Abstract
Objective
We conducted a comprehensive gene expression meta‐analysis in dermatomyositis (DM) muscle and skin tissues to identify shared disease‐relevant genes and pathways across tissues.
Methods
Six publicly available data sets from DM muscle and two from skin were identified. Meta‐analysis was performed by first processing data sets individually then cross‐study normalization and merging creating tissue‐specific gene expression matrices for subsequent analysis. Complementary single‐gene and network analyses using Significance Analysis of Microarrays (SAM) and Weighted Gene Co‐expression Network Analysis (WGCNA) were conducted to identify genes significantly associated with DM. Cell‐type enrichment was performed using xCell.
Results
There were 544 differentially expressed genes (FC ≥ 1.3, q < 0.05) in muscle and 300 in skin. There were 94 shared upregulated genes across tissues enriched in type I and II interferon (IFN) signaling and major histocompatibility complex (MHC) class I antigen‐processing pathways. In a network analysis, we identified eight significant gene modules in muscle and seven in skin. The most highly correlated modules were enriched in pathways consistent with the single‐gene analysis. Additional pathways uncovered by WGCNA included T‐cell activation and T‐cell receptor signaling. In the cell‐type enrichment analysis, both tissues were highly enriched in activated dendritic cells and M1 macrophages.
Conclusion
There is striking similarity in gene expression across DM target tissues with enrichment of type I and II IFN pathways, MHC class I antigen‐processing, T‐cell activation, and antigen‐presenting cells. These results suggest IFN‐γ may contribute to the global IFN signature in DM, and altered auto‐antigen presentation through the class I MHC pathway may be important in disease pathogenesis.
Introduction
Dermatomyositis (DM) is an idiopathic inflammatory myopathy (IIM) distinguished from other IIMs by pathognomonic skin rashes. Prior gene expression studies have identified a type I interferon (IFN) signature in the muscle, skin, and peripheral blood of patients with juvenile and adult DM 1, 2, 3, suggesting its potential role in disease pathogenesis. In addition, the type I IFN signature appears to correlate with disease activity 4, 5, and early data from a clinical trial of sifalimumab, an IFN‐α antagonist, demonstrated neutralization of the IFN signature with treatment and more clinical improvement in subjects with greater IFN reduction 6. DM also shares overlapping clinical features with other type I IFN‐mediated diseases, including systemic lupus erythematosus (SLE) and the monogenic interferonopathies, STING‐associated vasculopathy 7, and chronic atypical neutrophilic dermatosis (CANDLE) 8. These observations suggest that type I IFN is important in disease pathogenesis. However, whether dysregulated type I IFN signaling is the primary cause of disease or secondary to another immune‐ or nonimmune‐mediated mechanism remain unknown, and less is known about alternative immune pathways that may underlie the IFN response.
Recently, more attention has been paid to the role of type II IFN in IFN‐mediated diseases, including in SLE and DM. Type I IFN, which includes IFN‐α and IFN‐β, is primarily secreted by dendritic cells and macrophages and is involved in the innate and antiviral response, whereas type II IFN, or IFN‐γ, is secreted by T and NK cells and is important for linking innate and adaptive immune responses. Using a modular approach to deconstruct the IFN signature in SLE, Chiche et al identified three distinct IFN modules expressed in the peripheral blood of SLE patients, which were each activated in stepwise fashion 9. The first IFN module was stable over the disease course and consisted of genes induced primarily by type I IFN, whereas the second and third IFN modules correlated with skin and renal disease activity, and the genes in these modules were equally induced by both types I and II IFN, suggesting a role for type II IFN in SLE. In juvenile DM, IFN‐γ transcripts have been identified in the muscle tissue of 11 untreated patients and co‐localized with inflammatory infiltrates and T cells 10. However, the role of type II IFN in DM has not been thoroughly explored using unbiased, data‐driven methods.
The rarity of DM limits the feasibility of well‐powered studies, making data‐driven translational studies challenging. Computational methods allow researchers to integrate historical data sets from different research groups using methods that are robust to different assay technologies as an approach to overcome these challenges. Gene expression meta‐analysis is a strategy used to increase sample size by combining diverse data sets developed on different platforms, which increases the power to detect differentially expressed genes and discover new biological insights. Furthermore, studying gene expression in the target tissues of a disease strengthens the association of results when findings are shared between tissues and also allows for comparison of tissue‐specific gene expression. This approach has been applied to the study of gene expression in rheumatoid arthritis comparing synovial fluid and blood 11, as well as in antineutrophil cytoplasmic antibody–associated vasculitis comparing orbital tissue, peripheral blood leukocytes, and sinus brushings 12.
The purpose of this study was to conduct a comprehensive gene expression meta‐analysis leveraging publicly available microarray data sets to identify novel gene signatures expressed in DM muscle and skin using hypothesis‐free methods. Because skin and muscle disease may follow a discordant clinical course in DM, a secondary objective of this study was to compare gene signatures across tissues to determine if immune pathways are shared across tissues and identify unique immune pathways that might explain these clinical observations. Refining our knowledge of the dysregulated immune pathways in target tissues in DM is critical to advancing biomarker and drug development for this rare and understudied disease and challenging disease paradigms leading to new mechanistic hypotheses.
Methods
Gene expression meta‐analysis pipeline
An overview of the pipeline is presented in Figure 1. Publicly available microarray data sets from Gene Expression Omnibus (GEO) 13 were searched for the key words “myositis” and “dermatomyositis.” These samples were curated to include only subjects with dermatomyositis, including both adult and juvenile subtypes. All other myositis subtypes were excluded. Any samples annotated as inactive disease or nonlesional skin were excluded. Additional clinical covariates, including age, sex, treatment status, and duration of disease, were not available for all samples and thus, in order to avoid introducing false positives from imputation, were not included. Age was annotated for 93% of muscle samples, of which 44% were pediatric. All skin samples were from adult subjects. In muscle, sex was annotated in 77% of samples of which 88% were female, and in skin, sex was known for all samples, of which 66% were female.
Figure 1.
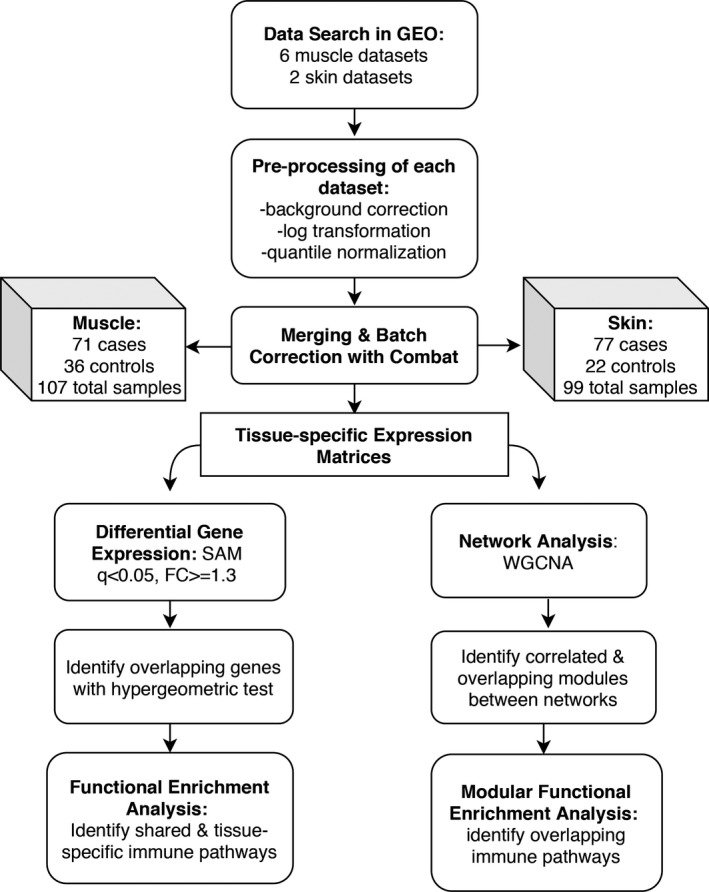
Gene expression meta‐analysis pipeline. Abbreviations: GEO, Gene Expression Omnibus; SAM, Significance Analysis of Microarrays; FC, fold change; DEG, differentially expressed genes; WGCNA: Weighted Gene Coexpression Network Analysis.
Raw data were downloaded for each data set and processed. The processing steps included background correction, log2 transformation, quantile normalization, and probe to gene mapping using R language 14 v3.5.2. We used the R package, SCAN.UPC 15 from Bioconductor 16 to process the data from Affymetrix platforms and the BrainArray database 17 v22 to map probes to Entrez gene IDs. For GSE32245 from the Stanford platform, we used the limma package v3 18 to perform processing steps and mapped probes to genes using GPL files. For GSE3307, the data from A and B arrays were merged using the mean intensities of common probes. Once processed, all muscle data sets were merged in accordance with the previously published pipeline by Hughey and Butte 19. During this step, the expression data were mean centered and reduced to the number of common probes across all data sets. Cross‐study normalization was performed with ComBat within sva package v3 20. Principle component analysis (PCA) plots and boxplots were used to evaluate for successful batch correction and to detect outliers using three standard deviations of the first two principal components. One sample, GSM799008 from GSE32245, was recognized as an outlier and removed from the analysis.
Differential gene expression: Significance Analysis of Microarrays
Significance Analysis of Microarrays (SAM), implemented in the R package siggenes v1.56 21, was used to determine differentially expressed genes between cases and controls in each tissue using a cutoff false discovery rate (FDR) 22 q value of less than 0.05 and fold change (FC) of 1.3 or more. Unsupervised hierarchical clustering using the Ward algorithm 23 was used to generate heatmaps. Significant overlap between up‐ and downregulated genes between muscle and skin was assessed using a hypergeometric test. Functional enrichment analysis of gene lists was carried out by overrepresentation analysis using the ReactomePA package v1.26 24, which detects enrichment of Reactome Database 25 pathway terms. The P values were calculated from the hypergeometric distribution and controlled for multiple comparisons using the Benjamini‐Hochberg method.
Network analysis: Weighted Gene Co‐Expression Network Analysis
In order to detect associations that were not identified on the single gene level, network analysis was performed using Weighted Gene Co‐Expression Network Analysis (WGCNA) v1.68 26, 27. Muscle and skin coexpression networks were each constructed using step‐by‐step signed network construction followed by module detection. In this context, a module is defined as a highly connected set of genes that are positively or negatively correlated. We then calculated the significance between the module eigengene, or first principal component in the module, and case/control status. To compare muscle and skin networks, we calculated the significance of the gene overlap between each muscle and skin module using a hypergeometric test adjusting P values for multiple comparisons. Additionally, network preservation statistics were computed with a module Z‐summary score with a Z‐summary greater than 10 considered to be strong evidence of preservation in the corresponding network and a score less than 10 but greater than 2 considered to be weak to moderate evidence of network preservation as previously described 28.
Within the significant modules, we identified hub genes. A hub gene is a highly connected gene likely to be of biological significance and defined as a gene significance (a measure of how biologically significant each gene is in the network) greater than 0.2 and module membership (how connected a gene is to all the other genes in the module) greater than 0.8 26. Module enrichment was evaluated using ReactomePA v1.26 and clusterProfiler R packages v3.10 29. Network visualization using the STRING v10 database 30 was used to assess evidence for protein‐protein interactions between these hub genes.
Cell‐type enrichment analysis
Enrichment of cell types was determined with xCell v1.1 31. Cell types were filtered to remove erythrocytes, epithelial cells, and stromal cells because they are not relevant to our analysis, resulting in total of 42 immune cell types. Significance assessment to determine the likelihood the cell type is in the mixture was calculated using predefined beta distribution parameters from random mixtures generated from the reference data sets in xCell 31. A t test was used to determine differential cell enrichment between cases and controls using a threshold of P < 0.01. The false discovery rate was controlled by the Benjamini‐Hochberg method. Significantly differentially enriched cell types were visualized with hierarchical clustering using the Ward algorithm.
The IFN signature
The upregulated gene lists identified by SAM were uploaded into Interferome v2.0 32 to determine whether genes were influenced by type I or type II IFN signaling. Interferome is a database of curated results from experiments assessing the IFN response in vitro and in vivo. For our purposes, the reference data set was filtered to only include prior experiments performed in vivo in human disease states.
Results
A total of six microarray data sets from muscle tissue and two from skin tissue were identified. After curation and removal of one outlier in the skin data set, a total of 107 samples from muscle and 99 samples from skin were included. The number of samples from cases and controls is shown in Figure 1. All studies 1, 3, 33, 34, 35 included are displayed in Supplementary Table 1. Studies were successfully merged and batch‐corrected as visualized by PCA plots displayed in Supplementary Figure S1. The expression matrices contained 11 703 common genes in the merged muscle data set and 17 418 common genes in the merged skin data set.
Single gene analysis reveals overlap of upregulated genes enriched in IFN responses
In the single‐gene analysis, 544 genes were significantly differentially expressed between cases and controls in muscle, of which 443 were upregulated and 101 were downregulated (FC ≥ 1.3, q < 0.05; Figure 2A). In skin, 300 genes were differentially expressed, of which 214 were upregulated and 86 were downregulated (Figure 2B). There was significant overlap by the hypergeometric test in the upregulated genes across skin and muscle with a total of 94 shared genes, P = 6.4xE−78 (see Supplementary Table 2 for the full gene list). Many of these genes were IFN‐stimulated genes (ISGs), including CXCL10, RSAD2, ISG15, IFI44L, IFIT3, IF44, and MX1 being the most strongly expressed across both tissues (Figure 3A). There was no significant overlap in repressed genes with only two shared genes, DDX3Y and RPS4Y1, both expressed on the Y chromosome. Enrichment of the 94 overlapping upregulated genes revealed several pathways involved in both innate and adaptive immune responses (Figure 3B). The most enriched terms, consistent with prior gene expression studies, were related to type I IFN signaling. However, several other themes emerged, including type II IFN signaling, class I mediated antigen‐processing and presentation, chemokine signaling, and complement activation.
Figure 2.
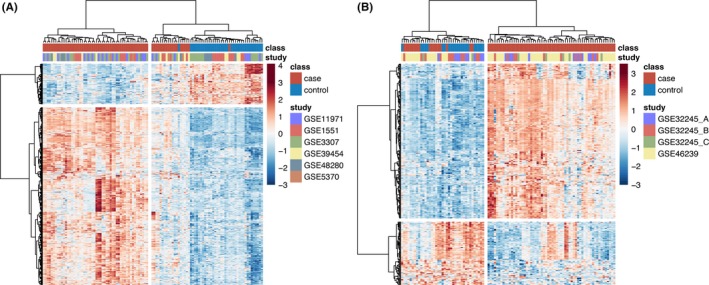
Heatmaps of differentially expressed genes identified by Significance Analysis of Microarrays (SAM). Hierarchical clustering of significant differentially expressed genes (DEGs) in muscle (A) and skin (B) using the cutoff of the false discovery rate P value < 0.05 and fold change ≥ 1.3 demonstrates clustering of cases and controls and that the majority of DEGs in both tissues are upregulated.
Figure 3.
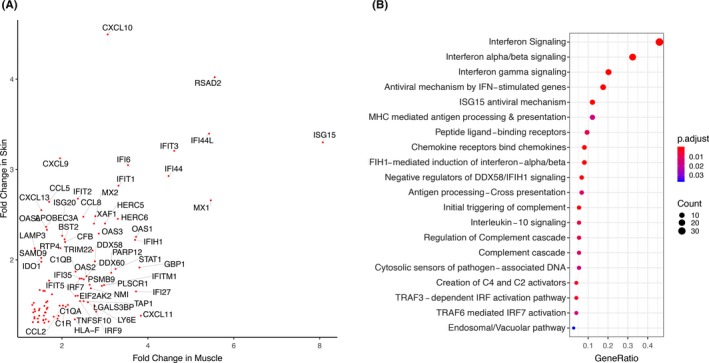
There are 94 overlapping genes in muscle and skin enriched in common immune pathways. A, Gene symbols of 94 overlapping differentially expressed genes plotted by fold change in muscle and skin. B, Enrichment of 94 overlapping genes by overrepresentation analysis using the Reactome Database. P values are calculated using a hypergeometric distribution and corrected for multiple comparisons by the Benjamini‐Hochberg method.
Analysis of the tissue‐specific upregulated gene lists revealed significant overlap in the enriched pathways of both gene sets as well. Significantly enriched terms in both gene lists were type I IFN signaling, type II IFN signaling, and terms related to class I major histocompatibility complex (MHC) antigen‐processing pathways. Complement activation and interleukin (IL) signaling were pathways uniquely enriched in skin, whereas neutrophil degranulation, extracellular matrix organization, collagen formation, and MHC class II antigen processing and presentation were terms uniquely enriched in muscle. A network map of the most enriched terms in each tissue and the differentially expressed genes identified by SAM is displayed in Figure 4, demonstrating the interconnectedness of these pathways on the gene level and emphasizing the network similarity between muscle and skin. Enrichment of downregulated gene lists revealed repression of glucose metabolism in muscle and repression of fatty acid and steroid metabolism in skin.
Figure 4.
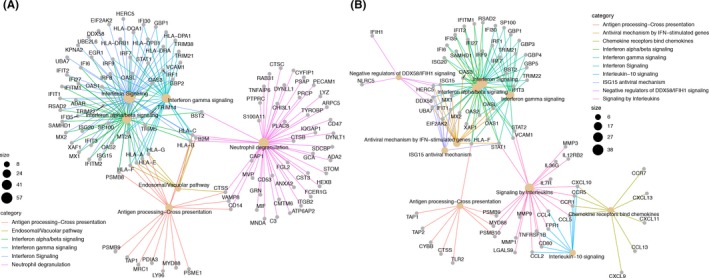
Network maps demonstrating the most enriched terms in each tissue, muscle (A) and skin (B), and the upregulated genes found to be differentially expressed acting in these pathways. These maps demonstrate the interrelatedness of many of these immune pathways in both tissues and highlight the similarity of genes and pathways across the two tissues.
Network analysis identifies overlapping gene modules between muscle and skin and enrichment of terms related to T‐cell activation
By applying WGCNA to the merged gene expression matrices from each tissue, we identified eight modules in muscle and seven modules in skin that met our significance threshold and were either positively or negatively correlated to case/control status (see Figure 5A). At least one module in each tissue‐specific network had significant overlap on the gene level assessed by a hypergeometric test with one or more modules in the other network, as indicated by the asterisks in Figure 5A. Hierarchical clustering of these gene modules revealed clustering of both upregulated and downregulated modules. A concentrated cluster of modules highly correlated with DM cases in both muscle and skin networks consisted of the M1 and M3 modules along with the S1 and S3 modules (denoted by the green box in Figure 5A). These modules were also the highly preserved modules by network preservation statistics 28 in both networks reaching Z‐summary scores greater than 10 (Supplementary Table 3).
Figure 5.
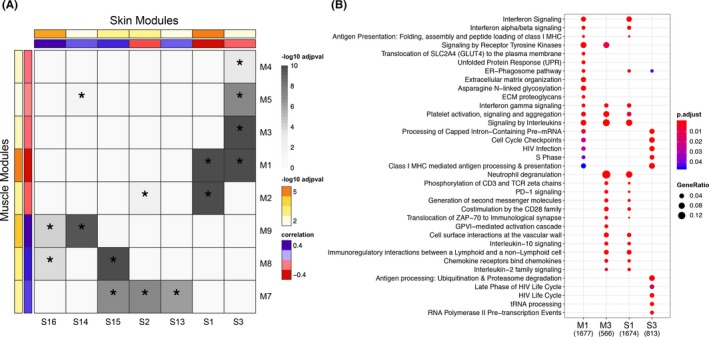
A, Heatmap demonstrating hierarchical clustering of significant modules identified by WGCNA in each tissue based on gene overlap. Correlation between module eigengene and disease status, sign*(correlation), is shown by the red to blue bar, where red represents modules most positively correlated with cases and blue represents modules negatively correlated with cases. The ‐log10 adjusted P value of this correlation is indicated by the yellow to orange bar. The degree of pairwise overlap in genes between muscle modules and skin modules computed using a hypergeometric test is denoted by grey coloring where modules with significant overlap in genes (P < 0.05) are denoted by an asterisk and the color range corresponds to –log10(P value) of the pairwise overlap. B, Enrichment of pathways from the Reactome database across a cluster of highly overlapping and interesting modules in each tissue (green box in pane A) demonstrating enrichment of shared pathways across tissues. P values are adjusted for comparisons across modules by the Benjamini‐Hochberg method.
Enrichment analysis of these specific modules (M1, M3, S1, and S3) using the Reactome Pathway database revealed enrichment of immune pathways relevant to disease that was consistent with those identified in the single‐gene analysis, including type I and II IFN signaling, class I MHC antigen processing and presentation, and chemokine signaling (Figure 5B). However, several additional pathways not seen on the single‐gene level were also enriched. Most notably, in the M3 and S1 modules, there were several pathways enriched relating to T‐cell activation, including CD28 co‐stimulation, IL‐2 signaling, and T‐cell receptor signaling events (Figure 5B). There were also signs of immunoregulatory signaling pathways, including PD‐1 signaling and IL‐10 signaling in both networks. Unique to the muscle network, the M1 module was also enriched in the unfolded protein response. Although there was no overlap of pathway enrichment in the single‐gene analysis, we did find overlap on the network level between the M8 and S6 module. These modules were both enriched in processes related to energy metabolism, including the citric acid cycle, respiratory electron transport chain, adenosine triphosphate synthesis, and fatty acid metabolism.
There were 615 hub genes in the muscle network and 271 hub genes in the skin network identified as meeting the criteria of module membership greater than 0.2 and gene significance greater than 0.8. The top hub genes in each network were from the M1 module in muscle as well as the S1 module in skin, consistent with the fact that these modules were most strongly correlated with case/control status. The 30 most significant hub genes are displayed in Supplementary Table 4. These genes are likely to be of high biological relevance within each network and include several of the same genes identified in the single‐gene analysis, including genes classically upregulated by type I IFN (STAT1, MX1, IFI44, ISG15), genes classically upregulated by type II IFN (CXCL10 and GBP1), genes involved in class I MHC antigen processing (HLA‐A, ‐B and ‐F, B2M, TAP1), and immunoproteasome genes (PSMB8 and PSMB9). There was strong evidence for interactions between the proteins encoded by these overexpressed genes in the STRING database, and network visualization for the top 50 hub genes in each network are included in Supplementary Figure S2.
Cell‐type enrichment analysis identifies antigen‐presenting cells as the most enriched cell type in both tissues
A total of 42 immune cell types were evaluated by cell‐type enrichment analysis using xCell. Nineteen of these cell types were significantly enriched in muscle, and 20 cell types were significantly enriched in skin. Hierarchical clustering showed separation of cases and controls, with cases being most significantly enriched in antigen‐presenting cells, including activated dendritic cells and other dendritic cell types and classically activated M1 macrophages (Figure 6). There was also enrichment of CD4+ and CD8+ T cells as well as gamma‐delta (γδ) T cells; however, there was no enrichment of B cells. Interestingly, regulatory T cells were enriched in cases in skin, whereas these cells were enriched in controls in muscle tissue.
Figure 6.
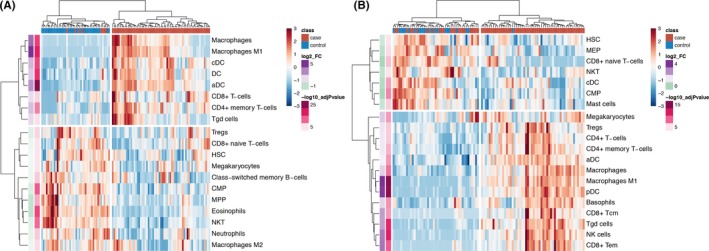
Heatmaps of cell‐type enrichment calculated using xCell demonstrates clustering of cases and controls based on cell types in both muscle (A) and skin (B). Enrichment was strongest for dendritic cells and M1 macrophages in both tissues.
The DM IFN signature is enriched for IFN‐stimulated genes induced by both type I and type II IFN
The upregulated gene lists for each tissue were uploaded to Interferome v2.0 to determine whether the ISGs identified in our gene lists were influenced by type I or type II IFN based on curated results of previous studies measuring the in vivo IFN response in human disease states. The majority of ISGs in both tissues were stimulated by both type I and type II IFN. In muscle,133 genes were classified as ISGs, of which 14 were exclusively stimulated by type I IFN, whereas, 119 were stimulated by both type I and type II IFN. A similar observation was seen in the skin gene list where 124 genes were classified as ISGs, of which only 4 were exclusively stimulated by type I IFN, whereas the other 120 were induced by both type I and type II IFN. Interestingly, two genes previously identified as markers to differentiate type II from type I IFN responses 36, GBP1 and GBP2, were significantly upregulated in our analysis: both GBP1 and GBP2 were upregulated in muscle, and GBP1 was upregulated in skin in the single‐gene analysis. In the network analysis, both GBP1 and GBP2 were highly coexpressed in the most significantly upregulated modules in both networks, providing further evidence that IFN‐γ may contribute to the global IFN signature in DM target tissues.
Discussion
Gene expression meta‐analysis is a powerful method for identifying new biological insights of rare, complex diseases that can be explored with future functional studies. This analysis demonstrates, through both single‐gene and network analyses, the presence of shared transcriptomic signatures across muscle and skin tissues in DM and simultaneous activation of innate and adaptive immune responses. Through an unbiased approach, we confirm the presence of a type I IFN signature and MHC class I antigen presentation across muscle and skin tissues, which is consistent with prior studies in DM muscle 35 and skin 3. We additionally show a type II IFN signature in both tissues and that the majority of the overexpressed ISGs are induced by both type I and type II IFNs. This result is consistent with a prior study that found that the majority of ISGs expressed in rheumatic diseases are induced by both IFN‐α and IFN‐γ in vitro 36. Furthermore, in the network analysis, the most significant modules in each tissue are enriched in terms related to T‐cell activation and T‐cell receptor signaling. Together, these findings suggest that IFN‐γ signaling might contribute to the global IFN signature in DM and that altered autoantigen presentation via the class I MHC pathway may play a role in disease propagation.
Through computational cell enrichment analysis, we also demonstrate an immune infiltrate in both tissues most enriched for antigen‐presenting cells, including activated dendritic cells and inflammatory M1 macrophages as well as CD4+ and CD8+ T cells. These results complement our gene expression findings and support the hypothesis that altered autoantigen presentation may be a key pathway in DM. Several lines of evidence from prior literature support this concept: i) there is early and widespread MHC class I presentation on muscle fibers and endothelial cells in myositis 37, ii) transgenic MHC class I presentation on muscle cells results in spontaneous development of myositis in a mouse model 38, and iii) there is increased expression of immunoproteasome subunits, dendritic cells, monocytes, and CD8+ T cells in peripheral blood and muscle biopsies of patients with myositis 39. Future immunohistochemistry and single‐cell analyses will be integral to help validate the infiltrating immune cell types and behaviors of these cells.
It remains debated whether muscle cells can serve as professional antigen‐presenting cells because it is unknown if they express co‐stimulatory molecules 40. However, it is possible that co‐stimulation could be provided by other antigen‐presenting cells in the correct inflammatory milieu. There are also compelling data that suggest mechanisms by which autoantigens are continuously generated, including enhanced expression of autoantigens in regenerating muscle fibers, suggesting damaged fibers may be the source of autoantigens in myositis 41. These authors also demonstrated that granzyme B, a proteolytic enzyme released by cytotoxic CD8+ T cells, cleaves many autoantigens in systemic autoimmune diseases, representing a possible cycle whereby effector cells continuously generate more autoantigens 42. Interestingly, the gene encoding granzyme B, GZMB, was a significant hub gene in the S1 module from the skin network.
Our finding of an IFN‐γ signature in DM using data‐driven methods is a novel finding. This result is supported by work from Wong et al, wherein both IFN‐β and IFN‐γ transcripts correlated most strongly with the IFN score in DM skin 3, and another recent study demonstrating IFN‐γ transcripts in muscle tissue from subjects with JDM 10. These results also highlight the difficulty of differentiating types I and II IFN pathways, especially in a highly inflammatory milieu, and highlight the significant overlap of these signaling pathways on downstream gene expression. In addition to enrichment of IFN‐γ signaling seen in the single‐gene and network analyses, we also found overexpression of genes classically induced by IFN‐γ, including CXCL9, CXCL10, GBP1, and GBP2 as well as the presence of M1 macrophages by cell enrichment, which are activated by IFN‐γ. Together, this evidence supports the model where IFN‐γ signaling contributes to the global IFN signature in DM. Because the majority of upregulated ISGs in our data have been shown to be stimulated by both type I and type II IFN in historical experiments, we hypothesize that these pathways converge and synergize in DM. An alternative hypothesis is that there is a dynamic IFN response in DM where type I IFN signaling is more important in initiating disease and type II IFN becomes more prominent during disease propagation. Future single‐cell studies will be informative in determining the cell types responsible for the IFN signature in DM.
In addition to innate and adaptive immune dysfunction, this unbiased analysis also provides evidence for metabolic dysfunction in DM. In the network analysis, there is evidence of ER stress with enrichment of the unfolded protein response, consistent with what has been shown in human myositis 37 and in a mouse model of myositis 43, providing further evidence that this may be an important pathway that causes intrinsic muscle damage in myositis. In both tissues, there is enrichment of metabolic pathways in the downregulated genes, including oxidative phosphorylation and either glycolysis (muscle) or fatty acid metabolism (skin), suggesting metabolic dysfunction in both tissues. The implications of this finding in skin is less clear, as repression of fatty acid metabolism appears to be a general finding among inflammatory skin disorders 3.
This study is strengthened by combining publicly available data sets across the most frequently affected tissues in DM—muscle and skin—which amounts to be the largest global study of gene expression across DM using this unbiased methodology. Although studies of peripheral blood may be useful for biomarker development, focusing on gene expression in target tissues is more informative for learning about disease pathophysiology. By applying an unbiased and network‐based analysis, we capture a robust signal of immune pathways overexpressed in DM and detect additional pathways at the systems level not possible to appreciate on the single‐gene level. This computational approach demonstrates how diverse data sets can be effectively combined to study rare autoimmune diseases such as DM.
There are several limitations to this study. The design is cross sectional, which only allows for a static view of gene expression. Although we demonstrate consistent strong signatures across tissues with identification of 94 overlapping genes and enrichment of overlapping immune pathways, the study population is inherently heterogeneous, and individual clinical data regarding possible confounders, including age, sex, disease duration, and treatment status are unavailable to include in the analysis. The ability to adjust our analysis for these covariates would further strengthen the validity of our analysis by normalizing some of the variation in the population and minimizing confounding. Additionally, gene expression is gauged from bulk tissues, which represent a heterogenous population of immune cells rather than the contribution of gene expression from individual immune cells. Because data on the degree of inflammatory infiltrate were unavailable, we were not able to determine whether our results were attributable to infiltrating immune cells versus tissue cells. Lastly, gene expression studies are a useful discovery tool, but the nature of this data type limits the conclusions that can be drawn with regards to biology without validating functional studies.
In conclusion, we demonstrate striking similarity in the muscle and skin gene expression of DM and presence of both type I and II IFN gene signatures, suggesting IFN‐γ signaling may be important in disease pathogenesis. Upregulation of genes involved in MHC class I antigen‐processing and presentation, T‐cell activation and presence of antigen‐presenting cells suggests altered autoantigen presentation and T‐cell cytotoxicity may be important pathways in disease propagation. Given the importance of the immunoproteasome in processing class I MHC antigens, immunoproteasome inhibition could represent a druggable pathway in DM, as others have suggested and mouse work supports 43, and will be an area of exciting future research. Longitudinal studies investigating changes in the type I and II specific IFN responses and the crosstalk between these pathways are also needed to clarify potential DM‐specific therapeutic targets. Advances in computational methods and genomic technologies will provide a powerful approach to further explore these pathways and understand their relevance in DM.
Author Contributions
All authors of this manuscript qualify for authorship by the following criteria:
Jessica Neely: 1a,b,c, 2 and 3
Dmitry Rychkov: 1a,b,c, 2 and 3
Manish Paranjpe: 1a,c, 2 and 3
Michael Waterfield: 1c, 2 and 3
Susan Kim: 1a,c, 2 and 3
Marina Sirota: 1 a,c, 2 and 3
Supporting information
Acknowledgments
The authors thank the original authors who produced the transcriptomic data utilized in this study and made it publicly available.
This study was funded by the Cure JM Foundation, a CARRA‐Arthritis Foundation grant, and by PREMIER, a National Institutes of Health, National Institute of Arthritis and Musculoskeletal and Skin Diseases, P30 Center for the Advancement of Precision Medicine in Rheumatology at University of California San Francisco (P30AR070155).
No potential conflicts of interest relevant to this article were reported.
Contributor Information
Jessica Neely, Email: Jessica.Neely@ucsf.edu.
Marina Sirota, Email: marina.sirota@ucsf.edu.
References
- 1. Greenberg SA, Pinkus JL, Pinkus GS, Burleson T, Sanoudou D, Tawil R, et al. Interferon‐α/β‐mediated innate immune mechanisms in dermatomyositis. Ann Neurol 2005;57:664–78. [DOI] [PubMed] [Google Scholar]
- 2. Baechler E, Bauer JW, Slattery CA, Ortmann WA, Espe KJ, Novitzke J, et al. An interferon signature in the peripheral blood of dermatomyositis patients is associated with disease activity. Mol Med 2007;13:59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wong D, Kea B, Pesich R, Higgs BW, Zhu W, Brown P, et al. Interferon and biologic signatures in dermatomyositis skin: specificity and heterogeneity across diseases. PLoS One 2012;7:e29161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Greenberg SA, Higgs BW, Morehouse C, Walsh RJ, Kong SW, Brohawn P, et al. Relationship between disease activity and type 1 interferon‐ and other cytokine‐inducible gene expression in blood in dermatomyositis and polymyositis. Genes Immun 2012;13:207–13. [DOI] [PubMed] [Google Scholar]
- 5. Huard C, Gullà SV, Bennett DV, Coyle AJ, Vleugels RA, Greenberg SA. Correlation of cutaneous disease activity with type 1 interferon gene signature and interferon β in dermatomyositis. Br J Dermatol 2017;176:1224–30. [DOI] [PubMed] [Google Scholar]
- 6. Higgs BW, Zhu W, Morehouse C, White WI, Brohawn P, Guo X, et al. A phase 1b clinical trial evaluating sifalimumab, an anti‐IFN‐α monoclonal antibody, shows target neutralisation of a type I IFN signature in blood of dermatomyositis and polymyositis patients. Ann Rheum Dis 2014;73:256–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu Y, Jesus AA, Marrero B, Yang D, Ramsey SE, Montealegre Sanchez GA, et al. Activated STING in a vascular and pulmonary syndrome. N Engl J Med 2014;371:507–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu Y, Ramot Y, Torrelo A, Paller AS, Si N, Babay S, et al. Mutations in proteasome subunit β type 8 cause chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature with evidence of genetic and phenotypic heterogeneity. Arthritis Rheum 2012;64:895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiche L, Jourde‐Chiche N, Whalen E, Presnell S, Gersuk V, et al. Modular transcriptional repertoire analyses of adults with systemic lupus erythematosus reveal distinct type I and type II interferon signatures. Arthritis Rheumatol 2014;66:1583–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moneta GM, Pires Marafon D, Marasco E, Rosina S, Verardo M, Fiorillo C, et al. Muscle expression of type I and type II interferons is increased in juvenile dermatomyositis and related to clinical and histological features. Arthritis Rheumatol 2019;71:1011–21. [DOI] [PubMed] [Google Scholar]
- 11. Afroz S, Giddaluru A, Vishwakarma S, Naz S, Khan AA, Khan N. A comprehensive gene expression meta‐analysis identifies novel immune signatures in rheumatoid arthritis patients. Front Immunol 2017;8:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Friedman MA, Choi D, Planck SR, Rosenbaum JT, Sibley CH. Gene expression pathways across multiple tissues in antineutrophil cytoplasmic antibody‐associated vasculitis reveal core pathways of disease pathology. J Rheumatol 2019;46:609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barrett T, Wihite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 2013;41:D991–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. The R Project for Statistical Computing. URL: http://www.r-project.org/.
- 15. Piccolo SR, Sun Y, Campbell JD, Lenburg ME, Bild AH, Johnson WE. A single‐sample microarray normalization method to facilitate personalized‐medicine workflows. Genomics 2012;100:337–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. Orchestrating high‐throughput genomic analysis with Bioconductor. Nat Methods 2015;12:115–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dai M. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids 2005;33:e175 10.1093/nar/gni179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. LIMMA powers differential expression analyses for RNA‐sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hughey JJ, Butte AJ. Robust meta‐analysis of gene expression using the elastic net. Nucleic Acids Res 2015;43:e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high‐throughput experiments. Bioinformatics 2012;28:882–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001;98:5116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goeman JJ, Solari A. Multiple hypothesis testing in genomics. Stat Med 2014;33:1946–78. [DOI] [PubMed] [Google Scholar]
- 23. Murtagh F, Legendre P. Ward's hierarchical agglomerative clustering method: which algorithms implement ward's criterion? [research article] J Classif 2014;31:274–95. [Google Scholar]
- 24. Yu G, He QY. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst 2016;12:477–9. [DOI] [PubMed] [Google Scholar]
- 25. Fabregat A, Sidiroupoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, et al. The Reactome pathway Knowledgebase. Nucleic Acids Res 2016;44:D481–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang B, Horvath S. A general framework for weighted gene co‐expression network analysis. Stat Appl Genet Mol Biol 2005;4:17. [DOI] [PubMed] [Google Scholar]
- 28. Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? [research support]. PLoS Comput Biol 2011;7:e1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta‐Cepas J, et al. STRING v10: protein‐protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017;18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, et al. Interferome v2. 0: an updated database of annotated interferon‐regulated genes. Nucleic Acids Res 2013;41:D1040–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reed AM, Collins EJ, Shock LP, Klapper DG, Frelinger JA. Diminished class II‐associated Ii peptide binding to the juvenile dermatomyositis HLA‐DQ α 1*0501/DQ β 1*0301 molecule. J Immunol 1997;159:6260–5. [PubMed] [Google Scholar]
- 34. Suárez‐Calvet X, Gallardo E, Nogales‐Gadea G, Querol L, Navas M, Díaz‐Manera J, et al. Altered RIG‐I/DDX58‐mediated innate immunity in dermatomyositis. J. Pathol 2014;233:258–68. [DOI] [PubMed] [Google Scholar]
- 35. Zhu W, Streicher K, Shen N, Higgs BW, Morehouse C, Greenlees L, et al. Genomic signatures characterize leukocyte infiltration in myositis muscles. BMC Med Genomics 2012;5:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hall JC, Casciola‐Rosen L, Berger AE, Kapsogeorgou EK, Cheadle C, Tzioufas AG, et al. Precise probes of type II interferon activity define the origin of interferon signatures in target tissues in rheumatic diseases. Proc Natl Acad Sci USA 2012;109:17609–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nagaraju K, Casciola‐Rosen L, Lundberg I, Rawat R, Cutting S, Thapyial R, et al. Activation of the endoplasmic reticulum stress response in autoimmune myositis: potential role in muscle fiber damage and dysfunction. Arthritis Rheum 2005;52:1824–35. [DOI] [PubMed] [Google Scholar]
- 38. Nagaraju K, Raben N, Loeffler L, Parker T, Rochon PJ, Lee E, et al. Conditional up‐regulation of MHC class I in skeletal muscle leads to self‐sustaining autoimmune myositis and myositis‐specific autoantibodies. Proc Natl Acad Sci USA 2000;97:9209–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ghannam K, Martinez‐Gamboa L, Spengler L, Krause S, Smiljanovic B, Bonin M, et al. Upregulation of immunoproteasome subunits in myositis indicates active inflammation with involvement of antigen presenting cells, CD8 T‐cells and IFNγ. PLoS One 2014;9:e104048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nagaraju K. Immunological capabilities of skeletal muscle cells. Acta Physiol Scand 2001;171:215–23. [DOI] [PubMed] [Google Scholar]
- 41. Casciola‐Rosen L, Nagaraju K, Plotz P, Wang K, Levine S, Gabrielson E, et al. Enhanced autoantigen expression in regenerating muscle cells in idiopathic inflammatory myopathy. J Exp Med 2005;201:591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Darrah E, Rosen A. Granzyme B cleavage of autoantigens in autoimmunity. Cell Death Differ 2010;17:624–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rayavarapu S, Coley W, Van der Meulen JH, Cakir E, Tappeta K, Kinder TB, et al. Activation of the ubiquitin proteasome pathway in a mouse model of inflammatory myopathy: a potential therapeutic target. Arthritis Rheum 2013;65:3248–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


