Abstract
Continuing crop domestication/redomestication and modification is a key determinant of the adaptation and fulfillment of the food requirements of an exploding global population under increasingly challenging conditions such as climate change and the reduction in arable lands. Monocotyledonous crops are not only responsible for approximately 70% of total global crop production, indicating their important roles in human life, but also the first crops to be challenged with the abovementioned hurdles; hence, monocot crops should be the first to be engineered and/or de novo domesticated/redomesticated. A long time has passed since the first green revolution; the world is again facing the challenge of feeding a predicted 9.7 billion people in 2050, since the decline in world hunger was reversed in 2015. One of the major lessons learned from the first green revolution is the importance of novel and advanced trait-carrying crop varieties that are ideally adapted to new agricultural practices. New plant breeding techniques (NPBTs), such as genome editing, could help us succeed in this mission to create novel and advanced crops. Considering the importance of NPBTs in crop genetic improvement, we attempt to summarize and discuss the latest progress with major approaches, such as site-directed mutagenesis using molecular scissors, base editors and especially homology-directed gene targeting (HGT), a very challenging but potentially highly precise genome modification approach in plants. We therefore suggest potential approaches for the improvement of practical HGT, focusing on monocots, and discuss a potential approach for the regulation of genome-edited products.
Keywords: Gene targeting (GT), Homology-directed repair (HDR), Homology-directed gene targeting (HGT), CRISPR/Cas, Targeted mutagenesis, Precision breeding, Monocots
Background
Status of Food Production Using Monocots
Most of the present important crop plants were domesticated approximately 10,000–13,000 years ago by our ancestors. The domestication of food crops forever changed human life from hunting-gathering groups to stationary living communities (Meyer and Purugganan 2013; Hickey et al. 2019). Among all food crops, cereals might have been the first to be artificially selected and intentionally planted for food (Meyer et al. 2012; Asano et al. 2011). The domestication process is still conducted though modern breeding techniques, which have completely changed the methods of selection and adaptation of crop traits (Meyer et al. 2012). The major monocots used as daily staples are rice, maize and wheat. In 2018–2019, the production of corn, wheat and rice accounted for approximately 70% of the total world crop production (FAO 2019a; USDA 2019). In the first half of the twentieth century, the world population increased rapidly and disproportionately to the increase in food production, leading to dire predictions for the second half of the century (Khush 2001). We now know that this large-scale famine did not happen due to the first green revolution (GR), which doubled cereal grain production within just 10 years of its beginning in the 1950s. Wheat and rice played major roles in the first GR, indicating the pivotal role of monocot crops in human life.
Food Requirements in 2050 Vs Present Production: a Major Challenge
The first GR accelerated world food production, which first reached 1 billion tons in 1950, but needed only 10 years to double that number by the use of high-yield varieties, chemical fertilizers and pesticides and the adoption of new cultivation methods involving irrigation systems (Khush 2001). Seventy years after the start of the first GR, the world is again facing the same challenge of feeding a much larger population. Unfortunately, the miracles of the first GR are now reaching their limits. The increases in the yield and production of food crops are slowing and will not meet the requirements of 9.7 billion people in 2050 in the present scenario (UN 2019); for more details, please review the Food and Agriculture Organization (FAO) report (2009) and its revised version published in 2012. A recent report from FAO detailed that the decline in world hunger had reversed in 2015 and that the number of hungry people is slowly increasing at present. As of 2018, over 820 million people are still living under the hunger line (FAO 2019b). It is worth noting that this conclusion was drawn considering the present situation of food production and agriculture, even with worldwide support for conventional and modern molecular-assisted breeding and smart agriculture practices (Ray et al. 2013). Obviously, world agriculture is now challenging, with many novel negative factors, such as more vigorous climate change, soil nutrition deficiency, and global sea level rise, but the other hurdles remain the same as before the first GR. This reality indicates that to cope with the challenges and to fulfill the food production demand, the world must accept and apply new technologies, especially new plant breeding techniques (NPBTs) (Lusser et al. 2011), for crop improvement and agricultural practices (Zaidi et al. 2019). Moreover, the world demands a second GR that can sustain and secure food production for mankind.
Introduction of NPBTs and Genome-Editing-Based Precision Breeding
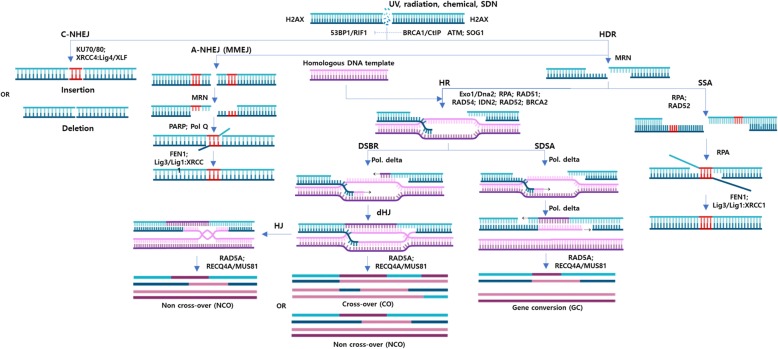
NPBTs, especially the recently emerged genome-editing technologies, offer various solutions to improve crop traits, such as (a) crops that can well adapt to environmental changes resulting in sustainable yield, (b) crops that efficiently use limited resources to produce more food and (c) crops with improved nutritional value. In general, genome editing is a two-stage process that includes (1) DNA damage generation such as single-stranded/double-stranded breaks (SSBs/DSBs) or nucleotide deaminations (for base editors) and (2) host cell repair of damaged sites. The repair process can be error-prone during the canonical nonhomologous end joining (C-NHEJ), alternative NHEJ (A-NHEJ), or single-stranded annealing (SSA) pathways that usually ligate DSB ends without the need for additional DNA templates (Fig. 1). The base excision repair (BER) or nucleotide excision repair (NER) pathways that cells use to fix damaged nucleotides such as deaminated ones may also be error-prone. Other repair pathways such as homologous recombination (HR) or oligonucleotide-directed mutagenesis (ODM) require the presence of homologous DNA donors to replicate genetic information and they have been shown to generate error-free products (Figs. 1 and 3b). Recently, Liu’s team published an exciting novel precision editing approach called ‘prime editing’ which used a reverse transcriptase (RT)-nCas9 (H840A) fusion to precisely add DNA modifications to specific sites (Fig. 3c) (Anzalone et al. 2019). Repair pathways in animals as well as plants have been extensively reviewed elsewhere (Belhaj et al. 2013; Hsu et al. 2014; Doudna and Charpentier 2014; Bortesi and Fischer 2015; Rees and Liu 2018). The NHEJ and BER/NER approaches are highly efficient in generating unpredictable error-prone products, while ODM, HGT and base editor (BE) have been considered as techniques for precision editing of genes in plants. However, in plants, the main obstacle to HGT applications is their extremely low efficacy (Paszkowski et al. 1988; Puchta et al. 1996). Many attempts have been made to improve plant HGT for practical applications. Important enhancements have been shown with CRISPR/Cas complexes with or without homologous donor template delivery and amplification by ssDNA replicons (Baltes et al. 2014; Cermak et al. 2015; Gil-Humanes et al. 2017; Wang et al. 2017). In this review, we summarize recent data regarding genome editing approaches in monocot plants with special focus on HGT and provide perspectives for monocot crop improvement and commercialization.
Fig. 1.
DSB repair pathways.1017In the C-NHEJ pathway, DSB formation induces binding to broken ends by KU70/80 heterodimers that subsequently recruit the DNA damage response kinase (DDK) complex such as DNA-PKcs in mammals. DDK then activates the 53BP1/RIF1 complex, which plays a role in shielding the broken ends from resection by antagonizing BRCA1/CtIP activity. DNA-PK also activates other KU-recruited proteins, such as XLF, XRCC4 and Lig4, for ligating the broken ends. In the HDR pathway, DSB formation induces cell cycle arrest initiated with the activation of ATM resulting from sensing a chromatin structure change. Activated monomeric ATM then phosphorylates the MRN complex and P53/SOG1, which regulates the cell cycle checkpoint and arrest. MRN activation supports end resection for HDR. Limited resection leads to MMEJ, and if a substantial level of resection is formed in the absence of a donor template, SSA is likely to be used for the repair. MMEJ requires PARP and Pol Q for its processes, and SSA requires the role of RAD52. Both MMEJ and SSA require the ssDNA flap endonuclease FEN1 and Lig3/Lig1:XRCC1 for ligating final products. Extensive resection of the broken ends is facilitated by Exonuclease 1 (Exo1) and/or Dna2. In the presence of donor template, the 3′ overhangs of resected ends could be protected by RPA binding and then recruiting RAD51 to the ssDNA with support and control by BRCA2. RAD51 binds to the resected ssDNA overhang, forming nucleoprotein filaments or presynaptic filaments. With the support of RAD54, the filament structure invades the donor template sequence and searches for and anneals to the complementary sequence; then, displacement loop (D-loop) formation occurs. Subsequently, the free 3′ OH end of the invaded ssDNA primes donor template-dependent DNA synthesis. This process determines the outcomes of HDR with several sub-pathways (DSBR with dHJ and SDSA) with the supportive activity of RAD5A, RECQ4A and MUS81. The DNA fragments and protein structures are not pictured to scale. The potential proteins involved in the processes of each pathway or sub-pathway are denoted adjacent to their approaching lines. XRCC: X-ray repair cross-complementing protein; XLF: XRCC4-like factor; Lig4: DNA ligase 4; PARP: poly-ADP-ribose polymerase; Pol Q: DNA polymerase theta
Fig. 3.
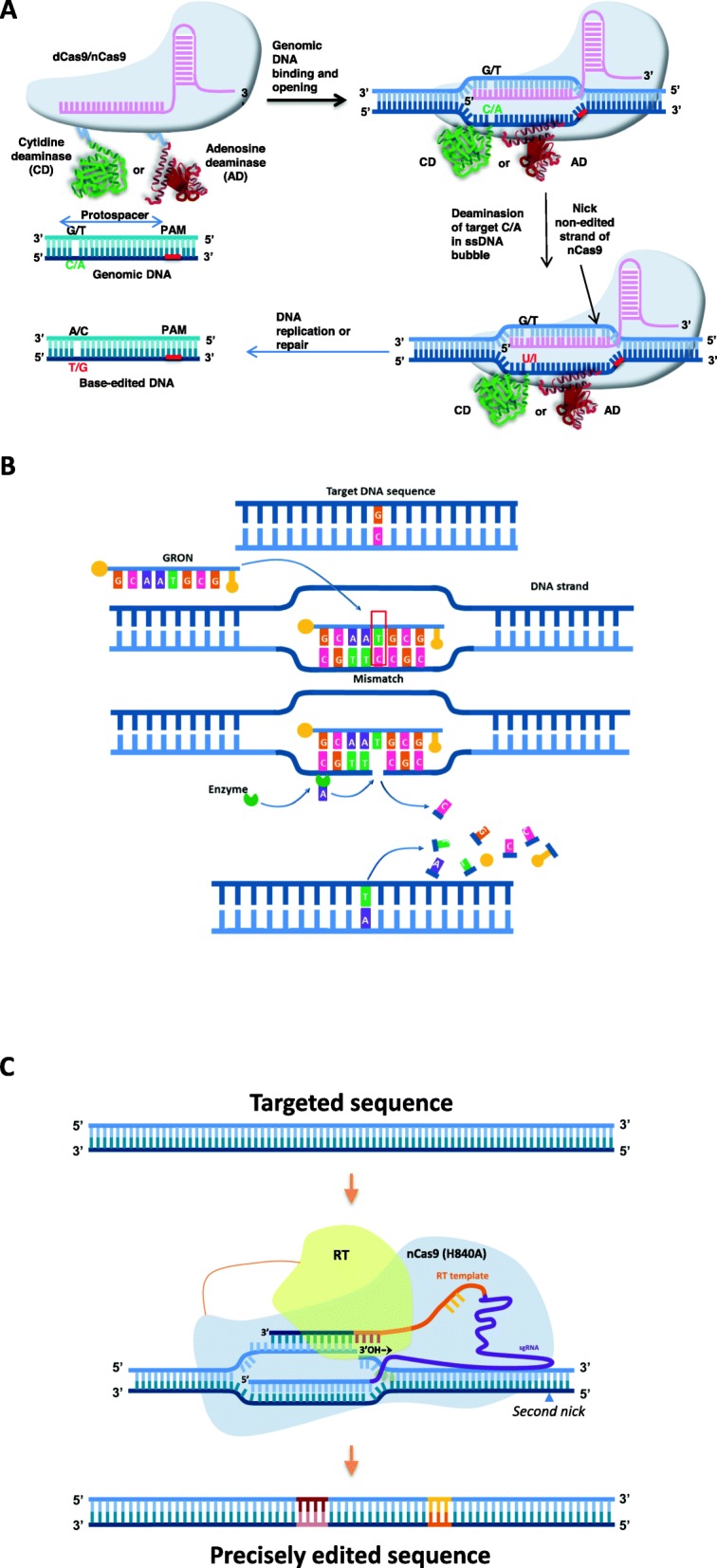
Non-DSB precise gene targeting approaches. a Base of approach editing. Cytosine Base Editors (CBEs) and Adenosine Base Editors (ABEs) are the two types of base editors that have been published so far. CBEs: Dead Cas9 (blue) binds to target C (green) via the RNA (pink) guide, which mediates the separation of local DNA strands. A tethered APOBEC1 (green) enzyme by cytosine deamination converts the single-stranded target C to U. The initial G: C is replaced by the A: T base pair at the target location through DNA repair or replication. ABEs: A hypothetical deoxyadenosine deaminase (red) and catalytically impaired nCas9 (Cas9 D10A nickase) bind target DNA in the RNA guide to expose a small bubble of single-stranded DNA that catalyzes the conversion of A to I within this bubble. b Oligonucleotide-directed mutagenesis process. A gene repair oligonucleotide (GRON), which contains designed modifications, is delivered and paired with the target DNA sequence. GRON creates a mismatch at the target site and triggers a DNA repair mechanism. DNA repair enzymes detect the mismatch and repair the target DNA sequence using GRON as a template. Once the repair process is completed during cell division and multiplication, the GRON is removed and degraded. The target sequence is modified with designed changes. The representative DNA fragments and protein structures are not pictured to scale. c Prime editing. Prime editor is a CRISPR/Cas complex developed by fusion of a reverse transcriptase (RT) to a C-terminal of nickase Cas9 (H840A) and a prime editing gRNA (pegRNA) with a 3 ‘extension that could bind to the 3 ‘nicked strands produced by the nCas9. When bound, the 3′-OH free nicked strand is used as a substratum for the RT to copy genetic information from the 3 ‘extension of pegRNA
Review
Genome Editing Technologies
Targeted Mutagenesis Using Molecular Scissors to Form DSBs
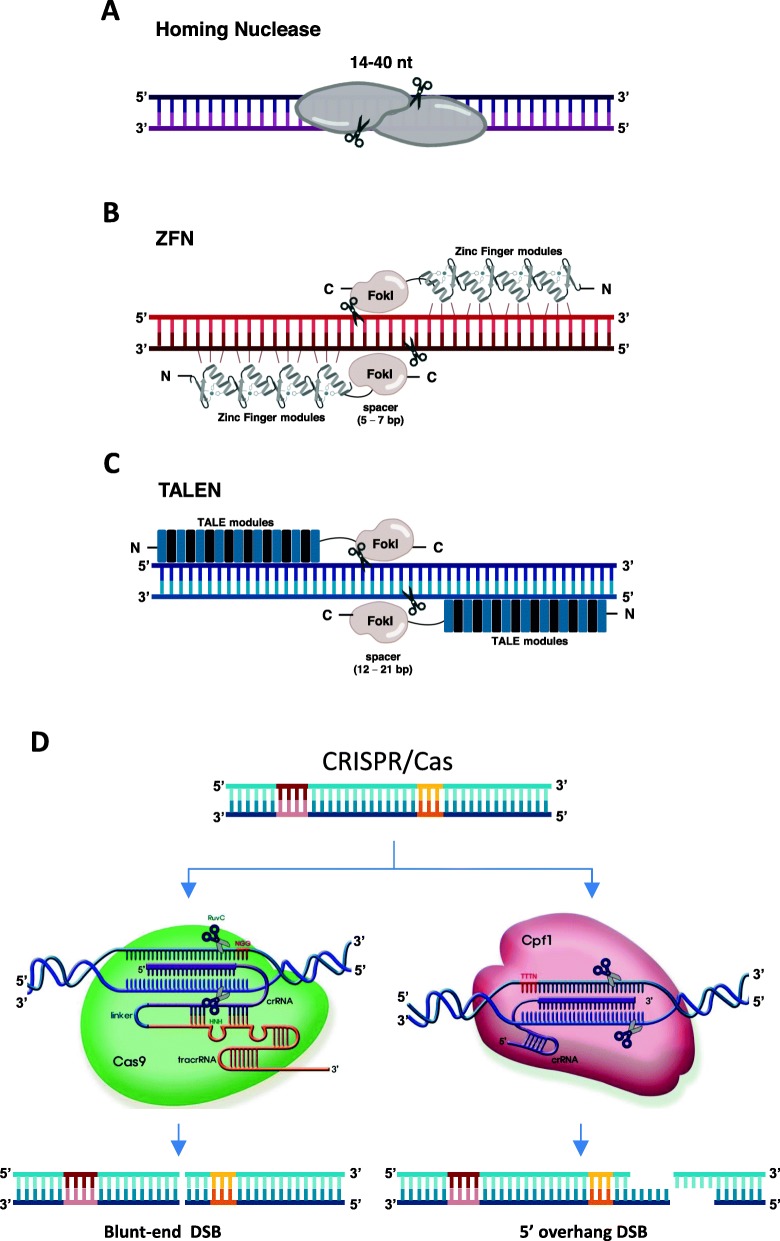
Since the discovery of restriction enzymes, the field of biotechnology has entered a new era of molecular engineering facilitated by recombinant DNA technology. Several generations of molecular scissors have been discovered, characterized and developed for DSB-based targeted genome mutagenesis. The technology has been improved from the long recognition sequence homing nucleases to protein-dependent DNA binding nucleases, such as zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), and ultimately to the 3rd generation RNA-guided molecular scissors CRISPR /Cas (Fig. 2). With the invention of target-specific synthetic molecular scissors, the specific modification of a gene of interest in a living organism has become possible. Consequently, there are several key factors involved in targeted mutagenesis induced by molecular scissors, including: 1) the ability to specifically recognize and bind to the targeted DNA sequence, 2) effective DSB formation, and 3) error-prone DSB repair.
Fig. 2.
Four generations of molecular scissors. The first, second and third generations of molecular scissors, Homing nuclease (a); ZFN (b); and TALEN (c), are characterized as nucleases relying on DNA binding domains to recognize DNA target sites. Homing nucleases recognize long DNA sequences of 14–40 bp with their DNA binding domains. A ZFN or TALEN is designed by connecting 3–6 zinc finger motifs or 17–20 TALE modules, respectively, for DNA binding and an endonuclease domain of FokI restriction enzyme for cutting. FokI works only in homodimer form, so usually one has to design pairs of ZFNs or TALENs to target a DNA site. FokI activity usually produces DSB with 4 nt overhangs. The fourth generation, CRISPR/Cas (d), is also the most powerful one; it uses guide RNA components to form active complexes, thereby interrogating and searching for target DNA sites based on Watson-Crick base pairing between the guide RNA and targeted strand. The DNA fragments and protein structures are not pictured to scale
The host’s repair of the DSB errors leads to error-free or error-prone outcomes depending on many factors, including the cell cycle state and the availability of homologous DNA templates at the damaged sites. In plant somatic cells, DSB repair by either of the two major pathways, homology-directed repair (HDR) or nonhomologous end joining (NHEJ), usually leads to either error-free or error-prone products. The majority of the error-prone products appear as insertion or deletion (indel) DNA mutations resulting from C-NHEJ or A-NHEJ (Fig. 1). A possibly lower portion of error-prone products may result from SSA repair in the absence of a homologous donor template and from Holliday junction resolution in the last steps of the double-stranded break repair (DSBR) subpathway if the DSB flanking sequences of the sister chromatids are not perfectly matched (Fig. 1). In this section, we briefly summarize the abovementioned molecular scissors. Extensive reviews of the same material can be found elsewhere (Carroll, 2011; Gaj et al., 2013).
Generation 0: Homing Nucleases
Homing nucleases are endonucleases (Mw < 40 kDa) that recognize long DNA sequences (14–40 nt) for their cutting activity (Fig. 2a). Homing nucleases can work alone as monomers or in pairs as homodimers (Chevalier and Stoddard 2001). Members of the LAGLIDADG homing endonucleases family such as I-CreI or I-SceI recognize targeted sequences of 22 bp and 18 bp respectively, thus allowing more specific targeting in the host cells (approximately once every 7 × 109 bp) (Jurica et al. 1998; Niu et al. 2008; Chevalier and Stoddard 2001; Jasin 1996). However, this feature also introduces limitations via the scarcity of targetable sites in the genomes of host cells. To compensate for this, researchers have engineered these nucleases for a wider range of binding and cutting sites or combinations of different homing nucleases to recognize multiple sites (Chevalier et al. 2002). Engineered homing nucleases often cleave correct sites as efficiently as wild-type nucleases (Chevalier and Stoddard 2001; Yang et al. 2009; Gao et al. 2010; D'Halluin et al. 2013). However, the engineering of homing nucleases for wider applications is still inefficient, laborious and time consuming.
Generations 1 and 2: Protein-Guided DSB Formation, ZFN and TALENs
ZFNs are derived from the discovery of the zinc finger, a finger-like DNA binding motif found in TFIIIA, a transcription factor from the eggs of Xenopus laevis (Miller et al. 1985). Its structure comprises 30 repetitive amino acid sequences and is stabilized by a zinc ion (Miller et al. 1985; Berg 1988). Berg (1988) suggested that the zinc finger protein structure might play a key role in the recognition of DNA sequences. ZFN was first developed in 1996 by fusing a nonspecific DNA cleavage domain of FokI, a type II-S restriction enzyme, to the C-terminal of the zinc finger motifs (Kim et al. 1996). Typically, three consecutive nucleotides can be specifically recognized by one zinc finger motif, and therefore, several connected zinc finger motifs fused to FokI can bind the target DNA of interest (Kim et al. 1996). ZFN is the first artificial restriction enzyme that recognizes desirable sites in the genome. Due to their binding specificity and dimerization-dependent FokI activity requirement, ZFNs were typically designed in pairs to recognize 9–18 bp using connected 3–6 zinc finger motifs on both the sense and antisense strands of the targeted sequences spaced by 5–7 bp between ZFNs (Kim et al. 1996; Bitinaite et al. 1998; Laity et al. 2001; Urnov et al. 2010) (Fig. 2b). Post cleavage, the DSB sites were recovered by DNA repair mechanisms that showed insertions or deletions at similar rates (Kim et al. 2013). However, for wider application of this technology, one should overcome the limitations of low editing efficiency (0–24%), elevated design and optimization cost, and high off-target possibility. Many efforts have been made to overcome these barriers. For example, to enhance the cleavage activity of the FokI cleavage domain, Gou and coworkers performed direct evolution to optimize a ZFN named ‘Sharkey’. Several approaches were tested to reduce the off-target effect, e.g., extending the recognition length by using more zinc finger modules (Pattanayak et al. 2011; Guo et al. 2010).
TALEN is the second-generation form of molecular scissors, discovered during studies of the plant immune system under attack and hijacking by pathogenic bacteria (Dangl and Jones 2001). AvrBs3, an effector protein secreted by the plant pathogen Xanthomonas campestris, is injected into host cells, thereby binding to the plant UPA-box gene and functioning as a transcription activator to modulate host cell gene expression for its efficient colonization (Kay et al. 2009). The causal agents secreted by Xanthomonas were identified and named transcription activator-like effectors (TALEs). TALEs have 33–35 amino acids that are highly conserved, except for those located at positions 12 and 13. These two hypervariable residues (namely, repeat-variable diresidues (RVDs)) are oriented toward the outside of the protein and play a key role in recognizing a specific nucleotide (Moscou and Bogdanove 2009). Common rules of RVD nucleotide recognition for binding were validated as NG for thymine; HD for cytosine; NN for guanine or adenine; and NI for adenine. The first TALENs were introduced by fusing a DNA binding TAL type III effector with a FokI cleavage domain Fig. 2c (Li et al. 2011). However, unlike ZFN, which recognizes 3 bp per zinc finger module, TALENs allow more precise recognition because each RVD of TALE can recognize only one nucleotide. TALENs were designed in pairs with a 12–21 nt distance between two binding sites for the highest cutting activity (Miller et al. 2011). The combination of the TAL effectors AvrXa7, PthXo1 and FokI was demonstrated to function as molecular scissors for cutting and hence modifying the binding sites of the TALs, subsequently resulting in resistance to rice blight disease (Li et al. 2011). The initial NN RVD repeat recognized either guanine or adenine, raising concerns about its specificity (Moscou and Bogdanove 2009). Ultimately, an NK RVD repeat that recognizes only guanine was discovered, fulfilling the specificity requirement for the TALEN molecular scissors (Miller et al. 2011).
One of the weak points of the TALEN approach is the large size of the binding domain, as every nucleotide requires a repeat of ~ 34 amino acids for binding. Thus, to assure high specificity for one TALEN binding to 20 nt, its DNA binding domain must be 680 amino acids long. In addition, assembly of the highly repeated modules was time consuming and laborious. Thus, a well-designed modular RVD repeat library was in high demand and was eventually developed (Zhang et al. 2011; Cermak et al. 2011; Kim et al. 2013). Another limitation of TALENs for practical applications is that they are sensitive to methylated cytosine, thereby preventing them from binding to the modified nucleotide efficiently. In an attempt to overcome the hurdle, the TALE domain was designed to contain the single asparagine RVD (N*) motif (N* refers to Asn instead of Asn-Gly), a base-recognition domain that could effectively bind to 5 ‘methylated cytosine. The engineered TALENs (N*) showed higher efficacies for genome editing in mammalian cells and rice (Valton et al. 2012; Kaya et al. 2017). TALENs have the advantages of high editing efficiency, low off-target activity and lower design cost than ZFNs and the drawbacks of difficult construction, no activity on methylated cytosines (Kim et al. 2013), and difficult introduction into cells owing to their large size (Kim and Ka 2015).
Generation 3: CRISPR/Cas
Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas) was shown to be a DNA interference-based defense machinery of prokaryotes such as bacteria and archaea against phage infection (Barrangou et al. 2007; Brouns et al. 2008). CRISPR/Cas systems were classified into two classes according to the number of complexity of their effector modules (Makarova et al. 2011; Makarova et al. 2015). Class 1 systems involve effector complexes formed by multiple subunits, whereas in class 2 systems, single multidomain proteins constitute the effector complexes. Furthermore, each class has been divided into several subtypes (class 1: types I, III and IV; and class 2: types II, V and VI) based on their effector architectures with unique signature proteins (Koonin and Makarova 2019). Almost all of the CRISPR/Cas systems used in genome engineering to date are from class 2 due to the simplicity of their effector modules (Additional file 1: Table S1). The most widely used CRISPR/Cas systems are Cas9 and Cas12a (Cpf1).
In the native CRISPR/Cas9 system, phage DNAs were shown to be cleaved by the Cas9 effector complex, which includes the Cas9 protein as a nuclease and a complexed RNA structure formed by a CRISPR RNA (crRNA) and a trans-activating CRISPR RNA (tracrRNA) as a probe. The two-component RNA secondary structure facilitates Cas9 assembly, searching and binding to dsDNA target sites by Watson-Crick complementarity to 19–21 nt of the 5′ end of the crRNA (protospacer) and subsequently cleaving both the strands of the dsDNA at the 3rd nucleotide proximal to a 5′-NGG-3′ protospacer-adjacent motif (PAM) site (Fig. 2d, CRISPR/Cas9). Originally, crRNA:tracrRNA required maturation from a precursor crRNA:tracrRNA by RNase III processing activity, making it more difficult to apply. In the first application of CRISPR/Cas9 for genome editing, the crRNA and tracrRNA were engineered to make a single guide RNA molecule by connecting the 3′ crRNA repeat and 5′ tracrRNA anti-repeat, thereby facilitating the use of the system (Jinek et al. 2012). The Cas9 protein remains inactive until it binds to a guide crRNA:tracrRNA structure. The guide RNA-bound Cas9 complex undergoes conformational changes and then stochastically searches for potential targets by PAM scanning and binding using the PAM-interacting motif. Then, the Cas-sgRNA complex again changes conformation, and the guide RNA sequence is used to pair with the sequence located upstream of the PAM via the Watson-Crick rule (Sternberg et al. 2014; Jinek et al. 2014; Zhu et al. 2019). The gRNA and its seed sequence (10-nucleotide RNA proximal to the NGG PAM) should be fully complemented for R-loop formation and to trigger Cas9 cleavage activities via its endonuclease domains (HNH and RuvC) (Jinek et al. 2012; Jiang et al. 2013; Hsu et al. 2013). The targeted and nontargeted strands of the dsDNA are cleaved by HNH and RuvC, respectively, generating mostly blunt ends (Fig. 2d) (Anders et al. 2014; Nishimasu et al. 2014). Nickase Cas9 (nCas9) that cuts either the targeted strand or nontargeted strand and dead Cas9 (dCas9) were also created by inactivating either the endonuclease domains or both domains for alternative gene editing and regulation.
Unlike Cas9, the Cpf1 system does not require a tracrRNA to mature the crRNA and to form an effector complex for its cleavage activity. The Cpf1 protein was also shown to process the precursor crRNA (Zetsche et al. 2015). After assembly, the Cpf1 effector complex recognizes a T-rich PAM for the initiation of binding and searching for target sites. Its seed sequence was illustrated to range from 1 to 10 nt proximal to the PAM (Kim et al. 2016). The Cpf1 protein has a Nuc nuclease domain that cleaves the target strand and a RuvC domain that cleaves the nontargeted strand (Schunder et al. 2013; Makarova and Koonin 2015; Stella et al. 2017). The nuclease domains cut the target dsDNAs at the 18th nt on the nontargeted strand and the 23rd nt on the targeted strand distal to the PAM, generating 5′ overhang ends (Fig. 2d) (Zetsche et al. 2015).
Precision Editing
Base Substitution
It is now well known that the majority of genetic diseases result from point mutations, but the potential DSB-based repair approaches for correcting these mutations are not applicable due to their inaccessibility and the unsuitability of the repair mechanisms (Cox et al. 2015; Hilton and Gersbach 2015). Therefore, a single-base-change technique is highly demanded and has been developed for at least transition fixation (C/G- > T/A or A/T- > G/C): the so-called cytosine base editors (CBEs) or adenosine base editors (ABEs) (Fig. 3a) (Gaudelli et al. 2017; Komor et al. 2016). The basal principle behind the technique is the fusion of dead or nickase Cas9 (d/nCas9) with a cytosine or adenosine deaminase and introduction of the editor complex to the targeted site by the CRISPR guide RNA structure. Deamination of C or A produces U or I, respectively, leading to lesion-by-pass replication and resulting in C/G- > T/A or A/T- > G/C transition, respectively. In addition, the Cas9-based CBEs and ABEs were shown to work in a framed window that was either narrow (13th to 17th nucleotides upstream of the 5′-NGG-3′ PAM) (Komor et al. 2016) or wide (4th to 20th nucleotides upstream of the 5′-NGG-3′ PAM) (Zong et al. 2018) at asymmetric frequency distributions (Additional file 2: Table S2) depending on the types of deaminase used. This fact raises the possibility of controllably and precisely editing every single base of interest by carefully calculating and evaluating the editing frequencies of base editors for a base of a given target. This could also help to avoid the possibility of bystander base changes and unintended off-targets (Gehrke et al. 2018).
Oligonucleotide-Directed Mutagenesis (ODM)
Oligonucleotide-directed mutagenesis (ODM) and rapid trait development system (RTDS) are two common names for an oligonucleotide-mediated targeted gene modification technique. This technique uses synthetic oligonucleotides or gene repair oligonucleotides (GRON), which function as a template for endogenous DNA repair to form a heterotriplex with a targeted genome site via homology binding using their sequences, which are identical to the site except at the intentionally modified nucleotide(s), thereby triggering gene conversion and resulting in specific base changes (Fig. 3b). The GRON itself is not inserted into the host genome, and site-directed nucleases or double-strand breaks are not required for this technique. Therefore, ODM was classified as one of the precise gene editing techniques (for review, see Sauer et al. 2016). The changes could be point mutations, multiple base changes, insertions or deletions. The GRON was subsequently degraded during cell divisions, and the modified gene retained its normal pattern of expression and stability within the genome (Sauer et al. 2016).
The first application of synthetic nucleotides was shown in yeast in 1988 (Moerschell et al. 1988) and then in mammalian cells for correction of a faulty human β-globin that causes sickle cell anemia in 1996 (Cole-Strauss 1996; Yoon et al. 1996). In plants, Beetham and coworkers used RNA/DNA chimeric molecules in a work known as the chimeraplasty approach to target tobacco acetoacetate synthase (ALS) (or aceto hydroxyl acid synthesis (AHAS)). Tobacco ALS is a biallelic gene (including alleles ALS1 and ALS2) due to its allotetraploid genome. Therefore, two chimeric ODM oligonucleotides were designed to engineer ALS1 and ALS2 as P196 (CCA) to CAA and to CTA, respectively. Particle bombardment of the oligos and subsequent selection on medium containing 200 ppb of chlorsulfuron revealed one out four ALS alleles with a Pro-196 (CCA) to Thr-196 (ACA) modification. The efficiency was two orders of magnitude higher than that of the control (Beetham et al. 1999). The ODM approach was also conducted in several studies in dicots, such as canola (Gocal 2015) and Arabidopsis (Sauer et al. 2016).
In monocots, ODM was used to target AHAS in maize (Zhu et al. 1999) and rice (Okuzaki and Toriyama 2004). In maize, nucleotide changes were induced at two sites, S621A (AGT to AAT) for imidazolinone and sulfonylurea herbicide resistance and P165A, mimicking the point mutation in tobacco in Beetham’s work (Beetham et al. 1999). The oligonucleotides were transformed into maize cells by bombardment and selected with either 7 μM imazethapyr for S612A or 20 bbp chlorsulfuron for P165A. The mutation frequencies were 1.0 × 10− 4 to 1.4 × 10− 4, approximately three orders of magnitude higher than that of spontaneous mutation and gene targeting by homologous recombination pathway in plants (Tong Zhu et al. 1999). In rice, three chimeric DNA/RNA oligonucleotides for targeted modification of ALS, P171A, W548 L and S627I, were introduced into rice calli by bombardment. Screening by herbicide selection (chlorsulfuron for P171A and bispyribac-sodium for W548 L and S627I) and Sanger sequencing identified independent transformants for both P171A and W548 L but not S627I at a frequency of 1 × 10− 4. The ODM approach was also demonstrated in a wheat system using a transient assay with GFP as a reporter. The authors claimed that using 2,4-D in osmotic media boosted the gene targeting efficiency and that the repair of point mutations had a higher frequency than that of single base deletions in immature wheat embryos (Dong et al. 2006).
ODM products have been considered non-GMOs in a number of countries, although not in the EU, due to the targeted point mutation mechanism and transgene-free outcome (Eriksson 2018). In 2011, the UK Advisory Committee of Releases into the Environment (ACRE) suggested that plants being developed by the ODM system should not be regulated as GMOs. Afterwards, the Federal Office of Consumer Protection and Food Safety of Germany decided that ODM products do not constitute GMOs in 2017. Based on its precise modification and the GMO regulation of this technology, ODM has potential for genome editing. However, its low efficiency is the main barrier for its application in research; thus, improving the editing frequency is essential. Recently, ODM and SDN have been combined to enhance the efficiency with the range of precise editing from 0.09% to 0.23% in an EPSPS target gene. This study also claimed that the transgene targeting efficiency of CRISPR/Cas9 was nearly 3 times higher than that of TALEN (Sauer et al. 2016).
HR-Based Gene Targeting
In 1988, gene targeting (GT) or HGT was first defined as modification of the host genome achieved by the integration of foreign DNA via the HR pathway (Paszkowski et al. 1988). This method provides a wide range of targeted genome modifications, such as precise insertion, deletion or replacement of a gene or an allele. In fact, HR is an ideal mechanism that can precisely repair DSBs during the S and G2 phases of the cell cycle, while homologous sequences (sister chromatids or donor templates) are available (Tamura et al. 2002). However, its low frequency in higher plants is still a hurdle for practical applications (Puchta 2005). The first application of HGT in a crop was the targeted knockout of the rice Waxy gene using positive/negative selection, which achieved approximately 1% frequency but also left a positive selection marker in the genome (Terada et al. 2002).
Since then, two other important achievements in the plant gene targeting field regarding frequency enhancement have come to light: (1) the key finding of on-target DSB roles (Puchta et al. 1993) and (2) methods to introduce high doses of autonomously homologous donor templates into targeted cells (Baltes et al. 2014). By inducing DSBs at a specific locus using the highly specific restriction enzyme I-Sce I, HDR efficiency can be enhanced from 10 to 100 times (Puchta et al. 1996). To further enhance the efficiency of HR for gene targeting, several approaches have been developed. First, site-specific nucleases such as ZFNs, TALENs and CRISPR/Cas systems are applied to induce double-strand breaks at the target sequence (Belhaj et al. 2013; Voytas 2013). The second approach takes advantage of the virus replicon system to increase the delivery ability and the number of donor templates; hence, the HGT efficiency is improved (Baltes et al. 2014). Apart from that, certain studies have demonstrated that overexpression of HR-involved genes or suppression of the NHEJ pathway led to improvement of HGT frequency (Endo et al. 2016; Qi et al. 2013; Shaked et al. 2005).
Prime Editing
RNAs were shown to involve in DSB repairs via non-templated or templated mechanisms in human and yeast cells (for more details, see review by Meers et al. 2016). In addition, Butt et al. (2017) successfully engineered the SpCas9 guide RNA scaffold called chimeric single-guide RNA (cgRNA) for acting as sgRNAs and repair templates in rice protoplast. The HGT rate for the replacement of two nucleotides of OsALS locus was shown to be as high as 16.88% of total mutations when plasmids carrying the CRISPR/Cas9 and cgRNA expression cassettes were transfected to the protoplast. Further, targeted insertion of 3xHA tag at the OsHDT701 locus using a cgRNA showed up to 4.69% of total mutations. However, the HGT rates were much lower when only cgRNA-SpCas9 ribonucleoprotein (RNP) complex was transfected while the mutation rates mediated by NHEJ were much higher (Butt et al. 2017). RNA transcripts were further validated as donor templates for HDR-mediated targeting OsALS locus using CRISPR/Cpf1 ribonucleoprotein (RNP) complex. Nonetheless, the HGT frequency obtained with ssRNA donor templates was at 0.07–0.13%, nearly ten folds lower compared to that of ssDNA donors (Li et al. 2019). To expand the use of RNA as templates for plant HGT approaches, further work needs to be done.
Recently, prime editing using guide RNA extensions for priming reverse transcription-mediated precise editing has been shown to be an excellent precision genome editing technique in mammalian cell lines. It would also be an excellent alternative for HGT with a shorter editing sequence coverage. Anzalone and colleagues tested variations in prime editing methods and demonstrated a wide range of specific genetic modifications, including 19 insertions up to 44 bp; 23 deletions up to 80 bp; 119 point mutations, including 83 transversions; and 18 hybrid edits at 12 human and mouse cell lines without explicit DSBs (Anzalone et al. 2019). The prime editor’s best version used a CRISPR / Cas complex developed by fusion of a reverse transcriptase (RT) to a C-terminal of nickase Cas9 (H840A) and a prime editing gRNA (pegRNA) with a 3 ‘extension that could bind to the 3 ‘nicked strands produced by the nCas9. When bound, the nicked strand’s free 3′-OH is used as a substratum for the RT to copy genetic information from pegRNA’s 3 ‘extension (Fig. 3c). If we design pegRNAs to produce modified nucleotides, they would be inserted into the genome during downstream repair processes. A second nick site present downstream of the first nick would support the retention of the de novo nucleotides introduced. (Fig. 3c) (Anzalone et al. 2019). Although prime editor has not yet been used in plant system, we expect this technology to have a bright future in plant genome editing, as plant HGT is much more challenging.
HGT in Monocots
HDR Mechanisms in Plants
One of the principal questions regarding cell response to DSBs is which repair consequences the cells favor: error-free or error-prone DNA products? In meiosis, error-prone crossing over (CO) or break-induced repair (BIR) (or even NHEJ) is preferred for creating genetic diversity by exchanging genetic information between parental chromosomes, a key factor for adaptation to environmental changes. However, we can expect an opposite situation in mitotic cells, which require genetic stability rather than diversity. In that case, should NHEJ be abolished from mitotic cells? The answer is absolutely not, and one of the key reasons may be the limitation of time, because a single DSB persistence may induce programmed cell death after a certain period of time (Nowsheen and Yang 2012). What can the cells do? NHEJ is so abundant and efficient in mending the broken ends. What can we expect from the bulky HDR apparatus?
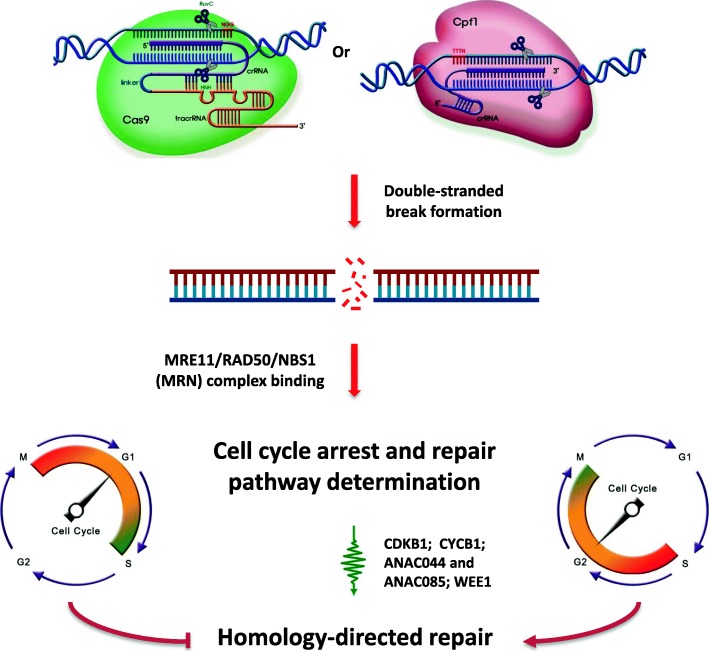
HDR has been extensively studied in yeasts and mammals for understanding the mechanisms of genetic diseases caused by DNA DSB damage. Most of the components of the plant HDR pathway are homologs of these known proteins (Schuermann et al. 2005), but the regulation of DSB responses in the kingdoms may be different (Yokota et al. 2005). Unlike in animal systems, HDR efficiency in plant somatic cells is extremely low (Szostak et al. 1983; Puchta et al. 1996) and very much dominated by NHEJ. Plant mitotic HDR is absent in the G1 phase and limited to S/G2, while NHEJ is active throughout the cell cycle (Fig. 4). The HDR pathway is determined by the presence of a sister chromatid as a homologous DNA donor, which is normally produced by replication in the S phase and remains present until the M phase. Even in these favorable cell cycle phases, the HDR pathway has to compete with the predominant NHEJ, and hence, it can be chosen in only certain conditions (Heyer et al. 2010; Voytas 2013; Jasin and Rothstein 2013). Therefore, a comprehensive knowledge of the conditions that favor HDR in plant somatic cells would offer key strategies in plant gene targeting for crop improvement.
Fig 4.
Homology-directed repair pathway determination and its favorable cell contexts. Activation of the MRN complex and P53/SOG1 triggers the activation of cell cycle checkpoint proteins such as CDKB1 (G2/M checkpoint) and CYCB1 (S phase checkpoint) or NAC-type transcription factors ANAC044 and ANAC085 (S/G2 checkpoints) or WEE1 kinase for cell cycle arrest
Sensing DSBs and Cell Cycle Arrest
In mammals, DSB formation induces cell cycle arrest, which is necessary to help the cell repair the damage in a reasonable time (Kastan and Bartek 2004). The process is initiated with the conformational changes of the ATM (ataxia telangiectasia mutated) homodimer resulting from sensing a chromatin structure change following DSB formation. Activation of human ATM by autophosphorylation of its serine 1981 disassociates its activated monomers (Bakkenist and Kastan 2003). Monomeric ATM then phosphorylates all the members of the MRE11 (meiotic recombination 11)/RAD50 (Radiation sensitive 50)/NBS1 (Nijmegen breakage syndrome 1) (MRN) complex, a DSB sensor holoenzyme, and is additionally phosphorylated by MRE11 (Lee and Paull 2005; Lamarche et al. 2010). Subsequently, ATM plays a central role in activating cell cycle checkpoint kinases and P53 and indirectly induces the suppression of cyclin-dependent kinases that ultimately leads to cell cycle arrest (Kastan and Bartek 2004; Harper and Elledge 2007; Yata and Esashi 2009). In Arabidopsis, SOG1 (SUPPRESSOR OF GAMMA RESPONSE 1), activated by ATM, is responsible for the regulation of multiple downstream proteins such as CDKB1 (G2/M checkpoint) and CYCB1 (S phase checkpoint), the NAC-type transcription factors ANAC044 and ANAC085 (S/G2 checkpoints) or WEE1 kinase for cell cycle arrest (Yoshiyama et al. 2013; Weimer et al. 2016; Takahashi et al. 2019; De Schutter et al. 2007).
HDR Pathway Determination
Post DSB formation, cell cycle arrest at S/G2 favors the essential condition for the HDR pathway (Fig. 4). In animals, the HDR pathway is determined by recruitment competition between KU70/80-DNA-PK and the MRN complex to the DSB ends and subsequent resection regulation by BRCA1/CtIP and 53BP1/RIF1, which favors HDR and NHEJ, respectively. However, only the Ku complex but not DNA-PK is conserved in plants (West et al. 2002; Tamura et al. 2002), suggesting an alternative regulation of activation by kinases in the plant kingdom. KU70 was shown to colocalize and interact with MRE11 in somatic cells and therefore was proposed to be a key player in the determination of the DSB repair pathway (Goedecke et al. 1999). Because the majority of DNA end binding proteins in a cell are KU70/80 (Gottlieb and Jackson 1993), NHEJ becomes dominant, and hence, HDR efficiency, especially in plant mitotic cells, is extremely low. Recently, it has been increasingly accepted that DSB end resection plays a key role in the determination of NHEJ- or HDR-mediated repair. NHEJ repair keeps the broken end resection in a limited range for its amendment, but HDR requires DSB end resection to produce 3′-protruding ends that are long enough for template annealing and replication of homologous genetic information. NHEJ resection length usually ranges from 0 to 14 bp, although very rare cases can be 25 bp and longer (Lieber 2010).
ATM-dependent phosphorylation of RAD50, NBS1 and MRE11 of the MRN complex plays an important role in DSB end resection and determines the ultimate repair pathway in an MRN-dependent manner. MRE11 acts as an endonuclease that nicks DNA upstream of the break and subsequently resects 3′- > 5′ toward the break, and then, the end is further resected by Endonuclease 1 and Dna 2 (Kijas et al. 2015). CtIP, activated by ATM, acts in concert with the MRN complex to enhance resection and HDR. CtIP physically interacts with the MRN complex and, more importantly, BRCA1 (Limbo et al. 2007), a protein that inactivates 53BP1 by dephosphorylation (Isono et al. 2017), thereby supporting DSB end resection for HDR determination. However, in a recent study, 53BP1 was shown to shield DSB ends from extensive resection, which might result in a strong bias toward RAD52-dependent error-prone SSA (Ochs et al. 2016). Broken end resection is also controlled by phosphorylated MRE11, which protects exonuclease 1 from extensive resection by phosphorylating it (Kijas et al. 2015). In Arabidopsis, PHF11 (plant homeodomain finger 11) plays roles in binding and suppressing RPA, thereby enhancing Exo1 resection (Gong et al. 2017). Furthermore, the resection coordination activity of MRE11 and CtIP/Ctp1 may inactivate KU70/80 and unload it from the broken ends. Meanwhile, a predefined resection length may deactivate the MRN complex and disassociate it from the ends (Langerak et al. 2011).
DSB Amendment by HDR
Once the HDR pathway is determined, in the presence of homologous DNA templates, HDR can occur through gene conversion or synthesis-dependent strand annealing (SDSA), single-stranded annealing (SSA) or crossover (CO, DSB repair (DSBR))/noncrossover (NCO) via double Holliday junction (dHj) formation (Holliday 1977). Only the former HDR subpathway can produce precise sequence products (Fig. 1). In plant somatic cells, SDSA was proven to be the major HDR mechanism to precisely repair damaged DNA (Szostak et al. 1983; Puchta et al. 1996; Voytas 2013). The differentiation of HDR subpathways has been well studied in yeasts and mammals but still remains a matter of investigation in higher plants. In the case of HDR, phosphorylation of H2AX histone protein by ATM or DNA-PKcs is important to open nucleosomes for strand annealing. As one H2AX is present for every 10 nucleosomes, efficient HDR requires relaxing up to thousands of base pairs (Lieber 2010). The resection of broken ends at a controllable length of 3′ ssDNA overhangs would favor RAD51-dependent SDSA repair. RPA binds to the resected ssDNAs to prevent the formation of a secondary loop for RAD51 loading. RAD51 loading is facilitated by BRCA2 through its BRC motif, which plays dual roles as an ssDNA-dsDNA junction binding protein as well as a RAD51 docking site provider (Seeliger et al. 2012; Heyer et al. 2010; Dray et al. 2006). The tight regulation of RAD51 loading and nucleofilament formation has been shown to involve a BRCA2-antagonistic protein called FIDGETIN-LIKE-1 (FIGL1) (Fernandes et al. 2018; Girard et al. 2015; Kumar et al. 2019). Extensive end resection with the involvement of Exonuclease 1 (Exo1) and/or Sgs1-Dna2 would lead to RPA disassociation facilitated by RAD52, which redirects to the error-prone SSA repair pathway (Heyer et al. 2010) (Fig. 1).
In Arabidopsis, INVOLVED IN DE NOVO2 (IDN2) was shown to help RAD51 loading by binding to RPA and unloading it from DSB ends (Liu et al. 2017). RAD51 binds to the resected ssDNA overhang, forming nucleoprotein filaments or presynaptic filaments. The filament structure invades the donor template sequence and then searches for and anneals to the complementary sequence; this process is followed by displacement loop (D-loop) formation (Rajanikant et al. 2008). RAD54 binds to and is required for supporting RAD51 strand invasion and annealing and for the disassociation of RAD51 afterward (Klutstein et al. 2008; Osakabe et al. 2006). RAD54 formed DNA repair foci in living Arabidopsis cells depending on ATM-SOG1 signaling and coincided with the formation of phosphorylated H2AX (Hirakawa et al. 2017). Subsequently, the free 3′ OH end of the invaded ssDNA primes donor template-dependent DNA synthesis. This process determines the outcomes of HDR with several subpathways (DSBR, dHJ and SDSA) depending on the type of DNA synthesis and resolution of the final products (Fig. 1). In the later stage of homologous template-dependent synthesis in somatic cells, the D-loop may be processed and reannealed by the activity of RAD5A, REC4Q and MUS81 (Mannuss et al. 2010; Hartung et al. 2006). Only SDSA can generate precise repair products and is favored in mitotic cells (Heyer et al. 2010; Puchta 2005).
HGT in Monocots
Plant gene targeting or HGT was defined by the homology-directed repair (HDR) of an endogenous gene by exogenously introduced homologous DNAs (Paszkowski et al. 1988). Obviously, the initial experiment obtained a very low frequency of homologous recombination (~ 10 − 4), indicating difficulty but feasibility in engineering plant genomes by site-specific gene targeting. Early in the 1990s, a transgenic approach using a preintroduced yeast mitochondrial I-SceI endonuclease as a DSB inducer was adopted in attempts to investigate the mechanisms of DSB repair in plants, especially the HDR pathway in plant somatic cells (Puchta et al. 1993; Fauser et al. 2012; Szostak et al. 1983). It became clear that the HDR pathway employing homologous DNA templates to precisely repair DSB-damaged DNAs occurred mainly via the SDSA mechanism (Fig. 1) with an extremely low efficiency. Nonetheless, the induced DSBs could improve HGT efficiency up to two orders of magnitude (Szostak et al. 1983; Puchta et al. 1996), a large step in plant gene targeting research. Recently, the emerging CRISPR/Cas systems, which have proven to be powerful molecular scissors for in vivo generation of site-specific DSBs, have revolutionized the plant gene targeting approach and brought hope for practical applications in crop improvement.
However, despite the application of flexible approaches (i.e., particle bombardment, protoplast transfection and Agrobacterium-mediated transformation) for the delivery and execution of HGT tools, gene targeting in most major crops is still challenging. As mentioned in the previous sections, most of our knowledge about the principal mechanisms of plant HDR has been taken from yeast and animal research studies, and some of those results are inconsistent with observations in the plant kingdom. Therefore, the plant genome engineering community should continuously focus on research to understand plant-specific factors involved in DSB repair, especially via the HDR pathway, the only approach providing precise gene targeting products. Using this background knowledge, one can propose approaches for improving gene targeting frequency. Two of the most important factors affecting gene targeting efficiency in plant somatic cells are 1) DSB formation at the targeted sites and 2) the number of homologous DNA templates available for the sites of breakage (Puchta et al. 1993; Puchta 2005; Townsend et al. 2009; Endo et al. 2016; Baltes et al. 2014).
Because most of the early studies focused on gene targeting in model dicot plants such as Arabidopsis, tobacco and tomato (for reviews, see (Voytas 2013; Puchta 2005), monocot gene targeting represented a large gap in the early reports, indicating major challenges in monocot gene targeting. In this section, we aim to summarize recent knowledge regarding gene targeting in the monocot plants that represent most of the major food crops for human beings. In addition, we discuss challenges and suggest potential solutions for improving gene targeting frequency in monocots.
HGT without Targeted DSBs
In vivo plant gene targeting without assisted selection was extremely low (Puchta and Hohn 1991; Paszkowski et al. 1988). The first targeted knockout of an endogenous “waxy” allele via HGT was successfully generated in rice at a 0.94% frequency by Terada and coworkers (2002) with an innovative positive (hygromycin phosphotransferase II (HptII)-based)/negative (using diphtheria toxin A (DT-A) subunit) selection method (Table 1). The frequency of the gene-targeted waxy and xyl (b1,2-xylosyltransferase) knockout alleles was further improved by the transformation frequency (Ozawa et al. 2012). The weak point of this strategy is the obligatory use of an associated marker gene; hence, the product is subject to GMO categorization. Therefore, Cre/loxP was applied to excise the marker from the gene-targeted allele (Terada et al., 2010; Dang et al. 2013). The approach was later successfully applied to functional genomic studies via tagging endogenous genes with visible marker(s) (Yamauchi et al. 2009; Moritoh et al. 2012; Ono et al. 2012; Tamaki et al. 2015). The positive/negative system using the DT-A subunit might have posed risks to dicots, because it has not been successfully applied in those plants. Therefore, an alternative positive/negative selection system was developed as an alternative, based on a caffeic acid O-methyltransferase (codA) D314A single-mutated version as the negative selection marker (Osakabe et al. 2014) or neomycin phosphotransferase II (NptII) (positive)/RNAi-based anti-NptII (negative) selection at much lower frequencies (Nishizawa-Yokoi et al. 2015b), which might be a result of less efficient G418 selection in rice. Nonetheless, the positive/negative selection strategy was shown to be unsuccessful in barley (Horvath et al. 2016), highlighting its extremely low efficiency in the absence of DSB and the high genome complexity of monocot gene targeting. In an herbicide-selection-based gene targeting experiment, Endo and coworkers successfully replaced the WT allele of rice ALS with the W548 L and S627I alleles and obtained homozygous T2 plants hypertolerant against an herbicide named bispyribac (BS). Under BS selection, gene targeting occurred at both loci at ~ 3% (Endo et al. 2007). The frequency of targeting OsALS for BS tolerance was enhanced to 6% by using the abovementioned HptII/DT-A selection system, and the selection marker was subsequently excised with the piggyBac system, which can remove a marker gene without leaving a DNA scar (Nishizawa-Yokoi et al. 2015a).
Table 1.
Major HGT studies in monocots
| No. | Gene/Allele | Monocot | HDR tool | Selection marker | HDR allele-associated marker | Homologous donor length (bp) (5′ arm + 3′ arm) | Gene targeting efficiency | Reference | |
|---|---|---|---|---|---|---|---|---|---|
| Gene targeting without targeted DSBs | |||||||||
| 1 | waxy (Granule-Bound Starch Synthase gene knockout) | rice | T-DNA | HptII (positive)/DT-A (negative) | HptII (positive) | 6300 + 6800 | 0.94% (per total surviving calli) | Terada et al. 2002 | |
| 2 | Acetolactate synthase (ALS) (W548 L and S627I) | rice | T-DNA | Bispyrobac-sodium (BS) herbicide (positive) | BS herbicide (positive) | 8092 | ~ 3% (per total surviving calli) | Endo et al. 2007 | |
| 3 | Alcohol dehydrogenase2 (Adh2) | rice | T-DNA | HptII (positive)/DT-A (negative) | HptII (positive) | 6200 + 6000 | 1.9% (per total surviving calli) | Terada et al. 2007 | |
| 4 | GUS-HptII tagging | rice | T-DNA | HptII (positive)/DT-A (negative) | HptII (positive) | 3100 + 3100 | ~ 5.3% (per total surviving calli) | Yamauchi et al. 2009 | |
| α-subunit of anthranilate synthase gene (OASA2) | rice | T-DNA | Trp-analog 5MT (positive) | Trp-analog 5MT (positive) | 7000 in total | 0.34% (per total surviving calli) | Saika et al. 2011 | ||
| 5 | Domains Rearranged Methylase 2 (OsDRM2), 70 bp targeted deletion | rice | T-DNA | HptII (positive)/DT-A (negative) | HptII (positive) | 3000 + 3100 | 1.9% (per total surviving calli) | Moritoh et al. 2012 | |
| OsDRM2, 3000 bp targeted deletion | rice | T-DNA | HptII (positive)/DT-A (negative) | HptII (positive) | 3000 + 3000 | 0.4% (per total surviving calli) | |||
| 6 | Repressor Of Silencing 1a (ROS1a) | rice | T-DNA | HptII (positive)/DT-A (negative) | HptII (positive) | 3000 + 3000 | 1.1% (per total surviving calli) | Ono et al. 2012 | |
| 7 | waxy | rice | T-DNA | HptII (positive)/DT-A (negative) | HptII (positive) | 6800 + 5700 | 1.5% (per total surviving calli) | Ozawa et al. 2012 | |
| β1,2-Xylosyltransferase (XylT) | rice | T-DNA | HptII (positive)/DT-A (negative) | HptII (positive) | 5500 + 5600 | 1.8% (per total surviving calli) | |||
| 8 | OsRac1 | rice | T-DNA | HptII (positive)/DT-A (negative), Cre/loxP-based removal of HptII | HptII (positive), Cre/loxP-based removal of HptII | 3000 + 3000 | ~ 5.3% (per total surviving calli) | Dang et al. 2013 | |
| Caffeic acid O-methyltransferase (CAOMT) | rice | T-DNA | HptII (positive)/CodA (negative) | HptII (positive) | 3700 + 6000 | ~ 12.1% (per total surviving calli) | Osakabe et al. 2014 | ||
| 9 | ALS (W548 L and S627I) | rice | T-DNA | BS herbicide (positive) and HptII (positive)/DT-A (negative), piggyBac-based removal of HptII | BS herbicide (positive) and HptII (positive), piggyBac-based removal of HptII | 8092 | ~ 6% (per total hygromycin-resistant calli) | Nishizawa-Yokoi et al. 2015a | |
| cleistogamy 1 (Oscly1) | rice | T-DNA | BS herbicide (positive) and HptII (positive)/DT-A (negative), piggyBac-based removal of HptII | BS herbicide (positive) and HptII (positive), piggyBac-based removal of HptII | 6000 | ~ 0.078% | |||
| 10 | waxy | rice | T-DNA | NptII (positive)/RNAi mediated anti-NptII (negative) | NptII (positive) | 5000 + 5100 | 0.26% (per total surviving calli) | Nishizawa-Yokoi et al. 2015b | |
| Glyoxalase I (OsGlb33) | rice | T-DNA | NptII (positive)/RNAi mediated anti-NptII (negative) | NptII (positive) | 5400 + 6200 | 0.21% (per total surviving calli) | |||
| Targeted DSB-based gene targeting | Site-specific Nuclease | ||||||||
| 11 | Inositol-1,3,4,5,6-pentakisphosphate 2-kinase (IPK) and phosphinothricin acetyltransferase (PAT) | Maize | Silicon carbide whiskers-mediated transformation | Bialaphos herbicide (positive) | Bialaphos herbicide (positive) | 815 + 815 | ? | Shukla et al. 2009 | Meganucleases |
| 12 | Synthetic DNA | Maize | T-DNA | NptII and GFP (positive) | NptII and GFP (positive) | 2992 + 1200 | 0.085% (True event per immature embryo) | Ayar et al. 2013 | ZFNs |
| OsPDS | rice | Bombardment | Transient | Transient | 23 + 37 (single stranded oligos) | 6.9% (2/29 clones of PCR product, restriction digestion for enrichment of HDR products | Shan et al. 2013 | CRISPR/Cas9 | |
| 13 | Pre-intergrated GFP | Barley | Bombardment | GFP/YFP fluorescence | GFP/YFP fluorescence | 196 + 334 | 2–3% (per total surviving calli) | Budhagatapalli et al. 2015 | TALENs |
| 14 | Acetolactate synthase (ALS) genes (ALS1 and ALS2) | Maize | Bombardment | Chlorsulfuron and bialaphos (positive selection of ALS HDR events). | bialaphos (positive selection of ALS HDR events). | 749 dsDNA donor and 127 single stranded oligo donor | 0.2% (dsDNA donor) and 0.35% (ssDNA donors) | Svitashev et al. 2015 | CRISPR/Cas9 |
| Upstream of the LIGULELESS1 (LIG1) gene | Bialaphos (positive) | Bialaphos (positive) | 1099 + 1035 | 0.7% (meganulease) and 2.5–4.1 (CRISPR/Cas9) | Meganuclease and CRISPR/Cas9 | ||||
| 15 | OsALS (W548 L and S627I) | rice | Bombardment | BS herbicide (positive) | BS herbicide (positive) | ? | 1.4–6.3% (per total hygromycin tolerant calli) | Li et al. 2016 | TALENs |
| 16 | OsALS (W548 L and S627I) | rice | Bombardment and T-DNA | Hygromycine and BS (positive) | BS (positive) | 100 + 46 (330 bp interval) | 7.0–25% (number of perfect HDR events per total calli) | Sun et al. 2016 | CRISPR/Cas9 |
| 17 | OsALS (W548 L and S627I) | rice | T-DNA | Hygromycine and BS (positive), lig4 background | BS (positive) | 329 + 635 | 0.147–1.000% (number of perfect HDR events per total calli) | Endo et al. 2016 | CRISPR/Cas9 |
| 18 | Nitrate transporter NRT1.1B | rice | Bombardment | Hygromycine (positive) | None | 100 + 100 | 6.72% | Li et al. 2018a | CRISPR/Cas9 |
| 19 | OsALS (W548 L and S627I) | rice | Bombardment | Hygromycine and BS (positive) | BS (positive) | 97 bp left arm only or 97 + 121 | 0.66% (only left homologous arm) and 1.22% (both arms) | Li et al. 2018b | CRISPR/Cpf1 |
| 20 | Wheat ubiquitin gene (TaUbi) | wheat | WDV replicon bombardment | GFP (positive) | GFP | 748 + 773 | 3.8% (per total protoplasts) and 5.74% (per total cells, normalized to scutela transformation efficiency) | Gil-Humanes et al. 2017 | CRISPR/Cas9 |
| Mildew Locus O (TaMLO) | wheat | WDV replicon bombardment | BFP (positive) | BFP | 674 + 647 | 6.4% (per total cells, normalized to scutela transformation efficiency) | |||
| 5-enolypyruvylshikimate-3-phosphate synthase (TaEPSPS) | wheat | WDV replicon bombardment | dsRED (positive) | dsRED | 210 + 646 | 4.7% (per total cells, normalized to scutela transformation efficiency) | |||
| TaUbi and TaMLO, multiplexed | wheat | WDV replicon bombardment | GFP and BFP (positive) | GFP and BFP | 748 + 773 (TaUbi) and 674 + 647 (TaMLO) | 1.1% (per total cells, normalized to scutela transformation efficiency) | |||
| TaUbi and TaEPSPS, multiplexed | wheat | WDV replicon bombardment | GFP and dsRED (positive) | GFP and dsRED | 748 + 773 (TaUbi) and 210 + 646 (TaEPSPS) | 0.4% (per total cells, normalized to scutela transformation efficiency) | |||
| 21 | OsACT and OsGST | Rice | WDV replicon T-DNA | GFP and kanamycin (positive) | GFP and kanamycin | 500 + 500 | Cas9 overexpressed background: 19.4% (OsACT) and 7.7% (OsGST); WT background 8.5% (OsACT) and 4.7% (OsGST) | Wang et al. 2017 | CRISPR/Cas9 |
Targeted DSB-Based HGT
DSBs induced at the gene targeting sites were shown to dramatically enhance efficiency by several orders of magnitude (Puchta et al. 1993). Since the introduced I-SceI meganuclease-mediated DSBs showed significant enhancement of gene targeting frequency, ectopic recombination was tested in maize and revealed remarkably higher efficiencies than the no-DSB strategies particle bombardment and Agrobacterium-mediated transformation (D'Halluin et al. 2008; Ayar et al. 2013). However, because the preintroduced homing nuclease targets a predefined sequence in the genome of the plant, gene targeting for a native gene/allele of interest in plant genomes still fell far short of expectations, and site-specific molecular scissors were in high demand. Bearing that in mind, researchers engineered ZFNs, zinc finger motifs for DNA binding fused to the type IIS endonuclease FokI, for efficiently and specifically generating DSBs in vivo (Kim et al. 1996) and obtained significant enhancement of gene targeting efficiency at native loci in Drosophila (~ 1.5%) (Bibikova et al. 2003) and human cells (~ 18%) (Urnov et al. 2005). Subsequently, ZFNs were applied to plant gene targeting and yielded an average of 17% HGT efficiency with a preintegrated GUS:NPTII reporter system in tobacco protoplasts (Wright et al. 2005). A similar strategy also obtained ~ 10% HGT efficiency in restoring a preintegrated defective herbicide-tolerance gene (Cai et al. 2009). For targeting multiple allelic loci, also acting as an herbicide-tolerance selection marker, the efficiency was several-fold lower at ~ 2% in tobacco (Townsend et al. 2009). In monocots, ZFN-based gene targeting was first shown to be efficient in maize via integrated insertion of an herbicide-tolerance gene as a selection marker into a native inositol-1,3,4,5,6-pentakisphosphate 2-kinase (IPK) gene (Shukla et al. 2009). Although ZFNs offered a great advantage over meganucleases in plant gene targeting, their design, validation and specificity optimization processes were extremely time consuming and laborious (Puchta and Hohn 2010).
TALENs, the second generation of sequence-specific nucleases, also used protein-based DNA binding domains for targeting sites of interest. Their highly specific and modular binding repeats offered an easier alternative for plant gene targeting. The first plant gene targeting events via HGT using TALENs were in tobacco calli regenerated from protoplasts at 3.5% efficiency without any selection marker (Zhang et al. 2013). Overall, 3.5% of calli showed HGT events without antibiotic selection; however, it is not clear how many protoplasts were used for transfection. The TALEN approach was first applied in monocots to demonstrate the feasibility of gene targeting and reached 2–3% post bombardment of leaves with TALENs plus donors (Budhagatapalli et al. 2015). In rice, a similar range (1.4–6.3%) of gene targeting frequencies was obtained with the OsALS herbicide-tolerance allele (Li et al. 2016).
With the advent of CRISPR/Cas, which revolutionized molecular scissors for DSB formation, plant gene targeting is in theory applicable to any gene/crop of interest due to the simplicity, flexibility and versatility of the system (Jinek et al. 2012; Zetsche et al. 2015). CRISPR/Cas tools have been adapted for wide use in genome engineering studies in various kingdoms, including Plantae (Jinek et al. 2012; Hsu et al. 2014; Barrangou and Doudna 2016). The first attempt to modify a monocot genome via HGT using CRISPR/Cas9 was shown in 2013 by Shan and coworkers. In a transient experiment, OsPDS was modified by HGT at a 6.9% frequency in rice protoplasts using CRISPR/Cas9 for DSB formation and single-stranded oligos as donor templates (Shan et al. 2013). Gene targeting in maize was shown with an efficiency comparison between Agrobacterium-mediated delivery and particle bombardment and between a meganuclease and CRISPR/Cas9 at two loci, ALS and LIG1 (Svitashev et al. 2015). Several herbicide-tolerant lines were obtained from the bombardment approach only, indicating a very low targeting efficiency in maize and the requirement of a high dose of donor template and editing tools for enhancing it. The herbicide-tolerance ALS allele was also used in another CRISPR/Cas9-based gene targeting work using a short dsDNA donor delivered as linearized or plasmid forms by bombardment or Agrobacterium. With hygromycin and BS herbicide for double selection, the total frequency of HGT events reached 22.5–25%. In detail, most of the HGT lines (42/52) obtained from bombardment showed a range of diversity, with mixtures of perfect W548 L and imperfect S627I. In Agrobacterium-mediated delivery, most HGT lines (30/40) were perfect but heterozygous, and the HGT alleles were co-located at the loci with NHEJ alleles (Sun et al. 2016). The highly chimeric HGT patterns indicate prolonged activity of CRISPR/Cas9; unsynchronized states of the cells used in the experiments and/or predominance of organogenesis during shoot formation post editing. Therefore, to synchronize DSB formation and HDR, Endo et al. (2016) used calli stably expressing SpCas9 and sequential transformation of sgRNAs and repair templates for OsALS gene targeting. The HGT frequency was too low, and it was difficult to obtain the target plants. However, when the DNA ligase 4 (LIG4) gene, a key player in the NHEJ pathway, was knocked out before targeting, up to a 1% frequency of gene targeting among the total herbicide-tolerant calli was observed, indicating competition between the NHEJ and HDR pathways (Endo et al. 2016). In an attempt to modify the nitrate transporter gene NRT1.1B using CRISPR/Cas9-based tools, Li and coworkers obtained 6.72% precise replacement of 4 SNPs in the gene sequence without an additional allele-associated selection marker (Li et al. 2018a). The gene-targeted lines might contain DNA insertions in their genome due to the high frequency of DNA integration of the bombardment system, but this possibility was not examined.
An alternative to the Cas9 system is Cpf1-based molecular scissors. The latter cut dsDNAs using a T-rich PAM for binding initiation and usually form 5′ overhangs at their distal ends relative to the PAM (Zetsche et al. 2015). CRISPR/Cpf1 was also used for gene targeting in monocots and showed precise SDSA-based gene replacement at the OsALS loci at comparable frequencies (0.66–1.22%) (Li et al. 2018b) to those of Cas9 systems (Endo et al. 2016).
Replicon-Based HDR
Because of the highly efficient replication of geminivirus genomes and their single-stranded DNA nature, these genomes have been used as perfect DNA template cargo for gene targeting in plants. Geminiviral genomic DNAs have been reconstructed to overexpress foreign proteins in plants at up to 80-fold higher levels than those of conventional T-DNA systems (Needham et al. 1998; Mor et al. 2003; Zhang and Mason 2006) due to their highly autonomous replication inside host nuclei and the ability to reprogram cells (Gutierrez 1999; Hanley-Bowdoin et al. 2013). Furthermore, Rep/RepA has been reported to promote a cell environment that is permissive for HR to stimulate the replication of viral DNA (Baltes et al. 2014). Interestingly, it has been reported that somatic HR is promoted by geminiviral infection (Richter et al. 2014). The above characteristics of geminiviral replicons have been shown to make them perfect delivery tools for introducing large amounts of homologous donor templates to plant nuclei. Likewise, the movement and coat proteins of a bean yellow dwarf virus (BeYDV)–based replicon were removed and replaced with Cas9 or TALEN to improve gene targeting in plants (Baltes et al. 2014; Butler et al. 2016; Cermak et al. 2015; Dahan-Meir et al. 2018).
In monocots, wheat dwarf virus (WDV) was first engineered for CRISPR/Cas9-based genome editing and gene targeting in wheat (Gil-Humanes et al. 2017). More importantly, this work showed the feasibility of multiplexed gene targeting of multiple homeoalleles of the wheat genome at a 1% frequency. A similar approach using WDV was also applied in rice for targeted insertion of GFP-2A-NPTII to the C terminals of ACT1 and GST genes in a Cas9-overexpressing WT background. The WDV replicon-based tools showed significantly higher targeted knock-in efficiencies than conventional T-DNA tools (Wang et al. 2017).
Present Challenges
Despite higher success rates in gene targeting in plants, most of the abovementioned cases required marker-associated or selectable loci, while the selection and regeneration of HGT events from edited cells are still challenging (Butler et al. 2016; Gil-Humanes et al. 2017; Hummel et al. 2018). The most effective delivery method for HGT tools was reported to be particle bombardment, with relatively high frequencies of gene targeting (see Table 1) due to the high doses of introduced donor DNAs, but it also resulted in multiple DNA integration and/or regeneration difficulties. In addition, compared to other delivery methods, such as Agrobacterium-mediated transformation, particle bombardment requires special equipment and costly consumables that are not widely available in every research laboratory. Agrobacterium-mediated transformation is a very common and cost-effective method for plant gene targeting, but it showed too low frequencies with conventional T-DNA cargos (Table 1). There has been one solution for delivery of high copy numbers of donor DNAs, without facilitating multiple DNA integration, using autonomous DNA replicons (Baltes et al. 2014; Cermak et al. 2015), but this technique is still challenging in monocots if not used in combination with bombardment (Wang et al. 2017) or with a stable Cas9-overexpressing background and selectable marker (Gil-Humanes et al. 2017). The frequencies were dramatically reduced if multiple allelic loci and/or polyploid plants were targeted (Table 1). Furthermore, the effective application of replicon cargos in plant gene targeting has been shown to be limited by their size (Baltes et al. 2014; Suarez-Lopez and Gutierrez 1997; Gil-Humanes et al. 2017). Therefore, plant gene targeting, especially in cases of nonselectable alleles, is still a matter of improvement.
Potential Solutions and Perspectives on Monocot HGT
To improve plant gene targeting frequency, understanding HDR mechanisms and finding optimal conditions for HDR are the most important subjects in the field. The initial data on DSB-based gene targeting led to an important conclusion that in plant somatic cells, the majority of HGT-based products were formed via the SDSA pathway (Fig. 1) (Puchta 1998; Voytas 2013; Vu et al. 2017; D'Halluin et al. 2008). Because it is well known that DSB formation is one of the key factors in gene targeting and that viral replicons are used as efficient delivery systems for HDR donor templates, we will discuss and propose only other factors regarding monocot gene targeting here.
The Role of Homologous Donor Templates
The initial experiments for understanding plant homologous recombination were mostly conducted in a transient manner using newly introduced homologous DNAs/plasmids in plant protoplasts/cells. Baur et al. (1990) reported extrachromosomal homologous recombinations between two plasmids in tobacco mesophyll protoplasts. The most favorable donor plasmids were in linearized forms that obtained 15- to 88-fold higher recombination efficiency and were proportional to homologous zone size. The closer the break sites were to homologous zones, the higher the recombination frequencies were (Table 2) (Baur et al. 1990). Puchta and Hohn also confirmed that the homologous zone sizes (456 bp to 1200 bp) have a direct correlation with extrachromosomal recombination frequencies in Nicotiana plumbaginifolia protoplasts. The frequency was significantly reduced when the homologous zone size was 456 bp or lower (Puchta and Hohn 1991). Single-stranded DNA templates were shown to be efficient substrates for extrachromosomal recombination because they could directly facilitate the initial annealing step between the donor and targeted DNAs. Double-stranded circular DNAs were the least efficient templates for the recombination mode (Bilang et al. 1992; de Groot et al. 1992).
Table 2.
Potential approaches for improvement of HGT in monocots shown in this review
| No. | Category | Approach | HGT mode | Tested in organism | HGT enhancement properties | References |
|---|---|---|---|---|---|---|
| 1 | homologous repair template | Homologous size and form | Extrachromosomal | Nicotiana tabacum | The longer the better; Linearized forms were much better | Baur et al. 1990 |
| 2 | homologous repair template | Homologous size and form | Extrachromosomal | Nicotiana plumbaginifolia | Homologous size was preferred to be more than 456 bp | Puchta and Hohn 1991 |
| 3 | homologous repair template | Homologous form | Extrachromosomal | Nicotiana tabacum | ssDNA was more preferred | Bilang et al. 1992; de Groot et al. 1992 |
| 4 | HDR-related genes | EcRecA overexpression | Intrachromosomal | Nicotiana tabacum | 10 folds-efficiency increase with nuclear localized EcRECA | Reiss et al. 1996 |
| 5 | HDR-related genes | RuvC overexpression | Somatic crossover; Extrachromosomal; intrachromosomal | Nicotiana tabacum | Somatic crossover (12 folds); intrachromosomal recombination (11 folds); and extrachromosomal recombination (56 folds) | Shalev et al. 1999 |
| 6 | HDR-related genes | yeast RAD54 overexpression | SDSA | Arabidopsis thaliana | 27-fold enhancement | Shaked et al. 2005 |
| 7 | NHEJ and HDR-related genes | RAD50 knockout | Intrachromosomal | Arabidopsis thaliana | 8–10 folds of SSA or intrachromosomal recombination | Gherbi et al. 2001 |
| 8 | NHEJ related genes | AtMLH1 knockout | Intrachromosomal | Arabidopsis thaliana | Knockout mutation reduced 72% HDR frequencies | Dion et al. 2007 |
| 9 | Chromatin modeling | AtFAS1 or AtFAS2 knockout | Interchromosomal | Arabidopsis thaliana | 40-fold enhancement of somatic homologous recombination | Endo et al. 2006 |
| 10 | Cell Cycle Synchronization/S phase | Hydroxyurea treatment | SDSA | Yarrowia lipolytica, Arxula adeninivorans, Saccharomyces cerevisiae, Kluyveromyces lactis and Pichia pastoris | 1.2- to 8-fold enhancement | Tsakraklides et al. 2015 |
| 11 | Cell Cycle Synchronization/S-G2 phases | Cas9 fused with N-terminal (110a.a) of human Gemini | SDSA | Human cell lines (HEK293T) | 72% enhancement of somatic homologous recombination | Gutschner et al. 2016 |
| 12 | Culture conditions | Polyamines (putrescine, spermidine and spermine) | SDSA | Mouse hair follicle model and Human cell lines (U2OS and HEK293) | Polyamines promoted RAD51 loading onto ssDNAs in in vitro assays | Lee et al. 2019 |
| 13 | NHEJ related genes | Suppression by chemical inhibitors | SDSA | Human cell lines (HEK293T) | KU70 and DNA ligase IV suppression by Scr7 obtained 4–5-fold increase of HR efficiency | Chu et al. 2015 |
| 14 | NHEJ related genes | Suppression by chemical inhibitors | SDSA | Human cell lines and mouse | DNA ligase IV suppression by Scr7 enhanced CRISPR/Cas9-based HDR frequency up to 19 folds | Maruyama et al. 2015 |
Positive-Negative Selection
Selection and/or regeneration of gene targeting transformants are critical to the success of the approach. The dual mode of selection strongly enhanced the possibility of obtaining gene targeting events in monocots (see Table 1), even without the involvement of the revolutionary CRISPR/Cas molecular scissors. The positive-negative selection system provides a large advantage in rice HGT and may help us improve crops by HGT (Terada et al. 2002). The hurdles in the removal of the associated positive selection markers have been solved by using a smart transposon-based excision system (Nishizawa-Yokoi et al. 2015a). It is exciting to combine the positive-negative selection system with the high DSB performance of CRISPR/Cas complexes for monocot gene targeting.
Overexpression of Genes Involved in the HDR Pathway
A good number of HDR-related protein homologs have been identified among prokaryotes and eukaryotes. Attempts have also been made to study and/or improve HDR in somatic cells by overexpressing the proteins in targeted organisms. We discuss these approaches in this section, thereby highlighting important points for the improvement of plant gene targeting frequency.
The Escherichia coli RecA protein (EcRecA) was shown to be involved in HR in this bacterium by facilitating ssDNA searching and annealing to its homologous DNA repair templates and subsequently exchanging and displacing the sequence (Radding 1981; Muniyappa et al. 1984; Chen et al. 2008). Overexpression of EcRecA in tobacco protoplasts enhanced the DNA repair efficiency 3-fold upon treatment with interstrand DNA crosslinking agent (mitomycin C) (Table 2). Intrachromosomal HR frequency was also shown to be 10 times higher in cells expressing the protein (Reiss et al. 1996). However, an SpCas9-EcRecA fusion was shown to enhance indel mutation via supporting the SSA repair mode (Fig. 1) and hence to suppress homology-directed gene conversion at 33% in mammalian cells (Lin et al. 2017). In contrast, Cai and coworkers showed a 1.7-fold increase in HGT frequency after cotransfection of the CRISPR/Cas9 complex and EcRecA into human embryonic kidney (HEK) 293FT cells (Cai et al. 2019). In E. coli, EcRecA acted in concert with the RuvC protein to resolve Holliday junctions in the late stage of DNA recombination (Iwasaki et al. 1991). By introducing RuvC into the nuclei of tobacco plants, Shalev and coworkers obtained strong enhancement of somatic crossover (12-fold), intrachromosomal recombination (11-fold), and extrachromosomal recombination (56-fold) (Shalev et al. 1999). This improvement may also be useful and applicable for DSB formation-based plant gene targeting approaches.
Activities of helicases have been shown in the initiation of homologous recombination. A transgenic approach using E. coli RecQ (EcRecQ) revealed positive effects on extrachromosomal recombination of a two-vector system cointroduced into rice leaves. The EcRecQ transient expression driven by a monocot-specific promoter induced a 4-fold increase in extrachromosomal gene targeting. The stimulation was much higher, at 20–40-fold in cases of stable EcRecQ expression (Li et al. 2004). This report confirmed the importance of helicase activities in HDR and suggested another potential approach for the enhancement of monocot HGT frequency.
As discussed earlier, RAD54 plays roles in concert with the activities of RAD51 during the HDR-mediated DSB amendment stage. Overexpression of yeast RAD54 in Arabidopsis was reported to increase gene targeting frequencies up to 27-fold, indicating the importance of strand invasion and/or chromatin remodeling in the HDR pathway (Shaked et al. 2005). The developmental stages of explants used for monocot gene targeting may differentially support the HDR pathway. The largest amount of recombination occurred in embryogenic cells, and this result was explained by the higher expression levels of OsRAD51 mRNA in the cells (Yang et al. 2010). Enhancement of the resection of the broken ends by overexpressing OsRecQl4 (BLM counterpart) and/or OsExo1 (Exo1 homolog) might positively support gene targeting in rice (Kwon et al. 2012).
Knockout of Genes Relating to the HDR Pathway
As discussed above, RAD50 plays a central role in the MRN/MRX complex for the resection of the broken ends of dsDNAs. Knockout mutations of RAD50 led to developmental lethality in mice (Roset et al. 2014) and suppression of gene targeting in moss (Kamisugi et al. 2012). Surprisingly, a homozygous rad50 KO A. thaliana showed hyperrecombination in somatic cells, as it supported 8- to 10-fold higher gene conversion frequencies of an inverted repeat substrate (Table 2) (Gherbi et al. 2001). This led to an important conclusion that MRN/MRX activities are required by NHEJ more than by HR. The data suggest a strategy that transiently suppresses plant RAD50 during a gene targeting experiment to achieve high frequencies.
Sequence divergence between homologous DNA templates and targeted loci has been shown to affect plant HGT frequency. The HGT frequencies were dramatically reduced by 4.1-, 9.6-, 11.7- or 20.3-fold when the levels of sequence divergence were increased by 0.5%, 2%, 4% or 9%, respectively. The sequence divergence might trigger a nucleotide mismatch repair (NMR) mechanism with the involvement of the NMR key protein AtMSH2 and hence disturb the HDR process (Li et al. 2006; Emmanuel et al. 2006). AtMLH1, a homolog of E. coli MutL that is involved in NMR, was shown to be required for homologous recombination and homeologous recombination. AtMHL1 mutation led to strong HDR reduction but a less severe reduction in homeologous recombination (Dion et al. 2007). The data indicate the potential for the regulation of MSH2 and/or MLH1 expression for the enhancement of HGT in monocots, especially when homologous DNA templates with obligate mismatches are used.
Chromosome accessibility is a key factor determining DSB formation and the subsequent repair of the broken DNAs. During replication or transcription, the chromatin is loosened, and the nucleosomes are opened for the assessment of related proteins involved in these processes. The Arabidopsis thaliana chromatin assembly factor 1 (CAF-1) complex involved in nucleosome assembly is formed by AtFAS1, AtFAS2 and AtMSI1 subunits. Endo and coworkers showed that knockout mutations of either AtFAS1 or AtFAS2 led to enhancement of somatic HR, potentially by 40-fold, thanks to the opening of nucleosomes for accessibility, cell cycle synchronization favoring HDR conditions, and high expression of HDR-related genes in the mutant backgrounds (Endo et al. 2006). The data suggest a potential enhancement of gene targeting via transient AtFAS1/2 knockdown by RNAi while introducing editing tools in somatic cells of monocots.
Another approach was tested in several studies that showed positive effects on HGT by suppressing important genes involved in the NHEJ pathway, such as KU70/80 or Lig4 (Nishizawa-Yokoi et al. 2012; Endo et al. 2016). This approach also showed a reduction in stable integration of T-DNA in the KU70/80 and Lig4 suppression conditions, suggesting a mechanism of T-DNA integration in the genome.
Favorable Tissue Culture Conditions for Gene Targeting
Polyamines accumulated in cells with induced DSBs and were subsequently shown to improve HGT by promoting RAD51-mediated DNA strand exchange. During in vitro assays, polyamines facilitated the capture of duplex DNA by the RAD51 presynaptic filament (Lee et al. 2019). Physical support of the substances may be a good approach for enhancing the activity of RAD51, a key protein in the SDSA subpathway for gene targeting in monocot somatic cells. Chemicals that suppress genes involved in the NHEJ pathway were used for testing HGT enhancement effects. Some chemicals inhibited DNA-PK (Robert et al. 2015) or KU70/80 or Lig4 (Table 2) (Chu et al. 2015; Maruyama et al. 2015), thereby enhancing HGT frequency in mammalian cell lines (Yu et al. 2015). It is still not clear whether we can achieve similar gene targeting enhancement in plants. Data obtained from our laboratory showed nearly no effects of SCR7 and/or RS-1 on tomato gene targeting using geminiviral replicons in combination with CRISPR/Cpf1 (unpublished data). Temperature is an important factor enhancing CRISPR/Cas9-based targeted mutagenesis in plants (LeBlanc et al. 2018) and CRISPR/Cpf1-based HDR in zebrafish and Xenopus by controlling genome accessibility (Moreno-Mateos et al. 2017). Recently, we re-engineered geminiviral replicon vectors in combination with CRISPR/Cpf1 and showed enhancement of HGT frequency at high temperatures and under lighting conditions (Vu et al. 2019).
Cell Cycle Synchronization
One of the reasons that HDR is limited to the S-G2 phases is the availability of sister chromatids to be used as donor templates. As a consequence, the majority of HDR genes might have evolved to be specifically expressed in these phases. The ideas are to artificially favor cellular conditions (S and G2 phases) in which HDR is more efficient and that limit NHEJ blocking of the targeted sites in other phases (M and G1), especially in the case of Cas9s, because they cut in the core sequences proximal to their PAMs. To that end, cell cycle synchronization at the S/G2 phase using chemical (hydroxyurea) or molecular approaches could be applied (Tsakraklides et al. 2015; Gutschner et al. 2016). Cas9 fused with the N-terminal (110a.a) end of human Gemini, a replication licensing factor that is a direct target of an M/G1-restricted E3 ubiquitin ligase for proteolysis, synchronized Cas9 expression in the S/G2 phase, thereby enhancing HGT up to 87% compared to only Cas9 (Table 2) (Gutschner et al. 2016).
In planta Gene Targeting
Gene targeting in maize may be performed during fertilization because it provides a permissive environment for sequence exchange by HGT (Djukanovic et al. 2006). In 2012, Fauser and coworkers demonstrated the feasibility of using the pre-integrated target, donor template and homing nuclease (I-SceI) in the planta gene targeting in Arabidopsis to correct the truncated GUS marker in the target with the remaining part located in the donor template. Crossing of the lines carrying homozygous target and donor alleles with a line expressing I-SceI obtained somatic GT events in the F1 generation that could be inherited in the F2 progeny at 6.8 × 10− 3 frequency (Fauser et al. 2012). Targeted mutagenesis using CRISPR/Cas9 has been shown to be a highly valuable in planta approach for crop improvement (Kelliher et al. 2019). These approaches could also be applied for CRISPR/Cas-based monocot gene targeting, and it would avoid the laborious, time consuming and complex tissue culture process. In planta gene targeting could reduce the mutation rate compared to the tissue culture system, which is accompanied by many mutations. However, the targeting tools should be redesigned to match the conditions (pollen-specific and/or ovule-specific) so that they work within the short time period of pollination and fertilization.
HDR-Based Monocot Events and Regulatory Aspects
Genome-edited crops, including those created by CRISPR/Cas-based targeted mutagenesis and HGT approaches with or without the uses of DNA cargos, are referred to as products of “new breeding techniques (NBTs)” (Laaninen 2016; Lusser et al. 2011) or “new genetic modification techniques (nGMs)” (Eckerstorfer et al. 2019). In most of these genome-editing events, foreign genetic editing tools could be excluded from the organisms after finishing their roles, except that exotic DNA sequence(s) need to be introduced to specific site(s) in their genome(s). Likewise, most of the genome-edited transformants could not be distinguished among other mutated crops generated by conventional mutagens or natural mutations (Friedrichs et al. 2019; Grohmann et al. 2019), and hence, they should not be regulated. The regulatory legislation seems to be more complicated for HGT events because they have been regulated either as non-GMOs or GMOs by the US, EU, Japan, Australia, and others (for an extensive review, see Eckerstorfer et al. 2019). In this section, we summarize and discuss the regulatory aspects of HGT crops, including monocots, as the critical hurdle for the commercialization of HGT crops. We would also attempt to propose a regulatory principle that could be useful for countries during the legislation process.
Current Status in Regulatory Policies for Genome-Edited Crops
The US is the leading country in the commercialization of GM crops to date, with 75 Mha of planted biotech crops in 2018 (ISAAA 2019). In the same year, the US was also the leading country to release policies for the regulation of genome-edited crops. The USDA announced that “Under its biotechnology regulations, USDA does not currently regulate or have any plans to regulate plants that could otherwise have been developed through traditional breeding techniques as long as they are developed without the use of a plant pest as the donor or vector and they are not themselves plant pests” (USDA_Press 2018). This means that genome modifications such as deletions, base substitutions and plant DNA modifications, being similar to those potentially generated by conventional cross-breeding, are all deregulated by USDA policies (NatPlants/Editorial 2018). In Japan, the Ministry of Environment released its final policy on environmental safety on Feb. 8, 2019. According to the decision, creating food items using genome editing is not considered to produce GMOs, under the conditions that any DNA from the nucleases required to edit the target organism are not left within the genome and the resulting gene edits could have also occurred naturally. The Japanese Ministry of Health, Labor and Welfare announced a nearly identical assessment with regard to food safety on March 27, 2019 (USDA/JA9050 2019). Brazil, Argentina, Canada, Chile and Colombia have decided to regulate genome-edited crops at similar levels to the US (Ledford 2019). The Australian government adopted a middle level of regulation because SDN-1 products would not be regulated (Mallapaty 2019). By contrast, on July 25, 2018, the European Court of Justice decided that genome-edited crops would be subject to the same rules as transgenic plants or animals (ECJ 2018). Other governments, including those of the Republic of Korea, China, Russia and India, are still making their determinations of how to regulate this technology.
SDN Declaration
In fact, according to the released ruling policies of the governments except the EU, not all genome-edited transformants are considered non-GM. In principle, the genome-edited crops were initially divided under the classification of the so-called site-directed nucleases (SDN) by the European Food Safety Authority (EFSA) in 2012: “In SDN-1 applications, only the SDNs are introduced into plant cells (stably or transiently), generating site-specific mutations by nonhomologous end-joining (NHEJ). In SDN-2 applications, homologous repair DNA (donor DNA) is introduced together with the SDN complex to create specific nucleotide sequence changes by homologous recombination (HR) or homology-directed repair (HDR). The SDN-2 technique can introduce substantial changes to the nucleotide sequences of the target gene but more precise changes according to the bioengineer’s plan. SDN-2 techniques can provide unlimited SNP alleles that can boost innovative crop breeding. In the SDN-3 technique, a large stretch of donor DNA (up to several kilobases) is introduced together with the SDN complex to target DNA insertion into a predefined genomic locus. The predefined locus may or may not have extensive similarity to the DNA to be inserted. The insertion can take place either by HR or by NHEJ. In the case of insertion by means of NHEJ, the technique is denominated the SDN-3–NHEJ technique” (EFSA 2012). This classification is now generally accepted as the basal information for genome-edited crop regulation. On August 20, 2018, Japan’s Ministry of Environment (MOE) released a draft of its regulatory policies, adding some detailed requirements for SDNs to be excluded from the Cartagena Protocol regulation (USDA/JA8064 2018). The levels of regulation are decided based on the presence/absence of foreign genetic carriers, the levels of modification and the natural existence of the modification in genome-edited organisms. From another point of view, they are assessed on a case-by-case basis (see Table 3). From the released regulations, it is now clear that HGT will be regulated as either non-GMO (some cases of SDN-2) or GMO (some cases of SDN-2 and SDN-3).
Table 3.
Gene- edited crop regulation status on the basis of SDN
| Category | EFSA-2012 Definition | The major contents of Regulation | |||
|---|---|---|---|---|---|
| USA | Japan | Australia | EU | ||
| SDN-1 | After the intended site-specific cleavage of the DNA in the genome, random mutation (base substitution, insertion, or deletion) occurring for one or a few bases as a natural repair mechanism | Excluded from regulation if the resulting plants are free of DNA from “plant pests” such as viruses or bacteria | When there are no transgenic genes and/or fragments of transgenic genes in the final product, however, the genome edited foods will not be considered to be foods derived from recombinant DNA technology, as long as, the DNA double-strand break induced by engineered restriction enzyme and following repair (i.e., mutation) is: a) base-pair deletion; b) substitution; c) naturally occurring gene deletion; and/or, d) concomitant insertion (mutation) of one to several base pairs. | Excluded from regulation | Regulated as GMO(s) |
| SDN-2 | Systematically induces mutation for one or a few bases by artificially synthesizing a short DNA fragment (template) that is homologous to the target base sequence and introducing it along with an artificial restriction enzyme at the time of cleaving. | Explicitly regulated | Regulated as GMO(s) | ||
| SDN-3 | Forms a special DNA fragment at a specific domain on the genome by introducing a long DNA fragment containing a gene of several thousand base pairs not originating from compatible same or related varieties (transgene) in a form sandwiched by sequences homologous to the target sequence. | Explicitly regulated | Regulated as GMO(s) | ||
| Effective date | 28 March 2018 | 27 March 2019 | 8 October 2019 | 25 July 2018 | |
The classification and regulatory considerations have created a major challenge for plant gene targeting approaches to be commercialized, even though their efficacy would be enhanced at a practical level. Gene targeting for modifying SNPs is deregulated by “relaxed” governments such as the US but not Australia. HGT products subject to the SDN-3 category, containing inserted sequence(s) that could not potentially form in nature, will all be regulated as transgenic products (Table 3).
A Regulatory Proposal for NBPT Products
Many governments seem to be trying to create sufficient oversight to protect the public interest and at the same time not create new obstacles to technical innovation. Genome-editing-based precision breeding is an innovative technology, but the technologies will evolve continuously. In particular, HDR-based precision breeding technologies are the most cutting-edge technology among genome-editing techniques because they can produce both precise SDN-1 and SDN-2/SDN-3 products. HDR-based precision breeding products are generally classified as SDN-2 or SDN-3, as they use a DNA donor template during the gene editing process and are thus regulated as GMO in Australia and Japan, with potential exceptions. Mechanical classification of HDR-based genome-edited products in the GMO category might pose the most unreasonable obstacle to this plant breeding innovation. In fact, HDR-based precision breeding can fulfill the long-awaited dream of breeders by precisely introducing beneficial gene alleles from crossable relatives without other trait compromises, such as linkage drag (mixing of targeted beneficial traits and unintended undesirable traits by linkage effects). What might be the solution? We must now again remind ourselves of the purpose of the regulation of biotechnology products. Regulations exist to prevent new products from harming human health or the environment. Many various technologies can be used to produce the same or similar, effectively indistinguishable, products to traditional breeding products; therefore, the consistent risk-based regulatory approach is to treat similar products identically. In this view, it is worth referring to the Canadian regulatory policy, which regulates only plants with novel traits (PNTs), irrespective of the technologies used (Ellens et al. 2019). According to Canadian regulation, some SDN-1 products or even chemically mutagenized products can be regulated. However, this regulatory policy provides more open opportunities to use various innovative technologies, including genome editing or GMO. In the end, the fruitfulness of NPBT crops will mainly depend on the level of regulation on NPBT products.
Conclusions
The development of novel traits for monocot crops is crucial for coping with future challenges in crop production for feeding approximately 10 billion people in 2050 (Hickey et al. 2019; Ray et al. 2013). The recent development of NPBT has paved ways for crop improvement to cope with difficult missions. Targeted mutagenesis approaches (Fig. 1, error-prone approaches, and Fig. 2) of NBPT have been gaining significant success in targeting a wide range of crop plants, including monocots, due to the ease and high frequencies of the revolutionized molecular scissors, especially the CRISPR/Cas complexes (Fig. 2 and Additional file 1: Table S1). Similar outcomes could also be expected with base-editing techniques (Fig. 3b and Additional file 2: Table S2). However, precision editing approaches such as ODM (Fig. 3a) and especially plant HGT approaches are still facing hurdles in practical application due to their low efficiencies and the complexities of editing event regeneration (Fig. 1, HR and SDSA approaches, and Table 1). A significant number of studies have been conducted to unveil the mechanisms of the HDR pathway (Fig. 1) and to overcome the obstacles in HGT frequency (Table 2). The major HGT support strategies to date are (1) appropriate selection of HGT events; (2) enhancement of DSB formation frequency and specificity at the targeted site; (3) high-dose delivery of homologous donors; (4) overexpression/interference of genes involved in HR/C-NHEJ; (5) chemical-based activation/suppression of genes involved in HR/C-NHEJ; and (6) chemical/biological-based cell cycle synchronization (Table 2). Although it is still not clear, we realize the potency of the in planta HGT approach, which helps to avoid the laborious and time-consuming tissue culture process. In addition, it may reduce unintended effects due to the high performance of the CRISPR/Cas system as well as genetic variation in callus-mediated plant regeneration. Appropriate applications of each of these strategies alone or in combination and with the use of CRISPR/Cas complexes may offer a better way to overcome the low efficiency and regeneration concerns. More work still needs to be done for practical customization of monocot crop traits using the HGT technique.
Recently, negative predictions about food production have forced several governments to accept NPBTs as the only way to sustain our future. Regulatory legislation has been more relaxed, with NPBT products produced by SDN-1 and SDN-2 (case-by-case) in the USA, Japan and Australia but not the EU. Countries with pending regulations include China, India, and the Republic of Korea. Based on this background and understanding, we have attempted to propose a few principles for upcoming regulatory policies for these countries.
Additional Files
Additional file 1: Table S1. Targeted mutagenesis-genome editing in monocots using CRISPR/Cas.
Additional file 2: Table S2. Major applications of base-editing approaches in plants.
Acknowledgments
Not applicable.
Abbreviations
- A-NHEJ
Alternative nonhomologous end joining
- BER
Base excision repair
- Cas
CRISPR-associated protein
- C-NHEJ
Canonical nonhomologous end joining
- CO
Crossover
- CRISPR
Clustered regularly interspaced short palindromic repeat
- dHj
Double Holliday junction
- DSBR
Double-stranded break repair
- DT-A
Diphtheria toxin A
- FAO
Food and Agriculture Organization
- GR
Green revolution
- GT
Gene targeting
- HDR
Homology-directed repair
- HGT
Homology-directed gene targeting
- HptII
Hygromycin phosphotransferase II
- HR
Homologous recombination
- MMEJ
Microhomology-mediated end joining
- NCO
Noncrossover
- NER
Nucleotide excision repair
- NPBTs
New plant breeding techniques
- NptII
Neomycin phosphotransferase II
- SDN
Site-directed nucleases
- SDSA
Synthesis-dependent strand annealing
- SSA
Single stranded annealing
- TALENs
Transcription activator-like effector nucleases
- ZFNs
Zinc-finger nucleases
Authors’ Contributions
All the authors contributed to writing the manuscript. Vu TV and Kim JY revised the manuscript. Kim JY finalized the manuscript. All the authors read and approved the final manuscript.
Funding
This work was supported by the National Research Foundation of Korea (Grant NRF 2017R1A4A1015515) and by the Next-Generation BioGreen 21 Program (SSAC, Grant PJ01322601), Rural Development Administration (RDA), Republic of Korea.
Availability of Data and Materials
Not applicable.
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12284-019-0355-1.
References
- Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513(7519):569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzalone Andrew V., Randolph Peyton B., Davis Jessie R., Sousa Alexander A., Koblan Luke W., Levy Jonathan M., Chen Peter J., Wilson Christopher, Newby Gregory A., Raguram Aditya, Liu David R. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576(7785):149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Yamasaki M, Takuno S, Miura K, Katagiri S, Ito T, Doi K, Wu J, Ebana K, Matsumoto T, Innan H, Kitano H, Ashikari M, Matsuoka M. Artificial selection for a green revolution gene during japonica rice domestication. Proc Natl Acad Sci U S A. 2011;108(27):11034–11039. doi: 10.1073/pnas.1019490108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayar A, Wehrkamp-Richter S, Laffaire JB, Le Goff S, Levy J, Chaignon S, Salmi H, Lepicard A, Sallaud C, Gallego ME, White CI, Paul W. Gene targeting in maize by somatic ectopic recombination. Plant Biotechnol J. 2013;11(3):305–314. doi: 10.1111/pbi.12014. [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421(6922):499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF. DNA replicons for plant genome engineering. Plant Cell. 2014;26(1):151–163. doi: 10.1105/tpc.113.119792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat Biotechnol. 2016;34(9):933–941. doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Baur M, Potrykus I, Paszkowski J. Intermolecular homologous recombination in plants. Mol Cell Biol. 1990;10(2):492–500. doi: 10.1128/mcb.10.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beetham PR, Kipp PB, Sawycky XL, Arntzen CJ, May GD. A tool for functional plant genomics: chimeric RNA/DNA oligonucleotides cause in vivo gene-specific mutations. Proc Natl Acad Sci U S A. 1999;96(15):8774–8778. doi: 10.1073/pnas.96.15.8774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V. Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods. 2013;9(1):39. doi: 10.1186/1746-4811-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JM. Proposed structure for the zinc-binding domains from transcription factor IIIA and related proteins. Proc Natl Acad Sci U S A. 1988;85(1):99–102. doi: 10.1073/pnas.85.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300(5620):764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- Bilang R, Peterhans A, Bogucki A, Paszkowski J. Single-stranded DNA as a recombination substrate in plants as assessed by stable and transient recombination assays. Mol Cell Biol. 1992;12(1):329–336. doi: 10.1128/mcb.12.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc Natl Acad Sci U S A. 1998;95(18):10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortesi L, Fischer R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv. 2015;33(1):41–52. doi: 10.1016/j.biotechadv.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321(5891):960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhagatapalli N, Rutten T, Gurushidze M, Kumlehn J, Hensel G. Targeted modification of gene function exploiting homology-directed repair of TALEN-mediated double-Strand breaks in barley. G3 (Bethesda) 2015;5(9):1857–1863. doi: 10.1534/g3.115.018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler NM, Baltes NJ, Voytas DF, Douches DS. Geminivirus-mediated genome editing in potato (Solanum tuberosum L.) using sequence-specific nucleases. Front Plant Sci. 2016;7:1045. doi: 10.3389/fpls.2016.01045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt H, Eid A, Ali Z, Atia MAM, Mokhtar MM, Hassan N, Lee CM, Bao G, Mahfouz MM. Efficient CRISPR/Cas9-mediated genome editing using a chimeric single-guide RNA molecule. Front Plant Sci. 2017;8:1441. doi: 10.3389/fpls.2017.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CQ, Doyon Y, Ainley WM, Miller JC, Dekelver RC, Moehle EA, Rock JM, Lee YL, Garrison R, Schulenberg L, Blue R, Worden A, Baker L, Faraji F, Zhang L, Holmes MC, Rebar EJ, Collingwood TN, Rubin-Wilson B, Gregory PD, Urnov FD, Petolino JF. Targeted transgene integration in plant cells using designed zinc finger nucleases. Plant Mol Biol. 2009;69(6):699–709. doi: 10.1007/s11103-008-9449-7. [DOI] [PubMed] [Google Scholar]
- Cai Y, Cheng T, Yao Y, Li X, Ma Y, Li L, Zhao H, Bao J, Zhang M, Qiu Z, Xue T. In vivo genome editing rescues photoreceptor degeneration via a Cas9/RecA-mediated homology-directed repair pathway. Sci Adv. 2019;5(4):eaav3335. doi: 10.1126/sciadv.aav3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188(4):773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Baltes NJ, Cegan R, Zhang Y, Voytas DF. High-frequency, precise modification of the tomato genome. Genome Biol. 2015;16:232. doi: 10.1186/s13059-015-0796-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39(12):e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453(7194):489–484. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- Chevalier BS, Kortemme T, Chadsey MS, Baker D, Monnat RJ, Stoddard BL. Design, activity, and structure of a highly specific artificial endonuclease. Mol Cell. 2002;10(4):895–905. doi: 10.1016/S1097-2765(02)00690-1. [DOI] [PubMed] [Google Scholar]
- Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001;29(18):3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kuhn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33(5):543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- Cole-Strauss A, Yoon K, Xiang Y, Byrne BC, Rice MC, Gryn J, Holloman WK, Kmiect EB. Correction of the mutation responsible for sickle cell Anemia by an RNA-DNA oligonucleotide. Science. 1996;273(5280):1386–1389. doi: 10.1126/science.273.5280.1386. [DOI] [PubMed] [Google Scholar]
- Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21(2):121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan-Meir T, Filler-Hayut S, Melamed-Bessudo C, Bocobza S, Czosnek H, Aharoni A, Levy AA. Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 2018;95(1):5–16. doi: 10.1111/tpj.13932. [DOI] [PubMed] [Google Scholar]
- Dang TT, Shimatani Z, Kawano Y, Terada R, Shimamoto K. Gene editing a constitutively active OsRac1 by homologous recombination-based gene targeting induces immune responses in rice. Plant Cell Physiol. 2013;54(12):2058–2070. doi: 10.1093/pcp/pct147. [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411(6839):826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- de Groot MJ, Offringa R, Does MP, Hooykaas PJ, van den Elzen PJ. Mechanisms of intermolecular homologous recombination in plants as studied with single- and double-stranded DNA molecules. Nucleic Acids Res. 1992;20(11):2785–2794. doi: 10.1093/nar/20.11.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schutter K, Joubes J, Cools T, Verkest A, Corellou F, Babiychuk E, Van Der Schueren E, Beeckman T, Kushnir S, Inze D, De Veylder L. Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell. 2007;19(1):211–225. doi: 10.1105/tpc.106.045047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Halluin K, Vanderstraeten C, Stals E, Cornelissen M, Ruiter R. Homologous recombination: a basis for targeted genome optimization in crop species such as maize. Plant Biotechnol J. 2008;6(1):93–102. doi: 10.1111/j.1467-7652.2007.00305.x. [DOI] [PubMed] [Google Scholar]
- D'Halluin K, Vanderstraeten C, Van Hulle J, Rosolowska J, Van Den Brande I, Pennewaert A, D'Hont K, Bossut M, Jantz D, Ruiter R, Broadhvest J. Targeted molecular trait stacking in cotton through targeted double-strand break induction. Plant Biotechnol J. 2013;11(8):933–941. doi: 10.1111/pbi.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion E, Li L, Jean M, Belzile F. An Arabidopsis MLH1 mutant exhibits reproductive defects and reveals a dual role for this gene in mitotic recombination. Plant J. 2007;51(3):431–440. doi: 10.1111/j.1365-313X.2007.03145.x. [DOI] [PubMed] [Google Scholar]
- Djukanovic V, Orczyk W, Gao H, Sun X, Garrett N, Zhen S, Gordon-Kamm W, Barton J, Lyznik LA. Gene conversion in transgenic maize plants expressing FLP/FRT and Cre/loxP site-specific recombination systems. Plant Biotechnol J. 2006;4(3):345–357. doi: 10.1111/j.1467-7652.2006.00186.x. [DOI] [PubMed] [Google Scholar]
- Dong C, Beetham P, Vincent K, Sharp P. Oligonucleotide-directed gene repair in wheat using a transient plasmid gene repair assay system. Plant Cell Rep. 2006;25(5):457–465. doi: 10.1007/s00299-005-0098-x. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Dray E, Siaud N, Dubois E, Doutriaux MP. Interaction between Arabidopsis Brca2 and its partners Rad51, Dmc1, and Dss1. Plant Physiol. 2006;140(3):1059–1069. doi: 10.1104/pp.105.075838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECJ . Judgment of the court (Grand Chamber) 2018. [PubMed] [Google Scholar]
- Eckerstorfer MF, Engelhard M, Heissenberger A, Simon S, Teichmann H. Plants developed by new genetic modification techniques-comparison of existing regulatory frameworks in the EU and non-EU countries. Front Bioeng Biotechnol. 2019;7:26. doi: 10.3389/fbioe.2019.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA Scientific opinion addressing the safety assessment of plants developed using zinc finger nuclease 3 and other site-directed nucleases with similar function. European food safety authority (EFSA) EFSA J. 2012;10(10):2943. doi: 10.2903/j.efsa.2012.2943. [DOI] [Google Scholar]
- Ellens KW, Levac D, Pearson C, Savoie A, Strand N, Louter J, Tibelius C. Canadian regulatory aspects of gene editing technologies. Transgenic Res. 2019;28(Suppl 2):165–168. doi: 10.1007/s11248-019-00153-2. [DOI] [PubMed] [Google Scholar]
- Emmanuel E, Yehuda E, Melamed-Bessudo C, Avivi-Ragolsky N, Levy AA. The role of AtMSH2 in homologous recombination in Arabidopsis thaliana. EMBO Rep. 2006;7(1):100–105. doi: 10.1038/sj.embor.7400577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Ishikawa Y, Osakabe K, Nakayama S, Kaya H, Araki T, Shibahara K, Abe K, Ichikawa H, Valentine L, Hohn B, Toki S. Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J. 2006;25(23):5579–5590. doi: 10.1038/sj.emboj.7601434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Mikami M, Toki S. Biallelic gene targeting in Rice. Plant Physiol. 2016;170(2):667–677. doi: 10.1104/pp.15.01663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Osakabe K, Ono K, Handa H, Shimizu T, Toki S. Molecular breeding of a novel herbicide-tolerant rice by gene targeting. Plant J. 2007;52(1):157–166. doi: 10.1111/j.1365-313X.2007.03230.x. [DOI] [PubMed] [Google Scholar]
- Eriksson D. The Swedish policy approach to directed mutagenesis in a European context. Physiol Plant. 2018;164(4):385–395. doi: 10.1111/ppl.12740. [DOI] [PubMed] [Google Scholar]
- FAO . Food Outlook - Biannual Report on Global Food Markets. . Rome Licence: CC BY-NC-SA 30 IGO. 2019. [Google Scholar]
- FAO . The state of food security nutrition in the world. 2019. [Google Scholar]
- Fauser F, Roth N, Pacher M, Ilg G, Sanchez-Fernandez R, Biesgen C, Puchta H. In planta gene targeting. Proc Natl Acad Sci U S A. 2012;109(19):7535–7540. doi: 10.1073/pnas.1202191109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes JB, Duhamel M, Seguela-Arnaud M, Froger N, Girard C, Choinard S, Solier V, De Winne N, De Jaeger G, Gevaert K, Andrey P, Grelon M, Guerois R, Kumar R, Mercier R. FIGL1 and its novel partner FLIP form a conserved complex that regulates homologous recombination. PLoS Genet. 2018;14(4):e1007317. doi: 10.1371/journal.pgen.1007317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichs S, Takasu Y, Kearns P, Dagallier B, Oshima R, Schofield J, Moreddu C. Meeting report of the OECD conference on “genome editing: applications in agriculture-implications for health, environment and regulation”. Transgenic Res. 2019;28(3–4):419–463. doi: 10.1007/s11248-019-00154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Smith J, Yang M, Jones S, Djukanovic V, Nicholson MG, et al. Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 2010;61(1):176–187. doi: 10.1111/j.1365-313X.2009.04041.x. [DOI] [PubMed] [Google Scholar]
- Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR. Programmable base editing of a*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;551(7681):464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke JM, Cervantes O, Clement MK, Wu Y, Zeng J, Bauer DE, Pinello L, Joung JK. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat Biotechnol. 2018;36(10):977–982. doi: 10.1038/nbt.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherbi H, Gallego ME, Jalut N, Lucht JM, Hohn B, White CI. Homologous recombination in planta is stimulated in the absence of Rad50. EMBO Rep. 2001;2(4):287–291. doi: 10.1093/embo-reports/kve069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Humanes J, Wang Y, Liang Z, Shan Q, Ozuna CV, Sanchez-Leon S, Baltes NJ, Starker C, Barro F, Gao C, Voytas DF. High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 2017;89(6):1251–1262. doi: 10.1111/tpj.13446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard C, Chelysheva L, Choinard S, Froger N, Macaisne N, Lemhemdi A, Mazel J, Crismani W, Mercier R. AAA-ATPase FIDGETIN-LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLoS Genet. 2015;11(7):e1005369. doi: 10.1371/journal.pgen.1005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocal G. Non-transgenic trait development in crop plants using oligo-directed mutagenesis: Cibus’ rapid trait development system. 2015. [Google Scholar]
- Goedecke W, Eijpe M, Offenberg HH, van Aalderen M, Heyting C. Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat Genet. 1999;23(2):194–198. doi: 10.1038/13821. [DOI] [PubMed] [Google Scholar]
- Gong Y, Handa N, Kowalczykowski SC, de Lange T. PHF11 promotes DSB resection, ATR signaling, and HR. Genes Dev. 2017;31(1):46–58. doi: 10.1101/gad.291807.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72(1):131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- Grohmann L, Keilwagen J, Duensing N, Dagand E, Hartung F, Wilhelm R, Bendiek J, Sprink T. Detection and identification of genome editing in plants: challenges and opportunities. Front Plant Sci. 2019;10:236. doi: 10.3389/fpls.2019.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Gaj T, Barbas CF., 3rd Directed evolution of an enhanced and highly efficient FokI cleavage domain for zinc finger nucleases. J Mol Biol. 2010;400(1):96–107. doi: 10.1016/j.jmb.2010.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C. Geminivirus DNA replication. Cell Mol Life Sci. 1999;56(3–4):313–329. doi: 10.1007/s000180050433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Haemmerle M, Genovese G, Draetta GF, Chin L. Post-translational regulation of Cas9 during G1 enhances homology-directed repair. Cell Rep. 2016;14(6):1555–1566. doi: 10.1016/j.celrep.2016.01.019. [DOI] [PubMed] [Google Scholar]
- Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S. Geminiviruses: masters at redirecting and reprogramming plant processes. Nat Rev Microbiol. 2013;11(11):777–788. doi: 10.1038/nrmicro3117. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28(5):739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Hartung F, Suer S, Bergmann T, Puchta H. The role of AtMUS81 in DNA repair and its genetic interaction with the helicase AtRecQ4A. Nucleic Acids Res. 2006;34(16):4438–4448. doi: 10.1093/nar/gkl576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey LT, N Hafeez A, Robinson H, Jackson SA, SCM L-B, Tester M, Gao C, Godwin ID, Hayes BJ, BBH W. Breeding crops to feed 10 billion. Nat Biotechnol. 2019;37(7):744–754. doi: 10.1038/s41587-019-0152-9. [DOI] [PubMed] [Google Scholar]
- Hilton IB, Gersbach CA. Enabling functional genomics with genome engineering. Genome Res. 2015;25(10):1442–1455. doi: 10.1101/gr.190124.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa T, Hasegawa J, White CI, Matsunaga S. RAD54 forms DNA repair foci in response to DNA damage in living plant cells. Plant J. 2017;90(2):372–382. doi: 10.1111/tpj.13499. [DOI] [PubMed] [Google Scholar]
- Holliday R. Recombination and meiosis. Philos Trans R Soc Lond Ser B Biol Sci. 1977;277(955):359–370. doi: 10.1098/rstb.1977.0024. [DOI] [PubMed] [Google Scholar]
- Horvath M, Steinbiss HH, Reiss B. Gene targeting without DSB induction is inefficient in barley. Front Plant Sci. 2016;7:1973. doi: 10.3389/fpls.2016.01973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel AW, Chauhan RD, Cermak T, Mutka AM, Vijayaraghavan A, Boyher A, Starker CG, Bart R, Voytas DF, Taylor NJ. Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnol J. 2018;16(7):1275–1282. doi: 10.1111/pbi.12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISAAA (2019) Global Status of Commercialized Biotech/GM Crops: 2018. Brief 54
- Isono M, Niimi A, Oike T, Hagiwara Y, Sato H, Sekine R, Yoshida Y, Isobe SY, Obuse C, Nishi R, Petricci E, Nakada S, Nakano T, Shibata A. BRCA1 directs the repair pathway to homologous recombination by promoting 53BP1 Dephosphorylation. Cell Rep. 2017;18(2):520–532. doi: 10.1016/j.celrep.2016.12.042. [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Takahagi M, Shiba T, Nakata A, Shinagawa H. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J. 1991;10(13):4381–4389. doi: 10.1002/j.1460-2075.1991.tb05016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 1996;12(6):224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol. 2013;5(11):a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31(3):233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, Kaplan M, Iavarone AT, Charpentier E, Nogales E, Doudna JA. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343(6176):1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Monnat RJ, Jr, Stoddard BL. DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-Cre I. Mol Cell. 1998;2(4):469–476. doi: 10.1016/S1097-2765(00)80146-X. [DOI] [PubMed] [Google Scholar]
- Kamisugi Y, Schaefer DG, Kozak J, Charlot F, Vrielynck N, Hola M, Angelis KJ, Cuming AC, Nogue F. MRE11 and RAD50, but not NBS1, are essential for gene targeting in the moss Physcomitrella patens. Nucleic Acids Res. 2012;40(8):3496–3510. doi: 10.1093/nar/gkr1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432(7015):316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Kay S, Hahn S, Marois E, Wieduwild R, Bonas U. Detailed analysis of the DNA recognition motifs of the Xanthomonas type III effectors AvrBs3 and AvrBs3Deltarep16. Plant J. 2009;59(6):859–871. doi: 10.1111/j.1365-313X.2009.03922.x. [DOI] [PubMed] [Google Scholar]
- Kaya H, Numa H, Nishizawa-Yokoi A, Toki S, Habu Y. DNA methylation affects the efficiency of transcription activator-like effector nucleases-mediated genome editing in rice. Front Plant Sci. 2017;8:302. doi: 10.3389/fpls.2017.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher T, Starr D, Su X, Tang G, Chen Z, Carter J, Wittich PE, Dong S, Green J, Burch E, McCuiston J, Gu W, Sun Y, Strebe T, Roberts J, Bate NJ, Que Q. One-step genome editing of elite crop germplasm during haploid induction. Nat Biotechnol. 2019;37(3):287–292. doi: 10.1038/s41587-019-0038-x. [DOI] [PubMed] [Google Scholar]
- Khush GS. Green revolution: the way forward. Nat Rev Genet. 2001;2(10):815–822. doi: 10.1038/35093585. [DOI] [PubMed] [Google Scholar]
- Kijas AW, Lim YC, Bolderson E, Cerosaletti K, Gatei M, Jakob B, Tobias F, Taucher-Scholz G, Gueven N, Oakley G, Concannon P, Wolvetang E, Khanna KK, Wiesmuller L, Lavin MF. ATM-dependent phosphorylation of MRE11 controls extent of resection during homology directed repair by signalling through exonuclease 1. Nucleic Acids Res. 2015;43(17):8352–8367. doi: 10.1093/nar/gkv754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim J, Hur JK, Been KW, Yoon SH, Kim JS. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol. 2016;34(8):863–868. doi: 10.1038/nbt.3609. [DOI] [PubMed] [Google Scholar]
- Kim Y, Kweon J, Kim A, Chon JK, Yoo JY, Kim HJ, Kim S, Lee C, Jeong E, Chung E, Kim D, Lee MS, Go EM, Song HJ, Kim H, Cho N, Bang D, Kim S, Kim JS. A library of TAL effector nucleases spanning the human genome. Nat Biotechnol. 2013;31(3):251–258. doi: 10.1038/nbt.2517. [DOI] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A. 1996;93(3):1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klutstein M, Shaked H, Sherman A, Avivi-Ragolsky N, Shema E, Zenvirth D, Levy AA, Simchen G. Functional conservation of the yeast and Arabidopsis RAD54-like genes. Genetics. 2008;178(4):2389–2397. doi: 10.1534/genetics.108.086777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS. Origins and evolution of CRISPR-Cas systems. Philos Trans R Soc Lond Ser B Biol Sci. 2019;374(1772):20180087. doi: 10.1098/rstb.2018.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Duhamel M, Coutant E, Ben-Nahia E, Mercier R. Antagonism between BRCA2 and FIGL1 regulates homologous recombination. Nucleic Acids Res. 2019;47(10):5170–5180. doi: 10.1093/nar/gkz225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YI, Abe K, Osakabe K, Endo M, Nishizawa-Yokoi A, Saika H, Shimada H, Toki S. Overexpression of OsRecQl4 and/or OsExo1 enhances DSB-induced homologous recombination in rice. Plant Cell Physiol. 2012;53(12):2142–2152. doi: 10.1093/pcp/pcs155. [DOI] [PubMed] [Google Scholar]
- Laaninen T. New plant-breeding techniques applicability of GM rules. EPRS PE. 2016;582:018. [Google Scholar]
- Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11(1):39–46. doi: 10.1016/S0959-440X(00)00167-6. [DOI] [PubMed] [Google Scholar]
- Lamarche BJ, Orazio NI, Weitzman MD. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010;584(17):3682–3695. doi: 10.1016/j.febslet.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerak P, Mejia-Ramirez E, Limbo O, Russell P. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 2011;7(9):e1002271. doi: 10.1371/journal.pgen.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc C, Zhang F, Mendez J, Lozano Y, Chatpar K, Irish VF, Jacob Y. Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J. 2018;93(2):377–386. doi: 10.1111/tpj.13782. [DOI] [PubMed] [Google Scholar]
- Ledford H. CRISPR conundrum: strict European court ruling leaves food-testing labs without a plan. Nature. 2019;572(7767):15. doi: 10.1038/d41586-019-02162-x. [DOI] [PubMed] [Google Scholar]
- Lee CY, Su GC, Huang WY, Ko MY, Yeh HY, Chang GD, Lin SJ, Chi P. Promotion of homology-directed DNA repair by polyamines. Nat Commun. 2019;10(1):65. doi: 10.1038/s41467-018-08011-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308(5721):551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Li HQ, Terada R, Li MR, Iida S. RecQ helicase enhances homologous recombination in plants. FEBS Lett. 2004;574(1–3):151–155. doi: 10.1016/j.febslet.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Li J, Zhang X, Sun Y, Zhang J, Du W, Guo X, Li S, Zhao Y, Xia L. Efficient allelic replacement in rice by gene editing: a case study of the NRT1.1B gene. J Integr Plant Biol. 2018;60(7):536–540. doi: 10.1111/jipb.12650. [DOI] [PubMed] [Google Scholar]
- Li L, Jean M, Belzile F. The impact of sequence divergence and DNA mismatch repair on homeologous recombination in Arabidopsis. Plant J. 2006;45(6):908–916. doi: 10.1111/j.1365-313X.2006.02657.x. [DOI] [PubMed] [Google Scholar]
- Li S, Li J, He Y, Xu M, Zhang J, Du W, Zhao Y, Xia L. Precise gene replacement in rice by RNA transcript-templated homologous recombination. Nat Biotechnol. 2019;37(4):445–450. doi: 10.1038/s41587-019-0065-7. [DOI] [PubMed] [Google Scholar]
- Li S, Li J, Zhang J, Du W, Fu J, Sutar S, Zhao Y, Xia L. Synthesis-dependent repair of Cpf1-induced double strand DNA breaks enables targeted gene replacement in rice. J Exp Bot. 2018;69(20):4715–4721. doi: 10.1093/jxb/ery245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011;39(1):359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Liu B, Chen CY, Yang B. TALEN-mediated homologous recombination produces site-directed DNA Base change and herbicide-resistant Rice. J Genet Genomics. 2016;43(5):297–305. doi: 10.1016/j.jgg.2016.03.005. [DOI] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell. 2007;28(1):134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Petersen TS, Jensen KT, Bolund L, Kuhn R, Luo Y. Fusion of SpCas9 to E. coli rec a protein enhances CRISPR-Cas9 mediated gene knockout in mammalian cells. J Biotechnol. 2017;247:42–49. doi: 10.1016/j.jbiotec.2017.02.024. [DOI] [PubMed] [Google Scholar]
- Liu M, Ba Z, Costa-Nunes P, Wei W, Li L, Kong F, Li Y, Chai J, Pontes O, Qi Y. IDN2 interacts with RPA and facilitates DNA double-Strand break repair by homologous recombination in Arabidopsis. Plant Cell. 2017;29(3):589–599. doi: 10.1105/tpc.16.00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusser M, Parisi C, Plan D, Rodriguez-Cerezo E. New plant breeding techniques: state-of-the-art and prospects for commercial development. Brussels: European Commission Joint Research Centre; 2011. [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9(6):467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV. Annotation and classification of CRISPR-Cas systems. Methods Mol Biol. 2015;1311:47–75. doi: 10.1007/978-1-4939-2687-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13(11):722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S (2019) Australian gene-editing rules adopt ‘middle ground’. Nature. 10.1038/d41586-019-01282-8 [DOI] [PubMed]
- Mannuss A, Dukowic-Schulze S, Suer S, Hartung F, Pacher M, Puchta H. RAD5A, RECQ4A, and MUS81 have specific functions in homologous recombination and define different pathways of DNA repair in Arabidopsis thaliana. Plant Cell. 2010;22(10):3318–3330. doi: 10.1105/tpc.110.078568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33(5):538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meers C, Keskin H, Storici F. DNA repair by RNA: templated, or not templated, that is the question. DNA Repair (Amst) 2016;44:17–21. doi: 10.1016/j.dnarep.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RS, DuVal AE, Jensen HR. Patterns and processes in crop domestication: an historical review and quantitative analysis of 203 global food crops. New Phytol. 2012;196(1):29–48. doi: 10.1111/j.1469-8137.2012.04253.x. [DOI] [PubMed] [Google Scholar]
- Meyer RS, Purugganan MD. Evolution of crop species: genetics of domestication and diversification. Nat Rev Genet. 2013;14(12):840–852. doi: 10.1038/nrg3605. [DOI] [PubMed] [Google Scholar]
- Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Moerschell RP, Tsunasawa S, Sherman F. Transformation of yeast with synthetic oligonucleotides. Proc Natl Acad Sci U S A. 1988;85(2):524–528. doi: 10.1073/pnas.85.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor TS, Moon YS, Palmer KE, Mason HS. Geminivirus vectors for high-level expression of foreign proteins in plant cells. Biotechnol Bioeng. 2003;81(4):430–437. doi: 10.1002/bit.10483. [DOI] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Fernandez JP, Rouet R, Vejnar CE, Lane MA, Mis E, Khokha MK, Doudna JA, Giraldez AJ. CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat Commun. 2017;8(1):2024. doi: 10.1038/s41467-017-01836-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritoh S, Eun CH, Ono A, Asao H, Okano Y, Yamaguchi K, Shimatani Z, Koizumi A, Terada R. Targeted disruption of an orthologue of DOMAINS REARRANGED METHYLASE 2, OsDRM2, impairs the growth of rice plants by abnormal DNA methylation. Plant J. 2012;71(1):85–98. doi: 10.1111/j.1365-313X.2012.04974.x. [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326(5959):1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Muniyappa K, Shaner SL, Tsang SS, Radding CM. Mechanism of the concerted action of recA protein and helix-destabilizing proteins in homologous recombination. Proc Natl Acad Sci U S A. 1984;81(9):2757–2761. doi: 10.1073/pnas.81.9.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NatPlants/Editorial A CRISPR definition of genetic modification. Nat Plants. 2018;4(5):233. doi: 10.1038/s41477-018-0158-1. [DOI] [PubMed] [Google Scholar]
- Needham PD, Atkinson RG, Morris BAM, Gardner RC, Gleave AP. GUS expression patterns from a tobacco yellow dwarf virus-based episomal vector. Plant Cell Rep. 1998;17(8):631–639. doi: 10.1007/s002990050456. [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A, Endo M, Ohtsuki N, Saika H, Toki S. Precision genome editing in plants via gene targeting and piggyBac-mediated marker excision. Plant J. 2015;81(1):160–168. doi: 10.1111/tpj.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A, Nonaka S, Osakabe K, Saika H, Toki S. A universal positive-negative selection system for gene targeting in plants combining an antibiotic resistance gene and its antisense RNA. Plant Physiol. 2015;169(1):362–370. doi: 10.1104/pp.15.00638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A, Nonaka S, Saika H, Kwon YI, Osakabe K, Toki S. Suppression of Ku70/80 or Lig4 leads to decreased stable transformation and enhanced homologous recombination in rice. New Phytol. 2012;196(4):1048–1059. doi: 10.1111/j.1469-8137.2012.04350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Tenney K, Li H, Gimble FS. Engineering variants of the I-SceI homing endonuclease with strand-specific and site-specific DNA-nicking activity. J Mol Biol. 2008;382(1):188–202. doi: 10.1016/j.jmb.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowsheen S, Yang ES. The intersection between DNA damage response and cell death pathways. Exp Oncol. 2012;34(3):243–254. [PMC free article] [PubMed] [Google Scholar]
- Ochs F, Somyajit K, Altmeyer M, Rask MB, Lukas J, Lukas C. 53BP1 fosters fidelity of homology-directed DNA repair. Nat Struct Mol Biol. 2016;23(8):714–721. doi: 10.1038/nsmb.3251. [DOI] [PubMed] [Google Scholar]
- Okuzaki A, Toriyama K. Chimeric RNA/DNA oligonucleotide-directed gene targeting in rice. Plant Cell Rep. 2004;22(7):509–512. doi: 10.1007/s00299-003-0698-2. [DOI] [PubMed] [Google Scholar]
- Ono A, Yamaguchi K, Fukada-Tanaka S, Terada R, Mitsui T, Iida S. A null mutation of ROS1a for DNA demethylation in rice is not transmittable to progeny. Plant J. 2012;71(4):564–574. doi: 10.1111/j.1365-313X.2012.05009.x. [DOI] [PubMed] [Google Scholar]
- Osakabe K, Abe K, Yoshioka T, Osakabe Y, Todoriki S, Ichikawa H, Hohn B, Toki S. Isolation and characterization of the RAD54 gene from Arabidopsis thaliana. Plant J. 2006;48(6):827–842. doi: 10.1111/j.1365-313X.2006.02927.x. [DOI] [PubMed] [Google Scholar]
- Osakabe K, Nishizawa-Yokoi A, Ohtsuki N, Osakabe Y, Toki S. A mutated cytosine deaminase gene, codA (D314A), as an efficient negative selection marker for gene targeting in rice. Plant Cell Physiol. 2014;55(3):658–665. doi: 10.1093/pcp/pct183. [DOI] [PubMed] [Google Scholar]
- Ozawa K, Wakasa Y, Ogo Y, Matsuo K, Kawahigashi H, Takaiwa F. Development of an efficient agrobacterium-mediated gene targeting system for rice and analysis of rice knockouts lacking granule-bound starch synthase (waxy) and beta1,2-xylosyltransferase. Plant Cell Physiol. 2012;53(4):755–761. doi: 10.1093/pcp/pcs016. [DOI] [PubMed] [Google Scholar]
- Paszkowski J, Baur M, Bogucki A, Potrykus I. Gene targeting in plants. EMBO J. 1988;7(13):4021–4026. doi: 10.1002/j.1460-2075.1988.tb03295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8(9):765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H. Repair of genomic double-strand breaks in somatic plant cells by one-sided invasion of homologous sequences. Plant J. 1998;13(3):331–339. doi: 10.1046/j.1365-313X.1998.00035.x. [DOI] [Google Scholar]
- Puchta H. The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J Exp Bot. 2005;56(409):1–14. doi: 10.1093/jxb/eri025. [DOI] [PubMed] [Google Scholar]
- Puchta H, Dujon B, Hohn B. Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 1993;21(22):5034–5040. doi: 10.1093/nar/21.22.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H, Dujon B, Hohn B. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc Natl Acad Sci U S A. 1996;93(10):5055–5060. doi: 10.1073/pnas.93.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H, Hohn B. A transient assay in plant cells reveals a positive correlation between extrachromosomal recombination rates and length of homologous overlap. Nucleic Acids Res. 1991;19(10):2693–2700. doi: 10.1093/nar/19.10.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H, Hohn B. Breaking news: plants mutate right on target. Proc Natl Acad Sci U S A. 2010;107(26):11657–11658. doi: 10.1073/pnas.1006364107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Zhang Y, Zhang F, Baller JA, Cleland SC, Ryu Y, Starker CG, Voytas DF. Increasing frequencies of site-specific mutagenesis and gene targeting in Arabidopsis by manipulating DNA repair pathways. Genome Res. 2013;23(3):547–554. doi: 10.1101/gr.145557.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radding CM. Recombination activities of E. coli recA protein. Cell. 1981;25(1):3–4. doi: 10.1016/0092-8674(81)90224-5. [DOI] [PubMed] [Google Scholar]
- Rajanikant C, Melzer M, Rao BJ, Sainis JK. Homologous recombination properties of OsRad51, a recombinase from rice. Plant Mol Biol. 2008;68(4–5):479–491. doi: 10.1007/s11103-008-9385-6. [DOI] [PubMed] [Google Scholar]
- Ray DK, Mueller ND, West PC, Foley JA. Yield trends are insufficient to double global crop production by 2050. PLoS One. 2013;8(6):e66428. doi: 10.1371/journal.pone.0066428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees HA, Liu DR. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet. 2018;19(12):770–788. doi: 10.1038/s41576-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss B, Klemm M, Kosak H, Schell J. RecA protein stimulates homologous recombination in plants. Proc Natl Acad Sci U S A. 1996;93(7):3094–3098. doi: 10.1073/pnas.93.7.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter KS, Kleinow T, Jeske H. Somatic homologous recombination in plants is promoted by a geminivirus in a tissue-selective manner. Virology. 2014;452-453:287–296. doi: 10.1016/j.virol.2014.01.024. [DOI] [PubMed] [Google Scholar]
- Robert F, Barbeau M, Ethier S, Dostie J, Pelletier J. Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Med. 2015;7:93. doi: 10.1186/s13073-015-0215-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roset R, Inagaki A, Hohl M, Brenet F, Lafrance-Vanasse J, Lange J, Scandura JM, Tainer JA, Keeney S, Petrini JH. The Rad50 hook domain regulates DNA damage signaling and tumorigenesis. Genes Dev. 2014;28(5):451–462. doi: 10.1101/gad.236745.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saika H, Oikawa A, Matsuda F, Onodera H, Saito K, Toki S. Application of gene targeting to designed mutation breeding of high-tryptophan rice. Plant Physiol. 2011;156(3):1269–1277. doi: 10.1104/pp.111.175778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer NJ, Mozoruk J, Miller RB, Warburg ZJ, Walker KA, Beetham PR, Schopke CR, Gocal GF. Oligonucleotide-directed mutagenesis for precision gene editing. Plant Biotechnol J. 2016;14(2):496–502. doi: 10.1111/pbi.12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuermann D, Molinier J, Fritsch O, Hohn B. The dual nature of homologous recombination in plants. Trends Genet. 2005;21(3):172–181. doi: 10.1016/j.tig.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Schunder E, Rydzewski K, Grunow R, Heuner K. First indication for a functional CRISPR/Cas system in Francisella tularensis. Int J Med Microbiol. 2013;303(2):51–60. doi: 10.1016/j.ijmm.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Seeliger K, Dukowic-Schulze S, Wurz-Wildersinn R, Pacher M, Puchta H. BRCA2 is a mediator of RAD51- and DMC1-facilitated homologous recombination in Arabidopsis thaliana. New Phytol. 2012;193(2):364–375. doi: 10.1111/j.1469-8137.2011.03947.x. [DOI] [PubMed] [Google Scholar]
- Shaked H, Melamed-Bessudo C, Levy AA. High-frequency gene targeting in Arabidopsis plants expressing the yeast RAD54 gene. Proc Natl Acad Sci U S A. 2005;102(34):12265–12269. doi: 10.1073/pnas.0502601102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalev G, Sitrit Y, Avivi-Ragolski N, Lichtenstein C, Levy AA. Stimulation of homologous recombination in plants by expression of the bacterial resolvase ruvC. Proc Natl Acad Sci U S A. 1999;96(13):7398–7402. doi: 10.1073/pnas.96.13.7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL, Gao C. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31(8):686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, Choi VM, Rock JM, Wu YY, Katibah GE, Zhifang G, McCaskill D, Simpson MA, Blakeslee B, Greenwalt SA, Butler HJ, Hinkley SJ, Zhang L, Rebar EJ, Gregory PD, Urnov FD. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009;459(7245):437–441. doi: 10.1038/nature07992. [DOI] [PubMed] [Google Scholar]
- Stella S, Alcon P, Montoya G. Structure of the Cpf1 endonuclease R-loop complex after target DNA cleavage. Nature. 2017;546(7659):559–563. doi: 10.1038/nature22398. [DOI] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507(7490):62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Lopez P, Gutierrez C. DNA replication of wheat dwarf geminivirus vectors: effects of origin structure and size. Virology. 1997;227(2):389–399. doi: 10.1006/viro.1996.8353. [DOI] [PubMed] [Google Scholar]
- Sun Y, Zhang X, Wu C, He Y, Ma Y, Hou H, Guo X, Du W, Zhao Y, Xia L. Engineering herbicide-resistant Rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mol Plant. 2016;9(4):628–631. doi: 10.1016/j.molp.2016.01.001. [DOI] [PubMed] [Google Scholar]
- Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, Cigan AM. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2015;169(2):931–945. doi: 10.1104/pp.15.00793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Ogita N, Takahashi T, Taniguchi S, Tanaka M, Seki M, Umeda M (2019) A regulatory module controlling stress-induced cell cycle arrest in Arabidopsis. Elife:8. 10.7554/eLife.43944 [DOI] [PMC free article] [PubMed]
- Tamaki S, Tsuji H, Matsumoto A, Fujita A, Shimatani Z, Terada R, Sakamoto T, Kurata T, Shimamoto K. FT-like proteins induce transposon silencing in the shoot apex during floral induction in rice. Proc Natl Acad Sci U S A. 2015;112(8):E901–E910. doi: 10.1073/pnas.1417623112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Adachi Y, Chiba K, Oguchi K, Takahashi H. Identification of Ku70 and Ku80 homologues in Arabidopsis thaliana: evidence for a role in the repair of DNA double-strand breaks. Plant J. 2002;29(6):771–781. doi: 10.1046/j.1365-313X.2002.01258.x. [DOI] [PubMed] [Google Scholar]
- Terada R, Johzuka-Hisatomi Y, Saitoh M, Asao H, Iida S. Gene targeting by homologous recombination as a biotechnological tool for rice functional genomics. Plant Physiol. 2007;144(2):846–856. doi: 10.1104/pp.107.095992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada R, Nagaharaa M, Furukawa F, Shimamoto M, Yamaguchi K, Iida S. Cre-loxP mediated marker elimination and gene reactivation at the waxy locus created in rice genome based on strong positive–negative selection. Japanese Society for Plant Cell and Molecular Biology (Plant Biotechnology) 2010;27:29–37. [Google Scholar]
- Terada R, Urawa H, Inagaki Y, Tsugane K, Iida S. Efficient gene targeting by homologous recombination in rice. Nat Biotechnol. 2002;20(10):1030–1034. doi: 10.1038/nbt737. [DOI] [PubMed] [Google Scholar]
- Tong Zhu DJP, Tagliani L, Clair GST, Baszczynski CL, Bowen B. Targeted manipulation of maize genes in vivo using chimeric. 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459(7245):442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakraklides V, Brevnova E, Stephanopoulos G, Shaw AJ. Improved gene targeting through cell cycle synchronization. PLoS One. 2015;10(7):e0133434. doi: 10.1371/journal.pone.0133434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UN (2019) World population prospects 2019. UN Department Global Communications. https://population.un.org/wpp/Publications/Files/WPP2019_Highlights.pdf. Accessed 25 Sept 2019.
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435(7042):646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- USDA . World agricultural production. 2019. pp. 8–19. [Google Scholar]
- USDA/JA8064 . Japan Holds Second Meeting to Discuss Genome Editing Technology. 2018. [Google Scholar]
- USDA/JA9050 . Japanese Health Ministry Finalizes Genome Edited Food Policy. 2019. [Google Scholar]
- USDA_Press . Secretary Perdue Issues USDA Statement on Plant Breeding Innovation. 2018. [Google Scholar]
- Valton J, Dupuy A, Daboussi F, Thomas S, Maréchal A, Macmaster R, et al. Overcoming transcription activator-like effector (TALE) DNA binding domain sensitivity to cytosine methylation. J Biol Chem. 2012;287(46):38427–38432. doi: 10.1074/jbc.C112.408864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas DF. Plant genome engineering with sequence-specific nucleases. Annu Rev Plant Biol. 2013;64:327–350. doi: 10.1146/annurev-arplant-042811-105552. [DOI] [PubMed] [Google Scholar]
- Vu GTH, Cao HX, Fauser F, Reiss B, Puchta H, Schubert I. Endogenous sequence patterns predispose the repair modes of CRISPR/Cas9-induced DNA double-stranded breaks in Arabidopsis thaliana. Plant J. 2017;92(1):57–67. doi: 10.1111/tpj.13634. [DOI] [PubMed] [Google Scholar]
- Vu TV, Sivankalyani V, Kim EJ, Tran MT, Kim J, Sung YW, Doan DTH, Kim JY (2019) Highly efficient homology-directed repair using transient CRISPR/Cpf1-geminiviral replicon in tomato. bioRxiv. 10.1101/521419 [DOI] [PMC free article] [PubMed]
- Wang M, Lu Y, Botella JR, Mao Y, Hua K, Zhu JK. Gene targeting by homology-directed repair in Rice using a Geminivirus-based CRISPR/Cas9 system. Mol Plant. 2017;10(7):1007–1010. doi: 10.1016/j.molp.2017.03.002. [DOI] [PubMed] [Google Scholar]
- Weimer AK, Biedermann S, Harashima H, Roodbarkelari F, Takahashi N, Foreman J, Guan Y, Pochon G, Heese M, Van Damme D, Sugimoto K, Koncz C, Doerner P, Umeda M, Schnittger A. The plant-specific CDKB1-CYCB1 complex mediates homologous recombination repair in Arabidopsis. EMBO J. 2016;35(19):2068–2086. doi: 10.15252/embj.201593083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West CE, Waterworth WM, Story GW, Sunderland PA, Jiang Q, Bray CM. Disruption of the Arabidopsis AtKu80 gene demonstrates an essential role for AtKu80 protein in efficient repair of DNA double-strand breaks in vivo. Plant J. 2002;31(4):517–528. doi: 10.1046/j.1365-313X.2002.01370.x. [DOI] [PubMed] [Google Scholar]
- Wright DA, Townsend JA, Winfrey RJ, Jr, Irwin PA, Rajagopal J, Lonosky PM, Hall BD, Jondle MD, Voytas DF. High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J. 2005;44(4):693–705. doi: 10.1111/j.1365-313X.2005.02551.x. [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Johzuka-Hisatomi Y, Fukada-Tanaka S, Terada R, Nakamura I, Iida S. Homologous recombination-mediated knock-in targeting of the MET1a gene for a maintenance DNA methyltransferase reproducibly reveals dosage-dependent spatiotemporal gene expression in rice. Plant J. 2009;60(2):386–396. doi: 10.1111/j.1365-313X.2009.03947.x. [DOI] [PubMed] [Google Scholar]
- Yang M, Djukanovic V, Stagg J, Lenderts B, Bidney D, Falco SC, Lyznik LA. Targeted mutagenesis in the progeny of maize transgenic plants. Plant Mol Biol. 2009;70(6):669–679. doi: 10.1007/s11103-009-9499-5. [DOI] [PubMed] [Google Scholar]
- Yang Z, Tang L, Li M, Chen L, Xu J, Wu G, Li H. Monitoring homologous recombination in rice (Oryza sativa L.) Mutat Res. 2010;691(1–2):55–63. doi: 10.1016/j.mrfmmm.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Yata K, Esashi F. Dual role of CDKs in DNA repair: to be, or not to be. DNA Repair (Amst) 2009;8(1):6–18. doi: 10.1016/j.dnarep.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Yokota Y, Shikazono N, Tanaka A, Hase Y, Funayama T, Wada S, Inoue M. Comparative radiation tolerance based on the induction of DNA double-strand breaks in tobacco BY-2 cells and CHO-K1 cells irradiated with gamma rays. Radiat Res. 2005;163(5):520–525. doi: 10.1667/RR3355. [DOI] [PubMed] [Google Scholar]
- Yoon K, Cole-Strauss A, Kmiec EB. Targeted gene correction of episomal DNA in mammalian cells mediated by a chimeric RNA.DNA oligonucleotide. Proc Natl Acad Sci U S A. 1996;93(5):2071–2076. doi: 10.1073/pnas.93.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama KO, Kobayashi J, Ogita N, Ueda M, Kimura S, Maki H, Umeda M. ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep. 2013;14(9):817–822. doi: 10.1038/embor.2013.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, Liu H, La Russa M, Xie M, Ding S, Qi LS. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16(2):142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SS, Vanderschuren H, Qaim M, Mahfouz MM, Kohli A, Mansoor S, Tester M. New plant breeding technologies for food security. Science. 2019;363(6434):1390–1391. doi: 10.1126/science.aav6316. [DOI] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29(2):149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Mason H. Bean yellow dwarf virus replicons for high-level transgene expression in transgenic plants and cell cultures. Biotechnol Bioeng. 2006;93(2):271–279. doi: 10.1002/bit.20695. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang F, Li X, Baller JA, Qi Y, Starker CG, Bogdanove AJ, Voytas DF. Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol. 2013;161(1):20–27. doi: 10.1104/pp.112.205179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Peterson DJ, Tagliani L, St Clair G, Baszczynski CL, Bowen B. Targeted manipulation of maize genes in vivo using chimeric RNA/DNA oligonucleotides. Proc Natl Acad Sci U S A. 1999;96(15):8768–8773. doi: 10.1073/pnas.96.15.8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Clarke R, Puppala AK, Chittori S, Merk A, Merrill BJ, Simonovic M, Subramaniam S. Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9. Nat Struct Mol Biol. 2019;26(8):679–685. doi: 10.1038/s41594-019-0258-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Yuan, Song Qianna, Li Chao, Jin Shuai, Zhang Dingbo, Wang Yanpeng, Qiu Jin-Long, Gao Caixia. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nature Biotechnology. 2018;36(10):950–953. doi: 10.1038/nbt.4261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Targeted mutagenesis-genome editing in monocots using CRISPR/Cas.
Additional file 2: Table S2. Major applications of base-editing approaches in plants.
Data Availability Statement
Not applicable.