Abstract
Purines, among most influential molecules, are reported to have essential biological function by regulating various cell types. A large number of studies have led to the discovery of many biological functions of the purine nucleotides such as ATP, ADP, and adenosine, as signaling molecules that engage G protein-coupled or ligand-gated ion channel receptors. The role of purines in the regulation of cellular functions at the gene or protein level has been well documented. With the advances in multiomics, including those from metabolomic and bioinformatic analyses, metabolic reprogramming was identified as a key mechanism involved in the regulation of cellular function under physiological or pathological conditions. Recent studies suggest that purines or purine-derived products contribute to important regulatory functions in many fundamental biological and pathological processes related to metabolic reprogramming. Therefore, this review summarizes the role and potential mechanism of purines in the regulation of metabolic reprogramming. In particular, the molecular mechanisms of extracellular purine- and intracellular purine-mediated metabolic regulation in various cells during disease development are discussed. In summary, our review provides an extensive resource for studying the regulatory role of purines in metabolic reprogramming and sheds light on the utilization of the corresponding peptides or proteins for disease diagnosis and therapy.
Keywords: Purines, Metabolic reprogramming, Target therapy, Signal transduction
Background
In 1924, a sharply distinct metabolism of tumor cells, the enhancement of glycolysis and decrease of the oxidative phosphorylation (OXPHOS), was firstly reported by Otto Warburg [1–3]. This type of tumor cell-specific metabolism favoring anaerobic over aerobic metabolism occurred even if environmental oxygen supply is abundant [1, 2, 4, 5]. This phenomenon was observed in different types of tumor cells and was later named the “Warburg effect” [6–9]. The mainstream view that has been established suggests that metabolic alterations contribute to the proliferation of tumor cells and are strictly programmed by a series of complicated pathways that remain unknown [4, 7, 10, 11]. Therefore, similar to transcriptional reprogramming, the concept of “metabolic reprogramming” was formed. Furthermore, the molecular characteristics of metabolic reprogramming have also been expanded to include changes in the activity of certain key enzymes in metabolic pathways, such as lipolysis and PPP, and metabolic regulators, including transporters of glucose or amino acids [12–14].
Recent studies confirmed that metabolic reprogramming is a key mechanism involved in physiological and pathological processes in cells via regulation of proliferation, differentiation, activation, and death [15–20]. Although it has been shown that metabolic reprogramming has an essential impact on cellular function, the molecular mechanism underlying the regulation of metabolic reprogramming remains unclear. This review summarizes the regulatory role of purines involved in metabolic reprogramming and in molecular mechanisms and provides a systematic perspective for further translational practice (Table 1).
Table. 1.
The current realm of metabolic reprogramming
| Cell type | Biological process | Metabolic alteration | Reference |
|---|---|---|---|
| Solid tumor cell | Proliferation, survival, and oncogenesis | Glycolysis, OXPHOS, PPP, glutaminolysis, and lipolysis | [7, 11] |
| Leukemia | Cell growth and autophagy | Glutaminolysis, glycolysis, and lipid metabolism | [8, 9] |
| Macrophages | Macrophages polarization | Glycolysis, l-arginine metabolism, glutaminolysis, OXPHOS, and lipolysis | [21, 22] |
| T cells | Activation and differentiation | Glycolysis, glutaminolysis, FAO, and PPP | [21, 23] |
| B cells | Activation, differentiation, and antibody production | Glycolysis, OXPHOS, and PPP | [21, 22, 24] |
| Dendritic cells | Activation | OXPHOS and glycolysis | [21] |
| MDSCs | Activation, differentiation, and inflammatory response | Amino acid metabolism, glycolysis, and FAO | [21, 25] |
| Neutrophils | Microbicidal functions | Glycolysis and PPP | [21] |
| Natural killer cells | Activation and killing function | Glycolysis and OXPHOS | [26] |
| Induced pluripotent stem cells | Generation of iPSCs from somatic cells | Glycolysis and OXPHOS | [27] |
| Mesenchymal stem cells (MSC) | Survival and activation | glycogen synthesis and glycolysis | [19, 28] |
| Hematopoietic stem cells (HSCs) | Lineage commitment, proliferation | Glucose metabolism, glutamine metabolism, FAO | [19, 29] |
| Erythrocytes | Oxygen release | Glycolysis | [30, 31] |
OXPHOS, oxidative phosphorylation; PPP, pentose phosphate pathway; FAO, fatty acids oxidation; MDSCs, myeloid-derived suppressor cells
The growing realm of metabolic reprogramming
The realm of metabolic reprogramming has been expanded to include not only tumor cells but also different, active cell types, including immune cells and stem cells, due to the development of omics such as proteomics, metabolomics, and metabolite tracking, including 18F-deoxyglucose positron emission tomography (FDG-PET) and metabolic flux analysis [6, 12, 19, 21, 29]. Considerable evidence has proven that metabolic reprogramming can regulate cell fate or commitment of stem cells via epigenetic regulation by metabolites, possibly contributing to the regenerative ability of stem cells [19, 27–29, 32–34]. The metabolic state of the organism affects the immune system through an altered fuel supply and thus the cellular metabolism used by immune cells. These affected components, in turn, regulate immune cell function, which we now know can subsequently regulate whole body metabolism such that an immunometabolic loop exists between systemic and cellular metabolism in the organism [35]. It has been reported that enhanced glycolysis and PPP are essential for the activation of T cells [21, 36]. Other immune cells, including B cells, macrophages, dendrite cells, and natural killer (NK) cells, play roles in the immune response with observed metabolic reprogramming [21–24, 26]. Of note, through regulation of the activation of immune regulatory cells such as regulatory T (Treg) cells and myeloid-derived suppressor cells (MDSCs), metabolic reprogramming might play a critical role in the self-regulation of the immune system [21, 25, 37, 38]. In addition, modulation of metabolic reprogramming for the treatments of autoimmune diseases and chronic inflammation has also increased expectations, especially for treatments of T cell–related disease [39, 40]. For tumor immunotherapy, metabolic rewiring has been reported to regulate therapeutic outcomes by significantly modulating tumor cells, tumor-associated immune cells, and tumor microenvironments [21, 41]. Several drugs targeting metabolic alterations have been utilized in preclinical or clinical trials [42]. For instance, metabolic reprogramming and current targets, such as programmed cell death protein-1 (PD-1), are being reevaluated. Studies have confirmed that metabolic alteration in fatty acid oxidation (FAO) or glycolysis is triggered by PD-1 ligation [43]. Additionally, hypoxia-induced factor (HIF), which is critically involved in metabolism, contributes to the induction of programmed cell death-ligand 1 (PD-L1) [44]. Therefore, metabolic reprogramming might start a revolution in the classic system of tumor pharmacology. Of note, our group discovered the critical role of metabolic reprogramming in erythrocyte function and hypoxia response, expanding the vision for metabolic reprogramming to include nonproliferating cells [30, 45]. Therefore, manipulating metabolic reprogramming could further help us develop novel targets for disease treatments.
The mechanism of metabolic reprogramming–mediated regulation of cellular processes largely depends on the sequential metabolites that alter metabolism. Current studies have revealed that small metabolites can be released into the extracellular microenvironment and function as ligands, for example, purines that bind to a matched receptor [30]. In addition, metabolic reprogramming provides energy and material pools that enable functional change through increased production of ATP and nucleic acid.
Currently, thousands of articles aimed at metabolic reprogramming are being published, greatly enriching the theoretical foundation of metabolic reprogramming. Undoubtedly, the concept and realm of “metabolic reprogramming” are still developing.
Overview of purines
The metabolism of purines
Biological purines are a series of small molecules with purine rings that exist inside or outside cells. Among all kinds of purines, the most frequently mentioned is probably adenosine-5′-triphosphate (ATP), which has been recognized as a critical metabolic molecule within the cell and was first described as an extracellular messenger by Burnstock in 1972 [46, 47]. Under normal conditions, ATP is produced from the gradual phosphorylation of ADP/adenosine monophosphate (AMP), forming a primary energy machine for biological processes [47]. Serving as the raw purinergic material of ADP/AMP/ATP, adenosine can be bypass degraded with the help of several enzymes including adenosine deaminase (ADA), S-adenosylhomocysteine hydrolase (SAHH), and adenosine kinase (ADK), leading to the generation of inosine, hypoxanthine, adenosylhomocysteine (AdoHcy), and AMP [48]. Additionally, intracellular purines can be utilized for the generation of cyclic purines such as cAMP, which is among the most famous second messengers [49].
Under stress, however, ATP tends to be released from injured or necrotic cells and subsequently degraded by four ectonucleotidase groups, namely, ectonucleoside triphosphate diphosphohydrolases (CD39), ecto-59-nucleotidase (CD73), ectonucleotide pyrophosphatase/phosphodiesterase (NPPs), and alkaline phosphatases, eventually leading to adenosine production, which serves as the major source of extracellular purines [50]. The other sources of extracellular adenosine involve direct secretion with the help of equilibrative nucleoside transporters (ENTs), which enable adenosine to travel bidirectionally [51]. Some of these extracellular purines, including ATP and adenosine, function as signal molecules and bind to exclusive receptors to complete their various regulatory functions [48].
The biological function of purines
The components and regulatory process of purines in cytoplasm differ from those in the extracellular microenvironment; therefore, herein, we elaborate on their different biological functions.
Extracellular purines
Extracellular purines mainly regulate cellular functions as a signaling ligand by engaging receptors on the cell surface. Purine receptors can be divided into two groups: P1 receptors for adenosine binding and P2 receptors for other pyrimidine nucleotide binding. The distribution of these receptors varies depending on different cell types, which determines the complex downstream regulatory network of the purinergic pathway [52]. Additionally, some of these receptors are coupled with G proteins and are capable of generating second messengers such as cyclic purines [53].
Adenosine signaling involves adenosine and four receptors and has a large impact on physiological homeostasis in many ways [48]. Under pathological conditions, the concentration of extracellular adenosine can elevate dramatically in response to stress or assist in repairing cell damage [48]. The adenosine receptor can be divided into four types (ADORA1, ADORA2A, ADORA2B, and ADORA3), which are all coupled with G proteins to function [54]. The role of the adenosine pathway may be tissue-specific because of the various distribution patterns of the adenosine receptor [48]. The adenosine pathway plays a beneficial role in cardiovascular protection; therefore, targeted drugs have been seriously investigated or already applied in clinical practice [55]. Adenosine-related research, in recent years, has focused on the cellular response to stress, hypoxia, immunity, inflammation, etc. [48, 55, 56].
ATP receptors can be subclassified according to their transduction mechanism (i.e., as metabotropic P2YRs or ionotropic P2XRs) [57]. The fast-acting P2XRs constitute a family of seven ligand-gated ion channel receptors (P2X1–7) that have a single physiological agonist (ATP). Composed of subunits of 379–595 amino acids in length, they share a similar tertiary topology with an intracellular NH2 and a more extended COOH terminus, a large extracellular loop responsible for ligand binding, and two transmembrane-spanning regions (TM1 and TM2) [58, 59]. Two hydrophobic transmembrane domains (M1 and M2) are separated by an ectodomain within which 10 cysteine residues form disulfide bridges [59]. TM1 is involved in channel gating, and a helix of TM2 forms a channel pore. Following ATP binding, P2XRs become permeable to Na+, K+, and Ca2+. Moreover, activation of a P2X7R induces formation of a large pore that enables the passage of molecules as large as 900 Da [52]. Unlike the P2XRs, P2Yrs belong to the G protein–coupled receptors and are not accessible exclusively to ATP. In fact, P2Yrs can be selectively engaged and activated by small metabolites including ATP, ADP, and UDP [60]. Based on current understanding, the main role of P2Yrs is thought to be involved in cardiovascular regulation and immunological regulation, where they are considered to be a potential drug target [61]. However, little research has been carried out.
In general, there are other types of purines that exist extracellularly, such as inosine and hypoxanthine, which mainly originate from the degradation of extracellular adenosine. Studies have shown nonexclusive binding between inosine and several types of receptors, such as GABAA receptors and adenosine receptors [62, 63]; however, little is known about the exclusive receptors for or the signaling pathways of inosine or hypoxanthine. Since little evidence has been generated to show an obvious relation between metabolic reprogramming and inosine or hypoxanthine, the extracellular purines discussed in this review mainly refer to ATP and adenosine.
Intracellular purines
In addition to extracellular purines, intracellular purines, which are mainly classified as either noncyclic purines or cyclic purines, have a vital and common regulatory role. There are some molecular sensors for intracellular purines, and their purine-dependent activation can initiate a signal transduction cascade.
Noncyclic purines exist both inside and outside cells and include adenosine, AMP, ADP, and ATP. Apart from the extracellular effect on receptors, AMP and ADP, in the past decade, were reported to regulate metabolism via their intracellular sensor, AMP-activated protein kinase (AMPK). During energy-depleting exercise, an increase of free AMP and ADP, which can directly bind to the γ regulatory subunits of AMPK, leads to a conformational change that activates AMPK via phosphorylation. The process can be elicited by even modest decreases in ATP production that result in relative increases in AMP or ADP [64]. In a coordinated response, AMPK increases metabolic output through nutrient utilization in addition to inhibiting anabolic pathways, thus enabling adaptive changes in growth, differentiation, and metabolism during energy deficiency [65]. In mammals, AMPK plays a general role in coordinating growth and metabolism and special roles in regulating metabolism in dedicated tissues such as fat, neuron, and muscle [66].
Small intracellular cyclic purines, including cyclic AMP (cAMP) and cyclic GMP (cGMP), have been well recognized as critical second messengers within cells. As the first-observed second messenger, cAMP was proven to be engaged in various physiological functions including memory formation, muscle contraction, and endocrine regulation, such as insulin signaling [67–69]. The effect of cAMP largely depends on the activation of cAMP receptor proteins (CRPs) or protein kinase A (PKA), a sensor of the concentration of intracellular cAMP levels that functions in downstream signaling to regulate the biological reactions or genetic expression within the cell [49, 70]. Similar to cAMP, cGMP interacts with some intracellular molecules such as protein kinase G (PKG) to fulfill downstream action [71]. In addition, cyclic purines have been revealed as activators of transcription factors or cation channels, indicating an epigenetic regulatory role of intracellular purines [69].
The integration of the metabolized purines and the regulatory pathway mediated by purines is illustrated in Fig. 1.
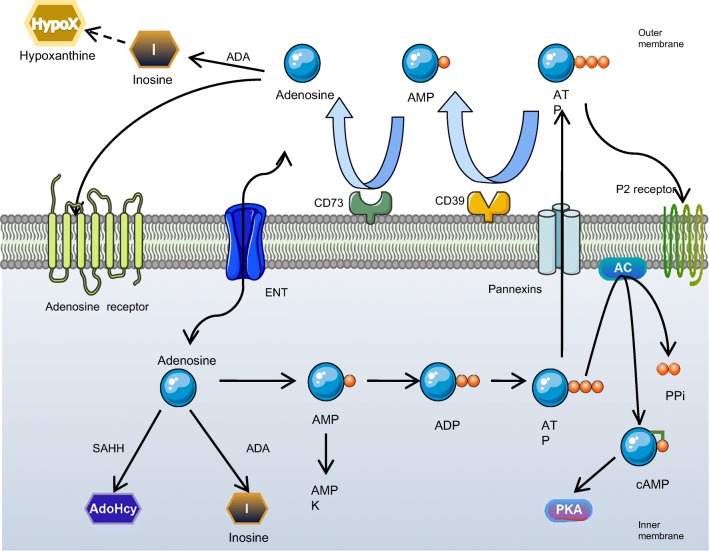
Fig. 1.
An integration of pathways and metabolism of biological purines: intracellular ATP is partly metabolized by adenylate cyclase (AC) to cAMP which could activate PKA. Cells release ATP through pannexin channels under stress. The accumulation of extracellular ATP is dephosphorylated to adenosine by 4 ecto-nucleotidases including CD39, CD73, NPPs, and alkaline phosphatases. Adenosine can further be metabolized by adenosine deaminase (ADA) to inosine (I) and hypoxanthine (HypoX) or function as a signaling molecule by activation of its adenosine receptors on multiple cell types. Once uptake by equilibrative nucleoside transporters (ENTs), adenosine is further metabolized by ADA to inosine, S-adenosylhomocytesine hydrolase (SAHH) to adenosylhomocysteine (AdoHcy) or adenosine kinase (ADK) to AMP, which could stimulate AMPK when [AMP]/[ATP] ratio escalates
Purine-mediated regulation of metabolic reprogramming
According to the classification of purines described above, in this part, the regulation of metabolic reprogramming by extracellular and intracellular purines is summarized.
Extracellular purines
Adenosine pathway
As mentioned above, the adenosine pathway is involved in cellular functions in response to stress, hypoxia, immunity, and inflammation [48, 56, 59]. These response functions, however, link closely to metabolic changes within the cell, and metabolic reprogramming is common under these conditions. The regulatory role of adenosine in metabolic reprogramming within inflammatory or immune cells has been confirmed, including the positive effect of adenosine/ADORA2 signaling in macrophage activation via increased fructose 2,6-bisphosphate and glycolytic flux [22, 72, 73]. Moreover, a series of studies by Sitkovsky and coworkers have demonstrated that adenosine/ADORA2A signaling contributes greatly to the suppression of T cells according to the tumor microenvironment, which is significantly regulated by metabolic reprogramming [31, 74–76]. Since the adenosine pathway can upregulate lipolysis and mobilize energy stores to cope with caloric deficiency, there is a high probability that metabolic reprogramming regulated by adenosine signaling plays a leading role in the progression of tumor cells in energy-deprived environments [77–79]. On the other hand, researchers have attempted to explore how metabolic reprogramming regulates the development of B cells and dendritic cells via the adenosine pathway. They hypothesized that the kind of impact that the adenosine pathway has on metabolic reprogramming of immune cells may explain the complicated mechanism of immune therapy resistance [21, 80]. In addition to contributing to the well-known understanding of metabolic reprogramming in mitotic cells, our group found a novel role for hypoxia-triggered adenosine signaling in erythroid physiology [30, 81, 82]. Using a translation study design and mature metabolomics protocols, we successfully confirmed that adenosine/ADORA2B/AMPK signaling triggers metabolic reprogramming, further inducing 2,3-BPG production in erythrocytes and providing materials for nucleotide synthesis in erythroid progenitor cells, thereby enhancing oxygen release and stress hematopoiesis, respectively, during oxygen crisis [30, 82]. This discovery led to the prospect of purine as a drug target or biomarker for nonmitotic cellular diseases. Intriguingly, these facts and hypotheses are almost accompanied by microenvironmental hypoxia, which has been proven to activate the adenosine pathway and promote metabolic reprogramming. Indeed, serving as the key factor in hypoxia metabolism, HIF might act as the bridge between adenosine signaling and metabolic reprogramming. According to the classification and working mechanism of HIF, the variable stability of HIF-1α and HIF-2α determines the effect of oxygen availability on transcription, leading to a change in biological processes [83]. Initially, studies were focused on the upregulation of purinergic receptors mediated by HIF [56]. However, recent studies have shown that both HIF-1α and HIF-2α can function downstream of purinergic signaling under hypoxic conditions [84–86]. More attention has been paid to HIF-1α owing to its ubiquitous distribution, although HIF-2α is expressed distinctively among tissues [87]. In addition, HIF-1α is reportedly regulated reciprocally by the adenosine pathway, and the downstream factors of HIF include a series of molecules that regulate metabolism, such as glycolytic enzymes and those involved in glucose transport [56, 88–91]. Therefore, we suspect that HIF, along with the adenosine pathway, might function as an indispensable component for the regulation of metabolic reprogramming.
However, the relationships between the adenosine pathway and metabolic reprogramming cannot be simply unidirectional. Apart from the regulatory effect that adenosine has on metabolism, metabolic reprogramming can also contribute to the generation of adenosine and eventually promote the process of adenosine/receptor/metabolic reprogramming [92, 93]. This kind of feedback can greatly amplify the downstream effect. For example, the activated adenosine pathway has been verified as a promoter of the regeneration of pancreatic β cells by inducing glycolysis [94], which potentially adds to the generation of ATP and subsequent adenosine. Thus, targeting the adenosine pathway through metabolic reprogramming would be highly efficient for treatment approaches.
ATP pathway
Considering that the concentration of ATP in the extracellular milieu is extremely low under physiological conditions (in the nanomolar range), even a small amount of ATP release can elicit a relatively strong signal against the background, characterizing ATP-based purinergic signaling as an extracellular messenger system with a high signal-to-noise ratio [95].
The cradle of knowledge about purinergic signaling was neurotransmission, for which ATP was long known to have a well-defined role as a neuromediator. During recent decades, our knowledge of how ATP signaling impacts brain pathology has increased considerably [96]. During a mild nonlethal ischemic episode, astrocytes exert neuroprotective actions via the release of ATP [97]. In addition to this, HIF-1α in astrocytes is reported to be upregulated dependent on ATP binding to P2X7R in response to mild ischemia [98]. Acting as the sensor of ischemic tolerance, P2X7R provides neuroprotection against subsequent severe ischemia [99]. These events are all associated with ATP/P2X7R/ HIF-1α signals in astrocytic activation, suggesting the importance of purinergic signaling for neuroprotection. For further understanding metabolic reprogramming mediated by purinergic signaling in the nervous system, the study of downstream pathways in the brain is urgently needed.
The more we learn about purinergic receptors, the more we realize that purinergic signaling extends well beyond the central or peripheral nervous systems, and in fact, its participation in inflammation and immune response might be even more relevant and pervasive. In these scenarios, P2X7R stands out as a leading player and a promising target for innovative therapeutics [53].
ATP plays a widely accepted role in self-defense, among plants and mammals, as one of the most ancient and conserved danger-associated molecular patterns (DAMPs). DAMPs include a unique class of molecules that are normally sequestered intracellularly and are released only upon cell damage. Once in the extracellular space, DAMPs elicit danger signals and activate the immune system by interacting with specific receptors. P2X7R is unanimously recognized as the main sensor for ATP during inflammation and the main trigger of pro-inflammatory effects [100–102]. Accordingly, its inhibitor, oxidized ATP, is an efficient immune suppressor for preventing transplant rejection [103, 104]. Following a pro-inflammatory response, activation of the inflammasome, a multiprotein complex, is required for an immune response [105]. In addition, negative regulation of GSK3β activity is an accepted mechanism by which P2X7R might contribute to T cell proliferation downstream of T lymphocyte activation [106, 107], since the expression of constitutively active GSK3β decreases T cell proliferation, differentiation, and survival [108, 109]. However, GSK3β is also reported to mediate metabolic reprogramming, specifically enhancing glycolysis and pyruvate metabolism, in TGF-β1-treated ARPE-19 cells [110]. These contradictory findings put us in a quandary such that we cannot exactly determine how GSK works in ATP/P2X7R-mediated metabolic reprogramming. On the other hand, P2X7R is linked to HIF-1α by a biunivocal relationship in which active P2X7R stimulates HIF-1α, but in turn, HIF-1α activation upregulates P2X7R expression [111]. Finally, it remains unclear whether the generation of immunological memory is impacted by the same pathway, but evidence has shown that P2X7R, indeed, plays a hitherto unsuspected and intrinsic role in supporting the generation of long-lived memory CD8+ T cells by driving their mitochondrial maintenance and metabolic reprogramming, in part, in an AMPK-activated manner [112].
The proliferative effect of P2X7R upregulation is described in human leukemic lymphocytes and human neuroblastoma cells [113–115], which relates to an enhanced mitochondrial energy state [116, 117]. P2X7R acts as a key determinant of tumor microenvironment composition due to its combined action on ectonucleotidases, ATP release, and immune cell infiltration [118, 119]. These findings characterize P2X7R as a promising therapeutic target in oncology. Small drug-like P2X7R antagonists have already been taken to clinical trial for select chronic inflammatory diseases. However, rather disappointing results regarding clinical efficacy were obtained [120–122]. The unsatisfactory outcomes may prompt the use of P2X7R-targeted biologics. Anti-P2X7R antibodies are currently under development [123, 124]. Better P2X7R modulators than small-drug agonists may be found.
Interestingly, recent research has demonstrated that ATP release from chemotherapy-treated tumor cells is a potent driver of the antitumor immune response through P2X7R activation on dendritic cells (DCs), which are induced to complete maturation and acquire full competence during antigen presentation [102, 125, 126]. In addition, chemotherapy treatment for AML may result in the induction of an immune suppressive microenvironment. This scenario, at least in part, depends on the local effects of ATP release from dying leukemia cells. And the released ATP may lead to an increase of tolerogenic DCs and leukemia-associated Tregs in a P2X7R-dependent manner [127]. ATP may directly elicit the Treg suppressive function or may be metabolized into adenosine, which enhances Treg activity [119, 128].
Arguably, P2X7R plays an essential role in the setting of chronic stress, which is known to contribute to depression in humans due to the priming of inflammation. In rodents exposed to chronic unpredictable stressors, increases in extracellular ATP release in the hippocampus were observed [129]. Furthermore, antagonistic action and genetic knockout of P2X7 prevented the development of depression-like behaviors in this model [129]. And a similar role for P2X7 has been observed in schizophrenia [130]. All the facts indicate that ATP and P2X7R can, again, be expected to have involvement in the development of this disease, given the strong evidence that purinergic signaling is increasingly involved during the pathology of psychiatric disorders [131, 132]. However, whether metabolic reprogramming participates in psychiatric conditions remains unclear.
In addition to the controversial interaction between biological processes and stimulated ATP-P2X7R signals, further studies reveal how P2X7R works to initiate and regulate metabolic reprogramming. The expression of P2X7R was found to enable better adaptability to unfavorable ambient conditions via upregulation of glycolysis (P2X7R-expressing cells overexpressing PFK and PDHK1) and intracellular glycogen stores (HEK293-P2X7 cells induce the expression of the ubiquitous glucose transporters Glut1 and G3PDH). P2X7R confers a substantial growth advantage by activating a series of important intracellular pathways that contribute to the generation of the energy and components required for the synthesis of amino acids, lipids, and nucleic acids [117].
As for P2YRs and metabolic reprogramming, Yvan first reported that P2YRs are involved in the inhibitory action of ATP on glucose transport [133]. More recently, P2Y6R activation by UDP or MRS2957 was also shown to increase GLUT4 translocation and glucose uptake in primary 3T3-L1 cells [134]. Under pathological conditions, P2YRs were reported to play a regulatory role in metabolic reprogramming in type 2 diabetes mellitus (T2DM) development. In pancreatic islets of humans and other species, P2YRs’ activation has been shown to stimulate insulin secretion in the glucose-dependent manner [135, 136].
An overview of current evidence about the regulatory effects of extracellular purines on metabolic reprogramming is shown in Table 2.
Table. 2.
Extracellular purines and metabolic reprogramming
| Receptors | Cell types | Biological process | Metabolic alterations | Reference |
|---|---|---|---|---|
| P1 receptors | ||||
|
ADORA2A ADORA2B |
Macrophage | Activation and inflammatory response | Glycolytic flux, glycolysis, and OXPHOS | [22, 71, 72] |
| Unknown | DCs, tumor-associated macrophages | Immunosuppressive function, pro-tumor effects | Glycolysis and OXPHOS | [38, 74] |
| ADORA2A | Brown adipocytes | Brown adipose tissue activation, thermogenesis | Lipolysis | [73] |
| ADORA2B | Erythrocytes | Oxygen release | Glycolytic flux and glycolysis | [30, 31] |
| ADORA2B | Hematopoietic progenitor cells | Stress hematopoiesis | Glycolytic flux and glycolysis | [77] |
| P2 receptors | ||||
| P2X7R | HEK293 cells | Cell growth | Glycolysis and glycogen stores | [92] |
| P2YRs | WA and 3T3-L1 cells | Unknown | Glycolytic flux and metabolism | [93, 94] |
OXPHOS, oxidative phosphorylation
Intracellular pathways
AMP pathways
One of the fundamental requirements of all cells is the ability to balance ATP generation and consumption. AMPK, regulated by the [AMP]/[ATP] ratio, is well established as a metabolic regulator [64, 65, 137]. As the [AMP]/[ATP] ratio increases, AMP binds to AMPKγ, which subsequently leads to conformational changes that enable phosphorylation of the α subunit by kinases upstream of AMPK, liver kinase B 1 (LKB1), and calcium/calmodulin-dependent protein kinase kinase-β (CaMKKβ), to activate AMPK [138, 139]. Falling energy levels lead to AMPK activation in an AMP-dependent manner, favoring ATP production by stimulating fatty acid oxidation (FAO), glucose uptake, and glycolysis, as well as by reducing ATP-consuming processes such as transcription and protein synthesis, depending on the tissues and the triggers [140].
In skeletal muscle during prolonged exercise, the AMP-AMPK pathway, which is necessary for efficient muscle contraction, promotes mitochondrial biogenesis and nutrient uptake [65]. In addition to influencing metabolite transport, the pathway also promotes FAO via phosphorylation and inactivation of the second isoform of acetyl-CoA carboxylase (ACC2) [65], leading to decreased malonyl-CoA generation and increased transport of fatty acids. This mechanism has been suggested to exist in multiple cell types, including adipocytes, macrophages, and hepatocytes [141, 142].
AMPK promotes systemic and cellular immunometabolism in an organism. Researchers demonstrated that AMPKα1 (an overriding AMPK isoform in T cells) is essential for Th1 and Th17 cell proliferation and primary T cell responses to viral and bacterial infections in vivo, indicating that AMPK-dependent regulation of metabolic homeostasis is a key modulator of T cell–mediated adaptive immunity [143]. Treatment with (aminooxy) acetic acid (AOA) may increase the phosphorylation of AMPKα in Th17 cells without affecting the mammalian/mechanistic target of rapamycin complex 1 (mTOR) activity [144]. In contrast, pentanoate, a physiologically abundant short-chain fatty acid, likely enhances glycolytic metabolism via the inhibition of AMPK in Th17 cells [145]. All these facts uncovered the complicated underlying mechanism of AMPK-modulating metabolism alterations of T helper cells and have contributed to proposed new perspectives and challenges. A study of AMPK-deficient CD8+ T cells showed that the generation of memory T cells is dependent on AMPK-induced upregulation of oxidative phosphorylation, mitochondrial biogenesis, and FAO [146, 147]. R. D. Michalek used in vitro analysis and showed that AMPK is stimulated in murine TCR-activated Tregs, thereby inhibiting mTOR and promoting lipid oxidation instead of glycolysis. Activated AMPK and targeted induction of lipid metabolism were shown to enhance the number of Tregs in vivo [148]. Collectively, AMPK is recognized as a checkpoint for activated T cells, regulating mRNA translation and glutamine-dependent mitochondrial metabolism to maintain T cell bioenergetics and viability [149]. However, loss of AMPK was found to cause only a slight disturbance in T cell homeostasis in LKB1−/−T cells [143, 150], suggesting that AMPK-dependent regulation of metabolic homeostasis is indeed a modulator of T cell–mediated adaptive immunity, but its specific role remains controversial. Similar to that of T cells, B cell metabolism is also reported to be linked to AMPK activation, which prevents the differentiation of unswitched memory B cells to plasmablasts but supports their differentiation into CD27−IgD− B cells by inhibiting mTOR [23]. Depletion of glucose can also appropriately limit macrophage and effector T cell–driven inflammation and concomitantly initiate M2 macrophage and Treg-dependent TGF-β secretion upon activation of AMPK signaling [151, 152].
High glycolytic flux, coupled with a decreased vascular supply, leads to profound glucose depletion within tumors [153]. Glucose depletion has a profound impact on the function of surrounding immune cells by favoring the growth and differentiation of Tregs, which have higher AMPK activity, lower glucose oxidation, and greater fatty acid oxidation to support energy homeostasis [151, 154, 155]. AMPK is found to have an effect on ovarian cancer [156]. Impaired mitochondrial respiration resulting from glucose deprivation and limited influx of glutamine is also involved in tumor-infiltrating T cells, making them dysfunctional and incapable of controlling malignant progression [157]. Stromal fibroblasts, which can sustain the viability of cancer cells and enhance malignant transformation, can also be reprogrammed by glucose depletion to support tumor growth [158]. Glucose depletion and subsequent AMPK activation in fibroblasts might activate autophagy, driving the secretion of nonessential amino acids (NEAAs) that are significantly depleted in tumor microenvironment [159, 160]. All these facts indicate that AMPK behalves a central role in the metabolic reprogramming of tumor.
Tightly linked to inflammation, tumor, and other pathological states, hypoxia induces ATP depletion and reactive oxygen species (ROS) accumulation [161], which directly activates AMPK and consequently mediates metabolic rewiring to promote survival during energy restriction through activation of catabolic pathways (glucose uptake, glycolysis, FAO, and gluconeogenesis) and suppression of anabolic pathways (protein, glycogen, and fatty acid synthesis) [65, 137, 162, 163]. AMPK is also responsible for the regulation of the redox state by alleviating glucose deprivation–induced NADPH deficiency via decreased FAS and increased FAO [137]. Of note, researchers thought it reasonable to propose that hypoxia could collaboratively drive cancer-related fibroblast function with glucose deprivation [158].
Recent studies revealed that the decline in the sensitivity and responsiveness of AMPK due to age had been shown to be a factor in many age-associated diseases, including cardiovascular diseases, metabolic syndrome, and even Alzheimer disease [164]. However, whether metabolic reprogramming has a role in these diseases is a subject of debate.
Therefore, we wondered how AMPK achieves its metabolism-altering effects. Phosphorylated AMPK could lead to the induction of cellular NAD+ levels, resulting in activation of sirtuin 1 (SIRT1)—an NAD+-dependent deacetylase that positively regulates PGC-1α such that mitochondrial energy metabolism and biogenesis are activated [139, 165, 166]. It is highly suspected that the regulatory role of AMPK is linked to downstream proteins such as mTOR, mitochondrial uncoupling protein 2 (UCP2), carnitine palmitoyltransferase 1C (CPT1C), and p53 [167, 168].
Studies have suggested that treatment with rapamycin, which inhibits mTOR and glycolysis, may be a useful strategy to restrict tumor growth by reversing metabolic reprogramming in tumor cells. However, studies have shown that untargeted rapamycin treatment also impairs the ability of tumor-infiltrating NK cells to kill tumor cells, similar to the effect of obesity [35].
As a highly expressed mitochondrial membrane protein in a variety of tumor cell types, UCP2 is capable of uncoupling mitochondrial respiration and protecting against ROS by reducing mitochondrial respiration [169, 170]. It also serves as a key regulator of energy metabolism that limits glucose oxidation while promoting the oxidation of alternative substrates such as glutamine and fatty acids [171–174]. Activation of AMPK has been shown to be involved in the upregulation of UCP2 expression [175, 176], whereas UCP2 overexpression has been shown to increase signaling from AMPK [177], although in both cases, the underlying mechanisms remain unclear. Based on established facts, activation of AMPK signaling in UCP2-overexpressing cells is associated with a downregulation of HIF expression [177], and hypoxia decreased UCP2 expression via HIF-1-mediated suppression of PPARγ [178]. A recent study clearly indicated that defects in MTO1, the mitochondrial tRNA modification enzyme, lead to reprogramming of cell metabolism mediated by the HIF-PPARγ-UCP2-AMPK axis. MTO1 fibroblasts exhibit HIF-1 activation, downregulation of PPARγ, UCP2, and inactivation of AMPK. Due to metabolic reprogramming, the utilization of fatty acids for the de novo synthesis of fatty acids and β-oxidation appears to be compromised, leading to the accumulation of lipid droplets in MTO1 fibroblasts. However, cells depleted of the GTPBP3 protein produce the opposite effect [179]. In addition, UCP2 has also been found to accommodate a metabolic shift from glucose oxidation to FAO and induce ketogenesis in an AMPK-related manner, resulting in neuron viability alteration and progressive neurodegeneration [180].
Theoretically, competing for AMPK binding sites via the use of an agonist could lead to metabolic turbulence in tumor cells. Metformin, the most widely prescribed T2DM drug, has been shown to modulate metabolism by acting as a mild inhibitor of Complex I in the respiratory chain [181]. Recently, metformin was recognized for its ability to induce ablation of miR-21-5p, leading to the activation of AMPK in addition to the inhibition of mTOR [182], thus showing therapeutic potential for breast cancer. However, Cristina Oliveras-Ferraros and colleagues established a preclinical model of estrogen-dependent MCF-7 cells that were adapted to grow chronically (> 10 months) in the presence of a graded, millimolar mixture of metformin. They found that breast cancer cells, in which metabolic reprogramming has been verified to be related to AMPK, showed acquired resistance to metformin [183, 184]. In other critical findings, metformin could suppress downstream signals such as those for the hypoxia-induced stabilization of HIF-1 by reprogramming oxygen metabolism in hepatocellular carcinoma and playing an AMPK-independent role in T cells [185, 186]. All the facts suggest that metformin may miss the target during clinical treatment by promoting acquired resistance and creating side effects. As another AMPK agonist, 5-aminimidazole-4-carboxamide1-β-D-ribofuranoside (AICAR) is a cell-permeable precursor to ZMP that simulates AMP to bind to the AMPKγ subunit [187]. Finally, a number of natural compounds, such as resveratrol and curcumin, reportedly activate AMPK and confer similar beneficial effects on metabolic reprogramming-related diseases, such as metformin AICAR [185, 188, 189], creating a bright future for the treatment of metabolic diseases.
An illustration of the AMP/AMPK pathway for regulating metabolic reprogramming is shown in Fig. 2.
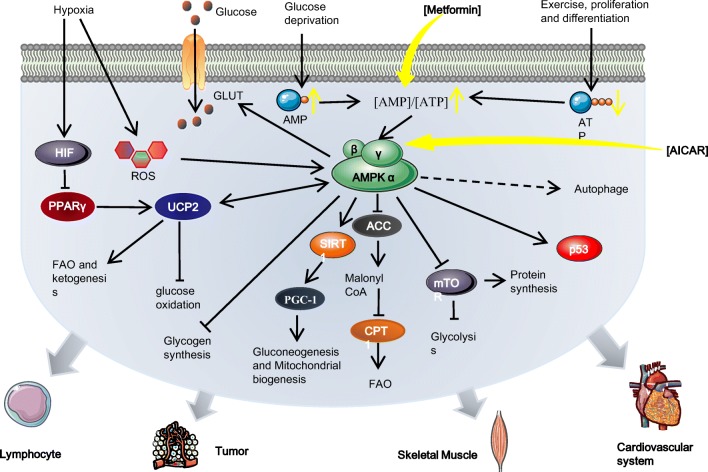
Fig. 2.
The regulatory role AMP plays in metabolic reprograming via activation of AMPK: under hypoxia, glucose deprivation and prolonged exercise in multiple tissues, the [AMP]/[ATP] ratio is elevated, thus initiating direct AMP binding to AMPKγ and stimulating AMPK. The activation of AMPK mediates metabolic reprogramming to promote cell survival during energy restriction by promoting catabolic pathways and suppressing anabolic pathways. Specifically, activated AMPK leads to the stimulation of SIRT1 which positively regulates PGC-1α and subsequent mitochondrial energy metabolism and biogenesis. AMPK activity can sustain glycolysis via the direct inhibition of mTOR. AMPK activation also promotes FAO via phosphorylation and inactivation of ACC2, which limits malonyl-CoA generation. Hypoxia elicits metabolic reprogramming via the accumulation of ROS or the HIF-PPARγ-UCP2-AMPK axis. UCP2 overexpression has been shown to increase signaling from AMPK, resulting in the limitation of glucose oxidation and promotion of FAO and ketogenesis. The AMPK agonist metformin activates AMPK by increasing [AMP]/[ATP] ratio. Another agonist, AICAR, simulates AMP to bind to AMPKγ
Intracellular cyclic purines
Regarding metabolism, the cAMP/PKA pathway has been confirmed to promote metabolic reprogramming in different cell types [190, 191], even in those of nonmammals such as Trypanosoma brucei [192]. Through metabolomics, proteomics, and other omics, Cheng and coworkers proposed that the metabolic reprogramming mediated by cAMP/PKA contributes to the adaptation of Escherichia coli to glycerol [193]. For mammals, the cAMP/PKA pathway enhances the uptake of glucose for metabolic rewiring, thus assisting in the prevention of hepatic steatosis [194, 195]. Additionally, cAMP-mediated metabolic effects may be closely linked to the impact of agmatine during calorie restriction [196]. Lu and coworkers managed to increase glucose uptake by upregulating GLUT expression in corticotropinoma after exposure of AtT-20 cells to 8-bromoadenosine 30,50-cyclic monophosphate (8-Br-cAMP, an analogue of cAMP) [194].
Some transcription factors might be responsible for cAMP-regulated metabolic reprogramming. For example, as an enzyme downstream from AMPK, SIRT1 could also be phosphorylated by PKA and thus induce modification during lipolysis [190]. It is considered a potential mechanism in the physiological thermogenic process. Furthermore, it is reported that cAMP-mediated HIF-1α activation could enhance the glycolysis of pancreatic β-cells and upregulate insulin secretion, creating a promising future for diabetes therapy treatments [197]. However, unlike the potential role of regulatory cAMP in metabolic reprogramming [71], little is known about whether cGMP participates in regulating metabolic reprogramming.
Although a seemingly similar mechanism, cyclic purines function intracellularly, and the various upstream pathways make their effects various and complicated. Together with downstream tissue-specific effector molecules and cellular heterogeneity, targeting cyclic purines might not be sufficient or efficient for metabolic reprogramming manipulation.
Conclusion
In summary, apart from the maintenance of homeostasis, purinergic signaling certainly plays a remarkable regulatory role in metabolic reprogramming. It is worth noting that purines could also be generated within metabolic reprogramming, forming a positive purine-metabolite reprogramming feedback pathway. Intriguingly, although both purinergic signaling and metabolic reprogramming may be ubiquitous, current studies of their relationships are focusing on some classic topics such as oncology or immunology. Therefore, more investigations about other popular themes such as autophagy and aging-related diseases might be required to expand the discussion about these relationships.
On the other hand, how underlying mechanisms express identical purinergic signaling function differently in different cellular metabolism pathways remains a perplexing issue. Due to heterogeneity, purine mediates metabolic reprogramming in various ways. Therefore, to clarify the specific pathways in certain cells and provide targets for diagnosis and treatment, new single-cell metabolomics analysis techniques are in urgent demand. Even though plenty of mysteries about the impact of purines on metabolic reprogramming have made the underlying mechanism complicated, advancements have revealed more surprises in solving some confusion with regard to immunotherapeutic resistance and hematopoiesis regulation. Because of the prosperity from and progress in drugs targeting purinergic signaling, metabolic reprogramming or both, we see a bright future for the translation of these advances.
A summary of the regulatory network of purines, metabolic reprogramming, and biological processes is illustrated in Fig. 3.
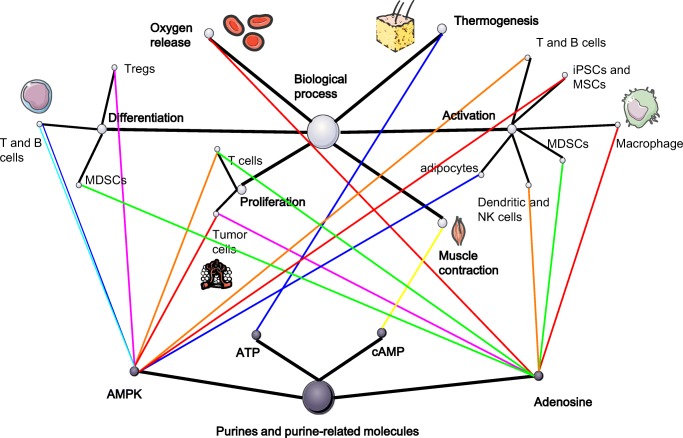
Fig. 3.
A summary of the regulatory network of purines, metabolic reprogramming, and biological process: the dark spots above represent the purines and the light spots represent the various biological process. Finally, the lines linking these two balls represent the metabolic pathways that are regulated by purines and result in the functional alteration. The types of metabolic pathways are represented by the colors of lines, which are illustrated in the lower right corner
Funding information
This work was financially supported by the grants from the Health and Family Planning Commission of Hunan Province (B20180855), Innovation Driven Planning of Central South University(2018CX028), and High-level Talent Planning of Xiangya Hospital.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Human participants and/or animals
This article does not contain any studies with human participants performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhenwei Tang and Wenrui Ye contributed equally to this work.
Contributor Information
Xiang Chen, Email: chenxiangck@126.com.
Hong Liu, Email: hongliu1014@csu.edu.cn.
References
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8(6):519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramaiah A. Pasteur effect and phosphofructokinase. Curr Top Cell Regul. 1974;8:297–345. doi: 10.1016/b978-0-12-152808-9.50014-6. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida GJ. Metabolic reprogramming: the emerging concept and associated therapeutic strategies. J Exp Clin Cancer Res. 2015;34(1):1–10. doi: 10.1186/s13046-015-0221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krebs HA. The Pasteur effect and the relations between respiration and fermentation. Essays Biochem. 1972;8(1):1. [PubMed] [Google Scholar]
- 6.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Libby Catherine J., Tran Anh Nhat, Scott Sarah E., Griguer Corinne, Hjelmeland Anita B. The pro-tumorigenic effects of metabolic alterations in glioblastoma including brain tumor initiating cells. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2018;1869(2):175–188. doi: 10.1016/j.bbcan.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herranz D, et al. Metabolic reprogramming induces resistance to anti-NOTCH1 therapies in acute lymphoblastic leukemia. Nat Med. 2015;21(10):1182. doi: 10.1038/nm.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rozovski U, et al. Metabolism pathways in chronic lymphocytic leukemia. Leuk Lymphoma. 2016;57(4):758–765. doi: 10.3109/10428194.2015.1106533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boroughs LK, Deberardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. 2015;17(4):351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aguilar E, et al. Metabolic reprogramming and dependencies associated with epithelial cancer stem cells independent of the epithelial-mesenchymal transition program. Stem Cells. 2016;34(5):1163. doi: 10.1002/stem.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wettersten HI, Hakimi AA, Morin D, Bianchi C, Johnstone ME, Donohoe DR, Trott JF, Aboud OA, Stirdivant S, Neri B, Wolfert R, Stewart B, Perego R, Hsieh JJ, Weiss RH. Grade-dependent metabolic reprogramming in kidney cancer revealed by combined proteomics and metabolomics analysis. Cancer Res. 2015;75(12):2541–2552. doi: 10.1158/0008-5472.CAN-14-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang HJ, et al. GLUT1 gene is a potential hypoxic marker in colorectal cancer patients. BMC Cancer. 2009;9(1):241. doi: 10.1186/1471-2407-9-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong J, Xiao D, Zhao Z, Ren P, Li C, Hu Y, Shi J, Su H, Wang L, Liu H, Li B, Gao P, Qing G. Epigenetic silencing of microRNA-137 enhances ASCT2 expression and tumor glutamine metabolism. Oncogenesis. 2017;6(7):e356. doi: 10.1038/oncsis.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deberardinis RJ, et al. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Søreide K, Sund M. Epidemiological-molecular evidence of metabolic reprogramming on proliferation, autophagy and cell signaling in pancreas cancer. Cancer Lett. 2015;356(2):281–288. doi: 10.1016/j.canlet.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 17.Sinclair LV, et al. Corrigendum: control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat Immunol. 2013;14(5):500–508. doi: 10.1038/ni.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng H. T cell exit from quiescence and differentiation into Th2 cells depend on raptor-mTORC1-mediated metabolic reprogramming. Immunity. 2013;39(6):1043–1056. doi: 10.1016/j.immuni.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryall JG, Cliff T, Dalton S, Sartorelli V. Metabolic reprogramming of stem cell epigenetics. Cell Stem Cell. 2015;17(6):651–662. doi: 10.1016/j.stem.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eng CH, Abraham RT. The autophagy conundrum in cancer: influence of tumorigenic metabolic reprogramming. Oncogene. 2011;30(47):4687–4696. doi: 10.1038/onc.2011.220. [DOI] [PubMed] [Google Scholar]
- 21.Subhra NBSP, Biswas K. Metabolic reprogramming of immune cells in cancer progression. Immunity. 2015;43(3):435. doi: 10.1016/j.immuni.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Kelly B, O'Neill LA. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 2015;25(7):771–784. doi: 10.1038/cr.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torigoe M, Iwata S (2017) Metabolic Reprogramming Commits Differentiation of Human CD27(+)IgD(+) B Cells to Plasmablasts or CD27(-)IgD(-) Cells. J Immunol 199(2):425–434 [DOI] [PubMed]
- 24.Deng J, et al. Homocysteine activates B cells via regulating PKM2-dependent metabolic reprogramming. J Immunol. 2016;198(1):170. doi: 10.4049/jimmunol.1600613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Khami Amir A., Rodriguez Paulo C., Ochoa Augusto C. Metabolic reprogramming of myeloid-derived suppressor cells (MDSC) in cancer. OncoImmunology. 2016;5(8):e1200771. doi: 10.1080/2162402X.2016.1200771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keating SE, Zaiatz-Bittencourt V, Loftus RM, Keane C, Brennan K, Finlay DK, Gardiner CM. Metabolic reprogramming supports IFN-γ production by CD56bright NK cells. J Immunol. 2016;196(6):2552–2560. doi: 10.4049/jimmunol.1501783. [DOI] [PubMed] [Google Scholar]
- 27.Hsu YC, Chen CT, Wei YH. Mitochondrial resetting and metabolic reprogramming in induced pluripotent stem cells and mitochondrial disease modeling. Biochim Biophys Acta. 2016;1860(4):686–693. doi: 10.1016/j.bbagen.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 28.Chen C, Tang Q, Zhang Y, Dai M, Jiang Y, Wang H, Yu M, Jing W, Tian W. Metabolic reprogramming by HIF-1 activation enhances survivability of human adipose-derived stem cells in ischaemic microenvironments. Cell Prolif. 2017;50(5):e12363. doi: 10.1111/cpr.12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oburoglu, et al. Glucose and glutamine metabolism regulate human hematopoietic stem cell lineage specification. Cell Stem Cell. 2014;15(2):169–184. doi: 10.1016/j.stem.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Liu H, Zhang Y, Wu H, D’Alessandro A, Yegutkin GG, Song A, Sun K, Li J, Cheng NY, Huang A, Edward Wen Y, Weng TT, Luo F, Nemkov T, Sun H, Kellems RE, Karmouty-Quintana H, Hansen KC, Zhao B, Subudhi AW, Jameson-van Houten S, Julian CG, Lovering AT, Eltzschig HK, Blackburn MR, Roach RC, Xia Y. Beneficial role of erythrocyte adenosine A2B receptor-mediated AMP-activated protein kinase activation in high-altitude hypoxia. Circulation. 2016;134(5):405–421. doi: 10.1161/CIRCULATIONAHA.116.021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kjaergaard J, et al. A 2A adenosine receptor gene deletion or synthetic A 2A antagonist liberate tumor-reactive CD8 + T cells from tumor-induced immunosuppression. J Immunol. 2018;201(2):782–791. doi: 10.4049/jimmunol.1700850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carey BW, et al. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2014;518(7539):413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryall JG, Dell’Orso S, Derfoul A, Juan A, Zare H, Feng X, Clermont D, Koulnis M, Gutierrez-Cruz G, Fulco M, Sartorelli V. The NAD(+)-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell Stem Cell. 2015;16(2):171–183. doi: 10.1016/j.stem.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shyhchang N, et al. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell. 2013;155(4):778–792. doi: 10.1016/j.cell.2013.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michelet Xavier, Dyck Lydia, Hogan Andrew, Loftus Roisin M., Duquette Danielle, Wei Kevin, Beyaz Semir, Tavakkoli Ali, Foley Cathriona, Donnelly Raymond, O’Farrelly Cliona, Raverdeau Mathilde, Vernon Ashley, Pettee William, O’Shea Donal, Nikolajczyk Barbara S., Mills Kingston H. G., Brenner Michael B., Finlay David, Lynch Lydia. Metabolic reprogramming of natural killer cells in obesity limits antitumor responses. Nature Immunology. 2018;19(12):1330–1340. doi: 10.1038/s41590-018-0251-7. [DOI] [PubMed] [Google Scholar]
- 36.Chang CH, Curtis JD, Maggi LB, Jr, Faubert B, Villarino AV, O’Sullivan D, Huang SCC, van der Windt GJW, Blagih J, Qiu J, Weber JD, Pearce EJ, Jones RG, Pearce EL. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153(6):1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerriets VA, Kishton RJ, Johnson MO, Cohen S, Siska PJ, Nichols AG, Warmoes MO, de Cubas AA, MacIver NJ, Locasale JW, Turka LA, Wells AD, Rathmell JC. Foxp3 and toll-like receptor signaling balance Treg cell anabolic metabolism for suppression. Nat Immunol. 2016;17(12):1459–1466. doi: 10.1038/ni.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sica A, Strauss L. Energy metabolism drives myeloid-derived suppressor cell differentiation and functions in pathology. J Leukoc Biol. 2017;102(2):325–334. doi: 10.1189/jlb.4MR1116-476R. [DOI] [PubMed] [Google Scholar]
- 39.Bettencourt IA, Powell JD. Targeting metabolism as a novel therapeutic approach to autoimmunity, inflammation, and transplantation. J Immunol. 2017;198(3):999. doi: 10.4049/jimmunol.1601318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Binger KJ, Côrtereal BF (2017) Immunometabolic Regulation of Interleukin-17-Producing T Helper Cells: Uncoupling New Targets for Autoimmunity. Front Immunol 8:311 [DOI] [PMC free article] [PubMed]
- 41.Huang L, Xu H, Peng G. TLR-mediated metabolic reprogramming in the tumor microenvironment: potential novel strategies for cancer immunotherapy. Cell Mol Immunol. 2018;15:428–437. doi: 10.1038/cmi.2018.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. J Clin Invest. 2013;123(9):3685–3692. doi: 10.1172/JCI69741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patsoukis N, Bardhan K, Chatterjee P, Sari D, Liu B, Bell LN, Karoly ED, Freeman GJ, Petkova V, Seth P, Li L, Boussiotis VA. PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat Commun. 2015;6:6692. doi: 10.1038/ncomms7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noman MZ, et al. PD-L1 is a novel direct target of HIF-1α, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J Exp Med. 2014;211(5):781–790. doi: 10.1084/jem.20131916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun K, et al. Sphingosine-1-phosphate promotes erythrocyte glycolysis and oxygen release for adaptation to high-altitude hypoxia. Nat Commun. 2016;7:12086. doi: 10.1038/ncomms12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burnstock G. Purinergic nerves. Pharmacol Rev. 1972;24(24):509–581. [PubMed] [Google Scholar]
- 47.Knowles JR. Enzyme-catalyzed phosphoryl transfer reactions. Annu Rev Biochem. 1980;49(1):877–919. doi: 10.1146/annurev.bi.49.070180.004305. [DOI] [PubMed] [Google Scholar]
- 48.Liu H, Xia Y. Beneficial and detrimental role of adenosine signaling in diseases and therapy. J Appl Physiol (1985) 2015;119(10):1173. doi: 10.1152/japplphysiol.00350.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mcdonough KA, Rodriguez A. The myriad roles of cyclic AMP in microbial pathogens: from signal to sword. Nat Rev Microbiol. 2012;10(1):27–38. doi: 10.1038/nrmicro2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferrari D, et al. Purinergic signaling during immune cell trafficking. Trends Immunol. 2016;37(6):399–411. doi: 10.1016/j.it.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 51.Antonioli L, et al. Regulation of enteric functions by adenosine: pathophysiological and pharmacological implications. Pharmacol Ther. 2008;120(3):233–253. doi: 10.1016/j.pharmthera.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 52.Surprenant A, Rassendren F, Kawashima E, North RA, Buell G. The cytolytic P2Z receptor for extracellular ATP identified as a P2X receptor (P2X7) Science. 1996;272(5262):735–738. doi: 10.1126/science.272.5262.735. [DOI] [PubMed] [Google Scholar]
- 53.Adinolfi E, et al. The P2X7 receptor: a main player in inflammation. Biochem Pharmacol. 2017;151:S0006295217307335. doi: 10.1016/j.bcp.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 54.Eltzschig HK. Extracellular adenosine signaling in molecular medicine. J Mol Med (Berl) 2013;91(2):141–146. doi: 10.1007/s00109-013-0999-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geldenhuys WJ, et al. Exploring adenosine receptor ligands: potential role in the treatment of cardiovascular diseases. Molecules. 2017;22(6):917. doi: 10.3390/molecules22060917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haskó G, Csóka B, Németh ZH, Vizi ES, Pacher P. A2B adenosine receptors in immunity and inflammation. Trends Immunol. 2009;30(6):263–270. doi: 10.1016/j.it.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abbracchio MP, Burnstock G. Purinoceptors: are there families of P2X and P2Y purinoceptors? Pharmacol Ther. 1994;64(3):445–475. doi: 10.1016/0163-7258(94)00048-4. [DOI] [PubMed] [Google Scholar]
- 58.Jarvis MF, Khakh BS. ATP-gated P2X cation-channels. Neuropharmacology. 2009;56(1):208–215. doi: 10.1016/j.neuropharm.2008.06.067. [DOI] [PubMed] [Google Scholar]
- 59.Idzko M, Ferrari D, Eltzschig HK. Nucleotide signalling during inflammation. Nature. 2014;509(7500):310–317. doi: 10.1038/nature13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cekic C, Linden J. Purinergic regulation of the immune system. Nat Rev Immunol. 2016;16(3):177–192. doi: 10.1038/nri.2016.4. [DOI] [PubMed] [Google Scholar]
- 61.Nishimura A, Sunggip C, Oda S, Numaga-Tomita T, Tsuda M, Nishida M. Purinergic P2Y receptors: molecular diversity and implications for treatment of cardiovascular diseases. Pharmacol Ther. 2017;180:113–128. doi: 10.1016/j.pharmthera.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 62.Welihinda AA, et al. Enhancement of inosine-mediated A 2A R signaling through positive allosteric modulation. Cell Signal. 2017;42:227–235. doi: 10.1016/j.cellsig.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yarom M, et al. Identification of inosine as an endogenous modulator for the benzodiazepine binding site of the GABAA receptors. J Biomed Sci. 1998;5(4):274–280. doi: 10.1007/BF02255859. [DOI] [PubMed] [Google Scholar]
- 64.Steinberg GR, Kemp BE. AMPK in health and disease. Physiol Rev. 2009;89(3):1025–1078. doi: 10.1152/physrev.00011.2008. [DOI] [PubMed] [Google Scholar]
- 65.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13(4):251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kahn BB, et al. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 2005;1(1):15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 67.Ricciarelli R, Fedele E. cAMP, cGMP and amyloid β: three ideal partners for memory formation. Trends Neurosci. 2018;41:255–266. doi: 10.1016/j.tins.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 68.Rasmussen H, Kelley G, Douglas JS. Interactions between Ca2+ and cAMP messenger system in regulation of airway smooth muscle contraction. Am J Phys. 1990;258(6 Pt 1):L279. doi: 10.1152/ajplung.1990.258.6.L279. [DOI] [PubMed] [Google Scholar]
- 69.Kresge N, Simoni RD, Hill RL. Earl W. Sutherland’s discovery of cyclic adenine monophosphate and the second messenger system. J Biol Chem. 2005;19(280):e39. [Google Scholar]
- 70.Skalhegg BS, Tasken K. Specificity in the cAMP/PKA signaling pathway. differential expression, regulation, and subcellular localization of subunits of PKA. Front Biosci. 2000;5(1):D678. doi: 10.2741/skalhegg. [DOI] [PubMed] [Google Scholar]
- 71.Buglioni Alessia, Burnett John C. New Pharmacological Strategies to Increase cGMP. Annual Review of Medicine. 2016;67(1):229–243. doi: 10.1146/annurev-med-052914-091923. [DOI] [PubMed] [Google Scholar]
- 72.Fan F, et al. Age-associated metabolic dysregulation in bone marrow-derived macrophages stimulated with lipopolysaccharide. Sci Rep. 2016;6:22637. doi: 10.1038/srep22637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruizgarcía A, et al. Cooperation of adenosine with macrophage Toll-4 receptor agonists leads to increased glycolytic flux through the enhanced expression of PFKFB3 gene. J Biol Chem. 2011;286(22):19247–19258. doi: 10.1074/jbc.M110.190298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hatfield SM, Sitkovsky M. A2A adenosine receptor antagonists to weaken the hypoxia-HIF-1α driven immunosuppression and improve immunotherapies of cancer. Curr Opin Pharmacol. 2016;29:90–96. doi: 10.1016/j.coph.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Akio O, et al. The development and immunosuppressive functions of CD4+CD25+FoxP3+regulatory T cells are under influence of the adenosine-A2A adenosine receptor pathway. Front Immunol. 2012;3(3):190–190. doi: 10.3389/fimmu.2012.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hatfield SM, et al. Immunological mechanisms of the antitumor effects of supplemental oxygenation. Sci Transl Med. 2015;7(277):277ra30. doi: 10.1126/scitranslmed.aaa1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gnad T, et al. Adenosine activates brown adipose tissue and recruits beige adipocytes via A2A receptors. Nature. 2014;516(7531):395–399. doi: 10.1038/nature13816. [DOI] [PubMed] [Google Scholar]
- 78.Ohta A. A metabolic immune checkpoint: adenosine in tumor microenvironment. Front Immunol. 2016;7(Pt 1):109. doi: 10.3389/fimmu.2016.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bantug GR, Galluzzi L, Kroemer G, Hess C. The spectrum of T cell metabolism in health and disease. Nat Rev Immunol. 2018;18(1):19–34. doi: 10.1038/nri.2017.99. [DOI] [PubMed] [Google Scholar]
- 80.Burrows N, Maxwell PH. Hypoxia and B cells. Exp Cell Res. 2017;356(2):197–203. doi: 10.1016/j.yexcr.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 81.Song A, Zhang Y, Han L, Yegutkin GG, Liu H, Sun K, D’Alessandro A, Li J, Karmouty-Quintana H, Iriyama T, Weng T, Zhao S, Wang W, Wu H, Nemkov T, Subudhi AW, Jameson-van Houten S, Julian CG, Lovering AT, Hansen KC, Zhang H, Bogdanov M, Dowhan W, Jin J, Kellems RE, Eltzschig HK, Blackburn M, Roach RC, Xia Y. Erythrocytes retain hypoxic adenosine response for faster acclimatization upon re-ascent. Nat Commun. 2017;8:14108. doi: 10.1038/ncomms14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu Hong, Zhang Yujin, D'Alessandro Angelo, Nemkov Travis, Couturier Jacob, Zhao Shushan, Hansen Kirk C, Lovering Andrew T., Roach Robert, Kellems Rodney E., Eltzschig Holger K., Blackburn Michael R., Xia Yang. Adenosine Signaling-Mediated Metabolic Reprogramming Regulates Erythropoiesis. Blood. 2016;128(22):2437–2437. [Google Scholar]
- 83.Palazon A, et al. HIF transcription factors, inflammation, and immunity. Immunity. 2014;41(4):518–528. doi: 10.1016/j.immuni.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stefania M, et al. A3 adenosine receptors modulate hypoxia-inducible factor-1alpha expression in human A375 melanoma cells. Neoplasia. 2005;7(10):894–903. doi: 10.1593/neo.05334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gessi S, et al. A 1 and a 3 adenosine receptors inhibit LPS-induced hypoxia-inducible factor-1 accumulation in murine astrocytes. Pharmacol Res. 2013;76(10):157–170. doi: 10.1016/j.phrs.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 86.Torres A, Erices JI, Sanchez F, Ehrenfeld P, Turchi L, Virolle T, Uribe D, Niechi I, Spichiger C, Rocha JD, Ramirez M, Salazar-Onfray F, San Martín R, Quezada C. Extracellular adenosine promotes cell migration/invasion of glioblastoma stem-like cells through A3 adenosine receptor activation under hypoxia. Cancer Lett. 2019;446:112–122. doi: 10.1016/j.canlet.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 87.Lagory EL, Giaccia AJ. The ever-expanding role of HIF in tumour and stromal biology. Nat Cell Biol. 2016;18(4):356. doi: 10.1038/ncb3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fraisl P, Aragonés J, Carmeliet P. Inhibition of oxygen sensors as a therapeutic strategy for ischaemic and inflammatory disease. Nat Rev Drug Discov. 2009;8(2):139. doi: 10.1038/nrd2761. [DOI] [PubMed] [Google Scholar]
- 89.Sitkovsky M, Lukashev D. Regulation of immune cells by local-tissue oxygen tension: HIF1 alpha and adenosine receptors. Nat Rev Immunol. 2005;5(9):712–721. doi: 10.1038/nri1685. [DOI] [PubMed] [Google Scholar]
- 90.Xie H, Simon MC. Oxygen availability and metabolic reprogramming in cancer. J Biol Chem. 2017;292(41):jbc.R117.799973. doi: 10.1074/jbc.R117.799973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen Changhu, Pore Nabendu, Behrooz Alireza, Ismail-Beigi Faramarz, Maity Amit. Regulation ofglut1mRNA by Hypoxia-inducible Factor-1. Journal of Biological Chemistry. 2000;276(12):9519–9525. doi: 10.1074/jbc.M010144200. [DOI] [PubMed] [Google Scholar]
- 92.Borg N, Alter C, Görldt N, Jacoby C, Ding Z, Steckel B, Quast C, Bönner F, Friebe D, Temme S, Flögel U, Schrader J. CD73 on T-cells orchestrates cardiac wound healing after myocardial infarction by purinergic metabolic reprogramming. Circulation. 2017;136:297–313. doi: 10.1161/CIRCULATIONAHA.116.023365. [DOI] [PubMed] [Google Scholar]
- 93.Wang X, Yang K, Xie Q, Wu Q, Mack SC, Shi Y, Kim LJY, Prager BC, Flavahan WA, Liu X, Singer M, Hubert CG, Miller TE, Zhou W, Huang Z, Fang X, Regev A, Suvà ML, Hwang TH, Locasale JW, Bao S, Rich JN. Purine synthesis promotes maintenance of brain tumor initiating cells in glioma. Nat Neurosci. 2017;20(5):661–673. doi: 10.1038/nn.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Andersson O, Adams BA, Yoo D, Ellis GC, Gut P, Anderson RM, German MS, Stainier DYR. Adenosine signaling promotes regeneration of pancreatic β cells in vivo. Cell Metab. 2012;15(6):885–894. doi: 10.1016/j.cmet.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yegutkin GG. Enzymes involved in metabolism of extracellular nucleotides and nucleosides: functional implications and measurement of activities. Crit Rev Biochem Mol Biol. 2014;49(6):473–497. doi: 10.3109/10409238.2014.953627. [DOI] [PubMed] [Google Scholar]
- 96.Jimenez-Mateos EM, Smith J, Nicke A, Engel T (2018) Regulation of P2X7 receptor expression and function in the brain. Brain Res Bull S0361-9230(18):30734–2 [DOI] [PubMed]
- 97.Liu HT, Sabirov RZ, Okada Y. Oxygen-glucose deprivation induces ATP release via maxi-anion channels in astrocytes. Purinergic Signal. 2008;4(2):147–154. doi: 10.1007/s11302-007-9077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hirayama Y, Koizumi S. Hypoxia-independent mechanisms of HIF-1α expression in astrocytes after ischemic preconditioning. Glia. 2017;65(3):523–530. doi: 10.1002/glia.23109. [DOI] [PubMed] [Google Scholar]
- 99.Lizhen L, et al. Protective role of reactive astrocytes in brain ischemia. J Cereb Blood Flow Metab. 2008;28(3):468–481. doi: 10.1038/sj.jcbfm.9600546. [DOI] [PubMed] [Google Scholar]
- 100.Ferrari D, Pizzirani C, Adinolfi E, Lemoli RM, Curti A, Idzko M, Panther E, di Virgilio F. The P2X7 receptor: a key player in IL-1 processing and release. J Immunol. 2006;176(7):3877–3883. doi: 10.4049/jimmunol.176.7.3877. [DOI] [PubMed] [Google Scholar]
- 101.Alessandra P, et al. ATP is released by monocytes stimulated with pathogen-sensing receptor ligands and induces IL-1beta and IL-18 secretion in an autocrine way. Proc Natl Acad Sci U S A. 2008;105(23):8067–8072. doi: 10.1073/pnas.0709684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ghiringhelli F, Apetoh LA. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1beta-dependent adaptive immunity against tumors. Nat Med. 2009;15(10):1170–1178. doi: 10.1038/nm.2028. [DOI] [PubMed] [Google Scholar]
- 103.Andrea V, et al. Effect of the purinergic inhibitor oxidized ATP in a model of islet allograft rejection. Diabetes. 2013;62(5):1665–1675. doi: 10.2337/db12-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vergani A, Tezza S, D’Addio F, Fotino C, Liu K, Niewczas M, Bassi R, Molano RD, Kleffel S, Petrelli A, Soleti A, Ammirati E, Frigerio M, Visner G, Grassi F, Ferrero ME, Corradi D, Abdi R, Ricordi C, Sayegh MH, Pileggi A, Fiorina P. Long-term heart transplant survival by targeting the ionotropic purinergic receptor P2X7. Circulation. 2013;127(4):463–475. doi: 10.1161/CIRCULATIONAHA.112.123653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Di VF. The therapeutic potential of modifying inflammasomes and NOD-like receptors. Pharmacol Rev. 2013;65(3):872–905. doi: 10.1124/pr.112.006171. [DOI] [PubMed] [Google Scholar]
- 106.Amoroso F, Capece M, Rotondo A, Cangelosi D, Ferracin M, Franceschini A, Raffaghello L, Pistoia V, Varesio L, Adinolfi E. The P2X7 receptor is a key modulator of the PI3K/GSK3β/VEGF signaling network: evidence in experimental neuroblastoma. Oncogene. 2015;34(41):5240–5251. doi: 10.1038/onc.2014.444. [DOI] [PubMed] [Google Scholar]
- 107.Gómez-Villafuertes R, et al. Ca2+/calmodulin-dependent kinase II signalling cascade mediates P2X7 receptor-dependent inhibition of neuritogenesis in neuroblastoma cells. FEBS J. 2010;276(18):5307–5325. doi: 10.1111/j.1742-4658.2009.07228.x. [DOI] [PubMed] [Google Scholar]
- 108.Beurel E, Michalek SM, Jope RS. Innate and adaptive immune responses regulated by glycogen synthase kinase-3 (GSK3) Trends Immunol. 2010;31(1):24–31. doi: 10.1016/j.it.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Martin M, et al. Toll-like receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat Immunol. 2005;6(8):777–784. doi: 10.1038/ni1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huang L, Zhang C, Su L, Song Z. GSK3β attenuates TGF-β1 induced epithelial–mesenchymal transition and metabolic alterations in ARPE-19 cells. Biochem Biophys Res Commun. 2017;486(3):744–751. doi: 10.1016/j.bbrc.2017.03.113. [DOI] [PubMed] [Google Scholar]
- 111.Tafani M, de Santis E, Coppola L, Perrone GA, Carnevale I, Russo A, Pucci B, Carpi A, Bizzarri M, Russo MA. Bridging hypoxia, inflammation and estrogen receptors in thyroid cancer progression. Biomed Pharmacother. 2014;68(1):1–5. doi: 10.1016/j.biopha.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 112.Borges da Silva Henrique, Beura Lalit K., Wang Haiguang, Hanse Eric A., Gore Reshma, Scott Milcah C., Walsh Daniel A., Block Katharine E., Fonseca Raissa, Yan Yan, Hippen Keli L., Blazar Bruce R., Masopust David, Kelekar Ameeta, Vulchanova Lucy, Hogquist Kristin A., Jameson Stephen C. The purinergic receptor P2RX7 directs metabolic fitness of long-lived memory CD8+ T cells. Nature. 2018;559(7713):264–268. doi: 10.1038/s41586-018-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Adinolfi E, et al. P2X7 receptor expression in evolutive and indolent forms of chronic B lymphocytic leukemia. Blood. 2002;99(2):706–708. doi: 10.1182/blood.v99.2.706. [DOI] [PubMed] [Google Scholar]
- 114.Raffaghello L, Chiozzi P, Falzoni S, di Virgilio F, Pistoia V. The P2X7 receptor sustains the growth of human neuroblastoma cells through a substance P-dependent mechanism. Cancer Res. 2006;66(2):907–914. doi: 10.1158/0008-5472.CAN-05-3185. [DOI] [PubMed] [Google Scholar]
- 115.Baricordi OR, Melchiorri L, Adinolfi E, Falzoni S, Chiozzi P, Buell G, di Virgilio F. Increased proliferation rate of lymphoid cells transfected with the P2X(7) ATP receptor. J Biol Chem. 1999;274(47):33206–33208. doi: 10.1074/jbc.274.47.33206. [DOI] [PubMed] [Google Scholar]
- 116.Adinolfi E, Callegari MG, Ferrari D, Bolognesi C, Minelli M, Wieckowski MR, Pinton P, Rizzuto R, di Virgilio F. Basal activation of the P2X7 ATP receptor elevates mitochondrial calcium and potential, increases cellular ATP levels, and promotes serum-independent growth. Mol Biol Cell. 2005;16(7):3260–3272. doi: 10.1091/mbc.E04-11-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Amoroso F, Falzoni S, Adinolfi E, Ferrari D, di Virgilio F. The P2X7 receptor is a key modulator of aerobic glycolysis. Cell Death Differ. 2012;3(8):e370. doi: 10.1038/cddis.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.De Marchi Elena, Orioli Elisa, Pegoraro Anna, Sangaletti Sabina, Portararo Paola, Curti Antonio, Colombo Mario Paolo, Di Virgilio Francesco, Adinolfi Elena. The P2X7 receptor modulates immune cells infiltration, ectonucleotidases expression and extracellular ATP levels in the tumor microenvironment. Oncogene. 2019;38(19):3636–3650. doi: 10.1038/s41388-019-0684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sara T, et al. Extracellular ATP exerts opposite effects on activated and regulatory CD4+ T cells via purinergic P2 receptor activation. J Immunol. 2012;189(3):1303–1310. doi: 10.4049/jimmunol.1103800. [DOI] [PubMed] [Google Scholar]
- 120.Di VF, et al. The P2X7 receptor in infection and inflammation. Immunity. 2017;47(1):15. doi: 10.1016/j.immuni.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 121.North RA. P2X receptors. Philos Trans R Soc Lond. 2016;371(1700):20150427. doi: 10.1098/rstb.2015.0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.De ME, et al. P2X7 receptor as a therapeutic target. Adv Protein Chem Struct Biol. 2016;104:39. doi: 10.1016/bs.apcsb.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 123.Danquah W, et al. Nanobodies that block gating of the P2X7 ion channel ameliorate inflammation. Sci Transl Med. 2016;8(366):366ra162. doi: 10.1126/scitranslmed.aaf8463. [DOI] [PubMed] [Google Scholar]
- 124.Gilbert SM, et al. A phase 1 clinical trial demonstrates nfP2X 7 targeted antibodies provide a novel, safe and tolerable topical therapy for BCC. Br J Dermatol. 2017;177(1):117–124. doi: 10.1111/bjd.15364. [DOI] [PubMed] [Google Scholar]
- 125.Laetitia A, et al. Tumor cell death and ATP release prime dendritic cells and efficient anticancer immunity. Cancer Res. 2010;70(3):855–858. doi: 10.1158/0008-5472.CAN-09-3566. [DOI] [PubMed] [Google Scholar]
- 126.Ma Y, et al. Anticancer chemotherapy-induced intratumoral recruitment and differentiation of antigen-presenting cells. Immunity. 2013;38(4):729–741. doi: 10.1016/j.immuni.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 127.Lecciso M, et al. ATP release from chemotherapy-treated dying leukemia cells elicits an immune suppressive effect by increasing regulatory T cells and tolerogenic dendritic cells. Front Immunol. 2017;8:1918. doi: 10.3389/fimmu.2017.01918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Giovanna B, et al. Expression of ectonucleotidase CD39 by Foxp3+ Treg cells: hydrolysis of extracellular ATP and immune suppression. Blood. 2007;110(4):1225–1232. doi: 10.1182/blood-2006-12-064527. [DOI] [PubMed] [Google Scholar]
- 129.Yue N, et al. Activation of P2X7 receptor and NLRP3 inflammasome assembly in hippocampal glial cells mediates chronic stress-induced depressive-like behaviors. J Neuroinflammation. 2017;14(1):102. doi: 10.1186/s12974-017-0865-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Laskaris LE, et al. Microglial activation and progressive brain changes in schizophrenia. Br J Pharmacol. 2016;173(4):666–680. doi: 10.1111/bph.13364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mondelli V, et al. Brain microglia in psychiatric disorders. Lancet Psychiatry. 2017;4(7):S2215036617301013. doi: 10.1016/S2215-0366(17)30101-3. [DOI] [PubMed] [Google Scholar]
- 132.Kesteren CFMGV, et al. Immune involvement in the pathogenesis of schizophrenia: a meta-analysis on postmortem brain studies. Transl Psychiatry. 2017;7(3):e1075. doi: 10.1038/tp.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fischer Y, Becker C, Löken C, et al. J Biol Chem. 1999;274(2):755–761. doi: 10.1074/jbc.274.2.755. [DOI] [PubMed] [Google Scholar]
- 134.Balasubramanian R, et al. Enhancement of glucose uptake in mouse skeletal muscle cells and adipocytes by P2Y6 receptor agonists. PLoS One. 2014;9(12):e116203–e116203. doi: 10.1371/journal.pone.0116203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hillairebuys D, et al. Stimulation of insulin secretion and improvement of glucose tolerance in rat and dog by the P2y-purinoceptor agonist, adenosine-5'-O-(2-thiodiphosphate) Br J Pharmacol. 1993;109(1):183–187. doi: 10.1111/j.1476-5381.1993.tb13551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fernandez-Alvarez J, Hillaire-Buys D, Loubatières-Mariani MM, Gomis R, Petit P. P2 receptor agonists stimulate insulin release from human pancreatic islets. Pancreas. 2001;22(1):69–71. doi: 10.1097/00006676-200101000-00012. [DOI] [PubMed] [Google Scholar]
- 137.Hardie DG. AMPK: positive and negative regulation, and its role in whole-body energy homeostasis. Curr Opin Cell Biol. 2015;33:1–7. doi: 10.1016/j.ceb.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 138.Sebbagh M, Santoni MJ, Hall B, Borg JP, Schwartz MA. Regulation of LKB1/STRAD localization and function by E-cadherin. Curr Biol. 2009;19(1):37–42. doi: 10.1016/j.cub.2008.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chiaranunt P, Ferrara JL, Byersdorfer CA. Rethinking the paradigm: how comparative studies on fatty acid oxidation inform our understanding of T cell metabolism. Mol Immunol. 2015;68(2):564–574. doi: 10.1016/j.molimm.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Horman S, Beauloye C, Vanoverschelde JL, Bertrand L. AMP-activated protein kinase in the control of cardiac metabolism and remodeling. Curr Heart Fail Rep. 2012;9(3):164–173. doi: 10.1007/s11897-012-0102-z. [DOI] [PubMed] [Google Scholar]
- 141.Steinberg GR, Schertzer JD. AMPK promotes macrophage fatty acid oxidative metabolism to mitigate inflammation: implications for diabetes and cardiovascular disease. Immunol Cell Biol. 2014;92(4):340–345. doi: 10.1038/icb.2014.11. [DOI] [PubMed] [Google Scholar]
- 142.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13(9):1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mayer A, et al. AMP-activated protein kinase regulates lymphocyte responses to metabolic stress but is largely dispensable for immune cell development and function. Eur J Immunol. 2008;38(4):948–956. doi: 10.1002/eji.200738045. [DOI] [PubMed] [Google Scholar]
- 144.Xu T, Stewart KM, Wang X, Liu K, Xie M, Ryu JK, Li K, Ma T, Wang H, Ni L, Zhu S, Cao N, Zhu D, Zhang Y, Akassoglou K, Dong C, Driggers EM, Ding S. Metabolic control of TH17 and induced Treg cell balance by an epigenetic mechanism. Nature. 2017;548(7666):228–233. doi: 10.1038/nature23475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Luu M, Pautz S, Kohl V, Singh R, Romero R, Lucas S, Hofmann J, Raifer H, Vachharajani N, Carrascosa LC, Lamp B, Nist A, Stiewe T, Shaul Y, Adhikary T, Zaiss MM, Lauth M, Steinhoff U, Visekruna A. The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat Commun. 2019;10(1):760. doi: 10.1038/s41467-019-08711-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Julia R, et al. AMPKα1: a glucose sensor that controls CD8 T-cell memory. Eur J Immunol. 2013;43(4):889–896. doi: 10.1002/eji.201243008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gj VDW, et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36(1):68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, Rathmell JC. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186(6):3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Grady SL, Hwang J, Vastag L, Rabinowitz JD, Shenk T. Herpes simplex virus 1 infection activates poly(ADP-ribose) polymerase and triggers the degradation of poly(ADP-ribose) Glycohydrolase. J Virol. 2012;86(15):8259–8268. doi: 10.1128/JVI.00495-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Maciver NJ, et al. The liver kinase B1 (LKB1) is a central regulator of T cell development, activation, and metabolism. J Immunol. 2011;187(8):4187–4198. doi: 10.4049/jimmunol.1100367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Angelin A, Gil-de-Gómez L, Dahiya S, Jiao J, Guo L, Levine MH, Wang Z, Quinn WJ, III, Kopinski PK, Wang L, Akimova T, Liu Y, Bhatti TR, Han R, Laskin BL, Baur JA, Blair IA, Wallace DC, Hancock WW, Beier UH. Foxp3 reprograms T cell metabolism to function in low-glucose, High-Lactate Environments. Cell Metab. 2017;25(6):1282–1293. doi: 10.1016/j.cmet.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rémi M, et al. AMPKα1 regulates macrophage skewing at the time of resolution of inflammation during skeletal muscle regeneration. Cell Metab. 2013;18(2):251–264. doi: 10.1016/j.cmet.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 153.Ho PC, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, Tsui YC, Cui G, Micevic G, Perales JC, Kleinstein SH, Abel ED, Insogna KL, Feske S, Locasale JW, Bosenberg MW, Rathmell JC, Kaech SM. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell. 2015;162(6):1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gualdoni GA, Mayer KA, Göschl L, Boucheron N, Ellmeier W, Zlabinger GJ. The AMP analog AICAR modulates the Treg/Th17 axis through enhancement of fatty acid oxidation. FASEB J. 2016;30(11):3800–3809. doi: 10.1096/fj.201600522R. [DOI] [PubMed] [Google Scholar]
- 155.Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, Rathmell JC. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186(6):3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sereni Maria Isabella, Baldelli Elisa, Gambara Guido, Ravaggi Antonella, Hodge K Alex, Alberts David S, Guillen-Rodriguez Jose M, Dong Ting, Memo Maurizio, Odicino Franco, Angioli Roberto, Liotta Lance A, Pecorelli Sergio L, Petricoin Emanuel F, Pierobon Mariaelena. Kinase-driven metabolic signalling as a predictor of response to carboplatin–paclitaxel adjuvant treatment in advanced ovarian cancers. British Journal of Cancer. 2017;117(4):494–502. doi: 10.1038/bjc.2017.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Song M, Sandoval TA, Chae CS, Chopra S, Tan C, Rutkowski MR, Raundhal M, Chaurio RA, Payne KK, Konrad C, Bettigole SE, Shin HR, Crowley MJP, Cerliani JP, Kossenkov AV, Motorykin I, Zhang S, Manfredi G, Zamarin D, Holcomb K, Rodriguez PC, Rabinovich GA, Conejo-Garcia JR, Glimcher LH, Cubillos-Ruiz JR. IRE1α–XBP1 controls T cell function in ovarian cancer by regulating mitochondrial activity. Nature. 2018;562(7727):423–428. doi: 10.1038/s41586-018-0597-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Schworer S, Vardhana SA, Thompson CB. Cancer metabolism drives a stromal regenerative response. Cell Metab. 2019;29(3):576–591. doi: 10.1016/j.cmet.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, Kremer D, Hwang RF, Witkiewicz AK, Ying H, Asara JM, Evans RM, Cantley LC, Lyssiotis CA, Kimmelman AC. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536(7617):479–483. doi: 10.1038/nature19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Katheder NS, Khezri R, O’Farrell F, Schultz SW, Jain A, Rahman MM, Schink KO, Theodossiou TA, Johansen T, Juhász G, Bilder D, Brech A, Stenmark H, Rusten TE. Microenvironmental autophagy promotes tumour growth. Nature. 2017;541(7637):417–420. doi: 10.1038/nature20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yang Z, et al. ROS signaling under metabolic stress: cross-talk between AMPK and AKT pathway. Mol Cancer. 2017;16(1):79. doi: 10.1186/s12943-017-0648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lin L, Huang H, Liao W, Ma H, Liu J, Wang L, Huang N, Liao Y, Liao W. MACC1 supports human gastric cancer growth under metabolic stress by enhancing the Warburg effect. Oncogene. 2015;34(21):2700–2710. doi: 10.1038/onc.2014.204. [DOI] [PubMed] [Google Scholar]
- 163.Yang T, et al. MACC1 mediates acetylcholine-induced invasion and migration by human gastric cancer cells. Oncotarget. 2016;7(14):18085–18094. doi: 10.18632/oncotarget.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kubben Nard, Misteli Tom. Shared molecular and cellular mechanisms of premature ageing and ageing-associated diseases. Nature Reviews Molecular Cell Biology. 2017;18(10):595–609. doi: 10.1038/nrm.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cantó C, et al. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 2010;11(3):213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Garciaroves PM, et al. Gain-of-function R225Q mutation in AMP-activated protein kinase gamma3 subunit increases mitochondrial biogenesis in glycolytic skeletal muscle. J Biol Chem. 2008;283(51):35724–35734. doi: 10.1074/jbc.M805078200. [DOI] [PubMed] [Google Scholar]
- 167.Maniam S, et al. Cofactor strap regulates oxidative phosphorylation and mitochondrial p53 activity through ATP synthase. Cell Death Differ. 2015;22(1):156–163. doi: 10.1038/cdd.2014.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sanchezmacedo N, et al. Depletion of the novel p53-target gene carnitine palmitoyltransferase 1C delays tumor growth in the neurofibromatosis type I tumor model. Cell Death Differ. 2013;20(4):659–668. doi: 10.1038/cdd.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Klingenberg Martin. Mechanism and evolution of the uncoupling protein of brown adipose tissue. Trends in Biochemical Sciences. 1990;15(3):108–112. doi: 10.1016/0968-0004(90)90194-g. [DOI] [PubMed] [Google Scholar]
- 170.Echtay KS, Roussel D, St-Pierre J, Jekabsons MB, Cadenas S, Stuart JA, Harper JA, Roebuck SJ, Morrison A, Pickering S, Clapham JC, Brand MD. Superoxide activates mitochondrial uncoupling proteins. Nature. 2002;415(6867):96–99. doi: 10.1038/415096a. [DOI] [PubMed] [Google Scholar]
- 171.Claire P, et al. Uncoupling protein-2 controls proliferation by promoting fatty acid oxidation and limiting glycolysis-derived pyruvate utilization. FASEB J. 2008;22(1):9–18. doi: 10.1096/fj.07-8945com. [DOI] [PubMed] [Google Scholar]
- 172.Alexandra K, et al. Loss of UCP2 attenuates mitochondrial dysfunction without altering ROS production and uncoupling activity. PLoS Genet. 2014;10(6):e1004385. doi: 10.1371/journal.pgen.1004385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jin Z, et al. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J. 2014;30(24):4860–4873. doi: 10.1038/emboj.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Vozza A, Parisi G, de Leonardis F, Lasorsa FM, Castegna A, Amorese D, Marmo R, Calcagnile VM, Palmieri L, Ricquier D, Paradies E, Scarcia P, Palmieri F, Bouillaud F, Fiermonte G. UCP2 transports C4 metabolites out of mitochondria, regulating glucose and glutamine oxidation. Proc Natl Acad Sci U S A. 2014;111(3):960–965. doi: 10.1073/pnas.1317400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhonglin X, et al. Upregulation of mitochondrial uncoupling protein-2 by the AMP-activated protein kinase in endothelial cells attenuates oxidative stress in diabetes. Diabetes. 2008;57(12):3222–3230. doi: 10.2337/db08-0610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 176.Marc F, et al. Short-term overexpression of a constitutively active form of AMP-activated protein kinase in the liver leads to mild hypoglycemia and fatty liver. Diabetes. 2005;54(5):1331–1339. doi: 10.2337/diabetes.54.5.1331. [DOI] [PubMed] [Google Scholar]
- 177.Pauline E, et al. Mitochondrial retrograde signaling mediated by UCP2 inhibits cancer cell proliferation and tumorigenesis. Cancer Res. 2014;74(14):3971–3982. doi: 10.1158/0008-5472.CAN-13-3383. [DOI] [PubMed] [Google Scholar]
- 178.Wang M, et al. Uncoupling protein 2 downregulation by hypoxia through repression of peroxisome proliferator-activated receptor γ promotes chemoresistance of non-small cell lung cancer. Oncotarget. 2017;8(5):8083. doi: 10.18632/oncotarget.14097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Boutoual R, et al. Defects in the mitochondrial-tRNA modification enzymes MTO1 and GTPBP3 promote different metabolic reprogramming through a HIF-PPARγ-UCP2-AMPK axis. Sci Rep. 2018;8(1):1163. doi: 10.1038/s41598-018-19587-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Szelechowski M, et al. Metabolic reprogramming in amyotrophic lateral sclerosis. Sci Rep. 2018;8(1):3953. doi: 10.1038/s41598-018-22318-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Daurio NA, et al. AMPK activation and metabolic reprogramming by tamoxifen through estrogen receptor-independent mechanisms suggests new uses for this therapeutic modality in cancer treatment. Cancer Res. 2016;76(11):3295. doi: 10.1158/0008-5472.CAN-15-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Pulito C, Mori F, Sacconi A, Goeman F, Ferraiuolo M, Pasanisi P, Campagnoli C, Berrino F, Fanciulli M, Ford RJ, Levrero M, Pediconi N, Ciuffreda L, Milella M, Steinberg GR, Cioce M, Muti P, Strano S, Blandino G. Metformin-induced ablation of microRNA 21-5p releases Sestrin-1 and CAB39L antitumoral activities. Cell Discov. 2017;3:17022. doi: 10.1038/celldisc.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lodi A, Woods SM, Ronen SM. Magnetic resonance-detectable metabolic consequences of MEK inhibition. NMR Biomed. 2014;27(6):700. doi: 10.1002/nbm.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Oliveras-Ferraros C, Vazquez-Martin A, Cuyàs E, COROMINAS-FAJA B, Rodríguez-Gallego E, Fernández-Arroyo S, Martin-Castillo B, Joven J, MENENDEZ MENENDEZ J. Acquired resistance to metformin in breast cancer cells triggers transcriptome reprogramming toward a degradome-related metastatic stem-like profile. Cell Cycle. 2014;13(7):1132–1144. doi: 10.4161/cc.27982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhou X, et al. Metformin suppresses hypoxia-induced stabilization of HIF-1α through reprogramming of oxygen metabolism in hepatocellular carcinoma. Oncotarget. 2016;7(1):873–884. doi: 10.18632/oncotarget.6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zarrouk M, et al. Adenosine-mono-phosphate-activated protein kinase-independent effects of metformin in T cells. PLoS One. 2014;9(9):e106710. doi: 10.1371/journal.pone.0106710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hardie DG. AMP-activated protein kinase as a drug target. Annu Rev Pharmacol Toxicol. 2007;47(1):185–210. doi: 10.1146/annurev.pharmtox.47.120505.105304. [DOI] [PubMed] [Google Scholar]
- 188.Popovics P, Frigo DE, Schally AV, Rick FG. Targeting the 5'-AMP-activated protein kinase and related metabolic pathways for the treatment of prostate cancer. Expert Opin Ther Targets. 2015;19(5):617–632. doi: 10.1517/14728222.2015.1005603. [DOI] [PubMed] [Google Scholar]
- 189.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444(7117):337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gerhart-Hines Z, et al. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD + Mol Cell. 2011;44(6):851–863. doi: 10.1016/j.molcel.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Maus M, Cuk M, Patel B, Lian J, Ouimet M, Kaufmann U, Yang J, Horvath R, Hornig-Do HT, Chrzanowska-Lightowlers ZM, Moore KJ, Cuervo AM, Feske S. Store-operated Ca2+ entry controls induction of lipolysis and the transcriptional reprogramming to lipid metabolism. Cell Metab. 2017;25(3):698–712. doi: 10.1016/j.cmet.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Gong KW, et al. cAMP-specific phosphodiesterase TbPDE1 is not essential in Trypanosoma brucei in culture or during midgut infection of tsetse flies. Mol Biochem Parasitol. 2001;116(2):229. doi: 10.1016/s0166-6851(01)00315-2. [DOI] [PubMed] [Google Scholar]
- 193.Cheng KK, Lee BS, Masuda T, Ito T, Ikeda K, Hirayama A, Deng L, Dong J, Shimizu K, Soga T, Tomita M, Palsson BO, Robert M. Global metabolic network reorganization by adaptive mutations allows fast growth of Escherichia coli on glycerol. Nat Commun. 2014;5(1):3233. doi: 10.1038/ncomms4233. [DOI] [PubMed] [Google Scholar]
- 194.Lu Jie, Montgomery Blake K., Chatain Grégoire P., Bugarini Alejandro, Zhang Qi, Wang Xiang, Edwards Nancy A., Ray-Chaudhury Abhik, Merrill Marsha J., Lonser Russell R., Chittiboina Prashant. Corticotropin releasing hormone can selectively stimulate glucose uptake in corticotropinoma via glucose transporter 1. Molecular and Cellular Endocrinology. 2018;470:105–114. doi: 10.1016/j.mce.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Tong T, Ryu SE, Min Y, de March CA, Bushdid C, Golebiowski J, Moon C, Park T. Olfactory receptor 10J5 responding to α-cedrene regulates hepatic steatosis via the cAMP–PKA pathway. Sci Rep. 2017;7(1):9471. doi: 10.1038/s41598-017-10379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Nissim I, et al. The molecular and metabolic influence of long term agmatine consumption. J Biol Chem. 2014;289(14):9710–9729. doi: 10.1074/jbc.M113.544726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Carlessi R, Chen Y, Rowlands J, Cruzat VF, Keane KN, Egan L, Mamotte C, Stokes R, Gunton JE, Bittencourt PIH, Newsholme P. GLP-1 receptor signalling promotes β-cell glucose metabolismviamTOR-dependent HIF-1α activation. Sci Rep. 2017;7(1):2661. doi: 10.1038/s41598-017-02838-2. [DOI] [PMC free article] [PubMed] [Google Scholar]