Abstract
Intercellular propagation of protein aggregation is emerging as a key mechanism in the progression of several neurodegenerative diseases, including Alzheimer's disease and frontotemporal dementia (FTD). However, we lack a systematic understanding of the cellular pathways controlling prion-like propagation of aggregation. To uncover such pathways, here we performed CRISPR interference (CRISPRi) screens in a human cell-based model of propagation of tau aggregation monitored by FRET. Our screens uncovered that knockdown of several components of the endosomal sorting complexes required for transport (ESCRT) machinery, including charged multivesicular body protein 6 (CHMP6), or CHMP2A in combination with CHMP2B (whose gene is linked to familial FTD), promote propagation of tau aggregation. We found that knocking down the genes encoding these proteins also causes damage to endolysosomal membranes, consistent with a role for the ESCRT pathway in endolysosomal membrane repair. Leakiness of the endolysosomal compartment significantly enhanced prion-like propagation of tau aggregation, likely by making tau seeds more available to pools of cytoplasmic tau. Together, these findings suggest that endolysosomal escape is a critical step in tau propagation in neurodegenerative diseases.
Keywords: Tau protein (Tau), protein aggregation, endosomal sorting complexes required for transport (ESCRT), endosome, lysosome, CRISPR/Cas, functional genomics, fluorescence resonance energy transfer (FRET), Membrane damage
Introduction
Neurodegenerative diseases are one of the most pressing challenges facing humanity. A formidable roadblock to the development of effective therapies is our incomplete understanding of the underlying molecular and cellular mechanisms. A major breakthrough was the discovery that scrapie, an infectious neurodegenerative disease, is caused by the cell-to-cell propagation of protein aggregates via “prion” forms of the protein (1). In this process, a prion seed converts healthy, native proteins to adopt an aggregated, prion conformer. More recently, findings from numerous, independent studies support the hypothesis that prion-like propagation also underlies common, noninfectious neurodegenerative diseases, such as Alzheimer's disease (AD)3 (recently reviewed in Ref. 2). However, the mechanisms that control aggregate uptake and propagation remain to be fully elucidated, especially in those diseases that involve cytoplasmic proteins. A systematic understanding of these mechanisms is important, both for the development of therapeutics and for furthering our understanding of why specific neuronal subtypes and brain regions are especially susceptible to specific diseases.
Of particular interest to us are the mechanisms controlling propagation of aggregated forms of the protein tau. Tau aggregation is one of the hallmarks of AD and the levels of aggregated tau correlate with cognitive deficits and neuronal loss (3–6). Beyond AD, tau aggregation also defines a number of other neurodegenerative diseases, collectively termed tauopathies, some of which are caused by familial point mutations in tau (7).
Propagation of tau aggregation can be modeled in cultured HEK293 cells that express fluorescently tagged versions of tau, as first established by the Diamond lab (8). In this system, addition of aggregated tau seeds to the culture medium causes the fluorescently tagged tau in the cells to convert from a diffuse, soluble form to aggregated puncta. This cell-based model has enabled the characterization of tau species with seeding activity from patient brains (9), and the creation of a minimal synthetic tau that retains seeding capability (10). Furthermore, cell-based models can also be used as a biosensor to detect and propagate distinct prion strains of tau from different tauopathies (11–13). Importantly, seeding of tau aggregation in the cell-based model is predictive of in vivo seeding in a mouse model (14).
In addition to their utility as “biosensors” for tau aggregates with prion properties, cell-based models can also be used to elucidate cellular pathways that control propagation of tau aggregation. Previous work from others and us leveraged cell-based models to uncover mechanisms that mediate tau uptake into cells (8, 15–17). In those studies, binding of tau to specific cell-surface heparan sulfate proteoglycans was found to mediate cellular uptake. These results were validated in human iPSC-derived neurons and mouse brain slices (16), supporting the physiological relevance of the cell-based model.
Although these studies established the mechanism for tau uptake, the downstream cellular pathways controlling propagation of tau aggregation have not been systematically characterized. We hypothesized that tau aggregation in the cytosol would be influenced by multiple cellular pathways, including those controlling trafficking of tau seeds through the endolysosomal pathway, localization of tau seeds to the cytosol, templated aggregation of soluble tau, and clearance of tau aggregates (Fig. 1A).
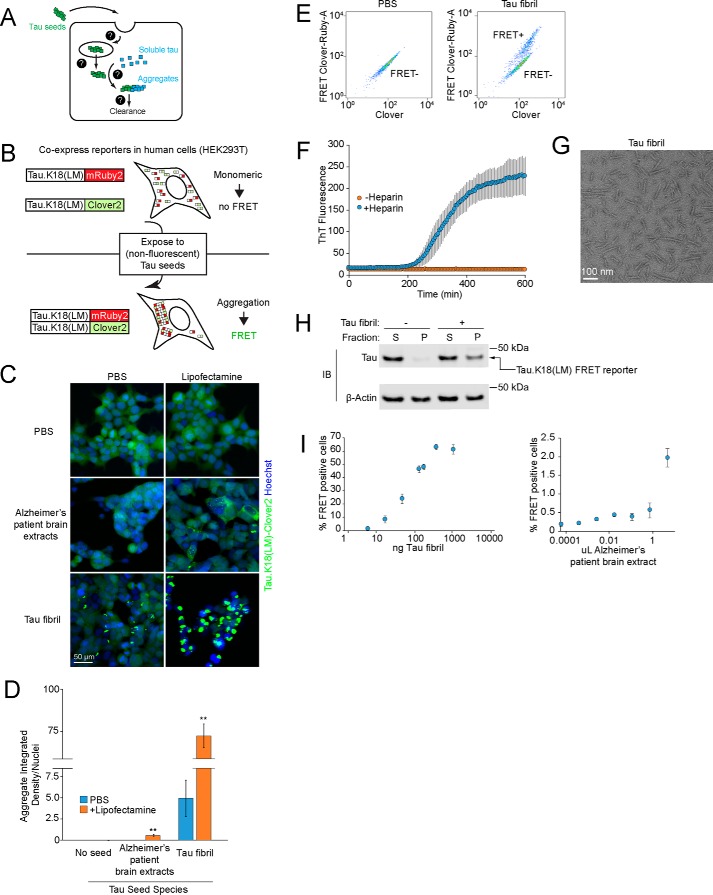
Figure 1.
Tau seeds induce tau aggregation in a FRET-based reporter cell line. A, overview of cellular processes that may control the prion-like tau propagation and aggregation. Question marks represent unknown cellular mechanisms. B, schematic representation of the FRET-based reporter assay to monitor tau aggregation in HEK293T cells. In the absence of tau seeds, fluorescently labeled tau.K18(LM) is monomeric. Exposure to nonfluorescent tau seeds induces aggregation of the reporter, which can be measured by the formation of tau aggregates by fluorescence microscopy or an increase in FRET intensity by flow cytometry. C, induction of fluorescent tau aggregates in a FRET reporter cell line. Representative images of cells treated with PBS (top row), Alzheimer's patient brain extracts after 5 days (second row), or fibrils of recombinant human 0N4R tau after 2 days (third row). For each tau seed, each condition is complexed with (right column) or without (left column) Lipofectamine. Nuclei were counter-stained with Hoechst 33342. D, comparison of intracellular fluorescent tau aggregates from images in C. Integrated density quantification of fluorescent tau aggregates seeded with various tau seeds complexed with (blue) or without (orange) Lipofectamine were quantified and divided by total nuclei per image. n = 3 technical replicates (with at least 50 nuclei per image), error bars represent mean ± S.D., *, p < 0.05; **, p < 0.01 (two-tailed Student's t test for comparison to PBS (no Lipofectamine) control for each tau seeding condition). E, representative flow cytometry plot of FRET reporter cells after 2 day treatment with PBS (left) or tau fibrils (right). F, incubation of recombinant 0N4R tau with heparin and constant agitation at 37 °C induces fibrillization. Fibrillization is monitored using an increase in thioflavin T fluorescence (excitation 440 nm, emission: 485 nm), which occurs in the presence (blue) of heparin (10 μg/ml), but not in the absence (orange) of heparin. Error bars represent mean ± S.D. from n = 3 technical replicates. G, representative negative stain electron micrograph of tau fibrils. H, lysates from FRET reporter cells treated with PBS or tau fibrils for 2 days were fractionated at 1000 × g into soluble (S) or pellet (P) fractions, and subjected to SDS-PAGE and immunoblotting using antibodies against tau and β-actin. I, quantification of % FRET-positive cells using flow cytometry across concentration ranges of tau fibrils (left) or human Alzheimer's patient brain extracts (right). Error bars represent mean ± S.D. for n = 3 technical replicates.
To uncover relevant cellular pathways downstream of tau uptake, we here combine our CRISPR interference-based genetic screening approach (18, 19) with a cell-based model of tau aggregation using fluorescence resonance energy transfer (FRET). Using this approach, we uncover endolysosomal escape of tau seeds as a critical step in the propagation of tau aggregation. Defects in the ESCRT machinery compromise endolysosomal integrity, thereby promoting the escape of tau seeds from endolysosomal compartments and accelerating subsequent templating of tau aggregation in the cytosol. These findings provide insight into the mechanisms of tau trafficking and suggest a source for new potential therapeutic targets.
Results
Cell-based model of prion-like propagation of tau aggregation
We established a cell-based model to monitor the prion-like propagation of tau aggregation in HEK293T cells. Such a model had previously been pioneered by the Diamond lab (8) and adapted for flow cytometry using a FRET-based strategy to monitor tau aggregation (20). In this FRET-based strategy, two versions of the tau repeat domain containing disease-associated P301L and V337M mutations are expressed as fusions with either the FRET donor CFP or the FRET acceptor YFP. When exposed to tau fibrils from recombinant or cell/brain-derived lysate sources, the CFP and YFP tags are brought into close proximity, enabling FRET. We generated a reporter line following a similar strategy (Fig. 1B). Instead of the CFP-YFP FRET pair, we used Clover2 and mRuby2, because proteins of this type had been shown to have a very high dynamic range for FRET, with a high Förster radius (21). We selected a monoclonal line for optimal expression and dynamic range of the FRET signal.
In the absence of seeding, these cells showed diffuse intracellular fluorescence without visible aggregates when monitored by fluorescence microscopy (Fig. 1, C and D) and they appeared as a single population when FRET levels are monitored by flow cytometry (Fig. 1E). In contrast, exposure of these cells to extracts from AD patient brains caused the tau reporter constructs to aggregate, as reflected by formation of fluorescent puncta (Fig. 1, C and D). However, seeded aggregation with brain-derived tau required co-incubation with Lipofectamine 2000 (here referred to as Lipofectamine) to achieve modest aggregation (Fig. 1, C and D), consistent with reports from other groups (12–14).
Our goal was to eliminate the use of Lipofectamine, because the use of a lipocationic carrier may bypass physiologically relevant uptake or trafficking pathways. Accordingly, we purified monomeric His6-tagged 0N4R human tau from Escherichia coli and induced fibrillization with heparin, which we monitored by an increase in thioflavin T fluorescence (Fig. 1F) and by negative stain EM (Fig. 1G). We found that treatment of our FRET reporter cells with these tau fibrils caused robust formation of aggregates, even in the absence of Lipofectamine. This activity was confirmed using multiple criteria, including formation of puncta by fluorescence microscopy (Fig. 1, C and D), appearance of a FRET-positive population by flow cytometry (Fig. 1E), and biochemical characterization of tau in the insoluble fraction (Fig. 1H). Finally, we tested the effects of increasing concentrations of tau fibrils on our FRET-based reporter. We found that His6-tagged fibrils robustly triggered tau aggregation in a dose-dependent manner across nearly 2 orders of magnitude in concentration in the absence of Lipofectamine, as quantified by the percentage of FRET-positive cells (Fig. 1I). Brain lysates also produced an increase in FRET-positive cells, although the magnitude was more modest. Together, these features make our FRET-based model suitable for use in a genetic screen to identify cellular factors that control prion-like propagation of tau aggregation.
Genetic screen to identify cellular factors that control prion-like propagation of tau aggregation
To identify cellular factors that control propagation of tau aggregation (Fig. 1A), we conducted a CRISPR interference (CRISPRi)-based genetic screen (Fig. 2A). First, we transduced the reporter cell line described above with a lentiviral expression construct for a catalytically inactive Cas9-BFP-KRAB (dCas9-BFP-KRAB) fusion protein. dCas9-BFP-KRAB can be directed by small guide RNAs (sgRNAs) to silence a gene of interest (22), enabling massively parallel genetic screens in mammalian cells (18). We then transduced the cells with pooled sgRNA libraries that target protein homeostasis factors, which we designed specifically for this study based on the rationale that protein homeostasis factors were likely to control or modulate tau aggregation and clearance. These libraries target 2,949 genes encoding genes that function in autophagy, protein folding, or the ubiquitin-proteasome system with at least five independent sgRNAs for each gene, plus 750 nontargeting control sgRNAs. Cells transduced with these libraries were exposed to recombinant tau fibrils at concentrations that would yield FRET-positive cells at 50% of the maximum percentage of FRET-positive cells (Fig. 1I), thereby maximizing the dynamic range for detecting cellular factors that either increase or decrease tau aggregation. FRET-negative and FRET-positive cell populations were separated by FACS, collecting sufficient numbers of cells from each population for an average ×1000 representation (cells per sgRNA elements in the library). Genomic DNA was isolated and the locus encoding the sgRNAs was PCR-amplified. Frequencies for each sgRNA in each population were determined by next generation sequencing. We evaluated genes for the effect their knockdown had on the formation of tau aggregates (Table S1) using our previously described bioinformatics pipeline (18, 23–25).
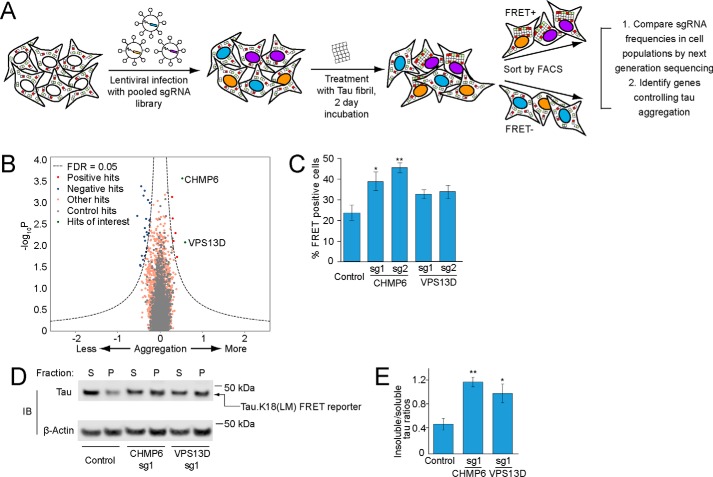
Figure 2.
CRISPRi screen for cellular factors controlling tau aggregation. A, strategy for pooled FRET-based CRISPRi screen. FRET reporter cells stably expressing the CRISPRi machinery (dCas9-BFP-KRAB) were transduced with pooled lentiviral expression libraries of sgRNAs targeting proteostasis genes. Following transduction and selection, cells were treated with tau fibrils and incubated for 2 days. Cells were detached and sorted into FRET-negative and -positive populations by FACS. sgRNA-encoding cassettes were amplified from genomic DNA of the cell populations and their frequencies were quantified using next generation sequencing to identify genes that control tau aggregation. B, volcano plot summarizing phenotypes and statistical significance (by our MAGeCK-iNC pipeline, see “Experimental procedures”) of the genes targeted by the sgRNA libraries. Nontargeting sgRNAs were randomly grouped into negative control “quasi-genes” (gray dots) to derive an empirical false discovery rate (FDR). Hit genes that passed an FDR < 0.05 threshold are shown in blue (knockdown decreases aggregation) or red (knockdown increases aggregation), other genes are shown in orange. Two hit genes of interest are shown in green and labeled. C–E, validation of hit genes CHMP6 and VPS13D. FRET reporter cells transduced with individual sgRNAs targeting two hit genes or a nontargeting control sgRNA, and 5 days after transduction treated for 2 days with tau fibrils. C, % of FRET-positive cells was quantified by flow cytometry. Error bars represent mean ± S.D. of n = 3 technical replicates. *, p < 0.05; **, p < 0.01 (two-tailed Student's t test for comparison to the nontargeting control sgRNA). D, representative immunoblot for the tau fluorescent protein construct in the soluble and insoluble fractions as in Fig. 1H. E, quantification of insoluble/soluble tau ratios from immunoblots in D. Error bars represent mean ± S.D. for n = 3 biological replicates. *, p < 0.05; **, p < 0.01 (two-tailed Student's t test for comparison to the nontargeting control sgRNA).
Two genes stood out for the strong enhancement of tau aggregation by their knockdown: CHMP6 and VPS13D (Fig. 2B). We decided to prioritize these two genes for further characterization, because both are related to genes implicated in neurodegenerative diseases.
CHMP6 is part of the Endosomal Sorting Complex Required for Transport (ESCRT)-III complex, which is required for numerous cellular processes involving membrane remodeling (26). Mutations in the ESCRT-III component CHMP2B cause familial frontotemporal lobar dementia (FTD) and have been shown to cause endolysosomal defects (27, 28).
The VPS13 protein family is comprised of four closely-related proteins, VPS13A-D (29). VPS13 family proteins are localized at various inter-organelle membrane contact sites and facilitates nonvesicular lipid transport (30, 31). Interestingly, mutations of VPS13D are associated with recessive ataxia (32). Previously, VPS13A and VPS13C mutations have been associated with a Huntington's-like syndrome (Chorea-acanthocytosis) (33, 34) and Parkinson's disease (35), respectively.
To confirm these screening hits, we cloned 2 individual sgRNAs each targeting CHMP6 and VPS13D, and confirmed target knockdown by qPCR (Table S2). Using these sgRNAs, we validated the effect of CHMP6 and, to a lesser extent, VPS13D knockdown on tau aggregation by flow cytometry (Fig. 2C) and biochemical solubility assay (Fig. 2, D and E).
CHMP6 knockdown accelerates tau aggregation following tau seed uptake
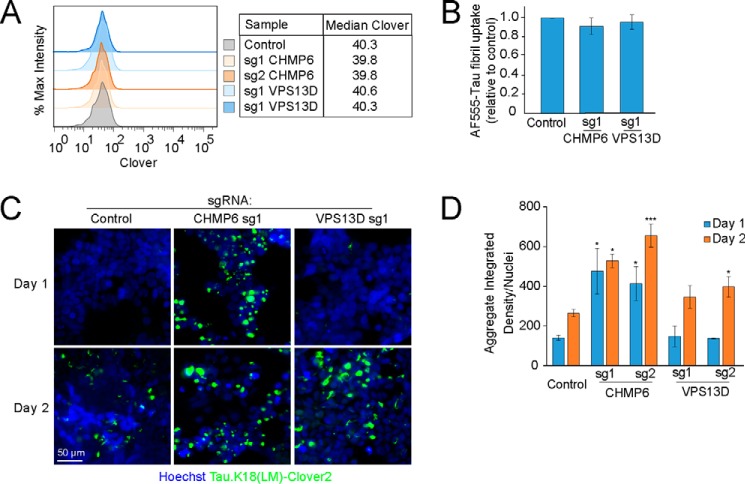
We next investigated the mechanism by which knockdown of CHMP6 and VPS13D might affect tau aggregation. First, we excluded the possibility that knockdown of these genes alters the levels of our tau reporter (Fig. 3A). Since we previously identified factors controlling cellular uptake of tau (16), we tested whether knockdown of CHMP6 and VPS13D impacted the uptake of tau fibrils. However, we found that knockdown of the genes did not impact tau fibril uptake (Fig. 3B), suggesting that their impact on tau aggregation is mediated downstream of seed uptake.
Figure 3.
CHMP6 knockdown accelerates the prion-like propagation of tau aggregation. FRET reporter cells or CRISPRi-HEK293T cells were transduced with individual sgRNAs targeting CHMP6 or VPS13D, or a nontargeting control sgRNA, and characterized for different phenotypes 5 days after transduction (A) Knockdown of CHMP6 and VPS13D does not impact steady-state levels of the tau-Clover2 construct in FRET reporter cells, as quantified by flow cytometry. B, individual gene knockdown does not impact uptake of tau fibrils. CRISPRi HEK293T cells were incubated with AF555-labeled tau fibrils for 1 h at 37 °C, stringently washed, and red fluorescence representing internalized tau fibrils was quantified by flow cytometry. The bar graph shows normalized fluorescence intensities and standard deviation of n = 3 technical replicates. C, CHMP6 knockdown accelerates prion-like propagation of tau aggregation. Representative fluorescence micrographs of the Tau.K18(LM)Clover2 reporter in cells 1 and 2 days after fibril addition are shown. Nuclei were counter-stained with Hoechst 33342. D, quantification of C. Tau aggregates were quantified by integrated density across the entire image and divided by total nuclei per image. Error bars represent mean ± S.D., where n = 3 images per condition (with at least 50 nuclei per image). *, p < 0.05; ***, p < 0.001 (two-tailed Student's t test for comparison to the values for nontargeting control sgRNA of the same day).
We next sought to evaluate whether knockdown of CHMP6 and VPS13D increased the rate of tau aggregation, or decreased the rate of tau aggregate clearance. To this end, we utilized high-content imaging analysis to track the fibril-induced aggregation of tau over time. As expected from the results in our primary screen, we observed increased levels of aggregates 48 h post-seeding with fibrils when either CHMP6 or VPS13D were knocked down (Fig. 3, C and D). Intriguingly, the timeline of tau aggregate formation was differentially affected by the two gene knockdowns. Although VPS13D knockdown did not cause a statistically significant increase in aggregates 24 h after treatment with tau fibrils compared with a nontargeting control sgRNA, CHMP6 knockdown promoted early aggregation 24 h post-seeding (Fig. 3, C and D). Interestingly, aggregate formation from 24 to 48 h post-seeding did not change substantially with CHMP6 knockdown, suggesting that the majority of soluble tau rapidly aggregates following treatment with tau fibrils in that background. Although we observe a rapid increase in aggregate formation in CHMP6 knockdown cells, this does not rule out an additional effect of CHMP6 knockdown on aggregate clearance. Given the intriguing acceleration of tau aggregation by CHMP6, and the comparatively weaker phenotype of VPS13D, we decided to focus our mechanistic studies on CHMP6.
Endolysosomal escape of tau seeds is rate-limiting for propagation of tau aggregation
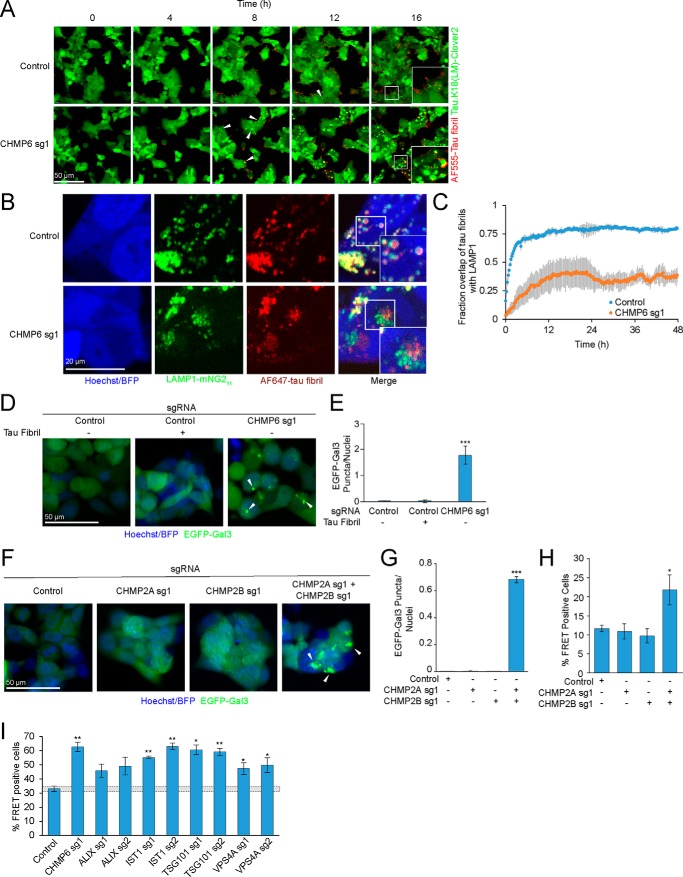
To investigate the mechanism by which CHMP6 knockdown accelerates seeded tau aggregation, we monitored fibril entry and aggregate formation simultaneously by longitudinal imaging in cells expressing a nontargeting control sgRNA compared with CHMP6 knockdown (Fig. 4A, Movies S1 and S2). In CHMP6 knockdown cells, large tau aggregates rapidly formed soon after tau fibrils localized to cells, within 12 h post-seeding. In control cells, by contrast, tau fibrils localized to control cells long before aggregates form. We confirmed that these fibril puncta partially colocalize with the late-endosome/lysosome markers LAMP1 and Rab7a, and frequently localize to the lumen of LAMP1- and Rab7a-positive compartments (Fig. 4B, Fig. S1A, Movies S3 and S4). (Large LAMP1 and Rab7a vesicles with visible lumina were observed also in the absence of fibrils, and therefore not induced by the fibrils themselves, Fig. S1B.) These results suggest that fibrils normally accumulate in endolysosomal compartments, where they do not encounter cytosolic tau. In CHMP6 knockdown cells, co-localization of fibrils with LAMP1-positive compartments was markedly reduced (Fig. 4, B and C, Movie S5). Therefore, CHMP6 knockdown seems to accelerate exit of fibrils from the endolysosomal pathway into the cytosol, where it can then seed aggregation of cytosolic tau.
Figure 4.
CHMP6 knockdown compromises endolysosomal membrane integrity. A, time-lapse microscopy of cell entry of tau-AF555 fibrils and resulting aggregation of the cytosolic tau-Clover2 construct. Representative images of CRISPRi-HEK293T cells expressing tau.K18(LM)-clover2 transduced with either nontargeting control (top) or CHMP6-targeting sgRNA (bottom) are shown. Corresponding movies are provided as Movie S1 (control sgRNA) and Movie S2 (CHMP6 sgRNA). B, representative fluorescence microscopy images of CRISPRi-HEK293T cells LAMP1-mNG11 endogenously labeled with the split-mNeonGreen system. Cells were transduced with nontargeting control (top) or CHMP6 sgRNA (bottom) and treated with AF647-tau fibrils and followed by automated time-lapse microscopy for 48 h. Images for the 12-h time point are shown here. Corresponding movies for 48-h time intervals are provided as Movie S4 (Control sgRNA) and Movies S5 (CHMP6 sg1). C, quantification of Tau fibril colocalization with LAMP1 from representative images and movies shown in B and Movies S4 and S5. Time represents start of treatment with tau fibrils and image acquisition. Error bars represent mean ± S.D. for n = 3 technical replicates (with at least 5 nuclei per image). D, CHMP6 knockdown causes endolysosomal vesicle damage. Representative fluorescence microscopy images of CRISPRi-HEK293T cells expressing an EGFP-Galectin3 (EGFP-Gal3) reporter transduced with either control (left) or CHMP6 (right) sgRNA. Nuclei were counter-stained with Hoechst 33342. E, quantification of EGFP-Gal3 puncta divided by number of nuclei in fluorescence microscopy images shown in D. Error bars represent mean ± S.D. for n = 3 technical replicates (with at least 50 nuclei per image). ***, p < 0.001 (two-tailed Student's t test for comparison to the nontargeting control sgRNA). F, knockdown of various ESCRT components increases tau aggregation. FRET reporter cells were transduced with individual sgRNAs targeting ESCRT components or a nontargeting control sgRNA, and 5 days after transduction treated for 2 days with tau fibrils. Error bars represent mean ± S.D. for n = 3 technical replicates. *, p < 0.05; **, p < 0.01 (two-tailed Student's t test for comparison to the nontargeting control sgRNA). G, simultaneous, but not individual, knockdown of CHMP2A and CHMP2B results in endolysosomal damage, monitored as in C. Nuclei were counter-stained with Hoechst 33342. Scale bar = 50 μm. H, quantification of EGFP-Gal3 puncta divided by nuclei in fluorescence microscopy images shown in G. Error bars represent mean ± S.D. for n = 3 technical replicates (with at least 50 nuclei per image). ***, p < 0.001 (two-tailed Student's t test for comparison to the nontargeting control sgRNA). I, simultaneous, but not individual, knockdown of CHMP2A and CHMP2B increases prion-like tau aggregation. % FRET-positive reporter cells transduced with sgRNAs as indicated 2 days after tau fibril treatment. Error bars represent mean ± S.D. where n = 3 technical replicates. *, p < 0.05 (two-tailed Student's t test for comparison to the nontargeting control sgRNA).
A mechanism underlying this CHMP6 phenotype is suggested by the recently reported role of the ESCRT machinery in the repair of endolysosomal membrane damage (36). We hypothesized that knocking down CHMP6 may compromise ESCRT-mediated membrane repair and facilitate tau fibril escape from damaged endolysosomes. We tested this hypothesis by monitoring the formation endolysosomal damage using a cytosolic GFP fusion of galectin 3 (GAL3), a lectin that binds β-galactosides and forms puncta when these sugars are exposed on damaged endolysosomes (37). Although tau fibrils themselves did not cause measurable endolysosomal damage based on our GAL3-GFP reporter (Fig. 4, D and E), knocking down CHMP6 indeed induced GAL3-GFP puncta, revealing endolysosomal damage. This demonstrates that CHMP6 plays a critical role in the maintenance of endolysosomal integrity.
CHMP6 was the only ESCRT protein that was a strong hit in our primary CRISPRi screen. However, targeted knockdown of several other ESCRT components, including members of the ESCRT-I complex (TSG101), the ESCRT-III complex (IST1), and the ESCRT-III–associated Vps4a promoted tau aggregation (Fig. 4F), pointing to a role for the ESCRT pathway in general, as opposed to a specialized role for CHMP6.
Other ESCRT proteins have several paralogues in the human genome, so we hypothesized that these paralogues may have been false-negatives in the CRISPRi screen because they have partially redundant functions. To test this hypothesis, we targeted CHMP2B, a gene with disease-associated mutations involved in familial frontotemporal lobar dementia (38). We hypothesized that its phenotype might have been masked by its close homolog, CHMP2A, which could partially compensate for a loss in CHMP2B function. Indeed, we found that simultaneous, but not individual, knockdown of CHMP2A and CHMP2B generated GAL3 puncta indicative of endolysosomal damage (Fig. 4, G and H) and likewise promoted prion-like propagation of tau aggregation (Fig. 4I). This finding supports our hypothesis that maintenance of endolysosomal membrane integrity by the broader ESCRT-machinery counteracts endolysosomal escape of tau seeds.
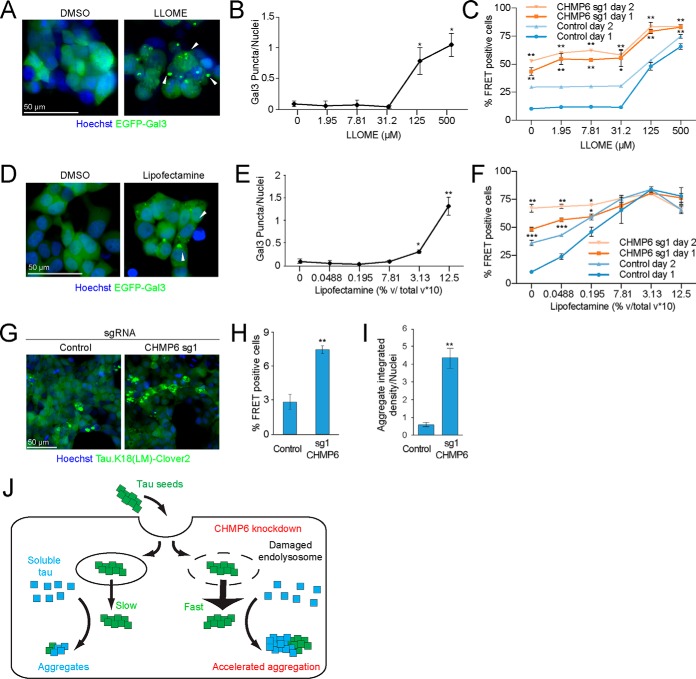
A key implication of this model is that endolysosomal damage may promote the prion-like propagation of tau aggregation. To test this concept, we treated cells with leucyl-leucyl-O-methyl ester (LLOME), a lysosomotropic compound that accumulates in acidified organelles and rapidly forms membranolytic polymers after cleavage by cathepsin C (36, 39, 40). We confirmed that LLOME damages endolysosomal membranes in our cell line using the GAL3-GFP reporter (Fig. 5, A and B). Interestingly, LLOME phenocopied CHMP6 knockdown in its acceleration of seeded tau aggregation only at concentrations where we observe endolysosomal membrane damage (Fig. 5C).
Figure 5.
Small molecules damage endolysosomal compartments and phenocopy the acceleration of the prion-like propagation of tau aggregation following CHMP6 knockdown. A, treatment with the lysosomotropic drug LLOME damages endolysosomal vesicles. Representative fluorescence microscopy images of CRISPRi-HEK293T cells expressing the EGFP-Gal3 reporter treated with DMSO (left) or 500 μm LLOME (right) for 24 h. B, quantification of EGFP-Gal3 puncta divided by number of nuclei in fluorescence microscopy images shown in A. Error bars represent mean ± S.D. for n = 3 technical replicates (with at least 50 nuclei per image). ***, p < 0.001 (two-tailed Student's t test for comparison to DMSO control). C, LLOME treatment accelerates the prion-like propagation of tau aggregation. % FRET-positive cells transduced with control (blue) or CHMP6 sgRNA (orange) were analyzed 24 (dark line) or 48 (light line) h following co-treatment with DMSO or increasing concentrations of LLOME and tau fibrils. Error bars represent mean ± S.D. for n = 3 technical replicates. *, p < 0.05; **, p < 0.01 (two-tailed Student's t test for comparison to the values for nontargeting control sgRNA of the same day). D, Lipofectamine treatment damages endolysosomal vesicles. Representative fluorescence microscopy images of CRISPRi-HEK293T cells expressing the EGFP-Gal3 reporter treated with DMSO (left) or 1.25% (v/v) Lipofectamine 2000 (right) for 6 h. E, quantification of EGFP-Gal3 puncta divided by number of nuclei in fluorescence microscopy images shown in D. Error bars represent mean ± S.D. for n = 3 technical replicates. ***, p < 0.001 (two-tailed Student's t test for comparison to no Lipofectamine treatment). F, Lipofectamine treatment accelerates the prion-like propagation of tau aggregation. % FRET-positive cells transduced with control (blue) or CHMP6 sgRNA (orange) were analyzed 24 (dark line) or 48 (light line) h following co-treatment with PBS or increasing concentrations of Lipofectamine 2000 and tau fibrils. Error bars represent mean ± S.D. where n = 3 technical replicates. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (two-tailed Student's t test for comparison to the values for nontargeting control sgRNA of the same day). G–I, CHMP6 knockdown increases the prion-like propagation tau aggregation when seeding with Alzheimer's patient brain extracts. G, representative images of FRET reporter cells transduced with control (left) or CHMP6 sgRNA (right) and treated with Alzheimer's patient brain extract after 5 days. H, quantification of images in G. Tau aggregates were quantified by integrated density across the entire image and divided by the total nuclei per image. Error bars represent mean ± S.D. for n = 3 images per condition (with at least 50 nuclei per image). **, p < 0.01 (two-tailed Student's t test for comparison to nontargeting control sgRNA) (I) % FRET-positive cells 5 days following treatment with Alzheimer's patient brain extract. Error bars represent mean ± S.D., where n = 3 technical replicates. **, p < 0.01 (two-tailed Student's t test for comparison to nontargeting control sgRNA). J, model for the increased tau aggregation via CHMP6 knockdown and generation of damaged endolysosomes.
As mentioned above, lipocationic reagents, such as Lipofectamine, are frequently used to deliver tau aggregates to cells for in vitro studies of prion-like propagation (13, 14, 41). Interestingly, these agents have previously been demonstrated to induce endolysosomal damage (42). Thus, lipocationic agents might facilitate cargo delivery and escape, in part, by causing endolysosomal membrane damage. Indeed, we found that GAL3 puncta indicative of endolysosomal damage are visible 24 h after treating cells with Lipofectamine (Fig. 5, D and E). Moreover, pre-treatment with Lipofectamine 6 h prior to seeding cells with tau fibrils significantly increased the formation of tau aggregates in a concentration-dependent manner, including at concentrations lower than the threshold required to induce a Gal3 reporter response (Fig. 5F). This suggests that Lipofectamine may assist in the prion-like spread of tau aggregates both by acting as a delivery vehicle and damaging endolysosomal membranes.
When we combined CHMP6 knockdown with either LLOME or Lipofectamine treatment, we found that the relative impact of CHMP6 knockdown on tau aggregate seeding was diminished in the presence of LLOME (Fig. 5C) or Lipofectamine (Fig. 5F), supporting the idea that CHMP6 knockdown and LLOME/Lipofectamine treatment promotes propagation of tau aggregation at least partially via the same mechanism.
Finally, we wanted to validate the CHMP6 phenotype using tau seeds derived from Alzheimer's patient brain-derived extracts. Indeed, we found that CHMP6 knockdown increased the rate of tau aggregation in our reporter line seeded with patient brain-derived tau by both microscopy (Fig. 5, G and H) and flow cytometry (Fig. 5I). Taken together, our results support endolysosomal escape of tau seeds as a rate-limiting step in our cell-based model of prion-like tau propagation. Propagation can be accelerated by compromising endolysosomal integrity either by directly damaging endolysosomes or by interfering with their repair through the ESCRT machinery (Fig. 5J).
Discussion
Using our CRISPRi-based genetic screening platform in a cell-based model of prion-like propagation of tau aggregation, we found that defects in the ESCRT machinery compromise the integrity of the endolysosomal pathway and thereby promote endolysosomal escape of tau seeds and accelerated propagation of tau aggregation. Although our observations were made in a cell-based model, it is intriguing to speculate that they are relevant for propagation of tau aggregation in the context of neurodegenerative diseases. Indeed, endolysosomal changes are among the first cellular symptoms in Alzheimer's disease (43), and have been postulated to be a central driver of pathogenesis in many neurodegenerative diseases (44–46). Furthermore, several risk genes for neurodegenerative diseases are thought to function in the endolysosomal pathway, including CHMP2B (38).
Although the ESCRT-III subunit CHMP6 was a top hit in our genetic screen, CHMP2B knockdown by itself did not have a major impact on endolysosomal integrity and the propagation of tau aggregation. This was likely the case because human cells express CHMP2A, a paralogue of CHMP2B, which can mostly compensate for CHMP2B in our cell-based model, combined knockdown of CHMP2A and CHMP2B phenocopied CHMP6 knockdown. CHMP6 does not have a paralogue in mammalian cells and is an essential gene, whereas CHMP2B is nonessential, based on the cancer dependence map (depmap.org)4 (47), and knockout mouse phenotypes (48). This provides a rationale for CHMP6 deficiency not being associated with neurodegenerative diseases, it may not be compatible with life. CHMP2B deficiency can be expected to cause a milder phenotype that is unmasked only later in life.
Many mechanisms have been proposed to explain the toxicity of tau aggregates. Intriguingly, tau aggregates can damage membranes in vitro (49), and may damage the endolysomal pathway in patient neurons. In combination with our findings, such a mechanism of toxicity would predict a “vicious circle” or feed-forward mechanism, in which tau aggregates would damage the endolysosomal pathway, thereby promoting their own propagation. They could also promote spreading of other aggregates, compatible with the co-occurrence of different protein pathologies, such as tau and α-synuclein, in many cases of neurodegenerative disease (50). However, in our HEK293T cell-based model, we did not find evidence of endolysosomal damage introduced by tau fibrils (Fig. 4, D and E), possibly due to differential susceptibility of different cell types to tau toxicity.
In summary, our results further support the concept that therapeutic strategies aimed at maintaining or restoring the function of the endolysosomal pathway or promoting its repair may be promising in neurodegenerative diseases. Future studies will be aimed at understanding mechanisms underlying the VPS13D phenotype, which seems distinct from the endolysosomal escape pathway controlled by CHMP6.
Although our genetic screen with libraries targeting protein homeostasis factors unexpectedly uncovered the ESCRT machinery in counteracting endolysosomal escape of tau, we had expected to find molecular chaperones or co-chaperones controlling tau aggregation among the top hits. Results obtained in vitro (51–54) and in vivo (55) suggest that specific chaperones and co-chaperones can strongly modulate tau aggregation, and are potential therapeutic targets for tauopathies (56). The fact that knockdown of individual chaperones did not have a major impact on tau aggregation could be due to redundancy in the chaperone network of cells. Future CRISPR activation (CRISPRa) screens have the potential to yield complementary results by over-expressing endogenous genes (18, 19) and may reveal relevant chaperones in the cellular context. Finally, future screens in iPSC-derived neurons using our recently developed platform (25) may reveal additional, neuron-specific pathways, and also uncover factors that underlie selective vulnerability of specific neuronal subtypes (57).
Experimental procedures
Preparation of extracts from Alzheimer's disease patient brains
The Alzheimer's disease brain sample was received from the Neurodegenerative Brain Bank of the UCSF Memory and Aging Center (UCSF/MAC). All research participants at UCSF/MAC undergo longitudinal clinical and imaging assessment. Upon death, the fresh brain was slabbed into 8–10–mm thick coronal slabs upon procurement. These slabs were alternately fixed, in 10% neutral buffered formalin for 72 h, or snap frozen. Twenty-six tissue blocks covering dementia-related regions of interest were dissected from the fixed slabs, and hematoxylin and eosin and immunohistochemical stains were applied following standard diagnostic procedures developed for patients with dementia (58, 59). Immunohistochemistry was performed using antibodies against TDP-43 (rabbit, 1:2000, Proteintech Group, Chicago, IL), hyperphosphorylated tau (CP-13, S202/T205, mouse, 1:250, courtesy of P. Davies), β-amyloid (1–16, mouse, clone DE2, 1:500, Millipore, Billerica, MA), and α-synuclein (LB509, mouse, 1:5000, courtesy of J. Trojanowski and V. Lee). All immunohistochemical runs included positive control sections to exclude technical factors as a cause of absent immunoreactivity. Neuropathological diagnosis followed currently accepted guidelines (60–64). For this study, a region from the parietal cortex containing a high amount of AD-tau pathology was sampled from a snap frozen block. A brain extract was prepared and phosphotungstate-insoluble fractions were purified as previously described (13). The extract was diluted in PBS, 1:40, in Dulbecco's PBS and flash frozen in liquid nitrogen and stored at −80 °C.
Purification, characterization, labeling, and fibrillization of recombinant tau
N-terminally His6 -tagged Human WT 0N4R tau protein was expressed in RosettaTM 2(DE3)-competent cells (MilliporeSigma, number 71400-3) essentially as previously described (51). Briefly, protein expression was induced with 200 μm isopropyl 1-thio-β-d-galactopyranoside for 3.5 h at 30 °C. Cells were lysed via a microfluidizer (Microfluidics catalog number M-100EH) followed by boiling of the lysate for 20 min. The clarified supernatant was subsequently dialyzed overnight into Buffer A (20 mm MES, pH 6.8, 50 mm NaCl, 1 mm EGTA, 1 mm MgCl2, 2 mm DTT, 0.1 mm phenylmethylsulfonyl fluoride) and purified by cation exchange using a HiTrap Capto SP ImpRes column (GE Healthcare catalog number 17546851) with elution buffer (Buffer A with 1 m NaCl). Fractions containing tau as determined by Coomassie-stained SDS-PAGE were dialyzed into PBS, concentrated with an Amicon Ultra-15 centrifugal 3-kDa MWCO filter (Millipore catalog number UFC900324), endotoxin purified using a Pierce high capacity endotoxin removal spin column (ThermoFisher catalog number 88274), filter sterilized using a Millex-GV syringe filter unit (Millipore catalog number SLGV033RB), and snap frozen in PBS at −80 °C. Aggregation was induced by incubating 10 μm tau (0.43 mg/ml) with 0.022 mg/ml of heparin (Fisher catalog number 9041-08-1, lot 177772) and shaken at 37 °C overnight in a shaker at 1200 rpm (VWR catalog number 12620-942).
To generate fluorescently labeled tau fibrils, 0.6 μl of 10 mg/ml of Alexa Fluor 555 (ThermoFisher catalog number A37571), 180 μl of 0.43 mg/ml of tau fibrils, and 19.4 μl of 1 m sodium bicarbonate were mixed at room temperature in the dark for 1 h. Labeled tau fibrils were subsequently purified from unlabeled dye with using a Zeba 7k MWCO spin desalting column (ThermoFisher catalog number 89882).
Tau fibrils were negatively stained with 0.75% uranyl formate (pH 5.5–6.0) on thin amorphous carbon-layered 400-mesh copper grids (Pelco catalog number 1GC400). Five μl of sample was applied to the grid for 20 s before taking the droplet off with Whatman paper, followed by two washes with 5 μl of ddH2O and three applications of 5 μl of uranyl formate removed by vacuum. Grids were imaged at room temperature using a Fei Tecnai 12 microscope operating at 120 kV. Images were acquired on a US 4000 CCD camera at ×66873 resulting in a sampling of 2.21 Å/pixel.
Plasmid and library design and construction
Plasmids for the FRET-based aggregation reporter were constructed by cloning a fusion of the K18 repeat domain of tau containing the P301L/V337M mutation (20) in-frame with C-terminal Clover2 (Addgene number 54711) or mRuby2 (Addgene number 54768 (21)), gifts from Michael Davidson, into the lentiviral expression vector pMK1200 (23) (Addgene number 84219) under the control of the constitutive EF1A promoter, to obtain pMK1253 or pMK1254, respectively. The K18-Scarlet-I reporter was constructed by cloning a fusion of the K18 construct from pMK1253 in-frame with the C-terminal mScarlet-I (Addgene number 98839, a gift from Dorus Gadella (65)) as well as replacing the EF1a promoter with a CAG promoter to obtain pJC49.
Pooled CRISPRi sgRNA libraries targeting human protein homeostasis genes were designed using our next-generation algorithm (66). SgRNA protospacers for these libraries are listed in Table S2. Oligonucleotide pools encoding the library were synthesized by Agilent, PCR amplified, and cloned into our optimized lentiviral sgRNA expression vector as previously described (18).
For generation of individual sgRNAs, pairs of oligonucleotides (IDT) were annealed and ligated into our optimized lentiviral sgRNA expression vector. For double sgRNA expression constructs, CHMP2B and CHMP2A targeting oligos were annealed and ligated into pMJ114 and pMJ179, and a double-sgRNA vector was generated from these as previously described (67). The fluorescent Gal3 reporter was PCR amplified from pEGFP-hGal3 (Addgene number 73080, a gift from Tamotsu Yoshimori) and Gibson cloned into pJC41, which uses the pMK1200 backbone (described above) and replaces the EF1a promoter with a CAG promoter.
For stable expression of the CRISPRi machinery, we modified our established lentiviral (d)Cas9 expression vectors (18) by replacing the SFFV promoter with a minimal ubiquitous chromatin opening element (UCOE) (68) upstream of the EF1α promoter, resulting in pMH0006 (UCOE-SFFV-dCas9-BFP-KRAB).
Cell culture, cell line generation, and treatment conditions
All cells were maintained in a tissue culture incubator (37 °C, 5% CO2) and checked regularly for mycoplasma contamination. HEK293T cells were cultured in DMEM supplemented with 10% fetal bovine serum (Seradigm catalog number 97068-085, lot 076B16), Pen/Strep (Life Technologies, catalog number 15140122), and l-glutamine (Life Technologies, catalog number 25030081).
To generate the FRET reporter line, HEK293T cells were infected with lentivirus from plasmids pMK1253 and pMK1254 and cells with the highest dynamic FRET signal 2 days after seeding with tau fibrils were selected. To introduce CRISPRi functionality, the cells were lentivirally transduced with pHR-SFFV-dCas9-BFP-KRAB (Addgene number 46911, a gift from Stanley Qi and Jonathan Weissman (22)), monoclonal cell lines were selected and CRISPRi activity was validated as previously described (25).
Cellular markers were endogenously labeled using the split-mNeonGreen2 system (69), following conditions described in Ref. 70. Briefly, synthetic guide RNAs (IDT, Alt-R reagents) were first complexed in vitro with purified Streptococcus pyogenes Cas9 protein (UC Berkeley Macrolab). Cas9/RNA complexes were then mixed with ssDNA oligo donors (IDT, Ultramer reagents) and nucleofected (Lonza catalog number AAF-1002B, Amaxa program CM-130) into HEK cells stably expressing SFFV-mNeonGreen21–10. Fluorescent cells were selected by flow cytometry (SONY Biotechnology catalog number SH800S). Sequences for CRISPR RNA and donors used are listed as follows: LAMP1 (C-term mNG11): crRNA sequence, 5′-GTGCACCAGGCTAGATAGTC-3′; donor oligonucleotide sequence, 5′-CCCAGAGAAAGGAACAGAGGCCCCTGCAGCTGCTGTGCCTGCGTGCACCAGGCTACATCATATCGGTAAAGGCCTTTTGCCACTCCTTGAAGTTGAGCTCGGTACCACTTCCTGGACCTTGAAACAAAACTTCCAATCCGCCACCGATAGTCTGGTAGCCTGCGTGACTCCTCTTCCTGCCGACGAGGTAGGCGATGAGG-3′; RAB7A (N-term mNG11): crRNA sequence, 5′-TAGTTTGAAGGATGACCTCT-3′; donor oligonucleotide sequence, 5′-TGTTTCCATCACACTCACAGTGATTTCTCCTTTTCCCCCTTTAGTTTGAAGGATGACCGAGCTCAACTTCAAGGAGTGGCAAAAGGCCTTTACCGATATGATGGGTGGCGGATTGGAAGTTTTGTTTCAAGGTCCAGGAAGTGGTACCTCTAGGAAGAAAGTGTTGCTGAAGGTTATCATCCTGGGAGATTCTGGGTAAG-3′.
To generate the LAMP1/K18-mScarletI CRISPR cells, the LAMP1-mNG11 cells were lentivirally transduced and sorted sequentially with pMH006 and pJC49. To generate CRISPRi-HEK293T cells that monitor EGFP-Gal3 damage or only generate tau.K18(LM)-Clover2 aggregates, CRISPRi-HEK293T cells were lentivirally transduced with pMK1253 and pJC41, and a polyclonal population was sorted by FACS.
Primary CRISPRi screen
For pooled screening of libraries, 7.5 million HEK293T cells were seeded into a 15-cm2 plate with complete DMEM on day 0. On day 1, 5 μg of lentiviral plasmid packaging mix (24) and 5 μg of pooled sgRNA library plasmid was transfected using Lipofectamine 2000 (ThermoFisher catalog number 11668019) and incubated for 2 days. On day 3, conditioned media was removed and filter sterilized using a Millex-GV syringe filter unit (Millipore catalog number SLGV033RB). Lentivirus was precipitated (Alstem catalog number VC100) according to manufacturer protocols and resuspended in complete DMEM. 20 million FRET reporter cells were added to lentivirus-containing media and seeded into a T175 flask. On day 4, media from the T175 was replaced with DMEM complete with 2.5 μg/ml of puromycin. On day 8, cells infected with pooled sgRNA libraries were trypsinized and replated into at 100 μl/well (25,000 cells/well) of several 96-well plates. In addition, 0.3 μl of 0.43 μg/μl of tau fibrils were added to each well. 48 h later, cells were trypsinized and sorted using an Aria II FACS cytometer into FRET-negative and FRET-positive populations. Genomic DNA was isolated using a Macherey-Nagel Blood L kit (Machery-Nagel catalog number 740954.20) and followed according to manufacturer protocols. SgRNA-encoding regions were then amplified and sequenced as previously published (18). Phenotypes and p values for each gene were calculated using our most recent bioinformatics pipeline (25). For genes targeted by more than one sgRNA library, values for the more significant phenotype were selected. Full results are listed in Table S1.
Secondary assays based on microscopy and flow cytometry
To monitor tau aggregation, FRET reporter cells were seeded (25,000 cells/well) into 100 μl/well in a 96-well black bottom plates (Greiner Bio-One number 655097) with 0.3 μl of 0.43 mg/ml of tau fibrils on day 1 and analyzed 24 or 48 h after seeding. For Alzheimer's patient brain extracts, 1.5 μl of extract, 0.375% total (v/v) Lipofectamine 2000 (ThermoFisher catalog number 11668019), and 7.85 μl of Opti-MEM (Thermo catalog number 31985062) were mixed and incubated at room temperature for 2 h. Lipofectamine-brain extract complexes were then added to cells previously plated in 100 μl (10,000 cells/well) for 6 h. Cells were analyzed 5 days after seeding. For analysis, cells were stained with Hoechst 33342 (ThermoFisher catalog number 5553141) at 1 μg/ml and analyzed by flow cytometry using a BD FACSCelesta or by fluorescence microscopy using an InCell 6000 (GE Healthcare catalog number 28-9938-51). Digital images were analyzed using CellProfiler by counting the integrated density of identified aggregates/nuclei and averaged between 3 images. Cells with sgRNA knockdown were similarly analyzed using a comparable protocol 5 days after transduction with individual sgRNA-encoding lentivirus.
For experiments measuring tau aggregation in the presence of inducers of endolysosmal damage, FRET reporter cells were seeded (25,000 cells/well) into 100 μl/well in a 96-well black bottom plate and treated with LLOME (Sigma catalog number L7393-500MG) at varying concentrations with 0.3 μl of 0.43 μg/μl of Tau fibrils. For treatment with Lipofectamine 2000, FRET reporter cells were seeded (25,000 cells/well) into 100 μl/well in a 96-well black bottom plate and treated with Lipofectamine 2000 at varying concentrations. Cells were then treated 0.3 μl of 0.43 μg/μl of Tau fibrils 6 h later. 24 or 48 h later after seeding, cells were stained with Hoechst 33342 (1 μg/ml) and analyzed by flow cytometry using a BD FACSCelesta or by fluorescence microscopy using an InCell 6000 GE Healthcare (catalog number 28-9938-51). Digital images were collected and analyzed using CellProfiler by quantifying the integrated density of identified aggregates and Hoechst-stained nuclei. In cases where CellProfiler was unable to identify nuclei, nuclei were counted interactively using ImageJ. Cells with sgRNA knockdown were similarly analyzed using a comparable protocol 5 days after transduction with individual sgRNA-encoding lentivirus.
To monitor tau fibril uptake, on day 0, CRISPRi-HEK293T cells previously transduced for 5 days with lentivirus expressing single sgRNAs were seeded (25,000 cells/well) into 100 μl/well in a 96-well plate. On day 1, cells were treated with 1 μl of 0.39 μg/μl of AF555-tau fibril for 1 h at 37 °C and collected for analysis by flow cytometry. Median mRuby2 values were calculated in FlowJo and averaged between 3 technical replicates.
To monitor tau.K18(LM)-Clover2 steady-state levels, on day 0, FRET reporter cells previously transduced for 5 days with lentivirus expressing single sgRNAs were seeded (25,000 cells/well) into 100 μl/well in a 96-well plate. On day 1, cells were collected for analysis by flow cytometry. Median Clover2 values were calculated in FlowJo and averaged between 3 technical replicates.
To monitor localization of AF555-labeled tau fibrils, HEK293T cells expressing Tau.K18(LM)-Clover2 were seeded (12,500 cells/well) into 100 μl/well in a 96-well black bottom plate (Greiner Bio-One number 655097) on day 0 in complete DMEM. On day 1, 0.3 μl of 0.39 μg/μl of AF555-tau fibrils were added to cell culture media and placed into an InCell 6000 (GE Healthcare catalog number 28-9938-51) incubator. Images were taken at 20-min intervals between incubations.
To monitor co-localization of tau fibrils with LAMP1, fluorescently labeled CRISPRi-HEK293T cells were seeded in glass-bottom 96-well plates (Cellvis number P96-1.5P) pre-coated with fibronectin (Roche catalog number 11051407001) at 15,000 cells/well in 150 μl of complete DMEM (including 10% fetal bovine serum). After incubation for 3 h to allow for cell adhesion, cells were treated with 0.11 μg of AF555-tau PFFs per well. 22 h post-treatment, cells were counterstained with Hoechst 33342 (0.5 μg/ml, 30 min at 37 °C) and imaged in complete DMEM without phenol red. Live-cell imaging was performed on a Dragonfly spinning disk instrument (Andor) at 37 °C in 5% CO2 atmosphere equipped with a ×63/1.47 NA objective (Leica) and an iXon Ultra 888 EMCCD camera (Andor), acquiring time-lapse datasets at 0.4Hz. Images were analyzed by thresholding LAMP1 and tau fibril images and masking pixels positive for LAMP1 and tau fibrils. Colocalization was calculated by dividing the total image intensity of the masked image with total image intensity of the thresholded tau fibrils. Cell profiler scripts are available at kampmannlab.ucsf.edu/resources.4
To monitor Gal3-EGFP puncta formation, CRISPRi-HEK293T cells expressing EGFP-Gal3 were seeded into 100 μl/well (25,000 cells/well) in a 96-well black bottom plates and treated with LLOME or Lipofectamine at varying concentrations. 24 h after seeding, cells were stained with Hoechst 33342 (1 μg/ml) and digital images were collected and analyzed by an InCell 6000 by counting EGFP-Gal3 puncta/nuclei and averaged between 3 images. Cells with sgRNA knockdown were similarly analyzed using a comparable protocol 5 days after transduction with individual sgRNA-encoding lentivirus and puromycin selection of transduced cells.
Cell fractionation and immunoblot
Cells were seeded into 3 ml at 250,000 cells/well in 6-well dishes with 4.8 μl of 0.43 mg/ml of tau fibrils, and harvested after 48 h by washing with PBS and releasing with 0.25% trypsin. Cells were resuspended with DMEM pre-warmed to 37 °C, spun down, and washed again with PBS. Cells were resuspended in 20 μl of PBS and lysed by flash freezing on dry ice and rapidly thawed at 42 °C. This step was repeated twice. The resulting lysate was spun at 1000 × g and the resulting supernatant was transferred to a new tube and respun to remove any carry-over insoluble material. The pellet was rinsed 3 times with PBS and resuspended to the corresponding volume of supernatant and briefly sonicated with a tip sonicator (Sonopuls 2070) for a brief 1-s pulse at 10% maximum intensity. Equivalent fractions of total volume for 100 ng of supernatant and resuspended pellet were boiled with SDS loading buffer (50 mm Tris-Cl, pH 6.8, 2% (2 w/v) SDS, 0.1% (w/v) bromphenol blue) and 10 mm DTT, subjected to SDS-PAGE on 4–12% BisTris polyacrylamide gels (ThermoFisher catalog number NP0322BOX) and transferred to nitrocellulose membranes. Primary antibodies against human tau (DAKO catalog number A0024) and β-actin (Cell Signaling catalog number 3700) were used to detect proteins. Blots were then incubated with secondary antibodies (Li-Cor catalog numbers 926-32213 and 926-68072) and imaged on the Odyssey Fc Imaging system (Li-Cor catalog number 2800). Digital images were processed and analyzed using Licor Image StudioTM software.
qRT-PCR
CRISPRi-HEK293T cells expressing a constitutive nontargeting or targeting sgRNA were collected by centrifugation at 1000 × g for 10 min, washed twice with ice-cold PBS, and processed for qPCR using a RNA purification kit (Zymo catalog number D7011). 500 ng of total RNA from each sample were reverse transcribed using SuperscriptTM III reverse transcriptase using an oligo(dT) primer (Invitrogen catalog number 18080044). The resulting cDNA was diluted 5-fold using 10 mm Tris (pH 8.0) and 0.67 μl of this dilution was used for each quantitative real-time PCR (qPCR). qPCR were set up using SensiMix 2x Mastermix (Bioline catalog number QT615-20) and oligonucleotides targeting genes of interest (IDT) in triplicate and run on QuantStudio 6 Flex (Applied Biosystems catalog number 4485694) using protocols according to the mastermix manufacturer's specifications. All reactions were normalized to an internal loading control (glyceraldehyde-3-phosphate dehydrogenase) and the sgRNA activity is expressed as knockdown efficiency. The qPCR primer sequences are listed in Table S2.
Author contributions
J. J. C. and M. K. conceptualization; J. J. C., R. T., J. Y. H., S. K. S., S.-A. M., M. Y. H., L. T. G., M. D. L., and M. K. resources; J. J. C., P. R., M. N., R. T., E. T., D. R. S., J. E. G., M. D. L., and M. K. formal analysis; J. J. C., D. L. N., P. R., M. N., E. T., J. Y. H., S. K. S., S.-A. M., L. T. G., M. D. L., and M. K. investigation; J. J. C. and M. K. methodology; J. J. C. and M. K. writing-original draft; J. J. C., D. L. N., P. R., M. N., R. T., E. T., J. Y. H., S. K. S., S.-A. M., D. R. S., L. T. G., J. E. G., M. D. L., and M. K. writing-review and editing; R. T. and M. K. software; D. R. S., L. T. G., J. E. G., and M. K. funding acquisition; M. K. supervision.
Supplementary Material
Acknowledgments
We thank Kathleen Keough and Nia Teerikorpi for contributing to preliminary studies. We thank all co-authors, and Adam Frost, Bryce Mendelsohn, Jay Debnath, Amanda Woerman, Kartika Widjaja, Avi Samelson, Nina Dräger, Emmy Li, Poornima Ramkumar, and other members of the Kampmann Lab for discussions and feedback on the manuscript. We thank Eric Chow and Derek Bogdanoff (UCSF Center for Advanced Technology) for support with next-generation sequencing and Sarah Elmes (UCSF Laboratory for Cell Analysis) for support with FACS. For human tissue samples, we thank Lea Grinberg, William Seeley, and the Neurodegenerative Disease Brain Bank at the University of California, San Francisco.
This work was supported by National Institutes of Health New Innovator Award Grant DP2 GM119139 (to M. K.), R01 AG062359 (to M. K.), R56 AG057528 (to M. K. and L. T. G.), U54 NS100717 (to M. K.), S10 OD021741 (supporting electron microscopy), R01 NS059690 (to J. E. G.), P30 CA082103 (supporting the UCSF Laboratory for Cell Analysis), P01AG019724 and P50AG023501 (supporting the UCSF Neurodegenerative Disease Brain Bank), a Paul G. Allen Distinguished Investigator Award (to M. K.), a Chan Zuckerberg Biohub Investigator Award (to M. K.), the Tau Consortium (to M. K., L. T. G., J. E. G. and the UCSF Neurodegenerative Disease Brain Bank), the Consortium for Frontotemporal Dementia Research (UCSF Neurodegenerative Disease Brain Bank), a QB3/Calico Longevity Postdoctoral Fellowship (to J. J. C.), an Alzheimer's Association Postdoctoral Fellowship (to J. J. C.), a National Defense Science & Engineering Graduate Fellowship (to S. K. S.), and an EMBO long-term postdoctoral fellowship ALTF 1193–2015 (to M. Y. H.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
This article contains Movies S1–S5, Tables S1 and S2, and Fig. S1.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- AD
- Alzheimer's disease
- CRISPRi
- CRISPR interference
- ESCRT
- endosomal sorting complexes required for transport
- LLOME
- leucyl-leucyl-O-methyl ester
- sgRNA
- single guide RNA
- FTD
- frontotemporal lobar dementia
- GAL3
- galectin 3
- UCOE
- ubiquitous chromatin opening element
- qRT
- quantitative RT
- YFP
- yellow fluorescent protein
- CFP
- cyan fluorescent protein
- BisTris
- [bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- DMEM
- Dulbecco's modified Eagle's medium
- iPSC
- induced pluripotent stem cell.
References
- 1. Prusiner S. B. (1982) Novel proteinaceous infectious particles cause scrapie. Science 216, 136–144 10.1126/science.6801762 [DOI] [PubMed] [Google Scholar]
- 2. Vaquer-Alicea J., and Diamond M. I. (2019) Propagation of protein aggregation in neurodegenerative diseases. Annu. Rev. Biochem. 88, 785–810 10.1146/annurev-biochem-061516-045049 [DOI] [PubMed] [Google Scholar]
- 3. Bejanin A., Schonhaut D. R., La Joie R., Kramer J. H., Baker S. L., Sosa N., Ayakta N., Cantwell A., Janabi M., Lauriola M., O'Neil J. P., Gorno-Tempini M. L., Miller Z. A., Rosen H. J., Miller B. L., Jagust W. J., and Rabinovici G. D. (2017) Tau pathology and neurodegeneration contribute to cognitive impairment in Alzheimer's disease. Brain 140, 3286–3300 10.1093/brain/awx243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berg L., McKeel D. W. Jr, Miller J. P., Storandt M., Rubin E. H., Morris J. C., Baty J., Coats M., Norton J., Goate A. M., Price J. L., Gearing M., Mirra S. S., and Saunders A. M. (1998) Clinicopathologic studies in cognitively healthy aging and Alzheimer's disease: relation of histologic markers to dementia severity, age, sex, and apolipoprotein E genotype. Arch. Neurol. 55, 326–335 10.1001/archneur.55.3.326 [DOI] [PubMed] [Google Scholar]
- 5. Giannakopoulos P., Herrmann F. R., Bussière T., Bouras C., Kövari E., Perl D. P., Morrison J. H., Gold G., and Hof P. R. (2003) Tangle and neuron numbers, but not amyloid load, predict cognitive status in Alzheimer's disease. Neurology 60, 1495–1500 10.1212/01.WNL.0000063311.58879.01 [DOI] [PubMed] [Google Scholar]
- 6. Gómez-Isla T., Hollister R., West H., Mui S., Growdon J. H., Petersen R. C., Parisi J. E., and Hyman B. T. (1997) Neuronal loss correlates with but exceeds neurofibrillary tangles in Alzheimer's disease. Ann. Neurol. 41, 17–24 10.1002/ana.410410106 [DOI] [PubMed] [Google Scholar]
- 7. Wang Y., and Mandelkow E. (2016) Tau in physiology and pathology. Nat. Rev. Neurosci. 17, 5–21 10.1038/nrg.2015.11 [DOI] [PubMed] [Google Scholar]
- 8. Frost B., Jacks R. L., and Diamond M. I. (2009) Propagation of tau misfolding from the outside to the inside of a cell. J. Biol. Chem. 284, 12845–12852 10.1074/jbc.M808759200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mirbaha H., Holmes B. B., Sanders D. W., Bieschke J., and Diamond M. I. (2015) Tau trimers are the minimal propagation unit spontaneously internalized to seed intracellular aggregation. J. Biol. Chem. 290, 14893–14903 10.1074/jbc.M115.652693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stöhr J., Wu H., Nick M., Wu Y., Bhate M., Condello C., Johnson N., Rodgers J., Lemmin T., Acharya S., Becker J., Robinson K., Kelly M. J. S., Gai F., Stubbs G., Prusiner S. B., and DeGrado W. F. (2017) A 31-residue peptide induces aggregation of tau's microtubule-binding region in cells. Nat. Chem. 9, 874–881 10.1038/nchem.2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaufman S. K., Sanders D. W., Thomas T. L., Ruchinskas A. J., Vaquer-Alicea J., Sharma A. M., Miller T. M., and Diamond M. I. (2016) Tau prion strains dictate patterns of cell pathology, progression rate, and regional vulnerability in vivo. Neuron 92, 796–812 10.1016/j.neuron.2016.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sanders D. W., Kaufman S. K., DeVos S. L., Sharma A. M., Mirbaha H., Li A., Barker S. J., Foley A. C., Thorpe J. R., Serpell L. C., Miller T. M., Grinberg L. T., Seeley W. W., and Diamond M. I. (2014) Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron 82, 1271–1288 10.1016/j.neuron.2014.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Woerman A. L., Aoyagi A., Patel S., Kazmi S. A., Lobach I., Grinberg L. T., McKee A. C., Seeley W. W., Olson S. H., and Prusiner S. B. (2016) Tau prions from Alzheimer's disease and chronic traumatic encephalopathy patients propagate in cultured cells. Proc. Natl. Acad. Sci. U.S.A. 113, E8187–E8196 10.1073/pnas.1616344113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holmes B. B., Furman J. L., Mahan T. E., Yamasaki T. R., Mirbaha H., Eades W. C., Belaygorod L., Cairns N. J., Holtzman D. M., and Diamond M. I. (2014) Proteopathic tau seeding predicts tauopathy in vivo. Proc. Natl. Acad. Sci. U.S.A. 111, E4376–E4385 10.1073/pnas.1411649111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Holmes B. B., DeVos S. L., Kfoury N., Li M., Jacks R., Yanamandra K., Ouidja M. O., Brodsky F. M., Marasa J., Bagchi D. P., Kotzbauer P. T., Miller T. M., Papy-Garcia D., and Diamond M. I. (2013) Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc. Natl. Acad. Sci. U.S.A. 110, E3138–E3147 10.1073/pnas.1301440110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rauch J. N., Chen J. J., Sorum A. W., Miller G. M., Sharf T., See S. K., Hsieh-Wilson L. C., Kampmann M., and Kosik K. S. (2018) Tau internalization is regulated by 6-O-sulfation on heparan sulfate proteoglycans (HSPGs). Sci. Rep. 8, 6382 10.1038/s41598-018-24904-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stopschinski B. E., Holmes B. B., Miller G. M., Manon V. A., Vaquer-Alicea J., Prueitt W. L., Hsieh-Wilson L. C., and Diamond M. I. (2018) Specific glycosaminoglycan chain length and sulfation patterns are required for cell uptake of tau versus α-synuclein and β-amyloid aggregates. J. Biol. Chem. 293, 10826–10840 10.1074/jbc.RA117.000378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gilbert L. A., Horlbeck M. A., Adamson B., Villalta J. E., Chen Y., Whitehead E. H., Guimaraes C., Panning B., Ploegh H. L., Bassik M. C., Qi L. S., Kampmann M., and Weissman J. S. (2014) Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661 10.1016/j.cell.2014.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kampmann M. (2018) CRISPRi and CRISPRa screens in mammalian cells for precision biology and medicine. ACS Chem. Biol. 13, 406–416 10.1021/acschembio.7b00657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kfoury N., Holmes B. B., Jiang H., Holtzman D. M., and Diamond M. I. (2012) Trans-cellular propagation of Tau aggregation by fibrillar species. J. Biol. Chem. 287, 19440–19451 10.1074/jbc.M112.346072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lam A. J., St-Pierre F., Gong Y., Marshall J. D., Cranfill P. J., Baird M. A., McKeown M. R., Wiedenmann J., Davidson M. W., Schnitzer M. J., Tsien R. Y., and Lin M. Z. (2012) Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods 9, 1005–1012 10.1038/nmeth.2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gilbert L. A., Larson M. H., Morsut L., Liu Z., Brar G. A., Torres S. E., Stern-Ginossar N., Brandman O., Whitehead E. H., Doudna J. A., Lim W. A., Weissman J. S., and Qi L. S. (2013) CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kampmann M., Bassik M. C., and Weissman J. S. (2013) Integrated platform for genome-wide screening and construction of high-density genetic interaction maps in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 110, E2317–E2326 10.1073/pnas.1307002110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kampmann M., Bassik M. C., and Weissman J. S. (2014) Functional genomics platform for pooled screening and generation of mammalian genetic interaction maps. Nat. Protocols 9, 1825–1847 10.1038/nprot.2014.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tian R., Gachechiladze M. A., Ludwig C. H., Laurie M. T., Hong J. Y., Nathaniel D., Prabhu A. V., Fernandopulle M. S., Patel R., Ward M. E., and Kampmann M. (2019) CRISPR-based platform for multimodal genetic screens in human iPSC-derived neurons. Neuron 10.1016/j.neuron.2019.07.014 10.1016/j.neuron.2019.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hurley J. H. (2015) ESCRTs are everywhere. EMBO J. 34, 2398–2407 10.15252/embj.201592484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clayton E. L., Mizielinska S., Edgar J. R., Nielsen T. T., Marshall S., Norona F. E., Robbins M., Damirji H., Holm I. E., Johannsen P., Nielsen J. E., Asante E. A., Collinge J., FReJA Consortium, and Isaacs A. M. (2015) Frontotemporal dementia caused by CHMP2B mutation is characterised by neuronal lysosomal storage pathology. Acta Neuropathol. 130, 511–523 10.1007/s00401-015-1475-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Urwin H., Authier A., Nielsen J. E., Metcalf D., Powell C., Froud K., Malcolm D. S., Holm I., Johannsen P., Brown J., Fisher E. M., van der Zee J., Bruyland M., FReJA Consortium, F. R., Van Broeckhoven C., Collinge J., Brandner S., Futter C., and Isaacs A. M. (2010) Disruption of endocytic trafficking in frontotemporal dementia with CHMP2B mutations. Hum. Mol. Genet. 19, 2228–2238 10.1093/hmg/ddq100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Velayos-Baeza A., Vettori A., Copley R. R., Dobson-Stone C., and Monaco A. P. (2004) Analysis of the human VPS13 gene family. Genomics 84, 536–549 10.1016/j.ygeno.2004.04.012 [DOI] [PubMed] [Google Scholar]
- 30. Kumar N., Leonzino M., Hancock-Cerutti W., Horenkamp F. A., Li P., Lees J. A., Wheeler H., Reinisch K. M., and De Camilli P. (2018) VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 217, 3625–3639 10.1083/jcb.201807019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yeshaw W. M., van der Zwaag M., Pinto F., Lahaye L. L., Faber A. I., Gómez-Sanchez R., Dolga A. M., Poland C., Monaco A. P., van IJzendoorn S. C., Grzeschik N. A., Velayos-Baeza A., and Sibon O. C. (2019) Human VPS13A is associated with multiple organelles and influences mitochondrial morphology and lipid droplet motility. eLife 8, e43561 10.7554/eLife.43561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seong E., Insolera R., Dulovic M., Kamsteeg E. J., Trinh J., Brüggemann N., Sandford E., Li S., Ozel A. B., Li J. Z., Jewett T., Kievit A. J. A., Münchau A., Shakkottai V., et al. (2018) Mutations in VPS13D lead to a new recessive ataxia with spasticity and mitochondrial defects. Ann. Neurol. 83, 1075–1088 10.1002/ana.25220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rampoldi L., Dobson-Stone C., Rubio J. P., Danek A., Chalmers R. M., Wood N. W., Verellen C., Ferrer X., Malandrini A., Fabrizi G. M., Brown R., Vance J., Pericak-Vance M., Rudolf G., Carrè S., Alonso E., Manfredi M., Németh A. H., and Monaco A. P. (2001) A conserved sorting-associated protein is mutant in chorea-acanthocytosis. Nat. Genet. 28, 119–120 10.1038/88821 [DOI] [PubMed] [Google Scholar]
- 34. Ueno S., Maruki Y., Nakamura M., Tomemori Y., Kamae K., Tanabe H., Yamashita Y., Matsuda S., Kaneko S., and Sano A. (2001) The gene encoding a newly discovered protein, chorein, is mutated in chorea-acanthocytosis. Nat. Genet. 28, 121–122 10.1038/88825 [DOI] [PubMed] [Google Scholar]
- 35. Lesage S., Drouet V., Majounie E., Deramecourt V., Jacoupy M., Nicolas A., Cormier-Dequaire F., Hassoun S. M., Pujol C., Ciura S., Erpapazoglou Z., Usenko T., Maurage C. A., Sahbatou M., Liebau S., et al. (2016) Loss of VPS13C function in autosomal-recessive Parkinsonism causes mitochondrial dysfunction and increases PINK1/Parkin-dependent mitophagy. Am. J. Hum. Genet. 98, 500–513 10.1016/j.ajhg.2016.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Skowyra M. L., Schlesinger P. H., Naismith T. V., and Hanson P. I. (2018) Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 360, eaar5078 10.1126/science.aar5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aits S., Kricker J., Liu B., Ellegaard A. M., Hämälistö S., Tvingsholm S., Corcelle-Termeau E., Högh S., Farkas T., Holm Jonassen A., Gromova I., Mortensen M., and Jäättelä M. (2015) Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy 11, 1408–1424 10.1080/15548627.2015.1063871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Skibinski G., Parkinson N. J., Brown J. M., Chakrabarti L., Lloyd S. L., Hummerich H., Nielsen J. E., Hodges J. R., Spillantini M. G., Thusgaard T., Brandner S., Brun A., Rossor M. N., Gade A., Johannsen P., et al. (2005) Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat. Genet. 37, 806–808 10.1038/ng1609 [DOI] [PubMed] [Google Scholar]
- 39. Maejima I., Takahashi A., Omori H., Kimura T., Takabatake Y., Saitoh T., Yamamoto A., Hamasaki M., Noda T., Isaka Y., and Yoshimori T. (2013) Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 32, 2336–2347 10.1038/emboj.2013.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thiele D. L., and Lipsky P. E. (1990) Mechanism of l-leucyl-l-leucine methyl ester-mediated killing of cytotoxic lymphocytes: dependence on a lysosomal thiol protease, dipeptidyl peptidase I, that is enriched in these cells. Proc. Natl. Acad. Sci. U.S.A. 87, 83–87 10.1073/pnas.87.1.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nonaka T., Watanabe S. T., Iwatsubo T., and Hasegawa M. (2010) Seeded aggregation and toxicity of α-synuclein and tau: cellular models of neurodegenerative diseases. J. Biol. Chem. 285, 34885–34898 10.1074/jbc.M110.148460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou X., and Huang L. (1994) DNA transfection mediated by cationic liposomes containing lipopolylysine: characterization and mechanism of action. Biochim. Biophys. Acta 1189, 195–203 10.1016/0005-2736(94)90066-3 [DOI] [PubMed] [Google Scholar]
- 43. Cataldo A. M., Peterhoff C. M., Troncoso J. C., Gomez-Isla T., Hyman B. T., and Nixon R. A. (2000) Endocytic pathway abnormalities precede amyloid β deposition in sporadic Alzheimer's disease and Down syndrome: differential effects of APOE genotype and presenilin mutations. Am. J. Pathol. 157, 277–286 10.1016/S0002-9440(10)64538-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hu Y. B., Dammer E. B., Ren R. J., and Wang G. (2015) The endosomal-lysosomal system: from acidification and cargo sorting to neurodegeneration. Transl. Neurodegener. 4, 18 10.1186/s40035-015-0041-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schreij A. M., Fon E. A., and McPherson P. S. (2016) Endocytic membrane trafficking and neurodegenerative disease. Cell Mol. Life Sci. 73, 1529–1545 10.1007/s00018-015-2105-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Small S. A., Simoes-Spassov S., Mayeux R., and Petsko G. A. (2017) Endosomal traffic jams represent a pathogenic hub and therapeutic target in Alzheimer's disease. Trends Neurosci. 40, 592–602 10.1016/j.tins.2017.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tsherniak A., Vazquez F., Montgomery P. G., Weir B. A., Kryukov G., Cowley G. S., Gill S., Harrington W. F., Pantel S., Krill-Burger J. M., Meyers R. M., Ali L., Goodale A., Lee Y., Jiang G., et al. (2017) Defining a cancer dependency map. Cell 170, 564–576.e16 10.1016/j.cell.2017.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bult C. J., Blake J. A., Smith C. L., Kadin J. A., Richardson J. E., and Mouse Genome Database Group (2019) Mouse Genome Database (MGD) 2019. Nucleic Acids Res. 47, D801–D806 10.1093/nar/gky1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ait-Bouziad N., Lv G., Mahul-Mellier A. L., Xiao S., Zorludemir G., Eliezer D., Walz T., and Lashuel H. A. (2017) Discovery and characterization of stable and toxic Tau/phospholipid oligomeric complexes. Nat. Commun. 8, 1678 10.1038/s41467-017-01575-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yan X., Uronen R. L., and Huttunen H. J. (2018) The interaction of α-synuclein and Tau: a molecular conspiracy in neurodegeneration? Semin. Cell Dev. Biol. 2018, S1084-9521(17)30389-0 10.1016/j.semcdb.2018.05.005 [DOI] [PubMed] [Google Scholar]
- 51. Mok S. A., Condello C., Freilich R., Gillies A., Arhar T., Oroz J., Kadavath H., Julien O., Assimon V. A., Rauch J. N., Dunyak B. M., Lee J., Tsai F. T. F., Wilson M. R., Zweckstetter M., Dickey C. A., and Gestwicki J. E. (2018) Mapping interactions with the chaperone network reveals factors that protect against tau aggregation. Nat. Struct. Mol. Biol. 25, 384–393 10.1038/s41594-018-0057-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kundel F., De S., Flagmeier P., Horrocks M. H., Kjaergaard M., Shammas S. L., Jackson S. E., Dobson C. M., and Klenerman D. (2018) Hsp70 inhibits the nucleation and elongation of Tau and sequesters Tau aggregates with high affinity. ACS Chem. Biol. 13, 636–646 10.1021/acschembio.7b01039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Voss K., Combs B., Patterson K. R., Binder L. I., and Gamblin T. C. (2012) Hsp70 alters tau function and aggregation in an isoform specific manner. Biochemistry 51, 888–898 10.1021/bi2018078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Baughman H. E. R., Clouser A. F., Klevit R. E., and Nath A. (2018) HspB1 and Hsc70 chaperones engage distinct tau species and have different inhibitory effects on amyloid formation. J. Biol. Chem. 293, 2687–2700 10.1074/jbc.M117.803411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Abisambra J. F., Blair L. J., Hill S. E., Jones J. R., Kraft C., Rogers J., Koren J. 3rd, Jinwal U. K., Lawson L., Johnson A. G., Wilcock D., O'Leary J. C., Jansen-West K., Muschol M., Golde T. E., Weeber E. J., Banko J., and Dickey C. A. (2010) Phosphorylation dynamics regulate Hsp27-mediated rescue of neuronal plasticity deficits in tau transgenic mice. J. Neurosci. 30, 15374–15382 10.1523/JNEUROSCI.3155-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Young Z. T., Mok S. A., and Gestwicki J. E. (2018) Therapeutic strategies for restoring Tau homeostasis. Cold Spring Harb. Perspect. Med. 8, a024612 10.1101/cshperspect.a024612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kampmann M. (2017) A CRISPR approach to neurodegenerative diseases. Trends Mol. Med. 23, 483–485 10.1016/j.molmed.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hyman B. T., and Trojanowski J. Q. (1997) Consensus recommendations for the postmortem diagnosis of Alzheimer disease from the National Institute on Aging and the Reagan Institute Working Group on diagnostic criteria for the neuropathological assessment of Alzheimer disease. J. Neuropathol. Exp. Neurol. 56, 1095–1097 10.1097/00005072-199710000-00002 [DOI] [PubMed] [Google Scholar]
- 59. Tartaglia M. C., Sidhu M., Laluz V., Racine C., Rabinovici G. D., Creighton K., Karydas A., Rademakers R., Huang E. J., Miller B. L., DeArmond S. J., and Seeley W. W. (2010) Sporadic corticobasal syndrome due to FTLD-TDP. Acta Neuropathol. 119, 365–374 10.1007/s00401-009-0605-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cairns N. J., Bigio E. H., Mackenzie I. R., Neumann M., Lee V. M., Hatanpaa K. J., White C. L. 3rd, Schneider J. A., Grinberg L. T., Halliday G., Duyckaerts C., Lowe J. S., Holm I. E., Tolnay M., Okamoto K., et al. (2007) Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the Consortium for Frontotemporal Lobar Degeneration. Acta Neuropathol. 114, 5–22 10.1007/s00401-007-0237-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dickson D. W., Bergeron C., Chin S. S., Duyckaerts C., Horoupian D., Ikeda K., Jellinger K., Lantos P. L., Lippa C. F., Mirra S. S., Tabaton M., Vonsattel J. P., Wakabayashi K., Litvan I., and Office of Rare Diseases of the National Institutes of Health (2002) Office of Rare Diseases neuropathologic criteria for corticobasal degeneration. J. Neuropathol. Exp. Neurol. 61, 935–946 10.1093/jnen/61.11.935 [DOI] [PubMed] [Google Scholar]
- 62. Hauw J. J., Daniel S. E., Dickson D., Horoupian D. S., Jellinger K., Lantos P. L., McKee A., Tabaton M., and Litvan I. (1994) Preliminary NINDS neuropathologic criteria for Steele-Richardson-Olszewski syndrome (progressive supranuclear palsy). Neurology 44, 2015–2019 10.1212/WNL.44.11.2015 [DOI] [PubMed] [Google Scholar]
- 63. McKeith I. G., Dickson D. W., Lowe J., Emre M., O'Brien J. T., Feldman H., Cummings J., Duda J. E., Lippa C., Perry E. K., Aarsland D., Arai H., Ballard C. G., Boeve B., Burn D. J., et al. (2005) Diagnosis and management of dementia with Lewy bodies: third report of the DLB Consortium. Neurology 65, 1863–1872 10.1212/01.wnl.0000187889.17253.b1 [DOI] [PubMed] [Google Scholar]
- 64. Montine T. J., Phelps C. H., Beach T. G., Bigio E. H., Cairns N. J., Dickson D. W., Duyckaerts C., Frosch M. P., Masliah E., Mirra S. S., Nelson P. T., Schneider J. A., Thal D. R., Trojanowski J. Q., et al. (2012) National Institute on Aging-Alzheimer's Association guidelines for the neuropathologic assessment of Alzheimer's disease: a practical approach. Acta Neuropathol. 123, 1–11 10.1007/s00401-011-0910-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mastop M., Bindels D. S., Shaner N. C., Postma M., Gadella T. W. J. Jr, and Goedhart J. (2017) Characterization of a spectrally diverse set of fluorescent proteins as FRET acceptors for mTurquoise2. Sci. Rep. 7, 11999 10.1038/s41598-017-12212-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Horlbeck M. A., Gilbert L. A., Villalta J. E., Adamson B., Pak R. A., Chen Y., Fields A. P., Park C. Y., Corn J. E., Kampmann M., and Weissman J. S. (2016) Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife 5, e19760 10.7554/eLife.19760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Adamson B., Norman T. M., Jost M., Cho M. Y., Nuñez J. K., Chen Y., Villalta J. E., Gilbert L. A., Horlbeck M. A., Hein M. Y., Pak R. A., Gray A. N., Gross C. A., Dixit A., Parnas O., Regev A., and Weissman J. S. (2016) A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167, 1867–1882.e21 10.1016/j.cell.2016.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Müller-Kuller U., Ackermann M., Kolodziej S., Brendel C., Fritsch J., Lachmann N., Kunkel H., Lausen J., Schambach A., Moritz T., and Grez M. (2015) A minimal ubiquitous chromatin opening element (UCOE) effectively prevents silencing of juxtaposed heterologous promoters by epigenetic remodeling in multipotent and pluripotent stem cells. Nucleic Acids Res. 43, 1577–1592 10.1093/nar/gkv019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Feng S., Sekine S., Pessino V., Li H., Leonetti M. D., and Huang B. (2017) Improved split fluorescent proteins for endogenous protein labeling. Nat. Commun. 8, 370 10.1038/s41467-017-00494-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Leonetti M. D., Sekine S., Kamiyama D., Weissman J. S., and Huang B. (2016) A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proc. Natl. Acad. Sci. U.S.A. 113, E3501–E3508 10.1073/pnas.1606731113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.