Abstract
The RNA world hypothesis assumes that life on Earth began with nucleotides that formed information‐carrying RNA oligomers able to self‐replicate. Prebiotic reactions leading to the contemporary nucleosides are now known, but their execution often requires specific starting materials and lengthy reaction sequences. It was therefore proposed that the RNA world was likely proceeded by a proto‐RNA world constructed from molecules that were likely present on the early Earth in greater abundance. Herein, we show that the prebiotic starting molecules bis‐urea (biuret) and tris‐urea (triuret) are able to directly react with ribose. The urea‐ribosides are remarkably stable because they are held together by a network of intramolecular, bifurcated hydrogen bonds. This even allowed the synthesis of phosphoramidite building blocks and incorporation of the units into RNA. Investigations of the nucleotides’ base‐pairing potential showed that triuret:G RNA base pairs closely resemble U:G wobble base pairs. Based on the probable abundance of urea on the early Earth, we postulate that urea‐containing RNA bases are good candidates for a proto‐RNA world.
Keywords: base pairing, origin of life, prebiotic chemistry, proto-RNA, urea
The prebiotic starting molecules bis‐urea (biuret) and tris‐urea (triuret) directly react with ribose. The urea‐ribosides are remarkably stable because of a network of intramolecular, bifurcated hydrogen bonds, which even allowed the synthesis of phosphoramidite building blocks and incorporation of the units into RNA. Urea‐containing RNA bases are therefore good candidates for a proto‐RNA world.
Introduction
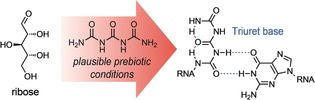
Urea, the bisamide of carbonic acid, is widely distributed in the biosphere and plays a fundamentally important role in the biosynthesis of proteins and the entire N‐cycle of organisms in general.1, 2 It is also believed to have formed early on the prebiotic Earth and before the process of chemical evolution that gave the centrally important molecules of life.3 Urea is a key starting molecule for many prebiotic chemical reactions,4, 5, 6, 7, 8, 9, 10, 11 and was the first organic compound synthesized from inorganic matter (ammonium cyanate) by the chemist Friedrich Wöhler in 1828.12 Wöhler's synthesis was the starting point of the field of organic chemistry and was, among others, essential to defeating the mainstream ideology of “vitalism”, which stated that organic matter contained a special vital force.13 Current theories about the origin of life are built upon the RNA world hypothesis, which predicts the early formation of information‐encoding RNA that was able to self‐replicate and that featured properties leading to their survival under early Earth conditions.14, 15, 16, 17 It is assumed that based on the processes of chemical evolution, more and more complex RNA and RNA–peptide structures were created that finally led to the emergence of life.18, 19 RNA and the constituting nucleosides that are needed to establish faithful replication of “genetic” information are, however, rather complex chemical structures. The problem of finding prebiotically plausible pathways to the canonical nucleosides (Figure 1 a; known as the nucleoside problem)20 led to the idea that RNA was potentially proceeded by a proto‐RNA that could more easily arise from prebiotically privileged starting materials.21 As a result, emerging discussions about the origin of life have often emphasized the significance of informational polymers that are simpler than RNA. A revolutionary study from Eschenmoser's group, for example, demonstrated that α‐threofuranosyl nucleic acid (TNA) is capable of forming antiparallel duplexes and can even pair with cDNA or RNA.22 TNA was later simplified to an acyclic polymer known as glycol nucleic acid (GNA),23, 24, 25 and various other XNA backbones have since been investigated.26 We know that formaldehyde and Ca(OH)2 can give sugars by the formose reaction,27, 28, 29 as first described by Butlerov.30 Although it remains unclear how exactly such a process could lead to the substantial accumulation of ribose on the early Earth, recent developments have led to significant improvements in the prebiotic synthesis of ribose from simpler aldoses.31, 32, 33 We also know that urea was one of the most likely nitrogen‐containing molecules present on the early Earth and that it can activate mineral phosphate to achieve phosphorylations.4, 5 As urea itself has even been shown to react directly with ribose under mildly acidic conditions,34 we asked whether the construction of informational base‐pairing systems based on the pyrolysis products of urea, biuret and triuret (Figure 1 b),35 would be possible.
Figure 1.
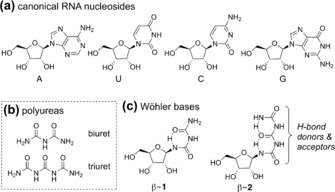
a) The chemical structures of the canonical RNA nucleosides. b) The chemical structures of biuret and triuret. c) Depiction of the urea‐based nucleosides with potentially stabilizing hydrogen bonds.
The fundamental chemical problem associated with this notion is that such nucleosides (Figure 1 c) should be highly prone to hydrolysis in an aqueous environment. Investigating the structures of potential urea bases, however, led us to discover that those containing biuret (β‐1) and triuret (β‐2) are highly stabile even in water, probably because of the intramolecular H‐bonds (Figure 1 c). The question of the possible prebiotic existence of urea (Wöhler) RNA is therefore directly associated with the question as to which extent these non‐covalent interactions protect from hydrolysis.
Results and Discussion
To investigate the formation of urea nucleosides under plausible prebiotic conditions, we mixed aqueous solutions of ribose with either biuret or triuret in the presence of boric acid and heated the mixture at 95 °C for 18 h, allowing the mixture to slowly dry down. Boric acid, which forms complexes with vicinal 1,2‐diols because of its high electron deficiency, was included for its known stabilizing36, 37, 38 and directing39, 40, 41 effects on ribose. The resulting solid was then taken up in dilute (100 mm) sodium carbonate buffer (pH 9.5) and heated again at 95 °C for 1 h (Figure 2).
Figure 2.
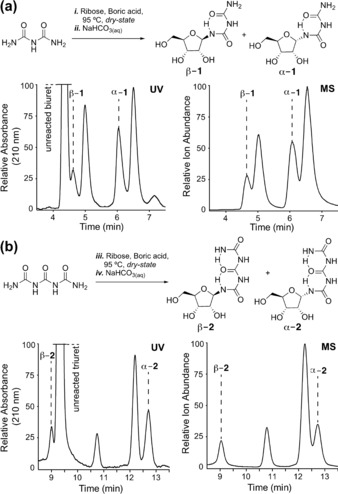
Reaction of a) biuret and b) triuret with ribose and analysis of the reaction mixture by HPLC‐MS, showing the successful formation of urea‐based nucleosides.
The mixture was subsequently analyzed by reverse‐phase HPLC‐MS. Gratifyingly, we noted significant formation of the corresponding nucleosides with both biuret and triuret. These wet–dry conditions mimic the intermittently concentrating environments that might be found on drying beaches or lagoons (or even today in Death Valley),42 as was recently discussed in relation to the prebiotic synthesis of canonical and non‐canonical purine nucleosides.39 For the biuret reaction, we detected formation of four major nucleoside products. Of those, two are the α‐ and β‐anomers of the ribofuranosides (α‐1 and β‐1). These structures were confirmed by independent synthesis of the α‐ and β‐anomers (see below) followed by co‐injection studies. The absolute stereochemical configurations of the α‐ and β‐anomers were confirmed by NOESY‐NMR spectroscopy (see the Supporting Information). The other two compounds detected in the HPLC‐MS experiment are likely the pyranosidic species (not further investigated). Similar but not identical data were obtained for triuret. Here too, we detected four reaction products of which two are the α‐ and β‐anomers of the ribofuranosides (α‐2 and β‐2), with the remaining compounds likely being the pyranosides again. We focused our initial studies on the ribofuranosides and noted to our surprise that both the biuret and the triuret species are quite stable. Both compounds can be kept for prolonged periods of time in an aqueous solution at neutral pH without signs of anomerization. This unusual stability of the biuret and triuret nucleosides prompted us to study if one could generate phosphoramidites and insert them into RNA. This would require the Wöhler nucleosides to survive even the nucleophilic reaction and deprotection conditions needed for solid‐phase RNA synthesis.
The synthesis of the phosphoramidite building blocks is shown in Scheme 1. We began with the 3′,5′‐silyl protection of 1‐azidoribose 3 to obtain 4, followed by TOM protection of the 2′‐OH group to give 5. Desilylation of the 3′,5′‐positions (affording compound 6) and subsequent DMTr protection of the 5′‐OH group furnished compound 7. We then protected the 3′‐OH group with an acetyl group to give 8 and reduced the azide by catalytic hydrogenation followed by reaction of the amine with trimethylsilylisocyanate. This provides the urea riboside 9. A second reaction with trichloroacetylisocyanate in pyridine followed by cleavage of the trichloroacetate group with basic alumina in methanol gave the biuret riboside 10 as a mixture of the α‐ and β‐isomers (β‐10 and α‐10), which we separated by column chromatography (SiO2, CH2Cl2/CH3OH 100:0→98:2). The unwanted α‐isomer (α‐10) isomerized upon heating in the presence of DBU, which gave a mixture of β‐10 and α‐10. Iterative anomerization allowed us to increase the total isolated yield of the β‐anomer (β‐10) for subsequent reactions. To obtain the triuret nucleoside 11, we repeated the trichloroacetylisocyanate reaction followed by basic alumina treatment using the pure β‐anomer of the biuret nucleoside (β‐10). Interestingly, we observed very little anomerization during these reactions (as monitored by TLC). The β‐triuret nucleoside 11 was therefore obtained in anomerically pure form and in 58 % yield. Both the β‐biuret (β‐10) and the β‐triuret (11) nucleosides were next converted into the corresponding phosphoramidites. To this end, we cleaved the 3′‐acetyl group with ammonia (even these conditions do not lead to hydrolysis of the glycosidic bond), and then treated the ensuing compounds 12 and 13 with bis(2‐cyanoethyl)‐N,N‐diisopropylphosphoramidite in the presence of diisopropylamine and tetrazole. This final step provided the phosphoramidite building blocks 14 and 15 in good yields of 70 % and 54 %, respectively.
Scheme 1.
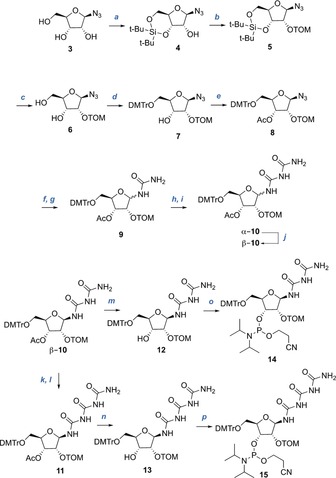
Phosphoramidite building block synthesis of the biuret and triuret nucleosides. Reagents and conditions: a) t‐Bu2Si(OTf)2, DMF, 0 °C, 1 h; b) i‐PrSiO(CH2)Cl, NaH, THF, 0 °C, overnight, 55 % over 2 steps; c) HF‐pyridine, pyridine, CH2Cl2, room temperature, 1 h, 61 %; d) DMTrCl, pyridine, room temperature, overnight, 77 %; e) Ac2O, DMAP, pyridine, room temperature, 2 h, 91 %; f) 10 % Pd/C, H2, THF, room temperature, 2 h, then g) TMS‐isocyanate, THF, room temperature, overnight, 76 % (mixture of diastereomers); h) trichloroacetylisocyanate, pyridine, THF, room temperature, 1 h, then i) Al2O3, MeOH, room temperature, 1 h, 90 % (d.r. α/β=1.6:1); j) DBU, THF, 50 °C, overnight, 28 % (96 % based on recovered starting material); k) trichloroacetylisocyanate, pyridine, THF, room temperature, 1 h, then l) Al2O3, MeOH, room temperature, 1 h, 58 %; m) NH3, MeOH, room temperature, 4 h, 74 %; n) NH3, MeOH, room temperature, 4 h, 76 %; o) bis(2‐cyanoethyl)‐N,N‐diisopropylphosphoramidite, diisopropylamine‐tetrazole, CH3CN, room temperature, overnight, 70 %; p) bis(2‐cyanoethyl)‐N,N‐diisopropylphosphoramidite, diisopropylamine‐tetrazole, CH3CN, room temperature, overnight, 54 %.
For the solid‐phase oligonucleotide synthesis, we used a standard ultra‐mild RNA synthesis protocol. The urea bases were coupled once for 20 min. After full assembly of the RNA strands using Pac chemistry conditions, we deprotected the RNA strands and cleaved them from the solid support with NH3 in methanol at room temperature. The silyl protecting groups were finally removed using HF‐TEA in DMSO at 65 °C. Figure 3 shows the crude HPLC chromatogram of the triuret‐containing RNA strand as an example, as well as the MALDI‐TOF mass spectrum obtained from purification of the major species. It is clearly evident that the RNA strands are efficiently produced and can be cleanly purified. Similar data were obtained for the biuret‐containing strands (Figure S7). It is remarkable that the urea bases survive the RNA synthesis conditions to give RNA strands in excellent purity.
Figure 3.
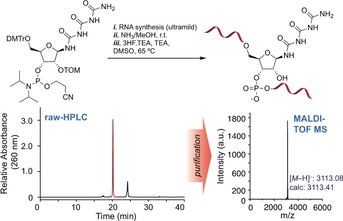
Preparatory HPLC and MALDI‐TOF mass data for an exemplary Tri‐containing RNA strand with the sequence 5′‐CUUACTriCUGA‐3′.
This stability allowed us to next investigate the pairing properties of the biuret and triuret bases using thermal melting curve studies (10 mm sodium phosphate, 150 mm NaCl, pH 7.0). If a candidate proto‐RNA were to have existed before the emergence of modern RNA, then it stands to reason that its bases needed to pair with the canonical nucleosides to allow a smooth evolutionary transition from proto‐RNA to RNA. For the measurements we prepared RNA strands with a C:G or a U:A base pair in a central position. We then exchanged the pyrimidine base C or U with the biuret (Bi) or triuret (Tri) base. The data are compiled in Figure 4 b. The unmodified RNA duplexes feature, as expected, rather high melting temperatures of 55 °C (C:G) and 49 °C (U:A). For all Bi:A/G/U/C base pairs, we measured much lower melting temperatures, which were in addition quite similar irrespective of the counterbase (between 28 °C and 31 °C). This shows that the biuret base does not prefer a particular counterbase and that base pairing is in general weak, if it occurs at all. For the larger triuret base, we noted significantly higher melting temperatures between 35 °C and 45 °C. In addition, we observed a clear base‐pairing preference with G. The melting temperature for the Tri:G base pair is around 5 °C higher than those for the others, which clearly points to a significant pairing selectivity. Analysis of the base pairing potential of the triuret base shows that it is in principle possible to form a wobble‐type base pair with G. We therefore tested an RNA duplex with a central U:G wobble base pair and observed indeed the same melting temperature (45 °C). Our hypothesis was further validated by the observation that triuret also forms a stable base pair with the structurally related base inosine (I), giving an almost identical melting temperature of 44 °C. To exclude that this is pure chance, we next prepared RNA strands with either two or even three consecutive triuret bases and paired this strand with a counterstrand containing either two or three central G bases. Comparative melting point studies showed exactly the same behavior between the U:G wobble and the Tri:G base pairs.
Figure 4.
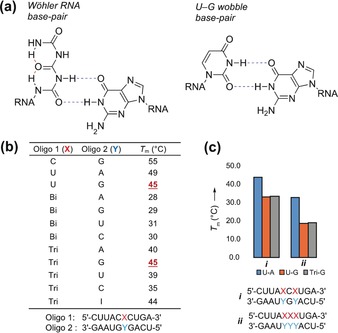
a) Chemical structure and base‐pairing properties of triuret and similarity between the triuret‐G base pair and a U‐G wobble base pair. b) Summary of T m analyses for oligonucleotides containing biuret and triuret. c) Summary of T m analyses for oligonucleotides containing more than one modified base or U‐G wobble base pair. Solutions were buffered with 10 mm sodium phosphate (pH 7) and 150 mm NaCl.
To gain deeper insight into the origin of the stability of the Tri:G base pairing, we prepared 8 mer palindromic RNA strands that were designed to form canonical C:G, wobble U:G, or Tri:G pairs at the central position (GGUXGACC, where X=C, U, or Tri) and analyzed their 2D 1H–1H NOESY spectra.43 The high chemical shift similarity in the fingerprint region of the spectra (Figure S17) confirms the same overall structure for the three oligonucleotides, namely formation of an A‐form double‐stranded RNA (Figure 5 c).
Figure 5.
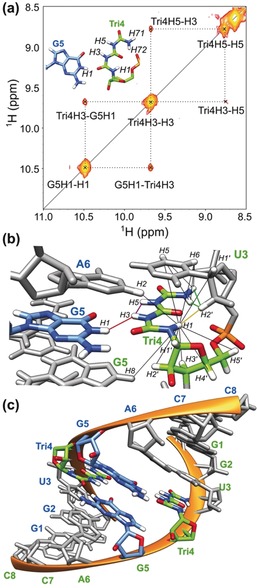
NMR analysis of the triuret:G base pairing. a) Excerpt from the 1H–1H NOESY spectrum (t mix=40 ms) of the dsRNA (GGUXGACC, where X=Tri) showing the inter‐strand cross‐peaks between the H1 imino proton of G5 and the H3 amide proton of Tri4. The inset shows a model for the triuret:G base paring. b) NOE contacts of the triuret base amide protons. Essential structure‐defining NOE contacts are highlighted for Tri4H1–U3H2′ (yellow), Tri4H71/H72–U3H2′ (green), G5H1–Tri4H3 (red), and Tri4H3–Tri4H5 (purple). Other observed NOE contacts are shown as black lines. c) Structural model of the GGUXGACC oligonucleotide showing the non‐canonical base pairing between G5 (blue) and Tri4 (green) bases.
The imino region of the NOESY spectra provides direct information on the hydrogen bond interactions between base pairs (Figure 5 a and Figure S16). The Tri base has five amide protons (H1, H3, H5, H71, and H72) and three carbonyl oxygen atoms that could potentially be involved in base pairing. Out of the five protons, H3 and H5 are partially or fully solvent‐exposed and thus they show strong exchange cross‐peaks at the water resonance (Figure S19). The number and intensity of cross‐peaks between the H1 proton of Tri and the sugar/base protons of the previous uridine base (U3) indicate that the H1 proton is located in a similar arrangement to the H5 proton of a U or a C or as the H8 proton of an A and a G base in canonical dsRNA structures; for example, we observed a strong NOE cross‐peak between Tri4H1 and U3H2′ (yellow line in Figure 5 b). Similarly, the terminal NH2 group of Tri shows strong cross‐peaks with the H2′ and H5 protons of U3 (green lines in Figure 5 c). Thus, the NOESY spectra suggest that both H1 and the NH2 group point towards the phosphate backbone. Direct evidence for Tri:G base pairing was obtained from a NOESY spectrum recorded with a relatively short (40 ms) mixing time (Figure 5 a). This experiment provides a clear cross‐peak between the H1 imino proton of G5 and the H3 amide proton of Tri of the opposite strand (red line in Figure 5 b). In addition, the TriH3 proton shows an intrabase cross‐peak with H5 (purple line in Figure 5 c). These, as well as a handful of other NOE cross‐peaks between the triuret base protons and the surrounding protons, support the structural model depicted in Figure 4 a (all other NOE distance restraints are depicted with black lines in Figure 5 b), with the triuret moiety clearly forming bifurcated hydrogen bonds.
To obtain a three‐dimensional model for the Tri:G base pair, we performed molecular modeling using the online software ROSIE44, 45 followed by NOE‐based structure calculations using the software CNSsolve.46, 47 During the structure calculations, only the conformation of the triuret base was altered while the phosphate backbone and all other bases were kept at their fixed position. Figure 5 c displays the obtained low‐energy structure model for the double‐stranded 8 mer RNA. It is clearly evident that the two Tri:G base pairs are well accommodated in the structure and that the extended network of hydrogen bonds within the Tri structure and between Tri and the opposite G establish the measured stability.
All of these results confirm that triuret, which is itself a condensation product of urea, is able to form a folded pseudobase that pairs with guanine. Finally, in order to further demonstrate the prebiotic plausibility of Wöhler RNA, we synthesized a homo‐RNA oligomer containing exclusively triuret bases (Figure 6 a). The only additional structural modification was the inclusion of a dye at the 5′‐end (Cy3), which was necessary to allow UV detection at 548 nm and therefore purification by HPLC. Remarkably, despite having five, in principle hydrolysable triuret bases in a row, the homo‐strand was bench‐stable both at room temperature as well as when subjected to the harsh conditions necessary for RNA deprotection and cleavage from the solid support. Figure 6 shows the crude HPLC chromatogram obtained from the material directly after its synthesis and the correct MALDI‐TOF mass spectrum, confirming the integrity of the material.
Figure 6.
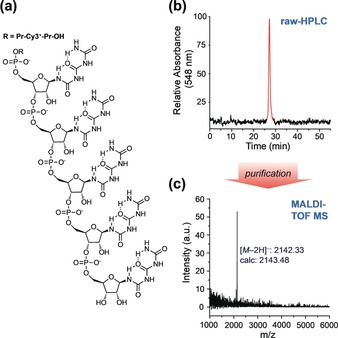
a) Depiction of the 5 mer triuret oligomer containing a Cy3‐fluorophore at the 5′‐end for better detectability. b) Crude HPLC chromatogram of the triuret oligomer and c) MALDI‐TOF mass spectrum of the triuret oligomer.
Conclusion
In summary, we have shown that biuret and triuret are able to condense directly with sugars (here ribose) to form stable bis‐ and tris‐urea nucleosides. Within an RNA strand, triuret is able to form stable wobble‐type base pairs with G as well as with the prebiotically relevant base inosine.48 As discussed, biuret and triuret are obtained upon pyrolysis of urea, one of the most likely building blocks available on the early Earth. Given that various tri‐, tetra‐, and pentose sugars are prebiotically accessible from glycoaldehyde,31, 32 which is itself accessible from either formaldehyde30 or HCN by ultraviolet irradiation,49 our discovery creates the prebiotically attractive possibility of generating information‐encoding oligomers whose key building blocks are derived of simple one‐carbon units. The chemistry described here now needs to be explored with sugars simpler than ribose. Discussed examples are threose‐22, 50 and glycol‐based23, 25 backbones.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
We thank the Deutsche Forschungsgemeinschaft for financial support via SFB1309, SPP1784, and CA275/11‐1. This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement no. EPiR 741912) and through a H2020 Marie Skłodowska‐Curie Action (LightDyNAmics, 765866). H.O. acknowledges support through a Marie Skłodowska‐Curie Individual Fellowship (752420) from the European Commission. P.R. acknowledges support from the Center for NanoScience and from the Fonds der Chemischen Industrie.
H. Okamura, A. Crisp, S. Hübner, S. Becker, P. Rovó, T. Carell, Angew. Chem. Int. Ed. 2019, 58, 18691.
Contributor Information
Dr. Hidenori Okamura, http://www.carellgroup.de.
Dr. Petra Rovó, Email: petra.rovo@lmu.de.
Prof. Dr. Thomas Carell, Email: thomas.carell@lmu.de.
References
- 1. G. Shambaugh III , Am. J. Clin. Nutr. 1977, 30, 2083–2087. [DOI] [PubMed] [Google Scholar]
- 2. Campbell J. W., Nature 1965, 208, 1299–1301. [DOI] [PubMed] [Google Scholar]
- 3. Schlesinger G., Miller S. L., J. Mol. Evol. 1983, 19, 383–390. [DOI] [PubMed] [Google Scholar]
- 4. Lohrmann R., Orgel L., Science 1971, 171, 490–494. [DOI] [PubMed] [Google Scholar]
- 5. Osterberg R., Orgel L., J. Mol. Evol. 1972, 1, 241–248. [DOI] [PubMed] [Google Scholar]
- 6. Powner M. W., Gerland B., Sutherland J. D., Nature 2009, 459, 239–242. [DOI] [PubMed] [Google Scholar]
- 7. Robertson M. P., Miller S. L., Nature 1995, 375, 772–774. [DOI] [PubMed] [Google Scholar]
- 8. Shapiro R., Proc. Natl. Acad. Sci. USA 1999, 96, 4396–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schneider C., Becker S., Okamura H., Crisp A., Amatov T., Stadlmeier M., Carell T., Angew. Chem. Int. Ed. 2018, 57, 5943–5946; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 6050–6054. [Google Scholar]
- 10. Becker S., Schneider C., Okamura H., Crisp A., Amatov T., Dejmek M., Carell T., Nat. Commun. 2018, 9, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hud N. V., Nat. Commun. 2018, 9, 5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wöhler F., Ann. Phys. 1828, 88, 253–256. [Google Scholar]
- 13. Ramberg P. J., Ambix 2000, 47, 170–195. [DOI] [PubMed] [Google Scholar]
- 14. Orgel L. E., Crit. Rev. Biochem. Mol. Biol. 2004, 39, 99–123. [DOI] [PubMed] [Google Scholar]
- 15. Crick F. H. C., J. Mol. Biol. 1968, 38, 367–379. [DOI] [PubMed] [Google Scholar]
- 16. Orgel L. E., J. Mol. Biol. 1968, 38, 381–393. [DOI] [PubMed] [Google Scholar]
- 17. Gilbert W., Nature 1986, 319, 618. [Google Scholar]
- 18. Joyce G. F., Nature 1989, 338, 217–224. [DOI] [PubMed] [Google Scholar]
- 19. Joyce G. F., PLOS Biol. 2012, 10, e1001323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hud N. V., Cafferty B. J., Krishnamurthy R., Williams L. D., Chem. Biol. 2013, 20, 466–474. [DOI] [PubMed] [Google Scholar]
- 21. Engelhart A. E., Hud N. V., Cold Spring Harbor Perspect. Biol. 2010, 2, a002196–a002196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schöning K.-U., Scholz P., Guntha S., Wu X., Krishnamurthy R., Eschenmoser A., Science 2000, 290, 1347–1351. [DOI] [PubMed] [Google Scholar]
- 23. Zhang L., Peritz A., Meggers E., J. Am. Chem. Soc. 2005, 127, 4174–4175. [DOI] [PubMed] [Google Scholar]
- 24. Karri P., Punna V., Kim K., Krishnamurthy R., Angew. Chem. Int. Ed. 2013, 52, 5840–5844; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 5952–5956. [Google Scholar]
- 25. Zhang S., Chaput J. C., Curr. Protoc. Nucleic Acid Chem. 2010, 42, 440.1–4.40.18. [DOI] [PubMed] [Google Scholar]
- 26. Anosova I., Kowal E. A., Dunn M. R., Chaput J. C., Van Horn W. D., Egli M., Nucleic Acids Res. 2016, 44, 1007–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Usami K., Okamoto A., Org. Biomol. Chem. 2017, 15, 8888–8893. [DOI] [PubMed] [Google Scholar]
- 28. Nitta S., Furukawa Y., Kakegawa T., Origins Life Evol. Biospheres 2016, 46, 189–202. [DOI] [PubMed] [Google Scholar]
- 29. Sagi V. N., Punna V., Hu F., Meher G., Krishnamurthy R., J. Am. Chem. Soc. 2012, 134, 3577–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Butlerow A., C. R. Hebd. Seances Acad. Sci. 1861, 53, 145–147. [Google Scholar]
- 31. Lamour S., Pallmann S., Haas M., Trapp O., Life 2019, 9, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kofoed J., Reymond J.-L., Darbre T., Org. Biomol. Chem. 2005, 3, 1850–1855. [DOI] [PubMed] [Google Scholar]
- 33. Oberhuber M., Joyce G. F., Angew. Chem. Int. Ed. 2005, 44, 7580–7583; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2005, 117, 7752–7755. [Google Scholar]
- 34. Björnberg O., Vodnala M., Domkin V., Hofer A., Rasmussen A., Andersen G., Piškur J., Nucleosides Nucleotides Nucleic Acids 2010, 29, 433–437. [DOI] [PubMed] [Google Scholar]
- 35. Chemat F., Poux M., Tetrahedron Lett. 2001, 42, 3693–3695. [Google Scholar]
- 36. Ricardo A., Carrigan M. A., Olcott A. N., Benner S. A., Science 2004, 303, 196–196. [DOI] [PubMed] [Google Scholar]
- 37. Scorei R., Cimpoiaşu V. M., Origins Life Evol. Biospheres 2006, 36, 1–11. [DOI] [PubMed] [Google Scholar]
- 38. Furukawa Y., Horiuchi M., Kakegawa T., Origins Life Evol. Biospheres 2013, 43, 353–361. [DOI] [PubMed] [Google Scholar]
- 39. Becker S., Thoma I., Deutsch A., Gehrke T., Mayer P., Zipse H., Carell T., Science 2016, 352, 833–836. [DOI] [PubMed] [Google Scholar]
- 40. Kim H.-J., Furukawa Y., Kakegawa T., Bita A., Scorei R., Benner S. A., Angew. Chem. Int. Ed. 2016, 55, 15816–15820; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 16048–16052. [Google Scholar]
- 41. Kim H.-J., Benner S. A., Proc. Natl. Acad. Sci. USA 2017, 114, 11315–11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kubecka P., Weather 2001, 56, 218–221. [Google Scholar]
- 43. McDowell J. A., Turner D. H., Biochemistry 1996, 35, 14077–14089. [DOI] [PubMed] [Google Scholar]
- 44. Das R., Karanicolas J., Baker D., Nat. Methods 2010, 7, 291–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lyskov S., Chou F.-C., Conchúir S. Ó., Der B. S., Drew K., Kuroda D., Xu J., Weitzner B. D., Renfrew P. D., Sripakdeevong P., Borgo B., Havranek J. J., Kuhlman B., Kortemme T., Bonneau R., Gray J. J., Das R., PLoS One 2013, 8, e63906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Acta Crystallogr. Sect. D 1998, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 47. Brunger A. T., Nat. Protoc. 2007, 2, 2728–2733. [DOI] [PubMed] [Google Scholar]
- 48. Kim S. C., O'Flaherty D. K., Zhou L., Lelyveld V. S., Szostak J. W., Proc. Natl. Acad. Sci. USA 2018, 115, 13318–13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ritson D., Sutherland J. D., Nat. Chem. 2012, 4, 895–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yu H., Zhang S., Chaput J. C., Nat. Chem. 2012, 4, 183–187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


