Abstract
Background
Drought is one of the most serious factors limiting plant growth and production. Sheepgrass can adapt well to various adverse conditions, including drought. However, during germination, sheepgrass young seedlings are sensitive to these adverse conditions. Therefore, the adaptability of seedlings is very important for plant survival, especially in plants that inhabit grasslands or the construction of artificial grassland.
Results
In this study, we found a sheepgrass MYB-related transcription factor, LcMYB2 that is up-regulated by drought stress and returns to a basal level after rewatering. The expression of LcMYB2 was mainly induced by osmotic stress and was localized to the nucleus. Furthermore, we demonstrate that LcMYB2 promoted seed germination and root growth under drought and ABA treatments. Additionally, we confirmed that LcMYB2 can regulate LcDREB2 expression in sheepgrass by binding to its promoter, and it activates the expression of the osmotic stress marker genes AtDREB2A, AtLEA14 and AtP5CS1 by directly binding to their promoters in transgenic Arabidopsis.
Conclusions
Based on these results, we propose that LcMYB2 improves plant drought stress tolerance by increasing the accumulation of osmoprotectants and promoting root growth. Therefore, LcMYB2 plays pivotal roles in plant responses to drought stress and is an important candidate for genetic manipulation to create drought-resistant crops, especially during seed germination.
Keywords: LcMYB2, Seed germination, Root growth, Osmoprotectant
Background
Seed germination and seedling establishment are the most critical stages in the life cycle of plants [1], also the germination and young seedlings establishment phases for most plants are the most sensitive to stressful conditions, such as drought. Drought has been known as the most important factor limiting plant growth and productivity, particularly in arid and semiarid regions. Thus, the research on seed germination and seedling establishment under drought stress is required not only for evaluating and breeding the ecological adaptation of the species, but also for developing effective strategies for restoration and application arid and semiarid grassland.
Drought typically has complex ramifications that impose hyperosmotic and ionic stresses on cells by disrupting the water potential gradient of the soil-root-plant-atmosphere continuum [2]. Plants, especially those constantly growing in severe environments, have evolved many strategies to cope with abiotic stress. Generally, plants adopt either stress tolerance or stress avoidance strategies. For example, establishing deeper roots, which helps roots absorb water from deeper underground in drought conditions, is an avoidance strategy [2, 3]. Controlling stomata aperture to reduce transpiration and accumulating osmoprotectants (e.g. proline or soluble sugars) are usually thought to be stress tolerance strategies [4–7].
The primary signal caused by drought is osmotic stress, which is a signal that overlaps with that of salt stress and increases the accumulation of phytohormone abscisic acid (ABA) [8, 9]. Endogenous ABA plays a critical role in regulating stomatal aperture and inducing the biosynthesis of osmolytes through signaling cascades [10–13]. ABA increases, it binds to soluble ABA receptors (PYR/PYL/RCAR), and together they bind to and inhibit plant protein phosphatases (PP2Cs) [14–17]. The inactive PP2Cs are released from SnRK2 kinases, activating them so that they phosphorylate and activate downstream transcription factors and effectors in ABA response pathways [2, 18, 19].
Many types of transcription factors, such as basic Leucine Zipper (bZIP); NAM, ATAF and CUC transcription factor (NAC); APETUAP2/Ethylene-Responsive-Element Binding Protein (AP2/ERF), and MYB DNA-binding domain protein (MYB), are involved in drought stress responses [20–22]. MYB transcription factors are reported to play a role in multiple functions, including metabolism, cell fate and identity, developmental processes and responses to biotic and abiotic stresses during the plant life cycle [23]. Members of the MYB transcription factor family are divided into four classes depending on the number of adjacent repeats (one, two, three or four) [23]. It has been reported that AtMYB60 and AtMYB61 regulate stomatal movement and plant drought tolerance in opposite manners [24, 25]. MYB96, which is involved in the development of lateral roots, regulates drought response by integrating ABA and auxin signals [26]. In addition, the overexpression of AtMYB44, StMYB1R-1, TaMYB33 or TaPIMP1 improved drought stress tolerance in transgenic plants through different mechanisms [27–30]. An important discovery, that a 366 bp-insertion including three MYB cis-elements in the promoter of ZmVPP1 confers drought-inducible expression to ZmVPP1 in the variant, indicates that MYB-type transcription factors have significant functions in drought response [31]. Sheepgrass, which is widely distributed in Eurasia, adapts well to drought, cold, saline and alkaline conditions [32, 33]. To explore the mechanism that underlies the abiotic stress tolerance of sheepgrass, several transcriptome analyses have been performed in the past few years [32–35], and several genes identified by transcriptome analyses have actually enhanced the abiotic stress tolerance of transgenic plants [36–42]. Although MYB transcription factors play pivotal roles in drought responses, there is still no report on MYB proteins from sheepgrass that elucidates their contributions to sheepgrass drought tolerance. In a previous study, we revealed that 15 MYB and MYB-related transcription factors that are involved in drought stress response [35]. A transcript (contig41859), named Leymus chinensis MYB DNA-binding domain protein 2 (LcMYB2) is largely induced by drought and was selected for further analysis. The transcript levels of LcMYB2 were enhanced under manninol, salt, ABA and cold treatments. The results of subcellular localization of 35S-LcMYB2-GFP and the distribution of the β-galactosidase activity of GAL4-BD-LcMYB2 indicate that LcMYB2 localizes to the nucleus and activates the transcription of lacZ. Chromatin immunoprecipitation (CHIP) analysis using anti-LcMYB antibodies showed that LcMYB2 can bind to the promoters of Leymus chinensis Dehydration Responsive Element Binding Protein 2(LcDREB2), Arabidopsis thaliana Dehydration Responsive Element Binding Protein 2A(AtDREB2A), Arabidopsis thaliana △1-Pyrroline-5-carboxylate synthetas (AtP5CS1) and Arabidopsis thaliana Late-embryogenesis-abundant protein (AtLEA14). Overexpressing LcMYB2 in A. thaliana promotes seed germination and enhances root growth under osmotic and ABA treatment and further increases soluble sugar and proline content with 300 mmol/L mannitol treatment. In addition, transgenic seedlings performed better than wild-type under natural drought stress. Taken together, these results indicated that LcMYB2 plays critical roles in the drought responses of sheepgrass through both avoidance and tolerance strategies. Furthermore, this work provides important information for understanding the intrinsic characteristics of sheepgrass drought tolerance and supplies an important candidate gene for improving drought stress tolerance with genetic engineering.
Results
LcMYB2 expression pattern analysis
Based on 454 high throughput sequencing and expression profile analyses of sheepgrass under drought stress, we found 15 MYB and MYB-related transcription factors that were responsive to changes of water content in plant tissues [32, 35]. Contig41859, which was up-regulated by drought stress and named LcMYB2, was a MYB-related transcription factor with unknown function that attracted our attention (Additional file 1: S1).
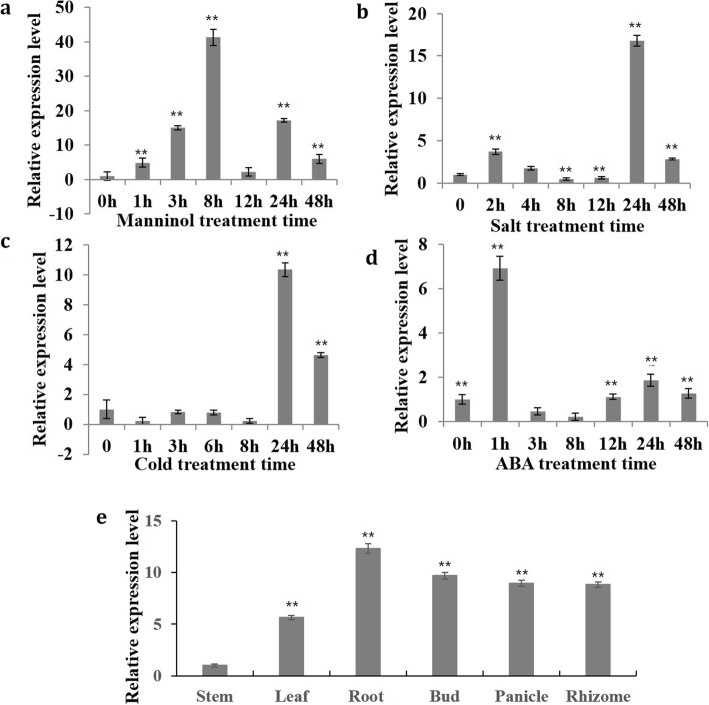
LcMYB2 is highly induced by 300 mM mannitol at the 8th hour after treatment (Fig. 1a), whereas it is relatively slower responding to salt and cold stress (24 h, Fig. 1b, c). However, it is quickly upregulated by ABA treatment (Fig. 1d). The maximal level of mRNA accumulation under mannitol treatment is higher than under salt, cold and ABA treatments, indicating that LcMYB2 mainly functions in response to osmotic stress. Furthermore, the expression level of LcMYB2 in different organs is also detected under normal growth conditions. The results show that LcMYB2 has the highest transcript level in roots (Fig. 1e). Based on these combined results, we predict that LcMYB2 is mainly responsible for the osmotic stress response in roots, which may benefit plants under drought stress.
Fig. 1.
The expression patterns of LcMYB2 under different treatments and its tissue-specific expression. a to d Expression of LcMYB2 in the seedlings of 8-week-old sheepgrass were treated with or without 300 mM mannitol, 400 mmol/L NaCl, cold and 100 μmol/L ABA treatments for 0, 1, 3, 8, 12, 24 h or 48 h after stress treatments. e LcMYB2 expression in stem, leaf, root, bud, panicle and rhizome of 1-year-old sheepgrass in flowering period. LcACTIN was used as a positive control for data normalization. Three independent replicates of measurements were performed for each time point, and the data are shown as the mean ± standard deviation (SD) (n = 3)
Isolation and sequence analysis of the LcMYB2
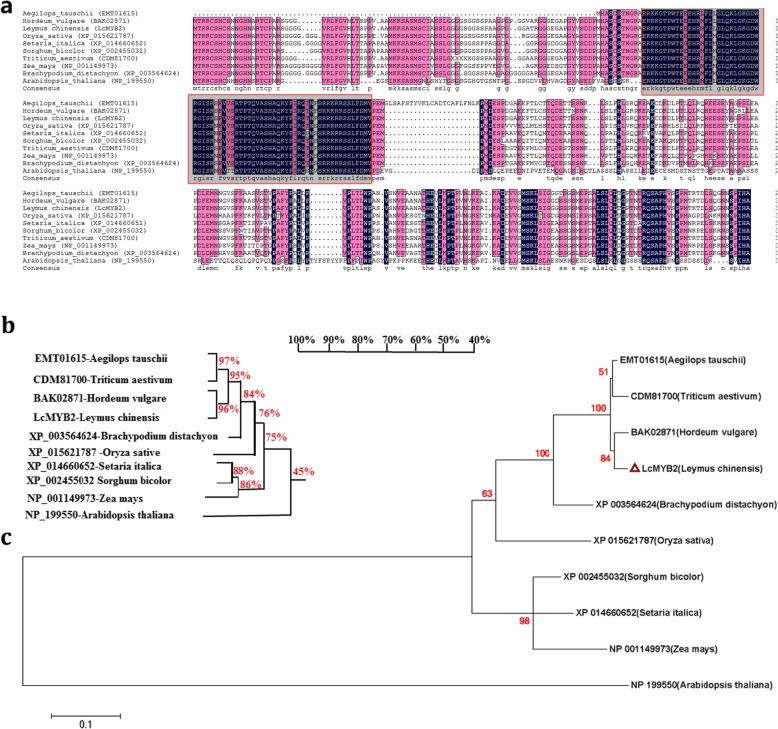
Putative full-length LcMYB2 was isolated from sheepgrass by Rapid-Amplification of cDNA Ends-PCR (RACE-PCR) and classical PCR based on the 454 high-throughput data (SRA065691; Additional file 2: S2). The length of LcMYB2 Open reading frame (ORF), region is 1092 bp, encoding 363 amino acids (GenBank: KY316376). The molecular mass of the putative protein is approximately 38.4 kDa, and its theoretical isoelectric point (pI) is 8.3 (predicted by DNAMAN 7.0). Multiple sequence alignment of LcMYB2 with its homologs shows that a conserved domain exists among these sequences (amino acids 101–171; Fig. 2a). Sequence similarity and phylogenetic analysis show that LcMYB2 forms a clade with BAK02871 (Hordeum vulgare), CDM81700 (Triticum aestivum) and EMT01615 (Aegilops tauschii) by a high nodal support values (Fig. 2b, c). However, the functions of these LcMYB2 homologs have not been reported thus far. The analysis of LcMYB2 biological function is of great significance for Leymus chinensis and closely related species Triticum aestivum and Aegilops tauschii homologs.
Fig. 2.
Multiple sequence alignment and phylogenic analysis of LcMYB2 with homologous sequences on NCBI. a Sequence alignment using DNAMAN; b Homology tree constructed with DNAMAN. The numbers on the branches indicate the similarity of the sequences; c Molecular phylogenetic analysis by the maximum likelihood method with MEGA6.0; the numbers on the branches indicate the percentage of bootstrap support from 1000 replicates; the branch length represents the divergence distance
Subcellular localization and transcription activity assay of LcMYB2
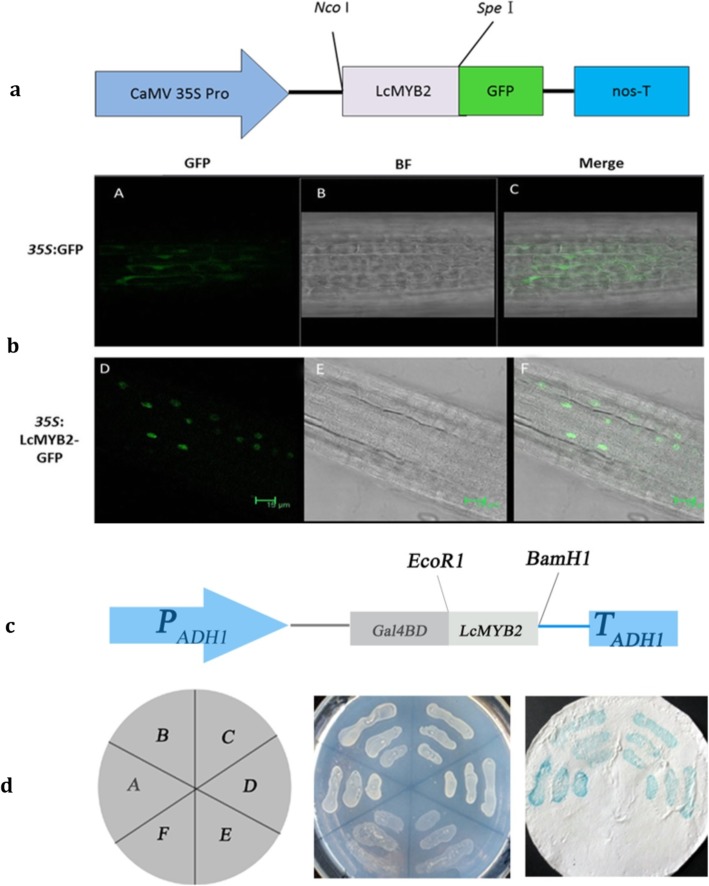
To determine the subcellular localization of LcMYB2, the ORF of LcMYB2 (without the TGA stop codon) was fused to a GFP reporter gene under the control of the CaMV 35S promoter (Fig. 3a). Recombinant CaMV35S::LcMYB2-GFP and CaMV35S::GFP were transformed into Arabidopsis separately by the floral dip method. Confocal microscopy showed that the GFP protein was localized throughout the whole cell, whereas the LcMYB2-GFP fusion protein was present only in the nucleus (Fig. 3b), suggesting that LcMYB2 is a nuclear-localized protein.
Fig. 3.
Subcellular localization and transcription activity of LcMYB2. (a) The LcMYB2-GFP construct; (b) Subcellular location of LcMYB2; A, D: GFP; B, E: bright field observation; C, F: merge; (c) The Gal4BD-LcMYB2 construct; (d) Transcription activity detected by X-gal staining, Left, The arrangement of yeast lines harboring different constructs, A and B are LcMYB2-containing yeast; C and D are positive controls; E and F are negative controls; Medium, The phenotype of yeast growing on SD/−His-Trp medium; Right, Beta-galactosidase activity assay of using X-gal
The transcriptional activation of LcMYB2 was tested using a yeast one-hybrid assay system. The LcMYB2 ORF was inserted at the 3′-end of GAL-BD under the control of PADH1 to form a BD-LcMYB2 fusion gene (Fig. 3c). The yeast strain AH109, harboring BD-LcMYB2 or BD-WRKY15 (positive controls), grew normally on SD/−His-Trp medium, whereas AH109 harboring only BD (negative control) did not grow. In β-galactosidase activity assays on Whatman filter paper, blue signal appeared in the regions where BD-LcMYB2 or BD-WRKY15-containing yeast were growing (Fig. 3d). Therefore, we suggest that LcMYB2 serves as a transcription activator and functions in the nucleus.
Performance of transgenic plants under osmotic stress, ABA treatment and natural drought treatment
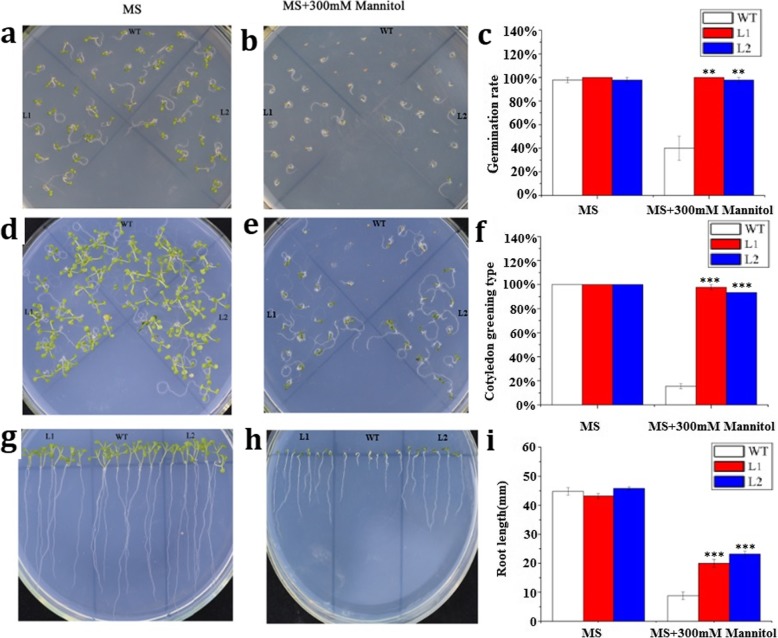
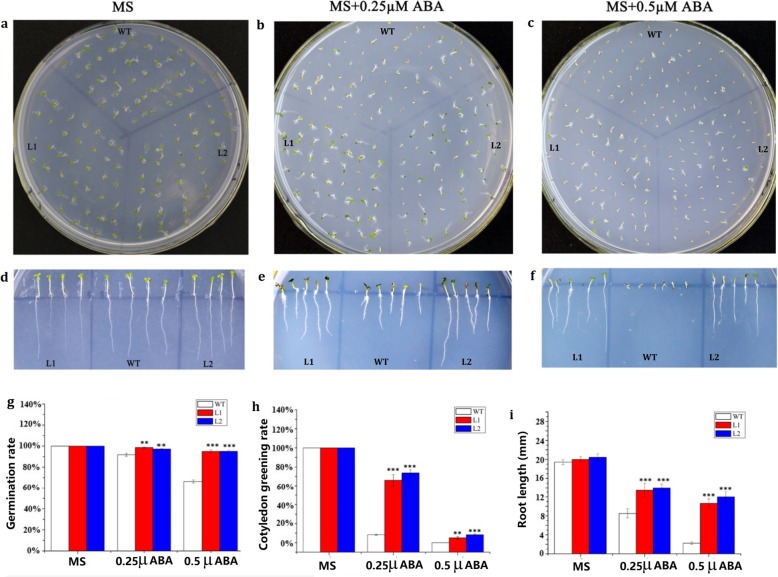
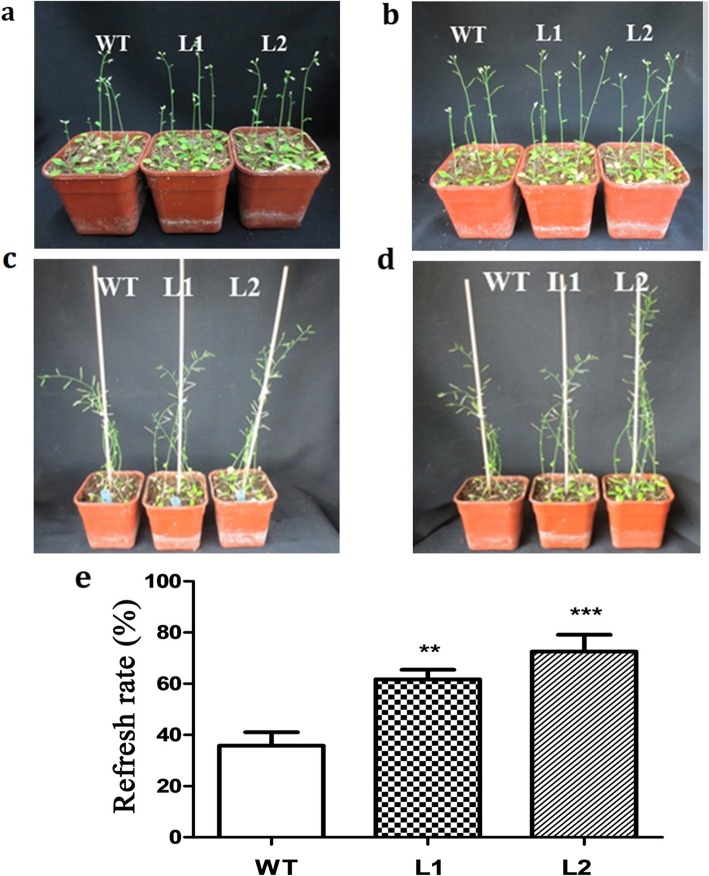
First, we probed the biological functions of LcMYB2 at the seed germination stage under osmotic or ABA treatment. Under normal conditions (Murashige-Skoog medium,MS), there were no significant differences between transgenic and wild-type seeds in germination rate, cotyledon greening rate or root length (Fig. 4a, d, g, c, f, i; Fig. 5a, d, g, h, i; Additional file 5: S5). Under treatment with 300 mmol/L mannitol, there were significant differences in germination rate (p < 0.01; Fig. 4b, c), and the cotyledon greening rate and root length had very significant differences (p < 0.001) between transgenic and wild-type seeds (Fig. 4e, f, h, i; Additional file 5: S5). Under treatment with 0.25 μmol/L ABA, the germination rate (p < 0.01), cotyledon greening rate (p < 0.001) and root length (p < 0.001) were significantly different between transgenic and wild-type seeds, and similar results were obtained with 0.5 μmol/L ABA treatment (Fig. 5b, c, e, f, g, h, i). Taken together, these data indicate that LcMYB2 can promote seed germination and root growth under osmotic stress and possibly via the ABA signaling pathway. In addition, the transgenic plants maintained green leaves longer under natural drought stress conditions and had a higher refresh rate after rewatering than did wild-type (Fig. 6).
Fig. 4.
Phenotypic and statistics analysis of LcMYB2 transgenic Arabidopsis under mannitol stress. a Germination on MS medium as control. Photos were taken on the 5th day; b Germination on MS containing 300 mM mannitol; c Statistics of germination rates; d Cotyledon greening rate on MS medium, Photos were taken on the 10th day; e Cotyledon greening rate on MS containing 300 mM mannitol; f Statistics of cotyledon greening; g Root growth on MS. Photos were taken on the 10th day; h Root growth on MS containing 300 mM mannitol; i Statistics of root length
Fig. 5.
Phenotypic and statistics analysis of LcMYB2 transgenic Arabidopsis under ABA stress. a Germination on MS medium as control; b Germination on MS containing 0.25 μM ABA; c Germination on MS containing 0.5 μM ABA; d Root length on MS medium; e Root length on MS containing 0.25 μM ABA; f Root length on MS containing 0.5 μM ABA; g Statistics of germination rates in different growth environments; h Statistics of cotyledon greening rates in different growth environments; i Statistics of root length in different growth environments. The germination and cotyledon greening rates were calculated halfway through the third day, and the photos of germination were taken on the 4th day after planting on medium. The photos of root growth were taken on the 5th day, and the root length was measured at the same time
Fig. 6.
Phenotypic and statistics analysis of LcMYB2 under natural drought. a Plants under natural drought stress for 21 days; b Plants under natural drought stress for 28 days; c Plants under natural drought stress for 42 days; d Plants 3 days after rewatering
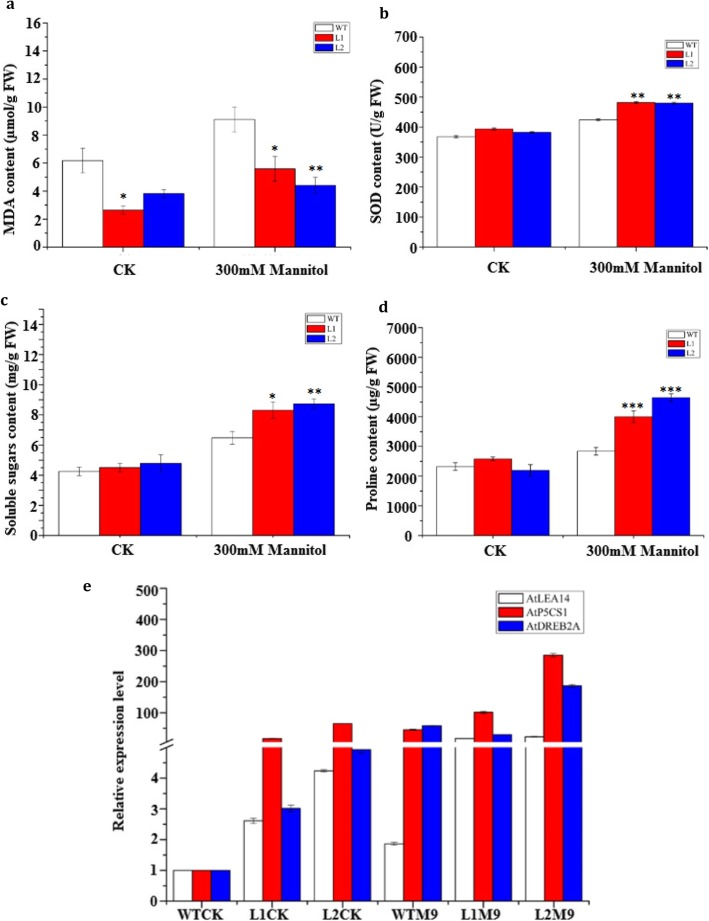
To investigate the physiological responses of transgenic and wild-type A. thaliana under osmotic stress, we irrigated 4-week-old seedlings with 300 mmol/L mannitol. Two days later, the malondialdehyde (MDA), Superoxide dismutase (SOD), soluble sugars and proline contents were measured. The results showed that the two transgenic lines overexpressing LcMYB2 accumulated greater amounts of SOD (p < 0.01), soluble sugars (p < 0.05/0.01) and proline (p < 0.001) than wild-type lines under 300 mmol/L mannitol treatment, whereas they had lower MDA content (Fig. 7a, b, c, d). The greater accumulation of proline and soluble sugars in the transgenic lines might provide extra protection to the cells under drought stress. The lower MDA and higher SOD content indicates that less damage occurred in the cells of transgenic plants. These results together suggest that LcMYB2 promotes osmotic stress resistance. The gene expression levels of AtDREB2A, AtP5CS1, and AtLEA14 were measured by Quantitative Real Time PCR (qPCR) on the 9th hour after treatment with 300 mmol/L mannitol. The expression levels of these genes were higher in transgenic plants than in wild-type plants both in the control check (CK) and in the treatment group (M9; Fig. 7e).
Fig. 7.
LcMYB2 significantly increased the accumulation of proline under mannitol stress. a MDA content of wild-type and transgenic Arabidopsis thaliana under control conditions and osmotic stress; b SOD content of wild-type and transgenic Arabidopsis thaliana under control conditions and osmotic stress; c Soluble sugar content of wild-type and transgenic Arabidopsis thaliana under control conditions and osmotic stress; d Proline content of wild-type and transgenic Arabidopsis thaliana under control conditions and osmotic stress; e Changes in gene expression of AtDREB2A, AtP5CS1 and AtLEA14. Physiological indices were measured with the wild-type and transgenic Arabidopsis thaliana exposed to 300 mM mannitol for 2 days
CHIP analysis
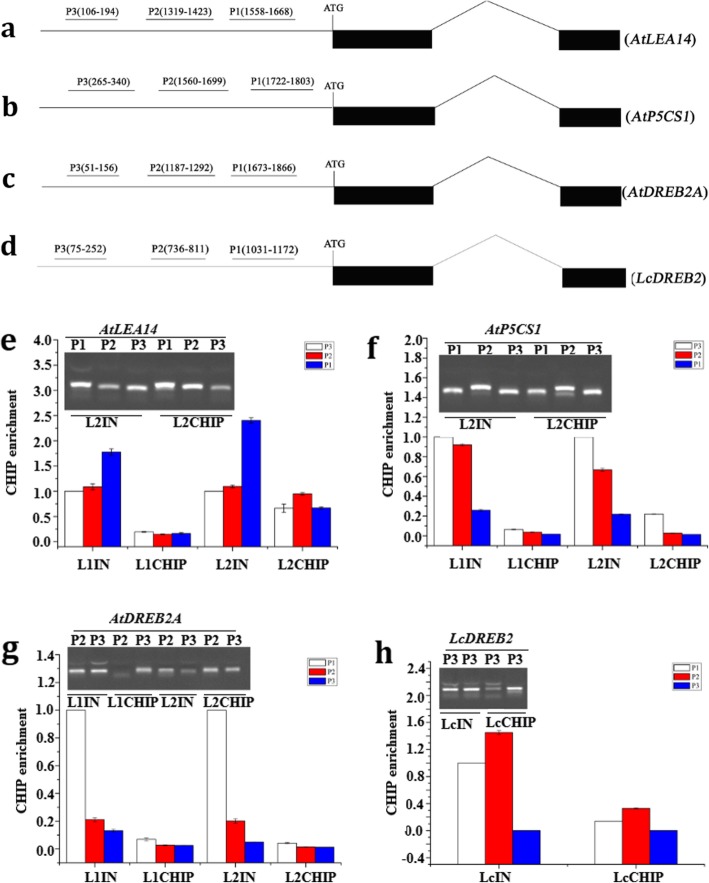
It has been shown that MYB proteins can recognize the motifs A/TAACCA and C/TAACG/TG [43]. Therefore, we analyzed the promoter sequences of AtLEA14, AtP5CS1, AtDREB2A and LcDREB2 using the sequences ~ 1500–2000 bp upstream of the predicted transcription start sites (TSSs), and several possible motifs were found in the putative promoter regions (Additional file 3: S3). We further confirmed our prediction with CHIP experiments, and the signals were detected in qPCR and universal PCR reactions with DNAs, released from proteins LICHIP, L2CHIP or LcCHIP, as templates. These results indicated that the LcMYB2 (or together with its interaction proteins) regulates the expression of AtLEA14, AtP5CS1, AtDREB2A and LcDREB2 (Fig. 8).
Fig. 8.
LcMYB2 binds to the promoter regions of targets, as revealed by CHIP. L1IN, L2IN, LcIN were the plant total nucleoproteins of transgenic Arabidopsis line 1, line 2 and Leymus chinensis leaf, while, L1CHIP, L2CHIP and LcCHIP were proteins that pulled down by antibody of LcMYB2 (anti-LcMYB2) from L1IN, L2IN, LcIN respectively. The DNAs released from these proteins were collected for PCR and qPCR analyses. a Schematic of the AtLEA14 gene; b Schematic of the AtP5CS1 gene; c Schematic of the AtDREB2A gene; d Schematic of the LcDREB2 gene; e The binding of LcMYB2 to the promoter region of AtLEA14. f: The binding of LcMYB2 to the promoter region of AtP5CS1; g The binding of LcMYB2 to the promoter region of AtDREB2A; h The binding of LcMYB2 to the promoter region of LcDREB2
Discussion
MYB and MYB-related transcription factors constitute a large family in plants and are involved in many biological processes, for example, secondary metabolism and responses to environmental factors [44–46]. Isolating and characterizing the functions of these genes provide a way to learn about plant-specific events at the transcriptional level. AtMYB2, a MYB-related protein, is induced by drought and by ABA; whereas BcMYB1 is rapidly and strongly induced by drought but only slightly by exogenous ABA, indicating that MYB proteins respond to environmental changes through both ABA-dependent and ABA-independent pathways [47–49]. LcMYB2 is induced to its maximal level at 1 hour after treatment with exogenous ABA and 8 hours after treatment with exogenous mannitol (Fig. 1a, d). However, the extent of LcMYB2 induction by mannitol is greater than by ABA, suggesting that LcMYB2 functions mainly in an ABA-independent pathway.
When facing drought stress, plants always adopt avoidance or resistance strategies to mitigate the negative effects of the stress. Rooting deeply is one of the avoidance strategies. For example, the introduction of the DEEPER ROOTING 1 (DRO1) gene into a shallow-rooting rice cultivar increased the downward growth of roots, and the resulting transgenic lines had higher yields under drought conditions [3]. Our results demonstrate that LcMYB2 is induced by mannitol and promotes root elongation at the germination stage and during seedling growth under osmotic stress and ABA treatment (Fig. 4 and Fig. 5). Therefore, LcMYB2 has the potential to enhance plant root growth to avoid drought stress.
Osmotic adjustment is usually thought to be one of the main mechanisms of resistance to drought or salt stress. Compatible osmolytes, such as proline, soluble sugars and LEA proteins, are often measured as critical physiological criteria to evaluate the tolerance of plants to abiotic stresses. Proline biosynthesis is catalyzed by P5CS1/2 and P5CR, and it is thought to protect subcellular structures and macromolecules under osmotic stress [50, 51]. Petunias overexpressing AtP5CS or OsP5CS accumulate more proline and appear to have drought tolerance [52]. In addition, soluble sugars, especially sucrose or trehalose, are correlated with the acquisition of desiccation tolerance and are thought to stabilize the membrane structure in dry environments [53, 54]. LEA proteins, first found in cotton, were shown to be up-regulated by drought stress in many species and function as compatible solutes to maintain the cellular structure in severe dehydration conditions [54–56]. Overexpression of some LEA protein-encoding genes confers enhanced drought tolerance in transgenic plants [57, 58]. Here, we found that LcMYB2 was induced by osmotic stress in sheepgrass (300mMmannitol); and A. thaliana overexpressing LcMYB2 accumulated more soluble sugars and free proline and expressed higher levels of AtLEA14, AtP5CS1 and AtDREB2A than wild-type A. thaliana seedlings under mannitol treatment (Fig. 7). In sheepgrass, many LEA protein-encoding genes and two P5CS-encoding genes were induced significantly by drought stress 35). Here, we demonstrate that LcMYB2 can bind to the promoter regions of AtLEA14 and AtP5CS1 (Fig. 8a, b, e, f). Therefore, we suggest that LcMYB2 functions in sheepgrass by elevating the content of osmoprotectants.
DREB proteins have been extensively studied and have been shown to improve drought tolerance in transgenic plants. AtDREB2A and AtDREB2B are strongly induced by dehydration stress in roots and stems, and constitutive expression of AtDREB2A results in significant drought stress tolerance [21, 59]. In addition, OsDREB2B is markedly induced by various stresses, and overexpressing OsDREB2B in A. thaliana or rice increases the expression of DREB2A target genes and improves transgenic plant drought stress tolerance [60, 61]. Previous studies have proposed that DREB proteins, such as DREB1 and DREB2, regulate low-temperature and drought-responsive genes by binding to the DRE/CTR elements through ABA-independent pathway [62, 63].
In sheepgrass, the highest transcript level of LcDREB2a occurs at the 12th hour under 20% PEG6000 treatment [42], whereas LcMYB2 transcript accumulation reaches the highest point at 8 h after 300 Mm mannitol treatment (Fig. 1a). Expression profile sequence analysis showed that both contig62249 (LcDREB2C/LcDREB2B/LcDREB2A) and contig41859 (LcMYB2) were up-regulated by drought stress and returned to basal levels after rewatering; however, the fold change of contig41859 was larger than that of contig62249 in response to drought stress (Additional file 1: S1). These results indicate that LcMYB2 is a possible transcription regulator upstream of LcDREB2. Therefore, we cloned the promoter sequence of LcDREB2 (~ 1500 bp upstream of the predicted transcription start site, Additional file 2: S2) and assayed the binding of LcMYB2 protein to this promoter region using CHIP analysis. Universal PCR and qPCR enrichment of CHIP DNA revealed that LcMYB2 can bind to the promoter regions of both AtDREB2A and LcDREB2 (Fig. 8c, d, g, h). Therefore, we propose that LcMYB2 improves drought tolerance by activating LcDREB2 in sheepgrass. Also, we find that LcMYB2 can bind to the promoters of sheepgrass LcDREB2 and Arabidopsis AtDREB2A, maybe they all have MYB binding elements in their promoters, which shows that the mechanism of response to stress in plants is conservative. Otherwise, pearson correlation analysis with differentially expressed genes DEGs of sheepgrass transcriptome data under drought stress [35] to gain novel insights on the mechanism of LcMYB2 on drought stress, the pearson correlation analysis results showed that LcMYB2 expression level was perfectly positively correlated with the expressions of typical stress response genes of LcLEA, LcDREB2c, LcMYB39, Peroxidase 56 under drought stress in sheepgrass (Additional file 6: S6), which provided evidence for the analysis of LcMYB2 function under drought stress. The pearson correlation analysis results about the expression levels of LcMYB2 with LcLEA, LcDREB2c, LcMYB39 in sheepgrass were mutually underlying with the results of CHIP-PCR on LcMYB2 binding the corresponding promoter elements of AtLEA14, AtP5CS1, AtDREB2A and LcDREB2. This research integrates the RNA-seq further analysis, CHIP-PCR and universal PCR results, and provides the evidence for understanding the function of LcMYB2 under drought stress.
With the development of the sheepgrass industry (artificial cultivation, natural grassland improvement, and artificial grassland establishment), the drought resistance of sheepgrass during the seed germination and seedling establishment stages in water-deficient areas is critical for propagation and is necessary for reaping economic and ecological benefits. Therefore, LcMYB2 is an important candidate for improving plant drought stress tolerance through genetic engineering.
Conclusions
In conclusion, we showed that a drought and osmotic-inducible transcription factor, LcMYB2, improves the drought and osmotic tolerance of plants by bingding the elements of promoters from the AtDREB2, AtLEA14, AtP5CS1 and LcDREB2 and regulating the transcription of drought-responsive genes to increase the accumulation of osmoprotectants (Fig. 9). These may be among the reasons underlying the tolerance of sheepgrass to drought-prone environments. Others, some the cis-elements of stress response genes from the monocotyledons and dicotyledonous plants are conserved in evolution, they both can be bound by the same trans-factors.
Fig. 9.
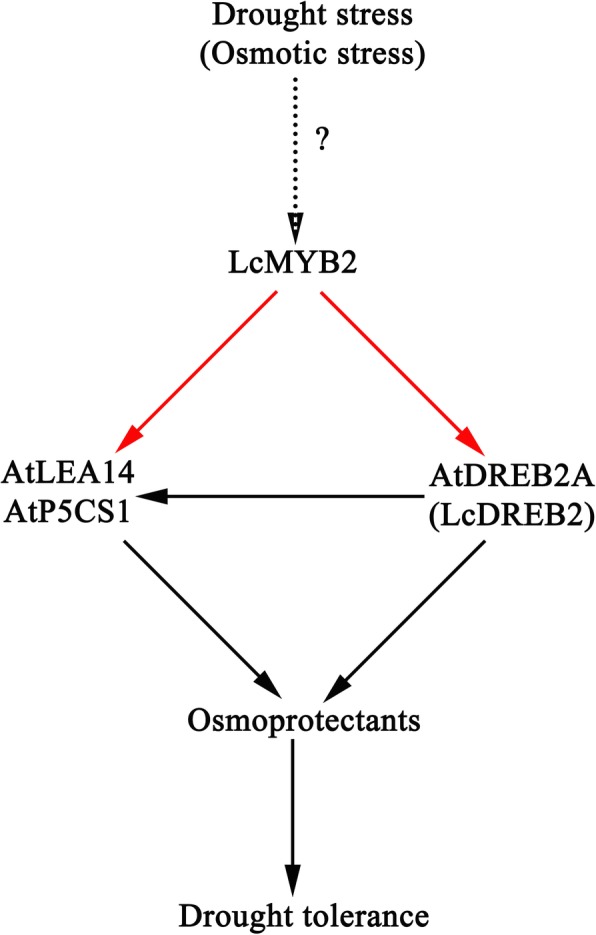
A hypothetical model of LcMYB2 function in transgenic plants
Methods
Plant materials and treatments
Sheepgrass (National certified variety Zhongke 1 from Institute of Botany, the Chinese Academy of Sciences, Beijing, China) was used for this experiment. The sheepgrass seedlings were grown in a plastic pot containing nutrition soil (Pindstrup Bubstrate, Denmark) and vermiculite (2:1, v/v) in the greenhouse at 27/23 °C, 16 h light/8 h dark for 8 weeks before treatments. Abiotic stresses were performed as follows: the plastic pots containing the seedlings were submerged in 100 μmol/L for ABA treatment, 300 mmol/L mannitol for osmotic stress and 400 mmol/L NaCl for salt stress. Seedlings were transferred to a growth chamber at 4 °C for cold stress. The seedlings were sampled at 0, 1, 3, 8, 12 or 24 h after stress treatments, immediately frozen in liquid nitrogen and stored at − 80 °C for RNA isolation. The abiotic stress experiments were excuted at least three times by different people to test the expression patterns of target genes. Stem, leaf, root, bud, panicle and rhizomes were also collected from 2-year-old sheepgrass seedlings for tissue-specific analysis.
Arabidopsis thaliana (A. thaliana; Columbia ecotype) seeds were surface-sterilized with 10% NaClO for 10 min, and then washed 5 times with sterile water. The sterilized seeds were planted on MS solid media (pH 5.8) for germination.
RNA isolation and expression pattern analysis of LcMYB2
Total RNA was extracted using Trizol reagent (TaKaRa, Dalian, China) according to the manufacturer’s instructions. First-strand cDNA was synthesized using the PrimeScript™ RT reagent Kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. qRT-PCR was carried out in triplicate according to the SYBR PremixExTaq™ protocol (TaKaRa, Dalian, China) on a LightCycler480 Real-Time PCR System (Roche, Rotkreuz, Switzerland) with the following program: 95 °C for 5 s and 68 °C for 30 s for 45 cycles. Tissue-specific expression of LcMYB2 was detected using semiquantitative PCR. All primers used in this research are listed in Additional file 4: S4.
Amplification and sequence analysis of LcMYB2
First-strand cDNA for amplification of the 5′ and 3′ ends of LcMYB2 was synthesized with SMARTer™ RACE cDNA Amplification Kit (Clontech, Palo Alto, CA) according to the manufacturer’s instructions. The gene-specific primer (GSP-5’RACE), designed according to the 454 high-throughput sequencing results, and universal primers (UPM) were used to amplify the 5′ end of LcMYB2. The putative full-length sequence of LcMYB2 was amplified using gene-specific primers (LcMYB2-F/R) designed according to the sequence assembled from the 5′-RACE and 454 sequencing results. The national center for biotechnology information (NCBI) database was searched for homologs of LcMYB2 using the BLASTX program. Multiple sequence alignment was executed in DNAMAN software (version 7.0) using the selected amino acid sequences. The phylogenetic relationships among the homologs were inferred using the maximum likelihood method based on the JTT matrix-based model in MEGA software version 6.0 [64, 65].
Subcellular localization and transcriptional activity assay of LcMYB2
The ORF of LcMYB2 was ligated into pCAMBIA1302 to form the LcMYB2-GFP fusion protein. The recombinant plasmid was introduced into Agrobacterium tumefaciens EHA105 using a freeze-thaw method and further transformed into A. thaliana using the floral dip method [66]. Positive seedlings were selected on solid MS containing 50 μg/L hygromycin (Roche) and further confirmed by PCR. T3 seeds of the transgenic plants were germinated on MS, and the GFP fluorescence in the roots was observed under a laser confocal scanning microscope (Leica TCS SP5). To assess LcMYB2 transcription activity, the ORF was inserted downstream of GAL-BD in the pBridge vector to obtain pBD-LcMYB2. The recombinant vectors were introduced into the yeast strain AH109, and positive transformants were selected on SD (−Trp) medium and confirmed by PCR. β-galactosidase activity was assayed according to the Yeast Protocols Handbook (Clontech).
Arabidopsis transformation
To reveal the biological function of LcMYB2, the ORF was fused to the p3301–121 vector (modified from vector pCAMBIA3301 and pBI121, donated by the Shen lab) under the control of the CaMV 35S promoter. The recombinant constructs were transformed into A. thaliana by Agrobacterium tumefaciens EHA105 using the floral dip method. Positive transgenic Arabidopsis seeds were screened on MS medium supplemented with 20 μg/L glufosinate ammonium and further confirmed by PCR using DNA extracted from the putative positive seedlings. T3 seeds were used for germination assays under different treatments.
Drought stress tolerance analysis of transgenic seedlings
To reveal the function of LcMYB2 at the germination stage, T3 seeds of transgenic and wild-type A. thaliana were planted on normal solid MS medium and solid MS medium supplemented mannitol (300 m mol/L) or ABA (0.25 μmol/L and 0.5 μmol/L). The Petri dishes were placed in a growth chamber at 22 °C with a 16 h / 8 h, light/dark photoperiod. Plants were photographed, and the germination rate, cotyledon greening rate and root length were and measured. Osmotic and ABA stress tolerance experiments were repeated at least three times.
To test the role of LcMYB2 at the seedling stage under drought conditions, 4-day seedlings were transplanted into a pot containing vermiculite and turfy soil (2:1, v/v) and grown in a growth chamber at 22 °C with a 16 h / 8 h, light/dark photoperiod for 1 week with sufficient water, then started to natural drought stress without water for the following 42 days. During the drought process, the soil water content was monitored every day in each pot. On the 42nd day, the seedlings were irrigated again, and survival rates were statistically analyzed after 3 days. There are 60 seedlings for drought stress experiment in each line (WT, L1, L2), respectively.
Measurement of lipid peroxidation and of proline and soluble sugar content
Four-week-old transgenic and WT A. thaliana seedlings were irrigated with 300 mmol/L mannitol. The leaves were sampled at 0 h and 9 h after treatment for gene expression analysis. Two days later, the leaves of transgenic and wild-type lines were harvested for physiological measurements. The level of MDAwas determined by a revised method described by Kramer et al. [67]. SOD content was measured with the nitro-blue tetrazolium (NBT) reduction method as previously described [68]. The contents of proline and soluble sugars were determined according to the protocols previously described by Shan et al. [69] and Bailey [70], respectively. Three replicates were carried out for each assay, and the variability was indicated with the standard error (SE).
Chromatin immunoprecipitation assays
Four-week-old seedlings of sheepgrass were treated with 300 mmol/L mannitol for 8 h and 24 h to induce the expression of LcMYB2, after which time, the samples were harvested. Six-week-old seedlings of A. thaliana overexpressing LcMYB2 (L1: line 1, L2: line 2) were tested. All samples were fixed with formaldehyde for CHIP analysis. Antibodies against LcMYB2 were prepared by Beijing Protein Innovation Co., Ltd. The EpiQuik Plant ChIP kit (Epigentek, Brooklyn, NY) was used for CHIPanalysis. CHIP DNA was detected with universal PCR (40 cycles of 95 °C, 30 S; 68 °C, 30 S) and qPCR (45 cycles of 95 °C, 5 S; 68 °C, 30 S). The method of comparing Ct values was adopted to analyze the qPCR CHIP efficiency.
Statistical analysis
Analysis of variance (ANOVA) and t-test were used to compare the differences between samples. *** indicates p-values < 0.001, ** indicates p-values < 0.01) and * indicates p-values < 0.05.
Supplementary information
Additional file 1. S1 MYB-related transcription factor with unknown function.
Additional file 2. S2. Amplification of full-length LcMYB2 and Amplification of LcDREB2 promoter sequence.
Additional file 3. S3 Promoter sequence used in CHIP experiment and Possible MYB recognition site.
Additional file 4. S4. Primers used in this research.
Additional file 5. S5. The root growth experiment.
Additional file 6. S6. Pearson correlation analysis between LcMYB2 and other genes.
Acknowledgements
We thank Dr. Xiaobing Dong supplied much contribution in materials collection and management.
Abbreviations
- ABA
Abscisic acid
- AP2/ERF
APETUAP2/Ethylene-Responsive-Element Binding Protein
- bZIP
Basic Leucine Zipper
- CHIP
Chromatin immunoprecipitation
- DREB
Dehydration Responsive Element Binding Protein
- LcMYB2
Leymus chinensis MYB DNA-binding domain protein 2
- LEA
Late-embryogenesis-abundant protein
- MS
Murashige-Skoog medium
- MYB
MYB DNA-binding domain protein 2
- NAC
NAM, ATAF and CUC transcription factor
- NCBI
The national center for biotechnology information
- ORF
Open reading frame;
- P5CS1
△1-Pyrroline-5-carboxylate synthetas
- PP2Cs
Plant protein phosphatases
- qPCR
Quantitative Real Time PCR
- RACE-PCR
Rapid-Amplification of cDNA Ends-PCR
Authors’ contributions
GL and LC conceived and designed the experiments. SH performed most of the experiments. PZ, XG and LC made substantial contributions to the data analysis and the manuscript writing. GL gave the final approval the manuscript. JJ, WY, ZL, SC, XL and DQ were involved in performing the experiments; all authors read and approved the final manuscript.
Funding
This work was supported by the National Basic Research Program of China (“973” program, 2014CB138704), the Project of Science and Technology Service Network Initiative of the Chinese Academy of Sciences (STS, KFJ-EW-STS-119), Kulun Banner Science and Technology Poverty Alleviation Project of Chinese Academy of Sciences, and the Science and Technology Major Project of Inner Mongolia Autonomous Region of China. The funders had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript, but just provide the financial support.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Pincang Zhao, Shenglin Hou and Xiufang Guo contributed equally to this work.
Contributor Information
Pincang Zhao, Email: zhaopincang@163.com.
Shenglin Hou, Email: shenglinhou@aliyun.com.
Xiufang Guo, Email: xiufguo@163.com.
Junting Jia, Email: jiajunting123@126.com.
Weiguang Yang, Email: anda580@163.com.
Zhujiang Liu, Email: liuzhujiang1986@163.com.
Shuangyan Chen, Email: sychen@ibcas.ac.cn.
Xiaoxia Li, Email: lixx2013@ibcas.ac.cn.
Dongmei Qi, Email: qidm@ibcas.ac.cn.
Gongshe Liu, Email: liugs@ibcas.ac.cn.
Liqin Cheng, Email: lqcheng@ibcas.ac.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12870-019-2159-2.
References
- 1.Baskin CC, Baskin JM. Seeds: ecology, biogeography, and evolution of dormancy and germination 2nd ed.[M]:AP; 2014.
- 2.Mickelbart MV, Hasegawa PM, Bailey-Serres J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat Rev Genet. 2015;16:237–251. doi: 10.1038/nrg3901. [DOI] [PubMed] [Google Scholar]
- 3.Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet. 2013;45:1097. doi: 10.1038/ng.2725. [DOI] [PubMed] [Google Scholar]
- 4.Chen THH, Murata N. Glycinebetaine: an effective protectant against abiotic stress in plants. Trends Plant Sci. 2008;13:499–505. doi: 10.1016/j.tplants.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Kollist H, Nuhkat M, Roelfsema MRG. Closing gaps: linking elements that control stomatal movement. New Phytol. 2014;203:44–62. doi: 10.1111/nph.12832. [DOI] [PubMed] [Google Scholar]
- 6.Valliyodan B, Nguyen HT. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr Opin Plant Biol. 2006;9:189–195. doi: 10.1016/j.pbi.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 7.Valluru R, Van den Ende W. Plant fructans in stress environments: emerging concepts and future prospects. J Exp Bot. 2008;59:2905–2916. doi: 10.1093/jxb/ern164. [DOI] [PubMed] [Google Scholar]
- 8.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu JK. Abiotic stress signaling and responses in plants. Cell. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci U S A. 2009;106:21425–21430. doi: 10.1073/pnas.0912021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Savoure A, Hua XJ, Bertauche N, VanMontagu M, Verbruggen N. Abscisic acid-independent and abscisic acid-dependent regulation of proline biosynthesis following cold and osmotic stresses in Arabidopsis thaliana. Mol Gen Genet. 1997;254:104–109. doi: 10.1007/s004380050397. [DOI] [PubMed] [Google Scholar]
- 12.Strizhov N, Abraham E, Okresz L, Blickling S, Zilberstein A, Schell J, et al. Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J. 1997;12:557–569. doi: 10.1046/j.1365-313x.1997.00557.x. [DOI] [PubMed] [Google Scholar]
- 13.Urano K, Maruyama K, Ogata Y, Morishita Y, Takeda M, Sakurai N, et al. Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J. 2009;57:1065–1078. doi: 10.1111/j.1365-313X.2008.03748.x. [DOI] [PubMed] [Google Scholar]
- 14.Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, et al. Regulators of PP2C phosphatase activity function as Abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 15.Miyakawa T, Fujita Y, Yamaguchi-Shinozaki K, Tanokura M. Structure and function of abscisic acid receptors. Trends Plant Sci. 2013;18:259–266. doi: 10.1016/j.tplants.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–U679. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- 17.Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, et al. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009;462:665–U143. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- 18.Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–U138. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends Plant Sci. 2010;15:395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 20.Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res. 2011;124:509–525. doi: 10.1007/s10265-011-0412-3. [DOI] [PubMed] [Google Scholar]
- 21.Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, et al. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell. 2006;18:1292–1309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu ZS, Chen M, Li LC, Ma YZ. Functions and application of the AP2/ERF transcription factor family in crop improvement. J Integr Plant Biol. 2011;53:570–585. doi: 10.1111/j.1744-7909.2011.01062.x. [DOI] [PubMed] [Google Scholar]
- 23.Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, et al. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol. 2005;15:1196–1200. doi: 10.1016/j.cub.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 25.Liang YK, Dubos C, Dodd IC, Holroyd GH, Hetherington AM, Campbell MM. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr Biol. 2005;15:1201–1206. doi: 10.1016/j.cub.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 26.Seo PJ, Xiang FN, Qiao M, Park JY, Lee YN, Kim SG, et al. The MYB96 transcription factor mediates Abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009;151:275–289. doi: 10.1104/pp.109.144220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, et al. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008;146:623–635. doi: 10.1104/pp.107.110981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin YX, Wang MC, Tian YC, He WX, Han L, Xia GM. Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis. Mol Biol Rep. 2012;39:7183–7192. doi: 10.1007/s11033-012-1550-y. [DOI] [PubMed] [Google Scholar]
- 29.Shin D, Moon SJ, Han S, Kim BG, Park SR, Lee SK, et al. Expression of StMYB1R-1, a novel potato single MYB-like domain transcription factor, increases drought Tolerance. Plant Physiol. 2011;155:421–432. doi: 10.1104/pp.110.163634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang ZY, Liu X, Wang XD, Zhou MP, Zhou XY, Ye XG, et al. An R2R3 MYB transcription factor in wheat, TaPIMP1, mediates host resistance to Bipolaris sorokiniana and drought stresses through regulation of defense- and stress-related genes. New Phytol. 2012;196:1155–1170. doi: 10.1111/j.1469-8137.2012.04353.x. [DOI] [PubMed] [Google Scholar]
- 31.Wang XL, Wang HW, Liu SX, Ferjani A, Li JS, Yan JB, et al. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat Genet. 2016;48:1233–1241. doi: 10.1038/ng.3636. [DOI] [PubMed] [Google Scholar]
- 32.Chen SY, Huang X, Yan XQ, Liang Y, Wang YZ, Li XF, et al. Transcriptome analysis in Sheepgrass (Leymus chinensis): a dominant perennial grass of the Eurasian steppe. PLoS One. 2013;8:11. doi: 10.1371/annotation/dd945f7c-c50b-461d-ab38-15e8b0966458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhai Junfeng, Dong Yuanyuan, Sun Yepeng, Wang Qi, Wang Nan, Wang Fawei, Liu Weican, Li Xiaowei, Chen Huan, Yao Na, Guan Lili, Chen Kai, Cui Xiyan, Yang Meiying, Li Haiyan. Discovery and Analysis of MicroRNAs in Leymus chinensis under Saline-Alkali and Drought Stress Using High-Throughput Sequencing. PLoS ONE. 2014;9(11):e105417. doi: 10.1371/journal.pone.0105417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Yepeng, Wang Fawei, Wang Nan, Dong Yuanyuan, Liu Qi, Zhao Lei, Chen Huan, Liu Weican, Yin Hailong, Zhang Xiaomei, Yuan Yanxi, Li Haiyan. Transcriptome Exploration in Leymus chinensis under Saline-Alkaline Treatment Using 454 Pyrosequencing. PLoS ONE. 2013;8(1):e53632. doi: 10.1371/journal.pone.0053632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao PC, Liu PP, Yuan GX, Jia JT, Li XX, Qi DM, et al. New insights on drought stress response by global investigation of gene expression changes in Sheepgrass (Leymus chinensis). Front Plant Sci. 2016;7:954. [DOI] [PMC free article] [PubMed]
- 36.Cheng LQ, Li XX, Huang X, Ma T, Liang Y, Ma XY, et al. Overexpression of sheepgrass R1-MYB transcription factor LcMYB1 confers salt tolerance in transgenic Arabidopsis. Plant Physiol Biochem. 2013;70:252–260. doi: 10.1016/j.plaphy.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 37.Gao Q, Li XX, Jia JT, Zhao PC, Liu PP, Liu ZJ, et al. Overexpression of a novel cold-responsive transcript factor LcFIN1 from sheepgrass enhances tolerance to low temperature stress in transgenic plants. Plant Biotechnol J. 2016;14:861–874. doi: 10.1111/pbi.12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu CQ, Gong P, Yin YY. China's water crisis needs more than words. Nature. 2011;470:307. doi: 10.1038/470307a. [DOI] [PubMed] [Google Scholar]
- 39.Li XX, Gao Q, Liang Y, Ma T, Cheng LQ, Qi DM, et al. A novel salt-induced gene from sheepgrass, LcSAIN2, enhances salt tolerance in transgenic Arabidopsis. Plant Physiol Biochem. 2013;64:52–59. doi: 10.1016/j.plaphy.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 40.Li Xiaoxia, Hou Shenglin, Gao Qiong, Zhao Pincang, Chen Shuangyan, Qi Dongmei, Lee Byung-Hyun, Cheng Liqin, Liu Gongshe. LcSAIN1, a Novel Salt-Induced Gene from SheepGrass, Confers Salt Stress Tolerance in Transgenic Arabidopsis and Rice. Plant and Cell Physiology. 2013;54(7):1172–1185. doi: 10.1093/pcp/pct069. [DOI] [PubMed] [Google Scholar]
- 41.Peng XJ, Ma XY, Fan WH, Su M, Cheng LQ, Alam I, et al. Improved drought and salt tolerance of Arabidopsis thaliana by transgenic expression of a novel DREB gene from Leymus chinensis. Plant Cell Rep. 2011;30:1493–1502. doi: 10.1007/s00299-011-1058-2. [DOI] [PubMed] [Google Scholar]
- 42.Peng XJ, Zhang LX, Zhang LX, Liu ZJ, Cheng LQ, Yang Y, et al. The transcriptional factor LcDREB2 cooperates with LcSAMDC2 to contribute to salt tolerance in Leymus chinensis. Plant Cell Tiss Org Cult. 2013;113:245–256. doi: 10.1007/s11240-012-0264-0. [DOI] [Google Scholar]
- 43.Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gatica-Arias A, Farag MA, Stanke M, Matousek J, Wessjohann L, Weber G. Flavonoid production in transgenic hop (Humulus lupulus L.) altered by PAP1/MYB75 from Arabidopsis thaliana L. Plant Cell Rep. 2012;31:111–119. doi: 10.1007/s00299-011-1144-5. [DOI] [PubMed] [Google Scholar]
- 45.Lotkowska ME, Tohge T, Fernie AR, Xue GP, Balazadeh S, Mueller-Roeber B. The Arabidopsis transcription factor MYB112 promotes anthocyanin formation during salinity and under high light stress. Plant Physiol. 2015;169:1862–1880. doi: 10.1104/pp.15.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren LJ, Hu ZL, Li YL, Zhang B, Zhang YJ, Tu Y, et al. Heterologous expression of BoPAP1 in tomato induces stamen specific anthocyanin accumulation and enhances tolerance to a long-term low temperature stress. J Plant Growth Regul. 2014;33:757–768. doi: 10.1007/s00344-014-9423-3. [DOI] [Google Scholar]
- 47.Chen BJ, Wang Y, Hu YL, Wu Q, Lin ZP. Cloning and characterization of a drought-inducible MYB gene from Boea crassifolia. Plant Sci. 2005;168:493–500. doi: 10.1016/j.plantsci.2004.09.013. [DOI] [Google Scholar]
- 48.Urao T, Noji M, YamaguchiShinozaki K, Shinozaki K. A transcriptional activation domain of ATMYB2, a drought-inducible Arabidopsis Myb-related protein. Plant J. 1996;10:1145–1148. doi: 10.1046/j.1365-313X.1996.10061145.x. [DOI] [PubMed] [Google Scholar]
- 49.Urao T, Yamaguchishinozaki K, Urao S, Shinozaki K. An Arabidopsis Myb homolog is induced by dehydration stress and its gene-product binds to the conserved Myb recognition sequence. Plant Cell. 1993;5:1529–1539. doi: 10.1105/tpc.5.11.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kishor PBK, Sangam S, Amrutha RN, Laxmi PS, Naidu KR, Rao KRSS, et al. Regulation of proline biosynthesis, degradation, uptake and transport in higher plants: its implications in plant growth and abiotic stress tolerance. Curr Sci. 2005;88:424–438. [Google Scholar]
- 51.Szabados L, Savoure A. Proline: a multifunctional amino acid. Trends Plant Sci. 2010;15:89–97. doi: 10.1016/j.tplants.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 52.Yamada M, Morishita H, Urano K, Shiozaki N, Yamaguchi-Shinozaki K, Shinozaki K, et al. Effects of free proline accumulation in petunias under drought stress. J Exp Bot. 2005;56:1975–1981. doi: 10.1093/jxb/eri195. [DOI] [PubMed] [Google Scholar]
- 53.Crowe JH, Hoekstra FA, Crowe LM. An hydrobiosis. Annu Rev Physiol. 1992;54:579–599. doi: 10.1146/annurev.ph.54.030192.003051. [DOI] [PubMed] [Google Scholar]
- 54.Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- 55.Dure L, Greenway SC, Galau GA. Developmental biochemistry of cottonseed embryogenesis and germination-changing messenger ribonucleic-acid populations as shown by invitro and invivo protein-synthesis. Biochemistry. 1981;20(14):4162–4168. doi: 10.1021/bi00517a033. [DOI] [PubMed] [Google Scholar]
- 56.Xu DP, Duan XL, Wang BY, Hong BM, Ho THD, Wu R. Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 1996;110:249–257. doi: 10.1104/pp.110.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao BZ, Huang YM, Tang N, Xiong LZ. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor Appl Genet. 2007;115:35–46. doi: 10.1007/s00122-007-0538-9. [DOI] [PubMed] [Google Scholar]
- 58.Goyal K, Walton LJ, Tunnacliffe A. LEA proteins prevent protein aggregation due to water stress. Biochem J. 2005;388:151–157. doi: 10.1042/BJ20041931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakashima K, Shinwari ZK, Sakuma Y, Seki M, Miura S, Shinozaki K, et al. Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol Biol. 2000;42:657–665. doi: 10.1023/A:1006321900483. [DOI] [PubMed] [Google Scholar]
- 60.Chen JQ, Meng XP, Zhang Y, Xia M, Wang XP. Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol Lett. 2008;30:2191–2198. doi: 10.1007/s10529-008-9811-5. [DOI] [PubMed] [Google Scholar]
- 61.Matsukura S, Mizoi J, Yoshida T, Todaka D, Ito Y, Maruyama K, et al. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol Genet Genomics. 2010;283:185–196. doi: 10.1007/s00438-009-0506-y. [DOI] [PubMed] [Google Scholar]
- 62.Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, et al. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci U S A. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 65.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clough SJ, Bent AF. Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 67.Kramer GF, Norman HA, Krizek DT, Mirecki RM. Influence of Uv-B radiation on polyamines, lipid-peroxidation and membrane-lipids in cucumber. Phytochemistry. 1991;30:2101–2108. doi: 10.1016/0031-9422(91)83595-C. [DOI] [Google Scholar]
- 68.Durak I, Yurtarslanl Z, Canbolat O, Akyol O. A methodological approach to superoxide-dismutase (sod) activity assay based on inhibition of Nitroblue Tetrazolium (Nbt) reduction. Clin Chim Acta. 1993;214:103–104. doi: 10.1016/0009-8981(93)90307-P. [DOI] [PubMed] [Google Scholar]
- 69.Shan DP, Huang JG, Yang YT, Guo YH, Wu CA, Yang GD, et al. Cotton GhDREB1 increases plant tolerance to low temperature and is negatively regulated by gibberellic acid. New Phytol. 2007;176:70–81. doi: 10.1111/j.1469-8137.2007.02160.x. [DOI] [PubMed] [Google Scholar]
- 70.Bailey RW. The reaction of pentoses with anthrone. Biochem J. 1958;68:669–672. doi: 10.1042/bj0680669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. S1 MYB-related transcription factor with unknown function.
Additional file 2. S2. Amplification of full-length LcMYB2 and Amplification of LcDREB2 promoter sequence.
Additional file 3. S3 Promoter sequence used in CHIP experiment and Possible MYB recognition site.
Additional file 4. S4. Primers used in this research.
Additional file 5. S5. The root growth experiment.
Additional file 6. S6. Pearson correlation analysis between LcMYB2 and other genes.
Data Availability Statement
Not applicable.