Summary
The transcriptional coactivator p300 is a histone lysine acetyltransferase that is typically recruited to transcriptional enhancers and regulates gene expression by acetylating chromatin. Here we show that p300 activation directly depends on the activation and oligomerisation status of transcription factor (TF) ligands. Using two model TFs, IRF3 and STAT1, we demonstrate that TF dimerization enables trans-autoacetylation of p300 in a highly conserved and intrinsically disordered autoinhibitory lysine-rich loop (AIL), resulting in HAT activation. We describe a p300 crystal structure in which the AIL invades the active site of a neighbouring HAT domain thus revealing a snap-shot of a trans-autoacetylation reaction intermediate. Substrate access to the active site involves rearrangement of an autoinhibitory RING domain. Our data explain how cellular signalling, TF activation and dimerization controls p300 activation thus explaining why gene transcription is associated with chromatin acetylation.
Keywords: P300, CBP, acetyltransferase, Chromatin, IRF3, STAT1, transcription factor, transcriptional regulation
Introduction
Signals that emanate from cellular receptors ultimately lead to changes in gene expression that drive cellular change and organismal development. Gene expression is typically controlled through the coordinated activity of DNA-binding transcription factors (TFs), chromatin regulators, and the general transcription machinery. For instance, in the innate immune system, pattern recognition receptors (PRRs) recognize various pathogen-associated molecular patterns (PAMPs)1. Once engaged by PAMPs, PRRs bind to adaptor proteins such as STING (stimulator of IFN genes). These adaptor proteins engage the latent DNA binding TF interferon (IFN) regulatory factor 3 (IRF3) and enable recruitment and activation of the non-canonical IKK kinase TBK11. TBK1 then phosphorylates IRF3 in a C-terminal motif, which results in removal of autoinhibition, dimerization and adaptor displacement2,3. Activated IRF3 dimers bind to p300/CBP (also known as KAT3B and KAT3A) to stimulate chromatin acetylation and gene expression of antiviral type I IFNs IFN-α and IFN-β3–5. Type I IFNs cytokines are secreted and bind to specific cell surface IFN receptors, which results in activation of Janus kinase/signal transducers and activators of transcription (JAK/STAT) signalling6. Activated, tyrosine phosphorylated STATs dimerise, translocate to the nucleus and bind to p300/CBP to stimulate transcription of IFN-stimulated genes (ISGs)7.
p300/CBP are known to interact with more than 400 binding partners including the basal transcription machinery8. The large protein interactome of p300/CBP results in near universal recruitment of these histone lysine acetyltransferases (HATs) to enhancers and p300 occupancy has been used to identify enhancers genome-wide9,10. CBP/p300 catalyse acetylation of histone H3K27, a modification that is considered an ‘activation’ mark11. However, recruitment of CBP/p300 does not always correlate with gene activation and is occasionally associated with repression12–16. A large number of chromatin regions that bind CBP/p300 therefore do not contain this canonical H3K27ac modification, indicating that HAT activity at such sites is blocked15,17. Thus a major challenge is to understand the mechanism that allows switching between inactive and active states of CBP/p300 on enhancers, and to causally link cellular signalling to the recruitment of CBP/p300, regulation of HAT activity and to the establishment of repressed, poised and active chromatin.
Here we have investigated how the activation and oligomerisation status of p300 TF ligands such as IRF3 and STAT1 impacts the catalytic activity of p300. We found that the kinase-activated and dimeric, but not inactive or monomeric variants of these TFs, support robust p300 HAT activation. We demonstrate that TF dimerization enables p300 trans-autoacetylation in a lysine-rich, intrinsically disordered autoinhibitory loop (AIL) in the HAT domain that serves as a ‘pseudo-substrate’ and is important for the regulation of p300 HAT activity18. A crystal structure of the core domain of p300 provides a snapshot of a potential trans-autoacetylation reaction intermediate in which the AIL projects into the active site of a neighbouring p300 molecule. As HAT activation is intimately linked to TF activation, these results causally relate cellular signalling to the activation and DNA targeting of a chromatin modifier and provide mechanistic insights into the long-standing and general correlation between an active, acetylated chromatin structure and gene transcription.
Results
Transcription factor dimerization activates p300
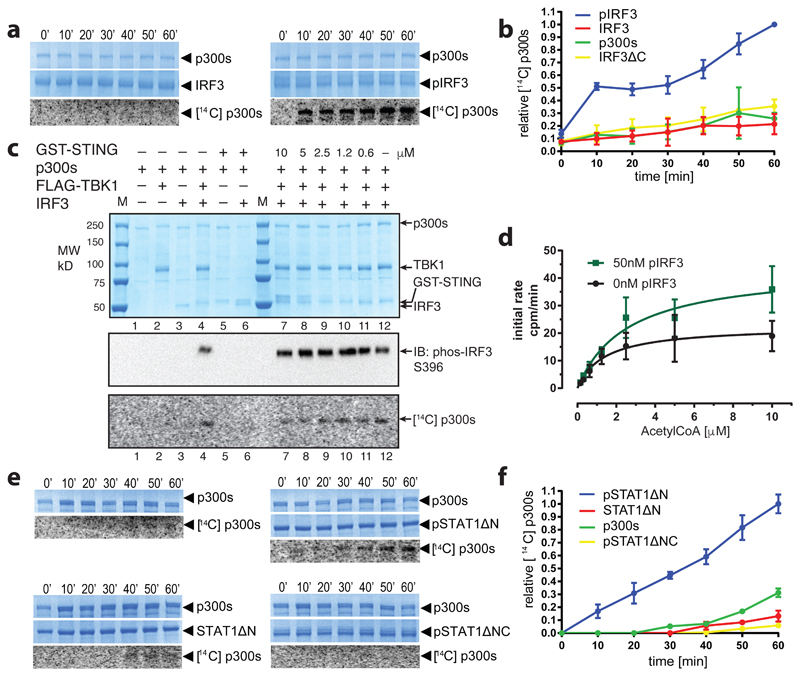
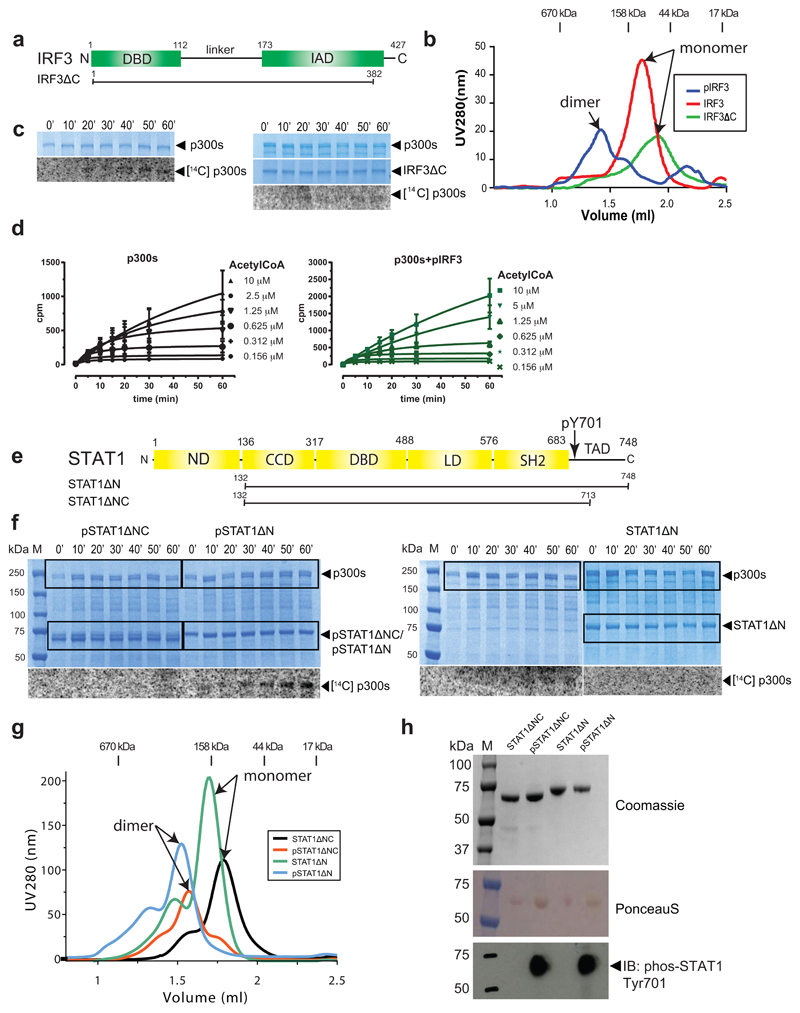
To explore if p300 is activated by signal-dependent IRF3 dimerization, we produced three recombinant IRF3 species: inactive monomers (IRF3), active IRF3 dimers by TBK1 phosphorylation (pIRF3) and a truncation mutant lacking the C-terminal autoinhibitory element (IRF3ΔC) (Extended Data Fig. 1a, b). Truncation of the C-terminal autoinhibitory element allows CBP/p300 binding but abolishes IRF3 dimerization19. We confirmed the oligomerisation status by gel filtration chromatography (Extended Data Fig. 1b), and investigated the impact of IRF3 activation and oligomerisation status on p300s autoacetylation in the presence of 14C acetyl-CoA. p300s spans from the TAZ1 to the NCBD/IBiD domain and contains a deletion of the flexible N- and C-terminal regions (Extended Data Fig. 4a). p300s autoacetylated slowly in the absence of IRF3 (Extended Data Fig. 1c). Inclusion of inactive, monomeric IRF3 or IRF3ΔC did not modify HAT activity (Fig. 1a and Extended Data Fig. 1c). In contrast, inclusion of active, TBK1-phosphorylated IRF3 dimers (pIRF3) resulted in a rapid burst of autoacetylation followed by a gradual increase of acetylated p300 (Fig. 1a, b). As IRF3ΔC did not support p300 HAT activation, IRF3 dimerization and not merely p300 binding is essential for HAT activation.
Figure 1. Transcription factor dimerization activates p300.
(a) p300s was incubated for the indicated times in the presence or absence of inactive, monomeric IRF3 or TBK1-phosphorylated, dimeric pIRF3. Samples were analysed by SDS-PAGE followed by Coomassie staining and autoradiography. Representative data of three independent experiments are shown. (b) Quantification of autoacetylation of p300s. (c) p300 is activated by TBK1-mediated IRF3 phosphorylation. p300s was incubated with recombinant GST-STING, TBK1 and IRF3 in the presence of ATP and [14C] acetyl-CoA. Top panel: Coomassie-stained SDS-PAGE gel. Middle panel: Analysis of IRF3 phosphorylation on S396 using immunoblotting. Bottom panel: autoradiography. Representative data of three independent experiments are shown. (d) HAT scintillation proximity assay. The degree of Histone H4 substrate acetylation was quantified using scintillation counting. (e) As in panel a but using inactive, monomeric STAT1ΔN or activated, dimeric pSTAT1ΔN. Activated, dimeric pSTAT1ΔNC lacking the C-terminal TAD did not stimulate p300s autoacetylation. Samples were analysed as in panel (a). Representative data of three independent experiments are shown. (f) Quantification of autoacetylation of p300s. Intensity values were normalized by dividing by the maximum autoacetylation signal obtained after 60 minutes. Error bars shown in panels (b), (d) and (f): Three independent experiments were performed and the mean value and error bars representing the standard deviation are shown. Data analysis and plotting was done with GraphPad Prism 7.0. For gel source data, see Supplementary Figure 1.
p300 HAT activation was directly dependent on TBK1-mediated phosphorylation of IRF3 on Ser396, a critical residue for IRF3 activation and dimerization2,3. Only when both TBK1 and IRF3 were included in the reaction did we observe p300 activation (Fig. 1c, Lane 4). We only observed a modest stimulatory effect of the adaptor protein STING (Fig. 1c, Lanes 7-12), likely due to the relatively high amounts of TBK1, which is already active and phosphorylates IRF3 even in the absence of STING20. We conclude that IRF3 phosphorylation by TBK1 and its dimerization are required for p300 HAT activation.
To analyse the impact of pIRF3 on p300 activation and histone substrate acetylation, we established a scintillation proximity HAT assay. We incubated saturating amounts of a biotinylated Histone H4 substrate peptide with p300s in the absence or presence of equimolar pIRF3 and increasing concentrations of [3H] acetyl-CoA (Fig. 1d). pIRF3 stimulated p300 histone substrate acetylation as determined by the increased rate of H4 acetylation obtained in the presence of pIRF3 (Vmax = 43.8 ± 5.3 cpm/min as compared to Vmax = 22.5 ± 2.8 cpm/min in the absence of pIRF3). These data indicate that pIRF3 not only stimulates p300 autoacetylation and activation, but also more efficient histone substrate acetylation.
We also investigated the effect of STAT1 on p300 activation. STATs are activated in response to cytokine receptor engagement and JAK kinase activation21. JAK-mediated STAT1 phosphorylation on tyrosine 701 induces dimerization and translocation to the nucleus where STAT1 binds to DNA elements to regulate gene expression. STAT1 contains a C-terminal transactivation domain (TAD) through which it interacts with CBP/p3007. A naturally occurring splice variant, STAT1β, lacks the TAD and acts in a dominant negative manner22. Structures of the active, STAT1 Tyr701-phosphorylated dimer bound to DNA as well as the STAT1 TAD bound to the TAZ2 domain of CBP have been determined previously23,24.
To understand the impact of STAT1 activation and oligomerisation status on p300 activity, we produced STAT1ΔN lacking the N-domain (ND) and STAT1ΔNX lacking the ND and TAD as non-phosphorylated monomers or as Tyr701-phosphorylated dimers (Extended Data Fig. 1e-h). We found that p300s autoacetylated slowly in the absence of STAT1, and that addition of non-phosphorylated, monomeric STAT1ΔN did not stimulate p300 autoacetylation beyond background levels (Fig. 1e, f). In contrast, addition of Tyr701-phosphorylated STAT1ΔN (pSTAT1ΔN) dimers to p300s resulted in a rapid increase of p300 autoacetylation. Activation required the C-terminal TAD of STAT1 as addition of a Tyr701-phosphorylated STAT1 dimer (pSTAT1ΔNC) lacking the TAD, did not stimulate p300 autoacetylation (Fig. 1e, f).
STAT1 dimerization, and not merely interaction with the TAZ2 domain, is required for activation of p300. Unphosphorylated, monomeric STAT1ΔN, which contains the TAD and is able to interact with the TAZ2 domain of CBP, did not stimulate p300 activity. Stimulation with STAT1 was however not as potent as compared to that of IRF3, possibly because our STAT1 preparation is unphosphorylated on Ser727, which is required for maximal gene activation25. Together, our data are in agreement with a model in which the AIL peptide serves as an intramolecular ‘pseudosubstrate’ and competitive HAT inhibitor18. Dimeric ligands such as pIRF3 and pSTAT1 allow p300 activation by bringing two molecules together to enable trans-autoacetylation of the AIL, which in turn relieves autoinhibition and enables more efficient entry of substrates into the HAT active site.
Structure of p300 adopts a AIL swap conformation
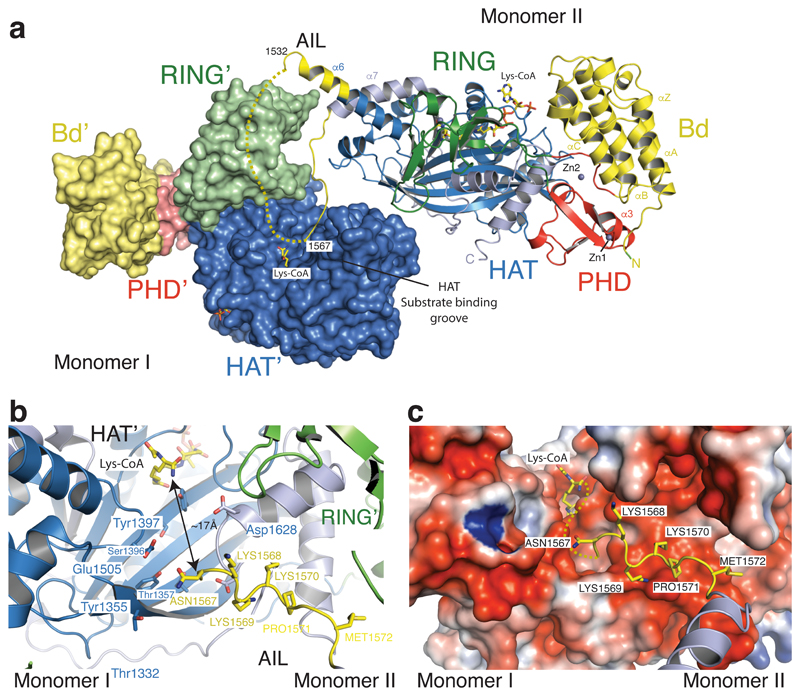
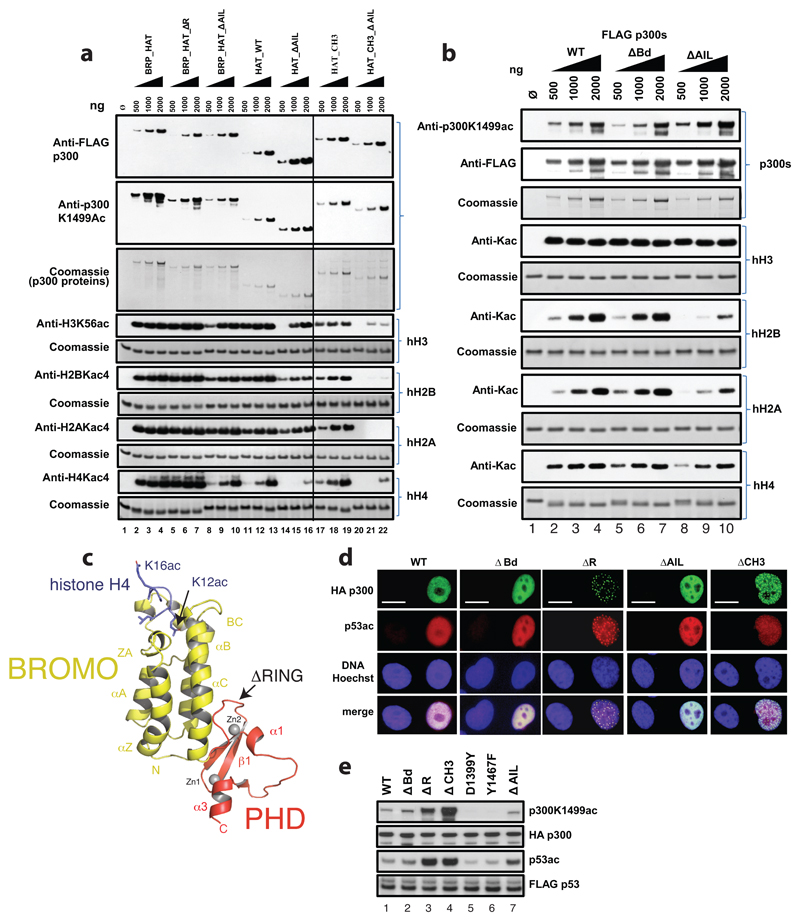
To further understand the role of the AIL in regulation of these structural transitions, we crystallized the hypoacetylated form of BRP-HAT containing the AIL. Crystals were obtained using a similar protocol as published previously26. Crystals diffracted to a minimal Bragg spacing of 3.1 Å and we determined the structure by molecular replacement (Extended Data Table 1). The crystal form contained four p300 molecules in the asymmetric unit (Extended Data Fig. 2). Structural comparison with our previous structure showed that the bromo-PHD-HAT domains overlay well on each other with a root–mean–square–deviation (RMSD) of ~ 1 Å. However the RING domains were not visible in the initial electron density map. Anomalous difference density maps showed a density peak for the zinc atom of the RING domain but not at the expected location. Manual repositioning allowed correct placement of the RING domains into the new position and refinement of the structure (Fig. 2a, Extended Data Fig. 3).
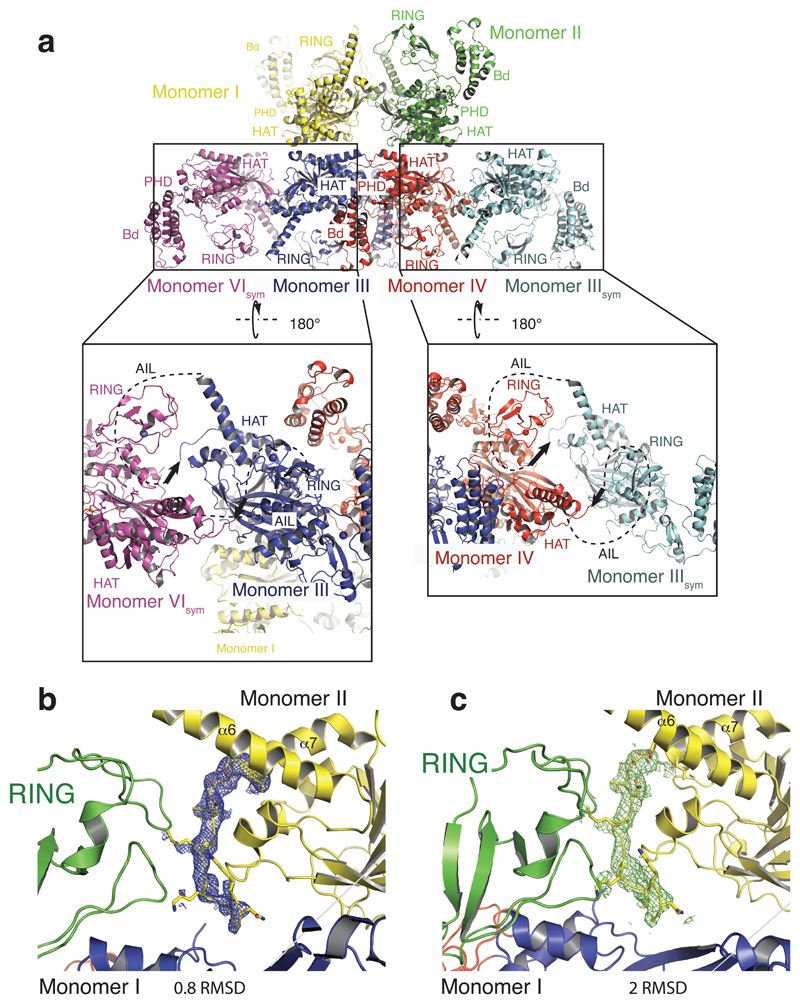
Figure 2. The structure of p300 adopts a AIL swap conformation.
(a) Monomer I is surface rendered and monomer II is shown as a cartoon. The AIL loop from monomer II is shown in yellow. The AIL lies near the HAT substrate binding groove of monomer I. A disordered segment of the AIL is shown as a dotted line. (b) Close up view of the residues of the AIL loop from monomer II and residues of monomer I in the substrate binding pocket. (c) Binding of the positively charged AIL is mediated by interactions with negatively charged residues in the HAT binding pocket.
The p300 molecules show an antiparallel arrangement of the BRP-HAT domains (Extended Data Fig. 2a). In this configuration, the HAT domains from two neighbouring molecules are closely apposed (Fig. 2a). In all protomers, AIL residues 1520–1532 adopt a helical extension of α6 which packs against the outwardly rotated RING domain of the neighbouring protomer (Fig. 2a). In monomer II, residues 1566–1581 extend away from the HAT domain and associate with the substrate binding pocket of the HAT domain in monomer I, ~17 Å away from the lysine substrate binding tunnel (Fig. 2b). The remainder of the AIL (residues 1532–1564) is disordered. In this conformation, positively charged residues K1568, K1569, K1570 project towards the highly electronegative substrate-binding pocket of the HAT domain in monomer I (Fig. 2c). As our SEC-MALLS data show that p300 is monomeric at low micromolar concentrations (see Extended Data Fig. 6), the AIL loop-swapped interactions do not appear to mediate formation of stable dimers, but may instead constitute more transient self-associations. Although the AIL is clearly flexible and the electron density over the exchanged region not visible in all protomers (Fig. 2b, c; Extended Data Fig. 2b, c), this arrangement supports the interpretation that at high concentrations and when in close proximity to each other, two p300 monomers can engage each other by a AIL loop–swap.
Structural rearrangement of the RING domain
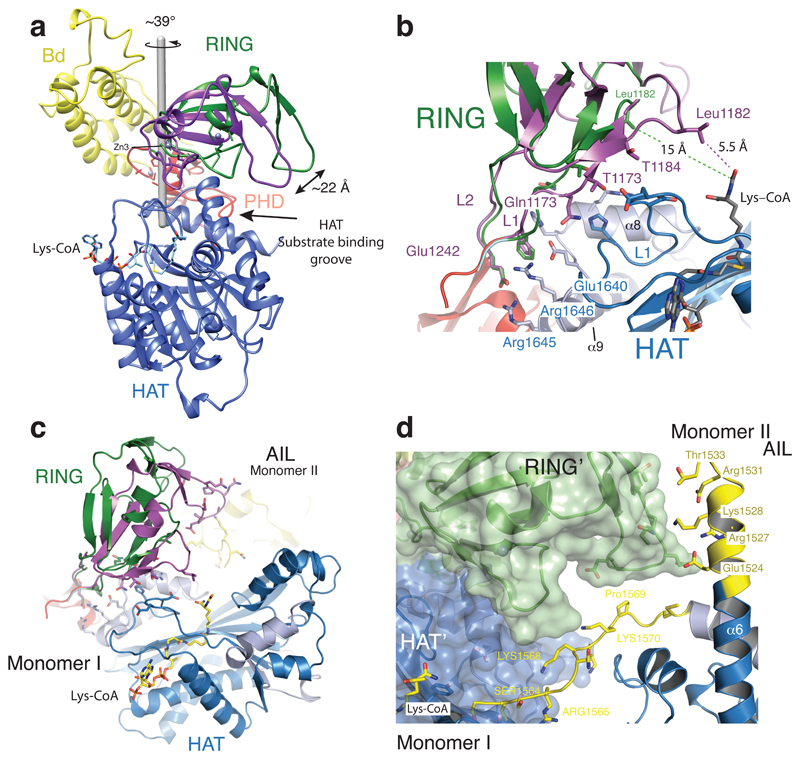
We previously proposed that active-site restriction by the RING domain is a negative regulatory mechanism for HAT activity26. A restricted active site is predicted to reduce the probability of substrates engaging with the active site by random diffusion and could thus be important in allowing for the regulation of acetylation by substrate recruitment. In agreement with this model, mutations that map to the structural framework that holds the RING domain in place result in HAT activation in cells26. In our current structure, the RING domain rotates by ~39° away from the HAT active site resulting in an overall displacement by ~22 Å as compared to the previously determined structure lacking the AIL (Fig. 3a). The axis of rotation is located perpendicular to the flexible loops L1 and L2 that connect the RING to the PHD domain.
Figure 3. Structural rearrangement of the RING domain.
(a) The RING domain (green) rotates ~39° resulting in a ~22 Å displacement away from the active site. The rotation axis is indicated as a grey rod. (b) In the loop-swap conformation, residues in the RING-HAT interface are disrupted thus resulting in a more open HAT active site. Leu1182 is positioned ~15Å away from the Lys-CoA inhibitor in the loop-swap conformation (green) but within 5.5Å in the absence of the loop swap (magenta). (c) Repositioning of the RING domain allows the AIL from monomer II to approach the HAT active site of monomer I. (d) Details of the interaction surface of the AIL from monomer II with the RING domain of monomer I.
The inward rotated conformation (magenta in Fig. 3a) is stabilized by interactions between Glu1242 of the RING domain and Arg1645 and Arg1646 of helix α9 of the HAT domain. In addition, Gln1173, Thr1174 and Thr1184 of the RING domain pack against the unusually long loop, L1 in the HAT domain that covers the CoA portion of the Lys-CoA inhibitor. As a result, Leu1182 resides within ~5.5 Å of the lysine moiety of Lys–CoA (Fig. 3b). This inward conformation of the RING domain thus restricts substrate access to the HAT domain: The incoming AIL from the neighbouring p300 monomer II would clash with the RING domain in the inward conformation (Fig. 3c).
In the outward rotated conformation, the interactions that attach the RING to the HAT domain are mostly disrupted (Fig. 3b). Leu1182 is positioned ~15 Å away from the substrate-binding site and the RING domain is cradled by the AIL extension of helix α6 of the neighbouring p300 molecule (monomer II residues 1524-1533; Fig. 3d). Despite shape complementarity, with a small buried surface area of ~320 Å2, the interface is predominantly polar, uncharacteristic of a typical protein-protein interface. However, this interaction could help to stabilize an outward rotated conformation of the RING domain and a more open HAT active site, apparently to enable AIL access and trans-acetylation.
Regulation of HAT activity by flanking domains
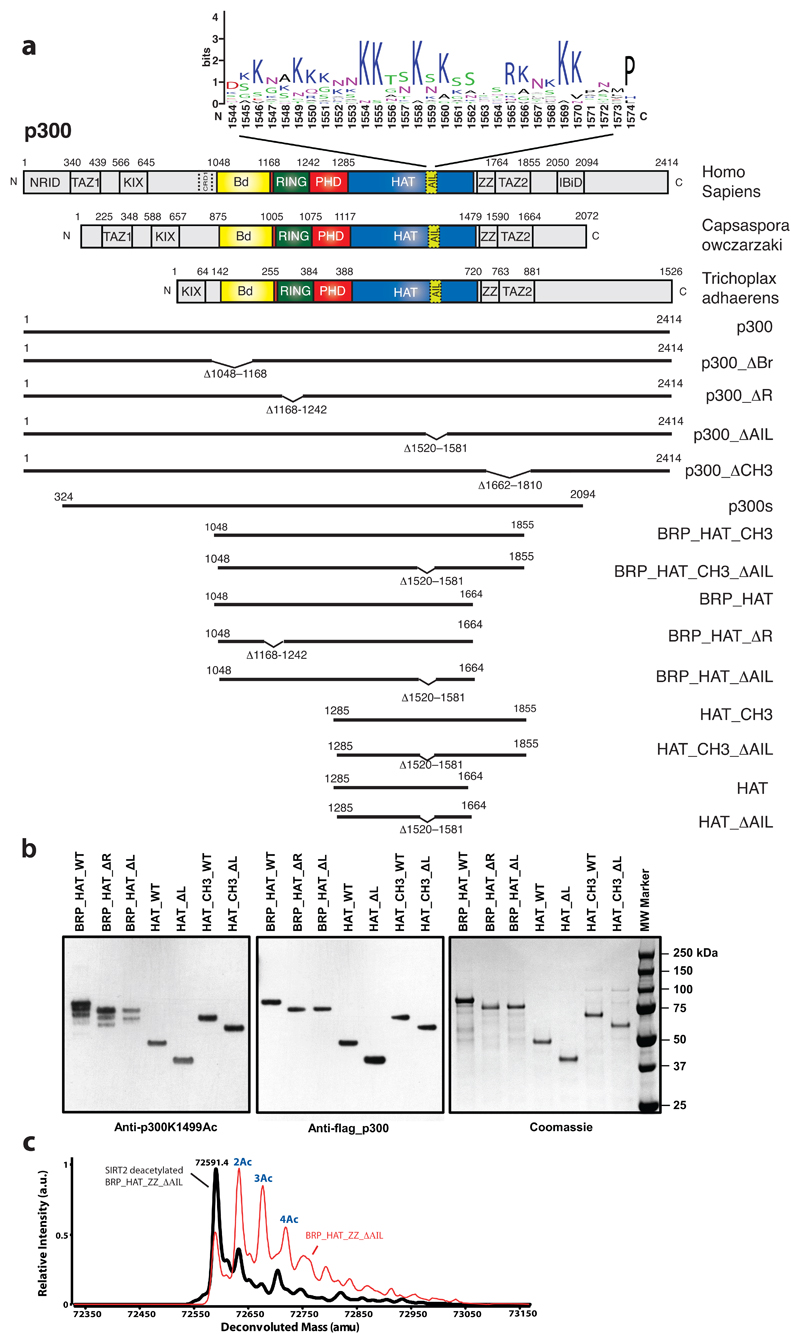
To systematically analyse the flanking domains, we generated a series of p300 constructs (Extended Data Fig. 4a) and analysed the impact on HAT activity in vitro and in cells. Overexpression of p300 generally resulted in hyperacetylated, active p300 variants (Extended Data Fig. 4b,c) which likely masks the functional role of structural elements potentially involved in autoinhibition of deacetylated p300. Deletion of the RING domain did not drastically alter auto- or histone acetylation (Extended Data Fig. 5a). This deletion did not adversely affect structural integrity of p300, as shown by a crystal structure of the BPΔR module containing this deletion (Extended Data Fig. 5c).
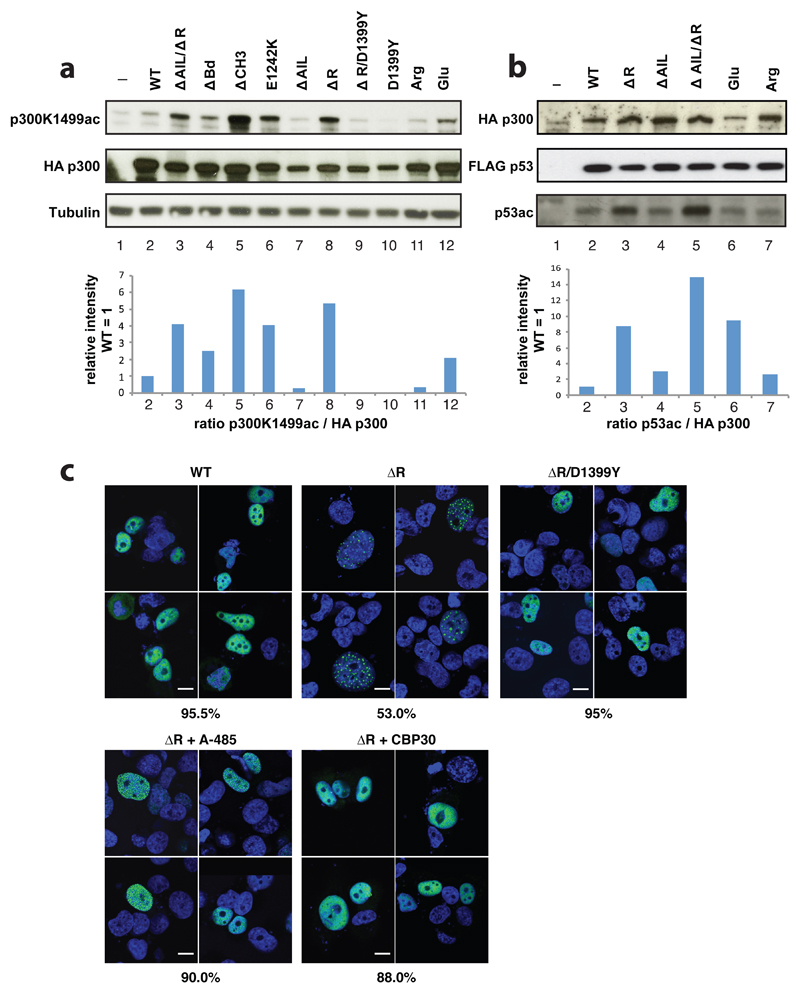
Deletion of the AIL in all constructs resulted in decreased histone acetylation but bromodomain deletion (ΔBd) did not impact HAT function (Extended Data Fig. 5a, b). Together, we agree with previous observations made for CBP that at least in the active, hyperacetylated state of the enzyme, RING deletion does not substantially impact HAT activity and that the p300 AIL positively contributes to substrate acetylation27. We next introduced mutations into full-length p300 and monitored their effect on p300 auto- and p53 acetylation upon transient co-overexpression in cells. Deletion of the RING and CH3 domains resulted in significantly increased p300 and p53 acetylation but deletion of the Bd or AIL had no major impact (Extended Data Fig. 5e). As expected, introduction of the catalytic mutants D1399Y or Y1467F abolished p300 or p53 acetylation (Extended Data Fig. 5e). Immunofluorescence analysis showed that wild-type p300 as well as a ΔBd and ΔAIL deletion were uniformly distributed in the nucleus but that the HAT activating p300 variants ΔRING and ΔCH3, formed nuclear foci that co-localized with p53 (Extended Data Fig. 5d). To validate these results, we analysed and confirmed the phenotype of p300 mutants and p53 acetylation in another cell line (Fig. 4a, b). In addition, we analysed p300 variants in which eleven lysine amino acids spanning amino acids 1546-1570 of the AIL were mutated to arginine or glutamate and found reduced or slightly increased p300 autoacetylation or p53 acetylation levels, respectively (Fig. 4a, b).
Figure 4. Regulation of HAT activity by flanking domains.
(a) Indicated variants of p300 were transiently co-transfected with p53 in COS cells and samples analysed by western blotting using the indicated antibodies. Bottom panel: quantification p300 K1499Ac signal. (b) Analysis of p53 acetylation. Bottom panel: quantification p53 acetylation signal. Representative data of three independent experiments are shown. For details on the mutants see Extendend Data Fig. 4a. Arg and Glu: lysine amino acids in the AIL segment spanning amino acids 1546-1570 were mutated to arginine or glutamate, respectively (c) H1299 cells were transfected with the indicated construct and analyzed by immunoflorescence using Anti-HA for p300 (green) and cell nuclei were stained with Hoechst (blue). Bottom panels: Cells were treated with the A-485 HAT or the CBP30 Bromodomain inhibitor. Percentage of cells showing the indicated phenotype (n=200 cells) is indicated below each panel. Scale bar, 10 μm. For gel source data, see Supplementary Figure 1.
As we observed formation of nuclear foci only with HAT activating variants, we hypothesized that hyperacetylation drives p300 to form biomolecular condensates in cells. Accordingly, introduction of a HAT inactivating D1399Y mutation into p300 ΔRING, treatment with the p300 HAT inhibitor A-48528 or with the CBP/p300 Bd inhibitor CBP30 greatly reduced foci formation (Fig. 4c). We therefore conclude that HAT activation drives biomolecular condensation of p300 in cells, apparently through Bd substrate engagement.
AIL acetylation and RING domain repositioning regulate a dynamic interaction with the p300 substrate binding pocket
We next sought to understand how the highly conserved and intrinsically disordered AIL segment contributes to regulation of the catalytic function of p300. The AIL spanning amino acid residues 1532-1567 is positively charged in the deacetylated state, with an estimated isoelectric point (pI) of 10.9, and net charge of 7 at neutral pH. In contrast, upon autoacetylation of residues spanning Lys1542-156029, we estimate a pI of 3.5 and a net charge of –2. As the proximal substrate binding groove of p300 is largely acidic (Fig. 3c), we hypothesized, in agreement with earlier predictions30, that a deacetylated AIL would engage the substrate binding site through electrostatic interactions, presumably to prevent access of positively charged lysine-containing substrates. Given the disordered nature of the AIL, this proposed interaction is expected to be highly dynamic27.
We tested this postulate through all-atom Monte Carlo simulations31. To make this approach tractable, our simulations held the backbone dihedral angles associated with the folded domains fixed, but all other degrees of freedom, including all backbone and side chain dihedral angles in the AIL were fully sampled. As a result, these simulations should be seen as assessing how the AIL interacts with the remainder of p300 given the observed crystal structure. Simulations were performed on the AIL in the deacetylated and acetylated state in the context of the p300 monomer. These simulations allowed us to interrogate how acetylation influenced the conformation and intra-molecular interactions of the AIL.
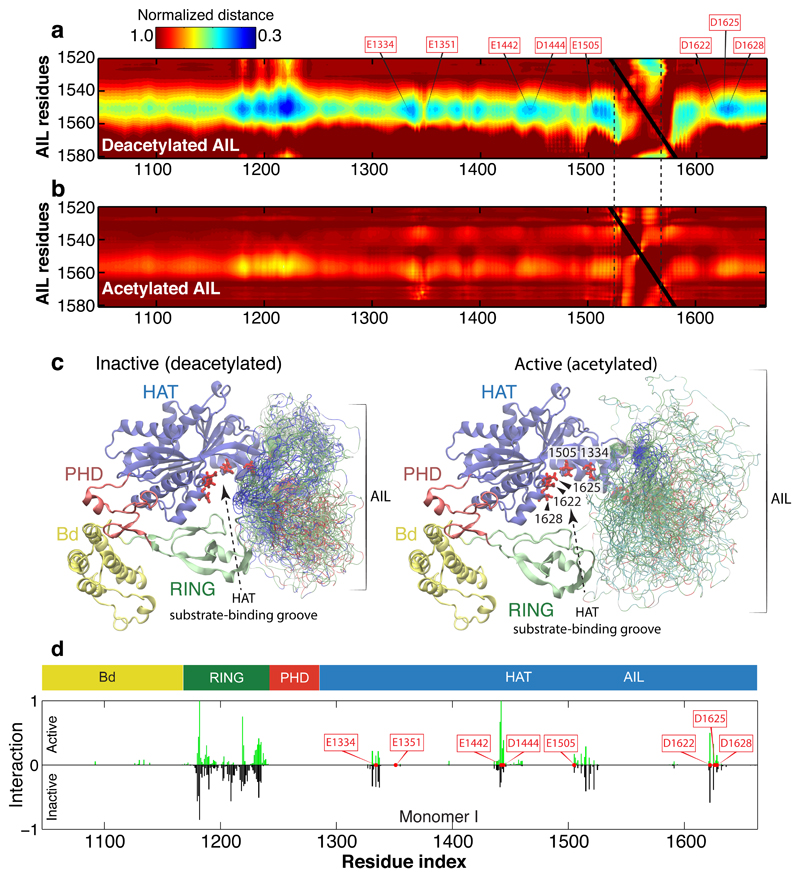
Simulations of the deacetylated AIL revealed the presence of extensive yet highly degenerate electrostatic interactions between the AIL and the RING domain and HAT substrate binding site. These interactions were quantifiable in terms of the normalized distances between pairs of amino acid residues (Fig. 5a and Supplementary Video 1). Lysine residues in the AIL dynamically associate through long-range electrostatic interactions with acidic residues (E1334, E1442, E1505, D1622, D1625 and D1628) in the p300 HAT substrate binding pocket (Fig. 5c). The importance of these residues for substrate acetylation has been shown previously32, and NMR data for CBP confirm that the AIL is intrinsically disordered in the deacetylated state27.
Figure 5. Acetylation of the AIL regulates dynamic interaction with the substrate binding pocket of p300.
(a) Normalized distance between the AIL and residues in the inactive monomer. Inter-residue distances were normalized by the distances expected if the AIL behaved as a self-avoiding random coil. Electrostatic interaction mediated by conserved lysine residues between K1542 and K1560 of the AIL and aspartic/glutamic acid residues around the active site of the HAT domain, as shown by the residues highlighted (E1334, E1351, E1442, D1444, E1505, D1622, D1625, and D1628). The extensive contacts between the AIL and the RING domain originates in part from the RING domain’s proximity to the AIL in its inhibitory conformation. (b) Normalized distance between the AIL and all residues in the active (acetylated) monomer. After acetylation, lysine-mediated electrostatic interactions are lost. (c) Representative conformations with the AIL shown as an ensemble for the inactive deacetylated monomer (left) and the active acetylated monomer (right). The Cα atoms of residues in the AIL are coloured according to charge: blue (positive), red (negative) and green (non-charged). The HAT substrate-binding groove is more exposed in the active acetylated state, due to both the relative position of the RING domain and the lack of preferential interactions by the AIL. (d) Inter-molecular interactions in the loop-swapped dimer between the AIL of one HAT and the adjacent subunit of the other. The adjacent subunit is either in the active (top) and inactive (bottom) conformation. In the active state, the AIL is able to directly engage with residues E1442 and E1444 from the adjacent HAT substrate binding groove, suggesting the position of the RING domain has a steric impact on the accessibility of the AIL.
In contrast, in the acetylated state, we found no interactions between the AIL and the substrate binding site (Fig. 5b and Supplementary Video 2). The acetylated AIL essentially behaved like a self-avoiding random coil without any strong biases for interaction with itself or with the surrounding folded domains, including the Bd. It has been proposed that the AIL of CBP, when acetylated on K1596 (K1558 in p300), engages the Bd intramolecularly thus competing with histone binding and negatively regulating substrate acetylation27. Isothermal calorimetry experiments showed highest binding affinity for multiacetylated peptides including diacetylated histone H3 K14acK18ac or H4 K12acK16ac peptides, generally following the pattern KacNNNKac (Extended Data Table 2). Monoacetylated peptides typically had weaker binding affinity. A crystal structure of the H4 K12acK16ac peptide bound to BPΔR (Extended Data Fig. 5c) confirmed the acetyllysine-specific binding mode. However, a AIL peptide acetylated on three lysines K1549, K1558, K1560, corresponding to some of the most highly acetylated residues in the AIL29, failed to bind to the BRP module. Thus our model is that the multiacetylated AIL is not a substrate for the Bd, presumably because of suboptimal spacing or sequence environment of the AIL Kac sites.
To understand how the RING domain influences the ability of substrates, including the AIL, to enter the active site of an adjacent p300 molecule, we performed simulations of the AIL in context of the loop-swapped dimer with a harmonic potential to maintain the AIL in the active site to assess potential inter-molecular interactions (Fig. 5d, Extended Data Fig. 6a). In the active RING conformation, the AIL is able to engage the substrate binding site. However, in the inactive conformation, the frequency of contacts between the AIL and the acidic active site residues E1442 and D1444, residues proximal to the lysine substrate binding tunnel, was reduced by 70-75% (Fig. 5d). Thus, in the inactive conformation the RING domain at least partially reduces catalytic activity by limiting accessibility of the active site to the AIL and other substrates.
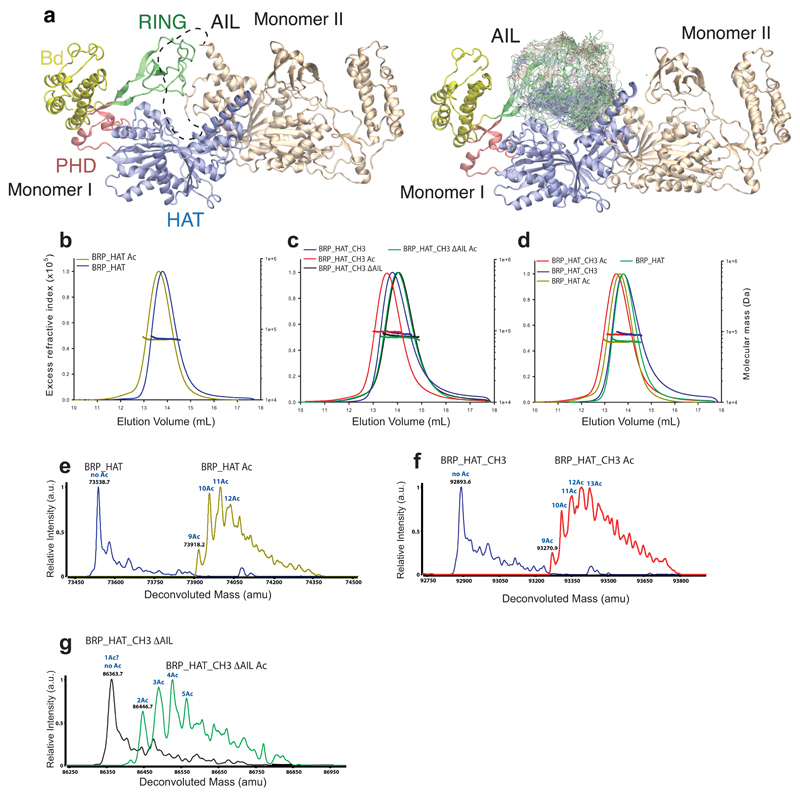
One prediction from our models is that the deacetylated form of p300 adopts a more compact conformation, due to dynamic engagement of the AIL with the HAT substrate-binding site, while the acetylated form adopts a more ‘open’ conformation (Fig. 5d). To test this possibility, we produced hypo- and hyperacetylated p300 variants (Extended Data Fig. 6e-g).
We analysed these preparations by multi-angle laser light scattering coupled to size exclusion chromatography (SEC-MALLS). All preparations were monomeric at the concentration tested (2 mg·ml-1) (Extended Data Fig. 6b-d, Extended Data Table 3). Hyperacetylation of p300 BRP-HAT resulted in a small decrease in the elution volume indicative of a larger hydrodynamic radius (Extended Data Fig. 6b). A similar result was obtained upon comparison of hyper- and hypoacetylated BRP-HAT-CH3 (Extended Data Fig. 6c). In contrast, a variant lacking the AIL showed no shift in the elution volume upon hyperacetylation (Extended Data Fig. 6c). Thus our data agree with the model that the catalytic p300 ‘core’ spanning the BRP-HAT-CH3 domains, adopts a compact conformation in the hypoacetylated state and that autoacetylation results in a more extended conformation.
Discussion
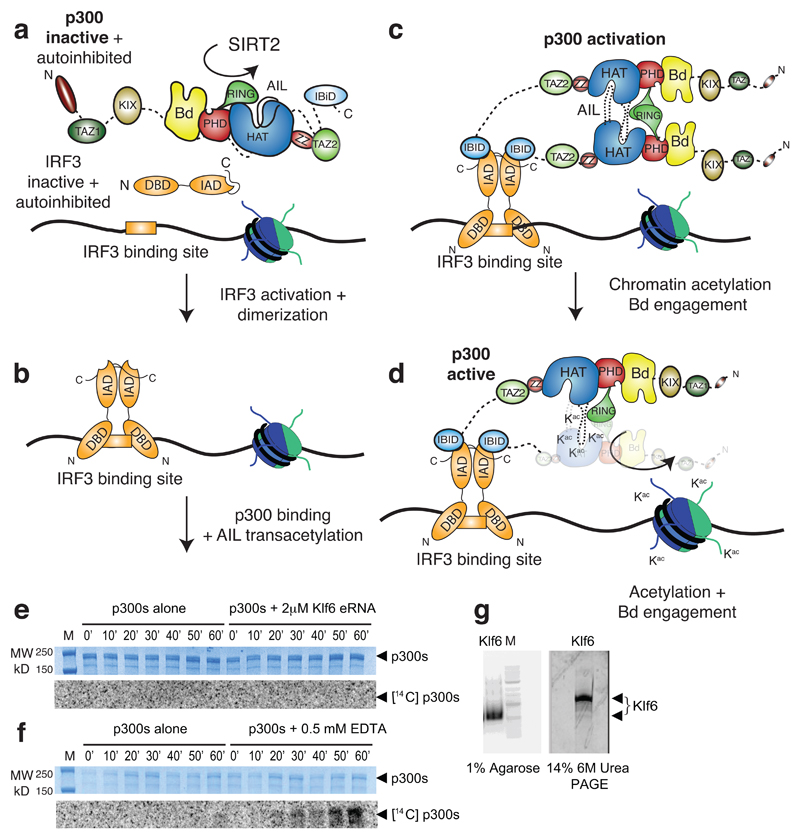
Our findings represent detailed mechanistic insights into how cellular signalling controls activity of a chromatin regulator. We propose a multi-step process for p300 HAT activation and signal transmission to chromatin (Extended Data Fig. 7a-d). In the basal state, the deacetylated AIL is expected to maintain an overall positively charged environment in close proximity to the enzyme’s active site, which prevents access of positively charged lysine-rich substrates. Direct access to the CoA binding tunnel and autoacetylation of the AIL in cis appears to be prohibited, in part due to positioning of the RING domain (Fig. 5d).
Cellular signalling initiates phosphorylation of TFs, such as IRF3 or STAT1, which results in their activation and dimerization. The activated, dimeric TFs are in their DNA-binding competent conformation and can engage two molecules of p300 in the nucleus thus increasing the likelihood of AIL disengagement from its inhibitory position in cis and capture in trans by a second p300 molecule. Association of two p300 molecules does not necessarily require precise stereospecific interactions between the structured domains as acetylation at multiple lysines in the AIL indicates a series of possible conformations in such transiently associating dimers. We predict that regulated oligomerisation uncouples recruitment from HAT activation which could explain why not all CBP/p300 recruitment events result in chromatin acetylation and gene activation12–17,33.
It has been proposed that enhancer RNA (eRNA) interacts with the AIL to regulate CBP HAT activity34. We have attempted to reproduce these results using Klf6, one of the most potent eRNAs reported34. We could not detect p300 HAT activation using up to equimolar amounts of Klf6 (Extended Data Fig. 7e, g). We note that Bose et al. purified CBP in buffer containing EDTA, which is detrimental to the structure of CBP/p300 due to the presence of multiple zinc-binding domains35. Unfolded CBP/p300 have a high tendency to aggregate, and to form non-specific interactions35. Paradoxically, as the HAT domain is not affected, inclusion of EDTA can have an ‘activating’ effect in biochemical assays, apparently due to such non-specific aggregation (Extended Data Fig. 7f). The detrimental effects of EDTA on the structure and function of CBP/p300 need to be taken into account in the interpretation of such data.
The ability of certain histone-modifying enzymes to bind to the PTM they generate has led to models where such enzymes might propagate modified chromatin domains by a positive-feedback loop36. According to this view histone PTMs and other chromatin modifications form an additional, DNA sequence-independent layer of the genome which is read out by enzymes that recognize these modifications to ‘epigenetically’ regulate genomic function36. An alternative view posits that histone PTMs ultimately depend on DNA sequence-dependent recruitment of chromatin modifiers and so do not necessarily form an independent ‘epigenetic’ layer of the genome8,37–39. The controversy has arisen because it has been difficult to disentangle, for most chromatin regulators, the relative contributions of DNA targeting and histone PTM substrate engagement to the overall chromatin modification reaction.
We show that regulation of p300 is linked to the activation and oligomerisation status of TF ligands and therefore conclude that specificity for p300-mediated chromatin acetylation arises mainly through TF-mediated and DNA sequence-dependent genome targeting. How then does the BRP ‘PTM reader’ module contribute to p300 function? While it is clear that the Bd can engage acetylated histone peptides and bind to hyperacetylated chromatin26,40, deletion or mutation of the Bd has no apparent effect on substrate acetylation26,41, has only minimal effects in a hematopoiesis model system42, and Bd inhibition does not adversely affect genome targeting of CBP43.
We favour the model that DNA binding provides the lead anchoring mechanism: Local hyperacetylation increases the binding valency by allowing Bd substrate engagement, which further helps to compartmentalize the biochemical reaction and contributes to signal maintenance 40. p300 HAT activating mutants form biomolecular condensates in cells when transiently overexpressed (Fig. 4c, Extended Data Fig. 5d). Treatment with a HAT or Bd inhibitor greatly reduces condensate formation, indicating that hyperacetylation and Bd-substrate engagement are critical in driving assembly. The formation of condensates, possibly through phase-separation, may provide a mechanism to enable signal integration on enhancers and transcriptional control44. It will be critical to disentangle cause-effect relationships of DNA targeting, chromatin modification and histone PTM substrate engagement of other chromatin regulators45–47.
Methods
Constructs
For cell-free protein expression, cDNA of p300 (NCBI reference sequence: NM_001429.3) variants were cloned into the pIVEX2.4d vector (Roche) with a N-terminal 6x His tag and a C-terminal FLAG tag. In the ΔR constructs, the RING domain encompassing residues 1169–1241 was replaced by Glycine amino acid residue linker. In the ΔAIL constructs, loop amino acid residues comprising residues 1520–1581 were replaced by the flexible linker sequence SGGSG. For E.coli expression, cDNA encoding residues 1048-1282, for the BRP or BPΔR were cloned into the vector pETM-33 (EMBL) with a TEV cleavable N-terminal glutathione S-transferase (GST) tag. p300 BRP_HAT variants were cloned into pFASTBAC1 (Thermo Fisher) and expressed in insect cells as shown earlier26. p300s constructs, spanning amino acid residues 324-2414, were cloned into pFASTBAC1 vector with an N-terminal FLAG tag. HA-tagged full-length p300 variants were cloned into pcDNA3.1 (Thermo Fisher). Point mutations were introduced by QuikChange mutagenesis (Agilent). Point mutations and nucleotide deletions carried out in p300FL (1-2414) or p300s (324-2094) were done through transfer vectors as described previously26. STAT1ΔN (136-748), STAT1ΔNC (136-713) and IRF3ΔC (1-382) with a C-terminal intein tag were cloned into the pTXB1 vector (New England Biolabs) using the restriction enzymes NdeI (STAT1) or NcoI (IRF3) and SpeI. IRF3 (1-427) with an N-terminal His-tag cleavable by TEV protease was cloned using the restriction enzymes NcoI and XhoI into the vector pETM-11 (EMBL). All constructs were confirmed by DNA sequencing.
Expression and Purification
Expression and purification of FLAG-tagged p300s constructs was done as described previously2. This method allows purification of p300s variants that are already preacetylated. Expression and purification of p300 BPR_HAT and SIRT2 were done as described in26. TBK1 was expressed in insect cells and purified as described previously20. Cell-free protein synthesis was done in a 50 μL reaction volume. Briefly, 10 μg mL-1 of His-p300 variants in pIVEX2.4d were added to a reaction mixture containing 1 mM amino acid mix, 0.8 mM rNTPs (guanosine-, uracil-, and cytidine- 5’ triphosphate ribonucleotides), 1.2 mM adenosine 5’-triphosphate, 55 mM HEPES, pH 7.5, 68 μM folinic acid, 0.64 mM cyclic adenosine monophosphate, 3.4 mM dithiothreitol, 27.5 mM ammonium acetate, 2 mM spermidine, 5 μM ZnCl2, 80 mM creatine phosphate, 208 mM potassium glutamate, 16 mM magnesium acetate, 250 μg mL-1 creatine kinase, 27 μg mL-1 T7 RNA polymerase, 0.175 μg mL-1 tRNA, and 67 μL mL-1 S30 E. coli bacterial extract. Incubation was carried out at 22 °C with agitation for 16 h. Proteins were purified using Ni-NTA chromatography (IMAC Sepharose 6 FF, GE healthcare) in buffer 1 (20 mM TRIS, pH 8.0, 300 mM NaCl, 1 mM DTT, 5 μM ZnCl2) containing Complete Protease Inhibitors EDTA-Free (Roche). The resin was washed with 20 CV of buffer 1 and the protein eluted with 5 CV buffer 1 containing 300 mM Imidazole. The protein was concentrated in a prewashed Amicon Ultra 0.5 ml Ultracel 10K Centrifugal filter (Molecular weight cut off = 10kDa; EMD Millipore). The protein was buffer exchanged into buffer 1 using 0.5 ml Zeba Spin desalting columns (Molecular weight cut off = 7kDa; Thermo Scientific), flash frozen in liquid N2 and stored at -80 °C.
For expression of GST-BRP and GST- BPΔR fusion proteins in E. coli BL21 (DE3), LB medium enriched with 100 μM ZnCl2 was used. Cell pellets were resuspended in buffer 1 containing Complete Protease Inhibitors EDTA-Free (Roche) and lysed by using a Microfluidizer (Microfluidics Corp., MA, USA). The lysate was clarified by centrifugation for 30 minutes at 39,000 g in a JA-25.5 rotor (Beckman) and applied to a Glutathione Sepharose 4 Fast Flow resin according to instructions by the manufacturer (GE Healthcare). The resin was washed with buffer 1 and incubated with His-tagged TEV protease (1:100 w/w) for 14-16 h at 4°C. Subtractive Ni-NTA chromatography (IMAC Sepharose 6 FF, GE Healthcare) was then employed to remove the residual His-tag and TEV protease. The untagged protein was further purified by gel filtration on a High Load 16/60 Superdex 75 column (GE Healthcare) equilibrated in 20 mM HEPES, pH 7.5, 300 mM NaCl, 0.5 mM TCEP and 5 μM ZnCl2. The final protein was concentrated to 15 mg/ml in a prewashed Amicon Ultra-15 Centrifugal filter (Molecular weight cut off = 10kDa; EMD Millipore), flash frozen in liquid N2 and stored at -80 °C.
The expression and purification of non-phosphorylated STAT1 variants (STAT1ΔN, STAT1ΔNC) and IRF3ΔC (1-382) was done in E.coli using the IMPACT expression system (New England Biolabs). For the expression of Y701 phosphorylated variants (pSTAT1ΔN, pSTAT1ΔNC), proteins were co-expressed with Elk receptor tyrosine kinase domain in E.coli BL21(DE3) TKB1 cells (Agilent). Cells were harvested by centrifugation and resuspended in buffer 2 (20 mM HEPES pH 7.5, 500 mM NaCl). The cells were lysed in a microfluidiser (Microfluidics Corp., MA, USA) and the soluble fraction was obtained by centrifugation for 30 minutes at 39,000 g in a JA-25.5 rotor (Beckman). The supernatant was first passed over chitin beads (New England Biolabs) and washed with buffer 2 for 10 column volumes. The protein was cleaved at 4°C for 16h in buffer 2 containing 50 mM DTT, eluted and further purified by gel filtration on a High Load 16/60 Superdex 200 column (GE Healthcare) equilibrated in buffer 2.
GST-STING, comprising the soluble cytoplasmic domain spanning amino acids 138-378, was expressed in E.coli BL21(DE3) at 37°C for 3 h. The cells were harvested by centrifugation and resuspended in buffer 3 (20 mM TRIS, pH 8.0, 300 mM NaCl, 1 mM DTT) containing Complete Protease Inhibitors EDTA-Free (Roche). The cells were lysed in a microfluidiser (Microfluidics Corp., MA, USA) and the soluble fraction was obtained by centrifugation as above. The supernatant was passed over equilibrated Glutathione Sepharose 4 Fast Flow resin according to instructions by the manufacturer (GE Healthcare). The resin was washed with buffer 3 and eluted with 10 mM reduced Glutathione in buffer 3. The protein was further purified by gel filtration on a High Load 16/60 Superdex 200 column (GE Healthcare) equilibrated in 20 mM HEPES, pH 7.5, 300 mM NaCl, 0.5 mM TCEP. The final protein was concentrated to 16 mg/ml in a prewashed Amicon Ultra-15 Centrifugal filter (Molecular weight cut off = 30 kDa; EMD Millipore), flash frozen in liquid N2 and stored at -80 °C.
IRF3 was expressed in E.coli BL21(DE3) at 18°C for 16 h. The cells were harvested by centrifugation and resuspended in buffer 2 containing 10 mM imidazole. The cells were lysed in a microfluidiser (Microfluidics Corp., MA, USA) and the soluble fraction was obtained by centrifugation as above. The supernatant was passed over Ni2+–conjugated IMAC sepharose resin (GE Healthcare) and washed with buffer 2 containing 20 mM imidazole. The protein was eluted in buffer 2 containing 500 mM imidazole and was further purified by gel filtration on a High Load 16/60 Superdex 200 column in buffer 2 containing 0.5 mM TCEP. IRF3 was phosphorylated in vitro at a 1:10 molar ratio TBK1:IRF3 (1mg/ml) in presence of 5 mM MgCl2 and 1 mM ATP. The reaction was incubated at 30°C for 1h and then for an additional 10h at 21°C. Phosphorylated IRF3 was further purified by size exclusion chromatography on a Superdex S200 16/60 column (GE Healthcare) in 20 mM HEPES, pH 7.5, 300 mM NaCl, 0.5 mM TCEP. The production of recombinant histones was done following standard procedures48.
Crystallization and structure determination
The p300 BRP_HAT construct comprising the AIL and the mutation Y1467F was deacetylated as done previously26. The protein at 4.5 mg ml-1 was incubated with a three-fold molar excess of the bi-substrate inhibitor Lys-CoA32 prior to crystallization. Crystals in the P21 space group were grown by hanging-drop vapor diffusion at 4 °C by mixing equal volumes of protein and crystallization solution containing 100 mM HEPES, pH 7.5, 18-22% polyethylene glycol 3350, 0.2 M NaCl. Crystals were cryoprotected in 20-25% ethylene glycol and drop frozen in liquid nitrogen. We collected native diffraction data to a minimum Bragg spacing of 3.1 Å resolution at the ESRF on beamline ID29 under a nitrogen gas stream at 100 K, at a wavelength of 1.282 Å. We processed the data with XDS (Extended Data Table 1). The structure of the p300 BRP_HAT was determined by molecular replacement using Phaser. There are four copies in the asymmetric unit and the RING domains were initially not visible in the electron density map and are partially disordered. Inspection of an anomalous difference map indicated peak density for the zinc ions and allowed positioning of the RING domain in the outward rotated conformation. A final model was produced by iterative rounds of manual model building in Coot and refinement using PHENIX. The final model contains residues 1045 -1664 with a deletion of residues 1534-1567 and was refined to a 3.1 Å resolution with an Rwork and an Rfree of 19% and 26%, respectively (Extended Data Table 1). Analysis of the refined structure by MolProbity showed that there are no residues in disallowed regions of the Ramachandran plot. The MolProbity all atom clash score was 1.91, placing the structure in the 100th percentile among structures refined at 3.1 Å resolution (N=2108).
The BPΔR construct at 15 mg ml-1 was mixed with 2 mM of a 11-mer histone peptide H4 (10-20) GLGKacGGAKacRHR (only the underlined amino acid sequence is visible in the electron density map) containing two acetylated Lysine residues at positions K12 and K16 (H4K12K16). Crystals in the P212121 space group were grown by hanging-drop vapor diffusion at 21°C by mixing equal volumes of protein and crystallization solution containing 1.6 M Ammonium Sulfate, 100 mM Bicine, pH 9.0. Crystals were cryoprotected in 20% ethylene glycol and drop frozen in liquid nitrogen. We collected native diffraction data to a minimum Bragg spacing of 2.5Å resolution at the ESRF on beamline ID29 under a nitrogen gas stream at 100K, at a wavelength of 1.0Å (Extended Data Table 1). Data processing, molecular replacement and refinement were done as indicated above. The final model contains two copies of the BPΔR module corresponding to residues 1049 -1279 of p300 in the asymmetric unit. As expected, replacement of the RING domain residues 1169-1241 by a single Glycine amino acid linker did not adversely affect the remainder of the BP module. Analysis of the refined structure by MolProbity showed that there are no residues in disallowed regions of the Ramachandran plot. The MolProbity all atom clash score was 0.97 placing the structure in the 100th percentile (N=6960).
Monte Carlo simulations
All-atom Monte Carlo simulations were performed using ABSINTH implicit solvent model and version 2 of the CAMPARI Monte Carlo simulation engine (http://campari.sourceforge.net)31. The initial AIL loop was constructed using MODELLER, and the complete set of backbone and side chain torsional angles were sampled for the AIL for which electron density was missing. Simulation analysis was performed with MDTraj and CTraj (http://pappulab.wustl.edu/CTraj.html)49. The backbone degrees of freedom of the folded domains were not sampled, while all amino acid side chains were fully sampledCAMPARI simulations explore conformational space through perturbation to the torsional angles (as opposed to Cartesian positions, as is typical for molecular dynamics). Consequently, a fully closed loop represents a major sampling challenge. To address this, we severed the covalent backbone bond between the N-terminal part of the AIL loop and the folded domain, and replaced this bond with a strong harmonic potential that recapitulates the distances constraint associated with the covalent bond. This allows moves to fully rotate the chain and dramatically improves the efficiency of conformational sampling.
We generated 5000 independent non-overlapping starting configurations and used a clustering approach to identify the most distinct 200 conformations. These were used as the starting conformations for full simulations. We ran 200 independent simulations of the deacetylated and acetylated p300 in the monomeric form, and 200 independent simulations of the loop-swapped p300 dimer in the active and inactive form (800 simulations total). Analysis was performed after an initial equilibration. Dimer simulations applied a harmonic potential between residue 1550 from the AIL and residue 1442 from the other monomer to maintain the AIL in the active site. This allowed us to directly compare active site accessibility of the AIL. For monomer simulations, no restraints were applied.
Each residue on the folded structure was evaluated for contacts with any residue in the AIL, and these contacts were summed to give an effective contact score. In this manner, the residues on the folded structure that most frequently interacted with any residue on the AIL were directly identified. Interaction was primarily of electrostatic nature, with residues E1334, E1444, E1505, D1622, D1625 and D1628 engaging in direct interactions. There are also extensive interactions between the AIL and the RING domain, although we cannot rule out that these interactions are driven by the harmonic applied to pull E1442 towards the active site. As might be expected, the AIL-RING interactions differed between the active and inactive conformation.
To assess interactions between the AIL and the folded domains in the monomer simulations scaling map analysis was performed. In this analysis, a simulation of the AIL as a true self-avoid random coil is performed to generate a reference state, and then the mean inter-residue distances obtained in the full simulations are normalized by the distances obtained from this reference. The self-avoiding random coil simulations are performed using an identical protocol to the full simulations, with the notable exception that all attractive interactions between atoms or solvation effects, instead using only the repulsive part of the Lennard-Jones potential. This ensures we generate a sequence and structure-specific self-avoiding random coil ensemble that provides a true reference state. Extensive details on the technical aspects associated with the generation of this reference state have been described previously50. The scaling maps allow us to easily identify local regions that engage in interactions that cause deviations from self-avoiding random coil behavior.
HAT assays
The standard autoacetylation HAT assay was done using 14C-acetyl-CoA (Perkin-Elmer). Autoacetylation of p300 was quantified by autoradiography after SDS-PAGE gel analysis. The p300s preparations were equilibrated in 1x HAT buffer (25 mM TRIS-HCl, pH 7.5, 100 mM NaCl, 1 mM DTT, 10% glycerol and 1x Complete EDTA-free protease inhibitor (Roche)) for 10 min at 30°C prior to initiation of the reaction by the addition of 200 μM 14C-acetyl-CoA for the indicated time points. For experiments containing IRF3 STAT1 or the eRNA Klf6, autoacetylation assays were performed at a fixed equimolar concentration (2 μM) of p300s and the indicated TF or Klf6. Assays were performed in triplicates with different batches of proteins and on different days. At the indicated time point, 5 μl of the reaction was quenched by addition of 5 μl of 2x SDS gel loading buffer followed by analysis on a 4-20% SDS-PAGE gel. Experiments shown in Fig. 1c were done in 1x kinase buffer (20 mM HEPES, pH 7.5, 250 mM NaCl, 20 mM β-glycerol phosphate, 1 mM sodium vanadate, 10 mM MgCl2, 1 mM DTT, 1 mM ATP and a mix of 20 μM [14C] acetyl-CoA and 80 μM cold acetyl-CoA (A2056, Sigma). 1 μM of p300s was incubated in the presence or absence of 1 μM IRF3, 2 μM TBK1 and 1 μM STING (lanes 1-6) or increasing amounts of STING as indicated (lanes 7-12). The gels analyzed by western blotting as indicated below or were fixed for 30 min in a solution containing 3% glycerol, 10% glacial acetic acid, 20% ethanol (v/v/v) in water. The gels soaked for 5 min in a solution containing 1% glycerol, 5% PEG8000 in water and were dried for 30 min using a Bio-Rad Gel Dryer and the radioactivity quantified on a phosphorimage analyser (Typhoon, GE Healthcare) followed by analysis using imageJ 1.8.0_11251.
A p300 HAT scintillation proximity assay (SPA) was designed similar to that described previously28. Briefly, as a substrate we used a synthetic histone H4 peptide containing 15 amino acids derived from the N-terminus of human H4 that was chemically attached to biotin with an amino hexanoic linker (Biotin-C6- GRGKGGKGLGKGGAK) (from peptid.de). The synthetic peptide was re-suspended in water and adjusted to pH 7.0 with concentrated NaOH.
A typical reaction contained p300s (50 nM), 12.5 μM biotinylated H4 peptide, acetyl-CoA (0.1 μM to 10 μM set at ~10x apparent Km) in 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 μM ZnCl2, 0.01% Tween-20, 0.1% BSA (w/v). For reactions containing pIRF3, 50 nM was added. 20μl of a 2x reaction mixture containing p300s, H4 peptide with and without pIRF3 was preincubated at 30°C for 5 min. The reaction was initiated by the addition of 20 μl of 2x acetyl-CoA containing a 1:3 mix of Tritiated [3H] acetyl-CoA (PerkinElmer; NET290050UC) with cold acetyl-CoA. Eg. for 10 μM final acetyl-CoA concentration, a mix of 5 μM [3H] acetyl-CoA and 15 μM cold acetyl-CoA (A2056, Sigma) was used. The reaction was quenched at the indicated time points by delivering 40 μl of the reaction mix into 120μl of 0.5N HCl in a FlashPlate Plus Streptavidin 96-well scintillant coated microplate (Perkin Elmer, SMP103001PK). The plate was incubated for 1h, and light emission was counted in a MicroBeta2 Scintillation Counter (Perkin Elmer) at 1 minute per well in the top count mode. Counts per minute (cpm) were plotted against acetyl-CoA concentration. Typical progress curves are shown in Extended Data Fig.1d. The initial rate was estimated by linear regression during the first 10 minutes of the reaction and plotted against acetyl-CoA concentration. All data were analyzed using GraphPad Prism 7.0.
For results shown in Fig. 4, acetylation reactions, were performed in acetylation reaction buffer HAT (25 mM TRIS-HCl, pH 7.5, 100 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 10 % glycerol and 1× Complete EDTA-free protease inhibitor (Roche)) with 50 μM Acetyl CoA (Sigma), 100 ng/ml TSA and 2 μg of purified histone octamer. Reactions were incubated 30 min at 30 °C and stopped by addition of 3x SDS gel loading buffer, then used for Coomassie staining and Immunoblotting.
Multi angle laser light scattering-size exclusion chromatography
Prior to SEC-MALLS runs, p300 variants were acetylated and deacetylated using p300 HAT or SIRT2 as described previously26. The reactions were analyzed by liquid chromatography-mass spectrometry (LC-MS) as done previously52. Size-exclusion chromatography was performed at a flow rate of 0.5 ml min-1 on a Superdex 200 Increase 10/300 GL column equilibrated in SEC-MALLS buffer (20 mM HEPES, 300mM NaCl, 5μM ZnCl2, 0.5mM TCEP) at 21 °C. A 50 μl sample of p300 at 2 mg ml-1 was injected onto the column and multi angle laser light scattering was recorded with a laser emitting at 690 nm using a DAWN-EOS detector (Wyatt TechnologyCorp. Santa Barbara, CA). The refractive index was measured using a RI2000 detector (Schambeck SFD). The molecular weight was calculated form differential refractive index measurements across the center of the elution peaks using the Debye model for protein using ASTRA software version 6.0.5.3.
In vitro eRNA transcription
eKlf6 eRNA corresponding to 496 nucleotides of the sense strand of human chr13:5802100-580259634, was produced by in vitro transcription from a pMA plasmid containing a eKlf6 insert synthesized by GeneArt Gene Synthesis (Thermo Fisher). 50 μg of pMA_Klf6 plasmid was linearized with 80 U of KpnI-HF in a final volume of 100 μl and incubated at 37 °C for 14-16h. The in vitro transcription reaction was done in a final volume of 1 ml, using 1x T7 buffer, T7 RNA Polymerase and 1 U of RNaseOUT Recombinant Ribonuclease Inhibitor (Thermo Fisher). After incubation for 2 h at 37°C, 0.5 U of TURBO™ DNase (2 U/μl) (Thermo Fisher) and 1μM CaCl2 was added to the reaction and incubated for 30 min at 37 °C. Following DNaseI treatment, 2 μl of a 30 mg/ml stock of proteinase K powder (Thermo Fisher), dissolved in proteinase K buffer (10 mM TRIS pH 7.5, 1 mM CaCl2, 40% Glycerol), was added and incubated for 45 min at 37 °C. Buffer was exchanged into 20 mM HEPES, pH 7.5, 300 mM NaCl, 0.5 mM TCEP using Amicon Ultra-0.5ml Centrifugal Filters (Molecular weight cut off = 3kDa; EMD Millipore). To further purify the RNA, 3 volumes of TRIzol (Thermo Fisher) was added to the RNA sample, followed by isopropanol precipitation. Purified Kfl6 RNA was resuspended in 20 mM HEPES, pH 7.5, 300 mM NaCl, 0.5mM TCEP. RNA was quantified using a Nanodrop spectrophotometer (Thermo Fisher). The quality of Klf6 was assessed by agarose gel electrophoresis in 1x TBE buffer or using denaturing 6M Urea 14% PAGE (Extended Data Fig. 7c).
Immunoblotting, Immunofluorescence and antibodies
We have used the following cell lines: COS and H1299. They are not on the list of commonly misidentified cell lines maintained by the International Cell Line Authentication Committee. COS cells were purchased from ATCC product reference: ATCC-CR1-1651; lot N°, 4171903. H1299 cells have been authenticated on December 6th 2016 by « LGC Standards: Cell Line Authentication Service". Method used: Cell lines were authenticated using Short Tandem Repeat (STR) analysis as described in 2012 in ANSI Standard (ASN-0002) Authentication of Human Cell Lines: Standardization of STR Profiling by the ATCC Standards Development Organization (SDO) as described in Int. J. Cancer. 2012 Nov 8. doi: 10.1002/Ijc.27931. Upon the reception of COS cells and after the authentication of H1299 cells, they have been expanded. Mycoplasma contamination test (MycoAlert Mycoplasma Detection Kit: Lonza cat N°: LT07-418) was performed and the Mycoplasma-free cells were frozen and kept in liquid nitrogen. After thawing they are kept in culture for 30 passages with a Mycoplasma contamination test after 15 passages. For immunoblotting, proteins were separated on 4–12% Bis-Tris SDS-PAGE gel (NuPAGE precast gel, Thermo Fisher) and transferred onto a nitrocellulose membrane (Hybond C+, GE Healthcare). Membranes were blocked with 5% skim milk in PBST buffer (PBS, 0.1% Tween-20) and probed with anti–p300 K1499ac rabbit polyclonal antibody (1:2,500 dilution; Cell Signaling, 4771), anti-Kac rabbit polyclonal antibody (1:2,500 dilution; Cell Signaling, 9441), anti-Flag mouse monoclonal antibody (1:2,500 dilution; Sigma, F1804), anti-HA rabbit polyclonal antibody (1:2500 dilution, Abcam, ab9110). For the detection of STAT1 or IRF3 phosphorylation, the membrane was blocked with 5% milk in PBST followed by overnight incubation at 4 °C with anti-phospho-Stat1 (Tyr701) Rabbit monoclonal antibody (1:2,500 dilution; Cell Signaling #9171) in PBST buffer containing 5% bovine serum albumin (BSA). For detection of IRF3 S396 phosphorylation, we used anti-phospho IRF3 S396 Rabbit monoclonal antibody (1:2.500 dilution; Cell Signaling #4947). Incubations were done as above. Membranes were washed extensively in PBST buffer before and after incubation with anti-rabbit or anti mouse HRP-conjugated secondary antibody (1:10,000 dilution; GE Healthcare, NA934 or NA931), and protein bands were visualized on film after the ECL reaction (ECL Prime, GE Healthcare). Immunofluorescence was done as described previously26 or as follows: 24h post-transfection, cells were treated with a DMSO control or with 2.5μM (final concentration) of HAT inhibitor A-485 (Phil Cole, Harvard Medical School) or CBP/p300 Bromodomain inhibitor (CBP30, Sigma # SML1133) dissolved in DMSO. The final DMSO concentration in all assays was 0.25%. After 24h, Cells were rinsed once with RNase-free PBS 1X. Next, cells were permeabilised in freshly made 0.2% Triton (SIGMA) buffer for 5 min and were then fixed in freshly made 4% formalin solution (SIGMA) for 10 min at room temperature. After three washes in RNase-free 1x PBS at room temperature, the cells were incubated in 5% skim milk in 1x PBS for 30 min and then probed with anti-HA high-affinity monoclonal antibody (1:100 dilution; Roche Applied Science, cat N°:11867423001) overnight at 4°C. Cells were washed extensively with 1x PBS before and after incubation with Alexa Fluor 488-conjugated secondary antibody (1:500 dilution; Invitrogen, Cat N°:A-11006) for 1h at 37°C. Cells were counterstained with Hoechst (250ng ml-1) and examined under a confocal laser scanning microscope (LSM510, Zeiss).
Extended Data
Extended Data Figure 1. The impact of IRF3 or STAT1 activation and oligomerisation on p300 autoacetylation.
(a) Domain structure of IRF3. Truncation construct used is shown below. (b) Size-exclusion chromatography of IRF3 variants. The red curve is for unphosphorylated IRF3. The blue chromatogram is for phosphorylated pIRF3. The green curve is C-terminally truncated IRF3ΔC. Representative data of three independent experiments are shown. (c) A constant amount of p300s (2 μM) was incubated alone or in the presence of C-terminally truncated IRF3ΔC (2 μM) for the indicated time points. Samples were analyzed by SDS-PAGE followed by Coomassie staining and autoradiography. (d) Progress curves of HAT scintillation proximity assay. Histone H4 substrate acetylation in the presence (green) or absence (black) of pIRF3 and varying concentrations of [3H] acetyl-CoA. The degree of Histone H4 substrate acetylation at different time points and the initial velocity (cpm/min) at the indicated acetyl-CoA concentrations were determined and plotted in Figure 1E. Three independent experiments were performed and the mean value and error bars representing the standard deviation are shown. (e) Domain structure of STAT1. Truncation constructs used are shown below. The Tyr701 phosphorylation site is indicated. (f) Uncropped images of SDS-PAGE gels shown in Figure 1D. 14C autoacetylation signal of p300s is shown below. (g) Size-exclusion chromatography of STAT1 variants. The black curve is for STAT1ΔNC. The green chromatogram is for STAT1ΔNC. The red curve is for Y701 phosphorylated pSTAT1ΔNC. The blue chromatogram is for Y701 phosphorylated pSTAT1ΔN. (h) SDS-PAGE analysis of STAT1 variants and analysis by western blotting. Coomassie staining of SDS-PAGE gel (top); PonceauS staining (middle) and western blot using anti Phospho-Stat1 (Tyr701). Representative data of three independent experiments are shown. For gel source data, see Supplementary Figure 1.
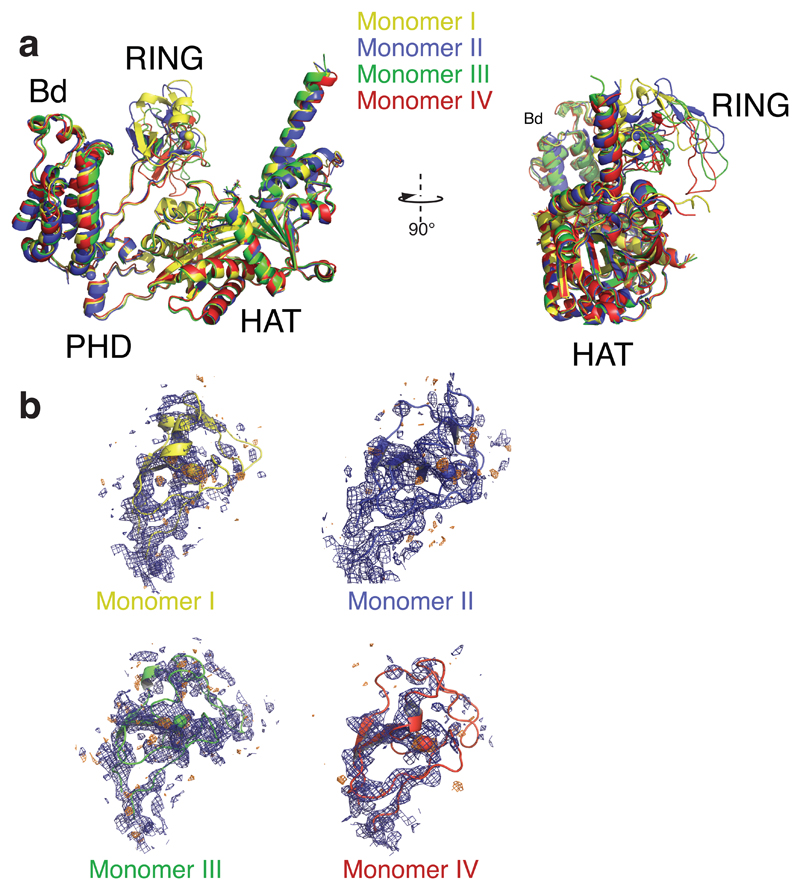
Extended Data Figure 2. Structural analysis of the RING domains.
(a) Superposition of the four p300 molecules (monomer I-IV) in the asymmetric crystallographic unit. While the Bromodomains (Bd), PHD and HAT domains superpose with a root mean square deviation of ~ 0.9 Å, the RING domains take multiple conformations, (b) 2Fo-Fc (blue mesh) as well as anomalous difference Fourier maps (orange mesh) for the four RING domains contoured around 1σ and 2.5σ, respectively.
Extended Data Figure 3. Crystal packing of the p300 core molecule.
(a) There are four p300 molecules (monomer I-IV) in the asymmetric crystallographic unit. The four molecules show an antiparallel arrangement of the BRP-HAT domains. As a result HAT domains from monomer I and II are closely apposed. Monomer III and IV engage monomer IVsym and monomer IIIsym, respectively, of a neighboring crystallographic unit, showing that all promoters are in a AIL-loop swap conformation. Black arrows indicate the direction of the AIL. The disordered segment of the AIL is shown as a black dotted line. (b) Electron density of the AIL. A 2Fo-Fc and (c) a Fo-Fc difference density omit map contoured at 0.8 or 2.0 RMSD (root means square deviation), respectively. Coloring as in Figure 3.
Extended Data Figure 4. Regulation of HAT activity by flanking domains.
(a) Domain structure of p300. Sequence conservation of the AIL is shown using WebLogo53. Constructs used are shown. (b) Analysis of in vitro expression of the indicated p300 variants. Purified proteins were analyzed for autoacetylation by immunoblotting with anti-p300 K1499 acetyl antibody (left panel), anti-FLAG antibody (middle) and Coomassie staining (right). Representative data of three independent experiments are shown. (c) Representative mass spec analysis of BRP_HAT_ZZ_ΔAIL following after in vitro expression (red curve) and after SIRT2 mediated deacetylation (black curve).
Extended Data Figure 5. Regulation of HAT activity by flanking domains.
(a) The AIL contributes to histone substrate acetylation of activated p300. The details of the constructs used are indicated in Extended Data Figure 4. Defined amounts of p300 variants were incubated with acetyl-CoA and the indicated histones prior to SDS-PAGE analysis followed by Coomassie staining and western blotting with the indicated antibodies. (b) The indicated amounts of purified p300s variants were incubated with histone octamers as in panel (a), followed by SDS-PAGE and immunoblot analysis with the indicated antibodies. Anti-Kac: pan-acetyl-lysine antibody. Representative data of three independent experiments are shown. (c) Crystal structure of the H4 K12acK16ac peptide bound to the BΔRP module containing an in frame RING deletion. Amino acid residues 1169-1241 were replace by a single Glycine residue. The deletion removes the RING domain (black arrow) and does not adversely affect structural integrity of the BΔRP module. (d) Indicated variants of p300 were co-expressed with p53 in H1299 cells and analyzed by immunofluorescence with the indicated antibodies or (e) by western blotting. Representative data of three independent experiments are shown. Scale bar, 10 μm.
Extended Data Figure 6. Autoacetylation changes the hydrodynamic properties of p300.
(a) Simulations of the AIL in context of the loop-swapped dimer. Left panel: Cartoon of the trajectory of the AIL (dashed line). Right panel: Representative conformations with the AIL Cα backbone atoms are coloured according to charge. (b) SEC-MALLS analysis of deacetylated (blue) and acetylated (yellow) p300 core. Note the decrease in elution volume upon acetylation. (c) SEC-MALLS analysis of deacetylated (blue), acetylated (red) BRP_HAT_CH3 and deacetylated (black) and acetylated BRP_HAT_CH3 ΔAIL (green). There is no increase in elution volume upon acetylation of theΔAIL construct. (d) Comparison of acetylated and deacetylated BRP_HAT and BRP_HAT_CH3. The deacetylated BRP_HAT (green) and deacetylated BRP_HAT_CH3 (blue) elute at the same position indicating a similar hydrodynamic radius. The acetylated BRP_HAT (yellow) and BRP_HAT_CH3 (red) elute at a larger elution volume. The normalized refractive index is plotted as a function of elution volume from an S200 column coupled to a MALLS detector. Calculated molecular masses are plotted as a function of volume for each eluted peak. The experiment was carried out at least three times with similar results. One representative example of each sample is shown. (e) Mass spectrometry analysis using electrospray ionization (ESI) of the BRP_HAT before (blue), and after (yellow) autoacetylation. The molecular mass and the number of acetylation events are indicated. (f) BRP_HAT_CH3 before (blue) and after (red) autoacetylation. (g) BRP_HAT_CH3_ΔAIL before (black) and after (green) autoacetylation.
Extended Data Figure 7. Molecular model and controls showing that p300 acetyltransferase activity is not stimulated by eRNA.
(a) p300 is maintained in the inactive state by deacetylases such as SIRT2. IRF3 is autoinhibited by a C-terminal segment in the IAD domain. (b) TBK1 phosphorylation activates and dimerises IRF3. The activated IRF3 dimer engages the IBID domain of p300. (c) Recruitment of two molecules of p300 results in trans-autoacetylation in the AIL loop and HAT activation. (d) Activated p300 can acetylate chromatin and engage acetylated substrates via the Bd. (e) A constant amount of p300s (2 μM) was incubated in [14C] acetyl-CoA alone or in the presence of 2 μM Klf6 eRNA for the indicated time points. Samples were analyzed by SDS-PAGE followed by Coomassie staining (top) and autoradiography (bottom). (f) As in (e) but in the presence of 0.5 mM EDTA. The experiment was carried out at least two times with consistency. One representative example is shown. (g) Quality control of Klf6 RNA. 3 μg Klf6 was deposited on a 1% Agarose gel or a 14% 6M Urea PAGE gel and detected by SYBR Safe stain. M: 100bp DNA ladder (NEB).
Extended Data Table 1. Data collection, phasing and refinement statistics.
* Data from 1 crystal. *Values in parentheses are for highest-resolution shell.
| BRP-HAT | BP∆R | |
|---|---|---|
| Data collection | ||
| Space group | P21 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 100.7, 146.6, 116.3 | 49.6, 83.7, 165.6 |
| α, β, γ (°) | 90, 91.7, 90 | 90, 90, 90 |
| Resolution (Å) | 50–3.10* | 42.7–2.50* |
| No. reflections | 106462 | 23996 |
| Rsym or Rmerge | 8.7 (89.7)* | 6.3 (141.3)* |
| l l σl | 7.48 (0.7)* | 7.6 (1.0)* |
| Completeness (%) | 99.0 (94.0)* | 97.29 (92.2)* |
| Redundancy | 1.9 (1.9)* | 9.1 (3.6)* |
| Refinement | ||
| Resolution (Å) | 50-3.1 | 42.7-2.5 |
| Rwork / Rfree | 0.19 / 0.26 | 0.24 / 0.27 |
| No. atoms | 19370 | 2769 |
| Protein | 19100 | 2693 |
| Lys-CoA ligand | 256 | – |
| Zinc | 14 | 4 |
| H4 K12AcK16Ac | – | 76 |
| B-factors (mean; Å2) | ||
| Proteins | 95.0 | 64.0 |
| RING domains | 164.8 | |
| Lys-CoA ligand | 69.2 | – |
| H4 K12AcK16Ac | – | 60.2 |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.013 | 0.004 |
| Bond angles (°) | 1.73 | 0.68 |
Extended Data Table 2. Thermodynamic analysis of the interaction between p300 BRP and histone peptides by ITC.
Mean and s.d. were determined from experiments performed in triplicate. Horizontal lines separate experiments involving different histone peptides or different protein constructs.
Histone peptide sequences: H3 (1-20) ARTKQTQRKSTGGKAPRKQL, H3 (11-30) TGGKAPRKQLATKASRKSAP, H4 (4-24) GKGGKGLGKGGAKRHRKVLRD.
AIL (Autoinhibitory loop peptide) SKNAKKKNNKKTSKNKSS (1545-1562). No binding was detected to non-acetylated peptides, or with a construct containing a mutation that abolishes acetyllysine binding (N1132A).
*binding stoichiometry
| Protein | Peptide (residues) | Kd (μM) | N* |
|---|---|---|---|
| BRP | H3 unmodified (1-20) | -- | No binding |
| H3 K4me1 (1-20) | -- | No binding | |
| H3 K4me3 (1-20) | -- | No binding | |
| H3 K9ac (1-20) | -- | No binding | |
| H3 K14ac (1-20) | 1761 ± 356 | 1.13 | |
| H3 K18ac (1-20) | large | 1.01 | |
| H3 K9acK14ac (1-20) | 578 ± 47 | 1.06 | |
| H3 K14acK18ac (1-20) | 104 ± 27 | 1.03 | |
| H3 K23ac (11-30) | -- | No binding | |
| H3 K18acK23ac (11-30) | -- | No binding | |
| H3 S10pho (1-20) | -- | No binding | |
| H3T11pho (1-20) | -- | No binding | |
| BRP | H4 unmodified (4-24) | -- | No binding |
| H4 K5ac (4-24) | -- | No binding | |
| H4 K8ac (4-24) | 58 ± 12 | 1.04 | |
| H4 K5acK8ac (4-24) | 828 ± 10 | 1.03 | |
| H4 K8acK12ac (4-24) | 90 ± 18 | 1.01 | |
| H4 K12ac (4-24) | 71 ± 14 | 1.05 | |
| H4 K16ac (4-24) | -- | No binding | |
| H4 K12K16diac (4-24) | 25 ± 5 | 0.99 | |
| H4 K20ac (1 -24) | 305 ± 6 | 1.00 | |
| H4 K16acK20ac (1-24) | 205 ± 10 | 1.23 | |
| H4 K5K8K12K16K20penta-ac (1-24) | 38 ± 4 | 1.07 | |
| H4 S1pho K5K8K12K16K20penta-ac (1-24) | 54 ± 2 | 1.12 | |
| BRP_N1131A | H4 K20ac (1-24) | -- | No binding |
| H4 K16acK20ac (1-24) | -- | No binding | |
| H4 K5K8K12K16K20penta-ac (1-24) | -- | No binding | |
| H4 S1pho K5K8K12K16K20penta-ac (1-24) | -- | No binding | |
| BRP | AIL (1545-1562) | -- | No binding |
| BRP | AIL K1549K1558K1560tri-ac (1545-1562) | -- | No binding |
Extended Data Table 3. Summary of SEC-MALLS and mass spectrometry experiments.
Column labelling: Molar masses determined by Mass spectrometry (MMMS), MMth the theoretical molar mass calculated from the appropriate primary sequences. Acetylation levels were estimated based on the mass differences as compared to the non-acetylated sample. MMSLS (Molar masses determined by SEC-MALLS) at a concentration of 2 mg·ml-1. All p300 constructs contained the mutation Y1467F. The experiment was carried out at least three times with consistency. Results from one representative example are shown. The mass and errors reported for SEC-MALLS are the weight average molar mass and residual standard deviations of the observed data from the fitted values calculated using ASTRA.
| Sample |
MMMS Da |
MMth Da |
Acetylation level |
MMSLS Da (2 mg·ml-1) |
|---|---|---|---|---|
| BRP_HAT | 73538 | 73538 | ~0 | 73380 ± 1.5% |
| BRP_HAT Acetyl | 73918 | 73538 | >8 | 71690 ± 1.6% |
| BRP_HAT_CH3 | 92893 | 92891 | ~0 | 92810 ± 2.0% |
| BRP_HAT_CH3 Acetyl | 93270 | 92891 | >9 | 90700 ± 2.0% |
| BRP_HAT_CH3 ΔAIL | 86363 | 86362 | ~0 | 84650 ± 2.2% |
| BRP_HAT_CH3 ΔAIL Acetyl | 86446 | 86362 | >2 | 80450 ± 1.7% |
Supplementary Material
Supplementary Information is available in the online version of the paper.
Acknowledgements
This work was supported by the grant 16-0280 from Worldwide Cancer Research. EO was supported by an EMBL Interdisciplinary Postdoctoral (EIPOD) fellowship. SR was supported by the Fondation ARC pour la recherche sur le Cancer and by the Fondation FINOVI. ASH is a postdoctoral fellow in the laboratory of Dr. Rohit V. Pappu (RVP) at Washington University in St. Louis. The computational work was supported by the Human Frontiers Science Program (grant RGP0034/2017 to RVP) and the St. Jude Collaborative Research Consortium on Membraneless Organelles (to RVP). We thank the staff at the ESRF beamlines BM29. We thank Luca Signor (Institut de Biologie Structurale (Grenoble)) for mass spectroscopy analysis. We thank Russel Vance (University of California, Berkeley, CA) for the plasmid encoding GST-STING and Philip A. Cole (Harvard Medical School) for the A-485 inhibitor. SK and DP were supported by ANR Episperm3 program. Additional support to SK was from: Fondation ARC ‘Canc’air’ project (RAC16042CLA), Plan Cancer (CH7-INS15B66) and Plan Cancer (ASC16012CSA) and the ‘Université Grenoble Alpes’ ANR-15-IDEX-02 LIFE and IDEX SYMER.
Footnotes
Data availability. Coordinates for the p300 core structure and BPΔR bound to a diacetylated histone H4 peptide are available from the Protein Data Bank under accession number 6GYR and 6GYT, respectively. Source data are available for Fig. 1b, f and Extended data Fig. 1d. Fig. 1d shows the initial velocities from reactions shown in Extended data Fig. 1d.
Author Contributions E.O. designed and performed most experiments, analysed and validated the data and revised the draft with assistance from S.R., Z.I., N.H and J.G. A.S.H. performed computational modelling and revised the draft. S. K. provided supervision, funding acquisition and commented on the draft. D.P. was involved in conceptualization, supervision, project administration, funding acquisition and wrote original and revised draft.
Author Information Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nature immunology. 2016;17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 2.Panne D, McWhirter SM, Maniatis T, Harrison SC. Interferon regulatory factor 3 is regulated by a dual phosphorylation-dependent switch. J Biol Chem. 2007;282:22816–22822. doi: 10.1074/jbc.M703019200. M703019200 [pii] [DOI] [PubMed] [Google Scholar]
- 3.Zhao B, et al. Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins. Proc Natl Acad Sci U S A. 2016;113:E3403–3412. doi: 10.1073/pnas.1603269113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parekh BS, Maniatis T. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-beta promoter. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- 5.Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. S0092-8674(07)00656-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stark GR, Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang JJ, et al. Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc Natl Acad Sci U S A. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bedford DC, Brindle PK. Is histone acetylation the most important physiological function for CBP and p300? Aging (Albany NY) 2012;4:247–255. doi: 10.18632/aging.100453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. ng1966 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. nature07730 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin Q, et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bedford DC, Kasper LH, Fukuyama T, Brindle PK. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics : official journal of the DNA Methylation Society. 2010;5:9–15. doi: 10.4161/epi.5.1.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao L, et al. Integrated genome-wide chromatin occupancy and expression analyses identify key myeloid pro-differentiation transcription factors repressed by Myb. Nucleic Acids Res. 2011;39:4664–4679. doi: 10.1093/nar/gkr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waltzer L, Bienz M. Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature. 1998;395:521–525. doi: 10.1038/26785. [DOI] [PubMed] [Google Scholar]
- 15.Holmqvist PH, Mannervik M. Genomic occupancy of the transcriptional co-activators p300 and CBP. Transcription. 2013;4:18–23. doi: 10.4161/trns.22601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasper LH, Qu C, Obenauer JC, McGoldrick DJ, Brindle PK. Genome-wide and single-cell analyses reveal a context dependent relationship between CBP recruitment and gene expression. Nucleic Acids Res. 2014;42:11363–11382. doi: 10.1093/nar/gku827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2010 doi: 10.1038/nature09692. nature09692 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson PR, et al. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol. 2004;11:308–315. doi: 10.1038/nsmb740. nsmb740 [pii] [DOI] [PubMed] [Google Scholar]
- 19.Qin BY, et al. Crystal structure of IRF-3 in complex with CBP. Structure. 2005;13:1269–1277. doi: 10.1016/j.str.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 20.Larabi A, et al. Crystal structure and mechanism of activation of TANK-binding kinase 1. Cell reports. 2013;3:734–746. doi: 10.1016/j.celrep.2013.01.034. [DOI] [PubMed] [Google Scholar]
- 21.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 22.Shuai K, Stark GR, Kerr IM, Darnell JE., Jr A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science. 1993;261:1744–1746. doi: 10.1126/science.7690989. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, et al. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. doi:S0092-8674(00)81443-9 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Wojciak JM, Martinez-Yamout MA, Dyson HJ, Wright PE. Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains. EMBO J. 2009;28:948–958. doi: 10.1038/emboj.2009.30. emboj200930 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 26.Delvecchio M, Gaucher J, Aguilar-Guerreri C, Ortega E, Panne D. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat Struct Mol Biol. 2013;21 doi: 10.1038/nsmb.2642. [DOI] [PubMed] [Google Scholar]
- 27.Park S, et al. Role of the CBP catalytic core in intramolecular SUMOylation and control of histone H3 acetylation. Proc Natl Acad Sci U S A. 2017;114:E5335–E5342. doi: 10.1073/pnas.1703105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lasko LM, et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature. 2017;550:128–132. doi: 10.1038/nature24028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karanam B, Jiang L, Wang L, Kelleher NL, Cole PA. Kinetic and mass spectrometric analysis of p300 histone acetyltransferase domain autoacetylation. J Biol Chem. 2006;281:40292–40301. doi: 10.1074/jbc.M608813200. [DOI] [PubMed] [Google Scholar]
- 30.Karanam B, et al. Multiple roles for acetylation in the interaction of p300 HAT with ATF-2. Biochemistry (Mosc) 2007;46:8207–8216. doi: 10.1021/bi7000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vitalis A, Pappu RV. ABSINTH: a new continuum solvation model for simulations of polypeptides in aqueous solutions. Journal of computational chemistry. 2009;30:673–699. doi: 10.1002/jcc.21005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, et al. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. 2008;451:846–850. doi: 10.1038/nature06546. nature06546 [pii] [DOI] [PubMed] [Google Scholar]
- 33.Soutoglou E, et al. Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity. EMBO J. 2001;20:1984–1992. doi: 10.1093/emboj/20.8.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bose DA, et al. RNA Binding to CBP Stimulates Histone Acetylation and Transcription. Cell. 2017;168:135–149 e122. doi: 10.1016/j.cell.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matt T, Martinez-Yamout MA, Dyson HJ, Wright PE. The CBP/p300 TAZ1 domain in its native state is not a binding partner of MDM2. Biochem J. 2004;381:685–691. doi: 10.1042/BJ20040564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nature reviews Genetics. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 37.Ptashne M. Epigenetics: core misconcept. Proc Natl Acad Sci U S A. 2013;110:7101–7103. doi: 10.1073/pnas.1305399110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rando OJ. Combinatorial complexity in chromatin structure and function: revisiting the histone code. Curr Opin Genet Dev. 2012;22:148–155. doi: 10.1016/j.gde.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen UT, et al. Accelerated chromatin biochemistry using DNA-barcoded nucleosome libraries. Nature methods. 2014;11:834–840. doi: 10.1038/nmeth.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rack JG, et al. The PHD finger of p300 Influences Its Ability to Acetylate Histone and Non-Histone Targets. J Mol Biol. 2014 doi: 10.1016/j.jmb.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 42.Kimbrel EA, et al. Systematic in vivo structure-function analysis of p300 in hematopoiesis. Blood. 2009;114:4804–4812. doi: 10.1182/blood-2009-04-217794. [DOI] [PubMed] [Google Scholar]
- 43.Picaud S, et al. Generation of a Selective Small Molecule Inhibitor of the CBP/p300 Bromodomain for Leukemia Therapy. Cancer Res. 2015;75:5106–5119. doi: 10.1158/0008-5472.CAN-15-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A Phase Separation Model for Transcriptional Control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coleman RT, Struhl G. Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene. Science. 2017;356 doi: 10.1126/science.aai8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laprell F, Finkl K, Muller J. Propagation of Polycomb-repressed chromatin requires sequence-specific recruitment to DNA. Science. 2017;356:85–88. doi: 10.1126/science.aai8266. [DOI] [PubMed] [Google Scholar]
- 47.Wang X, Moazed D. DNA sequence-dependent epigenetic inheritance of gene silencing and histone H3K9 methylation. Science. 2017;356:88–91. doi: 10.1126/science.aaj2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. doi:S0076-6879(99)04003-3 [pii] [DOI] [PubMed] [Google Scholar]
- 49.McGibbon RT, et al. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys J. 2015;109:1528–1532. doi: 10.1016/j.bpj.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holehouse AS, Garai K, Lyle N, Vitalis A, Pappu RV. Quantitative assessments of the distinct contributions of polypeptide backbone amides versus side chain groups to chain expansion via chemical denaturation. J Am Chem Soc. 2015;137:2984–2995. doi: 10.1021/ja512062h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaczmarska Z, et al. Structure of p300 in complex with acyl-CoA variants. Nature chemical biology. 2017;13:21–29. doi: 10.1038/nchembio.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.