Salmonella is an intracellular bacterium found in the gastrointestinal tract of mammalian, avian, and reptilian hosts. Mouse models have been extensively used to model in vivo distinct aspects of human Salmonella infections and have led to the identification of several host susceptibility genes. We have investigated the susceptibility of Collaborative Cross strains to intravenous infection with Salmonella enterica serovar Typhimurium as a model of human systemic invasive infection.
KEYWORDS: host resistance, immunophenotyping, QTL mapping, Salmonella
ABSTRACT
Salmonella is an intracellular bacterium found in the gastrointestinal tract of mammalian, avian, and reptilian hosts. Mouse models have been extensively used to model in vivo distinct aspects of human Salmonella infections and have led to the identification of several host susceptibility genes. We have investigated the susceptibility of Collaborative Cross strains to intravenous infection with Salmonella enterica serovar Typhimurium as a model of human systemic invasive infection. In this model, strain CC042/GeniUnc (CC042) mice displayed extreme susceptibility with very high bacterial loads and mortality. CC042 mice showed lower spleen weights and decreased splenocyte numbers before and after infection, affecting mostly CD8+ T cells, B cells, and all myeloid cell populations, compared with control C57BL/6J mice. CC042 mice also had lower thymus weights with a reduced total number of thymocytes and double-negative and double-positive (CD4+, CD8+) thymocytes compared to C57BL/6J mice. Analysis of bone marrow-resident hematopoietic progenitors showed a strong bias against lymphoid-primed multipotent progenitors. An F2 cross between CC042 and C57BL/6N mice identified two loci on chromosome 7 (Stsl6 and Stsl7) associated with differences in bacterial loads. In the Stsl7 region, CC042 carried a loss-of-function variant, unique to this strain, in the integrin alpha L (Itgal) gene, the causative role of which was confirmed by a quantitative complementation test. Notably, Itgal loss of function increased the susceptibility to S. Typhimurium in a (C57BL/6J × CC042)F1 mouse background but not in a C57BL/6J mouse inbred background. These results further emphasize the utility of the Collaborative Cross to identify new host genetic variants controlling susceptibility to infections and improve our understanding of the function of the Itgal gene.
INTRODUCTION
Salmonella enterica is a relatively common Gram-negative bacterium that is generally transmitted via the consumption of contaminated food or water (1). Infection with Salmonella can lead to a variety of pathologies, with worldwide health and economic costs. Human-restricted Salmonella serovars S. enterica serovar Typhi and S. Paratyphi result in typhoid fever, causing an estimated 190,000 deaths per year, and are typically observed in nations lacking adequate sanitation and clean drinking water programs (2, 3). Symptoms of typhoid include fever, abdominal pain, and general malaise (4). In contrast, nontyphoidal strains, such as S. Typhimurium, lead to 93.8 million cases of gastroenteritis annually (5). Symptoms of gastroenteritis involve diarrhea, vomiting, and nausea (1). In immunocompromised patients, nontyphoidal strains can also result in systemic and invasive infections involving bacteremia and sepsis (6).
The study of Salmonella in mouse models is typically conducted with S. Typhimurium, as it is known to induce systemic infections in mice similar to the bacteremia observed in immunocompromised patients (1). After systemic infection with S. Typhimurium, the bacteria are rapidly cleared from the bloodstream (within 2 h), followed by localization of approximately 10% of the inoculum within macrophages and polymorphonuclear cells of visceral organs, such as the spleen and liver, where it can replicate efficiently. In order to resolve the resulting systemic infection, the host must activate a robust innate and adaptive immune response (1, 7).
Many factors are known to be involved in the clinical outcomes and the ability of the host to clear Salmonella infection in both humans and mouse models. Factors include the bacterial strain, the dosage of infection, and the host immune status, microbiome, and genetic makeup (1, 6, 8, 9). Host genetics are increasingly being recognized as a crucial element involved in host susceptibility to infection. While many genes, such as those for Toll-like receptor 4 (TLR4), interleukin 12 (IL-12), and signal transducer and activator of transcription 4 (STAT4), have been implicated in the in vivo response to Salmonella, the complete cohort of genes involved has yet to be determined (8, 10–12). Nonetheless, establishing the genetic factors that influence disease is essential for the elucidation of host immune response pathways and will enable the identification of novel targets for therapeutic drug development. Recent genome-wide association studies have revealed the contribution of host genetic determinants of enteric fever and invasive nontyphoidal Salmonella infection in human populations (13–15).
One approach used for the detection of novel genes involved in complex traits, such as Salmonella susceptibility, utilizes a murine genetic reference population known as the Collaborative Cross (CC) (16). While traditional models tend to use highly homogeneous mouse populations, the CC has been designed to model the range of genetic variation of the human population (17). The CC is a panel of recombinant inbred mice derived from eight founder strains, including five laboratory strains and three wild-derived inbred strains (18), resulting in highly variable phenotypes. The genomes of the CC strains feature relatively well dispersed recombination sites and balanced allele origins from all eight founder strains (19), allowing for the genetic dissection of complex traits (20). Moreover, the CC serves as a platform to develop improved models of infectious disease and to map loci associated with variations in susceptibility to pathogens (21).
We previously utilized the CC to demonstrate that host genetic factors contribute to significant variations in Salmonella susceptibility (22). Following challenge of 35 CC strains with S. Typhimurium, we showed that the bacterial burdens of the spleen and liver were significantly different between strains (22). One strain in particular, known as CC042/GeniUnc (CC042), was shown to be extremely susceptible to S. Typhimurium infection, with greater than 1,000-fold higher numbers CFU being found in the spleen and liver of these mice compared to the numbers found in the highly susceptible C57BL/6J (B6) reference strain (22). It has been shown that a missense mutation in the solute carrier family 11, member 1, gene (Slc11a1) contributes to the high susceptibility of the C57BL/6J strain (23), and the CC042 strain inherited this missense mutation from the C57BL/6J founder strain (22). While the Slc11a1 mutation partially accounts for the high susceptibility of CC042 mice, other host genetic variants are required to explain the extreme CC042 phenotype and have yet to be identified. The current work reports on the characterization of the CC042 immunophenotype, the mapping of two loci associated with the susceptibility phenotype, and the identification of a causal variant. CC042 mice were found to have a primary immunodeficiency with alterations in spleen, thymus, and bone marrow hematopoietic cell populations. Quantitative trait locus mapping showed that the Salmonella susceptibility phenotype was controlled by at least two regions on chromosome 7 (Stsl6 and Stsl7). Within the Stsl7 locus, a de novo 15-bp deletion mutation in the intron 1 splice acceptor site of the integrin alpha L chain gene (Itgal), resulting in complete protein abrogation, was shown to increase susceptibility to Salmonella infection. This study provides a foundation for investigating the role of Itgal in the response to Salmonella and illustrates how the CC can serve to identify mechanisms of infectious and immunological traits.
RESULTS
Clinicopathological characterization of CC042 mice.
To characterize the Salmonella susceptibility observed in CC042 mice, clinicopathological parameters were evaluated in naive CC042 mice, and C57BL/6J mice were used as a reference, as they have a well-characterized immune phenotype prior to and during Salmonella infection. While C57BL/6J mice are typically considered susceptible to infection, they were previously shown to be more resistant to Salmonella Typhimurium infection than CC042 mice (22).
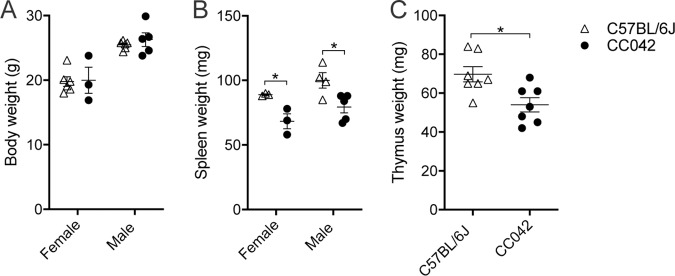
CC042 mice did not present any other overt visible phenotypes prior to infection, with the exception of a characteristic white head spot, most likely inherited from the WSB/EiJ CC founder parent (24). Age- and sex-matched CC042 mice had body weights comparable to those of the C57BL/6J mice (Fig. 1A). However, the CC042 mice displayed significantly smaller spleens and thymi, in terms of mass, than the C57BL/6J mice (Fig. 1B and C). Examination of hematological parameters in naive CC042 mice indicated total white blood cell and lymphocyte counts lower than those in the C57BL/6J mice (Table 1). CC042 mice presented a small but significant increase in the number of red blood cells, hemoglobin, and mean corpuscular hemoglobin concentration (MCHC) (Table 1). In addition, the splenic microarchitecture of CC042 mice was assessed using hematoxylin-eosin staining and was shown to present moderate extramedullary hematopoiesis with a prominent presence of megakaryocytes in comparison to the findings for C57BL/6J mice (Fig. 2A). Histopathological examination of the CC042 mouse liver and kidney was normal (data not shown). The extramedullary erythropoiesis may explain the increased numbers of circulating red blood cells (RBCs) and may be indicative of bone marrow failure.
FIG 1.
CC042 mice display reduced spleen and thymus size relative to body weight. Body (A), spleen (B), and thymus (C) weights are shown for C57BL/6J and CC042 naive mice. Only male mice were used for the calculation of thymus weight, and data were pooled from two experiments. The results in the graphs represent the mean ± SEM. Sidak’s multiple-comparison test (two-way analysis of variance) was used to analyze body (A) and spleen (B) weights, while Welch’s t test was used for thymus weight (C). *, P < 0.05.
TABLE 1.
Hematologic parameters in C57BL/6J and CC042 micea
| Parameter | Value for the following mice: |
P value (t test) | |
|---|---|---|---|
| C57BL/6J (n = 9) | CC042 (n = 9) | ||
| WBCs (109/liter) | 5.64 ± 0.41 | 4.33 ± 0.30 | 0.0205 |
| RBCs (1012/liter) | 9.13 ± 0.15 | 10.22 ± 0.39 | 0.0186 |
| Hemoglobin concn (g/liter) | 142.67 ± 1.21 | 163.11 ± 6.23 | 0.0054 |
| Hematocrit (liter/liter) | 0.46 ± 0.01 | 0.49 ± 0.02 | 0.1429 |
| MCV (fl) | 50.00 ± 0.53 | 47.78 ± 0.15 | 0.0009 |
| MCH (pg) | 15.66 ± 0.18 | 16.00 ± 0.09 | 0.1165 |
| MCHC (g/liter) | 313.11 ± 2.18 | 335.11 ± 2.37 | 0.0000 |
| Platelet count (109/liter) | 945.44 ± 66.90 | 808.33 ± 49.76 | 0.1196 |
| Neutrophils (%) | 6.11 ± 0.61 | 7.67 ± 0.82 | 0.1467 |
| Lymphocytes (%) | 87.67 ± 0.62 | 86.44 ± 1.21 | 0.3840 |
| Monocytes (%) | 5.00 ± 0.94 | 3.56 ± 0.77 | 0.2517 |
| Eosinophils (%) | 1.22 ± 0.36 | 2.33 ± 0.60 | 0.1334 |
| Neutrophils (109/liter) | 0.34 ± 0.05 | 0.32 ± 0.03 | 0.6839 |
| Lymphocytes (109/liter) | 4.96 ± 0.38 | 3.75 ± 0.27 | 0.0206 |
| Monocytes (109/liter) | 0.26 ± 0.04 | 0.16 ± 0.05 | 0.1370 |
| Eosinophils (109/liter) | 0.08 ± 0.02 | 0.09 ± 0.02 | 0.5630 |
Hematology data are for C57BL/6J and CC042 naive mice aged 14 to 16 weeks. Values indicate the mean ± SEM. WBCs, white blood cells; RBCs, red blood cells; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration.
FIG 2.
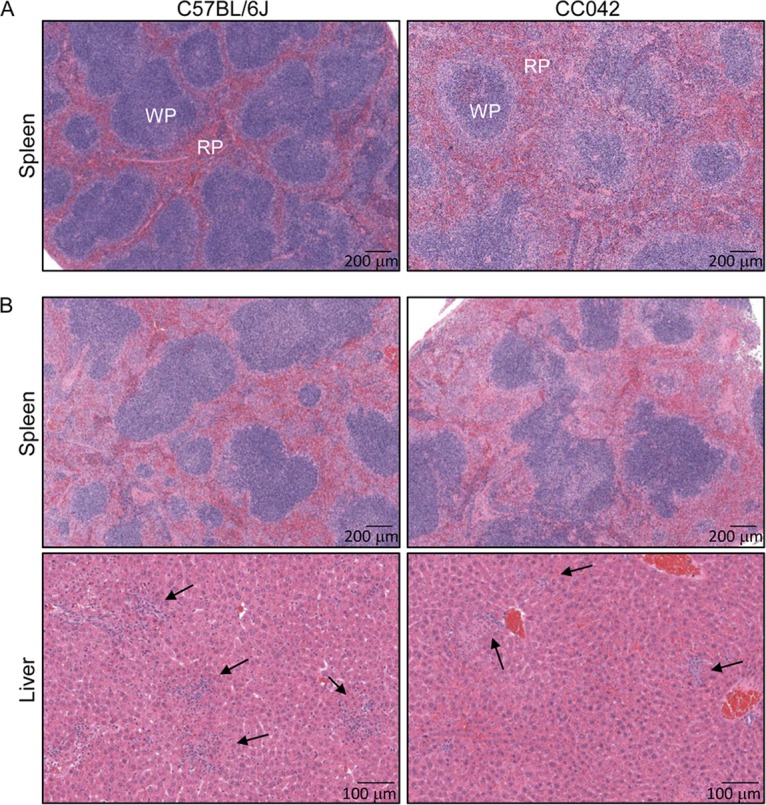
Pathological changes in the spleens and livers of infected C57BL/6J and CC042 mice. The images show C57BL/6J and CC042 mouse spleen sections stained with hematoxylin and eosin at day 0 after Salmonella infection (A) and spleen and liver sections stained with hematoxylin and eosin at day 3 after Salmonella infection (B) and are representative of those for 6 C57BL/6J mice and 6 CC042 mice. Foci of necrotic hepatocytes associated with histiocytes and neutrophils were seen in both C57BL/6J and CC042 mice but were smaller and less numerous in the CC042 mouse samples (2 to 4 foci per ×40-magnification field of view in CC042 mice compared to 4 to 5 foci in C57BL/6J mice). Arrows point to inflammatory foci. WP, white pulp; RP, red pulp.
CC042 mice display a primary immunodeficiency.
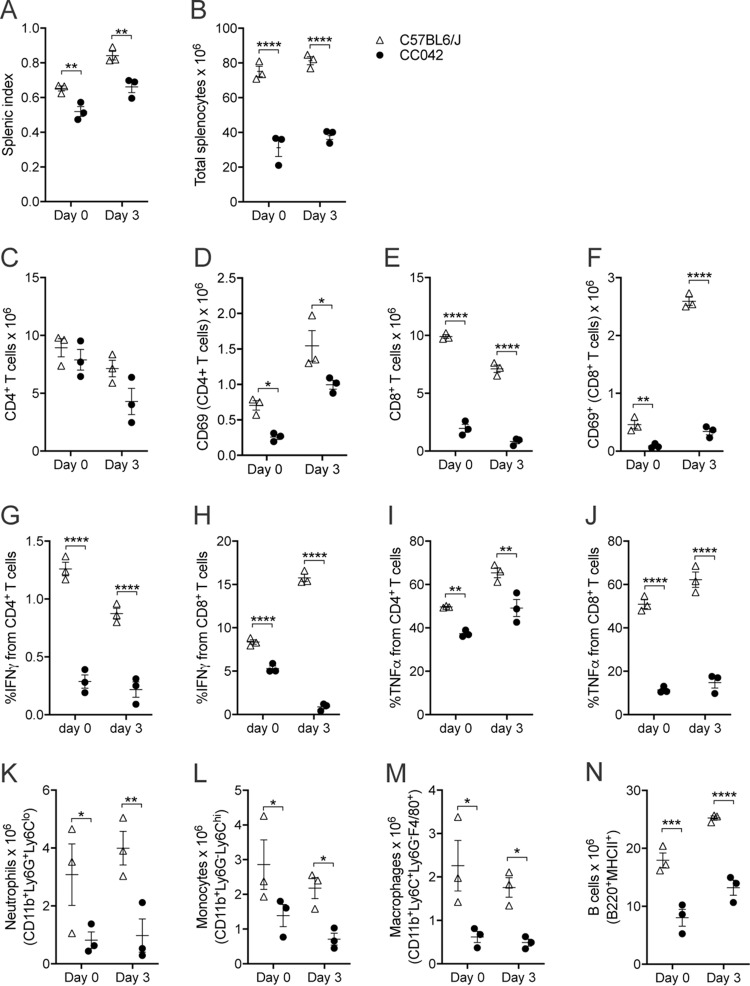
To identify if the reduced splenic size (Fig. 3A) observed in CC042 mice may be due to alterations in the cellular immune compartment, flow cytometry for lymphoid and myeloid cells was performed on the splenocytes of CC042 and C57BL/6J naive mice (the gating scheme is shown in Fig. S1a in the supplemental material). The mean total splenocyte counts in CC042 mice were significantly reduced to (31.2 ± 5.1) × 106 cells from the (75.1 ± 3.0) × 106 cells for the C57BL/6J mouse spleens (Fig. 3B). CC042 mice showed significant alterations in the splenic lymphoid compartment. Despite carrying similar numbers of splenic CD4+ T cells as C57BL/6J mice, CC042 mice displayed a reduction in the number of activated CD4+ T cells, as measured by CD69 expression (Fig. 3C and D). In addition, the numbers of splenic CD8+ T cells and activated CD69-expressing CD8+ T cells were also significantly reduced in CC042 mice compared to C57BL/6J mice (Fig. 3E and F). Further, CD4+ and CD8+ T cell effector function was assessed by measuring intracellular staining of gamma interferon (IFN-γ) and tumor necrosis factor alpha (TNF-α) after activation with anti-CD3 and anti-CD28. We observed diminished IFN-γ and TNF-α production from CC042 T cells both in naive mice and in the mice after infection (Fig. 3G to J). Reduced numbers of neutrophils, monocytes, macrophages, and B cells were also observed in CC042 mice prior to infection (Fig. 3K to N). Overall, CC042 mice presented an abnormal immunophenotype characterized by a reduced spleen size and a generalized reduction of spleen cells, affecting the lymphoid and myeloid compartments. This reduction in splenic immune cells together with the observed leukopenia indicates a potential defect in leukocyte development.
FIG 3.
CC042 mice have significantly reduced total splenocyte numbers. Flow cytometry analysis of C57BL/6J and CC042 mouse spleens was performed at day 0 (naive) and day 3 after Salmonella Typhimurium infection. (A and B) Splenic index (A) and total splenocyte count (B) for C57BL/6J and CC042 mice at day 0 and day 3 of infection. (C to F) Total cell counts and total CD69+ cells for CD4+ T cells (C and D) and CD8+ T cells (E and F). (G to J) Percentage of CD4+ T cells and CD8+ T cells producing IFN-γ (G and H) and TNF-α (I and J) for uninfected splenocytes and Salmonella-infected splenocytes at day 3 postinfection. (K to N) Total cell counts for neutrophils (K), monocytes (L), macrophages (M), and B cells (N). The results in the graphs show the mean ± SEM. Data are representative of those from six experiments in naive mice and three experiments at day 3 of infection. Cell populations were defined as follows: CD4+ T cells, TCRβ+ CD4+; CD8+ T cells, TCRβ+ CD8α+; neutrophils, CD11b+ Ly6G+ Ly6Clo; monocytes, CD11b+ Ly6G− Ly6Chi; macrophages, CD11b+ Ly6G− Ly6C+ F4/80+; and B cells, B220+ MHC-II+. Analysis was conducted using the Benjamini, Krieger, and Yekutieli correction for multiple testing (two-way analysis of variance), where significance is indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
CC042 mice have reduced thymic cellularity and altered T cell development.
To assess if the reduction in CD8+ T cells and total activated T cells in the spleens of CC042 mice may potentially be due to defective T cell maturation, flow cytometry of the thymus was carried out (the gating scheme is shown in Fig. S1b). In agreement with the previous observation that CC042 mice had a reduced thymus size, CC042 mice displayed a 2-fold reduction in total thymocyte counts (Fig. 4A). While the percentages of CD4+ and CD8+ T cells double positive for CD4 and CD8 (DP CD4+ and CD8+ T cells) and single positive for either CD4 or CD8 (SP CD4+ and SP CD8+ T cells) were comparable between CC042 and C57BL/6J mice due to the reduction in thymocyte numbers in CC042 mice, a significant decrease in the counts of T cells double negative for CD4 and CD8 (DN T cells), DP CD4+ and CD8+ T cells, and SP CD4+ or SP CD8+ T cells (25) was observed in CC042 mice (Fig. 4B and C). Further examination of the DN subset revealed significant alterations in CC042 mice (Fig. 4D and E). While no significant difference in the proportion of DN stage 1 (DN1) cells was observed, a significant reduction in the proportion of DN2 thymocytes was present in CC042 mouse thymi compared to the C57BL/6J mouse controls. Unexpectedly, the diminished proportion of DN2 cells did not result in the suppression of downstream subsets, as the fraction of DN3 thymocytes was comparable between the CC042 mice and the C57BL/6J mouse controls and a significantly increased proportion of DN4 cells was observed in CC042 mice compared to the C57BL/6J mouse controls. Given the reduction in total thymocytes, a significant decrease in DN1, DN2, DN3, and DN4 total cell counts was observed in CC042 mice compared to the C57BL/6J mouse controls. Overall, CC042 mice displayed a reduction in thymocyte numbers and altered progression of T cell precursors through maturation, specifically in the DN stages.
FIG 4.
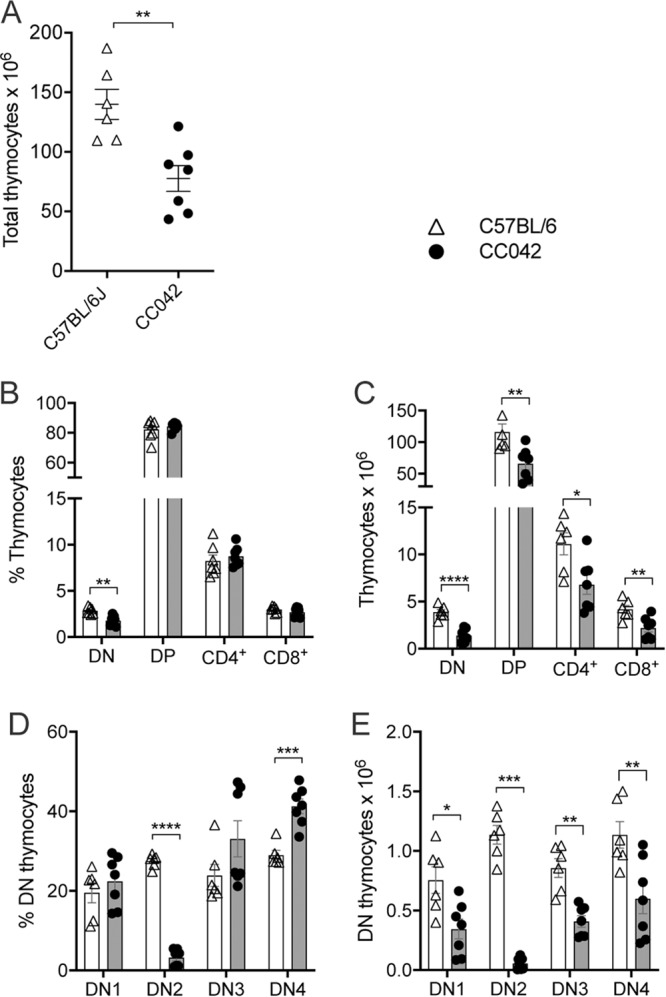
CC042 mice show significantly decreased total thymocyte numbers. (A) Total thymocyte counts for C57BL/6J and CC042 mice. (B to E) Percentage and total numbers of thymocytes by developmental stage analyzed via flow cytometry, where data are for 6 to 7 mice per genotype. The results in the graphs indicate the mean ± SEM for data pooled from two independent experiments. Thymocytes develop through double-negative (DN) stages (stages 1 to 4) to double-positive (DP) stages before splitting to either CD4+ (CD4+ SP) or CD8+ (CD8+ SP) T cell populations. Cell populations were gated as follows: DN, CD4− CD8a−; DP, CD4+ CD8a+; CD4+, CD4+ CD8a−; and CD8+, CD4− CD8a+. DN1, DN2, DN3, and DN4 subpopulations were gated from the DN population as follows: DN1, CD44+ CD25−; DN2, CD44+ CD25+; DN3, CD44− CD25+; and DN4, CD44− CD25−. Multiple t tests using the Holm-Sidak method were used to assess significance. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
CC042 mice show alterations in hematopoietic progenitor populations.
The observed reduction in peripheral blood leukocytes and the cellularity of the spleen and thymus in CC042 mice suggested that the hematopoietic development of these populations may be impaired. To investigate hematopoiesis in CC042 mice, flow cytometry (Table 2) of bone marrow hematopoietic progenitor populations was conducted.
TABLE 2.
Hematopoietic progenitor population surface markers
| Population | Markers |
|---|---|
| LSK | Lin− Sca1+ cKit+ |
| HSC | Lin− Sca1+ cKit+ CD150+ CD34− CD48− Flt3− |
| MPP1 | Lin− Sca1+ cKit+ CD150+ CD34+ CD48− Flt3− |
| MPP2 | Lin− Sca1+ cKit+ CD150+ CD34+ CD48+ Flt3− |
| MPP3 | Lin− Sca1+ cKit+ CD150− CD34+ CD48+ Flt3− |
| MPP4 | Lin− Sca1+ cKit+ CD150− CD34+ CD48+ Flt3+ |
| LKS− | Lin− cKit+ Sca1− |
| CMP | Lin− cKit+ Sca1− IL-7R− CD34+ CD16/CD32− |
| GMP | Lin− cKit+ Sca1− IL-7R− CD34+ CD16/CD32+ |
| MEP | Lin− cKit+ Sca1− IL-7R− CD34− CD16/CD32− |
| CLP | Lin− cKitlo Sca1lo IL-7R+ |
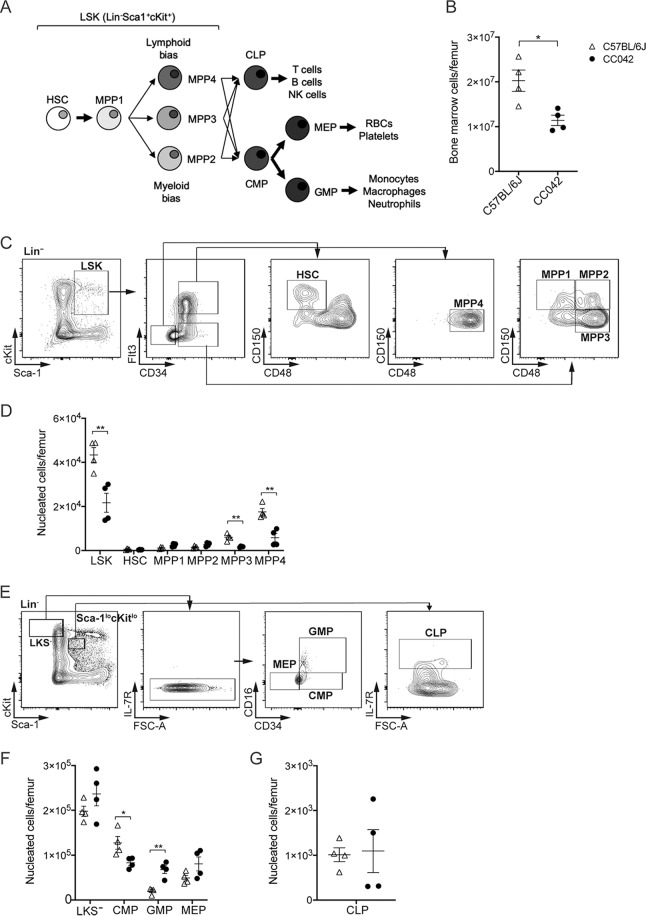
Bone marrow hematopoietic stem cells (HSCs) differentiate to multipotent progenitor (MPP) subpopulations, defined based on the expression of the CD150, CD34, CD48, and Flt3 cell surface markers (Table 2 and Fig. 5A) (26). CC042 mouse bone marrow contained fewer cells (Fig. 5B) and a reduced number of Lin− Sca-1+ c-Kit+ (LSK) cells than C57BL/6J mouse bone marrow, but the bone marrow of the two strains contained equivalent numbers of HSCs (Fig. 5C and D). However, examination of the LSK subsets revealed significant alterations in the proportions of MPP subpopulations. The CC042 mouse LSK compartment displayed a significant reduction in downstream MPP stage 3 (MPP3) and MPP4 cells in comparison to the numbers for the C57BL/6J mouse controls (Fig. 5C and D). The depletion of the MPP3 and MPP4 populations in CC042 mouse bone marrow suggests a maturation block in the progression from the MPP1 to the MPP3 and MPP4 cell stages.
FIG 5.
CC042 mice have altered bone marrow-resident hematopoietic progenitor populations. Flow cytometry analysis of hematopoietic progenitors in femoral bone marrow from C57BL/6J and CC042 mice. (A) Schematic diagram of the stages of hematopoietic stem cell differentiation. (B) Total number of bone marrow cells per femur averaged over the total number of cells collected for two femurs. (C) Gating scheme used to analyze Lin− Sca1+ cKit+ (LSK) cells. (D) Total LSK cells per femur by developmental stage. LSK cells progress through the HSC, MPP1, MPP2, MPP3, and MPP4 subsets. (E) Gating scheme used to analyze Lin− cKit+ Sca1− (LKS−) and common lymphoid progenitors (CLP). FSC-A, forward scatter area. (F) Total number of LKS− cells per femur grouped by progenitor stage. LKS− cells comprise common myeloid progenitors (CMP), granulocyte-macrophage progenitors (GMP), and megakaryocyte-erythroid progenitors (MEP). (G) Total number of CLPs per femur. The data in the graphs represent the mean ± SEM. Data are representative of those from three independent experiments. The cell populations are defined in Table 2. Significance was determined by Welch’s t test (B and G) and multiple t tests using the Holm-Sidak method (D and F). *, P < 0.05; **, P < 0.01.
MPP subpopulations give rise to common lymphoid progenitors (CLPs) and common myeloid progenitors (CMPs) (Fig. 5A and E) (27). CMPs form both megakaryocyte-erythroid progenitors (MEPs), which produce RBCs and platelets, and granulocyte-macrophage progenitors (GMPs) (27). CC042 mice showed a decreased number of CMP cells but an increase in the number of downstream GMPs compared to C57BL/6J mice (Fig. 5F). No significant difference in the number of CLPs was observed between CC042 and C57BL/6J mice (Fig. 5G). The maturational blocks observed upstream of MPP3, MPP4, and CMP could partially explain the reduced hematopoietic cell populations present in peripheral organs.
CC042 mice mount a distinct response to infection.
To characterize the impact of infection on the spleen and liver, histopathological examination of CC042 mouse spleens and livers was conducted 3 days after Salmonella infection. The livers of C57BL/6J mice harbored typical lesions for this strain, characterized by multifocal necrotic lesions with the aggregation of histiocytes and neutrophils typical of granulomas (Fig. 2B). The necrotic hepatocyte foci were found to be smaller and less abundant in CC042 mice (2 to 4 foci per ×40-magnification field of view) than in C57BL/6J mice (4 to 5 foci per field of view) (Fig. 2B). CC042 mouse spleens were also observed to have reduced neutrophil infiltration during infection compared to C57BL/6J mouse spleens (Fig. 2B). The increased megakaryocyte numbers and erythropoiesis observed in the spleens of uninfected CC042 mice (Fig. 2A) were also present in the spleens of infected CC042 mice (Fig. 2B). During infection, CC042 mice displayed a severe splenic immunodeficiency with an important reduction in the number of activated CD4+ T cells, CD8+ T cells, activated CD8+ T cells, neutrophils, monocytes, macrophages, and B cells compared to C57BL/6J mice (Fig. 3C to J). The reduction in CD8+ T cell counts combined with the generalized defect in total T cell activation and myeloid cell numbers may account for the observed increase in susceptibility to Salmonella infection.
A mutation within Itgal is responsible for the susceptibility of CC042 mice to Salmonella infection.
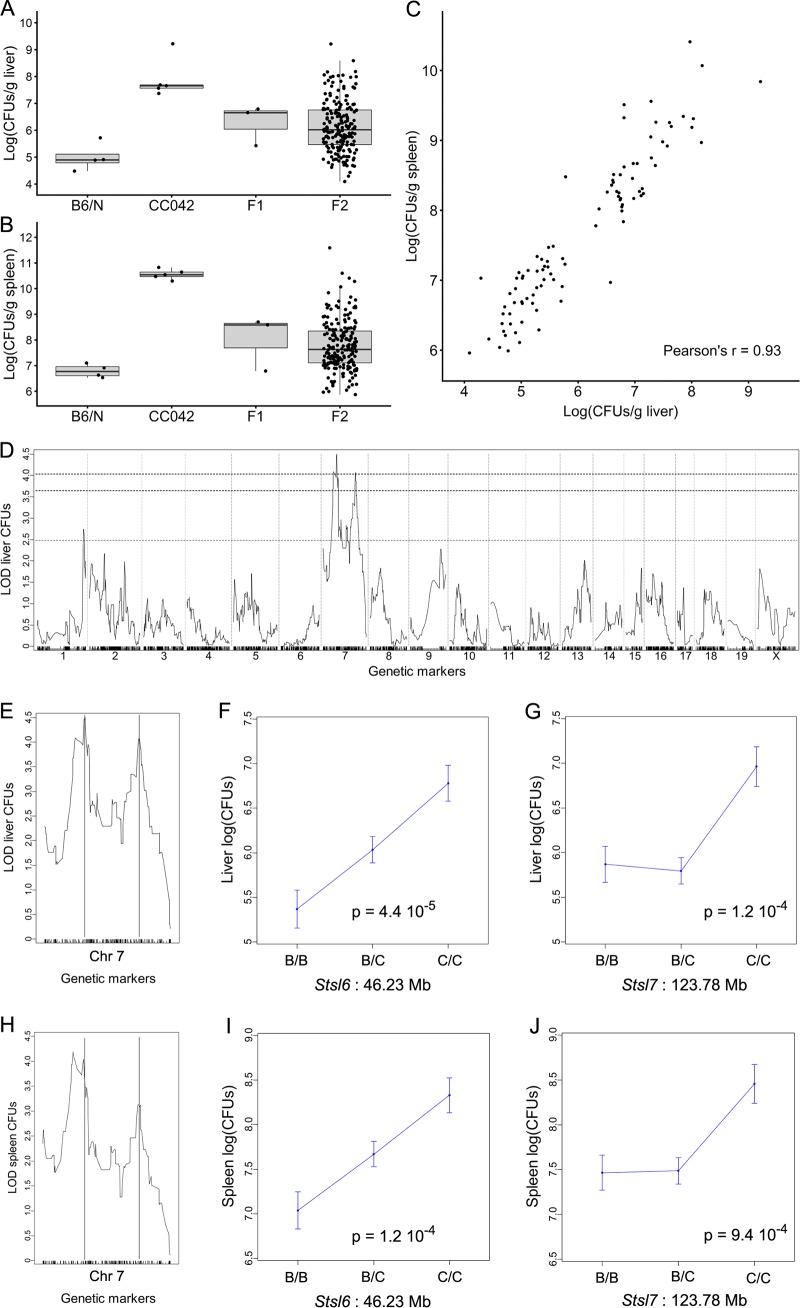
To identify the genes which, in addition to Slc11a1, control the extreme susceptibility to Salmonella Typhimurium of CC042 mice, we produced an F2 cross between CC042 mice and mice of the C57BL/6NCrl (B6N) strain, which is closely related to the C57BL/6J strain with the same Slc11a1 missense mutation. The C57BL/6N strain was separated from the C57BL/6J strain in 1951 (https://www.jax.org/strain/005304). Both strains have accumulated their own variants and are considered substrains. The number of genome-wide single nucleotide polymorphisms (SNPs) between the two C57BL/6 substrains is estimated to be ∼10,700, based on C57BL/6N sequencing data (28). This allows polymorphic markers to be found between the two substrains and the mapping of quantitative trait loci (QTL) to be performed even in regions where the CC042 genome is of C57BL/6J mouse origin. Infected F1 mice showed intermediate bacterial loads in the liver and spleen compared with the parental strains (Fig. 6A and B), while the bacterial loads in F2 mice spanned over the parental range. Ninety-four F2 mice with the highest or lowest liver bacterial loads were selected for QTL mapping. Since the bacterial loads in the liver and spleen were strongly correlated in the 94 selected individuals (Fig. 6C), QTL mapping was performed on liver bacterial loads.
FIG 6.
Susceptibility of CC042 mice to S. Typhimurium is controlled by two linked loci on chromosome 7. (A and B) Bacterial load in liver (A) and in spleen (B) at day 4 postinfection with S. Typhimurium in C57BL/6NCrl (n = 4), CC042 (n = 5), (C57BL/6NCrl × CC042)F1 (n = 3), and (C57BL/6NCrl × CC042)F2 (n = 196) mice. Bacterial loads in F2 mice spanned the values for the two parental strains. (C) Bacterial loads in the 94 individuals selected for genotyping, showing a strong correlation between the two organs (Pearson’s r = 0.93). (D) Genome-wide QTL mapping of the liver bacterial load identified two statistically significant peaks on chromosome 7. The horizontal dashed lines indicate the 0.05, 0.1, and 0.63 (top to bottom) significance thresholds estimated from 10,000 permutations. (E, H) QTL positions are indicated by vertical lines in liver (E) and in spleen (H). See Table 3 for details on each QTL. (F, G, I, J) The proximal Stsl6 QTL acted semidominantly on liver (F) and on spleen (I) bacterial loads, while the CC042-inherited allele at the distal Stsl7 QTL had a recessive mode of action both for liver (G) and for spleen (J). For both QTLs, the CC042-inherited allele was associated with an increased bacterial load. B, the B6 allele; C, the CC042 allele.
QTL mapping identified only two significant QTLs (at a 0.05 genome-wide significance level), and both of these were on chromosome 7 (Fig. 6D and E) and were named Salmonella Typhimurium susceptibility locus 6 (Stsl6) and Stsl7. Details for each QTL are given in Table 3. Stsl6 (peak position at 46.23 Mb) showed a semidominant mode of inheritance, with heterozygotes having a bacterial load intermediate between those of the two types of homozygotes (Fig. 6F). The CC042 allele at Stsl7 (peak position at 123.78 Mb) was recessive, and homozygotes had 10 times higher liver bacterial loads (Fig. 6G).
TABLE 3.
Summary of the significant QTLs identified in the (CC042 × B6/N)F2 crossa
| QTL | LOD | Peak position (Mb) | Peak linkage (cM) | SNP | 1.5-LOD drop interval (Mb) | Width (Mb) | Inheritance of susceptible allele | Variance (%) |
|---|---|---|---|---|---|---|---|---|
| Stsl6 | 4.50 | 46.23 | 29.66 | BackupUNC070497127 | 34.5–51.5 | 17 | Semidominant | 19.8 |
| Stsl7 | 4.06 | 123.78 | 67.42 | BackupJAX00654337 | 116.1–128.9 | 12.8 | Recessive | 18.0 |
Both loci are on chromosome 7, the significance level was a P value of 0.05, the susceptible allele was that of CC042, and the susceptible haplotype was that of the WSB/EiJ CC founder parent. cM, centimorgan.
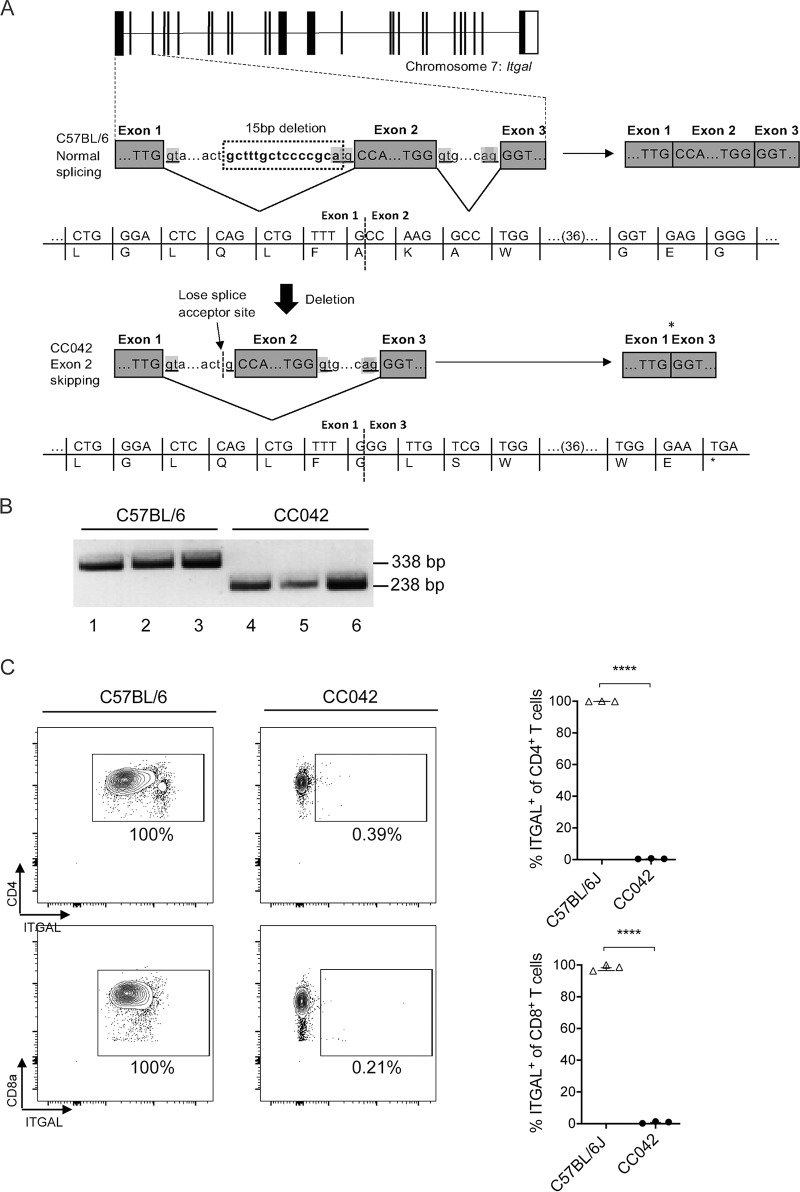
CC042 mice inherited the Stsl6 and Stsl7 QTL regions from the WSB founder (Fig. S2). Other CC strains also inherited WSB alleles in either or both QTL regions but had much lower bacterial loads than CC042 mice (Fig. S2). In particular, CC035 mice also carried WSB-derived alleles at Stsl6 and Stsl7. This result suggests that the susceptibility alleles present at these two loci in CC042 mice could result from de novo mutations which occurred during the development of the CC042 strain. These private variants have been previously identified by the sequencing of CC strains and are publicly available (29). Three private variants were identified in the Stsl6 confidence interval in genes Ush1c, Ccdc123, and 4930435C17Rik, and only one was identified within the Stsl7 interval in the integrin alpha L (Itgal) gene, also known as Cd11a. This variant is a 15-bp deletion located at the 3′ end of intron 1 and results in the disruption of the intron 1 splice acceptor site (Fig. 7A). We hypothesized that the intron 1 splice donor would attach to the intron 2 splice acceptor, resulting in the skipping of exon 2 during splicing. Joining exon 1 to exon 3 would create a frameshift and a downstream premature stop codon, resulting in a truncated form of the ITGAL protein.
FIG 7.
The Itgal mutation in CC042 mice resulted in the skipping of exon 2 and the absence of protein expression. (A) Schematic diagram illustrating the Itgal mutation in CC042 mice. A 15-bp deletion resulted in the loss of the intron 1 splice acceptor site, resulting in the skipping of exon 2 and the formation of a premature stop codon. (B) PCR analysis of Itgal cDNA from C57BL/6J and CC042 mice. Amplification of the region flanking exon 2 produces a 338-bp PCR product in C57BL/6J mice carrying the wild-type Itgal gene (lanes 1 to 3). A 238-bp PCR product was produced in CC042 mice and corresponds to a 100-bp deletion due to the skipping of exon 2 (lanes 4 to 6). (C) Flow cytometry analysis of ITGAL surface expression on CD4+ and CD8+ T cells. ITGAL gates were constructed using fluorescence-minus-one panels, and significance was calculated using Welch’s t test. ****, P < 0.0001. The results in the graphs represent the mean ± SEM.
To verify this prediction, reverse transcription and amplification of Itgal transcripts were conducted in C57BL/6J and CC042 cells using primers flanking exon 2. C57BL/6J cells were found to express a 338-bp transcript indicative of the inclusion of exon 2 (Fig. 7B). In contrast, CC042 cells produced a 238-bp transcript corresponding to the loss of the 100-bp-long exon 2. To confirm the predicted loss of the ITGAL protein, flow cytometry of splenic leukocytes labeled with a fluorescently conjugated anti-ITGAL antibody was conducted. As predicted, none of the CC042 leukocyte populations examined, including CD4+ and CD8+ T cells, expressed the ITGAL protein (Fig. 7C). In contrast, all C57BL/6J leukocytes examined expressed the ITGAL protein.
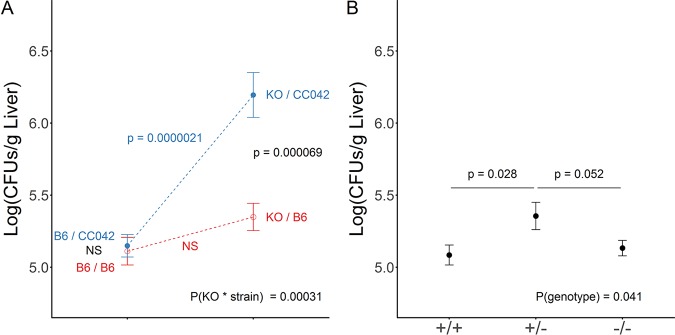
To confirm in vivo the role of the Itgal deletion in the extreme susceptibility of the CC042 strain to S. Typhimurium, we performed a quantitative complementation test. Compound heterozygous mice carrying a knockout (KO) Itgal allele (ItgalKO) and a CC042 mouse deleted Itgal allele (Itgaldel) had significantly higher liver bacterial loads (∼6.3 log CFU/g liver; Fig. 8A) than mice heterozygous for either of these two alleles (∼5.1 to 5.3 log CFU/g liver). Therefore, there was no significant difference between Itgal+/Itgal+ and Itgal+/ItgalKO mice in the B6 mouse background (Fig. 8A, red line), while the difference between Itgal+/Itgaldel and ItgalKO/Itgaldel mice was highly significant (Fig. 8A, blue line; P = 2.1 × 10−6). These results demonstrate that the Itgal deletion present in CC042 mice contributes to their increased susceptibility. Remarkably, the Itgal KO mutation had only a mild impact on the liver bacterial load in a B6 inbred background, with no significant difference between Itgal+/Itgal+ and ItgalKO/ItgalKO mice, but, unexpectedly, resulted in a slightly higher bacterial load in Itgal+/ItgalKO mice than in Itgal+/Itgal+ mice (Fig. 8B).
FIG 8.
The quantitative complementation test confirmed the role of the CC042 Itgal loss-of-function variant in susceptibility to S. Typhimurium. (A) Bacterial load in liver at day 4 postinfection with S. Typhimurium in C57BL/6J (B6/B6) (n = 23), (Itgal−/− × C57BL/6J)F1 (KO/B6) (n = 17), (C57BL/6J × CC042)F1 (B6/CC042) (n = 31), and (Itgal−/− × CC042)F1 (KO/CC042) (n = 20) mice determined with data pooled from two independent experiments. On the left are the results for mice which carry one B6 allele and either a B6 or a CC042 allele. On the right are the results for mice which carry one Itgal KO allele and either a B6 allele or a CC042 allele. (Itgal−/− × CC042)F1 (KO/CC042) mice showed bacterial loads higher by approximately 1 log CFU than the three other groups, indicating the absence of complementation between Itgal KO and CC042 alleles. Differences between groups were assessed by one-way analysis of variance. (B) The interaction between the genetic background (C57BL/6J or CC0042) and Itgal genotype (+/+ or −/−) was assessed by two-way analysis of variance. In an inbred C57BL/6J background, the Itgal genotype had only a mild impact on the susceptibility to S. Typhimurium (x axis, genotype at the Itgal locus; data are for 34, 17, and 23 +/+, +/−, and −/− mice, respectively; P values are from a pairwise Student's t test). NS, not significant.
DISCUSSION
The CC is a murine genetic reference population that was developed to reflect the genetic diversity and complexity of the human population (19). In a previous study, we utilized the CC to investigate the role of genetic factors on host susceptibility to Salmonella Typhimurium infection (22). The CC042 strain was identified as being highly susceptible to S. Typhimurium infection (22). In this study, we sought to characterize the immunophenotype of the CC042 strain and to identify the underlying genetic factors contributing to the extreme susceptibility phenotype. We demonstrated that CC042 mice display a generalized immunodeficiency with a reduced cellularity of the spleen, thymus, and bone marrow. CC042 mice also exhibited alterations in immune progenitor development in both the thymus and the bone marrow. Using genetic linkage mapping and in vivo complementation testing, we identified a de novo 15-bp deletion in intron 1 of the Itgal gene to be causatively linked to the Salmonella susceptibility phenotype in CC042 mice. This mutation results in exon 2 skipping and a premature stop codon.
The Itgal mutation identified in CC042 mice was demonstrated to result in the complete loss of ITGAL protein expression on all leukocytes examined. ITGAL is an integrin α-chain protein which, in conjunction with the ITGB2 β-chain (ITGB2 can dimerize with any of ITGAL, ITGAM, ITGAD, or ITGAX), forms the lymphocyte function-associated antigen 1 (LFA-1) integrin complex (30). The LFA-1 complex is expressed on all leukocytes and plays a critical role in various immunological functions (30). Circulating leukocytes utilize LFA-1 to bind to its cognate ligand, ICAM-1, expressed on endothelial cells (31, 32). Binding allows leukocytes to anchor in the bloodstream prior to extravasation into lymph nodes or inflamed tissues (31, 32). LFA-1 also enables stabilization of the immunological synapse during T cell activation, thus permitting prolonged T cell receptor (TCR)-major histocompatibility complex (MHC) interactions (31, 32). Given that LFA-1 plays a crucial role in leukocyte migration and T cell activation, it is unsurprising that the absence of ITGAL function may result in ineffective immune responses to Salmonella infection in CC042 mice.
CC042 mice displayed significant defects in splenic, thymic, and bone marrow immune cell populations. A study by Bose et al. similarly reported that Itgal−/− mice had reduced splenic and thymic cellularity but did not observe a reduction in bone marrow cell counts (33). Moreover, Itgal−/− mice or mice treated with anti-LFA-1 antibodies are consistently reported to have reductions in splenic T cells (33–35). Multiple reports specifically highlight defects in the CD8+ T cell subset in Itgal−/− mice in terms of numbers, activation, proliferation, and recruitment to infected tissues (34–36). CC042 mice were similarly observed to have a more significant impairment in CD8+ T cell populations than in CD4+ T cell populations. Defects in T cell activity may be due to the inability of T cells to sustain long-lived interactions with antigen-presenting cells (APCs). Itgal−/− T cells are noted to traffic at a higher rate through lymph nodes, while T cells expressing low levels of LFA-1 have been shown to only transiently form contacts with APCs, resulting in diminished expression of effector proteins, including IFN-γ (37, 38). This is consistent with our observation of a low level of expression of IFN-γ by CD4+ and CD8+ T cells after TCR stimulation in CC042 mice. Notably, both CC042 and Itgal−/− mice have previously been shown to display suppression of IFN-γ in lung tissue upon Mycobacterium tuberculosis infection (39, 40). Production of IFN-γ by T cells is crucial for the clearance of intracellular bacterial infections, such as those caused by Salmonella, as IFN-γ activates bacterial killing within infected macrophages (41, 42). Notably, IFN-γ production has been shown to be inducible by interferon-stimulated gene 15 (ISG15) through an ITGAL-dependent signaling mechanism (43). Thus, it is most likely that the immunosuppressed phenotype observed in CC042 mice in response to Salmonella may be due to reduced T cell activation and suppression of IFN-γ production via the ITGAL-dependent pathway.
Alterations in thymocyte maturation were also observed in CC042 mice, potentially explaining the corresponding reduction in peripheral T cell counts. ITGAL has been implicated in thymopoiesis, as Itgal−/− mice exhibit a general reduction in all thymocytes, while the use of anti-ITGAL antibodies has been shown to result in the impairment of double-positive CD4+/CD8+ and single-positive CD8+ T cell development (33, 44). Thymocyte defects may be due to alterations further upstream, as bone marrow hematopoiesis was also defective in CC042 mice. Hematopoietic progenitor populations have also been observed to be altered in Itgal−/− mice (33). In competitive reconstitution assays in irradiated mice, Itgal−/−-derived bone marrow progenitors were unable to compete against wild-type cells, presenting a role for ITGAL in hematopoietic cell generation (33). It should be noted that the mutation in Kitl, also inherited from WSB/EiJ mice, may contribute to the defects in hematopoietic progenitor generation.
Several animal studies have delineated a significant role for ITGAL in the response to infection. Itgal−/− mice have been reported to be highly susceptible to bacterial infection by both intracellular M. tuberculosis and extracellular Streptococcus pneumoniae, as characterized by reduced survival, increased bacterial burdens, and defects in leukocyte recruitment to infected tissues (39, 45). During M. tuberculosis infection, the absence of the ITGAL protein was shown to result in impaired containment of the bacteria, demonstrated by diffuse lung granuloma formation (39). Interestingly, upon M. tuberculosis infection, CC042 mice displayed significantly increased bacterial burdens in the lung and spleen, increased necrotic lung granuloma formation, and more rapid disease progression compared to C57BL/6J mice (40). This phenotype is likely partly explained by the Itgal deletion that we have reported here. Notably, in a recent study, Smith et al. reported their work on variants controlling susceptibility to M. tuberculosis in the Collaborative Cross and that the Itgal gene is involved in the extreme susceptibility of CC042 mice to M. tuberculosis (46). Studies utilizing anti-LFA-1 antibodies to neutralize ITGAL protein activity have shown that ITGAL function is necessary for the timely clearance of respiratory syncytial virus (RSV) and for the control of parasitemia during Trypanosoma cruzi infection (36, 47). In contrast, two separate studies have reported that Itgal−/− mice display increased resistance to Listeria monocytogenes infection (34, 48). Counterintuitively, resistance to L. monocytogenes occurs despite Itgal−/− mice displaying a significant reduction in CD8+ T cell numbers and lytic activity during L. monocytogenes infection (34). The discrepancy between the responses to L. monocytogenes and other pathogens may be due to differential methods of leukocyte recruitment in response to various inflammatory stimuli. One study found that neutrophil recruitment during S. Typhimurium and L. monocytogenes infection operates in ITGB2-dependent and -independent manners, respectively (49).
To our surprise, mice homozygous for the Itgal KO mutation on the B6 inbred background did not show an increased liver bacterial load, while increased susceptibility was observed in compound heterozygous mice on a (B6 × CC042)F1 background. The mild increase in the bacterial load (0.28 log CFU) in Itgal+/ItgalKO mice compared with Itgal+/Itgal+ mice has no simple explanation and would require confirmation experiments and further investigations. The major differences between the B6 mouse and B6 × CC042 mouse backgrounds suggest that the impact of ITGAL loss of function may depend on the genotype at other loci. There are in fact many examples where the phenotype induced by the inactivation of a gene is influenced by the genetic background (50, 51). One of the modifier loci could be Stsl6, which we identified in the F2 cross. While the comparison of the founder haplotypes in CC042 mice and other CC strains suggested that, like for Stsl7, the causative variant was likely a de novo mutation proper to CC042 mice, we failed to identify a candidate private variant for Stsl6 from the published data. Moreover, due to the linkage between these two loci, the F2 cross did not allow investigation of genetic interactions between the two loci. However, loci other than Stsl6 are likely genetic modifiers of the Itgal loss-of-function mutation and could explain the difference in phenotypes between the B6 and (B6 × CC042)F1 genetic backgrounds. Their identification could use a two-generation cross where all mice are homozygous for a deficient Itgal allele. The strong effect of the Itgal mutation observed in the (ItgalKO mouse × CC042)F1 mice suggests that the alleles of the CC042 mice at these modifiers act in a dominant fashion. Therefore, evaluating the susceptibility to S. Typhimurium of progeny from an (ItgalKO mouse × CC042)F1 × ItgalKO mouse backcross could identify genomic regions involved in the variable impact of the Itgal mutation. The identification of the causal genes would require combining positional information from genetic mapping with that from other approaches, including immunophenotyping of the ItgalKO and (ItgalKO mouse × CC042)F1 parents and analysis of gene expression in relevant tissues or cell types. One of the main advantages of the CC is the ability to model complex traits in genetically diverse populations (20), in particular, for studying host-pathogen interactions (21). The hope is that genetic factors uncovered through CC mouse studies may be applicable to studies of human disease (52). Indications that ITGAL may be important for responses to Salmonella infection in humans is supported by the finding that ITGAL is upregulated upon infection with S. Typhi in human volunteers (53). Moreover, ITGAL has been implicated in a range of inflammatory diseases in humans. A recent genome-wide association study identified an ulcerative colitis risk allele at the ITGAL locus resulting in the upregulation of ITGAL protein expression (54). ITGAL has also been shown to be upregulated in systemic lupus erythematosus, likely due to reported DNA hypomethylation at the ITGAL promoter (55, 56). Meanwhile, gene pathway analysis has uncovered a multiple sclerosis susceptibility gene network involving both ITGAL and its cognate receptor gene, ICAM1 (57). Furthermore, a deficiency of ITGAL’s dimerization partner, ITGB2, results in leukocyte adhesion deficiency I (LAD-I), which is classically characterized by recurrent bacterial infections (58). These studies suggest that overexpression of ITGAL may result in excessive activation of the immune system, resulting in inflammatory disease, while deficiency may result in immunosuppression.
Characterization of the CC042 mouse strain has provided a foundation for the future use of the CC in infection susceptibility studies. We have used this model to identify and characterize the novel Salmonella-susceptible CC042 strain and to identify Itgal as a gene critical in the Salmonella response pathway.
MATERIALS AND METHODS
Ethics statement and animals.
The animal experiments performed at McGill University were conducted in accordance with guidelines provided by the Canadian Council on Animal Care (CCAC). The guidelines included in the Guide to the Care and Use of Experimental Animals (59) were the choice of an appropriate endpoint in experiments using animals for research, teaching, and testing; the euthanasia of animals used in science; the husbandry of animals in science; laboratory animal facility characteristics, design, and development; and training of personnel working with animals in science. The animal use protocol was approved by the McGill University Facility Animal Care Committee (protocol number 5797). The animal experiments performed at the Institut Pasteur were conducted in compliance with French and European regulations and were approved by the Institut Pasteur Ethics Committee (project number 2014-0050) and authorized by the French Ministry of Research (decision number 8563).
CC042/GeniUnc (CC042) mice were originally generated through the CC at Geniad (60) in Australia and then maintained at the Institut Pasteur (Paris, France) and McGill University (Montréal, QC, Canada) under specific-pathogen-free conditions. C57BL/6J (B6) mice were purchased from The Jackson Laboratory (stock number 000664). The C57BL/6NCrl (B6N) mice used to generate (B6N × CC042)F2 mice were purchased from Charles River. The B6.129S7-Itgaltm1Bll/J (Itgal KO) mice used to perform complementation tests were purchased from The Jackson Laboratory (stock number 005257). F1 mice were obtained by crossing CC042 males with B6N females, and the F1 mice were intercrossed to produce F2 mice.
Salmonella Typhimurium infection.
The infectious dose was generated via culture of frozen S. Typhimurium strain SL1344 stocks in Trypticase soy broth at 37°C until an optical density at 600 nm of 0.1 to 0.2 was reached. To determine the specific bacterial concentration, the bacterial suspension was diluted in saline and plated on Trypticase soy agar (TSA). Using the resulting concentration, an inoculum of 1,000 CFU in 200 μl was administered to mice via intravenous injection into the caudal vein. All healthy mice aged between 7 and 12 weeks were included. Investigators were blind to the genotypes during the monitoring of Salmonella-infected mice. The dose was verified via serial dilution of the inoculum and bacterial culture on TSA. Following infection, the mice were monitored and assessed using body condition scoring. The mice were humanely euthanized at different time points after Salmonella infection for sample collection. To determine the bacterial loads, the spleen and liver were aseptically removed, weighed, and added to 0.9% saline. The organs were homogenized using a Polytron homogenizer (Kinematica, Bohemia, NY), serially diluted in phosphate-buffered saline (PBS), and cultured on TSA.
Hematology.
For complete blood count and white blood cell differential, blood samples were collected in EDTA tubes from mice aged 8 to 10 weeks. Analyses were performed at the Comparative Medicine Animal Resources Centre, McGill University.
Histology.
Tissues were collected and placed in 10% formalin at room temperature for 24 h to enable fixation. The organs were then transferred to a 70% ethanol solution at 4°C, followed by organ processing, embedding, and sectioning at the Goodman Cancer Research Center histology facility, McGill University. The resulting tissue sections were stained with hematoxylin and eosin prior to microscopic examination.
Flow cytometry analyses.
For flow cytometry analyses, whole spleens and thymi were aseptically collected, weighed, and transferred to 3 ml of phosphate-buffered saline (PBS). Tissues were mechanically dissociated using the back end of 3-ml syringes to pass them through a 70-μm-mesh-size strainer (Fisherbrand). The resulting cell suspensions were passed through a second 70-μm-mesh-size strainer and transferred to a 15-ml conical tube. The cells were spun at 1,400 rpm for 5 min at 4°C. The supernatant was discarded, and the pellets were resuspended in 10 ml (spleen) or 5 ml (thymus) of ammonium-chloride-potassium (ACK) lysis buffer at room temperature. The suspension was spun at 1,400 rpm for 5 min at 4°C, and the supernatant was discarded. The cell pellets were resuspended in 10 ml PBS and passed through a third cell strainer to generate single-cell suspensions. For bone marrow cell preparations, the femurs were aseptically collected. The bones were cleaned of flesh using a scalpel blade. The epiphyses of the femurs were cut, and a 25-gauge needle and syringe were used to pass 2 ml of PBS through the bone to displace the bone marrow. The bone marrow-containing medium was transferred to a 50-ml tube, and 2 ml of red blood cell lysis buffer (Sigma-Aldrich) was added. After 1 min of gentle mixing, 15 ml of PBS was added, the tubes were centrifuged for 7 min at 1,400 rpm, and the supernatant was discarded. The resulting cell pellet was resuspended in 2 ml of PBS.
The following monoclonal antibodies were used for flow cytometric analysis of spleen, thymus, and bone marrow samples: from Invitrogen, CD4-phycoerythrin (PE)-Cy7 (antibody GK1.5), CD11b-allophycocyanin (APC) (antibody M1/70), and Ly6C-PE (antibody HK1.4); from BioLegend, B220/CD45R-peridinin chlorophyll protein (PerCP)-Cy5.5 (antibody RA3-GB2), CD8α-APC-Cy7 (antibody 53-6.7), CD11a-Alexa Fluor 488 (antibody M17/4), CD49b-Pacific Blue (antibody DX5), c-Kit-PE-Cy7 (antibody 2B8), F4/80-PE-Cy7 (antibody BM8), Flt3-PE (antibody A2F10), interleukin 7 receptor (IL-7R)-PE (antibody A7R34), Ly6G-APC-Cy7 (antibody 1A8), Sca-1-APC-Cy7 (antibody D7), and TER119-PerCP-Cy5.5 (antibody TER-119); and from eBioscience, B220/CD45R-APC (antibody RA3-6B2), B220/CD45R-PerCP-Cy5.5 (antibody RA3-GB2), CD11b-PerCP-Cy5.5 (antibody M1/70), CD11c-eFluor450 (antibody N418), CD16/CD32-fluorescein isothiocyanate (FITC) (antibody 93), CD25-Alexa Fluor 488 (antibody eBio3C7), CD34-eFluor450 (antibody RAM34), CD44-PE (antibody IM7), CD48-HM48.1 (FITC), CD69-FITC (antibody H1-2F3), CD150-Alexa Fluor 647 (antibody mShad150), major histocompatibility complex class II MHC-II-FITC (antibody AF6-120.1), Sca-1-APC (antibody D7), and TCRβ-PE-Cy5 (antibody H57.597).
Live cells in cell suspensions prepared from spleen, thymus, and bone marrow were counted using a hematocytometer, plated in 96-well plates, and washed twice with PBS. The cells were stained with 1:400 Zombi Aqua Fixable viability dye (BioLegend) for 15 min at room temperature in the dark, followed by washing with PBS. Cells were incubated with antibody for 20 min at 4°C. The cells were fixed using Cytofix/Cytoperm solution (BD) according to the manufacturer’s directions, washed in PBS, and resuspended in PBS containing 2% fetal bovine serum (FBS). Cell acquisition was completed using a FACSCanto II (BD) or LSR Fortessa cell analyzer (BD) flow cytometer (McGill Flow Cytometry Core Facility) using FACSDiva software (BD). Compensation was completed using OneComp eBeads (Invitrogen), and data were analyzed using FlowJo software (version 10.0.8r1).
For cytokine intracellular staining, splenocytes were stimulated with precoated anti-CD3 (5 μg/ml; eBioscience) and soluble anti-CD28 (2 μg/ml; eBioscience) in the presence of a protein transport inhibitor cocktail (eBioscience) for 6 h at 37°C in 5% CO2 in complete RPMI (10% FBS, 1 mM sodium pyruvate, 1% nonessential amino acids, 1% penicillin-streptomycin, β-mercaptoethanol). Cells were incubated with the following antibodies: anti-CD4-PE (antibody GK1.5; Invitrogen), anti-CD8α-BV421 (antibody 53-6.7; BioLegend), and anti-TCRβ-FITC (antibody H57-597; eBioscience). Cells were then fixed and permeabilized per the manufacturer’s protocol (in Cytofix/Cytoperm solution; BD) and stained intracellularly with the antibodies anti-TNF-α-PerCP-Cy5.5 (antibody MPG-XT22; BioLegend) and anti-IFN-γ-APC (antibody XMG1.2; BioLegend) for 30 min at 4°C. Cells were acquired on an eight-color FACSCanto II flow cytometer using FACSDiva software (BD). The data were analyzed using FlowJo software (version 10.0.8r1). Doublets were removed by side scatter height versus side scatter width gating.
Bone marrow-derived macrophage (BMDM) cell culture, RNA extraction, reverse transcription, PCR, and gel electrophoresis.
Bone marrow was extracted from mouse femurs as described above (see “Flow cytometry analyses” above). The 2-ml cell suspension that was recovered was split into two petri dishes, and 7 ml of complete RPMI (RPMI, 10% FBS, 1% penicillin-streptomycin) and 4 ml of macrophage colony-stimulating factor (M-CSF) were added to each dish. The cells were incubated at 37°C in 5% CO2. At day 3 of culture, the supernatant was removed and replaced with 6 ml RPMI. The dishes were scraped, and 10 ml of RPMI and 8 ml of M-CSF were added. The suspension was split into 2 petri dishes. At day 6, the supernatant was removed and the cells were rinsed once in 5 ml PBS, before being collected in 5 ml of PBS. The cells were centrifuged for 7 min at 1,400 rpm. The supernatant was discarded, and the pellets were resuspended in the TRIzol reagent (catalog number 15596-018; Invitrogen) for RNA extraction. Reverse transcription of 2.5 μg of RNA was conducted using ABM 5× All-in-One RT MasterMix (catalog number G486; ABM Life Science Products) and amplification of the resulting cDNA was completed using a standard PCR and EasyTaq DNA polymerase (catalog number AP111-01; Transgen Biotech) (forward primer, 5′-CCATGCAAGAGAAGCCACCAT-3′; reverse primer, 5′-AGTGATAGAGGCCTCCCGTGT-3′). RNA was visualized using gel electrophoresis on 2% agarose gels.
F2 cross and QTL mapping.
One hundred ninety-six offspring from an F2 cross of CC042 and C57BL/6NCrl mice were infected with Salmonella Typhimurium and euthanized at day 4 postinfection. Spleen and liver bacterial loads were determined as described above. Ninety-four animals with extreme phenotypes (half with high bacterial loads and half with low bacterial loads) were selected for genotyping. SNP genotyping was performed by Neogen Inc. (Lincoln, NE), using the Mouse Universal Genotyping Array (MUGA) containing 7,800 SNPs (61). All statistical tests (Pearson’s R correlation coefficient, QTL mapping) were performed using R statistical software. QTL mapping on the liver bacterial load was performed by using the J/qtl interface 1.3.5 for the R/qtl software version running under R (version 3.2.2) (62). The significance thresholds of limit-of-detection (LOD) scores were estimated by 10,000 permutations of experimental data.
CC042 private variants in the QTL regions.
Most CC strains were recently sequenced (29), and data are publicly available, including the list of private variants specific to each CC strain. We retrieved the CC042 private variants localized within the Stsl6 and Stsl7 QTLs. Three CC042 private variants were present within Stsl6 (Ush1c, Ccdc123, 4930435C17Rik), and only one was present within Stsl7 (Itgal).
Itgal genotyping.
Amplification of the region containing the CC042 Itgal deletion was conducted using a standard PCR (forward primer, 5′-TGCTTGGGTGTAGGCAGCCTCA-3′; reverse primer, 5′-CTTCAATCTGCAAGACCTGGTA-3′). DNA amplicons were digested using FauI with CutSmart buffer (catalog number R0651S; New England BioLabs) for 4 h at 55°C. The reaction was stopped by incubating the samples at 80°C for 15 min. The digested DNA was run on a 1.5% agarose gel in Tris-borate-EDTA (TBE) buffer.
Quantitative complementation testing.
CC042 mice were crossed with C57BL/6J and Itgal KO mice to produce ItgalB6/ItgalCC042 and ItgalKO/ItgalCC042 genotypes on a B6 × CC042 genetic background. C57BL/6J mice were crossed with Itgal KO mice to produce the ItgalB6/ItgalKO genotype on a B6 mouse genetic background. Mice were infected with S. Typhimurium, and bacterial loads were determined at day 4 postinfection, as described above.
Data availability.
All relevant data that support the findings of this study are within the paper and the files in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
We thank Line Larivière and Hyejin Park for technical assistance and Patricia D’Arcy (McGill University) and Isabelle Lanctin and Tommy Penel (DTPS/C2RA-Central Animal Facility, Institut Pasteur) for mouse breeding and screening.
D.M. is the recipient of a Canadian Institutes of Health Research (CIHR) project grant (grant MOP133700) and a Natural Sciences and Engineering Research Council (NSERC) discovery grant (grant RGPIN-2017-04717). A.N. is a Canada Research Chair Tier II in Hematopoiesis and Lymphocyte Differentiation and the recipient of CIHR project grant PJT-153016 and NSERC discovery grant RGPIN-2016-05657. Funding had been provided to M.T. by an NSERC Undergraduate Student Research Award and the Fonds de Recherche du Québec Nature et Technologies and to J.K. by the McGill Faculty of Medicine Solvay Fellowship. The flow cytometry work/cell sorting was performed in the Flow Cytometry Core Facility for flow cytometry and single cell analysis of the Life Science Complex (McGill University) and was supported by funding from the Canadian Foundation for Innovation. This study was also supported by AgroParisTech (France) through a Ph.D. fellowship to J.Z.
We declare no competing financial interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Kang E, Crouse A, Chevallier L, Pontier SM, Alzahrani A, Silue N, Campbell-Valois FX, Montagutelli X, Gruenheid S, Malo D. 2018. Enterobacteria and host resistance to infection. Mamm Genome 29:558–576. doi: 10.1007/s00335-018-9749-4. [DOI] [PubMed] [Google Scholar]
- 2.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, et al. 2012. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radhakrishnan A, Als D, Mintz ED, Crump JA, Stanaway J, Breiman RF, Bhutta ZA. 2018. Introductory article on global burden and epidemiology of typhoid fever. Am J Trop Med Hyg 99:4–9. doi: 10.4269/ajtmh.18-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LaRock DL, Chaudhary A, Miller SI. 2015. Salmonellae interactions with host processes. Nat Rev Microbiol 13:191–205. doi: 10.1038/nrmicro3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM. 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 6.Dhanoa A, Fatt QK. 2009. Non-typhoidal Salmonella bacteraemia: epidemiology, clinical characteristics and its’ association with severe immunosuppression. Ann Clin Microbiol Antimicrob 8:15. doi: 10.1186/1476-0711-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dougan G, John V, Palmer S, Mastroeni P. 2011. Immunity to salmonellosis. Immunol Rev 240:196–210. doi: 10.1111/j.1600-065X.2010.00999.x. [DOI] [PubMed] [Google Scholar]
- 8.Gilchrist JJ, MacLennan CA, Hill A. 2015. Genetic susceptibility to invasive Salmonella disease. Nat Rev Immunol 15:452–463. doi: 10.1038/nri3858. [DOI] [PubMed] [Google Scholar]
- 9.Vogt SL, Finlay BB. 2017. Gut microbiota-mediated protection against diarrheal infections. J Travel Med 24:S39–S43. doi: 10.1093/jtm/taw086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eva MM, Yuki KE, Dauphinee SM, Schwartzentruber JA, Pyzik M, Paquet M, Lathrop M, Majewski J, Vidal SM, Malo D. 2014. Altered IFN-gamma-mediated immunity and transcriptional expression patterns in N-ethyl-N-nitrosourea-induced STAT4 mutants confer susceptibility to acute typhoid-like disease. J Immunol 192:259–270. doi: 10.4049/jimmunol.1301370. [DOI] [PubMed] [Google Scholar]
- 11.Mastroeni P, Harrison JA, Robinson JH, Clare S, Khan S, Maskell DJ, Dougan G, Hormaeche CE. 1998. Interleukin-12 is required for control of the growth of attenuated aromatic-compound-dependent salmonellae in BALB/c mice: role of gamma interferon and macrophage activation. Infect Immun 66:4767–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qureshi ST, Lariviere L, Leveque G, Clermont S, Moore KJ, Gros P, Malo D. 1999. Endotoxin-tolerant mice have mutations in Toll-like receptor 4 (Tlr4). J Exp Med 189:615–625. doi: 10.1084/jem.189.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunstan SJ, Hue NT, Han B, Li Z, Tram TTB, Sim KS, Parry CM, Chinh NT, Vinh H, Lan NPH, Thieu NTV, Vinh PV, Koirala S, Dongol S, Arjyal A, Karkey A, Shilpakar O, Dolecek C, Foo JN, Phuong LT, Lanh MN, Do T, Aung T, Hon DN, Teo YY, Hibberd ML, Anders KL, Okada Y, Raychaudhuri S, Simmons CP, Baker S, de Bakker PIW, Basnyat B, Hien TT, Farrar JJ, Khor CC. 2014. Variation at HLA-DRB1 is associated with resistance to enteric fever. Nat Genet 46:1333–1336. doi: 10.1038/ng.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alvarez MI, Glover LC, Luo P, Wang L, Theusch E, Oehlers SH, Walton EM, Tram TTB, Kuang Y-L, Rotter JI, McClean CM, Chinh NT, Medina MW, Tobin DM, Dunstan SJ, Ko DC. 2017. Human genetic variation in VAC14 regulates Salmonella invasion and typhoid fever through modulation of cholesterol. Proc Natl Acad Sci U S A 114:E7746–E7755. doi: 10.1073/pnas.1706070114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilchrist JJ, Rautanen A, Fairfax BP, Mills TC, Naranbhai V, Trochet H, Pirinen M, Muthumbi E, Mwarumba S, Njuguna P, Mturi N, Msefula CL, Gondwe EN, MacLennan JM, Chapman SJ, Molyneux ME, Knight JC, Spencer CCA, Williams TN, MacLennan CA, Scott JAG, Hill A. 2018. Risk of nontyphoidal Salmonella bacteraemia in African children is modified by STAT4. Nat Commun 9:1014. doi: 10.1038/s41467-017-02398-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgan AP, Welsh CE. 2015. Informatics resources for the Collaborative Cross and related mouse populations. Mamm Genome 26:521–539. doi: 10.1007/s00335-015-9581-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts A, Pardo-Manuel de Villena F, Wang W, McMillan L, Threadgill DW. 2007. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm Genome 18:473–481. doi: 10.1007/s00335-007-9045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, Bleich A, Bogue M, Broman KW, Buck KJ, Buckler E, Burmeister M, Chesler EJ, Cheverud JM, Clapcote S, Cook MN, Cox RD, Crabbe JC, Crusio WE, Darvasi A, Deschepper CF, Doerge RW, Farber CR, Forejt J, Gaile D, Garlow SJ, Geiger H, Gershenfeld H, Gordon T, Gu J, Gu W, de Haan G, Hayes NL, Heller C, Himmelbauer H, Hitzemann R, Hunter K, Hsu H-C, Iraqi FA, Ivandic B, Jacob HJ, Jansen RC, Jepsen KJ, Johnson DK, Johnson TE, Kempermann G, et al. 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet 36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- 19.Aylor DL, Valdar W, Foulds-Mathes W, Buus RJ, Verdugo RA, Baric RS, Ferris MT, Frelinger JA, Heise M, Frieman MB, Gralinski LE, Bell TA, Didion JD, Hua K, Nehrenberg DL, Powell CL, Steigerwalt J, Xie Y, Kelada SNP, Collins FS, Yang IV, Schwartz DA, Branstetter LA, Chesler EJ, Miller DR, Spence J, Liu EY, McMillan L, Sarkar A, Wang J, Wang W, Zhang Q, Broman KW, Korstanje R, Durrant C, Mott R, Iraqi FA, Pomp D, Threadgill D, Pardo-Manuel de Villena F, Churchill GA. 2011. Genetic analysis of complex traits in the emerging collaborative cross. Genome Res 21:1213–1222. doi: 10.1101/gr.111310.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saul MC, Philip VM, Reinholdt LG, Center for Systems Neurogenetics of Addiction, Chesler EJ. 2019. High-diversity mouse populations for complex traits. Trends Genet 35:501–514. doi: 10.1016/j.tig.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noll KE, Ferris MT, Heise MT. 2019. The Collaborative Cross: a systems genetics resource for studying host-pathogen interactions. Cell Host Microbe 25:484–498. doi: 10.1016/j.chom.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Malo D, Mott R, Panthier J-J, Montagutelli X, Jaubert J. 2018. Identification of new loci involved in the host susceptibility to Salmonella Typhimurium in Collaborative Cross mice. BMC Genomics 19:303. doi: 10.1186/s12864-018-4667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vidal S, Tremblay ML, Govoni G, Gauthier S, Sebastiani G, Malo D, Skamene E, Olivier M, Jothy S, Gros P. 1995. The Ity/Lsh/Bcg locus: natural resistance to infection with intracellular parasites is abrogated by disruption of the Nramp1 gene. J Exp Med 182:655–666. doi: 10.1084/jem.182.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z, Zhang X, Wang W. 2012. HTreeQA: using semi-perfect phylogeny trees in quantitative trait loci study on genotype data. G3 (Bethesda) 2:175–189. doi: 10.1534/g3.111.001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Germain RN. 2002. T-cell development and the CD4-CD8 lineage decision. Nat Rev Immunol 2:309–322. doi: 10.1038/nri798. [DOI] [PubMed] [Google Scholar]
- 26.Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, Offner S, Dunant CF, Eshkind L, Bockamp E, Lió P, Macdonald HR, Trumpp A. 2008. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 27.Cabezas-Wallscheid N, Klimmeck D, Hansson J, Lipka DB, Reyes A, Wang Q, Weichenhan D, Lier A, von Paleske L, Renders S, Wunsche P, Zeisberger P, Brocks D, Gu L, Herrmann C, Haas S, Essers MAG, Brors B, Eils R, Huber W, Milsom MD, Plass C, Krijgsveld J, Trumpp A. 2014. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell 15:507–522. doi: 10.1016/j.stem.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 28.Simon MM, Greenaway S, White JK, Fuchs H, Gailus-Durner V, Wells S, Sorg T, Wong K, Bedu E, Cartwright EJ, Dacquin R, Djebali S, Estabel J, Graw J, Ingham NJ, Jackson IJ, Lengeling A, Mandillo S, Marvel J, Meziane H, Preitner F, Puk O, Roux M, Adams DJ, Atkins S, Ayadi A, Becker L, Blake A, Brooker D, Cater H, Champy M-F, Combe R, Danecek P, di Fenza A, Gates H, Gerdin A-K, Golini E, Hancock JM, Hans W, Hölter SM, Hough T, Jurdic P, Keane TM, Morgan H, Müller W, Neff F, Nicholson G, Pasche B, Roberson L-A, Rozman J, et al. 2013. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol 14:R82. doi: 10.1186/gb-2013-14-7-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srivastava A, Morgan AP, Najarian ML, Sarsani VK, Sigmon JS, Shorter JR, Kashfeen A, McMullan RC, Williams LH, Giusti-Rodríguez P, Ferris MT, Sullivan P, Hock P, Miller DR, Bell TA, McMillan L, Churchill GA, de Villena F-M. 2017. Genomes of the mouse Collaborative Cross. Genetics 206:537–556. doi: 10.1534/genetics.116.198838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schittenhelm L, Hilkens CM, Morrison VL. 2017. beta2 integrins as regulators of dendritic cell, monocyte, and macrophage function. Front Immunol 8:1866. doi: 10.3389/fimmu.2017.01866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evans R, Patzak I, Svensson L, De Filippo K, Jones K, McDowall A, Hogg N. 2009. Integrins in immunity. J Cell Sci 122:215–225. doi: 10.1242/jcs.019117. [DOI] [PubMed] [Google Scholar]
- 32.Hogg N, Patzak I, Willenbrock F. 2011. The insider’s guide to leukocyte integrin signalling and function. Nat Rev Immunol 11:416–426. doi: 10.1038/nri2986. [DOI] [PubMed] [Google Scholar]
- 33.Bose TO, Colpitts SL, Pham QM, Puddington L, Lefrancois L. 2014. CD11a is essential for normal development of hematopoietic intermediates. J Immunol 193:2863–2872. doi: 10.4049/jimmunol.1301820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bose TO, Pham Q-M, Jellison ER, Mouries J, Ballantyne CM, Lefrançois L. 2013. CD11a regulates effector CD8 T cell differentiation and central memory development in response to infection with Listeria monocytogenes. Infect Immun 81:1140–1151. doi: 10.1128/IAI.00749-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Revilla C, Gonzalez AL, Conde C, Lopez-Hoyos M, Merino J. 1997. Treatment with anti-LFA-1 alpha monoclonal antibody selectively interferes with the maturation of CD4− 8+ thymocytes. Immunology 90:550–556. doi: 10.1046/j.1365-2567.1997.00183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rutigliano JA, Johnson TR, Hollinger TN, Fischer JE, Aung S, Graham BS. 2004. Treatment with anti-LFA-1 delays the CD8+ cytotoxic-T-lymphocyte response and viral clearance in mice with primary respiratory syncytial virus infection. J Virol 78:3014–3023. doi: 10.1128/jvi.78.6.3014-3023.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capece T, Walling BL, Lim K, Kim KD, Bae S, Chung HL, Topham DJ, Kim M. 2017. A novel intracellular pool of LFA-1 is critical for asymmetric CD8(+) T cell activation and differentiation. J Cell Biol 216:3817–3829. doi: 10.1083/jcb.201609072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reichardt P, Patzak I, Jones K, Etemire E, Gunzer M, Hogg N. 2013. A role for LFA-1 in delaying T-lymphocyte egress from lymph nodes. EMBO J 32:829–843. doi: 10.1038/emboj.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghosh S, Chackerian AA, Parker CM, Ballantyne CM, Behar SM. 2006. The LFA-1 adhesion molecule is required for protective immunity during pulmonary Mycobacterium tuberculosis infection. J Immunol 176:4914–4922. doi: 10.4049/jimmunol.176.8.4914. [DOI] [PubMed] [Google Scholar]
- 40.Smith CM, Proulx MK, Olive AJ, Laddy D, Mishra BB, Moss C, Gutierrez NM, Bellerose MM, Barreira-Silva P, Phuah JY, Baker RE, Behar SM, Kornfeld H, Evans TG, Beamer G, Sassetti CM. 2016. Tuberculosis susceptibility and vaccine protection are independently controlled by host genotype. mBio 7:e01516-16. doi: 10.1128/mBio.01516-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ingram JP, Brodsky IE, Balachandran S. 2017. Interferon-gamma in Salmonella pathogenesis: new tricks for an old dog. Cytokine 98:27–32. doi: 10.1016/j.cyto.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramirez-Alejo N, Santos-Argumedo L. 2014. Innate defects of the IL-12/IFN-gamma axis in susceptibility to infections by mycobacteria and salmonella. J Interferon Cytokine Res 34:307–317. doi: 10.1089/jir.2013.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swaim CD, Scott AF, Canadeo LA, Huibregtse JM. 2017. Extracellular ISG15 signals cytokine secretion through the LFA-1 integrin receptor. Mol Cell 68:581–590.e5. doi: 10.1016/j.molcel.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fine JS, Kruisbeek AM. 1991. The role of LFA-1/ICAM-1 interactions during murine T lymphocyte development. J Immunol 147:2852–2859. [PubMed] [Google Scholar]
- 45.Prince JE, Brayton CF, Fossett MC, Durand JA, Kaplan SL, Smith CW, Ballantyne CM. 2001. The differential roles of LFA-1 and Mac-1 in host defense against systemic infection with Streptococcus pneumoniae. J Immunol 166:7362–7369. doi: 10.4049/jimmunol.166.12.7362. [DOI] [PubMed] [Google Scholar]
- 46.Smith CM, Proulx MK, Lai R, Kiritsy MC, Bell TA, Hock P, Pardo-Manuel de Villena F, Ferris MT, Baker RE, Behar SM, Sassetti CM. 2019. Functionally overlapping variants control TB susceptibility in Collaborative Cross mice. bioRxiv doi: 10.1101/785725. [DOI] [PMC free article] [PubMed]
- 47.Ferreira CP, Cariste LM, Santos Virgilio FD, Moraschi BF, Monteiro CB, Vieira Machado AM, Gazzinelli RT, Bruna-Romero O, Menin Ruiz PL, Ribeiro DA, Lannes-Vieira J, Lopes MF, Rodrigues MM, de Vasconcelos J. 2017. LFA-1 mediates cytotoxicity and tissue migration of specific CD8(+) T cells after heterologous prime-boost vaccination against Trypanosoma cruzi infection. Front Immunol 8:1291. doi: 10.3389/fimmu.2017.01291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyamoto M, Emoto M, Emoto Y, Brinkmann V, Yoshizawa I, Seiler P, Aichele P, Kita E, Kaufmann S. 2003. Neutrophilia in LFA-1-deficient mice confers resistance to listeriosis: possible contribution of granulocyte-colony-stimulating factor and IL-17. J Immunol 170:5228–5234. doi: 10.4049/jimmunol.170.10.5228. [DOI] [PubMed] [Google Scholar]
- 49.Conlan JW, North RJ. 1994. Listeria monocytogenes, but not Salmonella typhimurium, elicits a CD18-independent mechanism of neutrophil extravasation into the murine peritoneal cavity. Infect Immun 62:2702–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Montagutelli X. 2000. Effect of the genetic background on the phenotype of mouse mutations. J Am Soc Nephrol 11(Suppl 16):S101–S105. [PubMed] [Google Scholar]
- 51.Nadeau JH. 2003. Modifier genes and protective alleles in humans and mice. Curr Opin Genet Dev 13:290–295. doi: 10.1016/s0959-437x(03)00061-3. [DOI] [PubMed] [Google Scholar]
- 52.Threadgill DW, Miller DR, Churchill GA, de Villena FP-M. 2011. The collaborative cross: a recombinant inbred mouse population for the systems genetic era. ILAR J 52:24–31. doi: 10.1093/ilar.52.1.24. [DOI] [PubMed] [Google Scholar]
- 53.McArthur MA, Fresnay S, Magder LS, Darton TC, Jones C, Waddington CS, Blohmke CJ, Dougan G, Angus B, Levine MM, Pollard AJ, Sztein MB. 2015. Activation of Salmonella Typhi-specific regulatory T cells in typhoid disease in a wild-type S. Typhi challenge model. PLoS Pathog 11:e1004914. doi: 10.1371/journal.ppat.1004914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Lange KM, Moutsianas L, Lee JC, Lamb CA, Luo Y, Kennedy NA, Jostins L, Rice DL, Gutierrez-Achury J, Ji SG, Heap G, Nimmo ER, Edwards C, Henderson P, Mowat C, Sanderson J, Satsangi J, Simmons A, Wilson DC, Tremelling M, Hart A, Mathew CG, Newman WG, Parkes M, Lees CW, Uhlig H, Hawkey C, Prescott NJ, Ahmad T, Mansfield JC, Anderson CA, Barrett JC. 2017. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet 49:256–261. doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu Q, Kaplan M, Ray D, Ray D, Zacharek S, Gutsch D, Richardson B. 2002. Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis Rheum 46:1282–1291. doi: 10.1002/art.10234. [DOI] [PubMed] [Google Scholar]
- 56.Balada E, Castro-Marrero J, Felip L, Ordi-Ros J, Vilardell-Tarrés M. 2014. Clinical and serological findings associated with the expression of ITGAL, PRF1, and CD70 in systemic lupus erythematosus. Clin Exp Rheumatol 32:113–116. [PubMed] [Google Scholar]
- 57.Damotte V, Guillot-Noel L, Patsopoulos NA, Madireddy L, El Behi M, De Jager PL, Baranzini SE, Cournu-Rebeix I, Fontaine B. 2014. A gene pathway analysis highlights the role of cellular adhesion molecules in multiple sclerosis susceptibility. Genes Immun 15:126–132. doi: 10.1038/gene.2013.70. [DOI] [PubMed] [Google Scholar]
- 58.Hanna S, Etzioni A. 2012. Leukocyte adhesion deficiencies. Ann N Y Acad Sci 1250:50–55. doi: 10.1111/j.1749-6632.2011.06389.x. [DOI] [PubMed] [Google Scholar]
- 59.Canadian Council on Animal Care. 1993. Guide to the care and use of experimental animals, vol 1, 2nd ed Canadian Council on Animal Care, Ottawa, ON, Canada. [Google Scholar]
- 60.Morahan G, Balmer L, Monley D. 2008. Establishment of “The Gene Mine”: a resource for rapid identification of complex trait genes. Mamm Genome 19:390–393. doi: 10.1007/s00335-008-9134-9. [DOI] [PubMed] [Google Scholar]
- 61.Morgan AP, Fu CP, Kao CY, Welsh CE, Didion JP, Yadgary L, Hyacinth L, Ferris MT, Bell TA, Miller DR, Giusti-Rodriguez P, Nonneman RJ, Cook KD, Whitmire JK, Gralinski LE, Keller M, Attie AD, Churchill GA, Petkov P, Sullivan PF, Brennan JR, McMillan L, Pardo-Manuel de Villena F. 2015. The mouse universal genotyping array: from substrains to subspecies. G3 (Bethesda) 6:263–279. doi: 10.1534/g3.115.022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Broman KW, Wu H, Sen S, Churchill GA. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data that support the findings of this study are within the paper and the files in the supplemental material.