Indeed, chemical analyses of depolymerized suberin in potato and Arabidopsis have identified ferulate, ω-hydroxyacids, and primary alcohols as signature products of FHT. The catalytic properties of FHT were initially characterized in extracts from potato tubers (Lotfy et al., 1994) and cell suspensions of tobacco (Lotfy et al., 1996). Later, in vitro recombinant FHT protein was shown to display feruloyl transferase activity and to transfer feruloyl to ω-hydroxyacids and primary alcohols. In FHT-RNAi potato plants and Arabidopsis ASFT/HHT mutants, ferulate and C18:1 ω-hydroxyacid are strongly reduced, which demonstrates the role of FHT (ASFT/HHT) in suberization (Gou et al., 2009; Molina et al., 2009; Serra et al., 2010). Here, the function of kiwifruit AchnFHT was elucidated in vitro and in vivo. AchnFHT encoded a FHT belonging to the BAHD acyltransferases and shared high similarity to the potato, tobacco, and Arabidopsis proteins (Fig. 1b). Recombinant AchnFHT protein displayed feruloyl transferase activity with the function of transferring feruloyl from feruloyl-CoA to ω-hydroxypalmitic acid and to 1-tetradecanol in vitro (Fig. 3). Transient overexpression of AchnFHT in N. benthamiana resulted in increased production of ferulate, primary alcohols, and ω-hydroxyacids (specifically C18:1 ω-hydroxyacid) (Fig. 4). In addition, pronounced increases in the three types of suberin monomers and in AchnFHT expression were detected in kiwifruit wound tissues (Fig. 7). Collectively, the results demonstrate that AchnFHT functions as a feruloyl transferase that is implicated in cross-linking aliphatic and aromatic domains, via conjugating feruloyl with primary alcohols and ω-hydroxyacids (especially C18:1 ω-hydroxyacid).
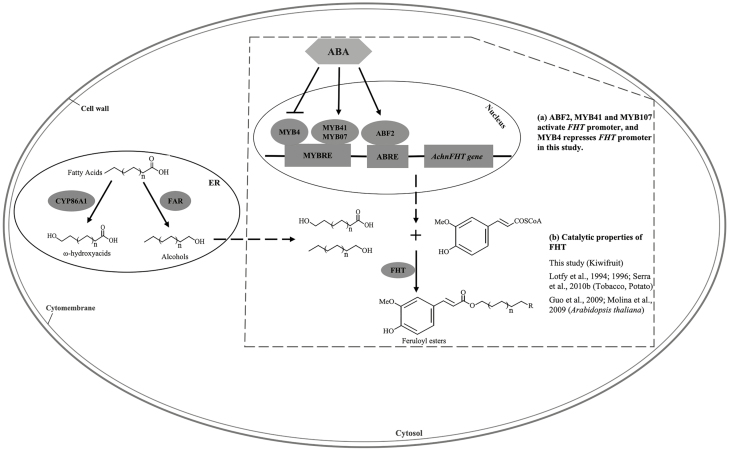
Apart from the transport and polymerization of suberin monomers, the subcellular organization of enzymes in the suberin biosynthesis pathway remains unclear (Beisson et al., 2012; Vishwanath et al., 2015). The endoplasmic reticulum (ER) has been identified as the place where the FHT substrates ω-hydroxyacids and fatty alcohols are biosynthesized (Höfer et al., 2008; Kosma et al., 2012); however, we found that the AchnFHT protein was distributed in the cytosol (Fig. 6). Therefore, it may be speculated that the ω-hydroxyacids and fatty alcohols were transported from the ER to the cytosol (Pollard et al., 2008) and were then catalysed by AchnFHT to form feruloyl esters before the monomers were exported to the apoplast (Fig. 8).
Fig. 8.
Schematic diagram of the regulatory mode of ABA-induced suberin monomer biosynthesis in kiwifruit. ABA induces production of AchnABF2, AchnMYB41, and AchnMYB107, which activate the downstream AchnFHT gene to produce feruloyl esters. ABA promotes suberin monomer biosynthesis via reducing the repression of the AchnFHT promoter by AchnMYB4. Results from this study are highlighted in the dashed box. ABRE, ABA-responsive element; MYBRE, MYB recognition element. (This figure is available in colour at JXB online.)
Several TFs implicated in regulation of suberin biosynthesis have been characterized in Arabidopsis (AtMYB41, AtMYB9, and AtMYB107), potato (StNAC103), and apple (MdMYB93) (Kosma et al., 2015; Lashbrooke et al., 2016; Legay et al., 2016; Verdaguer et al., 2016). Arabidopsis AtMYB41 is implicated in the ABA-mediated NaCl stress response and in suberin biosynthesis. Overexpression of AtMYB41 in Arabidopsis and N. benthamiana can induce suberin accumulation via up-regulation of suberin biosynthetic genes including ASFT, CYP86A1, KCS, and FAR (Kosma et al., 2015). However, Arabidopsis mutants of AtMYB107 display a noticeable reduction in suberin monomers and down-regulation of suberin biosynthetic genes (Lashbrooke et al., 2016). Arabidopsis AtABF2, which encodes bZIP protein, is a master transcriptional activator in ABA-mediated stress responses (Fujita et al., 2013; Yoshida et al., 2015). Kiwifruit AchnMYB41 and AchnMYB107 shared high homology with the N-terminal portions of their respective orthologues AtMYB41 and AtMYB107, which contain the R2R3 MYB domain (Fig. 2d, f), while AchnABF2 shared the conserved bZIP domain of AtABF2 (Fig. 2c). Indeed, transient overexpression of AchnMYB41, AchnMYB107, and AchnABF2 triggered the expression of many genes implicated in the aliphatic and phenolic pathways, including FHT, FHT2, CYP86A1, CYP86B1, FARs, KCSs, PAL2, and 4CL1 (Table 1), which cover the biosynthesis of central metabolites and advanced suberin building-blocks. The significant influence of AchnMYB41, AchnMYB107, and AchnABF2 on suberin biosynthetic genes was strongly supported by the accumulation of their products, ferulate, ω-hydroxyacids, and primary alcohols (Fig. 4). Moreover, their regulatory functions with regards to AchnFHT were also confirmed through yeast one-hybrid and dual-luciferase assays, which showed that these three TFs directly interacted with the gene promoter to activate the expression of AchnFHT (Fig. 5). AtMYB4 encodes a transcriptional repressor of C4H and 4CL to control hydroxycinnamic acid metabolism in Arabidopsis (Jin et al., 2000). Similarly, the kiwifruit homolog of AtMYB4, AchnMYB4, functioned as a repressor in the regulation of AchnFHT, inhibiting the AchnFHT promoter in a dual-luciferase assay, and it reduced the expression of FHT and the accumulation of ferulate, ω-hydroxyacids. and primary alcohols when it was overexpressed in N. benthamiana leaves. These results indicate that AchnABF2, AchnMYB41, and AchnMYB107 function as transcriptional activators but that AchnMYB4 acts as a repressor in regulating AchnFHT (and probably other suberin biosynthetic genes) to coordinate the biosynthesis of suberin monomers.
ABA has been shown to be induced upon wounding and to promote the wound suberization processes via up-regulation of suberin biosynthetic genes (Lulai et al., 2008; Han et al., 2018). By using FLD, an inhibitor of ABA biosynthesis, it has been unequivocally confirmed that ABA plays a key role in regulating wound-healing processes in potato tubers and tomato fruit (Lulai et al., 2008; Tao et al., 2016). The ABA-dependent signal transduction pathway in plants from the perception of the stress signal to gene expression involves different TFs including ABFs and MYBs (Agarwal and Jha, 2010). In our current study, the expression of the TF genes AchnABF2, AchnMYB41, and AchnMYB107 in wound tissues was induced by exogenous ABA treatment, but inhibited by FLD (Fig. 7). However, in contrast, expression of AchnMYB4 was increased and reduced by FLD and ABA, respectively. These results indicated that AchnABF2, AchnMYB4, AchnMYB41, and AchnMYB107 regulate AchnFHT expression in response to ABA. A model for the involvement of these four TFs in the regulation of AchnFHT in suberin biosynthesis via the ABA signaling pathway is presented in Fig. 8. AchnABF2, AchnMYB41, and AchnMYB107 are induced by ABA, and AchnFHT is then activated by the binding of the respective proteins (Fig. 8a). AchnFHT then catalyses the transfer of feruloyl to fatty ω-hydroxyacids and to fatty alcohols to form feruloyl esters (Fig. 8b). In contrast, ABA inhibits the expression of AchMYB4 to relieve the repression of the AchnFHT promoter that results from AchnMYB4, thereby promoting the biosynthesis of suberin monomers.
In conclusion, kiwifruit AchnFHT encoding a FHT is involved in suberin monomer biosynthesis, and a regulation scheme involving four transcription factors is proposed that acts via ABA signaling, possibly involving other suberin biosynthetic genes. AchnABF2, AchnMYB41, and AchnMYB107 play transcriptional activation roles in the regulation of AchnFHT, while AchnMYB4 works as a transcriptional repressor.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primer sequences used for quantitative RT-PCR.
Table S2. Primer sequences used for full-length amplification and vector construction.
Fig. S1. SDS-PAGE gel of the purified AchnFHT protein stained with Coomassie brilliant blue.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (31772365, 31972468) and the National Key Research and Development Program of China (2018YFD0401303).
References
- Agarwal PK, Jha B. 2010. Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biologia Plantarum 54, 201–212. [Google Scholar]
- Bernards MA, Razem FA. 2001. The poly(phenolic) domain of potato suberin: a non-lignin cell wall bio-polymer. Phytochemistry 57, 1115–1122. [DOI] [PubMed] [Google Scholar]
- Beisson F, Li-Beisson Y, Pollard M. 2012. Solving the puzzles of cutin and suberin polymer biosynthesis. Current Opinion in Plant Biology 15, 329–337. [DOI] [PubMed] [Google Scholar]
- Beuerle T, Pichersky E. 2002. Enzymatic synthesis and purification of aromatic coenzyme A esters. Analytical Biochemistry 302, 305–312. [DOI] [PubMed] [Google Scholar]
- Boher P, Serra O, Soler M, Molinas M, Figueras M. 2013. The potato suberin feruloyl transferase FHT which accumulates in the phellogen is induced by wounding and regulated by abscisic and salicylic acids. Journal of Experimental Botany 64, 3225–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagnon V, Diehl P, Benveniste I, Meyer D, Schaller H, Schreiber L, Franke R, Pinot F. 2009. CYP86B1 is required for very long chain omega-hydroxyacid and alpha, omega-dicarboxylic acid synthesis in root and seed suberin polyester. Plant Physiology 150, 1831–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBolt S, Scheible WR, Schrick K, et al. 2009. Mutations in UDP-glucose:sterol glucosyltransferase in Arabidopsis cause transparent testa phenotype and suberization defect in seeds. Plant Physiology 151, 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domergue F, Vishwanath SJ, Joubès J, et al. 2010. Three Arabidopsis fatty acyl-coenzyme A reductases, FAR1, FAR4, and FAR5, generate primary fatty alcohols associated with suberin deposition. Plant Physiology 153, 1539–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efetova M, Zeier J, Riederer M, Lee CW, Stingl N, Mueller M, Hartung W, Hedrich R, Deeken R. 2007. A central role of abscisic acid in drought stress protection of Agrobacterium-induced tumors on Arabidopsis. Plant Physiology 145, 853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sharkawy I, Manríquez D, Flores FB, Regad F, Bouzayen M, Latché A, Pech JC. 2005. Functional characterization of a melon alcohol acyl-transferase gene family involved in the biosynthesis of ester volatiles. Identification of the crucial role of a threonine residue for enzyme activity. Plant Molecular Biology 59, 345–362. [DOI] [PubMed] [Google Scholar]
- Fugate KK, Ribeiro WS, Lulai EC, Deckard EL, Finger FL. 2016. Cold temperature delays wound healing in postharvest sugarbeet roots. Frontiers in Plant Science 7, 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Yoshida T, Yamaguchi-Shinozaki K. 2013. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiologia Plantarum 147, 15–27. [DOI] [PubMed] [Google Scholar]
- Gao S, Gao J, Zhu X, Song Y, Li Z, Ren G, Zhou X, Kuai B. 2016. ABF2, ABF3, and ABF4 promote ABA-mediated chlorophyll degradation and leaf senescence by transcriptional activation of chlorophyll catabolic genes and senescence-associated genes in Arabidopsis. Molecular Plant 9, 1272–1285. [DOI] [PubMed] [Google Scholar]
- Gou JY, Yu XH, Liu CJ. 2009. A hydroxycinnamoyltransferase responsible for synthesizing suberin aromatics in Arabidopsis. Proceedings of the National Academy of Sciences, USA 106, 18855–18860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graça J, Cabral V, Santos S, Lamosa P, Serra O, Molinas M, Schreiber L, Kauder F, Franke R. 2015. Partial depolymerization of genetically modified potato tuber periderm reveals intermolecular linkages in suberin polyester. Phytochemistry 117, 209–219. [DOI] [PubMed] [Google Scholar]
- Han X, Lu W, Wei X, Li L, Mao L, Zhao Y. 2018. Proteomics analysis to understand the ABA stimulation of wound suberization in kiwifruit. Journal of Proteomics 173, 42–51. [DOI] [PubMed] [Google Scholar]
- Han X, Mao LC, Wei XP, Lu WJ. 2017. Stimulatory involvement of abscisic acid in wound suberization of postharvest kiwifruit. Scientia Horticulturae 224, 244–250. [Google Scholar]
- Höfer R, Briesen I, Beck M, Pinot F, Schreiber L, Franke R. 2008. The Arabidopsis cytochrome P450 CYP86A1 encodes a fatty acid omega-hydroxylase involved in suberin monomer biosynthesis. Journal of Experimental Botany 59, 2347–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsters M, Waele DD, Depicker A, Messens E, Schell J. 1978. Transfection and transformation of Agrobacterium tumefaciens. Molecular and General Genetics 163, 181–187. [DOI] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. 2000. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. The EMBO Journal 19, 6150–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Cai Q, Huang W, Dastmalchi K, Rigau J, Molinas M, Figueras M, Serra O, Stark RE. 2018. Potato native and wound periderms are differently affected by down-regulation of FHT, a suberin feruloyl transferase. Phytochemistry 147, 30–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosma DK, Molina I, Ohlrogge JB, Pollard M. 2012. Identification of an Arabidopsis fatty alcohol:caffeoyl-Coenzyme A acyltransferase required for the synthesis of alkyl hydroxycinnamates in root waxes. Plant Physiology 160, 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosma DK, Murmu J, Razeq FM, Santos P, Bourgault R, Molina I, Rowland O. 2015. AtMYB41 activates ectopic suberin synthesis and assembly in multiple plant species and cell types. The Plant Journal 80, 216–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz W, Johnson P, Mcknight S. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240, 1759–1764. [DOI] [PubMed] [Google Scholar]
- Lashbrooke J, Cohen H, Levy-Samocha D, et al. 2016. MYB107 and MYB9 homologs regulate suberin deposition in angiosperms. The Plant Cell 28, 2097–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legay S, Guerriero G, André C, Guignard C, Cocco E, Charton S, Boutry M, Rowland O, Hausman JF. 2016. MdMYB93 is a regulator of suberin deposition in russeted apple fruit skins. New Phytologist 212, 977–991. [DOI] [PubMed] [Google Scholar]
- Leide J, Hildebrandt U, Hartung W, Riederer M, Vogg G. 2012. Abscisic acid mediates the formation of a suberized stem scar tissue in tomato fruits. New Phytologist 194, 402–415. [DOI] [PubMed] [Google Scholar]
- Lotfy S, Javelle F, Negrel J. 1996. Purification and characterization of hydroxycinnamoyl-coenzyme A: ω-hydroxypalmitic acid O-hydroxycinnamoyltransferase from tobacco (Nicotiana tabacum L.) cell-suspension cultures. Planta 199, 475–480. [Google Scholar]
- Lotfy S, Negrel J, Javelle F. 1994. Formation of ω-feruloyloxypalmitic acid by an enzyme from wound-healing potato tuber discs. Phytochemistry 35, 1419–1424. [Google Scholar]
- Lulai EC, Suttle JC, Pederson SM. 2008. Regulatory involvement of abscisic acid in potato tuber wound-healing. Journal of Experimental Botany 59, 1175–1186. [DOI] [PubMed] [Google Scholar]
- Ma X, Koepke J, Panjikar S, Fritzsch G, Stöckigt J. 2005. Crystal structure of vinorine synthase, the first representative of the BAHD superfamily. The Journal of Biological Chemistry 280, 13576–13583. [DOI] [PubMed] [Google Scholar]
- Meshi T, Iwabuchi M. 1995. Plant transcription factors. Plant & Cell Physiology 36, 1405–1420. [PubMed] [Google Scholar]
- Min T, Yin XR, Shi YN, Luo ZR, Yao YC, Grierson D, Ferguson IB, Chen KS. 2012. Ethylene-responsive transcription factors interact with promoters of ADH and PDC involved in persimmon (Diospyros kaki) fruit de-astringency. Journal of Experimental Botany 63, 6393–6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina I, Li-Beisson Y, Beisson F, Ohlrogge JB, Pollard M. 2009. Identification of an Arabidopsis feruloyl-coenzyme A transferase required for suberin synthesis. Plant Physiology 151, 1317–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard M, Beisson F, Li Y, Ohlrogge JB. 2008. Building lipid barriers: biosynthesis of cutin and suberin. Trends in Plant Science 13, 236–246. [DOI] [PubMed] [Google Scholar]
- Ranathunge K, Schreiber L, Franke R. 2011. Suberin research in the genomics era—new interest for an old polymer. Plant Science 180, 399–413. [DOI] [PubMed] [Google Scholar]
- Reid KE, Olsson N, Schlosser J, Peng F, Lund ST. 2006. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biology 6, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra O, Hohn C, Franke R, Prat S, Molinas M, Figueras M. 2010. A feruloyl transferase involved in the biosynthesis of suberin and suberin-associated wax is required for maturation and sealing properties of potato periderm. The Plant Journal 62, 277–290. [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. 2001. The R2R3-MYB gene family in Arabidopsis thaliana. Current Opinion in Plant Biology 4, 447–456. [DOI] [PubMed] [Google Scholar]
- Tao XY, Mao LC, Li JY, Chen JX, Lu WJ, Huang S. 2016. Abscisic acid mediates wound-healing in harvested tomato fruit. Postharvest Biology and Technology 118, 128–133. [Google Scholar]
- Verdaguer R, Soler M, Serra O, Garrote A, Fernández S, Company-Arumí D, Anticó E, Molinas M, Figueras M. 2016. Silencing of the potato StNAC103 gene enhances the accumulation of suberin polyester and associated wax in tuber skin. Journal of Experimental Botany 67, 5415–5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwanath SJ, Delude C, Domergue F, Rowland O. 2015. Suberin: biosynthesis, regulation, and polymer assembly of a protective extracellular barrier. Plant Cell Reports 34, 573–586. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. The Plant Journal 33, 949–956. [DOI] [PubMed] [Google Scholar]
- Wei X, Mao L, Han X, Lu W, Xie D, Ren X, Zhao Y. 2018. High oxygen facilitates wound induction of suberin polyphenolics in kiwifruit. Journal of the Science of Food and Agriculture 98, 2223–2230. [DOI] [PubMed] [Google Scholar]
- Yan B, Stark RE. 2000. Biosynthesis, molecular structure, and domain architecture of potato suberin: a 13C NMR study using isotopically labeled precursors. Journal of Agricultural and Food Chemistry 48, 3298–3304. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Maruyama K, Mogami J, Todaka D, Shinozaki K, Yamaguchi-Shinozaki K. 2015. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant, Cell & Environment 38, 35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. 2010. AREB1, AREB2, and AREB3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. The Plant Journal 61, 672–685. [DOI] [PubMed] [Google Scholar]
- Zeng JK, Li X, Xu Q, Chen JY, Yin XR, Ferguson IB, Chen KS. 2015. EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnology Journal 13, 1325–1334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.