A single ATPase powers extension and retraction of a broadly distributed class of type IV pili.
Abstract
A widespread class of prokaryotic motors powered by secretion motor adenosine triphosphatases (ATPases) drives the dynamic extension and retraction of extracellular fibers, such as type IV pili (T4P). Among these, the tight adherence (tad) pili are critical for surface sensing and biofilm formation. As for most other motors belonging to this class, how tad pili retract despite lacking a dedicated retraction motor ATPase has remained a mystery. Here, we find that a bifunctional pilus motor ATPase, CpaF, drives both activities through adenosine 5′-triphosphate (ATP) hydrolysis. We show that mutations within CpaF result in a correlated reduction in the rates of extension and retraction that directly scales with decreased ATP hydrolysis and retraction force. Thus, a single motor ATPase drives the bidirectional processes of pilus fiber extension and retraction.
INTRODUCTION
The best-studied motor adenosine triphosphatases (ATPases) in eukaryotes are required for intracellular transport and motility, and the current dogma is that these motor proteins facilitate transport in a unidirectional manner (1, 2). Consistent with this, individual cells often have dozens of specialized motor ATPases to facilitate directional movement of specific cargo (1). Thus, motor ATPase specificity is hypothesized to be important for tight regulatory control over separate operations. In prokaryotes, the most widespread motor ATPases are secretion motor ATPases belonging to the PilT/VirB11 ATPase family that drive the polymerization of protein subunits into extracellular filaments including type II and type IV secretion systems (T2SS or T4SS), competence pili of Gram-positive bacteria, archaeal flagella, and T4P (3–5). Retraction of the fibers of T4P (3, 6–12) is critical for their function, as is also hypothesized for the T2SS (13) and Gram-positive competence pili (14). Despite its importance, the retraction mechanism has remained elusive for most of these systems, as they have a single motor ATPase, which is required for fiber extension (Table 1). Phylogenetic analysis suggests that the ancestor of these systems had a single ATPase (15). The T4aP and some T4bP represent an exception to this rule, since they have an antagonistic ATPase, derived through an early duplication of the extension motor ATPase gene (15), that drives fiber retraction (6, 10, 16). How do other fiber systems retract in the absence of a retraction ATPase?
Table 1. Most extracellular filaments that use related extension ATPases to power extension do not have an antagonistic retraction ATPase.
|
Extracellular filament type |
Extension ATPase |
Predicted retraction/ evidence of retraction |
Retraction ATPase present? |
| Type II secretion | GpsE | Yes | No |
| Gram-positive competence pili |
ComGA | Yes | No |
| Type IVa pili | PilB | Yes | Yes |
| Type IVb pili | Variable depending on system |
Yes | Sometimes |
| Type IVc tad pili | CpaF | Yes | No |
RESULTS
Here, we use the tad-type IVc pili (T4cP) as a model to elucidate the retraction mechanism for the systems that lack a retraction ATPase (fig. S1). We recently showed that the tad pili of Caulobacter crescentus can retract despite lacking a retraction ATPase orthologous gene (3). Analysis of a local database of fully sequenced bacterial genomes revealed that one-third (836 of 2554) encode an extension ATPase gene that is co-oriented with two platform protein genes, consistent with tad pilus operons (Fig. 1 and table S2) (17), highlighting that tad ATPases are both highly and broadly distributed even by this conservative metric. These ATPases appear to be frequently acquired through horizontal gene transfer (Fig. 1). Their broad distribution throughout the bacteria makes them an important target for mechanistic study. We hypothesized that the mechanism of retraction may be mediated by (i) an unidentified retraction ATPase encoded in trans, (ii) a bifunctional ATPase that powers both extension and retraction, or (iii) a process independent of adenosine 5′-triphosphate (ATP) hydrolysis.
Fig. 1. Tad-like ATPases are widely distributed.
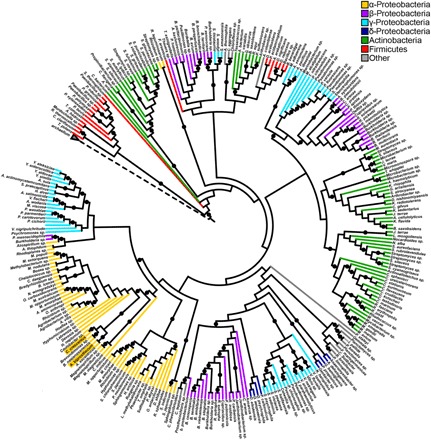
Rooted cladogram of 297 TadA-like ATPases and 3 archaellum ATPases generated by RAxML maximum-likelihood analysis. Branches are colored according to taxonomic class, and nodes with rapid bootstrap values greater than or equal to 85 are indicated by a black circle. The dashed node represents the archaellum ATPase clade. The accession numbers used to construct the tree can be found in table S2, and the tree can also be viewed at https://itol.embl.de/tree/684565227170291554769353.
To differentiate between these hypotheses, we first sought to identify potential unknown retraction ATPases. Retraction is hypothesized to be critical for bringing pilus-dependent phages in proximity to the bacterial cell envelope where phage entry occurs, and a retraction mutant should hypothetically be resistant to pilus-dependent phage infection. We, thus, performed transposon sequencing (Tn-seq) on mutant libraries infected with the pilus-dependent phage ɸCb5 (18) to identify genes required for phage infection. A previous study performed similar experiments using the pilus-dependent phage ɸCbK (19); however, we found that pilus retraction was not essential for ɸCbK infection (fig. S2). In contrast, ɸCb5 infection was prevented when pilus retraction was obstructed (fig. S2). As expected, Tn-seq of infected mutant libraries demonstrated that insertions in the tad pilus–encoding cpa genes resulted in increased phage resistance and, furthermore, revealed that no additional putative motor ATPase proteins outside of the pilus locus conferred increased phage resistance (fig. S3 and table S3).
To determine whether the single tad pilus motor CpaF may play a role in retraction, we used a sensitized, hyperpiliated strain of C. crescentus that has increased numbers of dynamic pili (movie S1) (20, 21). Pilus labeling (22) of a ∆cpaF-complemented mutant confirmed that the expression of the CpaF ATPase is required for pilus extension as shown previously (Fig. 2A) (21, 23). We next assessed the dynamics of CpaF localization during pilus extension and retraction. An mCherry-CpaF fusion (fig. S4) revealed that the ATPase localized to the base of extending or retracting pili equally (Fig. 2, B and C, and movie S2). In some events, we noticed that delocalization of mCherry-CpaF from the base of a retracting pilus fiber correlated with a pronounced reduction in retraction speed (Fig. 2D and movie S3), suggesting that CpaF may play a role in both pilus extension and retraction.
Fig. 2. CpaF is required for tad pilus synthesis and is localized during both pilus extension and retraction.
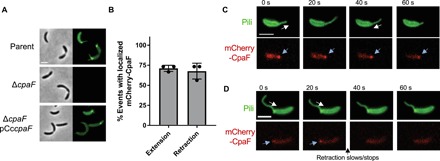
(A) Representative images of hyperpiliated CB13 pil-cys strains labeled with Alexa Fluor 488 maleimide (AF488-mal). (B) Quantification of pilus extension and retraction events with localized mCherry-CpaF. Data are from 15 extension, and retraction events are from three independent, biological replicates (n = 45 total extension and retraction events). Error bars show means + SD. (C) Representative time-lapse images of mCherry-CpaF localization during both pilus extension and retraction. (D) Representative time-lapse images of mCherry-CpaF delocalization during pilus retraction that correlates with halted retraction. Scale bars, 2 μm. White arrows indicate the direction of pilus movement (away from the cell body is extension, and toward the cell body is retraction), and blue arrows indicate mCherry-CpaF foci.
To distinguish between a bifunctional ATPase model and an ATP-independent model of retraction, we performed an unbiased genetic screen by selecting for retraction-deficient mutants. Because pili are important for adherence (24) and their retraction is critical for ɸCb5 infection, ultraviolet (UV)–mutagenized cells were exposed to ɸCb5 phage and resistant mutants were enriched for those that were still able to attach to surfaces. We then screened mutant isolates for their ability to make pili by fluorescence microscopy. Whole-genome sequencing revealed that several isolates contained mutations within the cpaF gene (cpaFF244LK245R and cpaFD310N), and Western blot analysis demonstrated that mutant proteins were still expressed (fig. S1 and S5). Homology-based modeling of CpaF to solved crystal structures (25) revealed high-confidence structural prediction matching a GspE archaeal T2SS ATPase (26), and mutations identified in CpaF mapped near the predicted ATPase active site of the protein (fig. S6). These results suggested a role for CpaF ATPase activity in pilus retraction, and we thus made an additional, targeted mutation (cpaFI355C) that was previously shown to reduce ATPase activity by half in a T4aP retraction ATPase (27), yet still allow for T4aP assembly when introduced to the extension ATPase (28). By fluorescence microscopy, these mutants had reduced extension and retraction rates (Fig. 3, A and B, and movies S4 to S6), with extension and retraction exhibiting a correlated reduction in rates for each mutation (fig. S7).
Fig. 3. CpaF is a bifunctional ATPase that drives extension and retraction of tad pili.
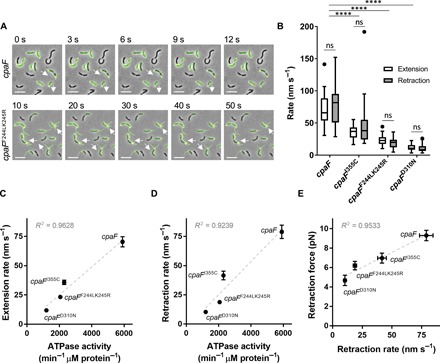
(A) Representative time-lapse images of indicated bNY30a pil-cys cpaF strains labeled with AF488-mal. White arrows show directionality of some active pili. Scale bars, 5 μm. (B) Quantification of extension and retraction rates in indicated strains. White boxes show extension rates, and gray boxes show retraction rates. Box and whisker plots show 5 to 95% confidence intervals. Data were collected from three independent, biological replicates. cpaF extension n = 30, retraction n = 30; cpaFI355C extension n = 30, retraction n = 30; cpaFF244LK245R extension n = 30, retraction n = 30; and cpaFD310N extension n = 30, retraction n = 30. Statistics were determined using Sidak’s multiple comparisons test. ****P < 0.0001. ns, not significant. (C and D) Correlated averages of extension (C) or retraction (D) rates from data shown in (B) and ATPase activity from in vitro ATPase assays. Error bars show SEM. ATPase activity was determined from three replicates of a coupled-enzyme assay, where ATPase activity is depicted as the change in NADH min−1 μM protein−1. (E) Correlated averages of retraction forces and retraction rates. Error bars show SEM. Retraction force measurements of indicated strains were determined from micropillar assays. cpaF n = 33, cpaFI355C n = 34, cpaFF244LK245R n = 34, and cpaFD310N n = 7.
The above mutations fall near the predicted ATPase active site. We therefore hypothesized that the reduction in extension and retraction rates is a result of altered ATP-hydrolyzing activity. ATP hydrolysis assays of mutant CpaF proteins revealed that they exhibited reduced ATP hydrolysis (Fig. 3, C and D). Furthermore, ATP hydrolysis was reduced by varying amounts in different mutants, and this reduction was highly correlated with the reduction in both extension and retraction rates. Together, these data support a model whereby CpaF is a bifunctional motor protein that drives both extension and retraction through ATP hydrolysis. In line with this, we hypothesized that if CpaF was the motor driving forceful pilus retraction, then retraction force should likewise be reduced in ATPase mutants. To measure retraction forces, we used a micropillar-based assay in which retracting pili bind to elastic micropillars and mediate micropillar bending, enabling force measurement (3, 29). cpaF point mutants exhibited reduced forces of retraction comparable and correlated to reductions of both ATPase activity and extension/retraction rates for individual mutants (Fig. 3E and fig. S8), demonstrating that ATP hydrolysis by CpaF drives force generation for tad pilus retraction.
While these data suggest that CpaF drives both pilus extension and retraction in C. crescentus, we sought to determine whether this mechanism extends to other tad pili. The tad pili of Asticcacaulis biprosthecum, an alphaproteobacterium that harbors bilateral stalks, fall into the same phylogenetic clade (Fig. 1) as those of C. crescentus. We labeled the tad pili in A. biprosthecum and showed that pilus synthesis in this species is also dependent on expression of CpaF (fig. S9). Unlike C. crescentus cells, fewer A. biprosthecum cells harbored dynamic pili, with the majority of piliated cells harboring static pilus fibers (Fig. 4, A and B, fig. S9, and movie S7). To test whether the differences in pili dynamics were due to differences in CpaF, we performed a cross-complementation experiment where we expressed either the C. crescentus cpaF (CccpaF) in a ΔcpaF mutant of A. biprosthecum or the A. biprosthecum cpaF (AbcpaF) in a ΔcpaF mutant of C. crescentus (Fig. 4A). Expressing A. biprosthecum CpaF in C. crescentus reduced the percentage of piliated cells that exhibited dynamic activity by approximately fourfold, while expressing C. crescentus CpaF in A. biprosthecum resulted in a fourfold increase in the percentage of cells harboring dynamic pili, supporting a role for CpaF in the control of tad pilus dynamic activity (Fig. 4B and movies S8 and S9). Alignment of CpaF protein sequences from either species revealed high sequence identity between the two proteins with the exception of the N-terminal region (fig. S10). We hypothesize that the N terminus has a regulatory role in controlling the dynamics of tad pili. Because expression of the C. crescentus motor ATPase increased the number of cells with active pili in A. biprosthecum, we used the CccpaF and CccpaFI355C alleles to determine whether decreased ATPase activity would result in reduced extension and retraction rates for this tad system as observed in C. crescentus. In line with the model that CpaF mediates both extension and retraction of tad pili, expression of the ATPase “slow” cpaFI355C allele resulted in proportional reductions in extension and retraction rates (Fig. 4C). The rate of retraction in A. biprosthecum expressing cpaFI355C was twofold higher than in C. crescentus, suggesting that other factors play a role in modulating rates of dynamic extension and retraction.
Fig. 4. Bifunctional ATPase activity of CpaF is conserved in other tad pilus systems.
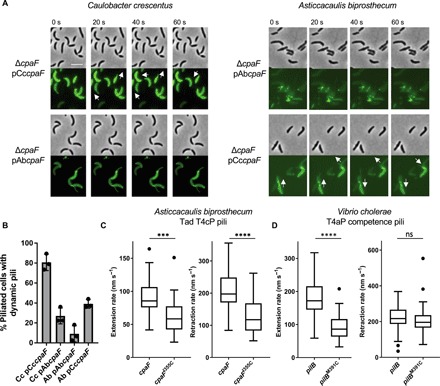
(A). Representative time-lapse images of indicated pil-cys strains labeled with AF488-mal that contain either their native CpaF motor protein or the cross-complement of the other species (Cc, C. crescentus; Ab, A. biprosthecum). White arrows show directionality of some active pili. Scale bar, 4 μm. Look-up tables were normalized for each species to the same values. (B) Quantification of the percentage of piliated cells with actively extending and retracting pili for indicated species shown in (A). Number of piliated cells quantified for each strain: Cc pCccpaF n = 163, Cc pAbcpaF n = 158, Ab pAbcpaF n = 103, and Ab pCccpaF n = 161. Bars show mean with SD, and each point represents one replicate. (C) Extension (left) and retraction (right) rates of A. biprosthecum–type IVc tad pili using Cc cpaF wild-type or I355C mutant alleles. Box and whisker plots show 5 to 95% confidence intervals. Number of events quantified: cpaF extension n = 30, retraction n = 30; cpaFI355C extension n = 29, retraction n = 27. (D) Extension (left) and retraction (right) rates of V. cholerae–type IVa pili that have either wild-type or a slow M391C mutant allele of the PilB extension ATPase. The PilT retraction ATPase in this system was left intact. Box and whisker plots show Tukey’s confidence intervals. Number of events quantified: pilB extension n = 67, retraction n = 69; pilBM391C extension n = 68, retraction n = 71. Statistics were determined using two-tailed t tests. ***P < 0.001, ****P < 0.0001.
The dynamic activity of T4aP and some T4bP is dependent on opposing ATPases that mediate either extension or retraction. We hypothesized that, in contrast to the CpaF mutations described above, slow mutations in the active site of the T4aP extension ATPase in these systems should have no effect on retraction rates. We tested this hypothesis using the T4aP of Vibrio cholerae, which exhibit dynamic extension and retraction important for natural transformation (12). These T4aP use an extension ATPase, PilB, or a retraction ATPase, PilT, to extend or retract (12, 30). We made a mutation (pilBM391C) in the ATP-hydrolyzing domain of PilB analogous to the cpaFI355C mutation (27). Consistent with the model that distinct ATPases drive either extension or retraction of T4aP, the slow mutation in the extension ATPase reduced the extension rate by half, while the retraction rate remained unchanged (Fig. 4D).
DISCUSSION
Our findings elucidate the mechanism of retraction in a class of broadly distributed dynamic extracellular filaments and provide insight into the evolution of molecular motors. Retraction of related fibers—which include the Gram-positive competence pili, T2SS, and other classes of T4P—is proposed to be essential for their function in fundamental bacterial behaviors including natural transformation, motility, secretion, and surface attachment. The majority of these nanomachines, as well as the conjugative T4SS F-pili that likewise dynamically extend and retract, have a single ATPase (31). Therefore, retraction may be mediated by a bifunctional motor protein in these systems, including the T4b V. cholerae toxin coregulated pili, where the insertion of a minor pilin is thought to initiate retraction (8). Evolution of a dedicated retraction ATPase in the T4aP and some T4bP may have enabled faster and more forceful retraction. In addition, this evolution may have provided tighter regulation of the extension-retraction switch or provided a deeper regulatory plasticity over pilus retraction frequency, which may otherwise be less flexible and hardwired through a sole biochemical input in single ATPase pilus systems.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions
Bacterial strains used in this study are listed in table S1. A hyperpiliated derivative of C. crescentus (20) was used throughout this study unless otherwise noted. C. crescentus strains were grown at 30°C in peptone yeast extract (PYE) medium (32) supplemented with kanamycin (5 μg/ml) where appropriate. V. cholerae strains were grown at 30°C in lysogeny broth (LB) medium. Escherichia coli DH5α (Bioline) was used for cloning, and E. coli Rosetta2 (Novagen) was used for CpaF overexpression. E. coli strains were grown in LB medium at 37°C supplemented with kanamycin (25 μg/ml) and chloramphenicol (20 μg/ml) where necessary.
Plasmids were transferred to C. crescentus by electroporation or conjugation with S-17 E. coli strains as described previously (33). In-frame deletion strains were constructed by double homologous recombination using pNPTS-derived plasmids as previously described (34). Briefly, plasmids were introduced to C. crescentus, and a two-step recombination was performed using kanamycin resistance selection followed by sucrose sensitivity selection. Complementation constructs were constructed using the low–copy number vector pMR10 with genes under control of the leaky lac promotor.
For construction of the pNPTS-derived plasmids, ~500–base pair (bp) flanking regions of DNA on either side of the desired mutations were amplified from bNY30a or A. biprosthecum C19 genomic DNA where appropriate. Point mutations were built into the R1 and F2 primers as indicated in table S1. Upstream regions were amplified using F1 and R1 primers, while downstream regions were amplified using F2 and R2 primers. The resulting amplified DNA was purified (QIAquick) and assembled into pNPTS138 that had been digested with restriction enzyme Eco RV (New England Biolabs) using the HiFi Assembly Master Mix (New England Biolabs). For the plasmid pNPTS138cpaF::mCherry-cpaF, ~500-bp upstream of the cpaF gene was amplified from bNY30a genomic DNA using the indicated F1 and R1 primers, the gene encoding mCherry was amplified from vector pRVCHYN-2 (35) using F2 and R2 primers, and the first ~500 bp of the cpaF gene was amplified from bNY30a genomic DNA using primers F3 and R3. A codon-optimized DNA sequence encoding a linker (GSAGSAAGSGEF) in-frame between the mCherry gene and the cpaF start codon was built into primers R2 and F3. All three DNA products were then assembled into pNPTS138 as described above.
For construction of pMR10-derived plasmids, primers pMR10CB13cpaFF and pMR10CB13cpaFR were used to amplify the cpaF gene from bNY30a genomic DNA, and pMR10AbcpaFF and pMR10AbcpaFR were used to amplify the cpaF gene from A. biprosthecum C19 genomic DNA. For CB13 cpaF point mutation alleles cpaFI355C, cpaFD310N, and cpaFF244LK245R, upstream regions of the mutation were amplified using primers pMR10CB13cpaFF + R1 primers, and downstream regions of the mutation were amplified using F2 + pMR10Cb13cpaFR primers as indicated in table S1, with the point mutations built into the R1 and F2 primers. The upstream and downstream DNA were purified (QIAquick) and then stitched together using splicing by overlap extension (SOE) polymerase chain reaction (PCR) (36). cpaF regions from CB13 were then digested using restriction enzymes Sac I (New England Biolabs) and Eco RI (New England Biolabs) followed by heat inactivation at 65°C for 20 min. The cpaF region from A. biprosthecum was digested using restriction enzymes Hind III (New England Biolabs) and Eco RI followed by heat inactivation at 80°C for 20 min. Digested products were then ligated into plasmid pMR10 that was digested with the same enzymes.
For construction of pET28-derived plasmids, primers pET28CB13cpaFF and pET28CB13cpaFR were used to amplify the cpaF gene from bNY30a genomic DNA. For CB13 cpaF point mutation alleles, up- and downstream regions were amplified as described above, with the point mutation built into the R1 and F2 primers and stitched together in the same way using SOE PCR. cpaF regions were then digested using restriction enzymes Nde I (New England Biolabs) and Eco RI followed by heat inactivation at 65°C for 20 min and ligated into pET28 that was digested with the same enzymes.
V. cholerae mutants were constructed by Multiplex Genome Editing by Natural Transformation and natural transformation as previously described (12, 37, 38) and derived from the El Tor isolate E7946 (39). In short, transforming DNA was constructed using SOE PCR and transformed or cotransformed into strains (37, 38).
Phylogenetic analysis
To determine which bacteria encode a Tad-like ATPase, a local database of 2554 completed bacterial genomes were annotated with the Pfam library using the software hmmer v.3.0 and an E value threshold of 1 × 10−10 (40, 41). The presence of a TadABC-like operon was established by the presence of three co-oriented open reading frames separated by no more than 500 bp that encode proteins containing domains found in the TadABC proteins. Specifically, TadB and TadC were identified by the presence of a single domain with at least two-third coverage of the T2SSF domain. TadA homologs were identified by the presence of a T2SSE domain. All genomes analyzed as well as the accession numbers identified as TadABC-like can be found in table S2. Of the 1184 TadABC-like operons that were identified, 297 were randomly selected for phylogenetic analysis, and three archaellum ATPases were used as an outgroup (table S2). The TadA and archaellum ATPase sequences were aligned using the default parameters of muscle version 3.8.31. The resulting alignment was used to generate a phylogenetic tree with RAxML version 8.1.3 set to perform 100 rapid bootstraps and subsequent maximum-likelihood search using the GAMMA model of rate heterogeneity and JTT substitution model (42). The resulting tree was visualized using the Interactive Tree of Life visualization software (43) and can also be viewed at https://itol.embl.de/tree/684565227170291554769353. The root was inferred as the branch separating the archaellum ATPases from the Tad-like ATPases.
Pilus labeling, microscopy, and analysis
In C. crescentus and A. biprosthecum, pili were labeled as described previously with some differences (3). Briefly, cells were grown to an OD600 (optical density at 600) of 0.15 to 0.3. Cells (100 μl) were incubated with Alexa Fluor 488 maleimide (AF488-mal) (25 μg/ml; Thermo Fisher Scientific) for 10 min at room temperature. Cells were then pelleted by centrifugation at 5200g for 1 min, washed once with 100 μl of PYE, and resuspended in a final volume of 5 to 10 μl of PYE before 1 μl of resuspended cells was spotted under a 1% agarose (SeaKem) PYE pad and imaged. Imaging was performed on a Nikon Ti2 microscope using a Plan Apo 60× objective, green fluorescent protein (GFP) and dsRed filter cubes, a Hamamatsu ORCA-Flash4.0 camera, and Nikon NIS Elements Imaging Software. mCherry-CpaF time lapses were performed at 10 s per frame, and analysis of localized mCherry-CpaF was performed manually using Nikon NIS Elements Analysis Software. Because cpaF mutants exhibited differences in rates of extension and retraction, rates were analyzed from strain YB9040 imaged at 3 s per frame, YB9202 imaged at 5 s per frame, YB9241 imaged at 10 s per frame, and YB9240 imaged at 20 s per frame to avoid phototoxicity and photobleaching while still capturing extension and retraction events. Rates were analyzed from strain YB9233 imaged at 3 s per frame and YB9258 imaged at 5 s per frame. Only extension and retraction events that lasted for multiple frames were analyzed. To calculate rates of retraction, the change in pilus length was manually measured using the Nikon NIS Elements Analysis Software and divided by the amount of time over which the length change occurred.
In V. cholerae, pili were labeled as described previously with some differences (12). To constitutively activate competence, the master competence regulator TfoX was ectopically overexpressed (via Ptac-tfoX), and quorum sensing was constitutively activated via deletion of luxO (37, 44). Strains were grown to late-log phase in LB Miller broth with 100 μM IPTG (isopropyl-β-d-thiogalactopyranoside) (to induce TfoX expression), 20 mM MgCl2, and 10 mM CaCl2. Approximately 108 colony-forming units (CFUs) of this culture were centrifuged at 18,000g for 1 min and then resuspended in instant ocean medium (7 g liter−1; Aquarium Systems) before labeling with AF488-mal (25 μg/ml) for 15 min in the dark. Labeled cells were centrifuged, washed twice, and resuspended using instant ocean medium. All imaging was performed under 0.2% Gelzan (Sigma) pads made with instant ocean medium. A Nikon Ti2 microscope with a Plan Apo 60× objective, a GFP filter cube, a Hamamatsu ORCA-Flash4.0 camera, and Nikon NIS Elements Imaging Software was used to image cell bodies and fluorescently labeled pili. To determine the rates of extension and retraction, labeled cells were imaged by time-lapse microscopy every second for 1 min. Extension and retraction events were manually calculated using measurement tools of the NIS Elements Analysis software. For extension and retraction rate calculations, only cells that began and completed extension or retraction events within a 1-min window were analyzed. Pili that were already extending or retracting when imaging began and/or pili that were shorter than 0.3 μm were not analyzed.
Genome-wide transposon mutagenesis coupled to deep sequencing (Tn-seq)
Transposon mutagenesis of C. crescentus NA1000 was done by intergeneric conjugation from E. coli S17-1 λpir harboring the himar1 derivative pMAR2xT7 (45). A Tn library of >200,000 gentamicin acid– and nalidixic acid–resistant clones was collected. NA1000::Tn(GentR) bank was grown overnight in PYE and then freshly restarted in PYE either in the absence or presence of bacteriophage φCb5 [at 103 multiplicity of infection (MOI)]. After 8 or 24 hours of incubation under agitation at 30°C, each cell culture was harvested, and chromosomal DNA was extracted. Genomic DNA was used to generate barcoded Tn-seq libraries and submitted to Illumina HiSeq 4000 sequencing (Fasteris SA). Tn insertion–specific reads (150 bp long) were sequenced using the himar-Tnseq2 primer (5′-AGACCGGGGACTTATCAGCCAACCTGT-3′). Specific reads attesting an integration of the transposon on a 5′-TA-3′–specific DNA locus were sorted (Rstudio_V1.1.442) from the tens of millions of reads generated by sequencing and then mapped (Map_with_Bowtie_for_Illumina_V1.1.2) to the C. crescentus NA1000 genome (NC_011916.1) using the Web-based analysis platform Galaxy (http://usegalaxy.org). Using Samtool_V0.1.18, BED file format encompassing the Tn insertion, coordinates were generated and then imported into SeqMonk V1.40.0 (www.bioinformatics.babraham.ac.uk/projects/) to assess the total number of Tn insertions per chromosome position (Tn insertion per millions of reads count) or per coding sequence (CDS). For CDS Tn insertion ratio calculation, SeqMonk datasets were exported into Microsoft Excel files (dataset S1) for further analyses, as described previously (46). Briefly, to circumvent ratio issues for a CDS Tn insertion value of 0 and CDS that does not share sufficient statistical Tn insertions, an average value of all CDS-Tn insertions normalized to the gene size was calculated, and 1% of this normalized value was used to correct each CDS-Tn insertion value.
Identification of mutants deficient in pilus retraction
To isolate mutants deficient in retraction, stationary phase cultures of the parent strain YB9034 were first mutagenized using UV light irradiation, resulting in at least 90% killing. Approximately 1 × 108 CFUs were then mixed with ɸCb5 phage at an MOI of 1 and added to 3 ml of PYE in a 12-well polystyrene plate and incubated at room temperature with overnight shaking at 150 rpm on an orbital shaker. The next morning, the medium was removed from the plates, and the wells were washed once with 3 ml of water to remove unattached cells. Three milliliters of PYE was added to each well, and plates were then incubated again with shaking at room temperature for 3.5 hours before they were washed again. Plates were then incubated with shaking for 3 days until turbidity was observed in the wells. The contents of each well were then struck out onto PYE plates to isolate for single colonies. Isolates were then imaged by microscopy for altered pilus phenotypes and selected for whole-genome sequencing.
Whole-genome sequencing and analysis of isolated mutants
DNA was extracted from mutant isolates using the Wizard Genomic DNA Purification Kit (Promega). Briefly, 1.5 ml of stationary phase cultures was centrifuged at 16,000g for 3 min to pellet cells. The supernatant was discarded, and pellets were resuspended in 200 μl of TES buffer [10 mM tris-HCl (pH 8), 20 mM EDTA, and 1% (w/v) sarkosyl]. Ribonuclease was added to the cell suspension at a final concentration of 0.05 mg/ml and incubated at 70°C for 10 min. Twenty micrograms of proteinase K was then added to the cell suspension and incubated at 70°C for 25 min. Thirty microliters of 7.5 M ammonium acetate was then added to the cells. One milliliter of resin provided in the kit was added to the cell suspension and then flushed through the Wizard Minicolumns. Two milliliters of column wash solution was then flushed through the columns to clean the DNA. Cleaned DNA was eluted using 80°C TE buffer [10 mM tris-HCl (pH 8) and 1 mM EDTA] and sent to the Center for Genomics and Bioinformatics core facility, Indiana University Bloomington for library preparation and next-generation sequencing analysis.
Five hundred nanograms of DNA was sheared on a Covaris E220 sonicator and used in library preparation using a NETFLEX Rapid DNA-Seq kit (Bio Scientific) according to the manufacturer’s protocol. For multiplexing, 2× 8-nucleotide dual-indexed adapters from the TruSeq RNA CD index kit (Illumina) were added to the libraries. The barcoded libraries were cleaned by double-sided bead cut with AMPure XP beads (Beckman Coulter), verified using a Qubit3 fluorometer (Thermo Fisher Scientific) and a 2200 TapeStation bioanalyzer (Agilent Technologies), and pooled. The pool was sequenced on NextSeq 500 (Illumina) with the NextSeq75 High Output v2 kit (Illumina), and paired-end 2× 38-bp reads were generated. The reads were demultiplexed using bcl2fastq (software version 2.20.0.422), and about 4 to 6 M reads were assigned to each library. Trimmomatic (47) (version 0.33; nondefault parameters) was used to trim reads of adapter and low-quality bases. Reads were mapped to the Caulobacter vibrioides strain CB13B1a (CP023315.3) with breseq (version 0.30.1) using default parameters (48). Custom python scripts were used to combine the breseq output and identify common variants across isolates for a given reference genome. Reads were also mapped to the reference genome with bowtie2 (49) and visualized with JBrowse (50) to look for complex rearrangements.
Phage sensitivity assays
Phage sensitivity assays on plates were performed by spotting dilutions of ɸCb5 phage onto lawns of growing C. crescentus strains. To make lawns, 200 μl of stationary phase cultures was mixed with 3 ml of top agar (0.5% agar in PYE) and spread over 1.5% PYE agar plates. After the top agar solidified, 5 μl of phage diluted in PYE was spotted on top. Plates were grown for 2 days at 30°C before imaging.
Phage sensitivity assays in tubes were performed by adding ~106 CFU of indicated strains with ~1011 plaque-forming units of ɸCb5 phage for an MOI of 105 in 3 ml of PYE where indicated. A final concentration of 500 μM methoxypolyethylene glycol maleimide, MW (molecular weight) 5000 (PEG5000-mal) (Sigma) was added to tubes where indicated to block pilus retraction as shown previously (3, 51). Cultures were then grown with shaking at 30°C overnight before imaging.
CpaF antibody production
For antibody production, an N-terminally tagged full-length version of CpaF from C. crescentus NA1000 was expressed and purified. Antibodies were raised in New Zealand white rabbits.
Western analysis
To compare the expression of cpaF alleles between strains, ~109 cells from exponential phase cultures were centrifuged for 3 min at 16,000g. The supernatant was discarded, pellets were resuspended in phosphate-buffered saline, and SDS loading buffer [62.5 mM tris-HCl (pH 6.8), 10% (v/v) glycerol, 2% (w/v) SDS, 0.05% (v/v) β-mercaptoethanol, and 0.0025% (w/v) Bromophenol blue] was added. Samples were heated to 100°C for 6 min and then separated on 10% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gels. Samples were transferred to nitrocellulose membranes and probed with affinity-purified α-CpaF antibody at 1:500 dilution in 5% (w/v) nonfat milk powder resuspended in 1× TTBS [20 mM tris-HCl (pH 7.6), 130 mM NaCl, and 0.05% (v/v) Tween 20] overnight. The nitrocellulose membrane was washed three times with 1× TTBS and then probed at a 1:20,000 dilution of horseradish peroxidase–conjugated goat α-rabbit antibody (Bio-Rad) in 5% nonfat milk solution for 1 hour. Membranes were developed with SuperSignal West Dura substrate (Thermo Fisher Scientific).
Force measurement using micropillars
Retraction forces were obtained by Polyacrylamide MicroPillars assays as described previously (29). Briefly, an equidistant (3 μm from center to center) polyacrylamide micropillar array with a spring constant of 25 ± 4 pN/μm was obtained by unmolding a silica mold at the center of a 25-mm-diameter round coverglass (Warner Instruments). To enable attachment of the pili to the pillars, the pillars were coated with a 0.01% poly-l-lysine solution (Sigma) covalently linked to the pillars by UV-activated sulfoSANPAH (Thermo Fisher Scientific) for 1 hour, followed by a 1-hour incubation with a water suspension of 0.2% (w/v) 0.02-μm carboxylate-modified beads (Molecular Probes). After 2 hours of treatment, the coverglass was set at the bottom center of an observation chamber (Invitrogen), and 500 μl of fresh PYE liquid medium supplemented with kanamycin (5 μg/ml) was added into the chamber. Mid-log phase bacteria culture (100 μl) was centrifuged at 7500 rpm for 1 min, the supernatant was removed, and the pellet was resuspended in 100 μl of fresh PYE medium containing kanamycin. Then, 25 μl of this cell suspension was added to the micropillar array. Subsequently, 10-Hz videos of the pillar tips movement were recorded with a 60× objective. Last, a combination of ImageJ plugin and MATLAB program (MathWorks Inc., Natick, MA) implementing a cross-correlation tracking of the pillars’ tip motion was used to analyze the videos and extract the maxima of the pillars’ deflections. Retraction forces were calculated on the basis of pillar calibration.
Expression and purification of His6-CB13CpaF and ATPase assays
Flasks (250 ml) containing 50-ml cultures of Rosetta2 expression strains carrying pET28 plasmids were grown with shaking at 37°C to exponential growth phase OD600 (0.5 to 0.7). Cultures were cooled down on ice for 20 min before 0.1 mM IPTG was added to induce protein expression. Induced cultures were incubated with shaking at 16°C overnight. The following day, 30 ml of cultures was centrifuged at 5000g for 10 min at 4°C to harvest cells. The supernatant was removed, and cells were resuspended in 10 ml of buffer A [20 mM tris-HCl (pH 8), 150 mM sodium citrate, and 5 mM 2-mercaptoethanol] with a protease inhibitor tablet (Pierce, EDTA-free). Cell suspensions were sonicated to lyse cells and then centrifuged at 100,000g to separate soluble and insoluble fractions. Cell lysates were then incubated at 4°C for 1 to 1.5 hours on 1 ml of Ni-NTA resin (Qiagen). The resin was then loaded into columns (Bio-Rad) and washed with 10 ml of cold buffer A. Proteins were eluted with 5 ml of elution buffer [20 mM tris-HCl (pH 8), 150 mM sodium citrate, 5 mM 2-mercaptoethanol, and 500 mM imidazole] and collected in 0.5-ml fractions. Fractions (2.5 ml) containing the highest protein content were pooled, and the imidazole was removed by running the 2.5-ml protein suspension on PD-10 size exclusion columns (GE Healthcare) followed by 3.5 ml of cold buffer A to elute proteins. Visual inspection of SDS-PAGE gels indicated that the proteins were >95% pure.
ATP hydrolysis activity was measured using a coupled-enzyme assay (52). Sixty microliters of ATPase buffer [50 mM tris-HCl (pH 8), 1 mM dithiothreitol, 90 mM NaCl, 10 mM MgOAc, bovine serum albumin (50 μg/ml), and 5% glycerol] containing 10 μg of protein was mixed with 60 μl of ATPase buffer supplemented with 10 mM ATP, 10 mM MgCl2, 1 mM phosphoenolpyruvate, 0.8 mM NADH (reduced form nicotinamide adenine dinucleotide), 0.6 U of pyruvate kinase, and 0.96 U of lactate dehydrogenase in a 96-well polystyrene plate and incubated at 30°C. Reactions were performed in triplicate, and absorbance at 340 nm was measured every minute for 1.5 hours. The slope of linear absorbance decay between 25 and 45 min was used to calculate the ATP hydrolysis activity for all samples. An NADH absorbance standard curve was used to calculate concentrations of NADH, and ATPase activity rates are reported as the change in nM of NADH min−1 μM protein−1. Background rates of NADH oxidation were subtracted by normalization of absorbance at 340 nm over time to no protein controls.
Statistics
Statistical significance was calculated using tests on Prism 7 software. Statistical differences between two groups were calculated using two-tailed Student’s t tests. Statistical tests between multiple groups were calculated using Sidak’s multiple comparisons test. Sample sizes were chosen on the basis of historical data, and no methods were used to predetermine sample size.
Supplementary Material
Acknowledgments
We thank S. Shaw, J. Shaevitz, D. Kearns, and C. Fuqua for the helpful discussion comments on the manuscript, and P. Caccamo for the hfsA deletion plasmid and advice on A. biprosthecum genetics. We also thank R. Snyder for the preliminary work on this project. We thank the Center for Genomics and Bioinformatics at Indiana University for whole-genome sequencing and single-nucleotide polymorphism mutant identification. Funding: This study was supported by grant R35GM122556 from the NIH and by a Canada 150 Research Chair in Bacterial Cell Biology to Y.V.B., by grant AI116566 from the NIH to N.B., by grants R35GM128674 and AI118863 from the NIH to A.B.D., and by NSF fellowships 1342962 to K.R.H. and C.K.E. Author contributions: C.K.E. designed and coordinated the overall study in consultation with Y.V.B. C.K.E. designed and performed the CB13 and A. biprosthecum experiments. N.B. designed and J.K. performed the micropillar experiments. A.B.D. designed and J.L.C. performed the V. cholerae experiments. K.R.H. performed the phylogenetic analysis. P.H.V. and G.P. designed and G.P. performed the Tn-seq experiments. All authors analyzed the data. C.K.E. and Y.V.B. wrote the manuscript. All authors contributed to the editing of the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/12/eaay2591/DC1
Table S1. Strains, plasmids, and primers used in this study.
Table S2. Strains used for phylogenetic analysis depicted in Fig. 1.
Table S3. Tn-seq data depicted in fig. S3.
Movie S1. C. crescentus bNY30a with labeled pili exhibiting dynamic cycles of extension and retraction.
Movie S2. C. crescentus bNY30a with labeled pili exhibiting localized mCherry-CpaF at the base of extending and retracting pili.
Movie S3. C. crescentus bNY30a with labeled pili exhibiting delocalization of mCherry-CpaF from the base of retracting pilus that coincides with cessation of retraction.
Movie S4. C. crescentus bNY30a expressing cpaFI355C with labeled pili exhibiting slowed pilus extension and retraction.
Movie S5. C. crescentus bNY30a expressing cpaFF244LK245R with labeled pili exhibiting slowed pilus extension and retraction.
Movie S6. C. crescentus bNY30a expressing cpaFD310N with labeled pili exhibiting slowed pilus extension and retraction.
Movie S7. A. biprosthecum expressing its own cpaF (AbcpaF) harboring nondynamic, labeled pili.
Movie S8. C. crescentus bNY30a expressing AbcpaF harboring mostly nondynamic, labeled pili.
Movie S9. A. biprosthecum expressing CccpaF with labeled pili exhibiting dynamic cycles of extension and retraction.
Fig. S1. The tad pilus structure and gene locus in C. crescentus CB13.
Fig. S2. ϕCb5 phage requires pili and their retraction for Caulobacter infection.
Fig. S3. Tn-seq experiments reveal that Tn insertions in the pilus operon improved growth fitness during ϕCb5 phage infection in C. crescentus NA1000.
Fig. S4. mCherry-CpaF is partially degraded.
Fig. S5. Mutant cpaF expression profiles.
Fig. S6. Mutations in cpaF fall into the ATPase active site of the protein.
Fig. S7. Extension and retraction rates of cpaF mutants are correlated.
Fig. S8. Forces of retraction are reduced and correlated with ATPase activity of cpaF mutants.
Fig. S9. A. biprosthecum CpaF is required for pilus synthesis.
Fig. S10. C. crescentus CB13 and A. biprosthecum CpaF ATPases are highly conserved except for a variable N-terminal region.
REFERENCES AND NOTES
- 1.Vale R. D., The molecular motor toolbox for intracellular transport. Cell 112, 467–480 (2003). [DOI] [PubMed] [Google Scholar]
- 2.J. Howard, Mechanics of Motor Proteins and the Cytoskeleton (Sinauer Associates, 2001). [Google Scholar]
- 3.Ellison C. K., Kan J., Dillard R. S., Kysela D. T., Ducret A., Berne C., Hampton C. M., Ke Z., Wright E. R., Biais N., Dalia A. B., Brun Y. V., Obstruction of pilus retraction stimulates bacterial surface sensing. Science 358, 535–538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCallum M., Tammam S., Khan A., Burrows L. L., Howell P. L., The molecular mechanism of the type IVa pilus motors. Nat. Commun. 8, 15091 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Planet P. J., Kachlany S. C., DeSalle R., Figurski D. H., Phylogeny of genes for secretion NTPases: Identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. U.S.A. 98, 2503–2508 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burrows L. L., Pseudomonas aeruginosa twitching motility: Type IV pili in action. Annu. Rev. Microbiol. 66, 493–520 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Craig L., Forest K. T., Maier B., Type IV pili: Dynamics, biophysics and functional consequences. Nat. Rev. Microbiol. 17, 429–440 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Ng D., Harn T., Altindal T., Kolappan S., Marles J. M., Lala R., Spielman I., Gao Y., Hauke C. A., Kovacikova G., Verjee Z., Taylor R. K., Biais N., Craig L., The Vibrio cholerae minor pilin TcpB initiates assembly and retraction of the toxin-coregulated pilus. PLOS Pathog. 12, e1006109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skerker J. M., Berg H. C., Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. U.S.A. 98, 6901–6904 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merz A. J., So M., Sheetz M. P., Pilus retraction powers bacterial twitching motility. Nature 407, 98–102 (2000). [DOI] [PubMed] [Google Scholar]
- 11.N. I. Abu-Lail, H. Beyenal, in Characterization of Biomaterials (Elsevier, 2013), pp. 207–253. [Google Scholar]
- 12.Ellison C. K., Dalia T. N., Vidal Ceballos A., Wang J. C.-Y., Biais N., Brun Y. V., Dalia A. B., Retraction of DNA-bound type IV competence pili initiates DNA uptake during natural transformation in Vibrio cholerae. Nat. Microbiol. 3, 773–780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korotkov K. V., Sandkvist M., Hol W. G. J., The type II secretion system: Biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 10, 336–351 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muschiol S., Balaban M., Normark S., Henriques-Normark B., Uptake of extracellular DNA: Competence induced pili in natural transformation of Streptococcus pneumoniae. Bioessays 37, 426–435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denise R., Abby S. S., Rocha E. P. C., Diversification of the type IV filament superfamily into machines for adhesion, protein secretion, DNA uptake, and motility. PLOS Biol. 17, e3000390 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craig L., Pique M. E., Tainer J. A., Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2, 363–378 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Tomich M., Planet P. J., Figurski D. H., The tad locus: Postcards from the widespread colonization island. Nat. Rev. Microbiol. 5, 363–375 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Bendis I., Shapiro L., Properties of Caulobacter ribonucleic acid bacteriophage φCb5. J. Virol. 6, 847–854 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christen M., Beusch C., Bösch Y., Cerletti D., Flores-Tinoco C. E., Del Medico L., Tschan F., Christen B., Quantitative selection analysis of bacteriophage φCbK susceptibility in Caulobacter crescentus. J. Mol. Biol. 428, 419–430 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Lagenaur C., Agabian N., Caulobacter crescentus pili: Structure and stage-specific expression. J. Bacteriol. 131, 340–346 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skerker J. M., Shapiro L., Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 19, 3223–3234 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellison C. K., Dalia T. N., Dalia A. B., Brun Y. V., Real-time microscopy and physical perturbation of bacterial pili using maleimide-conjugated molecules. Nat. Protoc. 14, 1803–1819 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhattacharjee M. K., Kachlany S. C., Fine D. H., Figurski D. H., Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J. Bacteriol. 183, 5927–5936 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bodenmiller D., Toh E., Brun Y. V., Development of surface adhesion in Caulobacter crescentus. J. Bacteriol. 186, 1438–1447 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. E., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamagata A., Tainer J. A., Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism. EMBO J. 26, 878–890 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hockenberry A. M., Hutchens D. M., Agellon A., So M., Attenuation of the Type IV pilus retraction motor influences Neisseria gonorrhoeae social and infection behavior. MBio 7, e01994-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Black W. P., Wang L., Jing X., Saldaña R. C., Li F., Scharf B. E., Schubot F. D., Yang Z., The type IV pilus assembly ATPase PilB functions as a signaling protein to regulate exopolysaccharide production in Myxococcus xanthus. Sci. Rep. 7, 7263 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biais N., Higashi D., So M., Ladoux B., Techniques to measure pilus retraction forces. Methods Mol. Biol. 799, 197–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seitz P., Blokesch M., DNA-uptake machinery of naturally competent Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 110, 17987–17992 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clarke M., Maddera L., Harris R. L., Silverman P. M., F-pili dynamics by live-cell imaging. Proc. Natl. Acad. Sci. U.S.A. 105, 17978–17981 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poindexter J. S., Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28, 231–295 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ely B., Genetics of Caulobacter crescentus. Methods Enzymol. 204, 372–384 (1991). [DOI] [PubMed] [Google Scholar]
- 34.Ried J. L., Collmer A., An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 57, 239–246 (1987). [DOI] [PubMed] [Google Scholar]
- 35.Thanbichler M., Iniesta A. A., Shapiro L., A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 35, e137 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bryksin A. V., Matsumura I., Overlap extension PCR cloning: A simple and reliable way to create recombinant plasmids. Biotechniques 48, 463–465 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dalia A. B., McDonough E., Camilli A., Multiplex genome editing by natural transformation. Proc. Natl. Acad. Sci. U.S.A. 111, 8937–8942 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dalia T. N., Yoon S. H., Galli E., Barre F.-X., Waters C. M., Dalia A. B., Enhancing multiplex genome editing by natural transformation (MuGENT) via inactivation of ssDNA exonucleases. Nucleic Acids Res. 45, 7527–7537 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller V. L., DiRita V. J., Mekalanos J. J., Identification of toxS, a regulatory gene whose product enhances toxR-mediated activation of the cholera toxin promoter. J. Bacteriol. 171, 1288–1293 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eddy S. R., Profile hidden Markov models. Bioinformatics 14, 755–763 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L., Potter S. C., Punta M., Qureshi M., Sangrador-Vegas A., Salazar G. A., Tate J., Bateman A., The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 44, D279–D285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Letunic I., Bork P., Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu J., Miller M. B., Vance R. E., Dziejman M., Bassler B. L., Mekalanos J. J., Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 99, 3129–3134 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liberati N. T., Urbach J. M., Miyata S., Lee D. G., Drenkard E., Wu G., Villanueva J., Wei T., Ausubel F. M., An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U.S.A. 103, 2833–2838 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murray S. M., Panis G., Fumeaux C., Viollier P. H., Howard M., Computational and genetic reduction of a cell cycle to its simplest, primordial components. PLOS Biol. 11, e1001749 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deatherage D. E., Barrick J. E., Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151, 165–188 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buels R., Yao E., Diesh C. M., Hayes R. D., Munoz-Torres M., Helt G., Goodstein D. M., Elsik C. G., Lewis S. E., Stein L., Holmes I. H., JBrowse: A dynamic web platform for genome visualization and analysis. Genome Biol. 17, 66 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ellison C. K., Rusch D. B., Brun Y. V., Flagellar mutants have reduced pilus synthesis in Caulobacter crescentus. J. Bacteriol. 201, e00031-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kiianitsa K., Solinger J. A., Heyer W.-D., NADH-coupled microplate photometric assay for kinetic studies of ATP-hydrolyzing enzymes with low and high specific activities. Anal. Biochem. 321, 266–271 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Evinger M., Agabian N., Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132, 294–301 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pate J. L., Ordal E. J., The fine structure of two unusual stalked bacteria. J. Cell Biol. 27, 133–150 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mignolet J., Panis G., Viollier P. H., More than a Tad: spatiotemporal control of Caulobacter pili. Curr. Opin. Microbiol. 42, 79–86 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera: A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Sievers F., Wilm A., Dineen D. G., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., Higgins D. G., Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/12/eaay2591/DC1
Table S1. Strains, plasmids, and primers used in this study.
Table S2. Strains used for phylogenetic analysis depicted in Fig. 1.
Table S3. Tn-seq data depicted in fig. S3.
Movie S1. C. crescentus bNY30a with labeled pili exhibiting dynamic cycles of extension and retraction.
Movie S2. C. crescentus bNY30a with labeled pili exhibiting localized mCherry-CpaF at the base of extending and retracting pili.
Movie S3. C. crescentus bNY30a with labeled pili exhibiting delocalization of mCherry-CpaF from the base of retracting pilus that coincides with cessation of retraction.
Movie S4. C. crescentus bNY30a expressing cpaFI355C with labeled pili exhibiting slowed pilus extension and retraction.
Movie S5. C. crescentus bNY30a expressing cpaFF244LK245R with labeled pili exhibiting slowed pilus extension and retraction.
Movie S6. C. crescentus bNY30a expressing cpaFD310N with labeled pili exhibiting slowed pilus extension and retraction.
Movie S7. A. biprosthecum expressing its own cpaF (AbcpaF) harboring nondynamic, labeled pili.
Movie S8. C. crescentus bNY30a expressing AbcpaF harboring mostly nondynamic, labeled pili.
Movie S9. A. biprosthecum expressing CccpaF with labeled pili exhibiting dynamic cycles of extension and retraction.
Fig. S1. The tad pilus structure and gene locus in C. crescentus CB13.
Fig. S2. ϕCb5 phage requires pili and their retraction for Caulobacter infection.
Fig. S3. Tn-seq experiments reveal that Tn insertions in the pilus operon improved growth fitness during ϕCb5 phage infection in C. crescentus NA1000.
Fig. S4. mCherry-CpaF is partially degraded.
Fig. S5. Mutant cpaF expression profiles.
Fig. S6. Mutations in cpaF fall into the ATPase active site of the protein.
Fig. S7. Extension and retraction rates of cpaF mutants are correlated.
Fig. S8. Forces of retraction are reduced and correlated with ATPase activity of cpaF mutants.
Fig. S9. A. biprosthecum CpaF is required for pilus synthesis.
Fig. S10. C. crescentus CB13 and A. biprosthecum CpaF ATPases are highly conserved except for a variable N-terminal region.


