Abstract
In contrast to the continuous increase in survival rates for many cancer entities, colorectal cancer (CRC) and pancreatic cancer are predicted to be ranked among the top 3 cancer-related deaths in the European Union by 2025. Especially, fighting metastasis still constitutes an obstacle to be overcome in CRC and pancreatic cancer. As described by Fearon and Vogelstein, the development of CRC is based on sequential mutations leading to the activation of proto-oncogenes and the inactivation of tumour suppressor genes. In pancreatic cancer, genetic alterations also attribute to tumour development and progression. Recent findings have identified new potentially important transcription factors in CRC, among those the activating transcription factor 2 (ATF2). ATF2 is a basic leucine zipper protein and is involved in physiological and developmental processes, as well as in tumorigenesis. The mutation burden of ATF2 in CRC and pancreatic cancer is rather negligible; however, previous studies in other tumours indicated that ATF2 expression level and subcellular localisation impact tumour progression and patient prognosis. In a tissue- and stimulus-dependent manner, ATF2 is activated by upstream kinases, dimerises and induces target gene expression. Dependent on its dimerisation partner, ATF2 homodimers or heterodimers bind to cAMP-response elements or activator protein 1 consensus motifs. Pioneering work has been performed in melanoma in which the dual role of ATF2 is best understood. Even though there is increasing interest in ATF2 recently, only little is known about its involvement in CRC and pancreatic cancer. In this review, we summarise the current understanding of the underestimated ‘cancer gene chameleon’ ATF2 in apoptosis, epithelial-to-mesenchymal transition and microRNA regulation and highlight its functions in CRC and pancreatic cancer. We further provide a novel ATF2 3D structure with key phosphorylation sites and an updated overview of all so-far available mouse models to study ATF2 in vivo.
Introduction
Cancer genes are historically classified as tumour suppressors that inhibit tumour development and balance proliferation under physiological conditions or as oncogenes that promote tumour initiation, progression and metastasis. In the last 10 years, there is growing evidence that genes might behave as tumour suppressors or oncogenes dependent on the genetic context, the cell type and the experimental stimulus (1). The term ‘cancer gene chameleon’ has been introduced to describe the functional antagonistic duality of such genes.
The activating transcription factor 2 (ATF2) is a member of the activator protein 1 (AP-1) transcription factor family of basic leucine zipper (bZIP) containing DNA-binding proteins. The N-terminal zinc finger region and the transactivation domain enable the transcriptional activity of ATF2, whereas the C-terminus organises its homodimerization and heterodimerization. Partners in heterodimers are JUN, FOS, CREB and MAF leading to the formation of the AP-1 transcription factor complex (2). ATF2 can also work as an epigenetic regulator by being a histone acetyltransferase (HAT) that specifically acetylates histones H2B and H4, triggering its own DNA-binding effectiveness (3). There are several reports about the role of ATF2 as a driver for tumour aggressiveness, whereas ATF2-mediated suppressive effects also are described (4). Its dual role in cancer is majorly dependent on its subcellular localisation (5). ATF2 regulates a plethora of target genes involved in proliferation, transformation, repair, inflammation and apoptosis (6). The protein has many phosphorylation sites that can be activated by different signalling pathways and trigger ATF2 specific functions (7). After cellular stress (e.g. UV radiation, hypoxia or inflammatory cytokines), ATF2 contributes to epithelial-to-mesenchymal transition (EMT), a transformation of epithelial cells into mesenchymal highly migrating cells, enabling tumour invasiveness (8–10). Furthermore, it can act as a DNA damage sensor (11). ATF2 function is best studied in melanoma (12–14). There are several somatic and tissue-specific Atf2 knockout (KO) mouse models for in vivo study of Atf2 function, and Atf2 loss has been mostly associated with lethality or degeneration (15–17). Pioneering work of the group of Ze´ev Ronai has shown that ATF2 loss might have oncogenic or tumour suppressive activity in melanoma development dependent on the mouse model that has been used (5). Thus, ATF2 needs to be more intensively studied to further exploit its potential as a therapeutic target.
The ATF2 cascade
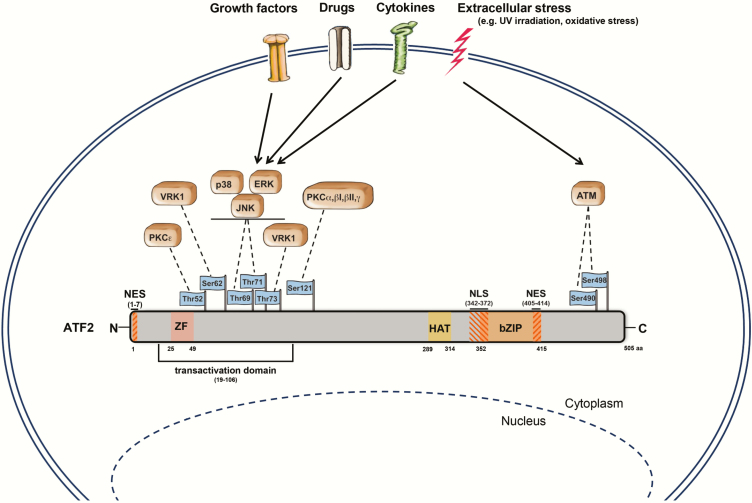
The ATF2 is located on chromosome 2q32 and is translated into a 505 amino acid (aa) large protein (18). ATF2 consists of multiple domains; the most prominent are the N-terminally located transactivation domain (aa 19–106), the zinc finger (ZF, aa 25–49), the bZIP domain (aa 352–415) and the nuclear localisation (aa 342–372) and nuclear export signals (aa 1–7, 405–414) (5,6,19) (Figure 1). Moreover, ATF2 harbours a HAT domain (aa 289–314), rendering it an epigenetic modulator that specifically acetylates histones H2B and H4 in vitro (3).
Figure 1.
Schematic model of ATF2 and its upstream kinases. ATF2 is a transcriptional activator of 505 aa length. This protein includes a ZF domain (aa 25–49), a transactivation domain (aa 19–106), a HAT domain (aa 289–314), a bZIP motif (aa 352–415), nuclear export signals (aa 1–7, aa 405–414) and a nuclear localisation signal (aa 342–372). Dependent on extracellular stress (inflammatory cytokines, oxidative stress, growth factors and UV/ionising irradiation) or drug treatment (e.g. retinoic acid and TPA), various upstream kinases (ATM, ERK, JNK, p38, VRK1 and PKC) phosphorylate ATF2 at its corresponding phosphorylation sites leading to its activation and nuclear translocation. Adapted from Kawasaki et al., Watson et al. and the UniProt database (https://www.uniprot.org/) (3,6,160). Figure was majorly drawn by Joerg Pekarsky (Department of Functional and Clinical Anatomy, Friedrich-Alexander University Erlangen-Nürnberg).
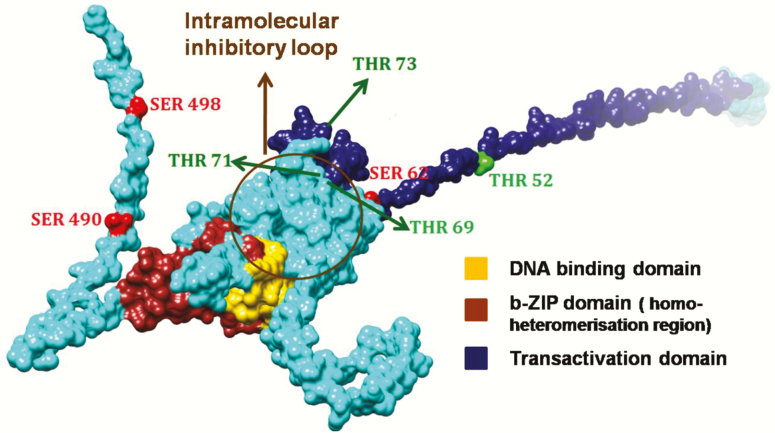
Under physiological conditions, ATF2 shows only low transactivation activity as its bZIP DNA-binding domain (C-terminus) interacts with the N-terminal activation domain forming an intramolecular inhibitory loop (Figure 2) (20). This inhibition is relieved (not depicted) in the presence of activating proteins, such as adenovirus E1A (21,22), hepatitis B virus (HBV) protein X (23) or human T-cell leukemia virus Type-1 (HTLV-I) protein Tax (24) and through phosphorylation at the transactivation domain (20,25,26). Upon activation, ATF2 is translocated into the nucleus where it either forms a homodimer (27) or heterodimer, either with intra-family proteins (ATF3, CRE-BPa or JDP2) or other bZIP proteins (28,29). Dependent on its dimerisation partner, ATF2 can bind to cAMP-response elements (CRE, 5’-TGACGTC-3’) or to the AP-1 binding motif together with c-JUN (TRE, 5’-TGAGTCA-3’) (30).
Figure 2.
Predicted structure model of ATF2 highlighting those phosphorylation sites discussed in this review. The phosphorylation sites of ATF2 are highlighted dependent on its residues: Thr residues (Thr52, Thr69, Thr71 and Thr73) were visualised in green, Ser residues (Ser62, Ser490 and Ser498) in red. Since the 3D structure of the entire ATF2 protein is not known, the structure of ATF2 was modelled using a combination of homology modelling and fold recognition approaches. The Nuclear magnetic resonance structure covering aa 19–56, a part of the transactivation domain (PDB ID: 1BH1A), the X-ray structures covering aa 354–414 (1T2K) were applied for homology remodelling using MODELLER 9v3 (161,162). The residual amino acids of the 505 aa protein were modelled using MUSTER (163). The best of 10 models was ranked, checked for stereochemistry and correct folding and was finally energy minimised and rendered using CHIMERA (164).
ATF2 and c-JUN form the AP-1 complex
The dimeric AP-1 transcription factor complex regulates stimuli-dependent cellular processes ranging from proliferation to differentiation. AP-1 dimers are composed of two sub-units, either of the bZIP protein family (ATF2, JUN, FOS, MAF, C/EBP) or of non-bZIP members (NF-κB, NFAT, SMAD). Due to the heterogeneity of dimerisation partners, AP-1 dimers bind to a plethora of promoters with either a heptameric TGAGTCA (e.g. JUN:FOS, JUN:JUN) or octameric TG/TACNTCA (e.g. ATF:JUN) binding motif (31).
ATF2 and c-JUN, both constitute bZIP transcription factors that are phosphorylated upon extracellular stimuli and build heterodimers to induce target gene expression. The best-studied and most prominent binding partner of ATF2 is c-JUN, both together forming the AP-1 complex (2,32). Other possible ATF2 binding partners in the AP-1 complex could be JUNB, JUND, CREB1, BRCA1, OCT1, NF1, JDP2, MAFA, PDX1, BETA2, NF-YA or NFAT (6). Apart from binding to CRE sites, the ATF2/c-JUN complex recognises and binds to the octameric AP-1 motif and, consequently, induces target gene expression (30). The formation of ATF2-c-JUN dimers is triggered by phosphorylation mediated by the upstream kinases extracellular signal-regulated kinase (ERK), c-JUN NH2-terminal kinase (JNK) and p38, leading to a stress-response target gene profile (33). JNK is the only mitogen-activated protein kinase (MAPK) that phosphorylates c-JUN (34). In contrast to ATF2, which is ubiquitously and abundantly expressed in a variety of tissues, the expression of c-JUN seems to be rather limited and more complexly regulated (31,35–39). Liu et al. have demonstrated that ATF2 harbours two nuclear localisation signals (NLS) and one nuclear export signal (NES). By stress-induced dimerisation with c-JUN, ATF2 is restrained in the nucleus and reinforces c-JUN gene expression (40).
Upstream signalling
The activation of ATF2 relies on multiple upstream kinases that target specific ATF2 phosphosites, thereby determining its transcriptional outcome and target gene signature (41–43). Figure 1 schematically depicts the most prominent and best-studied ATF2 phosphorylation sites. The various mechanisms of ATF2 upstream regulation and a concomitant wide array of ATF2-dependent target genes are presented in the sections below.
Mitogen-activated protein kinases (Thr69/Thr71)
Activated ATF2 mediates extracellular stimuli by induction of ATF2-dependent target genes. A plethora of stimuli (cytokines, growth factors, genotoxic agents and UV irradiation) activate ATF2 at its N-terminal aa Threonine 69 (Thr69) and 71 (Thr71) via MAPKs (39,44). It was shown that these kinases, such as p38, JNK and ERK, phosphorylate ATF2 at Thr69/Thr71 via a two-step mechanism, requiring Thr71 phosphorylation for subsequent Thr69 phosphorylation (33). This mode of activation, however, seems to be stimulus dependent and it is yet unclear to which extent these MAPKs cooperatively induce ATF2 phosphorylation (41,45). Together with SMAD1, ATF2 phosphorylation at Thr69/Thr71 results in the expression of p57KIP2 and, thereby, induces DNA damage response (46). In addition to Thr69/71, also Serine (Ser) 90 is phosphorylated upon UV radiation without effecting ATF2 transcriptional activity (47). In pancreatic β-cells, it was shown that Ca2+/calmodulin-dependent protein kinase IV (CaMKIV) phosphorylates ATF2 at Thr69, Thr71 and Thr73, independent of JNK and p38 signalling. Activated ATF2 binds to the insulin promoter and regulates its gene expression positively correlating with increasing calcium ion concentration (48).
ATF2 constitutes just one of several ATF family members and, therefore, it is not surprising that it shares significant homology with another ATF transcription factor. ATF7 depicts a major sequence homologue to ATF2, particularly within the transcriptional activation (N-terminus) and the DNA-binding domain (C-terminus) (49,50). Due to alternative splicing, a short variant of ATF7, named ATF7-4, was detected in the cytoplasm that is able to abrogate ATF2 transcriptional activity by inhibiting its phosphorylation at Thr71. Upon stimuli, ATF7-4 is phosphorylated leading to its poly-ubiquitination and 26S-proteasomal degradation. Thereby, an ATF7-4 bound kinase is released to phosphorylate ATF7/ATF2 at Thr53/71 allowing subsequent phosphorylation at Thr51/69 by p38, leading to the activation of ATF7/ATF2 (51).
Protein kinase C (Thr52/Ser121/Ser340/Ser367)
Another upstream kinase contributing to post-transcriptional modifications of ATF2 constitutes protein kinase C (PKC). Dependent on its isoforms, stimulus and cell types, PKC phosphorylates ATF2 at residues Thr52 (52), Ser121 (53), Ser340 or Ser367 (43).
PKCα phosphorylates ATF2 at Ser121 (54) after retinoic acid (RA) treatment leading to enhanced transcription of the ATF2 target gene c-JUN. The phosphorylation at Ser121 is also observed upon stimulation with 12-O-tetradecanoylphorbol-13-acetate (TPA) and induces ATF2 translocation into the nucleus. There, it binds to c-JUN and mediates gene expression of genes harbouring an AP-1 or CRE-like binding sequence (55).
Another PKC isoform, PKCε, mediates dual phosphorylation at Thr69/71. However, as shown in melanoma, these phosphorylated residues did not influence the subcellular localisation of ATF2 (52). In addition, PKCε was reported to phosphorylate ATF2 at Thr52. Previous investigations in squamous carcinomas and melanoma could already link high levels of PKCε to tumour promoting effects (56), whereas inhibition of this isoform sensitises tumour cells to apoptosis induced by TNF-related apoptosis-inducing ligand (TRAIL) (57). Phosphorylation at Thr52 keeps ATF2 in the nucleus where it functions as a classic transcription factor by binding to target genes involved in cell survival (52). However, under genotoxic stress, PKCε-mediated phosphorylation of ATF2 at Thr52 is abrogated and ATF2 is exported from the nucleus to the cytoplasm. There, ATF2 accumulates at the mitochondrial outer membrane and suppresses the formation of the voltage-dependent anion-selective channel protein 1 (VDAC1):hexokinase-1 (HK1) complex. This results in alterations in the mitochondrial membrane permeability and promotes apoptosis via cytochrome c release (58).
Vaccinia-related kinase 1 (Ser62/Thr73)
The human vaccinia-related kinase 1 (VRK1) constitutes a nuclear serine/threonine kinase that is involved in the regulation of p53 and cell division processes (59,60). VRK1 phosphorylates its substrate ATF2 mainly at Thr73 and Ser62 and, thereby, stabilises ATF2 and augments its intracellular levels (42). It was demonstrated that phosphorylation of N-terminal residues of ATF2 by VRK1 can coordinate with post-translational modifications mediated by JNK. This allows distinct ATF2 activation by different cellular stimuli and potentiates its transcriptional activity as phosphorylated ATF2 is prevented from ubiquitination and degradation (61).
Ataxia-telangiectasia mutated kinase (Ser490/Ser498)
The protein kinase ataxia-telangiectasia mutated (ATM) constitutes another kinase capable of phosphorylating ATF2. ATM is part of the PI3K-like protein kinase family and, following DNA double-strand breaks (DSB), phosphorylates its substrates at threonine and serine residues (62,63).
To date, a number of studies already implicated a potential role of ATF2 in DSB repair due to its role on chromatin remodelling and its associations with the HATs TIP49b and TIP60 (64–68). Following ionising radiation (IR), ATM phosphorylates ATF2 at its C-terminal residues Ser490 and Ser498 resulting in a transcriptionally independent DNA damage response. Moreover, ATF2 is then co-localised with γ–H2AX and IR-induced foci (IRIF) (69). Li et al. could confirm the significance of ATF2 as an ATM substrate by generating phospho-mutant mice for Ser490/498 (S472/S480 in mice) that could not be phosphorylated by ATM (11). When exposed to IR, these mice were more sensitive to DNA damage-induced cell death, suffered from multiple organ damage (intestine, liver), had a faster tumour development in p53 KO background and were more susceptible to tumour formation in a skin carcinogenesis model (11).
Epigenetic regulation by ATF2
Colorectal and pancreatic tumours arise due to the accumulation of mutations in driver genes (70,71) as best described for colorectal cancer (CRC) in the model of Fearon and Vogelstein (72). However, both cancer entities also harbour epigenetic imprints, such as DNA methylation of tumour suppressors and microRNAs (miRNAs) dysregulation driving their tumorigenic phenotype (73,74). Epigenetic events lead to altered accessibility of chromatin due to methylation (repressive), phosphorylation (activating) or acetylation (activating) (75). Thus, epigenetic changes constitute the driving force that directs gene activity or repression, leading to cell differentiation without altering the nucleotide sequence (76).
Kawasaki et al. showed that ATF2 also functions as a HAT that specifically acetylates histones H2B and H4 in vitro (3). Further, they revealed that phosphorylation of ATF2 at Thr69/Thr71 not only induced CRE-dependent transcription but also augmented its intrinsic HAT activity. As hypothesised by Kawasaki et al., ATF2 not only possess an intrinsic HAT activity but also associates with the E1A-associated protein p300, a transcriptional co-activator with a HAT domain (3). p300 and ATF2 are both parts of the differentiation regulatory factor (DRF) complex. Binding of p300 leads to acetylation of the ATF2 residues Lysine (Lys) 357 and 374, important residues in the bZIP DNA-binding domain. Since acetylation occurred in the bZIP domain, conformation changes could lead to altered ATF2 DNA-binding capacity and affect its intramolecular inhibition (77).
Epigenetic regulators of the KDM3 family of histone demethylases, i.e. KDM3A, KDM3B and JMJD1C, have been shown to demethylate H3K9me2 and to be associated with aggressive CRC, cancer stem cells and Wnt/β-catenin target gene expression (78). A study by Chen et al. could show that Jumonji domain containing 1C (JMJD1C) was significantly upregulated in CRC and associated with metastasis promoting effects through activation of ATF2 (79). Knockdown of JMJD1C-attenuated ATF2 transcriptional activity by modulating H3K9me2 and led to reduced metastasis. Since overexpression of ATF2 could reverse these effects, the authors postulated an oncogenic role for ATF2 in CRC (79).
ATF2 regulation by miRNAs
ATF2 expression was shown to be regulated by miRNAs (miRs) (Figure 3). miRNAs are small non-coding RNAs that regulate gene expression post-transcriptionally by binding to the 3’ untranslated regions (UTR) of their mRNA targets, leading to mRNA destabilisation or translation repression. Therefore, ordinarily, miRNAs coordinate gene expression negatively (80). In silico, over 200 miRNAs are predicted to target the 3´UTR of the ATF2 gene (miRWalk 3.0, P < 0.05; http://mirwalk.umm.uni-heidelberg.de/). In this review, we describe the ATF2/miRNA interactions that were experimentally demonstrated.
Figure 3.
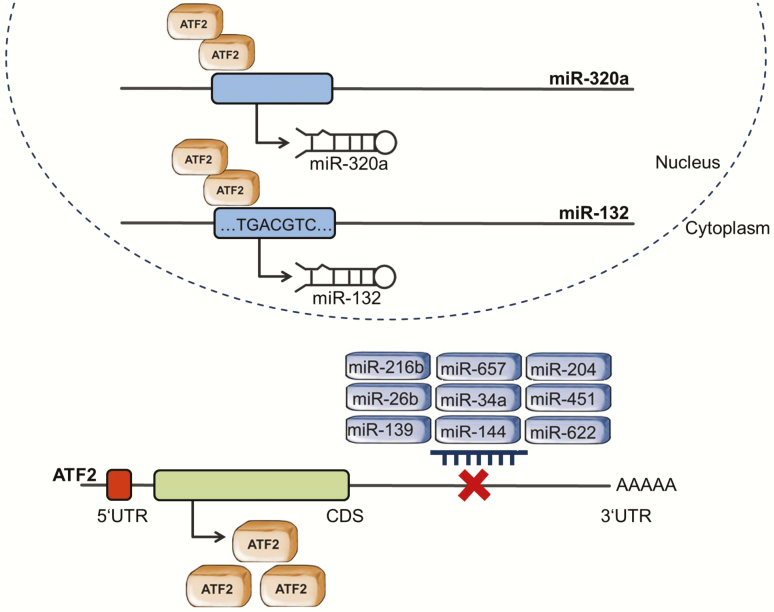
Schematic representation of ATF2 and miRNAs interaction. ATF2 and miRNAs constitute a regulatory network. ATF2 acts as a transcription factor regulating the expression of miRNAs, while its own expression is controlled by miRNAs. Overall, the miRWalk software shows that over 200 miRNAs are predicted to target the ATF2 promotor. Here, we summarise the ATF2/miRNA interactions that were experimentally validated.
ATF2 regulation by miRNAs was reported in several tumour types, such as lung, ovarian and renal cancer. However, until now, this modulation has been poorly studied in pancreatic and CRC. In CRC, Knowlton et al. observed that 13 miRNAs predicted to target ATF2 are dysregulated during cancer progression. miR-20a and miR-20b were upregulated at Stage III samples compared to normal tissue, while others, as miR-26a, miR-26b, miR-452 and miR-520a-3p, were downregulated in the same comparison. Although the authors did not analyse these interactions further, their results suggest that ATF2 expression can be regulated by miRNAs in CRC (81). To the best of our knowledge, the regulation of ATF2 by miRNAs in pancreatic cancer has not yet been reported.
In prostate cancer cells, ATF2 expression was shown to be controlled by miR-204. The authors demonstrated that this miRNA downregulation and ATF2 overexpression led to increased cell proliferation, migration and invasion (82). Another study with prostate and breast cancer cells showed that a miRNA sponge simultaneously inhibited the expression of the known oncogenic miRNAs miR-21, miR-155 and miR-221/-222 and, thereby increased ATF2 expression, a target of miR-21 (83).
Noteworthy, ATF2 expression was reported to be controlled by miRNAs after chemo- and radiotherapy. In different models, it was observed that antitumor treatment induced downregulation of distinct miRNAs leading to an upregulation of ATF2. For instance, in non-small cell lung carcinoma (NSCLC), radiotherapy reduced miR-144 levels in cancer tissue and cell lines. Expression of miR-144 was inversely correlated with ATF2 levels and associated with increased cell resistance. A luciferase assay demonstrated that miR-144 controls ATF2 expression by directly targeting its 3´UTR (84). Also, in NSCLC, cisplatin treatment altered the activation of the ATF2/c-JUN complex. The mentioned therapy reduced the levels of miR-216b, which targets c-JUN. Upregulation of c-JUN was associated with increased formation of the ATF2/c-JUN complex and apoptosis control (85). The activation of the ATF2/c-JUN complex was likewise modulated by miR-139 in ovarian cancer (86).
In comparison to therapy-sensitive cells, osteosarcoma, laryngeal and renal treatment-resistant cancer cells displayed reduced levels of miR-34a, miR-26b and miR-451, respectively. miR-26b and miR-451 were shown to directly regulate ATF2 through binding to its 3´UTR (87,88). On the other hand, miR-34a modulation of ATF2 was shown indirectly as overexpression of this miRNA resulted in increased ATF2 expression (89). Furthermore, miR-451 and miR-26b were also demonstrated to directly regulate ATF2 expression in hepatocellular carcinomas and lung cancer, respectively (90,91). Downregulation of miR-26b in CRC is associated with poor prognosis in patients as a decrease in miR-26b results in upregulation of its target ATF2 (92). Moreover, ATF2 expression is also regulated by miR-657 in myeloma cells after treatment with Spatholobus suberectus Dunn, a plant extract shown to induce apoptosis and cell cycle arrest (93). Therefore, miRNA regulation of ATF2 expression appears to be closely associated with ATF2 therapy-resistance functions. Finally, miR-622 was reported to regulate the expression of ATF2 in glioma cells. Noteworthy, the authors observed that the expression of miR-622 and ATF2 are inversely correlated in glioma specimens obtained from human patients (94).
miRNAs regulated by ATF2
ATF2 was also shown to regulate the expression of miRNAs. The transcription factor was reported to interact with miR-320a promotor and to induce its expression in cervical and colon cancer cells treated with ionising radiation. In the analysed cells, ATF2 cooperated with the ETS transcription factor ELK1 (ELK1) and the YY1 transcription factor (YY1) to activate miR-320a transcription. MiR-320 then directly inhibited X‑linked inhibitor of apoptosis (XIAP) expression, leading to apoptosis and inhibiting cell migration (95). Moreover, in gastric cancer cells, ATF2 can bind to a specific CRE site on miR-132 promotor, inducing its transcription (Figure 3). The overexpression of this miRNA suppressed CD44 and fibronectin 1 (FN1) expression, enabling lymphocytes to home and inducing apoptosis. MiR-132 expression further inhibited tumour cells migration and invasion (95,96).
Downstream targets of ATF2
The transcriptional functions of ATF2 display a broad spectrum of target genes due to its versatile dimerisation options, with c-JUN being the most prominent and best-studied binding partner. As Watson et al. excellently reviewed the transcriptional targets of ATF2, their review should be considered for detailed information on binding partners and corresponding stimuli of ATF2 target genes (6). Briefly, some of the transcriptional targets of ATF2 can be divided into the following categories (with the target genes in brackets): cell cycle (CCNA1, CCND1, GADD45, p21WAF1, RB1 and SERPINB5), immune and inflammatory responses (ELAM1, HBG1, IFNB1, IFNG, IL1B, IL23A, IL6, IL8 and TNFα), expression of AP-1 binding partners (ATF3 and JUN) and apoptosis regulation (ACHE, BCL2L1, CHOP, DP5 and TRAIL). Besides these, ATF2 regulates target gene expression of other intercellular and intracellular pathways, that is, PDGFRA, MMP2, PLAU or HSPA5 (2,5,6).
Interestingly, further ATF2 target genes in CRC and pancreatic cancer were identified. In CRC cells, ATF2 targets in a stimulus-dependent manner the expression of CDK4 (97), ITGA5 (9), MLCK (98), MRP2 (99), TGFB2 (100) and p57KIP2 (101). Gene expression profiling in CRC cells further revealed an ATF2 dependency of the non-canonical Wnt target genes PLOD2, HADH, LCOR and REEP1(102). In pancreatic cells, so far, only insulin was shown to be an ATF2 downstream target under Forskolin or UV treatment (103,104).
ATF2 in apoptosis
In a stimulus-, tissue- and cell type-dependent manner, ATF2 steers a plethora of target genes each contributing to a broad array of functions associated with ATF2. This review focuses on the dual role of ATF2 as tumour suppressor and oncogene and highlights its involvement in apoptosis.
Cells undergoing apoptosis, a programmed cell death, indispensably contribute to the balance of tissue homeostasis. Apoptosis can be induced by intracellular or extracellular events (e.g. oxidative stress and genotoxic agents) (105) and any dysfunctions in its mechanisms lead to imbalance and pathologic conditions (106). Preliminary stages of CRC evade the programmed cell death either by inhibition of apoptosis, by inactivation of apoptosis-inducing or by enhancing apoptosis-repressing genes (105), thereby contributing to the hallmarks of cancer (107). ATF2 as a transcription factor is capable of inducing or inhibiting apoptosis by inducing target gene expression of genes involved in the mechanisms of programmed cell death (e.g. ACHE, BCL2L1 or CHOP) (108–110).
Studies in various cancer entities have shown a dual role for ATF2, either supporting tumour growth and invasion (oncogene) or having tumour suppressive features (tumour suppressor). Studies in melanoma could even demonstrate that within one cancer entity, the function of ATF2 depends on its subcellular localisation: oncogenic ATF2 in the nucleus (where it is transcriptionally active); tumour suppressive ATF2 in the cytoplasm (13,111).
The oncogenic potential of ATF2 was already studied more than 20 years ago in a study conducted by Ronai et al. (112). They could show that ATF2 contributes to radiation resistance of human melanoma cells. However, ATF2 lacking its transactivation domain demonstrated higher sensitivity to irradiation by UV or X-rays. These effects might be attributable to the role of ATF2 in DNA repair (112).
Apart from melanoma, ATF2 was also investigated in pancreatic cancer revealing an upregulation of ATF2 in cancer tissues compared to adjacent non-tumour material (113). Silencing of ATF2 resulted in inhibition of proliferation, induction of apoptosis and rendered tumour cells susceptible to the anti-tumour drug gemcitabine (113). The oncogenic duality of ATF2 was further studied in bladder cancer where ATF2 activation was shown to be dependent on the androgen receptor (AR) with ATF2 activity being positively correlated to AR-induced urothelial tumorigenesis. Androgen was demonstrated to induce phosphorylation of ERK, an upstream kinase of ATF2, leading to the phosphorylation of ATF2 at Thr71 and its nuclear translocation (114). Moreover, Atf2 was shown to be indispensable for mouse skin tumour growth and progression since its downstream targets lead to cell proliferation and tumour progression(115,116).
In contrast, Maekawa et al. demonstrated the role of ATF2 as a tumour suppressor in breast cancer. They showed that a heterozygous loss of Atf2 in mice resulted in the development of mammary tumours after a long latency period majorly caused by a decreased expression of Atf2 target genes Maspin and Gadd45, both involved in the induction of apoptosis (117). In synovial sarcoma, the translocation fusion oncoprotein SS18-SSX serves as a scaffold to link ATF2 and the transcriptional co-repressor transducin-like enhancer of split 1 (TLE1). The presence of SS18-SSX disrupts ATF2 function by transcriptional downregulation of shared ATF2 target genes, whereas restoration of ATF2 and its target genes inhibits tumour growth and induces apoptosis (118).
Contrary to the study by Ronai et al. depicting its oncogenic potential (112), ATF2 also functions as a tumour suppressor in melanoma and its way of function seems to depend on its subcellular localisation. Melanoma cells treated with paclitaxel and vemurafenib demonstrated that these agents induced ATF2 translocation to the mitochondria and an ATF2-mediated release of Bcl-2-interacting mediator of cell death (BIM) and activation of VDAC1. This type of apoptosis is majorly induced by cytochrome c release after drug treatment (119).
Investigations of the role of ATF2 in apoptosis of head and neck squamous cell carcinoma (HNSCC) revealed a TNFα dependency of ATF2 phosphorylation, its translocation to the nucleus and the expression of target genes involved in apoptosis (120). In this study, decreased ATF2 levels did not influence in vivo tumour growth but led to chemo-resistance.
In oesophageal squamous epithelial cancer cells, ATF2 controls and induces the cell cycle regulator p21WAF1, and oxidative stress together with silencing of ATF2 decreased p21WAF1 expression and enhanced apoptosis (121).
A study using deoxycholic acid (DCA) treatment on rat pancreatic acinar cell line AR42J revealed inhibition of proliferation and an induction of apoptosis by targeting ATF2 (122). Moreover, phosphodiesterase-4 inhibitors (PDE4is) that have been previously used in respiratory diseases showed remarkable cytotoxic effects on pancreatic cancer cells. This PDE4is-induced apoptosis is mediated by ATF2 phosphorylation via p38 (123).
ATF2 not only seems to be involved in apoptosis in epithelial-derived cancers but also acts as a regulator of apoptosis in leukaemia. An et al. revealed that the anti-tumour drug selenite inhibits cancer cell growth by disruption of the JNK/ATF2 axis and the downregulation of ATF2-dependent cell cycle target genes (124).
Due to its omnipresent role in apoptosis, ATF2 might be of interest in drug design in clinical studies. Promising data on ATF2-derived peptides showed that silencing of transcriptional activity of ATF2 and induction of c-JUN activity inhibits melanoma growth and metastasis (125–127).
ATF2 and EMT
The EMT describes complex alterations in differentiation with a switch from an epithelial to a more invasive, mesenchymal phenotype that occurs during embryogenesis and tissue repair, as well as in tumour progression (128). The process of tumour progression is based on tumour cells gaining the ability to detach from the parental tumour mass, to migrate and invade surrounding tissues or basement membranes, to disseminate through the vascular system and to finally reattach at distant organs (metastasis) (129). Concomitantly to tumour progression, dedifferentiation of tumour cells at the invasive front (the area into which neoplastic tumour cells invade) is accompanied by distinct morphological changes. These altered phenotypic patterns resemble those of cells undergoing EMT: EMT cells show disrupted intercellular junctions and changed cell polarity and acquire a migratory and invasive phenotype (130,131).
Mesenchymal-like colorectal tumour cells that overexpress the EMT transcription factor TWIST1 showed an upregulation of AP-1, the complex between the key components c-JUN and ATF2 (9). TWIST1-induced cell invasion was triggered by AP-1 dependent integrin α5 induction, which plays a major role in EMT. ATF2 loss has been found together with high TWIST1 and low E-Cadherin expression in early disseminating aggressive Her2+ breast cancer cells (10).
Interestingly, EMT cells were characterised by a predominantly nuclear ATF2 expression pattern, which is associated with an oncogenic function of ATF2. In pancreatic tumour cells, the repressor of the AP-1 complex, Jun dimerization protein 2 (JDP2), was shown to inhibit EMT (132). Downregulation of JDP2 was associated with tumour metastasis in pancreatic cancer cells (133). In parallel, treatment with a p38 MAPK inhibitor resulted in a decreased TGFβ-mediated EMT via decreased p-ATF2 levels (8).
ATF2 in inflammation and metabolism
There is accumulating evidence that ATF2 is involved in inflammation (134). ATF2 expression has been shown to be triggered by hypoxia, reactive oxygen species and inflammatory cytokines majorly through phosphorylation on Thr69 and Thr71 residues. Phosphorylation at Thr69/71 by ERK, JNK and p38 MAPKs leads to nuclear accumulation of ATF2 (135). They reported that activated ATF2 is participating in microglia-mediated neuro-inflammation. It also triggers peripheral inflammation-associated diseases, such as arthritis or hepatitis (136). In Atf2 KO mice, there was a reduced level of inflammatory cytokines, such as IL6, IL1B or TNFα after stimulation with lipopolysaccharides (137). Vice versa, in lung cancer, IL8-mediated ATF2 activation promotes tumour invasiveness (138). By inhibiting the AP-1 transcriptional activity, the anti-inflammatory flavonoid Baicalein also showed a potent anti-angiogenic effect (139). Treatment of CRC cells with tolfenamic acid (TA), a non-steroidal anti-inflammatory drug with anti-cancer potential, revealed an ATF2-mediated induction of ATF3 expression. Upon TA treatment, ATF2 was phosphorylated by its upstream MAP kinases (JNK, ERK and p38) and, thereby, induced ATF3 transcription with ATF3 finally inducing apoptosis (140).
Vice versa, ATF2 itself can actively attenuate inflammation. In late-stage human melanoma LU1205 cells, ATF2 transcriptionally suppresses TNFα, leading to an Fas-mediated programmed cell death upon UVC radiation (141).
Furthermore, ATF2 seems to play a role in impaired glucose homeostasis and development of diabetes. ß-cells in the pancreatic islets of Langerhans produce and secrete insulin, thus maintaining glucose homeostasis. In pancreatic tumour cells, ATF2 interacts with chromatin-associated fumarase to provide a survival signal under glucose deprivation (142). Importantly, ATF2 is a necessary transactivator of insulin gene expression. Furthermore, it has been shown to contribute to diabetes by inducing abnormal upregulation of TNFα or IL1B expression in islet ß-cells (143,144).
ATF2 and prognosis
For some tumour entities, ATF2 expression could already be associated with patient prognosis. High expression of ATF2 correlated with aggressive clinic-pathological characteristics and predicted poor prognosis of renal cell carcinoma (RCC) patients (145). In RCC cells, ATF2 loss attenuated migratory capabilities and suppressed tumour invasion mostly by stabilising the epithelial phenotype (145). In NSCLC, high ATF2 levels were significantly associated with worse prognosis (146) and, recently, Zhou et al. demonstrated that overexpression of ARHGEF39 led to increased ATF2 levels by activating the Rac1-p38-ATF2 signalling pathway promoting tumour invasion and proliferation (147). Similarly, Zheng et al. showed that ATF2 activation triggers cell proliferation and lymph node metastasis in NSCLC (148). In extramammary Paget’s disease (EMPD), a rare malignancy of the skin, it was shown that p-ATF2 and p-STAT3 are both overexpressed and associated with the tumour stage: both markers displayed the highest expression levels in advanced EMPD (149).
A study in breast cancer demonstrated a tumour suppressive role of ATF2 as its loss was associated with enhanced anchorage-independent growth, downregulation of cell cycle and apoptosis regulating proteins and upregulation of EMT promoting proteins. Further, the authors could show that ATF2 was essential for the growth-inhibitory effects of tamoxifen in ER-positive cells and that phosphorylation of ATF2 at Thr71 could serve as a prognostic and predictive marker for response to tamoxifen (150).
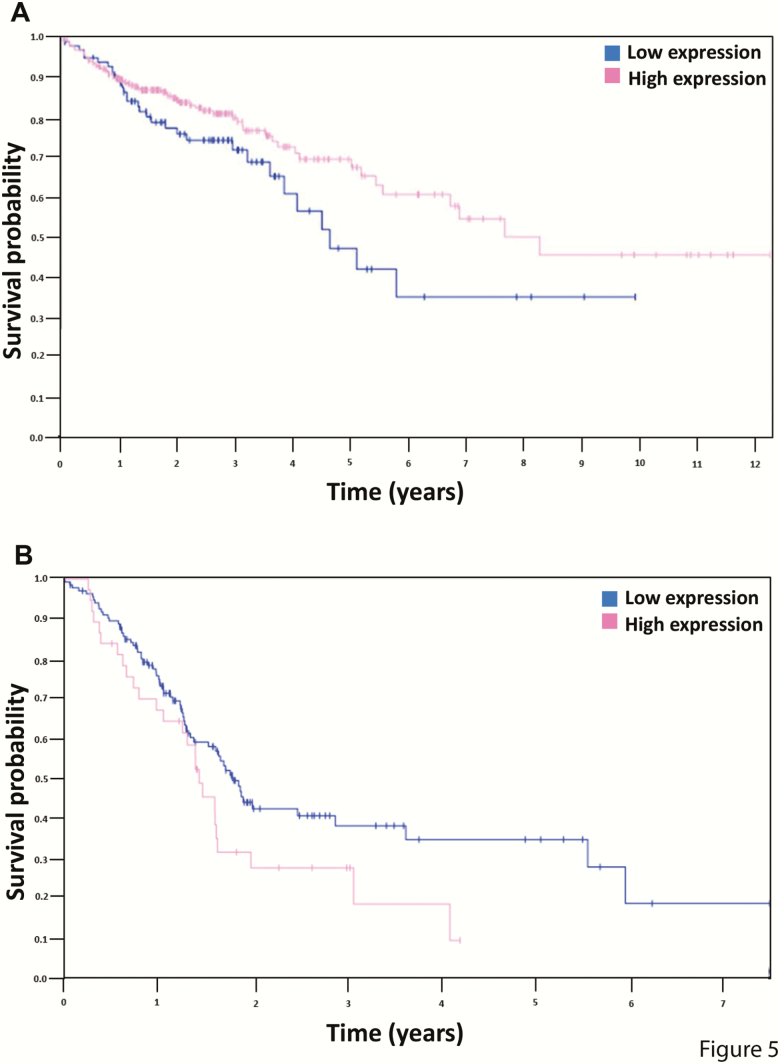
Although Pradhan et al. identified, in a global systems biology approach, ATF2 as an important transcription factor in CRC (151), only little is known about the role of ATF2 in CRC or pancreatic carcinogenesis in different stages of the disease or in chemo-resistance. Interestingly, in silico The Cancer Genome Atlas (TCGA) analysis (https://www.cbioportal.org/; Table 2) showed that mutations in the ATF2 gene seem to be negligibly low in CRC (n = 3504) and pancreatic cancer samples (n = 1057), with a somatic mutation frequency of 0.4% and 0.5%, respectively (Figure 4) (152,153). Although the literature has already proven the divergent role of ATF2 in cancer, it becomes more obvious when considering the ATF2 expression levels in CRC and pancreatic cancer. Analysis of patient survival from TCGA RNA-seq data showed in CRC (n = 438) that low ATF2 expression characterises a high-risk subgroup of patients (Figure 5A), whereas, in pancreatic cancer (n = 176), the situation is reversed with low ATF2 being associated with better patient survival (Figure 5B). Though these data sets revealed only marginally significant differences for low and high ATF2-expressing tumours, the prognostic value in CRC and pancreatic cancer seemed to be remarkably reversed.
Table 2.
Sequencing studies obtained from cBioPortal for CRC and pancreatic cancer to investigate the ATF2 mutation spectrum
| Study number | Cancer type | Study details | Number of samples | References |
|---|---|---|---|---|
| 1 | CRC | Whole-exome sequencing of 619 CRCs with clinicopathologic annotations | 619 | Giannakis et al. (165) |
| 2 | Colon cancer | Comprehensive profiling of 74 paired colon tumour-normal samples | 74 | Seshagiri et al. (166) |
| 3 | CRC | Whole-exome sequencing in 224 of the 276 colorectal carcinoma tumour/normal pairs | 276 | Cancer Genome Atlas Network (167) |
| 4 | CRC | Colorectal Adenocarcinoma TCGA Pan Cancer data | 594 | Liu et al. (168) |
| 5 | CRC | TCGA Colorectal Adenocarcinoma; raw data at the NCI | 640 | NA |
| 6 | CRC | Targeted sequencing of primary, metastatic and normal tissues from 69 colorectal adenocarcinoma patients | 138 | Brannon et al. (169) |
| 7 | CRC | Targeted sequencing of 1134 metastatic colorectal tumour/normal pairs | 1134 | Yaeger et al. (170) |
| 8 | CRC | Whole-exome sequencing of tumour/normal pairs from 29 African American colon cancers | 29 | Guda et al. (171) |
| 9 | Pancreatic cancer | Whole-exome sequencing of 23 surgically resected pancreatic carcinomas with acinar differentiation and their matched normals | 23 | Jiao et al. (172) |
| 10 | Pancreatic cancer | Whole-exome sequencing of 99 pancreatic samples and their matched normals | 99 | Biankin et al. (173) |
| 11 | Pancreatic cancer | Whole-genome and deep-exome sequencing analysis of 456 pancreatic ductal adenocarcinomas | 456 | Bailey et al. (174) |
| 12 | Pancreatic cancer | Pancreatic Adenocarcinoma TCGA PanCancer data. | 184 | Hoadley et al. (175) |
| 13 | Pancreatic cancer | TCGA Pancreatic Adenocarcinoma; raw data at the NCI | 186 | NA |
| 14 | Pancreatic cancer | Whole-exome sequencing of 109 micro-dissected pancreatic cancer cases and normal control tissue | 109 | Witkiewicz et al. (176) |
Figure 4.
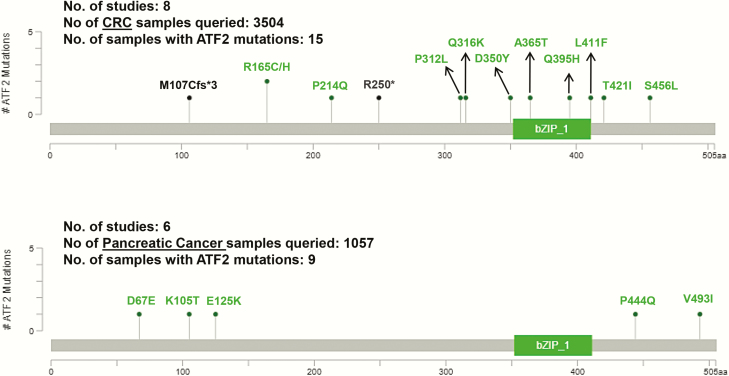
ATF2 mutation spectrum in colorectal and pancreatic cancer. In both CRC and pancreatic cancer, the occurrence of ATF2 mutations is negligibly low. Genomics data from 3504 samples (eight studies) in different stages of colorectal adenocarcinoma and CRC were queried from the cBioPortal; 13 tumours showed missense mutations (green colour) and 2 tumours had truncating mutations (black colour) in ATF2, which are depicted in the upper panel. Out of the 1057 pancreatic cancer samples queried (5 studies), 9 pancreatic tumours were found to have only missense mutations (green) in ATF2 which are depicted in the lower panel. Adapted from cBioportal, https://www.cbioportal.org/ (152,153).
Figure 5.
Kaplan–Meier plots of survival probability according to ATF2 expression in colon adenocarcinoma (A) and pancreatic cancer (B). (A) In colon adenocarcinomas (n = 438), the 5-year survival rate with high ATF2 expression was 69% (cut-off = 5.54, P-value = 0.06), whereas, in (B) pancreatic cancer (n = 176), the 5-year survival rate with high ATF2 expression was at 0% (cut-off = 10.33, P-value = 0.072). Based on the fragments per kilobase million (FPKM) value of each gene, patients were classified into two groups and the association between prognosis (survival) and gene expression (FPKM) was examined. The best expression cut-off refers the FPKM value that yields maximal difference with regard to survival between the two groups at the lowest log-rank P-value. Best expression cut-off was selected based on survival analysis. Image credit: adapted from The Human Protein Atlas, version 18.1 (www.proteinatlas.org, https://www.proteinatlas.org/ENSG00000115966-ATF2/pathology/tissue/colorectal+cancer/COAD, https://www.proteinatlas.org/ENSG00000115966-ATF2/pathology/tissue/pancreatic+cancer).
De Robertis et al. demonstrated that, in murine EphA2high cell populations of adenocarcinoma, ATF2 was significantly upregulated. Deregulation ATF2 was supposed as a novel prognostic and predictive target being associated with worse disease-free survival (DFS) in EphA2high patients already validated as an independent prognostic marker in Stage I–III CRC patients(92).
To our knowledge, the variable prognostic value of ATF2 has not yet been connected to any cause, but it might be reasonable that the acquisition of tumour samples (tumour centre and invasion front) and the general tumour heterogeneity might influence the overall relevance of ATF2 expression in cancer prognosis.
Mouse model to study ATF2 function
The Atf2 function has been assessed in multiple mouse models (Table 1). The first attempt to study Atf2 in mouse models produced hypomorphic variants by Atf2 deletion (15). The hypomorphic allele showed a severe phenotype in bone and cartilage development, resulting in hypochondrodysplasia and also neurological changes in the brain. Such findings shed first light on the importance of Atf2 in the regulation of development and physiology; however, the global view was limited by the fact that the allele still produced a partly functional protein. The approach to knock out Atf2 from the mouse genome resulted in more severe and lethal phenotype immediately after birth with 100% penetrance. The aspirated meconium was identified as the major cause of perinatal death in Atf2 KO pups (16).
Table 1.
ATF2-related mouse models
| Line | Principal investigator | Allele form | Repository | Phenotype | References |
|---|---|---|---|---|---|
| Atf2tm1Sis | Shunsuke Ishii | KO | Rikken | Meconium aspiration syndrome | Maekawa et al. (16) |
| Atf2tm1Glm | Laurie Glimcher | Hypomorphic | JAX | Hypochondroplasia, neurological abnormalities | Reimold et al. (15) |
| Atf2tm1a(EUCOMM)Hmgu | IMPC | KO first/cKO potential | EMMA | Under investigation | |
| Atf2tm1b(EUCOMM)Hmgu | IMPC | KO | EMMA | Under investigation | |
| Atf2tm1(T51A,T53A) | Nic Jonesa | Mutant | NA | Lethal with Atf7 KO | Breitwieser et al. (154) |
| Atf2tm1(S472A,S480A) | Ze’ev A. Ronaia | Mutant | NA | DNA damage response | Li et al. (11) |
| Atf2 Δ8,9 | Ze’ev A. Ronaia | Transcriptional inactive | NA | Shah et al. (13) | |
| Atf2flox/flox | Ze’ev A. Ronaia | Conditional KO | NA | Shah et al. (13) | |
| Atf2flox/flox;TyrcreERT2 | Ze’ev A. Ronaia | Melanocyte deleted | NA | Shah et al. (13) |
IMSR: International Mouse Strain Resource; IMPC: International Mouse Phenotyping Consortium.
aCorresponding author—not registered at IMSR.
The role of ATF2 in carcinogenesis has been suggested in multiple tumour models and also from patient data; however, severe lethal phenotypes hamper the study of tumorigenesis in Atf2 KO animals and, thus, alternative gene-targeting strategies were introduced. One approach was to study the biological function of Atf2 in living organisms by the production of mice with targeting functional domains and substitution mutations in critical sites for particular protein functions. In this case, two mouse models were designed. The first aimed to target important phosphorylation sites on the N-terminus of Atf2 in exon 3, Thr51 and Thr53 both substituted by alanine. This mouse model showed the same subset of perinatal lethal phenotype as previously published for Atf2 KO mice (16); however, if crossed to Atf7 KO, an even more severe embryonic lethal phenotype showed up, suggesting that Atf7, as a close homolog of Atf2, could compensate the Atf2 function related to these particular regulatory domains (154). Due to the lethal phenotype, this model is not suitable to study tumorigenesis in adult mice. Another substitution mouse mutant has been created in order to block phosphorylation sites close to the C-terminus in Ser472 and Ser480. Interestingly, such Atf2 mutant mouse does not promote any lethal phenotype as all mice are well viable. However, Atf2(S472A,S480A) mice have insufficient DNA damage response and are prone to spontaneous tumour formation. Worth mentioning is that ATF2 phosphorylation by ATM after DNA damage response is a p53-independent impaired expression of the cell cycle inhibitor p21, which in turn hampers cell cycle arrest in Atf2(S472A,S480A) cells after DNA damage. Interestingly, simultaneous mutation in p53 makes tumour formation more abundant than in single-mutated Atf2(S472A,S480A) or p53 KO mice (11). When the Atf2(S472A,S480A)-mutated mice were exposed to two-stage skin carcinogenesis protocols by 7,12-dimethylbenz(a)anthracene treatment (tumour inducer), the initiation dynamics and tumour number were higher than in control mice, suggesting that mutation in Atf2 is triggering the tumorigenic process (11).
A very specific problem arises when Atf2 function needs to be studied in specific tumour types since most of the targeted mutations are lethal just after birth or the ubiquitous expression of the mutated allele causes spontaneous tumour formation without control of any tissue specificity. In order to solve this problem and to study Atf2 effects in specific tissues or cell populations, the conditional allele, where the DNA-binding domain is flanked by LoxP sites, was produced (13). Inducible KO strategy majorly solved the issue with embryonic lethality and helped to establish genetic models to study the regulatory role of Atf2 in melanoma (13). The melanoma model was based on a NrasQ61K gain of function mutation on the Ink4a KO mouse genetic background. In this model, spontaneous melanoma appears in 30% of mice. The tissue specificity for Atf2 KO in melanocytes was achieved by using combination of conditional KO allele of Atf2, where the sequence for the DNA-binding domain is flanked by two LoxP sites and can be removed by Cre recombinase. As a Cre driver, the TyrCreERT2 (melanocyte-specific tyrosinase promoter-driven inducible Cre expression) was used. The Cre must be activated by hydroxyl-tamoxifen treatment, which allows precise temporal–spatial gene induction in the skin in a well-regulated time window (13). Remarkably, mutation of Atf2 in melanocytes had negative effects on melanoma promotion and only 5% of mice were diagnosed with melanoma compared to controls where 30% of mice were affected (13). There is a striking discrepancy from other types of tumours, where Atf2 mutations lead to cancer promotion and more severe tumour phenotypes (14,155,156). The Atf2 regulatory network in melanocytes relies on the negative regulation of expression of microphthalmia-associated transcription factor (MITF), which is responsible for the early phase of melanocyte transformation(13). However, a later mouse model of melanoma based on a different genetic background (BrafV600E/V600E; Pten–/–) showed opposite effects and the transcriptionally inactive form of Atf2Δ8,9 resulted in potentiation of melanoma formation and metastasis development (14). Remarkably, the Atf2 protein from Atf2Δ8,9 allele is in reading frame with all properties of the wild-type form with just an exception in the DNA-binding domain. A similar endogenous splicing variant, Atf2SV5, is also abundantly found in human melanomas. This might implicate that, beside transcription regulatory function, there is also a complex interacting network that, in non-DNA-binding mode, can cause readjustment of differentiation programs in (pro-) cancerogenic cells and enhance transformation capacity and tumour invasiveness even further (14).
From multiple melanoma models and also when compared to other tumour models, it is obvious that Atf2 itself can have dual effects on tumour development and invasiveness. The specificity of Atf2 action can be attributed to tissue-specific features and/or to genetic programs critical in carcinogenesis.
Xenograft models to study ATF2 in melanoma development
In order to study the role of ATF2 in melanoma development, xenograft models have been used with multiple mouse and human-derived melanoma cell lines. Their advantage is that cells can be genetically manipulated without genetic influence on the host. However, disadvantages might be that mouse cell lines used for isograft approaches might have different molecular signatures than patient-derived cells and vice versa, and the patient-derived cells must be grafted as xenografts into mice with compromised immune system. Despite all these obstacles, melanoma cell line grafts provided valuable molecular insights into the role of ATF2 in regulation of melanoma development and invasiveness. It has been shown that ATF2-mediated inhibition of Sox10 expression caused downregulation of MITF, which in turn led to a pronounced transformation phenotype of melanoma cells. This was supported by the fact that inhibition of ATF2 expression was able to reverse that effect (13). The role of PKCε on tumorigenic activity of ATF2 has been approached in isograft models showing that the ATF2 downstream cascade negatively influences fucosylation of the proteome in melanoma cells. Interestingly, the decrease of general protein fucosylation levels led to loss of cell adhesion and promotion of invasive behaviour. Moreover, if fucosylation was genetically restored in tumour cells, the invasive character of melanoma cells was decreased together with infiltration of primary tumours with natural killer cells (12).
The molecular pathways behind the diverse functions of ATF2 have mainly been discovered in melanoma models and this brought up new strategies on how to attenuate ATF2 effects in melanoma progression. Peptides derived from the ATF2 protein have been used in screenings to discover possible new pharmaceutical tools to target ATF2-dependent effects in melanoma cell invasiveness. The N-terminal peptide spanning aa 50–100 of ATF2 showed an active role in the sensitisation of melanoma cells to treatment and shifted cells into apoptosis (127). The molecular mechanism of the ATF2-derived peptide was identified as a simultaneous reduction of ATF2 transcription activity together with an enhancement of the JNK/JUN and JUND signalling pathway (126). The effect of the peptide was observed in sequestration of ATF2 from the nucleus into the cytosol, which overall resulted in inhibition of ATF2-dependent tumorigenic potential (157).
Xenografts also allowed the identification of novel drugs interfering with ATF2 biological functions. The pharmaceutical mimicking effect of the anti-oncogenic character of ATF2-derived peptides was identified in celastrol (CSL) and acetyl isogambogic acid compounds and constituted promising pharmacological tools to influence melanoma progression (158). The MAPK pathway activation with the small molecule MC3181 resulted in strong phosphorylation and activation of JNK pathways and triggered pro-apoptotic behaviour of ATF2 (159).
All models combining genetically modified mouse models, isografts of modified melanoma cells or patient-derived tumour cell lines support the view on ATF2 as an important tissue contextual regulator of melanoma development and invasiveness.
Outlook
It is worth analysing the prognostic value of ATF2 in different tumour types dependent on the stage and histological subtype. Historically, the divergence of ATF2 signalling has been first described in melanoma and the function of ATF2 is best understood in this tumour entity. Nevertheless, TCGA studies have shown a potential prognostic role of ATF2 also in CRC and pancreatic cancer. Thus, we hope that this review will motivate the research community to analyse the role of ATF2 also in other tumour types. The availability of a large range of Atf2 mutant mouse strains could be a suitable source for mechanistic functional studies. As a central link between DNA damage and MAPK signalling, ATF2 could be a potential therapeutic target. Due to its multiple regulation levels together with its action as a tumour suppressor or an oncogene, the therapeutic approach should be carefully determined in a tumour type-specific manner.
Funding
This article is partly based upon work from COST Action CA17118, supported by the European Cooperation in Science and Technology (COST; www.cost.eu). K.H., J.P. and R.S.S. were supported by grants from the Bavarian-Czech University Agency (BTHA-JC-2019-1) and the Manfred-Stolte-Stiftung (Bayreuth, Germany). K.H. was sponsored by a grant for the realization of equal opportunities for women in research and teaching by Bavarian state loans for the promotion of equality and by a grant of the German Academic Exchange Service. J.P. was supported by RVO 68378050 by the Academy of Sciences of the Czech Republic; LM2015040 Czech Centre for Phenogenomics by the Ministry of Education, Youth and Sports; CZ.02.1.01/0.0/0.0/16_013/0001789 Upgrade of the Czech Centre for Phenogenomics: developing towards translation research by the Ministry of Education, Youth and Sports and Education Research and Development Foundation; Z.1.05/1.1.00/02.0109 Biotechnology and Biomedicine Centre of the Academy of Sciences and Charles University in Vestec; CZ.1.05/2.1.00/19.0395 Higher quality and capacity for transgenic models by Ministry of Education, Youth and Sports and Education Research and Development Foundation.
References
- 1. Stepanenko A. A., Vassetzky Y. S. and Kavsan V. M (2013) Antagonistic functional duality of cancer genes. Gene, 529, 199–207. [DOI] [PubMed] [Google Scholar]
- 2. Lopez-Bergami P., Lau E. and Ronai Z (2010) Emerging roles of ATF2 and the dynamic AP1 network in cancer. Nat. Rev. Cancer, 10, 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kawasaki H., Schiltz L., Chiu R., Itakura K., Taira K., Nakatani Y. and Yokoyama K. K (2000) ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature, 405, 195–200. [DOI] [PubMed] [Google Scholar]
- 4. Bhoumik A. and Ronai Z (2008) ATF2: a transcription factor that elicits oncogenic or tumor suppressor activities. Cell Cycle, 7, 2341–2345. [DOI] [PubMed] [Google Scholar]
- 5. Lau E. and Ronai Z. A (2012) ATF2 - at the crossroad of nuclear and cytosolic functions. J. Cell Sci., 125, 2815–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watson G., Ronai Z. A. and Lau E (2017) ATF2, a paradigm of the multifaceted regulation of transcription factors in biology and disease. Pharmacol. Res., 119, 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gozdecka M. and Breitwieser W (2012) The roles of ATF2 (activating transcription factor 2) in tumorigenesis. Biochem. Soc. Trans., 40, 230–234. [DOI] [PubMed] [Google Scholar]
- 8. Bakin A. V., Rinehart C., Tomlinson A. K. and Arteaga C. L (2002) p38 mitogen-activated protein kinase is required for TGFbeta-mediated fibroblastic transdifferentiation and cell migration. J. Cell Sci., 115, 3193–3206. [DOI] [PubMed] [Google Scholar]
- 9. Nam E. H., Lee Y., Moon B., Lee J. W. and Kim S (2015) Twist1 and AP-1 cooperatively upregulate integrin α5 expression to induce invasion and the epithelial-mesenchymal transition. Carcinogenesis, 36, 327–337. [DOI] [PubMed] [Google Scholar]
- 10. Harper K. L., Sosa M. S., Entenberg D., et al. (2016) Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature, 540, 588–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li S., Ezhevsky S., Dewing A., et al. (2010) Radiation sensitivity and tumor susceptibility in ATM phospho-mutant ATF2 mice. Genes Cancer, 1, 316–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lau E., Feng Y., Claps G., et al. (2015) The transcription factor ATF2 promotes melanoma metastasis by suppressing protein fucosylation. Sci. Signal., 8, ra124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shah M., Bhoumik A., Goel V., et al. (2010) A role for ATF2 in regulating MITF and melanoma development. PLoS Genet., 6, e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Claps G., Cheli Y., Zhang T., et al. (2016) A transcriptionally inactive ATF2 variant drives melanomagenesis. Cell Rep., 15, 1884–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reimold A. M., Grusby M. J., Kosaras B., et al. (1996) Chondrodysplasia and neurological abnormalities in ATF-2-deficient mice. Nature, 379, 262–265. [DOI] [PubMed] [Google Scholar]
- 16. Maekawa T., Bernier F., Sato M., et al. (1999) Mouse ATF-2 null mutants display features of a severe type of meconium aspiration syndrome. J. Biol. Chem., 274, 17813–17819. [DOI] [PubMed] [Google Scholar]
- 17. Ackermann J., Ashton G., Lyons S., James D., Hornung J. P., Jones N. and Breitwieser W (2011) Loss of ATF2 function leads to cranial motoneuron degeneration during embryonic mouse development. PLoS One, 6, e19090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ozawa K., Sudo T., Soeda E., Yoshida M. C. and Ishii S (1991) Assignment of the human CREB2 (CRE-BP1) gene to 2q32. Genomics, 10, 1103–1104. [DOI] [PubMed] [Google Scholar]
- 19. Nagadoi A., Nakazawa K., Uda H., Okuno K., Maekawa T., Ishii S. and Nishimura Y (1999) Solution structure of the transactivation domain of ATF-2 comprising a zinc finger-like subdomain and a flexible subdomain. J. Mol. Biol., 287, 593–607. [DOI] [PubMed] [Google Scholar]
- 20. Li X. Y. and Green M. R (1996) Intramolecular inhibition of activating transcription factor-2 function by its DNA-binding domain. Genes Dev., 10, 517–527. [DOI] [PubMed] [Google Scholar]
- 21. Liu F. and Green M. R (1990) A specific member of the ATF transcription factor family can mediate transcription activation by the adenovirus E1a protein. Cell, 61, 1217–1224. [DOI] [PubMed] [Google Scholar]
- 22. Zu Y. L., Takamatsu Y., Zhao M. J., Maekawa T., Handa H. and Ishii S (1992) Transcriptional regulation by a point mutant of adenovirus-2 E1a product lacking DNA binding activity. J. Biol. Chem., 267, 20181–20187. [PubMed] [Google Scholar]
- 23. Maguire H. F., Hoeffler J. P. and Siddiqui A (1991) HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science, 252, 842–844. [DOI] [PubMed] [Google Scholar]
- 24. Wagner S. and Green M. R (1993) HTLV-I Tax protein stimulation of DNA binding of bZIP proteins by enhancing dimerization. Science, 262, 395–399. [DOI] [PubMed] [Google Scholar]
- 25. Livingstone C., Patel G. and Jones N (1995) ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J., 14, 1785–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sano Y., Tokitou F., Dai P., Maekawa T., Yamamoto T. and Ishii S (1998) CBP alleviates the intramolecular inhibition of ATF-2 function. J. Biol. Chem., 273, 29098–29105. [DOI] [PubMed] [Google Scholar]
- 27. Hai T. W., Liu F., Coukos W. J. and Green M. R (1989) Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev., 3, 2083–2090. [DOI] [PubMed] [Google Scholar]
- 28. Chatton B., Bocco J. L., Goetz J., Gaire M., Lutz Y. and Kedinger C (1994) Jun and Fos heterodimerize with ATFa, a member of the ATF/CREB family and modulate its transcriptional activity. Oncogene, 9, 375–385. [PubMed] [Google Scholar]
- 29. Vlahopoulos S. A., Logotheti S., Mikas D., Giarika A., Gorgoulis V. and Zoumpourlis V (2008) The role of ATF-2 in oncogenesis. Bioessays, 30, 314–327. [DOI] [PubMed] [Google Scholar]
- 30. Hai T. and Curran T (1991) Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc. Natl. Acad. Sci. USA, 88, 3720–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van Dam H. and Castellazzi M (2001) Distinct roles of Jun: Fos and Jun: ATF dimers in oncogenesis. Oncogene, 20, 2453–2464. [DOI] [PubMed] [Google Scholar]
- 32. Hayakawa J., Mittal S., Wang Y., et al. (2004) Identification of promoters bound by c-Jun/ATF2 during rapid large-scale gene activation following genotoxic stress. Mol. Cell, 16, 521–535. [DOI] [PubMed] [Google Scholar]
- 33. Ouwens D. M., de Ruiter N. D., van der Zon G. C., et al. (2002) Growth factors can activate ATF2 via a two-step mechanism: phosphorylation of Thr71 through the Ras-MEK-ERK pathway and of Thr69 through RalGDS-Src-p38. EMBO J., 21, 3782–3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chang L. and Karin M (2001) Mammalian MAP kinase signalling cascades. Nature, 410, 37–40. [DOI] [PubMed] [Google Scholar]
- 35. Angel P., Hattori K., Smeal T. and Karin M (1988) The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell, 55, 875–885. [DOI] [PubMed] [Google Scholar]
- 36. Takeda J., Maekawa T., Sudo T., Seino Y., Imura H., Saito N., Tanaka C. and Ishii S (1991) Expression of the CRE-BP1 transcriptional regulator binding to the cyclic AMP response element in central nervous system, regenerating liver, and human tumors. Oncogene, 6, 1009–1014. [PubMed] [Google Scholar]
- 37. Stein B., Angel P., van Dam H., Ponta H., Herrlich P., van der Eb A. and Rahmsdorf H. J (1992) Ultraviolet-radiation induced c-jun gene transcription: two AP-1 like binding sites mediate the response. Photochem. Photobiol., 55, 409–415. [DOI] [PubMed] [Google Scholar]
- 38. van Dam H., Duyndam M., Rottier R., Bosch A., de Vries-Smits L., Herrlich P., Zantema A., Angel P. and van der Eb A. J (1993) Heterodimer formation of cJun and ATF-2 is responsible for induction of c-jun by the 243 amino acid adenovirus E1A protein. EMBO J., 12, 479–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van Dam H., Wilhelm D., Herr I., Steffen A., Herrlich P. and Angel P (1995) ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J., 14, 1798–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu H., Deng X., Shyu Y. J., Li J. J., Taparowsky E. J. and Hu C. D (2006) Mutual regulation of c-Jun and ATF2 by transcriptional activation and subcellular localization. EMBO J., 25, 1058–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Baan B., van Dam H., van der Zon G. C., Maassen J. A. and Ouwens D. M (2006) The role of c-Jun N-terminal kinase, p38, and extracellular signal-regulated kinase in insulin-induced Thr69 and Thr71 phosphorylation of activating transcription factor 2. Mol. Endocrinol., 20, 1786–1795. [DOI] [PubMed] [Google Scholar]
- 42. Sevilla A., Santos C. R., Barcia R., Vega F. M. and Lazo P. A (2004) c-Jun phosphorylation by the human vaccinia-related kinase 1 (VRK1) and its cooperation with the N-terminal kinase of c-Jun (JNK). Oncogene, 23, 8950–8958. [DOI] [PubMed] [Google Scholar]
- 43. Sakurai A., Maekawa T., Sudo T., Ishii S. and Kishimoto A (1991) Phosphorylation of cAMP response element-binding protein, CRE-BP1, by cAMP-dependent protein kinase and protein kinase C. Biochem. Biophys. Res. Commun., 181, 629–635. [DOI] [PubMed] [Google Scholar]
- 44. Gupta S., Campbell D., Dérijard B. and Davis R. J (1995) Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science, 267, 389–393. [DOI] [PubMed] [Google Scholar]
- 45. Baan B., van der Zon G. C., Maassen J. A. and Ouwens D. M (2009) The nuclear appearance of ERK1/2 and p38 determines the sequential induction of ATF2-Thr71 and ATF2-Thr69 phosphorylation by serum in JNK-deficient cells. Mol. Cell. Endocrinol., 311, 94–100. [DOI] [PubMed] [Google Scholar]
- 46. Jia H., Cong Q., Chua J. F., et al. (2015) p57Kip2 is an unrecognized DNA damage response effector molecule that functions in tumor suppression and chemoresistance. Oncogene, 34, 3568–3581. [DOI] [PubMed] [Google Scholar]
- 47. Morton S., Davis R. J. and Cohen P (2004) Signalling pathways involved in multisite phosphorylation of the transcription factor ATF-2. FEBS Lett., 572, 177–183. [DOI] [PubMed] [Google Scholar]
- 48. Ban N., Yamada Y., Someya Y., et al. (2000) Activating transcription factor-2 is a positive regulator in CaM kinase IV-induced human insulin gene expression. Diabetes, 49, 1142–1148. [DOI] [PubMed] [Google Scholar]
- 49. Persengiev S. P. and Green M. R (2003) The role of ATF/CREB family members in cell growth, survival and apoptosis. Apoptosis, 8, 225–228. [DOI] [PubMed] [Google Scholar]
- 50. Hamard P. J., Dalbies-Tran R., Hauss C., Davidson I., Kedinger C. and Chatton B (2005) A functional interaction between ATF7 and TAF12 that is modulated by TAF4. Oncogene, 24, 3472–3483. [DOI] [PubMed] [Google Scholar]
- 51. Diring J., Camuzeaux B., Donzeau M., Vigneron M., Rosa-Calatrava M., Kedinger C. and Chatton B (2011) A cytoplasmic negative regulator isoform of ATF7 impairs ATF7 and ATF2 phosphorylation and transcriptional activity. PLoS One, 6, e23351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lau E., Kluger H., Varsano T., Lee K., Scheffler I., Rimm D. L., Ideker T. and Ronai Z. A (2012) PKCε promotes oncogenic functions of ATF2 in the nucleus while blocking its apoptotic function at mitochondria. Cell, 148, 543–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kawasaki H., Song J., Eckner R., Ugai H., Chiu R., Taira K., Shi Y., Jones N. and Yokoyama K. K (1998) p300 and ATF-2 are components of the DRF complex, which regulates retinoic acid- and E1A-mediated transcription of the c-jun gene in F9 cells. Genes Dev., 12, 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Maekawa T., Sakura H., Kanei-Ishii C., Sudo T., Yoshimura T., Fujisawa J., Yoshida M. and Ishii S (1989) Leucine zipper structure of the protein CRE-BP1 binding to the cyclic AMP response element in brain. EMBO J., 8, 2023–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yamasaki T., Takahashi A., Pan J., Yamaguchi N. and Yokoyama K. K (2009) Phosphorylation of activation transcription factor-2 at serine 121 by protein kinase C controls c-Jun-mediated activation of transcription. J. Biol. Chem., 284, 8567–8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sand J. M., Aziz M. H., Dreckschmidt N. E., Havighurst T. C., Kim K., Oberley T. D. and Verma A. K (2010) PKCepsilon overexpression, irrespective of genetic background, sensitizes skin to UVR-induced development of squamous-cell carcinomas. J. Invest. Dermatol., 130, 270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gillespie S., Zhang X. D. and Hersey P (2005) Variable expression of protein kinase C epsilon in human melanoma cells regulates sensitivity to TRAIL-induced apoptosis. Mol. Cancer Ther., 4, 668–676. [DOI] [PubMed] [Google Scholar]
- 58. Abu-Hamad S., Arbel N., Calo D., Arzoine L., Israelson A., Keinan N., Ben-Romano R., Friedman O. and Shoshan-Barmatz V (2009) The VDAC1 N-terminus is essential both for apoptosis and the protective effect of anti-apoptotic proteins. J. Cell Sci., 122, 1906–1916. [DOI] [PubMed] [Google Scholar]
- 59. Vega F. M., Sevilla A. and Lazo P. A (2004) p53 Stabilization and accumulation induced by human vaccinia-related kinase 1. Mol. Cell. Biol., 24, 10366–10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Valbuena A., Sanz-García M., López-Sánchez I., Vega F. M. and Lazo P. A (2011) Roles of VRK1 as a new player in the control of biological processes required for cell division. Cell. Signal., 23, 1267–1272. [DOI] [PubMed] [Google Scholar]
- 61. Fuchs S. Y., Tappin I. and Ronai Z (2000) Stability of the ATF2 transcription factor is regulated by phosphorylation and dephosphorylation. J. Biol. Chem., 275, 12560–12564. [DOI] [PubMed] [Google Scholar]
- 62. Paull T. T. (2015) Mechanisms of ATM activation. Annu. Rev. Biochem., 84, 711–738. [DOI] [PubMed] [Google Scholar]
- 63. Lovejoy C. A. and Cortez D (2009) Common mechanisms of PIKK regulation. DNA Repair (Amst)., 8, 1004–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cho S. G., Bhoumik A., Broday L., Ivanov V., Rosenstein B. and Ronai Z (2001) TIP49b, a regulator of activating transcription factor 2 response to stress and DNA damage. Mol. Cell. Biol., 21, 8398–8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ikura T., Ogryzko V. V., Grigoriev M., Groisman R., Wang J., Horikoshi M., Scully R., Qin J. and Nakatani Y (2000) Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell, 102, 463–473. [DOI] [PubMed] [Google Scholar]
- 66. Neuwald A. F., Aravind L., Spouge J. L. and Koonin E. V (1999) AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res., 9, 27–43. [PubMed] [Google Scholar]
- 67. Masson C., Menaa F., Pinon-Lataillade G., Frobert Y., Radicella J. P. and Angulo J. F (2001) Identification of KIN (KIN17), a human gene encoding a nuclear DNA-binding protein, as a novel component of the TP53-independent response to ionizing radiation. Radiat. Res., 156, 535–544. [DOI] [PubMed] [Google Scholar]
- 68. Kanemaki M., Kurokawa Y., Matsu-ura T., Makino Y., Masani A., Okazaki K., Morishita T. and Tamura T. A (1999) TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J. Biol. Chem., 274, 22437–22444. [DOI] [PubMed] [Google Scholar]
- 69. Bhoumik A., Takahashi S., Breitweiser W., Shiloh Y., Jones N. and Ronai Z (2005) ATM-dependent phosphorylation of ATF2 is required for the DNA damage response. Mol. Cell, 18, 577–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Raskov H., Pommergaard H. C., Burcharth J. and Rosenberg J (2014) Colorectal carcinogenesis–update and perspectives. World J. Gastroenterol., 20, 18151–18164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Maitra A. and Hruban R. H (2008) Pancreatic cancer. Annu. Rev. Pathol., 3, 157–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fearon E. R. and Vogelstein B (1990) A genetic model for colorectal tumorigenesis. Cell, 61, 759–767. [DOI] [PubMed] [Google Scholar]
- 73. Botla S. K., Savant S., Jandaghi P., et al. (2016) Early epigenetic downregulation of microRNA-192 expression promotes pancreatic cancer progression. Cancer Res., 76, 4149–4159. [DOI] [PubMed] [Google Scholar]
- 74. Lao V. V. and Grady W. M (2011) Epigenetics and colorectal cancer. Nat. Rev. Gastroenterol. Hepatol., 8, 686–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Raskov H., Soby J. H., Troelsen J., Bojesen R. D., and Gogenur I (2019) Driver gene mutations and epigenetics in colorectal cancer. Ann. Surg. [DOI] [PubMed] [Google Scholar]
- 76. Toh T. B., Lim J. J. and Chow E. K (2017) Epigenetics in cancer stem cells. Mol. Cancer, 16, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Karanam B., Wang L., Wang D., Liu X., Marmorstein R., Cotter R. and Cole P. A (2007) Multiple roles for acetylation in the interaction of p300 HAT with ATF-2. Biochemistry, 46, 8207–8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zarour L. R., Anand S., Billingsley K. G., et al. (2017) Colorectal cancer liver metastasis: evolving paradigms and future directions. Cell. Mol. Gastroenterol. Hepatol., 3, 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chen C., Aihemaiti M., Zhang X., Qu H., Sun Q. L., He Q. S. and Yu W. B (2018) Downregulation of histone demethylase JMJD1C inhibits colorectal cancer metastasis through targeting ATF2. Am. J. Cancer Res., 8, 852–865. [PMC free article] [PubMed] [Google Scholar]
- 80. Eichhorn S. W., Guo H., McGeary S. E., et al. (2014) mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell, 56, 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Knowlton D. L., Tang K., Henstock P. V. and Subramanian R. R (2011) miRNA alterations modify kinase activation in the IGF-1 pathway and correlate with colorectal cancer stage and progression in patients. J. Cancer, 2, 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhang S., Dong X., Ji T., Chen G. and Shan L (2017) Long non-coding RNA UCA1 promotes cell progression by acting as a competing endogenous RNA of ATF2 in prostate cancer. Am. J. Transl. Res., 9, 366–375. [PMC free article] [PubMed] [Google Scholar]
- 83. Jung J., Yeom C., Choi Y. S., Kim S., Lee E., Park M. J., Kang S. W., Kim S. B. and Chang S (2015) Simultaneous inhibition of multiple oncogenic miRNAs by a multi-potent microRNA sponge. Oncotarget, 6, 20370–20387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Song L., Peng L., Hua S., Li X., Ma L., Jie J., Chen D., Wang Y. and Li D (2018) miR-144-5p enhances the radiosensitivity of non-small-cell lung cancer cells via targeting ATF2. Biomed Res. Int., 2018, 5109497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Huang G., Pan J., Ye Z., Fang B., Cheng W. and Cao Z (2017) Overexpression of miR-216b sensitizes NSCLC cells to cisplatin-induced apoptosis by targeting c-Jun. Oncotarget, 8, 104206–104215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Jiang Y., Jiang J., Jia H., Qiao Z. and Zhang J (2018) Recovery of miR-139-5p in ovarian cancer reverses cisplatin resistance by targeting C-Jun. Cell. Physiol. Biochem., 51, 129–141. [DOI] [PubMed] [Google Scholar]
- 87. Tian L., Zhang J., Ren X., Liu X., Gao W., Zhang C., Sun Y. and Liu M (2017) Overexpression of miR-26b decreases the cisplatin-resistance in laryngeal cancer by targeting ATF2. Oncotarget, 8, 79023–79033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sun X., Lou L., Zhong K. and Wan L (2017) MicroRNA-451 regulates chemoresistance in renal cell carcinoma by targeting ATF-2 gene. Exp. Biol. Med. (Maywood), 242, 1299–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pu Y., Zhao F., Wang H. and Cai S (2017) MiR-34a-5p promotes multi-chemoresistance of osteosarcoma through down-regulation of the DLL1 gene. Sci. Rep., 7, 44218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lv G., Hu Z., Tie Y., Du J., Fu H., Gao X. and Zheng X (2014) MicroRNA-451 regulates activating transcription factor 2 expression and inhibits liver cancer cell migration. Oncol. Rep., 32, 1021–1028. [DOI] [PubMed] [Google Scholar]
- 91. Arora H., Qureshi R., Park A. K. and Park W. Y (2011) Coordinated regulation of ATF2 by miR-26b in γ-irradiated lung cancer cells. PLoS One, 6, e23802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. De Robertis M., Loiacono L., Fusilli C., et al. (2017) Dysregulation of EGFR pathway in EphA2 cell subpopulation significantly associates with poor prognosis in colorectal cancer. Clin. Cancer Res., 23, 159–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lim H. J., Park M. N., Kim C., Kang B., Song H.S., Lee H., Kim S.H., Shim B.S., and Kim B (2019) MiR-657/ATF2 signaling pathway has a critical role in Spatholobus suberectus Dunn extract-induced apoptosis in U266 and U937 cells. Cancers (Basel), 11, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zhang R., Luo H., Wang S., et al. (2015) MiR-622 suppresses proliferation, invasion and migration by directly targeting activating transcription factor 2 in glioma cells. J. Neurooncol., 121, 63–72. [DOI] [PubMed] [Google Scholar]
- 95. Hu Z., Tie Y., Lv G., Zhu J., Fu H. and Zheng X (2018) Transcriptional activation of miR-320a by ATF2, ELK1 and YY1 induces cancer cell apoptosis under ionizing radiation conditions. Int. J. Oncol., 53, 1691–1702. [DOI] [PubMed] [Google Scholar]
- 96. Liu F., Cheng Z., Li X., Li Y., Zhang H., Li J., Liu F., Xu H. and Li F (2017) A novel Pak1/ATF2/miR-132 signaling axis is involved in the hematogenous metastasis of gastric cancer cells. Mol. Ther. Nucleic Acids, 8, 370–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Xiao L., Rao J. N., Zou T., et al. (2007) Polyamines regulate the stability of activating transcription factor-2 mRNA through RNA-binding protein HuR in intestinal epithelial cells. Mol. Biol. Cell, 18, 4579–4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Al-Sadi R., Guo S., Ye D., Dokladny K., Alhmoud T., Ereifej L., Said H. M. and Ma T. Y (2013) Mechanism of IL-1β modulation of intestinal epithelial barrier involves p38 kinase and activating transcription factor-2 activation. J. Immunol., 190, 6596–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Arana M. R., Tocchetti G. N., Domizi P., et al. (2015) Coordinated induction of GST and MRP2 by cAMP in Caco-2 cells: role of protein kinase A signaling pathway and toxicological relevance. Toxicol. Appl. Pharmacol., 287, 178–190. [DOI] [PubMed] [Google Scholar]
- 100. Namachivayam K., MohanKumar K., Arbach D., Jagadeeswaran R., Jain S. K., Natarajan V., Mehta D., Jankov R. P. and Maheshwari A (2015) All-trans retinoic acid induces TGF-β2 in intestinal epithelial cells via RhoA- and p38α MAPK-mediated activation of the transcription factor ATF2. PLoS One, 10, e0134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Jia H., Cong Q., Chua J. F., et al. (2015) p57Kip2 is an unrecognized DNA damage response effector molecule that functions in tumor suppression and chemoresistance. Oncogene, 34, 3568–3581. [DOI] [PubMed] [Google Scholar]
- 102. Voloshanenko O., Schwartz U., Kranz D., Rauscher B., Linnebacher M., Augustin I. and Boutros M (2018) β-catenin-independent regulation of Wnt target genes by RoR2 and ATF2/ATF4 in colon cancer cells. Sci. Rep., 8, 3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Han S. I., Yasuda K. and Kataoka K (2011) ATF2 interacts with beta-cell-enriched transcription factors, MafA, Pdx1, and beta2, and activates insulin gene transcription. J. Biol. Chem., 286, 10449–10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Hay C. W., Ferguson L. A. and Docherty K (2007) ATF-2 stimulates the human insulin promoter through the conserved CRE2 sequence. Biochim. Biophys. Acta, 1769, 79–91. [DOI] [PubMed] [Google Scholar]
- 105. Stoian M., State N., Stoica V. and Radulian G (2014) Apoptosis in colorectal cancer. J. Med. Life, 7, 160–164. [PMC free article] [PubMed] [Google Scholar]
- 106. Watson A. J. (2004) Apoptosis and colorectal cancer. Gut, 53, 1701–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Hanahan D. and Weinberg R. A (2011) Hallmarks of cancer: the next generation. Cell, 144, 646–674. [DOI] [PubMed] [Google Scholar]
- 108. Zhang J. Y., Jiang H., Gao W., Wu J., Peng K., Shi Y. F. and Zhang X. J (2008) The JNK/AP1/ATF2 pathway is involved in H2O2-induced acetylcholinesterase expression during apoptosis. Cell. Mol. Life Sci., 65, 1435–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Salameh A., Galvagni F., Anselmi F., De Clemente C., Orlandini M. and Oliviero S (2010) Growth factor stimulation induces cell survival by c-Jun. ATF2-dependent activation of Bcl-XL. J. Biol. Chem., 285, 23096–23104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Averous J., Bruhat A., Jousse C., Carraro V., Thiel G. and Fafournoux P (2004) Induction of CHOP expression by amino acid limitation requires both ATF4 expression and ATF2 phosphorylation. J. Biol. Chem., 279, 5288–5297. [DOI] [PubMed] [Google Scholar]
- 111. Berger A. J., Kluger H. M., Li N., Kielhorn E., Halaban R., Ronai Z. and Rimm D. L (2003) Subcellular localization of activating transcription factor 2 in melanoma specimens predicts patient survival. Cancer Res., 63, 8103–8107. [PubMed] [Google Scholar]
- 112. Ronai Z., Yang Y. M., Fuchs S. Y., Adler V., Sardana M. and Herlyn M (1998) ATF2 confers radiation resistance to human melanoma cells. Oncogene, 16, 523–531. [DOI] [PubMed] [Google Scholar]
- 113. Li M., Wu X., Liu N., Li X., Meng F. and Song S (2017) Silencing of ATF2 inhibits growth of pancreatic cancer cells and enhances sensitivity to chemotherapy. Cell Biol. Int., 41, 599–610. [DOI] [PubMed] [Google Scholar]
- 114. Inoue S., Mizushima T., Ide H., Jiang G., Goto T., Nagata Y., Netto G. J. and Miyamoto H (2018) ATF2 promotes urothelial cancer outgrowth via cooperation with androgen receptor signaling. Endocr. Connect., 7, 1397–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Papassava P., Gorgoulis V. G., Papaevangeliou D., Vlahopoulos S., van Dam H. and Zoumpourlis V (2004) Overexpression of activating transcription factor-2 is required for tumor growth and progression in mouse skin tumors. Cancer Res., 64, 8573–8584. [DOI] [PubMed] [Google Scholar]
- 116. Zoumpourlis V., Papassava P., Linardopoulos S., Gillespie D., Balmain A. and Pintzas A (2000) High levels of phosphorylated c-Jun, Fra-1, Fra-2 and ATF-2 proteins correlate with malignant phenotypes in the multistage mouse skin carcinogenesis model. Oncogene, 19, 4011–4021. [DOI] [PubMed] [Google Scholar]
- 117. Maekawa T., Shinagawa T., Sano Y., et al. (2007) Reduced levels of ATF-2 predispose mice to mammary tumors. Mol. Cell. Biol., 27, 1730–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Su L., Sampaio A. V., Jones K. B., et al. (2012) Deconstruction of the SS18-SSX fusion oncoprotein complex: insights into disease etiology and therapeutics. Cancer Cell, 21, 333–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Gao Z., Shang Q., Liu Z., Deng C. and Guo C (2015) Mitochondrial ATF2 translocation contributes to apoptosis induction and BRAF inhibitor resistance in melanoma through the interaction of Bim with VDAC1. Oncotarget, 6, 36338–36353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Duffey D., Dolgilevich S., Razzouk S., Li L., Green R. and Gorti G. K (2011) Activating transcription factor-2 in survival mechanisms in head and neck carcinoma cells. Head Neck, 33, 1586–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Walluscheck D., Poehlmann A., Hartig R., et al. (2013) ATF2 knockdown reinforces oxidative stress-induced apoptosis in TE7 cancer cells. J. Cell. Mol. Med., 17, 976–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Zhang G., Zhang J., Shang D., Qi B. and Chen H (2015) Deoxycholic acid inhibited proliferation and induced apoptosis and necrosis by regulating the activity of transcription factors in rat pancreatic acinar cell line AR42J. In Vitro Cell. Dev. Biol. Anim., 51, 851–856. [DOI] [PubMed] [Google Scholar]
- 123. Mouratidis P. X., Colston K. W., Bartlett J. B., Muller G. W., Man H. W., Stirling D. and Dalgleish A. G (2009) Antiproliferative effects of CC-8062 and CC-8075 in pancreatic cancer cells. Pancreas, 38, 78–84. [DOI] [PubMed] [Google Scholar]
- 124. An J. J., Shi K. J., Wei W., et al. (2013) The ROS/JNK/ATF2 pathway mediates selenite-induced leukemia NB4 cell cycle arrest and apoptosis in vitro and in vivo. Cell Death Dis., 4, e973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Bhoumik A., Huang T. G., Ivanov V., Gangi L., Qiao R. F., Woo S. L., Chen S. H. and Ronai Z (2002) An ATF2-derived peptide sensitizes melanomas to apoptosis and inhibits their growth and metastasis. J. Clin. Invest., 110, 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Bhoumik A., Gangi L. and Ronai Z (2004) Inhibition of melanoma growth and metastasis by ATF2-derived peptides. Cancer Res., 64, 8222–8230. [DOI] [PubMed] [Google Scholar]
- 127. Bhoumik A., Ivanov V. and Ronai Z (2001) Activating transcription factor 2-derived peptides alter resistance of human tumor cell lines to ultraviolet irradiation and chemical treatment. Clin. Cancer Res., 7, 331–342. [PubMed] [Google Scholar]
- 128. Kalluri R. and Weinberg R. A (2009) The basics of epithelial-mesenchymal transition. J. Clin. Invest., 119, 1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Spaderna S., Schmalhofer O., Hlubek F., Berx G., Eger A., Merkel S., Jung A., Kirchner T. and Brabletz T (2006) A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology, 131, 830–840. [DOI] [PubMed] [Google Scholar]
- 130. Brabletz T., Jung A., Reu S., Porzner M., Hlubek F., Kunz-Schughart L. A., Knuechel R. and Kirchner T (2001) Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc. Natl. Acad. Sci. USA, 98, 10356–10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Gui T., Sun Y., Shimokado A. and Muragaki Y (2012) The roles of mitogen-activated protein kinase pathways in TGF-β-induced epithelial-mesenchymal transition. J. Signal Transduct., 2012, 289243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Xu Y., Liu Z. and Guo K (2012) The effect of JDP2 and ATF2 on the epithelial-mesenchymal transition of human pancreatic cancer cell lines. Pathol. Oncol. Res., 18, 571–577. [DOI] [PubMed] [Google Scholar]
- 133. Yuanhong X., Feng X., Qingchang L., Jianpeng F., Zhe L. and Kejian G (2010) Downregulation of AP-1 repressor JDP2 is associated with tumor metastasis and poor prognosis in patients with pancreatic carcinoma. Int. J. Biol. Markers, 25, 136–140. [DOI] [PubMed] [Google Scholar]
- 134. Yu T., Li Y. J., Bian A. H., et al. (2014) The regulatory role of activating transcription factor 2 in inflammation. Mediators Inflamm., 2014, 950472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Li M., Zhang D., Ge X., Zhu X., Zhou Y., Zhang Y., Peng X. and Shen A (2019) TRAF6-p38/JNK-ATF2 axis promotes microglial inflammatory activation. Exp. Cell Res., 376, 133–148. [DOI] [PubMed] [Google Scholar]
- 136. Hu G., Zhao X., Wang C., et al. (2017) MicroRNA-145 attenuates TNF-α-driven cartilage matrix degradation in osteoarthritis via direct suppression of MKK4. Cell Death Dis., 8, e3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Reimold A. M., Kim J., Finberg R. and Glimcher L. H (2001) Decreased immediate inflammatory gene induction in activating transcription factor-2 mutant mice. Int. Immunol., 13, 241–248. [DOI] [PubMed] [Google Scholar]
- 138. Desai S., Laskar S., and Pandey B. N (2014) Role of ATF-2 in regulation of epithelial–mesenchymal transition and radio-sensitivity of A549 cells mediated by secreted soluble factors. J Radiat Res., 55, i116–i117. [Google Scholar]
- 139. Huang Y., Miao Z., Hu Y., Yuan Y., Zhou Y., Wei L., Zhao K., Guo Q. and Lu N (2017) Baicalein reduces angiogenesis in the inflammatory microenvironment via inhibiting the expression of AP-1. Oncotarget, 8, 883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Lee S. H., Bahn J. H., Whitlock N. C. and Baek S. J (2010) Activating transcription factor 2 (ATF2) controls tolfenamic acid-induced ATF3 expression via MAP kinase pathways. Oncogene, 29, 5182–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Ivanov V. N. and Ronai Z (1999) Down-regulation of tumor necrosis factor alpha expression by activating transcription factor 2 increases UVC-induced apoptosis of late-stage melanoma cells. J. Biol. Chem., 274, 14079–14089. [DOI] [PubMed] [Google Scholar]
- 142. Wang T., Yu Q., Li J., et al. (2017) O-GlcNAcylation of fumarase maintains tumour growth under glucose deficiency. Nat. Cell Biol., 19, 833–843. [DOI] [PubMed] [Google Scholar]
- 143. Lawrence M. C., Naziruddin B., Levy M. F., Jackson A. and McGlynn K (2011) Calcineurin/nuclear factor of activated T cells and MAPK signaling induce TNF-{alpha} gene expression in pancreatic islet endocrine cells. J. Biol. Chem., 286, 1025–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Ammendrup A., Maillard A., Nielsen K., Aabenhus Andersen N., Serup P., Dragsbaek Madsen O., Mandrup-Poulsen T. and Bonny C (2000) The c-Jun amino-terminal kinase pathway is preferentially activated by interleukin-1 and controls apoptosis in differentiating pancreatic beta-cells. Diabetes, 49, 1468–1476. [DOI] [PubMed] [Google Scholar]
- 145. Wu D. S., Chen C., Wu Z. J., et al. (2016) ATF2 predicts poor prognosis and promotes malignant phenotypes in renal cell carcinoma. J. Exp. Clin. Cancer Res., 35, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. You Z., Zhou Y., Guo Y., Chen W., Chen S. and Wang X (2016) Activating transcription factor 2 expression mediates cell proliferation and is associated with poor prognosis in human non-small cell lung carcinoma. Oncol. Lett., 11, 760–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Zhou H., Cai L., Zhang X., Li A., Miao Y., Li Q., Qiu X. and Wang E (2018) ARHGEF39 promotes tumor progression via activation of Rac1/P38 MAPK/ATF2 signaling and predicts poor prognosis in non-small cell lung cancer patients. Lab. Invest., 98, 670–681. [DOI] [PubMed] [Google Scholar]
- 148. Zheng R., Liu Q., Wang T., Wang L. and Zhang Y (2018) FAM98A promotes proliferation of non-small cell lung cancer cells via the P38-ATF2 signaling pathway. Cancer Manag. Res., 10, 2269–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Chen S. Y., Takeuchi S., Moroi Y., et al. (2009) Concordant overexpression of phosphorylated ATF2 and STAT3 in extramammary Paget’s disease. J. Cutan. Pathol., 36, 402–408. [DOI] [PubMed] [Google Scholar]
- 150. Rudraraju B., Droog M., Abdel-Fatah T. M., et al. (2014) Phosphorylation of activating transcription factor-2 (ATF-2) within the activation domain is a key determinant of sensitivity to tamoxifen in breast cancer. Breast Cancer Res. Treat., 147, 295–309. [DOI] [PubMed] [Google Scholar]
- 151. Pradhan M. P., Prasad N. K. and Palakal M. J (2012) A systems biology approach to the global analysis of transcription factors in colorectal cancer. BMC Cancer, 12, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Cerami E., Gao J., Dogrusoz U., et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov., 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Gao J., Aksoy B. A., Dogrusoz U., et al. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal., 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Breitwieser W., Lyons S., Flenniken A. M., Ashton G., Bruder G., Willington M., Lacaud G., Kouskoff V. and Jones N (2007) Feedback regulation of p38 activity via ATF2 is essential for survival of embryonic liver cells. Genes Dev., 21, 2069–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Bhoumik A., Fichtman B., Derossi C., et al. (2008) Suppressor role of activating transcription factor 2 (ATF2) in skin cancer. Proc. Natl. Acad. Sci. USA, 105, 1674–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Gozdecka M., Lyons S., Kondo S., Taylor J., Li Y., Walczynski J., Thiel G., Breitwieser W. and Jones N (2014) JNK suppresses tumor formation via a gene-expression program mediated by ATF2. Cell Rep., 9, 1361–1374. [DOI] [PubMed] [Google Scholar]
- 157. Bhoumik A., Jones N. and Ronai Z (2004) Transcriptional switch by activating transcription factor 2-derived peptide sensitizes melanoma cells to apoptosis and inhibits their tumorigenicity. Proc. Natl. Acad. Sci. USA, 101, 4222–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Abbas S., Bhoumik A., Dahl R., Vasile S., Krajewski S., Cosford N. D. and Ronai Z. A (2007) Preclinical studies of celastrol and acetyl isogambogic acid in melanoma. Clin. Cancer Res., 13, 6769–6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Graziani G., Artuso S., De Luca A., et al. (2015) A new water soluble MAPK activator exerts antitumor activity in melanoma cells resistant to the BRAF inhibitor vemurafenib. Biochem. Pharmacol., 95, 16–27. [DOI] [PubMed] [Google Scholar]
- 160. The UniProt Consortium. (2019) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res., 47, D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Webb B. and Sali A (2016) Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics, 54, 5.6.1–5.6.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Sali A. and Blundell T. L (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol., 234, 779–815. [DOI] [PubMed] [Google Scholar]
- 163. Wu S. and Zhang Y (2008) MUSTER: improving protein sequence profile-profile alignments by using multiple sources of structure information. Proteins, 72, 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C. and Ferrin T. E (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem., 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- 165. Giannakis M., Mu X. J., Shukla S. A., et al. (2016) Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep., 15, 857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Seshagiri S., Stawiski E. W., Durinck S., et al. (2012) Recurrent R-spondin fusions in colon cancer. Nature, 488, 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Cancer Genome Atlas Network (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature, 487, 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Liu J., Lichtenberg T., Hoadley K. A., et al. ; Cancer Genome Atlas Research Network (2018) An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell, 173, 400–416.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Brannon A. R., Vakiani E., Sylvester B. E., et al. (2014) Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol., 15, 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Yaeger R., Chatila W. K., Lipsyc M. D., et al. (2018) Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell, 33, 125–136.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Guda K., Veigl M. L., Varadan V., et al. (2015) Novel recurrently mutated genes in African American colon cancers. Proc. Natl. Acad. Sci. USA, 112, 1149–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Jiao Y., Yonescu R., Offerhaus G. J., et al. (2014) Whole-exome sequencing of pancreatic neoplasms with acinar differentiation. J. Pathol., 232, 428–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Biankin A. V., Waddell N., Kassahn K. S., et al.; Australian Pancreatic Cancer Genome Initiative (2012) Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature, 491, 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Bailey P., Chang D. K., Nones K., et al. ; Australian Pancreatic Cancer Genome Initiative (2016) Genomic analyses identify molecular subtypes of pancreatic cancer. Nature, 531, 47–52. [DOI] [PubMed] [Google Scholar]
- 175. Hoadley K. A., Yau C., Hinoue T., et al.; Cancer Genome Atlas Network (2018) Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell, 173, 291–304.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Witkiewicz A. K., McMillan E. A., Balaji U., et al. (2015) Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun., 6, 6744. [DOI] [PMC free article] [PubMed] [Google Scholar]