Clostridioides (formerly known as Clostridium) difficile is the leading cause of hospital-acquired gastrointestinal infections in the United States and one of three urgent health care threats identified by the Centers for Disease Control and Prevention. C. difficile disease is mediated by the production of toxins that disrupt the epithelial barrier and cause a robust host inflammatory response.
KEYWORDS: Clostridioides difficile, Clostridium difficile, IL-17A, IL-22, IL-23, ILC3, neutrophils, Th17, type 3 immunity
ABSTRACT
Clostridioides (formerly known as Clostridium) difficile is the leading cause of hospital-acquired gastrointestinal infections in the United States and one of three urgent health care threats identified by the Centers for Disease Control and Prevention. C. difficile disease is mediated by the production of toxins that disrupt the epithelial barrier and cause a robust host inflammatory response. Studies in humans as well as animal models of disease have shown that the type of immune response generated against the infection dictates the outcome of disease, often irrespective of bacterial burden. Much of the focus on immunity during C. difficile infection (CDI) has been on type 3 immunity because of the established role for this arm of the immune system in other gastrointestinal inflammatory conditions such as inflammatory bowel disease (IBD). For example, interleukin-22 (IL-22) production by group 3 innate lymphoid cells (ILC3s) protects against pathobionts translocating across the epithelium during CDI. On the other hand, interleukin-17 (IL-17) production by Th17 cells increases CDI-associated mortality. Additionally, neutropenia has been associated with increased susceptibility to CDI in humans, but increased neutrophilia in mouse models correlates with host pathology. Taking the data together, these findings suggest dual roles for type 3 immune responses during infection. Here, we review the complex role of type 3 immunity during CDI and delineate what is known about innate and adaptive cellular immunity as well as the downstream effector cytokines known to be important during this infection.
INTRODUCTION
Clostridioides (formerly known as Clostridium) difficile is a spore-forming, Gram-positive, anaerobic bacterium that was identified as the cause of pseudomembranous colitis in 1978 (1, 2). C. difficile is the leading cause of hospital-acquired gastrointestinal infections in the United States (3). According to estimates by the Centers for Disease Control and Prevention, C. difficile causes almost 500,000 infections and 29,000 deaths in a single year in the United States alone (4). The annual cost of C. difficile infection (CDI) in the United States is estimated at almost 40 billion dollars (5–7). Several studies have reported significant increases in the prevalence and severity of CDI over the last 2 decades, and these increases are thought to be attributable to the emergence of hypervirulent C. difficile transferase (CDT)-expressing strains, including but not limited to ribotype 027 strains (also known as NAP1 strains) (8–10).
C. difficile causes disease in hosts with a perturbed gut microbiota usually due to the use of broad-spectrum antibiotics (6). Typically, the infection is acquired in hospital settings, although the incidence of community-acquired infections is also on the rise (11). Some reports suggest that although the majority of community-acquired infections are associated with antibiotic use, 30% to 35% of infected patients have no prior antibiotic exposure (11, 12).
CLOSTRIDIOIDES DIFFICILE BIOLOGY AND PATHOGENESIS
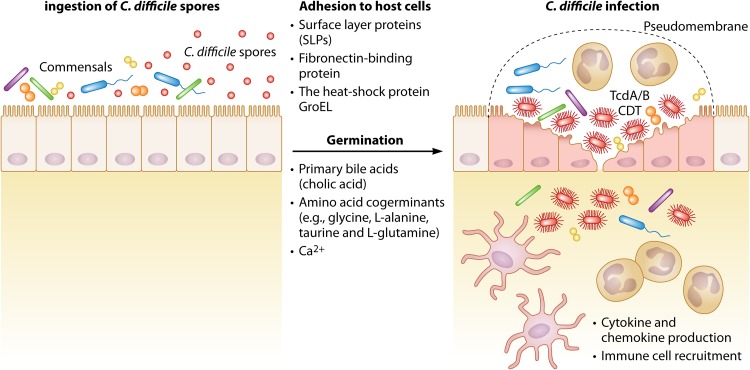
Since C. difficile is an obligate anaerobe, the vegetative form of this bacterium is unable to survive outside the host in an aerobic environment. Therefore, dissemination of C. difficile is mediated by dormant spores ingested through the oral-fecal route (13). Once ingested, these spores germinate and the vegetative bacteria cause disease by toxin production. Several signals have been shown to be important for spore germination, including bile acids, amino acids, and Ca2+ (Fig. 1). In humans, the two main primary bile acids are cholic acid and chenodeoxycholic acid. These bile acids are the end products of cholesterol metabolism, and although they are mostly reabsorbed and recycled to the liver, they can also be found in the large intestine. There, bile salt hydrolases expressed by many colon microbiota species convert these primary bile acids to secondary bile acids. The primary bile acid cholate is a known inducer of germination, whereas chenodeoxycholate inhibits germination (14). In a study of the effect of several secondary bile acids, Thanissery et al. showed that many secondary bile acids, namely, deoxycholate (DCA), lithocholate (LCA), ursodeoxycholate (UDCA), isodeoxycholate (iDCA), isolithocholate (iLCA), ω-muricholate (ωMCA), and hyodeoxycholate (HDCA), inhibited spore germination (15). C. difficile spores use the subtilisin-like receptor CspC pseudoprotease as the bile acid germinant receptor (13, 16).
FIG 1.
The pathogenesis of C. difficile. C. difficile spores are ingested by hosts with altered micriobiota, where the gut microbial community is perturbed, usually due to the use of antibiotics. Once the spores are in the large intestines, several signals, including the primary bile acid cholic acid, amino acid cogerminants such as glycine, l-alanine, taurine, and l-glutamine and Ca2+ ions, trigger germination. After germination, adhesion of vegetative cells is mediated by C. difficile’s surface layer proteins as well as the fibronectin-binding protein and the heat shock protein GroEL. C. difficile infection is mediated by the production of its main virulence factors, namely, toxins A and B (TcdA/B) and, in some strains, the binary toxin CDT. These toxins cause disruption of the actin cytoskeleton, epithelial cell rounding, and cell death. Production of damage-associated molecular patterns (DAMPs) and several cytokines and chemokines by epithelial cells leads to the recruitment of neutrophils and other immune cells. The influx of neutrophils, along with fibrin, mucin, and cellular debris, leads to the formation of pseudomembranes, which are characteristic of C. difficile colitis.
Bile acids are necessary for C. difficile germination, but they are not sufficient on their own. Other signals are needed for germination, including amino acid cogerminants. Glycine is the most effective cogerminant, although l-alanine, taurine, and l-glutamine are also good cogerminants (17). Recently, Shrestha et al. showed a role for CspA in recognition of these amino acid cogerminants (18). Finally, a role for Ca2+ has been described where in vitro media and ex vivo mouse ileal contents depleted for Ca2+ did not support C. difficile spore germination (19).
Once C. difficile spores germinate into vegetative bacteria, they adhere to epithelial cells and cause disease by producing toxin A (TcdA) and toxin B (TcdB) and, in some strains, a third toxin called CDT (20, 21). TcdA and TcdB are glucosyltransferases that inactivate Rho family GTPases, leading to the disruption of the actin cytoskeleton of epithelial cells, cell rounding, and cell death (22). CDT has been shown to enhance bacterial adhesion by inducing microtubule protrusions on host cells (23).
TcdA and TcdB are considered C. difficile’s main virulence factors, since strains that do not produce at least one of these toxins are avirulent (24). In addition to the toxins, however, C. difficile expresses other virulence factors that mediate colonization and adherence to epithelial cells. For example, all C. difficile strains express surface layer proteins (SLPs) that form the outermost layer of the bacterium. The surface layer of C. difficile is composed of two proteins (high-molecular-weight and low-molecular-weight proteins) that have been shown to be involved in adherence to host cells (25). Other C. difficile virulence factors include fibronectin-binding protein and the heat shock protein GroEL, both of which are thought to be involved in enhanced adhesion of C. difficile to host cells (24).
C. difficile colonization and adhesion to epithelial cells and toxin production lead to the upregulation of many proinflammatory cytokines and the recruitment of neutrophils and other inflammatory immune cells. The influx of neutrophils, along with fibrin, mucin, and cellular debris, causes the formation of the pseudomembranes that are characteristic of C. difficile colitis (Fig. 1). Clinical presentation of disease ranges from mild diarrhea to toxic megacolon and host mortality (26). The standard of care for treatment of severe CDI is vancomycin (27). While vancomycin treatment is effective therapy in most cases, recurrence affects 1 in 5 patients with CDI (4). Fecal microbial transplants (FMT) have proven successful in 80% to 90% of patients with recurrent infection, but the mechanism of protection and the long-term effects these transplants might have on the host are not fully understood (28, 29).
THE ROLE OF THE IMMUNE SYSTEM DURING CDI
Growing evidence has supported the idea that the type of immune response mounted by the host against CDI directly affects the severity and outcome of disease. Therefore, better understanding of protective and pathogenic immune responses during CDI will help identify new targets for future immunotherapies and preventative strategies. One of such studies that emphasized the importance of considering the host immune system in assessing CDI severity was a study that showed that biomarkers of intestinal inflammation such as CXCL5 and interleukin-8 (IL-8) are better predictors of the time to resolution of C. difficile diarrhea than bacterial burden (30, 31). Both CXCL5 and IL-8 bind to IL-8 receptor 2 and recruit and activate neutrophils. The levels of these biomarkers were elevated in diarrheal patients compared to controls, and the same biomarkers have been associated with severe CDI in human patients and mouse models (30, 31).
Data from other human and mouse studies support the notion that manipulating the immune response can modulate the severity of CDI independently of C. difficile burden or toxin production. For example, Buonomo et al. showed that pretreating mice with the type 2 cytokine interleukin 25 (IL-25) can protect mice against severe CDI via induction of eosinophils and downstream protection of epithelial integrity (32). In a study investigating a related mechanism, Cowardin et al. also showed that CDT expression by the hypervirulent R20291 strain of C. difficile causes severe CDI by inducing eosinophil apoptosis, leading to a decrease of eosinophil recruitment to the site of infection via a Toll-like receptor 2 (TLR2)-dependent mechanism (33). Both of those studies established a novel role for type 2 immunity in protection against CDI and have paved the way for future therapeutics that may target this pathway in patients with CDI.
Neutrophils, downstream of type 3 immune responses, have long been described as the hallmark of C. difficile infection (34–36). Type 3 immune responses include the activity of RORγt-positive (RORγt+) lymphocytes, which have the ability to produce IL-17A alone or with IL-22 as their signature cytokine(s) (37). These cells include CD4+ Th17 cells, CD8+ Tc17 cells, and group 3 innate lymphoid cells (ILC3s) (38–40). Because of the long-described association between CDI and neutrophilia downstream of type 3 immunity, many studies have interrogated this pathway and its role during infection. Here, we review innate and adaptive type 3 responses and describe their dual roles in the pathogenesis of C. difficile.
INNATE IMMUNITY AND GROUP 3 INNATE LYMPHOID CELLS
Innate lymphoid cells (ILCs) represent a subset of cells that share lymphoid cell morphologies but lack the somatically rearranged surface receptors expressed by T and B lymphocytes and therefore lack antigen specificity (41). Like T cell lymphocytes, noncytotoxic innate lymphoid cells are subdivided into three groups based on the transcription factors required for their development and the effector cytokines that they produce. Group 1 ILCs (ILC1s) express the transcription factor T-bet and produce gamma interferon (IFN-γ) and tumor necrosis factor alpha (TNF-α); group 2 ILCs (ILC2s) express the transcription factor GATA3 and produce IL-4, IL-5, IL-9, and IL-13; and group 3 ILCs (ILC3s) express the transcription factor RORγt and produce IL17A, IL-17F, IL-22, TNF-α, and granulocyte macrophage colony-stimulating factor (GM-CSF) (41–48). ILC3s play a role in defense against extracellular pathogens, including Citrobacter rodentium, Candida albicans, and Streptococcus pneumoniae (49–51). In addition, innate lymphoid cells have been implicated in the pathogenesis of inflammatory bowel disease (IBD). Crohn’s disease is associated with an increase in levels of IFN-γ-producing ILC1s, and T cell-independent mouse models of colitis exhibited an increase in IL-17 production by ILC3s (40, 52, 53). IL-17- and IL-22-producing ILC3s are also associated with psoriasis vulgaris (54). Taking the data together, ILCs share many effector cytokines and therefore many functions with T cell lymphocytes, including protection against infections, tissue healing and repair, and autoimmunity and inflammation. However, the main difference between ILCs and T cell lymphocytes is the lack of antigen specificity and immunological memory.
As with all lymphocytes, ILCs are derived from a common lymphoid progenitor (CLP) in the bone marrow. A cell population that is similar in phenotype to CLPs but that expresses the integrin α4β7 gives rise to all ILCs and NK cells but not T and B cells. Downstream of this cell type are two populations that express the transcriptional repressor Id2. Id2+ cells that do not express the transcription factor promyeloid leukemia zinc finger (PLZF) can give rise to lymphoid tissue inducer (LTi) cells. In contrast, Id2+ cells that coexpress PLZF are restricted to ILC1, ILC2, and ILC3 lineages (41). Once these cells differentiate in the bone marrow, they can be found in various tissues, including peripheral blood, the gut, lung, and skin, with different frequencies (55).
In the context of C. difficile infection, there are three reports assessing the roles of ILCs during CDI. First, Geiger et al. showed that Nfil3−/− mice, which have a deficit in all three ILC subsets, were more susceptible to infection with spores of C. difficile strain VPI 10463 (56). These mice lack bZIP transcription factor Nfil3, previously shown to be required for NK cell and ILC1 development. Geiger et al. showed that these mice also had significant deficiencies in intestinal ILC3s and lung and visceral adipose tissue ILC2s. They showed that Nfil3 is required for development of these ILCs but not necessarily for the maintenance of mature ILC3s since a conditional knockout (KO) of Nfil3 in mature NKp46+ ILC3s did not diminish their frequencies.
In a later study by the same group, Abt et al. investigated directly the protective or pathogenic role of each ILC subset during CDI (57). The authors used Rag2 common-gamma-chain double-knockout mice (Ragγc−/−), which lack all ILC subsets in addition to T and B cells. They showed that Ragγc−/− mice develop more-severe CDI than wild-type (WT) mice and Rag1−/− mice, lacking only T and B cells. Next, the authors showed that transferring ILCs is sufficient to protect Ragγc−/− mice from increased severity of disease. Furthermore, by using Rag1 IFN-γ double-knockout and Rag1 Tbet double-knockout mice, they demonstrated a role for T-bet+, IFN-γ-producing ILC1s in protection from CDI early during infection. The authors also observed that IL-22−/− mice had higher weight loss rates after infection with spores and significantly higher mortality after infection with a mixed spore/vegetative-cell inoculum. As the authors point out in the discussion of their results, the more profound effect of knocking out IL-22 on mortality during a vegetative infection might indicate that IL-22 is more important during the severe stages of CDI and sepsis caused by translocating pathobionts than in the early stages of C. difficile colonization after ingestion of spores. When IFN-γ was neutralized in the Rag1 IL-22 double-knockout mice, 100% of the mice succumbed to the infection, suggesting combined roles for IFN-γ-producing ILC1s and IL-22-producing ILC3s in providing early protection from CDI. Interestingly, the authors found that neutralizing IL-17A in Rag1−/− mice did not change the outcome of infection, suggesting that there was no role for IL-17A-producing ILCs in this model. Note that the use of Rag.IL-17, Rag.IFN-γ, and Rag.IL-22 double-knockout mice does not test exclusively the roles of these cytokines produced by ILCs, since subsets of other innate immune cells are capable of producing IFN-γ, IL-22, and IL-17A.
In support of the idea of a protective role for IL-22 in CDI, Hasegawa et al. found that CDI induces the production of IL-22 in the colon, liver, and lung (58). The authors found that CDI-induced upregulation of IL-22 is independent of RAG1, suggesting that innate immune cells, predominantly ILCs, represent the source of IL-22 (59). Hasegawa and colleagues found that IL-22−/− mice developed more-severe disease and that this increased disease severity was independent of adaptive and neutrophilic immune responses. Instead, severe disease was due to translocation of pathobionts across the damaged epithelium following CDI. The authors demonstrated elegantly that during CDI, increased IL-22 levels led to the induction of the complement C3, the deposition of C3 on the surface of pathobionts, and the downstream phagocytosis of these bacteria by neutrophils and macrophages. Sadighi Akha et al. reported no increased weight loss in mice treated with anti-IL-22 antibody after CDI (60). This contrasting result was perhaps due to the use of a neutralizing antibody against IL-22, rather than a knockout model, which could have meant less-efficient abrogation of IL-22 signaling. Nonetheless, Hasegawa et al. and Abt et al. independently showed a protective role for IL-22 during CDI (57, 58).
More recently, a third study, by Frisbee et al., of the role of ILCs during CDI described a protective role for ILC2s and eosinophils downstream of the type 2 cytokine interleukin 33 (IL-33) (61). Frisbee and colleagues showed that treating mice with an antibiotic cocktail decreased IL-33 protein levels in the cecum. On the other hand, pretreatment of mice with IL-33 was sufficient to protect antibiotic-treated mice from severe CDI. IL-33 treatment increased recruitment of ILC2s to the colon during CDI, and adoptive transfer of these ILC2s to naive Ragγc−/− mice was sufficient to protect them from severe disease.
ADAPTIVE IMMUNITY AND TH17 CELLS
Th17 cells were first described by Harrington et al., who found a subset of T cells that did not fit the dichotomy of Th1 and Th2 cells that was the dogma at the time (38). Differentiation of those cells was inhibited by IFN-γ and IL-4, but the absence of these cytokines and the presence of IL-23 led to the presence of a cell population that expressed the transcription factor RORγt and was capable of producing IL-17A (38). Since then, Th17 cells have been shown to produce IL-17F, IFN-γ, IL-21, IL-22, TNF-α, and GM-CSF. Through cytokine production, Th17 cells have downstream effects on neutrophil and mononuclear phagocyte recruitment as well as on antimicrobial peptide production by epithelial cells. While Th17 cells represent an important form of defense against extracellular bacteria and fungi, they have also been implicated in the pathogenesis of autoimmune diseases such as psoriasis and IBD (37, 48).
In contrast to the roles of innate and humoral responses in C. difficile infection, the role of CD4 T cells has been vastly understudied. Some evidence for the protective role that CD4 T cells might play against CDI comes from studies in HIV patients. Sanchez et al. reported higher rates of C. difficile infection in patients with advanced stages of HIV than in HIV-infected patients without AIDS (62). Similarly, Jha et al. found that in Indian HIV patients, the rate of C. difficile infection correlated with HIV seropositivity as well as with low CD4 T cell counts (63). Other studies reporting at least a 2-fold increase of CDI in AIDS patients and correlations with low CD4 T cell counts are reviewed nicely by Collini et al. (64).
In a mouse study of the protective role CD4 T cells might play in CDI, Johnston et al. observed that wild-type C57BL/6 mice challenged with C. difficile were protected from a subsequent challenge with the same C. difficile strain (65). They found that the rechallenged mice had protective colonic IgA and systemic IgG responses against C. difficile toxins A and B. To test the role of T cell help in generating protective humoral responses, the authors used CD4−/− mice, which lack the expression of the CD4 coreceptor, have a deficit in CD4 T cells, and lack IgG class switching. Upon rechallenge, CD4−/− mice were protected from CDI and this protection was long-lived. These mice generated IgA responses but no IgG response, as predicted. Interestingly, when the authors used MHCII−/− mice, which have a dramatic decrease in CD4 T cells and a deficiency in IgA and IgG class switching, those mice were not protected against rechallenge. The authors found no antitoxin antibodies in the sera or mucosa of these mice. Altogether, these data suggest that T cell help is needed in protection against recurrent CDI and that this protection correlates with antitoxin IgA or IgG antibody generation. As the authors point out, however, that study did not address the differences in the microbiota between the knockout and wild-type strains with respect to cohousing, 16S rRNA sequencing, or the use of littermate controls. These differences likely play a role in protection against rechallenge in some knockout strains, especially considering the drop in the bacterial burden in these mice. Another limitation was that the production of IgA antibody did not require CD4+ T cell help. The authors postulated that CD4− T cells might help with the generation of antitoxin IgA antibodies without providing data to support that hypothesis.
In another report, Ryan et al. investigated the ability of the immune system to recognize C. difficile surface layer proteins (SLPs) and the downstream cytokine profile (66). They found that pulsing bone marrow-derived dendritic cells (BMDCs) with SLPs led to the production of several cytokines such as IL-12p70, IL-23, IL-1β, TNF-α, and IL-10 and that this recognition was dependent on TLR4 signaling. They also observed that coculturing these BMDCs with CD4 T cells led to the induction of T helper responses and the production of IL-17, IFN-γ, and IL-4 by these T cells. Finally, the authors found that TLR4−/− and Myd88−/− mice developed more-severe disease than WT mice. However, they did not show directly in vivo that this phenotype was dependent on recognition of SLPs. This increase in the severity of disease in TLR4−/− mice was also unlikely to be dependent on CD4 T cells because differences in weight loss between TLR4−/− and WT mice are observed as early as day 1 of infection, before a T cell response against C. difficile can be generated. Nonetheless, that report described TLR4-dependent recognition of C. difficile proteins and downstream induction of T helper cells. Their results lead the way to study of the protective role of C. difficile-specific T cells in longer or recurrent models of disease.
On the other hand, there is also evidence that certain subsets of CD4 T cells play a pathogenic role during infection. In a study focused on the severity of C. difficile infection following treatment with the nonsteroidal anti-inflammatory drug (NSAID) indomethacin, Maseda et al. found that indomethacin-treated mice developed CDI that was more severe than that seen with untreated controls (67). This increased severity of disease was associated with increased numbers of neutrophils, CD4 T cells, and RORγt+ CD4 T cells in the colon lamina propria and peritoneum during infection. Indomethacin-treated mice also had higher levels of the type 3 cytokines IL-6 and IL-1β in colonic tissue than untreated controls. Finally, indomethacin treatment also led to alterations in the composition of the gut microbiota that the authors hypothesize might play a role in exacerbation of CDI severity in these mice. Although that study showed a correlation between enhanced Th17 responses in mice and more-severe CDI, it remains unclear whether Th17 cells alone are necessary and sufficient to exacerbate CDI severity in indomethacin-treated mice.
More recently, Saleh et al. studied the mechanism of increased severity of CDI in IBD patients (68). Using a dextran sulfate sodium (DSS) murine model of colitis, that study showed that DSS-treated mice developed more-severe CDI despite full recovery from DSS colitis. This increased severity of disease was not associated with enhanced C. difficile burden or toxin production. Furthermore, cohousing of DSS-treated mice with untreated controls to restore an intact microbiota alone was not sufficient to protect these mice from severe disease. Instead, severe disease in DSS mice was dependent on CD4+ T cells and depletion of CD4-expressing cells before infection protected these mice from severe CDI. Using adoptive-transfer studies, the authors showed that the presence of Th17 cells alone is sufficient to exacerbate the severity of CDI very early during infection. Additionally, blocking IL-17 signaling by either neutralizing the IL-17A cytokine or blocking the IL-17 receptor alpha provided protection against severe disease in the early stages of infection. Finally, in patients with CDI, serum levels of two type 3 cytokines IL-6 and IL-23 were associated with severe CDI. Patients with high levels of IL-6 in the serum were significantly more likely to succumb to the infection than those with low serum IL-6 levels. This is consistent with a previous report of higher IL-6 levels in the serum of 8 patients with severe CDI (69). Taking the data together, that study directly demonstrated that the presence of Th17 cells alone is sufficient to worsen the outcome of CDI, highlighting the importance of further studies into the role of these cells during disease.
The only study to look at human T cell responses to C. difficile infection was done by Yacyshyn et al. in 2014 (70). In that study, the authors used flow cytometry to characterize IL-17, IFN-γ, and FoxP3 expression in PBMCs of C. difficile patients. The patient cohort included 20 patients with initial C. difficile infection, 6 patients that developed recurrent disease during the study, 14 patients with known previous episodes of CDI, 20 inpatient controls, and 16 healthy controls. Many of the results obtained in that study failed to reach statistical significance, perhaps due to the small number of patients in each of the groups. Nonetheless, the authors observed that patients with CDI had slightly higher numbers of circulating IL-17+ CD4 T cells than healthy controls; however, there were no differences between CDI patients and inpatient controls with respect to those cells. The authors also made the qualitative observation that 5 of the 6 patients that developed recurrent disease during the course of the study had higher levels of IL-17+, IFN-γ+, and FoxP3+ T cells, and they postulated that the Th17 cells in these patients may have been coexpressing two or more of these proteins. However, as the authors note, the staining procedures were done separately for IL-17, IFN-γ, and FoxP3; thus, directly assessing coexpression was not feasible. Despite the limitations of that study, it does raise the issue of the role of Th17 cells, and perhaps of IFN-γ-producing Th17 cells, in C. difficile recurrence. Future studies with larger patient numbers and a deeper phenotypic characterization of T cell subsets are needed to address this fascinating issue. A role for CD8+ Tc17 cells during CDI has yet to be described.
EFFECTOR CYTOKINES AND DOWNSTREAM RESPONSES
IL-23.
Interleukin 23 (IL-23) is a cytokine that was first described by Oppmann et al. in 2000 (71). IL-23 shares the p40 subunit with IL-12 but has a unique p19 subunit that is distantly related to the p35 subunit of IL-12. Prior to the discovery of IL-23, it was thought that IL-12 played a major role during autoimmune diseases because many of the antibodies targeting IL-12 were against the shared p40 subunit. Since then, it has been shown that IL-23 plays a major role in the pathogenesis of inflammatory bowel disease (IBD) and other autoimmune diseases such as experimental autoimmune encephalomyelitis (EAE) (72). Polymorphisms in the IL-23R gene have been linked to IBD (73). Later, it was reported that IL-23 causes colitis in a T cell transfer model by directly acting on Th17 cells via the IL-23 receptor and promoting their survival as well as inducing the production of IL-17A and other Th17-derived cytokines (74). IL-23 has also been shown to play a role in T cell-independent colitis induced by infection with Helicobacter hepaticus. Buonocore et al. showed that induction of colitis in this model was dependent on a RORγt-expressing innate lymphoid cell subset (ILC3s) and the production of IL-17A and IFN-γ (40). In a separate report, Powell et al. (75) showed that IL-23 can promote IL-17A production by ILCs where T-bet signaling is lost and can induce more-severe colitis. Taking the data together, the studies showed an effect of IL-23 signaling on both T cell and ILC populations leading to the production of IL-17A and contributing to the pathogenesis of IBD.
Because of the long-appreciated role of the IL-23 axis in IBD, several studies have evaluated the involvement of this cytokine in C. difficile colitis. Buonomo et al. found higher levels of IL-23 in colonic tissue biopsy specimens isolated from C. difficile-positive patients than in biopsy specimens from C. difficile-negative patients (76). Similarly, Darkoh et al. found higher levels of IL-23, as well as of IL-8, in the stool of patients with C. difficile diarrhea than in the stool of other diarrheal patients (77).
In a mouse model of CDI, it was shown previously that abrogation of IL-23 signaling by a gene knockout or antibody neutralization protects mice from severe CDI (76). Using the same mouse genotype in two separate reports, McDermott et al. found that IL-23 knockout mice showed reduced recruitment of neutrophils and Ly6Chi monocytes to the colon and a slight but not statistically significant reduction in histopathological scores during infection (78, 79). The authors, however, did not assess the effect of knocking out IL-23 on survival or clinical signs of disease severity. Finally, in a study aimed at understanding the mechanism of IL-23 induction during CDI, Cowardin et al. found that C. difficile toxins A/B can act synergistically with pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) to activate the inflammasome, leading to production of IL-1β and downstream induction of IL-23 (80).
Taking the data together, the studies established that CDI induces IL-23 and that elevated IL-23 levels cause severe disease. None of the studies, however, described the cellular mechanism of increased severity of disease. More investigation is needed to evaluate whether this effect is T cell dependent or independent and which downstream signals and effector cells are required for exacerbation of CDI severity.
IL-17A.
IL-17A was first described in 1988 and was shown to induce inflammation by triggering the release of IL-6, IL-8, G-CSF, and IL-1β and other inflammatory cytokines by various target cell types (81). It was then shown that injection of IL-17A into the airway in vivo triggered neutrophil recruitment to the lung by the induction of CXC cytokines (82). Later, lymphocytes expressing the transcription factor RORγt, including Th17, ILC3 and Tc17 cells, were shown to produce IL-17A and the roles of these cells in bacterial and fungal infections as well as in autoimmune diseases started to be investigated.
In a study directly testing the role of IL-17 signaling during CDI, Tateda et al. reported enhanced survival in IL-17A and IL-17F double-knockout mice (IL-17 KO) (83). This enhanced survival correlated with the suppression of type 17 cytokines such as IL-1β and IL-6 as well as a decrease in neutrophil recruitment to the colon during infection. That study suggested that IL-17 signaling plays a pathogenic role during CDI, although the authors did not investigate the roles of IL-17A and IL-17F separately. A role for IL-17F during CDI has yet to be examined. Those findings are consistent with another study showing that blocking of IL-17RA, which binds to IL-17A and IL-17F, as well as neutralization of IL-17A protected susceptible mice from severe disease early during infection (68). Finally, another study also described an association with increased IL-17A and worsening of C. difficile disease. Wang et al. found that increased levels of IL-17A and IL-23 in the absence of IL-27 signaling led to increased mortality postinfection (84).
In a study that involved human C. difficile patients, Jafari et al. described the production of many Th17 cytokines, including IL-8, IL-6, IL-1β, IFN-γ, IL-17A, and IL-22, by colonic biopsy specimens from C. difficile-negative donors in response to stimulation with C. difficile strains R20291 and 630 (85). Yu et al. found elevated levels of IL-17A, IL-6, IL-1β, IL-8, and IFN-γ, among many other cytokines, in the serum of C. difficile patients compared to healthy controls, as might be expected in an inflamed mucosa (86). However, comparing the levels of these cytokines between patients with mild/moderate disease and those with severe disease, they did not find a correlation between high serum levels of type 3 cytokines and increased severity of disease. The authors concluded that severe disease was associated with a decrease in the IFN-γ/IL-17A ratio in the serum; however, their data suggest that this was strictly due to a decrease in IFN-γ levels in patients with severe CDI rather than to an increase in IL-17A levels. The absence of a correlation between the levels of any of the type 3 cytokines in serum and disease severity was perhaps due to the small number of patients (17–21). Indeed, a more recent study that included almost 400 patients per group reported higher serum IL-23 and IL-6 levels in patients with severe CDI (68). Assessment of the profiles of these cytokines in colonic tissue of CDI patients has yet to be performed and might reveal a correlation between high IL-17A levels and severe disease, which has yet to be described in human CDI.
Neutrophilic responses in the defense against C. difficile.
Further downstream of IL-23 signaling and the induction of IL-17A production by T cells and ILC3s, some studies have evaluated the role of neutrophils during CDI. IL-17A and other cytokines in the IL-17 family, including IL-17B, IL-17C, and IL17F, have been shown to promote neutrophil recruitment. These cytokines bind to receptors on immune cells (such as monocytes and macrophages) and nonimmune cells (such as epithelial and endothelial cells and fibroblasts). In response, these cells produce granulocyte colony-stimulating factor (G-CSF) and chemokines such as CXCL1, CXCL2, and CXCL5, leading to enhanced production of neutrophils in the bone marrow and recruitment to the tissue, respectively (87, 88).
In the context of CDI, studies have shown dual roles for neutrophils during infection. Reports that have suggested a protective role during infection include a study that found that in patients with leukemia, neutropenia was one of the factors associated with CDI occurrence (89). In another study of hematopoietic stem cell transplant patients, neutropenia was the only independent predictor of recurrent CDI (90). Mouse data that support this beneficial role for neutrophils come from a study of the effects of Toll-like receptor and MyD88 signaling during CDI. Jarchum et al. found that MyD88 signaling protects against severe CDI by recruiting neutrophils to the site of infection (91). Depletion of neutrophils in this model led to increased CDI-associated mortality. Finally, Hasegawa et al. (92) showed that loss of Nod1 signaling in response to CDI led to increased mortality due to defective CXCL1-dependent recruitment of neutrophils.
On the other hand, some studies found correlations between enhanced neutrophilic responses and increased severity of CDI. In general, leukocytosis and high white blood cell counts have been associated with increased CDI-induced mortality (93, 94). More specifically, in an analysis of fecal samples from children with and without symptomatic C. difficile diarrhea, elevated levels of the neutrophil recruiters CXCL5 and IL-8 correlated with persistent diarrhea. Furthermore, in diarrheal patients, time to diarrhea resolution was significantly increased in those with high CXCL5 and IL-8 levels in the stool (30, 31). Additionally, blocking neutrophil recruitment with an anti-CD18 antibody resulted in reduced tissue pathology following injection of C. difficile toxin A into rabbit ileal loops (34). Similarly, reduced neutrophil recruitment in the context of IL-23 blockade correlated with reduced histopathology in a mouse model of infection (78). A more comprehensive review of the dual roles of neutrophils during CDI was done by Jose and Madan (95).
CONCLUSIONS
In summary, C. difficile infections represent a major cause of infectious disease mortalities in the United States. Although antibiotic and FMT treatments exist for the infection, the high level of recurrence and the emergence of hypervirulent strains highlight the importance of developing new therapies. Targeting the immune system in many animal models has proven successful at preventing CDI-associated mortality, and many of these approaches focus on type 3 immune responses. Overall, the host immune system seems to represent a double-edged sword in the context of C. difficile infection (Fig. 2). The majority of studies investigated one cytokine or cell type in isolation. Given the plasticity of Th17 and ILC3s, studies that assess the production of one cytokine by these cells are limited in scope. A more comprehensive description of the phenotype and cytokine profile of type 3 immune cells, as well as other cell types, is required to fully understand which immune responses are beneficial and which are harmful during CDI.
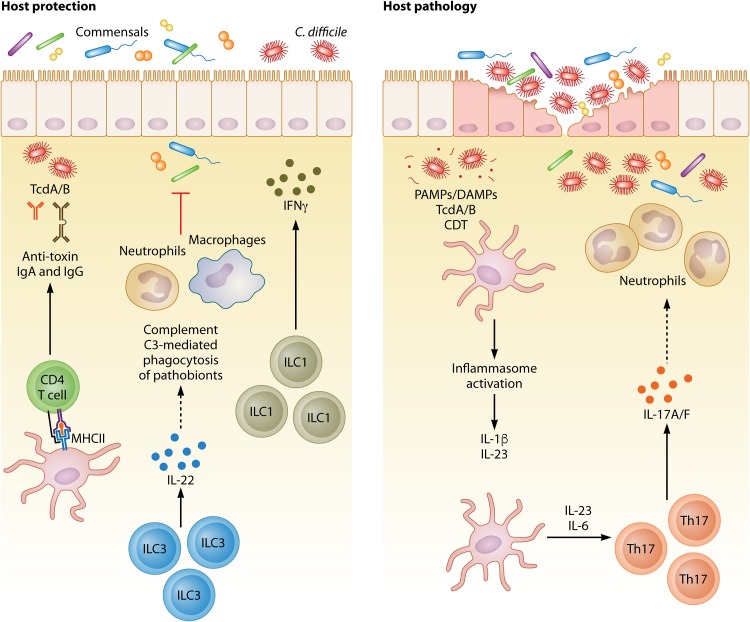
FIG 2.
Protective and pathogenic type 3 immunity during C. difficile infection. Several in vitro, in vivo, and human studies have assessed the role of type 3 immunity during CDI. The type and magnitude of this response directly influence the outcome of disease. Protective immune responses include CD4 T cell-dependent generation of IgA and IgG antibodies against C. difficile toxins, IFN-γ production by ILC1s, IL-22 production by ILC3s, and complement-mediated phagocytosis of translocating commensals downstream of IL-22. On the other hand, enhanced type 3 responses and excess inflammation can have off-target effects on the host and lead to increased pathology and mortality. For example, inflammasome activation and the downstream induction of IL-1β and IL-23 increased Th17 responses downstream of IL-6 and IL-23, and those responses, together with enhanced levels of neutrophilia, have been found to be associated with increased disease severity.
Many studies of CDI focus on the contributions of either the gut microbiota or the host immune system to C. difficile colonization and disease progression. However, few reports have described the interaction between the two and how that can control immune progression. For example, whereas we know that IFN-γ/IL-22-producing ILCs are protective and IL-17A-producing Th17 cells are pathogenic during CDI, little is known about the microbial and metabolic signals that drive the domination of one pathway over the other. Complete metagenomic and metabolomic studies of the gut microbiome coupled with comprehensive characterization of the downstream immune response would help improve connecting the microbiome to the host immune system, two important factors in C. difficile disease progression.
Further downstream of type 3 responses, the role of neutrophils during CDI has yet to be fully described. Evidence exists for both protective and pathogenic roles for neutrophils in disease. For example, blocking neutrophil recruitment downstream of Myd88 signaling leads to increased mortality. However, in humans, higher levels of the known neutrophil recruiters IL-8 and CXCL-5 correlate with increased mortality. There is evidence of heterogeneity in the neutrophil population in humans. Some of these neutrophil types include immature and mature neutrophils; immunosuppressive low-density neutrophils (LDNs), also known as granulocytic-myeloid-derived suppressor cells (G-MDSCs); proinflammatory LDNs, also known as low-density granulocytes (LDGs); and proinflammatory and immunosuppressive normal-density neutrophils (NDNs) (96). Future studies should describe functional differences in the neutrophils present at the site of infection and their prospective effects on disease outcome. This can include functional assays such as measurement of levels of myeloperoxidase, reactive oxygen species (ROS), or neutrophil extracellular trap (NET) formation as well as RNA sequencing and flow cytometry techniques to describe differences in the phenotype and function of distinct neutrophil populations that may be protective or pathogenic in different contexts.
Altogether, our understanding of immunity to CDI has continued to grow in the last few decades. However, a deeper understanding of cellular immunity is needed before these discoveries can be translated into therapies that harness the power of a patient’s own immune system to protect them against severe disease.
Biographies

Mahmoud M. Saleh, Ph.D., is a mucosal immunologist at the University of Virginia, Charlottesville, VA. He started his scientific career while studying biology and chemistry at Guilford College in Greensboro, NC. His undergraduate research was focused on the effects of bisphenol A (BPA) on the viability and reproduction of Caenorhabditis elegans. He then joined the University of Virginia’s Department of Microbiology, Immunology, and Cancer Biology, where he became interested in immune responses to Clostridioides difficile infection because of the clinical relevance and translational potential. His dissertation project described a novel mechanism by which Th17 cells can increase CDI-associated mortality and disease severity. His work was recognized by multiple awards, including the University of Virginia’s Department of Medicine award for an outstanding publication and awards for travel to multiple microbiology and mucosal immunology conferences. His goal is to continue to understand and modulate immune responses to enhance the outcome of inflammatory diseases.

William A. Petri, Jr., M.D., Ph.D., is Professor of Medicine in the Division of Infectious Diseases & International Health at the University of Virginia and Vice Chair of the Department of Medicine, where he practices internal medicine and the subspecialty of infectious diseases. His research is focused on the immunopathogenesis of diarrheal infections, including C. difficile infection, in a mouse model and in patients at UVA hospital.
REFERENCES
- 1.Bartlett JG, Chang TW, Gurwith M, Gorbach SL, Onderdonk AB. 1978. Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. N Engl J Med 298:531–534. doi: 10.1056/NEJM197803092981003. [DOI] [PubMed] [Google Scholar]
- 2.Oren A, Garrity GM. 2016. Notification of changes in taxonomic opinion previously published outside the IJSEM. Int J Syst Evol Microbiol 66:2469–2470. doi: 10.1099/ijsem.0.001150. [DOI] [PubMed] [Google Scholar]
- 3.Magill SS, O’Leary E, Janelle SJ, Thompson DL, Dumyati G, Nadle J, Wilson LE, Kainer MA, Lynfield R, Greissman S, Ray SM, Beldavs Z, Gross C, Bamberg W, Sievers M, Concannon C, Buhr N, Warnke L, Maloney M, Ocampo V, Brooks J, Oyewumi T, Sharmin S, Richards K, Rainbow J, Samper M, Hancock EB, Leaptrot D, Scalise E, Badrun F, Phelps R, Edwards JR. 2018. Changes in prevalence of health care-associated infections in U.S. hospitals. N Engl J Med 379:1732–1744. doi: 10.1056/NEJMoa1801550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lessa FC, Winston LG, McDonald LC, Emerging Infections Program C. difficile Surveillance Team. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:2369–2370. doi: 10.1056/NEJMc1505190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song X, Bartlett JG, Speck K, Naegeli A, Carroll K, Perl TM. 2008. Rising economic impact of Clostridium difficile-associated disease in adult hospitalized patient population. Infect Control Hosp Epidemiol 29:823–828. doi: 10.1086/588756. [DOI] [PubMed] [Google Scholar]
- 6.Ananthakrishnan AN. 2011. Clostridium difficile infection: epidemiology, risk factors and management. Nat Rev Gastroenterol Hepatol 8:17–26. doi: 10.1038/nrgastro.2010.190. [DOI] [PubMed] [Google Scholar]
- 7.Hopkins RJ, Wilson RB. 2018. Treatment of recurrent Clostridium difficile colitis: a narrative review. Gastroenterol Rep (Oxf) 6:21–28. doi: 10.1093/gastro/gox041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pepin J. 2004. Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. CMAJ 171:466–472. doi: 10.1503/cmaj.1041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bacci S, Mølbak K, Kjeldsen MK, Olsen K. 2011. Binary toxin and death after Clostridium difficile infection. Emerg Infect Dis 17:976–982. doi: 10.3201/eid1706.101483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucado J, Gould C, Elixhauser A. 2012. Clostridium difficile infections (CDI) in hospital stays, 2009: statistical brief #124. https://www.ncbi.nlm.nih.gov/pubmed/22574332. [PubMed]
- 11.Khanna S, Pardi DS, Aronson SL, Kammer PP, Orenstein R, St Sauver JL, Harmsen WS, Zinsmeister AR. 2012. The epidemiology of community-acquired Clostridium difficile infection: a population-based study. Am J Gastroenterol 107:89–95. doi: 10.1038/ajg.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chitnis AS, Holzbauer SM, Belflower RM, Winston LG, Bamberg WM, Lyons C, Farley MM, Dumyati GK, Wilson LE, Beldavs ZG, Dunn JR, Gould LH, MacCannell DR, Gerding DN, McDonald LC, Lessa FC. 2013. Epidemiology of community-associated Clostridium difficile infection, 2009 through 2011. JAMA Intern Med 173:1359. doi: 10.1001/jamainternmed.2013.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu D, Sorg JA, Sun X. 8 February 2018, posting date Clostridioides difficile biology: sporulation, germination, and corresponding therapies for C. difficile infection. Front Cell Infect Microbiol doi: 10.3389/fcimb.2018.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Britton RA, Young VB. 2012. Interaction between the intestinal microbiota and host in Clostridium difficile colonization resistance. Trends Microbiol 20:313–319. doi: 10.1016/j.tim.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thanissery R, Winston JA, Theriot CM. 2017. Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe 45:86–100. doi: 10.1016/j.anaerobe.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francis MB, Allen CA, Shrestha R, Sorg JA. 2013. Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS Pathog 9:e1003356. doi: 10.1371/journal.ppat.1003356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shrestha R, Sorg JA. 2018. Hierarchical recognition of amino acid co-germinants during Clostridioides difficile spore germination. Anaerobe 49:41–47. doi: 10.1016/j.anaerobe.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shrestha R, Cochran AM, Sorg JA. 2019. The requirement for co-germinants during Clostridium difficile spore germination is influenced by mutations in yabG and cspA. PLoS Pathog 15:e1007681. doi: 10.1371/journal.ppat.1007681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kochan TJ, Somers MJ, Kaiser AM, Shoshiev MS, Hagan AK, Hastie JL, Giordano NP, Smith AD, Schubert AM, Carlson PE, Hanna PC. 2017. Intestinal calcium and bile salts facilitate germination of Clostridium difficile spores. PLoS Pathog 13:e1006443. doi: 10.1371/journal.ppat.1006443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cowardin CA, Petri WA. 2014. Host recognition of Clostridium difficile and the innate immune response. Anaerobe 30:205–209. doi: 10.1016/j.anaerobe.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buonomo EL, Petri WA. 2016. The microbiota and immune response during Clostridium difficile infection. Anaerobe 41:79–84. doi: 10.1016/j.anaerobe.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chumbler NM, Farrow MA, Lapierre LA, Franklin JL, Haslam DB, Haslam D, Goldenring JR, Lacy DB. 2012. Clostridium difficile toxin B causes epithelial cell necrosis through an autoprocessing-independent mechanism. PLoS Pathog 8:e1003072. doi: 10.1371/journal.ppat.1003072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwan C, Stecher B, Tzivelekidis T, van Ham M, Rohde M, Hardt W-D, Wehland J, Aktories K. 2009. Clostridium difficile toxin CDT induces formation of microtubule-based protrusions and increases adherence of bacteria. PLoS Pathog 5:e1000626. doi: 10.1371/journal.ppat.1000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Awad MM, Johanesen PA, Carter GP, Rose E, Lyras D. 2014. Clostridium difficile virulence factors: insights into an anaerobic spore-forming pathogen. Gut Microbes 5:579–593. doi: 10.4161/19490976.2014.969632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calabi E, Calabi F, Phillips AD, Fairweather NF. 2002. Binding of Clostridium difficile surface layer proteins to gastrointestinal tissues. Infect Immun 70:5770–5778. doi: 10.1128/iai.70.10.5770-5778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poutanen SM, Simor AE. 2004. Clostridium difficile-associated diarrhea in adults. CMAJ 171:51–58. doi: 10.1503/cmaj.1031189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stevens VW, Nelson RE, Schwab-Daugherty EM, Khader K, Jones MM, Brown KA, Greene T, Croft LD, Neuhauser M, Glassman P, Goetz MB, Samore MH, Rubin MA. 2017. Comparative effectiveness of vancomycin and metronidazole for the prevention of recurrence and death in patients with Clostridium difficile infection. JAMA Intern Med 177:546. doi: 10.1001/jamainternmed.2016.9045. [DOI] [PubMed] [Google Scholar]
- 28.Van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman J, Tijssen JGP, Speelman P, Dijkgraaf MGW, Keller JJ. 2013. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 29.Leslie JL, Vendrov KC, Jenior ML, Young VB. 2019. The gut microbiota is associated with clearance of Clostridium difficile infection independent of adaptive immunity. mSphere 4:e00698-18. doi: 10.1128/mSphereDirect.00698-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El Feghaly RE, Stauber JL, Deych E, Gonzalez C, Tarr PI, Haslam DB. 2013. Markers of intestinal inflammation, not bacterial burden, correlate with clinical outcomes in Clostridium difficile infection. Clin Infect Dis 56:1713–1721. doi: 10.1093/cid/cit147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El Feghaly RE, Stauber JL, Tarr PI, Haslam DB. 2013. Intestinal inflammatory biomarkers and outcome in pediatric Clostridium difficile infections. J Pediatr 163:1697–1704.e2. doi: 10.1016/j.jpeds.2013.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buonomo EL, Cowardin CA, Wilson MG, Saleh MM, Pramoonjago P, Petri WA. 2016. Microbiota-regulated IL-25 increases eosinophil number to provide protection during Clostridium difficile infection. Cell Rep 16:432–443. doi: 10.1016/j.celrep.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cowardin CA, Buonomo EL, Saleh MM, Wilson MG, Burgess SL, Kuehne SA, Schwan C, Eichhoff AM, Koch-Nolte F, Lyras D, Aktories K, Minton NP, Petri WA. 2016. The binary toxin CDT enhances Clostridium difficile virulence by suppressing protective colonic eosinophilia. Nat Microbiol 1:16108. doi: 10.1038/nmicrobiol.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly CP, Becker S, Linevsky JK, Joshi MA, O'Keane JC, Dickey BF, LaMont JT, Pothoulakis C. 1994. Neutrophil recruitment in Clostridium difficile toxin A enteritis in the rabbit. J Clin Invest 93:1257–1265. doi: 10.1172/JCI117080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly CP, Pothoulakis C, LaMont JT. 1994. Clostridium difficile colitis. N Engl J Med 330:257–262. doi: 10.1056/NEJM199401273300406. [DOI] [PubMed] [Google Scholar]
- 36.Pépin J, Valiquette L, Gagnon S, Routhier S, Brazeau I. 2007. Outcomes of Clostridium difficile-associated disease treated with metronidazole or vancomycin before and after the emergence of NAP1/027. Am J Gastroenterol 102:2781–2788. doi: 10.1111/j.1572-0241.2007.01539.x. [DOI] [PubMed] [Google Scholar]
- 37.Annunziato F, Romagnani C, Romagnani S. 2015. The 3 major types of innate and adaptive cell-mediated effector immunity. J Allergy Clin Immunol 135:626–635. doi: 10.1016/j.jaci.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 38.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. 2005. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol 6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 39.Kondo T, Takata H, Matsuki F, Takiguchi M. 2009. Cutting edge: phenotypic characterization and differentiation of human CD8 + T cells producing IL-17. J Immunol 182:1794–1798. doi: 10.4049/jimmunol.0801347. [DOI] [PubMed] [Google Scholar]
- 40.Buonocore S, Ahern PP, Uhlig HH, Ivanov II, Littman DR, Maloy KJ, Powrie F. 2010. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature 464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Artis D, Spits H. 2015. The biology of innate lymphoid cells. Nature 517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 42.Klose CSN, Flach M, Möhle L, Rogell L, Hoyler T, Ebert K, Fabiunke C, Pfeifer D, Sexl V, Fonseca-Pereira D, Domingues RG, Veiga-Fernandes H, Arnold SJ, Busslinger M, Dunay IR, Tanriver Y, Diefenbach A. 2014. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell 157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 43.Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, Furusawa J, Ohtani M, Fujii H, Koyasu S. 2010. Innate production of TH2 cytokines by adipose tissue-associated c-Kit+Sca-1+ lymphoid cells. Nature 463:540–544. doi: 10.1038/nature08636. [DOI] [PubMed] [Google Scholar]
- 44.Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TKA, Bucks C, Kane CM, Fallon PG, Pannell R, Jolin HE, McKenzie A. 2010. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 464:1367–1370. doi: 10.1038/nature08900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price AE, Liang H-E, Sullivan BM, Reinhardt RL, Eisley CJ, Erle DJ, Locksley RM. 2010. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc Natl Acad Sci U S A 107:11489–11494. doi: 10.1073/pnas.1003988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JKM, Doherty JM, Mills JC, Colonna M. 2009. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature 457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cupedo T, Crellin NK, Papazian N, Rombouts EJ, Weijer K, Grogan JL, Fibbe WE, Cornelissen JJ, Spits H. 2009. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat Immunol 10:66–74. doi: 10.1038/ni.1668. [DOI] [PubMed] [Google Scholar]
- 48.Miossec P, Korn T, Kuchroo VK. 2009. Interleukin-17 and type 17 helper T cells. N Engl J Med 361:888–898. doi: 10.1056/NEJMra0707449. [DOI] [PubMed] [Google Scholar]
- 49.Satoh-Takayama N, Vosshenrich CAJ, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention J-J, Thiam K, Cerf-Bensussan N, Mandelboim O, Eberl G, Di Santo JP. 2008. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity 29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 50.Gladiator A, Wangler N, Trautwein-Weidner K, LeibundGut-Landmann S. 2013. Cutting edge: IL-17-secreting innate lymphoid cells are essential for host defense against fungal infection. J Immunol 190:521–525. doi: 10.4049/jimmunol.1202924. [DOI] [PubMed] [Google Scholar]
- 51.Van Maele L, Carnoy C, Cayet D, Ivanov S, Porte R, Deruy E, Chabalgoity JA, Renauld J-C, Eberl G, Benecke AG, Trottein F, Faveeuw C, Sirard J-C. 2014. Activation of type 3 innate lymphoid cells and interleukin 22 secretion in the lungs during Streptococcus pneumoniae infection. J Infect Dis 210:493–503. doi: 10.1093/infdis/jiu106. [DOI] [PubMed] [Google Scholar]
- 52.Bernink JH, Peters CP, Munneke M, Te Velde AA, Meijer SL, Weijer K, Hreggvidsdottir HS, Heinsbroek SE, Legrand N, Buskens CJ, Bemelman WA, Mjösberg JM, Spits H. 2013. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol 14:221–229. doi: 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
- 53.Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD, Cella M, Colonna M. 2013. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity 38:769–781. doi: 10.1016/j.immuni.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Villanova F, Flutter B, Tosi I, Grys K, Sreeneebus H, Perera GK, Chapman A, Smith CH, Di Meglio P, Nestle FO. 2014. Characterization of innate lymphoid cells in human skin and blood demonstrates increase of NKp44+ ILC3 in psoriasis. J Invest Dermatol 134:984–991. doi: 10.1038/jid.2013.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hazenberg MD, Spits H. 2014. Human innate lymphoid cells. Blood 124:700–709. doi: 10.1182/blood-2013-11-427781. [DOI] [PubMed] [Google Scholar]
- 56.Geiger TL, Abt MC, Gasteiger G, Firth MA, O’Connor MH, Geary CD, O’Sullivan TE, van den Brink MR, Pamer EG, Hanash AM, Sun JC. 2014. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J Exp Med 211:1723–1731. doi: 10.1084/jem.20140212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abt MC, Lewis BB, Caballero S, Xiong H, Carter RA, Sušac B, Ling L, Leiner I, Pamer EG. 2015. Innate immune defenses mediated by two ILC subsets are critical for protection against acute Clostridium difficile infection. Cell Host Microbe 18:27–37. doi: 10.1016/j.chom.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hasegawa M, Yada S, Liu MZ, Kamada N, Muñoz-Planillo R, Do N, Núñez G, Inohara N. 2014. Interleukin-22 regulates the complement system to promote resistance against pathobionts after pathogen-induced intestinal damage. Immunity 41:620–632. doi: 10.1016/j.immuni.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sonnenberg GF, Fouser LA, Artis D. 2011. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol 12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 60.Sadighi Akha AA, McDermott AJ, Theriot CM, Carlson PE, Frank CR, McDonald RA, Falkowski NR, Bergin IL, Young VB, Huffnagle GB. 2015. Interleukin-22 and CD160 play additive roles in the host mucosal response to Clostridium difficile infection in mice. Immunology 144:587–597. doi: 10.1111/imm.12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frisbee AL, Saleh MM, Young MK, Leslie JL, Simpson ME, Abhyankar MM, Cowardin CA, Ma JZ, Pramoonjago P, Turner SD, Liou AP, Buonomo EL, Petri WA. 2019. IL-33 drives group 2 innate lymphoid cell-mediated protection during Clostridium difficile infection. Nat Commun 10:2712. doi: 10.1038/s41467-019-10733-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanchez TH, Brooks JT, Sullivan PS, Juhasz M, Mintz E, Dworkin MS, Jones JL. 2005. Bacterial diarrhea in persons with HIV infection, United States, 1992–2002. Clin Infect Dis 41:1621–1627. doi: 10.1086/498027. [DOI] [PubMed] [Google Scholar]
- 63.Jha AK, Uppal B, Chadha S, Bhalla P, Ghosh R, Aggarwal P, Dewan R. 27 December 2012, posting date Clinical and microbiological profile of HIV/AIDS cases with diarrhea in North India. J Pathog doi: 10.1155/2012/971958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Collini PJ, Kuijper E, Dockrell DH. 2012. Clostridium difficile infection in patients with HIV/AIDS. Curr HIV/AIDS Rep 10:273–282. doi: 10.1007/s11904-013-0162-z. [DOI] [PubMed] [Google Scholar]
- 65.Johnston PF, Gerding DN, Knight KL. 2014. Protection from Clostridium difficile infection in CD4 T cell- and polymeric immunoglobulin receptor-deficient mice. Infect Immun 82:522–531. doi: 10.1128/IAI.01273-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ryan A, Lynch M, Smith SM, Amu S, Nel HJ, McCoy CE, Dowling JK, Draper E, O'Reilly V, McCarthy C, O'Brien J, Ní Eidhin D, O'Connell MJ, Keogh B, Morton CO, Rogers TR, Fallon PG, O'Neill LA, Kelleher D, Loscher CE. 2011. A role for TLR4 in Clostridium difficile infection and the recognition of surface layer proteins. PLoS Pathog 7:e1002076. doi: 10.1371/journal.ppat.1002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maseda D, Zackular JP, Trindade B, Kirk L, Roxas JL, Rogers LM, Washington MK, Du L, Koyama T, Viswanathan VK, Vedantam G, Schloss PD, Crofford LJ, Skaar EP, Aronoff DM. 2019. Nonsteroidal anti-inflammatory drugs alter the microbiota and exacerbate Clostridium difficile colitis while dysregulating the inflammatory response. mBio 10:e02282-18. doi: 10.1128/mBio.02282-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saleh MM, Frisbee AL, Leslie JL, Buonomo EL, Cowardin CA, Ma JZ, Simpson ME, Scully KW, Abhyankar MM, Petri WA. 2019. Colitis-induced Th17 cells increase the risk for severe subsequent Clostridium difficile infection. Cell Host Microbe 25:756–765.e5. doi: 10.1016/j.chom.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rao K, Erb-Downward JR, Walk ST, Micic D, Falkowski N, Santhosh K, Mogle JA, Ring C, Young VB, Huffnagle GB, Aronoff DM. 2014. The systemic inflammatory response to Clostridium difficile infection. PLoS One 9:e92578. doi: 10.1371/journal.pone.0092578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yacyshyn MB, Reddy TN, Plageman LR, Wu J, Hollar AR, Yacyshyn BR. 2014. Clostridium difficile recurrence is characterized by pro-inflammatory peripheral blood mononuclear cell (PBMC) phenotype. J Med Microbiol 63:1260–1273. doi: 10.1099/jmm.0.075382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oppmann B, Lesley R, Blom B, Timans JC, Xu Y, Hunte B, Vega F, Yu N, Wang J, Singh K, Zonin F, Vaisberg E, Churakova T, Liu M-r, Gorman D, Wagner J, Zurawski S, Liu Y-J, Abrams JS, Moore KW, Rennick D, de Waal-Malefyt R, Hannum C, Bazan JF, Kastelein RA. 2000. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity 13:715–725. doi: 10.1016/S1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 72.Ahern PP, Izcue A, Maloy KJ, Powrie F. 2008. The interleukin-23 axis in intestinal inflammation. Immunol Rev 226:147–159. doi: 10.1111/j.1600-065X.2008.00705.x. [DOI] [PubMed] [Google Scholar]
- 73.McGovern D, Powrie F. 2007. The IL23 axis plays a key role in the pathogenesis of IBD. Gut 56:1333–1336. doi: 10.1136/gut.2006.115402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ahern PP, Schiering C, Buonocore S, McGeachy MJ, Cua DJ, Maloy KJ, Powrie F. 2010. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity 33:279–288. doi: 10.1016/j.immuni.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Powell N, Walker AW, Stolarczyk E, Canavan JB, Gökmen MR, Marks E, Jackson I, Hashim A, Curtis MA, Jenner RG, Howard JK, Parkhill J, MacDonald TT, Lord GM. 2012. The transcription factor T-bet regulates intestinal inflammation mediated by interleukin-7 receptor+ innate lymphoid cells. Immunity 37:674–684. doi: 10.1016/j.immuni.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Buonomo EL, Madan R, Pramoonjago P, Li L, Okusa MD, Petri WA. 2013. Role of interleukin 23 signaling in Clostridium difficile colitis. J Infect Dis 208:917–920. doi: 10.1093/infdis/jit277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Darkoh C, Turnwald BP, Koo HL, Garey KW, Jiang Z-D, Aitken SL, DuPont HL. 2014. Colonic immunopathogenesis of Clostridium difficile infections. Clin Vaccine Immunol 21:509–517. doi: 10.1128/CVI.00770-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McDermott AJ, Falkowski NR, McDonald RA, Pandit CR, Young VB, Huffnagle GB. 2016. Interleukin-23 (IL-23), independent of IL-17 and IL-22, drives neutrophil recruitment and innate inflammation during Clostridium difficile colitis in mice. Immunology 147:114–124. doi: 10.1111/imm.12545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McDermott AJ, Falkowski NR, McDonald RA, Frank CR, Pandit CR, Young VB, Huffnagle GB. 2017. Role of interferon-γ and inflammatory monocytes in driving colonic inflammation during acute Clostridium difficile infection in mice. Immunology 150:468–477. doi: 10.1111/imm.12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cowardin CA, Kuehne SA, Buonomo EL, Marie CS, Minton NP, Petri WA. 2015. Inflammasome activation contributes to interleukin-23 production in response to Clostridium difficile. mBio 6:e02386-14. doi: 10.1128/mBio.02386-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sud V, Abboud A, Tohme S, Vodovotz Y, Simmons RL, Tsung A. 25 June 2018, posting date IL-17A—a regulator in acute inflammation: insights from in vitro, in vivo and in silico studies. Cytokine doi: 10.1016/j.cyto.2018.03.030. [DOI] [PubMed] [Google Scholar]
- 82.Laan M, Cui Z-H, Hoshino H, Lötvall J, Sjöstrand M, Gruenert DC, Skoogh B-E, Lindén A. 1999. Neutrophil recruitment by human IL-17 via CXC chemokine release in the airways. J Immunol 162:2347–2352. [PubMed] [Google Scholar]
- 83.Tateda K, Saji T, Ishii Y, Akasaka Y, Kimura S, Mori N, Kajiwara C, Nakagawa T. 2016. Endogenous IL-17 as a factor determining the severity of Clostridium difficile infection in mice. J Med Microbiol 65:821–827. doi: 10.1099/jmm.0.000273. [DOI] [PubMed] [Google Scholar]
- 84.Wang L, Cao J, Li C, Zhang L. 2018. IL-27/IL-27 receptor signaling provides protection in Clostridium difficile-induced colitis. J Infect Dis 217:198–207. doi: 10.1093/infdis/jix581. [DOI] [PubMed] [Google Scholar]
- 85.Jafari NV, Kuehne SA, Bryant CE, Elawad M, Wren BW, Minton NP, Allan E, Bajaj-Elliott M. 2013. Clostridium difficile modulates host innate immunity via toxin-independent and dependent mechanism(s). PLoS One 8:e69846. doi: 10.1371/journal.pone.0069846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu H, Chen K, Sun Y, Carter M, Garey KW, Savidge TC, Devaraj S, Tessier ME, von Rosenvinge EC, Kelly CP, Pasetti MF, Feng H. 4 August 2017, posting date Cytokines are markers of the Clostridium difficile-induced inflammatory response and predict disease severity. Clin Vaccine Immunol doi: 10.1128/CVI.00037-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iwakura Y, Ishigame H, Saijo S, Nakae S. 2011. Functional specialization of interleukin-17 family members. Immunity 34:149–162. doi: 10.1016/j.immuni.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 88.Matsuzaki G, Umemura M. 2018. Interleukin-17 family cytokines in protective immunity against infections: role of hematopoietic cell-derived and non-hematopoietic cell-derived interleukin-17s: IL-17 family cytokines in infections. Microbiol Immunol 62:1–13. doi: 10.1111/1348-0421.12560. [DOI] [PubMed] [Google Scholar]
- 89.Luo R, Greenberg A, Stone CD. 2015. Outcomes of Clostridium difficile infection in hospitalized leukemia patients: a nationwide analysis. Infect Control Hosp Epidemiol 36:794–801. doi: 10.1017/ice.2015.54. [DOI] [PubMed] [Google Scholar]
- 90.Huang AM, Marini BL, Frame D, Aronoff DM, Nagel JL. 2014. Risk factors for recurrent Clostridium difficile infection in hematopoietic stem cell transplant recipients. Transpl Infect Dis 16:744–750. doi: 10.1111/tid.12267. [DOI] [PubMed] [Google Scholar]
- 91.Jarchum I, Liu M, Shi C, Equinda M, Pamer EG. 2012. Critical role for MyD88-mediated neutrophil recruitment during Clostridium difficile colitis. Infect Immun 80:2989–2996. doi: 10.1128/IAI.00448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hasegawa M, Yamazaki T, Kamada N, Tawaratsumida K, Kim Y-G, Nunez G, Inohara N. 2011. Nucleotide-binding oligomerization domain 1 mediates recognition of Clostridium difficile and induces neutrophil recruitment and protection against the pathogen. J Immunol 186:4872–4880. doi: 10.4049/jimmunol.1003761. [DOI] [PubMed] [Google Scholar]
- 93.Bauer MP, Hensgens MPM, Miller MA, Gerding DN, Wilcox MH, Dale AP, Fawley WN, Kuijper EJ, Gorbach SL. 2012. Renal failure and leukocytosis are predictors of a complicated course of Clostridium difficile infection if measured on day of diagnosis. Clin Infect Dis 55:S149–S153. doi: 10.1093/cid/cis340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Solomon K, Martin AJ, O'Donoghue C, Chen X, Fenelon L, Fanning S, Kelly CP, Kyne L. 2013. Mortality in patients with Clostridium difficile infection correlates with host pro-inflammatory and humoral immune responses. J Med Microbiol 62:1453–1460. doi: 10.1099/jmm.0.058479-0. [DOI] [PubMed] [Google Scholar]
- 95.Jose S, Madan R. 2016. Neutrophil-mediated inflammation in the pathogenesis of Clostridium difficile infections. Anaerobe 41:85–90. doi: 10.1016/j.anaerobe.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ley K, Hoffman HM, Kubes P, Cassatella MA, Zychlinsky A, Hedrick CC, Catz SD. 2018. Neutrophils: new insights and open questions. Science Immunol 3:eaat4579. doi: 10.1126/sciimmunol.aat4579. [DOI] [PubMed] [Google Scholar]