Summary
Awns are stiff, hair‐like structures which grow from the lemmas of wheat (Triticum aestivum) and other grasses that contribute to photosynthesis and play a role in seed dispersal. Variation in awn length in domesticated wheat is controlled primarily by three major genes, most commonly the dominant awn suppressor Tipped1 (B1). This study identifies a transcription repressor responsible for awn inhibition at the B1 locus.
Association mapping was combined with analysis in biparental populations to delimit B1 to a distal region of 5AL colocalized with QTL for number of spikelets per spike, kernel weight, kernel length, and test weight.
Fine‐mapping located B1 to a region containing only two predicted genes, including C2H2 zinc finger transcriptional repressor TraesCS5A02G542800 upregulated in developing spikes of awnless individuals. Deletions encompassing this candidate gene were present in awned mutants of an awnless wheat. Sequence polymorphisms in the B1 coding region were not observed in diverse wheat germplasm whereas a nearby polymorphism was highly predictive of awn suppression.
Transcriptional repression by B1 is the major determinant of awn suppression in global wheat germplasm. It is associated with increased number of spikelets per spike and decreased kernel size.
Keywords: awns, B1 locus, fine mapping, positional cloning, wheat (Triticum aestivum), zinc finger protein
Introduction
Awns are stiff, hair‐like structures common in grass inflorescences. In Poaceae, awns emerge from the lemma of young spikelets at an early developmental stage. Awns are important for seed dispersal in wild relatives of wheat with a brittle rachis; spikelets attach to passing animals via barbs lining the awns. In addition, awns balance spikelets as they fall, and their expansion and contraction in response to changes in humidity drives them into the soil (Elbaum et al., 2007). Awns also may deter herbivores from ingesting heads (Grundbacher, 1963), making awn suppression important for developing forage cultivars (Cash et al., 2009). The absence of awns in rice is considered to be a key domestication trait facilitating grain harvest and storage (Toriba & Hirano, 2013), but the history of the evolution and spread of awn suppression in domesticated wheat is not well understood.
Major variation for awn length in domesticated wheats emerge from different combinations of three dominant genes: B1 (Tipped 1), B2 (Tipped 2) and Hd (Hooded) (McIntosh et al., 2014). On its own, the B1 awn suppressor produces an apically awnletted phenotype, characterized by short awns at the end of the spike but absent elsewhere (Watkins & Ellerton, 1940). The B2 allele reduces awn length most dramatically towards the top and bottom of the wheat spike, while the Hd allele reduces awn length consistently and can produce curved, ‘hooked’ awns (Watkins & Ellerton, 1940). Combination of these genes produces a nearly awnless or completely awnless phenotype (Yoshioka et al., 2017). None of the genes controlling awn length in wheat have been cloned, but mapping studies place Hd and B2 on the short arm of chromosome 4A and the long arm of chromosome 6B, respectively (Sourdille et al., 2002; Yoshioka et al., 2017). The B1 locus has a long history as a physical marker, and is located distal to the major genes controlling vernalization requirement (VRN‐A1) and the spelt head type (Q) on 5AL (Kato et al., 1998). Recent fine mapping has narrowed the B1 region to a 7.5 cM interval on the distal end of 5AL closely linked to marker BW8226_227 (Mackay et al., 2014). In Aegilops tauschii, the donor of the D‐genome in hexaploid wheat, an additional dominant awn suppressor Anathera (Antr) was located distally on 5DS (Nishijima et al., 2018). Deletion of the short arm of chromosome 3B in Chinese Spring also produces an awned phenotype, suggesting that further uncharacterized genes are involved in awn development (Ma et al., 2012).
Awn suppression in barley is controlled by two major genes: the homeobox gene Knox3 (Müller et al., 1995) underlying the Hooded (K) locus that replaces awns with sterile flowers, and a SHORT INTERNODES transcription factor (Yuo et al., 2012) underlying the short awn 2 (lks2) locus (Takahashi, 1955; Roig et al., 2004). In rice, awn suppression is an important domestication trait and is derived from mutations in genes involved in awn development: a helix‐loop‐helix protein Awn‐1 (An‐1), an auxin response factor OsETT2, a YABBY transcription factor DROOPING LEAF (DL), LONG AND BARBED AWN (LABA1) involved in cytokinin biosynthesis, and an EPIDERMAL PATTERNING FACTOR‐LIKE 1 (EPFL1) gene REGULATOR OF AWN ELONGATION 2 (RAE2) or GRAIN NUMBER, GRAIN LENGTH AND AWN DEVELOPMENT1 (GAD1) (Luo et al., 2013; Toriba & Hirano, 2013; Hua et al., 2015; Bessho‐Uehara et al., 2016; Jin et al., 2016). However, awn suppression in wheat does not appear to be related to these genes (Yoshioka et al., 2017).
Variation in awn length across modern wheat cultivars and landraces suggests that awns are variably adaptive in different environments. Previous studies support a role for awns in supplying photosynthate to developing grain of wheat and barley (Grundbacher, 1963; Kjack & Witters, 1974; Motzo & Giunta, 2002; Li et al., 2006; Tambussi et al., 2007; Ali et al., 2010; Maydup et al., 2014), and the location of awns on the wheat head facilitates movement of carbohydrates into kernels and positions them favorably for photosynthesis (Evans et al., 1972; Li et al., 2006; Ali et al., 2010). Awns may continue contributing to photosynthesis if leaves senesce early or are damaged by disease or drought (Tambussi et al., 2007), and the absence of awns can halve the rate of net ear photosynthesis (Evans et al., 1972). Potentially as a consequence of its silica coating, awn tissue tolerates water deficit better than other important photosynthetic tissues such as the flag leaf (Tambussi et al., 2005; Peleg et al., 2010). Besides increased area for photosynthesis, in warmer climates awns can play a role in cooling the wheat spike during grain fill (Motzo & Giunta, 2002). Awned wheats have been demonstrated to perform better in hotter climates or under water stress. The presence of awns is associated with smaller numbers of larger kernels (Rebetzke et al., 2016). Likewise in rice, wild‐type awned plants have larger kernels with a reduced number per panicle when compared to awnless GAD1 mutants (Jin et al., 2016).
Awnless or awnletted wheats are widely cultivated and comprise the dominant morphology in many parts of the world. Potential explanations for the prevalence of awn‐inhibited types is their association with a reduced incidence of pre‐harvest sprouting (King & Richards, 1984; Cao et al., 2016), use of wheat as forage and historical ease of harvest. In warm growing regions such as the southeastern U.S. where awnless varieties are historically dominant, the proportion of awned varieties has increased over the past two decades. Given the importance of awn status in selection of cultivars for local adaptation and end use, and the influence of awns on spike and kernel morphology, a better understanding of the genetic basis of awn suppression also should improve understanding of these processes. The present study investigated the relationship between awn suppression and kernel quality and spike morphology in association and biparental mapping populations. Fine‐mapping was combined with analysis of mutant lines and gene expression to identify a candidate gene responsible for awn suppression at the B1 locus. The companion paper to the present contribution by Huang et al. (2020) presents parallel identification of B1 and, through transcriptome analyses of awnletted B1 overexpressing plants, proposes possible pathways through which B1 may act for awn inhibition.
Materials and Methods
Genome‐wide association analyses (GWAS)
An association mapping panel of 640 elite soft winter wheat breeding lines was grown in two replications as 1 m rows spaced 30 cm apart at Raleigh, North Carolina during the 2016–2017 growing season. At heading, presence or absence of a fully awned phenotype was noted. These lines were entries in collaborative yield testing nurseries in the southeast soft wheat growing region of the United States, the Gulf Atlantic Wheat Nursery (GAWN) and the SunWheat Nursery, over a period of nine growing seasons. Association analyses were performed for grain yield and test weight using historical data available for the 640 entries. The GAWN and SunWheat yield trials were evaluated at one location in up to seven states from 2008 to 2016: Arkansas (Stuttgart or Marianna), Florida (Citra or Quincy), Georgia (Plains), Louisiana (Winnsboro), North Carolina (Kinston), Texas (Farmersville) and Virginia (Warsaw). Experimental designs at each environment were randomized complete block designs with one to three replications. Plot size was typical of yield trial plots for wheat in the region at a minimum of 1.3 m wide and 3.1 m long. The dataset was balanced for individual years, where the same set of genotypes was evaluated across different locations, and unbalanced between different years.
Mixed model analyses of grain yield and test weight data were performed as described in Sarinelli et al. (2019), except that the dataset in the present study was expanded to include the SunWheat nursery. Best linear unbiased estimates (BLUEs) of each genotype were calculated as the estimated genotypic effect plus overall mean and used as the response variable for association mapping.
Genotyping by sequencing (GBS; Elshire et al., 2011) was performed according to Poland et al. (2012), with 96 individual samples barcoded, pooled into a single library and sequenced on an Illumina HiSeq 2500. TASSEL5 GBS v2 pipeline v.5.2.35 (Glaubitz et al., 2014) was used to align raw reads to the International Wheat Genome Sequencing Consortium (IWGSC) RefSeqv1.0 assembly (https://wheat-urgi.versailles.inra.fr/Seq-Repository/Assemblies) using burrows‐wheeler aligner (BWA) v.0.7.12 and call single nucleotide polymorphisms (SNPs) (Li et al., 2009). The SNPs were filtered to retain samples with ≤ 20% missing data, ≥5% minor allele frequency and ≤ 10% of heterozygous calls per marker. Missing SNPs were imputed using Beagle (Browning & Browning, 2016).
The GWAS for awn status, grain yield and test weight were conducted in R v.3.3.1 (R Core Team, 2016) using the gapit package (Lipka et al., 2012). Population structure and relatedness between individuals were accounted for using the first three principal components of the genomic relationship matrix, determined using the GBS markers and the prcomp function in r v.3.3.1 (R Core Team 2016). Markers were declared significant based on the Bonferroni corrected P‐value at alpha = 0.01.
Biparental mapping populations
A population of 341 F5‐derived recombinant inbred lines (RILs) was developed from the cross of awned cultivar LA95135 with awnless SS‐MVP57 (LM population). LA95135 possesses the Rht‐D1b (dwarfing) allele, and the b1 allele for awns. SS‐MPV57 has the Rht‐D1a allele and the Ppd‐D1a allele conferring photoperiod insensitivity, as well as the Tipped1 (B1) allele for awn suppression. Awned (NIL37‐3) and awnless (NIL37‐14) sister lines were derived from a F5 plant of RIL37 heterozygous at B1 and homozygous for Rht‐D1b and Ppd‐D1b.
During the winter of 2016–2017, the LM population was grown in the glasshouse to evaluate spike morphology and kernel weight. Imbibed kernels from each RIL were placed in a cold chamber kept at 4°C for 8 wk and were transplanted into plastic cones (volume 0.7 l, diameter 6.5 cm and depth 25 cm) containing soil mix. Plants were grown in a completely randomized design with four replications in a glasshouse set to a 16 h : 8h, light : dark photoperiod and a 20°C/15°C (day/night) temperature. The primary tillers of each plant were used for evaluation of spike length, spikelet number per spike, kernel weight, and presence or absence of awns.
The LM RIL population was evaluated in the field at Raleigh, NC and Kinston, NC during the 2017–2018 season. The 341 RILs were grown in an incomplete block design with two replications at each location. The population was divided into five blocks, each consisting of 72 entries and the two parents of the population. Sister lines NIL37‐3 and NIL37‐14 were grown at Raleigh, NC in an experiment with two blocks of 25 replications each.
Plots consisted of 1 m rows spaced 30 cm apart. Six representative spikes from each row were harvested and the number of spikelets per spike recorded. Kernels were weighed and counted to determine kernel weight. Rows were hand‐harvested and threshed using a Vogel thresher. A 15‐ml sample of grain was cleaned and morphometric parameters (grain length, width, area and weight) were obtained using a MARVIN grain analyzer (GAT Sensorik GMBH, Neubrandenburg, Germany). An estimate of test weight was obtained by measuring the weight of grain in the 15‐ml sample. BLUEs were calculated for individual RILs using R/lme4, treating genotype as a fixed effect and all other terms as random (Bates et al., 2015).
The RIL population was genotyped and SNP identified using the GBS protocol as described above, except that missing data were not imputed. A linkage map was constructed with these data, KASP markers for major effect loci Rht‐D1 and Ppd‐D1, and the presence or absence of awns as a physical marker using R/qtl and R/asmap (Broman et al., 2003; Taylor & Butler, 2017). Quantitative trait loci (QTL) analysis was performed using composite interval mapping, with significance thresholds for an α = 0.05 determined using 1000 permutations.
Three F2 populations (referred to as G × M, G × S and L × S) were developed from crosses between awned breeding lines from the association mapping panel, GA06493‐13LE6 (G) and LA09264C‐P2 (L), with the awnletted cultivars SS‐MPV57 (M) and SS8641 (S). After heading, a total of 950 plants were evaluated for awn suppression. Fully awned plants were placed in one category, whereas awnletted or awnless plants were placed in a separate category.
Genomic DNA of the RIL and F2 populations was evaluated with KASP markers developed from sequences flanking GBS SNPs in the B1 region (Supporting Information Tables S1, S2) in the LM mapping population. In addition, polymorphisms were identified from exome capture data obtained using a previously described assay (Krasileva et al., 2017) for parental lines of mapping populations used in the USDA/IWYP‐WheatCAP project that included LA95135 and SS‐MPV57 (https://www.triticeaecap.org/qtl-cloning-projects). Linkage maps of the distal region of the long arm of chromosome 5A containing B1 and selected KASP markers for each F2 and the RIL population were developed using R/qtl (Broman et al., 2003).
Scaffold assemblies of awnless winter wheats with the B1 suppressor (Cadenza, Paragon, Robigus, and Claire) and the awned tetraploid wheat Kronos were downloaded from the Earlham Institute website (https://opendata.earlham.ac.uk/opendata/data/). Scaffolds containing portions of the B1 region in the awnless assemblies and Kronos were identified using blast and aligned to the IWSGC reference assembly RefSeqv1.0 of Chinese Spring using LAST (Kiełbasa et al., 2011). SNPs between the awned and awnless wheats for KASP marker design were identified with SAmtools (Li et al., 2009).
Analysis of mutant lines
Kernels of the awnless cv Brundage were treated using fast neutron irradiation with a center total dose of 7 Gy air at the McClellan Nuclear Radiation Center (McClellan, McClellan Park, CA, USA). M1 kernels were planted at the Parker Farm in Moscow, Idaho. The main tiller was harvested from each M1 plant and kernels were planted as 1 m rows in the 2017–2018 crop season. Rows were noted as being either awnless or segregating for presence of awns. Markers in the B1 region were evaluated on DNA isolated from one awnless plant and up to four awned plants from segregating rows. DNA samples of Chinese Spring and the Chinese Spring deletion line 5AL‐6 (TA4535‐6) missing the terminal 32% of 5AL were included as controls to evaluate genome specificity of markers. Progenies from the families with the smallest deletion around B1 were grown in the greenhouse in Raleigh in 2018 to confirm the awned phenotype.
Identification of haplotypes in diverse wheat germplasm
Genomic DNA from a panel of 455 winter and 1984 spring wheat accessions from the core collection of the USDA‐ARS National Small Grains Collection (NSGC) representing global diversity was evaluated using KASP markers developed from a 127 kb region flanking the B1 locus. Data on the presence or absence of awns for winter wheat accessions was gathered from the U.S. National Plant Germplasm System descriptor data (https://npgsweb.ars-grin.gov). The spring wheat accessions were grown as single 1 m rows at Raleigh, NC. At heading time, single tillers were selected for each accession, the presence or absence of awns was noted and genomic DNA was isolated from the flag leaf.
Sequencing the B1 region
The two parents of the RIL population and eight individuals representing diverse haplotypes for markers flanking B1 were selected for Sanger sequencing (Table S3). Amplification of the TraesCS5A02G542800 coding sequence and 7 kb of surrounding nonrepetitive sequence was performed with a nested PCR design to obtain specificity using NEB Longamp polymerase (New England Biolabs, Ipswich, MA, USA). The region was divided into 2.5 kb and 4.5 kb regions, and forward and reverse primers were developed for each region. Forward sequencing primers were designed every 500–800 bp for Sanger sequencing of the region. CodonCode Aligner software (CodonCode Corp., http://www.codoncode.com) was used to check base calls and assemble sequence reads into contigs.
Gene expression
To evaluate expression of candidate genes, tissue from immature inflorescences was collected from primary and secondary tillers of plants of LA95135 and SS‐MPV57, as well as the awned cultivar AGS 2000 (b1) and awnless cultivar Massey (B1). Individual samples from spikelets at similar stages were grouped for analysis to assess developmental variation in gene expression. RNA was isolated from plant tissue using the Zymopure RNA Extraction kit (Zymo Research, Irvine, CA, USA), and reverse transcribed with the ThermoFischer Reverse Transcription kit. The predicted exons of candidate genes TraesCS5A02G542700 and TraesCS5A02G542800 were aligned to similar sequences on chromosomes 4B and 4D to design genome‐specific primers for qPCR with an amplicon size of 100–150 bp and a T m of 60–62°C (Table S4). Quantitative PCR reactions were performed using a CFX384 real time PCR machine (Bio‐Rad Laboratories, Hercules, CA, USA) with Sybr green qPCR Master Mix (Applied Biosystems, Foster City, CA, USA). Reactions included primers for the candidate gene along with a β‐ACTIN control at an annealing temperature of 61°C (Table S4). There was a minimum of six awned and awnless samples at each developmental stage and three technical replications were performed per sample. Cq values were calculated for each replication using the Biorad CFX Maestro software and normalized to expression relative to the endogenous control β‐ACTIN with 2(ACTIN CT−TARGET CT).
As an additional evaluation of gene expression over the course of apical development, the sequences of genes in the B1 region were submitted to the WheatExp wheat expression database (https://wheat.pw.usda.gov/WheatExp/). β‐ACTIN primer sequences were also submitted to verify that its expression is consistent during apical development.
Results
B1 awn suppression is associated with test weight, spikelets per spike and kernel weight
Of the 640 lines evaluated in the eastern soft winter wheat panel, 58% had awns. Association mapping utilizing 14 567 GBS markers identified 30 markers significantly associated (adjusted P‐value < 0.01) with the presence or absence of awns (Table S5). The significant markers were located distally on the long arm of chromosome 5A, consistent with the location of the B1 awn suppressor (Fig. 1). Alignment of markers to IWGSC RefSeqv1.0 of Chinese Spring wheat placed the B1 locus in a 25 Mb region between 681 455 268 bp and 706 705 101 bp. A SNP located at 698 528 417 bp on chromosome 5A (5A28417) was highly significant (P‐value = 7 × 10−57) and co‐segregated with awn status in this set of lines. Association analysis of historical data for test weight and grain yield of the GAWN and SunWheat regional testing nurseries identified a significant association of test weight with SNP 5A28417 (P‐value = 4.1 × 10−7; Fig. 1a). Test weight is a measure of the weight of a standard volume of grain and is a general indicator of grain quality parameters such as kernel size and density. No significant markers for grain yield were identified.
Figure 1.
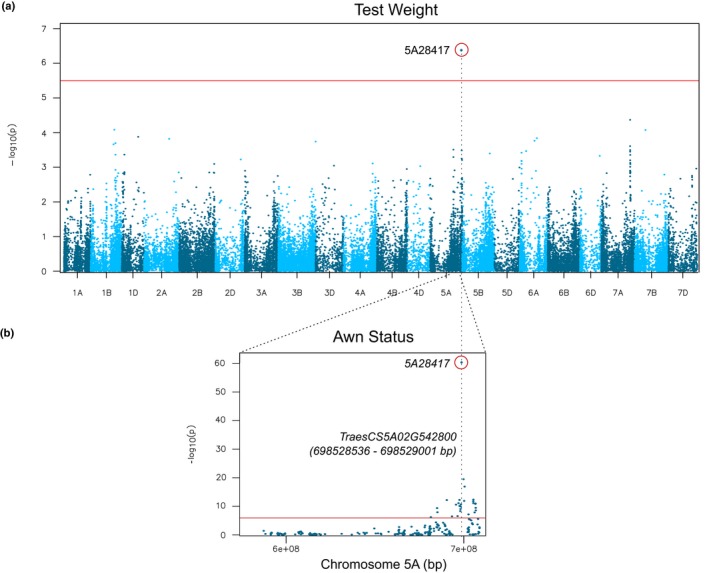
Association analysis identifies the candidate awn inhibition gene on wheat chromosome 5A. Genome‐wide association analysis for test weight (a) and awn status (b). The most significant marker awn status (5A28417) also is the most predictive marker for test weight and is located 219 bp upstream of candidate zinc finger TraesCS5A02G542800.
Analysis of the LM RIL population developed from the cross of fully awned cultivar LA95135 with awnless SS‐MPV57 indicated that awn inhibition was controlled by a single locus on chromosome 5A, co‐segregating with SNP 5A28417. Observed awn phenotypes in RILs having residual heterozygosity at B1 illustrated that the awn suppression was mostly dominant with short awns present on lemmas of apical spikelets of heterozygous plants (Fig. 2a).
Figure 2.
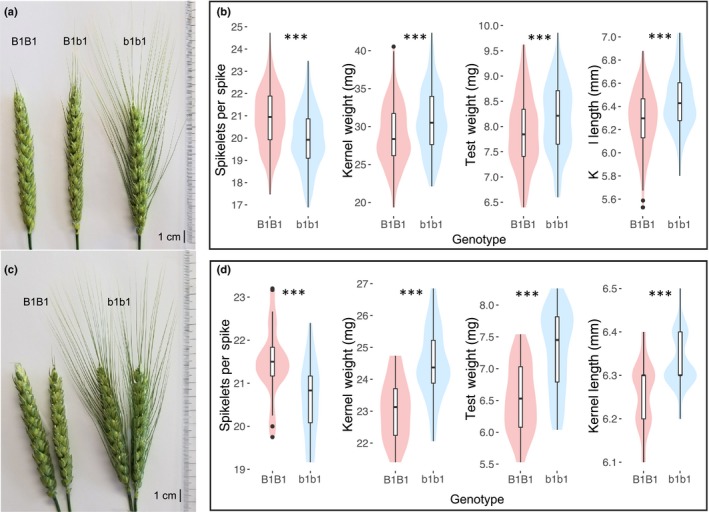
Differences observed between awned and awnless recombinant inbred lines (RIL) and near isogenic lines (NIL) of wheat. (a) Spikes from a segregating field row from the LA95135 × SS‐MPV57 (L × M) RIL population show the effect of the mostly dominant Tipped1 (B1) allele. (b) Comparison of best linear unbiased estimates calculated for spikelets per spike, kernel weight, test weight and kernel length indicate differences between awned vs awnless lines in the L × M RIL population. Significant difference are indicated (***,P < 0.001). (c) Spikes for HIF‐derived NIL families 37‐14 (left) and 37‐3 (right), differing at the B1 locus. (d) The B1 NILs also differ significantly (***,P < 0.001) for all four traits in a highly replicated field experiment in Raleigh, NC in 2018.
Multiple QTL for spikelets per spike and kernel morphometric traits were identified in the LM RIL population, including highly significant QTL associated with awn suppression at the B1 locus and with markers predictive of major effect photoperiod locus Ppd‐D1 and plant height gene Rht‐D1 (Fig. 3; Table S6). The Rht‐D1b semi‐dwarfing allele was associated with the largest effects, contributing to decreases in kernel weight, kernel width and kernel area (Table S6). The Ppd‐D1a allele for photoperiod insensitivity was also associated with decreased kernel weight in the greenhouse and a 2% and 6.1% decrease in number of spikelets per spike in the field and greenhouse, respectively. The presence or absence of awns, included in the genetic map as a physical marker, was significantly associated with QTL for number of spikelets per spike (LOD = 8.4), kernel weight (LOD = 6.5), kernel length (LOD = 9.5), and test weight (LOD = 5.6) (Fig. 3). In the greenhouse experiment, the presence of awns increased thousand kernel weight by 1.47 g (5.1%), and decreased spikelets per spike by 0.48 spikelets. In the field experiment, presence of awns increased kernel weight by 0.88 mg (3.2%), and decreased spikelets per spike by 0.37 spikelets (Fig. 2b). In addition, QTL at B1 were significantly associated with estimated test weight and kernel length in 2018 field data (Fig. 2b). Highly significant (P < 0.001) differences in mean spikelets per spike, kernel weight, estimated test weight, and kernel length were observed when awned and awnless F5‐derived sister lines from heterozygous inbred line RIL37 were evaluated in the 2018 field experiment (Fig. 2c,d). Most notably, awned line NIL37‐3 had 0.85 fewer spikelets per spike, a 6.3% increase in kernel weight, and an 11.6% greater estimated test weight compared with awnless sister line NIL37‐14.
Figure 3.
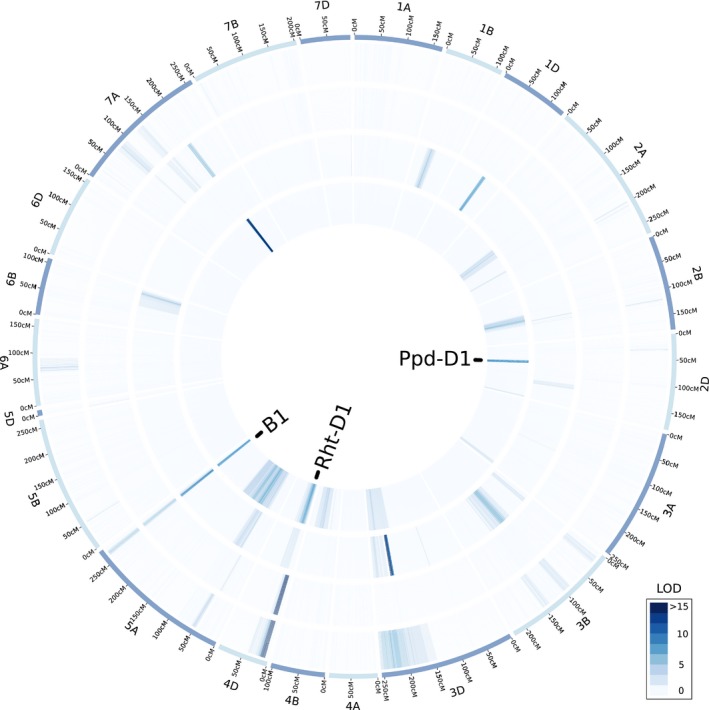
Quantitative trait loci (QTL) mapping in a wheat population segregating for Tipped1 (B1) awn suppression. Heatmap of logarithm of the odds (LOD) scores for composite interval mapping in the LA95135 × SS‐MPV57 recombinant inbred line (RIL) population of traits significantly impacted by presence or absence of awns. Traits from the outer to inner circle are thousand kernel weight, test weight, kernel length and spikelets per spike. Location of the Rht‐D1, Ppd‐D1 and B1 loci are noted in the center. Data on thousand kernel weight, test weight and kernel length were collected from the field in Raleigh, NC in 2018. Data on spikelets per spike were collected from the field in Raleigh and Kinston, NCa in 2018.
Fine mapping identifies B1 candidate genes
Significant SNP 5A28417 identified in the B1 QTL region was located 219 bp upstream of predicted gene model TraesCS5A02G542800 located on chromosome 5A from 698 528 636 bp to 698 529 001 bp in Chinese Spring RefSeqv1.0 (IWGSC 2018). The LM RIL population and three F2 populations developed from crosses between selected awned and awnless individuals from the association panel were genotyped for eight KASP markers targeting an 8.52‐Mb region flanking TraesCS5A02G542800 (Fig. 4; Table S2). In each population, an awnless phenotype was conferred by a single dominant allele co‐segregating with KASP marker 5A28417. Exome capture of the LA95135 and SS‐MPV57 parents of the LM RIL population did not reveal polymorphisms in the TraesCS5A02G542800 coding sequence. Polymorphisms identified in predicted genes proximal (TraesCS5A02G542600 and TraesCS5A02G5426700) and distal (TraesCS5A02G542900) were targeted for marker development. KASP marker 5A15019 targeted an A/G variant in intron five of predicted gene TraesCS5A02G542700 and BW8226_227 targeted a T/G polymorphism in exon two of TraesCS5A02G542600. Marker 5A30334 targeted a C/T variant in exon one of predicted gene TraesCS5A02G542900.
Figure 4.
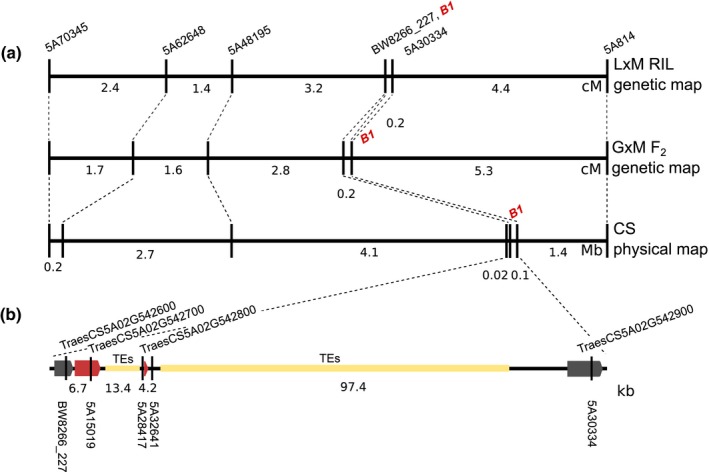
Fine mapping the Tipped1 (B1) locus in wheat. (a) Genetic distances in the B1 region calculated from the LA95135 × SS‐MPV57 (L × M) recombinant inbred line (RIL) population and F2 population GA06493‐13LE6 × SS‐MPV57 (G × M), compared to physical distances obtained using the International Wheat Genome Sequencing Consortium RefSeqv1.0 Chinese Spring reference genome. (b) The fine‐mapped B1 region in Chinese Spring containing candidate gene TraesCS5A02G542800 is shown relative to the genetic and physical maps. The B1 locus co‐segregates with single nucleotide polymorphism (SNP) markers 5A15019, 5A28417 and 5A32641. Genes co‐segregating with awn status are highlighted in red. Approximate positions of SNP markers are shown, including the most significant genotyping by sequencing marker in both the RIL population and association mapping results (5A28417), and the marker most predictive of awn status in global germplasm (5A32641).
Recombination within the biparental populations narrowed the genomic region to a 127‐kb region containing two predicted genes (Fig. 4). Recombination events observed between the awn phenotype and marker 5A30334 in the LM RIL population located the SNP in TraesCS5A02G542900 0.2 cM distal to B1 (Fig. 4a). However, this did not exclude potential causative variation in the TraesCS5A02G542900 promoter region. No recombination was observed between B1 and 5A28417, 5A15019 and BW8226_227 in the LM population. Of the 950 individuals evaluated from the three F2 populations, individual GM#101 from the cross between GA06493‐13LE6 and SS‐MPV57 was determined to be homozygous for BW8226_227 and heterozygous for 5A28417 and 5A15019. The awn phenotype segregated in a progeny test of 16 F3 plants derived from GM#101, placing the SNP in TraesCS5A02G542600 0.2 cM proximal to B1. Mackay et al. (2014) also identified BW8226_22 as associated with but proximal to B1, both in a MAGIC population and a diversity panel. These results narrowed candidate genes underlying B1 awn suppression to predicted genes TraesCS5A02G542700, TraesCS5A02G542800, and the promoter region of TraesCS5A02G542900.
Analysis of awned M2 plants of the awnless variety Brundage identified deletions on the distal part of 5AL encompassing the candidate genes (Table 1). Genomic DNA of awned and awnless M2 plants along with Chinese Spring and deletion line 5AL‐6 (TA4535‐6) missing the terminal 32% of 5AL was used to amplify 11 KASP markers targeting 5AL from 696 Mb to 706 Mb. For all markers, amplification was observed for wild‐type Brundage, and awnless plants from each M2 family and from Chinese Spring. No amplification was obtained for deletion line 5AL‐6, suggesting genome specificity of the primers. Deletions in the region were observed for all 53 awned plants selected from 17 segregating M2 families (Table 1). As expected with gamma irradiation, large deletions were observed with the majority of the M2 lines having lost > 7.5 Mb of the surrounding region. The smallest deletion was observed for mutant line Br‐187 (Fig. 5) and was estimated to be between 19.6 kb and 392.8 kb in size and included candidate genes TraesCS5A02G542700 and TraesCS5A02G542800. Amplification of markers 5A10592 and 5A30334 indicated that TraesCS5A02G542900 was not deleted from Br‐187, eliminating it as a candidate gene (Tables 1, S1). An awnletted phenotype was observed for the B1 hemizygous F1 from the cross of Br‐187 with awnletted cultivar NC‐Neuse (Fig. 5).
Table 1.
Marker analysis of awned mutants of awnless common wheat (Triticum aestivum) cv Brundage.
| Marker name | Positiona (bp) | WT | Number of M2 families | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 4 | 1 | 2 | 1 | 3 | 2 | 1 | |||
| 5A814 | 696993814 | + | − | + | − | + | − | + | + |
| 5A78871 | 697887148 | + | − | − | − | + | − | + | + |
| 5A208800 | 698208800 | + | − | − | − | + | − | + | + |
| 5A13057 | 698513057 | + | − | − | − | − | − | − | − |
| 5A15019 | 698515019 | + | − | − | − | − | − | − | − |
| 5A28417 | 698528417 | + | − | − | − | − | − | − | − |
| 5A32641 | 698532641 | + | − | − | − | − | − | − | − |
| 5A10592 | 698610592 | + | − | − | − | − | − | − | + |
| 5A30334 | 698630000 | + | − | − | − | − | − | − | + |
| 5A48195 | 702748195 | + | − | − | − | − | + | + | + |
| 5A70348 | 705570348 | + | − | − | + | − | + | + | + |
Presence (+) and absence (−) of amplification with markers flanking the Tipped1 (B1) locus in awned M2 families developed by fast neutron irradiation of the awnless cultivar Brundage (WT). Number of families (unique mutation events) for each haplotype is recorded. All reported haplotypes were validated in multiple individuals from each family. WT, wild‐type.
Position in bp on 5A pseudomolecule of the IWGSC RefSeqv1.0 Chinese Spring reference genome.
Figure 5.

Deletion of B1 in awned wheat mutants. Wild‐type Brundage wheat (Br‐WT), Brundage mutant line 187 (Br‐187) having the smallest deletion in the terminal part of chromosome 5AL surrounding TraesCS5A02G542800. The awnletted phenotype is observed in the F1 hybrid from the cross Br‐187 × NC‐Neuse (B1).
Evaluation of awned and awnless wheats in the exome capture dataset identified additional SNPs proximal to and within predicted gene TraesCS5A02G542700 annotated as universal stress protein family with protein kinase domain. Haplotypes were compared to sequence data of the orthologous gene TRIUR3_34498 from Triticum Urartu (https://www.ebi.ac.uk/ena/data/view/GCA_000347455.1), the awned progenitor of the A‐genome in wheat. An SNP unique to awned wheats and T. urartu predicted to create a mis‐sense mutation in exon 9 (5A16541) was targeted for development of a KASP assay. The winter wheat diversity panel was screened with the 5A16541 marker and previously developed marker for intronic SNP 5A15019. Of the 455 lines, 55 having the SS‐MPV57 (B1) 5A15019 allele and 99 individuals with the SS‐MPV57 5A16541 allele possess awns, suggesting that neither of these polymorphisms in TraesCS5A02G542700 underlie B1 awn suppression.
Characterization of B1 candidate gene TraesCS5A02G542800
The fine mapping narrowed the B1 locus to a 127‐kb region of mostly repetitive sequences in the Chinese Spring reference genome containing predicted genes TraesCS5A02G542700 and TraesCS5A02G542800. Marker analysis of wheat germplasm suggested that polymorphisms in predicted gene TraesCS5A02G542700 were not predictive of awn suppression. Thus, TraesCS5A02G542800, annotated as a 366‐bp single exon predicted C2H2 zinc finger transcription factor seemed a strong candidate for the dominant B1 awn suppressor. A predicted protein sequence was used to identify related genes in the Uniprot database. Although similar C2H2 zinc fingers in related grass species were identified, there are no publications characterizing these genes. The most similar characterized gene to TraesCS5A02G542800 is KNUCKLES (KNU) in Arabadopsis thaliana, a C2H2 zinc finger involved in floral development sharing conserved zinc finger and N‐terminal ethylene‐associated response motifs (EAR‐like) (Fig. 6).
Figure 6.
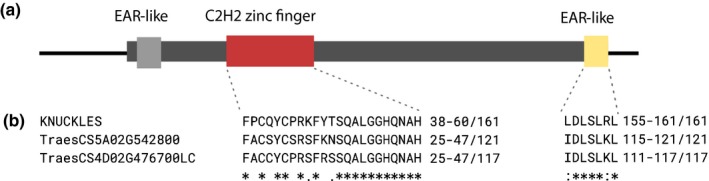
Wheat Tipped1 (B1) candidate is a C2H2 zinc finger protein with ethylene‐associated response (EAR) motif. (a) Gene model of TraesCS5A02G542800 with potential conserved domains. (b) Alignment of functional motifs from characterized Arabidopsis C2H2 zinc finger KNUCKLES with candidate B1 gene TraesCS5A02G542800 and its D‐genome homoeolog TraesCS4D02G476700LC. Cystedeine and histidine residues responsible for zinc finger binding and core plant zinc finger sequence QALGGH are conserved between the wheat genes and KNUCKLES (Takatsuji, 1999 from Payne et al., 2004.) A C‐terminal EAR‐like motif implicated in recruitment of histones in KNUCKLES is conserved (Hiratsu et al., 2002 from Payne et al., 2004). A potential N‐terminal EAR motif is not conserved.
Variation within the TraesCS5A02G542800 coding sequence was not found between the Chinese Spring refseq v.1.0, scaffold assemblies of winter wheats with the B1 suppressor (Cadenza, Paragon, Robigus and Claire) and the awned tetraploid wheat Kronos (https://opendata.earlham.ac.uk/opendata/data/). In Chinese Spring, the gene is surrounded by > 100 kb of repetitive elements, hindering assembly and comparison of the region in the scaffold assemblies. In the available B1 cultivar scaffolds, however, ≥ 10 kb of additional repetitive elements were observed proximal to the candidate gene used herein, suggesting insertion or removal of transposable elements since the divergence of the available B1 assemblies and Chinese Spring.
Gene B1 is located in a region of 5AL originating from a translocation with the 4A chromosome that occurred in the A‐genome diploid progenitor of common wheat (Dvorak et al., 2018). Nucleotide blast was used to identify two regions on chromosomes 4B and 4D syntenic to the fine‐mapped region. Sequences similar to the candidate C2H2 zinc finger were found in both regions, annotated as TraesCS4D02G476700LC on long arm of chromosome 4D (78% nucleotide identity), and unannotated on chromosome 4B (80% nucleotide identity). The 4BL sequence contains a frameshift mutation and is not annotated in either the high confidence or low confidence gene set of the IWGSC v.1.1 annotation (https://wheat-urgi.versailles.inra.fr/Seq-Repository/Annotations). TraesCS4D02G476700LC differs somewhat from TraesCS5A02G542800 in the region between the zinc finger and EAR‐like motifs, including a 10 amino acid insertion in the A‐genome from positions 90–99 (Fig. 6). Phylogenetic analysis by Huang et al. (2020) grouped B1 and its homoeologs with orthologous proteins of the progenitor species T. urartu and Aegilops tauschii.
Upregulation of TraesCS5A02G542800 is associated with B1 awn suppression
In data from WheatEXP, TraesCS5A02G542800 expression increases as the early inflorescence develops, and decreases after the emergence of the head (Fig. S1), whereas TraesCS4D02G476700LC increases in expression later in the development of the spike. The WheatFP browser (http://bar.utoronto.ca/efp_wheat/cgi-bin/efpWeb.cgi) (Winter et al., 2007; Ramírez‐González et al., 2018) was used to localize gene expression in an awned spring variety Azhurnaya. TraesCS5A02G5426800 is most highly expressed in the developing inflorescence, with some expression in the developing awns, ovaries and grain tissues, whereas TraesCS4D02G476700LC is mostly expressed later in spike development (Figs S2, S3). Similar analyses of predicted gene TraesCS5A02G5426700 indicate that this gene is expressed in most tissues, with higher expression in spikes after the development of awn tissue (Fig. S4).
Gene expression data from quantitative reverse trancription (qRT)‐PCR determined that TraesCS5A02G5426800 showed a seven‐fold average increase in gene expression (P < 1e‐7) in awnless plants compared to awned plants (Fig. 7), whereas TraesCS5A02G5426700 was expressed likewise in both awned and awnless individuals (Fig. S5). Developing inflorescences were grouped into four stages based on development (Fig. 7b). Within each group, expression of TraesCS5A02G5426800 was higher in awnless individuals, suggesting a mechanism of awn suppression associated with this gene.
Figure 7.
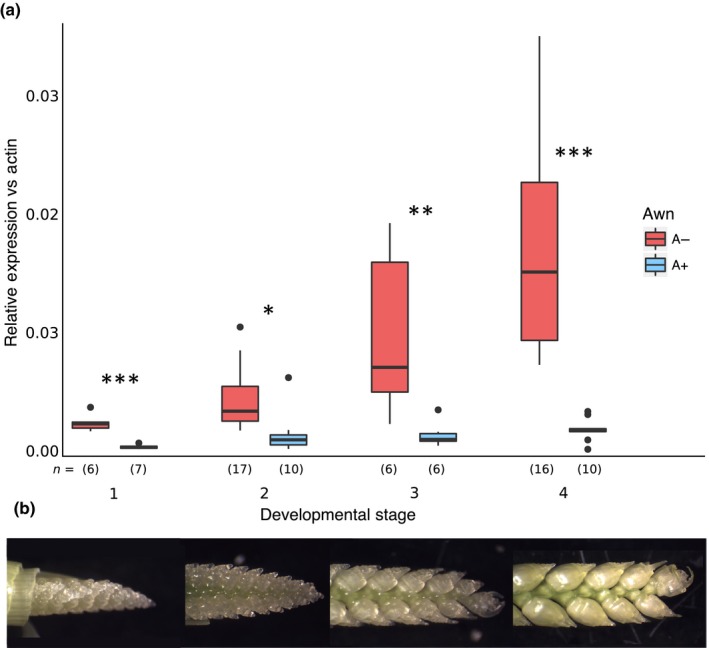
Expression of TraesCS5A02G542800 increases in spikes of awnless wheat. (a) Expression of TraesCS5A02G542800 in apical meristems of awned (blue, A+) and awnless (red, A−) wheat plants at different developmental stages where 1 = younger meristems; and 4 = older meristems. Expression is relative to the reference gene β‐ACTIN. Significant difference between awned and awnless individuals are indicated (***, P < 0.001; **, P < 0.01; *, P < 0.05). Number of biological replicates (n) is given per group, with three technical replications per biological replicate. (b) Representative spikes at each developmental stage.
Given the fine‐mapping of awn status to this interval, differences in expression imply the existence of cis‐regulatory variation either directly upstream in the promoter of TraesCS5A02G5426800 or in nearby enhancers or repressors. A 7‐kb region of nonrepetitive sequences surrounding the B1 candidate gene (3 kb upstream and 4 kb downstream) was sequenced in a set of 10 individuals, including LA95135, SS‐MVP57 and eight accessions of Triticum aestivum selected from the NSGC wheat core collection based on diversity in geographical origin and haplotypes of KASP markers in the region from 698.51 Mb to 698.62 Mb of 5AL (Table S4). No polymorphisms in the TraesCS5A02G5426800 coding sequence were observed, and sequence variation proximal to the gene in the promoter region (including marker 5A28417) was not predictive of the awn suppression phenotype. A 30‐bp deletion 4005 bp downstream of the TraesCS5A02G5426800 start codon was most predictive of awn suppression in this set. KASP marker 5A32641 designed around this deletion co‐segregated with B1 in the biparental mapping populations (Fig. 4).
Haplotype diversity in global wheat germplasm
Marker 5A32641 was highly predictive of awn suppression in 2439 winter and spring wheat accessions in the USDA NSGC core set. Of the 455 winter wheat accessions evaluated 57% were awnless, compared to 45% of 1984 spring wheat accessions (Table S7). The 30‐bp deletion distal to TraesCS5A02G5426800 was present in 98% of awnless winter wheat accessions and all but 59 of 696 awnless spring lines (92%). Awnless lines without the 30‐bp deletion downstream of TraesCS5A02G5426800 may possess either the Hd or B2 awn suppressors. Of the 1558 awned accessions, only 18 were homozygous for the deletion. Of these, 13 accessions are landraces from Sudan, Egypt and Oman, suggesting that they may share a rare genetic variant in either the candidate gene region or at the B1 target. Overall, these results suggest that the dominant B1 inhibitor is the primary determinant of awn suppression in wheat globally.
Eight haplotypes were identified when six SNP markers flanking B1 were examined in conjunction with marker 5A32641 (Table 2). Marker haplotype 1 (Hap1) present in the awned cultivar LA95135 and Hap8 present in awnless cultivar SS‐MPV57 were the most common, detected in 55% and 35% of accessions, respectively (Fig. 8). The majority of awnless wheats were categorized as having the B1‐associated Hap8. A small number of awnless spring wheat accessions from various geographic regions possessed Hap7 characterized by the 30‐bp deletion distal to the candidate B1 gene and differing from Hap8 at marker 5A28417 proximal to B1. By contrast, seven of the eight haplotypes were observed in awned accessions, with Hap1 being by far the most common. Higher haplotype diversity associated with the ancestral b1 allele is expected assuming that B1 originated and spread during or after the domestication of cultivated wheat.
Table 2.
Observed haplotypes near Tipped1 (B1) in diverse common wheat germplasm.
| Number of lines | Marker name | Allele | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Position (bp) | |||||||||
| 5A15019 | 5A28417 | 5A29396 | 5A32641 | 5A91913 | 5A613482 | 5A614919 | |||
| 698515019 | 698528417 | 698529396 | 698532641 | 698591913 | 698613482 | 698614919 | |||
| Hap1 | 1421 | A | A | A | A | A | A | A | b1 |
| Hap2 | 20 | A | B | A | A | A | A | A | b1 |
| Hap3 | 44 | B | B | A | A | B | A | B | b1 |
| Hap4 | 7 | B | B | A | A | B | B | A | b1 |
| Hap5 | 12 | B | B | A | A | B | B | B | b1 |
| Hap6 | 65 | B | B | C | A | B | B | C | b1 |
| Hap7 | 11 | B | A | B | B | B | B | B | B1 |
| Hap8 | 859 | B | B | B | B | B | B | B | B1 |
For each competitive allele specific PCR (KASP) marker, allele A is the LA95135 (b1) allele, allele B the SS‐MPV57 (B1) allele, and allele C a null allele. Position of SNPs on 5A pseudomolecule of International Wheat Genome Sequencing Consortium Chinese Spring RefSeqv1.0 are indicated below the marker name. The B1 allele associated with each haplotype is noted.
Figure 8.
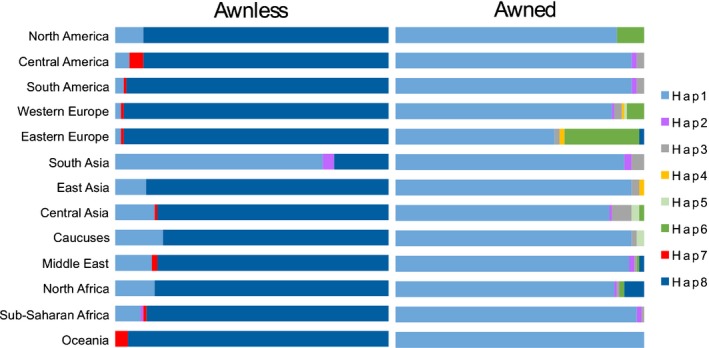
Distribution of haplotypes in the Tipped1 (B1) region in 2439 global wheat accessions. The majority of awn inhibited wheats possess B1‐associated Hap8 and Hap7, implicating B1 as the predominant determinant of awn suppression in all regions except South Asia. Hap1 that are awnless lines likely possess other awn‐inhibiting genes. Although Hap1 is the predominant haplotype associated with the b1 allele in awned accessions, rare haplotypes associated with geographical origin are observed. In Eastern Europe, Hap6 is a major haplotype associated with the presence of awns that is rare outside of Europe and North America. Lines from Central Asia contain a diverse set of haplotypes associated with the presence of awns. Bars are sized based on proportion awnless and awned accessions evaluated from each region.
Hap1 also was observed in awnless accessions that may possess the b1 allele and some combination of the Hd and B2 alleles. Of these lines, 67% originated from Central and South Asia, primarily Nepal and India, suggesting regional variation in control of awn suppression (Fig. 8). The greatest diversity in haplotypes was in accessions from Central Asia where all haplotypes except Hap4 were observed. Although all eight haplotypes were present in accessions from Western Europe, 95% of these accessions had either Hap1 or Hap8.
Discussion
Global variation for awn length in wheat suggests that the presence or absence of awns may be differentially adaptive to varying environments and production systems. Awns are known to contribute to yield in warmer, drier environments, cooling the wheat spike and supplying carbohydrates to developing grain. (Grundbacher, 1963; Kjack & Witters, 1974; Motzo & Giunta, 2002; Li et al., 2006; Tambussi et al., 2007; Ali et al., 2010; Maydup et al., 2014). A significant association of the Tipped1 (B1) awn suppressor with reduced test weight in the association panel used herein suggests that awns influence kernel size in winter wheat grown in the southeastern United States, although there remains the possibility that a tightly linked variant co‐segregating with B1 also is impacting kernel morphology. In the biparental cross of awned cultivar LA95135 with awnless SS‐MVP57 (LM) recombinant inbred line (RIL) population, quantitative trait loci (QTL) analysis confirmed that the presence of awns was associated with increased kernel weight and greater kernel length, but with decreased spikelets per spike. In rice, which has nonphotosynthetic awns, disabling GRAIN LENGTH AND AWN DEVELOPMENT1 (GAD1) suppresses awn elongation, but also decreases grain length while increasing grains per panicle (Jin et al., 2016). Thus, a current hypothesis is that decreased grain size in awnless wheats may also be due to changes in floret development associated with awn inhibition, rather than simply decreased availability of photosynthate.
Except in forage cultivars, the most economically important organ in wheat is the spike. As such, developing a better understanding of the gene networks that control spike development is critical for wheat breeders. Consistent with other studies, it was observed that awn suppression was associated with an increase in the number of spikelets per spike (Rebetzke et al., 2016). A major hindrance to increasing yield of modern wheat cultivars is the availability of sink tissues to fill with carbohydrates – increasing spikelet number is therefore one strategy to increase the number of grains harvested per unit area (Miralles & Slafer, 2007). The final number of spikelets in wheat is correlated with the duration of the reproductive growth period, and genes associated with flowering time in wheat can influence the number of spikelets per spike (Guo et al., 2018). In this QTL mapping study, the segregating Ppd‐D1 (photoperiod sensitivity) flowering time allele was associated with QTL for both spikelets per spike and flowering time, but the B1 region was associated only with spikelets per spike, indicating that B1 influences the number of rachis nodes through a different mechanism.
Association of B1 awn suppression with the upregulation of candidate gene TraesCS5A02G5426800 was identified in parallel by Huang et al. (2020) using RNA‐sequencing of bulked awned and awnless lines. In addition, they found that constitutive over‐expression of TraesCS5A02G5426800 produces an awnletted phenotype in transgenic plants produced from an awned wild‐type. The closest characterized genes to TraesCS5A02G5426800 as identified through protein blast are a set of zinc finger transcription factors in Arabidopsis, all of which contain conserved zinc finger and ethylene‐associated response motifs (EAR‐like) motif domains. This family of transcription factors are usually repressors; the zinc finger domain binds to the target sequence and the EAR‐like motif recruits histones which downregulate the target gene (Kagale & Rozwadowski, 2011). The best‐characterized member of this family, KNUCKLES (KNU), disrupts stem cell maintenance in Arabidopsis (Payne et al., 2004 ). The floral homeotic protein AGAMOUS (AG) upregulates KNU by binding to its promoter, with KNU in turn repressing the homeodomain protein WUSCHEL (WUS) until a specific developmental stage (Sun et al., 2009). WUS is responsible for the maintenance of stem cells in Arabidopsis, and overexpression of KNU pre‐emptively terminates floral meristem development (Sun et al., 2009). In this way, KNUCKLES represses growth of certain floral tissues to allow other floral tissues to develop at the same pace. If B1 plays a similar role in wheat, its overexpression suppressing developing awn tissue, this suggests a potential explanation for the dominance of the B1 allele. This is examined in the B1 companion paper to the present contribution, where constitutive overexpression of B1 suggests a role as a transcriptional repressor (Huang et al., 2020).
Haplotype analysis of global wheat germplasm confirms that B1 is the predominant source of awn suppression in hexaploid wheat. The B1 companion paper in this issue observed similar results in common wheat and identifies this gene as a source of awn suppression in tetraploid Triticum turgidum ssp. durum (Huang et al., 2020). Using six selected KASP assays, two dominant haplotypes were identified in the region of nonrepetitive sequence flanking B1, with a smaller number of lines having haplotypes associated with geographical regions and the greatest number of haplotypes being present in central Asia and Europe. In the companion paper, Huang et al. (2020) identified six B1‐like haplotypes when sequencing a 1961‐bp region flanking the B1 coding sequence and using a different set of markers near B1. They reported a 25‐bp deletion upstream of the candidate gene identified as being linked to B1 but not entirely predictive, and as this deletion is found in T. urartu it is unlikely to be causative. The present authors identified a 30‐bp deletion c. 4 kb downstream of the candidate gene that was not assayed by Huang et al. to be predictive of the B1 awn suppressor in nearly all lines in the present study. The marker 5A32641 assaying this deletion distinguished haplotypes 1–6 associated with b1 from haplotypes 7–8 associated with B1. Alignment of the regions in the B and D genomes syntenic to B1 reveal major variation in transposable elements both upstream and downstream of the candidate gene with expansion observed in the A genome. In addition, assemblies of the region in wheat lines carrying the B1 allele reveal further TE expansion in the region compared to lines carrying the b1 allele. Determining if the observed upregulation of B1 is a product of a binding site mutation, a by‐product of TE insertions, or some other causative polymorphism will require further work.
Wheat's large genome size, hexaploidy and long‐range linkage disequilibrium can make association mapping more challenging than in other species. Using next‐generation sequencing technologies, wheat geneticists have created communal resources that facilitate fine mapping and identification of candidate genes. The newly published wheat reference genome, tools that make it accessible to other researchers and technologies that take advantage of the reference genome (such as gene expression and exome capture datasets), were all used in the present study to reduce the time and cost of positional cloning. In parallel, Huang et al. (2020) also identified overexpression of TraesCS5A02G5426800 as underlying B1, through a combination of bulked segregant RNA‐seq and fine‐mapping in durum wheat, deletion mapping in UK bread wheats and transgenic validation. Cloning of B1 should allow for further experiments to identify the downstream targets and effects of this gene, and to explore its interaction with other developmental genes. Improved understanding of the relationship between awn development and control of spikelets per spike, grain number and grain size in wheat will provide insight into pathways that can be manipulated to increase grain yield. Characterization of B1 adds to our growing understanding of the gene networks underlying spike development in wheat and may help breeders produce varieties better adapted to local climate and end‐use.
Author contributions
ND, MG and GBG planned and designed the research; ND, MG, EL and MS performed experiments and collected phenotypes; ND, EL and PT analyzed genome‐wide sequencing data; QH and DF developed and phenotyped the mutant population; JPM and DM planted and maintained field experiments; AA, KJ and EA performed and analyzed exome capture data of parents; and ND, MG and GBG wrote the manuscript.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Expression of TraesCS5A02G542800 and TraesCS4D01G476700LC in different tissue.
Fig. S2 Expression patterns of TraesCS5A02G542800.
Fig. S3 Expression patterns of TraesCS4D01G476700LC.
Fig. S4 Expression patterns of TraesCS5A01G542700.
Fig. S5 Expression of TraesCS5A02G542700 in apical meristems of awned and awnless wheat.
Table S1 Positions and descriptions of KASP markers.
Table S2 Sequences of KASP markers.
Table S3 Haplotypes and awn status of sequenced lines.
Table S4 Sequences of quantitative reverse transcription (qRT‐)PCR markers.
Table S5 SNP significantly associated with awn suppression.
Table S6 QTL results for yield components in LA95135 × SS‐MPV57 population.
Table S7 Geographical distribution of B1 haplotypes.
Acknowledgements
The authors thank staff of the USDA‐ARS Plant Science Unit for assistance in genotyping and growing field trials, collaborating breeding programs for data from evaluation of GAWN and SunWheat yield trials, and the J. Dubcovsky lab at the University of California Davis for use of instrumentation for phenotyping grain traits. Support was provided by the Agriculture and Food Research Initiative Competitive Grant 67007‐25939 (WheatCAP‐IWYP) from the USDA NIF.
References
- Ali MA, Hussain M, Khan MI, Ali Z, Zulkiffal M, Anwar J, Sabir W, Zeeshan M. 2010. Source‐sink relationship between photosynthetic organs and grain yield attributes during grain filling stage in spring wheat (Triticum aestivum). International Journal of Agriculture and Biology 12: 509–515. [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S. 2015. Fitting linear mixed‐effects models using lme4. Journal of Statistical Software 67: 1–48. [Google Scholar]
- Bessho‐Uehara K, Wang DR, Furuta T, Minami A, Nagai K, Gamuyao R, Asano K, Angeles‐Shim RB, Shimizu Y, Ayano M et al 2016. Loss of function at RAE2, a previously unidentified EPFL, is required for awnlessness in cultivated Asian rice. Proceedings of the National Academy of Sciences, USA 113: 8969–8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen Ś, Churchill GA. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Browning BL, Browning SR. 2016. Genotype imputation with millions of reference samples. The American Journal of Human Genetics 98: 116–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Hayashi K, Tokui M, Mori M, Miura H, Onishi K. 2016. Detection of QTLs for traits associated with pre‐harvest sprouting resistance in bread wheat (Triticum aestivum L.). Breeding Science 66: 260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash SD, Bruckner PL, Wichman DM, Kephart KD, Berg JE, Hybner R, Hafla AN, Surber LMM, Boss DL, Carlson GR et al 2009. Registration of ‘Willow Creek’ forage wheat. Journal of Plant Registrations 3: 185–190. [Google Scholar]
- Dvorak J, Wang L, Zhu T, Jorgensen CM, Luo MC, Deal KR, Gu YQ, Gill BS, Distelfeld A, Devos KM, Qi P. 2018. Reassessment of the evolution of wheat chromosomes 4A, 5A, and 7B. Theoretical and Applied Genetics 131: 2451–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum R, Zaltzman L, Burgert I, Fratzl P. 2007. The role of wheat awns in the seed dispersal unit. Science 316: 884–886. [DOI] [PubMed] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE. 2011. A robust, simple genotyping‐by‐sequencing (GBS) approach for high diversity species. PLoS ONE 6: e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans LT, Bingham J, Jackson P, Sutherland J. 1972. Effect of awns and drought on the supply of photosynthate and its distribution within wheat ears. Annals of Applied Biology 70: 67–76. [Google Scholar]
- Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES. 2014. TASSEL‐GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS ONE 9: e90346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundbacher FJ. 1963. The physiological function of the cereal awn. Botanical Review 29: 366–381. [Google Scholar]
- Guo Z, Chen D, Röder MS, Ganal MW, Schnurbusch T. 2018. Genetic dissection of pre‐anthesis sub‐phase durations during the reproductive spike development of wheat. The Plant Journal 95: 909–918. [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Ohta M, Matsui K, Ohme-Takagi M. 2002. The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Letters 514: 351–354. [DOI] [PubMed] [Google Scholar]
- Hua L, Wang DR, Tan L, Fu Y, Liu F, Xiao L, Zhu Z, Fu Q, Sun X, Gu P et al 2015. LABA1, a domestication gene associated with long, barbed awns in wild rice. Plant Cell 27: 1875–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Zheng Q, Melchkart T, Bekkaoui Y, Konkin DJF, Kagale S, Martucci M, You FM, Clarke M, Adamski NM et al 2020. Dominant inhibition of awn development by a putative zinc‐finger transcriptional repressor expressed at the B1 locus in wheat. New Phytologist 225: 340–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium (IWGSC) , Appels R, Eversole K, Feuillet C, Keller B, Rogers J, Stein N, Pozniak CJ, Choulet F, Distelfeld A, Poland J, Ronen G. 2018. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361: eaar7191. [DOI] [PubMed] [Google Scholar]
- Jin J, Hua L, Zhu Z, Tan L, Zhao X, Zhang W, Liu F, Fu Y, Cai H, Sun X et al 2016. GAD1 encodes a secreted peptide that regulates grain number, grain length, and awn development in rice domestication. Plant Cell 28: 2453–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Rozwadowski K. 2011. EAR motif‐mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6: 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Miura H, Akiyama M, Kuroshima M, Sawada S. 1998. RFLP mapping of the three major genes, Vrn1, Q and B1, on the long arm of chromosome 5A of wheat. Euphytica 101: 91–95. [Google Scholar]
- Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. 2011. Adaptive seeds tame genomic sequence comparison. Genome Research 21: 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Richards RA. 1984. Water uptake in relation to pre‐harvest sprouting damage in wheat: ear characteristics. Australian Journal of Agricultural Research 35: 327–336. [Google Scholar]
- Kjack JL, Witters RE. 1974. Physiological activity of awns in isolines of atlas barley 1. Crop Science 14: 243–248. [Google Scholar]
- Krasileva KV, Vasquez‐Gross H, Howell T, Bailey P, Paraiso F, Clissold L, Simmonds J, Ramirez‐Gonzalez RH, Wang X, Borrill P et al 2017. Uncovering hidden variation in polyploid wheat. Proceedings National Academy Sciences, USA 114: E913–E921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and AMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang H, Li H, Zhang L, Teng N, Lin Q, Wang J, Kuang T, Li Z, Li B et al 2006. Awns play a dominant role in carbohydrate production during the grain‐filling stages in wheat (Triticum aestivum). Physiologia Plantarum 127: 701–709. [Google Scholar]
- Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z. 2012. GAPIT: genome association and prediction integrated tool. Bioinformatics 28: 2397–2399. [DOI] [PubMed] [Google Scholar]
- Luo J, Liu H, Zhou T, Gu B, Huang X, Shangguan Y, Zhu J, Li Y, Zhao Y, Wang Y et al 2013. An‐1 encodes a basic helix‐loop‐helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 25: 3360–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CY, Gao LY, Li N, Li XH, Ma WJ, Appels R, Yan YM. 2012. Proteomic analysis of albumins and globulins from wheat variety Chinese Spring and its fine deletion line 3BS‐8. International Journal of Molecular Sciences 13: 13398–13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay IJ, Bansept‐Basler P, Barber T, Bentley AR, Cockram J, Gosman N, Greenland AJ, Horsnell R, Howells R, O'Sullivan DM et al 2014. An eight‐parent multiparent advanced generation inter‐cross population for winter‐sown wheat: creation, properties, and validation. G3: Genes, Genomes, Genetics 4: 1603–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maydup ML, Antonietta M, Graciano C, Guiamet JJ, Tambussi EA. 2014. The contribution of the awns of bread wheat (Triticum aestivum L.) to grain filling: responses to water deficit and the effects of awns on ear temperature and hydraulic conductance. Field Crops Research 167: 102–111. [Google Scholar]
- McIntosh RA, Dubcovsky J, Rogers JW, Morris CF, Appels R, Xia X. 2014. Catalogue of gene symbols for wheat: 2013‐14 Supplement. Annual Wheat Newsletter 58. [Google Scholar]
- Miralles DJ, Slafer GA. 2007. Sink limitations to yield in wheat: how could it be reduced? Journal of Agricultural Science 145: 139–149. [Google Scholar]
- Motzo R, Giunta F. 2002. Awnedness affects grain yield and kernel weight in near‐isogenic lines of durum wheat. Australian Journal of Agricultural Research 53: 1285–1293. [Google Scholar]
- Müller KJ, Romano N, Gerstner O, Garcia‐Marotot F, Pozzi C, Salamini F, Rohde W. 1995. The barley Hooded mutation caused by a duplication in a homeobox gene intron. Nature 374: 727–730. [DOI] [PubMed] [Google Scholar]
- Nishijima R, Ikeda TM, Takumi S. 2018. Genetic mapping reveals a dominant awn‐inhibiting gene related to differentiation of the variety anathera in the wild diploid wheat Aegilops tauschii . Genetica 146: 75–84. [DOI] [PubMed] [Google Scholar]
- Payne T, Johnson SD, Koltunow AM. 2004. KNUCKLES (KNU) encodes a C2H2 zinc‐finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development 131: 3737–3749. [DOI] [PubMed] [Google Scholar]
- Peleg Z, Saranga Y, Fahima T, Aharoni A, Elbaum R. 2010. Genetic control over silica deposition in wheat awns. Physiologia Plantarum 140: 10–20. [DOI] [PubMed] [Google Scholar]
- Poland JA, Brown PJ, Sorrells ME, Jannink JL. 2012. Development of high‐density genetic maps for barley and wheat using a novel two‐enzyme genotyping‐by‐sequencing approach. PLoS ONE 7: e32253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team 2016. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; http://www.R-project.org [Google Scholar]
- Ramírez‐González RH, Borrill P, Lang D, Harrington SA, Brinton J, Venturini L, Davey M, Jacobs J, Van Ex F, Pasha A et al 2018. The transcriptional landscape of polyploid wheat. Science 361: eaar6089. [DOI] [PubMed] [Google Scholar]
- Rebetzke GJ, Bonnett DG, Reynolds MP. 2016. Awns reduce grain number to increase grain size and harvestable yield in irrigated and rainfed spring wheat. Journal of Experimental Botany 67: 2573–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig C, Pozzi C, Santi L, Müller J, Wang Y, Stile MR, Rossini L, Stanca M, Salamini F. 2004. Genetics of barley hooded suppression. Genetics 167: 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarinelli JM, Murphy JP, Tyagi P, Holland JB, Johnson JW, Mergoum M, Mason RE, Babar A, Harrison S, Sutton R et al 2019. Training population selection and use of fixed effects to optimize genomic predictions in a historical USA winter wheat panel. Theoretical and Applied Genetics 132: 1247–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourdille P, Cadalen T, Gay G, Gill B, Bernard M. 2002. Molecular and physical mapping of genes affecting awning in wheat. Plant Breeding 121: 320–324. [Google Scholar]
- Sun B, Xu Y, Ng KH, Ito T. 2009. A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes and Development 23: 1791–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi R. 1955. The origin and evolution of cultivated barley. Advances in Genetics 7: 227–266. [Google Scholar]
- Takatsuji H. 1999. Zinc-finger proteins: the classical zinc finger emerges in contemporary plant science. Plant Molecluar Biology 39: 1073–1078. [DOI] [PubMed] [Google Scholar]
- Tambussi EA, Bort J, Guiamet JJ, Nogués S, Araus JL. 2007. The photosynthetic role of ears in C3 cereals: metabolism, water use efficiency and contribution to grain yield. Critical Reviews in Plant Sciences 26: 1–16. [Google Scholar]
- Tambussi EA, Nogués S, Araus JL. 2005. Ear of durum wheat under water stress: water relations and photosynthetic metabolism. Planta 221: 446–458. [DOI] [PubMed] [Google Scholar]
- Taylor J, Butler D. 2017. R Package ASMap: efficient genetic linkage map construction and diagnosis. Journal of Statistical Software 79: 1–29.30220889 [Google Scholar]
- Toriba T, Hirano HY. 2013. The DROOPING LEAF and Os ETTIN 2 genes promote awn development in rice. The Plant Journal 77: 616–626. [DOI] [PubMed] [Google Scholar]
- Watkins AE, Ellerton S. 1940. Variation and genetics of the awn in Triticum . Journal of Genetics 40: 243–270. [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An “electronic fluorescent pictograph” browser for exploring and analyzing large‐scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka M, Lehisa JC, Ohno R, Kimura T, Enoki H, Nishimura S, Nasuda S, Takumi S. 2017. Three dominant awnless genes in common wheat: fine mapping, interaction and contribution to diversity in awn shape and length. PLoS ONE 12: e0176148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuo T, Yamashita Y, Kanamori H, Matsumoto T, Lundqvist U, Sato K, Ichii M, Jobling SA, Taketa S. 2012. A short internodes (SHI) family transcription factor gene regulates awn elongation and pistil morphology in barley. Journal of Experimental Botany 63: 5223–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Expression of TraesCS5A02G542800 and TraesCS4D01G476700LC in different tissue.
Fig. S2 Expression patterns of TraesCS5A02G542800.
Fig. S3 Expression patterns of TraesCS4D01G476700LC.
Fig. S4 Expression patterns of TraesCS5A01G542700.
Fig. S5 Expression of TraesCS5A02G542700 in apical meristems of awned and awnless wheat.
Table S1 Positions and descriptions of KASP markers.
Table S2 Sequences of KASP markers.
Table S3 Haplotypes and awn status of sequenced lines.
Table S4 Sequences of quantitative reverse transcription (qRT‐)PCR markers.
Table S5 SNP significantly associated with awn suppression.
Table S6 QTL results for yield components in LA95135 × SS‐MPV57 population.
Table S7 Geographical distribution of B1 haplotypes.


