Abstract
Background
Ovarian clear cell carcinoma (OCCC) is a histological subtype of epithelial ovarian cancer (EOC) with distinct pathological, biological, and molecular features. OCCCs are more resistant to conventional treatment regimen of EOC and have the worst stage-adjusted prognosis amongst EOC subtypes. As the OCCC incidence rate in Asian populations has significantly increased in recent decades, it is critical to elucidate its molecular features that could lead to OCCC-tailored therapeutic strategies.
Methods
Gene expression profiles of 222 OCCC were analyzed by hierarchical clustering and statistical analyses.
Findings
We identified two OCCC gene expression subtypes: EpiCC—epithelial-like, which is associated with early-stage disease, with a relatively higher rate of gene mutations in the SWI/SNF complex; and MesCC—mesenchymal-like, associated with late-stage and higher enrichment of immune-related pathway activity. Genetic, copy number and transcriptomic analyses showed that both EpiCC and MesCC carried OCCC-associated aberrations. The EpiCC/MesCC classification was reproducible in validation cohorts and OCCC cell lines. MesCC tumors had a poorer progression-free survival (PFS) than EpiCC tumors (HR: 3·0, p = 0·0006). Functional assays in cell lines showed that the MesCC subtype was more proliferative and more anoikis-resistant than the EpiCC. By applying the EpiCC/MesCC classification to the TCGA renal clear cell carcinoma cohort, our results indicated interoperability of the subtyping scheme, and revealed preferential drug response of MesCC to bevacizumab.
Interpretation
The EpiCC/MesCC classification shows promise for prognostic and therapeutic stratification in OCCC patients and warrants further investigation in the context of OCCC gene expression subtype-tailored treatment strategies.
Keywords: Ovarian cancer, Clear cell, Microarray gene expression, Molecular subtype
Research in context.
Evidence before this study
Ovarian clear cell carcinoma (OCCC) is a distinct histological subtype of epithelial ovarian cancer (EOC) that is associated with reduced sensitivity to chemotherapy and poorer outcomes than other EOC subtypes in advanced stages. There is hence a need to identify more effective treatments for this disease in order to improve outcomes for OCCC patients. Gene expression profiles of EOC have been shown to confer prognostic and possible therapeutic implications for patients but these studies have mainly focused on stratifying high grade serous ovarian cancer with a paucity of data on OCCC. Hence, the therapeutic and prognostic value of OCCC gene expression signatures remains largely unexplored.
Added value of this study
From an unsupervised analysis of 222 OCCC gene expression profiles, we identified two OCCC gene expression subtypes—EpiCC and MesCC. Each subtype was associated with distinct clinical features and outcomes with MesCC subtype having a significantly worse PFS than EpiCC subtype. The MesCC subtype was also significantly associated with more advanced stage OCCC (stage III/IV) whereas the EpiCC subtype was more frequently seen in stage I/II OCCCs. The subtypes were reproducible in two independent cohorts and in OCCC cell lines. Multi-omics analyses revealed that these two transcriptomic subtypes shared partial OCCC-associated molecular footprints. Applying the OCCC subtypes to a renal clear cell carcinoma cohort demonstrated interoperability of the subtyping scheme, and revealed preferential drug response of MesCC to bevacizumab.
Implication of all the available evidence
The association of EpiCC and MesCC subtypes in early and late stage OCCCs respectively suggests that specific biological features of OCCCs may determine the metastatic potential and hence subsequent outcomes for patients with this disease. Therapeutic agents tailored to each of the subtypes, e.g. the use of antiangiogenic drugs in MesCC, could help shape the treatment strategies urgently required to improve the dismal outcomes of advanced OCCC.
CRediT authorship contribution statement
Tuan Zea Tan: Data curation. Jieru Ye: Data curation. Yee Chung Vin: . Lim Diana: Data curation. Natalie Yan Li Ngoi: Data curation. Tan David Shao Peng: . Huang Ruby Yun-Ju: Formal analysis, Investigation.
Alt-text: Unlabelled box
1. Introduction
Ovarian clear cell carcinoma (OCCC), characterized by its clear cytoplasmic appearance, is the second most common histological subtype of epithelial ovarian cancer (EOC) after high grade serous ovarian cancers (HGSOC) in East Asian populations. OCCC prevalence varies across race and ethnicity: 6% in the America [1], 10% in Asian American [1], 15–27% in Japan and South Korea [2, 3], and 13% in Singapore [4]. Although OCCC is still considered a rare tumor type as defined by the Gynecologic Cancer InterGroup (GCIG), there has been a significant increase in the reported incidence rates of OCCC in American Asians [5], in Japan [3], and in Singapore [4]. OCCC frequently presents at an early stage among younger women and has the worst prognosis amongst EOC when adjusted for stage [6]. The poorer prognosis in OCCC could be attributed to the poor response rate to platinum-based chemotherapy, which stands at 11–27% and drops drastically to only 1–2% in recurrent settings [6, 7].
The relative chemoresistance of OCCCs has led to limited therapeutic options for patients with advanced stage or recurrent disease. There is thus an urgent need to identify more effective treatments for this disease in order to improve outcomes for OCCC patients. Endeavors have been made to stratify HGSOC patients into gene expression molecular subtypes (GEMS) [[8], [9]–10] and to reproduce the GEMS in OCCC [11]. Developing tailored therapeutic based on GEMS is an area of interest and is currently being explored in clinical trials [8, 12] like the VIP study (NCT03188159). Currently, a paucity of data exists for OCCC as only a few small (n < 50) studies have specifically studied GEMS in this disease.
In this study, we aggregated 222 OCCC gene expression profiles and have identified two GEMS in OCCC (epithelial/EpiCC, and mesenchymal/MesCC) by leveraging on a database previously compiled [13]. The two OCCC subtypes were reproducible not only in validation sets but also in OCCC cell lines. Importantly, the significance between EpiCC and MesCC in the risk for progression was apparent for the stage I disease. Both the EpiCC and MesCC subtypes shared OCCC-associated molecular footprints. In addition, we also discovered the interoperability of the OCCC subtypes in renal clear cell carcinoma (RCCC).
2. Materials and methods
2.1. Gene expression processing and discovery cohort compilation
Samples annotated as OCCC were extracted from CSIOVDB [13] and GSE65986 [14], RMA-normalized (Affymetrix Power Tool version 1.15.0; apt-probeset-summarize function with default parameters except –a rma-sketch, and library files from Affymetrix) and standardized (ComBat [15] with default parameters; cohort was used as batch information) to form the discovery cohort (n = 136; Suppl. Fig. 1). Principal component analysis was used to verify that the batch effect was not overwhelming in the merged dataset.
2.2. Validation cohorts compilation from public repository and the National University Hospital
Two validation cohorts were compiled: first, the pre-processed data of GSE73614 [11] was downloaded from GEO (accessed 2018, Feb). GSE73614 (n = 37) was used as validation cohort as it was hybridized on Agilent microarray platform as opposed to the Affymetrix microarray platform used in the discovery cohort; Second, 24 frozen archival OCCC tumors, and one cell samples from patient's ascites fluid from the Department of Obstetrics & Gynecology, National University of Singapore were collected according to protocols approved by the Institution Review Board from 2006 to 2017. The samples were additionally reviewed by a gynaecologic pathologist (DL) to verify that the tumors were primary to ovary and were pure OCCC. The samples showed typical morphology of OCCC according to WHO criteria: a malignant tumor composed of clear, eosinophilic and hobnail cells, displaying a combination of tubulocystic, papillary and solid patterns. The samples were then subjected to microarray profiling using Affymetrix GeneChip® Human Transcriptome Array 2.0 (Affymetrix, Inc., Santa Clara, CA). The data was first RMA-normalized (Affymetrix Power Tool version 1.15.0; apt-probeset-summarize function with default parameters except – a rma-sketch, and HTA2.0 r3 library files) and standardized (ComBat [15] with default parameters; cohort was used as batch information) with a subset of samples with confirmed OCCC histology from GSE69207 [13].
2.3. Molecular subtype identification and characterization
ConsensusClusterPlus [16] v1.44.0 (with default parameters settings except Euclidean distance, max K = 20, and 1000 permutations) in R v3.5.1 bioconductor v3.8 was employed to identify subtypes in the discovery cohort using the most varying genes (standard deviation > 0·9; yielded 1346 genes). Silhouette analysis was used to select core samples (silhouette width > +0·15; top 70% of samples) based on previous experiences [8, 13]. Differentially expressed genes identified in the core samples (SAM [17] v3.0 with default parameter settings) were used as gene expression signature (Suppl. Table S1). Pathway enrichment was analyzed by Enrichr [18] (last accessed: 2019 Mar). The MesCC score and the enrichment score of the pathways from Msigdb [19, 20] v6.1 were estimated using a Kolmogorov-Smirnov-based method [21] and GSVA [22] v1.28.0 (with default parameter settings except ssGSEA computation method was used), respectively. Immune infiltration was estimated using Cibersort (with default parameter settings) [23].
2.4. Mutation processing and analysis
Sample preparation and sequencing: targeted sequencing and processing were performed by ACT Genomics® (Taipei City, Taiwan). Genomic DNA was extracted from 25 FFPE tumor samples and eight OCCC lines. The gDNA integrity was investigated by Fragment AnalyzerTM (Advanced Analytical Technologies, Inc.) and Quantitative real-time PCR (qPCR). Extracted gDNA was amplified using 4 pools of primer pairs (Ion AmpliSeq Comprehensive Cancer Panel, Life Technologies) targeting coding exons of analyzed genes. Amplicons were ligated with barcoded adaptors using the Ion AmpliSeq Library Kit (Life Technologies). Barcoded libraries were subsequently conjugated with sequencing beads by emulsion PCR and enriched using Ion Chef system (Life Technologies) according to the Ion PI IC 200 protocol (Life Technologies). The quality and the quantity of amplified libraries were determined using the fragment analyzer (AATI) and Qubit (Invitrogen). Sequencing was performed on the Ion Proton sequencer using the Ion PI chip (Life Technologies) according to the manufacturer's protocol. Sequencing metrics were given in Suppl. Table S10.
Single Nucleotide Variant (SNV) calling: raw reads were mapped to the hg19 genome using the Ion Torrent Suite (version 5.2). Coverage depth was calculated using Torrent Coverage Analysis. SNV and short insertions/deletions (INDEL) were identified using the Torrent Variant Caller. The coverage was down-sampled to 4000 prior to annotation by Variant Effect Predictor (version 88) with COSMIC v.81 and 1000 Genomes Project Phase 3 database. Variants with coverage ≥ 25, allele frequency ≥ 5% and actionable variants with allele frequency ≥ 2% were retained.
Copy number variant identification: amplicons with read counts in the lowest 5th percentile of all detectable amplicons and amplicons with a coefficient of variation ≥ 0·3 were removed. The remaining amplicons from four different pools were normalized to correct the pool design bias. ONCOCNV [24] was applied on ACT Genomics in-house PBMCs (as baseline) and on the FFPE samples for the normalization of total amplicon number, amplicon GC content, amplicon length, and technology-related biases. The samples were then segmented with a gene-aware model.
Visualization: mutation profile was plotted with OncoPrinter in cBioPortal [25] v2.1.0.
2.5. Functional assays of ovarian cancer clear cell lines
The OCCC cell lines were sent for short tandem repeat (STR) assay to confirm their identities (Suppl. Table S11).
Population doubling time: cell line population doubling time was extracted from Matsumura et al. [26].
Wound healing: cells were seeded into culture inserts (#80209, Ibidi, Martinsried, Germany) and grown until 90% to 100% confluence. After 24 h, the inserts were removed and non-attached cells were washed away. Fresh L-15 media (#21083-027, Life Technologies, Waltham, Massachusetts, United States) with 20% FBS was added to the cells for time-lapse microscopy using Nikon Eclipse C1 Microscope Live-imaging system at 37 °C incubation. The images were then analyzed using NIS-Elements, Advance Research.
Anoikis resistance Annexin V/PI staining: 500,000 cells were seeded in each ultra-low attachment (ULA) 10-cm dish (#3263, Corning, Corning, New York, United States). After incubation at 37 °C, 5% CO2 for 48 to 72 h, the cells were collected and stained with Propidium iodide (#P4864, Sigma-Aldrich, St. Louis, Missouri, United States) and Annexin V (#A35122, Life Technologies) for 15 min at room temperature. The stained cells were then analyzed using BD LSR II FACS analyzer with proper gating.
Drug treatment: nine OCCC cell lines (JHOC9, KOC5C, KOC7C, OVISE, OVTOKO, RMG1, RMG2, RMG5 and TAYA) were tested for their sensitivity to cisplatin (#P3494, Sigma-Aldrich). Cells were seeded in 96-well plates at an optimal density, which was determined for each cell line that it would reach 80–90% confluency by the end of the assay. After overnight incubation, cells were treated with nine concentrations of each drug (four-fold serial dilution) for 48h. The percentage of the cell population responding to the drug relative to the un-treated/vehicle controls was measured using a CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay, following the manufacturer's recommendations (#G5430, Promega, Madison, Wisconsin, United States). Dose-response curves were plotted using GraphPad Prism version 5.04 (GraphPad Software, La Jolla, CA, USA), to derive GI50 (drug concentration for 50% growth inhibitory effects on cells) for each cell line in three independent experiments.
2.6. Statistical analysis
Statistical analyses were conducted using Matlab® R2016b version 9.1.0.960167, statistics and machine learning toolbox version 11.0 (MathWorks; Natick, MA, USA). Fisher's exact test or chi-squared test was applied for association analysis. Spearman correlation coefficient test was applied to assess the significance of correlation. Kaplan–Meier analyses were conducted using GraphPad Prism® version 5.04 (GraphPad Software, La Jolla, CA, USA) for patients with PFS (n = 103) or OS (n = 112) data from discovery and validation cohorts. Statistical significance of the Kaplan–Meier analysis was calculated by log-rank test. Cox regression analysis was performed using R v3.5.1 survival v2.42-6 package.
2.7. Data availability
The OCCC gene expression data from NUH cohort can be accessed online with the accession GEO: GSE129617.
3. Results
3.1. Epithelial and mesenchymal subtypes are found in ovarian clear cell carcinoma
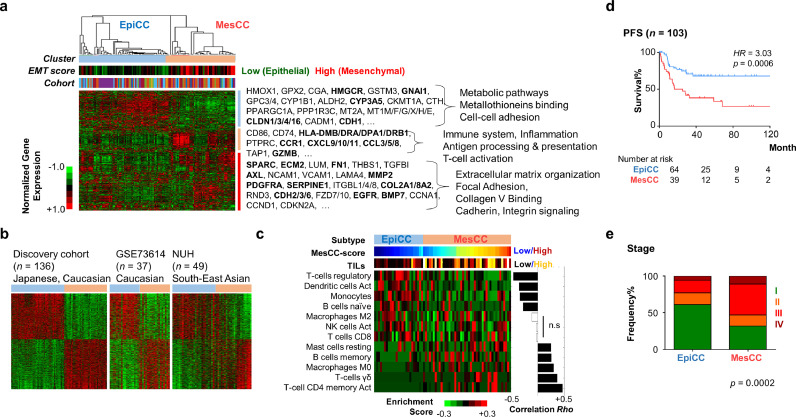
To identify gene expression molecular subtypes (GEMS) in OCCC, we compiled a discovery cohort of 136 OCCC transcriptomic profiles and performed consensus clustering (Suppl. Fig. S1; Materials and Methods), which yielded two dominant clusters (Fig. 1(a); Suppl. Fig. S2). One cluster had a lower epithelial–mesenchymal transition (EMT) score [21] and a higher expression of cell-cell adhesion genes (CDH1, CLDN3 and CLDN4) reported to be associated with a non-EMT phenotype, and was thus called epithelial OCCC (EpiCC) subtype. EpiCC showed enriched expression of metabolic and metallotheioneins binding pathways (HMGCR2, GNAI1, GPC3, and MT2A). The second cluster, which we denoted as mesenchymal OCCC (MesCC) subtype, had a higher EMT score and showed enriched expression of genes involved in extracellular matrix (ECM) organization, focal adhesion and collagen V binding such as SPARC, ECM2, FN1, COL2A1 and CDH2. MesCC also showed enriched expression in genes involved in immune-related genes such as genes in the antigen processing and presentation pathways (HLA-DMB, HLA-DRA, HLA-DPA1, CD74 and TAP1). Silhouette analysis indicated that EpiCC and MesCC were two distinct subtypes (Suppl. Fig. 3a). We used silhouette width to select core samples for the purpose of generating subtype signatures (Materials and Methods; Suppl. Table S1). Applying the subtype signatures, the EpiCC and MesCC were reproduced in two independent validation cohorts (Fig. 1(b)).
Fig. 1.
Ovarian clear cell carcinoma has two gene expression molecular subtypes. (a) Gene expression heatmap of consensus clusters identified in a discovery cohort of ovarian clear cell carcinoma (OCCC; n = 136). Color bars indicate cluster labels; epithelial–mesenchymal transition (EMT) score; and cohorts. Selected genes are listed and bold-faced. Selected pathway enriched was shown on the right. (b) Gene expression heatmap of OCCC subtype signature in discovery cohort (Japanese and Caucasian), and two validation cohorts (GSE73614; Caucasian; n = 37) and NUH (South-East Asian; n = 49). (c) Heatmap showing estimated enrichment score of various immune cell infiltration in the two OCCC subtypes identified in discovery cohort. Only samples with significant (p < 0.05) infiltration of any immune cell types are shown. MesCC score (blue = low, maroon = high) indicates degree of MesCC subtype within a tumor. Tumor infiltrating lymphocytes (TILs, sum of immune cells enrichment score) color bar indicates high infiltration in MesCC. Correlation Rho and significance were assessed by Spearman correlation coefficient test. (d) Kaplan–Meier analysis of PFS data from discovery and validation cohorts show that MesCC has poorer prognosis. p-Value is computed by log-rank test. (e) Bar plots showing frequency% (y-axis) in the EpiCC and MesCC (x-axis), the distribution of stage by combining data from discovery and validation cohorts. p-Value is computed by chi-square test. Color code: heatmap: green = low expression/score, red = high expression/score; Abbrev.: EpiCC, epithelial subtype of ovarian clear cell carcinoma; MesCC, mesenchymal subtype of ovarian clear cell carcinoma; Act., activated; PFS, progression-free survival; HR, hazard ratio; n.s., not significant.
To validate the association of MesCC subtype with the expression of immune-related genes, we performed pathway and immune cell infiltration analyses by correlating the pathway/immune cell infiltration scores with a MesCC score (Fig. 1(c), Suppl. Table S2, S3, Materials and Methods). Indeed, compared with EpiCC, MesCC had higher enrichment of immune-related pathways activity (Suppl. Table S2, S3) as well as tumor infiltrating lymphocytes (TIL; Fig. 1(c)). Interestingly, EpiCC and MesCC could be infiltrated by different immune cell types, as EpiCC showed enriched expression of genes associated with regulatory T-cells and activated dendritic cells whereas MesCC showed enriched expression of genes associated with CD4 memory and γδ T-cells (Fig. 1(c)). Genes associated with natural killer and CD8 T-cells, however, showed no correlation with the MesCC score. Apart from immune-related pathways, pathway analyses revealed enrichment of EMT, angiogenesis, TGFβ signaling, IL6_JAK_STAT3 signaling, PI3K-AKT signaling in MesCC (Suppl. Table S2). On the other hand, enrichment of catabolic and metabolic processes was found in EpiCC (Suppl. Table S2).
Next, as the sample sizes of each cohort were small, we performed clinico-pathological association by amalgamating the data from the discovery and validation cohorts to increase the statistical power. MesCC was associated with older OCCC patients with the median age of 58 (Mann–Whitney test, p = 0·0216) (Suppl. Fig. 3b). Using the samples with survival data, the MesCC subtype was significantly associated with worse prognosis in terms of progression-free survival (PFS) (Fig. 1(d); Hazard Ratio = 3·0, log-rank test, p = 0·0006) but there was no significant correlation of EpiCC and MesCC with overall survival (OS; Suppl. Fig. 3c). The trend for MesCC to have impact on survival was consistent when stratifying the cohort by disease stage. MesCC had poorer PFS and OS in both early and late stages, although statistical significances were not reached due to the small number of samples having reported events (Suppl. Fig. 3d, e) except for PFS in late stage. Univariate and multivariate Cox regression analyses further affirmed that the MesCC correlated with poor PFS and remained an independent prognosis factor when adjusted by age and stage (Suppl. Table S4). MesCC was significantly associated with stage III and IV whereas EpiCC was significantly associated with stage I and II (Fig. 1(e); Suppl. Fig. 3f). We repeated the analyses within each cohort and the results were largely consistent in at least two of the three cohorts (Suppl. Fig. 3g). In summary, OCCC could be classified into the good prognostic EpiCC subtype associated with low EMT status, early staged disease, and infiltration of regulatory T-cells and activated dendritic cells; and the poor prognostic MesCC subtype associated with high EMT status, advanced staged disease, and infiltrating CD4 memory and γδ T-cells.
3.2. EpiCC and MesCC harbor classical morphological features and molecular footprints of ovarian clear cell carcinoma
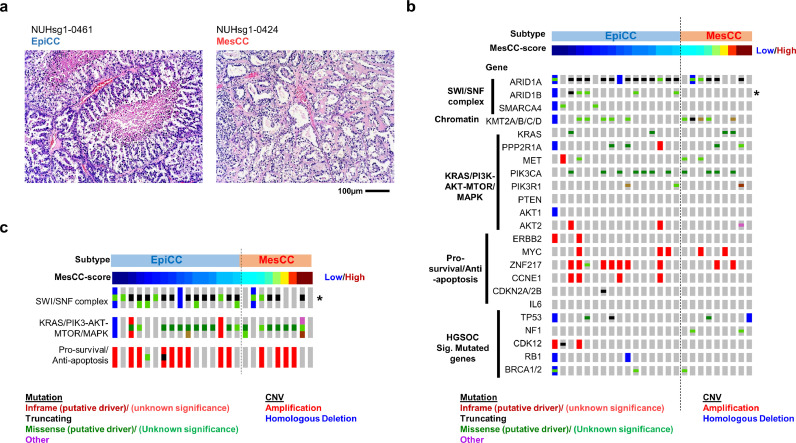
To confirm that these two subtypes were represented in true OCCC, we conducted a histological review of the OCCCs from NUH (n = 25). This showed that both EpiCC and MesCC subtypes displayed typical morphological features of clear cell carcinoma (Fig. 2(a)). Concordant with the histological review, the expression of markers CTH [27], HNF1B [28], and ZNF217 [2] indicated that both EpiCC and MesCC expressed higher levels of OCCC markers genes expression than non-OCCC histological subtypes (Suppl. Fig. 4a; Suppl. Table S5), with EpiCC having the highest expression. Similarly, using two reported OCCC gene expression signatures [29, 30], the enrichment scores of the signatures verified the identities of EpiCC and MesCC as OCCC (Suppl. Fig. 4b; Suppl. Table S5).
Fig. 2.
EpiCC and MesCC harbor classical ovarian clear cell carcinoma molecular footprint. (a) Hematoxylin and eosin stain of an EpiCC (left) and a MesCC (right) ovarian clear cell carcinoma (OCCC). Oncoplot of selected genes (b) and pathways (c). (Mut)ation (red = inframe mutation, black = truncating mutation, green = missense mutation, and purple = other mutation types) and copy number variation (CNV; red is amplification, blue is homologous deletion) in NUH cohort (n = 25) are shown. Top color bar indicates the OCCC subtype. Next color bars (blue = low, maroon = high) show MesCC score.
Next, we conducted targeted panel sequencing to investigate if EpiCC and MesCC harbored gene mutations or copy number variations (CNVs) commonly associated with OCCC (Fig. 2(b)). We curated a list of prevalent mutations and CNVs reported in OCCC [2, 7, 31, 32] (Suppl. Table S6). In 25 OCCC from the NUH cohort, we observed that both EpiCC and MesCC had a high prevalence of ARID1A (81% and 56%, respectively), and PIK3CA aberrations (44% in both). Our analysis detected E542 and E545 pathogenic recurrent mutation hotspots in PIK3CA, but no known pathogenic recurrent hotspots in ARID1A (Suppl. Table S7). There were only two recurrent mutations detected, R1461 and Q605 in ARID1A. Interestingly, there was no significant difference between EpiCC and MesCC in gene mutation and copy number aberration commonly associated with OCCC, except for ARID1B (Fisher exact test, p = 0·04). There was no known pathogenic or recurrent mutation detected in ARID1B (Suppl. Table S7). Of note, genes involved in the SWI/SNF complex were more prone to mutations in EpiCC (Fisher exact test, p = 0·0014; Fig. 2(c)). We also checked the prevalence of the significantly mutated genes in HGSOC reported by TCGA and the result indicated that EpiCC and MesCC had low incidences of HGSOC-associated mutations or CNVs [10] (0–12%; Fig. 2(b); Suppl. Table S6). Subsequently, we further explored whether there would be difference of the mutation signatures between EpiCC and MesCC. We estimated the mutation signatures for the 25 samples from NUH OCCC cohort by using the targeted sequencing results (Suppl. Fig. 4c; Materials and Methods). In agreement with the recent report on the mutation landscape stratified by the ovarian cancer histotypes [33], the NUH OCCC cohort exhibited primarily the aging and APOBEC mutation signatures. The former was enriched in MesCC (Fisher exact test, p = 0·058), and the later was enriched in EpiCC (Fisher exact test, p = 0·057), albeit this is not statistically significant. A larger study is needed to verify this findings. Altogether, these results presented evidence that EpiCC and MesCC, while exhibit shared OCCC-associated transcriptomic and genomic characteristics, they both harbor distinct molecular features which might reflect the functional differences in biology.
3.3. The EpiCC and MesCC classification is reproducible in OCCC cell lines
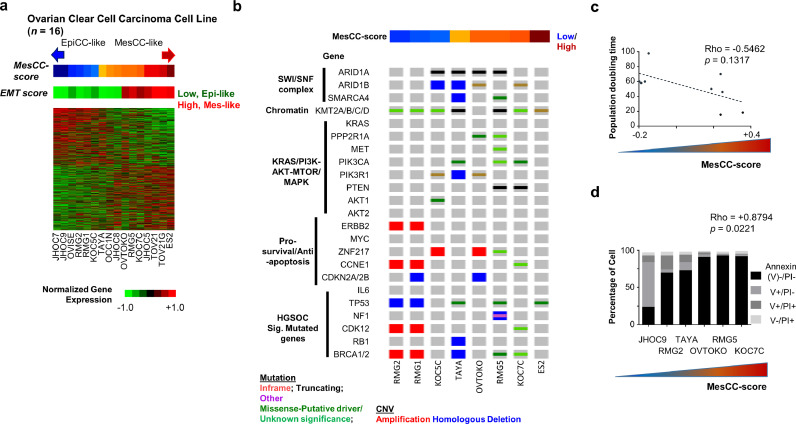
In order to have functional characterization of the EpiCC and MesCC, we projected the EpiCC/MesCC subtype signatures on a panel of 16 OCCC cell lines (Fig. 3(a)). Similarly, the OCCC cell lines could be grouped into EpiCC or MesCC. We subsequently checked the mutation and copy number profiles of the OCCC cell lines for OCCC-associated mutations/CNVs (Fig. 3(b); Suppl. Table S8). The OCCC cell lines had lower mutation rates in ARID1A (50%) and PIK3CA (37·5%) genes, and higher mutation rate (87·5%) in KMT2 genes (KMT2A/B/C; Suppl. Table S6) compared to the NUH OCCC cohort. In terms of HGSOC-associated mutations, the OCCC cell lines harbored relatively higher mutation rates than the OCCC clinical samples (0–37·5%; Suppl. Table S6). However, we noted that the OCCC cell lines had a higher rate of OCCC-associated mutations/CNVs than that of HGSOC. In addition, there was no significant mutation or copy number aberration correlated with EpiCC or MesCC, due to a limited number of samples.
Fig. 3.
EpiCC and MesCC are reproducible in ovarian clear cell cancer cell lines. (a) Gene expression heatmap (green = low, red = high) of EpiCC/MesCC signatures in 16 ovarian clear cell cancer (OCCC) cell lines. Color bars indicate MesCC score (blue = low, maroon = high); and epithelial–mesenchymal transition (EMT) score (green = low, red = high). (b) Oncoplot of selected genes (Mut)ation (red = inframe mutation, black = truncating mutation, green = missense mutation, and purple = other mutation types) and copy number variation (CNV; red is amplification, blue is homologous deletion). (c) Scatter plot of population doubling time (y-axis) and MesCC score (x-axis). Dashed line indicate linear regression fit. (d) Bar plot showing percentage of cell (y-axis) in different apoptotic states (Annexin V−/PI− = viable cells, black; V+/PI− = apoptotic cells, gray; V−/PI+ = necrotic cell, light gray; V+/PI+ = late apoptotic cells, dark gray) in different cell lines (x-axis) undergoing anoikis resistance assay. Correlations were computed using Spearman correlation coefficient test. For experiments in C and D, three replicates for each cell lines were used. Color bar indicates MesCC score.
The MesCC scores were annotated to each cell line to correlate with cellular functions which might explain the preferential aggressiveness. We first correlated the MesCC score of each cell line with its respective population doubling time (Fig. 3(c)). There was a trend of MesCC lines having a higher proliferation rate (Spearman correlation coefficient test, Rho = −0.5462, p = 0.1317). As the development of resistance to anoikis is a key step in EOC dissemination [34], we investigated the anoikis resistance of six OCCC cell lines (JHOC9, RMG2, TAYA, OVTOKO, RMG5, and KOC7C) by growing them on ultra-low attachment surface followed by Annexin V/PI staining (Fig. 3(d)). We observed a strong positive correlation of MesCC score with the number of anoikis-resistant cells (Spearman correlation coefficient test, Rho = +0·879, p = 0·022), affirming the aggressive nature of MesCC subtype. Alongside, the migration efficiencies of these cell lines were checked using the wound healing assay. Intriguingly, there was no significant correlation of EpiCC or MesCC subtype with relative cell migration (Suppl. Fig. 5a, b). In addition, we checked the cisplatin 50% inhibitory concentrations (IC50) of the cells and observed no correlation with the MesCC score (Spearman correlation coefficient test, p = 0·76; Suppl. Fig. 5c). Collectively, these data indicated that the EpiCC and MesCC GEMS appear to reflect the underlying ability of OCCC cells to withstand anoikis and proliferate, which may account for the increased prevalence of the MesCC signature in advanced stage disease and correspondingly poorer PFS.
3.4. The EpiCC and MesCC classification in RCCC
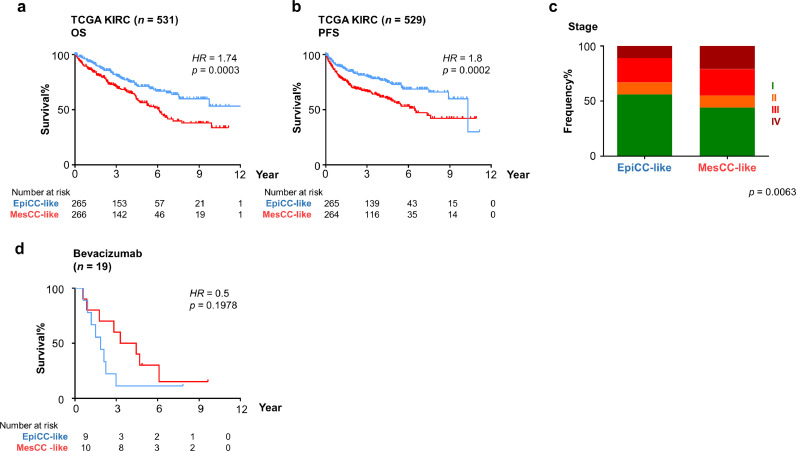
Renal clear cell carcinoma (RCCC) has been reported to share genomic similarities with the OCCC [28]. We therefore assessed the relevance of the EpiCC and MesCC classification in RCCC cohort of TCGA (TCGA KIRC) [35] (Fig. 4(a) and (b)). Similar to our observation in OCCC, the RCCC tumors annotated as the MesCC-like subtype had significantly poorer OS and PFS than EpiCC (HR= 1·74 and 1·8, respectively; log-rank test, p < 0·0005). In concordance with OCCC, the MesCC-like RCCC showed enrichment in late stage disease whereas EpiCC-like RCCC showed enrichment in stage I disease (Fig. 4(c); Chi-square test, p = 0·0063). Multivariate Cox regression analysis indicated the subtype classification is an independent prognostic factor in RCCC (Suppl. Table S9), consistent with the observation in OCCC (Suppl. Table S4). These results suggested interoperability of EpiCC/MesCC classification in OCCC and RCCC. Between the two subtypes, the MesCC-like RCCC patients were found to have improved clinical outcomes following treatment with bevacizumab but this was not significant (log-rank test, p = 0·19; Fig. 4(d)). This is nevertheless of interest since certain EOC GEMS has been reported to exhibit increased benefit with adjuvant bevacizumab [12]. These results suggest that the EpiCC/MesCC classification could be relevant in RCCC, and indicate the potential utility of antiangiogenic therapy in the context of OCCC with MesCC subtype.
Fig. 4.
Aggressiveness of MesCC is observed in renal clear cell carcinoma. Stratifying patients into EpiCC-like (blue) and MesCC-like (red) based on median of MesCC score, Kaplan–Meier analyses of (a) Overall survival (OS) and (b) progression-free survival (PFS) in TCGA renal clear cell carcinoma (KIRC; n = 534); (c) Bar plot showing the frequency% distribution (y-axis) in EpiCC-like and MesCC-like renal clear cell carcinoma (x-axis). Chi-square test is used to assess significance. (d) PFS in TCGA KIRC further stratified by treatment regimen containing bevacizumab. Log-rank tests are used to assess significance. Abbrev.: HR, Hazard ratio.
4. Discussion
In terms of pathology, clinical phenotype and molecular biology, OCCC represents a distinct subtype of EOC. The OCCC are associated with a poorer response rate to platinum-based chemotherapy in first-line and recurrent settings [6, 7], and worse prognosis in advanced stages compared with other EOC subtypes. Therapeutic options for OCCC patients are limited and the increasing incidence of OCCC [3–5] in certain Asian populations clearly delineates a critical need for more effective OCCC-specific clinical management. The classification of tumors by using GEMS may provide a viable strategy for therapeutic stratification but has not been well-characterized in OCCC. In this study, we have identified two molecular subtypes, EpiCC and MesCC, in 222 OCCC samples as well as in OCCC cell lines, and demonstrated that: (i) EpiCC are associated with a lower EMT score, lower disease stage and lower risk of progression; (ii) MesCC are associated with a higher EMT score, more advanced disease, and a greater propensity to progress. The association of poor prognosis with MesCC is concordant with the characteristics of EMT, which has been linked to metastasis, chemo-resistance and poor disease outcome [21]. Functionally, the MesCC cell lines showed a preferential ability to overcome anoikis and have increased proliferative capacity, which could explain the clinical phenotype of this OCCC molecular subtype.
By scrutinizing the genetic, copy number and transcriptomic profiles of the EpiCC and MesCC subtypes, we have verified that both subtypes harbor OCCC-associated molecular footprints such as high rates of ARID1A, and PIK3CA mutations; low rates of TP53, BRCA1/2 mutations; high rates of ZNF217 amplification; and elevated OCCC gene expression signatures scores. The observed rates of genetic alterations in EpiCC and MesCC samples are comparable to the rates reported in OCCC from other cohorts [7, 28, 31], thus validating the identities of that EpiCC and MesCC as OCCC. While both EpiCC and MesCC harbor OCCC-associated molecular alterations, EpiCC has a higher mutation rate in the SWI/SNF complex, suggesting that EpiCC and MesCC may be regulated differently at the epigenetic level. This warrants further investigation, especially for the development of therapeutics that target epigenetic aberrations.
From a therapeutic perspective, the fact that the MesCC signature was present in RCCC [28] and was associated with improved outcomes with the VEGF-targeting humanized monoclonal antibody bevacizumab suggests that this could also be a potential therapeutic strategy in MesCC. Several OCCC phase II clinical trials testing antiangiogenic drugs like nintedanib (NCT02866370), sunitinib malate (NCT00979992), and the immune checkpoint inhibitor durvalumab (NCT03405454) are underway. These compounds may be effective in MesCC given the high angiogenesis, IL6-JAK-STAT3 signaling [36], and immune signatures exhibited by this subtype and future trials testing the combination of these drugs in advanced OCCC are warranted. One question of interest is whether the intrinsic activation of the angiogenesis pathway also results into anoikis resistance in MesCC. Indeed, the angiogenesis factor ANGPTL4 has been reported to enhance anoikis resistance and tumor metastasis in head and neck squamous cell carcinoma [37]. Given that the genetic aberrations of MesCC remain similar to EpiCC, it is possible that perturbations in epigenetic regulation of the signaling pathways are the key determinants of these molecular subtypes.
Finally, as the majority of OCCC gene expression profiles utilized in this study are curated and re-analyzed from public repositories, our findings may be limited by the completeness, accuracy and quality of the samples collected. We cannot rule out the possibility of misdiagnoses but these cases should be minimal given that OCCC has a distinctive morphology. Another related limitation is the lack of data on the therapeutic regimen for each patient. This has hindered our analyses related to progression-free survival and therapeutic response, as the statistical power is low due to insufficient data. Similarly, the sample size in this study is still relatively small as compared to studies involving disease of higher prevalence. Therefore, the OCCC may be more heterogeneous than the two molecular subtypes we reported. More samples of OCCC and additional validation studies in future would be required to verify the observations uncovered in this study.
In conclusion, we have identified two distinct OCCC molecular subtypes that have significantly different PFS outcomes after first line treatment. We envisage that this subtyping scheme could provide a basis for future GEMS directed therapeutic strategies in an effort to improve outcomes for women with advanced and recurrent OCCC.
Declaration of competing interest
The other authors declared there is no conflict of interest.
Acknowledgments
This work was supported by National Research Foundation (NRF) Singapore and the Singapore Ministry of Health under its Research Centres of Excellence initiative to RYH; National Medical Research Council (NMRC) under its Center Grant scheme to National University Cancer Institute (NCIS), NR17NMR120OM to RYH; and charitable funding from the Pangestu Family to DSPT. RYH is supported by Yushan Young Scholar from Ministry of Education, Republic of China (Taiwan). DSPT is supported by the National Medical Research Council (NMRC) Singapore Clinician Scientist Award (NMRC/CSA-INV/0016/2017). The funding sources had no involvement in the study design, analysis or interpretation.
DSPT received research funding from AstraZeneca, Karyopharm Therapeutics, Bayer, and is on advisory boards of AstraZeneca, Roche, D3 Singapore, Tessa Therapeutics, Genmab, Bayer, MSD. DSPT received honoraria/travel support from AstraZeneca, Novartis, Roche, MSD, Bayer, Merck Serono. DSPT also receives charitable research funding from the Pangetsu Family.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.11.017.
Appendix. Supplementary materials
References
- 1.Torre L.A., Trabert B., DeSantis C.E., Miller K.D., Samimi G., Runowicz C.D. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68(4):284–296. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuzaki S., Yoshino K., Ueda Y., Matsuzaki S., Kakuda M., Okazawa A. Potential targets for ovarian clear cell carcinoma: a review of updates and future perspectives. Cancer Cell Int. 2015;15:117. doi: 10.1186/s12935-015-0267-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Machida H, Matsuo K, Yamagami W, Ebina Y, Kobayashi Y, Tabata T. Trends and characteristics of epithelial ovarian cancer in Japan between 2002 and 2015: a JSGO-JSOG joint study. Gynecol Oncol. 2019;153(3):589–596. doi: 10.1016/j.ygyno.2019.03.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tay S.K., Cheong M.A. Evidence for ethnic and environmental contributions to frequency of ovarian clear cell carcinoma. Aust N Z J Obstet Gynaecol. 2014;54(3):225–230. doi: 10.1111/ajo.12188. [DOI] [PubMed] [Google Scholar]
- 5.Park H.K., Ruterbusch J.J., Cote M.L. Recent trends in ovarian cancer incidence and relative survival in the United States by race/ethnicity and histologic subtypes. Cancer Epidemiol Biomarkers Prev. 2017;26(10):1511–1518. doi: 10.1158/1055-9965.EPI-17-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugiyama T., Kamura T., Kigawa J., Terakawa N., Kikuchi Y., Kita T. Clinical characteristics of clear cell carcinoma of the ovary: a distinct histologic type with poor prognosis and resistance to platinum-based chemotherapy. Cancer. 2000;88(11):2584–2589. [PubMed] [Google Scholar]
- 7.Mabuchi S., Sugiyama T., Kimura T. Clear cell carcinoma of the ovary: molecular insights and future therapeutic perspectives. J Gynecol Oncol. 2016;27(3):e31. doi: 10.3802/jgo.2016.27.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan T.Z., Miow Q.H., Huang R.Y., Wong M.K., Ye J., Lau J.A. Functional genomics identifies five distinct molecular subtypes with clinical relevance and pathways for growth control in epithelial ovarian cancer. EMBO Mol Med. 2013;5(7):1051–1066. doi: 10.1002/emmm.201201823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tothill R.W., Tinker A.V., George J., Brown R., Fox S.B., Lade S. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14(16):5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research N Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winterhoff B., Hamidi H., Wang C., Kalli K.R., Fridley B.L., Dering J. Molecular classification of high grade endometrioid and clear cell ovarian cancer using TCGA gene expression signatures. Gynecol Oncol. 2016;141(1):95–100. doi: 10.1016/j.ygyno.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kommoss S., Winterhoff B., Oberg A.L., Konecny G.E., Wang C., Riska S.M. Bevacizumab may differentially improve ovarian cancer outcome in patients with proliferative and mesenchymal molecular subtypes. Clin Cancer Res. 2017;23(14):3794–3801. doi: 10.1158/1078-0432.CCR-16-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan T.Z., Yang H., Ye J., Low J., Choolani M., Tan D.S. CSIOVDB: a microarray gene expression database of epithelial ovarian cancer subtype. Oncotarget. 2015;6(41):43843–43852. doi: 10.18632/oncotarget.5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lisowska K.M., Olbryt M., Dudaladava V., Pamula-Pilat J., Kujawa K., Grzybowska E. Gene expression analysis in ovarian cancer – faults and hints from DNA microarray study. Front Oncol. 2014;4:6. doi: 10.3389/fonc.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 16.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan T.Z., Miow Q.H., Miki Y., Noda T., Mori S., Huang R.Y. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol Med. 2014;6(10):1279–1293. doi: 10.15252/emmm.201404208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boeva V., Popova T., Lienard M., Toffoli S., Kamal M., Le Tourneau C. Multi-factor data normalization enables the detection of copy number aberrations in amplicon sequencing data. Bioinformatics. 2014;30(24):3443–3450. doi: 10.1093/bioinformatics/btu436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):1. doi: 10.1126/scisignal.2004088. pl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumura N., Huang Z., Mori S., Baba T., Fujii S., Konishi I. Epigenetic suppression of the TGF-beta pathway revealed by transcriptome profiling in ovarian cancer. Genome Res. 2011;21(1):74–82. doi: 10.1101/gr.108803.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cochrane D.R., Tessier-Cloutier B., Lawrence K.M., Nazeran T., Karnezis A.N., Salamanca C. Clear cell and endometrioid carcinomas: are their differences attributable to distinct cells of origin? J Pathol. 2017;243(1):26–36. doi: 10.1002/path.4934. [DOI] [PubMed] [Google Scholar]
- 28.Ji J.X., Wang Y.K., Cochrane D.R., Huntsman D.G. Clear cell carcinomas of the ovary and kidney: clarity through genomics. J Pathol. 2018;244(5):550–564. doi: 10.1002/path.5037. [DOI] [PubMed] [Google Scholar]
- 29.Yamaguchi K., Mandai M., Oura T., Matsumura N., Hamanishi J., Baba T. Identification of an ovarian clear cell carcinoma gene signature that reflects inherent disease biology and the carcinogenic processes. Oncogene. 2010;29(12):1741–1752. doi: 10.1038/onc.2009.470. [DOI] [PubMed] [Google Scholar]
- 30.Yanaihara N., Anglesio M.S., Ochiai K., Hirata Y., Saito M., Nagata C. Cytokine gene expression signature in ovarian clear cell carcinoma. Int J Oncol. 2012;41(3):1094–1100. doi: 10.3892/ijo.2012.1533. [DOI] [PubMed] [Google Scholar]
- 31.Murakami R., Matsumura N., Brown J.B., Higasa K., Tsutsumi T., Kamada M. Exome sequencing landscape analysis in ovarian clear cell carcinoma shed light on key chromosomal regions and mutation gene networks. Am J Pathol. 2017;187(10):2246–2258. doi: 10.1016/j.ajpath.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 32.Li M., Li H., Liu F., Bi R., Tu X., Chen L. Characterization of ovarian clear cell carcinoma using target drug-based molecular biomarkers: implications for personalized cancer therapy. J Ovarian Res. 2017;10(1):9. doi: 10.1186/s13048-017-0304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y.K., Bashashati A., Anglesio M.S., Cochrane D.R., Grewal D.S., Ha G. Genomic consequences of aberrant DNA repair mechanisms stratify ovarian cancer histotypes. Nat Genet. 2017;49(6):856–865. doi: 10.1038/ng.3849. [DOI] [PubMed] [Google Scholar]
- 34.Cai Q., Yan L., Xu Y. Anoikis resistance is a critical feature of highly aggressive ovarian cancer cells. Oncogene. 2015;34(25):3315–3324. doi: 10.1038/onc.2014.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cancer Genome Atlas Research N Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anglesio M.S., George J., Kulbe H., Friedlander M., Rischin D., Lemech C. IL6-STAT3-HIF signaling and therapeutic response to the angiogenesis inhibitor sunitinib in ovarian clear cell cancer. Clin Cancer Res. 2011;17(8):2538–2548. doi: 10.1158/1078-0432.CCR-10-3314. [DOI] [PubMed] [Google Scholar]
- 37.Liao Y.H., Chiang K.H., Shieh J.M., Huang C.R., Shen C.J., Huang W.C. Epidermal growth factor-induced ANGPTL4 enhances anoikis resistance and tumour metastasis in head and neck squamous cell carcinoma. Oncogene. 2017;36(16):2228–2242. doi: 10.1038/onc.2016.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The OCCC gene expression data from NUH cohort can be accessed online with the accession GEO: GSE129617.