Abstract
Bacteria and archaea use CRISPR-Cas adaptive immune systems to destroy complementary nucleic acids using RNAs derived from CRISPR loci. Here, we provide the first functional evidence for type IV CRISPR-Cas, demonstrating that the system from Pseudomonas aeruginosa strain PA83 mediates RNA-guided interference against a plasmid in vivo, both clearing the plasmid and inhibiting its uptake. This interference depends on the putative NTP-dependent helicase activity of Csf4/DinG.
Introduction
Prokaryotes use small non-coding RNAs derived from CRISPR arrays to guide Cas proteins to invasive complementary nucleic acids such as phage and plasmid DNA for destruction.1–5
Computational analyses have grouped CRISPR-Cas systems into two classes, six types, and at least 33 subtypes.6 The function of class 1, type IV systems, remains largely unknown, in part because this system is comparatively rare and considered a minimal CRISPR-Cas variant.6 All type IV systems contain genes predicted to encode a multi-subunit complex composed of large (csf1, cas8-like), backbone (csf2, cas7-like), and tail subunits (csf3, cas5-like). To date, type IV systems have been subdivided into IV-A and IV-B subtypes. Type IV-A systems contain genes that encode for a putative helicase (dinG) and a cas6-like endonuclease, whereas type IV-B system lack these genes and are often missing a CRISPR locus.6 It has been shown that the type IV-A Cas6 from Mahella australiensis and a Cas6-like homolog, Csf5 from Aromatoleum aromaticum, are involved in biogenesis of CRISPR-derived RNAs (crRNA).7,8 Additionally, the type IV-A proteins from A. aromaticum (AaCsf1, AaCsf2, AaCsf3, and AaCsf5) were shown to form a complex that assembles on a crRNA.8 It has been hypothesized that the type IV-A ribonucleoprotein complex enables RNA-guided target detection, the function of which remains unknown.8 Here, we investigate the type IV CRISPR-Cas systems in Pseudomonas aeruginosa and demonstrate that a type IV-A system mediates interference against plasmids in vivo.
Materials and Methods
Bioinformatic analysis
A Psi-Blast was performed on January 8, 2018, on the NCBI protein database with the seed WP_018940624.1 (Csf1), WP_013006553.1 (Csf2), and WP_018940624.1 (Csf3) of Thioalkalivibrio sp. K90Mix. Csf1, Csf2, and Csf3 returned 389, 113, and 737 hits, of which 38, 48, and 41 hits were from P. aeruginosa, respectively. We proceeded by downloading amino acid sequences solely from P. aeruginosa for later bioinformatic analysis. Each hit from the Csf1 and Csf3 searches were found in the Csf2 search. In addition, genes upstream and downstream of these hits were downloaded and compared by amino acid identity. Position relative to Csf2 hits was used to compare genes, revealing four conserved architectures. Each system, including intergenic regions and seven genes in each direction of Csf2, in a given architecture was saved for further analysis. In some of these systems, the downstream region of Csf3 contained putative CRISPR arrays already annotated, while the rest did not have annotation. An in-house script was written to search for non-identical repeats, with a search seed of the palindromic region CCCCGC or GCCGCC (available at https://github.com/adamcatching/type_IV_repeat_finder). By accounting for a Hamming distance up to three between nearby putative repeats, unannotated regions were found to have CRISPR arrays. This was assisted with the inference of direction, using the rate of divergence from the consensus sequence over the array to find the conserved orientation between systems. Spacers between repeats were declared to have originated from a Pseudomonas plasmid if they appeared at least once on an annotated plasmid. Spacers were used to search for protospacers that originated from phages/prophages or plasmids. This was done using BLASTn to find matches in prokaryotes, which were then determined to be either located on a plasmid or a prophage (using the online tool PHASTER9). To rule out that BLASTn hits were spacer sequences in CRISPR arrays, the protospacer was confirmed to be within a gene or, where intergenic, confirmed not to neighbor a csf gene.
Accession numbers
NZ_CP017294—P. aeruginosa PA83 plasmid unnamed1
| Annotation | Feature | GenPept | Genomic position |
|---|---|---|---|
| Putative ATP-dependent DNA helicase DinG | Csf4/DinG | PSA83_06667 | 299162..301341 |
| Hypothetical protein | Csf5/Cas6 | PSA83_06666 | 298385..299113 (complement) |
| CRISPR type AFERR-associated protein Csf1 | Csf1 | PSA83_06665 | 297670..298401 (complement) |
| CRISPR type AFERR-associated protein Csf2 | Csf2 | PSA83_06664 | 296619..297665 (complement) |
| CRISPR type AFERR-associated protein Csf3 | Csf3 | PSA83_06663 | 295957..296619 (complement) |
CRISPR array used in plasmid transformation efficiency and maintenance assays
We used the direct repeat found on the PA83 plasmid upstream of spacers that had hits to phage (5′-GTGTTCCCCGCATACGCGGGGGTGAACGG-3′). The CRISPR arrays used in our experiments contain a single spacer flanked by two of the same direct repeats. We evaluated two targeting spacers (TS1 and TS2) and one non-targeting spacer (NTS). The spacer sequences we used in our experiments were TS1: 5′-TGGAGCAACACCTGAAGGAAGGCTTGATGAGC-3′, TS2: 5′-CTCAACCGAGGGTGGTTTTGTCTA-3′, and NTS 5′-CTGAGTGTGATCGATGCCATCAGCGAAGGGCC-3′. The targeting spacers TS1 and TS2 target a sequence specific to the CAO1 gene from Oryza sativa (rice). These target sequences were picked for future experiments unrelated to this manuscript.
Plasmids
All P. aeruginosa genes and CRISPRs were synthesized by Twist Bioscience (San Francisco, CA). PaCsf1was codon optimized for synthesis (see DNA sequence in Supplementary Table S1). Primers are listed in Supplementary Table S1. csf1, csf2, and csf5/cas6 were placed on the same plasmid, with ribosome binding sites in between each gene, and initially obtained in the pTWIST CMV expression vector. The CMV enhancer was removed, and a T7 promoter was inserted upstream of the three gene polycistronic block via two separate mutagenesis reactions. The cassette was polymerase chain reaction (PCR) amplified and subcloned into pCDF using NcoI and PacI sites for origin of replication compatibility with other plasmids expressing the system. The CRISPR array with TS1, csf3, and csf4/dinG were synthesized and inserted into the pTWIST Amp High Copy expression vector. The CRISPR was PCR amplified and subcloned into multiple cloning site 1 of pACYC via EcoRI and SacI. csf3 and csf4/dinG were PCR amplified and subcloned into multiple cloning site 2 of pACYC containing the CRISPR via NdeI and EcoRV sites. CRISPR arrays with TS2 and NTS were subcloned into pACYC plasmids containing csf3 and csf4/dinG via EcoRI and SacI. To test each targeting spacer independently, TS1 and TS2 were tested with distinct vectors.
Plasmid transformation efficiency assay
Plasmid transformation efficiency assays were adapted from work previously described.10 In brief, Escherichia coli BL21-AI cells were co-transformed with pCDF-csf1-csf2-csf5/cas6 and pACYC-Pa83CRTS1-csf3-csf4/dinG to reconstitute the components of the PA83 type IV-A1 operon. The type IV system genes and CRISPR were placed on two different plasmids, as has been previously done to express multi-subunit class 1 systems.1,11,12 This strategy makes it easier to clone and later mutate systems containing multiple genes and a CRISPR locus, and reduces the overall size of expression plasmids. This cell line was made chemically competent using standard methods. This cell line was transformed with 25 ng of target or non-target plasmid. The target and non-target plasmid are in pET27b(+), and the non-target differs from the target in that it is truncated and does not contain the protospacer sequence. After transformation with the target or non-target plasmid, the cells were grown overnight at 37°C in 5 mL Luria–Bertani (LB) media supplemented with 1 mM IPTG, 0.2% L-arabinose, and 50, 25, and 50 μg/mL of kanamycin, chloramphenicol, and streptomycin, respectively, shaking at 200 rpm. Serially diluted cells were plated on LB agar plates supplemented with 1 mM IPTG, 0.2% l-arabinose, and 50, 25, and 50 μg/mL of kanamycin, chloramphenicol, and streptomycin respectively. The next day, colony-forming units were counted for analysis. The error bars represent standard error of the mean calculated from three independent transformations. The experiment was performed with three biological replicates that produced the same results.
Plasmid maintenance assay
Plasmid maintenance assays were adapted from work previously described.10,13 We transformed BL21-AI cells to generate individual cell lines expressing either the full or partial type IV-A operons. We transformed these cells lines with an equimolar mixture of target and non-target plasmid. After transformation, the cells were grown for 1 h at 37°C in 500 μL of LB media supplemented with 1 mM IPTG and 0.2% l-arabinose for induction, shaking at 200 rpm. Each experiment contained an uninduced control for comparison, where cells were grown solely in LB. Induced cells were plated on LB agar supplemented with 1 mM IPTG, 0.2% l-arabinose, and 50, 25, and 50 μg/mL of kanamycin, chloramphenicol, and streptomycin, respectively, and uninduced cells were plated on the same LB agar conditions without IPTG or l-arabinose. The next day, 48 colonies were randomly screened for the presence of target or non-target plasmid by colony PCR using forward (5′-GAGTTCTGGCTGGCTAGCC-3′) and reverse (5′-GGAATTGTGAGCGGATAACAA-3′) primers that amplify a 679 bp region of the target or 384 bp of the non-target plasmid. PCR reactions were separated by agarose gel electrophoresis and stained with ethidium bromide. The number of PCR products corresponding to the target and non-target were counted, and the data are expressed as a ratio of target to non-target bands. The error bars represent standard error of the mean calculated from three independent experiments. Values significantly below one indicate CRISPR interference. csf4/dinG mutants were made by site-directed mutagenesis using the pACYC-Pa83CRTS1-csf3-csf4/dinG plasmid as a template. All individual components of the type IV operon were removed from either pCDF-csf1-csf2-csf5/cas6 or pACYC-Pa83CRTS1-csf3-csf4/dinG plasmids via site-directed mutagenesis. All site-directed mutagenesis reactions were performed with the Q5® Site-Directed Mutagenesis Kit (New England BioLabs, Inc., Ipswich, MA). Primers are listed in Supplementary Table S1.
Results
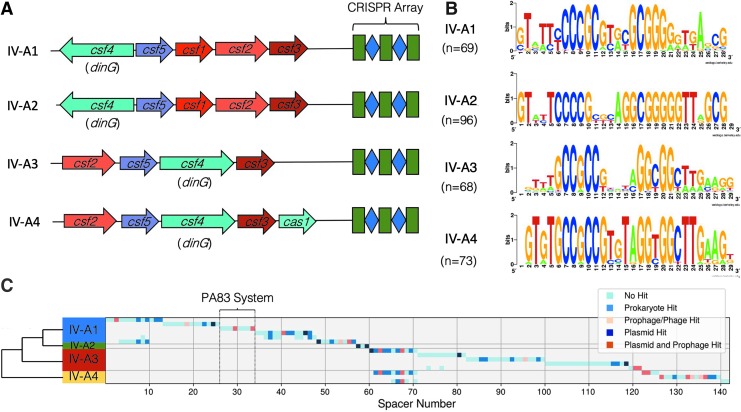
Using the type IV CRISPR-Cas system from Thioalkalivibrio sp. K90Mix14 as a seed, we searched P. aeruginosa genomes for putative type IV-A CRISPR-Cas systems using PSI-BLAST.15 We observed four variants of type IV-A systems based on amino acid sequence identity, gene arrangement, and CRISPR repeat sequence (Fig. 1A, Supplementary Fig. S1, and Supplementary Table S2). Variant systems were grouped based on the shared sequence identity between conserved genes (csf2, csf3, csf4, csf5/cas6; Supplementary Fig. S1). The architecture of the gene position and direction was conserved within each variant (Fig. 1A). Some variants had annotated CRISPR arrays. However, visual inspection of the region downstream of csf3 found unannotated palindromic repeats. An in-house script was written to take mutant repeats into account so that CRISPRs containing repeats that differ slightly in sequence from one another were found. Using Hamming distance of three mismatches between a system's repeats, a total of 335 repeats were found, with their respective consensus sequences shown in Figure 1B.
FIG. 1.
Type IV-A CRISPR-Cas systems found in Pseudomonas aeruginosa. (A) Classification of type IV-A CRISPR-Cas systems found in P. aeruginosa. Conserved architectures of these four variants are observed. All four variants include csf2, csf3, csf4/dinG, and csf5/cas6. Despite the similar architecture between type IV-A1 and type-IV-A2, they represent two separate groups based on differences in direct repeat sequences and amino acid divergence (Supplementary Fig. S1). (B) Consensus sequences for direct repeats found in type IV-A CRISPR arrays in P. aeruginosa. Palindromic regions are in the center of the repeat. (C) Table of spacers found in each system. Systems are grouped by variant, with group 2 consolidated, as all their CRISPR arrays are identical. Spacers are colored based on mapping results according to the legend. A dendrogram was generated based on representative csf2 sequences to illustrate the relationship between variants.
By identifying these repeats, a total of 195 spacers were found, of which 142 were unique (Fig. 1C). While redundant spacers were removed from consideration, all repeats were used to create consensus motifs (Fig. 1B). Out of the 142 spacers, 52 are complementary to putative protospacers. Twenty-five of these protospacers are found in prokaryotes without prophage or plasmid match, while 27 of the protospacers match either plasmids (n = 8), prophages (n = 5), or elements with signatures of both prophages and plasmids (n = 14; Fig. 1C and Supplementary Tables S3 and S4).
Given that type IV systems are often found on plasmids, we selected a plasmid-encoded system from P. aeruginosa strain PA83 as a model. This strain has a CRISPR array with a spacer matching a phage, suggesting it may be active. Because of the large number of anti-CRISPR proteins found in P. aeruginosa,16 we investigated the interference function heterologously in E. coli using distinct plasmid transformation efficiency and maintenance assays.
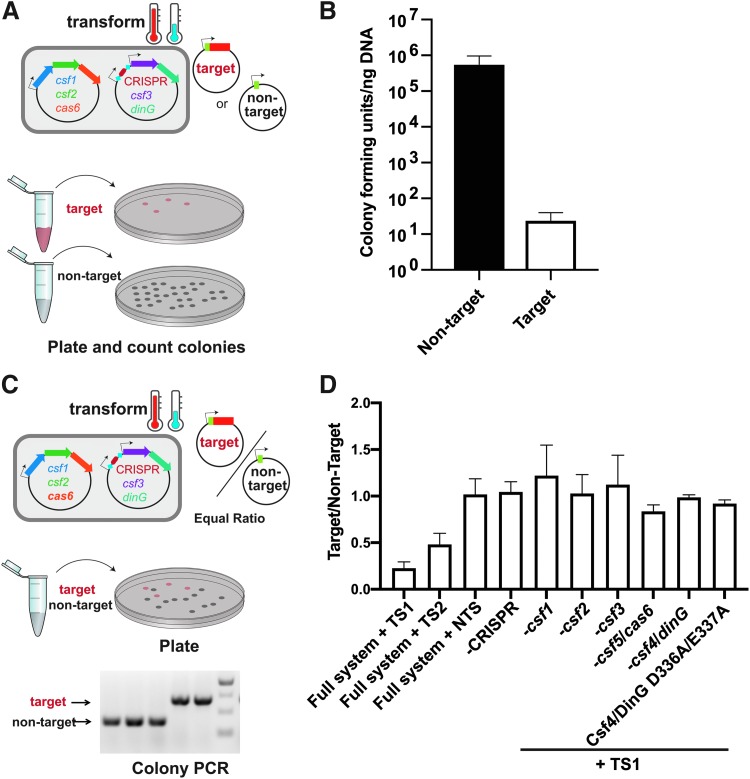
In the plasmid transformation efficiency assay, we compared the number of colonies generated when the cells harboring the full P. aeruginosa PA83 type IV-A1 operon were transformed with a target or non-target plasmid (Fig. 2A). Successful inhibition of transformation was observed with the target plasmid but not with the non-target plasmid, leading to around a four order of magnitude decrease in transformation efficiency (Fig. 2B).
FIG. 2.
The type IV-A1 CRISPR-Cas variant from P. aeruginosa PA83 mediates RNA-guided interference in vivo. (A) Plasmid transformation efficiency assay described in the methods section. Small arrows on plasmids indicate a T7 promoter. (B) A reduction in plasmid transformation efficiency was observed when cells harboring a CRISPR array with TS1, csf1, csf2, csf5/cas6, csf3, and csf4/dinG were transformed with the target compared to those transformed with the non-target plasmid. (C) Plasmid maintenance assay described in the Methods section. Small arrows on plasmids indicate a T7 promoter. (D) An interference defect is observed when any component is removed and when the Csf4/DinG DEAH-box is mutated (D336A/E337A). TS1 was used in each deletion line and the Csf4/DinG mutant. As each condition represents a different competent cell line, an uninduced control was always included (Supplementary Fig. S2). TS, targeting spacer; NTS, non-targeting spacer.
We next evaluated the interference requirements of the type IV-A1 operon from P. aeruginosa PA83 by systematically deleting each component and evaluating interference with plasmid maintenance assays (Fig. 2C). As each deletion required comparing different competent cell lines, we switched to plasmid maintenance assays to evaluate interference differences by PCR to reduce variation that could be attributed to differences between the competencies of prepared cell lines. In this experiment, equimolar amounts of plasmids with target or non-target sequences were mixed and transformed into cells expressing the type IV-A1 system. The ratio of colonies containing target versus non-target plasmid was assessed with colony PCR. A ratio less than one of target/non-target indicates interference (Fig. 2). An interference deficiency was observed when any component of the type IV-A1 operon was removed, including a point mutation in the genes encoding the putative DEAH-box helicase, Csf4/DinG (Fig. 2D and Supplementary Fig. S2). DEAH-box helicases are proteins that unwind double-stranded nucleic acids using the energy released by NTP hydrolysis.17 The putative activity of the DEAH-box of the Csf4/DinG helicase was inactivated by mutating the first two residues of the DEAH-box (D336A/E337A), and indeed, we found that these mutations resulted in an interference defect (Fig. 2D and Supplementary Fig. S2). The target and non-target plasmids both contain a T7 promoter upstream of a 5′-CTTTC-3′ sequence that lies adjacent to the protospacer. In the case of the target plasmid, this sequence is directly upstream of the protospacer. It is unclear at this time whether this is a protospacer adjacent motif utilized by the type IV-A1 system or if it is using another sequence in the vicinity of the protospacer. It is also uncertain whether DNA or RNA is targeted. However, the crRNA is not expected to base pair with the RNA transcribed from the target.
Discussion
Here, we provide the first functional evidence that a type IV CRISPR-Cas system mediates RNA-guided interference against plasmids in vivo and that this activity requires the putative NTP-dependent helicase activity of Csf4/DinG.
Although we show type IV CRISPR system interference requires the putative DinG helicase, the function of CRISPR-associated DinG proteins remains unclear. However, studies on DinG proteins that are not associated with CRISPR systems show that related DinG helicases are involved in recombinational DNA repair and the re-initiation of replication after DNA damage.18 Also, it has been shown that non-CRISPR–associated DinG unwinds R-and D-loops, forked substrates, and 5′ single-stranded overhangs with 5′ to 3′ polarity,18,19 allowing other proteins to access nucleic acid or modify nucleic acid structure. Additionally, there is evidence that non-CRISPR–associated DinG is recruited to disturbances in duplex DNA via changes in redox potential, and that in turn DinG recruits a nuclease to the disturbance site.20 The type IV–associated and non-CRISPR–associated DinG proteins are somewhat similar in sequence, suggesting the CRISPR-associated DinG protein may express similar functions. However, such activities remain to be confirmed in CRISPR-associated DinG proteins.
Unlike type I systems, which encode for the helicase-nuclease Cas3, the protein sequences of the type IV system appear not to contain any obvious nuclease domains. Thus, it is not clear how type IV-A systems protect against plasmid targets and whether the plasmid targets are cleaved. We speculate that similar to type I and type III systems, multi-subunit complexes composed of Csf proteins and a crRNA use the crRNA as a guide to bind complementary nucleic acid forming R-loops. We expect DinG is recruited to the R-loops and then that DinG either acts directly to destroy the plasmid through an unknown mechanism or recruits an endogenous nuclease to mediate RNA-guided interference. However, these hypotheses remain to be tested. As type IV-A systems lack a predicted effector nuclease and often lack a Cas1–Cas2 adaptation module, it has been reasoned that this CRISPR-Cas system cannot function as an independent adaptive immune system.21 Here, we show that a type IV-A system from P. aeruginosa is capable of interference. However, it remains unknown if type IV-A systems are capable of acquiring their own spacer sequences and whether DinG or the putative Csf crRNA complex are involved.
Here, we provide the first evidence that a type IV-A CRISPR-Cas system from P. aeruginosa mediates RNA-guided interference of plasmid DNA, and that such activity requires a putative multi-subunit crRNA-guided complex and a putative NTP-dependent helicase Csf4/DinG. However, many outstanding questions remain for this area of CRISPR-Cas biology. Do type IV-A systems act alone as distinct RNA-guided adaptive immune systems,21,22 or does interference require additional host factors? Do type IV-A systems associate with the acquisition machinery of other CRISPR-Cas systems to acquire immunity?8,21,22 Do type IV-A systems have roles apart from immunity altogether such as regulating gene expression or genome stability?
Conclusion
We conclude that the type IV-A1 system from P. aeruginosa strain PA83 mediates RNA-guided interference against plasmids in vivo. This activity requires all components of the type IV-A1 operon, including the accessory protein Csf4/DinG and its putative NTP-dependent helicase activity.
Supplementary Material
Acknowledgments
The authors wish to thank two anonymous reviewers for their helpful comments during manuscript editing.
Author Disclosure Statement
J.B.-D. is a scientific advisory board member of SNIPR Biome and Excision Biotherapeutics and a scientific advisory board member and co-founder of Acrigen Biosciences. No competing financial interests exist for the remaining authors.
Funding Information
Research in the Jackson Lab is supported by Utah State University RC grant, and New Faculty Start-up funding from the Department of Chemistry and Biochemistry, the Research and Graduate Studies Office, and the College of Science. The Bondy-Denomy Lab was supported by the UCSF Program for Breakthrough Biomedical Research funded in part by the Sandler Foundation, an NIH Director's Early Independence Award DP5-OD021344, and R01GM127489. A.C. was funded by the training grant T32GM008284.
Supplementary Material
References
- 1. Brouns SJJ, Jore MM, Lundgren M, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008;321:960–964. DOI: 10.1126/science.1159689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007;315:1709–1712. DOI: 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- 3. Barrangou R. Diversity of CRISPR-Cas immune systems and molecular machines. Genome Biol 2015;16:247 DOI: 10.1186/s13059-015-0816-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature 2015;526:55–61. DOI: 10.1038/nature15386 [DOI] [PubMed] [Google Scholar]
- 5. Jackson RN, van Erp PB, Sternberg SH, et al. Conformational regulation of CRISPR-associated nucleases. Curr Opin Microbiol 2017;37:110–119. DOI: 10.1016/j.mib.2017.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Makarova KS, Wolf YI, Koonin EV. Classification and nomenclature of CRISPR-Cas systems: where from here? CRISPR J 2018;1:325–336. DOI: 10.1089/crispr.2018.0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Taylor HN, Warner EE, Armbrust MJ, et al. Structural basis of Type IV CRISPR RNA biogenesis by a Cas6 endoribonuclease. RNA Biol 2019;16:1438–1447. DOI: 10.1080/15476286.2019.1634965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Özcan A, Pausch P, Linden A, et al. Type IV CRISPR RNA processing and effector complex formation in Aromatoleum aromaticum. Nat Microbiol 2019;4:89–96. DOI: 10.1038/s41564-018-0274-8 [DOI] [PubMed] [Google Scholar]
- 9. Arndt D, Grant JR, Marcu A, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 2016;44:W16–W21. DOI: 10.1093/nar/gkw387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Almendros C, Guzmán NM, Díez-Villaseñor C, et al. Target motifs affecting natural immunity by a constitutive CRISPR-Cas system in Escherichia coli. PLoS One 2012;7:e50797 DOI: 10.1371/journal.pone.0050797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jackson RN, Golden SM, van Erp PBG, et al. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science 2014;345:1473–1479. DOI: 10.1126/science.1256328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gasiunas G, Sinkunas T, Siksnys V. Molecular mechanisms of CRISPR-mediated microbial immunity. Cell Mol Life Sci 2014;71:449–465. DOI: 10.1007/s00018-013-1438-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Erp PBG, Jackson RN, Carter J, et al. Mechanism of CRISPR-RNA guided recognition of DNA targets in Escherichia coli. Nucleic Acids Res 2015;43:8381–8391. DOI: 10.1093/nar/gkv793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol 2017;37:67–78. DOI: 10.1016/j.mib.2017.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Altschul S. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997;25:3389–3402. DOI: 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Borges AL, Davidson AR, Bondy-Denomy J. The discovery, mechanisms, and evolutionary impact of anti-CRISPRs. Annu Rev Virol 2017;4:37–59. DOI: 10.1146/annurev-virology-101416-041616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Byrd AK, Raney KD. Superfamily 2 helicases. Front Biosci Landmark Ed 2012;17:2070–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Voloshin ON, Camerini-Otero RD. The DinG protein from Escherichia coli is a structure-specific helicase. J Biol Chem 2007;282:18437–18447. DOI: 10.1074/jbc.M700376200 [DOI] [PubMed] [Google Scholar]
- 19. Voloshin ON, Vanevski F, Khil PP, et al. Characterization of the DNA damage-inducible helicase DinG from Escherichia coli. J Biol Chem 2003;278:28284–28293. DOI: 10.1074/jbc.M301188200 [DOI] [PubMed] [Google Scholar]
- 20. Grodick MA, Segal HM, Zwang TJ, et al. DNA-mediated signaling by proteins with 4Fe–4S clusters is necessary for genomic integrity. J Am Chem Soc 2014;136:6470–6478. DOI: 10.1021/ja501973c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koonin EV, Krupovic M. Evolution of adaptive immunity from transposable elements combined with innate immune systems. Nat Rev Genet 2015;16:184–192. DOI: 10.1038/nrg3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Makarova KS, Koonin EV. Annotation and classification of CRISPR-Cas systems. CRISPR 2015;1311:47–75. DOI: 10.1007/978-1-4939-2687-9_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.