All animals are associated with microorganisms; hence, host-microbe interactions are of fundamental importance for life on earth. However, we know little about the molecular basis of these interactions. Therefore, we studied the deep-sea Riftia pachyptila symbiosis, a model association in which the tubeworm host is associated with only one phylotype of endosymbiotic bacteria and completely depends on this sulfur-oxidizing symbiont for nutrition. Using a metaproteomics approach, we identified both metabolic interaction processes, such as substrate transfer between the two partners, and interactions that serve to maintain the symbiotic balance, e.g., host efforts to control the symbiont population or symbiont strategies to modulate these host efforts. We suggest that these interactions are essential principles of mutualistic animal-microbe associations.
KEYWORDS: host-microbe interactions, symbiosis, holobiont, chemosynthesis, hydrothermal vents, metaproteomics
ABSTRACT
The deep-sea tubeworm Riftia pachyptila lacks a digestive system but completely relies on bacterial endosymbionts for nutrition. Although the symbiont has been studied in detail on the molecular level, such analyses were unavailable for the animal host, because sequence information was lacking. To identify host-symbiont interaction mechanisms, we therefore sequenced the Riftia transcriptome, which served as a basis for comparative metaproteomic analyses of symbiont-containing versus symbiont-free tissues, both under energy-rich and energy-limited conditions. Our results suggest that metabolic interactions include nutrient allocation from symbiont to host by symbiont digestion and substrate transfer to the symbiont by abundant host proteins. We furthermore propose that Riftia maintains its symbiont by protecting the bacteria from oxidative damage while also exerting symbiont population control. Eukaryote-like symbiont proteins might facilitate intracellular symbiont persistence. Energy limitation apparently leads to reduced symbiont biomass and increased symbiont digestion. Our study provides unprecedented insights into host-microbe interactions that shape this highly efficient symbiosis.
INTRODUCTION
All animals are associated with microorganisms (1–3), and consequently, mutualistic bacterium-animal symbioses play critical roles in the physiology, ecology, and evolution of animals, thereby shaping life on our planet. Many of these mutualistic symbioses are based on nutritional benefits for both partners. Symbionts supply their host with nutrients otherwise lacking in the host's diet, while the host in turn provides the symbionts with metabolites, shelter, and optimal growth conditions (4). To establish and stably maintain their alliance, the partners have to interact on the molecular level. The host's immune system needs to control the symbiont population without erasing it altogether (5), for example, by restricting the symbionts to certain organs and/or by downregulating its own immune response (reviewed in reference 6). Symbionts, on the other hand, often employ strategies resembling those of pathogens to colonize and persist in their host. For example, similar protein secretion systems are used by both symbionts and pathogens for interactions with the host (4, 7–9).
In many animals, host-microbe interactions are difficult to assess due to the high number of microbes potentially involved and the presence of long- and short-term associations, which are hard to distinguish (9). Therefore, low-complexity models are important to identify and characterize interaction mechanisms (10). Symbioses of marine invertebrates and their chemoautotrophic symbionts have emerged as suitable study systems. In these symbioses, animal hosts such as gutless annelids and bivalves are often tightly associated with one or a few symbiont types, which enable the eukaryotes to prevail in otherwise hostile environments (11). One of the most conspicuous representatives of these associations, and the first animal in which chemoautotrophic symbionts were discovered, is the giant tube worm Riftia pachyptila (short Riftia), which thrives around deep-sea hydrothermal vents of the East Pacific (12, 13). The host’s absolute dependency on its symbiont makes Riftia an ideal system to study beneficial host-microbe interactions in a mutualistic symbiosis.
The worm completely lacks a digestive system but instead receives all necessary nutrients from its chemosynthetic endosymbiont (12–15). The host in turn provides the endosymbiont with all necessary inorganic compounds for chemosynthesis (16). This association is remarkably productive: Riftia grows extraordinarily fast (>85-cm increase in tube length per year [17]) and reaches body lengths of up to 1.5 m (18).
The uncultured gammaproteobacterial Riftia symbiont, a single 16S rRNA phylotype tentatively named “Candidatus Endoriftia persephone” (19–21), densely populates bacteriocytes in the host trophosome, a specialized organ that fills most of the worm’s body cavity (14). The bacteria oxidize inorganic reduced compounds, such as hydrogen sulfide, to generate energy for carbon fixation (13, 22–26). The symbiont can store elemental sulfur, an intermediate of sulfide oxidation, in sulfur globules (27). Trophosome tissue containing large amounts of stored sulfur has a light yellowish color. During sulfide limitation, i.e., when energy availability is restricted due to low environmental sulfide concentrations, stored sulfur is consumed and the trophosome appears much darker (27–29). Thus, the energetic status of the symbiosis can be directly inferred from the color of the trophosome.
Riftia has been extensively studied, especially with respect to its anatomy, biochemistry, symbiont transmission, and substrate transfer between host, symbionts, and the environment (for examples, see references 24 and 29–32; see references 16 and 33 for reviews). The symbiont’s metabolism has been studied in detail as well (16), in particular by means of metagenomics and metaproteomics (19, 25, 34, 35). However, little is known about interactions between the two symbiotic partners and, particularly, about the proteins directly involved in these processes.
Our study aimed to illuminate the underlying mechanisms of host-symbiont interactions on the protein level. For this purpose, we employed a state-of-the-art global metaproteomics approach, which required comprehensive sequence data for both partners. While the genome of the Riftia symbiont was sequenced previously (19, 34), until now no such information was available for the host. Therefore, we sequenced the transcriptome of the Riftia host de novo. This enabled us to build a comprehensive protein database, which we used to compare protein abundance patterns in symbiont-containing and symbiont-free Riftia tissues. By comparing sulfur-rich and sulfur-depleted specimens, we furthermore examined the dynamics of host-symbiont interactions under high- and low-energy conditions. Our analysis sheds light on metabolite exchange processes between both partners, on the host’s symbiont maintenance strategies, and on the symbiont’s molecular mechanisms to persist inside the host.
RESULTS AND DISCUSSION
Interaction analysis of a chemosynthetic deep-sea symbiosis.
We sequenced the Riftia host transcriptome de novo and combined it with three existing symbiont genomes to create a comprehensive holobiont database for identification of Riftia host and symbiont proteins (see Materials and Methods). Our metaproteomic analysis included comparisons between symbiont-containing and symbiont-free tissues of specimens with light and dark trophosomes. As trophosome color and bacterial sulfur content are directly correlated (27, 28), samples from specimens with light and dark trophosomes will here be referred to as sulfur-rich (S-rich) and sulfur-depleted (S-depleted) samples, respectively. A fully replicated data set and stringent study design enabled us to find statistically significant differences in individual protein abundance between sample types as well as abundance differences between functional protein groups. For an overview of all identified proteins, see Text S1, section 1, and Fig. S1 in the supplemental material. We identified numerous molecular interaction processes (Fig. 1), including (i) metabolite exchange between host and symbiont, (ii) host strategies of symbiont maintenance, and (iii) symbiont mechanisms to persist inside the host. Furthermore, we found that (iv) sulfur availability affects symbiotic interactions in Riftia. Beyond the results presented here, our comprehensive metaproteome data sets and our newly established transcriptome-based Riftia host database (all available from the PRIDE archive; see below) also provide a valuable resource for future Riftia studies and microbe-eukaryote symbiosis research in general.
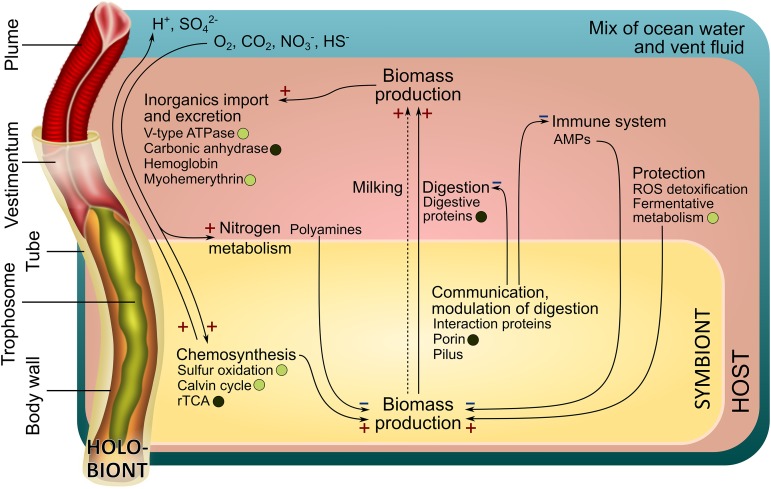
FIG 1.
Main interactions in the Riftia symbiosis. "HOST" refers to processes in Riftia host tissues, while "SYMBIONT" refers to processes in the bacterial endosymbiont. A plus sign indicates presumably stimulating interactions, and a minus sign indicates presumably inhibiting interactions. For example, host efforts that protect the symbiont population from oxidative stress, i.e., ROS detoxification and fermentative metabolism (on the right), can promote symbiont biomass production (+). In contrast, host immune system-related proteins and antimicrobial peptides (AMPs) may inhibit symbiont biomass production (−). Circles, where present, indicate that the respective proteins are more abundant in S-rich (energy-rich) specimens (light circles) or S-depleted (energy-limited) specimens (dark circles). The dashed arrow indicates putative transfer of small organic compounds "Milking"; see Text S1, section 3).
Supplementary Results and Discussion with supplemental figures. Download Text S1, PDF file, 1.9 MB (2.1MB, pdf) .
Copyright © 2019 Hinzke et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolite exchange between host and symbiont. (i) Riftia digests its symbionts for nutrition.
Our results suggest that the main mode of nutrient transfer from symbiont to host is the active digestion of symbiont cells, and that this process might involve endosome-like maturation of symbiont-containing vesicles. We detected a total of 113 host enzymes involved in protein, amino acid, and glycan degradation, as well as in glycolysis and fatty acid beta oxidation. Twenty-two of these proteins were significantly more abundant in trophosome samples than in the other tissues (Table 1). Overall, nearly all of the respective protein groups had higher abundances (i.e., higher organism-specific normalized spectral abundance factor values, or %orgNSAF) in the symbiont-bearing trophosome than in other tissues, both in S-rich and S-depleted specimens (Fig. 2). Many of the protein degradation-related proteins contain signal peptides and thus are likely either contained in lysosomes or secreted into the symbiont-containing vesicles to digest the symbiont cells (Table 1 and Table S1a).
TABLE 1.
Proteins which are putatively involved in symbiont digestion and which had significantly higher abundances in trophosome samples than in other tissues of S-rich and S-depleted specimens
| Accession | Description | Sig ina
: |
Secreted/membraneb | |
|---|---|---|---|---|
| S-rich troph | S-depl troph | |||
| Protein digestion | ||||
| Host_DN32373_c0_g1_i1::g.193014 | Cathepsin Z | x | x | M |
| Host_DN34261_c0_g1_i1::g.35886 | Cathepsin B | x | x | S |
| Host_DN38047_c1_g1_i1::g.177385 | Cathepsin Z | x | x | M |
| Host_DN41150_c0_g1_i1::g.101468 | Cathepsin L1 | x | x | S |
| Host_DN34118_c0_g1_i3::g.155432 | Digestive cysteine proteinase 2 | x | x | S |
| Host_DN39514_c3_g1_i1::g.201492 | Legumain | x | x | S |
| Host_DN34848_c0_g1_i1::g.215091 | Dipeptidyl peptidase 1 | o | x | S |
| Amino acid degradation | ||||
| Host_DN37934_c0_g3_i4::g.212722 | 4-Hydroxyphenylpyruvate dioxygenase | x | x | S |
| Host_DN35553_c0_g1_i1::g.72896 | Maleylacetoacetate isomerase | x | x | |
| Host_DN37934_c0_g3_i6::g.212725 | 4-Hydroxyphenylpyruvate dioxygenase | x | x | |
| Host_DN40417_c0_g1_i7::g.93374 | d-Aspartate oxidase | x | x | Possibly M |
| Host_DN41135_c1_g1_i1::g.101501 | Homogentisate 1,2-dioxygenase | x | x | |
| Host_DN39303_c6_g1_i3::g.66273 | Urocanate hydratase | x | x | |
| Host_DN37934_c0_g3_i11::g.212729 | 4-Hydroxyphenylpyruvate dioxygenase | o | x | |
| Host_DN39293_c0_g3_i16::g.11113 | Histidine ammonia-lyase | o | x | |
| Host_DN41135_c1_g1_i2::g.101503 | Homogentisate 1,2-dioxygenase | o | x | |
| Host_DN40306_c1_g4_i8::g.129962 | Aminoacylase-1 | o | x | |
| Glycan degradation | ||||
| Host_DN36692_c1_g2_i4::g.169924 | Lysosomal alpha-glucosidase | x | x | M/possibly S |
| Host_DN36692_c1_g2_i3::g.169923 | Glucoamylase 1 | o | x | |
| Host_DN37016_c0_g1_i1::g.156600 | Lysosomal alpha-mannosidase | o | x | S |
| Fatty acid beta oxidation | ||||
| Host_DN34874_c0_g1_i9::g.215370 | Propionyl-coenzyme A carboxylase beta chain, mitochondrial |
x | o | |
| Host_DN41664_c1_g5_i6::g.166806 | Peroxisomal bifunctional enzyme | o | x | |
Sig, Significance (x, significant; o, nonsignificant; false discovery rate, 0.05); troph, trophosome; S-depl, S depleted.
Subcellular localization (M, membrane-associated; S, secreted) was predicted using Phobius, TMHMM, and SignalP. Possibly M or S indicates localization prediction based on one tool only.
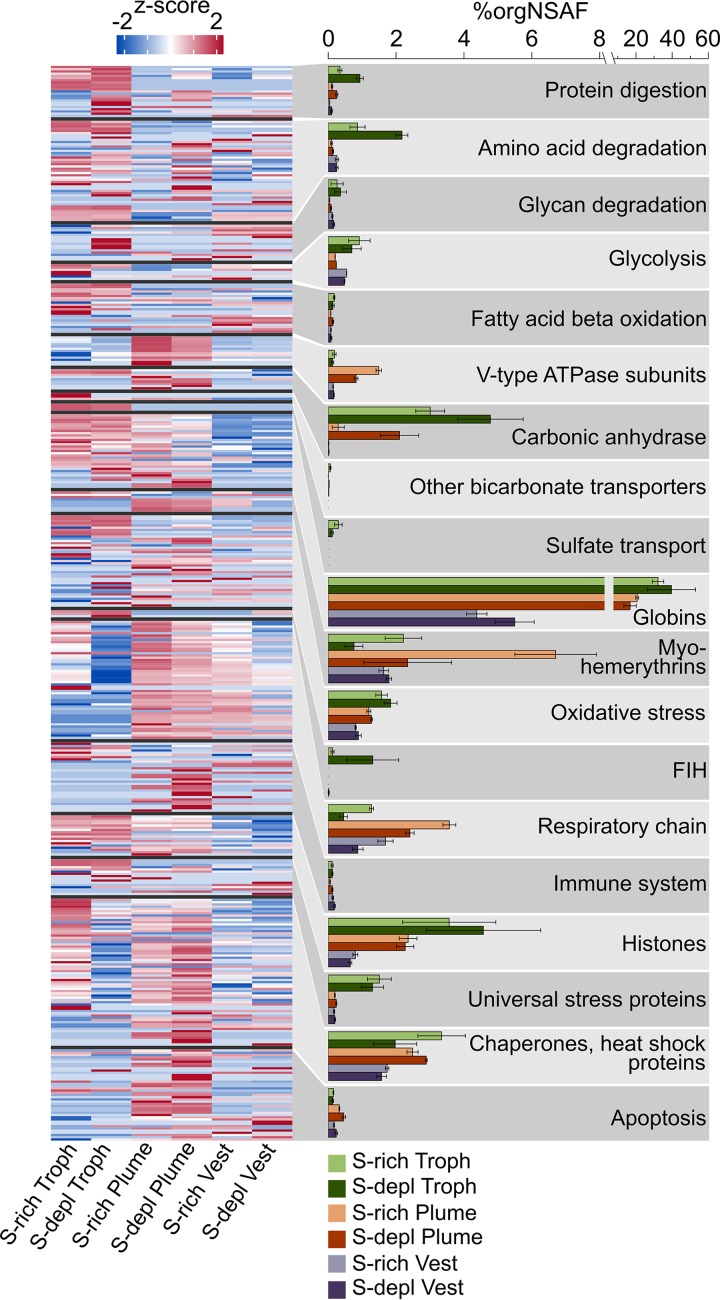
FIG 2.
Functional groups of selected Riftia host proteins and their relative abundances in tissue samples. The heatmap shows log-normalized, centered, and scaled protein abundances. The bar chart shows summed abundances in %orgNSAF (percent normalized spectral abundance factor per organism, i.e., of all host proteins) of all proteins in the respective category. Error bars indicate standard error of the mean. Note the different scaling in the right part of the x axis. The “Chaperones, heat shock proteins” category also includes chaperonins and Clp proteases. FIH, factor inhibiting hypoxia-inducible factor 1α. S-depl, S depleted. Vest, vestimentum. Troph, trophosome. For a list of all identified proteins and their abundances, see Table S1a. (Categories presented in this figure are labeled with X in Table S1a in the column labeled Figure 2. The table can be filtered for these categories.)
(a) All Riftia host proteins identified in this study. (b) Potential autophagy-related Riftia proteins. (c) Potential Riftia antimicrobial peptides. (d) Genomes and metagenomes used for SMART analysis of eukaryote-like protein structures. (e) Sampling dates, cruise number, and number of biological replicates of Riftia samples used in this study. (f) Number of proteins with significant abundance differences in pairwise comparisons of Riftia tissues. (g) All Riftia symbiont proteins identified in this study. (h) Hemerythrin and myohemerythrin isoforms in Riftia and other invertebrates used for alignment in Text S1, Fig. S3. (i) Carbonic anhydrase isoforms in Riftia as detected in this study and described in the literature. Download Table S1, XLSX file, 1.8 MB (1.8MB, xlsx) .
Copyright © 2019 Hinzke et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Our findings are in accordance with previous biochemical, autoradiographic, and microscopic studies, which suggested symbiont digestion in the Riftia trophosome (14, 36–38). Moreover, abundant degradative enzymes and symbiont digestion appear to be common in other mutualistic symbioses as well, including deep-sea mussels (39, 40), shallow-water clams (41, 42), and the gutless oligochaete Olavius algarvensis (43, 44).
Our metaproteome analysis suggests that symbiont digestion in Riftia involves maturation of symbiont-containing host vesicles in a process resembling the maturation of endosomes. Endosomes form after endocytosis of extracellular compounds and mature from early to late endosomes, which ultimately fuse with lysosomes (45). The endosome-associated proteins Rab5 and Rab7 showed significantly higher abundances in trophosome samples than in other host tissues (Table S1a). Rab5 and Rab7 localize to early and late stages, respectively, of endosomes and autophagosomes and are markers for these recycling-related organelles (45–47). The idea of symbiont degradation via an endosome-like maturation process in Riftia is additionally supported by our transmission electron microscopy (TEM) images of Riftia bacteriocytes (Fig. 3), which showed multilamellar bodies. These myelin-like structures can form in endosomes (48) and also during autophagic digestion and have, therefore, previously been attributed to autophagy in the Riftia trophosome (37). However, our results suggest that autophagy plays a less prominent role in symbiont digestion, as we detected only two autophagy-related proteins (Table S1b) in the trophosome metaproteome.
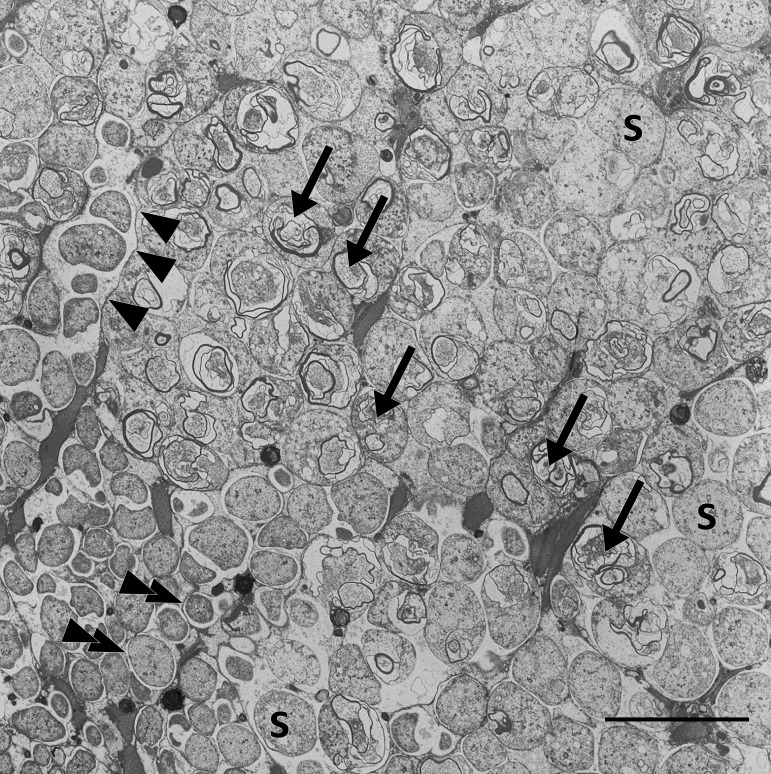
FIG 3.
Transmission electron micrograph of a Riftia trophosome tissue section. Within the lobular trophosome tissue, this section shows the median and peripheral zones of an individual lobule with host bacteriocytes containing intracellular coccoid symbionts (S) located in dedicated vesicles (arrowheads, bacteriocyte membrane; double arrowheads, vesicle membrane). While the lower left area of the image shows mostly intact symbiont cells, arrows in the central area point to symbiont cells in the state of digestion by the host, where cell degradation is indicated by the presence of lamellar bodies. Image brightness and contrast were adjusted for visual clarity. Scale bar, 10 μm.
Moreover, only 12 of 41 detected apoptosis-related Riftia proteins were identified in the trophosome, mostly with similar or significantly lower abundances than in other tissues, and caspases, the main apoptotic effectors, were not detected at all on the protein level in trophosome samples (see also Text S1, section 2). These results suggest that bacteriocyte cell death, which follows after symbiont digestion, probably does not involve apoptosis. This contradicts previous observations (37) but is in line with microscopic results, which did not indicate apoptosis in the trophosome (49). We therefore suggest that an alternative, nonapoptotic cell death mechanism exists in Riftia trophosomes. A nonapoptotic, nonautophagic cell death mechanism was recently described in pea aphid bacteriocytes (50). In the aphids, the proposed mechanism involved hypervacuolation of host bacteriocytes, which, however, was not observed in the Riftia trophosome. Also, in cancer cells, a caspase-independent (nonapoptotic) cell death mechanism was described, which involves the lysosomal protease cathepsin B (51), a putative regulator of lysosome production and autophagic activity (52). As cathepsin B was significantly more abundant in trophosome than in other Riftia tissues, we speculate that this protease, among other degradative enzymes, is involved in controlled cell death in the Riftia trophosome.
Besides symbiont digestion, a second mode of nutrient transfer, the release of small organic carbon compounds by intact symbionts (termed “milking”), was suggested to be present in Riftia (36, 53). Our calculated δ13C ratios might support this hypothesis (Text S1, section 3). However, as we did not detect dedicated symbiont exporters for organic acids or sugars on the proteome level, nutrient transfer by milking is probably less relevant for overall host nutrition than symbiont digestion.
(ii) Riftia dedicates a substantial part of its proteome to provisioning the symbionts with O2, sulfide, and CO2.
We found highly abundant and diverse globins, myohemerythrins, V-type ATPase subunits, and carbonic anhydrases in the host proteome (Fig. 2), indicating that Riftia dedicates a substantial part of its proteome to provisioning the symbiont with all necessary substrates for chemosynthesis.
Globins made up about one-third of all trophosomal host proteins and one-fifth of the total proteome in the plume (i.e., the worm’s symbiont-free gas exchange organ; Fig. 2), with extracellular hemoglobins being particularly abundant (in sum, 32 to 40 %orgNSAF in trophosome and 17 to 21% in plume samples). Riftia has three distinct extracellular hemoglobins composed of globin chains and, in the case of the hexagonal bilayer hemoglobin, globin linker chains (54–56). We detected several of these subunits, including isoforms that are (to our knowledge) hitherto undescribed (Table S1a). Riftia’s extracellular hemoglobins have been shown to bind both O2 and sulfide (56, 57; reviewed in references 58 and 59). Consequently, abundant hemoglobins in the highly vascularized plume would ensure efficient uptake of these compounds for transport to the symbionts (see Text S1, section 4, for more details on sulfur metabolism in the host). Moreover, reversible O2 and sulfide binding to abundant hemoglobins in the trophosome not only provides the bacteria with chemosynthetic substrates and prevents spontaneous sulfide oxidation but also protects the symbionts from oxygen (60). As suggested previously (23), Riftia symbionts are microaerophilic, i.e., sensitive to high oxygen levels. This idea is corroborated by the presence of several ROS-scavenging enzymes (superoxide dismutase, alkyl hydroperoxide reductase, and rubrerythrin) and cytochrome c oxidase cbb3 subunits in the symbiont metaproteome. cbb3 has a high affinity for oxygen and participates in microaerobic respiration (61). In addition to extracellular hemoglobins, we identified four low-abundance (0.002 to 0.084 %orgNSAF) globins that are probably intracellular and might store O2 (Text S1, section 5).
Besides hemoglobins, myohemerythrins were detected in all tissues, with particularly high abundances of 6.7 %orgNSAF in S-rich plumes. With their comparatively high oxygen-binding capacity (62), hemerythrins could facilitate oxygen uptake from the environment into the plume and are possibly also involved in O2 storage and intracellular transport in Riftia. Moreover, the abundance distribution of the nine detected myohemerythrins suggests a tissue-specific function (Text S1, section 6).
V-type ATPase subunits were found with highest total abundances of up to 1.5 %orgNSAF in Riftia plumes (Fig. 2), and almost all of the detected subunits were significantly more abundant or exclusively detected in the plumes. V-type ATPases have a pivotal function in regulating internal pH and CO2 uptake (63) and thus in symbiont provisioning. The high energy demand of V-type ATPase-dependent pH regulation could be met via a relatively higher respiration activity in the plume, as indicated by comparatively higher total abundances of respiratory chain proteins (Fig. 2), ATP synthase, and mitochondrial ribosomal proteins in this tissue. Additionally, carbonic anhydrase (CA), another important enzyme for CO2 uptake, was detected in all tissues. While we observed tissue-specific abundance patterns of individual CAs (Text S1, section 7 and Fig. S4), overall CA abundance was highest in the trophosome (Fig. 2). CA facilitates CO2 diffusion into the plume by converting it to HCO3− (63, 64) and likely back-converts the HCO3− to CO2 for fixation by the symbionts in the trophosome. Our analysis suggests that three of the Riftia CAs are membrane bound (Text S1, section 7) and could thus facilitate CO2 diffusion into bacteriocytes by converting HCO3− to CO2 on the bacteriocyte cell surface (65, 66). Transport of HCO3− to the bacteriocytes could be mediated by sodium bicarbonate exchangers, which we identified in trophosome and plume samples (Table S1a).
While carbon for fixation by the Riftia symbiont is likely mainly transported in the form of CO2/HCO3−, the host may additionally pre-fix CO2 into organic C4 compounds, which are then transported to the symbiont (67). We did identify host phosphoenolpyruvate carboxykinase and pyruvate carboxylase, which could be involved in this process (Text S1, section 8).
(iii) Riftia’s nitrogen metabolism depends less on the symbiont than previously assumed.
Riftia symbionts supply their host not only with carbon and energy sources but likely also with ammonium produced by bacterial nitrate reduction (Fig. 4 and Text S1, section 9). However, with regard to the subsequent metabolization of organic nitrogen, the host might be more self-sufficient than previously thought: previous biochemical analyses suggested that only the symbiont, but not the host, can de novo synthesize pyrimidines (68) and produce polyamines (69). In contrast to those studies, we found the multifunctional CAD protein (which combines the three enzyme functions carbamoyl-phosphate synthetase 2, aspartate transcarbamoylase, and dihydroorotase) in the Riftia host metatranscriptome, suggesting that the host can catalyze the first steps of pyrimidine synthesis. As we did not detect CAD protein on the protein level, expression levels and associated activities in the host are likely rather low, and most of the pyrimidine demand could be satisfied by digesting symbionts. In addition, we found key genes involved in polyamine synthesis in the host's metatranscriptome and also detected several of the respective proteins in the host's metaproteome (Fig. 4). Our results suggest that, while both Riftia symbiosis partners can synthesize spermidine, in fact only the host is able to generate spermine. Host spermidine synthase and spermine synthase were exclusively detected in trophosome samples in our study, suggesting that the polyamines produced by these proteins have a role in symbiont-host interactions. They could, for example, be involved in restricting the symbiont to its cell compartment, i.e., the bacteriocyte vesicle, as suggested for bacterial pathogens (Text S1, section 10). In addition, only the host seems to possess a full urea cycle and might degrade not only its own but also nitrogen-containing metabolites of the symbiont (Text S1, section 9). These results suggest that the symbiont provides the host with necessary metabolic energy and building blocks for biosynthesis but that the host has also retained key biosynthetic capacities for N-containing organic compounds.
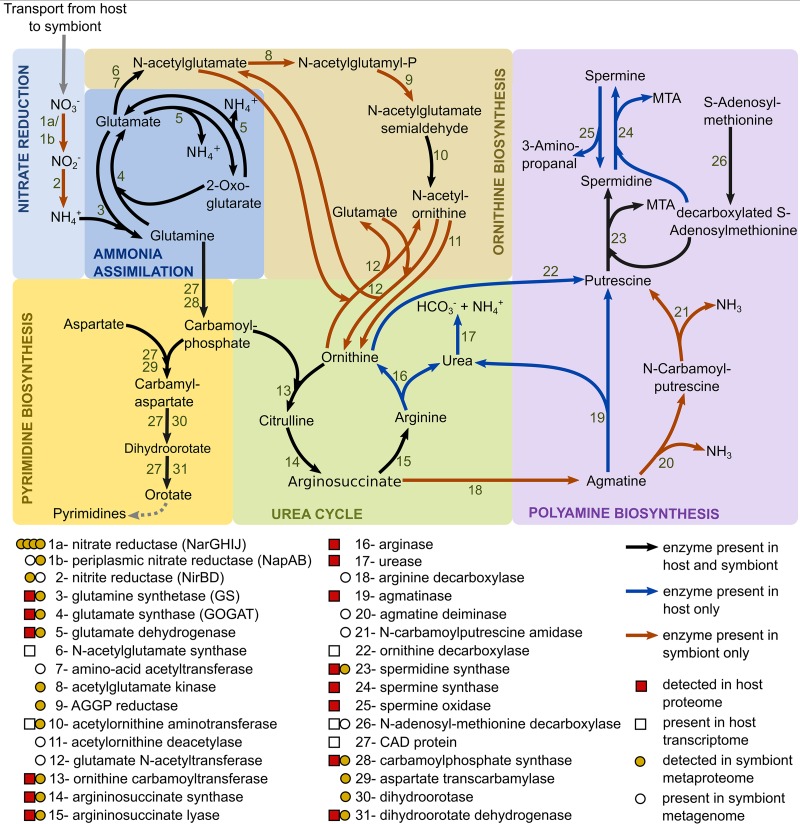
FIG 4.
Main nitrogen metabolic pathways in Riftia symbiosis. AGGP reductase, N-acetyl-gamma-glutamyl-phosphate reductase; CAD protein, multifunctional carbamoyl-phosphate synthetase 2, aspartate transcarbamoylase, and dihydroorotase protein; MTA, 5′-methylthioadenosine. Note that the symbiont might also be capable of nitrate respiration (25, 60), which is not depicted here.
Host strategies of symbiont maintenance.
(i) Riftia protects its symbiont from oxidative damage and may even generate hypoxic conditions in the trophosome. We found several reactive oxygen species (ROS)-scavenging enzymes (superoxide dismutase, peroxiredoxin, and glutathione S-transferase) as well as proteins indicative of anaerobic metabolism and universal stress proteins with significantly higher individual abundance and in higher total amounts (summed %orgNSAF) in the trophosome than in other tissues (Fig. 2 and Text S1, section 11). Riftia’s ROS-detoxifying enzymes probably protect not only the host but also the microaerophilic symbiont against ROS. Upregulation of host proteins involved in ROS detoxification was previously shown in the Wolbachia symbiosis (70, 71). Additionally, malate dehydrogenase was highly abundant in trophosomes. This enzyme is regularly observed in different invertebrates under anaerobic conditions (72) and is involved in maintaining redox balance during anaerobiosis (73). Therefore, the host might generate hypoxic conditions in the trophosome, as also indicated by the overall lower abundance of host respiratory chain proteins in trophosome than in other tissues of both S-rich and S-depleted specimens. We also detected hypoxia-inducible factor 1-alpha inhibitors (factor inhibiting HIF1a; FIH) almost exclusively in trophosome samples, which further supports the idea that free oxygen concentrations in the trophosome are low. This is in line with the high oxygen-binding capacity of Riftia hemoglobins (23, 60) and with the suggestion of fermentative metabolism under hypoxic and even normoxic conditions in Riftia based on biochemical results (74). Taken together, lower oxygen concentration in the trophosome, (partial) anaerobic host metabolism, and host ROS-detoxifying enzymes in this tissue would not only protect the symbionts from oxidative damage but also decrease the competition between the Riftia host and its symbionts for oxygen.
(ii) The Riftia immune system might be involved in symbiont population control. We detected several proteins that potentially are involved in a specific immune reaction of Riftia against its symbiont in the trophosome. Two bactericidal permeability-increasing proteins (BPIPs) were detected, one exclusively in the trophosome, the other only in the plume. BPIPs act specifically against Gram-negative bacteria, causing initial growth arrest and subsequent killing due to inner membrane damage (75). In Riftia, BPIPs could be involved in keeping the symbiont population under control, e.g., as part of the digestion process or by preventing the symbionts from leaving their intracellular host vesicles. Likewise, in the Vibrio-squid symbiosis, BPIPs have been implied in restricting the symbiont population to the light organ (76). In addition to BPIPs, a pathogen-related protein (PRP) was present in all replicates of S-rich trophosome but absent from all other tissues. In plants, PRPs accumulate during defense responses against pathogens (reviewed in reference 77). PRPs have also been described in nematodes (78) and humans (79), although their function remains elusive.
We also found that histones had overall higher abundance in Riftia trophosome than in other tissues. Four of these histones were significantly more abundant in trophosomes than in other tissues, and three additional histones were exclusively detected in trophosome samples (Table S1a). Besides being crucial for DNA interactions, histones and histone-derived peptides can have antimicrobial effects (80–82). A BLASTP search of the detected Riftia histones against the antimicrobial peptide (AMP) database APD3 (83) gave hits for four of the Riftia histones (Table S1c), stimulating the speculation that these histones have antimicrobial properties. While AMP-like histone-derived peptides in the plume might be involved in defense against environmental microbes, the high abundance of histones in the trophosome could point to a function in host-symbiont interaction. Host-derived AMPs could, for example, be involved in controlling the symbiont’s cell cycle. In their life cycle, the symbionts apparently differentiate from actively dividing stem cells into growing but nondividing larger cells (49). As various AMPs were shown to inhibit cell division or septum formation and to cause filamentous cell morphologies (reviewed in reference 84), we speculate that Riftia AMPs inhibit cell division as well, e.g., via interaction with the symbiont protein GroEL. Interaction between a host AMP and a symbiont GroEL has been proposed to lead to cell elongation of bacterial weevil symbionts (85). A role of histones and histone-derived peptides in immune system responses has been described or suggested in various other organisms, including catfish (80), Komodo dragons (86), toads (81), and humans (82).
Beyond these and a few other individual immunity-related proteins, we did not observe an overall higher abundance of host immune system proteins (such as lysozyme, complement system proteins, or peptidoglycan recognition proteins) in the trophosome than in symbiont-free tissues. This indicates that the host immune system does not play a major role in controlling symbiont population size. More likely, symbiont population control might to a large part be a result of digestion of symbionts (a “mowing” process), which effectively prevents the symbionts from escaping their compartments and/or overgrowing the host. Nevertheless, the immune system might be involved in phage protection and symbiont recognition during establishment of the symbiosis (Text S1, section 12).
Symbiont persistence mechanisms. (i) Eukaryote-like protein structures in the symbiont might be involved in host communication.
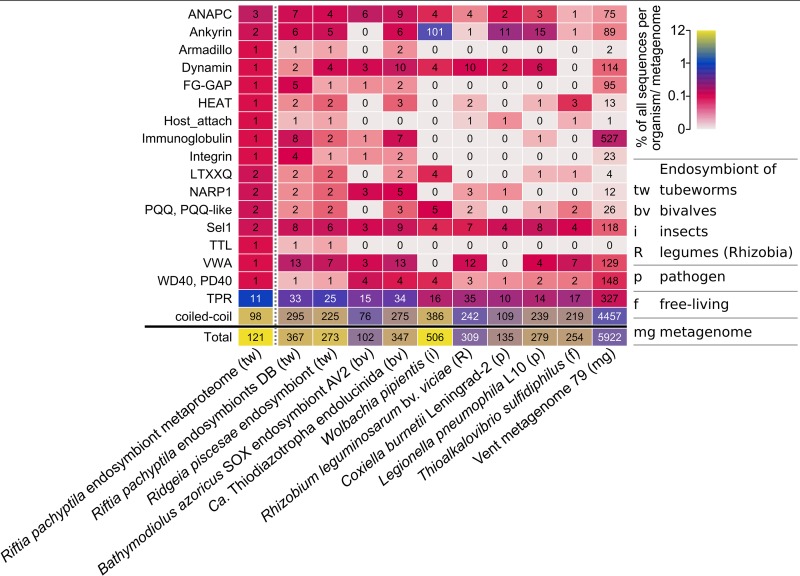
The metagenome of the Riftia symbiont “Ca. E. persephone” encodes several protein groups with possible roles in symbiont-host interactions, including eukaryote-like protein (ELP) structures, as revealed by our SMART analysis (Table S2). We detected more than 100 of these symbiont proteins in the trophosome samples (Fig. 5), which points to a symbiosis-relevant function.
FIG 5.
Selected domains with eukaryote-like structures and with putative functions in symbiont-host interactions in the Riftia symbiont and in selected other organisms and metagenomes. Color scale shows the percentage of genes/proteins containing the respective domain relative to all gene/protein sequences in this organism or metagenome. Numbers indicate the total number of genes/proteins containing the respective domain. For an overview of all analyzed organisms and domains, see Text S1, Fig. S5. For details on the organisms and communities, see Table S1d. The vent metagenome was sampled from hydrothermal vent fluid at a diffuse-flow vent site (Crab Spa) (137), which also houses Riftia. For further information about the selected protein groups, see Table S2. Riftia pachyptila endosymbiont metaproteome refers to the Riftia symbiont proteins detected in this study.
Domains and protein families with a putative role in host-symbiont interactions. Download Table S2, PDF file, 0.4 MB (414.9KB, pdf) .
Copyright © 2019 Hinzke et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Among the ELPs detected in the symbiont metaproteome were two ankyrin repeat-containing proteins, which contain a signal peptide and are therefore likely secreted (predicted by Phobius, http://phobius.sbc.su.se/). Ankyrin repeats were found to mediate protein-protein interactions (87). In the sponge Cymbastela concentrica, symbiont ankyrins were proposed to interact with the eukaryote’s phagocytosis system: the symbiont ankyrins were heterologously expressed in Escherichia coli and led to inhibition of phagocytosis by amoebae (88). Likewise, a secreted Legionella pneumophila ankyrin protein apparently interferes with host endosome maturation (89). The “Ca. E. persephone” ankyrin repeat-containing proteins therefore could directly interact with host proteins as well, e.g., to modulate endosome maturation and thus to interfere with symbiont digestion by the host. Similarly, proteins with tetratricopeptide repeat (TPR)/Sel1 domains, which we also detected in the “Ca. E. persephone” metaproteome, have been shown to impact phagocytosis by amoebae (90).
The Riftia symbiont furthermore encodes eukaryote-like proteins of the tubulin-tyrosine ligase family (TTL proteins). These proteins posttranslationally modify tubulin and thus interact with the eukaryotic cytoskeleton (91). We found one TTL protein in the “Ca. E. persephone” metaproteome. Other protein groups that are involved in protein-protein interactions in eukaryotes, e.g., with cytoskeletal proteins, and that we detected in “Ca. E. persephone” include armadillo repeat proteins (92) and HEAT repeat-containing proteins (93). As several of the protein structures analyzed here are also found in other mutualistic symbionts and pathogens (Text S1, section 13, and Table S2), it is conceivable that parallels exist between interaction processes of mutualistic and pathogenic associations and that the Riftia symbiont employs a strategy similar to that of pathogens to communicate with its host on the molecular level.
(ii) Symbiont membrane proteins may export effector proteins into host cells and lead to strain adaptation.
We detected various outer membrane-related proteins in the “Ca. E. persephone” proteome, including a porin (Sym_EGV52132.1), which was one of the most abundantly expressed symbiont proteins, and 12 type IV pilus (T4P) system proteins (PilQ, PilF, PilC, PilBTU, PilM, PilN, PilP, FimV, PilH, and PilY1). Five additional T4P structure proteins were encoded in the metagenome (pilVWXE and pilO). These proteins are in direct contact with the host cells and thus are likely involved in interactions between both symbiosis partners, including such processes that facilitate the symbiont’s persistence inside the host cells.
The abundant symbiont porins could transport effector molecules, e.g., to modulate digestion by the host. A role of porins in effector transport during symbiosis has been hypothesized for Vibrio fischeri OmpU, a channel protein that is important for symbiont recognition by the squid host (94).
The T4P system is a complex structure, which in Pseudomonas aeruginosa comprises more than 40 proteins, including structural and regulatory proteins (95). It can have several functions in different species: adhesion, secretion, and natural transformation (95–98). As the “Ca. E. persephone” T4P system likely is not involved in adhesion to host cells during symbiosis (although it might be during the initial infection), it could participate in protein secretion and/or natural transformation. The Riftia symbiont’s T4P system could export putative effector proteins (e.g., ankyrins and SET domain proteins; Text S1, sections 13 and 14) for host interactions. Interestingly, in the pathogen Francisella tularensis subsp. novicida, a T4P structure is involved in secretion of infection-moderating proteins (97).
Besides their putative function in effector protein export, symbiont membrane proteins may also lead to bacterial strain adaptation. The Riftia symbiont population is polyclonal, i.e., although there is only one 16S rRNA phylotype, this phylotype consists of several distinct strains (20). T4P system-mediated exchange of genetic material between different symbiont strains would add to this diversity in the symbiosis and might additionally enable exchange of symbiosis-related genes within the free-living “Ca. E. persephone” population. Natural transformation in symbionts has only recently been shown for V. fischeri in culture (99) and the earthworm symbiont Verminephrobacter eiseniae, which likely employs a T4P structure for DNA uptake (98). As microbial cell densities are comparatively high in eukaryote-prokaryote mutualisms, natural transformation in these systems might actually be more common than previously recognized. While mostly only one to three symbiont cells are located in one host vesicle, individual vesicles with up to 14 symbiont cells have also been reported (49), which might allow for exchange of genetic material. The proposed DNA uptake by the Riftia symbiont may not only facilitate exchange between symbiont strains but also promote horizontal gene transfer between host and symbiont, e.g., of eukaryote-like proteins. This hypothesis, as well as the speculation that “Ca. E. persephone” is capable of conjugation (Text S1, section 14), certainly warrant further investigations.
S availability affects symbiotic interactions in Riftia. (i) S-depleted Riftia hosts digest more symbionts than S-rich specimens.
We compared the metaproteomes of Riftia specimens with and without stored sulfur (i.e., energy-rich versus energy-depleted specimens; Text S1, Fig. S1) to examine how energy availability impacts symbiotic interactions (see Table S1f for total numbers of differentially abundant proteins). Metabolite transfer is apparently especially influenced by the energy regime: the host supposedly relies more on symbiont digestion in times of S shortage. Proteinaceous symbiont biomass was notably lower in S-depleted trophosomes (32%) than in S-rich trophosomes (58%) (Fig. 6). Simultaneously, overall abundances for several groups of host digestive enzymes were higher in S-starved trophosomes (Fig. 2), and a number of individual host proteins were significantly more abundant in these S-depleted samples, such as enzymes involved in protein digestion (including cathepsin B), amino acid degradation, the late endosome-related protein Rab7, and histones (Table S1a). One reason for this supposed increase in symbiont digestion in S-depleted trophosomes could be a lower nutritional value of the energy-depleted symbionts. S-depleted symbionts have lower abundances of enzymes involved in sulfur oxidation, probably due to lower S availability. Therefore, less energy might be available for biosynthesis under S depletion, rendering the symbiont less nutritious for the host. The animal would then, especially if S depletion is prolonged, have to rely on increased symbiont digestion in order to still satisfy its basal metabolic demands. Thus, S-depleted hosts may, despite increased symbiont digestion, have less energy available. This idea is supported by the observation that host proteins involved in the energy-generating glycolysis, tricarboxylic acid (TCA) cycle, respiratory chain, ATP synthesis, and biosynthetic pathways were less abundant in S-depleted trophosomes than in S-rich trophosomes. Concomitant with the postulated lower nutritional value of S-depleted symbionts, the Calvin cycle key enzyme RubisCO had an about 10-fold lower abundance in S-depleted symbionts. Abundance of the reverse TCA (rTCA) cycle key enzyme ATP citrate lyase (NCBI accession no. EGV51152.1), on the other hand, was slightly higher in S-depleted symbionts than in S-rich symbionts, albeit only 1.4-fold. Under S-depleted conditions, symbionts apparently rely relatively more on the rTCA cycle, which is more energy efficient than the Calvin cycle (35). The Calvin cycle could be used in addition to the rTCA cycle under favorable conditions to maximize carbon fixation. Moreover, symbiont enzymes involved in translation were overall more abundant in S-rich trophosomes than in S-depleted trophosomes. Less protein biosynthesis in S-depleted symbionts would not only impact the nutritional value of these symbionts but also directly decrease the proteinaceous symbiont biomass. The reason for the lower proteinaceous biomass of symbionts in S-depleted trophosomes is, therefore, probably 2-fold: the host digests more symbionts and the symbionts produce less biomass than in energy-rich trophosomes.
FIG 6.
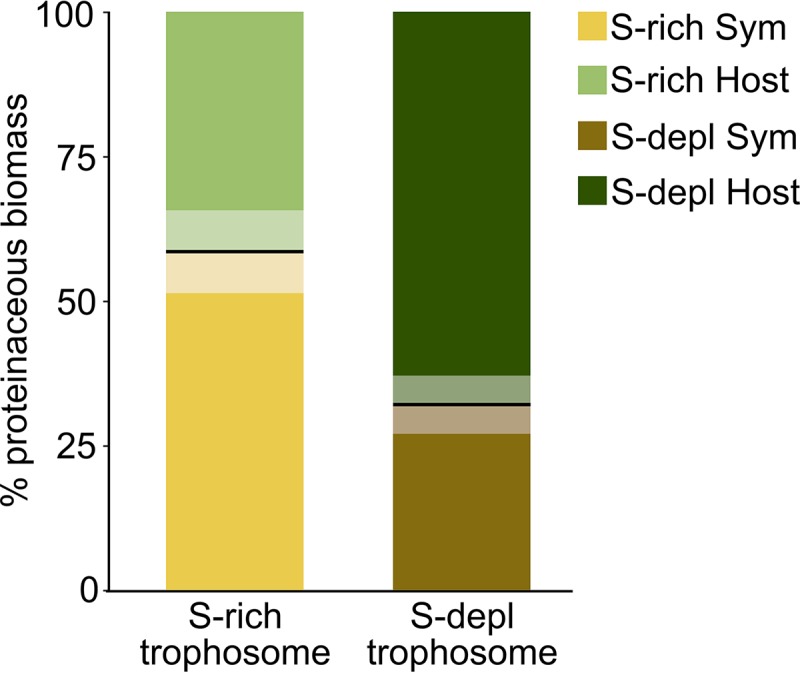
Percent proteinaceous biomass contributions of host and symbiont as calculated from the share of host and symbiont spectral counts in all spectral counts of the respective samples (127; see Materials and Methods for details). Boldface lines indicate the means, and semitransparent areas indicate standard error of the mean. Sym, symbiont; S-depl, S depleted.
These findings are in contrast to previous results (29), which showed no significant differences in autotrophic activity and symbiont abundance between Riftia specimens from high- versus low-sulfide habitats. Increased symbiont digestion may be a short-term adaptation to fluctuating environmental conditions, whereas under long-term low-S conditions the symbiosis might adapt by other means, e.g., by reduced growth rates. Decrease in symbiont abundance or total protein under energy-limiting conditions also has been noted in Bathymodiolus (100) and Codakia orbicularis bivalves (42) as well as in O. algarvensis oligochaetes (43). Thus, relying on the symbionts as a nutrient source also under unfavorable conditions appears to be a common symbiosis mechanism that would ensure survival of the host and a subset of the symbiont population, ultimately prolonging survival of the individual holobiont.
(ii) S availability influences CO2 uptake, pH regulation, and O2 regime in the Riftia host.
S-depleted hosts seem to invest relatively more biosynthetic capacities in CO2 uptake and less in pH regulation, and their trophosomes are supposedly less hypoxic than those of S-rich hosts (Text S1, sections 11 and 15). At the same time, S availability appeared to have little influence on non-symbiont-related processes in the host, as only very few (i.e., <10) individual proteins differed significantly in abundance between S-rich and S-depleted plume and vestimentum samples. This indicates that the host’s metabolism is very well buffered against changes in environmental conditions.
(iii) Higher digestion pressure might result in symbiont countermeasures.
A putative “Ca. E. persephone” dodecin was significantly more abundant in S-depleted Riftia specimens than in S-rich specimens. This protein might be involved in protecting the symbiont against oxygen and/or digestion stress (Text S1, section 14). A symbiont porin, which was also significantly more abundant in S-depleted specimens, might be involved in counteracting the supposedly higher digestion pressure (described above) (Text S1, section 14).
Conclusions.
To fully understand the biology of organisms, it is crucial to study them together with their symbiotic partners as holobionts (101). Given its low complexity, high specificity, and extreme dependence of the host on the symbiont, the association of Riftia and its bacterial partner serves as an excellent system to study mutualistic host-microbe interactions. While Riftia lives in a unique and remote environment, many of the interactions we identified, like symbiont digestion by the host, high host investment in substrate transfer to the symbiont, host-directed symbiont population control, and eukaryote-like symbiont proteins that could interact with the host's molecular machinery, seem to be critical in other symbiotic associations as well, including insects, mussels, and oligochaetes. These interactions might therefore represent common principles among evolutionarily diverse mutualistic animal-microbe associations.
Our study provides access to the Riftia host transcriptome and protein sequences and thus paves the way for future research on host-microbe interactions in Riftia and other systems. Promising research directions include the elucidation of protein functions, e.g., of Riftia immune system proteins and symbiont eukaryote-like proteins by heterologous gene expression and biochemical assays in model systems. Moreover, our work stimulates future in-depth studies of the molecular mechanisms involved in recognition of both partners during the initial infection of Riftia larvae by free-living symbionts. Putative differences between Riftia’s short- and long-term adaptation strategies in response to changing environmental conditions also warrant further investigation.
MATERIALS AND METHODS
Sampling.
Riftia samples were obtained during several research cruises in 2008, 2014, and 2017, with RV Atlantis, to the deep-sea hydrothermal vent fields on the East Pacific Rise at 9°50′N, 104°17′W. Riftia specimens were collected by the human occupied vehicle Alvin or the remotely operated vehicle Jason in approximately 2,500-m water depth. Specimens were kept at 4°C in cold seawater until dissection, which was performed within 4 h after recovery. Only healthy-looking specimens were used. Sampling dates for all Riftia tissue samples for proteomics, transcriptomics, and transmission electron microscopy (TEM) are summarized in Table S1e in the supplemental material. Different specimens were used for proteomics, transcriptomics, and TEM. Riftia specimens were dissected onboard, and samples from four different organs (here referred to as tissues) were stored at –80°C: the lamellae of the tentacular crown were shaved off to provide plume samples, trophosome samples were dissected from whole trophosome, body wall samples were retrieved and washed after removal of the trophosome, and vestimental samples were cut off from the lateral portions of the vestimentum. As trophosome color is directly correlated to the tissue’s elemental sulfur content (27, 28), we classified specimens as sulfur rich (S rich), S depleted, and medium S according to their trophosome color (yellow/light green, dark green/black, or medium green, respectively). To ensure comparability of the classifications, the same light source and dissection tray were used for all samples. Sulfur-rich and sulfur-depleted specimens were used for transcriptome sequencing and comparative metaproteomics, while specimens with medium sulfur content were only used for transcriptome sequencing (Table S1e).
Extraction of whole-tissue RNA.
RNA was extracted from a total of 22 tissue samples from 9 specimens with high, medium, and low trophosome sulfur content (6× trophosome, 6× body wall, 5× plume, 5× vestimentum) (Fig. 1). Tissue samples were homogenized by bead beating with lysing matrix D (MP Biomedicals) in 1 ml TRIzol (Thermo Fisher Scientific; 3 times at 6.5 m/s for 30 s, with 3 min of cooling on ice between steps). After 5 min of acclimatization to room temperature, samples were applied onto QIAShredder columns (Qiagen) and centrifuged (16,000 × g, 3 min, 4°C). Afterwards, RNA was isolated from the aqueous flowthrough according to the TRIzol extraction protocol, with the modification that samples were centrifuged for 20 min at 12,000 × g and 4°C for phase separation. Ten micrograms of glycogen was added for RNA precipitation. RNA was washed twice with 75% ethanol and purified using the Norgen RNA clean-up and concentration kit according to the manufacturer’s protocol A, including DNA removal with DNase (Qiagen). Quality of extracted RNA was assessed using NanoDrop (Thermo Fisher Scientific) and Bioanalyzer (Agilent) analyses.
Transcriptome sequencing and assembly. (i) Transcriptome sequencing.
Transcriptome sequencing was performed employing the TruSeq stranded mRNA [poly(A)-based] library protocol (Illumina) on a HiSeq 4000 (Illumina) according to the manufacturer’s guidelines.
(ii) Transcriptome assembly.
High-throughput paired-end Illumina sequencing resulted in an average of about 26 million reads per end per library (minimum of 16,045,121 reads per end, maximum of 31,318,532 reads per end; 95% confidence interval, 1,673,590). After demultiplexing and quality checking of reads in FastQC v0.11.5 (102), we trimmed low-quality bases and adapters with Trimmomatic v0.32 (103) using the settings ILLUMINACLIP:AllAdapters.fa:2:30:10 SLIDINGWINDOW:4:20 and LEADING:5 TRAILING:5 HEADCROP:15 MINLEN:75. Although bacterial mRNA does not possess a poly(A) tail, previous research has shown that bacterial reads can still be present in poly(A)-enriched RNA-sequencing libraries (104). To filter out potential symbiont contaminations from our host transcriptomes, we used the Bowtie 2 v2.2.9 aligner (105) in very-sensitive mode to map the quality-filtered paired-end reads against the published genomes of the endosymbionts of Riftia (Riftia1, NCBI locus tag prefix RIFP1SYM; Riftia2, locus tag prefix RIFP2SYM) and Tevnia jerichonana (34). Unmapped paired-end reads were subsequently extracted using SAMtools v1.4.1 (106). Potential environmental sequence contaminations from sample handling were excluded with DeconSeq v0.4.3 (107), using coverage and identity thresholds of 0.90 and 0.95, respectively. The decontaminated host reads were normalized, pooled, and assembled with Trinity v2.3.2 (108). To optimize the transcriptome assembly, we performed four different assemblies with different parameters and input files: (i) only paired reads, (ii) paired and unpaired reads, (iii) only paired reads plus Jaccard-clip option (to reduce chimeras), and (iv) paired and unpaired reads plus Jaccard-clip option.
To assess the completeness of the different assemblies, we compared our transcriptomes to the BUSCO v2.0 eukaryote and metazoan orthologous data sets (109). Overall, the best results in terms of transcriptome completeness and quality were obtained by the assembly approach using paired and unpaired reads plus the Jaccard-clip option (complete BUSCO, 99.0%) (Table S3). This data set was used for all further analyses.
Transcriptome completeness for the four different Riftia transcriptome assemblies based on the BUSCO eukaryote and metazoan datasets. Download Table S3, PDF file, 0.3 MB (333.7KB, pdf) .
Copyright © 2019 Hinzke et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(iii) ORF prediction.
TransDecoder v3.0.1 (110) was used to identify coding regions in the assembled transcripts. To improve open reading frame (ORF) prediction, we examined all candidate ORFs for homology to known proteins by searching the Swiss-Prot (http://www.uniprot.org) and Pfam (111) databases (downloaded 3 January 2017) with BLASTP (E value of 1e−05) (112) and HMMER3 (113), respectively. ORFs that were longer than 100 amino acids and/or had a database entry were retained. The FASTA headers of the TransDecoder output files were modified with a custom PERL script to include the BLASTP protein annotations.
Database generation.
A common database for protein identification of Riftia host and symbiont was generated. To this end, host protein sequences were clustered at 95% identity with CD-HIT v. 4.6 (114). For symbiont sequences, the three proteomes of the Riftia1 symbiont (NCBI PRJNA60889, ID 60889), Riftia2 symbiont (NCBI PRJNA60891, JGI 2600255285), and Tevnia symbiont (NCBI PRJNA60887, ID 60887) (34) were used. Riftia1 was used as basis for clustering the symbiont protein sequences with CD-Hit-2D (114). Subsequently, the combined symbiont database was clustered at 95% identity. Identifier prefixes were added to distinguish between host and symbiont sequences for Calis-p (115 and see below). Host and symbiont databases were concatenated, and the cRAP database containing common laboratory contaminants (116) was added. The final database contained 71,194 sequences (67,092 host and 3,986 symbiont protein sequences).
Proteomics sample preparation and analysis.
For metaproteomics analysis, we used three biological replicates per tissue (trophosome, vestimentum, and plume) and condition (specimens with S-rich and S-depleted trophosomes), which resulted in a total of 18 samples. Tissues were disrupted by bead beating for 45 s at 6.0 m/s with lysing matrix D tubes (MP Biomedicals) in SDT buffer (4% [wt/vol] sodium dodecyl sulfate [SDS], 100 mM Tris-HCl, pH 7.6, 0.1 M dithiothreitol [DTT]), followed by heating to 95°C for 10 min. Tryptic peptides were generated following the FASP protocol of Wiśniewski et al. (117), with minor modifications as described by Hamann et al. (118). Peptide concentrations were determined with the Pierce Micro BCA (bicinchoninic acid) assay (Thermo Scientific Pierce) according to the manufacturer’s instructions. The tryptic digest was desalted on-line during liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis.
All samples were analyzed by one-dimensional LC-MS/MS as described by Hinzke et al. (119), using 4-h gradients. Samples were analyzed in a randomized block design (120) and run in technical triplicates. Two technical replicate runs were acquired with a 50-cm analytical column, one with a 75-cm analytical column. To standardize the stable isotope fingerprinting (SIF) analysis (115), human hair was measured in technical duplicate alongside the Riftia samples in the replicate run using a 75-cm column.
Proteomics data evaluation. (i) Protein identification, quantification, and statistical analyses.
For protein identification, MS/MS spectra of combined technical triplicate runs were searched against the combined host and symbiont database using the Sequest HT node in Proteome Discoverer version 2.0.0.802 (Thermo Fisher Scientific) as described in Kleiner et al. (115). For protein abundance estimates, normalized spectral abundance factors (NSAFs) (121) were calculated per sample and organism (%orgNSAF) (122). Statistical evaluation was performed based on spectral counts using the edgeR package (123) in R (124). The edgeR package uses an overdispersed Poisson model for analysis of count data. Overdispersion is moderated across proteins using empirical Bayes methods, and differentially abundant proteins are detected using an overdispersion-adapted analog to Fisher’s exact test (123). We filtered for proteins with at least 10 spectral counts for host proteins and at least 5 spectral counts for symbiont proteins in at least three samples and employed a false discovery rate (FDR) of 0.05 to assign statistical significance to protein abundance differences. For graphical representation, heatmaps were generated with the R package ComplexHeatmaps (125) and intersection plots with the R package UpsetR (126). Protein biomasses of host and symbiont were calculated as described in Kleiner et al. (127). Spectral counts of all symbiont proteins and of all host proteins identified with at least two unique peptides were summed individually, and these two sums were divided by the sum of all spectral counts (host plus symbiont proteins with at least two unique peptides) and multiplied by 100 to give the percentage of proteinaceous biomass for host and symbiont.
δ13C values of Riftia symbiont and host were calculated from mass spectrometry data with Calis-p (115) using one technical replicate LC-MS/MS run (75-cm analytical column). Human hair was used as a reference material.
(ii) Protein annotations, functional characterization, and categorization.
Besides the annotations included in the database, proteins where further characterized using the online tools described in Table S4. Proteins were manually categorized into functional groups based on their annotations and on protein function information in the UniProt (128), NCBI (https://www.ncbi.nlm.nih.gov/), and InterPro (129) databases. We used the Transporter Automatic Annotation Pipeline (TransAAP) (http://www.membranetransport.org/transportDB2/TransAAP_login.html) of TransportDB2 (130) and TCDB (131) with gblast 2 (http://www.tcdb.org/labsoftware.php) to annotate transporters in the Riftia1 symbiont metagenome database. To detect potential antimicrobial peptides (AMPs) among the host proteins, we searched the detected host proteins against the antimicrobial peptide database APD3 (83) using BLASTP (112) in BLAST+ 2.7.1 (132). Results were filtered for identity of >75% and E value of <0.005. We screened the Riftia proteome for homologs of known autophagy-related Drosophila melanogaster proteins (as listed in reference 133) by BLAST searching (BLASTP [112] in BLAST+ 2.8.1 [132]) the Riftia host proteome against the respective Drosophila amino acid sequences (Table S1b).
Tools used to characterize Riftia host and symbiont proteins included in the combined Riftia host and symbiont database used in this study. Download Table S4, PDF file, 0.4 MB (383.4KB, pdf) .
Copyright © 2019 Hinzke et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(iii) SMART analysis of eukaryote-like and potential interaction domains.
We used the SMART tool (134) to screen the Riftia symbiont protein database for proteins and domains that could be involved in symbiont-host interactions. Structures that did not meet the threshold required by SMART were excluded, whereas overlapping features were included. We manually filtered the SMART annotations to find putative interaction-relevant structures based on Pfam and SMART database information. To compare the Riftia symbiont with other host-associated (mutualistic or pathogenic) and free-living organisms, we also included domains not present in the Riftia annotations but possibly relevant for host-bacterium interactions in other organisms based on the literature. All annotations we included are given in Table S2. The organisms we used for comparison and their associated proteome accession numbers can be found in Table S1d. Proteins with structures that did not pass the threshold criterion in SMART were removed.
(iv) Multiple-sequence alignments.
We used the alignment tool MUSCLE, provided by EMBL (https://www.ebi.ac.uk/Tools/msa/muscle/), for multiple-sequence alignment of protein sequences. Alignments were verified visually.
TEM.
The trophosome sample for TEM was fixed at room temperature for 1 h in fixative containing 4% paraformaldehyde, 1% glutaraldehyde, 10% sucrose in 50 mM HEPES (glutaraldehyde was added directly before use) and stored at 4°C. The sample was washed three times with washing buffer (100 mM cacodylate buffer [pH 7.0], 1 mM CaCl2, 0.09 M sucrose) for 10 min each step and treated with 1% osmium tetroxide in washing buffer for 1 h at room temperature. After three additional washing steps in washing buffer for 10 min each, the sample was dehydrated in a graded series of ethanol (30%, 50%, 70%, 90%, and 100%) on ice for 30 min each step. Afterwards, the material was subjected to stepwise infiltration with the acrylic resin LR White according to Hammerschmidt et al. (135). Sections were cut with a diamond knife on an ultramicrotome (Reichert Ultracut, Leica UK Ltd.), stained with 4% aqueous uranyl acetate for 5 min, and finally examined with a transmission electron microscope (LEO 906; Carl Zeiss Microscopy GmbH) at an acceleration voltage of 80 kV. The micrographs were edited using Adobe Photoshop CS6.
Data availability.
The mass spectrometry proteomics data and the combined host and symbiont database have been deposited to the ProteomeXchange Consortium via the PRIDE (136) partner repository with the data set identifier PXD012439. Transcriptomics raw data have been deposited to the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) with the BioProject accession number PRJNA534438 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA534438/).
ACKNOWLEDGMENTS
We thank the captains and crews of RV Atlantis, DSV Alvin, and ROV Jason for their excellent support during the cruises AT15-28, AT26-10, AT26-23, and AT37-12, which were funded through grants of the U.S. National Science Foundation. We are grateful to Ruby Ponnudurai for sampling, to Jana Matulla and Annette Meuche for excellent technical assistance, to Marc Strous for supporting this project by providing access to the proteomics equipment, to Xiaoli Dong for help with database annotations, and to Maryam Ataeian, Jackie Zorz, and Angela Kouris for help with MS measurements. Sandy Gerschler did preliminary SMART analyses. Marie-Katherin Zühlke provided helpful feedback on figure generation. Målin Tietjen and Lizbeth Sayavedra gave valuable input for RNA sample preparation.
This work was supported by the German Research Foundation DFG (grant MA 6346/2-1 to S.M., grant BR 5488/1-1 to C.B.), the German Academic Exchange Service DAAD (T.H.), a fellowship of the Institute of Marine Biotechnology Greifswald (T.H.), the Canada Foundation for Innovation, the Government of Alberta and the Natural Sciences and Engineering Research Council of Canada NSERC through a Banting Fellowship (M.K.), the U.S. National Science Foundation (grants OCE-0452333, OCE-1136727, OCE-1131095, and OCE-1559198 to S.M.S.), and the WHOI Investment in Science Fund (S.M.S.). P.R. was supported by a grant from the DFG CCGA Comprehensive Center for Genome Analysis, Kiel, and the DFG CRC1182 “Origin and Function of Metaorganisms.” R.H. and T.B.H.R. were supported by the DFG CRC1182 “Origin and Function of Metaorganisms,” subprojects B2, Z3, and INF.
Footnotes
Citation Hinzke T, Kleiner M, Breusing C, Felbeck H, Häsler R, Sievert SM, Schlüter R, Rosenstiel P, Reusch TBH, Schweder T, Markert S. 2019. Host-microbe interactions in the chemosynthetic Riftia pachyptila symbiosis. mBio 10:e02243-19. https://doi.org/10.1128/mBio.02243-19.
Contributor Information
Daniel Distel, Northeastern University.
Edward G. Ruby, University of Hawaii at Manoa.
REFERENCES
- 1.Bosch TCG, McFall-Ngai MJ. 2011. Metaorganisms as the new frontier. Zoology 114:185–190. doi: 10.1016/j.zool.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bang C, Dagan T, Deines P, Dubilier N, Duschl WJ, Fraune S, Hentschel U, Hirt H, Hülter N, Lachnit T, Picazo D, Pita L, Pogoreutz C, Rädecker N, Saad MM, Schmitz RA, Schulenburg H, Voolstra CR, Weiland-Bräuer N, Ziegler M, Bosch T. 2018. Metaorganisms in extreme environments: do microbes play a role in organismal adaptation? Zoology 127:1–19. doi: 10.1016/j.zool.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 3.McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A 110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moya A, Peretó J, Gil R, Latorre A. 2008. Learning how to live together: genomic insights into prokaryote–animal symbioses. Nat Rev Genet 9:218–229. doi: 10.1038/nrg2319. [DOI] [PubMed] [Google Scholar]
- 5.Feldhaar H, Gross R. 2009. Insects as hosts for mutualistic bacteria. Int J Med Microbiol 299:1–8. doi: 10.1016/j.ijmm.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nyholm SV, Graf J. 2012. Knowing your friends: invertebrate innate immunity fosters beneficial bacterial symbioses. Nat Rev Microbiol 10:815–827. doi: 10.1038/nrmicro2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hentschel U, Steinert M, Hacker J. 2000. Common molecular mechanisms of symbiosis and pathogenesis. Trends Microbiol 8:226–231. doi: 10.1016/s0966-842x(00)01758-3. [DOI] [PubMed] [Google Scholar]
- 8.Dale C, Moran NA. 2006. Molecular interactions between bacterial symbionts and their hosts. Cell 126:453–465. doi: 10.1016/j.cell.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 9.McFall-Ngai M. 2008. Are biologists in “future shock”? Symbiosis integrates biology across domains. Nat Rev Microbiol 6:789–792. doi: 10.1038/nrmicro1982. [DOI] [PubMed] [Google Scholar]
- 10.Webster NS. 2014. Cooperation, communication, and co-evolution: grand challenges in microbial symbiosis research. Front Microbiol 5:164. doi: 10.3389/fmicb.2014.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubilier N, Bergin C, Lott C. 2008. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat Rev Microbiol 6:725–740. doi: 10.1038/nrmicro1992. [DOI] [PubMed] [Google Scholar]
- 12.Felbeck H. 1981. Chemoautotrophic potential of the hydrothermal vent tube worm, Riftia pachyptila Jones (Vestimentifera). Science 213:336–338. doi: 10.1126/science.213.4505.336. [DOI] [PubMed] [Google Scholar]
- 13.Cavanaugh CM, Gardiner SL, Jones ML, Jannasch HW, Waterbury JB. 1981. Prokaryotic cells in the hydrothermal vent tube worm Riftia pachyptila Jones: possible chemoautotrophic symbionts. Science 213:340–342. doi: 10.1126/science.213.4505.340. [DOI] [PubMed] [Google Scholar]
- 14.Hand SC. 1987. Trophosome ultrastructure and the characterization of isolated bacteriocytes from invertebrate-sulfur bacteria symbioses. Biol Bull 173:260–276. doi: 10.2307/1541878. [DOI] [PubMed] [Google Scholar]
- 15.Felbeck H, Childress JJ, Somero GN. 1981. Calvin-Benson cycle and sulphide oxidation enzymes in animals from sulphide-rich habitats. Nature 293:291–293. doi: 10.1038/293291a0. [DOI] [Google Scholar]
- 16.Stewart FJ, Cavanaugh CM. 2005. Symbiosis of thioautotrophic bacteria with Riftia pachyptila, p 197–225. In Overmann J. (ed), Molecular basis of symbiosis. Progress in molecular and subcellular biology, vol 41 Springer, Berlin, Germany. [DOI] [PubMed] [Google Scholar]
- 17.Lutz RA, Shank TM, Fornari DJ, Haymon RM, Lilley MD, Von Damm KL, Desbruyeres D. 1994. Rapid growth at deep-sea vents. Nature 371:663–664. doi: 10.1038/371663a0. [DOI] [Google Scholar]
- 18.Jones ML. 1981. Riftia pachyptila Jones: observations on the vestimentiferan worm from the Galápagos Rift. Science 213:333–336. doi: 10.1126/science.213.4505.333. [DOI] [PubMed] [Google Scholar]
- 19.Robidart JC, Bench SR, Feldman RA, Novoradovsky A, Podell SB, Gaasterland T, Allen EE, Felbeck H. 2008. Metabolic versatility of the Riftia pachyptila endosymbiont revealed through metagenomics. Environ Microbiol 10:727–737. doi: 10.1111/j.1462-2920.2007.01496.x. [DOI] [PubMed] [Google Scholar]
- 20.Polzin J, Arevalo P, Nussbaumer T, Polz MF, Bright M. 2019. Polyclonal symbiont populations in hydrothermal vent tubeworms and the environment. Proc Biol Sci 286:20181281. doi: 10.1098/rspb.2018.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Distel DL, Lane DJ, Olsen GJ, Giovannoni SJ, Pace B, Pace NR, Stahl DA, Felbeck H. 1988. Sulfur-oxidizing bacterial endosymbionts: analysis of phylogeny and specificity by 16S rRNA sequences. J Bacteriol 170:2506–2510. doi: 10.1128/jb.170.6.2506-2510.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dover CLV. 2000. The ecology of deep-sea hydrothermal vents. Princeton University Press, Princeton, NJ. [Google Scholar]
- 23.Fisher CR, Childress JJ, Minnich E. 1989. Autotrophic carbon fixation by the chemoautotrophic symbionts of Riftia pachyptila. Biol Bull 177:372–385. doi: 10.2307/1541597. [DOI] [Google Scholar]
- 24.Robidart JC, Roque A, Song P, Girguis PR. 2011. Linking hydrothermal geochemistry to organismal physiology: physiological versatility in Riftia pachyptila from sedimented and basalt-hosted vents. PLoS One 6:e21692. doi: 10.1371/journal.pone.0021692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markert S, Gardebrecht A, Felbeck H, Sievert SM, Klose J, Becher D, Albrecht D, Thürmer A, Daniel R, Kleiner M, Hecker M, Schweder T. 2011. Status quo in physiological proteomics of the uncultured Riftia pachyptila endosymbiont. Proteomics 11:3106–3117. doi: 10.1002/pmic.201100059. [DOI] [PubMed] [Google Scholar]
- 26.Petersen JM, Zielinski FU, Pape T, Seifert R, Moraru C, Amann R, Hourdez S, Girguis PR, Wankel SD, Barbe V, Pelletier E, Fink D, Borowski C, Bach W, Dubilier N. 2011. Hydrogen is an energy source for hydrothermal vent symbioses. Nature 476:176–180. doi: 10.1038/nature10325. [DOI] [PubMed] [Google Scholar]
- 27.Pflugfelder B, Fisher CR, Bright M. 2005. The color of the trophosome: elemental sulfur distribution in the endosymbionts of Riftia pachyptila (Vestimentifera; Siboglinidae). Mar Biol 146:895–901. doi: 10.1007/s00227-004-1500-x. [DOI] [Google Scholar]
- 28.Wilmot DB Jr, Vetter RD. 1990. The bacterial symbiont from the hydrothermal vent tubeworm Riftia pachyptila is a sulfide specialist. Mar Biol 106:273–283. doi: 10.1007/BF01314811. [DOI] [Google Scholar]
- 29.Scott KM, Boller AJ, Dobrinski KP, Le Bris N. 2012. Response of hydrothermal vent vestimentiferan Riftia pachyptila to differences in habitat chemistry. Mar Biol 159:435–442. doi: 10.1007/s00227-011-1821-5. [DOI] [Google Scholar]
- 30.Liu H, Wang H, Cai S, Zhang H. 2017. A novel ω3-desaturase in the deep sea giant tubeworm Riftia pachyptila. Mar Biotechnol 19:345–350. doi: 10.1007/s10126-017-9753-9. [DOI] [PubMed] [Google Scholar]
- 31.Drozdov AL, Galkin SV. 2012. Morphology of gametes and insemination in the vestimentiferan Riftia pachyptila. OJMS 02:96–102. doi: 10.4236/ojms.2012.23013. [DOI] [Google Scholar]
- 32.Sanchez S, Hourdez S, Lallier FH. 2007. Identification of proteins involved in the functioning of Riftia pachyptila symbiosis by Subtractive Suppression Hybridization. BMC Genomics 8:337. doi: 10.1186/1471-2164-8-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bright M, Lallier FH. 2010. The biology of vestimentiferan tubeworms. Oceanogr Mar Biol 48:213–266. doi: 10.1201/EBK1439821169-c4. [DOI] [Google Scholar]
- 34.Gardebrecht A, Markert S, Sievert SM, Felbeck H, Thürmer A, Albrecht D, Wollherr A, Kabisch J, Le Bris N, Lehmann R, Daniel R, Liesegang H, Hecker M, Schweder T. 2012. Physiological homogeneity among the endosymbionts of Riftia pachyptila and Tevnia jerichonana revealed by proteogenomics. ISME J 6:766–776. doi: 10.1038/ismej.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Markert S, Arndt C, Felbeck H, Becher D, Sievert SM, Hügler M, Albrecht D, Robidart J, Bench S, Feldman RA, Hecker M, Schweder T. 2007. Physiological proteomics of the uncultured endosymbiont of Riftia pachyptila. Science 315:247–250. doi: 10.1126/science.1132913. [DOI] [PubMed] [Google Scholar]
- 36.Bright M, Keckeis H, Fisher CR. 2000. An autoradiographic examination of carbon fixation, transfer and utilization in the Riftia pachyptila symbiosis. Mar Biol 136:621–632. doi: 10.1007/s002270050722. [DOI] [Google Scholar]
- 37.Pflugfelder B, Cary SC, Bright M. 2009. Dynamics of cell proliferation and apoptosis reflect different life strategies in hydrothermal vent and cold seep vestimentiferan tubeworms. Cell Tissue Res 337:149–165. doi: 10.1007/s00441-009-0811-0. [DOI] [PubMed] [Google Scholar]
- 38.Boetius A, Felbeck H. 1995. Digestive enzymes in marine invertebrates from hydrothermal vents and other reducing environments. Mar Biol 122:105–113. doi: 10.1007/BF00349283. [DOI] [Google Scholar]
- 39.Ponnudurai R, Kleiner M, Sayavedra L, Petersen JM, Moche M, Otto A, Becher D, Takeuchi T, Satoh N, Dubilier N, Schweder T, Markert S. 2017. Metabolic and physiological interdependencies in the Bathymodiolus azoricus symbiosis. ISME J 11:463–477. doi: 10.1038/ismej.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Streams ME, Fisher CR, Fiala-Médioni A. 1997. Methanotrophic symbiont location and fate of carbon incorporated from methane in a hydrocarbon seep mussel. Mar Biol 129:465–476. doi: 10.1007/s002270050187. [DOI] [Google Scholar]
- 41.König S, Le Guyader H, Gros O. 2015. Thioautotrophic bacterial endosymbionts are degraded by enzymatic digestion during starvation: case study of two lucinids Codakia orbicularis and C. orbiculata. Microsc Res Tech 78:173–179. doi: 10.1002/jemt.22458. [DOI] [PubMed] [Google Scholar]
- 42.Caro A, Got P, Bouvy M, Troussellier M, Gros O. 2009. Effects of long-term starvation on a host bivalve (Codakia orbicularis, Lucinidae) and its symbiont population. Appl Environ Microbiol 75:3304–3313. doi: 10.1128/AEM.02659-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wippler J, Kleiner M, Lott C, Gruhl A, Abraham PE, Giannone RJ, Young JC, Hettich RL, Dubilier N. 2016. Transcriptomic and proteomic insights into innate immunity and adaptations to a symbiotic lifestyle in the gutless marine worm Olavius algarvensis. BMC Genomics 17:942. doi: 10.1186/s12864-016-3293-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woyke T, Teeling H, Ivanova NN, Huntemann M, Richter M, Gloeckner FO, Boffelli D, Anderson IJ, Barry KW, Shapiro HJ, Szeto E, Kyrpides NC, Mussmann M, Amann R, Bergin C, Ruehland C, Rubin EM, Dubilier N. 2006. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature 443:950–955. doi: 10.1038/nature05192. [DOI] [PubMed] [Google Scholar]
- 45.Hyttinen JMT, Niittykoski M, Salminen A, Kaarniranta K. 2013. Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim Biophys Acta 1833:503–510. doi: 10.1016/j.bbamcr.2012.11.018. [DOI] [PubMed] [Google Scholar]
- 46.Chavrier P, Parton R, Hauri H, Simons K, Zerial M. 1990. Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell 62:317–329. doi: 10.1016/0092-8674(90)90369-p. [DOI] [PubMed] [Google Scholar]
- 47.Vieira OV, Botelho RJ, Grinstein S. 2002. Phagosome maturation: aging gracefully. Biochem J 366:689–704. doi: 10.1042/BJ20020691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marchetti A, Mercanti V, Cornillon S, Alibaud L, Charette SJ, Cosson P. 2004. Formation of multivesicular endosomes in Dictyostelium. J Cell Sci 117:6053–6059. doi: 10.1242/jcs.01524. [DOI] [PubMed] [Google Scholar]
- 49.Bright M, Sorgo A. 2003. Ultrastructural reinvestigation of the trophosome in adults of Riftia pachyptila (Annelida, Siboglinidae). Invertebr Biol 122:345–366. doi: 10.1111/j.1744-7410.2003.tb00099.x. [DOI] [Google Scholar]
- 50.Simonet P, Gaget K, Balmand S, Ribeiro Lopes M, Parisot N, Buhler K, Duport G, Vulsteke V, Febvay G, Heddi A, Charles H, Callaerts P, Calevro F. 2018. Bacteriocyte cell death in the pea aphid/Buchnera symbiotic system. Proc Natl Acad Sci U S A 115:E1819–E1828. doi: 10.1073/pnas.1720237115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bröker LE, Huisman C, Span SW, Rodriguez A, Kruyt FA, Giaccone G. 2004. Cathepsin B mediates caspase-independent cell death induced by microtubule stabilizing agents in non-small cell lung cancer cells. Cancer Res 64:27–30. doi: 10.1158/0008-5472.can-03-3060. [DOI] [PubMed] [Google Scholar]
- 52.Man SM, Kanneganti T-D. 2016. Regulation of lysosomal dynamics and autophagy by CTSB/cathepsin B. Autophagy 12:2504–2505. doi: 10.1080/15548627.2016.1239679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Felbeck H, Jarchow J. 1998. Carbon release from purified chemoautotrophic bacterial symbionts of the hydrothermal vent tubeworm Riftia pachyptila. Physiol Zool 71:294–302. doi: 10.1086/515931. [DOI] [PubMed] [Google Scholar]
- 54.Zal F, Lallier FH, Green BN, Vinogradov SN, Toulmond A. 1996. The multi-hemoglobin system of the hydrothermal vent tube worm Riftia pachyptila. II. Complete polypeptide chain composition investigated by maximum entropy analysis of mass spectra. J Biol Chem 271:8875–8881. doi: 10.1074/jbc.271.15.8875. [DOI] [PubMed] [Google Scholar]
- 55.Zal F, Leize E, Lallier FH, Toulmond A, Van Dorsselaer A, Childress J. 1998. S-sulfohemoglobin and disulfide exchange: the mechanisms of sulfide binding by Riftia pachyptila hemoglobins. Proc Natl Acad Sci U S A 95:8997–9002. doi: 10.1073/pnas.95.15.8997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flores JF, Fisher CR, Carney SL, Green BN, Freytag JK, Schaeffer SW, Royer WE. 2005. Sulfide binding is mediated by zinc ions discovered in the crystal structure of a hydrothermal vent tubeworm hemoglobin. Proc Natl Acad Sci U S A 102:2713–2718. doi: 10.1073/pnas.0407455102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arp AJ, Childress JJ, Vetter RD. 1987. The sulphide-binding protein in the blood of the vestimentiferan tube-worm, Riftia pachyptila, is the extracellular haemoglobin. J Exp Biol 128:139–158. [Google Scholar]
- 58.Bailly X, Vinogradov S. 2005. The sulfide binding function of annelid hemoglobins: relic of an old biosystem? J Inorg Biochem 99:142–150. doi: 10.1016/j.jinorgbio.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 59.Hourdez S, Weber RE. 2005. Molecular and functional adaptations in deep-sea hemoglobins. J Inorg Biochem 99:130–141. doi: 10.1016/j.jinorgbio.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 60.Hentschel U, Felbeck H. 1993. Nitrate respiration in the hydrothermal vent tubeworm Riftia pachyptila. Nature 366:338–340. doi: 10.1038/366338a0. [DOI] [Google Scholar]
- 61.Pitcher RS, Brittain T, Watmough NJ. 2002. Cytochrome cbb3 oxidase and bacterial microaerobic metabolism. Biochem Soc Trans 30:653–658. doi: 10.1042/bst0300653. [DOI] [PubMed] [Google Scholar]
- 62.Mangum CP. 1992. Physiological function of the hemerythrins, p 173–192. In Mangum CP. (ed), Blood and tissue oxygen carriers. Advances in comparative and environmental physiology, vol 13 Springer, Berlin, Germany. [Google Scholar]
- 63.De Cian M-C, Andersen AC, Bailly X, Lallier FH. 2003. Expression and localization of carbonic anhydrase and ATPases in the symbiotic tubeworm Riftia pachyptila. J Exp Biol 206:399–409. doi: 10.1242/jeb.00074. [DOI] [PubMed] [Google Scholar]
- 64.Goffredi SK, Girguis PR, Childress JJ, Desaulniers NT. 1999. Physiological functioning of carbonic anhydrase in the hydrothermal vent tubeworm Riftia pachyptila. Biol Bull 196:257–264. doi: 10.2307/1542950. [DOI] [PubMed] [Google Scholar]
- 65.Sanchez S, Andersen AC, Hourdez S, Lallier FH. 2007. Identification, sequencing, and localization of a new carbonic anhydrase transcript from the hydrothermal vent tubeworm Riftia pachyptila. FEBS J 274:5311–5324. doi: 10.1111/j.1742-4658.2007.06050.x. [DOI] [PubMed] [Google Scholar]
- 66.De Cian M-C, Andersen AC, Toullec J-Y, Biegala I, Caprais J-C, Shillito B, Lallier FH. 2003. Isolated bacteriocyte cell suspensions from the hydrothermal-vent tubeworm Riftia pachyptila, a potent tool for cellular physiology in a chemoautotrophic symbiosis. Mar Biol 142:141–151. doi: 10.1007/s00227-002-0931-5. [DOI] [Google Scholar]
- 67.Felbeck H. 1985. CO2 fixation in the hydrothermal vent tube worm Riftia pachyptila (Jones). Physiol Zool 58:272–281. doi: 10.1086/physzool.58.3.30155998. [DOI] [Google Scholar]
- 68.Minic Z, Simon V, Penverne B, Gaill F, Hervé G. 2001. Contribution of the bacterial endosymbiont to the biosynthesis of pyrimidine nucleotides in the deep-sea tube worm Riftia pachyptila. J Biol Chem 276:23777–23784. doi: 10.1074/jbc.M102249200. [DOI] [PubMed] [Google Scholar]
- 69.Minic Z, Hervé G. 2003. Arginine metabolism in the deep sea tube worm Riftia pachyptila and its bacterial endosymbiont. J Biol Chem 278:40527–40533. doi: 10.1074/jbc.M307835200. [DOI] [PubMed] [Google Scholar]
- 70.Brennan LJ, Keddie BA, Braig HR, Harris HL. 2008. The endosymbiont Wolbachia pipientis induces the expression of host antioxidant proteins in an Aedes albopictus cell line. PLoS One 3:e2083. doi: 10.1371/journal.pone.0002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zug R, Hammerstein P. 2015. Wolbachia and the insect immune system: what reactive oxygen species can tell us about the mechanisms of Wolbachia-host interactions. Front Microbiol 6:1201. doi: 10.3389/fmicb.2015.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hourdez S, Lallier FH. 2007. Adaptations to hypoxia in hydrothermal-vent and cold-seep invertebrates. Rev Environ Sci Biotechnol 6:143–159. doi: 10.1007/s11157-006-9110-3. [DOI] [Google Scholar]
- 73.Fields JHA, Quinn JF. 1981. Some theoretical considerations on cytosolic redox balance during anaerobiosis in marine invertebrates. J Theor Biol 88:35–45. doi: 10.1016/0022-5193(81)90327-1. [DOI] [Google Scholar]
- 74.Arndt C, Schiedek D, Felbeck H. 1998. Metabolic responses of the hydrothermal vent tube worm Riftia pachyptila to severe hypoxia. Mar Ecol Prog Ser 174:151–158. doi: 10.3354/meps174151. [DOI] [Google Scholar]
- 75.Elsbach P, Weiss J. 1998. Role of the bactericidal/permeability-increasing protein in host defence. Curr Opin Immunol 10:45–49. doi: 10.1016/s0952-7915(98)80030-7. [DOI] [PubMed] [Google Scholar]
- 76.Chen F, Krasity BC, Peyer SM, Koehler S, Ruby EG, Zhang X, McFall-Ngai MJ. 2017. Bactericidal permeability-increasing proteins shape host-microbe interactions. mBio 8:e00040-17. doi: 10.1128/mBio.00040-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Van Loon CL, Van Strien EA. 1999. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol 55:85–97. doi: 10.1006/pmpp.1999.0213. [DOI] [Google Scholar]
- 78.Asojo OA, Goud G, Dhar K, Loukas A, Zhan B, Deumic V, Liu S, Borgstahl GEO, Hotez PJ. 2005. X-ray structure of Na-ASP-2, a pathogenesis-related-1 protein from the nematode parasite, Necator americanus, and a vaccine antigen for human hookworm infection. J Mol Biol 346:801–814. doi: 10.1016/j.jmb.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 79.Eberle HB, Serrano RL, Füllekrug J, Schlosser A, Lehmann WD, Lottspeich F, Kaloyanova D, Wieland FT, Helms JB. 2002. Identification and characterization of a novel human plant pathogenesis-related protein that localizes to lipid-enriched microdomains in the Golgi complex. J Cell Sci 115:827–838. [DOI] [PubMed] [Google Scholar]
- 80.Park IY, Park CB, Kim MS, Kim SC. 1998. Parasin I, an antimicrobial peptide derived from histone H2A in the cat fish, Parasilurus asotus. FEBS Lett 437:258–262. doi: 10.1016/S0014-5793(98)01238-1. [DOI] [PubMed] [Google Scholar]
- 81.Cho JH, Sung BH, Kim SC. 2009. Buforins: histone H2A-derived antimicrobial peptides from toad stomach. Biochim Biophys Acta 1788:1564–1569. doi: 10.1016/j.bbamem.2008.10.025. [DOI] [PubMed] [Google Scholar]
- 82.Rose F, Bailey K, Keyte JW, Chan WC, Greenwood D, Mahida YR. 1998. Potential role of epithelial cell-derived histone H1 proteins in innate antimicrobial defense in the human gastrointestinal tract. Infect Immun 66:3255–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang G, Li X, Wang Z. 2016. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res 44:D1087–D1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brogden KA. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 85.Login FH, Balmand S, Vallier A, Vincent-Monégat C, Vigneron A, Weiss-Gayet M, Rochat D, Heddi A. 2011. Antimicrobial peptides keep insect endosymbionts under control. Science 334:362–365. doi: 10.1126/science.1209728. [DOI] [PubMed] [Google Scholar]
- 86.Bishop BM, Juba ML, Russo PS, Devine M, Barksdale SM, Scott S, Settlage R, Michalak P, Gupta K, Vliet K, Schnur JM, van Hoek ML. 2017. Discovery of novel antimicrobial peptides from Varanus komodoensis (Komodo dragon) by large-scale analyses and de-novo-assisted sequencing using electron-transfer dissociation mass spectrometry. J Proteome Res 16:1470–1482. doi: 10.1021/acs.jproteome.6b00857. [DOI] [PubMed] [Google Scholar]
- 87.Li J, Mahajan A, Tsai M-D. 2006. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry 45:15168–15178. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- 88.Nguyen M, Liu M, Thomas T. 2014. Ankyrin-repeat proteins from sponge symbionts modulate amoebal phagocytosis. Mol Ecol 23:1635–1645. doi: 10.1111/mec.12384. [DOI] [PubMed] [Google Scholar]
- 89.Pan X, Lührmann A, Satoh A, Laskowski-Arce M, Roy CR. 2008. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320:1651–1654. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reynolds D, Thomas T. 2016. Evolution and function of eukaryotic-like proteins from sponge symbionts. Mol Ecol 25:5242–5253. doi: 10.1111/mec.13812. [DOI] [PubMed] [Google Scholar]
- 91.Prota AE, Magiera MM, Kuijpers M, Bargsten K, Frey D, Wieser M, Jaussi R, Hoogenraad CC, Kammerer RA, Janke C, Steinmetz MO. 2013. Structural basis of tubulin tyrosination by tubulin tyrosine ligase. J Cell Biol 200:259–270. doi: 10.1083/jcb.201211017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Coates JC. 2003. Armadillo repeat proteins: beyond the animal kingdom. Trends Cell Biol 13:463–471. doi: 10.1016/s0962-8924(03)00167-3. [DOI] [PubMed] [Google Scholar]
- 93.Yoshimura SH, Hirano T. 2016. HEAT repeats–versatile arrays of amphiphilic helices working in crowded environments? J Cell Sci 129:3963–3970. doi: 10.1242/jcs.185710. [DOI] [PubMed] [Google Scholar]
- 94.Nyholm SV, Stewart JJ, Ruby EG, McFall-Ngai MJ. 2009. Recognition between symbiotic Vibrio fischeri and the haemocytes of Euprymna scolopes. Environ Microbiol 11:483–493. doi: 10.1111/j.1462-2920.2008.01788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Leighton TL, Buensuceso RNC, Howell PL, Burrows LL. 2015. Biogenesis of Pseudomonas aeruginosa type IV pili and regulation of their function. Environ Microbiol 17:4148–4163. doi: 10.1111/1462-2920.12849. [DOI] [PubMed] [Google Scholar]
- 96.Stone BJ, Kwaik Y. 1999. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J Bacteriol 181:1395–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hager AJ, Bolton DL, Pelletier MR, Brittnacher MJ, Gallagher LA, Kaul R, Skerrett SJ, Miller SI, Guina T. 2006. Type IV pili-mediated secretion modulates Francisella virulence. Mol Microbiol 62:227–237. doi: 10.1111/j.1365-2958.2006.05365.x. [DOI] [PubMed] [Google Scholar]
- 98.Davidson SK, Dulla GF, Go RA, Stahl DA, Pinel N. 2014. Earthworm symbiont Verminephrobacter eiseniae mediates natural transformation within host egg capsules using type IV pili. Front Microbiol 5:546. doi: 10.3389/fmicb.2014.00546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pollack-Berti A, Wollenberg MS, Ruby EG. 2010. Natural transformation of Vibrio fischeri requires tfoX and tfoY. Environ Microbiol 12:2302–2311. doi: 10.1111/j.1462-2920.2010.02250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stewart FJ, Newton ILG, Cavanaugh CM. 2005. Chemosynthetic endosymbioses: adaptations to oxic-anoxic interfaces. Trends Microbiol 13:439–448. doi: 10.1016/j.tim.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 101.Gilbert SF, Sapp J, Tauber AI. 2012. A symbiotic view of life: we have never been individuals. Q Rev Biol 87:325–341. doi: 10.1086/668166. [DOI] [PubMed] [Google Scholar]
- 102.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 103.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Egas C, Pinheiro M, Gomes P, Barroso C, Bettencourt R. 2012. The transcriptome of Bathymodiolus azoricus gill reveals expression of genes from endosymbionts and free-living deep-sea bacteria. Mar Drugs 10:1765–1783. doi: 10.3390/md10081765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schmieder R, Edwards R. 2011. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS One 6:e17288. doi: 10.1371/journal.pone.0017288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren B, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 110.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 113.Eddy SR. 2009. A new generation of homology search tools based on probabilistic inference. Genome Inform 23:205–211. doi: 10.1142/9781848165632_0019. [DOI] [PubMed] [Google Scholar]
- 114.Huang Y, Niu B, Gao Y, Fu L, Li W. 2010. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kleiner M, Dong X, Hinzke T, Wippler J, Thorson E, Mayer B, Strous M. 2018. Metaproteomics method to determine carbon sources and assimilation pathways of species in microbial communities. Proc Natl Acad Sci U S A 115:E5576–E5584. doi: 10.1073/pnas.1722325115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.The Global Proteome Machine Organization. 2012. The global proteome machine: cRAP protein sequences. https://www.thegpm.org/crap/.
- 117.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. 2009. Universal sample preparation method for proteome analysis. Nat Methods 6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 118.Hamann E, Gruber-Vodicka H, Kleiner M, Tegetmeyer HE, Riedel D, Littmann S, Chen J, Milucka J, Viehweger B, Becker KW, Dong X, Stairs CW, Hinrichs K-U, Brown MW, Roger AJ, Strous M. 2016. Environmental Breviatea harbour mutualistic Arcobacter epibionts. Nature 534:254–258. doi: 10.1038/nature18297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hinzke T, Kouris A, Hughes R-A, Strous M, Kleiner M. 2019. More is not always better: evaluation of 1D and 2D-LC-MS/MS methods for metaproteomics. Front Microbiol 10:238. doi: 10.3389/fmicb.2019.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Oberg AL, Vitek O. 2009. Statistical design of quantitative mass spectrometry-based proteomic profiling experiments. J Proteome Res 8:2144–2156. doi: 10.1021/pr8010099. [DOI] [PubMed] [Google Scholar]
- 121.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. 2006. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res 5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 122.Mueller RS, Denef VJ, Kalnejais LH, Suttle KB, Thomas BC, Wilmes P, Smith RL, Nordstrom DK, McCleskey RB, Shah MB, VerBerkmoes NC, Hettich RL, Banfield JF. 2010. Ecological distribution and population physiology defined by proteomics in a natural microbial community. Mol Syst Biol 6:374. doi: 10.1038/msb.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.R Core Team. 2017. R: a language and environment for statistical computing. http://www.R-project.org/. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 125.Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 126.Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H. 2014. UpSet: visualization of intersecting sets. IEEE Trans Vis Comput Graph 20:1983–1992. doi: 10.1109/TVCG.2014.2346248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kleiner M, Thorson E, Sharp CE, Dong X, Liu D, Li C, Strous M. 2017. Assessing species biomass contributions in microbial communities via metaproteomics. Nat Commun 8:1558. doi: 10.1038/s41467-017-01544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.The UniProt Consortium. 2017. UniProt: the universal protein knowledgebase. Nucleic Acids Res 45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang H-Y, Dosztányi Z, El-Gebali S, Fraser M, Gough J, Haft D, Holliday GL, Huang H, Huang X, Letunic I, Lopez R, Lu S, Marchler-Bauer A, Mi H, Mistry J, Natale DA, Necci M, Nuka G, Orengo CA, Park Y, Pesseat S, Piovesan D, Potter SC, Rawlings ND, Redaschi N, Richardson L, Rivoire C, Sangrador-Vegas A, Sigrist C, Sillitoe I, Smithers B, Squizzato S, Sutton G, Thanki N, Thomas PD, Tosatto SCE, Wu CH, Xenarios I, Yeh L-S, Young S-Y, Mitchell AL. 2017. InterPro in 2017–beyond protein family and domain annotations. Nucleic Acids Res 45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Elbourne LDH, Tetu SG, Hassan KA, Paulsen IT. 2017. TransportDB 2.0: a database for exploring membrane transporters in sequenced genomes from all domains of life. Nucleic Acids Res 45:D320–D324. doi: 10.1093/nar/gkw1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Saier MH Jr, Reddy VS, Tsu BV, Ahmed MS, Li C, Moreno-Hagelsieb G. 2016. The transporter classification database (TCDB): recent advances. Nucleic Acids Res 44:D372–D379. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chang Y-Y, Neufeld TP. 2010. Autophagy takes flight in Drosophila. FEBS Lett 584:1342–1349. doi: 10.1016/j.febslet.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Letunic I, Bork P. 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46:D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hammerschmidt S, Wolff S, Hocke A, Rosseau S, Müller E, Rohde M. 2005. Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect Immun 73:4653–4667. doi: 10.1128/IAI.73.8.4653-4667.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu Q-W, Wang R, Hermjakob H. 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Labonté JM, Pachiadaki M, Fergusson E, McNichol J, Grosche A, Gulmann LK, Vetriani C, Sievert SM, Stepanauskas R. 2019. Single cell genomics-based analysis of gene content and expression of prophages in a diffuse-flow deep-sea hydrothermal system. Front Microbiol 10:1262. doi: 10.3389/fmicb.2019.01262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Results and Discussion with supplemental figures. Download Text S1, PDF file, 1.9 MB (2.1MB, pdf) .
Copyright © 2019 Hinzke et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(a) All Riftia host proteins identified in this study. (b) Potential autophagy-related Riftia proteins. (c) Potential Riftia antimicrobial peptides. (d) Genomes and metagenomes used for SMART analysis of eukaryote-like protein structures. (e) Sampling dates, cruise number, and number of biological replicates of Riftia samples used in this study. (f) Number of proteins with significant abundance differences in pairwise comparisons of Riftia tissues. (g) All Riftia symbiont proteins identified in this study. (h) Hemerythrin and myohemerythrin isoforms in Riftia and other invertebrates used for alignment in Text S1, Fig. S3. (i) Carbonic anhydrase isoforms in Riftia as detected in this study and described in the literature. Download Table S1, XLSX file, 1.8 MB (1.8MB, xlsx) .
Copyright © 2019 Hinzke et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Domains and protein families with a putative role in host-symbiont interactions. Download Table S2, PDF file, 0.4 MB (414.9KB, pdf) .
Copyright © 2019 Hinzke et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Transcriptome completeness for the four different Riftia transcriptome assemblies based on the BUSCO eukaryote and metazoan datasets. Download Table S3, PDF file, 0.3 MB (333.7KB, pdf) .
Copyright © 2019 Hinzke et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Tools used to characterize Riftia host and symbiont proteins included in the combined Riftia host and symbiont database used in this study. Download Table S4, PDF file, 0.4 MB (383.4KB, pdf) .
Copyright © 2019 Hinzke et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The mass spectrometry proteomics data and the combined host and symbiont database have been deposited to the ProteomeXchange Consortium via the PRIDE (136) partner repository with the data set identifier PXD012439. Transcriptomics raw data have been deposited to the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) with the BioProject accession number PRJNA534438 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA534438/).