Abstract
Aberrant DNA methylations have been reported to be significantly associated with lung squamous cell cancer (LUSC). The aim of this study was to investigate the DNA methylation-driven genes in LUSC by integrative bioinformatics analysis. In the present study, methylation-driven genes in LUSC were screened out, and survival analysis related to these genes was performed to confirm their value in prognostic assessment. Gene expression and methylation data were downloaded from The Cancer Genome Atlas (TCGA), and the MethylMix algorithm was used to identify methylation-driven genes. ConsensusPathDB was used to perform Gene Ontology and pathway enrichment analysis of methylation-driven genes. Survival analysis was performed to investigate the correlation with prognosis. In total, 52 differentially expressed methylation-driven genes were identified in LUSC and adjacent tissues. Survival analysis showed that DQX1, GPR75, STX12, and TRIM61 could serve as independent prognostic biomarkers. In addition, the combined methylation and gene expression survival analysis revealed that the combined expression level of the genes ALG1L, DQX1, and ZNF418 alone can be used as a prognostic marker or drug target. Methylation of four sites of gene ZNF418, four sites of ZNF701, two sites of DQX1, and four sites of DCAF4L2 was significantly associated with survival. The present study provides an important bioinformatic and relevant theoretical basis for subsequent early diagnosis and prognostic assessment of LUSC.
Keywords: lung squamous cell carcinoma, DNA methylation-driven genes, biomarkers, survival analysis
Introduction
Lung cancer is a common malignant tumor, and is the leading cause of cancer-related death in the United States and worldwide (1). The cancer occurs below the bronchi, and there are no typical clinical symptoms in the early stage. According to the characteristics of pathological tissues, lung cancer can be classified into two major categories: Small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). NSCLC is the most common subtype, accounting for >80% of all lung cancer cases (2). The lung squamous cell cancer (LUSC) subtype accounts for 25–30% of all lung cancers, and is mainly presented as central lung cancer (3). Studies have shown that squamous cell carcinoma is associated with smoking, and is common in men (1,2). However, not all smokers develop lung cancer. Only 10–20% of smokers develop lung cancer in their lifetime, which may be due to genetic differences in susceptibility (4). In recent years, great progress has been made in molecular targeted therapy for LUSC; however, further research is required. Unfortunately, some patients do not benefit from conventional chemotherapy and radiotherapy due to drug resistance and toxic effects. Studies have shown that changes in neoplasms at the molecular level occur earlier than various clinical features (5,6). Therefore, research focusing on the molecular level may be more conducive to the early diagnosis and treatment of cancer to improve the prognosis. New diagnostic markers and therapeutic targets are urgently needed to further improve the diagnosis and treatment of LUSC, and to reduce the fatality rate.
Although progress continues to be made in the treatment of non-squamous NSCLC, the needs of patients with squamous NSCLC remain unmet. With regard to targeted chemotherapy drugs, there has been a focus on non-squamous cell disease in terms of most regulatory approvals for advanced NSCLC, and updates to clinical practice guidelines. Although the progress that has been made with the therapeutic strategies of squamous NSCLC is limited, the identification of more effective treatments for this patient group is gaining momentum. Therefore, further research on LUSC will provide new directions for treatment.
With the use of the current advanced DNA methylation and RNA sequence research methods, great progress has also been made in research on the relationship between DNA methylation and gene expression during the onset and infiltration of neoplasms. In a study of the integrative analysis of DNA methylation and mRNA expression, Shi et al (7) revealed the function of epigenetic changes on LUSC. However, the mechanism contributing to oncogenesis is unclear, and in view of this, Gevaert (8) developed MethylMix, a new computational algorithm implemented in R software, to identify abnormal methylated genes and predict changes in transcription. The Cancer Genome Atlas (TCGA) (9), a well-known database on the cancer genome, provides a large amount of genetic information and clinical data, which can assist in understanding the clinical characteristics of molecular information.
In the present study, LUSC-related genes with abnormal methylation were identified in the TCGA database, and associated differential genes of abnormal methylation in LUSC were determined. Gene expression and abnormal methylation gene data of LUSC samples from the TCGA database were analyzed. Four candidate genes [DQX1 (DEAQ-box RNA dependent ATPase 1), GPR75 (probable G-protein coupled receptor 75), STX12 (syntaxin 12), and TRIM61 (putative tripartite motif-containing protein 61)] were identified from 52 methylation-driven genes (P<0.05), which may be independent prognostic biomarkers. Furthermore, the genes ALG1L (ALG1 chitobiosyldiphosphodolichol beta-mannosyltransferase like), DQX1, and ZNF418 (zinc finger protein 418) were confirmed to meaningfully predict prognosis by integrative survival analysis. In addition, a significant association between site methylation and survival was found.
Materials and methods
Data acquisition and preprocessing
Methylation and mRNA expression data of LUSC patients were downloaded from the TCGA database (https://portal.gdc.cancer.gov/) (accessed March, 2019) (10). The methylation data were obtained from 573 samples, including 69 normal samples and 504 cancer samples, and the mRNA expression data were obtained from 551 samples, including 49 normal samples and 502 LUSC samples. Clinical survival data were also included. The LIMMA 3.40.6 package (http://www.bioconductor.org/packages/release/bioc/html/limma.html) (11) in R language was employed to identify aberrant methylated genes and differentially expressed genes between lung cancer and normal tissues.
Identification of methylation-driven gene
The MethylMix (http://www.bioconductor.org/packages/release/bioc/html/MethyIMix.html), an algorithm implemented by R 3.5.2 software (http://www.rproject.org/) was used to analyze the correlation between gene methylation and gene expression and to screen methylation-driven genes. The following three data files are required: DNA methylation data for normal group; DNA methylation data from cancer group; and matched gene expression data for cancer group. Based on the MethylMix algorithm, the correlation between the level of methylation and gene expression was calculated. Next, genes significantly associated were identified and the hypomethylation and hypermethylation genes were also determined by the β-mixed model. Finally, methylation-driven genes were identified (8,12). P<0.05 and Cor<0.3 were the selected criteria for screening.
Gene Ontology (GO) enrichment analysis
ConsensusPathDB (http://cpdb.molgen.mpg.de/CPDB, version 34) is an online software integrating interaction networks in Homo sapiens, including binary and complex signaling, gene regulatory and drug-target interactions, as well as biochemical pathways (13,14). The methylation-driven genes list was submitted following the instructions on the website, the submit list was clicked and the data were downloaded. Finally, the GOplot R package was used to plot the enrichment results. P<0.05 was set as the cut-off criterion for GO enrichment analysis.
Pathway analysis
ConsensusPathDB was also used to perform pathway enrichment analysis of methylation-driven genes (14,15). The pathway enrichment analysis of methylation-driven genes was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.ad.jp/kegg/). The pathways were selected with P<0.05 as the cut-off criterion.
Survival analysis of driver genes and methylated sites
The methylation levels of driver genes were extracted, followed by Kaplan-Meier analysis using the survival analysis package in R software to compare the effects of different methylation levels of driver genes on survival (16). The P-value was obtained using the long-rank test. The level of gene methylation was then combined with gene expression data to analyze the combined effect on survival, which was also performed using the survival R package. In addition, the methylation information on the related sites of the methylation-driven genes was extracted, based on the downloaded methylation and clinical data of squamous cell carcinoma from the TCGA to further determine the value of the methylation-driven genes for prognostic evaluation. The survival curve was drawn using the survival R package. P<0.05 was considered to indicate a statistically significant value.
Results
Screening of methylation-driven genes
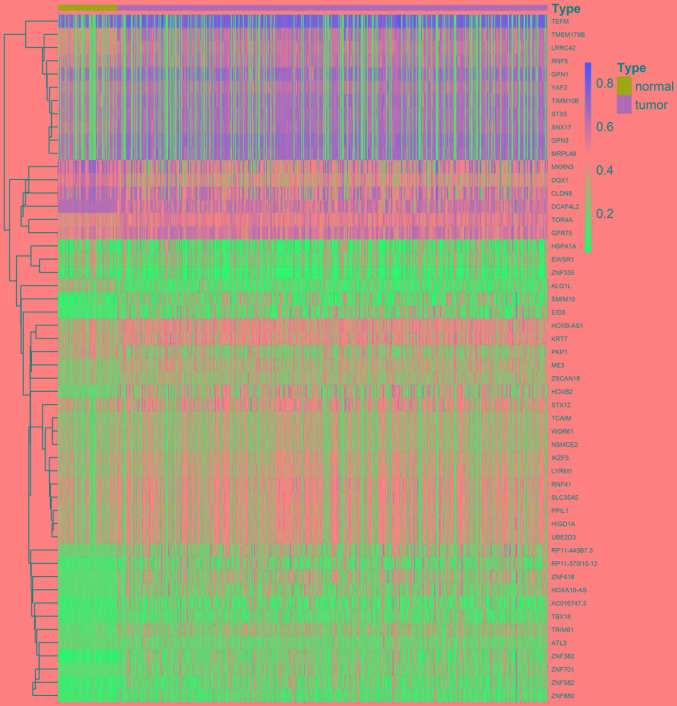
A total of 52 methylation-driven genes were identified by comparing the levels of methylation in tumor and normal tissues, and the genes were visualized using a heatmap (Fig. 1). The methylation level of 44 genes in the cancer group was higher compared with that in the normal group, and the methylation level of 8 genes was lower compared with that of the normal group. Five methylation-driven genes with the smallest P-values were selected to plot a distribution map of the degree of methylation (Fig. 2A-E). The distribution of the remainder of the genes is shown in Fig. S1.
Figure 1.
Heatmap of methylation-driven genes. The red color represents hypermethylation, whereas the blue color represents hypomethylation.
Figure 2.
Distribution map of methylation states of selective genes. Mixture models of ALG1L (A), ZNF418 (B), ZNF701 (C), DQX1 (D) and DCAF4L2 (E) are presented. The horizontal axis represents the methylation level, the vertical axis represents the number of samples, the histogram represents the methylation states in the tumor group, and the curve represents the methylation distribution trend of the tumor group. The black line at the top represents the methylation state in the non-tumor group.
Correlation analysis between gene methylation and expression
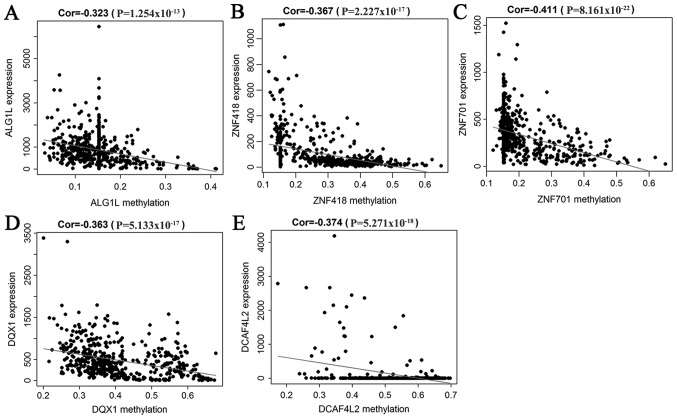
A correlation analysis between the methylation level of 52 methylation-driven genes and their expression was performed, which indicated that the level of methylation was negatively correlated with the respective expression of these genes. Five genes were selected to generate their scatterplots, and to estimate the correlation coefficient. These genes were the first 5 with the smallest P-values in the correlation test (Fig. 3A-E).
Figure 3.
Correlation between the expression level of selective genes and the degree of methylation. (A-E) Pearson's correlation analysis revealed a negative correlation between gene expression and methylation for (A) ALF1L, (B) ZNF418, (C) ZNF701, (D) DQX1 and (E) DCAF4L2. The abscissa represents the methylation level, and the ordinate represents gene expression. Cor represents the correlation coefficient of Pearson's analysis.
GO enrichment analysis
GO enrichment analysis was conducted using the ConsensusPathDB online software. Methylation-driven genes were enriched in ‘intracellular membrane-bounded organelle’, ‘Smc5-Smc6 complex’, and ‘SUMO ligase complex’ (Fig. 4).
Figure 4.
GO enrichment analysis of methylation-driven genes. Genes are on the left, pathways are on the right, and different pathways are presented in different colors. The color for each pathway is annotated below the ring diagram. Genes and their corresponding pathways are linked together. GO, Gene Ontology.
Pathway analysis
The pathway enrichment analysis is shown in Fig. 5, and was conducted in ConsensusPathDB online. In total, 13 pathways were enriched. These genes were significantly linked to ‘BARD1 signaling events’, ‘Nicotine Pathway (Dopaminergic Neuron)’ and ‘Pharmacodynamics’ (Fig. 5).
Figure 5.
Pathways of significant enrichment. Every circle represents the pathway, the size of the circle represents the number of genes enriched by the pathway, and each line represents the correlation between the pathways. The color of the circle represents the P-value. Edge width represents the percentage of genes with the same pathway, and the edge color represents genes from input with the same difference before the pathway.
Survival analysis of methylation-driven genes in LUSC
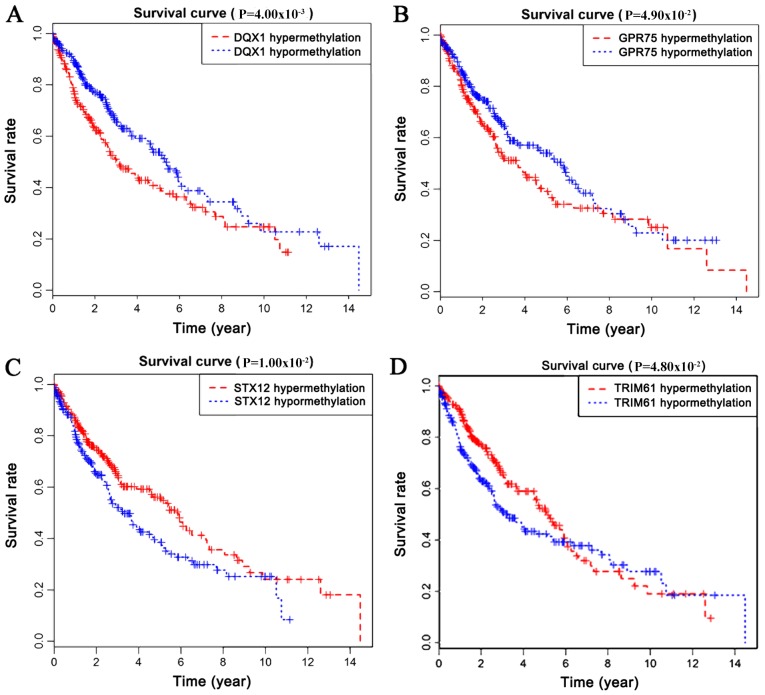
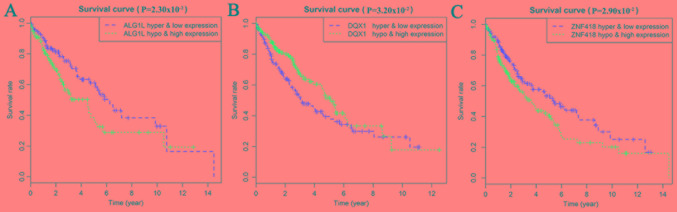
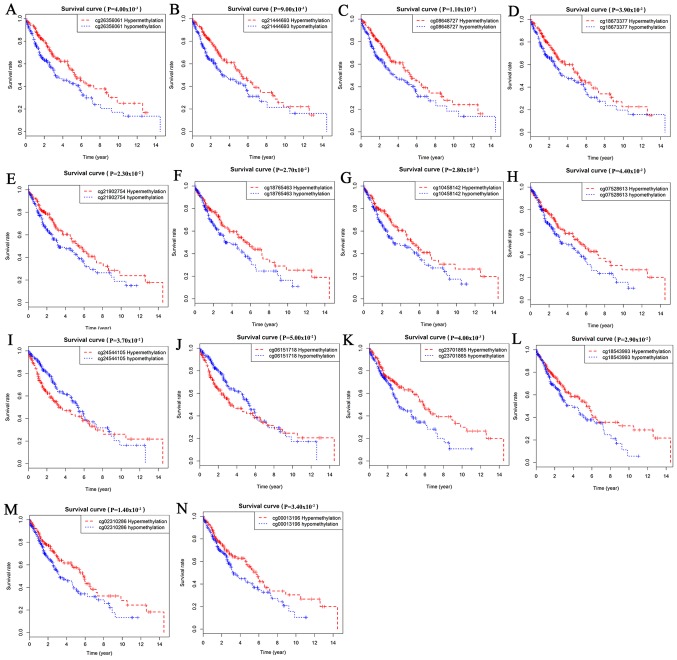
Survival analysis was statistically significant when the P-value was <0.05. By analyzing the degree of gene methylation, it was found that the genes DQX1, GPR75, STX12, and TRIM61 significantly affected the survival and prognosis of lung cancer patients (Fig. 6). Taking genes methylation and expression as comprehensive factors, it was confirmed that the genes ALG1L, DQX1, and ZNF418 were closely related to prognosis (Fig. 7). The methylation levels of multiple sites of ZNF418, ZNF701, DQX1 and DCAF4L2 were found to be correlated with patient survival (Fig. 8).
Figure 6.
(A-D) Survival curves related to the methylation levels of four genes. (A) DQX1; (B) GPR75; (C) STX12; (D) TRIM6.
Figure 7.
Survival curves related to the combination of gene expression and methylation level. (A) ALG1L; (B) DQX1; and (C) ZNF418.
Figure 8.
Survival curves related to methylation sites. (A-D) ZNF418; (E-H) ZNF701; (I and J) DQX1; and (K-N) DCAF4L2.
Discussion
LUSC, which usually has a poor prognosis, is one of the major subtypes of lung cancer, and is difficult to treat as patients tend to be older and have a higher incidence of comorbidities (17,18). The occurrence of lung cancer is an intricate biological process involving multiple steps and factors, and is closely related to changes in genetic information. Epigenetic changes are involved in important aspects of lung cancer development, and its regulatory mechanisms mainly involve DNA methylation, and non-coding RNA expression regulation and histone modification (19).
In recent years, significant advances have been made in the molecular biological mechanisms of LUSC, early diagnostic evaluation markers, as well as specific genetic alterations. DNA methylation is an epigenetic mechanism that leads to tumorigenesis, and has attracted a lot of attention from researchers. DNA methylation abnormalities have been found at the genome level in the majority of tumors, including lung cancer (20,21), and the frequency of CpG island hypermethylation in tumor cells is much higher compared with that of gene mutation (22). Abnormalities in DNA methylation often occur in the early stages of cancer, and persist throughout the development of the cancer. Methylation or demethylation may lead to tumor gene activation or inactivation of cancer suppressor genes. DNA methylation is considered to be a vitally important mechanism that causes cells to change from a normal to malignant state (23), and is the possible cause of tumor treatment tolerance (24). However, some studies have found that differentially methylated genes serve as potential cancer driver genes (25,26).
Previous research findings have verified that enhanced expression of genes caused by hypomethylation, and decreased expression caused by hypermethylation, serve an important role in the regulation and development of malignant carcinoma (27,28). Methylation is a major epigenetic modification of genomic DNA, and an important means of regulating genomic function. Although epigenetic modifications are reversible, they have great potential as effective therapeutic targets (29). Therefore, the detection and treatment of DNA methylation as a target will result in new strategies for the diagnosis and treatment of lung cancer. For example, Sugimoto et al (30) found that patients benefit from aberrant methylation of GRWD1 (glutamate rich WD repeat containing 1) in tumor development, due to activity of the GRWD1 gene being inhibited by its own methylation in tumor cells, whereas expression of the GRWD1 gene can benefit tumor cell growth. In contrast, Chen et al (31) found that hypermethylation of the AGTR1 promoter is more common in patients with LUSC. Ni et al (32) showed that the methylation of gene SHOX2 (short stature homeobox 1) is more pronounced in lung cancer, especially in LUSC, and is a potential non-invasive biomarker of lung cancer. A study by Guo et al (33) found that the hypermethylated state of WIF-1 gene, commonly found in NSCLC, is not only more likely to occur in squamous cell carcinoma, but its expression was also correlated with poor clinical prognosis. In a study on tumor and corresponding non-malignant lung tissue specimens, Kim et al (34) demonstrated that high methylation of the Wrap53α promoter predicts a worse prognosis in patients with borderline significance. Zhang et al (35) reported that PAX6 gene hypermethylation is an independent prognostic indicator, and is significantly correlated with an overall low survival rate of NSCLC, which may therefore be a potentially attractive biomarker for prognostic assessment in NSCLC patients. Thus, comprehensive functional and survival analysis of methylation-driven genes is able to provide a deeper understanding of their underlying mechanisms, and to identify novel strategies for lung cancer therapy.
In the present study, our aim was to investigate methylation-driven genes in patients with LUSC by analyzing data on gene methylation downloaded from TCGA, and to assess their relationship with prognosis. Fifty-two methylation-driven genes were identified via the MethylMix package of R software. Enrichment analysis was performed to investigate cellular functions and pathways significantly associated with these genes, and to further reveal the biological mechanism of these methylation-driven genes. Functional enrichment analysis revealed that these genes are mainly associated with ‘intracellular membrane-bounded organelle’, ‘Smc5-Smc6 complex’, and a variety of other functions, such as ‘SUMO ligase complex’. These genes were found to be involved in ‘BARD1 signaling events’, the ‘Nicotine Pathway (Dopaminergic Neuron)’, and ‘Pharmacodynamics’ by enrichment pathway analysis. These analyses revealed the function of these genes, and their relationships with each other.
Survival analysis confirmed that methylation of the genes DXQ1, GPR75, STX12 and TRIM61 is closely related to prognosis. Survival analysis for combined gene expression and methylation indicated that ALG1L, DQX1, and ZNF418 could serve as markers to predict prognosis, and may even be targets in future targeted therapy of LUSC. The methylation levels of multiple sites were shown to be correlated with patient survival.
A previous study found that ZNF418 is expressed in a variety of tissues, such as lung, pancreas, muscle, and heart (36). ZNF418 is significantly down-regulated in gastric carcinoma tissues (37). In the present study, integrated survival and gene expression and methylation analysis revealed that the gene ZNF418 tends to predict a poor prognosis in patients with LUSC. This suggests that ZNF418 may be involved in the mechanism of progression in LUSC. DCAF4L2 is a member of the WD-repeat domain-containing protein family, which is an intermediary of protein-protein interaction. The levels of DCAF4L2 have been previously reported to be elevated in lung cancer and in human colorectal cancer, leading to worse clinical staging, involving lymphatic and distant metastases. These findings confirm its oncogenic role (38). GPR75, a G protein-coupled receptor, is significantly hypermethylated in colorectal neoplasia (39). Consistently, the present study also showed that it is hypermethylated in LUSC, and this hypermethylation is associated with infaust outcome of LUSC. STX12 is a member of the syntaxin family of soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs). A previous study reported that STX12 is able to mediate tumor cell invasion (40). The present study has revealed that the expression of STX12 is negatively correlated with methylation, and hypermethylation of STX12 is associated with favorable clinical results in LUSC. These results are theoretically consistent with LUSC; however, further validation of our findings is needed in future studies.
The study of methylation-driven genes has important clinical significance for the early diagnosis and prognosis of lung cancer. Methylation-driven genes are expected to be identified as novel tumor markers for clinical application in the future. It is plausible that our study provides credible potential targets for the biological mechanism and clinical management of LUSC. However, further research is required to confirm our findings.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was funded by the Scientific Research Projects of Chongqing Municipal Health Bureau (2013-2-174), China.
Availability of data and materials
The datasets generated and/or analyzed in the current study are available from the corresponding author on reasonable request.
Authors' contributions
PH and QL drafted the manuscript, QL collected and organized the data, PH contributed to bioinformatics analysis by R software, and JX contributed to the research design. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemjabbar-Alaoui H, Hassan OU, Yang YW, Buchanan P. Lung cancer: Biology and treatment options. Biochim Biophys Acta. 2015;1856:189–210. doi: 10.1016/j.bbcan.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lorenzo-González M, Fernández-Villar A, Ruano-Ravina A. Disentangling tobacco-related lung cancer-genome-wide interaction study of smoking behavior and non-small cell lung cancer risk. J Thorac Dis. 2019;11:10–13. doi: 10.21037/jtd.2018.11.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jordan EJ, Kim HR, Arcila ME, Barron D, Chakravarty D, Gao J, Chang MT, Ni A, Kundra R, Jonsson P, et al. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov. 2017;7:596–609. doi: 10.1158/2159-8290.CD-16-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aoki MN, Amarante MK, de Oliveira CEC, Watanabe MAE. Biomarkers in non-small cell lung cancer: Perspectives of individualized targeted therapy. Anticancer Agents Med Chem. 2018;18:2070–2077. doi: 10.2174/1871520618666180827102101. [DOI] [PubMed] [Google Scholar]
- 7.Shi YX, Wang Y, Li X, Zhang W, Zhou HH, Yin JY, Liu ZQ. Genome-wide DNA methylation profiling reveals novel epigenetic signatures in squamous cell lung cancer. BMC Genomics. 2017;18:901. doi: 10.1186/s12864-017-4223-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gevaert O. MethylMix: An R package for identifying DNA methylation-driven genes. Bioinformatics. 2015;31:1839–1841. doi: 10.1093/bioinformatics/btv020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomczak K, Czerwinska P, Wiznerowicz M. The cancer genome atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, Staudt LM. Toward a shared vision for cancer genomic data. N Engl J Med. 2016;375:1109–1112. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Law CW, Alhamdoosh M, Su S, Dong X, Tian L, Smyth GK, Ritchie ME. RNA-seq analysis is easy as 1-2-3 with limma, Glimma and edgeR. Version 3. F1000Res. 2016;5 doi: 10.12688/f1000research.9005.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cedoz PL, Prunello M, Brennan K, Gevaert O. MethylMix 2.0: An R package for identifying DNA methylation genes. Bioinformatics. 2018;34:3044–3046. doi: 10.1093/bioinformatics/bty156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. ConsensusPathDB: Toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39((Database issue)):D712–D717. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamburov A, Stelzl U, Lehrach H, Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013;41((Database issue)):D793–D800. doi: 10.1093/nar/gks1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herwig R, Hardt C, Lienhard M, Kamburov A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat Protoc. 2016;11:1889–1907. doi: 10.1038/nprot.2016.117. [DOI] [PubMed] [Google Scholar]
- 16.Dudley WN, Wickham R, Coombs N. An introduction to survival statistics: Kaplan-meier analysis. J Adv Pract Oncol. 2016;7:91–100. doi: 10.6004/jadpro.2016.7.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langer CJ, Obasaju C, Bunn P, Bonomi P, Gandara D, Hirsch FR, Kim ES, Natale RB, Novello S, Paz-Ares L, et al. Incremental innovation and progress in advanced squamous cell lung cancer: Current status and future impact of treatment. J Thorac Oncol. 2016;11:2066–2081. doi: 10.1016/j.jtho.2016.08.138. [DOI] [PubMed] [Google Scholar]
- 18.Paes de Araújo R, Bertoni N, Seneda AL, Felix TF, Carvalho M, Lewis KE, Hasimoto ÉN, Beckmann M, Drigo SA, Reis PP, Mur LAJ. Defining metabolic rewiring in lung squamous cell carcinoma. Metabolites. 2019;9:E47. doi: 10.3390/metabo9030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim D, Kim DH. Epigenome-based precision medicine in lung cancer. Methods Mol Biol. 2018;1856:57–85. doi: 10.1007/978-1-4939-8751-1_4. [DOI] [PubMed] [Google Scholar]
- 20.Qazi TJ, Quan Z, Mir A, Qing H. Epigenetics in Alzheimer's disease: Perspective of DNA methylation. Mol Neurobiol. 2018;55:1026–1044. doi: 10.1007/s12035-016-0357-6. [DOI] [PubMed] [Google Scholar]
- 21.Pan Y, Liu G, Zhou F, Su B, Li Y. DNA methylation profiles in cancer diagnosis and therapeutics. Clin Exp Med. 2018;18:1–14. doi: 10.1007/s10238-017-0467-0. [DOI] [PubMed] [Google Scholar]
- 22.Schmiemann V, Böcking A, Kazimirek M, Onofre AS, Gabbert HE, Kappes R, Gerharz CD, Grote HJ. Methylation assay for the diagnosis of lung cancer on bronchial aspirates: A cohort study. Clin Cancer Res. 2005;11:7728–7734. doi: 10.1158/1078-0432.CCR-05-0999. [DOI] [PubMed] [Google Scholar]
- 23.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 24.Da Costa EM, McInnes G, Beaudry A, Raynal NJ. DNA methylation-targeted drugs. Cancer J. 2017;23:270–276. doi: 10.1097/PPO.0000000000000278. [DOI] [PubMed] [Google Scholar]
- 25.Manolakos A, Ochoa I, Venkat K, Goldsmith AJ, Gevaert O. CaMoDi: A new method for cancer module discovery. BMC Genomics. 2014;15(Suppl 10):S8. doi: 10.1186/1471-2164-15-S10-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gevaert O, Villalobos V, Sikic BI, Plevritis SK. Identification of ovarian cancer driver genes by using module network integration of multi-omics data. Interface Focus. 2013;3:20130013. doi: 10.1098/rsfs.2013.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 28.Duruisseaux M, Esteller M. Lung cancer epigenetics: From knowledge to applications. Semin Cancer Biol. 2018;51:116–128. doi: 10.1016/j.semcancer.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17:630–641. doi: 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]
- 30.Sugimoto N, Maehara K, Yoshida K, Yasukouchi S, Osano S, Watanabe S, Aizawa M, Yugawa T, Kiyono T, Kurumizaka H, et al. Cdt1-binding protein GRWD1 is a novel histone-binding protein that facilitates MCM loading through its influence on chromatin architecture. Nucleic Acids Res. 2015;43:5898–5911. doi: 10.1093/nar/gkv509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen R, Hong Q, Jiang J, Chen X, Jiang Z, Wang J, Liu S, Duan S, Shi S. AGTR1 promoter hypermethylation in lung squamous cell carcinoma but not in lung adenocarcinoma. Oncol Lett. 2017;14:4989–4994. doi: 10.3892/ol.2017.6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ni S, Ye M, Huang T. Short stature homeobox 2 methylation as a potential noninvasive biomarker in bronchial aspirates for lung cancer diagnosis. Oncotarget. 2017;8:61253–61263. doi: 10.18632/oncotarget.18056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo H, Zhou S, Tan L, Wu X, Wu Z, Ran R. Clinicopathological significance of WIF1 hypermethylation in NSCLC, a meta-analysis and literature review. Oncotarget. 2017;8:2550–2557. doi: 10.18632/oncotarget.13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim DS, Lee WK, Park JY. Promoter methylation of Wrap53α, an antisense transcript of p53, is associated with the poor prognosis of patients with non-small cell lung cancer. Oncol Lett. 2018;16:5823–5828. doi: 10.3892/ol.2018.9404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Yang X, Wang J, Liang T, Gu Y, Yang D. Down-regulation of PAX6 by promoter methylation is associated with poor prognosis in non small cell lung cancer. Int J Clin Exp Pathol. 2015;8:11452–11457. [PMC free article] [PubMed] [Google Scholar]
- 36.Dang DT, Pevsner J, Yang VW. The biology of the mammalian Krüppel-like family of transcription factors. Int J Biochem Cell Biol. 2000;32:1103–1121. doi: 10.1016/S1357-2725(00)00059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hui HX, Hu ZW, Jiang C, Wu J, Gao Y, Wang XW. ZNF418 overexpression protects against gastric carcinoma and prompts a good prognosis. Onco Targets Ther. 2018;11:2763–2770. doi: 10.2147/OTT.S160802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, Chen Y, Han J, Meng Q, Xi Q, Wu G, Zhang B. DCAF4L2 promotes colorectal cancer invasion and metastasis via mediating degradation of NFκb negative regulator PPM1B. Am J Transl Res. 2016;8:405–418. [PMC free article] [PubMed] [Google Scholar]
- 39.Ashktorab H, Daremipouran M, Goel A, Varma S, Leavitt R, Sun X, Brim H. DNA methylome profiling identifies novel methylated genes in African American patients with colorectal neoplasia. Epigenetics. 2014;9:503–512. doi: 10.4161/epi.27644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams KC, Coppolino MG. SNARE-dependent interaction of Src, EGFR and β1 integrin regulates invadopodia formation and tumor cell invasion. J Cell Sci. 2014;127:1712–1725. doi: 10.1242/jcs.134734. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed in the current study are available from the corresponding author on reasonable request.