Abstract
Yam (Dioscorea spp.) is an important food crop cultivated for its edible tubers in Cameroon. Surveys were conducted in Cameroon to determine the incidence and severity of yam mosaic disease and associated viruses in 124 yam farms in four agro-ecological zones in 2014 and 2016. Dioscorea rotundata, D. cayenensis, D. alata, D. Dumetorum and D. bulbifera were most frequently detected yam species in the fields. Symptoms of virus disease were observed on 81.5% of the farms surveyed and the disease incidence ranged from 0 to 96.7%, with an overall mean of 26.5%. Mean symptom severity estimated using a numerical rating scale of 1–5, ranged from 2 to 4.1, with an overall mean of 2.6. Representative set of leaf samples collected from farmers’ fields were tested for three viruses known to cause yam mosaic disease in West Africa, viz., Yam mosaic virus (YMV), Yam mild mosaic virus (YMMV) and Cucumber mosaic virus (CMV), using multiplex RT-PCR. YMV and YMMV were detected in 220 (37.2%) of the 591 samples tested and 75% of the farms surveyed. None of the samples tested positive to CMV. Phylogenetic analysis based on the coat protein sequencing of 27 YMV isolates clustered these isolates into three phylogenetic groups. This study demonstrated high prevalence of mosaic disease in yam fields and YMV as main causal agent. Knowledge generated in this study will be useful to augment diagnostic tools and yam mosaic disease control with a view to improve on yam production in Cameroon.
Electronic supplementary material
The online version of this article (10.1007/s13337-019-00552-3) contains supplementary material, which is available to authorized users.
Keywords: Yam virus disease surveys, Sequence diversity, Viral diseases, Virus diagnostics
Introduction
Yam (Dioscorea spp.) is a multi-species tuber-producing vine belonging to the family Dioscoreaceae. The genus includes about 603 domesticated and wild species distributed in tropical and subtropical areas of the world [5]. This vegetatively propagated crop is an important starchy staple and source of income for millions of people in the yam belt extending from Côte d’Ivoire to Cameroon, where over 67.3 million MT of the world’s estimated 73 million MT of yams are produced annually [9; FAOSTAT, 2017]. Nigeria is the highest producer with about 47.9 million MT, while Cameroon with its annual production of 648,407 MT is ranked 6th within the yam belt and 7th in the world behind Nigeria, Ghana, Côte d’Ivoire, Benin, Ethiopia and Togo [FAOSTAT, 2017]. Based on production quantities, yam is ranked 3rd after cassava and cocoyam/taro in Cameroon [18]. The most economically important yam species in Cameroon are D. rotundata, D. cayenensis, D. alata, and D. dumetorum, which are cultivated in all agro-ecological zones of the country [18, 20]. Yam cultivation in Cameroon has been increasing due to growing demand for this high value tuberous crop, as reflected in the expansion in surface area from 28,038 ha to 57,612 ha, and production from 262,610 MT to 648,407 MT from 2000 to 2017, respectively [FAOSTAT, 2017]. However, gains in productivity were marginal from 9.4 MT/ha in 2000 to 11.3 MT/ha in 2017. Yam production in Cameroon, like in other yam-producing countries in Sub-Saharan Africa [17], is hampered by many biotic constraints.
Diseases caused by viral pathogens are amongst the most significant as they were reported to reduce tuber yields by up to 40% [2, 17, 24]. In the lack of organized seed sector, farmers continue to reuse their saved seeds which are often infected by viruses. Reuse of the same seeds over generations and informal seed exchanges between farmers over the decades have likely contributed to widespread distribution and increase incidence of viral diseases in yam fields. Viruses infecting yams in the yam belt include, Yam mosaic virus (YMV, genus Potyvirus), Yam mild mosaic virus (YMMV, genus Potyvirus), Yam mild mottle virus (YMMV, genus Carmovirus), Cucumber mosaic virus (CMV, genus Cucumovirus), and several species of Dioscorea-infecting badnaviruses [6, 9, 13, 23]. Significant losses in yam tuber yields are inflicted by YMV and CMV infections in Côte d’Ivoire and Nigeria [2, 6, 11, 17].
Previous studies have reported the occurrence of YMV and yam badnaviruses in Cameroon [20, 22]; however, knowledge on their distribution and incidence is limited to the western highlands (Zone III) and the humid forest with monomodal rainfall (Zone IV) agro-ecological zones (AEZs). This study was undertaken with an objective to survey all the yam producing zones to generate information on the extent of virus spread and determine virus diversity, to augment diagnostic tools and control measures through ‘clean seed’ interventions in Cameroon.
Materials and methods
Survey area
Surveys were conducted in two seasons, in October 2014 and July 2016, in four of the five AEZs based on their importance in yam production: High Guinea Savannah (Adamawa Region) or Zone II; Western Highlands (West and North-West Regions) or Zone III; Humid Forest with monomodal rainfall (South-West and Littoral Regions) or Zone IV; and Humid forest with bimodal rainfall (South, East and Centre Regions) or Zone V (Figs. 1, 2). Surveys were not conducted in Sudano-Sahelian (or Zone I) as yam cultivation is limited or non-existent in this AEZ. A total of 124 farms were surveyed; 48 farms (12 per AEZ) in 2014 and 76 farms (19 per AEZ) in 2016 (Table 1; Figs. 1, 2). Geographic coordinates were recorded at each farm location.
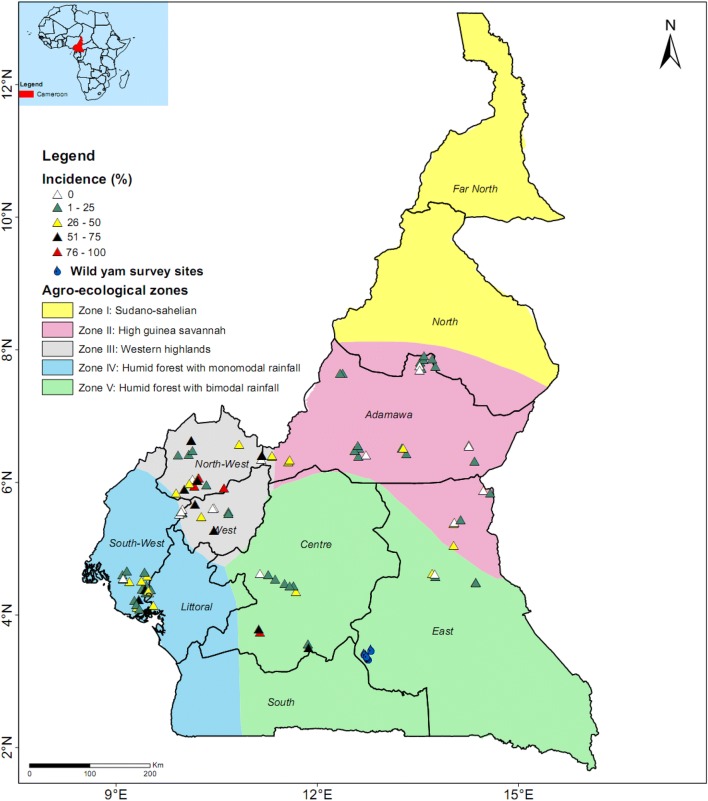
Fig. 1.
Percent virus disease incidence in the surveyed yam farms in Cameroon
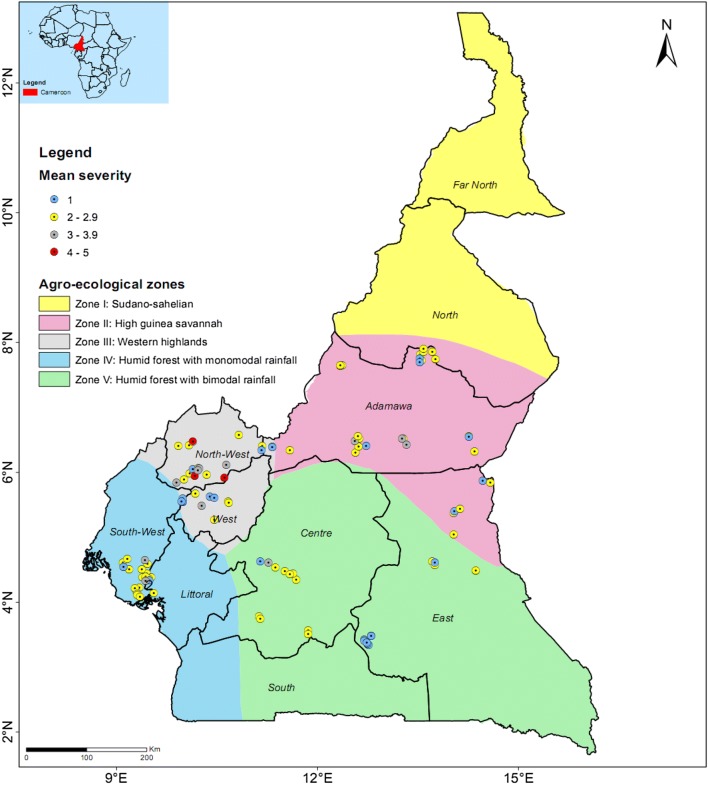
Fig. 2.
Mean virus disease severity assessed based on 1–5 rating scale in the order of increasing symptom severity in the surveyed yam fields in Cameroon
Table 1.
Yam viral disease incidence and severity across four agro-ecological zones in Cameroon in 2014 and 2016
| Agro-ecological zones | Farms surveyed | Mean incidence (%) | Mean severity (1–5 scale) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2014 | 2016 | Total | Range | 2014 | 2016 | Overall | Range | 2014 | 2016 | Overall | |
|
Zone II: high guinea savanna |
12 | 19 | 31 | 0–56.7 | 30.0b | 3.0b | 13.4c | 2–3.0 | 2.5ab | 2.4b | 2.4b |
|
Zone III: western highlands |
12 | 19 | 31 | 0–90.0 | 54.4a | 32.8a | 41.2a | 2–4.1 | 2.6a | 3.0a | 2.8a |
|
Zone IV: humid forest with monomodal rainfall |
12 | 19 | 31 | 0–70.0 | 36.7ab | 19.5a | 26.1b | 2–3.1 | 2.3b | 2.6b | 2.5b |
|
Zone V: humid forest with bimodal rainfall |
12 | 19 | 31 | 0–96.7 | 33.3b | 20.2a | 25.3bc | 2–3.0 | 2.5ab | 2.4b | 2.5b |
| F | – | – | 2.5 | 6.1 | 6.8 | – | 1.4 | 4.2 | 4.5 | ||
| Total | 48 | 76 | 124 | – | – | – | – | – | – | – | – |
Means followed by different letters within each column are significantly different from each other (Fisher’s LSD test, p < 0.05)
Sampling protocol and data recording
In each yam farm, 30 plants were examined by observing along the ‘X’ transects [25], with 15 plants per diagonal axis. Each plant was assessed for symptom type, and severity, and percent viral disease incidence per field was determined. Viral disease severity was scored using a numerical disease rating scale of 1–5, where: 1 = no visible symptoms; 2 = mild mosaic/mottling on most leaves but no leaf distortion; 3 = mild to moderate mosaic on most leaves, leaf narrowing and leaf distortion on about 1/3 of the plant (mostly in lower portion); 4 = severe mosaic on most leaves in about 2/3 of the plant, leaf distortion and mild to moderate stunting; 5 = severe mosaic, severe leaf distortion and severe stunting [28]. Depending on symptom types, up to 6 leaf samples per field were collected for virus testing. Where symptomatic plants were absent, non-symptomatic leaves were collected. The leaf samples were shipped to laboratory for virus testing.
Detection of viruses on leaf samples
All leaves were tested for YMV, YMMV and CMV using the multiplex-reverse transcription polymerase chain reaction (RT-PCR). Total nucleic acid was extracted from 100 mg of yam leaf tissue from each sample, using modified Cetyltrimethyl Ammonium Bromide (CTAB) method as previously reported by Abarshi et al. [1]. Multiplex RT-PCR was conducted using 100 ng/µl total nucleic acid in 12.5 µl reaction mixture, using a thermal cycler (SeeAmp Thermal Cycler, Seegene South Korea). The reaction mixture contained 2.5 µl of reaction buffer, 1.5 mM MgCl2, 0.3 U of Taq DNA polymerase, 12 U of M-MLV (Promega, USA), 0.2 mM dNTPs mix (New England Biolabs, USA), 0.1 µM of each CMV, 0.2 µM of each YMV, 0.36 µM of each YMMV oligonucleotide (IDT, Belgium) and 2 µl of diluted 1:50 (v/v) (100 ng/µl) total nucleic acid extract. Oligonucleotides used for nucleic acid amplification were YMMV-F: GGCACACATGCAAATGAATGC and YMMV-R: CACCAGTAGAGTGAACATAG for YMMV; CMV-F: GCCGTAAGCTGGATGGACAA and IITA CMV-R: CCGCTTGTGCGTTTAATGGCT for CMV; and YMV-F3x: GACAATGATGGACGGTGCGG and YMVB3x: GTTTGCCATCAAATCCAAACAT for YMV. The thermal cycling condition consisted of reverse transcriptase (RT) phase at 42 °C for 30 min, followed by one cycle of initial denaturation at 94 °C for 1 min, annealing at 54 °C for 2 min and extension at 72 °C for 3 min; 35 cycles at 94 °C for 1 min, 54 °C for 2 min, 72 °C for 1 min; and final extension at 72 °C for 5 min. PCR product was resolved in 2% agarose gel, pre-stained with EZ-Vision Bluelight DNA Dye (VWR, USA) and visualized using GelDoc (Biorad, USA). Samples with RT-PCR amplicons showing expected band sizes of 241, 330 and 520 bp, indicated positive results for YMMV, YMV and CMV, respectively (data not shown).
Sequencing and phylogenetic analyses of YMV and YMMV isolates
Viral nucleic acid extracts of 27 samples that tested positive to YMV and 2 samples positive to YMMV were selected for sequencing of full coat protein (CP) encoding segment (Supplementary Table 1). These samples represent wide area coverage and symptom types. RT-PCR reaction was performed in 50 µl reaction mixture as detailed before, except for using oligonucleotide primer pairs: YMV CP Bam-FP: AGAGGATCCGCAGATACACAGCCAGAT and YMV CP Eco-RP: ATCGAATTCCTACATACCTCTCATGCCCA, to amplify a 906 bp fragment corresponding to the CP encoding sequence of YMV; and the YMMV CP Bam-FP: CAGAGAGGATCCGCAAGTAAGGAACA and YMMV CP Eco-RP: TTGATCGAATTCCTAGATATTGCGCAC to amplify a 803 bp fragment corresponding to the CP of YMMV. RT-PCR was performed using thermal cycling conditions described earlier, and the amplified products were purified by ethanol precipitation and the products were used for sequencing in both orientations using amplicon-specific primers, at The Iowa State University DNA Facility (Iowa, USA). All the sequences were analyzed and edited manually using BioEdit Software version 7.2.1. The nucleotide sequences were compared with previously published YMV and YMMV CP sequences from Cameroon and other West African countries available in the NCBI GenBank, using the Clustal-W method [8], and phylogenetic relationships were reconstructed using the maximum likelihood (ML) method based on the Tamura–Nei model [26], conducted on MEGA7 [15]. Pairwise genetic distance between sequences was calculated using the maximum composite likelihood (MCL) approach.
Data analysis
Analysis of variance (ANOVA) and post hoc tests were conducted on the data with R statistical computing software, using the statistical software package agricolae [12]. Means were compared using the Fisher’s Least Significant Difference (LSD) test, at 95% confidence interval. Pearson’s product-moment correlation test was used to determine interrelationships among variables (between yam viral disease incidence and severity, viral disease incidence and altitude of sampling location, and viral disease severity and altitude of the sampling location), using the R statistical computing software package ggpubr [14].
Results
Prevalence, incidence and severity of yam viral diseases
In the 124 yam fields surveyed, D. rotundata was the most frequently encountered yam species followed by D. cayenensis, D. alata, D. dumetorum and D. bulbifera in both surveys (Table 2). Dioscorea cayenensis was the most frequently planted species in Zone III, whilst D. alata, D. cayenensis and D. rotundata were observed in all the four agro-ecological zones (Table 2). Virus disease symptoms were recorded in 46 (95.9%) of the 48 farms surveyed in 2014, and 55 (72.4%) of the 76 farms surveyed in 2016, with an overall prevalence of 81.5% for two years. A total of 3720 plants (1440 in 2014 and 2280 in 2016) were examined for symptoms. Virus disease incidence in decreasing order was recorded on D. cayenensis, D. rotundata, D. alata, D. bulbifera and D. dumetorum (Table 2). During the surveys a total of 591 leaf samples (287 in 2014 and 304 in 2016) were collected for virus testing. Eight symptom types were observed, including mosaic, leaf distortion, mottling, leaf bleaching, vein banding, shoe-stringing, stunting and leaf curling (Fig. 3, Table 2). The eight symptom types were observed more frequently on D. rotundata and D. cayenensis, followed by D. alata (Table 2). Only mosaic and leaf distortion were observed on both D. dumetorum and D. bulbifera.
Table 2.
Viral disease incidence and severity scores recorded in four agro-ecological zones and five yam species, during two surveys conducted in Cameroon in 2014 and 2016
| Agro-ecological zone | Yam species | Plants examined | Mean incidence (%) | Mean severity | Associated symptoms |
|---|---|---|---|---|---|
|
Zone II: high guinea savannah |
D. alata | 41 | 9.4 | 3 | M, Lb, Ld, Sh, St |
| D. bulbifera | 5 | 0 | 0 | – | |
| D. cayenensis | 24 | 41.2 | 2.9 | M, Mo, Ld, Sh, St | |
| D. dumetorum | 1 | 0 | 0 | – | |
| D. rotundata | 859 | 15.5 | 2.4 | M, Mo, Cu, Lb, Ld, Sh, St, Vb | |
| Total | 930 | 16.5 | 2.5 | ||
|
Zone III: western highlands |
D. alata | 62 | 3.8 | 2 | M, Mo, Ld, Sh |
| D. bulbifera | 11 | 4.6 | 3 | M, Ld | |
| D. cayenensis | 541 | 65.1 | 3 | M, Mo, Cu, Lb, Ld, Sh, Vb | |
| D. dumetorum | 112 | 3.8 | 2 | M, Ld, Ln | |
| D. rotundata | 204 | 32.2 | 2.5 | M, Mo, Cu, Lb, Ld, Sh, St, Vb | |
| Total | 930 | 43.6 | 3 | ||
| Zone IV: humid forest with monomodal rainfall | D. alata | 114 | 13.6 | 2.1 | M, Mo, Ld, St |
| D. bulbifera | 0 | 0 | 0 | – | |
| D. cayenensis | 9 | 50.0 | 3.3 | M, Mo, Ld, St | |
| D. dumetorum | 172 | 5.0 | 2.3 | M, Ld, St | |
| D. rotundata | 635 | 37.8 | 2.6 | M, Mo, Lb, Ld, Sh, St | |
| Total | 930 | 28.2 | 2.6 | ||
| Zone V: humid forest with bimodal rainfall | D. alata | 103 | 16.1 | 2.9 | M, Mo, Ld |
| D. bulbifera | 0 | 0 | 0 | – | |
| D. cayenensis | 292 | 53.4 | 2.7 | M, Mo, Cu, Lb, Ld, Sh, Vb | |
| D. dumetorum | 23 | 00 | 0 | Ld | |
| D. rotundata | 512 | 18.3 | 2.4 | M, Mo, Cu, Lb, Ld, Sh, Vb | |
| Total | 930 | 26.8 | 2.6 | ||
| Totals by species | D. alata | 320 | 11.2 | 2.4 | M, Mo, Lb, Ld, St |
| D. bulbifera | 16 | 4.6 | 3 | M, Ld | |
| D. cayenensis | 866 | 61.1 | 2.9 | M, Mo, Cu, Lb, Ld, Sh, St, Vb | |
| D. dumetorum | 308 | 3.6 | 2.2 | M, Ld, St | |
| D. rotundata | 2210 | 23.7 | 2.5 | M, Mo, Cu, Lb, Ld, Sh, St, Vb | |
| Overall | 3720 | 28.8 | 2.7 | ||
Yam viral disease symptoms: M = Mosaic, Mo = Mottling, Cu = Curling, Lb = Leaf bleaching, Ld = Leaf distortion, Sh = Shoe-stringing, St = Stunting, Vb = Vein-banding
Fig. 3.
Variety of viral disease symptoms observed on naturally infected yams in farmers’ fields in Cameroon. a Uninfected, b Stunting, mosaic and distortion, c Severe mosaic, d Shoe-stringing with mosaic, e Bleaching, f Leaf distortion, g Green-vein banding, h Mosaic with bleaching, and i Mild mottling
Virus disease incidence ranged from 0 to 96.7%, with an overall mean of 26.5% (Table 1). Of the 124 farms surveyed, virus disease incidence was 1–25% in 49 farms, 26–50% in 27 farms, 51–75% in 16 farms and > 75% in 9 farms (Fig. 1). Viral disease incidence was least in D. dumetorum (3.6%) and highest in D. cayenensis (61.1%) with a mean incidence of 28.8%, whilst mean viral disease severity was least in D. dumetorum (2.2) and highest in D. bulbifera (3.0) with and overall mean of 2.7 (Table 2). Average severity per farm ranged from 2 to 4.1, with an overall mean of 2.6 (Table 1). Average severity scores of ≥ 4 were recorded in four farms (Weh, Bambui, Bamenda upstation and Akum) on D. cayenensis, all of which are in Zone III (Fig. 2). Mean viral disease severity per AEZ was highest (p < 0.05) in Zone III (2.8), compared with other zones with 2.4–2.5 (Table 1). The disease incidence was highest (p < 0.05) in Zone III (41.2%), compared with Zone II (13.4%), Zone IV (26.1%), and Zone V (25.3%) (Table 1). Viral disease incidence scores of 50% or higher were recorded only on D. cayenensis, in Zones III (65.1%), IV (50%) and V (53.4%) in both surveys indicating that D. cayenensis is most susceptible of the five yam species observed in farms in Cameroon (Table 2).
Pearson’s product-moment correlation tests showed a positive correlation between yam viral disease incidence and severity (r = 0.57, p < 0.001), a weak but marginally not significant correlation between viral disease incidence and altitude (r = 0.16, p = 0.08) and no significant correlation between viral disease severity and altitude (r = 0.002, p = 0.99) (Table 3).
Table 3.
Correlation analysis of yam viral disease incidence, severity and altitude of sampling locations for 124 farms assessed in Cameroon
| Parameter 1 | Parameter 2 | Correlation coefficient (r) | p-value | Level of significance |
|---|---|---|---|---|
| Incidence | Altitude | 0.16 | 0.08 | ns |
| Severity | Altitude | 0.002 | 0.99 | ns |
| Incidence | Severity | 0.57 | 3.7e− 12 | ** |
ns no significant correlation
**Significant correlation at p < 01
Detection of YMV, YMMV and CMV
Of the 591 samples tested, 220 tested positive to YMV or YMMV. In total, 199 of these virus-positive samples showed symptoms and 21 were asymptomatic. YMV was detected in 174 symptomatic and 15 asymptomatic samples, YMMV was detected in 17 symptomatic and 6 asymptomatic samples, and YMV + YMMV was detected in 8 samples, all of which were symptomatic (Table 4). CMV was not detected in any of the samples. YMV and YMMV were detected in all the four agro-ecological zones, while mixed infection of the two viruses occurred only in Zone V (Table 5). In leaf samples from Zone II, III, IV and V, viruses were detected in 32.9, 37.3, 39.6 and 39.2%, respectively. YMV was detected in 29.5, 35.2, 35.6 and 27.7% while YMMV was detected in 3.4, 2.1, 4.0 and 6.1% of the samples from Zone II, III, IV and V, respectively (Table 5). Viruses were detected in 93 (75%) of the 124 farms sampled for testing (Table 5). Based on viruses tested, viral disease prevalence in decreasing order was 83.9% in Zone IV, 77.4% in Zone V, 74.2% in Zone III and 64.5% in Zone II. YMV and YMMV were detected in 88 (70.1%) and 20 (16.1%) farms, respectively. YMV and YMMV were detected in D. alata, D. cayenensis, D. dumetorum and D. rotundata, but not in D. bulbifera (Table 5). YMV, YMMV and mixed infection of both viruses were frequently associated with samples having mosaic symptoms (Table 4). In the leaves sampled from D. cayenensis, viruses were detected in 62.5, 48.9, 75 and 58.4% in Zone II, III, IV and V, respectively (Table 5).
Table 4.
Distribution of 220 virus-positive leaf samples from major yam-growing areas in Cameroon according to symptom types and three viruses tested in 2014 and 2016
| Symptoms associated with leaf samples | Virus-positive samples per symptom type | ||||
|---|---|---|---|---|---|
| YMV | YMMV | YMV + YMMV | CMV | aTotal | |
| Curling (Cu) | 8 | 1 | 0 | 0 | 9 |
| Leaf bleaching (Lb) | 3 | 0 | 0 | 0 | 3 |
| Leaf distortion (Ld) | 6 | 4 | 1 | 0 | 11 |
| Mosaic (M) | 146 | 8 | 7 | 0 | 161 |
| Mottling (Mo) | 5 | 3 | 0 | 0 | 8 |
| Shoe-stringing (Sh) | 5 | 1 | 0 | 0 | 6 |
| Vein-banding (Vb) | 1 | 0 | 0 | 0 | 1 |
| Total | 174 | 17 | 8 | 0 | 199 |
| Asymptomatic samples | 15 | 6 | 0 | 0 | 21 |
| Overall total | 189 | 23 | 8 | 0 | 220 |
aTotal number of virus-positives samples per symptom type
Table 5.
Distribution of three viruses detected in cultivated yams within four agro-ecological zones in Cameroon
| Agro-ecological zone | Yam species | Farms sampleda | Samples tested | Viruses detected (%) | ||||
|---|---|---|---|---|---|---|---|---|
| YMV | YMMV | CMV | YMV + YMMV | Total | ||||
|
Zone II: high guinea savannah |
D. alata | 8 | 25 | 12.5 | 0 | 0 | 37.5 | |
| D. cayenensis | 8 | 50 | 12.5 | 0 | 0 | 62.5 | ||
| D. dumetorum | 1 | 0 | 0 | 0 | 0 | 0 | ||
| D. rotundata | 132 | 28.8 | 2.3 | 0 | 0 | 31.1 | ||
| Total | 31(64.5) | 149 | 29.5 | 3.4 | 0 | 0 | 32.9 | |
| Zone III: western highlands | D. alata | 10 | 20 | 20 | 0 | 0 | 40 | |
| D. bulbifera | 4 | 0 | 0 | 0 | 0 | 0 | ||
| D. cayenensis | 88 | 47.7 | 1.1 | 0 | 0 | 48.9 | ||
| D. dumetorum | 18 | 0 | 0 | 0 | 0 | 0 | ||
| D. rotundata | 25 | 28 | 0 | 0 | 0 | 28 | ||
| Total | 31(74.2) | 145 | 35.2 | 2.1 | 0 | 0 | 37.3 | |
|
Zone IV: humid forest with monomodal rainfall |
D. alata | 17 | 11.8 | 0 | 0 | 0 | 11.8 | |
| D. bulbifera | 1 | 0 | 0 | 0 | 0 | 0 | ||
| D. cayenensis | 4 | 75 | 0 | 0 | 0 | 75 | ||
| D. dumetorum | 34 | 8.8 | 17.8 | 0 | 0 | 26.5 | ||
| D. rotundata | 93 | 48.4 | 0 | 0 | 0 | 48.4 | ||
| Total | 31(83.9) | 149 | 35.6 | 4.0 | 0 | 0 | 39.6 | |
|
Zone V: humid forest with bimodal rainfall |
D. alata | 22 | 27.3 | 13.6 | 0 | 0 | 40.9 | |
| D. cayenensis | 48 | 41.7 | 4.2 | 0 | 12.5 | 58.4 | ||
| D. dumetorum | 4 | 50 | 0 | 0 | 0 | 50 | ||
| D. rotundata | 74 | 17.6 | 5.4 | 0 | 2.7 | 25.7 | ||
| Total | 31(77.4) | 148 | 27.7 | 6.1 | 0 | 5.4 | 39.2 | |
| Total | 124(75) | 591 | 31.9 | 3.9 | 0 | 1.4 | 37.2 | |
aNumber of farms sampled (percentage of virus-positive farms given in parenthesis)
Molecular diversity of YMV and YMMV isolates detected in this study
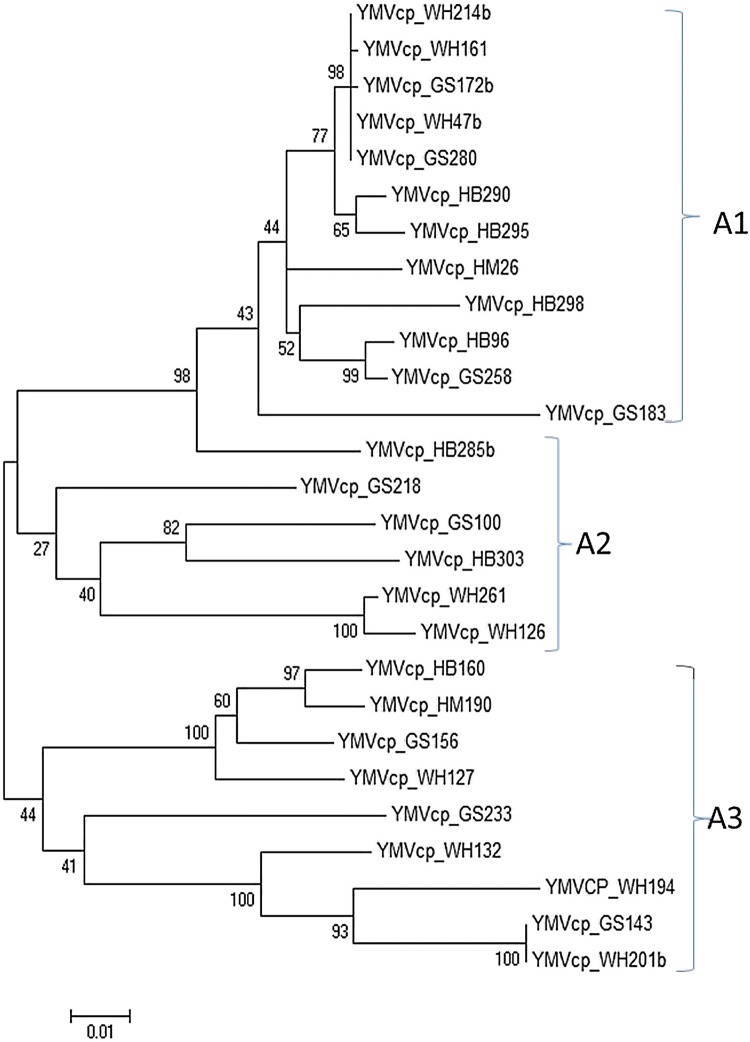
Phylogenetic analysis and pairwise comparison grouped the 27 YMV isolates sequenced in this study into three major clusters (A1, A2 and A3) as supported by low bootstrap values (< 50%; Fig. 4). However, clustering was not representative of any agro-ecological zone or yam species suggesting wide distribution of various YMV isolates due to inter-zonal movement of planting material and likely spread of viruses between plants by aphid vectors. The 13 YMV isolates in Cluster A1 were from zone II (4 isolates), III (3 isolates), IV (1 isolate) and V (5 isolates) and originated from 10 landraces of D. cayenensis and D. rotundata (Fig. 4, Supplementary Table 1). The five isolates in cluster A2 were identifiable with zone II (1 isolate), III (3 isolates) and V (1 isolate) and were isolated from D. cayenensis, D. rotundata and D. schimperiana. Also, the nine isolates in cluster A3 were identifiable with zone II (3 isolates), III (4 isolates), IV (1 isolate) and V (1 isolate) and were isolated from several landraces of D. cayenensis, D. rotundata and D. esculenta.
Fig. 4.
Unrooted phylogenetic tree constructed using the maximum-likelihood method with the aid of MEGA 7 software. The phylogenetic tree was constructed using the complete coat protein encoding gene of 27 YMV isolates detected in this study in Cameroon. Bootstrap values (1000 replicates) are shown at the branch nodes and branches corresponding to partitions reproduced in < 50% of bootstrap replicates are collapsed. The three groupings are indicated
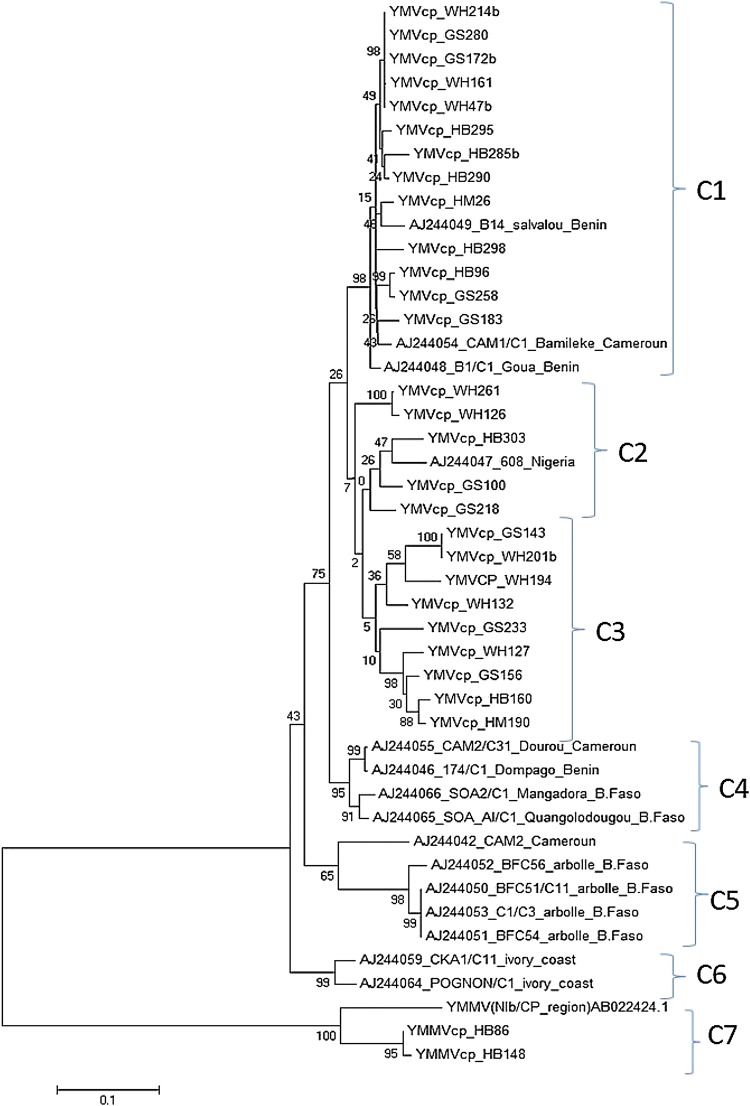
The 27 YMV and 2 YMMV isolates sequenced in this study were compared with 16 sequences retrieved from the NCBI GenBank database representative of Cameroon, and other West African countries. Phylogenetic analyses partitioned all 45 isolates into seven clusters (C1, C2, C3, C4, C5, C6 and C7) (Fig. 5). The topology of the phylogenetic tree did not change significantly, as the 27 YMV and 2 YMMV isolates detected in this study, still occurred in four clusters (C1, C2, C3 and C7). Of the isolates from the NCBI GenBank database, only AJ244049_Benin, AJ244054_Cameroon, and AJ244048_Benin occurred in cluster C1; AJ244047_Nigeria in cluster C2; and YMMV (Nib/CP_region) AB024424.1 in cluster C7 in association with virus isolates detected in this study (Fig. 5). Cluster C3 was exclusively constituted of YMV isolates detected in this study. The remaining 12 virus isolates from the NCBI GenBank separated from isolates detected in this study and were partitioned into three clusters: C4 (AJ244055_Cameroon, AJ244046_Benin, AJ244066 and AJ24406 from Burkina Faso); C5 (AJ244052, AJ244050, AJ244053 and AJ244051 from Burkina Faso); and C6 (AJ2044059_Côte d’Ivoire) (Fig. 5).
Fig. 5.
Rooted phylogenetic tree depicting evolutionary relationships among Yam mosaic virus (YMV) and Yam mild mosaic virus (YMMV) isolates characterized in Cameroon. The phylogenetic tree was constructed using the complete coat protein encoding gene of 27 YMV isolates, 2 YMMV isolates and 16 YMV isolates from NCBI GenBank and the tree was rooted using YMMV as out-group. The tree was constructed using the MEGA7 [20]. Bootstrap values (1000 replicates) are shown at the branch nodes and branches corresponding to partitions reproduced in < 50% of bootstrap replicates are collapsed
Discussion
Occurrence of YMV in Cameroon was reported previously [20], but YMMV detected in this study is the first report in Cameroon. The occurrence of these two viruses in all countries of the yam belt can be attributed to exchange of infected yam germplasm and transmission by aphid vectors [9]. CMV was not detected in yam, although its occurrence in some weed and vegetable hosts has been reported in Sub-Saharan Africa [10, 28]. YMV and YMMV were detected in all four agro-ecological zones, covering eight of the ten regions of Cameroon. A very high prevalence of YMV and YMMV was observed, confirming previous reports that these viruses are the most frequent potyviruses on yams in sub-Saharan Africa [10, 16]. Symptoms observed during the surveys in all agro-ecological zones included mosaic, leaf distortion, mottling, stunting, shoe-stringing and leaf bleaching which have been previously reported in Benin, Cameroon, Côte d’Ivoire, Ghana and Nigeria [19, 28]. This indicates that virus infection symptom development on yams is independent of agro-ecology in sub-Saharan Africa.
Viral disease incidence and severity were high across agro-ecological zones and yam species, with the highest incidence in Zone III, dominated by the cultivation of D. cayenensis landraces, which are probably more susceptible to yam viral diseases compared with other yam species encountered in the study. The lowest viral disease incidence occurred in Zone II, although the lowest severity did not occur in that agro-ecological zone. This observation suggests that perhaps the variety of D. rotundata called Bakokae which dominates in Zone II is more tolerant to virus infection, as previous reports have shown that some varieties of this yam species are resistant to viral infections [21]. This observation and the fact that both YMV and YMMV were detected in leaf samples without symptoms, further indicate that some yams in Cameroon are tolerant to virus infections. Extensive screening is needed to identity varieties that are resistant to viral infections in Cameroon, to provide genetic resources for yam breeding, to develop improved varieties and promote production.
None of the three viruses tested in the study was detected in some symptomatic leaves, indicating that the symptoms might have been due to other viruses not tested or other biotic and abiotic factors. Further characterization of such samples by deep sequencing of small RNAs is required to identify the occurrence of any other viruses with the samples. Moreover, some asymptomatic leaves tested positive for both viruses, indicating latent infection. Based on the foregoing we concluded that the absence or presence of symptoms alone cannot be used to accurately predict the occurrence of viral diseases on yams.
There was a significant positive correlation between viral disease incidence and severity, but there was absence of correlation between altitude and viral disease incidence or severity. This indicates that altitude does not have any influence on the incidence and severity of yam viral diseases in Cameroon.
The CP encoding gene sequence of 27 YMV isolates did correlate neither with geographical areas of origin nor their yam host species, as earlier reported [3], as isolates from different agro-ecological zones and yam host species clustered together in the same groups. Except for cluster A2 which contained isolates from Zones II, III, V, the other two clusters (A1 and A3) contained YMV isolates from all the four agro-ecological zones. Furthermore, except for one YMV isolate from D. esculenta and another from D. schimperiana, all other isolates were from the D. cayenensis/rotundata complex. This indicates that the D. cayenensis/rotundata complex is infected by several YMV strains as reported in previous studies conducted in Africa [4]. The high genetic variability amongst the YMV isolates in Cameroon may be due to intra-species recombination events [7], mutation caused by virus-yam host interaction, and virus transmission by insect vectors, particularly aphids [4]. The occurrence in the same cluster, of virus isolates from different agro-ecological zones and different landraces of the D. cayenensis/rotundata complex may be associated with local exchange of infected planting material amongst the different major yam production areas of the country.
The 15 YMV isolates retrieved from the NCBI GenBank database corresponded to the six African phylogenetic groups (I, II, III, IV, VII and IX) previously reported [4, 7]. When compared with these 15 isolates, phylogenetic analysis revealed that some of the YMV isolates detected in this study are associated to phylogenetic groups III and VII of the six groups previously reported in Africa [7]. Amongst the NCBI isolates two from Benin and one from Cameroon, all being members of phylogenetic group III clustered together with some isolates of this study in group C1. Also, the only member of Phylogenetic group VII, isolated from Nigeria clustered together with some YMV isolates of this study in group C2. This is an indication that YMV isolates of Cluster C1 and C2 are members of phylogenetic groups III and VII, reported in previous studies [7]. Nine YMV isolates formed a unique group together in cluster C3, suggesting a possible new phylogenetic group which may be associated with isolates from other parts of the world, that were not used in this study.
Further studies are needed to continue the improvement of knowledge on yam viral disease ecology and phylogenetic structure of viruses infecting yams in Cameroon. While our study was restricted to Cameroon, the findings could be relevant in all other countries in Central Africa and the Congo Basin, considering that the climate and geography of Cameroon, with its tropical, equatorial and mountain climates, is representative of much of Central Africa and the Congo basin [27].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank yam farmers for their cooperation and permission to survey their farms. We acknowledge the CGIAR Research Program on Roots, Tubers and Bananas (CRP-RTB) for financial support to this research work and PhD research fellowship granted to the first author. We also thank Mr. Samuel Nanga Nanga of IITA - Cameroon for preparing geographic maps.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Abarshi MM, Mohammed IU, Jeremiah SC, Legg JP, Kumar PL, Hillocks RJ, Maruthi MN. Multiplex RT-PCR assays for the detection of both RNA and DNA viruses infecting cassava and the common occurrence of mixed infections by two cassava streak brown streak viruses in east Africa. J Virol Methods. 2012;179:176–184. doi: 10.1016/j.jviromet.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 2.Adeniji MO, Shoyinka SA, Ikotun T, Asiedu R, Hughes Jd’A, Odu BO. Yield loss in guinea yam (Dioscorea rotundata Poir.) due to infection by Yam mosaic virus (YMV) genus potyvirus. Ife J Sci. 2012;14(2):237–244. [Google Scholar]
- 3.Aleman-Verdageur ME, Goudou-Urbino C, Dubern J, Beachy RN, Fauquet C. Analysis of the sequence diversity of the P1, HC, P3, Nib, and CP genomic regions of several yam mosaic potyvirus isolates: implications for the intra-species molecular diversity of potyviruses. J Gen Virol. 1997;78:1253–1264. doi: 10.1099/0022-1317-78-6-1253. [DOI] [PubMed] [Google Scholar]
- 4.Ayisah KD, Gumedzoe YMD. Genetic diversity among Yam mosaic virus (YMV) isolates infecting yam of the complex Dioscorea cayenensis-rotundata in Togo. Int J Biol Chem Sci. 2012;6(3):1090–1101. [Google Scholar]
- 5.Bhattacharjee R, Gedil M, Sartie A, Otoo E, Dumet D, Kikuno H, Kumar PL, Asiedu R. Dioscorea. In: Kole C, editor. Wild crop relatives—genomic and breeding resources, industrial crops. Berlin: Springer; 2011. pp. 71–96. [Google Scholar]
- 6.Bömer M, Turaki A, Silva G, Kumar PL, Seal SE. A sequence-independent strategy for amplification and characterization of episomal badnavirus sequences reveals three previously uncharacterized yam badnaviruses. Viruses. 2016;8:188. doi: 10.3390/v8070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bousalem M, Douzery EJP, Fargette D. High genetic diversity, distant phylogenetic relationships and intra-species recombination events among natural populations of Yam mosaic virus: a contribution to understanding potyvirus evolution. J Gen Virol. 2000;81:243–255. doi: 10.1099/0022-1317-81-1-243. [DOI] [PubMed] [Google Scholar]
- 8.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31(13):3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eni AO, Hughes Jd’A , Asiedu R, Rey MEC. Incidence and diversity of mixed viruses lower in yam tuber sprouts compared with field leaf samples: implications for virus-free planting material control strategy. Afr J Agric Res. 2013;8(23):3060–3067. doi: 10.5897/AJAR2013.6770. [DOI] [Google Scholar]
- 10.Eni AO, Kumar PL, Asiedu R, Alabi OJ, Naidu RA, Hughes Jd’A, Rey MEC. Characterization of cucumber mosaic virus isolated from yam (Dioscorea spp.) in West Africa. Afr J Biotechnol. 2013;12(22):3472–3480. doi: 10.5897/AJB2013.12303. [DOI] [Google Scholar]
- 11.Fauquet C, Thouvenel G C. In: Plant viral disease in the Ivory Coast. Documentation technique No 46, Edition de l’Ostrom, Paris. 1987. pp. 29.
- 12.Felipe M. agricolae: Statistical procedure for agricultural research. R package version 1.2-8; 2017. http://CRAN.R-project.org/package=agricolae.
- 13.Hughes Jd’A, Dongo L, Atiri GI. Viruses infecting cultivated yams (Dioscorea alata and Dioscorea rotundata) in Nigeria. Phytopathology. 1997;87:545. doi: 10.1094/PHYTO.1997.87.5.542. [DOI] [Google Scholar]
- 14.Kassambara A. ggpubr: ‘ggplot2’ Based Publication Ready Plots. R package version 1.2-8; 2018. https://CRAN.R-project.org/package=ggpubr.
- 15.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mignouna HD, Njukeng AP, Abang MM, Asiedu R. Inheritance of resistance to Yam mosaic virus, genus Potyvirus, in white yam (Dioscorea rotundata) Theor Appl Genet. 2001;103:1196–2000. doi: 10.1007/s001220100728. [DOI] [Google Scholar]
- 17.Mignouna D, Kumar PL, Coyne D, Bandyopadhyay R, Ortega-Beltran A, Bhattacharjee R, De Koeyer D. Identifying and managing plant health risks for key African crops: yam, taro and cocoyam. In: Neuenschwander P, Tamò M, editors. Critical issues in plant health: 50 years of research in African agriculture. Cambridge: Burleigh Dodds Science Publishing; 2019. p. 429. [Google Scholar]
- 18.Ngo-Ngwe MFS, Omokolo DN, Joly S. Evolution and phylogenetic diversity of yam species (Dioscorea spp.): implications for conservation and agricultural practices. PLOS One. 2015;10(12):e0145364. doi: 10.1371/journal.pone.0145364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Njukeng A P, Hughes J d’A, Atiri G I, Akpo E J A. Distribution of Yam mosaic virus in Nigeria. In: VIIIth International Plant Virus Epidemiology Symposium, Aschersleben, 2003; 12–17 May 2002.
- 20.Njukeng AP, Azeteh IN, Mbong GA. Survey of the incidence and distribution of two viruses infecting yam (Dioscorea spp.) in two agro-ecological zones in Cameroon. Int J Current Microbiol Appl Sci. 2014;3(4):1153–1166. [Google Scholar]
- 21.Odu BO, Asiedu R, Shoyinka SA, Hughes Jd’A. Analysis of resistance to Yam mosaic virus, genus Potyvirus in white Guinea yam (Dioscorea rotundata Poir) genotypes. J Agric Sci. 2011;56(1):1–13. [Google Scholar]
- 22.Offei SK. Virus and viral diseases of sub-Saharan Africa: analysis of responses to questionnaire by scientists in sub-Saharan Africa. In: Hughes Jd’A, Odu BO., editors. Proceedings of a conference organized by IITA. 2003. pp. 128–137. [Google Scholar]
- 23.Seal SE, Muller E. Molecular analysis of full-length sequence of a new yam badnavirus from Dioscorea sansibarensis. Adv Virol. 2007;152:809–815. doi: 10.1007/s00705-006-0888-7. [DOI] [PubMed] [Google Scholar]
- 24.Séka K, Etchian AO, Assiri PK, Toualy MNY, Diallo HA, Kouassi NK, Aké S. Yield loss caused by Yam mosaic virus (YMV) and Cucumber mosaic virus (CMV) on the varieties of Dioscorea spp. Int J Agron Agric Res. 2014;5(2):64–71. [Google Scholar]
- 25.Sseruwagi P, Sserubombue WS, Legg JP, Ndunguru J, Thresh JM. Methods of surveying the incidence and severity of cassava mosaic disease and whitefly vector populations on cassava in Africa: a review. Virus Res. 2004;100:129–142. doi: 10.1016/j.virusres.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 26.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 27.Tsalefac M, HiolHiol F, Mahe G, Laraque A, Sonwa DJ, Scholte P, Pokam W, Haensler A, Beyene T, Ludwig F, Mkankam FK, Manetsa DV, Ndjatsana M, Doumenge C. Climat de l’Afrique centrale: passé présent et futur. In: De Wasseige C, Tadoum M, Eba’a AR, Doumenge C, editors. Les forêt du bassin du Congo-Forêt et changement climatiques. Neufchâteau: Weyrich; 2015. pp. 37–52. [Google Scholar]
- 28.Yeyeh TMN, Diallo HA, Akinbade SA, Seka K, Kumar PL. Distribution, incidence and distribution of viral diseases of yam (Dioscorea spp.) in Côte d’Ivoire. Afr J Biotechnol. 2014;13(3):465–470. doi: 10.5897/AJB2013.13274. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.