Abstract
Market transparency is in strong demand by consumers, and the authentication of species is an important step for seafood traceability. In this study, a simple molecular strategy, COIBar–RFLP (cytochrome oxidase I barcode–restriction fragment length polymorphism), is proposed to unveil commercial fraud based on the practice of species substitution in the swordfish trade. In particular, COI barcoding allowed the identification of the species Prionace glauca, Mustelus mustelus, and Oxynotus centrina in slices labeled as Xiphias gladius. Furthermore, the enzymatic digestion of COI amplicons using the MboI restriction endonuclease allowed the simultaneous discrimination of the four species. Interestingly, an intraspecific differential MboI pattern was obtained for the swordfish samples. This pattern was useful to differentiate the two different clades revealed in this species by phylogenetic analyses using several molecular markers. These results indicate the need to strengthen regulations and define molecular tools for combating the occurrence of fraud along the seafood supply chain and show that COIBar–RFLP could become a standardized molecular tool to assess seafood authenticity.
Keywords: COIBar–RFLP (cytochrome oxidase I barcode–restriction fragment length polymorphism), seafood, fraud, DNA barcoding, food authenticity
1. Introduction
Swordfish fishery is one of the most important fishing activities in the Mediterranean Sea, in particular in South Italy. Quotas have been established to combat overfishing, and fisheries have been closed over several months to protect juveniles. According to recent data from the International Commission for the Conservation of Atlantic Tuna, Italy ranks the highest in terms of swordfish catches, which amount to 45% of the total allowable in the period 2003–2016 [1]. The highest demand for fish products in general, and swordfish in particular, occurs during summer, especially in restaurants [2] but also in local markets. As a result of the high demand, the price of these large pelagic fishes is on average higher than that of small fishes [3].
With the increase in demand and price, alimentary fraud potentially increases too. This can include food mislabeling, substitution, counterfeiting, misbranding, dilution, and adulteration [4]. The mislabeling of seafood can be harmful for health, in terms of economic loss, as well as for the loss of biodiversity it may cause in the case of illegal trade of threatened species. For these reasons, European regulations have focused on traceability and, in particular, on the mandatory declaration of the species present in a product on the product label [5,6,7,8,9,10].
Despite these adequate legislative tools, the number of cases of food fraud perpetrated in the fish trade in Europe and worldwide is increasing. The results of recent investigations on this phenomenon have shown that the percentage of mislabeling was around 30% of the total samples collected [11,12,13,14,15]. To address this problem, researchers have increasingly asserted the importance of using molecular tools based on DNA sequencing for detecting food fraud. The most common mitochondrial (mt) genes used for this purpose have been cytochrome b, 16S rRNA, and cytochrome oxidase I (COI). Other mtDNA targets, such as the mtDNA control region (CR) [16,17], which has been the most popular molecular marker used for genetic population structure studies [18,19,20,21,22,23,24], have seen limited use in fish and seafood species identification. In recent years, COI has been standardized as a barcode gene for species identification in several animal taxa [25,26,27,28,29,30,31,32,33] including fishes [34,35,36,37,38,39,40]. More specifically, the high number of COI-barcode fish sequences available in the large public gene sequence databases (BOLD and GenBank) [41,42] have made this gene the most highly used gene to clearly identify fish species and cases of mislabeling of seafood products [38,43,44,45,46,47,48,49,50,51,52,53,54]. However, in the context of seafood traceability, the main goal for the implementation of these analyses is to reduce the time it takes from sampling to obtaining gene sequencing results, as well as the costs of processing.
An already well-proven technique for the identification of species is polymerase chain reaction (PCR)–restriction fragment length polymorphism (RFLP), by which the PCR product of an amplified gene is cut with different restriction endonucleases to obtain a species-specific RFLP [55,56,57], useful for species authentication. In this regard, the combination of DNA barcoding of COI and the consolidated method of RFLP analysis (COIBar–RFLP, cytochrome oxidase I barcode–restriction fragment length polymorphism) has been successfully used to discriminate several fish species belonging to the Engraulidae, Merluccidae, Soleidae, and Acipenseridae families in processed seafood products [49,52,53,54,58]. It should be noted that the time and cost of execution of the COIBar–RFLP are lower than those of DNA sequencing (about 7 h and 10 euros per sample vs 24 h and 17 euros per sample, respectively).
Focusing on swordfish adulteration problems, the most commonly used species for fraudulent substitution are elasmobranches, including some species of shark. It should be noted that the market of shark meat is very wide for both fresh and frozen foods also in Italy, and cases of mislabeling have been frequently recorded for these products imported from all over the world [59,60,61].
Therefore, food fraud occurs due to an economic return when using shark meat. However, the substitution of a more valuable fish, such as swordfish, with shark meat leads to an even more serious fraud in economic terms. In the last decades, several studies have been carried out to detect the rate of mislabeling of different seafood products, and in some cases, shortfin mako (Isurus oxyrinchus) and blue shark (Prionace glauca) have been found to be sold as swordfish [36,37,62,63].
On the basis of the considerations above, the aim of this work is to extend the use of COIBar–RFLP to investigate the identity of swordfish products in the south of Italy and to discriminate swordfish (Xiphias gladius) from other fish species to detect fraudulent actions, such as species substitution, which represent the most common fraud in seafood. First, we sequenced the conventional COI barcode in a large number of samples collected in local fish markets and supermarkets, labeled as swordfish slices. Subsequently, the COIBar–RFLP procedure was applied on reference samples of the COI-barcoded species to obtain a species-specific restriction enzyme pattern. Finally, this pattern was used for swordfish slice authentication.
2. Materials and Methods
2.1. Sampling
Fresh and frozen slices of swordfish were acquired in 2010 and 2018 from local fish markets and supermarkets of south Italy for a total of 35 samples. Another 10 samples from the local harbor were collected and identified on the basis of morphological traits [64,65] and used to construct a reference COI-barcode library. The samples collected in 2010 had already been processed [37] and were used in this study only for the application of COIBar–RFLP. The remaining samples, preserved at room temperature in 1.5 mL labeled tubes filled with 95% ethanol, were processed for DNA barcoding and COIBar–RFLP (Table 1). DNA samples were deposited as vouchers at the Department of Biological, Geological, and Environmental Science, Section of Animal Biology, in Catania, Italy.
Table 1.
Samples examined in this study.
| Code | DNA Sample Voucher | Declared Species | GenBank Access N° | Species Matched by BLAST | Matched Accession From BLAST | % Identity With 100% Coverage |
|---|---|---|---|---|---|---|
| XIP1 | DBGES18-125 | Xiphias gladius * | JN083390 | Xiphias gladius | JN083390 | 100 |
| XIP2 | DBGES18-129 | Xiphias gladius * | MN447670 | Xiphias gladius | JN049558 | 99.71 |
| MUS1 | DBGES18-133 | Mustelus mustelus * | MN447688 | Mustelus mustelus | JN641215 | 99.39 |
| MUS2 | DBGES18-134 | Mustelus mustelus * | MN447689 | Mustelus mustelus | JN641214 | 99.24 |
| OXY1 | DBGES18-111 | Oxynotus centrina * | MN447691 | Oxynotus centrina | JF834320 | 98.96 |
| OXY2 | DBGES18-130 | Oxynotus centrina * | MN447692 | Oxynotus centrina | JF834320 | 98.66 |
| PRI1 | DBGES18-121 | Prionace glauca * | MN447694 | Prionace glauca | KJ146044 | 99.85 |
| PRI2 | DBGES18-138 | Prionace glauca * | MN447695 | Prionace glauca | MH719984 | 99.83 |
| SCY1 | DBGES18-144 | Scyliorhinus canicula * | MN457949 | Scyliorhinus canicula | KJ205311 | 99.70 |
| SCY2 | DBGES18-146 | Scyliorhinus canicula * | MN457950 | Scyliorhinus canicula | KJ205311 | 99.85 |
| X1 | DBGES10-045 | Xiphias gladius | JN083389 | Xiphias gladius | JN083389 | 100 |
| X2 | DBGES10-048 | Xiphias gladius | JN083387 | Xiphias gladius | JN083387 | 100 |
| X3 | DBGES10-049 | Xiphias gladius | JN049559 | Xiphias gladius | JN049559 | 100 |
| X4 | DBGES10-054 | Xiphias gladius | JN083397 | Xiphias gladius | JN083397 | 100 |
| X5 | DBGES10-055 | Xiphias gladius | JN083387 | Xiphias gladius | JN083387 | 100 |
| X6 | DBGES10-058 | Xiphias gladius | JN049559 | Xiphias gladius | JN049559 | 100 |
| X7 | DBGES10-059 | Xiphias gladius | JN049559 | Xiphias gladius | JN049559 | 100 |
| X8 | DBGES10-060 | Xiphias gladius | JN083387 | Xiphias gladius | JN083387 | 100 |
| X9 | DBGES10-068 | Xiphias gladius | JN083393 | Xiphias gladius | JN083393 | 100 |
| X10 | DBGES10-074 | Xiphias gladius | JN049559 | Xiphias gladius | JN049559 | 100 |
| X11 | DBGES10-077 | Xiphias gladius | JN083393 | Xiphias gladius | JN083393 | 100 |
| X12 | DBGES10-080 | Xiphias gladius | JN083387 | Xiphias gladius | JN083387 | 100 |
| X13 | DBGES10-083 | Xiphias gladius | MN447696 | Prionace glauca | MH719984 | 99.83 |
| X14 | DBGES10-089 | Xiphias gladius | JN083386 | Xiphias gladius | JN083386 | 100 |
| X15 | DBGES10-095 | Xiphias gladius | JN083387 | Xiphias gladius | JN083387 | 100 |
| Y1 | DBGES18-132 | Xiphias gladius | MN447671 | Xiphias gladius | JN049558 | 99.71 |
| Y2 | DBGES18-135 | Xiphias gladius | MN447672 | Xiphias gladius | JN083390 | 99.71 |
| Y3 | DBGES18-136 | Xiphias gladius | MN447673 | Xiphias gladius | JN083387 | 99.71 |
| Y4 | DBGES18-137 | Xiphias gladius | MN447674 | Xiphias gladius | JN049558 | 99.71 |
| Y5 | DBGES18-139 | Xiphias gladius | MN447675 | Xiphias gladius | JN083387 | 99.85 |
| Y6 | DBGES18-141 | Xiphias gladius | MN447676 | Xiphias gladius | JN083387 | 99.71 |
| Y7 | DBGES18-143 | Xiphias gladius | MN447690 | Mustelus mustelus | JN641214 | 99.39 |
| Y8 | DBGES18-144 | Xiphias gladius | MN447677 | Xiphias gladius | JN049558 | 99.85 |
| Y9 | DBGES18-151 | Xiphias gladius | MN447678 | Xiphias gladius | JN083387 | 99.71 |
| Y10 | DBGES18-152 | Xiphias gladius | MN447693 | Oxynotus centrina | JF834320 | 98.07 |
| Y11 | DBGES18-153 | Xiphias gladius | MN447679 | Xiphias gladius | JN083387 | 99.12 |
| Y12 | DBGES18-154 | Xiphias gladius | MN447680 | Xiphias gladius | JN049558 | 99.85 |
| Y13 | DBGES18-161 | Xiphias gladius | MN447681 | Xiphias gladius | JN083390 | 99.41 |
| Y14 | DBGES18-162 | Xiphias gladius | MN447682 | Xiphias gladius | JN083387 | 98.97 |
| Y15 | DBGES18-172 | Xiphias gladius | MN447697 | Prionace glauca | MH194484 | 99.39 |
| Y16 | DBGES18-173 | Xiphias gladius | MN447683 | Xiphias gladius | JN049558 | 99.27 |
| Y17 | DBGES18-174 | Xiphias gladius | MN447684 | Xiphias gladius | JN083387 | 99.85 |
| Y18 | DBGES18-175 | Xiphias gladius | MN447685 | Xiphias gladius | JN083390 | 99.27 |
| Y19 | DBGES18-182 | Xiphias gladius | MN447686 | Xiphias gladius | JN049558 | 99.71 |
| Y20 | DBGES18-187 | Xiphias gladius | MN447687 | Xiphias gladius | JN083387 | 99.56 |
DBGES (Department of Biological, Geological and Environmental Sciences). BLAST (Basic Local Alignment Search Tool). * = species of the samples identified by using morphological features.
2.2. DNA Barcoding
Genomic DNA was extracted from 25 mg of tissue using a commercial kit based on silica purification (DNeasy tissue kit, Qiagen, Hilden, Germany) following the manufacturer’s guidelines. All samples were analyzed by amplifying a portion of about 650 bases of the COI gene in a 20 μL reaction mixture also containing the M13 tailed primers (VF2_t1 and FishR2_t1) described in Ivanova et al. [66] to improve the sequencing quality of the PCR products and following the PCR conditions reported by Pappalardo et al. [54]. All PCR products were checked by 0.8% agarose gel electrophoresis, visualized with SYBR® Safe (Thermo Fisher, Waltham, MA USA), displayed through a Safe Imager TM 2.0 Blue Light Transilluminator (Thermo Fisher, Waltham, MA USA), and then purified with the QIAquick PCR purification kit (Qiagen, Hilden, Germany). Sanger sequencing, using M13 primers, was subsequently conducted by Genechron in both forward and reverse directions to generate the DNA barcodes [67].
The sequence chromatograms were checked visually and assembled. Multiple-sequence alignment was carried out by the online version of MAFFT v.7 [68]. Ambiguous sequences were trimmed, and primer sequences were cut. The sequences were carefully checked for the presence of nuclear mitochondrial pseudogenes or NUMTs (nuclear mitochondrial DNA sequences), which could be easily coamplified with orthologous mtDNA sequences [69]. The EMBOSS Transeq tool [70] was used to translate the nucleotide sequences to amino acids to check for premature stop codons and to verify that the open reading frames were maintained in the protein-coding locus. To confirm the identity of the amplified sequences, we conducted BLAST (Basic Local Alignment Search) searches in GenBank with default parameters [71]. All sequences obtained from the present study were published in the National Center for Biotechnology Information database (NCBI), and their GenBank accession numbers are reported in Table 1. After the BLAST search, six shark species sequences (HM909857, JF493927, KF899461, KI709900, JF493694, JN641217) downloaded from GenBank were added to our dataset to construct a phylogenetic tree. We used jModelTest v 2.1.10 [72] to select the best-fitting substitution model for our sequences according to the corrected Akaike information criterion. A maximum likelihood (ML) tree by using a GTR + I + G model was implemented in MEGA v 6.0 (Biodesign Institute, Arizona, MA, USA) [73]. The evaluation of the statistical confidence of nodes was based on 1000 non-parametric bootstrap replicates [74].
2.3. COIBar–RFLP
The selection of the most suitable restriction enzymes to discriminate swordfish from other shark species (Mustelus mustelus, L., 1758, Oxinotus centrina (L., 1758), P. glauca (L., 1758), Scyliorhinus canicula L., 1758) was performed through “Remap” [75]. The in silico analysis was preliminarily carried out using a total of 10 COI barcode sequences (of about 650 bases) of the examined species, downloaded from public databases (GenBank and BOLD) [41,42]. Five different restriction enzymes were tested to scan all validated sequences and to detect the expected size of the digested products: HpaII (C*CGG), HinfI (G*ANTC), MboI (*GATC), RsaI (GT*AC), and HindIII (A*AGCTT). Finally, a total of 49 COI sequences were analyzed by Remap to test for evidence of intraspecific variation at the recognition site of the restriction endonuclease suitable for simultaneous discrimination of the examined species (Figure 1, Table 2).
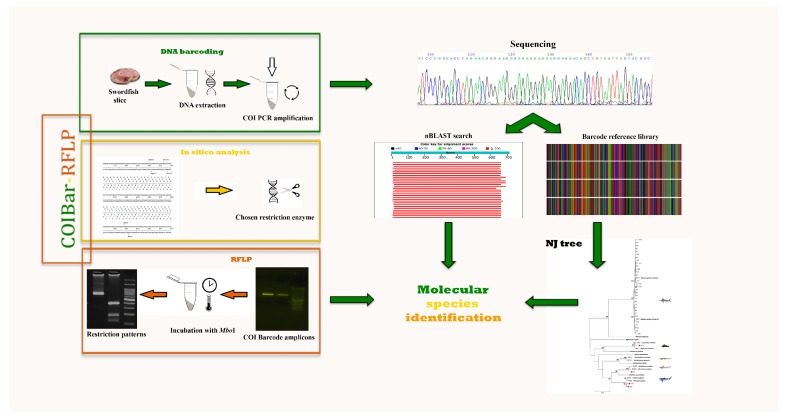
Figure 1.
Flow chart of COIBar–RFLP (cytochrome oxidase I barcode–restriction fragment length polymorphism) or species discrimination. DNA barcoding steps: DNA isolation from swordfish slices and barcode region PCR amplification. In silico analysis steps: search for an appropriate restriction enzyme. RFLP steps: incubation of barcode amplicons with MboI to obtain the COIBar–RFLP pattern. ML, maximum likelihood; nBLAST, nucleotide Basic Local Alignment Search Tool.
Table 2.
In silico analysis of swordfish and shark COI sequences scanned by Remap and using MboI as restriction endonuclease.
| Species | Sequence Number | Genbank Accession Number | Sequence Size (bases) | Restriction Fragment Size (base pair) |
|---|---|---|---|---|
| Xiphias gladius | 10 | JN083387 | 682 | ≈ 145 - 270 - 220 |
| JN083389 | 682 | ≈ 145 - 270 - 220 | ||
| JN049558 | 682 | ≈ 145 - 270 - 220 | ||
| JF952886 | 652 | ≈ 145 - 265 - 220 | ||
| HQ024928 | 652 | ≈ 145 - 265 - 220 | ||
| HQ024927 | 652 | ≈ 145 - 265 - 220 | ||
| KR086931 | 652 | ≈ 145 - 265 - 220 | ||
| GU324195 | 652 | ≈ 145 - 265 - 220 | ||
| DQ107625 | 655 | ≈ 145 - 265 - 200 | ||
| DQ107623 | 655 | ≈ 145 - 265 - 220 | ||
| Mustelus mustelus | 10 | JN641215 | 676 | ≈ 80 - 390 - 171 |
| JN641214 | 679 | ≈ 80 - 390 - 169 | ||
| JN641213 | 672 | ≈ 80 - 390 - 167 | ||
| JN641212 | 666 | ≈ 80 - 390 - 156 | ||
| JN641211 | 681 | ≈ 80 - 390 - 170 | ||
| KJ768265 | 652 | ≈ 80 - 390 - 142 | ||
| KJ768266 | 652 | ≈ 80 - 390 - 142 | ||
| JN641208 | 656 | ≈ 75 - 390 - 156 | ||
| JN641209 | 669 | ≈ 80 - 390 - 160 | ||
| JN641210 | 664 | ≈ 80 - 390 - 160 | ||
| Oxynotus centrina | 9 | KT307360 | 648 | ≈ 510 - 95 |
| KT307361 | 620 | ≈ 480 - 95 | ||
| KT307362 | 648 | ≈ 510 - 95 | ||
| KT307363 | 648 | ≈ 510 - 95 | ||
| KT307364 | 648 | ≈ 510 - 95 | ||
| JF834320 | 672 | ≈ 505 - 100 | ||
| KY176547 | 642 | ≈ 495 - 105 | ||
| GU805137 | 637 | ≈ 500 - 95 | ||
| GU805138 | 648 | ≈ 510 - 95 | ||
| Prionace glauca | 10 | JN312505 | 652 | ≈ 85 - 405 - 70 |
| JN312504 | 652 | ≈ 85 - 405 - 70 | ||
| JN312503 | 652 | ≈ 85 - 405 - 70 | ||
| KP193446 | 652 | ≈ 85 - 405 - 70 | ||
| KP193455 | 652 | ≈ 85 - 405 - 70 | ||
| KP193350 | 652 | ≈ 85 - 405 - 70 | ||
| KP193339 | 652 | ≈ 85 - 405 - 70 | ||
| KP193159 | 652 | ≈ 85 - 405 - 70 | ||
| KC015834 | 652 | ≈ 85 - 405 - 70 | ||
| KC015833 | 652 | ≈ 85 - 405 - 70 | ||
| Scyliorhinus canicula | 10 | JN641243 | 675 | ≈ 85 - 400 - 100 |
| JN641242 | 676 | ≈ 85 - 400 - 100 | ||
| JN641241 | 652 | ≈ 70 - 400 - 100 | ||
| JN641240 | 680 | ≈ 85 - 400 - 100 | ||
| JN641239 | 675 | ≈ 85 - 400 - 100 | ||
| JN641238 | 671 | ≈ 85 - 400 - 100 | ||
| JN641237 | 680 | ≈ 85 - 400 - 100 | ||
| JN641236 | 674 | ≈ 85 - 400 - 100 | ||
| JN641234 | 672 | ≈ 85 - 400 - 100 | ||
| JN641233 | 680 | ≈ 85 - 400 - 100 |
Afterwards, the COI-barcode PCR products obtained from X. gladius and shark samples were digested with the selected restriction enzymes. For each endonuclease, a 15 μL reaction volume containing 13 μL of unpurified PCR product, 1 μL of digestion buffer (1X), and 1 μL of each endonuclease (10 U each) was prepared. The reaction mixtures were incubated at an optimum temperature of 37 °C for 1 h. The digested amplicons were then separated on a 3% agarose gel using Trackit TM 100 bp DNA ladder (Invitrogen) as a size standard. The restriction pattern obtained from the validated samples was exploited to unequivocally identify the unknown commercial slices.
3. Results
3.1. DNA Barcoding
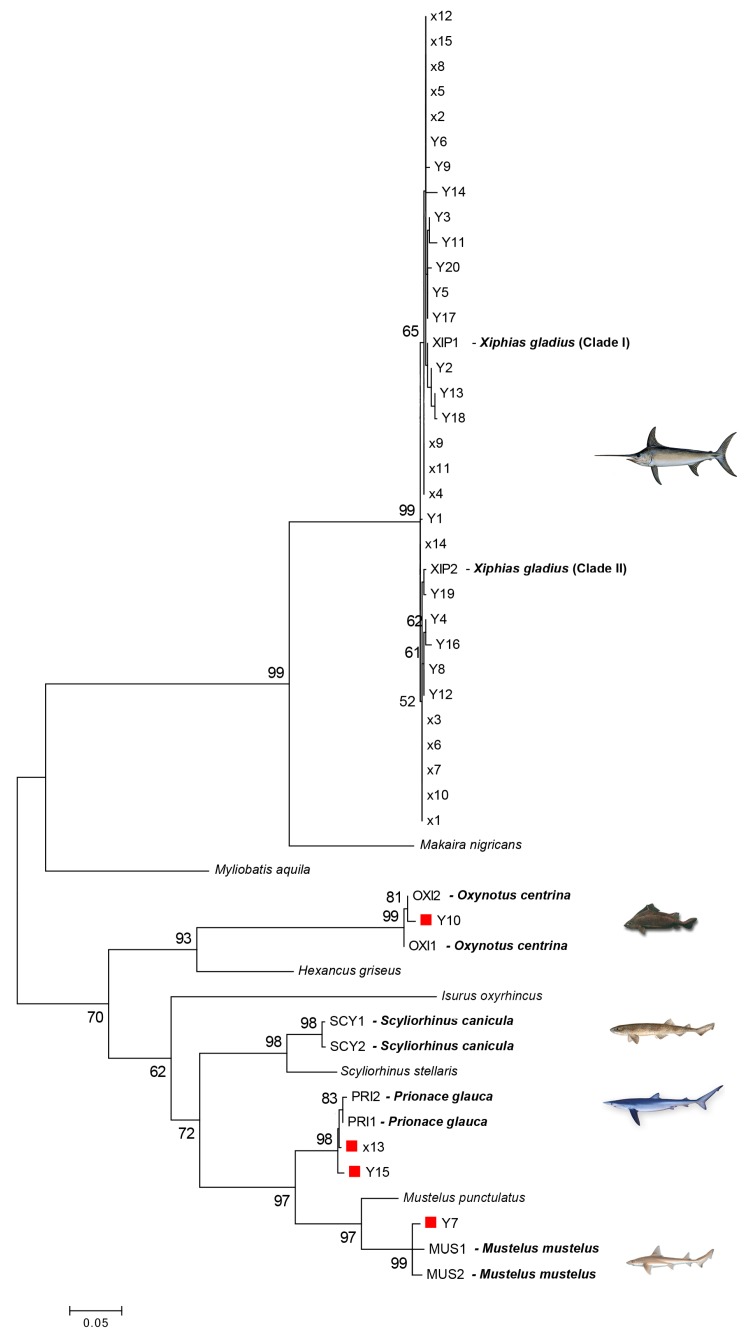
The length range of the obtained COI sequences was between 669 bases and 681 bases. Each of them was a functional mitochondrial sequence without stop codons. NUMTs generally smaller than 600 bases were not sequenced [71]. Five species were identified in all examined samples: X. gladius (Xiphiidae), P. glauca (Charcarinidae), M. mustelus (Triakidae), S. canicula (Scyliorinidae), and O. centrina (Oxynotidae). The sequences obtained from morphologically validated species were compared with the sequences retrieved from GenBank through a BLAST search. The identity percentage between the COI query sequences and their top-match sequences ranged from 98.07% to 100% (Table 1). The ML tree (Figure 2) showed the relationship between the sequences of several unidentified samples and the reference barcode sequences. High bootstrap values (>60%) supported the nodes connecting the sequences of the same species in the tree. The samples of X. gladius clustered into two main clades (named clade I and II), as already found by Pappalardo et al. [36,37]. Only one case of mislabeling (1 out of 15) was found in the samples examined in 2010 (6.7%), while 15% (3 out of 20) of mislabeling was found in the samples collected during 2018 (Table 1). Swordfish was substituted with P. glauca (2 products), M. mustelus (1 product), and O. centrina (1 product).
Figure 2.
Maximum likelihood (ML) tree showing the relationships of unknown samples sequences (X and Y) to validated reference barcode sequences. The numbers above the nodes represent bootstrap analyses after 1000 replicates. Bootstrap values greater than 60% are shown. The red square indicates swordfish mislabeled samples. Scale bar refers to a distance of 0.05 nucleotide substitutions per site.
3.2. COIBar–RFLP
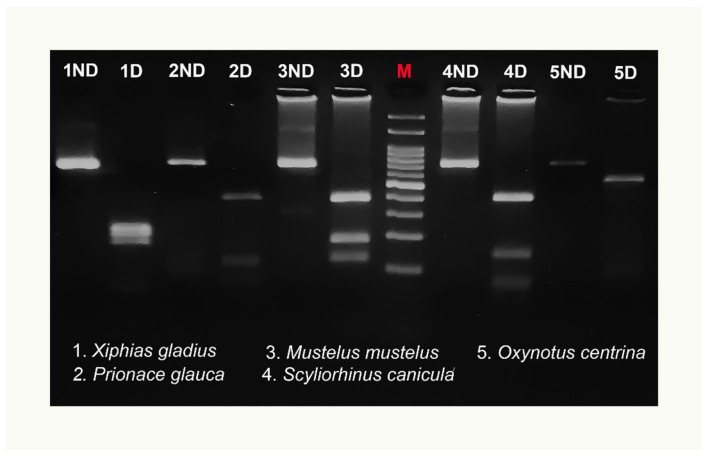
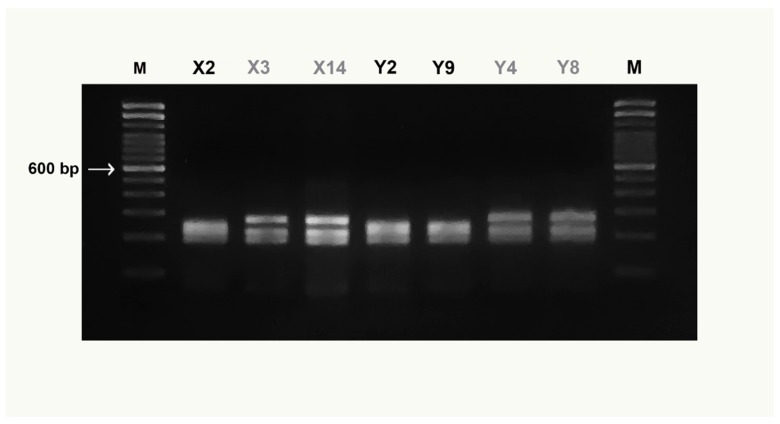
The preliminary in silico analysis using “Remap” showed that the MboI enzyme produced a species-specific pattern useful to discriminate simultaneously all examined species. No intraspecific variation of the MboI recognition sites was detected for any species tested by “Remap”, with the exception of the X. gladius digestion pattern (Table 2). Figure 3 highlights both the size of the undigested COI amplicon, of about 750 bp, and the MboI differential restriction pattern obtained for each species: one fragment of 510 bp was obtained for O. centrina; two fragments of 110 and 400 bp and of 150 and 400 bp were obtained, respectively, for P. glauca and S. canicula; finally, three fragments of 120, 180, and 390 bp were obtained for M. mustelus. The negative control is not shown in the figure. The enzymatic digestion of X. gladius amplicons produced two different patterns (Figure 4) corresponding to clades I and II, already described in this species. In particular, three fragments of 170, 220, and 240 bp were detected for clade I and three fragments of 170, 220, and 280 bp were found for clade II. On the basis of this intraspecific pattern, the swordfish sample shown in Figure 3 belongs to clade I.
Figure 3.
Example of COIBar–RFLP identification of swordfish and shark species on a 3% agarose gel by restriction by MboI of the cytochrome oxidase I amplicons. Bands smaller than 100 bp were not considered. The 5ND and 5D bands differ in intensity because they were obtained from two different PCR amplifications. ND = not digested, D = digested. M = molecular weight marker (100 bp DNA ladder, biotechrabbit GmbH, Berlin, Germany).
Figure 4.
COIBar–RFLP patterns obtained from MboI digestion of X. gladius samples. Clade I (black code), Clade II (grey code). M = molecular weight marker (Trackit ™ 100 bp DNA ladder, Invitrogen). For X and Y codes see Table 1.
4. Discussion
The results obtained in this study once again confirm the efficacy of COIBar–RFLP in discriminating fish species in commercial products and also highlight the fraudulent practice of species substitutions in seafood products, consisting in the use of less valuable shark species in place of swordfish. The MboI endonuclease restriction enzyme produced species-specific restriction patterns of the COI amplicons useful to differentiate X. gladius from shark species. Another interesting result proving the sensitivity of this methodology is the intraspecific differential MboI pattern obtained for the swordfish samples. This pattern was useful to discriminate the two different clades revealed in this species by phylogenetic analyses using several molecular markers [36,37,76,77,78]. COI DNA-barcoding showed that 15% of the swordfish samples purchased in local fish markets during 2018 was mislabeled, with an evident economic loss for the consumers. This percentage was at least two times higher than that recorded in 2010, demonstrating that despite the current European legislation focused on consumer protection against fraud, fraud remains frequent and widespread. In this context, there is no doubt that molecular tools are very useful and effective to fight commercial fraud and that DNA-based methods have become increasingly important for seafood authentication. However, while the practice of commercial fraud in the seafood market is a global concern, to date there is no standardized global methodology to expose this practice. Firstly, all states have not yet incorporated into their legislation the use of molecular methods to combat commercial fraud; this is true for Italy, for example. Secondly, significant differences among countries have been found in methods used by accredited laboratories for food authenticity [79]. Thirdly, together with the classic methods (protein- and DNA sequence-based methods), new and sophisticated methods are being developed to identify seafood species [80]. It is evident that the first two issues can be solved only by adopting a common global policy to fight food fraud. The European legislation, for example, could require, rather than only suggest, the application of DNA analysis in the context of seafood traceability [81], also indicating the most useful methodology to be used across European laboratories. In this regard, the features that molecular methods should have for a rapid authentication of species in seafood products can be debated. To be effective for routine activities carried out by local food safety and quality authorities, from the traceability of the catch to the labeling of the products, effectiveness in terms of cost and time-saving and correctness of species identification should be a priority. Among the classic methods, the protein-based methods, such as isoelectric focusing of sarcoplasmic proteins, are still used as official methods for fish species identification [82], but the DNA-sequencing methods, and the DNA-barcoding methodology in particular, have become more common in laboratories specialized in food authentication ([3] and literature therein). Increasingly, new methodologies are emerging for species identification, such as qPCR, DNA microarrays, high-resolution melting analysis, mass spectrometry, high-throughput sequencing, and the recently developed handheld testing devices [80], all of them suitable and effective in terms of cost and time consumption.
However, these new methodologies require, in some cases, extensive technical equipment and specific skills by the operators and need to be standardized for use as official methods. Furthermore, the application of these methods is limited to a few cases of species authentication, while wide databases of reference samples are needed for their validation as official methods. The methodological approach we propose, COIBar–RFLP, although it cannot substitute DNA sequencing in general, takes advantage of large databases of reference DNA sequences of fish species and of the positive results from several study cases for species of relevant commercial interest under various food matrices [49,52,53,54,58]. COIBar–RFLP successfully and simultaneously discriminated the fish species analyzed in these studies, through the banding pattern obtained after digestion with only one endonuclease restriction enzyme. This simple, robust, easy-to-perform, and cost-effective strategy can potentially cover a wide range of species and provide a versatile tool to monitor the mislabeling of fish products. However, it should be noted that poor enzyme storage, as well as the processing conditions, could compromise the advantages of the methodology in terms of expected time of processing and misleading results. In a recent investigation on the methodological approach performed in 45 European laboratories, Griffiths et al. [79] revealed that PCR–RFLP was used in 40% of the laboratories involved in seafood authentication; this result suggests that this method could become a standardized molecular tool to assess seafood authenticity.
5. Conclusions
The efficacy of COIBar–RFLP was tested for species authentication on slices labeled as swordfish. The illegal practice of species substitution was observed, with the species P. glauca, M. mustelus, and O. centrina being sold in place of swordfish. These results indicate the need to strengthen regulations and to define molecular tools to fight the occurrence of fraud along the seafood supply chain, from the traceability of the catch to the labeling of the products, and to achieve market transparency, which is highly demanded by the consumers. Finally, the future perspectives of COIBar–RFLP rest on the need to build a database of COI restriction patterns to be used for unequivocal species identifications.
Acknowledgments
We thanks the University of Catania for the economic support.
Author Contributions
A.M.P. and V.F. conceived and designed the experiments; A.M.P., A.R., and L.R. performed the experiments; A.M.P., C.F., and S.S. analyzed the data; A.M.P. and V.F. wrote the paper.
Funding
This work was supported by the Annual Research Plan 2016-2018 of the Department of Biological, Geological and Environmental Sciences, University of Catania (Grants #22722132134).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.SCRS Report . Report of the Standing Committee on Research and Statistics (SCRS) International Commission for the Conservation of Atlantic Tunas-Biennial Period 2016–2017. SCRS; Madrid, Spain: 2017. [Google Scholar]
- 2.ISMEA Osservatorio consumi extra-domestici. Analisi & Ricerche n. 3 2005. [(accessed on 25 June 2019)]; Available online: http://www.ismea.it.
- 3.Cosmina M., Demartini E., Gaviglio A., Mauracher C., Prestamburgo S., Trevisan G. Italian Consumers’ Attitudes Towards Small Palegic Fish. New Medit. 2012;1:52–57. [Google Scholar]
- 4.Reilly A. Overview of Food Fraud in the Fisheries Sector. FIAM; Rome, Italy: 2018. FAO In Fisheries and Aquaculture Circular. [Google Scholar]
- 5.European Union Regulation (EC) No. 178/2002 of the European parliament and of the Council of 28 January 2002 laying down the general principles and requirements of food law, establishing the European food safety authority and laying down procedures in matters of food safety. Off. J. Eur. Union. 2002;31:1–24. [Google Scholar]
- 6.European Union Regulation (EC) No. 1169/2011 of the European parliament and of the council of 25 October 2011 on the provision of food information to consumers. Off. J. Eur. Union. 2011;304:18–63. [Google Scholar]
- 7.European Union Regulation (EC) No. 1379/2013 of the European parliament and of the council of 11 December 2013 on the common organisation of the markets in fishery and aquaculture products. Off. J. Eur. Union. 2013;354:1–21. [Google Scholar]
- 8.Altalex IT Art 440. Codice Penale Italiano, Libro II Titolo VI Capo II. [(accessed on 10 April 2019)];2017 Available online: http://www.altalex.com/documents/news/2014/06/03/dei-delitti-contro-l-incolumita-pubblica.
- 9.Decreto Legislativo 27 Gennaio 1992, n. 109 Attuazione delle Direttive n. 89/395/CEE e n. 89/396/CEE Concernenti L’etichettatura, la Presentazione e la Pubblicità Dei prodotti Alimentari. [(accessed on 27 January 1992)]; Available online: http://www.comune.jesi.an.it/MV/leggi/dlvo109-92.htm.
- 10.Decreto Legislativo 6 settembre 2005, n. 206-codice del consumo, a norma dell’art. 7 della legge 29 luglio 2003, n. 229. [(accessed on 6 September 2005)]; Available online: https://www.camera.it/parlam/leggi/deleghe/testi/05206dl.htm.
- 11.Benard-Capelle J., Guillonneau V., Nouvian C., Fournier N., Le Loët K., Dettai A. Fish mislabeling in France: Substitution rates and retail types. PeerJ. 2015;2:e714. doi: 10.7717/peerj.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christiansen H., Dettai A., Heindler F.M., Collins M.A., Duhamel G., Hautecoeur M., Steinke D., Volckaert A.M., Van de Putte A.P. Diversity of mesopelagic fishes in the southern ocean—A phylogeographic perspective using DNA barcoding. Front. Ecol. Evol. 2018;6:120. doi: 10.3389/fevo.2018.00120. [DOI] [Google Scholar]
- 13.Do T.D., Choi T.J., Kim J., An H.E., Park Y.J., Karagozlu M.Z., Kim C.B. Assessment of marine fish mislabelling in South Korea’s markets by DNA barcoding. Food Control. 2019;100:53–57. doi: 10.1016/j.foodcont.2019.01.002. [DOI] [Google Scholar]
- 14.Garcia-Vazquez E., Perez J., Martinez J.L., Pardiñas A.F., Lopez B., Karaiskou N., Triantafyllidis A. High level of mislabelling in Spanish and Greek hake markets suggests the fraudulent introduction of African species. J. Agric. Food Chem. 2011;59:475–480. doi: 10.1021/jf103754r. [DOI] [PubMed] [Google Scholar]
- 15.Von Der Heyden S., Barendse J., Seebregts A.J., Matthee C.A. Misleading the masses: Detection of mislabelled and substituted frozen fish products in south Africa. ICES J. Mar. Sci. 2010;67:176–185. doi: 10.1093/icesjms/fsp222. [DOI] [Google Scholar]
- 16.Quinteiro J., Vidal R., Izquierdo M., Sotelo C.G., Chapela M.J., Pérez-Martín R.I., Rehbein H., Hold G.L., Russell V.J., Pryde S.E., et al. Identification of hake species (Merluccius genus) using sequencing and PCR-RFLP analysis of mitochondrial DNA control region sequences. J. Agric. Food Chem. 2001;49:5108–5114. doi: 10.1021/jf010421f. [DOI] [PubMed] [Google Scholar]
- 17.Kumar G., Kocour M., Kunal S.P. Mitochondrial DNA variation and phylogenetic relationships among five tune species based on sequencing of D-loop region. Mitochondr. DNA Part A. 2016;27:1976–1980. doi: 10.3109/19401736.2014.971313. [DOI] [PubMed] [Google Scholar]
- 18.Rocco L., Ferrito V., Costagliola D., Marsilio A., Pappalardo A.M., Stingo V., Tigano C. Genetic divergence among and within four Italian populations of Aphanius fasciatus (Teleostei, Cyprinodontiformes) Ital. J. Zool. 2007;74:371–379. doi: 10.1080/11250000701451225. [DOI] [Google Scholar]
- 19.Pappalardo A.M., Ferrito V., Messina A., Patarnello T., De Pinto V., Guarino F., Tigano C. Genetic structure of the killifish Aphanius fasciatus Nardo 1827 (Teleostei, Cyprinodontidae), results of mitochondrial DNA analysis. J. Fish Biol. 2008;72:1154–1173. doi: 10.1111/j.1095-8649.2007.01748.x. [DOI] [Google Scholar]
- 20.Ferrito V., Pappalardo A.M., Canapa A., Barucca M., Doadrio I., Olmo E., Tigano C. Mitochondrial phylogeography of the killifish Aphanius fasciatus (Teleostei, Cyprinodontidae) reveals highly divergent Mediterranean populations. Mar. Biol. 2013;160:3193–3208. doi: 10.1007/s00227-013-2307-4. [DOI] [Google Scholar]
- 21.Cuttitta A., Patti B., Maggio T., Quinci E.M., Pappalardo A.M., Ferrito V., De Pinto V., Torri M., Falco F., Nicosia A., et al. Larval population structure of Engraulis encrasicolus in the strait of Sicily as revealed by morphometric and genetic analyses. Fish Ocean. 2015;24:135–149. doi: 10.1111/fog.12098. [DOI] [Google Scholar]
- 22.Pedrosa-Gerasmio I.R., Agmata A.B., Santos M.D. Genetic diversity, population genetic structure, and demographic history of Auxis thazard (Perciformes), Selar crumenophthalmus (Perciformes), Rastrelliger kanagurta (Perciformes) and Sardinella lemuru (Clupeiformes) in Sulu-Celebes sea inferred by mitochondrial DNA sequences. Fish Res. 2015;162:64–74. [Google Scholar]
- 23.Machado V.N., Willis S.C., Teixeira A.S., Hrbek T., Farias I.P. Population genetic structure of the Amazonian black flannelmouth characin (Characiformes, Prochilodontidae: Prochilodus nigricans Spix & Agassiz, 1829): Contemporary and historical gene flow of a migratory and abundant fishery species. Environ. Biol. Fishes. 2017;100:1–16. [Google Scholar]
- 24.Duong T., Uy S., Chheng P., So N., Thi Tran T., Nguyen N.T., Pomeroy R., Egna H. Genetic diversity and structure of striped snakehead (Channa striata) in the lower Mekong basin: Implications for aquaculture and fisheries management. Fish Res. 2019;218:166–173. doi: 10.1016/j.fishres.2019.05.014. [DOI] [Google Scholar]
- 25.Hebert P.D.N., Cywinska A., Ball S., deWaard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hebert P.D.N., Ratnasingham S., deWaard J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergence, among closely related species. Proc. R. Soc. Lond. B. 2003;270:S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hebert P.D.N., Stoeckle M.Y., Zemlak T.S., Francis C.M. Identification of birds through DNA barcodes. PLoS Biol. 2004;2:e312. doi: 10.1371/journal.pbio.0020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hajibabaei M., Smith M.A., Janzen D.H., Rodriguez J.J., Whitfield J.B., Hebert P.D.N. A minimalist barcode can identify a specimen whose DNA is degraded. Mol. Ecol. Notes. 2006;6:959–964. doi: 10.1111/j.1471-8286.2006.01470.x. [DOI] [Google Scholar]
- 29.Hajibabaei M., Singer G.A.C., Hebert P.D.N., Hickey D.A. DNA barcoding: How it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet. 2007;23:167–172. doi: 10.1016/j.tig.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 30.Ward R.D., Holmes B.H., White W.T., Last P.R. DNA barcoding Australasian chondrichthyans: Results and potential uses in conservation. Mar. Fresh Res. 2008;59:57–71. doi: 10.1071/MF07148. [DOI] [Google Scholar]
- 31.Vitale D.G.M., Viscuso R., D’Urso V., Gibilras S., Sardella A., Marletta A., Pappalardo A.M. Morphostructural analysis of the male reproductive system and DNA barcoding in Balclutha brevis Lindberg 1954 (Homoptera, Cicadellidae) Micron. 2015;79:36–45. doi: 10.1016/j.micron.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Federico C., Lombardo D., La Porta N., Pappalardo A.M., Ferrito V., Lombardo F., Saccone S. Rapid molecular identification of necrophagous Diptera by means of variable-length of intron sequences in the wingless gene. J. Forensic Legal Med. 2018;56:66–72. doi: 10.1016/j.jflm.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 33.Conti E., Mulder C., Pappalardo A.M., Ferrito V., Costa G. How soil granulometry, temperature and water predict genetic differentiation in namibian Ariadna spiders and explain their behavior. Ecol. Evol. 2019;9:4382–4391. doi: 10.1002/ece3.4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward R.D., Zemlak T.S., Innes B.H., Last P.R., Hebert P.D.N. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. Lond. B. 2005;360:1847–1857. doi: 10.1098/rstb.2005.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steinke D., Zemlak T.S., Hebert P.D.N. Barcoding Nemo: DNA-based identifications for the ornamental fish trade. PLoS ONE. 2009;4:e6300. doi: 10.1371/journal.pone.0006300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pappalardo A.M., Guarino F., Reina S., Messina A., De Pinto V. Swordfish COI-DNA barcoding as a suitable tool for species and stock identification. FEBS J. 2011;278:440. [Google Scholar]
- 37.Pappalardo A.M., Guarino F., Reina S., Messina A., De Pinto V. Geographically widespread swordfish barcode stock identification: A case study of its application. PLoS ONE. 2011;6:e25516. doi: 10.1371/journal.pone.0025516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pappalardo A.M., Ferrito V. DNA barcoding species identification unveils mislabeling of processed flatfish products in southern Italy markets. Fish Res. 2015;164:153–158. doi: 10.1016/j.fishres.2014.11.004. [DOI] [Google Scholar]
- 39.Pappalardo A.M., Cuttitta A., Sardella A., Musco M., Maggio T., Patti B., Mazzola S., Ferrito V. DNA barcoding and COI sequence variation in Mediterranean lanternfishes larvae. Hydrobiologia. 2015;745:155–167. doi: 10.1007/s10750-014-2161-5. [DOI] [Google Scholar]
- 40.Pappalardo A.M., Federico C., Sabella G., Saccone S., Ferrito V. A COI nonsynonymous mutation as diagnostic tool for intraspecific discrimination in the European anchovy Engraulis encrasicolus (Linnaeus) PLoS ONE. 2015;10:e0143297. doi: 10.1371/journal.pone.0143297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Ostell J., Pruitt K.D., Sayers E.W. GenBank. Nucleic Acids Res. 2018;46:D41–D47. doi: 10.1093/nar/gkx1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ratnasingham S., Hebert P.D.N. BOLD: The barcode of life data system. Mol. Ecol. Notes. 2007;7:355–364. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dawnay N., Ogden R., McEwing R., Carvalho G.R., Thorpe R.S. Validation of the barcoding gene COI for use in forensic genetic species identification. Forensic Sci. Int. 2007;173:1–6. doi: 10.1016/j.forsciint.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 44.Espiñeira M., González-Lavín N., Vieites J.M., Santaclara F.J. Development of a method for the genetic identification of flatfish species on the basis of mitochondrial DNA sequences. J. Agric. Food Chem. 2008;56:8954–8961. doi: 10.1021/jf800570r. [DOI] [PubMed] [Google Scholar]
- 45.Cawthorn D.M., Steinman H.A., Corli Witthuhn R. Establishment of a mitochondrial DNA sequence database for the identification of fish species commercially available in south Africa. Mol. Ecol. Res. 2011;11:979–991. doi: 10.1111/j.1755-0998.2011.03039.x. [DOI] [PubMed] [Google Scholar]
- 46.Armani A., Castigliego L., Guidi A. Fish frauds: The DNA challenge. CAB reviews 071. Anim. Sci. Rev. 2012;7:227–238. [Google Scholar]
- 47.Armani A., Guardone L., La Castellana R., Gianfaldoni D., Guidi A., Castigliego L. DNA barcoding reveals commercial and health issues in ethnic seafood sold on the Italian market. Food Control. 2015;55:206–214. doi: 10.1016/j.foodcont.2015.02.030. [DOI] [Google Scholar]
- 48.Khaksar R., Carlson T., Schaffner D.W., Ghorashi M., Best D., Jandhyala S., Traverso J., Amini S. Unmasking seafood mislabeling in U.S. markets: DNA barcoding as a unique technology for food authentication and quality control. Food Control. 2015;56:71–76. doi: 10.1016/j.foodcont.2015.03.007. [DOI] [Google Scholar]
- 49.Ferrito V., Bertolino V., Pappalardo A.M. White fish authentication by COIBar-RFLP: Toward a common strategy for the rapid identification of species in convenience seafood. Food Control. 2016;70:130–137. doi: 10.1016/j.foodcont.2016.05.026. [DOI] [Google Scholar]
- 50.Nagalakshmi K., Annam P.K., Venkateshwarlu G., Pathakota G.B., Lakra W.S. Mislabelling in Indian seafood: An investigation using DNA barcoding. Food Control. 2016;59:196–200. doi: 10.1016/j.foodcont.2015.05.018. [DOI] [Google Scholar]
- 51.Ferrito V., Pappalardo A.M. Seafood species identification by DNA barcoding, a molecular tool for food traceability. Biodivers. J. 2017;8:65–72. [Google Scholar]
- 52.Pappalardo A.M., Copat C., Ferrito V., Grasso A., Ferrante M. Heavy metal content and molecular species identification in canned tuna: Insights into human food safety. Mol. Med. Rep. 2017;15:3430–3437. doi: 10.3892/mmr.2017.6376. [DOI] [PubMed] [Google Scholar]
- 53.Pappalardo A.M., Federico C., Saccone S., Ferrito V. Differential flatfish species detection by COIBar-RFLP in processed seafood products. Eur. Food Res. Technol. 2018;244:2191–2201. doi: 10.1007/s00217-018-3129-x. [DOI] [Google Scholar]
- 54.Pappalardo A.M., Petraccioli A., Capriglione T., Ferrito V. From fish eggs to fish name: Caviar species discrimination by COIBar-RFLP, an efficient molecular approach to detect fraud in the caviar trade. Molecules. 2019;24:2468. doi: 10.3390/molecules24132468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cocolin L., D’Agaro E., Manzano M., Lanari D., Comi G. Rapid PCR-RFLP method for the identification of marine fish fillets (seabass, seabream, umbrine, and dentex) J. Food Sci. 2000;65:1315–1317. doi: 10.1111/j.1365-2621.2000.tb10604.x. [DOI] [Google Scholar]
- 56.Hold G.L., Russell V.J., Pryde S.E., Rehbein H., Quinteiro J., Rey-Mendez M., Sotelo C.G., Pérez-Martin R.I., Santos A.T., Rosa C. Validation of a PCR-RFLP based method for the identification of salmon species in food products. Eur. Food Res. Technol. 2001;212:385–389. doi: 10.1007/s002170000237. [DOI] [Google Scholar]
- 57.Hsieh H.S., Chai T., Hwang D.F. Using the PCR-RFLP method to identify the species of different processed products of billfish meats. Food Control. 2007;18:369–374. doi: 10.1016/j.foodcont.2005.11.002. [DOI] [Google Scholar]
- 58.Pappalardo A.M., Ferrito V. A COIBar-RFLP strategy for the rapid detection of Engraulis encrasicolus in processed anchovy products. Food Control. 2015;57:385–392. doi: 10.1016/j.foodcont.2015.03.038. [DOI] [Google Scholar]
- 59.Dent F., Clarke S. State of the Global Market for Shark Products. Food and Agriculture Organization FAO; Rome, Italy: 2015. [Google Scholar]
- 60.Blanco M., Pérez-Martín R.I., Sotelo C.G. Identification of shark species in seafood products by forensically informative nucleotide sequencing (FINS) J. Agric. Food Chem. 2008;56:9868–9874. doi: 10.1021/jf8015128. [DOI] [PubMed] [Google Scholar]
- 61.Barbuto M., Galimberti A., Ferri E., Labra M., Malandra R., Galli P., Casiraghi M. DNA barcoding reveals fraudulent substitutions in shark seafood products: The Italian case of palombo (Mustelus spp.) Food Res. Int. 2010;43:376–381. doi: 10.1016/j.foodres.2009.10.009. [DOI] [Google Scholar]
- 62.Filonzi L., Chiesa S., Vaghi M., Nonnis Marzano F. Molecular barcoding reveals mislabelling of commercial fish products in Italy. Food Res. Int. 2010;43:1383–1388. doi: 10.1016/j.foodres.2010.04.016. [DOI] [Google Scholar]
- 63.Herrero B., Lago F.C., Vieites J.M., Espiñeira M. Authentication of swordfish (Xiphias gladius) by RT-PCR and FINS methodologies. Eur. Food Res. Technol. 2011;233:195–202. doi: 10.1007/s00217-011-1502-0. [DOI] [Google Scholar]
- 64.Tortonese E. Fauna d’Italia, ‘Osteichtyes’: Pesci ossei. Calderini Bologna. 1975;11:133–139. [Google Scholar]
- 65.Whitehead P.J.P., Bauchot M.L., Hureau J.C., Nielsen J., Tortonese E. Fishes of the North-Eastern Atlantic and the Mediterranean. UNESCO; Paris, France: 1986. [Google Scholar]
- 66.Ivanova N.V., Zemlak T.S., Hanner R.H., Hebert P.D.N. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes. 2007;7:544–548. doi: 10.1111/j.1471-8286.2007.01748.x. [DOI] [Google Scholar]
- 67.GENECHRON. [(accessed on 17 November 2018)]; Available online: http://www.genechron.it/index.php/sanger-sequencing.
- 68.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.TRANSEQ. [(accessed on 26 July 2019)]; Available online: http://www.ebi.ac.uk/Tools/st/emboss_transeq.
- 70.GENBANK. [(accessed on 12 September 2019)]; Available online: http://www.ncbi.nlm.nih.gov/genbank/
- 71.Zhang D.X., Hewitt G.M. Nuclear integrations: Challenges for mitochondrial DNA markers. Trends Ecol. Evol. 1996;11:247–251. doi: 10.1016/0169-5347(96)10031-8. [DOI] [PubMed] [Google Scholar]
- 72.Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 75.REMAP EMBOSS. [(accessed on 26 July 2019)]; Available online: http://emboss.bioinformatics.nl.
- 76.Alvarado-Bremer J.R., Baker A.J., Mejuto J. Mitochondrial DNA control region sequences indicate extensive mixing of swordfish (Xiphias gladius) populations in the Atlantic ocean. Can. J. Fish Aquat. Sci. 1995;52:1720–1732. doi: 10.1139/f95-764. [DOI] [Google Scholar]
- 77.Alvarado-Bremer J.R., Mejuto J., Greig T.W., Ely B. Global population structure of the swordfish (Xiphias gladius L.) as revealed by analysis of the mitochondrial DNA control region. J. Exp. Mar. Biol. Ecol. 1996;197:295–310. doi: 10.1016/0022-0981(95)00164-6. [DOI] [Google Scholar]
- 78.Bradman H., Grewe P., Appleton B. Direct comparison of mitochondrial markers for the analysis of swordfish population structure. Fish Res. 2011;109:95–99. doi: 10.1016/j.fishres.2011.01.022. [DOI] [Google Scholar]
- 79.Griffiths A.M., Sotelo C.G., Mendes R., Perez Martin R.I., Schröder U., Shorten M., Silva H.A., Verrez-Bagnis V., Mariani S. Current methods for seafood authenticity testing in Europe: Is there a need for harmonisation? Food Control. 2014;45:95–100. doi: 10.1016/j.foodcont.2014.04.020. [DOI] [Google Scholar]
- 80.Verrez-Bagnis V., Sotelo C.G., Mendes R., Silva H., Kappel K., Schröder U. Methods for seafood authenticity testing in Europe. In: Mérillon J.-M., Ramawat K.G., editors. Bioactive Molecules in Food, Reference Series in Phytochemistry. Springer; Berlin, Germany: 2018. pp. 1–55. [Google Scholar]
- 81.EUR-Lex Council Regulation (EC) No. 1224/2009 of 20 November. Off. J. Eur. Union. 2009;343:50. [Google Scholar]
- 82.Kappel K., Schröder U. Substitution of high-priced fish with low-priced species: Adulteration of common sole in German restaurants. Food Control. 2016;59:478–486. doi: 10.1016/j.foodcont.2015.06.024. [DOI] [Google Scholar]