Abstract
Adhesion G protein–coupled receptors (aGPCRs) represent a distinct family of GPCRs that regulate several developmental and physiological processes. Most aGPCRs undergo GPCR autoproteolysis-inducing domain–mediated protein cleavage, which produces a cryptic tethered agonist (termed Stachel (stinger)), and cleavage-dependent and -independent aGPCR signaling mechanisms have been described. aGPCR G1 (ADGRG1 or G protein–coupled receptor 56 (GPR56)) has pleiotropic functions in the development of multiple organ systems, which has broad implications for human diseases. To date, two natural GPR56 ligands, collagen III and tissue transglutaminase (TG2), and one small-molecule agonist, 3-α-acetoxydihydrodeoxygedunin (3-α-DOG), have been identified, in addition to a synthetic peptide, P19, that contains seven amino acids of the native Stachel sequence. However, the mechanisms by which these natural and small-molecule agonists signal through GPR56 remain unknown. Here we engineered a noncleavable receptor variant that retains signaling competence via the P19 peptide. We demonstrate that both natural and small-molecule agonists can activate only cleaved GPR56. Interestingly, TG2 required both receptor cleavage and the presence of a matrix protein, laminin, to activate GPR56, whereas collagen III and 3-α-DOG signaled without any cofactors. On the other hand, both TG2/laminin and collagen III activate the receptor by dissociating the N-terminal fragment from its C-terminal fragment, enabling activation by the Stachel sequence, whereas P19 and 3-α-DOG initiate downstream signaling without disengaging the N-terminal fragment from its C-terminal fragment. These findings deepen our understanding of how GPR56 signals via natural ligands, and a small-molecule agonist may be broadly applicable to other aGPCR family members.
Keywords: collagen, cell signaling, development, G protein–coupled receptor (GPCR), shedding, 3-α-DOG, ADGRG1/GPR56, GPCR autoproteolysis-inducing (GAIN) domain, tethered agonist, TG2
Introduction
Adhesion G protein–coupled receptors (aGPCRs)5 comprise a major class of adhesion receptors that mediate cell–cell and cell–extracellular matrix (ECM) interactions (1–4). Among the 33 members of the aGPCR class, several have emerged as critical regulators of development with important implications for human health and disease (1, 3, 4). Two structural characteristics distinguish aGPCRs from all other GPCRs. These include their large multidomain N termini with adhesive modules and their unique juxtamembrane GPCR autoproteolysis-inducing (GAIN) domain (1–5).
Most aGPCRs undergo GAIN domain–mediated autoproteolysis during protein maturation to generate an N-terminal fragment (NTF) containing adhesion domains and most of the GAIN domain and a C-terminal fragment (CTF) containing a cryptic tethered peptide agonist, called Stachel, that emanates extracellularly from the seven-transmembrane-spanning (7TM) domain (6, 7). The NTF and CTF remain noncovalently associated on the plasma membrane (1–5, 8). Currently, there are three models to describe aGPCR activity modulation: ligand binding dissociates the NTF from its CTF, exposing the tethered agonist to activate downstream signaling via heterotrimeric G proteins (6, 7, 9, 10); direct and transient interaction between the extracellular and 7TM domains alters downstream signaling in a Stachel-independent manner (11, 12); or ligand binding induces a conformation change that exposes the Stachel sequence and leads to receptor activation without cleavage and/or removal of the NTF (13). These models are unlikely to be mutually exclusive and could, in principle, occur in the same receptor.
ADGRG1 (GPR56) has critical functions in the development of several organ systems with broad implications for human diseases and their treatment (3, 14–31). Mutations in GPR56 cause a severe human brain malformation called bilateral frontoparietal polymicrogyria, characterized by cortical lamination defects, cerebellar hypoplasia, and central nervous system (CNS) hypomyelination (18, 32). In the CNS, GPR56 regulates embryonic brain development and postnatal CNS myelination (22, 26, 27, 33–37). Myelin is a multilayered glial membrane surrounding axons in the vertebrate nervous system. In the CNS, myelin is formed by specialized glial cells called oligodendrocytes (38). GPR56 promotes oligodendrocyte development and CNS myelination by coupling to Gα12/13 and leading to activation of the RhoA pathway (26, 27). In the peripheral nervous system, GPR56 regulates myelin formation and maintenance (23).
As a member of the aGPCR family, GPR56 undergoes GAIN domain–mediated cleavage to generate an NTF and a CTF, which remain noncovalently associated on the plasma membrane (8, 39). Current literature supports cleavage-dependent (7, 40) and cleavage-independent signaling mechanisms for GPR56 (11, 12). To date, there are two natural ligands, collagen III and tissue transglutaminase 2 (TG2), and one small molecule agonist, 3-α-acetoxydihydrodeoxygedunin (3-α-DOG), of GPR56. Collagen III is the ligand of GPR56 in the developing cerebral cortex and skeletal muscles (28, 35, 36, 41) and TG2 in melanoma cells (39) and oligodendrocytes (22). Additionally, a 19-amino-acid synthetic peptide (P19) that mimics the GPR56 Stachel sequence functions as a natural tethered agonist for GPR56 (42). However, it remains elusive whether these confirmed natural and small-molecule agonists of GPR56 require GAIN domain–mediated cleavage to activate G-protein signaling downstream of the receptor.
In this study, we show that P19 can equally activate both cleaved and noncleaved receptors, whereas the other three agonists require GAIN domain–mediated receptor autoproteolysis. Furthermore, we demonstrate that TG2 ligand action demands the presence of a matrix protein, laminin, thus mediating a tripartite form of signaling among GPR56, TG2, and matrix protein. Taken together, our results provide support for cleavage-dependent GPR56 activation by its natural and small-molecule agonists.
Results
GAIN domain–mediated cleavage is required for GPR56 activation by natural and small-molecule agonists
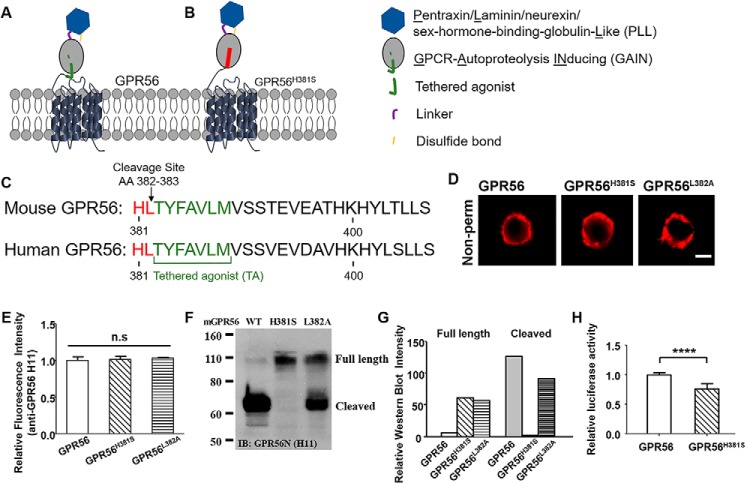
To investigate the requirement of GAIN domain–mediated autoproteolysis for GPR56 signaling, we first generated GPR56 mutants that abolish receptor cleavage but retain cell surface expression. Based on the published GAIN domain crystal structures (5), we made two missense mutants, H381S and L382A (Fig. 1, A–C). Immunostaining with an anti-mouse GPR56 mAb, H11 (35), showed that both mutant receptors had cell surface abundances that are comparable with the WT receptor (Fig. 1, D and E). Western blot analysis revealed that GPR56H381S completely lacks the cleaved NTF, whereas ∼ 56% of the GPR56L382A is cleaved (Fig. 1, F and G), showing that the GPR56H381S receptor is defective in undergoing GAIN domain–mediated cleavage. Therefore, we chose GPR56H381S for the remainder of the study. We next examined the basal activity of this mutant receptor compared with WT GPR56 using a serum response element (SRE)–luciferase assay and found that GPR56H381S mutant has significantly reduced basal activity (Fig. 1H). The lower basal activity associated with the GPR56H381S mutant is not the result of poor overall health of the transfected cells because there were comparable cell numbers over a 48-h period after transfection between cells transfected with WT and mutant receptors (Fig. S1).
Figure 1.
Generation of noncleavable mutant GPR56. A, schematic of GPR56, including the PLL, pentraxin/laminin/neurexin/sex-hormone-binding-globulin-Like domain (blue), cleavable GAIN domain (gray; tethered agonist, green), and 7-transmembrane domain. The GAIN domain can be cleaved between amino acids 382 and 383. B, noncleavable mutant GPR56H381S has no cleavage site in the GAIN domain. C, comparison of the tethered agonist regions of mouse and human GPR56. The tethered agonist (TA, green) is conserved between two species. The mutant sites of GPR56H381S and GPR56L382A are highlighted in red. The arrow indicates the cleavage site. D, expression of WT GPR56 and mutant GPR56 (GPR56H381S and GPR56L382A). Scale bar = 5 μm. E, no significant difference was observed among their expression. F and G, Western blotting of whole-cell lysates of cells expressing WT and mutant constructs. The presence of full-length GPR56 and cleaved fractions was determined with the GPR56 N-terminal antibody. IB, immunoblot. H, basal activity of WT and GPR56H381S constructs as measured by the SRE–luciferase reporter assay. Data are presented as mean ± S.D.; n = 3; ****, p < 0.0001; ns, not significant; two-tailed Student's t test.
We next investigated whether GAIN domain–mediated receptor cleavage is required for GPR56 activation by the known agonists collagen III (35), 3-α-DOG (42), and P19 (42), none of which require cofactors for receptor activation. Collagen III, P19, and 3-α-DOG all equally activated WT receptor–mediated downstream G protein–dependent signaling by SRE–luciferase assay (Fig. 2A). However, only P19 could elicit signaling from the GPR56H381S mutant receptor (Fig. 2). To further confirm this observation, we performed GTP-RhoA pulldown assays to measure levels of activated RhoA upon ligand stimulation. Treatment with 3-α-DOG, P19, and collagen III resulted in robust RhoA activation (Fig. 2, B and C). This result indicated that the GPR56H381S mutant receptor retained signaling competence but did not respond to natural or small-molecule agonist stimulation.
Figure 2.
GAIN domain cleavage is required for GPR56 activation. A, cells transfected with WT GPR56 and GPR56H381S were treated with 3-α-DOG (5 μm), P19 (20 μm), or collagen III (Col III, 50 nm) in an SRE–luciferase reporter assay. Data are normalized to the Renilla luciferase control and expressed as -fold over the signal obtained from cells transfected with SRE–luciferase only. Data are presented as mean ± S.D.; n = 3; *, p < 0.05; ***, p < 0.001; ****, p < 0.0001; two-tailed Student's t test. B and C, RhoA activation by treatment with 3-α-DOG (5 μm), P19 (20 μm), or collagen III (50 nm) on WT GPR56-expressing HEK293T cells. Data are presented as mean ± S.D.; n = 3; *, p < 0.05; **, p < 0.01; two-tailed Student's t test.
TG2-mediated GPR56 activation requires both receptor cleavage and the presence of laminin
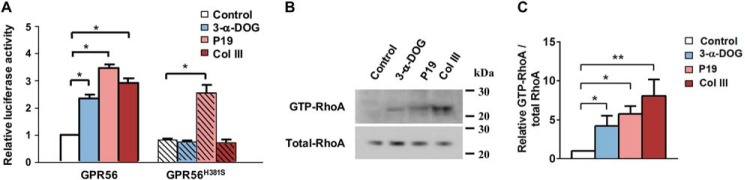
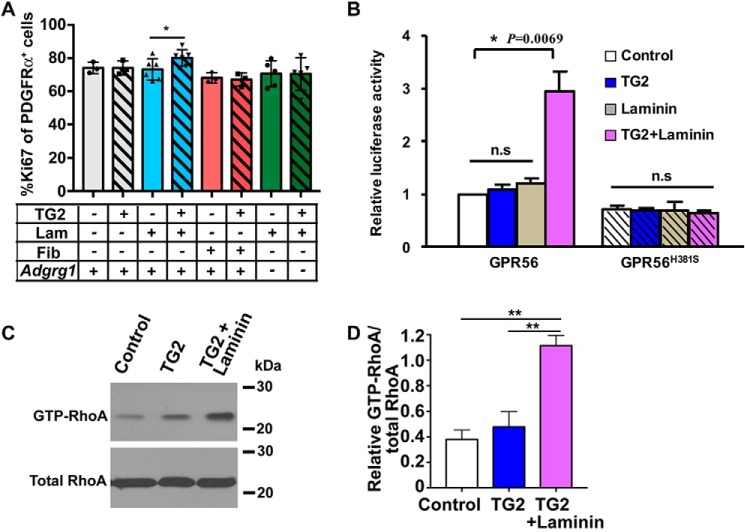
TG2 was first identified as a binding partner of GPR56 in melanoma cells, although the signaling consequences of this interaction were not revealed (39, 43). Very recently, we discovered that TG2 is also the ligand of oligodendrocyte-expressed GPR56 during CNS myelination by promoting OPC proliferation (22). We performed in vitro experiments using Gpr56+/+ and Gpr56−/− oligodendrocyte precursor cells (OPCs) and recombinant TG2 and found that TG2 can only augment the proliferation of OPCs in the presence of laminin (Fig. 3A). This effect was GPR56-dependent and highly specific because TG2 plus laminin had no effect on Gpr56−/− OPCs and fibronectin, another extracellular matrix protein that is known to interact with TG2, had no effect in this assay (Fig. 3A).
Figure 3.
TG2 requires the presence of the ECM protein laminin to activate GPR56. A, TG2 promotes oligodendrocyte precursor cell proliferation in an GPR56- and laminin (Lam)-dependent manner. B, cells transfected with WT GPR56 and GPR56H381S were treated with TG2 (200 nm), laminin (2 μg/ml), or TG2 plus laminin in an SRE–luciferase reporter assay. Data are normalized to Renilla luciferase control and expressed as -fold over the signal obtained from cells transfected with SRE–luciferase only. Data are presented as mean ± S.D.; n = 3; **, p < 0.01; ns, not significant; two-tailed Student's t test. C and D, RhoA activation by treatment with TG2 (200 nm) or TG2 plus laminin (2 μg/ml) of WT GPR56-expressing HEK293T cells. Data are presented as mean ± S.D.; n = 3; **, p < 0.01; two-tailed Student's t test. Fib, fibronectin.
To extend our characterization of TG2 as the ligand for GPR56, we investigated its ability to activate GPR56 downstream signaling in vitro on WT GPR56 and cleavage-deficient GPR56H381S using SRE–luciferase assays. We found that TG2 could activate WT GPR56 in the presence of laminin, whereas TG2 or laminin alone were not effective (Fig. 3B). Similarly, we only observed a robust elevation of GTP-RhoA upon dual stimulation of TG2 and laminin (Fig. 3, C and D). In contrast, the GPR56H381S receptor was not responsive to stimulation with TG2 and laminin together (Fig. 3B), indicating that GPR56 cleavage is required for TG2/laminin-mediated GPR56 activation.
Ligand-induced shedding of GPR56 NTF is required for downstream signaling
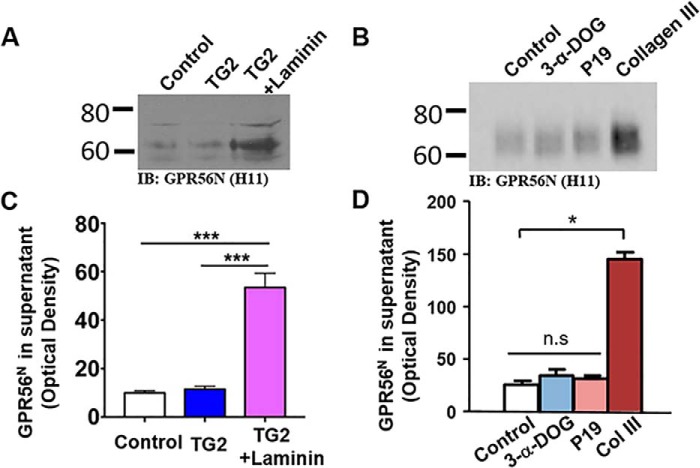
A potential explanation for the inability of TG2 binding alone to activate GPR56 is that this interaction is not sufficient to dissociate the NTF from its CTF. As a corollary, we hypothesized that GAIN domain–mediated cleavage was also required to generate a receptor permissive for dissociation of the NTF by TG2 plus laminin. To test this, we treated serum-starved, GPR56-transfected HEK293T cells with TG2 alone or TG2 plus laminin for 10 min. The cell pellets were used to measure GPR56-stimulated GTP-RhoA levels by RhoA pulldown assay (Fig. 3, C and D), and the supernatants were concentrated and subjected to Western blot analysis with the anti-GPR56 NTF antibody H11. Strikingly, we found that TG2 in the presence of laminin causes GPR56 NTF dissociation and shedding in conjunction with increased GTP-RhoA levels in the cell pellet, whereas TG2 alone was incapable of dissociating the GPR56 NTF and did not lead to increased GTP-RhoA levels (Figs. 4, A and C, and 3, C and D). We demonstrate that GPR56 mediates a tripartite interaction of receptor, ligand, and matrix protein and that ligand-induced shedding of GPR56 NTF is required for downstream signaling.
Figure 4.
Dissociation and shedding of GPR56 NTF. A and B, HEK293T Cells transfected with WT GPR56 were treated with TG2 (200 nm), TG2 plus laminin (2 μg/ml), 3-α-DOG (5 μm), P19 (20 μm), or collagen III (50 nm). Western blotting reveals the NTF of GPR56 in the supernatant. IB, immunoblot. C and D, quantification of GPR56 NTF in the supernatant. Data are presented as mean ± S.D.; n = 3; *, p < 0.05; ***, p < 0.001; ns, not significant; two-tailed Student's t test.
We next asked whether dissociation of the GPR56 NTF is a general mechanism for all of its known ligands. To test this, we performed similar NTF shedding experiments for 3-α-DOG, P19, and collagen III. Only collagen III treatment, but not 3-α-DOG and P19, resulted in increased levels of NTF in the supernatant (Fig. 4, B and D).
3-α-DOG is a partial orthosteric agonist of GPR56
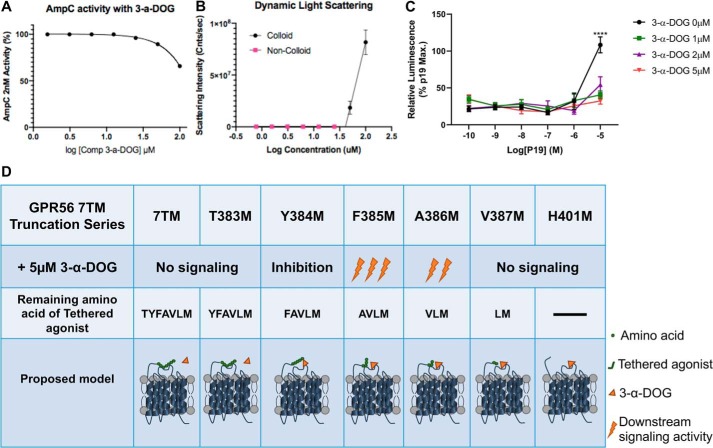
3-α-DOG is a small molecule that was first identified in a high-throughput screening assay (42). It activates truncated GPR56 7TM with an EC50 of ∼5 μm (42). Small-molecule aggregations or nonspecific inhibiting or activating proteins in vitro could be a major source of false positive results in drug discovery (44–46). Therefore, before exploring the 3-α-DOG activation mechanism, we examined whether 3-α-DOG is an aggregator using an AmpC β-lactamase enzymatic assay. AmpC is one of the most sensitive enzymes and has been studied extensively for aggregate-based inhibition (47). 2 nm AmpC β-lactamase was incubated with varying concentrations of 3-α-DOG (from 1.56 μm to 100 μm) for 5 min at room temperature. 3-α-DOG reduced AmpC activity to ∼65% only at the highest concentration of 100 μm (Fig. 5A). To investigate whether 3-α-DOG forms colloids, a dynamic light scattering assay was used (48). Different concentrations of 3-α-DOG were measured at a wavelength of 826.6 nm. 3-α-DOG scattered as an aggregate only at 50 and 100 nm (Fig. 5B). Taken together, these results suggest that 3-α-DOG is not an aggregator at any relevant concentration near its EC50.
Figure 5.
The activation mechanism of 3-α-DOG on GPR56. A, AmpC β-lactamase inhibition response curve with different concentrations of 3-α-DOG. B, critical aggregation concentration curve for 3-α-DOG by dynamic light scattering assay. C, 3-α-DOG inhibits P19 responses. SRE–luciferase reporter assay responses were normalized to the maximal P19 response in the absence of 3-α-DOG. Data are presented as mean ± S.D.; n = 3; ****, p < 0.0001; two-tailed Student's t test. D, summary of 3-α-DOG (orange triangle) activity on various truncated GPR56 7TM receptor (42). The seven-amino-acid Stachel is depicted as seven solid green circles (each represents one amino acid).
To investigate whether 3-α-DOG functions as either an orthosteric activator or an allosteric modulator, we performed a competition assay by examining the efficacy of P19 in the presence of 3-α-DOG. To our surprise, 3-α-DOG inhibits P19 activity at concentrations as low as 1 μm (Fig. 5C). It is worth noting that a previously published result demonstrated that 3-α-DOG requires the presence of three of the seven amino acids (VLM) in the Stachel sequence to activate the receptor (Fig. 5D) (42). Taken together, our findings support that 3-α-DOG is a partial orthosteric agonist that requires the presence of a partial Stachel sequence to activate the receptor.
Discussion
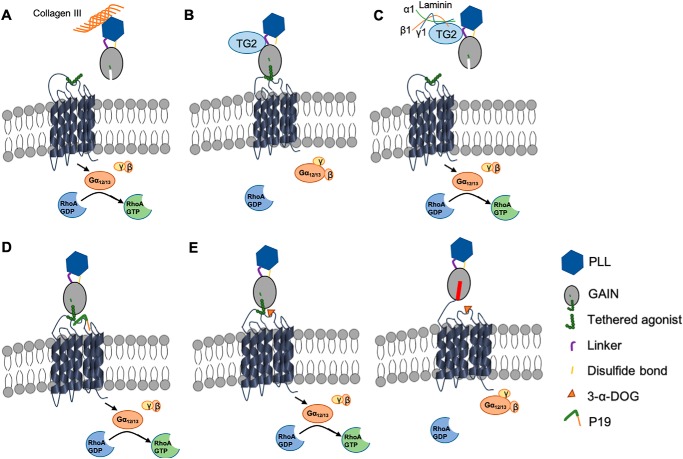
The evidence to date indicates aGPCR signaling modes that are either GAIN domain–mediated receptor cleavage–dependent or cleavage-independent (1, 3, 6, 7, 9, 11–13). However, most of the studies that were performed to arrive at these models did not test the influence of natural ligands or agonists. In this study, we investigated the signaling requirements of natural GPR56 ligands, a synthetic peptide agonist, and a small-molecule agonist. We focused on Gα12/13 and the RhoA pathway because this GPR56 signaling pathway is the only one to have been documented in vivo (22, 26, 27, 35) and in vitro (7, 41, 42). To investigate the requirement of receptor cleavage for its downstream signaling, we generated a cleavage-deficient receptor, GPR56H381S. We demonstrated that both natural ligands (collagen III and TG2) require receptor cleavage and activate GPR56 via a Stachel-dependent mechanism. Our previous work showed that collagen III facilitates RhoA activation by removing the GPR56 NTF from its CTF (40). Here we further demonstrate that collagen III requires GAIN domain–mediated receptor cleavage to initiate downstream signaling (Fig. 6A).
Figure 6.
Schematic of the GPR56 cleavage-dependent signaling pathway. A, collagen III binds to GPR56 NTF, causing conformational changes, including NTF shedding. Exposure of the Stachel tethered agonist results in 7TM domain activation. B, TG2 binds to GPR56 but fails to activate the receptor in the absence of the ECM protein laminin. C, combined binding of TG2 and laminin to GPR56 induces dissociation of GPR56 NTF from its CTF, allowing Stachel to initiate downstream signaling. D, P19 possesses the seven amino acids of the tethered agonist and activates downstream signaling. E, 3-α-DOG binds to part of the orthosteric site of the receptor and activates GPR56 when the other pocket of the ligand binding site is engaged by part of Stachel. Stachel of the noncleavable receptor GPR56H381S is buried by the NTF and therefore cannot be activated by 3-α-DOG.
Our results are consistent with published results. Using engineered truncated GPR56 CTFs that either possess or lack the Stachel sequence, Hall and co-workers (12) demonstrated that the Stachel sequence is required in a pathway-dependent manner; activation of SRE luciferase required the presence of the Stachel sequence, whereas transforming growth factor α shedding and activation of nuclear factor of activated T cells (NFAT) luciferase were Stachel-independent.
TG2 has been shown previously to be the GPR56 binding partner in melanoma cells (39). Recently, we showed, through a combined genetic and unbiased proteomic approach, that TG2 is the ligand of oligodendrocyte GPR56 (22). However, the downstream signaling of this binding remained largely unknown. This study elucidates a novel tripartite signaling model for GPR56, TG2, and laminin. TG2 has also been shown to interact directly with laminin (49). Here we show that TG2, in the presence of laminin, binds to the GPR56 NTF and dissociates the NTF from the CTF, allowing the endogenous GPR56-tethered peptide agonist to initiate G-protein signaling (Fig. 6, B and C). It is not possible to directly test the need for the Stachel sequence for GPR56 signaling because this will render a noncleavable receptor. Nevertheless, our data are very compelling in support of the Stachel-dependent signaling mechanism by demonstrating that shedding of the NTF leads to receptor activation (Figs. 3 and 4).
As P19 is a synthetic mimetic of the authentic tethered peptide agonist, it presumably engages the receptor orthosteric site, which is unaffected in both the cleaved and cleavage-deficient receptor (Fig. 6D). On the contrary, 3-α-DOG can only activate the cleaved receptor, unlike the synthetic peptide agonist P19 (Fig. 2). Combining the previously published results using a series of truncated GPR56 7TM constructs that contain various Stachel sequence lengths (Fig. 5D), our results support the notion that 3-α-DOG functions as a partial orthosteric agonist of GPR56 that requires the presence of part of the Stachel sequence (Fig. 6E). The crystal structure of GPR56 NTF confirms partial exposure of the Stachel sequence in cleaved GPR56 (50). We suspect that the noncleavable GPR56H381S receptor does not possess such exposure and therefore cannot be activated by 3-α-DOG (Fig. 6E).
Experimental procedures
Antibodies and other reagents
For (IHC) Immunohistochemistry and Western blot analyses, we used the following primary antibodies: mouse anti-GPR56 (H11, 1:200) (35) and mouse anti-RhoA (Cytoskeleton, catalog no. ARH03-A, 1:500). Secondary antibodies were goat anti-mouse Alexa 546 (ThermoFisher, 1:1000) and goat anti-mouse IgG-HRP (Sigma, catalog no. A4416, 1:3000). Recombinant laminin was purchased from Invitrogen, and P19 peptide was synthesized by Biomatik USA. The dual SRE–luciferase construct (50) was a kind gift from Dr. Demet Araç (University of Chicago).
Generation of constructs
Mouse GPR56 cDNA was cloned into the pCDNA3.1(+) vector as described previously (34); mutant GPR56H381S and GPR56L382A constructs were created by site-directed mutagenesis using the QuikChange II XL site-directed mutagenesis kit (Stratagene) as described previously (8). The primers for these mutageneses were as follows: H381S forward, 5′-catcctg cctgtgcaac TCcctgacct actttgcagt-3′; H381S reverse, 5′-actgcaaagt aggtcaggGA gttgcacagg caggatg-3′; L382A forward, 5′-cctg cctgtgcaac cacGCgacct actttgcagt gc-3′; L382A reverse, 5-gc actgcaaagt aggtcGCgtg gttgcacagg cagg-3′. These mutant constructs were confirmed by Sanger sequencing.
Cell surface immunofluorescence imaging and Western blotting
HEK293T cells were used for immunoblotting and immunocytochemistry experiments. For cell surface staining of GPR56, HEK293T cells were plated on poly-d-lysine coated coverglass and transfected with WT, H381S, or L382A mutant Gpr56 cDNA using Xfect transfection reagent (Clontech) and cultured for 48 h. The transfected cells were fixed in 2% (PFA) paraformaldehyde for 10 min without permeabilization and then incubated with a mouse anti-GPR56 (H11) antibody visualized by mouse Alexa 546. Images were captured by a confocal LSM 510 NLO system.
For Western blotting, transfected HEK293T cells were harvested 2 days after transfection and solubilized for 1 h at 4 °C in RIPA buffer (Boston BioProducts). The cell lysates were used for Western blotting with a standard Western blotting protocol.
OPC cultures
OPCs were isolated from mixed male and female P5–8 Gpr56+/+ or Gpr56−/− mouse forebrains as described previously (51, 52). Briefly, OPCs were purified by negatively selecting with mouse anti-Thy1.2 (Serotec, catalog no. MCA02R) and mouse anti-GalC (Millipore, catalog no. MAB342), followed by mouse anti-O4 (O4 hybridoma supernatant) for positive selection. After releasing OPCs from the O4 plate by trypsinization, cells were resuspended in proliferation medium containing (PDGF-AA) Platelet-derived growth factor composed of two A subunits and NT-3 (PeproTech). OPCs were plated on coverslips coated with poly-d-lysine before coating with laminin I (R&D Systems, catalog no. 3400-010-01) or fibronectin (Millipore, catalog no. 341668) as described previously (53). After 24 h, TG2 (2 nm) was added to the cultures and incubated for an additional 24 h before fixation with 4% PFA and staining for PDGFRα and Ki67.
TG2 activation assay
HEK 293T cells were transiently transfected with mouse Gpr56 cDNA. Twenty-four hours after transfection, the cells were subjected to serum starvation for 36 h, followed by addition of recombinant TG2 (200 nm) with or without recombinant laminin (2 μg/ml) or acetic acid as a control for 10 min. The conditioned media were harvested, filtered, and concentrated as described previously (8). Equal volumes of the concentrated media were used for Western blotting (40), whereas the cell pellets were used for the Rho-GTP pulldown assay.
Rho-GTP Rhotekin pulldown assay
The Rho-GTP pulldown assay was performed as described previously (35), using HEK293T cells transfected with mouse Gpr56 cDNAs. The transfected cells were stimulated with recombinant TG2 (200 nm) with or without recombinant laminin (2 μg/ml). The treated cells were lysed in 300 μl of ice-cold RIPA buffer containing protease inhibitor mixture (Boston BioProducts). Equal amounts of total lysate protein were incubated with 60 μg of GST-RBD beads at 4 °C for 90 min. The beads were washed twice with RIPA buffer and once with Tris-buffered saline. Bound Rho proteins were eluted by Laemmli sample buffer and detected by Western blotting using mouse monoclonal anti-RhoA antibody.
Luciferase reporter assays
HEK293T cells were seeded at 1 × 104 cells in 96-well plates in DMEM and 10% FBS 24 h prior to transfection. For each well, 20 ng of dual SRE–luciferase reporter plasmid and 2 ng of GPR56 (WT) or GPR56H381S (mutant) were transfected using Lipofectamine 2000 (Invitrogen). Six hours after transfection, cells underwent 16 h of serum starvation prior to treatment with various agonists. Cells were lysed and analyzed using the DualGlo® luciferase assay system (E2920, Promega, Madison, WI) according to the manufacturer's protocol. Luminescence was read using a Synergy-2 microplate reader (BioTek, Winooski, VT). All firefly luciferase data were normalized to the Renilla luciferase signal and expressed as -fold increase over the signal obtained from cells transfected with WT GPR56.
AmpC β-lactamase enzymatic assay
2 nm AmpC β-lactamase was incubated with varying concentrations of 3-α-DOG (from 1.56 μm to 100 μm) for 5 min at room temperature in 50 mm potassium Pi (pH 7.0). The reaction was initiated by adding 70 μm 7-β-Thien-2-yl-acetamido-3-[(4-nitro-3-carboxyphenyl)thiomethyl]-3-cephem-4-carboxylic acid (CENTA), a chromogenic β-lactamase substrate (Millipore Sigma, 219475-25MG), and the reaction progress was monitored at 405 nm by an HP8453a spectrophotometer over 150 s using UV-vis Chemstation software (Agilent Technologies).
Dynamic light scattering assay
3-α-DOG dilutions were delivered from concentrated DMSO stocks into filtered 50 mm potassium Pi buffer (pH 7.0), at room temperature to reach a final concentration of 1% DMSO. Measurements were made in triplicate for each concentration using a DynaPro Plate Reader II (Wyatt Technology) with a 75-milliwatt laser at a wavelength of 826.6 nm. Normalized scattercounts per second were obtained to measure critical aggregation concentration.
Statistical analysis
All data were analyzed using GraphPad Prism8 (San Diego, CA). Results are expressed as means ± S.D. An unpaired, two-sided Student's t test was used to compare two groups. p values of less than 0.05 were considered to indicate statistical significance. One-way ANOVA followed by Tukey's post hoc test was used to analyze multiple treatment condition experiments. Sample size was not predetermined by statistical methods but based on similar studies in the field.
Author contributions
B. Z., R. L., P. J., T. L., H. C. O., S. G., and P. L. data generation; B. Z., P. J., and P. L. formal analysis; B. Z. and X. P. validation; B. Z., P. L., K. R. M., and X. P. writing-review and editing; X. P. conceptualization; R. L., P. J., S. G., and X. P. writing-original draft; P. J., S. G., and K. R. M. software; P. J., S. G., and X. P. investigation; B. K. S. and X. P. supervision; X. P. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Demet Araç (University of Chicago) for sharing the dual SRE–luciferase construct and Dr. Gregory G. Tall for sharing recombinant TG2 and critical reading of this manuscript.
This research was supported in part by NINDS, National Institute of Health Grants R01 NS094164 (to X. P.), R01 NS108446 (to X. P.), R21NS108312 (to X. P.), and R01 NS079445 (to K. R. M.) and National Multiple Sclerosis Society Postdoctoral Fellowship Award FG 2063-A1/2 (to S. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Fig S1.
- aGPCR
- adhesion G protein–coupled receptor
- ECM
- extracellular matrix
- GAIN
- GPCR autoproteolysis-inducing
- NTF
- N-terminal fragment
- CTF
- C-terminal fragment
- TM
- transmembrane
- CNS
- central nervous system
- 3-α-DOG
- 3-α-acetoxydihydrodeoxygedunin
- SRE
- serum response element
- OPC
- oligodendrocyte precursor cell
- cDNA
- complementary DNA
- RIPA
- radioimmune precipitation assay.
References
- 1. Langenhan T., Piao X., and Monk K. R. (2016) Adhesion G protein-coupled receptors in nervous system development and disease. Nat. Rev. Neurosci. 17, 550–561 10.1038/nrn.2016.86 [DOI] [PubMed] [Google Scholar]
- 2. Langenhan T., Aust G., and Hamann J. (2013) Sticky signaling: adhesion class G protein-coupled receptors take the stage. Sci. Signal. 6, re3 [DOI] [PubMed] [Google Scholar]
- 3. Hamann J., Aust G., Araç D., Engel F. B., Formstone C., Fredriksson R., Hall R. A., Harty B. L., Kirchhoff C., Knapp B., Krishnan A., Liebscher I., Lin H. H., Martinelli D. C., Monk K. R., et al. (2015) International Union of Basic and Clinical Pharmacology: XCIV: adhesion G protein-coupled receptors. Pharmacol. Rev. 67, 338–367 10.1124/pr.114.009647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Folts C. J., Giera S., Li T., and Piao X. (2019) Adhesion G protein-coupled receptors as drug target for neurological diseases. Trends Pharmacol. Sci. 40, 278–293 10.1016/j.tips.2019.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Araç D., Boucard A. A., Bolliger M. F., Nguyen J., Soltis S. M., Südhof T. C., and Brunger A. T. (2012) A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 31, 1364–1378 10.1038/emboj.2012.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liebscher I., Schön J., Petersen S. C., Fischer L., Auerbach N., Demberg L. M., Mogha A., Cöster M., Simon K. U., Rothemund S., Monk K. R., and Schöneberg T. (2014) A tethered agonist within the ectodomain activates the adhesion G protein-coupled receptors GPR126 and GPR133. Cell Rep. 9, 2018–2026 10.1016/j.celrep.2014.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stoveken H. M., Hajduczok A. G., Xu L., and Tall G. G. (2015) Adhesion G protein-coupled receptors are activated by exposure of a cryptic tethered agonist. Proc. Natl. Acad. Sci. U.S.A. 112, 6194–6199 10.1073/pnas.1421785112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jin Z., Tietjen I., Bu L., Liu-Yesucevitz L., Gaur S. K., Walsh C. A., and Piao X. (2007) Disease-associated mutations affect GPR56 protein trafficking and cell surface expression. Hum. Mol. Genet. 16, 1972–1985 10.1093/hmg/ddm144 [DOI] [PubMed] [Google Scholar]
- 9. Demberg L. M., Winkler J., Wilde C., Simon K. U., Schön J., Rothemund S., Schöneberg T., Prömel S., and Liebscher I. (2017) Activation of adhesion G protein-coupled receptors: agonist specificity of Stachel sequence-derived peptides. J. Biol. Chem. 292, 4383–4394 10.1074/jbc.M116.763656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Müller A., Winkler J., Fiedler F., Sastradihardja T., Binder C., Schnabel R., Kungel J., Rothemund S., Hennig C., Schöneberg T., and Prömel S. (2015) Oriented cell division in the C. elegans embryo is coordinated by G-protein signaling dependent on the adhesion GPCR LAT-1. PLoS Genet. 11, e1005624 10.1371/journal.pgen.1005624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Salzman G. S., Zhang S., Gupta A., Koide A., Koide S., and Araç D. (2017) Stachel-independent modulation of GPR56/ADGRG1 signaling by synthetic ligands directed to its extracellular region. Proc. Natl. Acad. Sci. U.S.A. 114, 10095–10100 10.1073/pnas.1708810114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kishore A., Purcell R. H., Nassiri-Toosi Z., and Hall R. A. (2016) Stalk-dependent and stalk-independent signaling by the adhesion G protein-coupled receptors GPR56 (ADGRG1) and BAI1 (ADGRB1). J. Biol. Chem. 291, 3385–3394 10.1074/jbc.M115.689349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scholz N., Guan C., Nieberler M., Grotemeyer A., Maiellaro I., Gao S., Beck S., Pawlak M., Sauer M., Asan E., Rothemund S., Winkler J., Prömel S., Nagel G., Langenhan T., and Kittel R. J. (2017) Mechano-dependent signaling by latrophilin/CIRL quenches cAMP in proprioceptive neurons. Elife 6, e28360 10.7554/eLife.28360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moreno M., Pedrosa L., Paré L., Pineda E., Bejarano L., Martínez J., Balasubramaniyan V., Ezhilarasan R., Kallarackal N., Kim S. H., Wang J., Audia A., Conroy S., Marin M., Ribalta T., et al. (2017) GPR56/ADGRG1 inhibits mesenchymal differentiation and radioresistance in glioblastoma. Cell Rep. 21, 2183–2197 10.1016/j.celrep.2017.10.083 [DOI] [PubMed] [Google Scholar]
- 15. Daria D., Kirsten N., Muranyi A., Mulaw M., Ihme S., Kechter A., Hollnagel M., Bullinger L., Döhner K., Döhner H., Feuring-Buske M., and Buske C. (2016) GPR56 contributes to the development of acute myeloid leukemia in mice. Leukemia 30, 1734–1741 10.1038/leu.2016.76 [DOI] [PubMed] [Google Scholar]
- 16. Pabst C., Bergeron A., Lavallée V. P., Yeh J., Gendron P., Norddahl G. L., Krosl J., Boivin I., Deneault E., Simard J., Imren S., Boucher G., Eppert K., Herold T., Bohlander S. K., et al. (2016) GPR56 identifies primary human acute myeloid leukemia cells with high repopulating potential in vivo. Blood 127, 2018–2027 10.1182/blood-2015-11-683649 [DOI] [PubMed] [Google Scholar]
- 17. Solaimani Kartalaei P., Yamada-Inagawa T., Vink C. S., de Pater E., van der Linden R., Marks-Bluth J., van der Sloot A., van den Hout M., Yokomizo T., van Schaick-Solernó M. L., Delwel R., Pimanda J. E., van Ijcken W. F., and Dzierzak E. (2015) Whole-transcriptome analysis of endothelial to hematopoietic stem cell transition reveals a requirement for Gpr56 in HSC generation. J. Exp. Med. 212, 93–106 10.1084/jem.20140767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Piao X., Hill R. S., Bodell A., Chang B. S., Basel-Vanagaite L., Straussberg R., Dobyns W. B., Qasrawi B., Winter R. M., Innes A. M., Voit T., Ross M. E., Michaud J. L., Déscarie J. C., Barkovich A. J., and Walsh C. A. (2004) G protein-coupled receptor-dependent development of human frontal cortex. Science 303, 2033–2036 10.1126/science.1092780 [DOI] [PubMed] [Google Scholar]
- 19. Bahi-Buisson N., Poirier K., Boddaert N., Fallet-Bianco C., Specchio N., Bertini E., Caglayan O., Lascelles K., Elie C., Rambaud J., Baulac M., An I., Dias P., des Portes V., Moutard M. L., et al. (2010) GPR56-related bilateral frontoparietal polymicrogyria: further evidence for an overlap with the cobblestone complex. Brain 133, 3194–3209 10.1093/brain/awq259 [DOI] [PubMed] [Google Scholar]
- 20. Bae B. I., Tietjen I., Atabay K. D., Evrony G. D., Johnson M. B., Asare E., Wang P. P., Murayama A. Y., Im K., Lisgo S. N., Overman L., Šestan N., Chang B. S., Barkovich A. J., Grant P. E., et al. (2014) Evolutionarily dynamic alternative splicing of GPR56 regulates regional cerebral cortical patterning. Science 343, 764–768 10.1126/science.1244392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Olaniru O. E., Pingitore A., Giera S., Piao X., Castañera González R., Jones P. M., and Persaud S. J. (2018) The adhesion receptor GPR56 is activated by extracellular matrix collagen III to improve β-cell function. Cell Mol. Life Sci. 75, 4007–4019 10.1007/s00018-018-2846-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Giera S., Luo R., Ying Y., Ackerman S. D., Jeong S. J., Stoveken H. M., Folts C. J., Welsh C. A., Tall G. G., Stevens B., Monk K. R., and Piao X. (2018) Microglial transglutaminase-2 drives myelination and myelin repair via GPR56/ADGRG1 in oligodendrocyte precursor cells. Elife 7, e33385 10.7554/eLife.33385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ackerman S. D., Luo R., Poitelon Y., Mogha A., Harty B. L., D'Rozario M., Sanchez N. E., Lakkaraju A. K. K., Gamble P., Li J., Qu J., MacEwan M. R., Ray W. Z., Aguzzi A., Feltri M. L., et al. (2018) GPR56/ADGRG1 regulates development and maintenance of peripheral myelin. J. Exp. Med. 215, 941–961 10.1084/jem.20161714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chang G. W., Hsiao C. C., Peng Y. M., Vieira Braga F. A., Kragten N. A., Remmerswaal E. B., van de Garde M. D., Straussberg R., König G. M., Kostenis E., Knäuper V., Meyaard L., van Lier R. A., van Gisbergen K. P., Lin H. H., and Hamann J. (2016) The adhesion G protein-coupled receptor GPR56/ADGRG1 is an inhibitory receptor on human NK cells. Cell Rep. 15, 1757–1770 10.1016/j.celrep.2016.04.053 [DOI] [PubMed] [Google Scholar]
- 25. Holmfeldt P., Ganuza M., Marathe H., He B., Hall T., Kang G., Moen J., Pardieck J., Saulsberry A. C., Cico A., Gaut L., McGoldrick D., Finkelstein D., Tan K., and McKinney-Freeman S. (2016) Functional screen identifies regulators of murine hematopoietic stem cell repopulation. J. Exp. Med. 213, 433–449 10.1084/jem.20150806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ackerman S. D., Garcia C., Piao X., Gutmann D. H., and Monk K. R. (2015) The adhesion GPCR Gpr56 regulates oligodendrocyte development via interactions with Gα12/13 and RhoA. Nat. Commun 6, 6122 10.1038/ncomms7122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Giera S., Deng Y., Luo R., Ackerman S. D., Mogha A., Monk K. R., Ying Y., Jeong S. J., Makinodan M., Bialas A. R., Chang B. S., Stevens B., Corfas G., and Piao X. (2015) The adhesion G protein-coupled receptor GPR56 is a cell-autonomous regulator of oligodendrocyte development. Nat. Commun. 6, 6121 10.1038/ncomms7121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. White J. P., Wrann C. D., Rao R. R., Nair S. K., Jedrychowski M. P., You J. S., Martínez-Redondo V., Gygi S. P., Ruas J. L., Hornberger T. A., Wu Z., Glass D. J., Piao X., and Spiegelman B. M. (2014) G protein-coupled receptor 56 regulates mechanical overload-induced muscle hypertrophy. Proc. Natl. Acad. Sci. U.S.A. 111, 15756–15761 10.1073/pnas.1417898111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Saito Y., Kaneda K., Suekane A., Ichihara E., Nakahata S., Yamakawa N., Nagai K., Mizuno N., Kogawa K., Miura I., Itoh H., and Morishita K. (2013) Maintenance of the hematopoietic stem cell pool in bone marrow niches by EVI1-regulated GPR56. Leukemia 27, 1637–1649 10.1038/leu.2013.75 [DOI] [PubMed] [Google Scholar]
- 30. Chen G., Yang L., Begum S., and Xu L. (2010) GPR56 is essential for testis development and male fertility in mice. Dev. Dyn. 239, 3358–3367 10.1002/dvdy.22468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shashidhar S., Lorente G., Nagavarapu U., Nelson A., Kuo J., Cummins J., Nikolich K., Urfer R., and Foehr E. D. (2005) GPR56 is a GPCR that is overexpressed in gliomas and functions in tumor cell adhesion. Oncogene 24, 1673–1682 10.1038/sj.onc.1208395 [DOI] [PubMed] [Google Scholar]
- 32. Piao X., Chang B. S., Bodell A., Woods K., Benzeev B., Topcu M., Guerrini R., Goldberg-Stern H., Sztriha L., Dobyns W. B., Barkovich A. J., and Walsh C. A. (2005) Genotype-phenotype analysis of human frontoparietal polymicrogyria syndromes. Ann. Neurol 58, 680–687 10.1002/ana.20616 [DOI] [PubMed] [Google Scholar]
- 33. Jeong S. J., Luo R., Singer K., Giera S., Kreidberg J., Kiyozumi D., Shimono C., Sekiguchi K., and Piao X. (2013) GPR56 functions together with α3β1 integrin in regulating cerebral cortical development. PLoS ONE 8, e68781 10.1371/journal.pone.0068781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koirala S., Jin Z., Piao X., and Corfas G. (2009) GPR56-regulated granule cell adhesion is essential for rostral cerebellar development. J. Neurosci. 29, 7439–7449 10.1523/JNEUROSCI.1182-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luo R., Jeong S. J., Jin Z., Strokes N., Li S., and Piao X. (2011) G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc. Natl. Acad. Sci. U.S.A. 108, 12925–12930 10.1073/pnas.1104821108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jeong S. J., Li S., Luo R., Strokes N., and Piao X. (2012) Loss of Col3a1, the gene for Ehlers-Danlos syndrome type IV, results in neocortical dyslamination. PLoS ONE 7, e29767 10.1371/journal.pone.0029767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li G., Adesnik H., Li J., Long J., Nicoll R. A., Rubenstein J. L., and Pleasure S. J. (2008) Regional distribution of cortical interneurons and development of inhibitory tone are regulated by Cxcl12/Cxcr4 signaling. J. Neurosci. 28, 1085–1098 10.1523/JNEUROSCI.4602-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Emery B. (2010) Regulation of oligodendrocyte differentiation and myelination. Science 330, 779–782 10.1126/science.1190927 [DOI] [PubMed] [Google Scholar]
- 39. Xu L., Begum S., Hearn J. D., and Hynes R. O. (2006) GPR56, an atypical G protein-coupled receptor, binds tissue transglutaminase, TG2, and inhibits melanoma tumor growth and metastasis. Proc. Natl. Acad. Sci. U.S.A. 103, 9023–9028 10.1073/pnas.0602681103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Luo R., Jeong S. J., Yang A., Wen M., Saslowsky D. E., Lencer W. I., Araç D., and Piao X. (2014) Mechanism for adhesion G protein-coupled receptor GPR56-mediated RhoA activation induced by collagen III stimulation. PLoS ONE 9, e100043 10.1371/journal.pone.0100043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Luo R., Jin Z., Deng Y., Strokes N., and Piao X. (2012) Disease-associated mutations prevent GPR56-collagen III interaction. PLoS ONE 7, e29818 10.1371/journal.pone.0029818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stoveken H. M., Larsen S. D., Smrcka A. V., and Tall G. G. (2018) Gedunin- and Khivorin-derivatives are small-molecule partial agonists for adhesion G protein-coupled receptors GPR56/ADGRG1 and GPR114/ADGRG5. Mol. Pharmacol. 93, 477–488 10.1124/mol.117.111476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang L., Friedland S., Corson N., and Xu L. (2014) GPR56 inhibits melanoma growth by internalizing and degrading its ligand TG2. Cancer Res. 74, 1022–1031 10.1158/0008-5472.CAN-13-1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McGovern S. L., Helfand B. T., Feng B., and Shoichet B. K. (2003) A specific mechanism of nonspecific inhibition. J. Med. Chem. 46, 4265–4272 10.1021/jm030266r [DOI] [PubMed] [Google Scholar]
- 45. Aldrich C., Bertozzi C., Georg G. I., Kiessling L., Lindsley C., Liotta D., Merz K. M. Jr, Schepartz A., and Wang S. (2017) The ecstasy and agony of assay interference compounds. ACS Cent. Sci. 3, 143–147 10.1021/acscentsci.7b00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Irwin J. J., Duan D., Torosyan H., Doak A. K., Ziebart K. T., Sterling T., Tumanian G., and Shoichet B. K. (2015) An Aggregation advisor for ligand discovery. J. Med. Chem. 58, 7076–7087 10.1021/acs.jmedchem.5b01105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Feng B. Y., and Shoichet B. K. (2006) A detergent-based assay for the detection of promiscuous inhibitors. Nat. Protoc. 1, 550–553 10.1038/nprot.2006.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Coan K. E., and Shoichet B. K. (2008) Stoichiometry and physical chemistry of promiscuous aggregate-based inhibitors. J. Am. Chem. Soc. 130, 9606–9612 10.1021/ja802977h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Aeschlimann A. (1992) What is your diagnosis? Ruptured Baker's cyst. Schweiz. Rundsch. Med. Prax. 81, 677–678 [PubMed] [Google Scholar]
- 50. Salzman G. S., Ackerman S. D., Ding C., Koide A., Leon K., Luo R., Stoveken H. M., Fernandez C. G., Tall G. G., Piao X., Monk K. R., Koide S., and Araç D. (2016) Structural basis for regulation of GPR56/ADGRG1 by its alternatively spliced extracellular domains. Neuron 91, 1292–1304 10.1016/j.neuron.2016.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Watkins T. A., Emery B., Mulinyawe S., and Barres B. A. (2008) Distinct stages of myelination regulated by γ-secretase and astrocytes in a rapidly myelinating CNS coculture system. Neuron 60, 555–569 10.1016/j.neuron.2008.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang S., Sdrulla A., Johnson J. E., Yokota Y., and Barres B. A. (2001) A role for the helix-loop-helix protein Id2 in the control of oligodendrocyte development. Neuron 29, 603–614 10.1016/S0896-6273(01)00237-9 [DOI] [PubMed] [Google Scholar]
- 53. Dugas J. C., Tai Y. C., Speed T. P., Ngai J., and Barres B. A. (2006) Functional genomic analysis of oligodendrocyte differentiation. J. Neurosci. 26, 10967–10983 10.1523/JNEUROSCI.2572-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.