Summary
Plant pathogens are a significant challenge in agriculture despite our best efforts to combat them. One of the most effective and sustainable ways to manage plant pathogens is to use genetic modification (GM) and genome editing, expanding the breeder's toolkit. For use in the field, these solutions must be efficacious, with no negative effect on plant agronomy, and deployed thoughtfully. They must also not introduce a potential allergen or toxin. Expensive regulation of biotech crops is prohibitive for local solutions. With 11–30% average global yield losses and greater local impacts, tackling plant pathogens is an ethical imperative. We need to increase world food production by at least 60% using the same amount of land, by 2050. The time to act is now and we cannot afford to ignore the new solutions that GM provides to manage plant pathogens.
Keywords: biotechnology, food security, genetic modification, plant disease, plant pathogens, resistance
| Contents | ||
|---|---|---|
| Summary | 70 | |
| I. | Introduction | 70 |
| II. | Intervention based on pathogen recognition and effectors | 73 |
| III. | Intervention by modification of defence signalling and regulation | 74 |
| IV. | Intervention by targeting recessive traits/susceptibility genes | 75 |
| V. | Intervention via other dominant plant resistance genes | 75 |
| VI. | Intervention with antimicrobial peptides | 76 |
| VII. | Intervention using RNA interference | 77 |
| VIII. | Practical path to deployment | 77 |
| IX. | Path to market in Africa | 78 |
| X. | How to deploy resistance durably | 79 |
| XI. | Trends that shape the future | 80 |
| XII. | The time is now | 81 |
| Acknowledgements | 81 | |
| References | 81 | |
I. Introduction
From the earliest days of farming, plant disease and pests have been a critical challenge for farmers. Although mankind has split the atom, travelled to the moon and connected the world, plant pathogens continue to be a significant challenge to food security despite our best efforts to thwart them (Fig. 1). Estimates of average global losses to diseases and pests range from 11–30% (Oerke & Dehne, 2004; Savary et al., 2019). Importantly, crop losses are highest in regions that already suffer from food insecurity (Savary et al., 2019). Losses from diseases would be far worse without past steady advances in agricultural practices, including cultural controls, agrochemical use and plant breeding. However, we have learned that there are no ‘silver bullets’. An integrated approach is needed to combat plant diseases, combining the best technologies and practices that are available.
Figure 1.
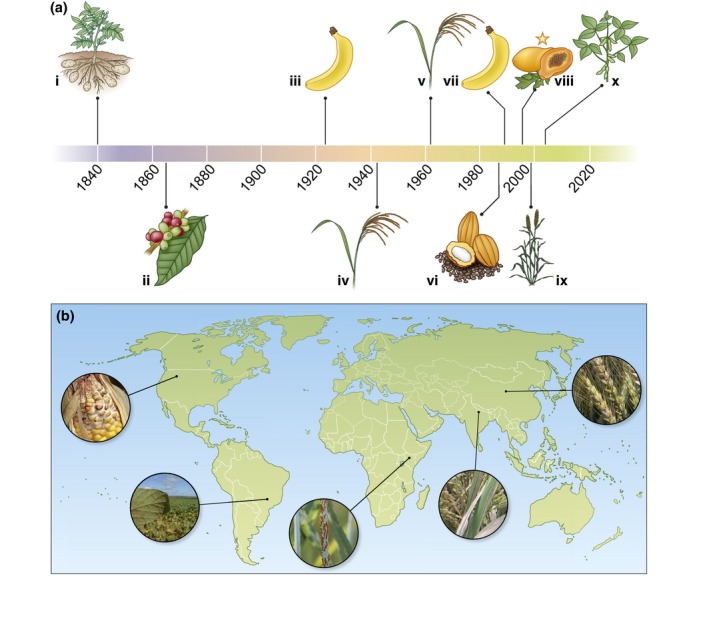
Major disease outbreaks in the last 150 yr and current critical disease challenges. (a) A timeline of major disease outbreaks: (i) Introduction of the oomycete Phytophthora infestans which causes potato late blight results in the Irish potato famine in which 1 million people die and 1.5 million people emigrate. (ii) The rust fungus Hemileia vastatrix wipes out the coffee crop in Sri Lanka; the British become tea drinkers. (iii) The vascular fungal pathogen causing Fusarium wilt of banana nearly wipes out the Gros Michel variety; the resistant Cavendish banana is adopted. (iv) The fungus Cochliobolus miyabeanus, which causes Brown spot disease of rice is a factor in the Great Bengal Famine in which 2 million people die of starvation. (v) Bacterial leaf blight of rice (Xanthomonas oryzae pv. oryzae) causes epidemics throughout Southeast Asia with yield losses up to 80%. (vi) Witches’ broom caused by the fungus Moniliophthora perniciosa is causing losses of up to 75% of annual cacao production in Brazil. (vii) The new Fusarium wilt isolate TR4 is identified and threatens Cavendish banana. (viii) Ringspot virus devastates the papaya industry in Hawaii; a GM variety is introduced that resists infection. (ix) A new race of the stem rust fungus Puccinia graminis (UG99) is spreading throughout Africa and the Middle East, threatening the world wheat supply. (x) Asian soybean rust caused by Phakopsora pachyrhizi reaches Brazil, costing growers US$2 billion annually in damages and control measures. (b) Examples of current disease challenges in major agricultural regions in the world that cause significant losses such as corn stalk and ear rots in the USA (4.15%), Soybean rust in Brazil (6.65%), Stem rust of wheat in sub‐Saharan Africa (8.89%), bacterial blight of rice in India (8.51%) and Fusarium head blight of wheat in China (8.75%). Source: Savary et al. (2019). Pictures: Gibberella zeae (corn ear rot) (photograph by Scot Adams, via Flickr, CC BY 2.0); Phakopsora pachyrhizi (Asian soybean rust) (photograph by Peter van Esse); Puccinia graminis f. sp. tritici (Wheat stem rust) (Photo by Yue Jin); Xanthomonas oryzae f. sp. oryzae (bacterial blight) (photograph provided by IRRI under creative commons licence); Fusarium graminearum (Fusarium head blight) (photograph by Gary C. Bergstrom, Cornell University, USA).
The benefits of an integrated approach can be seen in the management of stem rust in wheat, a disease that caused periodic costly epidemics in the USA between 1918 and 1960 (Pardey et al., 2013). Only the combined effort of cultural practices (removal of barberry, the sexual host of this pathogen), improved chemical control (development of demethylation inhibitor and quinone outside inhibitor fungicides) and an extensive breeding program spearheaded by Norman Borlaug have enabled the containment of this particular disease of wheat.
However, there are limitations to such efforts. Some pesticides are rapidly losing efficacy due to pathogen evolution, and their use faces increasingly strict regulations to minimize unwanted side effects (Geiger et al., 2010; Bolton et al., 2012; Lamichhane et al., 2015; Wieczorek et al., 2015; Godoy et al., 2016; Berger et al., 2017). Crop breeding can produce resistance to individual diseases, but it is challenging to select for genetic resistance against multiple diseases simultaneously while maintaining the strong performance traits of elite varieties. For example, wheat blast is an emerging disease that will require wheat breeders to select for blast resistance while maintaining resistance against stem rust (Islam et al., 2016). To make matters more complicated, new races of stem rust have emerged and must also be tackled to ensure the stability of the world's wheat supply (Singh et al., 2015). Finally, the introgression of a single resistance via classical breeding facilitates pathogen adaption to that resistance.
The disease issues of wheat are not an isolated example, and challenges such as these are becoming more frequent as global warming and increased global trade facilitate the spread of known and emerging pathogens (Bebber et al., 2014). Top of these issues is the fundamental reality that 821 million people do not have enough to eat (FAO et al., 2018). The world population is projected to reach nearly 10 billion in 2050 (United Nations, 2017). This forecast brings with it the associated need to increase world food production by at least 60% (Alexandratos & Bruinsma, 2012; United Nations, Department of Economic and Social Affairs, Population Division, 2017) With this development in mind, tackling plant pathogens is not a mere academic exercise but an ethical imperative that requires action.
One of the most effective and sustainable ways to manage plant pathogens is to use GM and genome editing to expand the genetic tools available to breeders. In this review, we present an inventory of the genetic disease solutions currently available for bacterial, viral, fungal and oomycete pathogens. We will highlight the success stories of the potential of GM technologies and will outline what is needed for the effective deployment and realisation of the benefits they offer. Examples of genetic disease solutions are listed in Table 1. We evaluate these examples in light of population growth and other challenges and describe the trends that will shape the future.
Table 1.
Examples of genetic disease solutions currently available for bacterial, viral, fungal and oomycete pathogens.
| Point of intervention | GM technology | Example | References |
|---|---|---|---|
| Pathogen perception | Interspecies transfer of PRRs | EF‐Tu receptor (EFR) | Lacombe et al. (2010); Schoonbeek et al. (2015); Schwessinger et al. (2015); Boschi et al. (2017); Kunwar et al. (2018) |
| Interspecies transfer of NLRs | Rpi‐Vnt1 | Foster et al. (2009); http://www.isaaa.org/ | |
| Bs2 | Horvath et al. (2012) | ||
| Modification of NLRs | Pikp‐1 | Maqbool et al. (2015) | |
| NLR protease trap | PBS1 kinase | Kim et al. (2016) | |
| NLR resurrection | NRCs (NLR helpers) | Wu et al. (2017) | |
| Pathogen effector binding | Deletion of effector targets | MAPK3K StVIK1 | Murphy et al. (2018) |
| Modification of effector binding sites | COI1 | Zhang et al. (2015) | |
| Deletion of effector binding sites | Os11N3/OsSWEET14 | Li et al. (2012) | |
| Addition of effector binding sites | Xa27 | Hummel et al. (2012) | |
| Defence signalling pathway | Altered expression of signalling components | NPR1 | Xu et al. (2017) |
| Altered expression of transcription factors | IPA1/OsSPL14 | Wang et al. (2018b) | |
| Recessive resistance alleles | Gene deletion | mlo | Kusch & Panstruga (2017) |
| Gene modification | bs5 | Iliescu et al. (2013) | |
| Dominant plant resistance proteins | Interspecies transfer of signalling components | PFLP | Huang et al. (2007); Namukwaya et al. (2012); J. N. Tripathy et al. (2014); Tang et al. (2001); Huang et al. (2004); Ger et al. (2014); Yip et al. (2007); Liau et al. (2003) |
| Transfer of detoxifying enzymes targeting pathogen toxins | Oxalate oxidase | Donaldson et al. (2001); Schneider et al. (2002); Hu et al. (2003); Dong et al. (2008); Walz et al. (2008); Partridge‐Telenko et al. (2011) | |
| Transfer of adult plant resistance (APR) alleles | Lr34 | Krattinger et al. (2016); Risk et al. (2013); Schnippenkoetter et al. (2017); Sucher et al. (2017); Rinaldo et al. (2017) | |
| Antimicrobial compound production | Transfer of antimicrobials from plants | Rs‐AFP defensin | Jha & Chattoo (2010); Li et al. (2011) |
| Transfer of antimicrobials from microorganisms or animals | Virus KP4 | Clausen et al. (2000); Schlaich et al. (2006); Quijano et al. (2016) | |
| Expression of synthetic antimicrobials | MsrA1 | Osusky et al. (2000); Rustagi et al. (2014) | |
| RNAi | Viral gene silencing through RNAi | Coat protein or replicase domain gene from Papaya ringspot virus | Fitch et al. (1992); Ferreira et al. (2002); Ye & Li (2010); http://www.isaa.org/ |
| AC1 from bean golden mosaic virus | Bonfim et al. (2007); http://www.isaaa/org | ||
| Coat protein gene from plum pox virus | Scorza et al. (2013); http://www.isaaa.org/ | ||
| Coat protein gene from potato virus Ya | Lawson et al. (1990); http://www.isaaa.org/ | ||
| Putative replicase domain or helicase domain gene from potato leaf roll virusb | Lawson et al. (2001); http://www.isaaa.org/ | ||
| Coat protein gene from cucumber mosaic cucumovirus, zucchini yellow mosaic potyvirus and watermelon mosaic potyvirus 2 | Tricoli et al. (1995); http://www.isaaa.org/ | ||
| Fungal and oomycete gene silencing through RNAi | HAM34 or CES1 gene of Bremia lactucae | Govindarajulu et al. (2015) |
Examples that are currently in the market are shown in bold.
NewLeaf Y® potato, no longer commercially available.
NewLeaf Plus® potato, no longer commercially available.
II. Intervention based on pathogen recognition and effectors
Research over the past 20 yr has led to an increasingly refined knowledge of the plant immune system and its surveillance capacity. It is able to distinguish ‘self’ from ‘nonself’ as well as perturbations of ‘self’ by monitoring the extracellular and intracellular environment (Jones & Dangl, 2006; Cook et al., 2015). However, pathogens can overcome this system in an evolutionary arms race, producing proteins and molecules called effectors that are used to suppress host immunity and manipulate the plant cell to facilitate colonisation (Cook et al., 2015; Uhse & Djamei, 2018). Effectors are secreted into the extracellular environment or delivered in an orchestrated way into the host. This process is often done via specialised mechanisms such as the type III secretion systems of bacteria, haustoria of fungi and oomycetes and the stylet of nematodes (Panstruga & Dodds, 2009; Galán et al., 2014; Bird et al., 2015; Espada et al., 2016; Deng et al., 2017; Lo Presti & Kahmann, 2017).
Plants have two main surveillance systems to detect pathogen incursions. One class of receptors, known as pattern‐recognition receptors (PRRs), monitors the extracellular environment for conserved pathogen molecules such as flagellin, the bacterial elongation factor Tu, and chitin (Gómez‐Gómez & Boller, 2000; Zipfel et al., 2006; Miya et al., 2007; Faulkner et al., 2013; Cao et al., 2014; Hind et al., 2016). This class also recognises extracellular effectors that increase pathogen virulence (Wang et al., 1996; Thomas et al., 1997; Rep et al., 2005; van den Burg et al., 2006; van Esse et al., 2007; Catanzariti et al., 2015; Pruitt et al., 2015), and has been recently reviewed (Boutrot & Zipfel, 2017).
Intracellular pathogen effectors are recognized by another class of receptors that make up a large family of proteins characterised structurally by a nucleotide binding site (NBS) and leucine‐rich repeats (LRR) that are known as NOD‐like receptors (NLR) proteins (Dodds & Rathjen, 2010; Jones et al., 2016). This large family is well characterised and can be divided into two major groups in plants by features at their N terminus: one set has a Toll/interleukin‐1 receptor‐like (TIR) domain and the other a coiled coil (CC) domain (Jones et al., 2016), which confer discrete signalling capacity. Some NLRs have integrated domains that resemble/contain effector targets such as heavy metal‐associated binding domains, WRKY domains and RPM1‐interacting protein 4 (RIN4) (Le Roux et al., 2015; Maqbool et al., 2015; Sarris et al., 2016). Finally, an additional layer of the NLR resistance network is emerging in Solanaceous plants, a clade of helper NLRs has been identified and that connect to several NLRs that detect pathogens (Wu et al., 2017).
In the ongoing evolutionary arms race, some pathogens use the plant's defences against itself by misdirecting the host immune system to produce an immune response to the wrong pathogen to maintain host susceptibility. For example, some bacterial pathogens hijack the Coronatine‐insensitive protein 1 (COI1) jasmonate receptor, rewiring defence responses to activate jasmonate responses and concomitantly suppress the more effective salicylic acid defence pathway (He et al., 2004). Similarly, the necrotrophic fungal pathogens Stagonospora nodorum and Pyrenophora tritici‐repentis activate an inappropriate cell death response benefiting the pathogen by triggering the NLR receptor Tsn1 (Faris et al., 2010).
Knowledge of the plant immune system has provided strategies to intervene at the point of pathogen perception. Extended or novel recognition capacity can be created in a number of ways, for example by introducing receptors from other plants with novel recognition specificity (Fig. 2a,b; Tai et al., 1999; Foster et al., 2009; Lacombe et al., 2010; J. N. Tripathy et al., 2014; Albert et al., 2015; Kawashima et al., 2016; Steuernagel et al., 2016; Witek et al., 2016; Ghislain et al., 2019); through modification of the integrated domains in NLRs that are targeted by the pathogen (Maqbool et al., 2015); or by reactivation of NLR genes disabled by effectors through the introduction of novel helper NLRs (Wu et al., 2017). Another original strategy is the design of the so‐called ‘NLR protease traps’. This strategy makes use of NLRs that can recognise the cleavage of plant proteins by specific pathogen proteases. This detection leads to a subsequent activation of immunity. Modification of the proteins monitored by such NLRs, such that the cleavage site will be targeted by a different pathogen protease, can broaden or alter the specificity of the plant's immune response (Kim et al., 2016).
Figure 2.

Success stories with different points of intervention: (a) The 3R potato contains three NLRs effective against Phytophthora infestans, which is present as a single mating type in Uganda and Kenya. (b) The cell‐surface EF‐Tu receptor (EFR) provides field level of resistance against the devastating tomato wilt pathogen Ralstonia solanacearum. (c) The Tomelo, genome‐edited tomato has resistance against powdery mildew due to modification of the mlo gene. (d) Heterologous expression of hypersensitive response‐assisting protein (Hrap) and plant ferredoxin‐like protein (Pflp) from sweet pepper provides field level resistance against Xanthomonas wilt disease in banana. (e) Overexpression of a virus coat protein in papaya provides commercial control against Papaya ringspot virus in Hawaii. In each case, the control plant(s) are on the left and the transgenic plants on the right. Pictures: photographs provided by (a) Marc Ghislain, © International Potato Center; (b) Dr Sanju Kunwar and Dr Mathews Paret, University of Florida; (c) Sophien Kamoun, The Sainsbury Laboratory. (d) Photograph reprinted by permission from Springer Nature Customer Service Centre GmbH: Springer Nature, Nature Biotechnology, field trial of Xanthomonas wilt disease‐resistant bananas in East Africa, (L. Tripathi et al., 2014). (e) Photograph provided by Dennis Gonsalves, republished with permission of the American Phytopathological Society, from Ferreira et al. (2002). Permission conveyed through Copyright Clearance Center, Inc.
Beyond strategies based on pathogen recognition, a growing understanding of effectors and their function has allowed interventions at the point of pathogen modulation of host responses. For example, knowledge of the plant targets of effector activity reveals which host components are manipulated to promote disease. This knowledge has been successfully applied to interfere with these points of vulnerability by removing them (Bozkurt et al., 2014; Boevink et al., 2016; Yang et al., 2016; Murphy et al., 2018) or replacing them with variants that are immune to effector action but retain the native function in the host (Zhang et al., 2015). For bacterial pathogens expressing transcription activator‐like (TAL) effectors that activate the expression of susceptibility genes in the host, resistance can be engineered by deletion of the TAL DNA binding sites in the promoter (Li et al., 2012; Jia et al., 2017). Another approach to engineer resistance to these bacterial pathogens is to add TAL effector binding sites to a cell‐death‐promoting (‘executor’) gene that is triggered by the TAL effectors present in common pathotypes (Hummel et al., 2012; Wang et al., 2018a).
Resistance of an entire plant species to all isolates of a microbial species is classically referred to as nonhost or species resistance. This nonhost resistance is brought about by physical factors, the plant immune system, and a general inability of the nonadapted pathogen to evade and/or disarm the plant's immune system (Nürnberger & Lipka, 2005). However, nonhost resistance does not represent a single phenomenon that can be used to engineer resistant crops. For example, most plant–pathogen systems cannot be neatly classified into the two extremes of host/nonhost systems (Bettgenhaeuser et al., 2014). In addition, there is no single mechanism behind nonhost resistance but various distinct and unique mechanisms (Cook et al., 2015). Therefore, the approaches that traditionally have been contained in the term nonhost resistance that are not perception‐related will be discussed in other sections of the review.
Resistance that is provided by NLRs and PRRs is robust, mechanistically well understood and, for NLRs, often results in strong immunity. There are clear advantages to working with the plant's innate immune system. Introduced receptors activate signalling pathways that are already in place in the plant. Importantly, activation of defences generally only occurs when a pathogen is perceived, minimising the cost to the plant overall. Furthermore, crop plants already contain hundreds of these receptors, therefore the likelihood that they are potential allergens or toxins is vanishingly small. Indeed, a late blight resistant potato containing an NLR receptor introduced from a wild relative is currently on the market in the USA (http://www.isaaa.org/). This is an important advance, but care must also be taken to deploy resistance genes durably; pathogens are extremely adaptive and single recognition specificities can be rapidly overcome by pathogen evolution.
III. Intervention by modification of defence signalling and regulation
Perception of pathogens by the plant's immune system is translated into defence responses through hormones, signalling pathways and changes in defence genes. The major hormones involved in plant defences are salicylic acid (SA), jasmonate (JA), and ethylene (ET). In addition, there is extensive crosstalk with essentially all other hormonal signalling pathways, including gibberellins, auxin, brassinolide, cytokinins, and abscisic acid (reviewed by Robert‐Seilaniantz et al., 2011; De Vleesschauwer et al., 2014; Berens et al., 2017). Most major signalling components seem to be conserved throughout angiosperms (Berens et al., 2017), with some variations in the details of signalling, crosstalk, and mode of defence against different types of pathogens (De Vleesschauwer et al., 2014; Berens et al., 2017). In general, SA primarily mediates resistance to biotrophic pathogens, while JA in concert with ET mediates resistance to necrotrophic pathogens. There is cross‐inhibition between SA and JA resulting in tradeoffs between resistance to biotrophs and necrotrophs. Constitutive induction of SA or JA signalling produces resistance to pathogens ordinarily controlled by these responses but produces pleiotropic effects on growth and yield.
One way to engineer resistance without causing such pleiotropic side effects is to tightly control the timing and location of gene expression. An example of this strategy is the use of the TL1‐binding factor 1 (TBF1) promoter and leader sequences. TBF1 contains two pathogen‐responsive upstream open reading frames to drive expression of either a constitutively active NLR protein or non‐expressor of pathogenesis‐related genes 1 (NPR1), a key regulator of SA response, in rice (Xu et al., 2017). The combined effects of transcriptional and translational control produced resistance to rice blast without a notable yield penalty.
A naturally occurring example of localised pathway overexpression is the quantitative resistance to biotrophic pathogens that is conferred by the loss of function of downy mildew resistance 6 (DMR6) in Arabidopsis (van Damme et al., 2008; Zeilmaker et al., 2015). DMR6 is widely conserved and encodes a salicylate‐5‐hydroxylase that is induced around pathogen infection sites (Zhang et al., 2017). Loss of DMR6 function presumably increases the local SA concentration at the infection site (Zeilmaker et al., 2015). This knowledge was used to engineer a loss‐of‐function allele of a DMR6 homologue in tomato. This allele resulted in a quantitative resistance to biotrophic pathogens (de Toledo Thomazella et al., 2016).
Defence responses are controlled by networks of transcriptional regulators (Tsuda & Somssich, 2015). Therefore, the overexpression of specific transcription factors is a potential strategy to engineer resistance if pleiotropic effects on yield can be avoided. One interesting case is the rice gene Ideal Plant Architecture 1 (IPA1)/OsSPL14 in which a natural allelic variant increased both yield and resistance to rice blast. Specific phosphorylation of the IPA1 protein in response to blast infection alters IPA1 binding specificity. This shift in specificity allows the protein to bind to and activate WRKY45, a defence regulatory transcription factor, providing quantitative resistance. By contrast, nonphosphorylated IPA1 promotes the expression of at least one yield‐related gene (Wang et al., 2018b). If this posttranslational regulation is conserved, IPA1 expression may be useful to control disease in other crops.
IV. Intervention by targeting recessive traits/susceptibility genes
Plant breeders have long been aware of recessive disease resistances, which have been identified in two ways, through mutagenesis and via breeding. With the onset of genome‐editing technologies, it is now possible to readily reconstitute recessive traits in other species. Many recessive traits can be generated by other methods in diploid crops, but genome editing opens up the possibility of reconstitution in polyploid crops such as wheat and potato. Most well understood recessive resistance traits remove or alter host factors needed for pathogen infection and hence are known as susceptibility genes. However, there are exceptions such as the dmr6 mutation discussed above that alters signalling pathways. Recessive resistance can be very broad and durable, as exemplified by the powdery mildew resistance conferred by the mildew resistance locus O (mlo) allele, which is effective in crops as diverse as apple, tomato, barley and wheat (Kusch & Panstruga, 2017). For the complex wheat genome, all three homoeoalleles of mlo were targeted simultaneously using genome‐editing techniques (Wang et al., 2014). Alleles of mlo that give strong resistance unfortunately also give strong pleiotropic phenotypes (Kusch & Panstruga, 2017). However, the mlo allele can now be easily modified with gene‐editing tools. This process could allow a more precise calibration between achieving mlo‐mediated resistance and minimising mlo‐mediated pleiotropic effects (Fig. 2c; Nekrasov et al., 2017). Still, care should be taken with mlo modification because the allele may result in enhanced susceptibility to other pathogens. Known examples are the necrotrophic fungi Magnaporthe oryzae, Fusarium graminearum and Ramularia collo‐cygni, which all are more virulent in hosts with an mlo background (Jarosch et al., 1999; Jansen et al., 2005; McGrann et al., 2014). This increased susceptibility may be particularly relevant in wheat, in which blast disease caused by Magnaporthe oryzae pathotype Triticum is a critical emerging pathogen (Islam et al., 2016).
Another widely deployed recessive resistance that has potential value as a genome‐editing target is potyvirus resistance mediated by variants of eukaryotic translation initiation factor 4E (eIF4E). This type of resistance was first observed in mutants of Arabidopsis thaliana that exhibited loss of susceptibility to tobacco etch virus (TEV; Potyvirus) due to a deficiency in the eIFiso4E gene, an isoform of eIF4E (Lellis et al., 2002). Similar to A. thaliana, eIF4E‐mediated resistance against potyviruses is found in several resistant crop cultivars including pepper (Capsicum annuum), lettuce (Lactuca sativa), and wild tomato (Solanum habrochaites) (Ruffel et al., 2002, 2005; Nicaise et al., 2003). However, the plasticity in editing eIF4E appears to be restricted, because simple knockouts often result either in severe pleiotropic effects or a lack of effect due to redundancy (Bastet et al., 2017). Therefore, editing of eIF4E may be more successful when guided by naturally existing allelic variation (Bastet et al., 2017). Another example of a naturally occurring recessive resistance allele is bacterial spot 5 (bs5), which was identified in pepper breeding populations as a Xanthomonas resistance locus (Jones et al., 2002). The basis of resistance is a six base pair deletion in Bs5, a CYSTM protein, resulting in a protein product that lacks two amino acids in a highly conserved domain (Iliescu et al., 2013). Knockout mutations of CYSTM proteins give rise to severe growth and reproduction defects (Albert et al., 2015). This situation suggests that the specific change in bs5 preserves other housekeeping functions and selectively interferes with pathogen action. Bs5 is widely conserved, raising the possibility that the bs5 phenotype may be recapitulated by creating the specific six base pair deletion in other plants susceptible to Xanthomonas, such as tomato.
Forward genetic approaches have yielded only a few targets for modification without incurring strong pleiotropic phenotypes in crops. Furthermore, recessive traits are typically not favoured by breeders, and therefore few have been molecularly characterised. The best and most widely deployed traits have been identified from nature. We therefore predict that the most effective recessive resistance traits will be those inspired by naturally occurring variants found in older breeding populations or wild relatives.
V. Intervention via other dominant plant resistance genes
V..1. Plant ferredoxin‐like protein and hypersensitive response‐assisting protein
Two interesting examples of plant proteins that confer disease resistance in various crops in a dominant fashion are plant ferredoxin‐like protein (PFLP) (Lin et al., 1997; Dayakar et al., 2003) and hypersensitive response‐assisting protein (HRAP) (Chen et al., 1998, 2000). Both proteins were isolated from sweet pepper (Capsicum annuum) and enhanced the production of reactive oxygen species and the hypersensitive response in reaction to harpins produced by Gram‐negative bacteria (Choi et al., 2013). HRAP may act in the extracellular space, where it could contribute to dissociation of harpins into active monomers or dimers, facilitating recognition by the plant (Chen et al., 1998, 2000). PFLP, formerly called amphipathic protein 1 (AP1), shows high similarity to ferredoxin proteins that function as electron carriers in photosynthetic tissues, where they are involved in many metabolic processes (Lin et al., 1997; Dayakar et al., 2003). Both PFLP and HRAP are effective against multiple bacterial pathogens when overexpressed in rice, banana, and other species (Tang et al., 2001; Ger et al., 2002, 2014; Liau et al., 2003; Huang et al., 2004, 2007; Pandey et al., 2005; Yip et al., 2007; Tripathi et al., 2010; Namukwaya et al., 2012; L. Tripathy et al., 2014). Field trials conducted in Uganda with PFLP‐ and HRAP‐expressing bananas indicated that both genes are highly effective against bacterial wilt caused by Xanthomonas campestris (Fig. 2d), while no negative effect on yield or plant morphology was observed (J. N. Tripathi et al., 2014, 2017). In addition, a bioinformatic approach did not reveal any potential allergenicity or toxicity associated with either of these proteins (Jin et al., 2017). A combination of PFLP or HRAP did not have a synergistic or additive effect, yet resistance in bananas that express both genes may be more durable (Muwonge et al., 2016).
PFLP and HRAP are valuable tools to engineer resistance to bacterial pathogens. The lack of mechanistic insights makes it difficult to predict what the full and long‐term effect of these proteins could be on plant health and agronomic performance. Additionally, the effect of overexpression of these genes on the performance of fungal, viral or oomycete pathogens has not been investigated. However, the urgent need to find a solution against bacterial wilt of banana, combined with successful field trials in which no negative effects were observed, argue for a staggered deployment combined with detailed monitoring of performance of HRAP and PFLP in the field.
V..2. Detoxification enzymes
Plant enzymes that neutralise fungal toxins can play a role in plant defences, and transfer of their genes can improve resistance (Johal & Briggs, 1992). For example, Fusarium head blight is a significant fungal disease of wheat, as well as a source of mycotoxins in food that can poison humans and animals. Expression of a barley UDP‐glucosyltransferase in wheat metabolises the Fusarium graminearum toxin deoxynivalenol to a less toxic derivative, leading to reduced symptoms of Fusarium head blight in the field (Li et al., 2015). Similarly, oxalic acid is a virulence factor for Sclerotinia sclerotiorum, and transfer of oxalate oxidase from wheat produces significant resistance to Sclerotinia in many species, including peanut, tomato, potato, oilseed rape and soybean (Donaldson et al., 2001; Schneider et al., 2002; Hu et al., 2003; Dong et al., 2008; Walz et al., 2008; Partridge‐Telenko et al., 2011).
V..3. Wheat adult plant resistance genes
The adult plant resistance (APR) or ‘slow rusting’ genes of wheat are another class of potentially transferable resistance genes. These genes produce dominant partial resistance to multiple biotrophic pathogens in mature plants but not in seedlings. Several APR genes are known, but only two, Lr34 and Lr67, have been cloned. Lr34 encodes an ATP‐binding cassette (ABC) transporter with an unknown substrate. The resistance allele in the D genome contains two specific mutations and is dominant over the other native Lr34 alleles in hexaploid wheat (Krattinger et al., 2009). Wheat lines carrying Lr34 are partially resistant to multiple biotrophic pathogens including stem rust, stripe rust, leaf rust and powdery mildew. As a consequence, Lr34 has been widely used in breeding. Similarly, the wheat Lr67 resistance gene is a specific dominant allele of a hexose transporter that provides resistance to multiple rusts and powdery mildew. The protein encoded by the Lr67 resistance allele is inactive in sugar transport, so it is likely to have a dominant negative effect (Moore et al., 2015). Introduction of the Lr34 resistance allele by transformation into rice (Krattinger et al., 2016), barley (Risk et al., 2013), sorghum (Schnippenkoetter et al., 2017), maize (Sucher et al., 2017) and durum wheat (Rinaldo et al., 2017) and of Lr67 to barley (Milne et al., 2018) also produced resistance to biotrophic pathogens. As for mlo, the mechanism by which resistance is triggered by Lr34 and Lr67 is poorly understood, although it is likely to involve the induction of biotic or abiotic stress responses that precondition the host to limit pathogen growth. Expression of these genes in some heterologous plants, for example Lr34 in barley (Risk et al., 2013), has produced deleterious effects while, in other cases for example Lr34 in durum wheat (Rinaldo et al., 2017), no obvious negative phenotypes were noted. Given the likely dominant negative mode of action of these proteins, relative quantities of wild‐type vs mutant proteins may need to be optimised in each system. This situation may also suggest that these types of resistances are more applicable to polyploid crops than diploid crops.
VI. Intervention with antimicrobial peptides
Over the past decades, antimicrobial peptides and proteins have received a lot of attention as potential tools to create disease‐resistant crops. Antimicrobials are produced by organisms across all kingdoms and are a part of their innate immune systems (Brogden, 2005). Their activity is quite diverse and includes destruction of fungal cell walls, membrane permeabilisation, transcriptional inhibition and ribosome inactivation (Dempsey et al., 1998; van der Biezen, 2001; Brogden, 2005). Crops have been designed that express or over‐express (1) plant‐derived compounds such as pathogenesis‐related (PR) proteins and defensins that are normally produced during the plant's defence response, (2) antimicrobial proteins or peptides derived from microorganisms or animal cells, or (3) synthetic peptides designed based on sequences of existing antimicrobial compounds (Dempsey et al., 1998; van der Biezen, 2001; Castro & Fontes, 2005; Montesinos, 2007; Ali et al., 2018). Unlike the success of crops expressing anti‐insecticidal proteins from Bacillus thuringensis (Bt) that have been commercialised in different countries around the world, no crops expressing antimicrobial proteins have been commercialised to date (http://www.isaaa.org/). Development of crops engineered to express antimicrobials is challenging as antimicrobial proteins can often have phytotoxic effects, lead to over‐activation of stress responses, resulting in undesired phenotypes such as negative yield impacts, or have adverse effects on human or animal health (Montesinos, 2007). However, careful design or selection of suitable antimicrobials, followed by assessment of the agronomic performances of the engineered crops as well as of the potential impact on human or animal health may yet yield potential new solutions to crop diseases.
VII. Intervention using RNA interference
RNA interference (RNAi) was first discovered in plants as a mechanism to recognise and defend against nonself nucleic acids. In addition to this defensive role, RNAi is a fundamental mechanism for the regulation of endogenous genes. Initiation of RNAi production occurs after double‐stranded RNA or endogenous microRNAs are processed by Dicer‐like proteins. The resulting small interfering (si)RNAs can be recruited by Argonaute (AGO) proteins that recognise and cleave complementary strands of RNA, resulting in gene silencing. RNAi‐based resistance can be engineered against many viruses by expressing ‘hairpin’ structures, double‐stranded RNA molecules that contain viral sequences, or simply by overexpressing dysfunctional viral genes (reviewed in Rosa et al., 2018). Moreover, a single double‐stranded RNA molecule can be processed into a variety of siRNAs and thereby effectively target several viruses using one hairpin construct. While viruses fight back with proteins that inhibit the silencing machinery of plants, the use of RNAi has nonetheless been validated as a powerful strategy to control many plant viruses (e.g. Lawson et al., 1990; Tricoli et al., 1995; Ferreira et al., 2002; Bonfim et al., 2007; Scorza et al., 2013), as well as nematodes (Huang et al., 2006) and insects (Baum et al., 2007; Bolognesi et al., 2012). The impact of RNAi technology deployed as a GM solution against viruses is powerfully demonstrated by the ‘Rainbow papaya’ (Fig. 2e). Introduction of the Rainbow papaya averted a collapse of the Hawaiian papaya industry from a severe outbreak of Papaya ringspot virus in the 1990s (Ferreira et al., 2002; Gonsalves et al., 2004). Since its introduction, 20 years ago, the GM trait introduced into Rainbow papaya has provided a sustainable and effective control of the virus. A similar GM trait has been used to engineer virus‐resistant squash, which has an even longer commercial history (Tricoli et al., 1995).
Following on these successes, RNAi has been explored as a strategy to control fungi and oomycetes as well, and initial patent applications for methods to control fungi using RNAi were made as early as 2006 (Roberts et al., 2007). Fungicide target genes in the pathogen are obvious candidates for this approach, as disruption is known to be lethal. Indeed, significant effects have been observed in Fusarium species by targeting the cytochrome P450, family 51 (Cyp51) genes that underlie the azole fungicide target sterol 14α‐demethylase with host‐induced gene silencing (HIGS) (Koch et al., 2013). Additional pathogen genes that have been targeted include pathogenicity factors, developmental genes and genes involved in metabolism. HIGS of a Verticillium hydrophobin gene resulted in strong resistance to V. dahliae in cotton (Zhang et al., 2016). Similarly, HIGS targeted to a cellulose gene and a highly expressed conserved gene of Bremia lactucae resulted in high levels of resistance to this pathogen in lettuce (Govindarajulu et al., 2015). More often, however, HIGS experiments produce quantitative effects, for example when targeting rust fungi (Panwar et al., 2013, 2018; Yin & Hulbert, 2018) and virulence factors of V. dahliae in tomato (Song & Thomma, 2018). Overall, HIGS seems to be quite effective against some pathogens (Govindarajulu et al., 2015; Wang et al., 2016) but ineffective against others (Kettles et al., 2018). However, there appears to be an apparent disconnect between the earliest publications and patent filings on HIGS a decade ago and practical examples of HIGS deployed in the field. This may suggest that, although effects are observed, they are not strong enough to provide field level solutions to many pathogens.
Until recently, it was unclear how small RNA molecules would be exchanged between host and pathogens. However, compelling evidence has shown that small RNAs are delivered to fungal pathogens via extracellular vesicles (Cai et al., 2018). A better understanding of this process in diverse plant−pathogen interactions may allow us to better optimise HIGS strategies to provide field‐relevant levels of resistance. In short, RNAi appears to be a promising additional control strategy in the arsenal of plant breeders against at least some pathogens. The modular nature of RNAi is especially suitable to multiplexing via synthetic biology approaches. In addition, RNAi strategies may be particularly relevant when no pathogen resistance can be identified in natural populations.
VIII. Practical path to deployment
After a solution against a crop disease is discovered in the laboratory, it must pass several further hurdles. The first of these hurdles is that it also must be effective in the field without reducing agronomic performance. Subsequently, a commercial development process requires the generation and evaluation of a large number of transgenic lines to choose a transgenic event that only has the specific and intended modifications. Once this rigorous vetting procedure has been completed, introgression of this event into commercial cultivars and development of a regulatory dossier is initiated (reviewed by Prado et al., 2014).
A genetically modified crop must meet regulatory approval in each country where it will be grown or imported. Regulatory requirements in different countries are not standardised, and this situation increases the complexity of the task (Prado et al., 2014). Costs are often prohibitive, with estimates for international product deregulation between US$7M and US$35M (Kalaitzandonakes et al., 2007; Phillips McDougall, 2011) out of a total estimated product development cost of US$136M (Phillips McDougall, 2011). A cost−benefit calculation is fundamental to determining the commercial practicality of different disease‐resistance solutions. As an example, Box 1 summarises the data needed to deregulate a transgenic disease‐resistant crop in the USA. In the USA, the Food and Drug Administration (US FDA) assesses evidence for the safety of any added protein and the substantial equivalence of the crop to its nontransgenic equivalent. The Environmental Protection Agency (US EPA) assesses the consumer safety and lack of environmental impact of any ‘plant incorporated protectant’. The United States Department of Agriculture (USDA) assesses the potential of the new plant to be a weed or plant pest. The level of evidence required for any of these points is determined by the relative risk of the introduced trait. As mentioned above, the first immune receptor has been deregulated in the US: the Rpi‐vnt1 receptor with effectivity against late blight of potato. In this case, the US EPA and FDA accepted arguments that the protein is present at vanishingly small amounts, is not a potential allergen, and is similar to proteins already consumed (Clark et al., 2014; FDA, 2015; EPA, 2017). Therefore, animal feeding studies and extensive biochemical analyses on purified protein, which would have been extremely difficult in the case of an NLR (Bushey et al., 2014), were not required. However, a hypothetical product that expressed high levels of an artificial antimicrobial protein without a history of safe consumption would require more extensive evidence for safety and have concomitantly higher regulatory costs. Given the costs, time, and risk involved in developing and deregulating GM crops, only very high‐value traits on broad acreage crops are currently commercially viable targets. Only a handful of crop diseases, for example soybean rust and potato late blight, meet this economic threshold.
Box 1. Regulatory authorities and scope of regulation of bioengineered crops in the United States (EPA, 2019; FDA, 2019; USDA, 2019).
1.
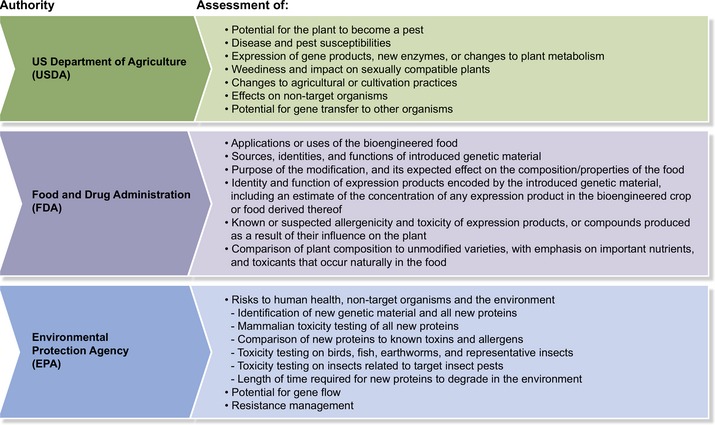
The USDA has recently released guidelines for the regulatory status of plants produced by gene editing, stating that certain classes of changes that could have been accomplished by traditional breeding are not subject to regulation if they were produced without plant pest sequences (that is not by Agrobacterium transformation). These changes include deletions, single nucleotide changes, and insertions of DNA from a sexually compatible relative (USDA, 2018). Although disease‐resistant food crops may still be subject to regulation by the FDA and EPA, this ruling drastically decreases the cost of bringing many types of disease‐resistant crops to market in the USA. By contrast with the scientifically based and pragmatic US guidelines, a recent ruling in the European Union states that all plants produced by gene editing are still subject to the same regulation as transgenic plants (Callaway, 2018). This effectively rules out the use of gene editing for any crop grown in or exported to Europe, robbing European growers of powerful solutions that could lead to more sustainable agriculture.
IX. Path to market in Africa
Africa is the continent where there is the greatest need and opportunity for agricultural growth, given the expected population growth and amount of unused arable land. Genetically modified or edited crops could play a significant role in helping Africa's agriculture to meet the needs of its growing population. Currently, adoption of GM crops in Africa is limited as they are commercially available only in Sudan (cotton) and South Africa (maize, cotton and soybean) (ISAAA, 2017). The adoption of GM crops in those countries has been successful, and acreage has increased steeply since they were first introduced (ISAAA, 2017). At present, several other countries in Africa have regulatory frameworks in place and are conducting field trials to prepare for general release when government policies allow. These countries are Ethiopia, Kenya, Uganda, Tanzania, Mozambique, Malawi, Swaziland, Cameroon, Nigeria, Ghana, and Burkina Faso (ISAAA, 2017). In Uganda, field trials are being conducted with potato expressing a stack of immune receptors providing protection against potato late blight disease, as well as bananas that are resistant to bacterial wilt (Fig. 2a; L. Tripathy et al., 2014; Ghislain et al., 2019).
Regulatory costs and time associated with the process can vary greatly and depend on the crop, the country, the developer and the inserted genes. Cost estimates for the development of a single GM variety (late blight resistant potato) in a developing country by a not‐for‐profit institution vary from US$1.4 million to US$1.6 million over 8–9 yr of review (Schiek et al., 2016). In many African countries, genome‐edited crops are expected to be regulated through the GM regulatory framework, similarly to the laws in Europe (ISAAA, 2017).
In Africa, as elsewhere, a second major barrier to advancing genetically engineered disease resistance is public concern about the safety of GM crops, despite an overwhelming body of evidence for the safety of these crops (National Academies of Science, Engineering and Medicine, 2016). This situation is unfortunate, given the potential for GM to address food losses caused by plant disease. This, in turn, can help to increase food production locally to accommodate a rapidly growing population. Africa's close ties to Europe influence its attitudes about GM crops, which are generally conservative and not based on scientific facts. Given the challenges that lie ahead, a shift to a scientific and pragmatic stance on the use of GM technology would be timely. The success of adoption of GM crops in Sudan and South Africa and the ongoing trials and safety assessments in other African countries might convince the public and politicians to open the doors to these molecular breeding approaches.
X. How to deploy resistance durably
It is clear that plant pathologists and breeders have uncovered a versatile arsenal of solutions to bring to bear against plant pathogens that offers great potential for global food security and sustainability. However, plant pathogens are highly adaptable and have much faster life cycles than their plant hosts, and therefore resistance conferred by most single genes or modes of action will be easily defeated. This reality is a key challenge for classical breeding, because durable resistance generally requires combinations of multiple resistance genes and quantitative trait loci (QTLs) at different locations in a genome. The problem is compounded by introgression of new resistances from non‐elite cultivars and wild relatives, which are often subject to yield loss due to linkage drag. Moreover, when a new disease or breeding goal appears, breeding for the new and existing traits becomes even more complex. Last, some important crop plants are notoriously difficult to breed, such as the tetraploid potato, sugarcane, and the (almost) sterile banana.
Genetic modification allows several dominant disease‐resistance genes to be introduced together in a single well‐characterised region of the genome overcoming many of these challenges. Critically, it is possible to introduce several dominant resistance traits into elite cultivars, polyploid crops, sterile plants and parental lines to be used in subsequent breeding efforts. Even if additional breeding is required, the key combined resistances will remain intact as a single locus. Unlike dominant resistance traits, recessive resistances present more of a challenge as they cannot be combined at a single locus, but genome editing in base breeding lines can accelerate the process of introducing these resistances.
Each resistance approach reviewed here took years of collaborative research effort. Many of the solutions were found by tapping into the large, but not unlimited, genetic diversity found in nature. It is therefore critical that thoughtful, durable deployment and stewardship of these hard‐won resources is achieved. The definition of durable resistance is fluid, and in each case is dependent on the strength of resistance required and the time that is needed for the resistance to hold (Brown, 2015). The question must be – ‘Does the combined solution work well enough and long enough?’
Given the requirement for clear resistance phenotypes in the field, many combined solutions will include the strong resistance conferred by NLR genes. Several factors influence the durability of combined NLR genes; major factors being the impact on virulence of the pathogen, the strength of the resistance, the exposure of a pathogen to an NLR, the total inoculum in the environment, and the capacity of the pathogen for sexual recombination (or lack thereof) (McDonald & Linde, 2002; Brown, 2015; Stam & McDonald, 2018). Although these factors are likely to play a role in the durability of each of the other resistance mechanisms reviewed here, the points of impact are likely to be different. Therefore, combining several modes of action will potentially result in resistance that is both effective and long lasting. For example, an NLR stack of Tm‐22 and Tm‐2 is predicted to be durable, as the two mutations in the movement protein of tomato mosaic virus that are required to overcome this resistance are predicted to disrupt function of the viral movement protein (Lanfermeijer et al., 2005). However, even greater durability may be achieved by combining these two genes with a different mode of action such as a hairpin RNAi construct.
Both the private and public sectors should pursue ever more durable ways of using the agricultural resources at hand. In the long run, a shift away from environmental and genetic uniformity in agricultural systems will result in a more durable status quo between crop and pathogens (McDonald & Stukenbrock, 2016). However, a critical assessment needs to be made on the timelines that this would take and at what cost to the efficiency and productivity of monoculture‐based agriculture this change would come. Compared with an average of 13 yr to deploy new transgenic lines, it can be debated whether an overhaul of the agricultural system before the population peak of 2050 is desirable or even possible. The pragmatic approach is to work with the best possible solutions that we have available today to ensure we will be in a position to deploy even better solutions later this century.
XI. Trends that shape the future
There are several trends that will affect the way in which we will design solutions and deploy traits. As exemplified in the paragraph above, it is important that several traits can be combined into one locus, preferably at a known location in the genome. This approach presents a unique technical challenge as cassettes need to be designed that contain multiple traits against one disease. An important trend therefore is the technical advance that is made to construct cassettes that contain multiple traits. Already, this is feasible to a certain extent, as has been demonstrated with gene stacks that contain three NLRs that recognise P. infestans (Ghislain et al., 2019, Fig. 2a) and a five R gene stack in wheat against wheat stem rust (Michael Ayliffe, personal communication). Although generating cassettes with multiple large inserts has traditionally been challenging, recent technical advances such as Gene Assembly in A grobacterium by Nucleic acid Transfer using Recombinase technologY (GAANTRY) has enabled the generation of stable cassettes with up to 10 genes with at total size of 28.5 kbp (Collier et al., 2018). Therefore, the generation of a cassette that can effectively target one or two key diseases is now technically feasible. As traits are dominant, combinations can subsequently be made via breeding. An example of what such a strategy may look like is the commercial maize line known by its trade name SmartStax™. To generate this line, four different biotech maize lines were crossed and resulted in the combination of six Bt genes and two herbicide tolerance genes, providing control for weeds and lepidopteran insects. Nonetheless, the ability to generate large stacks of combined traits will be a critical development over the coming years.
For gene stacks to be functional, the causal genes that underlie resistance must be identified. For many crops the reservoir of cloned resistance genes is still limited. However, the second trend is that new affordable sequencing technologies combined with bioinformatic approaches allow ever faster identification of causal resistance genes. This identification can now already be done, even in complex genomes such as wheat and potato and wild relatives of crops such as pigeonpea (Kawashima et al., 2016; Steuernagel et al., 2016; Witek et al., 2016). In addition, the ability to obtain a good quality reference genome assembly is now reduced to standard practice. With the ever‐dropping cost of sequencing and increase in processing power these approaches will soon become commonplace. This capability is important because it allows scientists to explore the rich genetic diversity of crop relatives. Nature has had millions of years to test and select resistance mechanisms, providing a wealth of potentially validated strategies. By making use of affordable, powerful sequencing capacity, wild germplasm can be mined for a distribution of resistance traits at the centre of origin. As many pathogens have co‐evolved with a wild progenitor species, a resistance trait against a specific disease that is overrepresented in the centre of origin of a wild progenitor may reflect that this trait is particularly effective with little cost to the host (Stam et al., 2017).
A third trend is the miniaturisation of sequencing technologies. Pathogen detection and analyses of the microbiome with a portable DNA sequencer has already successfully been executed (Hu et al., 2019). By the time that most solutions developed today will reach the field, such real‐time monitoring of pathogen populations in the field will be possible and likely standardised enough to be performed by growers or agronomists. A better understanding of pathogen population structure and dynamics may inform the best intervention strategy (genetic or other) against a given disease, for example via identification of key effectors.
No review would be complete without mentioning the fourth trend, which is the expanding use of genome‐editing tools. Genome editing can already be used to produce recessive traits, however as we set out in this review relatively few effective recessive targets have been identified. In addition, most targets that are simple knockouts have already been introduced via tilling, except in polyploid crops. Editing also provides the ability to precisely modify existing resistance genes or their expression, allowing the in situ conversion of a susceptible allele to a resistant one. Use of genome editing to integrate dominant resistance traits at a single locus will have even broader benefits, although it is important to note that this approach is already feasible using site‐specific recombination (SSR) systems (Srivastava & Thomson, 2016). However, the more efficient introduction of traits, or replacement of single traits in a stack may be accomplished via genome‐editing technologies (Rinaldo & Ayliffe, 2015). In addition, genes can be introduced anywhere in the genome. For instance, introducing new resistance genes next to already existing resistance loci could generate greater flexibility for the breeder. Gene stacks could be created and updated by precise addition and removal of genes. Finally, precise gene editing would introduce less ‘foreign’ DNA than Agrobacterium transformation, which may help deregulation in some countries. However, this is a legislative and not a scientific advantage.
A final trend that is developing in parallel is the rapid progress in protein structural biology techniques such as cryo‐EM. This will allow a better understanding of NLR and PRR function. Unlike the other trends described here, this trend has the capacity to be truly transformative in the way plant disease is tackled. The first step towards designing recognition specificity has already been made via the modification of HMA domains in NLRs with integrated domains (Maqbool et al., 2015). In addition, some NLR families can recognise multiple effectors from different pathogens via direct interaction (Saur et al., 2019). Unlike PRR proteins, how NLRs signal has been one of the long‐standing questions in plant pathology. However, two recent landmark publications have described the mechanism of activation for the A. thaliana HOP‐ACTIVATED RESISTANCE 1 (ZAR1) protein using cryo‐EM techniques (Wang et al., 2019a,2019b). All this information can be coupled to advances that are made in deep learning and synthetic biology, such those already used in drug discovery (Chen et al., 2018). This situation may enable scientists to develop recognition specificities for key pathogen effectors in the form of designer NLRs and PRRs.
XII. The time is now
We have in hand the means to thwart plant diseases that have plagued mankind since the dawn of agriculture. The genetic methods to combat disease reviewed here are more effective, environmentally friendly and safer than many current, common methods of control. We need to double our food production in 50 yr, and 70% of this increase needs to be achieved by adopting new technology. Therefore, we cannot ignore these approaches. However, almost none of the currently available GM solutions have reached growers, in large part due to consumer anxieties, even though the most ardent opponents of the technology ironically are the least informed about science and genetics (Fernbach et al., 2019), and the scientific consensus is that GM crops are as safe as those developed by classical breeding (National Academies of Sciences & Engineering and Medicine, 2016). Unfortunately, some legislators ignore the facts about GM safety and benefits, therefore blocking solutions that would benefit society broadly (Court of Justice of the European Union, 2018). Due to global trade, Europe's conservative attitude towards GM crops has affected agriculture worldwide, including those regions where GM crops could have great local benefits. To break this deadlock, interdisciplinary approaches that include social scientists need to be taken, and scientists should stay in dialogue with consumers and policy makers. It is up to this generation of scientists, seed companies, international agricultural organisations, and legislators to responsibly deploy the valuable and available genetic disease solutions to help reduce the footprint of agriculture on the planet while increasing its output.
Acknowledgements
This research was supported by grants from the 2Blades Foundation to The Sainsbury Laboratory. We acknowledge Yogesh Kumar Gupta, Diana Horvath, John Pitkin, Jonathan Jones and Sérgio Brommonschenkel for critically reading the manuscript. We acknowledge Yogesh Kumar Gupta for contributing the ggplot2 script for the world map in Fig. 1b. We acknowledge Matt Hayes and Casiana Vera Cruz for kindly providing pictures of Fusarium head blight and bacterial blight respectively in Fig. 1b. We acknowledge Marc Ghislain for kindly providing a picture of transgenic 3R potatoes in Fig. 2(a), Dr Sanju Kunwar and Dr Mathews Paret for kindly providing the field trial pictures with EF‐Tu receptor tomato in Fig. 2(b), Sophien Kamoun for kindly providing a picture of genome‐edited tomatoes in Fig. 2(c), and Dennis Gonsalves for kindly providing a picture of Rainbow papaya in Fig. 2(e). Fig 2(d) was reprinted by permission from Springer Nature Customer Service Centre GmbH: Springer Nature, Nature Biotechnology, Field trial of Xanthomonas wilt disease‐resistant bananas in East Africa (L. Tripathi et al., 2014). Fig. 2(e) was republished with permission of the American Phytopathological Society, from Ferreira et al. (2002). Permission conveyed through Copyright Clearance Center Inc.
References
- Albert I, Böhm H, Albert M, Feiler CE, Imkampe J, Wallmeroth N, Brancato C, Raaymakers TM, Oome S, Zhang H et al 2015. An RLP23‐SOBIR1‐BAK1 complex mediates NLP‐triggered immunity. Nature Plants 1: 15140. [DOI] [PubMed] [Google Scholar]
- Alexandratos N, Bruinsma J. 2012. World agriculture towards 2030/2050: the 2012 revision. ESA Working Paper No. 12 – 03 Rome, Italy: FAO. [Google Scholar]
- Ali S, Ganai BA, Kamili AN, Bhat AA, Mir ZA, Bhat JA, Tyagi A, Islam ST, Mushtaq M, Yadav P et al 2018. Pathogenesis‐related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiological Research 212–213: 29–37. [DOI] [PubMed] [Google Scholar]
- Bastet A, Robaglia C, Gallois JL. 2017. eIF4E resistance: natural variation should guide gene editing. Trends in Plant Science 22: 411–419. [DOI] [PubMed] [Google Scholar]
- Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, Johnson S, Plaetinck G, Munyikwa T, Pleau M et al 2007. Control of coleopteran insect pests through RNA interference. Nature Biotechnology 25: 1322–1326. [DOI] [PubMed] [Google Scholar]
- Bebber DP, Holmes T, Smith D, Gurr SJ. 2014. Economic and physical determinants of the global distributions of crop pests and pathogens. New Phytologist 202: 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berens ML, Berry HM, Mine A, Argueso CT, Tsuda K. 2017. Evolution of hormone signaling networks in plant defense. Annual Review of Phytopathology 55: 401–425. [DOI] [PubMed] [Google Scholar]
- Berger S, El Chazli Y, Babu AF, Coste AT. 2017. Azole resistance in Aspergillus fumigatus: a consequence of antifungal use in agriculture? Frontiers in Microbiology 8: 1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettgenhaeuser J, Gilbert B, Ayliffe M, Moscou MJ. 2014. Nonhost resistance to rust pathogens – a continuation of continua. Frontiers in Plant Science 5: 664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen EA. 2001. Quest for antimicrobial genes to engineer disease‐resistant crops. Trends in Plant Science 6: 89–91. [DOI] [PubMed] [Google Scholar]
- Bird DM, Jones JT, Opperman CH, Kikuchi T, Danchin EG. 2015. Signatures of adaptation to plant parasitism in nematode genomes. Parasitology 142: S71–S84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink PC, Wang X, McLellan H, He Q, Naqvi S, Armstrong MR, Zhang W, Hein I, Gilroy EM, Tian Z et al 2016. A Phytophthora infestans RXLR effector targets plant PP1c isoforms that promote late blight disease. Nature Communications 7: 10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognesi R, Ramaseshadri P, Anderson J, Bachman P, Clinton W, Flannagan R, Ilagan O, Lawrence C, Levine S, Moar W et al 2012. Characterizing the mechanism of action of double‐stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS ONE 7: e47534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton MD, Rivera‐Varas V, del Río Mendoza LE, Khan MFR, Secor GA. 2012. Efficacy of variable tetraconazole rates against Cercospora beticola isolates with differing in vitro sensitivities to DMI fungicides. Plant Disease 96: 1749–1756. [DOI] [PubMed] [Google Scholar]
- Bonfim K, Faria JC, Nogueira EOPL, Mendes E, Aragão FJL. 2007. RNAi‐mediated resistance to bean golden mosaic virus in genetically engineered common bean (Phaseolus vulgaris). Molecular Plant‐‐Microbe Interactions 20: 717–726. [DOI] [PubMed] [Google Scholar]
- Boschi F, Schvartzman C, Murchio S, Ferreira V, Siri MI, Galván GA, Smoker M, Stransfeld L, Zipfel C, Vilaró FL et al 2017. Enhanced bacterial wilt resistance in potato through expression of Arabidopsis EFR and introgression of quantitative resistance from Solanum commersonii . Frontiers in Plant Science 8: 1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F, Zipfel C. 2017. Function, discovery, and exploitation of plant pattern recognition receptors for broad‐spectrum disease resistance. Annual Review of Phytopathology 55: 257–286. [DOI] [PubMed] [Google Scholar]
- Bozkurt TO, Richardson A, Dagdas YF, Mongrand S, Kamoun S, Raffaele S. 2014. The plant membrane‐associated REMORIN1.3 accumulates in discrete perihaustorial domains and enhances susceptibility to Phytophthora infestans . Plant Physiology 165: 1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogden KA. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nature Reviews Microbiology 3: 238–250. [DOI] [PubMed] [Google Scholar]
- Brown JK. 2015. Durable resistance of crops to disease: a Darwinian perspective. Annual Review of Phytopathology 53: 513–539. [DOI] [PubMed] [Google Scholar]
- van den Burg HA, Harrison SJ, Joosten MHAJ, Vervoort J, de Wit PJGM. 2006. Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Molecular Plant‐‐Microbe Interactions 19: 1420–1430. [DOI] [PubMed] [Google Scholar]
- Bushey DF, Bannon GA, Delaney BF, Graser G, Hefford M, Jiang X, Lee TC, Madduri KM, Pariza M, Privalle LS et al 2014. Characteristics and safety assessment of intractable proteins in genetically modified crops. Regulatory Toxicology Pharmacology 69: 154–170. [DOI] [PubMed] [Google Scholar]
- Cai Q, Qiao L, Wang M, He B, Lin FM, Palmquist J, Huang SD, Jin H. 2018. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360: 1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. 2018. CRISPR plants now subject to tough GM laws in European Union. Nature 560: 16. [DOI] [PubMed] [Google Scholar]
- Cao Y, Liang Y, Tanaka K, Nguyen CT, Jedrzejczak RP, Joachimiak A, Stacey G. 2014. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin‐induced complex with related kinase CERK1. eLife 3: 1–19. doi: 10.7554/elife.03766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro MS, Fontes W. 2005. Plant defense and antimicrobial peptides. Protein and Peptide Letters 12: 13–18. [PubMed] [Google Scholar]
- Catanzariti AM, Lim GT, Jones DA. 2015. The tomato I‐3 gene: a novel gene for resistance to Fusarium wilt disease. New Phytologist 207: 106–118. [DOI] [PubMed] [Google Scholar]
- Chen C‐H, Lin H‐J, Feng T‐Y. 1998. An amphipathic protein from sweet pepper can dissociate harpinPss multimeric forms and intensify the harpinPss‐mediated hypersensitive response. Physiological and Molecular Plant Pathology 52: 139–149. [Google Scholar]
- Chen CH, Lin HJ, Ger MJ, Chow D, Feng TY. 2000. cDNA cloning and characterization of a plant protein that may be associated with the harpinPSS‐mediated hypersensitive response. Plant Molecular Biology 43: 429–438. [DOI] [PubMed] [Google Scholar]
- Chen H, Engkvist O, Wang Y, Olivecrona M, Blaschke T. 2018. The rise of deep learning in drug discovery. Drug Discovery Today 23: 1241–1250. [DOI] [PubMed] [Google Scholar]
- Choi MS, Kim W, Lee C, Oh CS. 2013. Harpins, multifunctional proteins secreted by Gram‐negative plant‐pathogenic bacteria. Molecular Plant‐‐Microbe Interactions 26: 1115–1122. [DOI] [PubMed] [Google Scholar]
- Clark P, Habig J, Ye J, Collinge S. 2014. Petition for determination of nonregulated status for Innate™ potatoes with late blight resistance, low acrylamide potential, reduced black spot, and lowered reducing sugars: Russet Burbank event W8 . USDA Petition no. 14‐093‐01p. [WWW document] URL https://www.aphis.usda.gov/brs/aphisdocs/14_09301p.pdf [accessed 15 January 2019].
- Clausen M, Kräuter R, Schachermayr G, Potrykus I, Sautter C. 2000. Antifungal activity of a virally encoded gene in transgenic wheat. Nature Biotechnology 18: 446–449. [DOI] [PubMed] [Google Scholar]
- Collier R, Thomson JG, Thilmony R. 2018. A versatile and robust Agrobacterium‐based gene stacking system generates high‐quality transgenic Arabidopsis plants. The Plant Journal 95: 573–583. [DOI] [PubMed] [Google Scholar]
- Cook DE, Mesarich CH, Thomma BP. 2015. Understanding plant immunity as a surveillance system to detect invasion. Annual Review of Phytopathology 53: 541–563. [DOI] [PubMed] [Google Scholar]
- Court of Justice of the European Union . 2018. Organisms obtained by mutagenensis are GMOs and are, in principle, subject to the obligations laid down by the GMO Directive . Press release no. 111/18. [WWW document] URL https://curia.europa.eu/jcms/upload/docs/application/pdf/2018-07/cp180111en.pdf [accessed 17 January 2019].
- van Damme M, Huibers RP, Elberse J, van den Ackerveken G. 2008. Arabidopsis DMR6 encodes a putative 2OG‐Fe(II) oxygenase that is defense‐associated but required for susceptibility to downy mildew. The Plant Journal 54: 785–793. [DOI] [PubMed] [Google Scholar]
- Dayakar BV, Lin HJ, Chen CH, Ger MJ, Lee BH, Pai CH, Chow D, Huang HE, Hwang SY, Chung MC et al 2003. Ferredoxin from sweet pepper (Capsicum annuum L.) intensifying harpinpss‐mediated hypersensitive response shows an enhanced production of active oxygen species (AOS). Plant Molecular Biology 51: 913–924. [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer D, Xu J, Höfte M. 2014. Making sense of hormone‐mediated defense networking: from rice to Arabidopsis . Frontiers in Plant Science 5. doi: 10.3389/fpls.2014.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey DA, Silva H, Klessig DF. 1998. Engineering disease and pest resistance in plants. Trends in Microbiology 6: 54–61. [DOI] [PubMed] [Google Scholar]
- Deng W, Marshall NC, Rowland JL, McCoy JM, Worrall LJ, Santos AS, Strynadka NCJ, Finlay BB. 2017. Assembly, structure, function and regulation of type III secretion systems. Nature Reviews Microbiology 15: 323–337. [DOI] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. 2010. Plant immunity: towards an integrated view of plant−pathogen interactions. Nature Reviews Genetics 11: 539–548. [DOI] [PubMed] [Google Scholar]
- Donaldson PA, Anderson T, Lane BG, Davidson AL, Simmonds DH. 2001. Soybean plants expressing an active oligomeric oxalate oxidase from the wheat gf‐2.8 (germin) gene are resistant to the oxalate‐secreting pathogen Sclerotina sclerotiorum . Physiological and Molecular Plant Pathology 59: 297–307. [Google Scholar]
- Dong X, Ji R, Guo X, Foster SJ, Chen H, Dong C, Liu Y, Hu Q, Liu S. 2008. Expressing a gene encoding wheat oxalate oxidase enhances resistance to Sclerotinia sclerotiorum in oilseed rape (Brassica napus). Planta 228: 331–340. [DOI] [PubMed] [Google Scholar]
- EPA . 2017. Final registration decision for the new active ingredient and food use of VNTl protein and the genetic material necessary for its production in potatoes . EPA‐HQ‐OPP‐2016‐0036 [WWW document] URL https://www.regulations.gov/document?D=epa-hq-opp-2016-0036-0013 [accessed 15 January 2019].
- EPA . 2019. EPA's regulation of biotechnology for use in pest management . [WWW document] URL https://www.epa.gov/regulation-biotechnology-under-tsca-and-fifra/epas-regulation-biotechnology-use-pest-management [accessed 5 February 2019].
- Espada M, Silva AC, Eves van den Akker S, Cock PJ, Mota M, Jones JT. 2016. Identification and characterization of parasitism genes from the pinewood nematode Bursaphelenchus xylophilus reveals a multilayered detoxification strategy. Molecular Plant Pathology 17: 286–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse HP, Bolton MD, Stergiopoulos I, de Wit PJGM, Thomma BPHJ. 2007. The chitin‐binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Molecular Plant‐‐Microbe Interactions 20: 1092–1101. [DOI] [PubMed] [Google Scholar]
- FAO, IFAD, UNICEF, WFP, WHO . 2018. The state of food security and nutrition in the world 2018. Building climate resilience for food security and nutrition Rome: FAO. [Google Scholar]
- Faris JD, Zhang Z, Lu H, Lu S, Reddy L, Cloutier S, Fellers JP, Meinhardt SW, Rasmussen JB, Xu SS et al 2010. A unique wheat disease resistance‐like gene governs effector‐triggered susceptibility to necrotrophic pathogens. Proceedings of the National Academy of Sciences, USA 107: 13544–13549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner C, Petutschnig E, Benitez‐Alfonso Y, Beck M, Robatzek S, Lipka V, Maule AJ. 2013. LYM2‐dependent chitin perception limits molecular flux via plasmodesmata. Proceedings of the National Academy of Sciences, USA 110: 9166–9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA . 2015. Biotechnology consultation – Note to file BNF no. 146 . [WWW document] URL https://www.fda.gov/Food/IngredientsPackagingLabeling/GEPlants/Submissions/ucm486681.htm [accessed 15 January 2019].
- FDA . 2019. Consultation procedures under FDA's 1992 Statement of Policy – Foods derived from new plant varieties . [WWW document] URL https://www.fda.gov/food/ingredients-additives-gras-packaging-guidance-documents-regulatory-information/consultation-procedures-under-fdas-1992-statement-policy-foods-derived-new-plant-varieties [accessed 12 May 2019].
- Fernbach PM, Light N, Scott SE, Inbar Y, Rozin P. 2019. Extreme opponents of genetically modified foods know the least but think they know the most. Nature Human Behaviour 3: 251–256. doi: 10.1038/s41562-018-0520-3. [DOI] [PubMed] [Google Scholar]
- Ferreira SA, Pitz KY, Manshardt R, Zee F, Fitch M, Gonsalves D. 2002. Virus coat protein transgenic papaya provides practical control of papaya ringspot virus in Hawaii. Plant Disease 86: 101–105. [DOI] [PubMed] [Google Scholar]
- Fitch MMM, Manshardt RM, Gonsalves D, Slightom JL, Sanford JC. 1992. Virus resistant papaya plants derived from tissues bombarded with the coat protein gene of papaya ringspot virus. Bio/Technology 10: 1466–1472. [Google Scholar]
- Foster SJ, Park TH, Pel M, Brigneti G, Sliwka J, Jagger L, van der Vossen E, Jones JDG. 2009. Rpi‐vnt1.1, a Tm‐2 2 homolog from Solanum venturii, confers resistance to potato late blight. Molecular Plant–Microbe Interactions 22: 589–600. [DOI] [PubMed] [Google Scholar]
- Galán JE, Lara‐Tejero M, Marlovits TC, Wagner S. 2014. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annual Review of Microbiology 68: 415–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger F, Bengtsson J, Berendse F, Weisser WW, Emmerson M, Morales MB, Ceryngier P, Liira J, Tscharntke T, Winqvist C et al 2010. Persistent negative effects of pesticides on biodiversity and biological control potential on European farmland. Basic and Applied Ecology 11: 97–105. [Google Scholar]
- Ger MJ, Chen CH, Hwang SY, Huang HE, Podile AR, Dayakar BV, Feng TY. 2002. Constitutive expression of hrap gene in transgenic tobacco plant enhances resistance against virulent bacterial pathogens by induction of a hypersensitive response. Molecular Plant‐‐Microbe Interactions 15: 764–773. [DOI] [PubMed] [Google Scholar]
- Ger MJ, Louh GY, Lin YH, Feng TY, Huang HE. 2014. Ectopically expressed sweet pepper ferredoxin PFLP enhances disease resistance to Pectobacterium carotovorum subsp. carotovorum affected by harpin and protease‐mediated hypersensitive response in Arabidopsis. Molecular Plant Pathology 15: 892–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M, Byarugaba AA, Magembe E, Njoroge A, Rivera C, Román ML, Tovar JC, Gamboa S, Forbes GA, Kreuze JF et al 2019. Stacking three late blight resistance genes from wild species directly into African highland potato varieties confers complete field resistance to local blight races. Plant Biotechnology Journal 17: 1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy CV, Seixas CDS, Soares RM, Marcelino‐Guimarães FC, Meyer MC, Costamilan LM. 2016. Asian soybean rust in Brazil: past, present, and future. Pesquisa Agropecuária Brasileira 51: 407–421. [Google Scholar]
- Gómez‐Gómez L, Boller T. 2000. FLS2: an LRR receptor‐like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis . Molecular Cell 5: 1003–1011. [DOI] [PubMed] [Google Scholar]
- Gonsalves C, Lee DR, Gonsalves D. 2004. Transgenic virus‐resistant papaya: the Hawaiian ‘Rainbow’ was rapidly adopted by farmers and is of major importance in Hawaii today. APSnet Features. doi: 10.1094/APSnetFeature-2004-0804. [DOI] [Google Scholar]
- Govindarajulu M, Epstein L, Wroblewski T, Michelmore RW. 2015. Host‐induced gene silencing inhibits the biotrophic pathogen causing downy mildew of lettuce. Plant Biotechnology Journal 13: 875–883. [DOI] [PubMed] [Google Scholar]
- He P, Chintamanani S, Chen Z, Zhu L, Kunkel BN, Alfano JR, Tang X, Zhou JM. 2004. Activation of a COI1‐dependent pathway in Arabidopsis by Pseudomonas syringae type III effectors and coronatine. The Plant Journal 37: 589–602. [DOI] [PubMed] [Google Scholar]
- Hind SR, Strickler SR, Boyle PC, Dunham DM, Bao Z, O'Doherty IM, Baccile JA, Hoki JS, Viox EG, Clarke CR et al 2016. Tomato receptor FLAGELLIN‐SENSING 3 binds flgII‐28 and activates the plant immune system. Nature Plants 2: 16128. [DOI] [PubMed] [Google Scholar]
- Horvath DM, Stall RE, Jones JB, Pauly MH, Vallad GE, Dahlbeck D, Staskawicz BJ, Scott JW. 2012. Transgenic resistance confers effective field level control of bacterial spot disease in tomato. PLoS ONE 7: e42036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Bidney DL, Yalpani N, Duvick JP, Crasta O, Folkerts O, Lu G. 2003. Overexpression of a gene encoding hydrogen peroxide‐generating oxalate oxidase evokes defense responses in sunflower. Plant Physiology 133: 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Green G, Milgate A, Stone E, Rathjen J, Schwessinger B. 2019. Pathogen detection and microbiome analysis of infected wheat using a portable DNA sequencer. BioRxiv. doi: 10.1101/429878. [DOI] [Google Scholar]
- Huang G, Allen R, Davis EL, Baum TJ, Hussey RS. 2006. Engineering broad root‐knot resistance in transgenic plants by RNAi silencing of a conserved and essential root‐knot nematode parasitism gene. Proceedings of the National Academy of Sciences, USA 103: 14302–14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HE, Ger MJ, Chen CY, Pandey AK, Yip MK, Chou HW, Feng TY. 2007. Disease resistance to bacterial pathogens affected by the amount of ferredoxin‐I protein in plants. Molecular Plant Pathology 8: 129–137. [DOI] [PubMed] [Google Scholar]
- Huang HE, Ger MJ, Yip MK, Chen CY, Pandey AK, Feng TY. 2004. A hypersensitive response was induced by virulent bacteria in transgenic tobacco plants overexpressing a plant ferredoxin‐like protein (PFLP). Physiological and Molecular Plant Pathology 64: 103–110. [Google Scholar]
- Hummel AW, Doyle EL, Bogdanove AJ. 2012. Addition of transcription activator‐like effector binding sites to a pathogen strain‐specific rice bacterial blight resistance gene makes it effective against additional strains and against bacterial leaf streak. New Phytologist 195: 883–893. [DOI] [PubMed] [Google Scholar]
- Iliescu EC, Balogh M, Szabo Z, Kiss GB. 2013. Identification of a Xanthomonas euvesicatoria resistance gene from pepper (Capsicum annuum) and method for generating plants with resistance . International (PCT) patent application WO/2014/068346A2, filed October 30, 2013.
- ISAAA . 2017. Global status of commercialized biotech/GM crops in 2017: Biotech crop adoption surges as economic benefits accumulate in 22 years . ISAAA Brief no. 53. Ithaca, NY: ISAAA. [Google Scholar]
- Islam MT, Croll D, Gladieux P, Soanes DM, Persoons A, Bhattacharjee P, Hossain MS, Gupta DR, Rahman MM, Mahboob MG et al 2016. Emergence of wheat blast in Bangladesh was caused by a South American lineage of Magnaporthe oryzae . BMC Biology 2016: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen C, von Wettstein D, Schäfer W, Kogel KH, Felk A, Maier FJ. 2005. Infection patterns in barley and wheat spikes inoculated with wild‐type and trichodiene synthase gene disrupted Fusarium graminearum . Proceedings of the National Academy of Sciences, USA 102: 16892–16897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosch B, Kogel KH, Schaffrath U. 1999. The ambivalence of the barley Mlo locus: mutations conferring resistance against powdery mildew (Blumeria graminis f. sp. hordei) enhance susceptibility to the rice blast fungus Magnaporthe grisea . Molecular Plant–Microbe Interactions 12: 508–514. [Google Scholar]
- Jha S, Chattoo BB. 2010. Expression of a plant defensin in rice confers resistance to fungal phytopathogens. Transgenic Research 19: 373–384. [DOI] [PubMed] [Google Scholar]
- Jia H, Zhang Y, Orbović V, Xu J, White FF, Jones JB, Wang N. 2017. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnology Journal 15: 817–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Goodman RE, Tetteh AO, Lu M, Tripathi L. 2017. Bioinformatics analysis to assess potential risks of allergenicity and toxicity of HRAP and PFLP proteins in genetically modified bananas resistant to Xanthomonas wilt disease. Food and Chemical Toxicology 109: 81–89. [DOI] [PubMed] [Google Scholar]
- Johal GS, Briggs SP. 1992. Reductase activity encoded by the HM1 disease resistance gene in maize. Science 258: 985–987. [DOI] [PubMed] [Google Scholar]
- Jones JB, Minsavage GV, Roberts PD, Johnson RR, Kousik CS, Subramanian S, Stall RE. 2002. A non‐hypersensitive resistance in pepper to the bacterial spot pathogen is associated with two recessive genes. Phytopathology 92: 273–277. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Jones JD, Vance RE, Dangl JL. 2016. Intracellular innate immune surveillance devices in plants and animals. Science 354: aaf6395. [DOI] [PubMed] [Google Scholar]
- Kalaitzandonakes N, Alston JM, Bradford KJ. 2007. Compliance costs for regulatory approval of new biotech crops. Nature Biotechnology 25: 509–511. [DOI] [PubMed] [Google Scholar]
- Kawashima CG, Guimarães GA, Nogueira SR, MacLean D, Cook DR, Steuernagel B, Baek J, Bouyioukos C, Do Vale Araújo Melo B, Tristão G et al 2016. A pigeonpea gene confers resistance to Asian soybean rust in soybean. Nature Biotechnology 34: 661–665. [DOI] [PubMed] [Google Scholar]
- Kettles GJ, Hofinger BJ, Hu P, Bayon C, Rudd JJ, Balmer D, Courbot M, Hammond‐Kosack KE, Scalliet G, Kanyuka K. 2018. Analysis of small RNA silencing in Zymoseptoria tritici – wheat interactions. BioRxiv. doi: 10.1101/501650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Qi D, Ashfield T, Helm M, Innes RW. 2016. Using decoys to expand the recognition specificity of a plant disease resistance protein. Science 351: 684–687. [DOI] [PubMed] [Google Scholar]
- Koch A, Kumar N, Weber L, Keller H, Imani J, Kogel KH. 2013. Host‐induced gene silencing of cytochrome P450 lanosterol C14α‐demethylase‐encoding genes confers strong resistance to Fusarium species. Proceedings of the National Academy of Sciences, USA 110: 19324–19329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta‐Espino J, McFadden H, Bossolini E, Selter LL, Keller B. 2009. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323: 1360–1363. [DOI] [PubMed] [Google Scholar]
- Krattinger SG, Sucher J, Selter LL, Chauhan H, Zhou B, Tang M, Upadhyaya NM, Mieulet D, Guiderdoni E, Weidenbach D et al 2016. The wheat durable, multipathogen resistance gene Lr34 confers partial blast resistance in rice. Plant Biotechnology Journal 14: 1261–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunwar S, Iriarte F, Fan Q, Evaristo da Silva E, Ritchie L, Nguyen NS, Freeman JH, Stall RE, Jones JB, Minsavage GV et al 2018. Transgenic expression of EFR and Bs2 genes for field management of bacterial wilt and bacterial spot of tomato. Phytopathology 108: 1402–1411. [DOI] [PubMed] [Google Scholar]
- Kusch S, Panstruga R. 2017. mlo‐based resistance: an apparently universal ‘weapon’ to defeat powdery mildew disease. Molecular Plant‐‐Microbe Interactions 30: 179–189. [DOI] [PubMed] [Google Scholar]
- Lacombe S, Rougon‐Cardoso A, Sherwood E, Peeters N, Dahlbeck D, van Esse HP, Smoker M, Rallapalli G, Thomma BP, Staskawicz B et al 2010. Interfamily transfer of a plant pattern‐recognition receptor confers broad‐spectrum bacterial resistance. Nature Biotechnology 28: 365–369. [DOI] [PubMed] [Google Scholar]
- Lamichhane JR, Dachbrodt‐Saaydeh S, Kudsk P, Messéan A. 2015. Toward a reduced reliance on conventional pesticides in European agriculture. Plant Disease 100: 10–24. [DOI] [PubMed] [Google Scholar]
- Lanfermeijer FC, Warmink J, Hille J. 2005. The products of the broken Tm‐2 and the durable Tm‐2 2 resistance genes from tomato differ in four amino acids. Journal of Experimental Botany 56: 2925–2933. [DOI] [PubMed] [Google Scholar]
- Lawson C, Kaniewski W, Haley L, Rozman R, Newell C, Sanders P, Tumer NE. 1990. Engineering resistance to mixed virus infection in a commercial potato cultivar: resistance to potato virus X and potato virus Y in transgenic Russet Burbank. Bio/Technology 8: 127–134. [DOI] [PubMed] [Google Scholar]
- Lawson EC, Weiss JD, Thomas PE, Kaniewski WK. 2001. NewLeaf Plus® Russet Burbank potatoes: replicase‐mediated resistance to potato leafroll virus. Molecular Breeding 7: 1–12. [Google Scholar]
- Le Roux C, Huet G, Jauneau A, Camborde L, Trémousaygue D, Kraut A, Zhou B, Levaillant M, Adachi H, Yoshioka H et al 2015. A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 161: 1074–1088. [DOI] [PubMed] [Google Scholar]
- Lellis AD, Kasschau KD, Whitham SA, Carrington JC. 2002. Loss‐of‐susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Current Biology 12: 1046–1051. [DOI] [PubMed] [Google Scholar]
- Li T, Liu B, Spalding MH, Weeks DP, Yang B. 2012. High‐efficiency TALEN‐based gene editing produces disease‐resistant rice. Nature Biotechnology 30: 390–392. [DOI] [PubMed] [Google Scholar]
- Li X, Shin S, Heinen S, Dill‐Macky R, Berthiller F, Nersesian N, Clemente T, McCormick S, Muehlbauer GJ. 2015. Transgenic wheat expressing a barley UDP‐glucosyltransferase detoxifies deoxynivalenol and provides high levels of resistance to Fusarium graminearum . Molecular Plant‐‐Microbe Interactions 28: 1237–1246. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhou M, Zhang Z, Ren L, Du L, Zhang B, Xu H, Xin Z. 2011. Expression of a radish defensin in transgenic wheat confers increased resistance to Fusarium graminearum and Rhizoctonia cerealis . Functional and Integrative Genomics 11: 63–70. [DOI] [PubMed] [Google Scholar]
- Liau CH, Lu JC, Prasad V, Hsiao HH, You SJ, Lee JT, Yang NS, Huang HE, Feng TY, Chen WH et al 2003. The sweet pepper ferredoxin‐like protein (pflp) conferred resistance against soft rot disease in Oncidium orchid. Transgenic Research 12: 329–336. [DOI] [PubMed] [Google Scholar]
- Lin H‐J, Cheng H‐Y, Chen C‐H, Huang H‐C, Feng T‐Y. 1997. Plant amphipathic proteins delay the hypersensitive response caused by harpinPss and Pseudomonas syringae pv. syringae . Physiological and Molecular Plant Pathology 51: 367–376. [Google Scholar]
- Lo Presti L, Kahmann R. 2017. How filamentous plant pathogen effectors are translocated to host cells. Current Opinion in Plant Biology 38: 19–24. [DOI] [PubMed] [Google Scholar]
- Maqbool A, Saitoh H, Franceschetti M, Stevenson CE, Uemura A, Kanzaki H, Kamoun S, Terauchi R, Banfield MJ. 2015. Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. eLife 4: e08709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald BA, Linde C. 2002. Pathogen population genetics, evolutionary potential, and durable resistance. Annual Review of Phytopathology 40: 349–379. [DOI] [PubMed] [Google Scholar]
- McDonald BA, Stukenbrock EH. 2016. Rapid emergence of pathogens in agro‐ecosystems: global threats to agricultural sustainability and food security. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 371: 20160026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougall P. 2011. The cost and time involved in the discovery, development and authorisation of a new plant biotechnology derived trait . [WWW document] URL https://croplife-r9qnrxt3qxgjra4.netdna-ssl.com/wp-content/uploads/2014/04/Getting-a-Biotech-Crop-to-Market-Phillips-McDougall-Study.pdf [accessed 15 January 2019].
- McGrann GR, Stavrinides A, Russell J, Corbitt MM, Booth A, Chartrain L, Thomas WT, Brown JK. 2014. A trade off between mlo resistance to powdery mildew and increased susceptibility of barley to a newly important disease, Ramularia leaf spot. Journal of Experimental Botany 65: 1025–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne RJ, Dibley KE, Schnippenkoetter WH, Mascher M, Lui AC, Wang L, Lo C, Ashton AR, Ryan PR, Lagudah E. 2018. The wheat Lr67 gene of the Sugar Transport Protein 13 family confers multipathogen resistance in barley. Plant Physiology 179: 1285–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. 2007. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proceedings of the National Academy of Sciences, USA 104: 19613–19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos E. 2007. Antimicrobial peptides and plant disease control. FEMS Microbiology Letters 270: 1–11. [DOI] [PubMed] [Google Scholar]
- Moore JW, Herrera‐Foessel S, Lan C, Schnippenkoetter W, Ayliffe M, Huerta‐Espino J, Lillemo M, Viccars L, Milne R, Periyannan S et al 2015. A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nature Genetics 47: 1494–1498. [DOI] [PubMed] [Google Scholar]
- Murphy F, He Q, Armstrong M, Giuliani LM, Boevink PC, Zhang W, Tian Z, Birch PRJ, Gilroy EM. 2018. The potato MAP3K StVIK is required for the Phytophthora infestans RXLR effector Pi17316 to promote disease. Plant Physiology 177: 398–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muwonge A, Kunert K, Tripathi L. 2016. Expressing stacked HRAP and PFLP genes in transgenic banana has no synergistic effect on resistance to Xanthomonas wilt disease. South African Journal of Botany 104: 125–133. [Google Scholar]
- Namukwaya B, Tripathi L, Tripathi JN, Arinaitwe G, Mukasa SB, Tushemereirwe WK. 2012. Transgenic banana expressing Pflp gene confers enhanced resistance to Xanthomonas wilt disease. Transgenic Research 21: 855–865. [DOI] [PubMed] [Google Scholar]
- National Academies of Sciences, Engineering and Medicine . 2016. Genetically engineered crops: experiences and prospects. Washington, DC, USA: The National Academies Press. [PubMed] [Google Scholar]
- Nekrasov V, Wang C, Win J, Lanz C, Weigel D, Kamoun S. 2017. Rapid generation of a transgene‐free powdery mildew resistant tomato by genome deletion. Scientific Reports 7: 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise V, German‐Retana S, Sanjuán R, Dubrana MP, Mazier M, Maisonneuve B, Candresse T, Caranta C, LeGall O. 2003. The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the Potyvirus Lettuce mosaic virus . Plant Physiology 132: 1272–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberger T, Lipka V. 2005. Non‐host resistance in plants: new insights into an old phenomenon. Molecular Plant Pathology 6: 335–345. [DOI] [PubMed] [Google Scholar]
- Oerke E‐C, Dehne H‐W. 2004. Safeguarding production—losses in major crops and the role of crop protection. Crop Protection 23: 275–285. [Google Scholar]
- Osusky M, Zhou G, Osuska L, Hancock RE, Kay WW, Misra S. 2000. Transgenic plants expressing cationic peptide chimeras exhibit broad‐spectrum resistance to phytopathogens. Nature Biotechnology 18: 1162–1166. [DOI] [PubMed] [Google Scholar]
- Pandey AK, Ger MJ, Huang HE, Yip MK, Zeng J, Feng TY. 2005. Expression of the hypersensitive response‐assisting protein in Arabidopsis results in harpin‐dependent hypersensitive cell death in response to Erwinia carotovora . Plant Molecular Biology 59: 771–780. [DOI] [PubMed] [Google Scholar]
- Panstruga R, Dodds PN. 2009. Terrific protein traffic: the mystery of effector protein delivery by filamentous plant pathogens. Science 324: 748–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panwar V, Jordan M, McCallum B, Bakkeren G. 2018. Host‐induced silencing of essential genes in Puccinia triticina through transgenic expression of RNAi sequences reduces severity of leaf rust infection in wheat. Plant Biotechnology Journal 16: 1013–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panwar V, McCallum B, Bakkeren G. 2013. Host‐induced gene silencing of wheat leaf rust fungus Puccinia triticina pathogenicity genes mediated by the barley stripe mosaic virus. Plant Molecular Biology 81: 595–608. [DOI] [PubMed] [Google Scholar]
- Pardey PG, Beddow JM, Kriticos DJ, Hurley TM, Park RF, Duveiller E, Sutherst RW, Burdon JJ, Hodson D. 2013. Right‐sizing stem‐rust research. Science 340: 147–148. [DOI] [PubMed] [Google Scholar]
- Partridge‐Telenko DE, Hu J, Livingstone DM, Shew BB, Phipps PM, Grabau EA. 2011. Sclerotinia blight resistance in Virginia‐type peanut transformed with a barley oxalate oxidase gene. Phytopathology 101: 786–793. [DOI] [PubMed] [Google Scholar]
- Prado JR, Segers G, Voelker T, Carson D, Dobert R, Phillips J, Cook K, Cornejo C, Monken J, Grapes L et al 2014. Genetically engineered crops: from idea to product. Annual Review of Plant Biology 65: 769–790. [DOI] [PubMed] [Google Scholar]
- Pruitt RN, Schwessinger B, Joe A, Thomas N, Liu F, Albert M, Robinson MR, Chan LJ, Luu DD, Chen H et al 2015. The rice immune receptor XA21 recognizes a tyrosine‐sulfated protein from a Gram‐negative bacterium. Science Advances 1: e1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quijano CD, Wichmann F, Schlaich T, Fammartino A, Huckauf J, Schmidt K, Unger C, Broer I, Sautter C. 2016. KP4 to control Ustilago tritici in wheat: enhanced greenhouse resistance to loose smut and changes in transcript abundance of pathogen related genes in infected KP4 plants. Biotechnology Reports (Amsterdam) 11: 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, Meijer M, Houterman PM, van der Does HC, Cornelissen BJ. 2005. Fusarium oxysporum evades I‐3‐mediated resistance without altering the matching avirulence gene. Molecular Plant‐‐Microbe Interactions 18: 15–23. [DOI] [PubMed] [Google Scholar]
- Rinaldo AR, Ayliffe M. 2015. Gene targeting and editing in crop plants: a new era of precision opportunities. Molecular Breeding 35: doi: 10.1007/s11032-015-0210-z. [DOI] [Google Scholar]
- Rinaldo A, Gilbert B, Boni R, Krattinger SG, Singh D, Park RF, Lagudah E, Ayliffe M. 2017. The Lr34 adult plant rust resistance gene provides seedling resistance in durum wheat without senescence. Plant Biotechnology Journal 15: 894–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risk JM, Selter LL, Chauhan H, Krattinger SG, Kumlehn J, Hensel G, Viccars LA, Richardson TM, Buesing G, Troller A et al 2013. The wheat Lr34 gene provides resistance against multiple fungal pathogens in barley. Plant Biotechnology Journal 11: 847–854. [DOI] [PubMed] [Google Scholar]
- Roberts JK, Pitkin JW, Adams TH. 2007. In planta RNAi control of fungi . US patent application 2008‐0022423 A1, filed February 1, 2007.
- Robert‐Seilaniantz A, Grant M, Jones JD. 2011. Hormone crosstalk in plant disease and defense: more than just jasmonate‐salicylate antagonism. Annual Review of Phytopathology 49: 317–343. [DOI] [PubMed] [Google Scholar]
- Rosa C, Kuo YW, Wuriyanghan H, Falk BW. 2018. RNA interference mechanisms and applications in plant pathology. Annual Review of Phytopathology 56: 581–610. [DOI] [PubMed] [Google Scholar]
- Ruffel S, Dussault MH, Palloix A, Moury B, Bendahmane A, Robaglia C, Caranta C. 2002. A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). The Plant Journal 32: 1067–1075. [DOI] [PubMed] [Google Scholar]
- Ruffel S, Gallois JL, Lesage ML, Caranta C. 2005. The recessive potyvirus resistance gene pot‐1 is the tomato orthologue of the pepper pvr2‐eIF4E gene. Molecular Genetics and Genomics 274: 346–353. [DOI] [PubMed] [Google Scholar]
- Rustagi A, Kumar D, Shekhar S, Yusuf MA, Misra S, Sarin NB. 2014. Transgenic Brassica juncea plants expressing MsrA1, a synthetic cationic antimicrobial peptide, exhibit resistance to fungal phytopathogens. Molecular Biotechnology 56: 535–545. [DOI] [PubMed] [Google Scholar]
- Sarris PF, Cevik V, Dagdas G, Jones JDG, Krasileva KV. 2016. Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biology 14: doi: 10.1186/s12915-016-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saur IM, Bauer S, Kracher B, Lu X, Franzeskakis L, Müller MC, Sabelleck B, Kümmel F, Panstruga R, Maekawa T et al 2019. Multiple pairs of allelic MLA immune receptor‐powdery mildew AVRA effectors argue for a direct recognition mechanism. eLife 8: pii: e44471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savary S, Willocquet L, Pethybridge SJ, Esker P, McRoberts N, Nelson A. 2019. The global burden of pathogens and pests on major food crops. Nature Ecology and Evolution 3: 430–439. [DOI] [PubMed] [Google Scholar]
- Schiek B, Hareau G, Baguma Y, Medakker A, Douches D, Shotkoski F, Ghislain M. 2016. Demystification of GM crop costs: releasing late blight resistant potato varieties as public goods in developing countries. International Journal of Biotechnology 14: 112. [Google Scholar]
- Schlaich T, Urbaniak BM, Malgras N, Ehler E, Birrer C, Meier L, Sautter C. 2006. Increased field resistance to Tilletia caries provided by a specific antifungal virus gene in genetically engineered wheat. Plant Biotechnology Journal 4: 63–75. [DOI] [PubMed] [Google Scholar]
- Schneider M, Droz E, Malnoë P, Chatot C, Bonnel E, Métraux J‐P. 2002. Transgenic potato plants expressing oxalate oxidase have increased resistance to oomycete and bacterial pathogens. Potato Research 45: 177–185. [Google Scholar]
- Schnippenkoetter W, Lo C, Liu G, Dibley K, Chan WL, White J, Milne R, Zwart A, Kwong E, Keller B et al 2017. The wheat Lr34 multipathogen resistance gene confers resistance to anthracnose and rust in sorghum. Plant Biotechnology Journal 15: 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoonbeek HJ, Wang HH, Stefanato FL, Craze M, Bowden S, Wallington E, Zipfel C, Ridout CJ. 2015. Arabidopsis EF‐Tu receptor enhances bacterial disease resistance in transgenic wheat. New Phytologist 206: 606–613. [DOI] [PubMed] [Google Scholar]
- Schwessinger B, Bahar O, Thomas N, Holton N, Nekrasov V, Ruan D, Canlas PE, Daudi A, Petzold CJ, Singan VR et al 2015. Transgenic expression of the dicotyledonous pattern recognition receptor EFR in rice leads to ligand‐dependent activation of defense responses. PLoS Pathogens 11: e1004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorza R, Callahan A, Dardick C, Ravelonandro M, Polak J, Malinowski T, Zagrai I, Cambra M, Kamenova I. 2013. Genetic engineering of plum pox virus resistance: ‘HoneySweet’ plum—from concept to product. Plant Cell, Tissue and Organ Culture 115: 1–12. [Google Scholar]
- Singh RP, Hodson DP, Jin Y, Lagudah ES, Ayliffe MA, Bhavani S, Rouse MN, Pretorius ZA, Szabo LJ, Huerta‐Espino J et al 2015. Emergence and spread of new races of wheat stem rust fungus: continued threat to food security and prospects of genetic control. Phytopathology 105: 872–884. [DOI] [PubMed] [Google Scholar]
- Song Y, Thomma BPHJ. 2018. Host‐induced gene silencing compromises Verticillium wilt in tomato and Arabidopsis. Molecular Plant Pathology 19: 77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava V, Thomson J. 2016. Gene stacking by recombinases. Plant Biotechnology Journal 14: 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam R, McDonald BA. 2018. When resistance gene pyramids are not durable—the role of pathogen diversity. Molecular Plant Pathology 19: 521–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam R, Silva‐Arias GA, Nosenko T, Scheikl D, Hörger AC, Stephan W, Haberer G, Tellier A. 2017. A small subset of NLR genes drives local adaptation to pathogens in wild tomato. BioRxiv: 1–50. doi: 10.1101/210559. [DOI] [Google Scholar]
- Steuernagel B, Periyannan SK, Hernandez‐Pinzon I, Witek K, Rouse MN, Yu G, Hatta A, Ayliffe M, Bariana H, Jones JD et al 2016. Rapid cloning of disease‐resistance genes in plants using mutagenesis and sequence capture. Nature Biotechnology 34: 652–655. [DOI] [PubMed] [Google Scholar]
- Sucher J, Boni R, Yang P, Rogowsky P, Büchner H, Kastner C, Kumlehn J, Krattinger SG, Keller B. 2017. The durable wheat disease resistance gene Lr34 confers common rust and northern corn leaf blight resistance in maize. Plant Biotechnology Journal 15: 489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai TH, Dahlbeck D, Clark ET, Gajiwala P, Pasion R, Whalen MC, Stall RE, Staskawicz BJ. 1999. Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proceedings of the National Academy of Sciences, USA 96: 14153–14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K, Sun X, Hu Q, Wu A, Lin C, Lin H, Twyman RM, Christou P, Feng T. 2001. Transgenic rice plants expressing the ferredoxin‐like protein (AP1) from sweet pepper show enhanced resistance to Xanthomonas oryzae pv. oryzae . Plant Science 160: 1035–1042. [DOI] [PubMed] [Google Scholar]
- Thomas CM, Jones DA, Parniske M, Harrison K, Balint‐Kurti PJ, Hatzixanthis K, Jones JD. 1997. Characterization of the tomato Cf‐4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf‐4 and Cf‐9. Plant Cell 9: 2209–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Toledo Thomazella DP, Brail Q, Dahlbeck D, Staskawicz BJ. 2016. CRISPR‐Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad‐spectrum disease resistance. BioRxiv. doi: 10.1101/064824. [DOI] [Google Scholar]
- Tricoli DM, Carney KJ, Russell PF, McMaster JR, Groff DW, Hadden KC, Himmel PT, Hubbard JP, Boeshore ML, Quemada HD. 1995. Field evaluation of transgenic squash containing single or multiple virus coat protein gene constructs for resistance to cucumber mosaic virus, watermelon mosaic virus 2, and zucchini yellow mosaic virus. Bio/Technology 13: 1458–1465. [Google Scholar]
- Tripathi JN, Lorenzen J, Bahar O, Ronald P, Tripathi L. 2014. Transgenic expression of the rice Xa21 pattern‐recognition receptor in banana (Musa sp.) confers resistance to Xanthomonas campestris pv. musacearum . Plant Biotechnology Journal 12: 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi L, Atkinson H, Roderick H, Kubiriba J, Tripathi JN. 2017. Genetically engineered bananas resistant to Xanthomonas wilt disease and nematodes. Food and Energy Security 6: 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi L, Mwaka H, Tripathi JN, Tushemereirwe WK. 2010. Expression of sweet pepper Hrap gene in banana enhances resistance to Xanthomonas campestris pv. musacearum . Molecular Plant Pathology 11: 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi L, Tripathi JN, Kiggundu A, Korie S, Shotkoski F, Tushemereirwe WK. 2014. Field trial of Xanthomonas wilt disease‐resistant bananas in East Africa. Nature Biotechnology 32: 868–870. [DOI] [PubMed] [Google Scholar]
- Tsuda K, Somssich IE. 2015. Transcriptional networks in plant immunity. New Phytologist 206: 932–947. [DOI] [PubMed] [Google Scholar]
- Uhse S, Djamei A. 2018. Effectors of plant‐colonizing fungi and beyond. PLoS Pathogens 14: e1006992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United Nations, Department of Economic and Social Affairs, Population Division . 2017. World population prospects: The 2017 revision, key findings and advance tables. Working Paper No. ESA/P/WP/248.
- USDA . 2018. Details on USDA plant breeding innovations . [WWW document] URL https://www.aphis.usda.gov/aphis/ourfocus/biotechnology/brs-news-and-information/2018_brs_news/pbi-details [accessed 11 January 2019].
- USDA . 2019. Coordinated framework: Roles of U.S. regulatory agencies . [WWW document] URL https://www.aphis.usda.gov/aphis/ourfocus/biotechnology/sa_regulations/ct_agency_framework_roles [accessed 5 February 2019].
- Walz A, Zingen‐Sell I, Loeffler M, Sauer M. 2008. Expression of an oxalate oxidase gene in tomato and severity of disease caused by Botrytis cinerea and Sclerotinia sclerotiorum . Plant Pathology 57: 453–458. [Google Scholar]
- Wang GL, Song WY, Ruan DL, Sideris S, Ronald PC. 1996. The cloned gene, Xa21, confers resistance to multiple Xanthomonas oryzae pv. oryzae isolates in transgenic plants. Molecular Plant–‐Microbe Interactions 9: 850–855. [DOI] [PubMed] [Google Scholar]
- Wang J, Hu M, Qi J, Han Z, Wang G, Qi Y, Wang HW, Zhou JM, Chai J. 2019a. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science. doi: 10.1126/science.aav5870. [DOI] [PubMed] [Google Scholar]
- Wang J, Hu M, Wu S, Qi J, Wang G, Han Z, Qi Y, Gao N, Wang HW, Zhou JM et al 2019b. Ligand‐triggered allosteric ADP release primes a plant NLR complex. Science 364: doi: 10.1126/science.aav5868. [DOI] [PubMed] [Google Scholar]
- Wang J, Zeng X, Tian D, Yang X, Wang L, Yin Z. 2018a. The pepper Bs4C proteins are localized to the endoplasmic reticulum (ER) membrane and confer disease resistance to bacterial blight in transgenic rice. Molecular Plant Pathology 19: 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhou L, Shi H, Chern M, Yu H, Yi H, He M, Yin J, Zhu X, Li Y et al 2018b. A single transcription factor promotes both yield and immunity in rice. Science 361: 1026–1028. [DOI] [PubMed] [Google Scholar]
- Wang M, Weiberg A, Lin FM, Thomma BP, Huang HD, Jin H. 2016. Bidirectional cross‐kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nature Plants 2: doi: 10.1038/nplants.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL. 2014. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nature Biotechnology 32: 947–951. [DOI] [PubMed] [Google Scholar]
- Wieczorek TM, Berg G, Semaškienė R, Mehl A, Sierotzki H, Stammler G, Justesen AF, Jørgensen LN. 2015. Impact of DMI and SDHI fungicides on disease control and CYP51 mutations in populations of Zymoseptoria tritici from Northern Europe. European Journal of Plant Pathology 143: 861–871. [Google Scholar]
- Witek K, Jupe F, Witek AI, Baker D, Clark MD, Jones JD. 2016. Accelerated cloning of a potato late blight‐resistance gene using RenSeq and SMRT sequencing. Nature Biotechnology 34: 656–660. [DOI] [PubMed] [Google Scholar]
- Wu CH, Abd‐El‐Haliem A, Bozkurt TO, Belhaj K, Terauchi R, Vossen JH, Kamoun S. 2017. NLR network mediates immunity to diverse plant pathogens. Proceedings of the National Academy of Sciences, USA 114: 8113–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Yuan M, Ai C, Liu L, Zhuang E, Karapetyan S, Wang S, Dong X. 2017. uORF‐mediated translation allows engineered plant disease resistance without fitness costs. Nature 545: 491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, McLellan H, Naqvi S, He Q, Boevink PC, Armstrong M, Giuliani LM, Zhang W, Tian Z, Zhan J et al 2016. Potato NPH3/RPT2‐Like Protein StNRL1, targeted by a Phytophthora infestans RXLR effector, is a susceptibility factor. Plant Physiology 171: 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, Li H. 2010. 20 years of transgenic research in China for resistance to papaya ringspot virus. Transgenic Plant Journal 4: 58–63. [Google Scholar]
- Yin C, Hulbert SH. 2018. Host‐induced gene silencing (HIGS) for elucidating Puccinia gene function in wheat. Methods in Molecular Biology 1848: 139–150. [DOI] [PubMed] [Google Scholar]
- Yip MK, Huang HE, Ger MJ, Chiu SH, Tsai YC, Lin CI, Feng TY. 2007. Production of soft rot resistant calla lily by expressing a ferredoxin‐like protein gene (pflp) in transgenic plants. Plant Cell Reports 26: 449–457. [DOI] [PubMed] [Google Scholar]
- Zeilmaker T, Ludwig NR, Elberse J, Seidl MF, Berke L, Van Doorn A, Schuurink RC, Snel B, Van den Ackerveken G. 2015. DOWNY MILDEW RESISTANT 6 and DMR6‐LIKE OXYGENASE 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. The Plant Journal 81: 210–222. [DOI] [PubMed] [Google Scholar]
- Zhang T, Jin Y, Zhao JH, Gao F, Zhou BJ, Fang YY, Guo HS. 2016. Host‐induced gene silencing of the target gene in fungal cells confers effective resistance to the cotton wilt disease pathogen Verticillium dahliae . Molecular Plant 9: 939–942. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yao J, Withers J, Xin XF, Banerjee R, Fariduddin Q, Nakamura Y, Nomura K, Howe GA, Boland W et al 2015. Host target modification as a strategy to counter pathogen hijacking of the jasmonate hormone receptor. Proceedings of the National Academy of Sciences, USA 112: 14354–14359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhao L, Zhao J, Li Y, Wang J, Guo R, Gan S, Liu CJ, Zhang K. 2017. S5H/DMR6 encodes a salicylic acid 5‐hydroxylase that fine‐tunes salicylic acid homeostasis. Plant Physiology 175: 1082–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, Boller T, Felix G. 2006. Perception of the bacterial PAMP EF‐Tu by the receptor EFR restricts Agrobacterium‐mediated transformation. Cell 125: 749–760. [DOI] [PubMed] [Google Scholar]


